Abstract
Tandem repeat elements such as the diverse class of satellite repeats occupy large parts of eukaryotic chromosomes, mostly at (peri)centromeric and (sub)telomeric regions1. Some elements, however, are located in euchromatic regions throughout the genome and were hypothesized to regulate gene expression in cis by modulating local chromatin structure, or in trans via repeat-derived transcripts2–4. Here we show that a satellite repeat in the mosquito Aedes aegypti promotes sequence-specific gene silencing via the expression of two PIWI-interacting RNAs (piRNAs). Whereas satellite repeats and piRNA sequences generally evolve extremely fast5–7, this locus was conserved for approximately 200 million years, suggesting a central function in mosquito biology. piRNA production commenced shortly after egg-laying, and inactivation of the more abundant of the piRNAs resulted in failure to degrade maternally provided transcripts and developmental arrest. Our results reveal a novel mechanism by which satellite repeats regulate global gene expression in trans via piRNA-mediated gene silencing that is essential for embryonic development.
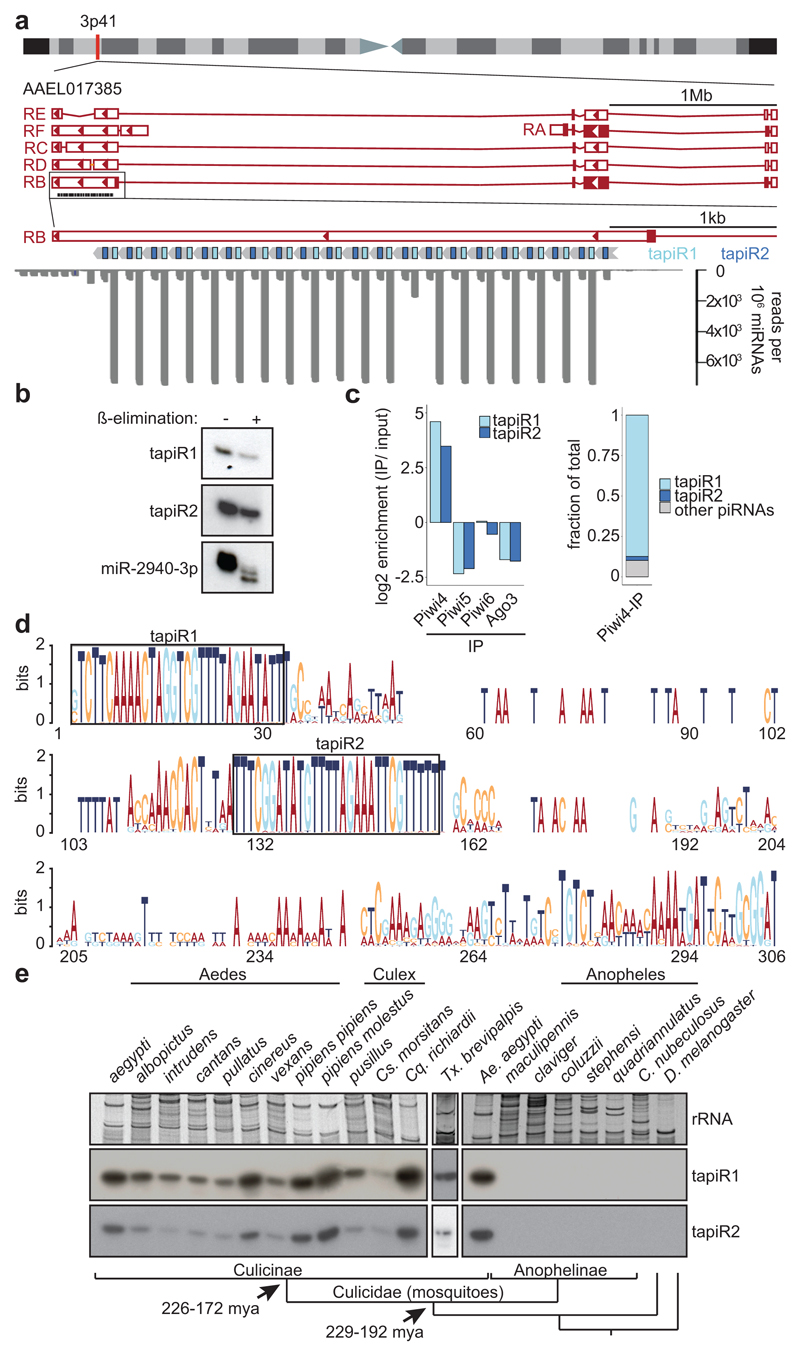
Satellite repeats comprise a substantial portion of eukaryotic genomes, but little is known about their functions. Many satellite repeats are actively transcribed, and some produce small interfering (si)RNAs required for heterochromatin formation and maintenance1. We analysed small RNAs in germline (ovaries) and somatic (carcasses) tissues of Ae. aegypti and in the somatic cell line Aag2. Although satellite repeats constitute <10% of the genome, they were not only covered by siRNAs, but also by PIWI-interacting (pi)RNAs (Extended Data Fig. 1A). piRNAs are thought to protect animal genomes from harmful parasitic elements like transposable elements (TEs)8. In contrast to the fruit fly Drosophila melanogaster8, piRNAs are not restricted to germline tissues in Ae. aegypti, and TE-derived piRNAs are underrepresented compared to TE abundance in the genome. Instead, satellite repeat-derived piRNAs were highly overrepresented especially in somatic tissues (Extended Data Fig. 1a). Intriguingly, 74% of reads in the soma, and 43% of reads in the germline, stem from only two individual sequences that map to a single repeat locus. This locus, ~3.5 kb in size, consists of 20 full repeats and one disrupted repeat unit organized in a head-to-tail array that overlaps with the 3’UTR of the gene of unknown function, AAEL017385 (Fig. 1a). These two highly abundant satellite-derived small RNAs were 30 and 29 nucleotides in size (Extended Data Fig. 1b) and resistant to β-elimination, suggesting that they are 2’-O-methylated at their 3’ end, a common feature of mature PIWI-bound piRNAs8 (Fig. 1b, Supplementary Fig. 1). We named these two sequences tapiR1 and 2 (tandem repeat-associated piRNA1/2).
Figure 1. An evolutionary conserved satellite repeat produces piRNAs that associate with Piwi4.
(a) Schematic representation of the current AAEL017385 annotation (AaegL5.1) and the tapiR satellite repeat locus (top), and small RNA read coverage in Aag2 cells (bottom). Filled boxes represent open reading frames. (b) Northern blot of tapiR1 and 2 upon ß-elimination in Aag2 cells. miR-2940-3p serves as positive control. (c) tapiR1 and 2 enrichment in the indicated PIWI immunoprecipitations (IP) (left), and fraction of total Piwi4-enriched reads (≥2-fold, right). (d) Sequence logo of all repeat monomers from Ae. aegypti, Ae. albopictus and Cx. quinquefasciatus. (e) Northern blot of tapiR1 and 2 in the indicated mosquito species and other insects. Ethidium bromide-stained rRNA serves as loading control. Phylogenetic relationships based on ref.10 are indicated in the bottom panel. Bar lengths are arbitrary.
Ae. Aegypti has an expanded PIWI gene family of seven PIWI genes (Piwi2-7 and Ago3)9. Based on the ubiquitous expression of tapiR1 and 2 in somatic and germline tissues (Extended Data Fig. 2a), we performed immunoprecipitation (IP) in Aag2 cells of the PIWI proteins that are expressed in both germline and soma (Piwi4-6 and Ago3)9. Both tapiR1 and 2 exclusively associated with Piwi4 and, in fact, most of the Piwi4-associated piRNAs comprised of tapiR1 (87%) and tapiR2 (2%) (Fig. 1c, Extended Data Fig. 2b,c, Supplementary Fig. 2a). Conversely, only knockdown of Piwi4, but not other PIWI or AGO-clade Argonaute genes, reduced tapiR1 and 2 levels (Extended Figure 2d,e, Supplementary Fig. 2b,c). Together, these data establish tapiR1 and 2 as PIWI-interacting RNAs.
Akin to piRNAs, satellite repeats evolve extremely fast and display high sequence divergence between species and can even be species-specific1,5,6. Hence, it is unexpected that the tapiR tandem repeat locus is conserved in the closely related Ae. albopictus and the more distantly related Culex quinquefasciatus (Extended Data Fig. 3a), while it is absent from the malaria vector Anopheles gambiae. The tandem repeat locus differed in the number of monomers across species, and the monomers exhibited substantial length and sequence divergence, both between species and between monomers within one species. However, the sequences encoding tapiR1 and 2 and a downstream T were by far more conserved than the overall monomer, suggesting that these sequences are under extensive selective constraints (Fig. 1d, Extended Data Fig. 3b-d). We analysed expression of the two piRNAs in different mosquitoes, including species for which no genome assembly is available. We detected both tapiR1 and 2 in all five tested genera of the Culicinae subfamily of mosquitoes (Fig. 1e, Extended Data Fig. 3e), but not in the Anophelinae subfamily, nor in a biting midge or Drosophila. These observations suggest that the locus evolved in the late Triassic after divergence of the Anophelinae and Culicinae subfamily of mosquitoes (229-192 mya10), but before further divergence of the culicine genera (226-172 mya10). This establishes the tapiR tandem repeat locus as one of the few ancient satellite repeats that have hitherto been described3,11–13. Deep evolutionary conservation suggests important and conserved functions for the locus and its associated piRNAs that are independent of the upstream AAEL017385 gene (Extended Data Fig. 3g-i, Supplementary Note 1).
We tested the potential for target silencing by tapiR1 in trans using a series of luciferase reporters. We validated that tapiR1 and 2 efficiently target RNAs harbouring a single, fully complementary target site in the 3’UTR (Fig. 2a). Addition of an antisense oligonucleotide (AO) complementary to tapiR1, but not tapiR2, relieved silencing of the tapiR1 reporter in a concentration-dependent manner, and vice versa (Extended Data Fig. 4a). These results confirm that tapiR-mediated silencing is sequence-specific, and that the AOs target the mature piRNAs and do not, or only to a minor extent, target the tapiR1/2 precursor. Additionally, tapiR AO treatment did not influence miRNA stability (Extended Data Fig. 4b). Unlike most miRNAs14, silencing was independent of the position of the target site in the mature mRNA (Fig. 2b, Extended Data Fig. 5a), yet was largely abolished for target sites in introns or a non-translated transcript expressed from a polymerase III promoter (Extended Data Fig. 5b). We noticed that firefly and Renilla luciferase genes, which we used as reporter and normalization control, respectively, contain potential tapiR1 target sites. Renilla luciferase is indeed potently suppressed by tapiR1, and firefly luciferase slightly (Extended Data Fig. 5c-e), yet, mutating these target sites did not affect our conclusions (Extended Data Fig. 5f,g).
Figure 2. tapiR1 silences target RNAs in trans through seed-mediated basepairing.
(a) Schematic representation of tapiR1 reporter constructs (left) and luciferase assay with the indicated reporters in Aag2 cells (right). (b-d) Representative luciferase assay of the indicated reporters, with (b) target sites at different positions in the RNA, or (c,d) target sites in the 3’ UTR with three consecutive mismatches (c), or increasing number of mismatches (d). (e) log2 expression of mRNAs and lncRNAs in tapiR1 or control AO treated Aag2 cells (average from three biological replicated). A pseudo-count of one was added to plot values of zero. Diagonal lines indicate a fold change of two. (f) RT-qPCR of tapiR1 target genes in AO treated Aag2 cells. Bars indicate mean ± s.d., and individual measurements (n=18 (a), n=6 (b-d), n=2 (f); see Supplementary Table 7).
To assess sequence requirements for tapiR1 targeting, we introduced mismatches in the piRNA target site. Three consecutive mismatches were tolerated unless they were located in the t1 to t9 region of the target (the nucleotides expected to basepair to piRNA positions 1 to 9) (Fig. 2c, Extended Data Fig. 6a), and single mismatches only impaired silencing at positions t3 to t7 (Extended Data Fig. 6a,b), reminiscent of a microRNA seed14 and comparable to the piRNA seed in C. elegans15,16.
Basepairing at the potential Argonaute cleavage site between nucleotides t10 and t11 was dispensable for silencing, as was a mismatch at t1, suggesting that the first nucleotide of tapiR1 is anchored in a binding pocket of Piwi4, similar to other Argonaute proteins17. Unexpectedly, a mismatch at position t2 was tolerated as well, but only when the rest of the target site was perfectly complementary (Extended Data Fig. 6c). Additionally, in contrast to C. elegans15, G:U wobble pairs were not allowed inside the seed and had the same effect as a mismatch at the same position (Extended Data Fig. 6d). Increasing numbers of mismatches at the 3’ end did not interfere with silencing when at least half of the piRNA could basepair with the target site (Fig. 2d), indicating that the 3’ part of the piRNA is not necessarily required, yet the seed region alone not sufficient, for silencing, a pattern that is more similar to target requirements of miRNAs than those of piRNAs studied thus far15,18–20.
Some satellite repeats can influence gene expression by modulation of the local chromatin environment in cis, or via siRNAs targeting of homologous repeat insertions2–4. In contrast, given the conservation of the tapiR repeat locus, the efficiency of target reporter silencing and the targeting requirements of tapiR1, we hypothesized that the tapiR locus has the potential to silence a broad range of remote genes in trans, and thus to regulate diverse cellular processes. To test this idea, we predicted tapiR1 target sites and verified that about half were sufficient to mediate silencing in luciferase reporter assays (Extended Data Fig. 7). We then blocked tapiR1-mediated silencing with the tapiR1 AO in Aag2 cells and assessed global gene expression by RNA-seq. Intriguingly, expression of 134 genes was strongly and significantly increased (Fig. 2e, Supplementary Table 1) and some transposons, though not globally affected, were upregulated as well (Extended Data Fig. 8a, Supplementary Table 2). We validated these results by RT-qPCR and confirmed that target gene expression was increased in a concentration-dependent manner upon tapiR1 AO treatment (Fig. 2f). As expected, knockdown of Piwi4, but not the miRNA Argonaute Ago1, led to derepression of target gene expression and reporters carrying cellular target sites (Extended Data Fig. 8b,c). Yet tapiR2 AO treatment did not relieve silencing of these target genes (Extended Data Fig. 8d), confirming again that the AOs are specific for the mature piRNAs. Our results indicate that tapiR1 directly and strongly regulates expression of a subset of cellular genes in a sequence-dependent manner, with only limited basepairing needed to mediate silencing. Yet, similar to miRNAs21, minimum free energy predictions of tapiR1/target RNA duplexes were not sufficient to define bona fide target genes (Extended Data Fig. 8e,f).
Satellite repeats are often expressed in a developmental or stage-specific manner22. In Ae. aegypti, tapiR1 and 2 were not maternally deposited and thus are absent during the first three hours of embryonic development before zygotic genome activation (ZGA) (Fig. 3a, Extended Data Fig. 9a), but could be detected in all subsequent life stages (Extended Data Fig. 9b,c). Likewise, Piwi4 is expressed in all analysed tissues, but is already present in early embryos (Extended Data Fig. 9d,e). After fertilization, embryonic development initially depends on maternally deposited information, but eventually developmental control is fully transferred to the zygotic genome (maternal-to-zygotic transition, MZT). This process is marked by the degradation of maternal transcripts, initially through maternal decay activities and later, after ZGA, also by zygotic components. One of the decay mechanisms involves zygotically-expressed miRNAs23. We hypothesized that tapiR1 is similarly involved in the zygotic degradation pathway and necessary for embryonic development. To test this idea, we injected either tapiR1-specific or control AOs into early pre-blastoderm embryos (Fig. 3b), and assessed their development using a discrete scoring scheme (Supplementary Fig. 2e). Strikingly, >90% of all tapiR1 AO-injected embryos were arrested early in development, whereas about half of all control embryos showed obvious signs of developmental progression (Fig. 3c). In accordance, only a small fraction of tapiR1 AO-injected embryos hatched (Fig. 3d), indicating that tapiR1 deficiency impedes development. RNA sequencing from tapiR1 or control AO-injected embryos revealed massive deregulation of cellular transcripts (Fig. 3e, Extended Data Fig. 9f, Supplementary Tables 3,4). We confirmed these results, and the dependency on Piwi4 for a selection of target genes by RT-qPCR upon tapiR1 AO and Piwi4 dsRNA injection (Extended Data Fig. 9g,h). Target genes with predicted tapiR1 target sites were more strongly upregulated than genes without target sites (Fig. 3f) and additionally, a subset of these genes was also found to be upregulated in tapiR1 AO-treated Aag2 cells (Extended Data Fig. 9i). These findings show that tapiR1 controls regulatory circuits by direct gene targeting in vivo, and that this function is essential for embryonic development, likely by promoting mRNA turnover of a subset of maternal transcripts. In line with this conclusion, confirmed target genes are down-regulated after the onset of tapiR1 expression (Fig. 3g), and tapiR1 targets are overrepresented in transcripts that are maternally provided and degraded during MZT (Fig. 3h).
Figure 3. tapiR1 promotes turnover of maternal transcripts and is essential for embryonic development.
(a) Northern blot of tapiR1 and 2 in Ae. aegypti embryos. Ethidium bromide-stained rRNA serves as loading control. (b) Outline of the experimental procedure for panels c-e. (c) Percent of embryos that reached the indicated developmental stages (Supplementary Fig. 2e) at 2.5 days post-AO injection (X2 test of independence: X2 (2, N = 521) = 105.05, p<2.2e-16). (d) Percent of hatched embryos at four days post-AO injection. Box-whisker-plot represent mean, first and third quartile, and maximum and minimum (n=6 (c), n=5 (d), Supplementary Table 7). (e) log2 RNA expression of genes in embryos injected with tapiR1 or control AO at 20.5 h post-injection (mean counts from five biological replicates plus a pseudo-count of one). Diagonal lines highlight a -fold change of two. (f) Cumulative distribution of log2 -fold changes of genes grouped by the minimum free energy (mfe) of predicted piRNA/target RNA duplexes. (g) Expression of tapiR1 target genes in embryos. RT-qPCR was performed on samples shown in (a). Abd-A is a gene not targeted by tapiR1 and serves as negative control. Bars indicate mean ± s.d. (n=2; see Supplementary Table 7). (h) Fraction of genes grouped by their expression pattern in early embryos.
Maternally-deposited piRNAs have been shown to promote degradation of nanos24 and other transcripts involved in germ cell development in Drosophila25. Yet, this was dependent on the existing pool of transposon-derived piRNAs, which, due to its large diversity, is ideal to be used to degrade a large number of transcripts. In contrast, we propose that, analogous to abundant miRNAs in other animals23, Culicinae mosquitoes have evolved a specific zygotic piRNA to destabilize a defined set of maternally-deposited transcripts in early embryonic development, the biogenesis and mechanism of which remains to be defined (Extended Data Fig. 10, Supplementary Fig. 2f,g, Supplementary Note 2). To our knowledge, this is the first demonstration of sequence-specific gene silencing by transcriptional products from a satellite repeat in trans, underlining the regulatory potential of tandemly repeated DNA.
Methods
Cell culture
Aedes aegypti Aag2 cells were cultured in Leibovitz's L-15 medium (Invitrogen) supplemented with 10% heat-inactivated Fetal Bovine Serum (PAA Laboratories), 2% Tryptose Phosphate Broth Solution (Sigma Aldrich), 1x MEM Non-Essential Amino Acids (Invitrogen) and 50 U/ml penicillin/ streptomycin (Invitrogen) at 25 °C. Aag2 cells are a widely-used non-clonal cell line of probably embryonic origin26 that expresses all somatic PIWI proteins and produces both primary and secondary piRNAs via ping-pong dependent amplification27,28. For all experiments, cells were seeded the day before and used at 70-80% confluency. Cells were regularly confirmed to be negative for mycoplasma contamination. The cell line was a gift from R. Andino, University of California, San Francisco.
Mosquito rearing
Injections of embryos for RNA sequencing and northern blotting presented in Fig. 3a,c-e,g were performed using a cell fusion agent virus-free, isofemale Aedes aegypti strain called Jane. This strain was derived from a field population originally sampled in the Muang District of Kamphaeng Phet Province, Thailand29, and reared for 26 generations at 28±1 °C, 75±5% relative humidity, 12:12 hour light-dark cycle. Embryos were hatched under low pressure for 30-60 min. Larvae were grown in dechlorinated tap water and fed fish food powder (Tetramin) every two days. Adults were maintained in cages with constant access to a 10% sucrose solution. Female mosquitoes were fed on commercial rabbit blood (BCL) through a membrane feeding system (Hemotek Ltd.) using pig intestine as membrane. For AO injections, female mosquitoes were transferred to 25 °C and 70% humidity for at least two days before forced to lay eggs, and embryos were then placed back to 28 °C immediately after the injection. For the time-course experiment in Fig. 3a,g, embryos were kept at 25 °C during the course of the experiment. All other injection experiments were conducted with Ae. aegypti Liverpool strain, reared at 28 ±1 °C, 70% humidity as described above, however, fed on human blood (Sanquin Blood Supply Foundation, Nijmegen, The Netherlands), and maintained at 28 °C throughout experiments.
Experiments presented in Extended Data Figs. 2a and 10a,b were performed with the Ae. aegypti Rockefeller strain, obtained from Bayer AG, Monheim, Germany. The mosquitoes were maintained at 27±1 °C with 12:12 hour light:dark cycle and 70% relative humidity, as described30. For the bloodfeeding experiment, mosquitoes were offered human blood (Sanquin Blood Supply Foundation, Nijmegen, The Netherlands) and five engorged females were selected and sacrificed at each of the indicated time points.
Insects used for tapiR northern blot analyses in Fig. 1f were either laboratory-reared or wild-caught species. Aedes aegypti (Liverpool strain), Culex pipiens pipiens, Cx. pipiens molestus, Toxorhynchites brevipalpis, Anopheles coluzzii, An. quadriannulatus, An. stephensi mosquitoes, Culicoides nubeculosus biting midges, and D. melanogaster fruit flies (genotype w1118) were laboratory strains, which were stored at -80 °C until use. Ae. albopictus, Ae. cantans, Ae. intrudens, Ae. pullatus, Ae. cinereus, Ae. vexans, Cx. pusillus, Culiseta morsitans, Coquillettidia richiardii, An. maculipennis, An. claviger, and An. coluzzii were wild-caught individuals collected in different regions in Italy, Sweden, or the Netherlands between July 2014 and June 201531. Species were identified at the species level, and stored at -20 °C for a maximum of two years.
Gene knockdown
Double-stranded RNA was generated by in vitro transcription of T7 promoter-flanked PCR products with T7 RNA polymerase. Primer sequences are given in Supplementary Table 5. The reaction was carried out at 37 °C for 3-4 h, then heated to 80 °C for 10 min and gradually cooled down to room temperature to facilitate dsRNA formation. The dsRNA was purified with the GeneElute Total RNA Miniprep Kit (Sigma Aldrich).
Aag2 cells were seeded in 24-well plates the day before the experiment and transfected with X-tremeGENE HP transfection reagent (Roche) according to the manufacturer’s instructions, using a ratio of 4 μL reagent per μg of dsRNA. The transfection medium was replaced after 3 h with fully supplemented Leibovitz-15 medium. For Extended Data Figs. 2d,e, 3f, and 8c, cells were harvested after 48 h; in other experiments, the knockdown was repeated 48 h after the first transfection and cells were then harvested after 24 h. Knockdown was confirmed by RT-qPCR. For the RNAi screen shown in Extended Data Fig. 10f, a luciferase reporter harbouring the tapiR1 target site of AAEL001555 in the 3’UTR, and a Renilla luciferase reporter were co-transfected with the dsRNA during the second transfection.
For injection of embryos with dsRNA, engorged female Ae. aegypti Liverpool mosquitoes were allowed to lay eggs for 45 min. Embryos were desiccated for 1.5 min, covered with Halocarbon oil (Sigma Aldrich) and injected with 500 ng/μL dsRNA with the Pneumatic PicoPump PV820 (World Precision Instruments) with 30 psi inject pressure. Injected embryos were then transferred to a wet Whatman paper and kept at 28 °C and 80% humidity for 21 h. Per experiment, 30-60 embryos were injected per condition.
RNA isolation
RNA from cells and mosquitoes was isolated with Isol-RNA lysis buffer (5PRIME) according to the manufacturer's instructions. Briefly, 200 μL chloroform was added to 1 mL lysis buffer, and centrifuged at 16,060 x g for 20 min at 4 °C. Isopropanol was added to the aqueous phase, followed by incubation on ice for at least one hour, and centrifugation at 16,060 x g for 10 min at 4 °C. The pellet was washed three to five times with 85% ethanol and dissolved in RNase-free water. RNA was quantified on a Nanodrop photospectrometer.
Periodate treatment and β-elimination
Total RNA was treated with 25 mM NaIO4 in a final concentration of 60 mM borax and 60 mM boric acid (pH 8.6) for 30 min at room temperature. In the control, NaIO4 was replaced by an equal volume of water. The reaction was quenched with glycerol and β-elimination was induced with a final concentration of 40 mM NaCl for 90 min at 45 °C. RNA was ethanol precipitated and analysed by northern blot.
Generation of antibodies
Custom-made antibodies (Eurogentec) against endogenous PIWI proteins have been described previously32. Briefly, rabbits were immunized with a mix of two unique peptides (Ago3: (C+)TSGADSSESDDKQSS, (C+)IIYKRKQRMSENIQF; Piwi4: (C+)HEGRGSPSSRPAYSS, (C+)HHRESSAGGRERSGN; Piwi5: (C+)DIVRSRPLDSKVVKQ, CANQGGNWRDNYKRAI; Piwi6: MADNPQEGSSGGRIR(+C), (C+)RGDHRQKPYDRPEQS). Sera were collected and purified against each peptide separately (antibodies purified against the peptides in bold were used). Specificity of the Piwi5 and Ago3 antibodies were validated previously32. Specificity of the Piwi4 and Piwi6 antibodies was confirmed by Western blotting of Aag2 cells stably expressing PTH (Protein A, TEV cleavage site, 6x His-tag)-tagged PIWI upon knockdown of the respective PIWI protein, or a control knockdown (dsRLuc) (Supplementary Fig. 2a).
Immunoprecipitation and western blotting
Aag2 cells were lysed with RIPA buffer (10 mM Tris-HCl, 150 mM NaCl, 0.5 mM EDTA, 0.1% SDS, 1% Triton-X-100, 10% DOC, 1x protease inhibitor cocktail), supplemented with 10% glycerol and stored at -80 °C until use. The IP was performed with custom-made antibodies against Piwi4-6 and Ago3 (1:10 dilution) at 4 °C for 4 h on rotation. Protein A/G Plus beads (Santa Cruz) were added at a dilution of 1:10 and then incubated overnight at 4 °C on rotation. Beads were washed 3 times with RIPA buffer, and half was used for RNA isolation and protein analysis each. For RNA extraction, beads were treated with proteinase K for 2 h at 55 °C and isolated with phenol-chloroform extraction. Equal amounts of RNA for input and IPs were then analysed by northern blotting. For western blotting, the IP samples were boiled in 2x Laemmli buffer for 10 min at 95 °C, separated on 7.5% SDS-polyacrylamide gels, and blotted on 0.2 μm nitrocellulose membranes (Bio-Rad) in a wet blot chamber on ice. Membranes were blocked for 1 h with 5% milk in PBS-T (137 mM NaCl, 12 mM phosphate, 2.7 mM KCl, pH 7.4, 0.1 % (v/v) Tween-20) and incubated with PIWI (dilution 1:1000) and tubulin primary antibodies (rat anti-tubulin alpha, MCA78G, 1:1000, Sanbio) overnight at 4 °C. The next day, membranes were washed 3 times with PBS-T and incubated with secondary antibodies conjugated to a fluorescence dye (IRDye 800CW conjugated goat anti-rabbit, 1:10,000, Li-Cor, and IRDye 680LT conjugated goat anti-rat, 1:10,000, Li-Cor) for 1 h at room temperature in the dark. After washing three times in PBS-T, signal was detected with the Odyssey-CLx Imaging system (Li-Cor). Alternatively, Ae. aegypti Liverpool embryos were bead-beaten in RIPA buffer supplemented with 1mM PMSF (Sigma Aldrich), and boiled in 1x Laemmli buffer. Western blotting was performed as described above, however, the membrane was blocked in 5% BSA in PBS-T, and incubated with pSer2 PolII (rabbit anti-RNA polymerase II CTD YSPTSPS (phosphor S2), ab5095, 1:1000, Abcam), and tubulin primary antibodies. Uncropped western blots can be found in Supplementary Fig. 1.
Northern blot
piRNAs were detected by northern blot analyses, as published33. Briefly, RNA was denatured at 80 °C for 2 min in Gel Loading Buffer II (Ambion) and size-separated on 0.5 x TBE (45 mM Tris-borate, 1 mM EDTA), 7 M urea, 15% denaturing polyacrylamide gels. RNA was then blotted on Hybond-NX nylon membranes (GE Healthcare) in a semi-dry blotting chamber for 45 min at 20 V and 4 °C and crosslinked to the membrane with EDC crosslinking solution (127 mM 1-methylimidazole (Sigma-Aldrich), 163 mM N-(3-dimethylaminopropyl)-N´-ethylcarbodiimide hydrochloride (Sigma-Aldrich), pH 8.0) at 60 °C for 2 h. Crosslinked membranes were pre-hybridized in ULTRAHyb-Oligo hybridization buffer (Thermo Scientific) for 1 h at 42 °C and probed with 32P 5' end-labelled DNA oligonucleotide probes overnight at 42 °C. Membranes were then washed with decreasing concentrations of SCC (300 mM NaCl, 30 mM sodium citrate, pH 7.0; 150 mM NaCl, 15 mM sodium citrate; 15 mM NaCl, 1.5 mM sodium citrate) and 0.1 % SDS, and exposed to Carestream BioMax XAR X-Ray films (Kodak). Probe sequences can be found in Supplementary Table 5.
To detect a putative tapiR precursor transcript, RNA was size-separated on a 0.5 x TBE, 7 M urea, 6% denaturing polyacrylamide gel, transferred to Hybond-N+ nylon membranes (GE Healthcare) in a semi-dry blotting chamber for 2 h at 200 mA at 4 °C, UV-crosslinked at 150 mJ, pre-hybridized in ULTRAHyb Ultrasensitive hybridization buffer (Thermo Scientific), and probed with 32P-labeled DNA probes overnight at 42 °C, washed as described above, exposed to a Storage Phosphor Screen GP (Kodak), and developed with the Amersham Typhoon 5 Biomolecular Imager (GE Healthcare). The probe was produced with the Amersham Rediprime DNA Labelling System II (GE Healthcare) from a PCR-amplified sequence of the tapiR locus spanning 1.5 repeat units (~450 nt) that was cloned with flanking T7 sites separated by EcoRI and ZraI restriction sites into the pUC19 vector.
Uncropped northern blots can be found in Supplementary Fig. 1.
Reporter cloning and luciferase assay
Reporters were constructed by cloning annealed and phosphorylated oligonucleotides with the indicated tapiR1 or control target sites in the pMT-GL3 vector34. This vector encodes the Photinus pyralis firefly luciferase (GL3) under a copper-inducible metallothionein promoter. Sense and antisense oligonucleotides (Sigma Aldrich) were annealed by heating to 80 °C, and gradually cooling down to room temperature, phosphorylated with T4 polynucleotide kinase (Roche) at 37 °C for 30 min, purified and then ligated into the pMT-GL3 vector. For cloning of 3’UTR reporters the target site was cloned into the PmeI and SacII restriction sites, while for the 5’UTR reporters the target site and an upstream BamHI site were cloned into NotI and XhoI restriction sites. ORF reporters were constructed by cloning a Kozak sequence followed by the first 45 nt of luciferase and the target site into XhoI and NcoI sites. For designing the intron reporters, the first intron of RpS7 (AAEL009496) was cloned behind the duplicated first 45 nt of luciferase in the pMT-GL3 vector, the original ATG of firefly luciferase was mutated, 3 stop codons were introduced at the 3’ end of the intron, and a BsaBI restriction site was inserted in the first third of the intron of by site directed mutagenesis. Afterwards, oligonucleotides encoding tapiR1 or control target sequences were inserted into BsaBI as described above. IRES-containing reporters were designed by cloning the 5’ UTR of cricket paralysis virus amplified from infected S2 cells into PmeI and SacII restriction sites of the pMT-GL3 vector with a mutated tapiR1 target site (ΔtapiR1) using the HD In-Fusion cloning kit (Takara). Afterwards oligonucleotides containing tapiR1 or control target sites were cloned into the SacII site in the 3’UTR as described above. RNAPIII reporters were constructed by cloning the Ae. aegypti U6 promoter and the GL3 3’UTR including different tapiR1 target sites and a series of six T’s as termination signal into pUC19 with the HD In-Fusion cloning kit (Takara). As normalization control, a part of GL3 ORF was cloned downstream of the U6 promoter. Sequences of the oligonucleotides are provided in Supplementary Table 5. Correct insertion of target sites was confirmed by Sanger sequencing for all clones.
Where indicated, mutated firefly or Renilla luciferase versions were used that harbour synonymous mutations destroying the predicted target sites for tapiR1 (firefly luciferase, nt 782: 5’-gagtcgtcttaatgtatagatttgaagaa-3’ mutated to 5’-gtgtcgtgcttatgtaccggttcgaggag-3’, and Renilla luciferase, nt 462 5’-tgaatggcctgatattgaagaa-3’ mutated to 5’-tgagtggccagatatcgaggag-3’; modified nucleotides in bold). Aag2 cells were seeded in 96-well plates the day before the experiment and transfected with 100 ng of the indicated plasmids and 100 ng pMT-Renilla34 per well, using 2 μL X-tremeGENE HP DNA transfection reagent per 1 μg plasmid DNA according to the manufacturer's instructions. Alternatively, 100 ng reporter plasmid and 100 ng pMT-Renilla were co-transfected with the indicated amounts of unlabelled, fully 2’O-methylated antisense RNA oligonucleotide using an additional amount of 4 μL X-tremeGENE HP DNA transfection reagent (Roche) per 1 μg oligonucleotide. Medium was replaced 3 h after reporter plasmid transfection with 0.5 mM CuSO4 in fully supplemented Leibovitz's L-15 medium to induce the metallothionein promoter. 24 h later, cells were lysed in 30 μL Passive lysis buffer (Promega) and activity of both luciferases was measured in 10 μL of the sample with the Dual Luciferase Reporter Assay system (Promega) on a Modulus Single Tube Reader (Turner Biosystems). Firefly luciferase was normalized to Renilla luciferase activity. For each construct, at least two to three independent clones were measured in triplicate wells to exclude clonal effects.
RT-qPCR
1 μg of total RNA was treated with DNase I (Ambion) for 45 min at 37 °C and reverse transcribed using the Taqman reverse transcription kit (Applied Biosystems) according to the manufacturer’s protocol. Real-time PCR was performed with the GoTag qPCR Master Mix (Promega) and measured on a LightCycler480 instrument (Roche) with 5 min initial denaturation and 45 cycles of 5 s denaturation at 95 °C, 10 s annealing at 60 °C and 20 s amplification at 72 °C. Starting fluorescence values of specific mRNAs were calculated with linear regression method of log fluorescence per cycle number and LinRegPCR program, version 2015.3, as described60.
Stem-loop RT-qPCR
Quantification of tapiR1 piRNA levels were performed similar to miRNA quantification35. Briefly, 100 ng of total RNA was reverse transcribed with 10 pmol stem-loop RT primer using 25 U Superscript II reverse transcriptase (Invitrogen) in 1x First Strand buffer, 0.33 mM dNTPs, and 2 U RNase inhibitors. cDNA was then measured by qPCR as described above.
3’ RACE, and 5’ RACE of slicer products
3' Rapid Amplification of cDNA Ends (3' RACE) was performed using the FirstChoice RLM-RACE Kit (Thermo Fischer Scientific) according to the manufacturer's instructions. Amplification products were separated on agarose gel, purified and Sanger sequenced. Slicer products were detected from 1 μg total RNA by 5’ RACE following the instructions, but without prior Calf Intestine Alkaline Phosphatase and Tobacco Acid Pyrophosphatase treatment. After amplification by PCR, RACE products in the size range of 150-250 nt were purified from agarose gel, cloned into pUC19 with HD In-Fusion cloning kit (Takara Bio) according to the manufacturer’s instructions, and sequenced by Sanger sequencing. Primer sequences can be found in Supplementary Table 5.
Antisense oligonucleotide treatment and injection
Aag2 cells were seeded in 24-well plates the day before the experiment. Cells were treated with 500 nM 5’Cy5-labelled, fully 2’O-methylated antisense RNA oligonucleotide in 530 μL medium with 4 μL X-tremeGENE HP DNA transfection reagent (Roche) per 1 μg oligonucleotide. Medium was replaced after 3 h and cells from three independent experiments were harvested 48 h after transfection and prepared for RNA sequencing (see below). For 5’ RACE, cells were treated with 200 nM antisense oligonucleotides together with 50 nM siRNA duplexes, and RNA was harvested 24 h later.
Ae. aegypti Jane embryos were injected with antisense oligonucleotides as described above. However, the engorged female mosquitoes were kept at 25 °C and 70% humidity and allowed to lay eggs for 45 min. Injection was performed with a FemtoJet 4x (Eppendorf) with 1200 hPa pressure. Injected embryos were then transferred to a wet Whatman paper and kept at 27 °C and 80% humidity for the indicated times. Per experiment, 50 to 150 embryos were injected per condition 1-2 h post-egg laying.
Scoring of embryo development and hatching
Injected embryos were allowed to develop for 2.5 days after injection on a moist Whatman paper and then fixed in 4% paraformaldehyde for 8 h to overnight. Afterwards, the pigment of the endochorion was bleached with Trpis solution36 (0.037 M sodium chlorite, 1.45 M acetic acid) for 24 to 48 h. Embryos were washed 5 times in PBS and images were taken with a EVOS FL imaging system (Thermo Fisher Scientific). Embryos with evident larval segmentation (head, fused thoracical elements and abdomen) were scored as fully developed and embryos without any evident structure of the ooplasm as undeveloped. Individuals that showed first signs of structural rearrangements of the ooplasm but did not complete larval segmentation were scored as intermediate (see Supplementary Fig. 2e). To avoid biases, the scoring was performed blindly. Six independent experiments were performed using the maximum number of embryos that was practically feasible.
Hatching rate of injected embryos was analysed at 4 days post-injection. Embryos were kept moist for two days and then allowed to slowly dry for the rest of the period. The embryos were transferred to water and then forced to hatch by applying negative pressure for a period of 30 min. The number of hatched L1 larvae was counted immediately afterwards. Five independent experiments were performed with numbers as practically feasible.
Poly(A) tail length (PAT) assay
Ligation-mediated (LM) PAT assay was performed as published37, and RACE PAT as published38. Superscript IV Reverse transcriptase (Invitrogen) was used for cDNA synthesis. RACE-PAT products were amplified by PCR with 3 min initial denaturation at 95 °C, 35 cycles of 30 s denaturation, 30 s annealing at 60 °C and 1 min extension at 72 °C, and final extension for 5 min, while PCR for LM-PAT was performed likewise, but with 2 min extension each cycle. PCR products were separated on agarose gel and stained with ethidium bromide. As control, Sindbis virus poly(A) tails were compared between in vitro transcribed RNA, produced as described previously39,40, and total RNA isolated from SINV-infected Aag2 cells.
Sequence logo
Repeat monomers from the satellite repeat loci in Ae. aegypti, Ae. albopictus, and Cx. quinquefasciatus were extracted manually from the current genome annotations obtained from VectorBase (Aedes aegypti Liverpool AaegL5, Aedes albopictus Foshan AaloF1, Culex quinquefasciatus Johannesburg CpipJ2). A repeat unit was defined as the sequence starting from the first tapiR1 nucleotide until one nucleotide upstream of the next tapiR1 sequence. Sequences were aligned using MAFFT (v7.397)41 (with options –genafpair –leavegappyregion --kimura 1 --maxiterate 1000 --retree 1) and the sequence logo was constructed with the R package ggseqlogo42.
Small RNA sequencing
Small RNAs from Aag2 cells (input) or PIWI immunoprecipitations were cloned with the TruSeq small RNA sample preparation kit (Illumina) according to the manufacturer’s instructions. For the input sample, size selected 19- to 33-nt small RNAs purified from polyacrylamide gel were used to construct the library as described43, whereas IP samples were not extracted from gel but isolated as described32. Libraries were sequenced on an Illumina HiSeq 4000 instrument by Plateforme GenomEast (Strasbourg, France). Sequenced libraries are available under BioProject number PRJNA594491.
mRNA sequencing
RNA was isolated from Aag2 cells 48 h after AO transfection (3 independent experiments), or from embryos at 20.5 h after AO injection (~50 embryos pooled per experiment from 5 independent experiments) with RNAsolv reagent following standard phenol-chloroform extraction. Polyadenylated RNAs were extracted and sequencing libraries were prepared using the TruSeq stranded mRNA Library Prep kit (Illumina) following the manufacturer’s instructions, and sequenced on an Illumina HiSeq 4000 instrument (2x50 bases). Sequenced libraries are available under BioProject number PRJNA482553.
Analysis of mRNA sequencing
Reads were mapped to the Ae. aegypti genome AaegL5 as provided by VectorBase (https://www.vectorbase.org) with STAR (version 2.5.2b)44 in 2-pass mode: first mapping was done for all samples (options: --readFilesCommand zcat --outSAMtype None --outSAMattrIHstart 0 --outSAMstrandField intronMotif), identified splice junctions were combined (junctions located on the mitochondrial genome were filtered out, as these are likely false positives), and this list of junctions was used in a second round of mapping (with –sjdbFileChrStartEnd), using default parameters as above. Reads were quantified with the additional option –quantMode GeneCounts to receive reads per gene. Alternatively, reads were quantified on TEfam transposon consensus sequences (https://tefam.biochem.vt.edu/tefam/get_fasta.php) with Salmon (v.0.8.2)45, default settings and libType set to “ISR”. Statistical and further downstream analyses were performed with DESeq246 from Bioconductor. Significance was tested at an FDR of 0.01 and a log2 -fold change of 0.5. tapiR1 target sites were predicted with the online tool from RNAHybrid47 with helix constraints from nucleotide two to seven, and no G:U wobble allowed in the seed. Predictions were made on the AaegL5.1 geneset as provided by VectorBase, and on TEfam transposon consensus sequences.
For Fig. 3h, publicly available sequencing datasets48 (accession numbers: SRR923702, SRR923826, SRR923837, SRR923853, SRR923704) were mapped and quantified as described above. Genes were categorized based on their expression in embryos at 0-2 h vs. 12-16 h post egg-laying. Genes not detected in the 0-2 h sample were defined as purely zygotic, and genes that did not increase or decrease by more than log2(0.5) as maternal stable. Genes that changed in expression by more than log2(0.5), log2(2), and log2(5) from 0-2 h to 12-16 h were categorized as maternal unstable fraction (decreased expression), or as genes that are maternally provided but are also transcribed by the zygote (increased expression). tapiR1 targets were defined as genes that were significantly upregulated at least two-fold in tapiR1 AO injected embryos and harbour a predicted tapiR1 target site (mfe <= -24).
A list of publicly available datasets that were used in this study is provided in Supplementary Table 6.
The code is available at https://github.com/RebeccaHalbach/Halbach_tapiR_2020.git.
Analysis of small RNA sequencing
3’ sequencing adapters (TGGAATTCTCGGGTGCCAAGG) were trimmed from the sequence reads with Cutadapt (version 1.14)49 and trimmed reads were mapped with Bowtie (version 0.12.7)50 to the Aedes aegypti LVP_AGWG genome sequence AaegL5.1 obtained from VectorBase with at most 1 mismatch. Reads that mapped to rRNAs or tRNAs were excluded from the analyses. Alternatively, 3’sequencing adapters ((NNN)TGGAATTCTCGGGTGCCAAGGC) and three random bases were trimmed from publicly available datasets from Ae. aegypti somatic and germline tissues51 (SRR5961503, SRR5961504, SRR5961505, SRR5961506) and then processed as described above. Oxidized libraries, IPs and input sample were normalized to the total number of mapped reads, all other libraries to the total number of miRNAs (in millions). piRNAs that were at least two-fold enriched in a PIWI-IP compared to the corresponding input sample and were present with at least 10 reads per million (rpm) in the IP sample were considered PIWI-bound. Mapping positions were overlapped with basefeatures and repeatfeatures retrieved from VectorBase and counted with bedtools52. Reads that mapped to two or more features were assigned to only one feature with the following hierarchy: open reading frames > non-coding RNAs (incl. lncRNAs, pseudogenes, snoRNAs, snRNAs, miRNAs) > LTR retrotransposons > Non-LTR retrotransposons (SINEs, LINEs, Penelope) > “Cut and paste” DNA transposons > other DNA transposons (Helitrons, MITEs) > satellite and tandem repeat features > DUST > other /unknown repeats. Accordingly, reads that mapped to a repeat feature and an intron or UTR were classified as repeat-derived, whereas all other reads mapping to introns or UTRs were considered as gene-derived. Positions not overlapping with any annotation were summarized as “other”. Results were then visualized with ggplot253, or Gviz54 in R.
The code is available at https://github.com/RebeccaHalbach/Halbach_tapiR_2020.git.
Extended Data
Extended Data Figure 1. Expression of piRNAs from a satellite repeat locus.
(a) Fraction of siRNAs and piRNAs mapping on genomic features in adult Ae. aegypti female ovaries (germline) or carcasses (soma). Small RNAs that overlapped multiple features were assigned to only one category (see Methods). The leftmost bar depicts the fraction of each feature category in the genome. (b) Read length distribution of tapiR1 and 2 in Aag2 cells, and adult germline and somatic tissues (β-eliminated, or untreated).
Extended Data Figure 2. tapiR1 and 2 are expressed in Ae. aegypti mosquitoes and associate with Piwi4.
(a,d,e) Northern blot of tapiR1 and 2 in different tissues of adult mosquitoes (a), upon dsRNA-mediated knockdown of individual PIWI genes (d), and upon knockdown of miRNA and siRNA pathway genes (e), or a control dsRNA treatment (dsFLuc and dsRLuc) in Aag2 cells. U6 snRNA or ethidium bromide-stained rRNA serves as loading controls. (b) Western blot analysis of the indicated PIWI proteins before (input) and after immunoprecipitation (IP) used for the small RNA northern blot of panel c. An IP with empty beads serves as negative control. Tubulin was used to control for non-specific binding. (c) Immunoprecipitation of the indicated PIWI proteins from Aag2 cells followed by northern blot analyses for tapiR1 and 2.
Extended Data Figure 3. Expression of tapiR1 is conserved in culicine mosquitoes and is independent of AAEL017385.
(a) Schematic representation of the tapiR satellite repeat locus in Ae. aegypti, Ae. albopictus and Cx. quinquefasciatus. Numbers indicate repeat lengths, and, for Cx. quinquefasciatus, lengths of deviating repeat monomers. (b-d) Sequence logos constructed from all individual tapiR repeat units in Ae. aegypti (b), Ae. albopictus (c), or Cx. quinquefasciatus (d). Gaps in the sequence logos mainly arise due to size heterogeneity in few repeat monomers. (e) Evolutionary relationships of dipterous genera based on ref.10. Bar lengths are arbitrary and do not reflect evolutionary distances. (f) Northern blot of tapiR1 in Aag2 cells treated with dsRNA targeting different transcripts of AAEL017385 (indicated in panel g), or, as control, firefly luciferase (FLuc). Ethidium bromide stained rRNA serves as loading control. (g) Top panel: Schematic representation of AAEL017385 and the tapiR satellite repeat locus. The primer used for 3’ RACE, and positions targeted by dsRNA in panel F are indicated with an arrow and wavy lines, respectively. Bottom panel: 3’ RACE analysis of AAEL017385. Indicated are sequences from the current AaegL5 genome annotation and RACE PCR products. The sequences of the 5’ terminal part tapiR1 and 2 repeats are highlighted with colours. (h) Northern blot of a potential tapiR1/2 precursor transcript. (i) RNA-seq read coverage of the tapiR repeat locus and AAEL017385 (top panel), and sashimi plot indicating spliced reads (bottom panel).
Extended Data Figure 4. Antisense oligonucleotides relieve tapiR1-mediated silencing.
(a) Luciferase assay of reporters with a fully complementary target site for tapiR1 (left panel) or tapiR2 (right panel) in the 3’UTR. Aag2 cells were co-transfected with the reporter and increasing amounts of a fully 2’O-methylated antisense tapiR RNA oligonucleotide (AO), or a control AO. Firefly luciferase activity was normalized to the activity of a co-transfected Renilla luciferase reporter. Indicated are mean, standard deviation and individual measurements from a representative experiment measured in triplicate wells. (b) Northern blot of tapiR1, tapiR2, and three different miRNAs in Aag2 cells upon treatment with the indicated concentrations of tapiR1, tapiR2, or control AO. Ethidium bromide-stained rRNA serves as loading control.
Extended Data Figure 5. Renilla luciferase contains a functional tapiR1 target site.
(a) Schematic representation of the different reporter constructs used in this study. pMT: metallothionein promoter; RNAPIII: RNA polymerase III reporter. (b) Representative northern blot (right panel) and quantification of RNAPIII reporters carrying the indicated tapiR1 target sites. Values are normalized to a non-targeted transfection control. Mean, standard deviation, and individual measures of three independent experiments (indicated with colours) quantified in triplicates are shown. The panels are split to reflect that the samples were loaded at different locations of the same gel. (c) Schematic representation of predicted tapiR1 target sites and minimum free energy of the indicated structures in the coding sequences of Renilla luciferase (RLuc) or firefly luciferase (FLuc). Numbers indicate the position of the targets relative to the first nucleotide in the ORFs. (d) Luciferase assay of Aag2 cells transfected with reporters carrying either a scrambled (scr) site or the predicted target site from firefly luciferase (left panel) or Renilla luciferase (right panel) from panel (c) in the 3’UTR of FLuc. (e) Luciferase activity of FLuc or RLuc constructs with synonymous mutations introduced into the predicted tapiR1 target site (ΔtapiR1 site) and the parental clones. (f) Luciferase assay of reporters carrying target sites for tapiR1 as indicated in panel c in the 3’UTR of either the parental firefly luciferase, or the ΔtapiR1 firefly luciferase version. (g) Reporter assay with luciferase carrying tapiR1 target sites with single mismatches in the 3’UTR as used in Extended Data Fig. 6b, using RLuc with a mutated tapiR1 target site (ΔtapiR1 site) for normalization. Left panel is a zoom to the x-axis of the right panel. Shown are mean, standard deviation and individual measurements from representative experiments performed with at least two different clones per construct, and each measured in triplicate wells.
Extended Data Figure 6. tapiR1 uses a G:U wobble sensitive seed sequence for target recognition.
(a) Schematic representation of the reporter constructs used in panel b and Fig. 2. Numbers indicate the position of the mismatch relative to the 5’ end of the piRNA. (b) Luciferase activity of reporters carrying a tapiR1 target site with single mismatches. (c) Luciferase activity of reporters with the tapiR1 target site from RLuc and indicated mismatches in the 3’UTR of FLuc (left panel). Predicted tapiR1/target RNA duplexes are presented in the right panel. (d) Luciferase activity of tapiR1 reporters carrying mismatches or G:U wobble basepairs at the indicated positions. Firefly luciferase activity was normalized to the activity of a co-transfected Renilla luciferase reporter to control for differences in transfection efficiencies. Data represent mean, standard deviation and individual measurements of representative experiments with two independent clones per construct and measured in triplicates wells.
Extended Data Figure 7. Validation of tapiR1 target genes.
(a) Predicted structures and minimum free energy of tapiR1/target RNA duplexes analysed in panel b. (b) Luciferase assay of reporters carrying the predicted target sites from panel a in the 3’UTR of firefly luciferase. Firefly luciferase activity was normalized to the activity of a co-transfected Renilla luciferase reporter to control for differences in transfection efficiencies. Indicated are mean, standard deviation and individual measurements from representative experiments performed with one to three independent clones per construct and measured in triplicate wells. (c) AAEL001741, AAEL017422, and AAEL000453 were annotated in the previous AaegL3 gene set, but not in the current AaegL5 gene set. Read coverage in tapiR1 AO and control AO treated Aag2 cells at these genomic regions suggests that these regions are actively transcribed but repressed by tapiR1. Red boxes indicate the positions of tapiR1 target sites.
Extended Data Figure 8. tapiR1/Piwi4 silence gene expression in Aag2 cells.
(a) log2 mRNA expression of transposable elements in Aag2 cells treated with a tapiR1-specific antisense oligonucleotide (AO) or control AO. Depicted are the means of three biological replicates. A pseudo-count of one was added to all values to plot values of zero. Diagonal lines represent a -fold change of two. Significance was tested at an FDR of 0.01 and a log2 -fold change of 0.5. (b) Luciferase assay of reporters harbouring different tapiR1 target sites (from Extended Data Fig. 7a) in the 3’UTR of firefly luciferase. Firefly luciferase activity was normalized to the activity of a co-transfected Renilla luciferase reporter to control for differences in transfection efficiencies. Data represent mean, standard deviation and individual measurements of representative experiments measured in triplicate wells. (c,d) RT-qPCR of tapiR1 target genes upon dsRNA-mediated knockdown of FLuc (control), Piwi4, or Ago1 in Aag2 cells (c), or after treatment with different concentrations of control, tapiR1, or tapiR2 AO (d). Depicted are mean, standard deviation, and individual measurements of a representative experiment as measured from triplicate wells (c), or from duplicate wells (d). Even skipped (eve) does not harbour a tapiR1 target site and serves as control. (e) Violin plot of log2 -fold changes in mRNA expression of all genes upon tapiR1 or control AO treatment in Aag2 cells (left) and mosquito embryos (right), either with or without predicted tapiR1 target site. (f) log2 -fold changes in RNA expression of genes upon treatment with tapiR1 or control AO in Aag2 cells (left) and mosquito embryos (right) plotted against the minimum free energy of predicted tapiR1/target RNA duplexes. Blue dots indicate target sites that were confirmed to be functional in luciferase reporter assays, and red dots indicate target sites that were not functional (see Extended Data Fig. 7b).
Extended Data Figure 9. tapiR1 regulates gene expression in mosquito embryos.
(a) Western blot analysis of phosphorylated RNA polymerase II (Ser2, middle panel) and Tubulin (bottom panel) in embryos at the indicated timepoints after egg-laying, and corresponding stem-loop RT-qPCR of tapiR1 measured in technical duplicates (top panel). (b,c) Northern blot of tapiR1 and 2 in developmental stages of Ae. aegypti mosquitoes (b), or at different time points after blood feeding (c). U6 snRNA (B) or ethidium bromide-stained rRNA (c) were analysed to verify equal loading. (d,e) RNA-seq read counts on Piw4 in the indicated adult tissues (d) or developmental stages (e). Libraries used for these analyses are listed in Supplementary Table 6. (f) log2 mRNA expression of transposable elements in embryos injected with tapiR1 or control AO. Mean counts of five biological replicates are shown. Significance was tested at an FDR of 0.01 and a log2 -fold change of 0.5. Diagonal lines indicate a -fold change of two. (g,h) RT-qPCR of the indicated tapiR1 target genes at 9 h after injection of tapiR1 or control AOs (g), or dsRNA-mediated knockdown of FLuc (control), Piwi4, or Ago1 (h) 21 h after injection in embryos. Mean, standard deviation, and individual measurements of a representative experiment are presented. Even skipped (eve) is not a tapiR1 target gene and serves as negative control. (i) Overlap of upregulated tapiR1 target genes (log2 -fold change ≥1, with a predicted target site with mfe ≤ 24) in Aag2 cells and Ae. aegypti embryos.
Extended Data Figure 10. tapiR1 regulates gene expression at a post-transcriptional level.
(a) Schematic representation of different modes of silencing of all three small RNA silencing pathways in Drosophila. (b) Top panel: Sequences of tapiR1, target gene AAEL026349, an siRNA targeting the gene at the same position, and Sanger sequencing results of 5’ RACE of Aag2 cells treated with tapiR1 AO and siRNAs. The tapiR1 target site is indicated in blue, RACE sequencing adapter in yellow, gene sequence in dark grey. The predicted slice site between nucleotides 10 and 11 is marked with a red vertical line. Bottom panel: summary of the results from 5’ RACE in the indicated conditions. Numbers refer to the number of sequenced clones with the 5’ RACE adapter ligated to the predicted slice site, and the total number of sequenced clones between brackets. (c) Small RNA coverage in Aag2 cells (unnormalized) and individual reads (direction of the arrow indicates the strand) on tapiR1 target genes. Red boxes indicate positions of tapiR1 target sites on the mRNA. (d) Schematic representation (top panel) and luciferase expression (bottom panels) of IRES-containing reporter constructs. Depicted are mean, standard deviation, and individual measurements of a representative experiment performed in triplicate wells with two different reporter clones. The bottom right panel illustrates firefly luciferase (FLuc) activity normalized to Renilla luciferase (RLuc); raw luciferase counts from the same experiment are shown in the left (FLuc) and middle (RLuc) panels. (e) Luciferase activity of a reporter harbouring the tapiR1 target site of AAEL001555 in the 3’UTR of FLuc upon dsRNA-mediated knockdown of the indicated genes. Symbols are color-coded according to the indicated RNA decay pathways. FLuc expression was normalized to RLuc expression to control for differences in transfection efficiencies and expressed relative to non-targeting control dsRNA (Sindbis virus dsRNA). Depicted are mean and standard deviation of one experiment performed in triplicates. Horizontal lines indicate a -fold change of 1 and 1.5. (f) AAEL008511 target gene expression as measured by RT-qPCR upon knockdown of the indicated genes. Primer sets located 5’ (upstream) and 3’ (downstream) to the tapiR1 target site were used for PCR. Mean, standard deviation, and individual measurements of one out of three experiments performed in triplicate are shown. The other two experiments are presented in Supplementary Fig. 2f,g. The horizontal line indicates a -fold change of two. (g) Schematic illustration of the two poly(A) tail length (PAT) assays and expected results of genes with increasing poly(A) tail lengths. LM-PAT, ligation mediated-PAT; RACE-PAT, Rapid amplification of cDNA ends-PAT. (h-j) Electrophoretic analysis with ethidium bromide-stained agarose gels of a LM-PAT assay (h), and RACE-PAT assay (i) of different tapiR1 target genes upon treatment with two concentrations of tapiR1 or control AO. As positive control, poly(A) tail length was measured from Sindbis virus (SINV) RNA in vitro transcribed from a plasmid, or from in infected Aag2 cells where the poly(A) tail is elongated during viral replication55 (j).
Supplementary Material
Acknowledgments
We thank past and current members of the laboratory for discussions. We are grateful to A.B. Crist and A. Baidaliuk (Institut Pasteur, Paris, France) for their help with mosquito rearing and embryo injections, and to C. Bourgouin and N. Puchot (Institut Pasteur, Paris, France) for assistance with the microinjection apparatus. We thank B. Dutilh (Radboud University Medical Center, Nijmegen, The Netherlands) for discussions about analyses of target site enrichment, and G.-J. van Gemert (Radboud University Medical Center, Nijmegen, The Netherlands) and M. Kristan (London School of Hygiene and Tropical Medicine, London, United Kingdom) for kindly providing mosquitoes. T. Möhlmann (Wageningen University, Wageningen, The Netherlands) is acknowledged for collecting wild-caught mosquito samples. The following reagent was provided by the NIH/NIAID Filariasis Research Reagent Resource Center for distribution by BEI Resources, NIAID, NIH: Aedes aegypti, Strain Black Eye Liverpool, Eggs, NR-48921. Sequencing was performed by the GenomEast platform, a member of the ‘France Génomique’ consortium (ANR-10-INBS-0009). This work is financially supported by a Consolidator Grant from the European Research Council under the European Union’s Seventh Framework Programme (grant number ERC CoG 615680) and a VICI grant from the Netherlands Organization for Scientific Research (grant number 016.VICI.170.090). A stay of R.H. at Pasteur Institute, Paris, France was supported by ERASMUS+.
Footnotes
Author contributions
R.H., P.M., and R.P.v.R designed the experiments and analysed the data. R.H. performed the computational analyses and most of the experiments, except for PIWI-IPs for small RNA sequencing (J.J. and E.T.), design and validation of PIWI antibodies (B.P.), and tissue isolations and blood feeding experiment (C.B.F.V. and C.J.K.). C.B.F.V. and C.J.K. provided wild-caught mosquito samples. I.R. assisted with the experiments, and S.H.M. and L.L. helped with optimizing embryo injections. R.H. and R.P.v.R. wrote the paper. All authors read and contributed to the manuscript.
Author information
The authors declare no competing financial interests.
Data and code availability
Raw sequence data is deposited in the NCBI Sequence Read Archive under BioProject numbers PRJNA482553 and PRJNA594491. The source code is available at https://github.com/RebeccaHalbach/Halbach_tapiR_2020.git.
References
- 1.Garrido-Ramos MA. Satellite DNA: An Evolving Topic. Genes (Basel) 2017;8 doi: 10.3390/genes8090230. pii: E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feliciello I, Akrap I, Ugarkovic D. Satellite DNA Modulates Gene Expression in the Beetle Tribolium castaneum after Heat Stress. PLoS Genet. 2015;11:e1005466. doi: 10.1371/journal.pgen.1005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li YX, Kirby ML. Coordinated and conserved expression of alphoid repeat and alphoid repeat-tagged coding sequences. Dev Dyn. 2003;228:72–81. doi: 10.1002/dvdy.10355. [DOI] [PubMed] [Google Scholar]
- 4.Pezer Z, Ugarkovic D. Satellite DNA-associated siRNAs as mediators of heat shock response in insects. RNA Biol. 2012;9:587–595. doi: 10.4161/rna.20019. [DOI] [PubMed] [Google Scholar]
- 5.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 6.Lau NC, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 7.Melters DP, et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013;14:R10. doi: 10.1186/gb-2013-14-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czech B, Hannon GJ. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem Sci. 2016;41:324–337. doi: 10.1016/j.tibs.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miesen P, Joosten J, van Rij RP. PIWIs Go Viral: Arbovirus-Derived piRNAs in Vector Mosquitoes. PLoS Pathog. 2016;12:e1006017. doi: 10.1371/journal.ppat.1006017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reidenbach KR, et al. Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC Evol Biol. 2009;9:298. doi: 10.1186/1471-2148-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plohl M, et al. Long-term conservation vs high sequence divergence: the case of an extraordinarily old satellite DNA in bivalve mollusks. Heredity (Edinb) 2010;104:543–551. doi: 10.1038/hdy.2009.141. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Lage A, Rodriguez-Farina F, Gonzalez-Tizon A, Mendez J. Origin and evolution of Mytilus mussel satellite DNAs. Genome. 2005;48:247–256. doi: 10.1139/g04-115. [DOI] [PubMed] [Google Scholar]
- 13.Chaves R, Ferreira D, Mendes-da-Silva A, Meles S, Adega F. FA-SAT Is an Old Satellite DNA Frozen in Several Bilateria Genomes. Genome Biol Evol. 2017;9:3073–3087. doi: 10.1093/gbe/evx212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D, et al. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science. 2018;359:587–592. doi: 10.1126/science.aao2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen EZ, et al. Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell. 2018;172:937–951. doi: 10.1016/j.cell.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto N, et al. Crystal Structure of Silkworm PIWI-Clade Argonaute Siwi Bound to piRNA. Cell. 2016;167:484–497. doi: 10.1016/j.cell.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Mohn F, Handler D, Brennecke J. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science. 2015;348:812–817. doi: 10.1126/science.aaa1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuter M, et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- 20.Goh WS, et al. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 2015;29:1032–1044. doi: 10.1101/gad.260455.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ugarkovic D. Functional elements residing within satellite DNAs. EMBO Rep. 2005;6:1035–1039. doi: 10.1038/sj.embor.7400558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vastenhouw NL, Cao WX, Lipshitz HD. The maternal-to-zygotic transition revisited. Development. 2019;146 doi: 10.1242/dev.161471. pii: dev161471. [DOI] [PubMed] [Google Scholar]
- 24.Rouget C, et al. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barckmann B, et al. Aubergine iCLIP Reveals piRNA-Dependent Decay of mRNAs Involved in Germ Cell Development in the Early Embryo. Cell Rep. 2015;12:1205–1216. doi: 10.1016/j.celrep.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan Q, Fallon AM. Small heat shock proteins distinguish between two mosquito species and confirm identity of their cell lines. Am J Trop Med Hyg. 1990;43:669–676. doi: 10.4269/ajtmh.1990.43.669. [DOI] [PubMed] [Google Scholar]
- 27.Vodovar N, et al. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS One. 2012;7:e30861. doi: 10.1371/journal.pone.0030861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miesen P, Girardi E, van Rij RP. Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res. 2015;43:6545–6556. doi: 10.1093/nar/gkv590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fansiri T, et al. Genetic mapping of specific interactions between Aedes aegypti mosquitoes and dengue viruses. PLoS Genet. 2013;9:e1003621. doi: 10.1371/journal.pgen.1003621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goertz GP, Vogels CBF, Geertsema C, Koenraadt CJM, Pijlman GP. Mosquito co-infection with Zika and chikungunya virus allows simultaneous transmission without affecting vector competence of Aedes aegypti. PLoS Negl Trop Dis. 2017;11:e0005654. doi: 10.1371/journal.pntd.0005654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohlmann TWR, et al. Community analysis of the abundance and diversity of mosquito species (Diptera: Culicidae) in three European countries at different latitudes. Parasit Vectors. 2017;10:510. doi: 10.1186/s13071-017-2481-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joosten J, et al. The Tudor protein Veneno assembles the ping-pong amplification complex that produces viral piRNAs in Aedes mosquitoes. Nucleic Acids Res. 2019;47:2546–2559. doi: 10.1093/nar/gky1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 34.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trpiš M. A new bleaching and decalcifying method for general use in zoology. Canadian J Zoology. 1970;48:892–893. [Google Scholar]
- 37.Murray EL, Schoenberg DR. Assays for determining poly(A) tail length and the polarity of mRNA decay in mammalian cells. Methods Enzymol. 2008;448:483–504. doi: 10.1016/S0076-6879(08)02624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salles FJ, Strickland S. Analysis of poly(A) tail lengths by PCR: the PAT assay. Methods Mol Biol. 1999;118:441–448. doi: 10.1385/1-59259-676-2:441. [DOI] [PubMed] [Google Scholar]
- 39.Hahn CS, Hahn YS, Braciale TJ, Rice CM. Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc Natl Acad Sci USA. 1992;89:2679–2683. doi: 10.1073/pnas.89.7.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Mierlo JT, et al. Novel Drosophila viruses encode host-specific suppressors of RNAi. PLoS Pathog. 2014;10:e1004256. doi: 10.1371/journal.ppat.1004256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagih O. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics. 2017;33:3645–3647. doi: 10.1093/bioinformatics/btx469. [DOI] [PubMed] [Google Scholar]
- 43.van Cleef KW, et al. Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res. 2014;42:8732–8744. doi: 10.1093/nar/gku528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akbari OS, et al. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 (Bethesda) 2013;3:1493–1509. doi: 10.1534/g3.113.006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011;17:3. 2011. [Google Scholar]
- 50.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis SH, et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat Ecol Evol. 2018;2:174–181. doi: 10.1038/s41559-017-0403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics. 2014;47:11 12 11–34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2016. [Google Scholar]
- 54.Hahne F, Ivanek R. Visualizing Genomic Data Using Gviz and Bioconductor. Methods Mol Biol. 2016;1418:335–351. doi: 10.1007/978-1-4939-3578-9_16. [DOI] [PubMed] [Google Scholar]
- 55.Frey TK, Strauss JH. Replication of Sindbis virus. VI. Poly(A) and poly(U) in virus-specific RNA species. Virology. 1978;86:494–506. doi: 10.1016/0042-6822(78)90088-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.