Abstract
Dopaminergic neurons (DANs) drive learning across the animal kingdom, but the upstream circuits that regulate their activity and thereby learning remain poorly understood. We provide the first synaptic-resolution connectome of the circuitry upstream of all DANs in a learning center, the mush-room body (MB) of Drosophila larva. We discover afferent sensory pathways and a large population of neurons that provide feedback from MB output neurons and link distinct memory systems (aversive and appetitive). We combine this with functional studies of DANs and their presynaptic partners and with comprehensive circuit modelling. We find that DANs compare convergent feedback from aversive and appetitive systems which enables the computation of integrated predictions that may improve future learning. Computational modelling reveals that the discovered feedback motifs increase model flexibility and performance on learning tasks. Our study provides the most detailed view to date of biological circuit motifs that support associative learning.
Introduction
To behave adaptively in an ever-changing environment, animals must be able to learn new associations between sensory cues (conditioned stimuli, CS) and rewards or punishments (aversive and appetitive unconditioned stimuli, US), and continuously update previous memories, depending on their relevance and reliability1–3.
Modulatory neurons (e.g. dopaminergic, DANs) convey information about rewards and punishments and provide the teaching signals for updating the valence associated with CS in learning circuits across the animal kingdom (e.g. the vertebrate basal ganglia1;4, or the insect mushroom body, MB3;5). The co-occurrence of CS and modulatory neuron activity tuned only to the received US can support simple associative memory formation6. To account for more complex behavioral phenomena, theories have been developed in which learning can be regulated by previously formed associations7;8. According to reinforcement learning theories, learning is driven by errors between predicted and actual US (prediction errors)7;8, which are thought to be represented by the activity of DANs1;4. Indeed, in many model organisms, the responses of modulatory neurons have been shown to be adaptive: in monkeys1, rodents4;9, and insects3;5;10;11. Despite recent progress3;4;9, the basic principles by which DAN activity is adaptively regulated and teaching signals are computed are not well understood.
A prerequisite for the adaptive regulation of modulatory neuron activity is convergence of afferent pathways that convey information about received US1;4 with feedback pathways that convey information about previous experience. A comprehensive synaptic-resolution connectivity map of the feedback circuits that regulate modulatory neurons would provide a basis for understanding how learning is adaptively regulated by prior learning, but it has previously been out of reach.
Insects, especially their larval stages, have small and compact brains that have recently become amenable to large-scale electron microscopy (EM) circuit mapping12;13. Both adult3;6;14 and larval15 insect stages possess a brain center essential for associative learning, the MB. The MB contains neurons called Kenyon Cells (KCs) that sparsely encode CS; MB modulatory neurons (collectively called MBINs) that provide the teaching signals; and MB output neurons (MBONs) whose activity represents learnt valences of stimuli3;6;14;15. In the Drosophila larva, most modulatory neurons are dopaminergic (DANs), some are octopaminergic (OANs), and some have unidentified neurotransmitters (simply called MBIN)15. Modulatory neurons and MBONs project axon terminals and dendrites, respectively, onto the KC axons in a tiled manner, defining MB compartments, in both adult3 and larval15 Drosophila. In adult Drosophila, it has been shown that co-activation of KCs and DANs reduces the strength of the KC-MBON synapse in that compartment3;16. Different compartments have been implicated in the formation of distinct types of memories, for example aversive and appetitive, or short- and long-term3;14;15;17;18. However, despite a good understanding of the structure and function of the core components of the MB in both adult3;6;14 and larval Drosophila15, the circuits presynaptic to modulatory neurons that regulate their activity have remained relatively uncharacterized.
We therefore reconstructed all neurons presynaptic to all modulatory neurons in an EM volume that spans the entire nervous system of a first instar Drosophila larva, in which we had previously reconstructed all the core components of the MB12. We also determined which individual modulatory neurons are activated by punishments and reconstructed their afferent US pathways from nociceptive and mechanosensory neurons. We characterized the neurotransmitter profiles of some of the neurons in the network and functionally confirmed some of the identified structural connections. Finally, we developed a model of the circuit constrained by the connectome, the neurotransmitter data, and the functional data and used it to explore the computational advantages offered by the newly discovered architectural motifs for performing distinct learning tasks.
Results
Larval MB modulatory neurons for aversive and appetitive memory formation
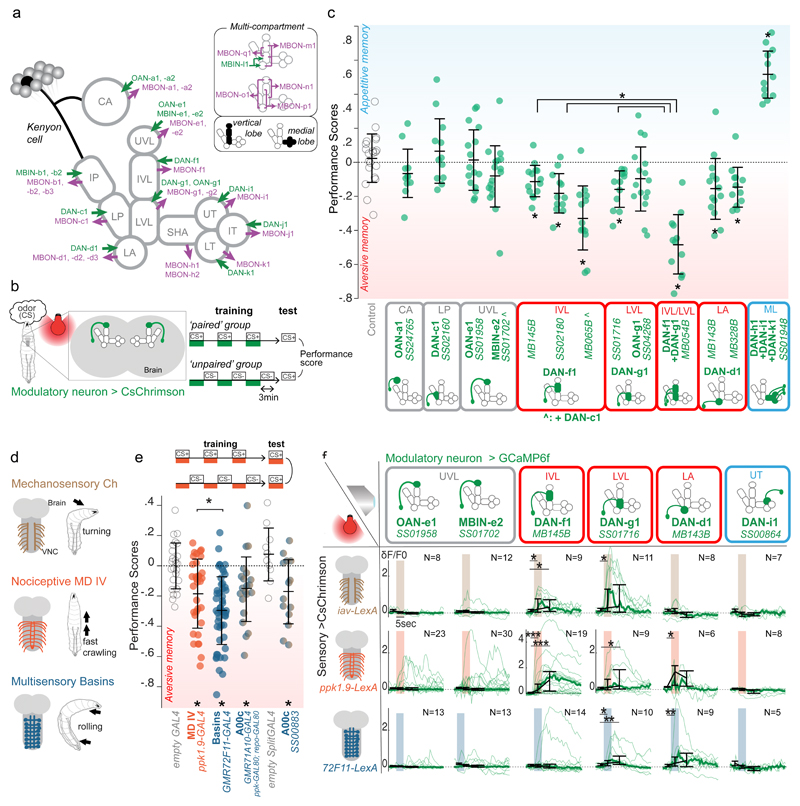
We begun with a functional characterisation of larval modulatory neurons and asked which ones signal punishment or reward. Activation of all DL-cluster DANs that target the vertical lobe, the lateral appendix, and the peduncle (with a broadly-expressing TH-GAL4 driver line) has been shown to induce aversive memory for paired odor19. Activation of individual larval PAM-cluster DANs that target the MB medial lobe has been shown to induce appetitive memory for paired odor20. However, the role of other individual modulatory neuron types was not known. We generated Split-GAL4 lines that drive expression selectively in one or two modulatory neurons per hemisphere (Fig. 1a, Extended Data 1, Supplementary Table).
Figure 1. Individual vertical lobe DANs can induce aversive memory and represent different kinds of punishments in Drosophila larva.
a Schematic diagram of the larval MB compartments with an example KC, all MBONs and modulatory neurons.
b Schematic diagram of the one-odor associative memory optogenetic training protocol. Individual performance score is the difference between odor preferences of larvae after ’paired’ or ’unpaired’ protocol.
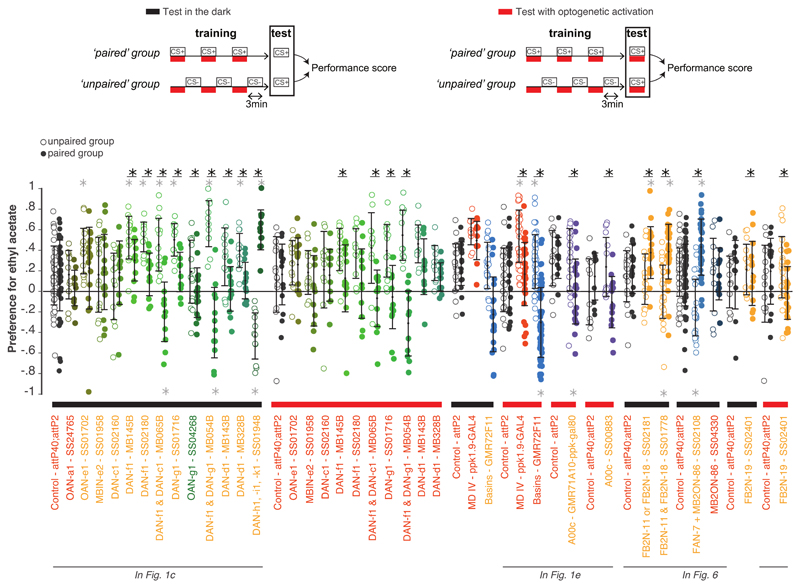
c Scores obtained with SplitGAL4-driven optogenetic activation of modulatory neurons in the training protocol shown in (b): Activating DANs of vertical (red) and medial (blue) lobe compartments induces aversive and appetitive memory, respectively. N=21,11,12,16,18,14,12,14,12,16,12,14,12,12 (see Supplementary SourceData_Figure1 for more details). Mean and standard deviations shown. *, **, ***, P<0.05, 0.01, 0.001, two-sided Mann-Whitney U test with Holm-Bonferroni correction in all legends unless otherwise stated.
d Different somatosensory neurons induce distinct innate escape responses23–25.
e Optogenetic activation of nociceptive neurons, Basins, or the Basin-target A00c induces aversive memory when paired with odor. (See Extended Data 2). N=25,33,52,21,14. Scores (computed as in b) are compared to the control group.
f ”Aversive DANs” respond to optogenetic activation of somatosensory neurons with specific tuning. Plots show calcium transients in selected modulatory neurons evoked by mechanosensory, nociceptive and Basin neurons activation. thin lines: averaged responses for one brain (3 repeats); thick lines: median across all animals. Black plots: median peak δF/F0 of individual curves in different time windows: 1 sec before, 1 sec during, and 2 sec following the stimulation. Error bars show the 25th and 75th percentile of peak δF/F0.
We then paired an odor (CS) with optogenetic activation of these modulatory neurons in a three-trial, one-odor, short-term memory associative memory paradigm (Fig. 1b).
We found that activation of DAN-f1 (innervating the intermediate vertical lobe), DAN-g1 (lower vertical lobe), or DAN-d1 (lateral appendix) established aversive memory for paired odor (Fig. 1c and Extended Data 2). In contrast, as previously reported15;20, activation of DANs that innervate the medial lobe led to the formation of an appetitive memory for paired odor (Fig. 1c and Extended Data 2). Thus, similar to findings in the adult fly3;17, larval DANs that innervate distinct lobes signal opposite valences.
Pairing of an odor with the activation of DAN-c1 (lower peduncle) or of the non-dopaminergic modulatory neurons induced neither appetitive nor aversive memory (Fig. 1c and Extended Data 2). Thus, our analysis revealed at least three functionally distinct classes of compartments in the larval MB: medial lobe compartments whose DANs can induce appetitive memory for paired odor; lateral appendix and lower and intermediate vertical lobe compartments whose DANs can induce aversive memory for paired odor; and others whose modulatory neurons were not sufficient to induce short-term memory (Fig. 1c).
Punishment encoding across larval MB modulatory neurons
Next, we asked whether there is any functional diversity within the population of DANs whose activation signals punishment.
Larvae sense multiple types of innately aversive somatosensory stimuli that evoke distinct types of innate avoidance and escape responses21–25 (Fig. 1d). Already the mildest of these punishments, vibration, that is transduced by mechanosensory neurons evokes a turning avoidance response and induces aversive memory for paired odor26. Fittingly, we found that optogenetic activation of nociceptive sensory neurons and of Basin interneurons (and their downstream A00c interneurons) that evokes more vigorous fast crawling and rolling escape, respectively, also induces aversive memory for paired odor (Fig. 1d-e and Extended Data 2).
We therefore asked how individual modulatory neurons respond to different punishment types by monitoring their calcium transients in response to optogenetic activation of specific somatosensory neurons. In each of the three DANs whose activation induced aversive memory for paired odor, we found reliable responses to at least two fictive punishment types. Each punishment type evoked reliable and statistically significant responses in at least two DANs, but each DAN’s tuning differs (Fig. 1f). Thus, these three DANs could combinatorially encode punishment type or salience.
For comparison, we also tested responses of a few modulatory neurons whose activation paired with odor did not induce short-term aversive memory and found they were not significantly activated by the fictive punishments (Fig. 1f).
EM reconstruction of all input neurons to MB modulatory neurons
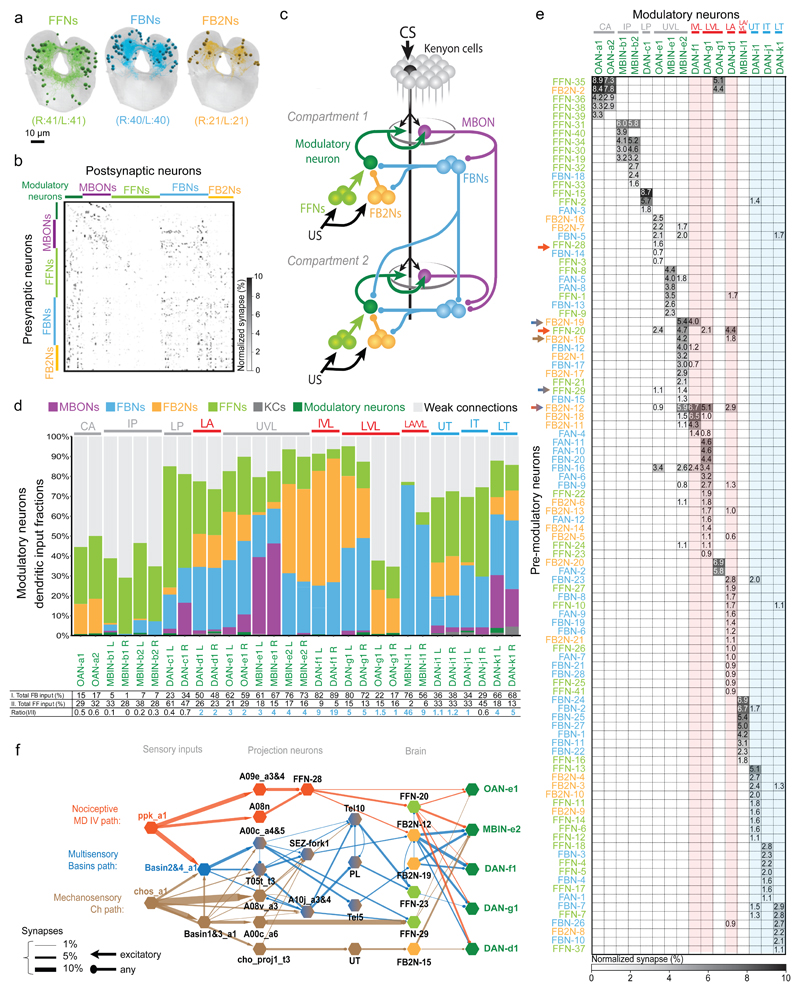
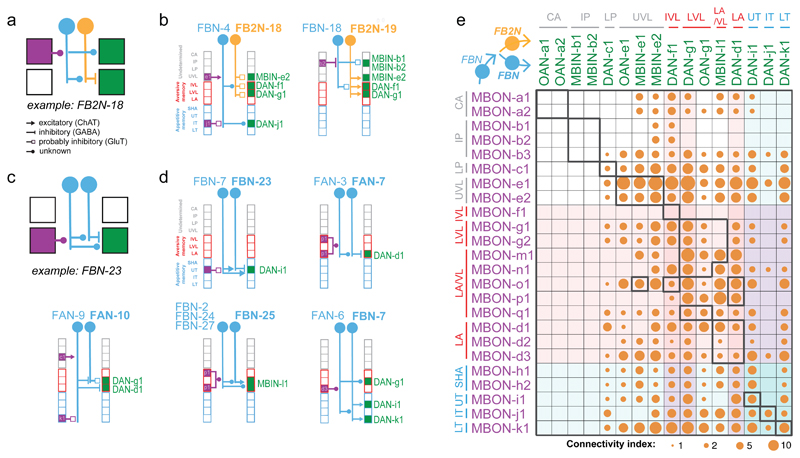
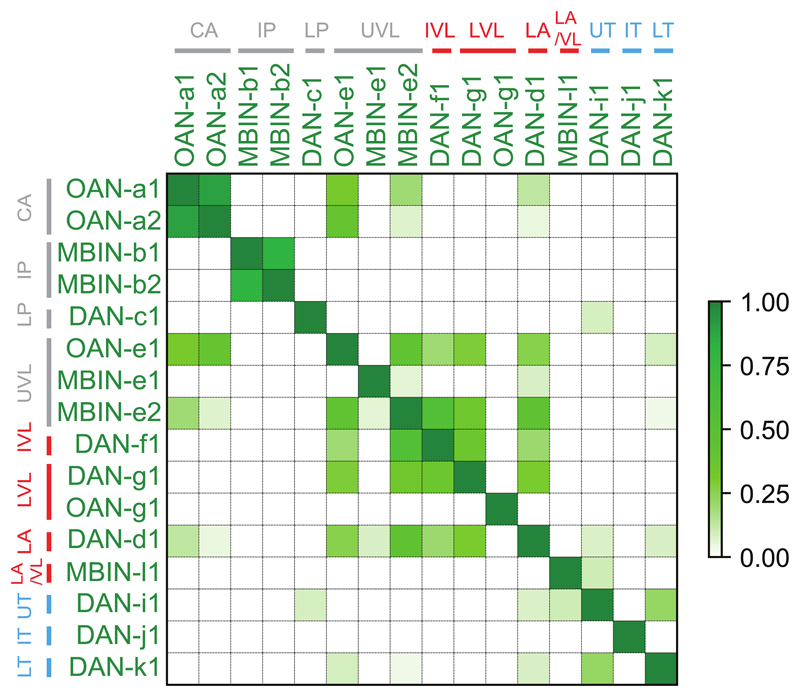
To provide a basis for understanding how the activity and function of modulatory neurons is regulated, we sought to comprehensively identify all the neurons presynaptic to them. We have previously reconstructed all of the KCs, MBONs, and modulatory neurons in an EM volume of a 1st instar larval nervous system12. Here we systematically reconstructed all neurons presynaptic to all modulatory neurons (i.e. pre-modulatory neurons) in the same EM volume (Fig. 2a-e). We identified 213 left-right homologous pairs and 5 unpaired pre-modulatory neurons. Out of these, 102 homologous pairs were reliably and strongly connected (see Material and Methods, Fig. 2a,b,e, Supplementary Adjacency Matrices 1-3, Supplementary Atlas). While the “other weakly connected partners” could also influence modulatory neuron activity, especially in combination with each other, we focus our study mainly on the 102 reliably and strongly connected partners.
Figure 2. Comprehensive EM reconstruction of pre-modulatory neurons reveals a multilayered recurrent architecture for regulating learning.
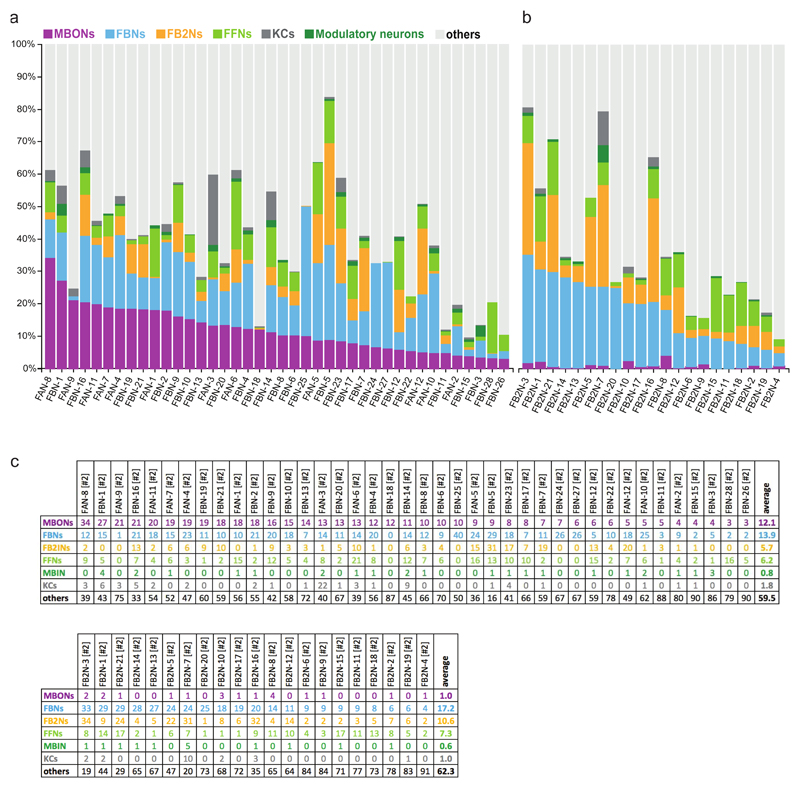
a Projections of EM reconstructions of 102 neuron pairs found to be strongly connected to modulatory neurons. The majority (61) relays inputs from MBONs: 40 FBNs pairs (light blue) ; and 21 FB2Ns (yellow). The remaining 41 are classified as FFNs (light green).
b Connectivity matrix showing normalized synaptic input (in %) each homologous pair of postsynaptic (columns) neurons receives from each pair of presynaptic (rows) neurons.
c Schematic wiring diagram of the Extended MB circuit.
d Fraction of total dendritic input each modulatory neuron receives from different neuron types. Most DANs receive more than half of their input from MBON feedback pathways, whereas most OANs receive most input from weakly connected partners. Some DANs extend dendritic arbors to the KCs which accounts for KCs dendritic input; KCs axonic inputs are described elsewhere (Eichler et al. 2017). Red and blue, aversive and appetitive memory compartments, respectively, in all legends.
Bottom: Percent of inputs onto modulatory neurons from (I.) MBON, FBN, and FB2N, (II.) FFNs, and their ratio. This ratio is greater than 1 in most cases.
e Connectivity matrix showing normalized synaptic input (expressed as % input) each modulatory neuron (columns) receives from each pre-modulatory neuron (rows).
Pre-modulatory neurons synapse onto a single or a few functionally related modulatory neuron(s) (See also Extended Data 3).
f US pathways converge with feedback pathways at modulatory neurons and at FB2Ns. Diagram shows the shortest identified US pathways from somatosensory neurons to vertical lobe modulatory neurons. Thickness of the arrow represents fraction of input.
We asked how the functional diversity of modulatory neurons relates to their input diversity. As expected, functionally distinct DANs receive inputs from distinct subsets of pre-modulatory neurons, and functionally similar DANs share a higher fraction of presynaptic partners with each other than with other DANs (Fig. 2e and Extended Data 3). Nevertheless, each modulatory neuron type, that is distinguishable based on the compartment it innervates, or based on neurotransmitter expression, receives input from a unique combination of neurons and thus potentially encodes a unique set of features.
Feedback neurons reveal a highly recurrent architecture for computing teaching signals
We aimed to characterize the pre-modulatory neurons based on the inputs they receive. We asked whether they convey information about previously formed memories (via feedback originating from MBONs), or about received US (via afferent input from sensory neurons), or both. Surprisingly, we found that the majority (61/102) of pre-modulatory neurons receive feedback input from MBONs (Fig. 2a-c, Extended Data 4a-c). 40 neuron pairs receive reliable direct input from MBONs, providing one-step feedback from MBONs to modulatory neurons (we call these one-step feedback neurons, FBNs, Fig. 2a-c, Extended Data 4a). Another 21 pre-modulatory neuron pairs receive reliable direct input from FBNs (but not MBONs) and provide two-step feedback from MBONs (two-step feedback neurons, FB2Ns, Fig. 2a-c, Extended Data 4b). The majority of FBNs also receive input from other FBNs, providing two-step, as well as one-step feedback (Fig. 2b-c, Extended Data 4a). The remaining pre-modulatory neurons do not receive reliable direct MBON or FBN input, so we classified them tentatively as “feedforward neurons” (FFNs, Fig. 2a-c).
To determine the likelihood that MBONs could influence modulatory neuron activity via the feedback pathways, we analyzed the fraction of total input that FBNs and FB2Ns receive from MBONs, and that modulatory neurons receive from feedback pathways. In previous studies we have demonstrated functional connections when neurons received 2% or more of their input from another neuron24;25. We found that FBNs receive on average 12% of their synaptic input from MBONs and 26% from MBONs and FBNs combined (Extended Data 4a,c). Similarly, FB2Ns receive on average 17% of their synaptic input from FBNs and 28% from FBNs and FB2Ns combined (Extended Data 4b-c). Based on these input fractions we expect that MBONs can significantly influence FBN and FB2N activity. Strikingly, we found that many modulatory neurons receive more than 50% of their total dendritic input from all feedback pathways combined (Fig. 2d). This suggests that modulatory neuron activity could be strongly influenced by MBON activity.
Multilevel convergence of afferent and feedback pathways
We investigated how the feedback pathways from MBONs converge with afferent pathways from US sensing neurons. We focused on the DANs that respond to somatosensory neuron activation (Fig. 1f) and asked whether they receive somatosensory and MBON input via distinct or overlapping pre-modulatory neurons. We had previously reconstructed all first-order neurons downstream of nociceptive and mechanosensory sensory neurons, and a subset of second- and third-order ones24;25. This enabled us to search for shortest pathways from these somatosensory neurons to modulatory neurons. While the pathways identified in this way represent only a subset of existing pathways, nevertheless, we were able to identify two-, three-, and four-step pathways from the somatosensory neurons to six different pre-modulatory neurons that target the vertical lobe and lateral appendix modulatory neurons: three FFNs and three FB2Ns (Fig. 2f, Supplementary Fig. 1, Supplementary Adjacency Matrix 1, Supplementary Atlas). Thus, the afferent US pathways converge with feedback pathways from MBONs at multiple levels, both onto the modulatory neurons themselves (via FFNs) and onto the pre-modulatory FB2Ns.
Modulatory neurons receive convergent one-step feedback from multiple MBONs innervating functionally distinct compartments
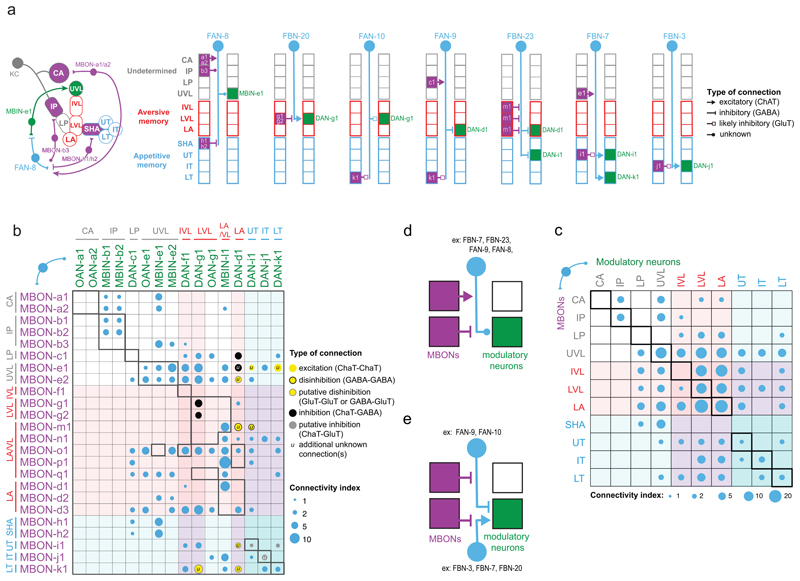
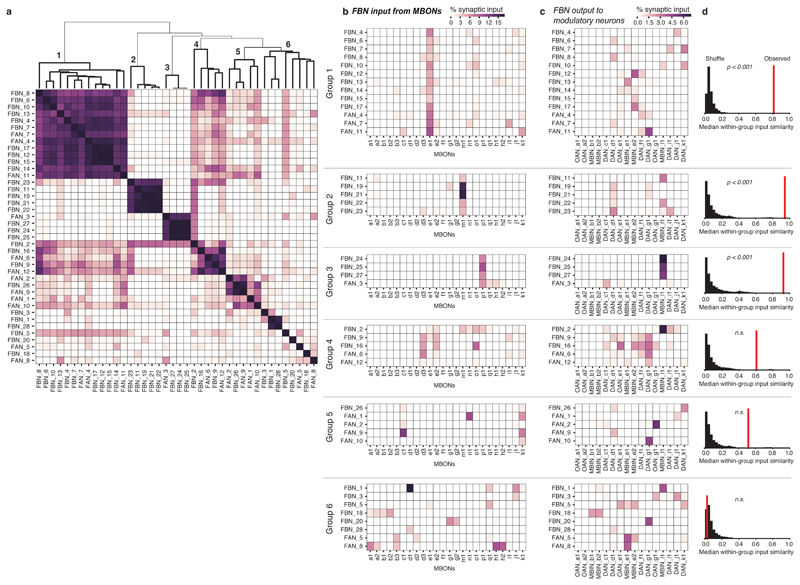
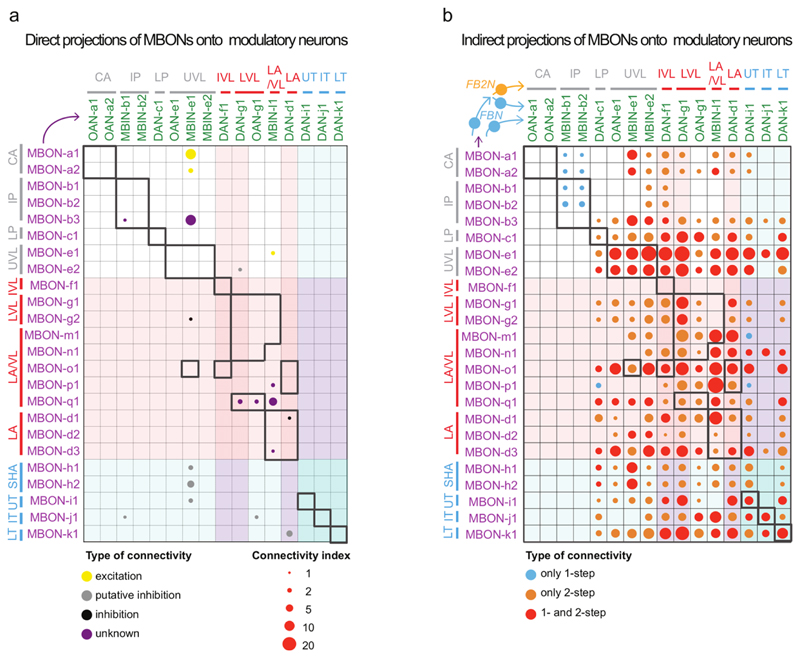
Next, we analyzed in more detail the types of one-step feedback motifs formed by FBNs (Fig. 2a-c, 3a-c, Extended Data 5-9, Supplementary Fig. 2a-b, Supplementary Adjacency Matrix 1, and Supplementary Atlas). We observed a surprising diversity of one-step FBNs that linked unique combinations of MBONs with unique combinations of modulatory neurons (See Fig. 3a for examples and Extended Data 5 for the complete dataset). Out of the 40 FBNs, 7 provide exclusively within-compartment feedback, and 13 provide exclusively cross-compartment feedback. Interestingly, many FBNs (17, Fig. 3a,d, Extended Data 5, Supplementary Fig. 2a) integrate input from MBONs of functionally distinct compartments (Fig. 1c, Extended Data 7). Almost all of these FBNs (at least 13/17) receive GABAergic (inhibitory) or glutamatergic (potentially also inhibitory27;28 in insects) input from MBONs innervating compartments, and cholinergic (excitatory) inputs from MBONs innervating other compartments (Fig. 3a,d, Extended Data 5, Supplementary Fig. 2a). The integration of inhibitory and excitatory inputs may enable these FBNs to more accurately read out the results of learning-induced plasticity by comparing its effects across compartments.
Figure 3. Modulatory neurons receive convergent one-step feedback from multiple MBONs from functionally distinct compartments.
a Diagram representing the connectivity of 7 example pairs of homologous FBNs. Boxes indicate separate MB compartments, with presynaptic MBON(s) on the left side, and postsynaptic modulatory neuron(s) on the right. When known (Extended Data 9 and Eichler et al. 2017), the neurotransmitter profiles of the MBONs and FBNs are indicated by the type of arrow. See Extended Data 5 for diagrams of each of the 40 FBNs.
b Connectivity matrix shows the one-step feedback connections between MBONs and modulatory neurons via FBNs, obtained by multiplying the MBON→FBN, and FBN→modulatory neuron normalized connectivity matrices. Connectivity indexes are the square roots of the matrix products. Colored circles indicate putative signs of connections if neurotransmitters of both MBONs and FBN(s) are known. Red and blue color shades, aversive and appetitive compartments, respectively. Pure within-compartment connections (excluding multi-compartment MBONs) are boxed in bold. The four pure within-compartment connections with known neurotransmitters are potentially inhibitory, in contrast to cross-compartment connections.
c Connectivity indexes from (b) pooled per compartment.
d-e Summary diagram of commonly observed convergence motifs. d, Many FBNs (at least 13) receive convergent inputs of opposite sign (i.e. excitatory and inhibitory) from functionally distinct compartments. e, DANs (e.g. DAN-g1 and DAN-i1) receive convergent inputs of opposite sign from functionally distinct compartments, via distinct one-step FBN pathways.
Not only do most FBNs receive input from multiple MBONs, but also most modulatory neurons receive input from multiple FBNs (Fig. 2e, 3a, Extended Data 6, Supplementary Fig. 2b). We therefore analyzed one-step connections from all MBONs to all modulatory neurons via FBNs. We found that most modulatory neurons receive one-step feedback from many MBONs and from each of the three functionally distinct regions of the MB: upper vertical lobe (unknown function), vertical lobe aversive memory compartments, and medial lobe appetitive memory compartments (Fig. 3b-c). This is in stark contrast to the direct connections from MBONs to modulatory neurons, which are sparse and connect few compartments (Extended Data 8a-b). Thus, the newly discovered FBNs greatly increase the connectivity between MBONs and modulatory neurons, enabling the output from functionally distinct regions of the MB to influence the activity of single modulatory neurons during memory formation.
A modulatory neuron receives inhibitory and excitatory feedback from compartments of opposite valence
To gain a better understanding of the way in which feedback motifs influence modulatory neuron activity, we were able to determine the neurotransmitter profiles of some of them (see Materials and Methods).
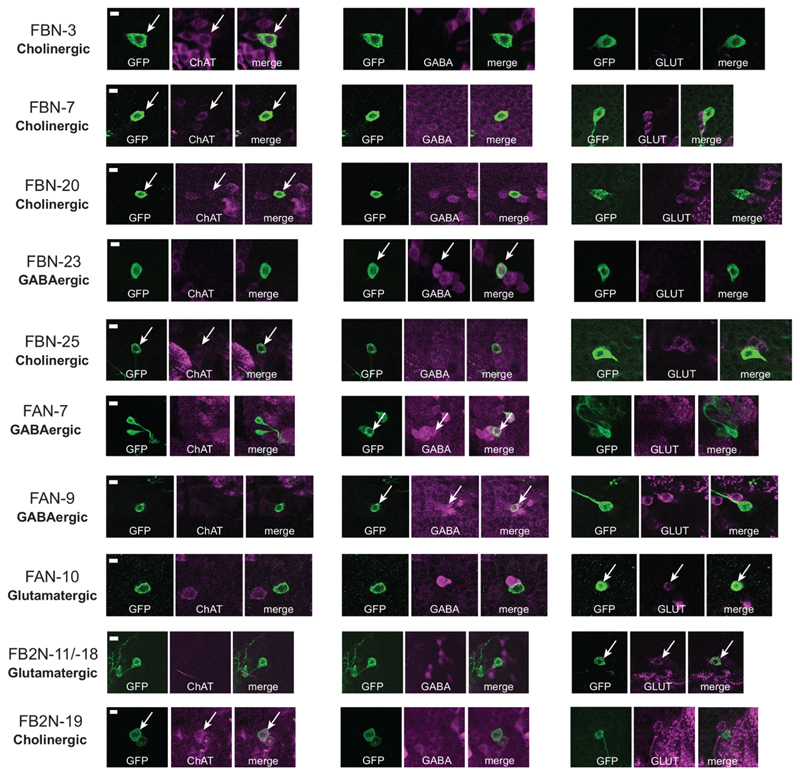
We found that four of the tested FBNs are cholinergic (i.e. excitatory), three are GABAergic (i.e. inhibitory), and one is glutamatergic (likely also inhibitory27;28, Fig. 3a and Extended Data 9).
For a few cases where we could identify the neurotransmitter profiles of both the MBON12 and the FBN in a one-step feedback connection, we attempted to predict the signs of these connections (Fig. 3b). Out of these, all of the true within-compartment feedback connections are potentially inhibitory (4/4), comprising a GABAergic or glutamatergic MBON and an excitatory FBN (Fig. 3b,e). In contrast, most of the (8/11) cross-compartment connections seem functionally excitatory, either disinhibitory (comprising an inhibitory MBON and an inhibitory FBN), or excitatory (comprising an excitatory MBON and an excitatory FBN, Fig. 3b and 3e). Furthermore, some modulatory neurons (e.g. DAN-g1 and DAN-i1) receive both potentially inhibitory feedback from their own compartment and potentially excitatory feedback from compartments of opposite valence (Fig. 3b,e).
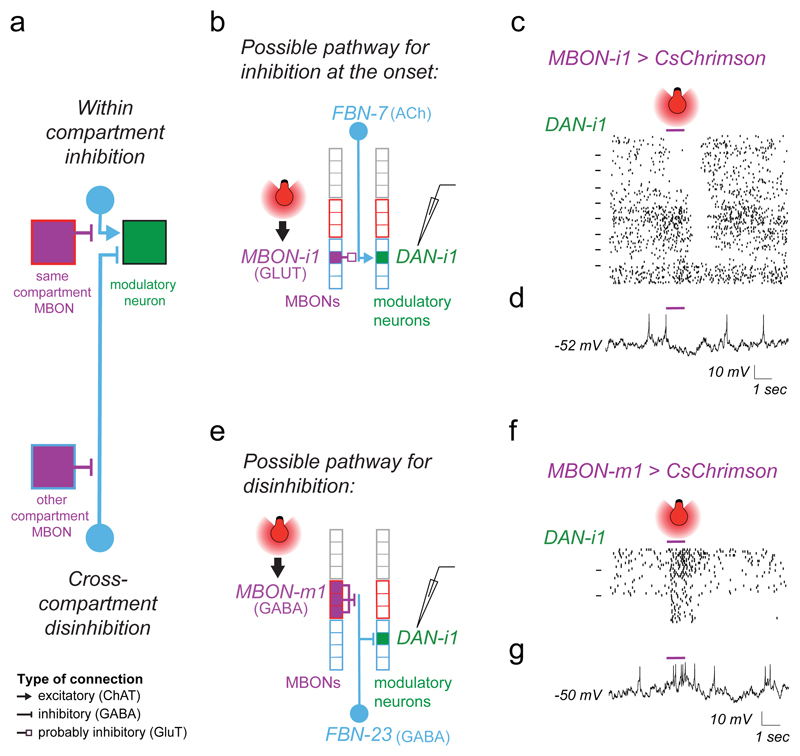
We wanted to functionally confirm the two types of predicted feedback connections onto the same DAN (Fig. 3e, 4a-g). DAN-i1 potentially receives inhibitory one-step feedback from MBON-i1 in its own compartment (Fig. 4a-b), and disinhibitory one-step feedback from MBON-m1 from compartments of opposite valence (Fig. 4a,e). Neither of these MBONs synapses directly onto DAN-i1. Furthermore, DAN-i1 receives two-step feedback from MBON-i1, but not from MBON-m1 (Fig. 5e). We activated these MBONs optogenetically while recording intracellularly from DAN-i1.
Figure 4. Functional inhibitory and excitatory feedbacks from compartments of opposite valence converge onto a DAN.
a Schematic diagram showing potentially inhibitory within-compartment and disinhibitory cross-compartment feedbacks converging onto a DAN. These two predicted connections are tested separately (b-d and e-g).
b The cholinergic FBN-7 downstream of the glutamatergic MBON-i1 could mediate inhibitory one-step within-compartment feedback onto DAN-i1.
c Whole-cell patch-clamp recording of DAN-i1 reveals inhibitory within-compartment feedback connection from MBON-i1 (purple bar). Action potentials from 180 traces from 9 animals are shown as raster plots. Dashes on the left separate distinct animals. We observed long-latency inhibitory responses in DAN-i1 at the onset (top 3/9 animals, 55.3 ± 17.3 ms, mean ± std) or offset (next 5/9 animals, 95.3 ± 43.5 ms) of MBON-i1 activation (purple bar)
d Example trace from (c) (more in Supplementary Fig. 3a).
e The GABAergic FBN-23 downstream of the GABAergic MBON-m1 could mediate disinhibitory cross-compartment one-step feedback onto DAN-i1.
f Whole-cell patch-clamp recording of DAN-i1 reveals a functionally excitatory cross-comparment connection from MBON-m1.
Action potentials from 30 traces from 3 animals are shown as raster plots. We observed a long-latency excitatory response in DAN-i1 at the onset of MBON-m1 activation (purple bar, 3/3 animals, 51.3 ± 7.7 ms). g Example trace from (f) (more in Supplementary Fig. 3d).
Figure 5. Two-step feedback from most MBONs to most modulatory neurons further increases inter-compartment connectivity.
a Schematic diagram of a two-step feedback motif involving an FBN (blue) and an inhibitory FB2N (yellow). The FBN provides one-step feedback to some compartments and two-step feedback to others via the FB2N. Arrowheads denote the type of synaptic connection in a-d.
b Two example two-step within-compartment feedback motifs involving FB2Ns with identified neurotransmitters.
c Schematic diagram of a two-step feedback motif involving two FBNs (blue) rather than an FBN and an FB2N. d Five example two-step within-compartment feedback motifs involving FBNs with identified neurotransmitters illustrate the diversity of two-step feedback connections.
e Most modulatory neurons receive two-step feedback from most MBONs. Connectivity matrix shows connections between MBONs and modulatory neurons via two-step feedback pathways, obtained by multiplying the MBON→FBN, FBN→FB2N/FBN and FB2N/FBN→modulatory neuron connectivity matrices. The connectivity indexes are the cubic root of the resulting matrix products.
Activating MBON-i1 evoked long-latency (55ms ± 17) inhibitory responses in DAN-i1 in 3/9 animals (Fig. 4c-d, Supplementary Fig. 3a), consistent with a polysynaptic connection mediated by an FBN (Fig. 3b and 4b). The inter-animal variability could be due to different baseline activity levels of the FBN (as illustrated in Supplementary Fig. 3b). In 5/9 animals (Fig. 4c and Supplementary Fig. 3a) we observed inhibitory responses to the offset of MBON-i1 activation only. These offset responses had a longer latency (95 ms ± 44) than the onset responses and could therefore be mediated by a longer two-step feedback pathway (See Supplementary Fig. 3c).
In contrast, we found that activating MBON-m1 evoked excitatory responses in DAN-i1 in 3/3 animals with a similar latency (47 ms ± 9) to the inhibitory responses evoked by MBON-i1 activation (Fig. 4e-g, Supplementary Fig. 3d).
In summary, we confirmed with physiological recording an inference we had made from structural connectivity and neurotransmitter information (Fig. 3e): functionally inhibitory and excitatory MBON connections from compartments of opposite valence converge onto the same DAN (Fig. 4a-g).
Two-step feedback further increases inter-compartment connectivity
We also analyzed the connections between all MBONs to all modulatory neurons via two-step pathways (Fig. 2a-c, 5a-e, Extended Data 4a-c, Supplementary Fig. 4, 5a-d, 6, Supplementary Adjacency Matrix 1, Supplementary Atlas). We found two-step feedback pathways from most MBONs to most modulatory neurons that further increase intercompartment connectivity (Fig. 5e, 5c, 5e and Extended Data 8b). We were able to determine neurotransmitter profiles for seven neurons that provide two-step feedback: three were cholinergic, two were GABAergic, and two were glutamatergic (Fig. 5b,d, Extended Data 9). In summary, we found a diverse set of two-step feedback motifs that could support within- and cross-compartment computations.
Feedback neurons can drive memory formation
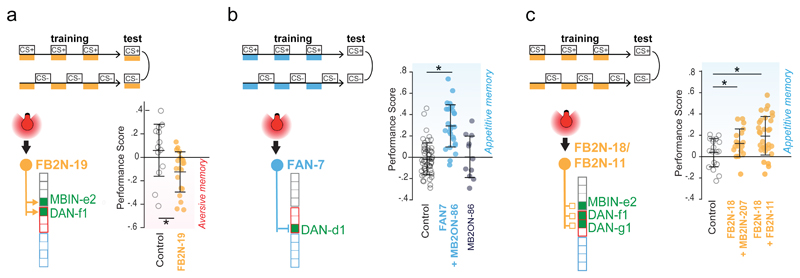
So far, we have shown that some indirect feedback connections from MBONs to DANs are functional (Fig. 4b-g). We also wanted to test whether the feedback neurons can sufficiently influence DAN activity to induce learning. We generated Split-GAL4 lines that drive expression selectively in one or very few neuron types for: a cholinergic FB2N, a glutamatergic FB2N, and a GABAergic FBN (Extended Data 1, 9, 10). These neurons project onto DANs whose activation can induce aversive memory for paired odor (Fig. 1c, Extended Data 2). We asked whether optogenetic activation of these neurons is sufficient to induce memory in our olfactory training paradigm (Fig. 6). We found that activation of the excitatory cholinergic FB2N induces aversive memory for paired odor (Fig. 6a, Extended Data 2), similar to activation of its postsynaptic DAN-f1 (Fig. 1c). Interestingly, pairing of an odor with the activation of the GABAergic FBN or the glutamatergic FB2N induces appetitive memory for paired odor (Fig. 6b-c, Extended Data 2a-b), opposite to activation of their postsynaptic DANs (Fig. 1c).
Figure 6. Feedback neurons can drive associative memory formation.
(a-c). We generated Split-GAL4 lines to target CsChrimson expression almost exclusively to single FBNs or FB2Ns that innervate vertical lobe DANs (See Extended Data 10). We optogenetically activated these neurons instead of a US in our associative memory paradigm (as in Fig. 1b-c). Horizontal lines, means and standard deviations. Statistics are as in Fig. 1c.
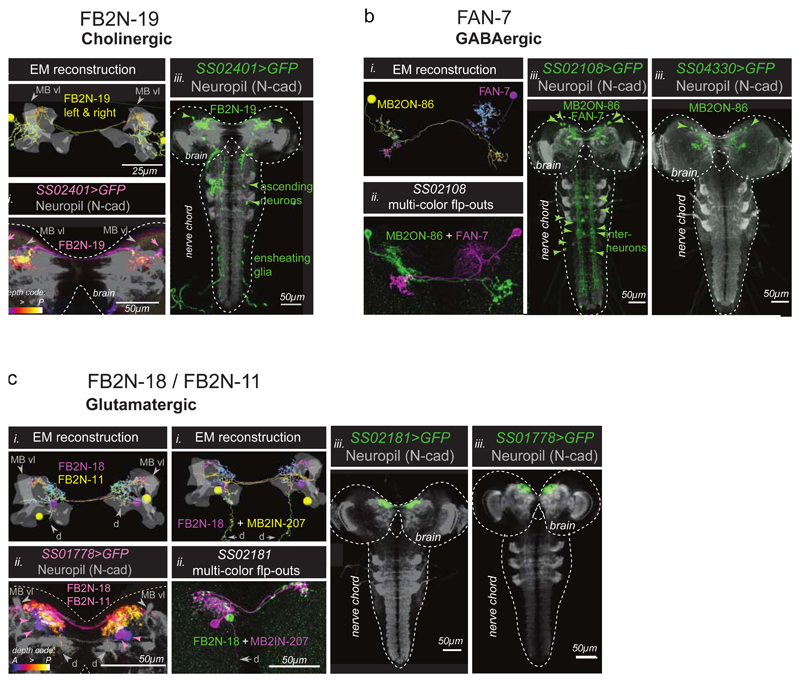
a, Activation of the excitatory FB2N-19 in the training paradigm induces aversive memory, same as the activation of its postsynaptic DAN-f1 (Fig. 1c). N=13,20
b, Activation of FAN-7 induces appetitive memory, opposite to its postsynaptic DAN-d1 (Fig. 1c). The SS02108-Split-GAL4 line drives expression in two brain neurons: the GABAergic FAN-7 and MB2ON-86, and a few somatosensory interneurons in the nerve cord (unlikely to evoke an appetitive memory). We therefore used the SS04330-Split-GAL4 line that drives expression specifically in MB2ON-86 as an additional control and found this neuron did not induce memory formation. N=52,27,11.
c, Activation of FB2N-18 with or without FB2N-11 induces appetitive memory, opposite to activation of their presynaptic DANs (Fig. 1c). The SS01778-Split-GAL4 line drives expression in the glutamatergic FB2N-18 and FB2N-11, which have very similar morphology and connectivity (Supplementary Fig. 3, 4b-d, and Extended Data 10). Both connect most strongly to DAN-f1 and weakly to MBIN-e2 (and DAN-g1 for FB2N-18). The SS02181-Split-GAL4 line drives expression in FB2N-18 and in MB2IN-207, a weakly connected pre-modulatory neuron (unlikely to be able to significantly influence modulatory neuron activity). N=18,18,31.
Connectivity-constrained model of the circuit reveals feedback neurons improve performance on complex learning tasks
To explore the computational consequences of the feedback neurons, we developed a model of the circuit constrained by i) the connectome, ii) the known neurotransmitter identities, and iii) the valences of compartments (Fig. 1c). The presence or absence of connections between MBONs, DANs, and feedback neurons in the model was determined by the connectome, and for neurons known to be excitatory or inhibitory, the signs of the connections were fixed to be consistent with this designation. We additionally used synapse counts from the connectome to set the initial strengths of model connections. Since synapse counts alone are unlikely to fully determine functional interactions, we then adjusted these connection strengths using gradient descent to optimize the network to perform a set of associative learning tasks29 (see Materials and Methods). Unlike in standard recurrent neural network models, we modeled ongoing modifications of KC to MBON connections using a synaptic plasticity rule that depends on the timing of KC and modulatory neuron activity consistent with experimental findings16.
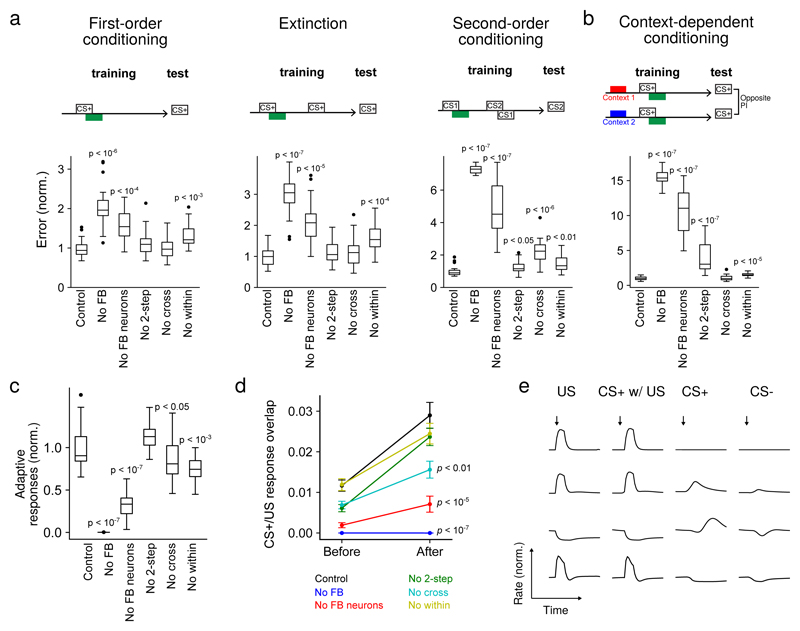
We assessed the contributions of different feedback pathways by repeating the optimization procedure for networks lacking such feedback and comparing their performance. Tasks included first-order conditioning and extinction which have been demonstrated in both larval15;30 and adult Drosophila3;31, and second-order and context-dependent conditioning which have so far been investigated only in adult32;33. In second-order conditioning, a reinforcement predicting conditioned stimulus is used to reinforce a second stimulus, while in context-dependent conditioning, the US valence depends on a previous contextual input.
We found that the performance on all tasks was significantly degraded in the absence of all feedback, including direct MBON feedback, one-step feedback via FBNs, and two-step feedback via FB2Ns and FBNs (Fig. 7a-b). The removal of the indirect feedback alone (with intact direct MBON feedback) also significantly degraded the performance on all tasks, with especially strong effects on the more complex tasks (Fig. 7a-b). Even the removal of two-step feedback alone significantly diminished performance on two of the more complex tasks (second-order conditioning and context-dependent conditioning), with a drastic effect on context-dependent conditioning (Fig. 7a-b). Thus, each additional feedback level improves the performance of the network when it is tested on challenging associative learning tasks.
Figure 7. Model reveals effects of feedback motifs on learning task performance.
a Normalized error (mean-squared difference between decoded and target valence, normalized to error for control networks) after optimizing models to perform first-order conditioning, extinction, and second-order conditioning. Error shown for six cases: Control: full network; No FB: networks in which feedback onto modulatory neurons, including direct MBON connections, is removed; No FB neurons: indirect FBN/FB2N feedback is removed but direct MBON connections are intact; No 2-step: only FB2Ns and all FBN-to-FBN connections are removed; No cross: indirect cross-compartment connections are removed; No within: indirect within-compartment connections are removed. Performance of model networks was assessed by optimizing n=20 networks independently. Performance measures were compared across network types and significance assessed with a two-sided Mann-Whitney U test (a-d). p-values represent a comparison to control networks after conditioning. Box: lower and upper quartiles, line: median, whiskers: range of the data up to 1.5 interquartile range, points: outliers of the whiskers range (a-c). b Similar to a, but for networks optimized to perform a context-dependent conditioning task.
c Adaptive response index for networks in a, defined as the magnitude of firing rate change to CS+ presentation before and after conditioning, averaged over modulatory neurons. Results are normalized by the index for control networks.
d Example responses of DANs from networks in a to US alone (US), CS+ paired with US following training (CS+ w/ US), CS+ alone after training (CS+), and CS prior to training (CS).
Row i: A DAN selective only to US.
Row ii: A DAN selective to US that acquires a CS+ response after conditioning.
Row iii, iv: DANs with ”prediction-error” like responses. CS+ responses are opposite in sign to US responses.
Row iii: A DAN inhibited by US and activated when US is omitted.
Row iv: A DAN excited by US and inhibited when US is omitted.
e CS+/US response overlap before and after conditioning. Overlap equals the dot product of the vectors of firing rate changes across the modulatory neuron population during CS+ and US presentations.
We also constructed networks lacking one- and two-step feedback within or across compartments. Removal of within-compartment feedback diminished performance on all tasks, while removal of cross-compartment communication substantially reduced performance for second-order conditioning (Fig. 7a-b). In total, each of the feedback categories revealed by the EM reconstruction may be important for associative learning paradigms that require computations such as prediction, prediction error, or context dependence.
Feedback neurons enable adaptive responses of modulatory neurons in the model
The high fraction of feedback input originating from MBONs onto modulatory neurons suggests that their activity could be adaptively regulated by prior learning. To test this idea, we computed an index that quantifies the mean change in modulatory neuron firing rates in response to CS+ (i.e. the CS that was paired with the US) presentations before and after conditioning. We found that this index is indeed substantially enhanced by the presence of feedback neurons (by FBNs and FB2Ns together, or even just FBNs alone, Fig. 7c). The optimized networks exhibit a diversity of adaptive modulatory neuron responses (some examples are shown in Fig. 7d).
After a CS/US pairing, many modulatory neurons acquired responses to CS+ that resemble their responses to the US that had been paired with that CS+ (Fig. 7e, 7d-ii). These responses were significantly attenuated in networks that lacked feedback, including those that lacked just indirect feedback, and just cross-compartment feedback (Fig. 7e). Such responses have been observed in modulatory neurons across the animal kingdom4;5;34, including adult Drosophila10;11. They are consistent with a computation of the valence that is predicted by the CS+ (i.e. a predicted value) and could drive the formation of an association between a novel CS and a CS+ during higher-order conditioning.
Some modulatory neurons acquired CS+ responses that were opposite in sign to their responses to that US and potentially represent prediction errors (Fig. 7d-iii, 7d-iv). Some of those appear to be activated by the omission of a predicted US whose valence is opposite that of the neuron’s preferred US (Fig. 7d-iii). Such responses potentially represent positive prediction errors and have been proposed to support extinction by inducing a parallel memory of opposite valence2;3;31. Consistent with this idea, in adult flies, DANs of opposite valence and direct cross-compartment MBON-to-DAN connections have been implicated in extinction3;31, but the role of indirect feedback pathways has not been investigated. In our model we find that removing indirect feedback significantly reduces performance of networks optimized to extinguish a previous association (Fig. 7a). Some modulatory neurons also showed prolonged inhibition in response to the omission of a predicted US whose valence is the same as the neuron’s preferred US (Fig. 7d-iv). Such responses potentially represent negative prediction errors1 and have been proposed to support extinction by erasing the memory formed by the activation of that modulatory neuron2. Thus, our model raises the possibility that extinction could be implemented via multiple mechanisms in this circuit2.
Discussion
Modulatory neurons (e.g. dopaminergic, DAN) are key components of higher-order circuits for adaptive behavioral control and they provide teaching signals that drive memory formation and updating1;3;4;9;15;31;35. Here, we provide the first synaptic-resolution connectivity map of a recurrent neural network that regulates the activity of modulatory neurons in a higher-order learning center, the Drosophila larval MB (Fig. 2a-f). We also functionally tested some of the newly identified structural pathways and developed a model of the circuit to explore the roles of the newly discovered architectural motifs in different learning tasks (Fig. 7a-e).
Feedback pathways enable adaptive regulation of learning by prior learning
We discovered a large population of 61 feedback neuron pairs that provide one- and/or two-step feedback from the MB output neurons (MBONs) to modulatory neurons (Fig. 2a-d, 3a-b and 5a-e). Strikingly, we found that many modulatory neurons receive more than 50% of their total dendritic input from feedback pathways (Fig. 2d). These results suggest that prior memories as represented by the pattern of MBON activity can strongly influence modulatory neuron activity.
Learning and memory systems in vertebrates9 and insects3;14;17;18 are often organized into distinct compartments implicated in forming distinct types of memories (e.g. aversive and appetitive or short- and long-term). Interestingly, we found that the majority of the discovered feedback pathways link distinct memory systems, suggesting that the MB functions as an interconnected ensemble during learning (Fig. 3b, 5e). Thus, prior memories formed about an odor in one compartment can influence the formation and updating of distinct types of memories about that odor in other compartments.
In adult Drosophila, functional connections between some MBONs and DANs31;35–41 have been reported, and some have been shown to play a role in short-term memory formation35;38;42, long-term memory consolidation37;40, extinction and re-consolidation3;31;39, or in synchronizing DAN ensemble activity in a context-dependent manner36. In some cases, direct MBON-to-DAN connections have been demonstrated31;35;37;39. While direct connections from several MBONs onto DANs exist in the larva12 (Extended Data 8a), we find that indirect connections via the feedback neurons account for a much larger fraction of a modulatory neuron’s dendritic input than direct MBON synapses (Fig. 2d). This suggests that adaptive DAN responses may be largely driven by such indirect feedback.
Some of the one-step within-compartment feedback motifs we found are analogous to the feedback motifs so far described for the DANs in the vertebrate midbrain4;43–47. While the diversity and the inputs of striatal feedback neurons have not yet been fully explored, in the future it will be interesting to determine whether many of the striatal feedback neurons also link distinct memory systems.
Circuit motifs for computing integrated predicted value signals across aversive and appetitive memory systems
The use of internal predictions can dramatically increase the flexibility of a learning system1;7;8. Our study reveals candidate circuit motifs that could compute integrated predicted value signals across appetitive and aversive memory systems. Thus, a prominent motif we identified is convergence of excitatory and inhibitory connections from MBONs from compartments of opposite valence onto DANs (Fig. 3b-e, Fig. 4a-g). In naïve animals, odor-evoked MBON excitation in all compartments is thought to be similar. However, associative learning selectively depresses conditioned odor drive to MBONs in compartments where modulatory neuron activation has been paired with the odor3;16. We propose that by comparing the conditioned odor-evoked MBON excitation in compartments of opposite valence via cross-compartment feedback connections, modulatory neurons compute an integrated predicted value signal across appetitive and aversive domains.
Convergence of feedback and US pathways could allow the computation of prediction errors
An important aspect of reinforcement learning theories is the idea that modulatory neurons compare predicted and actual US (to compute so-called prediction errors) and drive memory formation or extinction depending on the sign of the prediction error. While Drosophila modulatory neurons have not yet been directly shown to represent prediction errors, adult and larval Drosophila are capable of extinction3;30;31, and our study reveals candidate motifs that could support the comparison of expected and actual US. We found that modulatory neurons receive convergent input from feedback pathways from MBONs and from US pathways (Fig. 2d-f). Modulatory neurons could therefore potentially compute prediction errors by comparing inhibitory drive from the feedback pathways to the excitatory drive from the US pathways, or vice versa. Consistent with this idea, we observed in our model some DANs that are inhibited by US alone and activated by CS+ alone, or vice versa (Fig. 7d-iii and 7d-iv).
Our study also reveals that US pathways and feedback pathways converge at two levels: not only at the modulatory neurons themselves, but also at the two-step feedback neurons (FB2Ns (Fig. 2f). Actual and expected outcomes could therefore also be compared by FB2Ns. A recent study in the mouse VTA has found that some pre-DAN neurons encoded only actual, or only expected reward, while others encoded both variables4;43. Thus, both in vertebrates and in insects, comparing predicted and actual outcomes may be a complex computation involving multiple levels of integration that eventually converges onto an ensemble of modulatory neurons43.
Diversity of feedback inputs across modulatory neurons suggests a range of distinct and distributed teaching signals
An assumption in many reinforcement learning models is that all modulatory neurons receive a global scalar reward prediction error signal1. Here, we were able to analyze the comprehensive set of inputs of every individual uniquely identifiable modulatory neuron in a learning center. This revealed that each modulatory neuron receives a unique set of feedback inputs (Fig. 2e) that could enable each neuron to compute a unique set of features. Consistent with this, we observed a diversity of adaptive response types in the modulatory neurons in our model (Fig. 7d). This suggests that instead of computing a single global reward prediction error that is distributed to all modulatory neurons, the network uses a range of distinct compartmentalized and distributed teaching signals.
Multilevel and cross–compartment feedback increase performance and flexibility
Our connectivity and modeling studies revealed two architectural features of the circuit that provides input to the modulatory neurons that increase its performance and flexibility on learning tasks (Fig. 7a-b). The first is the multilevel feedback architecture that includes not only the previously known direct MBON feedback31;35;37;39, but also multiple levels of indirect feedback. The second is the extensive set of cross-compartment connections. Modeling suggests that these motifs support improved performance on complex tasks that require the computation of variables such as predictions, prediction errors, and context.
In summary, we present the first complete circuit diagram of a recurrent network that computes teaching signals in a biological system, providing insights into the architectural motifs that increase its computational power and flexibility. Our connectome-constrained model provides numerous predictions that can be tested in the future in a tractable model organism, for which genetic tools can be generated to monitor and manipulate individual neurons24;25;48. The connectome, together with the functional and modelling studies therefore provides exciting opportunities for elucidating the biological implementation of reinforcement learning algorithms.
Materials and Methods
Fly lines
In the main text and figures, short names are used to describe genotypes for clarity. See Supplementary Table for a complete list of full names of all driver lines and effectors. We used GAL4, Split-GAL4 lines to direct the expression of the red-shifted channel-rhodopsin 20XUAS-CsChrimson-mVenus49 (Bloomington Drosophila Stock Center BDSC 55134, gift of V. Jayaraman) or the Calcium indicator 20xUAS-IVS-GCaMP6f 50 in pairs of neurons or subsets of neurons. Split-GAL4 lines restrict expression of the effector to a few cells, under the double control of two enhancers (inserted in the attP2 and attP40 docking sites), selected by us or others in Janelia Research Campus (HHMI, VA, USA) based on their GAL4 expression pattern48;51;52.
Modulatory neurons (MBINs)
We used SS24765-Split-GAL4 to optogenetically activate OAN-a1 in the calyx. We generated SS02160-Split-GAL4 to activate DAN-c1 in the lower peduncle. For the vertical lobe, we generated SS01702-Split-GAL4 to activate or image calcium transients in MBIN-e2 (DAN-c1 was also covered by this line) and SS01958-Split-GAL4 to activate or image calcium transients in OAN-e1 in the UVL. We used SS02180-Split-GAL4, MB145B-Split-GAL4 (used for activation and calcium imaging, gift of G. Rubin and Y. Aso) and MB065B-Split-GAL453 (which also covered DAN-c1) to target DAN-f1 in the IVL. We used SS01716-Split-GAL420 to induce or image DAN-g1 activity in the LVL, and we generated SS04268-Split-GAL4 to activate OAN-g1, also in the LVL. MB054B-Split-GAL4 (gift of G. Rubin and Y. Aso) was also used to co-activate DAN-g1 and DAN-f1. We used two lines to target DAN-d1 in the lateral appendix: MB143B-Split-GAL4 (used for activation and calcium imaging) and MB328B-Split-GAL4 (both gifts of G. Rubin and Y. Aso). In the medial lobe, we generated a broad line SS01948-GAL4 which allows co-activation of DAN-h1, DAN-i1, DAN-k1, and sometimes DAN-j1. We also imaged calcium transients in DAN-i17 using the more specific GAL4 SS00864-Split-GAL4.
Neurons presynaptic to the modulatory neurons (MBINs)
We optogenetically activated multidendritic Class IV neurons (MD IV) with the driver line ppk-1.9-GAL48 (gift of D. Tracey); Basin interneurons with GMR72F11-GAL424; the ascending neuron A00c with GMR71A10-GAL424;54 crossed to ppk-GAL80 55, repo-GAL80 56 (to prevent expression in MD IV and glial cells, respectively). We also activated A00c with the more specific GAL4 line SS00883-Split-GAL4. We generated SS01778-Split-GAL4 and SS02181-Split-GAL4, which target FB2N-11 and/or FB2N-18. SS02108-Split-GAL4 targets FAN-7; SS02401-Split-GAL4 targets FB2N-19.
Control lines
As a control for the GAL4 lines inserted at the attP2 site, we used the empty control stock y w;;attP2 48;52 crossed to the effector line. As a control for Split-GAL4 lines with AD at attP40 and DBD at attP2, we used the empty stock y w;attP40;attP2 48;52 crossed to the effector line.
Lines for recording neuronal activity
Calcium transients in modulatory neurons were imaged using the following constructs to verify functional input of mechano-ch neurons: w; iav-LexA24 in attP40; 20xUAS-IVS-GCaMP6f 15.693 50 in attP2, 13XLexAop2-CsChrimson-tdTomato49 in VK00005. For Basins multisensory interneurons: w; GMR72F11-LexA52 in JK22C; 20xUAS-IVS-GCaMP6f 15.693 50 at attP2, 13XLexAop2-CsChrimson-tdTomato49 at VK00005. And for MD class IV nociceptive neurons: w; 13XLexAop2-CsChrimson-mVenus49 at attP40 (BDSC 55138); ppk-1kb-hs43-lexA-GAD10 at attP2, 20xUAS-IVS-GCaMP6f 50 at VK00005. All the effectors used in these stocks are a gift from V. Jayaraman. Transvection was tested by bathing some samples in 100 mM mecamylamine and observing the disappearance of responses to optogenetic stimulation (data not shown). If a response remained during mecamylamine application, the experiments were repeated using a spatially defined photo-stimulation using spatial light modulator (SLM) technology (see functional connectivity section for details of the procedure and the lines concerned).
For patch-clamp recording we crossed the genetic driver lines for MBON-m1 (SS02163-Split-GAL4) or for MBON-i1 (SS01726-Split-GAL4) to 58E02-LexAp65 at attP40 57; 13xLexAop2-IVS-GCaMP6f-p10 15.693 50 at VK00005 (BDSC 44276), 20xUAS-CsChrimson-mCherry 49 at su(Hw)attP1 in order to activate MBONs and visualize the medial lobe DANs (ML-DANs) for patch-clamping. Only data for DAN-i1, which was the most frequently hit by the recording pipette, as revealed by post hoc identification, are shown.
The reporter pJFRC29-10xUAS-IVS-myr::GFP-p10 58 at attP2 was used for immunostaining.
Learning experiments
Learning experiments were performed as previously described12;19;20. The larvae, with CsChrimson-expressing neurons, were reared in the dark at 25°C in food vials supplemented with 1:200 retinal. The experimenter selected two groups of 30 third-instar larvae and was blind to their specific genotype. The two groups underwent a training procedure involving odor and light exposures, either fully overlapping in time (paired group), or fully non-overlapping (unpaired group). The paired group was placed for 3 minutes on 4% agarose plates and exposed to constant red-light illumination (wavelength: 629 nm, power: 350 μW/cm2; except for ppk-1.9-GAL4, which targets neurons at the surface of the body and for which a light power of 35 μW/cm2 was used) paired with the presentation of 12 μl of odor ethyl-acetate (10−4 dilution in distilled water) absorbed on two filter papers located on the plate lid. These larvae were then transferred to a new plate with no odor and in the dark for 3 minutes. This paired training cycle was repeated three times in total. The unpaired group of larvae underwent odor presentation in the dark and red light without odor following the same protocol. The order of the sequence of presentation for odor and light stimulation (i.e. for paired group: half training protocols are in the sequence odor+/air-/odor+/air-/odor+/air- and half are air-/odor+/air-/odor+/air-/odor+, same logic for unpaired group) was alternated throughout all experiments. After a 3-minute test with odor presentation on one side of the plate lid, larvae were counted on the side of the odor, on the opposite side, and in the 1 cm-wide midline of the plate. Preference and performance indices were calculated as in a previous study59. Briefly, a preference index (PI) was first computed, for each group as: PI = [N (larvae on the odor side) - N (larvae on the no-odor side)]/N(total), N(total) includes larvae in the middle of the plate. Individual Learning Performance Score (LPS) was then computed for each pair of reciprocally trained group, as LPS = [PI (paired) – PI (unpaired)]/2. Positive scores indicate a larger proportion of larvae choosing the odor side in the paired group than in the unpaired group (i.e., appetitive olfactory memory), while negative scores indicate the reverse inequality (i.e., aversive olfactory memory). Because we tested larvae immediately after the last training trial, and less than 20 min after the first training trial, we assume the test reveals mainly short-term memory15. Experiments were performed en block for multiple genotypes with the same control line, as shown in the figures. The same genotype was tested over multiple days at random times of the day.
Circuit mapping and electron microscopy
We reconstructed neurons and annotated synapses in a single, complete central nervous system from a 6 hr old female [iso] Canton S G1 x w1118 [iso] 5905 larva, acquired with serial section transmission EM at a resolution of 3.8 x 3.8 x 50 nm, that was first published along with the detailed sample preparation protocol24. Briefly, the CNS was dissected and placed in 2% gluteraldehyde 0.1 M sodium cacodylate buffer (pH 7.4). An equal volume of 2% OsO4 was added and the larva was fixed with a Pelco BioWave microwave oven with 350-W, 375-W and 400-W pulses for 30 sec each, separated by 60-sec pauses, and followed by another round of microwaving but with 1% OsO4 solution in the same buffer. Next, samples were stained en bloc with 1% uranyl acetate in water and microwaved at 350 W for 3x3 30 sec with 60-sec pauses. Samples were dehydrated in an ethanol series, transferred to propylene oxide, and infiltrated and embedded with Epon resin. After sectioning the volume with a Leica UC6 ultramicrotome, sections were imaged semi-automatically with Leginon60 driving an FEI Spirit TEM (Hillsboro, OR), and then assembled with TrakEM261 using the elastic method62. The volume is available at https://l1em.catmaid.virtualflybrain.org/?pid=1.
To map the wiring diagram we used the web-based software CATMAID63, updated with a novel suite of neuron skeletonization and analysis tools 64, and applied the iterative reconstruction method64. All annotated synapses in this wiring diagram fulfill the four following criteria of mature synapses24;64: (1) There is a clearly visible T-bar or ribbon on at least two adjacent z-sections. (2) There are multiple vesicles immediately adjacent to the T-bar or ribbon. (3) There is a cleft between the presynaptic and the postsynaptic neurites, visible as a dark-light-dark parallel line. (4) There are postsynaptic densities, visible as dark staining at the cytoplasmic side of the postsynaptic membrane.
We validated the reconstructions as previously described24;64, a method successfully employed in multiple studies24;25;28;64–66. Briefly, in Drosophila, as in other insects, the gross morphology of many neurons is stereotyped and individual neurons are uniquely identifiable based on morphology66–68. Furthermore, the nervous system in insects is largely bilaterally symmetric and homologous, with mirror-symmetric neurons reproducibly found on the left and the right side of the animal. We therefore validated neuron reconstructions by independently reconstructing synaptic partners of homologous neurons on the left and right side of the nervous system. With this approach, we have previously estimated the false positive rate of synaptic contact detection to be 0.0167 (1 error per 60 synaptic contacts)19. Assuming the false positives are uncorrelated, for an n-synapse connection the probability that all n are wrong (and thus that the entire connection is a false positive) occurs at a rate of 0.0167n. Thus, the probability that a connection is a false positive reduces dramatically with the number of synaptic contacts contributing to that connection. Even for n = 2 synaptic contacts, the probability that a connection is not true is 0.00028 (once in every 3,586 two-synapse connections). We therefore consider ‘reliable’ connections those for which the connections between the left and right homologous neurons have at least 3 synapses each and their sum is at least 10. See24;64 for more details.
We also systematically asked what percentage of connections was conserved between left and right homologs, as a function of number of synapses in that connection. We did this for the 426 neurons that were presynaptic to MBINs on the left or the right. Thus, we found that if two neurons were connected with 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10 synapses on one hemisphere, the probability that homologous neurons are connected on the other hemisphere was 48%, 60%, 80%, 89%, 95%, 97%, 99%, 100%, 100%, and 100%, respectively. We also computed the fraction of input that each MBIN received from each presynaptic neuron and found that if a presynaptic neuron accounted for 1.6%, 2%, 3%, 4%, 5% of a postsynaptic neuron’s input the likelihood that the homologous neurons were connected on the other hemisphere was 95%, 99%, 100%, 100%, and 100%, respectively. Thus, numerically weak connections were not conserved, but numerically strong connections were 100% conserved. Interestingly, in previous studies we have demonstrated that the numerically strong connections in which the postsynaptic neuron receives at least 2% of input the presynaptic neuron were also functional24;25.
Thus, the numerically strong connections (that account for at least 2% of a postsynaptic neuron’s input) are reproducible between left and right hemispheres of the same individuals and across individuals, as evidenced by our functional connectivity experiments across individuals.
Immunostaining
Dissected brains were fixed in phosphate buffered saline (PBS, NaCl 137 mM, KCl 2.7 mM, Na2HPO4 8.1 mM, KH2PO4 1.5 mM, pH7.3) containing 4% paraformaldehyde (Merck) for 30-min at room temperature. After two 15-minute washes with PBT (PBS with 1% or 3% Triton X-100; Sigma-Aldrich), the brains were blocked with 5% normal goat serum (Vector Laboratories) in PBT and incubated for at least 24 hours with primary antibodies at 4°C. Before application of the secondary antibodies for at least 24 hours at 4°C or for 2 hours at room temperature, brains were washed several times with PBT. After that, brains were again washed with PBT, mounted in Vectashield (Vector Laboratories) and stored at 4°C in darkness. Images were taken with a Zeiss LSM 710M confocal microscope. The resulting image stacks were projected and analyzed with the image processing software Fiji69. Primary antibodies were used at the following dilutions: rabbit anti-GFP (cat# Af2020, Frontier Institute; 1:1000), chick anti-GFP (ab13970, abcam, 1:1000), rabbit anti-GABA (A2052, Sigma; 1:100), mouse anti-ChAT (ChAT4B1, DSHB Hybridoma Product deposited by P.M. Salvaterra; 1:50). Rabbit anti-DVGlut70 was diluted 1:1000. These antibodies were used in Drosophila larvae previously12;28. Secondary antibodies were used at the following dilutions: Alexa Fluor 568-conjugated goat anti-rabbit IgG (A-11036, Invitrogen Molecular Probes; 1:300), Alexa Fluor 633-conjugated goat anti-mouse IgG (A-21050, Invitrogen Molecular Probes; 1:300) and Alexa Fluor 488-conjugated goat anti-chicken IgG (A-11039, Invitrogen Molecular Probes; 1:300).
Identifying GAL4 lines that drive expression in modulatory neurons and their presynaptic partners
To identify GAL4 lines (listed in Supplementary Table) that drive expression in specific neurons, we performed single-cell FlpOut experiments (for FlpOut methodology see24;71) of many candidate GAL4 lines72. We generated high-resolution confocal image stacks of individual neuron morphology (multiple examples per cell type). Most MBONs and MBINs were uniquely identifiable based on the dendritic and axonal projection patterns (which MB compartment they project to and the shape of input or output arbor outside the MB). These were also compared to previously reported single-cell FlpOuts of dopaminergic and octopaminergic neurons in the larva54;59;73–75. For the neurons upstream of MBINs (FBNs/FANs/FB2Ns), we used morphology and cell body position to identify the lineage of the neuron. The precise shape and 3D location of dendritic and axonal projections were then examined and compared to all potential candidates in the lineage which have been fully reconstructed from the electron microscopy volume. In some cases, two neurons had very similar morphology at both light and EM level, and in such cases they also had nearly identical connectivity (e.g. FB2N-11 and FB2N-18).
Functional connectivity assays
Central nervous systems of third-instar larvae were dissected in a cold buffer containing 103 mM NaCl, 3 mM KCl, 5 mM TES, 26 mM NaHCO3, 1 mM NaH2PO4, 8 mM trehalose, 10 mM glucose, 2 mM CaCl2, 4 mM MgCl2 and adhered to poly-L-lysine (SIGMA, P1524) coated cover glass in small Sylgard (Dow Corning) plates.
For optogenetic activation, red illumination (617nm High-Power Lightguide Coupled LED Source, Mightex) was positioned above the sample to depolarize the axon terminal parts of the sensory neurons (MD IV or chordotonal) or the second order interneurons (Basins). Light stimulations were performed with 1 or 15 sec duration and in 40 and 600 cycles of laser on/off pulses of 20 msec/5 msec. Each preparation underwent three types of light stimulation of increasing power: ca. 390 μW/mm2, 920 μW/mm2 and 4.6 mW/mm2. Only the data for the highest light power during 1 sec is displayed (Fig.1f). The same stimulus was spaced with 30 sec for a total of three presentations in each scan. Each scan consisted in imaging dopaminergic neurons on a two-photon scanning microscope (Bruker) using a 60x 3 1.1 NA objective (Olympus). A mode-locked Ti:Sapphire laser (Chameleon Ultra II, Coherent) tuned to 925 nm was used for photo-activation of the GCaMP6f. Fluorescence was collected with photomultiplier tubes (Hamamatsu) after band-pass filtering. Images were acquired in line scanning mode (5.15 fps) for a single plane of the CNS. The same genotype was tested over multiple days at random times of the day. Data collection and analysis were not performed blind to the conditions of the experiments.
To overcome transvection observed between the transgenes at the attP40 landing site of the MB143B-Split-GAL4 line (targeting DAN-d1) crossed to w; 13XLexAop2-CsChrimson-mVenus49 in attP40; ppk-1kb-hs43-lexA-GAD10 in attP2, 20xUAS-IVS-GCaMP6f2 in VK00005, we used 3-dimension spatially defined photo-stimulation. MD IV neurons expressing CsChrimson were photo-activated by a holographic pattern generated by a two-photon 1040 nm laser (femtoTrain, Spectra-Physics) coupled to a phase-only SLM (Intelligent Imaging Innovations). GCaMP6f signal was imaged by a laser tuned to 925 nm (Insight DS+ Dual, Spectra-Physics). The optogenetic stimulations were 50 cycles of laser on/off pulses of 2 msec/18 msec, ranging from 1 to 1.5 mW/mm2. Off-target (equidistant from the Chrimson-expressing DAN-d1 neuron, but not targeting Chrimson-expressing MD IV neurons) and on-target stimulations were alternatively performed and the difference between transvection-only generated calcium signals and transvection + MD IV neuron activation-generated signal was computed and used as the fluorescence signal. DAN-d1 neurons were imaged at a frame rate of ca. 5 fps on a two-photon scanning microscope (Vivo, Intelligent Imaging Innovations) using a 25x 2 1.1 NA objective (Nikon).
For image analysis, image data were processed by Fiji software69 and analyzed using custom code in Matlab (The Mathworks, Inc). Specifically, we manually determine the regions of interest (ROIs) from maximum intensity projection of entire time series images, and measure the mean intensity. In all cases, changes in fluorescence were calculated relative to baseline fluorescence levels (F0) as determined by averaging over a period of at least 2 sec. just before the optogenetic stimulation. The δF/F0 values were calculated as δF/F0 = (Ft-F0)/F0, where Ft is the fluorescent mean value of a ROI in a given frame. Analyses were performed on the mean δF/F0 of the consecutive 3 stimulations.
Whole-cell patch-clamp recordings from DANs on optogenetic activation of MBONs
For recording, the isolated brain attached with VNC were dissected from third instar larvae in Baines external solution76, which contained (mM): 135 NaCl, 5 KCl,, 2 CaCl2.2H2O, 4 MgCl2.6H2O, 5 2-[(2-Hydroxy-1,1-bis(hydroxymethyl)ethyl)a ethanesulfonic acid, 5 N-[Tris(hydroxymethyl) methyl]–2-aminoethanesulfonic acid, and 36 sucrose. The pH was adjusted to 7.15 with NaOH, and osmolarity was 310-320 mOsm. The preparation was viewed with a 60x 1 NA water-immersion objective equipped with an Olympus microscopy (BX51WI; Olympus). GCaMP6f–labeled DANs were visualized with a 470-nm wavelength LED. The glial sheath above the targeted DANs was ruptured using 0.1% protease (Protease XIV; Sigma-Aldrich). Recording electrodes were pulled from thick-wall glass pipet (O.D. 1.5mm, I.D. 0.86mm) using P-97 puller (Sutter Instruments) and fire-polished to resistances of 10–15 Mω. The Baines intracellular solution76 contained (mM): 140 potassium gluconate, 5 KCl, 2 MgCl2.6H2O, 2 EGTA, 20 HEPES. The pH was adjusted to 7.4 with KOH, and the osmolarity was 280 mOsm. Biocytin was dissolved in intracellular solution at 0.5% for further post hoc morphological identification of recorded DANs. The data were acquired and processed using Digidata 1550, Multiclamp 700B, and Clampex 10.4 software (Molecular Devices). The recording was sampled at 20 kHz and filtered at 6 kHz under current-clamp mode. CsChrimson was activated by 617-nm wavelength LED.
DAN identification: After the electrophysiology recording, the preparation containing the VNC and brain was fixed in 4% paraformaldehyde in 0.1 M phosphate buffer saline (PBS) overnight at 4ºC, and then transferred to PBS until staining. After rinsing in PBS, the CNS preparations were placed in Streptavidin Alexa Fluor 647 (1:200) in PBS with 10% Triton X (overnight, room temperature). After rinsing, the preparations were dehydrated and mounted with DPX. The confocal images were captured with Zeiss 800 confocal laser microscope. Alexa Fluor 647 was excited with 633 nm-wavelength light, and mCherry-tagged CsChrimson neurons were excited with 567 nm-wavelength light.
Statistics
As most fluorescence and behavioral data were non-normally distributed (according to a Shapiro-Wilk test), we opted for non-parametric tests for paired comparisons. Similarly, for the model, we used a non-parametric test to compare performance. No statistical methods were used to pre-determine sample sizes but the sample sizes are similar to those reported in previous publications20;25;39.
For behavioral experiments, the performance scores obtained for each line tested in optogenetic reinforcement were compared to the ones of its corresponding empty line (i.e. w;;attP2 or w;attP40;attP2 for GAL4 or Split GAL4, respectively) using a non-parametric Mann-Whitney U test for independent sets of data. For multiple comparisons, the probability values were compared to a threshold of 0.05 adjusted with a Holm-Bonferroni correction to balance for Type I and Type II statistical errors, unless otherwise stated. Across GAL4 lines, comparisons of performance scores were done using the same methodology. Data were plotted using the Matlab script errorbarjitter, available at http://www.mathworks.com/matlabcentral/fileexchange/33658-errorbarjitter.
Fluorescence analyses were done using a non-parametric Wilcoxon test for paired comparisons between the maximum δF/F0 plus one standard deviation during 1 sec before photostimulation onset and the maximum δF/F0 at two time windows: during the 1 sec of the stimulation, and from 1 to 3 seconds after its onset.
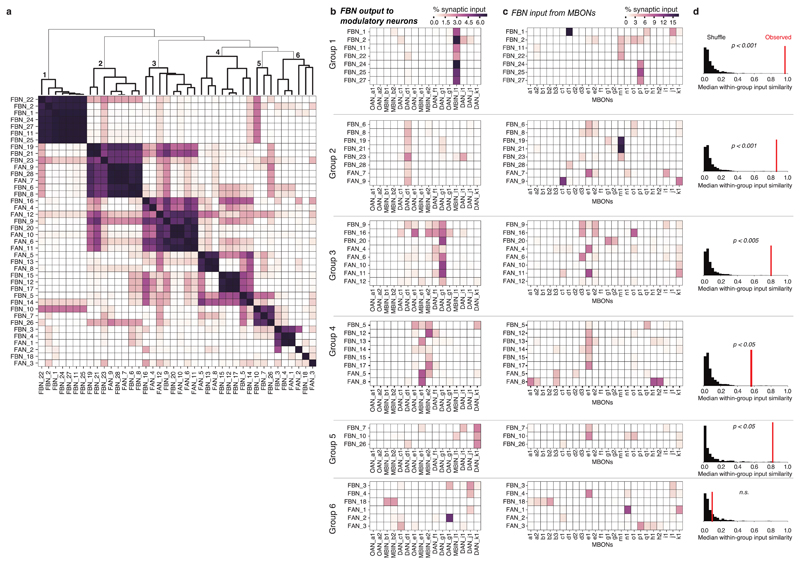
For the clustering analysis, we looked for clusters among FBNs/FANs based on the similarity of their synaptic partners separately for input and output. To find clusters based on synaptic inputs, we defined the similarity between a pair of FBN/FANs as the cosine similarity of the vector of inputs they receive from MBONs where the weight of a given connection is measured as the fraction of total input synapses on the postsynaptic neuron. Specifically, for vi and vj being the input vectors for FBN/FANs i and j, the similarity between them is defined Hierarchical clustering on the similarity matrix was done with Scipy using average linkage. We chose the top five clusters to highlight, which included all clearly differentiated groups of FBN/FANs. Clustering on the output patterns was done identically using the vectors of connectivity from FBN/FANs onto MBINs.
For the input-clustered groups, we assessed the similarity of the patterns of synaptic outputs and vice versa for the synaptic input patterns for output-clustered groups. We measured the overall group similarity as the median of all unique pairwise cosine similarities between neurons within the group. We used a permutation test to assess the significance of the observed similarities by randomizing the relationship between input pattern and output pattern for each FAN/FBN. For example, for each input-clustered group of size n, we randomly chose n output patterns and computed their median output similarity in the same way. A one-sided p-value was computed from the distribution of 10,000 random permutations with a Holm-Sidak correction for multiple comparisons across the groups.
Rate model of the MBON-i1-FBN-7-DAN-i1 one-step feedback motif
To illustrate the potential effects of different FBN-baselines we modeled the isolated MBON-i1-FBN-7-DAN-i1 feedback motif shown in Fig. 4b with rate equations where the output of neuron type (MBON, FBN, DAN), changed over time according to the equation where W was a matrix with positive and negative values corresponding to the direct interactions between neurons as shown in the circuit schematic of Fig. 4b, was a nonnegative tonic input into neuron i, is a stimulus input provided only to MBON, τi is a time constant, and parameters s, k and xh set the shape of the sigmoidal response. Equations were solved using ode45 in Matlab (The Mathworks, Inc). See Supplemental Fig. 3.
Connectivity-constrained model of the entire mushroom body with the feedback neurons Model dynamics
We constructed a recurrent network model of the larval MB containing MBONs, DANs and other feedback neurons. The network receives input from 70 KCs, and external cues, such as US. The normalized firing rate ri of neurons i is modeled as a threshold-linear function of its input:
| (1) |
where g represents positive rectification. Time is modeled in units of effective time constant (representing combined synaptic and membrane timescales). The connectivity matrix Wij is constrained using the EM reconstruction. The vector bi represents the static bias input to each neuron which determines its excitability, while Ii(t) represents time-varying external input. For MBONs, this includes external input from KCs,
KCs are initially silent, but during the presentation of an odor CS, the activity of a random fraction f of KCs is set to 1, leading to MBON activation. We assume all-to-all KC-to-MBON connectivity. At the beginning of each trial, weights WKC are initially set equal to their maximum value of 1/(NKCf), but are modified on each timestep according to a DAN-dependent synaptic plasticity rule. A weight W (t) from KC k to an MBON in compartment i evolves according to:
| (2) |
| (3) |
where di represents the level of dopamine in the compartment (a weighted sum of DAN inputs according to the DAN-to-MBON connectivity matrix), and rk represents the firing rate of the KC (note that modifications of weights onto MBONs depend only on KC and DAN activity). The terms and represent the firing rate rk and dopamine level di, respectively, low-pass filtered with time constant τ, which leads to an anti-Hebbian timing-dependent synaptic weight update in Equation 2. The second equation results in W(t) following these updates with a time constant of τW Equation 3. For simplicity, we assume that all modulatory neurons induce plasticity according to this rule.
Given the model dynamics described above, we use gradient descent optimization to find a set of network parameters that lead to good performance on the tasks we consider. After this optimization, KC-to-MBON weights are still time-varying quantities that evolve according to Equation 3, but all other parameters are fixed. Weights among DANs, MBONs, and feedback neurons are constrained by the EM reconstruction. Weight matrices are initialized using synapse counts from the EM data, scaled so that the ℓ2 norm of the inputs received by each neuron Only reliable connections, as defined previously, are included. Weights from neurons known to communicate using an inhibitory neurotransmitter, are then multiplied by −1. As optimization progresses, weights from neurons of known neurotransmitter identities are constrained to maintain a consistent sign by clipping at 0. Weights that were initialized to nonzero values rarely decayed to zero during optimization (7 ± 1% of weights, using a cutoff of 10% of each weight’s initial value to determine whether it has decayed). At the beginning of a trial, MBON rates are initialized to 0 while DAN and feedback neuron rates are initialized to 0.1. This promotes networks in which MBONs are primarily odor-driven, but some DANs and feedback neurons exhibit baseline levels of activity.
Tasks
Neuron i’s external input Ii(t) represents either KC input in the case of MBONs (as described above), or US or contextual signals (depending on the task) in the case of DANs and FB neurons. We assume that where WE is initialized as a random standard Gaussian variable and ej (t) = 0 or 1 depending on whether signal j is active. For most tasks, there are two signals (positive or negative US).
A linear readout of the MBONs determines the valence of the currently presented odor via where WM is initialized as a random Gaussian variable with variance 1/NMBON. Entries of WM corresponding to MBONs whose activation is known to produce approach or avoidance are constrained to be consistent with this sign.
Trials consist of 80 time units. In a first-order conditioning trial, a CS+ is presented for 3 time units starting randomly between t = 5 and t = 15, followed by a positive or negative US with a delay of 2 time units. A test CS+ presentation occurs between t = 65 and t = 75, and the system must output the appropriate valence of +1 or -1 depending on the US valence during this second presentation. For extinction, an additional CS+ presentation occurs randomly between t = 35 and t = 45, and the magnitude of the valence is halved for the final test CS+ presentation. For second-order conditioning, a new CS2 is presented at this time, followed by the original CS+, and the test occurs for CS2. Finally, for context-dependent conditioning, a contextual signal that determines the US valence is presented 3 time units prior to the first CS. At t = 30 and t = 60 firing rates are reset to their initial conditions to model an arbitrary time delay between CS presentations and preventing networks from using persistent activity, rather than synaptic plasticity, to maintain associations.
For networks trained on first-order conditioning, second-order conditioning, and extinction, training consists of random second-order conditioning and extinction trials (for which first-order conditioning is a subcomponent). On each trial, there is a 50% probability that one of the signals (e.g. the US) will be omitted, or a CS- odor will replace a CS+ odor, and the network report a valence of 0 in these cases, ensuring that only valid CS-US contingencies are learned.
Optimization
The weights of the network W, the external and readout weights WE, WM (described below), and the biases b are optimized using PyTorch using the RMSprop optimizer (www.pytorch.org). Optimization proceeds over 1500 epochs, each of which consists of a batch of 30 trials that are used to evaluate the loss function that is minimized through gradient descent. The loss is equal to the squared distance between the actual and target valence summed over timesteps, plus a regularization term for DAN activity. The regularization term equals which penalizes DAN activity that exceeds a baseline level of 0.1. This suppresses task-unrelated DAN activity and produces more realistic activity patterns in the DANs, but our results do not qualitatively change if this regularization is removed. We used a timestep of Δt = 0.5, although we verified that our qualitative results hold for smaller timesteps.
| Parameter | Notation | Value |
|---|---|---|
| KC coding level | f | 0.1 |
| Max. KC-to-MBON synaptic weight | wmax | 1/(NKCf) |
| Timing-dependent plasticity window | τ | 5 |
| Timescale of weight modifications | τW | 5 |
| Initial MBON rate | m0 | 0 |
| Initial DAN rate | d0 | 0.1 |
| Initial FB neuron rate | x0 | 0.1 |
| CS/US presentation length | Tstim | 3 |
| CS-US delay | ΔTUS | 2 |
| Trial length | T | 80 |
| Timestep | Δt | 0.5 |
| RMSprop learning rate | η | 0.002 |
| Batch size | B | 30 |
| Number of epochs | Nepochs | 1500 |
Extended Data
Extended Data Fig. 1. Expression patterns of Split-GAL4 lines.
Each panel shows a representative confocal maximum intensity projection (out of N=3) of the complete CNS of third-instar larvae (indicated by the dotted line in the first panel), with the neuropil labeled with anti-N-Cad antibody (blue) and the Split-GAL4line expression pattern revealed with UAS-myr-GFP (green). Arrowheads indicate cell bodies of identified neurons.
Extended Data Fig. 2. Detailed characterization of associative memories formed through different kinds of ”optogenetic punishments” or ”optogenetic rewards”.
Preference scores are shown for the trained odor, ethyl acetate, when it was paired (paired group, closed circles) or not paired (unpaired group, open circles) with optogenetic punishments or rewards. These Preference scores are used to compute the Learning Performance Scores as shown in Fig. 1c, 1e and 6a-c, additional testing conditions are also shown here. With some natural punishments, aversive memory is behaviorally expressed by trained Drosophila larvae only if the punishment is present at the moment of the test16. Here we assayed olfactory aversive memories in two ways: both with or without optogenetic punishment (red and black bars, respectively) during the retention test. Odor preference was decreased and increased, respectively, relative to genetic controls, after pairing the odor with the presence and absence of the following optogenetic punishments: co-activation of the aversive DAN-f1 and DAN-g1, co-activation of DAN-f1 and DAN-c1, or activation of Basins. Aversive memory formed by DAN activation (green) or by Basins activation (blue) was expressed to the same extent with or without the DANs activated during the retention test. However, memory evoked by the activation of nociceptive MD IV neurons (orange) or FB2N-19 (yellow) was fully expressed only if these neurons were active again during the retention test. Odor preference was increased and decreased, respectively, relative to genetic controls, after pairing the odor with the presence and absence of the following optogenetic rewards: the co-activation of DAN-h1, -i1, and -k1 (dark green); the activation of FB2N-18 and FB2N-11 (yellow), or activation of FAN-7 (blue-gray). Thus, both absence of odor in the unpaired group of animals, as well as the presence of odor in the paired group of animals can be associated with the activation of some DANs or some of their afferent neurons. These results suggest that presenting an odor unpaired with the activation of some of these DANs induces memory of opposite valence to the paired presentation. For other DANs or afferent neurons, only paired (e.g. A00c, purple) or only unpaired (e.g. the modulatory DAN-f1, the nociceptive MD IV sensory neuron, or FB2N-19) contingency significantly affected odor preference with respect to the control group. Either of these two observed types of effects can contribute to the negative or positive learning performance indexes plotted in Fig. 1c, 1e and 6a-c.
Sample sizes: N = 42, 11, 17, 16, 12, 14, 12, 13, 12, 16, 12, 14, 12, 12, 15, 14, 12, 11, 14, 13, 12, 14, 11, 11, 11, 18, 11, 20, 25, 33, 52,14, 21, 14, 14, 18, 18, 31, 52, 27, 11,11, 10, 13, 20 (control groups in bold). Mean and standard deviations are shown. Black *: p-value<Holm-Bonferroni-adjusted threshold for 0.05 from a two-sided Wilcoxon signed rank test comparison between paired and unpaired group. Grey *: p-value<Holm-Bonferroni-adjusted threshold for 0.05 from a two-sided Mann-Whitney U test comparison between the preference scores for a given group (paired or unpaired) and the preference scores (for paired or unpaired protocol, respectively) obtained by the control line shown on the left of each set of data. Exact p-values are available in SourceData_Figure1.xls.
Extended Data Fig. 3. Matrix of similarity between modulator neurons based on the amount of common input.
Similarity is obtained by counting the total number of inputs onto row modulatory neurons that are also inputs of column modulatory neurons, and dividing by the total number of inputs onto column modulatory neurons. An input here is a connection, consisting typically of many synapses, from a specific cell type onto the modulatory neuron. Inputs onto a modulatory neuron are considered if the pair of left and right neurons presynaptic to the pair of left and right modulatory neurons is each above a thresh-old of 1% (e.g. the presynaptic neuron makes 3 synapses onto a neuron with 300 postsynaptic sites) and the sum of both is over 3.3% (e.g. the sum of both connections is above 10 synapses for receiving neurons with 300 postsynaptic sites).
Interestingly, functionally similar DANs, whose activation leads to aversive memory for paired odors share a higher fraction of presynaptic partners with each other than with other DANs. By contrast some modulatory neurons that innervate the same compartment but express different neurotransmitters (e.g. OAN-g1 and DAN-g1) receive inputs from drastically different subsets of pre-modulatory neurons. Such modulatory neurons that innervate the same compartment could therefore be differentially recruited during learning. Interestingly though, OAN-e1 and MBIN-e2 in the UVL, whose activation paired with odor did not in-duce memory in our paradigm and that were not significantly activated by fictive punishments share a higher fraction of their input with the VL/LA DANs than with other modulatory neurons. This raises the possibility that the UVL modulatory neurons may be recruited by similar stimuli to the VL/LA DANs, but only in specific circumstances.
Extended Data Fig. 4. Input onto feedback neurons.
Figure shows the fractions of total dendritic input each pre-modulatory neuron (FBN, FB2N or FFN) receives from KCs, modulatory neurons, MBONs, FBNs, FB2Ns, FFNs, and from other non-MB neurons (others). a FBNs receive on average 12% of their inputs directly from MBONs and most of them also receive inputs from other FBNs, with an average of 26% from MBONs and other FBNs combined (see also Supplementary Figure 4a). b FB2Nsreceive inputs both from FBNs (on average 17%) and from other FB2Ns (on average 28% from FBNs and FB2Ns combined).Many feedback neurons also receive a significant fraction of input from other unknown neurons from other brain areas (other than MB), suggesting that the feedback about the learnt valences of stimuli is integrated with or modulated by other information. c Tables show percent of inputs onto FBNs (top) and FB2Ns (bottom) from MBONs, FBNs, FB2Ns, FFNs, modulatory neurons and Kenyon cells.
Extended Data Fig. 5. (related to Fig. 3a) Modulatory neurons receive convergent one-step feedback from multiple MBONs from functionally distinct compartments.
Connectivity of each of the 40 feedback neuron (FBN) pairs that provide one-step feedback from MBONs to DANs. Each diagram represents the connectivity of a single left-right pair of homologous FBNs. Each box indicates a separate compartment. Purple, compartment(s) of the presynaptic MBON(s). Green, compartment(s) of the postsynaptic modulatory neuron(s). FBNs are ordered according to the modulatory neuron they innervate, starting with peduncle modulatory neurons and ending with the medial lobe ones. Classical neurotransmitter profiles of the MBONs and FBNs are indicated by the arrow (cholinergic, excitatory connection), vertical line (GABAergic, inhibitory connection) or square (glutamatergic, probably also inhibitory connection) for the neurons for which they are known from immunostaining (For images, see Extended Data Fig. 9 for FBNs and ref.12 for MBONs), or by a circle when they are unknown. 7 FBNs provide exclusively within-compartment feedback. 13 FBNs provide exclusively cross-compartment feedback (named FANs, for feed-across). 8 FBNs synapse onto multiple modulatory neurons from multiple compartments. The largest class of FBNs (17) receives input from multiple MBONs, with the majority (at least 13) receiving input of potentially opposite sign from MBONs from functionally distinct compartments. More than a quarter of FBNs (at least 12) receive direct GABAergic(inhibitory) or glutamatergic (also potentially inhibitory) input from MBONs from one compartment and direct cholinergic(excitatory) input from MBONs from a functionally distinct compartments enabling them to compare the odor drive to these MBONs. Many DANs (DAN-f1, d1, i1, j1, and k1) receive potentially inhibitory (excitatory FBN downstream of an inhibitory MBON) one-step feedback from MBONs from one compartment and potentially disinhibitory (inhibitory FBN downstream of an inhibitory MBON) or excitatory (excitatory FBN downstream of an excitatory MBON) one-step feedback from MBONs from a functionally distinct compartment. A common pattern for the lobe DANs implicated in memory formation may be a likely inhibitory connection from an MBON from their own compartment and a likely disinhibitory connection from an MBON from a compartment of opposite valence (observed for both DAN-g1 and i1), that could enable these DANs to compare the odor drive to MBONs from compartments of opposite valence.
Extended Data Fig. 6. Clustering FBNs based on input from MBONs.
a Heat map of FBN similarity based on the pattern of FBN synaptic inputs from MBONs. The similarity between a pair of FBNs was computed as the cosine similarity between the vectors of their normalized synaptic inputs from all MBONs. Indices were ordered by agglomerative clustering with average linkage (dendrogram shown at top). We highlight six groups of FBNs defined by similarities in their input patterns (bold lines in dendrogram, numbered). b Heat maps showing patterns of input from MBONs onto FBNs for the input groups highlighted in a. In all cases, connectivity is measured in normalized synaptic input on the postsynaptic neuron. Most input groups receive dominant input from a single specific MBON (Groups 1,2, 5) or small group of MBONs (Groups 3 and 4), while Group 6 is not well-clustered and contains a variety of dissimilar input patterns. c Heat maps showing the patterns of synaptic output from FBNs to modulatory neurons for the input groups highlighted in a. d The observed similarity in the output patterns between FBNs within each group, compared to shuffled data. For each group clustered by input pattern, we computed the observed median of cosine similarity of the output vectors across all pairs of neurons (red line). In Groups 1,2, and 3, the neurons clustered by inputs had more similar output patterns than would be expected by chance. To determine significance, we compared the observed similarity to the distribution of the median cosine similarity for randomly permuted samples from the observed population of input vectors (black histograms, n=10000 randomized trials) with a one-sided permutation test. A Holm-Sidak correction was applied to p-values to correct for multiple comparisons. n.s.: p>0.05.
Extended Data Fig. 7. Clustering FBNs based on output onto modulatory neurons.
a Heat map of FBN similarity based on the pattern of FBN synaptic output across all modulatory neurons. The similarity between a pair of FBNs was computed as the cosine similarity between the vectors of normalized synaptic output onto all modulatory neurons. Indices were ordered by agglomerative clustering with average linkage (dendrogram shown at top). We highlight six groups of FBNs defined by similarities in their output patterns (bold lines in dendrogram, numbered). b Heat maps showing patterns of synaptic output from FBNs to modulatory neurons for output groups highlighted in a. Each group corresponds to several FBNs strongly targeting one or a small number of modulatory neurons, suggesting that some modulatory neurons are more strongly modulated than others. c Heat maps showing patterns of input onto FBNs from MBONs for the output groups highlighted in a. d The observed similarity in the input patterns between FBNs within each group, compared to shuffled data. For each group (as defined by output patterns), we computed the observed median of cosine similarity of the input vectors across all pairs of neurons (red line). In Groups 1–5, the neurons clustered by outputs had more input output patterns than would be expected by chance. To determine significance, we compared the observed similarity to the distribution of the median cosine similarity for randomly permuted samples from the observed population of input vectors (black histograms, n=10000 randomized trials) with a one-sided permutation test. A Holm-Sidak correction was applied to p-values to correct for multiple comparisons. n.s.: p>0.05.
Extended Data Fig. 8. Direct MBONs to modulatory neuron connectivity is very sparse, in contrast to the very dense connectivity via one-and two-step feedback pathways.
a Connectivity matrix showing normalized synaptic input (expressed as % input) each modulatory neuron (columns) receives from each MBON (rows). Only reliable connections for which the postsynaptic neuron receives at least 1% of input from the presynaptic neuron are shown. When the neurotransmitter of the MBON is known, the circle is color-coded to represent type of connection: excitatory (ChAT) or probably disinhibitory (GluT). Color shades represent the valence of the memory formed in a given compartment (red: aversive memory, blue: appetitive memory). True within-compartment feedback connections from an MBON that receives direct synaptic input from that modulatory neuron are boxed in bold. Very few modulatory neurons receive direct input from MBONs, in contrast to the dense connectivity between MBONs and modulatory neurons via the indirect one- and two-step feedback pathways (b). b Connectivity matrix showing indirect connections between MBONs and modulatory neurons via one-step and/or two-step feedback pathways. The matrix was obtained by summing the matrices from Fig. 3b and Fig. 5e. The color indicates the type of indirect connection existing between a given MBON and a given DAN. Bubble size represents a connectivity index computed as in Fig. 3b and Fig. 5e. A connectivity index of 1 or 10 means that for all connections comprising that indirect feedback pathway the presynaptic neuron accounts for 1% and 10% of input onto that postsynaptic neuron, respectively. One- and two-step feedback drastically increases the connectivity between MBONs and modulatory neurons, compared to direct connections (a).
Extended Data Fig. 9. Identification of neurotransmitters expressed in some FBNs/FB2Ns.
Neurotransmitter expression detected in neuron somata using antibody labelling. We identified GAL4 lines that drive gene expression in some of the FBN or FB2N neurons and used them to express GFP in these neurons. We stained central nervous systems with antibodies against GFP and either ChAT (choline acetyltransferase), GABA (gamma aminobutyric acid) or GLUT (vesicular glutamate transporter).Each row shows from left to right: the name of the individual neuron, anti-GFP (green), anti-ChAT (magenta), and both antibody stainings combined; anti-GFP (green), anti-GABA (magenta), and both antibody stainings combined; anti-GFP (green) and anti-GLUT (magenta), and both antibody stainings combined. Whether a cell is cholinergic, GABAergic or glutamatergic is listed at the beginning of each row under the neuron name. Images show confocal maximum intensity projections of specific neuronal cell bodies. At least two replicates were obtained per genotype. Scale bars: 5μm.
Extended Data Fig. 10. (related to Fig. 6) Identification of driver lines for EM-reconstructed Feedback neurons.
We were able to generate Split-GAL4 lines that drive expression in a single pair of neurons, or in very few cell types, for three different pairs of FBNs or FB2Ns that target VL DANs (a-c). We used these lines to optogenetically activate these neurons instead of a US during an associative learning paradigm (Fig. 6). i) Skeletons of specific feedback neurons reconstructed in the EM. Red dots, presynaptic sites. Blue dots, postsynaptic sites. Grey, mushroom body vertical lobe (MB vl) for reference. d, dendritic arbor. ii) Maximum intensity projections of confocal stacks of larval brains showing the same neurons visualized with reporters targeted using specific Split-GAL4 lines. For some lines multicolor flp-outs were used to visualize each neuron in a different color to facilitate identification and comparison with EM (N=1). Grey, neuropil visualized with N-cad. Dashed line, brain outline. iii) Maximum intensity projections of confocal stacks of the entire nervous system showing the complete expression pattern of each line revealed by driving UAS-myr-GFP. Grey, neuropil visualized with N-cad. Dashed line, nervous system outline. Representative image from N=3. a, The SS02401-Split-GAL4 line drives expression in FB2N-19 (i) in the brain (ii), and very weakly and stochastically (not reproducibly in all samples) in a few ascending neurons and ensheathing glia in the nerve chord (iii). b, The SS02108-Split-GAL4 line drives expression in FAN-7 and MB2ON-86 (i) in the brain visualized with multicolor flp-outs in (ii). Complete expression pattern of SS02108-Split-GAL4 visualized with UAS-myr-GFP shows additional expression in a few somatosensory interneurons in the nerve cord, called ladders, that mediate avoidance behavior and are hence unlikely to have a positive valence and evoke the appetitive memory observed in Fig.6b. We identified the SS04330-Split-GAL4 line as driving expression specifically in the MB2ON-86 neuron and used it as an additional control in Fig.6b. c, The SS01778-Split-GAL4 line drives expression in both FB2N-18 and FB2N-11, which have very similar morphology and very similar connectivity (Supplementary Figures 3 and 4b-d). The SS02181-Split-GAL4 line (ii shows multi-color flp-outs) drives expression in FB2N-18 and in MB2IN-207, one of the weakly connected pre-modulatory neurons from lineage DAMv12. Notice the ventrally projecting dendrite (d), a distinctive feature of MB2IN-207 neuron (i). UAS-myr-GFP expression patterns of the two lines show that they do not drive expression in any other neurons in the nerve cord (iii).
Supplementary Material
Acknowledgements
We thank Fly Light at HHMI Janelia Research Campus (JRC) for generating confocal images of the GAL4 lines, J. Simpson for sharing a 2-photon microscope, Y. Aso and G. Rubin, for sharing unpublished driver lines, V. Jayaraman for sharing unpublished versions of CsChrimson and GCamp6f, L. Feng and I. Andrade for help with circuit mapping from EM, K. Hibbard and JRC FLY Core, for generating some of the fly stocks, Fly EM at JRC for generating the EM volume, T. Saumweber for discussions, D. Bonnery for help with analyses, L. Abbott for helpful comments on the manuscript, Z. Zavala-Ruiz and the JRC Visiting Scientist Program and HHMI JRC for funding. M. Z. was also supported by the ERC consolidator grant RG95162 and Wellcome Trust Investigator Award RG86459. A.L.-K. was supported by the Burroughs Wellcome Foundation, the Gatsby Charitable Foundation, the Simons Collaboration on the Global Brain, and NSF award DBI-1707398. B.G. received grant support from the Deutsche Forschungsgemein-schaft Ge1091/4-1 and FOR 2705-TP2.
Footnotes
Code availability
Python code (used for the model in Fig.7) and Matlab code (used for Supplementary Fig. 3b) can be found in Supplementary_Software.zip. The Matlab code for the analysis of imaging and behavioral data are available from the corresponding author upon request.
Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Data availability
The source data and statistics for Figure 1 are available in SourceData_Figure1.xls. The electron microscopy volume of the whole central nervous system24 and the reconstructed neurons and synapses included in this paper are publicly available at https://l1em.catmaid.virtualflybrain.org/, accessible online thanks to the software CATMAID63;64–the same software used to reconstruct and analyze the neuronal circuits included here. Supplementary Adjacency MatriXx1.csv, 2.csv, and 3.csv contain all the connectivity information for all neurons from the Extended MB network discussed in the study. The light-microscopy image stacks of genetic driver lines are available from corresponding authors upon request.
Author contributions
C. E. conceived the study, designed and performed experiments and data analysis, and wrote the manuscript. A. F. performed circuit reconstruction and analysis, designed and performed experiments and data analysis, and wrote the manuscript. M. Z. and A. C. conceived the study, performed data analysis, and wrote the manuscript. A. L. K. developed the model and wrote the manuscript. M. W., C. M. S.-M. and J. V.-A. performed circuit reconstruction and analysis. M. S. performed patch-clamp recordings. R. A., K. E., and T. O. performed experiments. A. S. T. and B. G. contributed data analysis and experimental design. R. D. F. and J. W. T. contributed data and reagents.
Competing Interests Statement
The authors declare no competing interests.
References
- [1].Schultz W. Neuronal Reward and Decision Signals: From Theories to Data. Physiol Rev. 2015;95:853–951. doi: 10.1152/physrev.00023.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dunsmoor JE, Niv Y, Daw N, Phelps EA. Rethinking Extinction. Neuron. 2015;88:47–63. doi: 10.1016/j.neuron.2015.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cognigni P, Felsenberg J, Waddell S. Do the right thing: neural network mechanisms of memory formation, expression and update in drosophila. Current opinion in neurobiology. 2018;49:51–58. doi: 10.1016/j.conb.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Watabe-Uchida M, Eshel N, Uchida N. Neural Circuitry of Reward Prediction Error. Annu Rev Neurosci. 2017 doi: 10.1146/annurev-neuro-072116-031109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Menzel R. Searching for the memory trace in a mini-brain, the honeybee. Learn Mem. 2001;8:53–62. doi: 10.1101/lm.38801. [DOI] [PubMed] [Google Scholar]
- [6].Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- [7].Rescorla RA, Wagner AR. A theory of Pavlovian conditioning: Variations in the effectiveness of reinforcement and nonreinforcement. Classical Conditioning II: Current Research and Theory. 1972:64–99. [Google Scholar]
- [8].Sutton RS, Barto AG. Adaptive computation and machine learning. xviii. MIT Press; 1998. Reinforcement learning: an introduction; p. 322. [Google Scholar]
- [9].Watabe-Uchida M. The basal ganglia sensory system listens to prefrontal task needs. Neuron. 2019;103:353–355. doi: 10.1016/j.neuron.2019.07.018. [DOI] [PubMed] [Google Scholar]
- [10].Riemensperger T, Vller T, Stock P, Buchner E, Fiala A. Punishment prediction by dopaminergic neurons Drosophila. Current Biology. 2005;15:1953–1960. doi: 10.1016/j.cub.2005.09.042. [DOI] [PubMed] [Google Scholar]
- [11].Dylla KV, Raiser G, Galizia CG, Szyszka P. Trace Conditioning in Drosophila Induces Associative Plasticity in Mushroom Body Kenyon Cells and Dopaminergic Neurons. Front Neural Circuits. 2017;11:42. doi: 10.3389/fncir.2017.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Eichler K, et al. The complete connectome of a learning and memory centre in an insect brain. Nature. 2017;548:175. doi: 10.1038/nature23455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Takemura SY, et al. A connectome of a learning and memory center in the adult Drosophila brain. eLife. 2017;6 doi: 10.7554/eLife.26975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Davis RL. Traces of Drosophila memory. Neuron. 2011;70:8–19. doi: 10.1016/j.neuron.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Thum AS, Gerber B. Connectomics and function of a memory network: the mushroom body of larval Drosophila. Curr Opin Neurobiol. 2019;54:146–154. doi: 10.1016/j.conb.2018.10.007. [DOI] [PubMed] [Google Scholar]
- [16].Handler A, et al. Distinct Dopamine Receptor Pathways Underlie the Temporal Sensitivity of Associative Learning. Cell. 2019;178:60–75 e19. doi: 10.1016/j.cell.2019.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Waddell S. Reinforcement signalling in Drosophila; dopamine does it all after all. Curr Opin Neurobiol. 2013;23:324–329. doi: 10.1016/j.conb.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Aso Y, Rubin GM. Dopaminergic neurons write and update memories with cell-type-specific rules. eLife. 2016;5 doi: 10.7554/eLife.16135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Schroll C, et al. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol. 2006;16:1741–1747. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- [20].Saumweber T, et al. Functional architecture of reward learning in mushroom body extrinsic neurons of larval drosophila. Nature communications. 2018;9 doi: 10.1038/s41467-018-03130-1. 1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Caldwell JC, Miller MM, Wing S, Soll DR, Eberl DF. Dynamic analysis of larval locomotion in Drosophila chordotonal organ mutants. Proc Natl Acad Sci U S A. 2003;100:16053–16058. doi: 10.1073/pnas.2535546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tracey W, Wilson R, Laurent G, Benzer S. painless, a Drosophila gene essential for nociception. Cell. 2003;113:261–273. doi: 10.1016/s0092-8674(03)00272-1. [DOI] [PubMed] [Google Scholar]
- [23].Ohyama T, et al. High-throughput analysis of stimulus-evoked behaviors in Drosophila larva reveals multiple modality-specific escape strategies. PLoS One. 2013;8:e71706. doi: 10.1371/journal.pone.0071706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ohyama T, et al. A multilevel multimodal circuit enhances action selection in Drosophila. Nature. 2015;520:633–639. doi: 10.1038/nature14297. [DOI] [PubMed] [Google Scholar]
- [25].Jovanic T, et al. Competitive Disinhibition Mediates Behavioral Choice and Sequences in Drosophila. Cell. 2016;167:858–870 e19. doi: 10.1016/j.cell.2016.09.009. [DOI] [PubMed] [Google Scholar]
- [26].Eschbach C, et al. Associative learning between odorants and mechanosensory punishment in larval Drosophila. Journal of Experimental Biology. 2011;214:3897–3905. doi: 10.1242/jeb.060533. [DOI] [PubMed] [Google Scholar]
- [27].Liu WW, Wilson RI. Glutamate is an inhibitory neurotransmitter in the Drosophila olfactory system. Proc Natl Acad Sci U S A. 2013;110:10294–10299. doi: 10.1073/pnas.1220560110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fushiki A, et al. A circuit mechanism for the propagation of waves of muscle contraction in Drosophila. eLife. 2016;5 doi: 10.7554/eLife.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jiang L, Litwin-Kumar A. Models of heterogeneous dopamine signaling in an insect learning and memory center. bioRxiv. 2019 doi: 10.1371/journal.pcbi.1009205. 737064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mancini N, et al. Reversal learning in Drosophila larvae. Learning & Memory. 2019;26:424–435. doi: 10.1101/lm.049510.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Felsenberg J, et al. Integration of Parallel Opposing Memories Underlies Memory Extinction. Cell. 2018;175:709–722.e15. doi: 10.1016/j.cell.2018.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tabone CJ, de Belle JS. Second-order conditioning in Drosophila. Learn Mem. 2011;18:250–253. doi: 10.1101/lm.2035411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Brembs B, Wiener J. Context and occasion setting in Drosophila visual learning. Learn Mem. 2006;13:618–628. doi: 10.1101/lm.318606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schultz W, Dayan P, Montague PR. A neural substrate of prediction and reward. Science. 1997;275:1593–1599. doi: 10.1126/science.275.5306.1593. [DOI] [PubMed] [Google Scholar]
- [35].Zhao X, Lenek D, Dag U, Dickson BJ, Keleman K. Persistent activity in a recurrent circuit underlies courtship memory in Drosophila. eLife. 2018;7 doi: 10.7554/eLife.31425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cohn R, Morantte I, Ruta V. Coordinated and Compartmentalized Neuromodulation Shapes Sensory Processing in Drosophila. Cell. 2015;163:1742–1755. doi: 10.1016/j.cell.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ichinose T, et al. Reward signal in a recurrent circuit drives appetitive long-term memory formation. eLife. 2015;4:1–18. doi: 10.7554/eLife.10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ueoka Y, Hiroi M, Abe T, Tabata T. Suppression of a single pair of mushroom body output neurons in Drosophila triggers aversive associations. FEBS Open Bio. 2017;7:562–576. doi: 10.1002/2211-5463.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Felsenberg J, Barnstedt O, Cognigni P, Lin S, Waddell S. Re-evaluation of learned information in Drosophila. Nature. 2017;544:240–244. doi: 10.1038/nature21716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pavlowsky A, Schor J, Placais PY, Preat T. A GABAergic Feedback Shapes Dopaminergic Input on the Drosophila Mushroom Body to Promote Appetitive Long-Term Memory. Curr Biol. 2018;28:1783–1793.e4. doi: 10.1016/j.cub.2018.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].König C, Khalili A, Niewalda T, Gao S, Gerber B. An optogenetic analogue of second-order reinforcement in Drosophila. Biology Letters. 2019;15 doi: 10.1098/rsbl.2019.0084. 20190084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lyutova R, et al. Reward signaling in a recurrent circuit of dopaminergic neurons and peptidergic kenyon cells. Nature communications. 2019;10 doi: 10.1038/s41467-019-11092-1. 3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tian J, et al. Distributed and Mixed Information in Monosynaptic Inputs to Dopamine Neurons. Neuron. 2016;91:1374–1389. doi: 10.1016/j.neuron.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Beier KT, et al. Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell. 2015;162:622–634. doi: 10.1016/j.cell.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Haber SN, Fudge JL, McFarland NR. Striatonigrostriatal pathways in primates form an ascending spiral from the shell to the dorsolateral striatum. J Neurosci. 2000;20:2369–2382. doi: 10.1523/JNEUROSCI.20-06-02369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- [47].Menegas W, et al. Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. eLife. 2015;4:e10032. doi: 10.7554/eLife.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jenett A, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell reports. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Aso Y, et al. The neuronal architecture of the mushroom body provides a logic for associative learning. eLife. 2014;3:e04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Vogelstein JT, et al. Discovery of brainwide neural-behavioral maps via multiscale unsupervised structure learning. Science. 2014;344:386–392. doi: 10.1126/science.1250298. [DOI] [PubMed] [Google Scholar]
- [55].Yang CH, et al. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron. 2009;61:519–526. doi: 10.1016/j.neuron.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Awasaki T, Huang Y, O’Connor MB, Lee T. Glia instruct developmental neuronal remodeling through TGF-beta signaling. Nat Neurosci. 2011;14:821–823. doi: 10.1038/nn.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Liu C, et al. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature. 2012;488:512–516. doi: 10.1038/nature11304. [DOI] [PubMed] [Google Scholar]
- [58].Pfeiffer BD, Truman JW, Rubin GM. Using translational enhancers to increase transgene expression in Drosophila. Proc Natl Acad Sci U S A. 2012;109:6626–6631. doi: 10.1073/pnas.1204520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Rohwedder A, et al. Four Individually Identified Paired Dopamine Neurons Signal Reward in Larval Drosophila. Curr Biol. 2016;26:661–669. doi: 10.1016/j.cub.2016.01.012. [DOI] [PubMed] [Google Scholar]
- [60].Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- [61].Cardona A, et al. TrakEM2 software for neural circuit reconstruction. PLoS One. 2012;7:e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Saalfeld S, Fetter R, Cardona A, Tomancak P. Elastic volume reconstruction from series of ultra-thin microscopy sections. Nat Methods. 2012;9:717–720. doi: 10.1038/nmeth.2072. [DOI] [PubMed] [Google Scholar]
- [63].Saalfeld S, Cardona A, Hartenstein V, Tomancak P. CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics. 2009;25:1984–1986. doi: 10.1093/bioinformatics/btp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Schneider-Mizell CM, et al. Quantitative neuroanatomy for connectomics in Drosophila. eLife. 2016;5 doi: 10.7554/eLife.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Berck ME, et al. The wiring diagram of a glomerular olfactory system. eLife. 2016;5 doi: 10.7554/eLife.14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Goodman CS, Bate M, Spitzer NC. Embryonic development of identified neurons: origin and transformation of the H cell. J Neurosci. 1981;1:94–102. doi: 10.1523/JNEUROSCI.01-01-00094.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Bate M, Goodman CS, Spitzer NC. Embryonic development of identified neurons: segment-specific differences in the H cell homologues. J Neurosci. 1981;1:103–106. doi: 10.1523/JNEUROSCI.01-01-00103.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Costa M, Manton JD, Ostrovsky AD, Prohaska S, Jefferis GS. NBLAST: Rapid, Sensitive Comparison of Neuronal Structure and Construction of Neuron Family Databases. Neuron. 2016;91:293–311. doi: 10.1016/j.neuron.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Daniels RW, Gelfand MV, Collins CA, DiAntonio A. Visualizing glutamatergic cell bodies and synapses in Drosophila larval and adult CNS. J Comp Neurol. 2008;508:131–152. doi: 10.1002/cne.21670. [DOI] [PubMed] [Google Scholar]
- [71].Nern A, Pfeiffer BD, Rubin GM. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc Natl Acad Sci U S A. 2015;112:E2967–76. doi: 10.1073/pnas.1506763112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Li HH, et al. A GAL4 driver resource for developmental and behavioral studies on the larval CNS of Drosophila. Cell Rep. 2014;8:897–908. doi: 10.1016/j.celrep.2014.06.065. [DOI] [PubMed] [Google Scholar]
- [73].Selcho M, Pauls D, Han KA, Stocker RF, Thum AS. The role of dopamine in Drosophila larval classical olfactory conditioning. PLoS One. 2009;4:e5897. doi: 10.1371/journal.pone.0005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Selcho M, Pauls D, Huser A, Stocker RF, Thum AS. Characterization of the octopaminergic and tyraminergic neurons in the central brain of Drosophila larvae. J Comp Neurol. 2014;522:3485–3500. doi: 10.1002/cne.23616. [DOI] [PubMed] [Google Scholar]
- [75].Pauls D, Selcho M, Gendre N, Stocker RF, Thum AS. Drosophila larvae establish appetitive olfactory memories via mushroom body neurons of embryonic origin. The Journal of Neuroscience. 2010;30:10655–10666. doi: 10.1523/JNEUROSCI.1281-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Marley R, Baines RA. Dissection of third-instar Drosophila larvae for electrophysiological recording from neurons. Cold Spring Harb Protoc. 2011;2011 doi: 10.1101/pdb.prot065656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.