A systemic analysis identifies GROWTH-REGULATING FACTORS as DELLA-targeted transcription factors during cold stress in Arabidopsis.
Abstract
DELLA proteins are repressors of the gibberellin (GA) hormone signaling pathway that act mainly by regulating transcription factor activities in plants. GAs induce DELLA repressor protein degradation and thereby control a number of critical developmental processes as well as responses to stresses such as cold. The strong effect of cold temperatures on many physiological processes has rendered it difficult to assess, based on phenotypic criteria, the role of GA and DELLAs in plant growth during cold stress. Here, we uncover substantial differences in the GA transcriptomes between plants grown at ambient temperature (21°C) and plants exposed to cold stress (4°C) in Arabidopsis (Arabidopsis thaliana). We further identify over 250, to the largest extent previously unknown, DELLA-transcription factor interactions using the yeast two-hybrid system. By integrating both data sets, we reveal that most members of the nine-member GRF (GROWTH REGULATORY FACTOR) transcription factor family are DELLA interactors and, at the same time, that several GRF genes are targets of DELLA-modulated transcription after exposure to cold stress. We find that plants with altered GRF dosage are differentially sensitive to the manipulation of GA and hence DELLA levels, also after cold stress, and identify a subset of cold stress-responsive genes that qualify as targets of this DELLA-GRF regulatory module.
INTRODUCTION
In vascular plants, the phytohormones of the gibberellin (GA) family are associated most prominently with the promotion of germination, elongation growth, and flowering time (Ueguchi-Tanaka et al., 2007; Yoshida et al., 2014). GAs have, however, many other important but phenotypically less obvious biological functions in plant development as well as in stress responses (Claeys et al., 2014; Yoshida et al., 2014; Van De Velde et al., 2017). At the cellular level, GAs act by binding to GID1 (GIBBERELLIC ACID INSENSITIVE DWARF1) receptors, which induces the ubiquitylation and proteasomal degradation of DELLA repressors such as Arabidopsis (Arabidopsis thaliana) GAI (GIBBERELLIC ACID INSENSITIVE) and RGA (REPRESSOR-OF-ga1-3; Ueguchi-Tanaka et al., 2007; Willige et al., 2007; Yoshida et al., 2014). Through this mechanism, the strong DELLA-imposed growth repression of GA-deficient mutants can be relieved and growth can be normalized (Cao et al., 2005; Willige et al., 2007).
DELLAs regulate a number of structurally diverse transcription factors. DELLA-imposed repression of growth and development can often be explained as the consequence of DELLA interactions with specific transcription factors (de Lucas et al., 2008; Feng et al., 2008; Hou et al., 2010; Bai et al., 2012; Gallego-Bartolomé et al., 2012; Oh et al., 2012; Fukazawa et al., 2017; Van De Velde et al., 2017). In most cases, DELLA interactions result in the tethering of the transcription regulators away from their target promoters (de Lucas et al., 2008; Feng et al., 2008; Yoshida et al., 2014; Davière and Achard, 2016). Almost exclusively, these DELLA-transcription factor interactions have initially been identified with the help of the yeast two-hybrid system and were subsequently experimentally validated in planta (Claeys et al., 2014; Yoshida et al., 2014). In many organisms, DELLA function is encoded by a single gene, and Arabidopsis, which has five functionally redundant DELLAs, represents rather an exception (Gallego-Bartolomé et al., 2010; Van De Velde et al., 2017).
DELLAs belong to the GRAS protein family, which was defined after the cloning of GAI, RGA, and SCR (SCARECROW) from Arabidopsis (Pysh et al., 1999). DELLA family members can be distinguished from other GRAS proteins, such as SCR and SHORT ROOT, by their N-terminal DELLA and the neighboring VHYNP domains (Pysh et al., 1999). GRAS proteins without a DELLA domain have not been directly implicated in GA signaling, although there have been connections between GA-responsive growth and GRAS proteins through the strongly GA-regulated gene SCL3 (SCARECROW-LIKE3; Heo et al., 2011; Zhang et al., 2011; Yoshida and Ueguchi-Tanaka, 2014). Whether non-DELLA GRAS proteins interact with a similarly large number of transcription factors as DELLAs remains to be shown.
GAs and DELLAs regulate abiotic stress responses (Achard et al., 2007, 2008a, 2008b; Colebrook et al., 2014; Conti et al., 2014; Sakata et al., 2014). In relation to cold stress, the growth inhibitory effects after overexpression of CBFs (C-REPEAT BINDING FACTORs), important regulators of cold responses, are partially suppressed by GA application or in DELLA gene loss-of-function mutant backgrounds (Achard et al., 2008a). These phenotypes could, however, not be explained by direct physical DELLA-CBF interactions and, more recently, it was suggested that CBFs act by promoting DELLA stabilization through the activation of the GA catabolic gene GA2ox7 (GA2 OXIDASE7; Achard et al., 2008a; Zhou et al., 2017). Thus, the precise mechanism underlying the role of DELLAs in cold stress regulation remains to be elucidated.
GRFs (GROWTH-REGULATING FACTORs) form a nine-member transcription factor family in Arabidopsis with a role in growth control (e.g., of leaves and roots; Casadevall et al., 2013; Wang et al., 2013; Debernardi et al., 2014; Kim and Tsukaya, 2015; Omidbakhshfard et al., 2015; Rodriguez et al., 2015). GRFs are transcriptional activators that function together with GIFs (GRF INTERACTING FACTORs). In Arabidopsis, seven of the nine GRFs are regulated by the microRNA (miRNA) miR396 (Kim and Kende, 2004; Horiguchi et al., 2005; Lee et al., 2009; Wang et al., 2011; Hewezi et al., 2012; Kim and Tsukaya, 2015). In young leaves, GRF activity is restricted to the proximal proliferative zone of the leaf because miR396b is expressed in the distal part and antagonizes GRF expression (Liu et al., 2009; Rodriguez et al., 2010; Das Gupta and Nath, 2015; Beltramino et al., 2018). In adult leaves, miR396b expression spreads and restricts GRF activity in the entire organ (Beltramino et al., 2018). Plants overexpressing the miRNA-insensitive GRF5 or a miRNA-resistant variant of GRF3, rGRF3, have larger leaves than the wild type, while leaves of plants overexpressing miR396b are smaller (Horiguchi et al., 2005; Beltramino et al., 2018).
The strong general influence of cold temperatures on many physiological and biochemical processes has rendered it difficult to assess, based on phenotypic criteria, the contribution of DELLAs and GA to cold stress responses on plant growth. Furthermore, the multitude of cold response pathways makes it difficult to untangle molecular mechanistic relationships based on genetic studies (Ding et al., 2019). Here, we perform a dedicated molecular analysis to unravel the contribution of GAs and DELLAs to early cold stress responses. To this end, we establish the GA-modulated transcriptome after exposure of Arabidopsis seedlings to cold stress (4°C). We further identify transcription factors interacting with the DELLAs GAI and RGA in the yeast two-hybrid system using a comprehensive collection of 1956 Arabidopsis transcription regulators (Pruneda-Paz et al., 2014). By integrating both data sets, we isolate GRFs as DELLA-regulated transcription factors and unravel their role in GA-dependent plant growth control in the cold.
RESULTS
Cold Stress Promotes DELLA Accumulation and Influences GA Signaling and Biosynthesis
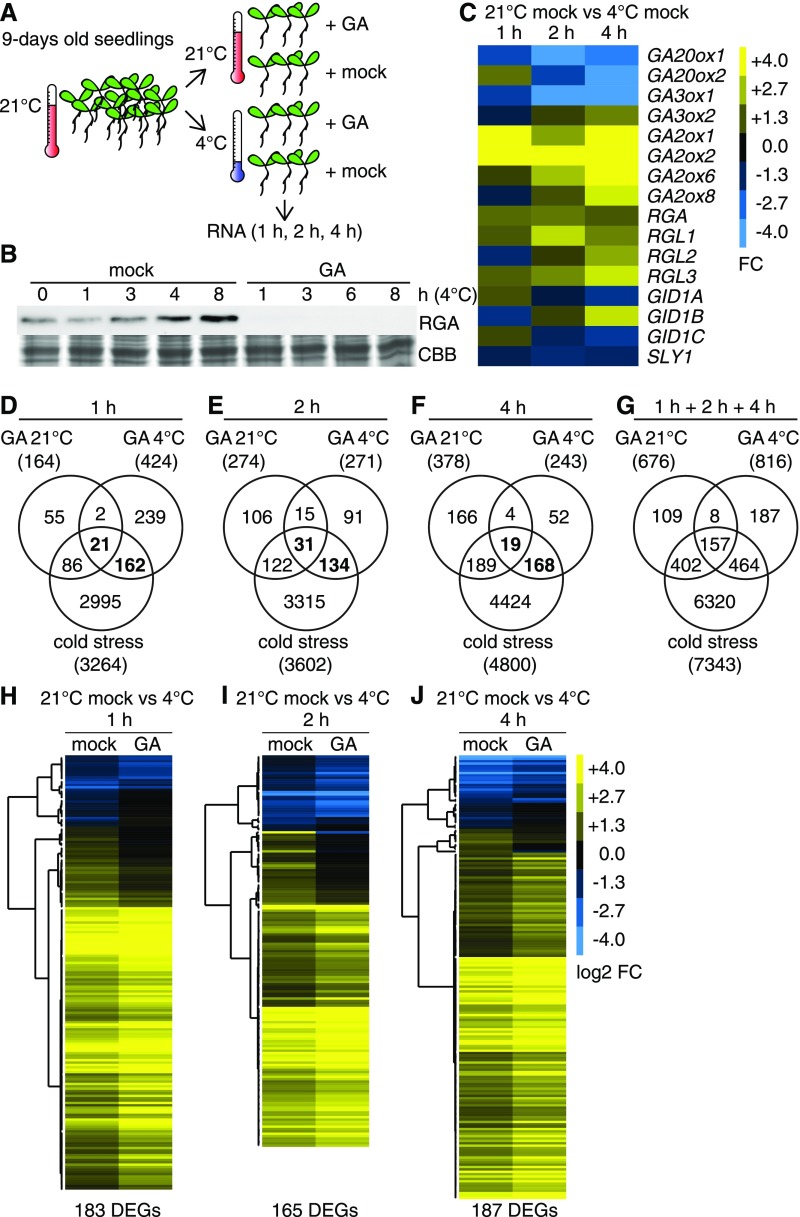
To test the effects of temperature on GA-regulated gene expression, seedlings grown at ambient temperature (21°C) for 9 d on solid medium were sprayed with 100 µM GA3 (GA) or a corresponding solvent control (mock) and immediately transferred to 4°C (cold stress) or kept at 21°C ambient temperature (Figure 1A). As previously reported, cold stress treatment led to an increase in RGA abundance over time but RGA was not detected in samples from GA-treated seedlings, which is an expected consequence of an efficient GA treatment (Figure 1B; Achard et al., 2008a). Using RNA collected from seedlings grown at 4 or 21°C for 1, 2, and 4 h, we performed RNA sequencing (RNA-seq) and examined the resulting data sets with regard to differential gene expression (Figures 1C to 1J; Supplemental Data Sets 1 and 2).
Figure 1.
GA Treatments Induce Substantially Different Transcriptome Changes in the Cold (4°C) and at Ambient (21°C) Temperature.
(A) Schematic representation of the experimental setup.
(B) Immunoblot analysis with an anti-RGA antibody and Coomassie Brilliant Blue (CBB)-stained gel of 9-d-old seedlings exposed to cold stress (4°C) in mock-treated and 100 µM GA3-treated (GA) samples for the specified periods of time.
(C) Heat map of the FC of GA biosynthesis and signaling genes after cold stress.
(D) to (G) Venn diagrams comparing differentially expressed gene sets in cold-stressed samples and GA-treated samples grown at ambient temperature and in the cold as specified. The total number of differentially expressed genes is provided in parentheses. Genes that are GA-regulated and cold stress-regulated are highlighted in boldface.
(H) to (J) Heat maps displaying the logarithmic FC (log2 FC) regulation of the cold- and GA-regulated genes as identified in (D) to (G) for the specified time points. DEGs, differentially expressed genes.
Our analysis of all Arabidopsis GA biosynthesis and signaling genes in the cold-treated samples revealed the downregulation of the GA anabolic genes GA20ox1, GA20ox2, and GA3ox1 and the upregulation of the catabolic GA2ox genes GA2ox1, GA2ox2, GA2ox6, and GA2ox8 (1.5 < fold change [FC], false discovery rate [FDR]-corrected P < 0.01; Figure 1C; Supplemental Data Set 2). These cold-dependent expression changes may be causal for the observed cold-dependent RGA accumulation, since they could result in decreased GA hormone levels (Figure 1B). Additionally, RGA (and more generally DELLA) accumulation could be explained by the observed cold stress-induced transcription of RGA and its paralogs RGA-LIKE1 (RGL1), RGL2, and RGL3 as well as by the downregulation of the GID1A and GID1C GA receptor genes and the SLY1 F-box protein gene (Figure 1C; Supplemental Data Set 2). Thus, cold stress affects GA biosynthesis and signaling genes in a manner that could explain cold stress-induced DELLA accumulation.
When we analyzed the efficiency of the cold stress treatment, we detected a total of 7343 differentially expressed cold stress-regulated genes, when summing up all cold stress-regulated genes from the three time points (1 + 2 + 4 h). This included established cold response genes (e.g., CBF1, CBF2, CBF3, RD29A, COR15A/B, and COR47) and thereby confirmed that the cold stress treatment had been efficient (Figures 1D to 1G; Supplemental Figure 1A; Supplemental Data Set 1; Gilmour et al., 2004; Eremina et al., 2016). In quantitative terms, the total number of cold stress-regulated transcripts, as determined in our analysis, was comparable to the number of genes identified in previous studies, which underlined the representative nature of the experimental setup (Jia et al., 2016; Zhao et al., 2016; Calixto et al., 2018).
When summed up over the three time points (1 + 2 + 4 h), 676 genes were differentially expressed after GA treatment at 21°C (1.5 < FC, FDR-corrected P < 0.01; Figure 1G). These included 18 genes that belonged to a list of 20 reference genes that had previously been identified as being consistently GA-regulated across several experiments (Figures 1D to 1G; Supplemental Figure 1B; Supplemental Data Sets 1 and 3; Claeys et al., 2014). Based on this observation and the fact that the abundance of the GA-labile RGA protein strongly decreased after the GA treatment (Figure 1B), we concluded that the GA treatment had been efficient.
Using the same filtering criteria, we identified a total of 816 GA-regulated genes (1 + 2 + 4 h) after exposure of the seedlings to the cold (1.5 < FC, FDR-corrected P < 0.01; Figure 1G). In comparison with the cold stress treatment, the quantitative effects of the GA treatment resulted in comparatively lower FC values (Supplemental Figure 2A). A principal component analysis performed with the data set revealed a substantially stronger effect of the cold stress treatment on all samples than of the GA treatment (Supplemental Figure 2B). Strikingly, the respective GA transcriptomes obtained from 4°C-grown and 21°C-grown plants differed substantially (Figures 1D to 1G; Supplemental Data Set 1). The overlap between the two gene sets (junction of the GA transcriptomes of 4°C-grown and 21°C-grown plants) was maximal at the 2-h time point, when 46 of 271 (17%; 4°C-grown seedlings) and 274 (17%; 21°C-grown seedlings) transcripts (Figure 1E), respectively, were shared between the GA-regulated gene sets at 4 and 21°C (Figures 1D to 1G; Supplemental Data Set 1). Only 165 (overlap of GA transcriptomes of 4°C-grown and 21°C-grown seedlings) of the 676 (21°C) and 816 (4°C) GA-regulated genes were GA-regulated at both temperatures (Figure 1G; Supplemental Figure 3; Supplemental Data Set 4). We thus concluded that GA and DELLAs control substantially different gene sets at ambient temperature and after exposure to cold stress.
At either temperature, 4 or 21°C, the majority of GA-regulated genes was only GA-responsive at one of the three time points (compare Figures 1D to 1F to Figure 1G; Supplemental Data Set 1). The respective data suggest that this was not a mere result of the applied analysis criteria, which may have led to the inclusion or exclusion of individual genes at a specific time point (Supplemental Figure 3; Supplemental Data Set 4). This was also supported by the principal component analysis, which separated the samples, in the first component, into 21°C-grown and 4°C-grown seedlings and, in the second component, according to the duration of the cold stress treatment (Supplemental Figure 2B).
When the GA- and cold-regulated gene sets were compared, only between 3.9% (4 h; 187 GA-regulated of 4800 cold-regulated genes; marked bold in Figure 1F) and 5.6% (1 h; 183 GA-regulated of 3264 cold-regulated genes; marked bold in Figure 1D) of the cold-regulated genes were also GA-regulated at 4°C (Figures 1D to 1G). The specific analysis of the GA- and cold-regulated genes revealed that GA treatments could modulate cold stress-induced gene expression in a positive or negative manner (Figure 1H; Supplemental Data Set 5). In summary, we found that the GA transcriptome at 4 and 21°C is largely specific for the respective temperature regime, that cold stress has a strong influence on the expression of GA pathway genes, and that the expression of a fraction of the cold stress-responsive genes is modulated by GA.
DELLAs Interact with a Broad Set of Transcription Factors
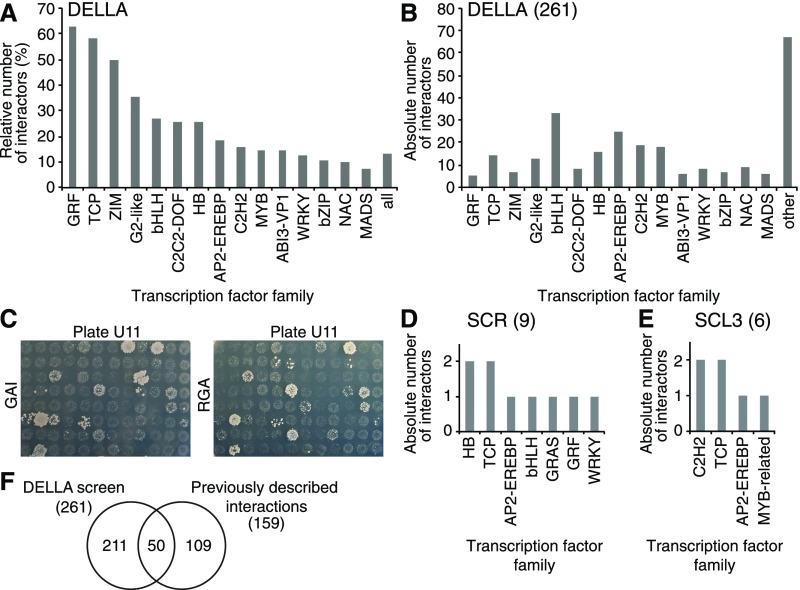
GA-modulated gene expression changes are the expected consequence of changes in DELLA-transcription factor interactions. To obtain an overview of the spectrum of DELLA-interacting transcription factors, we performed yeast two-hybrid interaction screens with RGA and GAI as baits and a collection of 1956 Arabidopsis transcription regulators as preys (Pruneda-Paz et al., 2014). Since full-length DELLA as baits are not compatible with yeast two-hybrid screening, N-terminally truncated versions of the regulators were used, as previously described by de Lucas et al. (2008). The screens identified 244 and 243 interactors of RGA and GAI, respectively (Supplemental Data Set 6). As expected based on the amino acid similarity of RGA and GAI and their known functional redundancy, both interactor sets overlapped substantially (226 or 86.6%) and, when combined, the screen yielded 261 DELLA interactors representing 13% of the transcription regulators in the available collection (Figures 2A to 2C; Supplemental Data Set 6; Gallego-Bartolomé et al., 2010; Pruneda-Paz et al., 2014). The interactors belonged to a total of 51 different transcription factor families, and there was no recognizable common denominator among these interactors in terms of their structure or the presence of known interaction domains (Figure 2B; Supplemental Data Set 6; Pruneda-Paz et al., 2014). In that regard, our results were comparable to previous analyses that had found that DELLAs interact with structurally diverse proteins (Marín-de la Rosa et al., 2014). Only 50 of the 261 interactors (19%) identified in our screen had previously been reported (Figure 2F; Supplemental Data Set 7; Van De Velde et al., 2017). On the one side, the screen retrieved 13 biologically validated DELLA interactions, including those with JASMONATE ZIM DOMAIN1 (JAZ1), INDETERMINATE proteins, and BRASSINAZOLE RESISTANT1 (Supplemental Data Set 7; Hou et al., 2010; Bai et al., 2012; Gallego-Bartolomé et al., 2012; Oh et al., 2012; Fukazawa et al., 2017; Van De Velde et al., 2017). On the other side, 109 previously validated interactions were not found in our screen, although clones for 65 of these 109 interactors were present in the available transcription factor collection, indicating that the complete DELLA-transcription factor interactome is even larger than identified by us. A complete list of all known DELLA interactors, based on our and previous analyses, is provided as Supplemental Data Set 7 (Van De Velde et al., 2017).
Figure 2.
Yeast Two-Hybrid Analyses Identify a Large Number of Structurally Diverse DELLA-Interacting Transcription Factors.
(A) and (B) Relative (A) and absolute (B) numbers of the 261 transcription factor interactions detected in the screens with GAI and RGA for the 15 most prominent transcription factor families. Sixty-seven interactors do not belong to the transcription factor families shown here. Both screens identified a largely overlapping set of interactions (86.6%).
(C) Photographs of one screening plate as a representative result for the GAI and RGA yeast two-hybrid baits, demonstrating the large overlap in the results obtained with the two DELLA bait proteins.
(D) and (E) Absolute numbers of the nine and six DELLA interactions detected in the screens with the non-DELLA GRAS proteins SCR and SCL3, respectively. The identities of the individual transcription factors, as well as an explanation of the transcription factor family nomenclature, are provided in Supplemental Data Set 6.
(F) Venn diagram showing the overlap between results of the DELLA interaction screen as described in this study and previoulsy described interactions.
Truncation of the RGA and GAI N termini for the yeast two-hybrid analysis rendered these DELLAs structurally close to the non-DELLA GRAS proteins SCR and SCL3 (Pysh et al., 1999). We therefore asked whether an equivalent screen with SCR and SCL3 would yield an interaction data set that was, in qualitative or quantitative terms, comparable to that of the DELLA proteins. However, SCR and SCL3 interacted only with nine and six proteins, respectively (Figures 2D and 2E). All but one of their interactors, APETALA2, was also present in the DELLA interactor set (Figures 2D and 2E; Supplemental Data Set 6). Thus, SCR and SCL3, at least according to our analysis, interact with a much smaller set of transcription factors than DELLAs.
DELLA-Interacting GRFs Are Transcribed in a GA- and Cold-Regulated Manner
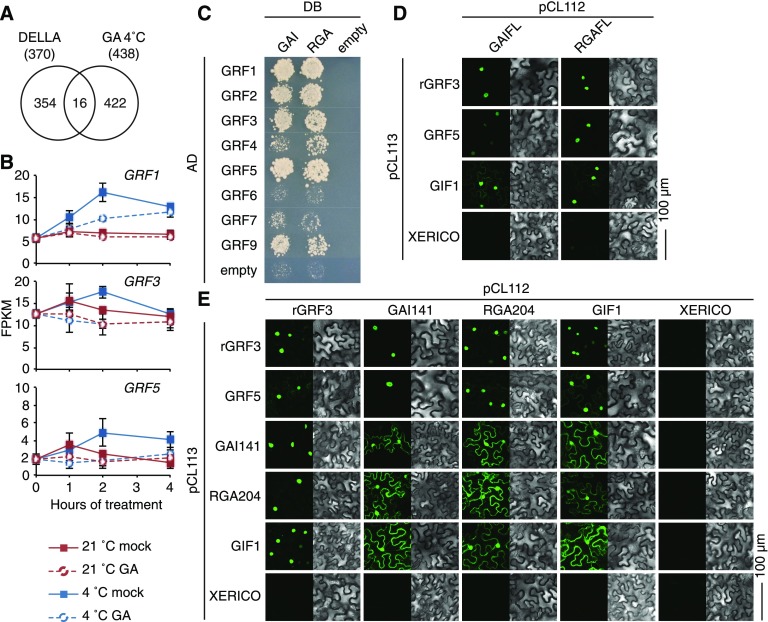
To identify transcription factors that could mediate GA-modulated cold stress responses, we examined the RNA-seq and interaction data sets for candidate genes. Since many transcription regulation events are subject to feedback regulation, we searched for DELLA-interacting transcription factors that were GA-regulated at the transcriptional level. The expression of 14 DELLA-interacting transcription factors, out of 16 DELLA-interacting proteins (including two non-transcription factor genes), was GA-modulated after cold stress and hence fulfilled this criterion (Figures 3A and 3B; Supplemental Figure 4A).
Figure 3.
GRF Transcription Factors Are DELLA Interactors and GRF Expression Is GA-Regulated in the Cold.
(A) Venn diagram showing the overlap between all 370 previously identified DELLA interactions and the 438 cold-regulated genes whose expression is GA- and cold-regulated.
(B) Graphs displaying fragments per kilobase million (FPKM) values of GRF gene expression of seedlings grown in ambient temperature (21°C) and in cold stress (4°C) in the absence (mock) and presence of 100 µM GA3. Shown are means and sd of three biological replicate samples from pooled seedling shoots.
(C) Targeted yeast two-hybrid interaction analysis between eight of the nine Arabidopsis GRF proteins and GAI as well as RGA together with the respective empty vector controls. AD, activation domain; DB, DNA-binding domain.
(D) and (E) Results of BiFC experiments performed in transiently transformed N. benthamiana leaf epidermal cells between full-length DELLA proteins (GAIFL and RGAFL; [D]) and N-terminally truncated DELLA proteins (GAI141 and RGA204; [E]) with interaction candidates as specified. XERICO is a biologically unrelated protein that serves as a negative control (Ko et al., 2006).
GRFs were particularly noteworthy in this regard for two main reasons. First, the cold stress-induced increases in transcript abundance of GRF1, GRF3, and GRF5 were attenuated in the presence of GA, indicating that cold stress-induced accumulation of DELLAs may contribute to this regulation (e.g., through DELLA-transcription factor interactions or by DELLAs promoting GRF expression; Figure 3B; Supplemental Figure 4B). At the same time, GRF genes did not respond, or responded less strongly, to GA when seedlings were grown at 21°C (Figure 3B; Supplemental Figure 4B). Second, five of the nine Arabidopsis GRFs interacted with DELLAs in our screen and, thereby, GRFs represented the transcription factor family with the proportionally largest fraction of DELLA interactors (Figures 2A and 3C). When we repeated the yeast two-hybrid screen in a GRF-targeted manner, we even found additional DELLA interactions with GRF2 and GRF4 (Figure 3C). Only GRF8 could not be analyzed because we failed to isolate a GRF8 cDNA in repeated attempts. Using bimolecular fluorescence complementation (BiFC), we confirmed the interactions between DELLAs and GRFs using N-terminally truncated as well as full-length DELLA variants in combination with the miRNA396-resistant rGRF3 and the miRNA396-nontargeted GRF5 (Figures 3D and 3E). In these experiments, the known GRF interactor GIF1 served as positive control and the unrelated RING E3 ubiquitin ligase XERICO served as negative control (Figures 3D and 3E; Kim and Kende, 2004; Ko et al., 2006). Interestingly, interactions between the truncated DELLA proteins with each other and with GIF1 were also detected in the cytoplasm, whereas they were strongly enriched in the nucleus when the DELLA interactions were tested against the GRFs. We concluded that GRF genes are upregulated after cold stress, that cold stress-induced DELLA accumulation positively regulates their expression, and that GRF proteins interact with DELLAs. GRFs thus qualify as components of a DELLA-regulated cold stress-modulated transcription module.
Leaf Growth Is Differentially Sensitive to GA and GRF Abundance
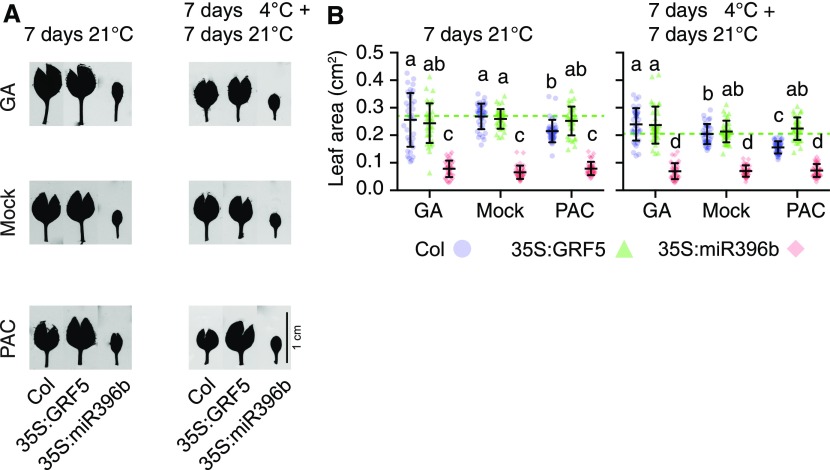
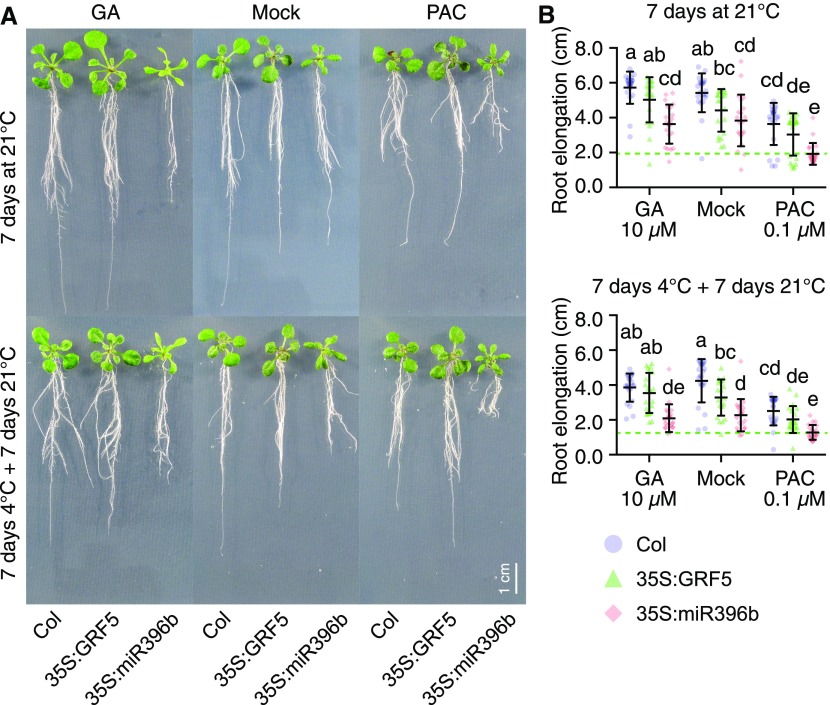
GRFs have been implicated in the control of cell proliferation and leaf growth (Kim et al., 2003; Horiguchi et al., 2005; Rodriguez et al., 2010). To examine the possible contribution of GA and DELLA abundance on GRF-dependent growth processes, we transferred 7-d-old wild-type and GRF5 overexpression seedlings (35S:GRF5) as well as seedlings overexpressing miRNA396b (35S:miRNA396b) to medium containing GA3 (10 µM), the GA biosynthesis inhibitor paclobutrazol (PAC; 0.1, 0.5, and 1.0 µM), or a mock solution. All seedlings were then grown for another 1 week at 21 or 4°C. Since plant growth arrested almost fully at 4°C, the respective seedlings were allowed to recover and grow for an additional 1 week at 21°C before the effects of the cold stress treatment on plant growth were assessed at the phenotype level.
GA treatments had no significant effect on leaf growth in the wild type and the 35S:GFR5 lines in these conditions (Figures 4A and 4B). PAC treatments, however, resulted in reduced leaf growth in these two genetic backgrounds and, interestingly, the relative effects of the PAC treatments were less pronounced in the GRF5 overexpressors than in the wild type (Figures 4A and 4B). Furthermore, the effect was more prominent when plants had been exposed to cold stress (Figures 4A and 4B). This observation would be in line with a scenario where high GRF levels in 35S:GFR5 seedlings outcompete the growth inhibitory effects of DELLAs that are the consequence of the PAC and/or the cold stress treatments. Along the same lines, the leaf area was strongly reduced in seedlings of the 35S:miRNA396b line, where GRFs are downregulated, and these seedlings did not respond to the GA or PAC treatment (Figures 4A and 4B). These findings not only supported previous reports that leaf growth was GRF-dependent but also invited the conclusion that GA and hence DELLAs regulate leaf growth via GRFs in our experimental conditions (Figures 4A and 4B; Rodriguez et al., 2010; Beltramino et al., 2018). These observations thus supported a role for DELLAs in the control of GRF dosage-dependent leaf growth. An extension of this model would suggest that high DELLA abundance, after PAC treatment or in cold-stressed plants, may mimic the growth behavior of 35S:miRNA396b seedlings. This model found support in our observation that leaf growth of the GRF5 overexpressor was partially insensitive to PAC inhibition compared with the wild type on medium containing 0.1 or 0.5 µM PAC and that the difference was fully inhibited at 1 µM PAC (Supplemental Figures 5A and 5B).
Figure 4.
GA Levels Differentially Modulate Leaf Area after Cold Stress in 35S:GRF5.
(A) Representative first true leaves of wild-type seedlings (Col) and of seedlings of transgenic lines overexpressing GRF5 (35S:GRF5), which is not targeted by miRNA396, or the GRF-targeting miRNA396b (35S:miR396b). Seedlings were germinated for 6 d on half strenth MS (without mock or hormone treatment) at ambient temperature. Seedlings were then transferred for 7 d to half strenth MS containing 10 µM GA3 (GA) or 0.1 µM PAC (PAC) or a corresponding mock solution for 7 d at 21°C. In a parallel setup, the seedlings were first exposed to a 7-d 4°C cold stress treatment, which fully arrests plant growth, followed by a 7-d recovery period at 21°C.
(B) Scatterplots of individual measurements (dots) of leaf areas from seedlings shown in (A). Also shown are means and sd of 16 seedlings. The dotted lines mark the means of the mock-treated wild-type sample and serve for orientation. Data sets with no statistical difference after ANOVA and Tukey’s HSD posthoc test fall into one group and are labeled with identical letters.
GRFs and DELLAs Control Cell Size and Cell Number
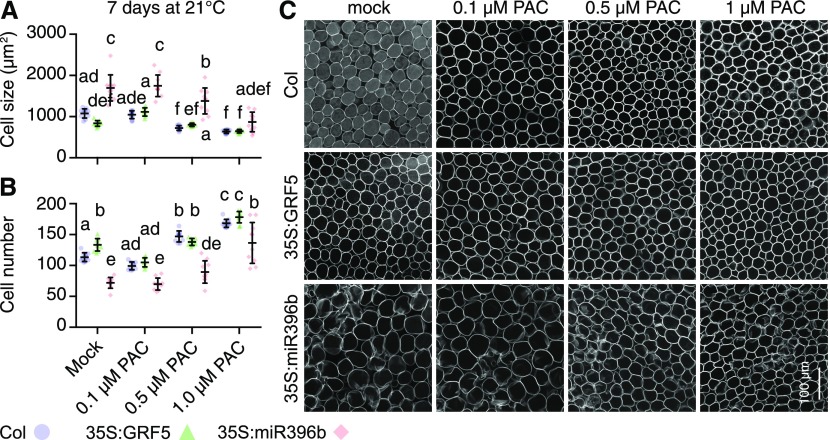
To understand whether the leaf growth differences observed were the result of differences in cell expansion or cell number, we measured cell size and cell number in a 400-µm × 400-µm region from images taken from the middle of the first true leaf of seedlings grown at 21°C on half-strength Murashige and Skoog medium (MS) for 4 d and subsequently on different concentrations of PAC for 7 d (Figure 5). In line with previous reports, we found that cell size in the miRNA396b overexpressor seedlings was significantly increased when compared with the wild type or the GRF5 overexpressor (Figures 5A and 5C; Horiguchi et al., 2005; Ferjani et al., 2007; Rodriguez et al., 2010). Conversely, we observed the corresponding changes in cell numbers (Figures 5B and 5C). In miRNA396b overexpressor seedlings with reduced GRF levels, we expected and found that the PAC treatment had a larger effect on the cell size phenotype than it had in the wild type and GRF5 overexpressors (Figures 5A and 5C). Conversely, cell number was reduced in the untreated miRNA396b overexpressors but, after PAC treatments, increased proportionally more strongly than in wild-type and 35S:GRF5 seedlings (Figures 5B and 5C). Cell size and cell number on different PAC concentrations were comparable between the wild-type and the 35S:GRF5 seedlings. On mock treatment, however, the GRF5 overexpressor had decreased cell size, and consequently more cells per region, indicating that the cells of the GRF5 overexpressor were not fully expanded in this condition (Figure 5). We concluded that the differences in leaf size between the wild type and the GRF5 overexpressor on 0.1 and 0.5 µM PAC may be the result of increased cell numbers in the 35S:GRF5 lines (Figure 4B).
Figure 5.
Cell Size Is Differentially Sensitive to PAC Treatments in Different GRF Genotypes.
(A) and (B) Scatterplots of individual measurements (dots) of cell size (A) and cell number (B) from 400-µm × 400-µm photographs taken from the first two true leaves from 10 seedlings, as shown in (C). Also shown are means and sd of measurements from 10 different leaves. Data sets with no statistical difference after ANOVA and Tukey’s HSD posthoc test fall into one group and are labeled with identical letters.
(C) Representative images of leaf palisade cells underlying the adaxial epidermis in the middle region of the leaf from plants with the specified genotypes and grown on medium supplemented with the specified concentrations of PAC.
Root Elongation and Development Are Hypersensitive to PAC Treatments in miRNA396b Overexpressor Lines
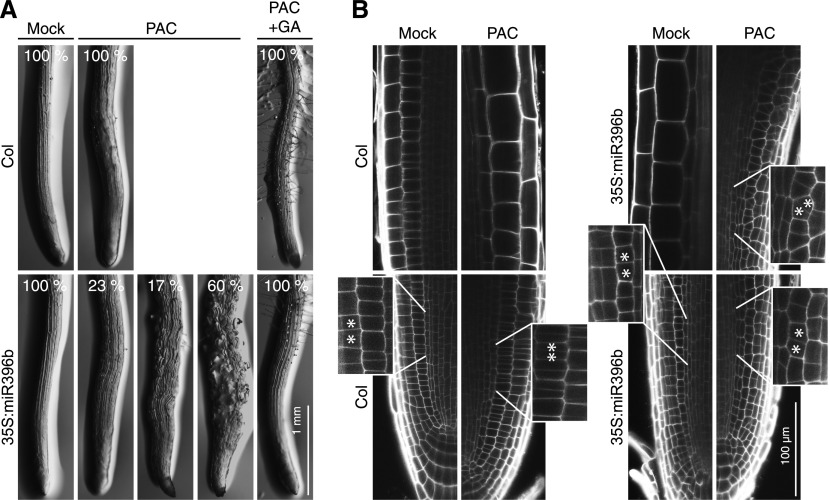
When we analyzed the effects of GA and PAC treatments on root elongation, we noted a strong sensitivity of the miRNA396b overexpressor line to the PAC treatment at ambient temperature and after cold stress treatment, which distinguished this genotype from the wild type and the 35S:GRF5 line (Figure 6; Supplemental Figure 5C). Most strikingly, however, we observed a strong impairment in primary root development in PAC-grown 35S:miRNA396b seedlings (Figure 7). There, we observed a high proportion of seedlings with disturbed root growth (17%) and, in most roots, we even noted the presence of bloated cells (60%; Figure 7A). These defects were fully suppressed when GA3 (10 µM) was applied together with PAC (5 µM), indicating that the PAC hypersensitivity of 35S:miRNA396b lines can be attributed to the effects of PAC on DELLAs (Figure 7A). Detailed examination revealed defects in tissue organization and differentiation (i.e., defects in the cell division planes in the root meristematic and elongation zones; Figure 7B). PAC and GA have not been reported to have similar effects on root development. We concluded that the reduction of GRFs may impair cellular interaction equilibria that, here, lead to effects on root growth and development.
Figure 6.
GA Abundance Modulates Root Elongation in Seedlings after Cold Stress in a GRF-Dependent Manner.
(A) Representative images of wild-type seedlings (Col) and of seedlings of transgenic lines overexpressing GRF5 (35S:GRF5), which is not targeted by miRNA396, or the GRF-targeting miRNA396b (35S:miR396b). Seedlings were germinated for 6 d on half strenth MS (without mock or hormone treatment) at ambient temperature. Seedlings were then transferred for 7 d to half strenth MS containing 10 µM GA3 (GA) or 0.1 µM PAC (PAC) or a corresponding mock solution for 7 d at 21°C. In a parallel setup, the seedlings were first exposed to a 7-d 4°C cold stress treatment, which fully arrests plant growth, followed by a 7-d recovery period at 21°C.
(B) Scatterplots of individual measurements (dots) of root elongation measurements of seedlings shown in (A). The dotted lines mark the means of the mock-treated wild-type sample and serve for orientation. Data sets with no statistical difference after ANOVA and Tukey’s HSD posthoc test fall into one group and are labeled with identical letters.
Figure 7.
miRNA396b Overexpression Lines Are Hypersensitive to PAC Treatments.
(A) Representative images of 13-d-old roots of wild-type and miRNA396b-overexpressing seedlings grown for 8 d on PAC (5 µM), PAC (5 µM) and GA3 (10 µM), and a corresponding mock solution. The relative abundances of seedling roots with the different phenotypes are provided in the images.
(B) Representative photographs of 10-d-old wild-type (Col) and 35S:miR396b roots grown for 5 d on mock or 5 μM PAC. Magnifications of epidermis and cortex cell layers showing cell division defects are shown in boxes.
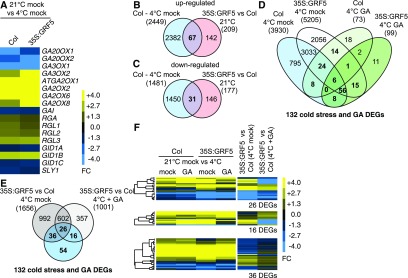
GRF5 Overexpression Alters the GA Transcriptome after Cold Stress
Our observation of a GRF-DELLA interaction at the molecular as well as the physiological level led to the expectation that cold- and GA-modulated transcription responses may be GRF-dependent. To test this hypothesis, we subjected wild-type and 35S:GRF5 seedlings to cold stress and GA treatment as described above (Figure 1A). We restricted the analysis to a single time point (4 h) because the differential GA-modulated GRF transcription was observed after a 2-h stress, and we assumed an additional 2 h for GRF translation (Figure 3B).
A principal component analysis of the data set (4-h time point) separated the samples according to the temperature (principal component 1, 36.4%) and the two contrasting genotypes (principal component 2, 14.8%; Supplemental Figure 6A). The RNA-seq data confirmed that GRF5 was strongly upregulated in 35S:GRF5 seedlings in all experimental conditions when compared with the wild type (Supplemental Figure 6B; Supplemental Data Sets 8 to 10). Furthermore, several GRF genes responded to the cold stress and the GA treatments in a similar manner to what we had observed in our initial RNA-seq experiment (Supplemental Figure 6C; Supplemental Data Set 9). Likewise, the transcription of GA biosynthesis and signaling genes in the wild type was regulated in a manner consistent with a negative feedback regulation that could be the consequence of a cold-imposed DELLA accumulation, as we had observed in the initial transcriptomics analysis (Figure 8A; Supplemental Data Set 9). Taken together, the similar expression behavior of marker genes from the GRF family and the GA pathway indicated that this RNA-seq data set is comparable to the first experiment described in this study (Figure 1).
Figure 8.
Genetic Modulation of GRF Levels Results in Changes of the Cold and the GA Transcriptomes.
(A) Heat map of the FC of GA biosynthesis and signaling genes after cold stress in the wild type (Col) and 35S:GRF5 (Supplemental Data Set 9).
(B) and (C) Venn diagrams identifying genes that are cold stress-regulated in the wild type and upregulated or downregulated in the GRF5 overexpressor grown at ambient temperatures, which may represent GRF targets during cold responses. The total numbers of differentially expressed genes are provided in parentheses (Supplemental Data Sets 10 and 11).
(D) Venn diagram comparing the cold stress-regulated transcriptomes of wild-type and GRF5 overexpressor seedlings and the GA-modulated cold transcriptomes of the two genotypes. Shown in boldface are 132 cold stress-regulated genes whose expression is GA modulated in the wild type and/or the GRF5 overexpression line, as further analyzed in (E) and (F). The total numbers of differentially expressed genes (DEGs) are provided in parentheses (Supplemental Data Set 10).
(E) Venn diagram comparing the 132 cold stress- and GA-modulated genes (D) with the differential cold stress and GA-modulated cold stress transcriptomes between GRF5 overexpression lines and the wild type (Supplemental Data Set 12).
(F) Heat maps of the FC of the 26, 16, and 36 differentially expressed genes identified in the comparisons between 35S:GRF5 and the wild-type (Col) transcriptomes in the cold. An expanded version of this heat map with gene identities is provided in Supplemental Figure 8.
In qualitative terms, the regulation of the GA pathway genes was not strongly affected in the 35S:GRF5 overexpression line (Supplemental Figure 6D; Supplemental Data Set 9). However, several GA pathway genes were differentially regulated after GRF5 overexpression, suggesting a role of GRFs in the regulation of GA biosynthesis, as had been previously suggested for a maize (Zea mays) GRF gene (Supplemental Figure 6D; Supplemental Data Set 9; Hewezi et al., 2012; Fina et al., 2017).
In view of our observation that DELLAs regulate GRFs, we analyzed these data based on several possible scenarios. Since GRF5 expression is cold stress-induced, we anticipated that the expression of genes that are differentially expressed after cold stress in the wild type may be regulated in 35S:GRF5 seedlings also at ambient temperature (Figures 8B and 8C; Supplemental Data Set 8). In support of this hypothesis, we identified 67 and 31 genes that were upregulated and downregulated after cold stress in the wild type among the 209 and 177 genes that were upregulated and downregulated in ambient temperature-grown 35S:GRF5 seedlings (Figures 8B and 8C; Supplemental Figure 7; Supplemental Data Set 8). Interestingly, known targets of the CBF regulon, such as COR15A, COR15B, RD29A, and KIN2, were among the genes upregulated in 35S:GRF5 at ambient temperatures, suggesting that GRF5 can independently regulate these normally cold stress-responsive genes (Supplemental Figure 7).
We further reasoned that the GA- and DELLA-regulated transcriptomes may change as a result of altered DELLA interactomes when GRF5, in the 35S:GRF5 line, is more abundant and can engage with more DELLA proteins. In line with this hypothesis, we observed that 35S:GRF5 seedlings responded in a substantially different manner to GA treatment and cold stress than the wild type (Figure 8D; Supplemental Data Set 10). Of 99 GA-regulated genes in cold-stressed 35S:GRF5 seedlings, only 9 genes overlapped with the 73 GA-regulated transcripts in cold-stressed wild-type seedlings (Figure 8D; Supplemental Data Set 10). At the same time, cold stress-induced transcription differed between the wild type (3930 genes) and the 35S:GRF5 overexpressors (5205 genes), but both genotypes also shared 3119 cold stress-regulated genes (Figure 8D; Supplemental Data Set 10). This indicated that proper cold stress response depends on GRF5 abundance. We thus concluded that GRF genes play a role in cold stress-induced gene expression and that GRF protein content may affect GA responses in the cold, which may be a direct consequence of GRF-DELLA interactions or the altered equilibrium between DELLAs and their interactions with other transcription factors.
To evaluate the differences in the GA responses in the two different GRF genotypes, we analyzed the 132 genes that were GA-regulated after cold stress in the wild type and/or the 35S:GRF5 line (Figure 8D, marked in bold). These genes could be classified according to three expression behaviors depending on the effects of the GA treatments in the 35S:GRF5 genotype. The expression of 26 differentially expressed genes was significantly regulated in the 35S:GRF5 overexpressor after cold stress and GA treatment (Figures 8E and 8F; Supplemental Figure 8A; Supplemental Data Sets 11 and 12A). The differential expression of 36 genes was normalized after GA treatment in the 35S:GRF5 overexpressor (Figures 8E and F; Supplemental Figure 8B; Supplemental Data Sets 11 and 12A). Finally, 16 differentially expressed genes were misregulated in the 35S:GRF5 overexpressor line only after GA treatment in the cold but not in the mock-treated cold-stressed samples (Figures 8E and F; Supplemental Figure 8C; Supplemental Data Sets 11 and 12A). In summary, we concluded that cold stress responses in the wild type are influenced by GRF5 levels, and possibly also other functionally redundant GRF factors not directly tested here, as well as by the interplay of GRFs with the GA pathway.
DISCUSSION
GAs and DELLA proteins play important roles in the plant’s responses to abiotic stress, including cold stress (Achard et al., 2008a; Colebrook et al., 2014). Cold stress affects many plant growth processes, but the general effects of cold temperature on biological molecules, on the metabolic activity of enzymes, and on the reactivity of signaling molecules have made it difficult to unravel the contribution of GA and DELLAs to cold stress based on phenotypic readouts. The accumulation of DELLAs in response to cold temperatures suggests that the DELLAs act early in the cold stress response pathway, and DELLA accumulation thereby precedes phenotypically apparent growth changes (Figure 1B). Therefore, the role of DELLAs in early cold stress response can be most accurately unraveled using molecular analyses. Here, we have examined the contribution of GA to cold-responsive growth by integrating transcriptomics and interactomics data sets (Figures 1 and 2). Based on our analyses, we identify DELLAs as regulators of GRFs in cold stress response (Figure 3).
First, we detected a profound role of cold stress in the transcription of several GA biosynthesis genes (Figure 1C). The observed reduction in GA3ox and GA20ox transcript abundance, combined with the upregulation of GA2ox genes, represents a layer of transcriptional regulation that could explain cold stress-induced DELLA accumulation as a result of reduced GA synthesis and increased GA catabolism (Figure 1B). Additionally, the down-regulation of GA signaling genes, particularly of GID1 GA receptor genes and the SLY1 F-box protein gene, could further argue for an attenuation of GA perception and DELLA degradation during cold stress due to reduced expression of these signaling compounds (Figure 1C). Less comprehensive but similar patterns of feedback regulation have previously been observed in response to GA treatments in ambient temperature-grown as well as stressed plants (Willige et al., 2007; Achard et al., 2008a; Colebrook et al., 2014). Our data thus support the existence of a cold stress-dependent feedback regulatory system that controls DELLA abundance through the regulation of GA biosynthesis and signaling genes.
When combining cold and GA treatments, we were able to assess the contribution of DELLAs to cold stress-induced gene expression (Figures 1D to 1J). We identified 816 genes that were GA-regulated after cold stress and noted that 165 genes, thus less than one-quarter of these genes, were also GA-regulated at ambient temperature (Figure 1G). DELLAs thus regulate substantially different gene sets at the two temperatures, at least in our experimental conditions, which should be the result of differential transcription factor regulation by DELLAs. A large fraction of the GA-regulated genes at 4°C, 621 of 816 genes, were also cold stress-regulated, indicating that the integration of DELLAs with the cold stress pathway plays an active role in the regulation of the transcriptional cold response (Figure 1G). Our analyses performed after 1, 2, and 4 h identified, for each time point, a comparable number of GA-regulated genes but, at the same time,revealed that there was only a restricted overlap between the individual GA-regulated transcriptomes (Figures 1D to 1G). This comparative analysis suggests that DELLAs act in a dynamic manner, possibly by interfering with the activity of different transcription factor sets during different stages of the cold stress response. Stress-dependent and tissue-specific effects of DELLAs on gene expression have previously been noted, and the identification of hundreds of DELLA-transcription factor interactions, as identified by us and others, form the molecular framework for these differential transcriptome responses, as will be discussed below (Cao et al., 2006; Zentella et al., 2007).
On the other side, and in view of the large number (7343) of cold stress-regulated genes in the wild type, our analyses uncover that the GA pathway regulates only a subset of the transcriptional responses of the cold response pathway (Figure 1G). Quantitative analysis further showed that DELLA interactions may have only modulating activity on the gene expression of the respective genes. This is already indirectly suggested by the comparatively minor effects of GA treatments on gene expression, in contrast to the strong gene expression differences observed after the cold treatment (Supplemental Figure 2). The cold stress-dependent regulation of most genes is generally moderately, and rarely strongly, regulated by DELLAs (Figures 1H to 1J).
Several transcription factors have been implicated in cold-responsive gene expression, and these may be targets of DELLA regulation. Foremost among them are the CBF transcription factors CBF1 to CBF3, whose overexpression induces phenotypes that can be suppressed by GA treatments or in della loss-of-function backgrounds (Achard et al., 2008a; Jia et al., 2016). As yet, there is no evidence that these effects are regulated by direct CBF-DELLA interactions. It is rather suggested that CBFs affect GA catabolism or that CBF activity is regulated by an indirect process involving competing interactions between DELLAs and JAZ proteins (Achard et al., 2008a; Zhou et al., 2017).
We identified over 250 DELLA-interacting transcription factors in a genome-wide transcription factor interaction screen (Figure 2). The two DELLA interactome data sets obtained with GAI and RGA overlapped substantially, which is consistent with the known biochemical redundancy between the Arabidopsis DELLA proteins (Gallego-Bartolomé et al., 2010). As already suggested by numerous biologically validated DELLA interactions and other large-scale screens, these DELLA interactors belong to structurally diverse transcription factor families (Claeys et al., 2014; Marín-de la Rosa et al., 2014). There was no recognizable theme that would allow rationalizing the fact that these interactors belonged to such diverse families and, in that, our study is no exception to the rule (Claeys et al., 2014; Marín-de la Rosa et al., 2014). At the same time, a substantial fraction of the already validated interactions was originally identified in the yeast two-hybrid system, and our analysis extends the list of candidate DELLA-regulated transcription factors substantially (Supplemental Data Set 7). Although we had no means of testing all individual interactions, we are confident, based on the previous reports, that the majority of these interactions will eventually be proven to be biologically relevant.
The question of whether there is specificity between these individual interactions, achieved through posttranslational regulation or tissue- and cell type-specific expression, or whether DELLAs simply function as global transcription regulators remains to be answered. There are examples in the published literature for cell type-specific interactions (e.g., in root hair-forming cells) but also for interactions with global regulators, such as the chromatin-remodeling proteins PICKLE and BRAHMA, which suggests that DELLAs may have a more general role in transcription regulation (Cao et al., 2006; Archacki et al., 2013; Sarnowska et al., 2013; Wild et al., 2016; Park et al., 2017). At the same time, the DELLA interactions identified here strongly outnumbered the number of interactions retrieved with non-DELLA GRAS family proteins, suggesting that the propensity to interact with transcription factors, at least in the yeast two-hybrid system, may be a specific feature of DELLA proteins (Figures 2D and 2E). The latter could be due to the specific primary amino acid sequence and the fact that RGA and GAI share high sequence identity (75%) but only restricted identity with SCR (38%) and with SCL3 (35%). Thus, besides the fact that GRAS proteins have a shared domain organization, the two classes of GRAS proteins differ at the amino acid level, which may influence their propensity of interacting with other proteins. Finally, we cannot rule out that the different behaviors are simply due to technical reasons related to the approach chosen.
To uncover interactors of DELLAs with a role in GA-regulated cold responses, we searched for DELLA-interacting transcription factors that were GA-regulated during cold stress at the transcriptional level. In this regard, GRF transcription factors were outstanding because at least six of the nine Arabidopsis GRFs interacted with DELLAs and because the transcription of three prominent family members, GRF1, GRF3, and GRF5, was GA-regulated in the cold (Figures 3B to 3E). Whereas we were able to verify these interactions in transiently transformed Nicotiana benthamiana leaf cells by BiFC, our other efforts to verify these interactions with biochemical methods failed, since we were unable to generate transgenic plants that allowed the detection of FLAG epitope-tagged or RFP-tagged GRFs and, consequently, GRF interactions (data not shown). However, while this work was in progress, others identified and biochemically verified the interaction between GRFs and DELLA in the context of a genetic study on nitrogen use efficiency in rice (Oryza sativa; Li et al., 2018). There, rice GRF4 was detected using a specific antibody, but also the instability of the protein in defined nutrient conditions was observed (Li et al., 2018). Thus, GRFs may be subject to different levels of regulation through miRNAs, DELLAs, and the proteasomal pathway (Hewezi et al., 2012; Omidbakhshfard et al., 2015; Li et al., 2018).
Using genotypes with high and low GRF levels, we could demonstrate that DELLAs and GAs interfere with GRF function at the level of leaf and root growth. Plants overexpressing miRNA396b, thus with strongly reduced GRF levels, were sensitized to elevated DELLA levels (Figures 4 to 7). Most dramatic was the change in root development associated with a GA-suppressible reduction in GA levels, as caused by PAC treatment of miR396b-overexpressing seedlings (Figure 7). Similar phenotypes were reported after root endodermis-specific expression of a stabilized form of GAI in Arabidopsis, and these phenotypes are also reminiscent of those of mutants with defects in microtubule organization (Ubeda-Tomás et al., 2009; Beck et al., 2011). GRFs have been implicated in cell division control and meristem formation, and the reduction of GRFs seems to severely disturb the interaction equilibrium for DELLAs in the root (Rodriguez et al., 2015; Ercoli et al., 2016). Overall, it became apparent from our various observations that the balance of GRF and DELLA dosage is essential for regulated plant development at the different stages assessed.
Through gene expression analyses, we have been trying to identify genes that are subject to DELLA-imposed GRF5 regulation. First of all, we found that several genes of the GA pathway, namely GA20ox1, GA2ox1, GA2ox8, RGL1, and RGL2, were more strongly regulated in the GRF5 overexpressor, which is in line with previous reports that GRFs target GA pathway genes, although the specific gene described in a previous study was not detected in our analysis (Supplemental Figure 6; Hewezi et al., 2012; Fina et al., 2017). Our gene expression analyses also find that the GRF5-DELLA interaction interferes with GRF-dependent gene regulation (Figure 8). The comparison of GRF gene regulation in the context of cold stress response and GA regulation reveals different scenarios where GRF5 acts as an activator or repressor of gene expression, as judged by the increased expression of the respective genes between the wild type and the GRF5 overexpressors at 4°C (Figure 8). At the same time, the presence and absence of DELLAs can influence the expression of these presumed GRF5 targets in a positive and negative manner. Further detailed analyses were not performed here, since we had no information about physiologically relevant target genes and we were, as mentioned above, not able to generate transgenic lines expressing GRFs that were amenable to such analyses.
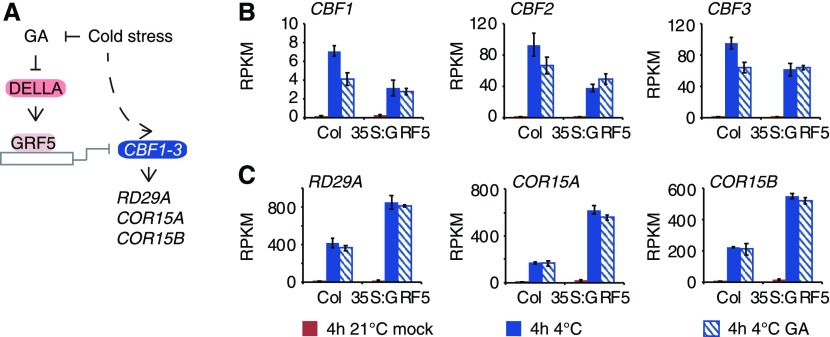
With great interest, we noted that the cold stress-dependent induction of all three CBF genes was attenuated in the 35S:GRF5 overexpression line, indicating that GRFs can repress CBF induction during cold stress (Figures 9A and 9B). At the same time, CBF expression was DELLA-dependent in the wild type, in line with a model where DELLAs act as coactivators of CBF expression (Figures 9A and 9B; Zhou et al., 2017). In GRF5 overexpressors, the DELLA-dependent effect was abolished, which could be a consequence of DELLA effects being outcompeted by increased GRF5 abundance (Figures 9A and 9B). What is more, also the expression of cold-regulated genes such as RD29A, COR15A, and COR15B, known CBF targets, followed the trend in CBF expression, although not necessarily the changes after GA treatment, which may be attributed to the underlying differential dynamics and stability of gene activation and repression, et cetera (Figures 9A and 9B). This regulation of cold stress-responsive genes is also in line with observations that cbf triple mutants still display residual induction of these genes, which should be due to the activity of other regulators (Jia et al., 2016; Zhao et al., 2016; Calixto et al., 2018). We, thus, uncover a new role of GRFs in cold stress-induced gene expression, apart from their other functions in ambient temperature-grown plants (Casadevall et al., 2013; Wang et al., 2013; Debernardi et al., 2014; Kim and Tsukaya, 2015; Omidbakhshfard et al., 2015; Rodriguez et al., 2015; Li et al., 2018; Zhang et al., 2018). Inversely, also the cold stress regulatory CBF genes have been shown to be implicated in developmental processes and, thus, GRFs and CBFs may also play together in processes unrelated to cold stress regulation (Jia et al., 2016).
Figure 9.
GRF5 Represses CBF Gene Expression in a GA-Modulated Manner.
(A) Schematic representation of the interactions in response to cold stress as determined in this study with a special emphasis on a possible crosstalk of the GA- and DELLA-dependent GRF5 regulation in the cold and cold stress-induced gene expression.
(B) and (C) Graphs displaying reads per kilobase of transcript per million mapped reads (RPKM) values of CBF1 to CBF3 (B) and CBF target (C) gene expression of seedlings grown in ambient temperature (21°C) or in cold stress (4°C) in the absence (mock) and presence of 100 µM GA3. Shown are means and sd of three biological replicate samples from pooled seedling shoots.
In summary, our study uncovers the identity of a GRF- and DELLA-modulated cold stress-responsive gene expression pathway that contributes to cold stress-responsive gene expression. GRFs can thus be added to the list of transcription factors regulating gene expression in the cold (Park et al., 2015). The identification of multiple other DELLA-interacting transcription factors and the biological characterization of these and other, previously reported, interactions will contribute in the future to a more detailed understanding of the GA pathway in cold-responsive gene expression.
METHODS
Biological Material
Wild-type, 35S:GRF5 (Horiguchi et al., 2005), and 35S:miR396b (Rodriguez et al., 2010) plants of the Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) were used for all experiments.
Molecular Cloning
DNA-binding domain yeast two-hybrid constructs of M5-truncated RGA (RGA204) and GAI (GAI141) were generated using Gateway-based cloning of PCR fragments obtained with the primers RGA-FW (5′-attB1-CGGAGTCAACTCGTTCTGTTATCCTGG-3′) and RGA-RV (5′-attB2-GTCAGTACGCCGCCGTCGA-3′) as well as GAI-FW (5′-attB1-CGAACGGCGTCGTGGAAACC-3′) and GAI-RV (5′-attB2-GCTAATTGGTGGAGAGTTTCCAAGCC-3′), respectively (Life Technologies). Activation domain (AD) yeast two-hybrid clones for GRF4 were obtained in the same manner from Col-0 cDNA with the primers 5′-attB1-CGATGGACTTGCAACTGAAAC-3′ and 5′-attB2-GTTAATGAAAAACTTGAGTAGAG-3′ and for GRF6 with 5′-attB1-CGATGGCTACAAGGATTCC-3′ and 5′-attB2-GTCAAATGAAGAGTGAAGTAG-3′. Yeast two-hybrid constructs for the remaining genes were retrieved from the transcription factor collection (Pruneda-Paz et al., 2014). SCR and SCL3 yeast two-hybrid constructs were previously published (Arabidopsis Interactome Mapping Consortium, 2011). For the BiFC experiment, the coding sequence of GRF5 was amplified with the primers 5′-attB1-CGATGATGAGTCTAAGTGGAAGTAG-3′ and 5′-attB2-GTTAGCTACCAGTGTCGAGTC-3′ and of GRF3 with 5′-attB1-CGATGGATTTGCAACTGAAAC-3′ and 5′-attB2-GTCAATGAAAGGCTTGTGTC-3′ from the respective AD vectors. The miRNA396-resistant version of GRF3, rGRF3, was obtained with mutation PCR with the primers 5′-GCACCGTGGCCGCAACAGGAGCCGTAAACCGGTCGAGACTCC-3′ and 5′-GTTGCATTGACGGTGGTTGGAGTCTCGACCGGTTTACGGCTCCTGTTG-3′ as described before (Hewezi et al., 2012). GIF1 was cloned with 5′-attB1-CGATGCAACAGCACCTGATG-3′ and 5′-attB2-GTCAATTCCCATCATCTGATG-3′. Entry vectors carrying the full-length sequences of RGA and GAI were previously published (Arabidopsis Interactome Mapping Consortium, 2011). An entry vector with the XERICO coding sequence was provided by Andrea Holzer (Technische Universität München).
Immunoblot Analysis
For immunoblot analyses, Arabidopsis Col-0 seeds were surface-sterilized, stratified for 2 d, and grown on half strenth MS agar for 9 d at 21°C under continuous light (110 to 150 μmol m−2 s−1; Osram L36W/840 Lumilux Cool White Hg) in a Sanyo growth chamber. Seedlings were then placed at 4°C under the same light conditions and spray-treated with 100 μM GA3 or a mock solution. Protein extracts were prepared from pooled whole seedlings, and immunoblot analyses were performed as previously described using an established anti-RGA rabbit antibody against the peptide KRDHHQFQGRLSNHGC (amino acids 2 to 16 of RGA; Eurogentec) and goat anti-rabbit IgG antibody, HRP-conjugate (catalog no.12-348, Sigma-Aldrich; Willige et al., 2007).
Transcriptomics
To obtain plant material for RNA-seq analysis, Arabidopsis Col-0 seeds were surface-sterilized, stratified for 2 d, and grown on half strenth MS agar for 9 d at 21°C under continuous light (110 to 150 μmol m−2 s−1; Osram L36W/840 Lumilux Cool White Hg) in a Sanyo growth chamber (Ewald). Seedlings were then spray-treated with 100 μM GA3 or a mock solution and placed at 21 or 4°C under continuous light (110 to 150 μmol m−2 s−1; Osram L36W/840 Lumilux Cool White Hg). Three biological replicates of seedling shoots, pooled from seedlings grown on three replicate plates per treatment, were collected after 0, 1, 2, and 4 h for each treatment. Total RNA was isolated using the NucleoSpin RNA Plant kit (Macherey-Nagel). DNA was removed by an on-column treatment with rDNase (Machery-Nagel). For library preparation and RNA sequencing, RNA was quantified with the KAPA library quantification kit (Illumina) and cDNA libraries were prepared using the TruSeq RNA Sample Preparation v2 kit (Illumina) following the manufacturer’s instructions. Clusters were generated and sequenced on eight-lane flow cells with the HiSeq 1000 platform (Illumina). The TruSeq Paired-End Cluster kit v3 (Illumina) and the TruSeq SBS kit v3 (Illumina) were used to generate 100-bp paired-end reads. The analysis of the raw sequences and the differential expression analysis were performed on the CLC Genomics Workbench v. 7.5.1 (Qiagen). Raw sequences were first quality trimmed (trim using quality scores, 0.05; maximum number of ambiguous nucleotides, 1) and then aligned and mapped to the Arabidopsis Col-0 (The Arabidopsis Information Resource 10) genome using the RNA-seq analysis tool with default settings (maximum number of hits for a read, 10; strand-specific, both; count paired reads as two, no; expression value, total counts; calculate RPKM for genes without transcripts, no; global alignment, no; autodetect paired distances, yes; similarity fraction, 0.8; length fraction, 0.8; mismatch cost, 2; insertion cost, 3; deletion cost, 3). Differential expression analysis was performed using the Empirical Analysis of DGE tool (total count filter cutoff, 5.0; estimate tagwise dispersions, yes; FDR corrected, yes). Genes with FDR-corrected P < 0.01 and FC > 1.5 were classified as differentially expressed genes. Heat maps were generated using the Cluster 3.0 and Treeview tools (de Hoon et al., 2004; Saldanha, 2004).
An independent RNA-seq experiment was performed to compare the responses to the 4°C and GA treatment using 9-d-old Col-0 wild-type and 35S:GRF5 transgenic seedlings (Horiguchi et al., 2005). Growth conditions and RNA preparation were as described above. Seedling shoot material, pooled from seedlings grown on three replicate plates per genotype and treatment, was harvested from the three genotypes after 4 h at 21°C (mock), 4°C (mock), and 4°C following 100 μM GA3 treatment. Library preparation and sequencing using Illumina technology was performed by GATC Biotech. After selection of polyadenylated mRNAs, strand-specific cDNA libraries were generated and sequenced, producing more than 25 million stranded 50-bp reads per sample. The raw sequence reads were quality trimmed (trim using quality scores, 0.05; maximum number of ambiguous nucleotides, 1) and aligned to The Arabidopsis Information Resource 10 using the RNA-seq analysis tool (mismatch cost, 2; insertion cost, 3; deletion cost, 3; length fraction, 0.8; similarity fraction, 0.8; global alignment, no; strand specific, reverse; maximum number of hits for a read, 10; count paired reads as two, no; expression value, total counts; calculate RPKM for genes without transcripts, no) of the CLC Genomics Workbench v. 10.1.1 (Qiagen). Differential expression analysis was performed using the Differential Expression for RNA-Seq tool (CLC Genomics Workbench v. 10.1.1; Qiagen). Differentially expressed genes were classified as for the previous experiment (FDR-corrected P < 0.01, FC > 1.5). Heat maps were generated using the Cluster 3.0 and Treeview tools (de Hoon et al., 2004; Saldanha, 2004).
Protein-Protein Interaction Assays
For yeast two-hybrid experiments, N-terminally truncated M5 versions of RGA and GAI and full-length coding sequences of SCR and SCL3 were cloned into pDEST-DB and screened by yeast mating against a previously published transcription factor collection of GAL4 AD fusion proteins in pDEST-AD (Pruneda-Paz et al., 2014). Yeast two-hybrid screening was done using previously described strains, plasmids, media, and transformation protocols (Altmann et al., 2018). For detection of AD autoactivators, the AD-fused transcription factor collection in MATa Y8800 yeast strains was mated with the empty pDEST-DB expressing MATα Y8930 and subsequently selected in the presence of 1 mM 3-amino-1,2,4-triazole (3-AT). The DELLA protein screens were performed twice, once with 1 mM 3-AT and a second time with 2 and 3 mM 3-AT for RGA and GAI, respectively. The SCR and SCL3 screens were performed once using 1 mM 3-AT. All positive transcription factor clones were confirmed by sequencing of the respective inserts.
In Planta Protein Interaction
BiFC assays were performed using transient transformation of Nicotiana benthamiana leaves with transformed Agrobacterium tumefaciens grown to OD600 = 0.4 as previously described (Bos et al., 2010). Fluorescence complementation was observed after 2 to 3 d with an Olympus FV1000 confocal microscope. The specified open reading frames were cloned for this purpose into the vectors pCL112 and pCL113 (Bos et al., 2010).
Physiological Experiments
For the quantification of leaf and root growth, seeds were surface-sterilized, stratified, and grown on half strenth MS 0.8% (w/v) agar plates in continuous white light (110 to 130 μmol m−2 s−1; Osram L36W/840 Lumilux Cool White Hg) in MLR-351 Sanyo growth chambers (Ewald) for 6 d before they were transferred to plates containing mock or 10 μΜ GA3 (Duchefa Biochemie) or 0.1, 0.5, or 1 μΜ PAC (Duchefa Biochemie). Seedlings and the first two true leaves of the seedlings were scanned after either 7 d at 21°C or 7 d at 4°C, where almost no growth was observed, and an additional 7 d at 21°C for recovery. Parameters were quantified with ImageJ. The experiment was repeated three times with similar results. In the case of the first repeat, no ambient temperature condition was included.
The cell size analysis was performed from 10 images taken from the midsections of true leaves 1 and 2 from 10 different seedling leaves. To this end, the palisade cell layer underlying the adaxial epidermis in the middle of the leaf blade between the vein and the leaf margin was photographed and analyzed after modified pseudo-Schiff-propidium iodide staining with an FV1000 confocal microscope (Olympus) as previously described by Horiguchi et al. (2005). The cell size analysis was performed once from a representative experiment. For the quantification of the 35S:miR396b root phenotype on PAC, Col-0 and 35S:miR396b seeds were surface-sterilized, stratified, and seedlings were grown on half strenth MS 0.8% agar plates for 5 d. Subsequently, seedlings were transferred on plates containing either mock, 5 μM PAC, or 5 μM PAC + 10 μΜ GA3. Root tips were imaged with an SZX16 stereoscope (Olympus) after 8 d of propidium iodide staining (Horiguchi et al., 2005). The frequency of the disorganized meristem phenotype was determined. For the microscopy visualization of the phenotype, Col-0 and 35S:miR396b seeds were surface-sterilized and grown for 5 d before they were transferred to medium containing either mock or 1 μΜ PAC. The photographs were taken after 8 d using propidium iodide staining with an FV1000 confocal microscope (Olympus).
The effects of the genotype and treatment on leaves and on root elongation were assessed with two-way ANOVA and Tukey’s HSD posthoc test (P < 0.05) with the R statistical package.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: GAI (AT1G14920), GIF1 (AT5G28640), GRF1 (AT2G22840), GRF2 (AT4G37740), GRF3 (AT2G36400), GRF4 (AT3G52910), GRF5 (AT3G13960), GRF6 (AT2G06200), GRF7 (AT5G53660), GRF9 (AT2G45480), RGA (AT2G01570), SCR (AT3G54220), SCL3 (AT1G50420), and XERICO (AT2G04240). The raw data of the RNA-seq experiments can be found at the National Center for Biotechnology Information Sequence Read Archive database under accession numbers SRP178244 and SRP158655 and Bioprojects PRJNA513856 and PRJNA487166.
Supplemental Data
Supplemental Figure 1. Expression of reference genes of cold and GA responses in the transcriptomics data set. (Supports Figure 1).
Supplemental Figure 2. The magnitude of cold stress-regulated gene expression is substantially higher than the magnitude of GA-regulated expression changes. (Supports Figure 1).
Supplemental Figure 3. GA modulates the expression of a common gene set comprising 165 genes at both experimental temperatures. (Supports Figure 1).
Supplemental Figure 4. Gene expression of DELLA-interacting proteins and GRF genes in ambient temperature and during cold stress with mock and GA treatment. (Supports Figure 3).
Supplemental Figure 5. GA levels modulate leaf area, root length and cell size in a GRF-dependent manner. (Supports Figure 4).
Supplemental Figure 6. GRF and GA pathway genes differentially respond to cold stress and GA treatments in 35S:GRF5. (Supports Figure 8).
Supplemental Figure 7. Cold stress-regulated genes are differentially regulated in 35S:GRF5 plants grown at ambient temperature. (Supports Figure 8).
Supplemental Figure 8. GRF5 overexpression affects cold and GA transcriptomes after cold stress. (Supports Figure 8).
Supplemental Data Set 1. Table of differentially expressed genes after GA and cold treatment.
Supplemental Data Set 2. Table of GA pathway genes and their transcript behavior in the RNA-seq experiment.
Supplemental Data Set 3. Table of known GA-regulated genes and their transcript behavior in the RNA-seq experiment.
Supplemental Data Set 4. Table of genes that were GA-regulated at both experimental temperatures.
Supplemental Data Set 5. Table of GA-modulated cold-regulated genes.
Supplemental Data Set 6. Table of the protein-protein interactions identified in the yeast two-hybrid screen.
Supplemental Data Set 7. Combined list of known and new DELLA-interacting proteins.
Supplemental Data Set 8. Table of differentially expressed genes after cold treatment in the wild type and in 35S:GRF5 at ambient temperature.
Supplemental Data Set 9. Table of GRF and GA pathway genes responding to cold stress and GA treatments.
Supplemental Data Set 10. Table of differentially expressed genes after cold and GA treatments in the wild type and 35S:GRF5.
Supplemental Data Set 11. Table of differentially expressed genes between 35S:GRF5 and the wild type at 4°C.
Supplemental Data Set 12. Table of GA-modulated cold stress-regulated genes differentially expressed between the wild type and 35S:GRF5.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dirk Inzé (VIB, Ghent, Belgium) for providing 35S:GRF5 transgenic seeds, Javier Palatnik (Instituto de Biología Molecular y Celular de Rosario, Rosario, Argentina) for providing 35S:miR396b transgenic seeds, Melina Altmann (Technische Universität München, Freising, Germany) for the SCR and SCL3 yeast two-hybrid vectors, as well as Stefan Engelhardt (Technische Universität München, Freising, Germany) for the BiFC vectors pCL112 and pCL113. This work was supported by the Deutsche Forschungsgemeinschaft (grant SFB924 to C.S.), the Federal Ministry of Education and Research (grant PLANT-KBBEIV:DELLA-STRESS), and the German-Israeli Foundation for Scientific Research and Development (grant I-1313-203.13/2015).
AUTHOR CONTRIBUTIONS
O.L. and C.S. designed research; O.L. performed all research; A.A. and P.F.-B. supported the yeast two-hybrid screen; O.L. and C.S. analyzed data and wrote the article.
References
- Achard P., Baghour M., Chapple A., Hedden P., Van Der Straeten D., Genschik P., Moritz T., Harberd N.P.(2007). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. USA 104: 6484–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Gong F., Cheminant S., Alioua M., Hedden P., Genschik P.(2008a). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Renou J.P., Berthomé R., Harberd N.P., Genschik P.(2008b). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18: 656–660. [DOI] [PubMed] [Google Scholar]
- Altmann M., Altmann S., Falter C., Falter-Braun P.(2018). High-quality yeast-2-hybrid interaction network mapping. Curr. Protoc. Plant Biol. 3: e20067. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archacki R., et al. (2013). BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in Arabidopsis. PLoS One 8: e58588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y.(2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Komis G., Ziemann A., Menzel D., Samaj J.(2011). Mitogen-activated protein kinase 4 is involved in the regulation of mitotic and cytokinetic microtubule transitions in Arabidopsis thaliana. New Phytol. 189: 1069–1083. [DOI] [PubMed] [Google Scholar]
- Beltramino M., Ercoli M.F., Debernardi J.M., Goldy C., Rojas A.M.L., Nota F., Alvarez M.E., Vercruyssen L., Inzé D., Palatnik J.F., Rodriguez R.E.(2018). Robust increase of leaf size by Arabidopsis thaliana GRF3-like transcription factors under different growth conditions. Sci. Rep. 8: 13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.I., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA 107: 9909–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto C.P.G., Guo W., James A.B., Tzioutziou N.A., Entizne J.C., Panter P.E., Knight H., Nimmo H.G., Zhang R., Brown J.W.S.(2018). Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell 30: 1424–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Cheng H., Wu W., Soo H.M., Peng J.(2006). Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 142: 509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Hussain A., Cheng H., Peng J.(2005). Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223: 105–113. [DOI] [PubMed] [Google Scholar]
- Casadevall R., Rodriguez R.E., Debernardi J.M., Palatnik J.F., Casati P.(2013). Repression of growth regulating factors by the microRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. Plant Cell 25: 3570–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H., De Bodt S., Inzé D.(2014). Gibberellins and DELLAs: Central nodes in growth regulatory networks. Trends Plant Sci. 19: 231–239. [DOI] [PubMed] [Google Scholar]
- Colebrook E.H., Thomas S.G., Phillips A.L., Hedden P.(2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217: 67–75. [DOI] [PubMed] [Google Scholar]
- Conti L., Nelis S., Zhang C., Woodcock A., Swarup R., Galbiati M., Tonelli C., Napier R., Hedden P., Bennett M., Sadanandom A.(2014). Small ubiquitin-like modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev. Cell 28: 102–110. [DOI] [PubMed] [Google Scholar]
- Das Gupta M., Nath U.(2015). Divergence in patterns of leaf growth polarity is associated with the expression divergence of miR396. Plant Cell 27: 2785–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière J.M., Achard P.(2016). A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant 9: 10–20. [DOI] [PubMed] [Google Scholar]
- Debernardi J.M., Mecchia M.A., Vercruyssen L., Smaczniak C., Kaufmann K., Inze D., Rodriguez R.E., Palatnik J.F.(2014). Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 79: 413–426. [DOI] [PubMed] [Google Scholar]
- de Hoon M.J., Imoto S., Nolan J., Miyano S.(2004). Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S.(2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Ding Y., Shi Y., Yang S.(2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222: 1690–1704. [DOI] [PubMed] [Google Scholar]
- Ercoli M.F., Rojas A.M., Debernardi J.M., Palatnik J.F., Rodriguez R.E.(2016). Control of cell proliferation and elongation by miR396. Plant Signal. Behav. 11: e1184809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremina M., Rozhon W., Poppenberger B.(2016). Hormonal control of cold stress responses in plants. Cell. Mol. Life Sci. 73: 797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Martinez C., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjani A., Horiguchi G., Yano S., Tsukaya H.(2007). Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol. 144: 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fina J., Casadevall R., AbdElgawad H., Prinsen E., Markakis M.N., Beemster G.T.S., Casati P.(2017). UV-B inhibits leaf growth through changes in growth regulating factors and gibberellin levels. Plant Physiol. 174: 1110–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J., Mori M., Watanabe S., Miyamoto C., Ito T., Takahashi Y.(2017). DELLA-GAF1 complex is a main component in gibberellin feedback regulation of GA20 oxidase 2. Plant Physiol. 175: 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadí D., Blázquez M.A.(2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 13446–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Marín J.A., Prat S., Blázquez M.A., Alabadí D.(2010). Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol. Biol. Evol. 27: 1247–1256. [DOI] [PubMed] [Google Scholar]
- Gilmour S.J., Fowler S.G., Thomashow M.F.(2004). Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54: 767–781. [DOI] [PubMed] [Google Scholar]
- Heo J.O., Chang K.S., Kim I.A., Lee M.H., Lee S.A., Song S.K., Lee M.M., Lim J.(2011). Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 108: 2166–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T., Maier T.R., Nettleton D., Baum T.J.(2012). The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol. 159: 321–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Kim G.T., Tsukaya H.(2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43: 68–78. [DOI] [PubMed] [Google Scholar]
- Hou X., Lee L.Y., Xia K., Yan Y., Yu H.(2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894. [DOI] [PubMed] [Google Scholar]
- Jia Y., Ding Y., Shi Y., Zhang X., Gong Z., Yang S.(2016). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 212: 345–353. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Choi D., Kende H.(2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36: 94–104. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Kende H.(2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 13374–13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Tsukaya H.(2015). Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J. Exp. Bot. 66: 6093–6107. [DOI] [PubMed] [Google Scholar]
- Ko J.H., Yang S.H., Han K.H.(2006). Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 47: 343–355. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Ko J.H., Lee S., Lee Y., Pak J.H., Kim J.H.(2009). The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151: 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Tian Y., Wu K., Ye Y., Yu J., Zhang J., Liu Q., Hu M., Li H., Tong Y., Harberd N.P., Fu X.(2018). Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Song Y., Chen Z., Yu D.(2009). Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 136: 223–236. [DOI] [PubMed] [Google Scholar]
- Marín-de la Rosa N., Sotillo B., Miskolczi P., Gibbs D.J., Vicente J., Carbonero P., Oñate-Sánchez L., Holdsworth M.J., Bhalerao R., Alabadí D., Blázquez M.A.(2014). Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol. 166: 1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y.(2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard M.A., Proost S., Fujikura U., Mueller-Roeber B.(2015). Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 8: 998–1010. [DOI] [PubMed] [Google Scholar]
- Park J., Oh D.H., Dassanayake M., Nguyen K.T., Ogas J., Choi G., Sun T.P.(2017). Gibberellin signaling requires chromatin remodeler PICKLE to promote vegetative growth and phase transitions. Plant Physiol. 173: 1463–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lee C.M., Doherty C.J., Gilmour S.J., Kim Y., Thomashow M.F.(2015). Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 82: 193–207. [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Nagel D.H., Kang S.E., Bonaldi K., Doherty C.J., Ravelo S., Galli M., Ecker J.R., Kay S.A.(2014). A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Rep. 8: 622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh L.D., Wysocka-Diller J.W., Camilleri C., Bouchez D., Benfey P.N.(1999). The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18: 111–119. [DOI] [PubMed] [Google Scholar]
- Rodriguez R.E., Ercoli M.F., Debernardi J.M., Breakfield N.W., Mecchia M.A., Sabatini M., Cools T., De Veylder L., Benfey P.N., Palatnik J.F.(2015). MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. Plant Cell 27: 3354–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R.E., Mecchia M.A., Debernardi J.M., Schommer C., Weigel D., Palatnik J.F.(2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata T., et al. (2014). Reduction of gibberellin by low temperature disrupts pollen development in rice. Plant Physiol. 164: 2011–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A.J.(2004). Java Treeview: Extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- Sarnowska E.A., et al. (2013). DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiol. 163: 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S., Federici F., Casimiro I., Beemster G.T., Bhalerao R., Swarup R., Doerner P., Haseloff J., Bennett M.J.(2009). Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19: 1194–1199. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Motoyuki A., Matsuoka M.(2007). Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58: 183–198. [DOI] [PubMed] [Google Scholar]
- Van De Velde K., Ruelens P., Geuten K., Rohde A., Van Der Straeten D.(2017). Exploiting DELLA signaling in cereals. Trends Plant Sci. 22: 880–893. [DOI] [PubMed] [Google Scholar]
- Wang L., Gu X., Xu D., Wang W., Wang H., Zeng M., Chang Z., Huang H., Cui X.(2011). miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J. Exp. Bot. 62: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Wang Q., Zhang B.(2013). Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 530: 26–32. [DOI] [PubMed] [Google Scholar]
- Wild M., Davière J.M., Regnault T., Sakvarelidze-Achard L., Carrera E., Lopez Diaz I., Cayrel A., Dubeaux G., Vert G., Achard P.(2016). Tissue-specific regulation of gibberellin signaling fine-tunes Arabidopsis iron-deficiency responses. Dev. Cell 37: 190–200. [DOI] [PubMed] [Google Scholar]
- Willige B.C., Ghosh S., Nill C., Zourelidou M., Dohmann E.M., Maier A., Schwechheimer C.(2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Ueguchi-Tanaka M.(2014). DELLA and SCL3 balance gibberellin feedback regulation by utilizing INDETERMINATE DOMAIN proteins as transcriptional scaffolds. Plant Signal. Behav. 9: e29726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Ueguchi-Tanaka M., Matsuoka M.(2014). Regulatory networks acted upon by the GID1-DELLA system after perceiving gibberellin. Enzymes 35: 1–25. [DOI] [PubMed] [Google Scholar]
- Zentella R., Zhang Z.L., Park M., Thomas S.G., Endo A., Murase K., Fleet C.M., Jikumaru Y., Nambara E., Kamiya Y., Sun T.P.(2007). Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Sun W., Singh R., Zheng Y., Cao Z., Li M., Lunde C., Hake S., Zhang Z.(2018). GRF-interacting factor1 regulates shoot architecture and meristem determinacy in maize. Plant Cell 30: 360–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.L., Ogawa M., Fleet C.M., Zentella R., Hu J., Heo J.O., Lim J., Kamiya Y., Yamaguchi S., Sun T.P.(2011). Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 2160–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Zhang Z., Xie S., Si T., Li Y., Zhu J.K.(2016). Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 171: 2744–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Chen H., Wei D., Ma H., Lin J.(2017). Arabidopsis CBF3 and DELLAs positively regulate each other in response to low temperature. Sci. Rep. 7: 39819. [DOI] [PMC free article] [PubMed] [Google Scholar]