HY5 promotes activation rather than repression of its direct targets, including SPA1, revealing a light-regulated feedback mechanism.
Abstract
The first exposure to light marks a crucial transition in plant development. This transition relies on the transcription factor HY5 controlling a complex downstream growth program. Despite its importance, its function in transcription remains unclear. Previous studies have generated lists of thousands of potential target genes and competing models of HY5 transcription regulation. In this work, we carry out detailed phenotypic and molecular analysis of constitutive activator and repressor HY5 fusion proteins. Using this strategy, we were able to filter out large numbers of genes that are unlikely to be direct targets, allowing us to eliminate several proposed models of HY5’s mechanism of action. We demonstrate that the primary activity of HY5 is promoting transcription and that this function relies on other, likely light-regulated, factors. In addition, this approach reveals a molecular feedback loop via the COP1/SPA E3 ubiquitin ligase complex, suggesting a mechanism that maintains low HY5 in the dark, primed for rapid accumulation to reprogram growth upon light exposure. Our strategy is broadly adaptable to the study of transcription factor activity. Lastly, we show that modulating this feedback loop can generate significant phenotypic diversity in both Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum).
INTRODUCTION
Plants continuously monitor the environment to tune development and optimize performance. They sense changes in factors such as temperature, water availability, gravity, nutrients, and many aspects of light, the latter of which are sensed by a diverse array of photoreceptors and feed into a variety of growth programs (Chen et al., 2004).
Deetiolation is one of the best-understood light-mediated developmental switches, activated very early in development, immediately following germination (Arsovski et al., 2012). When light is a limiting factor, the seedling will use etiolated growth, characterized by an elongated embryonic stem (hypocotyl) with the apical hook and tightly closed embryonic leaves (cotyledons) protecting the meristematic tissue. Rapid elongation during this stage allows the plant to quickly emerge from the soil and gain access to light. Upon sensing sufficient light, it transitions to deetiolated growth. This is characterized by a deceleration of hypocotyl growth, promotion of cotyledon expansion, and accumulation of chlorophyll. For the purposes of this article, we define photomorphogenesis as germination and growth in light, without a soil-like dark-growth stage. Deetiolation and photomorphogenesis are both triggered by photoreceptors transducing light signals to a set of key transcription factors (TFs) that change the expression of thousands of genes (Leivar et al., 2009).
One of these TFs, the basic domain/leucine zipper (bZIP) TF ELONGATED HYPOCOTYL 5 (HY5), is a master regulator of deetiolation and photomorphogenesis. hy5 was identified genetically in the now classic long-hypocotyl screen performed by Koornneef et al. (1980). Of the five loci identified in that study, hy5 was the only TF. This key factor has since been shown to control a network consisting of thousands of genes as plants modify their growth programs to fit the environment. Plants lacking HY5 are characterized by long hypocotyls under all light conditions, suggesting that HY5 responds to signals from several photoreceptor families to inhibit hypocotyl elongation (Koornneef et al., 1980; Chory, 1992). Studies of HY5 mutant and overexpression plants have implicated HY5 in regulating chlorophyll and anthocyanin biosynthesis, primary and lateral root development, shade and high-temperature responses, flowering time, and many other processes (Oyama et al., 1997; Ang et al., 1998; Holm et al., 2002; Andronis et al., 2008; Delker et al., 2014; Nozue et al., 2015; Gangappa and Botto, 2016). HY5 interacts with many light- and growth-related TFs (Holm et al., 2002; Datta et al., 2007; Singh et al., 2012; Jang et al., 2013; Abbas et al., 2014; Gangappa and Botto, 2016). It is worth noting that the transcription of most of these factors is also regulated by light.
HY5 is directly regulated at the protein level by another key control element in the light response: the E3 ubiquitin ligase complex composed of the CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) and SUPPRESSOR OF PHYA-105 (SPA) proteins (COP1/SPA). This complex degrades many photomorphogenesis-promoting factors in the dark and is deactivated upon light perception (Deng et al., 1992; Lau and Deng, 2012).
How HY5 regulates the transcription of its target genes remains an open question. Genomic studies have found that HY5 can bind to thousands of genes (Lee et al., 2007; Zhang et al., 2011; Kurihara et al., 2014). It has been characterized as a transcriptional activator, a repressor, or both (Ang et al., 1998; Lee et al., 2007; Ruckle et al., 2007; Kindgren et al., 2012; Delker et al., 2014; Gangappa and Botto, 2016; Norén et al., 2016; Xu et al., 2016; Gangappa and Kumar, 2017; Nawkar et al., 2017; Zhang et al., 2017). HY5 has also been reported to feed back to promote its own transcription (Abbas et al., 2014; Binkert et al., 2014). Complicating matters, in vitro work suggests that HY5 does not have its own activation or repression domain (Ang et al., 1998). Current hypotheses regarding HY5 activity include the following: (1) HY5 controls growth not only by its own DNA binding but by regulating the DNA binding of its interacting partners (Ram et al., 2014); (2) HY5 may have activator or repressor activity depending on the binding partner (Ruckle et al., 2007; Kindgren et al., 2012); and (3) HY5 may compete with other TFs for binding sites on DNA (Toledo-Ortiz et al., 2014; Gangappa and Kumar, 2017; Nawkar et al., 2017). These conflicting hypotheses thus require a new approach to clarify the exact molecular function of HY5.
Chromatin immunoprecipitation sequencing (ChIP-seq) and RNA sequencing (RNA-seq) experiments often end up with thousands of differentially expressed genes between genotypes or conditions. While this may provide an interesting mine for hypothesis generation, there is also the risk of false positives or indirect effects on transcription through downstream TF networks. Additionally, while many studies combine RNA-seq with ChIP-seq to find target genes, the multi-layered nature of transcription control in the context of epigenetic modifications, diverse signaling mechanisms, and higher-order protein complexes means that TF binding does not necessarily equal activity at a given locus (Eeckhoute et al., 2009; Schacht et al., 2014). Identifying the most likely direct targets thus requires the development of a higher-order filtering strategy to determine sites of physiologically relevant transcription regulation.
We generated chimeric constitutive repressor and activator forms of HY5 to understand the function of HY5 during early seedling development. We show that HY5 promotes the transcription of target genes. Using these new reagents, in combination with RNA-seq and ChIP-seq, we identify a high-confidence set of direct HY5 target genes as well as genes likely to be indirectly controlled by HY5-dependent transcriptional networks. We found SPA1, SPA4, and COP1 among the direct targets of HY5. We propose that this transcriptional control, coupled with COP1/SPA-mediated HY5 degradation in the dark, is a mechanism underlying the plant’s ability to quickly switch from etiolated to deetiolated growth upon light perception. In addition, we provide in planta evidence that HY5 lacks its own activation domain, supporting the hypothesis that the observed transcriptional activation is driven by preferential interaction with activating TFs. We propose that this multiplexed strategy may be generalizable to the study of TF function.
RESULTS
Chimeric HY5 Proteins Have Distinct Effects on Deetiolation
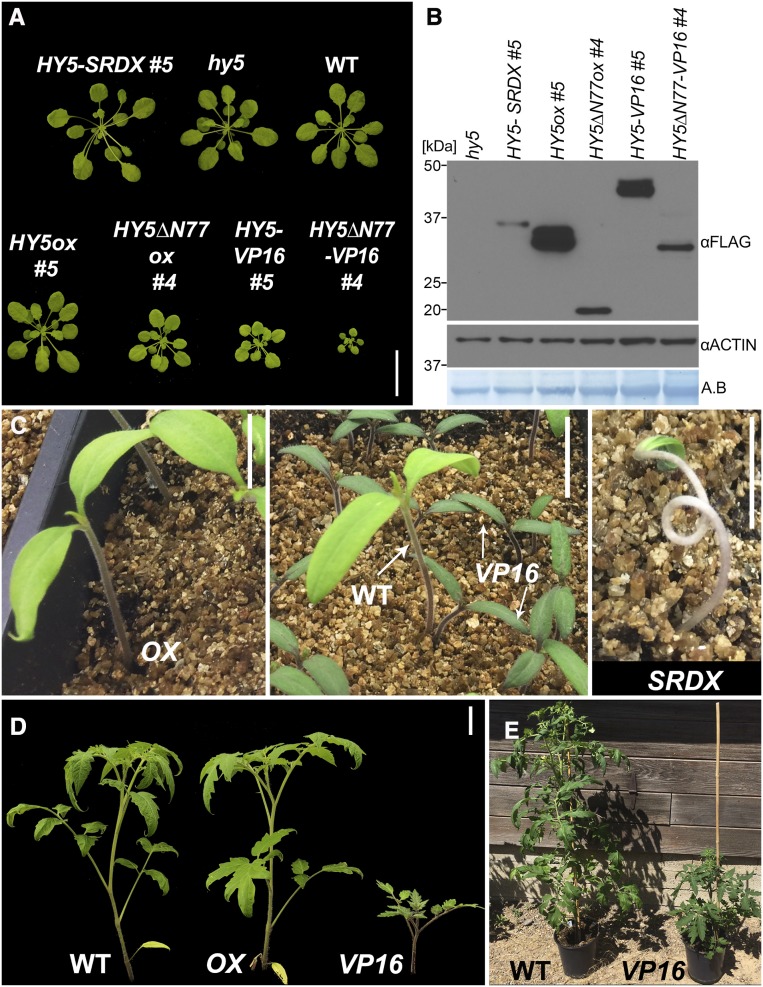
To determine the dominant transcriptional regulation activity of HY5 during deetiolation, we generated chimeric HY5 variants by adding a transcriptional silencing motif (EAR repressor motif of Arabidopsis [Arabidopsis thaliana] SUPERMAN gene [SRDX]; Ohta et al., 2001; Hiratsu et al., 2002) or the activation domain from VP16 (Triezenberg et al., 1988). The SRDX and VP16 domains have been shown to effectively repress or activate gene expression, respectively, when fused to plant TFs (Shani et al., 2009; Fujiwara et al., 2014). We expressed HY5-SRDX, HY5-VP16, and the native form of HY5 (HY5ox) with an N-terminal FLASH tag in the hy5 mutant background (Figure 1A; see Methods). We reasoned that the fusion proteins would specifically repress (HY5-SRDX) or activate (HY5-VP16) the transcription of HY5 direct targets. Since HY5 was shown to regulate its own promoter (Abbas et al., 2014; Binkert et al., 2014), we expressed these HY5 variants under the control of the constitutive 35S promoter. While overexpression under the control of this promoter may increase the false-positive rate, it provides a useful search space in our sorting strategy. Phenotypic and molecular observations in these lines were filtered using the differences between the Columbia-0 (Col-0) background control and the hy5 loss-of-function mutant as a primary benchmark.
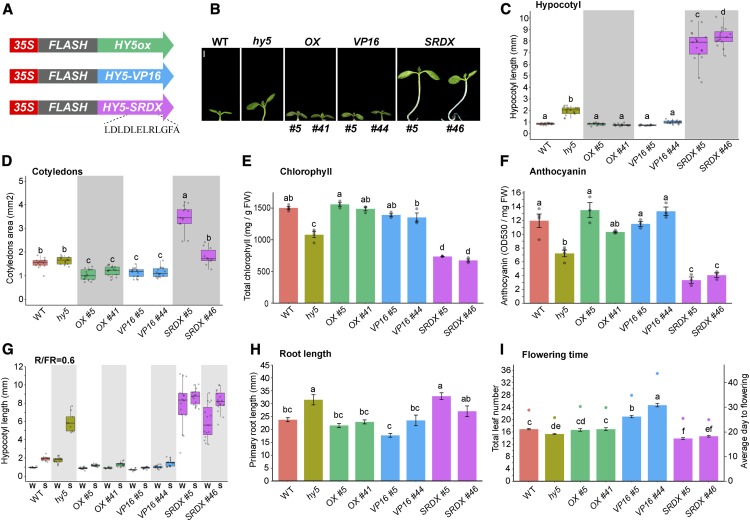
Figure 1.
Chimeric Activator and Repressor Forms of HY5 Phenotypically Recapitulate Overexpression and Loss-of-Function Mutants, Respectively.
(A) Schematic of the chimeric HY5 constructs expressed in hy5 mutant plants.
(B) Representative seedlings grown in continuous simulated white light (sWLc). Two independent transgenic lines from each construct are shown. Bar = 1 mm.
(C) to (I) Quantification of hypocotyl length (C), cotyledon area (D), total chlorophyll content (E), anthocyanin content (F), hypocotyl shade response (G), root length (H), and flowering time (I) of the genotypes shown in (B). Growth conditions were as follows. For (C) and (D), 6-d-old seedlings were grown in sWLc (∼100 µmol m−2 s−1; n = 13 to 17). For (E), 13-d-old seedlings were grown under growth light (100 µmol m−2 s−1) in long days (16 h of light/8 h of dark). For (F), seedlings were grown in constant growth light for 7 d and moved to high light (1080 µmol m−2 s−1) for 24 h. In (E) and (F), data represent means ± se (n = 3 to 4 biological replicates). For (G), seedlings were grown in sWLc (74 µmol m−2 s−1) for 3 d and moved to simulated shade (sWLc supplemented with far-red light [red:far-red light ratio = 0.6]) for an additional 3 d (n = 9 to 16). For (H), 7-d-old seedlings were grown in sWLc (74 µmol m−2 s−1); data represent means ± se (n = 7 to 12). For (I), total leaves at bolting (left y axis, bars) and days to bolting (right y axis, dots) under growth light in long days are shown. Data represent means ± se (n = 10 to 24). In the box plots in (C), (D), and (G), the boxes indicate the first and third quartiles and the whiskers indicate the minimum and maximum values, the black lines within the boxes indicate the median values, and gray dots mark the hypocotyl lengths of individual seedlings. FW, fresh weight; S, shade; W, sWLc. Different letters denote statistically equivalent groups (P < 0.05) among samples as assessed by one-way ANOVA and Tukey’s HSD. Transgene abbreviations are as follows: OX, 35S:FLASH-HY5ox; SRDX, 35S:HY5-FLASH-SRDX; VP16, 35S:FLASH-HY5-VP16.
We tested the effects of these fusion proteins on a series of hy5-related phenotypes during deetiolation, including hypocotyl elongation, cotyledon expansion, and chlorophyll content (as a proxy for chloroplast maturation). Expression of HY5ox and HY5-VP16 rescued the long hypocotyl of hy5 mutant seedlings, whereas HY5-SRDX caused a dramatic increase in hypocotyl length in continuous simulated white light (blue, red, and far-red LED lights) as well as monochromatic red, blue, and far-red light conditions (Figures 1B and 1C; Supplemental Figure 1). In addition, HY5-SRDX increased and HY5-VP16 decreased cotyledon size, and HY5-SRDX strongly reduced chlorophyll content, beyond that observed in the hy5 mutant (Figures 1D and 1E). Together, these data show that the constitutive transcriptional activator HY5-VP16 phenotypically follows the wild-type and HY5ox lines, while the constitutive repressor HY5-SRDX produces exaggerated phenotypes in the same direction as the hy5 loss-of-function mutant. This indicates that HY5 mainly works as an activator during deetiolation.
We then examined processes beyond deetiolation, finding that expression of HY5-VP16 and HY5ox rescued the hy5 mutant phenotypes in root growth, flowering time, shade-induced hypocotyl elongation, and anthocyanin level, while HY5-SRDX either did not change or exacerbated the mutant phenotype (the HY5-SRDX line 46 did not always follow this pattern due to its severe growth defect; Figures 1F to 1I; Supplemental Figures 2A and 2B). Recent studies have reported that HY5 is required as a transcriptional repressor to inhibit hypocotyl elongation at ambient temperature and that this activity is reduced in warm-temperature conditions (Delker et al., 2014; Gangappa and Kumar, 2017). Surprisingly, and contrary to the published model in which HY5 repression activity inhibits growth, we observed that seedlings expressing the HY5-SRDX constitutive repressor had longer hypocotyls than the controls, while those expressing HY5ox and HY5-VP16 had shorter hypocotyls at 20 and 28°C (Figures 2A and 2B). The phenotype of the chimeric HY5-SRDX suppressor suggests that releasing HY5 transcriptional repression activity is unlikely to be the major cause of the high temperature-mediated growth. To test this further, we examined the expression of the growth-related genes SMALL AUXIN UP RNA 19 (SAUR19), INDOLE-3-ACETIC ACID INDUCIBLE 19 (IAA19), and EXPANSIN A8 (EXP8), which were previously suggested to be repressed by HY5 (Jing et al., 2013; Delker et al., 2014; Gangappa and Kumar, 2017). We hypothesized that if HY5 directly regulates their expression, these transcripts will be reduced in HY5-SRDX and increased in HY5-VP16 transgenic lines. All three genes were upregulated in response to high temperature in wild-type and hy5 plants. However, the expression in HY5ox and HY5-VP16 plants was lower than in the wild type and that in HY5-SRDX plants was higher than in the wild type at both 20 and 28°C (Figures 2C to 2E). The observed repression of these growth-related genes is likely due to factors acting downstream of HY5 rather than to direct HY5 action. These results are in agreement with the phenotypes we observed and suggest that HY5 works as an activator in many processes.
Figure 2.
HY5 Transcriptional Repression Activity Is Unlikely To Be the Major Cause of High Temperature-Mediated Growth.
(A) Hypocotyl length in 20 and 28°C.
(B) Hypocotyl growth percentage (length in 28°C relative to the average length in 20°C).
In (A) and (B), seedlings were grown in 20°C long days (16 h of light/8 h of dark), white light (80 µmol m−2 s−1), for 4 d and then moved to 28°C or kept in 20°C for 3 d (n = 13 to 20). Different letters denote statistical differences (P < 0.05) among samples as assessed by one-way ANOVA and Tukey’s HSD.
(C) to (E) Expression of SAUR19, IAA19, and EXP8 in 4-d-old seedlings (line #5) grown in long days at 20°C and either moved to 28°C or kept at 20°C for an additional 6 h. Relative expression was assayed using RT-qPCR relative to the reference gene IPP2 and normalized to expression in the wild type at 20°C. Average values of three biological replicates per condition ± se are shown. Different letters denote statistical differences (P < 0.05) within the conditions as assessed by one-way ANOVA and Tukey’s HSD.
Identifying High-Confidence Direct Targets of HY5 Using Activator and Repressor Chimeras
All previous genomic studies searching for HY5 direct targets arrived at the same conclusion: HY5 can be both an activator and a repressor (Lee et al., 2007; Zhang et al., 2011; Kurihara et al., 2014). However, our results so far indicate that HY5 works mainly as an activator. To resolve this discrepancy, we performed RNA-seq and ChIP-seq with plants expressing our HY5 fusion proteins (using line number 5 from each construct). We reasoned that the expression of a direct HY5 target would respond to VP16 and SRDX by increasing or decreasing expression, respectively. These gene expression patterns could then be compared with those of HY5ox and hy5 and correlated with HY5 binding sites identified with ChIP-seq analysis, yielding a high-quality and high-confidence list of direct HY5 target genes.
Our ChIP-seq data recovered the ACGTG motif (bases 2 to 6 of the canonical G-box), the strongest binding motif for all HY5 fusion proteins, suggesting that the fusion of HY5 to VP16 or SRDX did not change the specificity of the DNA binding domain (Figures 3A and 3B).
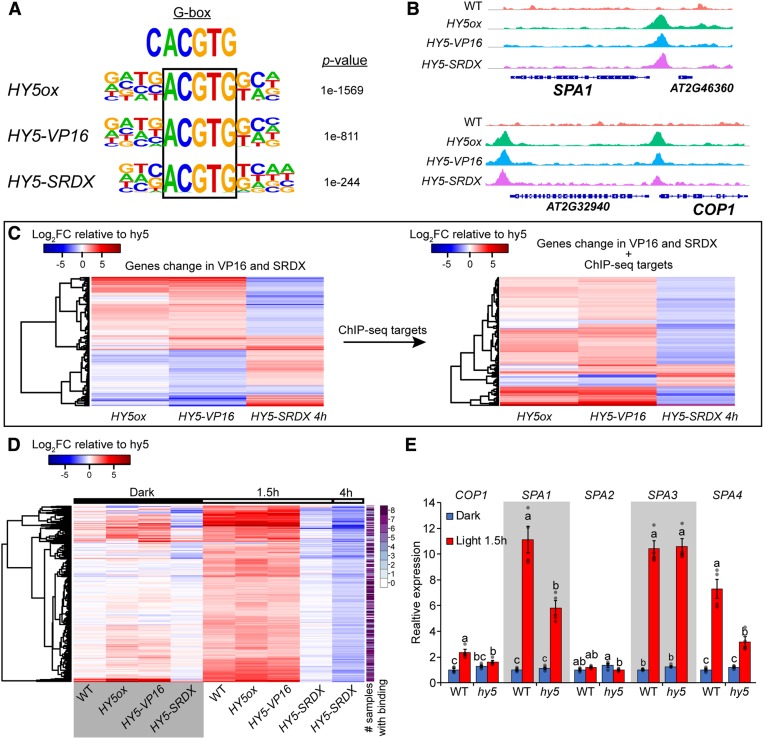
Figure 3.
HY5 Is a Transcriptional Activator during Deetiolation.
(A) De novo binding site motif enrichment from HY5ox, HY5-VP16, and HY5-SRDX ChIP-seq.
(B) Visualization of HY5 binding to the SPA1 and COP1 promoters.
(C) Heat map of differentially expressed genes relative to hy5 in HY5ox, HY5-VP16 after 1.5 h of continuous simulated white light (sWLc; 100 µmol m−2 s−1), and HY5-SRDX at 4 h of sWLc. Left panel: 1179 differentially expressed genes in HY5ox, HY5-VP16, and HY5-SRDX. Right panel: 127 genes from the list at left with HY5 binding peak (2000 bp upstream of the ATG or in the gene body) in all eight ChIP-seq samples.
(D) Heat map showing the 297 genes that are differentially expressed in the wild type and HY5ox, upregulated in HY5-VP16, and downregulated in HY5-SRDX compared with hy5. Samples are 3-d-old dark-grown seedlings after transfer to sWLc for the indicated times. The color scale represents the log2 fold change relative to hy5. The far-right column shows the number of ChIP-seq samples with HY5 binding at each locus.
(E) Expression of COP1, SPA1, SPA2, SPA3, and SPA4 in 3-d-old wild-type or hy5 seedlings grown in dark or dark plus 1.5 h of sWLc. Relative expression was assayed using RT-qPCR relative to the reference gene IPP2 and normalized to expression in the wild type in the dark. Average values of four biological replicates ± se are shown. Different letters denote statistical differences (P < 0.05) among samples as assessed by one-way ANOVA and Tukey’s HSD.
We then validated the transcriptional regulation activity of the chimeric HY5-VP16 and HY5-SRDX constructs by RNA-seq. There was no clear directional regulation of the differentially expressed genes (upregulated or downregulated) in HY5-VP16 and HY5-SRDX transgenic lines relative to hy5 mutant plants (Figure 3C, left; Supplemental Data Set 1). We filtered this list by selecting only the genes with HY5 peaks in all of our ChIP-seq samples (high-confidence HY5 binding sites). In this new list, 79% were upregulated in HY5-VP16 and downregulated in HY5-SRDX lines, indicating that HY5-VP16 activates and HY5-SRDX represses gene expression as expected (Figure 3C, right; Supplemental Data Set 1). Only 12 genes had the opposite response, and 10 of them were also downregulated in HY5ox. This strongly suggests that the binding at these loci is not the determining factor for expression and supports the hypothesis that these genes are being controlled by a repressor that is itself transcriptionally activated by HY5. The expression of the 15 remaining genes changed in the same direction in both HY5-SRDX and HY5-VP16 lines. Transcriptional regulation of this group is likely more complicated, possibly influenced by factors other than or farther downstream of HY5. Overall, the majority of gene expression profiles track well with the activator and repressor activity of VP16 and SRDX, respectively.
Having confirmed that the VP16 and SRDX fusion proteins regulate gene expression in the expected directions at the level of the whole transcriptome, we began our search for the crucial set of genes that are regulated directly by HY5. To increase the stringency and specificity of this analysis, we looked for genes that changed expression in hy5 and HY5ox (either up or down), were upregulated in HY5-VP16, and were downregulated in HY5-SRDX. A total of 297 genes satisfied these criteria (Figure 3D; Supplemental Data Set 2). All but two genes on this list were downregulated in hy5 compared with the wild type, suggesting that they are positively regulated by HY5. We found that 81% of the genes were bound by HY5 in at least four samples and only 3% lacked binding in all eight samples. We propose that these are the most likely direct HY5 targets in vivo. In contrast, previous studies searching for potential HY5 direct targets by comparing expression data with chromatin occupancy found that only 12% (Kurihara et al., 2014), 19% (Lee et al., 2007), or 43% (Zhang et al., 2011) of the differentially expressed genes in hy5 also had a HY5 ChIP peak.
The targets identified here represent both characterized and novel facets of HY5 physiology, including previously described HY5 targets, such as HY5-HOMOLOG, B-BOX (BBX22, BBX24, and BBX32), and LONG HYPOCOTYL IN FAR-RED (HFR1). Importantly, we filtered out growth-regulated genes previously suggested to be repressed by HY5, including SAUR19, IAA19, and EXP8. These three genes were upregulated in HY5-SRDX and downregulated in HY5-VP16, as shown earlier (Figures 2C to 2E; Supplemental Data Set 3). We consider it highly unlikely that these strong activator and repressor domains would act in an opposite role (activator becomes a repressor or repressor becomes an activator) at these specific loci, and therefore we conclude that they are probably regulated by factors downstream of HY5. In addition, we found that HY5 can directly regulate the expression of SCHLAFMÜTZE (SMZ) and SCHNARCHZAPFEN (SNZ), which repress FT to modify flowering time (Supplemental Figure 4; Mathieu et al., 2009). The mild early-flowering phenotype observed in hy5 and smz-2 snz-1 plants, and the strong late-flowering phenotypes of SMZ overexpression and HY5-VP16, are consistent with HY5 regulating flowering time through SMZ and SNZ. Of particular interest, we identified in our list of 297 HY5-regulated genes the E3 ubiquitin ligase complex components SPA1, SPA4, and COP1, which promote HY5 degradation (Figure 3D; Supplemental Data Set 2). We also observed HY5 on the promoters of SPA1, SPA3, SPA4, and COP1 (Figure 3B; Supplemental Figure 5A; Supplemental Data Set 2). The expression levels of COP1, SPA1, SPA3, and SPA4 were upregulated in the light relative to their expression in the dark and all except SPA3 showed weaker induction by light in hy5, suggesting a transcriptional/posttranslational feedback loop that fine-tunes HY5 abundance (Figure 3E; examined further in the following section). While the binding of HY5 to the COP1 promoter was previously described (Huang et al., 2012; Binkert et al., 2014; Xu et al., 2016), we wanted to confirm the binding to the SPAs in vitro. To this end, we tested HY5 activity on these promoters using a luciferase assay in the Drosophila S2 system. We confirmed HY5 transcriptional activation on the SPA1 promoter but not on the COP1, SPA3, or SPA4 promoters (Supplemental Figure 5B). This result suggests that either the binding of HY5 to the SPA1 promoter is stronger than the binding to the other promoters or that the COP1, SPA3, and SPA4 promoters are partially repressed in S2 cells. In addition, we found in a recently published ChIP-seq data set that HY5 binds to the promoters of SPA1, SPA3, SPA4, and COP1 when expressed under the control of its native promoter (Hajdu et al., 2018). These findings increase our confidence that HY5 binds to these promoters in vivo and that our results are most likely not an artifact of the 35S promoter driving the expression of our transgenes.
Our initial RNA-seq was done on whole seedlings, so we examined the contribution of organ-specific expression of the 297 HY5-regulated genes during a 12-h deetiolation time course in wild-type hypocotyls and cotyledons (Supplemental Figure 5C; Supplemental Data Set 4). The majority (96%) were upregulated, in at least one time point, in both the hypocotyl and the cotyledon. Thus, we conclude that HY5 promotes transcriptional activation nearly uniformly in multiple subregions of the plant.
HY5 Regulates the Expression of COP1/SPA Complex Components
Previous studies on hy5 mutants or overexpression of HY5 in wild-type seedlings did not describe modified growth patterns in the dark unless combined with hormone treatments or mutants that promote deetiolation in the dark (Ang and Deng, 1994; Ang et al., 1998; Alabadí et al., 2008; Stracke et al., 2010; Li and He, 2016; Shi et al., 2018). Consistent with these reports, our HY5ox lines did not show any notable phenotypes in etiolated seedlings. However, one of the 12 HY5-VP16 lines (#5) was partially deetiolated in the dark, with shorter stature and an open apical hook (Figures 4A to 4C; Supplemental Figure 1F). The HY5 protein and RNA levels in our HY5ox lines were higher than in HY5-VP16 or HY5-SRDX, and all chimeric proteins were similarly stabilized in the light (Supplemental Figure 6), suggesting that the dark phenotypes of HY5-VP16 #5 are due to activity rather than to expression level.
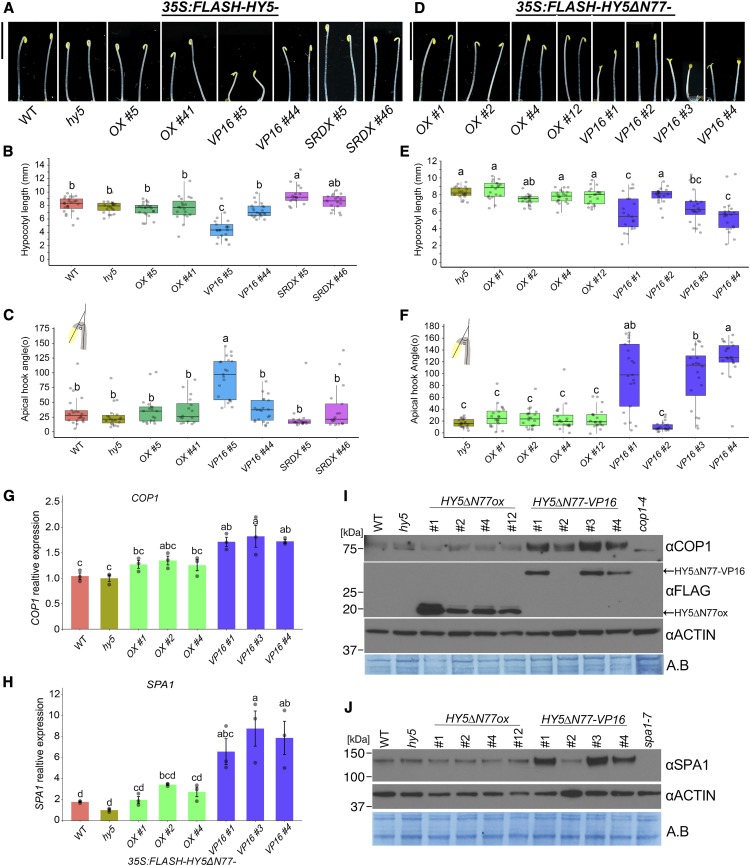
Figure 4.
Forcing Transcriptional Activation of HY5 Target Genes Promotes Deetiolation in the Dark and Promotes the Expression of COP1 and SPA1.
(A) and (D) Seedlings overexpressing full-length HY5 (A) or HY5 without the first 77 amino acids (HY5ΔN77 or HY5ΔN77-VP16; [D]) in the hy5 mutant background. Shown are representative seedlings grown in the dark for 3 d; HY5ΔN77-VP16 seedlings are heterozygotes, and others are homozygotes. Bars = 5 mm.
(B) and (E) Quantification of hypocotyl length of the seedlings shown in (A) and (D), respectively (n = 1423).
(C) and (F) Quantification of apical hook angle of the seedlings shown in (A) and (D), respectively (n = 1423).
(G) and (H) Expression of COP1 (G) and SPA1 (H) in 3-d-old dark-grown seedlings. Relative expression was assayed using RT-qPCR relative to the reference gene IPP2 and normalized to expression in hy5. Average values of three biological replicates per condition ± se are shown.
(I) and (J) Immunodetection of COP1 and HY5-FLAG (I) and SPA1 (J) protein levels from total protein extract of 3-d-old dark-grown seedlings. ACTIN and Amido Black (A.B.) are shown as loading controls.
In (G) to (J), the HY5∆N77-VP16 lines #1, #3, and #4 were segregating; therefore, we collected only seedlings with the deetiolated phenotype in the dark. Different letters denote statistical differences (P < 0.05) among samples as assessed by one-way ANOVA and Tukey’s HSD.
Two converging observations indicate that phenotypes induced by HY5-VP16 are highly sensitive to dosage or expression level. First, we noticed that many of the HY5-VP16 lines had a tendency to be silenced. Second, we found that HY5 directly promotes SPA1, SPA3, SPA4, and COP1 expression, possibly limiting HY5 protein accumulation (Figures 3B and 3E; Supplemental Data Set 2). These observations might explain the lack of dark phenotypes in medium or weakly expressing HY5ox or HY5-VP16 lines. We hypothesized that the COP1/SPA-mediated degradation may limit HY5-VP16 activity; therefore, we overexpressed HY5 lacking the COP1-interacting domain (HY5∆N77) with or without VP16 (Ang et al., 1998). Without this domain, HY5 cannot be degraded by COP1, thereby increasing its abundance and activity (Ang et al., 1998). None of the 10 HY5∆N77ox lines tested showed notable phenotypes in the dark; however, 5 out of 10 lines expressing HY5∆N77-VP16 showed a partly deetiolated phenotype in the dark (Figures 4D to 4F). In support of our interpretation, HY5∆N77-VP16 line #2 had very low expression of the chimeric protein and phenotypically resembled the wild type (Figures 4D to 4F and 4I; Supplemental Figure 7A). The partial deetiolation of HY5∆N77-VP16 suggests that HY5 may control its own degradation via COP1/SPA transcription. We therefore examined SPA1, SPA3, SPA4, and COP1 RNA levels in HY5∆N77-VP16 and HY5∆N77ox dark-grown seedlings. The expression of each of these components in the dark was higher in HY5∆N77-VP16 than in HY5∆N77ox, supporting our hypothesis that HY5 activates their transcription (Figures 4G and 4H; Supplemental Figures 7B and 7C). Consistent with this, we also found that COP1 and SPA1 protein levels were higher in HY5∆N77-VP16 than in HY5∆N77ox (Figures 4I and 4J; Supplemental Figures 7D and 7E). This result also suggests that HY5 depends on other TFs, potentially other targets of the COP1/SPA complex in the dark, to promote transcription. We did not observe any difference in gene expression between HY5-VP16 and HY5ox dark-grown seedlings in our RNA-seq data (Supplemental Figure 7F). While the expression of SPA1, SPA3, and SPA4 was higher than that of the wild type in HY5ox and HY5-VP16 etiolated seedlings, this degree of overexpression evidently was not enough to promote deetiolation. Together, these data indicate that the negative feedback loop between HY5 and COP1/SPAs can be uncoupled by disrupting the HY5-COP1 interaction.
HY5∆77-VP16 also had strong effects in adult plants compared with HY5∆N77ox or HY5-VP16. These plants were very small, with short petioles and small leaf blades, demonstrating the importance of tight regulation over HY5 in all stages of development (Figure 5A; Supplemental Figure 8A). There was a clear gradient in plant size and petiole length of the adult plants, from HY5∆N77-VP16 (the smallest) to HY5-SRDX (the largest), with HY5ox and hy5 being similar to the wild type. HY5∆N77ox was slightly smaller than the wild type but larger than HY5∆N77-VP16. While the protein levels of HY5∆N77 were lower than those of full-length HY5, in both native and VP16 constructs, the mature HY5∆N77 plants were smaller, indicating that HY5∆N77 is a hyperactive form of HY5. This is consistent with a previous study (Figures 5A and 5B; Supplemental Figure 8A; Ang et al., 1998). We therefore conclude that in mature plants, as in etiolated seedlings, HY5-dependent phenotypes are controlled by a HY5-COP1/SPA feedback loop.
Figure 5.
Dosage-Dependent HY5 Activity Controls Plant Size in Arabidopsis and Tomato.
(A) Arabidopsis plants of the indicated genotypes growing in short days (8 h of light/16 h of dark) for 42 d. Plants are as follows: 35S:FLASH-HY5-SRDX (HY5-SRDX), 35S:FLASH-HY5ox (HY5ox), 35S:FLASH-HY5ΔN77ox (HY5ΔN77ox), 35S:FLASH-HY5-VP16 (HY5-VP16), and 35S:FLASH-HY5ΔN77-VP16 (HY5ΔN77-VP16). Bar = 3 cm.
(B) HY5-FLASH protein levels from 7-d-old seedlings of the genotypes and growth conditions shown in (A), detected with an anti-FLAG antibody against FLAG-HY5 and FLAG-HY5ΔN77.
(C) Three-week-old tomato plants of the wild type, 35S:DOF-AtHY5ox (OX), 35S:DOF-AtHY5-VP16 (VP16), and 35S:DOF-AtHY5-SRDX (SRDX). DOF, HA-YFP-HA. Bars = 1.5 cm.
(D) Eight-week-old tomato plants. Bar = 5 cm.
(E) Mature tomato plants.
HY5 Transcriptional Control Activity Requires Additional Factors
HY5-VP16 has stronger phenotypic effects then HY5ox in the dark and at the adult stage, suggesting that HY5 is dependent on the expression of other proteins to activate transcription. To demonstrate this further, we compared HY5 activity in tobacco (Nicotiana benthamiana) with the ability of HY5 to activate gene expression in Drosophila S2 cells, which lack the plant proteins that interact with HY5 to form the activation complex. Using luciferase reporter assays, we tested HY5’s ability to activate a synthetic promoter made from four tandem repeats of the G-box (4xG-box), which HY5 has been shown to bind in vitro (Shi et al., 2018), and the CHALCONE SYNTHASE promoter (CHSp), which was previously reported to be a direct target promoter of HY5 (Ang et al., 1998). HY5 did not change the basal luciferase expression from either of the promoters in S2 cells, while HY5-VP16 strongly activated expression from both (Supplemental Figure 8B). Of interest, coexpression of HY5 and HY5-VP16 reduced luciferase activation from both promoters, supporting the hypothesis that HY5 lacks its own activation domain. By contrast, HY5 can weakly activate the expression of CHSp without VP16 in N.benthamiana leaves (Supplemental Figure 8C). This suggests that plant-specific transcriptional activators are required for HY5 activity.
Chimeric Arabidopsis HY5 Can Modulate Growth in Tomato
Early seedling establishment is crucial for survival in many crops. For example, tomato (Solanum lycopersicum) plants with long hypocotyls are quite fragile, which can lead to the loss of many seedlings during mechanical planting (Brigard et al., 2006). Conversely, early elongation may aid competition against weeds. HY5 plays key roles in regulating hypocotyl growth in tomato (Liu et al., 2004). Recent work in tomato has described strategies for generating phenotypic gradients (Rodriguez-Leal et al., 2017; Israeli et al., 2019). Given our observation of HY5-dependent size gradients in Arabidopsis, we tested the utility of chimeric Arabidopsis HY5 proteins with an N-terminal DOF (HA-YFP-HA) tag in generating phenotypic diversity in tomato plants. Similar to Arabidopsis, AtHY5-VP16 dramatically inhibited hypocotyl growth in tomato seedlings and generally slowed growth in later stages, resulting in bushy and dwarf plants (Figures 5C to 5E). Overexpression of AtHY5-SRDX, however, led to longer hypocotyls and seedling lethality. Interestingly, while it was recently shown that overexpression of the tomato HY5 (SlHY5) increased anthocyanin levels (Liu et al., 2018), AtHY5ox tomato plants did not visibly differ from the wild type. Together, these results suggest that Arabidopsis HY5 may be able to accomplish some, but not all, functions of tomato HY5. Nevertheless, the phenotypic changes we observed in AtHY5-VP16 and AtHY5-SRDX plants indicate that modification of HY5 activity can achieve dynamic control of growth. Fine-tuning temporal and spatial control over HY5 expression will be the next critical steps in optimizing this process.
DISCUSSION
HY5, a bZIP TF, plays a pivotal role in the transcriptional response of plants to changes in the local light conditions. Despite its importance to the biology of plants, HY5’s mechanism of action has been difficult to define. In this study, we used very stringent criteria to show that HY5 promotes transcriptional activation of its direct targets. Moreover, we define a negative feedback loop whereby HY5 promotes the expression of its negative regulators in the COP1/SPA complex. This use of a number of stringent criteria allowed us to filter out a large number of genes that were unlikely to be direct targets of HY5, thereby allowing us to eliminate several proposed models of HY5’s mechanism of action.
A Multiplex Strategy Using Constitutive Chimeras Defines Transcriptional Activity of HY5
Determining a causal relationship between TF binding and changes in gene expression is very challenging. In this study, we improve upon the existing RNA-seq/ChIP-seq pipeline. We directed HY5 to work as a repressor (HY5-SRDX) or as an activator (HY5-VP16) and combined RNA-seq and ChIP-seq with rigorous phenotypic analysis of these chimeric constructs. This approach allowed us to define high-confidence direct target genes and revealed that transcriptional activation is the major physiologically significant function of HY5. We identified a relatively small number of HY5 direct targets in comparison with previous studies (Lee et al., 2007; Zhang et al., 2011; Kurihara et al., 2014). This disparity is likely due to two reasons: (1) we collected our samples soon after the shift to light, while previous studies examined plants that had been grown in light for several days, which will lead to many indirect effects due to the phenotypic differences; and (2) the use of the chimeric HY5 constructs enabled us to filter out many indirect targets. In support of this, we found that ∼10% of our HY5 direct targets are TFs, indicating sequential layers of transcriptional control and potentially explaining the downregulation of genes that were thought to be direct targets of HY5.
Deeper examination of TF function will require that these methods be further refined. Our use of the constitutive 35S promoter was one solution to the problem of autoactivation by HY5. Nevertheless, the plants were sampled at steady state. This issue could be resolved using an inducible system to transiently express TFs of interest and examine short-time-scale changes in gene expression in a pulse-chase-like format. In the case of TFs that have strong intrinsic activation or repression activity, it may be necessary to identify and mutate these domains to an inactive form prior to fusing the protein to known activator and repressor domains in the strategy we have described here.
Refining Models of HY5 Function
Several models of HY5 function have been put forward in recent years, primarily relying on coanalysis of chromatin binding and gene expression. In light of our strict analysis, we examined areas of agreement and disagreement among these models. First, several groups have suggested that HY5 can act as a transcriptional repressor. This idea was supported by data suggesting that HY5 can compete with other TFs for DNA binding sites (Li et al., 2010; Xu et al., 2016) and that it can lead to downregulation of gene expression through interaction with chromatin-remodeling factors (Jing et al., 2013; Zhao et al., 2019). We cannot rule out the possibility that HY5 may contribute to transcriptional repression in some rare or specific cases. However, the contributions of these events to the tested phenotypes are not detectable in our conditions, and the proposed target genes examined in these studies do not respond in the predicted directions to the VP16 or SRDX fusion constructs.
It was also suggested that HY5 and PIFs work antagonistically and compete for binding at the same target genes, leading to opposing phenotypes during deetiolation at elevated temperature (Toledo-Ortiz et al., 2014; Gangappa and Kumar, 2017). However, we find only a few common targets between HY5 and PIFs, suggesting that this model is not sufficient to explain the combined functions of these TFs during deetiolation (Supplemental Figure 9). Both of these proposed models suggest that HY5 acts as a repressor; however, these are incompatible with our findings demonstrating the phenotypic and molecular requirements for HY5-based transcriptional activation. In addition, we demonstrated that the growth-related genes SAUR19, IAA19, and EXP8, which were shown to be directly activated by PIF4 and PIF5, are probably not directly regulated by HY5 (Figures 2C to 2E; Franklin et al., 2011; Hornitschek et al., 2012; Sun et al., 2013; Ma et al., 2016; Gangappa and Kumar, 2017). We therefore suggest that many of the cases in which HY5 is considered a repressor are in fact likely to be effects of the downstream transcriptional network. For example, HFR1, which represses PIF4 activity and was among our high-confidence HY5 direct targets, may account for the antagonistic activity of HY5 and PIF4.
Another potential interaction between HY5 and PIF activities is highlighted by the finding that COP1-SPA1 promotes PIF1 rapid degradation in the light and SPA1 directly phosphorylates PIF1 (Zhu et al., 2015; Paik et al., 2019). This is consistent with our identification of SPA1 as a high-confidence HY5 direct target. The observation that HY5∆77 is hyperactive relative to full-length HY5 provides some insight into this issue, in that the increased activity of degradation-resistant HY5∆77 leads to more SPA1. This increased population of SPA1 would then promote the degradation of PIF1, thus reducing PIF-dependent growth while maintaining the HY5 pool. We suggest a path in which, in wild-type plants, HY5 promotes the expression of SPA1 in the light, thus promoting degradation of PIF1 and providing a mechanism for maintaining a balance between HY5 and PIFs.
HY5 Requires Interacting Partners to Promote Gene Activation
Here, we show that HY5-VP16 and HY5ΔN77-VP16, but not HY5ox or HY5ΔN77ox, can promote deetiolation in the dark. Furthermore, HY5-VP16 activates reporter expression in insect cells and leads to phenotypic changes in tomato, while HY5 alone cannot. Taken together, these observations provide definitive evidence for the requirement of interacting partners for HY5 activity. Additionally, our finding that HY5 can promote deetiolation in the dark when stabilized and fused to an activation domain suggests that endogenous HY5-interacting transcriptional activators are expressed mainly in the light. This is consistent with a model in which HY5 directs the light-dependent growth program by binding target gene promoters, thereby marking them as targets for light-dependent transcriptional activators. Identification and characterization of the complexes that are needed for HY5 activity will be of future interest. It was shown that the human bZIP TFs FOS and JUN, together with the TF NFAT, form a complex that binds strongly to the DNA only when all three factors are present (Macián et al., 2000). HY5 may similarly require TFs from different families to form a complex that strongly binds to DNA and promotes transcription.
Feedback between HY5 and the COP1/SPA Complex Readies Seedlings for Deetiolation
A seedling’s ability to switch from etiolation mode to deetiolation upon light exposure is an important factor in determining its success in development. HY5 is a key activator of this process and, as such, is tightly regulated. We show here that HY5 directly promotes the expression of SPA1, SPA4, COP1 (as previously described), and to some extent SPA3 by directly binding to their promoters. As a result, accumulation of HY5 in the dark leads to an increase in the COP1/SPA complex, and thus its own degradation, and forms a negative feedback loop between HY5 and COP1/SPA, balancing the level of HY5. This negative feedback is consistent with a previous study showing that plants expressing VP16-HY5 are similar to the wild type (Stracke et al., 2010), as reaching an active threshold concentration while remaining vulnerable to COP1-mediated degradation is likely a rare event. By using HY5ΔN77-VP16, we were able to simultaneously uncouple this feedback loop and eliminate the need for HY5-interacting partners, thus demonstrating the potential detrimental effect of HY5 accumulation in the dark. For example, if a seedling switches to a deetiolated growth mode before emerging from the soil, it will never reach a light source, and if it activates the chloroplast maturation program prior to seeing light, it sensitizes itself to damaging reactive oxygen species when it is finally exposed (Seluzicki et al., 2017).
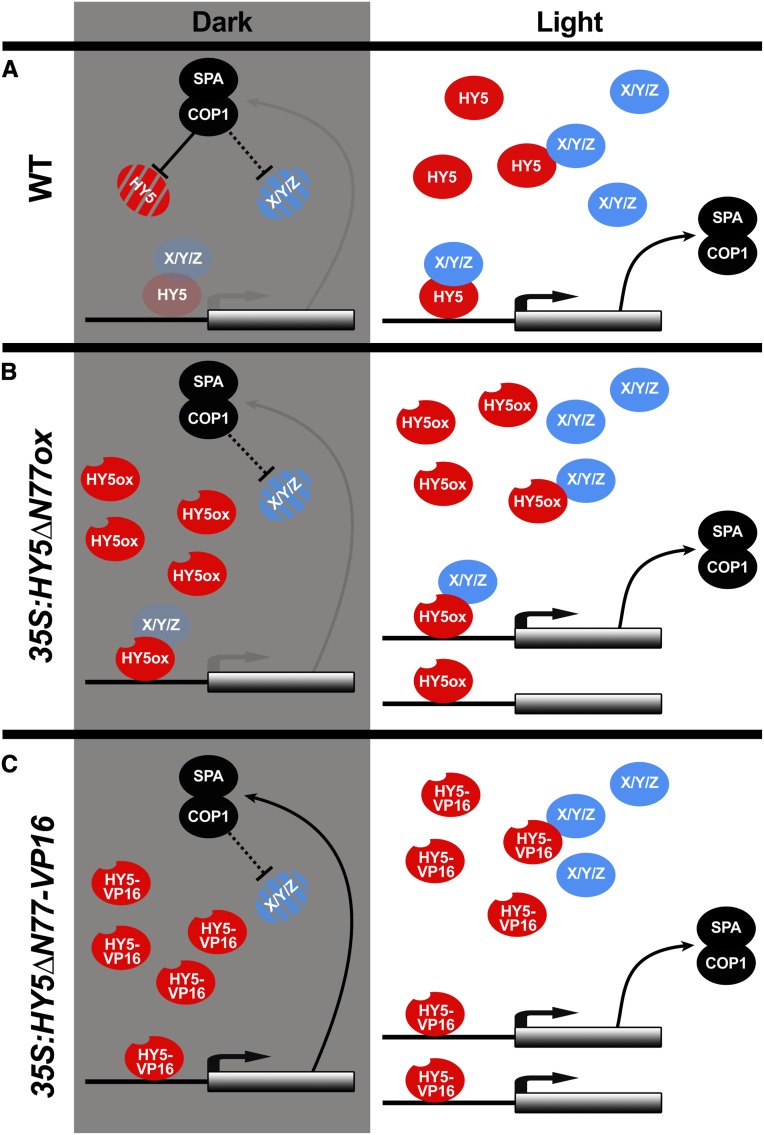
We propose a model, based on our results and the existing literature, in which HY5 is regulated at the transcript level by a number of factors including itself. At the protein level, HY5 is controlled by COP1/SPA, multiple photoreceptors, and the availability of other light-dependent HY5-interacting proteins (Figure 6; Ang et al., 1998; Lau and Deng, 2012; Zheng et al., 2013; Abbas et al., 2014; Binkert et al., 2014; Lu et al., 2015; Yang et al., 2018; Lau et al., 2019; Ponnu et al., 2019; Xu, 2019). In the dark, HY5 and its known partners are degraded, consistent with the observation that overexpression of HY5 alone cannot promote deetiolation. The negative feedback loop between HY5 and COP1/SPA in the dark restricts HY5 activity, limiting it to a small active pool. Upon exposure to light, the expression of COP1/SPA increases but their activity on HY5 is inhibited, allowing HY5 protein levels to rapidly increase, along with other interacting proteins. As they accumulate together, these proteins form complexes with HY5 that bind to DNA and activate gene expression. Importantly, none of the single or double mutants knocking out known HY5-interacting TF genes can mimic the hy5 long-hypocotyl phenotype in all light conditions (Holm et al., 2002; Datta et al., 2008; Kushwaha et al., 2008; Jang et al., 2013; Zhang et al., 2017). This suggests that HY5 acts through multiple parallel interactions with other TFs to run this light-dependent growth program.
Figure 6.
A HY5-COP1/SPA Feedback Loop Is a Light-Regulated Clutch.
(A) In wild-type dark-grown seedlings, HY5 can slightly activate the expression of its target genes. These genes include members of the COP1/SPA ubiquitin ligase complex, which promote HY5 degradation. The absence of the interacting proteins (X/Y/Z) limits HY5 activity. These interacting proteins are likely light-dependent at the level of transcription or protein stability, possibly by COP1/SPA. In the light, COP1 and SPA transcription increases, but the protein complex, which promotes HY5 degradation, becomes inactive, enabling HY5 and its interacting proteins to accumulate, interact, and activate the transcription of target genes.
(B) In the dark, HY5∆N77ox cannot increase the expression of its targets, since its interaction partners cannot accumulate. As interacting proteins accumulate in the light, however, HY5∆N77ox can function normally.
(C) Overexpression of HY5∆N77-VP16 can lead to gene activation in the dark, removing the requirement for interacting proteins and decoupling HY5 function from COP1/SPA activity.
Black lines indicate strong activity, and gray lines indicate very weak activity.
The identification of a HY5-COP1-SPA negative feedback loop reveals an intervention point for directed modulation of growth properties. Here, we show that by combinatorially directing transcription activity and enhancing protein stability, we can generate a phenotypic gradient in plant size. This raises another hypothesis, which remains to be tested, whereby the negative feedback is important for the maintenance of COP1/SPA levels during the day, becoming active at dusk and degrading HY5 and other proteins. We also show that chimeric HY5 proteins can modulate growth in tomato. We therefore propose that this feedback loop represents a distinct access point to tune desirable growth properties by modulating the strength of transcriptional activation and stability via the interaction with the COP1/SPA complex.
METHODS
Plant Materials, Growth Conditions, and Seedling Measurements
All Arabidopsis (Arabidopsis thaliana) plants used were in the Col-0 background. The mutants hy5, carrying a C→T mutation causing an early stop codon at the second amino acid Gln (Q2*; Lian et al., 2011; Chen et al., 2016), spa1-7 (Fittinghoff et al., 2006), and cop1-4 (McNellis et al., 1994) were described previously. Seeds were sterilized, stratified in the dark at 4°C, and germinated in constant white light for 1 h, or in the indicated light conditions (Supplemental Figure 10), on plates with 0.5× Linsmaier and Skoog medium (LS; Caisson Laboratories) with 0.8% (w/v) phyta-agar (Caisson Laboratories): long-day conditions were 16 h of light and 8 h of dark, and short-day conditions were 8 h of light and 16 h of dark. The temperature used was 21°C (unless otherwise stated). Hypocotyl length, apical hook angle, root length, and lateral root number assays were performed on T4 seedlings (except in Figures 4D to 4I, Supplemental Figure 1, and Supplemental Figures 7A to 7E, in which T3 seedlings used) grown vertically and scanned at the indicated times. For cotyledon size imaging, the cotyledons were cut and placed flat on plates with a thin layer of LS agar. Measurements were done using NIH ImageJ software (Schindelin et al., 2012).
Tomato (Solanum lycopersicum) strain M82 (sp) was germinated on soil in a growth chamber in long-day conditions under cool-white fluorescent light. Four-week-old seedlings were transferred to greenhouse conditions with natural daylength at 24 to 25°C.
DNA Constructs and Plant Transformation
The multisite Gateway system (Invitrogen) was used. To generate HY5ox, HY5-VP16, and HY5-SRDX gene constructs, full-length HY5 coding sequence (with or without stop codon) was PCR-amplified from Col-0 cDNA and recombined into pDONR-P2RP3. KpnI and XhoI were used to introduce the VP16 (PCR product) and SRDX (primers synthesized and annealed) coding sequences to the C terminus of HY5. HY5ΔN77 was generated by PCR from plasmids containing HY5ox and HY5-VP16 and cloned into pDONR-P2RP3. Different HY5 versions were cloned downstream of 2x35Spro in pDONR-P4P1R and FLASHv2 (the 2xStrepII-6xHis-protease cleavage site-3xFlag) in pDONR-P221, respectively. 35Spro:FLASH-HY5ox was recombined into the destination vector pH7m34GW; 35Spro:FLASH-HY5-VP16 into pK7m34GW; and 35Spro:FLASH-HY5-SRDX, 35Spro:FLASH-HY5ΔN77ox, and 35Spro:FLASH-HY5ΔN77-VP16 into pB7m34GW. hy5 mutant plants were transformed with Agrobacterium tumefaciens GV3101 carrying the various constructs using the floral dip method (Clough and Bent, 1998). Segregation analyses of antibiotic resistance were used to isolate single-insertion homozygous lines except for 35Spro:FLASH-HY5ΔN77-VP16, for which heterozygous lines were used.
For tomato transformation 2x35Spro in pDONR-P4P1R, HA-YFP-HA (DOF; Bürger et al., 2017) in pDONR-P221, and HY5ox, HY5-VP16, and HY5-SRDX in pDONR-P2RP3 were recombined into pK7m34GW. Cotyledon transformation in tomato was performed according to McCormick (1991). Primers used for cloning are detailed in Supplemental Data Set 5.
Immunoblot Analysis
Protein immunoblots were performed as described by Li et al. (2012), except a 4 to 12% Bis-Tris gel (Invitrogen) and semidry transfer (Thermo Fisher Scientific, Pierce G2 Fast Blotter) were used, except for SPA1, in which a wet transfer method was used. The primary antibodies used were αCOP1 1:250 (v/v; Balcerowicz et al., 2011; a gift from Ute Hoecker), αSPA1 1:5000 (v/v; Zhu et al., 2008; a gift from Xing-Wang Deng), αFLAG M2-HRP 1:5000 (v/v; Sigma-Aldrich, catalog no. A8592), αACTIN 1:75,000 (v/v; Sigma-Aldrich, catalog no. A0480), and αH3 1:5000 (v/v; Cell Signaling Technology, catalog no. 9715). Signal intensities were quantified using NIH ImageJ software (Schindelin et al., 2012).
RNA Analysis
For quantitative RT-qPCR, RNA was extracted using the RNeasy Micro Kit (Qiagen). cDNA synthesis was performed using a Maxima first-strand cDNA synthesis kit (Thermo Fisher Scientific) with 1 μg of RNA. RT-qPCR analysis was performed using a CFX384 Real-Time PCR Detection System (Bio-Rad), with Premix Ex Taq II (TaKaRa, catalog no. RR820A). Levels of mRNA were calculated relative to IPP2 as an internal control as described by Shleizer-Burko et al. (2011). Primers used for the RT-qPCR analysis are detailed in Supplemental Data Set 5.
RNA-Seq Experiments and Analysis
HY5 RNA-Seq
Line #5 was used for all constructs. Three biological replicates (two biological replicates collected for HY5-SRDX) of whole seedlings grown in the dark for 3 d or 3 d of dark plus 1.5 h in simulated white light (Supplemental Figure 10) were collected and snap-frozen in liquid nitrogen. Since we noticed that the effect of HY5-SRDX on hypocotyl growth is visible only after several hours of light (Supplemental Figure 3), we added the 4-h time point for HY5-SRDX and hy5, two biological replicates each. Dark samples were collected under safe green light. Total RNA was isolated using the RNeasy Micro Kit (Qiagen). Stranded mRNA-seq libraries were prepared using the Illumina TruSeq Stranded mRNA Library Prep Kit according to the manufacturer’s instructions. The library was sequenced at single-end 50 bp in the Illumina Hi-Seq 2500 System at the Salk Institute’s Next Generation Sequencing Core Facility. Raw reads were aligned to the The Arabidopsis Information resource (TAIR10) genome using TopHat (v2.0.11) with default parameters except that library type was set to first strand and intron length was set to 40 minimum, 2000 maximum (Kim et al., 2013). Mapped reads were counted by HTSeq (v0.6.0) with default parameters except “-m intersection-strict -s reverse” (Anders et al., 2015) and analyzed by edgeR (v3.16.5; Robinson et al., 2010). Only genes with counts-per-million values greater than 1 in at least two samples were kept for differential expression analysis. A design matrix was set up where each treatment (dark or light) for each genotype (hy5, wild type, HY5ox, HY5-VP16, and HY5-SRDX) at each time point (1.5 or 4 h) is one group, and a generalized linear model was fitted to the read counts with this design matrix. For differential gene expression, contrasts were set up between each of the genotypes and hy5 (at the relevant time point). Genes with P < 0.05 following Benjamini and Hochberg correction and log2 fold change < −0.5 or > 0.5 were considered differentially expressed. Lists of gene counts per million and fold change can be found in Supplemental Data Set 3.
Deetiolation RNA-Seq
Seedlings were grown vertically in the dark on 0.5× LS plates with nylon mash (100 μm, 44% open area, 40-inch-width ELKO filtering). After 3 d in the dark, the seedlings were moved to white light or kept in the dark, and at the indicated times, the roots were removed with a scalpel and two biological replicates of the remaining seedlings were collected into ice-cold 80% (v/v) acetone. On different days, the cotyledons and the hypocotyl were separated using a stereomicroscope as described by Park et al. (2012) and RNA was isolated as described above. Total RNA (500 ng) was used to prepare RNA-seq libraries, using the TruSeq Library prep kit-v2 (Illumina). Multiplexed libraries were sequenced on an Illumina Hi-Seq 2500. Raw reads were aligned to the TAIR10 genome using TopHat (v2.0.8b) with default parameters except that intron length was set to 40 minimum, 2000 maximum. Cuffdiff (v2.1.1) was used to calculate the expression change between the different time points in the light with the dark sample. Genes with false discovery rate < 0.05 in at least one time point were included in the heat map in Supplemental Figure 5C and Supplemental Data Set 4. We replace fragments per kilobase of transcript per million mapped reads values of 0 with 0.01.
ChIP-Seq Experiments and Analysis
Three biological replicates of HY5ox #5 and HY5-VP16 #5 and two for HY5-SRDX #5 of seedlings grown in the dark for 3 d and moved to simulated white light for 2.5 h were used for ChIP-seq analysis. The ChIP-seq experiment was performed as previously described by Kaufmann et al. (2010). Monoclonal αFLAG M2 antibody (Sigma-Aldrich #F1804) was used for immunoprecipitation. After elution, reversing cross-links, and DNA purification, Illumina TruSeq libraries were constructed according to the manufacturer’s protocols. A HY5ox_2 biological replicate was collected and processed for ChIP-seq separately. Multiplexed libraries were sequenced on an Illumina Hi-Seq 2500. Bowtie2 (v2.3.0) was used, with default parameters, to map sequencing reads to the TAIR10 genome (Langmead and Salzberg, 2012). HOMER (v4.9.1) Findpeaks script (Heinz et al., 2010) was used to call peaks using “-style factor -F 3 -P 0.0001 -L 2 -LP 0.001.” Immunoprecipitate from wild-type (Col-0) extract using anti-FLAG antibody was used as a background control. For HY5ox_2, immunoprecipitate from hy5 (collected and ChIP-seq together) was used as a background control, and for HY5ox_3 and HY5-VP16_3 Col-0_3, was used as a background control (collected and ChIP-seq together). The bedtools (v2.26.0) intersectBed function (Quinlan and Hall, 2010) was used to associate peaks with TAIR10 annotated genes within 2000 bp upstream of the transcription start site or with gene bodies. HOMER mergePeaks script was used to identify genes with binding in all six samples (Figure 3C, right). The full list of genes and peaks can be found in Supplemental Data Set 6. For motifs shown in Figure 3A, the peaks of the biological replicates were merged using HOMER mergePeaks script and then findMotifsGenome script used with default parameter except “-size 50.” Peaks were visualized using IGV browser (Thorvaldsdóttir et al., 2013).
Chlorophyll and Anthocyanin Measurements
For chlorophyll measurements, ∼50 mg of 13-d-old seedlings, grown in long-day conditions at 21°C, was collected. Fresh weights were measured after quick blotting to dry, and the tissue was immediately frozen in liquid nitrogen. After homogenization, the chlorophyll was extracted twice in 1 mL of ice-cold 80% acetone, and debris was removed by centrifugation at 10,000g for 20 min at 4°C. Chlorophyll was measured with a spectrophotometer, and levels were calculated according to Woodson et al. (2015). For anthocyanin measurements, 7-d-old plants, grown in constant light (100 µmol m−2 s−1) at 21°C, were transferred to high light (1080 µmol m−2 s−1) for 24 h. Anthocyanin was extracted and measured as described by Nakata and Ohme-Takagi (2014).
Dual-Luciferase Assay in S2 Cells
Schneider’s Drosophila S2 cells were maintained at high density in Schneider’s Drosophila medium (GIBCO) supplemented with 10% fetal bovine serum (GIBCO) and 10 mg/mL penicillin-streptomycin (Sigma-Aldrich). S2 cells at a density of 1 × 106 were plated on 24-well tissue culture plates. After 24 h, the cells were transfected using Effectene (Qiagen), according to the manufacturer’s instructions. In all transformations, 30 ng of pAC5.1:RENILLA was used as an internal transfection control, and empty pAC5.1/V5-HIS vector (Invitrogen) was added to a total of 220 ng of plasmid DNA in combination with 5 μL of Effectene. The Dual-Luciferase Reporter Assay System (Promega) was used according to the manufacturer’s instructions. Luciferase activity was measured with a TECAN Safire2 plate reader and normalized to the Renilla luciferase activity.
EcoRI and XhoI were used to introduce the HY5 and HY5-VP16 (using PCR on pDONR-P2RP3 plasmids containing HY5 or HY5-VP16) to PAC5.1/V5-HIS. XhoI and HindIII were used to introduce the HSP70 basal promoter, PCR-amplified from the CEX3:LUC plasmid (So et al., 2000; a gift from Ravi Allada), to create the pGL2-HSP70:LUC plasmid. The 4xG-box fragment was introduced by annealing two oligonucleotides and PCR-amplifying to introduce the pGL2-HSP70:LUC homology sequence. The CHS, COP1, SPA1, and SPA3 promoters were PCR-amplified from Col-0 DNA, and the SPA4 promoter (1450 bp upstream of the ATG) was synthesized (Genewiz), to introduce the pGL2-HSP70:LUC homology sequence. XhoI was used to open the pGL2-HSP70:LUC plasmid, and Gibson assembly was used to insert the different promoters.
Dual-Luciferase Assay in Nicotiana benthamiana
Leaves of N. benthamiana plants were coinfiltrated with A. tumefaciens cultures containing the effector and reporter constructs as described (Bürger et al., 2017). At 48 h after infiltration, 0.5-cm leaf discs were collected and the Dual-Luciferase Reporter Assay System (Promega) was used according to the manufacturer’s instructions using 500 μL of lysis buffer. Luciferase activity was measured with a TECAN Safire2 plate reader and normalized to Renilla luciferase activity. The 4xG-box promoter did not show any expression in N.benthamiana with any of our HY5 plasmids.
CHSp was PCR amplified and recombined into pDONR-P3P2. HY5, HY5-VP16, and GUS were PCR-amplified and recombined into pDONR-P1P4. The NOS terminator in pDONR-P4RP3R was used to get the destination vector 2X35S:HY5 or HY5-VP16 or GUS:-NosTer-CHSp:mini35:Firefly-35S:Renilla into pMDC-LRA plasmid (Li et al., 2014).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: HY5, AT5G11260; COP1, AT2G32950; SPA1, AT2G46340; SPA2, AT4G11110; SPA3, AT3G15354; SPA4, AT1G53090; SMZ, AT3G54990; SNZ, AT2G39250; SAUR19, AT5G18010; IAA19, AT3G15540; and EXP8, AT2G40610. RNA-seq and ChIP-seq data have been deposited into the Gene Expression Omnibus with the following accession numbers: HY5_RNA-seq GSE132859, HY5_ChIP-seq GSE132860, and De-etiolation_RNA-seq GSE132861. Genome browser tracks of the HY5 data set can be viewed at http://cactus2.salk.edu/aj2/pages/yburko/HY5_ChIP-seq_RNA-deq.php.
Supplemental Data
Supplemental Figure 1. Activity of chimeric HY5 proteins in different light conditions.
Supplemental Figure 2. Chimeric HY5 activators and repressors phenotypically recapitulates overexpression and loss-of-function mutants, respectively.
Supplemental Figure 3. Growth rates of HY5 transgenic plants during de-etiolation.
Supplemental Figure 4. HY5 positively regulates SMZ and SNZ.
Supplemental Figure 5. HY5 binding to the SPA promoters and the expression of HY5 regulated genes in hypocotyl and cotyledons during de-etiolation.
Supplemental Figure 6. Protein and RNA expression of HY5, HY5-VP16 and HY5-SRDX.
Supplemental Figure 7. HY5 activity in etiolated seedlings.
Supplemental Figure 8. HY5 activity in Drosophila S2 cells, N.benthamiana leaves and Arabidopsis.
Supplemental Figure 9. Venn diagram of the overlap between our HY5 direct target genes, PIF3, and PIF1,3,4,5.
Supplemental Figure 10. Light conditions.
Supplemental Data Set 1. List of VP16 and SRDX differentially expressed genes.
Supplemental Data Set 2. List of 297 HY5 regulated genes.
Supplemental Data Set 3. Complete HY5 RNA-seq data.
Supplemental Data Set 4. List of tissue-specific expression of HY5 target genes.
Supplemental Data Set 5. Primers used in this work.
Supplemental Data Set 6. Complete Chip-seq data.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Ute Hoecker (University of Cologne) for the α-COP1 and the spa1-7 seeds, Xing Wang Deng (Peking University) for α-SPA1, Ravi Allada (Northwestern University) for the CEX3:LUC plasmid, Alan Saghatelian (Salk institute) for the S2 cells, Huaming Chen (Salk institute) for help in setting the Anno-J browser, and Björn Willige, Carl Procko, Marco Burger (Salk institute), Naomi Ori (The Hebrew University of Jerusalem), and Xuelin Wu (Salk institute) for critical reading of the article. J.C. and J.R.E. are investigators of the Howard Hughes Medical Institute. This study was supported by the HHS|NIH|National Institute of General Medical Sciences (grant 5R35GM122604-02_05 to J.C.), the National Science Foundation (grant MCB-1024999 to J.R.E.), the Howard Hughes Medical Institute (to J.C. and J.R.E.), the European Molecular Biology Organization (grant ALTF 785-2013 to Y.B.), the United States-Israel Binational Agricultural Research and Development Fund (grant FI-488-13 to Y.B), by the Salk Pioneer Postdoctoral Endowment Fund (to M.Z and A.S.), and the Deutsche Forschungsgemeinschaft (research fellowship grant Za-730/1-1 to M.Z.). The Next Generation Sequencing Core Facility of the Salk Institute is supported by funding from the HHS|NIH|National Cancer Institute (grant CCSG: P30 014195), the Mary K. Chapman Foundation, and the Leona M. and Harry B. Helmsley Charitable Trust.
AUTHOR CONTRIBUTIONS
Y.B., A.S., and J.C. designed the research and wrote the article; J.C. and J.R.E. supervised; Y.B. performed most of the experiments and analyses with the following exceptions: Y.B. and M.Z. performed the Chip-seq and Y.B. and U.V.P. performed the hypocotyl- and cotyledon-specific RNA-seq.
Footnotes
Articles can be viewed without a subscription.
References
- Abbas N., Maurya J.P., Senapati D., Gangappa S.N., Chattopadhyay S.(2014). Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell 26: 1036–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D., Gallego-Bartolomé J., Orlando L., García-Cárcel L., Rubio V., Martínez C., Frigerio M., Iglesias-Pedraz J.M., Espinosa A., Deng X.W., Blázquez M.A.(2008). Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53: 324–335. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W.(2015). HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronis C., Barak S., Knowles S.M., Sugano S., Tobin E.M.(2008). The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Mol. Plant 1: 58–67. [DOI] [PubMed] [Google Scholar]
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W.(1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222. [DOI] [PubMed] [Google Scholar]
- Ang L.H., Deng X.W.(1994). Regulatory hierarchy of photomorphogenic loci: Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6: 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski A.A., Galstyan A., Guseman J.M., Nemhauser J.L.(2012). Photomorphogenesis. The Arabidopsis Book 10: e0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcerowicz M., Fittinghoff K., Wirthmueller L., Maier A., Fackendahl P., Fiene G., Koncz C., Hoecker U.(2011). Light exposure of Arabidopsis seedlings causes rapid de-stabilization as well as selective post-translational inactivation of the repressor of photomorphogenesis SPA2. Plant J. 65: 712–723. [DOI] [PubMed] [Google Scholar]
- Binkert M., Kozma-Bognár L., Terecskei K., De Veylder L., Nagy F., Ulm R.(2014). UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26: 4200–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigard J.P., Harkess R.L., Baldwin B.S.(2006). Tomato early seedling height control using a paclobutrazol seed soak. HortScience 41: 768–772. [Google Scholar]
- Bürger M., Willige B.C., Chory J.(2017). A hydrophobic anchor mechanism defines a deacetylase family that suppresses host response against YopJ effectors. Nat. Commun. 8: 2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C.(2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117. [DOI] [PubMed] [Google Scholar]
- Chen X., Yao Q., Gao X., Jiang C., Harberd N.P., Fu X.(2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26: 640–646. [DOI] [PubMed] [Google Scholar]
- Chory J.(1992). A genetic model for light-regulated seedling Arabidopsis. Development 115: 337–354. [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Datta S., Hettiarachchi C., Johansson H., Holm M.(2007). SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19: 3242–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Johansson H., Hettiarachchi C., Irigoyen M.L., Desai M., Rubio V., Holm M.(2008). LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C., et al. (2014). The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep. 9: 1983–1989. [DOI] [PubMed] [Google Scholar]
- Deng X.W., Matsui M., Wei N., Wagner D., Chu A.M., Feldmann K.A., Quail P.H.(1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71: 791–801. [DOI] [PubMed] [Google Scholar]
- Eeckhoute J., Métivier R., Salbert G.(2009). Defining specificity of transcription factor regulatory activities. J. Cell Sci. 122: 4027–4034. [DOI] [PubMed] [Google Scholar]
- Fittinghoff K., Laubinger S., Nixdorf M., Fackendahl P., Baumgardt R.L., Batschauer A., Hoecker U.(2006). Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J. 47: 577–590. [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Lee S.H., Patel D., Kumar S.V., Spartz A.K., Gu C., Ye S., Yu P., Breen G., Cohen J.D., Wigge P.A., Gray W.M.(2011). Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 108: 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S., Sakamoto S., Kigoshi K., Suzuki K., Ohme-Takagi M.(2014). VP16 fusion induces the multiple-knockout phenotype of redundant transcriptional repressors partly by Med25-independent mechanisms in Arabidopsis. FEBS Lett. 588: 3665–3672. [DOI] [PubMed] [Google Scholar]
- Gangappa S.N., Botto J.F.(2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant 9: 1353–1365. [DOI] [PubMed] [Google Scholar]
- Gangappa S.N., Kumar S.V.(2017). DET1 and HY5 control PIF4-mediated thermosensory elongation growth through distinct mechanisms. Cell Rep. 18: 344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu A., Dobos O., Domijan M., Bálint B., Nagy I., Nagy F., Kozma-Bognár L.(2018). ELONGATED HYPOCOTYL 5 mediates blue light signalling to the Arabidopsis circadian clock. Plant J. 96: 1242–1254. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.(2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Ohta M., Matsui K., Ohme-Takagi M.(2002). The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514: 351–354. [DOI] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J., Deng X.W.(2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C.(2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Huang X., Ouyang X., Yang P., Lau O.S., Li G., Li J., Chen H., Deng X.W.(2012). Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 24: 4590–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli A., Capua Y., Shwartz I., Tal L., Meir Z., Levy M., Bar M., Efroni I., Ori N. (2019). Multiple auxin-response regulators enable stability and variability in leaf development. Curr. Biol. 29: 1746–1759.e1745. [DOI] [PubMed]
- Jang I.C., Henriques R., Chua N.H.(2013). Three transcription factors, HFR1, LAF1 and HY5, regulate largely independent signaling pathways downstream of phytochrome A. Plant Cell Physiol. 54: 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y., Zhang D., Wang X., Tang W., Wang W., Huai J., Xu G., Chen D., Li Y., Lin R.(2013). Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25: 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C.(2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.(2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P., Norén L., López J.B., Shaikhali J., Strand A.(2012). Interplay between Heat Shock Protein 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Mol. Plant 5: 901–913. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Rolff E., Spruit C.J.P.(1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z. Pflanzenphysiol. 100: 147–160. [Google Scholar]
- Kurihara Y., Makita Y., Kawashima M., Hamasaki H., Yamamoto Y.Y., Matsui M.(2014). Next-generation sequencing of genomic DNA fragments bound to a transcription factor in vitro reveals its regulatory potential. Genes (Basel) 5: 1115–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha R., Singh A., Chattopadhyay S.(2008). Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 20: 1747–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L.(2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K., Podolec R., Chappuis R., Ulm R., Hothorn M.(2019). Plant photoreceptors and their signaling components compete for COP1 binding via VP peptide motifs. EMBO J. 38: e102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W.(2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17: 584–593. [DOI] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W.(2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H.(2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Gao S., Martinez C., He G., Zhou Z., Huang X., Lee J.H., Zhang H., Shen Y., Wang H., Deng X.W.(2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26: 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhang Q., Pedmale U.V., Nito K., Fu W., Lin L., Hazen S.P., Chory J.(2014). PIL1 participates in a negative feedback loop that regulates its own gene expression in response to shade. Mol. Plant 7: 1582–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.F., He J.X.(2016). BZR1 interacts with HY5 to mediate brassinosteroid- and light-regulated cotyledon opening in Arabidopsis in darkness. Mol. Plant 9: 113–125. [DOI] [PubMed] [Google Scholar]
- Lian H.L., He S.B., Zhang Y.C., Zhu D.M., Zhang J.Y., Jia K.P., Sun S.X., Li L., Yang H.Q.(2011). Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25: 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.C., Chi C., Jin L.J., Zhu J., Yu J.Q., Zhou Y.H.(2018). The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Environ. 41: 1762–1775. [DOI] [PubMed] [Google Scholar]
- Liu Y., Roof S., Ye Z., Barry C., van Tuinen A., Vrebalov J., Bowler C., Giovannoni J.(2004). Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc. Natl. Acad. Sci. USA 101: 9897–9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.D., Zhou C.M., Xu P.B., Luo Q., Lian H.L., Yang H.Q.(2015). Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8: 467–478. [DOI] [PubMed] [Google Scholar]
- Ma D., Li X., Guo Y., Chu J., Fang S., Yan C., Noel J.P., Liu H.(2016). Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 113: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macián F., García-Rodríguez C., Rao A.(2000). Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J. 19: 4783–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J., Yant L.J., Mürdter F., Küttner F., Schmid M.(2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S.(1991). Transformation of tomato with Agrobacterium tumefaciens In Plant Tissue Culture Manual: Supplement 7, Lindsey K., ed (Dordrecht, The Netherlands: Springer; ), pp. 311–319. [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W.(1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M., Ohme-Takagi M.(2014). Quantification of anthocyanin content. Bio Protoc. 4: e1098. [Google Scholar]
- Nawkar G.M., Kang C.H., Maibam P., Park J.H., Jung Y.J., Chae H.B., Chi Y.H., Jung I.J., Kim W.Y., Yun D.-J., Lee S.Y.(2017). HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: 2084–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norén L., Kindgren P., Stachula P., Rühl M., Eriksson M.E., Hurry V., Strand Å.(2016). Circadian and plastid signaling pathways are integrated to ensure correct expression of the CBF and COR genes during photoperiodic growth. Plant Physiol. 171: 1392–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K., Tat A.V., Kumar Devisetty U., Robinson M., Mumbach M.R., Ichihashi Y., Lekkala S., Maloof J.N.(2015). Shade avoidance components and pathways in adult plants revealed by phenotypic profiling. PLoS Genet. 11: e1004953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M.(2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K.(1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I., Chen F., Ngoc Pham V., Zhu L., Kim J.I., Huq E.(2019). A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat. Commun. 10: 4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Jiang K., Schatz M.C., Lippman Z.B.(2012). Rate of meristem maturation determines inflorescence architecture in tomato. Proc. Natl. Acad. Sci. USA 109: 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnu J., Riedel T., Penner E., Schrader A., Hoecker U.(2019). Cryptochrome 2 competes with COP1 substrates to repress COP1 ubiquitin ligase activity during Arabidopsis photomorphogenesis. Proc. Natl. Acad. Sci. USA 116: 27133–27141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M.(2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram H., Priya P., Jain M., Chattopadhyay S.(2014). Genome-wide DNA binding of GBF1 is modulated by its heterodimerizing protein partners, HY5 and HYH. Mol. Plant 7: 448–451. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K.(2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Leal D., Lemmon Z.H., Man J., Bartlett M.E., Lippman Z.B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171: 470–480.e478. [DOI] [PubMed]
- Ruckle M.E., DeMarco S.M., Larkin R.M.(2007). Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacht T., Oswald M., Eils R., Eichmüller S.B., König R.(2014). Estimating the activity of transcription factors by the effect on their target genes. Bioinformatics 30: i401–i407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluzicki A., Burko Y., Chory J.(2017). Dancing in the dark: Darkness as a signal in plants. Plant Cell Environ. 40: 2487–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E., Burko Y., Ben-Yaakov L., Berger Y., Amsellem Z., Goldshmidt A., Sharon E., Ori N.(2009). Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21: 3078–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Lyu M., Luo Y., Liu S., Li Y., He H., Wei N., Deng X.W., Zhong S.(2018). Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proc. Natl. Acad. Sci. USA 115: 6482–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shleizer-Burko S., Burko Y., Ben-Herzel O., Ori N.(2011). Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development 138: 695–704. [DOI] [PubMed] [Google Scholar]
- Singh A., Ram H., Abbas N., Chattopadhyay S.(2012). Molecular interactions of GBF1 with HY5 and HYH proteins during light-mediated seedling development in Arabidopsis thaliana. J. Biol. Chem. 287: 25995–26009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So W.V., Sarov-Blat L., Kotarski C.K., McDonald M.J., Allada R., Rosbash M.(2000). takeout, a novel Drosophila gene under circadian clock transcriptional regulation. Mol. Cell. Biol. 20: 6935–6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Favory J.J., Gruber H., Bartelniewoehner L., Bartels S., Binkert M., Funk M., Weisshaar B., Ulm R.(2010). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33: 88–103. [DOI] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Zhai Q., Li C.(2013). PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 25: 2102–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P.(2013). Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Johansson H., Lee K.P., Bou-Torrent J., Stewart K., Steel G., Rodríguez-Concepción M., Halliday K.J.(2014). The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triezenberg S.J., Kingsbury R.C., McKnight S.L.(1988). Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 2: 718–729. [DOI] [PubMed] [Google Scholar]
- Woodson J.D., Joens M.S., Sinson A.B., Gilkerson J., Salomé P.A., Weigel D., Fitzpatrick J.A., Chory J.(2015). Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350: 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D.(2019). COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol.. Available at: 10.1111/nph.16296. [DOI] [PubMed] [Google Scholar]
- Xu X., Chi W., Sun X., Feng P., Guo H., Li J., Lin R., Lu C., Wang H., Leister D., Zhang L.(2016). Convergence of light and chloroplast signals for de-etiolation through ABI4-HY5 and COP1. Nat. Plants 2: 16066. [DOI] [PubMed] [Google Scholar]
- Yang Y., Liang T., Zhang L., Shao K., Gu X., Shang R., Shi N., Li X., Zhang P., Liu H.(2018). UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat. Plants 4: 98–107. [DOI] [PubMed] [Google Scholar]
- Zhang H., He H., Wang X., Wang X., Yang X., Li L., Deng X.W.(2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65: 346–358. [DOI] [PubMed] [Google Scholar]
- Zhang X., Huai J., Shang F., Xu G., Tang W., Jing Y., Lin R.(2017). A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol. 174: 2487–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Peng T., Chen C.Y., Ji R., Gu D., Li T., Zhang D., Tu Y.T., Wu K., Liu X.(2019). HY5 interacts with the histone deacetylase HDA15 to repress hypocotyl cell elongation in photomorphogenesis. Plant Physiol. 180: 1450–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., et al. (2013). Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell 25: 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Maier A., Lee J.H., Laubinger S., Saijo Y., Wang H., Qu L.J., Hoecker U., Deng X.W.(2008). Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Bu Q., Xu X., Paik I., Huang X., Hoecker U., Deng X.W., Huq E.(2015). CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat. Commun. 6: 7245. [DOI] [PubMed] [Google Scholar]