Reactive oxygen species produced in the chloroplast phosphorylate a key translation initiation factor, thereby sensitizing cytosolic protein synthesis to the metabolic status of the chloroplast.
Abstract
Cytosolic mRNA translation is subject to global and mRNA-specific controls. Phosphorylation of the translation initiation factor eIF2α anchors a reversible regulatory switch that represses cytosolic translation globally. The stress-responsive GCN2 kinase is the only known kinase for eIF2α serine 56 in Arabidopsis (Arabidopsis thaliana). Here, we show that conditions that generate reactive oxygen species (ROS) in the chloroplast, including dark-light transitions, high light, and the herbicide methyl viologen, rapidly activated GCN2 kinase, whereas mitochondrial and endoplasmic reticulum stress did not. GCN2 activation was light dependent and mitigated by photosynthesis inhibitors and ROS quenchers. Accordingly, the seedling growth of multiple Arabidopsis gcn2 mutants was retarded under excess light conditions, implicating the GCN2-eIF2α pathway in responses to light and associated ROS. Once activated, GCN2 kinase preferentially suppressed the ribosome loading of mRNAs for functions such as mitochondrial ATP synthesis, the chloroplast thylakoids, vesicle trafficking, and translation. The gcn2 mutant overaccumulated transcripts functionally related to abiotic stress, including oxidative stress, as well as innate immune responses. Accordingly, gcn2 displayed defects in immune priming by the fungal elicitor, chitin. Therefore, we provide evidence that reactive oxygen species produced by the photosynthetic apparatus help activate the highly conserved GCN2 kinase, leading to eIF2α phosphorylation and thus affecting the status of the cytosolic protein synthesis apparatus.
Introduction
Reactive oxygen species (ROS) are byproducts of plant cellular metabolism and also serve as versatile secondary messengers (Foyer and Noctor, 2016; Mignolet-Spruyt et al., 2016; Choudhury et al., 2017; Mullineaux et al., 2018). Plants tightly regulate the ROS balance between production and scavenging through various enzymatic and nonenzymatic mechanisms (Das and Roychoudhury, 2014; Foyer and Noctor, 2016; Mittler, 2017). As ROS act as signaling molecules, these mechanisms function in a variety of physiological and developmental programs, including root development, the pathogen-induced hypersensitive response, and stomatal closure (Camejo et al., 2016; Tsukagoshi, 2016; Ehonen et al., 2019). These normal programs are quite often perturbed by a variety of external biotic and abiotic cues, which make ROS cytotoxic agents. The resulting adverse effects include oxidative damage to nucleic acids, carbohydrates, and especially lipids and proteins (Jacques et al., 2013; Demidchik, 2015).
Being both beneficial and deleterious, ROS production in plants is carefully compartmentalized to the apoplast, endoplasmic reticulum (ER), chloroplasts, peroxisomes, and mitochondria. Under active photosynthesis conditions, chloroplasts are the major producers of ROS (Schmitt et al., 2014; Czarnocka and Karpiński, 2018). Arabidopsis (Arabidopsis thaliana) plants adapted to low-light conditions and exposed to sudden high-light intensities experience photo-oxidative stress resulting from excess excitation energy, where the amount of absorbed light energy exceeds the photosynthetic capacity (Mateo et al., 2004; Li et al., 2009; Muñoz and Munné-Bosch, 2018). Under excess light, singlet oxygen (1O2) and superoxide anion (O2−1) are overproduced at PSII and PSI, respectively, leading to higher levels of hydrogen peroxide (H2O2; Asada, 2006; Mubarakshina and Ivanov, 2010). When high light elevates organellar ROS production beyond the capacity for their detoxification, this triggers stress-adaptive reprogramming via retrograde signaling to the nucleus (Dietz et al., 2016; Foyer and Noctor, 2016; Mignolet-Spruyt et al., 2016; Crisp et al., 2017).
Most of the work to dissect the roles of ROS on gene expression has focused on transcriptional control (Gadjev et al., 2006; Vaahtera et al., 2014), which is inherently slower than regulation at the level of translation. Thus, rapidly changing light and ROS levels may also act on translation to elicit a nimbler response in gene expression. Light is known to stimulate cytosolic translation (Tang et al., 2003; Juntawong and Bailey-Serres, 2012; Liu et al., 2012; Missra et al., 2015; Merchante et al., 2017), while the effect of ROS on cytosolic translation has received less attention (Khandal et al., 2009; Benina et al., 2015; Moore et al., 2016).
In mammals and yeast, translational control in response to a diverse range of stresses converges on the phosphorylation of the translation initiation factor eIF2α (eIF2α-P), a regulatory process performed by a family of up to four different kinases. Upon phosphorylation, eIF2α-P binds to eIF2B and inhibits its guanine nucleotide exchange activity, leading to a depletion of the ternary complex (eIF2-GTP-tRNA(i)Met) and, ultimately, a decline in translation initiation and protein synthesis (Wek, 2018). In plants, the only known kinase for eIF2α is GCN2 (GENERAL CONTROL NONDEREPRESSIBLE2), which responds to diverse stress stimuli, including inhibitors of amino acid and purine biosynthesis, wounding, UV light, hormones, and bacterial infection by phosphorylating eIF2α (Akbudak et al., 2006; Lageix et al., 2008; Zhang et al., 2008; Sesma et al., 2017; Izquierdo et al., 2018; Liu et al., 2019; Llabata et al., 2019). GCN2 is activated through its binding of various uncharged tRNAs, which accumulate during amino acid starvation and other stress conditions (Dong et al., 2000; Anda et al., 2017; Li et al., 2013).
In this study, we show that GCN2 kinase is activated rapidly under excess light stress and H2O2-mediated oxidative stress in Arabidopsis. This increase in GCN2 kinase activity and eIF2α-P levels specifically occurs in response to a signal emanating from the chloroplast, as it could be controlled by the application of various plastidic redox modulators. Moreover, eIF2α phosphorylation in response to herbicide stress is strictly light dependent. Seedlings with mutations in GCN2 grew more slowly than the wild type under continuous- or high-light stress. These mutants were also defective in the global reorganization of the translatome that occurs after GCN2 is activated by herbicides. Comparative transcriptome analysis between wild-type and gcn2 plants under herbicide stress revealed significant changes in the levels of mRNAs involved in the response to pathogens as well as abiotic stresses, including ROS metabolism. In summary, this study reveals that ROS emanating from the photosynthetic machinery feed back to the cytosolic protein synthesis apparatus to balance overall energy and metabolic resources with cellular demands.
Results
GCN2 Kinase Activity Is Activated by Excess Light
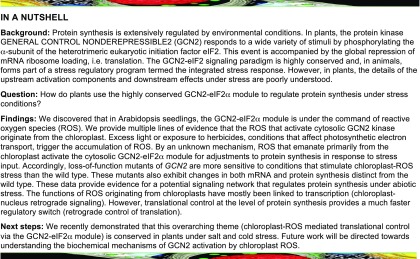
The activity of GCN2 kinase can be determined by monitoring the phosphorylation status of its primary substrate, the eukaryotic translation initiation factor eIF2α. GCN2 is activated by a number of environmental signals, including herbicides that inhibit amino acid biosynthesis, UV light, and salicylic acid (Lageix et al., 2008; Zhang et al., 2008). To determine if GCN2 is activated by light, we exposed 24-h dark-adapted seedlings to white light (80 µE m−2s−1). After dark adaptation, the eIF2α-P level (as determined by immunoblotting) declined to the level detected in unstimulated plants (Figures 1A and 1B). White light treatment induced intense eIF2α phosphorylation within 2 h in dark-adapted wild-type plants, but not in the gcn2 mutant (Figures 1A and 1B), indicating that GCN2 kinase is responsive to light. eIF2α-P production was fluence rate dependent and rapidly increased (after 30 min) in response to low to moderate fluence rates (Figure 1C). By contrast, heat shock at 37°C did not trigger an increase in GCN2 activity and instead suppressed the basal level of eIF2α-P in the light (Supplemental Figure 1). Under normal long-day light-dark cycle conditions, i.e., 80 µE m−2s−1 without prior dark adaptation, eIF2α-P levels rose during the light period and then declined early during the dark period (Figures 1D and 1E). These results indicate that GCN2 kinase is activated by white light and that responses to other signals should be interpreted in light of the diel dynamics of eIF2α-P. Once phosphorylated under high light (780 µE m−2s−1), eIF2α-P levels remained elevated for over 72 h (Supplemental Figure 2), indicating little adaptation to the high-light condition. By contrast, the light-triggered eIF2α-P dissipated within 6 h after shifting plants to darkness (Supplemental Figure 3). Because GCN2 responded most strongly to a sudden transition of dark-adapted plants to light of moderate or high fluence rate, we conclude that GCN2 is activated by excess light.
Figure 1.
Excess Light Triggers GCN2-Dependent eIF2α Phosphorylation in a Dose-Dependent Manner.
(A) Schematic of the light regimen. Seedlings were grown under a 16 h light 8 h dark cycle, followed by a 24 h dark acclimation starting at ZT2. The red arrow at T = 0 indicates the beginning of excess light treatment and the start of sampling.
(B) Immunoblot showing the time course of eIF2α phosphorylation in 14-d-old wild-type Ler-0 (Wt(Ler)) and gcn2-1 (gcn2) seedlings subjected to excess light stress (white light) as described in (A). Top, Probed with phosphospecific antibody against eIF2α-P (38 kDa). A partially cropped band at the top represents nonspecific binding of the antibody. Middle, Rubisco large subunit (∼55 kDa) as a loading control after Ponceau S staining of the blot. Bottom, Probed with antibody against eIF2α (38 kDa). +, arbitrary amount of total protein extract from glyphosate-treated wild-type seedlings indicating unphosphorylated (eIF2α) or phosphorylated (eIF2α-P) protein; (10, 30, 120) sampling time in min; M, molecular weight marker; MW, molecular weight.
(C) Fluence rate dependence of eIF2α phosphorylation in wild-type seedlings subjected to 2, 25, 80, and 200 µE m−2s−1 (µE) white light after dark acclimation as described in (A) and (B).
(D) Diel time course of eIF2α phosphorylation over 48 h in 14-d-old wild-type seedlings grown under a 16-h light (80 µE m−2s−1) and 8-h dark cycle.
(E) Quantification of the diel time course of the eIF2α phosphorylation signal from immunoblots as shown in (D). Dark periods are shaded in gray. Error bars represent SE of the mean of three independent biological replicates.
GCN2 Is Activated by ROS
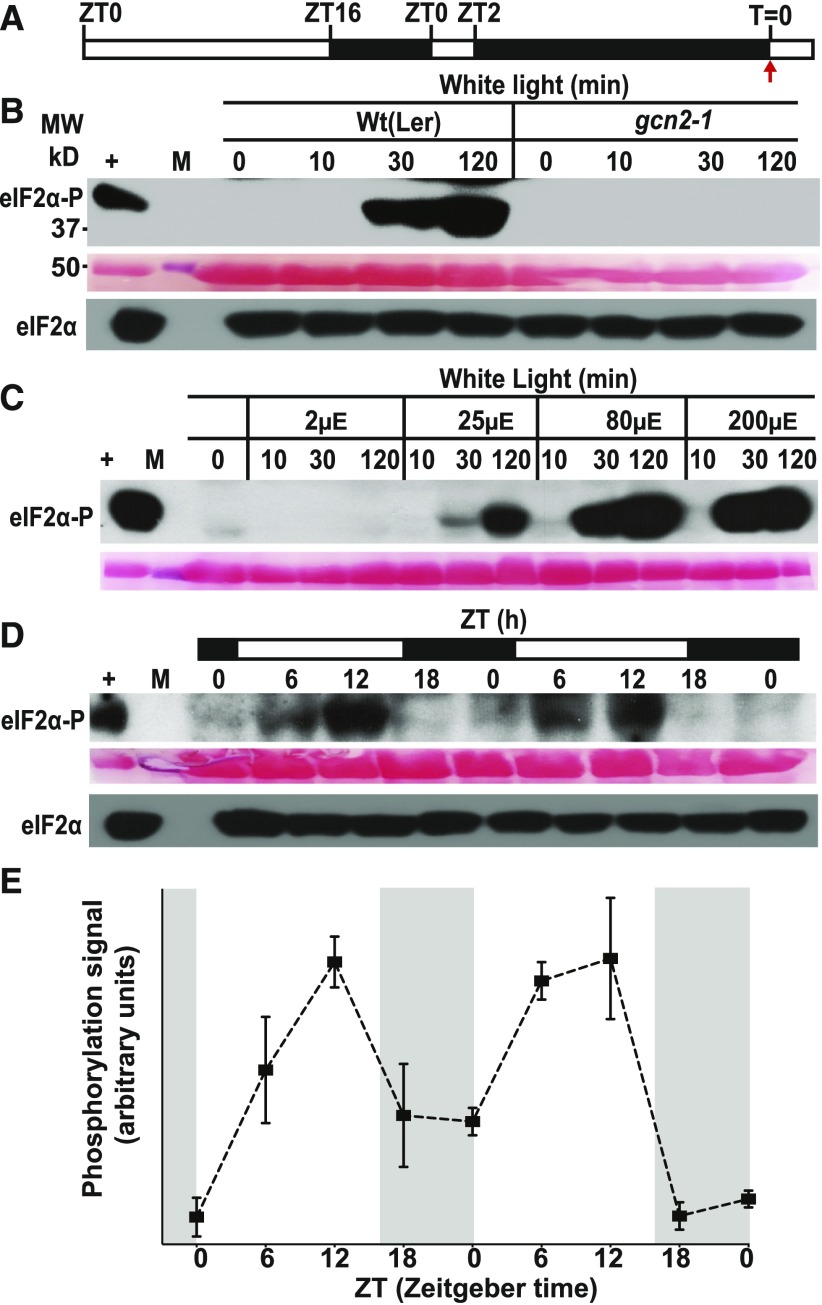
Because excess light leads to the rapid accumulation of ROS such as hydrogen peroxide, we tested whether GCN2 could be activated solely by H2O2. Indeed, ectopic H2O2 treatments in the dark resulted in a dose-dependent activation of GCN2 as early as 10 min, and this induction was reduced, although still detectable, at doses as low as 1 mM (Figures 2A to 2C). We confirmed that exposing dark-adapted seedlings to light triggered ROS accumulation in seedling leaves, as shown in situ with the ROS-sensitive dye 2′,7′-dichlorofluorescein diacetate (H2DCFDA; Figure 2D).
Figure 2.
Effect of Hydrogen Peroxide on GCN2 Activity and Microscopic Examination of ROS in Response to Photosynthetic Inhibitors.
(A) Schematic of the light regimen. Seedlings were grown under a 16-h light and 8-h dark cycle, dark-acclimated for 24 h, and sprayed with H2O2 in the dark starting at T = 0 (red arrow).
(B) Time course of eIF2α phosphorylation in 14-d-old wild-type Ler-0 (Wt(Ler)) and gcn2-1 seedlings (gcn2-1) treated with 10 mM H2O2 for 0, 10, 30, and 120 min as described in (A). For details, see legend to Figure 1.
(C) Treatment of wild-type seedlings with 1, 10, and 100 mM H2O2 after dark acclimation as described in (A).
(D) Representative images of 14-d-old wild-type leaves stained with the ROS-sensitive dye H2DCFDA after 24 h dark acclimation as in (A). T0, dark acclimated control; EL, exposed to 80 µE m−2s−1 white light for 30 min. DCMU+EL, treated with 8 µM DCMU 30 min prior to exposure to 80 µE m−2s−1 light. H2O2, treated with 10 mM H2O2 in the dark for 30 min. Oxidized H2DCFDA is represented in green and chlorophyll in magenta, and the superimposed view yields a white color. Scale bars are 25 µm.
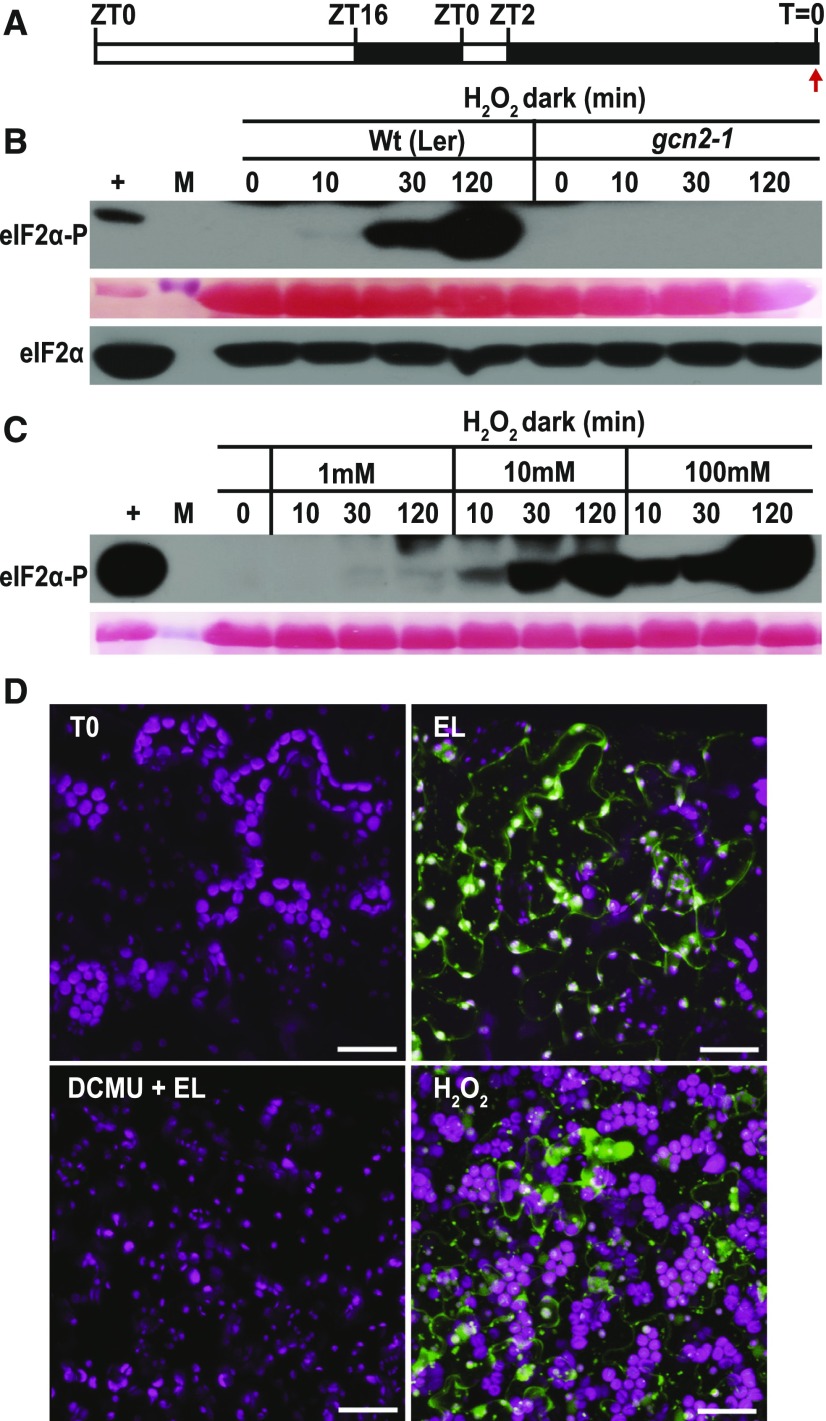
To gain deeper insights into the identity of the ROS (H2O2, 1O2, O2−1) that can activate GCN2, we analyzed eIF2α phosphorylation in response to specific ROS inducers. The conditional fluorescent (flu) mutant generates singlet oxygen (1O2) in plastids within 1 min upon a shift from prolonged darkness to light (Meskauskiene et al., 2001; op den Camp et al., 2003). Indeed, upon dark-to-light shift, flu1-1 plants showed more rapid (<5 min) eIF2α phosphorylation than wild type, and the basal level of phosphorylation was elevated as well (Figure 3A). 1O2 is rapidly converted to H2O2 in a nonenzymatic reaction utilizing either plastoquinone (PQ) or plastoquinol (PQH2), and accordingly, flu accumulates H2O2 as well as superoxide (Mubarakshina and Ivanov, 2010; Kim and Apel, 2013; Khorobrykh et al., 2015). Interestingly, the hypersensitive activation of GCN2 in flu1-1 was suppressed in seedlings pretreated with the herbicide 3-(3,4-dichlorophenyl)-1,1-dimethyl urea (DCMU), which blocks electron transport between PSII and PQ (Figure 3B). Thus, chloroplast singlet oxygen can serve as the trigger for GCN2 kinase, notwithstanding that singlet oxygen may act via hydrogen peroxide.
Figure 3.
flu Mutants Show Accelerated or Elevated GCN2 Activation by Photosynthetic ROS.
(A) Time course of eIF2α phosphorylation in rosette stage leaves of wild-type (Wt(Ler)) and flu1-1 plants. Plants were grown under continuous light (cLL), dark acclimated for 16 h (dark), and re-exposed to light at 80 µE m−2s−1 (dark to white light). Note that eIF2α-P levels are elevated within 5 min in the flu1-1 mutant. gcn2-1 is included as a negative control.
(B) eIF2α phosphorylation in flu1-1 and Wt(Ler) control seedlings after 12 d in continuous light, followed by 24 h dark acclimation and re-exposure to light (80 µE m−2s−1). Seedlings were sprayed with DMSO (mock) or 8 µM DCMU 30 min prior to light exposure. Time = 0 was sampled right before the light treatment. The difference in response times may be due to the different developmental stages, i.e., rosette leaves (A) versus seedlings (B). For details, see legend to Figure 1.
Herbicides Rely on Photosynthetic H2O2 to Activate GCN2
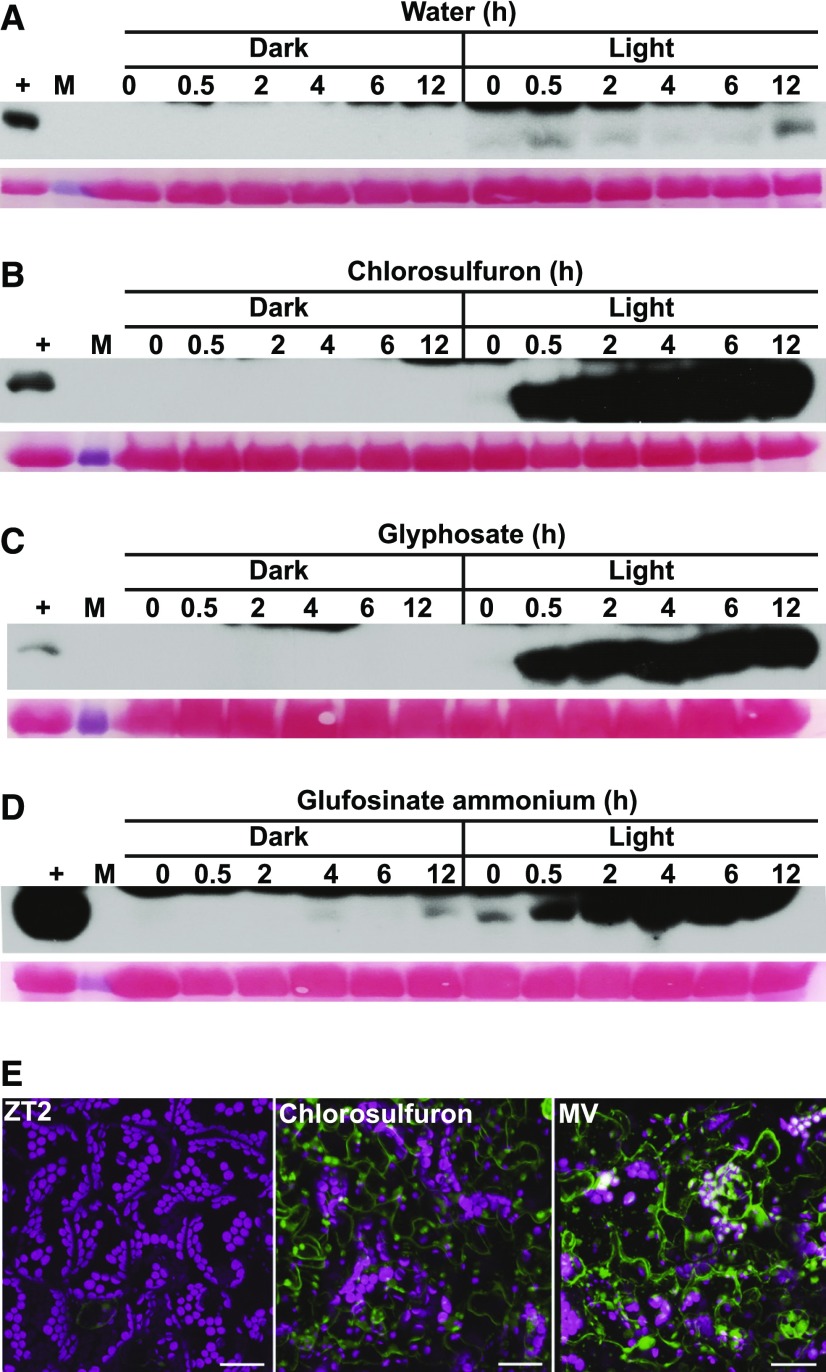
GCN2 is activated by uncharged tRNAs in organisms including plants (Wek et al., 1995; Li et al., 2013), and treatments with herbicides that inhibit amino acid biosynthesis activate GCN2 kinase (Lageix et al., 2008; Zhang et al., 2008). We tested three compounds that inhibit different branch points in amino acid biosynthesis for their effects on GCN2: chlorosulfuron for branched-chain amino acids, glyphosate for aromatic amino acids, and glufosinate for glutamine synthetase. Surprisingly, all three inhibitors required light to activate GCN2 kinase (Figures 4A to 4D). These data suggest that GCN2 cannot be activated solely by herbicides raising the levels of uncharged tRNAs, yet it requires another signal that is light dependent in our hands. We propose that ROS function as a light-dependent signal that initiates GCN2 activity. Consistent with this notion, treatment with the herbicide chlorosulfuron led to ROS accumulation in the leaf (Figure 4E), as do glyphosate and glufosinate treatment (Faus et al., 2015; Takano et al., 2019). Moreover, treatment with the antioxidant ascorbate delayed the activation of GCN2 kinase by excess light (Supplemental Figure 4).
Figure 4.
Herbicide-Induced GCN2 Activation Requires Light.
(A) to (D) Time course of eIF2α phosphorylation in 14-d-old wild-type Ler-0 seedlings treated with (A) water only, (B) 0.6 µM chlorosulfuron, (C) 150 µM glyphosate, (D) 15 µg/mL glufosinate ammonium under light and dark conditions. For treatments in the dark and details about the blots, see legend to Figure 1.
(E) Chlorosulfuron triggers ROS accumulation in the light, as visualized with H2DCFDA. 14-d-old wild-type leaves were sampled at ZT2 (left) and treated with 0.6 µM chlorosulfuron for 1 h starting at ZT2 (chlorosulfuron). MV, treated with 20 µM methyl viologen for 30 min. For details, see legend to Figure 2D. Scale bars are 25 µm.
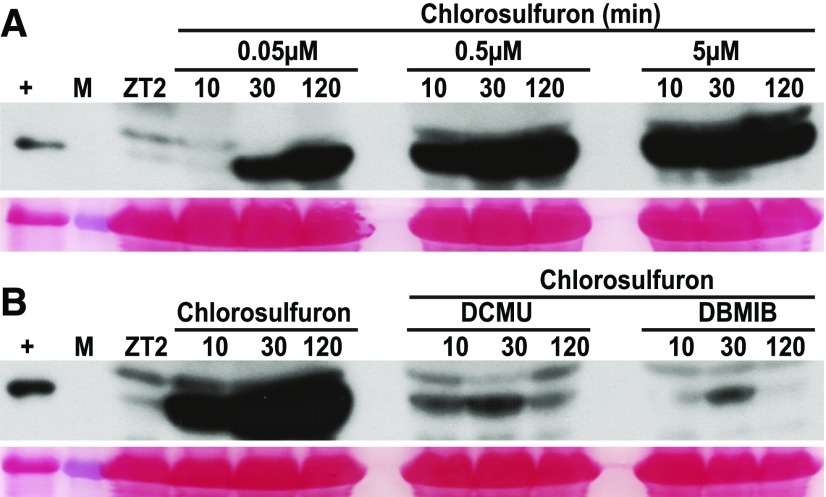
To further test the hypothesis that chlorosulfuron activates GCN2 by impinging on the balance of ROS metabolism in the chloroplast, we pretreated wild-type seedlings with photosynthetic inhibitors that block electron flux through the PQ/PQH2 pool prior to chlorosulfuron treatment. Specifically, application of the PQ oxidizer DCMU or the PQ reducer 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB) both suppressed the effect of a near-saturating dose of chlorosulfuron (Figures 5A and 5B). Taken together, these findings suggest that ROS, possibly in concert with uncharged tRNAs, activate GCN2 kinase.
Figure 5.
Photosynthetic Inhibitors Attenuate Chlorosulfuron-Triggered GCN2 Activation.
eIF2α phosphorylation in long-day-grown 14-d-old wild-type Ler-0 seedlings. Treatments with herbicides started at ZT2 in the light.
(A) Dose response for chlorosulfuron over a time course of up to 120 min.
(B) eIF2α phosphorylation triggered by chlorosulfuron (0.5 µM) was suppressed by pretreatment with 8 µM of DCMU or 16 µM of 2,5-Dibromo-6-isopropyl-3-methyl-1,4-benzoquinone (DBMIB). Seedlings were sprayed with DCMU and DBMIB 30 min prior to chlorosulfuron treatment. For details, see legend to Figure 1.
GCN2 Supports Seedling Growth under Excess Light Conditions
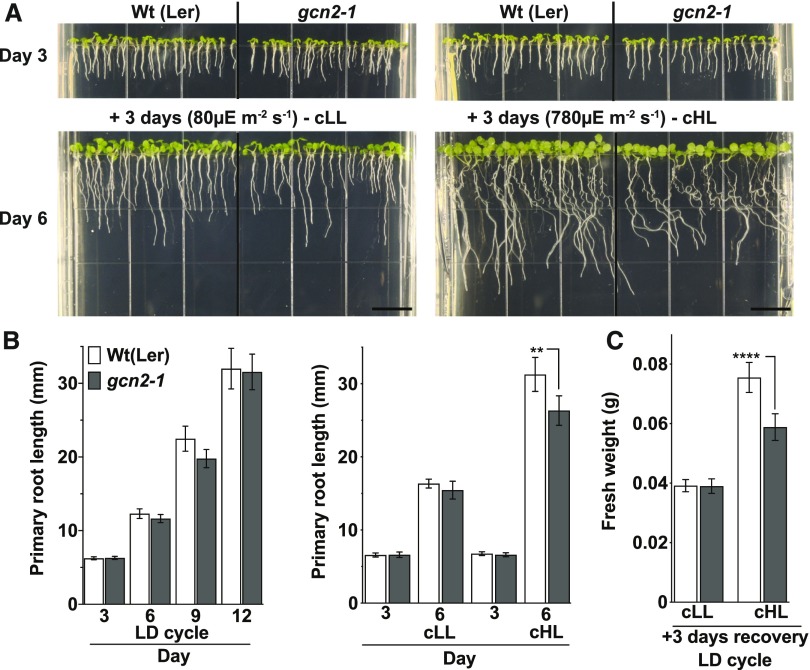
Under normal laboratory conditions, gcn2 mutants display few phenotypic abnormalities (Lageix et al., 2008; Zhang et al., 2008; Faus et al., 2015; Liu et al., 2019; Llabata et al., 2019). However, after exposure to 3 d of continuous high light, the growth of gcn2-1 seedlings was retarded compared with wild type, specifically in terms of root length and overall fresh weight (Figures 6A to 6C), whereas gcn2-1 seedlings grown in a regular day-night cycle were normal. A complementation line expressing genomic GCN2 under the control of its native promoter in the gcn2-1 background (Lageix et al., 2008) showed recovery from growth retardation under high light (Supplemental Figure 5). Two independent gcn2 alleles in the Columbia (Col-0) ecotype background (Supplemental Figures 6A to 6D) also had shorter roots after 3 d of continuous high light compared to the wild type and, following recovery in normal light, gcn2-2 showed lower fresh weight than the wild type (Supplemental Figures 7A to 7C and 8). However, other than gcn2-1, these Col-0 alleles also had shorter roots than the wild type under continuous normal light intensity (+3 days continuous light; Supplemental Figures 7 and 8), suggesting that the gcn2 phenotype is ecotype dependent. Taken together, given that three independent gcn2 loss-of-function alleles had similar whole-seedling phenotypes, we conclude that GCN2 kinase plays a physiological role in adaptation to excess light.
Figure 6.
Loss of GCN2 Renders Increased Sensitivity to High Light.
(A) Top, 3-d-old wild-type Ler-0 (Wt(Ler)) and gcn2-1 (gcn2-1) seedlings grown under a long-day photoperiod on plant medium supplemented with 0.1% sucrose. Bottom, The same seedlings after 3 d of additional continuous light at 80 µE m−2s−1 (cLL, left) or continuous high light at 780 µE m−2s−1 (cHL, right). Scale bars are 10 mm.
(B) Left, Primary root length of wild-type and gcn2-1 seedlings grown under a 16-h light and 8-h dark (long day, LD) cycle. Right, Root length of wild type and gcn2-1 as shown in (A).
(C) Fresh weights of wild type and gcn2-1 seedlings from (B) after an additional 3 d of recovery under a long-day (LD) photoperiod after the cLL or cHL treatment. Error bars represent SD of four biological replicates with n > 12 ([B], left) or n > 150 ([B], right; and [C]) per experiment (Welch’s t test, **P < 0.005; ****P < 0.0001).
What are the defects of gcn2 at the level of cellular physiology? The gcn2-1 mutants had no dramatic differences in PSII efficiency from the wild type during exposure to continuous high light, except for a slightly reduced quantum yield overall (Supplemental Figure 9). Moreover, in our hands, the gcn2-1 mutants accumulated as much ROS as wild type in the light, when GCN2 kinase is active (dark-to-light shift; Supplemental Figure 10). This result stands in contrast with previously published data, where gcn2-1 plants accumulated less ROS than wild type in response to treatment with the herbicide glyphosate (Faus et al., 2015). The gcn2-1 mutant also did not exhibit any difference in survival or overt phenotypes when light-grown seedlings were exposed to a concentration series of H2O2 (1 to 1000 mM) or to the herbicide methyl viologen (MV, paraquat), which leads to rapid superoxide-mediated hyperaccumulation of H2O2 in chloroplasts (Fujii et al., 1990). A drop in polysome loading was observed when wild-type plants were treated with H2O2; however, this drop was not abrogated in gcn2-1 (Supplemental Figure 11). Therefore, although GCN2 responds to ROS and helps the plant acclimate to ROS-producing conditions such as high light, it appears to regulate processes other than photosynthesis or H2O2 levels.
GCN2 Is Activated by ROS from the Chloroplast but Not Other Organelles
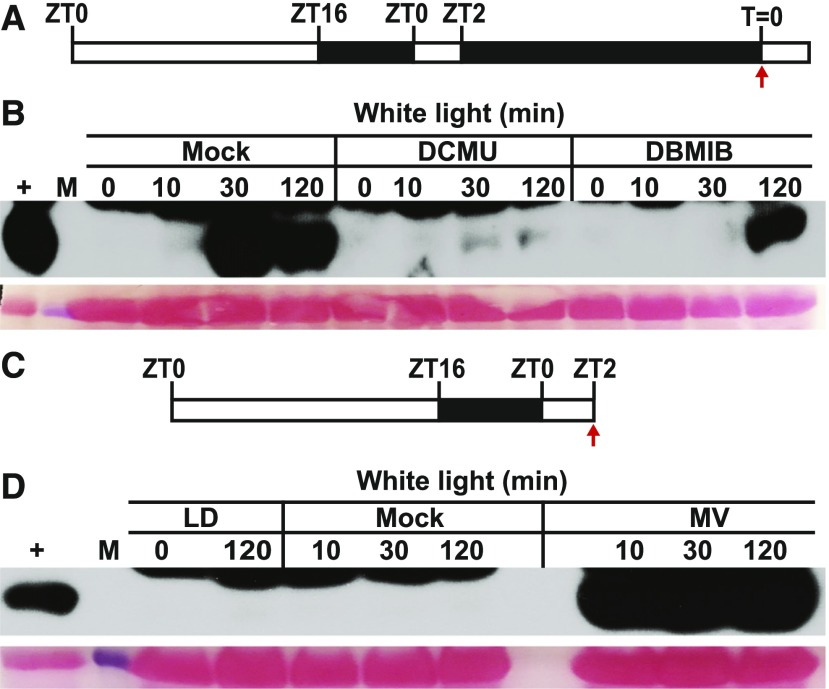
We hypothesized that the activation of GCN2 by light involves ROS produced by photosynthesis in the chloroplast. Strikingly, both DCMU and DBMIB suppressed GCN2 kinase activity in the light (Figures 7A and 7B). These results indicate that excess light activates GCN2 through photosynthetic electron transfer and that the activation signal likely originates from the PQH2 pool. The reason that DBMIB also inhibited GCN2, even though DBMIB should push PQ toward the ROS-producing PQH2 form, may lie in the overall reduction in photosynthetic electron flow and hence ROS production when DBMIB is present. In keeping with the hypothesis that photosynthetic ROS activates GCN2, norflurazon, an inhibitor of carotenoid biosynthesis, rapidly activated GCN2. The effect of norflurazon was kept in check by pretreatment with DCMU or DBMIB (Supplemental Figure 12). Similarly, MV triggered intense GCN2 activity within 10 min (Figures 7C and 7D). As expected, MV stimulated (Figure 4E) and DCMU suppressed ROS production by light in situ (Figure 2D). MV did not activate GCN2 under dark conditions (Supplemental Figure 13). However, MV significantly lowered the PSII quantum yield of gcn2-1 compared to wild-type plants (Supplemental Figure 14), suggesting a deficiency in recovery from ROS stress in the gcn2-1 mutant, which may potentially underlie the root growth defect evident under high light.
Figure 7.
Photosynthetic Inhibitors Modulate eIF2α Phosphorylation in the Light.
(A) Schematic of the light regimen consisting of a long-day photoperiod followed by 24 h dark acclimation and the start time of light treatment (80 µE m−2s−1) and sampling (T = 0).
(B) Time course of eIF2α phosphorylation in 14-d-old wild-type Ler-0 seedlings after lights-on. Thirty minutes prior to light exposure, seedlings were sprayed with either DMSO (mock), 8 µM DCMU, or 16 µM DBMIB.
(C) Schematic of the light regimen for (D).
(D) eIF2α phosphorylation starting at ZT2 (T = 0) either left untreated (LD) or treated with water up to 120 min (mock) or treated with 20 µM MV. For details, see legend to Figure 1.
Plants that overexpress a chloroplast-targeted glycolate oxidase (GO), a peroxisomal enzyme, provoke plastid-specific H2O2 responses in Arabidopsis (Fahnenstich et al., 2008). GO-overexpressing seedlings of three independent lines were dark adapted and then exposed to light. These seedlings appeared to show slightly accelerated and sustained GCN2 activation compared to the corresponding wild-type Col-0 (Supplemental Figures 15A and 15B), which could be suppressed with DCMU (Supplemental Figure 15B), which is consistent with elevated ROS accumulation in the GO-overexpressing seedlings (Fahnenstich et al., 2008). Taken together, these observations support the notion that light-induced production of H2O2 in chloroplasts serves as an activation signal for GCN2.
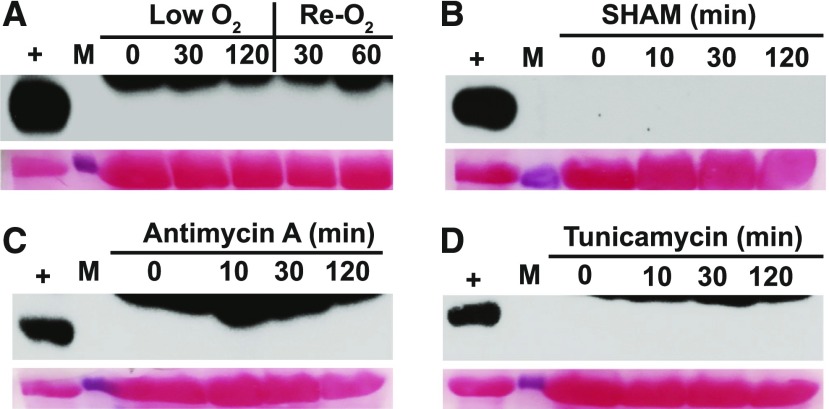
Next, we asked whether GCN2 could be activated by ROS generated from organelles other than the chloroplast. We treated hypoxic seedlings with oxygen, and we treated normal seedlings with the mitochondrial electron transport inhibitor antimycin-A (Maxwell et al., 1999) and the alternative oxidase inhibitor salicylhydroxamic acid (Stonebloom et al., 2012), both of which trigger mitochondrial ROS accumulation in the dark (Supplemental Figure 16). Neither of these treatments activated GCN2. Likewise, tunicamycin, an inhibitor of protein folding in the ER that can trigger ROS accumulation (Supplemental Figure 16; Ozgur et al., 2014), was also inactive toward GCN2 in the dark (Figures 8A to 8D). Thus, in our hands, ROS originating from the plastid were active in stimulating GCN2 kinase activity, while our attempts to stimulate ROS-induced activation of GCN2 from other cellular locales did not have the same effect.
Figure 8.
eIF2α Phosphorylation Is Specific to ROS from the Chloroplast.
14-d-old wild-type Ler-0 seedlings were dark-acclimated for 24 h and treated as follows to induce ROS stress from the mitochondria or ER.
(A) Hypoxia (low O2) with argon gas followed by re-oxygenation (Re-O2) in air.
(B) Sprayed with 200 µM salicylhydroxamic acid (SHAM), an inhibitor of alternative oxidase.
(C) Sprayed with 50 µM antimycin A, a mitochondrial complex III inhibitor.
(D) Sprayed with 5 µg/mL tunicamycin to induce ER stress. For details, see legend to Figure 1.
How Does GCN2 Regulate Ribosome Loading of mRNA?
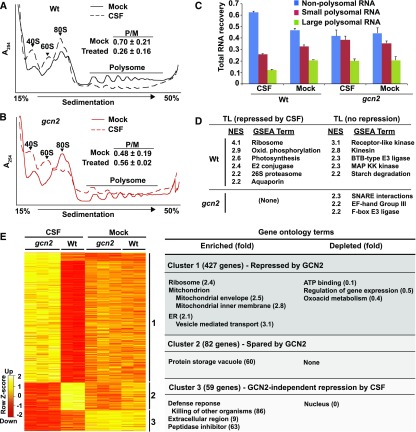
To investigate the consequence of GCN2 kinase activation at the translational level, we activated GCN2 in light-grown seedlings using chlorosulfuron (CSF). Cell extracts from wild-type and gcn2 seedling shoots were fractionated on Suc gradients to separate polysomal from nonpolysomal RNAs. As expected, GCN2 globally inhibited polysome loading in response to the brief (2 h) herbicide treatment (Figures 9A to 9C; Lageix et al., 2008; Zhang et al., 2008). Following microarray hybridizations of nonpolysomal, small polysomal, and large polysomal mRNAs, we calculated an ad hoc translation state for each mRNA (Missra et al., 2015) in response to each of the four treatments (i.e., ± GCN2 and ± CSF). We identified differentially translated mRNAs (Figure 9E; Supplemental Table 1). Wild-type and gcn2-1 plants had similar translation states under control conditions (Figure 9E), in keeping with the lack of kinase activity and the weak phenotypic differences. The mRNAs with changes in translation states in response to herbicide fell into three different clusters. Cluster 1 was the largest and was preferentially repressed by active GCN2 kinase. This cluster was enriched for mRNAs for proteins from three cellular locales, ER and vesicle trafficking, ribosomes, and mitochondrial membrane complexes, particularly proton-coupled ATP synthesis, as revealed by gene ontology analysis. Interestingly, ATP binding proteins, which include many kinases, were strongly depleted from this cluster, indicating that their mRNAs remained preferentially ribosome loaded. Cluster 2 encoded mRNAs that were spared from the translational repression by active GCN2 and was enriched for few functional terms. Cluster 3 included proteins that were translationally repressed by the herbicide in a GCN2-independent manner. This cluster was enriched for extracellular proteins and defense responses and was strikingly depleted for nuclear functions (Figure 9E). For a more sensitive functional analysis that surveys the entire translatome rather than just the few mRNAs passing a stringent false discovery rate, we turned to gene set enrichment analysis (GSEA; Figure 9D; Subramanian et al., 2005). GSEA of the herbicide response strongly confirmed some of the earlier trends (repressed, ribosome and mitochondrial oxidative phosphorylation; spared from repression, kinases) but also revealed new trends, especially in the protein turnover category, as well as translational repression of photosynthesis (thylakoid) functions. Thus, in general, core metabolic machinery was especially strongly repressed by GCN2, while regulators such as kinases and E3 ligases were relatively protected from translational repression.
Figure 9.
GCN2-Induced Regulation of the mRNA Translation State.
Cell extracts from Arabidopsis seedling shoots treated with 0.5 µM chlorosulfuron (2 h) were fractionated over Suc gradients to separate polysomal from nonpolysomal RNAs.
(A) and (B) UV light absorbance profiles of polysome gradients. Upon activation by CSF, GCN2 kinase inhibits polysome loading in the wild type but not in gcn2. The ratio of polysomes (P) to monosomes (M) is indicated with SE from three replicates.
(C) Gradient fractions were pooled into nonpolysomal (NP), small (SP), and large polysomal (LP) RNA pools, respectively. The histogram shows the average RNA recovered with SE.
(D) Gene set enrichment analysis of mRNA translation state. The differential translations states of 13,551 mRNAs were rank-ordered for each of two pairwise comparisons (wild type ± CSF, gcn2 ± CSF). Next, 257 gene sets harboring between 15 and 500 members were examined for a biased distribution along the rank-ordered mRNAs. Gene sets with a bias passing a family-wise error rate < 0.05 are listed with their normalized enrichment score (NES) where 0 equals no enrichment.
(E) The NP, SP, and LP RNA samples were processed for microarray gene expression profiling. Limma with FDR correction (P < 0.05) identified 568 differentially translated genes. Translation state values are displayed as a Z score. Three major clusters were identified and analyzed for functional enrichment or depletion using topGO (for details, see Supplemental Table 1).
To interpret these data properly, we note that GCN2 kinase causes a global reduction in polysome loading in wild-type plants (Figure 9A); this global reduction was masked during the subsequent translatome analysis, because the array hybridization was performed with equal amounts of total RNA per sample. We then reanalyzed the wild-type translatome by unmasking the global translational repression by herbicide treatment in the wild type (Supplemental Table 2). This analysis further confirmed the major results of gene ontology analysis from clustering and GSEA, yet it also uncovered additional details that link the molecular physiology of the GCN2 pathway with functional categories at the level of translational control. For example, defense responses were relatively protected from translational repression. Given that in animals and yeast GCN2 controls amino acid homeostasis, it is interesting that functional terms related to protein recycling appear among mRNAs that are preferentially translationally repressed, whereas terms related to amino acid metabolism characterize mRNAs that are spared from translational repression.
GCN2 Remodels the Transcriptome, Implicating GCN2 in Plant Responses to Stress
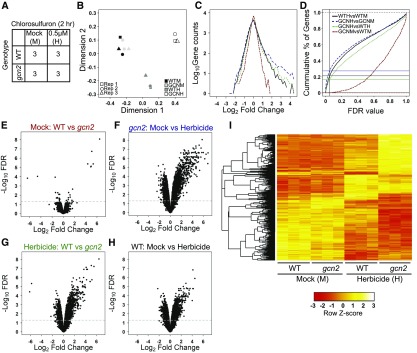
Although GCN2’s primary known role is as a translational regulator, one would expect that alterations in translation would result in alterations in mRNA transcript levels. Because analyzing transcript levels may shed additional light on the physiological role of GCN2, we also analyzed the transcriptome of GCN2 in response to CSF (Figure 10A; Supplemental Data Set 1). The four sets of triplicate data separated well in a multidimensional scaling plot (Figure 10B). In the absence of herbicide, wild-type and gcn2 seedlings had nearly identical transcript levels (Figures 10C to 10E), in keeping with the nearly indistinguishable whole-plant phenotypes. The response to herbicide was dramatic and was biased toward upregulation in both the wild type and gcn2 (Figures 10C, 10F, and 10H), as expected given that the herbicide treatment was short compared to the average lifetime of mRNAs. Many individual genes responded more strongly in the gcn2 mutant (blue traces) than in the wild type (black traces; Figures 10C, 10D, and 10F to 10I). Hence, the gcn2 mutant is hypersensitive to herbicide stimulus.
Figure 10.
Transcriptome Analysis of gcn2 Seedlings Treated with the Herbicide Chlorosulfuron.
(A) Schematic of the experiment. “3” indicates three biological replicates. WT, wild type.
(B) Gene expression data displayed by multidimensional scaling. Replicates are identified by a square, circle, and triangle. The plot was produced using limma’s plot MDS function on all the data. GCNH, gcn2 herbicide; GCNM, gcn2 mock; WTH, wild-type herbicide; WTM, wild-type mock.
(C) Line histogram of the number of differentially expressed genes for the four pairwise comparisons described in (D). See (D) for color codes. GCNH, gcn2 herbicide; GCNM, gcn2 mock; WTH, wild-type herbicide; WTM, wild-type mock.
(D) Line graphs of the number of genes deemed differentially expressed as a function of the false discovery rate for the four pairwise comparisons. An FDR of 0.05 is marked by the vertical line. The data were corrected for false discovery by multiple comparisons using the Benjamini-Hochberg method. Red, mock-treated gcn2 versus mock-treated wild type. Green, herbicide-treated gcn2 versus wild type. Black, wild type mock versus wild type herbicide. Blue, gcn2 mock versus gcn2 herbicide. The horizontal lines highlight where each curve intercepts FDR = 0.05.
(E) to (H) Scatterplots (“volcano plots”) of the fold-changes in mRNA level versus the false-discovery rate (FDR) for the four pairwise comparisons in (D). The horizontal line marks FDR = 0.05. WT, wild type.
(I) Cluster analysis of the four experimental transcriptomes. Included are 4632 genes (probe sets) that showed significant differences in mRNA level in both a one-way ANOVA and any pairwise comparison (limma FDR < 0.05). The clustering tree is shown on the left. WT, wild type.
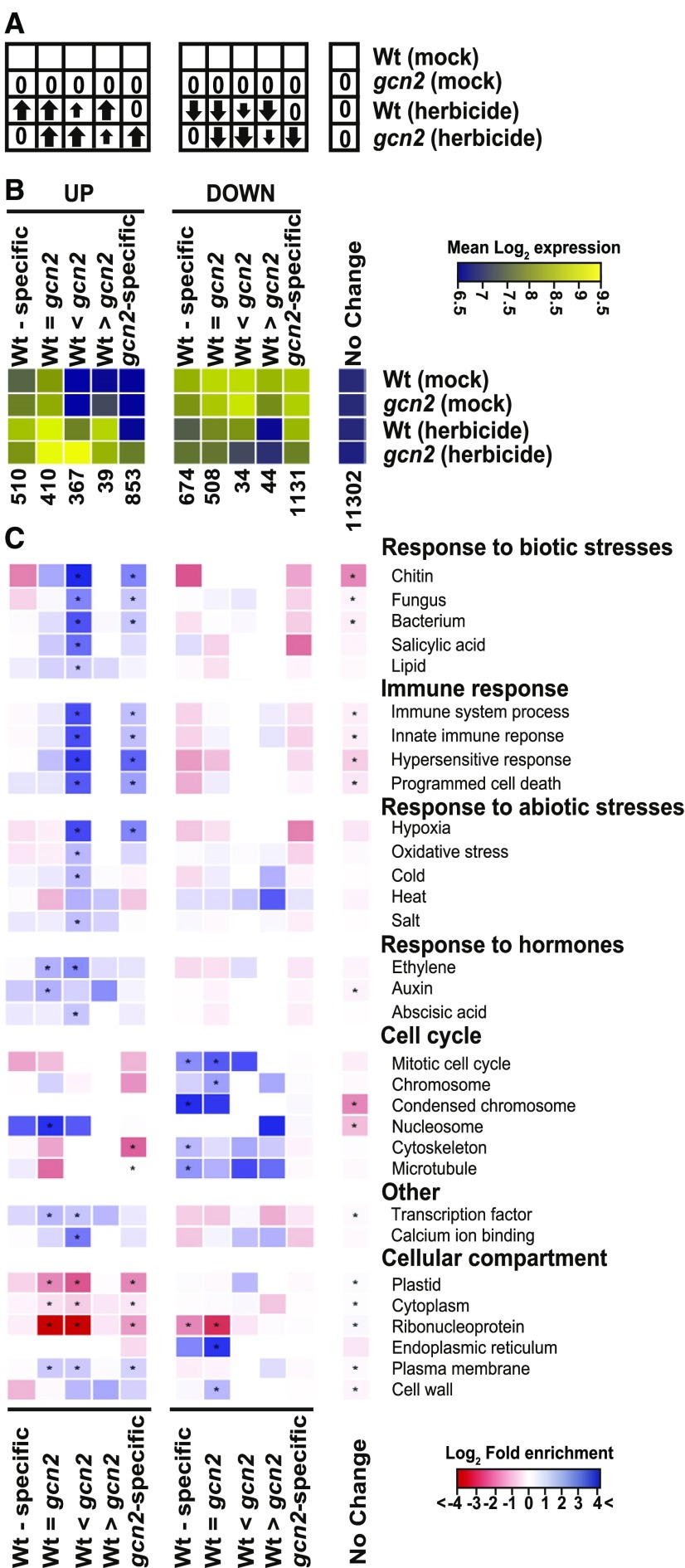
Although unsupervised clustering of the transcriptome response revealed clearly delineated response types (Figure 10I), we decided to bin genes according to 11 predefined filters (Figure 11) for a finer-grained, biologically motivated classification. The herbicide response was strongly affected by the GCN2 genotype, ranging from genes that only responded in wild type and not in gcn2 (wild type specific) all the way to genes that only responded in gcn2 but not in the wild type (gcn2-specific; Figures 11A and 11B). We were most interested in the mRNAs whose response was exaggerated in gcn2 (columns 3 and 5 in Figures 11B and 11C), indicating that wild-type GCN2 tempers their response to herbicide. Among these, the upregulated genes were biased toward the term “response to stimulus” (201/367 in column 3), especially biotic defense and immune responses, as well as many abiotic stresses including oxidative stress. This group includes genes for ROS signaling, such as the famous RbohD isoform of the NADPH oxidase responsible for the respiratory burst. The strongest enrichment was seen for “response to chitin,” a gene ontology term that contains numerous transcription factors. Thus, herbicide treatment triggered a defense response, which was attenuated by active GCN2 kinase.
Figure 11.
Gene Ontology Analysis of the GCN2-Responsive Transcriptome.
(A) Cartoon illustrating the filter being applied under each of 11 bins. An arrow indicates whether transcript levels are up or down or unchanged (0) compared to mock-treated wild-type Ler-0 (Wt) as a reference. The small arrow represents a lesser response.
(B) Heatmap of the average mRNA levels in each of the 11 bins in (A). The color scale shows the mean log2-transformed hybridization signals per bin on the Affymetrix gcRMA scale. The number of genes in each bin is indicated. Note that genes unresponsive to herbicide typically have lower mRNA levels than herbicide-responsive genes. Wt, wild type.
(C) All 11 bins were analyzed by gene ontology with topGO with correction for multiple testing. The color-coded heatmap displays the fold-enrichment of a given gene ontology term in each bin over what is expected by chance. Blue hues denote enriched terms, and red hues are depleted terms. Cells where the enrichment/depletion passes FDR < 0.05 are labeled with an *. Wt, wild type.
GCN2 Mediates Chitin Priming
Exposure to chitin primes an innate immune response that protects plants against the bacterial pathogen Pseudomonas syringae DC3000 (Zipfel and Robatzek, 2010). Given that “response to chitin” and related immune responses were the most prominent functional categories among the mRNAs that are upregulated in the gcn2-1 mutant, we examined whether gcn2-1 plants respond differently to chitin. Interestingly, while defense priming by chitin was observed in wild-type Landsberg erecta (Ler-0), this pattern-triggered immunity (PTI) response was significantly reduced in gcn2-1 (Supplemental Figure 17), indicating that GCN2 kinase plays a role in PTI (to some extent) and the activation of innate immune signaling. On one hand, the loss of priming by chitin in gcn2-1 is somewhat surprising because transcript levels in the “response to chitin” category were elevated rather than reduced in gcn2-1 treated with herbicide (Figure 11C). However, this observation suggests that the response of gcn2-1 to herbicide triggers a complex series of events that ultimately link GCN2 kinase activity with cellular signaling processes associated with stress, including biotic stress responses.
Discussion
Extensive studies in animals and yeast have demonstrated that GCN2 kinase mediates global translational repression in response to environmental signals by phosphorylating its primary substrate, eIF2α. In these organisms, GCN2 kinase is activated by uncharged tRNAs (Wek et al., 1995; Dong et al., 2000; Anda et al., 2017). The GCN2 pathway appears to be substantially conserved in plants. GCN2 kinase is activated by inhibitors of amino acid biosynthesis such as the herbicides CSF, glyphosate, and glufosinate, it can be activated by uncharged tRNAs in vitro, and it is activated in a mutant with a defect in Cys biosynthesis (Lageix et al., 2008; Zhang et al., 2008; Li et al., 2013; Dong et al., 2017). GCN2 kinase phosphorylates eIF2α (Lageix et al., 2008; Zhang et al., 2008), and the inhibitors of amino acid biosynthesis that activate GCN2 kinase indeed reduce overall ribosome loading of mRNAs (Lageix et al., 2008).
GCN2 Activation by Light and ROS
The true nature of the biochemical signal that activates GCN2 in planta remains unclear. Herbicides are synthetic activators of GCN2. GCN2 kinase is activated by numerous other agents, including UV light, wounding, the ethylene precursor 1-ACC, and the endogenous defense signals salicylic acid and methyl-jasmonate (Lageix et al., 2008), but it is unclear whether these signals are the authentic activators of GCN2 in nature. Here, we demonstrated that GCN2 kinase is also activated by light in a dosage-dependent fashion. This activation was observed most clearly in 24-h-dark-adapted plants, where GCN2 activity became undetectable within 6 h of dark acclimation and then increased rapidly upon exposure to excess light stress. Some activation of GCN2 was also evident in plants grown under a regular long-day light-dark cycle. Importantly, even the traditional activation of GCN2 by herbicides is strictly light dependent. These observations suggest that, besides uncharged tRNAs as a ligand for the kinase, a second, light-dependent signal is required for GCN2 activation.
It is unclear which biochemical sequence of events causes excess light to activate GCN2. The light-dark pattern of GCN2 activation coincides with the redox rhythm. Our experiments suggest that an ROS produced in the chloroplast during photosynthesis is a key intermediate in this process, although we cannot rule out the possibility that uncharged tRNAs may also be required. First, GCN2 is activated rapidly by ROS, by the application of hydrogen peroxide ectopically, and even more rapidly by MV, which triggers superoxide and hydrogen peroxide production from PSI. Internal triggers of plastidic ROS, i.e., plastidic expression of GO, the application of norflurazon, and exposure of flu1 mutants to excess light, also stimulate GCN2 activity. Surprisingly, heat stress, which also triggers plastidic ROS production (Czarnocka and Karpiński, 2018), suppressed eIF2α-P formation, possibly due to secondary effects of heat on GCN2 or phosphatase activity. In keeping with a role for ROS, the activation of GCN2 by excess light can be suppressed by treatment with ascorbate. This activation can also be repressed by manipulating the redox status of the photosynthetic apparatus with electron transport inhibitors, suggesting that a plastidic ROS functions as a source of the signal that activates GCN2. Chemical or reoxygenation treatments that stimulate ROS production in the mitochondria (Stonebloom et al., 2012; Paradiso et al., 2016; Cui et al., 2019) did not activate GCN2 at the time periods tested, nor did tunicamycin, a trigger of ER stress well known to promote eIF2α-P accumulation in animals.
ROS produced in the chloroplast can regulate translation in the cytosol (Khandal et al., 2009), with methyl-jasmonate functioning as a potential intermediate. Our work suggests that this pathway may operate in part via GCN2 kinase. The finding that methyl-jasmonate can activate GCN2 (Lageix et al., 2008) supports this hypothesis. However, in our hands, ROS also repressed translation in a GCN2-independent manner. These observations suggest that H2O2 regulates cytosolic translation via multiple pathways. A similar situation exists in fission yeast (Schizosaccharomyces pombe), where H2O2 stimulates GCN2 kinase and represses translation, but its effect on translation is at least partially GCN2 independent (e.g., Knutsen et al., 2015). These comparisons suggest that ROS-mediated activation of GCN2 kinase in Arabidopsis is evidence of a pan-eukaryotic phenomenon. The mechanism by which ROS activate GCN2 kinase in plants remains unknown, and it also remains unresolved in every other organism, including fission yeast, budding yeast (Saccharomyces cerevisiae), and mammals, where ROS-induced activation of eIF2α kinases has been observed over the past 15 years (Shenton et al., 2006; Anda et al., 2017).
The Effect of GCN2 on the Transcriptome
Given that the overt growth defects in the gcn2 mutant were mild, we analyzed its transcriptome to identify the consequences of GCN2 activation. We decided to stimulate GCN2 kinase activity with herbicide treatment rather than ROS because (1) ROS have numerous biochemical targets and trigger multiple signaling pathways, and (2) ROS accumulate in a nonuniform pattern across tissues. By contrast, CSF specifically inhibits branched-chain amino acid synthesis and suppresses bulk ribosome loading in a GCN2-dependent manner (Lageix et al., 2008), whereas hydrogen peroxide did not, under our conditions. Similar to data by Faus et al. (2015), in the absence of herbicide, the wild-type and gcn2 transcriptomes were essentially the same. Once GCN2 kinase is activated by herbicide, two roles can be distinguished. The first of these is GCN2 as an attenuator of transcript levels: among all the herbicide-inducible mRNAs, GCN2 attenuated more than half, either partially or completely. Many of these mRNAs are functionally associated with responses to biotic or abiotic stresses (Figure 11C, columns 3 and 5). The second role of GCN2 kinase is GCN2 as a repressor/activator: after treatment with herbicide, the levels of many mRNAs became repressed, often with the help of GCN2 (Figure 11C, columns 6 and 9). The functional classification of this cohort indicated that GCN2 also modulates the effect of herbicide on the cell cycle. Taken together, it is noteworthy that a substantial portion of the transcriptome-wide response to herbicide appears to be mediated by GCN2 kinase. It is also worth considering that our findings suggest GCN2 kinase functions to attenuate a stress response in Arabidopsis rather than mediating an active, integrated stress response, as is the case in mammals.
Several functional categories were strongly depleted among the herbicide-responsive mRNAs, namely those associated with the plastid as well as ribonucleoprotein complexes, i.e., translation and ribosome biogenesis. Given that CSF inhibits amino acid synthesis in the chloroplast, one might have expected a robust response in favor of chloroplast-bound proteins. However, this was not the case. Previously, Faus and coworkers compared the transcriptomes of wild type and gcn2-1 seedlings after 6 h of glyphosate treatment (Faus et al., 2015) compared to 2 h of CSF in this study. Both experiments showed a downregulation of cell-cycle-related functions. At 6 h of treatment, photosynthesis-related functions were clearly repressed by glyphosate treatment, and more so in wild type than gcn2, in contrast to our results; perhaps the photosynthesis response is a late response to herbicide that is amplified by GCN2 activity. Finally, in our hands, the defense response was exacerbated in gcn2, while in Faus et al. (2015)’s study, it was weakened in gcn2; this suggests that the defense response is more rapid and transient in gcn2 than in wild type.
The Effects of GCN2 on the Translatome
In animals and budding yeast, the activation of GCN2 boosts the ribosome loading of certain basic leucine zipper-type transcription factor mRNAs that harbor upstream open reading frames in their 5′ untranslated region. GCN2’s translational effects on mRNAs such as Activating transcription factor 4 and GCN4 and the resulting transcriptional changes are known as the integrated stress response and the general amino acid control, respectively (Wek, 2018). By comparison, in fission yeast, a nonhomologous but functionally analogous mRNA for a GATA transcription factor was only recently identified (Duncan et al., 2018). Although certain basic leucine zippers and several other classes of transcription factor mRNAs harbor upstream open reading frames in Arabidopsis (von Arnim et al., 2014), these mRNAs did not appear to be in any way poised for elevated translation once GCN2 was activated. This result suggests that GCN2 and eIF2α-P affect translation differently in plants compared to yeast and animals, but eventually they tie a stress signal, in our case herbicide, to a change in transcript level.
The most striking transcript-level effect upon loss of GCN2 function is an exaggerated defense response. This effect might be due to the loss of translational control in gcn2, specifically because translation of specific regulatory mRNAs remains high in the presence of the herbicide stress. Because the translation state of thousands of mRNAs is affected in the mutant, including transcriptional regulators (Supplemental Data Set 2, cluster 1), it is not possible to attribute this effect to a single mRNA. More likely, the hypersensitivity of transcript levels in gcn2 might be mediated by the aggregate of alterations at the level of GCN2-mediated ribosome loading. However, other explanations cannot be ruled out, such as signaling functions of GCN2 beyond eIF2α and translational control.
The Physiological Role of GCN2 Kinase
One conundrum surrounding plant GCN2 is that gcn2 mutants have rather mild phenotypes under favorable laboratory conditions (e.g., Faus et al., 2015; Liu et al., 2019; Llabata et al., 2019) and a near-normal transcriptome. Although GCN2 kinase can be activated by numerous treatments, very few of these conditions cause a maladaptive phenotype in gcn2 mutants, lending urgency to the question of which signals activate GCN2 in the wild.
Our data add to the evidence that GCN2 is part of the biotic defense response. First, GCN2 is activated by signals that arise during regular defense responses, such as salicylic acid, jasmonic acid, ethylene (Lageix et al., 2008), and reactive oxygen (Figure 2B). GCN2 is also activated in response to some but not all bacterial infections (Izquierdo et al., 2018; Liu et al., 2019). Second, the transcriptome of gcn2 overexpresses mRNAs from defense-related gene ontology terms such as “response to chitin” and the oxidative burst. Thus, the presence of GCN2 can attenuate the defense response at the transcriptional level, which is akin to what is observed during the preinvasive stages of bacterial infection (Liu et al., 2019). Third, GCN2 also preferentially affects the translation of mRNAs in the secretory system, and the secretory system is responsible for the secretion of many defense-related proteins, such as peptidases and chitinases. Finally, gcn2 plants had a defect in the priming of their bacterial defense response by chitin (Supplemental Figure 17) but were more resistant to unprimed infection by P. syringae than the wild type (Liu et al., 2019). In summary, these findings bolster the view that GCN2 is an integral part of the defense response against pathogens.
Here, we demonstrated that GCN2 kinase is not only activated by excess light, but gcn2 mutants also have a mild growth deficiency under prolonged continuous or high light, depending on the ecotype. Seedlings that had been transferred from the dark to light also trended toward a GCN2-mediated reduction in polysome loading (Supplemental Figure 18). These data support the notion that adaptation to excess light is part of the functional portfolio of GCN2 kinase.
In conclusion, this work establishes that the pan-eukaryotic protein kinase GCN2 is activated by ROS in a plant. While the activation of GCN2 by ROS has been observed in other organisms, our work now implicates ROS produced in a specific organelle, the chloroplast, in the activation of GCN2 and subsequent phosphorylation of a key translation initiation factor. Because ROS in the chloroplast originate from the photosynthetic apparatus, and because the active GCN2 kinase is capable of regulating cytosolic translation, this process represents a mechanism for the chloroplast to communicate information directly to the protein synthesis machinery of the cytosol. Although we do not discount the notion that ROS regulate cytosolic translation by yet other pathways, the phenotypic growth deficiencies of gcn2 plants exposed to high light suggest that GCN2 kinase is physiologically relevant under ROS-producing conditions and that GCN2-mediated eIF2α-P is part of the regular cellular homeostasis mechanisms whereby plants adapt to naturally changing environmental conditions.
METHODS
Plant Materials and Growth Conditions
The plant lines used in this study include Arabidopsis (Arabidopsis thaliana) ecotype Ler-0 and Col-0. The homozygous gcn2-1 mutant is Genetrap line GT8359 (Zhang et al., 2008). The GCN2 T-DNA insertion alleles gcn2-2 (SALK_032196; Faus et al., 2018) and a new allele with a T-DNA insertion in exon 15 (SALK_129334.2; Alonso et al., 2003), henceforth called gcn2-3, were obtained from the Arabidopsis Biological Resource Center (ABRC) and characterized by PCR genotyping. The gcn2-1;GCN2 line is the gcn2-1 mutant complemented with a genomic GCN2 gene under the control of its native promoter (Lageix et al., 2008). The flu1-1 mutant is in the Ler-0 background (Meskauskiene et al., 2001), and the GO overexpressing plants are in the Col-0 background (Fahnenstich et al., 2008).
Seeds were sterilized and stratified at 4°C for 2 d. The seeds were germinated on half strength Murashige and Skoog (MS) salt plant medium (MP Biomedicals, cat. no. 2,633,024) solidified with 0.65% Phytoagar (Bioworld, cat. no. 40100072-2). Standard conditions are a long-day cycle of 16 h white light (Philips F17T8/TL741 17 Watt; 80 ± 10 µE m−2s−1)/8 h dark at 22°C and 50% humidity. Unless stated otherwise, seedlings were grown on medium without Suc. Genomic DNA was extracted from 2-week-old seedlings using a GeneJET Plant Genomic DNA purification kit (Thermo Fisher Scientific, cat. no. K0791) as per manufacturer’s instructions, and PCR was performed with gene-specific GCN2 primers (GCN2-F1, 5′-AATTCGCCAAATTGTGGAAG-3′; GCN2-R1, 5′-ATAAGCAAATGACAGGTCCG-3′ for gcn2-2 and GCN2-F2, 5′- TAAGTTCCCCTGTGTCCCAC-3′; GCN2-R2, 5′-ACTTGGAGACATCAAACGCC-3′ for gcn2-3) and a left border T-DNA primer (LBa1, 5′-TGGTTCACGTAGTGGGCCATCG-3′). The predicted T-DNA insertion site was verified by DNA sequencing. Like gcn2-1, gcn2-2 and gcn2-3 homozygotes were unable to phosphorylate eIF2α.
Stress Treatments
For stress treatments performed in the dark, 2-week-old seedlings were dark acclimated for 24 h starting 2 h after lights-on (Zeitgeber time 2, ZT2). Dark samples were collected under a green safe light. For treatment with H2O2 (Sigma Aldrich cat. no. H1009), antimycin A (Alfa Aesar, cat. no. AAJ63522LB0), salicylhydroxamic acid (Thermo Fisher Scientific, cat. no. C132620050), tunicamycin (Sigma Aldrich, cat. no. T7765), and herbicide, dark-acclimated seedlings were sprayed six to eight times with the respective reagents from a distance of 4 inches. Seedlings were mock-treated with water only or with 0.1% DMSO, as appropriate. For heat and cold stress, plates containing dark-acclimated seedlings were shifted to 37°C or 4°C, respectively, in the dark for the desired times. Hypoxia (i.e., low O2) stress was administered in the dark using argon gas, and re-oxygenation treatment, also in the dark, was performed as previously described by Lokdarshi et al. (2016).
For stress treatments that involved a dark-to-light shift, 2-week-old seedlings grown on horizontal plates were dark-acclimated for 24 h starting at ZT2. A sample for time zero (T0) was collected under a green safe light, and plates were exposed to different white light intensities for the desired times. For chemical pretreatments with DCMU (Thermo Fisher Scientific, cat. no. D2425), DBMIB (Thermo Fisher Scientific, cat. no. 271993), CSF (Sigma Aldrich, cat. no. 34,322), glyphosate (Bioworld, cat. no. 30632003-1), and glufosinate ammonium, seedlings were treated (i.e., sprayed) with reagents or mock control (DMSO or water) 30 min prior to the end of the 24-h dark acclimation. MV and norflurazon were applied by spraying with 20 μM MV or 50 μM norflurazon, respectively. For pretreatment with ascorbate, seedlings were grown for 2 weeks on medium supplemented with 0.5 mM ascorbate. Other reagent concentrations are listed in the figure legends.
For pathogen-associated molecular pattern-triggered immunity (PTI) priming, 4-week-old Arabidopsis plants were hand-infiltrated with flg22 (1 μM) or chitin ([GlcNAc]7; 1 μM) and incubated at room temperature (∼22°C) for 24 h. After 24 h, pathogen-associated molecular pattern-infiltrated leaves were hand-infiltrated with Pst DC3000 strains harboring the vector pVSP61. For in planta bacterial growth enumeration, Pst DC3000 strains were inoculated into the leaves using a needleless syringe at a final concentration of 2 × 105 CFU/mL. Samples were collected at 0 and 96 h postinoculation (hpi). All pathogen-inoculation experiments were performed at least three times with three technical replicates per experiment. Statistical analysis of bacterial growth was performed by one-way ANOVA using GraphPad Software (Prism; Supplemental File).
Phenotype Characterization and High-Light Treatment
For phenotypic characterization under high light, wild-type and gcn2 seedlings were germinated on half strength MS medium containing 0.1% Suc under a long-day cycle of 16 h white light (80 ± 10 µE m−2s−1)/8 h dark at 22°C. 3- or 4-d-old vertically grown seedlings were transferred to half strength MS medium without Suc, and the plates were placed under continuous high light (780 ± 10 µE m−2s−1) at 18°C starting at ZT4 for 3 consecutive days. For mock treatments, plates were transferred to continuous light (80 ± 10 µE m−2s−1) at 22°C. To compensate for any increase in plate temperature from the high-intensity fluorescent tube lights (6000 lumens, Super Bright White; model HyperBC-4F-605, UL no. E472253), chamber temperature was adjusted to 18°C for the period of high-light treatment. Additionally, to avoid exposing the roots to high-light levels, the vertical plates were covered with black paper and the position of the plates was adjusted to focus light treatment primarily on the shoots. For recovery treatments, both mock- and high-light-exposed plates were transferred to a long day cycle of 16 h white (80 ± 10 µE m−2s−1)/8 h dark at 22°C for 3 d prior to fresh weight measurements.
Root Length, Fresh Weight Measurements, and Statistical Analysis
Photographs of vertically grown seedlings at day 3 postgermination and after stress treatments were taken with a digital camera (Canon). The primary root length was measured using ImageJ (ver. 1.41; http://rsb.info.nih.gov/ij/index.html). For fresh weight measurements, seedlings were weighed on an analytical balance. Statistical tests (Supplemental File) were performed using GraphPad Prism (ver. 7.0a; GraphPad Software).
Protein Extraction and Immunoblot Analysis
Total protein extraction was performed as described previously by Zhang et al. (2008). In brief, 2-week-old whole seedlings were harvested after the desired treatments and flash frozen in liquid nitrogen. For total protein extraction, seedlings were ground using a plastic pestle in a 1.5-mL tube with ice-cold extraction buffer containing 25 mM Tris-HCl (pH 7.5), 75 mM NaCl, 5% (v/v) glycerol, 0.05% (v/v) Nonidet P-40, 0.5 mM EDTA, 0.5 mM EGTA, 2 mM DTT, and 2% (w/v) insoluble polyvinylpyrrolidone (Sigma Aldrich P-6755) supplemented with 1× protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific; cat. no. PIA32959). Total protein content was quantified by Bradford assay (Thermo Fisher Scientific, cat. no. 23,236).
For immunoblot analysis, 50 μg of total protein was separated on a 12% (w/v) SDS-PAGE gel and electroblotted onto a polyvinylidene fluoride membrane. After 1 h of blocking at 22°C with TBST buffer (1× Tris-buffered saline [pH7.6], 0.1% Tween-20, 10% non-fat dry milk, and 0.2% BSA), the membrane was incubated for 48 h at 4°C with polyclonal rabbit phospho-eIF2α antibody (Cell Signaling, cat. no. 9712S) diluted to 1:5000 in 1× TBST with 5% BSA. Following washing with 1× TBST, 10 min each for 15 repeats, the membrane was incubated with a 1:2000 dilution of horseradish peroxidase conjugated anti-rabbit IgG (Vector Labs, cat. no. PI-1000) in blocking buffer for 1 h at room temperature. After washing with 1× TBST, 10 min each for 15 repeats, chemiluminescence was performed (WesternBright Quantum, Advansta) as per manufacturer’s protocol. For immunoblot analysis with polyclonal rabbit eIF2α antibody (a gift from Karen Browning, University of Texas, Austin), 5 μg of total protein was resolved by 12% (w/v) SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane. Blocking and incubation with antibodies were performed as described in Dennis et al. (2009), followed by chemiluminescent detection (Lokdarshi et al., 2016).
Polysome Profiling and Protein Fractionation
Two-week-old seedlings were flash frozen in liquid N2 and stored at −80°C prior to polysome profiling. Whole seedlings were ground in liquid N2, and 0.5 g of tissue powder was resuspended in 1 mL of polysome extraction buffer (200 mM Tris-HCl, pH 8.4, 50 mM KCl, 25 mM MgCl2, 1% deoxycholic acid, 2% polyoxyethylene 10 tridecyl ether, 50 μg/mL cycloheximide and 40 U/mL RNase inhibitor [Promega, cat. no. N2115]) and centrifuged at 13,000g for 5 min at 4°C. One milliliter of the supernatant was layered onto a 10-mL 15 to 50% linear gradient prepared using a Hoefer gradient maker and centrifuged at 35,000 rpm (Beckmann SW 41 Ti) for 3.5 h at 4°C. Absorbance at 254 nm was recorded using an ISCO UA 5 absorbance/fluorescence monitor, and individual data points were extracted using DATA acquisition software (DATAQ Instruments). Polysome-to-monosome ratios were calculated as described (Enganti et al., 2018).
ROS Localization and Microscopic Techniques
For subcellular detection of ROS in Arabidopsis leaves, 2-week-old stress-exposed seedlings were submerged in 15 μM H2DCFDA (Thermo Fisher Scientific, cat. no. D339) for 10 to 12 min in the dark. ROS detection was performed on a Leica SP8 laser scanning confocal microscope using the HeNe laser in the Advanced Microscopy and Imaging Facility at the University of Tennessee, Knoxville. The excitation filter was set to 488 nm, and the emission filter was set to 500 to 550 nm for H2DCFDA (emission maximum of 517 to 527 nm) and 660 to 690 nm for chlorophyll autofluorescence. Post processing of confocal Z-stack images was performed using ImageJ (ver. 1.4; http://rsb.info.nih.gov/ij/index.html).
Hydrogen Peroxide Quantification
H2O2 measurements were performed using an Amplex Red kit (Thermo Fisher Scientific, cat. no. A22188). In brief, 30 mg of 2-week-old seedling leaf tissue was flash frozen in liquid N2 and ground with a plastic pestle to a homogeneous powder. The pulverized tissue was resuspended in 100 μL of sterile 1× PBS and centrifuged at 17,000g 4°C for 2 min, and the supernatant was used for H2O2 measurements as per manufacturer’s protocol. Relative fluorescence was measured on a POLARstar OPTIMA plate reader (BMG LABTECH) with an excitation filter at 535 nm and emission filter at 600 nm.
Photosynthetic Efficiency Measurement
The maximum quantum yield of PSII [Qymax = Fv/Fm] was measured on a FluorCam 800MF (Photon Systems Instruments) as per manufacturer’s instructions and modifications from Murchie and Lawson (2013). In brief, 16 h white light/8 h dark grown 3-d-old wild-type Ler-0 and gcn2-1 seedlings were dark adapted for 2 min (Fo) prior to applying a saturating pulse of 1800 µE m-2s−1 for 0.8 s (Fm). Variable fluorescence (Fv) was calculated as the difference between Fo and Fm to obtain the maximum quantum yield [Fv/Fm].
Transcriptome Analysis
For microarray analysis, seedlings were grown as described (Missra et al., 2015). Eleven-day-old wild-type or gcn2-1 seedlings were treated around ZT 2 by dipping in a 0.5-µM CSF solution in water or in a mock treatment for 2 min. Treated seedling shoots were harvested 2 h after treatment by freezing in liquid nitrogen and scraping off the aerial tissue with a metal spatula. Total RNA extraction and purification as well as Affymetrix GeneChip Arabidopsis ATH1 Genome Array hybridization and scanning were performed as described (Missra and von Arnim, 2014). Each treatment/genotype combination was performed in triplicate at different times, resulting in four treatment/genotype combinations and a total of 12 microarrays.
The microarrays were analyzed in R (ver. 3.5.0; R Core Team, 2018) and normalized using the gcrma package (ver. 2.52.0; Wu et al., 2018) with default parameters. The mas5calls function was used to create present, marginal, or absent calls from the normalized data. Probes were filtered by mas5 calls, only keeping probes that expressed only present values in all three replicates of at least one condition. Probes for chloroplastic, mitochondrial, and control genes as well as probes with 0 variance across all 12 microarrays were also removed. Differentially expressed genes (DEGs) were identified using limma (version 3.36.2; Ritchie et al., 2015; Phipson et al., 2016). DEGs were identified using a factorial design with terms included to account for a replicate batch effect in the data. DEG results were corrected for multiple comparisons using FDR. A one-way ANOVA using limma’s topTableF ftest was used to find genes that had equal means between conditions for future filtering; the P values were corrected by the Benjamini-Hochberg method. This helped to define a set of nondifferentially expressed genes.
Following differential expression analysis, the intersection of genes found to be significant in the one-way ANOVA and limma’s factorial design were then divided according to expression patterns. The first group contained genes with a fold change of less than or equal to 1.3 or did not have an FDR < 0.05 and were considered not differentially expressed. The second group contained genes with a fold change greater than 1.3 and FDR < 0.05. This second group was further subdivided into seven gene sets where differential gene expression occurred: (1) only in the wild type; (2) only in the gcn2 mutant; (3 to 5) upregulated or downregulated in both genotypes with subgroups (3) wild type = gcn2 (i.e., |wild type-gcn2| < 1.3 fold change), (4) gcn2 > wild type, and (5) wild type > gcn2; (6) opposite differential expression patterns in wild type and gcn2 (e.g., upregulated in wild type and downregulated in gcn2 or vice versa); and (7) differentially expressed between wild type and gcn2 under control but not herbicide-treated conditions. These seven subgroups were reduced to five by pooling closely related groups and subdivided again into sets containing only upregulated and downregulated genes (total of 11 groups, one no differential expression, five upregulated, five downregulated).
The replicate heat map was created with the hclust package in R using Pearson’s R-square as distances and the complete algorithm. In the heat map, the expression level (Xijk) of gene i, in replicate j under condition k is displayed as a Z-score with:
Gene ontology analysis was performed on the 15 gene sets using the topGO package (version 2.32.0; Alexa and Rahnenfuhrer, 2016) and revision 46,221 (http://viewvc.geneontology.org/viewvc/GO-SVN/trunk/gene-associations/gene_association.tair.gz?revision=46221) of the TAIR gene association files from the gene ontology consortium. Only genes measured as expressed (15,934 genes) were used as the gene universe. topGO was run with node size 1, and FDR P value adjustment using a custom script and the classic Fisher, parent-child, and weight01 algorithms. Packages were obtained from CRAN or Bioconductor (version 3.7; Huber et al., 2015).
Translatome Analysis
After cell extracts from seedling shoots were subjected to sucrose gradient fractionation, we generated pools of nonpolysomal (NP), small polysomal (SP), and large polysomal (LP) RNAs. The RNAs were processed for ATH1 Affymetrix RNA hybridization to measure the abundance of each mRNA in each fraction. Hybridization signals were extracted and normalized using the standard gcRMA algorithm. Subsequently, a translation state (TL) was calculated for each mRNA, essentially as described by Missra et al. (2015) as follows:
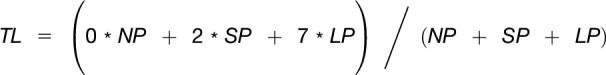 |
From the wild-type data set, one of the three replicates was eliminated due to excessive variance compared with the other two replicates; we assert that this variance is due to a technical flaw in the experiment. We initially did not adjust the data to account for the global loss of polysome loading in herbicide-treated wild type (Figure 9; Supplemental Table 1). In a subsequent, alternate analysis, a scaling factor (Kawaguchi et al., 2004) was applied to the data from herbicide-treated wild type in order to reflect the global repression of translation that was evident after RNA isolation. This was done by reducing the translation states of wild type treated with CSF herbicide by the global drop in ribosome loading, averaged from the biological replicates (Supplemental Table 2). Differentially translated mRNAs were identified by limma with FDR correction (P < 0.05). These mRNAs were clustered by hierarchical clustering with Pearson correlation, the preferred cluster number was settled upon by silhouette score analysis, and TL values were displayed as a Z-score (the fold difference from the average divided by the standard deviation across each of four treatments). Clusters were analyzed for functional enrichment using topGO. Pairwise comparisons were analyzed using the “preranked” function in the GSEA suite (Subramanian et al., 2005).
Accession Numbers
Sequence data for this article can be found in the GenBank/EMBL libraries under AGI locus identifiers At3g59410 (GCN2) and At5g05470/At2g40290 (eIF2α). Microarray data were deposited at NCBI under GEO accession number GSE52117.
Supplemental Data
Supplemental Figure 1. Heat stress suppresses eIF2α phosphorylation
Supplemental Figure 2. eIF2α phosphorylation under high light
Supplemental Figure 3. Dark acclimation suppresses eIF2α phosphorylation
Supplemental Figure 4. Ascorbate delays GCN2 activation in the light
Supplemental Figure 5. Complementation with GCN2 helps in recovery from high light
Supplemental Figure 6. Characterization of the gcn2-2 and gcn2-3 T-DNA insertion alleles of GCN2
Supplemental Figure 7. The gcn2-2 allele in the Col-0 background shows increased sensitivity to continuous-light and high-light treatments
Supplemental Figure 8. The gcn2 mutant alleles in the Col-0 background show increased sensitivity to continuous or high light
Supplemental Figure 9. GCN2 has minimal effects on PSII quantum yield (Fv/Fm)
Supplemental Figure 10. Dark-to-light shift modulates hydrogen peroxide levels
Supplemental Figure 11. Hydrogen peroxide treatment represses translation to similar levels in gcn2-1 and wild-type plants.
Supplemental Figure 12. Norflurazon induces GCN2 activity in a manner dependent on photosynthesis.
Supplemental Figure 13. Activation of GCN2 by MV requires light.
Supplemental Figure 14. Effect of MV on photosynthetic efficiency of wild type and gcn2-1.
Supplemental Figure 15. Ectopic GO augments GCN2 activation by photosynthetic ROS.
Supplemental Figure 16. Treatments intended to induce ROS accumulation in mitochondria and ER cause accumulation of H2O2.
Supplemental Figure 17. GCN2 imparts bacterial resistance by chitin-inducible priming.
Supplemental Figure 18. gcn2 and wild-type plants show similar ribosome RNA profiles upon dark adaptation and after return to light.
Supplemental Table 1. Gene ontology analysis of translatome data.
Supplemental Table 2. Global repression of translation by GCN2 kinase.
Supplemental Data Set 1. Transcriptome data.
Supplemental Data Set 2. Translation state data.
Supplemental File. ANOVA tables.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
GSEA Gramene: Gene set enrichment analysis
GSEA Araport: Gene set enrichment analysis
PTI Gramene: Pattern triggered immunity
PTI Araport: Pattern triggered immunity
Acknowledgments
We thank Karen Browning for the gift of antibody against eIF2α. We also thank Chanhong Kim for sharing flu1-1 seeds, Veronica G. Maurino for the GO overexpressors, and Jean-Marc Deragon for the gcn2-1 strain complemented with a wild-type GCN2 gene. This work was supported by the National Science Foundation (grants IOS-1456988 and NSF-MCB 1546402 to A.G.v.A.), the Donald L. Akers Jr. Faculty Enrichment Fellowship (to A.G.v.A), the National Science Foundation (grant IOS-1557437 to B.D.), and the National Institutes of General Medical Sciences (grant 1R01GM125743 to B.D.).
AUTHOR CONTRIBUTIONS
A.L., J.G., S.K.C., R.A.U.C., M.S., B.D., and A.G.v.A. designed the research; A.L., J.G., S.K.C., R.A.U.C., P.W.M., M.L., and M.S. performed the research; A.L., J.G., S.K.C., R.A.U.C., M.S., B.D., and A.G.v.A. analyzed data; A.L., B.D., and A.G.v.A. wrote the article.
References
- Alexa A., Rahnenfuhrer J. (2016). topGO: Enrichment analysis for gene ontology. R package version 2.26.0; http://www.mpi-sb.mpg.de/alexa. (Accessed October 30, 2018) [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Anda S., Zach R., Grallert B.(2017). Activation of Gcn2 in response to different stresses. PLoS One 12: e0182143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbudak N., Tezcan H., Akbudak B., Seniz V.(2006). The effect of harpin protein on plant growth parameters, leaf chlorophyll, leaf colour and percentage rotten fruit of pepper plants inoculated with Botrytis cinerea. Sci. Hortic. (Amsterdam) 109: 107–112. [Google Scholar]
- Asada K.(2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141: 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benina M., Ribeiro D.M., Gechev T.S., Mueller-Roeber B., Schippers J.H.(2015). A cell type-specific view on the translation of mRNAs from ROS-responsive genes upon paraquat treatment of Arabidopsis thaliana leaves. Plant Cell Environ. 38: 349–363. [DOI] [PubMed] [Google Scholar]
- Camejo D., Guzmán-Cedeño Á., Moreno A.(2016). Reactive oxygen species, essential molecules, during plant-pathogen interactions. Plant Physiol. Biochem. 103: 10–23. [DOI] [PubMed] [Google Scholar]
- Choudhury F.K., Rivero R.M., Blumwald E., Mittler R.(2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90: 856–867. [DOI] [PubMed] [Google Scholar]
- Crisp P.A., Ganguly D.R., Smith A.B., Murray K.D., Estavillo G.M., Searle I., Ford E., Bogdanović O., Lister R., Borevitz J.O., Eichten S.R., Pogson B.J.(2017). Rapid recovery gene downregulation during excess-light stress and recovery in Arabidopsis. Plant Cell 29: 1836–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F., Brosché M., Shapiguzov A., He X.-Q., Vainonen J.P., Leppälä J., Trotta A., Kangasjärvi S., Salojärvi J., Kangasjärvi J., Overmyer K.(2019). Interaction of methyl viologen-induced chloroplast and mitochondrial signalling in Arabidopsis. Free Radic. Biol. Med. 134: 555–566. [DOI] [PubMed] [Google Scholar]
- Czarnocka W., Karpiński S.(2018). Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 122: 4–20. [DOI] [PubMed] [Google Scholar]
- Das K., Roychoudhury A.(2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2: 53. [Google Scholar]
- Demidchik V.(2015). Mechanisms of oxidative stress in plants: From classical chemistry to cell biology. Environ. Exp. Bot. 109: 212–228. [Google Scholar]
- Dennis M.D., Person M.D., Browning K.S.(2009). Phosphorylation of plant translation initiation factors by CK2 enhances the in vitro interaction of multifactor complex components. J. Biol. Chem. 284: 20615–20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K.J., Turkan I., Krieger-Liszkay A.(2016). Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol. 171: 1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A.G.(2000). Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 6: 269–279. [DOI] [PubMed] [Google Scholar]
- Dong Y., et al. (2017). Sulfur availability regulates plant growth via glucose-TOR signaling. Nat. Commun. 8: 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Duncan C.D.S., Rodríguez-López M., Ruis P., Bähler J., Mata J.(2018). General amino acid control in fission yeast is regulated by a nonconserved transcription factor, with functions analogous to Gcn4/Atf4. Proc. Natl. Acad. Sci. USA 115: E1829–E1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehonen S., Yarmolinsky D., Kollist H., Kangasjärvi J.(2019). Reactive oxygen species, photosynthesis, and environment in the regulation of stomata. Antioxid. Redox Signal. 30: 1220–1237. [DOI] [PubMed] [Google Scholar]
- Enganti R., Cho S.K., Toperzer J.D., Urquidi-Camacho R.A., Cakir O.S., Ray A.P., Abraham P.E., Hettich R.L., von Arnim A.G.(2018). Phosphorylation of ribosomal protein RPS6 integrates light signals and circadian clock signals. Front. Plant Sci. 8: 2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahnenstich H., Scarpeci T.E., Valle E.M., Flügge U.I., Maurino V.G.(2008). Generation of hydrogen peroxide in chloroplasts of Arabidopsis overexpressing glycolate oxidase as an inducible system to study oxidative stress. Plant Physiol. 148: 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faus I., Niñoles R., Kesari V., Llabata P., Tam E., Nebauer S.G., Santiago J., Hauser M.T., Gadea J.(2018). Arabidopsis ILITHYIA protein is necessary for proper chloroplast biogenesis and root development independent of eIF2α phosphorylation. J. Plant Physiol. 224–225: 173–182. [DOI] [PubMed] [Google Scholar]
- Faus I., Zabalza A., Santiago J., Nebauer S.G., Royuela M., Serrano R., Gadea J.(2015). Protein kinase GCN2 mediates responses to glyphosate in Arabidopsis. BMC Plant Biol. 15: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G.(2016). Stress-triggered redox signalling: What’s in pROSpect? Plant Cell Environ. 39: 951–964. [DOI] [PubMed] [Google Scholar]
- Fujii T., Yokoyama E., Inoue K., Sakurai H.(1990). The sites of electron donation of photosystem I to methyl viologen. Biochim. Biophys. Acta Bioenerg. 1015: 41–48. [Google Scholar]
- Gadjev I., Vanderauwera S., Gechev T.S., Laloi C., Minkov I.N., Shulaev V., Apel K., Inzé D., Mittler R., Van Breusegem F.(2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141: 436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T., et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo Y., Kulasekaran S., Benito P., López B., Marcos R., Cascón T., Hamberg M., Castresana C.(2018). Arabidopsis nonresponding to oxylipins locus NOXY7 encodes a yeast GCN1 homolog that mediates noncanonical translation regulation and stress adaptation. Plant Cell Environ. 41: 1438–1452. [DOI] [PubMed] [Google Scholar]
- Jacques S., Ghesquière B., Van Breusegem F., Gevaert K.(2013). Plant proteins under oxidative attack. Proteomics 13: 932–940. [DOI] [PubMed] [Google Scholar]
- Juntawong P., Bailey-Serres J.(2012). Dynamic light regulation of translation status in Arabidopsis thaliana. Front. Plant Sci. 3: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R., Girke T., Bray E.A., Bailey-Serres J.(2004). Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 38: 823–839. [DOI] [PubMed] [Google Scholar]
- Khandal D., Samol I., Buhr F., Pollmann S., Schmidt H., Clemens S., Reinbothe S., Reinbothe C.(2009). Singlet oxygen-dependent translational control in the tigrina-d.12 mutant of barley. Proc. Natl. Acad. Sci. USA 106: 13112–13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorobrykh S.A., Karonen M., Tyystjärvi E.(2015). Experimental evidence suggesting that H2O2 is produced within the thylakoid membrane in a reaction between plastoquinol and singlet oxygen. FEBS Lett. 589: 779–786. [DOI] [PubMed] [Google Scholar]
- Kim C., Apel K.(2013). 1O2-mediated and EXECUTER-dependent retrograde plastid-to-nucleus signaling in norflurazon-treated seedlings of Arabidopsis thaliana. Mol. Plant 6: 1580–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen J.H., Rødland G.E., Bøe C.A., Håland T.W., Sunnerhagen P., Grallert B., Boye E.(2015). Stress-induced inhibition of translation independently of eIF2α phosphorylation. J. Cell Sci. 128: 4420–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageix S., Lanet E., Pouch-Pélissier M.N., Espagnol M.C., Robaglia C., Deragon J.M., Pélissier T.(2008). Arabidopsis eIF2α kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC Plant Biol. 8: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.W., AuYeung W.K., Lam H.M.(2013). The GCN2 homologue in Arabidopsis thaliana interacts with uncharged tRNA and uses Arabidopsis eIF2α molecules as direct substrates. Plant Biol (Stuttg) 15: 13–18. [DOI] [PubMed] [Google Scholar]
- Li Z., Wakao S., Fischer B.B., Niyogi K.K.(2009). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60: 239–260. [DOI] [PubMed] [Google Scholar]
- Liu M.J., Wu S.H., Chen H.M., Wu S.H.(2012). Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol. Syst. Biol. 8: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Afrin T., Pajerowska-Mukhtar K.M.(2019). Arabidopsis GCN2 kinase contributes to ABA homeostasis and stomatal immunity. Commun Biol 2: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llabata P., Richter J., Faus I., Słomiňska-Durdasiak K., Zeh L.H., Gadea J., Hauser M.T.(2019). Involvement of the eIF2α kinase GCN2 in UV-B responses. Front. Plant Sci. 10: 1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokdarshi A., Conner W.C., McClintock C., Li T., Roberts D.M.(2016). Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol. 170: 1046–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo A., Mühlenbock P., Rustérucci C., Chang C.C., Miszalski Z., Karpinska B., Parker J.E., Mullineaux P.M., Karpinski S.(2004). LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 136: 2818–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell D.P., Wang Y., McIntosh L.(1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 96: 8271–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C., Stepanova A.N., Alonso J.M.(2017). Translation regulation in plants: An interesting past, an exciting present and a promising future. Plant J. 90: 628–653. [DOI] [PubMed] [Google Scholar]
- Meskauskiene R., Nater M., Goslings D., Kessler F., op den Camp R., Apel K.(2001). FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98: 12826–12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignolet-Spruyt L., Xu E., Idänheimo N., Hoeberichts F.A., Mühlenbock P., Brosché M., Van Breusegem F., Kangasjärvi J.(2016). Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 67: 3831–3844. [DOI] [PubMed] [Google Scholar]
- Missra A., Ernest B., Lohoff T., Jia Q., Satterlee J., Ke K., von Arnim A.G.(2015). The circadian clock modulates global daily cycles of mRNA ribosome loading. Plant Cell 27: 2582–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missra A., von Arnim A.G.(2014). Analysis of mRNA translation states in Arabidopsis over the diurnal cycle by polysome microarray. Methods Mol. Biol. 1158: 157–174. [DOI] [PubMed] [Google Scholar]
- Mittler R.(2017). ROS are good. Trends Plant Sci. 22: 11–19. [DOI] [PubMed] [Google Scholar]
- Moore M., Gossmann N., Dietz K.J.(2016). Redox regulation of cytosolic translation in plants. Trends Plant Sci. 21: 388–397. [DOI] [PubMed] [Google Scholar]
- Mubarakshina M.M., Ivanov B.N.(2010). The production and scavenging of reactive oxygen species in the plastoquinone pool of chloroplast thylakoid membranes. Physiol. Plant. 140: 103–110. [DOI] [PubMed] [Google Scholar]
- Mullineaux P.M., Exposito-Rodriguez M., Laissue P.P., Smirnoff N.(2018). ROS-dependent signalling pathways in plants and algae exposed to high light: Comparisons with other eukaryotes. Free Radic. Biol. Med. 122: 52–64. [DOI] [PubMed] [Google Scholar]
- Muñoz P., Munné-Bosch S.(2018). Photo-oxidative stress during leaf, flower and fruit development. Plant Physiol. 176: 1004–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie E.H., Lawson T.(2013). Chlorophyll fluorescence analysis: A guide to good practice and understanding some new applications. J. Exp. Bot. 64: 3983–3998. [DOI] [PubMed] [Google Scholar]
- op den Camp, R.G., Przybyla, D., Ochsenbein, C., Laloi, C., Kim, C., Danon, A., Wagner, D., Hideg, E., Göbel, C., Feussner, I., Nater, M., Apel, K. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332. [DOI] [PMC free article] [PubMed]
- Ozgur R., Turkan I., Uzilday B., Sekmen A.H.(2014). Endoplasmic reticulum stress triggers ROS signalling, changes the redox state, and regulates the antioxidant defence of Arabidopsis thaliana. J. Exp. Bot. 65: 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradiso A., Caretto S., Leone A., Bove A., Nisi R., De Gara L.(2016). ROS production and scavenging under anoxia and re-oxygenation in Arabidopsis cells: A balance between redox signaling and impairment. Front. Plant Sci. 7: 1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B., Lee S., Majewski I.J., Alexander W.S., Smyth G.K.(2016). Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 10: 946–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2018). R: A language and environment for statistical computing. (Vienna, Austria: R Foundation for Statistical Computing). Available at: http://www.R-project.org. [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.(2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt F.J., Renger G., Friedrich T., Kreslavski V.D., Zharmukhamedov S.K., Los D.A., Kuznetsov V.V., Allakhverdiev S.I.(2014). Reactive oxygen species: Re-evaluation of generation, monitoring and role in stress-signaling in phototrophic organisms. Biochim. Biophys. Acta 1837: 835–848. [DOI] [PubMed] [Google Scholar]
- Sesma A., Castresana C., Castellano M.M.(2017). Regulation of translation by TOR, eIF4E and eIF2α in plants: Current knowledge, challenges and future perspectives. Front. Plant Sci. 8: 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton D., Smirnova J.B., Selley J.N., Carroll K., Hubbard S.J., Pavitt G.D., Ashe M.P., Grant C.M.(2006). Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 281: 29011–29021. [DOI] [PubMed] [Google Scholar]
- Stonebloom S., Brunkard J.O., Cheung A.C., Jiang K., Feldman L., Zambryski P.(2012). Redox states of plastids and mitochondria differentially regulate intercellular transport via plasmodesmata. Plant Physiol. 158: 190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P.(2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano H.K., Beffa R., Preston C., Westra P., Dayan F.E.(2019). Reactive oxygen species trigger the fast action of glufosinate. Planta 249: 1837–1849. [DOI] [PubMed] [Google Scholar]
- Tang L., Bhat S., Petracek M.E.(2003). Light control of nuclear gene mRNA abundance and translation in tobacco. Plant Physiol. 133: 1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H.(2016). Control of root growth and development by reactive oxygen species. Curr. Opin. Plant Biol. 29: 57–63. [DOI] [PubMed] [Google Scholar]
- Vaahtera L., Brosché M., Wrzaczek M., Kangasjärvi J.(2014). Specificity in ROS signaling and transcript signatures. Antioxid. Redox Signal. 21: 1422–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim A.G., Jia Q., Vaughn J.N.(2014). Regulation of plant translation by upstream open reading frames. Plant Sci. 214: 1–12. [DOI] [PubMed] [Google Scholar]
- Wek R.C.(2018). Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb. Perspect. Biol. 10: a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek S.A., Zhu S., Wek R.C.(1995). The histidyl-tRNA synthetase-related sequence in the eIF-2 α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 15: 4497–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Irizarry R., MacDonald J., Gentry J. (2018). gcrma: Background adjustment using sequence information. R package version 2.54.0. [Google Scholar]
- Zhang Y., Wang Y., Kanyuka K., Parry M.A., Powers S.J., Halford N.G.(2008). GCN2-dependent phosphorylation of eukaryotic translation initiation factor-2α in Arabidopsis. J. Exp. Bot. 59: 3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S.(2010). Pathogen-associated molecular pattern-triggered immunity: Veni, vidi...? Plant Physiol. 154: 551–554. [DOI] [PMC free article] [PubMed] [Google Scholar]