Unlike in Arabidopsis, loss of function of Polymerase IV in the Brassicaceae species Capsella rubella causes microspore arrest, revealing an important role for Polymerase IV in pollen development.
Abstract
In Arabidopsis (Arabidopsis thaliana), DNA-dependent RNA polymerase IV (Pol IV) is required for the formation of transposable element (TE)-derived small RNA transcripts. These transcripts are processed by DICER-LIKE3 into 24-nucleotide small interfering RNAs (siRNAs) that guide RNA-directed DNA methylation. In the pollen grain, Pol IV is also required for the accumulation of 21/22-nucleotide epigenetically activated siRNAs, which likely silence TEs via post-transcriptional mechanisms. Despite this proposed role of Pol IV, its loss of function in Arabidopsis does not cause a discernible pollen defect. Here, we show that the knockout of NRPD1, encoding the largest subunit of Pol IV, in the Brassicaceae species Capsella (Capsella rubella), caused postmeiotic arrest of pollen development at the microspore stage. As in Arabidopsis, all TE-derived siRNAs were depleted in Capsella nrpd1 microspores. In the wild-type background, the same TEs produced 21/22-nucleotide and 24-nucleotide siRNAs; these processes required Pol IV activity. Arrest of Capsella nrpd1 microspores was accompanied by the deregulation of genes targeted by Pol IV-dependent siRNAs. TEs were much closer to genes in Capsella compared with Arabidopsis, perhaps explaining the essential role of Pol IV in pollen development in Capsella. Our discovery that Pol IV is functionally required in Capsella microspores emphasizes the relevance of investigating different plant models.
INTRODUCTION
In flowering plants, male and female gametes are the products of a multistep process that starts from a cell undergoing a meiotic division, resulting in haploid spores that divide mitotically to form multicellular gamete-containing gametophytes. The pollen grain corresponds to the male gametophyte and forms after two mitotic divisions of the haploid microspore. The first mitotic division generates the large vegetative cell and a small generative cell that after another mitotic division will give rise to the two sperm cells. The second mitotic division can occur before pollen germination, as in Arabidopsis (Arabidopsis thaliana), or during pollen germination (Berger and Twell, 2011). Unlike in the male lineage, where all microspores will develop into pollen, in most flowering plants, only one megaspore survives and mitotically divides to give rise to the seven-celled female gametophyte, containing the two female gametes, the egg cell and the central cell (Tekleyohans et al., 2017). In contrast to animals, where germ cells separate early from the somatic lineage, plant germ cells originate from differentiated cells that acquire the competence to undergo meiotic divisions (Schmidt et al., 2015)
The formation of male and female plant gametes is associated with the partial resetting of epigenetic marks, which likely serves to achieve meiotic competence (Borges and Martienssen, 2013; Baroux and Autran, 2015; Borg and Berger, 2015). Epigenetic modifications can be applied directly on the DNA in the form of DNA methylation or on histones, the proteins that package DNA into nucleosomes. The specific type of modification and its position on the genomic locus define the transcriptional outcome. DNA methylation is generally (but not always) a repressive modification and is used to silence transposable elements (TEs) and genes during specific stages of plant development. In plants, DNA methylation can occur in the CG, CHG, and CHH contexts (where H corresponds to A, T, or C) and is established and maintained by different DNA methyltransferases. Methylation in the CG context is maintained by METHYLTRANSFERASE1, while the maintenance of CHG methylation requires CHROMOMETHYLASE3 (CMT3) and to a lesser extent CMT2 (Zhang et al., 2018). The RNA-directed DNA methylation (RdDM) pathway maintains CHH methylation by recruiting DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2). This pathway requires the plant-specific DNA-dependent RNA polymerases (Pol) IV and V (Xie et al., 2004; Herr et al., 2005; Onodera et al., 2005). Pol IV generates small (∼30–40-nucleotide) transcripts that are converted into double-stranded RNA by the action of RNA-DEPENDENT RNA POLYMERASE2 (RDR2; Blevins et al., 2015; Li et al., 2015; Zhai et al., 2015a). These double-stranded RNAs are processed into 23- and 24-nucleotide small interfering RNAs (siRNAs) by DICER-LIKE3 (DCL3; Xie et al., 2004; Singh et al., 2019). The 24-nucleotide siRNAs preferentially associate with ARGONAUTE4 and guide DRM2 to its targets by associating with Pol V transcripts (Cao and Jacobsen, 2002; Zilberman et al., 2003; Wierzbicki et al., 2009). Recent studies further uncovered that Pol IV is required for the accumulation of 21/22-nucleotide epigenetically activated siRNAs (easiRNAs) in pollen. Their biogenesis is triggered by microRNAs (miRNAs) such as miRNA845b and requires DCL2 and DCL4 (Borges et al., 2018). In addition, Pol IV establishes a hybridization barrier between plants of different ploidy grades (Borges et al., 2018; Martinez et al., 2018; Satyaki and Gehring, 2019).
Pollen formation in Arabidopsis is associated with the reprogramming of CHH methylation. There is a strong reduction in CHH methylation during meiosis, which is followed by a restoration of CHH methylation in the pollen vegetative cell and a locus-specific restoration in sperm (Calarco et al., 2012; Ibarra et al., 2012; Walker et al., 2018). Nevertheless, CHH methylation is not completely erased during meiosis, and locus-specific CHH methylation was shown to be of functional relevance for meiosis (Walker et al., 2018). In Arabidopsis, the accumulation of meiosis-specific small RNAs (sRNAs) depends on Pol IV (Huang et al., 2019). Meiotic defects have been reported in the RdDM pathway mutants rdr2, drm2, and ago4, albeit at low frequency in the Columbia (Col) accession (Oliver et al., 2016; Walker et al., 2018). In addition, a relaxation of heterochromatin occurs in the vegetative cell as a consequence of histone H1 depletion, which allows the DNA demethylase DEMETER (DME) to access and demethylate TEs in the vegetative cell (Slotkin et al., 2009; He et al., 2019). Demethylated TEs in the vegetative cell generate siRNAs that can move to sperm cells and may serve to enforce TE silencing in sperm (Ibarra et al., 2012; Martínez et al., 2016; Kim et al., 2019). Nevertheless, the functional relevance of enhanced TE silencing in sperm by mobile siRNAs remains to be demonstrated, since the loss of DME function in pollen causes a pollen germination defect but not a seed defect (Schoft et al., 2011).
Arabidopsis differs from many other species by its low repeat content of ∼24% of the 135-Mb genome (Maumus and Quesneville, 2014), which may explain its apparent tolerance to the lack of Pol IV and other RdDM components. Consistent with this notion, species with higher repeat and TE load suffer from reproductive defects upon the loss or impaired function of Pol IV; these defects range from sterility in tomato (Solanum lycopersicum; Gouil and Baulcombe, 2016) to maternal seed defects in Brassica rapa (Grover et al., 2018). Like Arabidopsis, the closely related Brassicaceae species Capsella (Capsella rubella) can reproduce by selfing; however, because of its recent transition to selfing (30,000–100,000 years ago), it has maintained high numbers of TEs and almost half of the 219-Mb genome is repetitive, with many TEs being located in the vicinity of genes (Foxe et al., 2009; Guo et al., 2009; Slotte et al., 2013; Niu et al., 2019). By contrast, Arabidopsis could reproduce by selfing ∼500,000 years ago and experienced a strong reduction in TE number (de la Chaux et al., 2012).
Given the different TE contents in both species, we hypothesized that the loss of function of Pol IV would have a stronger impact on Capsella compared with Arabidopsis, possibly resembling the defects in B. rapa (Grover et al., 2018). In this study, to test this hypothesis, we generated a loss-of-function allele in the Capsella Pol IV subunit NRPD1. We found that loss of NRPD1 function in Capsella caused impaired male fertility due to the postmeiotic arrest of pollen at the microspore stage. Wild-type microspores accumulated Pol IV-dependent siRNAs ranging from 21- to 24-nucleotides in size, suggesting that the formation of easiRNAs is initiated during or after meiosis. In Capsella and Arabidopsis microspores, 21/22-nucleotide and 24-nucleotide siRNAs were generated from the same TE loci, suggesting that Pol IV produces the precursors for both types of siRNAs. Consistently, the loss of DCL3 in Arabidopsis caused increased formation of 21/22-nucleotide siRNAs, supporting the idea that different DCLs compete for the same double-stranded RNA precursor molecule. Microspore arrest in Capsella nrpd1 mutant plants correlated with a substantially stronger deregulation of genes compared with Arabidopsis nrpd1 microspores, including genes encoding known regulators of pollen development. These findings indicate that Pol IV in Capsella generates siRNAs ranging from 21- to 24-nucleotides in size with important functional roles in pollen development.
RESULTS
Loss of Pol IV Function Causes Microspore Arrest in Capsella
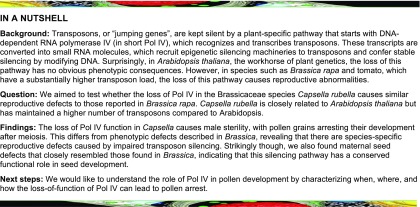
To explore the functional role of Pol IV in Capsella, we generated a knockout mutant in the NRPD1 subunit-encoding gene of Pol IV using CRISPR/Cas9 (hereafter referred to as Cr nrpd1; Figure 1A). The induced deletion caused a frame shift leading to a stop codon after 385 amino acids, resulting in the production of a truncated protein without the catalytic site (Onodera et al., 2005) that is likely a functional null allele. Like the Arabidopsis nrpd1 mutant (referred to as At nrpd1), TE-derived 24- nucleotide siRNAs were abolished in Cr nrpd1 leaves (Figure 1B; Wierzbicki et al., 2012), which was associated with a strong reduction in CHH methylation levels over TEs (Figure 1C). These results reveal that Pol IV has a conserved function in siRNA biogenesis and is required for RdDM in Capsella and Arabidopsis.
Figure 1.
Disruption of NRPD1 in Capsella Impairs 24-nucleotide siRNA Formation and RdDM.
(A) Deleted genomic region in Capsella NRPD1 at 1634 to 2108 bp (genomic sequence). Target 1 (T1) and target 2 (T2) sequences of CRISPR/Cas9 are indicated. AA, amino acids; CDS, coding sequence.
(B) Profile of TE-derived siRNAs in Capsella wild-type (wt) and nrpd1 leaves. RPM, reads per million mapped reads.
(C) DNA methylation levels at TEs in Capsella wild-type and nrpd1 leaves.
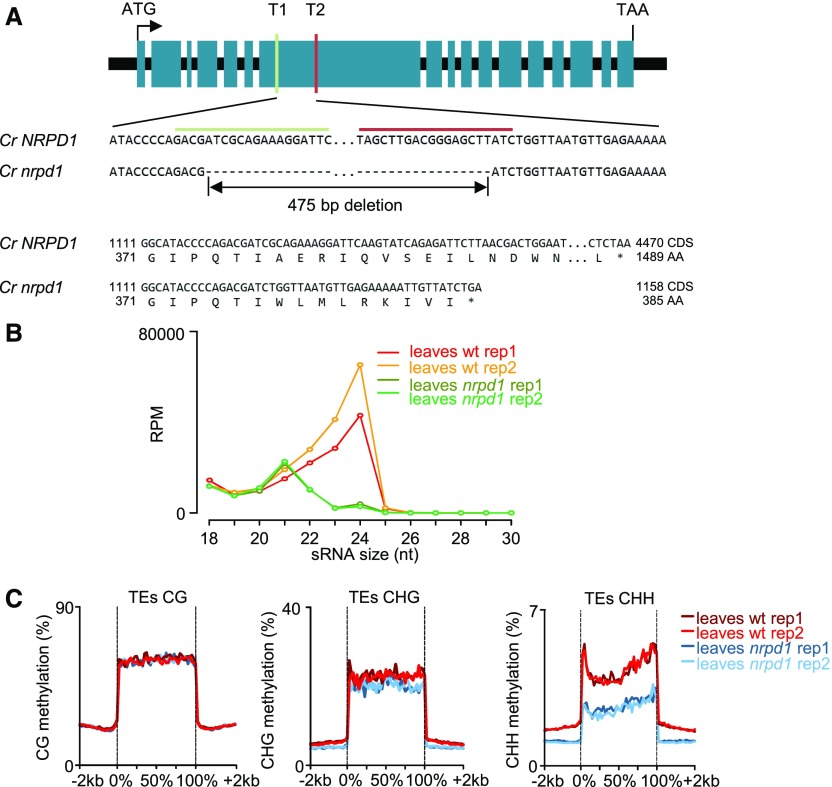
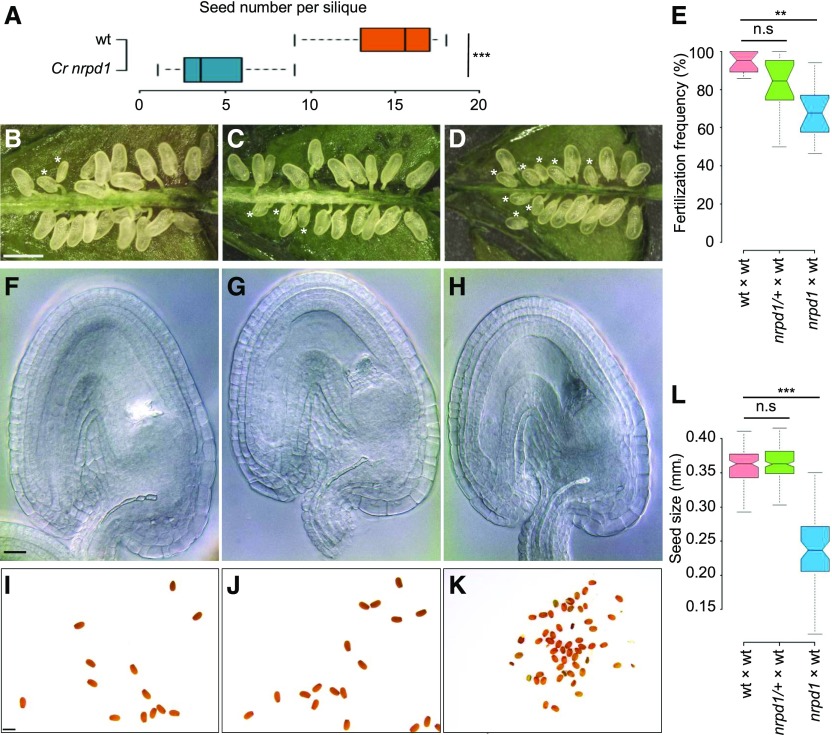
Strikingly, homozygous Cr nrpd1 plants had strongly reduced seed set (Figure 2A): on average, Cr nrpd1 siliques contained only three seeds, corresponding to ∼25% of normal wild-type seed set. The male fertility of Cr nrpd1 was strongly impaired, with most pollen arrested after meiosis at the microspore stage (Figures 3A to 3K). Only homozygous Cr nrpd1 mutants were impaired in pollen development, while heterozygous Cr nrpd1 plants were completely fertile and pollen development was normal (Figures 3C, 3G, and 3K), indicating that the loss of Pol IV function affects a stage prior to microspore development, or alternatively, tapetum development. Observation of cross sections of microsporangia confirmed that Cr nrpd1 mainly formed arrested microspores, with ∼20% of the pollen grains were able to complete development (Figure 3L). There were no obvious differences in tapetum development and degradation in Cr nrpd1 compared with the wild type (Figures 3M to 3T).
Figure 2.
Loss of NRPD1 in Capsella Affects Female Fertility and Causes a Reduction in Seed Size.
(A) Total seed numbers per silique in wild-type (wt; 11 siliques) and Cr nrpd1 mutant (24 siliques) plants. ***, P < 0.001 (Student’s t test). Boxes show medians and the interquartile range, and error bars show the full range excluding outliers.
(B) to (D) Siliques at 2 d after pollination from wt × wt (B), nrpd1/+ × wt (C), and nrpd1 × wt (D) crosses. Asterisks mark unfertilized ovules. Bar = 1 mm.
(E) Fertilization frequency in the indicated crosses. Sixteen siliques per cross combination were analyzed. **, P < 0.01 (Student’s t test). Boxes show medians and the interquartile range, and error bars show the full range excluding outliers. n.s., not significant.
(F) to (H) Ovules at 2 d after emasculation of wt (F), nrpd1/+ (G), and nrpd1 (H) plants. Bar = 20 μm.
(I) to (K) Seeds harvested from wt × wt (I), nrpd1/+ × wt (J), and nrpd1 × wt (K) crosses. Bar = 1 mm.
(L) Seed size of mature seeds derived from wt × wt (n = 120), nrpd1/+ × wt (n = 149), and nrpd1 × wt (n = 64) crosses. ***, P < 0.001 (Student’s t test). Boxes show medians and the interquartile range, and error bars show the full range excluding outliers.
Figure 3.
Cr nrpd1 Pollen Arrest at the Microspore Stage.
(A) to (H) Bright-field ([A]–[D]) and corresponding DAPI ([E]–[H]) staining of manually dissected pollen from anthers at stage 12/13. Pollen of the wild type ([A] and [E]), Cr nrpd1 homozygotes ([B] and [F]), Cr nrpd1 heterozygotes ([C] and [G]), and a complemented line ([D] and [H]) is shown. Bar = 50 μm.
(I) to (K) Confocal images of DAPI-stained pollen of the wild type (I), Cr nrpd1 homozygotes (J), and Cr nrpd1 heterozygotes (K). Bar = 5 μm.
(L) Percentage of mature pollen (MP) in anthers dissected at stage 12/13 from the wild type (wt), Cr nrpd1 homozygotes (nrpd1), Cr nrpd1 heterozygotes (nrpd1/+), and a complemented line (compl.). Pollen was dissected from two individual plants per genotype. Error bars represent sd. Numbers of pollen grains counted in each genotype are shown above the bars.
(M) to (T) Microsporangia cross sections stained with toluidine blue at anther stages 8 ([M] and [O]), 11 ([Q] and [S]), and 12 ([R] and [T]) of the wild type ([M], [Q], and [R]) and Cr nrpd1 ([O], [S], and [T]). Insets in (M) and (O) are shown enlarged in (N) and (P), respectively. Bar for (M), (O), and (Q) to (T) = 50 μm; bar for (N) and (P) = 50 μm.
Consistent with previous work showing a maternal effect of nrpd1 mutants in B. rapa (Grover et al., 2018), ∼30% of ovules of homozygous Cr nrpd1 remained unfertilized after pollination with wild-type pollen, while there was no statistically significant decrease in fertility in heterozygous Cr nrpd1 (Figures 2B to 2E). Inspection of Cr nrpd1 ovules did not reveal obvious abnormalities: wild-type and Cr nrpd1 ovules contained both an egg cell and unfused polar nuclei at 2 d after emasculation (Figures 2F to 2H). Furthermore, Cr nrpd1 homozygous, but not heterozygous, plants had strongly reduced seed size after self-fertilization or pollination with wild-type pollen (Figures 2I to 2L), revealing a maternal effect on ovule and seed development.
Complementation of Cr nrpd1 with the Arabidopsis NRPD1 genomic sequence under the control of the constitutive RPS5A promoter (Weijers et al., 2001) fully restored pollen development in the T1 generation of transgenic plants (Figures 3D, 3H, and 3L), confirming that the pollen defect is a consequence of impaired Pol IV function and that NRPD1 is functionally conserved in Arabidopsis and Capsella.
Meiotic abnormalities at low frequency were previously reported for mutants of the RdDM pathway in Arabidopsis (Oliver et al., 2016; Walker et al., 2018). However, we did not detect abnormal chromosome segregation during male meiosis in Cr nrpd1 (Supplemental Figure 1), and inspection of meiotic products revealed the formation of tetrads (Supplemental Figure 2), indicating that the pollen arrest after meiosis is not due to a defect in chromosome segregation.
Pol IV-Dependent Silencing of TEs in Capsella Microspores
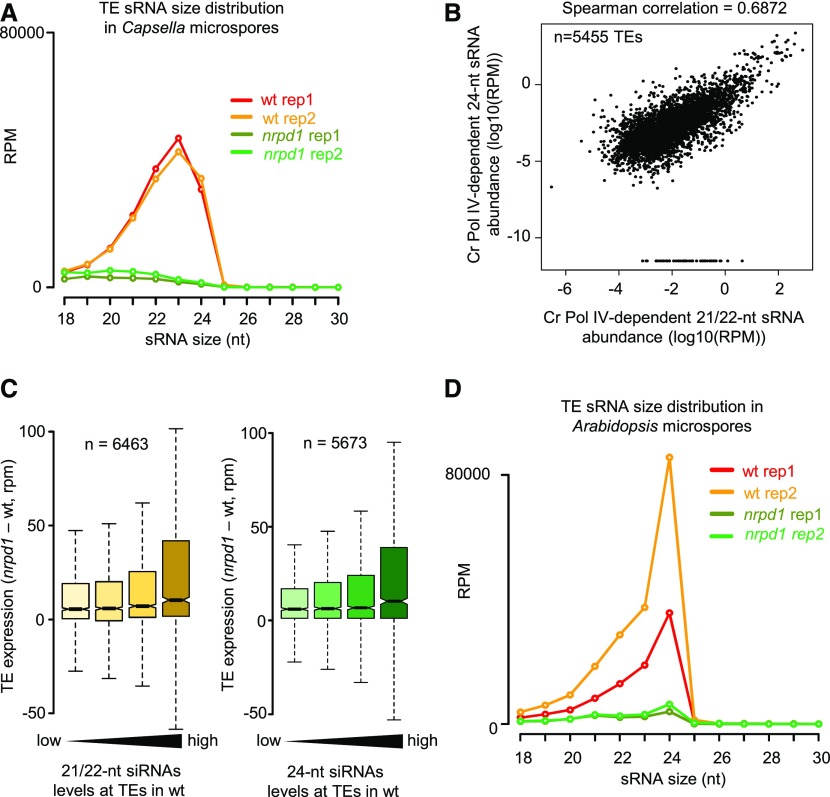
The arrest of Cr nrpd1 pollen at the microspore stage prompted us to compare sRNAs of wild-type and Cr nrpd1 microspores. We enriched for microspores using a Percoll gradient following previously established procedures (Dupl’áková et al., 2016). On average, the purity of the fractions was 84% (Supplemental Figure 3A). Sequencing of isolated sRNAs revealed that TE-derived siRNAs in the 21- to 24-nucleotide size range were abolished in Cr nrpd1 microspores (Figure 4A; Supplemental Figure 4A). As previously described for Arabidopsis meiocytes (Huang et al., 2019), we observed a strong accumulation of 23-nucleotide siRNAs in Capsella microspores (Figures 4A and 5A), which clearly differed from leaves, where 24-nucleotide TE-derived siRNAs were predominantly detected (Figure 1B). Nearly all TE loci generating 21/22-nucleotide siRNAs also formed 24-nucleotide siRNAs (Figure 4B). To investigate the functional role of Pol IV-dependent siRNAs in TE silencing, we isolated RNA from wild-type and mutant microspores and performed RNA sequencing (RNA-seq) analysis. Those TEs that lost 21/22- and 24-nucleotide siRNAs had higher transcript levels in Cr nrpd1 microspores compared with the wild type (Figure 4C), revealing a role for Pol IV-dependent siRNAs in TE silencing in microspores. Together, we conclude that Pol IV-dependent TE-derived siRNAs in the 21- to 24-nucleotide size range are present in microspores and are required for TE silencing. Which size class is required for TE silencing, and the pathway(s) involved, remain to be determined. This finding is consistent with the recent proposal that pollen easiRNAs are produced during or shortly after meiosis (Borges et al., 2018).
Figure 4.
Pol IV Is Required for 21- to 24-nucleotide siRNA Production in Capsella Microspores.
(A) Profile of TE-derived siRNAs in wild-type (wt) and nrpd1 Capsella microspores. RPM, reads per million mapped reads.
(B) Abundance of TE-derived Capsella Pol IV-dependent 21/22-nucleotide siRNAs and 24-nucleotide siRNAs in Capsella microspores. Values are indicated as the log10 of the average RPM of both libraries. Each dot represents one TE for a total of 5455 TEs. A pseudocount of 0.0001 was added to all zero values. The correlation has been tested by a Spearman test (correlation coefficient of 0.6872).
(C) Loss of 21/22-nucleotide and 24-nucleotide siRNAs at TEs is associated with increased transcript levels of TEs in Cr nrpd1 microspores. Increasing accumulation of siRNAs over TEs is plotted from low to high levels of accumulation. The bars from low to high represent quartiles. Only TEs with more siRNAs in the wild type than in Cr nrpd1 are represented. Differences between the first and last categories are significant (P = 3.4e-13 and 1.4e-9, respectively, Wilcoxon test). Boxes show medians and the interquartile range, and error bars show the full range excluding outliers.
(D) sRNA profile of TE-derived siRNAs from Arabidopsis wild-type and nrpd1 microspores.
Figure 5.
Meiocytes, Microspores, and Mature Pollen Grain Accumulate Overlapping Sets of siRNAs.
(A) TE-derived siRNA distribution in Arabidopsis meiocytes of the indicated genetic backgrounds (data from Huang et al., 2019). RPM, reads per million mapped reads; wt, wild type.
(B) TE-derived siRNA distribution in Arabidopsis pollen grains of the indicated genetic backgrounds.
(C) UpSet plot showing the overlap of TEs accumulating 21/22-nucleotide siRNAs or 24-nucleotide siRNAs in Arabidopsis microspores and meiocytes (data from Huang et al., 2019).
(D) UpSet plot showing the overlap of TEs accumulating 21/22-nucleotide siRNAs or 24-nucleotide siRNAs in Arabidopsis microspores and mature pollen grain (MPG; data from Martinez et al., 2018).
Pol IV-Dependent siRNAs Also Accumulate in Arabidopsis Microspores
Pol IV generates small (30 to 40 nucleotides) transcripts that are converted into 23- or 24-nucleotide siRNAs by the action of DCL3 (Blevins et al., 2015; Zhai et al., 2015a; Yang et al., 2016). To genetically dissect the Pol IV-dependent pathway leading to the formation of 21- to 24-nucleotide siRNAs, we tested for the presence of similar siRNAs in Arabidopsis microspores.
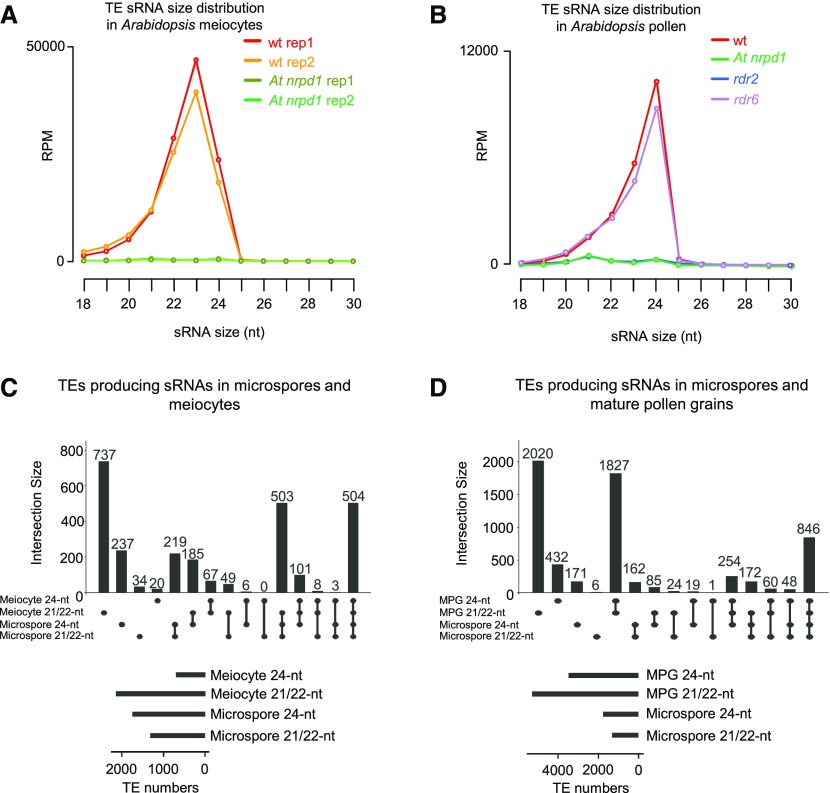
We sequenced sRNAs from Arabidopsis wild-type and nrpd1 microspores that had been enriched to 90% following the same procedures as applied for Capsella (Supplemental Figures 3B and 4B). As in Capsella microspores, Arabidopsis microspores accumulated TE-derived 21- to 24-nucleotide siRNAs, which were abolished in At nrpd1 microspores (Figure 4D). Thus, 21/22-nucleotide TE-derived siRNAs are already present in microspores and their formation depends on Pol IV, strongly supporting the idea that the biogenesis of easiRNAs present in mature pollen starts at an earlier stage, most likely during meiosis. Consistently, microspores and meiocytes as well as microspores and mature pollen grains share a large number of loci generating Pol IV-dependent siRNAs (Figures 5A to 5D).
To investigate the functional requirement for Pol IV-derived siRNAs in TE silencing, we correlated the levels of TEs producing Pol IV-dependent 21/22-nucleotide and 24-nucleotide siRNAs to TE expression changes in At nrpd1, a mutant that lacks Pol IV function. We found a significant association between Pol IV-dependent siRNA levels and expression change of TEs in At nrpd1 microspores (Figure 6A). These results, which are similar to the findings for Capsella (Figure 4C), reveal that TE-derived siRNAs are involved in the repression of TE expression in microspores.
Figure 6.
Pol IV/RDR2 Generate Templates for 21- to 24-nucleotide siRNAs.
(A) Loss of Pol IV-dependent 21/22-nucleotide easiRNAs is associated with increased transcript levels of TEs in Arabidopsis microspores. Increasing accumulation of siRNAs over TEs is plotted from low to high levels of accumulation. The bars from low to high represent quartiles. In both plots, siRNA levels at TEs in the wild type (wt) increase from left to right in quantiles. Differences between the first and last categories are significant (P = 2.6e-10 and 1.5 e-14, respectively, Wilcoxon test). Boxes show medians and the interquartile range, and error bars show the full range excluding outliers. RPM, reads per million mapped reads.
(B) Abundance of At Pol IV-dependent 21/22-nucleotide siRNAs and 24-nucleotide siRNAs at TEs in Arabidopsis microspores. Values are indicated as log10 of the average RPM of both libraries. Each dot represents one TE for a total of 1504 TEs. A pseudocount of 0.0001 was added to all zero values. The correlation has been tested by a Spearman test (correlation coefficient of 0.7686).
(C) Plots showing the distribution of the ratio of the number of reads mapped against the positive strand to the total number of mapped reads. Left plots show analysis for 21/22-nucleotide reads and right plots for 24-nucleotide reads.
(D) TE-derived siRNA distribution in inflorescences of rdr2 (left panel; data from Zhai et al., 2015a) and rdr6 (right panel; data from Panda et al., 2016).
(E) Average total 21/22-nucleotide or 24-nucleotide reads mapping against TEs in wild-type or dcl3 libraries (data from Li et al., 2015). Reads were normalized to show RPM values. Errors bars represent sd of two biological replicates.
(F) Average total 21/22-nucleotide reads mapping against miRNAs in wild-type or dcl3 libraries (data from Li et al., 2015). Reads were normalized to show RPM values. Errors bars represent sd of two biological replicates.
Pol IV is usually associated with 24-nucleotide siRNAs through the RdDM pathway, and its strong effect on the production of 21/22-nucleotide siRNAs in pollen is thus unexpected. One hypothesis is that Pol IV transcripts are direct precursors of 21/22-nucleotide siRNAs. If true, we would expect that TE-derived 21/22-nucleotide and 24-nucleotide siRNAs in microspores should arise from the same genomic loci. Consistently, as in Capsella microspores (Figure 4B), nearly all TE loci generating 21/22-nucleotide siRNAs also formed 24-nucleotide siRNAs (Figure 6B). Visualizing the individual reads in a genome browser showed that all read sizes accumulated along the same loci (Supplemental Figure 5). Taken together, these data indicate that TE-derived 21/22-nucleotide siRNAs and 24-nucleotide siRNAs are produced from the same loci in a Pol IV-dependent manner and that at least a subset of these siRNAs are able to repress TEs in both Capsella and Arabidopsis.
TE-Derived siRNA Production in Microspores Requires RDR2 Activity
TE-derived siRNAs were produced from both DNA strands (Figure 6C; Supplemental Figure 5), suggesting that an RNA-dependent RNA polymerase is involved in the production of the double-stranded RNAs used as templates for microspore TE-derived siRNA production.
Among the three RDRs with known functions in Arabidopsis, RDR2 is tightly associated with Pol IV (Li et al., 2015; Zhai et al., 2015a), and RDR6 has been shown to affect the production of some easiRNAs (Creasey et al., 2014; Martinez et al., 2018). To assess the potential involvement of RDR2 and RDR6 in TE-derived siRNA production in microspores, we analyzed publicly available sRNA sequencing data from rdr2 and rdr6 inflorescences (Zhai et al., 2015a; Panda et al., 2016). The sRNA pattern of wild-type and At nrpd1 inflorescence tissues was comparable to that of microspores (Figures 4D and 6D), indicating that the siRNAs identified in rdr2 and rdr6 inflorescences are comparable to those in microspores. While rdr6 showed a normal distribution pattern of TE-derived siRNAs, in rdr2 inflorescences, the accumulation of TE-derived siRNAs was abolished (Figure 6D), indicating that RDR2 is likely involved in TE siRNA biogenesis. We also generated siRNA profiles from rdr2 and rdr6 pollen (Figure 5B), which confirm the data obtained from inflorescences and reveal that RDR2, but not RDR6, is required for the generation of 21/22-nucleotide siRNAs. These results reinforce the idea that 21/22-nucleotide and 24-nucleotide TE-derived siRNAs present in pollen are processed from a double-stranded RNA produced by Pol IV and RDR2.
DICERs Producing TE-Derived siRNAs Compete for the Same Double-Stranded RNA Template
Our data suggest that TE-derived siRNAs of different size classes are derived from double-stranded RNAs produced by Pol IV/RDR2 (Figure 6B). We hypothesized that different size classes of siRNAs are produced due to different DICERs competing for the same double-stranded RNA template, as has been shown to occur upon disruption of DCL3 function (Gasciolli et al., 2005; Henderson et al., 2006; Kasschau et al., 2007; Bond and Baulcombe, 2015). If true, the impairment of DCL3 should increase the proportion of Pol IV-dependent 21/22-nucleotide siRNAs accumulating over defined loci.
To test this idea, we used publicly available sRNA sequencing data from dcl3 inflorescences (Li et al., 2015). We quantified the number of normalized 21/22-nucleotide siRNAs and 24-nucleotide siRNAs mapped against TEs in wild-type and dcl3 inflorescences. In the wild type, 24-nucleotide siRNAs were the most abundant siRNAs, exceeding the level of 21/22-nucleotide siRNAs by nearly sevenfold (Figure 6E). In dcl3, the abundance of 21/22-nucleotide siRNAs was highly increased, while 24-nucleotide siRNAs were depleted (Figure 6E). To rule out the possibility that the increased abundance of 21/22-nucleotide siRNAs is due to a normalization artifact, we performed the same analysis with miRNAs. As DCL3 is not involved in the 21/22-nucleotide miRNA pathway, any changes observed in dcl3 would be suggestive of a normalization artifact. The abundance of 21/22-nucleotide miRNAs in the wild type and dcl3 was highly similar (Figure 6F), strongly supporting the notion that the observed increase in 21/22-nucleotide siRNA levels in dcl3 inflorescences is not due to a normalization problem. These results indicate that there is indeed a competition between DCL3 and other DCLs for the same double-stranded RNA precursor, which is in agreement with previous findings (Gasciolli et al., 2005; Henderson et al., 2006; Kasschau et al., 2007; Bond and Baulcombe, 2015).
TE-Derived siRNAs Are Highly Enriched at COPIA95 in Capsella Microspores
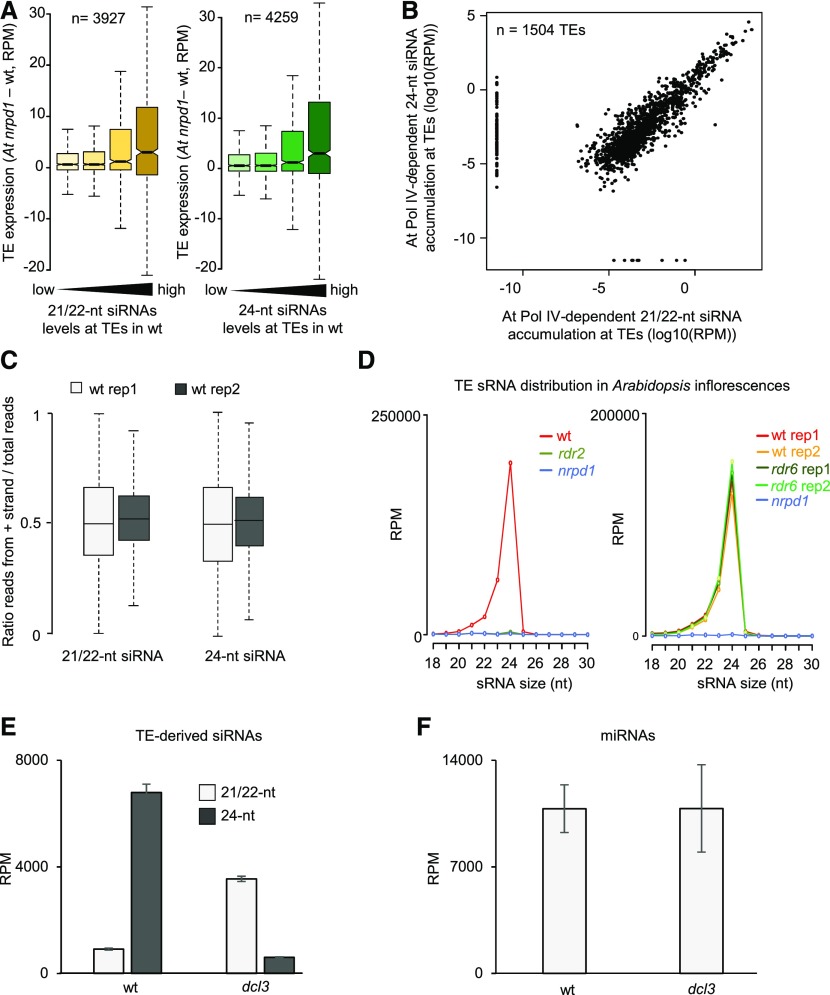
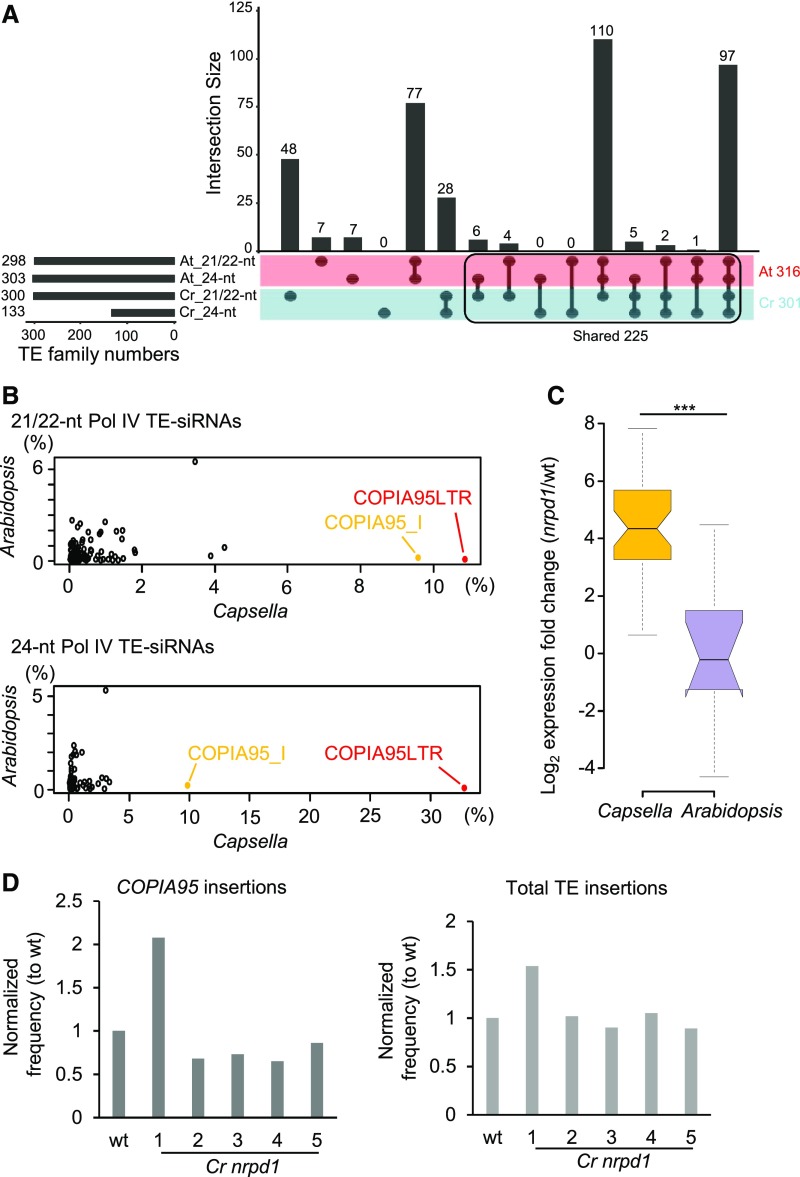
To investigate whether Pol IV-dependent siRNAs target similar loci in Arabidopsis and Capsella, we identified sRNA reads mapping to both genomes, which were then mapped to 527 previously reported Arabidopsis TE consensus sequences (Repbase; Bao et al., 2015). A similar number of TE families accumulated Pol IV-dependent siRNAs (21/22-nucleotide and/or 24-nucleotide siRNAs) in Arabidopsis (n = 316; Figure 7A, red color) and Capsella (n = 301; Figure 7A, cyan color) microspores, 225 of which (Figure 7A, black frame) were common to both species (Figure 7A). There were substantially fewer TE families forming 24-nucleotide siRNAs in Capsella microspores (133) compared with Arabidopsis (303), but the majority of those overlapped between both species (103). Nearly all TE families forming 24-nucleotide siRNAs also formed 21/22-nucleotide siRNAs (94.1% [285 out of 303 TE families] in Arabidopsis and 99.2% [132 out of 133 TE families] in Capsella; Figure 7A), supporting the idea that 21/22-nucleotide and 24-nucleotide siRNAs are derived from the same TE loci in microspores.
Figure 7.
COPIA95-siRNAs Are Highly Enriched in Capsella Microspores.
(A) UpSet plots of TE families accumulating Pol IV-dependent 21/22-nucleotide and 24-nucleotide siRNAs in Arabidopsis (At) and Capsella (Cr).
(B) Proportions of Pol IV-dependent 21/22-nucleotide and 24-nucleotide siRNAs accumulating at specific TE consensus sequences in relation to all TE-siRNAs. Reads mapping to COPIA95 long-terminal repeats (LTR) and internal (I) sequences are highlighted in red and yellow, respectively.
(C) Log2 expression fold change of mRNAs for COPIA95 elements in nrpd1 mutant microspores of Arabidopsis and Capsella compared with the corresponding wild type (wt). ***, P < 0.001 (Wilcoxon test). Boxes show medians and the interquartile range, and error bars show the full range excluding outliers.
(D) Relative number of COPIA95 insertions (left panel) and total TE insertions (right panel) compared with the corresponding wild-type control in five progeny of homozygous Cr nrpd1.
To investigate the specificity of TE-derived siRNAs in Arabidopsis and Capsella microspores, we calculated the proportion of siRNAs targeting specific TE families. Strikingly, nearly 20% of 21/22-nucleotide siRNAs and more than 40% of 24-nucleotide siRNAs were derived from the COPIA95 family (COPIA95 long-terminal repeats and internal) in Capsella microspores, while only 0.3% of both siRNA classes were derived from COPIA95 in Arabidopsis microspores (Figure 7B). We identified 17 and 70 TEs accumulating COPIA95-derived Pol IV-dependent siRNAs in Arabidopsis and Capsella, respectively, indicating that the COPIA95 TE family expanded in Capsella (Supplemental Data Set 1). The prominent targeting of COPIA95 in Capsella microspores by Pol IV-dependent siRNAs prompted the question of whether the loss of Pol IV function may cause increased expression and transposition of COPIA95. Indeed, COPIA95 was highly upregulated in Cr nrpd1 microspores but remained silenced in At nrpd1 microspores (Figure 7C). To test whether increased expression caused heritable transposition, we performed whole-genome sequencing of five homozygous Cr nrpd1 mutants derived from homozygous Cr nrpd1 parental plants. We mapped genomic reads to COPIA95 elements and found that one of the five tested mutants had a twofold increase in the number of COPIA95 elements (Figure 7D). To verify this result, we used the Transposable Element Polymorphism IDentification (TEPID) algorithm to identify de novo insertions (Stuart et al., 2016). Indeed, we detected increased numbers of insertions in the same mutant compared with the wild type (Figure 7D). This result supports the idea that Pol IV is required to prevent TE remobilization in Capsella and is, in particular, required to silence COPIA95.
Loss of Pol IV Causes Transcriptional Changes in Microspores
To explore the cause for postmeiotic arrest of Capsella microspores, we compared the transcriptome changes in At nrpd1 and Cr nrpd1 microspores. A comparable number of genes were upregulated (log2 fold change > 1, P < 0.05) in nrpd1 microspores of both species. However, approximately twice as many genes were downregulated (log2 fold change < −1, P < 0.05) in Cr nrpd1 microspores compared with At nrpd1 (Supplemental Figures 6A and 6B; Supplemental Data Set 2). While there was no significant overlap between downregulated genes in Cr nrpd1 and At nrpd1 microspores (Supplemental Figure 6B), there was a significant overlap of upregulated genes in Cr nrpd1 and At nrpd1. These genes were significantly enriched in Gene Ontology (GO) terms related to stimulus response, cell wall organization, and defense responses (Supplemental Figure 6C).
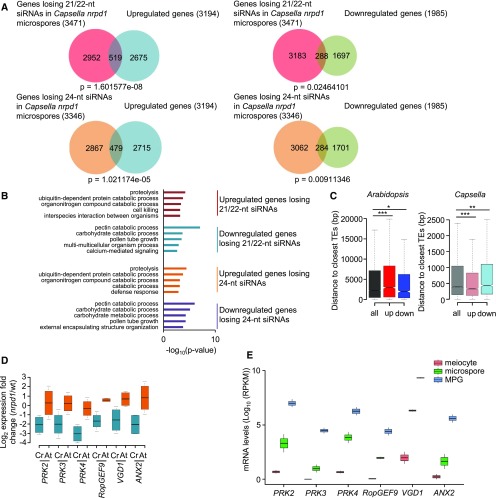
We tested whether deregulated genes in Cr nrpd1 and At nrpd1 microspores were targeted by 21/22-nucleotide or 24-nucleotide siRNAs. We found a significant overlap between upregulated genes and downregulated genes, with genes losing 21/22-nucleotide and 24-nucleotide siRNAs in Cr nrpd1 (Figure 8A; Supplemental Data Set 3). By contrast, in At nrpd1, only downregulated genes significantly overlapped with genes losing 21/22-nucleotide and 24-nucleotide siRNAs (Supplemental Figure 6D; Supplemental Data Set 4). Upregulated genes losing 21/22-nucleotide or 24-nucleotide siRNAs in Cr nrpd1 had functional roles in proteolysis and catabolic processes, cell killing, and interspecies organismal interactions (Figure 8B). The distance of TEs to neighboring genes was significantly shorter in Capsella compared with Arabidopsis, regardless of their direction of deregulation (Figure 8C; Wilcoxon test, P < 2e-15). Nevertheless, upregulated genes in Capsella had an even shorter distance to neighboring TEs than nonderegulated or downregulated genes, suggesting that neighboring TEs have an impact on gene expression in Cr nrpd1 microspores. However, there was no preference for COPIA95 among the TEs that were close to deregulated genes (P = 1, hypergeometric test).
Figure 8.
Deregulated Genes Differ in Arabidopsis and Capsella nrpd1 Mutant Microspores.
(A) Venn diagrams showing the overlap of deregulated genes (log2 fold change > 1, P < 0.05) in nrpd1 microspores of Capsella and genes losing 21/22-nucleotide and 24-nucleotide siRNAs at 2-kb upstream and downstream regions and gene bodies (log2 fold change < −1, P < 0.05) in Capsella nrpd1 microspores.
(B) Enriched GO terms for biological processes of intersected genes losing siRNAs and deregulated genes in Capsella nrpd1 microspores. The top 5 GO terms (P < 0.01) of each analysis are shown.
(C) Distance of Arabidopsis and Capsella genes to the closest TEs. all, all genes; down, significantly downregulated genes; up, significantly upregulated genes. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Wilcoxon test). Boxes show medians and the interquartile range, and error bars show the full range excluding outliers.
(D) Log2 expression fold change of PRK2, PRK3, PRK4, RopGEF9, VGD1, and ANX2 genes in nrpd1 microspores compared with the wild type (wt) in Capsella (Cr) and Arabidopsis (At). Boxes show medians and the interquartile range, and error bars show the full range.
(E) mRNA levels of PRK2, PRK3, PRK4, RopGEF9, VGD1, and ANX2 in Arabidopsis wild-type meiocytes, microspores, and mature pollen grain (MPG). +1 was added to all values to avoid negative log10 values. Boxes show medians and the interquartile range, and error bars show the full range. RPKM, reads per kilobase per million mapped reads.
Interestingly, downregulated genes associated with the loss of 21/22-nucleotide and 24-nucleotide siRNAs in Cr nrpd1 were enriched for genes involved in pollination; among these were known regulators of pollen tube growth such as VANGUARD1 (VGD1), ANXUR2 (ANX2), and RopGEF9 (Figure 8D; Supplemental Figure 7; Jiang et al., 2005; Zhang and McCormick, 2007; Boisson-Dernier et al., 2009). We also identified homologs of pollen receptor kinase-encoding genes PRK2-4 among downregulated genes losing 21/22-nucleotide and 24-nucleotide siRNAs (Figure 8D; Supplemental Figure 7). While there was also a significant overlap of downregulated genes in At nrpd1 with genes losing siRNAs (Supplemental Figure 6D), these genes were not enriched for genes involved in pollination (Supplemental Figure 6E), and the aforementioned genes were not deregulated in At nrpd1 (Figure 8D). The affected PRKs have partially redundant functions in pollen tube growth and the perception of female attractant peptides (Chang et al., 2013; Takeuchi and Higashiyama, 2016). Importantly, RNAi-mediated knockdown of PiPRK1, a PRK homolog in Petunia inflata, causes microspore arrest (Lee et al., 1996), suggesting that the reduced expression of PRKs may contribute to microspore arrest in Cr nrpd1. All genes were highly induced during the transition from microspore to mature pollen formation (Figure 8E), suggesting that their expression is required to ensure viable pollen formation, a hypothesis that remains to be tested.
DISCUSSION
In this study, we demonstrated that the loss of Pol IV function causes arrested microspore development and has a maternal effect on ovule and seed development in Capsella. Therefore, Cr nrpd1 mutants strongly differ from At nrpd1 mutants, which lack obvious reproductive abnormalities (Mosher et al., 2009). Mutations in NRPD1, NRPE1, and RDR2 have a maternal effect on seed development in B. rapa, while no defect in pollen development was reported for these mutants (Grover et al., 2018). The mutation in B. rapa NRPD1 was not a null allele; however, the mutation in RDR2 completely abolished the production of 24-nucleotide siRNAs (Grover et al., 2018), indicating that this mutant was a functional null mutant for RDR2. Since the loss of Cr NRPD1 function had a similar molecular effect to mutations in Arabidopsis and B. rapa NRPD1 (depletion of 24-nucleotide siRNAs and CHH methylation; Wierzbicki et al., 2012; Panda et al., 2016; Grover et al., 2018) and the Cr nrpd1 mutant could be complemented with the Arabidopsis NRPD1 sequence, we conclude that the molecular function of Pol IV is likely conserved between the three species but the targets differ. Interestingly, the loss of Pol IV function in tomato also causes sterility, but the cause for this phenotype remains to be explored (Gouil and Baulcombe, 2016). The microspore arrest in Cr nrpd1 might be a consequence of TEs being in close proximity to essential regulators of microspore development, such as PRK genes, or genes that cause microspore arrest upon overexpression. The distance of TEs to neighboring genes is substantially larger in Arabidopsis compared with Capsella, supporting this notion.
We observed a heritable remobilization of the COPIA95 element in progeny of the Cr nrpd1 mutant. This finding is consistent with the notion that this element is preferentially targeted by Pol IV-generated siRNAs in Capsella and strongly activated in Cr nrpd1 microspores. Interestingly, in Arabidopsis, the COPIA element ONSEN also undergoes transgenerational retrotransposition in nrpd1 after heat treatment, and new ONSEN insertions differ between siblings derived from a single plant (Ito et al., 2011). Since the amplification of COPIA95 was only observed in one of five tested Cr nrpd1 progeny, it is unlikely that the consistently observed microspore arrest is connected to TE remobilization. Alternatively, it is possible that those microspores only survived when TE remobilization did not occur or occurred at low frequency.
Cr nrpd1 microspores were completely depleted of TE-derived siRNAs, including 21/22-nucleotide siRNAs, which are usually not associated with Pol IV (Xie et al., 2004; Blevins et al., 2015; Zhai et al., 2015a). A similar depletion of 21/22-nucleotide siRNAs (easiRNAs) was previously reported in mature pollen grains of At nrpd1 mutants (Borges et al., 2018; Martinez et al., 2018). The biogenesis of easiRNAs is thought to be a consequence of reduced heterochromatin formation in the vegetative cell and the resulting TE activation (Slotkin et al., 2009; Creasey et al., 2014). Based on genetic data, Borges et al. (2018) proposed that easiRNA biogenesis occurs earlier (i.e., during or soon after meiosis). Our data reveal that 21/22-nucleotide TE-derived siRNAs are already present in microspores and, given their similarity to meiocyte siRNAs (Huang et al., 2019), are likely generated before or during meiosis.
In rice (Oryza sativa) and maize (Zea mays), highly abundant 21-nucleotide phased siRNAs (phasiRNAs) accumulate in premeiotic anthers, and 24-nucleotide phasiRNAs are enriched in meiotic stage anthers (Johnson et al., 2009; Komiya et al., 2014; Zhai et al., 2015b). The 21-nucleotide phasiRNAs are important for male fertility in rice, and the disruption of 24-nucleotide phasiRNA production yields conditional male sterility in maize (Fan et al., 2016; Teng et al., 2018). The biogenesis of premeiotic and meiotic phasiRNAs in maize appears to occur in the tapetum rather than in meiocytes, where they accumulate (Zhai et al., 2015b). The production of 24-nucleotide phasiRNAs depends on miR2275, a pathway that is widely present in the eudicots but missing in the Brassicaceae (Xia et al., 2019), suggesting evolutionary divergence of the functional role of phasiRNAs during pollen development. Cross sections did not reveal obvious tapetal defects in Cr nrpd1, indicating that microspore arrest in Cr nrpd1 is not due to a tapetal defect.
The strong dependency of TE-derived siRNA accumulation on Pol IV suggests that Pol IV transcripts are the precursors of all sizes of TE-derived siRNAs in microspores. In the absence of DCL3, other DCL proteins (DCL1, DCL2, and DCL4) are able to process Pol IV transcripts into 21- or 22-nucleotide siRNAs (Gasciolli et al., 2005; Henderson et al., 2006; Kasschau et al., 2007; Bond and Baulcombe, 2015). We thus propose that before or during meiosis, Pol IV transcripts are targeted by other DCLs in addition to DCL3, explaining why all sizes of Pol IV-dependent siRNAs are derived from the same TE loci.
In Arabidopsis siliques, a nucleus-localized form of DCL4 targets Pol IV transcripts and generates 21-nucleotide siRNAs (Pumplin et al., 2016). The abundance of these 21-nucleotide Pol IV-derived siRNAs was nevertheless low, contrasting to the high abundance in microspores. Perhaps the disruption of the nuclear envelope during meiosis allows cytoplasmic DCLs to gain access to Pol IV/RDR2 transcripts. This notion implies that meiosis is the trigger of Pol IV-dependent 21- to 24-nucleotide siRNA production, which is consistent with our genetic data. Not mutually exclusive with this scenario is the possibility that 22-nucleotide siRNAs produced during meiosis trigger secondary 21/22-nucleotide siRNA production in mature pollen grains by targeting TE transcripts expressed in the vegetative cells of pollen (Slotkin et al., 2009). This amplification of the signal by the canonical post-transcriptional gene silencing (PTGS) pathway (Martínez de Alba et al., 2013) should result in highly abundant 21/22-nucleotide siRNAs in mature pollen, which is in agreement with published siRNA profiles of pollen (Borges et al., 2018; Martinez et al., 2018).
We demonstrated that Pol IV-dependent siRNAs are required to silence TEs in microspores. This could be achieved by the canonical and noncanonical RdDM pathway involving 21/22-nucleotide siRNAs (Cuerda-Gil and Slotkin, 2016) and 24-nucleotide siRNAs or, alternatively, by the PTGS pathway. CHH methylation levels are low in meiocytes but are higher in microspores and in the vegetative cells of pollen (Walker et al., 2018). Nevertheless, CHH methylation levels in microspores are very low (Calarco et al., 2012), making it more likely that TE silencing in microspores and subsequently vegetative cells is achieved by PTGS; this notion is consistent with the high accumulation of 21/22-nucleotide siRNAs in mature pollen.
Recent work from our and other groups revealed that disrupting NRPD1 suppresses the hybridization barrier between plants of different ploidy levels (Martinez et al., 2018; Satyaki and Gehring, 2019). However, while Martinez et al. (2018) did not find a suppressive effect when using mutants in RdDM components such as RDR2 and NRPE1, Satyaki and Gehring (2019) found that such mutants suppressed hybrid seed failure. The difference between these studies lies in the use of tetraploid RdDM mutants by Satyaki and Gehring (2019), while RdDM mutants introgressed into omission of second division1 (osd1) were used by Martinez et al. (2018). The mutation of OSD1 suppresses the second meiotic division, leading to unreduced gamete formation (d’Erfurth et al., 2009). Here, we showed that RDR2 is required for easiRNA biogenesis, suggesting that the loss of easiRNAs is not sufficient to suppress the triploid block induced by the osd1 mutation. An important difference between osd1 and tetraploid plants is the ploidy of the genome at the beginning of meiosis, which is diploid and tetraploid, respectively. This difference can have a strong impact, since tetraploid plants undergo DNA methylation changes leading to the production of stable epialleles (Mittelsten Scheid et al., 2003). Biochemical feedback loops link DNA methylation to repressive histone 3 Lys-9 dimethylation that (via the homeodomain protein SHH1) recruit Pol IV (Law et al., 2013; Zhang et al., 2013). Since easiRNAs are generated during meiosis, it is thus possible that the requirement of RdDM activity for easiRNA formation and ploidy barriers may be different depending of the initial ploidy of the plants. If true, the signal establishing the triploid block depends on Pol IV but only indirectly on RdDM, suggesting that both pathways can be separated, as previously proposed for maize endosperm (Erhard et al., 2013).
In summary, our study in Capsella uncovered a functional requirement for Pol IV in microspores, highlighting the notion that Pol IV-dependent siRNA formation occurs earlier than previously hypothesized (Slotkin et al., 2009). We showed that Pol IV generates the precursors for 21- to 24-nucleotide siRNAs, perhaps because different DCLs are able to access Pol IV transcripts during meiosis. Our study highlights the relevance of investigating different plant models to gain novel insights into the molecular control of developmental processes.
METHODS
Plant Growth and Material
Mutants for Arabidopsis (Arabidopsis thaliana) alleles nrpd1-3 (Salk_128428) and dcl3-1 (Salk_005512) were previously described by Xie et al. (2004) and Pontier et al. (2005). For all experiments using Arabidopsis, the Col-0 accession was used as the wild type, while for Capsella (Capsella rubella), accession Cr1GR1 was used.
Seeds of Arabidopsis and Capsella were surface-sterilized in 5% commercial bleach and 0.01% Tween 20 for 10 min and washed three times in sterile distilled, deionized water. Seeds were sown on half-strength Murashige and Skoog medium (0.43% [w/v] Murashige and Skoog salts, 0.8% [w/v] bacto agar, 0.19% [w/v] MES hydrate, and 1% [w/v] Suc). After stratification for 2 d at 4°C, the plates were transferred to a growth chamber (16 h of light/8 h of dark, 110 μmol s−1 m−2, 21°C, 70% humidity). After 10 d, the seedlings were transferred to soil and grown in a growth chamber under a 16-h-light/8-h-dark cycle with light intensity of 150 μE from an Osram FQ 24W/840 HQ Constant Lumilux Cool White light source at 21°C and 70% humidity. Capsella plants were grown in a growth chamber under the same light conditions and light/dark cycle but at 18°C and 60% humidity.
Generation of Plasmids and Transgenic Plants
The Web tool CRISPR-P (http://cbi.hzau.edu.cn/cgi-bin/CRISPR) was used to design the sgRNAs for knocking out Capsella NRPD1 (Carubv10019657m; Lei et al., 2014). Sequence information for the primers containing the two sgRNA sequences is listed in Supplemental Table 1. The primers were used to amplify the fragment including Target1-sgRNA-scaffold-U6-terminator-U6-29promoter-Target2 using plasmid DT1T2-PCR as template (Wang et al., 2015). The amplified fragment was digested with BsaI and inserted into pHEE401E containing an egg cell-specific promoter-driven Cas9 cassette as previously described by Wang et al. (2015).
The pHEE401E-NRPD1-T1T2 construct was transformed into Agrobacterium tumefaciens (GV3101), and bacteria containing the plasmid were used to transform Capsella accession Cr1GR1 by floral dip (Clough and Bent, 1998). The genomic sequence of Arabidopsis NRPD1 with the stop codon was amplified from Col-0 genomic DNA and cloned into pDONR221 (Invitrogen). After being confirmed by sequencing, the fragment was inserted into pB7WG2 in which the Cauliflower mosaic virus 35S promoter was replaced by the 1.6-kb promoter sequence of RPS5A (Weijers et al., 2001).
Microscopy
Capsella inflorescences were harvested and fixed in 3:1 ethanol:acetic acid solution. Pollen grains were manually dissected from stage 12 and 13 anthers and stained with 4′,6-diamidino-2-phenylindole (DAPI; 1 μg/mL) as previously described by Brownfield et al. (2015). The slides were observed with an Axio Scope.A1 (Zeiss) and a 7800 confocal microscope (Zeiss).
To generate sections, Capsella inflorescences were harvested and fixed in FAA solution (50% [v/v] ethanol, 5% [v/v] acetic acid, and 4% [v/v] formaldehyde) and embedded using a Historesin Embedding Kit (702,218,500; Leica). Three-micrometer sections were prepared using an HM 355 S microtome (Microm) with glass knives. Sections were stained with 0.1% (w/v) toluidine blue for 1 min, washed five times with distilled water, air dried, and observed with an Axio Scope (Zeiss).
Microspore Extraction
The different pollen stages were extracted on a Percoll gradient following previously published procedures (Dupl’áková et al., 2016). The purity of each fraction was assessed by Alexander and DAPI staining.
RNA-Seq and sRNA-Seq
RNA of Arabidopsis microspores was isolated using TRIzol following the manufacturer’s protocol (Thermo Fisher Scientific, catalog no. 15596018). Purified RNA was treated with DNase I (Thermo Fisher Scientific, catalog no. EN0521) and loaded on a 15% TBE-urea polyacrylamide gel. RNA 15- to 27-nucleotides in size was retrieved and eluted by crushing the gel in PAGE elution buffer (1 M Tris, pH 7.5, 2.5 M NaOAc, and 0.5 M EDTA, pH 8), followed by overnight incubation and a new TRIzol extraction.
Capsella leaves were ground with liquid nitrogen, and 100 mg of fine powder from each sample was used for RNA isolation. Capsella microspores were ground in a precooled mortar with Lysis/Binding Solution from a mirVana miRNA isolation kit. Both long RNAs (>200 nucleotides) and short RNAs (<200 nucleotides) were isolated from leaves and microspores according to the manufacturer’s protocol (mirVana miRNA Isolation Kit, AM1560). Size selection of sRNAs was performed as described above.
For RNA-seq analysis, total RNA was treated using a Poly(A) mRNA Magnetic Isolation Module kit (New England Biolabs [NEB], catalog no.E7490). Libraries were prepared from the resulting mRNA with a NEBNext Ultra II kit (NEB, catalog no. E7770S). sRNA-seq libraries were generated with a NEBNext Multiplex Small RNA kit (NEB, catalog no. E7300S). RNA-seq libraries and sRNA-seq libraries were sequenced at the SciLife Laboratory and Novogene on a HiSeqX in 150-bp paired-end mode or an Illumina HiSeq2000 in 50-bp single-end mode, respectively.
Bisulfite Sequencing
Leaves of 6 to 10 wild-type and nrpd1 Capsella plants were pooled as one replicate. Genomic DNA was extracted using a MagJET Plant Genomic DNA Kit (K2761). Bisulfite conversion and library preparation were done as previously described (Moreno-Romero et al., 2016). Libraries were sequenced at the SciLife Laboratory on an Illumina HiSeq2000 in 125-bp paired-end mode.
DNA Sequencing
Genomic DNA was isolated from the leaves of one Capsella wild-type plant and five nrpd1 mutants using a MagJET Plant Genomic DNA Kit (K2761). Libraries were generated using a NEBNext Ultra II DNA Library Prep Kit for Illumina and sequenced at Novogene on an Illumina HiSeqX in 150-bp paired-end mode.
Bioinformatic Analysis
For sRNA data, adapters were removed from 50-bp-long single-end sRNA reads in each library. The resulting 18- to 30-bp-long reads were mapped to the respective reference genomes using bowtie (-v 0–best). All reads mapping to chloroplast and mitochondria and to structural noncoding RNAs (tRNAs, snRNAs, rRNAs, or snoRNAs) were removed. Mapped reads from both biological replicates were pooled together, sorted into two categories (21/22 nucleotides and 24 nucleotides long), and remapped to the same reference masked genome mentioned above using ShortStack (–mismatches 0–mmap f; Johnson et al., 2016) in order to improve the localization of sRNAs mapping to multiple genomic locations. We normalized the alignments by converting coverage values to reads per million values. TE-siRNAs were defined as siRNAs that overlap with annotated TEs. TEs accumulating 20 or more reads in the merged wild-type libraries were considered to be TE-producing siRNA loci. TEs losing siRNAs in nrpd1 were defined as those having less than 5% of reads left in nrpd1 compared with wild-type samples. To identify genes losing siRNAs in nrpd1 microspores, we determined siRNA coverage over the genomic loci plus 2-kb upstream and downstream regions and calculated differences from wild-type microspores using the Bioconductor RankProd Package (Hong et al., 2006; log2 fold change < −1, P < 0.05). For RNA analysis, for each biological replicate, reads were mapped to the Arabidopsis or Capsella reference genome using TopHat v2.1.0 (Trapnell et al., 2009) in single-end mode. Gene and TE expression was normalized to reads per kilobase per million mapped reads using GFOLD (Feng et al., 2012). For Capsella, the Capsella v1.0 annotated genome was used as a reference (Slotte et al., 2013; https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Crubella), which was also used as a reference in all Capsella analyses described in this article. For Arabidopsis, the TAIR10 annotation was used. Expression levels for each condition were calculated using the mean of the expression values in both biological replicates. Differentially regulated genes and TEs across the two replicates were detected using the rank product method, as implemented in the Bioconductor RankProd Package (Hong et al., 2006). For DNA methylation analysis, reads of each pair were split in 50-bp-long fragments and mapped in single-end mode using Bismark (Krueger and Andrews, 2011). Duplicated reads (aligning to the same genomic position) were eliminated, and methylation levels for each condition were calculated by averaging the replicates.
To estimate the number and identity of sRNA reads mapping to COPIA95 TEs in Capsella, 21/22-nucleotide and 24-nucleotide sRNA reads were first mapped to a consensus reference fasta file for Arabidopsis TEs available at Repbase (https://www.girinst.org/repbase/update/index.html; Jurka et al., 2005) using bowtie (-v 2 -m 3–best–strata). Reads mapping to COPIA95 TEs were remapped to the Capsella reference genome with ShortStack (–mismatches 0 –mmap f; Johnson et al., 2016) and normalized using the coverage values of single-copy genes.
New TE insertions in Capsella were identified using TEPID (Stuart et al., 2016) in pair-end mode based on the sequenced genomes of five Cr nrpd1 mutants and the corresponding wild type.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: AtNRPD1 (AT1G63020), CrNRPD1 (LOC17895349), DCL3 (AT3G43920), RDR2 (AT4G11130), RDR6 (AT3G49500), VGD1 (AT2G47040), ANX2 (AT5G28680), RopGEF9 (AT4G13240), PRK2 (AT2G07040), PRK3 (AT3G42880), and PRK4 (AT3G20190). The sequencing data generated in this study are available in the Gene Expression Omnibus under accession number GSE129744. Supplemental Table 2 summarizes all sequencing data generated in this study.
Supplemental Data
Supplemental Figure 1. Meiosis is not affected in Capsella nrpd1.
Supplemental Figure 2. Tetrad formation is normal in Cr nrpd1.
Supplemental Figure 3. Average purity of Capsella and Arabidopsis microspore extractions.
Supplemental Figure 4. Profiles of total sRNAs in Capsella and Arabidopsis microspores.
Supplemental Figure 5. Example of four loci producing Pol IV-dependent siRNAs in Arabidopsis.
Supplemental Figure 6. Deregulated genes in Arabidopsis nrpd1 mutant microspores.
Supplemental Figure 7. Representative pollen developmental genes accumulating 21/22-nucleotide and 24-nucleotide siRNAs in microspores.
Supplemental Table 1. Primer list.
Supplemental Table 2. Quality of sequencing samples.
Supplemental Data Set 1. COPIA95 elements accumulating Pol IV-dependent siRNAs in Capsella and Arabidopsis.
Supplemental Data Set 2. Up- and downregulated genes in Arabidopsis and Capsella nrpd1 microspores.
Supplemental Data Set 3. Up- and downregulated genes in Capsella microspores overlapping with regions losing 21/22- or 24-nucleotide siRNAs in Cr nrpd1 microspores.
Supplemental Data Set 4. Up-and downregulated genes in Arabidopsis microspores overlapping with regions losing 21/22- or 24-nucleotide siRNAs in At nrpd1 microspores.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We are grateful to Cecilia Wärdig for technical assistance. Sequencing was performed by the SNP&SEQ Technology Platform, Science for Life Laboratory at Uppsala University, a national infrastructure supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. This research was supported by the Swedish Research Councils VR and Formas (grants 2016-00961 and 2017-04119 to C.K.), the Knut and Alice Wallenberg Foundation (grant 2012.0087 to C.K.), and the Göran Gustafsson Foundation for Research in Natural Sciences and Medicine (to C.K.). Research in the Martienssen laboratory was supported by the U.S. National Institutes of Health (grant R01 GM067014) and by the Howard Hughes Medical Institute. Support was also provided by the Cold Spring Harbor Laboratory Shared Resources (funded in part by the Cancer Center Support Grant 5PP30CA045508).
AUTHOR CONTRIBUTIONS
Z.W., N.B., and C.K. performed the experimental design; Z.W., N.B., J.Y., and F.B. performed experiments; G.M. advised on experimental work; R.A.M. contributed experimental data; Z.W., N.B., J.S.-G., and C.K. wrote the article; all authors read and commented on the article.
Footnotes
Articles can be viewed without a subscription.
References
- Bao W., Kojima K.K., Kohany O. (2015). Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C., Autran D. (2015). Chromatin dynamics during cellular differentiation in the female reproductive lineage of flowering plants. Plant J. 83: 160–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F., Twell D. (2011). Germline specification and function in plants. Annu. Rev. Plant Biol. 62: 461–484. [DOI] [PubMed] [Google Scholar]
- Blevins T., Podicheti R., Mishra V., Marasco M., Wang J., Rusch D., Tang H., Pikaard C.S. (2015). Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4: e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A., Roy S., Kritsas K., Grobei M.A., Jaciubek M., Schroeder J.I., Grossniklaus U. (2009). Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136: 3279–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond D.M., Baulcombe D.C. (2015). Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: 917–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M., Berger F. (2015). Chromatin remodelling during male gametophyte development. Plant J. 83: 177–188. [DOI] [PubMed] [Google Scholar]
- Borges F., Martienssen R.A. (2013). Establishing epigenetic variation during genome reprogramming. RNA Biol. 10: 490–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F., Parent J.S., van Ex F., Wolff P., Martínez G., Köhler C., Martienssen R.A. (2018). Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat. Genet. 50: 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield L., Yi J., Jiang H., Minina E.A., Twell D., Köhler C. (2015). Organelles maintain spindle position in plant meiosis. Nat. Commun. 6: 6492. [DOI] [PubMed] [Google Scholar]
- Calarco J.P., Borges F., Donoghue M.T., Van Ex F., Jullien P.E., Lopes T., Gardner R., Berger F., Feijó J.A., Becker J.D., Martienssen R.A. (2012). Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. (2002). Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12: 1138–1144. [DOI] [PubMed] [Google Scholar]
- Chang F., Gu Y., Ma H., Yang Z. (2013). AtPRK2 promotes ROP1 activation via RopGEFs in the control of polarized pollen tube growth. Mol. Plant 6: 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Creasey K.M., Zhai J., Borges F., Van Ex F., Regulski M., Meyers B.C., Martienssen R.A. (2014). miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuerda-Gil D., Slotkin R.K. (2016). Corrigendum: Non-canonical RNA-directed DNA methylation. Nat. Plants 3: 16211. [DOI] [PubMed] [Google Scholar]
- de la Chaux N., Tsuchimatsu T., Shimizu K.K., Wagner A. (2012). The predominantly selfing plant Arabidopsis thaliana experienced a recent reduction in transposable element abundance compared to its outcrossing relative Arabidopsis lyrata. Mob. DNA 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth I., Jolivet S., Froger N., Catrice O., Novatchkova M., Mercier R. (2009). Turning meiosis into mitosis. PLoS Biol. 7: e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupl’áková N., Dobrev P.I., Reňák D., Honys D. (2016). Rapid separation of Arabidopsis male gametophyte developmental stages using a Percoll gradient. Nat. Protoc. 11: 1817–1832. [DOI] [PubMed] [Google Scholar]
- Erhard K.F. Jr., Parkinson S.E., Gross S.M., Barbour J.E., Lim J.P., Hollick J.B. (2013). Maize RNA polymerase IV defines trans-generational epigenetic variation. Plant Cell 25: 808–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., et al. (2016). PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc. Natl. Acad. Sci. USA 113: 15144–15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Meyer C.A., Wang Q., Liu J.S., Liu S., Zhang Y. (2012). GFOLD: A generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 28: 2782–2788. [DOI] [PubMed] [Google Scholar]
- Foxe J.P., Slotte T., Stahl E.A., Neuffer B., Hurka H., Wright S.I. (2009). Recent speciation associated with the evolution of selfing in Capsella. Proc. Natl. Acad. Sci. USA 106: 5241–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15: 1494–1500. [DOI] [PubMed] [Google Scholar]
- Gouil Q., Baulcombe D.C. (2016). DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet. 12: e1006526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover J.W., Kendall T., Baten A., Burgess D., Freeling M., King G.J., Mosher R.A. (2018). Maternal components of RNA-directed DNA methylation are required for seed development in Brassica rapa. Plant J. 94: 575–582. [DOI] [PubMed] [Google Scholar]
- Guo Y.L., Bechsgaard J.S., Slotte T., Neuffer B., Lascoux M., Weigel D., Schierup M.H. (2009). Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc. Natl. Acad. Sci. USA 106: 5246–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Vickers M., Zhang J., Feng X. (2019). Natural depletion of histone H1 in sex cells causes DNA demethylation, heterochromatin decondensation and transposon activation. eLife 8: e42530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Zhang X., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38: 721–725. [DOI] [PubMed] [Google Scholar]
- Herr A.J., Jensen M.B., Dalmay T., Baulcombe D.C. (2005). RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120. [DOI] [PubMed] [Google Scholar]
- Hong F., Breitling R., McEntee C.W., Wittner B.S., Nemhauser J.L., Chory J. (2006). RankProd: A Bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22: 2825–2827. [DOI] [PubMed] [Google Scholar]
- Huang J., Wang C., Wang H., Lu P., Zheng B., Ma H., Copenhaver G.P., Wang Y. (2019). Meiocyte-specific and AtSPO11-1-dependent small RNAs and their association with meiotic gene expression and recombination. Plant Cell 31: 444–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra C.A., et al. (2012). Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Gaubert H., Bucher E., Mirouze M., Vaillant I., Paszkowski J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472: 115–119. [DOI] [PubMed] [Google Scholar]
- Jiang L., Yang S.L., Xie L.F., Puah C.S., Zhang X.Q., Yang W.C., Sundaresan V., Ye D. (2005). VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17: 584–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C., Kasprzewska A., Tennessen K., Fernandes J., Nan G.L., Walbot V., Sundaresan V., Vance V., Bowman L.H. (2009). Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19: 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N.R., Yeoh J.M., Coruh C., Axtell M.J. (2016). Improved placement of multi-mapping small RNAs. G3 (Bethesda) 6: 2103–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. (2005). Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110: 462–467. [DOI] [PubMed] [Google Scholar]
- Kasschau K.D., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.Y., Ono A., Scholten S., Kinoshita T., Zilberman D., Okamoto T., Fischer R.L. (2019). DNA demethylation by ROS1a in rice vegetative cells promotes methylation in sperm. Proc. Natl. Acad. Sci. USA 116: 9652–9657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R., Ohyanagi H., Niihama M., Watanabe T., Nakano M., Kurata N., Nonomura K. (2014). Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 78: 385–397. [DOI] [PubMed] [Google Scholar]
- Krueger F., Andrews S.R. (2011). Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Du J., Hale C.J., Feng S., Krajewski K., Palanca A.M., Strahl B.D., Patel D.J., Jacobsen S.E. (2013). Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Karunanandaa B., McCubbin A., Gilroy S., Kao T. (1996). PRK1, a receptor-like kinase of Petunia inflata, is essential for postmeiotic development of pollen. Plant J. 9: 613–624. [Google Scholar]
- Lei Y., Lu L., Liu H.Y., Li S., Xing F., Chen L.L. (2014). CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7: 1494–1496. [DOI] [PubMed] [Google Scholar]
- Li S., Vandivier L.E., Tu B., Gao L., Won S.Y., Li S., Zheng B., Gregory B.D., Chen X. (2015). Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 25: 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez G., Panda K., Köhler C., Slotkin R.K. (2016). Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat. Plants 2: 16030. [DOI] [PubMed] [Google Scholar]
- Martinez G., Wolff P., Wang Z., Moreno-Romero J., Santos-González J., Conze L.L., DeFraia C., Slotkin R.K., Köhler C. (2018). Paternal easiRNAs regulate parental genome dosage in Arabidopsis. Nat. Genet. 50: 193–198. [DOI] [PubMed] [Google Scholar]
- Martínez de Alba A.E., Elvira-Matelot E., Vaucheret H. (2013). Gene silencing in plants: A diversity of pathways. Biochim. Biophys. Acta 1829: 1300–1308. [DOI] [PubMed] [Google Scholar]
- Maumus F., Quesneville H. (2014). Ancestral repeats have shaped epigenome and genome composition for millions of years in Arabidopsis thaliana. Nat. Commun. 5: 4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelsten Scheid O., Afsar K., Paszkowski J. (2003). Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat. Genet. 34: 450–454. [DOI] [PubMed] [Google Scholar]
- Moreno-Romero J., Jiang H., Santos-González J., Köhler C. (2016). Parental epigenetic asymmetry of PRC2-mediated histone modifications in the Arabidopsis endosperm. EMBO J. 35: 1298–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R.A., Melnyk C.W., Kelly K.A., Dunn R.M., Studholme D.J., Baulcombe D.C. (2009). Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286. [DOI] [PubMed] [Google Scholar]
- Niu X.M., Xu Y.C., Li Z.W., Bian Y.T., Hou X.H., Chen J.F., Zou Y.P., Jiang J., Wu Q., Ge S., Balasubramanian S., Guo Y.L. (2019). Transposable elements drive rapid phenotypic variation in Capsella rubella. Proc. Natl. Acad. Sci. USA 116: 6908–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver C., Santos J.L., Pradillo M. (2016). Accurate chromosome segregation at first meiotic division requires ago4, a protein involved in RNA-dependent DNA methylation in Arabidopsis thaliana. Genetics 204: 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y., Haag J.R., Ream T., Costa Nunes P., Pontes O., Pikaard C.S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622. [DOI] [PubMed] [Google Scholar]
- Panda K., Ji L., Neumann D.A., Daron J., Schmitz R.J., Slotkin R.K. (2016). Full-length autonomous transposable elements are preferentially targeted by expression-dependent forms of RNA-directed DNA methylation. Genome Biol. 17: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., Hakimi M.A., Lerbs-Mache S., Colot V., Lagrange T. (2005). Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19: 2030–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N., Sarazin A., Jullien P.E., Bologna N.G., Oberlin S., Voinnet O. (2016). DNA methylation influences the expression of DICER-LIKE4 isoforms, which encode proteins of alternative localization and function. Plant Cell 28: 2786–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyaki P.R.V., Gehring M. (2019). Paternally acting canonical RNA-directed DNA methylation pathway genes sensitize Arabidopsis endosperm to paternal genome dosage. Plant Cell 31: 1563–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Schmid M.W., Grossniklaus U. (2015). Plant germline formation: Common concepts and developmental flexibility in sexual and asexual reproduction. Development 142: 229–241. [DOI] [PubMed] [Google Scholar]
- Schoft V.K., Chumak N., Choi Y., Hannon M., Garcia-Aguilar M., Machlicova A., Slusarz L., Mosiolek M., Park J.S., Park G.T., Fischer R.L., Tamaru H. (2011). Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc. Natl. Acad. Sci. USA 108: 8042–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Mishra V., Wang F., Huang H.Y., Pikaard C.S. (2019). Reaction mechanisms of Pol IV, RDR2, and DCL3 drive RNA channeling in the siRNA-directed DNA methylation pathway. Mol. Cell 75: 576–589.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., et al. (2013). The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45: 831–835. [DOI] [PubMed] [Google Scholar]
- Stuart T., Eichten S.R., Cahn J., Karpievitch Y.V., Borevitz J.O., Lister R. (2016). Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. eLife 5: e20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H., Higashiyama T. (2016). Tip-localized receptors control pollen tube growth and LURE sensing in Arabidopsis. Nature 531: 245–248. [DOI] [PubMed] [Google Scholar]
- Tekleyohans D.G., Nakel T., Groß-Hardt R. (2017). Patterning the female gametophyte of flowering plants. Plant Physiol. 173: 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng C., Zhang H., Hammond R., Huang K., Meyers B.C., Walbot V. (2018). Dicer-like5 deficiency confers temperature-sensitive male sterility in maize. bioRxiv. Available at: 10.1101/498410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J., Gao H., Zhang J., Aldridge B., Vickers M., Higgins J.D., Feng X. (2018). Sexual-lineage-specific DNA methylation regulates meiosis in Arabidopsis. Nat. Genet. 50: 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.P., Xing H.L., Dong L., Zhang H.Y., Han C.Y., Wang X.C., Chen Q.J. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Geldner N., Offringa R., Jürgens G. (2001). Seed development: Early paternal gene activity in Arabidopsis. Nature 414: 709–710. [DOI] [PubMed] [Google Scholar]
- Wierzbicki A.T., Cocklin R., Mayampurath A., Lister R., Rowley M.J., Gregory B.D., Ecker J.R., Tang H., Pikaard C.S. (2012). Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 26: 1825–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Ream T.S., Haag J.R., Pikaard C.S. (2009). RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41: 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R., Chen C., Pokhrel S., Ma W., Huang K., Patel P., Wang F., Xu J., Liu Z., Li J., Meyers B.C. (2019). 24-nt reproductive phasiRNAs are broadly present in angiosperms. Nat. Commun. 10: 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.L., Zhang G., Tang K., Li J., Yang L., Huang H., Zhang H., Zhu J.K. (2016). Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res. 26: 1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., et al. (2015a). A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., Zhang H., Arikit S., Huang K., Nan G.L., Walbot V., Meyers B.C. (2015b). Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc. Natl. Acad. Sci. USA 112: 3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lang Z., Zhu J.K. (2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19: 489–506. [DOI] [PubMed] [Google Scholar]
- Zhang H., et al. (2013). DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc. Natl. Acad. Sci. USA 110: 8290–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., McCormick S. (2007). A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18830–18835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719. [DOI] [PubMed] [Google Scholar]