CRK2 associates with and activates RBOHD to trigger MAMP-induced ROS production, revealing a regulatory mechanism for plant NADPH oxidases through phosphorylation of the C terminus.
Abstract
Reactive oxygen species (ROS) are important messengers in eukaryotic organisms, and their production is tightly controlled. Active extracellular ROS production by NADPH oxidases in plants is triggered by receptor-like protein kinase-dependent signaling networks. Here, we show that CYSTEINE-RICH RLK2 (CRK2) kinase activity is required for plant growth and CRK2 exists in a preformed complex with the NADPH oxidase RESPIRATORY BURST OXIDASE HOMOLOG D (RBOHD) in Arabidopsis (Arabidopsis thaliana). Functional CRK2 is required for the full elicitor-induced ROS burst, and consequently the crk2 mutant is impaired in defense against the bacterial pathogen Pseudomonas syringae pv tomato DC3000. Our work demonstrates that CRK2 regulates plant innate immunity. We identified in vitro CRK2-dependent phosphorylation sites in the C-terminal region of RBOHD. Phosphorylation of S703 RBOHD is enhanced upon flg22 treatment, and substitution of S703 with Ala reduced ROS production in Arabidopsis. Phylogenetic analysis suggests that phospho-sites in the C-terminal region of RBOHD are conserved throughout the plant lineage and between animals and plants. We propose that regulation of NADPH oxidase activity by phosphorylation of the C-terminal region might be an ancient mechanism and that CRK2 is an important element in regulating microbe-associated molecular pattern-triggered ROS production.
INTRODUCTION
Plants are continuously confronted with stimuli from the surrounding environment, including abiotic cues and invading pathogens. Plant cells also perceive a plethora of signals from neighboring cells and distant tissues. Numerous plasma membrane proteins are involved in the meticulous monitoring and transduction of signals for inter- and intracellular communication. A common early feature of many cellular responses to various environmental changes involves the production of reactive oxygen species (ROS; Kimura et al., 2017; Waszczak et al., 2018). While ROS are an inevitable byproduct of aerobic metabolism and their unrestricted accumulation can have deleterious consequences (Waszczak et al., 2018), ROS are also ubiquitous signaling molecules in plants and animals (Suzuki et al., 2011; Waszczak et al., 2018). Eukaryotic cells produce ROS in several subcellular compartments as well as the extracellular space, in plants referred to as the apoplast (Kimura et al., 2017; Waszczak et al., 2018). A major component in the production of extracellular ROS is the evolutionarily conserved NADPH oxidase (NOX) family (Meitzler et al., 2014; Kimura et al., 2017). NOX-dependent ROS production is involved in regulation of immune functions, cell growth, and apoptosis in animals and plants (Jiménez-Quesada et al., 2016; Waszczak et al., 2018).
Plant NOXs, referred to as respiratory burst oxidase homologs (RBOHs), have been identified as homologs of phagocyte gp91phox/NOX2, which contains six transmembrane helices and a C-terminal NADPH- and FAD-binding cytoplasmic region (Torres et al., 2002). Unlike gp91phox/NOX2, RBOHs contain an additional N-terminal region with Ca2+ binding EF hands, similar to nonphagocytic NOXs, such as NOX5 (Suzuki et al., 2011). RBOH activity is strictly controlled to avoid damaging consequences of unrestricted ROS production (Suzuki et al., 2011). Arabidopsis (Arabidopsis thaliana) RBOHD is the best-characterized RBOH and is involved in biotic and abiotic stress responses (Torres et al., 2002; Lee et al., 2013, 2018). The N-terminal region of RBOHD is phosphorylated by a variety of protein kinases, including receptor-like cytoplasmic kinases (Ogasawara et al., 2008; Kimura et al., 2012; Dubiella et al., 2013; Kadota et al., 2014; Li et al., 2014; Zhang et al., 2018; Han et al., 2019; Kaya et al., 2019), for example BOTRYTIS-INDUCED KINASE 1 (BIK1; Kadota et al., 2014; Li et al., 2014). While previous research has suggested a predominant role of phosphorylation of the N-terminal region for regulation of RBOH, phosphorylation of the C-terminal region is important for the regulation of human gp91phox/NOX2 and NOX5 (Jagnandan et al., 2007; Raad et al., 2009). NADPH- and FAD-binding sites in C terminus are highly conserved in NOXs and RBOHs, but it is unclear whether the C terminus of plant RBOHs could also be a target for regulation of the ROS producing activity.
Apoplastic RBOH-dependent ROS production is a common response to the activation of receptor-like protein kinase (RLK; Shiu and Bleecker, 2001) signaling, in particular following perception of microbe-associated molecular patterns (MAMPs) or damage-associated molecular patterns (DAMPs; Couto and Zipfel, 2016; Kimura et al., 2017). However, the roles of the so-called ROS burst and its integration into RLK-triggered signaling networks are as yet unclear (Kimura et al., 2017). A large group of RLKs in plants is formed by the cysteine-rich RLKs (CRKs; Vaattovaara et al., 2019). The extracellular region of CRKs harbors two copies of the domain of unknown function 26 (DUF26), but the molecular function of the CRK ectodomain remains unknown (Vaattovaara et al., 2019). CRKs have been linked to ROS signaling (Idänheimo et al., 2014; Bourdais et al., 2015; Yeh et al., 2015; Yadeta et al., 2017) and cell death (Bourdais et al., 2015; Yadeta et al., 2017). CRKs are important signaling elements in plant development and biotic and abiotic stress responses (Chen et al., 2004; Acharya et al., 2007; Wrzaczek et al., 2010; Tanaka et al., 2012; Idänheimo et al., 2014; Bourdais et al., 2015; Yeh et al., 2015; Chern et al., 2016; Yadeta et al., 2017; Hunter et al., 2019).
Here, we characterize the role of CRK2 in immune signaling in response to MAMP perception. CRK2 exists in a preformed complex with RBOHD. CRK2 controls the activity of RBOHD, and functional CRK2 is required for full MAMP-induced ROS production. Importantly, we show that CRK2 phosphorylates the C-terminal region of RBOHD and modulates the ROS-production activity of RBOHD in vivo. Our results lead us to propose a novel mechanism for the regulation of RBOHD activity through phosphorylation of the C-terminal region and highlight a critical role for CRK2 in the precise control of the ROS burst in response to biotic stress.
RESULTS
CRK2 Kinase Activity Is Important for Plant Development
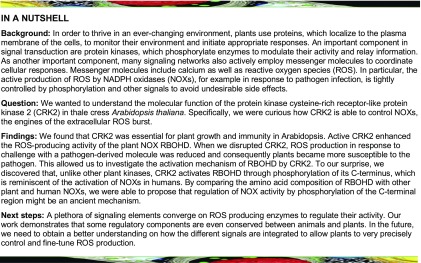
CRK2 has been previously implicated in stress responses and development in Arabidopsis (Bourdais et al., 2015). CRK2 is a typical CRK with an N-terminal signal peptide, extracellular region containing two DUF26 domains, transmembrane region, and intracellular protein kinase domain (Figure 1A). The crk2 mutant was smaller than wild-type (Columbia-0 [Col-0]) plants (Figure 1B; Bourdais et al., 2015) and displayed significantly reduced fresh (Figure 1C) and dry weight (Supplemental Figure 1A). Overaccumulation of the plant hormone salicylic acid (SA) often leads to a reduction of plant size, but SA levels were not significantly different between crk2 and wild-type plants (Supplemental Figure 1B). Expression of yellow fluorescent protein (YFP)-tagged CRK2 under the control of the CRK2 promoter (CRK2pro:CRK2-YFP) in crk2 background-restored plant growth (Figures 1B, 1C, and Supplemental Figure 1A). Substitution of the ATP binding Lys (K) at position 353 with Glu (E; CRK2K353E) or the Asp (D) at position 450 in the catalytic domain VIb (Stone and Walker, 1995) with Asn (N; CRK2D450N) abated the kinase activity of CRK2 in vitro (Hunter et al., 2019). Kinase-dead CRK2K353E-YFP or CRK2D450N-YFP under control of the CRK2 promoter was expressed in leaves and roots and displayed the same subcellular localization as wild-type CRK2-YFP (Supplemental Figures 1C and 1D) but failed to restore the growth defect of crk2 (Figures 1B, 1C, and Supplemental Figure 1A). The amino acid substitutions did not reduce the abundance of kinase-dead CRK2-YFP (Supplemental Figure 1E). In summary, our results show that CRK2 is important for proper plant growth, and its kinase activity is crucial for this function.
Figure 1.
CRK2 Kinase Activity Is Required for Plant Growth.
(A) Schematic representation of CRK2 structure. SP, Signal peptide (amino acids 1 to 29); DUF26-A (amino acids 39 to 132); DUF26-B (amino acids 146 to 243); TM, transmembrane domain (amino acids 261 to 283); and kinase domain (AAs 325 to 601).
(B) Representative pictures of 21-d-old plants of Col-0, crk2, CRK2pro:CRK2-YFP/crk2, CRK2pro:CRK2K353E-YFP/crk2, and CRK2pro:CRK2D450N-YFP/crk2 plants. Bar = 1 cm.
(C) Box plot shows the fresh weight of 21-day-old plants (n = 10). Differences between Col-0 and transgenic lines were evaluated with one-way ANOVA with Tukey-Kramer honestly significant difference (HSD) post-hoc test, *** P < 0.001; ns, not statistically significant. The experiment was repeated three times with similar results.
CRK2 Is Required for MAMP-Triggered Responses and Resistance to Pseudomonas syringae pv tomato DC3000
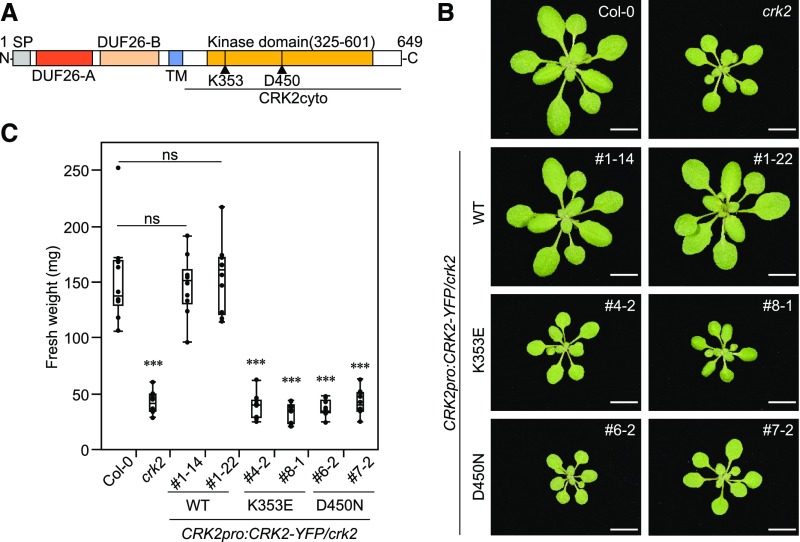
Previous results suggested that ROS production triggered by flg22, a MAMP derived from bacterial flagella, is reduced in crk2 (Bourdais et al., 2015). Therefore, we tested the role of CRK2 in MAMP-induced ROS production in detail. ROS production triggered by flg22 was reduced in crk2 and reintroduction of CRK2-YFP into the mutant background restored ROS production to the same levels as in Col-0 (Figure 2A). The flg22-induced ROS production in plants expressing CRK2D450N-YFP was comparable to crk2 (Figure 2B). Transcriptional upregulation of flg22 responsive genes (FLG22-INDUCED RECEPTOR-LIKE KINASE 1 [FRK1] and NDR1/HIN1-LIKE 10 [NHL10]) showed that MAMP perception was not impaired in crk2 (Supplemental Figures 2A and B). To test whether the reduced response of crk2 to flg22 was accompanied by altered pathogen susceptibility, we measured growth of the hemibiotrophic bacterial pathogen Pseudomonas syringae pv tomato DC3000 (Pto DC3000). The crk2 mutant was significantly more susceptible to the virulent pathogen compared to Col-0 (Figure 2C). CRK2-YFP but not the kinase-dead CRK2D450N-YFP rescued the pathogen susceptibility of crk2 (Figure 2C). ROS production induced by chitin (Supplemental Figure 2C) or pep1 (Supplemental Figure 2D) was also reduced in crk2 compared to Col-0, suggesting that the reduced MAMP- or DAMP-triggered ROS production in crk2 is a general response and not specific to flg22. MAMP-triggered ROS production is also an important element in pathogen-triggered stomatal closure. Therefore, we tested whether the reduced flg22-triggered ROS production in crk2 also coincided with altered stomatal closure. The crk2 mutant displayed slightly reduced stomatal aperture compared to wild-type plants under control conditions. However, treatment with flg22-triggered stomatal closure in wild-type but not in crk2 plants (Supplemental Figure 2E).
Figure 2.
CRK2 Regulates flg22-Triggered ROS Production and Resistance to a Virulent Bacterial Pathogen.
(A) and (B) flg22-induced ROS production in Col-0, crk2, and CRK2pro:CRK2-YFP/crk2 or CRK2pro:CRK2D450N-YFP/crk2. Leaf discs from 28-d-old plants were treated with 200 nM flg22 and ROS production was measured. Box plot shows cumulative ROS production over 40 min (top right).
(A) Values represent mean ± sem of n ≥ 16. Differences compared with Col-0 were evaluated with one-way ANOVA with Tukey-Kramer HSD, ***P < 0.001; ns, not statistically significant.
(B) Values represent the mean ± sem of n ≥ 19. Differences compared with Col-0 were evaluated with one-way ANOVA with Tukey-Kramer HSD, * P < 0.05, *** P < 0.001.
(C) Quantitative analysis of bacterial growth in Col-0, crk2, and CRK2pro:CRK2-YFP/crk2 or CRK2pro:CRK2D450N-YFP/crk2 following syringe infiltration with Pto DC3000 (1 × 105 CFU/mL). Box plot shows the numbers of bacteria in leaf discs after infiltration (0 day postinfiltration [0 DPI], n = 3) or 2 DPI (n = 6). Each data point contains four leaf discs. Letters indicate significant differences at P < 0.05 (one-way ANOVA with Tukey-Kramer HSD).
(A) to (C) The experiment was repeated three times with similar results.
To further investigate the role of CRK2 in flg22-triggered responses, we assessed Ca2+ signaling, MAPK activation, and callose deposition in crk2. Application of flg22 resulted in a rapid increase in cytosolic Ca2+ ([Ca2+]cyto) levels in wild-type plants, which express the Förster resonance energy transfer (FRET)-based Ca2+-sensor YCNano-65 (Choi et al., 2014; Lenglet et al., 2017; Toyota et al., 2018). This response was reduced in the YCNano65/crk2 background (Supplemental Figures 2F and G). Interestingly, flg22-dependent MAPK activation (Supplemental Figure 2H) was more pronounced in crk2 compared to Col-0. Callose deposition was enhanced in crk2 compared to Col-0 after 30 min of treatment with flg22 (Supplemental Figures 2I and 2J). However, flg22-triggered callose deposition was similar in Col-0 and crk2 after 12 h (Supplemental Figure 2I). Taken together, CRK2 is an essential component for mounting immune responses against Pto DC3000 in Arabidopsis, modulating extracellular ROS production, stomatal closure, Ca2+ influx, MAPK activation and early callose deposition.
CRK2 Interacts with RBOHD and Controls ROS Production
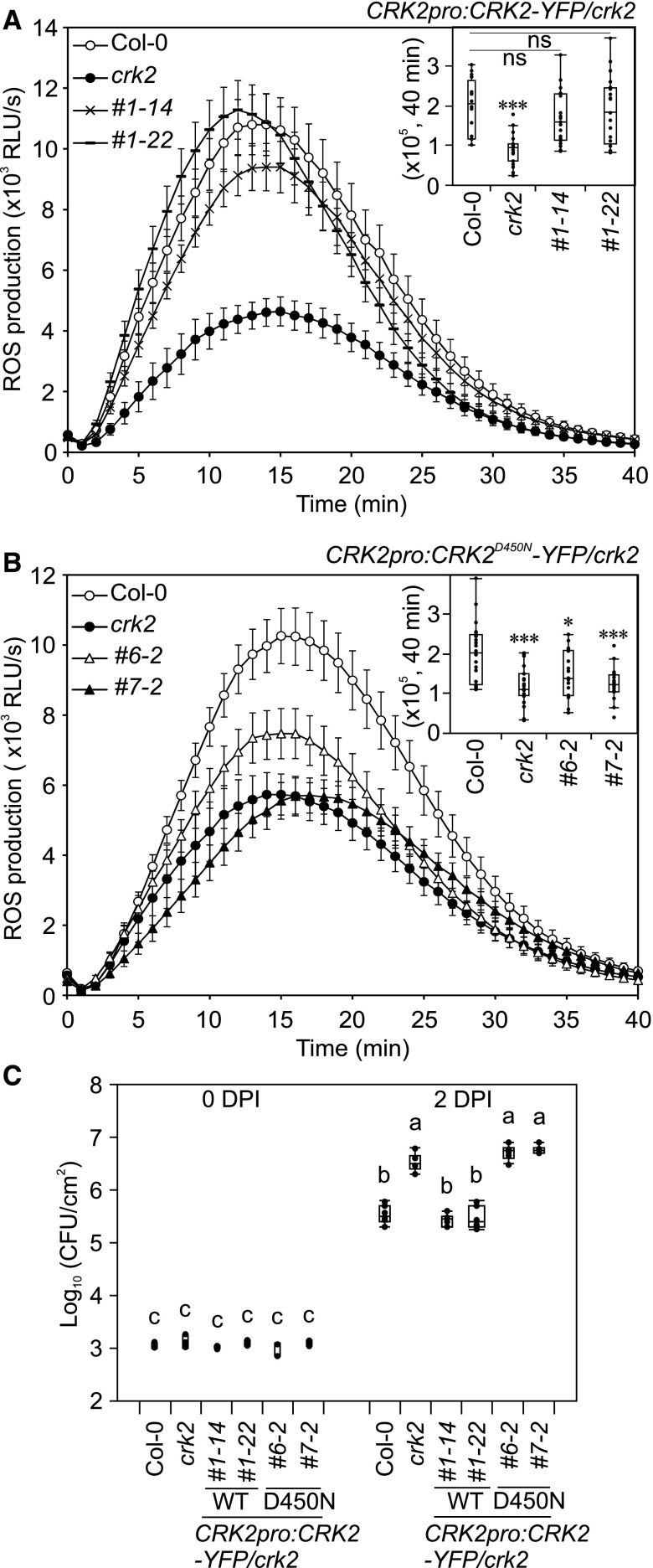
RBOHD is the main source of MAMP/DAMP-induced extracellular ROS production (Couto and Zipfel, 2016; Kimura et al., 2017) and flg22-, pep1-, and chitin-induced ROS production was significantly reduced in crk2 (Figure 2; Supplemental Figure 2). RBOH proteins, including RBOHD, are synergistically activated by protein phosphorylation and Ca2+ binding to EF-hand motifs in the N-terminal region (Kaya et al., 2019). Given the importance of CRK2 kinase activity in MAMP-induced ROS production, we investigated whether CRK2 could activate RBOHD. To test this, we used human embryonic kidney 293T (HEK293T) cells, a human cell culture that produces minimal amounts of extracellular ROS due to a lack of expression of endogenous NOXs (Ogasawara et al., 2008). HEK293T cells were cotransfected with 3FLAG-RBOHD and CRK2-3Myc or 3Myc-GFP as a control. Subsequently, RBOHD-mediated extracellular ROS production was measured by luminol-amplified chemiluminescence. Despite equal 3FLAG-RBOHD protein levels (Supplemental Figure 3A), ROS production in cells cotransfected with CRK2-3Myc and 3FLAG-RBOHD was strongly elevated compared to cells cotransfected with 3FLAG-RBOHD and 3Myc-GFP (Figure 3A). Cotransfection with the inactive variant CRK2D450N-3Myc did not enhance ROS production by 3FLAG-RBOHD compared to cotransfection with CRK2-3Myc (Figure 3A). Transfection of CRK2-3Myc in the absence of 3FLAG-RBOHD did not induce ROS production in HEK293T cells (Figure 3A).
Figure 3.
CRK2 Interacts with RBOHD.
(A) ROS production of RBOHD-expressing HEK293T cells. 3FLAG-RBOHD was transiently coexpressed with either 3Myc-GFP or CRK2 (wild type or D450N)-3Myc. After 30 min, 1 µM ionomycin was added to the medium (black arrow). Values represent mean ± sem of n = 3. E.V., Empty vector. The experiment was repeated three times with similar results.
(B) and (C) Coimmunoprecipitation analysis of interaction between RBOHD and CRK2. CRK2-YFP was immuno-precipitated using anti-GFP beads followed by immunoblotting with anti-RBOHD and anti-GFP antibodies. FLAG-RBOHD, 105 kD; CRK2-YFP, 99.9 kD; and YFP-6Myc, 36.7 kD.
(B) 35S:FLAG-RBOHD/rbohD × 35S:CRK2-YFP/Col-0 (F1) and 35S:FLAG-RBOHD/rbohD × 35S:YFP-6Myc/Col-0 (F1) plants. The experiment was repeated three times with similar results.
(C) 35S:FLAG-RBOHD/35S:CRK2-YFP/rbohD plants with 1 µM flg22 treatment. M, protein molecular marker; *, unspecific signal. Total protein from rbohD was used for immunoblot of input as a negative control.
(D) Schematic representation of RBOHD structure. EF hands (AAs 257 to 329); TM, transmembrane domains (AAs 374 to 605); FAD, FAD binding domain (AAs 613 to 730); NADPH, NADPH binding domain (AAs 736 to 904); RBOHD/N, RBOHD N-terminal region (AAs 1 to 376); RBOHD/C, RBOHD C-terminal region (AAs 606 to 922); C1, RBOHD/C1 (AAs 606 to 741); C2, RBOHD/C2 (AAs 696 to 831); C3, RBOHD/C3 (AAs 787 to 922).
(E) In vitro pull-down analysis of direct interaction between RBOHD and CRK2. MBP, 6His-MBP-RBOHD/N, and 6His-MBP-RBOHD/C were incubated with 6His-GST-CRK2cyto and pull-down with GST followed by immunoblotting with anti-6His and anti-MBP antibodies. 6His-GST-CRK2cyto, 68.5 kD; 6His-MBP-RBOHD/N, 84.7 kD; 6His-MBP-RBOHD/C, 78.4 kD; MBP, 50.8 kD. The experiment was repeated two times with similar results.
Since RBOHD can also be activated by Ca2+, HEK293T cells were treated with ionomycin, a Ca2+ ionophore that induces a rise in cytosolic Ca2+ levels. Ionomycin-induced transient ROS production (Δdelta ROS, ROST=30 to ROST=31) in CRK2-3Myc and 3FLAG-RBOHD cotransfected cells was not different from 3Myc-GFP and 3FLAG-RBOHD cotransfected cells (Figure 3A). Enhancement of RBOHD activity by CRK2 was not dependent on Ca2+ influx as the elevated basal ROS production activity of RBOHD cotransfected with CRK2-3Myc (ROST=0 to ROST=30 in Figure 3A) was also observed when using Ca2+-free assay buffer (Supplemental Figures 3B and C). These results suggest that CRK2-3Myc enhanced the basal ROS-producing activity of 3FLAG-RBOHD in HEK293T cells, uncoupling it from Ca2+ dependence.
The Arabidopsis genome encodes 10 RBOHs (Kaya et al., 2019). To test whether CRK2 specifically activates RBOHD, CRK2-3Myc was cotransfected with 3FLAG-RBOHF and 3FLAG-RBOHC into HEK293T cells. CRK2-3Myc enhanced basal ROS-producing activity of RBOHC and RBOHF in HEK293T cells similarly to RBOHD (Supplemental Figures 3D to 3G). However, while the basal ROS production activity (ROST=5) of RBOHD and RBOHF was elevated ∼10-fold, the basal activity of RBOHC was only elevated threefold.
To investigate whether CRK2 and RBOHD interact in planta, we performed coimmunoprecipitation assays using rbohD plants expressing 35S:CRK2-YFP and 35S:FLAG-RBOHD. CRK2-YFP was immunoprecipitated using an anti-GFP antibody coupled to magnetic beads, and copurified RBOHD was detected using a RBOHD-specific antibody. RBOHD copurified with CRK2 (Figure 3B) and treatment of plants with flg22 did not alter the interaction of CRK2 with RBOHD (Figure 3C). To analyze this interaction in more detail, we performed in vitro interaction assays between the cytosolic region of CRK2 (Figure 1A) and the cytosolic N-terminal and C-terminal regions of RBOHD (Figure 3D). Recombinant RBOHD/N or RBOHD/C tagged with 6His and maltose binding protein (MBP; 6His-MBP-RBOHD/N, 6His-MBP-RBOHD/C) or MBP were incubated with the cytosolic region of CRK2, which contains the kinase domain (CRK2cyto), tagged with 6His and glutathione S-transferase (GST; 6His-GST-CRK2cyto) and glutathione sepharose beads. GST pull-down assay showed that 6His-GST-CRK2cyto interacted in vitro with 6His-MBP-RBOHD/N but intriguingly also with 6His-MBP-RBOHD/C (Figure 3E). In summary, our results suggest that CRK2 is capable of direct interaction with RBOHD. CRK2 and RBOHD form a complex, which exists independently of flg22 perception in planta, in contrast to many other RLK-containing complexes that are formed in response to ligand binding.
CRK2 Phosphorylates RBOHD In Vitro
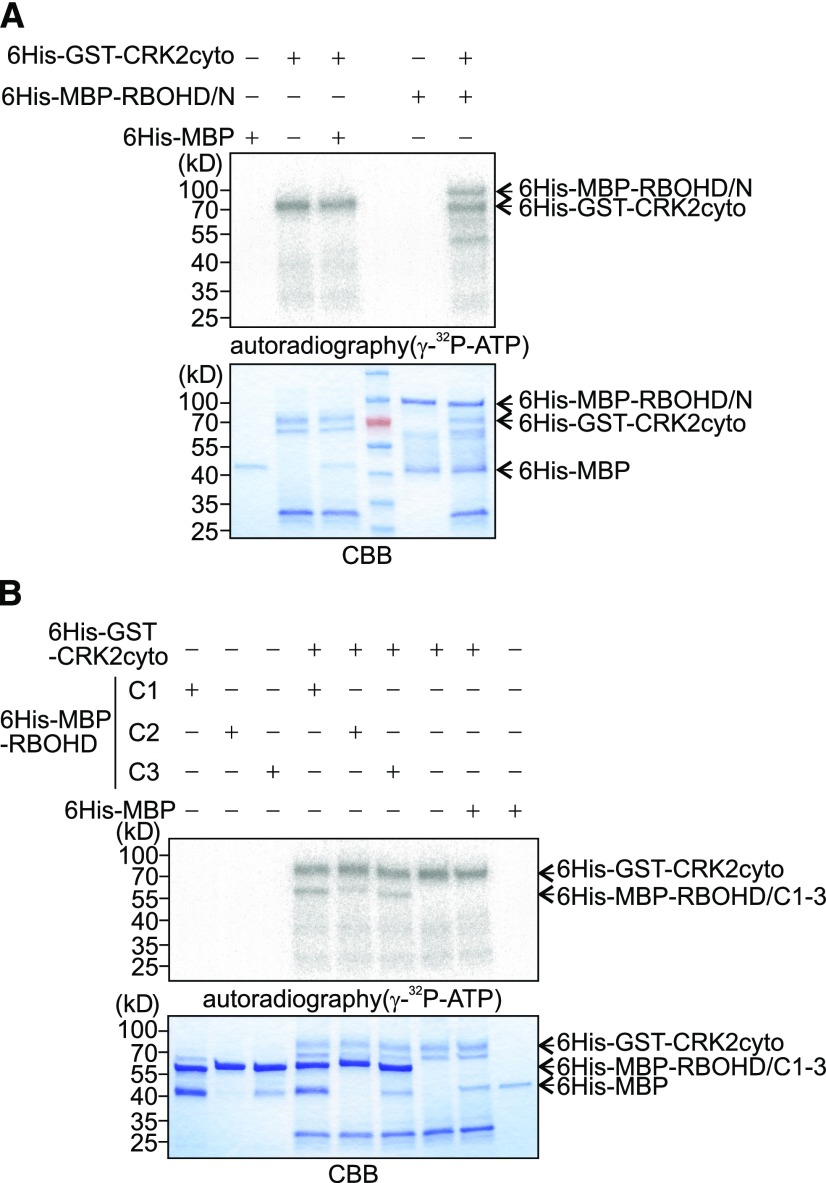
The kinase activity of CRK2 was essential for the full flg22-triggered ROS burst in planta as well as for enhancing ROS production by RBOHD in HEK293T cells. Therefore, we tested whether CRK2 could phosphorylate RBOHD in vitro. Recombinant 6His-GST-CRK2cyto and 6His-MBP tagged RBOHD cytosolic regions (Figures 1A and 3D) were produced in Escherichia coli and affinity purified. The 6His-GST-CRK2cyto phosphorylated 6His-MBP-RBOHD/N but not MBP (Figure 4A). Because of the similar molecular weight of 6His-GST-CRK2cyto (68.5 kD) and 6His-MBP-RBOHD/C (78.4 kD), RBOHD/C was divided into three overlapping fragments (C1, C2, and C3; Figure 3D). The results showed that the C1 and C3 fragments of 6His-MBP-RBOHD were preferentially phosphorylated by 6His-GST-CRK2cyto, while C2 displayed considerably weaker phosphorylation (Figure 4B). Mass spectrometric analysis of in-gel trypsin- or Lys-C-digested peptides identified in vitro RBOHD phosphorylation sites targeted by CRK2cyto (Supplemental Table 1). In the N-terminal region of RBOHD, two sites targeted by CRK2 were identified (S8 and S39), while three sites (S611, S703, S862) were identified in the C-terminal region. Taken together, our results show that the N- and C-terminal regions of RBOHD are phosphorylated by CRK2 in vitro.
Figure 4.
CRK2 Phosphorylates the Cytosolic Regions of RBOHD In Vitro.
(A) and (B) Autophosphorylation and transphosphorylation were visualized with [γ-32P] ATP and autoradiography (top). Input proteins were stained with Coomassie Brilliant Blue (CBB; bottom). Experiments were repeated three times with similar results. 6His-GST-CRK2cyto, 68.5 kD; 6His-MBP-RBOHD/N, 84.7 kD; 6His-MBP-RBOHD/C1, 57.9 kD; RBOHD/C2, 57.8 kD; RBOHD/C3, 58.4 kD; 6His-MBP, 44.3 kD.
(A) In vitro transphosphorylation of 6His-MBP-RBOHD N terminus by 6His-GST-CRK2cyto. 6His-MBP-RBOHD/N or 6His-MBP was incubated with 6His-GST-CRK2cyto in kinase buffer.
(B) In vitro transphosphorylation of 6His-MBP-RBOHD C terminus by 6His-GST-CRK2cyto. 6His-MBP-RBOHD/C1, RBOHD/C2, RBOHD/C3, or 6His-MBP was incubated with 6His-GST-CRK2cyto in kinase buffer.
CRK2 Regulates RBOHD via Phosphorylation of S703 and S862
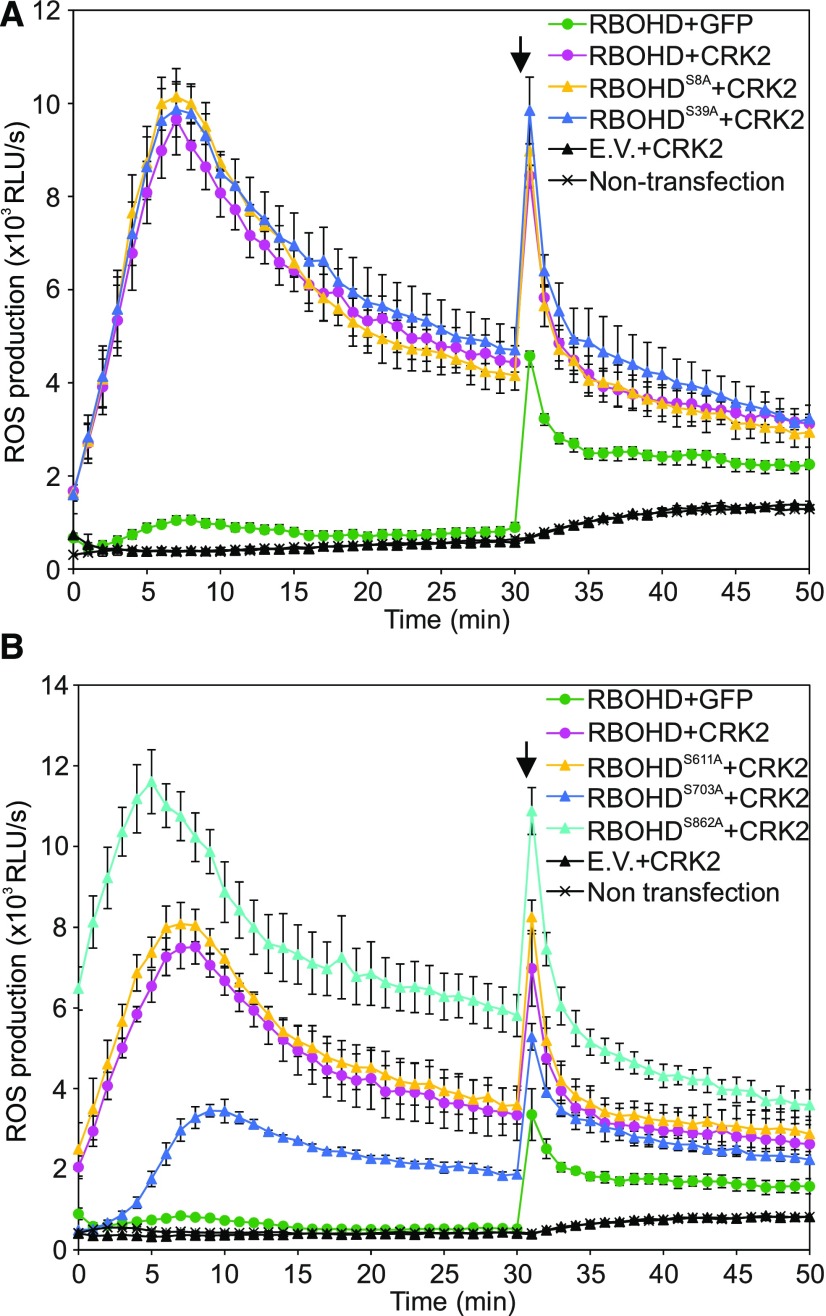
Among the RBOHD phospho-sites targeted by CRK2 in vitro, S8 and S39 have been previously described to be phosphorylated by SIK1 (Zhang et al., 2018) and BIK1 (Kadota et al., 2014; Li et al., 2014). S703 has been reported to be phosphorylated upon xylanase treatment, but no responsible kinase was identified (Benschop et al., 2007), while phosphorylation of S611 and S862 has not been described so far. In order to test whether the identified phospho-sites in RBOHD were important for the regulation of RBOHD activity, the residues S8, S39, S611, S703, and S862 were substituted with Ala to make them nonphosphorylatable. Wild-type RBOHD and phospho-site mutant constructs were transfected into HEK293T cells together with CRK2. Amino acid substitutions did not affect RBOHD protein levels (Supplemental Figures 4A and B). Substitution of S8 or S39 in the N-terminal cytoplasmic region of RBOHD did not impact ROS-producing activity compared to the wild-type protein when cotransfected with CRK2 (Figure 5A). The 3FLAG-RBOHDS703A and CRK2-3Myc cotransfected cells showed reduced basal ROS production as compared to 3FLAG-RBOHD and CRK2-3Myc (Figure 5B), suggesting that S703 could be a positive regulatory site for RBOHD activity. In contrast to 3FLAG-RBOHDS703A, HEK293T cells expressing 3FLAG-RBOHDS862A and CRK2-3Myc exhibited higher basal ROS production compared to 3FLAG-RBOHD and CRK2-3Myc (Figure 5B), suggesting that S862 could act as a negative regulatory site. ROS production of 3FLAG-RBOHDS611A cotransfected with CRK2-3Myc was similar to 3FLAG-RBOHD, suggesting no regulatory role of this single site. Mutation of S703 or S862 of RBOHD did not impair Ca2+-dependent activation of ROS production of RBOHD itself without CRK2 cotransfection (Supplemental Figures 4C and D). Taken together, our results suggest that the phospho-sites in the C-terminal cytoplasmic region of RBOHD could be crucial for fine-tuning ROS production activity in HEK293T cells.
Figure 5.
CRK2 Modulates the ROS-Production Activity of RBOHD via Phosphorylation of the C Terminus in HEK293T Cells.
(A) and (B) Effect of mutations of CRK2-dependent in vitro phosphorylation sites in the N-terminal (A) or C-terminal (B) cytosolic region of RBOHD. After 30 min, 1 µM ionomycin was added to the medium to promote Ca2+ influx (black arrow). Values represent mean ± sem of n = 3. E.V., Empty vector. The experiment was repeated three times with similar results.
(A) 3FLAG-RBOHD (wild type, S8A, or S39A) was transiently coexpressed with either 3Myc-GFP or CRK2-3Myc in HEK293T cells.
(B) 3FLAG-RBOHD (wild type, S611A, S703A, or S862A) were transiently coexpressed with either 3Myc-GFP or CRK2-3Myc in HEK293T cells.
Phosphorylation of S703 of RBOHD Modulates flg22-Induced ROS Production In Planta
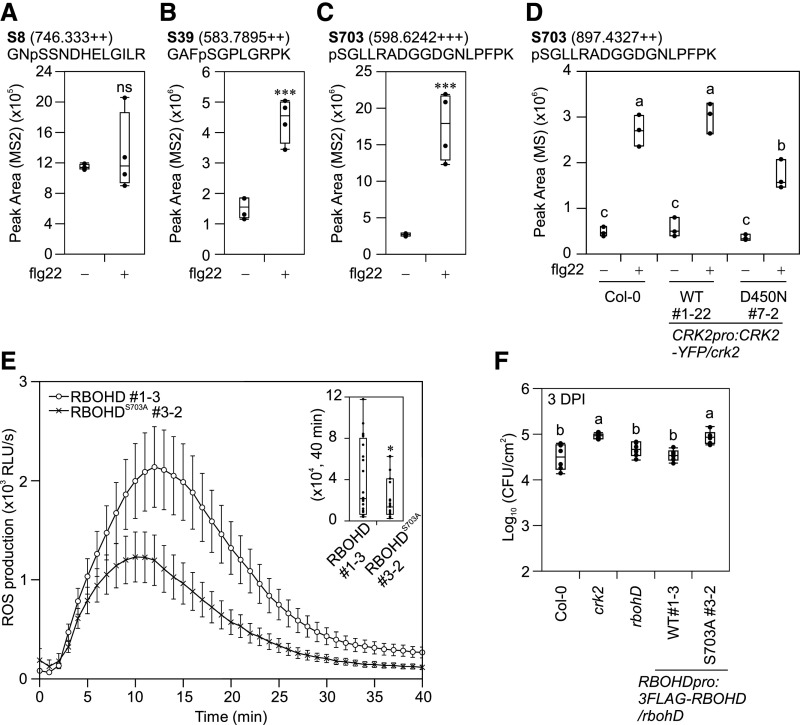
To investigate whether RBOHD phosphorylation sites targeted by CRK2 in vitro were also phosphorylated upon flg22-treatment in planta, we performed targeted phosphoproteomic analyses of Col-0 seedlings treated with flg22 for 5 min. Phosphorylation of S8 was not significantly induced by flg22-treatment (Figure 6A; Supplemental Figures 5A and S5B), while S39 phosphorylation was strongly enhanced (Figure 6B and Supplemental Figure S5C). Phosphorylation of S611 and S862 could not be evaluated, as trypsin or Lys-C digestion resulted in phospho-site-containing peptides of inappropriate length for liquid chromatography-mass spectrometry (LC-MS)-based targeted analyses. However, phosphorylation of S703, which was targeted by CRK2 in vitro, and mutation of which to Ala reduced ROS production in HEK293T cells, was significantly enhanced upon flg22 treatment (Figure 6C; Supplemental Figure 5D). In agreement with previous studies, phosphorylation of S163, S343, and S347 in RBOHD (Supplemental Figures 5E to 5G), as well as dual phosphorylation of the TEY-motif in the MAPKs MPK3, MPK6, and MPK11 (Supplemental Figures 5H to 5J), were enhanced by flg22-treatment (Kadota et al., 2015).
Figure 6.
RBOHD S703 Is Involved in Regulation of flg22-Induced ROS Production.
(A) to (C) Quantification of RBOHD phosphorylation in Col-0 upon flg22 treatment. 12-d-old seedlings were treated with water (−) or 1 µM flg22 (+) for 5 min. Total proteins were digested with trypsin (S8 and S39) or Lys-C (S703) and phosphopeptides were enriched, and then selected phosphopeptides were quantified by LC-MS/MS. Box plots show MS2 fragment peak ion areas of indicated phosphopeptides (n = 4). Differences between water- or flg22-treated samples were evaluated with one-way ANOVA with Tukey-Kramer HSD, ***P < 0.001; ns, not statistically significant.
(A) RBOHD S8 residue.
(B) RBOHD S39 residue.
(C) RBOHD S703 residue.
(D) Quantification of RBOHD S703 phosphorylation in Col-0, CRK2pro:CRK2-YFP/crk2 #1-22, and CRK2pro:CRK2D450N-YFP/crk2 #7-2 upon flg22 treatment. Leaf discs from 28-d-old plants were treated with water (−) or 200 nM flg22 (+) for 5 min. Total proteins were digested with Lys-C and phosphopeptides were enriched, and then selected phosphopeptides were quantified by LC-MS/MS. Box plots show MS2 fragment peak ion areas of indicated phosphopeptides (n = 3). Each data point contains 14 leaf discs. Letters indicate significant differences at P < 0.05 (one-way ANOVA with Tukey-Kramer HSD).
(E) flg22-induced ROS production in RBOHDpro:3FLAG-RBOHD/rbohD #1-3 and RBOHDpro:3FLAG-RBOHDS703A/rbohD #3-2. Leaf discs from 28-d-old plants were treated with 200 nM flg22. Box plot shows cumulative ROS production over 40 min (top right). Values represent mean ± sem of n ≥ 23. Difference between lines was evaluated with one-way ANOVA with Tukey-Kramer HSD, *P < 0.05. The experiment was repeated three times with similar results.
(F) Quantitative analysis of bacterial growth in Col-0, crk2, rbohD, RBOHDpro:3FLAG-RBOHD/rbohD #1-3 and RBOHDpro:3FLAG-RBOHDS703A/rbohD #3-2 following spray inoculation with Pto DC3000 (1 × 105 CFU/mL). Box plot shows the numbers of bacteria in leaf discs 3 d post-inoculation (3 DPI; n = 6). Each data point contains four leaf discs. Letters indicate significant differences at P < 0.05 (one-way ANOVA with Tukey-Kramer HSD).
To test whether phosphorylation of S703 in the C-terminal region of RBOHD was mediated by CRK2 in planta, we used leaf discs of CRK2pro:CRK2 (wild type or D450N)-YFP/crk2. The basal and flg22-induced phosphorylation of S8 and S39 were not significantly altered in CRK2pro:CRK2-YFP/crk2 or the kinase-dead variant (Supplemental Figures 5K to 5M), suggesting that CRK2 might not be the main kinase phosphorylating RBOHD S8 and S39 in planta. However, flg22-induced phosphorylation of S703 in RBOHD was significantly reduced in kinase-dead CRK2pro:CRK2(D450N)-YFP/crk2 plants compared CRK2pro:CRK2-YFP/crk2 (Figure 6D). These results suggest that CRK2 is responsible, at least partially, for flg22-induced phosphorylation of S703 in planta.
To investigate whether phosphorylation of S703 in the C-terminal region of RBOHD also impacts RBOHD-dependent ROS production in planta, we generated transgenic plants expressing RBOHD or RBOHDS703A under the control of the RBOHD promoter (RBOHDpro:3FLAG-RBOHD) in the rbohD background. The phospho-site mutations did not alter growth or development compared with the 3FLAG-RBOHD-expressing plants (Supplemental Figure 6A). Protein amounts of 3FLAG-RBOHD or 3FLAG-RBOHDS703A were comparable in the transgenic plant lines (Supplemental Figures 6B and 6C). ROS production triggered by flg22 was significantly reduced in 3FLAG-RBOHDS703A plants compared with 3FLAG-RBOHD plants (Figure 6E; Supplemental Figure 6D). The results suggest that phosphorylation of S703 in the C terminus of RBOHD is important for full flg22-triggered ROS production also in planta. Notably, expression of 3FLAG-RBOHDS703A in rbohD mutant background impaired flg22-triggered stomatal closure (Supplemental Figure 6E) and consequently enhanced susceptibility to PtoDC3000 after spray infection (Figure 6F). In summary, our results suggest phosphorylation of RBOHD S703 is an important regulatory element of plant immune responses.
C-Terminal Phosphorylation Sites Are Conserved in Plant and Animal NOXs
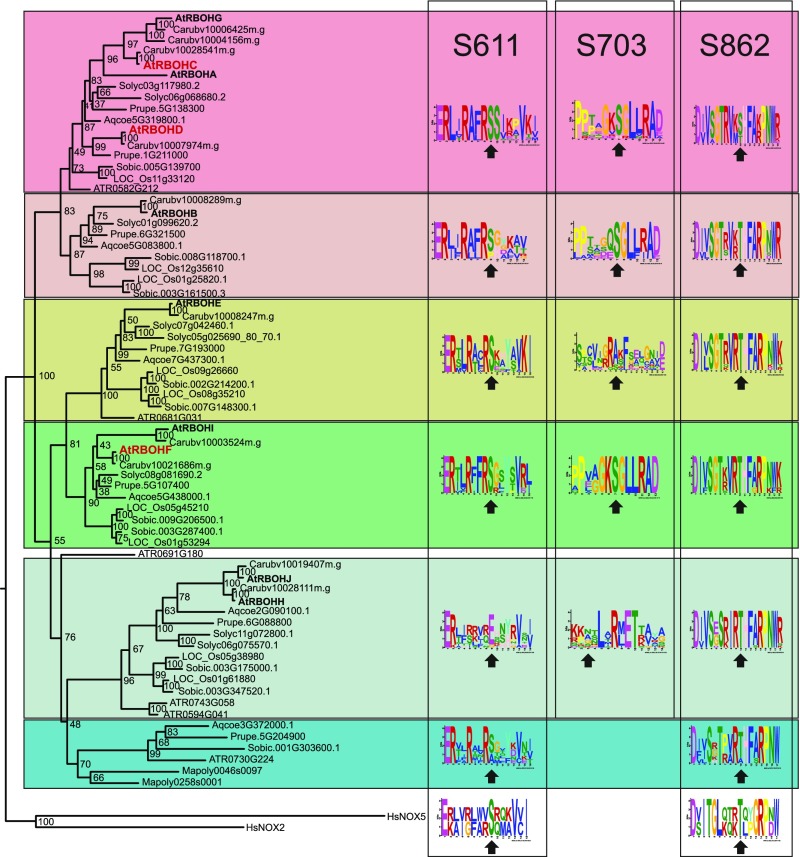
Since little is known about control of RBOH activity via its C terminus, we investigated whether regulation through S703 was unique to RBOHD or conserved in other RBOHs. We constructed a phylogenetic tree of RBOHs from plant genomes representing major branches of the plant lineages (Figure 7). Plant RBOHs form a monophyletic group, which is separated from human NOX2 and NOX5. The C-terminal region of plant NOXs contains a high number of serines and threonines (Supplemental Table 2). Furthermore, conservation of the amino acid sequence surrounding a phosphorylation site may be required for recognition of the site by the kinase (Miller and Turk, 2016; Trost et al., 2016). Accordingly, the sites corresponding to RBOHD S611, S703, and S862 exhibited conservation of amino acid motifs surrounding the phosphorylation site.
Figure 7.
Phosphorylation Sites in the C-Terminal Region Are Conserved in Plants and Animals.
Phylogenetic tree showing that plant RBOHs form a single clade that is parallel to the NADPH oxidases NOX2 and NOX5β from Homo sapiens. The tree was constructed using RAxML from a MAFFT alignment, 1000 bootstraps were calculated with RAxML. The full sequence alignment can be found in Wasabi at http://was.bi?id=p_nwi1. Plant species included were Arabidopsis (At), Capsella rubella (Carub), Prunus persica (Prupe), Solanum lycopersicum (Solyc), Aquilegia coerulea (Aqcoe), Oryza sativa (LOC_Os), Sorghum bicolor (Sobic), Amborella trichopoda (Atr), and Marchantia polymorpha (Mapoly). Numbers of phospho-sites in the meme figures represent the position of the amino acid in RBOHD from Arabidopsis. Arrows indicate the position of the phospho-site (S or T) or corresponding amino acid.
The phospho-sites in the C-terminal region displayed strong conservation throughout the plant RBOH clade. S703 was conserved in eight of the 10 RBOHs from Arabidopsis, but not in RBOHH and RBOHJ (Figure 7). In the clade containing AtRBOHE, the sequence motif harboring S703 was far less conserved compared to other clades. Intriguingly, AtRBOHE itself as well as its homolog from the Brassicaceae Capsella rubella contains a Ser corresponding to S703 of RBOHD, while the other members of the clade do not possess a Ser or Thr at this position. The phospho-sites S862, which may be involved in negative regulation based on experiments in HEK293T cells (Figure 5B), as well as S611 were strongly conserved in all plant RBOHs. Intriguingly, the sequence motifs harboring S611 and S862 are conserved even in human NOX2 and NOX5 (Figure 7). The C-terminal region binds FAD and NADPH. Therefore, it may underlie strong evolutionary constraints to conserve these binding properties. This may be also reflected in the strong conservation of C-terminal phospho-sites and their sequence context in plant and animal NOXs alike.
DISCUSSION
CRKs are a large group of RLKs involved in biotic and abiotic stress signaling in Arabidopsis (Bourdais et al., 2015). We have previously shown that flg22-triggered extracellular ROS production is altered in several crk mutants (Bourdais et al., 2015). In particular CRK2, a member of the basal clade of CRKs (Vaattovaara et al., 2019), has been highlighted since crk2 displays striking phenotypes (Bourdais et al., 2015), including reduced rosette size and diminished flg22-induced ROS production. Functional CRK2 restored the reduced rosette size (Figure 1) as well as the MAMP-triggered ROS burst (Figure 2). In addition to their roles in stress responses, extracellular ROS have also been implicated in leaf cell expansion (Schmidt et al., 2016). For example, the rbohD rbohF double mutant displays reduced rosette size (Torres et al., 2002). Overexpression of CRKs has been associated with increased SA accumulation (Chen et al., 2004; Acharya et al., 2007), which could potentially explain the growth phenotype of crk2. However, since SA levels were unaltered in the loss-of-function mutant crk2 (Supplemental Figure 1B), its smaller size may rather be a consequence of impaired ROS production (Figure 2). This is supported by the observation that CRK2 enhanced the activity of RBOHD and RBOHF in HEK293T cells (Figure 3A; Supplemental Figure 3F). Alternatively, other proteins interacting with or substrates of CRK2 might be involved in the regulation of plant growth.
In line with reduced MAMP- and DAMP-induced ROS production (Figures 2A and 2B; Supplemental Figures 2C and 2D), crk2 was more susceptible to the virulent bacterial pathogen Pto DC3000 (Figure 2C), suggesting that CRK2-mediated ROS production was essential to effectively counteract pathogen infection. Also, other flg22-induced defense responses were altered in crk2, including reduced changes in cytosolic Ca2+ and reduced stomatal closure and MAPK activation (Supplemental Figures 2E to 2H). Ca2+ is an important element in the activation of RBOH. Moreover, ROS also triggers Ca2+ fluxes in plants and contributes to stomatal closure (Kimura et al., 2017). Thus, the diminished increase of cytosolic Ca2+ and stomatal closure in crk2 may be a consequence of the impaired flg22-induced ROS production. Also, callose deposition (Caillaud et al., 2014; Ellinger et al., 2014) has been previously linked to ROS production (Couto and Zipfel, 2016) and CRK2 interacts with callose synthases, phosphorylates CALLOSE SYNTHASE 1 (CALS1) in vitro and salt-induced callose deposition is reduced in crk2 (Hunter et al., 2019). While flg22-triggered callose deposition in crk2 was indistinguishable from Col-0 after 12 h of treatment (Supplemental Figures 2I and 2J), crk2 exhibited more callose deposition after 30 min treatment with flg22 compared to Col-0. Interestingly, CRK2 forms clusters at the plasma membrane in response to flg22 treatment, and ROS is required for this process (Hunter et al., 2019). It is not clear how these clusters are integrated with the regulation of RBOHD activity, but it might serve to connect RBOHD-dependent ROS production with callose deposition. Another important element in response to biotic and abiotic cues is the activation of MAPK cascades (Bigeard et al., 2015; Boudsocq et al., 2015), and earlier reports suggest a bifurcation to ROS burst and MAPK activation in defense signaling following MAMP perception (Zhang et al., 2007; Yeh et al., 2016). CRK2 could be involved in balancing MAMP-induced ROS signaling pathways and MAPK signaling, but the mechanisms are still unclear.
Our results suggest that CRK2 participates in the control of ROS production via interaction with RBOHD. Intriguingly, CRK2 existed in a preformed complex with RBOHD in planta independent of MAMP treatment (Figure 3C), while many other RLK protein complexes are formed upon signal perception. The observation that, similar to flg22-triggered ROS production, also chitin-induced ROS production was reduced in crk2 suggests that CRK2 might not act on MAMP receptor complexes but rather on the regulation of ROS production itself. Phosphorylation of the C terminus is critical for the regulation of human NOXs. Phosphorylation of the NOX2 C terminus by protein kinase C enhances assembly of the multimeric NOX2 complex and its activity, whereas phosphorylation by ATM kinase inhibits NOX2 activity (Raad et al., 2009; Beaumel et al., 2017). NOX5 activity is regulated by Ca2+ binding to EF hands in the N terminus (Bánfi et al., 2004), but NOX5 is also activated by phosphorylation of the C terminus by protein kinase C α or calcium/calmodulin-dependent kinase II (Pandey et al., 2011; Chen et al., 2014). Although the C-terminal catalytic domain of RBOHs is highly conserved in plants and animals, the N terminus has been considered as important for activation of the ROS-production activity. Accordingly, multiple phospho-sites (S8, S39, S133, S148, S163, S339, S343, and S347) in the N-terminal region have been reported. Intriguingly, CRK2cyto interacted with and phosphorylated the RBOHD C-terminal region at S611, S703, and S862 (Figures 3E and 4B; Supplemental Table 1). Phosphorylation of S703 upon xylanase treatment has been reported (Benschop et al., 2007) but not linked with other MAMPs or modulation of ROS production. Treatment with flg22 enhanced phosphorylation of S703 in Arabidopsis (Figures 6C and 6D; Supplemental Figure 5D). Mutation S703A in RBOHD led to reduced CRK2-dependent RBOHD activity in HEK293T cells (Figure 5B) and decreased flg22-induced ROS production in Arabidopsis (Figure 6E; Supplemental Figure 6D). The flg22-induced phosphorylation of S703 was diminished in CRK2D450N-YFP/crk2 plants compared to the wild type (Figure 6D). These results suggest that phosphorylation of the RBOHD C terminus at S703 by CRK2 contributes to the regulation of MAMP-induced ROS production. However, other kinases than CRK2 might also target S703 in RBOHD since flg22-induced phosphorylation of S703 in CRK2D450N-YFP/crk2 plants was significantly reduced but not completely abolished. Phosphorylation sites in the C terminus are highly conserved among RBOHs (Figure 7), suggesting that phosphorylation of the C-terminal region could be a general feature of plant NOXs. Remarkably, two putative RBOHD phospho-sites, S611 and S862, were identified even in the human NOXs NOX2 and NOX5 (Figure 7).
Substitution of RBOHD S862 to Ala resulted in enhanced ROS-producing activity in HEK293T cells. However, substitution of RBOHD S611 to Ala, similarly to RBOHD S39A, did not alter ROS production in HEK293T cells (Figure 5). RBOHD S39A also did not affect flg22-induced ROS production but phospho-mimic S39D enhanced the ROS production and phosphorylation of S39 is enhanced by MAMP treatment in planta (Kadota et al., 2014; Figure 6B;Supplemental Figure 5C). These results suggest the importance to determine the phosphorylation status of S611 and S862 in planta in the future. RBOHD can also be regulated by Cys S-nitrosylation in the C terminus (Yun et al., 2011), but it is unclear how this modification is integrated with other regulatory mechanisms. Taken together, our results suggest that phosphorylation of the C-terminal region of plant NOXs is strongly conserved and important for controlling ROS production.
Several protein kinases phosphorylate RBOHD N terminus and regulate the activity including receptor-like cytoplasmic kinases (Kadota et al., 2014; Li et al., 2014; Lin et al., 2015), MAP4Ks (Zhang et al., 2018), CPKs (Dubiella et al., 2013), and RLKs (Chen et al., 2017), but how is regulation by phosphorylation of the N- and C-terminal regions coordinated? BIK1 activates RBOHD by phosphorylation (Kadota et al., 2014; Li et al., 2014), and ROS production in bik1 is reduced to a similar extent as in crk2. However, decreased flg22-induced ROS production in crk2 was not due to lower BIK1 transcript abundance (Supplemental Figure 7). BIK1 homologs, AVRPPHB SUSCEPTIBLE1 (PBS1) and AVRPPHB SUSCEPTIBLE1-LIKE (PBL) kinases, contribute to the regulation of RBOH activity and ROS production is progressively reduced in double mutants of bik1 with its homologs (Lin et al., 2015; Zhang et al., 2018). CRK2 and BIK1 might synergistically regulate ROS production, but we were unable to obtain a double mutant between bik1 and crk2 (Supplemental Table 3). Therefore, we propose that at least one of these components is essentially required. BIK1 has previously been shown to interact with other kinases including, CRKs (Lee et al., 2017), but interaction with CRK2 has not been investigated. BIK1 and CRK2 are likely highly coordinated in order to precisely control ROS production in response to environmental stimuli (Figure 8). Like CRKs, RBOHs are involved in diverse processes in stress responses and also plant development, and it is conceivable that different CRKs regulate the diverse set of RBOH proteins in various cellular contexts potentially via phosphorylation of the C-terminal region.
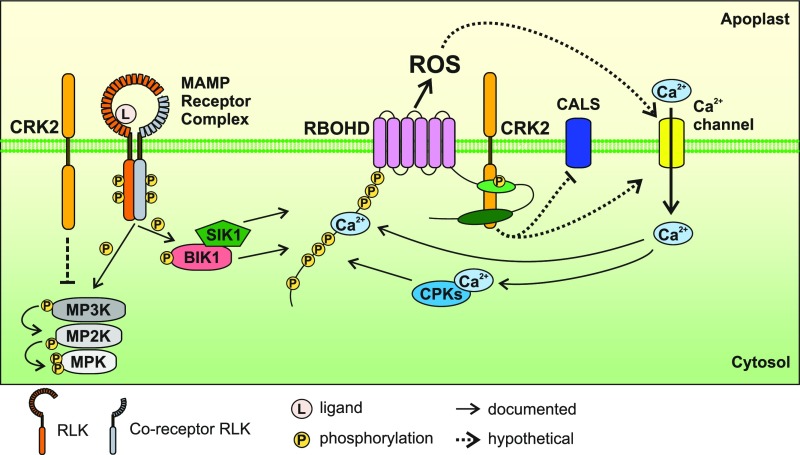
Figure 8.
Schematic Model for MAMP-Triggered RBOHD Activation.
MAMPs are recognized by MAMP receptor complexes. RBOHD N terminus is phosphorylated by BIK1 and SIK1, and apoplastic ROS production is induced. Apoplastic ROS production by RBOHD leads to Ca2+ influx into the cytosol. Ca2+ binding to RBOHD N terminus and to CPKs leads to Ca2+-dependent activation of RBOHD. We found that CRK2 also contributes to the activation of RBOHD via phosphorylation of its C terminus at S703. CRK2 can also mediate inhibition of MAPK activation and callose deposition via CALS after MAMP perception. MPK, mitogen-activated protein kinase; MP2K, MPKK; MP3K, MPKKK.
In summary, we propose that CRK2 is a central element in orchestrating the extracellular ROS burst and in mediating the balance between different defense responses. The full complexity and integration of the regulatory components controlling RBOH activity is still a topic of much speculation (Kimura et al., 2017). The diversity of regulators converging at RBOHs reflects the prominent role of apoplastic ROS in signal transduction, while strict control of their activity is required to circumnavigate oxidative damage. We suggest that RBOHD is regulated by phosphorylation of the C-terminal region to complement regulatory mechanisms targeting the N terminus (Figure 8). Based on the conservation of amino acid motifs in the C terminus of NOXs harboring Ser and Thr residues (Figure 7), we propose that this mode of regulation could be evolutionarily conserved in plants and animals. In the future, it will be interesting to investigate how CRK-mediated phosphorylation of the RBOH C terminus is integrated in the diverse processes that incorporate extracellular ROS.
METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants used in this study include Col-0, crk2 (Bourdais et al., 2015), rbohD (Torres et al., 2002), fls2 (Zipfel et al., 2004), bik1 (Veronese et al., 2006), 35S:FLAG-RBOHD/rbohD (Kadota et al., 2014), 35S:CRK2-YFP/Col-0, and 35S:YFP-6Myc/Col-0 (Hunter et al., 2019). To generate crk2/bik1 double mutant, crk2 and bik1 single-mutant plants were crossed. F1, F2, and F3 progenies were analyzed by PCR. F2 and F3 seeds were obtained by self-pollination. Primers are listed in Supplemental Table 4.
Seeds were sterilized by 70% (v/v) ethanol 2% (v/v) Triton X-100 for 5 min and washed three times with 99% (v/v) ethanol. Surface sterilized seeds were sown on 1× or 0.5-strength Murashige and Skoog (MS) medium containing 1% (w/v) Suc and subsequently stratified for 2 to 4 d in the dark at 4°C. Plants were grown in growth chambers (Panasonic, #MLR-352-PE) under 12 h light/12 h dark (22°C/18°C) at 145 µmol m−2 s−1 (Panasonic FL40SS ENW/37). After 10 d, seedlings were transferred to soil and grown in growth rooms under the following conditions: 12 h light/12 h dark (23°C/19°C) at 220 to 280 µmol m−2 s−1 (Osram L 36W/865 Lumilux Cool Daylight or Osram L 58W/865 Lumilux Cool Daylight), relative humidity 50 to 60%, unless otherwise stated.
For SA quantification, seeds were sown in commercial soil with perlite (Hasselfors Garden Yrkeskvalitet S-jord). Seeds were stratified for 3 d at 4°C, then grown for a week in long-day conditions (16 h light, 22°C, 40% humidity/8 h dark, 18°C, 60% humidity) in 150 μmol m−2 s−1 photon flux density with a light source Philips Master TL5 HE 21W/830. Seedlings were then transplanted into fresh soil and grown for 18 d more before sampling.
Cell Culture and Transfection
HEK293T cells (ATCC, catalog no. CRL-3216) were maintained at 37°C in 5% (v/v) CO2 in Dulbecco’s Modified Eagle’s Medium nutrient mixture Ham’s F-12 (Sigma, catalog no. D8062) supplemented with 10% (v/v) fetal bovine serum (Gibco, catalog no. 26140-079). Cells were transfected with pcDNA3.1 and pEF1 vectors using GeneJuice transfection regent (Merck Millipore, catalog no. 70967-3) according to the manufacturer’s instructions.
Plasmid Construction
CRK2 and RBOHD constructs for Arabidopsis were generated through MultiSite Gateway technology (Invitrogen). To generate pBm43GW-CRK2pro:CRK2-Venus (YFP)-3AT for crk2 complementation lines, the coding region of CRK2 or kinase-dead mutants (K353E or D450N) were recombined into pENTR/D-TOPO vector (Invitrogen). pDONRP4P1R/zeo-CRK2pro, pDONR/zeo-CRK2 (or pENTR/D-TOPO-CRK2 kinase-dead mutant), and p2R3a-VenusYFP-3AT were recombined with pBm43GW. To generate pHm43GW-pRBOHD:3FLAG-RBOHD-nosT, the coding region of 3FLAG-RBOHD was amplified by PCR from pcDNA3.1-3FLAG-RBOHD and cloned into pDONR/zeo vector (Invitrogen). The promoter region of RBOHD was amplified by PCR from pBin19g-pRBOHD:3FLAG-RBOHD and cloned into pDONRP4P1R/zeo vector (Invitrogen). pDONRP4P1R/zeo-RBOHDpro, pDONR/zeo-3FLAG-RBOHD, and p2R3a-nosT were recombined with pHm43GW. Single amino acid substitution mutants of CRK2 and RBOHD were generated by point-mutant primers and the mega-primer PCR method. pBm43GW-CRK2pro:CRK2-YFP-3AT and pHm43GW-pRBOHD:3FLAG-RBOHD-nosT constructs were transformed into crk2 and rbohD plants, respectively, by Agrobacterium tumefaciens strain GV3101 (pSoup)-mediated floral dipping (Clough and Bent, 1998). p2R3a-Venus(YFP)-3AT, p2R3a-nosT, pBm43GW and pHm43GW (Siligato et al., 2016), pBin19g-pRBOHD:3FLAG-RBOHD (Kadota et al., 2014), pcDNA3.1-3FLAG-RBOHD (Kaya et al., 2019), pDONR/zeo-CRK2, and pDONRP4P1R/zeo-CRK2pro (Hunter et al., 2019) have been described previously.
6His-GST-CRK2cyto and 6His-MBP-RBOHD/C constructs for recombinant proteins were generated by using In-Fusion technology (Clontech). The coding regions of CRK2cyto (wild type, K353E, and D450N) and RBOHD/C (full-length, C1, C2, and C3) were amplified by PCR and cloned into pOPINK (Addgene, catalog no. 41143) or pOPINM (Addgene, catalog no. 26044) vectors. pOPINM-RBOHD/N was previously described by Kadota et al. (2014).
For HEK293T cell experiments, pEF1-MCS-3Myc (BamHI-NotI-3Myc-stop fragment was inserted between KpnI and XbaI sites of pEF1/myc-His B vector [Invitrogen]) was generated. To generate pEF1-CRK2 (wild type or D450N)-3Myc, the codon-optimized coding sequence of Kozak-CRK2 (wild type or D450N) was cloned between BamHI and NotI sites of pEF1-MCS-3Myc. To generate pcDNA3.1-3FLAG-RBOHD mutant constructs, the coding regions of RBOHD (S8A, S39A, S611A, S703A, or S862A) were cloned into the BamHI site of pcDNA3.1-3FLAG-MCS (Kozak-3FLAG-BamHI-EcoRV-stop fragment was inserted between NheI and KpnI sites of pcDNA3.1(−) vector [Invitrogen]). Amino acid substituted mutants of CRK2 and RBOHD were generated by point-mutant primers and the mega-primer PCR method. pEF1-3Myc-GFP (Kawarazaki et al., 2013), pcDNA3.1-3FLAG-RBOHD, pcDNA3.1-3FLAG-RBOHC, pcDNA3.1-3FLAG-RBOHF, and pcDNA3.1-3FLAG-MCS were previously described by Kaya et al. (2019). Primer sequences are listed in Supplemental Table 4.
Subcellular Protein Localization
Fluorescent images were obtained using a TCS SP5 II HCS confocal microscope (Leica). For investigation of CRK2-YFP localization, 514 nm excitation and 525 to 590 nm detection range were used.
ROS Measurements
Leaf discs were collected using a cork borer from 4-week-old Arabidopsis plants and floated overnight in sterile distilled water in 96-well plates under continuous light (17 µmol m−2 s−1, light-emitting diode [LED]) at room temperature. On the following day, water was replaced with assay buffer containing 34 mg/L luminol sodium salt (Sigma Aldrich, catalog no. A4685), 20 mg/L horse radish peroxidase (Fujifilm Wako, catalog no. 169-10791), 200 nM flg22 (GenScript), 200 µg/mL Chitin (Sigma Aldrich, catalog no. C9752), or 1 µM AtPep1 (ATKVKAKQRGKEKVSSGRPGQHN, synthesized by Synpeptide). Luminescence was measured for 1 s every 1 min at room temperature using GloMax-Multi+Detection System (Promega). ROS production was expressed in relative luminescence units.
The ROS-producing activity of RBOHs in HEK293T cells was measured as previously described by Kimura et al. (2012). Two days after transfection, medium was removed and cells were gently washed with 1× Hanks’ Balanced Salt Solution (Gibco, catalog nos. 14025-092 or 14175-095). Measurements were started after addition of the assay buffer containing 250 µM lunimol sodium salt and 66.7 mg/L horseradish peroxidase. After 30 min measurement, 1 µM ionomycin (Calbiochem, catalog no. 407952) was added. Chemiluminescence was measured for 1 s every 1 min at 37°C using a GloMax-Multi+Detection System. ROS production was expressed in relative luminescence units. Expressed proteins were detected by immunoblotting with anti-FLAG (Sigma Aldrich, catalog no. F1804), anti-cMyc (Fujifilm Wako, catalog no. 017-2187), anti-β-actin (Sigma Aldrich, catalog no. A5316), and IRDye800CW anti-mouse IgG (LI-COR, catalog no. 926-32210) antibodies.
Bacterial Growth Assay
To quantify bacterial growth on 4-week-old plants infected with the virulent Pto DC3000 (Whalen et al., 1991), growth curve assays were performed as previously described by Wrzaczek et al. (2007). Spray infections were performed as previously described (Yao et al., 2013).
Stomatal Aperture
Measurements of stomatal aperture were performed as previously described by Kadota et al. (2014) with minor modifications. Leaves were cut at the petiole from 3-week-old plants and incubated in water for 2 h. Samples were then incubated overnight in stomatal opening buffer (50 mM KCl, 10 µM CaCl2, 0.01% [v/v] Tween20, 10 mM MES, pH 6.15) to induce stomatal opening. For treatments, 5 µM flg22 was added to the buffer for 2 h prior to imaging. Brightfield images were obtained using a TCS SP5 II HCS confocal microscope (Leica).
Ca2+ Imaging
Calcium imaging with YCNano-65-expressing plants was performed as previously described by Choi et al. (2014), Lenglet et al. (2017), and Toyota et al. (2018). In brief, YCNano-65 was visualized by a fluorescence stereo microscope (Nikon) with a 1× objective lens (Nikon), image-splitting optics (Hamamatsu Photonics), and a sCMOS camera (Hamamatsu Photonics). To excite YCNano-65, a mercury lamp (Nikon), a 436/20-nm excitation filter (Chroma) and a 455 nm dichroic mirror (Chroma) were used. The fluorescent signal from YCNano-65 was separated by a 515 nm dichroic mirror (Chroma) equipped in the image splitting optics. The resultant cyan fluorescent protein (CFP) and YFP (FRET) signals passed independently through a 480/40 nm and 535/30 nm emission filters, respectively (Chroma). A pair of the CFP and FRET images was simultaneously acquired every 4 s with the sCMOS camera using NIS-Elements imaging software (Nikon). A 7-d-old seedling was filled with ∼50 μL of 1 µM flg22 or 1× MS 1% (w/v) Suc liquid media. A region of interest was placed on the adaxial surface of cotyledons to analyze both CFP and FRET signals. The FRET/CFP ratio was calculated by the 6D imaging plug-in modules in NIS-Elements imaging software (Nikon).
MAPK Assay
MAPK assays were performed as previously described (Yadeta et al., 2017). In brief, 4-week-old Arabidopsis plants were sprayed with 10 µM flg22 with 0.025% (v/v) Silwett L-77. Leaf samples were ground in liquid nitrogen and sand. Extraction buffer (50 mM HEPES [pH 7.4], 50 mM NaCl, 10 mM EDTA, 0.2% [v/v] Triron X-100, 1% [v/v] protease inhibitor cocktail [Sigma, catalog no. P9599], 1% [v/v] Halt phosphatase inhibitor cocktail [Thermo Fisher Scientific, catalog no. 78428]) was added (2 mL/g plant powder). Samples were incubated at 4°C for 30 min and centrifuged at 12,000g, 4°C for 10 min. The supernatant was used for immunoblotting with anti-Phospho-p44/42 MAPK (Cell Signaling Technology, catalog no. 4370) and IRDye800CW anti-rabbit IgG (LI-COR, catalog no. 926-32211) antibodies.
Callose Staining
Callose staining was performed as previously described by Hunter et al. (2019).
RT-qPCR
Col-0, crk2, and fls2 seedlings were grown on MS 1% (w/v) Suc agar plate for 5 d and were transferred into MS 1% (w/v) Suc liquid media and grown for 5 d. Plants were incubated with 1 µM flg22 for 30 min, 1 h, and 3 h, respectively. Plants were ground in liquid nitrogen and total RNA was extracted using the GeneJET Plant RNA purification kit (Thermo Fisher Scientific, catalog no. K0802). Total RNA was treated with DNase I (Thermo Fisher Scientific, #EN0525), and cDNA was synthesized with Maxima H Minus Reverse Transcriptase (Thermo Fisher Scientific, catalog no. EP0751). qPCR analysis was performed with CFX real-time PCR (Bio-Rad) using 5× HOT FIREPol EvaGreen qPCR mix plus ROX (Soils Biodyne). SAND, TIP41, and YLS8 were used as reference genes for normalization. Relative expression was calculated with qBase+ (Biogazelle; https://www.qbaseplus.com/). Primers are listed in Supplemental Table 4.
Phytohormone Analysis
SA was analyzed from adult rosettes as previously described by Forcat et al. (2008) with minor modifications. Rosettes were flash-frozen in liquid nitrogen and freeze-dried for 24 h. Rosettes were ground in a mortar before weighing in to 2-mL tubes. About 6 mg aliquots of freeze-dried material were further homogenized by shaking with 5-mm stainless steel beads in a Tissue Lyser II (Qiagen) for 2 min at 25 Hz. Shaking was repeated after addition of 400 μL extraction buffer (10% [v/v] methanol, 1% [v/v] acetic acid) with internal standard (28 ng Salicylic-d4 Acid; CDN Isotopes). Samples were then incubated on ice for 30 min and centrifuged for 10 min at 16,000g and 4°C. Supernatants were centrifuged three times to remove all debris before liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis. The chromatographic separation was performed using an Acquity UHPLC thermo system (Waters) equipped with a Cortecs C18 column (2.7 μm, 2.1 × 100 mm; Waters). The solvent gradient (acetonitrile [ACN] /water with 0.1% [v/v] formic acid each) was adapted to a total run time of 7 min: 0 to 4 min 20% (v/v) to 95% (v/v) ACN, 4 to 5 min 95% (v/v) ACN, 5 to 7 min 95% (v/v) to 20% (v/v) ACN; flow rate 0.4 mL/min. For hormone identification and quantification, a tandem mass spectrometric system Xevo TQ-XS, triple quadrupole mass analyzer with a ZSpray ESI function (Waters) was used. Mass transitions were SA 137 > 93, D4-SA 141 > 97.
Protein Extraction and Coimmunoprecipitation
Coimmunoprecipitation was performed as previously described by Kadota et al. (2016). Homozygous 35S:FLAG-RBOHD/rbohD was crossed with homozygous 35S:CRK2-YFP/Col-0 or 35S:YFP-6Myc/Col-0. 35S:FLAG-RBOHD/35S:CRK2-YFP/rbohD F3 plants were selected by kanamycin resistance (homozygous FLAG-RBOHD insertion) and PCR (homozygous rbohD T-DNA insertion). F1 and F3 plants were grown on MS 1% (w/v) Suc agar plate for 7 d and were transferred into MS 1% (w/v) Suc liquid media and grown for 8 to 10 days. F3 plants were incubated in water or 1 µM flg22 for 10 min or 30 min after vacuum application for 2 min. Plants were ground in liquid nitrogen and sand. Extraction buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10% [v/v] glycerol, 5 mM DTT, 1% [v/v] protease inhibitor cocktail [Sigma Aldrich, P9599], 2% [v/v] IGEPAL CA630, 1 mM Na2MoO42H2O, 2.5 mM NaF, 1.5 mM activated sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride) was added at 1.5 to 2 mL/g fresh weight. Samples were incubated at 4°C for 1 h and centrifuged at 15,000g, 4°C for 20 min. Supernatants were adjusted to 5 mg/mL protein concentration and incubated for 1 h at 4°C with 100 μL of anti-GFP magnetic beads (Miltenyi Biotec, catalog no. 130-091-125). Bound proteins were analyzed by immunoblotting with anti-GFP (Invitrogen, catalog no. A11122), anti-RBOHD (Agrisera, catalog no. AS15-2962), and IRDye800CW anti-rabbit IgG (LI-COR, #926-32211) antibodies.
To detect CRK2-YFP, total protein was extracted from 13-day-old CRK2pro:CRK2 (wild type, K353E, or D450N)/crk2 T3 homozygous plants with the same extraction buffer and analyzed by immunoblotting with anti-GFP (Invitrogen, catalog no. A11122) and IRDye800CW anti-rabbit IgG (LI-COR, catalog no. 926-32211) antibodies. GFP signal intensity was quantified by ImageJ and normalized by Coomassie Brilliant Blue staining intensity.
To detect 3FLAG-RBOHD, total protein was extracted from 14-d-old RBOHDpro:3FLAG-RBOHD (wild type or S703A)/rbohD T3 homozygous plants with the same extraction buffer and analyzed by immunoblotting with anti-FLAG (Sigma Aldrich, catalog no. F1804 and IRDye800CW anti-mouse IgG (LI-COR, catalog no. 926-32210) antibodies.
Protein Purification from E. coli
Cytosolic regions of CRK2 were expressed in Escherichia coli Lemo21. Cytosolic regions of RBOHD and MBP were expressed in E. coli BL21. GST-tagged recombinant proteins were purified using glutathione sepharose 4B (GE Healthcare, catalog no. 17-0756-01) according to manufacturer’s instructions. MBP-tagged proteins were purified using amylose resin (New England Biolabs, catalog no. E8021S) according to manufacturer’s instructions.
In Vitro Pull-Down
6His-GST-CRK2cyto, 6His-MBP-RBOHD/N, 6His-MBP-RBOHD/C, and MBP were incubated with glutathione Sepharose 4B in the pull-down buffer (20 mM HEPES, 50 mM KCl, 5 mM MgCl2, 1% [v/v] Tween 20, 1 mM DTT, and 100 µM phenylmethylsulfonyl fluoride) at 4°C for 1 h. The glutathione sepharose 4B was washed four times with the pull-down buffer and eluted with 10 mM reduced gluthatione. The mixture was analyzed by immunoblotting anti-6× His (Invitrogen, catalog no. MA1-135), anti-MBP (Santa Cruz Biotechnology, catalog no. sc-13564), and IRDy800CW anti-mouse IgG antibodies.
In Vitro Kinase Assay
Purified recombinant proteins were incubated with [γ-32P] ATP for 30 min at room temperature in the kinase assay buffer (50 mM HEPES [pH 7.4], 1 mM DTT, 10 mM MgCl2, 0.6 mM unlabeled ATP). The mixture was subsequently separated by SDS-PAGE, and autoradiography was detected by FLA-5100 image analyzer (Fujifilm, Japan). For identification of in vitro phosphorylation sites by LC-electrospray ionization (ESI)-MS/MS, 1.5 mM unlabeled ATP was used in the kinase buffer. The proteins were separated by SDS-PAGE, followed by Coomassie Brilliant Blue staining and were digested by trypsin (Thermo Fisher Scientific, catalog no. 90057) or Lys-C (Thermo Fisher Scientific, catalog no. 90051).
Identification of In Vitro Phosphorylation Sites of RBOHD by LC-Electrospray Ionization-MS/MS
Trypsin or Lys-C-digested protein samples were analyzed by a Q Exactive mass spectrometer (Thermo Fisher Scientific) connected to Easy NanoLC 1000 (Thermo Fisher Scientific). Peptides were first loaded on a trapping column and subsequently separated inline on a 15-cm C18 column (75 μm × 15 cm, ReproSil-Pur 5 μm 200 Å C18-AQ, Dr. Maisch HPLC). The mobile phase consisted of water with 0.1% (v/v) formic acid (solvent A) or acetonitrile/water (80:20 [v/v]) with 0.1% (v/v) formic acid (solvent B). A linear 10-min gradient from 8 to 42% (v/v) B was used to elute peptides.
Mass spectrometry data was acquired automatically by using Xcalibur 3.1 software (Thermo Fisher Scientific). An information-dependent acquisition method consisted of an Orbitrap mass spectrometry survey scan of mass range 300 to 2000 m/z (mass-to-charge ratio) followed by higher-energy collisional dissociation (HCD) fragmentation for 10 most intense peptide ions. Raw data was searched for protein identification by Proteome Discoverer (version 2.2; http://www.matrixscience.com/blog/mascot-workflows-in-proteome-discoverer.html) connected to in-house Mascot (v. 2.6.1; https://www.thermofisher.com/fi/en/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/multi-omics-data-analysis/proteome-discoverer-software.html) server. Phosphorylation site locations were validated using phosphoRS algorithm (Taus et al., 2011). A SwissProt database (https://www.uniprot.org/) with a taxonomy filter Arabidopsis was used. Two missed cleavages were allowed. Peptide mass tolerance ± 10 ppm and fragment mass tolerance ± 0.02 D were used. Carbamidomethyl (C) was set as a fixed modification and Met oxidation, acetylation of protein N terminus, and phosphorylation of Ser and Thr were included as variable modifications. Only peptides with a false discovery rate of 0.01 were used.
Targeted (Phospho) Peptide Analysis
Plant Treatment
Arabidopsis seeds were sterilized by incubating with 1.5% (w/v) NaClO 0.02% (v/v) Triton X-100 solution for 5 min and vernalized at 4°C for 2 d. Sterilized seeds were germinated and grown in liquid culture on 6-well plates (30 seeds/well) in MGRL medium with 0.1% (w/v) Suc (2 mL/well; Fujiwara et al., 1992) at 23°C under continuous light (100 µmol m−2 s−1, LED) in a Percival growth chamber. Plates with 11-d-old seedlings were transferred from the growth chamber to a workbench and kept overnight for acclimatization before treatments. Seedlings were treated with either 1 µM flg22 or sterile water for 5 min, after which seedlings were immediately collected and flash-frozen in liquid nitrogen and stored at −80°C. Leaf discs were collected using a cork borer from 4-week-old Arabidopsis plants and floated overnight in sterile distilled water in Petri dishes under continuous light (17 µmol m−2 s−1, LED) at room temperature. On the following day, water was replaced with new sterile distilled water or 200 nM flg22. After 5 min incubation, leaf discs were immediately collected and flash-frozen in liquid nitrogen and stored at −80°C.
Phosphopeptide Enrichment
Frozen seedlings or leaf discs were disrupted using a Retsch mill (5 min, 30 Hz), and 500 μL urea extraction buffer (8 M urea in 100 mM Tris, pH 8.5, 20 µL/mL phosphatase inhibitor cocktail 3 [Sigma Aldrich, catalog no.P0044], 20 µL/mL phosphatase inhibitor cocktail 2 [Sigma Aldrich, catalog no. P5726], 5 mM DTT) was added to the disrupted frozen powders, mixed briefly, and incubated at room temperature (RT) for 30 min. After centrifugation at 15,000g for 10 min, supernatants were transferred to fresh tubes. Protein concentrations were determined using a Pierce 660-nm protein assay (Thermo Fisher Scientific). Extracts with 500 µg of protein were alkylated with 14 mM chloroacetamide at RT for 30 min in the dark, chloroacetamide was quenched by addition of 1/200 sample volume 1 M DTT. Samples were diluted 1:8 with 0.1 M Tris, pH 8.5, and 1 mM CaCl2 and were digested overnight at RT either with 5 µg trypsin or 5 µg Lys-C. Digestion reaction was terminated by addition of trifluoroacetic acid (TFA; 0.1% [v/v] final concentration), and peptides were desalted using C18 SepPaks (1-cc cartridge, 100 mg [WAT023590]). In brief, SepPaks were conditioned using methanol (1 mL), buffer B (80% [v/v] acetonitrile, 0.1% [v/v] TFA; 1 mL), and buffer A (0.1% [v/v] TFA; 2 mL). Samples were loaded by gravity flow, washed with buffer A (1 × 1 mL, 1 × 2 mL) and eluted with buffer B (2 × 400 µL). Forty microliters of eluates were kept separately to measure nonphosphopeptides, and the rest were used for further phosphopeptide enrichment. Phosphopeptide enrichment was performed by hydroxy acid-modified metal-oxide chromatography using titania as previously described, with minor modifications, by Nakagami (2014) and Sugiyama et al. (2007).
LC-MS/MS Data Acquisition
Samples were analyzed using an EASY-nLC 1200 (Thermo Fisher) coupled to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific). Peptides were separated on 16-cm frit-less silica emitters (New Objective, 0.75-µm inner diameter), packed in-house with reversed-phase ReproSil-Pur C18 AQ 1.9 µm resin (Dr. Maisch). Peptides were loaded on the column and eluted for 115 min using a segmented linear gradient of 5 to 95% (v/v) solvent B (0 min, 5% B; 0 to 5 min, 5% B; 5 to 65 min, 20% B; 65 to 90 min, 35% B; 90 to 100 min, 55% B; 100 to 105 min, 95% B; 105 to 115 min, 95% B; solvent A [0% ACN, 0.1% (v/v) formic acid]; solvent B [80% (v/v) ACN, 0.1% (v/v) formic acid]) at a flow rate of 300 nL/min. Mass spectra were acquired using a targeted (parallel reaction monitoring [PRM]) approach. The acquisition method consisted of a full-scan method combined with a nonscheduled PRM method. The 16 targeted precursor ions were selected based on the results of a data-dependent acquisition peptide search of phospho-enriched samples in Skyline (version 4.2.0.x; https://skyline.ms; MacLean et al., 2010). Mass spectrometry spectra were acquired in the Orbitrap analyzer with a mass range of 300 to 1750 m/z at a resolution of 70,000 full width at half maximum (FWHM) and a target value of 3 × 106 ions, followed by MS/MS acquisition for the 16 targeted precursors. Precursors were selected with an isolation window of 2.0 m/z. HCD fragmentation was performed at a normalized collision energy of 27. MS/MS spectra were acquired with a target value of 2 × 105 ions at a resolution of 17,500 FWHM, a maximum injection time of 120 ms, and a fixed first mass of m/z 100.
Mass Spectrometry Data Analysis
Raw data from PRM acquisition were processed using MaxQuant software (version 1.5.7.4; http://www.maxquant.org/; Cox and Mann, 2008). MS/MS spectra were searched by the Andromeda search engine against a combined database containing the sequences from Arabidopsis (TAIR10_pep_20101214; ftp://ftp.arabidopsis.org/home/tair/Proteins/TAIR10_protein_lists/) and sequences of 248 common contaminant proteins and decoy sequences. Trypsin specificity was required, and a maximum of two missed cleavages allowed. Minimal peptide length was set to seven amino acids. Carbamidomethylation of Cys residues was set as fixed, and phosphorylation of Ser, Thr and Tyr, oxidation of Met, and protein N-terminal acetylation was set as variable modifications. The match between runs option was disabled. Peptide-spectrum matches and proteins were retained if they were below a false discovery rate of 1% in both cases. The “msms.txt” output from MaxQuant was further analyzed using Skyline in PRM mode. Trypsin specificity was required and a maximum of two missed cleavages allowed. Minimal and maximum peptide lengths were set to seven and 25 amino acids, respectively. Carbamidomethylation of Cys, phosphorylation of Ser, Thr and Tyr, oxidation of Met, and protein N-terminal acetylation were set as modifications. Results were filtered for precursor charges of 2 and 3, and b- and y-ions with ion charges of +1 and +2. Product ions were set to “from ion 1 to last ion.” All chromatograms were inspected manually and peak integration was corrected for best representation of mass spectrometry 2 signals. Peak area data was exported and further processed. The Skyline documents containing the data for the targeted phophoproteomics experiments have been uploaded to Panorama Public and can be obtained from https://panoramaweb.org/RBOHDphosphorylation.url. Raw data have been deposited to the ProteomeXchange Consortium via the Panorama partner repository with the data set identifier PXD013525 (http://proteomecentral.proteomexchange.org/cgi/GetData set?ID=PXD013525).
Phylogenetic Analysis
Sequences for plant RBOH genes were extracted from public genome databases and manually curated. Sequences were aligned using MAFFT version 7.407 (Katoh and Standley, 2013) using iterative refinement (L-INS-i). This alignment was used to infer a phylogenetic maximum likelihood tree using RAxML version 8.1.3 (Stamatakis, 2014) with the JTT or DAYHOFF substitution models (which resulted in identical phylogenetic trees). Thousand bootstrap replicates were calculated using RAxML. Human NOX2 and NOX5β were used as outgroup to root the phylogenetic tree of plant NOXs. The sequence alignment of plant RBOHs, human NOX2 and NOX5β can be viewed on the Veidenberg et al. (2016) web server (http://was.bi?id=p_nwi1) and the tree as a nexus file is available as the Supplemental File. Sequence motifs were analyzed using the MEME suite (Bailey et al., 2009).
Statistical Analysis
Statistical analyses were performed with JMP Pro13 (SAS, https://www.jmp.com/). One-way ANOVA results are in Supplemental Table 5.
Accession Numbers
Phylogenetic tree of human and plant NOXs with bootstrap information for 1000 replicates and corresponding sequence alignment has been deposited on Wasabi (http://was.bi?id=p_nwi1). Data for the targeted phophoproteomics experiments have been uploaded to Panorama Public (https://panoramaweb.org/RBOHDphosphorylation.url). Raw data have been deposited to the ProteomeXchange Consortium via the Panorama partner repository with the dataset identifier PXD013525 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD013525). Materials used in the experimental work are available from the authors upon request.
Supplemental Data
Supplemental Figure 1. Complementation of crk2 with CRK2pro:CRK2-YFP
Supplemental Figure 2. MAMP-triggered ROS production and molecular responses in crk2
Supplemental Figure 3. CRK2 modulates the ROS-producing activity of RBOHC, D, and F in HEK293T cells
Supplemental Figure 4. ROS production activity of RBOHD S703A and S862A in HEK293T cells
Supplemental Figure 5. Quantification of RBOHD and MPK phosphorylation upon flg22 treatment
Supplemental Figure 6. RBOHD S703 is involved in regulation of flg22-induced ROS production
Supplemental Figure 7. Reduced ROS production in crk2 is not due to lower expression of BIK1
Supplemental Table 1. In vitro phosphorylation sites of 6His-MBP-RBOHDcyto by 6His-GST-CRK2cyto
Supplemental Table 2. The numbers of Ser (S) and Thre (T) residues in C terminus of NOXs in plants and animals
Supplemental Table 3. Progeny of CRK2/crk2 BIK1/bik1 parent and CRK2/crk2 bik1/bik1 parent
Supplemental Table 4. Primer sequences
Supplemental Table 5. One-way ANOVA results
Supplemental File. Nexus file of the alignment corresponding to the phylogenetic tree in Figure 7
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
The authors would like to thank Julia Krasensky-Wrzaczek and Alexey Shapiguzov (University of Helsinki, Finland) for critical comments on the article. We thank Tuomas Puukko, Nghia Le Tri, Simon Schmitz, Denis Owczarek, Jan-Niklas Weber (University of Helsinki, Finland), and Jiaqi Wang (Saitama University, Japan) for technical assistance and Riccardo Siligato for the Gateway Multisite vector system. L.V. and T.H. thank Trude Johansen (Norwegian University of Science and Technology, Norway) for technical assistance in LC-MS/MS-based hormone analysis. We thank Yasuhiro Kadota (RIKEN Yokohama, Japan) and Cyril Zipfel (University of Zurich, Switzerland) for 35S:FLAG-RBOHD/rbohD seeds and pOPINM-RBOHD/N and pBin19g-pRBOHD:3FLAG-RBOHD plasmids. We thank Simon Gilroy (University of Wisconsin) for YCNano65 seeds. The pcDNA3.1-3FLAG-RBOHD and pcDNA3.1-3FLAG-MCS plasmids were provided by Kazuyuki Kuchitsu (Tokyo University of Science, Japan). Microscopy imaging was performed at the Light Microscopy Unit, Institute of Biotechnology, University of Helsinki. Mass spectrometry analyses were performed at the Turku Proteomics Facility, supported by Biocenter Finland. This work was supported by the Academy of Finland (grants 275632, 283139, 312498, and 323917 to M.W.), the University of Helsinki (three–year fund allocation to M.W.), the Finnish Cultural Foundation (grant numbers 00170046 and 00181379 to L.V.), KAKENHI (grants 17H05007, 18H04775, and 18H05491 to M.T.), and the Max-Planck-Gesellschaft (to H.N.). K.H., S.K., A.V., and M.W. are members of the Centre of Excellence in the Molecular Biology of Primary Producers (2014 to 2019) funded by the Academy of Finland (grants 271832 and 307335).
AUTHOR CONTRIBUTIONS
S.K., K.H., H.N., and M.W. conceived and designed the project. S.K., K.H., L.V., H.C.T., M.C., A.V., A.R., L.M., M.M., T.W., M.T., and M.W. carried out experiments. S.K., K.H., L.V., A.V., A.R., T.H., M.T., and M.W. analyzed the data. A.H., S.C.S., and H.N. designed and performed targeted mass spectrometry analysis and analyzed the data. S.K. and M.W. wrote the article. All authors read and contributed to the final article.
Footnotes
Articles can be viewed without a subscription.
References
- Acharya B.R., Raina S., Maqbool S.B., Jagadeeswaran G., Mosher S.L., Appel H.M., Schultz J.C., Klessig D.F., Raina R.(2007). Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 50: 488–499. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.(2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37: W202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bánfi B., Tirone F., Durussel I., Knisz J., Moskwa P., Molnár G.Z., Krause K.H., Cox J.A.(2004). Mechanism of Ca2+ activation of the NADPH oxidase 5 (NOX5). J. Biol. Chem. 279: 18583–18591. [DOI] [PubMed] [Google Scholar]
- Beaumel S., Picciocchi A., Debeurme F., Vivès C., Hesse A.M., Ferro M., Grunwald D., Stieglitz H., Thepchatri P., Smith S.M.E., Fieschi F., Stasia M.J.(2017). Down-regulation of NOX2 activity in phagocytes mediated by ATM-kinase dependent phosphorylation. Free Radic. Biol. Med. 113: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop J.J., Mohammed S., O’Flaherty M., Heck A.J., Slijper M., Menke F.L.(2007). Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6: 1198–1214. [DOI] [PubMed] [Google Scholar]
- Bigeard J., Colcombet J., Hirt H.(2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8: 521–539. [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Danquah A., de Zélicourt A., Hirt H., Colcombet J.(2015). Plant MAPK cascades: Just rapid signaling modules? Plant Signal. Behav. 10: e1062197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdais G., et al. ; CRK Consortium (2015). Large-scale phenomics identifies primary and fine-tuning roles for CRKs in responses related to oxidative stress. PLoS Genet. 11: e1005373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud M.C., Wirthmueller L., Sklenar J., Findlay K., Piquerez S.J., Jones A.M., Robatzek S., Jones J.D., Faulkner C.(2014). The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 10: e1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Cao Y., Li H., Kim D., Ahsan N., Thelen J., Stacey G.(2017). Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat. Commun. 8: 2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Yu Y., Haigh S., Johnson J., Lucas R., Stepp D.W., Fulton D.J.(2014). Regulation of NADPH oxidase 5 by protein kinase C isoforms. PLoS One 9: e88405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Fan B., Du L., Chen Z.(2004). Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol. Biol. 56: 271–283. [DOI] [PubMed] [Google Scholar]
- Chern M., Xu Q., Bart R.S., Bai W., Ruan D., Sze-To W.H., Canlas P.E., Jain R., Chen X., Ronald P.C.(2016). A genetic screen identifies a requirement for cysteine-rich-receptor-like kinases in rice NH1 (OsNPR1)-mediated immunity. PLoS Genet. 12: e1006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.G., Toyota M., Kim S.H., Hilleary R., Gilroy S.(2014). Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA 111: 6497–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Couto D., Zipfel C.(2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16: 537–552. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M.(2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26: 1367–1372. [DOI] [PubMed] [Google Scholar]
- Dubiella U., Seybold H., Durian G., Komander E., Lassig R., Witte C.P., Schulze W.X., Romeis T.(2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 110: 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger D., Sode B., Falter C., Voigt C.A.(2014). Resistance of callose synthase activity to free fatty acid inhibition as an indicator of Fusarium head blight resistance in wheat. Plant Signal. Behav. 9: e28982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcat S., Bennett M.H., Mansfield J.W., Grant M.R.(2008). A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Hirai M.Y., Chino M., Komeda Y., Naito S.(1992). Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 99: 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.P., Köster P., Drerup M.M., Scholz M., Li S., Edel K.H., Hashimoto K., Kuchitsu K., Hippler M., Kudla J.(2019). Fine-tuning of RBOHF activity is achieved by differential phosphorylation and Ca2+ binding. New Phytol. 221: 1935–1949. [DOI] [PubMed] [Google Scholar]
- Hunter K., Kimura S., Rokka A., Tran H.C., Toyota M., Kukkonen J.P., Wrzaczek M.(2019). CRK2 enhances salt tolerance by regulating callose deposition in connection with PLDα1. Plant Physiol. 180: 2004–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idänheimo N., Gauthier A., Salojärvi J., Siligato R., Brosché M., Kollist H., Mähönen A.P., Kangasjärvi J., Wrzaczek M.(2014). The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem. Biophys. Res. Commun. 445: 457–462. [DOI] [PubMed] [Google Scholar]
- Jagnandan D., Church J.E., Banfi B., Stuehr D.J., Marrero M.B., Fulton D.J.(2007). Novel mechanism of activation of NADPH oxidase 5: Calcium sensitization via phosphorylation. J. Biol. Chem. 282: 6494–6507. [DOI] [PubMed] [Google Scholar]
- Jiménez-Quesada M.J., Traverso J.A., Alché J.(2016). NADPH oxidase-dependent superoxide production in plant reproductive tissues. Front. Plant Sci. 7: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Macho A.P., Zipfel C.(2016). Immunoprecipitation of plasma membrane receptor-like kinases for identification of phosphorylation sites and associated proteins. Methods Mol. Biol. 1363: 133–144. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K., Zipfel C.(2015). Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56: 1472–1480. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D., Shirasu K., Menke F., Jones A., Zipfel C.(2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54: 43–55. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M.(2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawarazaki T., Kimura S., Iizuka A., Hanamata S., Nibori H., Michikawa M., Imai A., Abe M., Kaya H., Kuchitsu K.(2013). A low temperature-inducible protein AtSRC2 enhances the ROS-producing activity of NADPH oxidase AtRbohF. Biochim. Biophys. Acta 1833: 2775–2780. [DOI] [PubMed] [Google Scholar]
- Kaya H., et al. (2019). Comparative analysis of the reactive oxygen species-producing enzymatic activity of Arabidopsis NADPH oxidases. Plant J. 98: 291–300. [DOI] [PubMed] [Google Scholar]
- Kimura S., Kaya H., Kawarazaki T., Hiraoka G., Senzaki E., Michikawa M., Kuchitsu K.(2012). Protein phosphorylation is a prerequisite for the Ca2+-dependent activation of Arabidopsis NADPH oxidases and may function as a trigger for the positive feedback regulation of Ca2+ and reactive oxygen species. Biochim. Biophys. Acta 1823: 398–405. [DOI] [PubMed] [Google Scholar]
- Kimura S., Waszczak C., Hunter K., Wrzaczek M.(2017). Bound by fate: The role of reactive oxygen species in receptor-like kinase signaling. Plant Cell 29: 638–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.S.K., Kim Y.C., Kwon S.J., Ryu C.M., Park O.K.(2017). The Arabidopsis cysteine-rich receptor-like kinase CRK36 regulates immunity through interaction with the cytoplasmic kinase BIK1. Front. Plant Sci. 8: 1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Rubio M.C., Alassimone J., Geldner N.(2013). A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412. [DOI] [PubMed] [Google Scholar]
- Lee Y., et al. (2018). A lignin molecular brace controls precision processing of cell walls critical for surface integrity in Arabidopsis. Cell 173: 1468–1480.e9. [DOI] [PubMed] [Google Scholar]
- Lenglet A., Jaślan D., Toyota M., Mueller M., Müller T., Schönknecht G., Marten I., Gilroy S., Hedrich R., Farmer E.E.(2017). Control of basal jasmonate signalling and defence through modulation of intracellular cation flux capacity. New Phytol. 216: 1161–1169. [DOI] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y., Chen S., Zhou J.M.(2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338. [DOI] [PubMed] [Google Scholar]
- Lin Z.J., Liebrand T.W., Yadeta K.A., Coaker G.(2015). PBL13 is a serine/threonine protein kinase that negatively regulates Arabidopsis immune responses. Plant Physiol. 169: 2950–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J.(2010). Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26: 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitzler J.L., Antony S., Wu Y., Juhasz A., Liu H., Jiang G., Lu J., Roy K., Doroshow J.H.(2014). NADPH oxidases: A perspective on reactive oxygen species production in tumor biology. Antioxid. Redox Signal. 20: 2873–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.J., Turk B.E.(2016). Rapid identification of protein kinase phosphorylation site motifs using combinatorial peptide libraries. Methods Mol. Biol. 1360: 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H.(2014). StageTip-based HAMMOC, an efficient and inexpensive phosphopeptide enrichment method for plant shotgun phosphoproteomics. Methods Mol. Biol. 1072: 595–607. [DOI] [PubMed] [Google Scholar]
- Ogasawara Y., et al. (2008). Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J. Biol. Chem. 283: 8885–8892. [DOI] [PubMed] [Google Scholar]
- Pandey D., Gratton J.P., Rafikov R., Black S.M., Fulton D.J.(2011). Calcium/calmodulin-dependent kinase II mediates the phosphorylation and activation of NADPH oxidase 5. Mol. Pharmacol. 80: 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raad H., Paclet M.H., Boussetta T., Kroviarski Y., Morel F., Quinn M.T., Gougerot-Pocidalo M.A., Dang P.M., El-Benna J.(2009). Regulation of the phagocyte NADPH oxidase activity: Phosphorylation of gp91phox/NOX2 by protein kinase C enhances its diaphorase activity and binding to Rac2, p67phox, and p47phox. FASEB J. 23: 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Kunkowska A.B., Schippers J.H.(2016). Role of reactive oxygen species during cell expansion in leaves. Plant Physiol. 172: 2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B.(2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siligato R., et al. (2016). MultiSite gateway-compatible cell type-specific gene-inducible system for plants. Plant Physiol. 170: 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A.(2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J.M., Walker J.C.(1995). Plant protein kinase families and signal transduction. Plant Physiol. 108: 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N., Masuda T., Shinoda K., Nakamura A., Tomita M., Ishihama Y.(2007). Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol. Cell. Proteomics 6: 1103–1109. [DOI] [PubMed] [Google Scholar]
- Suzuki N., Miller G., Morales J., Shulaev V., Torres M.A., Mittler R.(2011). Respiratory burst oxidases: The engines of ROS signaling. Curr. Opin. Plant Biol. 14: 691–699. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Osakabe Y., Katsura S., Mizuno S., Maruyama K., Kusakabe K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K.(2012). Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J. 70: 599–613. [DOI] [PubMed] [Google Scholar]
- Taus T., Köcher T., Pichler P., Paschke C., Schmidt A., Henrich C., Mechtler K.(2011). Universal and Confident Phosphorylation Site Localization Using phosphoRS. J Proteome Res 10: 5354–5362. [DOI] [PubMed] [Google Scholar]
- Torres M.A., Dangl J.L., Jones J.D.(2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M., Spencer D., Sawai-Toyota S., Jiaqi W., Zhang T., Koo A.J., Howe G.A., Gilroy S.(2018). Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Trost B., Kusalik A., Napper S.(2016). Computational analysis of the predicted evolutionary conservation of human phosphorylation sites. PLoS One 11: e0152809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaattovaara A., Brandt B., Rajaraman S., Safronov O., Veidenberg A., Luklová M., Kangasjärvi J., Löytynoja A., Hothorn M., Salojärvi J., Wrzaczek M.(2019). Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun Biol 2: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veidenberg A., Medlar A., Löytynoja A.(2016). Wasabi: An integrated platform for evolutionary sequence analysis and data visualization. Mol. Biol. Evol. 33: 1126–1130. [DOI] [PubMed] [Google Scholar]
- Veronese P., Nakagami H., Bluhm B., Abuqamar S., Chen X., Salmeron J., Dietrich R.A., Hirt H., Mengiste T.(2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszczak C., Carmody M., Kangasjärvi J.(2018). Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 69: 209–236. [DOI] [PubMed] [Google Scholar]
- Whalen M.C., Innes R.W., Bent A.F., Staskawicz B.J.(1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M., Brosché M., Salojärvi J., Kangasjärvi S., Idänheimo N., Mersmann S., Robatzek S., Karpiński S., Karpińska B., Kangasjärvi J.(2010). Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 10: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M., Rozhon W., Jonak C.(2007). A Proteasome-regulated glycogen synthase kinase-3 modulates disease response in plants. J. Biol. Chem. 282: 5249–5255. [DOI] [PubMed] [Google Scholar]
- Yadeta K.A., Elmore J.M., Creer A.Y., Feng B., Franco J.Y., Rufian J.S., He P., Phinney B., Coaker G.(2017). A cysteine-rich protein kinase associates with a membrane immune complex and the cysteine residues are required for cell death. Plant Physiol. 173: 771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Withers J., He S.Y.(2013). Pseudomonas syringae infection assays in Arabidopsis. Methods Mol. Biol. 1011: 63–81. [DOI] [PubMed] [Google Scholar]
- Yeh Y.H., Chang Y.H., Huang P.Y., Huang J.B., Zimmerli L.(2015). Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front. Plant Sci. 6: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y.H., Panzeri D., Kadota Y., Huang Y.C., Huang P.Y., Tao C.N., Roux M., Chien H.C., Chin T.C., Chu P.W., Zipfel C., Zimmerli L.(2016). The Arabidopsis malectin-like/LRR-RLK IOS1 is critical for BAK1-dependent and BAK1-independent pattern-triggered immunity. Plant Cell 28: 1701–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B.W., Feechan A., Yin M., Saidi N.B., Le Bihan T., Yu M., Moore J.W., Kang J.G., Kwon E., Spoel S.H., Pallas J.A., Loake G.J.(2011). S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478: 264–268. [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang M., et al. (2018). The MAP4 kinase SIK1 ensures robust extracellular ROS burst and antibacterial immunity in plants. Cell Host Microbe 24: 379–391.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D., Felix G., Boller T.(2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767. [DOI] [PubMed] [Google Scholar]