Arabidopsis KETCH1, an importin β, promotes the nuclear accumulation of ribosomal proteins and contributes to both male and female gametogenesis, likely by maintaining translational efficiency.
Abstract
Male and female gametophytes are generated from micro- or megaspore mother cells through consecutive meiotic and mitotic cell divisions. Defects in these divisions often result in gametophytic lethality. Gametophytic lethality was also reported when genes encoding ribosome-related proteins were mutated. Although numerous ribosomal proteins (RPs) have been identified in plants based on homology with their yeast and metazoan counterparts, how RPs are regulated, e.g., through dynamic subcellular targeting, is unknown. We report here that an Arabidopsis (Arabidopsis thaliana) importin β, KETCH1 (karyopherin enabling the transport of the cytoplasmic HYL1), is critical for gametogenesis. Karyopherins are molecular chaperones mediating nucleocytoplasmic protein transport. However, the role of KETCH1 during gametogenesis is independent of HYPONASTIC LEAVES 1 (HYL1), a previously reported KETCH1 cargo. Instead, KETCH1 interacts with several RPs and is critical for the nuclear accumulation of RPL27a, whose mutations caused similar gametophytic defects. We further showed that knocking down KETCH1 caused reduced ribosome biogenesis and translational capacity, which may trigger the arrest of mitotic cell cycle progression and lead to gametophytic lethality.
Introduction
Development of gametophytes is critical for plant reproduction. In angiosperms, megagametogenesis (Drews and Yadegari, 2002) and microgametogenesis (McCormick, 1993, 2004) produce female and male gametophytes, respectively. During megagametogenesis, meiosis of a megaspore mother cell (MMC) produces four megaspores, among which only one survives as a functional megaspore (FM). The FM undergoes three rounds of mitosis and cellularization to develop into an embryo sac, i.e., the female gametophyte (Drews and Yadegari, 2002). During microgametogenesis, meiosis of a microspore mother cell gives rise to a tetrad of microspores. After release from the tetrad, each microspore goes through an asymmetric cell division, referred to as pollen mitosis I (PMI), to produce a bicellular microspore containing a generative cell and a vegetative nucleus. The generative cell then undergoes another mitotic event, called pollen mitosis II, to produce two sperm cells enclosed in pollen together with the vegetative nucleus (McCormick, 1993, 2004).
Mutations of mitotic cell cycle regulators often impair gametogenesis (Liu and Qu, 2008). Cyclin-dependent kinase (CDK)-activating kinases CDKD;1 and CDKD;3 are essential for preserving mitotic activity in Arabidopsis (Arabidopsis thaliana), and double mutants with defects in both of these proteins showed gametophytic lethality due to defective mitosis (Takatsuka et al., 2015). Similarly, the triple mutant of CDK, cdka;1/+;cdkb1;1;cdkb1;2, was defective in male and female gametogenesis due to mitotic division defects (Nowack et al., 2012). Two RING-finger E3 ligases, RING-H1 group F1a (RHF1a) and RHF2a, interact and target a CDK inhibitor ICK4/KRP6 for degradation (Liu et al., 2008). Mutants of the E3 ligase pair resulted in interphase arrest of the mitotic cell cycle at the microspore stage of pollen development and at female gametogenesis (FG) stage 1 of embryo sac development, leading to male and female gametophytic lethality (Liu et al., 2008).
The other major class of genes whose mutations may cause gametophytic lethality encodes proteins involved in ribosome biogenesis. In Arabidopsis, RIBOSOMAL PROTEIN L10 (RPL10; Imai et al., 2008; Falcone Ferreyra et al., 2010, 2013), RPL27a (Szakonyi and Byrne, 2011; Zsögön et al., 2014), SLOW WALKER1 (SWA1; Shi et al., 2005), SWA2 (Li et al., 2009), and two DEAD-box RNA helicases, AtRH36 (Huang et al., 2010) and SWA3 (Liu et al., 2010), are likely required for the biogenesis of rRNAs, the export of preribosomes from the nucleus, or the biogenesis of ribosomes. Mutations at these genes all resulted in mitotic arrest during gametogenesis (Shi et al., 2005; Imai et al., 2008; Li et al., 2009; Falcone Ferreyra et al., 2010, 2013; Huang et al., 2010; Liu et al., 2010). Although these results demonstrated the importance of RPs and their associated proteins, how these proteins are regulated, such as via dynamic targeting, is poorly understood.
Karyopherins, more often known as importins or exportins, are molecular chaperones mediating the nucleocytoplasmic transport of proteins (Meier and Brkljacic, 2009; Tamura and Hara-Nishimura, 2014). Classic nucleocytoplasmic transport relies on importin α for recognizing nuclear localization signals (NLSs) and importin β for interacting with the nuclear pore complex (NPC) and Ran-GTP (Tamura and Hara-Nishimura, 2014). However, importin βs play diverse roles, such as in the assembly of the mitotic spindle, in nuclear membrane formation, in microRNA activities, and in maintaining protein stability (Harel and Forbes, 2004; Wang et al., 2011; Tamura and Hara-Nishimura, 2014; Cui et al., 2016; Liu et al., 2019).
We report here that an Arabidopsis importin β, KETCH1 (KARYOPHERIN ENABLING THE TRANSPORT OF THE CYTOPLASMICHYL1), is critical for male gametogenesis and FG. Functional loss of KETCH1 caused the arrest of male gametophytic development at PMI and of female gametophytic development at FG1, suggesting a key role of KETCH1 in mitotic progression during gametogenesis. We demonstrate that the role of KETCH1 during gametogenesis is independent of HYPONASTIC LEAVES 1 (HYL1), the previously reported KETCH1 cargo. We show that KETCH1 interacts with a few ribosomal proteins (RPs), including RPL27a, whose nuclear accumulation depends on KETCH1 and whose mutations caused similar gametophytic defects. Knocking down KETCH1 resulted in reduced translational capacity, which may trigger mitotic arrest.
Results
Functional Loss of KETCH1 Compromises Male and Female Transmission
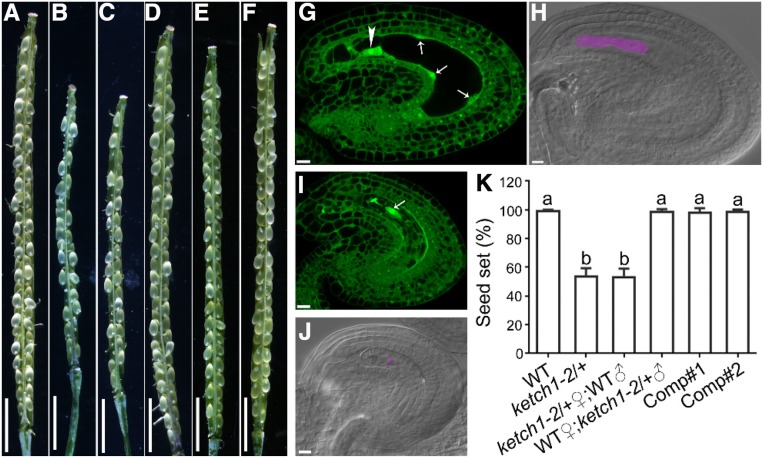
Arabidopsis KETCH1 encodes an importin β (Tamura and Hara-Nishimura, 2014). It was reported previously that mutations of KETCH1 resulted in embryo lethality, and thus no homozygous mutants of KETCH1 could be obtained (Zhang et al., 2017). However, compared to the full seed set in the wild type (Figures 1A and 1K), the heterozygous mutant of KETCH1, ketch1-2/+, contained around 47% tiny and wrinkled ovules (Figures 1B and 1K), much higher than 25% as expected for embryo lethality. As there were a higher number of aborted seeds, we decided to investigate if the mutation affected gametophyte development. Wild-type ovules at 24 h after pollination (HAP) were fertilized and showed developing embryo and endosperm by whole-mount clearing and optical sections of pistils (Figures 1G and 1H). By contrast, the small and wrinkled ovules in ketch1-2/+ were unfertilized, containing irregularly distributed nuclei (Figures 1I and 1J), suggesting a female gametophytic defect of ketch1-2. Indeed, when ketch1-2/+ was used as the female parent in reciprocal crosses with wild type, seed set was severely reduced (Figures 1C, 1D, and 1K), further supporting a role of KETCH1 in female gametophytic functionality. By analyzing the progenies of such crosses, we determined that both male and female gametophytic transmissions of ketch1-2 were severely reduced (Table 1), suggesting that KETCH1 is critical for male gametogenesis and FG, in addition to its roles in embryogenesis (Zhang et al., 2017).
Figure 1.
Reduced Seed Set in ketch1/+ Was Due to Female Gametophytic Defect.
(A) to (F) A representative silique from self-fertilized wild type (A) or ketch1-2/+ (B), from ketch1-2/+ pistil pollinated with wild-type pollen (C) or wild-type pistil pollinated with ketch1-2/+ pollen (D) or from two complementation lines, i.e., ProUBQ10:KETCH1-YFP;ketch1-2 (E) and (F).
(G) to (J) Optical sections (G) and (I) or whole-mount clearing (H) and (J) of wild-type (G) and (H) or ketch1-2 (I) and (J) ovules pollinated with wild-type pollen at 24 HAP. Arrows in (G) point at initiating peripheral endosperm; the arrowhead in (G) points at the nucleus of the zygote; the arrow in (I) points at the single nucleus in the embryo sac. Region highlighted in lilac is an elongated zygote in wild type (H) or the single nucleus in the abnormal embryo sac of ketch1-2 (J).
(K) Seed set. Results shown are means ± SD (n = 50 to 100). Means with different letters indicate significant difference (one-way ANOVA, Tukey’s multiple comparison test, P < 0.05). Bars = 2 mm for (A) to (F); 10 µm for (G) to (J).
Table 1. Transmission of Both Male and Female Is Defective by KETCH1 Loss of Function.
| Parents | Genotype of Progenies | ||||
|---|---|---|---|---|---|
| KETCH1 | ketch1-2/+ | ketch1-2 | Observed Ratio | Expected Ratio | |
| ♀ketch1-2/+ ×♂wild type | 105 | 60 | 0 | 1:0.57a | 1:1 |
| ♀wild type ×♂ketch1-2/+ | 60 | 30 | 0 | 1:0.50a | 1:1 |
| ♀ketch1-2/+ ×♂ketch1-2/+ | 77 | 115 | 0 | 1:1.49:0b | 1:2:1 |
Significantly different from the expected segregation ratio 1:1 (χ2 < χ20.05,1).
Significantly different from the expected segregation ratio 1:2:1 (χ2 < χ20.05,2).
Male Gametophytic Development Is Defective in KETCH1 Loss-of-Function Mutants
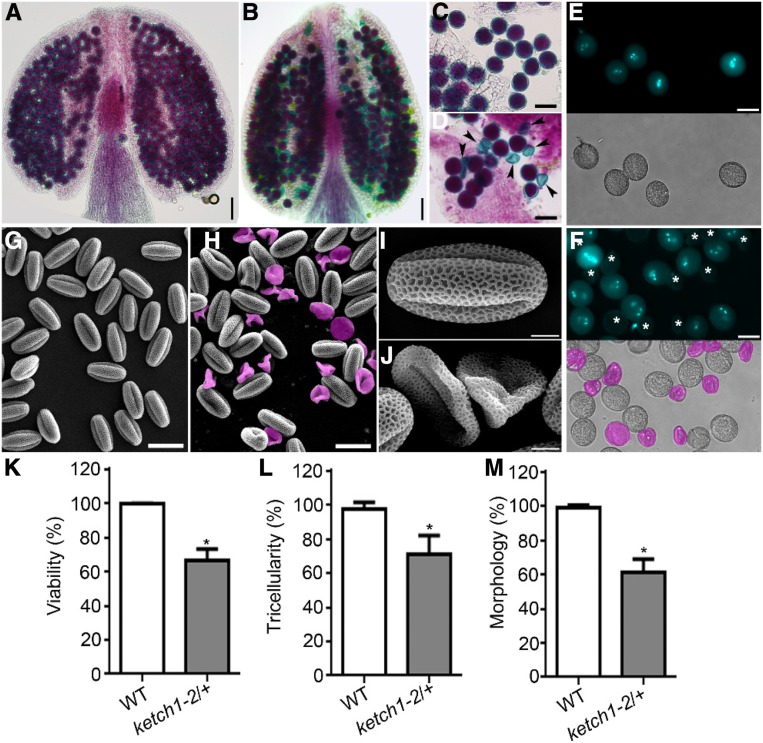
To determine the cause of reduced male transmission (Table 1), we performed Alexander staining to analyze cytoplasmic viability, DAPI staining to examine nuclear structure, and SEMs to examine pollen morphology. Compared to wild-type pollen grains that were stained purple by Alexander dye (Figures 2A, 2C, and 2K), 33.9% pollen from the ketch1-2/+ plants were light blue (Figures 2B, 2D, and 2K), indicating pollen abortion. In wild type, mature pollen grains stained with DAPI showed two sperm nuclei and one vegetative nucleus (Figures 2E, 2K, and 2L), whereas 30% pollen from the ketch1-2/+ plants failed to show nuclei at all (Figures 2F, 2K, and 2L). Finally, SEMs of the wild type showed oval-shaped pollen grains (Figures 2G, 2I, and 2M), whereas 32% pollen from ketch1-2/+ were wrinkled or collapsed (Figures 2H, 2J, and 2M). These results suggest that KETCH1 is critical for the development of male gametophytes, i.e., pollen.
Figure 2.
Pollen Development Was Defective in KETCH1 Loss-of-Function Mutants.
(A) to (D) Alexander staining of a representative anther (A) and (B) or mature pollen grains (C) and (D) from wild type (A and C) or ketch1-2/+ (B) and (D). Arrowheads point at aborted pollen grains.
(E) and (F) DAPI staining of mature pollen grains from wild type (E) or ketch1-2/+ (F). Bright-field (BF) images are shown at the bottom of corresponding fluorescent images. Aborted pollen grains are labeled by asterisks (in the fluorescent image) or in pink (in the BF image).
(G) to (J) Scanning electron micrographs (SEMs) of mature pollen from wild type (G) and (I) or ketch1-2/+ (H) and (J). Aborted pollen grains are highlighted in pink.
(K) to (M) Percentage of viable pollen by Alexander staining (K) of pollen with tricellular structure by DAPI staining (L) or of oval-shaped pollen by SEM (M). Results shown are means ± SD (n = 50 to 100). Asterisks indicate a significant difference of ketch1-2/+ from wild type (t test, P < 0.05). Bars = 50 µm for (A) and (B); 25 µm for (C), (D), (G), and (H); 20 µm for (E) and (F); 5 µm for (I) and (J).
To provide further evidence that ketch1-2 pollen was defective, we introduced quartet1 (qrt1) into ketch1-2/+. In qrt1, the four pollen grains of microsporogenesis remain fused and thus make genetic analysis of a heterozygous plant easier (Francis et al., 2006). In qrt1, the majority of tetrads had four or three pollen tubes during in vitro germination assays (Supplemental Figure 1). By contrast, the majority of tetrads in ketch1-2/+;qrt1 only had one or two pollen tubes (Supplemental Figure 1). These results suggest that functional loss of KETCH1 compromises gametophytic pollen development.
KETCH1 Loss of Function Impairs PMI
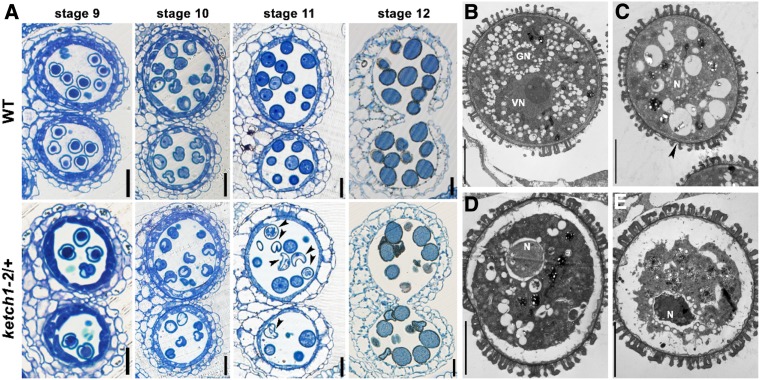
To determine at which stage the development of ketch1-2 pollen started to be defective, we analyzed anthers at different developmental stages by plastic embedding and semithin transverse sections. Anther development in Arabidopsis is classified into 14 stages (Sanders et al., 1999), which correspond to specific pollen developmental stages (Borg et al., 2009). Pollen development seems comparable between wild type and ketch1-2/+ before stage 10 (Figure 3A), i.e., the stage of unicellular microspore (Borg et al., 2009). At stage 11, the unicellular microspore in wild type undergoes one round of mitosis, i.e., PMI, to produce a bicellular microspore (Borg et al., 2009). This event accompanied the conversion of large vacuoles into numerous small ones in wild type (Figure 3A), as previously reported (Yamamoto et al., 2003). By contrast, some microspores in ketch1-2/+ pollen were deformed, containing large vacuoles at stage 11 (Figure 3A). At maturation, debris of aborted pollen grains was detected in ketch1-2/+ anther, alongside normally developed ones (Figure 3A). To confirm the results by plastic sections, we examined floral buds containing unicellular or bicellular microspores (Supplemental Figure 2) for optical sections by confocal laser scanning microscopy (CLSM) as described (Zhang et al., 2018). Consistent with the results obtained by plastic sections, CLSM showed microspores containing large vacuoles at stage 11 in ketch1-2/+ anthers in contrast to those of wild type (Supplemental Figure 2).
Figure 3.
KETCH1 Loss-of-Function Compromised PMI during Pollen Development.
(A) Representative semithin transverse sections of developing anthers at stage 9, stage 10, stage 11, or stage 12 from wild type or ketch1-2/+. Arrowheads point at aborted microspores.
(B) to (E) Representative TEM sections of ketch1-2/+ anthers at stage 11, showing a wild-type-looking microspore (B) and defective microspores at early (C), middle (D), or late (E) stages. Arrowhead in (C) points at invaginated PM. N, Nucleus; GN, generative nucleus; VN, vegetative nucleus. Bars = 20 µm for (A); 5 µm for (B) to (E).
To provide a detailed histological analysis of pollen developmental defects in ketch1-2/+, we performed ultrastructural analysis of ketch1-2/+ anthers using transmission electron microscopy (TEM). At stage 11, a portion of microspores in ketch1-2/+ anthers showed normal intracellular morphology, i.e., contained a vegetative nucleus, a generative cell nucleus, and numerous small vacuoles (Figure 3B). Others, however, showed similar defects, albeit to a different extent (Figures 3C to 3E). These microspores contained only one nucleus and few large vacuoles (Figures 3C to 3E). At early stages, the PM of these microspores was invaginated and wrinkled (Figure 3C). In these microspores, the single nucleus was abnormally surrounded by vacuolar structures (Figure 3C). Later on, defects appeared more obvious, such that the PM was detached from cell wall (Figure 3D) and finally degenerated together with the cytoplasm and the nucleus (Figure 3E). Results from TEM indicate that ketch1-2 microspores are defective during PMI, which is detrimental to pollen development.
KETCH1 Loss-of-Function Results in the Mitotic Arrest of FMs
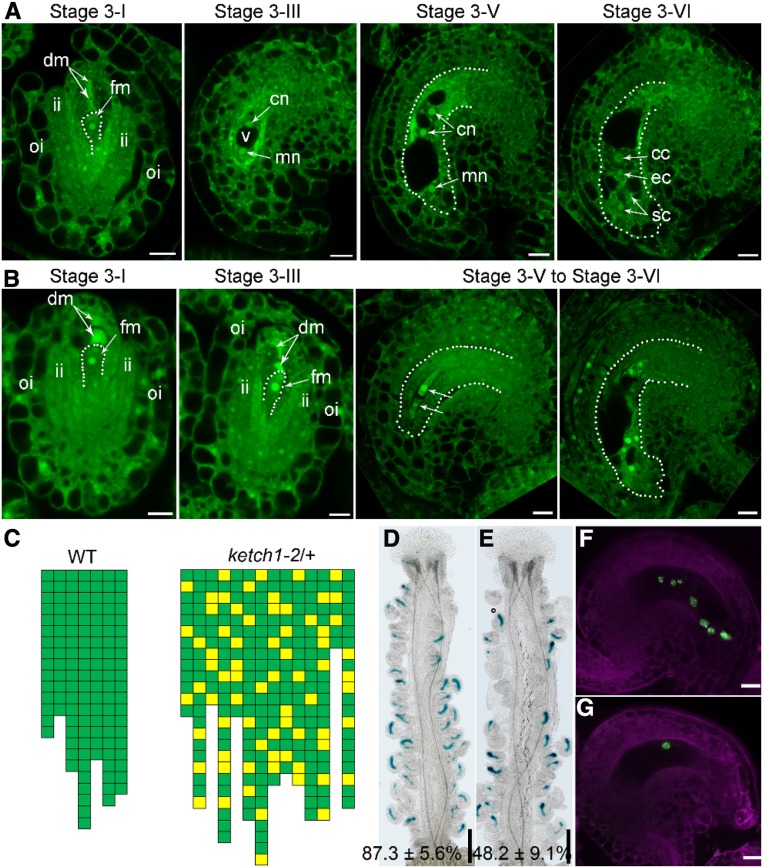
To characterize the defects in ketch1-2 female gametophytes (Table 1), we examined ketch1-2/+ ovules at different developmental stages by optical sections through CLSM (Wang et al., 2016). In wild type, FM persisted among the three degenerating sisters from MMC at stage 3-I (Figure 4A). At this stage, ketch1-2 was comparable to wild type (Figures 4A and 4B). Later on, FM underwent three rounds of mitotic cell division to produce the embryo sac (Figure 4A). At maturation, an embryo sac contains a central cell, an egg cell, and two synergid cells (Figure 4A). By contrast, almost 30% ovules in ketch1-2/+ did not contain a mature embryo sac (Figure 4B). Although all stage 3-I ovules in ketch1-2/+ contained a FM, FM in some ovules failed to go through the first mitotic division (Figure 4B). As a consequence, these ovules at stage 3-III did not contain two nuclei as in wild type (Figures 4A and 4C) but one FM (Figures 4B and 4C). In these ovules, integument growth was not affected and thus provided the information for developmental stages (Figure 4B). As a consequence of failed mitosis of FM, in 30% mature ovules of ketch1-2/+, the embryo sac contained irregular numbers of nuclei, ranging from one to six (Figures 4B and 4C). Indeed, by introducing an egg cell-specific reporter construct ProDD45:β-glucuronidase (GUS; Steffen et al., 2007; Wang et al., 2016), we found that a portion of mature ovules in ketch1-2/+ pistils did not show GUS signals (Figure 4E) in comparison to that in wild type (Figure 4D). In addition, the embryo sac-specific promoter-driven nucleus-targeted yellow fluorescent protein (YFP), i.e., ProES1:NLS-YFP (Wang et al., 2017), showed seven nuclei in wild-type ovules (Figure 4F). By contrast, 36 out of 156 ovules from ProES1:NLS-YFP;ketch1-2/+ showed only one nucleus (Figure 4G). These results show that KETCH1 is critical for the mitotic cell division of FM during megagametogenesis.
Figure 4.
KETCH1 Loss of Function Resulted in the Defective Development of Female Gametophytes.
(A) and (B) CLSMs of representative wild-type (A) or ketch1-2 (B) ovules during development. Dotted lines either illustrate functional megaspore (FM) of stage 3-I ovules or the embryo sac of ovules at other stages. cc, Central cell; cn, chalazal nucleus; dm, degenerating megaspore; ec, egg cell; fm, functional megaspore; ii, inner integument; mn, micropylar nucleus; oi, outer integument; sc, synergid cell.
(C) Quantification of embryo sac development based on CLSM of mature ovules. Each column represents one mature pistil; the number of cubes in each column indicates the number of countable ovules; green cubes indicate embryo sacs with normal seven-nucleus structure, whereas yellow cubes indicate abnormal embryo sacs as shown in (B). The positions of yellow squares indicate their positions in the pistils.
(D) and (E) Representative histochemical GUS staining of pistils from ProDD45:GUS (D) or from ProDD45:GUS;ketch1-2/+ transgenic plants (E). Values shown at the bottom (% of GUS-positive ovules) are means ± SD (n = 20). The ketch1-2/+ mutant is significantly different from wild type (t test, P < 0.05).
(F) and (G) Overlaid CLSM images of lysotracker red (magenta)-stained ProES1:NLS-YFP (F) or ProES1:NLS-YFP; ketch1-2/+ transgenic plants (G). Bars = 200 µm for (D) and (E); 10 µm for (A), (B), (F), and (G).
To provide further support of the roles KETCH1 plays in gametogenesis, we generated transgenic lines down-regulating KETCH1 specifically during gametogenesis using a gametophytic-specific promoter ProGPR1 (Yang et al., 2017). The expression of KETCH1-RNAi by ProGPR1 caused pollen abortion and reduced female fertility (Supplemental Figure 3), just like those by KETCH1 loss of function.
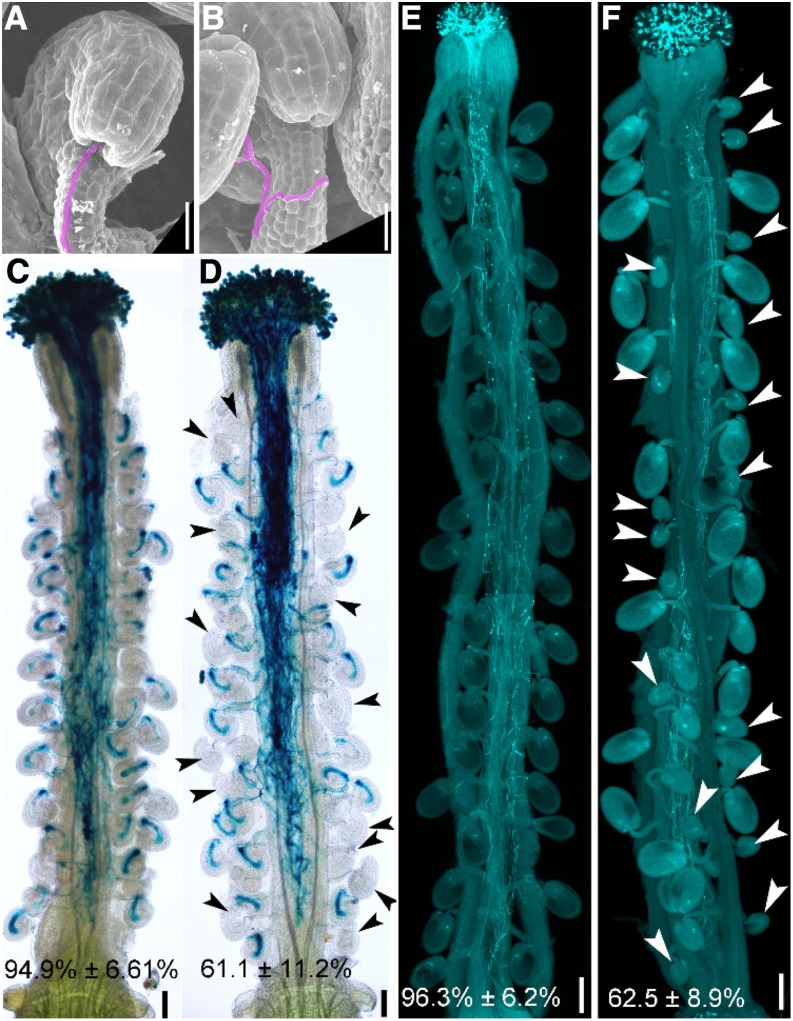
We suspected that the reduced seed set in the heterozygous ketch1-2/+ was due to defective embryo sacs. To test this hypothesis, we pollinated wild-type or ketch1-2/+ pistils by ProLAT52:GUS or by wild-type pollen and examined pistils at 12 HAP or 48 HAP, respectively. At 12 HAP, the majority of wild-type ovules were targeted by one pollen tube, as shown by SEM (Figure 5A) or by histochemical GUS staining of pistils (Figure 5C). In comparison, around 35% ovules in ketch1-2/+ failed to attract a pollen tube (Figure 5D). In these ovules, a pollen tube exited the transmission track and grew along the funiculus (Figure 5B). However, it failed to enter the micropyle (Figures 5B and 5D). At 48 HAP, wild-type ovules were fertilized and showed rapid size increase (Figure 5E). By contrast, more than 35% ovules in ketch1-2/+ remained tiny due to failed fertilization (Figure 5F). These results demonstrate that defective embryo sac development by KETCH1 loss of function compromises pollen tube guidance and thus causes reduced female fertility.
Figure 5.
KETCH1 Loss of Function Compromised Pollen Tube Guidance.
(A) and (B) SEMs of a wild-type (A) or a ketch1-2/+ ovule (B) pollinated with wild-type pollen at 12 HAP. Pollen tubes are shown in lilac.
(C) and (D) Histochemical GUS staining of a wild-type (C) or a ketch1-2/+ pistil (D) pollinated with ProLAT52:GUS pollen at 12 HAP. Two overlapping high-magnification images were taken for one pistil. The images were then overlaid with Photoshop (Adobe) to show the whole pistil. Arrowheads point at the ovules not targeted by a pollen tube.
(E) and (F) Aniline blue staining of a wild-type (E) or a ketch1-2/+ pistil (F) pollinated with wild-type pollen at 48 HAP. Arrowheads in (F) point at unfertilized ovules. Results shown at the bottom of (C) to (F) are means ± SD (n = 20). The ketch1-2/+ mutant is significantly different from wild type (t test, P < 0.05). Bars = 20 µm for (A) and (B); 100 µm for (C) and (D); 200 µm for (E) and (F).
Expression and Subcellular Localization of KETCH1
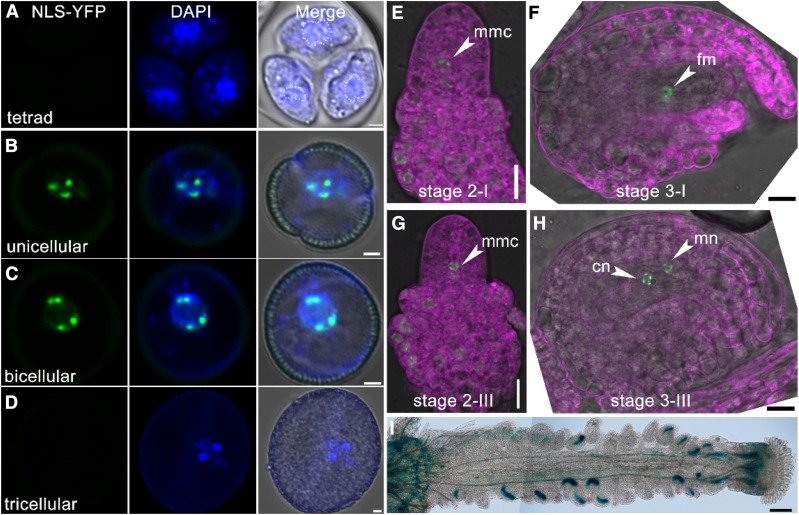
KETCH1 is highly expressed in reproductive tissues such as inflorescence, ovules, and pollen by quantitative RT-PCRs (Supplemental Figure 4). Analysis of KETCH1 promoter-driven GUS reporter lines, ProKETCH1:GUS, verified the enriched expression of KETCH1 in reproductive tissues/cells (Supplemental Figure 4). To provide further evidence that KETCH1 was critical for PMI during pollen development and mitosis of FM, we examined its expression specifically in these processes. A KETCH1 promoter-driven nuclear-localized YFP (ProKETCH1:NLS-YFP) was generated, and CLSM imaging was performed with 10 lines of ProKETCH1:NLS-YFP transgenic plants. During pollen development, signals were detected strongly in unicellular and bicellular microspores (Figures 6B and 6C) but hardly at the tetrad stage (Figure 6A) or in tricellular pollen (Figure 6D). During ovule development, signals were first detected in MMC and initiating integument cells (Figure 6E). At stage 2-III, signals were detected in FM in addition to integument (Figure 6G). At later stages, the promoter of KETCH1 was active in the nuclei of developing embryo sac (Figures 6F, 6H, and 6I). These results show that KETCH1 is expressed during gametogenesis.
Figure 6.
KETCH1 Is Expressed in Reproductive Cells.
(A) to (H) CLSM of ProKETCH1:NLS-YFP in developing microspores at tetrad (A), unicellular (B), bicellular (C), tricellular microspores (D), and ovules at stage 2-I (E), stage 2-III (G), stage 3-I (F), and stage 3-III ovules (H). Dotted lines in (A) illustrate vegetative nuclei. Images in (E) to (H) are merges of the RFP (for lysotracker red staining, in magenta), YFP (for NLS-YFP, green), and transmission channels.
(I) Histochemical GUS staining of a mature ProKETCH1:GUS pistil. cn, Chalazal nucleus; fm, functional megaspore; mmc, megaspore mother cell; mn, micropylar nucleus. Two overlapping high-magnification images were taken for one pistil. The images were then overlaid with Photoshop (Adobe) to show the whole pistil. Bars = 2 µm for (A) to (D), 10 µm for (E) to (H), 100 µm for (I).
To examine the subcellular localization of KETCH1, we generated ProUBQ10:KETCH1-YFP. The YFP translational fusion driven by ProUBQ10 rescued the male and female gametophytic defects of ketch1-2 (Figures 1E, 1F, and 1K; Supplemental Figure 5), indicating that GFP fusion did not interfere with the functionality of KETCH1. By CLSM, it was shown that KETCH1 was distributed both in the nucleus and in the cytoplasm either in ovules, in microspores, or in roots (Supplemental Figure 6), as reported (Zhang et al., 2017).
Functional Loss of HYL1 Did Not Affect Gametogenesis
KETCH1 was reported to mediate the nucleocytoplasmic transport of HYPONASTIC LEAVES 1 (HYL1; Zhang et al., 2017), a miRNA regulator (Lu and Fedoroff, 2000; Vazquez et al., 2004). Although the null mutant of HYL1, i.e., hyl1-2, was reported to show reduced fertility (Vazquez et al., 2004), both male and female gametophytic transmission of hyl1-2 were comparable to those of wild type (Supplemental Table 1), suggesting that compromised nucleocytoplasmic transport of HYL1 is not responsible for KETCH1 function during gametogenesis.
KETCH1 Interacts with RPs
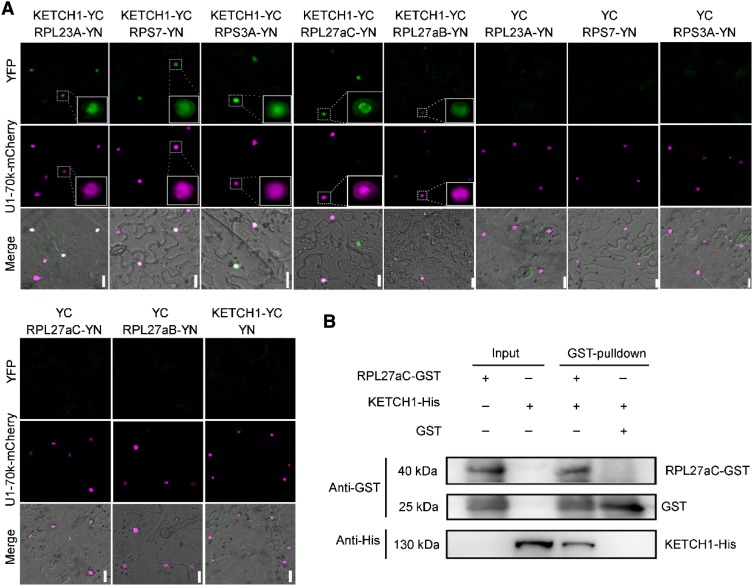
Because KETCH1 is an importin, the roles it plays are most likely conferred by its cargos. Phylogenetic analysis indicated that Arabidopsis KETCH1 is closely related to mammalian importin 5 (IPO5; Zhang et al., 2017), which mediates the nuclear import of several RPs in mammals (Jäkel and Görlich, 1998). We first examined whether KETCH1 interacts with these RP homologs by bimolecular fluorescence complementation (BiFC). Among the RPs whose homologs are cargos of mammalian IPO5, RPL23A, RPS7, RPS3A, and RPL27a, but not RPL5 or RPL23a, interacted with KETCH1 in the nucleus (Figure 7A; Supplemental Figure 7).
Figure 7.
KETCH1 Interacts with Several RPs.
(A) BiFC assays showing the interaction between KETCH1 and several RPs, including RPL23A, RPS7, RPS3A, RPL27aC, and RPL27aB. U1-70k-mCherry was used to label the nucleus and to mark transformed cells. Positive interactions were highlighted as insets. BiFC-positive signals are shown in green while U1-70k-mCherry is in magenta. From top to bottom, the YFP channel, the RFP channel, merges of the YFP, RFP, and transmission channels. For each combination, 30 pavement cells from five tobacco infiltrated leaves were examined with the same results.
(B) In vitro pull-down showing that KETCH1 interacts with RPL27aC. Experiments were performed three times with similar results. Bars = 20 µm.
Because RPL27a is required for both gametogenesis and embryogenesis, whose mutants showed similar, albeit less severe, phenotypes as ketch1-2 (Szakonyi and Byrne, 2011; Zsögön et al., 2014), we further verified its interaction with KETCH1 by in vitro pull-down assays (Figure 7B). The interaction between KETCH1 and RPL27a is specific because importin β4 (IMB4), a close homolog of KETCH1 (Tamura and Hara-Nishimura, 2014) and recently shown to regulate the nucleocytoplasmic shuttling of the transcription coactivator GRF-INTERACTING FACTOR1 (GIF1) in integument cells (Liu et al., 2019), did not interact with RPL27a (Supplemental Figure 7), and nor did GIF1 interact with KETCH1 (Supplemental Figure 7). These results suggest a pairwise specificity between importins and their cargos.
Nuclear Accumulation of RP RPL27a Requires KETCH1
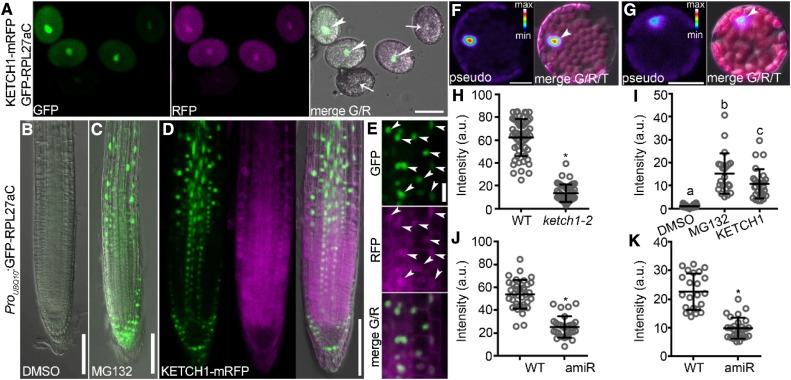
To determine whether KETCH1 mediates the nuclear accumulation of RPL27a, as would be expected if it fulfils the role of an importin, we introduced ProUBQ10:GFP-RPL27aC into ProUBQ10:KETCH1-mRFP/+;ketch1-2, in which pollen grains with red fluorescent protein (RFP) signals are comparable to those of wild type, whereas pollen grains without RFP signals are of the ketch1-2 genotype. We examined pollen grains from the ProUBQ10:GFP-RPL27aC;ProUBQ10:KETCH1-mRFP/+;ketch1-2 plants. In all pollen grains with clear RFP signals, there were strong GFP signals mostly in the nucleus (Figure 8A). By contrast, pollen grains without detectable RFP signals contained significantly reduced GFP signals (Figures 8A and 8H). GFP signals, i.e., RPL27a levels, are significantly higher when coexpressed with KETCH1-mRFP, suggesting that the nuclear accumulation of RPL27a depends on KETCH1.
Figure 8.
KETCH1 Positively Regulates the Nuclear Accumulation of RPL27a.
(A) Pollen grains from the ProUBQ10:GFP-RPL27aC;ProUBQ10:KETCH1-mRFP/-;ketch1-2 plants. Arrowheads point at the nuclei of pollen grains expressing GFP-RPL27aC (green) and KETCH1-mRFP (magenta); arrows point at the nuclei of pollen grains with weak or undetectable signals of GFP-RPL27aC and KETCH1-mRFP. Images are Z-stacks of eight confocal sections.
(B) and (C) A representative primary root from the ProUBQ10:GFP-RPL27aC plants treated with either DMSO (B) or MG132 (C). The images shown are merges of the GFP and transmission channels.
(D) A representative primary root from the ProUBQ10:GFP-RPL27aC;ProUBQ10:KETCH1-mRFP plants. From left to right, CLSM of the GFP channel, the RFP channel, and merges of the GFP, RFP, and transmission channels. Confocal imaging of the GFP channel was performed with the same settings for (B) to (D).
(E) Closeup of the images in (D). From top to bottom, CLSM of the GFP channel, the RFP channel, and the merge of the GFP and RFP channels. Arrowheads point to the nuclei with both GFP and RFP signals.
(F) and (G) A leaf protoplast from the ProUBQ10:GFP-RPL27aC (F) or ProUBQ10:GFP-RPL27aC;Pro35S:amiR-KETCH1 plants (G). Confocal imaging was performed with the same settings. Left, CLSM of the GFP channel; right, the merge of the GFP channel, autofluorescence from chlorophyll, and the transmission channel. For GFP signals, intensities are represented in pseudo-color, covering the full range of measured values within each data set (min to max). Arrowheads point at the nuclei.
(H) Intensity of nuclear-associated GFP in pollen grains expressing both KETCH1-mRFP and GFP-RPL27aC (wild type [WT]) or those without clear RFP signals (ketch1-2).
(I) Intensity of nuclear-associated GFP in the ProUBQ10:GFP-RPL27aC transgenic roots, either treated with DMSO, with MG132, or coexpressing KETCH1.
(J) and (K) Intensity of nuclear-associated GFP in leaf protoplasts (J) or leaf abaxial epidermal cells (K) of the ProUBQ10:GFP-RPL27aC (WT) or ProUBQ10:GFP-RPL27aC;Pro35S:amiR-KETCH1 (amiR) transgenic plants. a.u. represents arbitrary fluorescence unit. Results in (H) to (K) are means ± SD (n > 23). Asterisks in (H), (J), (K) indicate significant difference (t test, P < 0.05). Different letters in (I) indicate significantly different groups (one-way ANOVA, Tukey’s multiple comparisons test, t < 0.05). Bars = 20 µm for (A), (E), (F), and (G), 100 µm for (B) to (D).
Interestingly, in the meristem and elongating zones of the ProUBQ10:GFP-RPL27aC transgenic roots, GFP signals could only be detected when MG132, an inhibitor for the 26S proteasome, was added to the medium (Figures 8B, 8C, and 8I). Coexpressing KETCH1-mRFP by ProUBQ10 caused the same effect as MG132 (Figures 8D, 8E, and 8I), indicating that KETCH1 prevents 26S proteasome-mediated RPL27a degradation.
Although the homozygous ketch1-2 mutant cannot be obtained due to male and female gametophytic defects (Table 1), Pro35S:amiR-KETCH1 plants (amiR) that were generated previously showed reduced leaf size and morphology (Zhang et al., 2017), similar to that shown by the mutants of several RP-coding genes (Byrne, 2009; Rosado et al., 2012; Carroll, 2013). We thus used the amiR plants (Zhang et al., 2017) to examine whether down-regulating KETCH1 affected the nuclear accumulation of RPL27a also in leaves. By examining the abaxial epidermal cells of Pro35S:amiR-KETCH1;ProUBQ10:GFP-RPL27aC leaves, we confirmed that down-regulating KETCH1 significantly reduced the nuclear accumulation of RPL27a in leaf protoplasts and pavement cells (Figures 8F, 8G, 8J, and 8K), similar to that in pollen grains. Two other RPs that interact with KETCH1, i.e., RPL23A and RPS3A, were also significantly reduced in their nuclear accumulation, similar to that of RPL27a (Supplemental Figure 8). By contrast, GIF1, the interactor and cargo of IMB4 (Liu et al., 2019), did not differ in its nuclear accumulation between wild type and Pro35S:amiR-KETCH1 (Supplemental Figure 8). These results showed that KETCH1 positively regulates the nuclear accumulation of its interacting RPs.
Down-regulating KETCH1 Caused Reduced Ribosome Biogenesis and Translational Efficiency
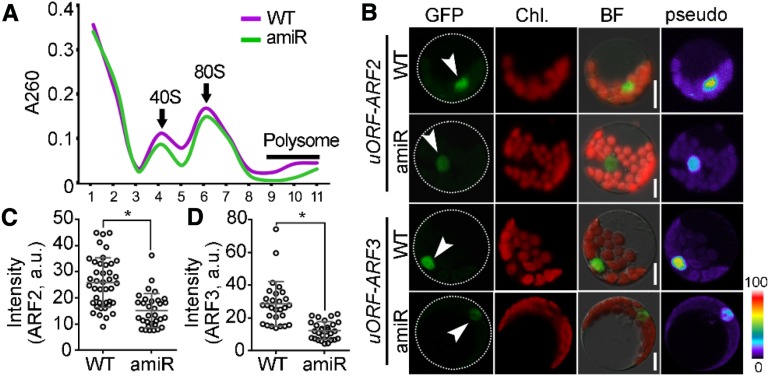
Because KETCH1 is required for the nuclear accumulation of its interacting RPs (Figure 8; Supplemental Figure 8), we wondered whether ribosome biogenesis and translational capacity would be affected by KETCH1. To test this hypothesis, we first performed polysome profiling assays in the Pro35S:amiR-KETCH1 plants to determine whether down-regulating KETCH1 could affect ribosome biogenesis. Indeed, the Pro35S:amiR-KETCH1 plants showed a clear reduction in both 40S and 80S subunit intensity peaks to those in the wild type (Figure 9A), suggesting reduced ribosome biogenesis by down-regulating KETCH1.
Figure 9.
Down-regulating KETCH1 Reduced Ribosomal Biogenesis and Translational Efficiency.
(A) Polysome profiling assay with sucrose density gradient. The OD260 absorption (A260) was monitored together with fractionation. The fractions containing 40S, 80S of ribosome, and polysomes in wild type (WT) or Pro35S:amiR-KETCH1 (amiR) are indicated. Three biological replicates were performed, and similar results were obtained.
(B) CLSM of a GFP-translational fusion of ARF2 or ARF3 from wild-type or Pro35S:amiR-KETCH1 protoplasts transformed with ProUBQ10:uORF-ARF2-GFP or ProUBQ10:uORF-ARF3-GFP. Dotted lines illustrate protoplast silhouettes and arrowheads point at the nuclei.
(C) and (D) GFP intensity in the nucleus for ARF2 (C) or ARF3 (D) from wild-type or Pro35S:amiR-KETCH1 protoplasts transformed with ProUBQ10:uORF-ARF2-GFP or ProUBQ10:uORF-ARF3-GFP. a.u., Arbitrary fluorescence unit. Results are means ± SD (n > 28) from two batches of transformation events. Asterisks indicate significant difference (t test, P < 0.05). Bars = 10 µm.
Next, we examined the translational capacity in the Pro35S:amiR-KETCH1 plants. Genes with an upstream open reading frame (uORF) located on their 5′ leader sequences are sensitive to translational efficiency (Fernandez et al., 2005), among which AUXIN RESPONSE FACTOR2 (ARF2) and ARF3 were shown to be affected translationally in the mutants of RPs (Rosado et al., 2012). We thus generated a transgenic line expressing a GFP translational fusion of ARF2 and ARF3 with their corresponding uORF into either wild-type or the Pro35S:amiR-KETCH1 protoplasts. The promoter ProUBQ10 was used to exclude the influence of transcription (Supplemental Figure 9). As shown in Figure 9, translational repression, as indicated by reduced GFP signals for either ARF2 or ARF3 with uORF, was detected in the Pro35S:amiR-KETCH1 protoplasts compared with their respective controls in wild type (Figures 9B to 9D). By contrast, GFP-fusion proteins translated from ARF2 or ARF3 without uORF did not significantly differ between wild type and amiR-KETCH1 (Supplemental Figure 9). These results suggest that KETCH1 positively regulates ribosome biogenesis and translational efficiency, likely by maintaining the nuclear accumulation of RPs.
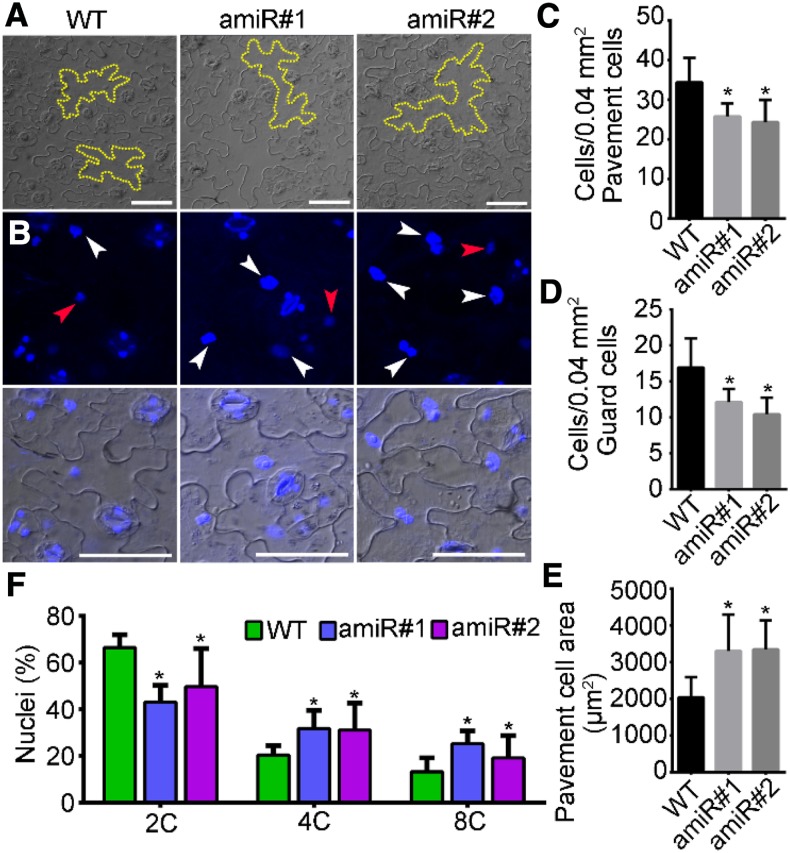
Because functional loss of KETCH1 resulted in mitotic arrest during gametogenesis (Figures 3 and 4) and ribosome biogenesis is involved in cell cycle progression in yeast and mammals (Dez and Tollervey, 2004; Donati et al., 2012), we considered the possibility that KETCH1 may indirectly mediate cell cycle progression since it positively regulates ribosome biogenesis via RPs. To test this hypothesis, we examined cell size and contents of leaf epidermal cells. Compared to those of wild type, Pro35S:amiR-KETCH1 contained larger pavement cells (Figures 10A and 10E) and reduced cell density (Figures 10C and 10D), as would be expected if cell cycle progression is delayed (Yang et al., 2019). 4′,6-diamidino-2-phenylindole (DAPI) staining of leaf epidermal cells further showed that a larger percentage of cells from the Pro35S:amiR-KETCH1 plants showed either 4C or 8C DNA content than those from wild type (Figure 10F). These results indicate that the expression of amiR-KETCH1 compromised cell cycle progression.
Figure 10.
The Expression of amiR-KETCH1 Compromised Cell Cycle Progression.
(A) Pavement cells from the wild type (WT) and two lines of Pro35S:amiR-KETCH1 (amiR#1 and amiR#2) after tissue clearing. Dotted lines illustrate the morphology of pavement cells.
(B) CLSM of DAPI-stained leaf epidermal cells from wild type, amiR#1, and amiR#2. Red arrowheads point at 2C nuclei, whereas white arrowheads points at nuclei with 4C or 8C (DNA contents).
(C) and (E) Quantification of the density (C) and (D) and the size (E) of leaf epidermal cells. Results shown are means ± SD (n = 10). Asterisks indicate significant difference from that of wild type (t test, P < 0.05).
(F) Percentage of nuclei either with 2C, 4C, or 8C DNA content. Results shown are means ± SE (n = 3). Asterisks indicate significant difference from that of wild type (t test, P < 0.05). More than 100 cells from three leaves of three individual plants were analyzed. Bars = 50 µm for (A) and (B).
Discussion
In this report, we demonstrate that KETCH1 is critical for both male gametogenesis and FG. Although KETCH1 is also required for embryogenesis (Zhang et al., 2017), several lines of evidence supported a critical role of KETCH1 in gametogenesis. First, male and female gametophytic transmission of ketch1-2 was severely reduced (Table 1). Second, reciprocal crosses using the heterozygous ketch1-2/+ as the female parent resulted in reduced seed set, similar to that of self-fertilized ketch1-2/+ (Figure 1). Third, the development of pollen and embryo sac were defective (Figures 2 to 4). Defective embryo sac development caused reduced pollen tube attraction, leading to partial female sterility (Figure 5). KETCH1 is expressed throughout male gametogenesis and FG (Figure 6), consistent with its roles in these processes.
The developmental function of KETCH1 is presumably performed through regulating its cargoes. Although HYL1 was shown to be a cargo of KETCH1 (Zhang et al., 2017), reciprocal crosses together with segregation ratio analysis of hyl1-2, a null mutant of HYL1, indicated that male and female gametophytic transmission was not affected in hyl1-2 (Supplemental Table 1). Instead, we show that RPL27a and a few other RPs interact specifically with KETCH1 (Figure 7; Supplemental Figure 7). Furthermore, the nuclear accumulation of RPL27a was significantly reduced in pollen grains and other cell types of ketch1-2 or by down-regulating KETCH1 (Figure 8; Supplemental Figure 8), suggesting that RPL27a and likely other RPs with which it interacts are also cargos of KETCH1. Among these KETCH1-interacting RPs, RPL27a is critical for gametophytic transmission (Szakonyi and Byrne, 2011; Zsögön et al., 2014), whose reduced nuclear accumulation in ketch1-2 may have contributed to its gametophytic lethality. On the other hand, down-regulating KETCH1 constitutively caused altered leaf morphology (Zhang et al., 2017), similar to that caused by mutations at RPL27aC or RPL23A (Degenhardt and Bonham-Smith, 2008; Szakonyi and Byrne, 2011), hinting at a genetic link between KETCH1 and its interacting RPs.
The interaction with KETCH1 may protect RPs from 26S proteasome-mediated degradation. In mammals, RPs, such as RPL27a, are rapidly degraded in the nucleus to balance a certain rate of ribosome-subunit production (Lam et al., 2007). Such a degradation depends on the 26S proteasome (Sung et al., 2016). Our data suggest a similar regulation in plants. Inhibiting the 26S proteasome by MG132 significantly increased the protein level of RPL27a, which was otherwise undetectable (Figure 8). Coexpression of KETCH1 showed the same effect as did MG132 on RPL27a (Figure 8), suggesting a protective role of KETCH1 through interaction. It has been demonstrated recently that IMB4 positively regulates the turnover of a Kinesin-4 (Ganguly et al., 2018), suggesting that importin-mediated protein stability against the 26S proteasome may be a common theme in plant cells.
Both the large and small subunits of the eukaryotic ribosome assemble in the nucleus (Byrne, 2009). Thus, reduced nuclear accumulation of RPs would compromise ribosomal biogenesis. This was indeed the case when KETCH1 was down-regulated (Figure 9). Consequently, translational capacity, indicated by the expression of uORF-containing ARF2 and ARF3, was reduced (Figure 9). It was proposed that cells might monitor their translational capacity to determine cell cycle progression, whereas mutations affecting ribosome biogenesis would prevent cells at the G1/S boundary, leading to mitotic cell cycle arrest (Dez and Tollervey, 2004; Donati et al., 2012). In addition, a study in mammals indicated that a delay of cell cycle progression may occur before any detectable difference on ribosome number or translation when an rRNA processing factor was depleted (Bernstein et al., 2007), suggesting that cells sense some aspect of ribosome biogenesis in order to control cell cycle progression. Because male gametogenesis and FG were arrested right before mitosis by KETCH1 loss of function (Figures 3 and 4), it was a tempting thought that reduced nuclear accumulation of RPs in ketch1-2 results in the arrest of mitotic cell division during gametogenesis, which was supported by the observation that the amiR-KETCH1 plants show reduced mitotic division in leaf epidermal cells (Figure 10). Whether this reduction was due to a general reduction of protein translation or to some specific component(s) merits further investigation.
A large portion of proteins are required in the nucleus, sometimes dynamically, once they have been synthesized in the cytoplasm. By contrast, only dozens of importins are encoded in a genome such as that of Arabidopsis (Tamura and Hara-Nishimura, 2014). Thus, the cargo specificity of importins has often been questioned. However, our data unequivocally demonstrated pairwise specificity between importins and cargos (Figure 7; Supplemental Figure 7). That one importin regulates multiple cargos, such as KETCH1 for HYL1 and several RPs, is a solution for this dilemma. Identifying consensus motifs that are responsible for the interaction with individual importins, if possible, will be a rewarding effort in the future.
METHODS
Plant Growth and Transformation
The T-DNA insertion line SALK_050129 (ketch1-2) was obtained from the Arabidopsis Biological Resource Center (www.arabidopsis.org). The Arabidopsis (Arabidopsis thaliana) Columbia-0 ecotype was used as the wild type. Plants were grown in nutrient-rich soil in greenhouse with normal light conditions (90 μmol/m2/s) at a long-day cycle (16 h light/8 h dark) at 22°C as described by Zhou et al. (2013). Stable transgenic plants were selected on half-strength Murashige and Skoog medium supplemented with 30 µg/mL Basta salts (Sigma-Aldrich) or 25 µg/ml hygromycin (Roche). Other materials, including hyl1-2 (Lu and Fedoroff, 2000), Pro35S:amiR-KETCH1 (Zhang et al., 2017), ProLAT52:GUS (Li et al., 2013), ProDD45:GUS (Wang et al., 2016), and ProES1:NLS-YFP (Wang et al., 2017), were described previously.
DNA Manipulation
All constructs were generated using the Gateway technology (Invitrogen). All entry vectors were generated in the pENTR/D/TOPO vector (Invitrogen). The promoter for KETCH1 was cloned with the primer pair ZP4211/ZP4212 from Columbia-0 genomic DNA. ProKETCH1 containing the 2135-bp sequence upstream of its start codon was introduced into the destination vectors GW:NLS-YFP (Wang et al., 2013) and GW:GUS (Zhou et al., 2013). The full-length coding sequence of KETCH1, RPL27aC, RPL27aB, RPS3A, RPL23A, RPS7, RPL5, and RPL23a was cloned using the primer pairs ZP4213/ZP4214, ZP7265/ZP7266, ZP8964/ZP8965, ZP8948/ZP8949, ZP7524/ZP7525, ZP8942/ZP8943, ZP8944/ZP8945, and ZP7504/ZP7505, respectively. The uORF for ARF2 or ARF3 (Rosado et al., 2012) was cloned using the primer pairs ZP9771/ZP9772 and ZP9773/ZP9774, respectively. The full-length coding sequence of ARF2 or ARF3 without uORF was cloned using the primer pairs ZP9772/ZP11174 and ZP9774/ZP11175, respectively. The entry vectors were used in LR reactions with the destination vector ProUBQ10:GW-YFP to generate ProUBQ10:ARF2-YFP or ProUBQ10:ARF3-YFP.
To generate the destination vector used in in planta protein expression, ProUBQ10:GW-YFP, ProUBQ10:GW-RFP, Pro35S of the destination vector Pro35S:GW-YFP, or Pro35S:GW-RFP (Karimi et al., 2002) was replaced with ProUBQ10, which was amplified with the primer pair ZP510/ZP511 and digested with HindIII/SpeI. Expression vectors were generated by LR reactions using Gateway LR Clonase II (Invitrogen).
For the KETCH1-RNAi construct, a 490-bp fragment of the KETCH1 coding sequence (from 1 bp to 490 bp starting from the start codon) was amplified with the primer pair ZP5338/ZP5339. The resulting PCR products were subcloned into the RNAi vector pTCK303 (Guo et al., 2010) to obtain the Pro35S:KETCH1-RNAi construct. The construct ProGPR1:KETCH1-RNAi was generated by cloning the promoter of GPR1 (ProGPR1; Yang et al., 2017) with ZP9474/ZP9475 and replacing Pro35S in Pro35S:KETCH1-RNAi.
Constructs used in BiFC assays were generated with entry vectors and the destination vector pSITE::cEYFP-C1 or pSITE-nEYFP-C1 (Martin et al., 2009) by LR reactions. BiFC constructs for IMB4 or GIF1 were described by Liu et al. (2019). For constructs used for protein expression in the pull-down assay, the coding sequence of RPL27aC was inserted into pGEX4T-1 to obtain a vector expressing the recombinant protein glutathione S-transferase (GST)-RPL27aC while the coding sequence of KETCH1 was inserted into pET30a to obtain a vector expressing the recombinant protein 6× His-KETCH1.
All PCR amplifications were performed using Phusion hot-start high-fidelity DNA polymerase using the annealing temperature and extension times recommended by the manufacturer (Finnzyme). All entry vectors were sequenced, and sequences were analyzed using VectorNTI (Invitrogen). The Bioneer PCR purification kit and the Bioneer Spin miniprep kit were used for PCR product recovery and plasmid DNA extraction, respectively. All primers are listed in Supplemental Table 2.
RNA Extraction, RT-PCRs, and qPCRs
The genotypes of ketch1-2/+ progenies were determined by PCRs using the following primers: ZP4148/ZP4149 for KETCH1 and ZP1/ZP4148 for ketch1-2. The genotype of qrt1 was determined by examining mature pollen.
Total RNAs were isolated using a Qiagen RNeasy plant mini kit according to the manufacturer’s instructions. Oligo(dT)-primed cDNAs were synthesized using SuperScript III reverse transcriptase with on-column DNase digestion (Invitrogen). For RT-PCR analysis of complementation lines, the endogenous or exogenous KETCH1 was amplified with the primer pair ZP5720/ZP4214 or ZP11/ZP4148, respectively. Primers to amplify ACTIN2 were as described by Zhou et al. (2013).
The qPCRs were performed with the Bio-Rad CFX96 real-time system using SYBR green real-time PCR master mix (Toyobo) as described by Zhou et al. (2013). Primers used in qPCRs were ZP4494/ZP4495 for KETCH1, ZP7994/ZP7995 for RPL27aC, ZP9772/GFP-F for ARF2-GFP, and ZP9774/GFP-F for ARF3-GFP. Primers for GAPDH and ACTIN2 in qPCRs were as described by Zhou et al. (2013). All primers are listed in Supplemental Table 2.
Phenotypic Analysis
Methods to analyze pollen development, including Alexander staining, DAPI staining, or SEM, were performed as described by Li et al. (2013) and Feng et al. (2016). Semithin sections and TEM of developing anthers were performed as described by Xie et al. (2014), Feng et al. (2017a), and Zhang et al. (2018). Arabidopsis in vitro pollen tube growth was performed as described by Boavida and McCormick (2007). Whole-mount ovule clearing, optical sections of developing flowers, and SEM were performed as described by Wang et al. (2016, 2017) and Liu et al. (2019). Histochemical GUS analysis of ProLAT52:GUS-pollinated pistils and aniline blue staining of pollinated pistils were performed as described by Li et al. (2013) and Feng et al. (2018). Measurement of epidermal cell size and density was conducted as described by Horváth et al. (2006). Cells were photographed from at least three different positions of a leaf, and on average 50 cells were analyzed per genotype. Cell outlines were traced and parameters such as cell area, perimeter, and shape factor were calculated with ImageJ (http://rsbweb.nih.gov/ij/). The analysis of DNA content was described previously by Yang et al. (2019). In brief, leaves were dissected and fixed in 70% ethanol for 3 h and then incubated in DAPI staining solution for at least 20 min before imaging. The total integrated density of DAPI fluorescence from selected nuclei was measured.
BiFC Assays
BiFC assays, in which a P19 protein was used to suppress gene silencing (Park et al., 2014), were performed as described by Li et al. (2018b) and Liu et al. (2019). U1-70K-mCherry was used to label the nucleus as described by Wang et al. (2012). Confocal imaging was performed 48 h after tobacco (Nicotiana tabacum) leaf infiltration.
Protein Biochemical Assays
For the purification of recombinant proteins in in vitro pull-down assays, pET30a-KETCH1 and GST-RPL27aC were transformed into BL21 competent cells (DE3). The BL21 cells were grown at 37°C in Lurani-Bertani medium in the presence of antibiotics (100 μg/mL kanamycin) to an OD600 of 0.6 to 0.8. Protein expression was induced by adding isopropyl-β-D-1-thiogalactopyranosid to a final concentration of 0.5 mM. Cells were further incubated in a horizontal shaker with slow shaking overnight at 16°C before pellet collection by centrifugation at 13,800g for 10 min at 4°C. Cell pellets were resuspended and lysed by sonification on ice in a lysis buffer (20 mM Tris·HCl, pH 7.5, 300 mM NaCl, 2 mM phenylmethylsulfonyl fluoride). Supernatant was centrifuged at 16,000g for 30 min at 4°C to completely remove cell debris. The 6× His-KETCH1 or GST-RPL27aC recombinant proteins were purified using an Ni-NTA column or glutathione agarose beads, respectively (Zhang et al., 2017).
In vitro pull-down assays were performed as described by Wang et al. (2016). Approximately 1 μg each of GST or GST-RPL27aC proteins was bound to 45 μL of glutathione sepharose beads for 1 h at 4°C. Bound beads were incubated with 1 μg of 6× His-KETCH1 proteins for another 1 h at 4°C. After three washes with 1× PBS buffer, the beads were eluted with 40 μL of 10 mM glutathione in 50 mM Tris (pH 8.0), mixed with a loading buffer, and then boiled at 100°C for 10 min. Approximately 15 μL of each boiled sample was used for SDS-PAGE and protein gel-blot analysis with an anti-His antibody (Beyotime, cat. no. AF5060, 1:1000 dilution) and an anti-GST antibody (Beyotime, cat. No. AF0174, 1:1000 dilution).
Polysome Profiling
Polysome profiling was performed as described by Li et al. (2018a). In brief, leaves from 15-d-old wild-type or Pro35S:amiR-KETCH1 plants were ground in liquid nitrogen, followed by resuspension in polysome extraction buffer. The extract was loaded onto a 10% to 60% Suc gradient and spun in a Hitachi CP100WX/CR22N at 30,000 rpm for 4 h at 4°C. Eleven fractions were collected into centrifugal tubes. The 40S and 80S of ribosome and polysomes were quantified by OD260 absorbance profile.
Analysis of DNA Content
For DAPI staining of nuclei, abaxial epidermal cells of the third true leaf from 3 week after germination plants were used for analysis. Areas of epidermis were chosen near the center of the leaf but away from veins. The analysis of DNA content was described previously (Yang et al., 2019). In brief, leaves were dissected and fixed in 70% ethanol for 3 h, and then incubated in DAPI staining solution for at least 20 min before imaging. The total integrated density of DAPI fluorescence from selected nuclei was measured.
Protoplast Preparation
Arabidopsis protoplasts were prepared according to a previous report (Wang et al., 2017). Three independent experiments involving at least 50 protoplasts were conducted to ensure reproducibility of the results.
Fluorescence Microscopy and Pharmacological Treatment
Staining of tissues with FM4-64 (Wang et al., 2013; Zhang et al., 2015; Chai et al., 2016; Feng et al., 2017b; Wan et al., 2017b) or Lysotracker red (Wang et al., 2016) was as described. For MG132 treatment, plant materials were treated with 100 µM MG132 for 0.5 h before examination. Fluorescent images were captured using a LSM 880 CLSM (Zeiss) with a 40/1.3 oil objective. YFP-RFP double-labeled plant materials were captured alternately using line-switching with the multitrack function (514 nm for YFP and 561 nm for RFP). Fluorescence was detected using a 520- to 550-nm band-pass filter for YFP or a 575- to 650-nm band-pass filter for RFP. Images were processed with an LSM image processing software (Zeiss). For the quantification of fluorescence intensity between nuclear and cytoplasmic fractions, a region of interest of the same size was defined either in the nucleus (Nuc) or cytoplasm (Cyt) within a pollen tube or protoplast. The ratio of fluorescence intensity between the nuclear and cytoplasmic ROS (Chai et al., 2016; Wan et al., 2017a) was calculated using ImageJ (http://rsbweb.nih.gov/ij/).
Statistical Analysis
Quantification data were analyzed using GraphPad Prism 6.02 (www.graphpad.com/scientific-software/prism/). All statistical analyses, one-way ANOVA (Tukey’s multiple comparisons test), and t test were performed with built-in analysis tools and parameters.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are At5g62000 for ARF2; At2g33860 for ARF3; At5g19820 for KETCH1/IMP3; At1g09700 for HYL1; At4g27640 for IMB4; At5g28640 for GIF1/AN3; At3g23860 for GPR1; At1g70600 for RPL27aC; At1g23290 for RPL27aB; At1g04480 for RPL23A; At5g39740 for RPL5; At2g39460 for RPL23a; At4g34670 for RPS3A; At3g02560 for RPS7.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. In vitro pollen germination was reduced by KETCH1 loss of function
Supplemental Figure 2. KETCH1 loss of function compromised PMI during pollen development
Supplemental Figure 3. Knocking down KETCH1 by ProGPR1:KETCH1-RNAi mimicked the fertility defects of ketch1-2/+
Supplemental Figure 4. KETCH1 is constitutively expressed
Supplemental Figure 5. Complementation of ketch1-2 by ProUBQ10:KETCH1-YFP
Supplemental Figure 6. KETCH1 is distributed both in the cytoplasm and in the nucleus
Supplemental Figure 7. The interaction between KETCH1 and RPL27a is specific
Supplemental Figure 8. KETCH1 positively regulates the nuclear accumulation of its interacting RPs
Supplemental Figure 9. ARFs without uORF did not show significant difference of translational efficiency between wild type and amiR-KETCH1
Supplemental Table 1. Male and female transmission of hyl1-2 was comparable to that of wild type
Supplemental Table 2. Oligos used in this study
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Yijun Qi for hyl1-2, the ABRC for ketch1-2, and Feng Yu for the help with polysome profiling. This work was supported by Natural Science Foundation of China (31871422 and 31625003 to Y.Z.; 31771558 and 31970332 to S.L.). Y.Z.’s laboratory is partially supported by the Tai-Shan Scholar Program by Shandong Provincial Government.
AUTHOR CONTRIBUTIONS
S.L. and Y.Z. conceived and supervised the project; F.X. and C.-Y.D. performed the experiments with the assistance by H.-H.L. and J.-H.W.; Z.-H.Z. contributed materials; Y.Z., S.L., F.X., and C.-Y.D. designed the experiments and analyzed the data; Y.Z. wrote the article with contributions of all the authors.
References
- Bernstein K.A., Bleichert F., Bean J.M., Cross F.R., Baserga S.J.(2007). Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol. Biol. Cell 18: 953–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boavida L.C., McCormick S.(2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52: 570–582. [DOI] [PubMed] [Google Scholar]
- Borg M., Brownfield L., Twell D.(2009). Male gametophyte development: A molecular perspective. J. Exp. Bot. 60: 1465–1478. [DOI] [PubMed] [Google Scholar]
- Byrne M.E.(2009). A role for the ribosome in development. Trends Plant Sci. 14: 512–519. [DOI] [PubMed] [Google Scholar]
- Carroll A.J.(2013). The Arabidopsis cytosolic ribosomal proteome: from form to function. Front Plant Sci 4: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai S., Ge F.R., Feng Q.N., Li S., Zhang Y.(2016). PLURIPETALA mediates ROP2 localization and stability in parallel to SCN1 but synergistically with TIP1 in root hairs. Plant J. 86: 413–425. [DOI] [PubMed] [Google Scholar]
- Cui Y., Fang X., Qi Y.(2016). TRANSPORTIN1 promotes the association of microRNA with ARGONAUTE1 in Arabidopsis. Plant Cell 28: 2576–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt R.F., Bonham-Smith P.C.(2008). Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol. 147: 128–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C., Tollervey D.(2004). Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 7: 631–637. [DOI] [PubMed] [Google Scholar]
- Donati G., Montanaro L., Derenzini M.(2012). Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res. 72: 1602–1607. [DOI] [PubMed] [Google Scholar]
- Drews G.N., Yadegari R.(2002). Development and function of the angiosperm female gametophyte. Annu. Rev. Genet. 36: 99–124. [DOI] [PubMed] [Google Scholar]
- Falcone Ferreyra M.L., Casadevall R., Luciani M.D., Pezza A., Casati P.(2013). New evidence for differential roles of l10 ribosomal proteins from Arabidopsis. Plant Physiol. 163: 378–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone Ferreyra M.L., Pezza A., Biarc J., Burlingame A.L., Casati P.(2010). Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress. Plant Physiol. 153: 1878–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C., Wang J.G., Liu H.H., Li S., Zhang Y.(2017a). Arabidopsis adaptor protein 1G is critical for pollen development. J. Integr. Plant Biol. 59: 594–599. [DOI] [PubMed] [Google Scholar]
- Feng Q.N., Kang H., Song S.J., Ge F.R., Zhang Y.L., Li E., Li S., Zhang Y.(2016). Arabidopsis RhoGDIs are critical for cellular homeostasis of pollen tubes. Plant Physiol. 170: 841–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q.N., Liang X., Li S., Zhang Y.(2018). The ADAPTOR PROTEIN-3 complex mediates pollen tube growth by coordinating vacuolar targeting and organization. Plant Physiol. 177: 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q.N., Zhang Y., Li S.(2017b). Tonoplast targeting of VHA-a3 relies on a Rab5-mediated but Rab7-independent vacuolar trafficking route. J. Integr. Plant Biol. 59: 230–233. [DOI] [PubMed] [Google Scholar]
- Fernandez J., Yaman I., Huang C., Liu H., Lopez A.B., Komar A.A., Caprara M.G., Merrick W.C., Snider M.D., Kaufman R.J., Lamers W.H., Hatzoglou M.(2005). Ribosome stalling regulates IRES-mediated translation in eukaryotes, a parallel to prokaryotic attenuation. Mol. Cell 17: 405–416. [DOI] [PubMed] [Google Scholar]
- Francis K.E., Lam S.Y., Copenhaver G.P.(2006). Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol. 142: 1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A., DeMott L., Zhu C., McClosky D.D., Anderson C.T., Dixit R.(2018). Importin-β directly regulates the motor activity and turnover of a kinesin-4. Dev. Cell 44: 642–651.e5. [DOI] [PubMed] [Google Scholar]
- Guo J., Wang F., Song J., Sun W., Zhang X.S.(2010). The expression of Orysa;CycB1;1 is essential for endosperm formation and causes embryo enlargement in rice. Planta 231: 293–303. [DOI] [PubMed] [Google Scholar]
- Harel A., Forbes D.J.(2004). Importin beta: conducting a much larger cellular symphony. Mol. Cell 16: 319–330. [DOI] [PubMed] [Google Scholar]
- Horváth B.M., Magyar Z., Zhang Y., Hamburger A.W., Bakó L., Visser R.G., Bachem C.W., Bögre L.(2006). EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 25: 4909–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.K., Huang L.F., Huang J.J., Wu S.J., Yeh C.H., Lu C.A.(2010). A DEAD-box protein, AtRH36, is essential for female gametophyte development and is involved in rRNA biogenesis in Arabidopsis. Plant Cell Physiol. 51: 694–706. [DOI] [PubMed] [Google Scholar]
- Imai A., Komura M., Kawano E., Kuwashiro Y., Takahashi T.(2008). A semi-dominant mutation in the ribosomal protein L10 gene suppresses the dwarf phenotype of the acl5 mutant in Arabidopsis thaliana. Plant J. 56: 881–890. [DOI] [PubMed] [Google Scholar]
- Jäkel S., Görlich D.(1998). Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17: 4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A.(2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Lam Y.W., Lamond A.I., Mann M., Andersen J.S.(2007). Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 17: 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Liu X., Qiang X., Li X., Li X., Zhu S., Wang L., Wang Y., Liao H., Luan S., Yu F.(2018a). EBP1 nuclear accumulation negatively feeds back on FERONIA-mediated RALF1 signaling. PLoS Biol. 16: e2006340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Cui Y., Ge F.R., Chai S., Zhang W.T., Feng Q.N., Jiang L., Li S., Zhang Y.(2018b). AGC1.5 kinase phosphorylates RopGEFs to control pollen tube growth. Mol. Plant 11: 1198–1209. [DOI] [PubMed] [Google Scholar]
- Li N., Yuan L., Liu N., Shi D., Li X., Tang Z., Liu J., Sundaresan V., Yang W.C.(2009). SLOW WALKER2, a NOC1/MAK21 homologue, is essential for coordinated cell cycle progression during female gametophyte development in Arabidopsis. Plant Physiol. 151: 1486–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ge F.R., Xu M., Zhao X.Y., Huang G.Q., Zhou L.Z., Wang J.G., Kombrink A., McCormick S., Zhang X.S., Zhang Y.(2013). Arabidopsis COBRA-LIKE 10, a GPI-anchored protein, mediates directional growth of pollen tubes. Plant J. 74: 486–497. [DOI] [PubMed] [Google Scholar]
- Liu H.H., Xiong F., Duan C.Y., Wu Y.N., Zhang Y., Li S.(2019). Importin β4 mediates nuclear import of GRF-interacting factors to control ovule development in Arabidopsis. Plant Physiol. 179: 1080–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Qu L.J.(2008). Meiotic and mitotic cell cycle mutants involved in gametophyte development in Arabidopsis. Mol. Plant 1: 564–574. [DOI] [PubMed] [Google Scholar]
- Liu J., et al. (2008). Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20: 1538–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Shi D.Q., Yuan L., Liu J., Yang W.C.(2010). SLOW WALKER3, encoding a putative DEAD-box RNA helicase, is essential for female gametogenesis in Arabidopsis. J. Integr. Plant Biol. 52: 817–828. [DOI] [PubMed] [Google Scholar]
- Lu C., Fedoroff N.(2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12: 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K., Kopperud K., Chakrabarty R., Banerjee R., Brooks R., Goodin M.M.(2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59: 150–162. [DOI] [PubMed] [Google Scholar]
- McCormick S.(1993). Male gametophyte development. Plant Cell 5: 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S.(2004). Control of male gametophyte development. Plant Cell 16 (Suppl): S142–S153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I., Brkljacic J.(2009). The nuclear pore and plant development. Curr. Opin. Plant Biol. 12: 87–95. [DOI] [PubMed] [Google Scholar]
- Nowack M.K., Harashima H., Dissmeyer N., Zhao X., Bouyer D., Weimer A.K., De Winter F., Yang F., Schnittger A.(2012). Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev. Cell 22: 1030–1040. [DOI] [PubMed] [Google Scholar]
- Park S.J., Jiang K., Tal L., Yichie Y., Gar O., Zamir D., Eshed Y., Lippman Z.B.(2014). Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 46: 1337–1342. [DOI] [PubMed] [Google Scholar]
- Rosado A., Li R., van de Ven W., Hsu E., Raikhel N.V.(2012). Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc. Natl. Acad. Sci. USA 109: 19537–19544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P.M., Bui A.Q., Weterings K., McIntire K.N., Hsu Y.-C., Lee P.Y., Truong M.T., Beals T.P., Goldberg R.B.(1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322. [Google Scholar]
- Shi D.Q., Liu J., Xiang Y.H., Ye D., Sundaresan V., Yang W.C.(2005). SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17: 2340–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen J.G., Kang I.H., Macfarlane J., Drews G.N.(2007). Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51: 281–292. [DOI] [PubMed] [Google Scholar]
- Sung M.K., Reitsma J.M., Sweredoski M.J., Hess S., Deshaies R.J.(2016). Ribosomal proteins produced in excess are degraded by the ubiquitin-proteasome system. Mol. Biol. Cell 27: 2642–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakonyi D., Byrne M.E.(2011). Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J. 65: 269–281. [DOI] [PubMed] [Google Scholar]
- Takatsuka H., Umeda-Hara C., Umeda M.(2015). Cyclin-dependent kinase-activating kinases CDKD;1 and CDKD;3 are essential for preserving mitotic activity in Arabidopsis thaliana. Plant J. 82: 1004–1017. [DOI] [PubMed] [Google Scholar]
- Tamura K., Hara-Nishimura I.(2014). Functional insights of nucleocytoplasmic transport in plants. Front Plant Sci 5: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Gasciolli V., Crété P., Vaucheret H.(2004). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14: 346–351. [DOI] [PubMed] [Google Scholar]
- Wan Z.Y., Chai S., Ge F.R., Feng Q.N., Zhang Y., Li S.(2017b). Arabidopsis PROTEIN S-ACYL TRANSFERASE4 mediates root hair growth. Plant J. 90: 249–260. [DOI] [PubMed] [Google Scholar]
- Wan Z.Y., Zhang Y., Li S.(2017a). Protein S-acyl transferase 4 controls nucleus position during root hair tip growth. Plant Signal. Behav. 12: e1311438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.G., Feng C., Liu H.H., Feng Q.N., Li S., Zhang Y.(2017). AP1G mediates vacuolar acidification during synergid-controlled pollen tube reception. Proc. Natl. Acad. Sci. USA 114: E4877–E4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.G., Feng C., Liu H.H., Ge F.R., Li S., Li H.J., Zhang Y.(2016). HAPLESS13-mediated trafficking of STRUBBELIG is critical for ovule development in Arabidopsis. PLoS Genet. 12: e1006269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.G., Li S., Zhao X.Y., Zhou L.Z., Huang G.Q., Feng C., Zhang Y.(2013). HAPLESS13, the Arabidopsis μ1 adaptin, is essential for protein sorting at the trans-Golgi network/early endosome. Plant Physiol. 162: 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Ye R., Xin Y., Fang X., Li C., Shi H., Zhou X., Qi Y.(2011). An importin β protein negatively regulates microRNA activity in Arabidopsis. Plant Cell 23: 3565–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. (2012). SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24: 3278–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H.T., Wan Z.Y., Li S., Zhang Y.(2014). Spatiotemporal production of reactive oxygen species by NADPH oxidase is critical for tapetal programmed cell death and pollen development in Arabidopsis. Plant Cell 26: 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Nishimura M., Hara-Nishimura I., Noguchi T.(2003). Behavior of vacuoles during microspore and pollen development in Arabidopsis thaliana. Plant Cell Physiol. 44: 1192–1201. [DOI] [PubMed] [Google Scholar]
- Yang K., Zhu L., Wang H., Jiang M., Xiao C., Hu X., Vanneste S., Dong J., Le J.(2019). A conserved but plant-specific CDK-mediated regulation of DNA replication protein A2 in the precise control of stomatal terminal division. Proc. Natl. Acad. Sci. USA 116: 18126–18131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Zhang Q., Zhao K., Luo Q., Bao S., Liu H., Men S.(2017). The Arabidopsis GPR1 gene negatively affects pollen germination, pollen tube growth, and gametophyte senescence. Int. J. Mol. Sci. 18: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.T., Li E., Guo Y.K., Yu S.X., Wan Z.Y., Ma T., Li S., Hirano T., Sato M.H., Zhang Y.(2018). Arabidopsis VAC14 is critical for pollen development through mediating vacuolar organization. Plant Physiol. 177: 1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.L., Li E., Feng Q.N., Zhao X.Y., Ge F.R., Zhang Y., Li S.(2015). Protein palmitoylation is critical for the polar growth of root hairs in Arabidopsis. BMC Plant Biol. 15: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Guo X., Ge C., Ma Z., Jiang M., Li T., Koiwa H., Yang S.W., Zhang X.(2017). KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: 4011–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L.Z., Li S., Feng Q.N., Zhang Y.L., Zhao X., Zeng Y.L., Wang H., Jiang L., Zhang Y.(2013). Protein S-ACYL Transferase10 is critical for development and salt tolerance in Arabidopsis. Plant Cell 25: 1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsögön A., Szakonyi D., Shi X., Byrne M.E.(2014). Ribosomal protein RPL27a promotes female gametophyte development in a dose-dependent manner. Plant Physiol. 165: 1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]