The cyclin-dependent kinase CDKG1 stabilizes recombination intermediates during male meiosis and DNA damage-induced somatic homologous recombination.
Abstract
The Arabidopsis (Arabidopsis thaliana) cyclin-dependent kinase G1 (CDKG1) is necessary for recombination and synapsis during male meiosis at high ambient temperature. In the cdkg1-1 mutant, synapsis is impaired and there is a dramatic reduction in the number of class I crossovers, resulting in univalents at metaphase I and pollen sterility. Here, we demonstrate that CDKG1 is necessary for the processing of recombination intermediates in the canonical ZMM recombination pathway and that loss of CDKG1 results in increased class II crossovers. While synapsis and events associated with class I crossovers are severely compromised in a cdkg1-1 mutant, they can be restored by increasing the number of recombination intermediates in the double cdkg1-1 fancm-1 mutant. Despite this, recombination intermediates are not correctly resolved, leading to the formation of chromosome aggregates at metaphase I. Our results show that CDKG1 acts early in the recombination process and is necessary to stabilize recombination intermediates. Finally, we show that the effect on recombination is not restricted to meiosis and that CDKG1 is also required for normal levels of DNA damage-induced homologous recombination in somatic tissues.
INTRODUCTION
During sexual reproduction, the genomic complement of diploid cells is halved by a highly specialized nuclear division, called meiosis, to produce haploid gametes. In meiotic prophase I, homologous chromosomes pair, synapse, and recombine. These three processes are intimately linked, and defects in any one often lead to reduced fertility due to nondisjunction. Meiotic prophase I is divided into five substages: leptotene, zygotene, pachytene, diplotene, and diakinesis. During leptotene, recombination is initiated by the formation of double strand breaks (DSBs) catalyzed by the highly conserved topoisomerase Sporulation-deficient11 (SPO11), followed by the invasion of single strand DSB ends into homologous sequences, creating a D-loop mediated by the recombinases Disrupted Meiotic cDNA1 (DMC1) and Radiation sensitive51 (RAD51; Bishop et al., 1992; Sung and Robberson, 1995; Keeney et al., 1997). Following D-loop formation, recombination-associated DNA synthesis occurs using the complementary strand of the invaded duplex as a template. If the D-loops are extended, they can give rise to a unique heteroduplex DNA configuration called a double Holliday junction. The mode of resolution of this joint molecule determines the outcome either as a crossover (CO) or as a noncrossover (NCO; Allers and Lichten, 2001).
The resolution of stabilized double Holliday junctions into COs is dependent on the activity of ZMM proteins including MutS homologue4 (MSH4), MSH5, and Human Enhancer of Invasion10 (HEI10; Higgins et al., 2004, 2008b; Chelysheva et al., 2012). These ZMM proteins are responsible for the formation of interference-sensitive class I COs that account for 80 to 90% of meiotic COs (Basu-Roy et al., 2013). A number of additional pathways repair DSBs that are not metabolized by the ZMM proteins. In most cases, these DSB repair pathways resolve joint molecules as NCOs, but they also contribute a small number of COs, mostly mediated by either MMS and UV sensitive81 (MUS81; Berchowitz et al., 2007; Higgins et al., 2008a) or Fanconi anemia D2 (FANCD2; Kurzbauer et al., 2018). These class II COs are insensitive to interference and usually make up 10 to 20% of the total CO number (Housworth and Stahl, 2003; Basu-Roy et al., 2013).
In Arabidopsis (Arabidopsis thaliana), between ∼150 and 250 DSBs are formed, but only ∼10 are processed as COs (Mercier et al., 2015). The rest are processed as NCOs via synthesis-dependent strand annealing, dissolution of joint molecules, or by other mechanisms. This implies that there are mechanisms designating which DSBs become resolved as COs and that there are inhibitory mechanisms preventing resolution of DSBs into COs at the remaining sites. While the factors involved in CO designation remain elusive, recent studies have identified anti-CO factors that include the DNA helicases RECQ4A and RECQ4B, Topoisomerase3α, RECQ Mediated Genome Instability1 (RMI), the DNA helicase FANCM and its cofactors MHF1 and MFH2 (FANCM interacting histone-fold protein), and the AAA-ATPase Fidgetin-like1 (FIGL1) and its interacting protein FIDGETIN-LIKE-1 INTERACTING PROTEIN (FLIP; Crismani et al., 2012; Dangel et al., 2014; Girard et al., 2014, 2015; Seguela-Arnaud et al., 2015, 2017; Fernandes et al., 2018; Kurzbauer et al., 2018; Serra et al., 2018).
The DNA helicase FANCM was first identified in Arabidopsis in a reverse genetic screen for factors suppressing CO formation (Crismani et al., 2012). In humans, FANCM is a core component of the Fanconi Anemia network that preserves genome stability by promoting the resolution of inter-strand cross-links (Kottemann and Smogorzewska, 2013). The FANCM protein contains an N-terminal helicase domain and a C-terminal nuclease domain (Whitby, 2010). The helicase activity is necessary for its role as an anti-CO protein as mutations in conserved residues within the helicase domain increase CO formation (Crismani et al., 2012). In Arabidopsis, the absence of FANCM increases CO frequency by threefold, and the extra COs are formed by the MUS81-dependent class II pathway (Crismani et al., 2012). In an independent study, FANCM was also found to be required for DSB-induced somatic homologous recombination (HR; Knoll et al., 2012). It was proposed that FANCM acts by unwinding the D-loop recombination intermediates and channelling them through an NCO pathway. In the absence of FANCM, these recombination intermediates would be processed through the class II recombination pathway. This implies that the recombination sites on which FANCM acts are distinct from those processed by the ZMM-dependent class I pathway.
Concomitant with the recombination process is the formation of the synaptonemal complex (SC). SC formation is initiated early in meiosis between homologous chromosomes. The axial element proteins Asynaptic1 (ASY1), REC8, and ASY3 form a protein axis where the chromatin loops from sister chromatids are anchored in linear arrays (Mercier et al., 2015; Lambing et al., 2017). The central element protein ZYP1 polymerizes between the tracks of axial element proteins, spatially aligning the homologous chromosomes in close register. After CO formation is complete, the SC dissociates and homologous chromosomes are kept together at CO sites.
There is a close interaction between SC formation and CO designation, but how this interaction happens and which proteins are involved remains unclear. Several observations suggest that recombination and SC formation are tightly linked. In most organisms, including Arabidopsis, SC formation is dependent on DSB formation and strand invasion as atspo11-1, atrad51-1, and dmc1 mutants defective in DSB formation or repair fail to synapse (Couteau et al., 1999; Li et al., 2004). Additionally, in some organisms, the number of SC initiation sites (SICs) is directly related with the number of late recombination nodules (Gray and Cohen, 2016). Finally, central element proteins have been shown to have a role in CO formation (Colaiácovo et al., 2003; Jang et al., 2003; Couteau et al., 2004; Zickler, 2006). Interestingly, unlike in other species, Arabidopsis ZMM proteins are not necessary for SC formation (Higgins et al., 2004, 2005, 2008b), indicating that once strand invasion has occurred SC polymerization can progress.
Although many of the components of the recombination and pairing machinery have been identified, their specific role and those of other associated molecules in these processes is not yet fully understood. One of those components is the cyclin-dependent kinase G1 (CDKG1) that belongs to the same kinase group as mammalian CDK10/11 (Doonan and Kitsios, 2009). CDKG1 is also related to the kinases present in the Ph1 locus in wheat (Triticum aestivum) that controls pairing and recombination between homologous chromosomes (Greer et al., 2012), and we previously showed that CDKG1 was necessary for normal levels of COs and synapsis in male meiosis at high ambient temperature (Zheng et al., 2014). While the formation of DSBs was not affected in the cdkg1-1 loss-of-function mutant, the number of COs was drastically reduced, causing plant sterility. In addition, loading of the axial elements of the SC was normal, but the polymerization of the central element protein ZYP1 was reduced in the cdkg1-1 mutant. As ZMM mutants undergo full synapsis (Higgins et al., 2004, 2008b; Mercier et al., 2005; Chelysheva et al., 2012; Wang et al., 2012), it is possible that CDKG1 acts upstream of the ZMM proteins.
To address how CDKG1 affects recombination and synapsis, we examined the genetic interactions between cdkg1-1 and a variety of mutants that have characterized defects in different stages of the CO determination and formation process. We show that, while synapsis is severely compromised in a cdkg1-1 mutant, it is restored by increasing the number of recombination intermediates in a double cdkg1-1 fancm-1 mutant. The restoration of synapsis in the double mutant is accompanied by the restoration of the wild-type numbers of HEI10 and MLH1 foci, indicating that synapsis is sufficient for normal class I CO formation in the absence of CDKG1. Thus, we infer that the synapsis defect in the single cdkg1-1 mutant results from a lack of suitable recombination precursors rather than the recombination defect being due to a lack of synapsis. In addition, the cdkg1-1 mutation is able to partially rescue the phenotype of the ZMM mutant, msh5-2, suggesting that in the absence of CDKG1 early intermediates not resolved by the ZMM proteins are processed by the class II processing pathways. Together, our results indicate that CDKG1 is necessary for the processing of recombination intermediates into the class I CO pathway.
RESULTS
Loss of CDKG1 Increases Class II COs
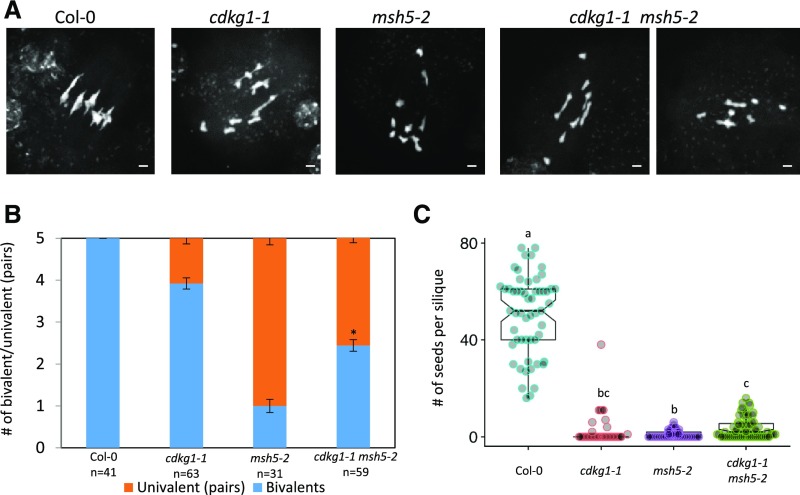
Our previous work showed that the cdkg1-1 mutant retains a low level of class I COs (2.5 ± 2.4 per cell; Zheng et al., 2014). To further investigate the role of CDKG in class I DSB repair, the cdkg1-1 mutant was crossed with the previously described class I CO mutant msh5-2 (Higgins et al., 2008b). In the msh5-2 mutant, class I CO formation is compromised and bivalent formation is dramatically reduced (from five bivalents in the wild type Columbia-0 [Col-0] to 1.1 ± 0.99 in the msh5-2 mutant; Figures 1A and 1B), as was reported previously (Higgins et al., 2008b). The cdkg1-1 msh5-2 double mutant showed a significant increase in bivalent formation at metaphase I (1.1 ± 0.99 bivalents in the msh5-2 mutant and 2.5 ± 0.95 in the cdkg1 msh5-2 double mutant; two-tailed t test, P < 0.001; Figures 1A and 1B) and a concomitant significant increase in seed set (Figure 1C).
Figure 1.
The cdkg1-1 Mutation Increases Bivalent Formation in the msh5-2 Mutant Background.
(A) DAPI-stained metaphase I spreads. Bar = 2 µm.
(B) Ratio of bivalent to univalent pairs present at metaphase I. Error bars represent average ± sd, and n indicates the number of metaphases counted for each genotype. Asterisk indicates that the bivalent number in the cdkg1-1 msh5-2 mutant is significantly different from the single msh5-2 mutant for P < 0.001, two-tailed t test.
(C) Fertility counts in the wild type (Col-0) and indicated mutants. Graphs show mean and interquartile range as well as the actual seed counts. For each genotype, at least 30 siliques from three independent plants were counted. Superscript letters indicate the significance groups for P < 0.001 calculated using ANOVA, with post hoc pairwise t tests using nonpooled sd and Bonferroni correction.
As the class I CO pathway is compromised by the absence of MSH5, the increased bivalent number in the double cdkg1-1 msh5-2 must be generated by increased class II COs. This suggests that loss of CDKG1 increases class II CO formation. If this were the case, we would also expect to see more class II COs in the cdkg1-1 single mutant compared with Col-0. To further test this possibility, we used a modeling approach to identify the best fit values of class II COs in the three mutant contexts (i.e., cdkg1-1, msh5-2, and cdkg1-1 msh5-2). Specifically, we compared bivalent counts observed experimentally with those observed in simulations where the number of class II COs varied from 0.5 to 7 per meiosis.
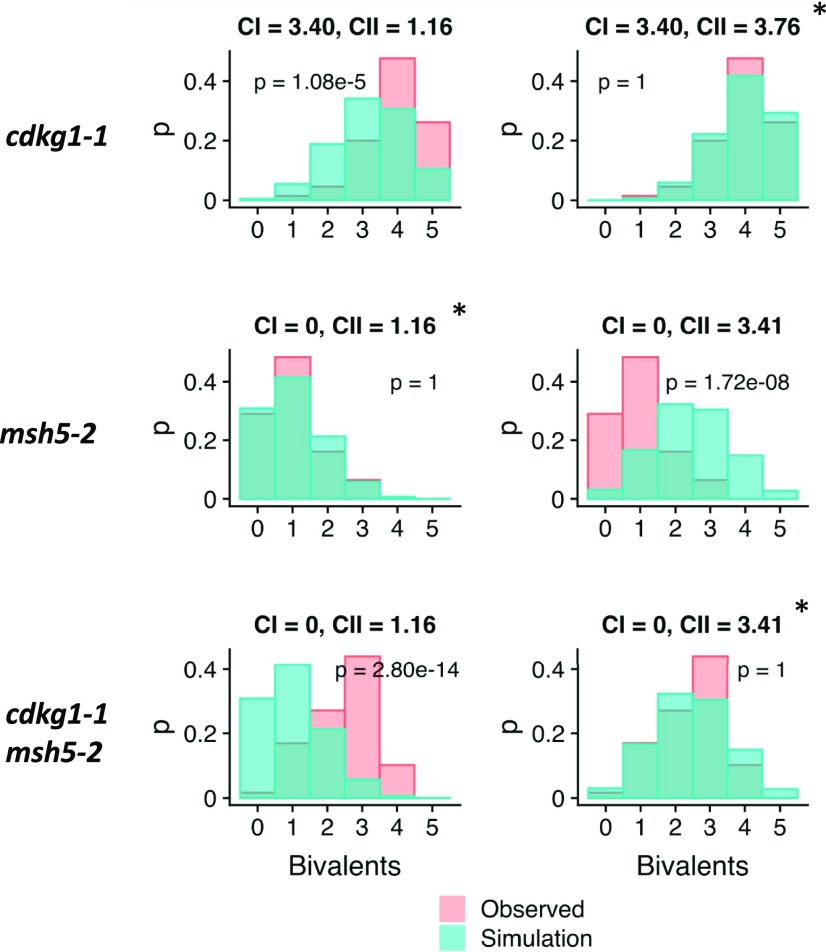
For simulations, we used the beam-film model of CO patterning (Zhang et al., 2014; White et al., 2017), with parameter values that were previously determined for the Arabidopsis wild-type male meiosis (Lloyd and Jenczewski, 2019); the class I CO maturation parameter (M) was adjusted to reflect the differing number of class I COs in the respective lines. For msh5-2, the best fit value was 1.16 class II COs; the best fit bivalent distribution was identical to that experimentally observed for msh5-2 but differed significantly from that of the cdkg1-1 msh5-2 double mutant (Figure 2). For the double cdkg1-1 msh5-2 mutant the best fit value was 3.41 class II COs per meiosis; the best fit bivalent distribution (3.41 class II COs) was no different from that experimentally observed for cdkg1-1 msh5-2 (Figure 2) but differed significantly from that observed experimentally for the single msh5-2 mutant (Figure 2). For the single cdkg1-1, the best fit value was 3.76 class II COs per meiosis; the best fit bivalent distribution (3.76 class II COs) was no different from that experimentally observed for cdkg1-1 (Figure 2) and equivalent to that determined for cdkg1-1 msh5-2 (Supplemental Figure 1). By contrast, distributions from simulations assuming 1.16 class II COs per meiosis (i.e., the number of class II COs derived from our msh5-2 observations) differed significantly from bivalent distributions observed for cdkg1-1 (Figure 2). The same results were obtained assuming 1.5 class II COs per meiosis (Supplemental Table 1), that is, the number of class II COs per meiosis reported in numerous previous studies. Together, these results suggest that loss of CDKG1 results in an approximately threefold increase in the number of class II COs in both the presence and absence of MSH5.
Figure 2.
Bivalent Distribution Comparisons for Simulated and Experimentally Observed Meioses.
Bivalent distributions for observed (pink) and simulated (blue) meioses for cdkg1-1, msh5-2, and cdkg1-1 msh5-2. For simulations, the number of class I (CI) COs was fixed based on experimental observations, and the number of class II (CII) COs varied from 0.5 to 7 per meiosis. Bivalent distributions of best fit simulations are shown (*). In addition, bivalent distributions for cdkg1-1 and cdkg1-1 msh5-2 are compared with those from simulated meiosis with 1.16 CII COs (the best fit value for msh5-2), and the msh5-2 bivalent distribution is compared with simulated meiosis with 3.41 CII COs (the best fit for cdkg1-1 msh5-2). P-values are Bonferroni-corrected values derived from two-sample Kolmogorov–Smirnov tests.
The Extra Class II COs Present in the cdkg1-1 Mutant Are Not MUS81 Dependent
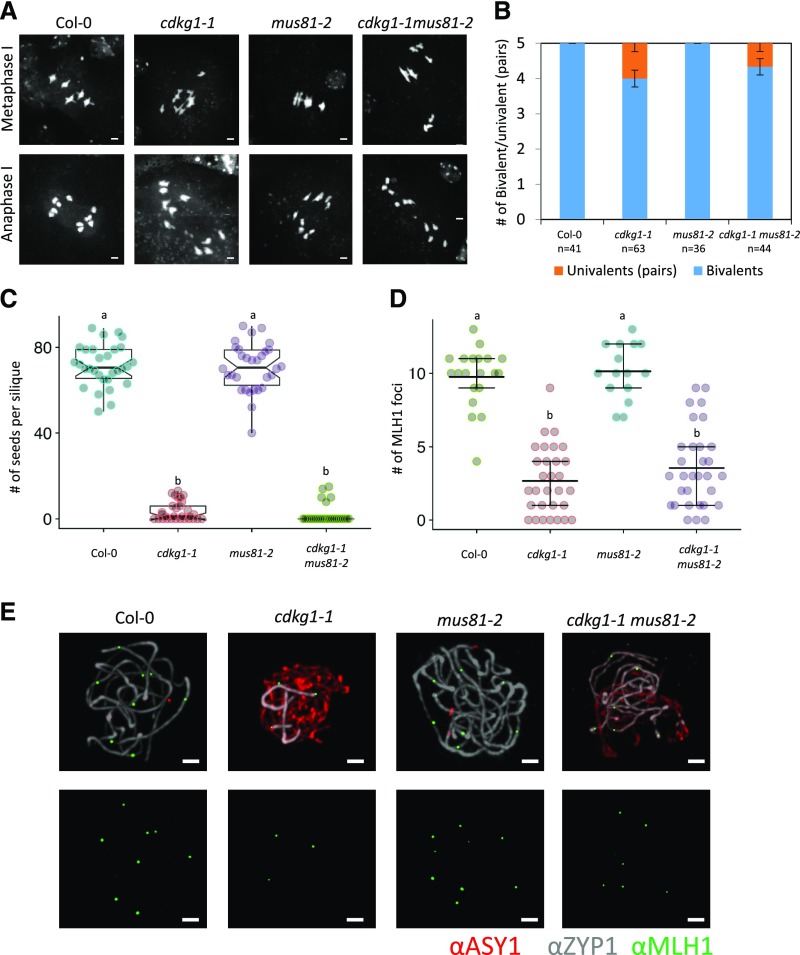
To establish whether the extra COs arise from a MUS81-dependent pathway, we crossed the cdkg1-1 mutant with the mus81-2 mutant. While bivalent formation and seed set are normal in the mus81-2 mutant, the double cdkg1-1 mus81-2 mutant shows similar bivalent formation and seed set to that observed in the single cdkg1-1 mutant (Figures 3A to 3C). This decreased bivalent formation is accompanied by reduced numbers of class I COs as detected by the presence of MLH1 foci (Figures 3D and 3E). In the double cdkg1-1 mus81-2 mutant, the number of MLH1 foci is reduced to levels comparable to that of the single cdkg1-1 mutant (2.7 ± 2.3 in cdkg1-1, n = 30 and 3.6 ± 2.7 in cdkg1-1 mus81-2, n = 31; P = 1 ANOVA, post hoc pairwise t test with nonpooled sd and Bonferroni correction; Figures 3D and 3E), while the single mus81-2 mutant has the wild-type levels of class I COs (9.75 ± 2.03, n = 20 in Col-0 and 10.1 ± 1.9, n = 15 in mus81-2; P = 1, ANOVA, post hoc pairwise t test with nonpooled sd and Bonferroni correction; Figures 3D and 3E).
Figure 3.
The Meiotic Phenotype of the Double cdkg1-1 mus81-2 Mutant Is Similar to the Single cdkg1-1 Mutant.
(A) DAPI-stained metaphase I spreads of the indicated mutants. Bar = 2 µm.
(B) Ratio of bivalent to univalent pairs present at metaphase I. Error bars represent average ± sd, and n indicates the number of metaphases counted for each genotype.
(C) Fertility counts in the indicated mutants. Graphs show mean and interquartile range as well as the actual seed counts. For each genotype at least 30 siliques from three independent plants were counted. Superscript letters indicate the significance groups for P < 0.001 calculated using ANOVA, with post hoc pairwise t tests using nonpooled sd and Bonferroni correction.
(D) Number of MLH1 foci observed in the indicated mutants. Graphs show mean and interquartile range as well as the actual foci counts. Superscript letters indicate the significance groups for P < 0.001 calculated using ANOVA, with post hoc pairwise t tests using nonpooled sd and Bonferroni correction.
(E) Immunolocalization of the class I CO marker protein MLH1 in the indicated mutants. The axial element protein ASY1 is labeled in red, the central element protein ZYP1 in gray, and MLH1 foci in green (top panels). The MLH1 channel (green) is also shown separately (bottom panels). Images represent maximum projections of Z-stacks. Bar = 2 µm.
Synapsis is also defective in the double cdkg1-1mus81-2 mutant. At pachytene, in both Col-0 and the mus81-2 mutant, full synapsis is observed with all the homologous chromosomes paired and a continuous signal of the central element protein ZYP1 being observed along the chromosome axis (Figure 3E). In both the single cdkg1-1 mutant and the double cdkg1-1 mus81-2 mutant, synapsis is impaired (Figure 3E). As the double cdkg1-1 mus81-2 mutant forms the same number of bivalents as the single cdkg1-1 mutant, we conclude that the extra class II COs are formed through a MUS81-independent pathway.
Synapsis and CO Formation Are Restored in the cdkg1-1 fancm-1 Double Mutant
From the ∼100 DSBs that enter the class II pathways, the majority are processed into NCOs by the activity of anti-recombinases (Mercier et al., 2015). One of these is the helicase FANCM that directs these recombination intermediates toward NCO repair or sister chromatid events. To determine the interaction between CDKG1 and the NCO pathways, we crossed the cdkg1-1 mutant with the well-described fancm-1 mutant and examined homologous chromosome pairing and CO formation in the double cdkg1-1 fancm-1 mutant.
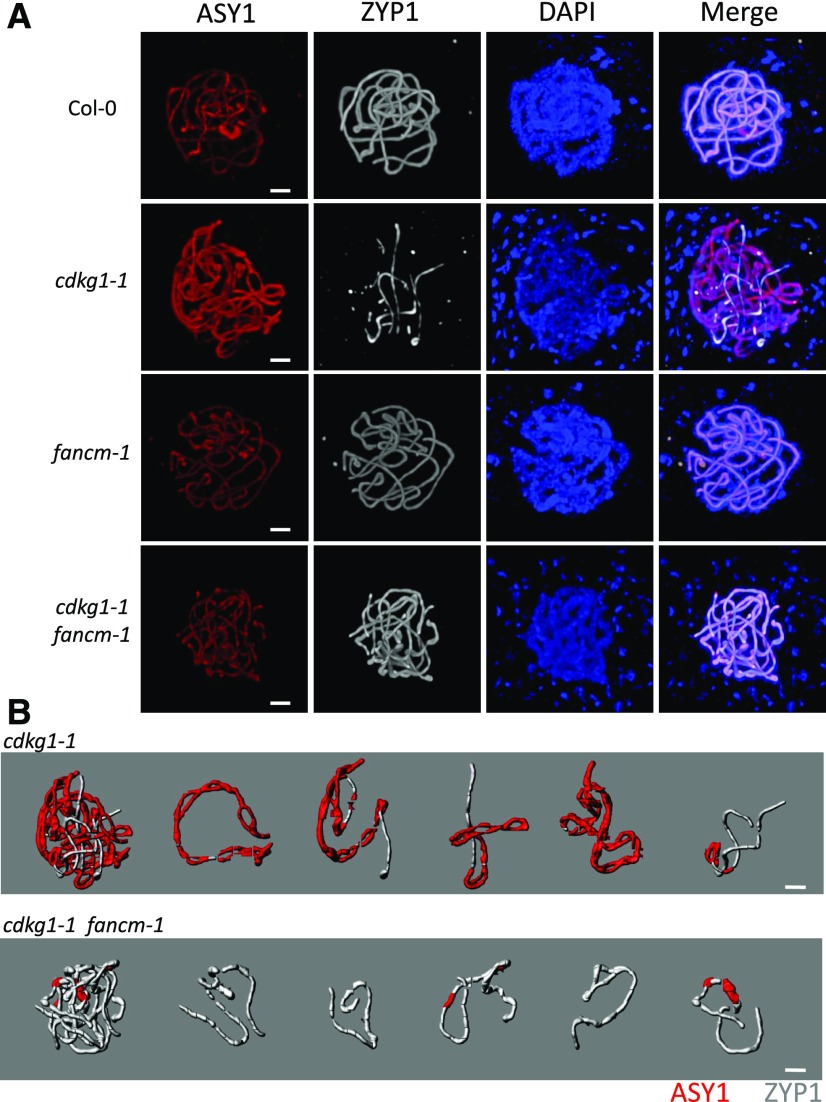
In Col-0 and single fancm-1 mutant cells at pachytene, full homologous chromosome synapsis is observed as determined by ZYP1 loading (Figure 4A). In the single cdkg1-1 mutant, as we have reported previously (Zheng et al., 2014), ZYP1 loading is dramatically reduced (Figure 4A) and the five bivalents fail to fully synapse (Figures 4A and 4B). Strikingly, in the double cdkg1-1 fancm-1 mutant, synapsis is fully restored (Figure 4A), and five nearly fully synapsed bivalents can be computationally isolated (Figure 4B). Using the presence of ASY1 to measure the total SC length and ZYP1 to quantify the synapsed regions, we calculated that in the cdkg1-1 fancm-1 double mutant the average synapsis is 96% (range, 71 to 100%; sd = 0.075; n = 10 cells). The average synapsis previously reported for the single cdkg1-1 mutant is 29.92% (range, 0 to 73.76%; sd = 0.185; n = 10 cells), showing that in the absence of both CDKG1 and FANCM, ZYP1 is able to load onto the chromosome axis at normal levels.
Figure 4.
Chromosome Synapsis Is Restored in the cdkg1-1 fancm-1 Double Mutant.
(A) Immunolocalization of the axial element protein ASY1 (red) and the central element protein ZYP1 (gray) in pachytene nuclei of Col-0, cdkg1-1, fancm-1, and cdkg-1-1 fancm-1 mutant plants. DNA is counterstained with DAPI (blue), and a merge of all channels is shown. Images represent maximum projections of Z-stacks. Bar = 2 µm.
(B) Three-dimensional reconstruction of a whole nucleus and individual bivalents from a cdkg1-1 pachytene-like nucleus and a cdkg1-1 fancm-1 pachytene nucleus. The nuclei were processed using Imaris software, and each bivalent pair was isolated and false colored. Unpaired regions are marked by the presence of ASY1 (red) and paired regions by the presence of ZYP1 (gray). Bar = 2 µm.
This observation could be explained by restoration of ZMM-associated synaptic initiation complexes in the double cdkg1-1 fancm-1 mutant or if the additional class II recombination intermediates (introduced by the fancm-1 mutation) are able to initiate synapsis. To distinguish between these two possibilities, we determined the localization of ZMM proteins MLH1 and HEI10.
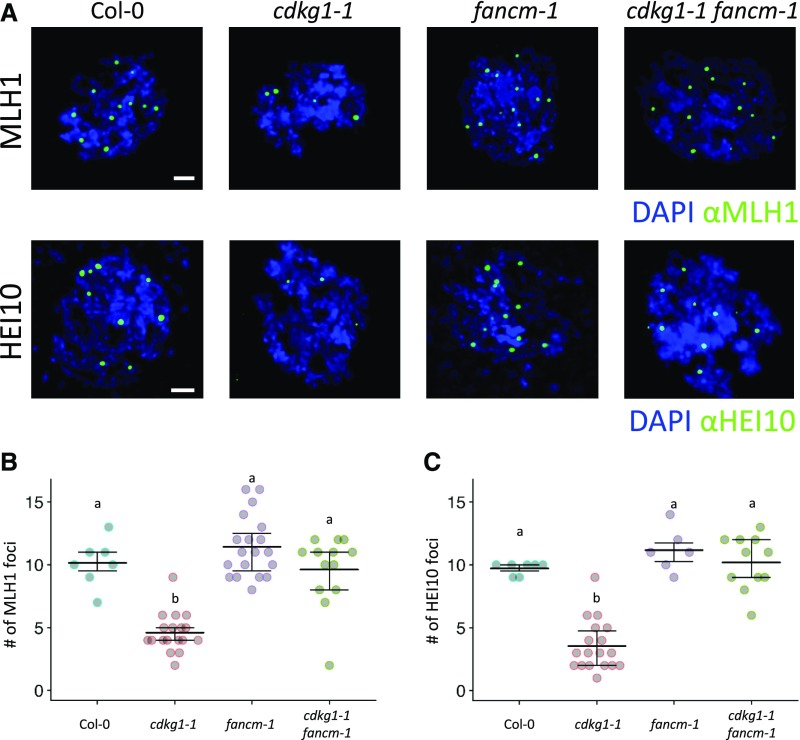
In late pachytene, the MLH1 protein localizes to mature recombination sites, and the number of MLH1 foci observed corresponds to the number of class I COs. In the fancm-1 mutant, we observe the wild-type levels of MLH1 foci (10.14 ± 1.87, n = 7 in Col-0 and 11.42 ± 2.43, n = 19 in fancm-1; P = 1, ANOVA, post hoc pairwise t test with nonpooled sd and Bonferroni correction), while in the cdkg1-1 drastically reduced numbers of MLH1 foci were observed (4.61 ± 1.5, n = 18; P < 0.001, ANOVA, post hoc pairwise t test with nonpooled sd and Bonferroni correction), as previously described by Zheng et al. (2014; Figures 5A and 5B). Interestingly, in the cdkg1-1 fancm-1 double mutant, we observed the wild-type levels of MLH1 (9.62 ± 2.82, n = 13; P = 1, ANOVA, post hoc pairwise t test with nonpooled sd and Bonferroni correction; Figures 5A and 5B). These data suggest that the presence of an increased number of recombination intermediates caused by the absence of FANCM is sufficient to restore class I CO-associated events.
Figure 5.
Class I CO Formation Is Restored in the cdkg1-1 fancm-1 Double Mutant.
(A) Immunolocalization of the class I CO marker proteins (green) MLH1 (top panel) and HEI10 (bottom panel) in diplotene nuclei of Col-0, cdkg1-1, fancm-1, and cdkg-1-1 fancm-1 mutant plants. DNA is counterstained with DAPI (blue). Images represent maximum projections of Z-stacks. Bar = 2 µm.
(B) and (C) Number of MLH1 (B) and HEI10 (C) foci observed in the indicated mutants. Graphs show mean and interquartile range as well as the actual foci counts. Superscript letters indicate the significance groups for P < 0.001, calculated using ANOVA, with post hoc pairwise t tests using nonpooled sd and Bonferroni correction.
Similar results were observed for HEI10 localization in late pachytene and diplotene cells. While the cdkg1-1 single mutant has fewer HEI10 foci, the wild-type levels are restored in the double cdkg1-1 fancm-1 mutant (Col-0: 9.9 ± 0.64, n = 7; cdkg1-1: 3.41 ± 1.91*, n = 17; fancm-1, 11.17 ± 1.5, n = 6; cdkg1-1 fancm-1, 9.9 ± 1.92, n = 10; *P < 0.001, ANOVA, post hoc pairwise t test with nonpooled sd and Bonferroni correction; Figures 5A and 5C). Besides marking mature class I CO sites later in meiosis, the ZMM protein HEI10 also marks early recombination intermediates in leptotene/zygotene (Chelysheva et al., 2012). We observed a reduced number of HEI10 foci in cdkg1-1 in leptotene and early zygotene cells (Col-0: 76 ± 11.01, n = 5; cdkg1-1: 21.58 ± 12.09, n = 12; P < 0.001, ANOVA, post hoc pairwise t test with nonpooled sd and Bonferroni correction), indicating that fewer class I recombination intermediates are present in this mutant, but this defect is restored in a cdkg1-1 fancm-1 double mutant (cdkg1-1 fancm-1: 67.11 ± 22.61, n = 9; P = 1; ANOVA, post hoc pairwise t test with nonpooled sd and Bonferroni correction; Supplemental Figure 2).
In addition, under our immunolocalization conditions and as seen by others (Hurel et al., 2018; Li et al., 2018), HEI10 also localizes to synapsed chromosome axes, emphasizing that the partial synapsis observed in a single cdkg1-1 mutant is restored in a cdkg1-1fancm-1 double mutant (Supplemental Figure 2).
Taken together, these data suggest that the increased number of ZMM-bound early recombination intermediates in the double cdkg1-1 fancm-1 mutant are able to restore synapsis and class I CO defects caused by the absence of CDKG1.
Recombination Intermediates Are Not Correctly Resolved in the cdkg1-1 fancm-1 Double Mutant
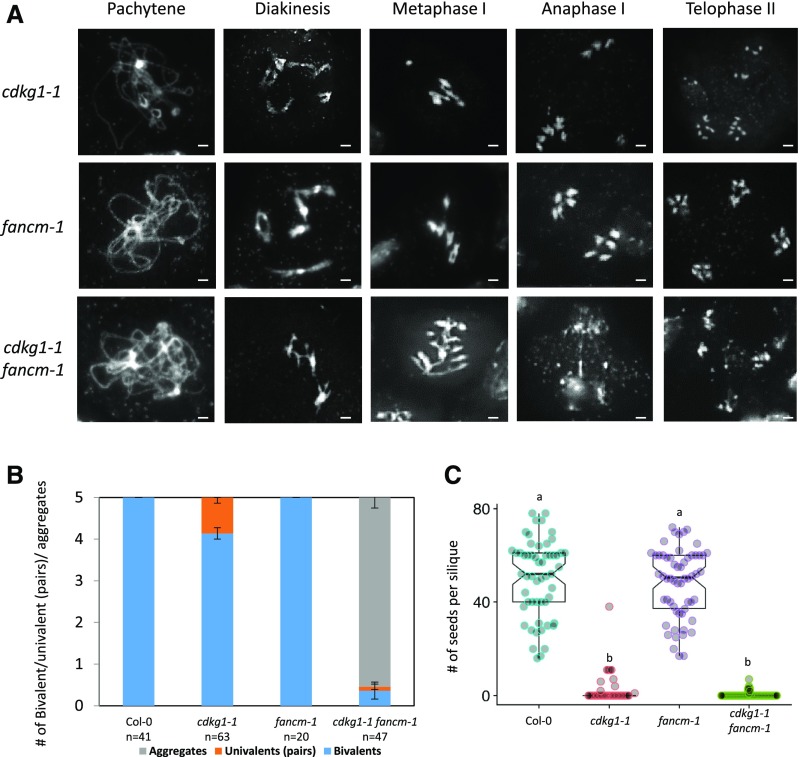
Although synapsis is restored in a cdkg1-1 fancm-1 double mutant, meiotic defects appear in late prophase. In the single fancm-1 mutant, full synapsis was observed at pachytene, five bivalents observed at metaphase I, and four meiotic products with five chromosomes each observed at telophase II (Figure 6A). In the cdkg1-1 fancm-1 double mutant, synapsis progressed normally, but chromosome aggregates (without fragmentation) were always observed at diakinesis and metaphase I (Figures 6A and 6B), suggesting that breaks are not correctly repaired. In addition, chromosome bridges and fragmentation were observed in anaphase I, resulting in the production of unbalanced meiotic products at telophase II (Figure 6A) and plant sterility as measured by seed set (Figure 6C).
Figure 6.
CDKG1 Is Necessary for the Resolution of Recombination Intermediates in the fancm-1 Mutant.
(A) DAPI-stained chromosome spreads of different meiotic stages in the cdkg1-1, fancm-1, and cdkg1-1 fancm-1 mutant as indicated. While in the single mutant five bivalents are observed at metaphase I, in the double mutant chromosome aggregates are present at metaphase I and chromosome bridges observed at anaphase I. Bar = 2 µm.
(B) Ratio of bivalent, univalent pairs and chromosome aggregates present at metaphase I. Error bars represent average ± sd, and n indicates the number of metaphases counted for each genotype.
(C) Fertility counts in the indicated mutants. Graphs show mean and interquartile range as well as the actual seed counts. For each genotype at least 30 siliques from three independent plants were counted. Superscript letters indicate the significance groups for P < 0.001, calculated using ANOVA, with post hoc pairwise t tests using nonpooled sd and Bonferroni correction.
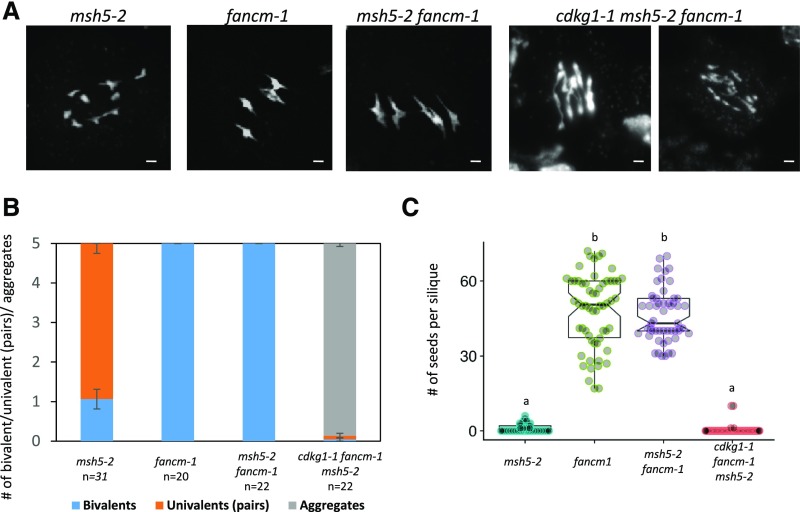
To confirm that the aberrant recombination intermediates in a cdkg1-1 fancm-1 double mutant occur via class II pathways rather than the class I pathway, we produced a triple cdkg1-1 fancm-1 msh5-2 mutant. As can be seen in Figure 7 and as has been previously reported by Crismani et al. (2012), the fancm-1 mutation restores bivalent formation and fertility in a msh5-2 mutant. However, in the triple cdkg1-1 fancm-1 msh5-2 mutant, recombination intermediates are not correctly resolved, resulting in the formation of chromosome aggregates at metaphase I, linkage between chromosomes, and chromosome breakage at anaphase I (Figures 7A and 7B). Thus, the meiotic behavior of the triple mutant mirrors that of the cdkg1-1 fancm-1 double mutant, and like the double cdkg1-1 fancm-1 mutant the triple mutants are sterile (Figure 7C). These results indicate that events leading to metaphase I chromosome aggregates occur when both CDKG1 and FANCM are absent and are independent of the status of the class I CO pathway.
Figure 7.
In the Absence of CDKG1, the fancm-1 Mutation Is Not Able to Rescue the msh5-2 Phenotype.
(A) DAPI-stained metaphase I spreads. Bar = 2 µm.
(B) Ratio of bivalent, univalent pairs and chromosome aggregates present at metaphase I. Error bars represent average ± sd, and n indicates the number of metaphases counted for each genotype.
(C) Fertility counts in the indicated mutants. Graphs show mean and interquartile range as well as the actual seed counts. For each genotype at least 30 siliques from three independent plants were counted. Superscript letters indicate the significance groups for P < 0.001, calculated using ANOVA, with post hoc pairwise t tests using nonpooled sd and Bonferroni correction.
CDKG1 Is Necessary for HR in Somatic Tissues
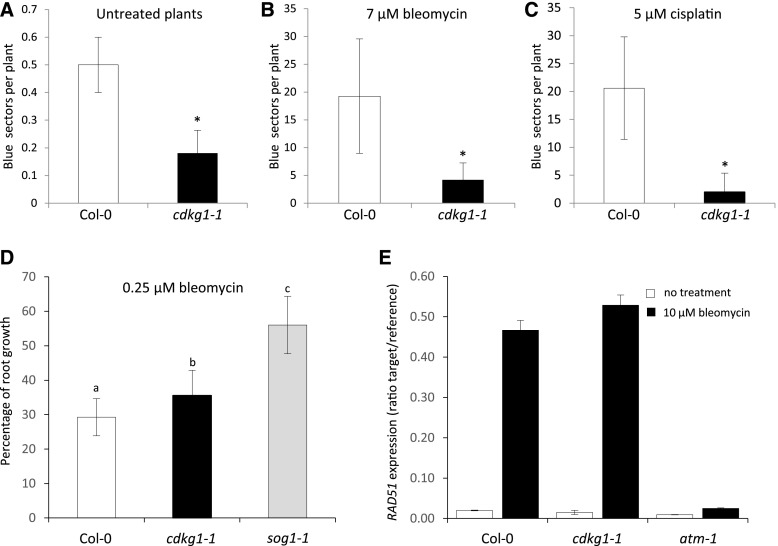
Our data suggest that CDKG1 is required for progression of the major recombination pathways in meiotic cells, raising the possibility that, like many other proteins (e.g., FANCM and RECQ4; Hartung et al., 2006; Knoll and Puchta, 2011), it plays a similar role in somatic cells. To test whether CDKG1 was also necessary for DNA repair in somatic cells, we crossed the cdkg1-1 mutant with the HR reporter line IC9 (Molinier et al., 2004). When DNA damage is induced by genotoxic agents, those lesions repaired through the HR pathway are detected by the presence of blue sectors in the IC9 line. In untreated plants, the number of spontaneous recombination events is significantly reduced in the cdkg1-1 mutant compared to the control IC9 line (0.5 ± 0.1 sectors in the IC9 line versus 0.2 ± 0.1 sectors in the IC9 cdkg1-1; P < 0.001; Figure 8A). In addition, the rate of HR DNA repair in the cdkg1-1 mutant was significantly reduced when plants were treated with bleomycin (from 19.2 ± 10.3 to 4.2 ± 3.0 blue sectors per plant; P < 0.001) and cisplatin treatments (from 20.6 ± 9.2 to 2 ± 3.4 blue sectors per plant; P < 0.001; Figures 8B and 8C). Surprisingly, cdkg1-1 plants, like Col-0 plants, were sensitive to root growth inhibition by bleomycin in contrast to the sog-1 mutant, which fails to respond to DNA damage and is less sensitive to root growth inhibition by bleomycin (Figure 8D; Yoshiyama et al., 2009). This suggests that most DNA breaks are repaired by a non–HR-dependent pathway. In support of this, we observed that the expression of genes involved in the HR pathway including RAD51, is not altered in the cdkg1-1 mutant compared to the wild type Col-0 (Figure 8E), while in the DNA repair-deficient mutant atm-1 expression of these genes is not induced in response to bleomycin treatment (Figure 8E; Garcia et al., 2003; Cimprich and Cortez, 2008; Culligan and Britt, 2008).
Figure 8.
Rates of Somatic HR Are Reduced in the cdkg1-1 Mutant.
(A) Spontaneous recombination rates in Col-0 and the cdkg1-1 mutant. Graphs represent averages ± sd for 10 plants. Asterisks indicate values that are significantly different for P < 0.001, using two-tailed t test.
(B) and (C) Recombination rates for Col-0 and the cdkg1-1 mutant in the presence of 7 µM bleomycin (B) or 5 µM cisplatin (C). Graphs represent averages ± sd for 10 plants.
(D) Percentage of root growth in media containing 0.25 µM bleomycin for Col-0, cdkg1-1, and sog1-1 seedlings compared with growth in media with no bleomycin. Graphs represent averages ± sd for thirty 8-d-old seedlings. Superscript letters indicate the significance groups for P < 0.001 calculated using ANOVA, with post hoc pairwise t tests using nonpooled sd and Bonferroni correction.
(E) Expression of the HR repair pathway gene RAD51 in Col-0, the cdkg1-1 mutant, and atm-1 control 8-d-old seedlings, treated or not with 10 µM bleomycin for 3 h, as determined by qPCR.
Taken together, these data suggest that CDKG1 has a global role in HR-based DNA repair in somatic and meiotic cells promoting the orderly progression of canonical HR pathways.
DISCUSSION
Cyclin-dependent kinases and their cognate cyclins have been shown to have important roles during meiosis and recombination in many organisms (Trovesi et al., 2013; Gómez-Escoda and Wu, 2017; Wijnker et al., 2019; Yang et al., 2020). We have previously demonstrated that CDKG1 is necessary for CO formation and homologous chromosome synapsis at high ambient temperature during male meiosis in Arabidopsis (Zheng et al., 2014). Here, we describe a role for CDKG1 in the stabilization of meiotic and somatic recombination intermediates.
CDKG1 Promotes the Stability of Early Recombination Intermediates
In the cdkg1-1 mutant, early events such as DSB formation and the loading of the strand invasion and exchange proteins RAD51 and DMC1 were found to be normal (Zheng et al., 2014). Despite this, synapsis and CO formation were drastically reduced, suggesting that CDKG1 acts downstream of these early recombination events. After strand invasion and D-loop stabilization, ∼100 sites are designated as potential class I COs and marked by the presence of ZMM proteins, including HEI10. As prophase progresses, the number of HEI10 foci decreases, until by late pachytene all remaining HEI10 foci colocalize with MLH1 and mark sites of future class I COs (Chelysheva et al., 2012). In the absence of the ZMM protein MSH5, class I COs are not formed, and the recombination intermediates are resolved as NCOs, resulting in the presence of univalents at metaphase I and consequent sterility (Higgins et al., 2008b; Lu et al., 2008). Unexpectedly, we observed increased CO number and seed set in the double cdkg1-1 msh5-2 mutant compared to the single msh5-2 mutant. As the class I CO pathway is defective in the double mutant, the extra COs must come from a ZMM-independent pathway. The presence of additional class II COs in both the single cdkg1-1 and the double cdkg1-1msh5-2 mutant is also supported by the results obtained when bivalent distributions were simulated. While for the single msh5-2 mutant the best fit bivalent number was obtained for 1.16 class II COs per meiosis, for the cdkg1-1 and cdkg1-1 msh5-2 mutants the best fit was observed for 3.76 and 3.41 class II COs, respectively, per meiosis. In yeast, mutations of the early recombination protein MLH2 and its interacting partner Mer3 also improve spore viability of msh4 mutants, and the authors suggest that this might be due to an increased capacity of the double mutants to make COs (Duroc et al., 2017). Indeed, the double msh4mlh2 and msh5mlh2 mutants show increased CO frequency over specific genetic intervals (Abdullah et al., 2004). The MLH1-Mer3 complex preferentially recognizes D-loops and DNA branched structures, and it is thought to act by stopping D-loop extension (Duroc et al., 2017). Furthermore, this activity seems to be independent of the fate of the D-loop intermediates, akin to what we observe in the cdkg1-1 mutant.
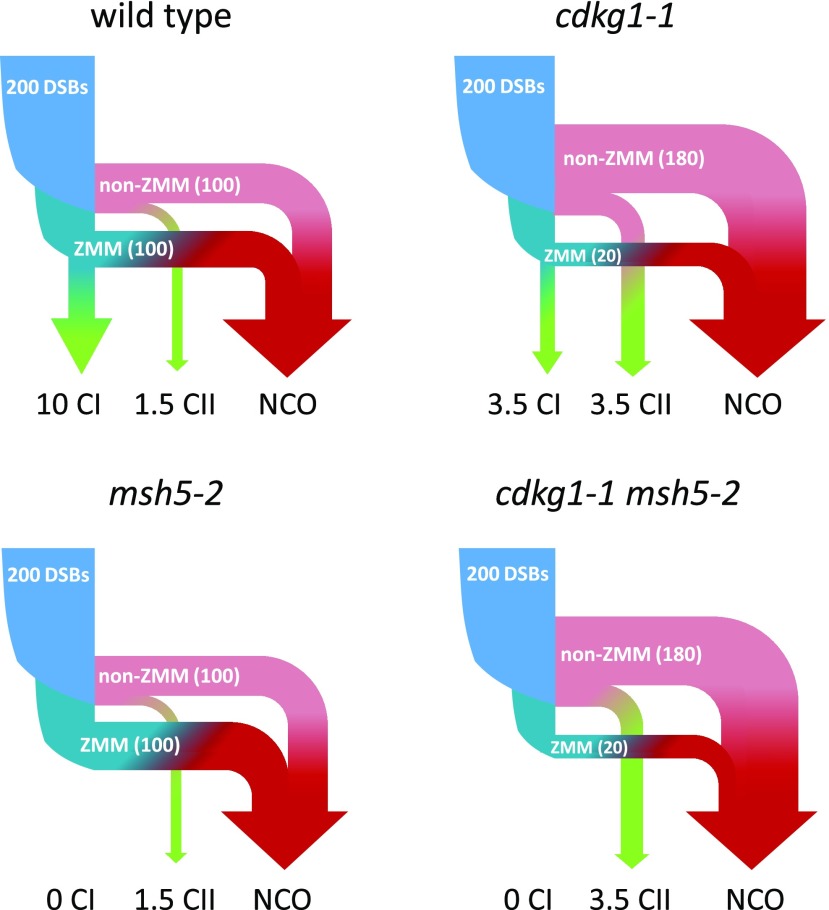
Where then do these additional class II COs come from? One possibility is that in the absence of CDKG1, a number of the recombination intermediates that would normally be destined for the class I pathway are repaired via the DSB processing pathways operating in parallel. In the wild type, of the ∼200 DSBs initiated in early prophase (Chelysheva et al., 2012; Choi et al., 2013; Girard et al., 2015; Séguéla-Arnaud et al., 2015) approximately half are thought to be processed by the ZMM proteins (Higgins et al., 2004, 2008a; Chelysheva et al., 2012), with the remaining breaks processed by parallel non-ZMM pathways, for example, via MUS81 or FANCD2 (Figure 9; Kurzbauer et al., 2018). A small percentage of the joint molecules processed by these secondary pathways are eventually resolved as class II COs (Mercier et al., 2015). Channelling more intermediates through non-ZMM pathways in a cdkg1-1 mutant could therefore result in additional class II COs, together with an equivalent decrease in the number of stable early class I intermediates (Figure 9). This possibility is directly supported by our observation of reduced HEI10 foci during leptotene in the cdkg1-1 mutant, indicating fewer DSBs being processed by the ZMM proteins in the absence of CDKG1. Given that the wild-type numbers of DSBs occur in a cdkg1-1 mutant (Zheng et al., 2014), a large number of intermediates that would normally be processed by ZMM proteins in the wild type must be processed by other pathways in a cdkg1-1 mutant (Figure 9). Another possible explanation is that in the absence of CDKG1, joint molecules that enter the class II pathways have a greater likelihood of being repaired as COs. In either situation, the recombination intermediates are processed by a MUS81-independent pathway, likely involving FANCD2 (Kurzbauer et al., 2018), as the double cdkg1-1mus81-2 mutant behaves as the single cdkg1-1 mutant.
Figure 9.
A Model for the Impact of cdkg1-1 and msh5-2 Mutations on the Class I (CI) CO, Class II (CII) CO, and Non-crossover (NCO) Pathways in Arabidopsis.
Arrow thickness relates to the proportion of the initial DSBs going through each pathway. Numbers indicate the number of DSBs channeled to each pathway. DSBs: ∼200 DMC1/RAD51/γH2AX foci are observed in early meiotic prophase (Choi et al., 2013; Girard et al., 2015; Séguéla-Arnaud et al., 2015; Xue et al., 2018). ZMM intermediates: ∼100 MSH4/MSH5/HEI10 foci are observed in the wild-type leptotene/zygotene (Higgins et al., 2004, 2008a; Chelysheva et al., 2012). In cdkg1-1, we observe ∼20 HEI10 foci in leptotene/zygotene. CI, class I CO.
A greater reliance on the DSB processing pathways that operate in parallel to the class I CO pathway is also suggested by the presence of chromosome aggregates at metaphase in a cdkg1-1 fancm-1 double mutant. The helicase FANCM is an integral component of the meiotic DSB processing machinery and promotes dissolution of joint molecules that have the potential to become class II COs, diverting them toward NCO or inter-sister repair. One interpretation of the observed chromosome aggregates in a double cdkg1-1 fancm-1 mutant (but not the respective single mutants) is that aberrant joint molecules form in the absence of CDKG1 and that FANCM is required to resolve these toxic recombination intermediates. The reciprocal interpretation is also possible, that is, that aberrant joint molecules form in the absence of FANCM that require CDKG1 for resolution. Further experimentation will be required to distinguish between these (or other) possibilities.
Elevated Numbers of Recombination Intermediates Restore Synapsis in the cdkg1-1 Mutant
Despite the chromosome abnormalities present at metaphase I, full homologous chromosome synapsis is observed in the double cdkg1-1 fancm-1 mutant. One of the main consequences of a fancm mutation is the persistence of a large number of recombination intermediates. In the absence of CDKG1, these recombination intermediates are able to act as synaptic initiation sites resulting in full synapsis in the cdkg1-1 fancm-1 mutant. By extension, this implies that the synapsis defect in a cdkg1-1 mutant is due to a deficit in the number of recombination intermediates able to act as synaptic initiation sites, rather than a problem in SC assembly or maintenance.
In some organisms, such as budding yeast, CO formation is necessary for full synapsis to occur (Zickler and Kleckner, 2016) but, in Arabidopsis, after the early recombination events, the SC is able to form even in the absence of ZMM proteins and consequently reduced CO formation (Higgins et al., 2004, 2008b; Mercier et al., 2005; Jackson et al., 2006; Macaisne et al., 2008; Chelysheva et al., 2012), while defects in DSB formation, strand invasion, and D-loop formation and stabilization do lead to incomplete synapsis in Arabidopsis as illustrated by the dmc1, atrad51, and atxrcc3 mutants (Couteau et al., 1999; Bleuyard and White, 2004; Li et al., 2004). One possible explanation for this difference is that plants have long chromosomes containing numerous synapsis initiation sites that are not CO-designated sites so loss of COs would have less effect on SC nucleation and polymerization (Zickler and Kleckner, 2016). This could help explain the phenotype of the single cdkg1-1 mutant. In the single cdkg1-1 mutant, the number of SICs is reduced, possibly due to the reduced stability of recombination intermediates, resulting in fewer HEI10 foci in leptotene, incomplete synapsis, and consequently reduced numbers of class I COs.
In the cdkg1-1 fancm-1 mutant, more recombination intermediates are maintained and can act as SIC sites. This is in agreement with synapsis initiation occurring early in plants, at the time of strand exchange and D-loop formation.
The Localization of Class I CO Markers Is Restored in a Double cdkg1-1 fancm-1 Mutant
In addition to the recovery of synapsis in the double cdkg1-1 fancm-1 mutant, there is also recovery of class I CO events as marked by the presence of HEI10 and MLH1. The recovery of HEI10 foci early in leptotene and zygotene and the normal levels of HEI10 and MLH1 foci at pachytene and diplotene suggest normal numbers of recombination intermediates can be processed by the ZMM pathway in the absence of both CDKG1 and FANCM. How the lack of FANCM restores the ZMM pathway when CDKG1 is absent is unclear. One possibility is that CDKG1 normally protects ZMM intermediates from dissolution by FANCM and that in the absence of both proteins these intermediates persist. Another possibility is that the additional class II CO sites, due to the fancm-1 mutation, may stabilize inter-homologue associations allowing ZMM intermediates to persist.
Despite the fact that normal levels of ZMM-associated foci are observed during pachytene and diplotene in cdkg1-1 fancm-1 double mutants, it is not possible to ascertain whether these sites mature as full CO events, as chromosome entanglements are observed at metaphase I. Unresolved ZMM intermediates are not required for the formation of chromosome aggregates, however, as aggregates are also observed in a cdkg1-1 fancm-1 msh5-2 triple mutant, where class I COs and their precursors are absent. The chromosome aggregate phenotype is reminiscent of that observed in fancm-1 mus81-2 double mutants (Crismani et al., 2012). In this mutant, unresolved class II recombination intermediates persist, leading to the formation of chromosome entanglements at metaphase I (Crismani et al., 2012). MUS81 has a well-described role in the resolution of Holliday junctions (Osman et al., 2003), and there is also support for CDKs having a role in CO maturation (Palmer et al., 2019).
In wheat–rye (Secale cereale) hybrids, which possess no true homologous chromosomes, only an average of 0.58 chiasmata per cell are observed at metaphase I, despite an average of ∼20 MLH1 foci being observed at diplotene (Martín et al., 2014). In this context, the ability of the MLH1 marked foci to mature as chiasmata is suppressed by the Ph1 locus, which contains a cluster of defective CDKs closely related to the CDKG group in Arabidopsis (Griffiths et al., 2006; Zheng et al., 2014). Ph1 is thought to negatively regulate CDK activity, leading the authors to suggest that CDK activity promotes maturation of MLH1 sites in wheat–rye hybrids (Martín et al., 2014).
A Wider Role for CDKG1 in Recombination Pathways in Arabidopsis
Our data indicate that in addition to its meiotic role, CDKG1 is also involved in somatic DNA repair. In the absence of CDKG1, we observed a reduction in the rate of HR DNA repair after spontaneous and induced DNA lesions. This reduction occurred after treatment with bleomycin, which induces DSBs, and also with cisplatin, which is thought to cause DNA cross-links (Brabec, 2002), suggesting that CDKG1 promotes DNA repair from both types of lesions. This contrasts with what is observed for FANCM and RECQ4A mutants. While FANCM is required for DSB-induced DNA repair, it suppresses spontaneous and cisplatin-induced HR (Knoll et al., 2012). RECQ4A, on the other hand, is thought to suppress the repair of defects arising during DNA replication and not the ones caused by DSB formation (Hartung et al., 2006). These observations again suggest that CDKG1 is important during the early stages of DNA repair, likely by helping to stabilize recombination intermediates, and show that CDKG1 acts independently of other anti-CO proteins during the recombination process. Although HR DNA repair was reduced in the cdkg1-1 mutant, we did not observe increased sensitivity to genotoxic agents or altered expression of DNA repair genes, indicating that despite the fact that HR is suppressed in the cdkg1-1 mutant, DNA damage is still repaired via alternative pathways.
A Model for the Role of CDKG1 during Male Meiosis
Our data indicate that CDKG1 is important for the formation and/or stabilization of both meiotic and somatic recombination intermediates. In the cdkg1-1 mutant, the stability of early recombination intermediates normally processed by the class I CO pathway is compromised, resulting in reduced HEI10 foci in leptotene and zygotene, reduced synapsis, and decreased levels of class I COs. As DSB formation appears normal in a cdkg1-1 mutant, our results suggest that the DSBs not captured by the class I recombination pathway are processed by parallel DNA repair pathways. This is accompanied by the formation of additional class II COs in both the presence and absence of an otherwise functioning ZMM pathway (Figure 9). This contrasts with what is observed in ZMM mutants, where recombination intermediates destined to be processed by the ZMM proteins are resolved as NCOs. We suggest therefore that CDKG1 acts upstream of the ZMM proteins and helps stabilize or maintain intermediates for processing by the ZMM recombination pathway.
In addition to its meiotic role, it is clear that CDKG1 also has wider functions including the role in promoting somatic DNA repair by HR described here. Moreover, CDKG1 affects splicing in somatic tissues (Cavallari et al., 2018), and in anthers, the loss of CDKG1 results in the incorrect splicing of the callose synthase gene CalS5, impaired callose synthesis, and abnormal pollen cell wall formation (Huang et al., 2013). This likely contributes to the low seed set observed in cdkg1-1 mutants, despite a relatively modest reduction in bivalent number. While the possibility that CDKG1 fine-tunes recombination outcomes during male meiosis by regulating the splicing of meiotic genes is an attractive hypothesis, alternative possibilities include the direct phosphorylation of meiotic proteins or their immediate regulators, leading to changes in activity or levels. However, the underlying molecular mechanisms remain obscure and merit further investigation.
METHODS
Plant Material and Growth Conditions
Plants were grown in growth rooms under long-day conditions (16-h-light/8-h-dark cycle, light intensity of 150 µmol m−2 s−1, provided by Sylvania 840 lamps) with the temperature set at 23°C. The mutant lines used in this study have previously been described; cdkg1-1, SALK_075762; (Zheng et al., 2014), mus81-2 (SALK_107515; Higgins et al., 2008a), msh5-2 (SALK_026553; Higgins et al., 2008b), fancm-1 (Crismani et al., 2012), and IC9 (Molinier et al., 2004). Homozygous lines for each mutant were used for crosses and double and triple mutants identified by PCR-based screening (the full list of primers used for genotyping can be found in Supplemental Table 3). For seed count experiments, all plants were grown at the same time under the same conditions, and at least 30 siliques from three different plants were counted per genotype. Statistical significance was calculated using ANOVA, with post hoc pairwise t tests using nonpooled sd and Bonferroni correction (full ANOVA results are given in Supplemental Table 4).
Meiotic Spread Preparation
Whole inflorescences from Col-0 and mutant plants were fixed in 3:1 ethanol:acetic acid and stored at 4°C. Flower buds in the size range 0.3 to 0.9 mm were used to prepare the squashes as described previously (Jenkins and Hasterok, 2007). Slides were then stained with 4′,6-diamidino-2-phenylindole (DAPI) for observation. The bivalent/univalent ratio was determined for at least 20 metaphases per genotype; exact numbers are shown in the respective figures. Statistical significance was calculated using a two-tailed Student’s t test.
Immunolabeling Arabidopsis Pollen Mother Cells
Meiocites from Col-0 and mutant plants where embedded in acrylamide to preserve their three-dimensional structure and used for the immunolocalization studies as described previously (Phillips et al., 2010), with the following modifications. Buds of Arabidopsis (Arabidopsis thaliana) plants in the prophase I meiotic stage (size range, 0.3 to 0.9 mm) were isolated into buffer A and fixed in 2% (w/v) paraformaldehyde for 30 min. After 2 × 10-min washes in buffer A, the buds were macerated with a brass rod in buffer A + 1% (v/v) Lipsol, and the suspension was then embedded in acrylamide. Embedded meiocytes were blocked and incubated with primary antibody solution for 24 to 36 h. The primary antibodies used were α-ASY1 (rat, 1:500; Armstrong et al., 2002), α-ZYP1 (guinea pig, 1:500; Higgins et al., 2005), α-MLH1 (rabbit, 1:250; Chelysheva et al., 2010), and α-HEI10 (rabbit, 1:250; Chelysheva et al., 2012). Secondary antibodies (Alexa Fluor 488 chicken anti-rat [A21470], Alexa Flour 546 donkey anti-rabbit [A10040], and Alexa Flour 633 goat anti–guinea pig [A21105], all from Molecular Probes) were used at 1:500 dilution and incubated overnight. Images were acquired using a TCS SP5II microscope or an SP8 confocal microscope (Leica). Images represent deconvolved maximum projection of Z-stacks. For images obtained with the SP5II microscope, the Z-stacks were deconvolved using AutoQuant ×2 (Media Cybernetics), and for the SP8 microscope using the built-in Lightning software (Leica Microsystems). The same deconvolution parameters were applied for every image. Bivalent tracking and analysis were performed using Imaris 7.3 (Bitplane).
Simulating CO Formation
Meiosis was modeled using the beam-film model of CO patterning (Zhang et al., 2014; White et al., 2017). Parameter values previously determined for the Arabidopsis wild-type male meiosis were applied for each of the five chromosomes (Lloyd and Jenczewski 2019). Class I and class II COs were simulated independently for 10,000 bivalents for each of the five chromosomes and then combined to obtain final CO positions for 10,000 meioses. For each meiosis, the number of bivalents was the number of chromosomes that received at least one CO of either class. The class I CO M value was adjusted to reflect the differing number of MLH1 foci (i.e., class I COs) observed in the respective lines (msh5-2 and msh5-2 cdkg1-1, M = 0, i.e., 100% reduction in class I COs; cdkg1-1 M = 0.354, i.e., 64.6% reduction in class I COs). The proportion of DSBs that become class II COs (T2Prob) was varied from 30% of wild-type values to 450% of wild-type values, corresponding to a range of 0.5 to 7 class II COs per meiosis. Complete parameter values used in simulations are provided in Supplemental Table 2. Best fit values for class II COs were obtained by comparing the average number of bivalents for each simulation round to the average number of bivalents observed cytologically. P-values were calculated using Bonferroni corrected values derived from two-sample Kolmogorov–Smirnov tests.
HR Assays
HR assays were performed as described previously (Hartung et al., 2007). Briefly, the 7-d-old wild-type Col-0 and cdkg1-1 mutant seedlings carrying the IC9 reporter construct (Molinier et al., 2004) were transferred to liquid media containing 7 µM bleomycin, 5 µM cisplatin, or solvent control and incubated in the light at 22°C for 8 d. The β-glucuronidase staining reaction was done at 37°C for 2 d, and pigment extraction was done using 70% (v/v) ethanol. The number of blue sectors per plant was counted under a binocular microscope. For the root growth assays, plants were germinated at 22°C in media containing 0.25 µM bleomycin or solvent control. Root growth was measured after 8 d. For each treatment, 30 seedlings were used, and the experiment repeated three times with similar results. Statistical significance was calculated using ANOVA, with post hoc pairwise t tests using nonpooled sd and Bonferroni correction.
qPCR
Total RNA was extracted from seedlings treated with 10 µM bleomycin or solvent control for 3 h using the RNeasy Plant Mini kit (Qiagen). Total mRNA (1 µg) was used to generate cDNA using the SuperScript III First-strand Synthesis kit (Invitrogen). qPCRs were performed using the LightCycler 480 system (Roche). Typically, 10 ng of cDNA was used in a 20 μL reaction containing 0.25 µM of each primer and 10 μL of LightCycler 480 SYBR Green I Master. Each sample was analyzed in triplicate using Rad51-specific primers as described by Wang et al. (2014), and Actin2 expression was used as a reference (Supplemental Table 3). Data were analyzed using the LightCycler 480 software.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CDKG1 (At5g63370), MSH5 (At3g20475), MUS81 (At4g30870), and FANCM (At1g35530).
Supplemental Data
Supplemental Figure 1. Equivalent numbers of class II COs are modeled for cdkg1-1 and cdkg1-1 msh5-2.
Supplemental Figure 2. HEI10 localization in leptotene, zygotene, pachytene and diplotene nuclei of Col-0, cdkg1-1, fancm-1 and cdkg-1-1 fancm-1 mutant plants as indicated.
Supplemental Table 1. Comparisons of experimental and simulated bivalent distributions.
Supplemental Table 2. Parameter values used in beam-film simulations.
Supplemental Table 3. List of primers used in this study.
Supplemental Table 4. Statistical analysis tables.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Raphael Mercier for kindly providing the fancm-1, msh5-2, and msh5-2 fancm-1 mutants; Holger Puchta for the HR recombination marker IC9 line; and Mathilde Grelon for providing the MLH1 and HEI10 antibodies. Special thanks to Glyn Jenkins, Kim Osman, and Eugenio Sanchez-Moran for many helpful discussions. This work was funded by the Biotechnology and Biological Sciences Research Council (grants BB/M009459/1 and BB/CSP1730/1), by a Marie Curie COFUND grant (663830-AU-110 to A.L.), by the National Science Centre Poland (grant 2014/14/M/NZ2/00519 to A.B.), and by the EU Erasmus+ Programme (to F.T. and D.D.).
AUTHOR CONTRIBUTIONS
C.N., A.L., D.W.P., and J.H.D. designed the experiments; C.N., D.D., A.B., and F.T. performed the experiments; C.N., D.W.P., and A.L. analyzed the data; C.N., A.L., D.W.P., and J.H.D. wrote the article. All authors read and approved the article.
Footnotes
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Abdullah M.F.F., Hoffmann E.R., Cotton V.E., Borts R.H.(2004). A role for the MutL homologue MLH2 in controlling heteroduplex formation and in regulating between two different crossover pathways in budding yeast. Cytogenet. Genome Res. 107: 180–190. [DOI] [PubMed] [Google Scholar]
- Allers T., Lichten M.(2001). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Caryl A.P., Jones G.H., Franklin F.C.H.(2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115: 3645–3655. [DOI] [PubMed] [Google Scholar]
- Basu-Roy S., Gauthier F., Giraut L., Mézard C., Falque M., Martin O.C.(2013). Hot regions of noninterfering crossovers coexist with a nonuniformly interfering pathway in Arabidopsis thaliana. Genetics 195: 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz L.E., Francis K.E., Bey A.L., Copenhaver G.P.(2007). The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D.K., Park D., Xu L., Kleckner N.(1992). DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456. [DOI] [PubMed] [Google Scholar]
- Bleuyard J.-Y., White C.I.(2004). The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J. 23: 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabec V.(2002). DNA modifications by antitumor platinum and ruthenium compounds: their recognition and repair In Progress in Nucleic Acid Research and Molecular Biology, Moldave K., ed (New York: Academic Press; ), pp. 1–68. [DOI] [PubMed] [Google Scholar]
- Cavallari N., Nibau C., Fuchs A., Dadarou D., Barta A., Doonan J.H.(2018). The cyclin-dependent kinase G group defines a thermo-sensitive alternative splicing circuit modulating the expression of Arabidopsis ATU2AF65A. Plant J. 94: 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L., Grandont L., Vrielynck N., le Guin S., Mercier R., Grelon M.(2010). An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: Immunodetection of cohesins, histones and MLH1. Cytogenet. Genome Res. 129: 143–153. [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Vezon D., Chambon A., Gendrot G., Pereira L., Lemhemdi A., Vrielynck N., Le Guin S., Novatchkova M., Grelon M.(2012). The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8: e1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Zhao X., Kelly K.A., Venn O., Higgins J.D., Yelina N.E., Hardcastle T.J., Ziolkowski P.A., Copenhaver G.P., Franklin F.C.H., McVean G., Henderson I.R.(2013). Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45: 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich K.A., Cortez D.(2008). ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiácovo M.P., MacQueen A.J., Martinez-Perez E., McDonald K., Adamo A., La Volpe A., Villeneuve A.M.(2003). Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474. [DOI] [PubMed] [Google Scholar]
- Couteau F., Belzile F., Horlow C., Grandjean O., Vezon D., Doutriaux M.P.(1999). Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., Nabeshima K., Villeneuve A., Zetka M.(2004). A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 14: 585–592. [DOI] [PubMed] [Google Scholar]
- Crismani W., Girard C., Froger N., Pradillo M., Santos J.L., Chelysheva L., Copenhaver G.P., Horlow C., Mercier R.(2012). FANCM limits meiotic crossovers. Science 336: 1588–1590. [DOI] [PubMed] [Google Scholar]
- Culligan K.M., Britt A.B.(2008). Both ATM and ATR promote the efficient and accurate processing of programmed meiotic double-strand breaks. Plant J. 55: 629–638. [DOI] [PubMed] [Google Scholar]
- Dangel N.J., Knoll A., Puchta H.(2014). MHF1 plays Fanconi anaemia complementation group M protein (FANCM)-dependent and FANCM-independent roles in DNA repair and homologous recombination in plants. Plant J. 78: 822–833. [DOI] [PubMed] [Google Scholar]
- Doonan J.H., Kitsios G.(2009). Functional evolution of cyclin-dependent kinases. Mol. Biotechnol. 42: 14–29. [DOI] [PubMed] [Google Scholar]
- Duroc Y., et al. (2017). Concerted action of the MutLβ heterodimer and Mer3 helicase regulates the global extent of meiotic gene conversion. eLife 6: e21900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J.B., et al. (2018). FIGL1 and its novel partner FLIP form a conserved complex that regulates homologous recombination. PLoS Genet. 14: e1007317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V., Bruchet H., Camescasse D., Granier F., Bouchez D., Tissier A.(2003). AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C., Chelysheva L., Choinard S., Froger N., Macaisne N., Lehmemdi A., Mazel J., Crismani W., Mercier R.(2015). AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 11: e1005448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C., Crismani W., Froger N., Mazel J., Lemhemdi A., Horlow C., Mercier R.(2014). FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res. 42: 9087–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Escoda B., Wu P.J.(2017). Roles of CDK and DDK in genome duplication and maintenance: meiotic singularities. Genes (Basel) 8: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S., Cohen P.E.(2016). Control of meiotic crossovers: From double-strand break formation to designation. Annu. Rev. Genet. 50: 175–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E., Martín A.C., Pendle A., Colas I., Jones A.M.E., Moore G., Shaw P.(2012). The Ph1 locus suppresses Cdk2-type activity during premeiosis and meiosis in wheat. Plant Cell 24: 152–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S., Sharp R., Foote T.N., Bertin I., Wanous M., Reader S., Colas I., Moore G.(2006). Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752. [DOI] [PubMed] [Google Scholar]
- Hartung F., Suer S., Bergmann T., Puchta H.(2006). The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 34: 4438–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Puchta H.(2007). Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18836–18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Armstrong S.J., Franklin F.C.H., Jones G.H.(2004). The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 18: 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Buckling E.F., Franklin F.C.H., Jones G.H.(2008a). Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54: 152–162. [DOI] [PubMed] [Google Scholar]
- Higgins J.D., Sanchez-Moran E., Armstrong S.J., Jones G.H., Franklin F.C.H.(2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19: 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.D., Vignard J., Mercier R., Pugh A.G., Franklin F.C.H., Jones G.H.(2008b). AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 55: 28–39. [DOI] [PubMed] [Google Scholar]
- Housworth E.A., Stahl F.W.(2003). Crossover interference in humans. Am. J. Hum. Genet. 73: 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.-Y., Niu J., Sun M.-X., Zhu J., Gao J.-F., Yang J., Zhou Q., Yang Z.-N.(2013). CYCLIN-DEPENDENT KINASE G1 is associated with the spliceosome to regulate CALLOSE SYNTHASE5 splicing and pollen wall formation in Arabidopsis. Plant Cell 25: 637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurel A., Phillips D., Vrielynck N., Mézard C., Grelon M., Christophorou N.(2018). A cytological approach to studying meiotic recombination and chromosome dynamics in Arabidopsis thaliana male meiocytes in three dimensions. Plant J. 95: 385–396. [DOI] [PubMed] [Google Scholar]
- Jackson N., Sanchez-Moran E., Buckling E., Armstrong S.J., Jones G.H., Franklin F.C.H.(2006). Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J. 25: 1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J.K., Sherizen D.E., Bhagat R., Manheim E.A., McKim K.S.(2003). Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 116: 3069–3077. [DOI] [PubMed] [Google Scholar]
- Jenkins G., Hasterok R.(2007). BAC ‘landing’ on chromosomes of Brachypodium distachyon for comparative genome alignment. Nat. Protoc. 2: 88–98. [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C.N., Kleckner N.(1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Knoll A., Higgins J.D., Seeliger K., Reha S.J., Dangel N.J., Bauknecht M., Schröpfer S., Franklin F.C.H., Puchta H.(2012). The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell 24: 1448–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A., Puchta H.(2011). The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants. J. Exp. Bot. 62: 1565–1579. [DOI] [PubMed] [Google Scholar]
- Kottemann M.C., Smogorzewska A.(2013). Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493: 356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzbauer M.T., Pradillo M., Kerzendorfer C., Sims J., Ladurner R., Oliver C., Janisiw M.P., Mosiolek M., Schweizer D., Copenhaver G.P., Schlögelhofer P.(2018). Arabidopsis thaliana FANCD2 promotes meiotic crossover formation. Plant Cell 30: 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambing C., Franklin F.C.H., Wang C.R.(2017). Understanding and manipulating meiotic recombination in plants. Plant Physiol. 173: 1530–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Chen C., Markmann-Mulisch U., Timofejeva L., Schmelzer E., Ma H., Reiss B.(2004). The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. USA 101: 10596–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Qin B., Shen Y., Zhang F., Liu C., You H., Du G., Tang D., Cheng Z.(2018). HEIP1 regulates crossover formation during meiosis in rice. Proc. Natl. Acad. Sci. USA 115: 10810–10815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A., Jenczewski E.(2019). Modelling sex-specific crossover patterning in Arabidopsis. Genetics 211: 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Liu X., An L., Zhang W., Sun J., Pei H., Meng H., Fan Y., Zhang C.(2008). The Arabidopsis MutS homolog AtMSH5 is required for normal meiosis. Cell Res. 18: 589–599. [DOI] [PubMed] [Google Scholar]
- Macaisne N., Novatchkova M., Peirera L., Vezon D., Jolivet S., Froger N., Chelysheva L., Grelon M., Mercier R.(2008). SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr. Biol. 18: 1432–1437. [DOI] [PubMed] [Google Scholar]
- Martín A.C., Shaw P., Phillips D., Reader S., Moore G.(2014). Licensing MLH1 sites for crossover during meiosis. Nat. Commun. 5: 4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R., Jolivet S., Vezon D., Huppe E., Chelysheva L., Giovanni M., Nogué F., Doutriaux M.P., Horlow C., Grelon M., Mézard C.(2005). Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3, whereas the other one is not. Curr. Biol. 15: 692–701. [DOI] [PubMed] [Google Scholar]
- Mercier R., Mezard C., Jenczewski E., Macaisne N., Grelon M.(2015). The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66: 297–327. [DOI] [PubMed] [Google Scholar]
- Molinier J., Ries G., Bonhoeffer S., Hohn B.(2004). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16: 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F., Dixon J., Doe C.L., Whitby M.C.(2003). Generating crossovers by resolution of nicked Holliday junctions: A role for Mus81-Eme1 in meiosis. Mol. Cell 12: 761–774. [DOI] [PubMed] [Google Scholar]
- Palmer N., Talib S.Z.A., Kaldis P.(2019). Diverse roles for CDK-associated activity during spermatogenesis. FEBS Lett. 593: 2925–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D., Nibau C., Ramsay L., Waugh R., Jenkins G.(2010). Development of a molecular cytogenetic recombination assay for barley. Cytogenet. Genome Res. 129: 154–161. [DOI] [PubMed] [Google Scholar]
- Séguéla-Arnaud M., Choinard S., Larchevêque C., Girard C., Froger N., Crismani W., Mercier R.(2017). RMI1 and TOP3α limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res. 45: 1860–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séguéla-Arnaud M., et al. (2015). Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl. Acad. Sci. USA 112: 4713–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra H., Lambing C., Griffin C.H., Topp S.D., Nageswaran D.C., Underwood C.J., Ziolkowski P.A., Séguéla-Arnaud M., Fernandes J.B., Mercier R., Henderson I.R.(2018). Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis. Proc. Natl. Acad. Sci. USA 115: 2437–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Robberson D.L.(1995). DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82: 453–461. [DOI] [PubMed] [Google Scholar]
- Trovesi C., Manfrini N., Falcettoni M., Longhese M.P.(2013). Regulation of the DNA damage response by cyclin-dependent kinases. J. Mol. Biol. 425: 4756–4766. [DOI] [PubMed] [Google Scholar]
- Wang K., Wang M., Tang D., Shen Y., Miao C., Hu Q., Lu T., Cheng Z.(2012). The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 8: e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xiao R., Wang H., Cheng Z., Li W., Zhu G., Wang Y., Ma H.(2014). The Arabidopsis RAD51 paralogs RAD51B, RAD51D and XRCC2 play partially redundant roles in somatic DNA repair and gene regulation. New Phytol. 201: 292–304. [DOI] [PubMed] [Google Scholar]
- Whitby M.C.(2010). The FANCM family of DNA helicases/translocases. DNA Repair (Amst.) 9: 224–236. [DOI] [PubMed] [Google Scholar]
- White M.A., Wang S., Zhang L., Kleckner N.(2017). Quantitative modeling and automated analysis of meiotic recombination. Methods Mol. Biol. 1471: 305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnker E., Harashima H., Müller K., Parra-Nuñez P., de Snoo C.B., van de Belt J., Dissmeyer N., Bayer M., Pradillo M., Schnittger A.(2019). The Cdk1/Cdk2 homolog CDKA;1 controls the recombination landscape in Arabidopsis. Proc. Natl. Acad. Sci. USA 116: 12534–12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M., Wang J., Jiang L., Wang M., Wolfe S., Pawlowski W.P., Wang Y., He Y.(2018). The number of meiotic double-strand breaks influences crossover distribution in Arabidopsis. Plant Cell 30: 2628–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., et al. (2020). The Arabidopsis Cdk1/Cdk2 homolog CDKA;1 controls chromosome axis assembly during plant meiosis. EMBO J. 39: e101625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama K., Conklin P.A., Huefner N.D., Britt A.B.(2009). Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl. Acad. Sci. USA 106: 12843–12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liang Z., Hutchinson J., Kleckner N.(2014). Crossover patterning by the beam-film model: analysis and implications. PLoS Genet. 10: e1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T., Nibau C., Phillips D.W., Jenkins G., Armstrong S.J., Doonan J.H.(2014). CDKG1 protein kinase is essential for synapsis and male meiosis at high ambient temperature in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 111: 2182–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D.(2006). From early homologue recognition to synaptonemal complex formation. Chromosoma 115: 158–174. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N.(2016). A few of our favorite things: Pairing, the bouquet, crossover interference and evolution of meiosis. Semin. Cell Dev. Biol. 54: 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]