Transit-peptide motifs that specifically enhance protein import into root leucoplasts demonstrate tissue-specific regulation of plastid protein import.
Abstract
Plastids differentiate into various functional types (chloroplasts, leucoplasts, chromoplasts, etc.) that have distinct proteomes depending on the specific tissue. Most plastid proteins are encoded by the nuclear genome, synthesized as higher molecular mass preproteins with an N-terminal transit peptide, and then posttranslationally imported from the cytosol. Evidence for tissue-specific regulation of import into plastids, and subsequent modulation of plastid proteomes, has been lacking. We quantified protein import into isolated pea (Pisum sativum) leaf chloroplasts and root leucoplasts and identified two transit-peptide motifs that specifically enhance preprotein import into root leucoplasts. Using a plastid preprotein expressed in both leaves and roots of stable transgenic plants, we showed that losing one of the leucoplast motifs interfered with its function in root leucoplasts but had no effect on its function in leaf chloroplasts. We assembled a list of all Arabidopsis (Arabidopsis thaliana) plastid preproteins encoded by recently duplicated genes and show that, within a duplicated preprotein pair, the isoform bearing the leucoplast motif usually has greater root protein abundance. Our findings represent a clear demonstration of tissue-specific regulation of organelle protein import and suggest that it operates by selective evolutionary retention of transit-peptide motifs, which enhances import into specific plastid types.
INTRODUCTION
Most plastid proteins are encoded by the nuclear genome and are synthesized in the cytosol. Proper import of proteins from the cytosol is essential for plastid biogenesis and plant development. Most nucleus-encoded plastid proteins are synthesized as higher molecular mass preproteins with N-terminal transit peptides, which are necessary and sufficient to direct the import of preproteins into plastids. Plastids develop into different functional types in a tissue-specific manner. For example, they develop into chloroplasts in green leaves for photosynthesis, into chromoplasts in orange-colored fruits for carotenoid accumulation, and into leucoplasts in tubers and roots for nutrient storage (Sakamoto et al., 2008; Jarvis and López-Juez, 2013). Plastids in different tissues have distinct proteomes in order to conduct their specialized functions. Although plastid proteomes are mostly determined by tissue-specific gene expression (Kleffmann et al., 2004), accumulating evidence suggests that preprotein import from the cytosol is part of the regulatory process (Jarvis and López-Juez, 2013; Chu and Li, 2018).
Several studies have suggested that preproteins have preferences for different plastid types. Using the ratio of precursor to mature protein after import as an approximate indication of import efficiency, it was previously shown that preproteins of ribulose 1,5-bisphosphate carboxylase (prRBCS) were imported preferentially into chloroplasts isolated from pea (Pisum sativum) leaves, whereas two other preproteins were imported equally well into those chloroplasts or into leucoplasts isolated from castor bean (Ricinus communis) seeds (Wan et al., 1996). Moreover, it has also been shown that prRBCS could not be imported into pea leucoplasts at all, whereas two other nonphotosynthetic preproteins could be imported (Yan et al., 2006). Plastid selectivity was shown to be determined by the transit peptide of each preprotein, because swapping the transit peptides resulted in a switch of respective plastid preference (Wan et al., 1996; Yan et al., 2006). Furthermore, only the transit peptide of the nonphotosynthetic ferredoxin III and FtsZ preproteins, but not that of prRBCS, directs the import of GFP into wheat (Triticum aestivum) endosperm leucoplasts (Primavesi et al., 2008). However, it is not known whether plastid selectivity is a general phenomenon or is only restricted to a few preproteins. Nor is it known how transit peptides determine the selectivity.
Transit-peptide sequences and lengths are very diverse. A few sequence features, including an enrichment of Ser and Ala for Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) transit peptides, respectively, has been found (Zybailov et al., 2008). Also, the N terminus of transit peptides has a higher percentage of uncharged amino acids (Ivey et al., 2000), although conflicting results have also been presented (Rial et al., 2000). A loosely conserved FGLK motif was found in several transit peptides (Karlin-Neumann and Tobin, 1986) but not in other transit peptides (von Heijne et al., 1989). Interestingly, it has been shown that if transit peptides are first divided into multiple subgroups, conserved sequence motifs can be identified within each subgroup (Lee et al., 2008). Detailed analyses of the prRBCS transit peptide have also identified several functional motifs for general import (Lee et al., 2006) and for Toc159 dependency (Lee et al., 2009) of this transit peptide. Chloroplast-type selectivity has been studied in Bienertia sinuspersici, which performs C4 photosynthesis within single cells using different subcellular domains to two separate types of chloroplasts. The two types of chloroplasts accumulate different sets of nucleus-encoded proteins to perform their specific C4- and C3-related functions. It has been shown that information for selecting specific chloroplast types also resides in the transit peptide of individual preproteins. It was further shown that the C-terminal one-third of the transit peptide of Pi dikinase, the key enzyme needed for generating phosphoenolpyruvate in C4 chloroplasts, is important for targeting proteins to the C4 chloroplasts (Wimmer et al., 2017).
In this study, we used a quantitative system (Chu and Li, 2015) to compare chloroplast and leucoplast import and investigated whether particular transit-peptide motifs are required for plastid selectivity. We identified two motifs that specifically conferred high import efficiency into root leucoplasts. We then produced transgenic Arabidopsis plants and found that one of the motifs indeed functioned specifically in roots. Finally, taking advantage of recent genome duplications in higher plants, as well as the whole-genome transcriptome and proteome data of Arabidopsis roots and leaves, we show, for transit peptides of homologous preproteins, that the isoform possessing extra leucoplast import-enhancing motifs usually has greater protein abundance in roots. Our results demonstrate that specific transit-peptide motifs are part of the mechanism determining tissue-specific plastid proteomes.
RESULTS
prFibrillin 1B, but Not Its Homolog prPGL35, Has High Import Efficiency into Leucoplasts
We previously optimized a system of protein import into isolated pea root leucoplasts (Chu and Li, 2015). The purity of the leucoplast preparation was verified by immunoblots (Supplemental Figure 1), and we have shown that a mitochondrial preprotein could not be imported into this leucoplast preparation (Chu and Li, 2015). Our optimization increased import efficiency and allows quantitative comparisons of protein import between leaf chloroplasts and root leucoplasts. That study also identified several preproteins with very high leucoplast import efficiencies (Chu and Li, 2015). Theoretically, identification of motifs necessary for efficient leucoplast import can be achieved by mutagenesis of their transit peptides or by swapping or fusing fragments between two transit peptides that have high and low import efficiencies. However, transit peptides exhibit extreme sequence and length diversity (von Heijne et al., 1989; von Heijne and Nishikawa, 1991; Zybailov et al., 2008), and the order of functional domains in different transit peptides may vary (Li and Teng, 2013). When domain boundaries are not known, Ala scanning or deletion may disrupt domain integrity. Moreover, swapping or fusion of two unrelated transit peptides may abolish import or produce results that are hard to interpret due to differing motif arrangements among transit peptides (Li and Teng, 2013). One way to bypass these problems is to find two homologous preproteins with similar sequences but very different leucoplast import efficiencies. Given their high sequence similarity, these two transit peptides most likely have the same domain order and structural framework, and sequence discrepancies between them would most likely represent the motifs responsible for divergent leucoplast import efficiencies.
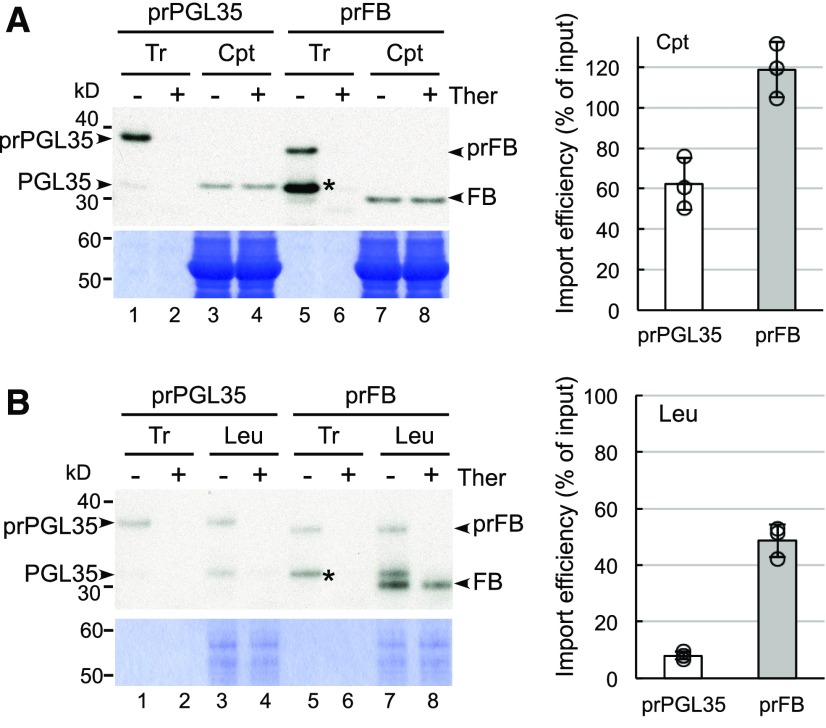
We screened our lab collection of preproteins for those having closely related homologs and compared the leucoplast import of pairs of homologs. We found one such candidate pair of preproteins: plastoglobulin 35 (AT4G04020; prPGL35) and Fibrillin 1B (AT4G22240; prFB). The amino acid sequences of these two preproteins are 80% identical, and even their transit-peptide sequences exhibit a 52% identity (Supplemental Figure 2A). When imported into isolated pea chloroplasts and leucoplasts, [35S]Met-prPGL35 and [35S]Met-prFB were processed into ∼32- and 30-kD mature proteins, respectively (Figure 1). Thermolysin treatment of chloroplasts and leucoplasts after import confirmed that these mature proteins localized within plastids (Figures 1A and 1B, lanes 4 and 8). We quantified the amounts of imported PGL35 and FB after thermolysin treatments. Both prPGL35 and prFB exhibited high import efficiency into chloroplasts, with more than 50% of the supplied preproteins localizing in chloroplasts (Figure 1A; import efficiency is represented by the percentage of supplied preproteins that were imported). However, whereas prFB exhibited 50% import efficiency into leucoplasts, import of prPGL35 into leucoplasts was very limited (Figure 1B). Therefore, it is likely that some transit-peptide sequence motifs that are present in prFB, but not in prPGL35, enable higher import efficiency into leucoplasts.
Figure 1.
Both prPGL35 and prFB Are Efficiently Imported into Chloroplasts but Only prFB Is Efficiently Imported into Leucoplasts.
(A) Import into chloroplasts (Cpt) isolated from pea leaves.
(B) Import into leucoplasts (Leu) isolated from pea roots.
Isolated plastids were incubated with in vitro-translated [35S]Met-prPGL35 or [35S]Met-prFB under import conditions for 10 min. After import, half of the plastids were further treated with thermolysin (Ther). Intact plastids were reisolated and analyzed by SDS-PAGE, Coomassie blue staining, and fluorography. An equal amount of protein was loaded in all lanes containing plastids on the same gel. Fluorographs and Coomassie blue staining of the same gels are shown. Imported mature proteins in the thermolysin-treated samples were quantified, and the import efficiencies were calculated and are shown on the right. Import efficiency was defined as the percentage of [35S]Met-labeled preproteins that was found as mature proteins in plastids. The values have been corrected for the difference in Met numbers between precursor and mature forms. Data shown are means ± sd of three independent experiments for chloroplasts (A) and four independent experiments for leucoplasts (B). Tr, 50% input for chloroplasts (A) and 10% input for leucoplasts (B). Asterisks indicate the internal initiation product of prFB from the Met at position 42.
Synthesis of prFB by in vitro translation produced a smaller protein (Figures 1A and 1B; Supplemental Figures 2B and 2C, asterisks) that resulted from translation initiation from the Met at residue 42 (Supplemental Figure 2A, arrowhead). Although this smaller protein was not imported into leucoplasts (Figure 1B, compare lanes 7 and 8), it was very similar in size to mature FB and could only be removed by thermolysin treatment of intact leucoplasts. To avoid the thermolysin treatment step when assessing a large number of mutant constructs in our subsequent experiments, we mutated the Met at residue 42 to Val, creating the prFB-M42V mutant. Production of the smaller protein was eliminated in prFB-M42V, and the mutant had the same chloroplast and leucoplast import efficiencies as wild-type prFB (Supplemental Figures 2B and 2C). Subsequent experiments were performed using the prFB-M42V variant.
Two Motifs of the prFB Transit Peptide Are Sufficient to Increase the Leucoplast Import Efficiency of prPGL35
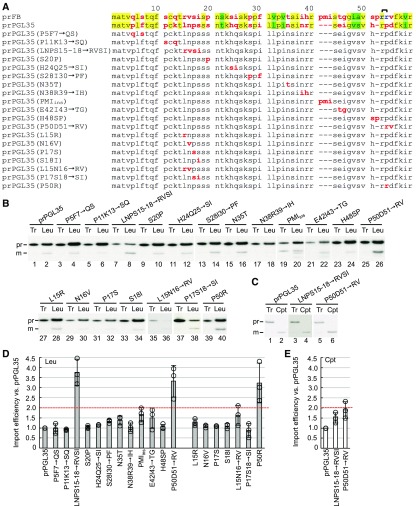
To test if the prFB transit peptide harbors motifs that can specifically confer high leucoplast import efficiency, we generated a series of mutants in which we replaced regions of the prPGL35 transit peptide that differ from prFB with the corresponding prFB sequence (Figure 2A). Import of all prPGL35 transit-peptide mutants was tested in isolated leucoplasts (Figure 2B). The results showed that two regions of the prFB transit peptide—Arg15-Val16-Ser17-Ile18 and Arg54-Val55—increased more than twofold the import of prPGL35 into leucoplasts (the LNPS15-18→RVSI and P50D51→RV mutants in Figure 2B, lanes 8 and 26, and in Figure 2D). For LNPS15-18→RVSI, we performed single- and double-residue replacements to identify the critical residues, but none of the single- or double-residue replacements had the same effect as replacing all four residues (Figure 2B, lanes 27 to 38, and Figure 2D).
Figure 2.
The LNPS15-18→RVSI, P50D51→RV, and P50R Mutations Are Sufficient to Increase the Import of prPGL35 into Leucoplasts More Than Twofold.
(A) Transit-peptide sequences of prFB, prPGL35, and prPGL35 mutants. The twin-positive motif is marked with a bracket at positions 53 and 54. Transit peptide length was predicted according to Köhler et al. (2015).
(B) Import of [35S]Met-prPGL35 and its transit-peptide mutants into leucoplasts (Leu). Isolated leucoplasts were incubated with in vitro-translated [35S]Met-prPGL35 or its mutants under import conditions for 10 min. Tr, 10% input.
(C) Import of [35S]Met-prPGL35 and two transit-peptide mutants into chloroplasts (Cpt). Isolated chloroplasts were incubated with in vitro-translated [35S]Met-prPGL35 or its mutants under import conditions for 10 min. Tr, 50% input.
In (B) and (C), pr and m indicate precursor and mature forms, respectively.
(D) and (E) Import efficiencies of preproteins shown in (B) and (C), respectively. Values were calculated and normalized to those of wild-type prPGL35. Data shown are means ± sd of three independent experiments.
For P50D51→RV, we noticed that the residue immediately preceding the replacement is Arg, so the replacement restored a motif with two consecutive Arg residues that is present in prFB but absent from prPGL35 (Figure 2A, bracket). A motif constituting two consecutive positive charges was previously found in all preproteins that are preferentially imported into old chloroplasts (Teng et al., 2012). Therefore, we tested if we could increase prPGL35 leucoplast import efficiency simply by restoring this RR motif. Indeed, when Pro-50 of prPGL35 was replaced with Arg (the P50R mutant), leucoplast import efficiency was increased to the same level as that of the P50D51→RV mutant (Figure 2B, lanes 26 and 40, and Figure 2D). We also tested the import efficiencies into chloroplasts of the LNPS15-18→RVSI and P50D51→RV mutants. Although the mutants also presented enhanced import efficiencies into chloroplasts, they were not as pronounced as the enhancement for leucoplasts (Figures 2C and 2E). Import time-course experiments were further performed with the wild-type prPGL35 and the LNPS15-18→RVSI and P50D51→RV mutants (Supplemental Figure 3). For leucoplasts, the two mutants imported with greater rates and exhibited threefold to fourfold increases in import efficiency compared with the wild type at the end of a 30-min import (Supplemental Figure 3B). For chloroplasts, the two mutants also had increased import rates, but all three preproteins had similar import efficiency at the end of import (Supplemental Figure 3D). These results show that the RVSI and twin-positive motifs are sufficient to enhance the import of prPGL35 into leucoplasts.
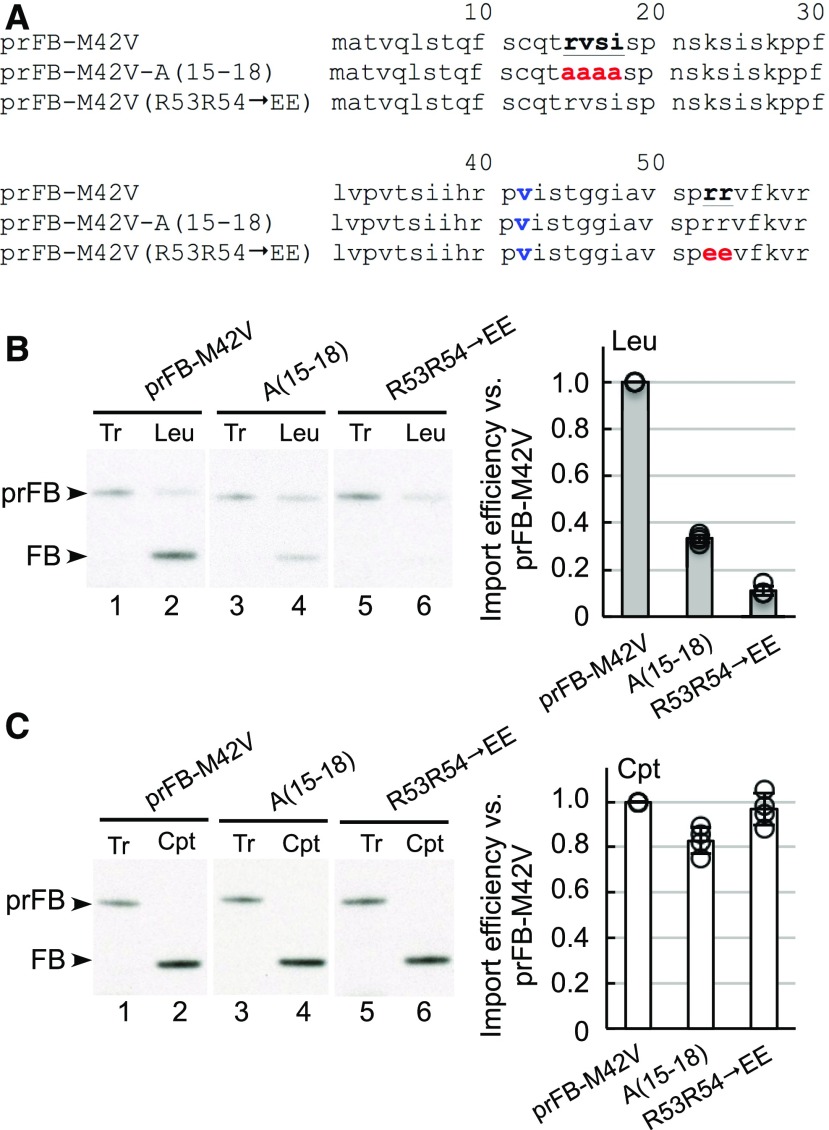
Next, we investigated if the RVSI and twin-positive motifs are necessary for efficient leucoplast import of prFB. As explained above, we used the prFB-M42V variant for mutagenesis. When we mutated the RVSI motif to four Ala residues (Figure 3A), the resulting A(15-18) mutant exhibited severely reduced import efficiency into leucoplasts but its import efficiency into chloroplasts was comparable to that of wild-type prFB-M42V (Figures 3B and 3C). Similarly, when we mutated the twin-positive motif to two Glu residues (the R53R54→EE mutant), import into leucoplasts was also severely reduced but import into chloroplasts was not affected (Figures 3B and 3C). These data show that both motifs are necessary for efficient prFB import specifically into leucoplasts.
Figure 3.
The Twin-Positive and RVSI Motifs Are Specifically Important for prFB Import into Leucoplasts.
(A) The transit-peptide sequences of prFB-M42V and its mutants. The RVSI and twin-positive motifs are denoted in boldface letters and underlined, and the Ala and Glu replacing them in the mutants are colored red. The Val replacing the Met at residue 42 is colored blue.
(B) and (C) Import of prFB-M42V and its mutants into leucoplasts and chloroplasts. [35S]Met preproteins were incubated with isolated leucoplasts (Leu) and chloroplasts (Cpt), respectively, under import conditions for 10 min. Intact plastids were reisolated and analyzed by SDS-PAGE. Import efficiencies were calculated and normalized to those of prFB-M42V. Tr, 20 and 50% input for leucoplasts (B) and chloroplasts (C), respectively. Data shown are means ± sd of four independent experiments.
The Twin-Positive Motif Is Also Necessary for Efficient Import of prTic40 and prcpHsc70-1 into Leucoplasts
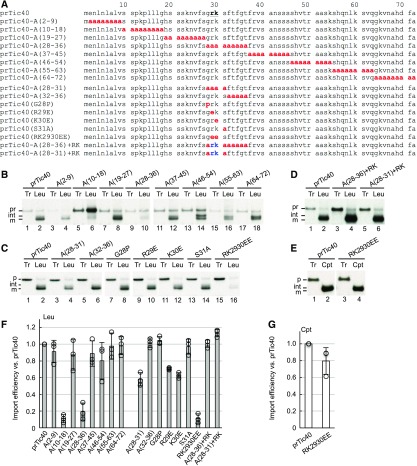
Previously, we showed that the preprotein prTic40 exhibits very high import efficiency into leucoplasts (Chu and Li, 2015). Although prTic40 does not have a homolog, a previous study generated Ala scanning mutants of the prTic40 transit peptide (Figure 4A; Teng et al., 2012). Therefore, we analyzed the import of these prTic40 transit-peptide mutants into leucoplasts to identify motifs necessary for efficient leucoplast import of prTic40. Two mutants, A(10-18) and A(28-36), exhibited severely reduced import into leucoplasts (Figure 4B, lanes 6 and 10, and Figure 4F). Previously, we had shown that the A(10-18) mutant was also not imported into chloroplasts (Teng et al., 2012). Therefore, the A(10-18) mutation disrupts a motif that is essential for general import into both chloroplasts and leucoplasts. Accordingly, we focused on the prTic40-A(28-36) mutant and two additional mutants in which we substituted either the N- or C-terminal halves of residues 28 to 36 with Ala: A(28-31) and A(32-36). We found that the A(28-31) mutant, but not the A(32-36) mutant, had a reduced leucoplast import efficiency relative to wild-type prTic40 (Figure 4C, lanes 4 and 6, and Figure 4F).
Figure 4.
The Twin-Positive Motif Is Necessary for Efficient Import of prTic40 into Leucoplasts.
(A) Transit-peptide sequences of prTic40 and its mutants. The twin-positive motif at positions 29 and 30 is shown in boldface letters and underlined. Mutated residues are colored red. The twin-positive motif added back to the mutants are colored blue.
(B) to (E) Import of [35S]Met-prTic40 and its mutants into leucoplasts ([B] to [D]) and chloroplasts (E). [35S]Met preproteins were incubated with isolated leucoplasts (Leu) or chloroplasts (Cpt) under import conditions for 25 min. Intact plastids were reisolated and analyzed by SDS-PAGE. Tr, 30% input. pr, int, and m indicate the precursor, intermediate, and mature forms of prTic40, respectively.
(F) and (G) Import efficiencies of each mutant shown in (B) to (E) were calculated and normalized to those of wild-type prTic40. Data shown are means ± sd of three independent experiments.
We then individually substituted residues 28 to 31 with an amino acid that has very different characteristics from the original amino acid. Substitution of Gly-28 to Pro and Ser-31 to Ala did not affect the import of prTic40 into leucoplasts (Figure 4C, lanes 8 and 14). However, substitution of positively charged Arg-29 or Lys-30 with Glu reduced the import of prTic40 into leucoplasts, and when we substituted both residues with Glu the import efficiency of the prTic40(RK2930EE) mutant decreased by almost 80% relative to wild-type prTic40 (Figure 4C, lanes 10, 12, and 16, and Figure 4F). Moreover, when the two positively charged residues were added back into the A(28-36) and A(28-31) mutants, their leucoplast import efficiency was fully restored (Figure 4D, lanes 4 and 6, and Figure 4F). These data indicate that the twin-positive motif is critical for high-efficiency import of prTic40 into leucoplasts. Import efficiency of prTic40(RK2930EE) into chloroplasts was reduced by ∼20% relative to wild-type prTic40 (i.e., not nearly as severe as seen for leucoplasts; Figures 4E and 4G). Thus, similar to our findings for prFB, the twin-positive motif is much more important for leucoplast import of prTic40 than it is for chloroplast import. Our previous results have shown that this twin-positive motif in prTic40 is also important for prTic40 import into old chloroplasts (Teng et al., 2012).
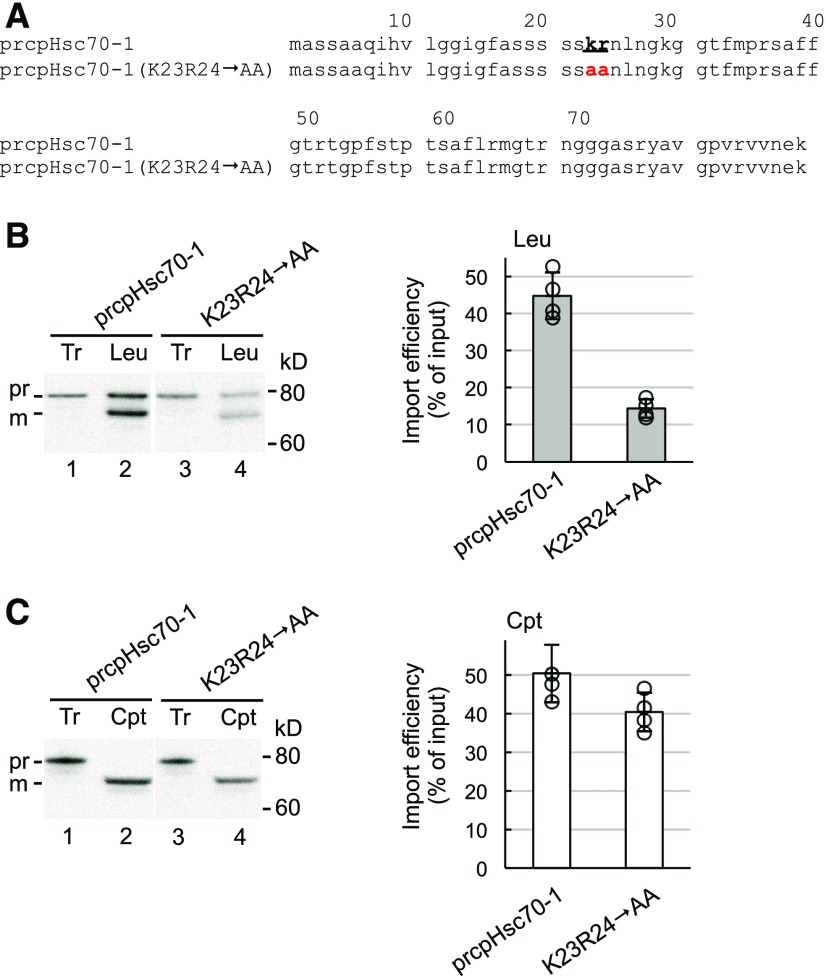
We further verified the importance of the twin-positive motif by examining another preprotein. We screened our lab collection of preproteins and found that Arabidopsis preprotein prcpHsc70-1 also imported very well into leucoplasts, with a leucoplast import efficiency of 45% that is comparable to that of prFB (Figure 5B). It also possesses a single twin-positive motif in its transit peptide (Lys23-Arg24; Figure 5A). Therefore, we mutated that motif to two Ala residues, creating the K23R24→AA mutant. Compared with wild-type prcpHsc70-1, import of the K23R24→AA mutant into leucoplasts was severely reduced, whereas its import into chloroplasts was only slightly affected (Figures 5B and 5C, lane 4), echoing the results observed for prFB and prTic40. We further performed leucoplast binding experiments in the absence of ATP to investigate if the twin-positive motif is important for preprotein binding to leucoplasts using the K23R24→AA mutant of cpHsc70 and the RK2930EE mutant of prTic40 (Supplemental Figure 4). For both mutants, their binding to leucoplasts was significantly reduced compared with their corresponding wild type, suggesting that the twin-positive motif is important for preprotein binding to leucoplasts.
Figure 5.
The Twin-Positive Motif Is Necessary for Efficient Import of prcpHsc70-1 into Leucoplasts.
(A) Transit-peptide sequences of prcpHsc70-1 and the K23R24→AA mutant. The twin-positive motif at positions 23 and 24 is denoted in boldface letters and underlined. The Ala residues replacing the twin-positive motif are colored red.
(B) and (C) Import of [35S]Met-prcpHsc70-1 and the K23R24→AA mutant into leucoplasts (B) and chloroplasts (C), respectively. [35S]Met preproteins were incubated with isolated leucoplasts (Leu) or chloroplasts (Cpt) under import conditions for 10 min. Intact plastids were reisolated and analyzed by SDS-PAGE. The import efficiency of each preprotein was calculated. Data shown are means ± sd of four independent experiments. Tr, 12.5 and 25% input for leucoplast (B) or chloroplast (C) experiments, respectively. pr and m indicate precursor and mature forms, respectively.
The Twin-Positive Motif Is Specifically Important for the Functions of cpHsc70-1 in Roots
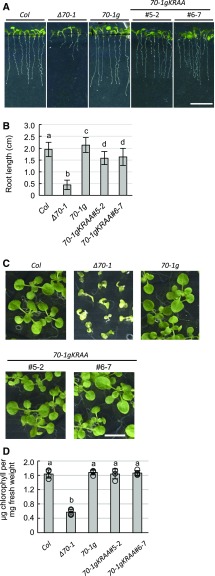
Arabidopsis cpHsc70-1 is one of a family of plastid Hsp70 proteins that are important for various aspects of plastid biogenesis, including serving as a motor for driving protein import into plastids (Schroda et al., 1999; Liu et al., 2007, 2014; Shi and Theg, 2010; Su and Li, 2010). A knockout mutant of cpHsc70-1, Δcphsc70-1, exhibits variegated cotyledons, malformed leaves, reduced chlorophylls, small size, and retarded root growth, particularly after heat shock treatments of germinating seeds (Supplemental Figure 5D; Su and Li, 2008). These phenotypes indicate that cpHsc70-1 is important for plastid functions in both leaves and roots. We found that the K23R24→AA transit-peptide mutation impaired the import of prcpHsc70-1 into isolated leucoplasts but not chloroplasts (Figure 5). To test the effect of that mutation in planta, we introduced the K23R24→AA mutation into a cpHsc70-1 genomic fragment, which was shown to complement the Δcphsc70-1 mutant (Su and Li, 2008), and transformed the mutated genomic fragment into Δcphsc70-1 plants. We obtained several independent transgenic plants and selected plants homozygous for a single insertion of the mutant transgene. Hereafter, the transformants are designated as 70-1gKRAA, and Δcphsc70-1 complemented with the wild-type cpHsc70-1 genomic fragment (Su and Li, 2008) is designated as 70-1g. We selected two representative 70-1gKRAA lines— nos. 5-2 and 6-7 (Supplemental Figure 5)—for further characterizations.
To analyze the effect of the K23R24→AA mutation, we heat-shocked seeds of various genotypes, germinated them, and then measured root length and leaf chlorophyll content. As shown in Figures 6A and 6B, roots of 70-1gKRAA plants were longer than those of Δcphsc70-1 mutant plants but were significantly shorter than roots of wild-type Columbia (Col) and 70-1g plants. These data indicate that the K23R24→AA transit-peptide mutation resulted in incomplete complementation of the Δcphsc70-1 defect in roots, possibly due to defective import of the K23R24→AA mutant into root leucoplasts. By comparison, whereas leaves of Δcphsc70-1 plants exhibited extremely retarded growth and reduced chlorophylls, leaves of the 70-1gKRAA plants were indistinguishable from those of 70-1g and Col plants and had similar amounts of chlorophyll. Thus, the K23R24→AA transit-peptide mutation did not interfere with the functions of cpHsc70-1 in leaves and the twin-positive motif is specifically important for root leucoplast import, both in isolated plastids and in planta.
Figure 6.
The Twin-Positive Motif Is Specifically Important for cpHsc70-1 Functions in Roots.
(A) and (C) Seeds of Col, Δcphsc70-1 (Δ70-1), 70-1g, and two independent lines of 70-1gKRAA (#5-2 and #6-7) were heat-shock-treated and grown for 7 d (A) or 14 d (C) on MS-agar medium. Bars = 1 cm.
(B) Quantification of the root lengths of plants shown in (A). Data shown are means ± sd (n ≥ 86 plants). Values with the same letter do not differ significantly according to one-way ANOVA (Supplemental Data Set 5) with Tukey’s multiple comparison test (P < 0.0001).
(D) Quantification of the chlorophyll contents of plants shown in (C). Data shown are means ± sd (n = 4 independent experiments). Values with the same letter do not differ significantly according to one-way ANOVA (Supplemental Data Set 5) with Tukey’s multiple comparison test (P < 0.0001).
Presence of the Twin-Positive Motif Is Associated with Higher Expression in Roots
We have shown that the twin-positive motif is necessary for efficient import of three preproteins into root leucoplasts. Next, we investigated if there is a general association between the presence of that motif in a transit peptide and greater protein abundance in roots. Such an association would further support that the twin-positive motif facilitates protein transport into root leucoplasts. However, the twin-positive motif is unlikely to be sufficient to mediate leucoplast import on its own, and its efficacy most likely depends on the context of the transit peptide in which it resides. It would be futile to compare many unrelated sequences and then try to correlate the presence of twin-positive motifs with levels of root expression. However, for homologous proteins with similar overall sequences and domain structures, if only one isoform has a twin-positive motif, then that isoform would be expected to have greater protein abundance in roots if the twin-positive motif truly facilitates protein import into root leucoplasts.
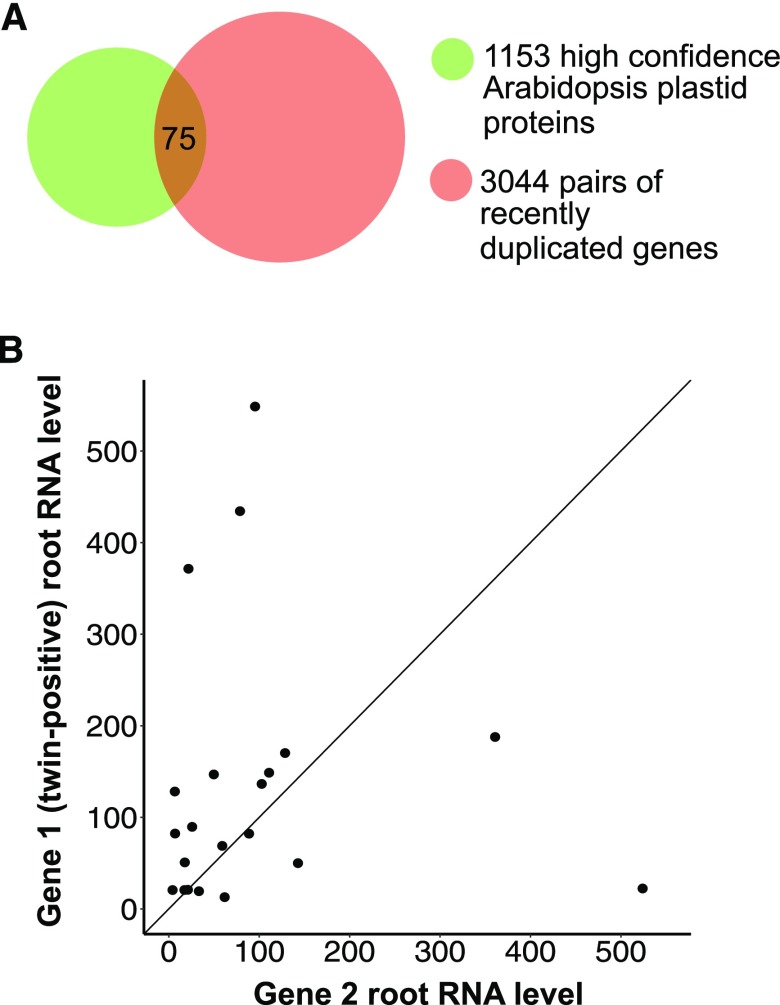
Accordingly, we systematically searched for homologous preproteins. In the plant lineage leading to Arabidopsis, the two most recent gene duplication events generated about 3000 pairs of duplicated genes in the Arabidopsis genome (Blanc et al., 2003). Some of these genes were duplicated as recently as 20 million years ago, so sequence similarity between duplicated genes is very high. We cross-referenced this list of 3044 pairs of duplicated genes (Supplemental Data Set 1; Blanc et al., 2003) with a list of 1153 high-confidence Arabidopsis plastid proteins (Bischof et al., 2011) and identified 75 pairs of plastid proteins encoded by recently duplicated genes (Figure 7A; Supplemental Data Set 2). We used the first 60 amino acids of their sequences to represent the transit peptide region of each preprotein because it has been estimated that the average length of the transit peptides of Arabidopsis chloroplast preproteins is 53.6 amino acids (Zybailov et al., 2008). Root protein levels for these 75 pairs of proteins were then downloaded from AtProteome (Supplemental Data Set 2; Baerenfaller et al., 2008). If the twin-positive motif is truly important for increasing import into root leucoplasts, then we anticipated that after the gene duplication event and over the course of subsequent evolution, the motif should be preferentially retained in the isoform with greater root protein abundance.
Figure 7.
Isoforms Possessing Extra Twin-Positive Motifs Have Significantly Higher RNA Expression in Roots.
(A) Venn diagram of the group of high-confidence Arabidopsis plastid proteins (1153 proteins) and the group of proteins encoded by recently duplicated gene pairs (3044 pairs), resulting in identification of 75 pairs of plastid preproteins encoded by recently duplicated genes.
(B) Among homologous gene pairs, the isoform with extra twin-positive motifs has significantly higher RNA expression in roots. RNA expression data were downloaded from the Arabidopsis eFP website. A Wilcoxon signed-rank test was performed (V = 156 and P < 0.05; V corresponds to the sum of ranks assigned to the differences with positive sign.).
Of the 75 preprotein pairs we identified, the twin-positive motif did not differ between the two isoforms in some pairs, some pairs comprised isoforms each with unique twin-positive motifs, and, in 22 pairs, only one of the isoforms possessed extra twin-positive motifs not found in its paralog (Table 1). Of these latter 22 pairs, root protein data were available for 19 pairs. For 14 of these 19 pairs, the isoform harboring extra twin-positive motifs exhibited higher protein levels in roots than its paralog (Table 1), supporting our hypothesis that the twin-positive motif is important for protein import into root leucoplasts. For two of the remaining five pairs that did not fit our hypothesis, the isoform possessing extra twin-positive motifs had a higher root RNA level but a lower protein level than its paralog. We directly tested the import of these two preprotein pairs into isolated leucoplasts. In both cases, the isoform with extra twin-positive motifs presented higher leucoplast import efficiency than its respective paralog (Supplemental Figure 6). Thus, for these two preprotein pairs, the isoform with extra twin-positive motifs was still imported more efficiently into root leucoplasts than its paralog, but the final steady state protein level may be under additional postimport control. Hence, for most preprotein pairs, the presence of twin-positive motifs is associated with greater protein abundance in roots or with more efficient root leucoplast import. Although our sample size is not large because the transit-peptide sequences of most of the 75 recently duplicated preprotein pairs are still very similar, we believe that the ratio of 16 pairs out of 19 represents strong evidence supporting the importance of the twin-positive motif in root leucoplast protein abundance.
Table 1. List of Recently Duplicated Arabidopsis Gene Pairs Encoding Plastid Preproteins in Which Only One of the Isoforms (the One at Top) Possesses Extra Twin-Positive Motifs.
| No. | Gene | Transit Peptide Sequence (First 60 Amino Acids of the N Terminus) | Root Protein (Log2)a | Association | Root RNA | Leaf RNA |
|---|---|---|---|---|---|---|
| 1 | AT2G30950.1 | MAASSACLVGNGLSVNTTTKQRLSKHFSGRQTSFSSVIRTSKVNVVKASLDGKKKQEGRR———– | −2.14 | Yes | 82.30 | 1632.73 |
| AT1G06430.1 | MAASSACLLGNGLSVYTT-KQRFQK–LG——-LDRTSKVTVVKASLD-EKKHEGRRGFFKLLLGNAA | – | 6.95 | 150.96 | ||
| 2 | AT1G17050.1 | MMMSCRNIDLGTSVLDHSCSSSSTSRRFLFGNSSKTVCMIGGRS-CVGNLVFLRRDLATCR—– | −1.41 | Yes | 50.76 | 151.48 |
| AT1G78510.1 | MMTSCRNIDLGTMMMACGCG—–RR-QFPSLAKTVCKFTSSNRSYGGLVGSCKAVPTKSKEISL | −3.32 | 17.70 | 53.73 | ||
| 3 | AT1G55490.1 | MASTFTATSSIGSMVAPNGHKSDKKLISKLSSSSFGRRQSVCPRPRRSSSAIVCAAKELH—- | −2.76 | Yes | 187.73 | 2437.78 |
| AT3G13470.1 | MASTFTATSSLGSLLAPNAIKLSS–ATSISSSSFGRRHNVCV–RRSRPAIVCAAKELHFNKD | −3.72 | 360.90 | 1454.63 | ||
| 4 | AT2G47400.1 | MTTIAAAGLNVATPRVVVRPVAR—VLGPVRLNYPWKFGS—–MKRMVVVKATSEGEISEKVEKS | −1.37 | Yes | 128.19 | 2174.71 |
| AT3G62410.1 | MATIAT-GLNIATQRVFVTSENRPVCLAGPVHLNNSWNLGSRTTNRMMKLQPIKAAPEGGI——- | – | 6.68 | 496.4 | ||
| 5 | AT3G22890.1 | MASMAAVLSK-TPFLSQPLTKSSPNSDLPFAAVSFPSKSLRR–RVGSIRAGLIAPDGGKLVE | 0.33 | Yes | 434.31 | 957.01 |
| AT4G14680.1 | MASMSTVFPKPTSFISQPLTKS-HKSDSVTTSISFPSNSKTRSLRTISVRAGLIEPDGGKL– | – | 78.73 | 216.16 | ||
| 6 | AT5G24650.1 | –MGKDGEGDKKR—ETMAVMSLMKDQQNPIQQFQVKFKEIETGFKSWLSKQKLPVEAAVVTAM | 0.74 | Yes | 170.20 | 364.01 |
| AT3G49560.1 | MVVGGGGEGDQKRSSGEMMAMASLFNDQQNPIQQFQVKFKEVETNFKTWLSKQSIPVEAA—– | −0.37 | 128.60 | 453.28 | ||
| 7 | AT3G05020.1 | MATQFSASVSLQTSCLATT-RISFQKPALISNHGKTN-LSFNLRRSIPSRRLSVSCAAKQET | 0.98 | Yes | 371.42 | 129.6 |
| AT5G27200.1 | MATSFCSSISMQAPFSATTTRFCLNKQATIFNNEKTNNLSFSLRRLMPAR-LAVSCAVKQE- | – | 21.75 | 8.18 | ||
| 8 | AT3G11630.1 | –MASVASS–TTLISSPSSRVFPAKSSLSSPSVSFLRTLSSPSASAS-LRSGFARRSSLSSTSRR | 1.47 | Yes | 548.53 | 2728.13 |
| AT5G06290.1 | MSMASIASSSSTTLLSS-SRVLLPSKSSLLSPTVSFPRIIPSSSASSSSLCSGFSSLGSLTT—- | −1.61 | 95.41 | 1446.05 | ||
| 9 | AT3G02730.1 | MPLSLRLSPSPTAL–SPTT—-GGFGPSRKQCRIPYSGVPTTKIGFCS—–LDSRKRGDSSVVRCSL- | −2.17 | Yes | 20.68 | 925.86 |
| AT5G16400.1 | MPLSLRLAPSPTSFRYSPITSTGAGGFSPVKQHCRIPNSGVAT-KIGFCSGGGGVLDSGRR | −2.27 | 17.33 | 485.15 | ||
| 10 | AT2G45440.1 | MAALKGYGLCSMDSALQFPCPKLFNSYKRRSSKWVSPKAAVVPNFHLPMRSLEVKNRTNT- | −1.83 | Yes | 148.71 | 254.88 |
| AT3G60880.1 | MSALKNYGLISIDSALHFPRSNQLQSYKR-NAKWVSPIAAVVPNFHLPMRSLEDKNRTNTD | – | 110.91 | 101.45 | ||
| 11 | AT3G29200.1 | MEASLLMRSSCCSSAIGGFFDHRRELSTSTPISTLLPLPSTKSSFSVRCSLPQPSKPRSG- | −0.83 | Yes | 82.15 | 144.48 |
| AT1G69370.1 | MEAKLLKPAFYNSPNLN-LTNSSRLISRLSIWNDKSKVGLSSGSLFLRLSAASPIRYSRGL | −2.69 | 88.73 | 162.94 | ||
| 12 | AT3G47060.1 | MTTTFEFLQPRIHG—FATCCSSNSLLYSKASRFFNDRCRVYRQNPNRFVSN-SITLPLQKKQ | −4.01 | Yes | 49.98 | 74.64 |
| AT5G58870.1 | MTS-IELLSPLIHDKFRFSTCCSTSSLLYLHASSFFRDRSFGFRQNPNRFVSNSSIQLPQS— | – | 142.86 | 456.78 | ||
| 13 | AT1G31230.1 | MPVVSLAKVVTSPAVAG-DLAVRVPFIYGKRLVSNRVSFGKLRRRSCIG-QCVRSELQSPRV | 0.49 | Yes | 136.48 | 196.58 |
| AT4G19710.2 | MATLKPSFTVSPPNSNPIRFGSFPPQCFLRVPKPRRLILPRFRKTTGGGGGLIRCEL–PDF | −1.34 | 102.78 | 215.33 | ||
| 14 | AT1G15500.1 | MEGLIQTRGILSLPAKPIGVRRLLQPSHGLKQRLFTT—NLPALSLSSNGHKKFQAFQQIPL | −0.12 | Yes | 159.78b | 354.58b |
| AT1G80300.1 | MEAVIQTRGLLSLPTKPIGVRSQLQPSHGLKQRLFAAKPRNLHGLSLSFNGHKKFQTFEP— | −0.19 | 159.78b | 354.58b | ||
| 15 | AT1G14410.1 | MSQLLSTPLMAVN———SNPRFLSSSSVLVTGGFAVKRHGFALKPTTKTVKLFSVKSRQTDYFE | −1.03 | Noc | 89.60 | 261.13 |
| AT2G02740.1 | MSQLLSSPPMAVFSKTFINHKFSDARFLSSHSILTSGGFAGK–IIPLKPTAR-LKL-TVKSRQ—– | −0.83 | 25.81 | 103.56 | ||
| 16 | AT2G37400.1 | MESLGKLQLHQQPNHLSFTHFSSS-FPKKPSSFSLRSLPRSTSSFKCISIKASSSKSQDSK— | – | Noc | 68.85 | 95.36 |
| AT3G53560.1 | MESLGKLQLHHQPFHLSFTHTSSSTFPKN—-LFKSSIQPISSLKSASIKASSSKFQNSITPL | −3.01 | 59.13 | 479.76 | ||
| 17 | AT1G77060.1 | MSMLMAVKTTSLCCSSLNLTASPTFRRNPRAARLVNPTARIQTRFHRLIEEQGIVLMPGC– | – | No | 22.30 | 210.06 |
| AT1G21440.1 | MSMLMAAKSTSLFSS–NPTISAKIGQNPRGVRSVYPTVRMQSRVHRLIEEQGAVLIPGVYD | 1.24 | 524.05 | 326.13 | ||
| 18 | AT3G25740.1 | —–MLQKISQS–ISLCN—GDQFKPLIYLAGAPTNFISSPLSGKKKSSSLRIKRIQQLQSTLEDRI | – | No | 19.34 | 26.80 |
| AT1G13270.1 | MASSVFLSSFSSSSSLQLCSSFHGEYLAPSRCFLGAPVTSSSLSLSGKKNSYSPRQFHVS———- | −3.01 | 33.33 | 478.56 | ||
| 19 | AT3G23790.1 | M-ASTSLGASILVSHCSSAPEFQVSGMRLVFGYKAFGCRTSRRGFR-VRCESKIQEKELRRCS | – | No | 20.90 | 28.86 |
| AT4G14070.1 | MQIRLKPDYSFFIASSSTMASTSSLGPSTLLSYGSPSRQFPDFGFRLISGHESVRIPSFRR– | −3.98 | 21.10 | 115.58 | ||
| 20 | AT5G47110.1 | MSISMALFSPPISSS-LQNPNLIPKISTSLLSTKRFSLISVPRASSDNGTTSP—-VVEIPKPA | – | 20.63 | 840.26 | |
| AT4G17600.1 | —-MALFSPPISSSSLQNPNFIPKFSFSLLSSNRFSLLSVTRASSDSGSTSPTAAVSVEAPEP- | – | 4.16 | 1218.96 | ||
| 21 | AT4G39030.1 | MLIKSQRLTLFSPLLSKTRRIPVNSHQTLVA-ESVITRRTLGAITATPSFHKNPVVIRRRI | – | 146.83 | 52.11 | |
| AT2G21340.1 | MQIQCKTLTFTVSSIPCNPKLPFPSSLTLRSWNPSFPSFRSSAVSGPKSSLKLNRFLRNC- | – | 49.80 | 224.11 | ||
| 22 | AT1G18170.1 | MANLFTATAP-FLSLSKP–FTKTASYHQCYASS-SNPPEPESSSPPPPPPPPQPLASQQKRKK | – | 12.83 | 98.83 | |
| AT1G73655.1 | MATLFTATVPSHHRFVSPSQHPKQSLLSQSLSVTFTENPQPTAVVTLQEQQLTDWITSPV—- | – | 61.78 | 542.31 |
Twin-positive motifs are denoted with boldface underlined letters. The root and leaf RNA (fluorescence intensity, arbitrary units) and the root protein data were downloaded from the Arabidopsis eFP Browser (Winter et al., 2007) and AtProteome (Baerenfaller et al., 2008) databases, respectively. “Yes” in the Association column indicates that the isoform with extra twin-positive motifs also has higher root protein abundance. Blank cells in the Association column indicate no data. The sequence alignment was produced using ClustalW with default parameters and refined manually.
Dashes in this column indicate not detected.
The same probe set was used for the two genes.
The isoform with extra twin-positive motifs also has higher leucoplast import, as shown in Supplemental Figure 6.
We also analyzed the correlation between the possession of extra twin-positive motifs and higher RNA levels in roots for the 22 pairs of paralogs listed in Table 1. Root RNA expression data were downloaded from the Arabidopsis eFP browser (Winter et al., 2007). For one of these pairs, the same probe set was used for the two genes (Table 1), so their expression could not be distinguished. A Wilcoxon signed-rank test was performed on the root RNA levels of the remaining 21 pairs, which showed that isoforms with extra twin-positive motifs (denoted as Gene 1 in Figure 7B) exhibited significantly higher root RNA levels than their paralogs (Figure 7B; V = 156 and P < 0.05; V corresponds to the sum of ranks assigned to the differences with positive sign). In comparison, when we conducted the same analysis on leaf RNA levels, no significant difference was found between the two groups (Supplemental Figure 7; V = 137 and P = 0.23 by Wilcoxon signed-rank test).
DISCUSSION
Taking advantage of the ability to isolate plastids from different tissues and knowledge of recent genome duplications in higher plants, we performed quantitative import assays, functional analyses in transgenic plants, and systematic transit-peptide sequence comparisons to identify and demonstrate the importance of the twin-positive motif for root leucoplast import. Our results reveal a tissue-specific modulation of plastid protein import executed via specific transit-peptide motifs. Analyses of the recently duplicated preproteins also suggest an association between higher RNA expression in roots and the possession of extra twin-positive motifs in transit peptides. This suggests that, after gene duplication, the gene exhibiting higher root RNA expression gained or retained twin-positive motifs leading to more efficient import into root leucoplasts. Therefore, there is a correlation between RNA expression levels, transit-peptide motifs, and protein abundance, supporting the notion that protein import from the cytosol helps determine tissue-specific plastid proteomes.
The RVSI motif in the prFB transit peptide is sufficient to increase more than threefold the import of prPGL35 into leucoplasts. It is also necessary for efficient import of prFB into leucoplasts. Unfortunately, we were unable to further establish which of the residues of that motif are most critical. No other preproteins encoded by the 75 pairs of recently duplicated genes we examined contain this motif within their first 60 amino acids. Further analyses are required to reveal how this motif exerts its function, whether any of its residues can be substituted by amino acids of similar properties, or if this motif requires other sequence elements in the transit peptides of prFB and prPGL35 to function.
Among all organellar targeting signals, plastid transit peptides may exhibit the greatest diversity in sequence and length (von Heijne and Nishikawa, 1991). Apart from a general enrichment of Ser and a deficiency of acidic amino acids, no consensus sequence motifs have been identified thus far (von Heijne et al., 1989; Zybailov et al., 2008). Previously, we presented a multi-selection and multi-order hypothesis to suggest that this sequence diversity may be due in part to embedded regulatory motifs (Li and Teng, 2013). We hypothesized that each transit peptide represents a selective assembly of functional motifs and that the order of the motifs may differ among transit peptides, explaining why no consensus sequences or domain structures have been identified through linear sequence comparisons of all transit peptides as a group. We also hypothesized that specific transit-peptide motifs exist that can increase import preferentially into particular plastid types. The combined effect of all motifs in a transit peptide would then determine the preference of each transit peptide for a particular plastid type. The results we present herein provide support for our multi-selection and multi-order hypothesis by demonstrating the existence of motifs that specifically enhance import into root leucoplasts. The existence of tissue-specific import regulation also suggests that caution should be exercised when comparing protein import and transit-peptide mutagenesis data generated with plastids isolated from different tissues (e.g., onion [Allium cepa] epidermal cells or rice calli).
Previously, we showed that the twin-positive motif is important for preferential preprotein import into old chloroplasts of pea seedlings (Teng et al., 2012). However, there are no large proteomic data sets in which young and old leaves are compared. Furthermore, it is also difficult to precisely correlate leaf age of pea seedlings with the age of Arabidopsis seedlings, for which all transcriptomic and proteomic data have been generated. Here, we have shown that the twin-positive motif is also important for efficient import into root leucoplasts. Tissue-specific expression patterns of proteins are more likely to be conserved between pea and Arabidopsis, so we could take advantage of the available quantitative root proteomic and transcriptomic data of Arabidopsis to perform our systematic analyses. Of note, not all proteins that are preferentially imported into old chloroplasts show efficient import into root leucoplasts, and vice versa. For example, the preprotein prL11 is preferentially imported into old chloroplasts but its import into leucoplasts is poor (Supplemental Figure 8), whereas preprotein prCpn10-2 is imported efficiently into leucoplasts but its import into chloroplasts is independent of chloroplast age (Teng et al., 2012; Chu and Li, 2015). It is possible that preferential import into leucoplasts and old chloroplasts both require multiple motifs that interact with multiple factors.
Interestingly, in Arabidopsis, several major translocon components, including Toc159, Toc33, Tic22, and Tic20, are encoded by multigene families, and different family members have different RNA and protein expression levels in roots and leaves (Supplemental Data Set 3). Genetic and biochemical evidence also indicates that Arabidopsis Toc159 and Toc33 are more important for preprotein import into leaf chloroplasts and that Toc132/Toc120 and Toc34 may be more important for preprotein import into root leucoplasts (Yu and Li, 2001; Constan et al., 2004; Ivanova et al., 2004; Kubis et al., 2004; Dutta et al., 2014; for review, see Chu and Li, 2018). Therefore, TOC complexes with the composition of Toc132/Toc120 and Toc34 may be part of the system in leucoplasts recognizing the twin-positive motif. The possible presence of factors recognizing plastid-type-specific transit-peptide motifs also fully agrees with data showing that plastid-type transition involves remodeling of the TOC complex. The proteins SP1 and SP2 act in facilitating degradation of TOC proteins and are important for remodeling the TOC machinery and for interconversion between different plastid types (Ling et al., 2012, 2019). It would be interesting to identify all the factors important for deciphering the twin-positive and RVSI motifs and to study how the synthesis and degradation of these factors are controlled in a tissue-specific manner.
It has been proposed that the mitochondrial inner membrane ∆Ψ imposes selection pressure for a higher abundance of Arg residues in mitochondrial presequences. To avoid mistargeting to mitochondria, plastid transit peptides generally have fewer Arg residues than mitochondrial presequences (Garg and Gould, 2016). A domain containing multiple Arg and hydrophobic residues located at the N-terminal region of mitochondrial presequences has also been shown to be critical for conferring targeting specificity to mitochondria (Lee et al., 2019). Interestingly, all of the twin-positive motifs we analyzed and almost all of the additional twin-positive motifs identified in the recently duplicated genes are located in the middle and C-terminal regions of transit peptides. It is possible that, in roots and older leaves, the mitochondrial ∆Ψ is not as high as in young leaves. The chance of a more positively charged sequence, located in regions other than the N terminus, being mistargeted to mitochondria is therefore reduced. Twin-positive motifs are then employed to increase import efficiency into plastids in roots and older leaves.
METHODS
Identification of Recently Duplicated Genes Encoding Plastid Preproteins
To generate a list of recently duplicated genes encoding plastid preproteins, we performed a Venn diagram analysis of the 1153 high-confidence Arabidopsis (Arabidopsis thaliana) plastid proteins (Bischof et al., 2011) and the 3044 recently duplicated gene pairs (Blanc et al., 2003; Supplemental Data Set 1), which initially generated 96 pairs of genes. We manually checked pairs for which the polypeptide lengths differed by more than 10% and then removed those that clearly had deletions in the transit-peptide or mature region of one of the genes. If one of the genes encoded a protein that has been experimentally shown to be non-plastid-localized, these pairs were also excluded.
Plant Growth and Plastid Isolation
Pea (Pisum sativum cv Green Arrow; De Bruyn Seed Co.) seedlings were grown at 20°C on vermiculite for 4 d in the dark for leucoplast isolation or for 7 d under a 12-h photoperiod with a light intensity of ∼150 μmol m−2 s−1 for chloroplast isolation. Leucoplasts were prepared from pea roots as previously described by Chu and Li (2015), except that pea roots were washed twice with the homogenization buffer (50 mM Tricine-KOH, pH 7.9, 330 mM sorbitol, 1 mM MgCl2, and 2 mM EDTA) and then homogenized with homogenization buffer containing 1% (w/v) BSA and reducing agents (2 mM ascorbic acid, 0.1 mM DTT, and 1.2 mM glutathione). Chloroplasts were isolated from pea leaves as described by Perry et al. (1991), except that 2 mM ascorbic acid, 0.1 mM DTT, and 1.2 mM glutathione were added to the grinding buffer used for homogenization. Isolated chloroplasts were adjusted to 1 mg chlorophyll/mL in import buffer (50 mM HEPES-KOH, pH 8.0, and 330 mM sorbitol). The isolated plastids were assayed using a bicinchoninic acid kit (Thermo Fisher Scientific) to determine the protein concentration. For each import reaction, 500 μg of chloroplast proteins or 114 μg of leucoplast proteins was used.
Wild-type and mutant Arabidopsis used in this study were in the Col ecotype. The Δcphsc70-1 knockout mutant and the genomic fragment complemented line (70-1g) were previously described by Su and Li, 2008). Surface-sterilized seeds of Arabidopsis were sown on 0.3% (w/v) Gelrite-solidified Murashige and Skoog (MS) medium containing Gamborg’s B5 vitamins and 0.5% (w/v) Suc. After a 3-d cold stratification period, seeds were grown in growth chambers under a 16-h photoperiod with a light intensity ∼80 μmol m−2 s−1 at 23°C. Production of the anti-Toc75 (Tu et al., 2004), anti-Tic110 (Tu et al., 2004), and anti-porin (Teng et al., 2012) antibodies was described previously, and they were used at a 1:2000 dilution.
Plasmid Construction and Plant Transformation
The leaf cDNA pools of Arabidopsis (Col ecotype) were used as templates to amplify the regions encoding prPGL35 (AT4G04020) and prFB (AT4G22240) with specific primers (Supplemental Data Set 4), and these regions were cloned into the HindIII/PstI of pSP72. Since the Met residues were located only in the transit peptide region of prPGL35 and prFB, two extra Met residues were introduced into the 3′ end of the cDNA before the stop codon. Transit-peptide mutants of prPGL35 and prFB were generated using a QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies) with specific primers (Supplemental Data Set 4), and all sequences were confirmed by sequencing. Plasmids encoding prTic40, prTic40 transit peptide mutants, and prcpHsc70-1 have been previously described by Teng et al. (2012).
The 70-1gKRAA genomic fragment was generated from the pGEM-T-cpHsc70-1g (Su and Li, 2008) using a QuikChange II Site-Directed Mutagenesis Kit and specific primers (Supplemental Data Set 4) and used to replace the original genomic fragment in pCambia1390/cpHsc70-1g (Su and Li, 2008). After confirming the sequences, the resulting plasmid was named pCambia1390-cpHsc70-1g-K23R24→AA and transformed into Agrobacterium tumefaciens GV3101. The Δcphsc70-1 mutant plants were infected by the floral spray method (Chung et al., 2000). Two independent transformations were performed. Four independent T1 transgenic lines were obtained and characterized. Transgenic plants harboring the 70-1gKRAA transgene were screened on MS medium containing 50 mg L−1 hygromycin. The genotype of transgenic plants was confirmed using specific primers as described (Supplemental Data Set 4; Su and Li, 2008) The 70-1gKRAA transgenic lines were further confirmed by PCR using specific primers (Supplemental Figure 5; Supplemental Data Set 4). Seeds of independent transgenic lines were collected, and lines homozygous for a single transgene insertion were identified based on the antibiotic-resistant ratio in the T2 and T3 generations. Experiments were performed using plants of the T3 generation.
Protein Import and Postimport Analyses
All preproteins were translated in vitro by TNT Coupled Wheat Germ Extract or Reticulocyte Lysate Systems (Promega) in the presence of [35S]Met and incubated with isolated plastids in the presence of 3 mM ATP at room temperature. Import was stopped by transferring reactions to a new tube containing 1 mL of cold import buffer. The plastids were pelleted at 3000 relative centrifugal force and 4°C for 6 min and resuspended in 200 μL of cold import buffer. Intact leucoplasts were reisolated by underlaying with 1 mL of 10% (v/v) Percoll, and intact chloroplasts were reisolated by loading onto 1 mL of 40% (v/v) Percoll. Both were centrifuged in a swinging-bucket rotor at 2900 relative centrifugal force and 4°C for 6 min. The plastids were washed once with import buffer. Thermolysin treatments of in vitro-translated products and plastids after import were performed as previously described by Perry et al. (1991), and intact plastids were reisolated as described above. Protein concentrations of the plastid samples were measured with the bicinchoninic acid kit (Thermo Fisher Scientific). Samples were analyzed by SDS-PAGE using the NuPAGE gel system (Invitrogen), exposed to x-ray films with TranScreen-LE intensifying screens (Carestream Health), and quantified using a phosphoimager (Typhoon FLA 9000; GE Healthcare Life Sciences).
Heat-Shock Treatments and Measurements of Root Lengths and Chlorophylls
Heat-shock treatments of imbibed seeds were performed as described (Su and Li, 2008). Briefly, sterilized Arabidopsis seeds were stratified at 4°C for 3 d and then plated on MS-agar medium with 0.5% (w/v) Suc. Plates were sealed with plastic electric tape and heated in a 44.5°C water bath in the dark for 2.5 h and then cooled down to room temperature by floating on tap water for 15 min. For root growth assays, plates were placed vertically and seeds were allowed to grow for 7 d. Root lengths were measured using photographs of seedlings and the software ImageJ (National Institutes of Health). For chlorophyll assays, plates were placed horizontally and seeds were allowed to grow for 14 d. Total chlorophyll was determined as described (Lichtenthaler, 1987).
Accession Numbers
Accession numbers are as follows: PGL35 (AT4G04020), FB (AT4G22240), Tic40 (AY157668), cpHsc70-1 (AT4G24280), L11 (AT1G32990), Toc75 (X83767), Tic110 (AT1G06950), and porin (AT3G01280).
Supplemental Data
Supplemental Figure 1. Immunoblotting analyses of isolated-leucoplast and total-root protein extracts.
Supplemental Figure 2. Sequence alignment of prFB and prPGL35 and the import behaviors of prFB-M42V into chloroplasts and leucoplasts.
Supplemental Figure 3. Import time-course experiments of prPGL35 and its transit peptide mutants into chloroplasts and leucoplasts.
Supplemental Figure 4. The twin-positive motif in the transit peptides of prTic40 and prcpHsc70-1 is important for leucoplast binding.
Supplemental Figure 5. Generation of the 70-1g and 70-1gKRAA transgenic lines.
Supplemental Figure 6. The isoforms with extra twin-positive motifs have higher leucoplast import efficiency.
Supplemental Figure 7. Isoforms possessing extra twin-positive motifs do not have significantly higher RNA expression in leaves.
Supplemental Figure 8. Preprotein prL11, which shows preference for old chloroplasts as reported by Teng et al., 2012, does not have high leucoplast import efficiency.
Supplemental Data Set 1. 3044 pairs of recently duplicated Arabidopsis genes.
Supplemental Data Set 2. 75 pairs of recently duplicated Arabidopsis genes encoding plastid preproteins.
Supplemental Data Set 3. RNA and protein levels of TOC and TIC family members.
Supplemental Data Set 4. Sequences of primers used to generate plasmid constructs or verify genotypes of transgenic plants used in this study.
Supplemental Data Set 5. ANOVA parameters for the statistical analyses in Figure 6.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Shin-Han Shiu (Michigan State University) for the list of recently duplicated genes, the Institute of Molecular Biology (Academia Sinica) Bioinformatics Core for assistance in sequence downloading and analyses, Jun-Yi Leu (Institute of Molecular Biology, Academia Sinica) for supporting K.S., and John O’Brien for English editing. This work was supported by the Ministry of Science and Technology of Taiwan (grant MOST 108-2321-B-001-021 to H.m.L.) and from the Academia Sinica of Taiwan.
AUTHOR CONTRIBUTIONS
C.-C.C. and H.-m.L. designed the research; C.-C.C. performed most of the experiments; K.S. analyzed the transit-peptide sequences of 75 pairs of homologous preproteins and the correlation between the presence of the twin-positive motif and RNA and protein expression levels; C.-C.C. and H.-m.L. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Baerenfaller K., Grossmann J., Grobei M.A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S.(2008). Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941. [DOI] [PubMed] [Google Scholar]
- Bischof S., Baerenfaller K., Wildhaber T., Troesch R., Vidi P.A., Roschitzki B., Hirsch-Hoffmann M., Hennig L., Kessler F., Gruissem W., Baginsky S.(2011). Plastid proteome assembly without Toc159: Photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins. Plant Cell 23: 3911–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Hokamp K., Wolfe K.H.(2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.C., Li H.M.(2015). Protein import into isolated pea root leucoplasts. Front. Plant Sci. 6: 690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.C., Li H.M.(2018). Developmental regulation of protein import into plastids. Photosynth. Res. 138: 327–334. [DOI] [PubMed] [Google Scholar]
- Chung M.H., Chen M.K., Pan S.M.(2000). Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Res. 9: 471–476. [DOI] [PubMed] [Google Scholar]
- Constan D., Patel R., Keegstra K., Jarvis P.(2004). An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J. 38: 93–106. [DOI] [PubMed] [Google Scholar]
- Dutta S., Teresinski H.J., Smith M.D.(2014). A split-ubiquitin yeast two-hybrid screen to examine the substrate specificity of atToc159 and atToc132, two Arabidopsis chloroplast preprotein import receptors. PLoS One 9: e95026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S.G., Gould S.B.(2016). The role of charge in protein targeting evolution. Trends Cell Biol. 26: 894–905. [DOI] [PubMed] [Google Scholar]
- Ivanova Y., Smith M.D., Chen K., Schnell D.J.(2004). Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol. Biol. Cell 15: 3379–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey R.A. III, Subramanian C., Bruce B.D.(2000). Identification of a Hsp70 recognition domain within the Rubisco small subunit transit peptide. Plant Physiol. 122: 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P., López-Juez E.(2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14: 787–802. [DOI] [PubMed] [Google Scholar]
- Karlin-Neumann G.A., Tobin E.M.(1986). Transit peptides of nuclear-encoded chloroplast proteins share a common amino acid framework. EMBO J. 5: 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffmann T., Russenberger D., von Zychlinski A., Christopher W., Sjölander K., Gruissem W., Baginsky S.(2004). The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14: 354–362. [DOI] [PubMed] [Google Scholar]
- Köhler D., Montandon C., Hause G., Majovsky P., Kessler F., Baginsky S., Agne B.(2015). Characterization of chloroplast protein import without Tic56, a component of the 1-megadalton translocon at the inner envelope membrane of chloroplasts. Plant Physiol. 167: 972–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S., Patel R., Combe J., Bédard J., Kovacheva S., Lilley K., Biehl A., Leister D., Ríos G., Koncz C., Jarvis P.(2004). Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16: 2059–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Kim J.K., Lee S., Choi S., Kim S., Hwang I.(2008). Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell 20: 1603–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Lee S., Lee G.J., Lee K.H., Kim S., Cheong G.W., Hwang I.(2006). Functional characterization of sequence motifs in the transit peptide of Arabidopsis small subunit of Rubisco. Plant Physiol. 140: 466–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Lee S., Lee J., Woo S., Razzak M.A., Vitale A., Hwang I.(2019). Molecular mechanism of the specificity of protein import into chloroplasts and mitochondria in plant cells. Mol. Plant 12: 951–966. [DOI] [PubMed] [Google Scholar]
- Lee D.W., Lee S., Oh Y.J., Hwang I.(2009). Multiple sequence motifs in the Rubisco small subunit transit peptide independently contribute to Toc159-dependent import of proteins into chloroplasts. Plant Physiol. 151: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.M., Teng Y.S.(2013). Transit peptide design and plastid import regulation. Trends Plant Sci. 18: 360–366. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H.(1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148: 350–382. [Google Scholar]
- Ling Q., Broad W., Trösch R., Töpel M., Demiral Sert T., Lymperopoulos P., Baldwin A., Jarvis R.P.(2019). Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science 363: eaav4467. [DOI] [PubMed] [Google Scholar]
- Ling Q., Huang W., Baldwin A., Jarvis P.(2012). Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338: 655–659. [DOI] [PubMed] [Google Scholar]
- Liu C., Willmund F., Golecki J.R., Cacace S., Hess B., Markert C., Schroda M.(2007). The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas. Plant J. 50: 265–277. [DOI] [PubMed] [Google Scholar]
- Liu L., McNeilage R.T., Shi L.X., Theg S.M.(2014). ATP requirement for chloroplast protein import is set by the Km for ATP hydrolysis of stromal Hsp70 in Physcomitrella patens. Plant Cell 26: 1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S.E., Buvinger W.E., Bennett J., Keegstra K.(1991). Synthetic analogues of a transit peptide inhibit binding or translocation of chloroplastic precursor proteins. J. Biol. Chem. 266: 11882–11889. [PubMed] [Google Scholar]
- Primavesi L.F., Wu H., Mudd E.A., Day A., Jones H.D.(2008). Visualisation of plastids in endosperm, pollen and roots of transgenic wheat expressing modified GFP fused to transit peptides from wheat SSU RubisCO, rice FtsZ and maize ferredoxin III proteins. Transgenic Res. 17: 529–543. [DOI] [PubMed] [Google Scholar]
- Rial D.V., Arakaki A.K., Ceccarelli E.A.(2000). Interaction of the targeting sequence of chloroplast precursors with Hsp70 molecular chaperones. Eur. J. Biochem. 267: 6239–6248. [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Miyagishima S.Y., Jarvis P.(2008). Chloroplast biogenesis: Control of plastid development, protein import, division and inheritance. The Arabidopsis Book 6: e0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M., Vallon O., Wollman F.A., Beck C.F.(1999). A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M.(2010). A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22: 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P.H., Li H.M.(2008). Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol. 146: 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P.H., Li H.M.(2010). Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y.S., Chan P.T., Li H.M.(2012). Differential age-dependent import regulation by signal peptides. PLoS Biol. 10: e1001416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S.L., Chen L.J., Smith M.D., Su Y.S., Schnell D.J., Li H.M.(2004). Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell 16: 2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G., Nishikawa K.(1991). Chloroplast transit peptides: The perfect random coil? FEBS Lett. 278: 1–3. [DOI] [PubMed] [Google Scholar]
- von Heijne G., Steppuhn J., Herrmann R.G.(1989). Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem. 180: 535–545. [DOI] [PubMed] [Google Scholar]
- Wan J., Blakeley S.D., Dennis D.T., Ko K.(1996). Transit peptides play a major role in the preferential import of proteins into leucoplasts and chloroplasts. J. Biol. Chem. 271: 31227–31233. [DOI] [PubMed] [Google Scholar]
- Wimmer D., Bohnhorst P., Shekhar V., Hwang I., Offermann S.(2017). Transit peptide elements mediate selective protein targeting to two different types of chloroplasts in the single-cell C4 species Bienertia sinuspersici. Sci. Rep. 7: 41187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J.(2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Khan S., Hase T., Emes M.J., Bowsher C.G.(2006). Differential uptake of photosynthetic and non-photosynthetic proteins by pea root plastids. FEBS Lett. 580: 6509–6512. [DOI] [PubMed] [Google Scholar]
- Yu T.-S., Li H.(2001). Chloroplast protein translocon components atToc159 and atToc33 are not essential for chloroplast biogenesis in guard cells and root cells. Plant Physiol. 127: 90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K.J.(2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]