JAZ repressors of the jasmonate pathway directly interact with and suppress RHD6 and RSL1 transcription factors to integrate jasmonate signaling and root hair development in Arabidopsis.
Abstract
Root hairs arise from trichoblasts and are crucial for plant anchorage, nutrient acquisition, and environmental interactions. The phytohormone jasmonate is known to regulate root hair development in Arabidopsis (Arabidopsis thaliana), but little is known about the molecular mechanism underlying jasmonate modulation in this process. Here, we show that the application of exogenous jasmonate significantly stimulated root hair elongation, but, on the contrary, blocking the perception or signaling of jasmonate resulted in defective root hairs. Jasmonate consistently elevated the expression levels of several crucial genes positively involved in root hair growth. Mechanistic investigation revealed that JASMONATE ZIM-DOMAIN (JAZ) proteins, critical repressors of jasmonate signaling, physically interacted with ROOT HAIR DEFECTIVE 6 (RHD6) and RHD6 LIKE1 (RSL1), two transcription factors that are essential for root hair development. JAZ proteins inhibited the transcriptional function of RHD6 and interfered with the interaction of RHD6 with RSL1. Genetic analysis indicated that jasmonate promoted root hair growth in a RHD6/RSL1-dependent manner. Moreover, overexpression of RHD6 largely rescued the root hair defects of JAZ-accumulating plants. Collectively, our study reveals a key signaling module in which JAZ repressors of the jasmonate pathway directly modulate RHD6 and RSL1 transcription factors to integrate jasmonate signaling and the root hair developmental process.
INTRODUCTION
Root hairs are tubular polarized extensions of root epidermal cells that greatly increase the root surface area, facilitating the acquisition of nutrients, environmental interactions, and plant anchorage. In Arabidopsis (Arabidopsis thaliana), root epidermal cell types are determined in a position-dependent manner (Ishida et al., 2008). Root epidermal cells that overlie a single cortical cell will develop as non-hair (N) cells, whereas root cells that are in contact with two underlying cortical cells will produce a root hair (H; Dolan et al., 1994; Galway et al., 1994; Ishida et al., 2008; Schiefelbein et al., 2009). Previous studies using genetic and molecular approaches identified several components that influence the root epidermal cell fate (Schiefelbein et al., 2009; Wei and Li, 2018). Among these components, the R2R3-type MYB protein WEREWOLF (WER) forms a transcription factor complex with basic helix-loop-helix (bHLH) proteins GLABRA3 (GL3)/ENHANCER OF GLABRA3 (EGL3), and the WD40 protein TRANSPARENT TESTA GLABRA1 (TTG1) to modulate the expression levels of downstream related genes such as GL2 (Galway et al., 1994; Masucci and Schiefelbein, 1996; Lee and Schiefelbein, 1999; Payne et al., 2000; Bernhardt et al., 2003, 2005; Zhang et al., 2003; Ryu et al., 2005; Kang et al., 2009; Schiefelbein et al., 2009; Song et al., 2011a). GL2, a homeodomain–Leu zipper transcription factor, is crucial for maintaining the N cell fate and repressing root hair development (Rerie et al., 1994; Di Cristina et al., 1996; Masucci and Schiefelbein, 1996).
A modulatory network for root hair growth that functions downstream of GL2 consists of a subset of bHLH transcription factors (Bruex et al., 2012; Pires et al., 2013; Lin et al., 2015). Among them, ROOT HAIR DEFECTIVE 6 (RHD6), a Class I member of the bHLH Group VIII subfamily, plays a crucial positive role in the regulation of root hair elongation (Masucci and Schiefelbein, 1994; Menand et al., 2007; Bruex et al., 2012). RHD6 is expressed explicitly in H cells and promotes their development; the loss-of-function rhd6 mutant plants have fewer root hairs than wild type (Masucci and Schiefelbein, 1994). Similarly, RHD6 LIKE1 (RSL1), the closest homolog of RHD6, has a partially redundant role with that of RHD6 in stimulating root hair growth (Menand et al., 2007). RHD6 and RSL1 both upregulate the expression of four Class II members of the Group VIII subfamily, including RSL2, RSL3, RSL4, and RSL5, which are positively involved in mediating root hair elongation redundantly (Yi et al., 2010; Pires et al., 2013). Simultaneous disruption of RSL4 and RSL2 results in rsl4 rsl2 double mutants with no visible root hairs (Yi et al., 2010). Recent studies have shown that the RSL4 and RSL2 transcription factors are required for root hair growth induced by phytohormones (e.g., auxin, cytokinin, or ethylene) and nutrient (e.g., phosphorus or nitrogen) deficiency (Yi et al., 2010; Datta et al., 2015; Zhang et al., 2016; Feng et al., 2017; Mangano et al., 2017).
The plant hormone jasmonate is ubiquitous in the plant kingdom and essential for multiple physiological processes (Wasternack and Hause, 2013; Chini et al., 2016; Hu et al., 2017; Huang et al., 2017; Zhang et al., 2017; Guo et al., 2018a; Howe et al., 2018). Jasmonate is perceived by a coreceptor complex consisting of the F-box protein CORONATINE-INSENSITIVE1 (COI1) and JASMONATE ZIM-DOMAIN (JAZ) proteins (Xie et al., 1998; Chini et al., 2007; Thines et al., 2007; Yan et al., 2009; Sheard et al., 2010). JAZ coreceptors are repressors of jasmonate signaling and interact with a wide variety of transcription factors (Chini et al., 2007; Thines et al., 2007; Cheng et al., 2009; Fernández-Calvo, 2011; Niu et al., 2011; Kazan and Manners, 2013; Schweizer et al., 2013; Guo et al., 2018b). Upon jasmonate perception, JAZ repressors are degraded via the SCFCOI1-26S proteasome pathway (Chini et al., 2007; Thines et al., 2007; Yan et al., 2009), subsequently releasing downstream transcription factors to activate jasmonate signaling. Deficiencies in the biosynthesis or perception of jasmonate attenuate several developmental processes in Arabidopsis, such as trichome initiation and stamen development (Sanders et al., 2000; Stintzi and Browse, 2000; Balbi and Devoto, 2008; Qi et al., 2011, 2015; Song et al., 2011b; Chini et al., 2016; Guo et al., 2018b; Han et al., 2018). Moreover, jasmonate has been reported to promote root hair growth (Zhu et al., 2006, 2011). However, the molecular mechanisms underlying the stimulation of root hair development by jasmonate remain elusive.
In this study, we undertook a genetic and molecular approach to investigate the role and underlying mechanism of jasmonate in modulating root hair growth. We initially confirmed that exogenous application of methyl jasmonate (MeJA) significantly increased the length of root hairs. We also discovered that the endogenous COI1/JAZ-mediated jasmonate signaling pathway is critical for root hair development—disruption of the COI1 receptor or accumulation of a JAZ repressor (e.g., JAZ4 or JAZ8) decreased root hair length. Further investigation of the detailed mechanism revealed that several JAZ repressors interacted with RHD6 and RSL1, two bHLH transcription factors crucial for root hair development. JAZ proteins repressed the transcriptional function of RHD6 and interfered with the interaction between RHD6 and RSL1. Phenotypic analysis showed that jasmonate promoted root hair growth in a manner dependent on RHD6 and RSL1. It also showed that overexpressing RHD6 largely rescued the root hair-defective phenotypes of coi1-2 and JAZ8-ΔJas-9 plants. Taken together, our study provides a mechanistic understanding of how JAZ repressors directly regulate RHD6/RSL1 transcription factors to integrate jasmonate signaling and root hair development in Arabidopsis.
RESULTS
COI1/JAZ-Mediated Jasmonate Signaling Is Critical for Root Hair Elongation
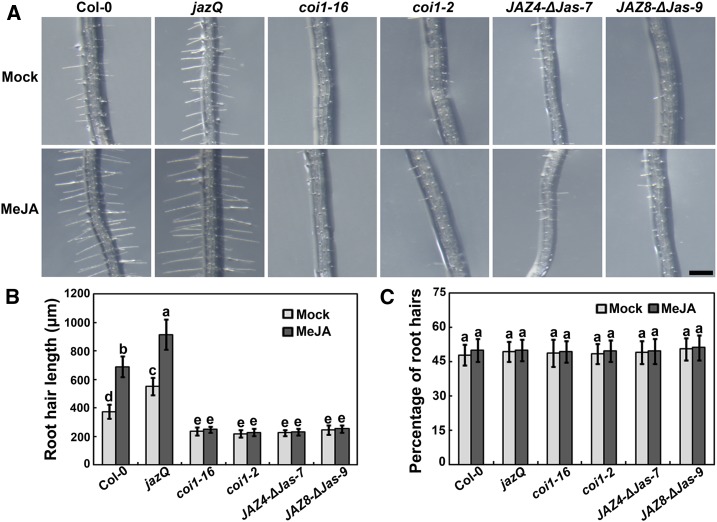
Previous studies have shown that exogenous application of MeJA stimulates root hair development in Arabidopsis (Zhu et al., 2006, 2011). To investigate the molecular mechanisms underlying how jasmonate promotes root hair development, we first confirmed the regulatory role of jasmonate using MeJA. Similar to previous studies (Zhu et al., 2006, 2011), root hair elongation was significantly enhanced in wild-type plants after the application of 20 µM MeJA (Figures 1A and 1B). We also examined the percentage of visible roots hairs in total epidermal cells and found that the percentage was not affected by MeJA in wild-type plants (Figure 1C). To better understand the function of jasmonate in root hair growth, we analyzed whether crucial components of endogenous jasmonate signaling modulate root hair initiation and elongation. The F-box protein COI1 is the jasmonate receptor and positively regulates jasmonate responses (Xie et al., 1998; Yan et al., 2009). As shown in Figures 1A and 1B, the root hairs of coi1-16 and coi1-2, two partial COI1 mutants, were much shorter than those of wild type and were completely unresponsive to MeJA. JAZ proteins are critical repressors of jasmonate signaling and directly link jasmonate perception with transcriptional changes (Chini et al., 2007; Thines et al., 2007). To investigate whether JAZ repressors are also involved in root hair growth, we generated transgenic plants (JAZ4-ΔJas-7 or JAZ8-ΔJas-9) overexpressing JAZ4 or JAZ8 with a deletion of the Jas domain that is required for the degradation of JAZ proteins (Chini et al., 2007; Thines et al., 2007), which are insensitive to MeJA treatment (Supplemental Figure 1). Further analysis showed that root hair elongation was dramatically compromised in JAZ4-ΔJas-7 and JAZ8-ΔJas-9 transgenic plants (Figures 1A and 1B). In contrast, the quintuple mutant jazQ, which lacks five JAZ transcriptional repressors (Campos et al., 2016), displayed root hairs much longer than those of wild type with or without MeJA induction (Figures 1A and 1B). Taken together, these results demonstrate that the COI1/JAZ-mediated endogenous jasmonate signaling is required for root hair elongation in Arabidopsis.
Figure 1.
Root-Hair Phenotypes of Jasmonate-Related Mutants and Transgenic Plants.
(A) Images of root hairs of untreated Col-0, jazQ, coi1-16, coi1-2, JAZ4-ΔJas-7 and JAZ8-ΔJas-9 control plants (Mock) and plants treated with methyl jasmonate (MeJA). Experiments were performed more than three times with similar results by analyzing different batches of seeds. Bar = 500 μm.
(B) and (C) Quantitative analysis of root hair length and percentage of visible hair cells in total epidermal cells in response to MeJA treatment. Seeds were first germinated and grown for 4 d on half-strength MS medium, and then were transferred to half-strength MS medium supplemented with 20 μM MeJA for 2 d before the examination. Values are means ± sd (n = 12 roots). Bars with different letters are significantly different from each other (P < 0.05). Data were analyzed by ANOVA.
Jasmonate Upregulates Several Root Hair-Related bHLH Transcription Factors and Their Targets
Recent studies have demonstrated that a series of bHLH family proteins, including the transcription factors RHD6 and RSL, play essential roles in modulating root hair growth (Yi et al., 2010; Bruex et al., 2012; Pires et al., 2013; Lin et al., 2015). Because jasmonate is also involved in this process, we queried whether it mediates the expression of those bHLH factors and their downstream targets, such as ROOT HAIR SPECIFIC10 (RHS10), EXPANSIN A7 (EXPA7), and LEU-RICH REPEAT/EXTENSIN1 (LRX1). To test this, we performed quantitative real-time PCR (RT-qPCR) analyses to detect their expression levels in wild-type seedlings with or without MeJA treatment. As shown in Figure 2, among those bHLH genes analyzed, RHD6, RSL1, RSL2, RSL4, and RSL5 were upregulated by MeJA treatment in wild-type plants. Similarly, transcripts of RHS10, EXP7, and LRX1 accumulated at higher levels in MeJA-treated wild-type seedlings (Figure 2). To verify the regulatory effect of jasmonate on those genes, we examined their expression in coi1-2, JAZ8-ΔJas-9, and jazQ plants. The basal expression level of several root hair-related genes was reduced in coi1-2 and JAZ8-ΔJas-9 compared with that in wild-type plants (Figure 2). Furthermore, transcript levels of most genes analyzed were significantly decreased in coi1-2 and JAZ8-ΔJas-9 upon MeJA treatment in comparison with wild type (Figure 2). In contrast, the quintuple mutant jazQ accumulated significantly more transcripts of those genes compared with the wild type either with or without MeJA induction (Figure 2). These results show that jasmonate stimulates the expression of several root hair-associated bHLH transcription factors and their downstream targets.
Figure 2.
Expression of Root Hair-Related bHLH Transcription Factors and Their Targets in Response to MeJA.
Seeds of Col-0, jazQ, coi1-2, and JAZ8-ΔJas-9 were germinated and grown for 4 d on half-strength MS medium and then were transferred to half-strength MS medium with 20 μM MeJA for 2 d. Total RNA was extracted from at least three batches of roots as biological replicates, and at least three technical replicates were analyzed for each sample. Error bars show SD from three independent biological replicates. Expression of ACTIN2 was used as a normalization control. Bars with different letters are significantly different from each other (P < 0.05). Data were analyzed by ANOVA.
JAZ Proteins Physically Interact with RHD6 and RSL1
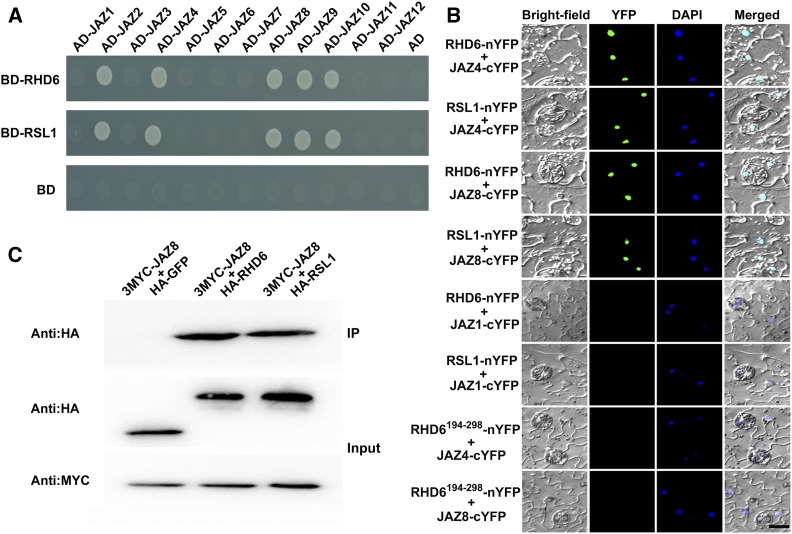
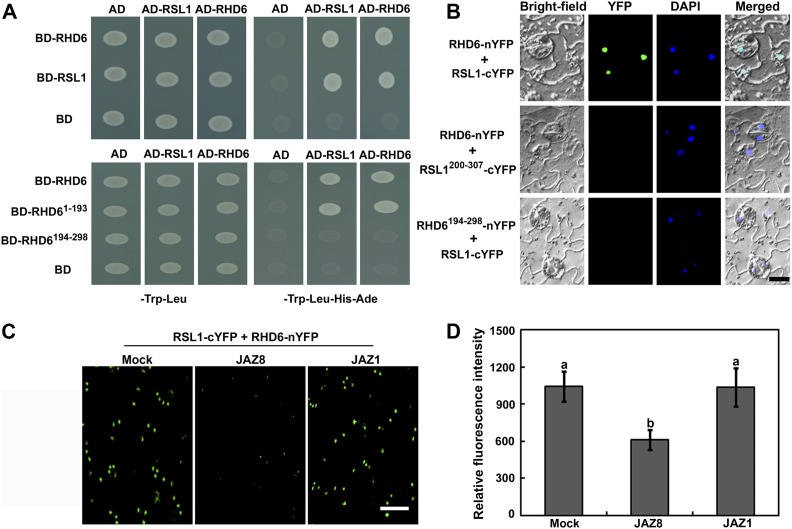
JAZ proteins are crucial negative modulators of jasmonate signaling (Chini et al., 2007; Thines et al., 2007). Recently, JAZ repressors were shown to physically associate with several transcription factors to regulate various jasmonate responses (Kazan and Manners, 2013; Schweizer et al., 2013; Guo et al., 2018b). As RHD6 and RSL1 are essential for root hair elongation and responsiveness to jasmonate treatment (Menand et al., 2007; Figure 2), we hypothesized that they might also interact with JAZ proteins. To test this, we first used the yeast two-hybrid system to identify possible interactions between full-length RHD6 and JAZ proteins. As shown in Figure 3A, RHD6 strongly interacted with JAZ2, JAZ4, JAZ8, JAZ9, and JAZ10 in yeast. Moreover, we also detected possible physical associations of JAZ proteins with RSL1 and found that the same JAZ proteins exhibited interactions with RSL1 in yeast (Figure 3A).
Figure 3.
Physical Interactions of JAZ Proteins with RHD6 and RSL1 Factors.
(A) Yeast two-hybrid assays. Interaction is indicated by the ability of cells to grow on dropout medium lacking Leu, Trp, His, and Ade and containing 3 mM 3-aminotriazole for 2 d after plating. pGBKT7 (BD) and pGADT7 (AD) were used as negative controls. BD, DNA binding domain; AD, Activation domain.
(B) BiFC analyses. Fluorescence was observed in the nucleus of transformed tobacco cells, resulting from the complementation of RHD6-nYFP (or RSL1-nYFP) with JAZ4-cYFP (or JAZ8-cYFP). Nuclei are indicated by DAPI staining. Bar = 15 μm.
(C) Co-immunoprecipitation analyses. MYC-JAZ8 fusion proteins were immunoprecipitated using an anti-MYC antibody (catalog no. A7470, Sigma-Aldrich; 1:250), and the coimmunoprecipitated protein was detected using an anti-HA antibody (catalog no. H9658, Sigma-Aldrich; 1:10000). Protein input for MYC-JAZ8 in immunoprecipitated complexes is also shown. Experiments were repeated three times with similar results. IP, immunoprecipitation.
To characterize which region(s) of JAZ proteins are required to interact with RHD6 and RSL1, we performed additional directed yeast two-hybrid analysis. JAZ4 and JAZ8 were each divided into the N-terminal fragment (JAZ41-176 or JAZ81-74) and the C-terminal fragment containing a Jas domain (JAZ4177-310 or JAZ875-131). As shown in Supplemental Figure 2, deletion of the C-terminal portion of JAZ4 or JAZ8 abolished the interactions with RHD6 and RSL1 in yeast. In contrast, deletion of the N-terminal residues of JAZ4 or JAZ8 did not affect their associations. These results demonstrate that the C-terminal region of JAZ4 or JAZ8 is essential for the interactions. Similarly, we identified fragments of RHD6 and RSL1 that were necessary for physical associations with JAZ proteins. RHD6 and RSL1 were each divided into the N-terminal region (RHD61-193 or RSL11-200) and the C-terminal portion including the bHLH DNA binding domain (RHD6194-298 or RSL1200-307). The N-terminal fragments of RHD6 and RSL1 exhibited interactions with JAZ4 and JAZ8 in yeast, whereas removal of the N-terminal part eliminated these interactions (Supplemental Figure 3). Further mapping revealed that the amino acids 120 to 193 of RHD6 (RHD6120-193) and amino acids 120 to 200 of RSL1 (RSL1120-200) are sufficient for the interactions (Supplemental Figure 3).
To corroborate that RHD6 and RSL1 are interacting partners of JAZ proteins, we performed in vivo bimolecular fluorescence complementation (BiFC) assays in tobacco (Nicotiana benthamiana). The N-terminal fragment of yellow fluorescent protein (nYFP), driven by the cauliflower mosaic virus (CaMV) 35S promoter (Pro35S), was fused with RHD6 and RSL1 to produce RHD6-nYFP and RSL1-nYFP. Next, JAZ4 and JAZ8 proteins were fused with the C-terminal fragment of YFP (cYFP) to generate JAZ4-cYFP and JAZ8-cYFP. When JAZ4-cYFP or JAZ8-cYFP was coexpressed with RHD6-nYFP or RSL1-nYFP in tobacco leaves, strong YFP fluorescence was detected in the nuclei of transformed cells, as revealed by staining with 4',6-diamidino-2-phenylindole (DAPI; Figure 3B). Moreover, the YFP signal was also observed in tobacco leaves when JAZ4-cYFP or JAZ8-cYFP was coexpressed with the N-terminal domain of RHD6 or RSL1 fused with nYFP (RHD61–193-nYFP or RSL11–200-nYFP; Supplemental Figure 4). No obvious fluorescence was observed in negative controls when JAZ1-cYFP was coexpressed with RHD6-nYFP, RSL1-nYFP, or when RHD6194–298-nYFP (the C-terminal domain of RHD6 fused with nYFP) was coexpressed with JAZ4-cYFP or JAZ8-cYFP. In addition to BiFC assays, co-immunoprecipitation assays provided further in vivo evidence for the association between JAZ8 protein and RHD6 or RSL1 (Figure 3C). Based on these experiments, we conclude that RHD6 and RSL1 physically interact with several JAZ proteins, suggesting that they may act as direct targets of JAZ repressors.
RHD6 and RSL1 Are Essential for Jasmonate-Promoted Root Hair Development
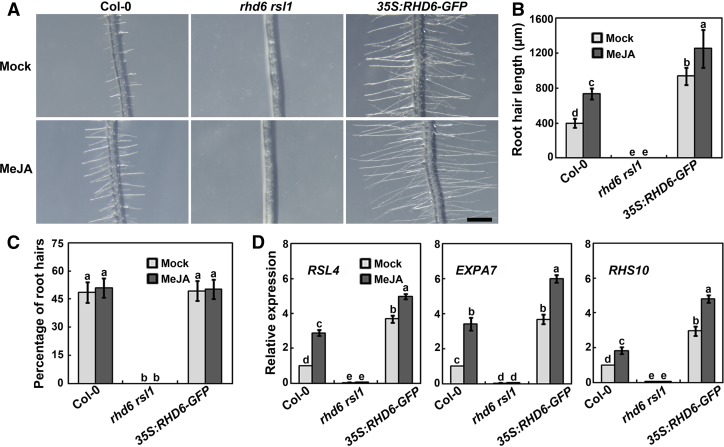
The RHD6 and RSL1 transcription factors positively regulate root hair growth in Arabidopsis (Masucci and Schiefelbein, 1994; Menand et al., 2007). Transgenic plants overexpressing RHD6 (Pro35S:RHD6-GFP) have noticeably longer root hairs, whereas the roots of rhd6 rsl1 double mutants are hairless (Menand et al., 2007). Having ascertained that JAZ repressors physically interact with RHD6 and RSL1, we followed up by asking whether jasmonate modulates root hair growth via these two factors. To test this, we investigated the responses of wild type, rhd6 rsl1 mutants, and Pro35S:RHD6-GFP plants to MeJA treatment. As expected, the root hair elongation of wild-type seedlings was obviously enhanced when treated with MeJA, although the percentage of visible roots hairs in total epidermal cells did not increase (Figures 4A to 4C). However, MeJA-induced root hair development was severely disrupted in rhd6 rsl1 mutants. As shown in Figures 4A and 4B, no root hairs were visible in rhd6 rsl1 mutants following the application of MeJA. Consistent with the phenotype, the basal and MeJA-induced expression of several root hair-related genes, such as RSL4, EXPA7, and RHS10, was dramatically decreased in rhd6 rsl1 mutants compared with wild type (Figure 4D). In contrast, root hairs of Pro35S:RHD6-GFP plants were much longer than those of wild type with or without MeJA stimulation (Figures 4A and 4B). Consistently, those genes analyzed exhibited much higher expression levels in Pro35S:RHD6-GFP plants in comparison with wild type (Figure 4D). These findings demonstrate that RHD6 and RSL1 transcription factors are necessary for jasmonate-stimulated root hair growth.
Figure 4.
RHD6 and RSL1 Are Required for Jasmonate-Stimulated Root Hair Development.
(A) Images of root hairs of Col-0, rhd6 rsl1, and Pro35S:RHD6-GFP upon mock or MeJA treatment. Experiments were performed more than three times with similar results. Bar = 500 μm.
(B) and (C) Quantitative analysis of the root hair length (B) and percentage of visible hair cells (C) in total epidermal cells in response to MeJA treatment. Seeds were germinated and grown for 4 d on half-strength MS medium and were then transferred to half-strength MS medium supplemented with 20 μM MeJA for 2 d before examination. Values are means ± sd (n = 12 roots).
(D) Expression of several root hair-related genes. Seeds of Col-0, rhd6 rsl1, and Pro35S:RHD6-GFP were germinated and grown for 4 d on half-strength MS medium, and then were transferred to half-strength MS medium supplemented with 0 (Mock) or 20 μM MeJA and grown for an additional 2 d before analysis. Error bars show SD from three independent biological replicates. Bars with different letters are significantly different from each other (P < 0.05). Data were analyzed by ANOVA.
Overexpression of RHD6 Largely Rescues the Root Hair Defects of coi1-2 Mutants and JAZ8-ΔJas-9 Transgenic Plants
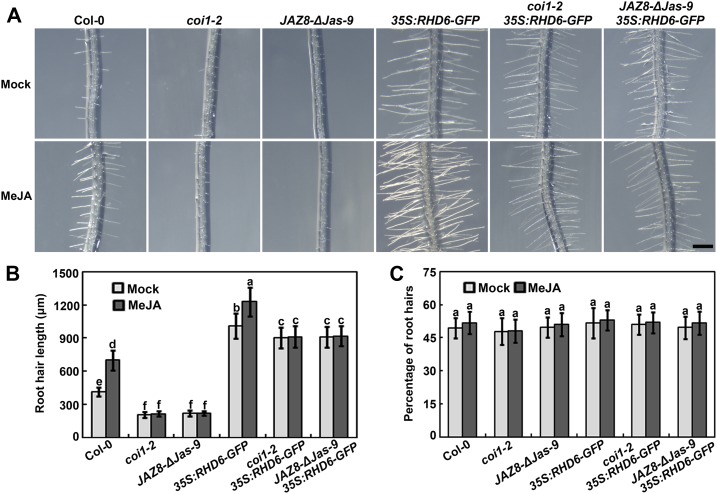
JAZ proteins repress jasmonate signaling by interacting with multiple transcription factors, and they overaccumulate in coi1 mutants, where they inhibit jasmonate responses (Chini et al., 2007; Thines et al., 2007; Katsir et al., 2008; Fonseca et al., 2009; Guo et al., 2018b). Because JAZ repressors physically associate with RHD6 and RSL1, we hypothesized that JAZ proteins might attenuate the function of these two factors, thus conferring the root hair-defective phenotype onto coi1 mutants. To verify this, we investigated whether overexpression of RHD6 could rescue the root hair defect of coi1-2. The coi1-2 mutant was genetically crossed with Pro35S:RHD6-GFP plants to generate coi1-2/Pro35S:RHD6-GFP. The root hair phenotype of coi1-2, Pro35S:RHD6-GFP, and coi1-2/Pro35S:RHD6-GFP was examined in response to MeJA. As expected, the root hair length, not the percentage of visible roots hairs in epidermis, was noticeably decreased in coi1-2 and was insensitive to MeJA stimulation (Figure 5). However, the phenotype of coi1-2/Pro35S:RHD6-GFP plants partially mimicked the root hair phenotype of Pro35S:RHD6-GFP, although their root hairs were shorter than those in the wild-type background. These results demonstrate that the transgenic expression of RHD6 compensates for the root hair development defect in coi1-2. To further dissect the genetic relationship between JAZ repressors and the RHD6 factor, we tested whether RHD6 overexpression could restore the phenotype of JAZ8-ΔJas-9 plants. Overexpression of RHD6 also largely rescued the root hair defect of JAZ8-ΔJas-9 plants with or without MeJA induction (Figures 5A and 5B). Our results imply that RHD6 functions downstream of COI1 and JAZ proteins to positively modulate jasmonate-promoted root hair development.
Figure 5.
Overexpression of RHD6 Partially Rescues the Root Hair Defects of coi1-2 and JAZ8-ΔJas-9.
(A) Images of root hairs of coi1-2 and related mutants or transgenic plants treated with (MeJA) or without (Mock) methyl jasmonate. Experiments were performed more than three times with similar results by analyzing different batches of seeds. Bar = 500 μm.
(B) and (C) Quantitative analysis of the root hair length (B) and percentage of visible hair cells in total epidermal cells (C) in response to MeJA treatment. Seeds were germinated and grown for 4 d on half-strength MS medium, and then were transferred to half-strength MS medium supplemented with 20 μM MeJA for an additional 2 d before examination. Values are means ± sd (n = 12 roots). Bars with different letters are significantly different from each other (P < 0.05). Data were analyzed by ANOVA.
JAZ Proteins Interfere with the Transcriptional Function of RHD6
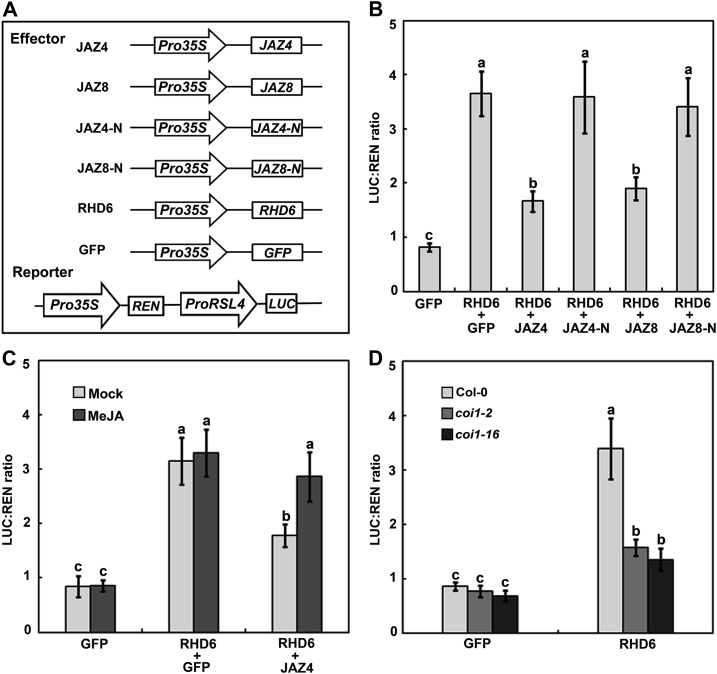
Having demonstrated that RHD6 and RSL1 transcription factors interact with JAZ proteins and are required for jasmonate-stimulated root hair growth, we further examined the regulatory effect of JAZ proteins on these factors. We initially used a dual-luciferase (LUC) reporter approach to analyze the regulation of JAZ proteins on the transcriptional function of RHD6 in wild-type Arabidopsis mesophyll protoplasts (Yoo et al., 2007). The effector constructs contained a GFP, JAZ4, JAZ8, or RHD6 gene driven by the CaMV 35S promoter (Figure 6A). Because the RSL4 gene is a direct target of RHD6 (Yi et al., 2010), its promoter (ProRSL4) was fused to LUC to produce a reporter construct (Figure 6A). Predictably, the expression of RHD6 dramatically elevated LUC expression driven by the RSL4 promoter (ProRSL4; Figure 6B; Feng et al., 2017). In contrast, the coexpression of RHD6 with JAZ4 or JAZ8 decreased LUC expression compared with the expression of RHD6 with GFP (Figure 6B). Furthermore, we coexpressed of RHD6 with the N-terminal fragment of JAZ4 (JAZ4-N, amino acids 1 to 176) or JAZ8 (JAZ8-N, amino acids 1 to 74), but no obvious difference of LUC expression was observed in comparison with expression of RHD6 with GFP (Figure 6B). These results suggest that JAZ proteins inhibit the transcriptional function of RHD6 and the inhibition depends on their physical interaction.
Figure 6.
JAZ Proteins Affect the Transcriptional Function of RHD6.
(A) Schematic of the effectors and reporters used in the transient transactivation assays. Pro35S, CaMV 35S promoter; ProRSL4, RSL4 promoter.
(B) Transient dual-luciferase reporter assays. JAZ4 and JAZ8, not JAZ4-N (amino acids 1 to 176) and JAZ8-N (amino acids 1 to 74), interfere with RHD6 to modulate the expression of RSL4. Error bars indicate SD from three biological replicates using different batches of plants.
(C) Transient dual-luciferase reporter assays. 100 μM MeJA attenuates the inhibition of JAZ4 on RHD6 transcription. Error bars indicate SD from three independent biological replicates using different batches of plants.
(D) Transient transcriptional activity assays. Activation of the RSL4 promoter by RHD6 is decreased in the coi1-2 and coi1-16. Error bars indicate SD from five independent biological replicates using different batches of plants. Bars with different letters are significantly different from each other (P < 0.05). Data were analyzed by ANOVA.
Previous studies revealed that the exogenous application of jasmonate induces the degradation of JAZ proteins (Chini et al., 2007; Thines et al., 2007). To verify the effect of JAZ proteins on the transcriptional role of RHD6, we investigated whether MeJA-induced JAZ protein degradation could attenuate the repression of JAZ proteins on RHD6. When the accumulation level of JAZ4 protein was reduced upon MeJA treatment, high-level LUC expression driven by ProRSL4 was detected in JAZ4-expressing wild-type Arabidopsis mesophyll protoplasts. (Figure 6C; Supplemental Figure 5). These findings imply that jasmonate reverses the interference of JAZ4 on the transcriptional function of RHD6. To gain a better understanding of whether the accumulation of endogenous JAZ proteins affects the transcriptional function of RHD6, we compared the abilities of RHD6 to stimulate downstream RSL4 in mesophyll protoplasts of wild type and the coi1-2 or coi1-16 mutant, which highly accumulates JAZ proteins. As shown in Figure 6D, an apparently reduced LUC expression driven by the RSL4 promoter was observed in RHD6-expressing coi1-2 or coi1-16 protoplasts compared with wild-type protoplasts. These results further support the notion that JAZ proteins interfere with the transcriptional function of RHD6 to modulate downstream targets.
RHD6 Interacts with RSL1, and JAZ8 Affects This Interaction
Dimerization of transcription factors is often a crucial regulatory mechanism in signal transduction (Klemm et al., 1998). Because RHD6 and RSL1 positively regulate root hair development redundantly, we tested whether they physically associate with themselves or each other to form homo- or heterodimers. To investigate this possibility, we used the yeast two-hybrid system to assay possible interactions. As shown in Figure 7A, RHD6 and RSL1 strongly interacted with both themselves and each other in yeast. Further analysis showed that the entire N-terminal fragment of RHD6 and the full-length of RSL1 were essential for these interactions with themselves and each other (Figure 7A; Supplemental Figure 6).
Figure 7.
JAZ8 Represses the Interaction between RHD6 and RSL1.
(A) Yeast two-hybrid assays showing the interaction between RHD6 and RSL1, and that the N-terminal part of RHD6 (RHD61-193) is responsible for the interaction. Yeast cells were plated onto dropout medium lacking Leu, Trp, His, and Ade and containing 3 mM 3-aminotriazole and observed after 2 d. BD, DNA binding domain; AD, Activation domain.
(B) BiFC analyses showing that RHD6 interacts with RSL1 in plant cells. Fluorescence was observed in nuclei of transformed tobacco cells, resulting from the complementation of RHD6-nYFP with RSL1-cYFP. No signal was obtained from negative controls in which RHD6-nYFP was coexpressed with RSL1200-307-cYFP (the C-terminal domain of RSL1 fused with cYFP), or RHD6194–298-nYFP (the C-terminal domain of RHD6 fused with nYFP) was coexpressed with RSL1-cYFP. Nuclei are indicated by DAPI staining. Bar = 15 μm.
(C) BiFC analyses showing that JAZ8 interferes with the interaction between RHD6 and RSL1. Similar results were obtained from at least three independent replicates. Bar = 30 μm.
(D) Quantitative analysis of YFP fluorescence intensity in (C). Fifty independent fluorescent spots were valued for fluorescence intensity. Error bars represent SD. Bars with different letters are significantly different from each other (P < 0.05). Data were analyzed by ANOVA.
To verify whether RHD6 and RSL1 transcription factors interact in plant cells, we performed BiFC assays. Full-length and truncated RSL1 proteins were ligated with the C-terminal YFP fragment driven by the CaMV 35S promoter to generate RSL1-cYFP and RSL1200-307-cYFP. When RHD6-nYFP or RHD61–193-nYFP was coexpressed with RSL1-cYFP in tobacco, strong YFP fluorescence was observed in the nuclei of transformed cells, as revealed by staining with DAPI (Figure 7B; Supplemental Figure 7). No fluorescence was observed in negative controls (Figure 7B; Supplemental Figure 7). These results demonstrate that RHD6 and RSL1 can interact to form a heterodimer. Because JAZ repressors directly bind RHD6 and RSL1 transcription factors (Figure 3), we performed BiFC assays to further investigate the effect of JAZ8 protein on the interaction of RHD6 with RSL1. When JAZ8 was coexpressed with RHD6-nYFP and RSL1-cYFP in tobacco, the YFP fluorescence signal was dramatically decreased (Figures 7C and D). As a control, the coexpression of JAZ1 with RHD6-nYFP and RSL1-cYFP did not affect the fluorescence intensity (Figures 7C and D). These results suggest that JAZ8 interferes with the interaction of RHD6 with RSL1.
DISCUSSION
The phytohormone jasmonate is a critical signal that regulates a wide range of physiological processes such as root elongation, male fertility, trichome development, anthocyanin accumulation, and stress responses (Wasternack and Hause, 2013; Chini et al., 2016; Huang et al., 2017; Hu et al., 2017; Zhang et al., 2017; Guo et al., 2018b; Howe et al., 2018). Jasmonate has also been reported to promote root hair development in Arabidopsis (Zhu et al., 2006, 2011); however, the detailed molecular mechanism remains largely unclear. In this study, we investigated the role of jasmonate and demonstrated that treatment with exogenous jasmonate (such as MeJA) has a stimulatory effect on root hair elongation, although the percentage of visible roots hairs in total epidermal cells did not increase in the presence of MeJA. Consistent with previous studies (Zhu et al., 2006, 2011), we found that wild-type seedlings had much longer root hairs in the presence of MeJA (Figures 1A and 1B). Consistently, expression analysis showed that transcripts of several genes positively related to root hair growth increased upon MeJA treatment (Figure 2). However, blocking endogenous jasmonate signaling attenuated root hair development. Disruption of the COI1 receptor or accumulation of the JAZ repressor resulted in a decrease of root hair length (Figures 1A and 1B). Collectively, our findings provide evidence that jasmonate positively regulates root hair elongation in Arabidopsis.
Jasmonate signal transduction involves a profound transcriptional reprogramming of cellular genetic programs associated with a complex interplay among negative and positive components (e.g., JAZ proteins and downstream transcription factors). JAZ repressors exhibit interactions with multiple transcription factors and interfere with their functions to suppress jasmonate signaling. The MYC transcription factors MYC2, MYC3, and MYC4 are the most extensively studied JAZ binding factors and mediate a subset of jasmonate processes, including the inhibition of root elongation and defense responses (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007; Fernández-Calvo et al., 2011). Recently, we demonstrated that JAZ proteins physically associate with INDUCER OF CBF EXPRESSION1 (ICE1) and ICE2 transcription factors to repress cold acclimation and freezing tolerance (Hu et al., 2013, 2017). Moreover, several crucial transcriptional modulators have been characterized as direct targets of JAZ proteins, including MYB24, GL3, ETHYLENE INSENSITIVE3 (EIN3), WRKY57, FILAMENTOUS FLOWER (FIL), and TARGET OF EAT1 (TOE1), which regulate male fertility, anthocyanin accumulation, stress responses, and flowering (Fernández-Calvo, 2011; Qi et al., 2011; Song et al., 2011b; Zhu, 2011; Jiang et al., 2014; Boter et al., 2015; Zhai et al., 2015). Despite the discovery of diverse transcription factors for jasmonate responses, the exact transcriptional regulation mechanisms underlying jasmonate-signaled processes, such as root hair elongation, remain to be elucidated. Further investigation of physical associations between JAZ proteins and critical regulators of other developmental processes will enhance our understanding of jasmonate signaling networks.
In this study, we found that several JAZ proteins physically interact with RHD6 and RSL1 transcription factors (Figure 3). RHD6 and RSL1 belong to the bHLH transcription factor family and contain a bHLH DNA binding domain. The N-terminal amino acids 120 to 193 of RHD6 (RHD6120-193) and amino acids 120 to 200 of RSL1 (RSL1120-200) are responsible for the interactions with JAZ4 and JAZ8 (Supplemental Figure 3). Interestingly, Fernández-Calvo et al. (2011) revealed that the interactions of MYC transcription factors with JAZ proteins also occur through their N-terminal regions. Further mapping in their study identified a conserved fragment, known as the JAZ Interaction Domain (JID), critical for the interactions with JAZ proteins. After BLAST analysis, we found that the RHD6120-193 and RSL1120-200 regions share several conserved residues with the JID domain of MYC transcription factors (Supplemental Figure 8), suggesting that RHD6 and RSL1 might also have a JID domain for the interactions with JAZ proteins in their N terminus. Dimerization of transcription factors is a crucial mechanism in signal transduction (Klemm et al., 1998). In this study, we found that RHD6 and RSL1 displayed interactions with themselves and each other to form homo- and heterodimers (Figures 7A and B). Further analysis demonstrated that the N-terminal fragment of RHD6 (RHD61-193) is sufficient for the formation of dimers (Supplemental Figures 6 and 7), implying that the C-terminal bHLH domain is dispensable for this process. Similarly, Cui et al. 2016 showed that the bHLH domain is also not essential for the dimerization of DYSFUNCTIONL TAPETUM1 (DYT1) with itself and its close homologs (such as bHLH089 and AMS). Feller et al. (2006) demonstrated that an ACT-like domain, not the bHLH domain, mediates the dimerization of several plant bHLH transcription factors. These findings show the involvement of other domains in dimerization of bHLH transcription factors, and further investigation may shed light on the distinct regulatory mechanisms of bHLH factors.
Our experimental results showed that the root hair development of rhd6 rsl1 was not responsive to jasmonate induction (Figure 4), indicating that jasmonate-stimulated root hair growth requires RHD6 and RSL1. We also observed that overexpression of RHD6 largely rescued the root hair defects of coi1-2 and JAZ8-ΔJas-9 plants (Figures 5A and 5B). Moreover, JAZ proteins repressed the transcriptional function of RHD6 (Figure 6) and interfered with the physical association of RHD6 and RSL1 (Figures 7C and 7D). These findings support the notion that RHD6 and RSL1 act as targets of JAZ repressors to modulate root hair elongation. Our study thus reveals a critical signaling module in which JAZ repressors of the jasmonate pathway directly regulate RHD6 and RSL1 transcription factors to integrate jasmonate signaling with the root-hair developmental process. Considering that the MYC2, MYC3, and MYC4 transcription factors regulate diverse aspects of jasmonate responses (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007; Fernández-Calvo et al., 2011; Chini et al., 2016; Howe et al., 2018), we investigated whether these master regulators are also involved in root hair development. Interestingly, the root hair length of the loss-of-function myc2-1 single mutant and myc2 myc3 myc4 triple mutant (Fernández-Calvo et al., 2011; Han et al., 2018) was comparable with its wild-type counterpart (Supplemental Figure 9). This observation suggests that these MYC transcription factors may not mediate jasmonate-stimulated root hair elongation. Consistently, no direct interaction between MYC transcription factors and RHD6 or RSL1 was observed in yeast (Supplemental Figure 10).
Exogenous application of jasmonate (e.g., MeJA) upregulated the expression levels of genes encoding RHD6 and several RSL transcription factors in wild-type seedlings (Figure 2). Several downstream target genes of these factors were responsive to MeJA treatment (Figure 2). In contrast, the basal and jasmonate-induced expression levels of these factors and their targets was decreased in coi1-2 and JAZ8-ΔJas-9 (Figure 2). Thus, the promotion of root hair elongation by jasmonate can be partially achieved through activating the transcription of RHD6 and RSL transcription factors. Taken together with the fact that JAZ proteins interact with and repress RHD6 and RSL1 (Figures 3 and 6), it is possible that jasmonate signaling exhibits dual regulatory effects on RHD6/RSL1-mediated root hair development. Similarly, Qi et al. (2011) showed that JAZ proteins directly bind and interfere with several MYB (e.g., MYB75 and GL1) and bHLH transcription factors (e.g., TRANSPARENT TESTA8 [TT8] and GL3) to regulate anthocyanin accumulation and trichome initiation. Their expression analyses also revealed that jasmonate induces the expression of those factors (Qi et al., 2011). Earlier studies found that the JAZ-interacting MYC2 transcription factor is rapidly induced by jasmonate (Chung et al., 2008; Pauwels et al., 2008). Based on the findings described above, we speculate that these dual regulations of downstream transcription factors by the jasmonate signal may be adaptive mechanisms to establish appropriate jasmonate signaling, thereby ensuring optimization for the growth and survival of plants.
Feng et al. (2017) demonstrated that the ethylene-activated EIN3 transcription factor physically interacts with RHD6 to form a transcriptional complex. The biochemical and genetic analysis in their study revealed that EIN3 and RHD6 directly coactivate the expression of RSL4 and other related genes to promote root hair elongation. These results indicate that EIN3 acts as a critical component in the RHD6/RSL-mediated root hair developmental process. Interestingly, EIN3 and its homolog EIN3-LIKE1 (EIL1) were also identified in another study as direct downstream targets of JAZ repressors in jasmonate responses (Zhu et al., 2011). Jasmonate-induced root hair growth is largely absent in ein3 eil1 double mutants. Consequently, jasmonate signaling may positively modulate RHD6/RSL-mediated root hair growth partially through EIN3 and EIL1. Based on the phenotypes of double mutants rhd6 rsl1 and ein3 eil1 in response to MeJA induction (Figure 4; Zhu et al., 2011), we speculate that RHD6/RSL1 factors play a dominant role in the regulation of jasmonate-stimulated root hair growth. To better understand the molecular mechanism of jasmonate-signaled root hair elongation in Arabidopsis, we propose the following simplified model involving RHD6/RSL and EIN3/EIL1 (Figure 8): when the concentration of jasmonate is elevated, the COI1–JAZ coreceptor perceives jasmonate and JAZ repressors are degraded via the SCFCOI1-26S proteasome pathway (Chini et al., 2007; Thines et al., 2007; Yan et al., 2009; Sheard et al., 2010). The degradation of JAZ proteins subsequently releases RHD6/RSL1 and EIN3/EIL1 transcription factors to form a transcriptional complex, which coordinately regulates the expression of genes essential for root hair growth (Figures 3 and 7; Zhu et al., 2011; Feng et al., 2017). Additionally, jasmonate signaling activates the expression of RHD6 and several RSL factors, thus enhancing root hair elongation (Figure 2).
Figure 8.
A Simplified Model for Jasmonate-Stimulated Root Hair Development in Arabidopsis.
When the concentration of jasmonate is elevated, the COI1–JAZ coreceptor perceives jasmonate and JAZ repressors are degraded. The degradation of JAZ proteins subsequently releases RHD6/RSL1 and EIN3/EIL1 transcription factors to form a transcriptional complex, which coordinately regulates the expression of genes essential for root hair development.
METHODS
Materials and Plant Growth Conditions
The phytohormone MeJA was purchased from Sigma-Aldrich. Taq DNA polymerases were obtained from Takara Biotechnology (Dalian, China), and other common chemicals were from Shanghai Sangong (Shanghai, China). Anti-MYC (catalog no. A7470, Sigma-Aldrich) and anti-HA (catalog no. H9658, Sigma-Aldrich) antibodies were purchased from Sigma-Aldrich. The wild-type and mutant Arabidopsis (Arabidopsis thaliana) plants used in this study were in the Columbia (Col-0) genetic background. The mutants coi1-2 (Xu et al., 2002), coi1-16 (Ellis and Turner, 2002), JazQ (Campos et al., 2016), and myc2 myc3 myc4 (Fernández-Calvo et al., 2011) were described previously. rhd6 rsl1 and transgenic Pro35S:RHD6-GFP plants were gifts from Hongwei Guo (Southern University of Science and Technology, Shenzhen, China). To generate JAZ4-ΔJas-7 and JAZ8-ΔJas-9 transgenic plants, the full-length cDNAs of JAZ4 and JAZ8 with the Jas domain (critical for the degradation of JAZ proteins) of each deleted were cloned into the binary vector pOCA30 in the sense orientation driven by the CaMV 35S promoter (Han et al., 2018). coi1-2/Pro35S:RHD6-GFP and JAZ8-ΔJas-9/Pro35S:RHD6-GFP were generated by genetic crossing using standard techniques. Arabidopsis seeds were sterilized (20% [v/v] bleach for 15 min) then sown on half-strength Murashige and Skoog (MS) medium and kept at 4°C for 3 d before germination. Plants were grown in an artificial growth chamber at 22°C under a 16-h-light (100 mE m−2 s−1, white fluorescent bulbs, full-spectrum light)/8-h-dark photoperiod.
MeJA Treatment
MeJA was dissolved in 10% (v/v) ethanol as a 10-mM stock solution. Arabidopsis seeds were first germinated and grown for 4 d on half-strength MS medium, and then were transferred to half-strength MS medium supplemented with 20 μM MeJA for 2 d. In the mock treatment, an equal volume of 10% (v/v) ethanol was added to the medium.
Microscopy
All root hairs shown in the same root range (the first 2 mm of the zone of maturation) were counted under an SZX16 microscope (Olympus) as described by Schnall and Quatrano (1992). A total of 12 similarly sized roots were counted. To measure hair length, both sides of the root in the same range were measured. Olympus software (Moon et al., 2019) was used to measure hair length. To assay the number of root hair cells and hairless cells in the root epidermis, at least 12 seedlings (6-d-old) were observed (Lee and Schiefelbein, 2002). In each root, seven adjacent epidermal cells from the same cell file were scored, and 28 cells from two hair cell files and the adjacent two hairless cell files were counted. Four trials were performed for each genotype with or without MeJA treatment. A cell was scored as a root hair cell if any protrusion was visible, regardless of the root hair length. Mean root hair length and percentage of visible hair cells in total epidermal cells were compared using analysis of variance (ANOVA).
RNA Extraction and RT-qPCR
Total RNA (for each sample, roots were collected) was extracted from roots using the TRIzol reagent (Invitrogen), and qRT-PCR was performed as described by Han et al. (2018). Briefly, 1 μg DNase-treated root total RNA was reverse-transcribed in a 20-μL reaction volume using Moloney murine leukemia virus reverse transcriptase (Fermentas) with oligo (dT) 18 primers. Then, 1 μL cDNA was used in a qRT-PCR reaction with the SYBR Premix Ex Taq kit (Takara) on a Roche Light Cycler 480 real-time PCR machine. RNA was extracted from at least three batches of roots as biological replicates for qRT-PCR analysis and at least three technical replicates were analyzed for each sample. Changes in expression of the target gene were calculated relative to the expression of ACTIN2 (AT3G18780) gene. The gene-specific primers used for transcript analysis are listed in the Supplemental Data Set.
Yeast Two-Hybrid Assays
To analyze the physical interactions of RHD6 and RSL1 with JAZ proteins, the full-length RHD6 and RSL1 coding sequences (CDS), respectively, were cloned into the bait vector pGBKT7 (Clontech) to produce BD-RHD6 and BD-RSL1, which contain the Gal4 DNA binding domain (BD). Full-length CDS of JAZ were inserted into pGADT7 (Clontech) to generate prey vectors (AD-JAZ) containing the Gal4 activation domain (AD). To identify specific regions responsible for the interactions, a variety of truncated forms of RHD6 or RSL1 were cloned into pGBKT7, and truncated forms of JAZ4 or JAZ8 were cloned into pGADT7. To analyze whether RHD6 and RSL1 interact with themselves or each other to form dimers, full-length CDS of RHD6 and RSL1 were also cloned into pGADT7. Yeast two-hybrid assays were performed as described previously (Hu et al., 2013). Briefly, the bait and prey vectors were cotransformed into the yeast strain AH109 (Yu et al., 2010), and interactions were indicated by the ability of cells to grow on dropout medium lacking Leu, Trp, His, and Ade and containing 3 mM 3-aminotriazole.
BiFC Assays
The cDNA sequences of the N-terminal 173-amino acid–enhanced YFP (nYFP) and C-terminal 64-amino acid YFP (cYFP) fragments were PCR amplified and individually cloned into tagging pFGC5941 plasmids to produce pFGC-nYFP and pFGC-cYFP, respectively (Kim et al., 2008). Full-length or truncated RHD6 was inserted into pFGC-nYFP to generate an N-terminal in-frame fusion with nYFP (RHD6-nYFP, RHD61-193-nYFPor RHD6194-298-nYFP). Similarly, full-length or truncated RSL1 was fused with nYFP or cYFP to generate RSL1-nYFP, RSL1-cYFP, RSL11-200-cYFP, or RSL1200-307-cYFP. Full-length JAZ1, JAZ4, or JAZ8 was inserted into pFGC-cYFP to generate a C-terminal in-frame fusion with cYFP (JAZ1-cYFP, JAZ4-cYFP, or JAZ8-cYFP). The resulting plasmids were transformed into Agrobacterium tumefaciens (strain EHA105; Pan et al., 2018), and the infiltration of wild tobacco (Nicotiana benthamiana) leaves was performed as described previously (Hu et al., 2019). The experiments were performed at least four times using different batches of plants; for each biological replicate, more than 12 wild tobacco plants were infiltrated and more than 600 cells were examined. Infected leaves were analyzed 48 h after infiltration. YFP and DAPI fluorescence were examined under a confocal laser-scanning microscope (Olympus). The primers used for cloning are listed in the Supplemental Data Set.
Co-immunoprecipitation Assays
Full-length CDS of RHD6, RSL1, and GFP were individually cloned into tagging plasmids with MYC or HA tag sequences in the sense orientation driven by the CaMV 35S promoter (Hu et al., 2013). MYC-fused JAZ8 and HA-fused RHD6 or HA-fused RSL1 were then coexpressed transiently in N. benthamiana. Infected leaves were sectioned 48 h after infiltration. Co-immunoprecipitation assays were performed using total leaf protein extracts as described previously (Hu et al., 2013). Briefly, MYC-fused JAZ8 were immunoprecipitated using an anti-MYC antibody (catalog no. A7470, Sigma-Aldrich; 1:250), and the coimmunoprecipitated protein was then detected using an anti-HA antibody (catalog no. H9658, Sigma-Aldrich; 1:10000). The primers used for cloning are listed in the Supplemental Data Set.
Transient Transcriptional Activation Assays
Full-length CDS of RHD6, JAZ8, JAZ4 (with a HA tag), and GFP (with a HA tag) were individually amplified and cloned into the pGreenII 62-SK vector (Biovector Science Lab) driven by the CaMV 35S promoter as effectors. The N-terminal fragment of JAZ4 (JAZ4-N, amino acids 1 to 176) and JAZ8 (JAZ8-N, amino acids 1 to 74) were also amplified and cloned into pGreenII 62-SK. The putative promoter sequences of the RSL4 were amplified by PCR and cloned into the pGreenII 0800-LUC vector (Biovector Science Lab) as the reporter (Hellens et al., 2005). Combinations of plasmids were transformed into Arabidopsis leaf mesophyll protoplasts according to the Sheen laboratory protocol (Sheen, 2001). Transfected cells were cultured for 16 to 18 h with or without 100 μM MeJA treatment (for 10 h), and then relative LUC activity was analyzed using a Dual-Luciferase Reporter Assay system (Promega), which measured the activities of firefly LUC and the internal control Renilla reniformis LUC (REN). The primers used for cloning are listed in the Supplemental Data Set.
Statistical Analysis
Statistical analysis was performed by ANOVA or Student's t test. The results are shown in the Supplemental File.
Accession Numbers
Arabidopsis Genome Initiative identification numbers for the genes discussed in this article are as follows: COI1, AT2G39940; JAZ1, AT1G19180; JAZ2, AT1G74950; JAZ3, AT3G17860; JAZ4, AT1G48500; JAZ5, AT1G17380; JAZ6, AT1G72450; JAZ7, AT2G34600; JAZ8, AT1G30135; JAZ9, AT1G70700; JAZ10, AT5G13220; JAZ11, AT3G43440; JAZ12, AT5G20900; RSL1, AT5G37800; RSL2, AT4G33880; RSL3,AT2G14760; RSL4, AT1G27740; RSL5, AT5G43175; RHD6, AT1G66470; LRX1, AT1G12040; RHS10, AT1G70460; EXPA7, AT1G12560; and ACTIN2, AT3G18780.
Supplemental Data
Supplemental Figure 1. Identification of JAZ4-ΔJas and JAZ8-ΔJas transgenic plants.
Supplemental Figure 2. The C-terminal region of JAZ4 or JAZ8 is involved in the interactions with RHD6 and RSL1.
Supplemental Figure 3. The N-terminal part of RHD6 or RSL1 is responsible for the interactions with JAZ4 and JAZ8.
Supplemental Figure 4. BiFC assay shows that the N-terminal part of RHD6 or RSL1 interacts with JAZ4 and JAZ8 in N. benthamiana.
Supplemental Figure 5. Accumulation of JAZ4 in Arabidopsis leaf mesophyll protoplasts.
Supplemental Figure 6. The entire N-terminal fragment of RHD6 and the full-length of RSL1 are necessary for the interactions with themselves and each other in yeast.
Supplemental Figure 7. BiFC assay shows that the N-terminal part of RHD6 interacts with itself and RSL1 in N. benthamiana.
Supplemental Figure 8. Alignment of the JAZ-interaction domain (JID) of RHD6 and related transcription factors.
Supplemental Figure 9. Root-hair phenotypes of myc2-1 and myc2 myc3 myc4 mutants.
Supplemental Figure 10. Yeast two-hybrid assay analysis of the interactions between RHD6 and MYC transcription factors.
Supplemental Data Set. Primers used for cloning and qRT-PCR.
Supplemental File. ANOVA and Student's t test tables.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
RSL Gramene: ROOT HAIR DEFECTIVE-SIX LIKE
RSL Araport: ROOT HAIR DEFECTIVE-SIX LIKE
RHD6 Gramene: ROOT HAIR DEFECTIVE SIX
RHD6 Araport: ROOT HAIR DEFECTIVE SIX
Acknowledgments
We thank Daoxin Xie (Tsinghua University), Gregg A. Howe (Michigan State University), Roberto Solano (Campus Universidad Autónoma), Hongwei Guo (Southern University of Science and Technology), Xiaoya Chen (Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences), Zhixiang Chen (Purdue University), and Feng Yu (Hunan University) for sharing research materials. We also thank Sirui Zhu (Hunan University) and the Central Laboratory of Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences for technical support. This work was supported by the Natural Natural Science Foundation of China (NSFC) (grants 31670281, 31870258, and 31922009 to Y.H.), the Applied Basic Research Foundation of Yunnan Province (Applied and Basic Research Foundation of Yunnan Provincial Science and Technology Commission) (grants 2017FA005 and 2019FI006 to Y.H.), the Youth Innovation Promotion Association of the Chinese Academy of Sciences (Youth Innovation Promotion Association CAS) (to Y.H.), Yunnan High Level Talents Special Support Plan (grant YNWR-QNBJ-2018-075 to Y.H.), and Candidates of the Young and Middle Aged Academic Leaders of Yunnan Province (grant 2017HB067 to Y.H.).
AUTHOR CONTRIBUTIONS
X.H. and Y.H. designed this study, interpreted data, and wrote the article; X.H., M.Z., and M.Y. performed experiments; all authors read and approved the final article.
References
- Balbi V., Devoto A.(2008). Jasmonate signalling network in Arabidopsis thaliana: Crucial regulatory nodes and new physiological scenarios. New Phytol. 177: 301–318. [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Lee M.M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J.(2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439. [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Zhao M., Gonzalez A., Lloyd A., Schiefelbein J.(2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298. [DOI] [PubMed] [Google Scholar]
- Boter M., Golz J.F., Giménez-Ibañez S., Fernandez-Barbero G., Franco-Zorrilla J.M., Solano R.(2015). FILAMENTOUS FLOWER is a direct target of JAZ3 and modulates responses to jasmonate. Plant Cell 27: 3160–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S.(2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruex A., Kainkaryam R.M., Wieckowski Y., Kang Y.H., Bernhardt C., Xia Y., Zheng X., Wang J.Y., Lee M.M., Benfey P., Woolf P.J., Schiefelbein J.(2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M.L., Yoshida Y., Major I.T., de Oliveira Ferreira D., Weraduwage S.M., Froehlich J.E., Johnson B.F., Kramer D.M., Jander G., Sharkey T.D., Howe G.A.(2016). Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 7: 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J.(2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R.(2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Chini A., Gimenez-Ibanez S., Goossens A., Solano R.(2016). Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 33: 147–156. [DOI] [PubMed] [Google Scholar]
- Chung H.S., Koo A.J., Gao X., Jayanty S., Thines B., Jones A.D., Howe G.A.(2008). Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146: 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., You C., Zhu E., Huang Q., Ma H., Chang F.(2016). Feedback regulation of DYT1 by interactions with downstream bHLH factors promotes DYT1 nuclear localization and anther development. Plant Cell 28: 1078–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Prescott H., Dolan L.(2015). Intensity of a pulse of RSL4 transcription factor synthesis determines Arabidopsis root hair cell size. Nat. Plants 1: 15138. [DOI] [PubMed] [Google Scholar]
- Di Cristina M., Sessa G., Dolan L., Linstead P., Baima S., Ruberti I., Morelli G.(1996). The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 10: 393–402. [DOI] [PubMed] [Google Scholar]
- Dolan L., Duckett C.M., Grierson C., Linstead P.J., Schneider K., Lawson E., Dean C., Poethig S., Roberts K.(1994). Clonal relations and patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474. [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K.(2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C., Turner J.G.(2002). A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215: 549–556. [DOI] [PubMed] [Google Scholar]
- Feller A., Hernandez J.M., Grotewold E.(2006). An ACT-like domain participates in the dimerization of several plant basic-helix-loop-helix transcription factors. J. Biol. Chem. 281: 28964–28974. [DOI] [PubMed] [Google Scholar]
- Feng Y., Xu P., Li B., Li P., Wen X., An F., Gong Y., Xin Y., Zhu Z., Wang Y., Guo H.(2017). Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: 13834–13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chico J.M., Solano R.(2009). The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12: 539–547. [DOI] [PubMed] [Google Scholar]
- Galway M.E., Masucci J.D., Lloyd A.M., Walbot V., Davis R.W., Schiefelbein J.W.(1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166: 740–754. [DOI] [PubMed] [Google Scholar]
- Guo Q., Major I.T., Howe G.A.(2018a). Resolution of growth-defense conflict: mechanistic insights from jasmonate signaling. Curr. Opin. Plant Biol. 44: 72–81. [DOI] [PubMed] [Google Scholar]
- Guo Q., Yoshida Y., Major I.T., Wang K., Sugimoto K., Kapali G., Havko N.E., Benning C., Howe G.A.(2018b). JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 115: E10768–E10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Hu Y., Zhang G., Jiang Y., Chen X., Yu D.(2018). Jasmonate negatively regulates stomatal development in Arabidopsis cotyledons. Plant Physiol. 176: 2871–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A.(2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A., Major I.T., Koo A.J.(2018). Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant Biol. 69: 387–415. [DOI] [PubMed] [Google Scholar]
- Hu Y., Han X., Yang M., Zhang M., Pan J., Yu D.(2019). The transcription factor INDUCER OF CBF EXPRESSION1 interacts with ABSCISIC ACID INSENSITIVE5 and DELLA proteins to fine-tune abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 31: 1520–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Jiang L., Wang F., Yu D.(2013). Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25: 2907–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Jiang Y., Han X., Wang H., Pan J., Yu D.(2017). Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 68: 1361–1369. [DOI] [PubMed] [Google Scholar]
- Huang H., Liu B., Liu L., Song S.(2017). Jasmonate action in plant growth and development. J. Exp. Bot. 68: 1349–1359. [DOI] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T.(2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59: 365–386. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Liang G., Yang S., Yu D.(2014). Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26: 230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.H., Kirik V., Hulskamp M., Nam K.H., Hagely K., Lee M.M., Schiefelbein J.(2009). The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21: 1080–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Chung H.S., Koo A.J., Howe G.A.(2008). Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11: 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M.(2013). MYC2: The master in action. Mol. Plant 6: 686–703. [DOI] [PubMed] [Google Scholar]
- Kim K.C., Lai Z., Fan B., Chen Z.(2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20: 2357–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm J.D., Schreiber S.L., Crabtree G.R.(1998). Dimerization as a regulatory mechanism in signal transduction. Annu. Rev. Immunol. 16: 569–592. [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J.(1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483. [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J.(2002). Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14: 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Ohashi Y., Kato M., Tsuge T., Gu H., Qu L.J., Aoyama T.(2015). GLABRA2 directly suppresses basic helix-loop-helix transcription factor genes with diverse functions in root hair development. Plant Cell 27: 2894–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R.(2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano S., et al. (2017). Molecular link between auxin and ROS-mediated polar growth. Proc. Natl. Acad. Sci. USA 114: 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J.D., Schiefelbein J.W.(1994). The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an Auxin- and Ethylene-associated Process. Plant Physiol. 106: 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J.D., Schiefelbein J.W.(1996). Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8: 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B., Yi K., Jouannic S., Hoffmann L., Ryan E., Linstead P., Schaefer D.G., Dolan L.(2007). An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480. [DOI] [PubMed] [Google Scholar]
- Moon S., et al. (2019). RSL class II transcription factors guide the nuclear localization of RHL1 to regulate root hair development. Plant Physiol. 179: 558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Figueroa P., Browse J.(2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62: 2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Wang H., Hu Y., Yu D.(2018). Arabidopsis VQ18 and VQ26 proteins interact with ABI5 transcription factor to negatively modulate ABA response during seed germination. Plant J. 95: 529–544. [DOI] [PubMed] [Google Scholar]
- Pauwels L., Morreel K., De Witte E., Lammertyn F., Van Montagu M., Boerjan W., Inzé D., Goossens A.(2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 105: 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C.T., Zhang F., Lloyd A.M.(2000). GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires N.D., Yi K., Breuninger H., Catarino B., Menand B., Dolan L.(2013). Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc. Natl. Acad. Sci. USA 110: 9571–9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Huang H., Song S., Xie D.(2015). Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 27: 1620–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D.(2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie W.G., Feldmann K.A., Marks M.D.(1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8: 1388–1399. [DOI] [PubMed] [Google Scholar]
- Ryu K.H., Kang Y.H., Park Y.H., Hwang I., Schiefelbein J., Lee M.M.(2005). The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development 132: 4765–4775. [DOI] [PubMed] [Google Scholar]
- Sanders P.M., Lee P.Y., Biesgen C., Boone J.D., Beals T.P., Weiler E.W., Goldberg R.B.(2000). The arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J., Kwak S.H., Wieckowski Y., Barron C., Bruex A.(2009). The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. J. Exp. Bot. 60: 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnall J.A., Quatrano R.S.(1992). Abscisic acid elicits the water-stress response in root hairs of Arabidopsis thaliana. Plant Physiol. 100: 216–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F., Fernández-Calvo P., Zander M., Diez-Diaz M., Fonseca S., Glauser G., Lewsey M.G., Ecker J.R., Solano R., Reymond P.(2013). Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25: 3117–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J.(2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127: 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Song S.K., Ryu K.H., Kang Y.H., Song J.H., Cho Y.H., Yoo S.D., Schiefelbein J., Lee M.M.(2011a). Cell fate in the Arabidopsis root epidermis is determined by competition between WEREWOLF and CAPRICE. Plant Physiol. 157: 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D.(2011b). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Browse J.(2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97: 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J.(2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Wasternack C., Hause B.(2013). Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Li J.(2018). Receptor-like protein kinases: Key regulators controlling root hair development in Arabidopsis thaliana. J. Integr. Plant Biol. 60: 841–850. [DOI] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G.(1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D.(2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D.(2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K., Menand B., Bell E., Dolan L.(2010). A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 42: 264–267. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J.(2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu F., et al. (2010). ANK6, a mitochondrial ankyrin repeat protein, is required for male-female gamete recognition in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 22332–22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q., Zhang X., Wu F., Feng H., Deng L., Xu L., Zhang M., Wang Q., Li C.(2015). Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 27: 2814–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A.(2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhang F., Melotto M., Yao J., He S.Y.(2017). Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 68: 1371–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Huang L., Yan A., Liu Y., Liu B., Yu C., Zhang A., Schiefelbein J., Gan Y.(2016). Multiple phytohormones promote root hair elongation by regulating a similar set of genes in the root epidermis in Arabidopsis. J. Exp. Bot. 67: 6363–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Gan L., Shen Z., Xia K.(2006). Interactions between jasmonates and ethylene in the regulation of root hair development in Arabidopsis. J. Exp. Bot. 57: 1299–1308. [DOI] [PubMed] [Google Scholar]
- Zhu Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 12539–12544. [DOI] [PMC free article] [PubMed] [Google Scholar]