The OPR protein MTHI1 is a major actor in the biogenesis of chloroplast ATP synthase in green algae that coregulates the expression of AtpH and AtpI, the two subunits of the proton channel.
Abstract
In the green alga Chlamydomonas (Chlamydomonas reinhardtii), chloroplast gene expression is tightly regulated posttranscriptionally by gene-specific trans-acting protein factors. Here, we report the identification of the octotricopeptide repeat protein MTHI1, which is critical for the biogenesis of chloroplast ATP synthase oligomycin-sensitive chloroplast coupling factor. Unlike most trans-acting factors characterized so far in Chlamydomonas, which control the expression of a single gene, MTHI1 targets two distinct transcripts: it is required for the accumulation and translation of atpH mRNA, encoding a subunit of the selective proton channel, but it also enhances the translation of atpI mRNA, which encodes the other subunit of the channel. MTHI1 targets the 5′ untranslated regions of both the atpH and atpI genes. Coimmunoprecipitation and small RNA sequencing revealed that MTHI1 binds specifically a sequence highly conserved among Chlorophyceae and the Ulvale clade of Ulvophyceae at the 5′ end of triphosphorylated atpH mRNA. A very similar sequence, located ∼60 nucleotides upstream of the atpI initiation codon, was also found in some Chlorophyceae and Ulvale algae species and is essential for atpI mRNA translation in Chlamydomonas. Such a dual-targeted trans-acting factor provides a means to coregulate the expression of the two proton hemi-channels.
INTRODUCTION
In chloroplasts, photosynthetic energy conversion is performed by oligomeric protein complexes comprising subunits of dual genetic origin. Indeed, due to the extensive gene transfer from the cyanobacterial ancestor of chloroplasts to the nucleus of the host cell, only some subunits of the photosynthetic apparatus are still organelle encoded, whereas others are expressed in the nucleo-cytosol and then imported into organelles. Thus, the assembly of photosynthetic protein complexes requires tight cooperation between two genetic compartments to avoid the wasteful or even deleterious accumulation of unassembled subunits. The first level of coordination between the two genetic compartments involves a plethora of nucleus-encoded factors that tightly control each posttranscriptional step of chloroplast gene expression: processing, trimming, splicing, editing, stabilization, translation activation, and decay of chloroplast RNAs (reviewed by Barkan and Goldschmidt-Clermont, 2000; Schmitz-Linneweber and Small, 2008; Woodson and Chory, 2008; Germain et al., 2013; Zoschke and Bock, 2018). Thanks to this nuclear control of chloroplast gene expression that emerged after endosymbiosis, gene expression remains proportional in the chloroplast and nucleo-cytosol, despite the huge imbalance in gene copy number, which may differ by as many as four orders of magnitude. In the unicellular green alga Chlamydomonas (Chlamydomonas reinhardtii), nucleus-encoded factors primarily belong to two major functional classes: the M factors involved in chloroplast mRNA maturation and stabilization and the T factors required for mRNA translation activation (Choquet and Wollman, 2002). Most of these factors belong to helical repeat protein families, such as PentatricoPeptide Repeat (PPR), Half A Tetratricopeptide repeat (HAT), mitochondrial TERmination Factor (mTERF), and octatricopeptide repeat (OPR) proteins (reviewed by Barkan and Small, 2014; Hammani et al., 2014). These proteins comprise tandem repeats of simple structural motifs that fold into antiparallel α helices and stack onto each other to form a concave surface well suited to interact with RNA molecules. Each repeat contacts one specific nucleotide via amino acids at determined positions, thereby allowing sequence-specific recognition. While the PPR family has greatly expanded in land plants, with more than 450 members in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), it remains limited in green algae (14 PPR proteins in C. reinhardtii; Tourasse et al., 2013), which instead express numerous OPR proteins (>125 in C. reinhardtiis versus only 1 in Arabidopsis).
Beside this nuclear control of chloroplast gene expression, other fine-tuning regulatory mechanisms set the synthesis of the individual subunits of a photosynthetic protein to the stoichiometry required for their functional assembly, as shown by the pleiotropic loss of all subunits of a complex in any mutant lacking expression of one of its major subunits. Two major mechanisms account for this concerted accumulation in Chlamydomonas (reviewed by Choquet and Vallon, 2000). Some subunits, particularly those encoded in the nucleus, are expressed normally but rapidly degraded when they cannot assemble, while many chloroplast-encoded subunits of the photosynthetic apparatus show assembly-dependent regulation of their synthesis, a process known as the control by epistasy of synthesis (CES) process (Choquet and Wollman, 2009). In the absence of their assembly partners, the rate of synthesis of CES subunits is dramatically reduced. In most cases, the CES process relies on negative feedback mediated by the unassembled CES subunit on its own translation (Choquet et al., 1988, 2003; Wostrikoff et al., 2004; Minai et al., 2006; Wostrikoff and Stern, 2007; Choquet and Wollman, 2009). However, the CES processes that control the biogenesis of the CF1 sector of ATP synthase present atypical features, accounting for the 3:3:1 uneven stoichiometry in the synthesis of the α, β, and γ subunits, respectively (Drapier et al., 2007).
Little is known about the mechanisms that ensure the 1:1:14:1 accumulation of the AtpF, AtpG, AtpH, and AtpI subunits, respectively, of the oligomycin-sensitive chloroplast coupling factor (CFo). The acetate-requiring ac46 mutant, which was isolated more than half a century ago (Levine, 1960), was later characterized as defective in photosynthesis due to a single nuclear mutation (Levine and Goodenough, 1970). The ac46 mutant does not express the chloroplast-encoded AtpH subunit (Lemaire and Wollman, 1989b) because it does not accumulate the monocistronic atpH mRNA (Majeran et al., 2001). Beside defective expression of AtpH, this mutant also shows strongly reduced synthesis of AtpI, another chloroplast-encoded CFo subunit (Lemaire and Wollman, 1989b), that, together with the tetradecameric ring of AtpH subunits, forms the membrane-embedded proton channel. The mutation thus affects the MTHI1 gene, whose product is required for the Maturation/stability and Translation of the atpH and atpI mRNAs, and the ac46 mutation was renamed mthi1-1. The coupled expression of AtpH and AtpI was possibly indicative of a CES relationship. In the mitochondria of the yeast Saccharomyces cerevisiae, mutants lacking expression of Atp9p, the mitochondrion-encoded counterpart of AtpH, show reduced synthesis of Atp6p and Atp8p, the former being the mitochondrial equivalent of AtpI (Jean-Francois et al., 1986; Ooi et al., 1987; Payne et al., 1991; Bietenhader et al., 2012). Together, these results prompted us to investigate the expression of the atpH and atpI genes in mthi1 mutants.
In this study, we characterized the MTHI1 OPR protein, a protein that is critical for the biogenesis of the chloroplast ATP synthase CFo. We showed that this protein controls the accumulation and translation of atpH mRNA, encoding a subunit of the selective proton channel, and enhances the translation of atpI mRNA, which encodes the other subunit of the channel. Finally, we identified the nucleotide targets of MTHI1.
RESULTS
Lack of MTHI1 Leads to the Reduced Accumulation and Translation of atpI mRNA
We recovered a photosynthetic mutant, kindly provided by Rachel Dent, generated by insertional mutagenesis with a paromomycin resistance cassette (Dent et al., 2005), originally called CAL014.01.38. This mutant shows the same phenotype as mthi1-1, lacks atpH mRNA, hence AtpH synthesis (Figure 1C) and accumulation of all subunits of the ATP synthase complex (Figure 1B). In addition, it shows a strongly reduced synthesis of AtpI in 14C pulse-labeling experiments (Figure 1C). Therefore, we renamed this mutant mthi1-2.
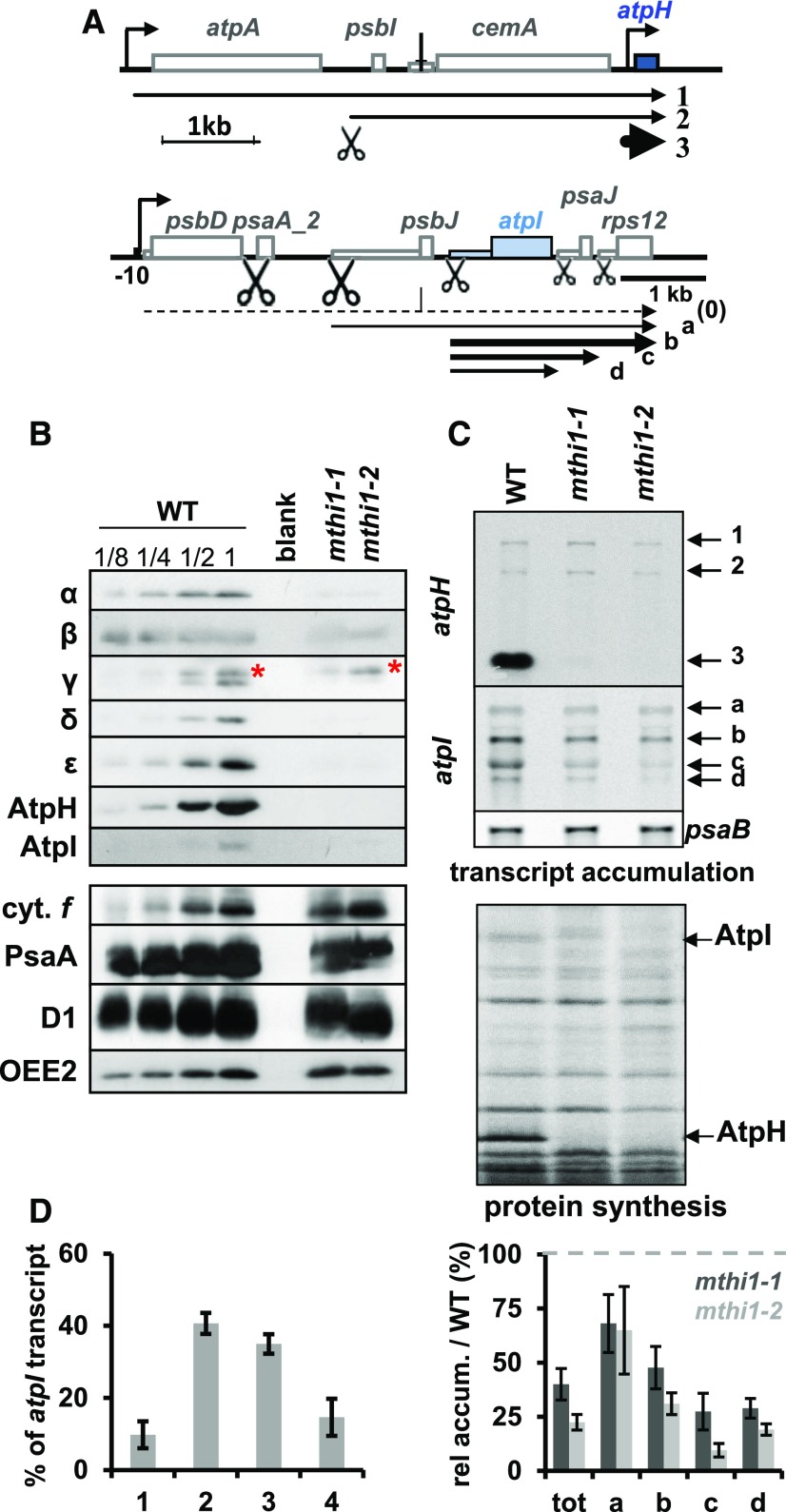
Figure 1.
Phenotypes of the mthi1 Mutants.
(A) Schematic representation of the atpH (top) and atpI (bottom) transcription units. CDSs are shown as thick rectangles, while 5′ UTRs are depicted as thin rectangles. Bent arrows represent promoters. The major transcripts detected in (C) with probes specific to atpH or atpI are indicated. (0) represents a precursor transcript that cannot be observed in the wild type because it is efficiently processed but can be detected in psaA trans-splicing mutants (Choquet et al., 1988). Scissors indicate the positions of processing events, whose efficiency is symbolized by their size.
(B) Pleiotropic loss of ATP synthase subunits in the mthi1 mutants. Total cell extracts of wild type (WT; a dilution series is shown) and the two mthi1 mutant strains were probed with antibodies against the proteins indicated on the left. The accumulation of all ATP synthase subunits was dramatically reduced in the two mutant strains, while that of cytochrome f (cyt. f), PsaA, and D1 and OEE2 (which were used as proxies for the abundance of the cytochrome b6f complex, PSI, and PSII, respectively) was unaffected. The red asterisk points to a cross-reaction of the antibody, preserved in the mutant strains, against the γ subunit of mitochondrial ATP synthase.
(C) (Top) Accumulation of the atpH and atpI transcripts in the same strains, assessed by RNA gel blots. The psaB transcript is provided as a loading control. (Bottom) Rate of translation of ATP synthase subunits in the same strains, assessed by 5′ pulse-labeling experiment in the presence of 14C acetate (5 µCi mL−1) and cycloheximide, an inhibitor of cytosolic translation (10 µg mL−1). The positions of the AtpI and AtpH subunits are indicated. WT, wild type.
(D) Quantification of atpI transcripts amount (±se) in wild-type (WT) and mutant strains estimated from RNA gel blots similar to the representative blot shown in (C). (Left) Relative accumulation (rel accum.) of the four atpI-containing transcripts in the wild type, expressed as the percentage of the total amount (tot) of atpI transcript. (Right) Relative abundance of each atpI transcript, and the sum of them, compared with that of the same band in the wild type (set to 100, symbolized by a gray dashed line) in the two mutants (dark gray, mthi1-1; light gray, mthi1-2; n = 4).
The reduced synthesis of AtpI prompted us to monitor the accumulation of its transcript in mthi1 mutants. The atpI gene belongs to a polycistronic transcription unit that comprises a gene encoding the PSII D2 protein (psbD), the second exon of psaA (apoprotein of complex Chlorophyll-Protein I), psbJ (PSII subunit), atpI, psaJ (PSI subunit), and ribosomal protein Rps12 (rps12; Figure 1A; Cavaiuolo et al., 2017). As reported previously by Liu et al. (1989), Rymarquis et al. (2006), and Jalal et al. (2015) and illustrated in Figures 1A and 1C, the wild type displays four major atpI transcripts, including the psbJ-atpI-psaJ-rps12, atpI-psaJ-rps12, atpI-psaJ, and atpI tetra-, tri-, di-, and monocistronic transcripts. The tri- and dicistronic transcripts account for 75% of the atpI-containing mRNAs. In mthi1 mutants, the accumulation of atpI transcripts was reduced by ∼60% and that of the di- and monocistronic transcripts was reduced by ∼85% and ∼75%, respectively (Figures 1C and 1D).
To explore whether this reduced transcript level was responsible for the reduced synthesis of AtpI in mthi1 mutants, we compared the loading of atpI transcripts on polysomes in the wild type and in three strains lacking AtpH expression: ΔatpH, an atpH deletion strain (Table 1 lists strains constructed in this study); mthi1-1; and mthi1-2 (Figure 2A). Free mRNAs and dissociated 50S and 30S ribosome subunits are found in the light fractions (6 to 10) of Suc gradients, while transcripts found in the heavy fractions (1 to 5) correspond to polysomes of increasing sizes (Minai et al., 2006; Eberhard et al., 2011). The distribution of psbD mRNA, whose expression is unrelated to ATP synthase biogenesis, was unchanged in the three mutant strains and the wild type, with a peak centered on fractions 4 and 5. The distribution of the four atpI transcripts was similar in the wild type and ΔatpH strain, with a peak centered on fraction 4. In mthi1 mutants, the distribution of the tetra- and tricistronic transcripts was similar to that in the wild type, likely because these transcripts comprise three and two open reading frames, respectively, in addition to the atpI coding sequence (CDS). In stark contrast, the two smaller transcripts were virtually absent in fractions 1 to 5 and were mostly found in fractions 7 to 9 (Figure 2A). Their exclusion from polysomal fractions indicates that the reduced synthesis of AtpI was not due to the reduced accumulation of atpI transcripts nor to an increased and rapid proteolytic disposal of AtpI in the absence of its assembly partner AtpH, but rather to a severely impaired translation of atpI transcripts in mthi1 mutants.
Table 1. Transformations Performed in this Study.
| Chloroplast Transformation | ||
|---|---|---|
| Plasmid | Recipient straina | Transformed strainb |
| pKrΔatpH | Wild type | ΔatpHc,d |
| mthi1-1 | ΔH mthi1-1c,f,g | |
| mthi1-2 | ΔH mthi1-2c,f,g | |
| pKrΔatpI | Wild type | ΔatpIc,e |
| mthi1-2 | ΔΙ mthi1f | |
| MTHI1-HA | ΔI MTHI1-HAc,f | |
| ΔatpHd | ΔH/Ic,f | |
| ΔH mthi1d | ΔH/I mthi1c,f | |
| pKr5′psaA-atpIh | Wild type | aAdI,c, f, h |
| mthi1-2 | mthi1-2 {aAI}a,f,g | |
| patpIStKr | ΔatpId | atpIStc,g |
| patpICtKr | ΔatpId | atpICtc,g |
| pKrdIfh | Wild type | dIfc,f,g,h |
| mthi1-1 | mthi1-1{dIf}c,f,g | |
| ΔatpHd | {ΔH, dIf}c,f,g | |
| ΔatpId | {ΔI, dIf}c,f,g | |
| ΔH/Id | {ΔH/I, dIf}c,f,g | |
| aAdId | {aAdI, dIf}c,f,g | |
| pKrdIfΔ1 | aAdId | Δ1e,f |
| pKrdIfΔ2 | Δ2e,f | |
| pKrdIfΔ3 | Δ3e,f | |
| pKrdIfΔ4 | Δ4e,f | |
| pKrdIfΔT | Δτe,g | |
| pKrdIAf | Wild type | dIAfe,g |
| pKrdHf | Wild type | dHfc,f,g,h |
| mthi1-1 | mthi1-1 {dHf}c,f,g | |
| ΔatpHd | {ΔH, dHf}c,f,g | |
| ΔatpId | {ΔΙ, dHf}c,f,g | |
| pKrdHf | ΔH/Id | {ΔH/I, dHf}c,f,g |
| pWFdHK | Wild type | dHKc,f,g |
| pWFdIK | Wild type | dIKc,f,g |
| pGatpH Kr | Wild type | pGatpHc,g |
| mthi1-2 | mthi1 {pGatpH}c,g | |
| aAdId | {aAdI, pGatpH}c,g | |
| mthi1 {aAdI}d | mthi1 {aAdI, pGatpHc,g | |
| patpHCt | Wild type | atpHCtc,g |
| patpHM | Wild type | atpHMc,g |
| P5′AatpH | Wild type | 5′AatpHc,g |
| Nuclear Transformation | ||
| Plasmidi | Recipient strainj | Transformed strainj |
| gMTHI1-HA | mthi1-1 | MTHI1-HA (g clones) |
| mthi1-2 | MTHI1-HA (g clones) | |
| cMTHI1-HA | mthi1-1 | MTHI1-HA (c clones) |
| mthi1-2 | MTHI1-HA (c clones) | |
| gMTHI1-HA_ΔC | mthi1-1 | ΔCg clones |
All recipient strains were spectinomycin sensitive. Transformed strains were selected based on resistance to spectinomycin (100 µg·mL−1) under low light (5 µE·m−2·s−1) and subcloned in darkness on TAP-spectinomycin (500 µg mL−1) until they reached homoplasmy.
Transformed strains are named based on their genotype. By convention, the chloroplast genotype is indicated in curly brackets for strains containing more than one mutation and follows, when required, the nuclear genotype.
These strains were initially selected based on spectinomycin resistance due to the presence of the recycling spectinomycin resistance cassette (Kr). Once homoplasmic with respect to the ATP synthase mutation, they were grown on TAP medium for several generations to allow for the spontaneous loss of the recycling cassette, according to Fischer et al. (1996), but not that of the ATP synthase transgene.
They therefore became spectinomycin sensitive again and could be used as a recipient strain in a new round of transformation experiments based on selection for spectinomycin resistance.
Homoplasmy was deduced from the loss of phototrophic growth capacity.
Homoplasmy was assessed by RNA gel-blot experiments.
Homoplasmy was assessed by RFLP of specific PCR products.
Chimeras are named as follows: the first two letters indicate the origin of the 5′ UTR, based on the nomenclature for chloroplast genes in Chlamydomonas (the first letter indicates the complex: A for PSI - psa-, B for PSII – psb-, C for cytochrome b6f, D for ATP synthase, R for Rubisco; the second letter indicates the gene whose 5′ UTR was borrowed: i.e., for aA for the 5′ UTR of psaA). The next two letters indicate the CDS used in the chimera, based on the same nomenclature. For historical reasons, the petA CDS is designated as f for cytochrome f, instead of cA, and the aadA CDS is designated as K. Unless required, the 3′ UTR is not mentioned and is usually that following the CDS, or the 3′rbcL UTR downstream of the aadA CDS. Thus, the full description of the dHf chimera would be atpH 5′UTR-petA CDS-petA 3′UTR, inserted at the petA locus, in replacement of the endogenous petA gene. The aAdI chimera comprises the psaA 5′UTR-atpI CDS-atpI 3UTR chimera, substituting the endogenous atpI gene at the atpI locus. A schematic map of all chimeras is also provided in the figures.
Plasmid DNA was linearized before transformation upstream of the MTHI1 gene by XbaI.
All recipient strains were nonphotosynthetic, and transformants were selected based on photoautotrophy on minimal medium (Harris, 1989) under high light (100 µE m−2 s−1).
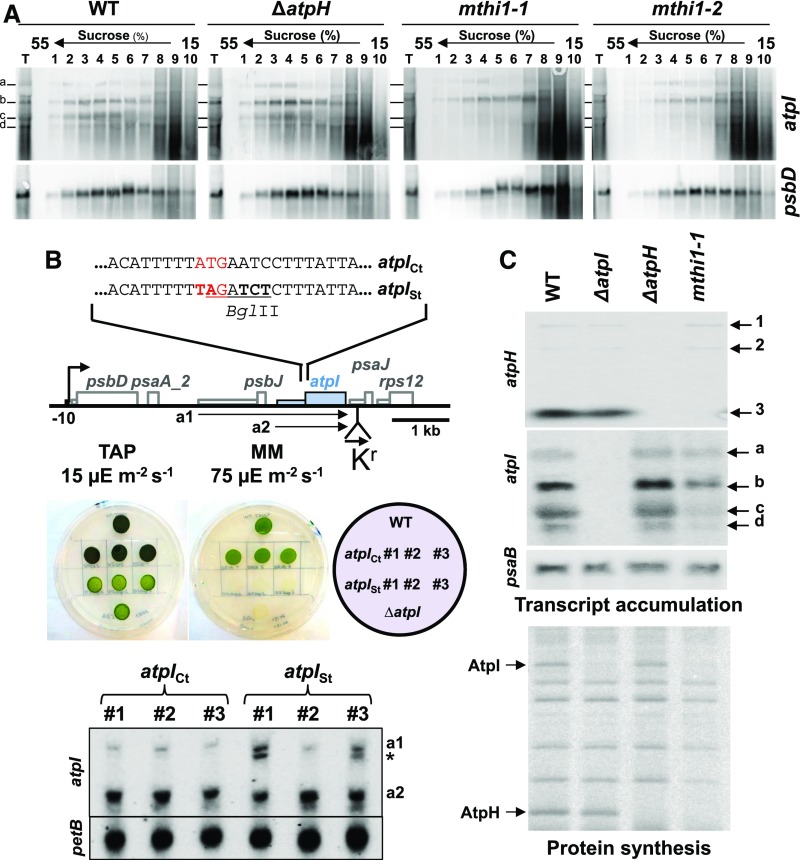
Figure 2.
The MTHI1 Factor Controls the Translation of atpI mRNA.
(A) Loading of atpI mRNAs on polysomes. Solubilized whole-cell extracts (T) from wild-type (WT), ΔatpH, and the two mthi1 mutant strains pretreated for 10 min with CAP (200 µg mL−1) were loaded on Suc gradients. After ultracentrifugation, 10 fractions were collected, and the transcripts present in each fraction were analyzed by RNA gel blots using the probes indicated on the right.
(B) Defective atpI mRNA translation is not responsible for its decreased abundance in mthi1 mutants. (Top) Schematic representation of the changes introduced into the atpI gene. Mutated nucleotides are shown in bold: they change the initiation codon (written in red) to a stop codon and introduced a BglII restriction fragment length polymorphism marker (underlined). (Middle) Phototrophic growth of the atpISt and atpICt strains assessed on minimal medium (devoid of acetate) under 75-µE m−2 s−1 light. Three independent transformants are shown. The growth of the strain on TAP medium (15 µE m−2 s−1) as well as the growth of the wild type (WT) and the ΔatpI strain are shown as controls. (Bottom) Accumulation, assessed by RNA gel blots, of atpI transcripts schematically depicted in (A) in a control strain bearing the aadA cassette alone and in strains bearing the aadA cassette associated with the untranslatable atpISt gene. Three independent transformants are shown for each construct. Because of the polar effect of the aadA cassette, cotranscripts with atpJ and/or rps12 cannot be observed. The origin of the transcripts indicated by an asterisk (*) is unknown. petB is provided as a loading control.
(C) atpH and atpI gene expression in the wild-type, ΔatpH, ΔatpI, and mthi1-1 strains. (Top) Accumulation of the atpH and atpI transcripts, assessed by RNA gel blots. psaB is provided as a loading control. (Bottom) Rate of translation of the atpH and atpI transcripts in the same strains, assessed as in Figure 1B by pulse labeling experiments. The positions of the AtpI and AtpH subunits are indicated. WT, wild type.
To further assess the relationship between atpI transcript accumulation and translation, we constructed an untranslatable version of the atpI gene, atpISt, whose initiation codon was replaced by an amber stop codon (Figure 2B). This mutated atpI gene, like all chimeric and mutated genes used in this study (Table 1), was associated with an aminoglycosyl adenine transferase (aadA) cassette to select transformants for spectinomycin resistance. After transformation, it replaced the endogenous atpI gene. Because they did not synthesize the AtpI subunit, the transformants were unable to perform phototrophic growth (Figure 2B). However, the mutated atpI transcripts accumulated to the same levels as those in control strains transformed with an unmodified atpI gene that was just associated with the aadA cassette (Figure 2B). The reduced accumulation of atpI mRNA in mthi1 mutants is not due to impaired translation but to the lack of MTHI1 that therefore not only activates the translation of atpI mRNA but also contributes to its stabilization.
AtpI and AtpH Are Synthesized Independently
The reduced translation of atpI transcripts in mthi1 mutants could be explained in two ways: either MTHI1 is a bifunctional protein required for the stable accumulation of atpH mRNA and for the translation of the atpI transcript, or AtpI is a CES subunit that requires the presence of AtpH to be synthesized at sustained rates, as in yeast. The similar loading of atpI transcripts on polysomes in wild-type and ΔatpH strains (Figure 2A) strongly argues against the latter hypothesis. As a further challenge, we compared the translation of atpH and atpI mRNAs by pulse-labeling experiments in strains mthi1-1, ΔatpH, and ΔatpI. While the synthesis of AtpI was strongly reduced in the mthi1-1 strain, it was similar in the ΔatpH and wild-type strains (Figure 2C). Conversely, AtpH was synthesized at the same level in the wild-type and ΔatpI strains. The two subunits are thus synthesized independently, ruling out a CES relationship and indicating that MTHI1 controls the expression of two different genes, unlike most M or T factors studied so far in Chlamydomonas.
The MTHI1 Factor Targets the 5′ UTR of atpI
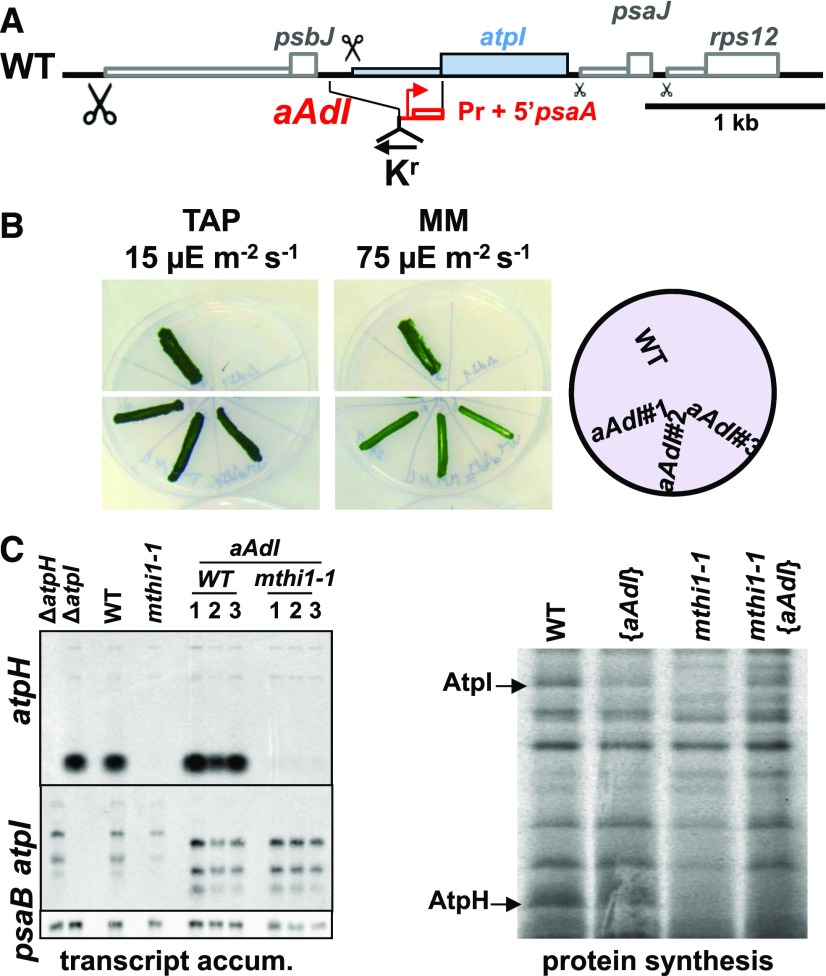
We studied the role of MTHI1 in atpI gene expression using chimeric genes. We first constructed a chimeric atpI gene in which the atpI 5′ untranslated region (UTR) was replaced by the promoter and 5′ UTR of the psaA gene (Figure 3A). After transformation, this aAdI chimera (see footnote h in Table 1 for the chimera nomenclature) replaced the endogenous atpI gene in the wild-type and mthi1-1 recipient strains. In the wild-type background, this gene was expressed at a level sufficient to sustain phototrophy (Figure 3B). When introduced into the mthi1-1 recipient strain, it did not restore phototrophy in transformants that still lacked atpH mRNA accumulation. However, pulse-labeling experiments showed that the synthesis of the AtpI subunit was restored (Figure 3C). The downregulation of atpI mRNA translation in the absence of MTHI1 thus depends on the 5′ UTR of atpI.
Figure 3.
The MTHI1 Factor Targets the atpI 5′ UTR.
(A) Schematic representation of the aAdI chimera, where the atpI 5′ UTR had been replaced by the promoter and 5′ UTRs of the psaA gene. The position of the recycling aadA cassette (Kr), inserted in reverse orientation with respect to atpH, is shown. WT, wild type.
(B) Photoautotrophic growth of the aAdI strain, assessed as in Figure 2B. WT, wild type.
(C) (Left) atpH and atpI transcript accumulation (transcript accum.) in the wild-type (WT) and mthi1-1 strains transformed by the aAdI construct, whose transcript is shorter than the endogenous atpI transcript due to the small size of the psaA 5′ UTR. The recipient strains are shown as well as the ΔatpH and ΔatpI strains for controls. Three independent transformants are shown for each genetic background. The psaB transcript is provided as a loading control. (Right) Rate of AtpH and AtpI synthesis in the wild-type and mthi1-1 strains and in the corresponding strains transformed by the aAdI construct, assessed as in Figure 1B by pulse-labeling experiments.
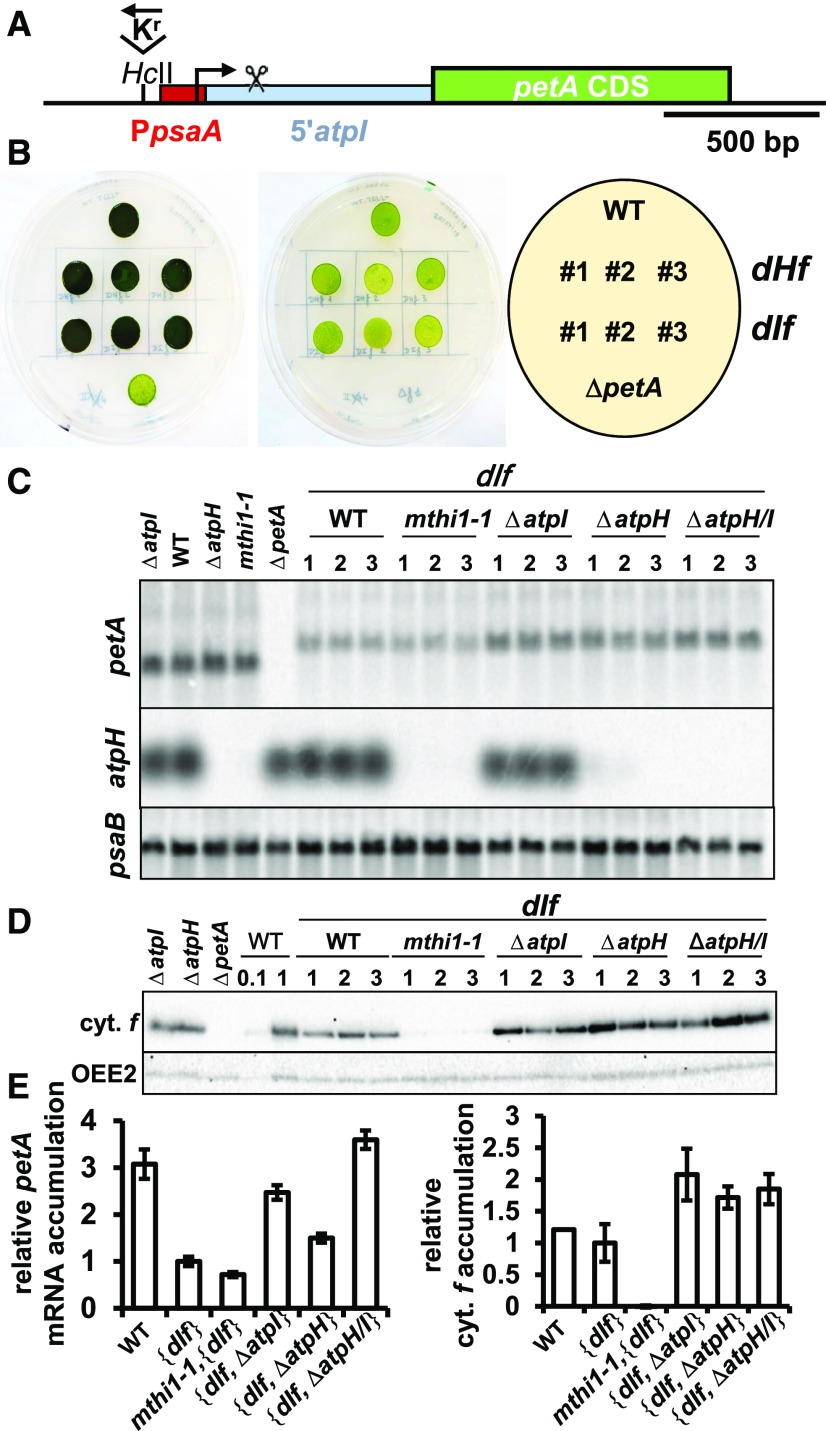
In another chimera, dIf, the atpI 5′ UTR was fused in frame to the petA (cytochrome f) CDS, previously shown to be a convenient reporter gene (Wostrikoff et al., 2004). Since the atpI 5′ UTR is thus far uncharacterized, we first determined its length (493 nucleotides) by 5′ RNA ligase-mediated rapid amplification of cDNA ends (Supplemental Figure 1A). Furthermore, as the atpI gene is part of a polycistronic unit, with no indication of a dedicated promoter (Supplemental Figure 1B; Cavaiuolo et al., 2017), we placed the psaA promoter upstream of the atpI 5′ UTR (Figure 4A). After transformation, the dIf chimera replaced the endogenous petA gene in the wild-type, mthi1-1, ΔatpH, ΔatpI, and ΔatpH/I strains.
Figure 4.
The atpI 5′ UTR Is Sufficient to Confer MTHI1-Dependent Translation to a Reporter Gene.
(A) Schematic map of the dIf construct inserted instead of the endogenous petA gene. The red rectangle indicates the psaA promoter region placed upstream of the psbJ-atpI intergenic fragment (in light blue), which was long enough to include the atpI processing site. The scissors above the intergenic region indicate the position of the 5′ end of the processed atpI mRNA. The position of the recycling selection cassette, upstream of the chimeric petA gene and in reverse orientation with respect to this latter, is shown.
(B) Photoautotrophic growth of the dHf (Figure 5) and dIf chimeric strains (three independent transformants) assessed as in Figure 2B. The growth of the wild-type (WT) and the ΔpetA strains are shown as controls.
(C) Accumulation, assessed by RNA gel blots, of the chimeric petA transcript, introduced by transformation of the chloroplast genome in the wild-type (WT), mthi1-1, ΔatpH, ΔatpI, and ΔatpH/I recipient strains. Untransformed recipient strains are shown on the left, as well as a ΔpetA strain for comparison. Three independent transformants are shown for each genetic context. The accumulation of atpH mRNA in the same strains is also shown, while that of the psaB mRNA is provided as a loading control.
(D) Accumulation of cytochrome f (cyt. f) in the same strains, assessed by immunoblots (loading control, OEE2). WT, wild type.
(E) Quantification (±se) of the petA transcript (left) and cytochrome f (cyt. f; right) in transformed strains shows a competition between the chimera and the endogenous atpI gene for the expression of 5′atpI-driven genes. Value for the dIf transcript in the wild-type (WT) recipient strain is set to 1 (n = 6).
Transformants derived from the wild-type strain grew on minimal medium (Figure 4B): the atpI 5′ UTR can drive cytochrome f synthesis at levels sufficient to sustain phototrophic growth. However, the accumulation of both the chimeric transcript and its cytochrome f gene product were lower than those of the endogenous petA gene. When introduced into the mthi1-1 strain, the accumulation of the chimeric transcript was further reduced, but only trace amounts of cytochrome f accumulated: the atpI 5′ UTR thus confers an MTHI1-dependent rate of translation to a reporter CDS. Similar results were obtained using the heterologous aadA CDS as a reporter (Supplemental Figure 2).
Most interestingly, the expression of the 5′atpI-petA chimera increased in the deletion strains ΔatpH, ΔatpI, and ΔatpH/I compared with the wild type background (Figures 4C to 4E). In the ΔatpH strain, the accumulation of the chimeric transcript increased 1.5-fold compared with the wild type background but remained lower than that of the endogenous petA gene, while the accumulation of its gene product was higher than that of the endogenous cytochrome f. The expression of the atpH and atpI genes thus relies on a common factor present in limiting amounts, possibly MTHI1. The accumulation of the chimeric mRNA further increased (by 2.5-fold) when the atpI gene was deleted and was also increased in a strain deleted for both atpH and atpI genes. Therefore, the chimeric and endogenous atpI transcripts compete for the binding of some factors in limiting amounts.
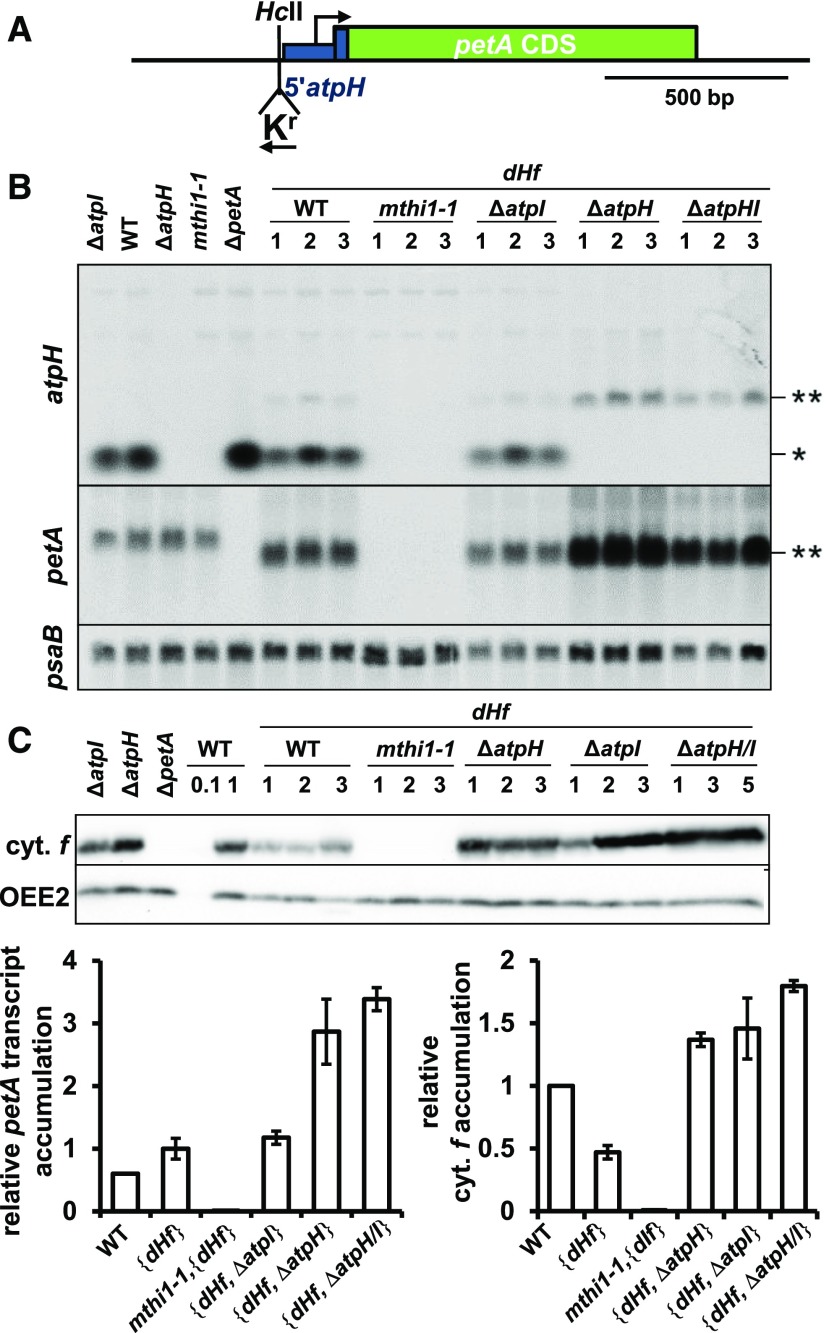
The MTHI1 Factor Targets the atpH 5′ UTR to Stabilize the Transcript and Activate Its Translation
We also identified the target of MTHI1 within atpH mRNA. The dHf chimeric gene, comprising the atpH promoter and 5′ UTR fused in frame to the petA CDS (Figure 5A), was introduced by transformation into the chloroplast genome of the wild-type, mthi1-1, ΔatpH, ΔatpI, and ΔatpH-atpI recipient strains. In the wild-type background, transformants were phototrophic: the atpH 5′ UTR allows cytochrome f to be expressed (Figure 4B). The dHf chimeric transcript accumulated to 150% of the level of endogenous petA transcript, but its protein product was two times less abundant than the endogenous cytochrome f (Figures 5B to 5D). In the mthi1-1 background, the chimeric petA mRNA did not accumulate and cytochrome f was absent (Figures 5B and 5C).
Figure 5.
The MTHI1 Factor Targets the atpH 5′UTR.
(A) Schematic representation of the dHf chimera, with the position of the recycling aadA cassette (in reverse orientation with respect to the petA gene) shown. The blue thick rectangle represents the first 25 nucleotides of the atpH CDS fused in frame with the petA CDS, which was added to the construct to improve the expression of the chimera.
(B) Accumulation of the atpH and petA transcripts, assessed by RNA gel blots, in the wild-type (WT), mthi1-1, ΔatpH, ΔatpI, and ΔatpH/I strains carrying the dHf chimera instead of the endogenous petA gene. Unstransformed wild-type, ΔatpH, ΔatpI, ΔpetA, and mthi1-1 strains are shown as controls. Asterisk indicates the position of atpH mRNA, while the double asterisk points to a cross-reaction of the probe that comprises the atpH 5′ UTR with the dHf chimeric transcript. Three independent transformants are shown for each genetic context. The psaB transcript is provided as a loading control.
(C) Cytochrome f (cyt. f) accumulation in the same strains, assessed by immunoblots, with OEE2 as a loading control. WT, wild type.
(D) Quantification of the relative accumulation (±se) of the petA transcript (left) and cytochrome f (cyt. f, right) in the same strains. Values for dHf transformed in the wild-type (WT) strain are set to 1 (n = 6).
Deletion of the atpH gene increased the expression of the chimera at the transcript and cytochrome f levels by 3- and 1.5-fold, respectively, compared to the wild-type background (Figures 5B to 5D). The chimeric transcript competes with the endogenous atpH mRNA for MTHI1 binding. Deletion of the atpI gene only moderately increased the accumulation of the chimeric transcript but stimulated its translation, while the simultaneous deletion of the two genes increased the accumulation of both the chimeric transcript and its cytochrome f gene product. Again, these observations were confirmed using the aadA CDS as a heterologous reporter gene (Supplemental Figure 3). We noted that a dicistronic petA-aadA transcript accumulated to the same level in the four progeny, but it was not expressed in the mthi1 offspring (Supplemental Figures 3B and 3C), suggesting that MTHI1 might also be required for the translation of atpH mRNA.
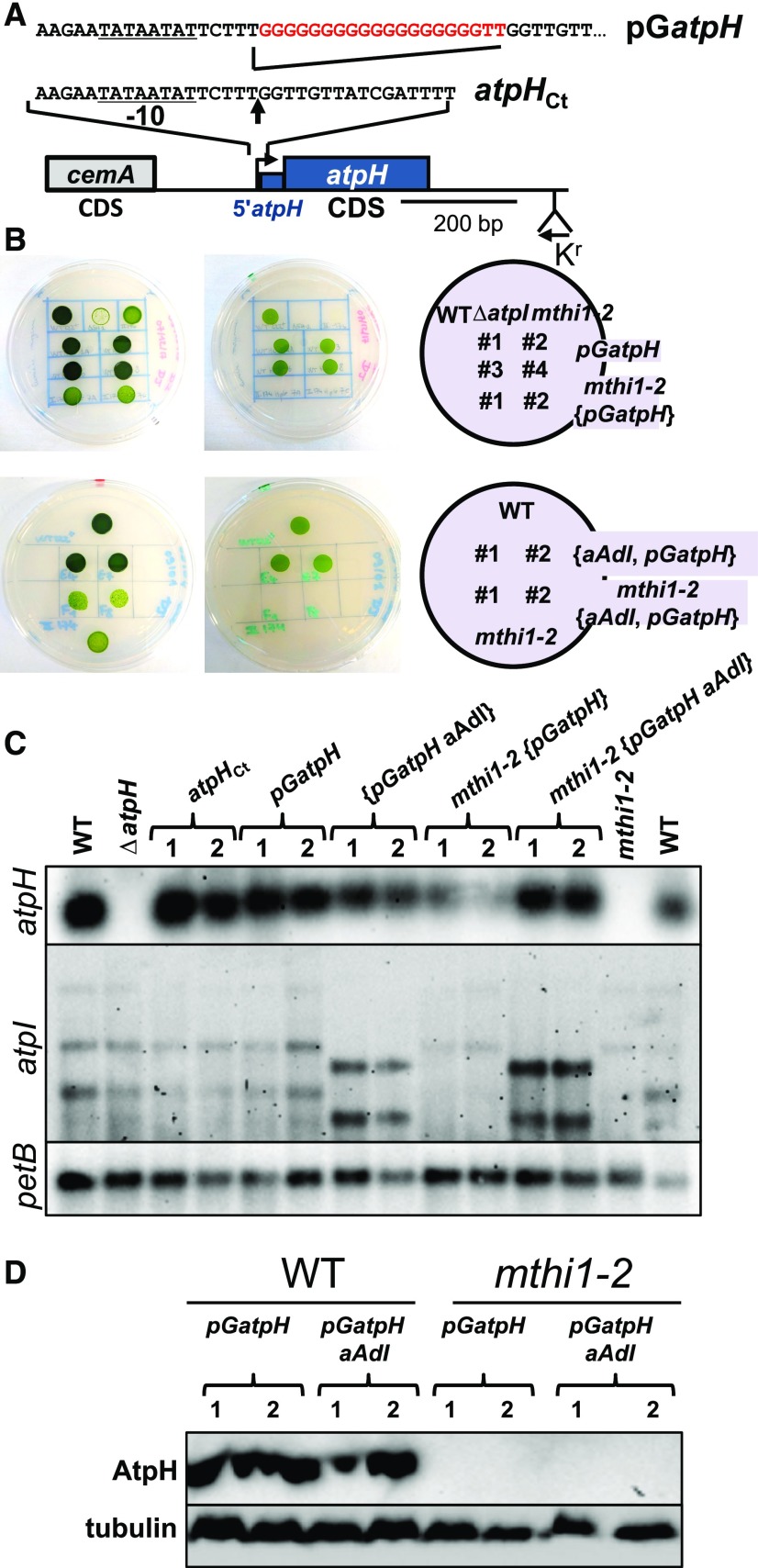
To address this point, we constructed a modified atpH gene whose transcript is stabilized independently of the presence of MTHI1 thanks to the insertion of a poly(G) cage immediately after the atpH transcription start site, a very stable secondary structure impeding the progression of 5′ → 3′ exoribonucleases (Vreken and Raué, 1992; Drager et al., 1996, 1998). This modified pGatpH gene (Figure 6A) replaced the endogenous atpH gene in the wild-type and mthi1-2 strains, and we monitored its expression in the transformants. Transformants recovered from the wild-type strain were phototrophic (Figure 6B) and accumulated similar amounts of atpH transcript (Figure 6C) and AtpH (Figure 6D) to the control strain: the poly(G) cage at the beginning of the atpH transcript did not prevent its translation. Transformants derived from the mthi1-2 strain recovered atpH mRNA accumulation, although at reduced levels, but were nevertheless unable to perform phototrophic growth. These transformants lacked accumulation of the AtpH subunit (Figure 6D), likely because the synthesis of the AtpI subunit was still impaired in the mthi1 background. To overcome this issue, we replaced the atpI gene of the mthi1-2 {pGatpH} strain with its chimeric aAdI version, whose expression does not depend on the presence of MTHI1 (Figures 2B and 2C). Despite the restored expression of AtpI, the mthi1-2 {aAdI, pGatpH} transformants were still unable to perform photosynthetic growth and lacked accumulation of the AtpH subunit (Figures 6B and 6D). Thus, beside stabilizing atpH mRNA, MTHI1 is also required for its translational activation.
Figure 6.
MTHI1 Is Required for the Translation of the atpH Gene.
(A) Schematic map of the pGatpH construct with a close-up view of the region surrounding the atpH transcription start site, indicated by a vertical arrow, where the poly(G) tract was inserted. The atpH promoter is underlined, and the position of the recycling aadA cassette is shown. A construct carrying the selection cassette at the same position but devoid of the poly(G) insertion was used as a control (atpHCt). To avoid any polar effect on the expression of the downstream located atpF gene (cotranscribed with atpH), all experiments were performed after excision of the recycling aadA cassette.
(B) Phototrophic growth of the pGatpH, mthi1-2 {pGatpH}, {aAdI pGatpH}, and mthi1-2 {aAdI pGatpH} strains (two independent transformants each) assessed as in Figure 2B. Growth of the wild-type (WT) and the mthi1-2 and ΔatpI strains are shown as controls.
(C) Accumulation, assessed by RNA gel blots, of the atpH and atpI transcripts in the wild-type (WT) strain transformed by the atpHCt and pGatpH constructs and in the mthi1-2 strain transformed with the pGatpH gene. The aAdI construct then replaced the endogenous atpI gene in the resulting pGatpH and mthi1-2 {pGatpH} strains. Two independent transformants are shown for each genetic background. The petB transcript is provided as a loading control.
(D) Accumulation of the AtpH subunit, assessed by immunoblots, in strains expressing the poly(G) construct. Tubulin is provided as a loading control.
Characterization of the MTHI1 Protein
We cloned the MTHI1 gene by complementing an mthi1-1, arg7, cw15 strain with an indexed library of cosmids (details in Supplemental Figure 4). Evidence that the MTHI1 gene actually corresponds to gene model Cre17.g734564 came from the complementation of both mthi1-1 and mthi1-2 mutations by an expressed sequence tag (EST) clone (AV629671) obtained from Kazusa DNA Research Institute. This chromosomic localization, however, is erroneous. Indeed, the ac46 mutant (mthi1-1) has been previously mapped to the complementation group XVI/XVII, which was later shown to correspond to chromosome 15 (Dutcher et al., 1991, Kathir et al., 2003). Crosses confirmed that the mthi1-1 mutation was linked to the CytC1 molecular marker on chromosome 15. It is of note that the MTHI1 gene was localized on chromosome 15 in version 4.0 of the Chlamydomonas genome and has been moved to chromosome 17 in version 5.5. Sequencing, using appropriate primers, of the EST clone and comparison with the genomic scaffold showed that the EST clone contained the full-length coding sequence of MTHI1 as an in-frame stop codon is located six nucleotides upstream of the initiation codon and that the MTHI1 gene is composed of 11 exons. A poly(A) tail was found 424 bp downstream of the stop codon and 15 nucleotides downstream of the TGTAA polyadenylation consensus signal (Silflow, 1998).
Sequencing of the MTHI1 region in the mutants revealed that the translation initiation codon was substituted by an AUU opale stop codon in strain mthi1-1, while the insertion of a C residue after codon 138 led to premature translation abortion after codon 188 in strain mthi1-2 (Supplemental Figure 5B).
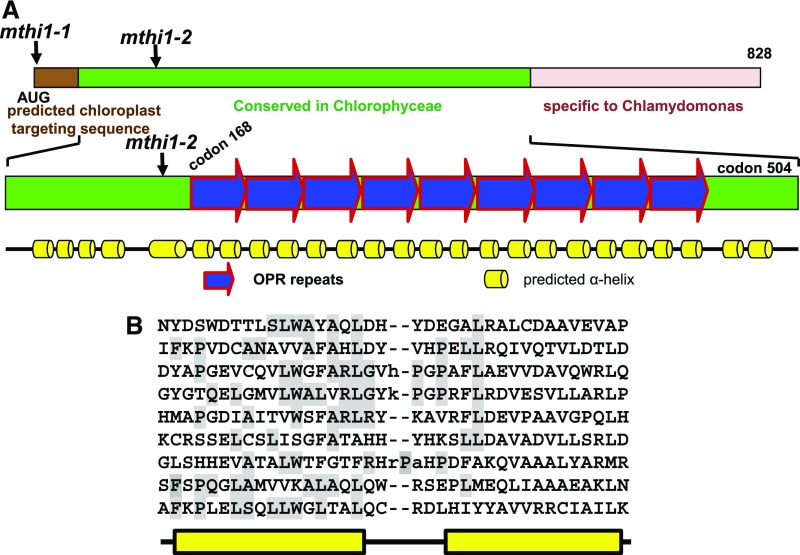
The MTHI1 gene encodes a protein of 828 amino acid residues (Figure 7A; Supplemental Figure 5C) predicted to be targeted to the chloroplast by the Predalgo and TargetP programs (Tardif et al., 2012; Almagro Armenteros et al., 2019). Prediction of secondary structure using Scratch protein predictor software (http://scratch.proteomics.ics.uci.edu/) suggested that the mature MTHI1 protein potentially comprises two different domains. Following a predicted chloroplast targeting peptide of 48 residues, the N-terminal domain (up to residue 566) contains pairs of α helices (Figure 7A; Supplemental Figure 5C), nine of which are typical OPR repeats (Figure 7B). The C-terminal domain mainly harbors coiled-coil or intrinsically disordered sequences with no obvious motifs but several stretches of Ala and Gln residues (Supplemental Figure 5C), as in other Chlamydomonas M and T factors (Boudreau et al., 2000; Auchincloss et al., 2002; Raynaud et al., 2007).
Figure 7.
The MTHI1 Protein.
(A) and (B) Schematic representation of the MTHI1 protein. The positions of the two mthi1 mutations are shown. (A) In the top diagram, the brown rectangle depicts the chloroplast transit peptide, as predicted by the ChloroP program. The green rectangle indicates the region of the protein conserved in other Chlorophyceae species (Supplemental Figure 6), and the pink rectangle represents a rapidly evolving and disordered region. The bottom diagram shows the predicted secondary structure of the conserved region. Blue arrows represent the OPR repeats, whose sequence is shown in (B), with the amino acid residues obeying the OPR consensus shaded in gray.
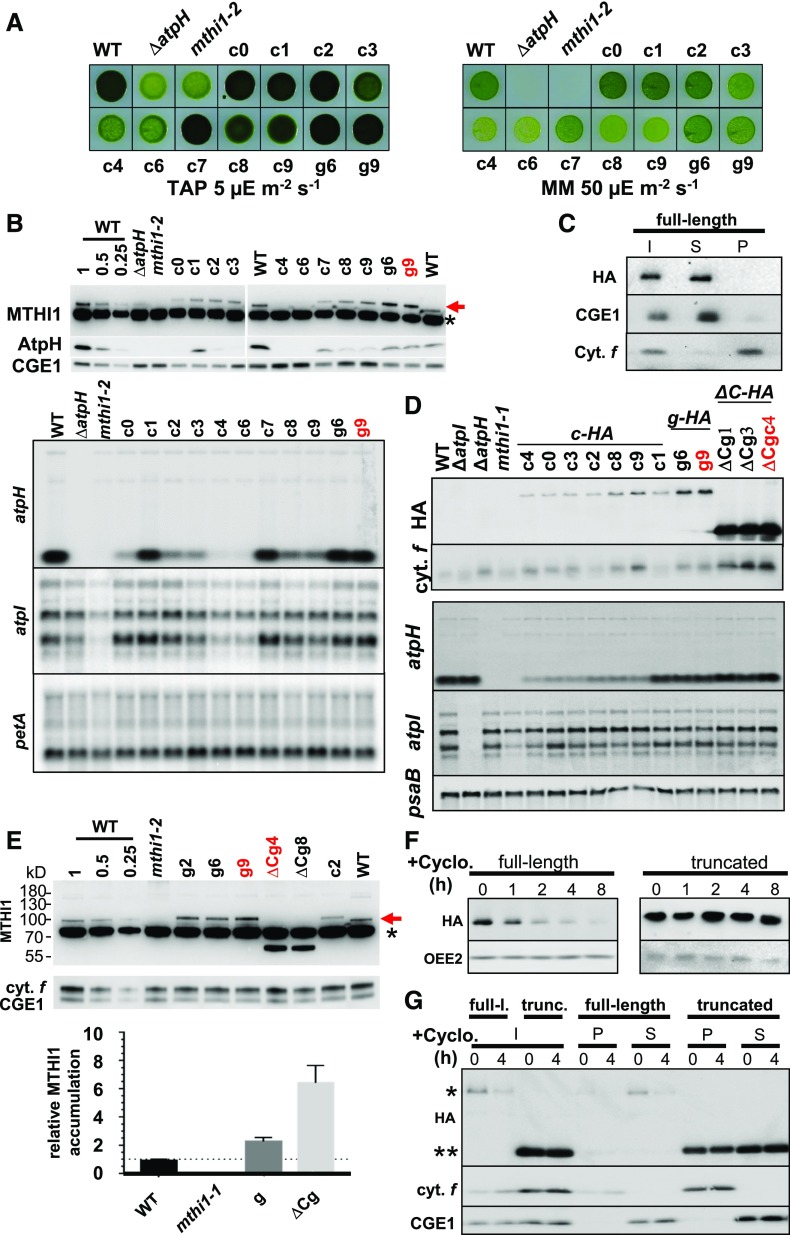
BLAST searches revealed orthologs of MTHI1 in green algae (Supplemental Figure 6). The region of similarity was restricted to the N-terminal, OPR-containing part of the protein, while the C-terminal tail was highly variable in length and sequence, even between the most closely related species. Thus, fusing a hemagglutinin (HA) tag for immuno-detection at the C terminus of MTHI1 should not be deleterious for its function. Indeed, we could still complement the mthi1-1 mutation with a tagged version of MTHI1 when the tag was inserted in genomic (g transformant) or cDNA (c transformant) constructs (Figure 8A). The tagged genomic construct, including 4280 bp upstream of the translation initiation codon, that is, presumably the whole MTHI1 promoter, allowed for greater accumulation of the tagged protein than the tagged cDNA construct (compared clones g6 and g9 with clones c in Figure 8B).
Figure 8.
Complementation of the mthi1-1 Mutant Strain.
(A) Complementation of the mthi1 strain with a tagged version of the MTHI1 gene, either the tagged cDNA (c clones) or the genomic construct (g clones), restores phototrophy, as assessed by plating the cells on minimal medium (MM) plates as in Figure 2B. The growth of the wild-type (WT), ΔatpH, and mthi1-2 strains is shown as a control.
(B) Accumulation of the MTHI1 protein (red arrow), either endogenous or tagged, the AtpH subunit (top), and the atpH and atpI transcripts (bottom) detected by immunoblots in the same strains with an antibody against the MTHI1 protein. Note the larger size of the tagged protein compared with the endogenous protein due to the insertion of the triple HA tag. CGE1and cytochrome f or petA mRNA are shown as the respective loading controls in protein and RNA gel blots. The name of the clone used for further analysis of MTHI1 in the next figures is written in red (asterisk, cross-contaminant).
(C) MTHI1 is a soluble protein. Cellular extract (I) from the complemented strain g9 was separated into soluble S and insoluble P fractions by ultracentrifugation and equal volumes of each fraction were probed with antibodies against the HA tag and against GrpE and cytochrome f (Cyt. f) as controls for the purity of the fractions.
(D) The C-terminal domain of MTHI1 is dispensable for its function. Accumulation of the tagged MTHI1 protein, probed with an antibody against the HA tag, and the atpH and atpI transcripts, assessed by RNA gel blots, in mthi1-1 strain complemented with the tagged versions of the MTHI1 gene, either the tagged cDNA (c-HA), the genomic construct (g-HA), or its C-terminally truncated version (ΔC-HA). All transformants were selected based on recovery of phototrophy on minimal medium plates. Overaccumulation of the truncated MTHI1 protein does not lead to an increased abundance of the atpH transcripts. Cytochrome f (cyt. f) and psaB are shown as the respective loading controls in the protein and RNA gel blots. The names of the clones used for further analysis of MTHI1 in the next figures are written in red. WT, wild type.
(E) Deletion of the C-terminal domain results in higher abundance of MTHI1. (Top) Accumulation of MTHI1 protein (red arrow) in mthi1-1 strains complemented with either the tagged MTHI1 cDNA (c), the tagged genomic construct (g), or its C-terminally truncated version (ΔCg), probed with an antibody against the MTHI1 protein. The name of the clones used for further analysis of MTHI1 is written in red (asterisk, cross-contaminant). WT, wild type. (Bottom) Quantification of MTHI1 accumulation (±se), estimated from immunoblots similar to the representative one shown in the top panel, in the strains shown in top panel, normalized to that of cytochrome f (cyt. f) and compared with the accumulation of MTHI1 in wild-type (WT) cells (set to 1). Error bars represent se (n = 3).
(F) The C-terminal domain of MTHI1 contributes to its high turnover. Stability of full-length (full-l.) MTHI1 or of its C-terminally truncated (trunc.) version, assessed by immunoblots in a culture treated with cycloheximide (+Cyclo.) for the indicated times. Accumulation of OEE2 in the same samples is shown as a loading control.
(G) Differential solubility of the full-length MTHI1 protein and its C-terminal truncated version. Cellular extracts of transformants expressing the full-length (*) and truncated (**) versions of the tagged MTHI1 protein, treated with cycloheximide (+Cyclo.) for 0 or 4 h (I, left), were fractionated into soluble (S) and membrane (P) fractions and analyzed as in (D). Distributions of CGE1 and cytochrome f (cyt. f) are shown to assess the purity of the fractions.
We overexpressed the MTHI1 protein and raised an antibody against the mature protein to compare the accumulation of the tagged protein in transformants with that of the endogenous protein in the wild type. Despite the higher accumulation of MTHI1 in clones transformed with the genomic construct, atpH mRNA was not more highly accumulated than in the wild type (Figures 8B and 8E). Therefore, either the C-terminal tag somehow decreases protein activity, or other factors limit the abundance of atpH mRNA. As expected based on the requirement of MTHI1 for the accumulation of atpH mRNA, the levels of MTHI1 and atpH mRNA were correlated (Figures 8B and 8D): the clones (e.g., c4 or c6) that accumulated less MTHI1, which was undetectable using the antibody against the whole protein (Figure 8B) but detectable using the antibody against the HA tag (c4 in Figure 8E), also accumulated atpH mRNA to levels just above the detection threshold. However, these clones were able to grow phototrophically (Figure 8A), confirming some restoration of ATP synthase.
We used one clone complemented with the tagged genomic construct (g9) to study the intra-organelle localization of MTHI1 and found that it was exclusively soluble (Figure 8C).
The C-Terminal Tail Is Dispensable for the Main Function of the MTHI1 Factor
The poor conservation of the C-terminal domain raised the question of its function. We thus constructed a truncated version of the gene encoding a protein lacking residues 573 to 797, that is, the most C-terminal domain, but still containing the HA tag. This truncated MTHI1 could still complement the mthi1-1 mutation. As shown in Figures 8D and 8E, the truncated protein accumulated to much higher levels than the full-length protein, but it did not proportionally increase the abundance of atpH or atpI mRNAs. Therefore, either part of the truncated MTHI1 protein was not involved in mRNA stabilization or other factors became limiting. To investigate the origin of this differential accumulation, we compared the stability of the full-length and truncated MTHI1 by following their decay in complemented strains incubated with cycloheximide, an inhibitor of cytosolic translation (Figure 8F). MTHI1 was short lived, with a half-life of ∼1 h. Most interestingly, its truncated version remained stable over the 8-h period of the experiment: the C terminus tail apparently controls the half-life of the whole protein. We repeated fractionation experiments on these complemented strains that were treated with cycloheximide for 4 h. In total cell extracts, the level of MTHI1-HA was strongly reduced upon cycloheximide treatment, while that of its truncated version was insensitive to the antibiotic. After fractionation into soluble and insoluble fractions, the full-length MTHI1 was almost exclusively found in the soluble fraction. By contrast, significant levels of its truncated version were found in the pellet, most likely as large aggregates that fell down during ultracentrifugation. Both the aggregated and soluble populations were stable over a 4-h period (Figure 8G).
MTHI1 Belongs to a Large Complex That Also Contains atpH and/or atpI mRNA
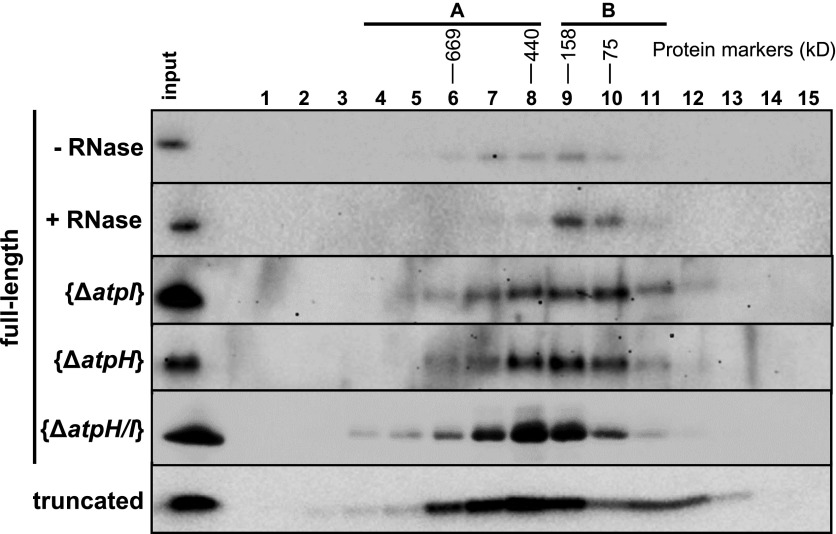
We used size exclusion chromatography to investigate whether MTHI1 belongs to a high molecular mass complex, as do almost all trans-acting factors studied so far (Boudreau et al., 2000; Vaistij et al., 2000b; Auchincloss et al., 2002; Dauvillée et al., 2003; Perron et al., 2004; Schwarz et al., 2007; Johnson et al., 2010; Boulouis et al., 2011). Soluble extract from clone g9 was fractionated on a Superose 6 column, a column that is optimal for separating protein complexes in the 5- to 5000-kD range. As shown in Figure 9, MTHI1 belongs to complexes ranging from 75 (fraction 10) to >700 kD (fraction 5), peaking in fractions 8 and 9 (150 to 450 kD). As no special care was taken to preserve the integrity of the RNA moiety, RNAs, if retained by MTHI1, were probably restricted to fragments closely surrounding its binding site and only accounted for a minor increase in molecular mass. When the supernatant was treated with RNase prior to loading on the column, MTHI1 presented a sharper distribution in slightly lighter fractions 9 and 10, which is consistent with a monomeric state. Thus, mRNAs appear to be responsible for the distribution of MTHI1 in high molecular mass ribonucleoprotein complexes. We analyzed the distribution of MTHI1 in complemented strains lacking atpH mRNA, atpI mRNA, or both. Upon deletion of either atpH or atpI, the distribution of MTHI1 remained unchanged and centered on fractions 8 to 10. The deletion of both atpH and atpI shifted the distribution of MTHI1 to larger complexes centered on fraction 8 but extending to still heavier fractions. Therefore, in the absence of its two RNA targets, MTHI1 undergoes conformational changes, possibly forming aggregates. A similar behavior had been already reported for the nucleus-encoded trans-acting factors Maturation/stability and Translation of complex C PetA subunit mRNA (MCA1 and TCA1, respectively) in the absence of their target petA mRNA (Boulouis et al., 2011). This behavior, however, is opposite that observed upon RNase treatment. Aggregation of MTHI1 in the absence of its RNA target was corroborated by the distribution pattern of the truncated MTHI1. Partially found in the pellet after ultracentrifugation, MTHI1 presented a bimodal distribution, with the first peak in fractions 11 and 12, likely corresponding to monomers, and a broad peak in fractions 2 to 8, with a maximum in fraction 8.
Figure 9.
MTHI1 Belongs to a High Molecular Weight Complex That Interacts with atpH and atpI Transcripts.
Soluble extracts listed at the left of the figure were fractionated on a Superose 6 10/300 HR column and probed with an antibody against the HA tag. Molecular masses of the complexes found in each fraction were estimated by comparing with standards of the High Molecular Weight Gel Filtration Calibration Kit (GE Healthcare).
MTHI1 Interacts with atpH mRNA
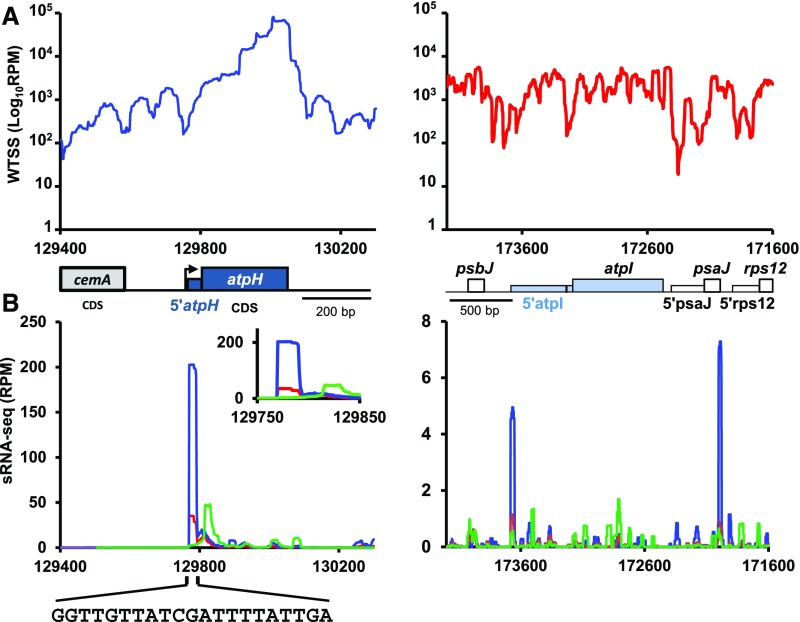
To investigate the interaction of MTHI1 with the atpH and atpI transcripts, we sequenced the small RNA fraction (size range, 11 to 44 nucleotides; sRNA-seq) of RNA samples prepared from the wild-type or mthi1-1 strains, since the interaction of an M factor on its target transcript tends to generate of a footprint that pinpoints its binding site (Ruwe and Schmitz-Linneweber, 2012; Zhelyazkova et al., 2012; Cavaiuolo et al., 2017). Figure 10A shows the normalized coverage of RNAs along the atpH and atpI loci. Figure 10B shows the coverage of sRNAs (11 to 44 nucleotides) over the same loci. A prominent peak of sRNAs ∼19 to 21 nucleotides long with a sharp 5′ end was found in the wild type at the very beginning of atpH mRNA. This peak was only seen after treatment of the RNA samples with RNA polyphosphatase (RPP), an enzyme that removes the pyrophosphate moiety of triphosphorylated transcription products. Most, if not all, monocistronic atpH mRNA is thus transcribed from the atpH promoter and does not result from the processing of the large precursor transcribed from the atpA promoter. In the mthi1-1 mutant, the amplitude of this peak was reduced more than fivefold (Figure 10B).
Figure 10.
Transcriptional Profiles of the atpH and atpI Genes.
(A) Coverage, normalized as RPM (log scale) of pooled the bidirectional and directional wild-type whole transcriptome shotgun sequencing (WTSS) along the atpH (left) and atpI (right) loci. Positions of the relevant genes and 5′ UTR are shown below. The black bar in the atpI 5′ UTR shows the position of the MTHI1 target (see below). Redrawn from the data in Cavaiuolo et al. (2017).
(B) sRNA mapping to the 5′ end of atpH mRNA is the footprint of MTHI1. Coverage, normalized as RPM, of pooled sRNA-seq along the same loci: the mock- (green) versus RPP-treated (blue) wild-type sRNA-seq libraries compared with RPP-treated libraries of the mthi1-1 mutant (red). Coverage is averaged over two biological replicates (libraries prepared from two independent RNA samples). Only reads mapping to the coding strand are shown. The inset for atpH shows a close-up view of the atpH 5′ UTR, and the sequence of the atpH footprint is shown. For atpI, a close-up view of the 5′ UTR (coding strand only) is shown in Supplemental Figure 9A. Note the very different values on y axes of the two graphs.
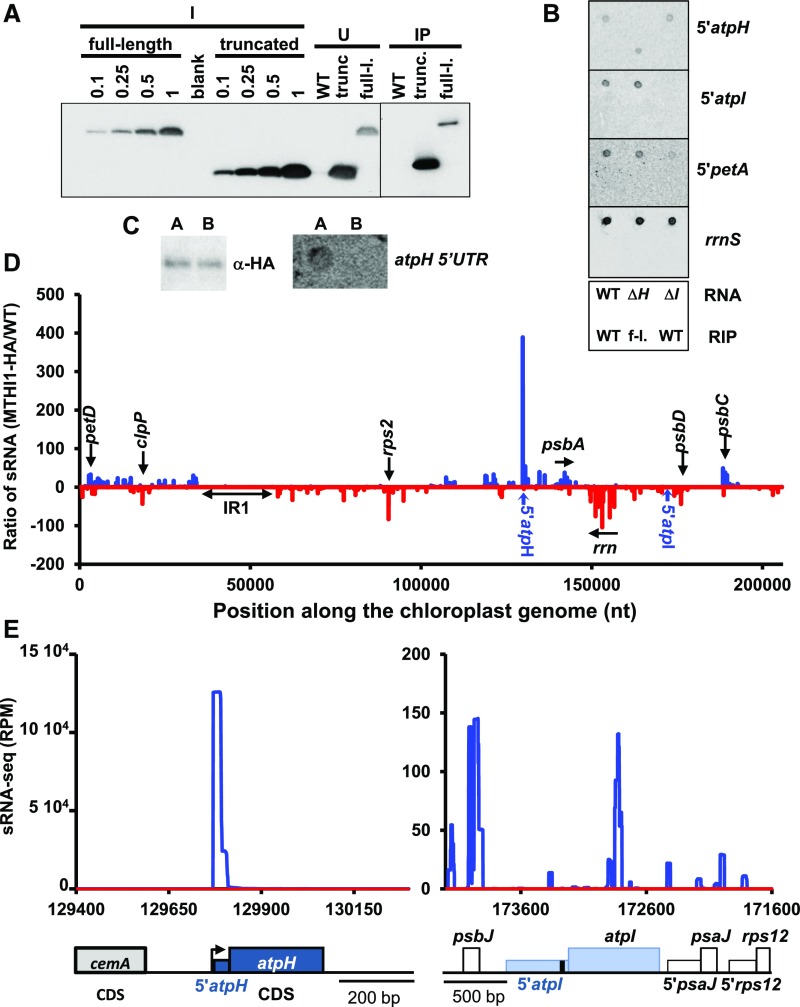
We subjected the g9 strain complemented with the tagged version of MTHI1 to immunoprecipitation with an antibody against the HA tag (Figure 11A). RNAs extracted from the pulled down material were monitored by dot-blot analysis (Figure 11B). We observed a signal with a probe specific for atpH 5′ UTR, but not with rrnS (chloroplastic 16S rRNA) or 5′petA probes used as specificity controls, nor when the same procedure was applied to the wild type (devoid of HA-tagged MTHI1). Thus, MTHI1 interacts specifically (either directly or indirectly) with the atpH 5′ UTR. By contrast, no signal was detected with a 5′atpI probe.
Figure 11.
The MTHI1 Protein Interacts Specifically with the atpH 5′ UTR.
(A) The full-length (full-l.) and truncated (trunc.) versions of the MTHI1 protein were immunoprecipitated from a soluble cellular extract with an antibody against the HA tag (U, unbound). Immunoprecipitation of a cellular extract from the wild-type (WT) strain is shown as a negative control. The apparent slower migration of the immunoprecipitated proteins in due to a smiling effect in the migration of the gel from which the composite figure (indicated by a vertical line) was made.
(B) RNA extracted from immunoprecipitates was analyzed by dot blot hybridized to the probes indicated on the right. The bottom panel shows the positions of the samples on the filter. Top line: RNA extracted from the wild-type (WT), ΔatpH, and ΔatpI strains (without immunoprecipitation), as a control for the specificity of the probes. Bottom line: immunoprecipitated RNA from the wild-type and from a complemented strain expressing the tagged MTHI1 (g9). f-l., full length.
(C) Fractions from size exclusion chromatography of a cellular extract from a strain expressing the full-length MTHI1 (first line in Figure 9), indicated by the bars A (fractions 4 to 8) and B (9 to 11), were pooled, immunoprecipitated with an antibody against the HA tag, and analyzed with the same antibody for the MTHI1 content of the immunoprecipitated fractions. Their RNA was extracted and analyzed by dot blot with a probe specific of the atpH 5′ UTR, which detected a (weak) signal in pooled fractions A, further analyzed by deep sRNA-seq.
(D) Ratio of normalized sRNA coverage in MTHI1 RIP samples. Differential enrichment was calculated as the ratio of the coverage at each nucleotide position in the MTHI1-HA sample to that in the wild-type control sample (+1). Blue curve, sRNAs mapping to the + strand; red curve, sRNA mapping to the − strand. Most enriched genome positions are shown on the graph, as well as the position of the inverted repeat (atpH and atpI 5′ UTRs).
(E) Coverage of immunoprecipitated RNA (normalized as RPM) over the atpH and atpI loci, schematically depicted at the bottom of the panel. The black bar in the atpI 5′ UTR shows the position of the MTHI1 target (see below). Blue curve, MTHI1-RIP sample; red curve, WT-RIP sample (negative control). A close-up view of the atpI 5′ UTR is shown in Supplemental Figure 7B. Note the very different values of the y axes in the two graphs.
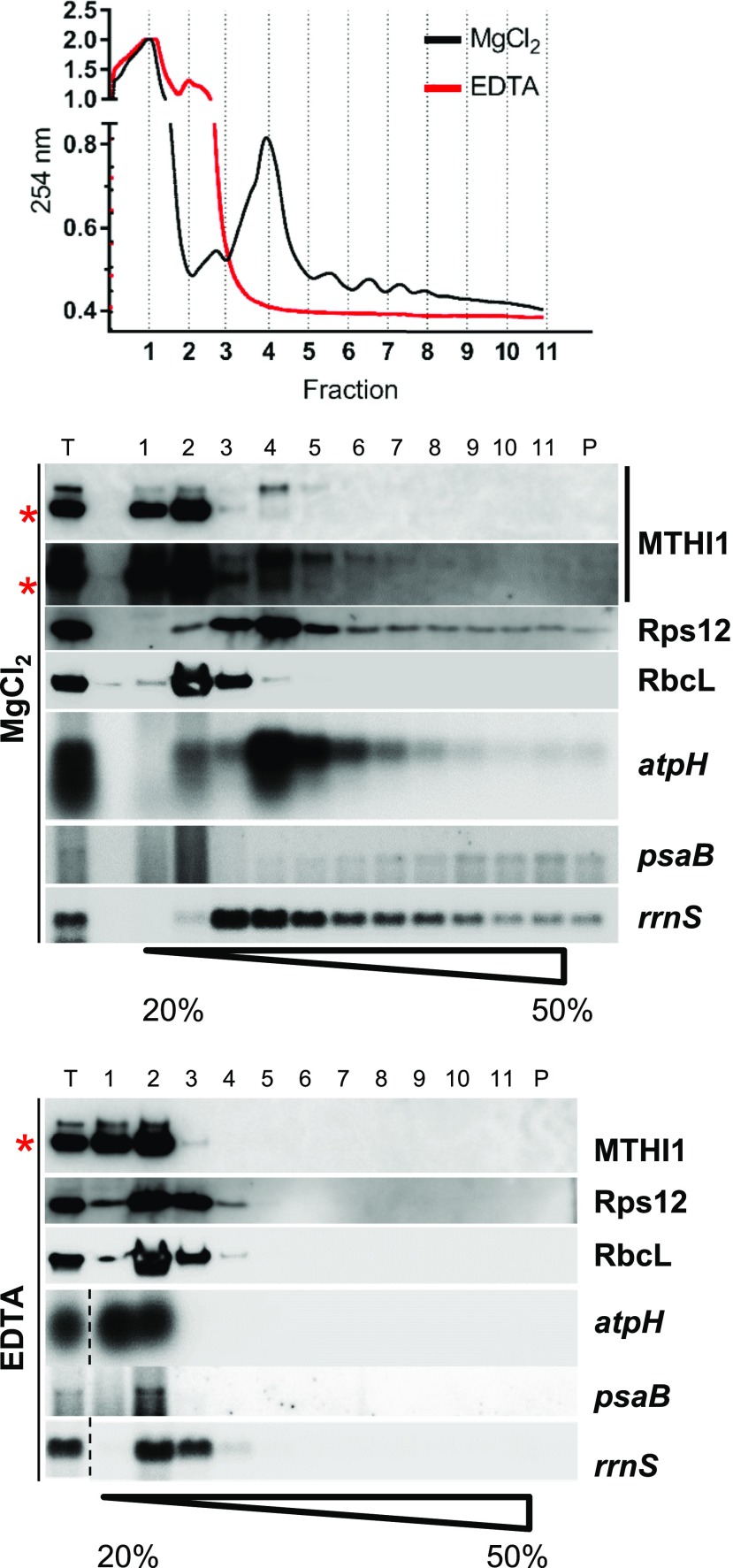
We sequenced sRNAs from the immunoprecipitated samples. To increase specificity, the MTHI1 complexes were purified by size exclusion chromatography. MTHI1-HA was then immunoprecipitated independently from pooled fractions 3 to 8 and 9 and 10 (Figure 9). Only RNAs extracted from fractions 3 to 8 gave rise to a 5′atpH signal in the dot-blot experiment (Figure 11C) and were used for library construction. Since atpH mRNA is triphosphorylated (Figure 10B; Cavaiuolo et al., 2017), all samples were treated with RPP prior to library construction. Figure 11D displays the ratio of normalized sRNA coverage (expressed as reads per million [RPM]) in the strain complemented with the tagged MTHI1 versus that in the wild type, plotted along the chloroplast genome. In the MTHI1-HA sample, sRNAs were strongly enriched at the 5′ end of the atpH 5′ UTR, as better shown in Figure 11E. By contrast, sRNAs were not enriched around the atpI 5′ UTR compared with the wild-type sample (Figures 11D and 11E), which is in agreement with the absence of atpI signal in the RNA immunoprecipitation (RIP) experiments (Figure 11B). However, in contrast to the lack of signal in dot blots hybridized to an rrnS probe, sRNAs mapping to the ribosomal operon were somehow enriched in the MTHI1-HA RIP library (Supplemental Figure 7C). To solve this discrepancy, we looked to the possible association of MTHI1 to ribosomes along a Suc gradient (Figure 12). As atpH contains a short CDS, it only accommodates a limited number of ribosomes and does not migrate deep in the gradient. The distribution of MTHI1 paralleled that of atpH mRNA: both were found in polysomal fractions 4 to 8, as shown by the UV light absorbance profile and by their dissociation upon EDTA treatment. Thus, MTHI1 remains bound to the atpH transcript when engaged in translation.
Figure 12.
MTHI1 Interacts with Polysomes.
The top panel shows the UV light absorbance profile along a Suc gradient, the middle panel shows the distribution of MTHI1, Rps12, and RbcL proteins and atpH, psaB, and rrnS (16S rRNA) transcripts in the wild-type cells, assessed by immunoblots, and the bottom panel shows the distribution of the same proteins and transcripts in samples treated with EDTA to dissociate the ribosomes. For the gradient in the presence of MgCl2, an overexposed blot immuno-decorated with the antibody against the MTHI1 protein is shown. T represents the total protein and RNA extracts. Note that atpH mRNA, encoding a short polypeptide, is not heavily loaded with ribosomes and does not penetrate deep in the gradient (red asterisk, cross-contamination).
Identification of the Targets of MTHI1
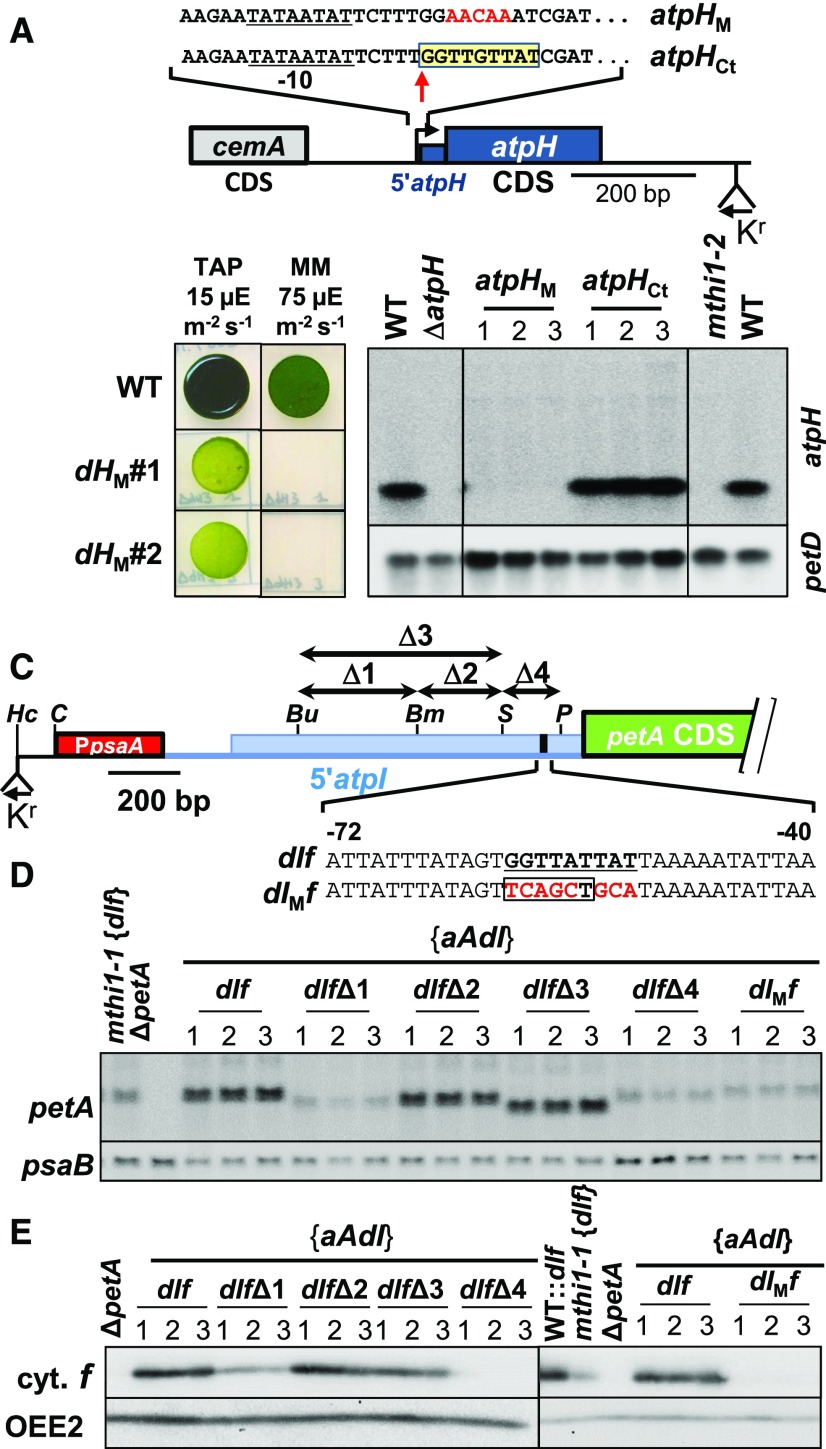
To gain more information about the target of the nine OPR repeats-containing MTHI1 protein, we investigated the conservation of the small (40-bp) atpH 5′ UTR, which is well conserved among green algae. Indeed, the first nine nucleotides (GGTTGTTAT) of the atpH transcript were strongly conserved in almost all Chlorophyceae, Pedinophyceae, and the Ulvale clade of Ulvophyceae (Supplemental Figure 8A; Supplemental Data Set), but not in Trebouxiophyceae or Prasinophytes. To test whether this sequence, which is often localized downstream of a putative Pribnow-10 box, corresponds to the target of MTHI1, we mutated it into the poorly related GGAACAAAT sequence (Figure 13A). After introducing this mutated gene into the chloroplast genome, the transformants lost phototrophy and failed to accumulate the atpH transcript (Figure 13B), suggesting that MTHI1 could no longer bind to and protect the transcript.
Figure 13.
Validation of the Putative MTHI1 Targets.
(A) Schematic map of the atpHM construct with a close-up view of the region of the MTHI1 binding site, highlighted in a yellow box. Mutated nucleotides are written in red. The atpH transcription start site is indicated by a vertical arrow. The atpH promoter is underlined, and the position of the recycling aadA cassette is shown. The control construct (atpHCt) carries the selection cassette but no mutation in the atpH gene.
(B) (Left) Phototrophic growth of the atpHM strain (two independent transformants), assessed as in Figure 2B. Growth of the wild type (WT) is shown as a control. MM, minimal medium. (Right) Accumulation of the atpH transcript in the wild type transformed by the atpHCt and atpHM constructs, assessed by RNA gel blots. Three independent transformants are shown for each genetic background. The petD transcript is provided as a loading control.
(C) Schematic representation of the 5′atpI 5′ UTR in the mutant dIf series.
The red rectangle represents the psaA promoter region, and the blue line shows the psbJ-atpI intergenic fragment inserted in the construct (larger than the atpI 5′ UTR, to allow the processing of the chimeric transcript). The blue rectangle symbolizes the processed atpI 5′ UTR, with the target of MTHI1 shown in black. Relevant restriction sites Bu (Bsu36I), Bm, (BsmI), S (SnaBI), P (PflMI), and Hc (HincII, where the selection cassette was inserted) are indicated. Arrows above the map indicate the position of the deletions, while the bottom insert shows the mutation introduced in the MTHI1 binding site (underlined) in the dIMf strain, with mutated nucleotides shown in red. A PvuII site introduced as a restriction fragment length polymorphism marker is boxed.
(D) Accumulation of the chimeric petA transcript in the {aAdI} strain transformed with the indicated dIf variants, assessed by RNA gel blots. Three independent transformants are shown for each construct. psaB mRNA is shown as a loading control.
(E) Accumulation of the chimeric cytochrome f (cyt. f) in the same strains, and in the {aAdI}, mthi1-1 {dIf}, and ΔpetA strains as controls, assessed by immunoblots. Immuno-detection of OEE2 is provided as a loading control.
Since MTHI1 also targets the atpI transcript, we searched for occurrence of this motif in the atpI 5′ UTR. In contrast to the atpH 5′ UTR, the long atpI 5′ UTR (493 bp in C. reinhardtii) is not conserved in Chlorophyta, except in some Chlorophyceae, Pedinophyceae, and in the Ulvale clade, for a stretch of ∼60 nucleotides upstream of the initiation codon (Supplemental Figure 8B; Supplemental Data Set). Strikingly, this conserved stretch starts by a GGTT(A/G)TTAT motif. We tested its significance by introducing deletions or mutations in the atpI 5′ UTR (Figure 13C). To facilitate the characterization of the resulting mutants, mutations were introduced in the atpI 5′ UTR of the dIf reporter gene (Figure 4A). Moreover, to avoid recombination between the 5′ UTRs of the endogenous atpI gene and the chimeras, the latter were introduced in the chloroplast genome of the {aAdI} strain, which lacks the atpI 5′ UTR (Figure 3A). A deletion of 168 bp in the atpI 5′ UTR (Δ1) strongly decreased the accumulation of the chimeric transcript (Figure 13D) and its cytochrome f gene product (Figure 13E). The deletion of the next 129 bp, either alone (Δ2) or together with the upstream 168 nucleotides (Δ3), did not alter the accumulation of the chimeric mRNA nor its gene product (Figures 13D and 13E), suggesting that antagonistic regulatory elements at the beginning and in the middle of the atpI 5′ UTR fine-tune the expression of the atpI gene, as already observed in other 5′ UTRs (Costanzo and Fox, 1993; Sakamoto et al., 1994). Deletion of 86 bp encompassing the GGTTATTAT motif (Δ4) decreased the accumulation of the chimeric transcript, although it remained more abundant than in strains carrying the Δ1 deletion, but this deletion totally abolished its translation. Mutation of this motif to TCAGCTGCA, leaving the rest of the UTR unaltered, led to the same decreased accumulation of the chimeric transcript as in the mthi1 mutants and prevented cytochrome f expression, confirming its importance for atpI mRNA translation.
DISCUSSION
MTHI1 Is a Major Player in CFo Biogenesis
Here, we show that MTHI1 plays a dual role in controlling the expression of AtpH and AtpI, the two subunits of the selective proton channel, and is therefore a major player in the biogenesis of the CFo sector of chloroplast ATP synthase.
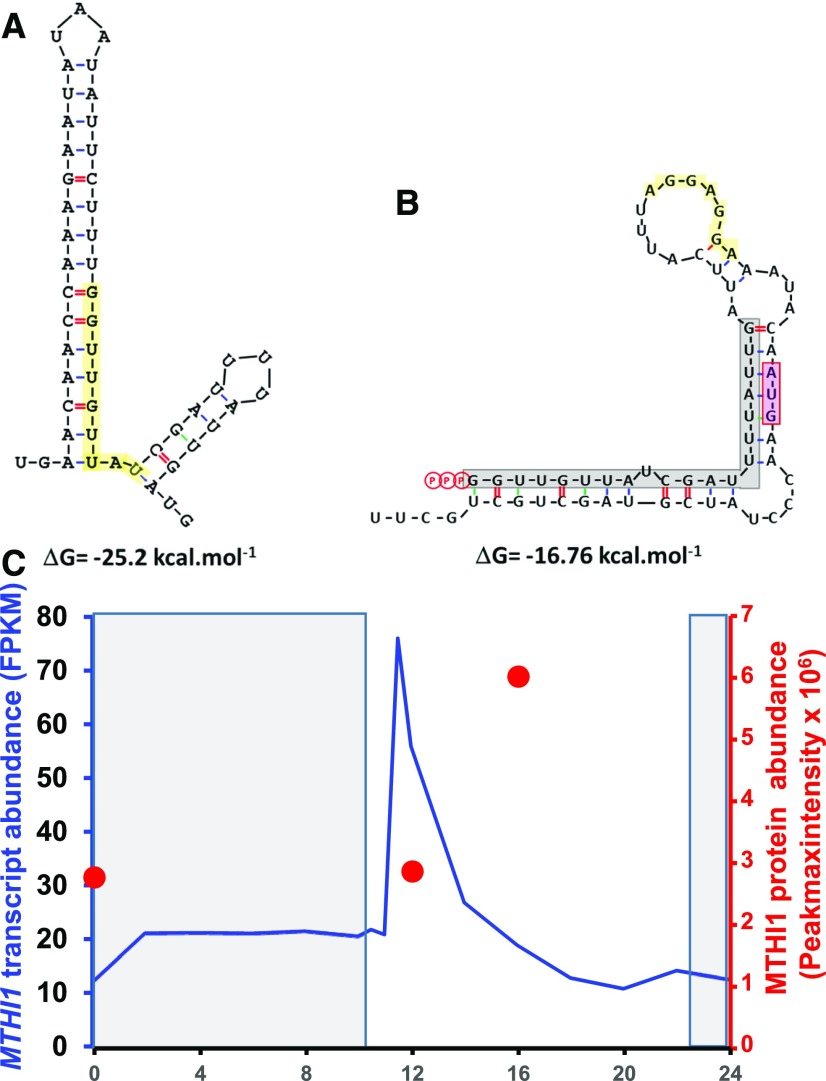
MTHI1 is required for the stable accumulation of the monocistronic atpH mRNA. Being an OPR protein, MTHI1 likely binds directly to its RNA target, as was shown for the OPR factor TAB1 (Rahire et al., 2012). Like PPR proteins in plants (reviewed by Barkan and Small, 2014), in Chlamydomonas, OPR proteins are involved in all posttranscriptional steps of chloroplast gene expression: maturation/stabilization (Murakami et al., 2005; Kleinknecht et al., 2014; Wang et al., 2016; Cavaiuolo et al., 2017; Viola et al., 2019), translation activation (Auchincloss et al., 2002; Eberhard et al., 2011; Rahire et al., 2012; Lefebvre-Legendre et al., 2015), and splicing (Rivier et al., 2001; Balczun et al., 2005; Merendino et al., 2006; Marx et al., 2015; Reifschneider et al., 2016). Like other M factors (Loiselay et al., 2008; Wang et al., 2015; Cavaiuolo et al., 2017), MTHI1 binds to the very 5′ end of its target mRNA to protect it from 5′ → 3′ exonucleases, whose action can alternatively be impaired by the addition of a poly(G) cage at the beginning of the transcript. The nine OPR repeat-containing MTHI1 protein interacts with the first nine nucleotides (GGTTGTTAT) of atpH mRNA, which is highly conserved among Chlorophyceae, Pedinophyceae, Nephroselmidophyceae, and in the Ulvale clade (Sun et al., 2016) of the Ulvophyceae class of green algae and whose mutation prevents atpH mRNA accumulation. This interaction results in a specific footprint in atpH mRNA that can be coimmunoprecipitated with MTHI1 protein and that is highly reduced, although not abolished, in the mthi1-1 mutant. Whether this is due to the leakiness of the mthi1-1 mutant (which reverts to some extent when plated on minimal medium), to a low affinity of other OPR proteins for atpH mRNA, or to an intrinsic stability of triphosphorylated transcripts that are poor substrates for 5′ → 3′ exonucleases (Richards et al., 2011; Luciano et al., 2012; Foley et al., 2015) remains to be determined. The monocistronic atpH mRNA is transcribed from its own promoter and does not result from the processing of precursors transcribed from the atpA promoter. Although atpH mRNA precursors accumulate to the wild-type levels in mthi1 mutants, they could not translate the AtpH subunit in the absence of the atpH translational activator MTHI1. In the wild type, the AtpH subunit is probably not synthesized from these precursors either, despite the presence of MTHI1, as the target of MTHI1 is sequestered within a stable secondary structure (Figure 14A), likely preventing the binding of OPR proteins, as do secondary structures for PPR proteins (Kindgren et al., 2015; Zoschke et al., 2016; Miranda et al., 2017; McDermott et al., 2018).
Figure 14.
Modulation of MTHI1 Action.
(A) Secondary structure sequestering the MTHI binding site in the precursor RNA transcribed from the atpA promoter. Lowest energy structure calculated at 25°C by RNA Folding Form (M-Fold: http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/rna-form1.cgi; Zuker, 2003) for the region surrounding the atpH 5′ end in the precursor transcript initiated at the atpA promoter. The MTHI1 binding site is shaded in yellow.
(B) Secondary structure of the atpH 5′ UTR in the dHf chimera, sequestering the initiation codon. Lowest energy structure calculated at 25°C by M-Fold for transcribed atpH sequences inserted upstream of the petA gene in chimera dHf. The footprint of MTHI1 is shaded in gray, the atpH initiation codon is shaded in pink, and the Shine-Dalgarno sequence is shaded in yellow.
(C) Variations in MTHI1 transcript and protein accumulation over the circadian cycle. Redrawn from the data in Strenkert et al. (2019). The dark period is indicated by the shaded area. Blue line shows the accumulation of the MTHI1 transcript over time (expressed as fragments per kilobase million [FPKM]); the red dots show the accumulation of the MTHI1 protein at the indicated time points (expressed as Peakmaxintensity).
The fate of trans-acting factors during translation remains poorly understood. Most trans-acting factors that have been studied are not found in polysomal fractions (Boudreau et al., 2000; Auchincloss et al., 2002; Dauvillée et al., 2003; Schwarz et al., 2007; Viola et al., 2019). Either their association does not resist the polysome preparation procedure or they dissociate from their target mRNA upon translation, raising the question of the stability of translated mRNAs (Kato et al., 2006; Viola et al., 2019). MTHI1, however, remains associated with atpH mRNA loaded on polysomes, while sRNAs derived from the ribosomal cluster were enriched in the MTHI1 RIP samples (even if rrnS signal was not observed in dot blots). This unique behavior may favor the re-initiation of atpH mRNA translation, whose rate of translation in exponentially growing cells is higher than that of most other photosynthetic transcripts.
MTHI1 also contributes to the stabilization of atpI mRNA while strongly enhancing its translation. However, we did not detect any specific footprint within the atpI 5′ UTR, nor did we find evidence for a binding of MTHI1 that would resist RIP experiments, whether analyzed by dot blot or deep sequencing. Similarly, we previously failed to observe a footprint diagnostic of an interaction of the translational activator TCA1 with its target, the 5′ UTR of petA (Cavaiuolo et al., 2017), despite experimental evidence that TCA1 interacts with this RNA region (Loiselay et al., 2008; Boulouis et al., 2011). It is likely that T factors (here MTHI1) interact only transiently with their target transcript (here the atpI 5′ UTR) to promote translation. However, mutating the putative MTHI1 binding site within the atpI 5′ UTR destabilized the 5′atpI-petA chimeric transcript as in the mthi1 mutants and totally prevented the synthesis of a reporter protein, highlighting its importance for the expression of the atpI gene.
PPR10 and MTHI1: An Example of Convergent Evolution
The mode of action of Chlamydomonas MTHI1 strikingly resembles that of the maize (Zea mays) PPR10 protein, even though the two proteins are not evolutionarily related, as they belong to different protein families (OPR versus PPR). PPR10 targets the atpI-atpH intergenic region to stabilize the transcripts of these adjacent and cotranscribed genes by, respectively, protecting them from 3′ → 5′ and 5′ → 3′ exonucleases (Pfalz et al., 2009). The binding of PPR10 generates a footprint matching the overlapping ends of the atpI and atpH transcripts (Zhelyazkova et al., 2012). In addition, PPR10 activates the translation of atpH mRNA by opening a secondary structure that would otherwise sequester the Shine-Dalgarno sequence (Prikryl et al., 2011). Similarly, MTHI1 may activate the translation of atpH mRNA by opening a secondary structure sequestering the atpH initiation codon (Figure 14B). However, unlike MTHI1, PPR10 is not involved in the translation activation of atpI mRNA (Zoschke et al., 2013).
The Two Target Genes of MTHI1 Are Widely Separated in the Chloroplast Genome
Since MTHI1 targets two genes that are widely separated in the chloroplast genome, it appears unusual compared to other factors characterized so far in Chlamydomonas. These factors target a single chloroplast transcript to allow its stable accumulation (Kuchka et al., 1989; Drapier et al., 1992; Drager et al., 1998; Boudreau et al., 2000; Loiselay et al., 2008; Johnson et al., 2010; Wang et al., 2015; Cavaiuolo et al., 2017) or to activate its translation (Rochaix et al., 1989; Stampacchia et al., 1997; Wostrikoff et al., 2001; Auchincloss et al., 2002; Dauvillée et al., 2003; Raynaud et al., 2007; Schwarz et al., 2007; Eberhard et al., 2011; Lefebvre-Legendre et al., 2015; Cavaiuolo et al., 2017). A recent genome-wide ribosome profiling study performed on the Chlamydomonas nac2 (mbd1-nac2) mutant, which is defective in the accumulation of psbD mRNA, only detected very limited changes in chloroplast gene expression, most of which were attributed to PSII deficiency rather than to the absence of Nuclear Affecting Chloroplast2 (NAC2) per se (Trösch et al., 2018). The only exception so far is the factor Maturation/stability of the psbB mRNA (MBB1), which is required for the stable accumulation of psbB mRNA, encoding CP47, a core antenna complex of PSII. MBB1 is also required for the correct processing and translation of the cotranscribed psbH mRNA, encoding another PSII subunit (Monod et al., 1994; Vaistij et al., 2000a, 2000b; Loizeau et al., 2014). In both cases, these bifunctional factors target two subunits that tightly interact in the assembled complex (Komenda et al., 2005; Boehm et al., 2011; Murphy et al., 2019), whose synthesis is highly interdependent in other organisms (Jean-Francois et al., 1986; Ooi et al., 1987; Payne et al., 1991; Komenda, 2005; Bietenhader et al., 2012). Such bifunctional factors would thus provide a mechanism alternative to the CES process to coregulate the expression of closely interacting subunits.
The landscape of the nuclear control of chloroplast gene expression in Chlamydomonas appears to be widely different from that in vascular plants. The trans-acting factors in land plants show a looser specificity. When targeting a polycistronic transcript, these factors may recognize both the 3′ end of the upstream transcript and the overlapping 5′ end of the downstream transcript (Pfalz et al., 2009; Zhelyazkova et al., 2012). Moreover, they often bind to similar sequences in different transcription units, often encoding subunits of different protein complexes. The maize protein Chloroplast RNA Processing1 (CRP1) activates the translation of both petA and psaC (4Fe-4S centers-containing subunit of PSI) transcripts and is also required for the processing of petB and petD (cytochrome b6 and PetD subunits of the cytochrome b6f complex, respectively) monocistronic RNAs in maize as in Arabidopsis (Barkan et al., 1994; Fisk et al., 1999; Schmitz-Linneweber et al., 2005; Ferrari et al., 2017). In addition to its role in atpI and atpH expression, the maize PPR10 protein also controls the accumulation of the monocistronic transcripts of the adjacent rpl23 and psaJ genes (Pfalz et al., 2009; Prikryl et al., 2011; Zhelyazkova et al., 2012). A recent genome-wide ribosome profiling study revealed an even more complex situation by highlighting the unexpected versatility of several PPR proteins in plants, since PPR10 also stabilizes the monocistronic psaI mRNA, while Proton Gradient Regulation3 (PGR3) binds to the rpl14-rps8 (ribosomal subunits Rps14 and Rps8, respectively) intergenic region to stabilize rpl14 mRNA at its 3′ end and to stimulate rps8 translation (Rojas et al., 2018).
The Paradoxical Specificity of Trans-Acting Factors in Chlamydomonas
The high specificity of trans-acting factors in Chlamydomonas appears paradoxical, since, for example, the GTT(G/A)TTAT target of MTHI1 is not restricted to atpH or atpI mRNA but is found several times in the chloroplast transcriptome of C. reinhardtii: 3 times for GTTGTTAT, in atpH, rpoC2, and rpoC1 (RNA Polymerase subunits β′′ and β′, respectively) transcripts; 2 times for GGTTATTAT, in atpI and rps3 (ribosomal subunit Rps3) transcripts; and 11 times for the more degenerate GGTTNTTAT motif. However, these extra motifs do not lead to footprints or sRNA enrichment in MTHI1-RIP samples, suggesting that the affinity of MTHI1 for its GGTTGTTAT target remains moderate and requires additional determinants, presently unknown, for its strong interaction with the atpH 5′ UTR. This interaction leads to the formation of an abundant footprint, whereas that with a very similar motif in the atpI 5′ UTR does not. This is unlikely to result from a differential affinity of MTHI1 for its two targets: changing GGTTGTTAT to GGTTATTAT did not modify the accumulation of the atpH transcript or lead to notable changes in cytochrome f expression from the dHf chimera (Supplemental Figure 9).
The correlated abundances of MTHI1 and atpH mRNA in a series of transformants argues for MTHI1 being a limiting factor for the expression of atpH. The stimulated expression of the dHf and dIf chimera in the absence of the atpI or atpH genes, respectively, suggests that the two genes share some common factors, MTHI1 being a likely candidate. However, the deletion of the abundant atpH mRNA, stoichiometrically bound to its stabilization factor, should release much more MTHI1 protein than the deletion of the atpI gene, whose mRNA, which is 10-fold less abundant (Cavaiuolo et al., 2017), interacts only transiently with its translational activator. Still, deleting the atpI gene stimulates the expression of the dIf chimera much more strongly than deleting atpH. Therefore, another factor(s) specific to the 5′ UTR of atpI mRNA likely limits atpI expression.
The interaction of several factors assembled in a complex on a target 5′ UTR may, despite the moderate specificity/affinity of each of them for its target, lead to a strong cooperative interaction that is much more stable than that between any two components taken separately. An atpH-specific factor interacting with both the atpH 5′ UTR and MTHI1 could tether it on the atpH 5′ end, but not on other occurrences of the same motif. A weak affinity of an atpI-specific factor for MTHI1 may similarly result in a transient, but still specific, interaction with the atpI 5′ UTR. Such cooperative interactions prevail for the few chloroplast genes whose expression has been studied in detail in Chlamydomonas. MCA1 and TCA1 are strictly required for the accumulation and translation of the petA transcript, respectively. Nevertheless, the translation of this transcript decreases 10-fold in the absence of MCA1, while its stability decreases by 85% in the absence of TCA1 (Wostrikoff et al., 2001; Raynaud et al., 2007; Loiselay et al., 2008). The two factors form a ternary complex with petA mRNA (Boulouis et al., 2011), and the absence of any of them weakens the interaction between the other two. Similarly, NAC2, the stabilization factor of the psbD transcript, recruits the 40-kD RNA Binding protein (RB40) to activate the translation of psbD mRNA, despite the poor specificity of RB40 for U-rich regions (Schwarz et al., 2007). Finally, MDA1 and TDA1, which are required for the accumulation and translation of atpA mRNA, also form a complex assembled onto the atpA mRNA (Viola et al., 2019).
Such a dually footed mechanism could favor the high plasticity of nucleo-chloroplastic interactions observed in Chlorophyceae: despite a mutation in its target, a trans-acting factor would, through its interaction with other factors, remain in contact with it, allowing the selection of compensatory mutations over time. It also helps to understand the recycling of M factors: once the target mRNA is degraded, the complex will dissociate, and due to the moderate affinity of the M factor for its target, the footprint sRNA will be released, rather than trapped, allowing the protein to interact with newly synthesized mRNAs.
The Coregulation of atpH and atpI: An Ancestral Situation
The joint control of atpH and atpI expression can be traced back during evolution. In Escherichia coli, the unc operon organization facilitates the concerted expression of ATP synthase subunits, even if additional translational controls are required to set their contrasted stoichiometry. Cyanobacteria, including Gloeomargarita lithophora, the extant free-living cyanobacterium most closely related to the ancestor of chloroplasts (Ponce-Toledo et al., 2017), partially retained this gene organization, with ATP synthase subunits now encoded by two distinct operons: atpI-atpH-atpG-atpF-atpD-atpA-atpC and atpB-atpE (for the sake of clarity, cyanobacterial genes are named here as their chloroplast counterparts, rather than by their true name; e.g., the atpE locus of Gloeomargarita encoding subunit C [AtpH] is nevertheless named atpH). This ancestral organization was largely preserved in Archeplastidia: while the genes encoding subunits γ, δ, AtpI, and ATPG may have been relocated to the nucleus in some species, those remaining in the chloroplast still belong to two transcription units: (atpI)-atpH-(atpG)-atpF-(atpD)-atpA and atpB-atpE. A notable exception is the Chlorophyceae in which atp genes are shuffled around the chloroplast genome (Supplemental Data Set), raising the question of their coregulation.
In the Ulvale clade of Ulvophyceae and in Pedinophyceae, the atpI and atpH genes, although adjacent on the chloroplast genome, share a sequence similar to the MTHI1 binding site in their 5′ UTRs (Supplemental Figure 8A and 8B; Supplemental Data Set). This suggests an ancestral situation that placed the expression of the two genes under the control of an ortholog of MTHI1, paving the way for their separation in Chlorophyceae. This sequence possibly appeared early during evolution in the common ancestor of Pedinophyceae, Ulvales, and Chlorophyceae, together with the appearance of an efficient processing system that in green algae chloroplasts generates independent monocistronic transcripts from polycistronic transcription units, which are remnants of the ancestral cyanobacterial operons.
MTHI1 Is Conserved in Chlorophyceae
In Chlorophyceae, the conservation of the MTHI1 target goes along with the conservation of the MTHI1 protein, since all sequenced genomes except Coelastrella encode an ortholog of MTHI1. The region of similarity is restricted to the OPR-containing, N-terminal part of the protein. Even this conserved region evolves rapidly, with multiple species-specific insertions, some of which interrupt the OPR repeats (Supplemental Figure 6). Strikingly, the two Ulvale genomes presently available each encode an OPR protein with nine OPR repeats (Supplemental Figure 6), which are the mutual best hits of CrMTHI1 and are predicted (based on a preliminary version of the OPR code) to recognize the GGTTGTTAT sequence. These OPR are shorter than their Chlorophycean orthologs, as they lack the disordered C-terminal extension.
Downstream of this conserved region, all chlorophycean MTHI1 orthologs possess a C-terminal tail, rich in stretches of identical residues (mostly Ala, Ser, Gln, and Arg) that is predicted to essentially be a random coil. These tails are not conserved in sequence or in length (Supplemental Figure 6A) and do not show similarity to other proteins in databases, suggesting that they have no specific functions. Indeed, in Chlamydomonas, the tail appears to be dispensable for the major function of the protein, as are the N-terminal tails of TCA1 (Raynaud et al., 2007), NAC2 (Boudreau et al., 2000), RNA processing trans-splicing of psaA1 (RAA1; Merendino et al., 2006), and TDA1 (Eberhard et al., 2011) and the C-terminal tail of Maturation/stability of ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) subunit RbcL (MRL1; Johnson et al., 2010). These tails could result from the introduction of junk GC-rich DNA within permissive regions of the genes or from the loss of stop codons upon mutations in GC-rich regions, progressively extending the CDS. However, while the full-length MTHI1 factor is short lived with a half-life of ∼1 h, its C-terminal truncated version is stable (>8 h). Perhaps these tails, a common feature of trans-acting factors in Chlorophyceae, modulate the stability of the proteins, although the proteolytic process controlled by these tails is unknown.
The accumulation of the short-lived, full-length MTHI1 protein thus depends on changes in the abundance of the MTHI1 transcript, as occurs over the circadian cycle (Figure 14C). MTHI1, a limiting factor for the expression of atpH and atpI, would couple the expression of these genes to that of the nucleus-encoded ATP synthase subunits, whose transcripts show a similar pattern of expression (Figure 8F in Zones et al., 2015). Thus, MTHI1 behaves as a genuine regulator of the biogenesis of ATP synthase.
METHODS
Strains, Media, Culture Conditions, and Chemicals
Wild-type t222+ (derived from 137c: nit1 nit2), mutants, and transformed strains of Chlamydomonas (Chlamydomonas reinhardtii) were grown at 25°C in Tris-acetate-phosphate (TAP) medium, pH 7.2 (Harris, 1989), under continuous light (5 to 10 μE m−2 s−1; white light-emitting diode, whose emission spectrum is shown in Supplemental Figure 10) unless otherwise specified. Crosses were performed according to Harris (1989).
Nucleic Acid Manipulations: DNA and Constructs
Standard nucleic acid manipulations were performed according to Sambrook et al. (1989). The primers used in this study are listed in the Supplemental Table. All DNA constructs were sequenced before transformation in Chlamydomonas.
DNA Constructs
Plasmids p-520 and p-70, containing a 7.8 kb PstI fragment of the chloroplast genome encompassing the 3′ end of atpA, psbI, cemA, atpH, atpF, and rps11 cloned in the Bluescript pBSKS+ vector and a 4.9-kb EcoR1 fragment of the chloroplast genome encompassing the psbJ, atpI, psaJ, and rps12 genes cloned into pUC8, respectively, were obtained from the Chlamydomonas Resource Center (http://chlamycollection.org/).
Deletion of the atpH Gene
To remove unwanted restriction sites, plasmid p-520 was first digested by SacI and NcoI, blunted with T4 DNA polymerase (pol), and religated on itself. Next, it was digested with PacI and XhoI, blunted with T4 DNA pol, and religated on itself to yield plasmid p-520Sh that only contains a 4916-bp insert.
A 789 bp DNA fragment upstream of the atpH CDS was amplified from template p-520 using primers cemA_RI and atpHDel, digested by EcoRI and EcoRV, and cloned into plasmid p-520Sh digested by the same enzymes to create plasmid pΔatpH. The recycling psaA-driven aadA cassette (Boulouis et al., 2015), excised from plasmid p5′aA-aadA485 by digestion with SacI and XhoI, was cloned into plasmid pΔatpH, digested by the same enzymes (restriction sites introduced when designing primer atpH_del) to yield plasmid pKrΔatpH.
Deletion of the atpI Gene
A 1013-bp DNA fragment was amplified by two-step megaprime PCR (Higuchi, 1990): primers psbJ_FW/atpIDel_RV and atpI_RV/atpIDel_FW allowed the amplification from plasmid p-70 of two partially overlapping amplicons that were mixed and used as templates in a third PCR with the external primers psbJ_FW and atpI_RV. In the final amplicon, the whole atpI 5′ UTR and CDS were deleted and replaced by a short multiple cloning site. After digestion by ClaI and HpaI, this amplicon was cloned into plasmid p-70 digested with the same enzymes, yielding plasmid pΔatpI. The recycling psaA-aadA cassette, excised from plasmid p5′aA-aadA485 by digestion with SacI and XhoI, was cloned into plasmid pΔatpI, digested by the same enzymes (restriction sites introduced when designing primer atpIDel_FW and atpIDel_RV) to yield plasmid pKrΔatpI.
Construction of Reporter Genes
5′atpH-Driven Reporter Genes
The atpH promoter and 5′ UTR, PCR amplified from the template plasmid p-520 using primers atpHprom and atpHATG, were digested by EcoRV and NcoI and cloned into the pWFaAK vector digested by the same enzymes to yield plasmid pWFdHK.
The promoter and a slightly extended 5′ UTR of atpH were similarly amplified using primers PCR atpHprom and atpHATG2, digested by HincII and NcoI and cloned into plasmid paAf (Wostrikoff et al., 2004), digested by the same enzyme to yield plasmid pdHf. The recycling psaA-driven aadA cassette, excised from plasmid p5′aA-aadA485 by digestion with SacI and KpnI and blunted with T4 DNA pol, was cloned into plasmid pdHf digested with HincII to yield plasmid pKrdHf.
pGatpH
The pGatpH construct was created by a two-step PCR procedure, using the external primers cemA-FW and atp_RV and template plasmid p-520Sh. The final amplicon, carrying the poly(G) track (909 bp), was digested with EcoRI and EcoRV and cloned into plasmid p-520Sh digested with the same enzymes to create plasmid patpH-pG. The recycling 5′aA-aadA485 was then cloned into plasmid patpH-pG digested with EcoRV to yield plasmid pKrpGatpH-pG.
5′atpI-Driven Reporter Genes
The atpI 5′ UTR was amplified from the template plasmid p-70 using oligonucleotides atpIATG and atpI5′FWprom. The resulting 637-bp amplicon was digested by NsiI and NcoI and cloned into vectors paAf or pWFaAK digested with the same enzymes to yield plasmid pdIf or pWFdIK, respectively. In these constructs, the ClaI-NsiI fragment from the psaA 5′ UTR and promoter regions provides a promoter to drive the expression of the promoter-less atpI 5′ UTR. The recycling psaA-aadA cassette, excised from plasmid p5′aA-aadA485 by digestion with SacI and KpnI and blunted with T4 DNA pol, was then cloned into plasmid pdIf digested with HincII to yield plasmid pKrdIf.
Reporter Genes Driven by Modified atpI 5′ UTRs
Plasmid pKrdIf was digested with either Bsu96I and BsmI, BsmI and SnaBI, Bsu96I and SnaBI, or SnaBI and PflMI, blunted by T4 DNA pol treatment, and religated on itself to yield plasmids pKrdIfΔ1, pKrdIfΔ2, pKrdIfΔ3, and pKrdIfΔ4, respectively. The putative target of MTHI1 within the atpI 5′ UTR was also modified by megaprime PCR: primers atpI5′_FW/atpITar_RV and atpITar_FW/atpI5′_RV allowed the amplification from plasmid p-70 of two partially overlapping amplicons that were mixed and used as templates in a third PCR with the external primers atpI5′_FW and atpI5′_RV. This 996-bp final amplicon was digested with SnaBI and PmlI and cloned into the pKrdIf vector digested by the same enzymes to create plasmid pKrdIfΔT.
5′psaA-Driven atpI
To remove unwanted restriction sites, the p-70 vector was cut with ClaI and NdeI, blunted with Klenow enzyme, and religated on itself to yield plasmid p-70_CN. This plasmid was then digested with EcoRI and SexAI, filled with Klenow, and religated on itself to generate plasmid p-70Sh.
The atpI 5′ UTR was then deleted from this plasmid by a two-step megaprime PCR procedure: primers psbJ_FW/atpIChim_RV and atpI-Chim_FW/atpI5′_RV allowed the amplification from plasmid p-70 of two partially overlapping amplicons that were mixed and used as templates in a third PCR with the external primers psbJ_FW and atpI5′_RV. This 743-bp amplicon was digested with KpnI and BstBI and cloned into the p-70Sh vector digested with the same enzymes to create plasmid patpIΔ5′.
To generate plasmid p5′psaA-atpIKr, the promoter and 5′ UTR of the psaA gene were amplified from the template plasmid ps1A1 (Kück et al., 1987) using primers psaAprom and psaAATG. The resulting 270-bp fragment amplicon was digested with ClaI and NcoI and cloned into vector patpIΔ5′, digested with the same enzymes to yield plasmid p5′psaA-atpI. This plasmid was digested with SmaI (a restriction site introduced in the psaAprom primer) and ligated with the recycling 5′aA-aadA cassette to yield plasmidepKr5′psaA-atpI.
AtpISt
An untranslatable version of the atpI gene was constructed by a two-step megaprime PCR procedure, using the external primers atpI_FW and atpI_RV2 and the mutagenic primers atpISt_FW and atpISt_RV on the template plasmid p-70Sh. The final 1069-bp amplicon was digested with KpnI and Bsu36I, and the resulting 654-bp fragment was cloned into p-70Sh digested with the same enzymes to create patpISt. The recycling 5′aA-aadA resistance cassette was then cloned into the HpaI site of this vector to yield plasmid patpIStKr.
MTHI1 Constructs
We constructed a vector encompassing the genomic sequence of the MTHI1 gene by digesting the 21H4 cosmid by EcoRV and XhoI, isolating the 10679-bp subfragment that was cloned into pBluescriptII SK− digested by XhoI and AleI to create plasmid pgMTHI1. A triple HA tag was fused to the C terminus of the protein by megaprime PCR, using the mutagenic primers MTHI1HAFW and MTHI1HARV and the external primers MTHI1FW5 and MTHI1RV5. The resulting 1252-bp amplicon was digested with SfrI and SpeI and cloned into the pgMTHI1 vector digested by the same enzymes to create plasmid pgMTHI1-HA. To remove most of the C-terminal domain of the protein but keep the triple HA tag, a 921-bp PCR product was amplified from template pgMTHI1-HA with primers MTHI1DelC_FW and MTHI1DelC_RV, digested by HindIII and SrfI, and cloned into plasmid pgMTHI1 digested by the same enzymes to generate plasmid pgMTHI1ΔC.
The AV629671 EST clone containing a full-length cDNA cloned into the pBluescriptII SK− vector, was obtained from the Kazusa DNA Research Institute (Asamizu et al., 2000). The triple HA tag was introduced in this plasmid by cloning the 350-bp fragment recovered from the digestion of the above-mentioned 1252-bp PCR fragment with FspAI and Bst1107I into the AV629671 vector digested by the same enzymes to yield plasmid pcMTHI1-HA.
MTHI1 Recoding
The MTHI1 CDS (Cre17.g734564.t1.1) was codon optimized for expression in Escherichia coli (ec) and synthesized by GeneCust (Supplemental Figure 11). The synthesized EcMTHI1 gene lacks the first 147 bp and contains instead 5′-ATGGCGATTGCAATTGGAATTCAT-3′ that is derived from the bacterial araB gene (ECK0064). EcMTHI1 was cloned into vector pET28a (Novagen) using the NcoI and HindIII restriction sites to yield plasmid pET28a-EcMTHI1. To introduce the hexa-His tag at the N terminus, primers EcMTHI1-F and EcMTHI1-R were used to amplify a 573-bp DNA fragment from plasmid pET28a-ecMTHI1, digested with NcoI and AgeI, and cloned into pET28a-ecMTHI1 digested with the same enzymes using the NEBuilder HighFidelity DNA Assembly Cloning (New England Biolabs) strategy, resulting in pET28a-6His-ecMTHI11.
RNA Isolation and Analysis
RNA extraction and RNA gel-blot analysis were performed as described previously (Drapier et al., 2002) with 33P-labeled probes derived from CDSs (Eberhard et al., 2002). Transcript accumulation was quantified from PhosphorImager scans of the blots, as described by Choquet et al. (2003). In Figures 2B and 6C, probes amplified with primers listed in the Supplemental Table were digoxigenin-labeled using digoxigenin-dUTP, the anti-digoxigenin Fab fragment, and CDP Star reagent (Roche). Signals were acquired in a ChemiTouch imaging system (Bio-Rad) and analyzed with ImageLab 3.0 software (Bio-Rad). Transcriptomic analyses were performed as described in Cavaiuolo et al. (2017). In Figure 2, polysome analyses were performed as described in Minai et al. (2006) and Eberhard et al. (2011). In Figure 12, we used a modified protocol adapted from Trösch et al. (2018) as follows. Cell cultures were grown to mid-logarithmic phase (2 to 3 × 106 cells mL−1) and supplemented with 100 µg mL−1 chloramphenicol 15 min before harvesting. Cell pellets were resuspended in polysome buffer (20 mM Tris, pH 8.0, 25 mM KCl, 50 mM β-mercaptoethanol, 0.5 mg mL−1 heparin, 100 μg mL−1 chloramphenicol, 0.2 M Suc, 1% [v/v] Triton X-100, and 1× Protease Inhibitor Cocktail [Roche]), with or without MgCl2 (25 mM). The cells were broken with a French press, and cell lysates were centrifuged at 10,000g for 15 min at 4°C to remove cell debris. EDTA samples were prepared without MgCl2 and supplemented with 20 mM EDTA. MgCl2 and EDTA supernatants were loaded on a 20 to 50% (w/v) continuous Suc gradient. The 20 and 50% Suc solutions were prepared in a buffer containing 20 mM Tris, pH 8.0, 25 mM KCl, 5 mM β-mercaptoethanol, 0.5 mg mL−1 heparin, and 100 μg mL−1 chloramphenicol supplemented with 25 mM MgCl2 or 1 mM EDTA. Suc gradients were centrifuged at 38,000 rpm for 150 min in a SW41 Ti rotor (Beckman). Eleven fractions were collected, and the pellet was resuspended in 1.1 mL of solution containing 5 mM EDTA and 0.1% SDS.
Transformation Experiments
Chloroplast transformation was performed by tungsten particle bombardment (Boynton et al., 1988) as described in Kuras and Wollman (1994) using a home-made helium gun. Transformants (listed in Table 1) were selected on TAP-Spec (100 µg mL−1) and subcloned on TAP-Spec (500 µg mL−1) until they reached homoplasmy, which was assessed as described in Table 1. For each transformation, at least four independent transformants were analyzed. Phenotypic variations between independent transformants proved negligible.
Nuclear transformation of mthi1 strains was performed by electroporation, as described by Raynaud et al. (2007), with the following parameters: 10 mF/1200 V·cm−1. Transformants were selected based on phototrophy on minimal medium (Harris, 1989) under high light (150 µE·m−2·s−1).
Protein Preparation, Separation, and Analysis
14C pulse-labeling experiments in the presence of cycloheximide (10 µg mL−1) and protein isolation, separation, and immunoblot analyses were performed on exponentially growing cells (2 to 3 × 106 cells·mL−1) as described previously (Kuras and Wollman, 1994). Immunoblots were repeated at least twice and performed on three independent transformants. Cell extracts, loaded on an equal chlorophyll basis, were analyzed by SDS-PAGE (12 to 18% acrylamide gradients and 8 M urea). At least three biological replicates were performed for each experiment. Proteins were detected by enhanced chemiluminescence. Primary antibodies, diluted 100,000-fold (antibodies against cytochrome f, D1, and PsaA), 50,000-fold (CF1β, tubulin subunit α), 10,000-fold (AtpH, CGE1, RbcL, Rps12, and ATP synthase subunit γ), 5000-fold (ATP synthase subunits δ, ε, and AtpI), and 2500-fold (AadA), were revealed by horseradish peroxidase–conjugated antibodies against rabbit IgG (no. W401B, Promega). Antibodies against the PSII Oxygene Evolving Enhancer2 (OEE2) subunit from the PSII oxygen-evolving complex, the β-subunit of F1/CF1, cytochrome f, and CGE1 have been described by de Vitry et al. (1989), Atteia et al. (1992), Lemaire and Wollman (1989a), Kuras and Wollman (1994), and Schroda et al. (2001), respectively. Antibody against Rps12 was kindly provided by S. Ramundo (Ramundo et al., 2013). Antibodies against D1 (no. AS05 084), PsaA (no. AS06 172), RbcL (no. AS03 037), AadA (no. AS09 580), and the ATP synthase subunits γ (no. AS08 312), δ (no. AS10 1590), ε (no. AS10 1586), AtpH (no. AS09 591), and AtpI (no. AS10 1583) were purchased from Agrisera and used according to the manufacturer’s instructions. Antibody against the α subunit of tubulin was purchased from Sigma-Aldrich (MABT868). MTHI1-HA was detected by enhanced chemiluminescence using monoclonal anti-HA.11 (no. MMS-101R, Covance) antibodies and horseradish peroxidase–conjugated antibody against mouse IgG (no. W402B, Promega). When required, protein accumulation (normalized to that of OEE2) or β F1 (mitochondrial ATP synthase subunit β), as internal standards, was quantified from ChemiTouch (Bio-Rad) scans of the membrane using ImageLab 3.0 software. For immuno-chase, cytosolic translation was arrested by supplementing cells grown in TAP medium (2 to 3 × 106 cells mL−1) with cycloheximide (final concentration, 10 µg mL−1) at t = 0, and aliquots were taken at the indicated time points.
Gel Filtration Experiments on Soluble Cellular Extracts
Size exclusion chromatography was performed according to Boulouis et al. (2011), with minor modifications. Cells from a 600-mL culture (2 to 3 × 106 cells·mL−1) were centrifuged, resuspended in 3 mL of breaking buffer (5 mM Hepes-KOH, pH 7.8, 20 mM KCl, 10% [v/v] glycerol, 0.5 g·L−1 heparin, and 5× Roche protease inhibitors in diethylpyrocarbonate-treated water), broken with a French press at 6000 psi, and centrifuged at 346,000g for 20 min to pellet membranes, debris, and unbroken cells. Next, 500 μL of the supernatant was loaded onto a Superose 6 10/300 HR column (GE Healthcare). Chromatography was performed on a Biologic DuoFlow chromatography system (Bio-Rad), and protein elution (monitored on the UV light channel of the QuadTec device) was performed at a rate of 200 µL·min−1 at 4°C with a buffer containing 80 mM Tricine-KOH, pH 7.8, 200 mM KCl, 20 mM ε-aminocaproic acid, and 0.1× Roche protease inhibitors. Sixteen 1-mL fractions, eluted 16 mL after void volume (8 mL), were collected and concentrated by centrifugation on Amicon Ultra-15 filter units (cutoff, 30 kD) at 4500g for 20 min. Fraction volumes were adjusted to 100 µL, out of which 20 μL were loaded on 8% acrylamide gels containing 8 M urea. Fraction 16 (lower molecular mass) lacked protein and was not loaded on the gels. For RNase treatment, stromal preparations prepared in breaking buffer lacking heparin were incubated at 4°C with 2500 U·mL−1 RNase A and 625 U mL−1 RNase I for 45 min under gentle and continuous shaking, prior to loading on the column. For further analysis by coimmunoprecipitation, the indicated fractions were pooled, concentrated on an Amicon Ultra-15 filter, and adjusted to 1 mL with lysis buffer before coimmunoprecipitation.
Coimmunoprecipitations
Coimmunoprecipitations were performed according to Boulouis et al. (2011), with minor modifications. Cells from a 400-mL culture (2 × 106 cells·mL−1) were centrifuged, resuspended in 2 mL of lysis buffer (20 mM Hepes-KOH, pH 7.2, 150 mM NaCl, 10 mM KCl, 1 mM MgCl2, 10% [v/v] glycerol, and 2× Roche protease inhibitors in diethylpyrocarbonate-treated water), broken by a French press at 6000 psi, and centrifuged at 34,000g for 30 min to pellet membranes and debris. Next, 500 μL of supernatant supplemented with 0.2% (v/v) Tween 20 was incubated for 1 h at 4°C in the presence of 20 μL of anti–HA-tag magnetic beads (Medical Biological Laboratories International) that were pre-equilibrated with lysis buffer supplemented with 0.2% (v/v) Tween 20. The beads were then washed three times with washing buffer (150 mM NaCl, 20 mM Hepes-KOH, pH 7.2, 10% [v/v] glycerol, 0.2% [v/v] Tween 20, and 1× Roche protease inhibitors) and twice more with 10 mM Tris-HCl, pH 7.5. Bound proteins were detached by boiling the beads for 2 min in the presence of 2% (w/v) SDS, while for RNA purification, immunoprecipitation beads were resuspended in 250 μL of AE buffer (50 mM Na-acetate pH 5.2, and 10 mM EDTA) and extracted with phenol/chloroform/isoamyl alcohol (25:24:1 [v/v]) prior to ethanol precipitation in the presence of 2 μL of GlycoBlue (Invitrogen).
Two-Step Centrifugation Procedure
Cells from a 400-mL culture (2 to 3 × 106 cells·mL−1) were centrifuged, resuspended in breaking buffer (final volume of 4 mL), broken by a French press (6000 psi), and centrifuged at 2100g for 5 min to remove unbroken cells, starch, and large debris. One milliliter of the supernatant (input [I]) was ultracentrifuged at 272,000g for 30 min. The supernatant (S) was recovered and the pellet (P) was resuspended in 1 mL of breaking buffer. After spectroscopic determination of chlorophyll concentration in the input fraction, equal volumes of the S and P samples were loaded onto a gel and analyzed by immunoblotting.
MTHI1 Overexpression, Purification, and Immunization
For antibody generation, MTHI1 was overexpressed in E. coli BL21 for 16 h at 15°C and purified by nickel–nitrilotriacetic acid agarose under denaturing conditions as described previously (Supplemental Figure 12; Muranaka et al., 2016). After elution, the fractions were concentrated on Amicon Ultra 50-kD filters units and washed several times with 6 M urea and 20 mM Tris-HCl, pH 8. For antibody generation, 0.5 mg of purified protein was used for an 88-d rabbit immunization protocol (Covalab).
Accession Numbers
Sequence data from this article can be found in the National Center for Biotechnology Information Database under the accession numbers indicated in Supplemental Data Set for the atpH and atpI 5′ UTRs and for the MTHI1 CDSs; petA (FJ423446.1); psbD (X04147.1); OEE2 (M15187.1); atpA (X60298.1); CGE1 (AAK96224.1); Rps12 (AAC16329.1); RbcL (ASF83644.1); PsaB (P09144.4); rrnS (J01395.1); aadA (MG052656.1); atpB (M13704.1); PsaA (1310243A); PsbA (1102190A).
Supplemental Data
Supplemental Figure 1. The atpI transcript has a long 5′UTR but no dedicated promoter.
Supplemental Figure 2. MTHI1 is required for the translation of 5′atpI-driven genes.
Supplemental Figure 3. MTHI1 targets the atpH 5′UTR.
Supplemental Figure 4. Cloning of the MTHI1 gene.
Supplemental Figure 5. The MTHI1 locus.
Supplemental Figure 6. Conservation of the MTHI1 sequence among Chlorophyceae.
Supplemental Figure 7. sRNA coverage (normalized as RPM) over the atpI 5′UTR and along the inverted repeat.
Supplemental Figure 8. Conservation of the MTHI1 target in atpH and atpI 5′UTRs.
Supplemental Figure 9. Changing GGTTGTTAT to GGTTATTAT does not affect the expression of 5′atpH-driven genes.
Supplemental Figure 10. Emission spectrum of the white LED lights used to grow Chlamydomonas.
Supplemental Figure 11. Sequence of the recoded MTHI1 gene.
Supplemental Figure 12. Purification and quantification of the recombinant EcMTHI1.
Supplemental Table. Oligonucleotides used in this work.
Supplemental Data Set. Chlorophyta species analyzed for conservation of the atpH and atpI 5′UTRs.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Rachel Dent and Silvia Ramundo for their kind gifts of the CAL014.01.38 mutant and the α-Rps12 antibody, respectively; Jean-David Rochaix for kindly providing the indexed cosmid library; and all members of Unité Mixte de Recherche 7141 for stimulating discussions and/or critical reading of the article. This work was supported by the Centre National de la Recherche Scientifique and Sorbonne Université (basic support to Unité Mixte de Recherche 7141), by the Agence Nationale de la Recherche (ChloroMitoCES: BLAN-NT09_451610 and ChloroRNP: ANR-13-BSV7-0001-01), and by the “Initiative d’Excellence” program (grant “DYNAMO,” ANR-11-LABX-0011-01).
AUTHOR CONTRIBUTIONS
Y.C. designed research. S.-I.O., M.C., D.J., R.K., M.R., S.E., D.D., and Y.C. performed research; M.C. contributed to bioinformatics analysis; F.-A.W., S.-I.O., and Y.C. analyzed data; S.-I.O., F.-A.W., and Y.C. wrote the article.
References
- Almagro Armenteros J.J., Salvatore M., Emanuelsson O., Winther O., von Heijne G., Elofsson A., Nielsen H.(2019). Detecting sequence signals in targeting peptides using deep learning. Life Sci Alliance 2: e201900429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E.E., Miura K., Kucho K., Inoue Y., Fukuzawa H., Ohyama K., Nakamura Y., Tabata S.(2000). Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA res. 7: 305–307. [DOI] [PubMed] [Google Scholar]
- Atteia A., de Vitry C., Pierre Y., Popot J.L.(1992). Identification of mitochondrial proteins in membrane preparations from Chlamydomonas reinhardtii. J. Biol. Chem. 267: 226–234. [PubMed] [Google Scholar]
- Auchincloss A.H., Zerges W., Perron K., Girard-Bascou J., Rochaix J.-D.(2002). Characterization of Tbc2, a nucleus-encoded factor specifically required for translation of the chloroplast psbC mRNA in Chlamydomonas reinhardtii. J. Cell Biol. 157: 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczun C., Bunse A., Hahn D., Bennoun P., Nickelsen J., Kück U.(2005). Two adjacent nuclear genes are required for functional complementation of a chloroplast trans-splicing mutant from Chlamydomonas reinhardtii. Plant J. 43: 636–648. [DOI] [PubMed] [Google Scholar]
- Barkan A., Goldschmidt-Clermont M.(2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559–572. [DOI] [PubMed] [Google Scholar]
- Barkan A., Small I.(2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65: 415–442. [DOI] [PubMed] [Google Scholar]
- Barkan A., Walker M., Nolasco M., Johnson D.(1994). A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 13: 3170–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M., Romero E., Reisinger V., Yu J., Komenda J., Eichacker L.A., Dekker J.P., Nixon P.J.(2011). Investigating the early stages of photosystem II assembly in Synechocystis sp. PCC 6803: Isolation of CP47 and CP43 complexes. J. Biol. Chem. 286: 14812–14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bietenhader M., et al. (2012). Experimental relocation of the mitochondrial ATP9 gene to the nucleus reveals forces underlying mitochondrial genome evolution. PLoS Genet. 8: e1002876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau E., Nickelsen J., Lemaire S.D., Ossenbühl F., Rochaix J.-D.(2000). The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 19: 3366–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulouis A., Drapier D., Razafimanantsoa H., Wostrikoff K., Tourasse N.J., Pascal K., Girard-Bascou J., Vallon O., Wollman F.A., Choquet Y.(2015). Spontaneous dominant mutations in chlamydomonas highlight ongoing evolution by gene diversification. Plant Cell 27: 984–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulouis A., Raynaud C., Bujaldon S., Aznar A., Wollman F.-A., Choquet Y.(2011). The nucleus-encoded trans-acting factor MCA1 plays a critical role in the regulation of cytochrome f synthesis in Chlamydomonas chloroplasts. Plant Cell 23: 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J.E., et al. (1988). Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240: 1534–1538. [DOI] [PubMed] [Google Scholar]
- Cavaiuolo M., Kuras R., Wollman F.-A., Choquet Y., Vallon O.(2017). Small RNA profiling in Chlamydomonas: Insights into chloroplast RNA metabolism. Nucleic Acids Res. 45: 10783–10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Goldschmidt-Clermont M., Girard-Bascou J., Kück U., Bennoun P., Rochaix J.-D.(1988). Mutant phenotypes support a trans-splicing mechanism for the expression of the tripartite psaA gene in the C. reinhardtii chloroplast. Cell 52: 903–913. [DOI] [PubMed] [Google Scholar]
- Choquet Y., Vallon O.(2000). Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie 82: 615–634. [DOI] [PubMed] [Google Scholar]
- Choquet Y., Wollman F.-A.(2002). Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 529: 39–42. [DOI] [PubMed] [Google Scholar]
- Choquet Y., Wollman F.-A. (2009). The CES process In Chlamydomonas Source Book, Harris E.E., Stern D.B., and Witman G.B., eds, 2nd ed, Volume Volume 2 (New York, London, Amsterdam: Academic Press, Elsevier; ), pp. 1027–1064. [Google Scholar]
- Choquet Y., Zito F., Wostrikoff K., Wollman F.-A.(2003). Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell 15: 1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M.C., Fox T.D.(1993). Suppression of a defect in the 5′ untranslated leader of mitochondrial COX3 mRNA by a mutation affecting an mRNA-specific translational activator protein. Mol. Cell. Biol. 13: 4806–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée D., Stampacchia O., Girard-Bascou J., Rochaix J.-D.(2003). Tab2 is a novel conserved RNA binding protein required for translation of the chloroplast psaB mRNA. EMBO J. 22: 6378–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R.M., Haglund C.M., Chin B.L., Kobayashi M.C., Niyogi K.K.(2005). Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 137: 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vitry C., Olive J., Drapier D., Recouvreur M., Wollman F.-A.(1989). Posttranslational events leading to the assembly of photosystem II protein complex: A study using photosynthesis mutants from Chlamydomonas reinhardtii. J. Cell Biol. 109: 991–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drager R.G., Girard-Bascou J., Choquet Y., Kindle K.L., Stern D.B.(1998). In vivo evidence for 5′-->3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J. 13: 85–96. [DOI] [PubMed] [Google Scholar]
- Drager R.G., Zeidler M., Simpson C.L., Stern D.B.(1996). A chloroplast transcript lacking the 3′ inverted repeat is degraded by 3′-->5′ exoribonuclease activity. RNA 2: 652–663. [PMC free article] [PubMed] [Google Scholar]
- Drapier D., Girard-Bascou J., Wollman F.-A.(1992). Evidence for nuclear control of the expression of the atpA and atpB chloroplast genes in Chlamydomonas. Plant Cell 4: 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapier D., Girard-Bascou J., Stern D.B., Wollman F.-A.(2002). A dominant nuclear mutation in Chlamydomonas identifies a factor controlling chloroplast mRNA stability by acting on the coding region of the atpA transcript. Plant J. 31: 687–697. [DOI] [PubMed] [Google Scholar]
- Drapier D., Rimbault B., Vallon O., Wollman F.-A., Choquet Y.(2007). Intertwined translational regulations set uneven stoichiometry of chloroplast ATP synthase subunits. EMBO J. 26: 3581–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S.K., Power J., Galloway R.E., Porter M.E.(1991). Reappraisal of the genetic map of Chlamydomonas reinhardtii. J. Hered. 82: 295–301. [DOI] [PubMed] [Google Scholar]
- Eberhard S., Drapier D., Wollman F.-A.(2002). Searching limiting steps in the expression of chloroplast-encoded proteins: Relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 31: 149–160. [DOI] [PubMed] [Google Scholar]
- Eberhard S., Loiselay C., Drapier D., Bujaldon S., Girard-Bascou J., Kuras R., Choquet Y., Wollman F.-A.(2011). Dual functions of the nucleus-encoded factor TDA1 in trapping and translation activation of atpA transcripts in Chlamydomonas reinhardtii chloroplasts. Plant J. 67: 1055–1066. [DOI] [PubMed] [Google Scholar]
- Ferrari R., Tadini L., Moratti F., Lehniger M.K., Costa A., Rossi F., Colombo M., Masiero S., Schmitz-Linneweber C., Pesaresi P.(2017). CRP1 Protein: (Dis)similarities between Arabidopsis thaliana and Zea mays. Front. Plant Sci. 8: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N., Stampacchia O., Redding K., Rochaix J.D.(1996). Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 251: 373–380. [DOI] [PubMed] [Google Scholar]
- Fisk D.G., Walker M.B., Barkan A.(1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18: 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley P.L., Hsieh P.K., Luciano D.J., Belasco J.G.(2015). Specificity and evolutionary conservation of the Escherichia coli RNA pyrophosphohydrolase RppH. J. Biol. Chem. 290: 9478–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain A., Hotto A.M., Barkan A., Stern D.B.(2013). RNA processing and decay in plastids. Wiley Interdiscip. Rev. RNA 4: 295–316. [DOI] [PubMed] [Google Scholar]
- Hammani K., Bonnard G., Bouchoucha A., Gobert A., Pinker F., Salinas T., Giegé P.(2014). Helical repeats modular proteins are major players for organelle gene expression. Biochimie 100: 141–150. [DOI] [PubMed] [Google Scholar]
- Harris E.H.(1989). The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use. (San Diego: Academic Press; ). [DOI] [PubMed] [Google Scholar]
- Higuchi R.(1990). Recombinant PCR In PCR Protocols: A Guide to Methods and Application. (London, New York: Academic Press; ). [Google Scholar]
- Jalal A., Schwarz C., Schmitz-Linneweber C., Vallon O., Nickelsen J., Bohne A.V.(2015). A small multifunctional pentatricopeptide repeat protein in the chloroplast of Chlamydomonas reinhardtii. Mol. Plant 8: 412–426. [DOI] [PubMed] [Google Scholar]
- Jean-Francois M.J., Lukins H.B., Marzuki S.(1986). Post-transcriptional defects in the synthesis of the mitochondrial H+-ATPase subunit 6 in yeast mutants with lesions in the subunit 9 structural gene. Biochim. Biophys. Acta 868: 178–182. [Google Scholar]
- Johnson X., Wostrikoff K., Finazzi G., Kuras R., Schwarz C., Bujaldon S., Nickelsen J., Stern D.B., Wollman F.-A., Vallon O.(2010). MRL1, a conserved Pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22: 234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathir P., LaVoie M., Brazelton W.J., Haas N.A., Lefebvre P.A., Silflow C.D.(2003). Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell 2: 362–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K., Ishikura K., Kasai S., Shinmyo A.(2006). Efficient translation destabilizes transcripts in chloroplasts of Chlamydomonas reinhardtii. J. Biosci. Bioeng. 101: 471–477. [DOI] [PubMed] [Google Scholar]
- Kindgren P., Yap A., Bond C.S., Small I.(2015). Predictable alteration of sequence recognition by RNA editing factors from Arabidopsis. Plant Cell 27: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinknecht L., Wang F., Stübe R., Philippar K., Nickelsen J., Bohne A.V.(2014). RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation. Plant Cell 26: 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J.(2005). Autotrophic cells of the Synechocystis psbH deletion mutant are deficient in synthesis of CP47 and accumulate inactive PS II core complexes. Photosynth. Res. 85: 161–167. [DOI] [PubMed] [Google Scholar]
- Komenda J., Tichý M., Eichacker L.A.(2005). The PsbH protein is associated with the inner antenna CP47 and facilitates D1 processing and incorporation into PSII in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 46: 1477–1483. [DOI] [PubMed] [Google Scholar]
- Kuchka M.R., Goldschmidt-Clermont M., van Dillewijn J., Rochaix J.-D.(1989). Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PS II. Cell 58: 869–876. [DOI] [PubMed] [Google Scholar]
- Kück U., Choquet Y., Schneider M., Dron M., Bennoun P.(1987). Structural and transcription analysis of two homologous genes for the P700 chlorophyll a-apoproteins in Chlamydomonas reinhardii: Evidence for in vivo trans-splicing. EMBO J. 6: 2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras R., Wollman F.-A.(1994). The assembly of cytochrome b6/f complexes: An approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 13: 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre-Legendre L., Choquet Y., Kuras R., Loubéry S., Douchi D., Goldschmidt-Clermont M.(2015). A nucleus-encoded chloroplast protein regulated by iron availability governs expression of the photosystem I subunit PsaA in Chlamydomonas reinhardtii. Plant Physiol. 167: 1527–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire C., Wollman F.-A.(1989a). The chloroplast ATP synthase in Chlamydomonas reinhardtii. I. Characterization of its nine constitutive subunits. J. Biol. Chem. 264: 10228–10234. [PubMed] [Google Scholar]
- Lemaire C., Wollman F.-A.(1989b). The chloroplast ATP synthase in Chlamydomonas reinhardtii. II. Biochemical studies on its biogenesis using mutants defective in photophosphorylation. J. Biol. Chem. 264: 10235–10242. [PubMed] [Google Scholar]
- Levine R.P.(1960). Genetic control of photosynthesis in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 46: 972–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine R.P., Goodenough U.W.(1970). The genetics of photosynthesis and of the chloroplast in Chlamydomonas reinhardi. Annu. Rev. Genet. 4: 397–408. [DOI] [PubMed] [Google Scholar]
- Liu X.Q., Gillham N.W., Boynton J.E.(1989). Chloroplast ribosomal protein gene rps12 of Chlamydomonas reinhardtii. Wild-type sequence, mutation to streptomycin resistance and dependence, and function in Escherichia coli. J. Biol. Chem. 264: 16100–16108. [PubMed] [Google Scholar]
- Loiselay C., Gumpel N.J., Girard-Bascou J., Watson A.T., Purton S., Wollman F.-A., Choquet Y.(2008). Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol. 28: 5529–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loizeau K., Qu Y., Depp S., Fiechter V., Ruwe H., Lefebvre-Legendre L., Schmitz-Linneweber C., Goldschmidt-Clermont M.(2014). Small RNAs reveal two target sites of the RNA-maturation factor Mbb1 in the chloroplast of Chlamydomonas. Nucleic Acids Res. 42: 3286–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano D.J., Hui M.P., Deana A., Foley P.L., Belasco K.J., Belasco J.G.(2012). Differential control of the rate of 5′-end-dependent mRNA degradation in Escherichia coli. J. Bacteriol. 194: 6233–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott J.J., Civic B., Barkan A.(2018). Effects of RNA structure and salt concentration on the affinity and kinetics of interactions between pentatricopeptide repeat proteins and their RNA ligands. PLoS One 13: e0209713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Olive J., Drapier D., Vallon O., Wollman F.-A.(2001). The light sensitivity of ATP synthase mutants of Chlamydomonas reinhardtii. Plant Physiol. 126: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx C., Wünsch C., Kück U.(2015). The octatricopeptide repeat protein Raa8 is required for chloroplast trans splicing. Eukaryot. Cell 14: 998–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino L., Perron K., Rahire M., Howald I., Rochaix J.-D., Goldschmidt-Clermont M.(2006). A novel multifunctional factor involved in trans-splicing of chloroplast introns in Chlamydomonas. Nucleic Acids Res. 34: 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minai L., Wostrikoff K., Wollman F.-A., Choquet Y.(2006). Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 18: 159–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda R.G., Rojas M., Montgomery M.P., Gribbin K.P., Barkan A.(2017). RNA-binding specificity landscape of the pentatricopeptide repeat protein PPR10. RNA 23: 586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod C., Takahashi Y., Goldschmidt-Clermont M., Rochaix J.-D.(1994). The chloroplast ycf8 open reading frame encodes a photosystem II polypeptide which maintains photosynthetic activity under adverse growth conditions. EMBO J. 13: 2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S., Kuehnle K., Stern D.B.(2005). A spontaneous tRNA suppressor of a mutation in the Chlamydomonas reinhardtii nuclear MCD1 gene required for stability of the chloroplast petD mRNA. Nucleic Acids Res. 33: 3372–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranaka L.S., Rütgers M., Bujaldon S., Heublein A., Geimer S., Wollman F.-A., Schroda M.(2016). TEF30 interacts with Photosystem II monomers and is involved in the repair of photodamaged photosystem II in Chlamydomonas reinhardtii. Plant Physiol. 170: 821–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy B.J., Klusch N., Langer J., Mills D.J., Yildiz Ö., Kühlbrandt W.(2019). Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F1-Fo coupling. Science 364: eaaw9128. [DOI] [PubMed] [Google Scholar]
- Ooi B.G., Lukins H.B., Linnane A.W., Nagley P.(1987). Biogenesis of mitochondria: A mutation in the 5′-untranslated region of yeast mitochondrial oli1 mRNA leading to impairment in translation of subunit 9 of the mitochondrial ATPase complex. Nucleic Acids Res. 15: 1965–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne M.J., Schweizer E., Lukins H.B.(1991). Properties of two nuclear pet mutants affecting expression of the mitochondrial oli1 gene of Saccharomyces cerevisiae. Curr. Genet. 19: 343–351. [DOI] [PubMed] [Google Scholar]
- Perron K., Goldschmidt-Clermont M., Rochaix J.-D.(2004). A multiprotein complex involved in chloroplast group II intron splicing. RNA 10: 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J., Bayraktar O.A., Prikryl J., Barkan A.(2009). Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28: 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce-Toledo R.I., Deschamps P., López-García P., Zivanovic Y., Benzerara K., Moreira D.(2017). An early-branching freshwater cyanobacterium at the origin of plastids. Curr. Biol. 27: 386–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J., Rojas M., Schuster G., Barkan A.(2011). Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. USA 108: 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahire M., Laroche F., Cerutti L., Rochaix J.-D.(2012). Identification of an OPR protein involved in the translation initiation of the PsaB subunit of photosystem I. Plant J. 72: 652–661. [DOI] [PubMed] [Google Scholar]
- Ramundo S., Rahire M., Schaad O., Rochaix J.D.(2013). Repression of essential chloroplast genes reveals new signaling pathways and regulatory feedback loops in chlamydomonas. Plant Cell 25: 167–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C., Loiselay C., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F.-A., Choquet Y.(2007). Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl. Acad. Sci. USA 104: 9093–9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifschneider O., Marx C., Jacobs J., Kollipara L., Sickmann A., Wolters D., Kück U.(2016). A Ribonucleoprotein supercomplex involved in trans-splicing of organelle group II introns. J. Biol. Chem. 291: 23330–23342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J., Liu Q., Pellegrini O., Celesnik H., Yao S., Bechhofer D.H., Condon C., Belasco J.G.(2011). An RNA pyrophosphohydrolase triggers 5′-exonucleolytic degradation of mRNA in Bacillus subtilis. Mol. Cell 43: 940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier C., Goldschmidt-Clermont M., Rochaix J.D.(2001). Identification of an RNA-protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 20: 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J.-D., Kuchka M., Mayfield S., Schirmer-Rahire M., Girard-Bascou J., Bennoun P.(1989). Nuclear and chloroplast mutations affect the synthesis or stability of the chloroplast psbC gene product in Chlamydomonas reinhardtii. EMBO J. 8: 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M., Ruwe H., Miranda R.G., Zoschke R., Hase N., Schmitz-Linneweber C., Barkan A.(2018). Unexpected functional versatility of the pentatricopeptide repeat proteins PGR3, PPR5 and PPR10. Nucleic Acids Res. 46: 10448–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwe H., Schmitz-Linneweber C.(2012). Short non-coding RNA fragments accumulating in chloroplasts: Footprints of RNA binding proteins? Nucleic Acids Res. 40: 3106–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymarquis L.A., Higgs D.C., Stern D.B.(2006). Nuclear suppressors define three factors that participate in both 5′ and 3′ end processing of mRNAs in Chlamydomonas chloroplasts. Plant J. 46: 448–461. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T.(1989). Molecular Cloning. (Cold Spring Harbor: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Sakamoto W., Chen X., Kindle K.L., Stern D.B.(1994). Function of the Chlamydomonas reinhardtii petd 5′ untranslated region in regulating the accumulation of subunit IV of the cytochrome b6/f complex. Plant J. 6: 503–512. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Small I.(2008). Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 13: 663–670. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R., Barkan A.(2005). RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M., Vallon O., Whitelegge J.P., Beck C.F., Wollman F.-A.(2001). The chloroplastic GrpE homolog of Chlamydomonas: Two isoforms generated by differential splicing. Plant Cell 13: 2823–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz C., Elles I., Kortmann J., Piotrowski M., Nickelsen J.(2007). Synthesis of the D2 protein of photosystem II in Chlamydomonas is controlled by a high molecular mass complex containing the RNA stabilization factor Nac2 and the translational activator RBP40. Plant Cell 19: 3627–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow C.D.(1998). Organization of the nuclear genome In The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas, Rochaix J.-D., Goldschmidt-Clermont M., and Merchant S., eds (Dordrecht: Kluwer Academic Publishers; ), pp. 25–40. [Google Scholar]
- Stampacchia O., Girard-Bascou J., Zanasco J.L., Zerges W., Bennoun P., Rochaix J.-D.(1997). A nuclear-encoded function essential for translation of the chloroplast psaB mRNA in chlamydomonas. Plant Cell 9: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strenkert D., Schmollinger S., Gallaher S.D., Salomé P.A., Purvine S.O., Nicora C.D., Mettler-Altmann T., Soubeyrand E., Weber A.P.M., Lipton M.S., Basset G.J., Merchant S.S.(2019). Multiomics resolution of molecular events during a day in the life of Chlamydomonas. Proc. Natl. Acad. Sci. USA 116: 2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Fang L., Zhang Z., Chang X., Penny D., Zhong B.(2016). Chloroplast phylogenomic inference of green algae relationships. Sci. Rep. 6: 20528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif M., Atteia A., Specht M., Cogne G., Rolland N., Brugière S., Hippler M., Ferro M., Bruley C., Peltier G., Vallon O., Cournac L.(2012). PredAlgo: A new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 29: 3625–3639. [DOI] [PubMed] [Google Scholar]
- Tourasse N.J., Choquet Y., Vallon O.(2013). PPR proteins of green algae. RNA Biol. 10: 1526–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trösch R., Barahimipour R., Gao Y., Badillo-Corona J.A., Gotsmann V.L., Zimmer D., Mühlhaus T., Zoschke R., Willmund F.(2018). Commonalities and differences of chloroplast translation in a green alga and land plants. Nat. Plants 4: 564–575. [DOI] [PubMed] [Google Scholar]
- Vaistij F.E., Boudreau E., Lemaire S.D., Goldschmidt-Clermont M., Rochaix J.-D.(2000b). Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 97: 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij F.E., Goldschmidt-Clermont M., Wostrikoff K., Rochaix J.-D.(2000a). Stability determinants in the chloroplast psbB/T/H mRNAs of Chlamydomonas reinhardtii. Plant J. 21: 469–482. [DOI] [PubMed] [Google Scholar]
- Viola S., Cavaiuolo M., Drapier D., Eberhard S., Vallon O., Wollman F.-A., Choquet Y.(2019). MDA1, a nucleus-encoded factor involved in the stabilization and processing of the atpA transcript in the chloroplast of Chlamydomonas. Plant J. 98: 1033–1047. [DOI] [PubMed] [Google Scholar]
- Vreken P., Raué H.A.(1992). The rate-limiting step in yeast PGK1 mRNA degradation is an endonucleolytic cleavage in the 3′-terminal part of the coding region. Mol. Cell. Biol. 12: 2986–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Johnson X., Cavaiuolo M., Bohne A.V., Nickelsen J., Vallon O.(2015). Two Chlamydomonas OPR proteins stabilize chloroplast mRNAs encoding small subunits of photosystem II and cytochrome b6 f. Plant J. 82: 861–873. [DOI] [PubMed] [Google Scholar]
- Wang G., Li X., Wang Z.(2016). APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44 (D1): D1087–D1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J.-D., Chory J.(2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9: 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K., Choquet Y., Wollman F.-A., Girard-Bascou J.(2001). TCA1, a single nuclear-encoded translational activator specific for petA mRNA in Chlamydomonas reinhardtii chloroplast. Genetics 159: 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K., Girard-Bascou J., Wollman F.-A., Choquet Y.(2004). Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J. 23: 2696–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K., Stern D.(2007). Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA 104: 6466–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P., Hammani K., Rojas M., Voelker R., Vargas-Suárez M., Börner T., Barkan A.(2012). Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 40: 3092–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zones J.M., Blaby I.K., Merchant S.S., Umen J.G.(2015). High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27: 2743–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Bock R.(2018). Chloroplast translation: Structural and functional organization, operational control, and regulation. Plant Cell 30: 745–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Watkins K.P., Barkan A.(2013). A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell 25: 2265–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Watkins K.P., Miranda R.G., Barkan A.(2016). The PPR-SMR protein PPR53 enhances the stability and translation of specific chloroplast RNAs in maize. Plant J. 85: 594–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M.(2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]