Viral βC1 protein disrupts the ATG3-GAPC interaction and thereby induces autophagy.
Abstract
Autophagy plays an important role in plant–pathogen interactions. Several pathogens including viruses induce autophagy in plants, but the underpinning mechanism remains largely unclear. Furthermore, in virus–plant interactions, viral factor(s) that induce autophagy have yet to be identified. Here, we report that the βC1 protein of Cotton leaf curl Multan betasatellite (CLCuMuB) interacts with cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPC), a negative autophagic regulator, to induce autophagy in Nicotiana benthamiana. CLCuMuB βC1 bound to GAPCs and disrupted the interaction between GAPCs and autophagy-related protein 3 (ATG3). A mutant βC1 protein (βC13A) in which I45, Y48, and I53 were all substituted with Ala (A), had a dramatically reduced binding capacity with GAPCs, failed to disrupt the GAPCs-ATG3 interactions and failed to induce autophagy. Furthermore, mutant virus carrying βC13A showed increased symptoms and viral DNA accumulation associated with decreased autophagy in plants. These results suggest that CLCuMuB βC1 activates autophagy by disrupting GAPCs–ATG3 interactions.
INTRODUCTION
Autophagy is a highly conserved mechanism that leads to the degradation and recycling of damaged or unwanted intracellular materials under stress conditions or during specific developmental processes (Klionsky and Codogno, 2013; Yin et al., 2016). Three major types of autophagy, that is, macroautophagy, microautopahgy, and chaperone-mediated autophagy, occur in eukaryotic cells (Massey et al., 2004; Klionsky, 2005). Macroautophagy (hereafter referred to as autophagy) is mediated by autophagosome, a de novo–formed double-membrane vesicle. The outer membrane of a mature autophagosome fuses with the lysosome (in mammals) or vacuole (in yeast and plants) to release the sequestered cargo for breakdown by acid hydrolases (Mizushima et al., 2008; Ohsumi, 2001).
In mammalian system, autophagy acts as a defense mechanism against some intracellular invading pathogens, including viruses (Boyle and Randow, 2013; Randow and Youle, 2014; Paul and Münz, 2016). Increasing evidence shows that viruses manipulate or hijack the autophagy pathway to promote pathogenesis, demonstrating the pivotal role of autophagy in the arms race between hosts and viruses (Shoji-Kawata and Levine, 2009; Dong and Levine, 2013). In plants, autophagy plays an important role in disease resistance or susceptibility to various pathogens (Han et al., 2011; Ismayil et al., 2019). It contributes to hypersensitive response cell death, but restricts the spread of programmed cell death beyond the initial infection site (Liu et al., 2005; Patel and Dinesh-Kumar, 2008; Hofius et al., 2009; Yoshimoto et al., 2009). Cytoplasmic glyceraldehyde-3-phosphat dehydrogenases (GAPCs) interact with autophagy-related protein 3 (ATG3) directly and negatively regulates autophagy and immunity. Silencing of GAPCs reduced Tobacco mosaic virus accumulation in N gene-containing plants and enhanced N gene–mediated cell death (Han et al., 2015).
Autophagy has also recently been shown to represent an antiviral mechanism in plants (Nakahara et al., 2012; Hafrén et al., 2017, 2018; Haxim et al., 2017; Li et al., 2018). In addition, autophagy may directly or indirectly promote virus infection (Hafrén et al., 2017; Fu et al., 2018).
Given that autophagy has an important role in host–virus interactions, it is not surprising that some viruses have evolved strategies that modulate host autophagy for their own benefit. For instance, Turnip mosaic virus (TuMV) antagonizes NBR1-dependent autophagy during infection, thereby limiting its antiviral capacity. NBR1-independent bulk autophagy prevents premature plant death, extending the lifespan of virus reservoirs and particle production (Hafrén et al., 2018). Cauliflower mosaic virus (CaMV) P6 protein disrupts the interaction between viral P4 and host NBR1 to protect viral replication factory inclusions from autophagic degradation (Hafrén et al., 2017). Barley stripe mosaic virus (BSMV) γb interferes with the interaction of ATG7 with ATG8 in a competitive manner to suppress autophagy, thereby promoting viral infection (Yang et al., 2018). Viral proteins can also promote autophagic degradation of components in RNA silencing pathway (Derrien et al., 2012; Cheng and Wang, 2016).
Betasatellites, such as Cotton leaf curl Multan betasatellite (CLCuMuB), are approximately half the size of the DNA genomes of their helper begomoviruses. They require the helper begomoviruses for replication and movement in plants and only encode a single multifunctional pathogenicity βC1 protein (Sattar et al., 2013). CLCuMuB βC1 functions as a viral suppressor of RNA silencing (VSR; Amin et al., 2011). CLCuMuB βC1 enhances the accumulation of its helper virus CLCuMuV and is required for the induction of viral symptoms in plants (Jia et al., 2016). Geminivirus βC1 can also subvert ubiquitination to assist their helper viruses to infect plants (Jia et al., 2016; Shen et al., 2016). We reported that autophagic machinery targets and degrades viral βC1 through its interaction with autophagy-related protein 8 (ATG8; Haxim et al., 2017). Although we know that CLCuMuV infection activates autophagy and enhances the autophagic flux (Haxim et al., 2017), the viral factor responsible for autophagy activation and the underlying mechanism are unknown.
On the other hand, it is well-established that various plant pathogens can trigger autophagy (Liu et al., 2005; Hofius et al., 2009; Hafrén et al., 2017; Haxim et al., 2017; Li et al., 2018). However, in plants only one bacterial and one fungal protein have been shown to be responsible for autophagic activation, but the underlying mechanism(s) remain(s) to be characterized (Dagdas et al., 2016; Üstün et al., 2018). In addition, no viral protein has been reported to activate autophagy in plants. In this study, we demonstrate that CLCuMuB βC1 activates autophagy by interfering with the interaction between NbGAPCs and NbATG3.
RESULTS
CLCuMuB βC1 Activates Plant Autophagy
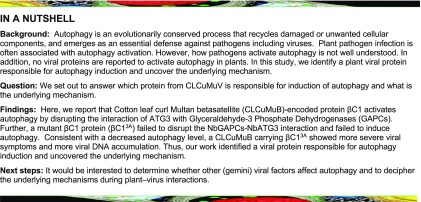
We previously showed that CLCuMuV infection induces autophagy but did not identify any viral protein responsible for this induction (Haxim et al., 2017). Here, we investigated whether βC1 affected autophagy in Nicotiana benthamiana. To achieve this, we used Cyan Fluorescent Protein (CFP)–tagged NbATG8f (CFP-NbATG8f) as a marker to visualize autophagosomes in the cytoplasm as well as autophagic bodies in the vacuole (Wang et al., 2013). In plants expressing HA-tagged βC1 (HA-βC1), numerous CFP-labeled autophagic bodies were observed compared to plants expressing the HA-tagged C-terminal luciferase (HA-cLUC) control (Figure 1A). The relative autophagy level in HA-βC1 expressing plants was 2- to 3-fold greater than the control (Figure 1B). Transmission electron microscopy confirmed βC1-mediated autophagic induction, and showed that overexpression of HA-βC1 increased the number of autophagic structures compared to the control (Figures 1C and 1D). In addition, HA-βC1 expression reduced the accumulation of the autophagy substrate, Joka 2/NBR1 (see below). These results indicated that CLCuMuB βC1 induces autophagy in N. benthamiana.
Figure 1.
CLCuMuB βC1 Induces Autophagy.
(A) Representative confocal microscopy images of autophagic activity detected by the specific autophagy marker CFP-NbATG8f in N. benthamiana leaves infiltrated with HA-βC1and HA-cLUC. Autophagosomes and autophagic bodies are revealed as CFP-positive puncta in mesophyll cells. CFP-NbATG8f fusion proteins are in cyan, and chloroplasts are in red. Bars = 20 mm.
(B) Quantification of the CFP-NbATG8f-labeled autophagic puncta/ cell from (A). More than 500 mesophyll cells for each treatment were used for the quantification. Relative autophagic activity in HA-cLUC-infected plants was normalized to control plants, which was set to 1.0. Values represent means ± se from three independent experiments; t test used for analyses, *P < 0.05.
(C) Representative transmission electron microscope (TEM) images of autophagic structures. Ultrastructure of autophagic bodies was observed in the vacuoles (indicated by V) of mesophyll cells of HA-cLUC control and plants infected with HA-βC1.
(D) Autophagosome-like structures from (C) were quantified. At least 30 cells for each treatment were used for the quantification. Relative autophagic activity in HA-cLUC–infected plants was normalized to control plants, which was set to 1.0. Values represent means ± se from three independent experiments; t test used for analyses, *P < 0.05.
CLCuMuB βC1 Interacts with NbGAPCs
Since CLCuMuB βC1 interacts with the autophagy-related protein ATG8 (Haxim et al., 2017), we investigated whether βC1-mediated autophagic induction was dependent on the βC1-ATG8 interaction. For this, we tested whether βC1V32A, a βC1 point mutant that cannot interact with ATG8, can induce autophagy in plants. Expression of HA-βC1 or HA-βC1V32A resulted in a similar level of CFP-labeled autophagic bodies (Supplemental Figure 1), revealing that βC1-mediated autophagic induction does not depend on its interaction with ATG8.
To identify the mechanism underlying activation of autophagy by βC1, we used affinity purification using GFP-Trap followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis to identify plant proteins that interact with βC1 (see Methods for details). When LC-MS/MS polypeptide profiles were searched against N. benthamiana protein databases, we identified many potential βC1-interacting proteins including autophagy-related protein 8 (ATG8). The ATG8-βC1 interaction has been reported (Haxim et al., 2017), suggesting that our screen is successful. Particularly, the partial sequence of cytosolic GAPC was identified with a high score (Supplemental Figure 2). Interestingly, this peptide sequence exists in all three N. benthamiana GAPC homologues (NbGAPC1–3; Han et al., 2015).
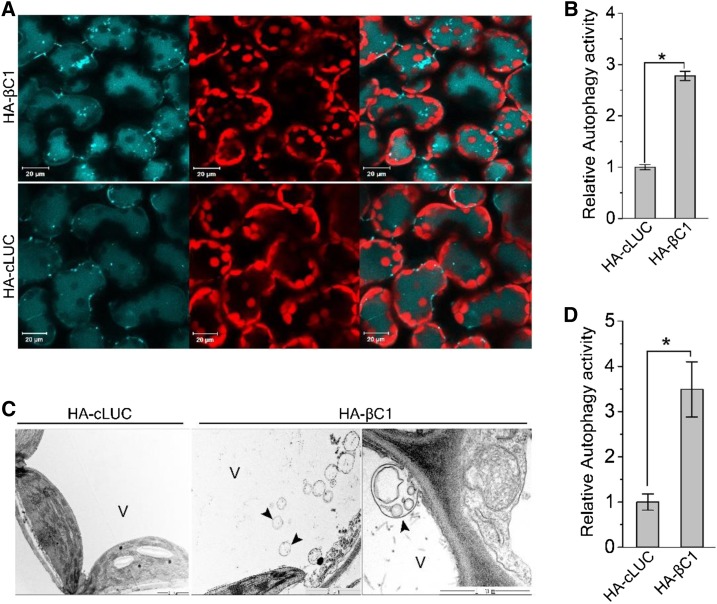
We first validated the interaction between GAPCs and βC1 in plants using a firefly Luciferase Complementation Imaging (LCI) assay (Chen et al., 2008). N-terminal luciferase-tagged βC1 (nLUC-βC1) was coexpressed with NbGAPC1-cLUC, NbGAPC2-cLUC, or NbGAPC3-cLUC in N. benthamiana. Positive signals were detected when βC1 was combined with all three NbGAPCs (Figure 2A), due to the reconstitution of the luciferase activity by βC1-NbGAPCs interactions. We also detected the signals in the positive control (NbATG3-nLUC/NbGAPC1-cLUC). However, no signal was detected in any of the negative controls (HA-cLUC + nLUC-βC1; Figure 2A). We further confirmed the interaction of βC1 with NbGAPCs by coimmunoprecipitation (Co-IP) assays. HA-βC1 coimmunoprecipitated with both NbGAPC1-GFP and NbGAPC2-GFP fusion proteins. However, HA-cLUC did not coimmunoprecipitate with NbGAPC1-GFP or NbGAPC2-GFP (Figure 2B).
Figure 2.
CLCuMuB βC1 Protein Interacts with NbGAPCs.
(A) LCI assays showed that βC1 interacts with NbGAPCs in plants. The image shows luminescence of a N. benthamiana leaf agroinfiltrated with nLUC-βC1 (left) or the negative control nLUC-HA (right) with the cLUC-tagged GAPC1, GAPC2, GAPC3, or a negative control cLUC. The positive control is GAPC1-cLUC and NbATG3-nLUC. Experiments were repeated three times with similar results.
(B) CLCuMuB βC1 protein co-immunoprecipitated with NbGAPCs. Total protein extracts were immunoprecipitated with anti-GFP beads and followed by immunoblotting (IB) using anti-GFP or anti-HA antibodies.
(C) The BiFC assay showed the interaction between βC1 and NbGAPCs. Cells were photographed at 48 hpi using a confocal laser scanning microscope. Scale bar = 50 μm.
(D) βC1 and NbGAPCs are localized in the soluble fraction. The total protein (T) extracted from leaves expressing NbGAPC- GFP and HA-βC1 was fractionated into soluble (S) and membrane (M) fractions by ultracentrifugation at 100,000g. Fractions were analyzed by immunoblot using antibodies against GFP, HA, and H+-ATPase (PM marker).
To identify the subcellular localization of the βC1-NbGAPCs interactions, citrine yellow fluorescent protein (YFP)–based Bimolecular Fluorescence Complementation (BiFC) assays (Han et al., 2015) were performed. βC1 was fused with the N-terminal domain of YFP (nYFP) to generate nYFP-βC1, and GAPC1 and GAPC2 were tagged with the C-terminal domain of YFP (cYFP) to generate NbGAPC1-cYFP and NbGAPC2-cYFP, respectively. nYFP-βC1 and 35S-nYFP were transiently coexpressed with NbGAPC1-cYFP or NbGAPC2-cYFP in N. benthamiana. Positive interactions (yellow fluorescence) were observed for nYFP-βC1 with both NbGAPC1-cYFP and NbGAPC2-cYFP, and such interactions occurred in the cytoplasm of leaf cells (Figure 2C). To confirm the specific localization of this interaction, we performed cell fractionation and immunoblotting assays. Total protein extracts from leaves coexpressing HA-βC1 and NbGAPC1-GFP were separated into soluble and microsomal membrane fractions by ultracentrifugation. Both βC1 and NbGAPC1 were mostly detected in the soluble fraction (Figure 2D).
Together, these results suggest that CLCuMuB βC1 specifically interacts with NbGAPCs.
CLCuMuB βC1 Interferes with the Interaction between NbGAPCs and NbATG3
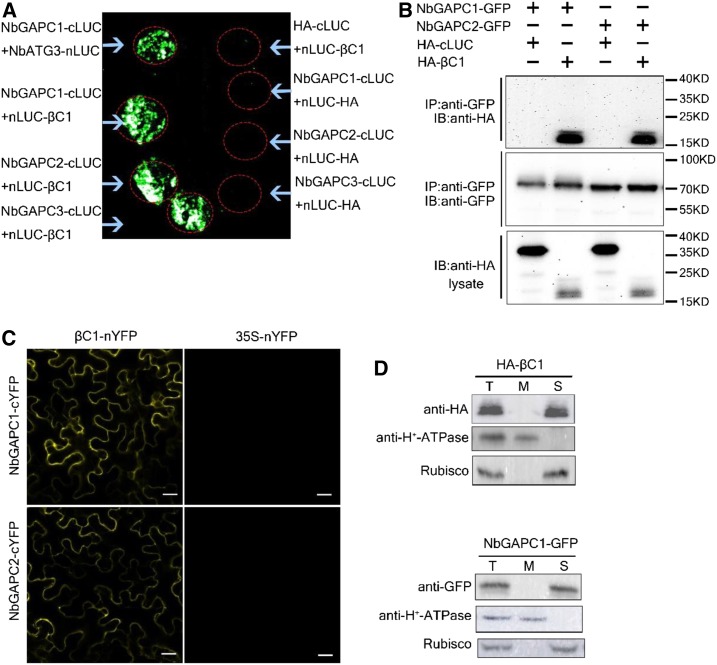
GAPCs interact with autophagy-related protein 3 (ATG3) to negatively regulate autophagy in plants (Han et al., 2015), whereas CLCuMuB βC1 induces autophagy and interacts with NbGAPCs as shown above. Based on these observations, we hypothesized that βC1 could interfere with the interaction of ATG3 with GAPCs to relieve the negative regulatory role of GAPCs on autophagy. To test this hypothesis, we examined the effect of βC1 on ATG3-GAPC interactions using BiFC assays. For this, we coexpressed NbATG3-nYFP and NbGAPC1-cYFP, or NbGAPC2-cYFP with HA-βC1 or HA-cLUC in N. benthamiana leaves. Stronger signals were detected in tissue expressing NbATG3YFP and NbGAPC1-cYFP or NbGAPC2-cYFP in the presence of HA-cLUC than HA-βC1 (Figures 3A and 3B). The protein levels of NbGAPC1-cYFP, NbGAPC2-cYFP, and NbATG3-nYFP were similar between the two groups (Figure 3C).
Figure 3.
CLCuMuB βC1 Disrupts the Interaction between NbGAPCs and NbATG3.
(A) A confocal image of BiFC assays shows that CLCuMuB βC1 interfered with the interaction between NbGAPCs and NbATG3. Photographs were taken 48 hpi post inoculation with HA-βC1or HA-cLUC. Scale bar = 200 μm.
(B) BiFC intensity (means ± SEM, n = 4) was quantified by YFP fluorescence. Relative BiFC intensity was normalized to the control. The raw data were analyzed by a two-sample t test to assess the significance level P ≤ 0.05 (*).
(C) Immunoblot analyses of BiFC construct combinations from the same experiment as in (A). The protein levels of cYFP-NbGAPC1/cYFP-NbGAPC2 and NbATG3-nYFP were monitored using a polyclonal GFP antibody (Huaxin Bochuang). The polyvinylidene difluoride (PVDF) membrane was stained with Ponceaux to visualize the large subunit of ribulose- 1,5-bisphosphate as a loading control.
(D) Co-IP assays show that CLCuMuB βC1 interferes with the interaction between NbGAPCs and NbATG3. Total protein extracts were immunoprecipitated with anti-GFP beads and monitored by immunoblotting (IB) using anti-Myc antibody. Protein levels were assessed using anti-GFP, anti-Myc, or anti-HA. The PVDF membrane was stained with Ponceaux to visualize the large subunit of ribulose-1,5-bisphosphate as a loading control.
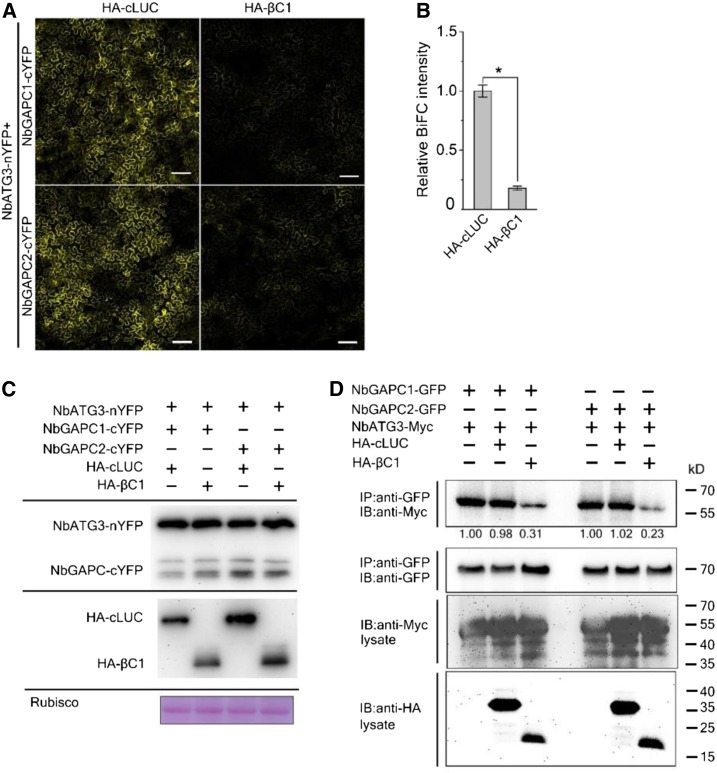
We further confirmed these results using a co-IP assay. For this, we coexpressed NbATG3-Myc with NbGAPC1-GFP or NbGAPC2-GFP in the presence of HA-βC1 or HA-cLUC in N. benthamiana. Coexpression of HA-βC1 reduced the amount of NbATG3 coimmunoprecipitated by NbGAPC1-GFP or NbGAPC2-GFP when compared to HA-cLUC (Figure 3D). Further, we performed a competitive pull-down assay to test the interference of different amounts of βC1 on ATG3-GAPC interactions. NbATG3-Myc, NbGAPC1-GFP, and HA-βC1 were expressed in N. benthamiana, respectively. NbGAPC1-GFP was trapped through GFP-Trap agarose, incubated with same amount of NbATG3-Myc and with gradient dilutions (1, 1/2, 1/4, 1/8, 1/16) of HA-βC1. Less NbATG3-Myc was pull-downed by NbGAPC1-GFP in the presence of HA-βC1 in a dose-dependent manner (Supplemental Figure 3).
Together, these results suggest that βC1 disrupts the interaction between NbGAPCs and NbATG3 by binding to NbGAPCs.
The βC1-GAPC Interaction is Important for βC1-Induced Autophagy
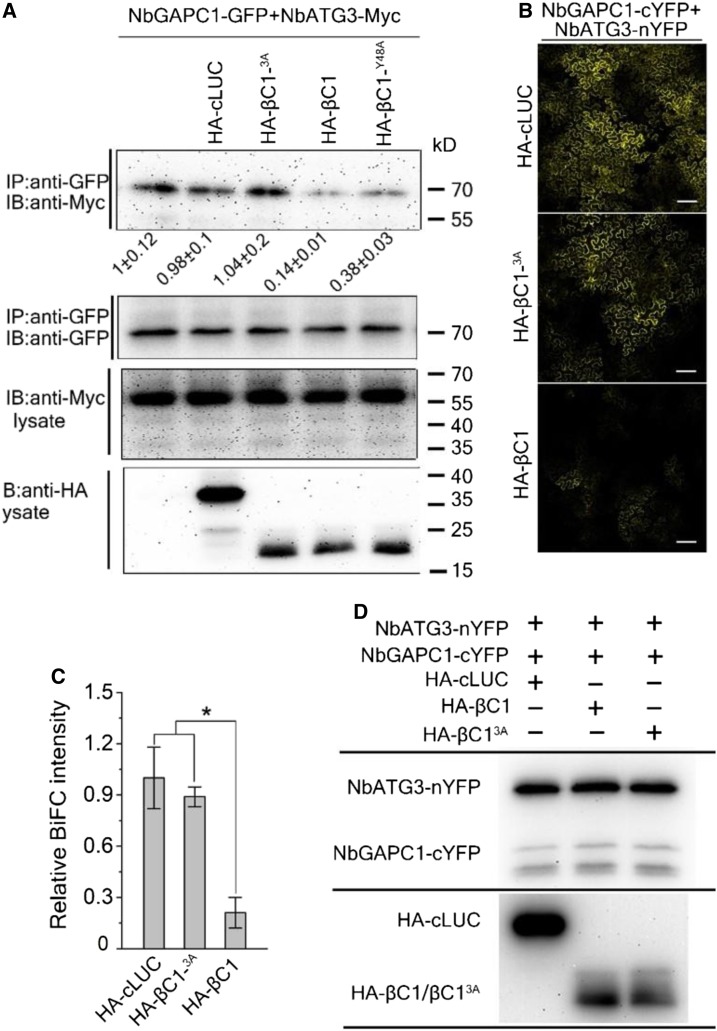
To explore the potential role of the βC1-NbGAPCs interaction in autophagy activation, we used βC1 deletion mutants to characterize the region required for binding to NbGAPCs in a yeast two-hybrid assay. These experiments identified a 13-amino acid motif from residue 40 to 53 of βC1 that is necessary for its interaction with NbGAPC1 (Supplemental Figure 4). Further, co-IP assays revealed that Ala substitutions at residues 45 IIe (βC1I45A), 48 Tyr (βC1Y48A), and 53 IIe (βC1I53A) decreased βC1 interaction with NbGAPC1 (Supplemental Figure 5). However, other βC1 mutants (V32A, Y42A, Y50A, E51A) still interacted with NbGAPC1. We also constructed double or triple mutants and found that a triple βC1 mutant (βC1I45A/Y48A/I53A, hereafter βC13A) dramatically decreased the βC1-NbGAPC1 interaction (Supplemental Figure 6). Importantly, the βC13A mutant localization was similar to that of wild-type βC1 (Supplemental Figure 7).
Given that βC13A interacts weakly with NbGAPC1, we hypothesized that the βC13A mutant may not interfere with the interaction between NbGAPC1 and NbATG3. To test this, we performed a co-IP assay in which NbGAPC1-GFP was coexpressed with NbATG3-Myc in the presence of HA-βC1 or HA-βC13A in N. benthamiana. Less NbATG3-Myc was coimmunoprecipitated by NbGAPC1-GFP in the presence of HA-βC1 but not with HA-βC1 3A compared to that in the presence of the HA-nLUC control (Figure 4A). We also tested the effect of several other mutations on the ATG3-GAPC interaction. Mutations (I45A, Y48A, I53A) in βC1 have little effect on the ATG3-GAPC interaction, whereas a strong effect was observed with the Y42A mutation (Supplemental Figure 8; Figure 4A). BiFC assays confirmed that, in contrast to wild-type βC1, βC13A failed to disrupt the ATG3-GAPC interactions (Figures 4B and 4C). The protein levels of NbGAPC1-cYFP, NbGAPC2-cYFP, and NbATG3-nYFP were similar in the BiFC assays (Figure 4D).
Figure 4.
βC1-GAPCs Interaction Is Important for Disrupting the ATG3-GAPCs Interaction in Plants.
(A) Co-IP assay shows that βC1, but not its mutants βC13A and βC1Y48A, interferes with the interaction between NbGAPCs and NbATG3. Total protein extracts were immunoprecipitated with anti-GFP beads and monitored by immunoblotting (IB) using an anti-Myc antibody. Protein levels were assessed using anti-GFP, anti-Myc, or anti-HA antibodies. Values represent means ± se from three independent experiments.
(B) Confocal image of BiFC assays show that βC1, but not βC13A, interferes with the interaction between NbGAPCs and NbATG3. Photographs were taken at 48 hpi. Scale bar = 200 μm.
(C) BiFC intensity (means ± SEM, n = 4) was quantified by YFP fluorescence. Relative BiFC intensity was normalized to the control. The raw data were analyzed by two-sample t tests to show the significance level of P ≤ 0.05 (*).
(D) Immunoblot analyses of BiFC construct combinations from the same experiments as in (B). The protein level of cYFP-NbGAPC1 and NbATG3-nYFP were assessed with the polyclonal GFP antibody (Huaxin Bochuang). The PVDF membrane was stained with Ponceaux to visualize the large subunit of ribulose-1,5-bisphosphate as a loading control.
Because the natural host of CLCuMuV is cotton, we tested the interaction between βC1 and cotton (Gossypium barbadense [Xinhai no. 21]) GAPC (GhGAPC). Indeed, βC1 also bound to GhGAPC and disrupted the interaction between GhGAPC and cotton ATG3 (GhATG3). Co-IP assays showed that HA-βC1, but not cLUC, was coimmunoprecipitated with GhGAPC-GFP fusion protein. Further, HA-βC13A dramatically decreased the βC1-GhGAPC interaction (Supplemental Figure 9A). Moreover, less GhATG3-Myc was coimmunoprecipitated by GhGAPC-GFP in the presence of HA-βC1 but not with HA-βC13A (Supplemental Figure 9B).
These results indicated that the interaction of βC1 with GAPC is critical for disruption of the interaction of ATG3 with GAPC.
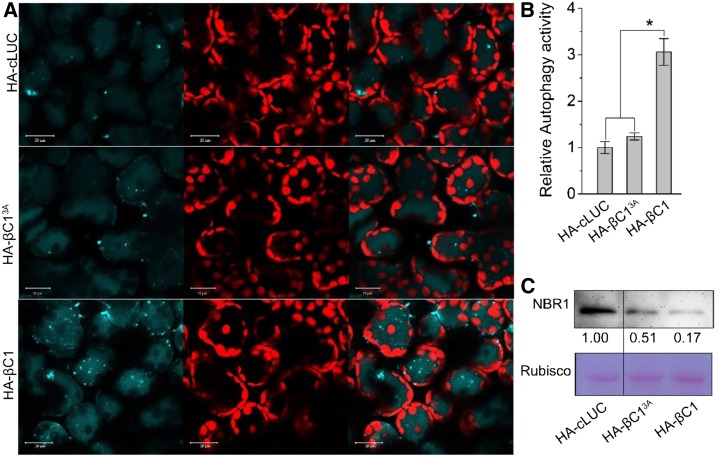
In addition, we examined the role of the βC1-NbGAPCs interaction in βC1-mediated autophagy induction using the βC13A mutant. Few autophagic bodies were observed in leaf tissue agroinfiltrated with HA-βC13A or the HA-cLUC control. By contrast, a high level of autophagic bodies accumulated in leaf tissue expressing HA-βC1 (Figures 5A and 5B). We also measured the autophagy flux by quantifying the autophagy substrate Joka2/NBR1, which is degraded in the vacuole in an autophagy-dependent manner (Xu et al., 2017). HA-cLUC or HA-βC13A overexpressing plants accumulated more Joka2/NBR1 than HA-βC1 overexpressing plants (Figure 5C), suggesting that βC1 but not βC13A accelerated autophagy flux and autophagy activity.
Figure 5.
Viral βC1, but not βC13A, Is Able to Activate Autophagy.
(A) Representative confocal microscopy images of dynamic autophagic activity revealed by the specific autophagy marker CFP-NbATG8f in plants infiltrated with HA-cLUC, HA-βC13A, or HA-βC1. Autophagosomes and autophagic bodies are revealed as CFP-positive puncta in mesophyll cells. CFP-NbATG8f fusion proteins are in cyan, and chloroplasts are in red. Bars = 20 mm.
(B) Quantification of the CFP-NbATG8f-labeled autophagic puncta/cell from (A). More than 500 mesophyll cells for each treatment were used for the quantification. Relative autophagic activity in HA-cLUC-infiltrated plants was normalized to control plants, which was set to 1.0. Values represent means ± se from three independent experiments; t test used for analyses, P ≤ 0.05 (*).
(C) Immunoblot assays showed that NbJoka2 protein level was reduced due to the increased autophagy flux in HA-βC1 plants. NbJoka2 was detected with anti-NBR1 polyclonal antibody.
These data suggest that the CLCuMuB βC1-GAPCs interaction is important for βC1-mediated autophagy induction.
CLCuMuB βC1 Induces Autophagy Independent of its VSR Activity
Since CLCuMuB βC1 also suppresses gene silencing, we investigated whether βC1-mediated autophagy induction is dependent on its activity as viral suppressor of RNA silencing (VSR). We compared the VSR activity of wild-type βC1 and the βC13A mutant by coexpressing them with GFP. Analysis of the agroinfiltrated plants under UV illumination at 4 dpi showed that leaf tissues expressing both HA-βC1 and HA-βC13A proteins showed similar GFP fluorescence. The immunoblot results indicate that VSR activity was not affected in mutant βC13A protein (Supplemental Figure 10). These results indicate that βC1-activated autophagy is independent of its VSR activity.
Disruption of βC1 Binding to GAPCs Reduces CLCuMuV-Induced Autophagy and Enhances Viral Infection
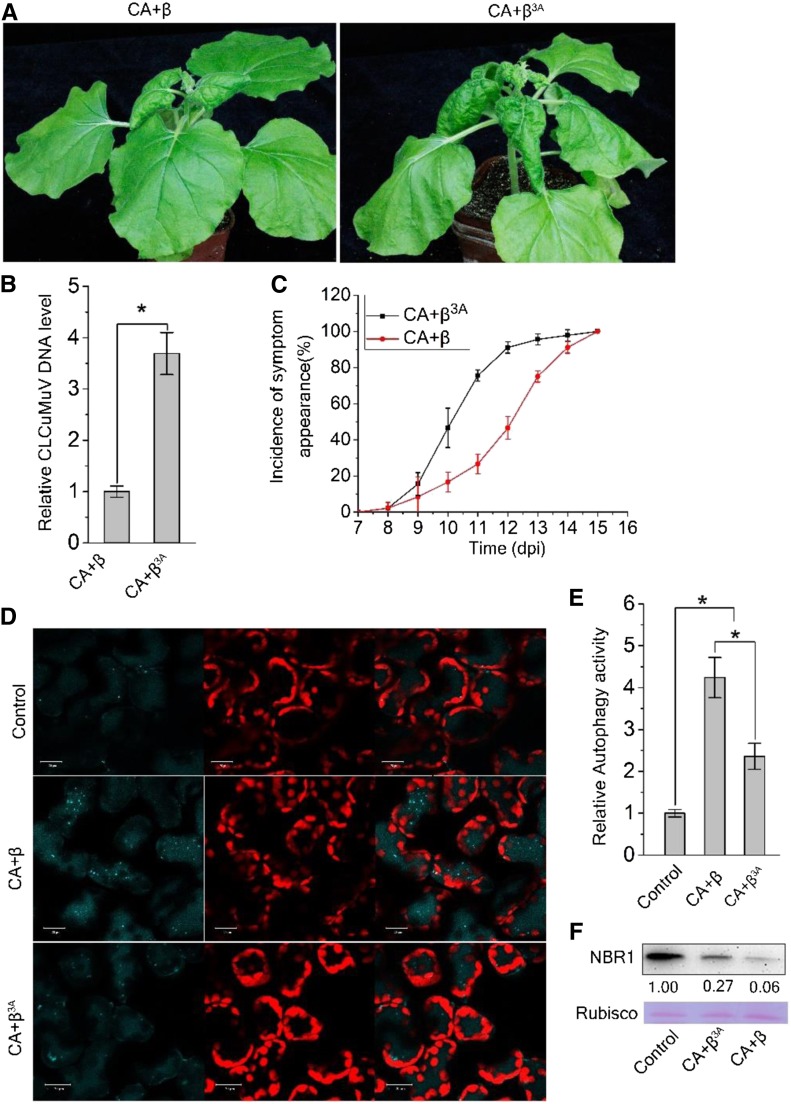
To explore the role of the βC1-NbGAPCs interaction during CLCuMuV (CA) infection, we generated a CLCuMuB mutant (β3A) virus by replacing βC1 with the βC13A mutant and inoculated the mutant virus (CA+β3A) or wild-type virus (CA+β) onto N. benthamiana leaves. First, we tested the GAPC-ATG3 interaction during viral infection and found that less NbATG3-Myc was coimmunoprecipitated by NbGAPC1-GFP in plants infected with wild-type virus (CA+β) than mutant virus (CA+β3A; Supplemental Figure 11). At 12 days post inoculation (dpi), the CA+β3A mutant virus caused more severe leaf curling symptoms compared to wild-type CA+β virus (Figure 6A), and viral DNA accumulated 4-fold higher in plants infected with CA+β3A mutant compared to CA+β wild-type (Figure 6B). The leaf curling symptoms caused by CA+β3A appeared 2 d earlier than those caused by CA+β (Figure 6C). Since βC13A has a much lesser ability to bind to NbGAPC1 and induce autophagy than βC1, we hypothesize that the enhanced viral infection of this mutant virus is due to decreased autophagic activation. We tested the autophagy level of CA+β3A-infected plants using the CFP-NbATG8f marker. Less CFP-labeled autophagic bodies were seen in CA+β3A-infected plants than in CA+β-infected plants (Figures 6D and 6E). We also measured the autophagy flux in CA+β3A or CA+β infected plants using Joka2/NBR1 as an indicator. CA+β3A-infected plants accumulated more Joka2/NBR1 than CA+β-infected plants (Figure 6F). Interestingly, noninfected control plants accumulated more Joka2/NBR1 and less CFP-labeled autophagic bodies than both CA+β- and CA+β3A-infected plants (Figures 6D to 6F).
Figure 6.
Disruption of βC1 Binding to GAPCs Reduces CLCuMuV-Induced Autophagy and Enhances Viral Infection.
(A) Mutant CLCuMuB (β3A), which encodes a mutant βC13A, caused more severe viral symptoms than wild-type CLCuMuB (β) when co-infected with CLCuMuV (CA). The photographs were taken at 12 dpi. A 3A point mutation in βC1 (βC13A) eliminates its interaction with NbGAPC.
(B) Relative accumulation of CLCuMuV DNA. Quantitative PCR analysis of the V1 gene sequence of CLCuMuV was used to determine viral DNA levels. Values represent means ± se from three independent experiments; t test used for analyses, P ≤ 0.05 (*).
(C) The incidence of symptom appearance at different time points of post infection. Symptom was indicated as the appearance of curled leaves caused by the infection with CA+ β or CA+β3A.
(D) βC13A-carrying virus has reduced induction of autophagy activity. Representative confocal microscopy images of dynamic autophagic activity revealed by the specific autophagy marker CFP-NbATG8f in plants infected with Control, CA+ β, CA+β3A.
(E) Quantification of the CFP-NbATG8f-labeled autophagic puncta/cell from (D). More than 500 mesophyll cells for each treatment were used for the quantification. Relative autophagic activity in noninfected plants was normalized to that of control plants, which was set to 1.0. Values represent means ± se from three independent experiments. P ≤ 0.05 (*).
(F) Protein gel blot assays showed the NbJoka2 protein level in CA+ β or CA+β3A infected plants. NbJoka2 was detected with anti-NBR1 polyclonal antibody.
We also confirmed that CA+β3A mutant virus is more infectious than wild-type CA+β virus in cotton plants. At 17 dpi, 60% of CA+β3A, but only 20% of CA+β infiltrated plants, showed disease symptoms. Further, CA+β3A caused more severe leaf curling symptoms in cotton plants than wild-type CA+β (Supplemental Figure 12A), and viral DNA accumulated 5-fold higher in plants infected with CA+β3A mutant than CA+β wild-type (Supplemental Figure 12B).
These results suggest that disruption of βC1 binding to GAPCs reduces virus-induced autophagy and enhances virus infection.
DISCUSSION
In this study, we established that CLCuMuB βC1 induces autophagy by interacting with the negative autophagy regulator GAPCs to disrupt the interaction between NbGAPCs and NbATG3. We identify a plant viral protein responsible for the autophagy induction and uncover the underlying mechanistic basis of pathogen-mediated autophagy activation in plants.
Autophagy has been reported to participate in plant–pathogen interactions. Pathogens have evolved the ability to hijack autophagy and degrade host defense components for their own benefits. For instance, the VSR protein P0 of Polerovirus is thought to mediate autophagic degradation of ARGONAUTE 1, a component of the cellular RNAi-based antiviral defense machinery (Derrien et al., 2012). Similarly, the VSR protein VPg of Turnip mosaic virus is also reported to mediate the degradation of the cellular RNAi-based antiviral defense component suppressor of gene silencing 3 (SGS3), partially via autophagy (Cheng and Wang, 2016). The VSR protein βC1 of Tomato yellow leaf curl China virus can mediate autophagic degradation of SGS3 through the calmodulin-like protein rgsCAM (Li et al., 2017). The movement protein of Rice stripe virus (RSV) induces autophagic degradation of remorin and overcomes remorin-mediated inhibition of viral movement by binding to and interfering with the S-acylation of remorin, thus facilitating virus infection (Fu et al., 2018). In these cases, viruses hijack autophagy to target host defense-related factors for degradation. However, host autophagy can also target viral virulence factors for degradation. The VSR protein 2b of Cucumber mosaic virus (CMV) is thought to be indirectly targeted by autophagy for its degradation through its binding to host ATG8 binding protein rgs‐CaM (Nakahara et al., 2012). In this study, we show that CLCuMuB βC1 induces autophagy. Since CLCuMuB βC1 is directly targeted by autophagy for its degradation through its direct binding to ATG8 (Haxim et al., 2017), it seems that CLCuMuB βC1 activates autophagy and thereby degrades the viral effector itself.
Pathogen effectors can also suppress host autophagy and promote infection in plants. Phytophthora infestans effector PexRD54 antagonizes a host selective autophagy cargo receptor Joak2/NBR1 by depleting Joka2/NBR1 out of ATG8 complexes through its binding to ATG8, which interferes with Joka2/NBR1-mediated plant defense (Dagdas et al., 2016). CaMV P6 protein is reported to inhibit autophagy (Zvereva et al., 2016) and protect viral replication factory inclusions from autophagic degradation (Hafrén et al., 2017). TuMV VPg and 6K2 proteins inhibit NBR1-dependent autophagy antiviral defense by blocking the degradation of viral protein HCpro (Hafrén et al., 2018). BSMV-γb inhibit the interaction between ATG7 and ATG8 to suppress autophagy and promote viral infection (Yang et al., 2018). In contrast to these pathogen effectors that antagonize or inhibit plant autophagy, both CLCuMuV infection and CLCuMuB βC1 activate autophagy (Haxim et al., 2017; this study). Considering TuMV not only hijacks autophagy to degrade plant antiviral protein SGS3 (Cheng and Wang, 2016) but also inhibits autophagy to block the degradation of viral effector (Li et al., 2017), it will be interesting to determine whether there are CLCuMuV protein(s) that suppress autophagy antiviral defense.
Host autophagy can also be induced during plant–pathogen interactions (Liu et al., 2005; Hofius et al., 2009; Hafrén et al., 2017; Haxim et al., 2017). To date, only two pathogen effectors, PexRD54 from Phytophthora infestans and HopM1 from Pseudomonas, have been shown to activate autophagy (Dagdas et al., 2016; Üstün et al., 2018). Interestingly, no viral effectors have been shown to activate autophagy, even though autophagy is activated by the infection of several viruses, including CaMV, TuMV, and geminiviruses (Hafrén et al., 2017; Haxim et al., 2017; Li et al., 2018). More importantly, the mechanism underlying activation of autophagy during plant–pathogen interactions is generally unknown in plants. In this study, we reveal that CLCuMuB βC1 activates autophagy by interfering with the interaction between NbGAPCs with NbATG3 through its binding to NbGAPCs. In this study, we also determined that the autophagy level in mutant CA+β3A virus-infected plants is higher than that in noninfected control plants. It is possible that other viral factors except βC1 could also induce autophagy.
Geminiviral βC1 is a multifunctional pathogenicity protein. βC1 is crucial for producing viral disease symptoms and enhancing viral DNA accumulation (Briddon et al., 2001; Jia et al., 2016), and complements the movement function of some bipartite begomoviruses (Saeed et al., 2007). βC1 can also subvert ubiquitination to enhance virus infection (Jia et al., 2016). Furthermore, βC1 plays important roles in suppressing posttranscriptional gene silencing (PTGS; Amin et al., 2011). In this study, we showed that the mutant protein βC13A, which cannot induce autophagy, has a similar PTGS suppression activity to wild-type βC1 (Supplemental Figure 8). This indicates that CLCuMuB βC1 protein induces autophagy independent of its VSR functionality.
In summary, we describe a first viral factor that is able to activate plant autophagy and the underlying activation mechanism. CLCuMuB βC1 functions as a viral autophagy activator by interfering with the interaction between GAPCs with ATG3 through its interaction with GAPCs. Interestingly, CLCuMuB βC1 is targeted and degraded by autophagy through its interaction with ATG8, and autophagy functions as an antiviral mechanism against CLCuMuV in plants (Haxim et al., 2017). Strikingly, the mutant CA+ β3A virus that is unable to induce autophagy causes more severe viral symptoms and increases viral DNA accumulation to a greater extent than wild-type virus (Figure 6A), consistent with the antiviral role of autophagy (Haxim et al., 2017). Since CLCuMuB βC1 is a viral virulence factor and is targeted by autophagy for degradation, CLCuMuB βC1-mediated autophagy may reduce its viral virulence and enable successful infection during the plant–virus coevolution. In addition, it is also possible that CLCuMuB βC1-mediated autophagy helps host cell survival for better viral propagation and transmission. Indeed, autophagy is reported to extend plant lifespan and to play a proviral role during some virus infections (Hafrén et al., 2017).
METHODS
Plant Materials, Growth Conditions, and Viral Inoculation
Nicotiana benthamiana and Gossypium barbadense (Xinhai no. 21; cotton) were grown in a growth chamber at 25°C with a 16-h–light/8-h–dark cycle, 3000 to 5000 Lumens. For viral inoculation, cotton seedlings at the two-cotyledon stage were vacuum infiltrated with Agrobacterium GV3101 (OD600 = 0.8) containing CA+β or CA+β3A, respectively.
Plasmid Constructs
The full-length infectious clones of CLCuMuV (GQ924756.1) and CLCuMuB (GQ906588.1) were described (Yin et al., 2019).Vectors NbGAPC1-GFP, NbGAPC2-GFP, NbGAPC1-cYFP, NbGAPC2-cYFP, cLUC-NbGAPC1, cLUC-NbGAPC2, cLUC-NbGAPC3, NbATG3-nYFP, and NbATG3-Myc were previously described by Han et al. (2015). DNA fragments of GhGAPC were PCR amplified and cloned into the LIC-pGFP vector to generate GhGAPC-GFP. Full-length GhATG3 was PCR amplified and cloned into the LIC-pMyc vector to generate GhATG3-Myc. Vectors HA-βC1, HA-cLUC, nYFP-βC1, and nLUC-βC1 were previously described by Haxim et al. (2017). HA-βC13A and other HA-βC1 mutations were obtained by overlapping PCR, and then cloned between the duplicated CaMV 35S promoter and the NOS terminator of pJG045, a pCAMBIA1300-based T-DNA vector (Chen et al., 2017). CLCuMuB-βC13A was generated by replacing CLCuMuB βC1 with βC13A by overlapping PCR. Primers sequences used for plasmid construction in this study are listed in Supplemental Table 1.
Confocal Microscopy and TEM
Agrobacterium tumefaciens GV3101 harboring the autophagy marker CFP-NbATG8f was infiltrated into leaves. About 48 h later, 20 mM protease inhibitor E-64d was infiltrated and 8 h later the samples were imaged using a Zeiss LSM 710 three-channel microscope with an excitation wavelength of 405 nm, and the emission was captured at 454 to 581 nm. TEM observation was performed as previously described by Wang et al. (2013).
Mass Spectrometry Analysis
Total protein was extracted as previously described by Han et al. (2015) from N. benthamiana leaves infected with GFP-βC1 or GFP constructs. Total protein extracts were incubated with GFP-Trap_A beads (ChromoTek). After washing, the purified GFP-tagged βC1 proteins were denatured at 98°C, separated on a SDS-PAGE gel (12%), and visualized by silver staining. Protein bands were excised and in-gel–digested with trypsin (Promega), and the peptides were extracted twice with 1% (v/v) trifluoroacetic acid in 50% (v/v) acetonitrile aqueous solution for 30 min. All the peptides were subjected to LC-MS/MS analysis as previously described by Han et al. (2015).
LCI Assays
LCI assays were performed as described (Ismayil et al., 2018). All combinations tested were agro-infiltrated into N. benthamiana leaves. The leaves were detached at 48 hpi, sprayed with 1 mM luciferin, and observed under a low-light cooled charge-coupled device imaging apparatus (iXon; Andor Technology). Photographs were taken 5 min after exposure to luciferin.
Protein Analyses and Co-IP
For protein analysis, total protein extracts were prepared from N. benthamiana leaves and separated by SDS-PAGE for immunoblot analysis with the indicated antibodies as described by Zheng et al. (2019). For co-IP assays, total proteins from N. benthamiana leaves (∼1 g tissue/sample) were extracted in ice-cold immunoprecipitation buffer (10% [v/v] glycerol, 25 mM Tris, pH 7.5, 150 mM NaCl, 1× protease inhibitor cocktail [Roche], and 0.15% [v/v] Nonidet P-40). Protein extracts were incubated with GFP-Trap_A (ChromoTek) beads for 3 h at 4°C. The precipitates were washed four times with ice-cold immune precipitation buffer at 4°C and were analyzed on immunoblots using anti-Myc (Abmart), anti-HA (Cell Signaling Technology), or anti-GFP (ChromoTek) antibodies with dilutions of 1:3000.
Cell Fractionation Assay
Cell fractionation and immunoblotting assays were conducted as previously described by Chen et al. (2017).
BiFC Assays
Citrine YFP-based BiFC was performed as described by Wang et al. (2018). The experimental group and corresponding control group were inoculated on the same leaf to reduce differences of expression conditions. Live plant imaging was performed using an LSM710 confocal microscope (Zeiss). Enhanced citrine YFP-derived fluorescence was acquired using a 514-nm laser and emission 519- to 587-nm filters. Images were analyzed with ZEN 2012 Light Edition.
Yeast Two-Hybrid Screen and Interaction Assays
For the yeast two-hybrid interaction assay, CLCuMuB βC1 was amplified by PCR and cloned into the yeast vector pYL302 (Du et al., 2013) to generate the LexA DNA binding domain containing bait vectors. NbGAPC1 was amplified and cloned into the B42 activation domain–containing vector pSAH20b. The interaction assay and yeast two-hybrid screen was performed as described (Wang et al., 2019).
Statistical Analysis
The data were expressed as mean ± SEM from three independent experiments. Statistical analysis was performed using the Student’s t test with *P < 0.05.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: CLCuMuV (EF465535), CLCuMuB (EF465536), Nb-GAPC1 (KM986323), Nb-GAPC2 (KM986324), and Nb-GAPC3 (KM986325), Gh-GAPC (XM_016837483.1), Gh-ATG3 (XP_016682025.1).
Supplemental Data
Supplemental Figure 1. CLCuMuB βC1V32A induces autophagy.
Supplemental Figure 2. Representative tandem mass spectrum (MS/MS spectrum) for a peptide from GAPC protein.
Supplemental Figure 3. βC1 interferes with the interaction between NbGAPCs and NbATG3.
Supplemental Figure 4. Amino acids 40–53 of βC1 are involved in its binding to NbGAPC1.
Supplemental Figure 5. βC1 mutants co-immunoprecipitated with NbGAPC1.
Supplemental Figure 6. Triple mutation (βC13A) dramatically decreased the βC1–NbGAPC1 interaction.
Supplemental Figure 7. Subcellular localization of CLCuMuB βC1 and βC13A.
Supplemental Figure 8. Effect of several other βC1 mutants on the ATG3–NbGAPC interaction.
Supplemental Figure 9. CLCuMuB βC1 binds to GhGAPC and disrupts the interaction between GhGAPC and GhATG3.
Supplemental Figure 10. Both βC1 and βC13A can reverse PTGS of GFP.
Supplemental Figure 11. The GAPC–ATG3 interaction during viral infection.
Supplemental Figure 12. Disruption of βC1 binding to GAPC enhances CLCuMuV infection in cotton.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. S. P. Dinesh-Kumar for revising the manuscript and Dr. Jian Ye (Chinese Academy of Sciences) for kindly providing the cotton seeds. This work was supported by the Chinese Ministry of Science and Technology | Department of S and T for Social Development (grant 2017YFA0503401), the National Natural Science Foundation of China (NSFC; grants 31920103013, 31901848, 31530059, 31421001, and 31872636), and the National Transgenic Program of China (grants 2019ZX08009-003 and 2019ZX08005-001).
Author Contributions
A.I. and Y.L. initiated the project, designed the experiments, analyzed the data, and wrote the article; A.I., M.Y., Y. Haxim, Yunjing Wang, J.L., L.H., Yan Wang, and X.Z. performed the experiments; X.W., U.N., Y. Hong, and L.H.-.B. were involved in analysis of data and helped write the article.
Footnotes
Articles can be viewed without a subscription.
References
- Amin I., Hussain K., Akbergenov R., Yadav J.S., Qazi J., Mansoor S., Hohn T., Fauquet C.M., Briddon R.W.(2011). Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-betasatellite complex. Mol. Plant Microbe Interact. 24: 973–983. [DOI] [PubMed] [Google Scholar]
- Boyle K.B., Randow F.(2013). The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Curr. Opin. Microbiol. 16: 339–348. [DOI] [PubMed] [Google Scholar]
- Briddon R.W., Mansoor S., Bedford I.D., Pinner M.S., Saunders K., Stanley J., Zafar Y., Malik K.A., Markham P.G.(2001). Identification of dna components required for induction of cotton leaf curl disease. Virology 285: 234–243. [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.-M.J.-M.(2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Liu D., Niu X., Wang J., Qian L., Han L., Liu N., Zhao J., Hong Y., Liu Y.(2017). Antiviral resistance protein Tm-22 functions on the plasma membrane. Plant Physiol. 173: 2399–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Wang A.(2016). The potyvirus silencing suppressor protein VPg mediates degradation of SGS3 via ubiquitination and autophagy pathways. J. Virol. 91: e01478–e01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagdas Y.F., et al. (2016). An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife 5: e10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien B., Baumberger N., Schepetilnikov M., Viotti C., De Cillia J., Ziegler-Graff V., Isono E., Schumacher K., Genschik P.(2012). Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA 109: 15942–15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Levine B.(2013). Autophagy and viruses: Adversaries or allies? J. Innate Immun. 5: 480–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Zhao J., Chen T., Liu Q., Zhang H., Wang Y., Hong Y., Xiao F., Zhang L., Shen Q., Liu Y.(2013). Type I J-domain NbMIP1 proteins are required for both Tobacco mosaic virus infection and plant innate immunity. PLoS Pathog. 9: e1003659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S., Xu Y., Li C., Li Y., Wu J., Zhou X.(2018). Rice stripe virus interferes with S-acylation of remorin and induces its autophagic degradation to facilitate virus infection. Mol. Plant 11: 269–287. [DOI] [PubMed] [Google Scholar]
- Hafrén A., Macia J.-L., Love A.J., Milner J.J., Drucker M., Hofius D.(2017). Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 114: E2026–E2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafrén A., Üstün S., Hochmuth A., Svenning S., Johansen T., Hofius D.(2018). Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol. 176: 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Wang Y., Zheng X., Jia Q., Zhao J., Bai F., Hong Y., Liu Y.(2015). Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 27: 1316–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Yu B., Wang Y., Liu Y.(2011). Role of plant autophagy in stress response. Protein Cell 2: 784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxim Y., et al. (2017). Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 6: e23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H.T., Mattsson O., Jørgensen L.B., Jones J.D.G., Mundy J., Petersen M.(2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783. [DOI] [PubMed] [Google Scholar]
- Ismayil, et al. (2018). Cotton Leaf Curl Multan virus C4 protein suppresses both transcriptional and post-transcriptional gene silencing by interacting with SAM synthetase. PLoS Pathog. 14: e1007282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismayil A., Yang M., Liu Y.(2019). Role of autophagy during plant-virus interactions. Semin. Cell Dev. Biol. S1084-9521: 30225–30228. [DOI] [PubMed] [Google Scholar]
- Jia Q., et al. (2016). CLCuMuB βC1 subverts ubiquitination by interacting with NbSKP1s to enhance geminivirus infection in Nicotiana benthamiana. PLoS Pathog. 12: e1005668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J.(2005). The molecular machinery of autophagy: Unanswered questions. J. Cell Sci. 118: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Codogno P.(2013). The mechanism and physiological function of macroautophagy. J. Innate Immun. 5: 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Zhang C., Li Y., Wu G., Hou X., Zhou X., Wang A.(2018). Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat. Commun. 9: 1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Zhao N., Li Z., Xu X., Wang Y., Yang X., Liu S.S., Wang A., Zhou X.(2017). A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog. 13: e1006213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Tallóczy Z., Levine B., Dinesh-Kumar S.P.(2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577. [DOI] [PubMed] [Google Scholar]
- Massey A., Kiffin R., Cuervo A.M.(2004). Pathophysiology of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 36: 2420–2434. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J.(2008). Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara K.S., et al. (2012). Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA 109: 10113–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y.(2001). Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2: 211–216. [DOI] [PubMed] [Google Scholar]
- Patel S., Dinesh-Kumar S.P.(2008). Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4: 20–27. [DOI] [PubMed] [Google Scholar]
- Paul P., Münz C.(2016). Autophagy and mammalian viruses: Roles in immune response, viral replication, and beyond. Adv. Virus Res. 95: 149–195. [DOI] [PubMed] [Google Scholar]
- Randow F., Youle R.J.(2014). Self and nonself: How autophagy targets mitochondria and bacteria. Cell Host Microbe 15: 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M., Zafar Y., Randles J.W., Rezaian M.A.(2007). A monopartite begomovirus-associated DNA beta satellite substitutes for the DNA B of a bipartite begomovirus to permit systemic infection. J. Gen. Virol. 88: 2881–2889. [DOI] [PubMed] [Google Scholar]
- Sattar M.N., Kvarnheden A., Saeed M., Briddon R.W.(2013). Cotton leaf curl disease - an emerging threat to cotton production worldwide. J. Gen. Virol. 94: 695–710. [DOI] [PubMed] [Google Scholar]
- Shen Q., Hu T., Bao M., Cao L., Zhang H., Song F., Xie Q., Zhou X.(2016). Tobacco RING E3 ligase NtRFP1 mediates ubiquitination and proteasomal degradation of a geminivirus-encoded βC1. Mol. Plant 9: 911–925. [DOI] [PubMed] [Google Scholar]
- Shoji-Kawata S., Levine B.(2009). Autophagy, antiviral immunity, and viral countermeasures. Biochim. Biophys. Acta 1793: 1478–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S., Hafrén A., Liu Q., Marshall R.S., Minina E.A., Bozhkov P.V., Vierstra R.D., Hofius D.(2018). Bacteria exploit autophagy for proteasome degradation and enhanced virulence in plants. Plant Cell 30: 668–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Xie K., Xu G., Zhou H., Guo Q., Wu J., Liao Z., Liu N., Wang Y., Liu Y.(2018). Plant G proteins interact with endoplasmic reticulum luminal protein receptors to regulate endoplasmic reticulum retrieval. J. Integr. Plant Biol. 60: 541–561. [DOI] [PubMed] [Google Scholar]
- Wang Y., et al. (2019). Geminiviral V2 protein suppresses transcriptional gene silencing through interaction with AGO4. J. Virol. 93: e01675-e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yu B., Zhao J., Guo J., Li Y., Han S., Huang L., Du Y., Hong Y., Tang D., Liu Y.(2013). Autophagy contributes to leaf starch degradation. Plant Cell 25: 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Wang S., Han S., Xie K., Wang Y., Li J., Liu Y.(2017). Plant Bax Inhibitor-1 interacts with ATG6 to regulate autophagy and programmed cell death. Autophagy 13: 1161–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Zhang Y., Xie X., Yue N., Li J., Wang X.B., Han C., Yu J., Liu Y., Li D.(2018). Barley stripe mosaic virus γb protein subverts autophagy to promote viral infection by disrupting the ATG7–ATG8 interaction. Plant Cell 30: 1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K., Han T., Xie K., Zhao J., Song J., Liu Y.(2019). Engineer complete resistance to Cotton Leaf Curl Multan virus by the CRISPR/Cas9 system in Nicotiana benthamiana. Phytopathol. Res. 1: 9. [Google Scholar]
- Yin Z., Pascual C., Klionsky D.J.(2016). Autophagy: Machinery and regulation. Microb. Cell 3: 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K.(2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Wu M., Li X., Cao J., Li J., Wang J., Huang S., Liu Y., Wang Y.(2019). Actin filaments are dispensable for bulk autophagy in plants. Autophagy 15: 2126–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvereva A.S., Golyaev V., Turco S., Gubaeva E.G., Rajeswaran R., Schepetilnikov M.V., Srour O., Ryabova L.A., Boller T., Pooggin M.M.(2016). Viral protein suppresses oxidative burst and salicylic acid-dependent autophagy and facilitates bacterial growth on virus-infected plants. New Phytol. 211: 1020–1034. [DOI] [PubMed] [Google Scholar]