Starch degradation in guard cells is induced by the phytohormone brassinosteroid and the redox signal hydrogen peroxide to promote stomatal opening.
Abstract
Starch is the major storage carbohydrate in plants and functions in buffering carbon and energy availability for plant fitness with challenging environmental conditions. The timing and extent of starch degradation appear to be determined by diverse hormonal and environmental signals; however, our understanding of the regulation of starch metabolism is fragmentary. Here, we demonstrate that the phytohormone brassinosteroid (BR) and redox signal hydrogen peroxide (H2O2) induce the breakdown of starch in guard cells, which promotes stomatal opening. The BR-insensitive mutant bri1-116 accumulated high levels of starch in guard cells, impairing stomatal opening in response to light. The gain-of-function mutant bzr1-1D suppressed the starch excess phenotype of bri1-116, thereby promoting stomatal opening. BRASSINAZOLE-RESISTANT1 (BZR1) interacts with the basic leucine zipper transcription factor G-BOX BINDING FACTOR2 (GBF2) to promote the expression of β-AMYLASE1 (BAM1), which is responsible for starch degradation in guard cells. H2O2 induces BZR1 oxidation, enhancing the interaction between BZR1 and GBF2 to increase BAM1 transcription. Mutations in BAM1 lead to starch accumulation and reduce the effects of BR and H2O2 on stomatal opening. Overall, this study uncovers the critical roles of BR and H2O2 in regulating guard cell starch metabolism and stomatal opening.
INTRODUCTION
Starch is the major storage carbohydrate in plants and is composed of Glc residues linked to each other by α-1,4-linkages with occasional α-1,6-branches (Zeeman et al., 2010). Starch plays important roles in plant fitness under challenging environmental conditions, as its degradation in response to several physiological demands mainly related to nocturnal, stress, and germination events provides ample carbon, energy, and carbon-derived metabolites (Thalmann and Santelia, 2017). Starch degradation is catalyzed by a series of enzymes including α-amylase, β-amylase, glucan water dikinase, phosphoglucan water dikinase, phosphoglucan phosphatase, and isoamylase (Streb and Zeeman, 2012). Transcripts of genes involved in starch degradation have diurnal patterns that are at least partly driven by the circadian clock to ensure that the rate of starch breakdown correlates with the anticipated length of the night. Alteration of the photoperiod leads to rapid changes in the expression levels of these genes and in the rates of starch degradation within one day/night cycle (Streb and Zeeman, 2012). Many enzymes involved in starch degradation are regulated by the thiol-based redox modification (Skryhan et al., 2018). The precise regulation of the genes encoding starch breakdown at the transcriptional and posttranscriptional levels allows for the coordination of energy demands and plant growth under diverse situations.
Guard cells, which flank the stomata in plant leaves, undergo volume adjustments to regulate the stomatal movement that is essential for gas exchange and water evaporation during photosynthesis and transpiration (Shimazaki et al., 2007). Unlike the starch in mesophyll cells that is synthesized during the day and degraded at night, the starch in guard cells starts being synthesized 1 h after the onset of the day and continues into the middle of the night, when it starts to slowly degrade; by dawn, about half of the starch in guard cells has been degraded. Upon light exposure, starch in guard cells is rapidly degraded within 1 h (Santelia and Lawson, 2016; Daloso et al., 2017). The transitory starch breakdown in guard cells might provide organic acids and sugars to increase guard cell turgor pressure and promote stomatal opening. β-Amylase1 (BAM1) and α-amylase3 (AMY3) are preferentially and highly expressed in guard cells and are responsible for the majority of starch degradation. The bam1 amy3 double knockout mutants display a guard cell–specific starch excess phenotype and fail to open stomata efficiently in response to light (Horrer et al., 2016). BAM1 is expressed in guard cells to control the stomatal movement, but it is also activated in mesophyll cells under drought and osmotic stress conditions to contribute to leaf starch degradation to obtain sugars and sugar-derived osmolytes. Mutant plants lacking BAM1 are impaired in Pro accumulation and change their osmotic tolerance (Valerio et al., 2011; Thalmann et al., 2016; Zanella et al., 2016). These results suggest that BAM1-mediated transitory leaf starch breakdown plays an important role in stomatal opening and in plant fitness under osmotic stress.
Brassinosteroids (BRs) are a group of plant steroid hormones that regulate a wide range of plant growth and development processes as well as plant responses to biotic and abiotic stresses (Clouse and Sasse, 1998). BR activation of the BRASSINOSTEROID-INSENSITIVE1 (BRI1) receptor kinase initiates a phosphorylation cascade that leads to the activation of the core transcription factors BRASSINAZOLE-RESISTANT1 (BZR1) and BRI1-EMS-SUPPRESSOR1 (BES1) to control the expression of target genes for BR responses (Ye et al., 2011; Wang et al., 2012; Chaiwanon et al., 2016). BR has been reported to play important roles in stomatal movement (Rajasekaran and Blake, 1999; Haubrick et al., 2006; Xia et al., 2014; Shi et al., 2015; Ha et al., 2016; Inoue et al., 2017; Singh et al., 2017; Ha et al., 2018; Kim et al., 2018). BR treatments promote stomatal closure and inhibit stomatal opening in Vicia faba and the application of BR to Pinus banksiana seedlings inhibited stomatal closure under water stress (Rajasekaran and Blake, 1999; Haubrick et al., 2006). In Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum), BR was found to induce stomatal opening at low concentrations of BR but to promote stomatal closure at high concentrations of BR (Xia et al., 2014; Inoue et al., 2017; Kim et al., 2018). The BR-deficient mutant dwarf5-7 exhibited impaired stomatal opening in response to blue light and reduced K+ accumulation by decreasing the expression of inward-rectifying K+ channel genes including POTASSIUM CHANNEL IN ARABIDOPSIS THALIANA1 (KAT1), ARABIDOPSIS POTASSIUM TRANSPORTER1 (AKT1), and ARABIDOPSIS POTASSIUM TRANSPORTER2 (AKT2). Application of BR restored the reduced stomatal opening phenotype in dwarf5-7 and in another BR biosynthesis mutant det2-1, but not in the BR receptor mutant bri1-6, suggesting that BR at the cellular level promotes stomatal opening (Inoue et al., 2017). However, BR also promotes stomatal closure through the phosphorylation and activation of OPEN STOMATA1 (OST1) in a BR-activated kinase CDG1-LIKE1 (CDL1)–dependent manner (Kim et al., 2018). Hydrogen peroxide (H2O2) is a rate-limiting second messenger in BR-mediated stomatal movement. In tomato, treatment with less than 0.1 µM 24-epibrassinolide triggered the transient production of H2O2 and changed the redox status of glutathione in guard cells, thereby leading to stomatal opening. Conversely, treatment with 1 µM 24-epibrassinolide induced the accumulation of H2O2, promoting stomatal closure (Xia et al., 2014).
In this study, we showed that the transitory starch breakdown in guard cells is necessary for BR-induced stomatal opening. BZR1 directly interacts with the basic leucine zipper (bZIP) transcription factor G-BOX BINDING FACTOR2 (GBF2) to synergistically promote the expression of BAM1 that then induces guard cell starch degradation and stomatal opening. The interaction between BZR1 and GBF2 is increased by H2O2 through the oxidative modification of BZR1. Our findings illustrate that the BZR1–GBF2 interaction regulates a transcriptional network that controls starch degradation in guard cells, enabling BR- and H2O2-interdependent regulation of stomatal opening.
RESULTS
Starch Degradation in Guard Cells Is Required for BR- and/or BZR1-Promoted Stomatal Opening
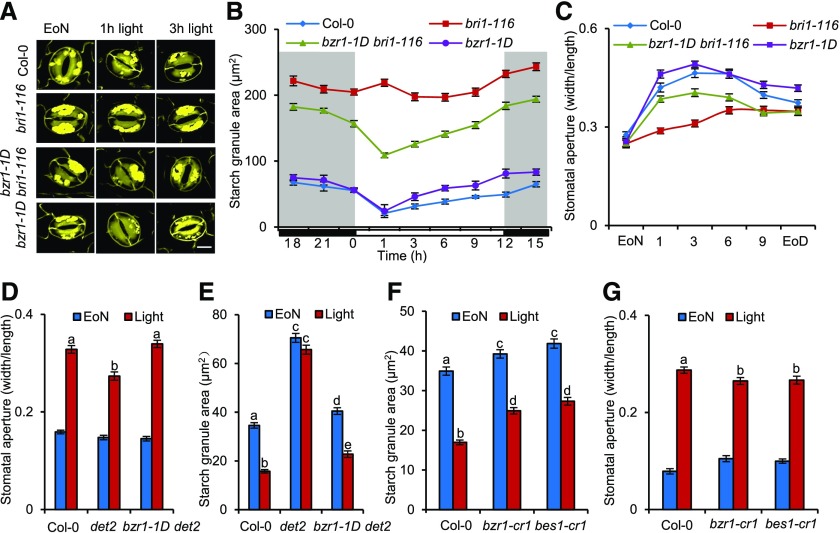
The transitory starch degradation in guard cells plays an important role in stomatal opening (Horrer et al., 2016). The phytohormone BR has been reported to promote stomatal opening at low concentrations (Xia et al., 2014; Inoue et al., 2017). To determine whether starch metabolism in guard cells is involved in BR-mediated stomatal opening, we analyzed the starch contents in the guard cells of the Arabidopsis wild-type Columbia-0 (Col-0) and BR-related mutants using a newly developed starch staining method in which starch granules in individual guard cells are labeled by periodic acid and fluorophore propidium iodide and then quantitatively analyzed with high-resolution confocal microscopy (Flütsch et al., 2018). In wild-type plants, starch started to be synthesized 1 h after light exposure, and its synthesis continued until midnight, when the content of starch peaked. Starch then began to slowly degrade during late night and rapidly degraded within 1 h upon light exposure (Figures 1A and 1B). The changing patterns of guard cell starch levels in the wild-type plants in our experimental conditions were consistent with previously published data (Horrer et al., 2016; Flütsch et al., 2018), but the absolute starch granule area per guard cell in this study was larger than that reported in previously published literature, which may be due to a number of different factors, such as the difference in the plant growth conditions, age of leaves, and staining time. In the BR-insensitive mutant bri1-116 and the BR-deficient mutant det2, the starch content of guard cells was significantly higher than that in the wild-type plants and slowly decreased after light exposure. The high levels of starch in guard cells of bri1-116 and det2 were partially suppressed by the dominant gain-of-function mutant bzr1-1D (Figures 1A to 1E), in which the mutant BZR1 protein is constitutively active through a strong interaction with PROTEIN PHOSPHATASE 2A (PP2A; Wang et al., 2002; Yin et al., 2002; Tang et al., 2011). To confirm the results of the pseudo-Schiff propidium iodide (mPS-PI) staining method, the iodine staining method, which is a routine technology for the detection of starch granules in plants, was used to analyze the starch levels in different BR mutants. Consistent with the mPS-PI staining results, the starch granules occupied a substantially larger area in the guard cells of bri1-116 and det2 than in those of the wild-type plants, and this excess starch phenotype of bri1-116 and det2 is partly rescued by bzr1-1D (Supplemental Figure 1A). However, the starch granules in the leaves of bri1-116 and det2 at the end of day were less accumulated than those in the wild-type plants, and these decreased-starch phenotypes were not rescued by bzr1-1D (Supplemental Figures 1B and 1C). These results suggest that BR promotes starch breakdown in guard cells through a BZR1-dependent pathway, while BR induces starch accumulation in leaves through a BZR1-independent pathway.
Figure 1.
Starch Degradation in Guard Cells Is Required for BR- or BZR1-Mediated Stomatal Opening.
(A) to (C) Dynamic changes of starch granules within guard cells and stomatal apertures in the leaves of the wild-type Col-0, bri1-116, bzr1-1D, and bzr1-1D bri1-116 plants at the indicated time points under 12-h-light/12-h-dark cycles. The cotyledons of 11-d-old seedlings of Col-0 and the indicated mutants were harvested at the indicated time points and immediately fixed in buffer for starch quantification ([A] and [B]) or used to directly measure the stomatal apertures (C). The starch granules in guard cells or the ratio of stomatal aperture width to length from at least 100 guard cells from 10 to 15 leaves of eight different plants were measured using ImageJ software. Three independent biological repeats were performed. Error bars indicate sd (n = 3 biological repeats). Bar = 20 µm. EoD, end of day.
(D) to (G) Quantification of the starch granules and stomatal apertures in guard cells of intact leaves of the wild type and different mutants at the EoN or after 1 h of light illumination. The rosette leaves of 5-week-old Col-0 and different mutants were harvested at the indicated time points and immediately fixed in buffer for starch quantification ([E] and [F]) or fixed using a Tape-Arabidopsis Sandwich method for stomatal aperture measurement ([D] and [G]). The guard cell starch granules or the ratio of stomatal aperture width to length from at least 100 guard cells from 10 to 15 leaves of eight different plants were measured using ImageJ software. Three independent biological repeats were performed. Error bar indicates sd (n = 3 biological repeats). Different letters above bars indicate statistically significant differences between samples (two-way, [E] and [F], or one-way, ([D] and [G]), ANOVA followed by a post hoc Tukey test, P < 0.05).
To investigate the roles of guard cell starch degradation in BR-mediated stomatal opening, we examined the stomatal apertures in intact leaves of BR-related mutants. The bri1-116 and det2 mutants displayed reduced stomatal openings in response to light, while the additional bzr1-1D mutation partially suppressed the stomatal response of bri1-116 and det2 (Figures 1C and 1D).
To further confirm that BR and BZR1 promote stomatal opening, we analyzed the stomatal apertures of BR-related mutants using the conventional stomatal opening technology in detached leaves. The BR-deficient mutants rot3-1 and det2 and the BR-insensitive mutant bri1-301 exhibited impaired stomatal opening in response to blue light, whereas BR signal-enhancing materials including bzr1-1D, bes1-D, bin2 bil1 bil2, and ProBZR1:bzr1-1D-CFP showed larger stomatal aperture than their corresponding wild types (Supplemental Figures 2A to 2E). Consistent with the data from intact leaves, the gain-of-function mutant bzr1-1D completely restored the stomatal opening phenotype of det2 (Supplemental Figure 2B). Furthermore, we showed that two independent sets of BZR1 and BES1 loss-of-function mutants displayed decreased starch degradation ratios in guard cells, reduced stomatal opening phenotypes, and slightly reduced BR sensitivity (Figures 1F and 1G; Supplemental Figures 2F and 3A to 3L). These results suggested that BZR1 and BES1 positively regulate guard cell starch degradation and stomatal opening.
H2O2 Is Required for BR-Promoted Guard Cell Starch Degradation
The activities of several enzymes involved in starch breakdown have been reported to be regulated by redox modification (Skryhan et al., 2018). H2O2 is considered to be a major signaling molecule that mediates redox modification in plants due to its remarkable stability within cells (half-time of 10−3 s) and rapid and reversible oxidation of target proteins (García-Santamarina et al., 2014). It is also required for the action of BR in the regulation of cell elongation, root stem cell maintenance, and stomatal closure (Xia et al., 2014; Tian et al., 2018).
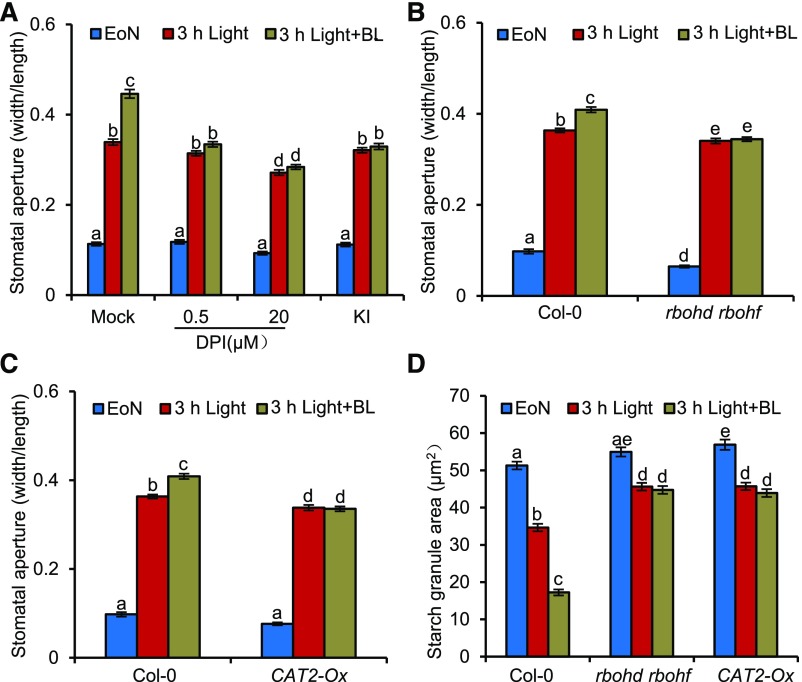
To examine whether H2O2 is involved in BR-mediated guard cell starch degradation and stomatal opening, we analyzed how BR induces stomatal opening in the H2O2-deficient plant materials. The rosette leaves of 4-week-old Col-0 were transferred to half-strength Murashige and Skoog (MS) liquid medium for overnight incubation in the dark, and then brassinolide (BL, the most active BR), H2O2, or mock solution was added to medium at the end of night (EoN) and illuminated with white light for 3 h. Fluorescein diacetate staining assays (Lawrence et al., 2018) showed that most of the guard cells in this experimental condition are alive and responsive to BR and H2O2 (Supplemental Figures 4A and 4B). BR treatment significantly promoted stomatal opening in the wild-type control plants; cotreatment with diphenyleneiodonium (DPI) and potassium iodide (KI), which reduced the accumulation of H2O2 in plants, strongly counteracted BR effects to promote stomatal opening (Figure 2A). Consistent with these results, the respiratory-burst oxidase homologue NADPH oxidase mutant, rbohd rbohf double mutants, and catalase overexpression plants (CAT2-Ox) failed to effectively open their stomata in response to BR (Figures 2B and 2C). BR treatments significantly induced the guard cell starch degradation in the wild-type plants, but not in rbohd rbohf and CAT2-Ox plants (Figure 2D). These results indicated that H2O2 plays an essential role in BR-promoted guard cell starch degradation and stomatal opening.
Figure 2.
H2O2 Is Required for BR-Mediated Guard Cell Starch Degradation and Stomatal Opening.
(A) DPI and KI treatment reduced the BR effects on stomatal opening. The rosette leaves of 4-week-old Col-0 were transferred to half strength MS liquid medium containing the indicated concentrations of DPI or 1 mM KI and incubated overnight in the dark. Next, 100 nM BL or 80% ethanol solution was added to the medium at the EoN and then illuminated with white light for 3 h.
(B) to (D) Quantification of stomatal aperture ([B] and [C]) and starch granules (D) in guard cells of intact leaves of the wild-type, rbohd rbohf, and CAT2-Ox plants with or without BR treatment. The rosette leaves of the 4-week-old wild-type Col-0, rbohd rbohf, and CAT2-Ox were transferred to half strength MS liquid medium and incubated overnight in the dark. Next, 100 nM BL or 80% ethanol solution was added to the medium at the EoN, and samples were illuminated with white light for 3 h.
The ratio of stomatal aperture width to length or guard cell starch granules from at least 100 guard cells from 10 to 15 leaves of eight different plants were measured using ImageJ software. Three independent biological repeats were performed. Error bars indicate sd (n = 3 biological repeats). Different letters above the bars indicate statistically significant differences between samples (two-way ANOVA followed by a post hoc Tukey test, P < 0.05).
H2O2 Induces Stomatal Opening at Low Concentrations
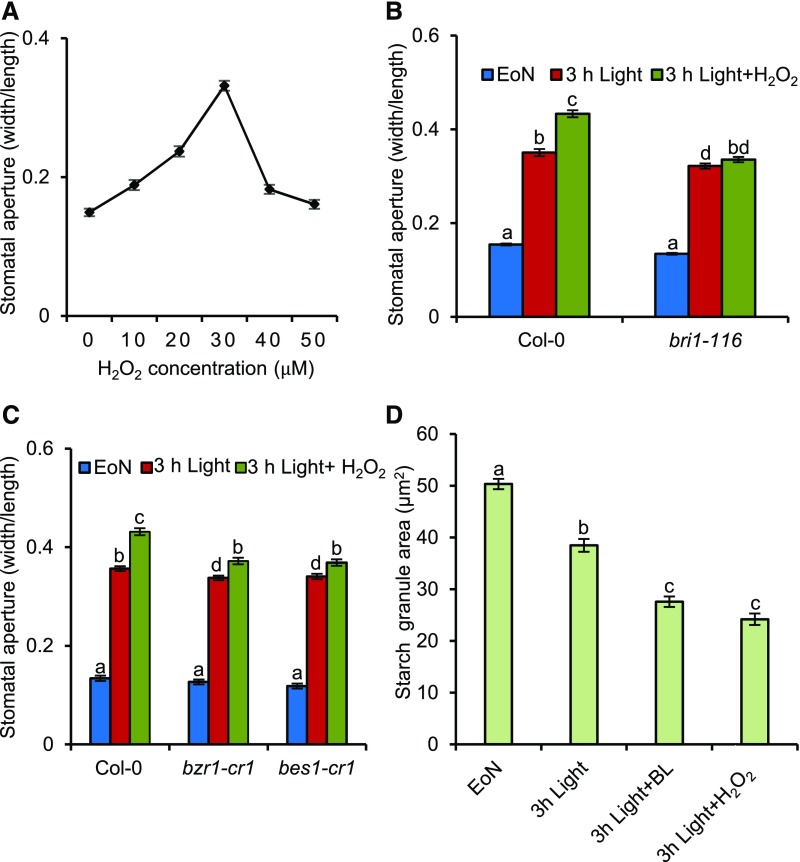
Reactive oxygen species (ROS) homeostasis in plants has been reported to exhibit time-of-day–specific changes, and H2O2 production peaks at noon and reaches trough levels at midnight (Lai et al., 2012). Here, we showed that the levels of H2O2 in guard cells of leaves was low at the EoN and then quickly increased after light illumination, consistent with the increased stomatal apertures of leaves (Supplemental Figure 5A). BR and H2O2 treatment both significantly induced the accumulation of H2O2 in guard cells (Supplemental Figure 5B). In addition, the stomatal apertures of rbohd rbohf and CAT2-Ox were smaller than those of the wild type under our experimental conditions. These results indicated that H2O2 might promote stomatal opening at the cellular level (Figures 2B and 2C). To confirm this hypothesis, we analyzed the stomatal apertures of the wild-type plants treated with low concentrations of H2O2. Stomatal opening was first promoted at 10 µM H2O2, with the most significant effects at 30 µM; however, stomatal opening was inhibited with increasing H2O2 concentrations (Figure 3A). To evaluate the roles of BR in H2O2-promoted stomatal opening, we investigated the H2O2 response in BR-related mutants. The effects of H2O2 on the stomatal opening in the BR-insensitive mutants bri1-116 and bri1-301 and the BR-deficient mutant det2 were significantly reduced, suggesting that BR is required for H2O2-induced stomatal opening (Figure 3B; Supplemental Figures 6A and 6B). The major transcription factor BZR1 integrates the BR and H2O2 signals to regulate cell elongation and root stem cell maintenance (Tian et al., 2018). To determine whether BZR1 is involved in H2O2-mediated stomatal opening, we analyzed the H2O2 responses in the BZR1 knockout mutant. The results showed that H2O2 induces stomatal opening in the wild-type plants but has weak effects on bzr1-cr1 and bes1-cr1 mutants (Figure 3C). The starch contents in the leaves of rbohd rbohf and CAT2-Ox were not significantly different compared with the wild type, but H2O2 treatment, similar to BR treatment, dramatically induced starch degradation in guard cells (Figure 3D; Supplemental Figures 7A and 7B). These results indicated that BR and H2O2 converge at transcription factor BZR1 to promote stomatal opening by inducing guard cell starch degradation.
Figure 3.
H2O2 Induces Stomatal Opening at Low Concentrations.
(A) H2O2 promotes stomatal opening at low concentrations. The rosette leaves of 4-week-old Col-0 were transferred to half strength MS liquid medium, incubated overnight in the dark, and then added to different concentrations of H2O2 and incubated for 3 h in darkness. The leaves were then harvested, and stomatal apertures were measured.
(B) to (D) Quantification of stomatal apertures ([B] and [C]) and starch granules (D) in guard cells of intact leaves of the wild type and different mutants with or without H2O2 and BR treatment. The rosette leaves of the 4-week-old wild-type Col-0, bzr1-cr1, and bes1-cr1 plants were transferred to half strength MS liquid medium and incubated overnight in the dark. Next, 30 µM H2O2, 100 nM BL, or 80% ethanol solution was added to the medium at the EoN and illuminated with white light for 3 h.
The ratio of stomatal aperture width to length or guard cell starch granules from 100 cells of 15 leaves of eight different plants were measured using ImageJ software. Three independent biological repeats were performed. Error bars indicate sd (n = 3 biological repeats). Different letters above the bars indicate statistically significant differences between the samples (two-way ANOVA, [B] and [C], or one-way ANOVA [D] followed by a post hoc Tukey test, P < 0.05).
H2O2 Enhances the Interaction between BZR1 and GBF2
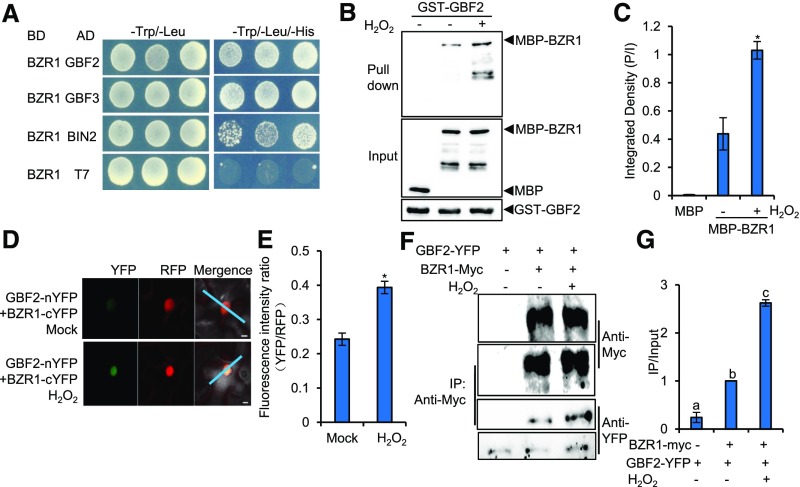
Considering that BR and H2O2 interdependently promote stomatal opening, and our previous findings that H2O2 positively regulates BR signaling by oxidative modification of BZR1 to increase its binding affinity to transcription factors PIF4 and ARF6 (Tian et al., 2018), we hypothesized that H2O2 might regulate stomatal opening by promoting the interaction between BZR1 and some transcription factors that have important roles in stomatal opening. To test this hypothesis, we performed a yeast two-hybrid screen to identify the transcription factors that interact with BZR1 and then analyzed the functions of these putative BZR1-interacting proteins in stomatal opening using reverse genetic approaches. Among these putative BZR1-interacting transcription factors, GBF2 shows the potential function of regulating stomatal movement. GBF2 belongs to the G group of bZIP transcription factors that play important functions in a plethora of processes related to plant development, environmental signaling, and stress response (Schütze et al., 2008). Therefore, GBF2 was selected for further in-depth study.
To determine whether GBF2 is involved in BR- and H2O2-mediated stomatal opening, we first repeated the yeast two-hybrid assays to confirm the interaction between BZR1 and GBF2. The results showed that BZR1 interacted with GBF2 and its homologous protein GBF3 in yeast (Figure 4A). We also performed glutathione S-transferase (GST) pull-down assays to test the direct interaction between BZR1 and GBF2 in vitro. As shown in Figure 4B, GST-GBF2 could pull down maltose binding protein (MBP)–BZR1, but could not pull down MBP alone. The H2O2 treatment dramatically increased the binding affinity of BZR1 to GBF2 (Figure 4C). To test whether BZR1 interacts with GBF2 in plants and whether H2O2 regulates this interaction, a ratiometric bimolecular fluorescence complementation assay (Hecker. et al., 2015) was performed. In this assay, BZR1 and GBF2 were simultaneously cloned into a single vector backbone that contained a monomeric red fluorescent protein driven by the 35S promoter as an internal marker for expression control and ratiometric analysis. The ratiometric bimolecular fluorescence complementation results showed that BZR1 interacted with GBF2 in the epidermal cells of tobacco (Nicotiana tabacum) leaves, and this interaction was significantly enhanced by the H2O2 treatment (Figures 4D and 4E). Based on Arabidopsis microarray data displayed in the eFP browser (Winter et al., 2007), BZR1 and GBF2 displayed similar ubiquitous expression patterns in whole plants, and they were also highly expressed in the guard cells of leaves (Supplemental Figures 8A and 8B). We further analyzed the tissue expression patterns of BZR1 and GBF2 using the ProBZR1:BZR1-YFP and ProGBF2:GBF2-YFP transgenic plants. BZR1 and GBF2 were both expressed in the pavement cells, stomatal lineage cells, and guard cells of epidermal leaves (Supplemental Figure 8C). Coimmunoprecipitation (CoIP) assays were used to further verify the interaction between BZR1 and GBF2 in plants. The results showed that BZR1 interacted with GBF2 in plants and that H2O2 enhanced this interaction (Figure 4F and 4G). Together, these results demonstrate that H2O2 enhances the interaction between BZR1 and GBF2 in vivo and in vitro.
Figure 4.
H2O2 Enhances the Interaction between BZR1 and GBF2 in Vivo and in Vitro.
(A) BZR1 interacts with GBF2, GBF3, and BIN2 in yeast.
(B) and (C) In vitro pull-down assays showed that H2O2 enhanced the interaction between BZR1 and GBF2. Panel (C) shows the quantification of pull-down assays (normalized to input) displayed in (B). P/I, ratio of immunoblot image intensities between pull down and input. Error bars represent the sd of three independent experiments. *P < 0.05, as determined by a Student’s t test.
(D) and (E) H2O2 enhanced the interaction between BZR1 and GBF2 in tobacco leaves. (D) The fluorescent signals of YFP (BiFC) and RFP (reference) were determined along a line drawn on confocal images using ImageJ software. Error bars in (E) indicate sd (n = 100 images). *P < 0.05, as determined by a Student’s t test. Bar = 10 μm.
(F) and (G) H2O2 increased the interaction between BZR1 and GBF2 in plants. Plants expressing BZR1-Myc from the native BZR1 promoter and GBF2-YFP from the native GBF2 promoter or plants expressing only GBF2-YFP were treated with water (mock) or 100 μM H2O2 for 1 h before CoIP. The CoIP experiments were performed using Myc-Trap agarose beads, and the immunoblots were probed using anti-Myc and anti-YFP antibodies. Panel (G) shows the quantification of CoIP assays (normalized to input) displayed in (F). IP/Input, ratio of immunoblot image intensities between IP and input. Error bars represent the sd of three independent experiments. Different letters above the bars indicate statistically significant differences between the samples (one-way ANOVA followed by a post hoc Tukey test, P < 0.05). IP, immunoprecipitation.
BZR1 and GBF2 Synergistically Promote Guard Cell Starch Degradation and Stomatal Opening
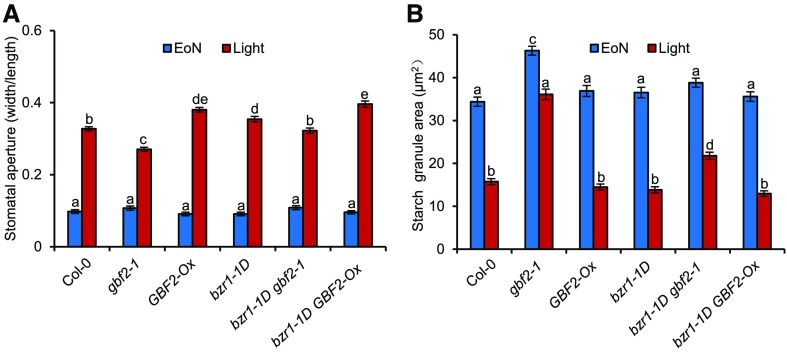
To characterize the function of GBF2 in stomatal movement, we analyzed the stomatal apertures of a series of GBF2 transgenic plants, including the GBF2-knockout mutant gbf2-1 (Salk_031900) that harbors a T-DNA insertion in the 5′ untranslated region (UTR) of GBF2 (Supplemental Figure 9A), GBF2 complementation lines (ProGBF2:GBF2-YFP/gbf2-1) with genomic GBF2 fused to a yellow fluorescent protein (YFP) driven by the native GBF2 promoter transgenically expressed in the gbf2-1 background, and GBF2 overexpression lines (GBF2-Ox) that were generated by the constitutive expression of the GBF2 coding sequence under the control of the cauliflower mosaic virus 35S promoter (Supplemental Figure 9B). Stomatal aperture in gbf2-1 exhibited a dramatic reduction, while GBF2 complementation lines restored the opening of stomatal pores to levels similar to that of the wild-type plants, indicating that the impaired light-induced stomatal opening of gbf2-1 was caused by GBF2 loss of function. By contrast, GBF2-Ox displayed significantly larger stomatal apertures than the wild-type plants (Figure 5A; Supplemental Figure 9C). These results demonstrate that GBF2 plays a positive role in stomatal opening.
Figure 5.
BZR1 and GBF2 Synergistically Promote Guard Cell Starch Degradation and Stomatal Opening.
(A) BZR1 and GBF2 synergistically promoted stomatal aperture in intact leaves of the 4-week-old wild-type and the indicated mutant plants at the EoN and 1 h after white light exposure, as determined by light microscopy and digital image processing.
(B) Quantification of starch content in guard cells of intact leaves of the 4-week-old wild type and mutants at the EoN and 1 h after light exposure. Error bars indicate sd (n = 100 cells).
The ratio of stomatal aperture width to length and guard cell starch granules from at least 100 guard cells from 10 to 15 leaves of eight different plants were measured using ImageJ software. Three independent biological repeats were performed. Error bar indicates sd (n = 3 biological repeats). Different letters above the bars indicate statistically significant differences between samples (two-way ANOVA followed by a post hoc Tukey test, P < 0.05).
Given the physical association of GBF2 and BZR1, we assessed whether the biological function of BZR1 requires that of GBF2. We crossed the BZR1 gain-of-function mutant bzr1-1D with the GBF2 loss-of-function gbf2-1, and the GBF2 gain-of-function GBF2-Ox. Stomatal aperture measurements showed that gbf2-1 suppressed and GBF2-Ox enhanced the stomatal opening phenotype of bzr1-1D, indicating that GBF2 is essential for BZR1 promotion of stomatal opening (Figure 5A). The BR and H2O2 treatments significantly induced stomatal opening in the wild type Col-0 plants but had weak effects on gbf2-1, suggesting that GBF2 plays an important role in BR- and H2O2-mediated stomatal opening (Supplemental Figures 9D and 9E). Hypocotyl elongation assays are routinely used to determine the contribution of GBF2 to the BR response. gbf2-1 and GBF2-Ox both had normal responses to BR and to the BR biosynthesis inhibitor propiconazole (PPZ) (Supplemental Figures 10A to 10D); furthermore, there were no growth differences (Supplemental Figure 10E), indicating that GBF2 is specifically required for BZR1-mediated stomatal opening, but not for BZR1-induced cell elongation.
Starch breakdown in guard cell plays important roles in stomatal opening (Horrer et al., 2016). To determine whether BZR1 and GBF2 promote stomatal opening by regulating starch degradation in guard cells, we measured the guard cell starch content in BZR1- and GBF2-related materials. GBF2-Ox and bzr1-1D mutants both showed normal guard cell starch degradation under light exposure. However, the mutation of GBF2 in both the wild-type plants and in the bzr1-1D background impaired the rapid degradation of guard cell starch in response to light (Figure 5B). These results indicate that BZR1 and GBF2 synergistically promote the degradation of starch in guard cells that, in turn, induces stomatal opening.
BZR1 and GBF2 Directly Regulate BAM1 Expression
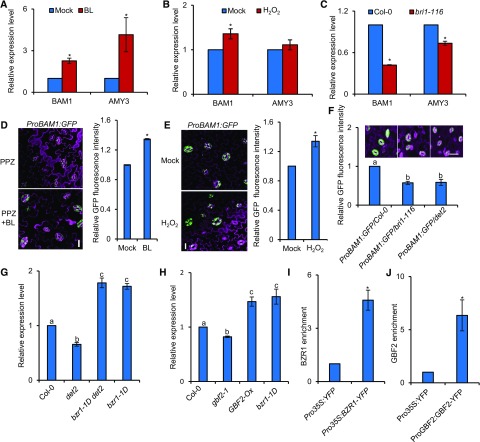
To decipher the molecular mechanism through which BR and H2O2 regulate starch levels in guard cells, we examined the expression of genes involved in guard cell starch degradation in response to BR and H2O2 using RT-qPCR with enriched epidermal cells. The enzymes BAM1 and AMY3 are preferentially expressed in guard cells under standard conditions and are responsible for the degradation of starch in guard cells (Horrer et al., 2016). BR and H2O2 treatment both significantly increased the transcript levels of BAM1. The expression of BAM1 was downregulated in the bri1-116 mutants (Figures 6A to 6C). To verify the effects of BR and H2O2 on the accumulation of BAM1, we monitored the fluorescent signal of ProBAM1:GFP transgenic plants. Consistent with the previously published results that the ProBAM1:GFP promoter-reporter was specifically expressed in the guard cells of epidermal leaves (Valerio et al., 2011), BR and H2O2 treatments both increased the fluorescence intensity of ProBAM1:GFP (Figures 6D and 6E; Supplemental Figures 11A and 11B). The fluorescence intensity of ProBAM1:GFP was partially reduced in bri1-116 and det2 mutants (Figure 6F). We further showed that the expression of BAM1 was reduced in bri1-116, det2, and gbf2-1 but activated in bzr1-1D (Figures 6G and 6H; Supplemental Figures 12A and 12B). To assess whether BZR1 and GBF2 directly bind to the BAM1 promoter and regulate its transcription, we then performed chromatin immunoprecipitation (ChIP)–qPCR experiments using Pro35S:BZR1-YFP and ProGBF2:GBF2-YFP transgenic plants. A high enrichment of BZR1 and GBF2 was observed in the promoter of BAM1, indicating that BAM1 is the direct target of BZR1 and GBF2 (Figures 6I and 6J). Together, these results suggest that BZR1 and GBF2 directly induce BAM1 expression to promote starch breakdown in guard cells.
Figure 6.
BZR1 and GBF2 Directly Control BAM1 Expression.
(A) to (C) RT-qPCR analysis of the effects of BR and H2O2 on the expression of BAM1 in guard cell–enriched epidermal fragments of the 4-week-old wild-type Col-0 treated with or without 100 nM BL (A), 30 µM H2O2 (B) for 3 h, or in bri1-116 mutants (C). PP2A was used as an internal control. Error bars indicate sd of three biological repeats. *P < 0.05, as determined by a Student’s t test.
(D) Confocal images of ProBAM1:GFP in the epidermis of leaves. The ProBAM1:GFP transgenic plants were grown in half strength MS medium with 2 µM PPZ for 11 d and then treated with 80% ethanol solution (mock) or 100 nM BL for 3 h. GFP signal intensity was analyzed using ImageJ software. Error bars indicate sd (n = 100 images). *P < 0.05, as determined by a Student’s t test. Bar = 20 µm.
(E) Confocal images of ProBAM1:GFP in the epidermis of leaves of 4-week-old plants treated with or without 30 µM H2O2 for 3 h. GFP signal intensity was analyzed by ImageJ software. Error bars indicate sd (n = 100 images). *P < 0.05, as determined by a Student’s t test. Bar, 20 µm.
(F) Plants expressing the ProBAM1:GFP transgene displayed low GFP fluorescence signals in the Col-0, bri1-116, and det2 background. GFP signal intensity was analyzed using ImageJ software. Error bars indicate sd of three biological repeats. Differnt letters above bars indicate statistically significant differences between samples (one-way ANOVA followed by a post hoc Tukey test, P<0.05). Bar = 20 µm.
(G) and (H) RT-qPCR analysis of BAM1 expression in guard cell–enriched epidermal fragments of 4-week-old BR-related mutants (G) and GBF2-related plants (H). PP2A was used as an internal control. Error bars indicate sd of three biological repeats. Different letters above bars indicate statistically significant differences between samples (one-way ANOVA followed by a post hoc Tukey test, P < 0.05).
(I) and (J) ChIP-qPCR showed that BZR1 and GBF2 bind to the promoter of BAM1. ChIP was performed using GFP-Trap followed by qPCR analysis. The level of BZR1 or GBF2 binding was calculated as the ratio between BZR1-YFP or GBF2-YFP and the Pro35S:YFP control, normalized to that of the control gene PP2A. Error bars indicate sd of three biological repeats. *P < 0.05, as determined by a Student’s t test..
Mutation in BAM1 Attenuates the Positive Effects of BR and H2O2 on Stomatal Opening
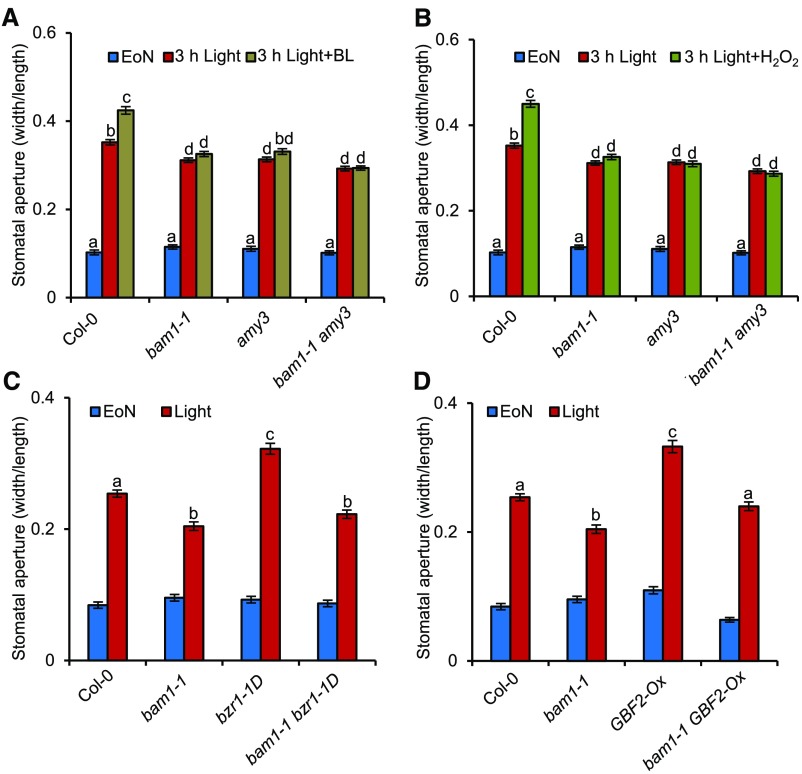
To determine whether BAM1-mediated guard cell starch degradation is involved in the BR- or H2O2-mediated stomatal opening process, we analyzed the effects of BR and H2O2 on bam1-1 mutants. Because of the important role of AMY3 in the starch degradation in guard cells, amy3 single mutant and bam1-1 amy3 double mutants were also used to analyze the effects of guard cell starch degradation on BR- and H2O2-induced stomatal opening processes. BR and H2O2 could effectively induce stomatal opening in wild-type plants, but not in bam1-1 and bam1-1 amy3 mutants, and had a weak effect on the amy3 single mutant (Figures 7A and 7B). A mutation in BAM1 also prevented bzr1-1D and GBF2-Ox from degrading starch to induce stomatal opening (Figures 7C and 7D). The BR-induced hypocotyl elongation assays showed that bam1-1 and amy3 mutants exhibited normal responses to BR and the BR biosynthesis inhibitor PPZ (Supplemental Figures 13A and 13B). These results indicate that BAM1 and AMY3 specifically contribute to the BR and H2O2 response in guard cells, but not to cell elongation in the hypocotyl.
Figure 7.
Mutation in BAM1 Attenuates the Promoting Effects of BR and H2O2 on Stomatal Opening.
(A) and (B) The bam1-1 mutant is less sensitive to BR (A) and H2O2 (B) in the stomatal opening process. The rosette leaves of the 4-week-old wild-type Col-0, bam1-1, amy3, and bam1-1 amy3 were transferred to half strength MS liquid medium and incubated overnight in darkness. Next, 30 µM H2O2, 100 nM BL, or 80% ethanol solution was added to the medium at EoN, and the leaves were illuminated with white light for 3 h.
(C) and (D) Mutation in BAM1 partly suppressed the stomatal aperture phenotype of bzr1-1D (C) and GBF2-Ox (D). The rosette leaves of 4-week-old Col-0 and different mutants were harvested at 1 h after white light exposure in the morning to measure the stomatal apertures.
The ratio of stomatal aperture width to length from at least 100 guard cells from 10 to 15 leaves of eight different plants were measured using ImageJ software. Three independent biological repeats were performed. Error bars indicate sd (n = 3 biological repeats). Different letters above the bars indicate statistically significant differences between samples (two-way, [A] and [B], or one-way, [C] and [D]) ANOVA followed by a post hoc Tukey test, P < 0.05).
DISCUSSION
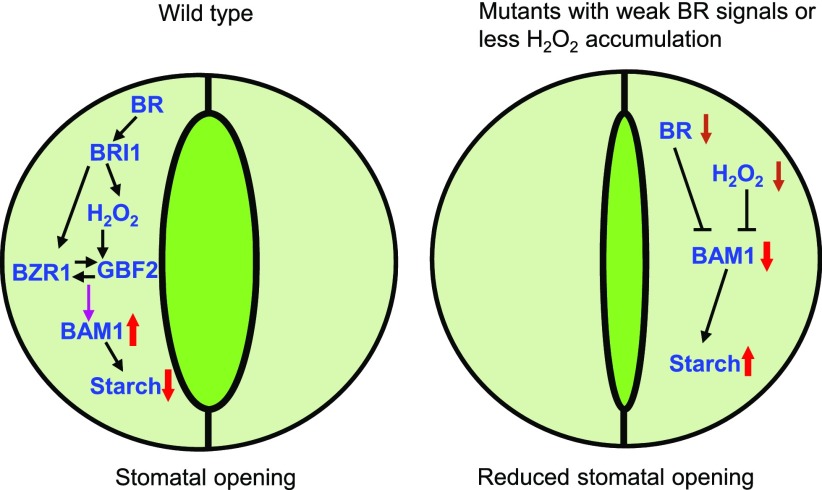
Starch is a major storage carbohydrate widely synthesized in plants and serves as an important carbon storage molecule for plant metabolism and growth under changing environmental conditions (Zeeman et al., 2010; Thalmann and Santelia, 2017). The enzymes involved in starch metabolism have been characterized in vascular plants, but how environmental signals and phytohormones regulate the activities of these enzymes remains unclear (Streb and Zeeman, 2012). In this study, we demonstrate that BR and H2O2 interdependently promote starch degradation in guard cells, which promotes stomatal opening. In wild-type plants, binding of BR to the receptor kinase BRI1 triggers the accumulation of H2O2 that induces BZR1 oxidation and enhances the interaction between BZR1 and GBF2, which directly promotes the expression of BAM1, inducing guard cell starch degradation and stomatal opening. By contrast, in the mutants with weak BR signals or reduced H2O2 content, the decreased expression levels of BAM1 lead to the accumulation of starch in guard cells and reduced stomatal opening (Figure 8).
Figure 8.
A Model for BR and H2O2 Promotion of Starch Degradation in Guard Cells to Induce Stomatal Opening.
In the wild-type plants, binding of BR to the receptor kinase BRI1 triggers H2O2 accumulation, which induces BZR1 oxidation and enhances the interaction between BZR1 and GBF2, and induces the expression of BAM1 to degrade starch and promote stomatal opening. Mutants with weak BR signals or reduced H2O2 accumulation have low expression levels of BAM1 and high levels of starch in guard cells, which results in reduced stomatal apertures. Black lines, posttranscriptional regulation; purple lines, transcriptional regulation; red arrows pointing up, upregulation; red arrows pointing down, downregulation; brown arrows pointing down, plant with weak BR signals or less H2O2 accumulation.
The transitory starch breakdown in the chloroplast of guard cells plays important roles in stomatal opening (Horrer et al., 2016). Our work provides several lines of evidence indicating that starch degradation in guard cells is critical for BR- and/or H2O2-promoted stomatal opening. First, the opposite effects of BR-hyperresponse and -hyporesponse mutants on guard cell starch degradation provide strong evidence for a correlation between BR-induced stomatal opening and starch breakdown in the chloroplast of guard cells. The BR-insensitive mutant bri1-116 and the BR-deficient mutant det2 contained high levels of starch in the guard cells and showed a slow starch-degraded ratio under light exposure, and both mutants displayed impaired stomatal opening in response to light. The high levels of starch in guard cells and impaired stomatal opening phenotypes of bri1-116 and det2 were suppressed by the gain-of-function mutant bzr1-1D, suggesting that BR could induce guard cell starch degradation and stomatal opening through BZR1-mediated regulation of key enzymes for starch degradation in guard cells, such as BAM1. However, the starch contents and starch degradation ratio in the guard cells of bzr1-1D are similar to those in the wild type, which may be due to the similar transcriptional activity of BZR1 in the guard cells of the wild type and bzr1-1D. When treated with BR or H2O2, the transcriptional activity of BZR1 is increased by promoting the nuclear localization of BZR1 or inducing the oxidative modification of BZR1, respectively, and then resulting in the increased expression of BAM1, guard cell starch degradation, and stomatal opening. Mutations in BAM1 and AMY3, which are required for starch degradation in guard cells, interfered with BR- or H2O2-induced stomatal opening, and counteracted the stomatal aperture phenotype of bzr1-1D. Finally, the direct induction of the β-amylase gene BAM1 by BZR1 and its partner GBF2 indicated that there was a specific mechanism for BR regulation of stomatal opening by promoting guard cell starch degradation.
BR, as a type of growth-promoting hormone, has been reported to enhance the photosynthetic rate in several plant species, and especially in plants stressed by various abiotic stress factors, but its molecular mechanism remains unclear (Wu et al., 2008; Jiang et al., 2012; Li et al., 2016). Here, we showed that BR promotes stomatal opening by inducing the starch degradation in guard cells, which may result in increased stomatal conductance and CO2 assimilation to increase photosynthetic rate. Consequently, overexpression of the BR biosynthetic gene DWF4 or BR receptor BRI1 induces stomatal opening, enhances photosynthesis, and promotes plant growth and development. The growth defect of BR-deficient mutants or BR-insensitive mutants may be partially due to the impaired stomatal opening and the reduced photosynthesis efficiency. In support of this, the levels of starch, which is a major product of photosynthesis, were significantly lower in the leaves of bri1-116 and det2 plants than in those of wild-type plants. However, the sizes of guard cells in BR-deficient mutants or BR-insensitive mutants are similar to those in wild-type plants (Inoue et al. 2017), suggesting that BR regulates the development of guard cells and other cells through different pathways.
BR plays dual roles in modulating stomatal movement, as low concentrations of BR promote stomatal opening, while high concentrations of BR induce stomatal closure. In this study, BR-deficient mutants exhibited small stomatal aperture, whereas BR signal-enhancing mutants such as bin2 bil1 bil2, bzr1-1D, and bes1-D showed large stomatal aperture.
We also established that BZR1 interacts with GBF2 to induce the expression of BAM1 and promote guard cell starch degradation, thereby promoting stomatal opening. Plants with mutation in BAM1 contained more starch in their guard cells and their stomata were more tightly closed than those of the wild-type plants. The bam1-1 mutant exhibited hyposensitivity to BR and H2O2 with regard to stomatal opening but showed a normal BR and H2O2 response with regard to stomatal closure, indicating that BAM1-mediated guard cell starch degradation specifically functions in stomatal opening, but not in stomatal closure (Figures 7A and 7B; Supplemental Figures 14A to 14C).
However, the molecular mechanism by which BAM1-mediated guard cell starch degradation promotes stomatal opening is unclear. One possibility is that guard cell starch degradation produces the malate2− that is an important counterion for K+ (Daloso et al., 2017; Thalmann and Santelia, 2017). The accumulation of malate2− and K+ reduces the water potential, which promotes the inflow of water and then induces the opening of the stomatal pore. A previous study has shown that the decreased expression of the inward-rectifying K+ channel gene KAT1 in the BR-deficient mutant dwarf5-7 resulted in reduced K+ levels and inhibition of stomatal opening (Inoue et al. 2017). Here, we showed that the expression of BAM1 is significantly decreased in the BR-insensitive and -deficient mutants, which led to the accumulation of starch and may produce less malate2− in guard cells. Together these results indicate that K+ and its counter ion malate2+ are simultaneously reduced in BR-deficient mutants through repressing the expression of KAT1 and BAM1, which caused the decreased water potential and impaired stomatal opening.
Kim et al. (2018) recently reported that exogenous BR treatment induces stomatal closure via the interaction between BR-associated CDL1 and abscisic acid–associated OST1. Both CDL1 and OST1 are required for BR- and abscisic acid–induced stomatal closure, as CDL1 and OST1 phosphorylate each other, activating downstream components to induce stomatal closure (Kim et al., 2018). However, under our experiment conditions, we found that CDL1 and OST1 are not involved in BR-mediated stomatal opening (Supplemental Figures 15A to 15D). The results obtained here indicated that BR regulates stomatal opening and closure through different mechanisms: it promotes stomatal opening by activating BAM1 to induce guard cell starch degradation but promotes stomatal closure by enhancing the kinase activity of CDL1 and OST1.
BR and blue light are key signals that regulate many aspects of plant growth and development throughout the plant life cycle (De Wit et al., 2016). Stomatal pores, surrounded by pairs of guard cells on the surfaces of plant leaves, regulate CO2 uptake and water loss, thereby affecting the efficiency of photosynthesis and transpiration (Murata et al., 2015). BR and blue light both play important roles in stomatal movement. Phototropins act as major blue light receptors for stomatal opening. Blue light induces the autophosphorylation of phototropins and initiates light signal transduction to finally activate a plasma membrane H+-ATPase and promote stomatal opening (Shimazaki et al., 2007). Blue light induces the guard cell starch degradation through a phototropin-dependent signaling pathway (Horrer et al., 2016).
In this study, we showed that the BR-insensitive mutant bri1-116 and BR-deficient mutant det2 both accumulated high levels of starch in guard cells and displayed impaired stomatal opening under blue light illumination and that these abnormal phenotypes of bri1-116 and det2 were suppressed by bzr1-1D, suggesting that BR and BZR1 maybe also be involved in the blue light–regulated guard cell starch degradation and stomatal movement. Recently, several groups reported that BR and blue light antagonistically regulate seedling photomorphogenesis through the interaction between the blue light photoreceptor cryptochromes and transcription factors BZR1, BES1, and HBI1 (Wang et al., 2018a, 2018b; He et al., 2019). CRY1 and CRY2 physically interact with these transcription factors in a blue light–dependent manner, inhibit their DNA binding ability and transcriptional activity, and further regulate BR signal transduction and plant photomorphogenesis (Wang et al., 2018a, 2018b; He et al., 2019). In addition to CRY1 and CRY2, other photoreceptors including the red light receptor phytochrome B and UV-B light receptor UVR8 all interact with BZR1/BES1 to regulate the activity of BZR1/BES1 (Liang et al., 2018; Wang et al., 2018b; Dong et al., 2019; He et al., 2019; Wu et al., 2019). Whether the blue light photoreceptors PHOT1 and PHOT2 interact with BZR1 and BES1 to regulate stomatal movement should be investigated further.
H2O2 was initially considered to be a toxic by-product of oxidative plant aerobic metabolism that causes irreversible damage to cells but now is recognized as an important signaling molecule that regulates a wide range of plant growth, development, and stress responses, especially in stomatal closure (Xia et al., 2015; Mittler, 2017). Drought stress induces the biosynthesis of abscisic acid in guard cells, which activates the protein kinase OST1 to induce the phosphorylation of respiratory burst oxidase homolog protein F (RBOHF) to produce O2− that is converted to H2O2 by superoxide dismutase, and then the accumulated H2O2 promotes stomatal closure (Qi et al., 2018; Han et al., 2019). Hao et al. (2012) reported that ATP as an important signaling molecule could promote stomatal opening, but this promotion was impaired in the null mutant of NADPH oxidase, rbohd rbohf, suggesting that H2O2 plays critical roles in the stomatal opening process. In this study, we confirmed that H2O2 positively regulates stomatal opening by several lines of evidence. First, the rbohd rbohf mutant and CAT2-Ox transgenic lines both displayed smaller stomatal apertures than the wild types and failed to efficiently open stomata in response to BR treatment. Second, exogenous application of H2O2 (<30 µM) significantly induced stomatal opening in a dose-dependent manner. Third, we showed that the guard cell starch degradation ratio in the rbohd rbohf and CAT2-Ox plants was dramatically lower than that in the wild type. These results demonstrated that H2O2 has a double effect on stomatal movement, that is, it promotes stomatal opening at low concentrations but induces stomatal closure at high concentrations, making it challenging to measure the absolute H2O2 levels in guard cells; therefore, it is difficult to estimate the concentration ranges of H2O2 that promote stomatal opening and closure in plants. The reason why H2O2 has opposing effects on stomatal movement remains unclear. One possibility is that H2O2 oppositely regulates the activity of BAM1 at different concentrations to regulate stomatal movement. Low concentrations of H2O2 enhanced the interaction between BZR1 and GBF2 to induce the expression of BAM1 and guard cell starch degradation to promote stomatal opening. By contrast, high concentrations of H2O2 inhibited the β-amylase activity of BAM1 by oxidative modification that has been demonstrated by several groups using a well-defined in vitro system (Sparla et al., 2006; Valerio et al., 2011). The regulation of BAM1 at the transcriptional level together with the reversible oxidation of key Cys residues at the posttranscription levels orchestrates the precise regulation of starch contents and stomatal apertures downstream of H2O2 and diverse hormonal and environmental signals.
In Arabidopsis, it has been shown that loss-of-function mutants in the BR biosynthesis or signaling pathways have an enhanced tolerance to drought, while the gain-of-function mutant bes1-D exhibited lower drought tolerance than the wild-type plants (Chen et al., 2017; Nolan et al., 2017; Ye et al., 2017). BES1 interacts with the drought-induced transcription factor RESPONSIVE TO DESICCATION26 (RD26) to negatively regulate the expression of drought-responsive genes and inhibit drought tolerance (Ye et al., 2017). In addition, BES1 has been reported to interact with the group III WRKY transcription factor WRKY54 to cooperatively regulate the expression of BR-regulated and dehydration-responsive genes, thereby controlling plant growth and drought tolerance (Chen et al., 2017). Here, we showed that BR promoted stomatal opening by increasing the activity of BAM1 to trigger the degradation of starch. The drought tolerance of the bam1-1 mutant is significantly higher than that of wild-type plants due to its reduced stomatal aperture and impaired starch breakdown (Valerio et al., 2011; Thalmann et al., 2016; Zanella et al., 2016). Transcriptomic analysis showed that the expression of BAM1 and AMY3 is regulated by RD26 and WRKY54, indicating that BZR1 and BES1 interact with multiple transcription factors to finely regulate the expression of BAM1 to control the contents of starch in guard cells (Chen et al., 2017; Ye et al., 2017). These results indicate that the drought-tolerant phenotype of the BR-deficient or -insensitive mutants might be at least partially due to the lower expression of BAM1 and impaired guard cell starch breakdown, which lead to reduced stomatal opening and water loss.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) Col-0 ecotype was used as the wild-type plant for all transgenic plants and knockout mutants, except bin2 bil1 bil2 in the Wassilewskija background and bes1-D in Enkheim-2 background (Yin et al., 2002; Yan et al., 2009). Arabidopsis plants were grown in a greenhouse with white light at 150 µmol·m−2·s−1 and 50% RH under a 12-h-light/12-h-dark cycle at 22 to 24°C for general growth and seed harvesting. Alternatively, plants were grown in a growth chamber with light-emitting diode light (RXZ, Ningbo Jiangnan Instruments), with a total photon fluorescence rate of 100 µmol·m−2·s−1. The T-DNA insertion knockout mutants bam1-1 (Salk_039895), amy3 (Sail_613_D12), and gbf2-1 (Salk_031900) were obtained from the Arabidopsis Biological Resource Center. All the other single and multiple mutants used in this study were described previously: bzr1-1D, bri1-116, bzr1-1D bri1-116, bri1-301, det2, bzr1-1D det2, bes1-ko, bin2 bil1 bil2, and rot3-1 (Li et al., 1996; Tsuge et al., 1996; Li and Chory, 1997; Wang et al., 2002; Xu et al., 2008; Yan et al., 2009). For hypocotyl length measurement, the seedlings were scanned by a K708 scanner (BenQ), and their hypocotyl lengths were measured using ImageJ software (National Institutes of Health; http://rsbweb.nih.gov/ij/).
Plasmid and Transgenic Plants
Full-length coding sequences of BZR1 and GBF2 were amplified by PCR and cloned into pENTR/SD/D-TOPO vectors (Thermo Fisher Scientific). Each entry clone was subcloned into Gateway-compatible destination vectors including pGCBDT7 (GAL4BD-X), pGCADT7 (GAL4AD-X), pDEST15 (N-GST), pMAL2CGW (N-MBP), pX-nYFP (Pro35S:C-nYFP), pX-cYFP (Pro35S:C-cYFP), and pX-YFP (Pro35S:C-YFP). Oligo primers used for cloning are listed in Supplemental Data Set 1. All binary vector constructs were introduced into Agrobacterium tumefaciens (strain GV3101) and transformed into Col-0 or mutants plants using the floral dip method (Clough and Bent, 1998).
Generation of the CRISPR/Cas9 Mutant
Two fragments of guide RNA were selected for BZR1 and BES1, respectively, and cloned into the CRISPR/Cas9 vector ProYAO:hSpCas9, as described previously (Yan et al., 2015). The constructs were then transformed into Col-0 plants using the Agrobacterium-mediated floral dip method (Clough and Bent, 1998). The transgenic lines were confirmed by PCR and sequencing to determine whether the targeted regions contained the desired mutations. The sequence information of targets and primers is listed in Supplemental Data Set 1.
Guard Cell Starch Quantification
The quantification of starch in guard cells was performed following previously described procedures (Horrer et al., 2016; Flütsch et al., 2018). Briefly, epidermal peels of the sixth rosette leaf or cotyledon of the wild type and various mutants were harvested at the indicated time points and immediately fixed in 50% (v/v) methanol and 10% (v/v) acetic acid at least for 12 h at 4°C in the dark. Samples were first destained by incubation in 80% (v/v) ethanol at 65°C for 5 to 10 min and, after removing the ethanol, they were repeatedly washed with water. Starch granules were stained with a mPS-PI staining method. Samples were treated with 1% (v/v) periodic acid at room temperature for 40 min. After rinsing with water, samples were incubated in Schiff reagent with propidium iodide (100 mM sodium metabisulfite and 0.15 N HCl; propidium iodide to a final concentration of 100 µg/mL was freshly added) for 1 to 2 h or until plants were visibly stained at room temperature. The samples were destained in water and then transferred into chloral hydrate solution (4 g of chloral hydrate, 1 mL of glycerol, and 2 mL of water) on microscopy slides. After incubation in the dark for 24 h, samples were fixed using Hoyer’s solution (30 g of gum arabic, 200 g of chloral hydrate, 20 g of glycerol, and 50 mL of water). Microscopy observations were performed under an LSM700 laser scanning confocal microscope (Zeiss). The excitation wavelength was 488 nm, and the emission was collected between 610 and 640 nm. The area of starch granules from at least 100 guard cells from 10 to 15 leaves of eight different plants was measured using ImageJ software. Three independent biological repeats were performed. Statistically significant differences were determined by two-way analysis of variance (ANOVA) followed by a post hoc Tukey test, considering P < 0.05 as significant (Supplemental Data Set 2).
Stomatal Bioassay
For determining stomatal aperture in intact leaves, rosette leaves of 4- to 5-week-old plants or cotyledons of 11-d-old seedlings from Col-0 and the indicated mutant plants were cut from plants at the EoN, or at 1, 3, 6, 9, and 12 h after light illumination, and quickly fixed using a Tape-Arabidopsis Sandwich method (Ibata et al., 2013; Lawrence et al., 2018). The upper epidermal surface was stabilized by affixing a strip of Time tape (Medi Pore Surgical tape, Ultraporous Hypollergenic), while the lower epidermal surface was affixed to a strip of Magic tape (Scotch tape, Chenguang). The Time tape was then carefully pulled away, leaving the lower epidermis sticking onto the Magic tape, and then stomata were imaged immediately by microscopy examination (Ibata et al., 2013; Lawrence et al., 2018).
To determine the stomatal aperture in detached leaves, the rosette leaves of 4- to 5-week-old plants were preincubated in assay buffer (10 mM KCl, 7.5 mM iminodiacetic acid, and 10 mM MES) in the dark 2.5 h before starting illumination with blue light (25 µmol·m−2·s−1) and red light (50 µmol·m−2·s−1) for 2 h. To analyze the effects of BR and H2O2 on stomatal movement, the rosette leaves of 4-week-old Col-0 or different mutants were transferred to half strength MS liquid medium with or without different concentrations of DPI or 1 mM KI for overnight incubation in the dark, and then different concentrations of H2O2 or 100 nM BL were added to the medium at the EoN and illuminated with white light for 3 h. The stomatal apertures from 100 cells of 15 leaves of eight different plants were measured using ImageJ software. Three independent biological repeats were performed. Error bars indicate sd. Different letters above the bars indicate statistically significant differences between the samples (two-way ANOVA, followed by a post hoc Tukey test, P < 0.05).
Yeast Two-Hybrid Screening
The full-length cDNA of BZR1 was cloned into the pBD-GAL4 vector and transformed into yeast strain AH109. The resulting yeast cells were then transformed with plasmid DNAs derived from an Arabidopsis yeast two-hybrid cDNA library CD4-22 ordered from Arabidopsis Biological Resource Center. More than 10 million yeast transformants were screened for growth on medium lacking His but containing 50 mM 3-aminotriazol. Recovered clones were then assayed for LacZ activity using a filter-lift assay. The HIS3+LacZ+ yeast cells were cultured, and the prey plasmids were isolated and then transformed into Escherichia coli for sequencing.
In Vitro Pull-Down Assays
GBF2 fused to GST was purified from bacteria that contains the GST-GBF2 construct using glutathione beads (GE Healthcare). BZR1 fused to MBP was purified using amylose resin (New England Biolabs). Glutathione beads containing 1 µg of GST-GBF2 were incubated with 1 µg of MBP or MBP-BZR1 as indicated in the pull-down buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 1 mM EDTA) with or without 1 mM H2O2 at 4°C for 1 h, and the beads were washed 10 times with wash buffer (20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 0.5% Triton X-100, and 1 mM EDTA). The proteins were eluted from beads by boiling in 50 μL of 2× SDS sample buffer and separated on a 8% SDS-PAGE gel. Gel blots were analyzed using anti-MBP (catalog no. E8038L, New England Biolabs; 1:5000 dilution).
CoIP Assays
Plants expressing BZR1-Myc from the native BZR1 promoter and GBF2-YFP from the native GBF2 promoter or plants expressing only GBF2-YFP were grown in half strength MS medium containing 1% Suc for 12 d and then treated with or without 100 μM H2O2 for 1 h. Plant materials were harvested and ground in liquid nitrogen and then extracted in lysis buffer containing 20 mM HEPES-KOH, pH 7.5, 40 mM KCl, 1 mM EDTA, 0.5% Triton X-100, and 1× protease inhibitor (Sigma-Aldrich). After centrifugation with 12,000g for 10 min at 4°C, the supernatant was incubated with Myc-Trap agarose beads (Chromotek) at 4°C for 1 h, and the beads were washed four times using wash buffer (20 mM HEPES-KOH, pH 7.5, 40 mM KCl, 1 mM EDTA, 300 mM NaCl, and 1% Triton X-100). The proteins were eluted from the beads by boiling with 2× SDS sample buffer, analyzed by SDS-PAGE, and immunoblotted with anti-YFP (1:5000 dilution) and anti-Myc (catalog no. M4439, Sigma-Aldrich; 1:5000 dilution) antibodies.
Ratiometric Bimolecular Fluorescence Complementation Assays
Full-length BZR1 or GBF2 genes were amplified by PCR and cloned into the pDONR221-P1P4 or pDONR221-P3P2 vector, respectively, using the BP recombination reaction (Invitrogen). The two-in-one LR reaction was performed with destination vector pBiFCt-2in1-NN and different pDONR221 vectors. Agrobacterial suspensions containing Pro35S:GBF2-nYFP-Pro35S:RFP-Pro35S:BZR1-cYFP constructs were injected into tobacco (Nicotiana tabacum) leaves. The transfected plants were kept in the greenhouse for at least 36 h at 22°C, and fluorescent signals were visualized using an LSM700 laser scanning confocal microscope (Zeiss). The signal intensities of YFP and red fluorescent protein (RFP) were determined by ImageJ software from at least 100 confocal images. Statistically significant differences were determined by one-way ANOVA, considering P < 0.05 as significant.
RNA Extraction from Epidermal Peels and Reverse Transcriptase qPCR Analysis
Guard cell–enriched fragments were isolated for RNA extraction following the procedure described previously (Bauer et al., 2013; Jalakas et al., 2017), with some modifications. Fully developed leaves from the 4- to 5-week-old wild-type plants or the indicated mutants were harvested after irradiating with white light (100 µmol·m−2·s−1) for 3 h. The central vein was removed with a razor blade and then blended in cold water with a handful of crushed ice for 0.5 to 1 min. The homogenate was passed through a 200-μm nylon mesh. The mesh was washed with cold water and the epidermal peels were transferred to a blender cup. The washes were repeated three times. Viability of guard cells was confirmed by microscopy (DP73 digital microscope camera, Olympus) analysis. The collected material was subsequently frozen in liquid nitrogen and ground in a mortar. Then, 100 mg of fine powder of guard cell–enriched epidermal peels was used to extract RNA with the TRIzol RNA extraction kit (TransGen). The first-strand cDNAs were synthesized using RevertAid reverse transcriptase (Thermo Fisher Scientific) and used as RT-PCR templates. Quantitative PCR analyses were performed on a CFX connect real-time PCR detection system (Bio-Rad) using a SYBR Green reagent (Roche) with gene-specific primers (Supplemental Data Set 1).
ChIP
Immunoprecipitation was performed following a previously described protocol (Tian et al., 2018), using 12-d-old seedlings of Pro35S:BZR1-YFP, ProGBF2:GBF2-YFP, and Pro35S:YFP transgenic plants. An affinity-purified anti-GFP polyclonal antibody was used for immunoprecipitation. The ChIP products were analyzed by qPCR, and the fold enrichment was calculated as the ratio between Pro35S:BZR1-YFP, ProGBF2:GBF2-YFP, and Pro35S:YFP and then normalized by the PP2A (At1G13320) gene, which was used as an internal control. The ChIP experiments were performed with three biological replicates from which the means and sds were calculated.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: GBF2 (At4G01120), BAM1 (At3G23920), AMY3 (At1G69830), BZR1 (At1G75080), BES1 (At1G19350), BRI1 (At4G39400), BIN2 (At1G06390), DET2 (At2G38050), ROT3 (At4G36380), CAT2 (At4G35090), RBOHD (At5G47910), RBOHF (At1G64060).
Supplemental Data
Supplemental Figure 1. Comparison the starch contents in guard cells and leaves of wild type and indicated mutants.
Supplemental Figure 2. BR and BZR1 promote stomatal opening in detached leaves.
Supplemental Figure 3. BZR1 and BES1 promote guard cell starch degradation and stomatal opening.
Supplemental Figure 4. Viability assay of guard cells in the detached leaves of wild-type plants.
Supplemental Figure 5. H2DCFDA staining for H2O2 in the guard cells of Col-0 treated with or without BL and H2O2.
Supplemental Figure 6. BR is required for H2O2-induced stomatal opening.
Supplemental Figure 7. Starch content in total leaves of wild type and ROS-related mutants.
Supplemental Figure 8. The Expression Patterns of BZR1 and GBF2 in plants.
Supplemental Figure 9. GBF2 is required for BR- and H2O2-mediated stomatal opening.
Supplemental Figure 10. GBF2 is not involved in BR-induced cell elongation.
Supplemental Figure 11. BR and H2O2 promote the expression of BAM1.
Supplemental Figure 12. BZR1 and GBF2 promote the expression of BAM1.
Supplemental Figure 13. BAM1 and AMY3 are not Required for BR-induced cell elongation.
Supplemental Figure 14. The bam1 mutants displayed the normal stomatal closure in response to BR, H2O2 and ABA.
Supplemental Figure 15. The OST1-CDL1 module is not involved in BR- and H2O2-induced stomatal opening.
Supplemental Data Set 1. Oligonucleotide sequences used in this study.
Supplemental Data Set 2. ANOVA analysis in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Shuhua Yang and Tae-wuk Kim for providing ost1-3, OST1-Ox, and cdl1 seeds. We thank Haiyan Yu and Xiaomin Zhao from the Analysis and Testing Center of State Key Laboratory of Microbial Technology, Shandong University, for assistance with the laser scanning confocal microscopy. This work was supported by grants from the Natural Science Foundation of Shandong Province (grants ZR2018ZC0334 and JQ201708), the Science and Technology Department of Shandong Province (grant 2019LZGC015), and the National Natural Science Foundation of China (grants 31600199, 31670284, 31970306, and 31870262).
AUTHOR CONTRIBUTIONS
J.-G.L., M.F., and M.-Y.B. designed experiments; J.-G.L., M.F., W.H., Y.T., L.-G.C., and Y.S. conducted experiments; and M.F. and M.-Y.B. wrote the article.
References
- Bauer H., et al. (2013). The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr. Biol. 23: 53–57. [DOI] [PubMed] [Google Scholar]
- Chaiwanon J., Wang W., Zhu J.Y., Oh E., Wang Z.Y.(2016). Information integration and communication in plant growth regulation. Cell 164: 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Nolan T.M., Ye H., Zhang M., Tong H., Xin P., Chu J., Chu C., Li Z., Yin Y.(2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29: 1425–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Clouse S.D., Sasse J.M.(1998). BRASSINOSTEROIDS: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 427–451. [DOI] [PubMed] [Google Scholar]
- Daloso D.M., Medeiros D.B., Dos Anjos L., Yoshida T., Araújo W.L., Fernie A.R.(2017). Metabolism within the specialized guard cells of plants. New Phytol. 216: 1018–1033. [DOI] [PubMed] [Google Scholar]
- De Wit M., Galvão V.C., Fankhauser C.(2016). Light-mediated hormonal regulation of plant growth and development. Annu. Rev. Plant Biol. 67: 513–537. [DOI] [PubMed] [Google Scholar]
- Dong H., Liu J., He G., Liu P., Sun J.(2019). Photoexcited phytochrome B interacts with BZR1 to repress brassinosteroid signaling in Arabidopsis. J. Integr. Plant Biol.. [DOI] [PubMed] [Google Scholar]
- Flütsch S., Distefano L., Santelia D.(2018). Quantification of starch in guard cells of Arabidopsis thaliana. Bio Protoc. 8: e2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Santamarina S., Boronat S., Hidalgo E.(2014). Reversible cysteine oxidation in hydrogen peroxide sensing and signal transduction. Biochemistry 53: 2560–2580. [DOI] [PubMed] [Google Scholar]
- Ha Y., Shang Y., Nam K.H.(2016). Brassinosteroids modulate ABA-induced stomatal closure in Arabidopsis. J. Exp. Bot. 67: 6297–6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha Y.M., Shang Y., Yang D., Nam K.H.(2018). Brassinosteroid reduces ABA accumulation leading to the inhibition of ABA-induced stomatal closure. Biochem. Biophys. Res. Commun. 504: 143–148. [DOI] [PubMed] [Google Scholar]
- Han J.P., Köster P., Drerup M.M., Scholz M., Li S., Edel K.H., Hashimoto K., Kuchitsu K., Hippler M., Kudla J.(2019). Fine-tuning of RBOHF activity is achieved by differential phosphorylation and Ca2+ binding. New Phytol. 221: 1935–1949. [DOI] [PubMed] [Google Scholar]
- Hao L.H., Wang W.X., Chen C., Wang Y.F., Liu T., Li X., Shang Z.L.(2012). Extracellular ATP promotes stomatal opening of Arabidopsis thaliana through heterotrimeric G protein α subunit and reactive oxygen species. Mol. Plant 5: 852–864. [DOI] [PubMed] [Google Scholar]
- Haubrick L.L., Torsethaugen G., Assmann S.A.(2006). Effect of brassinolide, alone and in concert with abscisic acid, on control of stomatal aperture and potassium currents of Vicia faba guard cell protoplasts. Physiol. Plant. 128: 134–143. [Google Scholar]
- He G., Liu J., Dong H., Sun J.(2019). The blue-light receptor CRY1 interacts with BZR1 and BIN2 to modulate the phosphorylation and nuclear function of BZR1 in repressing BR signaling in Arabidopsis. Mol. Plant 12: 689–703. [DOI] [PubMed] [Google Scholar]
- Hecker A., Wallmeroth N., Peter S., Blatt M.R., Harter K., Grefen C.(2015). Binary 2in1 vectors improve in planta (co)localization and dynamic protein interaction studies. Plant Physiol. 168: 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrer D., Flütsch S., Pazmino D., Matthews J.S., Thalmann M., Nigro A., Leonhardt N., Lawson T., Santelia D.(2016). Blue light induces a distinct starch degradation pathway in guard cells for stomatal opening. Curr. Biol. 26: 362–370. [DOI] [PubMed] [Google Scholar]
- Ibata H., Nagatani A., Mochizuki N.(2013). Perforated-tape epidermal detachment (PED): A simple and rapid method for isolating epidermal peels from specific areas of Arabidopsis leaves. Plant Biotechnol. 30: 497–502. [Google Scholar]
- Inoue S.I., Iwashita N., Takahashi Y., Gotoh E., Okuma E., Hayashi M., Tabata R., Takemiya A., Murata Y., Doi M., Kinoshita T., Shimazaki K.I.(2017). Brassinosteroid involvement in Arabidopsis thaliana stomatal opening. Plant Cell Physiol. 58: 1048–1058. [DOI] [PubMed] [Google Scholar]
- Jalakas P., Yarmolinsky D., Kollist H., Brosche M.(2017). Isolation of guard-cell enriched tissue for RNA extraction. Bio Protoc. 7: e2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.P., Cheng F., Zhou Y.H., Xia X.J., Mao W.H., Shi K., Chen Z.X., Yu J.Q.(2012). Brassinosteroid-induced CO(2) assimilation is associated with increased stability of redox-sensitive photosynthetic enzymes in the chloroplasts in cucumber plants. Biochem. Biophys. Res. Commun. 426: 390–394. [DOI] [PubMed] [Google Scholar]
- Kim T.W., Youn J.H., Park T.K., Kim E.J., Park C.H., Wang Z.Y., Kim S.K., Kim T.W.(2018). OST1 activation by the brassinosteroid-regulated kinase CDG1-LIKE1 in stomatal closure. Plant Cell 30: 1848–1863. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lai A.G., Doherty C.J., Mueller-Roeber B., Kay S.A., Schippers J.H., Dijkwel P.P.(2012). CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. USA 109: 17129–17134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence S. II, Pang Q., Kong W., Chen S.(2018). Stomata tape-peel: An improved method for guard cell sample preparation. J. Vis. Exp. 137: e57422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J.(1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938. [DOI] [PubMed] [Google Scholar]
- Li J., Nagpal P., Vitart V., McMorris T.C., Chory J.(1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401. [DOI] [PubMed] [Google Scholar]
- Li X.J., Guo X., Zhou Y.H., Shi K., Zhou J., Yu J.Q., Xia X.J.(2016). Overexpression of a brassinosteroid biosynthetic gene Dwarf enhances photosynthetic capacity through activation of Calvin cycle enzymes in tomato. BMC Plant Biol. 16: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T., Mei S., Shi C., Yang Y., Peng Y., Ma L., Wang F., Li X., Huang X., Yin Y., Liu H.(2018). UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev. Cell 44: 512–523. [DOI] [PubMed] [Google Scholar]
- Mittler R.(2017). ROS are good. Trends Plant Sci. 22: 11–19. [DOI] [PubMed] [Google Scholar]
- Murata Y., Mori I.C., Munemasa S.(2015). Diverse stomatal signaling and the signal integration mechanism. Annu. Rev. Plant Biol. 66: 369–392. [DOI] [PubMed] [Google Scholar]
- Nolan T.M., Brennan B., Yang M., Chen J., Zhang M., Li Z., Wang X., Bassham D.C., Walley J., Yin Y.(2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Song C.P., Wang B., Zhou J., Kangasjärvi J., Zhu J.K., Gong Z.(2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 60: 805–826. [DOI] [PubMed] [Google Scholar]
- Rajasekaran L.R., Blake T.J.(1999). New plant growth regulators protect photosynthesis and enhance growth under drought of Jack Pine seedlings. J. Plant Growth Regul. 18: 175–181. [DOI] [PubMed] [Google Scholar]
- Santelia D., Lawson T.(2016). Rethinking guard cell metabolism. Plant Physiol. 172: 1371–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze K., Harter K., Chaban C.(2008). Post-translational regulation of plant bZIP factors. Trends Plant Sci. 13: 247–255. [DOI] [PubMed] [Google Scholar]
- Shi C., Qi C., Ren H., Huang A., Hei S., She X.(2015). Ethylene mediates brassinosteroid-induced stomatal closure via Galpha protein-activated hydrogen peroxide and nitric oxide production in Arabidopsis. Plant J. 82: 280–301. [DOI] [PubMed] [Google Scholar]
- Shimazaki K., Doi M., Assmann S.M., Kinoshita T.(2007). Light regulation of stomatal movement. Annu. Rev. Plant Biol. 58: 219–247. [DOI] [PubMed] [Google Scholar]
- Singh R., Parihar P., Singh S., Mishra R.K., Singh V.P., Prasad S.M.(2017). Reactive oxygen species signaling and stomatal movement: Current updates and future perspectives. Redox Biol. 11: 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryhan K., Gurrieri L., Sparla F., Trost P., Blennow A.(2018). Redox regulation of starch metabolism. Front. Plant Sci. 9: 1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparla F., Costa A., Lo Schiavo F., Pupillo P., Trost P.(2006). Redox regulation of a novel plastid-targeted beta-amylase of Arabidopsis. Plant Physiol. 141: 840–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S., Zeeman S.C.(2012). Starch metabolism in Arabidopsis. Arabidopsis Book 10: e0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann M., Pazmino D., Seung D., Horrer D., Nigro A., Meier T., Kölling K., Pfeifhofer H.W., Zeeman S.C., Santelia D.(2016). Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell 28: 1860–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann M., Santelia D.(2017). Starch as a determinant of plant fitness under abiotic stress. New Phytol. 214: 943–951. [DOI] [PubMed] [Google Scholar]
- Tian Y., et al. (2018). Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat. Commun. 9: 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge T., Tsukaya H., Uchimiya H.(1996). Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122: 1589–1600. [DOI] [PubMed] [Google Scholar]
- Valerio C., Costa A., Marri L., Issakidis-Bourguet E., Pupillo P., Trost P., Sparla F.(2011). Thioredoxin-regulated beta-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. J. Exp. Bot. 62: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Li L., Xu P., Lian H., Wang W., Xu F., Mao Z., Zhang T., Yang H.(2018a). CRY1 interacts directly with HBI1 to regulate its transcriptional activity and photomorphogenesis in Arabidopsis. J. Exp. Bot. 69: 3867–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., et al. (2018b). Photoexcited CRYPTOCHROME1 interacts with dephosphorylated BES1 to regulate brassinosteroid signaling and photomorphogenesis in Arabidopsis. Plant Cell 30: 1989–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Bai M.Y., Oh E., Zhu J.Y.(2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46: 701–724. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J.(2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J.(2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.Y., et al. (2008). Brassinosteroids regulate grain filling in rice. Plant Cell 20: 2130–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Wang W., Xu P., Pan J., Zhang T., Li Y., Li G., Yang H., Lian H.(2019). phyB interacts with BES1 to regulate brassinosteroid signaling in Arabidopsis. Plant Cell Physiol. 60: 353–366. [DOI] [PubMed] [Google Scholar]
- Xia X.J., Gao C.J., Song L.X., Zhou Y.H., Shi K., Yu J.Q.(2014). Role of H2O2 dynamics in brassinosteroid-induced stomatal closure and opening in Solanum lycopersicum. Plant Cell Environ. 37: 2036–2050. [DOI] [PubMed] [Google Scholar]
- Xia X.J., Zhou Y.H., Shi K., Zhou J., Foyer C.H., Yu J.Q.(2015). Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 66: 2839–2856. [DOI] [PubMed] [Google Scholar]
- Xu W., Huang J., Li B., Li J., Wang Y.(2008). Is kinase activity essential for biological functions of BRI1? Cell Res. 18: 472–478. [DOI] [PubMed] [Google Scholar]
- Yan L., Wei S., Wu Y., Hu R., Li H., Yang W., Xie Q.(2015). High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8: 1820–1823. [DOI] [PubMed] [Google Scholar]
- Yan Z., Zhao J., Peng P., Chihara R.K., Li J.(2009). BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 150: 710–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Li L., Yin Y.(2011). Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J. Integr. Plant Biol. 53: 455–468. [DOI] [PubMed] [Google Scholar]
- Ye H., et al. (2017). RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 8: 14573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J.(2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Zanella M., Borghi G.L., Pirone C., Thalmann M., Pazmino D., Costa A., Santelia D., Trost P., Sparla F.(2016). β-Amylase 1 (BAM1) degrades transitory starch to sustain proline biosynthesis during drought stress. J. Exp. Bot. 67: 1819–1826. [DOI] [PubMed] [Google Scholar]
- Zeeman S.C., Kossmann J., Smith A.M.(2010). Starch: Its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61: 209–234. [DOI] [PubMed] [Google Scholar]