Analysis of the gene expression atlas of Selaginella moellendorffii reveals transcriptional programs that arose in vascular plants over the course of one billion of years of evolution.
Abstract
Selaginella moellendorffii is a representative of the lycophyte lineage that is studied to understand the evolution of land plant traits such as the vasculature, leaves, stems, roots, and secondary metabolism. However, only a few studies have investigated the expression and transcriptional coordination of Selaginella genes, precluding us from understanding the evolution of the transcriptional programs behind these traits. We present a gene expression atlas comprising all major organs, tissue types, and the diurnal gene expression profiles for S. moellendorffii. We show that the transcriptional gene module responsible for the biosynthesis of lignocellulose evolved in the ancestor of vascular plants and pinpoint the duplication and subfunctionalization events that generated multiple gene modules involved in the biosynthesis of various cell wall types. We demonstrate how secondary metabolism is transcriptionally coordinated and integrated with other cellular pathways. Finally, we identify root-specific genes and show that the evolution of roots did not coincide with an increased appearance of gene families, suggesting that the development of new organs does not coincide with increased fixation of new gene functions. Our updated database at conekt.plant.tools represents a valuable resource for studying the evolution of genes, gene families, transcriptomes, and functional gene modules in the Archaeplastida kingdom.
INTRODUCTION
The lycophyte Selaginella moellendorffii is an important model organism for plant evolutionary studies and comparative genomics, as it represents lycophytes, which, together with the euphyllophytes, are one of the two existing lineages that diverged from the first vascular plants. This plant is characterized by adaxial sporangia, a root system (roots and rhizophores), a vasculature, and leaves with a single vein, called microphylls. S. moellendorffii has a genome size of only ∼100 Mbp, which is one of the smallest reported plant genomes (Banks et al., 2011). Lycophytes appeared ∼400 million years (Myr) ago during the Silurian period and were particularly abundant during the Devonian to mid-Carboniferous period, peaking around 310 Myr ago in Euramerican coal swamps (Phillips and DiMichele, 1992). During the Carboniferous period, many lycophytes were populating Pennsylvanian forests, and their remains now exist as coal. As 70% of the biomass responsible for the Bashkirian and Moscovian coal formations in Euramerica came from lycophytes (Manfroi et al., 2012), they represent an important source of fossil fuels. Despite their decreasing abundance at the end of the Carboniferous period, the impact of lycophytes on the climate was significant, as their role as carbon sinks most likely contributed to the dramatic decline in atmospheric CO2 (Crowley and Berner, 2001).
Lycophytes produce spores for reproduction that are dispersed by wind and water. One means to increase spore dispersal and outcompete neighbors in light capture and thus facilitate the colonization of land is to increase the height of the sporophyte. This required the evolution of specialized vasculature (xylem and phloem) for the transport of nutrients, water, and various signaling molecules and the reinforcement of cell walls by lignification (Kenrick and Strullu-Derrien, 2014). This lignification, in turn, was associated with the evolution of the phenylpropanoid metabolic pathway, which synthesizes and polymerizes the heterogeneous aromatic lignin in the xylem cell walls (Li and Chapple, 2010; Tohge et al., 2013)
The evolution of roots provided water and structural support to the larger sporophytes. Despite their morphological and evolutionary diversity, all roots have the capacity to acquire and transport water and nutrients, grow in a downward direction, and form branch roots, and they all possess a root cap (set of terminal protective cells), a root apical meristem (a self-sustaining stem cell population), and a radial organization of cell types (from the outermost epidermal tissue to the innermost vascular tissue). Root morphology in extant plants and fossil records currently suggests that roots evolved several times independently during vascular plant evolution (Kenrick and Crane, 1997; Raven and Edwards, 2001; Kenrick and Strullu-Derrien, 2014; Hetherington and Dolan, 2019)
In addition to the presence of vasculature and roots, land plants collectively produce more than 200,000 secondary metabolites via complex biosynthetic pathways (Hartmann, 2007). These secondary metabolites are not directly required for growth, reproduction, and development but serve to provide defense against microorganisms and UV protection and to attract pollinators. Many secondary metabolites, including those that impart colors and flavors to plants, are lineage specific and play specialized roles in unique ecological niches (Weng et al., 2012). Selaginella contains numerous secondary metabolites, such as phenolics, alkaloids, flavonoids, lignans, selaginellins, and terpenoids (Weng and Noel, 2013). Many species of Selaginella have been used as traditional medicines for hundreds of years. In Colombia, Selaginella articulata is used to treat snakebites and neutralize Bothrops atrox venom (Otero et al., 2000). In India, Selaginella bryopteris is one of the plants proposed to be Sanjeevani—one that infuses life—due to its medicinal properties (Sah et al., 2005). While the medicinal uses of Selaginella are anecdotal, the efficacy of Selaginella metabolites has been investigated using modern approaches (Chen et al., 2005). For example, uncinoside A and uncinoside B biflavonoids have potent antiviral activities against respiratory syncytial virus (Ma et al., 2003), while biflavonoids from Selaginella tamariscina inhibit the production of nitric oxide and prostaglandins (Pokharel et al., 2006; Woo et al., 2006), which are important for the pathogenesis of some cancers (Lala and Chakraborty, 2001). In addition, biflavone ginkgetin from S. moellendorffii selectively induces apoptosis in ovarian and cervical cancer cells (Su et al., 2000). These findings show that Selaginella metabolites and their biosynthetic pathways merit more investigation.
To unravel the evolution of various organs and metabolic pathways, it is necessary to understand the functions of the underlying gene products and to compare these functions across species that possess or lack these organs and pathways (Hansen et al., 2014; Rhee and Mutwil, 2014; Ferrari et al., 2018). Recent studies have identified tissue-specific genes in roots, stems, leaves, shoot meristems, and root meristems in S. moellendorffii (Frank et al., 2015; Zhu et al., 2017; Mello et al., 2019). Furthermore, comparative analyses with early divergent model species have been performed to understand the molecular basis of the pluripotent cells of the meristems (Frank et al., 2015). However, the experimental functional characterization of even one gene can take years, and in silico prediction is currently the only large-scale approach to exploring gene function in S. moellendorffii. Classical comparative genomic approaches based solely on gene sequences are useful but have shortcomings, as they cannot readily reveal which genes work together in a pathway, i.e. form a functional gene module (Hartwell et al., 1999). Consequently, to study the evolution of new traits, we need to integrate the classical genomic approaches with predicted functional gene modules, which can be identified by expression and coexpression analysis (Ruprecht et al., 2011, 2016, 2017; Lampugnani et al., 2019). Coexpression analysis is based on the guilt-by-association principle, which states that genes with similar expression patterns across organs, developmental stages, and biotic as well as abiotic stresses tend to be involved in the same or closely related biological processes (Usadel et al., 2009; Lee et al., 2010; Hansen et al., 2014; Rhee and Mutwil, 2014; Proost and Mutwil, 2016; Ruprecht et al., 2017). Coexpression analyses have been applied to successfully identify genes involved in cyclic electron flow (Takabayashi et al., 2009), cell division (Takahashi et al., 2008), drought sensitivity, and lateral root development (Lee et al., 2010), plant viability (Mutwil et al., 2010), seed germination (Bassel et al., 2011), shade avoidance (Jiménez-Gómez et al., 2010), and many other processes (Stuart et al., 2003; Yu et al., 2003; Persson et al., 2005; Alejandro et al., 2012; Itkin et al., 2013; Radivojac et al., 2013; Rhee and Mutwil, 2014; Proost and Mutwil, 2017; Sibout et al., 2017; Zhao et al., 2016).
In this study, to enable such comparative genomic, transcriptomic, and coexpression approaches for S. moellendorffii, we generated a comprehensive RNA sequencing-based (RNA-seq) expression atlas, which captures the transcriptomes of the plant major organs. By uploading the expression data to the CoNekT-Plants database, conekt.plant.tools, we provide the plant community with advanced comparative transcriptomic analyses for algae and land plants. We use the database to reveal the predicted functional gene modules involved in the biosynthesis of lignified cell walls and various secondary metabolites. We also provide examples of comparative analysis of the different cell wall types and root-specific transcriptomes. Thus, the updated CoNekT-Plants database constitutes a valuable resource for studying transcriptional programs over 1 billion of years of evolution in the Archaeplastida kingdom.
RESULTS
Expression Atlas Databases for S. moellendorffii
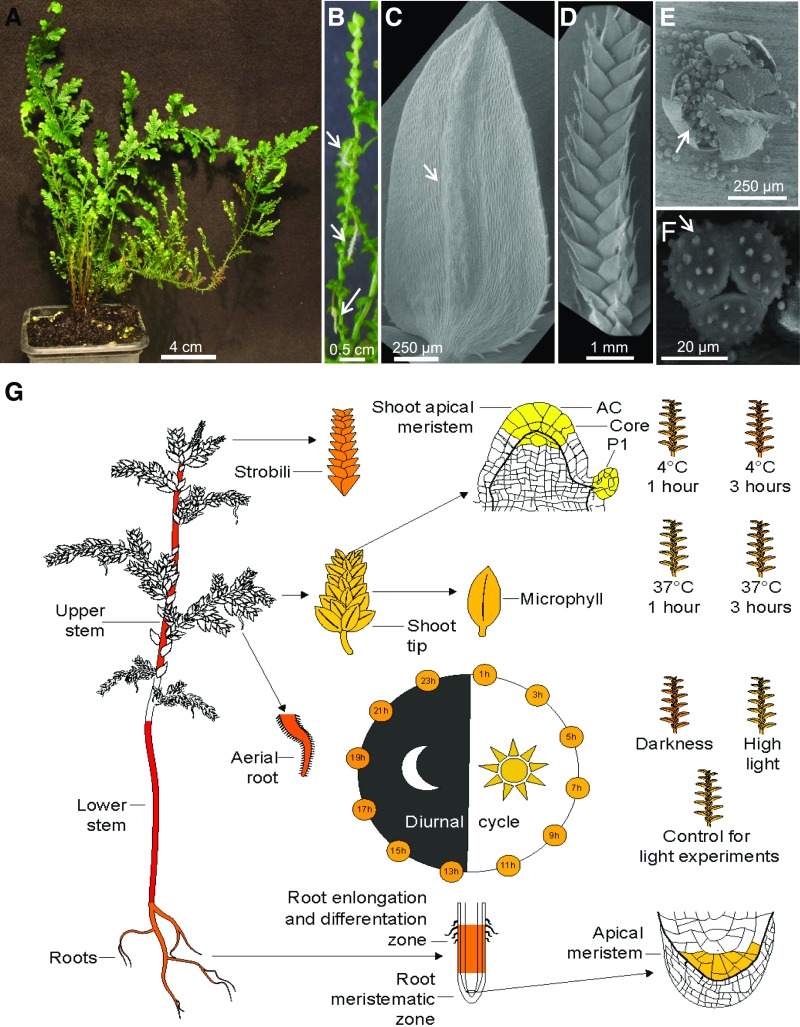
The typical S. moellendorffii plants used in this study (Figure 1A) contained aerial root-bearing rhizophores (Figure 1B), which produce roots with typical characteristics such as root hairs and root caps (Mello et al., 2019). The major photosynthetic organs of S. moellendorffii are single-veined leaves called microphylls (Figure 1C), which are covered with stomata (Supplemental Figure 1A, red arrows) and contain serrated epidermal cells (Supplemental Figure 1A, green arrow). The reproductive organs are represented by sporangia-bearing structures known as strobili (Figure 1D), which are found at the tips of shoots and contain male micro- and female mega-sporangia. Figures 1E and 1F show microsporangia with microspore tetrads; Supplemental Figure 1B shows strobili without sporophylls (Banks et al., 2011). The microphyll structures are found along the stem, shoots (Supplemental Figure 1C), and strobili (Supplemental Figures 1D and 1E).
Figure 1.
Tissues and Organs Used to Generate the S. moellendorffii Expression Atlas.
(A) A typical S. moellendorffii plant used in this study.
(B) Stem with aerial roots (indicated by arrows).
(C) to (G) SEM images of Selaginella. (C) Microphyll, with the vein indicated by an arrow. (D) Strobilus. (E) Crushed microsporangium with released microspore tetrads. (F) Microspore tetrad, with three microspores shown. (G) Expression profile of the C3′H gene from S. moellendorffii (PACid_15423174) in Selaginella eFP Browser. Expression values in the sampled organs, stress, and diurnal conditions are indicated by a color gradient. Yellow indicates no detectable expression, while red signifies the highest expression.
We assembled a gene expression atlas for S. moellendorffii by combining newly generated and publicly available RNA-seq samples from major organs, tissues, stress conditions, and a diurnal cycle study (Table 1; Supplemental Data Set 1 and 2). The sample clustering dendrogram showed the expected clustering of replicates, similar tissues, and organs (Supplemental Figure 2). We provide two options to visualize gene expression data: (1) an S. moellendorffii eFP Browser (http://bar.utoronto.ca/efp_selaginella/cgi-bin/efpWeb.cgi; Winter et al., 2007), providing a graphical anatogram that color-codes the different organs and conditions based on the average expression level of a gene of interest, and (2) the CoNekT-Plants database (www.conekt.plant.tools; Proost and Mutwil, 2018), where the expression level of each sample is shown based on condition and tissue.
Table 1. Samples Used to Generate the Expression Atlas.
| Sample Description | Number of Samples | Source |
|---|---|---|
| Whole plant | 2 | E-MTAB-4374 |
| Roots | 2 | This study |
| Aerial roots | 2 | This study |
| Roots, elongation and differentiating zone | 3 | E-GEOD-64665 |
| Roots, meristematic zone | 3 | E-GEOD-64665 |
| Strobili | 2 | This study |
| Shoot tips | 2 | This study |
| Stem, top 5 cm | 2 | This study |
| Stem, bottom 5 cm | 2 | This study |
| Microphylls | 2 | This study and SRX2779511 |
| Shoot apical meristem, AC | 2 | E-MTAB-4374 |
| Shoot apical meristem, Core | 2 | E-MTAB-4374 |
| Shoot apical meristem, P1 | 2 | E-MTAB-4374 |
| 4°C, 1 h | 2 | This study |
| 4°C, 3 h | 2 | This study |
| 37°C, 1 h | 2 | This study |
| 37°C, 3 h | 2 | This study |
| Control (of dark and HL) | 1 | This study |
| Dark, 48 h | 2 | This study |
| High light, 200 µE for 12 h | 2 | This study |
| Diurnal expression, aerial tissue sampled every 2 h | 39 | E-MTAB-7188 |
Aerial roots, adventitious roots originating at the tips of rhizophores; strobili, sporangia-bearing structures along the stem.
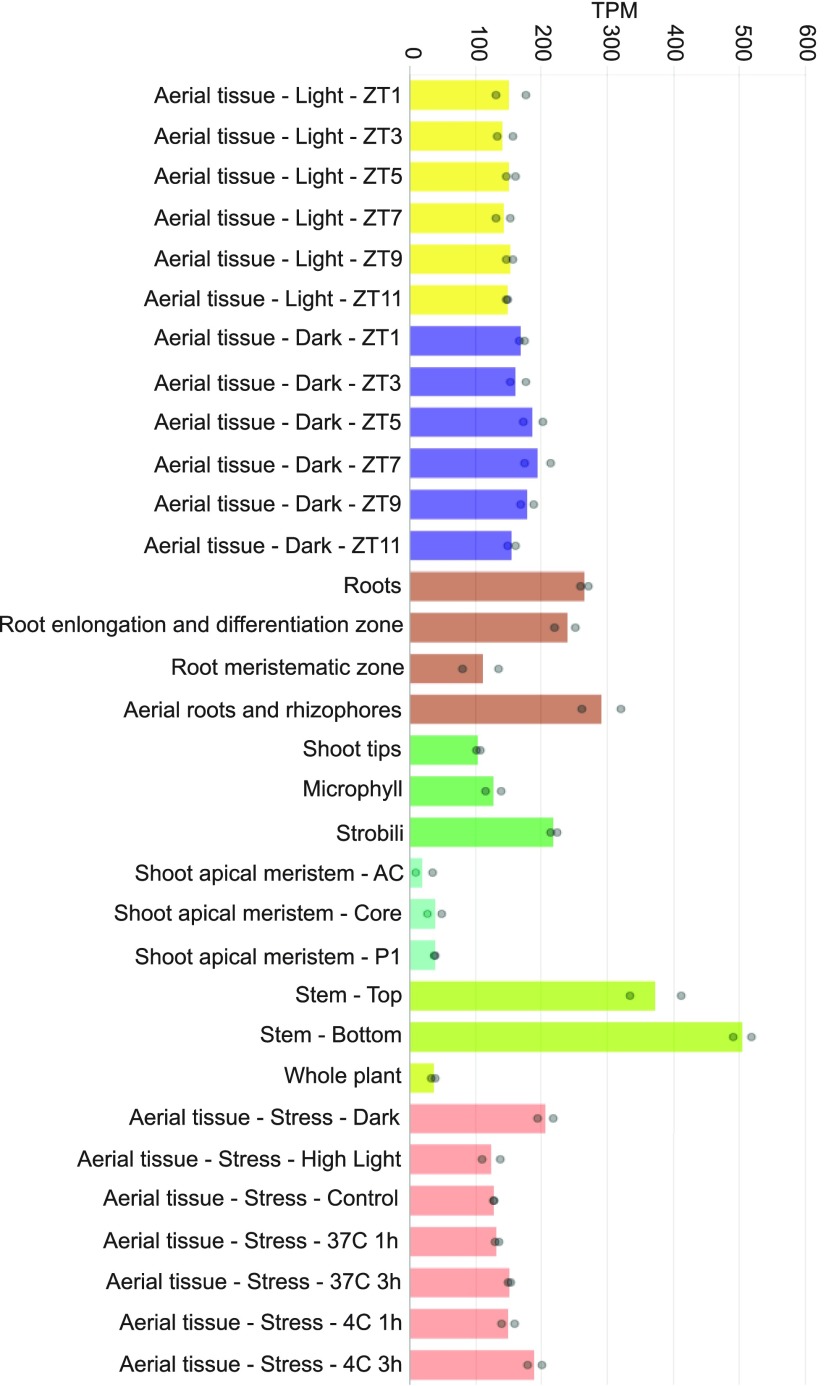
Since S. moellendorffii is a representative vascular plant, as an example, we present the expression profile of a p-coumarate 3-hydroxylase (C3′H) gene (Smo271465; PACid_15423174) involved in the first steps of lignin biosynthesis (Ralph et al., 2006). As expected of a gene involved in vasculature biogenesis, it is highly expressed in the stem and roots, as indicated by the red color in the eFP Browser representation (Figure 1G) and by brown and green bars in CoNekT (Figure 2).
Figure 2.
Expression Profile of the C3′H Gene from S. moellendorffii (PACid_15423174) in CoNekT-Plants.
The samples are shown on the x axis (at the top), while the TPM expression value is indicated on the y axis. The bars represent the average expression values and are color-coded according to the organ or experiment type. The dots indicate the minimum and maximum expression values of the replicates.
Coexpression Network Analysis of Lignin Biosynthesis Reveals the Association of Cellulose and Lignin Biosynthesis
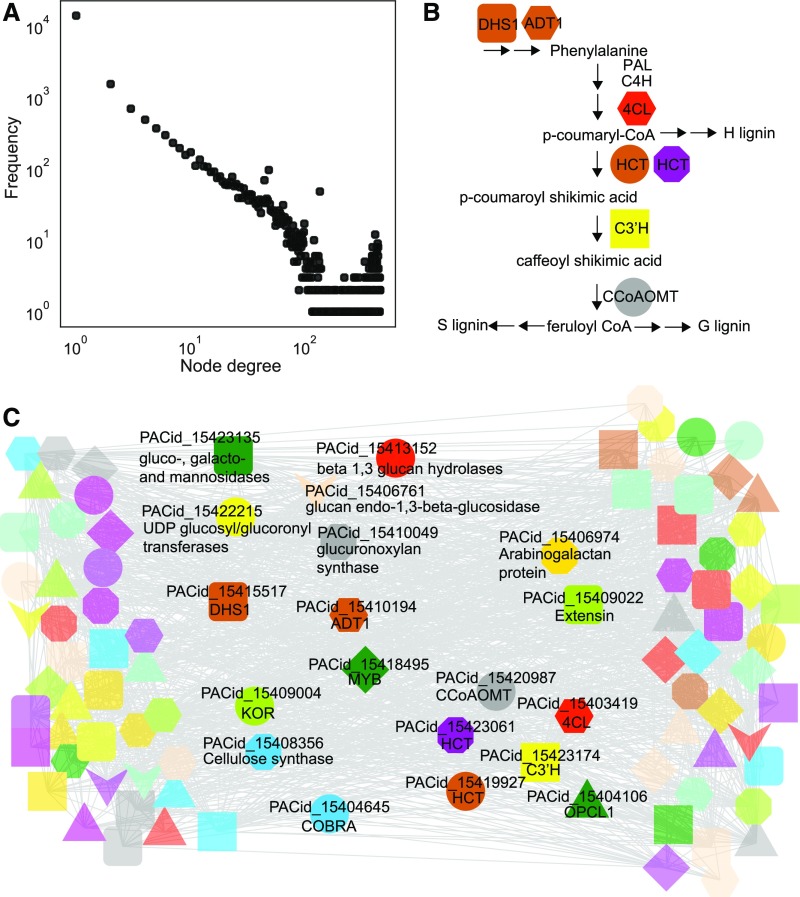
While only a small portion of real-world networks are scale-free (Broido and Clauset, 2019), coexpression networks tend to show scale-free topology (Mutwil et al., 2010). Scale-free networks show a power-law behavior, where only a few genes are connected (correlated) to many genes, while the majority of genes show only a few connections (Barabási and Bonabeau, 2003). Scale-free topology is thought to ensure that the network remains mostly unaltered in case of disruptive mutations and is thus an evolved property that ensures robustness against genetic and environmental perturbations (Barabási and Oltvai, 2004). To demonstrate that our expression atlas can generate biologically meaningful coexpression networks, we investigated whether the data can produce a typical scale-free network. The coexpression network of S. moellendorffii showed a scale-free topology, as plotting the number of connections a gene has (node degree) against the frequency of this association produced a negative slope (Figure 3A), confirming the scale-free topology of our network and suggesting the biological validity of our expression data.
Figure 3.
Coexpression Network of S. moellendorffii.
(A) Power-law plot obtained from S. moellendorffii expression data. The x axis shows the node degree (number of coexpression connections of a gene), while the y axis indicates the frequency of a degree. The two axes are log10-transformed.
(B) Simplified pathway of lignin biosynthesis. Colored nodes indicate orthogroups of the involved enzymes. Enzymes without colored nodes, such as PAL and C4H, are enzymes not found in the C3′H module.
(C) Coexpression network of S. moellendorffii C3′H gene PACid_15423174 (yellow square). Nodes indicate genes, edges connect coexpressed genes. Node colors/shapes indicate which genes belong to the same orthogroup.
Since lignocellulosic cell walls are the main characteristics of the plant vasculature, we decided to investigate the biosynthetic module of lignin in Selaginella. Lignin is produced by the Phe/Tyr pathway through the formation of lignin monomers followed by their polymerization in the cell wall (Liu et al., 2018). The three monolignols that are incorporated into the lignin polymer are guaiacyl (G), syringyl (S), and p-hydroxyphenyl (H; Vanholme et al., 2010), which polymerize into G-lignin, S-lignin, and H-lignin, respectively (Figure 3B). Although the appearance of S-lignin was thought to be specific to angiosperms due to its absence in conifers and ferns, S-lignin is found in Selaginella (Weng et al., 2008), suggesting the independent evolution of S-lignin in lycophytes and angiosperms.
The coexpression network neighborhood of the C3′H gene (Smo271465; PACid_15423174) contained genes encoding enzymes involved in the early stages of lignin biosynthesis (Figure 3C; Weng et al., 2011). The enzymes are 4-coumarate-CoA ligase (red hexagon, Figure 3C), hydroxycinnamoyl-CoA shikimate/quinatehydroxycinnamoyl transferase (represented by two genes in two different orthogroups), and caffeoyl-CoA O-methyltransferase (gray circle, Figure 3C). We also identified genes for two enzymes involved in the biosynthesis of Phe DHS1 (3-DEOXY-D-ARABINO-HEPTULOSONATE 7-PHOSPHATE SYNTHASE1; Vanholme et al., 2010) and ADT (AROGENATE DEHYDRATASE1; Cho et al., 2007), suggesting that lignin and Phe biosynthesis are coupled, likely to coordinate the high demand for this amino acid in lignin biosynthesis. In addition, we identified key genes of cell wall biosynthesis such as cellulose synthase, COBRA, and KORRIGAN (KOR; Mutwil et al., 2008; McFarlane et al., 2014), various cell wall proteins (arabinogalactan protein, extensin), and various polysaccharide-acting enzymes. Furthermore, we identified a member of the MYB transcription factor family that is, among other pathways, known to regulate vasculature formation (Figure 3C, green diamond; Nakano et al., 2015). The sequence annotation of the S. moellendorffii MYB indicates that it has a best hit to Arabidopsis (Arabidopsis thaliana) AtMYB55, which increased lignin content when overexpressed in rice (Oryza sativa; Koshiba et al., 2017). Importantly, similar modules containing genes involved in lignin, Phe, cell wall biosynthesis, and MYB regulators have been identified in Arabidopsis and Brachypodium distachyon (Sibout et al., 2017), suggesting that the conserved module shown in Figure 3C indeed evolved in vascular plants and is thus at least 400 million years old.
Expression and Phylogenetic Analyses Reveal the Duplication and Subfunctionalization of Cell Wall Biosynthesis Modules
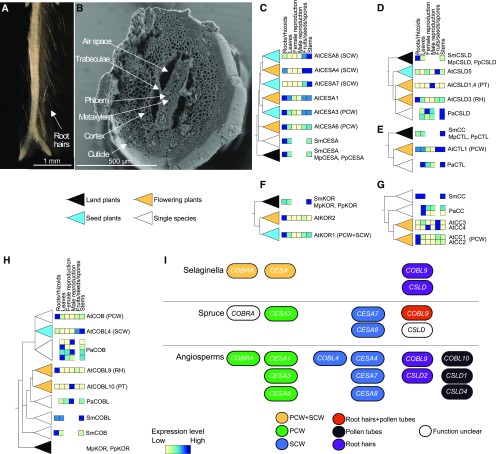
The composition of cell walls evolved to accommodate the different niches of land plants, with variations not only across different species but also among different organs within the same species (Knox, 2008). Arabidopsis contains at least four cell wall types: (1) the ubiquitous primary cell wall (PCW), (2) the secondary cell wall present in the vasculature (SCW), and the tip growing cell walls of (3) root hairs (RHs) and (4) pollen tubes (PTs; Ruprecht et al., 2016). Since S. moellendorffii contains PCW, bifurcating roots with root hairs (Figure 4A; Otreba and Gola, 2011), vasculature with SCW (Figure 4B), and lignified cortex, xylem, and phloem (Weng et al., 2011), it provides a unique reference to study the evolution of these four types of modules. To this end, we analyzed the phylogenetic trees and gene expression of the six major gene families involved in cellulosic cell wall biosynthesis: (1) cellulose synthases (CesA) that have subfunctionalized to play roles in PCW and SCW biosynthesis (Figure 4C; McFarlane et al., 2014), (2) cellulose synthase-like D genes (CslD) involved in tip growth (Figure 4D; Bernal et al., 2008), (3) CHITINASE-LIKE (CTL; Figure 4E), (4) KORRIGAN (KOR; Figure 4F), (5) COMPANION OF CELLULOSE SYNTHASE (CC; Figure 4G), and (6) COBRA (COB). To investigate whether members of these gene families have subfunctionalized to biosynthesize PCW, SCW, PTs, or RHs, we took advantage of the specific expression profiles of genes involved in PCW (typically ubiquitous expression), SCW (typically expressed in stems and roots), PTs (expressed in male reproduction), and RHs (expressed in roots/rhizoids). To perform such a comparison, we used the View comparative expression as heatmap/row normalized function on the page of each orthogroup (e.g., https://conekt.sbs.ntu.edu.sg/family/view/413).
Figure 4.
Phylogenetic and Expression Analysis of Cell Wall Biosynthesis Genes.
(A) S. moellendorffii bifurcating root tip, with RHs.
(B) SEM image of S. moellendorffii stem, with the major tissue types indicated. Phylogenetic trees from CoNekT-Plants for (C) cellulose synthases (CesAs), (D) cellulose synthase-like D (CslDs), (E) chitinase-like (CTL), (F) korrigan (KOR), (G) companion of cellulose synthase (CC), and (H) COBRA. For brevity, only the representative genes from Arabidopsis (genes starting with At), S. moellendorffii (Sm), spruce (Pa), and rice (Os) are shown. The color of the triangles indicates the oldest lineage found in the clade. For example, orange indicates that only flowering plant genes are found in a clade. The colored boxes to the left of the gene identifiers correspond to the expression in major organs, where light color indicates low expression, while dark color shows high gene expression. Genes involved in PCW, SCW, RHs, and PTs are indicated.
(I) Proposed model of the evolution of genes involved in PCW+SCW (orange nodes), PCW (green nodes), SCW (blue nodes), RH+PT (red nodes), RH (purple nodes), PT (black nodes) in Selaginella, spruce, and angiosperms. White CslD and COBRA node indicates spruce genes that could not be assigned to a functional clade due to node support value < 80.
The CesA family contains multiple nonredundant copies in angiosperms, where PCW CesAs are typically ubiquitously expressed, while SCW CesAs show specific expression in roots and stems (Ruprecht et al., 2016). Studies in Arabidopsis have shown that AtCESA1, AtCESA3, and AtCESA6 are needed for PCW formation, while AtCESA4, AtCESA7, and AtCESA8 are essential for SCW formation (McFarlane et al., 2014). Conversely, mutant AtCESA2, AtCESA5, and AtCESA9 phenotypes are mild and only lethal when combined with AtCESA6 knockout, while the function of AtCESA10 is yet unclear (McFarlane et al., 2014).
We observed that S. moellendorffii CesA genes (Figure 4C; Supplemental Figure 3) are found in the same clade as CESAs from the liverwort Marchantia polymorpha and the moss Physcomitrella patens (black triangle, Figure 4C) and in a clade that cannot be assigned to either PCW or SCW (white triangle, Figure 4C; Supplemental Figure 3). However, in contrast to the distinctive expression profiles of PCW and SCW CESAs from Arabidopsis, all Selaginella CESAs show a ubiquitous expression pattern in roots, leaves, and stems. This finding suggests that S. moellendorffii CesAs are part of one module that produces both PCW and SCW. Conversely, in seed plants, the CesAs duplicated and subfunctionalized to produce PCW (CesA3) and SCW (CesA7 and CesA8), and in flowering plants, the PCW CesAs further expanded to produce CesA1, CesA3, and CesA6, while SCW CesAs expanded to produce CesA4, CesA7, and CesA8 (Figure 4C; Supplemental Figure 3).
The CslD family is essential for tip growth of RHs and PTs. We detected one copy of CslD in S. moellendorffii expressed in roots (Figure 4D; Supplemental Figure 4), suggesting it functions in RH growth (Figure 4A). Conversely, in flowering plants, two clades of CslD genes were formed, where each clade shows either specific expression in roots (RH CslD3 clade) or the male gametophyte (PT CslD1 clade). The ubiquitously expressed CslD5 duplicated in the ancestor of seed plants (blue triangle, Figure 4D) and is involved in cell plate formation (Gu et al., 2016). In the ancestor of angiosperms, the PT CslDs further duplicated and subfunctionalized into nonredundant CslD1 and CslD4 (Figure 4D; Supplemental Figure 4; Bernal et al., 2008). The CslDs from spruce (Picea abies) could not be assigned to either PT or RH clade, and their ubiquitous expression profile does not allow us to infer their function (Figure 4D, white triangle).
While we did not observe any evidence for subfunctionalization for CTLs (Figure 4E; Supplemental Figure 5), KORs (Figure 4F; Supplemental Figure 6), or CCs (Figure 4G; Supplemental Figure 7), the COBRA family showed a distinctive duplication in the ancestor of flowering plants to produce RH-specific COBRA-like (Figure 4H, ATCOBL9, orange triangle; Ringli et al., 2005) and PT-specific COBRA-like clades (AtCOBL10; orange triangle, Figure 4H; Supplemental Figure 8; Li et al., 2013). Interestingly, these two clades are associated with genes from spruce and S. moellendorffii, suggesting that the COBRA family has duplicated in the ancestor of vascular plants to support anisotropic (RH and PT) and isotropic (PCW and SCW) growth. The presence of only one spruce gene in the anisotropic clade indicates that the spruce COBRA gene is involved in the biosynthesis of both PTs and RHs. The poor branch support for the isotropic clade does not allow us to infer whether the PCW and SCW COBRAs duplicated in the ancestor of seed or flowering plants (Figure 4H; Supplemental Figure 8; branch support < 80), but the presence of the S. moellendorffii COBRA clade (SmCOB; white triangle, Figure 4H) suggests that the duplication took place after emergence of vascular plants.
Taken together, based on the phylogenetic trees and the expression profiles that we obtained from CoNekT-Plants, we propose a model describing the duplication and subfunctionalization events of the cell wall modules (Figure 4I). Due to poor node support, it is unclear whether the CslDs and COBRAs duplicated in the ancestor of seed plants or flowering plants. While S. moellendorffii contains both PCWs and SCWs, these two cell wall types are biosynthesized by one type of CesAs and COBRAs. The PCW/SCW subfunctionalization of CesAs took place in the ancestor of seed plants, and the CesAs further subfunctionalized into at least three nonredundant isoforms (PCW, CesA1, CesA3, CesA6, and SCW, CesA4, CesA7, CesA8) in the ancestor of flowering plants. Since S. moellendorffii only contains RHs, only one anisotropic growth-supporting COBRA (COBL9) is found in lycophytes, while the corresponding COBRA in spruce is likely involved in the biosynthesis of PTs and RHs, which have dedicated genes in flowering plants (RH, COBL9, and PT, COBL10).
Analysis of Functional Modules in S. moellendorffii and the Archaeplastida Kingdom
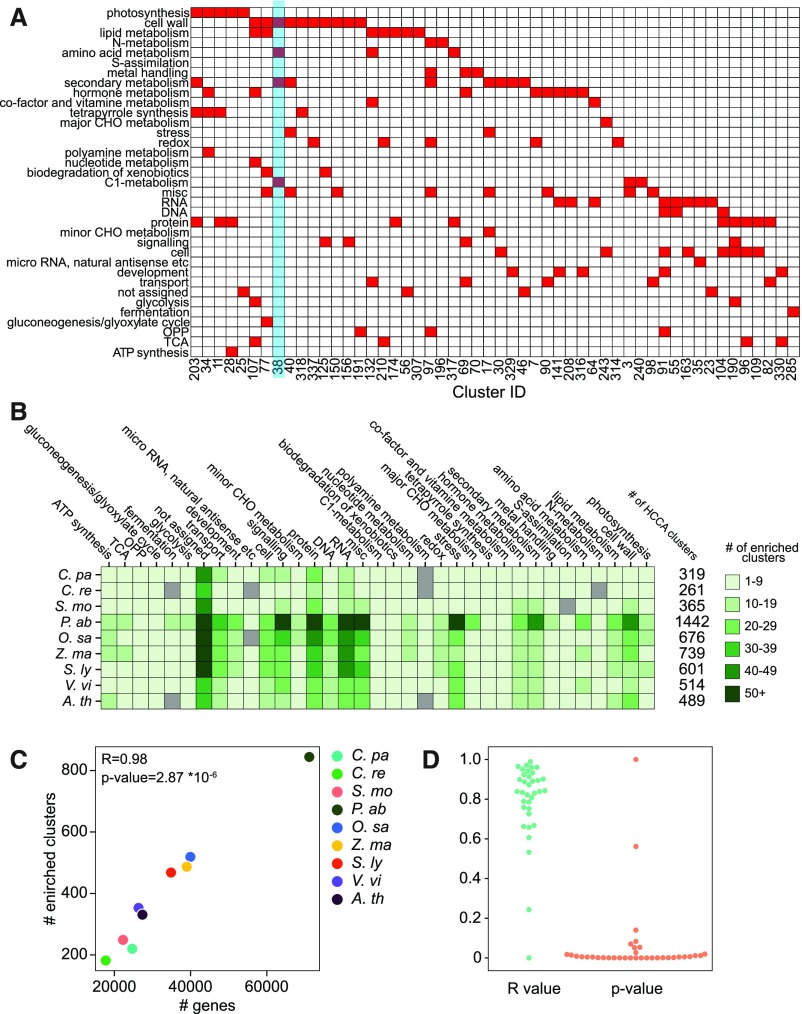
Coexpression networks contain clusters of highly connected genes (modules), which represent functionally related genes (Aoki et al., 2007; Mutwil et al., 2010; Rhee and Mutwil, 2014). We used the heuristic cluster chiseling algorithm (HCCA; Mutwil et al., 2010) to identify 365 clusters of coexpressed genes in S. moellendorffii (Supplemental Data Set 3) and investigated their biological functions by enrichment analysis of the MapMan functional bins. We found that 155 clusters were significantly enriched (false discovery rate [FDR] adjusted P value < 0.05) for at least one biological process (Figure 5; Supplemental Figure 9). All the biological processes captured by MapMan were enriched in at least one cluster, with the exception of S-assimilation (Figure 5; Supplemental Figure 9). Most of the processes, such as photosynthesis, cell wall (comprising various cell wall-acting pathways), not assigned (unknown), protein (biosynthesis, degradation, and modification), RNA (regulation of transcription), misc (various enzymes), and others were enriched in more than one cluster, suggesting that multiple functional modules are involved in these processes. Moreover, various processes were found in the same cluster, such as cell wall, amino acid metabolism, secondary metabolism, and C1 metabolism (Figure 5A; Supplemental Figure 9, blue rectangle). This is in line with the observed coordination of lignin and cell wall biosynthesis (Figure 3; Zhou et al., 2009) and suggests that these and other processes are transcriptionally coupled.
Figure 5.
Comparative Analyses of Biological Functions of the Coexpression Clusters.
(A) HCCA clusters of S. moellendorffii. The columns show cluster ID, while the rows correspond to MapMan bins that represent biological processes. Clusters enriched for a MapMan bin (FDR adjusted P value < 0.05) are indicated by red cells. Only 52 out of 365 clusters are shown in this figure, for brevity.
(B) The number of enriched bins found in the HCCA clusters of CoNekT-Plants species. The species are shown in rows, while the bins are indicated in the columns. The six shades of green indicate the number of clusters assigned to a bin.
(C) Relationship between the number of genes (y axis) and the number of HCCA clusters enriched for at least one MapMan bin (y axis) for each of the nine species in the CoNekT database (indicated by colored points). The Pearson r value and the resulting P value are indicated.
(D) Correlation between the number of genes assigned to a bin and the number of clusters enriched for the bin. Each point represents a MapMan bin. Blue points correspond to r values, while the orange points correspond to P values.
To investigate whether S. moellendorffii shows a typical number of functional modules, we plotted the number of functionally enriched clusters for the other species in CoNekT-Plants, which contains two algae (Cyanophora paradoxa, Chlamydomonas reinhardtii) and seven land plants (S. moellendorffii, Norway spruce, rice [O. sativa], maize [Zea mays], tomato [Solanum lycopersicum], grapevine [Vitis vinifera], and Arabidopsis). Despite a marked increase in the number of clusters in seed plants, the nine species show a similar distribution of enriched clusters for most processes (Figure 5B). The vascular plant-specific expansion includes misc (miscellaneous, containing various enzymes), hormone metabolism, secondary metabolism, and cell wall. This is in line with evolutionary studies that revealed that hormone metabolism and secondary metabolism appeared to provide chemical defense and responses to dehydration when plants started to colonize the land (Rensing et al., 2008; Banks et al., 2011; Wink, 2015).
The increased number of clusters can be explained by a simple linear relationship between the number of genes and the enriched clusters belonging to any biological process. For example, the number of genes in a species is positively correlated with the genome-wide number of clusters enriched for any biological process (Figure 5C). This positive correlation is seen for almost all biological processes, e.g., the number of genes assigned to a cell wall bin is strongly correlated with the number of its enriched clusters. The few exceptions to this rule (7 out of 35 bins) are involved in various aspects of primary metabolism (Figure 5D; Supplemental Data Set 4). These results demonstrate that the increased number of genes results in new coexpression clusters, possibly harboring novel functions.
Secondary Metabolic Pathways Are Coregulated with Specific Biological Processes
The colonization of land by plants was mediated by the development of new organs, biosynthetic pathways, and compounds. These innovations comprised novel tissues and organs such as the vasculature, roots, and flowers and various mechanisms to resist desiccation and light stress, which helped the plants establish sophisticated intracellular communication and interact with other organisms for defense and reproduction (Ehrlich and Raven, 1964; Hartmann, 2007; Chen et al., 2011; Pichersky and Lewinsohn, 2011; Chae et al., 2014). The latter innovations required the establishment and expansion of gene families involved in hormone biosynthesis and secondary metabolism (Figure 5B; Rensing et al., 2008; Banks et al., 2011).
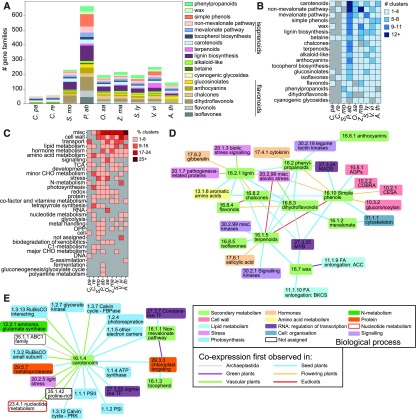
We first analyzed the number of gene families that take part in the various secondary metabolic pathways (Figure 6A; Supplemental Data Set 5). As previously observed (Rensing et al., 2008), we noticed an ∼5-fold increase in the number and diversity of families associated with secondary metabolism in vascular plants compared with algae (Figure 6A). Except for spruce, which showed an exceptionally high number of gene families (663) associated with secondary metabolism, all vascular plants showed ∼200 gene families involved in secondary metabolism (Figure 6A). The higher number of secondary metabolite gene families in spruce can, at least partly, be explained by the more than twofold higher number of total genes compared with Arabidopsis (71,158 genes versus 27,382 genes, respectively).
Figure 6.
Analysis of the Coexpression Patterns of Clusters Involved in Secondary Metabolism.
(A) The number of gene families (y axis) assigned to the different secondary metabolite MapMan bins in the species found in CoNekT-Plants (x axis).
(B) The number of HCCA clusters enriched for secondary metabolite bins, indicated by four shades of blue. Gray (e.g., terpenoids in algae) indicates that no clusters for a specific bin in a species were found.
(C) Co-occurrence analysis of secondary metabolite and other bins. The percentage of non-secondary metabolite bins enriched in clusters enriched for secondary metabolite bins. The four shades of burgundy indicate the percentage, while gray cells show that a given bin is not enriched with secondary metabolite bins.
(D) Bin co-occurrence network. Nodes represent MapMan bins color-coded according to the type, while edges connect bins that were enriched in the same cluster in at least three species. The edges indicate the oldest phylostratum where the bins co-occurred in a cluster.
(E) Bin co-occurrence network for the photosynthesis-related processes.
To identify the secondary biosynthetic modules in the Archaeplastida kingdom, we calculated which bins belonging to the secondary metabolism category are enriched in the HCCA clusters (Figure 6B). While only a few clusters were enriched for any secondary bins in the algae C. paradoxa and C. reinhardtii, vascular plants contained clusters enriched for nearly all secondary metabolites (Figure 6B). In line with the observation that the number of genes is correlated with the number of enriched clusters, spruce showed the highest number of clusters enriched for secondary metabolism (Figure 6B). Interestingly, we observed a species-specific expansion of distinct secondary metabolite pathways, such as lignin biosynthesis (phenylpropanoids) and dihydroflavonols in S. moellendorffii, terpenoids in V. vinifera, and glucosinolates in Arabidopsis (Figure 6B; Supplemental Figure 10). Spruce showed a distinct expansion for carotenoid and nonmevalonate pathway clusters, which can be attributed to the high amounts of photoprotective xanthophylls produced in needles and the high emissions of isoprenoids from spruce (Tegischer et al., 2002).
Next, we investigated how diverse biological processes are coexpressed with secondary metabolic pathways. As exemplified by lignin and cellulose biosynthesis (Figure 3C), functionally related processes can be transcriptionally coordinated (Figure 5A; Mutwil et al., 2011). To explore how secondary metabolism is integrated with other biological pathways, we investigated which MapMan bins tend to be coenriched with the secondary metabolism bin in the HCCA clusters. Figure 6C shows that the bin misc was most frequently coenriched with secondary metabolism. This bin includes several enzymes necessary for secondary metabolite biosynthesis, such as glutathione S transferases for the biosynthesis of sulfur-containing secondary metabolites (Czerniawski and Bednarek, 2018), cytochrome P450 for lignin biosynthesis (Vanholme et al., 2010), and O-methyltransferases involved in the biosynthesis of phenylpropanoids, flavonoids, and alkaloids (Lam et al., 2007). The other five most frequently coenriched bins are cell wall, transport, lipid metabolism, hormone metabolism, and amino acid metabolism (Figure 6C), suggesting that secondary metabolic pathways are tightly associated with these processes.
To reveal biologically relevant associations of secondary metabolism with the processes shown in Figure 6C, we created a network, where the nodes represent processes and the edges connect processes present in the same cluster in at least three species (Figure 6D; Supplemental Data Set 6 shows the association of all processes in all species). The color of the edges indicates when the two processes were first observed to be associated. Most of the associations appeared in seed plants (Figures 6D and 6E), which can be explained by the large number of HCCA clusters found in spruce (Figures 6A and 6B). The network shows that cell wall is transcriptionally associated with simple phenols, the building blocks of lignin. Lignin is in turn connected to stress responses, which is in line with the observed changes in lignin deposition upon biotic and abiotic stresses (Moura et al., 2010) such as cold (Gindl et al., 2000), drought stress (Hu et al., 2009), and mechanical stress (Soltani et al., 2006). Interestingly, secondary metabolite bins such as simple phenols, chalcones, dihydroflavonons, and phenylpropanoids were coenriched with MYB and MADS transcription factors (Figure 6D), which is in line with the known regulatory role of these transcription factors in these processes (Liu et al., 2015). Moreover, we observed a large association between carotenoids and the photosynthesis and carbon fixation machinery (Figure 6E), which is in agreement with the role of these pigments in promoting photosynthesis and photoprotection (Young, 1991; Ostroumov et al., 2013). The carotenoid bin was also associated with sigma-like transcription factors, which regulate the expression of photosynthesis genes (Chi et al., 2015). These and numerous other associations demonstrate the biologically meaningful integration of secondary metabolism with the cellular machinery. In addition, they can be used to further understand the wiring of biological processes in response to the environment.
Expression and Phylostratigraphic Analyses Reveal a Highly Conserved Root Transcriptome
Roots provide anchorage to the soil and efficient water and nutrient acquisition in land plants. In contrast to the tip-growing rhizoids in early, nonvascular land plants, all vascular plants possess roots that include a root cap, an apical meristem, and a radial cell type organization (Kenrick and Strullu-Derrien, 2014). Despite the differences observed between vascular plants regarding root morphology and architecture, the root transcriptomes are highly conserved, which suggests a unique, or highly convergent, origin of roots (Huang and Schiefelbein, 2015).
To further study the evolution of roots, we analyzed root transcriptomes using the “specific profile” feature in CoNekT-Plants, which can identify genes specifically expressed in certain organs or under certain conditions (https://conekt.sbs.ntu.edu.sg/search/specific/profiles). CoNekT also includes tools that allow the expression of conserved homologs to be extracted in different organs, tissues, and conditions for pairs (i.e., maximum two) of species (at https://conekt.sbs.ntu.edu.sg/specificity_comparison/). However, to identify the conserved root-specific transcriptome in more than two species, we extracted root-specific genes for each species using the “specific profile” feature and performed a custom meta-analysis. To detect expression specificity, CoNekT uses a specificity measure (SPM) that ranges between 0 (gene is not expressed in a given sample) and 1 (gene is exclusively expressed in the sample; Xiao et al., 2010). Using SPM > 0.85 cutoff and selecting Roots/rhizoid as the target organ, we obtained 4314, 12,177, 4316, 4185, and 3247 genes for S. moellendorffii, O. sativa, Z. mays, S. lycopersicum, and Arabidopsis, respectively (Supplemental Data Set 7). Note that since no rhizoids were collected for S. moellendorffii, the Root/rhizoid target organ corresponds to roots only. As CoNekT-Plants does not contain information about root samples for spruce and V. vinifera, they were omitted from the analysis. We hypothesized that the root-specific gene families emerged at a specific period in plant evolution. To investigate this notion, we performed phylostratigraphic analysis, where the age of a gene family is determined based on its presence in the most basal, i.e., earliest, plant (Ruprecht et al., 2017). The different phylostrata are “Archaeplastida,” “green plants,” “vascular plants,” “seed plants,” “flowering plants,” “monocots,” “eudicots,” and “species-specific.” For example, a gene family containing S. moellendorffii genes and Arabidopsis genes would be assigned to the “vascular plants” phylostratum, while a family containing S. lycopersicum genes and Arabidopsis genes would be assigned to the “eudicot” phylostratum.
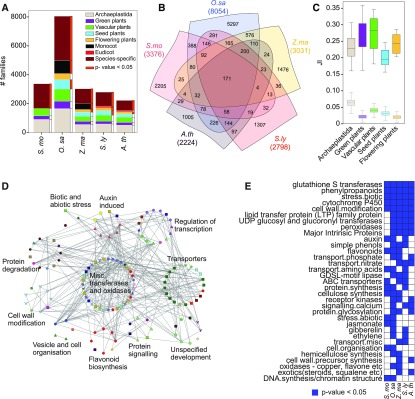
Our phylostratigraphic analysis revealed a quite similar distribution of the different phylostrata in the root-specific transcriptomes among the five different species (Figure 7A). Enrichment analysis showed that the “vascular plants” phylostratum gene families were not overrepresented in the roots (Figure 7A, green bars; Supplemental Data Set 8). Conversely, the “species-specific” phylostratum, which represents gene families that are only found in one species (Supplemental Data Set 7), was significantly overrepresented in most of the plants (Figure 7A, dark-orange bars; Supplemental Data Set 8). These results suggest that the evolution of the root organ did not require the acquisition of novel genetic material. Conversely, the significant enrichment (FDR adjusted P value < 0.05) of gene families of the “species-specific” phylostratum suggests that the different root morphologies observed in these plants required the generation of novel functions based on new genetic material (Figure 7A; Huang and Schiefelbein, 2015).
Figure 7.
Comparative Analysis of the Root Transcriptomes of S. moellendorffii and Flowering Plants.
(A) The number of root-specific gene families whose members are expressed in roots of five different plant species. The x axis indicates the species, while the y axis shows the number of gene families. The color of the bars indicates the phylostrata of the gene families.
(B) Venn diagram showing the overlap of the root-specific gene families in the five plants.
(C) Boxplots showing the distribution of JI values (y axis), capturing the overlap of gene families of genes specifically expressed in roots. The central line and the bounds of the box represent the median, the first, and the third quartile, respectively. The whiskers represent the minimum and maximum values of the observations. The gene families are divided into phylostrata (x axis), from oldest (Archaeplastida) to youngest (flowering plants). The transparent boxplots below represent the JI values calculated by random sampling of an equal number of genes from a phylostratum.
(D) Co-expression network of S. moellendorffii genes that belong to a family that contains members specifically expressed in roots in the five analyzed species. The genes (nodes) are grouped according to biological function.
(E) Blue cells indicate the MapMan bins that are significantly enriched (FDR adjusted P value < 0.05) among the root-specific genes. Bins that are enriched in at least two species are shown.
Next, we investigated the similarities of the root transcriptomes in the five vascular plants by analyzing the overlap of the root-specific gene families. While the majority of the families are species specific, a considerable number of families are shared across all five vascular plants (171 families), flowering plants (200 families), monocots (576 families), and eudicots (97 families), suggesting a further elaboration of the root transcriptome in the different lineages (Figure 7B). We then investigated how the different phylostrata contribute to the similarities of the root transcriptomes by calculating the Jaccard Index (JI) between the five phylostrata in all five species pairs. The JI ranges from 0 (no gene families assigned to a given phylostratum are in common) to 1 (all gene families are in common). Despite the differences in the JI values across the phylostrata, all five phylostrata contributed to the similarity of the root transcriptomes (Figure 7C), and the JI values were larger than the values that would be produced by chance (FDR adjusted P value < 0.05; Figure 7C, transparent box plots). This further supports the notion that the evolution of roots did not coincide with a punctuated appearance of novel genetic material.
To investigate the role of the 171 core gene families that are root specific in all vascular plants (Supplemental Data Set 7), we extracted S. moellendorffii root-specific genes belonging to these families and used CoNekT-Plants to create a custom network (https://conekt.sbs.ntu.edu.sg/custom_network/). Functional analysis of this network revealed groups of genes involved in root biology, such as transferases and oxidases (Hemsley et al., 2005; Lo et al., 2008; Li et al., 2017), transporters to mediate root signaling and nutrient uptake (Javot and Maurel, 2002; Little et al., 2005), auxin-induced genes important for lateral root growth (Casimiro et al., 2001; Grieneisen et al., 2007), and genes involved in the regulation of transcription, protein degradation, cell wall modification, and flavonoid biosynthesis (Tohge and Fernie, 2016). Functional enrichment analysis of the root-specific genes in all five species revealed bins corresponding to glutathione S transferase, phenylpropanoids, biotic stress, cytochrome P450, various transporters, cell wall modification, hormone metabolism, and others (Figure 7E). Thus, these genes and gene families constitute prime candidates to study root molecular biology in vascular plants.
DISCUSSION
S. moellendorffii, a model organism for lycophytes and one of the oldest extant vascular plants, is invaluable for investigating the evolution of land plants and vascular plants. In this study, we produced and collected expression data from several organs, tissue types, and environmental stresses to expand the eFP Browser (http://bar.utoronto.ca/efp_selaginella/cgi-bin/efpWeb.cgi; Figure 1G) and the CoNekT-Plants database (conekt.plant.tools; Figure 2) with data from this species. This comprehensive expression atlas can be used to predict gene function and perform comparative analyses among S. moellendorffii and the other eight species representing different families in the database.
We first investigated the conservation of the gene module involved in lignocellulose biosynthesis in S. moellendorffii. This module contains genes involved in Phe, lignin and cellulose biosynthesis, various polysaccharide-acting enzymes, cell wall structural proteins, and MYB transcription factor that putatively regulate the expression of these genes (Figure 3; Rao and Dixon, 2018). Importantly, a similar module has been observed in Arabidopsis and B. distachyon (Sibout et al., 2017), indicating that the lignocellulosic module is at least 400 million years old and is most likely present in a similar form in all vascular plants. As second-generation biofuels from lignocellulosic biomass are based on the products of this module (Somerville et al., 2010), a better understanding of the genes found in this module, their products, and the underlying regulatory networks will enable the improvement of lignocellulose in all crops.
All angiosperms possess PCWs, SCWs, tip-growing PTs, and RHs. While Arabidopsis contains four gene modules dedicated to each cell wall type (Ruprecht et al., 2016), phylogenetic and expression analysis of cell wall-related genes revealed that S. moellendorffii contains one gene module that biosynthesizes PCW and SCW (Figure 4). This supports the previous suggestion that the diversification of CesAs for PCW and SCW biosynthesis was not required for the evolution of vascular tissue (Harholt et al., 2012). The duplication and subfunctionalization events of the CesA, COBRA, and CslD families in the ancestor of seed and flowering plants produced multiple, nonredundant copies of each gene. What purpose are these gene and gene module duplications serving? Perhaps the duplication of the cell wall module relieved the multitasking strain (adaptive conflict) imposed on the ancestral Selaginella module and allowed for the further specialization and improvement of PCW and SCW in seed plants. Alternatively, these duplications may serve no purpose beyond duplicating these genes. For example, CslD2 and CslD3 are a product of an Arabidopsis-specific duplication (Supplemental Figure 4), and both are needed to produce normal RHs (Bernal et al., 2008), indicating that these genes underwent a duplication-degeneration-complementation-type (DCC) functional divergence (De Smet and Van de Peer, 2012). Since gene modules are composed of genes (Ruprecht et al., 2016), it is likely that the same duplication types that govern genes (e.g., DDC, specialization, neofunctionalization, gene sharing) can govern duplications of gene modules.
We observed a linear relationship between the number of genes assigned to a process and the number of clusters enriched for the process (Figure 5D). This relationship is sufficient to explain the observed increase in clusters enriched for processes such as cell wall, secondary metabolism, hormone metabolism, and miscellaneous in the vascular plants (Figure 5B). Interestingly, we detected clusters enriched for, e.g. hormone metabolism in the two algae C. paradoxa and C. reinhardtii. While these single-cellular organisms do not have or indeed require a hormonal system, they might contain precursors for these pathways. For example, enzymes involved in the biosynthesis of lignin precursors are found in C. paradoxa (Labeeuw et al., 2015), and the complete pathway appeared in the moss P. patens (Xu et al., 2009). These examples suggest that many of the pathways unique to land plants are likely a result of extension and subfunctionalization of ancient pathways present in algae.
Secondary metabolism dramatically expanded in land plants (Figures 6A and 6B), raising the question of how these new pathways were integrated into the existing cellular machinery. For example, lignin and cellulose synthesis are coordinated on the level of gene expression (Figure 3) and biosynthesis (Mutwil et al., 2009). Our analysis revealed that secondary metabolic pathways are often transcriptionally coordinated with relevant pathways (e.g. light stress response with carotenoids, biotic stress with lignin; Figures 6D and 6E), suggesting that the wiring of these pathways is evolutionarily advantageous.
The root transcriptome is conserved in vascular plants, indicating that a common molecular program is used to produce roots (Huang and Schiefelbein, 2015). This conservation is remarkable, as the lycophyte and euphyllophyte lineages are thought to have evolved roots independently based on fossil evidence showing early euphyllophytes without roots at a time when lycophytes already possessed them (Kenrick and Crane, 1997; Raven and Edwards, 2001; Hetherington and Dolan, 2019). The exact origin of roots is thus still under debate. The two possible, not mutually exclusive theories are (1) the existence of a primitive root developmental program in the common ancestor of vascular plants and/or (2) parallel recruitment of largely similar developmental programs independently in the lycophyte and euphyllophyte lineages. Our analysis revealed that the root transcriptome is not enriched for gene families that predate the appearance of roots but is instead enriched for gene families that appeared after the roots were established (Figures 7A and 7C). This suggests that root evolution is an ongoing selective process, which could also explain the large variations in root morphology and architecture observed in different species. Similar analyses for leaves, flowers, and other organs and tissues will shed more light on the question of whether the evolution of plant structures followed the same trajectory as in roots.
METHODS
Experimental Growth Conditions and Sampling
Selaginella moellendorffii was obtained from Evermoss (http://www.evermoss.de). The plants used for the organ expression atlas and temperature stress treatment were grown at 24°C under a 16 h light (90 μmol photons m−2 s−1 with a wavelength of 380 to 800 nm [Master TL5; Philips])/8 h dark photoperiod in soil, while the plants used for high-light and dark-stress treatment were grown on modified Hoagland solution (0.063 mM FeEDTA; 0.5 mM KH2PO4; 2.5 mM KNO3; 2 mM Ca(NO3)2 × 4H2O; 1 mM MgSO4 × 7H2O; 50.14 µM H3BO3; 9.25 µM MnCl2 × 4H2O; 1 µM ZnCl2; 1 µM CuCl2; 0.5 µM Na2MoO4 × 2H2O, pH 5.8, 0.8% [w/w] agar) at 24°C under a 12 h light (70 μmol photons m−2 s−1)/12 h dark period before being subjected to prolonged-darkness (48 h) and high-light (150 μmol photons m−2 s−1) treatments. Temperature-stressed plants were exposed to 4°C (cold stress) or 37°C (heat stress) for 1 or 3 h. The sampled organs were flash-frozen in liquid nitrogen and ground with a mortar and pestle. RNA was extracted from the samples using a Plant RNeasy plant kit (Qiagen) according to the manufacturer’s instructions. The integrity of the RNA was assessed using an RNA nano chip on an Agilent Bioanalyzer 2100; all samples showed RNA integrity values higher than the recommended thresholds. cDNA libraries were prepared from total RNA using poly-A enrichment and sequenced on the Illumina-HiSeq2500/4000 systems at the Beijing Genomics Institute. Scanning electron microscope (SEM) images were produced using freshly harvested and N2-frozen plant samples in a tabletop Hitachi TM3030.
Estimating Gene Expression from RNA-seq Data
The raw reads were mapped to the coding sequences (CDS) with Kallisto (Bray et al., 2016) to obtain transcripts per million (TPM) gene expression values. Publicly available RNA-seq data were downloaded from ENA (Silvester et al., 2018). The CDSs were downloaded from Phytozome (Banks et al., 2011; Goodstein et al., 2012). On average, 55% of the reads mapped to the CDSs (Supplemental Data Set 1). Hierarchical clustering of the samples revealed the expected clustering, where related samples were found in the same clade. The TPM values were combined into a TPM expression matrix (Supplemental Data Set 2). Due to the generation of a more updated diurnal atlas for S. moellendorffii (Ferrari et al., 2019), the eight samples generated capturing diurnal gene expression were excluded from the expression profile but were still used to generate the coexpression network (Supplemental Data Set 2).
Adding S. moellendorffii to the CoNekT-Plants Database
The normalized TPM matrix was uploaded in CoNekT-Plants together with CDS (Banks et al., 2011; Goodstein et al., 2012), annotation (Mercator, standard settings; Lohse et al., 2014), and protein domain information (InterProScan 5.32-71.0; Jones et al., 2014). The gene families were detected with OrthoFinder 1.1.8 (Emms and Kelly, 2015) with default parameters and Diamond (Buchfink et al., 2015). CoNekT-plants now includes data from nine species: Arabidopsis (Arabidopsis thaliana), Vitis vinifera, Solanum lycopersicum, Zea mays, Oryza sativa, Picea abies, S. moellendorfii, Chlamydomonas reinhardtii, and Cyanophora paradoxa.
Functional Enrichment Analysis of Clusters and Root Transcriptomes
CoNekT-Plants constructs coexpression networks using the highest reciprocal rank metric, where an edge cutoff of highest reciprocal rank < 100 is used to connect genes (Proost and Mutwil, 2017, 2018). The groups of densely connected genes (clusters) were inferred by CoNekT-Plants using the HCCA (Mutwil et al., 2010). The HCCA clusters were downloaded from CoNekT-Plants (Supplemental Data Set 3 for Selaginella clusters). For each cluster, we counted the number of times a specific biological process was represented. The observed distribution of biological processes was compared with a permuted distribution obtained by sampling an equal number of genes from the total pool of a species’ genes for 10,000 permutations. The empirical P values obtained were FDR corrected by the Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995). The functional enrichment of clusters was performed using the annotation from Mercator (Lohse et al., 2014) for first-level bins (Figures 5A and 5B), secondary metabolism bins (Figures 6A and 6B), and third-level bins (Figures 6D and 6E). Genes and gene families involved in secondary metabolism were defined using the annotation from Mercator, where bin 16 indicates secondary metabolism genes (Lohse et al., 2014; Supplemental Data Set 5).
CoNekT-Plants uses measures such as SPM, Tau, and entropy to define the specificity of expression of genes. Typically, SPM ∼0.8, high Tau value, and low entropy value indicate that a gene is expressed in a specific organ, tissue, or sample. To identify root-specific genes, we selected tissue specificity method root sample, with an SPM cutoff of 0.85. The functional enrichment of the root transcriptomes was performed by comparing the observed distribution of MapMan bins within the root-specific genes to a random distribution obtained by sampling an equal number of genes from the total pool of the species for 10,000 permutations. The empirical P values were FDR corrected (Figures 6D, 6E, and E7E).
Phylogenetic Analysis and Tree Construction
To study the phylogeny of cell wall modules, we entered protein sequences from the orthogroups containing AtCESAs, AtCSLDs, AtCTLs, AtKORs, AtCCs, and AtCOBLs into the PLAZA Interactive Phylogenetics Module (Van Bel et al., 2018). To help root the trees, the bryophytes Marchantia polymorpha and Physcomitrella patens were added to the analysis by selecting them in the species panel. The tool performed multisequence alignment with MUSCLE version 1.5.5 (Edgar, 2004), followed by trimming of poorly conserved positions. Phylogenetic tree construction was performed with the maximum likelihood method IQ-Tree (Nguyen et al., 2015), which detects the best-fitting protein model from the following widely used models: JTT, LG, WAG, Blosum62, VT, and Dayhoff. The program was run with the following commands: ‘iqtree-omp -st AA -s $MSA_FILE_PATH -pre $NEWICK_FILE_PATH -nt 1 -bb 1000 -mset JTT,LG,WAG,Blosum62,VT,Dayhoff -mfreq F -mrate R’. A thousand rounds of ultraparametric bootstrapping were run, and IQ-Tree used the FreeRate model to rate heterogeneity across sites. The selected models for CC, CTL, COB, KOR, CESA, and CSLD families by the IQ-Tree algorhithm were WAG+F+R3, WAG+F+R3, LG+F+R5, LG+F+R4, LG+F+R4, and LG+F+R5, respectively. The trees were visualized and rooted using TreeGraph2 (Stöver and Müller, 2010), and the node-branching pattern and node support ≥80 were used to manually identify the subfamilies. Supplemental Figures 3 to 8 contain the trees with node support values. The trees were rooted by selecting clades containing M. polymorpha and/or P. patens.
Phylostratigraphic Analysis of Root Transcriptomes
The phylostratum of a family was assessed by identifying the oldest clade found in the family (Ruprecht et al., 2017). To test whether a specific phylostratum was enriched in the root transcriptome, we randomly sampled (without replacement) the number of observed root-enriched gene families 10,000 times. The empirical P values were obtained by calculating whether the observed number of gene families for each phylostratum was larger than the number obtained from the 10,000 sampling procedure. The P values were FDR corrected (Figure 7A, orange bars; Supplemental Data Set 8).
To calculate whether a given phylostratum is enriched among the conserved families between two species, we used the JI, which represents the number of shared families between a species pair. For each phylostratum, the observed JI was compared with the JI obtained by shuffling the family-phylostratum assignment 10,000 times (Figure 7C).
Accession Numbers
The raw sequencing data are available from the EBI under accession number E-MTAB-8217. Nexus files with sequence alignments and trees are available from the Dryad Digital Repository (https://datadryad.org/stash) under doi:10.5061/dryad.1jwstqjr0.
Supplemental Data
Supplemental Figure 1. Scanning electron microscopy of Selaginella organs and tissues
Supplemental Figure 2. Sample relationship dendrogram of the 91 samples
Supplemental Figure 3. Phylogenetic trees of the CesA genes
Supplemental Figure 4. Phylogenetic trees of the CslD genes
Supplemental Figure 5. Phylogenetic trees of the CTL genes
Supplemental Figure 6. Phylogenetic trees of the KOR genes
Supplemental Figure 7. Phylogenetic trees of the CC genes
Supplemental Figure 8. Phylogenetic trees of the COBRA genes
Supplemental Figure 9. HCCA clusters of S. moellendorffii
Supplemental Figure 10. The relative abundance of secondary metabolite gene families
Supplemental Data Set 1. Kallisto mapping statistics
Supplemental Data Set 2. TPM normalized expression matrix
Supplemental Data Set 3. HCCA clusters of S. moellendorffii
Supplemental Data Set 4. Correlation analysis of functionally enriched clusters and number of genes assigned to a specific biological process
Supplemental Data Set 5. Gene families containing genes assigned to secondary metabolism-related MapMan bins
Supplemental Data Set 6. Association matrix of biological processes
Supplemental Data Set 7. Gene families specifically expressed in Roots/rhizoids in S. moellendorffii, O. sativa, Z. mays, S. lycopersicum, and Arabidopsis
Supplemental Data Set 8. Phylostrata enrichment of root-specific genes
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Nikola Winter for her help with taking the SEM images. Funding for this work was provided by the Max Planck Society (to C.F., B.O.H., F.K., A.F., T.T., and M.M.) and Nanyang Technological University (to M.M.).
AUTHOR CONTRIBUTIONS
M.M. conceived the project; C.F. and B.O.H. performed the experiments; C.F., D.S., and M.M. processed the data and performed bioinformatic analyses; A.P., E.E., and N.J.P. provided eFP browser assistance; F.K. helped with acquiring the SEM images; T.T. and A.F. provided feedback on the article and experimental design. C.F. and M.M. wrote the article with help from all the authors.
References
- Alejandro S., Lee Y., Tohge T., Sudre D., Osorio S., Park J., Bovet L., Lee Y., Geldner N., Fernie A.R., Martinoia E. (2012). AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 22: 1207–1212. [DOI] [PubMed] [Google Scholar]
- Aoki K., Ogata Y., Shibata D. (2007). Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 48: 381–390. [DOI] [PubMed] [Google Scholar]
- Banks J.A., et al. (2011). The selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A.-L., Bonabeau E. (2003). Scale-free networks. Sci. Am. 288: 60–69. [DOI] [PubMed] [Google Scholar]
- Barabási A.L., Oltvai Z.N. (2004). Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 5: 101–113. [DOI] [PubMed] [Google Scholar]
- Bassel G.W., Lan H., Glaab E., Gibbs D.J., Gerjets T., Krasnogor N., Bonner A.J., Holdsworth M.J., Provart N.J. (2011). Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc. Natl. Acad. Sci. USA 108: 9709–9714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57: 289–300. [Google Scholar]
- Bernal A.J., Yoo C.M., Mutwil M., Jensen J.K., Hou G., Blaukopf C., Sørensen I., Blancaflor E.B., Scheller H.V., Willats W.G.T. (2008). Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol. 148: 1238–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34: 525–527. [DOI] [PubMed] [Google Scholar]
- Broido A.D., Clauset A. (2019). Scale-free networks are rare. Nat. Commun. 10: 1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D. (2015). Fast and sensitive protein alignment using DIAMOND. Nat Methods 12: 59–60. [DOI] [PubMed] [Google Scholar]
- Casimiro I., Marchant A., Bhalerao R.P., Beeckman T., Dhooge S., Swarup R., Graham N., Inzé D., Sandberg G., Casero P.J., Bennett M. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae L., Kim T., Nilo-Poyanco R., Rhee S.Y. (2014). Genomic signatures of specialized metabolism in plants. Science 344: 510–513. [DOI] [PubMed] [Google Scholar]
- Chen F., Tholl D., Bohlmann J., Pichersky E. (2011). The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66: 212–229. [DOI] [PubMed] [Google Scholar]
- Chen K., Plumb G.W., Bennett R.N., Bao Y. (2005). Antioxidant activities of extracts from five anti-viral medicinal plants. J. Ethnopharmacol. 96: 201–205. [DOI] [PubMed] [Google Scholar]
- Chi W., He B., Mao J., Jiang J., Zhang L. (2015). Plastid sigma factors: Their individual functions and regulation in transcription. Biochim. Biophys. Acta 1847: 770–778. [DOI] [PubMed] [Google Scholar]
- Cho M.H., et al. (2007). Phenylalanine biosynthesis in Arabidopsis thaliana. Identification and characterization of arogenate dehydratases. J. Biol. Chem. 282: 30827–30835. [DOI] [PubMed] [Google Scholar]
- Crowley T.J., Berner R.A. (2001). CO2 and climate change. Science 292: 870–872. [DOI] [PubMed] [Google Scholar]
- Czerniawski P., Bednarek P. (2018). Glutathione S-transferases in the biosynthesis of sulfur-containing secondary metabolites in brassicaceae plants. Front. Plant Sci. 9: 1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R., Van de Peer Y. (2012). Redundancy and rewiring of genetic networks following genome-wide duplication events. Curr. Opin. Plant Biol. 15: 168–176. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich P.R., Raven P.H. (1964). Butterflies and plants: A study in coevolution. Evolution 18: 586–608. [Google Scholar]
- Emms D.M., Kelly S. (2015). OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari C., Proost S., Janowski M., Becker J., Nikoloski Z., Bhattacharya D., Price D., Tohge T., Bar-Even A., Fernie A., Stitt M., Mutwil M. (2019). Kingdom-wide comparison reveals the evolution of diurnal gene expression in Archaeplastida. Nat. Commun. 10: 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari C., Proost S., Ruprecht C., Mutwil M. (2018). PhytoNet: Comparative co-expression network analyses across phytoplankton and land plants. Nucleic Acids Res. 46 (W1): W76–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M.H., Edwards M.B., Schultz E.R., McKain M.R., Fei Z., Sørensen I., Rose J.K.C., Scanlon M.J. (2015). Dissecting the molecular signatures of apical cell-type shoot meristems from two ancient land plant lineages. New Phytol. 207: 893–904. [DOI] [PubMed] [Google Scholar]
- Gindl W., Grabner M., Wimmer R. (2000). The influence of temperature on latewood lignin content in treeline Norway spruce compared with maximum density and ring width. Trees (Berl. West) 14: 409–414. [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40: D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen V.A., Xu J., Marée A.F.M., Hogeweg P., Scheres B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013. [DOI] [PubMed] [Google Scholar]
- Gu F., Bringmann M., Combs J.R., Yang J., Bergmann D.C., Nielsen E. (2016). Arabidopsis CSLD5 functions in cell plate formation in a cell cycle-dependent manner. Plant Cell 28: 1722–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen B.O., Vaid N., Musialak-Lange M., Janowski M., Mutwil M. (2014). Elucidating gene function and function evolution through comparison of co-expression networks of plants. Front. Plant Sci. 5: 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J., Sørensen I., Fangel J., Roberts A., Willats W.G.T., Scheller H.V., Petersen B.L., Banks J.A., Ulvskov P. (2012). The glycosyltransferase repertoire of the spikemoss Selaginella moellendorffii and a comparative study of its cell wall. PLoS One 7: e35846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann T. (2007). From waste products to ecochemicals: Fifty years research of plant secondary metabolism. Phytochemistry 68: 2831–2846. [DOI] [PubMed] [Google Scholar]
- Hartwell L.H., Hopfield J.J., Leibler S., Murray A.W. (1999). From molecular to modular cell biology. Nature 402 (6761 Suppl): C47–C52. [DOI] [PubMed] [Google Scholar]
- Hemsley P.A., Kemp A.C., Grierson C.S. (2005). The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis. Plant Cell 17: 2554–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington A.J., Dolan L. (2019). Rhynie chert fossils demonstrate the independent origin and gradual evolution of lycophyte roots. Curr. Opin. Plant Biol. 47: 119–126. [DOI] [PubMed] [Google Scholar]
- Hu Y., Li W.C., Xu Y.Q., Li G.J., Liao Y., Fu F.L. (2009). Differential expression of candidate genes for lignin biosynthesis under drought stress in maize leaves. J. Appl. Genet. 50: 213–223. [DOI] [PubMed] [Google Scholar]
- Huang L., Schiefelbein J. (2015). Conserved gene expression programs in developing roots from diverse plants. Plant Cell 27: 2119–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M., et al. (2013). Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341: 175–179. [DOI] [PubMed] [Google Scholar]
- Javot H., Maurel C. (2002). The role of aquaporins in root water uptake. Ann. Bot. 90: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Gómez J.M., Wallace A.D., Maloof J.N. (2010). Network analysis identifies ELF3 as a QTL for the shade avoidance response in Arabidopsis. PLoS Genet. 6: e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., et al. (2014). InterProScan 5: Genome-scale protein function classification. Bioinformatics 30: 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P., Crane P.R. (1997). The origin and early evolution of plants on land. Nature 389: 33–39. [Google Scholar]
- Kenrick P., Strullu-Derrien C. (2014). The origin and early evolution of roots. Plant Physiol. 166: 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox J.P. (2008). Revealing the structural and functional diversity of plant cell walls. Curr. Opin. Plant Biol. 11: 308–313. [DOI] [PubMed] [Google Scholar]
- Koshiba T., Yamamoto N., Tobimatsu Y., Yamamura M., Suzuki S., Hattori T., Mukai M., Noda S., Shibata D., Sakamoto M., Umezawa T. (2017). MYB-mediated upregulation of lignin biosynthesis in Oryza sativa towards biomass refinery. Plant Biotechnol (Tokyo) 34: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeeuw L., Martone P.T., Boucher Y., Case R.J. (2015). Ancient origin of the biosynthesis of lignin precursors. Biol. Direct 10: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lala P.K., Chakraborty C. (2001). Role of nitric oxide in carcinogenesis and tumour progression. Lancet Oncol. 2: 149–156. [DOI] [PubMed] [Google Scholar]
- Lam K.C., Ibrahim R.K., Behdad B., Dayanandan S. (2007). Structure, function, and evolution of plant O-methyltransferases. Genome 50: 1001–1013. [DOI] [PubMed] [Google Scholar]
- Lampugnani E.R., Flores-Sandoval E., Tan Q.W., Mutwil M., Bowman J.L., Persson S. (2019). Cellulose synthesis: Central components and their evolutionary relationships. Trends Plant Sci. 24: 402–412. [DOI] [PubMed] [Google Scholar]
- Lee I., Ambaru B., Thakkar P., Marcotte E.M., Rhee S.Y. (2010). Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat. Biotechnol. 28: 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ge F.R., Xu M., Zhao X.Y., Huang G.Q., Zhou L.Z., Wang J.G., Kombrink A., McCormick S., Zhang X.S., Zhang Y. (2013). Arabidopsis COBRA-LIKE 10, a GPI-anchored protein, mediates directional growth of pollen tubes. Plant J. 74: 486–497. [DOI] [PubMed] [Google Scholar]
- Li W.Y., Chen B.X., Chen Z.J., Gao Y.T., Chen Z., Liu J. (2017). Reactive oxygen species generated by NADPH oxidases promote radicle protrusion and root elongation during rice seed germination. Int. J. Mol. Sci. 18: E110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Chapple C. (2010). Understanding lignification: Challenges beyond monolignol biosynthesis. Plant Physiol. 154: 449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little D.Y., Rao H., Oliva S., Daniel-Vedele F., Krapp A., Malamy J.E. (2005). The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Natl. Acad. Sci. USA 102: 13693–13698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Osbourn A., Ma P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8: 689–708. [DOI] [PubMed] [Google Scholar]
- Liu Q., Luo L., Zheng L. (2018). Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 19: E335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S.F., Yang S.Y., Chen K.T., Hsing Y.I., Zeevaart J.A.D., Chen L.J., Yu S.M. (2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Nagel A., Herter T., May P., Schroda M., Zrenner R., Tohge T., Fernie A.R., Stitt M., Usadel B. (2014). Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 37: 1250–1258. [DOI] [PubMed] [Google Scholar]
- Ma L.Y., Ma S.C., Wei F., Lin R.C., But P.P.H., Lee S.H.S., Lee S.F. (2003). Uncinoside A and B, two new antiviral chromone glycosides from Selaginella uncinata. Chem. Pharm. Bull. (Tokyo) 51: 1264–1267. [DOI] [PubMed] [Google Scholar]
- Manfroi J., Jasper A., Guerra-Sommer M., Uhl D. (2012). Sub-arborescent lycophytes in coal-bearing strata from the artinskian (Early Permian/Cisuralian) of the Santa Catarina coalfield (Paraná Basin, SC, Brazil). Revista de Brasileira Paleontologia 15: 135–140. [Google Scholar]
- McFarlane H.E., Döring A., Persson S. (2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65: 69–94. [DOI] [PubMed] [Google Scholar]
- Mello A., Efroni I., Rahni R., Birnbaum K.D. (2019). The Selaginella rhizophore has a unique transcriptional identity compared with root and shoot meristems. New Phytol. 222: 882–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura J.C.M.S., Bonine C.A.V., de Oliveira Fernandes Viana J., Dornelas M.C., Mazzafera P. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52: 360–376. [DOI] [PubMed] [Google Scholar]
- Mutwil M., Debolt S., Persson S. (2008). Cellulose synthesis: A complex complex. Curr. Opin. Plant Biol. 11: 252–257. [DOI] [PubMed] [Google Scholar]
- Mutwil M., Klie S., Tohge T., Giorgi F.M., Wilkins O., Campbell M.M., Fernie A.R., Usadel B., Nikoloski Z., Persson S. (2011). PlaNet: Combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 23: 895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M., Ruprecht C., Giorgi F.M., Bringmann M., Usadel B., Persson S. (2009). Transcriptional wiring of cell wall-related genes in Arabidopsis. Mol. Plant 2: 1015–1024. [DOI] [PubMed] [Google Scholar]
- Mutwil M., Usadel B., Schütte M., Loraine A., Ebenhöh O., Persson S. (2010). Assembly of an interactive correlation network for the Arabidopsis genome using a novel heuristic clustering algorithm. Plant Physiol. 152: 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y., Yamaguchi M., Endo H., Rejab N.A., Ohtani M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroumov E.E., Mulvaney R.M., Cogdell R.J., Scholes G.D. (2013). Broadband 2D electronic spectroscopy reveals a carotenoid dark state in purple bacteria. Science 340: 52–56. [DOI] [PubMed] [Google Scholar]
- Otero R., et al. (2000). Snakebites and ethnobotany in the northwest region of Colombia: Part I: traditional use of plants. J. Ethnopharmacol. 71: 493–504. [DOI] [PubMed] [Google Scholar]
- Otreba P., Gola E.M. (2011). Specific intercalary growth of rhizophores and roots in Selaginella kraussiana (Selaginellaceae) is related to unique dichotomous branching. Flora Morphol. Distrib. Funct. Ecol. Plants 206: 227–232. [Google Scholar]
- Persson S., Wei H., Milne J., Page G.P., Somerville C.R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. USA 102: 8633–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips T.L., DiMichele W.A. (1992). Comparative ecology and life-history biology of arborescent lycopsids in late carboniferous swamps of Euramerica. Ann. Mo. Bot. Gard. 79: 560–588. [Google Scholar]
- Pichersky E., Lewinsohn E. (2011). Convergent evolution in plant specialized metabolism. Annu. Rev. Plant Biol. 62: 549–566. [DOI] [PubMed] [Google Scholar]
- Pokharel Y.R., Yang J.W., Kim J.Y., Oh H.W., Jeong H.G., Woo E.R., Kang K.W. (2006). Potent inhibition of the inductions of inducible nitric oxide synthase and cyclooxygenase-2 by taiwaniaflavone. Nitric Oxide 15: 217–225. [DOI] [PubMed] [Google Scholar]
- Proost S., Mutwil M. (2016). Tools of the trade: Studying molecular networks in plants. Curr. Opin. Plant Biol. 30: 143–150. [DOI] [PubMed] [Google Scholar]
- Proost S., Mutwil M. (2017). PlaNet: Comparative co-expression network analyses for plants. Methods Mol. Biol. 1533: 213–227. [DOI] [PubMed] [Google Scholar]
- Proost S., Mutwil M. (2018). CoNekT: An open-source framework for comparative genomic and transcriptomic network analyses. Nucleic Acids Res. 46 (W1): W133–W140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P., et al. (2013). A large-scale evaluation of computational protein function prediction. Nat. Methods 10: 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J., Akiyama T., Kim H., Lu F., Schatz P.F., Marita J.M., Ralph S.A., Reddy M.S.S., Chen F., Dixon R.A. (2006). Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J. Biol. Chem. 281: 8843–8853. [DOI] [PubMed] [Google Scholar]
- Rao X., Dixon R.A. (2018). Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front. Plant Sci. 9: 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven J.A., Edwards D. (2001). Roots: Evolutionary origins and biogeochemical significance. J. Exp. Bot. 52: 381–401. [DOI] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69. [DOI] [PubMed] [Google Scholar]
- Rhee S.Y., Mutwil M. (2014). Towards revealing the functions of all genes in plants. Trends Plant Sci. 19: 212–221. [DOI] [PubMed] [Google Scholar]
- Ringli C., Baumberger N., Keller B. (2005). The Arabidopsis root hair mutants der2-der9 are affected at different stages of root hair development. Plant Cell Physiol. 46: 1046–1053. [DOI] [PubMed] [Google Scholar]
- Ruprecht C., Mendrinna A., Tohge T., Sampathkumar A., Klie S., Fernie A.R., Nikoloski Z., Persson S., Mutwil M. (2016). Famnet: A framework to identify multiplied modules driving pathway expansion in plants. Plant Physiol. 170: 1878–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht C., Mutwil M., Saxe F., Eder M., Nikoloski Z., Persson S. (2011). Large-scale co-expression approach to dissect secondary cell wall formation across plant species. Front. Plant Sci. 2: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht C., Proost S., Hernandez-Coronado M., Ortiz-Ramirez C., Lang D., Rensing S.A., Becker J.D., Vandepoele K., Mutwil M. (2017). Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules. Plant J. 90: 447–465. [DOI] [PubMed] [Google Scholar]
- Sah N.K., Singh S.N.P., Sahdev S., Banerji S., Jha V., Khan Z., Hasnain S.E. (2005). Indian herb ‘Sanjeevani’ (Selaginella bryopteris) can promote growth and protect against heat shock and apoptotic activities of ultra violet and oxidative stress. J. Biosci. 30: 499–505. [DOI] [PubMed] [Google Scholar]
- Sibout R., et al. (2017). Expression atlas and comparative coexpression network analyses reveal important genes involved in the formation of lignified cell wall in Brachypodium distachyon. New Phytol. 215: 1009–1025. [DOI] [PubMed] [Google Scholar]
- Silvester N., et al. (2018). The European nucleotide archive in 2017. Nucleic Acids Res. 46 (D1): D36–D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltani B.M., Ehlting J., Hamberger B., Douglas C.J. (2006). Multiple cis-regulatory elements regulate distinct and complex patterns of developmental and wound-induced expression of Arabidopsis thaliana 4CL gene family members. Planta 224: 1226–1238. [DOI] [PubMed] [Google Scholar]
- Somerville C., Youngs H., Taylor C., Davis S.C., Long S.P. (2010). Feedstocks for lignocellulosic biofuels. Science 329: 790–792. [DOI] [PubMed] [Google Scholar]
- Stöver B.C., Müller K.F. (2010). TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart J.M., Segal E., Koller D., Kim S.K. (2003). A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255. [DOI] [PubMed] [Google Scholar]
- Su Y., Sun C.M., Chuang H.H., Chang P.T. (2000). Studies on the cytotoxic mechanisms of ginkgetin in a human ovarian adenocarcinoma cell line. Naunyn Schmiedebergs Arch. Pharmacol. 362: 82–90. [DOI] [PubMed] [Google Scholar]
- Takabayashi A., Ishikawa N., Obayashi T., Ishida S., Obokata J., Endo T., Sato F. (2009). Three novel subunits of Arabidopsis chloroplastic NAD(P)H dehydrogenase identified by bioinformatic and reverse genetic approaches. Plant J. 57: 207–219. [DOI] [PubMed] [Google Scholar]
- Takahashi N., Lammens T., Boudolf V., Maes S., Yoshizumi T., De Jaeger G., Witters E., Inzé D., De Veylder L. (2008). The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J. 27: 1840–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegischer K., Tausz M., Wieser G., Grill D. (2002). Tree- and needle-age-dependent variations in antioxidants and photoprotective pigments in Norway spruce needles at the alpine timberline. Tree Physiol. 22: 591–596. [DOI] [PubMed] [Google Scholar]
- Tohge T., Fernie A.R. (2016). Specialized metabolites of the flavonol class mediate root phototropism and growth. Mol. Plant 9: 1554–1555. [DOI] [PubMed] [Google Scholar]
- Tohge T., Watanabe M., Hoefgen R., Fernie A.R. (2013). The evolution of phenylpropanoid metabolism in the green lineage. Crit. Rev. Biochem. Mol. Biol. 48: 123–152. [DOI] [PubMed] [Google Scholar]
- Usadel B., Obayashi T., Mutwil M., Giorgi F.M., Bassel G.W., Tanimoto M., Chow A., Steinhauser D., Persson S., Provart N.J. (2009). Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ. 32: 1633–1651. [DOI] [PubMed] [Google Scholar]
- Van Bel M., Diels T., Vancaester E., Kreft L., Botzki A., Van de Peer Y., Coppens F., Vandepoele K. (2018). PLAZA 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 46 (D1): D1190–D1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153: 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J.K., Akiyama T., Ralph J., Chapple C. (2011). Independent recruitment of an O-methyltransferase for syringyl lignin biosynthesis in Selaginella moellendorffii. Plant Cell 23: 2708–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J.K., Li X., Stout J., Chapple C. (2008). Independent origins of syringyl lignin in vascular plants. Proc. Natl. Acad. Sci. USA 105: 7887–7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J.K., Noel J.P. (2013). Chemodiversity in Selaginella: A reference system for parallel and convergent metabolic evolution in terrestrial plants. Front. Plant Sci. 4: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J.K., Philippe R.N., Noel J.P. (2012). The rise of chemodiversity in plants. Science 336: 1667–1670. [DOI] [PubMed] [Google Scholar]
- Wink M. (2015). Evolution of secondary metabolism in plants In Ecological Biochemistry: Environmental and Interspecies Interactions, Krauss G.-J., and Nies D.H., eds (Hoboken, NJ: Wiley-Blackwell; ), pp. 38–48. [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo E.R., Pokharel Y.R., Yang J.W., Lee S.Y., Kang K.W. (2006). Inhibition of nuclear factor-kappaB activation by 2′,8′′-biapigenin. Biol. Pharm. Bull. 29: 976–980. [DOI] [PubMed] [Google Scholar]
- Xiao S.J., Zhang C., Zou Q., Ji Z.L. (2010). TiSGeD: A database for tissue-specific genes. Bioinformatics 26: 1273–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., et al. (2009). Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinformatics 10 (Suppl 11): S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.J. (1991). The photoprotective role of carotenoids in higher plants. Physiol. Plant. 83: 702–708. [Google Scholar]
- Yu H., Luscombe N.M., Qian J., Gerstein M. (2003). Genomic analysis of gene expression relationships in transcriptional regulatory networks. Trends Genet. 19: 422–427. [DOI] [PubMed] [Google Scholar]
- Zhao J., He Q., Chen G., Wang L., Jin B. (2016). Regulation of non-coding RNAs in heat stress responses of plants. Front. Plant Sci. 7: 1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Lee C., Zhong R., Ye Z.H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Chen L., Zhang C., Hao P., Jing X., Li X. (2017). Global transcriptome analysis reveals extensive gene remodeling, alternative splicing and differential transcription profiles in non-seed vascular plant Selaginella moellendorffii. BMC Genomics 18 (Suppl 1): 1042. [DOI] [PMC free article] [PubMed] [Google Scholar]