Abstract
With the goal of charting plant transcriptional regulatory maps (i.e. transcription factors (TFs), cis-elements and interactions between them), we have upgraded the TF-centred database PlantTFDB (http://planttfdb.cbi.pku.edu.cn/) to a plant regulatory data and analysis platform PlantRegMap (http://plantregmap.cbi.pku.edu.cn/) over the past three years. In this version, we updated the annotations for the previously collected TFs and set up a new section, ‘extended TF repertoires’ (TFext), to allow users prompt access to the TF repertoires of newly sequenced species. In addition to our regular TF updates, we are dedicated to updating the data on cis-elements and functional interactions between TFs and cis-elements. We established genome-wide conservation landscapes for 63 representative plants and then developed an algorithm, FunTFBS, to screen for functional regulatory elements and interactions by coupling the base-varied binding affinities of TFs with the evolutionary footprints on their binding sites. Using the FunTFBS algorithm and the conservation landscapes, we further identified over 20 million functional TF binding sites (TFBSs) and two million functional interactions for 21 346 TFs, charting the functional regulatory maps of these 63 plants. These resources are publicly available at PlantRegMap (http://plantregmap.cbi.pku.edu.cn/) and a cloud-based mirror (http://plantregmap.gao-lab.org/), providing the plant research community with valuable resources for decoding plant transcriptional regulatory systems.
INTRODUCTION
Transcription factors (TFs) control gene expression by binding to specific cis-elements, which play essential roles in plant development and stress responses. Systematic identification of TFs, regulatory elements and functional interactions between them would greatly facilitate further mechanistic investigation (1,2). In the past decade, we have been dedicated to constructing a plant TF knowledge base (PlantTFDB) through identifying and annotating the genomic TF repertoires of 165 species covering the main lineages of green plants (3–6), and this resource has been widely used by the community. With TF binding motifs throughout the genome determined by experiments in plants (7,8) and in silico-mapped in 156 plants (6), directly scanning the TF binding motifs in the promoters of putative target genes is becoming a promising option. As prediction from direct scanning yields a rather high false positive rate, additional data such as DNase-seq footprints (9,10) and conserved elements (11–16) have been incorporated to screen for functional TFBSs. However, these data are available in only a few model plants (10,17), and conserved-element-based methods are still confounded by evolutionary constraints on other functional elements other than TF binding (18), hindering the systematic charting of transcriptional regulatory maps across the plant kingdom.
Comparisons of multiple related genomes with substantial divergence are widely used to detect evolutionary constraints and further identify functional elements (17,19–21). The availability of over 100 plant genomes provides a unique opportunity to calculate genome-wide evolutionary footprints and further infer plant functional regulatory maps. Here, we established the first genome-wide conservation landscapes for 63 representative plants and developed an algorithm for screening for functional transcriptional regulatory elements by coupling the base-varied binding affinities of TFs with the evolutionary footprints of their binding sites. Over 20 million functional TFBSs and 2 million functional interactions for 21 346 TFs were identified accordingly, charting the regulatory maps of these 63 plants. In addition, in response to the ever-increasing number of plant genomes, we introduced a new section, ‘extended TF repertoires’ (TFext), to enable users to access the TF repertoires of newly sequenced plants as soon as possible.
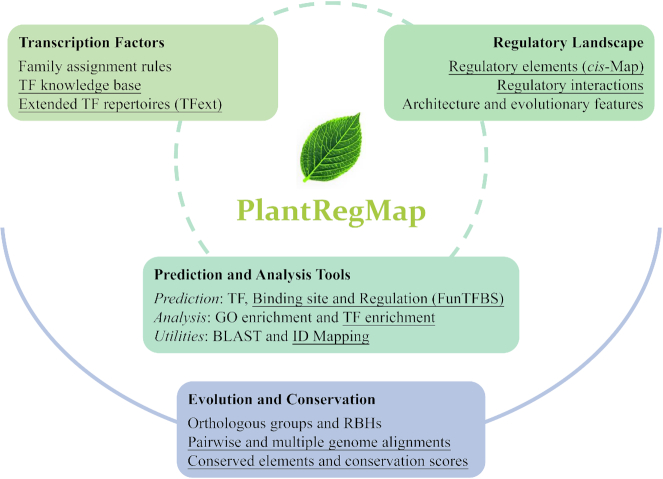
The PlantTFDB (i.e. plant TF knowledge base and TFext), conservation landscapes, regulatory landscapes and the set of prediction and analysis tools constitute an integrated plant regulatory data and analysis platform PlantRegMap (http://plantregmap.cbi.pku.edu.cn/, Figure 1) with a mirror on the cloud server (http://plantregmap.gao-lab.org/), providing the plant community with valuable resources for decoding plant transcriptional regulatory systems and genome sequences.
Figure 1.
The framework of PlantRegMap. The newly released/updated resources are marked with underlines (e.g. ‘Regulatory elements’).
RESULTS AND DISCUSSION
Updated annotations for previously collected TFs and extended TF repertoires
High-quality annotations for TFs (e.g. expert-curated description) is crucial for users to become familiar with the research status of TFs of interest and provides important clues for further study. Through extensively collecting expert-curated descriptions on expression, regulation and function as well as corresponding references for TFs from various public resources (22–24), we greatly improved the coverage of the collected TFs with such knowledge-based annotations (Table 1) and take another step towards constructing a TF knowledge base.
Table 1.
Summary of the update of expert-curated descriptions and references for the TF repertoires of 165 species
| PlantTFDB v4 | PlantTFDB v5 | |||||
|---|---|---|---|---|---|---|
| Type | Species | TF | Entry | Species | TF | Entry |
| Expression | 14 | 1 211 | 1 526 | 165 | 113 810 | 150 836 |
| Regulation | 7 | 620 | 620 | 161 | 65 726 | 66 721 |
| Function | 66 | 4 221 | 9 755 | 165 | 162 151 | 176 151 |
| References | 110 | 40 701 | 79 670 | 165 | 170 527 | 737 506 |
In addition to updating the previously collected TFs of 165 species with high-quality annotations, as the number of plant species with genome sequences is growing dramatically, we also created a new section, ‘extended TF repertoires’ (TFext, http://planttfdb.cbi.pku.edu.cn/index_ext.php), scheduled to update every six months, for users to access the TF repertoires of newly sequenced species quickly. Currently, it includes the TF repertoires of 52 plants (Supplementary Table S1) collected from multiple public resources (22,25–28). TFs collected in this section are provided with most essential information, including basic information, signature domain, CDS and protein sequences, nuclear localization signal and the corresponding best hit in A. thaliana. Similar to UniProtKB/TrEMBL (29), the records in TFext are taken as ‘precursors’ of those in the TF knowledge base and will be incorporated into the TF knowledge base after being curated with additional functional and evolutionary annotations such as expression profiles, multiple-species comparison as well as corresponding literature references during the regular update cycle of PlantRegMap.
Establishment of conservation landscapes in 63 plants
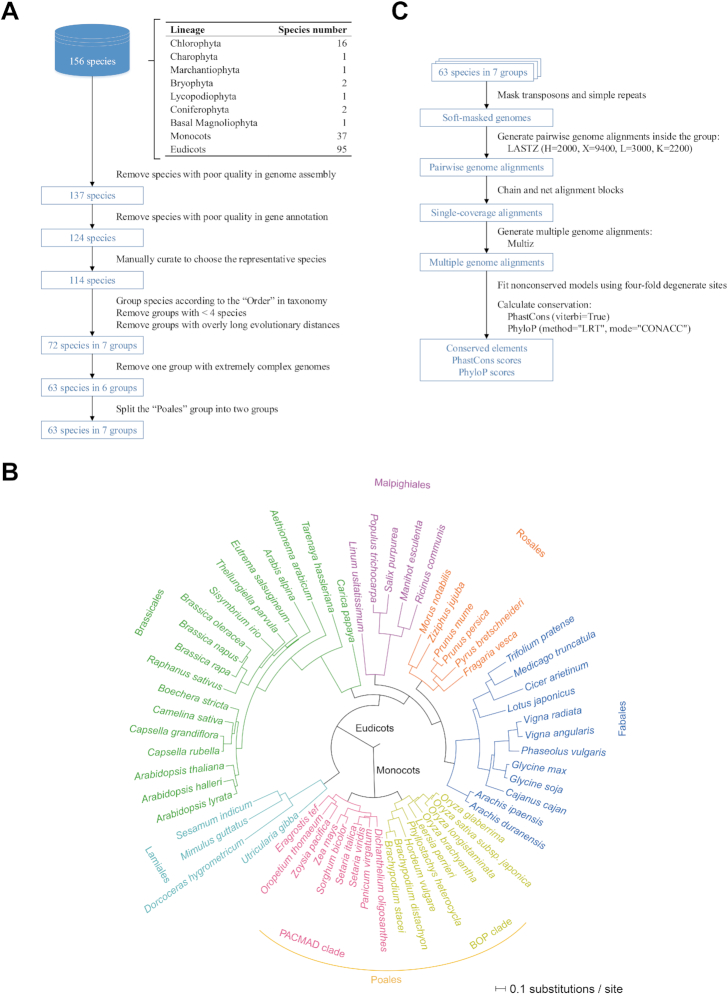
A high-quality, genome-wide conservation landscape is essential for detecting functional elements in genomic sequences (17,19–21). After considering both the number of species in a group and the divergence time inside the group, we chose 63 representative species and grouped them into seven groups according to taxonomy (30) (Figure 2A and B and Supplementary Table S2; see Supplementary Text for more details about species selection) with divergence times varying from 37 million years ago (MYA) (the PACMAD clade in Poales) to 106 MYA (Rosales) (Figure 3A). Following the established protocols (12,31) and an assessment of the LASTZ (32) parameters (Supplementary Figure S1 and Supplementary Text), we further generated 63 multiple genome alignments using each species as a reference and then detected evolutionary constraints (Figure 2C and Supplementary Text). Finally, we identified over 67 million conserved elements and calculated the base-by-base conservation scores (PhastCons and PhyloP) for over 22 billion base pairs, covering approximately 66% of the genome sequences and establishing the first conservation landscapes for the main lineages of angiosperms (Figures 2B and 3A and Supplementary Tables S3 and S4).
Figure 2.
The workflow for establishing conservation landscapes in 63 plants. (A) The workflow for choosing and grouping species for conservation analyses. (B) The phylogenetic tree for 63 species in seven groups, where the branch length within each group is shown as the substitution number per site at 4-fold degenerate sites. (C) The workflow for calculating conservation.
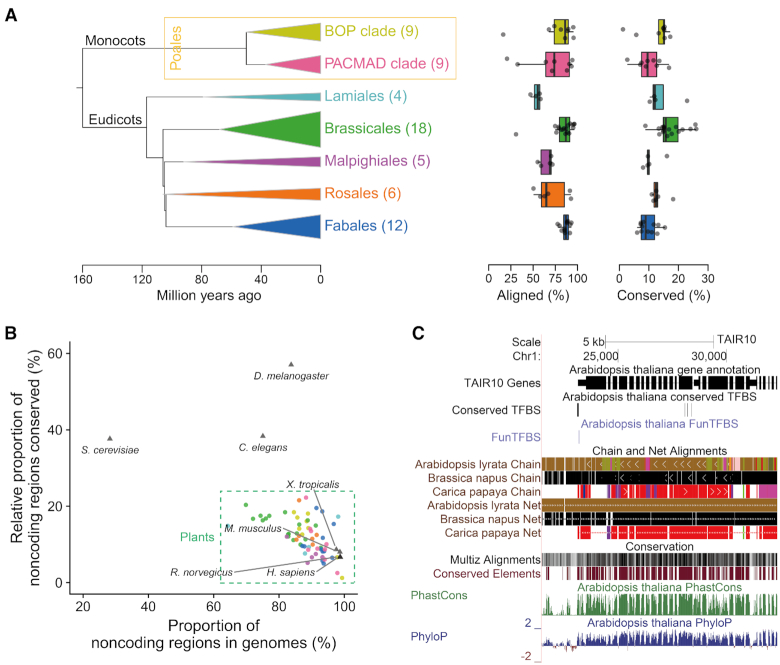
Figure 3.
The snapshot of conservation landscapes in 63 plants. (A) Summary of the genomes aligned and conserved for each group. Left: Phylogenetic tree showing the evolutionary relationships among groups. The width of the triangle corresponds to the maximum divergence time of the involved species, and the height corresponds to the species number (as shown in brackets) in each group. Middle and right: The box plots show the percentage of genomes aligned (middle) and conserved (right) for each group. (B) The proportion of noncoding regions in the genomes and their relative conservation ratios (proportion of noncoding regions conserved / proportion of coding regions conserved) for plants (63 species), animals (Homo sapiens, Mus musculus, Rattus norvegicus, Xenopus tropicalis, Drosophila melanogaster and Caenorhabditis elegans) and yeast (Saccharomyces cerevisiae). Each data point represents a species, and its colour represents the group. (C) Screenshot of the genome alignment and conservation resources in the genome browser. ‘Chain and Net Alignments’ are higher-level processing of the basic pairwise genome alignments (38), and ‘Multiz Alignment’ is the multiple genome alignment assembled by Multiz (39). Conserved elements and conservation scores (PhastCons score and PhyloP score) are calculated by PHAST (40) with fourfold degenerate sites as the neutral model.
For these seven groups, ∼54–87% of the genomes were aligned together, and at least 10–17% of the genomes were under evolutionary constraints (Figure 3A and Supplementary Tables S3 and S4). Compared with a previous study in A. thaliana (17), which used nine species in Brassicales to calculate conservation scores, our work which included more representative species (18 versus 9), aligned 20.49% (18.13 Mb) more genome sequences and detected 100.96% (4.84 Mb) more conserved noncoding regions with a higher accuracy (Supplementary Figure S2). Plants present a higher conserved ratio in the noncoding regions than vertebrates (i.e. Xenopus tropicalis, Mus musculus, Rattus norvegicus and Homo sapiens) but a lower ratio than organisms with a lower proportion of noncoding sequences in their genomes, such as fruit flies, worms and yeast (Figure 3B). Lineages undergoing more rounds of whole-genome duplication (e.g. Fabales) or sudden genome expansion (e.g. the PACMAD clade in Poales) show lower conservation ratios in their genomes, likely due to genomic degeneration after polyploidization or a lack of homologs in close species after sudden expansion. Notably, the conserved unannotated genomic regions (UGRs) in A. thaliana present a larger proportion covered by transcriptional signals (27% versus 10%) and a higher expression level than the nonconserved UGRs (Supplementary Figure S3), suggesting that many genes or functional elements remain to be decoded, even in the most well-annotated plant genome. Conservation landscapes in humans and fruit flies have been widely used to illustrate their genome sequences (20,21); thus, our conservation landscapes of 63 plants provide the research community with a unique chance to decode plant genomes. For users to conveniently access the conservation data, we have set up a genome browser (http://plantregmap.cbi.pku.edu.cn.org/cis-map.php) for users to visualize and decode plant genomes (Figure 3C).
Screening functional TFBSs by coupling the base-varied binding affinities of TFs with their evolutionary footprints
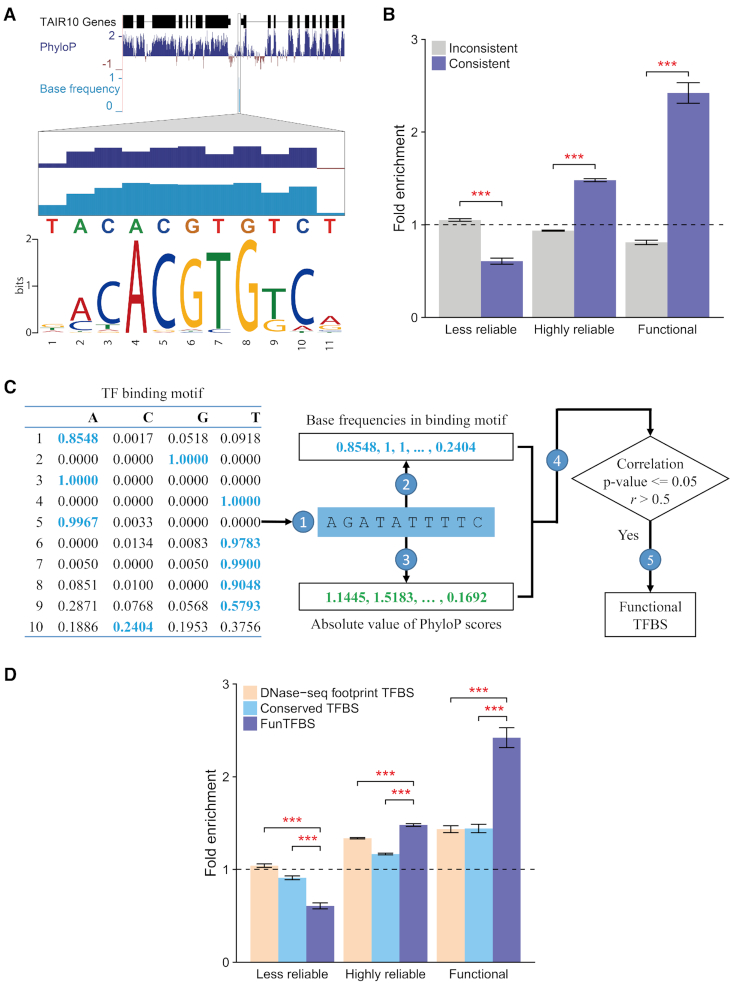
The establishment of conservation landscapes in the main lineages of angiosperms paves the way for systematic identification of functional TFBSs. However, other functional elements (such as noncoding RNAs and stem regions in RNA structures) may also contribute to the conservation of promoter sequences (18) (Supplementary Figure S4), confounding the use of algorithms that depend on conserved elements to screen for functional interactions. As the mutations of different base pairs on the TFBSs have different effects on the binding of TFs, we speculated that the base-varied binding affinity (base frequencies in the binding motifs) of the TF binding motifs would yield a consistent base-varied evolutionary constraint on the functional TFBSs (Figure 4A). To determine whether this feature could distinguish functional TFBSs from nonfunctional ones, we first generated an evaluation dataset by classifying the TFBSs identified from 124 ChIP-seq experiments for 21 TFs (33) into three classes: ‘Less reliable’, ‘Highly reliable’ and ‘Functional’. The ‘Less reliable’ and ‘Highly reliable’ TFBSs represent the TFBSs with low and high consistency among replicates, respectively, and the ‘Functional’ TFBSs are the ‘Highly reliable’ TFBSs that are further supported by expression data (see Supplementary Text for more details). A method with a higher screening efficiency would result in a lower percentage of TFBSs being ‘Less reliable’ but a higher percentage of TFBSs being ‘Highly reliable’ and ‘Functional’. Compared with the TFBSs whose conservation scores are inconsistent with their binding affinities, the consistent TFBSs are depleted in the ‘Less reliable’ TFBSs and enriched in the ‘Highly reliable’ TFBSs, particularly the ‘Functional’ ones (Figure 4B). Consistently, the functional and nonfunctional regulations were distinguished effectively (Supplementary Figure S5), suggesting that this feature allows screening of functional TFBSs and regulations.
Figure 4.
Screening for functional regulatory interactions by coupling the base-varied binding affinities of transcription factors (TFs) and consistent evolutionary constraints on their binding sites (TFBSs). (A) An example illustrating the consistency between the based-varied binding affinities (base frequency in the binding motifs) of the TFs and the evolutionary constraints on a functional TFBS. (B) Enrichment analysis of the TFBSs showing consistency or inconsistency between conservation scores and base frequencies in motifs in the ‘Less reliable’, ‘Highly reliable’, and ‘Functional’ categories of the evaluation dataset. The percentage of the TFBSs, directly scanned by motifs, included in each group of the evaluation dataset was used as the background. (Error bar indicates the standard deviation of the average fold change from 1 000 subsamples; the P-value was calculated based on the results from the 1000 subsamples, *** P-value < 0.001.) (C) An example workflow of the FunTFBS tool used to screen for functional transcription factor binding sites (TFBSs). For a TFBS candidate, the genomic sequences are extracted  , and the base frequency in the binding motifs
, and the base frequency in the binding motifs  and the PhyloP score
and the PhyloP score  for each base of the TFBS are calculated. Then, the Pearson correlation coefficient between the base frequencies in binding motifs and absolute value of PhyloP scores is calculated
for each base of the TFBS are calculated. Then, the Pearson correlation coefficient between the base frequencies in binding motifs and absolute value of PhyloP scores is calculated  , and only the TFBS candidates with a significant correlation (P-value ≤ 0.05) and correlation coefficient greater than 0.5 are kept as functional TFBSs
, and only the TFBS candidates with a significant correlation (P-value ≤ 0.05) and correlation coefficient greater than 0.5 are kept as functional TFBSs  . (D) Enrichment analysis of the TFBSs using the TFBSs screened by DNase-seq footprints (DNase-seq footprint TFBS), conserved elements (Conserved TFBS), and evolutionary footprints (FunTFBS).
. (D) Enrichment analysis of the TFBSs using the TFBSs screened by DNase-seq footprints (DNase-seq footprint TFBS), conserved elements (Conserved TFBS), and evolutionary footprints (FunTFBS).
Employing this feature, we developed an algorithm called FunTFBS to screen for functional TFBSs by identifying putative TFBSs whose conservation scores present a significant and strong correlation with the base frequencies in the binding motifs of TFs and to infer their functional regulatory interactions (Figure 4C and Supplementary Text). To determine whether our algorithm showed higher precision for functional TFBSs than the other motif-based methods, we first compared FunTFBS with the existing DNase-seq footprint-based and conserved-element-based methods using the evaluation dataset mentioned above. Our algorithm presented 42% and 33% decreases in the percentage of screened TFBSs that were designated ‘Less reliable’ TFBSs, but it presented 68% and 67% increases in the percentage of screened TFBSs that were designated ‘Functional’ TFBSs compared to the DNase-seq footprint-based and conserved-element-based methods, respectively (Figure 4D), suggesting that our algorithm can more efficiently screen for functional TFBSs.
We then assessed the precision of our algorithm in inferring transcriptional regulatory interactions based on experimentally validated interactions from the Arabidopsis transcriptional regulatory map (ATRM) (2). Our algorithm showed a 95–146% increase in the percentage of edges that were supported by the functional regulatory interactions in the ATRM compared with the DNase-seq footprint-based and conserved-element-based methods (Supplementary Figure S6), indicating the superiority of FunTFBS in inferring functional regulatory interactions. We further assessed the performance of our algorithm based on two other indexes: the percentage of regulatory pairs that coexist in the same biological process and the percentage of regulatory pairs that are highly correlated in expression (34), where higher numbers in the two indexes represent higher-quality interactions. Our algorithm showed the highest percentage of TFs and their targets coexisting in the two indexes (20–22% and 20–39% increases compared with the other two methods, respectively) (Supplementary Figures S7 and S8), further confirming the superiority of FunTFBS in screening for functional regulatory interactions.
Given the fact that A. thaliana, as the most popular model plant, has the most abundant experimentally validated, high-quality data on gene regulation, we performed most of evaluations in A. thaliana. Meanwhile, we further assessed the performance of FunTFBS in Glycine max, Oryza sativa and Arabidopsis lyrate based on TF ChIP-seq peaks downloaded from PCBase (35), and found that the TF binding sites screened by FunTFBS are significantly enriched in the corresponding ChIP peak regions compared with those screened by the conserved-element-based method (Supplementary Figure S9), suggesting the application potentials of FunTFBS in plants other than A. thaliana.
Functional regulatory maps in 63 plants
After confirming the superiority of FunTFBS in screening for functional regulatory interactions, we employed this method with integrated genomic TF binding motifs in 63 plants (6). Finally, we identified 21 997 501 functional TFBSs in 63 plant genomes, of which 2 493 577 are located in the gene promoter regions (TSS −500 bp to + 100 bp). Based on whether (at least) a functional TFBS of a TF presents at the promoter of a gene (if so, a regulatory interaction will be assigned between the TF and the gene), we further inferred 2 196 397 regulatory interactions for 21 346 TFs (Supplementary Table S5), charting the functional regulatory maps for the main lineages of angiosperms.
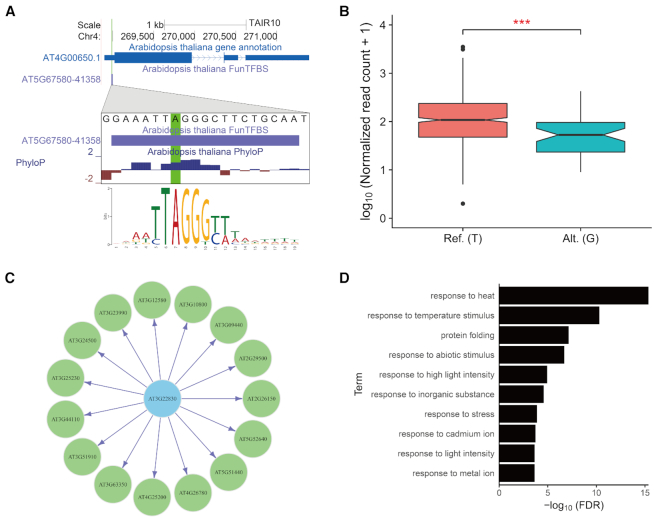
Our identified functional TFBSs are significantly enriched in expression quantitative trait loci (eQTLs) (Supplementary Figure S10), offering a unique chance to unveil the molecular mechanisms that underlie genetic variation and gene expression alteration. For example, according to the 1 203 transcriptomes from the 1001 Arabidopsis genomes project (36), one substitution (Chr4:268990 A>T) is associated with lower expression of AT4G00650 (Figure 5B), a major gene for variation in flowering time. By browsing our functional TFBSs, we found a TF (AT5G67580) that could bind to that position, and an A to T substitution would weaken its binding (Figure 5A), shedding light on the putative molecular mechanism. Moreover, the functional regulations also provide insight into the function of the TFs. For example, the target genes of a TF (AT3G22830) (Figure 5C) are enriched in ‘response to heat’ (Figure 5D), a biological process that corresponds well to the reported ‘heat stress response’ function of the TF (AT3G22830) (37).
Figure 5.
The application of FunTFBS for regulatory mechanism inference. (A and B) An eQTL (A to T substitution, highlighted in green) located in the TFBS of AT5G67580 predicted by FunTFBS (A) and the significant difference in expression of its target gene (B) (Wilcoxon rank sum test, *** P-value < 0.001). (C) The transcriptional regulatory network consisting of AT3G22830 and its target genes predicted by FunTFBS. (D) Enriched GO terms for the target genes of AT3G22830. Only the top 10 significant terms are shown.
A set of online tools for the prediction and analysis of transcriptional regulation
In the previous version, we have set up multiple online tools for transcriptional regulation prediction and analysis (6), which greatly facilitate exploration of the functional mechanisms of plant transcriptional regulatory systems by plant biologists. Here, we set up two novel online servers for ID mapping and functional TFBS screening and updated two existing servers for regulation prediction and analyses with newly released resources in this work.
ID mapping: Inconsistency between user-provided IDs and PlantRegMap-supported IDs (e.g. genome annotation IDs, UniProt AC/ID, Entrez Gene IDs and symbols) hinders users from using the sets of prediction and analyzing tools at PlantRegMap. Thus, this tool is set up to convert user-provided IDs to the PlantRegMap-supported IDs based on BLAST reciprocal best hits (RBHs).
FunTFBS: As mentioned above, this tool is employed to screen for functional TFBSs by coupling base-varied binding affinities of TFs and consistently evolutionary constraints on TFBSs.
Regulation Prediction: This tool infers regulatory interactions between TFs and input genes and finds over-represented upstream TFs for the input gene list. In this version, users can further refine the predicted regulatory interactions using the conserved elements and FunTFBS released in this work by simply choosing the output option.
TF enrichment: This tool enables users to find enriched upstream regulators for the input gene list based on pre-calculated regulations. In this version, functional regulations screened by conserved elements and FunTFBS are added to optimize the tool.
Resource availability
All the data released in this work (Table 2) can be freely accessed and downloaded at PlantRegMap (http://plantregmap.cbi.pku.edu.cn/), which includes PlantTFDB (i.e. TF knowledge base and Extended TF repertoires), conservation landscapes, regulatory maps and sets of prediction and analysis tools (Figure 1). In addition, we set up a mirror for PlantRegMap at the cloud server (http://plantregmap.gao-lab.org/) to provide continuous, high-quality service.
Table 2.
The resources released/updated in this work and their accessibility
| Module | Description | Content | URL |
|---|---|---|---|
| PlantTFDB | A portal for users to access the TF repertoires and corresponding annotations in plants. | TF knowledge base Extended TF repertoires (TFext) |
http://planttfdb.cbi.pku.edu.cn/index.php
http://planttfdb.cbi.pku.edu.cn/index_ext.php |
| cis-Map | A genome browser set up for users to browse genome alignments, conservation data and regulatory elements. | Pairwise genome alignments Multiple genome alignments Conserved elements PhastCons scores PhyloP scores Conserved TFBS FunTFBS |
http://plantregmap.cbi.pku.edu.cn/cis-map.php |
| Network | A portal for users to retrieve and visualize regulations. | Regulations inferred by conserved TFBS and FunTFBS |
http://plantregmap.cbi.pku.edu.cn/network.php |
| Tools | A set of online tools for ID mapping, functional TFBS screening, regulation prediction and TF enrichment analysis. | ID mapping FunTFBS |
http://plantregmap.cbi.pku.edu.cn/id_mapping.php
http://plantregmap.cbi.pku.edu.cn/funtfbs.php |
| Regulation prediction | http://plantregmap.cbi.pku.edu.cn/regulation_prediction.php | ||
| TF enrichment | http://plantregmap.cbi.pku.edu.cn/tf_enrichment.php | ||
| Download | A portal for users to download genome alignments, conservation data, regulatory elements and regulations. | Download individual files via HTTP Batch download via FTP |
http://plantregmap.cbi.pku.edu.cn/download.php
ftp://ftp.cbi.pku.edu.cn/pub/database/PlantRegMap/ |
CONCLUSION
In this work, we first updated the annotations for previously collected TFs and set up a new section, ‘extended TF repertoires’ (TFext), to promptly release the TF repertoires of newly sequenced species. Moreover, after the establishment of the first genome-wide conservation landscapes of 63 representative plants, we developed a more efficient algorithm to screen for functional TFBSs by coupling the base-varied binding affinities of the TFs and their evolutionary footprints. Using this algorithm, we systematically screened for functional TFBSs and regulatory interactions in 63 plants, charting functional regulatory maps for the main lineages of angiosperms. We believe that these resources will advance the understanding of plant transcriptional regulatory systems and allow further decoding of plant genome sequences.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Joint Genome Institute for the genome assemblies of unpublished species. The analyses were performed on the Computing Platform of the Center for Life Sciences of Peking University, and we thank Dr Fangjin Chen and Ting Fang for their assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [1470330]; China 863 Program [2015AA020108]; State Key Laboratory of Protein and Plant Gene Research; Beijing Advanced Innovation Center for Genomics (ICG) at Peking University; China Postdoctoral Science Foundation Grants [2014M560017 and 2015T80015 to J.J.]; Postdoctoral Fellowship from Peking University and Peking-Tsinghua Center for Life Sciences [to J.J.]. Funding for open access charge: State Key Laboratory of Protein and Plant Gene Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Taylor-Teeples M., Lin L., de Lucas M., Turco G., Toal T.W., Gaudinier A., Young N.F., Trabucco G.M., Veling M.T., Lamothe R. et al.. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature. 2015; 517:571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jin J., He K., Tang X., Li Z., Lv L., Zhao Y., Luo J., Gao G.. An arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors. Mol. Biol. Evol. 2015; 32:1767–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guo A.Y., Chen X., Gao G., Zhang H., Zhu Q.H., Liu X.C., Zhong Y.F., Gu X., He K., Luo J.. PlantTFDB: a comprehensive plant transcription factor database. Nucleic Acids Res. 2008; 36:D966–D969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang H., Jin J., Tang L., Zhao Y., Gu X., Gao G., Luo J.. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011; 39:D1114–D1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jin J., Zhang H., Kong L., Gao G., Luo J.. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014; 42:D1182–D1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jin J., Tian F., Yang D.C., Meng Y.Q., Kong L., Luo J., Gao G.. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017; 45:D1040–D1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weirauch M.T., Yang A., Albu M., Cote A.G., Montenegro-Montero A., Drewe P., Najafabadi H.S., Lambert S.A., Mann I., Cook K. et al.. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014; 158:1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O’Malley R.C., Huang S.S., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R.. Cistrome and Epicistrome features shape the regulatory DNA landscape. Cell. 2016; 165:1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neph S., Vierstra J., Stergachis A.B., Reynolds A.P., Haugen E., Vernot B., Thurman R.E., John S., Sandstrom R., Johnson A.K. et al.. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012; 489:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sullivan A.M., Arsovski A.A., Lempe J., Bubb K.L., Weirauch M.T., Sabo P.J., Sandstrom R., Thurman R.E., Neph S., Reynolds A.P. et al.. Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep. 2014; 8:2015–2030. [DOI] [PubMed] [Google Scholar]
- 11. Baxter L., Jironkin A., Hickman R., Moore J., Barrington C., Krusche P., Dyer N.P., Buchanan-Wollaston V., Tiskin A., Beynon J. et al.. Conserved noncoding sequences highlight shared components of regulatory networks in dicotyledonous plants. Plant Cell. 2012; 24:3949–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hupalo D., Kern A.D.. Conservation and functional element discovery in 20 angiosperm plant genomes. Mol. Biol. Evol. 2013; 30:1729–1744. [DOI] [PubMed] [Google Scholar]
- 13. Van de Velde J., Heyndrickx K.S., Vandepoele K.. Inference of transcriptional networks in Arabidopsis through conserved noncoding sequence analysis. Plant Cell. 2014; 26:2729–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burgess D., Freeling M.. The most deeply conserved noncoding sequences in plants serve similar functions to those in vertebrates despite large differences in evolutionary rates. Plant Cell. 2014; 26:946–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burgess D.G., Xu J., Freeling M.. Advances in understanding cis regulation of the plant gene with an emphasis on comparative genomics. Curr. Opin. Plant Biol. 2015; 27:141–147. [DOI] [PubMed] [Google Scholar]
- 16. Van de Velde J., Van Bel M., Vaneechoutte D., Vandepoele K.. A collection of conserved noncoding sequences to study gene regulation in flowering plants. Plant Physiol. 2016; 171:2586–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haudry A., Platts A.E., Vello E., Hoen D.R., Leclercq M., Williamson R.J., Forczek E., Joly-Lopez Z., Steffen J.G., Hazzouri K.M. et al.. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat. Genet. 2013; 45:891–898. [DOI] [PubMed] [Google Scholar]
- 18. Wang Y., Fan X., Lin F., He G., Terzaghi W., Zhu D., Deng X.W.. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:10359–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S.. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003; 423:241–254. [DOI] [PubMed] [Google Scholar]
- 20. Stark A., Lin M.F., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N. et al.. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007; 450:219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindblad-Toh K., Garber M., Zuk O., Lin M.F., Parker B.J., Washietl S., Kheradpour P., Ernst J., Jordan G., Mauceli E. et al.. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011; 478:476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sayers E.W., Agarwala R., Bolton E.E., Brister J.R., Canese K., Clark K., Connor R., Fiorini N., Funk K., Hefferon T. et al.. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019; 47:D23–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maglott D., Ostell J., Pruitt K.D., Tatusova T.. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011; 39:D52–D57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N. et al.. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012; 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cunningham F., Achuthan P., Akanni W., Allen J., Amode M.R., Armean I.M., Bennett R., Bhai J., Billis K., Boddu S. et al.. Ensembl 2019. Nucleic Acids Res. 2019; 47:D745–D751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jung S., Lee T., Cheng C.H., Buble K., Zheng P., Yu J., Humann J., Ficklin S.P., Gasic K., Scott K. et al.. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019; 47:D1137–D1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hirakawa H., Sumitomo K., Hisamatsu T., Nagano S., Shirasawa K., Higuchi Y., Kusaba M., Koshioka M., Nakano Y., Yagi M. et al.. De novo whole-genome assembly in Chrysanthemum seticuspe, a model species of Chrysanthemums, and its application to genetic and gene discovery analysis. DNA Res. 2019; 26:195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bairoch A., Apweiler R.. The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. Nucleic Acids Res. 1999; 27:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012; 40:D136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S. et al.. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005; 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harris R.S. Improved pairwise alignment of genomic DNA. 2007; The Pennsylvania State University; Ph.D. Thesis. [Google Scholar]
- 33. Song L., Huang S.C., Wise A., Castanon R., Nery J.R., Chen H., Watanabe M., Thomas J., Bar-Joseph Z., Ecker J.R.. A transcription factor hierarchy defines an environmental stress response network. Science. 2016; 354:aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang X., Wei X., Thijssen B., Das J., Lipkin S.M., Yu H.. Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat. Biotechnol. 2012; 30:159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chow C.N., Lee T.Y., Hung Y.C., Li G.Z., Tseng K.C., Liu Y.H., Kuo P.L., Zheng H.Q., Chang W.C.. PlantPAN3.0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019; 47:D1155–D1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. 1001 Genomes Consortium 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016; 166:481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang Y.C., Niu C.Y., Yang C.R., Jinn T.L.. The heat stress factor HSFA6b connects ABA signaling and ABA-Mediated heat responses. Plant Physiol. 2016; 172:1182–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Casper J., Zweig A.S., Villarreal C., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Karolchik D. et al.. The UCSC genome browser database: 2018 update. Nucleic Acids Res. 2018; 46:D762–D769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D. et al.. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004; 14:708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hubisz M.J., Pollard K.S., Siepel A.. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief. Bioinform. 2011; 12:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.