Abstract
Allosteric regulation is one of the most direct and efficient ways to fine-tune protein function; it is induced by the binding of a ligand at an allosteric site that is topographically distinct from an orthosteric site. The Allosteric Database (ASD, available online at http://mdl.shsmu.edu.cn/ASD) was developed ten years ago to provide comprehensive information related to allosteric regulation. In recent years, allosteric regulation has received great attention in biological research, bioengineering, and drug discovery, leading to the emergence of entire allosteric landscapes as allosteromes. To facilitate research from the perspective of the allosterome, in ASD 2019, novel features were curated as follows: (i) >10 000 potential allosteric sites of human proteins were deposited for allosteric drug discovery; (ii) 7 human allosterome maps, including protease and ion channel maps, were built to reveal allosteric evolution within families; (iii) 1312 somatic missense mutations at allosteric sites were collected from patient samples from 33 cancer types and (iv) 1493 pharmacophores extracted from allosteric sites were provided for modulator screening. Over the past ten years, the ASD has become a central resource for studying allosteric regulation and will play more important roles in both target identification and allosteric drug discovery in the future.
INTRODUCTION
The concept of allostery, or allosteric regulation, proposed in the 1960s, describes the existence of indirect coupling between two topographically and spatially distinct types of binding sites: allosteric and orthosteric sites (1,2). The binding of effectors (e.g. small molecules, ions, and DNA/RNA) to an allosteric site enables a protein to propagate perturbations from an allosteric site to an orthosteric site, ultimately achieving exquisite control of the protein's functional activities (3–7). Compared to the relatively conserved orthosteric sites in homologous protein families, the targeting of structurally less conserved allosteric sites by allosteric modulators confers the advantages of increased specificity and reduced side effects (8–14). In this context, the identification of allosteric modulators for a desired target is the subject of intense drug discovery efforts (15–20).
Despite the pharmacological merits of allosteric modulators, their discovery has faced a long-standing conundrum because of the difficulty in identifying allosteric sites via the experimental determination of allosteric protein-modulator complexes (21). Remarkably, recent computational advances in the detection of allosteric sites have significantly facilitated the identification of allosteric modulators for related targets (22–29), including sirtuin 6 (SIRT6) (15), signal transducer and activator of transcription 3 (STAT3) (30), glutathione peroxidase 4 (GPX4) (31) and cathepsin K (32). Based on the many computational allosteric approaches contributing to practical applications, we have proposed that allosteric modulator discovery has evolved from the realm of serendipity to structure-based drug design (33). Therefore, it is necessary to comprehensively annotate potential allosteric sites in known human allosteric proteins with both computational predictions and experimental determinations. These sites are useful for virtual screening and subsequent medicinal chemistry optimization of allosteric modulators.
As for orthosteric drugs, somatically acquired mutations at allosteric sites, referred to as allosteric mutations, may confer resistance to allosteric therapeutics (34–36). For example, single point mutations (A337V, P465S, V468F, and I502L) at the allosteric myristoyl binding site in the C-lobe of BCR-ABL kinase cause insensitivity to the allosteric inhibitor ABL001 in chronic myeloid leukemia (37). Analogously, the gatekeeper V211D mutation in the allosteric site of the N-lobe of MEK1 confers resistance to the allosteric inhibitor binimetinib in BRAF K601E-associated colon cancer (38). As such, it is advisable to systematically annotate known allosteric mutations from clinical samples. A comprehensive understanding of allosteric mutations, particularly clinically related variations, thus broadens the scope of available drug targets.
We have created a large, comprehensive database, the Allosteric Database (ASD), that has provided comprehensive information focusing on allosteric regulation since 2009 (39–41). Over the past decade, it has been visited >63 000 times from >100 countries around the world. ASD has garnered high acclaim from the scientific community (19,20,42) and has become a valuable resource for versatile allosteric applications. The fruitful results include the construction of benchmarking datasets for high-quality allosteric sites (ASBench (43)), the prediction of allosteric sites (e.g. Allosite (44) /AllositePro (45), AlloPred (46), PARS(47), CavityPlus (48), CryptoSite (49), pocketZebra (50), PocketDB (51), ALLO (52), ExProSE (53)), the assessment of allosteric interaction (Alloscore (54)), the exploration of allosteric communication (e.g. MCPath (55), AlloSigMA (26)), the analysis of allosteric mutations and evolution (e.g. AlloDriver (27), AlloMAPS (25), Trans-Omics (56)), and the screening of allosteric modulators (AlloFinder (30)). Such knowledge has promoted an upsurge in allosteric research, leading to the emergence of the framework of the allosterome. To further understand the landscape of the allosterome, in ASD 2019, we provide new allosteric features, including (i) the introduction of computational prediction of the locations of potential allosteric sites for known human allosteric proteins, (ii) the addition of allosteric mutations at allosteric sites of human proteins through large-scale exploration of somatic mutations in clinical samples across 33 cancer types, (iii) the construction of 7 new human allosterome maps and (iv) the development of allosteric site-based pharmacophore models. In addition, allosteric data such as experimentally determined allosteric proteins, sites, and modulators as well as allosteric-related diseases and allosteric drugs in different phases of clinical trials have also been updated and curated. Overall, the current ASD is useful for achieving a comprehensive understanding of the allosterome, facilitating the investigation of allosteric regulatory mechanisms and evolution and the discovery of allosteric drugs.
DATABASE GROWTH AND STATISTICS
New allosteric molecules and features, including structures, affinities, sites, functions, related diseases and external links, were collected using the methods described in our previous publications (39–41). According to the statistical data for ASD 2019 provided in Table 1, the known allosteric proteins have experienced a rapid increase over the past three years. Currently, 1949 allosteric proteins from various species are archived in the ASD, representing an increase of more than 32% (476 new proteins) since the last version. The allosteric phenomenon has been widely observed in 22 different protein categories defined by UniProt keyword ontology (57), and the growth of the data on allosteric proteins has been highly concentrated in the following categories: ion channels (from 93 to 154), GPCRs (from 114 to 169), kinases (from 201 to 249), and transferases (kinases excluded, from 21 to 119), which play key roles in disease therapies and biological pathways (3,4). Findings of additional classes of proteins have also confirmed the intrinsic properties of allostery, such as 147 hydrolases (proteases and phosphatases excluded), 126 oxidoreductases, 122 transcription factors, 120 transporters (ion channels excluded) and 87 lyases. The allosteric proteins in ASD 2019 come from 274 different species, and nearly 75% of the records belong to humans (43%) and bacterial species (31%) (see Figure 1). Notably, more than half of the 476 new proteins are from humans (244), and the acceleration of human record accumulation has contributed greatly to allosterome development. In addition to proteins, ASD 2019 contains 82070 allosteric modulators, composed of 31376 activators, 37471 inhibitors and 14622 regulators. Among the 10532 new curated modulators, 94% (9891) are exogenous allosteric lead compounds for human targets developed in the last three years, indicating a significant acceleration of the progress of allosteric drug discovery.
Table 1.
Data statistics for allosteric proteins and modulators in the updated ASD 2019
| Data category | ASD 2019 | ASD 2016 |
|---|---|---|
| Number of all proteins a | 1949 | 1473 |
| Number of kinases | 249 | 201 |
| Number of transferases | 185 | 144 |
| Number of GPCRs | 169 | 114 |
| Number of ion channels | 154 | 93 |
| Number of hydrolases | 147 | 117 |
| Number of oxidoreductases | 126 | 97 |
| Number of transcription factors | 122 | 94 |
| Number of transporters | 120 | 103 |
| Number of proteases | 103 | 78 |
| Number of lyases | 87 | 69 |
| Number of other proteinsb | 487 | 363 |
| Number of all modulators | 82 070 | 71 538 |
| Number of protein-modulator interactions | 89 554 | 75 462 |
| Number of crystal/NMR protein structures | 26 363 | 11 683 |
| Number of protein-modulator complex structures | 2542 | 1930 |
| Number of potential allosteric sites | 10 081 | 0 |
| Number of allosterome maps | 9 | 2 |
| Number of allosteric mutations | 1312 | 0 |
| Statistics of allosteric drug dataset | ||
| Number of allosteric drugs | 538 | 49 |
| Number of allosteric drug targets | 95 | 20 |
| Number of allosteric target-drug interactions | 638 | 56 |
| Number of allosteric-related diseases | 5983 | 3350 |
aIn the definition of the classifications for allosteric proteins in ASD 2019, several subtypes have been removed from the enzyme categories, including transferases (kinases excluded), hydrolases (proteases and phosphatases excluded), transporters (ion channels excluded), and receptors (nuclear receptors excluded).
bProtein categories including >50 proteins are displayed, while others are included in the ‘other proteins’.
Figure 1.
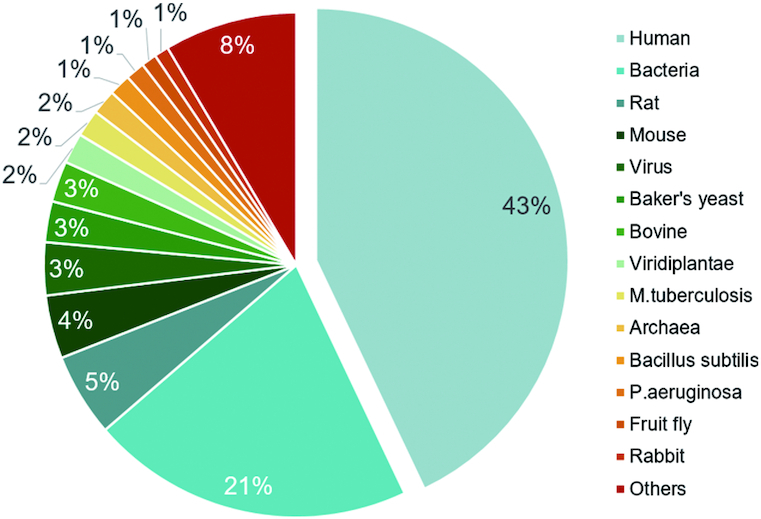
Statistics for allosteric proteins in ASD 2019.
In addition to these allosteric molecules, the data on their allosteric features and annotations are expanded in ASD 2019, including an ∼19% increase in allosteric interactions (from 75462 to 89554), an ∼32% increase in allosteric protein-modulator complexes (from 1930 to 2542), an ∼79% increase in related diseases (from 3350 to 5983), a 126% increase in protein structures (from 11683 to 26363), and a nearly 10-fold increase in allosteric chemicals in different phases of drug development (i.e. allosteric drugs, from 49 to 538). These allosteric drugs are well organized in the ‘ALLO-DRUG’ category under the FEATURES menu and classified into five phases: preclinical (451), phase I (23), phase II (36), phase III (9) and approved (19). Further analysis revealed that the targets associated with allosteric drugs include not only the traditional ion channels, GPCRs, and kinases but also many new types of proteins, including transporters, oxidoreductases, ligases, etc. These consistently rich data illustrate the ongoing expansion of the landscapes of the allosterome.
NEW FEATURES AND FUNCTIONS
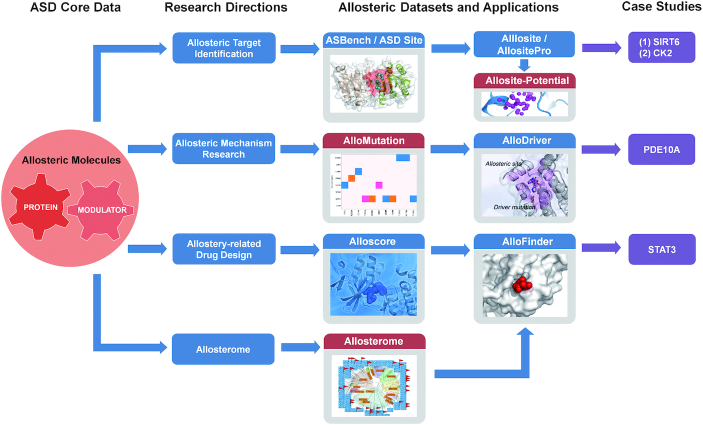
To further investigate the landscapes of the allosterome, novel allosteric features have been developed and organized in ASD 2019, including ‘ALLOSITE-POTENTIAL’ for potential allosteric sites in human proteins, ‘ALLOSTEROME’ for the evolution of allostery in a protein family, ‘ALLO-MUTATION’ for allosteric mutations in cancers, and ‘ALLO-DRUG’ for allosteric modulators in the preclinical and clinical phases. In addition to these data, two allosteric tools, ‘ALLO-PHARM’ and ‘ALLO-PATHWAY’, are integrated into the ‘TOOLS’ menu of the ASD, providing computational protocols for screening allosteric modulators and detecting allosteric pathways from orthosteric sites to allosteric sites. Figure 2 shows the workflow of the major ASD components or their combinations applied to allosteric drug design, target identification, mechanism research and allosterome analysis. The details of each new feature are described below.
Figure 2.
The workflow of the ASD major components or their combinations used for allosteric target identification, allosteric mechanism research, allostery-related drug design, and allosterome analysis. (i) The high-quality allosteric site benchmarking dataset (ASBench) derived from the ASD can be used to develop computational allosteric site prediction methods such as Allosite and AllositePro. The real cases include the identification of the SIRT6 allosteric activators MDL-800 and MDL-801 and a novel CK2α allosteric site. The Allosite-Potential datasets constructed by AllositePro could also provide an effective way for allosteric site identification throughout the human proteome. (ii) The allosteric mutation dataset in the ASD can be used to develop methods to predict allosteric driver mutations. The real cases are the identification of the allosteric L1143F driver mutation in human protein tyrosine phosphatase receptor type K (PTPRK) and the allosteric P360A driver mutation in human phosphodiesterase 10A (PDE10A). (iii) The allosterome maps can be used for allosteric evolutionary analysis of an allosteric site (an allosteric modulator) within its protein family (known allosteric modulators). (iv) The AlloFinder-based platform that integrates allosteric site prediction (AllositePro), allosteric interaction evaluation (Alloscore), and allosteric evolutionary analysis (Allosterome) can be used to automatically screen for allosteric modulators for a given target. A real case is the identification of a STAT3 allosteric inhibitor, K116. The new features in ASD 2019 are highlighted with red color.
POTENTIAL ALLOSTERIC SITES
Allosteric sites tend to be under lower sequence-conservation pressure and exhibit more structural diversity than conserved orthosteric sites, endowing allosteric modulators with the potential to achieve higher specificity, fewer side effects and lower toxicity (43–45,47,58). However, the slow pace of experimental validation seriously hinders the disclosure of the entire allosterome; allosteric sites have been identified in only a few proteins. Therefore, a large-scale dataset containing the potential allosteric sites of proteins is highly desirable and was therefore built in the current release. In ASD 2019, 10081 potential allosteric sites predicted from 4013 human proteins were constructed using our AllositePro method (45) (see Supporting Information) and are available under ‘ALLOSITE-POTENTIAL’ in the ‘FEATURES’ menu. These sites may be selectively shown through a combination of checkbox filters and protein subclass tree maps. Clicking the selected human protein causes an interactive page to open, and this page provides the details of potential allosteric sites, including protein information, a 3D representation window, a site selection panel, and a grid showing site properties. The proteins in the ALLOSITE-POTENTIAL dataset account for 75% of human proteins with PDB structures (59) and are widely distributed in all protein categories. These potential allosteric sites could provide an effective resource leading to the identification of allosteric regulation throughout the human proteome and be useful for allosteric drug discovery.
ALLOSTEROME MAPS
Owing to remarkable advances in structural biology and chemical biology in the past three years, an increasing number of allosteric sites have been determined by experimental crystallographic methods. These experimentally determined allosteric sites can be used to construct allosterome maps of protein families to illuminate the evolution of allosteric regulation (7). In ASD 2019, seven new human allosterome maps, including ‘Ion Channel’, ‘Protease’, ‘Phosphatase’, ‘Nuclear Receptor’, ‘Transferase’, ‘Transporter’, and ‘Hydrolase’ maps, were built to reveal the biological evolution of allosteric sites in a family; the ‘GPCR’ and ‘Kinase’ allosterome maps were also thoroughly updated by adding recently identified members (see Table 2). During the construction of the maps, all family annotations were collected from UniProt (57), GPCRDB (60), the Nuclear Receptor Resource (61), the Enzyme database (62), IUPHAR/BPS Pharmacology (63) and Wikipedia (https://en.wikipedia.org/), and the maps were constructed based on the multiple sequence alignment and dendrogram methods described previously (41).
Table 2.
Summary and statistics of the allosterome module in ASD 2019
| Number of human allosteric proteins | |||
|---|---|---|---|
| Allosterome categorya | All | Has modulators | Has complexes |
| Protein kinase allosterome | 113 | 85 | 30 |
| GPCR allosterome | 114 | 108 | 12 |
| Ion channel allosterome | 94 | 79 | 4 |
| Hydrolase allosterome | 66 | 52 | 16 |
| Transferase allosterome | 54 | 40 | 9 |
| Transporter allosterome | 53 | 34 | 7 |
| Protease allosterome | 45 | 34 | 13 |
| Phosphatase allosterome | 19 | 14 | 5 |
| Nuclear receptor allosterome | 17 | 12 | 4 |
aIn the definition of the classifications for allosteric proteins in ASD 2019, several subtypes have been removed from the enzyme categories, including transferases (kinases excluded), hydrolases (proteases and phosphatases excluded) and transporters (ion channels excluded).
The allosterome maps are organized in a grid under the ‘ALLOSTEROME’ feature in the ASD (Figure 2 and Table 2). Clicking on one map causes the details of the allosterome to be shown in a new tab, and the allosteric proteins with modulators and complexes are well annotated with white circle and red flag symbols, respectively. In nearly all the maps (except the transporter map), the percentage of allosteric proteins with modulators reached >70%, with particularly high percentages being achieved for the ‘GPCR’ (95%) and ‘Ion Channel’ allosteromes (84%). In four maps, >10 allosteric proteins with allosteric complexes are marked, including 30 kinases, 16 hydrolases, 13 proteases and 12 GPCRs. For each family, these allosteric sites actually cluster into different locations throughout the subfamilies. For example, 12 allosteric sites in human GPCRs show 3 different locations according to structural superimposition and 5 allosteric sites in ion channels are located in 4 different regions. The ‘ALLOSTEROME’ feature has systematically recorded allosteric protein accumulation from decades of allosterome development and offers the possibility of predicting the locations of novel sites based on known sites from the same family.
ALLOSTERIC MUTATIONS
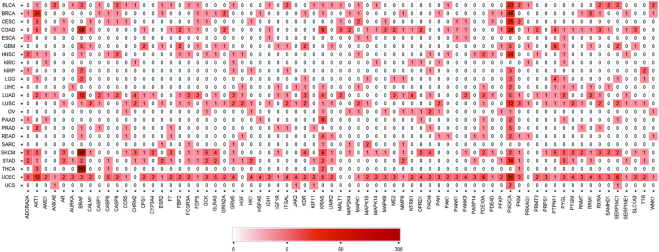
Mutations around an allosteric site may trap a protein in either an active or inactive conformation, and this structural disruption of uncontrolled proteins can perturb downstream signaling pathways and elicit multiple physiological and pathological conditions, including cancer (7). The increasing number of clinical mutations discovered from next-generation sequencing (NGS) data provides us with an unprecedented opportunity to systematically examine allosteric regulation related to disease conditions and promotes the development of small datasets and novel resources in this new field (24–27). To help biologists and chemists to easily explore allosteric mutations validated from NGS samples, in ASD 2019, the high-quality ‘Allo-Mutation’ dataset was established under the ‘FEATURES’ menu. Applying our previous workflow (24,27) to the most recent data on allosteric complexes and NGS samples, 1312 somatic missense mutations located in the allosteric sites of 133 human proteins were extracted from >100 000 patient samples from TCGA (64) and COSMIC (65) across 33 cancer types. Each allosteric mutation in a human protein from one kind of TCGA cancer is defined as a mutation record. The frequency of variants at allosteric sites for the allosteric proteins in each cancer type is depicted as a heatmap in Figure 3. For instance, allosteric mutations in BRAF present the highest frequency in THCA, SKCM and COAD samples, and allosteric mutations in PIK3CA have also been identified in numerous kinds of cancers. This module could be of great help for not only understanding the allosteric mechanism but also identifying allosteric targets in precision medicine.
Figure 3.
Heatmap displaying the frequency of mutations at the allosteric sites of human proteins in each cancer type. For each gene or each cancer type, records with a total frequency >6 are displayed in this heatmap.
ALLOSTERIC TOOLS
To facilitate studies of allosteric mechanisms and drug discovery, six specified allosteric computational tools are integrated under the ‘TOOLS’ menu of the ASD. ‘Allo-Pharm’ was built for allosteric target prediction by reverse searching of query molecules against allosteric pharmacophore datasets generated from allosteric complexes and ligand-free receptors. This structure-based pharmacophore tool provides an ensemble of spatially distributed chemical features that are essential for specific target-ligand binding (see Supporting Information). In total, 1493 pharmacophore models were extracted from 1374 allosteric protein-modulator complex structures, and 267 pharmacophore models were generated from unique allosteric pockets in 226 proteins via a pocket-based method (66–68). These pharmacophores of allosteric sites will be valuable for the high-throughput virtual screening of allosteric modulators. ‘Allo-Pathway’ is a tool for analyzing the allosteric communication pathway through revised dynamic network analysis (see Supporting Information). The allosteric pathways predicted by this tool could support an understanding of the roles of allostery in biological processes and signal transduction networks. In addition, AlloDriver (27) for allosteric mutagenesis prediction, AlloFinder (30) for the investigation of allosteric mechanisms, AllositePro (45) for allosteric site identification, and AlloScore (54) for the evaluation of allosteric interactions are integrated together in ASD 2019. Collectively, all of the above methods are expected to expedite the discovery of allosteric drugs and research on allosteric mechanisms.
DISCUSSION AND CONCLUSION
The discovery and development of first-in-class drugs is emerging as a focal point of the pharmaceutical industry (69,70). The development of an efficient strategy for discovering first-in-class drugs is therefore an area of intensive research. Allosteric drugs that exhibit a unique mechanism of action by targeting structurally diverse allosteric sites provide an innovative and promising opportunity for treating human diseases (11,12).
However, allosteric drug discovery is fraught with obstacles, and the greatest challenge is the effectiveness of using experimental methods to identify bona fide allosteric sites for a therapeutic target. As a supplement to experimental methods, computational predictions of allosteric sites in proteins made using the ASD data can expedite allosteric drug discovery. In particular, many of the computational predictions have been validated by experimental observations. For example, based on the Allosite-based allosteric site prediction (15), we recently discovered the first-in-class cellularly active SIRT6 allosteric activators MDL-800 and MDL-801 bound to the computationally predicted allosteric site and performed further crystallographic determinations. Through the AlloFinder-based platform (30) that integrates allosteric site prediction (AllositePro, an updated version of Allosite) (45), allosteric interacton evaluation (Alloscore) (54), and allosteric evolutionary analysis (Allosterome), we also recently identified a STAT3 allosteric inhibitor, K116. These findings strongly support the notion that computational allosteric methods have played an important role in structure-based drug design (23,33). In this study, we predicted a total of 10 081 potential allosteric sites for 4013 human allosteric proteins using AllositePro (45). This novel dataset of potential allosteric sites represents a breakthrough attributable to ASD 2019, because it is a valuable resource providing an attractive starting point for structure-based drug design at previously unexplored allosteric sites.
Allosteric mutations cause another challenge for allosteric drug discovery because of the notorious difficulty of identifying the locations of these mutation sites in proteins, as they may, for example, occur in allosteric ligand binding sites or allosteric communication pathways. The ‘Allo-Mutation’ dataset in ASD 2019, consisting of 1312 somatic missense mutations located in the allosteric sites of 133 human proteins, is useful for not only developing computational predictions of allosteric mutations but also uncovering new clinical disease targets. Such is the case for AlloDriver, which was recently developed based on this dataset (27). AlloDriver identified an L1143F driver mutation at the allosteric site of human protein tyrosine phosphatase receptor type K (PTPRK) from samples obtained from head and neck squamous cell carcinoma (HNSC) patients, which may expand the potential of PTPRK as a therapeutic target for HNSC (27). Additionally, the AlloMAPS database was built based on the theoretical prediction of allosteric effects of mutations on protein structure and function (25). Thus, this novel ‘Allo-Mutation’ dataset represents another breakthrough attributable to ASD 2019, because it is important for the identification of clinically relevant allosteric driver mutations that can expand the available therapeutic targets as well as for the investigation of allosteric communication pathways.
It has been suggested that allosteric sites are under lower evolutionary pressure than orthosteric sites. Notably, we have newly constructed human allosterome maps of seven protein families in ASD 2019, in addition to the update of two previous allosterome (GPCR and protein kinase) maps in ASD 2016. This ‘ALLOSTEROME’ dataset in ASD 2019 represents a useful improvement, which can reveal the specificity of an allosteric site in its protein family as well as trace the origin and evolution of allosteric regulation in protein families. Moreover, this information can also aid in the development of allosteric modulators that exploit differences in allosteric sites to achieve improved selectivity.
In summary, we anticipate that the new features in ASD 2019 will become a useful tool for allosteric modulator screening, allosteric mutation and evolutionary analysis, and allosteric mechanism research.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [91753117, 21778037, 81721004, 81603023, in part]; Shanghai Sailing Program [16YF1406500]; Shanghai Health and Family Planning Commission [20184Y0268]; Subgrant [2017YFC0908502] of the Chinese National Precise Medical Research key project [2017YFC0908500]; Innovation Program of Shanghai Municipal Education Commission [2019-01-07-00-01-E00036]; Shanghai Science and Technology Innovation [19431901600]; Shanghai Health and Family Planning System Excellent Subject Leader and Excellent Young Medical Talents Training Program [2018BR12], Subgrant [2018ZX09711002-001-006] of the National Science & Technology Major Project ‘Key New Drug Creation and Manufacturing Program’ [2018ZX09711002]; Natural Science Foundation of Shanghai Municipal Commission of Health and Family Planning [20164Y0033]; Shanghai Natural Science Foundation [16ZR1449300]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Changeux J.P. The concept of allosteric modulation: an overview. Drug Discov. Today Technol. 2013; 10:e223–e228. [DOI] [PubMed] [Google Scholar]
- 2. Liu J., Nussinov R.. Allostery: an overview of its history, concepts, methods, and applications. PLoS Comput. Biol. 2016; 12:e1004966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goodey N.M., Benkovic S.J.. Allosteric regulation and catalysis emerge via a common route. Nat. Chem. Biol. 2008; 4:474–482. [DOI] [PubMed] [Google Scholar]
- 4. Nussinov R., Tsai C.J., Liu J.. Principles of allosteric interactions in cell signaling. J. Am. Chem. Soc. 2014; 136:17692–17701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Good M., Tang G., Singleton J., Remenyi A., Lim W.A.. The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell. 2009; 136:1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Good M.C., Zalatan J.G., Lim W.A.. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011; 332:680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nussinov R., Tsai C.J.. Allostery in disease and in drug discovery. Cell. 2013; 153:293–305. [DOI] [PubMed] [Google Scholar]
- 8. Lu S., Ji M., Ni D., Zhang J.. Discovery of hidden allosteric sites as novel targets for allosteric drug design. Drug Discov. Today. 2018; 23:359–365. [DOI] [PubMed] [Google Scholar]
- 9. Nussinov R., Tsai C.J., Csermely P.. Allo-network drugs: harnessing allostery in cellular networks. Trends Pharmacol. Sci. 2011; 32:686–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nussinov R., Tsai C.J.. The different ways through which specificity works in orthosteric and allosteric drugs. Curr. Pharm. Des. 2012; 18:1311–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu S., Li S., Zhang J.. Harnessing allostery: a novel approach to drug discovery. Med. Res. Rev. 2014; 34:1242–1285. [DOI] [PubMed] [Google Scholar]
- 12. Wu P., Clausen M.H., Nielsen T.E.. Allosteric small-molecule kinase inhibitors. Pharmacol. Ther. 2015; 156:59–68. [DOI] [PubMed] [Google Scholar]
- 13. Clark A.C. Caspase allostery and conformational selection. Chem Rev. 2016; 116:6666–6706. [DOI] [PubMed] [Google Scholar]
- 14. Yu Z.H., Zhang Z.Y.. Regulatory mechanisms and novel therapeutic targeting strategies for protein tyrosine phosphatases. Chem Rev. 2018; 118:1069–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang Z., Zhao J., Deng W., Chen Y., Shang J., Song K., Zhang L., Wang C., Lu S., Yang X. et al.. Identification of a cellularly active SIRT6 allosteric activator. Nat. Chem. Biol. 2018; 14:1118–1126. [DOI] [PubMed] [Google Scholar]
- 16. Lu S., Zhang J.. Small molecule allosteric modulators of G-protein-coupled receptors: drug-target interactions. J. Med. Chem. 2019; 62:24–45. [DOI] [PubMed] [Google Scholar]
- 17. Chai H., Cheng X., Zhou B., Zhao L., Lin X., Huang D., Lu W., Lv H., Tang F., Zhang Q. et al.. Structure-based discovery of a subtype-selective inhibitor targeting a transient receptor potential vanilloid channel. J. Med. Chem. 2019; 62:1373–1384. [DOI] [PubMed] [Google Scholar]
- 18. Ni D., Li X., He X., Zhang H., Zhang J., Lu S.. Drugging K-Ras(G12C) through covalent inhibitors: Mission possible. Pharmacol. Ther. 2019; 202:1–17. [DOI] [PubMed] [Google Scholar]
- 19. Hauser A.S., Attwood M.M., Rask-Andersen M., Schioth H.B., Gloriam D.E.. Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov. 2017; 16:829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Changeux J.P., Christopoulos A.. Allosteric modulation as a unifying mechanism for receptor function and regulation. Cell. 2016; 166:1084–1102. [DOI] [PubMed] [Google Scholar]
- 21. Hardy J.A., Wells J.A.. Searching for new allosteric sites in enzymes. Curr. Opin. Struct. Biol. 2004; 14:706–715. [DOI] [PubMed] [Google Scholar]
- 22. Lu S., Huang W., Zhang J.. Recent computational advances in the identification of allosteric sites in proteins. Drug Discov. Today. 2014; 19:1595–1600. [DOI] [PubMed] [Google Scholar]
- 23. Lu S., Shen Q., Zhang J.. Allosteric methods and their applications: facilitating the discovery of allosteric drugs and the investigation of allosteric mechanisms. Acc. Chem. Res. 2019; 52:492–500. [DOI] [PubMed] [Google Scholar]
- 24. Shen Q., Cheng F., Song H., Lu W., Zhao J., An X., Liu M., Chen G., Zhao Z., Zhang J.. Proteome-Scale investigation of protein allosteric regulation perturbed by somatic mutations in 7,000 cancer genomes. Am. J. Hum. Genet. 2017; 100:5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tan Z.W., Tee W.V., Guarnera E., Booth L., Berezovsky I.N.. AlloMAPS: allosteric mutation analysis and polymorphism of signaling database. Nucleic Acids Res. 2019; 47:D265–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guarnera E., Tan Z.W., Zheng Z., Berezovsky I.N.. AlloSigMA: allosteric signaling and mutation analysis server. Bioinformatics. 2017; 33:3996–3998. [DOI] [PubMed] [Google Scholar]
- 27. Song K., Li Q., Gao W., Lu S., Shen Q., Liu X., Wu Y., Wang B., Lin H., Chen G. et al.. AlloDriver: a method for the identification and analysis of cancer driver targets. Nucleic Acids Res. 2019; 47:W315–W321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wodak S.J., Paci E., Dokholyan N.V., Berezovsky I.N., Horovitz A., Li J., Hilser V.J., Bahar I., Karanicolas J., Stock G. et al.. Allostery in its many disguises: from theory to applications. Structure. 2019; 27:566–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schueler-Furman O., Wodak S.J.. Computational approaches to investigating allostery. Curr. Opin. Struct. Biol. 2016; 41:159–171. [DOI] [PubMed] [Google Scholar]
- 30. Huang M., Song K., Liu X., Lu S., Shen Q., Wang R., Gao J., Hong Y., Li Q., Ni D. et al.. AlloFinder: a strategy for allosteric modulator discovery and allosterome analyses. Nucleic Acids Res. 2018; 46:W451–W458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li C., Deng X., Zhang W., Xie X., Conrad M., Liu Y., Angeli J.P.F., Lai L.. Novel allosteric activators for ferroptosis regulator glutathione peroxidase 4. J. Med. Chem. 2019; 62:266–275. [DOI] [PubMed] [Google Scholar]
- 32. Novinec M., Korenc M., Caflisch A., Ranganathan R., Lenarcic B., Baici A.. A novel allosteric mechanism in the cysteine peptidase cathepsin K discovered by computational methods. Nat. Commun. 2014; 5:3287. [DOI] [PubMed] [Google Scholar]
- 33. Lu S., He X., Ni D., Zhang J.. Allosteric modulator discovery: from serendipity to structure-based design. J. Med. Chem. 2019; 62:6405–6421. [DOI] [PubMed] [Google Scholar]
- 34. Agnello S., Brand M., Chellat M.F., Gazzola S., Riedl R.. A structural view on medicinal chemistry strategies against drug resistance. Angew. Chem. Int. Ed. Engl. 2019; 58:3300–3345. [DOI] [PubMed] [Google Scholar]
- 35. Shah N.H., Kuriyan J.. Understanding molecular mechanisms in cell signaling through natural and artificial sequence variation. Nat. Struct. Mol. Biol. 2019; 26:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kolch W., Kiel C.. From oncogenic mutation to dynamic code. Science. 2018; 361:844–845. [DOI] [PubMed] [Google Scholar]
- 37. Wylie A.A., Schoepfer J., Jahnke W., Cowan-Jacob S.W., Loo A., Furet P., Marzinzik A.L., Pelle X., Donovan J., Zhu W. et al.. The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature. 2017; 543:733–737. [DOI] [PubMed] [Google Scholar]
- 38. Gao Y., Maria A., Na N., Paula A.D.C., Gorelick A.N., Hechtman J.F., Carson J., Lefkowitz R.A., Weigelt B., Taylor B.S. et al.. V211D mutation in MEK1 causes resistance to MEK inhibitors in colon cancer. Cancer Discov. 2019; 9:1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang Z., Zhu L., Cao Y., Wu G., Liu X., Chen Y., Wang Q., Shi T., Zhao Y., Wang Y. et al.. ASD: a comprehensive database of allosteric proteins and modulators. Nucleic Acids Res. 2011; 39:D663–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang Z., Mou L., Shen Q., Lu S., Li C., Liu X., Wang G., Li S., Geng L., Liu Y. et al.. ASD v2.0: updated content and novel features focusing on allosteric regulation. Nucleic Acids Res. 2014; 42:D510–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shen Q., Wang G., Li S., Liu X., Lu S., Chen Z., Song K., Yan J., Geng L., Huang Z. et al.. ASD v3.0: unraveling allosteric regulation with structural mechanisms and biological networks. Nucleic Acids Res. 2016; 44:D527–D535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chang R.L., Andrews K., Kim D., Li Z., Godzik A., Palsson B.O.. Structural systems biology evaluation of metabolic thermotolerance in Escherichia coli. Science. 2013; 340:1220–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang W., Wang G., Shen Q., Liu X., Lu S., Geng L., Huang Z., Zhang J.. ASBench: benchmarking sets for allosteric discovery. Bioinformatics. 2015; 31:2598–2600. [DOI] [PubMed] [Google Scholar]
- 44. Huang W., Lu S., Huang Z., Liu X., Mou L., Luo Y., Zhao Y., Liu Y., Chen Z., Hou T. et al.. Allosite: a method for predicting allosteric sites. Bioinformatics. 2013; 29:2357–2359. [DOI] [PubMed] [Google Scholar]
- 45. Song K., Liu X., Huang W., Lu S., Shen Q., Zhang L., Zhang J.. Improved method for the identification and validation of allosteric sites. J. Chem. Inf. Model. 2017; 57:2358–2363. [DOI] [PubMed] [Google Scholar]
- 46. Greener J.G., Sternberg M.J.. AlloPred: prediction of allosteric pockets on proteins using normal mode perturbation analysis. BMC Bioinformatics. 2015; 16:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Panjkovich A., Daura X.. PARS: a web server for the prediction of protein allosteric and regulatory sites. Bioinformatics. 2014; 30:1314–1315. [DOI] [PubMed] [Google Scholar]
- 48. Xu Y., Wang S., Hu Q., Gao S., Ma X., Zhang W., Shen Y., Chen F., Lai L., Pei J.. CavityPlus: a web server for protein cavity detection with pharmacophore modelling, allosteric site identification and covalent ligand binding ability prediction. Nucleic Acids Res. 2018; 46:W374–W379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cimermancic P., Weinkam P., Rettenmaier T.J., Bichmann L., Keedy D.A., Woldeyes R.A., Schneidman-Duhovny D., Demerdash O.N., Mitchell J.C., Wells J.A. et al.. CryptoSite: Expanding the druggable proteome by characterization and prediction of cryptic binding sites. J. Mol. Biol. 2016; 428:709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Suplatov D., Kirilin E., Arbatsky M., Takhaveev V., Svedas V.. pocketZebra: a web-server for automated selection and classification of subfamily-specific binding sites by bioinformatic analysis of diverse protein families. Nucleic Acids Res. 2014; 42:W344–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bhagavat R., Sankar S., Srinivasan N., Chandra N.. An augmented pocketome: detection and analysis of small-molecule binding pockets in proteins of known 3D structure. Structure. 2018; 26:499–512. [DOI] [PubMed] [Google Scholar]
- 52. Akbar R., Helms V.. ALLO: A tool to discriminate and prioritize allosteric pockets. Chem. Biol. Drug Des. 2018; 91:845–853. [DOI] [PubMed] [Google Scholar]
- 53. Greener J.G., Filippis I., Sternberg M.J.E.. Predicting protein dynamics and allostery using multi-protein atomic distance constraints. Structure. 2017; 25:546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li S., Shen Q., Su M., Liu X., Lu S., Chen Z., Wang R., Zhang J.. Alloscore: a method for predicting allosteric ligand-protein interactions. Bioinformatics. 2016; 32:1574–1576. [DOI] [PubMed] [Google Scholar]
- 55. Kaya C., Armutlulu A., Ekesan S., Haliloglu T.. MCPath: Monte Carlo path generation approach to predict likely allosteric pathways and functional residues. Nucleic Acids Res. 2013; 41:W249–W255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yugi K., Kubota H., Hatano A., Kuroda S.. Trans-omics: how to reconstruct biochemical networks across multiple ‘Omic’ layers. Trends Biotechnol. 2016; 34:276–290. [DOI] [PubMed] [Google Scholar]
- 57. UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Panjkovich A., Daura X.. Exploiting protein flexibility to predict the location of allosteric sites. BMC Bioinformatics. 2012; 13:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Isberg V., Mordalski S., Munk C., Rataj K., Harpsoe K., Hauser A.S., Vroling B., Bojarski A.J., Vriend G., Gloriam D.E.. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res. 2016; 44:D356–D364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martinez E., Moore D.D., Keller E., Pearce D., Vanden Heuvel J.P., Robinson V., Gottlieb B., MacDonald P., Simons S. Jr, Sanchez E. et al.. The Nuclear Receptor Resource: a growing family. Nucleic Acids Res. 1998; 26:239–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000; 28:304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Harding S.D., Sharman J.L., Faccenda E., Southan C., Pawson A.J., Ireland S., Gray A.J.G., Bruce L., Alexander S.P.H., Anderton S. et al.. The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018; 46:D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tomczak K., Czerwinska P., Wiznerowicz M.. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. (Pozn.). 2015; 19:A68–A77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E. et al.. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019; 47:D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Koes D.R., Camacho C.J.. ZINCPharmer: pharmacophore search of the ZINC database. Nucleic Acids Res. 2012; 40:W409–W414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sunseri J., Koes D.R.. Pharmit: interactive exploration of chemical space. Nucleic Acids Res. 2016; 44:W442–W448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang X., Shen Y., Wang S., Li S., Zhang W., Liu X., Lai L., Pei J., Li H.. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017; 45:W356–W360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Eder J., Sedrani R., Wiesmann C.. The discovery of first-in-class drugs: origins and evolution. Nat. Rev. Drug Discov. 2014; 13:577–587. [DOI] [PubMed] [Google Scholar]
- 70. Lu S., Ni D., Wang C., He X., Lin H., Wang Z., Zhang J.. Deactivation pathway of Ras GTPase underlies conformational substates as targets for drug design. ACS Catal. 2019; 9:7188–7196. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.