Abstract
ProteomicsDB (https://www.ProteomicsDB.org) started as a protein-centric in-memory database for the exploration of large collections of quantitative mass spectrometry-based proteomics data. The data types and contents grew over time to include RNA-Seq expression data, drug-target interactions and cell line viability data. In this manuscript, we summarize new developments since the previous update that was published in Nucleic Acids Research in 2017. Over the past two years, we have enriched the data content by additional datasets and extended the platform to support protein turnover data. Another important new addition is that ProteomicsDB now supports the storage and visualization of data collected from other organisms, exemplified by Arabidopsis thaliana. Due to the generic design of ProteomicsDB, all analytical features available for the original human resource seamlessly transfer to other organisms. Furthermore, we introduce a new service in ProteomicsDB which allows users to upload their own expression datasets and analyze them alongside with data stored in ProteomicsDB. Initially, users will be able to make use of this feature in the interactive heat map functionality as well as the drug sensitivity prediction, but ultimately will be able to use all analytical features of ProteomicsDB in this way.
INTRODUCTION
ProteomicsDB (https://www.ProteomicsDB.org) is an in-memory database initially developed for the exploration of large quantities of quantitative human mass spectrometry-based proteomics data including the first draft of the human proteome (1). Among many features, it allows the real-time exploration and retrieval of protein abundance values across different tissues, cell lines, and body fluids via interactive expression heat maps and body maps. Today, ProteomicsDB supports multiple use cases across different disciplines and covering a wide range of data (2). For instance, tandem mass spectra, peptide identifications and peptide proteotypicity values can be used as starting points to develop targeted mass spectrometry assays. Because of the recent incorporation of a large amount of reference spectra from the ProteomeTools project (3,4) as well as spectra predicted by the artificial intelligence Prosit (5), both experimental and reference spectra can be used for assay development and to validate the identification of so far unobserved, or in fact any proteins. The integration of phenotypic data allows the exploration of the dose-dependent effect of drugs of interest (e.g. clinically approved drugs) on multiple cell lines (6–9). The dynamic identifier mapping in ProteomicsDB allows the integration of transcriptomics data from e.g. the Human Protein Atlas project (10) and Bgee (11), and thus facilitates the automated integration of different data sources within ProteomicsDB. This, in turn, allows the development of new tools. A wide range of drug-target interaction data can be visualized in ProteomicsDB as well, which enables the exploration of combination treatments in a dose-dependent protein-drug interaction graph in-silico.
ProteomicsDB is becoming an increasingly valuable resource in (proteomic) life science research, evidenced by the increasing number of external resources linking to ProteomicsDB, such as UniProt (12) and GeneCards (13), as well as resources making use of our application programming interface (API) to show e.g. protein expression information, as done by OmniPathDB (14) and Gene Info eXtension (GIX) (15).
In this version, we expanded the data content of ProteomicsDB by including additional publically available as well as in-house generated proteomic and transcriptomic studies. Furthermore, we expanded the drug-target interaction data now covering ∼1500 kinase inhibitors and tool compounds. The cell line viability data were enriched with an additional large dataset (16) now covering >20 000 drugs against 1500 cell lines. We further increased the amount of protein property information that is stored in ProteomicsDB, such as 13 000 melting points of proteins obtained by thermal proteome profiling (17). In addition, we expanded the biochemical assays section to include protein turnover data with synthesis and degradation curves for >6000 proteins. We further increased the number of reference tandem mass spectra in ProteomicsDB to >5 million from synthetic peptides and 40 million from predictions, which, in total, are represented by 3 billion fragment ions.
RESULTS
Overview
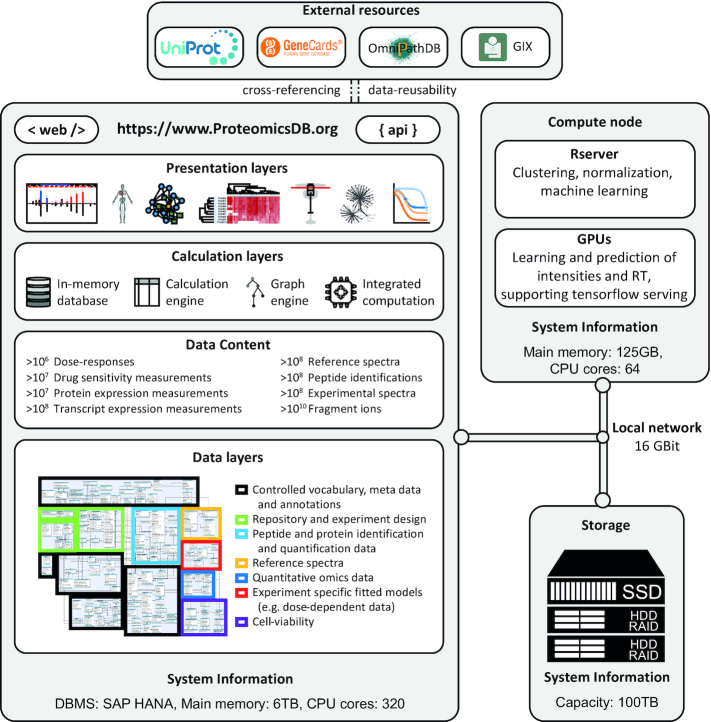
ProteomicsDB aims to provide real-time analytical functions to users, including computationally challenging tasks. For this purpose, ProteomicsDB was carefully designed and organized (Figure 1). It consists of a production unit, a computing unit, and a storage unit, all intra-connected via a 16Gbit local network. The production unit hosts the production server as well as the entire development and testing environment. The computing unit is one machine with a fully dockerized environment which currently handles two main tasks. First, an R server that handles R-procedures from ProteomicsDB such as the clustering available in the heat map. Second, a docker container with various services handling requests to our deep learning tool Prosit which is connected to two NVIDIA P100 GPU cards.
Figure 1.
The architecture of ProteomicsDB. The production unit hosts the SAP HANA in-memory database management system which involves three of the presented layers: the data layers, data content and the calculation layers. Parts of the calculation layers are shared between the production unit and the compute node, such as the clustering and correlation procedures for the interactive expression heat map which are calculated by the Rserver. Part of the data content is stored in the network storage unit, so that data are always available throughout the network if needed. The entire infrastructure is intra-connected via a 16 Gbit bandwidth local network that enables rapid communication and data transfer between units.
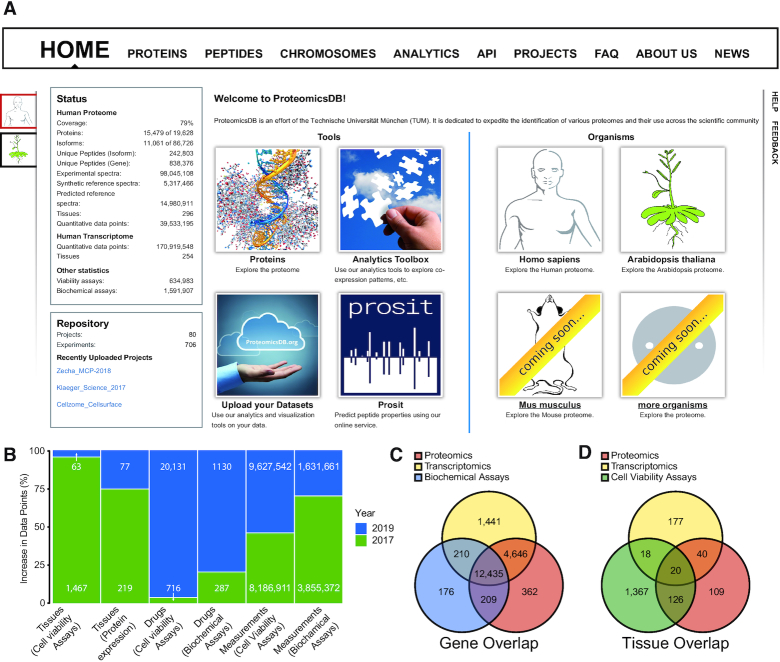
Over the past two years, the user interface and data content of ProteomicsDB were updated to accommodate new requirements such as hosting data from other organisms. Figure 2A shows the changes that were made to the front page such that users can select the organism of interest. Parts of the webpage have been renamed to be more generic and cover every organism, such as the ‘Human Proteins’ tab, which was renamed to ‘Proteins’. The front page statistics lists new information about the quantity of the data that is available for the chosen organism, including information about tissue coverage, quantitative multi-omics expression values, biochemical assay measurements as well as cell viability measurements. The main pane of the front page was redesigned to show the main features of the platform. It is now split into two sections. The left section provides direct links to the protein centric visualizations, the analytics toolbox, the new feature to upload custom data and a link to Prosit. The right section includes links that trigger the selection of the corresponding organism. To make organism selection available throughout the web interface, we additionally adjusted the left sidebar to show one icon per available organism. The ‘Feedback’ button that that was previously located in that position was transferred to the right pane below the ‘Help’ button. In light of these changes, all internal procedures and endpoints (e.g. API) were adjusted to support the new data types and organisms.
Figure 2.
Additions to ProteomicsDB. (A) The front page of ProteomicsDB has been adjusted to host new organisms as well as provide information about the quantity of the different data types that are stored in the database. (B) Barplot depicting the proportion and absolute number of data points added to ProteomicsDB (in blue) since the previous update manuscript in 2017 (green). (C) Venn diagram showing the number and overlap of genes for which proteomics, transcriptomics or biochemical assay data is available in ProteomicsDB. (D) Venn diagram showing the number and overlap of tissues (as well as cell lines and body fluids) for which the respective data types are available in ProteomicsDB.
Figure 2B depicts the data expansion in ProteomicsDB since 2017, grouped by categories. By re-analyzing and uploading more publically available proteomics studies, we increased the tissue coverage of ProteomicsDB by ∼70 human tissues and cell lines (+∼30%), to a total of almost 300 tissues and cell lines. The broader coverage of biological systems has direct impact on visualizations like the human body map or expression heat map. The plethora of data in ProteomicsDB allows not only the further online exploration of the proteome and its properties but also enables the development of new tools integrating different omics data sources. Currently, human proteomics and transcriptomics data are available for ∼17 000 genes and ∼60 tissues (Figure 2C, D). This large overlap enabled the implementation of a new missing value imputation approach which makes use of transcriptomics or proteomics data to estimate the presence and abundance of protein or RNA not covered in individual data sets. For ∼13 000 proteins, additional information derived from other biochemical assays such as melting behavior or synthesis or degradation curves are available. By integrating additional publicly available datasets, the overlap at the tissue- and protein level will increase further over the next years and eventually cover all the >1000 (cancer) cell lines for which we already have cell viability data. This, in turn, will aid the development of a better understanding of the molecular factors that govern the life of a particular cell.
New biochemical assay data, covering more protein properties
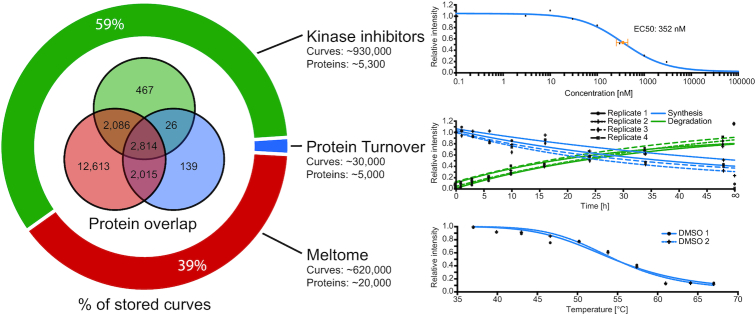
In addition to importing additional expression profile datasets, we further extended our biochemical assay portal by integrating the results of three additional studies covering target information of small molecule kinase inhibitors, melting (thermal aggregation) behavior of proteins and turnover data. First, in order to extend knowledge on druggable protein kinases (18), we imported ∼500 000 kinase inhibitor dose-response curves (Figure 3) covering 243 kinase inhibitors that are either approved for use or are in clinical trials (18) and ∼1300 tool compounds targeting kinases (unpublished). This data gives users a broader coverage and thus more options to select inhibitors to study a particular protein kinase. Various learnings might arise from such analysis, such as assessing the repurposing potential of clinical kinase inhibitors. Moreover, users can discover an appropriate molecule/inhibitor with respect to potency and selectivity to study the function of a particular kinase (19). Another use case is to identify inhibitors which share the same target(s) but have different off-targets, which can be used to identify and study the core signaling pathway of the shared target(s) or general on-target effects (18). In addition, the biochemical assay data and tools provided in ProteomicsDB (e.g. Inhibitor potency/selectivity analysis) can be used to discover new lead compounds for medicinal chemistry programs targeting a specific kinase of interest (20,21). The dose-response curves can be explored in the ‘Biochemical assay’ tab of the protein details view. This view allows users to filter the data by different properties, so that only compounds that fit the desired criteria will be displayed. For all curves, full experimental designs are stored for the users to browse and explore. For dose-response curves that belong to studies that are not published yet, the curve information is available but the experimental design, although fully imported, will only be shown when these studies are published. Second, the meltome data of ProteomicsDB was enriched with another study that covers the protein melting properties for many organisms (unpublished). Therefore, users can more thoroughly study the effect of temperature on selected proteins. We now cover the melting properties of ∼13 000 human proteins. ProteomicsDB thus provides an extensive resource and data-driven guidance on which temperature range should be used for e.g. a thermal shift assay or which temperature would be suitable for an isothermal dose response assay (ITDR). Third, we introduced a new assay type in the ‘Biochemical Assay’ tab which covers data from protein turnover measurements (synthesis and degradation). Users can obtain the half-life time of proteins of interest to assess their stability (22). This data can support the analysis of the mode of action of drugs (23) and might provide additional avenues into understanding the effectiveness of drugs in light of the stability of on- or off-target proteins (18). In total, ∼20 000 proteins (including isoforms) are covered by at least one and ∼3000 by all three biochemical assay types, providing potentially valuable insight into additional aspects of a protein's life cycle. As ProteomicsDB visualizes every curve (accessible via the ‘Biochemical assay’ tab in the ‘Protein Details’ view), users can assess the quality of each individual curve and underlying data points themselves.
Figure 3.
New biochemical assay data. The pie chart on the left shows the distribution of biochemical assay data available for three different applications. The Venn diagram inside the pie chart shows the overlap of proteins for which biochemical assay data of the respective type is available. The diagrams on the right show exemplary fitted curves for each biochemical assay type, accompanied by the number of curves and proteins that each assay covers.
Upload and online analysis of user expression data
Uploading expression profiles
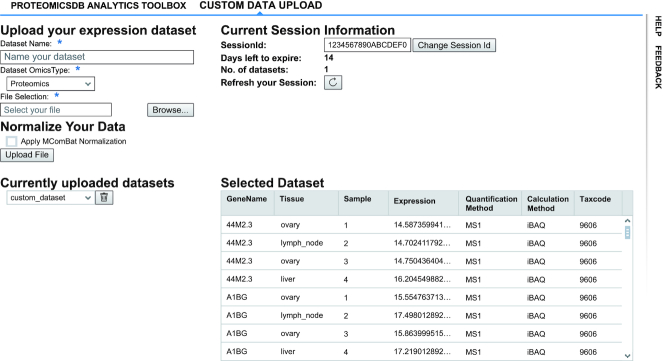
ProteomicsDB’s ability to interconnect and cross-reference data from various sources is one of its core features. However, this was so far only possible for data already stored in ProteomicsDB, limiting its usefulness for the interpretation of data acquired in a user laboratory. In order to fill this gap, we implemented a new feature called ‘Custom User Data Upload’ (Figure 4). Here, users can temporarily upload their expression profiles and optionally normalize them to the data stored in ProteomicsDB. On upload of a dataset, a temporary session is created in the database which can be accessed by a unique session ID. This session will automatically expire after 14 days, which will result in the permanent and not recoverable deletion of all corresponding data unless the user chooses to extend this period. Users can save and use their session ID to load their session to any other computer or browser. Data stored in such sessions are available via ODATA (https://www.odata.org) services within ProteomicsDB and will ultimately allow the integration into any existing analytical pipeline.
Figure 4.
Custom data analysis area of ProteomicsDB. The ‘Custom Data Upload’ tab enables users to upload their own expression datasets temporarily to ProteomicsDB. The datasets are session-specific so that no other user has access to this uploaded data.
The first use case we highlight is the comparison of custom expression data to expression data stored in ProteomicsDB. For this to be successful, we highly recommend making use of the normalization feature available upon upload. The uploaded expression profiles are normalized via MComBat (24) using the total sum normalized proteomics expression values of ProteomicsDB as a reference set. Because MComBat normalization depends on the calculation of a mean and variance for any given protein, only datasets with three or more samples can be normalized using this method. Every uploaded dataset has to adhere to a pre-defined comma-separated format (.csv files) where each row must provide the following information. (i) A gene name—HGNC symbol as the identifier, which will help us associate the uploaded proteins to the ones stored in ProteomicsDB and enable cross-dataset comparisons. (ii) A tissue or cell line name representing the origin of the measured sample, which will be used for visualizations. (iii) A sample name, which is important to separate samples with the same tissue of origin especially for the normalization step, as samples with the same sample and tissue/cell line name will be automatically aggregated as there is no way to separate them. (iv) The expression value of the corresponding protein in the sample in log10 scale, accompanied by the quantification and calculation method that was used, which will help with further comparisons of matching in-ProteomicsDB data. (v) The taxonomy code of each sample, which will allow dataset separation based on the selected organism, a feature which is discussed below. A detailed documentation on how to use this functionality as well as on the data upload format, can be found by clicking the ‘Help’ button that accompanies every view in ProteomicsDB (Figure 4).
Use of analytical tools on uploaded datasets
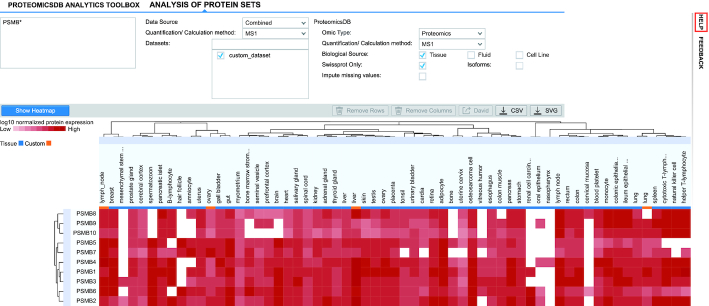
By uploading an expression dataset, back-end procedures take care of the data modelling and transformation, so that they are compatible to existing tools with no major differences to the data available in ProteomicsDB. The first tool making use of this is the interactive expression heat map. The heat map allows interactive visualization of expression patterns of multiple groups of proteins. Upon upload, users can choose a data source and focus their analysis on either data from ProteomicsDB, their own datasets noted as ‘User Data’ or the integration of both, noted as ‘Combined’. Because the heat map automatically aggregates tissues, duplicated tissue names provided in the custom dataset will appear as one column. The automatic mapping enables users to use all functionalities of the heat map, such as direct links to the ProteomicsDB’s protein summary views and perform GO enrichment analysis on the selected proteins. The ‘Combined’ option allows users to compare their data to data stored in ProteomicsDB. They can further allow a comparison of some or all datasets that they have uploaded to the in-database data. Users should expect that uploaded datasets that were not subjected to normalization during uploading, will clustered together. If the normalization step was enabled, then user samples should cluster with tissues or cell lines that have similar expression profiles in ProteomicsDB, ideally from the same origin. Figure 4 shows such an example where a custom dataset was co-clustered with data stored in ProteomicsDB. Some of the uploaded expression profiles of cell lines co-cluster with the respective cell lines stored in ProteomicsDB (here lung and liver samples). There are cases though (here ovary) that cluster with other tissues (here uterus). This feature enables users to find the closest cell lines for which ProteomicsDB contains, e.g. phenotypic information and explore compounds that may be effective in user cell lines.
Extended heat map features—missing values imputation
ProteomicsDB stores a large collection of transcriptomics expression profiles alongside the respective proteomic profiles. Having access to expression data from both sources and to the automatic mapping using the built-in Resource Identifier Relation Model, ProteomicsDB is able to perform data-driven missing value imputation using either data type. Especially proteomics data (depending on the depth of measurement) can show a large number of missing values. Data selected for imputation might come from different projects for both omics types. Even projects of the same omics type might differ in the distribution of their expression values. This phenomenon is commonly referred to as ‘batch effect’ and results in additional variance by the fact that we aggregate data across multiple ‘batches’. Here, the term ‘batch’ refers to experiments processed in one laboratory over a short time period using the same technological platform (25). We performed intra-omics normalization and batch effect correction using ComBat (26). Next, we apply MComBat (24) to perform inter-omics correction of systematic differences. MComBat, in contrast to ComBat, allows selecting a reference dataset so that all other datasets will be normalized based on the reference. Transcriptomics data are then transferred to the same scale of the proteomics expression data. Previous experiments showed that the correlation across all tissues between mRNA and protein expression data is higher with than without such an adjustment (27). Finally, we implemented the mRNA-guided missing value imputation method, described in (27). For this purpose, we train linear regression models and extrapolate protein abundance from transcriptomics abundance. To validate the performance of the generated models, we created artificial missing values in a random subset of the protein expression data that are stored in ProteomicsDB. We then used our models to extrapolate the protein abundances and compared them to two other common missing value imputation strategies: (a) replacing missing values with the minimum protein abundance of the corresponding sample and (b) random sampling from the corresponding sample's protein abundance distribution, as the created missing values originate from the whole abundance distribution. The mRNA-guided missing value imputation method showed the best correlation to the measured values (Supplementary Figure S1) which is why we implemented it. The entire procedure, from data normalization to training the regression model is performed by the R server (Figure 1). This is possible because the SAP HANA in-memory database management system supports direct connections to the R-server via proper adapters. Missing value imputation is available in the interactive heat map (Figure 5) and can be activated by the respective button. Once activated, and only if matching expression profiles are available, the model trained above and the adjusted transcriptomics expression data are used to fill in missing values in the protein expression matrix. The authors point out that missing value imputation can lead to issues and should therefore be carefully considered and evaluated on a case by case basis. Especially in the case of mRNA-guided missing value imputation, it becomes less accurate if the RNA dataset or protein expression data has a limited number of samples. Moreover, not all missing values can be imputed if RNASeq matching data is missing.
Figure 5.
Combined interactive expression heat map. User datasets can be clustered along with data stored in ProteomicsDB for a combined analysis. User datasets (marked in orange) that were normalized using MComBat subsequent to upload, cluster close to samples in ProteomicsDB (in blue) that were generated from the same or similar tissues or cell types.
Drug sensitivity prediction for proteomic profiles
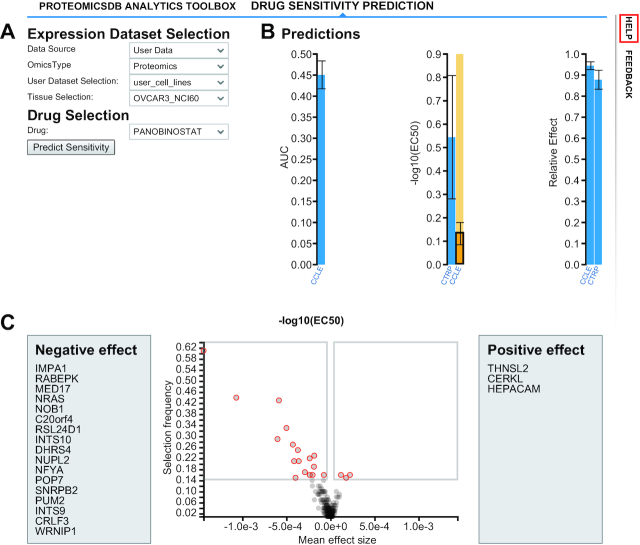
ProteomicsDB already covers a lot of phenotypic drug sensitivity information (Figure 2B) and to the best of our knowledge, no other platform exists which shows the full dose response curves across multiple resources including filters to the extent as ProteomicsDB’s cell viability viewer does. However, the list of cell lines for which this data is available is necessarily incomplete and likely entirely unavailable or impossible to generate if cells lines were derived from say patient tissue in a particular laboratory. In order to obtain an estimate of the susceptibility of such cell lines to drugs, without performing an experiment, ProteomicsDB provides a tool to model and estimate drug sensitivity, based on expression profiles. Recent proteome profiling of the NCI60 (28) and the CRC65 (27) cancer cell line panels, and an additional panel of 20 breast cancer cell lines (29) showed that protein signatures can predict drug sensitivity or resistance. On this basis, we implemented elastic net regression (30) in ProteomicsDB to model drug sensitivity as a function of quantitative protein expression profiles. This functionality can be used in the ‘Drug Sensitivity Prediction’ view (Figure 6). Here, users can select from a variety of tissues and cell lines whose proteomic profiles are stored in ProteomicsDB. Next, a drug or compound can be selected to check for its effect on the selected cell line (Figure 6A). Figure 6B shows the result of the prediction as bar plots - one for each predicted feature (area under the curve, pEC50, relative effect). Error bars show the range of the predictions of all bootstraps of the corresponding model. Each drug in ProteomicsDB might be accompanied by multiple models (multiple bars in each bar plot), because the drug may have been used in more than one drug sensitivity screen which was imported into ProteomicsDB (max. 4). It is important to point out that each model includes a certain set of predictor-proteins. If the sample on which a user wants to predict drug sensitivity does not contain some of the required proteins, prediction from some models is not possible. Selecting a bar of any bar plot generates a volcano plot (Figure 6C), which shows information for the interpretation of the trained model. The x-axis shows how strong the expression of a particular protein is associated with drug sensitivity or resistance, analogous to a correlation. The y-axis shows the number of bootstrap models contained the particular protein as a predictor, when training the elastic net model. Proteins that appear in the top left and right areas of the volcano plot (Figure 6C) are frequently selected from the models as predictors, as they have a high positive or negative correlation with drug sensitivity or resistance and can, therefore, represent potential biomarkers. Instead of predicting drug sensitivity on tissues or cell lines from ProteomicsDB, users also have the option to use this functionality on their own datasets, uploaded using the ‘Custom User Data Upload’ tab. Predictions can be applied to all user datasets, although it is highly recommended to use normalization upon uploading, as the models were trained on data stored in ProteomicsDB and expect values from the same or similar expression distributions.
Figure 6.
Drug sensitivity prediction. (A) Prediction is enabled for both, data stored in ProteomicsDB or user uploaded datasets. (B) This view visualizes the predicted sensitivity of a chosen cell line to a chosen drug expressed by area under the curve (AUC, left bar), the negative log of the effective concentration of the drug (EC50, middle bars) and the relative (cell killing) effect (right bars). If more than one bar is shown, more than one training data set was available for the particular drug and either one or several predictions are shown. (C). Each dot in the volcano plot, represents a protein that is associated to drug sensitivity or resistance on the basis of the elastic net model generated during training.
Real-time analytics and visualization for any organism
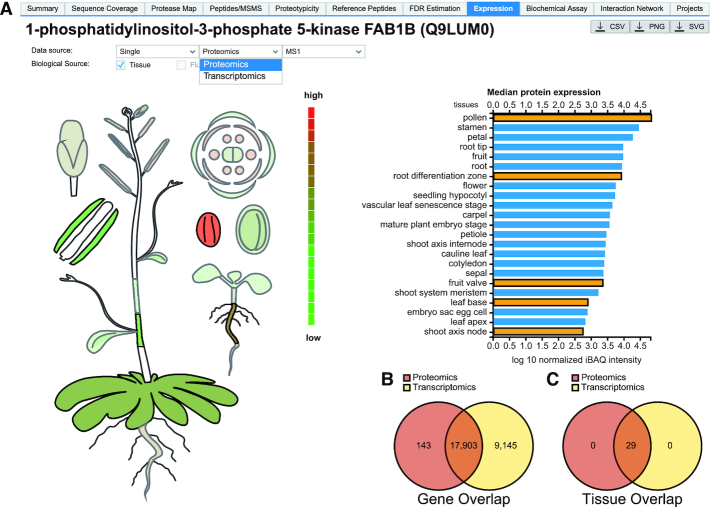
ProteomicsDB was initially developed for the exploration of the human proteome. As a result, every database view and endpoint was designed without explicit support for multiple organisms. In order to support the storage, handling and visualization of data from multiple organisms, all layers of ProteomicsDB (Figure 1) required modifications and extensive testing. In the new version presented here, we modified all backend procedures to support querying of data for a specific taxonomy. The API endpoints were modified to require a taxcode in order to respond with the desired data. With this functionality in place, we prepared the database and the data models to support and handle the protein sequence space of any organism. Similarly, the user interface was modified to support the visualization of data from a selected organism. Users can change the selected organisms by using the respective icons on the left hand side of each view, or directly on the front page of ProteomicsDB (Figure 2A). For the protein expression visualization, new interactive body maps for Arabidopsis thaliana and Mus musculus were generated (Figure 7A, Supplementary Figure S2) and function in the same way as the human body map.
Figure 7.
ProteomicsDB as a multi-organism and multi-omics platform. (A) Proteome or transcriptome expression data are visualized in the tissues of a chosen organism (left) and numerical expression data (medians in case multiple samples of the same tissue are available) are shown on the right for each tissue the protein was found in. Tissue bars selected by users turn orange and the respective tissue is highlighted on the body map on the left view projects the tissue aggregated omics expression values to the corresponding organism's body map. (B) Venn diagram is showing the overlap of gene-level data available for proteomics and transcriptomics for Arabidopsis thaliana. (C) Venn diagram showing the overlap of tissues for which proteomics and transcriptomics expression values are available in ProteomicsDB.
To bring Arabidopsis thaliana into ProteomicsDB, we downloaded, processed and imported the protein sequence space from UniProt, following the same mechanism as for human proteins. Upon import, appropriate decoy sequences were created for every protease, to allow false discovery (FDR) estimation by the picked FDR approach already implemented in ProteomicsDB (31). We furthermore imported the Plant Ontology (PO) (32) to be able to make use of ontologies for the different plant tissues. This step was not necessary for Mus musculus, since the Brenda Tissue Ontology (BTO) (33) that was previously imported into ProteomicsDB to support the analysis of human proteins covers any mammalian tissue. To complete the protein information and meta-data panel, we downloaded and imported protein domain information from SMART (34) using their RESTful API and GO annotations using the QuickGo-API of the European Bioinformatics Institute (EBI). Protein-protein interactions and functional pathway information were downloaded from STRING (35) and KEGG (36), respectively. The latter data were processed and transformed for import into our triple-store data model, which allows the automatic mapping of the respective STRING and KEGG identifiers to the corresponding UniProt accessions and our internal protein identifiers. With the meta-data imported, the proteomics and transcriptomics expression profiles for Arabidopsis thaliana were imported. The project covers 30 different tissues, including a tissue-derived cell line that was derived from callus tissue. Because of the generic design of ProteomicsDB, any analytical view (e.g. heat map) will work without further modifications for any other organism. However, due to the limited datasets available for phenotypic drug responses (and the respective drug targets), other views do not show any A. thaliana or M. musculus data yet.
As mentioned before, we have imported >5 million reference spectra acquired from synthetic human peptides in the ProteomeTools project. As a next step, we imported more than 10 million Prosit-predicted peptide spectra, in three different charge states and 3 different collision energies. By chance, these spectra also represent 70 000 peptides from Arabidopsis thaliana because their sequences are identical in either organism. In addition, we added predicted spectra for all peptides present in the experimental data set. Thus, akin to the human case, these reference spectra can be used to validate peptide identifications in experimental data using the mirror spectrum viewer integrated in ProteomicsDB. First, these are directly accessible in the ‘Peptides/MSMS’ tab of the ‘Protein Details’ view, where users can validate or invalidate i.e. one hit wonders (proteins which are only identified by a single peptide/spectrum), and more generally validate proteins/peptides in case the user wants confirmation that the protein is actually present in the sample of a project and consequently in a cell line or tissue in ProteomicsDB. Since ProteomicsDB contains up to 14 different types of reference spectra (11 fragmentation settings from ProteomeTools and 3 normalized collision energies from Prosit) as indicated in the list of available reference spectra, users can select the optimal match (37). Second, in the ‘Reference Peptides’ tab, where users can browse ProteomeTools and Prosit spectra for e.g. designing targeted mass spectrometric assays. The two separate views exist because for some proteins, no experimental spectra of endogenous proteins might be available, while many reference spectra might be available because the ProteomeTools synthesized all meaningful peptides for a hitherto unobserved protein. For proteins where experimental data from endogenous proteins is available, users can take experimental proteotypicity of peptides into account and thus rationalize which peptide to choose for an assay. Additionally, this view can be used to compare spectra created by different fragmentation methods and, more importantly, different collision energies to optimize their targeted assays for collision energies which generate desired fragment ions (e.g. highly intense and high m/z ions). Furthermore, spectra can now be downloaded in the mirrored spectrum viewer as msp-files. Finally, as mentioned above, ProteomicsDB is also ready to support Mus musculus data. However, the selection of mouse in ProteomicsDB will only be enabled once the data has been published.
FUTURE DIRECTIONS
The continuous updates introduced over the last years have transformed ProteomicsDB into a multi-omics resource for life science research covering proteomic and transcriptomic expression, pathway, protein-protein and protein-drug interactions, and cell viability data (Supplementary Figure S3). Many aspects of ProteomicsDB are already respecting the FAIR principles (38). For example, e.g. findability (F) is supported by unique identifiers, accessibility (A) via API endpoints including meta-data and reusability (R) by way of multiple online services taking advantage of ProteomicsDB’s API endpoints. However, more efforts are currently made to transform ProteomicsDB into a fully FAIR resource, e.g. by extending the API to allow access to all data stored in ProteomicsDB. One particular strength of ProteomicsDB is its versatile mapping service allowing the seamless connection between different data types. This enables subsequent modelling and data mining to further evolve ProteomicsDB from an information database to a knowledge platform. Along these lines, we plan to extend our analytical toolbox such that scientists in life science research can directly benefit from the wealth of data stored in ProteomicsDB. Here, we show the first steps into this direction by extending the toolbox as well as enabling users to upload their own expression data. Combined with ProteomicsDB’s flexible infrastructure, this will provide ease of use for data analysis, interpretation and machine learning capabilities not accessible to every laboratory or scientist. For this purpose, we are also planning to further extend the data content of ProteomicsDB to include, e.g. protein structures integrated with drug–target affinity data (20) or develop tools which allow the prediction of the target spaces of kinase inhibitors (39).
Two more extensions are planned that will allow the further integration and exploitation of reference spectra. The first one is to use synthetic or predicted reference spectra to systematically validate and assess the confidence of experimental data by evaluating their spectral similarity. As shown earlier, the integration of intensity information can lead to drastic improvements in either the number of identified peptides or the ability to differentiate correct from incorrect matches (5). Especially the latter will help to increase the confidence of each peptide identification and thus also increase the quality of identification and quantification results stored in ProteomicsDB. The second extension is the implementation of a smart tool which will allow users to build targeted assays based on data stored in ProteomicsDB as described.
Ultimately, the collected data and generated knowledge should culminate in actionable hypotheses. These may drive the design of laboratory experiments or eventually aid decision making in patient care. One way how ProteomicsDB could be used for the latter is by providing tools that assist molecular tumor boards. We plan to provide pipelines where researchers and clinicians will be able to upload the protein profiles of patient samples in a fully anonymized fashion and have in-depth bioinformatic analysis reports returned, spiked with a wide range of information including, e.g. protein and RNA abundance levels, biomarkers that predict sensitivity or resistance, potential off-label uses based on approved kinase inhibitors as well as general sample characterization, classification or origin identification based on similarities of molecular fingerprints.
DATA AVAILABILITY
ProteomicsDB is available at https://www.ProteomicsDB.org.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank all members of the Kuster laboratory for fruitful discussions and technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Science Foundation [SFB924, SFB1309, SFB1321]; German Federal Ministry of Education and Research (BMBF) [031L0008A, 031L0168]; SAP. Funding for open access charge: BMBF [031L0168].
Conflict of interest statement. T.S., S.G. and M.F. are founders and shareholders of msAId, which operates in the field of proteomics. M.W. and B.K. are founders and shareholders of OmicScouts and msAId, which operate in the field of proteomics. They have no operational role in the company. S.G., H.-C.E. and S.A. are employees of SAP SE. Neither company affiliation had any influence on the results presented in this study.
REFERENCES
- 1. Wilhelm M., Schlegl J., Hahne H., Gholami A.M., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H. et al.. Mass-spectrometry-based draft of the human proteome. Nature. 2014; 509:582–587. [DOI] [PubMed] [Google Scholar]
- 2. Schmidt T., Samaras P., Frejno M., Gessulat S., Barnert M., Kienegger H., Krcmar H., Schlegl J., Ehrlich H.C., Aiche S. et al.. ProteomicsDB. Nucleic Acids Res. 2018; 46:D1271–D1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zolg D.P., Wilhelm M., Schmidt T., Medard G., Zerweck J., Knaute T., Wenschuh H., Reimer U., Schnatbaum K., Kuster B.. ProteomeTools: Systematic characterization of 21 Post-translational protein modifications by liquid chromatography tandem mass spectrometry (LC-MS/MS) using synthetic peptides. Mol. Cell. Proteomics. 2018; 17:1850–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zolg D.P., Wilhelm M., Schnatbaum K., Zerweck J., Knaute T., Delanghe B., Bailey D.J., Gessulat S., Ehrlich H.C., Weininger M. et al.. Building ProteomeTools based on a complete synthetic human proteome. Nat. Methods. 2017; 14:259–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gessulat S., Schmidt T., Zolg D.P., Samaras P., Schnatbaum K., Zerweck J., Knaute T., Rechenberger J., Delanghe B., Huhmer A. et al.. Prosit: proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat. Methods. 2019; 16:509–518. [DOI] [PubMed] [Google Scholar]
- 6. Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H. et al.. A landscape of pharmacogenomic interactions in cancer. Cell. 2016; 166:740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rees M.G., Seashore-Ludlow B., Cheah J.H., Adams D.J., Price E.V., Gill S., Javaid S., Coletti M.E., Jones V.L., Bodycombe N.E. et al.. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2016; 12:109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Medico E., Russo M., Picco G., Cancelliere C., Valtorta E., Corti G., Buscarino M., Isella C., Lamba S., Martinoglio B. et al.. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat. Commun. 2015; 6:7002. [DOI] [PubMed] [Google Scholar]
- 9. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D. et al.. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S. et al.. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010; 28:1248–1250. [DOI] [PubMed] [Google Scholar]
- 11. Komljenovic A., Roux J., Wollbrett J., Robinson-Rechavi M., Bastian F.B.. BgeeDB, an R package for retrieval of curated expression datasets and for gene list expression localization enrichment tests [version 2; peer review: 2 approved, 1 approved with reservations]. F1000Res. 2018; 5:2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y. et al.. The GeneCards Suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics. 2016; 54:1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- 14. Turei D., Korcsmaros T., Saez-Rodriguez J.. OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods. 2016; 13:966–967. [DOI] [PubMed] [Google Scholar]
- 15. Knight J.D.R., Samavarchi-Tehrani P., Tyers M., Gingras A.C.. Gene Information eXtension (GIX): effortless retrieval of gene product information on any website. Nat. Methods. 2019; 16:665–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monga M., Sausville E.A.. Developmental therapeutics program at the NCI: molecular target and drug discovery process. Leukemia. 2002; 16:520–526. [DOI] [PubMed] [Google Scholar]
- 17. Savitski M.M., Reinhard F.B., Franken H., Werner T., Savitski M.F., Eberhard D., Martinez Molina D., Jafari R., Dovega R.B., Klaeger S. et al.. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science. 2014; 346:1255784. [DOI] [PubMed] [Google Scholar]
- 18. Klaeger S., Heinzlmeir S., Wilhelm M., Polzer H., Vick B., Koenig P.A., Reinecke M., Ruprecht B., Petzoldt S., Meng C. et al.. The target landscape of clinical kinase drugs. Science. 2017; 358:eaan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koch H., Busto M.E., Kramer K., Medard G., Kuster B.. Chemical proteomics uncovers EPHA2 as a mechanism of acquired resistance to small molecule EGFR kinase inhibition. J. Proteome Res. 2015; 14:2617–2625. [DOI] [PubMed] [Google Scholar]
- 20. Heinzlmeir S., Kudlinzki D., Sreeramulu S., Klaeger S., Gande S.L., Linhard V., Wilhelm M., Qiao H., Helm D., Ruprecht B. et al.. Chemical proteomics and structural biology define EPHA2 inhibition by clinical kinase drugs. ACS Chem. Biol. 2016; 11:3400–3411. [DOI] [PubMed] [Google Scholar]
- 21. Heinzlmeir S., Lohse J., Treiber T., Kudlinzki D., Linhard V., Gande S.L., Sreeramulu S., Saxena K., Liu X., Wilhelm M. et al.. Chemoproteomics-Aided medicinal chemistry for the discovery of EPHA2 inhibitors. Chem. Med. Chem. 2017; 12:999–1011. [DOI] [PubMed] [Google Scholar]
- 22. Zecha J., Meng C., Zolg D.P., Samaras P., Wilhelm M., Kuster B.. Peptide level turnover measurements enable the study of proteoform dynamics. Mol. Cell. Proteomics. 2018; 17:974–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Savitski M.M., Zinn N., Faelth-Savitski M., Poeckel D., Gade S., Becher I., Muelbaier M., Wagner A.J., Strohmer K., Werner T. et al.. Multiplexed proteome dynamics profiling reveals mechanisms controlling protein homeostasis. Cell. 2018; 173:260–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stein C.K., Qu P., Epstein J., Buros A., Rosenthal A., Crowley J., Morgan G., Barlogie B.. Removing batch effects from purified plasma cell gene expression microarrays with modified ComBat. BMC Bioinformatics. 2015; 16:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen C., Grennan K., Badner J., Zhang D., Gershon E., Jin L., Liu C.. Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PLoS One. 2011; 6:e17238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson W.E., Li C., Rabinovic A.. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007; 8:118–127. [DOI] [PubMed] [Google Scholar]
- 27. Frejno M., Zenezini Chiozzi R., Wilhelm M., Koch H., Zheng R., Klaeger S., Ruprecht B., Meng C., Kramer K., Jarzab A. et al.. Pharmacoproteomic characterisation of human colon and rectal cancer. Mol. Syst. Biol. 2017; 13:951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gholami A.M., Hahne H., Wu Z., Auer F.J., Meng C., Wilhelm M., Kuster B.. Global proteome analysis of the NCI-60 cell line panel. Cell Rep. 2013; 4:609–620. [DOI] [PubMed] [Google Scholar]
- 29. Lawrence R.T., Perez E.M., Hernandez D., Miller C.P., Haas K.M., Irie H.Y., Lee S.I., Blau C.A., Villen J.. The proteomic landscape of triple-negative breast cancer. Cell Rep. 2015; 11:630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zou H., Hastie T.. Regularization and variable selection via the elastic net. J. R. Stat. Soc.: B (Stat. Methodol.). 2005; 67:301–320. [Google Scholar]
- 31. Savitski M.M., Wilhelm M., Hahne H., Kuster B., Bantscheff M.. A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol. Cell. Proteomics. 2015; 14:2394–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walls R.L., Cooper L., Elser J., Gandolfo M.A., Mungall C.J., Smith B., Stevenson D.W., Jaiswal P.. The plant ontology facilitates comparisons of plant development stages across species. Front. Plant Sci. 2019; 10:631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gremse M., Chang A., Schomburg I., Grote A., Scheer M., Ebeling C., Schomburg D.. The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011; 39:D507–D513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Letunic I., Bork P.. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018; 46:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P. et al.. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019; 47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kanehisa M., Goto S.. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zolg D.P., Wilhelm M., Yu P., Knaute T., Zerweck J., Wenschuh H., Reimer U., Schnatbaum K., Kuster B.. PROCAL: a set of 40 peptide standards for retention time indexing, column performance monitoring, and collision energy calibration. Proteomics. 2017; 17:1700263. [DOI] [PubMed] [Google Scholar]
- 38. Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.W., da Silva Santos L.B., Bourne P.E. et al.. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li X., Li Z., Wu X., Xiong Z., Yang T., Fu Z., Liu X., Tan X., Zhong F., Wan X. et al.. Deep learning enhancing kinome-wide polypharmacology profiling: model construction and experiment validation. J. Med. Chem. 2019; doi:10.1021/acs.jmedchem.9b00855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ProteomicsDB is available at https://www.ProteomicsDB.org.