Abstract
Intron retention (IR) is a form of alternative splicing that has long been neglected in mammalian systems although it has been studied for decades in non-mammalian species such as plants, fungi, insects and viruses. It was generally assumed that mis-splicing, leading to the retention of introns, would have no physiological consequence other than reducing gene expression by nonsense-mediated decay. Relatively recent landmark discoveries have highlighted the pivotal role that IR serves in normal and disease-related human biology. Significant technical hurdles have been overcome, thereby enabling the robust detection and quantification of IR. Still, relatively little is known about the cis- and trans-acting modulators controlling this phenomenon. The fate of an intron to be, or not to be, retained in the mature transcript is the direct result of the influence exerted by numerous intrinsic and extrinsic factors at multiple levels of regulation. These factors have altered current biological paradigms and provided unexpected insights into the transcriptional landscape. In this review, we discuss the regulators of IR and methods to identify them. Our focus is primarily on mammals, however, we broaden the scope to non-mammalian organisms in which IR has been shown to be biologically relevant.
INTRODUCTION
The original assumption that one gene encodes only one polypeptide (1,2) was appended with the discovery of introns and mRNA splicing by Richard J. Roberts and Philip A. Sharp more than four decades ago (3,4). Considered for a long time as ‘junk DNA’, introns are now recognised as being central to the regulation of gene expression. Indeed, recent studies have unravelled a new facet of processed introns where they play an important role in regulating cell growth in yeast under stress conditions (5,6). The discovery of alternative splicing (AS), a mechanism of variable precursor mRNA processing, has changed ideas of how genes and proteins are defined. In this process, coding and non-coding gene fragments are alternatively skipped and joined, thus significantly enhancing both transcriptomic and proteomic complexity. Recent breakthroughs in high-throughput sequencing have shown that more than 95% of human multi-exonic genes are subject to AS and produce at least two alternative isoforms, demonstrating the central role of AS in normal biology (7–9).
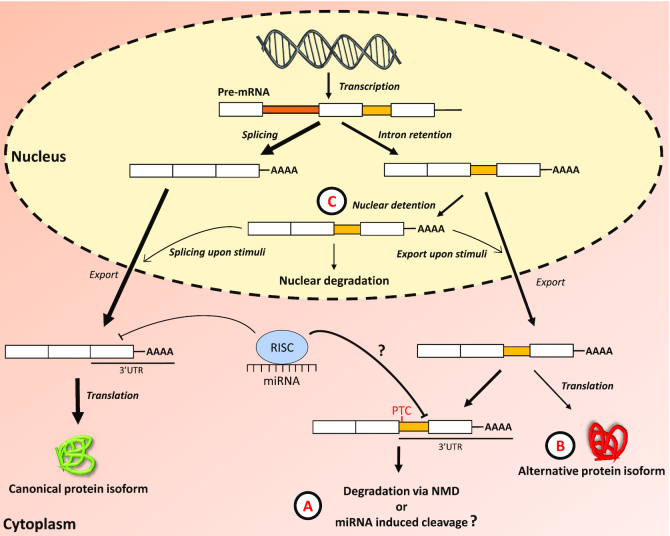
The main archetypes of AS are the cassette-type alternative exon usage, alternative 5′ or 3′ splice sites, mutually exclusive exons and intron retention (IR) (10,11). Unlike the other forms of AS, IR suffered from the misconception that it results from a malfunctioning spliceosome and associated factors. Consequently, IR has been relatively ignored in mammalian systems until recently. IR is characterized by the inclusion of one or more introns in mature mRNA transcripts, which can lead to diverse fates (Figure 1). Many introns contain in-frame premature termination codons (PTCs), leading to the detection and nonsense-mediated decay (NMD) of intron-containing transcripts through the cytoplasmic surveillance machinery (12–14). They may also be degraded via microRNA-induced mRNA cleavage, yet this mechanism remains to be experimentally validated (Figure 1A). Interaction with ribosomal subunits can lead to the translation of IR transcripts to generate alternative protein isoforms with novel functions (Figure 1B). Another class of intron-retained transcripts, called detained introns (ID), remain in the nucleus. ID transcripts can be degraded by a mechanism independent of NMD, where components of the nuclear RNA surveillance machinery, including the nuclear pore-associated protein Tpr and the exosome complex, are required (15). Alternatively, ID transcripts can be stored in the nucleus and rapidly exported to the cytoplasm upon specific stimuli (Figure 1C).
Figure 1.
The retention of an intronic sequence within the mature mRNA molecule can lead to multiple distinct fates. (A) IR transcripts are exported to the cytoplasm where they can interact with the ribosomal machinery, thus triggering their degradation via nonsense-mediated decay (NMD) if a premature termination codon (PTC) is encountered. The degradation of IR transcripts may also occur via interaction with the miRNA-RISC complex as retained introns located in the 3′ UTR of mature transcripts increase the number of miRNA binding sites. (B) The interaction with the ribosomal machinery can also lead to the production of alternative protein isoforms with novel biological functions. (C) IR transcripts can also be detained in the nucleus (ID), thus preventing the export of mRNAs and inhibiting translation. Detained IR transcripts may be degraded by nucleases, or are exported to the cytoplasm as fully spliced mRNAs or IR transcripts upon specific stimuli. Legend: question mark ?—degradation of IR transcripts via miRNA induced cleavage remains to be validated experimentally, RISC—RNA-induced silencing complex, miRNA—microRNA.
High-throughput RNA sequencing coupled with advances in bioinformatics algorithms to detect IR have enabled scientists to evaluate incidences of IR across species. While it has been shown that IR affects ∼80% of protein coding genes in humans (16), Braunschweig et al. compared IR occurrences across 11 vertebrate species and found that 50–75% of multi-exonic genes are affected in these species (17). Beyond that, IR is also widespread in fungi, insects, viruses and it represents the most frequent form of AS in plants (18,19). In contrast to humans where exon skipping is the most prevalent form of AS (20,21), IR occurs in 47% of all AS events in rice (22) and constitutes approximately two thirds of all AS events in Arabidopsis (23). The diverse fates of plant IR, similar to those demonstrated in animals (Figure 1), and its physiological importance have been recently reviewed elsewhere (24–26). Notably, most intron-retaining mRNA transcripts in plants do not contain PTCs and thus escape NMD (27). This indicates that introns are retained to fulfill a specific function in plants, for example, they are playing key roles in normal development and under stress conditions (28,29). Chaudhary et al. (24,26) recently proposed that plants employ AS to buffer against the stress-responsive transcriptome. IR would help by reducing the metabolic cost of translating newly synthesized transcripts and by selectively producing protein isoforms required for adaptation to varied stress conditions. As most intron-containing transcripts are sequestered in the nucleus under a particular stress or developmental stage in plants (30), the alteration of the transcriptional landscape by IR would directly influence the proteome composition under stress conditions. IR also plays a regulatory role during wheat growth. Pectin is an important component for cell wall remodelling during normal plant growth or following stress responses. Pectin methyl esterase inhibitor (PMEI) proteins control pectin activity in a tissue- or organ-specific manner. IR occurs in two of the PMEI genes to maintain an appropriate level of processed transcripts during flower development and pollen formation (31). Yet, the mechanisms contributing to the high incidence of IR in plants remain elusive.
In the single cell transcriptome of Schizosaccharomyces pombe, which contains over 2200 intron-containing-genes, IR is the dominant type of AS during meiosis. IR events appear to be co-associated (rather than mutually exclusive), suggesting coordinated IR regulation of meiosis in S. pombe (32). In Saccharomyces cerevisiae, orchestrated IR occurs during the transition from vegetative growth to sporulation as 13 meiosis-specific introns are incompletely spliced during exponential growth in rich media (33). Post-transcriptional regulation of the transition from mitosis to meiosis via IR is essential for yeast in order to maintain active growth. In Drosophila melanogaster, the generation of a bona fide protein translated from the Rieske Iron Sulphur (RFeSP) protein locus is a direct consequence of IR. Upon retention of the second intron within the RFeSP mRNA, the resulting novel protein accumulates in the mitochondrial compartment and lacks the iron sulphur domain that is otherwise present in the canonical isoform. However, it has been suggested that this alternative protein isoform, which is missing the functional domain, is not able to positively regulate mitochondrial respiration but would instead antagonise the function of the canonical RFeSP protein (34). IR has also been shown to be a key process in Human Immunodeficiency Virus (HIV) replication. Indeed, HIV encodes a viral accessory protein Rev, which is involved in the export and expression of many of the HIV mRNA species. Rev binds preferentially to unspliced viral RNAs to create a ribonucleoprotein complex. This complex, which recruits the host factor Exportin-1, allows the export of intact viral intron-containing RNAs to reach the cytoplasm for translation and virus packaging (35). IR is also widespread during parasite differentiation, which was shown in analyses of the intron-rich genomes of apicomplexan parasites. Additionally, IR prevents translation of stage specific isoforms of glycolytic enzymes in Toxoplasma gondii (36).
Although the relevance of IR has been known for decades in non-mammalian organisms, it has gained increased attention in recent years as its fundamental physiological importance in normal mouse and human biology and disease has been defined. The phenomenon of IR has emerged as an unexpected generator of variability in gene expression and transcriptomic diversity in various stages of development and in cell differentiation in mammals, e.g. in haematopoiesis (15,37–39). In human erythropoiesis, for example, an analysis of the RNA processing program has revealed the existence of abundant developmentally-dynamic IR events. Induction of high IR levels by splicing factors was suggested as a mechanism in late erythroblasts to modulate splicing events and to regulate gene expression (38). IR-coupled NMD also occurs during granulocyte differentiation in mice and humans, whereby groups of functionally related genes are co-regulated (39). For example, the expression of the nuclear lamina gene Lmnb1 is reduced due to increased levels of IR at the terminal stage of granulopoiesis triggering NMD of mature mRNA transcripts.
Different frequencies of IR observed between cell types further support its role as a mechanism to fine-tune gene expression. For example, this phenomenon is less frequent in muscle and embryonic stem cells (17) whereas there is a higher incidence rate of IR in neural and immune cell types. In these cells, IR facilitates the response to external stimuli that must occur rapidly to allow protein synthesis within a shorter timeframe (40,41) than what is required for de novo transcription and protein synthesis. During the differentiation of embryonic stem cells into neural progenitors, the up-regulation of genes with neuron-specific functions and down-regulation of genes involved in cell cycle progression is mediated by IR (17). IR has also been reported to play key roles in the response to stress. Under hypoxic conditions for example, IR becomes the predominant form of alternative splicing in tumour cells and leads to a reduction of HDAC6 and TP53BP1 expression, two proteins involved in cytotoxic response regulation and DNA repair (42). IR is also the most enriched form of AS during spermatogenesis. Stable ID transcripts have longer half-lives than constitutively spliced transcripts and are recruited to polyribosomes days after synthesis. This observation highlights the pivotal role of IR in the temporal expression of specific genes (43).
Aberrant IR has been reported in various diseases as a result of germline or somatic mutations at splice junctions. Mutations that induce mis-splicing can result in partial or complete IR (reviewed elsewhere, (44)) and potentially inactivate tumour suppressor genes in diverse cancers (45). In addition, IR characterizes the transcriptomes of many primary cancers (45–47). IR, generated by aberrant splicing, can produce abnormal transcripts that are translated into novel peptides. These peptides are recognized by the immune system (via MHC I) and potentially represent a source of tumour neoepitopes (48). Moreover, cancer-associated inactivating mutations in the spliceosome and associated factors might explain the consistently observed increase in IR levels. An acquired defect of the splicing machinery would induce a general increase of retained introns, especially the ones displaying weak splice sites (49). However, an analysis of the Cancer Genome Atlas (TCGA, (50)) tumour transcriptomes (tumour versus adjacent normal tissue) revealed a frequent global increase in IR levels despite an absence of mutations in the splicing machinery (46). This observation suggests that IR is not only regulated by a single elementary mechanism but rather a complex combination of cis- and trans-acting factors which orchestrate its regulation.
It is crucial to identify regulators of IR in order to provide approaches to control pathogenic IR changes. This review summarizes our current understanding on how IR is regulated in mammals and particularly in human. While emphasis has been placed on characteristics of mammalian intron-retaining transcripts and trans-acting modulators of IR, examples in non-mammalian organisms have been provided to highlight the diverse biological functions of IR. Experimental and computational approaches to identify the regulators of IR are detailed in the final section.
INTRINSIC FEATURES OF INTRON RETENTION
Sequence features
Certain sequence features are associated with an increase in the likelihood that an intron will be retained (Table 1) and the characterization of such features has been examined in many global gene regulation studies (16,17,46,51). In these studies, the cis-acting sequence features influencing the retention of certain introns include the presence of weaker splice sites at the 5′ and 3′ ends of the intron, hampering the ability of the spliceosome to recognise introns that would otherwise be spliced (52–54). Studies on specific IR events in human genes have shown that the excision of highly retained introns could be increased by strengthening the sub-optimal splice sites flanking the retained introns (55,56). Splice site mutation experiments in Drosophilia and Schizosaccharomyces have also demonstrated that mis-recognition of a single splice site could lead to IR (57,58). IR has also been shown to be associated with weak splice sites in plants, fungi and protists (19). Put simply, a decrease in splice site strength leads to higher relative frequencies of IR. While such correlations provide a conspicuous and intuitive justification of IR, they are not sufficient to be considered as sole contributors.
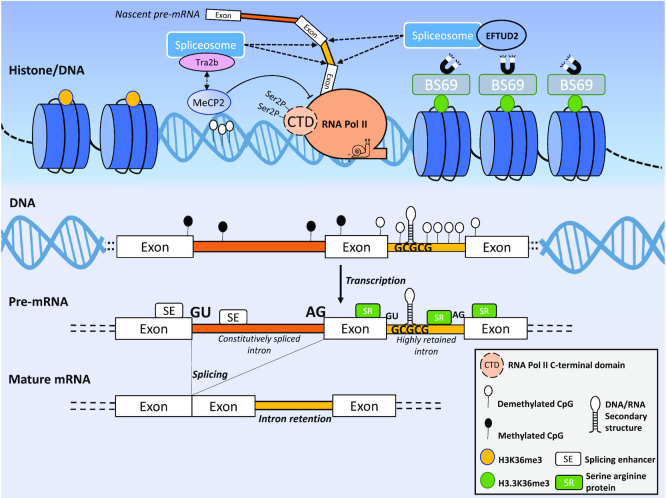
Table 1.
IR characteristics displayed in Figure 2 that positively regulate IR
| Level | Features | Contribution to IR | References |
|---|---|---|---|
| Histone/DNA | H3.3K36me3 Histone modification (magnets) | Chromatin-bound BS69 (via H3.3K36me3-mediated recruitment) interacts with snRNPs including EFTUD2, a component of the U5 small ribonucleoprotein complex, and destabilizes the spliceosome complex. BS69 has also been shown to repress RNA Pol II elongation | (77,88–90) |
| Slow RNA Pol II elongation (snail) | Impaired recognition and splicing of constitutive introns | (17,78,163) | |
| DNA | High CpG density/Reduced CpG methylation | Impaired binding of MeCP2 and recruitment of splicing factors to mRNAs | (78,81,84) |
| High intronic GC content | Generates DNA secondary structures that increase pausing of RNA Pol II over retained introns | (17,51,59,63) | |
| RNA | Weak splice site(s) | Less effective recognition of canonical splicing sites | (17,53,54) |
| Enrichment of RNA associated proteins binding sites in retained intron and flanking exon(s) | Binding of splicing repressors/IR enhancers | (70–72,94,95) | |
| Short intronic length | Reduce the availability of alternative splice sites and motifs for the binding of splicing factors | (17,51,53,54) | |
| High intronic GC content | Generates secondary structures that reduce the binding of RNA associated proteins/splicing enhancers | (17,51,59) |
An additional feature associated with retained introns is their higher GC content compared to constitutively spliced introns. The repetition of GC across the intronic sequence could be partially responsible for the retention of certain introns. Indeed, a study carried out by Sznajder and coworkers (59) in human tissues and peripheral blood lymphocytes showed that GC microsatellite expansion, which are predicted to form highly stable RNA secondary structures (hairpins and G-quadruplexes (60–62)), could induce IR in a variety of diseases. They also noted a positional bias on the location of the GC repeats within introns towards splice sites (within 0.07–0.8 kb). Therefore, the presence of these RNA structure induced by the GC-rich microsatellite expansions would have an inhibitory effect on splicing of a given host intron by preventing the binding of trans-acting factors (Figure 2; Table 1) (59). Furthermore, Veloso and co-workers (63) have demonstrated that an increase in GC content could negatively correlate with RNA Pol II elongation in human cells (Extrinsic features and trans-acting regulators of intron retention, RNA pol II elongation). These GC-rich sequences are likely to form stable DNA secondary structures which could explain the observed pausing of RNA Pol II over retained introns. Additionally, the presence of ultraconserved sequences in insects like Drosophilia seems to be an important factor in IR regulation. These ultraconserved sequences appear to occur primarily in intergenic and intronic sequences, and at intron–exon junctions. In the homothorax (hth) gene for example, where intron-retained hth transcripts constitute the majority of the total hth steady-state RNA pool during Drosophila embryogenesis, an ultraconserved sequence was found. The sequence spans an internal exon–intron junction (with the majority located in the intron) and is predicted to form a thermodynamically stable stem–loop RNA structure. This putative hairpin structure, which forms around the donor splice site in the hth transcripts, would block the U1 RNP complex from accessing the splice site, resulting in IR (64).
Figure 2.
IR is regulated at multiple levels. The upper panel (dark blue gradient) shows the histone/DNA modifications known to modulate IR. The lower panel (light blue gradient) displays epigenetic and sequence features of IR. Features that positively regulate IR are presented in Table 1.
Retained introns are generally shorter than non-retained introns in vertebrates and plants (17,51,54). In rice for example, the average size of retained introns is about 183 bp, which is significantly shorter than the mean size of introns (470 bp) (22). A similar trend is observed in the ciliate Tetrahymena thermophila, where IR is the dominant form of AS. The average intron length is 80 bp whereas introns that undergo cassette-exon inclusion or skipping show the largest average length (279 bp) (65). McGuire and co-workers found the same strong correlation between intron length and the prevalence of splice variation in fungi and protists (19). In all the 23 species examined where IR is most prevalent form of AS, shorter introns (<200 bp) were significantly more recognized by intron definition, where splice sites on either side of an intron are recognised as a unit. These observations demonstrate that IR might be related to commonly-shared features, including short intron length and higher GC content in some species. These features indicate possible involvement of intron definition as the preferred mechanism of splicing resulting in IR. Additionally, a short intron may have a lower chance of containing motifs for the binding of splicing factors that would otherwise lead to the splicing of the intron. A longer intron would also have a greater probability of containing alternative splice site(s) that would induce its partial splicing/retention and therefore would not be detected/considered as a (fully) retained intron per se. However, it is important to emphasize that the documented sequence features of retained introns are not always consistent between organisms. Notably, sequence feature analysis of retained and constitutively-spliced introns in Arabidopsis thaliana has shown that most retained introns display weaker 5′ sites, less PTC occurrence and lower GC content (66), diverging from the aforementioned organisms where the GC density is high in retained introns. These observations lead to the conclusion that the molecular mechanisms underlying IR regulation cannot be attributed exclusively to the sequence features described heretofore.
Motifs of cis-acting regulatory elements
In order to identify a general mechanism that regulates IR, over 2500 publicly available RNA-sequencing datasets were analysed and 1000 each of the most frequently and least frequently retained introns were identified (16). Notably, an enrichment for serine-arginine (SR) protein binding sites was observed in retained introns. Using the ENCODE (Encyclopedia of DNA Elements, (67)) database of SR protein knockdown experiments in HepG2 cells to determine the effect on IR, an enrichment of IR transcripts in SR protein-depleted HEPG2 cells was detected (16). SR protein binding sites are not specific to IR events in cancer but are also present in normal cells. Intron-bound SR proteins are likely to facilitate splicing, thereby inhibiting IR (Figure 2; Table 1). Retention of specific introns can be induced by RNA-binding proteins such as hnRNPLL. Indeed, the binding of hnRNPLL in specific exons of the Ptprc gene encoding the CD45 mRNA transcript can trigger the retention of introns surrounding the exons-bound in hnRNPLL (68). In addition, the splicing regulator CUG binding protein (CUG-BP), which is elevated in myotonic dystrophy type 1 (DM1) striated muscle, binds to the ClC-1 pre-mRNA and can induce the retention of intron 2 by binding to a U/G-rich motif causing myotonia in DM1 (69). These observations suggest that IR can also be induced by specific RNA-binding proteins and that the regulation of these RNA binding proteins, notably in diseases (70–72) may modulate the level of particular IR events by binding specific motifs in introns and their flanking exons (Figure 2; Table 1; Extrinsic features and trans-acting regulators of intron retention, splicing factors). An observation that could partially explain the differences in IR frequencies between vertebrates and plants, is the presence of transposable elements within retained introns (73). In the plant species Gossypium raimondii, where IR constitutes 40% of all AS events, transposable elements are present in 43% of the retained introns while their frequency is only 2.9% in all introns. These transposable element insertions are often near the 3′ splice site of the intron and might impact on the efficiency of intron splicing by decreasing RNA secondary structure flexibility. These elements also alter the distribution of branch points from preferred positions which might be an important factor of IR in plants.
In addition, an analysis of IR events and their conservation across diverse vertebrate species revealed an important and intriguing feature of retained introns (51). Namely, intron-retaining genes harbor a larger number of microRNA (miRNA) binding sites in their 3′ untranslated region (UTR), augmented by the presence of longer 3′ UTR sequences in human, mouse, dog, chicken and zebrafish. MiRNA target prediction algorithms also revealed a significantly higher density of putative miRNA binding sites in sequences of retained introns compared to non-retained introns in these species (51). Furthermore, miRNA-intron interactions in Arabidopsis and rice genes have been validated using public degradome sequencing data. Interestingly, double-stranded RNAs could be generated from specific cleaved intron remnants. These double-stranded RNAs could then be processed into siRNAs by the activity of Dicer-like 1 and 3 and be incorporated into Argonaute-associated silencing complexes to potentially cleave mature mRNA targets (74). These results indicate that IR-mediated decay and miRNA-induced translational repression may be complementary mechanisms orchestrating post-transcriptional gene expression control. Further, IR transcripts could potentially function as miRNA ‘sponges’ to indirectly regulate other transcripts by modulating the available pool of miRNAs. The biological relevance of this mechanism of gene regulation, whereby retained introns act as miRNA sponges to fine tune gene expression, requires experimental validation.
Conservation of IR features
Improved understanding of the conservation of IR features across species (17,51) and cancers (46) has refined the definition of IR and provided insights into its regulation. The comprehensive Cancer Genome Atlas (TCGA, (50)) comprises genomic data and key genomic changes in 33 different types of cancers, which has facilitated the analysis of IR features. Dvinge and Bradley (46) performed a large-scale analysis to identify differences in RNA splicing events between tumour and normal control samples across 16 distinct types of cancer. Their most striking observation was that IR was the most differentially altered form of AS, thus the abundance of IR transcripts in cancer cells may significantly contribute to transcriptomic diversity. Almost all the cancers displayed a marked increase in IR levels when compared to normal controls with the sole exception of breast cancer which exhibited a decrease in IR compared to normal breast tissue. They revealed the intrinsic sequence features of IR in these 16 types of cancers were consistent with previously observed common IR features (Intrinsic features of intron retention, sequence features). These data confirm that the likelihood of IR is partly determined by intrinsic features.
In exploring online databases in seven equivalent tissues from eleven vertebrate species, Braunschweig et al. (17) provided insight into the conservation of IR features. Although IR patterns have evolutionarily diverged in the analysed vertebrate tissues, functional conservation has been demonstrated in specific tissues (e.g. cerebellum and brain) compared to other tissues analysed. Conservation of such features was independently confirmed in a deep transcriptomic analysis of highly purified neutrophilic granulocytes from five vertebrate species (human, dog, mouse, chicken and zebrafish) spanning 430 million years of evolution (51). The features include the presence of weaker splice sites, a higher GC content, a shorter intronic length, and a consistent trend for the retained introns to be located near the 3′ termini of the gene body. In this in-depth phylogenetic exploration of IR, some unanticipated regulatory characteristics of this phenomenon were uncovered. For example, intron-retaining genes are transcriptionally co-regulated from bidirectional promoters and retained introns exhibit a higher density of miRNA binding sites (51).
EXTRINSIC FEATURES AND trans-acting REGULATORS OF INTRON RETENTION
Epigenetic factors
Epigenetic changes including DNA methylation and histone modifications are known to regulate alternative splicing (75–79). Relevant to IR, retained introns typically exhibit a higher density of CpG dinucleotides compared to non-retained introns (78). This observation prompted a recent investigation into the role of DNA methylation changes in IR control. Reduced DNA methylation near splice junctions and within introns was observed in diverse normal and neoplastic cells including blood, neurones, embryos, colon and fibroblasts (78). Similar findings have been reported in normal and malignant breast tissue (80,81).
Mechanistically, altered DNA methylation regulates IR by modulating the functions of trans-acting proteins involved in splicing control. Most splicing events occur co-transcriptionally (82,83), whereby proteins that recognize DNA methylation may be recruited to nascent mRNAs to regulate their processing. This observation indicates that reduced DNA methylation levels abrogate binding of methyl-CpG binding protein 2 (MeCP2) to splice junctions in the DNA and mRNA (78) (Figure 2; Table 1). As MeCP2 is involved in the recruitment of splicing factors to mRNAs (78,84,85), decreased MeCP2 binding interrupts the recruitment of splicing factors including Tra2b. In this scenario, IR occurs as a consequence of inefficient splicing. Binding of MeCP2 to other splicing factors including Srsf2, U1-70K, Srsf6, Srsf10, Srsf1, Srsf3, Srsf4, Srsf7, Tra2a were identified using immunoprecipitation coupled with mass spectrometry (78). Others have reported the interaction between MeCP2 and hnRNPs, SRSF4, SRSF6, SRSF7, PRPF3 and YB-1 (84–86). Thus, it is plausible that IR events mediated by reduced MeCP2 binding to splice junctions, consequent to decreased DNA methylation near splice junctions, are regulated via inefficient recruitment of one or more of these splicing factors. Yet, it is important to mention that in yeast species like Saccharomyces cerevisiae and Schizosaccharomyces pombe, cytosine methylation is absent (87) but IR is widespread. Thus, DNA methylation does not play a fundamental role in IR regulation in these species. Once again, these observations highlight the certainty that the molecular mechanisms underlying IR regulation cannot be attributed exclusively to one unique regulatory mechanism.
Post-translational modifications of lysine 36 in histone 3, particularly H3K36me3, have consistently been associated with IR in plants and mammals (77–90). In one study, ablation of the gene encoding the H3K36me3 methylase, SETD2, dramatically increased IR in human kidney tumours (88). Surprisingly, the opposite was observed in colon cancers where SETD2 deficient intestinal cells exhibit a marked decrease in IR (90) (Figure 2; Table 1). In rice, the depletion of SDG725, a plant-specific H3K36 methyltransferase, alters dramatically the patterns of IR by affecting >4700 genes (89). Increased IR at the 5′ region but decreased IR in the 3′ region of genes were observed following SDG725 knock-down. Interestingly, H3K36me2/me3 levels were also increased genome-wide, which is consistent with the results in animals, where the levels of these modifications coincide with position-specific alterations of IR. Previous studies have reported that H3K36me3 regulates the recruitment of either a splicing enhancer or a repressor (91,92). The recognition of H3K36me3-interacting proteins that differentially bind tissue- and position-specific intron-retaining transcripts may shed further light on IR programming. However, a detailed mechanistic explanation of tissue- and position-specific patterns of H3K36me3 regulation of IR remains to be determined.
One protein that regulates IR via H3K36me3 is BS69/ZMYND11. This protein binds preferentially to a variant of H3K36me3, called H3.3K36me3 which is known to associate with chromatin (77) (Figure 2; Table 1). Importantly for splicing regulation, chromatin-bound BS69/ZMYND11 interacts directly with EFTUD2, a component of the U5 small ribonucleoprotein complex (77). Knockdown of BS69/ZMYND11 markedly reduces IR in vitro, confirming its role in potentiating IR. This process occurs via H3.3K36me3 inhibition of the interaction of EFTUD2 with other components of the splicing machinery. Knockdown of SETD2 leads to a similar decrease in overall IR events (77). Thus, BS69/ZMYND11 regulation of IR is dependent on its interaction with H3.3K36me3. The decrease in IR reported recently in SETD2-deficient colon cancers (90) may also be regulated via the H3.3K36me3/BS69/EFTUD2 axis. However, this mechanism of IR regulation affects approximately one-fifth of all IR events associated with H3K36me3 (77), indicating that other ‘reader proteins’ may regulate IR through their interaction with this histone mark. Chromatin accessibility and IR have been associated in plants (93). In Arabidopsis and rice for instance, analysis of DNAse I-seq, a powerful experimental tool for genome-wide interrogation of chromatin accessibility, has shown that IR events are highly enriched in DNase I hypersensitive sites in both species. Mechanistically, as splicing is known to occur co-transcriptionally and is influenced by the speed of transcription, retained introns displaying a more open chromatin conformation would give less time for the spliceosomal machinery to process these introns. Besides, DNA-binding proteins that are associated with IR can also be affected by the structural conformation of chromatin (93).
Splicing factors
The spliceosomal complex comprises a number of core subunits: U1, U2, U4, U5, U6 and over 200 auxiliary proteins. While disruption to any of these proteins may cause IR consequent to splicing perturbation, the depletion of specific spliceosome proteins has been shown to have pronounced effects. Using mRNA sequencing coupled with mass spectrometry, a strong association between increased IR and decreased expression of splicing factors within the U1 and U2 subunits was observed during granulocyte differentiation (39). This finding indicates that IR maybe regulated by small nuclear ribonucleoprotein (snRNP) complexes responsible for the recognition of exon-intron boundaries. However, this phenomenon may be cell type or RNA transcript-specific as IR has been shown to be regulated by changes involving the U4, U5, U6 complexes that assemble post exon recognition in cancer cell lines (77). Individual knockdown of splicing factors including U2AF1, PCBP1, PCBP2, Tra2b, QKI, TIA-1 and PTBP1 have also been shown to promote IR (16,94,95). Proteins like SRSF1 and SRSF7 also have functions relevant to processes post IR including triggering NMD (96), suggesting that they may also regulate the outcome of IR.
In neoplastic cells, mutations in splicing factors have been consistently associated with aberrant IR. Mutations and knockdown of ZRSR2 increase the retention of U12-type introns in mRNA transcripts in blood cells studied from individuals with the myelodysplastic syndrome (97). Mutation of U2AF1 also leads to increased IR in acute myeloid leukaemia (98). Surprisingly, mutations in SF3B1 lead to reduced IR in acute myeloid leukaemia, which can be explained partly by the ability of SF3B1 mutant cells to utilise a cryptic alternative 3′ splice site to produce shorter non intron-retaining isoforms (99). Based on TCGA data (50), reduced IR in breast and lung cancers is associated with mutation of the splicing repressor, RBM10 (100). A study in uveal melanoma also revealed reduced IR of a gene encoding multidrug resistance-associated protein, ABCC5, in SF3B1 mutated compared to SF3B1 wild-type cancers (101). The mechanism by which SF3B1-mutant uveal melanoma cells promote more efficient splicing remains to be determined. It may involve an altered splicing program via cryptic splice site usage or inhibition of a splicing repressor similar to other cancers. Overall, these data in malignant cells support the importance of optimal splicing factor activities in preventing aberrant IR.
RNA Pol II elongation
A slower RNA Pol II elongation rate has been associated with more efficient splice site recognition, which promotes exon inclusion (102) (Figure 2; Table 1). As a corollary of this, a faster RNA Pol II elongation rate reflected by decreased RNA Pol II density in introns might be a characteristic of IR. However, the accumulation of the elongation-associated RNAPolII-Ser2P in retained introns, which is a hallmark of inefficient splicing factor recruitment, has been previously reported (78) (Figure 2). This indicates that slower RNA Pol II elongation is unlikely to facilitate better recognition and splicing of introns. Rather, accumulation of RNA Pol II may be indicative of sub-optimal recruitment of splicing factors, thereby promoting IR (Intrinsic features of intron retention, sequence features).
TOOLS TO INVESTIGATE REGULATORS OF IR
Experimental approaches used to study regulators of IR
In the context of experimental approaches for studying regulators of IR, it is essential to first establish methods for the identification and quantification of IR. Not least are some of the regulatory elements intrinsic to retained introns themselves. Alternative splicing events are routinely identified using RNA sequencing. For accurate IR calling, optimized sample and library preparation protocols are essential (103). This can be achieved, for example, by making sure nascent RNA and DNA contamination is eliminated via poly-A enrichment and DNase I treatment, respectively. For short-read protocols stranded paired-end sequencing is the preferred method and a high sequencing depth is crucial for reliable and reproducible quantification of IR (103). Using bulk RNA sequencing, specific sequential and structural characteristics associated with retained introns and their host genes have been revealed in several studies (37,39,51) (Intrinsic features of intron retention; Extrinsic features and trans-acting regulators of intron retention ).
IR is a low-frequency transcription event and its detection requires adequate read coverage and depth. Hence, accurate detection of IR from single cell sequencing is limited. For the quantification of known IR events, RNA Capture sequencing (CaptureSeq) (104) could provide a medium-throughput alternative to qRT-PCR-based IR validation and quantification. CaptureSeq uses a custom panel of oligonucleotide probes designed to bind complementary sequences specific to transcripts of interest. As a result, the sequence depth of targeted transcripts is markedly increased for the same total number of sequenced reads.
Intronic subsequences, such as identifier elements, can direct their subcellular localization and thus act as intrinsic regulators of IR-mediated mRNA localization (105–107). Regulators of the widespread phenomenon of nuclear intron detention are largely unknown and thus require further exploration (108). Isolation of subcellular compartments prior to RNAseq-based IR calling is essential to determine the sub-cellular localization of intron-retaining transcripts (39). A low-throughput alternative to subcellular fractionation/RNAseq are RNA-based in situ hybridization techniques (e.g. RNA ISH or smFISH) (109,110).
Apart from IR identification, quantification and localization, next generation sequencing techniques provide unique opportunities to shed light into intrinsic and extrinsic regulators of IR as well as other forms of alternative splicing. For example, long-read sequencing protocols, such as PacBio Single-Molecule, Real-Time (SMRT) Sequencing or Oxford Nanopore sequencing, are attractive techniques to study whole transcript isoforms (111–113). The genomic context and the impact of DNA mutations, such as single nucleotide variants near splice sites, can be studied using genome profiling via whole genome or whole exome sequencing (45,114).
For the high-throughput analysis of epigenetic regulators of IR the same toolbox can be used as for the analysis of epigenetically driven gene regulation. Methylated DNA immunoprecipitation sequencing (MeDIP) using monoclonal antibodies specific to 5-methylcytidine (5mC) followed by microarray analysis (MeDIP-Chip) or direct sequencing (MeDIP-Seq) has been used as a valuable tool to map methylated DNA on a genomic scale. However, whole genome bisulphite sequencing (WGBS) has the capability to resolve the methylation status of cytosines at single base resolution. WGBS can be used to show that IR can be regulated by differential DNA methylation in promyelocytes and granulocytes (39). Low-throughput techniques, such as methylation-specific PCR (115) may be used for validation purposes.
Nucleosome Occupancy and Methylome sequencing (NOME-Seq) is a derivative WGBS technique that is used to determine the footprint of nucleosome positioning. In brief, native chromatin is treated with the GpC methyltransferase M.CviPl prior to DNA sodium bisulfite treatment and WGBS. M.CviPl methylates GpC sites that are not bound by nucleosomes (116). However, the most widely used method to determine genome-wide nucleosome occupancy nowadays is the assay for transposase-accessible chromatin using sequencing (ATAC-seq) (117) for its simple and fast sample preparation, and lower DNA input requirements (118).
Iannone et al. described differences in nucleosome density around alternatively spliced exons (119). Nucleosome occupancy around retained introns, however, has not been systematically investigated to date. Chromatin structure-dependent regulation of alternative splicing has been previously described in the context of transcription factor CTCF-mediated chromatin loop formation (120). Methods for open chromatin profiling include DNase-seq (93) and its successors FAIRE-Seq, ATAC-seq, and NicE-seq (121). Chromatin immunoprecipitation sequencing (ChIP-seq) can be used to elucidate the impact of histone modifications on IR. Using this approach Wei et al. have demonstrated that SDG725, a plant-specific H3K36 methylase, mediates position-specific IR in rice (77,89).
To investigate the overall chromatin 3D configuration and chromatin interactions 3C, 4C, 5C-seq, Hi-C and Capture-C are being used. The role of the chromatin organising protein CTCF in alternative splicing has been studied along with data of various histone modifications (ChIP-seq, Hi-C and 4C data) to show that alternative exon usage can be regulated by CTCF-dependent chromatin organisation (120).
Using RNA sequencing data from splicing factor knockdown experiments available in ENCODE (encodeproject.org), it has been shown that knockdown of SR family proteins triggers a dramatic increase in IR, which suggests that many IR events depend on multiple splicing factors (RNA binding proteins, RBP), such as TIA1, SRSF1/7, U2AF2, PCBP1/2, and PTBP1 (16) (Extrinsic features and trans-acting regulators of intron retention, splicing factors). Footprinting of RBP binding can be conducted using RNA immunoprecipitation (low throughput) or variants of RNA cross-linking immunoprecipitation sequencing (high-throughput), such as HITS-CLIP, PAR-CLIP and iCLIP (122, 123). As indicated, ENCODE has continuously made CLIP-seq datasets available to the research community and thus provides a valuable and growing resource to mine for trans-acting regulators of IR and other forms of alternative splicing.
Computational methods to investigate regulators of IR
Custom computational pipelines are essential not just for IR detection (103) but also for the identification and analysis of IR regulators. The reliable identification of IR events in RNA sequencing data starts with the sample and library preparation steps, and sequencing protocol as discussed in the previous section. The computational analysis of RNA sequencing data includes rigorous quality control, transcript identification and quantification, followed by alternative splicing analysis. These analysis steps have recently been reviewed by Conesa et al. (124). IR calling and quantification approaches, however, differ from general alternative splicing analyses. In this context, bioinformatics challenges in investigating intronic regions and pitfalls associated with the analysis of IR from short-read sequencing data as well as limitations of long-read sequencing approaches have been recently discussed elsewhere (103). RNA sequencing-based IR detection/quantification software has not been systematically benchmarked to date, however, an overview of available tools is provided in Table 2.
Table 2.
Overview of IR detection/quantification algorithms
| Tool/Resource | Purpose/method | Website | Reference |
|---|---|---|---|
| IRFinder | Detecting IR from RNA-Seq experiments | github.com/williamritchie/IRFinder | (16) |
| Kma | R package for IR detection | github.com/pachterlab/kma | (125) |
| MISO | (Differential) gene isoform expression analysis; determines intronic percent spliced in (PSI) levels | genes.mit.edu/burgelab/miso | (127) |
| rMATS | Differential AS analysis | rnaseq-mats.sourceforge.net | (134) |
| spliceR | AS identification/quantification | bioconductor.org/packages/spliceR | (164) |
| IntEREst | IR quantification | github.com/gacatag/IntEREst | (126) |
| Psichomics | AS quantification and analysis | bioconductor.org/packages/psichomics | (135) |
| Whippet | Fast AS detection and quantification algorithm | github.com/timbitz/Whippet.jl | (165) |
| SUPPA2 | Fast differential splicing analysis | github.com/comprna/SUPPA | (136) |
| MAJIQ | Detection and quantification of local splicing variations from RNA-Seq data | majiq.biociphers.org | (166) |
| VAST-TOOLS | Toolset for profiling and comparing AS events in RNA-Seq data | github.com/vastgroup/vast-tools | (137) |
| ASTALAVISTA | AS quantification and analysis | astalavista.sammeth.net | (138) |
| JUM | AS quantification and analysis | github.com/qqwang-berkeley/JUM | (141) |
| SpliceHunter | AS quantification and analysis | bitbucket.org/canzar/splicehunter | (32) |
The many challenges in IR identification and quantification and how to overcome them were summarized in a recent review article (103). In brief, risk factors in the identification and quantification of IR events include transcriptional ‘noise’ introduced by DNA contamination or unprocessed pre-mRNA transcripts. IRFinder has a build-in routine that detects DNA contamination by calculating the ratio of the number of reads that map to intergenic regions to the number that maps to coding regions (16). To minimize the possibility of pre-mRNA contamination, sequencing libraries must be enriched for polyadenylated RNA. IRFinder detects experiments for which the library was not enriched for mature mRNA transcripts by counting the number of reads that map to a list of non-polyadenylated genes (small nucleolar RNAs and histone genes). Accurate IR estimation can also be affected by low or highly variable coverage in both intronic and exonic regions. This can be caused for example by repetitive sequences, such as Long and Short Interspersed Nuclear Elements (LINEs and SINEs), DNA transposons, tandem and low complexity repeat sequences. kma has coverage filters in place to identify and remove introns with highly variable coverage (125). IntEREst provides the possibility to exclude repeat regions from the analysis based on a user-provided table of repeat coordinates (126). IRFinder too has a routine that determines regions of poor unique mappability. Poorly mappable regions are then excluded from the measurable intron area (16).
IR event detection can be performed by conceptually different approaches (i) the splice-junction-only approach; (ii) the coverage-based approach; or (iii) both approaches combined (103). While bioinformatics software has been developed to assess alternative splicing from high-throughput transcriptomics data using either of these approaches (Table 2), only the three aforementioned tools have considered specific peculiarities important for IR event detection and quantification (16,125,126). To thoroughly identify IR events, it is of course necessary to first define what is an intron. For that purpose, IRFinder includes tools for preparing a reference genome, while introns are extracted from a given gtf file. Regions between two exons in any transcript are considered introns, while regions within introns covered by a non-intron feature (e.g. snoRNAs or miRNAs) are excluded (16). The R package IntEREst (Intron–Exon Retention Estimator) provides a dedicated function for preparing a reference genome with the option to collapse all isoforms of a gene to avoid assigning reads mapping to any alternatively skipped exons to their overlapping introns (126). With kma, intronic coordinates can be determined based on a genome reference (FASTA file) and feature file (GTF). kma ensures that none of the overlapping isoforms contain an exon in what is defined as intronic inclusion regions (125). kma adds a small region of the neighbouring exons to the intron coordinates to include reads spanning the intron–exon junctions for intron expression quantification.
The key metric that defines the ratio of transcripts retaining an intron to the total number of transcripts of a certain gene isoform is referred to as the IR-ratio by IRFinder or percentage spliced-in (PSI or Ψ) by other tools (127). The IR-ratio is the ratio between intronic abundance and intronic abundance plus exonic abundance; where the exonic abundance refers to the number of read fragments spliced across the intron and the intronic abundance is the median number of reads that map to an intron. IRFinder excludes overlapping features as well as the highest and lowest 30% of values from the intronic abundance and normalizes both the exonic and intronic abundance for feature length (16). kma can be used with existing transcript quantification methods, such as Bowtie (128) or eXpress (129) to determine the intron abundance in transcripts per million (TPM) or Fragments Per Kilobase of transcript per Million mapped reads (FPKM). kma computes Ψ as the ratio between intron expression and expression of the overlapping transcripts plus the intron expression (125). IntEREst quantifies intron expression using the FPKM metrics adapted for intron length and the total number of introns in a gene. IntEREst calculates the relative intron inclusion level (Ψ) based on the number of reads mapped to introns divided by the number of reads spanning the intron (or mapping exons flanking the intron) (126).
To determine regulators of IR, it is crucial to first determine differential IR events. For that purpose, several statistical approaches have been implemented: IRFinder has an integrated method to analyse digital transcript profile data with the Audic and Claverie test (130). However, this method is suitable only for small sample sizes (between 1 and 3 replicates) (16). For larger sample sizes the IRFinder output can be passed on to the R Bioconductor package DESeq2 (131), which fits the count data to a negative binomial generalized linear model and employs Wald statistics to determine differential IR events. Similarly, IntEREst (126) uses functions from established RNA sequencing analysis tools for differential IR analysis including edgeR (132), DEXSeq (133) and DESeq (131). rMATS uses the binomial distribution for modelling the estimation uncertainty in individual replicates and the normal distribution for modelling the variability among replicates based on inclusion read counts, skipping read counts, and intron inclusion levels (134). A likelihood-ratio test is then used to determine whether the difference between the mean inclusion levels is smaller than or equal to the user-defined threshold. Other investigators assume a non-normal distribution of percent spliced-in (PSI) values (similar or identical to IR ratios) and therefore provide the non-parametric Wilcoxon rank-sum, Kruskal–Wallis rank-sum and Fligner–Killeen tests in their psichomics R package together with a selection of multiple-testing correction methods (135). In the Python package SUPPA2 for differential splicing analysis, Trincado et al. (136) consider two distributions: one for the difference between PSI values amongst biological replicates and one for the different PSI values between conditions together with the average abundance of the transcripts in transcripts per million. P values of selected alternative splicing events are computed based on their empirical cumulative distribution function over |deltaPSI|. SUPPA2 includes the Benjamini–Hochberg method for multiple testing correction (136). VAST-TOOLs uses Bayesian inference followed by differential analysis of posterior distributions on PSI values (IR ratios). The posterior distributions are estimated using maximum-likelihood fitting (137).
Although there are no species or clade-specific software tools for alternative splicing analysis, some are more commonly used in plants and others in vertebrate species. Neither IRFinder nor kma were tested in non-vertebrate species, however, the principles of IR detection and quantification remain the same independent of the clade. The developers of IntEREst confirmed this assumption by successfully testing their software both in human and plant samples. Earlier analyses of AS in plants and other non-vertebrates used expressed sequence tags from shotgun sequencing experiments, which clearly lack behind the single nucleotide resolution that deep transcriptome sequencing offers. ASTALAVISTA is a tool that was used by many plant biologists for alternative splicing analyses (138) but can be used for other clades as well.
Nevertheless, a reliable and reproducible IR analysis in any species depends on well-curated genome annotations. The quality of genome annotations typically increases the more widely an organism is studied. In phylogenetic analyses of IR, it is therefore important to consider differences in annotation quality. Examples for quality transcriptome annotation efforts in plants are the Arabidopsis thaliana Reference Transcript Dataset (139) and the Gossypium austral full-length transcriptome atlas (140). However, similar quality reference transcriptomes have yet to be generated for other non-mammalian species. One approach to account for differences in annotation quality is to generate de novo exon–intron structures from the same number of random reads for each sample (8). Wang and Rio have recently developed the tool JUM (junction usage model) for alternative splicing analysis that addresses the problem of poorly annotated genomes by using ‘split’ reads. In this case reads that cannot be completely mapped to one location in the genome are viewed to achieve an annotation-free analysis of alternative splicing (including IR) in metazoan transcriptomes (141). A tool that was developed as part of a transcriptome analysis of meiosis in fission yeast is SpliceHunter (32), which harnesses full-length transcript sequences produced by long-read sequencing technologies to identify alternatively spliced isoforms. SpliceHunter can be used for other species as well.
Systematic analysis of IR events in RNA sequencing data has revealed conserved intrinsic features of IR regulation in retained introns and intron-retaining transcripts (17, 51). A plethora of tool collections, software repositories, or code libraries (e.g. bedtools – bedtools.readthedocs.io; BioPython – biopython.org, BioPerl – bioperl.org, Bioconductor – bioconductor.org) are available for the analysis of recurring sequence or structural features in and around retained introns. These include nucleotide or dinucleotide frequencies, intron length, locus and conservation. The maximum entropy model of short sequence motifs proposed by Yeo and Burge can be used to estimate the strengths of donor and acceptor sites in retained and non-retained introns (142). Computational analysis of epigenomic IR regulation can be performed analogous to the analysis of epigenomic gene expression regulation by shifting focus to donor and acceptor splice sites rather than transcription start and termination sites. Methods for the analysis of epigenomics data including DNA methylation (e.g. WGBS) (143), histone modification (e.g. ChIP-seq) (144), chromatin structure (3C-based technologies, MNase-seq, DNase-seq, FAIRE-seq, ATAC-seq) (118) data have been critically reviewed before. However, in order to acquire a holistic grasp of IR regulatory mechanisms, integrative ‘omics’ approaches involving these experimental methods should be pursued. Methods for multi-omics data integration and associated challenges have been discussed in recent reviews (145–147).
Modelling splicing regulation
The computational prediction of IR events has not been attempted to date, however multiple machine learning approaches have been proposed to predict exon usage. For example, a Bayesian neural network was used to identify the ‘splicing code’, which is comprised of hundreds of RNA sequence and structural features (including cis- elements described in literature) and predicts tissue-specific changes in alternative splicing (exon usage) (148). Following this, a deep neural network approach achieved an enhanced performance in predicting alternative splicing patterns (149). Based on deep learning and other machine learning approaches a number of tools and algorithms were recently developed that predict cryptic splicing caused by putative genetic variants and their role in rare genetic disorders (150–152).
However, all the above-mentioned studies focus on exonic splicing and primarily include cis-acting splicing regulators. An algorithm or tool that predicts IR is currently missing but should be within reach given the recent advances in exon splicing prediction and the identification of mechanisms of IR regulation. While machine learning has been used to predict alternative splicing, systems biology approaches are employed to study the dynamics of splicing regulatory networks using stochastic or deterministic modelling formalisms. Network modelling of splicing regulation has recently been discussed in the context of bioinformatics challenges in determining the effects of epigenetic modifications on alternative splicing (153). Splicing regulatory networks include cis- and trans-acting regulators of alternative splicing and their respective splicing targets (153). For example, a Bayesian network approach was used to predict the target network of the neuron-specific factor Nova in the mouse brain, comprising ∼700 alternative splicing events (154). Similarly, a kinetic model of co-transcriptional alternative splicing was used to predict that transcriptional elongation rates may affect splicing outcomes (155).
CONCLUSION AND PERSPECTIVE
IR is regulated at multiple levels, with specific molecular mechanisms awaiting further clarification
The phenomenon of IR is a conserved, orchestrated mechanism which is widespread across taxa. It plays a pivotal role in fine-tuning gene expression at the post-transcriptional level (44). Nonetheless, an understanding of the regulation of intron splicing is still far from complete and key players of IR regulation remain to be uncovered. For instance, the presence of a wide range of small RNAs (including miRNAs, splice-site RNAs, etc.) might directly affect IR by interacting with nascent pre-mRNA at the splice junction. The binding of these small RNAs adjacent to splice sites would prevent recognition of the intron as a prelude to its removal by the spliceosome. Indeed, during mouse granulopoiesis the level of IR increases upon differentiation (39) and that nuclear-enrichment of miRNAs is associated with hematopoietic differentiation (156). It is possible that the level of IR could be directly modulated by the nuclear localization of these miRNAs.
In addition, RNA editing can lead to alternative splicing either directly (157) or indirectly as in the case of the alternative splicing regulator Nova1, in which protein stability is increased through an amino acid substitution enabled by A-to-I editing (158). Evidence suggests that the double-stranded RNA-specific adenosine deaminase (ADAR2) can even modulate its own expression by editing an AA dinucleotide to an AI dinucleotide. The inosine is recognized by the splicing machinery as guanosine. ADAR2 is thus creating an alternative 3′ acceptor site in its own pre-mRNA (159). Most A-to-I editing sites reside in introns and 3′ UTR sequences. Alternative splice sites created by intronic RNA editing results in partial intron inclusion in mature mRNA transcripts (157).
‘Yet to be discovered’ roles of IR
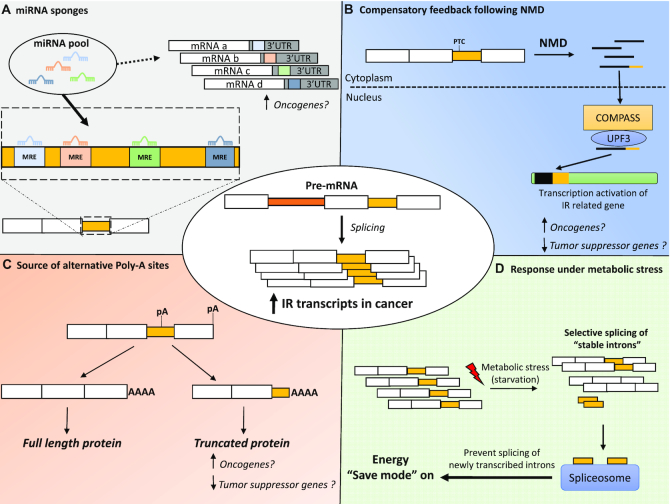
While many aspects of IR regulation remain uncertain, the inventory of the roles of IR in normal and disease biology is steadily expanding. The genetic paradox recently uncovered where, in a context of a gene knockout, a molecular mechanism activates the transcription of genes related to the inactivated gene has opened new possibilities in the potential roles of IR (160,161). Indeed, this genetic compensation mechanism is specifically triggered when mutation generating PTCs result in the degradation of the transcript via NMD. Transcripts retaining introns often contain one or more PTCs, which then initiate their degradation by NMD. Thus, PTC-containing intron-retaining transcripts could potentially trigger the up-regulatory feedback response known as nonsense-induced transcriptional compensation (Figure 3B). IR could also be a powerful asset under stress conditions (e.g. starvation). Indeed, as mentioned in the introductory section, recent studies (5,6) have proposed a mechanism in yeast whereby spliced introns may ‘clutter up’ the spliceosome apparatus, thus preventing it from splicing newly transcribed introns and expending energy under starvation conditions. Additionally, processed introns would also prevent the expression of ribosomal protein genes thereby decreasing protein production. In nutrient-poor environments, intron-retaining transcripts could swiftly provide a source of stable introns to interact with the spliceosome and reduce energy consumption (Figure 3D). Furthermore, new candidate tumour-suppressor genes that are inactivated by intronic polyadenylation in leukemia have been described (162). Thus, a new intriguing facet of IR could be its role to act as a source of alternative poly-adenylation sites (Figure 3C).
Figure 3.
The ‘yet to be discovered’ roles of IR and possible implications for cancer. (A) Acting as competing endogenous RNA or miRNA sponges, retained introns harbouring MREs might divert miRNAs away from their canonical target. MRE, miRNA response element; UTR, untranslated region. (B) Compensatory feedback after degradation of PTC-containing IR transcript via NMD. NMD – nonsense mediated decay; COMPASS – Complex Proteins Associated with Set1; Upf3 – nonsense-mediated mRNA decay protein 3. (C) Acting as an alternative source of polyadenylation sites to generate truncated protein isoforms. pA – polyadenylation site. (D) Source of stable introns interacting with the spliceosome wherein cancer cells survive under starvation conditions.
In cancer where IR is dysregulated (up-regulated in most cancers analysed by Dvinge et al. (46)), the consequences of the variation of IR level might be more dramatic than previously anticipated. Indeed, as portrayed in Figure 3, the accumulation of IR transcripts could exacerbate the effect of the ‘yet to be discovered roles of IR’. For instance, an increase of IR transcripts could have a greater ‘sponging’ effect on miRNAs, which could relieve oncogenes from miRNA-mediated suppression (Figure 3A). In addition, intron-containing transcripts, potentially harbouring alternative polyadenylation sites, could generate truncated proteins with oncogenic activity (Figure 3C). Furthermore, in order to divide indefinitely, cancer cells must pace nutrient intake. Cancer cells may adapt and survive in an environment deprived of nutrients and the up-regulation of IR transcripts, which would provide a rapid source of stable introns interacting with the spliceosome, might be a way for cancer cells to thrive even under starvation conditions (Figure 3D).
CONCLUDING REMARKS
Further studies into IR regulation and its diverse roles in normal and disease biology are needed in order to shed light on fundamental aspects of regulatory RNA biology. With technological advances and new computational approaches additional roles of IR will be uncovered, for example in lineage commitment and cell differentiation enabled through single-cell sequencing. In conclusion, the world of alternative splicing has expanded to accommodate a changing paradigm that places intron splicing as an important regulatory mechanism. From an intronic view of its RNA context, the question remains as to what complex machinery determines the fate of introns.
ACKNOWLEDGEMENTS
We thank the funding agencies for research and fellowship support.
FUNDING
National Health and Medical Research Council Project Grants 1080530, 1061906, 1128175 and 1129901 to J.E.J.R. and J.J.-L.W. and 1126306 to J.J.-L.W.; NSW Genomics Collaborative Grant (to J.E.J.R. and J.J.-L.W.); Cure the Future (to J.E.J.R.); an anonymous foundation (to J.E.J.R.); U.S. and J.J.-L.W. hold Fellowships from the Cancer Institute New South Wales; financial support was provided by Tour de Cure to C.G.B. and J.E.J.R; Tour de Rocks project support (to C.G.B.); Cancer Council NSW project grants [RG11-11 to J.E.J.R. and J.J-L.W.]. Funding for open access charge: An anonymous foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Beadle G.W., Tatum E.L.. Genetic control of biochemical reactions in neurospora. Proc. Natl. Acad. Sci. U.S.A. 1941; 27:499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beadle G.W., Tatum E.L.. Neurospora. ii. methods of producing and detecting mutations concerned with nutritional requirements. Am. J. Bot. 1945; 32:678–686. [Google Scholar]
- 3. Chow L.C., Gelinas R.E., Broker T.R., Roberts R.J.. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. 1977. Rev. Med. Virol. 2000; 10:362–371. [PubMed] [Google Scholar]
- 4. Berget S.M., Moore C., Sharp P.A.. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. U.S.A. 1977; 74:3171–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morgan J.T., Fink G.R., Bartel D.P.. Excised linear introns regulate growth in yeast. Nature. 2019; 565:606–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parenteau J., Maignon L., Berthoumieux M., Catala M., Gagnon V., Abou Elela S.. Introns are mediators of cell response to starvation. Nature. 2019; 565:612–617. [DOI] [PubMed] [Google Scholar]
- 7. Merkin J., Russell C., Chen P., Burge C.B.. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science. 2012; 338:1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barbosa-Morais N.L., Irimia M., Pan Q., Xiong H.Y., Gueroussov S., Lee L.J., Slobodeniuc V., Kutter C., Watt S., Colak R. et al.. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012; 338:1587–1593. [DOI] [PubMed] [Google Scholar]
- 9. Nilsen T.W., Graveley B.R.. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010; 463:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nilsen T.W. The spliceosome: The most complex macromolecular machine in the cell. Bioessays. 2003; 25:1147–1149. [DOI] [PubMed] [Google Scholar]
- 11. Keren H., Lev-Maor G., Ast G.. Alternative splicing and evolution: Diversification, exon definition and function. Nat. Rev. Genet. 2010; 11:345–355. [DOI] [PubMed] [Google Scholar]
- 12. Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E.. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007; 446:926–929. [DOI] [PubMed] [Google Scholar]
- 13. Eom T., Zhang C., Wang H., Lay K., Fak J., Noebels J.L., Darnell R.B.. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. Elife. 2013; 2:e00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng S., Gray E.E., Chawla G., Porse B.T., O’Dell T.J., Black D.L.. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat. Neurosci. 2012; 15:381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yap K., Lim Z.Q., Khandelia P., Friedman B., Makeyev E.V.. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012; 26:1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Middleton R., Gao D., Thomas A., Singh B., Au A., Wong J.J.-., Bomane A., Cosson B., Eyras E., Rasko J.E.J. et al.. IRFinder: Assessing the impact of intron retention on mammalian gene expression. Genome Biol. 2017; 18:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Braunschweig U., Barbosa-Morais N., Pan Q., Nachman E.N., Alipanahi B., Gonatopoulos-Pournatzis T., Frey B., Irimia M., Blencowe B.J.. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014; 24:1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim E., Magen A., Ast G.. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007; 35:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McGuire A.M., Pearson M.D., Neafsey D.E., Galagan J.E.. Cross-kingdom patterns of alternative splicing and splice recognition. Genome Biol. 2008; 9:R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sultan M., Schulz M.H., Richard H., Magen A., Klingenhoff A., Scherf M., Seifert M., Borodina T., Soldatov A., Parkhomchuk D. et al.. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008; 321:956–960. [DOI] [PubMed] [Google Scholar]
- 21. Wang Z., Burge C.B.. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008; 14:802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang G., Guo G., Hu X., Zhang Y., Li Q., Li R., Zhuang R., Lu Z., He Z., Fang X. et al.. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res. 2010; 20:646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang B., Brendel V.. Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:7175–7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chaudhary S., Khokhar W., Jabre I., Reddy A.S.N., Byrne L.J., Wilson C.M., Syed N.H.. Alternative splicing and protein diversity: Plants versus animals. Front. Plant Sci. 2019; 10:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jabre I., Reddy A.S.N., Kalyna M., Chaudhary S., Khokhar W., Byrne L.J., Wilson C.M., Syed N.H.. Does co-transcriptional regulation of alternative splicing mediate plant stress responses. Nucleic Acids Res. 2019; 47:2716–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chaudhary S., Jabre I., Reddy A.S.N., Staiger D., Syed N.H.. Perspective on alternative splicing and proteome complexity in plants. Trends Plant Sci. 2019; 24:496–506. [DOI] [PubMed] [Google Scholar]
- 27. Kalyna M., Simpson C.G., Syed N.H., Lewandowska D., Marquez Y., Kusenda B., Marshall J., Fuller J., Cardle L., McNicol J. et al.. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in arabidopsis. Nucleic Acids Res. 2012; 40:2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moran Y., Weinberger H., Reitzel A.M., Sullivan J.C., Kahn R., Gordon D., Finnerty J.R., Gurevitz M.. Intron retention as a posttranscriptional regulatory mechanism of neurotoxin expression at early life stages of the starlet anemone nematostella vectensis. J. Mol. Biol. 2008; 380:437–443. [DOI] [PubMed] [Google Scholar]
- 29. Boothby T.C., Zipper R.S., van der Weele Corine M., Wolniak S.M.. Removal of retained introns regulates translation in the rapidly developing gametophyte of marsilea vestita. Dev. Cell. 2013; 24:517–529. [DOI] [PubMed] [Google Scholar]
- 30. Göhring J., Jacak J., Barta A.. Imaging of endogenous messenger RNA splice variants in living cells reveals nuclear retention of transcripts inaccessible to nonsense-mediated decay in arabidopsis. Plant Cell. 2014; 26:754–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rocchi V., Janni M., Bellincampi D., Giardina T., D’Ovidio R.. Intron retention regulates the expression of pectin methyl esterase inhibitor (pmei) genes during wheat growth and development. Plant Biol. (Stuttg). 2012; 14:365–373. [DOI] [PubMed] [Google Scholar]
- 32. Kuang Z., Boeke J.D., Canzar S.. The dynamic landscape of fission yeast meiosis alternative-splice isoforms. Genome Res. 2017; 27:145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Juneau K., Palm C., Miranda M., Davis R.W.. High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc. Natl Acad. Sci. U.S.A. 2007; 104:1522–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gontijo A.M., Miguela V., Whiting M.F., Woodruff R.C., Dominguez M.. Intron retention in the drosophila melanogaster rieske iron sulphur protein gene generated a new protein. Nat. Commun. 2011; 2:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fernandes J.D., Booth D.S., Frankel A.D.. A structurally plastic ribonucleoprotein complex mediates post-transcriptional gene regulation in HIV-1. Wiley Interdiscip. Rev. RNA. 2016; 7:470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lunghi M., Spano F., Magini A., Emiliani C., Carruthers V.B., Di Cristina M.. Alternative splicing mechanisms orchestrating post-transcriptional gene expression: Intron retention and the intron-rich genome of apicomplexan parasites. Curr. Genet. 2016; 62:31–38. [DOI] [PubMed] [Google Scholar]
- 37. Edwards C.R., Ritchie W., Wong J.J., Schmitz U., Middleton R., An X., Mohandas N., Rasko J.E., Blobel G.A.. A dynamic intron retention program in the mammalian megakaryocyte and erythrocyte lineages. Blood. 2016; 127:e2–e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pimentel H., Parra M., Gee S.L., Mohandas N., Pachter L., Conboy J.G.. A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2016; 44:838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wong J.J., Ritchie W., Ebner O.A., Selbach M., Wong J.W., Huang Y., Gao D., Pinello N., Gonzalez M., Baidya K. et al.. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013; 154:583–595. [DOI] [PubMed] [Google Scholar]
- 40. Mauger O., Lemoine F., Scheiffele P.. Targeted intron retention and excision for rapid gene regulation in response to neuronal activity. Neuron. 2016; 92:1266–1278. [DOI] [PubMed] [Google Scholar]
- 41. Ni T., Yang W., Han M., Zhang Y., Shen T., Nie H., Zhou Z., Dai Y., Yang Y., Liu P. et al.. Global intron retention mediated gene regulation during CD4+ T cell activation. Nucleic Acids Res. 2016; 44:6817–6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Memon D., Dawson K., Smowton C.S., Xing W., Dive C., Miller C.J.. Hypoxia-driven splicing into noncoding isoforms regulates the DNA damage response. NPJ Genome Med. 2016; 1:16020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Naro C., Jolly A., Di Persio S., Bielli P., Setterblad N., Alberdi A.J., Vicini E., Geremia R., De la Grange P., Sette C.. An orchestrated intron retention program in meiosis controls timely usage of transcripts during germ cell differentiation. Dev. Cell. 2017; 41:8–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wong J.J.-., Au A.Y.M., Ritchie W., Rasko J.E.J.. Intron retention in mRNA: No longer nonsense: known and putative roles of intron retention in normal and disease biology. Bioessays. 2016; 38:41–49. [DOI] [PubMed] [Google Scholar]
- 45. Jung H., Lee D., Lee J., Park D., Kim Y.J., Park W.Y., Hong D., Park P.J., Lee E.. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat. Genet. 2015; 47:1242–1248. [DOI] [PubMed] [Google Scholar]
- 46. Dvinge H., Bradley R.K.. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015; 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pearson H., Daouda T., Granados D.P., Durette C., Bonneil E., Courcelles M., Rodenbrock A., Laverdure J., Côté C., Mader S. et al.. MHC class I-associated peptides derive from selective regions of the human genome. J. Clin. Invest. 2016; 126:4690–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smart A.C., Margolis C.A., Pimentel H., He M.X., Miao D., Adeegbe D., Fugmann T., Wong K., Van Allen E.M.. Intron retention is a source of neoepitopes in cancer. Nat. Biotechnol. 2018; 36:1056–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hubert C.G., Bradley R.K., Ding Y., Toledo C.M., Herman J., Skutt-Kakaria K., Girard E.J., Davison J., Berndt J., Corrin P. et al.. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev. 2013; 27:1032–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M.. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013; 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schmitz U., Pinello N., Jia F., Alasmari S., Ritchie W., Keightley M.C., Shini S., Lieschke G.J., Wong J.J., Rasko J.E.J.. Intron retention enhances gene regulatory complexity in vertebrates. Genome Biol. 2017; 18:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stamm S., Zhu J., Nakai K., Stoilov P., Stoss O., Zhang M.Q.. An alternative-exon database and its statistical analysis. DNA Cell Biol. 2000; 19:739–756. [DOI] [PubMed] [Google Scholar]
- 53. Galante P.A.F., Sakabe N.J., Kirschbaum-Slager N., de Souza S.J.. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004; 10:757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sakabe N.J., de Souza S.J.. Sequence features responsible for intron retention in human. BMC Genomics. 2007; 8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Romano M., Marcucci R., Baralle F.E.. Splicing of constitutive upstream introns is essential for the recognition of intra-exonic suboptimal splice sites in the thrombopoietin gene. Nucleic Acids Res. 2001; 29:886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dirksen W.P., Sun Q., Rottman F.M.. Multiple splicing signals control alternative intron retention of bovine growth hormone pre-mRNA. J. Biol. Chem. 1995; 270:5346–5352. [DOI] [PubMed] [Google Scholar]
- 57. Talerico M., Berget S.M.. Intron definition in splicing of small drosophila introns. Mol. Cell Biol. 1994; 14:3434–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Romfo C.M., Alvarez C.J., van Heeckeren W.J., Webb C.J., Wise J.A.. Evidence for splice site pairing via intron definition in schizosaccharomyces pombe. Mol. Cell Biol. 2000; 20:7955–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sznajder ŁJ., Thomas J.D., Carrell E.M., Reid T., McFarland K.N., Cleary J.D., Oliveira R., Nutter C.A., Bhatt K., Sobczak K. et al.. Intron retention induced by microsatellite expansions as a disease biomarker. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:4234–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ciesiolka A., Jazurek M., Drazkowska K., Krzyzosiak W.J.. Structural characteristics of simple RNA repeats associated with disease and their deleterious protein interactions. Front Cell Neurosci. 2017; 11:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Handa V., Yeh H.J.C., McPhie P., Usdin K.. The AUUCU repeats responsible for spinocerebellar ataxia type 10 form unusual RNA hairpins. J. Biol. Chem. 2005; 280:29340–29345. [DOI] [PubMed] [Google Scholar]
- 62. Park H., González ÀL., Yildirim I., Tran T., Lohman J.R., Fang P., Guo M., Disney M.D.. Crystallographic and computational analyses of AUUCU repeating RNA that causes spinocerebellar ataxia type 10 (SCA10). Biochemistry. 2015; 54:3851–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Veloso A., Kirkconnell K.S., Magnuson B., Biewen B., Paulsen M.T., Wilson T.E., Ljungman M.. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res. 2014; 24:896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Glazov E.A., Pheasant M., McGraw E.A., Bejerano G., Mattick J.S.. Ultraconserved elements in insect genomes: A highly conserved intronic sequence implicated in the control of homothorax mRNA splicing. Genome Res. 2005; 15:800–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xiong J., Lu X., Zhou Z., Chang Y., Yuan D., Tian M., Zhou Z., Wang L., Fu C., Orias E. et al.. Transcriptome analysis of the model protozoan, tetrahymena thermophila, using deep RNA sequencing. PLoS One. 2012; 7:e30630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cui Y., Zhang C., Cai M.. Prediction and feature analysis of intron retention events in plant genome. Comput. Biol. Chem. 2017; 68:219–223. [DOI] [PubMed] [Google Scholar]
- 67. The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cho V., Mei Y., Sanny A., Chan S., Enders A., Bertram E.M., Tan A., Goodnow C.C., Andrews T.D.. The RNA-binding protein hnRNPLL induces a T cell alternative splicing program delineated by differential intron retention in polyadenylated RNA. Genome Biol. 2014; 15:R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Charlet-B N., Savkur R.S., Singh G., Philips A.V., Grice E.A., Cooper T.A.. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002; 10:45–53. [DOI] [PubMed] [Google Scholar]
- 70. Del Gatto-Konczak F., Bourgeois C.F., Le Guiner C., Kister L., Gesnel M.C., Stévenin J., Breathnach R.. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell Biol. 2000; 20:6287–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Linares A.J., Lin C., Damianov A., Adams K.L., Novitch B.G., Black D.L.. The splicing regulator PTBP1 controls the activity of the transcription factor Pbx1 during neuronal differentiation. Elife. 2015; 4:e09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rogelj B., Easton L.E., Bogu G.K., Stanton L.W., Rot G., Curk T., Zupan B., Sugimoto Y., Modic M., Haberman N. et al.. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci. Rep. 2012; 2:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li Q., Xiao G., Zhu Y.. Single-nucleotide resolution mapping of the gossypium raimondii transcriptome reveals a new mechanism for alternative splicing of introns. Mol. Plant. 2014; 7:829–840. [DOI] [PubMed] [Google Scholar]
- 74. Meng Y., Shao C., Ma X., Wang H.. Introns targeted by plant microRNAs: A possible novel mechanism of gene regulation. Rice (N Y). 2013; 6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S.. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011; 479:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Maunakea A.K., Chepelev I., Cui K., Zhao K.. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013; 23:1256–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Guo R., Zheng L., Park J., Lv R., Chen H., Jiao F., Xu W., Mu S., Wen H., Qiu J. et al.. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell. 2014; 56:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wong J.J., Gao D., Nguyen T.V., Kwok C.T., van Geldermalsen M., Middleton R., Pinello N., Thoeng A., Nagarajah R., Holst J.. Intron retention is regulated by altered MeCP2-mediated splicing factor recruitment. Nat. Commun. 2017; 8:15134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gao D., Pinello N., Nguyen T.V., Thoeng A., Nagarajah R., Holst J., Rasko J.E.J., Wong J.J.L.. DNA methylation/hydroxymethylation regulate gene expression and alternative splicing during terminal granulopoiesis. Epigenomics. 2018; 11:95–109. [DOI] [PubMed] [Google Scholar]
- 80. Gascard P., Bilenky M., Sigaroudinia M., Zhao J., Li L., Carles A., Delaney A., Tam A., Kamoh B., Cho S. et al.. Epigenetic and transcriptional determinants of the human breast. Nat. Commun. 2015; 6:6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kim D., Shivakumar M., Han S., Sinclair M.S., Lee Y., Zheng Y., Olopade O.I., Kim D., Lee Y.. Population-dependent intron retention and DNA methylation in breast cancer. Mol. Cancer Res. 2018; 16:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Oesterreich F.C., Herzel L., Straube K., Hujer K., Howard J., Neugebauer K.M.. Splicing of nascent RNA coincides with intron exit from RNA polymerase II. Cell. 2016; 165:372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Herzel L., Neugebauer K.M.. Quantification of co-transcriptional splicing from RNA-seq data. Methods. 2015; 85:36–43. [DOI] [PubMed] [Google Scholar]
- 84. Long S., Ooi J., Yau P., Jones P.. A brain-derived MeCP2 complex supports a role for MeCP2 in RNA processing. Biosci. Rep. 2011; 31:333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Young J.I., Hong E.P., Castle J.C., Crespo-Barreto J., Bowman A.B., Rose M.F., Kang D., Richman R., Johnson J.M., Berget S. et al.. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:17551–17558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tsujimura K., Irie K., Nakashima H., Egashira Y., Fukao Y., Fujiwara M., Itoh M., Uesaka M., Imamura T., Nakahata Y. et al.. miR-199a links MeCP2 with mTOR signaling and its dysregulation leads to rett syndrome phenotypes. Cell Rep. 2015; 12:1887–1901. [DOI] [PubMed] [Google Scholar]
- 87. Capuano F., Mülleder M., Kok R., Blom H.J., Ralser M.. Cytosine DNA methylation is found in drosophila melanogaster but absent in saccharomyces cerevisiae, schizosaccharomyces pombe, and other yeast species. Anal. Chem. 2014; 86:3697–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Simon J.M., Hacker K.E., Singh D., Brannon A.R., Parker J.S., Weiser M., Ho T.H., Kuan P., Jonasch E., Furey T.S. et al.. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res. 2014; 24:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wei G., Liu K., Shen T., Shi J., Liu B., Han M., Peng M., Fu H., Song Y., Zhu J. et al.. Position-specific intron retention is mediated by the histone methyltransferase SDG725. BMC Biol. 2018; 16:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yuan H., Li N., Fu D., Ren J., Hui J., Peng J., Liu Y., Qiu T., Jiang M., Pan Q. et al.. Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. J. Clin. Invest. 2017; 127:3375–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Luco R.F., Pan Q., Tominaga K., Blencowe B.J., Pereira-Smith O., Misteli T.. Regulation of alternative splicing by histone modifications. Science. 2010; 327:996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pradeepa M.M., Sutherland H.G., Ule J., Grimes G.R., Bickmore W.A.. Psip1/ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLos Genet. 2012; 8:e1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ullah F., Hamilton M., Reddy A.S.N., Ben-Hur A.. Exploring the relationship between intron retention and chromatin accessibility in plants. BMC Genomics. 2018; 19:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. de Bruin R.G., Shiue L., Prins J., de Boer H.C., Singh A., Fagg W.S., van Gils J.M., Duijs J.M., Katzman S., Kraaijeveld A.O. et al.. Quaking promotes monocyte differentiation into pro-atherogenic macrophages by controlling pre-mRNA splicing and gene expression. Nat. Commun. 2016; 7:10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Dichmann D., Walentek P., Harland R.. The alternative splicing regulator Tra2b is required for somitogenesis and regulates splicing of an inhibitory Wnt11b isoform. Cell Rep. 2015; 10:527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Twyffels L., Gueydan C., Kruys V.. Shuttling SR proteins: More than splicing factors. FEBS J. 2011; 278:3246–3255. [DOI] [PubMed] [Google Scholar]
- 97. Madan V., Kanojia D., Li J., Okamoto R., Sato-Otsubo A., Kohlmann A., Sanada M., Grossmann V., Sundaresan J., Shiraishi Y. et al.. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun. 2015; 6:6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ilagan J.O., Ramakrishnan A., Hayes B., Murphy M.E., Zebari A.S., Bradley P., Bradley R.K.. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2014; 25:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pellagatti A., Armstrong R.N., Steeples V., Sharma E., Repapi E., Singh S., Sanchi A., Radujkovic A., Horn P., Dolatshad H. et al.. Impact of spliceosome mutations on RNA splicing in myelodysplasia: Dysregulated genes/pathways and clinical associations. Blood. 2018; 132:1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Seiler M., Peng S., Agrawal A.A., Palacino J., Teng T., Zhu P., Smith P.G., Caesar-Johnson S., Demchok J.A., Felau I. et al.. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 2018; 23:28–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Furney S.J., Pedersen M., Gentien D., Dumont A.G., Rapinat A., Desjardins L., Turajlic S., Piperno-Neumann S., de l.G., Roman-Roman S. et al.. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013; 3:1122–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. de l.M., Alonso C.R., Kadener S., Fededa J.P., Blaustein M., Pelisch F., Cramer P., Bentley D., Kornblihtt A.R.. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003; 12:525–532. [DOI] [PubMed] [Google Scholar]
- 103. Vanichkina D.P., Schmitz U., Wong J.J., Rasko J.E.J.. Challenges in defining the role of intron retention in normal biology and disease. Semin. Cell Dev. Biol. 2017; 75:40–49. [DOI] [PubMed] [Google Scholar]
- 104. Mercer T.R., Clark M.B., Crawford J., Brunck M.E., Gerhardt D.J., Taft R.J., Nielsen L.K., Dinger M.E., Mattick J.S.. Targeted sequencing for gene discovery and quantification using RNA CaptureSeq. Nat. Protoc. 2014; 9:989–1009. [DOI] [PubMed] [Google Scholar]
- 105. Buckley P.T., Khaladkar M., Kim J., Eberwine J.. Cytoplasmic intron retention, function, splicing, and the sentinel RNA hypothesis. Wiley Interdiscip. Rev. RNA. 2014; 5:223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Buckley P.T., Lee M.T., Sul J.Y., Miyashiro K.Y., Bell T.J., Fisher S.A., Kim J., Eberwine J.. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron. 2011; 69:877–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ortiz R., Georgieva M.V., Gutierrez S., Pedraza N., Fernandez-Moya S., Gallego C.. Recruitment of Staufen2 enhances dendritic localization of an intron-containing CaMKIIalpha mRNA. Cell Rep. 2017; 20:13–20. [DOI] [PubMed] [Google Scholar]
- 108. Boutz P.L., Bhutkar A., Sharp P.A.. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015; 29:63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bell T.J., Miyashiro K.Y., Sul J.Y., Buckley P.T., Lee M.T., McCullough R., Jochems J., Kim J., Cantor C.R., Parsons T.D. et al.. Intron retention facilitates splice variant diversity in calcium-activated big potassium channel populations. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21152–21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Urbanek M.O., Krzyzosiak W.J.. Discriminating RNA variants with single-molecule allele-specific FISH. Mutat. Res. 2017; 773:230–241. [DOI] [PubMed] [Google Scholar]
- 111. Rhoads A., Au K.F.. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics. 2015; 13:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Byrne A., Beaudin A.E., Olsen H.E., Jain M., Cole C., Palmer T., DuBois R.M., Forsberg E.C., Akeson M., Vollmers C.. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 2017; 8:16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang B., Tseng E., Regulski M., Clark T.A., Hon T., Jiao Y., Lu Z., Olson A., Stein J.C., Ware D.. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 2016; 7:11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Maselli R.A., Arredondo J., Nguyen J., Lara M., Ng F., Ngo M., Pham J.M., Yi Q., Stajich J.M., McDonald K. et al.. Exome sequencing detection of two untranslated GFPT1 mutations in a family with limb-girdle myasthenia. Clin. Genet. 2014; 85:166–171. [DOI] [PubMed] [Google Scholar]
- 115. Herman J.G., Graff J.R., Myohanen S., Nelkin B.D., Baylin S.B.. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Kelly T.K., Liu Y., Lay F.D., Liang G., Berman B.P., Jones P.A.. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012; 22:2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J.. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013; 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Chang P., Gohain M., Yen M.R., Chen P.Y.. Computational methods for assessing chromatin hierarchy. Comput. Struct. Biotechnol. J. 2018; 16:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Iannone C., Pohl A., Papasaikas P., Soronellas D., Vicent G.P., Beato M., ValcaRcel J.. Relationship between nucleosome positioning and progesterone-induced alternative splicing in breast cancer cells. RNA. 2015; 21:360–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ruiz-Velasco M., Kumar M., Lai M.C., Bhat P., Solis-Pinson A., Reyes A., Kleinsorg S., Noh K.M., Gibson T.J., Zaugg J.B.. CTCF-mediated chromatin loops between promoter and gene body regulate alternative splicing across individuals. Cell Syst. 2017; 5:62–637. [DOI] [PubMed] [Google Scholar]
- 121. Ponnaluri V.K.C., Zhang G., Esteve P.O., Spracklin G., Sian S., Xu S.Y., Benoukraf T., Pradhan S.. NicE-seq: High resolution open chromatin profiling. Genome Biol. 2017; 18:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lagier-Tourenne C., Polymenidou M., Hutt K.R., Vu A.Q., Baughn M., Huelga S.C., Clutario K.M., Ling S.C., Liang T.Y., Mazur C. et al.. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 2012; 15:1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Bergeron D., Pal G., Beaulieu Y.B., Chabot B., Bachand F.. Regulated intron retention and nuclear pre-mRNA decay contribute to PABPN1 autoregulation. Mol. Cell Biol. 2015; 35:2503–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szczesniak M.W., Gaffney D.J., Elo L.L., Zhang X. et al.. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016; 17:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Pimentel H., Conboy J.G., Pachter L.. Keep me around: intron retention detection and analysis. 2015; 02 October 2015, preprint: not peer reviewedhttp://arxiv.org/abs/1510.00696.
- 126. Oghabian A., Greco D., Frilander M.J.. IntEREst: Intron-exon retention estimator. BMC Bioinformatics. 2018; 19:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Katz Y., Wang E.T., Airoldi E.M., Burge C.B.. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010; 7:1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Langmead B., Salzberg S.L.. Fast gapped-read alignment with bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Roberts A., Pachter L.. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods. 2013; 10:71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Audic S., Claverie J.M.. The significance of digital gene expression profiles. Genome Res. 1997; 7:986–995. [DOI] [PubMed] [Google Scholar]
- 131. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Anders S., Reyes A., Huber W.. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012; 22:2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y.. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Saraiva-Agostinho N., Barbosa-Morais N.. Psichomics: Graphical application for alternative splicing quantification and analysis. Nucleic Acids Res. 2019; 47:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Trincado J.L., Entizne J.C., Hysenaj G., Singh B., Skalic M., Elliott D.J., Eyras E.. SUPPA2: Fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018; 19:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Tapial J., Ha K.C.H., Sterne-Weiler T., Gohr A., Braunschweig U., Hermoso-Pulido A., Quesnel-Vallieres M., Permanyer J., Sodaei R., Marquez Y. et al.. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017; 27:1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Foissac S., Sammeth M.. ASTALAVISTA: Dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 2007; 35:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Zhang R., Calixto C.P.G., Marquez Y., Venhuizen P., Tzioutziou N.A., Guo W., Spensley M., Entizne J.C., Lewandowska D., Ten Have S. et al.. A high quality arabidopsis transcriptome for accurate transcript-level analysis of alternative splicing. Nucleic Acids Res. 2017; 45:5061–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Feng S., Xu M., Liu F., Cui C., Zhou B.. Reconstruction of the full-length transcriptome atlas using PacBio iso-seq provides insight into the alternative splicing in gossypium australe. BMC Plant Biol. 2019; 19:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Wang Q., Rio D.C.. JUM is a computational method for comprehensive annotation-free analysis of alternative pre-mRNA splicing patterns. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E8181–E8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Yeo G., Burge C.B.. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004; 11:377–394. [DOI] [PubMed] [Google Scholar]
- 143. Bock C. Analysing and interpreting DNA methylation data. Nat. Rev. Genet. 2012; 13:705–719. [DOI] [PubMed] [Google Scholar]
- 144. Bailey T., Krajewski P., Ladunga I., Lefebvre C., Li Q., Liu T., Madrigal P., Taslim C., Zhang J.. Practical guidelines for the comprehensive analysis of ChIP-seq data. PLoS Comput. Biol. 2013; 9:e1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Gomez-Cabrero D., Abugessaisa I., Maier D., Teschendorff A., Merkenschlager M., Gisel A., Ballestar E., Bongcam-Rudloff E., Conesa A., Tegner J.. Data integration in the era of omics: Current and future challenges. BMC Syst. Biol. 2014; 8:I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Qin J., Yan B., Hu Y., Wang P., Wang J.. Applications of integrative OMICs approaches to gene regulation studies. Quant. Biol. 2016; 4:283–301. [Google Scholar]
- 147. Huang S., Chaudhary K., Garmire L.X.. More is better: recent progress in multi-omics data integration methods. Front. Genet. 2017; 8:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Barash Y., Calarco J.A., Gao W., Pan Q., Wang X., Shai O., Blencowe B.J., Frey B.J.. Deciphering the splicing code. Nature. 2010; 465:53–59. [DOI] [PubMed] [Google Scholar]
- 149. Leung M.K., Xiong H.Y., Lee L.J., Frey B.J.. Deep learning of the tissue-regulated splicing code. Bioinformatics. 2014; 30:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K., Hua Y., Gueroussov S., Najafabadi H.S., Hughes T.R. et al.. RNA splicing. the human splicing code reveals new insights into the genetic determinants of disease. Science. 2015; 347:1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B. et al.. Predicting splicing from primary sequence with deep learning. Cell. 2019; 176:535–548. [DOI] [PubMed] [Google Scholar]
- 152. Baeza-Centurion P., Minana B., Schmiedel J.M., Valcarcel J., Lehner B.. Combinatorial genetics reveals a scaling law for the effects of mutations on splicing. Cell. 2019; 176:549–563. [DOI] [PubMed] [Google Scholar]
- 153. Pacini C., Koziol M.J.. Bioinformatics challenges and perspectives when studying the effect of epigenetic modifications on alternative splicing. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2018; 373:1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Zhang C., Frias M.A., Mele A., Ruggiu M., Eom T., Marney C.B., Wang H., Licatalosi D.D., Fak J.J., Darnell R.B.. Integrative modeling defines the nova splicing-regulatory network and its combinatorial controls. Science. 2010; 329:439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Davis-Turak J., Johnson T.L., Hoffmann A.. Mathematical modeling identifies potential gene structure determinants of co-transcriptional control of alternative pre-mRNA splicing. Nucleic Acids Res. 2018; 46:10598–10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Wong J.J.L., Ritchie W., Gao D., Lau K.A., Gonzalez M., Choudhary A., Taft R.J., Rasko J.E.J., Holst J.. Identification of nuclear-enriched miRNAs during mouse granulopoiesis. J. Hematol. Oncol. 2014; 7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016; 17:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Irimia M., Denuc A., Ferran J.L., Pernaute B., Puelles L., Roy S.W., Garcia-Fernàndez J., Marfany G.. Evolutionarily conserved A-to-I editing increases protein stability of the alternative splicing factor Nova1. RNA Biol. 2012; 9:12–21. [DOI] [PubMed] [Google Scholar]
- 159. Rueter S.M., Dawson T.R., Emeson R.B.. Regulation of alternative splicing by RNA editing. Nature. 1999; 399:75–80. [DOI] [PubMed] [Google Scholar]
- 160. Ma Z., Zhu P., Shi H., Guo L., Zhang Q., Chen Y., Chen S., Zhang Z., Peng J., Chen J.. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature. 2019; 568:259–263. [DOI] [PubMed] [Google Scholar]
- 161. El-Brolosy M.A., Kontarakis Z., Rossi A., Kuenne C., Günther S., Fukuda N., Kikhi K., Boezio G.L.M., Takacs C.M., Lai S. et al.. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019; 568:193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Lee S., Singh I., Tisdale S., Abdel-Wahab O., Leslie C.S., Mayr C.. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature. 2018; 561:127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Fong N., Saldi T., Sheridan R.M., Cortazar M.A., Bentley D.L.. RNA pol II dynamics modulate co-transcriptional chromatin modification, CTD phosphorylation, and transcriptional direction. Mol. Cell. 2017; 66:546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Vitting-Seerup K., Porse B.T., Sandelin A., Waage J.. spliceR: an R package for classification of alternative splicing and prediction of coding potential from RNA-seq data. BMC Bioinformatics. 2014; 15:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Sterne-Weiler T., Weatheritt R.J., Best A.J., Ha K.C.H., Blencowe B.J.. Efficient and accurate quantitative profiling of alternative splicing patterns of any complexity on a laptop. Mol. Cell. 2018; 72:187–200. [DOI] [PubMed] [Google Scholar]
- 166. Norton S.S., Vaquero-Garcia J., Lahens N.F., Grant G.R., Barash Y.. Outlier detection for improved differential splicing quantification from RNA-seq experiments with replicates. Bioinformatics. 2018; 34:1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]