Abstract
The Gasterosteidae fish family hosts several species that are important models for eco-evolutionary, genetic, and genomic research. In particular, a wealth of genetic and genomic data has been generated for the three-spined stickleback (Gasterosteus aculeatus), the “ecology’s supermodel,” whereas the genomic resources for the nine-spined stickleback (Pungitius pungitius) have remained relatively scarce. Here, we report a high-quality chromosome-level genome assembly of P. pungitius consisting of 5,303 contigs (N50 = 1.2 Mbp) with a total size of 521 Mbp. These contigs were mapped to 21 linkage groups using a high-density linkage map, yielding a final assembly with 98.5% BUSCO completeness. A total of 25,062 protein-coding genes were annotated, and about 23% of the assembly was found to consist of repetitive elements. A comprehensive analysis of repetitive elements uncovered centromere-specific tandem repeats and provided insights into the evolution of retrotransposons. A multigene phylogenetic analysis inferred a divergence time of about 26 million years ago (Ma) between nine- and three-spined sticklebacks, which is far older than the commonly assumed estimate of 13 Ma. Compared with the three-spined stickleback, we identified an additional duplication of several genes in the hemoglobin cluster. Sequencing data from populations adapted to different environments indicated potential copy number variations in hemoglobin genes. Furthermore, genome-wide synteny comparisons between three- and nine-spined sticklebacks identified chromosomal rearrangements underlying the karyotypic differences between the two species. The high-quality chromosome-scale assembly of the nine-spined stickleback genome obtained with long-read sequencing technology provides a crucial resource for comparative and population genomic investigations of stickleback fishes and teleosts.
Keywords: genome assembly, stickleback, Pungitius pungitius, comparative genomics
Introduction
The small teleost fishes of the Gasterosteidae family have served as important model systems in ecology and evolutionary biology, and in the study of adaptation in particular (Wootton 1976, 1984; Bell and Foster 1994; Östlund-Nilsson et al. 2006; Von Hippel 2010). The most well-known member of this family is the three-spined stickleback (Gasterosteus aculeatus Linnaeus, 1758), also known as a supermodel in ecology and evolutionary biology (Gibson 2005). The genome assembly of the three-spined stickleback has been available since 2006 and was formally published in 2012 (Jones et al. 2012), making the species an attractive model system for studying genomic architecture of ecologically important traits, as well as for population genomic studies in general. The nine-spined stickleback (Pungitius pungitius Linnaeus, 1758), suggested to have diverged from the three-spined stickleback at least 13 Ma (Bell et al. 2009), is the next most frequently utilized model species from the Gasterostidae family. It has recently gained recognition as an especially interesting model system for comparative investigations of adaptive evolution (Shapiro et al. 2006; Herczeg et al. 2009; Shikano et al. 2013; Raeymaekers et al. 2017), sex chromosome evolution (Shikano et al. 2011; Dixon et al. 2019; Natri et al. 2019), and study of adaptive divergence in the face of strong genetic drift (Merilä 2013; Karhunen et al. 2014). Although the nine- and three-spined sticklebacks share very similar circumpolar distribution ranges and similar habitat requirements (Wootton 1984), the former shows a far greater degree of genetic population structuring than the latter (Shikano et al. 2010; DeFaveri et al. 2011; cf. Guo et al. 2019 vs. Fang et al. 2018). This, together with the fact that the genus Pungitius appears to be more specious than the genus Gasterosteus (8–10 vs. 3 species, respectively; Eschmeyer 2015), suggests that there are likely important differences in processes and forces governing differentiation between each of the two species. Hence, this species pair provides an interesting model system for comparative and population genomic investigations aiming to disentangle the relative importance of factors influencing processes of population differentiation and speciation.
Although genomic resources for the three-spined stickleback, including an annotated reference genome (Jones et al. 2012), are well-developed, those for the nine-spined stickleback are rather less developed, typically relying on the three-spined stickleback reference genome. The recently published ultra-high density linkage map (Rastas et al. 2016; Li et al. 2018) and a draft version of the nine-spined stickleback genome based on short-read sequencing technology (Nelson and Cresko 2018) are important developments in this regard. However, short-read technology-based draft assemblies are often of limited utility when dealing with fairly large and complex vertebrate genomes. Therefore, a high-quality genome assembly based on long-read technologies for the nine-spined stickleback would provide a valuable resource for comparative and population genomic studies of stickleback fishes.
High-contiguity chromosome-level assemblies obtained from long-range information are vital for resolving large repetitive regions and providing robust insights into genome and chromosome evolution. Furthermore, a particularly high degree of divergence in karyotype characterized by varied chromosomal morphology and diploid numbers in sticklebacks has been previously noted (Chen and Reisman 1970; Ocalewicz et al. 2011; Urton et al. 2011). The three- and nine-spined sticklebacks have a diploid chromosome number (2n) of 42, and the number of arms (NF) of 58 and 70, respectively, whereas the four-spined (Apeltes quadracus) and brook stickleback (Culaea inconstans) karyotypes have 23 pairs of chromosomes. Hence, the 2n of the nine-spined stickleback is more similar to that of three-spined stickleback than to those of the more closely related four-spined and brook sticklebacks. Thus, the exact ancestral karyotype of sticklebacks is not well understood in relation to the phylogeny (Kawahara et al. 2009) of the family.
Here, we present the first chromosome-level genome assembly of the nine-spined stickleback. The high-coverage long-read PacBio sequencing integrated with an ultra-dense linkage map yielded a high-quality contiguous assembly ordered into 21 pseudo-chromosomes. Using this new resource, we provide a comprehensive analysis of repetitive elements including centromeric repeats in the nine-spined stickleback genome. We also describe a recent duplication in the hemoglobin gene cluster and show that this region could potentially involve frequent copy number variations (CNVs) in the species. Utilizing our chromosome-scale assembly, we identify and pin-point structural variations potentially explaining the divergent karyotypes of the three- and nine-spined sticklebacks.
Materials and Methods
Sampling, DNA Extraction, and Sequencing
The sequenced male individual was caught April 28, 2014 from Pyöreälampi pond from northwestern Finland (66°15″N, 29°26″E). This small (<5 ha surface area) isolated pond was selected as the source because the level of genetic variability in this pond is very low, as revealed by earlier population genetic studies (Shikano et al. 2010). Genomic DNA was extracted from muscle tissue using phenol–chloroform method and fragmented to 20 kb size. All libraries were size selected using BluePippin (4–7 kb) and sequenced on PacBio RSII in a total of 86 SMRT cells (63 SMRT cells with P6v1/C4 chemistry, 23 SMRT cells with P6v2/C4 chemistry). For short-read sequencing, paired-end sequencing Illumina HiSeq2000 (rapid run 2 × 250 nt) was performed for the same individual.
Linkage Map Construction
Three F2-generation interpopulation crosses between pond and marine nine-spined sticklebacks were used as linkage mapping populations. Each of them consisted of first crossing an adult marine nine-spined stickleback female from Southern Finland (Helsinki, 60°13′N, 25°11′E) to a pond male from three different populations viz. Rytilampi, Finland, 66°23′N, 29°19′E; Pyöreälampi (Finland) 66°15′N, 29°26′E; and Bynästjärnen (Sweden) 64°27′N; 19°26′E in 2006, 2011, and 2012, respectively. After the F1 generations fish had matured, F2 generations were created by single full-sib mating within each hybrid cross. Fish in both parental generations and the resulting F2 offspring in the three crosses were RAD sequenced as described earlier for two of the crosses in Rastas et al. (2016).
The linkage mapping followed the Lep-MAP3 (LM3) pipeline (Rastas 2017). First, the individual fastq files were mapped to the contig assembly using BWA-MEM (v 0.7.10) (Li 2013) and sorted bam files were created using samtools (v 1.3.1) (Li et al. 2009). Second, the LM3 pipeline (samtools mpileup and custom scripts) was used to produce proper data for mapping, following with ParentCall2 module with XLimit = 2 parameter to call markers from autosomes and the sex chromosome. Third, Filtering2 module was run on the data with dataTolerance = 0.001 parameter to remove markers segregating in non-Mendelian fashion (1:1,000 by chance). SeparateChromosomes2 was then run with lodLimit = 75 finding 21 (major) linkage groups. These group names were mapped to chromosome names used in Rastas et al. (2016). After this, JoinSingles2All was run with lodLimit = 60 and lodDifference = 10 to add more markers into linkage groups. After these steps the maps had over 89,000 markers assigned to 21 chromosomes.
In the next step, the OrderMarker2 module was run on each chromosome twice with parameters informativeMask = 13 and 23, removing markers informative only for the female or male parent, respectively. The reason for constructing two maps was to reduce the uncertainty in map position caused by markers informative only for one but different parent (having no direct information between each other). The orders were inspected with the LMPlot module and Marey maps were made using custom R scripts.
Genome Assembly
The raw reads were error-corrected using the hierarchical genome assembly process and assembled using Celera assembler. Genome assembly was performed using Celera assembler 8.2, followed by polishing using Quiver (Chin et al. 2013), yielding 5,303 scaffolds with a total size of 522 Mbp. Quality checked Illumina HiSeq2500 reads were then mapped to the contigs using BWA-MEM (v0.7.10) (Li 2013) and the alignment was used to polish the PacBio assembly using Pilon (v 1.9) (Walker et al. 2014). Validation of the assembly in terms of its completeness was performed by searching for core eukaryotic orthologous genes using BUSCO (v3.0.1) (Simão et al. 2015). The contigs consisting of mitochondrial genome sequences were discarded owing to misassembly, and we then used the mitochondrial sequence described in Guo et al. (2016) as the reference sequence. The Illumina reads mapping to the mitochondrial genome were then extracted and variants were inferred using samtools (Li et al. 2009) mpileup. A consensus mitochondrial genome sequence was generated using GATK (DePristo et al. 2011) FastaAlternateReferenceMaker and added to the assembly.
Anchoring Contigs to the Linkage Map
Each marker in the linkage map had a coordinate in the contig assembly, and this information could be used directly to anchor contigs into pseudo-chromosomes. Each contig was placed based on most abundant linkage group in the markers. For the contigs where multiple SNPs supported different linkage groups, based on the number of such matches, the contig was either broken or assigned to the linkage group with largest number of hits. The median map position for each contig was computed and it was used to approximate place contigs within pseudo-chromosomes. A gap of 200 bases was inserted in between the anchored contigs. Contigs with only one marker were not anchored, except for the X chromosome region where typical contig lengths were shorter. The exact location and orientation of contigs and chimeric contigs were further fixed by manually inspecting the Marey maps. The recombination rate was estimated as the derivative of a nondecreasing spline function fitted to the Marey map using module cobs (He and Ng 1999) in R. The R code to estimate recombination rate is included in the supplement.
Transcriptome Assembly
The RNA-seq data from brain and liver for four individuals (two female and two male samples), all sampled from Pyöreälampi pond from northwestern Finland (66°15″N; 29°26″E) were generated. The cDNA libraries and sequencing were done by BGI Hongkong Co., Limited. Sequencing was performed on the Illumina HiSeq2000 platform with 90 bp paired-end strategy. The read data were evaluated for quality using FastQC and pooled to assemble using Trinity package (v2.0.6) (Grabherr et al. 2011; Emms and Kelly 2015). In silico normalization was performed on the reads prior to the assembly (Haas et al. 2013). Using default parameters, the Trinity de novo pipeline resulted in 255,469 transcripts with a CEGMA completeness of around 89% (complete match) to 100% (partial match). Transcript abundance estimates were obtained using the RSEM method implemented within the Trinity package. The assembled transcripts were then filtered based on a FPKM (fragments per kilobase of transcript per million fragments sequenced) threshold of 0.05, resulting in 123,174 transcripts.
Repeat Annotation
Repetitive sequences in the P. pungitius genome assembly were identified using both de novo and homology methods. The de novo repeat identification was performed using Repeat Modeler (v1.0.8, http://www.repeatmasker.org/RepeatModeler) and Transposon PSI (http://transposonpsi.sourceforge.net/). The sequences were combined and clustered using USEARCH (v9.2.64) (Edgar 2010) at a threshold of 80%. In addition, full-length LTR sequences were identified using LTR_finder (Xu and Wang 2007) and LTRharvest (Ellinghaus et al. 2008), and were combined using LTR_retriever (Ou and Jiang 2018). The sequences were classified using RepeatClassifier (a part of Repeatmodeler package), TEclass, Censor, and Dfam database (Wheeler et al. 2012). RepeatMasker (v 4.0.7) was used to annotate the identified repeat elements on the assembly.
Genome Annotation
The annotation was done on the repeat-masked genome following a two-pass approach using MAKER2 (v 2.31.9) pipeline (Holt and Yandell 2011). The first round used Genemark-ES (v 2.3e) (Lomsadze et al. 2005) for ab initio prediction of genes and SNAP model trained on CEGMA genes. The de novo transcriptome assembly, UniProt/SwissProt database (UniProt Consortium 2015) and G. aculeatus CDS sequences were used as evidence sets for the prediction of gene models. For the second round, SNAP (v 20131129) (Korf 2004) and AUGUSTUS (v 3.2.2) (Stanke et al. 2008) were trained on the gene model predicted from the first pass. Functional annotation was performed using BlastP against UniProt proteins with an E-value threshold of 1e-5, and InterProScan (v 5.4-47) (Jones et al. 2014) was used for domain annotation. The resulting gene models were filtered to retain those with AED value of 0.5 or less, having PFAM annotations and significant hits to known proteins against UniProt DB (E-value 1e-5).
Tandem Repeat Analysis
To determine the sequence and structure of centromeric repeat sequence, a random sample of 500,000 PacBio subreads were extracted. These were processed to retain only sequences with length greater than 1,000 bp and less than 5% Ns, and low complexity regions were masked using DUST filter as done in Melters et al. (2013). Tandem repeat finder (v4.0.7) (Benson 1999) was run on the resulting sequences. Sequences greater than 100 bp and occupying more than 80% of the read length were retained as putative centromeric repeats. The most abundant representative repeat sequence of 178 bp length was then aligned to the centromeric repeat sequence in the three-spined stickleback (GacCEN, accession KT321856 [Cech and Peichel 2015] using PRANK [Löytynoja and Goldman 2005]). To survey STRs in the genome, Phobos (version 3.3.12) was run to determine repeats up to unit size of 6, with otherwise default settings. The telomeric repeat arrays were identified by filtering Phobos tandem repeat output to include greater than 10 copies of repeat.
Identification of Orthogroups and Phylogenetic Analysis
The protein sequences from zebrafish (GRCz10, Ensembl release 89), Atlantic cod (gadMor2, Tørresen et al. 2017), platyfish (Xipmac4.4.2, Ensembl release 89), Nile tilapia (GCF_001858045.1_ASM185804v2), medaka (MEDAKA1, Ensembl release 89), tetraodon (TETRAODON 8.0, Ensembl release 89), fugu (FUGU5), three-spined stickleback (Glazer et al. 2015), and nine-spined stickleback were used for orthologous group analysis using OrthoFinder (v1.0.6) (Emms and Kelly 2015). Clustering into orthologous gene families was done based on best reciprocal BLAST hits resulting from an all-vs.-all BLAST with E-value threshold of 1e-5.
To perform phylogenetic analysis, single copy orthologs were extracted and further filtered to only retain complete BUSCO proteins (based on BUSCO Actinopterygii odb9). The protein alignments were then converted to codon alignment using pal2nal (Suyama et al. 2006), trimmed using gblocks and further filtered to remove the third codon position. First, BEAST analysis (version 2.5) was done (Bouckaert et al. 2019) using bModelTest (Bouckaert and Drummond 2017) and relaxed clock model. The Yule tree model was used with MRCA age calibrations added according to the divergence estimates in Betancur-R et al. (2013) for all the nodes except the sticklebacks and divergence time was estimated for the sticklebacks. The Markov-chain Monte Carlo (MCMC) analysis was run using a chain length of 10 million logging every 10,000th step. Tracer (v1.6.0) (Rambaut et al. 2018) was used to verify that the effective sample size is above 200 for all model parameters, indicating convergence of the MCMC analysis. TreeAnnotator (v2.4.8) was then used to generate a maximum clade credibility tree with median node heights.
To further check the robustness of the obtained estimates, we applied rigorous filters on the orthogroups. For this, BEAST analysis with the same parameters as mentioned above was performed on all the individual 2,691 gene sets. From the resulting trees, all those that did not support monophyly of the two stickleback species were eliminated. Second, to eliminate potential misalignments and paralogy, the protein alignments were inspected with a set of stringent thresholds to remove alignments with outlier-like values of bit score, alignment length, and gap open. This set of orthogroups was further filtered based on clock-likeness by excluding alignments for which a high standard deviation of the uncorrelated lognormal (Drummond et al. 2006) molecular clock was inferred in the single-gene BEAST analyses (a threshold of 0.1 for UCLDstddev was chosen). The remaining 778 gene alignments were concatenated to form a supermatrix and BEAST analysis was then rerun with this supermatrix using similar settings as above. MCMC analysis was performed using chain length of 1 billion, logging every 1,000 steps with a burn-in of ∼25%. The run was monitored using Tracer and TreeAnnotator was then used to generate maximum clade credibility tree. The tree resulting from the concatenated alignments of 778 genes was used as an input to gene family analysis using CAFE (v 3) (Han et al. 2013). Cafeerror was run for error model estimation and then CAFE was run with a global lambda estimation using the error model.
Hemoglobin Gene Cluster Analysis and Population Analysis
Hemoglobin genes from three-spined stickleback and zebrafish were used to query the nine-spined stickleback genome assembly using TBlastN. The coordinates of the obtained hits were used to extract sequences corresponding to the MN and LA cluster in the assembly. To predict genes in this region, GENSCAN was used with the human model. The predicted protein sequences were aligned using MUSCLE and maximum likelihood trees were generated in MEGA. To calculate read depth across the region, the Illumina reads were first mapped using BWA-MEM, bedtools was then used to calculate coverage per base.
Whole genome sequencing of five individuals each from Pyöreälampi, Finland, and Levin Navolok Bay, Russia, was done using Illumina HiSeq X 150PE, to 10× coverage (BGI Hong Kong). Reads were mapped to the reference genome with BWA-MEM (v. 0.7.15) and realigned around gaps with GATK IndelRealigner (v. 3.7). Duplicate reads were marked, and site-wise sequencing coverage computed with samtools (v. 1.9) markdup and depth, respectively. From these coverage counts, the mean coverage was computed for different repeat element classes, for all coding exons (totaling 205,048 after excluding LG12 which is the sex chromosome) and for the individual genes (coding exons only) within the MN hemoglobin cluster. The coverage for the repeat element classes and the hemoglobin genes were normalized by the mean sequencing coverage over all coding exons.
Synteny Analysis
Large-scale gene order synteny between G. aculeatus and P. pungitius was identified using MCScanX (v1). The collinear blocks with conserved gene order were identified using BlastP with E-value 1e-5 and match size of 10. The same was repeated using the Medaka (MEDAKA1, Ensembl release 89) assembly.
Results
Genome Assembly and Validation
We sequenced a male P. pungitius individual using the PacBio RSII platform yielding ∼110× of genome coverage. The error-corrected reads were assembled using Celera assembler (Miller et al. 2008) followed by polishing with Quiver (Chin et al. 2013). The assembly improvement was done by mapping Illumina HiSeq2500 reads to the polished assembly using Pilon (Miller et al. 2008; Walker et al. 2014). This resulted in an initial assembly consisting of 5,305 contigs with a total size of 521 Mb and an N50 of 1.2 Mb (table 1). The total assembly size is close to the genome size estimates of about 550–650 Mbp in other stickleback species (Hinegardner and Rosen 1972; Vinogradov 1998). The assembly size of the nine-spined stickleback, however, is higher than that of the current size of three-spined stickleback genome (∼460 Mbp, Glazer et al. 2015 and Peichel et al. 2017), this could result due to the underrepresentation of repetitive regions in the three-spined stickleback assembly.
Table 1.
Assembly Statistics
| Raw Assembly | Anchored Assembly | |
|---|---|---|
| Number of contigs | 5,303 | 4,939a |
| Total size of contigs | 521,182,237 | 521,203,469 |
| Longest contigs | 9,719,887 | 32,096,348 |
| N50 scaffold | 1,233,545 | 17,578,551 |
| Assembly validation | ||
| Complete BUSCOs | 97.1 % | |
| Complete single-copy BUSCOs | 4269 (93%) | |
| Complete duplicated BUSCOs | 183 (4%) | |
| Fragmented BUSCOs | 62 (1.4%) | |
| Missing BUSCOs | 68 (1.5%) | |
| Total BUSCO groups searched | 4,584 | |
21 LGs and unplaced contigs.
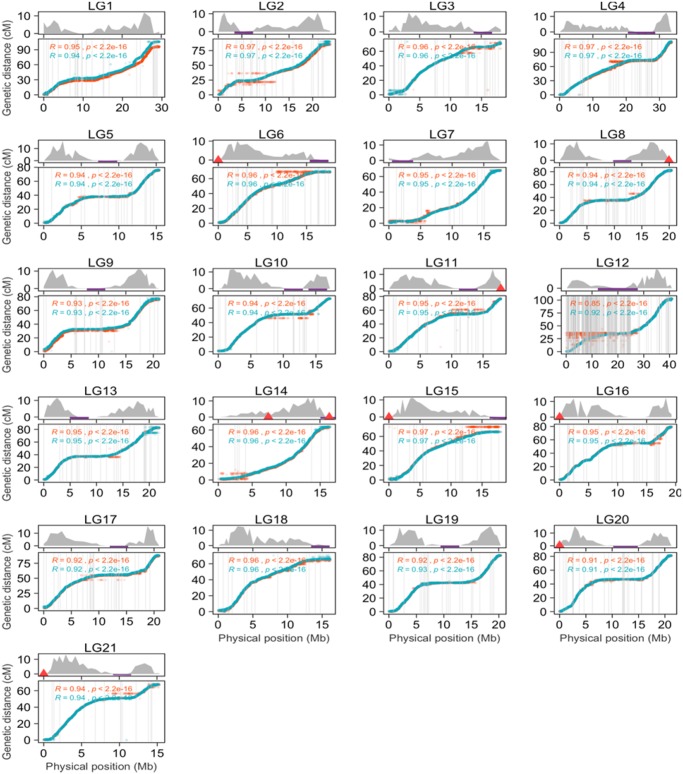
A high-density linkage map of almost 90,000 markers and 1,000 individuals was constructed and used to order and orient the majority of the assembled contigs. Utilizing this map, we placed 686 contigs, comprising ∼444 Mb (∼85%) of the assembly, into 21 pseudo-chromosomes. The largest assembled pseudo-chromosome LG12, which is also the sex chromosome of Eastern European lineage of P. pungitius (Shapiro et al. 2009; Rastas et al. 2016; Natri et al. 2019), is of size ∼40.9 Mbp. To validate the placement of the contigs, we inspected the collinearity between the physical and linkage maps. A high degree of correspondence was observed between the marker order in the linkage map and the assembled pseudo-chromosomes (average r = 0.95; fig. 1). Consistent with expectations, there was a general monotonic increase between the physical and genetic maps along most of the pseudo-chromosomes, except for some regions corresponding to lower recombination rates (fig. 1). Further validation of the assembly was performed by assessing the genome completeness using BUSCO (Simão et al. 2015). The results indicated high contiguity with 98.5% of BUSCO genes found complete or fragmented in the assembly (table 1 and supplementary fig. S1, Supplementary Material online).
Fig. 1.
—Concordance of the assembly with linkage map. The plots represent the correspondence between genetic (cM; y axis) and physical (Mb; x axis) positions of the markers on each of the pseudo-chromosomes (bottom panels). The turquoise points correspond to the sex averaged map. The orange points are map positions from a technical replicate using a different subset of markers (see Materials and Methods). The gray lines represent the contig borders and the maximum value on x axis corresponds to the size of the pseudo-chromosomes in the assembly. The corresponding recombination rates (cM/Mb) are plotted along the length of each pseudo-chromosome (top panel). The potential telomeric regions in the assembly are marked with red triangles and the purple lines represent locations of the identified centromere-associated tandem repeat in the nine-spined stickleback genome.
Genome Annotation
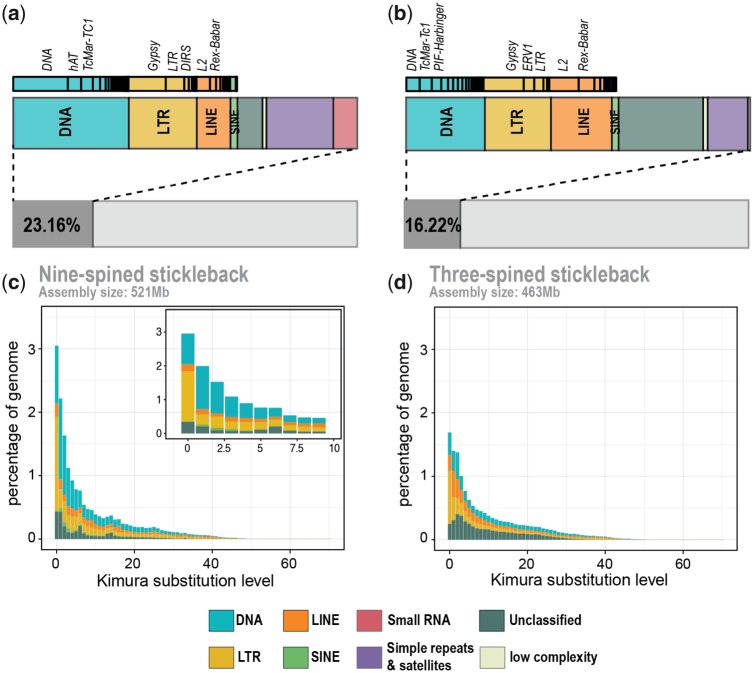
We constructed a nine-spined stickleback-specific repeat library using de novo and homology-based approaches (see Materials and Methods). Redundant sequences were removed, and the remaining sequences were classified (see Materials and Methods). Sequences with hits to UniProt/SwissProt database (UniProt Consortium 2015) were removed and the remaining 1,450 sequences were then combined with teleost repeat sequences from Repbase (Bao et al. 2015). Repeat masking using the custom-made repeat library identified 23.16% of the genome assembly as repetitive. A combination of evidence-based and ab initio gene predictions followed by filtering based on “annotation edit distance” (AED) score and presence of PFAM domains resulted in 25,062 high confidence gene models. Of these, 22,925 reside on the pseudo-chromosomes and the remaining 2,137 on unplaced contigs.
Genome-Wide Characterization of Transposable Elements
Analysis of repetitive elements in the nine-spined stickleback assembly revealed that the genome comprises 23.22% of repetitive sequences, with 6.91% of DNA transposons, ∼4.60% of long terminal repeat (LTR) retrotransposons and 2.28% of LINE and 0.5% of SINE elements (fig. 2a). To facilitate comparison, we created a three-spined stickleback-specific repeat library using the same approach, utilizing the genome assembly generated in Glazer et al. (2015). Using this set of repeats, we classified 16.22% of the three-spined stickleback genome as repetitive, with DNA transposons accounting for 3.73%, LTRs for 3.13% and LINE and SINE elements comprising 2.76% and 0.32%, respectively. This estimate for three-spined stickleback is close to that obtained in other studies (Chalopin et al. 2015; Gao et al. 2016). Similar to earlier observations, the three-spined stickleback genome was found to have no particular predominance of transposable element families and a similar lack of dominance was observed for the nine-spined stickleback, although DNA transposons were slightly more abundant than LTR elements. Furthermore, the diversity of the repeat families is fairly similar in the two species, whereas proportions vary. The abundance of different categories of repetitive elements in the two species is shown in figure 2a. The assembly quality and possible collapse of repetitive sequences naturally affect repeat annotation and thus the overall lower proportion of repeats in the three-spined stickleback genome is likely an underestimation. We also note the general lack of accumulation of repeat families in the nine-spined stickleback genome, supported by the observation that most repeat families seem to have recent activity (fig. 2c). This is consistent with a previous study of non-LTR retrotransposons in the three-spined stickleback and suggests that active DNA loss leads to lower accumulation (Blass et al. 2012).
Fig. 2.
—Transposable elements in stickleback genomes: (a) and (b) represent the fraction of the assembly comprising of repetitive elements in nine- and three-spined stickleback (Glazer et al. 2015) assemblies, respectively. Repeat landscapes representing the divergence of the repeat sequences to the consensus are represented for nine- and three-spined stickleback genomes in (c) and (d), respectively. The inset plot in (c) zooms into the youngest TEs in the nine-spined stickleback assembly.
To study the activity of transposable elements, we estimated the sequence divergence between the repeat copies as a proxy for their age. We found that LTRs are enriched in the youngest category and thus seem to have been recently active. In small populations, selection against repeats and elimination of accumulated repeat copies loses power and active repeat families may rapidly expand in size (Lynch and Conery 2003). To look for such patterns in the reference individual and the respective population, we sequenced (to 10× coverage) five additional individuals from the same small pond population, Pyöreälampi, Finland (FIN-PYO), and five individuals from a large marine population, Levin Navolok Bay, Russia (RUS-LEV), from which the pond population has, most probably, been derived (supplementary fig. S2, Supplementary Material online). We found that, some of the repeat families have significantly higher mean normalized read depths in the pond individuals than in the marine individuals, consistent with weakened selection against repeats in the small pond population (Lynch and Conery 2003). The most notable differences, 24.0% and 31.1% increases, were seen in LTRs and its Gypsy subfamily (one-sided Welch t-test: P = 6.0e-06 and P = 3.0e-06). The mean normalized coverage for other repeat families was 0.903–1.087, close to the expected coverage of one (see Materials and Methods) and demonstrating that the assembly represents the true number of repeats well, whereas the coverage for LTRs, and specifically Gypsy elements, were 1.804 and 2.219, respectively. Although these numbers indicate recent activity of the particular repeat families in the genome, they might also thus be underrepresented in the assembly.
Characterization of Short Tandem Repeats
Short tandem repeats (STRs) are ubiquitous elements comprising of tandemly repeated units of length 1–6 bp (Tóth et al. 2000). STRs are implicated in various facets of genome evolution like gene regulation and chromatin organization (Kashi et al. 1997; Li et al. 2002). Although a major portion of these repeats are known to reside in the noncoding intronic and intergenic regions of eukaryotic genomes, certain types of STRs have been known to occur in coding regions. The STRs in coding regions are predominantly tri-nucleotide (multiples of three) repeats owing to the strong selection to maintain the reading frame (Metzgar et al. 2000). We surveyed the STR abundances in the nine-spined stickleback genome using Phobos. STRs of lower unit size appeared to be more prevalent than those with larger unit sizes, with the dinucleotide repeats being overall the most abundant class of STRs in the assembly (supplementary fig. S3a, Supplementary Material online). The proportions of STRs of different motif sizes were fairly identical with slight variation among the pseudo-chromosomes (supplementary fig. S3b, Supplementary Material online). However, the nonrandom distribution in different genomic regions is apparent with the overall relative abundance of STRs in intronic and intergenic regions being many folds higher than in genes (supplementary fig. S3c, Supplementary Material online). As expected, the repeats of unit size three, six, and nine were relatively more abundant in the genic regions whereas the dinucleotide repeats were far more abundant in the noncoding regions (supplementary fig. S3c, Supplementary Material online). Among the dinucleotide repeats, AC was the most abundant motif, followed by AG. AGG was the most abundant triplet repeat motif across all regions (supplementary fig. S4, Supplementary Material online). In exonic regions, this was followed by the AGC and CCG motifs, both of which were relatively underrepresented in the intronic/intergenic regions. In addition, the AAT motif was fairly abundant in intergenic and intronic regions but rare in the coding regions (supplementary fig. S4, Supplementary Material online). These findings are in agreement with observations from other eukaryotic genomes (Stallings 1994; Tóth et al. 2000; Li et al. 2004). In addition, we also looked for telomeric tandem repeats characterized by a typical conserved G-rich hexamer motif TTAGGG. Large telomeric arrays were found in LG8, 11, 14, and 21 comprising 975.33, 272.17, 1,663, and 1,033.17 copies of the telomeric repeats, respectively (marked in fig. 1). LG14 contains a second, smaller, repeat array in the central region consisting of about 15 copies of telomeric hexamer (fig. 1). This could be an error or a remnant from earlier rearrangements.
Characterizing Centromeric Repeats in the Nine-Spined Stickleback Genome
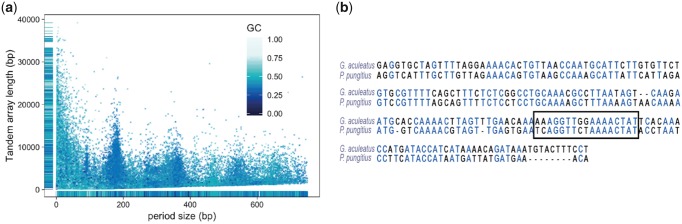
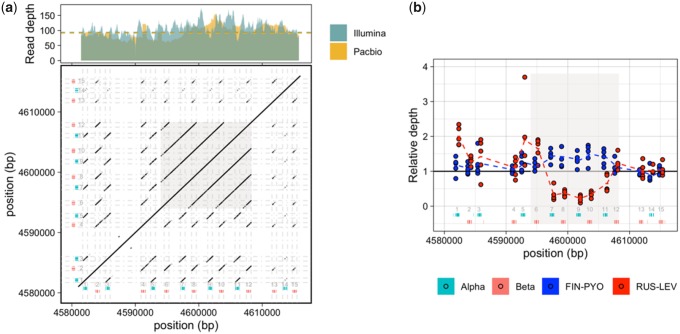
A substantial portion of eukaryotic genomes comprised satellite repeats. Large satellite tandem repeat arrays are often observed in the heterochromatin, including centromeric and pericentromeric regions, and frequently constitute the most abundant tandem repeat category in genome assemblies (Melters et al. 2013). Although the exact role of these repeat elements in structure and function of centromeres is not well understood, they are thought to be vital for various processes such as chromosome segregation, proper pairing of homologous chromosomes, and packaging of centromeric DNA (Plohl et al. 2008). These highly repetitive structures of heterochromatin are still a major impediment to proper genome assembly and mapping of such regions. Our long-read-based assembly allows insights into the sequence composition of these regions. Using the nine-spined stickleback-specific repeat library (fig. 2a), ∼18.4% of the assembly was annotated as known repetitive elements and the rest remained unclassified. The predominant repeat among the unclassified sequences occupied up to ∼2.2% of the assembly and accounted for about 45% of the bases masked by the unclassified repeats in the library. This repeat consisted of tandem repeat units with sizes of 176–180 and ∼360 bp. The low GC content of the sequence along with the distinct monomer size, often associated with centromeric satellites, warranted further analysis. To characterize the centromeric repeat monomer size and abundance, we performed a search for tandem repeats on a randomly selected subset of ∼500,000 PacBio subreads using Tandem Repeat Finder (Benson 1999). The results were parsed using a method similar to that used by Melters et al. (2013). Specifically, we retained only the shortest monomer representing the repeats longer than 50 bp and covering a minimum of 80% of the read length. The resulting repeats show a clear peak around a monomer length of ∼178 bp (fig. 3a). Furthermore, the AT-rich 178 bp monomer is organized into dimers (∼350 bp) and trimers (∼530 bp), as apparent from the distinct peaks (fig. 3a).
Fig. 3.
—Distribution of tandem repeats in the nine-spined stickleback assembly. (a) The x axis represents period size of the tandem repeat units and the y axis represents array size (period size × number of repeats). The points are colored based on GC content. (b). Alignment of the nine-spined stickleback putative centromeric repeat to the centromeric repeat sequence in the three-spined stickleback [GacCEN, accession KT321856 {Cech and Peichel 2015}; identity 61.2%]. Identical bases are represented in blue. The CENP-B box in three-spined stickleback is marked with a dashed rectangle.
Although functionally conserved, owing to the rapid evolution of the satellite DNA, there is often a significant sequence divergence in centromeric repeats among related species, with sequence similarity detectable only in species that have diverged within ∼50 Myr (Melters et al. 2013). In three-spined stickleback, an AT-rich 186 bp repeat was identified and confirmed to occur in centromeric constrictions (Cech and Peichel 2015). The alignment of the centromere-associated repeat sequence in nine-spined stickleback to that of the three-spined stickleback centromeric repeat (GacCEN), showed a considerable sequence similarity (∼61%) and particularly in the CENP-B box region (fig. 3b).
Furthermore, one of the distinguishing features of centromeric and pericentromeric regions is the apparent suppression of recombination. Thus, we investigated the positions of the representative centromeric repeat relative to the recombination rates along the pseudo-chromosomes. The identified repeat indeed consistently corresponded to regions of low recombination, marking pericentromeric regions (fig. 1, top panels). The only exception to this, the second array of centromeric-associated repeats close to the telomere of LG10 (fig. 1) consisted of smaller number of hits and probably represents a mistake in the assembly or misplacement of contigs due the ambiguity in the linkage map in the region.
Genomic Features, Transposable Elements, and Recombination Rates
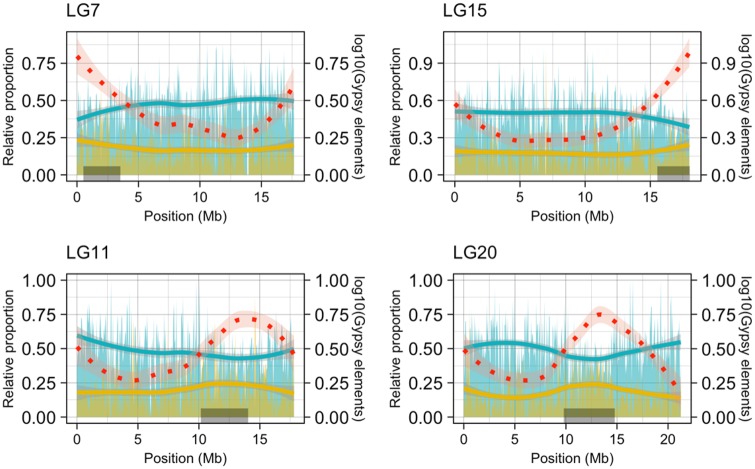
To understand how various genomic features vary relative to each other, we analyzed the distribution of GC content, gene density, transposable elements, and recombination rate along the assembled pseudo-chromosomes. In line with expectations, GC content and gene density were generally reduced in areas of low recombination, whereas TEs were enriched. To access the global trend of this variation, we computed correlations of recombination rates with GC content, gene density, and repeat density (supplementary figs. S5 and S6, Supplementary Material online). Indeed, GC content shows a significant positive correlation with recombination rate (r = 0.44, P value <2.2e-16) and transposable element density shows a strong negative correlation (r = −0.41, P <2.2e-16), whereas the gene density is only weakly positively correlated with the recombination rate (r = 0.061, P = 4.1e-05). Interestingly, TE density also shows a negative correlation with the density of STRs (r = −0.35, P <2.2e-16). Furthermore, the approximated pericentromeric region for each of the pseudo-chromosomes was defined based on the location of the inferred centromeric tandem repeat and reduction of recombination rates (for LG1 and LG16, we used only the region of low recombination). Using these compartments for comparison, we found a significant increase in GC content and gene density outside pericentromeric regions and a significant enrichment of TEs in the pericentromeric regions (supplementary fig. S6, Supplementary Material online). Apart from the general enrichment of TEs in the pericentromeric regions, we compared the relative proportions of LTR and DNA elements to the total TE content for each bin across each of the pseudo-chromosomes to look for enrichment of specific classes of TEs. Overall, LTRs, specifically gypsy elements, are consistently more enriched in pericentromeric regions than outside of them (Wilcoxon P <2.2e-16) and thus are likely associated with centromeric regions in the nine-spined stickleback chromosomes (fig. 4). Next, we defined pericentromeric regions in the three-spined stickleback chromosomes using locations of the GacCEN repeat (Cech and Peichel 2015) and including a 2 Mb flanking region on either side of the repeat. A similar increase in relative proportions of LTR elements was observed in the pericentromeric regions of the three-spined stickleback chromosomes (supplementary fig. S7, Supplementary Material online).
Fig. 4.
—Examples of relative proportions of LTR and DNA transposons along nine-spined stickleback pseudo-chromosomes. Distribution of relative proportions of LTR (yellow) and DNA TEs (blue) to total repeat content per 50 kb bin along selected pseudo-chromosomes (LG7, 11, 15, and 20). The red dotted line represents the log10 value of absolute abundance of LTR-gypsy elements across the pseudo-chromosomes. The gray rectangles (at the bottom) depict the pericentromeric region.
Gene-Based Phylogeny and Gene Family Evolution
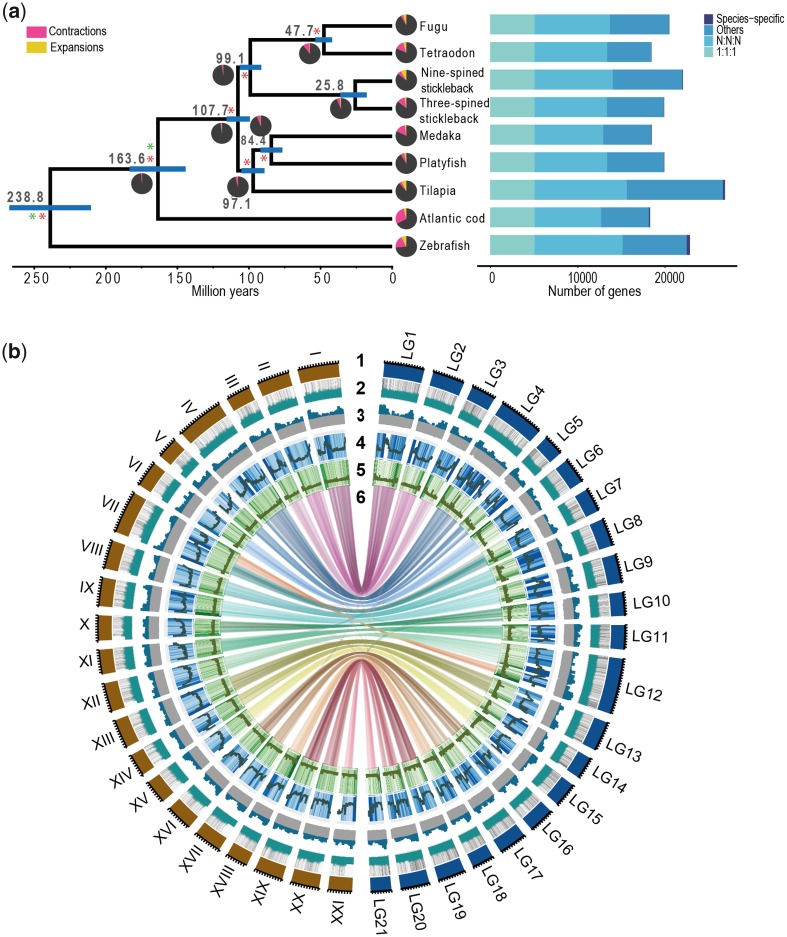
To assess global gene content evolution, we compared the inferred protein-coding gene set with proteins from eight other teleost species including zebrafish, Atlantic cod, platyfish, medaka, fugu, tilapia, tetraodon and three-spined stickleback. A total of 17,976 orthogroups were inferred using Orthofinder (Emms and Kelly 2015), of which 9,811 are present in all nine species and 5,098 were single-copy orthologs in the nine species. The two stickleback species share a set of 71 core orthogroups.
To infer the divergence times among the nine species, we retained 2,691 single-copy orthologs that were also a part of BUSCO Actinopterygii (odb9) genes set. The resulting sequences were concatenated into a supermatrix of 2,085,162 bp that was used for phylogenetic reconstruction and divergence time estimation with BEAST2 (Bouckaert et al. 2014). The phylogeny of the nine species was time calibrated by placing age constraints according to the timeline of teleost evolution inferred by Betancur-R et al. (2013). These age constraints were placed on all internal nodes except the divergence of the two stickleback species. Using this approach, the divergence time between nine- and three-spined sticklebacks was estimated to be around 27.1 Ma, with a 95% highest posterior density interval (HPD) of 25.5–28.8 Ma. To assess the robustness of the estimate, we performed separate BEAST analyses for each of the 2,691 individual gene alignments. This resulted in a distribution of divergence time estimates with the median and mean of 23.0 and 26.6 Ma respectively.
Although the BUSCO genes should appear as single copy in all species, the full data set may contain nonhomologous sequences. To reduce the error from these, we applied a set of rigorous filters. First, we removed 24 gene trees that did not support monophyly of the three- and nine-spined sticklebacks. These genes were excluded from further analyses because, given the long timescales separating sticklebacks from other species included in the phylogeny, their apparent non-monophyly is more likely to result from low phylogenetic signal than from processes like incomplete lineage sorting. Second, to avoid errors due to paralogy and misalignments, we performed stringent filtering of the protein alignments generated by Orthofinder and retained only the 825 high-confidence orthogroups (see Materials and Methods). Finally, we discarded genes not evolving in a clock-like manner. By selecting the remaining genes conforming, at least to some extent, to the molecular-clock assumption (Zuckerkandl and Pauling 1962), greater precision of divergence time estimates can be expected. In addition, as unidentified paralog sequences would likely increase the inferred rate variation, the selection of clock-like genes also further reduces the probability of paralogs remaining in the alignments. The above filtering steps resulted in a total of 778 orthogroups and a concatenated alignment of 548,248 bp. This concatenated alignment was analyzed with BEAST under the same settings as the full supermatrix. Using the concatenated data set, the divergence time between nine- and three-spined stickleback was estimated to be around 25.8 Ma (95% HPD 18.9–35.04 Ma, fig. 5a) whereas the separate analysis of 778 gene alignments gave median and mean divergence time estimates of 23.1 and ∼25.7 Ma, respectively (supplementary fig. S8a, Supplementary Material online). To assess the impact of the calibration points, we repeated the analysis of 778 gene alignments with only two calibration points (indicated by green asterisks in fig. 5a). The median estimate from this analysis was 23.9 Ma (mean of 27.1 Ma, supplementary fig. S8b, Supplementary Material online).
Fig. 5.
—Evolutionary and comparative genomic analysis. (a) Phylogenetic tree using orthogroups inferred from nine teleost species. The number of gene families expanded and contracted has been indicated in the pie diagrams in red and yellow, respectively. (b) Circos plot representing gene-level synteny between the nine- (right) and three-spined stickleback (left) genome assemblies. Tracks: 1, nine- and three-spined stickleback chromosomes; 2, gene density; 3, GC content; 4, TE density; 5, tandem repeat density; 6, links of synteny (defined by 10 collinear genes) between the two species.
Identification of a Recent Tandem Duplication in the Hemoglobin MN Cluster
Synteny analysis using MCScanX (represented as ribbons in fig. 5b) was further utilized to identify tandem duplicates across the assembly. Interestingly, one such duplication was indicated in the hemoglobin cluster in LG11. In teleost fish, hemoglobin genes occur in two distinct unlinked clusters, called MN and LA clusters, located on different chromosomes (Opazo et al. 2013). In the three-spined stickleback, the MN cluster comprising 11 alpha and beta genes, is present in chromosome 11, and LA cluster with two genes is located in chromosome 5. We therefore investigated the hemoglobin repertoire in nine-spined stickleback, using alpha and beta globin genes from three-spined stickleback and zebrafish as query sequences. In line with the expectation from three-spined stickleback genome, the MN and LA clusters were found in LG11 and LG5 of the nine-spined stickleback assembly. Although the arrangement of alpha (Hba) and beta (Hbb) hemoglobin genes on opposite strands in MN cluster is conserved between the species, we found four more alpha and beta globin genes (leading to 15 in total) in the nine-spined than in the three-spined stickleback genome. The entire set of genes was present in a single contig of the raw nine-spined stickleback assembly. To investigate for lineage-specific duplications in the nine-spined stickleback genome, we first excluded the possibility of misassembly by mapping and computing the depth of Illumina reads across the genes. We then performed a self-alignment of the hemoglobin cluster in the nine-spined stickleback and found high internal similarity in the region, extending outside exons and into intergenic regions (fig. 6a). This pattern suggests recent, en bloc duplications likely comprised of multiple Hba and Hbb genes. To estimate the relative age of the gene duplications, we inferred a phylogenetic tree from the predicted protein sequences (supplementary fig. S9, Supplementary Material online). The genes comprising the putative duplication block form a distinct clade next to the syntenic genes in the three-spined stickleback. As the high sequence identity indicates a very recent duplication event, we decided to study the region in more detail using population genomic data. For this, we used the data from five sequenced individuals from both the Pyöreälampi pond (Finland) population and the ancestral marine population from Levin Navolok Bay in the White Sea (Russia). The normalized mean sequencing coverages for individual hemoglobin genes show that the duplication region is fixed in the pond but missing, or present at low frequency, in the ancestral marine population (fig. 6b). This suggests that the duplication has happened since the split of the two populations at most 8,000 years ago, after the retreat of the ice sheet in northeastern Fennoscandia (Shikano et al. 2010; Bruneaux et al. 2013; Wang et al. 2015). Interestingly, other Hb genes within the same cluster (e.g., genes 1, 3, 5, and 6 in fig. 6a) show higher coverage in the marine population and thus additional duplications of individual hemoglobin genes may be relatively frequent.
Fig. 6.
—Analysis of hemoglobin MN cluster. (a) Dotplot (bottom) representing self-alignment of the MN cluster region of the hemoglobin in nine-spined stickleback. Top: Mapped read depth across the MN cluster of hemoglobin in nine-spined stickleback. (b) Relative read depth for five individuals each from nine-spined stickleback populations FIN-PYO (Pyöreälampi pond, Finland) and RUS-LEV (Levin Navolok Bay, Russia). The dashed lines connect the means of the read depth for each of the hemoglobin genes.
Chromosome Evolution in Sticklebacks
Although most teleosts have a karyotype comprising 24 pairs of chromosomes, three fusion events involving chromosomes 1, 4, and 7 were found to likely result in the reduced karyotype of the three-spined stickleback (Amores et al. 2014). Based on our macrosynteny analysis, the fusion of chromosomes 1 and 4 is distinctly detected in the nine-spined stickleback genome with 1:1 synteny correspondence across chromosome arms when compared with the corresponding ortholog in medaka (supplementary fig. S10, Supplementary Material online).
Generally, a high degree of overall genomic collinearity is observed between the nine- and three-spined stickleback chromosomes (fig. 5b). The translocation of the three-spined stickleback chromosome 7 arm to the sex chromosome (LG12) of nine-spined stickleback (Shikano et al. 2013; Rastas 2017) was the only major inter-chromosomal rearrangements observed. However, a considerable number of intra-chromosomal rearrangements were observed. Such a lack of gene order conservation was earlier described by comparing linkage maps between the two species (Rastas et al. 2016).
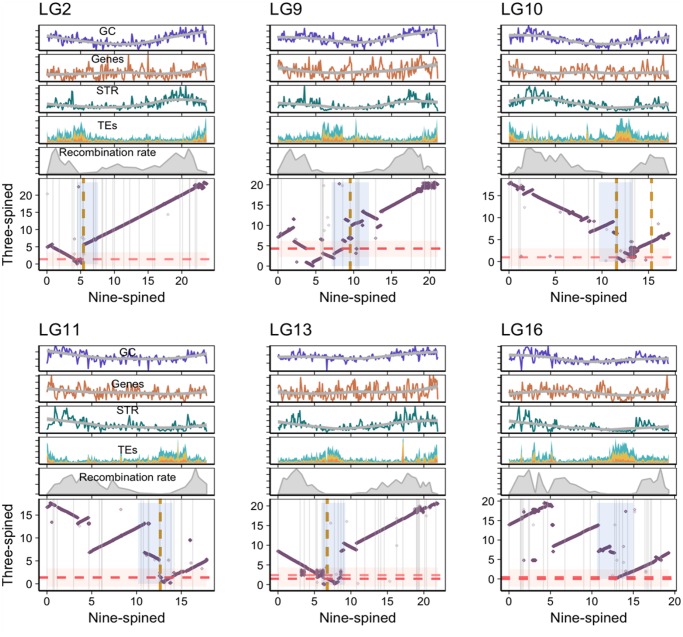
Both 2n and NF (number of chromosome arms) vary considerably across sticklebacks, with nine-spined stickleback having a 2n of 42 with highest NF of 70 among stickleback species with a distinctly larger number of metacentric and submetacentric chromosomes than the three-spined stickleback (Ocalewicz et al. 2011). A comparison of cytogenetic data of the four- and three-spined sticklebacks revealed various rearrangements leading to the differences in NF between the two stickleback species (Urton et al. 2011). A common mechanism to achieve an increase in NF is by pericentric inversions, as these retain 2n whereas increasing NF. Between the three-spined and four-spined sticklebacks, six pericentric inversions, involving chromosomes 1, 3, 8, 17, 20, and 21, have been observed (Urton et al. 2011). First, we attempted to investigate the synteny relationships within these chromosomes to infer potential rearrangements in the nine-spined stickleback chromosomes. The pericentric inversion of chromosome 1 (acrocentric in four-spined stickleback and metacentric in three-spined stickleback) seems specific to the four-spined stickleback karyotype because the chromosome 1 seems to be syntenic along the entire length (and metacentric) between three- and nine-spined sticklebacks. The pericentromeric inversions on the chromosomes orthologous to three-spined stickleback chromosomes 8 and 20 are likely shared by four- and nine-spined sticklebacks, both of which seem metacentric, thus reflecting likely rearrangements in the common ancestor of four- and nine-spined sticklebacks (supplementary fig. S11, Supplementary Material online). We also observe a pericentric inversion in LG21, whereas the rearrangements in LG17 and LG3 are rather unclear (supplementary fig. S11, Supplementary Material online). Because the nine-spined stickleback karyotype has a larger number of bi-armed chromosomes than that of the three-spined stickleback, we looked for additional pericentromeric inversions explaining this difference. First, we inferred karyotypes based on the pericentromeric regions defined in the assembly and looked for chromosomes that differ in morphology between the three- and nine-spined sticklebacks. Indeed, we find that LG2, 10, 11, 13, and LG16 of the nine-spined stickleback harbor potential NF-increasing inversions leading to different chromosomal morphologies in the two species (fig. 7). We further confirmed these rearrangements based on synteny with corresponding medaka chromosomes (supplementary fig. S12, Supplementary Material online). Further rearrangements and inversions in other chromosomes, with unclear indications of NF-increasing chromosome morphology changes, are presented in supplementary figure S13, Supplementary Material online.
Fig. 7.
— Conserved synteny between three-spined and nine-spined stickleback for chromosomes (LG2, 9–11, 13, and 16) potentially involving inversions that have led to divergent chromosomal morphology between three-and nine-spined sticklebacks. Top: The distribution (per 100 kb bins) of GC content, gene density, STR density, TE density (yellow, LTR; blue, DNA; orange, LINE; green, SINE) and recombination rates along the chromosomes. Bottom: Alignment of nine- (x axis) and three-spined (y axis) stickleback chromosomes. The shaded areas represent the putative pericentromeric region in the two genomes. The yellow and red dashed lines represent location of centromeric satellite repeats in nine-spined and three-spined stickleback, respectively.
Discussion
We have generated and described a high-quality chromosome-level genome assembly of the nine-spined stickleback. The raw assembly using PacBio long reads yielded 5,305 contigs and 686 of these were anchored into 21 pseudo-chromosomes with the aid of a high-resolution linkage map, representing 85% of the assembly size. About one-fourth of the nine-spined stickleback genome was found to consist of repetitive sequences. The completeness of the assembly enabled in-depth investigation of these highly repetitive regions of the genome.
In eukaryotic chromosomes, centromeric heterochromatin is often known to be dominated by satellite DNA consisting of large homogenized tandemly arranged arrays of monomeric repeat sequences (Melters et al. 2013). Our survey of tandem repeats in the assembly identified a high-copy 178 bp long centromere-associated repeat organized in such large arrays. This length of identified repeat is consistent with the length of 150–180 bp required to wrap around a single nucleosome unit (Henikoff 2001). Although this repeat sequence has not been validated through cytogenetic methods, it shares a high similarity to the centromeric repeat found in three-spined stickleback using cytogenetic methods (Cech and Peichel 2015).
Furthermore, transposable elements were found to be non-uniformly distributed along the length of all pseudo-chromosomes. Indeed, strong purifying selection against deleterious insertions of TEs in gene-rich regions of high recombination has been one of the proposed models to explain heterogeneity in TE distribution along chromosomes (Bartolomé et al. 2002). Consistent with this expectation, heterochromatic pericentromeric regions, marked by suppression of recombination, had consistently low GC content, were gene-poor and enriched with high copy numbers of TEs as compared with the GC-rich euchromatic region. Furthermore, the relationship between STRs and transposable elements has been subject to debate. Preferential accumulation of STRs outside TE-rich regions has been documented in several species (Morgante et al. 2002; Guo et al. 2009). However, in some species this negative correlation has been shown to be specific to certain chromosomes (Li et al. 2017). In our data, we consistently found a significantly lower proportion of STRs in the pericentromeric regions, and the negative correlation of the STRs with TEs held true for all individual pseudo-chromosomes. We also found a consistent relative enrichment of LTR elements (as compared with DNA elements) in the pericentromeric regions in both the nine- and three-spined stickleback genomes. The tendency of retrotransposon accumulation in centromeric regions and their potential contribution to the evolution of centromeres has been described earlier in many species (Kent et al. 2017). Furthermore, we found that the locations of the identified centromere-associated repeat were closely associated with the regions of reduced recombination rates. Chromatin immunoprecipitation sequencing along with fluorescence in situ hybridization could be useful in confirming the sequence composition (as performed for three-spined stickleback by Cech and Peichel 2015) and abundance of these centromere-specific repeats, as well as to gain insights into the repeat organization in functional centromeres of the nine-spined stickleback.
We estimated the age of divergence between the three- and nine-spined sticklebacks to be 26.6 Ma. The estimate is consistent with fossil evidence showing that the three-spined stickleback had diverged from nine-spined stickleback, and also from its congener, the blackspotted stickleback (Gasterosteuswheatlandi), at least ∼13 Ma (Bell et al. 2009). Our age estimate is considerably larger than a previous estimate that placed the same divergence at 15.86 Ma based on inferred substitution rates for mitochondrial DNA (Aldenhoven et al. 2010) and has been commonly used to as a calibration for phylogenetic analyses in sticklebacks (Teacher et al. 2011; Nelson and Cresko 2018). However, the accuracy of divergence time estimates from mitochondrial DNA has generally been questioned (Brandley et al. 2011; Zheng et al. 2011; Mulcahy et al. 2012).
A recent likelihood-based phylogeny, including representatives of Gasterosteiform species (Rabosky et al. 2018) that reported the divergence time between nine- and three-spined sticklebacks to be 29.6 Ma further corroborates our results, as does similar analysis in Fang et al. (2020). The phylogeny also dates the divergence of G. wheatlandi, closer to three-spined stickleback, at 14.3 Ma, consistent with the 13 Ma oldest three-spine stickleback fossil. Considering that the relative age of nine-spined stickleback to that of the G. wheatlandi is about twice the age from the phylogeny inferred in Rabosky et al. (2018), the age of the nine-spined stickleback divergence is expected to be at least on the order of 25–30 Ma. Thus, our estimate, being fairly robust to different filtering methods applied, supports that the divergence time between the nine- and three-spined sticklebacks occurred earlier than previously believed.
Rearrangements involving chromosomal inversions and translocation are believed to drive speciation by creating reproductive isolation (Faria and Navarro 2010). The rapid evolution of stickleback karyotypes, as apparent from the diversity in the number of chromosomes and chromosome arms, as well as the sex chromosome systems, has attracted considerable interest (Chen and Reisman 1970; Ross et al. 2009; Ocalewicz et al. 2011; Urton et al. 2011; Natri et al. 2013). Based on the pericentromeric regions defined using the centromere-associated repeats and reduced recombination rates, we inspected the pericentromeric rearrangements reported between three- and four-spined sticklebacks, and investigated rearrangements potentially implicated in the difference in chromosomal morphologies between nine-spined and three-spined sticklebacks. The nine-spined stickleback has the highest number of chromosome arms among the sticklebacks (Ocalewicz et al. 2011). Pericentric inversions around the centromere often drive such increases in NF. Using the karyotype derived from the assembled pseudo-chromosomes, we could highlight pericentric inversion events that potentially are responsible for some of the differences in chromosomal morphologies between three-spined and nine-spined sticklebacks. Although the results show the potential of karyotypic deductions using a high-quality genome, a cytogenetic validation of these rearrangements would further improve such deductions.
The diversity in the hemoglobin gene family has been well studied in many vertebrate species. In three-spined stickleback, a high internal similarity between genes in the MN cluster, potentially harboring a recent en bloc duplication comprising of Hba–Hbb or Hba–Hbb–Hba–Hbb has been suggested earlier (Opazo et al. 2013). Interestingly, we identified a recent duplication in the MN hemoglobin cluster, leading to a total of 15 hemoglobin alpha and beta genes in the nine-spined stickleback assembly, in contrast to 11 annotated hemoglobin genes in the MN cluster of the three-spined stickleback assembly. Multiplicity of hemoglobin genes has often been implicated in the ability of tolerating a wide range of environmental stressors (Borza et al. 2009; Opazo et al. 2013; Baalsrud et al. 2017). The observed difference in the hemoglobin cluster between the three- and nine-spined sticklebacks could thus be of potential biological interest if it is associated with the differing ability to tolerate lower oxygen levels in the two species (Lewis et al. 1972). However, the possibility that this difference in gene copy numbers in the two species could partly stem from the differences in the quality of assemblies cannot be excluded. The long-read data in our assembly potentially resolved some of the highly identical regions arising from very recent duplications, which might not be represented in three-spined stickleback genome assembly. Our analysis of the MN cluster in nine-spined stickleback showed a larger region of high internal similarity and populated by repetitive elements. The duplication could thus be a result of nonallelic homologous recombination owing to the presence of repetitive elements. Highly similar regions of segmental duplications are also often a source of genomic rearrangements due to high frequency of possible misalignments (Stankiewicz and Lupski 2002). The higher tendency of rearrangements in such regions is linked with high occurrences of CNVs (Perry et al. 2008). In line with this, our analyses of population samples revealed extensive copy number variation in hemoglobin genes even between closely related populations, suggesting that duplications of individual hemoglobin genes may be of frequent occurrence in the nine-spined sticklebacks inhabiting small water bodies with varying oxygen levels.
Conclusions
Chromosome-scale whole genome assemblies are a critical resource for elucidating genomic underpinnings and evolutionary forces driving variation in genome structure and organization among different species. With the advent of long-read sequencing technologies, complex genomes have been assembled to a fairly high quality owing to improved resolution of complex and repetitive regions. The new chromosome-scale assembly of the nine-spined stickleback genome, including detailed analyses of repetitive regions, provides a valuable resource to comparative genomic studies, as well as a solid template for population genomic studies of stickleback fishes. Furthermore, new phylogenetic analyses based on a large number of protein coding genes support the notion that the divergence of the three- and nine-spined sticklebacks took place about 26 Ma, that is, much earlier than the minimum estimate suggested by the fossil record. Finally, the results regarding extensive copy-number variation in hemoglobin genes, even among populations diverged less than 8,000 years ago, suggests that these genes will deserve further attention in studies seeking to understand the genetic underpinnings of local adaptation in stickleback fishes.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the Oulanka Biological Station, and Pia Saarinen in particular, for help in obtaining the individual used to generate the genome assembly. Kirsi Kähkönen and Ave Tooming-Klunderud are thanked for their help with DNA-extractions and sequencing. PacBio sequencing was performed at the Norwegian Sequencing Centre (NSC) and sequencing (Illumina) of population samples at BGI Hong Kong and Institute of Biotechnology, University of Helsinki. Our research was supported by grants from the Academy of Finland (250435, 263722, 265211, and 1307943 to J.M.), Helsinki Institute for Life Sciences (HiLIFE; H9701-11-109105 to J.M.), and Marie Curie Intra-European Fellowship within the 7th European Community Framework Programme under REA grant agreement PIEF-GA-2013- 624073 (F.C.F.C. and J.M.). M.M. was supported by the Norwegian Research Council (FRIPRO 275869). S.V. and K.S.J. were supported by UiO through a Strategic Research Initiative. We acknowledge the Abel computing Cluster (Norwegian metacenter for High Performance Computing [NOTUR] and the University of Oslo) operated by the Department for Research Computing at USIT, the University of Oslo IT-department as well as the University of Helsinki and ELIXIR Finland node hosted at CSC—IT Center for Science for the ICT resources. We also thank Joost Raeymaekers for providing helpful comments on the manuscript.
Data deposition: This project has been deposited at ENA under the accessions PRJEB33823 (genome assembly and raw sequencing reads for reference individual) and PRJEB33474 (whole genome sequenced individuals from Finland and Russia).
Literature Cited
- Aldenhoven JT, Miller MA, Corneli PS, Shapiro MD.. 2010. Phylogeography of ninespine sticklebacks (Pungitius pungitius) in North America: glacial refugia and the origins of adaptive traits. Mol Ecol. 19(18):4061–4076. [DOI] [PubMed] [Google Scholar]
- Amores A, et al. 2014. A RAD-tag genetic map for the platyfish (Xiphophorus maculatus) reveals mechanisms of karyotype evolution among teleost fish. Genetics 197(2):625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baalsrud HT, et al. 2017. Evolution of hemoglobin genes in codfishes influenced by ocean depth. Sci Rep. 7(1):7956.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé C, Maside X, Charlesworth B.. 2002. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol Biol Evol. 19(6):926–937. [DOI] [PubMed] [Google Scholar]
- Bell MA, Foster SA.. 1994. The evolutionary biology of the threespine stickleback. Oxford University Press. [Google Scholar]
- Bell MA, Stewart JD, Park PJ.. 2009. The world’s oldest fossil threespine stickleback fish. Copeia 2009(2):256–265. [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur-R R, et al. 2013. The tree of life and a new classification of bony fishes. PLoS Curr. 5. doi: 10.1371/currents.tol.53ba26640df0ccaee75bb 165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blass E, Bell M, Boissinot S.. 2012. Accumulation and rapid decay of non-LTR retrotransposons in the genome of the three-spine stickleback. Genome Biol Evol. 4(5):687–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borza T, Stone C, Gamperl AK, Bowman S.. 2009. Atlantic cod (Gadus morhua) hemoglobin genes: multiplicity and polymorphism. BMC Genet. 10(1):51.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, et al. 2014. BEAST2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10(4):e1003537.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, et al. 2019. BEAST2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 15(4):e1006650.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert RR, Drummond AJ.. 2017. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol Biol. 17(1):42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandley MC, et al. 2011. accommodating heterogenous rates of evolution in molecular divergence dating methods: an example using intercontinental dispersal of Plestiodon (Eumeces) lizards. Syst Biol. 60(1):3–15. [DOI] [PubMed] [Google Scholar]
- Bruneaux M, et al. 2013. Molecular evolutionary and population genomic analysis of the nine-spined stickleback using a modified restriction-site-associated DNA tag approach. Mol Ecol. 22(3):565–582. [DOI] [PubMed] [Google Scholar]
- Cech JN, Peichel CL.. 2015. Identification of the centromeric repeat in the threespine stickleback fish (Gasterosteus aculeatus). Chromosome Res. 23(4):767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalopin D, Naville M, Plard F, Galiana D, Volff J-N.. 2015. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol Evol. 7(2):567.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TR, Reisman HM.. 1970. A comparative chromosome study of the North American species of sticklebacks (Teleostei: Gasterosteidae). Cytogenet Genome Res. 9:321–332. [DOI] [PubMed] [Google Scholar]
- Chin C-S, et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10(6):563–569. [DOI] [PubMed] [Google Scholar]
- DeFaveri J, Shikano T, Shimada Y, Goto A, Merilä J.. 2011. Global analysis of genes involved in freshwater adaptation in threespine sticklebacks (Gasterosteus aculeatus). Evolution 65(6):1800–1807. [DOI] [PubMed] [Google Scholar]
- DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon G, Kitano J, Kirkpatrick M.. 2019. The origin of a new sex chromosome by introgression between two stickleback fishes. Mol Biol Evol. 36(1):28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A.. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4(5):e88.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U.. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16(1):157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschmeyer WN, editor. 2015. Catalog of fishes, genera, species, references. Available from: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.as
- Fang B, Merilä J, Ribeiro F, Alexandre CM, Momigliano P.. 2018. Worldwide phylogeny of three-spined sticklebacks. Mol Phylogenet Evol. 127:613–625. [DOI] [PubMed] [Google Scholar]
- Fang B, Momigliano P, Matschiner M, Merilä J. 2020. Estimating uncertainty in divergence times among three-spined stickleback clades using the multispecies coalescent. Mol Phylogenet Evol. https://doi.org/10.1016/j.ympev.2019.106646. [DOI] [PubMed] [Google Scholar]
- Faria R, Navarro A.. 2010. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol Evol. 25(11):660–669. [DOI] [PubMed] [Google Scholar]
- Gao B, et al. 2016. The contribution of transposable elements to size variations between four teleost genomes. Mob DNA 7:4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. 2005. The synthesis and evolution of a supermodel. Science 307(5717):1890–1891. [DOI] [PubMed] [Google Scholar]
- Glazer AM, Killingbeck EE, Mitros T, Rokhsar DS, Miller CT.. 2015. Genome assembly improvement and mapping convergently evolved skeletal traits in sticklebacks with genotyping-by-sequencing. G3 5:1463–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, et al. 2019. A phylogenomic perspective on diversity, hybridization and evolutionary affinities in the stickleback genus Pungitius. Mol Ecol. 28(17):4046–4064. [DOI] [PubMed] [Google Scholar]
- Guo B, Toli E-A, Merilä J.. 2016. Complete mitochondrial genome of the nine-spined stickleback Pungitius pungitius (Gasterosteiformes, Gasterosteidae). Mitochondrial DNA B 1(1):72–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W-J, Ling J, Li P.. 2009. Consensus features of microsatellite distribution: microsatellite contents are universally correlated with recombination rates and are preferentially depressed by centromeres in multicellular eukaryotic genomes. Genomics 93(4):323–331. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Thomas GWC, Lugo-Martinez J, Hahn MW.. 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 30(8):1987–1997. [DOI] [PubMed] [Google Scholar]
- He X, Ng P.. 1999. COBS: qualitatively constrained smoothing via linear programming. Comput Stat. 14(3):315. [Google Scholar]
- Henikoff S. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293(5532):1098–1102. [DOI] [PubMed] [Google Scholar]
- Herczeg G, Gonda A, Merilä J.. 2009. Evolution of gigantism in nine-spined sticklebacks. Evolution 63(12):3190–3200. [DOI] [PubMed] [Google Scholar]
- Hinegardner R, Rosen DE.. 1972. Cellular DNA content and the evolution of teleostean fishes. Am Nat. 106(951):621–644. [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12(1):491.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484(7392):55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhunen M, Ovaskainen O, Herczeg G, Merilä J.. 2014. Bringing habitat information into statistical tests of local adaptation in quantitative traits: a case study of nine-spined sticklebacks. Evolution 68(2):559–568. [DOI] [PubMed] [Google Scholar]
- Kashi Y, King D, Soller M.. 1997. Simple sequence repeats as a source of quantitative genetic variation. Trends Genet. 13(2):74–78. [DOI] [PubMed] [Google Scholar]
- Kawahara R, Miya M, Mabuchi K, Near TJ, Nishida M.. 2009. Stickleback phylogenies resolved: evidence from mitochondrial genomes and 11 nuclear genes. Mol Phylogenet Evol. 50(2):401–404. [DOI] [PubMed] [Google Scholar]
- Kent TV, Uzunović J, Wright SI.. 2017. Coevolution between transposable elements and recombination. Philos Trans R Soc Lond B Biol Sci. 372:pii: 20160458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5(1):59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DB, Walkey M, Dartnall H.. 1972. Some effects of low oxygen tensions on the distribution of the three-spined stickleback Gasterosteus aculeatus L. and the nine-spined stickleback Pungitius pungitius (L.). J Fish Biol. 4(1):103–108. [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN]. Available from: http://arxiv.org/abs/1303.3997
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-C, Korol AB, Fahima T, Beiles A, Nevo E.. 2002. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol. 11(12):2453–2465. [DOI] [PubMed] [Google Scholar]
- Li Y-C, Korol AB, Fahima T, Nevo E.. 2004. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol. 21(6):991–1007. [DOI] [PubMed] [Google Scholar]
- Li Z, et al. 2017. Genome-wide mapping and characterization of microsatellites in the swamp eel genome. Sci Rep. 7(1):3157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kemppainen P, Rastas P, Merilä J.. 2018. Linkage disequilibrium clustering-based approach for association mapping with tightly linked genomewide data. Mol Ecol Resour. 18(4):809–824. [DOI] [PubMed] [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M.. 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33(20):6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N.. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 102:10557–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2003. The origins of genome complexity. Science 302(5649):1401–1404. [DOI] [PubMed] [Google Scholar]
- Melters DP, et al. 2013. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14(1):R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merilä J. 2013. Lakes and ponds as model systems to study parallel evolution. J Limnol. 73:33–45. [Google Scholar]
- Metzgar D, Bytof J, Wills C.. 2000. Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 10(1):72–80. [PMC free article] [PubMed] [Google Scholar]
- Miller JR, et al. 2008. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 24(24):2818–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M, Hanafey M, Powell W.. 2002. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet. 30(2):194–200. [DOI] [PubMed] [Google Scholar]
- Mulcahy DG, et al. 2012. Estimating divergence dates and evaluating dating methods using phylogenomic and mitochondrial data in squamate reptiles. Mol Phylogenet Evol. 65(3):974–991. [DOI] [PubMed] [Google Scholar]
- Natri HM, Merilä J, Shikano T.. 2019. The evolution of sex determination associated with a chromosomal inversion. Nat Commun. 10(1):145.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natri HM, Shikano T, Merilä J.. 2013. Progressive recombination suppression and differentiation in recently evolved neo-sex chromosomes. Mol Biol Evol. 30(5):1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson TC, Cresko WA.. 2018. Ancient genomic variation underlies repeated ecological adaptation in young stickleback populations. Evol Lett. 2(1):9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocalewicz K, Woznicki P, Furgala-Selezniow G, Jankun M.. 2011. Chromosomal location of Ag/CMA3-NORs, 5S rDNA and telomeric repeats in two stickleback species. Ital J Zool. 78(1):12–19. [Google Scholar]
- Opazo JC, Butts GT, Nery MF, Storz JF, Hoffmann FG.. 2013. Whole-genome duplication and the functional diversification of teleost fish hemoglobins. Mol Biol Evol. 30(1):140–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östlund-Nilsson S, Mayer I, Huntingford FA.. 2006. Biology of the three-spined stickleback. CRC Press. [Google Scholar]
- Ou S, Jiang N.. 2018. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176(2):1410–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel CL, Sullivan ST, Liachko I, White MA.. 2017. Improvement of the threespine stickleback genome using a Hi-C-based proximity-guided assembly. J Hered. 108(6):693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, et al. 2008. Copy number variation and evolution in humans and chimpanzees. Genome Res. 18(11):1698–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plohl M, Luchetti A, Mestrović N, Mantovani B.. 2008. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 409(1–2):72–82. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, et al. 2018. An inverse latitudinal gradient in speciation rate for marine fishes. Nature 559(7714):392–395. [DOI] [PubMed] [Google Scholar]
- Raeymaekers JAM, et al. 2017. Adaptive and non-adaptive divergence in a common landscape. Nat Commun. 8(1):267.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA.. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 67(5):901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastas P. 2017. Lep-MAP3: robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics 33(23):3726–3732. [DOI] [PubMed] [Google Scholar]
- Rastas P, Calboli FCF, Guo B, Shikano T, Merilä J.. 2016. Construction of ultradense linkage maps with Lep-MAP2: stickleback F2 recombinant crosses as an example. Genome Biol Evol. 8(1):78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL.. 2009. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae). PLoS Genet. 5(2):e1000391.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Bell MA, Kingsley DM.. 2006. Parallel genetic origins of pelvic reduction in vertebrates. Proc Natl Acad Sci U S A. 103(37):13753–13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, et al. 2009. The genetic architecture of skeletal convergence and sex determination in ninespine sticklebacks. Curr Biol. 19(13):1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikano T, Laine VN, Herczeg G, Vilkki J, Merilä J.. 2013. Genetic architecture of parallel pelvic reduction in ninespine sticklebacks. G3 3:1833–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikano T, Natri HM, Shimada Y, Merilä J.. 2011. High degree of sex chromosome differentiation in stickleback fishes. BMC Genomics 12(1):474.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikano T, Shimada Y, Herczeg G, Merilä J.. 2010. History vs. habitat type: explaining the genetic structure of European nine-spined stickleback (Pungitius pungitius) populations. Mol Ecol. 19(6):1147–1161. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Stallings RL. 1994. Distribution of trinucleotide microsatellites in different categories of mammalian genomic sequence: implications for human genetic diseases. Genomics 21(1):116–121. [DOI] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D.. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24(5):637–644. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR.. 2002. Genome architecture, rearrangements and genomic disorders. Trends Genet. 18(2):74–82. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server):W609–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teacher AGF, Shikano T, Karjalainen ME, Merilä J.. 2011. Phylogeography and genetic structuring of European nine-spined sticklebacks (Pungitius pungitius)—mitochondrial DNA evidence. PLoS One 6(5):e19476.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tørresen OK, et al. 2017. An improved genome assembly uncovers prolific tandem repeats in Atlantic cod. BMC Genomics 18(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth G, Gáspári Z, Jurka J.. 2000. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 10(7):967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43:D204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urton JR, McCann SR, Peichel CL.. 2011. Karyotype differentiation between two stickleback species (Gasterosteidae). Cytogenet Genome Res. 135(2):150.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. 1998. Genome size and GC-percent in vertebrates as determined by flow cytometry: the triangular relationship. Cytometry 31(2):100–109. [DOI] [PubMed] [Google Scholar]
- Von Hippel F. 2010. Tinbergen’s legacy in behaviour: sixty years of landmark stickleback papers. BRILL.
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Shikano T, Persat H, Merilä J.. 2015. Mitochondrial phylogeography and cryptic divergence in the stickleback genus Pungitius. J Biogeogr. 42(12):2334–2348. [Google Scholar]
- Wheeler TJ, et al. 2012. Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res. 41(D1):D70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton RJ. 1976. Biology of the sticklebacks. London: Croom Helm.
- Wootton RJ. 1984. A Functional biology of the sticklebacks. London: Croom Helm.
- Xu Z, Wang H.. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35(Web Server):W265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Peng R, Kuro-o M, Zeng X.. 2011. Exploring patterns and extent of bias in estimating divergence time from mitochondrial DNA sequence data in a particular lineage: a case study of salamanders (Order Caudata). Mol Biol Evol. 28(9):2521–2535. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. 1962. Molecular disease, evolution, and genic heterogeneity. Horizons in Biochemistry, New York: Academic, pp 189–222.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.