Abstract
Bacterial ribosome biogenesis and translation occur in the same cellular compartment. Therefore, a biochemical gate-keeping step is required to prevent error-prone immature ribosomes from engaging in protein synthesis. Here, we provide evidence for a previously unknown quality control mechanism in which the abundant ribosome assembly factor, RbfA, suppresses protein synthesis by immature Escherichia coli 30S subunits. After 30S maturation, RbfA is displaced by initiation factor 3 (IF3), which promotes translation initiation. Genetic interactions between RbfA and IF3 show that RbfA release by IF3 is important during logarithmic growth as well as during stress encountered during stationary phase, low nutrition, low temperature, and antibiotics. By gating the transition from 30S biogenesis to translation initiation, RbfA and IF3 maintain the fidelity of bacterial protein synthesis.
INTRODUCTION
Unlike yeast and other eukaryotes, ribosome biogenesis in bacteria occurs in the same cell compartment as protein synthesis (1,2). Although bacteria lack membrane compartments, they must nevertheless encode robust biochemical checkpoints that gate the transition from ribosome biogenesis to translation. A barrier that prevents immature ribosomes from entering protein synthesis is essential for all cells, because translation by immature ribosomal subunits is inefficient and error-prone (3–9). How bacteria prevent immature ribosomes from initiating translation is not well understood. It is also not known whether the same checkpoint mechanism operates during logarithmic growth and during poor growth when immature subunits accumulate (2).
In yeast, 40S ribosome assembly factors act as fidelity checkpoints at the last stages of pre-40S maturation prior to translation initiation (10,11), through the formation of 80S-like complexes. These late assembly factors mask regions of the pre-40S ribosome that are recognized by translation initiation factors. A similar quality control step has not been clearly demarcated in bacteria (12,13). The binding sites of bacterial assembly factors also overlap the binding sites of translation initiation factors IF1, IF2 and IF3 (12–14), however, suggesting that bacterial assembly factors may also prevent translation initiation by immature subunits. Although several 30S assembly factors are known to act at the end of 30S biogenesis, it is unclear which of these, if any, directly block translation initiation.
Ribosome binding factor A (RbfA) is a strong candidate for the last gatekeeper in 30S biogenesis. The most abundant 30S subunit assembly factor, RbfA’s role in biogenesis was discovered because its overexpression suppressed genetic defects in pre-16S processing (15–17), whereas rbfA deletion impaired 30S biogenesis at low temperatures (18,19). A low-resolution cryo-electron microscopy structure of a 30S•RbfA complex showed that RbfA displaces the top of 16S helix (h) 44 and h45, rendering the 30S•RbfA complex unsuitable for joining with 50S subunits (12). Distortion of the decoding site explained why RbfA associated with pre-30S assembly intermediates and mature 30S subunits, but not with 70S ribosomes or polysomes (15,20,21).
The exclusion of RbfA from 70S ribosomes indicates that RbfA must be released before 30S subunits can initiate translation. RbfA is known to be released from mature 30S subunits by the GTPase RsgA (YjeQ) (20). In current models, GTP hydrolysis induces a conformational change within RsgA that promotes the release of RbfA and RsgA (22). Dissociation of RbfA and RsgA allows 16S helices h44 and h45 to dock with the 30S platform, rendering the 30S subunit suitable for translation (13,20,22,23).
Despite the well-characterized activity of RsgA GTPase, several observations suggested to us that additional Escherichia coli proteins displace RbfA from 30S ribosomes. First, RsgA is nonessential, and the level of RsgA is ∼10-fold less than the amount of RbfA during logarithmic growth (16). Second, it is not known what prevents RbfA from rebinding recycled 30S subunits. Additionally, RsgA’s GTPase activity is inhibited by the alarmone (p)ppGpp (24), which accumulates during stationary phase (25,26). This observation implies that E. coli employs a second RbfA-release factor under adverse conditions.
To test this possibility, we surveyed ribosome-associated proteins for their ability to displace RbfA. Among the proteins tested, IF3 was uniquely able to release RbfA from fully mature 30S subunits but not from immature pre-30S complexes. We also found that RbfA inhibits protein synthesis by pre-30S subunits in the presence of IF3, suggesting that RbfA acts as a gatekeeper to prevent premature entry of pre-30S subunits into the translation cycle. Biochemical and genetics results further showed that IF3 is essential for displacing RbfA during stationary phase, at lower temperature, and under antibiotics stress. Altogether, the results demonstrate that RbfA and IF3 enforce the barrier between ribosome biogenesis and translation, creating a checkpoint that is sensitive to the quality of the 30S decoding site.
MATERIALS AND METHODS
Strains and culture conditions
Strains are listed in the Supplementary Table S1. All bacterial strains were grown in LB media unless stated otherwise. Media were supplemented with antibiotics (100 μg/ml ampicillin, 25 μg/ml kanamycin, 10 μg/ml tetracycline and 100 μg/ml kasugamycin) as required.
Growth analysis: E. coli strains JK382 (parental) and JK378 (infC362) were transformed with either p15BHA over-expressing RbfA (15,27) or with empty vector (pSE420), and were grown overnight at 37°C in LB media supplemented with 10 μg/ml tetracycline plus 100 μg/ml ampicillin. These strains were sub-cultured in fresh media containing 50 μg/ml ampicillin and the optical density (OD600nm) was recorded at the indicated intervals. For growth analysis under nutrient limited conditions, cells were grown overnight as above and sub-cultured into minimal media: 1× MOPS, 2 mM K2HPO4, 0.1 μg/ml thiamine, 0.4% glucose, supplemented with antibiotics as above as previously reported (28).
Plate assays
For plate assays, a single colony from a freshly prepared plate was re-streaked on plates containing LB agar and antibiotics, and incubated at either 37°C or 18°C. Plates were imaged after 1–3 days. For the rbfA complementation assay, p15B-RbfA-A2C-HA was transformed into BX41 (ΔrbfA) cells. MRE600, BX41 and (BX41/ p15B-RbfA-A2C-HA) strains were grown on LB agar plates with no antibiotics, 25 μg/ml kanamycin and 100 μg/ml ampicillin plus 25 μg/ml kanamycin, respectively, in duplicates. Plates were grown at 37°C or 22°C, and imaged after 1–2 days. For antibiotics sensitivity, 5 μl of serially diluted culture of the indicated strains was spotted on LB-agar plates containing gentamycin (0.1 μg/ml), streptomycin (1 μg/ml), chloramphenicol (1 μg/ml), paromomycin (1 μg/ml), colistin (1 μg/ml), kanamycin (2 μg/ml) or neomycin (2 μg/ml). Plates were incubated overnight at 37°C or 24 h at 18°C and imaged.
Plasmids
Plasmids are listed in the Supplementary Table S1. QuikChange mutagenesis and domain separation was carried out in pProEx-HTb-IF3 to obtain pProEx-HTb-IF3-K110L (primers- IF3-K110L-F 5′AGGTATTACTCCGCAGCCTGATTC3′ and IF3-K110L-R 5′GGAGTAATACCTGATAGTCGCCTTC3′) and pProEx-HTb-IF3-Nter (primers- IF3-Nter-F 5′TAACTCGAGGCATGCGG3′ and IF3-Nter-R 5′GATAACTTTTTGCTTTTTCTTCTG3′) and pProEx-HTb-IF3-Cter (primers- IF3-Cter-F 5′CAGGTTAAGGAAATTAAATTCCG3′ and IF3-Cter-R 5′AATGGATCCCATGGCG3′). QuikChange mutagenesis was also carried out on plasmid p15BHA to obtain p15B-RbfA-A2C-HA (primers- p15BHA-F 5′GAATAAACCATGGATGTGCAAAGAATTTGGTCGCC3′ and p15BHA-R 5′GGCGACCAAATTCTTTGCACATCCATGGTTTATTC3′) for the RbfA complementation assay.
Ribosome purification
Mature ribosomes were purified from MRE600 following the protocol of (29). Near mature 30S (nm30S) subunits were isolated from JW0050-3 (ΔksgA) and TPR201 cells following the same protocol, except that the cell lysis was performed in an ethanol-dry ice bath. Pre-30S subunits were purified as follows: BX41 (ΔrbfA) cells were grown in 5 ml LB at 37°C for 9 h with shaking at 250 rpm. A 2 ml aliquot was transferred to 200 ml LB supplemented with 25 μg/ml kanamycin and grown overnight at 37°C with shaking at 250 rpm. 50 ml of this culture was then transferred to two flasks each containing 2 l LB supplemented with 25 μg/ml kanamycin. The cultures were grown at 37°C until O.D.600 ∼0.7, after which the cultures were shifted to a shaking incubator pre-cooled at 17°C and grown until O.D.600 ∼1.2 (5). The cultures were stored at 4°C for 30 min before pelleting cells at 4000 × g for 10 min at 4°C. The cell pellets were washed once with 10 mM Tris–HCl pH 7.5, 15 mM MgCl2 and re-pelleted as above and stored at −20°C until needed. The pellets were resuspended in 15 ml lysis buffer containing 10 mM Tris–HCl pH 7.5, 15 mM MgCl2 and 1 mg/ml lysozyme (hen egg white, Sigma) (freshly prepared). The cells were incubated for 5 min with intermittent vortexing, frozen in an ethanol-dry ice bath, and allowed to thaw at room temperature. Cell lysates were cleared twice by centrifugation at 20 000 × g for 20 min at 4°C. 10–40% sucrose gradients in 60 mM Tris–HCl pH 7.5, 30 mM MgCl2, 300 mM NH4Cl, 6 mM DTT, 3 mM β-mercaptoethanol were prepared in SW28 rotor tubes (Beckman Coulter) using a gradient master (BioComp). An equal quantity of cleared lysate (OD260nm ∼ 100) was layered onto each gradient and spun at 25 000 rpm for 2 h at 4°C (SW28 rotor; Beckman Coulter). The fraction corresponding to pre-30S particles was collected using a fractionator (BioComp) and analyzed for the presence of 17S rRNA on a 1.5% agarose-TAE gel. The protein component was analyzed by 4–20% SDS-PAGE (Bio-Rad). Pooled fractions were concentrated using 100 kDa MWCO ultrafiltration tubes (Millipore) and exchanged 3–5 times with buffer A (20 mM Tris–HCl pH 7.5, 40 mM NH4Cl, 60 mM KCl, 10 mM MgCl2). Aliquots were snap frozen in liquid nitrogen and stored at −80°C until further use.
Protein purification and fluorescence labeling
Escherichia coli proteins S1, RbfA, RsgA, IF1, IF2 and IF3 (WT, mutants and domains) with histidine tags were purified from BL21(DE3) cells harboring the respective plasmids listed in the Supplementary Table S1. Over-expression was induced with 1 mM IPTG at 37°C. Pellets were lysed in lysis buffer (50 mM Tris–HCl pH7.5, 500 mM NaCl) using sonication, and cell debris was removed by centrifugation at 20 000 × g for 20 min. Cleared lysates were passed through a 5 ml HiTrap Ni-column (GE Healthcare) pre-equilibrated with lysis buffer. The column was washed with 25 column volumes of lysis buffer containing 30 mM imidazole and 25 column volumes of lysis buffer with 30 mM imidazole plus 1 M NaCl. His-tagged protein was eluted using lysis buffer plus 500 mM imidazole. Protein-containing fractions were pooled and dialyzed in 50 mM Tris–HCl pH 7.5, 300 mM KCl, 10 mM MgCl2, 6 mM β-mercaptoethanol, 10% glycerol after the purity was checked by SDS PAGE. For fluorophore labeling, the protein of interest containing a single cysteine was dialyzed overnight at 4°C against 80 mM HEPES-K+ pH 7.5, 1 M KCl, 1 mM TCEP. A maleimide conjugate of Cy5 or Cy3 dye (GE Healthcare) was dissolved in anhydrous DMSO (Molecular Probes), mixed with the protein of interest following the manufacturer's instruction and incubated for 2–3 h at room temperature in the dark. Reactions were quenched with 6 mM β–mercaptoethanol, and the labeled protein was re-purified through Ni-column, concentrated in 3 kDa MWCO filtration units (Millipore) and exchanged with dialysis buffer as described above. Subsequently, concentrated proteins were dialyzed a second time against a large excess of dialysis buffer, aliquoted, snap frozen and stored at –80°C. The labeling efficiency and concentration were checked by UV absorption (Nanodrop; Thermo Scientific). IF3 was used before and after removing the histidine tag by TEV protease.
Ultracentrifugation pelleting assay and ultrafiltration assay
200 nM pre-30S or 30S subunits was incubated with 400 nM Cy5-RbfA in buffer A with 3 mM DTT (Buffer A*) for 15 min at 37°C (300 μl total volume is sufficient for six pelleting assays). Unbound Cy5-RbfA was removed by passing the reaction mixture through a 100 kDa MWCO filtration unit for 5 min at 10 000 × g. The filter was washed twice with 200–400 μl buffer A* by centrifugation as above and the complexes were recovered by centrifugation at 1000 × g following the manufacturer's instructions. To monitor the release of Cy5-RbfA in a pelleting assay (30,31), the pre-30S or 30S•Cy5-RbfA complex (50 μl) was incubated with excess IF1/IF2/IF3/mRNA/S1 (individually or in combination) in buffer A* for 15 min at 37°C in a 100 μl reaction volume. 10 μl of this mixture was saved as ‘input’ and 90 μl was separated through a 2 ml 1.1 M sucrose cushion in buffer A by ultracentrifugation in a SW50 rotor (Beckman Coulter). The pellet was dissolved in 30 μl buffer A*, and 10 μl of the input and pellet samples was mixed with 2 μl 2× Tricine loading dye and boiled for 2 min at 95°C and resolved by 4–20% SDS PAGE. Gels were scanned for Cy5 signal using a Typhoon imager (GE Healthcare) and quantified using ImageQuant before staining with Coomassie blue. For monitoring release with a filtration assay as previously described (20,30), unbound Cy5-RbfA was removed from the release reaction mixture by passing the reaction mixtures again through a 100 kDa MWCO filtration unit as above, and input and retentate was further processed similar to pelleting assay.
30S assembly intermediates were prepared by mixing 100 nM 17S rRNA (transcribed) with purified recombinant primary assembly r-proteins (S4, S7, S8, S15, S17 and S20) or primary plus secondary r-proteins (S4, S7, S8, S15, S17 and S20 plus S6, S16, S9, S13, S18 and S19) (200 nM) as previously described (32,33) in HKM20 buffer (80 mM HEPES-K+ pH 7.8, 330 mM KCl and 20 mM MgCl2) at 42°C for 1 h. 17S rRNA or native 30S subunits was also incubated with Cy5-RbfA separately as controls. The reaction mixtures were then layered onto 1.1 M sucrose cushions and analyzed as described above.
Primer extension
Primer extension on total RNA extracted from E. coli (BW25113) and BX41 (ΔrbfA) or rRNA purified from 30S fractions was performed using primers 5′ labeled with either 32P-or Cy5. The 16S 5′ end was analyzed with primer 161 (5′GCGGTATTAGCTACCGT3′) or primer 323 (5′AGTCTGGACCGTGTCTC3′) as described previously (5). For checking the methylation of 16S A1518 and A1519, 5′-32P-labeled primer 1523 (5′GGAGGTGATCCAACCGC3′) (34) was used on rRNA purified from immature and mature 30S subunits. In all cases, the rRNA was purified using RNeasy kit (Qiagen).
In vitro translation assay
Purified pre-30S, nm30S or 30S subunits and native 50S subunits were incubated with or without 3 μM RbfA and combined with in vitro translation components (PURExpress; New England Biolabs). Each reaction mixture (15–20 μl) was supplemented with 1 μl 35S methionine (1 μCi, Perkin Elmer) and 1 μl RNase inhibitor (40 U, Promega) and incubated at 37°C for 2 h in an incubator. The reaction was stopped by placing tubes on ice for 5 min, followed by the addition of 2 μl 2× Tricine loading dye. 8–10 μl of each reaction was loaded without boiling onto a 4–20% SDS PAGE. The gel was then dried under vacuum, exposed overnight against a phosphor screen and imaged using a Typhoon scanner.
3H-uridine pulse labeling and polysome profiling
BW25113 cells were first grown overnight in LB at 37°C with shaking at 250 rpm, then transferred to fresh 5 ml LB at 37°C with shaking at 250 rpm. After 3 h or after 10 h, 50 μl 3H-uridine (50 μCi) was added to the culture, which was allowed to grow for another 9 min, before the addition of 25 μg/ml chloramphenicol for 1 min. Cells were harvested, washed with cold 10 mM Tris–HCl pH 7.5 and 15 mM MgCl2, and the pellets were stored at –80°C. To analyze polysome profiles, cell lysate was prepared as above. Equal amounts of cleared lysate containing total ribosomes (OD260 nm ∼ 20) was separated through 10–40% sucrose gradient in the buffer as above for 2.5 h at 35 000 rpm in a SW41 rotor (Beckman Coulter). The gradients were analyzed using a fractionator (Biocomp) and the A260 nm recorded. Fractions (250 μl) covering the entire gradient were collected in 96-well plate and 3H-uridine incorporation was measured by liquid scintillation counting (Beckman). rRNA analysis was performed using primer extension as described above.
Western blot
Western blot analysis was performed as previously described (20,21). Briefly, reaction mixtures (0.5–1 μg), cell lysates (40–60 μg) or 30S fractions (5–10 μg) (concentrated using 3 kDa MWCO filtration tubes) from polysome profiling experiments were separated by 4–20% SDS PAGE and transferred to a PVDF membrane (Novex) and probed with polyclonal antibodies against IF3. Antibody binding was detected using a secondary goat anti-rabbit WesternDot-655 antibody (ThermoFisher), following the manufacturer's protocol. Membranes were imaged using a Typhoon scanner.
Native PAGE experiments
Co-localization assays were performed by incubating Cy5-RbfA and Cy3-IF3, together or separately, with or without pre-30S (ΔrbfA), 30S (MRE600), nm30S (ΔksgA) or nm30S (TPR201) subunits (100 nM each reactant) in buffer A* for 15 min at 37°C in a 10 μl reaction mixture. 2 μl native loading dye (50% sucrose, 0.02% bromophenol blue) was added before loading 8 μl on a native 4% PAGE (acrylamide: bis-acrylamide 37.5:1, Bio-Rad). The gel was run at ∼15.5°C for ∼45 min in 1× THEM buffer (56 mM HEPES-K+, 34 mM Tris–HCl pH 7.5, 5 mM MgCl2, 0.1 mM EDTA) before scanning with 532 nm excitation (Cy3) and an emission filter at 610 nm for FRET (Typhoon). The relative FRET efficiency Erel = (IAD – IA)/(IAD + ID) was calculated from (35), where 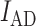 is the intensity of the acceptor in the presence of the donor (i.e. FRET), IA is the intensity of the acceptor in the absence of donor (i.e. direct excitation), and
is the intensity of the acceptor in the presence of the donor (i.e. FRET), IA is the intensity of the acceptor in the absence of donor (i.e. direct excitation), and 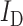 is the intensity of the donor. The gels were also imaged with 633 nm excitation (Cy5).
is the intensity of the donor. The gels were also imaged with 633 nm excitation (Cy5).
For mapping the presence of 5′ leader and 3′ trailer in pre-30S(ΔrbfA) and nm30SΔksgA and TPR201subunits, 10 nM purified rRNA was annealed with 15 nM Cy5-labeled primer-5′end (5′CTAGAGAGACTTGGTATTCATTTTTCGTCTTGCGACG3′), primer-3′end (5′TGTGTGAGCACTGCAAAGAACGCTTTAAGG3′) or primer-161 in a buffer containing 80 mM HEPES-K+ and 330 mM KCl at 70°C for 5 min followed by another incubation at 25°C for 5 min in a thermocycler. Subsequently, 10 mM MgCl2 was added to these reactions to stabilize the interaction between RNA and DNA oligomer. 2 μl native loading dye was added before samples were loaded onto a native 4% polyacrylamide gel. The gel was resolved, imaged and analyzed as above.
To determine the equilibrium dissociation constant (Kd) of Cy5-RbfA binding with 30S subunits, 100 nM Cy5-RbfA was incubated with increasing concentrations of 30S subunits (0–2 μM) in buffer A* for 15 min at 37°C. The relative Kd of unlabeled and Cy5-labeled RbfA (Krel) was determined by competition: 40 nM Cy5-RbfA was mixed with increasing concentrations of unlabeled RbfA (0–4 μM) and 30S subunits (0.17 μM). Samples were incubated for 1 hour at 37°C. In both cases, samples were resolved by native PAGE and quantified as above. Kd and Krel were obtained by fitting the fraction of bound Cy5-RbfA to quadratic expressions of the binding equilibria, as previously described (36).
For binding of RbfA with the 16S rRNA 5′ domain, an extended 5′domain RNA was transcribed and annealed with Cy3-SA5 oligonucleotide as previously described (37,38). The 5′ domain•Cy3-SA5 complex was incubated with Cy5-RbfA, with or without 5′ domain binding ribosomal proteins (S4, S17, S20 and S16), at 37°C for 15 min in HKM20 buffer. For measuring binding of RbfA with 16S (native) or 17S rRNA (transcribed), increasing concentrations of 16S (10–100 nM) or 17S (20–100 nM) rRNA was incubated with 100 nM Cy5-RbfA at 37°C for 15 min in HKM20 buffer. The complexes were resolved and analyzed as described above.
For binding of RbfA with the 30S translation initiation complex (30SIC), 30S subunits were first incubated at 42°C for 30 min in an initiation complex buffer (ICB) containing 50 mM Tris–HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2 as previously described (39). The initiation complex was formed by combining 100 nM 30S subunits, 500 nM mRNA (sodB), 300 nM fMet-tRNAfmet, 300 nM each IF1, IF2 and IF3, and 300 nM GTP in ICB at 37°C for 15 min. Subsequently, 100 nM Cy5-RbfA was incubated with 50 nM initiation complex in ICB at 37°C for 15 min. 2 μl native loading dye was added and the reaction mixtures were resolved and analyzed as described above.
RESULTS
The multi-functional initiation factor IF3 releases RbfA from 30S subunits
We employed an ultracentrifugation pelleting assay and an ultrafiltration assay (20,30,31) to investigate the binding and release of RbfA from E. coli ribosomal 30S subunits under various conditions. In order to distinguish RbfA from ribosomal proteins (r-proteins) of similar size, we fluorescently labeled RbfA with Cy5 at position 2 (Supplementary Figure S1A), which is exposed in the NMR structure of RbfA (40) and is not essential for RbfA function (Supplementary Figure S1B). Fluorescent labeling enabled accurate quantitation of bound and free RbfA and showed that Cy5-RbfA and unlabeled RbfA bind with similar (twofold) affinity (Supplementary Figure S1C and D).
In each pelleting assay or filtration assay, 30S•Cy5-RbfA complexes were incubated with translation initiation factors or mRNA, and the bound Cy5-RbfA was separated from the unbound protein by pelleting the reaction mixture through a sucrose cushion or by filtration. In accordance with previous data (20), we observed that RbfA formed a stable complex with mature 30S subunits (Kd = 120 ± 20 nM) in the absence of competitors (Supplementary Figure S2A).
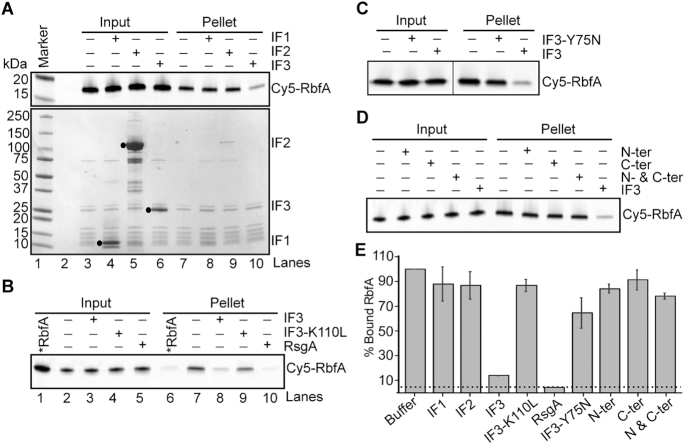
We next tested whether components of the translation initiation complex could displace RbfA. Filtration and pelleting assays showed that neither r-protein bS1, which acts during translation initiation, nor sodB mRNA could displace RbfA (Supplementary Figure S2B, C). RbfA was unchanged in the presence of either IF1 or IF2 (Figure 1A, lanes 8 and 9 and Figure 1E). By contrast, we observed that RbfA was dissociated from 30S subunits in the presence of 4 μM IF3, as demonstrated by a >80% decrease in the Cy5 fluorescence intensity in the pelleted fraction after ultracentrifugation (Figure 1A, lane 10 and Figure 1E). The addition of IF1 or IF2 to reactions with IF3 did not affect the ability of IF3 to promote dissociation of RbfA from 30S subunits, suggesting that IF3 acts alone on the 30S•RbfA complex (Supplementary Figure S2A). We confirmed that the ability of IF3 to remove RbfA depends on its association with the 30S subunit, since IF3-K110L, which does not bind 30S ribosomes (41), was not able to remove RbfA (Figure 1B, lane 9 and Figure 1E). To confirm that the RbfA release promoted by IF3 is meaningful, we compared this reaction to the known RbfA release by the RsgA GTPase. RsgA was more efficient at RbfA release; in the presence of 5 μM GTP, 500 nM RsgA was as effective as 4 μM IF3 (Figure 1B, lane 10 and Figure 1E). However, IF3 levels are typically ∼20 times higher than RsgA levels during logarithmic growth; for every 100 30S subunits the approximate copy numbers are 100:20:8–10:1 30S:IF3:RbfA:RsgA (16,17,42,43). Thus, these experiments suggested that IF3 provides an equally likely mechanism as RsgA for removing RbfA from 30S subunits during logarithmic growth.
Figure 1.
Full-length IF3 is required for RbfA release from 30S ribosomes. (A) Results of pelleting assay showing the release of Cy5-RbfA from 30S•Cy5-RbfA complexes in the presence of initiation factors. Only IF3 was able to displace RbfA from 30S subunits (lane 10). A complex of mature 30S (200 nM) and Cy5-RbfA (400 nM) was formed and unbound Cy5-RbfA was removed by filtration (input) before complexes were challenged with initiation factors (4 μM). Proteins were resolved by 4–20% SDS PAGE. Top panel, Cy5-RbfA fluorescence; bottom panel, Coomassie stain. Initiation factors (input) are indicated with black dots. (B) Pelleting assay showing that a non-binding IF3 mutant (IF3-K110L) cannot release of RbfA from 30S subunits (lane 9). RsgA (500 nM) and GTP (5 μM) was used as a positive control (lane 10). *RbfA (lanes 1 and 6); Cy5-RbfA only. (C) Results of pelleting assay showing that a mutation in the linker region of IF3 (IF3-Y75N) reduces Cy5-RbfA release. (D) Separated N- and C-terminal domains of IF3 were used alone or in combination. (E) Fraction of bound Cy5-RbfA in pellets from panels (A)–(D). Bars indicate mean and s.d., n = 3 independent trials. Dotted line indicates ∼5% Cy5-RbfA background in reactions lacking 30S subunits.
Conformational change of full length IF3 is required for RbfA release
IF3 is a 180 amino acid protein with two globular domains separated by a flexible linker. The C-terminal domain, which performs many of the functions of full-length IF3 including selection of the correct start codon (44), can bind the 30S subunit alone. The isolated N-terminal domain does not bind 30S subunits, but is nonetheless required for wild-type growth (45). Upon binding to 30S subunits, the N- and C-terminal domains of IF3 can adopt extended, intermediate, and compact conformations, owing to the dynamics of the inter-domain linker that are essential for IF3′s function in translation initiation (46).
Given these properties of IF3, we asked whether the conformational dynamics of IF3 is needed to promote the release of RbfA from 30S subunits. We used the IF3 mutation Y75N, which is located in the highly conserved linker region and is known to be critical for the start codon selection activity of IF3 (27,47). This mutation does not affect IF3 binding to 30S subunits, but it disfavors the extended conformation of IF3 (46). We found that the Y75N mutation reduced the amount of RbfA released compared to WT IF3 (Figure 1C and E), suggesting that IF3′s ability to fluctuate into the extended conformation is required for optimal dissociation of RbfA from 30S subunits. To completely abrogate the conformational dynamics, we separated IF3 into N-terminal and C-terminal fragments and tested their ability to remove RbfA from 30S subunits (Figure 1D and E). As expected, the separated domains individually or in combination failed to remove RbfA from 30S subunits, suggesting that conformational changes of the full-length IF3 are important for displacing RbfA. Moreover, that both domains of IF3 are required to release RbfA indicates a role for the N-terminal domain of IF3 in displacing assembly factors from newly formed or recycled 30S subunits.
Timing the release of RbfA during 30S subunit biogenesis
To identify at which step RbfA acts during 30S biogenesis, we sought to determine the influence of 30S subunit composition on RbfA release by IF3. We found that RbfA does not bind with naked 16S 5′ domain RNA, 16S or 17S rRNA, nor with early 30S assembly intermediates reconstituted in vitro (Supplementary Figure S3A, B, and C), consistent with previous studies showing that RbfA acts during late 30S subunit assembly or maturation (12,15,20,21).
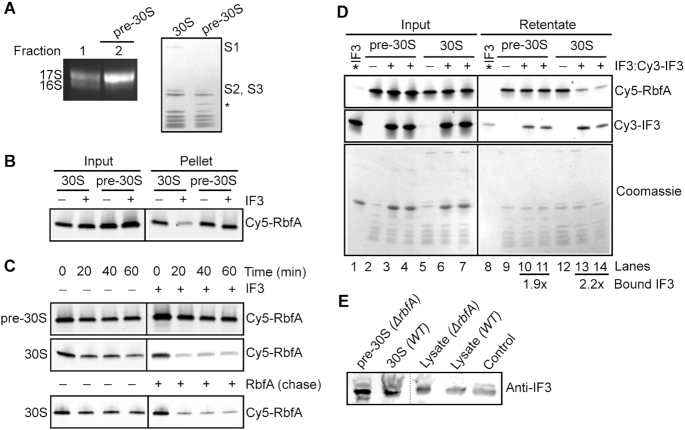
Late 30S assembly intermediates were purified from a △rbfA strain grown at low temperature that causes pre-30S complexes to accumulate (5). These pre-30S particles contain unprocessed 17S pre-rRNA and lack some r-proteins (Figure 2A). Cy5-RbfA was allowed to bind pre-30S particles as before, then challenged with IF3, followed by ultracentrifugation. Although RbfA readily binds pre-30S particles, IF3 was not able to promote the release of RbfA from these complexes. Thus, IF3 preferentially releases RbfA from mature 30S subunits (Figure 2B), as shown previously for RsgA (20).
Figure 2.
IF3 cannot release RbfA from immature pre-30S subunits. (A) Composition of pre-30S ΔrbfA particles. Left, 1.5% agarose-TAE gel showing the rRNA profile. Fraction 2 containing >90% 17S pre-rRNA (pre-30S) was used for further assays. Right, 4–20% SDS PAGE comparing proteins from mature 30S and pre-30S ΔrbfA particles. Proteins S1, S2 and S3 are missing in the pre-30S particles. The identity of the extra band (*) is not known. (B) Pelleting assay; Cy5-RbfA (400 nM) was complexed with mature 30S or pre-30S subunits (200 nM) and challenged with IF3 (4 μM) as in Figure 1. (C) Kinetics of Cy5-RbfA dissociation from pre-30S and 30S subunits. Top, with or without 4 μM IF3; bottom, with or without 4 μM unlabeled RbfA. After excess Cy5-RbfA was removed from Cy5-RbfA•30S complexes by filtration (‘0 min’), complexes were filtered a second time to remove free Cy5-RbfA after an additional 20–60 min at 37°C. The 0 min samples that were filtered once contain ∼30% more Cy5-RbfA than the samples that were filtered twice. (D) Binding of Cy5-RbfA and Cy3-IF3 to pre-30S or 30S subunits, as in C. Top panel, Cy5-RbfA; middle panel, Cy3-IF3; lower panel, Coomassie stain. *IF3 (lanes 1 and 8); Cy3-IF3 only. Average fold change in bound IF3 over *IF3 background (lane 8) was 1.9 ± 0.15 in lanes 10 and 11 and 2.2 ± 0.9 in lanes 13 and 14; n = 2. Added IF3 was a mixture of 80% unlabeled IF3 and 20% Cy3-IF3. Cy5-RbfA was scanned with 600 PMT voltage, whereas Cy3-IF3 (Input) with 400 PMT voltage and Cy3-IF3 (retentate) with 500 PMT voltage. (E) Anti-IF3 western blot showing the presence of IF3 in the pre-30S and 30S fractions from BX41 (ΔrbfA) and BW25113 (WT) compared to ΔrbfA and WT lysates and purified IF3 (control).
To understand why RbfA could not be released from pre-30S subunits, we compared the lifetimes of RbfA bound to pre-30S and mature 30S subunits by removing free RbfA by ultrafiltration at various times. Figure 2C shows that the half-life of the pre-30S•RbfA complex was >60 min as observed previously (20), and was unaffected by the presence of IF3. Whereas, the amount of RbfA bound to mature 30S subunits significantly decreased after 20 min in the presence of IF3. These experiments suggested that the structure of the pre-30S particles either stabilizes RbfA binding so that it cannot be removed by IF3, or prevents IF3 from acting on the complex.
To assess whether IF3 actively displaces RbfA from 30S subunits, a chase experiment was performed with excess unlabeled RbfA (Figure 2C, lower panel). The results showed that unlabeled RbfA chases Cy5-RbfA out of the bound fraction, implying that IF3 (and also RsgA) could capture 30S subunits after RbfA has dissociated. Since the time resolution of this experiment is 20 min, however, we cannot rule out active release of RbfA by either IF3 or RsgA at shorter times that are more relevant for cellular translation.
To test whether IF3 fails to release RbfA from pre-30S complexes because IF3 cannot bind them, we monitored the release of Cy5-RbfA in the presence of Cy3-IF3, followed by detection of the Cy3 signal to determine whether IF3 was retained with the 30S or pre-30S complexes after filtration. IF3 was able to bind pre-30S complexes, suggesting that either conformational changes in the linker of IF3 are compromised on pre-30S subunits, or that these conformational changes occur but cannot compete RbfA away from pre-30S subunits (Figure 2D, lanes 10 and 11). In a similar experiment with mature 30S subunits, IF3 remained bound after removing RbfA (Figure 2D, lanes 13 and 14). Thus, IF3 can bind both mature and immature 30S complexes, but cannot displace RbfA from pre-30S particles.
We sought to determine if IF3 binds pre-30S particles in vivo by analyzing purified pre-30S or 30S samples for the presence of IF3 by western blotting (Figure 2E). The complexes were fractionated by sucrose gradient sedimentation and validated by measuring rRNA and protein composition (as in Figure 2A). We found that IF3 co-purifies with pre-30S subunits in an △rbfA strain. Additionally, IF3 was previously detected by Western blotting in pre-30S particles in a △ybeY strain (6) and by mass spectrometry in pre-30S particles containing 16S mutations that block folding of a helix junction (48) (Supplementary Figure S4). Together, these data confirmed that the interaction of IF3 with pre-30S particles is physiologically measurable. Furthermore, pre-30S subunits have been found in 70S and polysome fractions of △rbfA, △rimM, △ybeY and △rpsO strains (3,5,6), suggesting that some pre-30S complexes are competent for translation and therefore must interact with IF3.
RbfA release marks the transition from ribosome biogenesis to translation initiation
Methylation of 16S A1518 and A1519 in h45 by the conserved RNA methylase KsgA is thought to be one of the last steps in the 30S subunit biogenesis preceding translation initiation (21,49,50). Therefore, we wondered if h45 methylation promotes RbfA release by IF3. First, we used primer extension to confirm that pre-30S complexes purified from an ΔrbfA strain are methylated by KsgA, as indicated by the presence of a reverse transcription stop at residue A1519 (Figure 3A, lane 4). This showed that RbfA is not required for h45 methylation, consistent with the proposal that RbfA acts downstream of KsgA (21).
Figure 3.
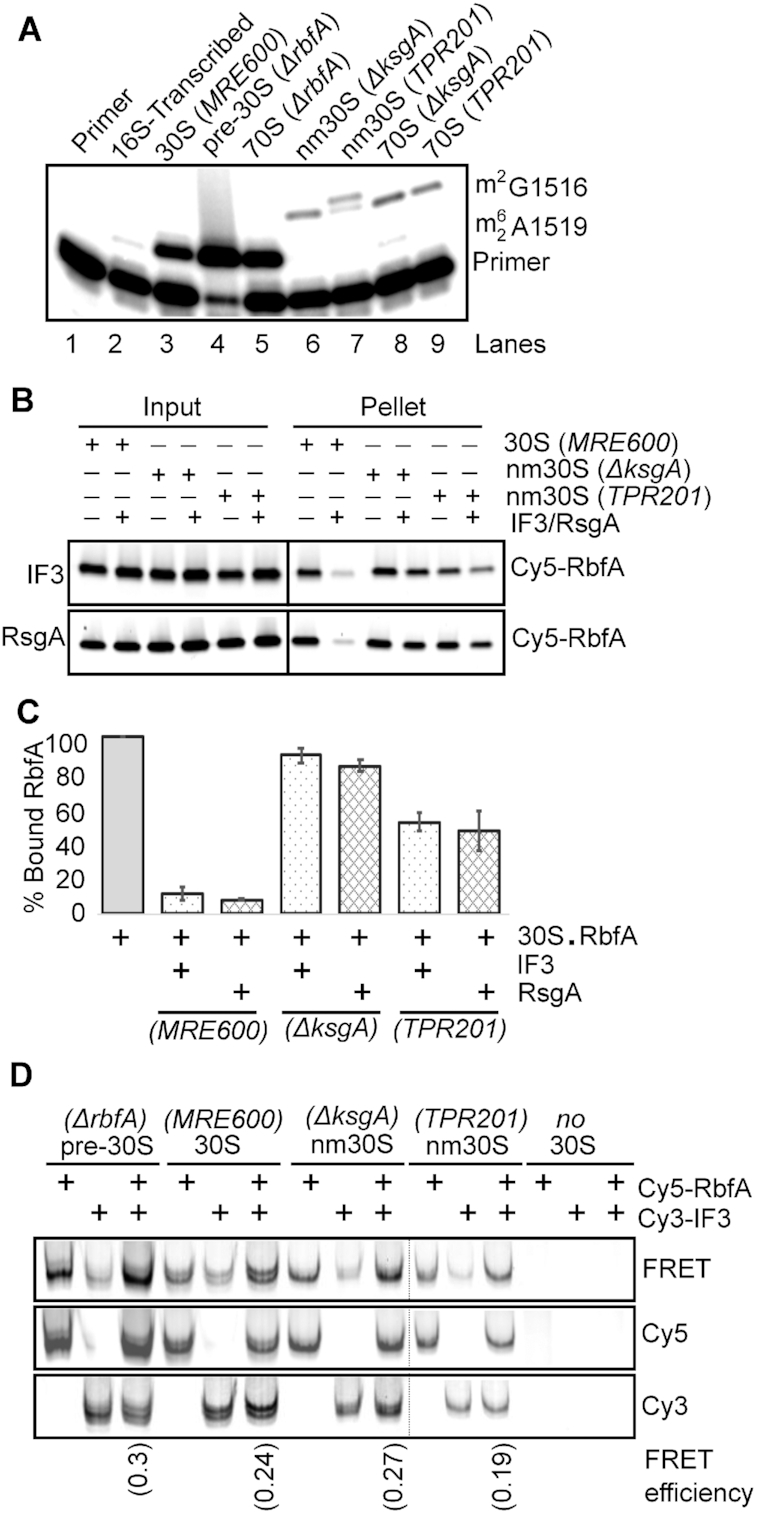
Methylation of 30S subunits is important for RbfA release. (A) Methylation of 16S A1519 in pre-30S and 30S subunits was measured by primer extension (see Materials and Methods). Pre-30S subunits from BX41 (ΔrbfA) were methylated by KsgA (lane 4); nm30S subunits from ΔksgA cells (JW0050-3) and TPR201 cells bearing an inactive ksgA allele were unmethylated (lanes 6 and 7). (B) Pelleting assay showing that neither IF3 (top) nor RsgA (bottom) promote Cy5-RbfA release from nm30SΔksgA subunits and promote only partial release from nm30STPR201 subunits. (C) % Bound RbfA relative to buffer control as in panel B; mean and s.d.; n = 2. (D) Native PAGE showing that Cy5-RbfA and Cy3-IF3 can bind pre-30S or 30S subunits simultaneously. 30S complexes were purified from the strains shown and incubated with Cy5-RbfA and Cy3-IF3 before native PAGE. Panels show the same gel scanned with Cy3 excitation (bottom), Cy5 excitation (middle), and FRET to Cy5 upon Cy3 excitation (top). The FRET efficiencies are indicated at the bottom.
Next, to determine whether h45 methylation is required for RbfA release, we purified unmethylated near-mature 30S (nm30S) subunits from two strains lacking KsgA activity: a ΔksgA deletion (51) and TPR201, which harbors a catalytically dead ksgA allele (52). nm30S subunits purified from these strains were not methylated at A1519 (Figure 3A, lanes 6 and 7), although they contain the full complement of r-proteins (21). Pelleting assays showed that RbfA was able to bind to these nm30S particles, but its release by IF3 was compromised. IF3 was unable to promote release of RbfA from nm30S subunits from the ΔksgA strain, and only partially able to release RbfA from TPR201 nm30S subunits (Figure 3B, upper panel and Figure 3C). The absence of h45 methylation also reduced the ability of RsgA to release RbfA from both types of nm30S subunits (Figure 3B, lower panel and Figure 3C). Thus, while h45 methylation by KsgA is not essential for RbfA recruitment, it is important for RbfA release by IF3 and RsgA.
We next examined the difference in IF3 binding to nm30S subunits to determine if this could explain IF3′s reduced ability to promote the release of RbfA from these subunits. nm30S subunits from ΔksgA and TPR201 strains were incubated with Cy5-RbfA and Cy3-IF3 and then subjected to native PAGE (Figure 3D). We observed colocalization of Cy3 and Cy5 fluorescence in the native gel for all of the 30S complexes tested, indicating that IF3 and RbfA can both bind to methylated and non-methylated 30S subunits. Furthermore, we observed FRET from Cy3-IF3 to Cy5-RbfA (EFRET ∼ 0.2–0.3), indicating that IF3 and RbfA can bind 30S, nm30S or pre-30S complexes at the same time. IF3 interacted less tightly with unmethylated nm30S subunits than with 30SWT subunits, however, judging from the intensities of individual protein bands. Connolly and Culver (2013) also showed that IF3 binds less tightly to nm30S subunits from ΔksgA cells, supporting our observation. Together, these data show that KsgA methylation of H45 enhances IF3 binding, and that methylation is important for the efficient removal of RbfA from 30S subunits. These results explain why RbfA overexpression is toxic to cells lacking KsgA (21), because RbfA cannot be released from nm30S subunits.
RbfA suppresses translation by pre-30S ribosomes
We previously observed that pre-30S ribosomes are partially active in protein synthesis and enter the polysome fraction in an E. coli strain lacking RbfA (5), supporting the idea that RbfA normally prevents translation by immature subunits. To test this, we compared the activity of pre-30S (ΔrbfA), nm30S (ΔksgA and TPR201) and mature 30S subunits in an in vitro translation assay using purified components (53) in the presence or absence of excess RbfA (Figure 4A). Pre-30SΔrbfAsubunits were intrinsically less active (29 ± 0.3%) than mature 30S subunits, consistent with previous work (5). In contrast, nm30S subunits had normal activity. Importantly, the addition of RbfA substantially inhibited translation by pre-30SΔrbfA and nm30S (ΔksgA and TPR201) complexes, while it did not inhibit translation by WT 30SMRE600 subunits (Figure 4A). Increasing the concentration of IF3 in the translation assay did not improve the translation efficiency for any of the 30S complexes tested. We concluded that IF3′s inability to displace RbfA from immature particles (pre-30SΔrbfA, nm30SΔksgA and nm30STPR201), even when IF3 was provided in excess, markedly decreased the activity of these subunits. Other proteins associated with pre-30S subunits, Era, RsgA, and Hfq, did not inhibit translation by pre-30S subunits (Supplementary Figure S5), indicating that this gate-keeping function is specific to RbfA.
Figure 4.
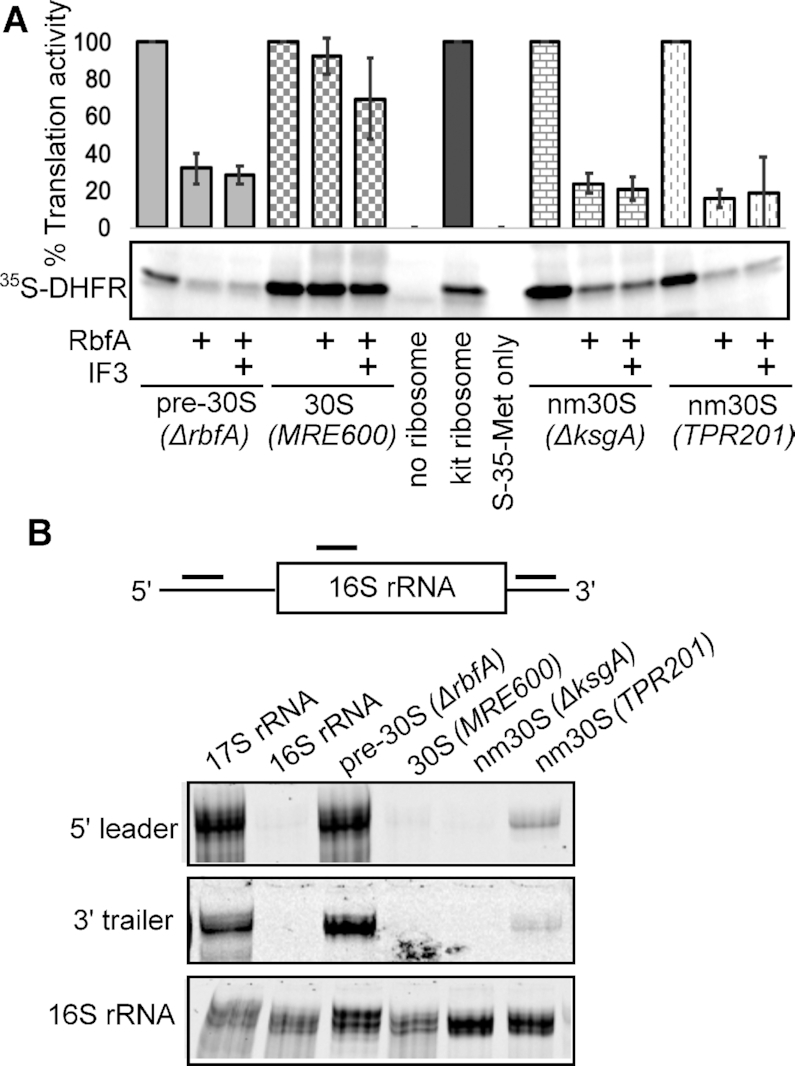
Translation initiation requires RbfA release from 30S subunits. (A) In vitro translation by pre-30S (methylated), nm30S (unmethylated) and mature 30S subunits in the presence of RbfA or RbfA plus IF3. Top, average and s.d., n = 2, relative to no RbfA controls for each complex. Bottom, 35S-labeled DHFR product. Immature or unmethylated subunits are active in protein synthesis but are inhibited by RbfA. Mature 30S subunits were unaffected by RbfA, presumably because RbfA was removed by IF3. (B) 16S rRNA processing. Binding of anti-sense oligomers to RNA from 30S complexes was detected by native 4% PAGE.
Accurate processing of the 5′ leader and 3′ trailer play a crucial role in determining the fate of immature 30S subunits for maturation or degradation (54). For example, defective processing of the 3′ trailer is a hallmark of misfolded intermediates, and triggers their decay. We probed the rRNA purified from pre-30SΔrbfA and nm30SΔksgA and TPR201 subunits with Cy5-labeled DNA oligomers (Figure 4B, scheme) to assess whether processing correlates with RbfA release. The rRNA from pre-30SΔrbfAsubunits hybridized with probes against the 5′ leader and the 3′ trailer, whereas nm30S subunits were fully processed in ΔksgA and ≥90% processed in TPR201 cells (Figure 4B). Since IF3 cannot release RbfA from any of these immature 30S complexes, these data suggest that the competition between RbfA and IF3 senses a range of perturbations to the decoding center that arise from incomplete assembly or a lack of h45 methylation.
RbfA and IF3 are genetically linked
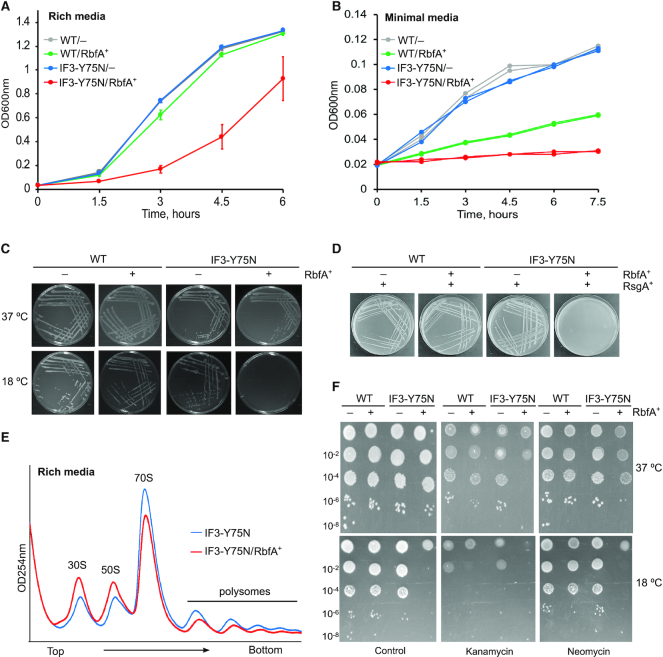
The results above show that IF3 can displace RbfA from mature 30S subunits in vitro, but not from immature subunits. To determine if this activity of IF3 is important in vivo, we looked for a genetic interaction between RbfA and IF3 under a variety of growth conditions. Since IF3 is essential, we used the non-lethal IF3-Y75N mutation (infC362; (27)), which is fortuitously defective in its ability to release RbfA from 30S subunits (Figure 1C). Figure 5A shows that the infC362 strain grows similarly to the parental E. coli strain. When these strains were transformed with a plasmid expressing RbfA (p15BHA), however, leaky expression of RbfA in the absence of IPTG inhibited the growth of cells expressing IF3-Y75N but not cells expressing wild type IF3 (Figure 5A). This level of RbfA expression is enough to complement an E. coli ΔrbfA strain (15), suggesting that RbfA and IF3 are genetically linked. The leaky expression of RbfA was also toxic when the infC362 cells were grown in minimal media (Figure 5B), which slows ribosome biogenesis and makes cells more dependent on ribosome assembly factors. The genetic interaction between IF3 and RbfA was confirmed by plate assays, which also showed that RbfA over-expression induces the cold sensitivity of infC362 cells (Figure 5C). Basal RbfA overexpression was even more toxic in IF3-Y75N ΔrsgA double mutant strain (Supplementary Figure S6A), whereas, it did not substantially alter growth of the ΔrsgA strain ((20) and Supplementary Figure S6B).
Figure 5.
Genetic interaction between RbfA and IF3. (A) Growth of E. coli strains in rich LB media at 37°C (OD600nm). WT, parental; IF3-Y75N, infC362; –, empty vector; and RbfA+, + p15BHA (RbfA overexpression). Mean and s.d., n = 4 biological replicates. (B) Growth of strains in (A) in minimal MOPS media (pH 7.2) supplemented with 0.4% glucose at 37°C; n = 2. (C) Growth of strains as in (A) on LB agar media plates at 37°C and 18°C; n = 3. (D) WT and IF3-Y75N cells harboring empty vector or p15BHA were transformed with pD421-rsgA for RsgA overexpression. Plates were incubated overnight at 37°C. Single colonies were subsequently streaked on a fresh plate. No transformation was observed for the IF3-Y75N/RbfA+/RsgA+ strain, n = 2. (E) Polysome profiles of strains infC362 (blue) and infC362/p15BHA (red), grown under similar conditions as (A) and collected at OD600nm ∼0.2. Experiment was performed in duplicate. (F) Serial dilutions of the indicated strains were spotted on LB-agar containing sublethal concentrations of kanamycin (2 μg/ml) or neomycin (2 μg/ml). Ampicillin (100 μg/ml) was added to all plates to maintain the plasmid; n = 2.
We next asked whether RsgA could compensate for the inability of IF3 to displace RbfA, by overexpressing RsgA in infC362 cells either alone, or together with RbfA. Surprisingly, not only did RsgA not rescue the toxicity of RbfA expression in infC362 cells, but dual over-expression of RsgA and RbfA was lethal in the IF3-Y75N background (Figure 5D). Over-expression of either protein alone, or dual over-expression of RsgA and RbfA, were not lethal in the presence of WT IF3. These results strongly suggested that RsgA and IF3 do not act redundantly to remove RbfA from 30S ribosomes, but function separately in 30S biogenesis.
To pinpoint the reason for RbfA toxicity in the IF3-Y75N background, the polysome profile of infC362/p15BHA strain was compared with infC362 strain. Figure 5E shows that excess RbfA reduced the size of the 70S and polysome peaks, with a concomitant increase in the amounts of free 30S and 50S ribosome subunits. This is consistent with an inability of IF3-Y75N to displace RbfA, which in turn stabilizes free 30S subunits. This reduction in the numbers of active ribosomes was not due to a defect in 30S biogenesis because we did not observe a defect in 16S rRNA 5′ processing as judged by primer extension (Supplementary Figure S7).
Finally, we tested the sensitivity of these strains to sublethal concentrations of aminoglycoside antibiotics kanamycin and neomycin. We found that infC362 cells overexpressing RbfA were more sensitive to kanamycin and neomycin at 37°C (Figure 5F, upper panel), which increased when cells were grown at 18°C (Figure 5F, lower panel). Both of these antibiotics interact with 30S subunits and promote misreading of mRNA (55), a function shared by the IF3-Y75N protein, and they may also interfere with the binding of IF3 (56). We concluded that wild type IF3 plays an important role in removing RbfA from 30S subunits, which becomes even more important at low temperature, low nutrient conditions, and when stressed by aminoglycoside antibiotics.
IF3 is likely the predominant pathway for RbfA release during stationary phase
Since the genetic interaction of IF3 and RbfA is stronger in low nutrient conditions, we reasoned that IF3 may be the dominant factor that releases RbfA during stationary phase, when the GTPase activity of RsgA is inhibited by (p)ppGpp (24). To test this possibility, we measured RbfA release in a mixture containing combinations of RsgA, GTP, ppGpp and IF3. We found that ppGpp inhibits the release activity of RsgA by ∼55% (Figure 6A, lane 10 and Figure 6B), yet did not inhibit IF3′s ability to release RbfA from 30S subunits (Figure 6A, lanes 11 and 12 and Figure 6B). These data indicate that IF3 can still displace RbfA when ppGpp levels are high enough to inhibit RsgA, as is the case during stationary phase (57).
Figure 6.
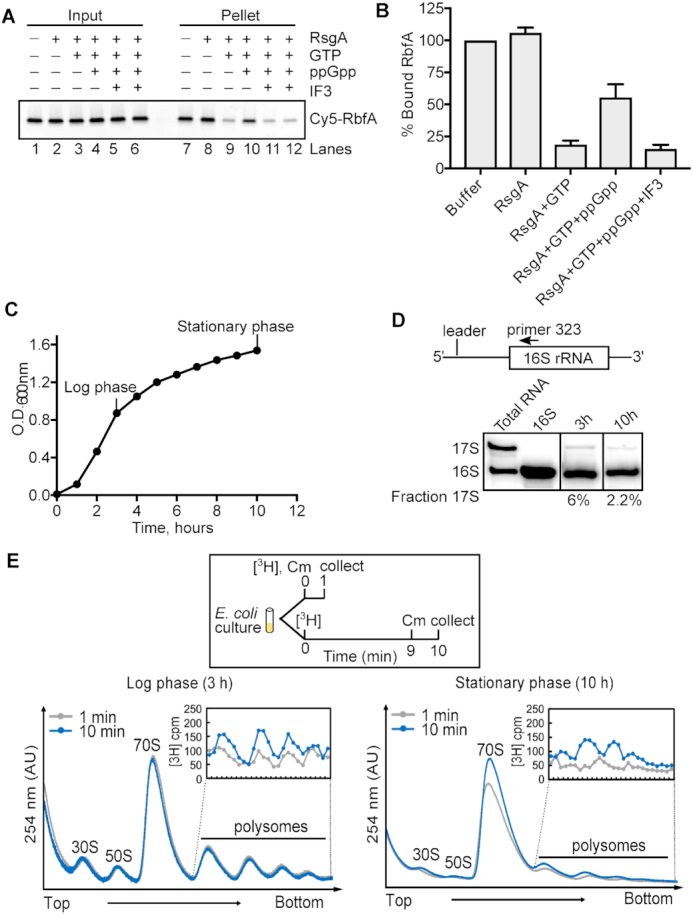
IF3 releases RbfA from 30S ribosomes during stationary phase. (A) Pelleting assay showing that RbfA release by 200 nM RsgA, 50 μM GTP is inhibited by 1.5 mM ppGpp (lane 10). 5 μM IF3 can release RbfA from 30S subunits under this condition (lanes 11 and 12). (B) Quantification of experiments performed in (A). Mean and s.d., n = 2. (C) Growth of WT E. coli (BW25113) in liquid LB medium. (D) Primer extension on total RNA from cells in (C) at 3 h and 10 h detects unprocessed 17S pre-rRNA as a proxy of ribosome biogenesis. (E) Ribosome biogenesis during log phase (3 h, left panel) and stationary phase (10 h, right panel) from tritium labeled cells. Cells were harvested for polysome analysis at 1 min (gray) and 10 min (blue) after the addition of 3H-uridine and 1 min after treatment with chloramphenicol (Cm). Insets: 3H-uridine in polysomes.
Because ribosome biogenesis is sharply reduced in stationary phase or when nutrients are limiting (58–61), we asked whether a checkpoint for biogenesis is needed under these conditions. To measure the amount of ribosome biogenesis during stationary phase, we purified total rRNA from cells in mid-log or stationary phase (Figure 6C) and measured the amount of unprocessed 17S pre-rRNA by primer extension (5,62). Immature 17S rRNA was present during both growth stages tested, although the proportion of immature rRNA was three times lower during stationary phase (2% versus 6%; Figure 6D). We next tracked de novo ribosome synthesis by pulse labeling the rRNA with 3H-uridine in cells in log phase (3 h, OD600 nm ∼ 0.6–0.8) and stationary phase (10 h, OD600 nm ∼1.6–1.8), to determine whether newly synthesized rRNA is assembled into mature ribosomes (Figure 6E). The numbers of ribosomes and polysomes are expected to be lower in stationary phase (63). Nevertheless, after 10 min, newly formed tritiated ribosomal subunits were observed in the polysome fractions from cells in stationary phase, indicating that ribosome biogenesis continues even when nutrients are limiting (Figure 6E).
DISCUSSION
The results presented here show that RbfA, the most abundant 30S subunit assembly factor in E. coli, inhibits translation by 30S subunits unless it is displaced during formation of the 30S initiation complex. Because RbfA is only released from 30S subunits when assembly and maturation are complete, we suggest that the opposing effects of RbfA and IF3 on pre-30S particles set up a checkpoint between 30S biogenesis and protein synthesis (Figure 7). RbfA unfolds 16S h44 (decoding center), whereas IF3 stabilizes h44, explaining how this checkpoint senses the assembly status of the 30S active site (Supplementary Figure S8). After 30S maturation, a hand-off from RbfA to IF3 (30S•RbfA→30S•IF3) leads to formation of a 30S translation initiation complex.
Figure 7.
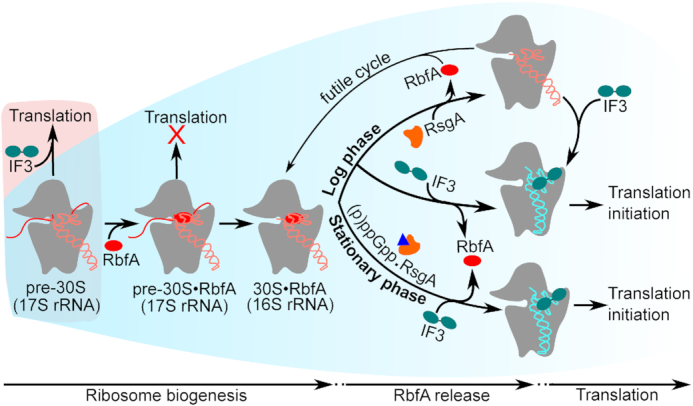
Model for RbfA quality control of 30S maturation. Under normal growth conditions (blue background), RbfA strongly interacts with immature pre-30S particles and prevents their entry into translation. Both RsgA and IF3 release RbfA from mature 30S subunits. When RsgA releases RbfA, 16S h44 and h45 can fluctuate into an undocked state (5,67), and free 30S subunits can again bind to RbfA, leading to a futile cycle (top). By contrast, when IF3 releases RbfA from mature 30S subunits, IF3 remains bound and ready to form a translation initiation complex (30SIC; middle). During stationary phase, RsgA is inhibited by (p)ppGpp (blue triangle) but RbfA is still released by IF3, allowing new subunits to initiate translation (bottom). Under stress (red background), when the pre-30S level exceeds the amount of RbfA, pre-30S complexes can enter translation.
RbfA as a gatekeeper for 30S ribosomes
During active growth conditions, bacteria must ensure the fidelity of translation by preventing the entry of pre-30S particles into translation, which are thought to be inefficient and error-prone (3–9). We found that RbfA directly inhibits translation by immature pre-30S particles (Figure 4A), providing a gatekeeping mechanism. Congruent with this role, RbfA forms stable complexes with pre-30S subunits (Figure 2B, C) that are not displaced by either RsgA (20) or IF3 (Figure 3B, C). In a cryo-EM structure of the RbfA•30S complex, RbfA dislocates 16S h45 and the top of h44 (12). These changes would prevent initiator tRNA binding and the formation of bridges B2a and B3 with the 50S subunit. One possibility is that RbfA binding tests the stability of interactions around the 30S decoding center, which is the last region of the 30S subunit to fold. This would explain why RbfA release is sensitive to KsgA methylation of A1518 and A1519, which stabilizes tertiary interactions between h45 and h44 (22,64). In yeast, late assembly factors were also suggested to directly block the premature entry of pre-40S complexes into the translation cycle (10).
Mechanism of RbfA release by IF3
Our observation that RbfA does not interact with the 30S pre-initiation complex (30SIC) (Supplementary Figure S3D, lane 3) suggests that RbfA is released before or during initiation of translation. In our experiments, only IF3 was able to release RbfA from mature 30S subunits (Figure 1 and S2). However, IF2 was previously reported to genetically interact with RbfA (19), consistent with reported effects of initiator tRNA mutations on 30S biogenesis (65).
Although our results don’t reveal how IF3 displaces RbfA from mature 30S ribosomes, cryo-EM and single molecule FRET findings suggest a plausible pathway for RbfA dissociation (Supplementary Figure S8). Cryo-EM structures showed that IF3 initially interacts with the 30S subunit through its N-terminal domain which binds the platform near uS11 (14,46). This interaction anchors IF3 to 30S subunits and allows for a conformational change within the linker region that extends the C-terminal domain towards the top of h44, where it reinforces docking of h44 and h45 and stabilizes the tRNA anti-codon in the P-site (14,46,66). Together these observations suggest that IF3 and RbfA exert opposite and mutually exclusive effects on docking and folding of h44 and h45, which explain how IF3 binding favors RbfA dissociation or prevents RbfA rebinding, and why this requires both domains of IF3. In-cell footprinting indicated that h44 is at least partially unfolded in free 30S subunits (67) and completely undocked in pre-30S complexes (5), supporting the notion that this region of the 30S ribosome can fluctuate between docked and undocked conformations.
Like IF3, RsgA GTPase also stabilizes the docked conformation of h44 and h45, and directly or indirectly promotes the release of RbfA (13,20,22,23). An important difference is that RsgA itself dissociates from the 30S complex upon GTP hydrolysis. By contrast, IF3 remains bound to mature 30S subunits as part of the initiation complex, and could be physiologically more important for preventing RbfA rebinding and toxicity. This idea is further supported by the fact that the N-terminal region of RsgA, which is important for RbfA release, is weakly conserved or absent from many bacteria (23).
RbfA release by IF3 increases stress tolerance
Our data indicate that RbfA release by IF3 is even more critical in suboptimal conditions such as low temperature, poor nutrients, or antibiotics (Figure 5). Ribosome biogenesis continues during stationary phase, albeit slowly (Figure 6D and E). However, as the GTPase activity of RsgA is inhibited by rising levels of (p)ppGpp (Figure 6A and B), IF3 is increasingly needed to remove RbfA from newly made 30S subunits. RsgA has been shown to promote 70S ribosome dissociation into 30S and 50S subunits (13,68), which may explain why inhibition of RsgA by (p)ppGpp is advantageous to the cell. Similarly, when RsgA and RbfA are both over-expressed, over-active RsgA may create an excess of free 30S subunits that are captured by RbfA, in a quasi-reversal of 30S biogenesis. If IF3 cannot reactivate these RbfA•30S complexes, the protein synthesis capacity is fatally depleted. During adaptation to low temperature and starvation, 70S ribosomes are protected by forming 100S hibernation complexes (69,70). Interestingly, IF3 is also known to compete with hibernation factors for binding to ribosomes following termination, suggesting a competition between 30S recycling and hibernation (71,72). It will be interesting to see which of these pathways is more important for maintaining cell survival under stress conditions.
Translation using pre-30S subunits is a survival strategy under stress
When pre-30S particles accumulate during stress, they can participate in translation to ensure survival. The results of our in vitro translation assays support previous reports that pre-30S complexes can enter the polysome pool in the absence of RbfA or when 30S assembly has stalled (3,5,6). We suggest that when the accumulation of pre-30S subunits exceeds RbfA concentrations, free pre-30S complexes which are competent can enable translation under different adverse growth conditions (Figure 7).
Together, our data support a model in which RbfA normally prevents the premature entry of pre-30S complexes into translation by exploiting the instability of 16S rRNA interactions within the immature decoding site (Figure 7). After 30S maturation, RbfA binding is destabilized, and RsgA or IF3 can release RbfA to keep up with the demand for new 30S subunits during logarithmic growth. Since IF3 remains bound with mature 30S subunits after RbfA release, the release of RbfA by IF3 marks end of ribosome biogenesis and beginning of translation initiation. Finally, our results hint that RbfA and IF3 also verify the quality of recycled mature 30S subunits, by preventing their disassembly by biogenesis factors.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Julie Brunelle, Mollie Rappé, Himani Galagali, and Laura Lessen for technical help, Drs Gloria Culver, Ruben Gonzalez, Rachel Green, Hyouta Himeno, Eric D. Brown, John Wertz, Isabella Möll, Harry Noller, and Umesh Varshney for the gifts of reagents and strains, Drs Balasubrahmanyam Addepalli and Patrick Limbach for the assistance with mass spectrometry, and Arthur Korman, Allen Buskirk, Yumeng Hao, Margaret Rodgers, Joaquin Ortega and Sean Connolly for helpful discussion and comments on the manuscript.
Author contributions: I.M.S. conceived the project, performed experiments, analyzed and interpreted the results and wrote the manuscript. S.A.W. helped design the project, interpret the results and edit the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01 GM60819 to S.A.W.]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Klinge S., Woolford J.L.. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019; 20:116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shajani Z., Sykes M.T., Williamson J.R.. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 2011; 80:501–526. [DOI] [PubMed] [Google Scholar]
- 3. Roy-Chaudhuri B., Kirthi N., Culver G.M.. Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4567–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andrade J.M., dos Santos R.F., Chelysheva I., Ignatova Z., Arraiano C.M.. The RNA‐binding protein Hfq is important for ribosome biogenesis and affects translation fidelity. EMBO J. 2018; 37:e97631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. ClatterbuckSoper S., Dator R., Limbach P., Woodson S.. InVivo X-Ray footprinting of pre-30S ribosomes reveals chaperone-dependent remodeling of late assembly intermediates. Mol. Cell. 2013; 52:506–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davies B.W., Köhrer C., Jacob A.I., Simmons L.A., Zhu J., Aleman L.M., RajBhandary U.L., Walker G.C.. Role of escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol. Microbiol. 2010; 78:506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cole S.E., LaRiviere F.J., Merrikh C.N., Moore M.J.. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol. Cell. 2009; 34:440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Soudet J., Gélugne J.P., Belhabich-Baumas K., Caizergues-Ferrer M., Mougin A.. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 2010; 29:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sharma H., Anand B.. Ribosome assembly defects subvert initiation Factor3 mediated scrutiny of bona fide start signal. Nucleic Acids Res. 2019; doi:10.1093/nar/gkz825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Strunk B.S., Loucks C.R., Su M., Vashisth H., Cheng S., Schilling J., Brooks C.L., Karbstein K., Skiniotis G.. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011; 333:1449–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Strunk B.S., Novak M.N., Young C.L., Karbstein K.. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012; 150:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Datta P.P., Wilson D.N., Kawazoe M., Swami N.K., Kaminishi T., Sharma M.R., Booth T.M., Takemoto C., Fucini P., Yokoyama S. et al.. Structural aspects of RbfA action during small ribosomal subunit assembly. Mol. Cell. 2007; 28:434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo Q., Yuan Y., Xu Y., Feng B., Liu L., Chen K., Sun M., Yang Z., Lei J., Gao N.. Structural basis for the function of a small GTPase RsgA on the 30S ribosomal subunit maturation revealed by cryoelectron microscopy. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13100–13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hussain T., Llácer J.L., Wimberly B.T., Kieft J.S., Ramakrishnan V.. Large-scale movements of IF3 and tRNA during bacterial translation initiation. Cell. 2016; 167:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dammel C.S., Noller H.F.. Suppression of a cold-sensitive mutation in 16s rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev. 1995; 9:626–637. [DOI] [PubMed] [Google Scholar]
- 16. Thurlow B., Davis J.H., Leong V., Moraes T.F., Williamson J.R., Ortega J.. Binding properties of YjeQ (RsgA), RbfA, RimM and Era to assembly intermediates of the 30S subunit. Nucleic Acids Res. 2016; 34:9918–9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gibbs M.R., Moon K.-M., Chen M., Balakrishnan R., Foster L.J., Fredrick K.. Conserved GTPase LepA (Elongation Factor 4) functions in biogenesis of the 30S subunit of the 70S ribosome. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xia B., Ke H., Shinde U., Inouye M.. The role of RbfA in 16 S rRNA processing and cell growth at low temperature in Escherichia coli. J. Mol. Biol. 2003; 332:575–584. [DOI] [PubMed] [Google Scholar]
- 19. Jones P.G., Inouye M.. RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol. Microbiol. 1996; 21:1207–1218. [DOI] [PubMed] [Google Scholar]
- 20. Goto S., Kato S., Kimura T., Muto A., Himeno H.. RsgA releases RbfA from 30S ribosome during a late stage of ribosome biosynthesis. EMBO J. 2011; 30:104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Connolly K., Culver G.. Overexpression of RbfA in the absence of the KsgA checkpoint results in impaired translation initiation. Mol. Microbiol. 2013; 87:968–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pedro Lopez-Alonso J., Kaminishi T., Kikuchi T., Hirata Y., Iturrioz I., Dhimole N., Schedlbauer A., Hase Y., Goto S., Kurita D. et al.. RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket. Nucleic Acids Res. 2017; 45:6945–6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Razi A., Guarné A., Ortega J.. The cryo-EM structure of YjeQ bound to the 30S subunit suggests a fidelity checkpoint function for this protein in ribosome assembly. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E3396–E3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Corrigan R.M., Bellows L.E., Wood A., Gründling A.. ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E1710–E1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peterson C.N., Mandel M.J., Silhavy T.J.. Escherichia coli starvation diets: Essential nutrients weigh in distinctly. J. Bacteriol. 2005; 187:7549–7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Potrykus K., Cashel M.. (p)ppGpp: Still Magical?. Annu. Rev. Microbiol. 2008; 62:35–51. [DOI] [PubMed] [Google Scholar]
- 27. Sussman J.K., Simons E.L., Simons R.W.. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol. 1996; 21:347–360. [DOI] [PubMed] [Google Scholar]
- 28. Mechold U., Murphy H., Brown L., Cashel M.. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of RelSeq, the Rel/Spo enzyme from streptococcus equisimilis. J. Bacteriol. 2002; 184:2878–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spedding G. Spedding G. In Ribosomes and Protein Synthesis: A practical Approach. 1990; NY: IRL Press, Oxford University Press. [Google Scholar]
- 30. Jeganathan A., Razi A., Thurlow B., Ortega J.. The C-terminal helix in the YjeQ zinc-finger domain catalyzes the release of RbfA during 30S ribosome subunit assembly. RNA. 2015; 21:1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shoemaker C.J., Green R.. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:E1392–E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Culver G.M., Noller H.F.. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA. 1999; 5:832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Traub P., Nomura M.. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl. Acad. Sci. U.S.A. 2006; 59:777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seistrup K.H., Rose S., Birkedal U., Nielsen H., Huber H., Douthwaite S.. Bypassing rRNA methylation by RsmA/Dim1during ribosome maturation in the hyperthermophilic archaeon Nanoarchaeum equitans. Nucleic Acids Res. 2017; 45:2007–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sabanayagam C.R., Eid J.S., Meller A.. High-throughput scanning confocal microscope for single molecule analysis. Appl. Phys. Lett. 2004; 84:1216–1218. [Google Scholar]
- 36. Bellur D.I., Woodson S.A.. A minimized rRNA-binding site for ribosomal protein S4 and its implications for 30S assembly. Nucleic Acids Res. 2009; 37:1886–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim H., Abeysirigunawarden S.C., Chen K., Mayerle M., Ragunathan K., Luthey-Schulten Z., Ha T., Woodson S.A.. Protein-guided RNA dynamics during early ribosome assembly. Nature. 2014; 506:334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abeysirigunawardena S.C., Kim H., Lai J., Ragunathan K., Rappé M.C., Luthey-Schulten Z., Ha T., Woodson S.A.. Evolution of protein-coupled RNA dynamics during hierarchical assembly of ribosomal complexes. Nat. Commun. 2017; 8:492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Julián P., Milon P., Agirrezabala X., Lasso G., Gil D., Rodnina M. V., Valle M.. The cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol. 2011; 9:e1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang Y.J., Swapna G.V.T., Rajan P.K., Ke H., Xia B., Shukla K., Inouye M., Montelione G.T.. Solution NMR structure of ribosome-binding factor A (RbfA), a cold-shock adaptation protein from Escherichia coli. J. Mol. Biol. 2003; 327:521–536. [DOI] [PubMed] [Google Scholar]
- 41. De Bellis D., Liveris D., Schwartz I., Goss D., Ringquist S.. Structure-function analysis of Escherichia coli translation initiation factor IF3: Tyrosine 107 and lysine 110 are required for ribosome binding. Biochemistry. 1992; 31:11984–11990. [DOI] [PubMed] [Google Scholar]
- 42. Howe J.G., Hershey J.W.B.. Initiation factor and ribosome levels are coordinately controlled in Escherichia coli growing at different rates. J. Biol. Chem. 1983; 258:1954–1959. [PubMed] [Google Scholar]
- 43. Liveris D., Klotsky R.A., Schwartz I.. Growth rate regulation of translation initiation factor IF3 biosynthesis in Escherichia coli. J. Bacteriol. 1991; 173:3888–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Petrelli D., La Teana A., Garofalo C., Spurio R., Pon C.L., Gualerzi C.O.. Translation initiation factor IF3: Two domains, five functions, one mechanism. EMBO J. 2001; 20:4560–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ayyub S., Dobriyal D., Varshney U.. Contributions of the N-terminal and C-terminal domains of initiation factor 3 towards its functions in the fidelity of initiation and anti-association of the ribosomal subunits. J. Bacteriol. 2017; 199:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Elvekrog M.M., Gonzalez R.L.. Conformational selection of translation initiation factor 3 signals proper substrate selection. Nat. Struct. Mol. Biol. 2013; 20:628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maar D., Liveris D., Sussman J.K., Ringquist S., Moll I., Heredia N., Kil A., Bläsi U., Schwartz I., Simons R.W.. A single mutation in the IF3 N-terminal domain perturbs the fidelity of translation initiation at three levels. J. Mol. Biol. 2008; 383:937–944. [DOI] [PubMed] [Google Scholar]
- 48. Sharma I.M., Rappé M.C., Addepalli B., Grabow W.W., Zhuang Z., Abeysirigunawardena S.C., Limbach P.A., Jaeger L., Woodson S.A.. A metastable rRNA junction essential for bacterial 30S biogenesis. Nucleic Acids Res. 2018; 46:5182–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Connolly K., Rife J.P., Culver G.. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol. Microbiol. 2008; 70:1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thammana P., Held W.A.. Methylation of 16S RNA during ribosome assembly in vitro. Nature. 1974; 251:682–686. [DOI] [PubMed] [Google Scholar]
- 51. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H.. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006; 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Andrésson Ó.S., Davies J.E.. Some properties of the ribosomal RNA methyltransferase encoded by ksgA and the polarity of ksgA transcription. MGG Mol. Gen. Genet. 1980; 179:217–222. [DOI] [PubMed] [Google Scholar]
- 53. Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., Ueda T.. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001; 19:751–755. [DOI] [PubMed] [Google Scholar]
- 54. Jacob A.I., Köhrer C., Davies B.W., RajBhandary U.L., Walker G.C.. Conserved Bacterial RNase YbeY Plays Key Roles in 70S Ribosome Quality Control and 16S rRNA Maturation. Mol. Cell. 2013; 49:427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Davies J., Gorini L., Davis B.D.. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1965; 1:93–106. [PubMed] [Google Scholar]
- 56. Chulluncuy R., Espiche C., Nakamoto J., Fabbretti A., Milón P.. Conformational response of 30S-bound IF3 to a-site binders streptomycin and kanamycin. Antibiotics. 2016; 5:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kriel A., Bittner A.N., Kim S.H., Liu K., Tehranchi A.K., Zou W.Y., Rendon S., Chen R., Tu B.P., Wang J.D.. Direct regulation of GTP homeostasis by (p)ppGpp: A critical component of viability and stress resistance. Mol. Cell. 2012; 48:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gourse R.L., Gaal T., Bartlett M.S., Appleman J.A., Ross W.. rRNA Transcription and growth rate–dependent regulation of ribosome synthesis in Escherichia Coli. Annu. Rev. Microbiol. 2002; 50:645–677. [DOI] [PubMed] [Google Scholar]
- 59. Booth I. Escherichia coli and Salmonella typhimurium. Cellular and Molecular Biology, Vol. 1 (of2). Trends Biochem. Sci. 1988; 13:493–494. [Google Scholar]
- 60. Murray H.D., Schneider D.A., Gourse R.L.. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol. Cell. 2003; 12:125–134. [DOI] [PubMed] [Google Scholar]
- 61. Dennis P.P., Bremer H.. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus. 2008; 3:doi:10.1128/ ecosal.5.2.3. [DOI] [PubMed] [Google Scholar]
- 62. Gupta N., Culver G.M.. Multiple in vivo pathways for Escherichia coli small ribosomal subunit assembly occur on one pre-rRNA. Nat. Struct. Mol. Biol. 2014; 21:937–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Reeve C.A., Amy P.S., Matin A.. Role of protein synthesis in the survival of carbon-starved Escherichia coli K-12. J. Bacteriol. 1984; 160:1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Boehringer D., O’Farrell H.C., Rife J.P., Ban N.. Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis. J. Biol. Chem. 2012; 287:10453–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shetty S., Varshney U.. An evolutionarily conserved element in initiator tRNAs prompts ultimate steps in ribosome maturation. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E6126–E6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. López-Alonso J.P., Fabbretti A., Kaminishi T., Iturrioz I., Brandi L., Gil-Carton D., Gualerzi C.O., Fucini P., Connell S.R.. Structure of a 30S pre-initiation complex stalled by GE81112 reveals structural parallels in bacterial and eukaryotic protein synthesis initiation pathways. Nucleic Acids Res. 2017; 45:2179–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. McGinnis J.L., Liu Q., Lavender C.A., Devaraj A., McClory S.P., Fredrick K., Weeks K.M.. In-cell SHAPE reveals that free 30S ribosome subunits are in the inactive state. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:2425–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Himeno H., Hanawa-Suetsugu K., Kimura T., Takagi K., Sugiyama W., Shirata S., Mikami T., Odagiri F., Osanai Y., Watanabe D. et al.. A novel GTPase activated by the small subunit of ribosome. Nucleic Acids Res. 2004; 32:5303–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wada A., Mikkola R., Kurland C.G., Ishihama A.. Growth phase-coupled changes of the ribosome profile in natural isolates and laboratory strains of Escherichia coli. J. Bacteriol. 2000; 182:2893–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Starosta A.L., Lassak J., Jung K., Wilson D.N.. The bacterial translation stress response. FEMS Microbiol. Rev. 2014; 38:1172–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Seshadri A., Varshney U.. Mechanism of recycling of post-termination ribosomal complexes in eubacteria: A new role of initiation factor 3. J. Biosci. 2006; 31:281–289. [DOI] [PubMed] [Google Scholar]
- 72. Yoshida H., Ueta M., Maki Y., Sakai A., Wada A.. Activities of Escherichia coli ribosomes in IF3 and RMF change to prepare 100S ribosome formation on entering the stationary growth phase. Genes Cells. 2009; 14:271–280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.