Abstract
Endogenous retroviruses (ERVs) contribute to ∼10 percent of the mouse genome. They are often silenced in differentiated somatic cells but differentially expressed at various embryonic developmental stages. A minority of mouse embryonic stem cells (ESCs), like 2-cell cleavage embryos, highly express ERV MERVL. However, the role of ERVs and mechanism of their activation in these cells are still poorly understood. In this study, we investigated the regulation and function of the stage-specific expressed ERVs, with a particular focus on the totipotency marker MT2/MERVL. We show that the transcription factor Zscan4c functions as an activator of MT2/MERVL and 2-cell/4-cell embryo genes. Zinc finger domains of Zscan4c play an important role in this process. In addition, Zscan4c interacts with MT2 and regulates MT2-nearby 2-cell/4-cell genes through promoting enhancer activity of MT2. Furthermore, MT2 activation is accompanied by enhanced H3K4me1, H3K27ac, and H3K14ac deposition on MT2. Zscan4c also interacts with GBAF chromatin remodelling complex through SCAN domain to further activate MT2 enhancer activity. Taken together, we delineate a previously unrecognized regulatory axis that Zscan4c interacts with and activates MT2/MERVL loci and their nearby genes through epigenetic regulation.
INTRODUCTION
Endogenous retroviruses (ERVs) cover ∼10% of the mouse and human genome (1). Most of them are silenced to protect cells from abnormal gene activation and potential mutations (2,3). In mammals, the repression mechanism and function of ERVs are now extensively investigated in various tissues and cells such as brain (4) and embryonic stem cells (ESCs) (5,6). ERVs are known to be repressed by KRAB-zinc finger proteins that mediate heterochromatin formation involving H3K9me2/3 and DNA methylation (5,7–11). Multiple proteins are involved in this process including H3.3, Atrx, Setdb1, G9a, Kdm1a, Chaf1 and Kap1 (9,12–16). However, a minority of ERVs are still expressed in sporadic ESCs. For instance, the ERV3 family member MERVL is expressed in the 2-cell stage embryo and a minority of ESCs (17). Once being expressed, ERVs participate in the regulation of host gene expression (18,19) and may provide new transcription factor binding sites and function as enhancers or promoters to drive gene expression (20). These ERVs also can donate new recombination hotspots and Ctcf binding sites to reorganize chromatin structure (21). If the expressed ERVs retain protein-coding capacity, they may activate viral restriction pathways in ESCs to prevent external viral infection (22).
Distinct ERVs are expressed dynamically during early embryogenesis (23). This requires flexible epigenetic regulation to allow appropriate expression of ERVs. Therefore, it is important to decipher the mechanism behind the dynamic expression of ERVs in early embryos and the function of ERVs. Recently, transcription factor Dux was found to be activated during zygotic gene activation and in 2-cell embryos (24–26) and it is critical to the expression of MERVL and 2-cell embryo genes during development and in ESCs (24–26). Retroelement long interspersed nuclear element 1 (LINE1) was also discovered to be essential for early mouse embryo development due to its role in maintaining chromatins in an open state (27). It remains elusive whether any other players are involved in the activation of MERVL and 2-cell embryo genes.
Here, we sought to further analyse published single cell RNA-seq data (28,29) in order to investigate the potential regulatory mechanism of ERV transcription in pre-implantation mouse embryos. We identified ERV groups that were uniquely expressed in each developmental stage of mouse preimplantation embryos. We further found Zscan4c as an activator of MT2/MERVL and 2-cell/4-cell genes in ESCs. We demonstrate that Zscan4c overexpression is associated with augmented H3K27ac, H3K4me1 and H3K14ac deposition on MT2 (MERVL LTR) and Zscan4c interacts with GBAF (GLTSCR1L- and BRD9-containing BAF) chromatin remodelling complex to activate MT2 enhancer activity, which subsequently promotes the expression of its nearby 2-cell/4-cell genes. Taken together, these findings extend our understanding of ERV regulation in early embryogenesis and demonstrate that Zscan4c plays a key role in regulating MERVL and 2-cell/4-cell genes in ESCs.
MATERIALS AND METHODS
Cell culture, inhibitor treatment and transfection
Mouse J1 ESCs were cultured under 5% CO2 at 37°C on plates coated with 0.2% gelatin (G1890, Sigma), in Dulbecco's modified Eagle's medium (SH30243.01m Hyclone) that was supplemented with 15% fetal bovine serum (FBS) (SH30070.03, Hyclone), 10 ng/ml mouse LIF (Z03077, GeneScript), 1% Penicillin–Streptomycin (P1400, Solarbio), 1% GlutaMAX (G0200, Solarbio), 1% nonessential amino acids (Gibco) and 0.1 mM β-mercaptoethanol (M3148-250, Sigma). Cells were passaged every 2–3 days using 0.25% trypsin–EDTA (25200-072, Gibco). 293T cells were cultured under 5% CO2 at 37°C, in Dulbecco's modified Eagle's medium (12100-046, Gibco) that was supplemented with 10% FBS (04-001-1A, Biological Industries) and 1% Penicillin–Streptomycin (P1400, Solarbio). Cells were passaged every 1.5–2 days using 0.25% trypsin–EDTA (25200-072, Gibco). For inhibitor treatment, cells were treated with 1 μM 5-Azacytidine (5-Aza) (A1907, APExBIO) for 24 h or 20 μM BI-7273 (T6783, TargetMol) for 48 h at 24 h post-passage. For transfection, 293T cells were transfected at 60% confluency with Polyjet (SL100688, SignaGen) according to the manufacturer's instructions.
Immunostaining and flow cytometry analysis
ESCs were fixed in ice-cold 80% ethanol. After permeabilization in 1% Triton X-100 for 30 min on ice, cells were blocked in 1% bovine serum albumin for 30 min. Immunostaining was performed using anti-MERVL-gag antibody (A-2801, EpiGentek). The secondary antibody used was Alexa Fluor 594-conjugated goat anti-rabbit antibody (ZSGB-BIO). Immunostained samples were analysed by flow cytometry on LSR Fortessa (BD Biosciences).
Chromatin immunoprecipitation (ChIP)
ChIP was done according to a published protocol (30). Approximately, 2 × 107 ESCs were harvested for ChIP. Briefly, ESCs were cross-linked with 1% (w/v) formaldehyde for 10 min at room temperature and quenched in 200 mM glycine. Cells were washed with chilled tris-buffered saline (TBS) containing EDTA, scraped off the plates and collected by centrifugation. The collected cell pellet was lysed in buffer containing 0.25% Triton X-100 and protease inhibitors. Subsequently, chromatin pellet was collected and sonicated. Sample was pre-cleared with protein G beads at 4°C for 2 h and immunoprecipitated with anti-Flag antibody (M20008-M, Abmart), anti-H3K4me1 antibody (ab176877, Abcam), anti-H3K27ac antibody (ab177178, Abcam), anti-H3K14ac antibody (ab52946, Abcam), anti-Brg1 antibody (#49360, Cell Signalling Technology), anti-Brd9 antibody (#71232, Cell Signalling Technology) at 4°C overnight. Immunoprecipitated chromatins were subsequently eluted and decrosslinked. The immunoprecipitated DNA was purified with phenol:chloroform and analysed by qPCR.
Chromatin immunoprecipitation coupled with high throughput sequencing (ChIP-seq) data analysis
DNA from ChIP was extracted and at least 2 ng ChIP DNA was used for ChIP-seq library preparation. The ChIP-seq sequencing libraries were prepared with kits from Novogene Corporation. Each library was subsequently pair-end sequenced by Novogene Corporation (Tianjin, China), resulting in pair-ended 25 million reads (150 bp). The adapter sequences of ChIP-seq data were removed with Cutadapt (31) and reads with Phred score <5 were excluded for analysis. Bowtie2 (32) was used to map reads to mm10 with the parameter ‘–very-sensitive’. ChIP-seq signal enrichment was obtained by bamCompare from Deeptools (33) with parameter ‘–scaleFactorsMethodreadCount –minMappingQuality 1 –operation ratio –pseudocount 1’. ChIP signal heatmap and line plot were also generated by Deeptools (33). Macs2 (34) was used to call Zscan4c binding peaks with parameter ‘--keep-dup auto -q 0.1'. MT2 and MERVL-int centre information were inferred from RepeatMasker (35,36). MEME-ChIP (37) was used to analyze Zscan4 binding motif with the parameter ‘-meme-mod zoops -meme-minw 4 -meme-maxw 10 -meme-nmotifs 10 -dreme-e 0.05 -centrimo-score 5.0 -centrimo-ethresh 10.0’, and sequences of top 2000 Zscan4c binding peaks were used as input. For Zscan4c function prediction, binding and expression target analysis (BETA) (38) was used to integrate Zscan4c ChIP data and Zscan4c overexpression data. ChIP-seq signal was mapped to MT2/MERVL consensus (GenBank ID: Y12713.1) (39) and visualized on Integrative Genomics Viewer (IGV) (40). For location statistics of MT2 motif and Dux motif, MT2 annotation on mm10 was first extracted from RepeatMasker. ‘vmatchPattern’ function from R package Biostrings was used to determine the locations of Zscan4c binding motif and Dux binding motif on MT2.
Whole genome bisulfite-sequencing (WGBS) data analysis
For WGBS data processing, all the WGBS reads were first processed with Trim Galore (0.4.4) to trim adaptor and low-quality reads. Adaptor-trimmed reads were then mapped to MT2 consensus sequence from Dfam (41) by bismark (v0.20.0) (42), and duplicated reads were depleted by deduplicate_bismark (v0.20.0), and bismark_methylation_extractor (v0.20.0) was used to get final DNA methylation level.
Generation of Zscan4c-related ESC cell lines
For the establishment of Zscan4c-depleted cell lines, two shRNAs targeting Zscan4c were transfected into J1 ESCs and selected in 2 μg/ml puromycin for ∼7 days. The sequences of shRNAs against Zscan4c are: GCGTTCAATTAGCAGACCA (Control shRNA), GAGTGAATTGCTTTGTGTC (Zscan4c shRNA1), CAGAAGCCTGGCATTCCCT (Zscan4c shRNA2). For the generation of Zscan4c overexpression cell line, J1 ESCs were transfected with 2 μg pCMV-Zscan4c vector or empty vector were selected in 500 μg/ml geneticin for ∼14 days. For the generation of Flag-Zscan4c overexpression cell line, J1 ESCs were transfected with 1 μg PB-Zscan4-Flag and 0.5 μg Pbase (PB-Zscan4-Flag:Pbase = 2:1) or empty vector were selected in 500 μg/ml hygromycin B for ∼14 days. Drug-resistant clones were picked to establish stable cell lines. For the generation of Zscan4c SCAN domain, internal domain, or zinc finger domain overexpression cell line, J1 ESCs that were transfected with 1 μg pCAG-3HA-Zscan4c-zinc finger domains, pCAG-3HA-Zscan4c-internal domain, pCAG-3HA-Zscan4c SCAN domain or empty vector were selected in 500 μg/ml hygromycin B for ∼14 days.
DNA-protein pull-down assay
DNA-protein pull-down was performed as previously described (43). In brief, 293T cells were transfected with HA-Zscan4c overexpression vector. Two days after transfection, 293T cells were lysed by sonication in lysis buffer containing 50 mM Tris–HCl pH8.0, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.5% NP-40 and protease inhibitors for the preparation of nuclear exact. Nuclear extracts were precleared with Streptavidin MagBeads (GenScript, L00424) for 1 h. After that, the precleared supernatant was incubated with 1 μg biotinylated double-stranded oligonucleotide and poly(dI-dC) on a rotating shaker at 4°C for 4 h. DNA-bound proteins were collected by incubating with Streptavidin MagBeads at 4°C for 1 h. Then the beads were washed five times with PBS, and the bound proteins were released from beads by boiling the beads in SDS loading buffer. Western blot was performed to detect the proteins with specific antibodies. The sequences of biotinylated double-strand oligonucleotides used in the pull-down assay are listed in Supplementary Table S1.
Luciferase assay
Mouse ESCs were transfected with pGL4.23 empty vector or vector containing MT2 LTR region of MERVL cloned from 2C::tdTomato reporter (17). MT2 reporter construct (200 ng) was transfected into each well of J1 ESCs in a 24-well plate together with 10 ng of pCMV-Renilla. After transfection, the cells were cultured in medium for 48 h. The medium was changed one day after transfection. For the generation of mutant Luciferase reporter, the region corresponding to 162–259 bp on MT2 consensus was deleted from MT2. For constructing rescued mutant Luciferase construct, Zscan4c binding motif (ATTAAAGGTGTGTT) was inserted back to mutant Luciferase reporter. For Luciferase assay after Brd9 inhibitor BI7273 treatment, one day after transfection, cells were treated with DMSO or 20 μM BI7273 (T6783, TargetMol) for 40 h. Luciferase and Renilla activity were determined by Dual-Luciferase Reporter Assay System (#E1910, Promega) according to the manufacturer's instructions.
Bisulphite sequencing
Approximately, 2 × 106 ESCs were harvested to extract genomic DNA, which was treated using the EpiTect Bisulphite Kit (QIAGEN) as described. DNA fragments for sequencing were obtained by PCR and inserted into pEASY-T1 vector. The sequences of primers are: TTGAAAGTGTTGGTGGATTAATAAGT (Forward), ATAAATTCCAAAAAATTAAATCTCC (Reverse). Five or more clones were obtained from J1 ESCs overexpressing Zscan4c and control ESCs.
Methylated DNA immunoprecipitation (MeDIP)
MeDIP was done using the SimpleDIP Methylated DNA immunoprecipitation (MeDIP) Kit (Cell Signalling Technology) according to the manufacturer's instructions. Two million ESCs were harvested to extract genomic DNA, which was sonicated to ∼500 bp DNA fragments. Sonicated genomic DNA (1 μg) was used for each immunoprecipitation or input. Rabbit anti-5mC antibody (#28692, Cell Signalling Technology) was used to immunoprecipitate methylated DNA. DNA was eluted from the antibody/protein G beads by 150 μl elution buffer for 30 min at 65°C after washing with 1 ml immunoprecipitation buffer for four times. Finally, the immunoprecipitated DNA and input DNA were purified with phenol:chloroform and analysed by qPCR.
Protein co-immunoprecipitation (co-IP)
J1 ESCs expressing Flag-Zscan4c and control J1 ESCs were used for Brg1-Zscan4c co-IP. For Brd9-Zscan4c co-IP, 293T cells were co-transfected with Zscan4c-Flag/pCAG-3HA or Zscan4-Flag/pCAG-3HA-Brd9. Cells were washed with pre-cooled PBS and lysed in 1 ml pre-cooled lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 20 mM KCl, 1.5 mM MgCl2, 1% glycine, 0.5% Triton-100 and protease inhibitor cocktail). The cell lysate was pre-cleared and immunoprecipitated with anti-HA magnetic beads (b26202, Bimake) or anti-Flag magnetic beads (b26102, Bimake). Beads were washed 3 times with lysis buffer and twice with wash buffer (50 mM Tris [pH 8.0], 300 mM NaCl, 1 mM EDTA, 1% Triton-100). Proteins were eluted in elution buffer (50 mM Tris [pH 7.5], 10 mM EDTA, 1% SDS) and loaded onto 10% SDS-PAGE together with 1.25% input.
Reverse transcription and quantitative real-time PCR (RT-qPCR)
Total RNA was extracted with Trizon Reagent (CW0580, CWBIO) and subjected to DNase I (EN0521, Thermo Scientific) treatment. The concentration of total RNA was determined by NanoDrop 2000 (Thermo Scientific). Then 1 μg RNA was subjected to reverse transcription with Transcriptor First Strand cDNA Synthesis Kit (4897030001-RCH, Roche) and Oligo-dT primers. The resulting cDNA was quantified by RT-qPCR analysis with Hieff qPCR SYBR Green Master Mix (H97410, Yeasen) in a QuantStudio 6 real-time PCR system (Life Technologies). Primers for qPCR analysis are listed in Supplementary Table S1.
RNA-seq and data analysis
Total RNA was extracted from control ESCs, Zscan4c-overexpressed ESCs and Zscan4c- depleted ESCs in Trizon reagent (CWBIO) and treated with DNase I (Thermo Scientific) treatment. Total RNA (4 μg) was poly(A) enriched with oligo(T) beads and RNA libraries were prepared for sequencing using standard Illumina protocols. Each library was sequenced for >40 million reads by GENEWIZ, Inc. (Suzhou, China). The adapter sequences of RNA-seq data and low-quality reads with Phred score <5 were excluded with Cutadapt (31) before further processing. Hisat2 (44) was used to map RNA-seq data to mm10 with parameter - k 20. Genes were annotated according to the Ensembl database. Transposable element annotations were from UCSC Genome Browser (RepeatMasker). Reads were counted using featureCounts (45) with parameter ‘-O -M -fraction’ to retain multi-mapped reads. TE repClass, LTR repFamily, LTR repName were considered as metafeatures and were counted together. TEtranscripts (46) with parameter ‘–mode multi’ was used to measure differentially expressed ERVs. For weighted co-expression network analysis (WGCNA), ERVs with high expression level (mean cpm > 1) were used in WGCNA (47) to construct a co-expression network. Cpm matrix with log2 transformed was considered as input data. Soft thresholding power 16 and signed networks were used. Other steps were carried out according to published procedures (47).
Genes with expression fold change >1.5 and adjusted P value <0.05 from DEseq2 results were used for gene ontology (GO) analysis. GO analysis was carried out with DAVID Bioinformatics Resources as described (48,49). The GSEA software (Gene Set Enrichment Analysis) (50) was used to analyse Zscan4c overexpression transcriptome profile. For mapping the expression of mouse Dux repeat units, we mapped RNA-seq and ChIP-seq reads directly to the mouse Dux repeat (AM398147.1) according to the method proposed by Erckersley-Maslin et al. (51).
Western blot
Cells were lysed in buffer containing 3% SDS, 30% glycerol, 150 mM Tris–HCl (pH7) and 5% β-mercaptoethanol. Protein extracts (30 μg) were loaded to a 10% SDS-PAGE gel and transferred to polyvinylidenefluoride (PVDF) membrane. Afterwards, the PVDF membranes were blocked in 5% milk for 2 h at room temperature and incubated with anti-Zscan4c antibody (1:5000 dilution, AB4340, Merck), β-Actin (1:5000 dilution, KM9001, Sungenebiotech), anti-Flag antibody (1:5000 dilution, 30503ES60, Yeasen), anti-HA antibody (1:5000 dilution, 30701ES60, Yeasen), anti-Brg1 antibody (1:2500 dilution, #49360, Cell Signalling Technology) or anti-Brd9 antibody (1:2500 dilution, #71232, Cell Signalling Technology) overnight at 4°C and subsequently incubated with goat anti-rabbit IgG-HRP (1:1000 dilution, sc-2004, Santa Cruz) or anti-mouse m-IgGκ BP-HRP (1:1000 dilution, sc-516102, Santa Cruz) for 90 min at room temperature. Membranes were treated with Immobilon Western HRP substrate (WBKLS0100, Merck) and images were taken by Chemiluminescence Imaging System (Tanon).
RESULTS
Dynamic expression of ERVs in early mouse embryos
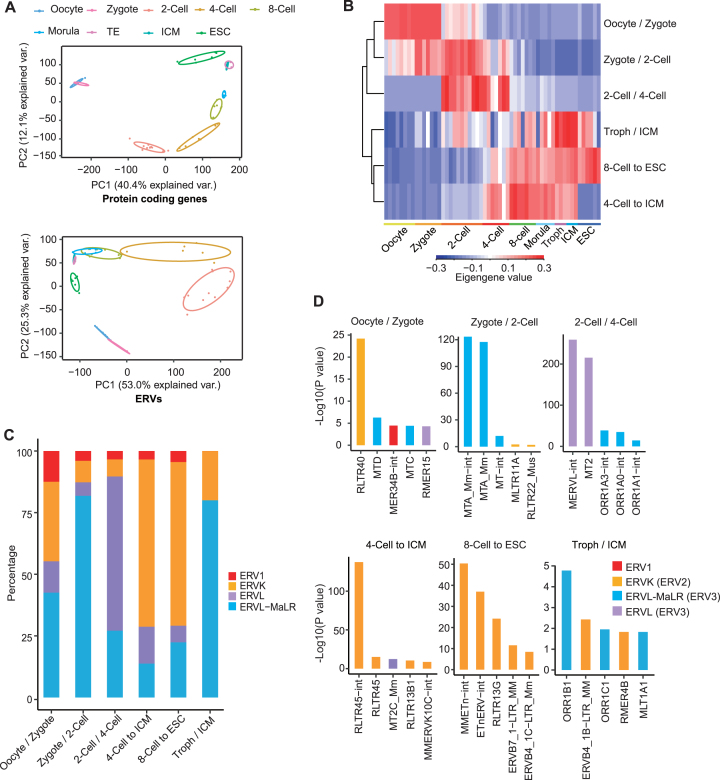
To investigate the expression pattern of transposable elements (TEs) in mouse preimplantation embryo, we analysed the differentially expressed TEs based on published single cell RNA-seq data (28). We first examined expression of ERVs, LINEs, short interspersed nuclear elements (SINEs) and DNA transposons in pre-implantation embryos, and found that ERVs exhibited strongest expression dynamics across different developmental stages among TEs (Supplementary Figure S1A and B). Similar to protein coding genes, principal component analysis (PCA) based on ERVs alone can distinguish different developmental stages (Figure 1A). To address which groups of ERVs are expressed at each different developmental stage, we performed weighted gene co-expression network analysis (WGCNA) of ERVs. This approach revealed 11 modules of ERVs with distinct expression patterns during early embryogenesis (Figure 1B; Supplementary Figure S1C). Out of 11 modules, 6 modules exhibited the same stage-specific expression during embryogenesis according to both RNA-seq datasets (28,29) we analysed (Figure 1B; Supplementary Figure S1D). ERVs from 3 modules (oocyte/zygote, zygote/2-cell, 2-cell/4-cell) demonstrated strong expression in totipotent cells (zygote to 4-cell) whereas the other three modules were expressed in both early embryos and ICM/ESCs (Figure 1B; Supplementary Figure S1D). Most modules mainly consisted of one or two families of ERVs (Figure 1C). The expressed ERVs in zygote/2-cell module were different from those in the oocyte/zygote module, implying that these ERVs in zygote/2-cell may be activated during zygotic activation (Figure 1D). ERV3 family (ERVL-MaLR and ERVL) member MTA was most highly enriched in the zygote/2-cell stage specific ERV module (Figure 1D). Another member of ERV3, MT2/MERVL was enriched in the module expressed in both 2-cell and 4-cell embryos (Figure 1D; Supplementary Table S2 and 3). Besides the ERV3 family (ERVL and ERVL-MaLR), a minority of ERV1 and ERVK were also expressed in 2-cell embryos (Figure 1C; Supplementary Figure S1E). For example, RLTR14 (ERV1) and RLTR19/RMER6-int (ERVK) were specifically expressed at 2-cell stage marked by MT2/MERVL (Supplementary Figure S1E). One or two repeat families were usually found in each module, demonstrating their specific expression in comparison to other repeats (Figure 1D; Supplementary Figure S1F; Supplementary Tables S2 and S3). Collectively, these results suggest that different families of ERV members are expressed in a stage-specific manner in the pre-implantation mouse embryos.
Figure 1.
Analysis of ERV expression dynamics in early embryogenesis by WGCNA. (A) Principal component analysis of embryo samples according to mRNA expression of protein coding genes and ERV expression. The selected ERV loci were separated from protein coding genes and were filtered by low expression (mean cpm < 1). Var., variation. (B) Hierarchical cluster of ERV modules using eigengene values. ERV modules were identified by WGCNA. Only modules with embryonic expression specificity in both datasets (28, 29) were presented. Troph, trophectoderm; ICM, inner cell mass. (C) The ERV repeat family composition of 6 stage-specified modules presented in percentage of all enriched ERVs in each module. (D) WGCNA enrichment of ERVs in six most stage-specified modules. P values were determined by Fisher's exact test.
Zscan4c activates MT2/MERVL expression
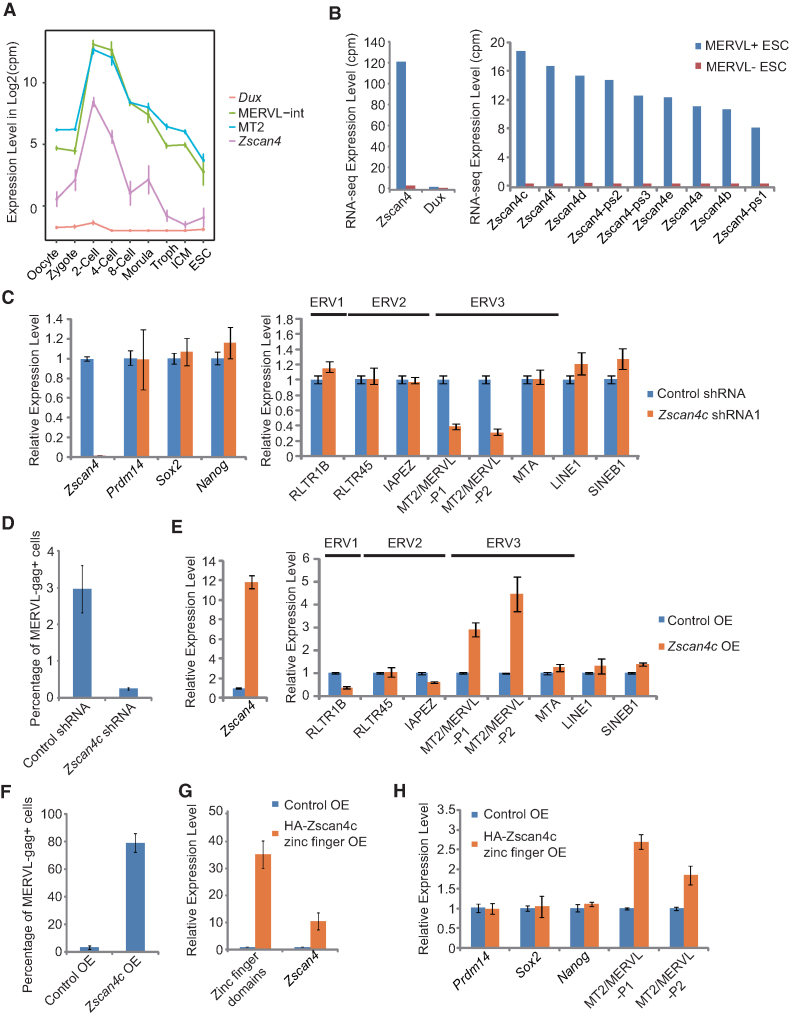
ERV3 family member MT2/MERVL marks totipotent 2-cell/4-cell embryos (Figure 1D) and a subset of 2-cell like ESCs (17). Recently, transcription factor Dux was discovered to be a crucial activator of 2-cell genes and MERVL (24–26). Our analysis and previous studies (25,26) showed that MT2/MERVL was not only expressed in 2-cell embryo, also in 4-cell embryos (Figure 2A). In addition, given that Dux is only highly expressed in early 2-cell stage mouse embryos (25,26), we speculated that there were other factor(s) essential to the activation of MT2/MERVL in 2-cell and 4-cell embryos in addition to Dux. Transcription factor Zscan4 cluster genes were newly discovered to be a marker of the intermediate state of 2-cell like ESCs (52). Similar to MT2 and MERVL, Zscan4 was also expressed at 2-cell/4-cell stages (Figure 2A). Moreover, Zscan4 expression was higher than Dux at transcript level in MERVL-gag+ 2-cell like ESCs (Figure 2B). Among expressed Zscan4 cluster genes, Zscan4c demonstrated the highest expression in 2-cell like ESCs (Figure 2B). Hence, we examined the possibility for Zscan4c as an activator of MT2/MERVL. Depletion of Zscan4c with two independent shRNAs led to a decreased expression of MT2/MERVL, without affecting the expression of pluripotency genes (Prdm14, Nanog and Sox2), other ERVs (RLTR1B, RLTR45, IAPEZ and MTA) or TEs (LINE1 and SINE B1) (Figure 2C; Supplementary Figure S2A and B). Depletion of Zscan4c also decreased MERVL-gag+ 2-cell like population (Figure 2D). In contrast, the overexpression of Zscan4c led to the upregulation of MT2/MERVL, but not other repeat sequences (RLTR1B, RLTR45, IAPEZ and MTA, LINE1 and SINE B1) (Figure 2E), while the expression of pluripotency genes (Prdm14 and Sox2) remained unchanged except for Nanog being slightly upregulated (Supplementary Figure S2C and D). Overexpression of Zscan4c similarly caused ∼80% ESCs to become MERVL-gag+ (Figure 2F). Moreover, overexpression of zinc finger domains (putative DNA binding domains) of Zscan4c alone, but not other domains of Zscan4c, activated MT2/MERVL in ESCs (Figure 2G and H; Supplementary Figure S2E–H). Intriguingly, overexpression of zinc finger domains also promoted Zscan4c expression (Figure 2G). Together, these results indicate that Zscan4c specifically activates MT2/MERVL expression in ESCs and its zinc finger domains play an important role in this process.
Figure 2.
The role of Zscan4c in activating of MT2/MERVL expression. (A) Expression of MT2/MERVL, Zscan4c and Dux during early embryogenesis according to published single cell RNA-seq data (28). (B) Expression of total Zscan4 gene cluster members and Dux in MERVL+ and MERVL- ESCs according to published RNA-seq data (17). (C) qPCR analysis of the expression of Zscan4c and retroelements after Zscan4c depletion in mESCs. P1, primer set1; P2, primer set 2. Biological-triplicate data (n = 3 dishes) are presented as mean ± s.e.m. (D) Flow cytometry analysis of MERVL-gag+ population of ESCs expressing control shRNA or Zscan4c shRNA. Biological triplicate data (n = 3 dishes) are presented as mean ± s.e.m. (E) qPCR analysis of the expression of Zscan4c and retroelements after Zscan4c overexpression in mESCs. P1, primer set1; P2, primer set 2. Biological triplicate data (n = 3 dishes) are presented as mean ± s.e.m. (F) Flow cytometry analysis of MERVL-gag+ population of ESCs after Zscan4c overexpression. Biological triplicate data (n = 3 dishes) are presented as mean ± s.e.m. (G) Expression of zinc finger domains of Zscan4c and full length Zscan4c determined by qPCR after overexpression of Zscan4c zinc finger domains. (H) Expression of pluripotency markers (Prdm14, Sox2 and Nanog) and MT2/MERVL determined by qPCR after overexpression of Zscan4c zinc finger domains. P1, primer set1; P2, primer set 2. Biological-triplicate data (n = 3 extracts) are presented as mean ± s.e.m. OE: overexpression.
Genome-wide activation of MT2/MERVL loci and 2-cell/4-cell embryo genes by Zscan4c
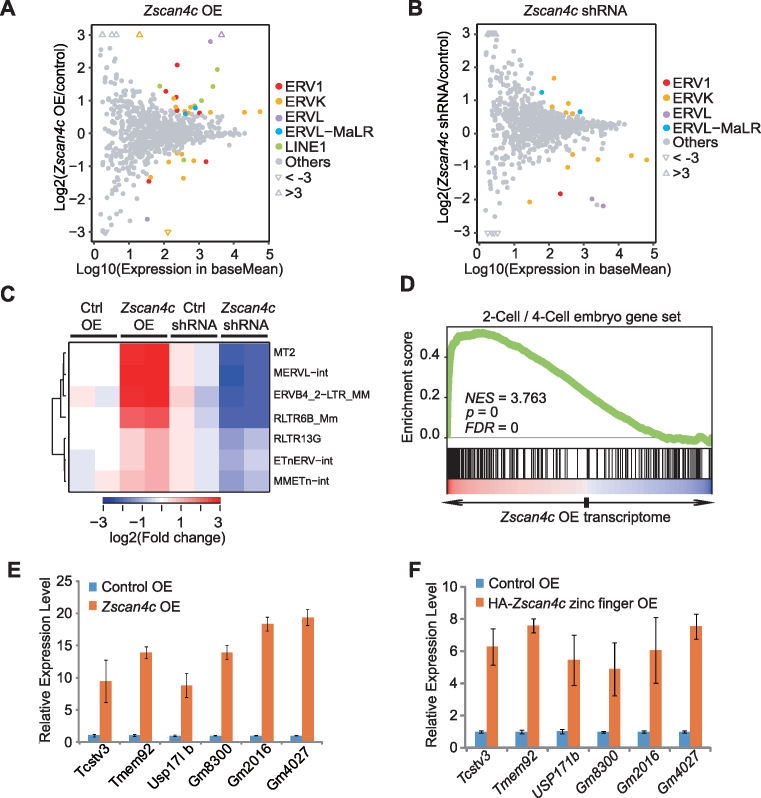
To identify genome-wide regulatory targets of Zscan4c, we performed RNA-seq analysis after Zscan4c overexpression and depletion in ESCs. RNA-seq results revealed that ERVL family members were strongly upregulated in Zscan4c-overexpressed ESCs (Figure 3A; Supplementary Table S4 and S5) and most downregulated after Zscan4c depletion in comparison to ERVs in other families (Figure 3B; Supplementary Table S6 and S7). Furthermore, if we overlapped upregulated ERVs in Zscan4c-overexpressed ESCs with those downregulated ERVs in Zscan4c-depleted ESCs, the enrichment of both MERVL-int and MT2 (MERVL LTR) was revealed (Figure 3C; Supplementary Table S4 and S6). Besides ERVs, Zscan4c overexpression or depletion also affected the expression of protein coding genes (Supplementary Tables S8–S11). After Zscan4c overexpression, GO terms related to the regulation of transcription, chromatin modification, cell cycle, and cell proliferation were enriched in upregulated genes (Supplementary Figure S3A) whereas GO terms related to metabolic processes were enriched in downregulated genes (Supplementary Figure S3B). In agreement with Zscan4c overexpression results, depletion of Zscan4c in ESCs resulted in enrichment of GO terms related to the regulation of transcription and cell proliferation in downregulated genes, while GO terms related to differentiation and development were enriched in upregulated genes (Supplementary Figure S3C and D). These implicate a role of Zscan4c in regulating gene transcription in ESCs. Similar to the effects of Dux overexpression in ESCs (24–26), Zscan4c overexpression also led to the enrichment of gene sets associated with 2-cell/4-cell embryo development (Figure 3D; Supplementary Figure S3E). We defined 170 genes that were upregulated after Zscan4c overexpression and downregulated after Zscan4c depletion at the same time as Zscan4c-activated genes. Among these 170 Zscan4c-activated genes, 56 genes (33%) were expressed specifically in 2-cell/4-cell embryos (Supplementary Figure S3F). Our qPCR results verified the upregulation of 2-cell/4-cell embryo genes after Zscan4c overexpression (Figure 3E). These Zscan4c-targeted 2-cell/4-cell genes were also upregulated after overexpression of Zscan4c zinc finger domains (Figure 3F). Together, these results implicate that Zscan4c activates MT2/MERVL and 2-cell/4-cell cleavage embryo genes in ESCs.
Figure 3.
Transcriptome analysis of Zscan4c-regulated retroelements and genes after Zscan4c overexpression or depletion. (A, B) Scatter diagrams of TE expression change after Zscan4c overexpression (OE) (A) and depletion by shRNA (B). TEtranscript results were used to plot these diagrams. Coloured dots indicate retroelements with significant expression change (Wald test, FDR adjusted P value < 0.05). Triangles represent ERVs with fold change >3. (C) Expression heatmap of selected retroelements after Zscan4c depletion and Zscan4c overexpression. Those selected TEs were upregulated after Zscan4c overexpression and downregulated after Zscan4c depletion (fold change > 1.5; Wald test, FDR adjusted P < 0.05). (D) Gene set enrichment analysis (GSEA) indicating that upregulated genes after Zscan4c overexpression were highly enriched in the 2-cell/4-cell embryo gene set. Red, upregulated genes; blue, downregulated genes. (E) qPCR analysis of the expression of 2-cell/4-cell embryo genes after Zscan4c overexpression in ESCs. (F) qPCR analysis of the expression of 2-cell/4-cell embryo genes after overexpression of Zscan4c zinc finger domains in ESCs. Biological-triplicate data (n = 3 extracts) are presented as mean ± s.e.m. Expression level of the control sample was set as 1. OE, overexpression.
Zscan4c mainly binds to enhancer regions to regulate gene expression
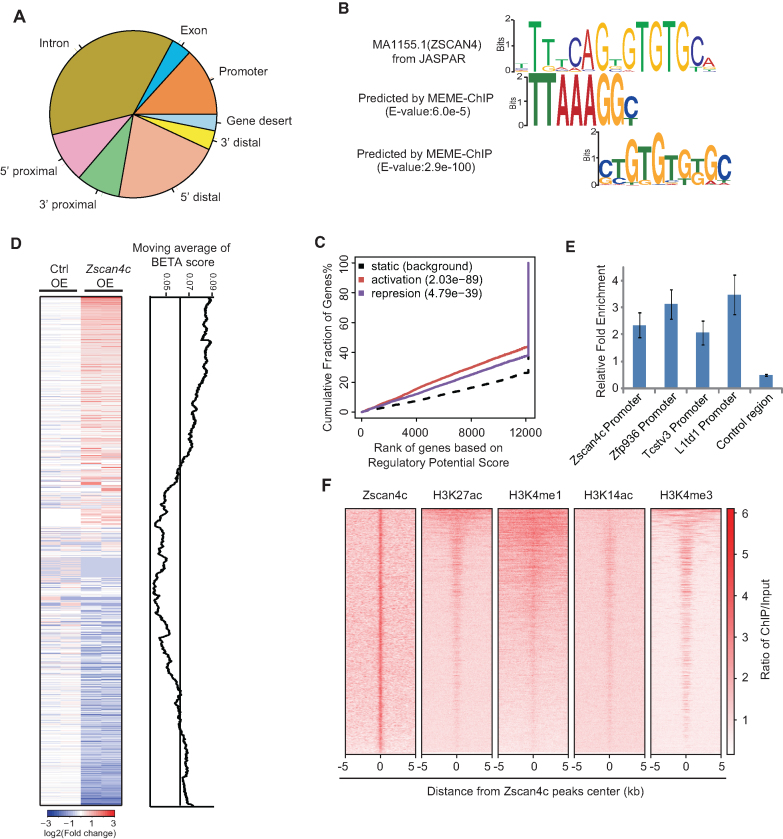
To further gain insight into the role of Zscan4c in activating MT2/MERVL and 2-cell/4-cell gene expression, we performed Zscan4c ChIP-seq after Zscan4c overexpression. Only 13.3% of Zscan4c binding sites were on gene promoter regions whereas most of the Zscan4c binding sites were on intergenic (46%) and intronic (36.8%) regions (Figure 4A; Supplementary Figure S4A). We performed DNA binding motif analysis with MEME-ChIP (37) to identify DNA motifs enriched at Zscan4c binding regions. Two motifs similar to human ZSCAN4 motif were identified (Figure 4B). Both of these motifs were mostly distributed distant from promoter regions (Supplementary Figure S4B). Through the combined analysis of RNA-seq data after Zscan4c overexpression and Zscan4c ChIP-seq data by binding and expression target analysis (BETA) (38), we found that Zscan4c could both activate and repress gene expression in ESCs (Figure 4C and D). The genes that were bound and activated by Zscan4c were related to transcription regulation, mRNA processing and stem cell population maintenance (Supplementary Figure S4C) whereas those genes directly repressed by Zscan4c were mainly enriched of genes important to metabolic processes and ion transport (Supplementary Figure S4D). Moreover, the expression activation effect of Zscan4c was more prominent than its repression role (P = 2.03e−89 for activation; P = 4.79e−39 for repression) (Figure 4C and D). Therefore, we mainly focused on studying activation role of Zscan4c on the gene expression. Our ChIP-qPCR results further verified the interaction of Zscan4c with the promoter regions of its activated genes (Figure 4E). By examining the published histone modifications (53–55) enriched around Zscan4c binding peaks, we found that Zscan4c peaks were enriched of histone marks related to enhancer (H3K27ac, H3K4me1 and H3K14ac) and promoter histone mark H3K4me3 (Figure 4F). However, Zscan4c was depleted from the transcription start site (TSS) region which is highly enriched of H3K4me3 (Supplementary Figure S4E). Based on these results, we conclude that Zscan4c interacts mainly with enhancer regions.
Figure 4.
Genome-wide binding profile of overexpressed Zscan4c on ESC genome. (A) Locations of Zscan4c peaks relative to the nearest transcription units (Promoter, 2 kb around transcriptional start sites; 5′ proximal, 2–10 kb upstream of gene; 5′ distal, 10–100 kb upstream of gene; 3′ proximal, 0–10 kb downstream of gene; 3′ distal, 10–100 kb downstream of gene; Gene desert, > 100 kb away from the nearest gene). (B) Zscan4c-recognized DNA motifs identified by MEME-ChIP and enriched on MT2. MEME Suite was used to determine E value of motif. (C) Prediction of activating/repressive function of Zscan4c by BETA according to Zscan4c ChIP-seq data and RNA-seq data after Zscan4c overexpression. Red line, upregulated genes; purple line, downregulated genes; dashed line, the non-differentially expressed genes as background. Genes were cumulated by the rank on the basis of the regulatory potential score from high to low. P values that represented the significance of the distributions of upregulated or downregulated genes were compared with the non-differentially expressed genes by the Kolmogorov–Smirnov test. (D) Genome-wide association of Zscan4c regulatory potential score with gene expression change after Zscan4c overexpression. Regulatory potential score was predicted by BETA minus function. Genes were ranked by expression in log2 (ratio of Zscan4c OE/Control OE) on heatmap on the left. The line plot (right) shows the corresponding BETA scores (moving averages) of genes. (E) ChIP-qPCR analysis of Zscan4c binding on gene promoters. ChIP-qPCR data was normalized to input and that of the control region. Biological-triplicate data (n = 3 extracts) are presented as mean ± s.e.m. (F) ChIP-seq enrichment heatmap of Zscan4c, H3K27ac, H3K4me1, H3K14ac, H3K4me3 signal on Zscan4c binding peaks. The ChIP-seq signal was calculated as the ratio of normalized reads relative to input.
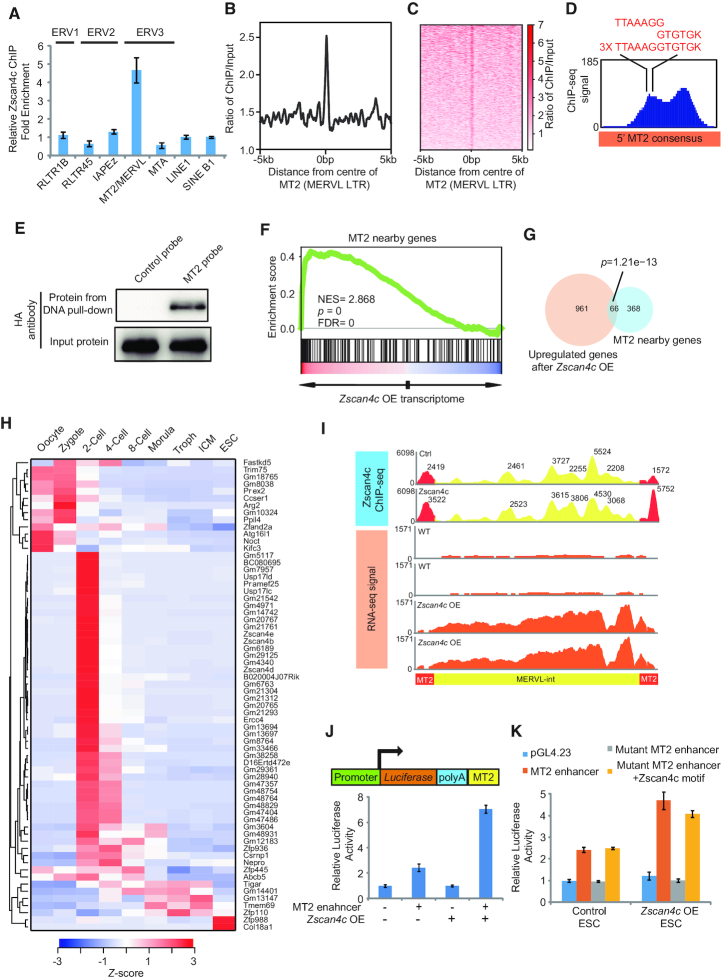
Zscan4c directly binds to MT2/MERVL to regulate 2-cell gene expression
Interestingly, the majority of Zscan4c binding peaks (58.7%) were mainly located on TEs (Supplementary Figure S5A), implying a role of Zscan4c in regulating TE expression. To test whether Zscan4c directly regulates MT2/MERVL, we first used ChIP-qPCR to validate the interaction of Zscan4c with MT2/MERVL. Indeed, Zscan4c directly interacted with MT2/MERVL instead of other ERVs and retroelements (Figure 5A). Notably, we only observed the enrichment of Zscan4c binding on MT2 and MERVL-int (Figure 5B and C; Supplementary Figure S5B and C), indicating a direct role of Zscan4c in activating MT2/MERVL expression. In addition, we further found Zscan4c peaks present on 5′ MT2 of MERVL consensus (Figure 5D). The motifs we identified were fused into one longer motif on MT2 (Figure 5D). All three copies of the fused motifs located in the centre of one Zscan4c peak on MT2 (Figure 5D). The Zscan4c binding motif is separated from the Dux motif yet frequently found together on MT2 elements (Supplementary Figure S5D). Our DNA-protein pull-down experiment showed that only MT2 probe corresponding to Zscan4c ChIP-seq peak region (Figure 5D), but not control probe, could successfully pull down full-length Zscan4c protein and zinc finger domains of Zscan4c (Figure 5E; Supplementary Figure S5E). This further confirmed that Zscan4c directly interacted with MT2 sequence. In addition, regions around MT2 and MERVL were enriched of upregulated genes after Zscan4c overexpression (Figure 5F; Supplementary Figure S5F). MT2 and MERVL nearby genes significantly overlapped with upregulated genes after Zscan4c overexpression (P = 1.21e–13 for MT2 nearby genes; P = 2.73e–4 for MERVL-int nearby genes) (Figure 5G; Supplementary Figure S5G). Among overlapped genes, most of them were expressed prior to the 4-cell stage (Figure 5H; Supplementary Figure S5H), implying that Zscan4c may activate 2-cell/4-cell embryo genes through MT2/MERVL. Moreover, enrichment of Zscan4c binding was detected at MT2 (Figure 5B and C) and flanking MERVL annotated regions (Supplementary Figure S5B and C), consistent with the fact that MT2 is the LTR of MERVL elements. Furthermore, mapping Zscan4c binding profile on MT2/MERVL consensus revealed the enrichment of Zscan4c binding only on MT2 region but not on MERVL-int (Figure 5I). However, expression of whole MT2/MERVL consensus was upregulated after Zscan4c overexpression (Figure 5I), implying that the Zscan4c may activate MERVL-int through binding to MT2 alone. The potential role of MT2 in activating Zscan4c-targeted 2-cell/4-cell embryo genes was exemplified by the case for Gm13697, from which MT2 located within 500 bp and it was activated after Zscan4c overexpression (Supplementary Figure S5I). Since the expression of MT2-nearby genes was activated after Zscan4c overexpression, we hypothesized that MT2 may act as an enhancer. We inserted MT2 to the downstream of Luciferase reporter gene and found a modest 2-fold upregulation of Luciferase activity after insertion of MT2 (Figure 5J), probably due to low expression of Zscan4 cluster genes in ESCs. The enhancer activity of MT2 was further elevated to 6-fold after Zscan4c overexpression (Figure 5J). Since there were three repeats of Zscan4c binding motifs on MT2, we deleted a large region (162–259 bp on MT2 consensus) containing all motifs to examine the role of these motifs. After deletion, MT2 enhancer activity was completely abolished (Figure 5K). Furthermore, inserting one copy of fused Zscan4c binding motif (TTAAAGGTGTGK) back to the mutant reporter completely rescued MT2 enhancer activity (Figure 5K), confirming that the Zscan4c binding motifs are critical to MT2 function. During embryogenesis, MT2 was also marked by enhancer histone mark H3K27ac (Supplementary Figure S5J), which peaked at 2-cell stage according to the analysis of published ChIP-seq data (56). The enhancer role of MT2 was further supported by the notion that expression of MT2-nearby genes was significantly higher in 2-cell embryos than in embryos of other stages (Supplementary Figure S5K). Taken together, these findings suggest that Zscan4c activates MT2/MERVL via direct binding to MT2 loci and activates 2-cell/4-cell embryo genes through regulating enhancer activity of MT2.
Figure 5.
Zscan4c interacts with MT2 and regulates nearby genes. (A) ChIP-qPCR analysis of Zscan4c binding on different retroelements. ChIP-qPCR data was normalized to input and that of LINE1. Biological triplicate data (n = 3 extracts) are presented as mean ± s.e.m. (B, C) Zscan4c binding profile plot (B) and enrichment heatmap (C) around the centre of MT2 sequence. The ChIP-seq signal was calculated as the ratio of normalized reads relative to input. (D) Zscan4c binding signal on MT2 and position of Zscan4c motif on MT2. Red sequences highlight the sequence of Zscan4c binding motif identified on MT2. (E) DNA pull-down analysis of the DNA binding activity of Zscan4c. DNA pull-down was done with 293T cells overexpressing HA-tagged Zscan4c. Probe with scrambled sequence was used as control. (F) MT2 nearby genes were enriched in upregulated genes after Zscan4c overexpression. Genes within 10 kb from MT2 locus were considered to be near MT2. Red, upregulated genes; blue, downregulated genes. (G) Overlap between upregulated genes after Zscan4c overexpression and MT2 10 kb nearby genes. P value was calculated by Fisher's exact test. (H) Expression heatmap of overlapped genes between upregulated genes after Zscan4c overexpression and genes within 10 kb from MT2. Troph, trophectoderm; ICM, inner cell mass. Average expression level of each developmental stage was determined and presented as Z-score across different stages. (I) Density plot of Zscan4c ChIP-seq signal and RNA-seq expression of MT2/MERVL consensus sequence from GeneBank (ID: Y12713.1). Density plot was plotted by IGV with ChIP-seq and RNA-seq bam files. Yellow region, ChIP-seq signal on MERVL-int; red region, ChIP-seq signal on MT2. ChIP input was included as control. Value of individual peak was highlighted. (J) Luciferase assay analysis of enhancer activity of MERVL LTR(MT2) after Zscan4c overexpression. MT2+, MT2 at the downstream of Luciferase gene; MT2–, reporter without MT2. Zscan4c OE +, Zscan4c overexpression; Zscan4c OE–, control vector overexpression. Biological-triplicate data (n = 3 extracts) are presented as mean ± s.e.m. (K) Luciferase assay analysis enhancer activity of wild type MT2, MT2 with mutant Zscan4c binding motif, and mutant MT2 with inserted Zscan4c binding motif after Zscan4c overexpression. Biological-triplicate data (n = 3 extracts) are presented as mean ± s.e.m.
Activation of MT2 is associated with augmented deposition of H3K4me1, H3K27ac, and H3K14ac on MT2
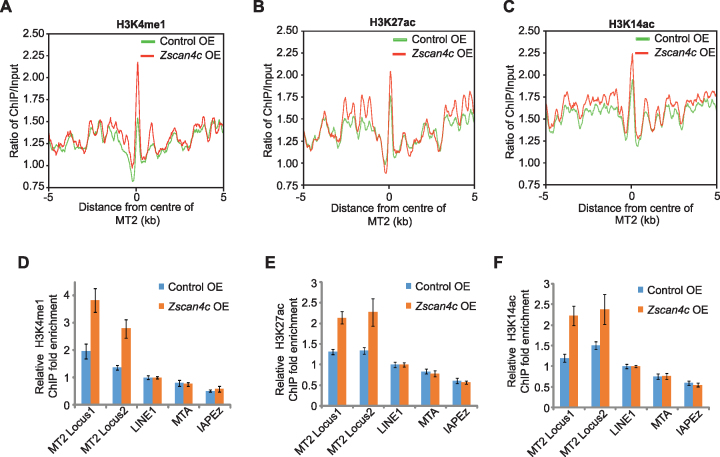
A previous study has shown that Zscan4c mediated gene expression change through degrading Uhrf1 and Dnmt1 proteins, and this subsequently led to the global DNA demethylation in ESCs (57,58). Hence, we checked whether MT2 was activated by Zscan4c through the same pathway. MeDIP results showed that there was indeed a decreased DNA methylation on MERVL after Zscan4c overexpression (Supplementary Figure S6A). Bisulphite sequencing confirmed the reduction of MT2 methylation after Zscan4c overexpression (Supplementary Figure S6B). However, Zscan4c could further promote MT2/MERVL expression after 5-Aza inhibition of DNA methylation in Zscan4c-overexpressed ESCs (Supplementary Figure S6C). These data suggest that Zscan4c may activate MT2 in a DNA-methylation independent manner as well. DNA methylation continuously decreased from zygote to ICM (29) (Supplementary Figure S6D) but chromatin at MT2 only became open at 2-cell/4-cell stage according to published ATAC-seq data (59) (Supplementary Figure S6E), suggesting the presence of factors other than DNA methylation in controlling MT2 activity during early embryogenesis. Enhancer regions are typically marked by H3K27ac, H3K4me1 and H3K14ac. Therefore, we asked whether the MT2 activation was associated with altered deposition of these histone marks on MT2. ChIP-seq of H3K27ac, H3K4me1 and H3K14ac revealed the increment of these histone marks on MT2 (Figure 6A–C). Our ChIP-qPCR results verified that H3K4me1, H3K27ac and H3K14ac were significantly enhanced on specific locus of MT2 (Figure 6D–F). We further examined whether the inhibition of H3K4me1 deposition affected MT2/MERVL expression. Knockout of H3K4me1 methyltransferase Mll3/Mll4 (54) resulted in reduced H3K4me1 deposition and abolished MT2/MERVL expression in ESCs (Supplementary Figure S6F–H). MT2-nearby regions were enriched of genes that were downregulated after the loss of H3K4me1 (Supplementary Figure S6I). The role of H3K27ac and H3K14ac was supported by the upregulation of MT2/MERVL after depletion of Hdac family members according to published RNA-seq data (60) (Supplementary Figure S6J). Together, these results demonstrate that MT2 activation after Zscan4c overexpression is associated with increased deposition of H3K4me1, H3K27ac and H3K14ac, which may contribute to the subsequent MT2 activation.
Figure 6.
Epigenetic regulation of MT2/MERVL after Zscan4c overexpression. (A–C) H3K4me1 (A), H3K27ac (B), and H3K14ac (C) profile plot around the centre of MT2 sequences in control ESCs (Control OE) and Zscan4c-overexpressed ESCs (Zscan4c OE). The ChIP-seq signal was calculated as the ratio of normalized reads relative to input. (D–F) ChIP-qPCR analysis of H3K4me1 (D), H3K27ac (E) and H3K14ac (F) enrichment on specific locus of MT2 in control ESCs and Zscan4c-overexpressed ESCs. LINE1, MTA and IAPEz were included as control. ChIP-qPCR data was normalized to input and that of LINE1. Biological-triplicate data (n = 3 extracts) are presented as mean ± s.e.m.
GBAF chromatin remodeling complex is important to MT2 activation in Zscan4c-overexpressed ESCs
Brg1 chromatin-remodeling complex, which is related to gene activation, was associated with Zscan4c (61). H3K4me1 also augments the recruitment of BAF complex to enhancers (62). Our co-immunoprecipitation experiment confirmed the interaction between Zscan4c and Brg1 in Zscan4c-overexpressed ESCs (Figure 7A). Recently, a non-canonical BAF complex GBAF complex was found to recognize H3K27ac and modulate enhancer activity (63,64). Thus, we speculated that the BAF complex that interacted with Zscan4c and regulated MT2 might be GBAF complex. We inspected the interaction of GBAF specific member Brd9 with Zscan4c by co-expressing HA-Brd9 and Flag-Zscan4c in 293T cells. Our co-immunoprecipitation results demonstrated that Brd9 interacted with Zscan4c (Supplementary Figure S7A). We could also confirm these results in ESCs (Figure 7A; Supplementary Figure S7B). We further investigated which domain of Zscan4c was implicated in the interaction between Zscan4c and Brd9/Brg1. We overexpressed HA-tagged individual domain of Zscan4c in ESCs and performed a co-immunoprecipitation experiment with HA antibody. Only SCAN domain of Zscan4c but not other domains of Zscan4c interacted with Brg1 and Brd9 (Figure 7B–D). In addition, Brd9 and Brg1 were weakly enriched at regions bound by Zscan4c (Figure 7E). However, more Brg1 and Brd9 were recruited to MT2 loci after Zscan4c overexpression (Figure 7F). Next, we examined whether MT2 enhancer activity and MERVL expression were affected by Brd9 activity. Inhibition of Brd9 activity by BI-7273 decreased Luciferase signal, demonstrating that Brd9 activity is important to MT2 enhancer activity (Figure 7G). Furthermore, inhibition of Brd9 by BI-7273 led to reduced expression of MERVL only in Zscan4c-overexpressed ESCs, but not in wild type ESCs (Supplementary Figure S7C). These results highlight an important role of Zscan4c-associated GBAF complex in activating MT2 enhancer activity.
Figure 7.
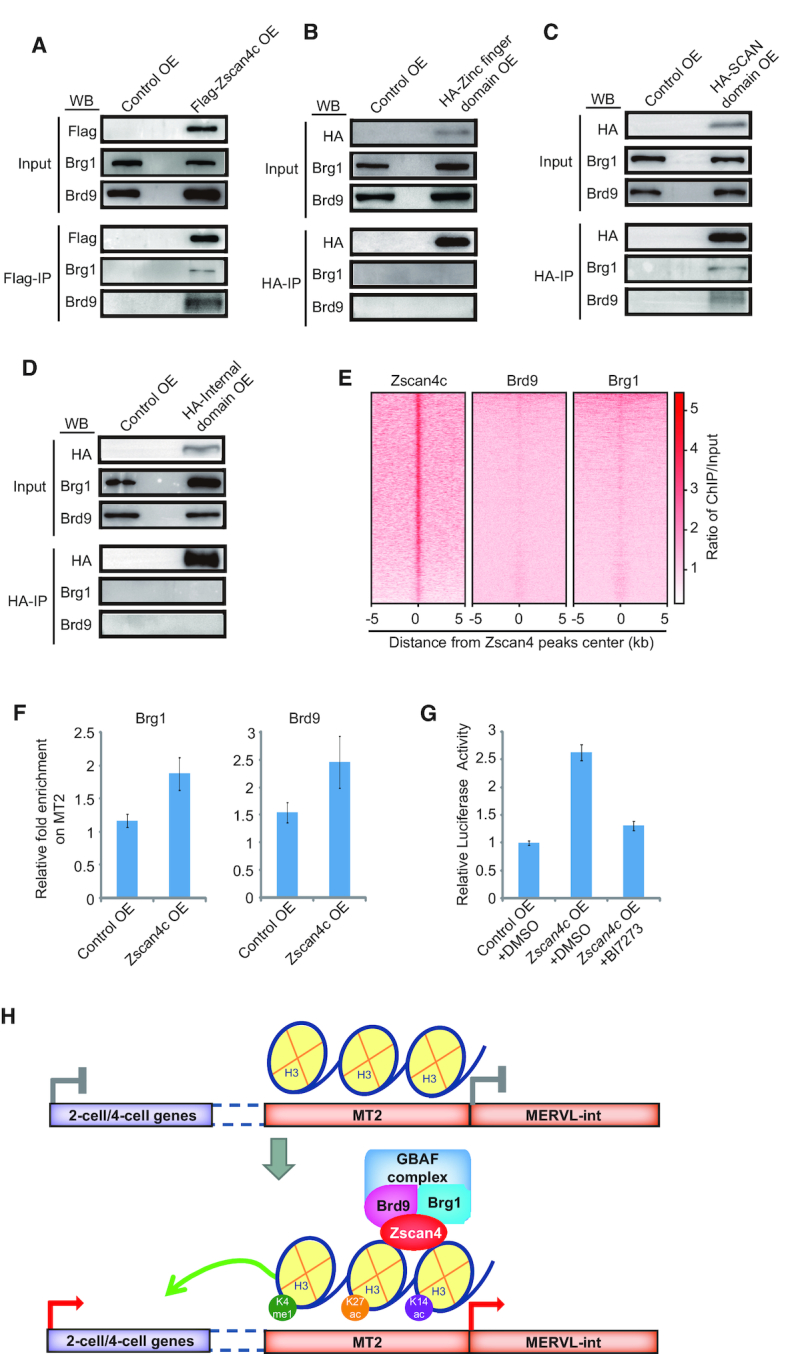
Role of Zscan4-associated GABF complex in MT2/MERVL activation. (A) Western blot analysis of Flag-Zscan4c/Brg1/Brd9 co-immunoprecipitation in Flag-Zscan4c overexpressed ESCs or in control vector overexpressed ESCs. IP, immunoprecipitation. IP was done with anti-Flag antibody. 1.25% input was loaded as control. (B–D) Western blot analysis of Brg1/Bd9 co-immunoprecipitation with HA-zinc finger domains (B), HA-SCAN domain (C), and HA-internal domain (D) of Zscan4c. IP was done with anti-HA antibody and separated by SDS/PAGE. 1.25% input was loaded as control. (E) Enrichment heatmap of Brd9 and Brg1 around the binding regions of Zscan4c. The ChIP-seq signal was calculated as the ratio of normalized reads relative to input. (F) ChIP-qPCR analysis of Brg1 and Brd9 enrichment on MT2 in control ESCs (Control OE) and Zscan4c-overexpressed (Zscan4c OE) ESCs. Biological triplicate data were presented as mean ± s.e.m. (G) Relative Luciferase activity of vector containing MT2 in control ESCs (Control OE) and Zscan4c-overexpressed (Zscan4c OE) ESCs treated with inhibitor of Brd9 (BI7273). DMSO treated samples were included as control. Biological triplicate data were presented as mean ± s.e.m. (H) Schematic plot of Zscan4c function in activating MT2/MERVL and 2-cell/4-cell cleavage genes. In the absence of Zscan4c, MT2 enhancer activity is not activated and MT2 nearby 2-cell/4-cell genes are silenced. In the presence of Zscan4c, Zscan4c binds to MT2, interacts with GBAF complex member Brg1 and Brd9, and augments H3K4me1, H3K27ac, and H3K14ac deposition on MT2/MERVL, thus activates MT2 which drives the expression of MERVL and enhances the expression of 2-cell/4-cell genes.
DISCUSSION
In summary, we proposed a model that activation of MT2/MERVL by Zscan4c is associated with augmented H3K4me1, H3K27ac and H3K14ac deposition on these loci and recruitment of GBAF complex (Figure 7H). Subsequently, Zscan4c-targeted MT2 enhancer regions further activate the expression of nearby genes in 2-cell/4-cell embryos (Figure 7H). Interestingly, ERVs demonstrate developmental stage-specific expression patterns (Figure 1D). In human, HERVK family member LTR14B is enriched at 2-cell stage while HERVL (MLT2A1) is enriched in 4-cell/8-cell embryos (23,65). However, in mouse, ERV3 (ERVL and ERVL-MaLR) members are enriched in 2-cell and 4-cell embryos (Figure 1D). These suggest that human and mouse express different groups of ERVs during early embryogenesis, which may have been exapted by the host for gene regulatory roles. It is also noticed that certain individual repeat loci demonstrated expression patterns different from those of other members in the same family, probably due to mutations and recombination during evolution. Previously, ERVs have been shown to function as enhancers and alternative promoters for protein coding genes (20,66). Therefore, the distinct expression pattern of ERVs should be mediated by the transcription regulatory network in each developmental stage. It was shown that Nanog mediated expression activation of LTR7 in human ESCs (23). Recently, it was found that KLF family transcription factors activated embryonic genome activation (EGA) and naive ESC specific enhancer ERVs during human embryogenesis (67). In counter of KLF transcription factor's action, KRAB-zinc finger proteins act to repress TE-derived enhancers and function as their tissue specific regulators (67). Dux expressed during EGA is important to activate EGA genes and promote MERVL expression in mouse 2-cell embryos (24–26). Similar to Dux, Zscan4c is also expressed in 2-cell/4-cell embryos (Figure 2A). However, the exact function of stage-specific expressed transcription factors in early embryogenesis and how they mediate ERV expression are less well understood. It is intriguing to look into how the stage-specific expressed transcription factors cooperate to mediate distinct expression pattern of ERVs during early embryogenesis.
The 2-cell/4-cell embryos represent a totipotent state which can generate an entire organism. However, the key activators of this state have yet to be found until recently when Dux and Dppa2/4 were discovered as central transcription regulators of zygotic activation genes in 2-cell embryos (24–26,51,68). LINE1 was also found to be critical to the global chromatin accessibility in early mouse embryos (27). Here, we identified an important role of Zscan4c and MT2/MERVL in regulating 2-cell/ 4-cell genes in ESCs. Previously, it was found that ESC developmental potency was enhanced by increasing Zscan4 activation frequency (69). Zscan4 also enhances reprogramming efficiency and facilitates the activation of pre-implantation genes (70). Our finding that Zscan4c activates 2-cell/4-cell embryo genes in ESCs is consistent with these results. However, we cannot exclude the possibility that Zscan4c may behave differently between ESCs and 2-cell/4-cell embryos. The role of Zscan4c may be different in 2-cell/4-cell embryos due to different developmental potency and chromatin state. Zscan4 is expressed at a much higher level in 2-cell/4-cell embryos than in ESCs (Figure 2A). The expression level of Zscan4 in 2-cell/4-cell embryos may be even higher than in Zscan4c-overexpressed ESCs (Figure 2A and 2E). In addition, the global epigenetic environment and gene expression are also different in 2-cell/4-cell embryos than in ESCs. Chromatin on MT2 is more open in 2-cell/4-cell embryos (Figure S6E) and other factors activating MT2/MERVL may also express highly in 2-cell/4-cell embryos. All these factors may influence MT2/MERVL expression. To further confirm the role of Zscan4c in early embryos, direct examination of Zscan4c binding on MT2 by ChIP-seq and single cell RNA-seq after Zscan4c depletion in 2-cell embryos will be helpful. It was found that knockout of Dux in early embryos only caused minor defects in EGA process and live mouse pups can still be born after Dux loss (71,72), although overexpression of Dux promotes 2-cell gene expression (24–26). In comparison to Dux, depletion of Zscan4 delays embryo development from 2-cell stage to 4-cell stage and ∼28% embryos completely failed to enter into 4-cell stage after Zscan4 suppression (73). Lack of Zscan4 also led to complete failure in blastocyst outgrowth and birth of live pups (73). These discoveries highlight the importance of Zscan4 family genes in early embryos. Since Dux, Dppa2/4 and Zscan4c all can activate the expression of MERVL and 2-cell/4-cell genes in ESCs (24–26,51,68,74), we speculate that multiple transcription factors, rather than a particular transcription factor, cooperate to mediate EGA process and safeguard the progress of early embryo development, although the extent of contribution to EGA may be different for each individual factor.
It was noteworthy that the population of MERVL-gag+ 2-cell like ESCs increased to ∼80% after Zscan4c overexpression (Figure 2F). This change after Zscan4c overexpression was accompanied by the alternation of ESC epigenetic states on MT2 (Figure 6A–C). Additionally, knockout of Mll3/4 led to the downregulation of MT2/MERVL (Supplementary Figure S6F–I), suggesting that H3K4me1 is central to MERVL expression. In addition, depletion of Hdac family members, which catalyze histone deacetylation, also resulted in increased MT2/MERVL expression (Supplementary Figure S6J), consistent with the finding that histone acetylation is also an important factor regulating expression of MT2/MERVL and 2-cell genes (75). These results reflected a direct participation of histone mark change in activating MT2/MERVL after Zscan4c overexpression. However, we cannot exclude the possibility that conversion of ESCs to 2C-like state may further strengthen the epigenetic change of MT2/MERVL region, given that the expression of other MT2/MERVL activators such as Dux and Dppa2/4 (24–26,51,74) were also upregulated after Zscan4c overexpression. These factors may, in turn, strengthen the action of Zscan4c on MT2.
Our DNA pull-down results supported the direct binding of Zscan4c on MT2 DNA (Figure 5E). Zinc finger domains of Zscan4c also demonstrate direct DNA binding activity (Supplementary Figure S5E) in regulating the expression of MT2/MERVL. Consistent with DNA pull-down results, ChIP-seq results revealed that DNA motif recognized by Zscan4c was located in the centre of the one Zscan4c peak on MT2 (Figure 5D) and this motif is important for Zscan4c to activate MT2 activity. The presence of a second Zscan4c peak implies that Zscan4c may recognize other motif(s) besides the ones we identified. These findings reinforced that direct binding on MT2 is important to the function of Zscan4c. Among the Zscan4c-activated genes, the upregulated expression of Dppa2/4, as well as Dux, were observed after Zscan4c overexpression (Supplementary Table S8). Dppa2/4 and Dux are also shown to activate Zscan4 and MT2/MERVL (24–26,51). However, Zscan4c does not directly bind to Dux promoter region (data not shown), suggesting an indirect activation of Dux through Dppa2/4 by Zscan4c. These data reflect that Zscan4c, Dppa2/4 and Dux may reinforce each other's expression and form a positive feedback loop to strengthen 2-cell like state of ESCs.
MT2/MERVL belongs to ERV3 (ERVL sub-family), whose silencing is mainly mediated by H3K9me2 and the protein complex responsible for its deposition (76). Zscan4c expression was shown to be related to global erasure of DNA methylation due to degradation of Uhrf1 and Dnmt1 whereas SCAN domain of Zscan4c is essential for the degradation (57,58). However, knockout of Dnmt1 in ESCs does not affect MERVL expression (77). In addition, zinc finger domains of Zscan4c can activate MT2/MERVL without SCAN domain (Figure 2G and H). These data imply that MERVL is not activated by Zscan4c mediated-global DNA methylation alone. In term of MERVL activation, we found that both enhancer epigenetic marks and GBAF complex were essential to activate MT2/MERVL expression. The BAF complex was also found to be important to H3K4me1 and H3K27ac deposition (63,64,78). Although zinc finger domains alone can activate MT2/MERVL expression, the impact of overexpressed zinc finger domains on MERVL expression was weaker than that of full-length Zscan4c (Figure 2E and H). These suggest that SCAN domain may further enhance the role of Zscan4c zinc fingers in activating MT2/MERVL and 2-cell/4-cell embryo genes. This is consistent with the fact that only SCAN domain of Zscan4c interacts with Brg1 and Brd9 (Figure 7B–D). The zinc finger domain of Zscan4c may activate MT2/MERVL through other mechanisms. One possible mechanism by which zinc finger domains may act through is activating Zscan4c expression, which may in turn activate MT2/MERVL. This is consistent with our finding that Zscan4c also binds to and activates its own gene (Figure 4E) (79). Hence, the epigenetic marks and Zscan4c-associated GBAF complex may work together to augment MT2/MERVL activity. In concordance with these data, histone deacetylation mediated by Pspc1 and Tet2 silences MERVL expression (80). These results support that multiple transcriptional and epigenetic regulatory pathways are required to work together to regulate ERVs.
Both MT2/MERVL and Zscan4c are expressed in 2-cell/4-cell cleavage stage embryos. It is intriguing to notice that Zscan4c is known to mediate telomere lengthening (81), which starts in cleavage stage embryos (82). Human DUX gene locates at the sub-telomeric region of chromosome 4q (83). The location of mouse Dux gene is still unclear. If mouse Dux also locates at the same region as human DUX, Zscan4c may regulate Dux through its action on telomere elongation. Hence, it will be necessary to confirm the detailed location of mouse Dux and investigate whether telomere also plays a role in regulating ERVs and EGA genes during development.
In conclusion, our study demonstrates that the expression of stage-specific ERV transcripts in mouse early embryos and Zscan4c activates enhancer activity of MERVL LTR MT2 to promote the expression of its nearby genes in 2-cell/4-cell embryos though mediating H3K27ac, H3K4me1, and H3K14ac deposition on MT2 and recruitment of GBAF complex. Future research will enable us to identify more regulators of ERVs and characterize the detailed mechanism through which these ERVs and regulators cooperate together in early development and diseases.
DATA AVAILABILITY
High-throughput sequencing data that generated from this study have been deposited in Gene Expression Omnibus under GSE120998 for RNA-seq and GSE125238 for ChIP-seq. Data analysed in this study are available in Gene Expression Omnibus under accession GSE53386 (28), GSE72784 (56), GSE108087 (60), GSE74055 (54), GSE90893 (55), GSE31284 (53), GSE66581 (59), GSE97778 (29) and GSE111264 (64).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Weiwei Zhai from the Genome Institute of Singapore for helpful discussions. We acknowledge Novogene and GENEWIZ China for their high-throughput sequencing services.
Author contributions: X.L., L.L. and W.Z. conceived the project; W.Z, Y.Z., J.G. and Y. S. preformed the bioinformatics analysis; F.C., R.C. and D.X. performed most experiments; J.Y. and R.G. established Zscan4c overexpression and knockdown cell lines; X.Z. did inhibitor treatment and flow cytometry analysis; X.L., L.L. and W.Z. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2018YFA0107000]; National Natural Science Foundation of China [31671352 to X.L., 31430052, 31571546 to L.L.]; Natural Science Foundation of Tianjin City [15JCZDJC65600 to X.L.]; Fundamental Research Funds for the Central Universities and the National Thousand Young Talents Plan.
Conflict of interest statement. None declared.
REFERENCES
- 1. Stocking C., Kozak C.A.. Murine endogenous retroviruses. Cell Mol. Life Sci. 2008; 65:3383–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burns K.H. Transposable elements in cancer. Nat. Rev. Cancer. 2017; 17:415–424. [DOI] [PubMed] [Google Scholar]
- 3. Babaian A., Mager D.L.. Endogenous retroviral promoter exaptation in human cancer. Mob. DNA. 2016; 7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brattas P.L., Jonsson M.E., Fasching L., Nelander Wahlestedt J., Shahsavani M., Falk R., Falk A., Jern P., Parmar M., Jakobsson J.. TRIM28 controls a gene regulatory network based on endogenous retroviruses in human neural progenitor cells. Cell Rep. 2017; 18:1–11. [DOI] [PubMed] [Google Scholar]
- 5. Gautam P., Yu T., Loh Y.H.. Regulation of ERVs in pluripotent stem cells and reprogramming. Curr. Opin. Genet. Dev. 2017; 46:194–201. [DOI] [PubMed] [Google Scholar]
- 6. Robbez-Masson L., Rowe H.M.. Retrotransposons shape species-specific embryonic stem cell gene expression. Retrovirology. 2015; 12:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang P., Wang Y., Macfarlan T.S.. The role of KRAB-ZFPs in transposable element repression and mammalian evolution. Trends Genet. 2017; 33:871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karimi M.M., Goyal P., Maksakova I.A., Bilenky M., Leung D., Tang J.X., Shinkai Y., Mager D.L., Jones S., Hirst M. et al.. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011; 8:676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He Q., Kim H., Huang R., Lu W., Tang M., Shi F., Yang D., Zhang X., Huang J., Liu D. et al.. The Daxx/Atrx complex protects tandem repetitive elements during dna hypomethylation by promoting H3K9 trimethylation. Cell Stem Cell. 2015; 17:273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leung D., Du T., Wagner U., Xie W., Lee A.Y., Goyal P., Li Y., Szulwach K.E., Jin P., Lorincz M.C. et al.. Regulation of DNA methylation turnover at LTR retrotransposons and imprinted loci by the histone methyltransferase Setdb1. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:6690–6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Imbeault M., Helleboid P.Y., Trono D.. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. 2017; 543:550–554. [DOI] [PubMed] [Google Scholar]
- 12. Elsasser S.J., Noh K.M., Diaz N., Allis C.D., Banaszynski L.A.. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015; 522:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collins P.L., Kyle K.E., Egawa T., Shinkai Y., Oltz E.M.. The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:8367–8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maksakova I.A., Thompson P.J., Goyal P., Jones S.J., Singh P.B., Karimi M.M., Lorincz M.C.. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenet. Chromatin. 2013; 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang B.X., El Farran C.A., Guo H.C., Yu T., Fang H.T., Wang H.F., Seah Y.F., Goh G.Y., Neo S.P. et al.. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell. 2015; 163:230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Macfarlan T.S., Gifford W.D., Agarwal S., Driscoll S., Lettieri K., Wang J., Andrews S.E., Franco L., Rosenfeld M.G., Ren B. et al.. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011; 25:594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L.. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012; 487:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bulut-Karslioglu A., De La Rosa-Velazquez I.A., Ramirez F., Barenboim M., Onishi-Seebacher M., Arand J., Galan C., Winter G.E., Engist B., Gerle B. et al.. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell. 2014; 55:277–290. [DOI] [PubMed] [Google Scholar]
- 19. Kunarso G., Chia N.Y., Jeyakani J., Hwang C., Lu X., Chan Y.S., Ng H.H., Bourque G.. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010; 42:631–634. [DOI] [PubMed] [Google Scholar]
- 20. Wang J., Xie G., Singh M., Ghanbarian A.T., Rasko T., Szvetnik A., Cai H., Besser D., Prigione A., Fuchs N.V. et al.. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014; 516:405–409. [DOI] [PubMed] [Google Scholar]
- 21. Rebollo R., Miceli-Royer K., Zhang Y., Farivar S., Gagnier L., Mager D.L.. Epigenetic interplay between mouse endogenous retroviruses and host genes. Genome Biol. 2012; 13:R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grow E.J., Flynn R.A., Chavez S.L., Bayless N.L., Wossidlo M., Wesche D.J., Martin L., Ware C.B., Blish C.A., Chang H.Y. et al.. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015; 522:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goke J., Lu X., Chan Y.S., Ng H.H., Ly L.H., Sachs F., Szczerbinska I.. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015; 16:135–141. [DOI] [PubMed] [Google Scholar]
- 24. Whiddon J.L., Langford A.T., Wong C.J., Zhong J.W., Tapscott S.J.. Conservation and innovation in the DUX4-family gene network. Nat. Genet. 2017; 49:935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hendrickson P.G., Dorais J.A., Grow E.J., Whiddon J.L., Lim J.W., Wike C.L., Weaver B.D., Pflueger C., Emery B.R., Wilcox A.L. et al.. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017; 49:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Iaco A., Planet E., Coluccio A., Verp S., Duc J., Trono D.. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017; 49:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jachowicz J.W., Bing X., Pontabry J., Boskovic A., Rando O.J., Torres-Padilla M.E.. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 2017; 49:1502–1510. [DOI] [PubMed] [Google Scholar]
- 28. Fan X., Zhang X., Wu X., Guo H., Hu Y., Tang F., Huang Y.. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015; 16:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang C., Liu X., Gao Y., Yang L., Li C., Liu W., Chen C., Kou X., Zhao Y., Chen J. et al.. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 2018; 20:620–631. [DOI] [PubMed] [Google Scholar]
- 30. Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J. et al.. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006; 38:431–440. [DOI] [PubMed] [Google Scholar]
- 31. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011; 17:3. [Google Scholar]
- 32. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T.. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014; 42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lindner I., Rubin D., Helwig U., Nitz I., Hampe J., Schreiber S., Schrezenmeir J., Doring F.. The L513S polymorphism in medium-chain acyl-CoA synthetase 2 (MACS2) is associated with risk factors of the metabolic syndrome in a Caucasian study population. Mol. Nutr. Food Res. 2006; 50:270–274. [DOI] [PubMed] [Google Scholar]
- 35. Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics. 2004; doi:10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 36. Bedell J.A., Korf I., Gish W.. MaskerAid: a performance enhancement to RepeatMasker. Bioinformatics. 2000; 16:1040–1041. [DOI] [PubMed] [Google Scholar]
- 37. Machanick P., Bailey T.L.. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011; 27:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang S., Sun H., Ma J., Zang C., Wang C., Wang J., Tang Q., Meyer C.A., Zhang Y., Liu X.S.. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc. 2013; 8:2502–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benit L., De Parseval N., Casella J.F., Callebaut I., Cordonnier A., Heidmann T.. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol. 1997; 71:5652–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hubley R., Finn R.D., Clements J., Eddy S.R., Jones T.A., Bao W., Smit A.F., Wheeler T.J.. The Dfam database of repetitive DNA families. Nucleic Acids Res. 2016; 44:D81–D89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krueger F., Andrews S.R.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang R.H., Yu H., Deng C.X.. A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:17108–17113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 46. Jin Y., Tam O.H., Paniagua E., Hammell M.. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics. 2015; 31:3593–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Langfelder P., Horvath S.. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008; 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 49. Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C. et al.. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007; 35:W169–W175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Eckersley-Maslin M., Alda-Catalinas C., Blotenburg M., Kreibich E., Krueger C., Reik W.. Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev. 2019; 33:194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rodriguez-Terrones D., Gaume X., Ishiuchi T., Weiss A., Kopp A., Kruse K., Penning A., Vaquerizas J.M., Brino L., Torres-Padilla M.E.. A molecular roadmap for the emergence of early-embryonic-like cells in culture. Nat. Genet. 2018; 50:106–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Karmodiya K., Krebs A.R., Oulad-Abdelghani M., Kimura H., Tora L.. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012; 13:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yan J., Chen S.A., Local A., Liu T., Qiu Y., Dorighi K.M., Preissl S., Rivera C.M., Wang C., Ye Z. et al.. Histone H3 lysine 4 monomethylation modulates long-range chromatin interactions at enhancers. Cell Res. 2018; 28:204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J., Plath K.. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017; 168:442–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dahl J.A., Jung I., Aanes H., Greggains G.D., Manaf A., Lerdrup M., Li G., Kuan S., Li B., Lee A.Y. et al.. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016; 537:548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dan J., Rousseau P., Hardikar S., Veland N., Wong J., Autexier C., Chen T.. Zscan4 inhibits maintenance DNA methylation to facilitate telomere elongation in mouse embryonic stem cells. Cell Rep. 2017; 20:1936–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eckersley-Maslin M.A., Svensson V., Krueger C., Stubbs T.M., Giehr P., Krueger F., Miragaia R.J., Kyriakopoulos C., Berrens R.V., Milagre I. et al.. MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs. Cell Rep. 2016; 17:179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu J., Huang B., Chen H., Yin Q., Liu Y., Xiang Y., Zhang B., Liu B., Wang Q., Xia W. et al.. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016; 534:652–657. [DOI] [PubMed] [Google Scholar]
- 60. He J., Fu X., Zhang M., He F., Li W., Abdul M.M., Zhou J., Sun L., Chang C., Li Y. et al.. Transposable elements are regulated by context-specific patterns of chromatin marks in mouse embryonic stem cells. Nat. Commun. 2019; 10:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Akiyama T., Xin L., Oda M., Sharov A.A., Amano M., Piao Y., Cadet J.S., Dudekula D.B., Qian Y., Wang W. et al.. Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res. 2015; 22:307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Local A., Huang H., Albuquerque C.P., Singh N., Lee A.Y., Wang W., Wang C., Hsia J.E., Shiau A.K., Ge K. et al.. Identification of H3K4me1-associated proteins at mammalian enhancers. Nat. Genet. 2018; 50:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jefimov K., Alcaraz N., Kloet S., Varv S., Aastedatter Sakya S., Dalager Vaagenso C., Vermeulen M., Aasland R., Andersson R.. The GBAF chromatin remodeling complex binds H3K27ac and mediates enhancer transcription. 2018; 10.1101/445148, 17 October 2018, preprint: not peer reviewed. [DOI]
- 64. Gatchalian J., Malik S., Ho J., Lee D.S., Kelso T.W.R., Shokhirev M.N., Dixon J.R., Hargreaves D.C.. A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat. Commun. 2018; 9:5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu L., Leng L., Liu C., Lu C., Yuan Y., Wu L., Gong F., Zhang S., Wei X., Wang M. et al.. An integrated chromatin accessibility and transcriptome landscape of human pre-implantation embryos. Nat. Commun. 2019; 10:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fort A., Hashimoto K., Yamada D., Salimullah M., Keya C.A., Saxena A., Bonetti A., Voineagu I., Bertin N., Kratz A. et al.. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat. Genet. 2014; 46:558–566. [DOI] [PubMed] [Google Scholar]
- 67. Pontis J., Planet E., Offner S., Turelli P., Duc J., Coudray A., Theunissen T.W., Jaenisch R., Trono D.. Hominoid-specific transposable elements and KZFPs facilitate human embryonic genome activation and control transcription in naive human ESCs. Cell Stem Cell. 2019; 24:724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yan Y.L., Zhang C., Hao J., Wang X.L., Ming J., Mi L., Na J., Hu X., Wang Y.. DPPA2/4 and SUMO E3 ligase PIAS4 opposingly regulate zygotic transcriptional program. PLoS Biol. 2019; 17:e3000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Amano T., Hirata T., Falco G., Monti M., Sharova L.V., Amano M., Sheer S., Hoang H.G., Piao Y., Stagg C.A. et al.. Zscan4 restores the developmental potency of embryonic stem cells. Nat. Commun. 2013; 4:1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hirata T., Amano T., Nakatake Y., Amano M., Piao Y., Hoang H.G., Ko M.S.. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci. Rep. 2012; 2:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen Z., Zhang Y.. Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat. Genet. 2019; 51:947–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Iaco A.D., Verp S., Offner S., Trono D.. DUX is a non-essential synchronizer of zygotic genome activation. 2019; 10.1101/569434, 06 March 2019, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- 73. Falco G., Lee S.L., Stanghellini I., Bassey U.C., Hamatani T., Ko M.S.. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 2007; 307:539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. De Iaco A., Coudray A., Duc J., Trono D.. DPPA2 and DPPA4 are necessary to establish a 2C-like state in mouse embryonic stem cells. EMBO Rep. 2019; 20:e47382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dan J., Yang J., Liu Y., Xiao A., Liu L.. Roles for histone acetylation in regulation of telomere elongation and two-cell state in mouse ES cells. J. Cell Physiol. 2015; 230:2337–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Leung D.C., Dong K.B., Maksakova I.A., Goyal P., Appanah R., Lee S., Tachibana M., Shinkai Y., Lehnertz B., Mager D.L. et al.. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:5718–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sharif J., Endo T.A., Nakayama M., Karimi M.M., Shimada M., Katsuyama K., Goyal P., Brind’Amour J., Sun M.A., Sun Z. et al.. Activation of endogenous retroviruses in dnmt1(-/-) ESCs involves disruption of SETDB1-mediated repression by NP95 binding to hemimethylated DNA. Cell Stem Cell. 2016; 19:81–94. [DOI] [PubMed] [Google Scholar]
- 78. Alver B.H., Kim K.H., Lu P., Wang X., Manchester H.E., Wang W., Haswell J.R., Park P.J., Roberts C.W.. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat. Commun. 2017; 8:14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dan J., Liu Y., Liu N., Chiourea M., Okuka M., Wu T., Ye X., Mou C., Wang L., Yin Y. et al.. Rif1 maintains telomere length homeostasis of ESCs by mediating heterochromatin silencing. Dev. Cell. 2014; 29:7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Guallar D., Bi X., Pardavila J.A., Huang X., Saenz C., Shi X., Zhou H., Faiola F., Ding J., Haruehanroengra P. et al.. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet. 2018; 50:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zalzman M., Falco G., Sharova L.V., Nishiyama A., Thomas M., Lee S.L., Stagg C.A., Hoang H.G., Yang H.T., Indig F.E. et al.. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010; 464:858–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu L., Bailey S.M., Okuka M., Munoz P., Li C., Zhou L., Wu C., Czerwiec E., Sandler L., Seyfang A. et al.. Telomere lengthening early in development. Nat. Cell Biol. 2007; 9:1436–1441. [DOI] [PubMed] [Google Scholar]
- 83. Leidenroth A., Clapp J., Mitchell L.M., Coneyworth D., Dearden F.L., Iannuzzi L., Hewitt J.E.. Evolution of DUX gene macrosatellites in placental mammals. Chromosoma. 2012; 121:489–497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput sequencing data that generated from this study have been deposited in Gene Expression Omnibus under GSE120998 for RNA-seq and GSE125238 for ChIP-seq. Data analysed in this study are available in Gene Expression Omnibus under accession GSE53386 (28), GSE72784 (56), GSE108087 (60), GSE74055 (54), GSE90893 (55), GSE31284 (53), GSE66581 (59), GSE97778 (29) and GSE111264 (64).