Abstract
TFBSshape (https://tfbsshape.usc.edu) is a motif database for analyzing structural profiles of transcription factor binding sites (TFBSs). The main rationale for this database is to be able to derive mechanistic insights in protein–DNA readout modes from sequencing data without available structures. We extended the quantity and dimensionality of TFBSshape, from mostly in vitro to in vivo binding and from unmethylated to methylated DNA. This new release of TFBSshape improves its functionality and launches a responsive and user-friendly web interface for easy access to the data. The current expansion includes new entries from the most recent collections of transcription factors (TFs) from the JASPAR and UniPROBE databases, methylated TFBSs derived from in vitro high-throughput EpiSELEX-seq binding assays and in vivo methylated TFBSs from the MeDReaders database. TFBSshape content has increased to 2428 structural profiles for 1900 TFs from 39 different species. The structural profiles for each TFBS entry now include 13 shape features and minor groove electrostatic potential for standard DNA and four shape features for methylated DNA. We improved the flexibility and accuracy for the shape-based alignment of TFBSs and designed new tools to compare methylated and unmethylated structural profiles of TFs and methods to derive DNA shape-preserving nucleotide mutations in TFBSs.
INTRODUCTION
A mechanistic understanding of transcriptional regulation and other cellular processes requires a structural characterization of transcription factor (TF)–DNA binding properties. TF–DNA binding preferences are commonly described as consensus sequence represented by a position weight matrix (PWM) (1,2) and visualized as motif logo (3,4). Traditional PWM-based methods assume that each nucleotide independently contributes to TF–DNA binding; however, this does not hold generally for every DNA-binding protein (5–7). One way of encoding interdependencies between nucleotide positions is k-mer models (8,9) with dinucleotide containing PWMs being the simplest description that cover exclusively interactions between adjacent base pairs (10,11). An alternative representation of interdependencies between base pairs is the three-dimensional (3D) DNA structure (12,13), resulting from physical interactions such as inter-base pair stacking and other interactions between nucleotide positions within a TF binding site (TFBS).
Several studies demonstrated that the readout of 3D DNA structure is an important component of the binding specificity of TFs (14–17) and downstream gene expression (18). In some cases, TFs recognize low-affinity binding sites with less pronounced sequence motifs (19,20) or tend to bind to DNA even in the absence of the sequence motif both in vitro (21) and in vivo (22). The initial version of TFBSshape (23) characterized the structural profile of binding sites using four DNA shape features, including helix twist (HelT), minor groove width (MGW), propeller twist (ProT) and Roll, all of which are considered important structural properties for TF–DNA readout mechanisms (12,24). However, since the original publication of TFBSshape (23), we have derived nine additional DNA shape features (25) and one biophysical feature, namely minor groove electrostatic potential (EP) (26), extending the mechanistic description of TF–DNA recognition (13,25,27). The new release of TFBSshape provides 14 DNA feature profiles for each dataset of TFs, comprised of 13 shape features and EP. The DNA shape features include six inter-base pair parameters (HelT, Rise, Roll, Shift, Slide and Tilt), six intra-base pair parameters (Buckle, Opening, ProT, Shear, Stagger and Stretch) and MGW.
Emerging evidence reveals that the DNA binding of some TFs is sensitive to DNA methylation at TFBSs both in vitro and in vivo (28–34). Several studies show that aberrant methylation patterns on DNA lead to human disease and cancer (35,36). CpG methylation is the most frequent DNA modification, where a methyl group is added at the major groove edge of the cytosine base. This not only changes the chemical signature of C/G base pairs but also alters the DNA structure by slightly widening the major groove due to the addition of a bulky methyl group and, in turn, narrowing the minor groove (36). We observed that such structural effects of DNA methylation are dependent on the sequence context (37). Methylation-induced changes in 3D DNA structure were found to explain the methylation-dependent cleavage rate of DNase I (38) and binding affinity of human Pbx–Hox heterodimers (37). Thus, it becomes essential to understand the structural readout mechanisms underlying the recognition through DNA shape changes due to CpG methylation. The new release of TFBSshape provides four shape features (HelT, MGW, ProT and Roll) for the structural profiles of methylated TFBSs derived from an in vitro high-throughput binding assay, EpiSELEX-seq (28), and a motif database for methylated TFBSs, MeDReaders (39). EpiSELEX-seq probes the sensitivity of TF binding to methylated DNA. MeDReaders integrates whole genome bisulfite sequencing (WGBS) and ChIP-seq data in multiple cell lines.
An increasing number of studies have demonstrated that mechanisms for TF recognition of specific DNA sequences involve an interplay between DNA base and shape readout (40). To independently probe the importance of these readout mechanisms, researchers either design mutations of the cognate binding sites or disrupt certain DNA shape signatures (41,42). The current TFBSshape release introduces a new tool for designing novel binding sites, by preserving either DNA shape or nucleotide sequence while varying the other feature group, which can provide systematic mutation design for downstream experiments. This release also provides a new shape alignment tool to align structural profiles based on ensemble binding sites for the investigation of possible shape readout mechanisms.
DATABASE EXPANSIONS
Increased collection of DNA structural profiles for unmethylated DNA
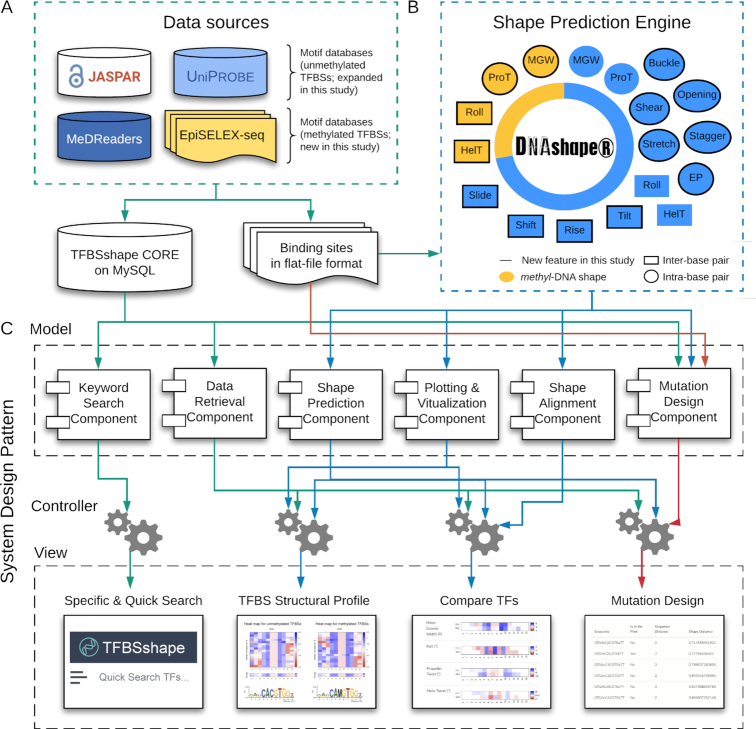
The TFBSshape database provides DNA structural profiles for the TFBS sequences collected from various data sources (Figure 1A). For the current version of the TFBSshape database, we added and updated the collection of DNA structural profiles based on the latest TFBS sequences obtained from JASPAR (43) and UniPROBE (44), the two main databases incorporated in the original version of TFBSshape. New content covers 1243 structural profiles for 1091 TFs in JASPAR (a 235% increase) and 886 structural profiles for 627 TFs in UniPROBE (a 141% increase) (Table 1). The current version of TFBSshape has been updated with the latest version of JASPAR 2020 (43) and with the continuously updated UniPROBE database (http://thebrain.bwh.harvard.edu/uniprobe/).
Figure 1.
Schematic overview of the architecture and key functionality of TFBSshape. (A) TFBSshape CORE collects TF information derived from two motif databases for unmethylated DNA, JASPAR (43) and UniPROBE (44), a database for in vivo binding to methylated DNA, MeDReaders (39), and a dataset obtained from the high-throughput binding assay, EpiSELEX-seq (28). The corresponding TFBS sequences were extracted from the aforementioned data sources and stored in flat-file format. (B) TFBSshape uses DNAshapeR (45) as a DNA shape prediction engine to generate structural profiles of TFBSs. The current version of TFBSshape predicts and analyzes 18 DNA features, including 14 features for unmethylated DNA (inter-base pair parameters HelT, Rise, Roll, Shift, Slide and Tilt, intra-base pair parameters Buckle, Opening, ProT, Shear, Stagger and Stretch, MGW and EP) and four features for methylated DNA (HelT, MGW, ProT and Roll). Among these features, 14 of them were added in the current version of TFBSshape. (C) We implemented the TFBSshape interface with a Model–View–Controller architectural pattern. The Model layer consists of multiple reusable and extendable components that are responsible for specific tasks such as searching and retrieving information from the database, performing DNA shape predictions, calculating similarities between vectors and generating statistical plots. The Controller layer handles business logics and prepares necessary data by calling the Model components to respond to the user's need. The View layer renders the final web page with the prediction results and presents them to the user through the browser. The Model and View layers are independent; thus, changes to one layer will not affect the functionality of the other layer.
Table 1.
Overview of the number of TFBS and TF datasets in the current version of the TFBSshape database
| TFBS datasets | TF datasets | |||
|---|---|---|---|---|
| Database sources | Original Version | Current Version | Original Version | Current Version |
| JASPAR | 371 | 1243 | 371 | 1091 |
| UniPROBE | 368 | 886 | 361 | 627 |
| MeDReaders | NA | 292 | NA | 175 |
| EpiSELEX-seq | NA | 7 | NA | 7 |
| Total | 739 | 2428 | 732 | 1900 |
Integration of motif databases for methylated DNA
In this release, we have expanded the dimensionality of the TFBSshape database by providing DNA structural profiles for methylated TFBSs obtained from EpiSELEX-seq binding experiments (28) and the recently published MeDReaders database for methylated motifs (39) (Figure 1A). EpiSELEX-seq probes the sensitivity of TF binding to DNA with 5-methylcytosine (5mC) in vitro using massively parallel sequencing (28). This method has investigated seven binding profiles for six TFs, including three human bZIP proteins and three human Pbx–Hox complexes. MeDReaders applied in silico approaches to predict methylated and unmethylated motifs of 175 TFs by incorporating WGBS and ChIP-seq datasets, providing unified access to most TFs that involved methylation-associated binding events in vivo (39). We analyzed 292 structural profiles of these 175 TFs for TFBSs with both high and low methylation levels of CpG sites derived from MeDReaders, which includes six human cell lines/tissues and one mouse cell line/tissue (Table 1).
Summary of total data collection
TFBSshape now provides DNA structural profiles for TFBSs from four data sources (JASPAR, UniPROBE, EpiSELEX-seq and MeDReaders), rather than just JASPAR and UniPROBE as previously used, and contains unmethylated and methylated DNA involved in in vitro and in vivo binding. In total, the current version of the TFBSshape database holds 2428 DNA structural profiles for 1900 TFs from 39 different species, representing a 229% increase compared to its original version (Table 1).
Additional shape and biophysical features for unmethylated DNA
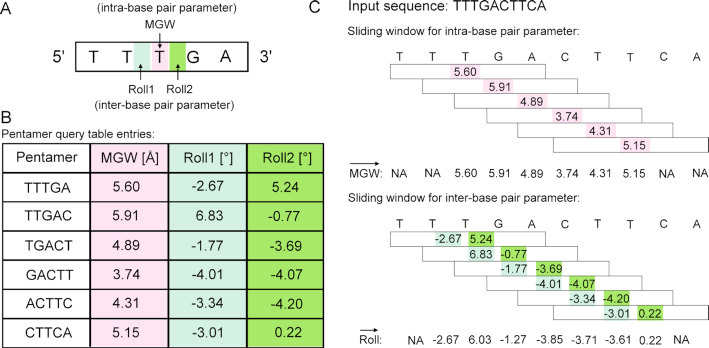
TFBSshape calculates DNA shape features for qualitative and quantitative analysis to improve the mechanistic understanding of TF–DNA recognition. The current version of TFBSshape analyzes nine additional shape features and one biophysical feature of DNA for each set of TFBS sequences, expanding the original set of four DNA shape features to a total set of 14 features, including six intra-base pair features (Buckle, Opening, ProT, Shear, Stagger and Stretch), six inter-base pair features (HelT, Rise, Roll, Shift, Slide and Tilt), MGW and EP. The feature values were predicted using our R/Bioconductor package DNAshapeR (45), where DNA shape features are derived from data mining of trajectories from all-atom Monte Carlo (MC) simulations for DNA fragments of different nucleotide sequences ranging 12–27 base pairs in length capturing all 512 unique pentamers in diverse sequence contexts (24). These MC simulations used a set of collective and internal variables (46) and the AMBER force field (47) for DNA fragments, explicit sodium counter ions (48) and a distant-dependent sigmoidal function to describe the solvent implicitly (49). Average values for each shape feature assigned to each of the 512 unique pentamers were calculated from equilibrated MC simulation trajectories using the Curves algorithm (50) and compiled into a pentamer query table for high-throughput prediction (24) (Figure 2). These DNA shape predictions have previously been validated using available experimental structures and hydroxyl radical cleavage measurements (24,51). EP values were calculated at the center of the minor groove within each approximate base-pair plane by solving the nonlinear Poisson–Boltzmann equation at physiological ionic strength using the DelPhi program (52) and a previously described protocol (15) for average DNA structures originating from MC simulations (26) (Figure 1B). The current version of TFBSshape provides qualitative illustrations for the 14 features of unmethylated DNA in heat maps with the option for downloading quantitative data for further analysis.
Figure 2.
Schematic illustration of the pentamer model for high-throughput prediction of DNA shape. (A) A pentamer model was used to characterize and predict DNA shape features for either one intra-base pair parameter (e.g. MGW, in pink) or two inter-base pair parameters (e.g. Roll, in light and dark green). The intra-base pair parameter specifies the relative location of the bases within a base pair, or in the case of MGW is defined with respect to a base-pair plane, while the inter-base pair parameter indicates the relative location of two adjacent base pairs, or refers to a base-pair step (24). (B) A sliding-pentamer window was used to mine the prediction results from MC simulations and, in turn, generate a query table of average DNA shape features of each pentamer (45). (C) The pentamer query table integrated with a sliding-pentamer window can be used to predict shape features for a given DNA sequence of any length in a high-throughput manner. For predicting intra-base pair parameters (e.g. MGW), each sliding step assigns a shape prediction for the central base pair. For predicting inter-base pair parameters (e.g. Roll), each sliding step assigns a shape prediction for two central base-pair steps. The overlapping values arising from two adjacent pentamers at the same nucleotide position will then be averaged. The sliding-window approach will result in a feature vector (12).
Introduction of shape features for methylated DNA
TFBSshape provides DNA shape features for DNA sequences that contain CpG dinucleotides, and the new release offers an alternative approach to determine how the intrinsic shape of methylated DNA affects TF binding. Recently, we developed a high-throughput method, methyl-DNAshape (37), for predicting the shape features of methylated DNA, including HelT, MGW, ProT and Roll. The current version of TFBSshape uses methyl-DNAshape (37) for deriving shape profiles for methylated DNA sequences. The methyl-DNAshape approach (37) uses MC simulations of DNA fragments with methylated CpG dinucleotides embedded in diverse sequence contexts. The MC simulation protocol is identical to the one described for unmethylated DNA fragments with the exception of 5-methylcytosine replacing cytosine in all occurring CpG base-pair steps. The methyl-DNAshape predictions were previously validated based on available experimental structures with CpG methylation (37). The MC simulations and limited data for validation restrict the current approach to CpG methylation despite cytosine methylation in other sequence contexts in plants (53) and at CpA dinucleotides in neurons (54,55).
The nucleotide sequences for which the new edition of TFBSshape provides CpG methylated shape profiles were obtained from MeDReaders (39) and EpiSELEX-seq data (28), which are represented qualitatively as heat maps and available as quantitative data for downloading. Since EpiSELEX-seq datasets provide paired methylated and unmethylated TFBS sequences, we were able to compare shape changes between methylated and unmethylated TFBSs directly from the binding data. For those datasets that only contain unmethylated TFBSs, such as JASPAR (43) and UniPROBE (44), we performed in silico CpG methylation on unmethylated DNA and predicted shape features of in silico methylated DNA fragments. Similarly, we compared the shape changes between these unmethylated and in silico methylated TFBS sequences, aiming to provide insights regarding the effects of methylated DNA on the binding of a TF even though in vitro or in vivo assays for this effect are unavailable.
NEW DATABASE FEATURES
Illustration and comparison of DNA shape profiles for individual TF dataset
TFBSshape now provides two search functions. Similar to the first release of TFBSshape, the user can specify the search criteria for four individual databases by selecting ‘Search TFs’ in the navigation bar on the left of the web page. In the current version of TFBSshape, a quick search bar can be found on the top of each page of the TFBSshape interface, allowing the user to search for any TF of interest across all four databases (Figure 3A and B). When expanding the details of the selected TF, the resulting web page displays background information about the selected TF on the upper panel and 14 shape feature profiles in the sliding menu on the lower panel (Figure 3C).
Figure 3.
Overview of the new TFBSshape web interface for displaying TFBS structural profiles. (A) The user can filter the TF dataset(s) of interest through the quick or advanced search functions. (B) The responsive table lists the search results. (C) The resulting page consists of background information and 14 DNA feature profiles displayed in the sliding menu for the structural profile for MAX TFBS sequences derived from UniPROBE (UP00060). (D) The MGW section comprises three data columns for unmethylated TFBS sequences, methylated TFBS sequences and the comparison of those two sets of sequences. Each of the first two columns contains three illustrations, including a heat map demonstrating predicted MGW feature profiles for individual sequences, an average heat map for all sequences and a DNA logo representing the PWM calculated using the WebLogo tool (4). The third column displays the differences in MGW distributions between unmethylated and methylated TFBSs with respect to each nucleotide position (ΔMGW). The difference between two MGW distributions in the center of the binding site is significant based on a one-sample statistical t-test.
Each shape feature profile consists of three data columns for unmethylated DNA, methylated DNA and a comparison of the results from the first two columns (Figure 3D). The first two data columns contain three illustrations, including shape feature heat maps for each individual sequence, average heat maps for each shape parameter and the motif logo representing the PWM calculated using TFBS sequence information. The third column demonstrates the comparison by presenting box plots along with a statistical one-sample t-test to compare the mean of the shape changes, for example ΔMGW, between the sets of unmethylated and methylated TFBS sequences. This comparison determines whether the shape profile is significantly altered when introducing 5mC methylation at CpG dinucleotides (Figure 3D).
The illustrations of the first data column are derived from the unmethylated TFBS sequences from JASPAR, UniPROBE and EpiSELEX-seq, while the illustrations of the second data column are derived from the methylated TFBS sequences from EpiSELEX-seq and MeDReaders. For EpiSELEX-seq (28), TFBSshape predicts the shape features on paired unmethylated and methylated TFBS sequences. However, since JASPAR (43) and UniPROBE (44) do not have methylated TFBS sequences for comparison, TFBSshape performs in silico methylation on unmethylated TFBS sequences and predicts their shape features. For MeDReaders (39), TFBSshape predicts the shape features for TFBS sequences with both high and low methylation levels. In some cases, the CpG-containing TFBS is not the optimal binding site, and thus, the comparison of changes due to the methylation might be difficult to see in a box plot when considering the entire set of TFBSs, including sequences without CpG dinucleotides (Supplementary Figure S1A). Therefore, TFBSshape provides an additional box plot that only compares the TFBS sequences with CpG and MpG (where MpG represents a CpG dinucleotide with the cytosines on both strands methylated at their C5 positions) (Supplementary Figure S1B).
Shape alignment for comparison of DNA shape profiles of two TF datasets
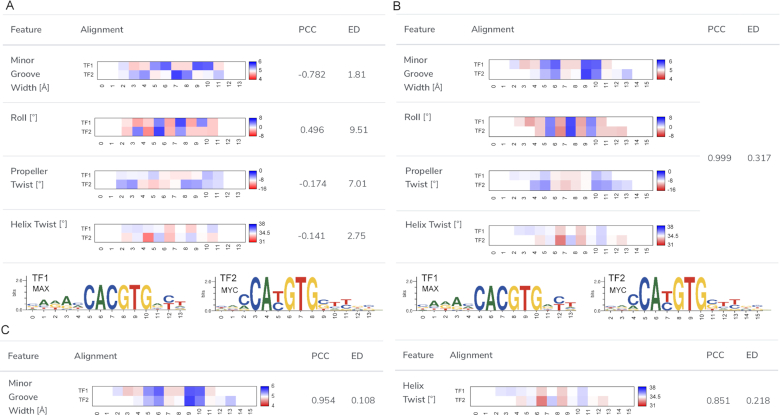
The original version of TFBSshape provided an interface for comparing two TFBS shape profiles from the database. However, with this interface, the user previously needed to specify the alignment of the two TF motifs by setting the reference positions for the compared datasets. This manual setting had two shortcomings. First, it was inconvenient for the user to manually repeat the comparison process, especially when prior knowledge was lacking or the alignment of the two shape profiles was ambiguous. Second, the setting only considered the alignment at the DNA sequence level; therefore, the mechanistic similarity in terms of shape of two TF binding profiles might have been overlooked. TFBSshape now offers a new function ‘Align by Shape’ to automatically determine the best alignment based on the selected shape features. Since the lengths and positions of compared TFBSs might vary within the sequences, a comparison without alignment results in low Pearson correlation coefficients (PCCs) and large Euclidean distances (EDs) among all shape features (Figure 4A). The new release of TFBSshape allows the user to select the shape features for the basis of alignment. According to the selection, TFBSshape calculates all possible combinations of alignments and displays results that are represented by the best PCC value and visualizes quantitative comparisons of average heat maps for the DNA shape features (Figure 4B). This tool can be used not only to determine the best alignment but also to investigate possible binding mechanisms through cross comparisons with similarities based on the selection of different shape features (Figure 4C).
Figure 4.
An example for a TFBSshape comparison of DNA shape preferences of two TFBS datasets using the shape alignment function. (A) The user can select the shape features used for calculating the best alignment from the checkbox at the shape alignment interface. Before the shape alignment, the comparison of the homologous TFs MAX from human (MA0058.1) and MYC from mouse (MA0147.2) from JASPAR demonstrates low Pearson correlation coefficients (PCCs) and large Euclidean distances (EDs) due to the poor alignment. The corresponding sequence motif logos with nucleotide positions numbered according to the alignment for the two TFs are shown in the bottom panel. (B) In this example, four shape features, including HelT, MGW, ProT and Roll, were selected. Using the chosen shape features, the best alignment was calculated and the average heat maps as well as the corresponding PCC and ED for the four DNA shape features are shown. Compared to the initial alignment, this shape alignment is a significant improvement in terms of PCC and ED. (C) The shape alignment tool can be used to compare the similarity between two TFs referring to different shape features. For example, the similarity for MGW is higher than the one for HelT.
Mutation design
The current version of TFBSshape introduces a mutation design tool to generate DNA sequences that preserve either DNA shape or DNA sequence features. For any given wild-type sequence 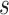 bound by a TF, the user can specify
bound by a TF, the user can specify 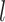 base pairs in lower case that are intended to be mutated. If at most
base pairs in lower case that are intended to be mutated. If at most 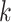 base pairs are expected to be mutated, there are
base pairs are expected to be mutated, there are 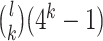 possible mutations. The distance between wild-type sequence
possible mutations. The distance between wild-type sequence 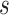 and each mutated sequence
and each mutated sequence 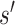 is determined by the similarity between the two strings of DNA sequences. This is calculated as Levenshtein distance
is determined by the similarity between the two strings of DNA sequences. This is calculated as Levenshtein distance 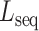 that counts the number of deletions, insertions or substitutions required to transform sequence
that counts the number of deletions, insertions or substitutions required to transform sequence 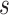 to
to 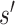 . Furthermore, to calculate DNA shape distances, shape profiles regarding four shape features (HelT, MGW, ProT and Roll) or user-selected shape features for
. Furthermore, to calculate DNA shape distances, shape profiles regarding four shape features (HelT, MGW, ProT and Roll) or user-selected shape features for 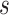 and
and 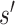 are first derived with DNAshapeR (45). The shape features are normalized between 0 and 1 using min–max normalization with the global minimum and maximum values retrieved from the DNAshape pentamer query table. The normalized shape features are then concatenated as vector
are first derived with DNAshapeR (45). The shape features are normalized between 0 and 1 using min–max normalization with the global minimum and maximum values retrieved from the DNAshape pentamer query table. The normalized shape features are then concatenated as vector 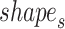 and
and 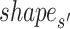 , respectively. The distance between these two vectors is represented as Euclidean distance
, respectively. The distance between these two vectors is represented as Euclidean distance 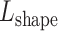 . DNA sequence and shape distances for all mutations are then sorted among all possible mutations (Supplementary Figure S2). The user can set thresholds for
. DNA sequence and shape distances for all mutations are then sorted among all possible mutations (Supplementary Figure S2). The user can set thresholds for 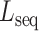 and
and 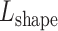 to obtain a list of desired mutations. For instance, a mutation with high
to obtain a list of desired mutations. For instance, a mutation with high 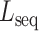 and low
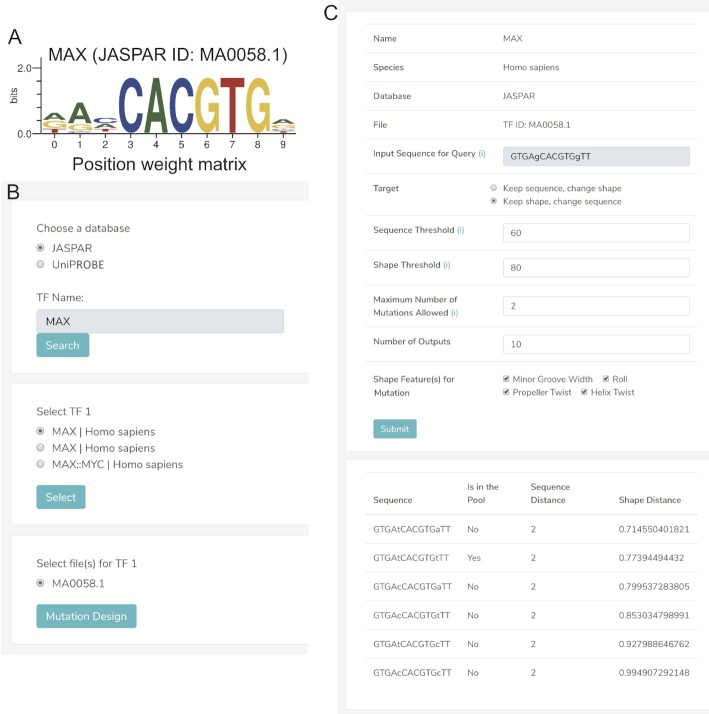
and low 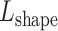 values changes predominantly DNA sequence while preserving most shape features. Moreover, whether a mutated sequence was previously detected by binding assays such as SELEX-seq and PBM will be indicated in the resulting table on the web page, so that the user can choose an alternative binding target of the same TF (Figure 5A–C). This function will assist researchers to design experiments to investigate the independent role of base and shape readout.
values changes predominantly DNA sequence while preserving most shape features. Moreover, whether a mutated sequence was previously detected by binding assays such as SELEX-seq and PBM will be indicated in the resulting table on the web page, so that the user can choose an alternative binding target of the same TF (Figure 5A–C). This function will assist researchers to design experiments to investigate the independent role of base and shape readout.
Figure 5.
Mutation design interface. In this example, we aimed to design mutations for a wild-type sequence ‘GTGAgCACGTGgTT’, which is bound by the TF MAX (only bases in lower case will be mutated). (A) MAX is a member of the basic helix-loop-helix (bHLH) family of TFs that binds to the E-box ‘CACGTG’ core binding site and refines its binding specificity through structural readout of the flanking regions (23). (B) A responsive window lists the available binding site data for MAX in TFBSshape. Here, the MAX binding profile (MA0058.1) in JASPAR is selected. (C) A detailed page lists options for mutation design and candidates. Note that ‘GTGAtCACGTGtTT’ could be a good candidate if the user is interested in mutations that preserve structural patterns in the flanking regions of the E-box since this mutation was previously detected as bound sequence (MA0058.1). See Supplementary Data for more details on the algorithm.
Web interface
We completely redesigned the TFBSshape web interface to meet modern web design standards. The implementation follows the Model–View–Controller architecture pattern for improving scalability, flexibility and extensibility (Figure 1C). The Model component handles data derived from heterogeneous sources from MySQL databases and flat experimental data files and implements the core functionality of the system, such as calculating DNA shape features and distances between two sequences, performing statistical analysis and generating various plots. The View component, which is the primary user interface component, provides multiple and synchronized views to present the information as well as interact with the user. Using Bootstrap as a front-end template engine in combination with HTML and JavaScript improves the visibility and usability of our functionality and enhances browsing and searching. The Model and View components are independent and loosely coupled with each other, thus supporting parallel development and simplifying updating or integration of new databases. The Controller component manages the application logic and acts as a mediator between the Model and View components, tightly coupling the independent components, which ensures consistent as well as flexible architecture. Moreover, we substantially increased the speed of displaying the structural profiles by precalculation of shape features. Finally, we introduced semantic URLs to facilitate external links to TFBSshape's detailed pages of individual profiles.
CONCLUSIONS AND FUTURE EXPANSIONS
The new version of TFBSshape has greatly increased the quantity and dimensionality of the available structural profiles in the database. The update includes the most recently released TF binding data from the motif databases, JASPAR 2020 (43) and UniPROBE (44), and incorporates the methylated TFBS sequences from the in vivo methylation database MeDReaders (39) and in vitro EpiSELEX-seq experiments (28). The original four shape features in the original version of TFBSshape (23) have been expanded to 14 features, which can be used, for example, to differentiate DNA binding specificities that are not apparent from nucleotide sequence alone. TFBSshape also introduces four shape features for methylated DNA that can be used to uncover mechanistic insights into the effect of methylation on local DNA structure in TF–DNA binding by comparing the structural profiles of unmethylated and methylated TFBSs. Moreover, the current version of TFBSshape provides new functions of mutation design and shape alignment. Finally, the new web interface provides an improved user experience through a modern web design with a Model–View–Controller architecture. In the future, it would be useful to include other types of DNA modification, such as 5-hydroxymethylcytosines or different methylated forms of bases (56–58), once the binding data and DNA shape prediction methods are available. The architecture of the current version of TFBSshape enables adding other TF motif databases such as UniBind (59) and MethMotif (60) and new TF–DNA binding profiles or new large-scale TF–DNA binding assays in the future.
Supplementary Material
ACKNOWLEDGEMENTS
A collaboration of the authors with Harmen Bussemaker on DNA methylation readout initiated some of the added functionalities of TFBSshape. The authors thank all current members of the Rohs laboratory for feedback and valuable input. The authors acknowledge Lin Yang and Tianyin Zhou for conceiving initial concepts of TFBSshape (23) and DNAshape (24), Satyanarayan Rao for participation in planning stages of the project, contributions to the shape alignment approach and development of methyl-DNAshape (37), and Jinsen Li for deriving the expanded set of 13 DNA shape features (25). The authors also thank Luigi Manna for administrating the server hosting TFBSshape, and Anthony Mathelier, Wyeth Wasserman and the JASPAR 2020 team (43) for providing data entries prior to their publication.
Notes
Present address: Beibei Xin, State Key Laboratory of Agrobiotechnology and National Maize Improvement Center, Department of Plant Genetics and Breeding, China Agricultural University, Beijing 100193, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01GM106056, R01HG003008 (in part), and R35GM130376 to R.R.]; the USC-Taiwan Postdoctoral Fellowship Program [to T.P.C.]; the Rose Hills Foundation [to N.M.]; the Human Frontier Science Program [RGP0021/2018 to R.R.]. Funding for open access charge: National Institutes of Health [R35GM130376].
Conflict of interest statement. None declared.
REFERENCES
- 1. Stormo G.D. DNA binding sites: representation and discovery. Bioinformatics. 2000; 16:16–23. [DOI] [PubMed] [Google Scholar]
- 2. Stormo G.D. Modeling the specificity of protein–DNA interactions. Quant. Biol. 2013; 1:115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schneider T.D., Stephens R.M.. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990; 18:6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benos P.V., Bulyk M.L., Stormo G.D.. Additivity in protein–DNA interactions: how good an approximation is it?. Nucleic Acids Res. 2002; 30:4442–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eggeling R., Roos T., Myllymaki P., Grosse I.. Inferring intra-motif dependencies of DNA binding sites from ChIP-seq data. BMC Bioinformatics. 2015; 16:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sharon E., Lubliner S., Segal E.. A feature-based approach to modeling protein–DNA interactions. PLoS Comput. Biol. 2008; 4:e1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kahara J., Lahdesmaki H.. Evaluating a linear k-mer model for protein–DNA interactions using high-throughput SELEX data. BMC Bioinformatics. 2013; 14(Suppl. 10):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Annala M., Laurila K., Lahdesmaki H., Nykter M.. A linear model for transcription factor binding affinity prediction in protein binding microarrays. PLoS One. 2011; 6:e20059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao Y., Ruan S., Pandey M., Stormo G.D.. Improved models for transcription factor binding site identification using nonindependent interactions. Genetics. 2012; 191:781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Siddharthan R. Dinucleotide weight matrices for predicting transcription factor binding sites: generalizing the position weight matrix. PLoS One. 2010; 5:e9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou T., Shen N., Yang L., Abe N., Horton J., Mann R.S., Bussemaker H.J., Gordân R., Rohs R.. Quantitative modeling of transcription factor binding specificities using DNA shape. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:4654–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang L., Orenstein Y., Jolma A., Yin Y., Taipale J., Shamir R., Rohs R.. Transcription factor family-specific DNA shape readout revealed by quantitative specificity models. Mol. Syst. Biol. 2017; 13:910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gordân R., Shen N., Dror I., Zhou T., Horton J., Rohs R., Bulyk M.L.. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Rep. 2013; 3:1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B.. The role of DNA shape in protein–DNA recognition. Nature. 2009; 461:1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Joshi R., Passner J.M., Rohs R., Jain R., Sosinsky A., Crickmore M.A., Jacob V., Aggarwal A.K., Honig B., Mann R.S.. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007; 131:530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rohs R., Jin X., West S.M., Joshi R., Honig B., Mann R.S.. Origins of specificity in protein–DNA recognition. Annu. Rev. Biochem. 2010; 79:233–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peng P.C., Sinha S.. Quantitative modeling of gene expression using DNA shape features of binding sites. Nucleic Acids Res. 2016; 44:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crocker J., Abe N., Rinaldi L., McGregor A.P., Frankel N., Wang S., Alsawadi A., Valenti P., Plaza S., Payre F. et al.. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell. 2015; 160:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Crocker J., Stern D.L.. Functional regulatory evolution outside of the minimal even-skipped stripe 2 enhancer. Development. 2017; 144:3095–3101. [DOI] [PubMed] [Google Scholar]
- 21. Pal S., Hoinka J., Przytycka T.M.. Co-SELECT reveals sequence non-specific contribution of DNA shape to transcription factor binding in vitro. Nucleic Acids Res. 2019; 47:6632–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Samee M.A.H., Bruneau B.G., Pollard K.S.. A de novo shape motif discovery algorithm reveals preferences of transcription factors for DNA shape beyond sequence motifs. Cell Syst. 2019; 8:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang L., Zhou T., Dror I., Mathelier A., Wasserman W.W., Gordân R., Rohs R.. TFBSshape: a motif database for DNA shape features of transcription factor binding sites. Nucleic Acids Res. 2014; 42:D148–D155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou T., Yang L., Lu Y., Dror I., Dantas Machado A.C., Ghane T., Di Felice R., Rohs R.. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013; 41:W56–W62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li J., Sagendorf J.M., Chiu T.P., Pasi M., Perez A., Rohs R.. Expanding the repertoire of DNA shape features for genome-scale studies of transcription factor binding. Nucleic Acids Res. 2017; 45:12877–12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chiu T.P., Rao S., Mann R.S., Honig B., Rohs R.. Genome-wide prediction of minor-groove electrostatic potential enables biophysical modeling of protein–DNA binding. Nucleic Acids Res. 2017; 45:12565–12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mathelier A., Xin B., Chiu T.P., Yang L., Rohs R., Wasserman W.W.. DNA shape features improve transcription factor binding site predictions in vivo. Cell Syst. 2016; 3:278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kribelbauer J.F., Laptenko O., Chen S., Martini G.D., Freed-Pastor W.A., Prives C., Mann R.S., Bussemaker H.J.. Quantitative analysis of the DNA methylation sensitivity of transcription factor complexes. Cell Rep. 2017; 19:2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., Das P.K., Kivioja T., Dave K., Zhong F. et al.. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017; 356:eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mann I.K., Chatterjee R., Zhao J., He X., Weirauch M.T., Hughes T.R., Vinson C.. CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res. 2013; 23:988–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tillo D., Ray S., Syed K.S., Gaylor M.R., He X., Wang J., Assad N., Durell S.R., Porollo A., Weirauch M.T. et al.. The Epstein-Barr virus B-ZIP protein Zta recognizes specific DNA sequences containing 5-methylcytosine and 5-hydroxymethylcytosine. Biochemistry. 2017; 56:6200–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zuo Z., Roy B., Chang Y.K., Granas D., Stormo G.D.. Measuring quantitative effects of methylation on transcription factor–DNA binding affinity. Sci. Adv. 2017; 3:eaao1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hu S., Wan J., Su Y., Song Q., Zeng Y., Nguyen H.N., Shin J., Cox E., Rho H.S., Woodard C. et al.. DNA methylation presents distinct binding sites for human transcription factors. eLife. 2013; 2:e00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R.. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016; 165:1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baylin S.B., Jones P.A.. A decade of exploring the cancer epigenome—biological and translational implications. Nat. Rev. Cancer. 2011; 11:726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dantas Machado A.C., Zhou T., Rao S., Goel P., Rastogi C., Lazarovici A., Bussemaker H.J., Rohs R.. Evolving insights on how cytosine methylation affects protein–DNA binding. Brief. Funct. Genomics. 2015; 14:61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rao S., Chiu T.P., Kribelbauer J.F., Mann R.S., Bussemaker H.J., Rohs R.. Systematic prediction of DNA shape changes due to CpG methylation explains epigenetic effects on protein–DNA binding. Epigenet. Chromatin. 2018; 11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lazarovici A., Zhou T., Shafer A., Dantas Machado A.C., Riley T.R., Sandstrom R., Sabo P.J., Lu Y., Rohs R., Stamatoyannopoulos J.A. et al.. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:6376–6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang G., Luo X., Wang J., Wan J., Xia S., Zhu H., Qian J., Wang Y.. MeDReaders: a database for transcription factors that bind to methylated DNA. Nucleic Acids Res. 2018; 46:D146–D151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Slattery M., Zhou T., Yang L., Dantas Machado A.C., Gordân R., Rohs R.. Absence of a simple code: how transcription factors read the genome. Trends Biochem. Sci. 2014; 39:381–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang X., Zhou T., Wunderlich Z., Maurano M.T., DePace A.H., Nuzhdin S.V., Rohs R.. Analysis of Genetic Variation Indicates DNA Shape Involvement in Purifying Selection. Mol. Biol. Evol. 2018; 35:1958–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Al-Zyoud W.A., Hynson R.M., Ganuelas L.A., Coster A.C., Duff A.P., Baker M.A., Stewart A.G., Giannoulatou E., Ho J.W., Gaus K. et al.. Binding of transcription factor GabR to DNA requires recognition of DNA shape at a location distinct from its cognate binding site. Nucleic Acids Res. 2016; 44:1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fornes O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A., Modi B.P., Correard S., Gheorghe M., Baranašić D. et al.. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic. Acids Res. 2019; doi:10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hume M.A., Barrera L.A., Gisselbrecht S.S., Bulyk M.L.. UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein–DNA interactions. Nucleic Acids Res. 2015; 43:D117–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chiu T.P., Comoglio F., Zhou T., Yang L., Paro R., Rohs R.. DNAshapeR: an R/Bioconductor package for DNA shape prediction and feature encoding. Bioinformatics. 2016; 32:1211–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sklenar H., Wüstner D., Rohs R.. Using internal and collective variables in Monte Carlo simulations of nucleic acid structures: chain breakage/closure algorithm and associated Jacobians. J. Comput. Chem. 2006; 27:309–315. [DOI] [PubMed] [Google Scholar]
- 47. Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A.. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995; 117:5179–5197. [Google Scholar]
- 48. Rohs R., Sklenar H., Shakked Z.. Structural and energetic origins of sequence-specific DNA bending: Monte Carlo simulations of papillomavirus E2–DNA binding sites. Structure. 2005; 13:1499–1509. [DOI] [PubMed] [Google Scholar]
- 49. Rohs R., Etchebest C., Lavery R.. Unraveling proteins: a molecular mechanics study. Biophys. J. 1999; 76:2760–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lavery R., Sklenar H.. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 1989; 6:655–667. [DOI] [PubMed] [Google Scholar]
- 51. Azad R.N., Zafiropoulos D., Ober D., Jiang Y., Chiu T.P., Sagendorf J.M., Rohs R., Tullius T.D.. Experimental maps of DNA structure at nucleotide resolution distinguish intrinsic from protein-induced DNA deformations. Nucleic Acids Res. 2018; 46:2636–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Honig B., Nicholls A.. Classical electrostatics in biology and chemistry. Science. 1995; 268:1144–1149. [DOI] [PubMed] [Google Scholar]
- 53. Zhang H., Lang Z., Zhu J.K.. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018; 19:489–506. [DOI] [PubMed] [Google Scholar]
- 54. Luo C., Keown C.L., Kurihara L., Zhou J., He Y., Li J., Castanon R., Lucero J., Nery J.R., Sandoval J.P. et al.. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science. 2017; 357:600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M. et al.. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Spruijt C.G., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W., Bauer C., Munzel M., Wagner M., Muller M., Khan F. et al.. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013; 152:1146–1159. [DOI] [PubMed] [Google Scholar]
- 57. Iurlaro M., Ficz G., Oxley D., Raiber E.A., Bachman M., Booth M.J., Andrews S., Balasubramanian S., Reik W.. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 2013; 14:R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kinde B., Gabel H.W., Gilbert C.S., Griffith E.C., Greenberg M.E.. Reading the unique DNA methylation landscape of the brain: non-CpG methylation, hydroxymethylation, and MeCP2. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:6800–6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gheorghe M., Sandve G.K., Khan A., Cheneby J., Ballester B., Mathelier A.. A map of direct TF–DNA interactions in the human genome. Nucleic Acids Res. 2019; 47:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xuan Lin Q.X., Sian S., An O., Thieffry D., Jha S., Benoukraf T.. MethMotif: an integrative cell specific database of transcription factor binding motifs coupled with DNA methylation profiles. Nucleic Acids Res. 2019; 47:D145–D154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.