Abstract
Fungal tRNA ligase (Trl1) rectifies RNA breaks with 2′,3′-cyclic-PO4 and 5′-OH termini. Trl1 consists of three catalytic modules: an N-terminal ligase (LIG) domain; a central polynucleotide kinase (KIN) domain; and a C-terminal cyclic phosphodiesterase (CPD) domain. Trl1 enzymes found in all human fungal pathogens are untapped targets for antifungal drug discovery. Here we report a 1.9 Å crystal structure of Trl1 KIN-CPD from the pathogenic fungus Candida albicans, which adopts an extended conformation in which separate KIN and CPD domains are connected by an unstructured linker. CPD belongs to the 2H phosphotransferase superfamily by dint of its conserved central concave β sheet and interactions of its dual HxT motif histidines and threonines with phosphate in the active site. Additional active site motifs conserved among the fungal CPD clade of 2H enzymes are identified. We present structures of the Candida Trl1 KIN domain at 1.5 to 2.0 Å resolution—as apoenzyme and in complexes with GTP•Mg2+, IDP•PO4, and dGDP•PO4—that highlight conformational switches in the G-loop (which recognizes the guanine base) and lid-loop (poised over the nucleotide phosphates) that accompany nucleotide binding.
INTRODUCTION
Invasive fungal infections, particularly Candidiasis and Aspergillosis, are a major cause of morbidity and mortality in individuals with neutropenia following cancer chemotherapy or hematopoietic stem cell transplantation. The recent emergence of Candida auris as a multi-drug-resistant invasive fungal pathogen in hospitalized patients is especially worrisome (1). The development of more effective (and less toxic) drugs for treatment of fungal infections hinges on defining new targets for antifungal drug discovery. We regard the tRNA splicing enzymes Trl1 (tRNA ligase) and Tpt1 (tRNA 2′-phosphotransferase) as attractive antifungal targets.
Fungal tRNA ligase Trl1 is an essential agent in the repair of RNA breaks with 2′,3′-cyclic phosphate and 5′-OH ends that are formed during tRNA splicing and the unfolded protein response (2,3). Trl1 executes three RNA repair reactions performed by three autonomous catalytic domains: (i) the 2′,3′-cyclic phosphate (>p) end is converted to a 3′-OH,2′-PO4 by a C-terminal cyclic phosphodiesterase (CPD) module; (ii) the 5′-OH end is phosphorylated by a central GTP-dependent polynucleotide kinase (KIN) module; and (iii) the 3′-OH,2′-PO4 and 5′-PO4 ends are joined by an N-terminal ATP-dependent RNA ligase (LIG) module to form a 2′-PO4, 3′-5′ phosphodiester splice junction (4–10). To complete the pathway, the essential enzyme Tpt1 transfers the 2′-PO4 from the splice junction to NAD+ to form ADP-ribose 1″–2″ cyclic phosphate and nicotinamide via two unique steps (11–17). First, NAD+ reacts with the tRNA 2′-PO4 to expel nicotinamide and generate a 2′-phospho-ADP-ribosylated RNA intermediate. Then, transesterification of the ADP-ribose 2″-O to the tRNA 2′-PO4 displaces the tRNA product and generates ADP-ribose 1″–2″ cyclic phosphate.
Fungal Trl1 enzymes are potential therapeutic targets because their domain structures and biochemical mechanisms are unique compared to the RtcB-type tRNA repair systems elaborated by metazoa, archaea, and many bacteria (18–26). RtcB is a GTP-dependent RNA ligase that splices 3′-PO4 and 5′-OH ends via a different chemical mechanism vis-à-vis Trl1. RtcB is absent from the proteomes of most fungi and mammalian proteomes have no homologs of the LIG domain of fungal Trl1. Trl1 LIG is distinguished from other ATP-dependent polynucleotide ligases by virtue of its stringent requirement for a terminal 2′-PO4 in order to seal 3′-OH and 5′-PO4 ends, a property that could be exploited for LIG inhibition. There is no 5′ kinase step in the RtcB pathway of RNA repair. Whereas polynucleotide kinases are widely distributed in nature, the KIN domains of fungal Trl1 enzymes are unique in that they have a strong preference for GTP as the phosphate donor (6,9,10,27,28). An ortholog of the Trl1 CPD domain is present in mammals in the form of CNP, a 2′,3′-cyclic nucleotide phosphodiesterase that is an abundant constituent of brain myelin (29). Mammalian CNP can substitute for Trl1 CPD as a tRNA splicing enzyme in vivo in yeast (30), yet its role in mammalian RNA metabolism is uncharted. CNP is unlikely to be essential for mammalian cell function generally, insofar as CNP-null mouse is viable to adulthood (31). [Adult CNP-null mice develop a neurodegenerative disorder despite maintaining normal myelin morphology. It is unclear whether this pathology is caused by a lack of CNP protein or CNP enzymatic activity.] A Tpt1 ortholog is also present in mammals, notwithstanding that mammalian tRNA ligation by RtcB does not generate a 2′-PO4 splice junction. Whereas it is not obvious what reactions Tpt1 performs in mammals, it is clear that Tpt1 is not essential for mammalian physiology, i.e., genetic ablation of mouse Tpt1 has no phenotypic consequences (32).
The above considerations highlight all four enzymatic activities in fungal tRNA splicing as potentially vulnerable to inhibition with selective toxicity for fungi versus mammalian hosts. To strengthen the case for tRNA splicing as an antifungal drug target, we aim to attain atomic structures of the tRNA splicing enzymes and elucidate the basis for their unique biochemical specificities. To that end, we recently reported the crystal structures of: (i) Trl1 LIG from the fungus Chaetomium thermophilum as the covalent LIG–AMP•Mg2+ intermediate and the LIG•ATP•(Mg2+)2 Michaelis complex (33); (ii) Trl1 KIN from the human pathogen Candida albicans in a complex with GDP•Mg2+, thereby revealing a ‘G-loop’ motif as the structural determinant of GTP specificity (28); and (iii) the Tpt1 ortholog from Clostridium thermocellum in a product-mimetic complex with ADP-ribose-1″-phosphate in the NAD+ site and a pAp moiety in the RNA site that gave insights to how Tpt1 recognizes a 2′-PO4 RNA splice junction (34).
In the present study, we report the crystal structure of the two-domain KIN-CPD ‘end-healing’ component of Candida albicans Trl1 with a phosphate anion in the CPD active site. This is the first structure available for a Trl1 CPD enzyme and it highlights features shared with other 2H phosphoesterases as well as active site motifs specific to the fungal Trl1 CPD clade. We also report a series of structures of the Candida Trl1 KIN domain – as apoenzyme and in complexes with GTP, IDP, and dGDP – that underscore conformational switches in the G-loop (which recognizes the guanine base) and lid-loop (poised over the nucleotide phosphates) in response to nucleotide binding.
MATERIALS AND METHODS
CalTrl1 domain purifications
Recombinant Candida Trl1(401-832) [KIN-CPD] and Trl1(401-636)-D445N [KIN-D445N] proteins were purified from soluble extracts of IPTG-induced Escherichia coli BL21(DE3) pET28b-His10Smt3-Trl1 strains by sequential nickel-affinity, tag cleavage, tag removal by second nickel-affinity, and gel filtration steps (10,28). The peak Superdex 200 fractions were concentrated by centrifugal ultrafiltration to 9.5 mg/ml (for KIN-CPD) or 8.5 mg/ml (for KIN) in buffer containing 20 mM HEPES-NaOH, pH 7.5, 200 mM NaCl, 1 mM DTT, 5% glycerol. Protein concentrations were determined by using the BioRad dye reagent with bovine serum albumin as the standard.
Crystallization and structure determination of KIN-CPD
CalTrl1 KIN-CPD crystals were grown at 22°C by sitting drop vapor diffusion. A 1 μl solution of 0.18 mM KIN-CPD, 10 mM MgCl2, 2 mM GTP and 2 mM 2′-AMP was mixed with an equal volume of precipitant solution containing 0.2 M potassium dihydrogen phosphate, 20% PEG3350. Crystals appeared overnight and increased in size over four days, after which they were transferred to precipitant solution containing 25% glycerol prior to freezing. X-ray diffraction data were collected from a single crystal at the Advanced Photon Source beamline 24ID-C. Indexing and merging of the diffraction data were performed in HKL2000 (35). KIN-CPD crystallized in space group P21 with one protomer in the asymmetric unit. The structure was solved after initial molecular replacement with Phenix Phaser-MR (36) using as a search model the previously reported Candida KIN domain structure (pdb 5U32) (28). The electron density was improved by solvent flattening with Solomon (37) and the CPD domain was traced using Phenix Autobuild (36). Iterative model building into electron density was performed with O (38). Refinement was accomplished with Phenix.Refine (36). Data collection and refinement statistics are summarized in Supplementary Table S1.
Crystallization and structure determination of KIN apoenzyme
KIN crystals were grown at 22°C by hanging drop vapor diffusion after mixing a solution of 250 μM KIN-D445N with an equal volume of precipitant solution containing 0.3 M sodium dihydrogen phosphate, 20% PEG3350. Crystals appearing overnight were harvested and transferred to precipitant solution containing 25% glycerol prior to freezing. X-ray diffraction data were collected from a single crystal at the Advanced Photon Source beamline 24ID-E. KIN apoenzyme crystallized in space group C2221 with two protomers in the asymmetric unit. The structure was solved by molecular replacement. There was no nucleotide in the active site; rather a phosphate anion was present at the position occupied by the GDP β phosphate in pdb 5U32.
Crystallization and structure determination of KIN complexes with various nucleotides
Crystals were grown at 22°C by hanging drop vapor diffusion after mixing a solution of 250 μM KIN-D445N, 10 mM MgCl2, and 2 mM GTP, ITP or dGTP with an equal volume of precipitant solution containing 0.15 M sodium dihydrogen phosphate, 20% PEG3350. KIN+GTP crystals appearing overnight were harvested immediately and transferred to precipitant solution containing 25% glycerol, 20 mM MgCl2, and 1 mM GTP prior to freezing. KIN+ITP and KIN+dGTP crystals that were harvested after 4–5 days were transferred to precipitant solutions containing 25% glycerol, 20 mM MgCl2 and 1 mM ITP or 1 mM dGTP prior to freezing. All of the KIN+NTP crystals were in space group P212121 with one protomer in the asymmetric unit. The structures were solved by molecular replacement. Refinement statistics are compiled in Supplementary Table S1. The KIN+GTP crystal that was harvested after overnight growth contained GTP and Mg2+ in the active site. All other KIN+NTP crystals that were harvested after 4 to 5 days contained an NDP in the active site. Additional KIN+GTP crystals were grown by sitting drop vapor diffusion by mixing a solution of 250 μM KIN-D445N, 10 mM MgCl2, 2 mM GTP and 300 μM 5′-OH DNA oligonucleotide d(CGCTTC) with an equal volume of a precipitant solution containing 0.1 M MES, pH 5.0, 30% PEG6000. The crystals were harvested after 4–5 days and transferred to precipitant solution containing 25% glycerol prior to freezing. The refined structure at 1.7 Å resolution (Supplementary Table S1) contained GDP and Mg2+ in the active site but no DNA oligonucleotide. This KIN•GDP•Mg2+ structure did not differ significantly from that reported for wild-type KIN (28) but was notable for its higher resolution (1.7 Å versus 2.2 Å previously).
S. cerevisiae trl1Δ complementation
We tested trl1Δ complementation by plasmid shuffle in a S. cerevisiae trl1Δ p(CEN URA3 TRL1) strain (6), which is unable to grow on medium containing 0.75 mg/ml FOA (5-fluoroorotic acid), a drug that selects against the URA3 plasmid. trl1Δ cells were transfected with CEN LEU2 plasmids harboring wild-type or mutated CalTRL1 under the control of the yeast TPI1 promoter (10). Individual Leu+ transformants were selected and streaked on agar medium containing FOA. CalTRL1 alleles that failed to give rise to FOA-resistant colonies after incubation for 8 days at 20°C, 30°C and 37°C were deemed to be lethal. Individual FOA-resistant CalTRL1 colonies were grown to mid-log phase in YPD broth and adjusted to A600 of 0.1. Aliquots (3 μl) of serial 10-fold dilutions were spotted to YPD agar plates, which were incubated for 2 days at 30°C and 37°C.
RESULTS
Structure of Candida albicans Trl1 KIN-CPD
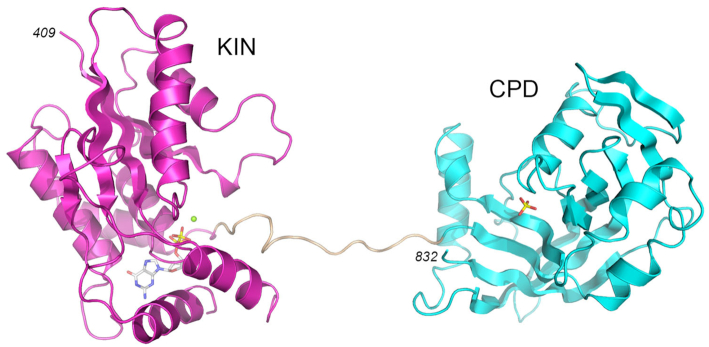
C. albicans Trl1 is an 832-amino acid polypeptide composed of N-terminal ligase (LIG), central kinase (KIN), and C-terminal cyclic phosphodiesterase (CPD) domains. We showed previously that an RNA with 5′-OH and 2′,3′-cyclic phosphate ends could be spliced in vitro by reaction with a mixture of recombinant LIG (aa 1–400) and KIN-CPD (aa 401–832) proteins (28). Here we grew crystals of the Candida KIN-CPD protein by sitting drop vapor diffusion after mixing a sample of the protein solution containing KIN-CPD, GTP (KIN phosphate donor), MgCl2 (KIN cofactor), and 2′-AMP (CPD product mimetic) with an equal volume of precipitant solution containing 200 mM potassium dihydrogen phosphate, 30% PEG3350. The crystals belonged to space group P21 and contained one KIN-CPD protomer in the asymmetric unit. The refined model of KIN-CPD at 1.95 Å resolution (Rwork/Rfree 0.185/0.221; Supplementary Table S1) comprised a KIN domain (aa 409–630) connected by an extended 13-aa linker to a CPD domain (aa 644–832) (Figure 1A and Supplementary Figure S1). The KIN domain had GDP and Mg2+ bound in the active site. The CPD domain had a phosphate anion in its active site (Figure 1A and Supplementary Figure S1). Amino acid sequence alignment of the KIN-CPD components of tRNA ligases from diverse fungal taxa indicates that the lengths of this linker segment are variable (13–40 aa) and the linker primary structure is not conserved (8). There were no contacts between the KIN and CPD modules within a single KIN-CPD polypeptide. The interactions of the KIN-CPD protomer with three symmetry related protein molecules in the crystal lattice are shown in Supplementary Figure S2. Analysis of the three interfaces in PISA returned a Complex Formation Significance Score of 0.000, implying that these interfaces are the result of crystal packing only. We regard this crystal packing interface as being of no physiological significance given that KIN-CPD is a monomer in solution (28). The structure of the KIN domain of KIN-CPD is highly similar to the structure of the isolated Candida Trl1 KIN domain in complex with GDP•Mg2+ (28).
Figure 1.
Structure of Candida Trl1 KIN-CPD. The tertiary structure is shown as a cartoon model with the KIN domain in magenta, CPD domain in cyan, and the interdomain linker in beige. GDP and magnesium in the KIN active site are depicted as a stick model and a green sphere, respectively. Phosphate anion in the CPD active site is shown as a stick model.
Overview of the Trl1 CPD domain structure
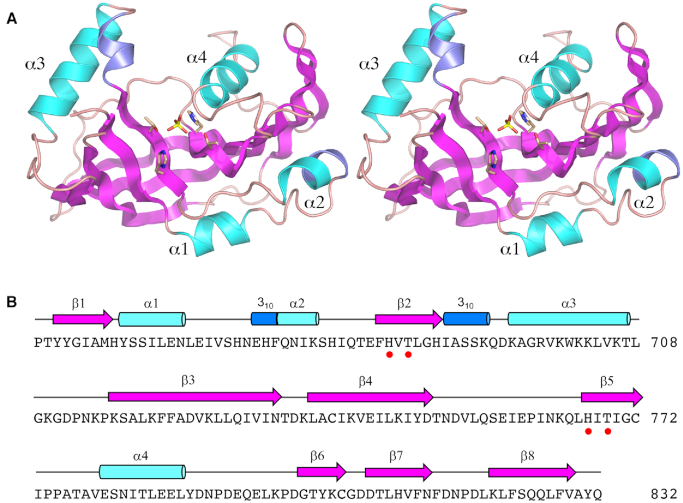
The core CPD tertiary structure (shown in stereo in Figure 2A) consists of eight β strands that form a central anti-parallel β sheet. Flanking the β sheet are four α helices and two 310 helices. The secondary structure elements are aligned to the CPD primary structure in Figure 2B. The signature 2H phosphoesterase family motifs – 682HVT684 and 767HIT769 – are located in strands β2 and β5. The two histidines and two threonines coordinate a phosphate anion in the active site. (Although we had preincubated KIN-CPD with the CPD product-like molecule 2′-AMP prior to crystallization, it was apparently out-competed for occupancy of the CPD active site by the 200 mM phosphate in the precipitant solution used to grow the crystals).
Figure 2.
Structure of the CPD domain. (A) Stereo view of the CPD tertiary structure, depicted as a cartoon model with magenta β strands, cyan α helices (numbered sequentially), and blue 310 helices. The phosphate anion in the active site and the histidine and threonine side chains that coordinate the phosphate are rendered as stick models. (B) Secondary structure elements (colored as in panel A) are displayed above the CPD primary structure. The histidine and threonine residues in the signature HxT motifs are denoted by red dots.
A DALI search (39) with the CPD structure retrieved other 2H phosphoesterase enzymes with varying extents of structural homology. The top hits were mammalian CNP (pdb 2YDD; Z score 13.8; rmsd of 3.0 Å at 150 Cα positions; 10% amino acid identity) and goldfish CNP (pdb 2I3E; Z score 12.3; rmsd of 2.6 Å at 149 Cα positions; 10% amino acid identity) (40,41). The next best homolog was E. coli ThpR (42), a biochemically validated RNA 2′,3′-cyclic phosphodiesterase enzyme (pdb 4QAK; Z score 6.1; rmsd of 3.5 Å at 111 Cα positions; 8% amino acid identity). Other hits with lower Z cores included: rotavirus VP3 (43), an RNA 2′,5′-phosphodiesterase that degrades the innate immune signaling molecule 2′–5′ oligoadenylate (pdb 5AF2; Z score 5.4; rmsd of 3.6 Å at 98 Cα positions; 16% amino acid identity); and Arabidopsis ADP-ribose 1″–2″-cyclic phosphodiesterase (44), an enzyme that hydrolyzes the product of the Tpt1 reaction (pdb 1FSI; Z score 4.9; rmsd of 4.3 Å at 108 Cα positions; 8% amino acid identity). The primary structure conservation between Candida Trl1 CPD and the other 2H phosphoesterases is low and mostly localized to the 2H catalytic motifs.
CPD active site
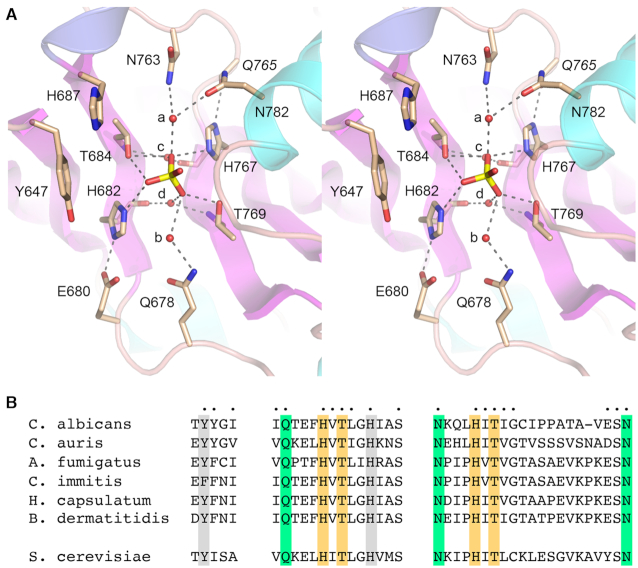
Figure 3A shows a detailed stereo view of the CPD active site. The antiparallel β2 and β5 strands (containing the HxT motifs) and their preceding loops are arranged with pseudo-two-fold symmetry around the phosphate anion in the center of the active site. The enzymic contacts to the phosphate reflect this two-fold symmetry. The phosphate oxygens receive hydrogen bonds from His682-Nϵ, Thr684-Oγ, His767-Nϵ and Thr769-Oγ. The tautomeric state of the two histidines, whereby histidine-Nϵ engages the phosphate, is ensured by hydrogen bonding of the histidine-Nδ to the amino acid two positions upstream of the histidine: to the Glu680 carboxylate in the case of His682 and to the Gln765 main chain carbonyl in the case of His767 (Figure 3A). The phosphate oxygens are also engaged to four water molecules (labeled a, b, c, and d in Figure 3A) that bridge to the enzyme. Waters a and b are coordinated to the side chain amide Nδ atoms of the residues four positions upstream of the catalytic histidines: Asn763 for water a and Gln678 for water b (Figure 3A). Water a makes an additional contact to Asn782 in helix α5. Waters c and d are engaged pseudo-symmetrically to the phosphate via bridging interactions with the HxT histidine main chain carbonyls and the threonine main chain amides (Figure 3A).
Figure 3.
CPD active site. (A) Stereo view of the CPD active site highlighting CPD interactions with the phosphate anion. Amino acids are shown as stick models with beige carbons. Waters are denoted by red spheres. Atomic contacts are indicated by dashed lines. (B) Conservation of three active site motifs among Trl1 CPD domains from six species of human pathogenic fungi: Candida albicans, Candida auris, Aspergillus fumigatus, Coccidioides immitis, Histoplasma capsulatum, and Blastomyces dermatitidis. Positions of amino acid side chain identity or similarity in all six proteins are indicated by dots above the C. albicans sequence. The phosphate-binding histidines and threonines are highlighted in gold shading. Amino acids making water-bridged contacts to the phosphate are in green shading. Two aromatic residues imputed to interact with the terminal nucleoside are shaded gray. The corresponding motifs in S. cerevisiae Trl1 CPD are shown at bottom.
Whereas our CPD structure does not show the position and trajectory of the substrate RNA 3′ end, we can glean some insights via comparison to the structure of E. coli ThpR in complex with 2′-AMP (42). The ThpR active site aligns very well with that of Candida Trl1 CPD, notwithstanding that the overall folds of the two proteins have significant differences. The HxT histidines and threonines in ThpR superimpose on the CPD HxT motifs. The upstream HxT motif in ThpR contacts the AMP 3′-OH (the leaving group in the CPD reaction), suggesting that the upstream catalytic histidine acts as a general acid; we envision that His682 plays a similar role in Trl1 CPD. Of note, the ThpR HxT histidine main chain carbonyls and threonine main chain amides coordinate a pair of phosphate-neighboring water molecules equivalent to waters c and d in our CPD structure. A key distinction is that CPD lacks a counterpart of ThpR Arg130, which makes a bidentate contact to the 2′-AMP phosphate and is proposed to stabilize the transition-state of the ThpR CPD reaction (42). Mutating this arginine to alanine abolished ThpR CPD activity (42). In the case of Trl1 CPD, there is no positively charged functional group near the active site phosphate other than the two histidines. We presume that HxT interactions with the scissile terminal phosphate suffice for catalysis of 3′ end healing during fungal tRNA splicing. The ThpR structure taught us that two aromatic amino acids – Phe8 and Phe48—pack against the nucleoside and nucleobase of 2′-AMP. The equivalent (and superimposable) residues in Trl1 CPD are Tyr647 (in strand β1) and His687 (in strand β2) (Figure 3A).
Distinctive fungal CPD motifs
The Candida albicans Trl1 CPD structure highlights active site constituents clustered in three motifs that are broadly conserved among other human fungal pathogens that are prevalent in the US, including Candida auris, Aspergillus fumigatus, Coccidioides immitis (the agent of Valley Fever), Histoplasma capsulatum, and Blastomyces dermatitis (Figure 3B). In addition to the active site histidines and threonines (shaded gold in Figure 3B), the conserved fungal residues include: the equivalents of Gln678, Asn763 and Asn782 (shaded green in Figure 3B) that make water-bridged phosphate contacts; and the Tyr/Phe and His equivalents of Tyr647 and His687 that we suggest interact with the terminal RNA nucleoside (shaded gray in Figure 3B). The shaded amino acids are conserved in Trl1 from the non-pathogenic yeast Saccharomyces cerevisiae (Figure 3B). Such conservation within the fungal clade may bode well for Trl1 CPD inhibitor discovery.
New structures of Candida Trl1 KIN in GTP-bound and nucleotide-free states
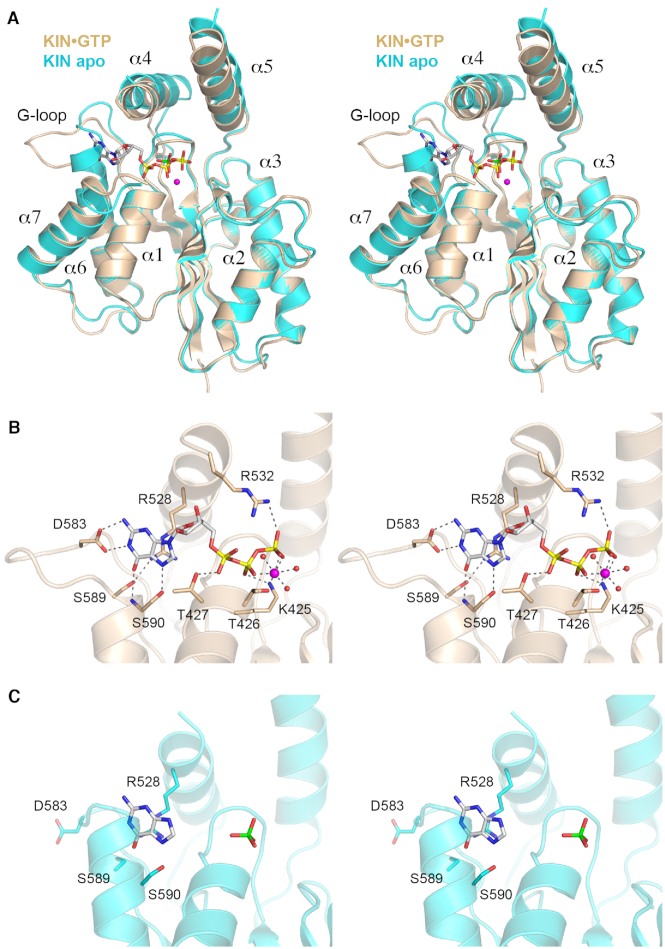
Our initial structure of the isolated KIN domain obtained from crystals grown from a mixture of KIN, GTP, and magnesium had GDP•Mg+ in the kinase active site (28), as did the KIN domain of KIN-CPD crystallized here in the presence of GTP and magnesium. This suggested that the γ phosphate of the added GTP was hydrolyzed during crystal growth. Alternatively, the KIN donor site was pre-filled with GDP during enzyme production in E. coli and GDP remained tightly bound during the KIN purification. To explore this issue, we grew crystals of a mutant kinase (KIN-D445N) from a mixture of KIN-D445N, GTP and magnesium. The mutant is inactive as a kinase because the Asp445 general base that deprotonates the 5′-OH RNA end for its attack on the GTP γ phosphate cannot perform this function when changed to an isosteric asparagine (28). The KIN+GTP crystals were in space group P212121 and had one KIN protomer in the asymmetric unit. The structure was refined to a resolution of 1.85 Å (Rwork/Rfree 0.170/0.215) (Supplementary Table S1). The electron density clearly revealed the presence of GTP and magnesium in the kinase active site (Supplementary Figure S3). The tertiary structure of the KIN-GTP complex is shown in Figure 4A. We also grew crystals of KIN-D445N in the absence of added nucleotide. The apoenzyme crystallized in space group C2221 and had two KIN protomers in the asymmetric unit. The apoenzyme structure was refined to a resolution of 1.98 Å (Rwork/Rfree 0.193/0.236). Whereas neither apoenzyme protomer had a nucleotide in the active site, both protomers had a phosphate anion in the active site (derived from the 300 mM phosphate in the precipitant solution used for crystallization) at the position corresponding to the β phosphate of GTP or GDP. Figure 4A shows a superposition of the KIN apoenzyme B protomer on the KIN•GTP structure. Pairwise alignment of the apo and GTP-bound KIN domains in DALI returned a Z score of 31.8 with a rmsd of 1.5 Å at 208 Cα positions.
Figure 4.
Structures of the KIN•GTP•Mg2+ complex and KIN apoenzyme. (A) Stereo view of the superimposed tertiary structures of the KIN•GTP•Mg2+ complex (colored gold) and the KIN apoenzyme (colored cyan). The seven α helices flanking the central β sheet are numbered sequentially. GDP is depicted as a stick model with yellow phosphorus atoms. Magnesium is depicted as a magenta sphere. The phosphate anion in the KIN apoenzyme is shown as a stick model with a green phosphorus atom. (B) Stereo view of the KIN active site highlighting interactions with GTP•Mg2+. Amino acids are shown as stick models with beige carbons. Magnesium and waters are denoted by magenta and red spheres, respectively. Atomic contacts are indicated by dashed lines. (C) Stereo view of the equivalent active site region of the KIN apoenzyme, highlighting a conformational switch in the G-loop segment.
The KIN fold comprises a central 5-strand β sheet surrounded by seven α helices (Figure 4A). The GTP phosphates are nestled into the P-loop (422GCGKTT427) (Figure 4A and B). The α4 and α5 helices form a ‘lid’ over the GTP phosphates, wherein the lid-loop that connects the lid helices is disordered, in both the KIN apoenzyme and KIN•GTP•Mg2+ structures (Figure 4A) as it was in the KIN•GDP•Mg2+ structure reported previously (28). Comparison of the KIN apoenzyme and KIN•GTP•Mg2+ structures reveals shifts in the positions of the lid helices (α4 and α5) and of α7 that accompany nucleotide binding (Figure 4A). Most striking, however, is the complete remodeling of the conformation of the G-loop element in apoenzyme versus KIN•GTP. The upshot of this difference (described below) is that the apoenzyme structure is incompatible with GTP binding and thus a conformational switch is necessary to mobilize and properly position the NTP specificity determinants resident in the G-loop.
KIN•GTP active site
Figure 4B shows a detailed stereo view of the atomic interactions with the GTP nucleotide in the KIN active site. As in the previous KIN•GDP•Mg2+ structure (28), the guanine nucleobase is engaged by a network of hydrogen bonds from G-loop amino acids Asp583 (to guanine N1 and N2 from the Asp carboxylate) and Ser590 (to guanine O6 and N7, from the main chain amide and Oγ, respectively). The Arg528 side chain of the lid α4 helix makes a π-cation stack on the guanine base. Arg528 also makes a hydrogen bond to the G-loop Ser589 side chain. There are no enzymic contacts to the ribose sugar in the KIN•GTP•Mg2+ complex. Main chain amide nitrogens of the 422GCGKTT427 P-loop donate hydrogen bonds to the α and β phosphates of GTP. The Thr427 hydroxyl donates a hydrogen bond to the GTP α phosphate. The Lys425 side chain makes a bifurcated ionic interaction with the β and γ phosphates. Arg528 of the lid-loop also engages the GTP γ phosphate (Figure 4B). Arg528 was disordered in the previous KIN•GDP structure (28) and is also not visualized in the present KIN apoenzyme structure (Figure 4C), suggesting that this arginine at the start of the lid-loop becomes ordered when it can engage the γ phosphate. Two of the sites in the octahedral coordination complex of the Mg2+ ion are occupied by nonbridging β and γ phosphate oxygens of GTP. The other sites are filled by the Thr426 Oγ atom and three waters (Figure 4B). These features of the GTP•Mg2+ substrate complex in the phosphate donor site suggest a mechanism whereby Lys425, Arg532, and the metal complex orient the γ phosphate for nucleophilic attack by the polynucleotide 5′-OH and stabilize the extra negative charge developed in the pentacoordinate transition state during an associative in-line phosphoryl transfer.
Conformational switch in the G-loop
A stereo view of the phosphate donor site fold in the KIN apoenzyme structure is shown in Figure 4C, onto which the guanine base from the KIN•GTP•Mg2+ structure is superimposed. The 579KLSKDENSLKSS590 segment that comprises the G-loop in the nucleotide-bound KIN structures includes only two amino acids that adopt a secondary structure: Ser589 and Ser590 at the outset of the α6 helix. By contrast, in the KIN apoenzyme, the α6 helix is extended proximally into the erstwhile G-loop so that the helix initiates with 585NS—i.e. four amino acids upstream of its onset in the GTP/GDP-bound state—and the conformation/trajectory of the residual preceding loop is completely different (Figure 4A). Thereby, the Asp583 side chain that confers GTP specificity is pointed away from the guanosine binding site and the extended α6 helix occludes the site occupied by the guanine base in the KIN•GTP•Mg2+ structure (Figure 4C). We surmise that a conformational switch entailing unfolding of the proximal α6 helix segment accompanies, and is necessary for, engagement of the GTP phosphate donor.
KIN structures in complex with inosine and deoxyguanosine nucleotides
By testing a series of purine ribonucleoside triphosphate analogs as substrates for HORNAp phosphorylation by Candida Trl1 KIN (28), it was found that inosine triphosphate (ITP) was the most effective substrate, implying that the exocyclic 2-NH2 moiety of GTP is not an essential determinant of nucleobase recognition per se. By contrast, 2-aminopurine triphosphate was a much less effective substrate for the kinase reaction. These results highlight the importance of the 6-oxo atom of guanine for phosphate donor function. To better understand base recognition by Candida KIN, we grew crystals from a mixture of KIN-D445N, ITP, and magnesium. The KIN + ITP crystals were in space group P212121 and had one KIN protomer in the asymmetric unit. The structure was refined to a resolution of 1.53 Å (Rwork/Rfree 0.174/0.208) (Supplementary Table S1). The active site contained inosine diphosphate (IDP) and a phosphate anion but did not include a magnesium ion (Figure 5A). We surmise that the input ITP ligand was hydrolyzed to IDP in crystallo [see Discussion for why this might be] and that the metal ion dissociated from the enzyme; the retention of a phosphate anion adjacent to, but clearly distinct from, the IDP β phosphate is presumably driven by the presence of 150 mM phosphate in the precipitant solution used to grow the crystals. In the KIN•IDP structure, Arg528 of the lid α4 helix makes a π-cation stack on the inosine base and Ser590 of the G-loop makes hydrogen bonds to the inosine N7 and O6 atoms (Figure 5A), just as in the KIN•GTP•Mg2+ structure (Figure 3B). However, the G-loop has shifted slightly (by 3.5 Å at the Asn585 Cα atom and 1.7 Å at the Asp583 Cα atom) and the Asp583 side chain now makes a single hydrogen bond to the inosine N1 atom (Figure 5A). The P-loop Lys425 side chain makes a bifurcated contact to the IDP β phosphate and the phosphate anion, analogous to the β–γ phosphate-bridging contact seen in the KIN•GTP•Mg2+ complex. Absent a metal, the P-loop Thr426 side chain donates a hydrogen bond to the IDP β phosphate. A notable feature of the KIN•IDP structure is that the 532RGDNHQSIK540 lid-loop peptide is visible and in contact with the IDP nucleotide and phosphate anion in a surprising fashion (Figure 5A). In particular, Arg532, which contacts the γ phosphate in the GTP•Mg2+ complex, is now oriented in the opposite direction and makes no contacts. The main chain carbonyls of Arg532 and Gly533 make hydrogen bonds to the IDP ribose 3′-OH. The Asp534 carboxylate is within close hydrogen-bonding distance of the IDP β phosphate (Figure 5A), counter to the expectation that there should be charge repulsion between these two moieties. The plausible explanation is that pH of the precipitant solution used to grow the crystals is 4.7. At this acidic pH, the IDP β phosphate is expected to be a monoanion and the Asp534 carboxylate is likely to be protonated. His536 makes bifurcated hydrogen bonds to the phosphate.
Figure 5.
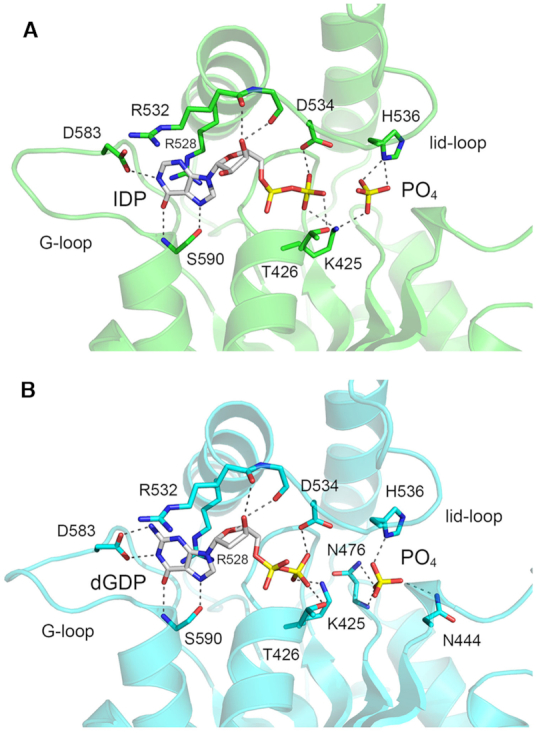
Structures of KIN in complexes with IDP and dGDP. The atomic interactions with IDP (panel A; KIN domain colored green) and dGDP (panel B; KIN domain colored cyan) and phosphate anions near the nucleotide β phosphates are indicated by dashed lines. The lid-loop is visible in both structures.
We also grew crystals from a mixture of KIN-D445N, dGTP, and magnesium under the same conditions used for the inosine nucleotide experiment. The structure was refined to a resolution of 1.7 Å (Rwork/Rfree 0.180/0.209) (Supplementary Table S1). The active site contained dGDP and a phosphate anion, but no magnesium (Figure 5B). The G-loop conformation and nucleobase contacts in the KIN•dGDP structure were identical to that of the KIN•GTP•Mg2+ structure. The lid-loop was visible and made contacts to dGDP similar to those seen in the KIN•IDP structure. The phosphate anion in the KIN•GDP complex was coordinated to His536, Asn476, and Asn444 (Figure 5B).
Mutational analysis of Candida Trl1 KIN
The C. albicans422GCGKTT427 P-loop and 532RGDNHQSIK540 lid-loop sequences in the KIN domain are well conserved among fungal Trl1 proteins (8). The corresponding sequences in S. cerevisiae Trl1 are 401GCGKTT406 and 511RGNNHQSIK519, respectively. Previous mutational analysis of the S. cerevisiae Trl1 P-loop showed that single-alanine substitutions for Lys404 and Thr405 had no effect on vegetative growth and little effect on GTP-dependent RNA kinase activity in vitro, whereas a K404A-T405A double-mutation was lethal and abolished GTP-dependent RNA kinase activity (6). By contrast a single alanine mutation of the P-loop Lys425 residue of C. albicans Trl1 was lethal in vivo (as gauged by trl1Δ complementation in S. cerevisiae) and eliminated GTP-dependent RNA kinase activity in vitro (28). Alanine scanning of the S. cerevisiae Trl1 lid-loop showed that Arg511 and His515 are essential for Trl1 function in vivo (6,8). Replacing Arg511 with lysine or glutamine was lethal in vivo, as was replacing His515 with either glutamine or asparagine (8). By contrast, alanine substitutions for S. cerevisiae lid-loop residues Asn513, Asn514, Gln516 and Ser517 had no apparent effect on cell growth (8). In light of the divergent effects of the P-loop lysine mutation on Saccharomyces versus Candida Trl1 and the conformational variability of the lid-loop in the available suite of Candida Trl1 KIN structures, we mutated the Candida P-loop threonines adjacent to Lys425 and we mutated components of the Candida lid loop. We changed P-loop Thr426 (which coordinates the magnesium cofactor in the GTP•Mg2+ complex) and Thr427 (which engages the α phosphate in all of the nucleotide-bound KIN structures) to alanine. We introduced alanine in lieu of lid-loop residues Arg532 (which contacts the γ phosphate in the GTP•Mg2+ complex), Asp534 (which contacts the β phosphate in the IDP and dGDP complexes), Asn535, His536 (which contacts the phosphate anion in the IDP and dGDP complexes), and Gln537. These alanine changes were made in the context of the full-length Candida Trl1 protein encoded on a CEN LEU2 plasmid under the transcriptional control of the S. cerevisiae TPI1 promoter (28). We then tested the CalTRL1-Ala alleles by plasmid shuffle for complementation of an S. cerevisiae trl1Δ p[CEN URA3 SceTRL1] strain (6).
The Candida T426A allele was lethal in vivo, i.e., Leu+trl1Δ CalTRL1-T426A transformants were unable to form colonies after incubation for 8 days at 20, 30, or 37°C on agar medium containing 5-fluoroorotic acid (FOA), a drug that selects against the p[CEN URA3 SceTRL1] plasmid. Thus, the metal coordinating P-loop threonine is essential for Candida Tr1 kinase activity in vivo. The other seven alleles yielded viable FOA-resistant trl1Δ CalTRL1-Ala isolates. These strains were tested by serial dilution for growth on YPD agar medium at 30°C and 37°C in parallel with a trl1Δ wild-type CalTRL1 control. The P-loop T427A and lid-loop N476A, D534A, N535A, H536A and Q537A mutant strains grew as well as wild-type at both temperatures, as gauged by colony size (Figure 6). By contrast, the R532A strain was severely defective for growth, forming pinpoint colonies at 30°C and 37°C (Figure 6). The importance of Arg532 for Candida Trl1 activity is in keeping with our inferences from the KIN•GTP•Mg2+ structure that the contacts of Arg532 with the GTP γ phosphate are on-pathway in the kinase reaction. Conversely, the dispensability of Asp534 for Trl1 function suggests that its β phosphate contacts in the IDP and dGDP complexes are off-pathway.
Figure 6.
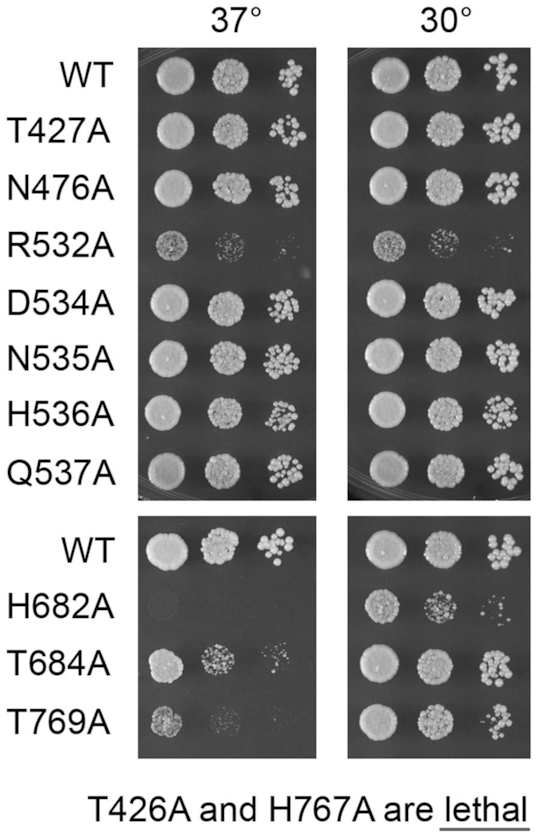
Effects of KIN and CPD mutations on Candida Trl1 activity in vivo. S. cerevisiae trl1Δ p[CEN LEU2 CalTRL1] strains with CalTRL1 alleles as specified on the left were grown in YPD liquid culture at 30°C and adjusted to the same A600. Serial 10-fold dilutions were spotted to YPD agar and incubated at the 30°C and 37°C. Photographs of the plates are shown. Lethal CalTRL1-Ala alleles that failed to complement trl1Δ are indicated at the bottom.
Mutational analysis of Candida Trl1 CPD
The His682, Thr684, His767 and Thr769 residues of the signature HxT motifs that engage the active site phosphate were replaced individually by alanine in the context of full-length Candida Trl1 and the CalTrl1-Ala mutants were tested for trl1Δ complementation in S. cerevisiae. The H767A mutation was lethal. The H682A strain was sick at 30°C and unable to grow at 37°C (Figure 6). The T684A and T769A strains grew fairly well at 30°C but displayed ts growth defects at 37°C, with T769A being less fit than T684A (Figure 6). The effects of mutating the Candida HxT histidines were in accord with previous findings for S. cerevisiae Trl1 and the plant tRNA ligase AtRNL (a Trl1 ortholog that complements S. cerevisiae trl1Δ), i.e. changing the proximal HxT histidine to alanine elicited a ts growth phenotype whereas alanine mutation of the distal histidine was lethal (6,8). Different phenotypes were seen for the SceTrl1 and AtRNL HxT threonine-to-alanine mutations vis-à-vis the Candida enzyme: the proximal threonine mutation in SceTrl1 and AtRNL was lethal while the distal threonine SceTrl1 and AtRNL mutant strains grew as well as wild-type at 30°C and 37°C (8).
DISCUSSION
The KIN-CPD structure presented here is, to our knowledge, the first instance of an atomic structure of the RNA 2′,3′-cyclic phosphodiesterase component of fungal tRNA ligase. With this accomplished, we now have exemplary structures in hand for all four enzymes that catalyze RNA repair during fungal tRNA splicing (28,33,34). The present study also extends our knowledge of the NTP donor specificity of the RNA kinase module of fungal tRNA ligase and reveals conformational changes in the G-loop and lid-loop of the KIN domain triggered by guanine nucleotide binding. Our findings bolster the case for tRNA splicing as an antifungal drug target.
CPD structure
It has been thought for some time, on the basis of its tandem HxT motifs, that the CPD domains of fungal and plant tRNA ligases belong to the 2H phosphoesterase superfamily (45). This classification was fortified by the findings that certain mutations of the signature HxT motifs drastically reduce CPD activity in vitro and are lethal in vivo when tested for complementation of S. cerevisiae trl1Δ (6,8,9,46). The crystal structure of Candida Trl1 CPD affirms its membership in the 2H superfamily by virtue of the conservation of the fold of the central concave β sheet and the atomic interactions of the HxT motif histidines and threonines with a phosphate anion in the active site, which we regard as a mimetic of the scissile RNA 2′,3′-cyclic phosphate. That said, the degree of amino acid identity between Trl1 CPD and other 2H family members is low and the Trl1 CPD diverges from other 2H enzymes with respect to structural elements outside the active site. On the other hand, we discern three active site motifs in the C. albicans CPD that are well conserved in the CPD domains of tRNA ligases from multiple other human fungal pathogens. Conserved residues in these motifs make water-mediated contacts to the phosphate in the CPD crystal structure or are imputed to interact with the terminal nucleoside of the RNA substrate. The array of hydrogen-bonding residues and several aromatic residues in the CPD active site (and the relative paucity of charged basic and acid residues) could provide a pocket for the binding of a small-molecule CPD inhibitor.
A striking feature of the KIN-CPD structure is that it adopts an extended conformation in which the KIN and CPD domains are far apart and connected by an unstructured peptide linker. It is possible that this particular extended conformation is a matter of crystal packing and that the domain arrangement of KIN-CPD in solution is flexible about the linker peptide. Although the KIN-CPD structure does not suggest a specific and stable interface between KIN and CPD, we do not exclude the prospect that an interdomain interface exists in Trl1 that is dependent on the presence of the LIG domain. A previous study employing bifunctional lysine chemical crosslinking agents to probe at low resolution the fold of the CPD domain of Coccidioides immitis Trl1 (10) identified a network of four lysine-lysine crosslinks involving four amino acids (Lys635, Lys693, Lys703 and Lys707 in C. immitis CPD) that are conserved in C. albicans CPD (as Lys642 in the linker; Lys692 in the second 310 helix; Lys702 in helix α3; and Lys706 in helix α3). These residues are indeed in physical proximity in the Candida CPD tertiary structure (Supplementary Figure S4) at atomic distances (9–12 Å) consistent with the chemical crosslinking results.
KIN substrate recognition and conformational switches
We suggested previously (28) that the peculiar G-loop and guanine-binding pocket of Candida KIN present a plausible target for a small molecule that could selectively inhibit the fungal Trl1 kinase without affecting the many other P-loop superfamily phosphotransferases that do not share the guanine specificity of the fungal Trl1 kinase domains. The structures solved presently of KIN in complexes with deoxyguanosine and inosine nucleotides underscore that: (i) the enzyme does not discriminate between ribose and deoxyribose sugars in the phosphate donor; (ii) the exocyclic –NH2 of guanine is not essential for occupancy of the G-loop by inosine, which still makes a hydrogen bond from N1 to G-loop Asp583; and (iii) the purine O6 atom is the key determinant of substrate recognition via its hydrogen bonding interactions with Ser590, which is essential for KIN activity (28). The notable finding here was that in the structure of the nucleotide-free KIN apoenzyme, the conformation of the erstwhile G-loop is rearranged such that the guanine-binding pocket is occluded. We surmise that the ordered N-terminal segment of the α6 helix of the apoenzyme must become disordered upon GTP binding and the preceding loop is reshaped accordingly to form the G-loop pocket. This situation raises the prospect that a small molecule that binds to the apo state of the G-loop segment might act as conformational trap to inhibit the kinase.
The present suite of nucleotide-bound KIN structures also highlights the conformational flexibility of the lid including: (i) a ‘closing’ movement of the lid α4 and α5 helices over the nucleotide in the transition from apoenzyme to GTP•Mg2+ complex; and (ii) conformational variability of the lid-loop ranging from: entirely disordered (apoenzyme and GDP•Mg2+ complexes); to a state in which Arg532 at the margin of the disordered loop interacts with the GTP γ phosphate (GTP•Mg2+ complex); to a situation in which the loop is visible in the magnesium-free dGDP and IDP complexes while making contacts to these nucleotides that we suspect are off-pathway (owing to the acidic crystallization conditions that favor the atypical Asp534 hydrogen bond to the NDP β phosphate). Mutagenesis of the KIN domain lid-loop fortifies our view that the interactions of Arg532 with the GTP γ phosphate in the GTP•Mg2+ complex are indeed on-pathway, i.e. an R532A mutation compromises (but does not abolish) Trl1 function in vivo. The corresponding P-loop arginine in the kinase domain of the bacterial RNA repair enzyme Pnkp makes analogous contact to the γ phosphate of its NTP substrate; mutating this Pnkp arginine (Arg120) to alanine reduced the kinase specific activity to 42% of the wild-type level (47). Comparing the effects of mutations at equivalent positions in the KIN domains of S. cerevisiae and C. albicans Trl1 enzymes indicates that the contacts of the individual loop and lid-loop amino acids can vary in their importance. For example, whereas the P-loop lysine and metal-binding threonine are both individually essential in C. albicans Trl1 (ref. 28 and Figure 6), equivalent single-alanine P-loop mutants in S. cerevisiae Trl1 are viable – though a double-mutation of Lys-Thr to Ala-Ala is lethal in S. cerevisiae (6). Alanines in lieu of the lid-loop arginine and histidine side chains are lethal in S. cerevisiae; the equivalent arginine mutant in C. albicans Trl1 is sick but alive and the histidine mutation is apparently benign. The take-home lesson is that structure-activity relations are not precisely transferrable between fungal Trl1 kinase orthologs.
Our use of the KIN general acid mutant D445N in the present structural studies was prompted by a desire to avoid hydrolysis of the input NTP substrate that was evident in the initial crystallization of wild-type KIN in the presence of GTP and magnesium (28). This maneuver clearly did not prevent NTP hydrolysis, insofar as several of the structures determined here had NDP in the KIN active site. It is conceivable that NTP hydrolysis by the KIN in the presence of magnesium is influenced by the acidic condition that favored successful crystallization. However the key parameter anent NTP hydrolysis seems to be the duration of the crystallization experiment: to wit, all of the crystals that contained NDP in the active site were harvested after 4–5 days, whereas the GTP•Mg2+ structure was obtained from a crystal that was harvested immediately after overnight growth. Notwithstanding that the electron density map clearly reveals the GTP•Mg2+ complex, we suspect that a fraction of the KIN enzymes in the crystal may have hydrolyzed GTP to GDP, insofar as the B factor of the GTP γ phosphorus atom (=51) was higher than the B factors of the α and β phosphorus atoms (=25).
The available LIG, KIN, and CPD structures of fungal tRNA ligase each have mechanistically instructive ligands in their respective active sites. The outstanding questions at this point concern how each catalytic module recognizes the specific RNA substrate on which it acts: RNA>p for CPD; HORNA for KIN, and pRNA2′p for LIG. Our attempts to date to crystallize KIN-D445N in the presence of GTP, magnesium, and a 5′-OH oligonucleotide to capture a Michaelis complex of the kinase reaction (a strategy that was successful with Clostridium thermocellum polynucleotide kinase; 48) have not been fruitful, insofar as the conditions that led to crystals (none of which contained the oligonucleotide) were the same as those that yielded KIN•nucleotide complexes, i.e., it is likely that high concentrations of phosphate and/or acidic pH were inimical to binding of the nucleic acid substrate to KIN (or of 2′-AMP to CPD). Still, the available Trl1 domain structures do suggest that it might be possible to inhibit multiple enzymatic steps in the fungal tRNA splicing pathway, which, if achieved, would reduce the risk of the emergence of drug resistance. The case for Trl1 as a drug target for Candidiasis is bolstered by the recent identification of TRL1 as an essential gene in C. albicans (49).
DATA AVAILABILITY
Structural coordinates have been deposited in Protein Data Bank under accession codes 6U05 (KIN-CPD), 6U00 (KIN-D455N-apo), 6U03 (KIN-D445N•GTP•Mg), 6TZX (KIN-D445N-IDP), 6TZ0 (KIN-D445N•dGDP) and 6TZM (KIN-D445N•GDP•Mg).
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R35-GM126945 to S.S.]; Geoffrey Beene Cancer Research Center (to S.S.); Deutsche Forschungsgemeinschaft [394320208 to A.B.]; MSKCC structural biology core laboratory is supported by National Cancer Institute [P30-CA008748]; X-ray diffraction data were collected at synchrotron facilities supported by grants and contracts from the National Institutes of Health [P41GM103403, HEI-S10RR029205]; Department of Energy [DE-AC02-06CH11357]. Funding for open access charge: National Institutes of Health [R35-GM126945].
Conflict of interest statement. None declared.
REFERENCES
- 1. Montoya M.C., Moye-Rowley W.S., Krysan D.J.. Candida auris: the canary in the mine of antifungal drug resistance. ACS Infect. Dis. 2019; 5:1487–1492. [DOI] [PubMed] [Google Scholar]
- 2. Abelson J., Trotta C.R., Li H.. tRNA splicing. J. Biol. Chem. 1998; 273:12685–12688. [DOI] [PubMed] [Google Scholar]
- 3. Sidrauski C., Cox J.S., Walter P.. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996; 87:405–413. [DOI] [PubMed] [Google Scholar]
- 4. Greer C.L., Peebles C.L., Gegenheimer P., Abelson J.. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983; 32:537–546. [DOI] [PubMed] [Google Scholar]
- 5. Apostol B.L., Westaway S.K., Abelson J., Greer C.L.. Deletion analysis of a multifunctional yeast tRNA ligase polypeptide: identification of essential and dispensable functional domains. J. Biol. Chem. 1991; 266:7445–7455. [PubMed] [Google Scholar]
- 6. Sawaya R., Schwer B., Shuman S.. Genetic and biochemical analysis of the functional domains of yeast tRNA ligase. J. Biol. Chem. 2003; 278:43298–43398. [DOI] [PubMed] [Google Scholar]
- 7. Wang L.K., Shuman S.. Structure-function analysis of yeast tRNA ligase. RNA. 2005; 11:966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang L.K, Schwer B., Englert M., Beier H., Shuman S.. Structure-function analysis of the kinase-CPD domain of yeast tRNA ligase (Trl1) and requirements for complementation of tRNA splicing by a plant Trl1 homolog. Nucleic Acids Res. 2006; 34:517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Remus B.S., Shuman S.. Distinctive kinetics and substrate specificities of plant and fungal tRNA ligases. RNA. 2014; 20:462–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Remus B.S., Schwer B., Shuman S.. Characterization of the tRNA ligases of pathogenic fungi Aspergillus fumigatus and Coccidioidesimmitis. RNA. 2016; 22:1500–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Culver G.M., McCraith S.M., Zillman M., Kierzek R., Michaud N., LaReau R.D., Turner D.H., Phizicky E.M.. An NAD derivative produced during transfer RNA splicing: ADP-ribose 1“-2” cyclic phosphate. Science. 1993; 261:206–208. [DOI] [PubMed] [Google Scholar]
- 12. Culver G.M., McCraith S.M., Consaul S.A., Stanford D.R., Phizicky E.M.. A 2′-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J. Biol. Chem. 1997; 272:13203–13210. [DOI] [PubMed] [Google Scholar]
- 13. Spinelli S.L., Consaul S.A., Phizicky E.M.. A conditional lethal yeast phosphotransferase mutant accumulates tRNA with a 2′-phosphate and an unmodified base at the splice junction. RNA. 1997; 3:1388–1400. [PMC free article] [PubMed] [Google Scholar]
- 14. Spinelli S.L., Kierzek R., Turner D.H., Phizicky E.M.. Transient ADP-ribosylation of a 2′-phosphate implicated in its removal from ligated tRNA during splicing in yeast. J. Biol. Chem. 1999; 274:2637–2644. [DOI] [PubMed] [Google Scholar]
- 15. Steiger M.A., Kierzek R., Turner D.H., Phizicky E.M.. Substrate recognition by a yeast 2′-phosphotransferase involved in tRNA splicing and its Escherichia coli homolog. Biochemistry. 2001; 40:14098–14105. [DOI] [PubMed] [Google Scholar]
- 16. Steiger M.A., Jackman J.E., Phizicky E.M.. Analysis of 2′-phosphotransferase (Tpt1p) from Saccharomyces cerevisiae: evidence for a conserved two-step reaction mechanism. RNA. 2005; 11:99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Munir A., Abdullahu L., Damha M.J., Shuman S.. Two-step mechanism and step-arrest mutants of Runellaslithyformis NAD+-dependent tRNA 2′-phosphotransferase Tpt1. RNA. 2018; 24:1144–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tanaka N., Shuman S. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 2011; 286:7727–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tanaka N., Chakravarty A.K., Maughan B., Shuman S.. A novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligation reactions. J. Biol. Chem. 2011; 286:43134–43143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tanaka N., Meineke B., Shuman S.. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J. Biol. Chem. 2011; 286:30253–30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chakravarty A.K., Subbotin R., Chait BT., Shuman S.. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:6072–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maughan W.P., Shuman S.. Distinct contributions of enzymic functional groups to the 2′,3′-cyclic phosphodiesterase, 3′-phosphate guanylylation, and 3′-ppG/5′-OH ligation steps of the Escherichia coli RtcB nucleic acid splicing pathway. J. Bacteriol. 2016; 198:1294–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Englert M., Xia S., Okada C., Nakamura A., Tanavde V., Yao M., Eom S.H., Koningsberg W.H., Söll D., Wang J.. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:15235–15240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Desai K.K., Bingman C.A., Phillips G.N., Raines R.T.. Structure of the noncanonical RNA ligase RtcB reveal the mechanism of histidine guanylylation. Biochemistry. 2013; 52:2518–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Popow J., Englert M., Weitzer S., Schleiffer A., Mierzwa B., Mechtler K., Trowitzsch S., Will C.L., Lürhmann R., Söll D. et al.. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011; 331:760–764. [DOI] [PubMed] [Google Scholar]
- 26. Kosmaczewski S.G., Edwards T.J., Han S.M., Eckwahl M.J., Meyer B.I., Peach S., Hesselberth J.R., Wolin S.L., Hammarlund M.. The RtcB RNA ligase is an essential component of the metazoan unfolded protein response. EMBO Rep. 2014; 15:1278–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Westaway S.K., Belford H.G., Apostol B.L., Abelson J., Greer C.L.. Novel activity of a yeast ligase deletion polypeptide: evidence for GTP-dependent tRNA splicing. J. Biol. Chem. 1993; 268:2435–2443. [PubMed] [Google Scholar]
- 28. Remus B.S., Goldgur Y., Shuman S.. Structural basis for the GTP specificity of the RNA kinase domain of fungal tRNA ligase. Nucleic Acids Res. 2017; 45:12945–12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Braun P.E., Lee J., Gravel M.. 2′,3′-Cyclic nucleotide 3′-phosphodiesterase: structure, biology, and function. Myelin Biol. Disorders. 2004; 1:499–522. [Google Scholar]
- 30. Schwer B., Aronova A., Ramirez A., Braun P., Shuman S.. Mammalian 2′,3′ cyclic nucleotide phosphodiesterase (CNP) can function as a tRNA splicing enzyme in vivo. RNA. 2008; 14:204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lappe-Siefke C., Goebbels S., Gravel M., Nicksh E., Lee J., Braun P.E., Griffiths I.R., Nave K.A.. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat. Genet. 2003; 33:366–374. [DOI] [PubMed] [Google Scholar]
- 32. Harding H.P., Lackey J.G., Hsu H.C., Zhang Y., Deng J., Xu R.M., Damha M.J., Ron D.. An intact unfolded protein response in Trpt1 knockout mice reveals phylogenic divergence in pathways for RNA ligation. RNA. 2008; 14:225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Banerjee A., Ghosh S., Goldgur Y., Shuman S.. Structure and two-metal mechanism of fungal tRNA ligase. Nucleic Acids Res. 2019; 47:1428–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Banerjee A., Munir A., Abdullahu L., Damha M.J., Goldgur Y., Shuman S.. Structure of tRNA splicing enzyme Tpt1 illuminates the mechanism of RNA 2′-PO4 recognition and ADP-ribosylation. Nat. Comm. 2019; 10:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 36. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010; D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abrahams J.P., Leslie A.G.W.. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Cryst. 1996; D52:30–42. [DOI] [PubMed] [Google Scholar]
- 38. Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M.. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991; A47:110–119. [DOI] [PubMed] [Google Scholar]
- 39. Holm L., Kaariainen S., Rosenstrom P., Schenkel A.. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008; 24:1780–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Myllykoski M., Raasakka A., Han H., Kursula P.. Myelin 2′,3′-cyclic nucleotide 3′-phosphodiesterase: active-site ligand binding and molecular conformation. PLoS ONE. 2012; 7:e32336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kozlov G., Denisov A.Y., Pomerantseva E., Gravel M., Braun P.E., Gehring K.. Solution structure of the catalytic domain of RICH protein from goldfish. FEBS J. 2007; 274:1600–1609. [DOI] [PubMed] [Google Scholar]
- 42. Remus B.S., Jacewicz A., Shuman S.. Structure and mechanism of E. coli RNA 2′,3′-cyclic phosphodiesterase. RNA. 2014; 20:1697–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brandmann T., Jinek M.. Crystal structure of the C-terminal 2′,5′-phosphodiesterase domain of group A rotavirus protein VP3. Proteins. 2015; 83:997–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hofmann A., Zdanov A., Genschik P., Ruvinov S., Filipowicz W., Wlodawer A.. Structure and mechanism of activity of the cyclic phosphodiesterase of Appr>p, a product of the tRNA splicing reaction. EMBO J. 2000; 19:6207–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mazumder R, Iyer L.M., Vasudevan S., Aravind L.. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res. 2002; 30:5229–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Remus B.S., Shuman S.. A kinetic framework for tRNA ligase and enforcement of a 2′-phosphate requirement for ligation highlights the design logic of an RNA repair machine. RNA. 2013; 19:659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Das U., Wang L.K., Smith P., Shuman S.. Structural and biochemical analysis of the phosphate donor specificity of the polynucleotide kinase component of the bacterial Pnkp•Hen1 RNA repair system. Biochemistry. 2013; 52:4734–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Das U., Wang L.K., Smith P., Jacewicz A., Shuman S.. Structures of bacterial polynucleotide kinase in a Michaelis complex with GTP•Mg2+ and 5′-OH oligonucleotide and a product complex with GDP•Mg2+ and 5′-PO4 oligonucleotide reveal a mechanism of general acid-base catalysis and the determinants of phosphoacceptor recognition. Nucleic Acids Res. 2014; 42:1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Segal E.S., Gritsenko V., Levitan A., Yadav B., Dror N., Steenwyk J.L., Silberberg Y., Meilich K., Rokas A., Gow N.A. et al.. Gene essentiality analyzed by in vivo transposon mutagenesis and machine learning in a stable haploid isolate of Candidaalbicans. mBio. 2018; 9:e02048-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural coordinates have been deposited in Protein Data Bank under accession codes 6U05 (KIN-CPD), 6U00 (KIN-D455N-apo), 6U03 (KIN-D445N•GTP•Mg), 6TZX (KIN-D445N-IDP), 6TZ0 (KIN-D445N•dGDP) and 6TZM (KIN-D445N•GDP•Mg).