Abstract
Exon skipping (ES) is reported to be the most common alternative splicing event due to loss of functional domains/sites or shifting of the open reading frame (ORF), leading to a variety of human diseases and considered therapeutic targets. To date, systematic and intensive annotations of ES events based on the skipped exon units in cancer and normal tissues are not available. Here, we built ExonSkipDB, the ES annotation database available at https://ccsm.uth.edu/ExonSkipDB/, aiming to provide a resource and reference for functional annotation of ES events in multiple cancer and tissues to identify therapeutically targetable genes in individual exon units. We collected 14 272 genes that have 90 616 and 89 845 ES events across 33 cancer types and 31 normal tissues from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx). For the ES events, we performed multiple functional annotations. These include ORF assignment of exon skipped transcript, studies of lost protein functional features due to ES events, and studies of exon skipping events associated with mutations and methylations based on multi-omics evidence. ExonSkipDB will be a unique resource for cancer and drug research communities to identify therapeutically targetable exon skipping events.
INTRODUCTION
Accumulated evidence has shown that the disruption of alternative splicing (AS) contributes to human diseases (1). Among the typical patterns of alternative splicing, exon skipping (ES) is the most common event (2). Since ES results in the loss of functional domains/sites or frame shifting of the open reading frame (ORF), skipped exons have been used as therapeutic targets (3–8). For example, MET has lost the binding site of E3 ubiquitin ligase CBL through exon 14 skipping event (9), resulting in an enhanced expression level of MET. MET amplification drives the proliferation of tumor cells. Multiple tyrosine kinase inhibitors, such as crizotinib, cabozantinib and capmatinib, have been used to treat patients with MET exon 14 skipping (10). Another example is the dystrophin gene (DMD) in Duchenne muscular dystrophy (DMD), a progressive neuromuscular disorder. To restore dystrophin protein functions that were lost due to frame shifting through ES, multi-exon skipping antisense oligonucleotides (ASOs) have been used for DMD treatment (11). Therefore, systematic identification and intensive analyses of ES events in pan-cancer and healthy tissues will provide important insights into disease mechanisms and identify novel cancer type-specific therapeutic targets.
With the recent exponential growth of cancer genomic and other biomedical data, several studies have analyzed alternative splicing events in multiple cancer or tissue types (e.g. pan-cancer studies) (12,13). There exist several web tools providing pan-cancer AS annotations such as TCGA SpliceSeq (14), TSVdb (15) and CAS-viewer (16). However, these studies focused on the identification of alternative splicing events and visualization of isoform structures based on known gene structures. Furthermore, these studies did not present detailed functional impacts of AS events. So far, a systematic and intensive annotation of ES events based on the skipped exon units in cancer and normal tissues regarding their functional impacts has not been available. Here, we built ExonSkipDB, the ES annotation database, aiming to provide resources and references for functional annotation of ES events in cancer to identify therapeutically targetable exons.
To achieve this goal, we first collected ES event information of total 14 272 genes from 33 cancer types and 31 normal tissues of the Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) from Kahles et al.’s study (12). These genes contain 90 616 and 89 845 ES events of TCGA and GTEx, respectively. For these ∼14K genes, we performed functional annotations including investigation of the open reading frame (ORF) for individual ES events and lost protein functional features of in-frame ES events based-on the canonical transcript sequences and amino acid sequences, respectively. We also analyzed mutations and methylations associated with ES events. Users can access a variety of annotations, including gene summaries, gene structures such as gene isoform and ES events from TCGA and GTEx, landscape of percent spliced in (PSI) and isoform abundances across multiple human tissues and cancers, ORF annotation, analysis of lost protein functional features, skipped exon region mutations with coverage depth and differential PSIs, sQTL and sQTM analyses, and ES gene-related drugs and diseases. This paper introduces ExonSkipDB, the web interface, and its applications, aiming to provide resources or references for functional annotation of exon skipping events in cancer. ExonSkipDB will be a unique resource for cancer and drug research communities to identify disease-associated exon skipping events.
DATABASE OVERVIEW
Through multiple annotations of the ExonSkipDB, users can get help from the following aspects. (i) Comparing ES events with PSIs and isoform abundance between pan-cancer and healthy human tissues can help detect potential cancer or cancer type-specific ES events. Through comparing 90 616 and 89 845 ES events from TCGA and GTEx, we identified 121 cancer-specific in-frame ES events-related genes. (ii) Analysis of the lost protein features of exon skipping events will help to deepen understanding the detailed loss-of-functional effects of tumorigenic ES events. We have identified 2427 and 249 in-frame genes that have lost protein functional domains and DNA-binding domain, respectively. One hundred forty-two genes lost specific amino acid sites such as the site required for ligand-induced CBL-mediated ubiquitination in MET exon 14. (iii) ORF annotation of skipped exons classifies translational and non-translational ES events and provides potential candidates to be targeted by antisense oligonucleotides to restore lost protein functions. Among ∼14 000 genes with ES events in cancer, there were 8667 and 9623 ES genes of in-frame and frame-shift ORFs, respectively. Among 9623 genes with the frame-shifted ES events, 453 genes can be targeted by the drugs from IUPHAR database. (iv) RNA-seq data studies, such as coverage depth check, differences in PSI value between mutated and non-mutated samples, and split read abundance evidence by sashimi plots, can identify mutation-associated ES events. One hundred twenty-three genes with non-synonymous mutations around skipped exons showed decreased expression at the skipped exons with differential PSI values compared to non-mutated samples of same cancer type. Furthermore, we performed manual curation of PubMed articles for 711 genes with recurrent splice-site or nonsense mutations. (v) Based on the involvement of methylation in differentiating elongation rate of Pol II (17), we analyzed the association between splicing and methylation (ES-specific splicing Quantitative Trait Methylation, sQTM). Through systematic survival analysis of sQTM pairs, we identified 50 prognostic cases. In addition, we identified 1534 approved drugs for targeting 1715 ES genes, and 7324 genes reported to be associated with 5039 different types of diseases in DrugBank (18) and DisGeNet (19), respectively. Figure 1 is an overview of ES annotations using MET as a representative example. All entries and annotation data are available for browsing and downloading on the ExonSkipDB web site (https://ccsm.uth.edu/ExonSkipDB).
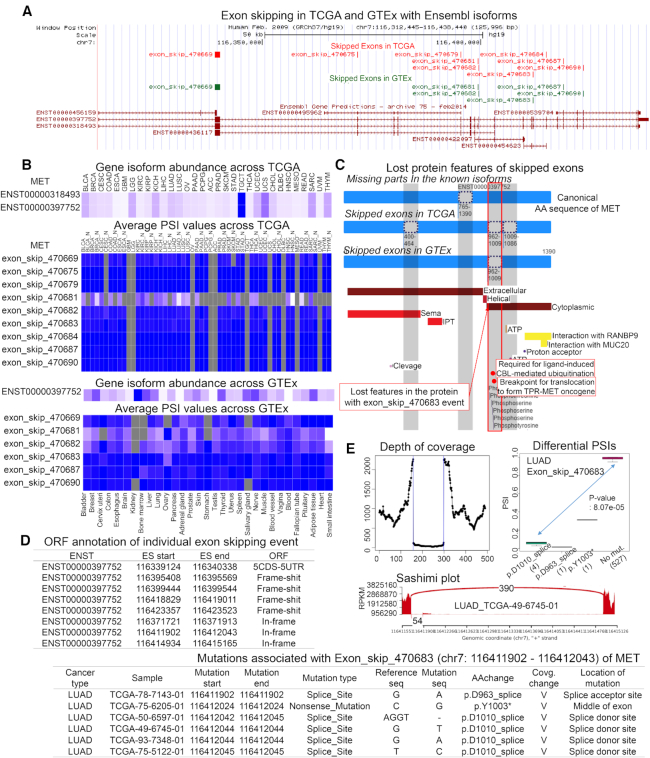
Figure 1.
Overview of ExonSkipDB. (A) The genomic structures of exon skipping events of TCGA and GTEx across reference gene model. (B) Abundance of isoforms and PSI values of individual exon skipping events across TCGA and GTEx. (C) The protein functional features based on the canonical protein sequence. The grey color shaded regions correspond to individual exon skipping events. (D) The analyzed ORF information of individual exon skipping events based on the canonical transcript sequence. (E) RNA-seq evidence for the mutation-associated exon skipping event. Consistent evidence through the depth of coverage, differential PSI values between mutated and non-mutated samples, and sashimi plots have identified 136 exon skipping events that have associations with mutations.
DATA INTEGRATION AND ANNOTATIONS
Exon skipping information
We downloaded the exon skipping event information of 8,705 patients of TCGA’s 33 cancer types and over 3,000 normal samples from GTEx's 31 different tissues (Supplementary Tables S1 and S2) from Kahles et al.’s study (12). Only the exon skipping events with conserved loci among the six splice-sites of three exons were used in this study. These exons are involved in exon skipping such as ‘upstream exon’, ‘skipped exon’ and ‘downstream exon’. Of these six sites, four sites are the acceptor site of the upstream exon, the acceptor and donor sites of the skipped exon, and the donor site of the downstream exon. Based on the GENCODE v19 annotation reference genome (20), we obtained 90 616 and 89 845 annotated gene structure-based exon skipping events from TCGA and GTEx, respectively.
Open reading frame (ORF) annotation
For specific exon skipping events based on the GENCODE v19 (20) (Supplementary Tables S3 and S4), we examined the open reading frames of major isoform transcript sequences. When both of the nucleotide start and end positions of exon skipping were located inside of the coding region (CDS) and the number of transcript sequences after exclusion of the skipped exon sequence was a multiple of three, then we reported such exon skipped gene isoform as ‘in-frame’. When one or two nucleotide insertions are present, we reported such transcripts as ‘frame-shift’.
Retention analysis of 39 protein features from UniProt
Firstly, we created the exon skipped transcript sequence based on the canonical transcript sequence. Then this sequence was aligned with non-redundant protein sequence database using BLASTX (21) and the mapped proteins with 100 percent identify were selected. Through this process, we obtained the loci of skipped exon on the genomic, transcriptomic, and protein sequences for the ES genes of TCGA and GTEx (Supplementary Tables S5 and S6). We searched the retention of 39 protein features of UniProt (22) at the canonical amino acid sequence level with skipped exon loci information, including 6 molecule processing features, 13 region features, 4 site features, 6 amino acid modification features, 2 natural variation features, 5 experimental info features and 3 secondary structure features. The lost protein functional features due to ES are listed in Supplementary Tables S7 and S8. Detailed information about all of the protein features is in UniProt.
Identifying mutations associated with exon skipping events
According to Wimmer et al. and Anna and Monika (23,24), there are five types of splicing mutations. Among these, exonic mutations creating new splicing site or leading splicing enhancer (ESE) disruption, and canonical splice site at the exon-intron boundaries can effect in improper splicing and exon skipping event. On the other hands, intronic mutations in the branch point or polypyrimidine tract, or new intronic splice site mutations can lead exon skipping or include intron fragment. In our annotation, we only could use exonic mutations since the variants of TCGA are not based on whole genome sequencing, but based on whole exon sequencing data sets. So, we tried to identify exonic mutations inducing exon skipping event including splice site mutations. For other types of exonic non-synonymous mutations, we delved into and searched for relevant publications to determine which coding region mutations may influence or induce exon skipping events. We have collected evidence of ES-induced mutations, including not only splice site mutations, but also nonsense and frameshift indel mutations from the previous studies (25–27; see 3–5). Therefore, to identify candidate mutations that might induce the exon skipping events, we overlapped the genomic coordinates of skipped exons with three types of non-synonymous mutations, including splice-site, nonsense, and frame-shift mutations from TCGA (Supplementary Table S9). Using genomeCoverageBed of bedtools (28), we drew the coverage depth plots based on the transcript nucleotide sequences of the three exons involved in exon skipping events from the bam files of RNA-seq data of TCGA samples. If the difference of mean read count between the middle exon (alternatively spliced, skipped exon) and two neighbor exons (constitutively spliced exon) is great than 10.0, we remained these cases for next step. It resulted 3554 exon skipping events among 2402 genes in 1886 samples. Next, we chose the cases where the PSI values in the mutated samples are different from the ones in the non-mutated samples as the final candidates. Since there are not many recurrent ES events, we applied the average PSI value as the threshold. If the PSIs of the mutated sample is less than 0.3 and the PSI of the non-mutated sample is >0.7, then we selected those ES events as potential mutation-associated ES events. Finally, we obtained 136 ES events from 123 genes that presented differences in coverage depth and PSIs between non-mutated and mutated samples (Supplementary Table S10). For these cases, we also provided sashimi plots to show additional evidence from the quantified split reads across ∼3000 cancer samples (29).
Exon skipping specific splicing quantitative trait loci (sQTL) and methylation (sQTM) analysis
For the ES-specific sQTL, we adopted sQTL analysis results from TCGA (12). There were 901 167 possible sQTL candidate single-nucleotide polymorphisms (SNPs) in 9909 genes across 33 cancer types. By integrating these loci with the coordinates of skipped exons, 4124 SNPs from 2254 ES genes were identified. For ES specific sQTM analysis, we first selected 2750 exon skipping events. These events have PSI values between 0.1 and 0.9 in at least 10 samples across 8088 pan-cancer samples and have methylation data in the same samples. The Illumina Human Methylation 450 data of these samples across 32 cancer types were downloaded from TCGA. Typically, a beta value greater than or equal to 0.6 is considered to be fully methylated, and a beta value less than or equal to 0.2 is considered to be fully unmethylated. Therefore, to calculate the association with PSI values, we have used the beta values between 0.2 and 0.6. We calculated the beta value for the region between the first exon's start site and the middle exon's end site. The average beta value of 1231 upstream CpG regions for individual exon skipping events in pan-cancer (Supplementary Table S11) was obtained. For the 8088 samples with methylation data, we investigated the effect of DNA methylation on exon skipping events using a linear regression approach, ultra-fast eQTL analysis via large matrix operations (MatrixEQTL) (30). We calculated the correlation p-value between the exon skipping event and its upstream region methylation, and the pair with a p-value less than 1.0 × E−5 were selected as a cis-sQTM pair. To identify the pathogenic sQTM, we performed the same analysis per cancer type and chose significant cis-sQTM pairs. For these significant cis-sQTMs, we performed the survival analysis using the survival package in R for the Kaplan–Meier method and log-rank test (Supplementary Table S12).
Drug and disease information
Drug-target interactions (DTIs) were extracted from DrugBank (18) (April 2018, version 5.1.0) and duplicated DTI pairs were excluded. All drugs were grouped using Anatomical Therapeutic Chemical (ATC) classification system codes. Disease-genetic information was extracted from a database of gene-disease associations (DisGeNet, June 2018, version 4.0) (19).
Manual curation of PubMed articles
For the 711 genes with recurrent splice-sites or nonsense mutations in more than two samples, PubMed's literature query was performed on June 2019 using the search expression applied to each gene. Taking MET as an example, it is ‘((MET [Title/Abstract]) AND exon [Title/Abstract]) AND skip [Title/Abstract])’. After a manual review of the abstracts, we found that there was 46 documentary evidence supporting exon skipping events of 9 genes (Supplementary Table S13).
Database architecture
The ExonSkipDB system is based on a three-tier architecture: client, server, and database. It includes a user-friendly web interface, a Perl's DBI module, and a MySQL database. This database was developed in MySQL 3.23 with the MyISAM storage engine.
WEB INTERFACE AND ANALYSIS RESULTS
ES event gene structure browser and ES event heatmaps provide potential disease-specific ES events
In order to understand the landscape of exon skipping events, co-localization information with known gene isoform structures is essential. For this, we mapped ES events based on gene structure using the custom track of UCSC genome browser (31). An example image of MET with the gene isoforms and skipped exons based on RefGene (32) and Ensembl (33) gene models is shown in Figure 1A. Through this image, the relationship between known isoforms and exon skipping events can be easily understood by users. In addition, the landscape of gene isoform abundance and PSI values across TCGA cancers and GTEx tissues in the heat maps allow users to gain insight into the disease- or tissue-specific exon skipping events (Figure 1B). For example, MET has three cancer-specific exon skipping events, such as ‘exon_skip_470675’, ‘exon_skip_470679’ and ‘exon_skip_470684’._In addition to ‘exon_skip_470679’, other ES events are predicted to be in-frame events based on the canonical transcript sequence (ENST00000397752). The ‘exon_skip_470675’ is located from 400 to 464 of the canonical protein amino acid sequence (P08581). From the lost protein feature analysis image in Figure 1C, this exon skipping event might lose the sema domain (PROSITE-ProRule: PRU00352 (34)). On the other hand, ‘exon_skip_470684’, anticipated to be located between 1009 to 1086 of the canonical amino acids, might lose protein kinase domain (PROSITE-ProRule: PRU00159) and the breakpoint for translocation to form TPR-MET oncogene. Furthermore, by comparing the ES genes with in-frame ORF between TCGA and GTEx, 121 cancer-specific ES genes were identified. Among the 121 genes, there were one of the cancer gene census genes, three kinases, four transcription factors, two tumor suppressors, and 14 IUPHAR drug targets (Supplementary Figure S1), including ARG2, AQP6, CDC6, DUSP9, ENPEP, GABPA, GABRA3, GPRC6A, GRM7, HTR3A, KCNU1, KLF12, NEK2, NR6A1, QRFPR, RET, SLC6A19, SLC6A3, STK17A, and TMPRSS11A. For each of these genes, the users can obtain a detailed annotation of exon skipping events (Supplementary Figure S2). As shown here, the gene structure images and other annotations of ExonSkipDB will help users identify disease-specific exon skipping events.
ORF analysis of individual exon skipping events will be helpful for screening potential therapeutic candidates
The open reading frame of the skipped exons is very important for assessing the translation capabilities of individual exon skipped transcripts into the effective proteins. We annotated the ORFs of individual ES events based on GENCODE v19. Out of 14 272 genes with ES events, we identified 8667 and 9623 genes with in-frame and frame-shift ORFs, respectively. Among the 9623 genes with frame-shifted ES events, 453 genes can be pharmacologically targeted according to the information from IUPHAR database. By performing over-representation analysis using Gene Set Enrichment Analysis (GSEA) (35), we found that the 181 and 173 genes are involved in ‘ion transport’ and ‘transmembrane transport’ biological pathways, respectively (Supplementary Table S14). These transcripts with frame-shifted ES events cannot be translated into normal proteins. If a critical functional domain is lost or partially expressed due to this frame-shift and causes disease, then we can consider the use of antisense oligonucleotide-based therapy to restore the lost function as seen in the exon skipping therapy for Duchenne muscular dystrophy (DMD) (36). For the genes with the in-frame ORF ES events, we systematically studied the retention of protein functional features. For example, MET has eight exon skipping events. We aligned these events on the Ensembl gene model of ENST00000397752 (Figure 1D). Among these, three and four were annotated as in-frame and frame-shift ORF ES events, respectively. As shown in Figure 1C, the transcripts with in-frame ES event can be translated into proteins; however, some functional features of these proteins will be lost. Obviously, ExonSkipDB’s ORF annotation will be helpful for users to identify translational ES events and targetable ES events for the antisense oligonucleotides (AS)s treatment.
Lost protein functional features might be caused by ES events
As mentioned above, the lost protein features of ES events are important for evaluating the functional effects of the related genes on tumorigenesis or disease development. Figure 1C shows an annotation of the lost protein functional features for the skipped exons of MET in the known transcripts and all in-frame ES events based on the canonical protein sequence. Through translating individual ES events at the amino acid sequence level and localizing all the protein functional features of UniProt, the users can obtain a positional relationship of the ES events and functional features. For example, MET has four in-frame ES events, which are shaded with grey in the protein feature image. Three of the four are skipped exons and one is a missed exon in the known gene isoform structure. Specifically, MET has a unique tumorigenesis site called ‘Required for ligand-induced CBL-mediated ubiquitination’. As shown in this example, the users can identify lost protein functional features of ES genes of interest.
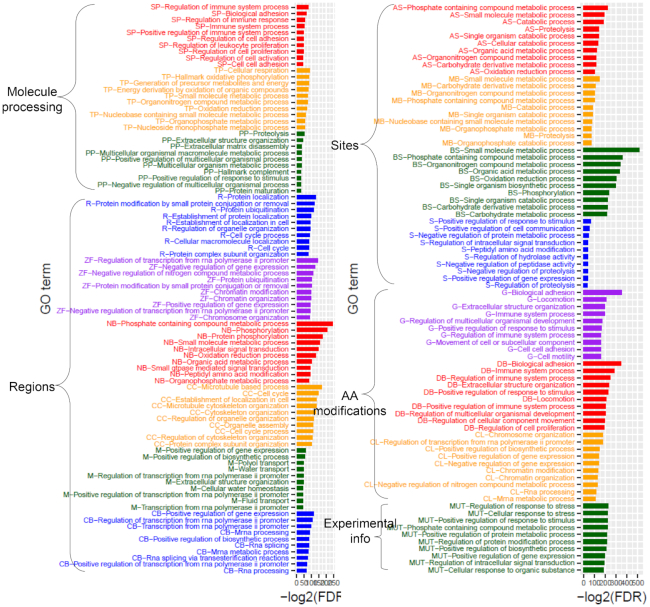
The overall statistics of all lost protein functional features of the 14 272 ES genes are listed in Table 1. For individual lost protein feature categories, we did over-representation analysis using Gene Set Enrichment Analysis (GSEA) to determine if the number of genes in each category is >30 and <700. This is a modified range from GSEA’s default range (25–500). There are 18 feature categories in total (Figure 2). We then found that the genes that lost the ‘molecule processing’ subsection were enriched in biological processes related to the ‘regulation of immune system process’, ‘cellular respiration’, and ‘proteolysis’. The genes that lost the ‘regions’ subsection were enriched in ‘protein localization’, ‘regulation of transcription from RNA polymerase II promoter’, ‘microtubule-based process’ and ‘positive regulation of gene expression’. Specifically, the genes that lost ‘compositional bias’ features were enriched in ‘positive regulation of gene expression’ as described as important in gene expression regulations in many reports (37–39). The genes that lost the ‘amino acid modifications’ subsection were enriched in the ‘biological adhesion’ process, consistent with previous studies about the needs of amino acid modifications for improving cell adhesions (40,41). Interestingly, the genes that lost the ‘sites’ subsection were enriched in the ‘small molecular metabolic processes’. Of 554 genes that lost this subsection, 118 genes are drug targets according to the International Union of Basic and Clinical Pharmacology (IUPHAR) (42). We identified 66 out of 118 genes that can be targeted by 136 approved drugs from DrugBank and 120 drugs are small molecule drug types. In addition, we found that 142 genes lost specific amino acid sites, such as ‘the site required for ligand-induced CBL-mediated ubiquitination’ in MET exon 14 skipping event. Overall, there were 21, 30, 16, 23, 23 and 4 sites of breakpoint, cleavage, important, inhibition, interaction, and required sites according to UniProt feature description, respectively. For 20 genes overlapped with important group genes out of 121 cancer-specific ES genes, we focused on studying the lost protein functional features (Supplementary Table S15). These 20 genes lost extracellular membrane domain, transmembrane domain, protein kinase domain, nuclear receptor, tyrosine–protein phosphatase, so on. Changes in the functional features due to these ES events can serve as a good resource for further validation of ES-caused tumorigenesis.
Table 1.
Number of in-frame ES event genes per lost protein functional features
| Subsection | Content | # in-frame exon skipped genes |
|---|---|---|
| Molecule processing | Initiator methionine | 13 |
| Signal peptide | 93 | |
| Transit peptide | 31 | |
| Propeptide | 45 | |
| Chain | 7044 | |
| Peptide | 8 | |
| Regions | Topological domain | 1243 |
| Transmembrane | 847 | |
| Intramembrane | 21 | |
| Domain | 2427 | |
| Repeat | 596 | |
| Calcium binding | 25 | |
| Zinc finger | 193 | |
| DNA binding | 21 | |
| Nucleotide binding | 269 | |
| Region | 1055 | |
| Coiled coil | 458 | |
| Motif | 192 | |
| Compositional bias | 497 | |
| Sites | Active site | 240 |
| Metal binding | 237 | |
| Binding site | 378 | |
| Site | 142 | |
| Amino acid modifications | Modified residue | 1374 |
| Lipidation | 18 | |
| Glycosylation | 624 | |
| Disulfide bond | 647 | |
| Cross-link | 234 | |
| Natural variations | Alternative sequence | 3123 |
| Natural variant | 2080 | |
| Experimental info | Mutagenesis | 665 |
| Sequence conflict | 1482 | |
| Secondary structure | Helix | 1750 |
| Turn | 952 | |
| Beta strand | 1623 |
Figure 2.
The overrepresentation enrichment analysis of ES genes per lost protein feature categories. For individual protein functional feature categories, which were lost in ES genes due to exons kipping, we have investigated the enriched genes in the biological processes. The abbreviations of categories are following with alphabetical order; Active site (AS), Binding site (BS), Coiled coil (CC), Compositional bias (CB), Cross-link (CL), Disulfide bond (DB), Glycosylation (G), Metal binding (MB), Motif (MB), Mutagenesis (MUT), Nucleotide binding (NB), Propeptide (PP), Repeat (R), Signal peptide (SP), Site (SP), Transit peptide (TP) and Zinc finger (ZF). In the left top panel, SP, TP, and PP belong to the UniProt's protein feature subsection of ‘Molecule processing’, and R, ZF, NB, CC, M and CB belong to the ‘Regions’ subsection. In the right top panel, AS, MS, BS and S belong to the ‘Sites’ subsection. G, DB and CL belong to the subsection of ‘Amino acid modifications’. Lastly, Mutagenesis (M) belongs to the ‘Experimental info’ subsection.
Non-synonymous mutation-associated exon skipping events
As we have seen from the mutation-induced exon skipping examples (such as MET exon 14 and DMD exon skipping), screening mutations associated with exon skipping events are a good approach to identify novel therapeutic targets. By overlapping genomic coordinates of somatic mutations including splice-site, nonsense, and frame-shift mutations (see Materials and Methods for the reason why we focused on these mutation types), and ES loci, we identified 11 204 genes with 75 130 non-synonymous mutations in 35 714 ES events. For 7139 cancer samples with non-synonymous mutations and available bam files, we plotted their coverage depth along the three exons of each ES event. Among them, we selected 136 ES events in 123 genes as shown in Table 2. These events have decreased coverage depth at the skipped exon and differential in PSI values between mutated and non-mutated samples (see Methods). Taking MET as an example, as shown in Figure 1E, in a LUAD sample with ‘p.D1010_splice’ mutation, the read coverage depth of the skipped exon is decreased by more than 1000.0 reads. Second, there is a difference in the distribution of PSI values between the samples with mutations and wild-type. Furthermore, the sashimi plot showed almost of the split reads were connected between the two flanking exons. Of the 123 mutation candidate genes associated with exon skipping, 8 ES events (7 genes) have more than 2 mutated samples in each cancer type. These ES events are IRF3, MET, PIK3R1, PTEN, SCRIB, TP53 and YLPM1. In addition, PLEC and RB1 have more than two mutated samples in pan-cancer.
Table 2.
Exon skipping events associated with mutations
| Gene | Cancer type | ESID | Gene | Cancer type | ESID | Gene | Cancer type | ESID |
|---|---|---|---|---|---|---|---|---|
| AASS | BRCA | exon_skip_479254 | HNRNPC | READ | exon_skip_111139 | RASA1 | HNSC | exon_skip_436351 |
| ACADVL | HNSC | exon_skip_148534 | IRF3 | SKCM | exon_skip_320801 | RB1 | LGG | exon_skip_100319 |
| ACE2 | LIHC | exon_skip_514046 | IRF3 | LIHC | exon_skip_320801 | RB1 | LUSC | exon_skip_100319 |
| ACSL1 | BRCA | exon_skip_433327 | KCTD10 | STAD | exon_skip_96034 | RB1 | SKCM | exon_skip_100319 |
| ACTR5 | HNSC | exon_skip_351842 | KDM4C | LIHC | exon_skip_495073 | RB1 | SARC | exon_skip_100320 |
| AK9 | SKCM | exon_skip_461739 | LAMTOR4 | CESC | exon_skip_468895 | RB1 | SKCM | exon_skip_100321 |
| AP1G2 | KIRP | exon_skip_111822 | LPCAT1 | COAD | exon_skip_440961 | RBBP7 | BRCA | exon_skip_514095 |
| AP1G2 | UCS | exon_skip_111839 | LRRC37B | TGCT | exon_skip_150743 | RBL2 | LIHC | exon_skip_136487 |
| ARAP1 | HNSC | exon_skip_75409 | LRRC49 | LUSC | exon_skip_123285 | RBM10 | LUAD | exon_skip_509946 |
| ARAP2 | STAD | exon_skip_428975 | LZTFL1 | SKCM | exon_skip_382808 | RBM11 | CESC | exon_skip_358911 |
| ARHGAP4 | CESC | exon_skip_517379 | MAOB | LIHC | exon_skip_514541 | RBM23 | KIRP | exon_skip_111436 |
| ARSA | ESCA | exon_skip_370946 | MAP4K4 | SKCM | exon_skip_328236 | RBM27 | SKCM | exon_skip_438439 |
| ATF7IP | LUAD | exon_skip_80383 | MCTP2 | SKCM | exon_skip_125176 | RWDD2B | LUAD | exon_skip_361480 |
| ATXN7L1 | BRCA | exon_skip_478799 | MET | LUAD | exon_skip_470683 | SAAL1 | LIHC | exon_skip_69749 |
| AXIN1 | LIHC | exon_skip_140168 | MFN1 | BRCA | exon_skip_379593 | SAFB | LUSC | exon_skip_301206 |
| BAP1 | UVM | exon_skip_384821 | MFSD11 | BRCA | exon_skip_156579 | SCRIB | STAD | exon_skip_493826 |
| BCHE | LIHC | exon_skip_389735 | MIB2 | LIHC | exon_skip_292 | SEMA6C | LUAD | exon_skip_30610 |
| BRCC3 | READ | exon_skip_513411 | MPP1 | LIHC | exon_skip_517757 | SIK3 | BRCA | exon_skip_77623 |
| BSDC1 | SKCM | exon_skip_24351 | MPP1 | LIHC | exon_skip_517758 | SLC44A1 | LUSC | exon_skip_497929 |
| BSDC1 | SKCM | exon_skip_24356 | MSH2 | STAD | exon_skip_325448 | SLC6A9 | BRCA | exon_skip_25867 |
| C16orf70 | OV | exon_skip_137511 | MSI2 | SARC | exon_skip_154718 | SLCO2A1 | LUAD | exon_skip_388682 |
| C20orf96 | LGG | exon_skip_354191 | MYO9B | LUAD | exon_skip_303297 | SMARCA1 | KIRP | exon_skip_516648 |
| CCDC125 | SKCM | exon_skip_442509 | NF1 | PCPG | exon_skip_150648 | SMARCA2 | KIRC | exon_skip_494778 |
| CCDC90B | HNSC | exon_skip_76335 | NF2 | CHOL | exon_skip_364330 | SMARCC1 | HNSC | exon_skip_383264 |
| CD44 | SKCM | exon_skip_57862 | NSFL1C | LUAD | exon_skip_354315 | SNRNP200 | KIRP | exon_skip_341775 |
| CDH1 | BRCA | exon_skip_138094 | OCIAD1 | LGG | exon_skip_423495 | SPCS1 | HNSC | exon_skip_375123 |
| CHD3 | SKCM | exon_skip_148936 | OCRL | LUSC | exon_skip_512323 | STAG2 | GBM | exon_skip_512303 |
| CHEK2 | LUSC | exon_skip_368202 | ODF2 | PAAD | exon_skip_499697 | SUPT3H | SKCM | exon_skip_459890 |
| CNOT6 | LUAD | exon_skip_440379 | PACRGL | LUSC | exon_skip_422774 | TBL1X | UCEC | exon_skip_509098 |
| COL13A1 | LUAD | exon_skip_42139 | PEA15 | LGG | exon_skip_12856 | TBX3 | STAD | exon_skip_96977 |
| COL7A1 | LUSC | exon_skip_383673 | PELI2 | LUSC | exon_skip_106790 | TCTN2 | THCA | exon_skip_88383 |
| COPS3 | SKCM | exon_skip_287002 | PGLYRP2 | LIHC | exon_skip_316122 | TEP1 | LUAD | exon_skip_110964 |
| CREBBP | CESC | exon_skip_141389 | PHKA2 | LUAD | exon_skip_514168 | TERF1 | OV | exon_skip_484059 |
| CTCF | BRCA | exon_skip_137805 | PHKA2 | BRCA | exon_skip_514195 | TNFRSF10A | PAAD | exon_skip_488782 |
| DIAPH3 | SKCM | exon_skip_103986 | PIK3R1 | SKCM | exon_skip_435166 | TP53 | OV | exon_skip_286364 |
| DLG3 | STAD | exon_skip_511069 | PIK3R1 | COAD | exon_skip_435167 | TP53 | OV | exon_skip_286376 |
| DSG2 | LUAD | exon_skip_296226 | PLEC | LUSC | exon_skip_494000 | TP53 | UCS | exon_skip_286379 |
| EEA1 | STAD | exon_skip_95294 | PLEC | SKCM | exon_skip_494000 | TP53 | BRCA | exon_skip_286384 |
| EPB41L4A | LUAD | exon_skip_443476 | PLOD3 | LUSC | exon_skip_478451 | TP53 | KICH | exon_skip_286388 |
| FARP2 | ESCA | exon_skip_336006 | PPP6C | SKCM | exon_skip_506902 | TRAPPC2L | BRCA | exon_skip_139393 |
| FAS | DLBC | exon_skip_43777 | PROM1 | BRCA | exon_skip_428630 | TULP4 | BRCA | exon_skip_455061 |
| FN1 | SKCM | exon_skip_346530 | PTCH1 | ESCA | exon_skip_505102 | UBE2D3 | LIHC | exon_skip_431479 |
| GCSAM | DLBC | exon_skip_386471 | PTEN | COAD | exon_skip_43702 | UBE2E1 | CESC | exon_skip_372157 |
| GLRX3 | LUAD | exon_skip_46116 | PTEN | CESC | exon_skip_43707 | USP40 | LUAD | exon_skip_347441 |
| GNB5 | LUAD | exon_skip_127162 | PTEN | SKCM | exon_skip_43707 | YIF1A | STAD | exon_skip_74538 |
| GPR143 | SKCM | exon_skip_513862 | PTEN | BRCA | exon_skip_43714 | YLPM1 | SKCM | exon_skip_108215 |
| HAUS6 | STAD | exon_skip_502793 | RABL3 | LUSC | exon_skip_386919 | ZNF512B | LUAD | exon_skip_358876 |
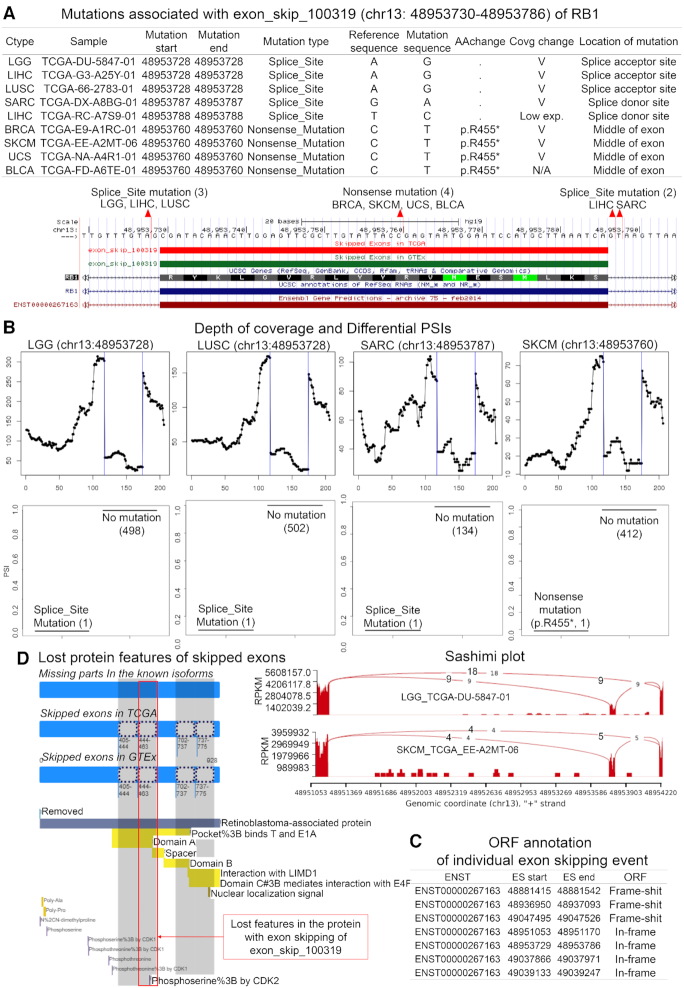
After manually curating the evidence of ES-associated mutation in these 9 genes, we identified a novel mutation-associated exon skipping event of RB1 (RB transcriptional corepressor 1), as shown in Figure 3. RB1 is a negative regulator of the cell cycle. The RB1 protein also stabilizes constitutive heterochromatin to maintain overall chromatin structure. The active hypo-phosphorylated RB1 protein binds to transcription factor E2F1 (32). There are 9 samples with five splice-site and nonsense mutations in ‘exon_skip_100319’ ES event of RB1 (Figure 3A). Three of them are located in the splice acceptor or donor sites of the skipped exon. The nonsense mutation is p.R455* of RB1. Samples with these mutations showed different PSI values than the non-mutated ones, and the coverage depth decreased consistently (Figure 3B). Since this ES event was predicted to create an in-frame ORF transcript (Figure 3C), annotation of lost protein features due to this in-frame ORF ES event can be achieved with ExonSkipDB (Figure 3D). This ES event seems to cause a loss of a portion of the ‘pocket’ (retinoblastoma associated protein A domain) between 444 to 463 amino acid sequence. In addition, RB1 exon skipping event caused by mutations in the splice site may result in the loss of normal RB1 function through binding and inhibiting E2F-DP dimer formation (E2 promoter-binding-protein-dimerization partner), thereby restricting cell replication.
Figure 3.
Splice-site mutation associated exon skipping event in RB1. Among 10 recurrent ES events, RB1 showed consistent expression change and differential PSI values with non-mutated samples in multiple cancer types. (A) Mutations associated with exon skipping event (ESID: exon_skip_100319) in RB1 and mutation location in the gene structure. (B) RNA-seq evidence for the mutation associated exon skipping event. Consistent evidence through the depth of coverage, differential PSI values between mutated and non-mutated samples, and sashimi plots. (C) The analyzed ORF information of individual exon skipping events based on the canonical transcript sequence. (D) The protein functional features based on the canonical protein sequence.
Another candidate is interferon regulatory factor 3 (IRF3). As shown in Supplementary Figure S3, ‘exon_skip_320801’ ES event location covers partial of 5′-UTR and CDS. Two LIHC (Liver Hepatocellular Carcinoma) and three SKCM (Skin Cutaneous Melanoma) samples had ‘Frame_shift_Indel' or ‘Nonsense' mutations in this skipped exons. These samples showed decreased read coverage depth and differential PSI values between mutated and non-mutated samples. Overall translation rates were also affected by characteristics of the 5′-UTR, including length and start-site consensus sequences, the presence of secondary structure, upstream AUGs, upstream open reading frames (uORFs) and internal ribosome entry sites (IRES) (43,44). In addition, 5′-UTRs can contain sequences that function as binding sites for regulatory proteins (45). In this context, we have searched binding sites by the known transcription factors of IRF3 (such as AP1, ATF2, NF-kappaB, NF-kappaB1, p300, p53 and STAT1) in this ES location (chr19:50164706–50165585) from GeneCards (46) using ConTra v3, a tool for identifying transcription factor binding sites (47). As shown in Supplementary Figure S4, we predicted three and one binding sites of the p300 and ATF2 proteins in the partial 5′-UTR of ‘exon_skip_32080’ ES event. Furthermore, analysis of the lost protein functional features showing in Supplementary Figure S3C indicates the skipped exon may lead to the loss of the ‘HERC5 binding site’ in the CSD portion. In summary, loss of functional and regulatory domains caused by this ES event may result in IRF3 dysfunction. These findings require further experimental validation. ExonSkipDB can also provide ES event located mutation information for public cell lines by integrating data from Cancer Cell Line Encyclopedia (CCLE) (48), In total, we found that 1570 cell-lines have overlaps with 243 628 ES-region mutations of TCGA samples across 12 368 genes. This information can be downloaded from the ExonSkipDB web site.
Exon skipping associated sQTLs and sQTMs
SNPs can alter splicing or alternative splicing by altering the sequence-specific binding affinity of the splicing factors to the pre-mRNAs, resulting splicing quantitative trait locus (sQTL) (49). To explore the association of exon skipping with sQTLs and sQTMs, we downloaded 901 167 sQTL candidate SNPs in 9909 genes of 33 cancer types from TCGA (12). By integrating these sQTLs with skipped exon loci, we identified 4124 SNPs located in 4298 skipped exons of 2254 genes. Of these, 51 SNPs in 50 genes were completely overlapped with SNVs in 40 TCGA samples. These genes are mainly involved in ‘regulation of cell proliferation’, ‘positive regulation of protein metabolic process’, and ‘phosphorylation’ (Supplementary Table S16).
DNA methylation was originally thought only affecting transcription. Emerging evidence shows that methylation also regulates alternative splicing of RNA (17). Three proteins are known to transmit DNA methylation-encoded information to splicing machinery through different mechanisms, including CTCF, methyl-CpG binding protein 2 (MeCP2), and HP1. CTCF can bind to DNA and reduce the elongation rate of Pol II, which facilitates exon inclusion. In this process, CTCF binding can be inhibited by DNA methylation. Therefore, DNA methylation can result in exon exclusion (50). Second, the binding of MeCP2 to the methylated DNA can reduce the elongation rate of Pol II, enhancing exon inclusion (51). Another mechanism involves in the formation of protein bridges, through which DNA methylation leads to histone modification of H3K9me3. At the chromatin level, HP1 proteins can accumulate on the regulated alternative exons and then HP1-bound splicing factors can exert their regulatory role on these alternative exons (52). More underlying mechanisms are remained to be identified. Since TCGA has DNA methylation and RNA-Seq data for the matched samples, we explored the regulatory mechanisms of DNA methylation on alternative splicing. PSI values measure how efficiently the sequences of interest are spliced into transcripts (53). We downloaded PSI values from the studies conducted by Kahles et al. (12) and calculated the average beta values of CpG methylations in the upstream region covering the two upstream exons in three ES-involved exons. We then used MatrixEQTL to identify significantly associated methylation-ES pairs in pan-cancer. The same approach was also used to identify cancer type-specific ES-specific sQTMs and prognostic candidate pairs. We set the threshold of significant sQTM pairs with a p-value less than 1.0 × e−5.
Using this approach, we identified 513 significant sQTM pairs of 434 genes. Among these sQTMs, 292 and 218 sQTM pairs had negative and positive correlations with exon skipping events, respectively. We analyzed the overrepresented biological processes (Supplementary Table S17) for these sQTM genes. The positively correlated genes were enriched in the process of ‘positive regulation of response to stimulus or cell communications’. The negatively correlated genes were actively involved in ‘macromolecular complex assembly’, ‘regulation of transcription from RNA polymerase II promoter’, ‘positive regulation of gene expression’, and ‘cellular response to stress’. This different roles in the cells might depend on the methylation locations across genes. We also examined the overlap between somatic mutation loci and 513 sQTMs’ exons. There were 192 TCGA samples with 140 mutations in the skipped exons of 125 ES genes. Interestingly, these somatically mutated sQTM genes were involved in the same biological processes as the 40 genes with mutated sQTLs (Supplementary Table S18). In addition, 77 exon skipping events (out of 513 sQTM exons) of the 67 genes were common in more than 2 cancer types. On the other hand, 146 exon skipping events in 137 genes were cancer type-specific sQTMs pairs with mutual exclusivity. For the PSI and beta values of these sQTMs, we generated t-distributed stochastic neighbor embedding (t-SNE) plots (see the statistics page of the ExonSkipDB web site). These images show the dispersion of each value in pan-cancer with a cancer type-specific distribution. For all sQTM pairs, we performed survival analysis and identified 50 prognostic sQTMs. One of the top-ranked prognostic sQTM genes was deoxythymidylate kinase (DTYMK) (Supplementary Figure S5). This gene was reported as a potential marker for brain tumors in cerebrospinal fluid (54).
ES gene-related FDA approved-drugs and human diseases
The pharmacological information related to the ES genes were obtained from DrugBank (18). Our analysis indicates that 1801 proteins encoded by ES genes are targeted by 1676 drugs. Among these, 1534 (91.50%) are FDA-approved drugs targeting 1715 proteins. 750, 142, 104, 93, 74 and 67 drugs targeted ‘cellular receptors’, ‘channels’, ‘transporters’, ‘synthases’, ‘kinases’, and ‘growth factors’, respectively. Related disease category shows the related disease information for each gene from a database of gene-disease associations (DisGeNet) (19). We found that the identified 7324 exon skipping events-related genes were associated with 5039 different types of diseases based on the published data searches. Of these, 955 genes were associated with 711 different syndromes.
DISCUSSION AND FUTURE DIRECTION
ExonSkipDB is the first database to systematically annotate the function of exon skipping events across human pan-cancer and multiple human tissues. To serve the broad biomedical research communities, we will continually update and curate exon skipping events by routinely checking newly published alternative splicing data sets for human diseases, including neurodegenerative diseases such as Alzheimer's disease. To make better use of ExonSkipDB, we will add the analysis results for the exon skipping events from over 1000 cell-lines in CCLE, as well as data sets for multiple mouse tissues. We will continue to expand our research and improve our approach to find clinically significant exon skip events and downstream target genes. The user-friendly website provides researchers with multiple annotations and facilitates a comprehensive functional study of exon skipping. Therefore, ExonSkipDB will be useful resource for many researchers in the fields of pathology, cancer genomics and precision medicine, and pharmaceutical and therapeutic research.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Center for Computational Systems Medicine (CCSM) for valuable discussion. We also thank Talissa Chapin for improving the English of the manuscript and web site.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01GM123037, U01AR069395-01A1, R01CA241930 to X.Z.]; The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Funding for open access charge: Dr & Mrs Carl V. Vartian Chair Professorship Funds to Dr Zhou from the University of Texas Health Science Center at Houston.
Conflict of interest statement. None declared.
REFERENCES
- 1. Tazi J., Bakkour N., Stamm S.. Alternative splicing and disease. Biochim. Biophys. Acta. 2009; 1792:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Florea L., Song L., Salzberg S.L.. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues [version 2; peer review: 2 approved]. F1000Research. 2013; 2:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barny I., Perrault I., Michel C., Soussan M., Goudin N., Rio M., Thomas S., Attie-Bitach T., Hamel C., Dollfus H. et al.. Basal exon skipping and nonsense-associated altered splicing allows bypassing complete CEP290 loss-of-function in individuals with unusually mild retinal disease. Hum. Mol. Genet. 2018; 27:2689–2702. [DOI] [PubMed] [Google Scholar]
- 4. Takeuchi Y., Mishima E., Shima H., Akiyama Y., Suzuki C., Suzuki T., Kobayashi T., Suzuki Y., Nakayama T., Takeshima Y. et al.. Exonic mutations in the SLC12A3 gene cause exon skipping and premature termination in Gitelman syndrome. J. Am. Soc. Nephrol.: JASN. 2015; 26:271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oda H., Sato T., Kunishima S., Nakagawa K., Izawa K., Hiejima E., Kawai T., Yasumi T., Doi H., Katamura K. et al.. Exon skipping causes atypical phenotypes associated with a loss-of-function mutation in FLNA by restoring its protein function. Eur. J. Hum. Genet.: EJHG. 2016; 24:408–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Han S., Miller J.E., Byun S., Kim D., Risacher S.L., Saykin A.J., Lee Y., Nho K. for Alzheimer's Disease Neuroimaging, I. . Identification of exon skipping events associated with Alzheimer's disease in the human hippocampus. BMC Med. Genet. 2019; 12:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ramsbottom S.A., Molinari E., Srivastava S., Silberman F., Henry C., Alkanderi S., Devlin L.A., White K., Steel D.H., Saunier S. et al.. Targeted exon skipping of a CEP290 mutation rescues Joubert syndrome phenotypes in vitro and in a murine model. Proc. Natl Acad. Sci. U.S.A. 2018; 115:12489–12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Niks E.H., Aartsma-Rus A.. Exon skipping: a first in class strategy for Duchenne muscular dystrophy. Expert Opin. Biol. Ther. 2017; 17:225–236. [DOI] [PubMed] [Google Scholar]
- 9. Awad M.M., Oxnard G.R., Jackman D.M., Savukoski D.O., Hall D., Shivdasani P., Heng J.C., Dahlberg S.E., Janne P.A., Verma S. et al.. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J. Clin. Oncol. 2016; 34:721–730. [DOI] [PubMed] [Google Scholar]
- 10. Reungwetwattana T., Liang Y., Zhu V., Ou S.I.. The race to target MET exon 14 skipping alterations in non-small cell lung cancer: the why, the how, the who, the unknown, and the inevitable. Lung Cancer. 2017; 103:27–37. [DOI] [PubMed] [Google Scholar]
- 11. Aartsma-Rus A., van Ommen G.J.. Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications. RNA. 2007; 13:1609–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kahles A., Lehmann K.V., Toussaint N.C., Huser M., Stark S.G., Sachsenberg T., Stegle O., Kohlbacher O., Sander C. Cancer Genome Atlas Research, N. et al.. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell. 2018; 34:211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mele M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M., Young T.R., Goldmann J.M., Pervouchine D.D., Sullivan T.J. et al.. Human genomics. the human transcriptome across tissues and individuals. Science. 2015; 348:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu H.Y., Peng Z.G., He R.Q., Luo B., Ma J., Hu X.H., Dang Y.W., Chen G., Pan S.L.. Prognostic index of aberrant mRNA splicing profiling acts as a predictive indicator for hepatocellular carcinoma based on TCGA SpliceSeq data. Int. J. Oncol. 2019; 55:425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun W., Duan T., Ye P., Chen K., Zhang G., Lai M., Zhang H.. TSVdb: a web-tool for TCGA splicing variants analysis. BMC Genomics. 2018; 19:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han S., Kim D., Kim Y., Choi K., Miller J.E., Kim D., Lee Y.. CAS-viewer: web-based tool for splicing-guided integrative analysis of multi-omics cancer data. BMC Med. Genet. 2018; 11:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lev Maor G., Yearim A., Ast G.. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015; 31:274–280. [DOI] [PubMed] [Google Scholar]
- 18. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. et al.. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pinero J., Bravo A., Queralt-Rosinach N., Gutierrez-Sacristan A., Deu-Pons J., Centeno E., Garcia-Garcia J., Sanz F., Furlong L.I.. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017; 45:D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J. et al.. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019; 47:D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wimmer K., Roca X., Beiglbock H., Callens T., Etzler J., Rao A.R., Krainer A.R., Fonatsch C., Messiaen L.. Extensive in silico analysis of NF1 splicing defects uncovers determinants for splicing outcome upon 5′ splice-site disruption. Hum. Mutat. 2007; 28:599–612. [DOI] [PubMed] [Google Scholar]
- 24. Anna A., Monika G.. Splicing mutations in human genetic disorders: examples, detection, and confirmation. J. Appl. Genet. 2018; 59:253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu H.X., Cartegni L., Zhang M.Q., Krainer A.R.. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat. Genet. 2001; 27:55–58. [DOI] [PubMed] [Google Scholar]
- 26. Littink K.W., Pott J.W., Collin R.W., Kroes H.Y., Verheij J.B., Blokland E.A., de Castro Miro M., Hoyng C.B., Klaver C.C., Koenekoop R.K. et al.. A novel nonsense mutation in CEP290 induces exon skipping and leads to a relatively mild retinal phenotype. Invest. Ophthalmol. Vis. Sci. 2010; 51:3646–3652. [DOI] [PubMed] [Google Scholar]
- 27. Lalonde S., Stone O.A., Lessard S., Lavertu A., Desjardins J., Beaudoin M., Rivas M., Stainier D.Y.R., Lettre G.. Frameshift indels introduced by genome editing can lead to in-frame exon skipping. PLoS One. 2017; 12:e0178700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quinlan A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinformatics. 2014; 47:doi:10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katz Y., Wang E.T., Silterra J., Schwartz S., Wong B., Thorvaldsdottir H., Robinson J.T., Mesirov J.P., Airoldi E.M., Burge C.B.. Quantitative visualization of alternative exon expression from RNA-seq data. Bioinformatics. 2015; 31:2400–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shabalin A.A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012; 28:1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Giron C.G. et al.. Ensembl 2018. Nucleic Acids Res. 2018; 46:D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sigrist C.J., de Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A., Bougueleret L., Xenarios I.. New and continuing developments at PROSITE. Nucleic Acids Res. 2013; 41:D344–D347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aoki Y., Yokota T., Wood M.J.. Development of multiexon skipping antisense oligonucleotide therapy for Duchenne muscular dystrophy. BioMed. Res. Int. 2013; 2013:402369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Quax T.E., Claassens N.J., Soll D., van der Oost J.. Codon bias as a means to fine-tune gene expression. Mol. Cell. 2015; 59:149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vetsigian K., Goldenfeld N.. Genome rhetoric and the emergence of compositional bias. Proc. Natl Acad. Sci. U.S.A. 2009; 106:215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hajjari M., Khoshnevisan A., Behmanesh M.. Compositional features are potentially involved in the regulation of gene expression of tumor suppressor genes in human tissues. Gene. 2014; 553:126–129. [DOI] [PubMed] [Google Scholar]
- 40. Sankaran S., Cavatorta E., Huskens J., Jonkheijm P.. Cell adhesion on RGD-displaying knottins with varying numbers of tryptophan amino acids to tune the affinity for assembly on Cucurbit[8]uril surfaces. Langmuir. 2017; 33:8813–8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huettner N., Dargaville T.R., Forget A.. Discovering cell-adhesion peptides in tissue engineering: beyond RGD. Trends Biotechnol. 2018; 36:372–383. [DOI] [PubMed] [Google Scholar]
- 42. Harding S.D., Sharman J.L., Faccenda E., Southan C., Pawson A.J., Ireland S., Gray A.J.G., Bruce L., Alexander S.P.H., Anderton S. et al.. The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018; 46:D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gray N.K., Wickens M.. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 1998; 14:399–458. [DOI] [PubMed] [Google Scholar]
- 44. Mignone F., Gissi C., Liuni S., Pesole G.. Untranslated regions of mRNAs. Genome Biol. 2002; 3:REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wickens M., Anderson P., Jackson R.J.. Life and death in the cytoplasm: messages from the 3′ end. Curr. Opin. Genet. Dev. 1997; 7:220–232. [DOI] [PubMed] [Google Scholar]
- 46. Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y. et al.. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics. 2016; 54:1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- 47. Kreft L., Soete A., Hulpiau P., Botzki A., Saeys Y., De Bleser P.. ConTra v3: a tool to identify transcription factor binding sites across species, update 2017. Nucleic Acids Res. 2017; 45:W490–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. 3rd, Barretina J., Gelfand E.T., Bielski C.M., Li H. et al.. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019; 569:503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qu W., Gurdziel K., Pique-Regi R., Ruden D.M.. Identification of Splicing Quantitative Trait Loci (sQTL) in Drosophila melanogaster with Developmental Lead (Pb(2+)) Exposure. Front. Genet. 2017; 8:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S.. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011; 479:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Young J.I., Hong E.P., Castle J.C., Crespo-Barreto J., Bowman A.B., Rose M.F., Kang D., Richman R., Johnson J.M., Berget S. et al.. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc. Natl Acad. Sci. U.S.A. 2005; 102:17551–17558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Piacentini L., Fanti L., Negri R., Del Vescovo V., Fatica A., Altieri F., Pimpinelli S.. Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLos Genet. 2009; 5:e1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schafer S., Miao K., Benson C.C., Heinig M., Cook S.A., Hubner N.. Alternative splicing signatures in RNA-seq data: percent spliced in (PSI). Curr. Protoc. Hum. Genet. 2015; 87:doi:10.1002/0471142905.hg1116s87. [DOI] [PubMed] [Google Scholar]
- 54. Gronowitz J.S., Kallander C.F., Hagberg H., Persson L.. Deoxythymidine-kinase in cerebrospinal fluid: a new potential ‘marker’ for brain tumours. Acta Neurochir. (Wien). 1984; 73:1–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.