Abstract
The volume of biological, chemical and functional data deposited in the public domain is growing rapidly, thanks to next generation sequencing and highly-automated screening technologies. These datasets represent invaluable resources for drug discovery, particularly for less studied neglected disease pathogens. To leverage these datasets, smart and intensive data integration is required to guide computational inferences across diverse organisms. The TDR Targets chemogenomics resource integrates genomic data from human pathogens and model organisms along with information on bioactive compounds and their annotated activities. This report highlights the latest updates on the available data and functionality in TDR Targets 6. Based on chemogenomic network models providing links between inhibitors and targets, the database now incorporates network-driven target prioritizations, and novel visualizations of network subgraphs displaying chemical- and target-similarity neighborhoods along with associated target-compound bioactivity links. Available data can be browsed and queried through a new user interface, that allow users to perform prioritizations of protein targets and chemical inhibitors. As such, TDR Targets now facilitates the investigation of drug repurposing against pathogen targets, which can potentially help in identifying candidate targets for bioactive compounds with previously unknown targets. TDR Targets is available at https://tdrtargets.org.
INTRODUCTION
Neglected tropical diseases (NTDs) disproportionately affect ∼1.5 billion people in low income and developing countries, where they are a leading cause for life-years lost to disability and premature death (1). Historically, the lack of involvement from the pharmaceutical industry, in combination with limited investment in public health research programs in affected countries, resulted in a deficiency of available drugs to effectively control a majority of these diseases (2). Moreover, drugs currently in use to treat these diseases are often compromised in terms of cost, difficulties in administration, efficacy, drug resistance, or safety profiles.
Drug discovery is a time-consuming and expensive process (3,4). For NTDs in particular, drug discovery programs need to survive long enough through pervasive funding shortages to make it into subsequent clinical trials (5). In this context, a strategic approach for NTD drug discovery is drug repositioning (6), which may help lower costs by facilitating regulatory approvals in early trials for drugs that have already undergone clinical research for other diseases and/or indications and failed for reasons other than safety (6). In addition, if the scope of drug repurposing is broadened to include drugs and bioactive compounds from research on non-human organisms, it can also lead to the identification of at least new chemical tools for probing the function of targets and pathways in human pathogens. Thus, by leveraging the vast amounts of data available from well-funded research programs on human diseases and model organisms, the drug discovery landscape on NTDs gets a positive boost (7).
Computational strategies are becoming ever more essential in translational drug discovery, both in academia and in the pharmaceutical industry. Smart, intensive integration of the increasing volumes of data generated during all phases of drug discovery is already enabling key challenges of the process to be addressed (8). Since its introduction, the TDR Targets database has been a reliable resource for neglected diseases researchers to access chemogenomics data for drug target prioritization and drug repurposing on neglected diseases. Introduced in 2008 (9), this open access resource allowed researchers to find novel protein targets and chemical inhibitors, and prioritize them for aiding drug development for NTD pathogens. TDR Targets makes use of publicly available genome-wide functional datasets to allow users to find and prioritize targets based on their knowledge of the biology of their pathogen of interest, and nature of the disease (10,11). This is implemented by a flexible, user-based target selection (using filtering criteria) and ranking (using criteria-specific weighting) (12,13).
Here, we describe the upgrades to the underlying datasets and functionality in the TDR Targets resource, accumulated since its previous publication in 2012 (13). The new TDR Targets release (v6.1, abbreviated TDR6 in this paper) integrates pathogen specific genomic information with functional data (e.g. expression, orthology-based relationships, essentiality) from a selection of organisms, along with bioactive compounds data (chemical structure, property and bioactivity/target information); all of which can be queried and browsed through the web application. All queries can be saved to a personal stash by registered users and published through the web application to maximize collaboration opportunities. Prioritized lists of targets can be exported for further off-line analysis. Full details of all novel features can be found in the release notes (https://tdrtargets.org/releases). This report presents a full walkthrough of the web application, its novel features, and examples to illustrate use cases.
OVERVIEW AND ORGANIZATION OF TDR TARGETS
As in previous releases of TDR Targets, TDR6 is also organized into two main sections: Targets and Compounds. The Targets section of the database contains genome-wide data for 20 human pathogens, and allows users to run queries and prioritizations of protein targets based on a number of features and data relevant to drug discovery (see Table 1). The compounds section of the database contains information on >2 million bioactive compounds, and allows queries based on the chemical properties of the compounds and their annotated bioactivities (see Table 2).
Table 1.
Available target queries in TDR targets
| Query group | Pathogens for which data is available | Data types available for querying |
|---|---|---|
| Names & Annotations | All | Gene identifiers and functional annotations (EC numbers, GO terms, Pfam domains, metabolic pathway mappings) |
| Protein Features | All | MW, isoelectric point, presence of predicted signal peptide, trans-membrane segments and glycosylphosphatidylinositol (GPI) anchors. |
| Structural Information | All | Availability of 3D structures in PDB; availability of structural models in Modbase |
| Gene expression | Plasmodium spp.; Leishmania spp.; Trypanosoma spp.; Mycobacterium tuberculosis; Echinococcus multilocularis; Entamoeba histolytica; Toxoplasma gondii | Gene expression data from pathogen life cycle stages and/or experimental conditions that are relevant to drug discovery. |
| Phylogenetic information | All | Filter targets using simplified ‘present/absent’ in other species criteria, based on ortholog group information. Includes model organisms (human) and other related pathogens. |
| Essentiality | C. elegans (model for helminths); E. coli (model for bacteria); S. cerevisiae (model for eukaryotic pathogens); Trypanosoma brucei; Mycobacterium tuberculosis; Toxoplasma gondii; Plasmodium berghei | Ortholog-based inference of essentiality of genes in life cycle stages and/or experimental conditions relevant to drug discovery. Integrated from selected genome-wide gene disruption (e.g. transposon, CRISPR/Cas) and knockdown (e.g. RNAi) datasets in pathogens and model organisms. |
| Target Validation Data | Schistosoma mansoni; Leishmania major; Trypanosoma cruzi; Trypanosoma brucei; Mycobacterium leprae; Mycobacterium tuberculosis; Plasmodium falciparum | Manually curated data on target validation credentials (genetic, chemical and/or pharmacological, observed phenotypes) |
| Druggability | All | Precedent for successful chemical modulation of target activity or function. Summarized into a druggability score calculated from the network model (see main text) |
| Assayability | All | Available biochemical assays for protein targets (mapping based on EC numbers) |
| Bibliographic references | All | Filter targets based on available publications |
Table 2.
Available compound queries in TDR targets
| Query group | Data types available for querying |
|---|---|
| Text-based searches | |
| Names & Annotations | Compound names or synonyms; Database identifiers (e.g. ChEMBL, PubChem); InCHI and InCHI key identifiers |
| Chemical Properties | Molecular weight; LogP octanol/water partition coefficient; number of H donors and acceptors, number of flexible bonds and number of matching Ro5 (Lipinski) |
| Compound formula | Search by compounds containing a specific number (e.g. 3) of defined atoms (e.g. Cl, F, Br, N) |
| Bioactivity | Text search on assay descriptions; numerical search for values in assays (e.g. IC50 < 5 μM) |
| Orphan compounds | Search for compounds that have bioactivity reports in whole-organism or whole-cell assays but lack target and mechanism information (orphans inhibitor/drugs) |
| Compounds with targets | Find compounds that have target information and mechanism based assays |
| Structure-based searches | |
| Compound similarity | Draw/paste compound or fragment 2D structure and search for similar compounds. Search is based on matching of chemical fingerprints |
| Compound substructure | Draw/paste compound or fragment 2D structure and search for compounds in the database that contain the query fragment. |
NEW FEATURES IN TDR TARGETS 6
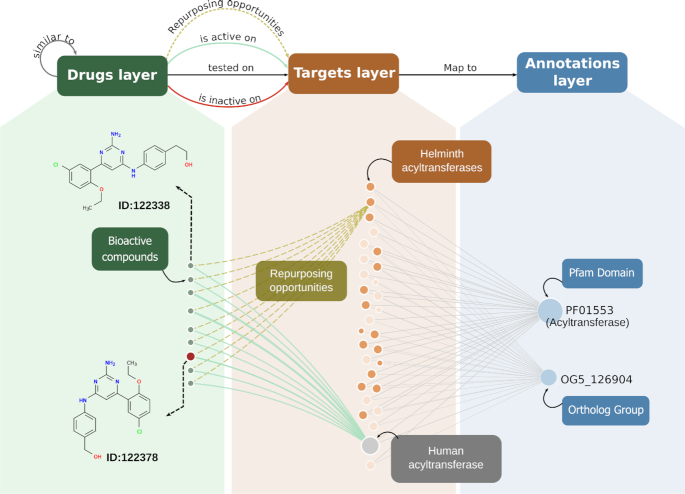
We have recently reported an integrative network model (14) where all genome-scale datasets available in TDR Targets (protein targets), chemical information (bioactive compounds) and their relations (bioactivity of compounds in target-based assays) were linked into a multilayered graph. In TDR6, this network model has been updated by integrating new datasets (described below). This model incorporates links between targets and bioactive compounds derived from manual curation of published bioactivity assays (i.e. direct links between targets and chemical compounds), as well as from computed relations (target-target links, and compound-compound links) based on protein annotations (Pfam domains, ortholog groups) and chemical similarity. A key aspect of these links in the multilayer-network model is that they enable the fast exploration and visualization of the neighborhood around selected targets and/or bioactive compounds. This allows users to explore compounds linked to targets, inspect the chemical similarity neighborhood around bioactive compounds, and visualize these data in a user friendly and comprehensive manner (see Figure 1).
Figure 1.
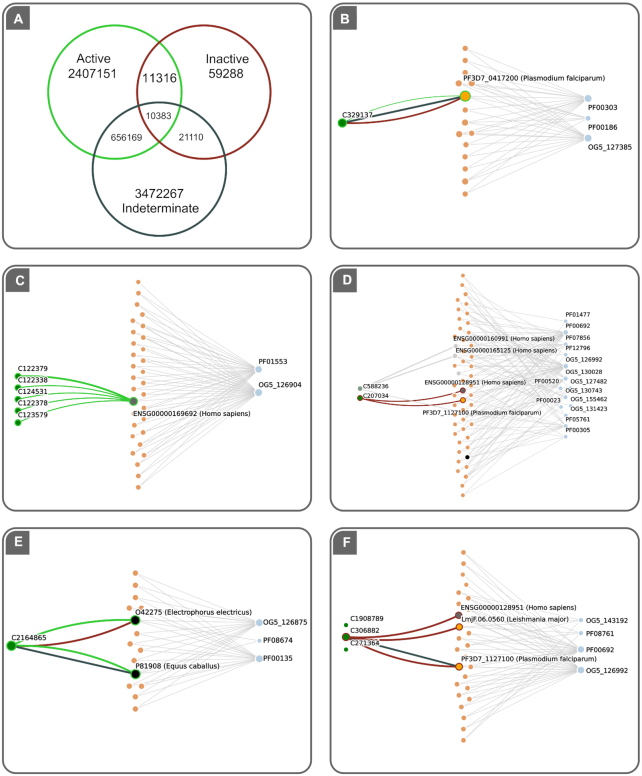
Schematic network model in TDR6 and sub-graph visualizations: Compound or target pages now display a subgraph visualization containing objects in the network vicinity of the selected entity. These graphs are built from a three-layer complex network graph. A schematic of this graph is presented at the top. Both target and compound subgraphs are always arranged as follows: Compound nodes (green) are connected to targets (orange = pathogens; gray = non-pathogens) through bioactivity edges. These links depict positive (green), neutral (gray) or negative (red) assay outcomes. Finally, targets map to a set of functional affiliations (annotations, blue nodes). In the example, the graph shows a set of known inhibitors for a human acyltransferase (15). These bioactivities (all positive) are drawn as green links between the compounds (green nodes) and the target (grey nodes for non pathogens, orange nodes for pathogens). The graph highlights the repurposing opportunities for helminth acyltransferases (dashed lines, added manually for this figure), based on shared annotations with the known druggable human target. The red node in the drug layer indicates the selected compound. Node sizes are determined by the number of connections in the network (degree), whereas bioactivity link widths (edges) are related to the cumulative number of experimental evidence for a given drug-target pair (number of assays).
With these updates, TDR6 now gives users the following functionalities: (i) network-driven whole-genome target prioritizations, (ii) exploration of drug repurposing; and iii) the exploration of candidate targets for orphan compounds. These use cases are possible by a number of precomputed network-based features such as a novel Network-Druggability Score (NDS). By associating a quantitative metric to targets based on the enrichment of bioactive compounds on closely connected network nodes, this score facilitate classification of targets into Druggability Groups (DGs), which are available to users in database queries.
The network model is also the basis for precomputed Network-Driven Prioritizations (NDPs) which can be queried by users and are also used internally by TDR6 to select connected targets and compounds for display in the newly developed network visualizations (see below). When starting from a compound of interest TDR6 uses the precomputed prioritizations of candidate targets to aid users in the navigation of the target space around the compound (and vice versa when starting from a target of interest). By providing these precomputed enrichment metrics and rankings the database now facilitates the discovery of new drug–target associations. Besides these new precomputed NDPs, users can prioritize targets using the same functionality as in previous TDR Targets releases.
This release also includes several data upgrades, namely the inclusion of 22 new genomes (20 new pathogens and 2 new model organisms), and extensive updates to chemical and bioactivity data among others. The improved and versatile user interface, together with data updates renew TDR Targets’ commitment to provide an integrated and powerful tool for exploring genomic and chemical data in the context of neglected tropical diseases.
USING TDR TARGETS 6
Whole-genome target prioritizations
The network model (14) is the base for the new druggability score, which is a network-derived metric that is related to the enrichment in bioactive compounds for a given target (NDS, ‘network druggability score’). NDSs are available for all Tier 1 organisms, which can be queried, and used to weight queries to filter (in or out) targets in user defined customized prioritization pipelines. As further explained in the network integration details, for each organism, targets were classified into five Druggability Groups (DG), from lowest (DG1) to highest scoring (DG5), according to their performance in the network prioritizations.
As in previous versions of TDR Targets, users can combine different datasets simply by running individual queries on different data types and combining them at the history page (9,10,12,13). This is useful when, for example, users would like to include additional data types to druggability-based prioritizations, such as those relying on gene expression in relevant life cycle stages, or those providing information on fitness/lethality of targets (essentiality).
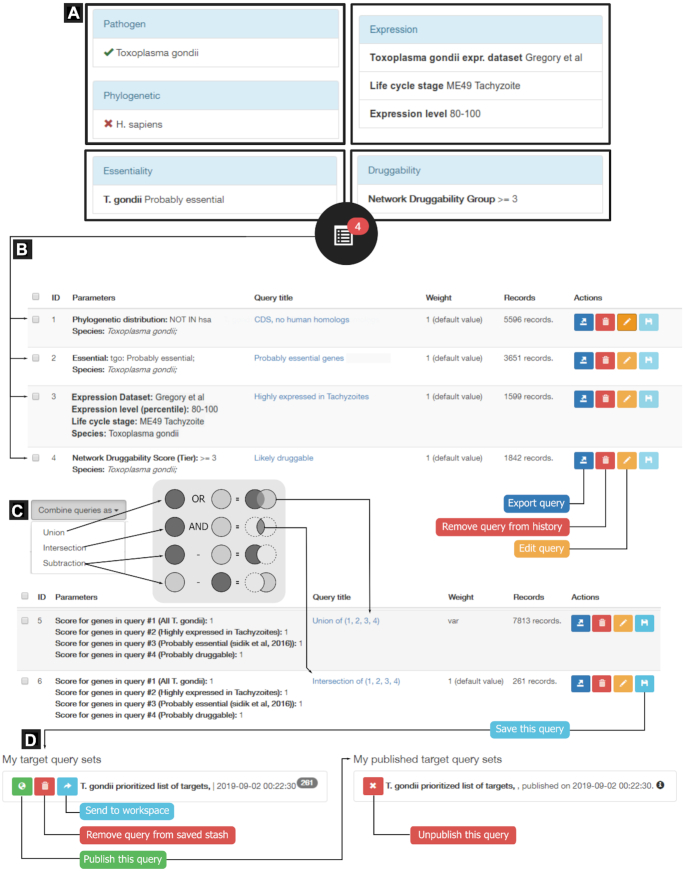
As an example, we present here a prioritization example using Toxoplasma gondii as the pathogen of interest. T. gondii is an apicomplexan parasite often used as a model to investigate the biology underlying several human and animal diseases (15). The search strategy is summarized in Figure 2. The query was started by searching for all T. gondii targets, and filtering out those targets with homologs in humans (to select only parasite-specific targets). Next, we selected candidate essential genes based on fitness profiles during infection of human fibroblasts (16); and also selected genes highly expressed in tachyzoites (replicative stage of T. gondii) by querying for genes in the top 80–100 percentile of RNAseq transcript abundance (17). These selections were combined with the network druggability rankings. For this we considered genes in druggability groups 3, 4 or 5 (DG ≥ 3) (see Figure 2). The figure shows all queries and their results as seen in the History page, and the operations performed when combining queries (union, intersection). The final list of ranked targets based on these criteria has been made public and is available in the TDR Targets section of posted lists.
Figure 2.
Target prioritization example strategy for T. gondii. The composite image shows (A) the query terms used to find T. gondii targets that have no homologs in humans, that are highly expressed in the virulent tachyzoite stage of the parasite during human cell infection, that are probably essential and are likely druggable according to Network Druggability Score. (B) Summary of queries performed at the ‘Targets’ page, showing how these queries appear in the ‘History’ page, where they can be reviewed and transformed. In-line query management buttons allow selected actions (remove, rename, export) on result-sets. (C) Query combinations allow users to execute union, intersect or subtract actions on queries with and from each other. Examples of union and intersection actions are shown. (D) Queries can be saved to a private stash (My query sets) from where they can be sent back to the workspace (to perform additional query operations) or shared publicly with other TDR Targets users (My published target query sets).
Drug repurposing strategies using query transformations
The druggability query in TDR6 allows users to select targets with known or predicted inhibitors/drugs. Information on targets with known drugs come from literature curation, whereas predicted (indirect) associations of targets with inhibitors/drugs are obtained through calculations of sequence similarity or orthology (to known druggable targets), or through network-supported inferences (14). All these methods are implemented in TDR6. Hence, when users filter a gene set based on druggability, they limit the selection to highly ranked targets, which should provide a rich source of drug repurposing opportunities.
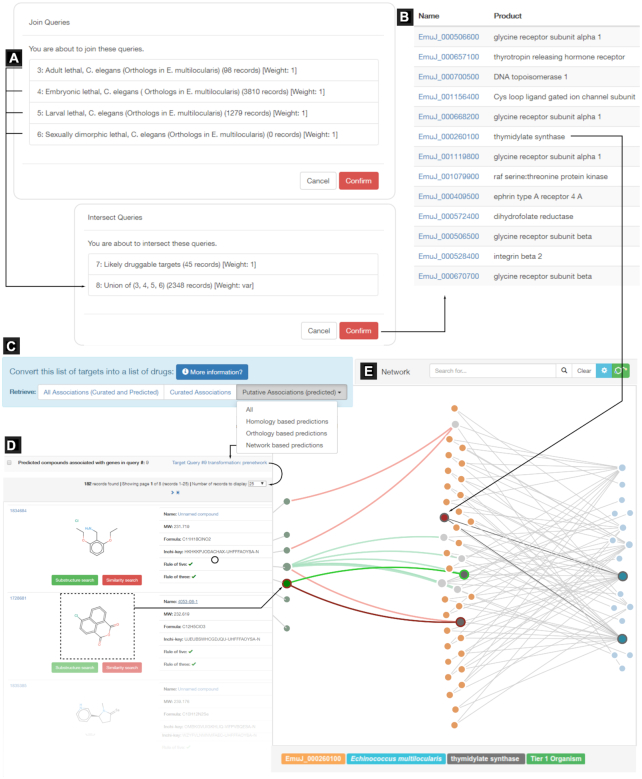
To showcase the utility of TDR6 in this area we show how to look for candidate drugs for repurposing for Echinococcus multilocularis (the causative agent of Alveolar Echinococcosis). This is shown in Figure 3. The process is similar to the one described previously for T. gondii, but in this query strategy we did not rule out human homologs, and have used C. elegans RNAi lethality datasets as a proxy for nematode essentiality. As a result, we obtained a whole-genome prioritization for E. multilocularis. Next, applying a druggability-based filter to this query, we have narrowed the gene selection to a handful of genes. The user may manually inspect the selected targets to find out which drugs were listed through indirect associations. Target pages will display all associated compounds in the druggability section, classified according to the source of the inference. For network driven inferences, the score for every compound proposed will appear both as a list and as a rank plot, to quickly identify promising candidates. Alternatively, to minimize manual inspection, the list of genes (i.e. the query itself) can be easily converted to a list of associated drugs by clicking on the ‘Convert this query’ buttons at the top of query results pages. This functionality provides a rapid way to get started on creating a screening library for a set of targets. Query transformations can be based on curated (known drugs for a set a targets), predicted (computed associations to drugs) or both. In all three approaches, the inhibitors/drugs associated with known druggable targets are transitively associated with the genes in the list. Figure 3 summarizes the prioritization strategy, the query conversion of gene list to compounds, and an example of the sub-graph visualization available from the compound page of a repurposing hit. Currently these conversions are run in the background and results appear in the History section of the website when done (users are also alerted by email).
Figure 3.
Drug repurposing opportunities for E. multilocularis using query transformations. A target prioritization scheme for Echinococcus multilocularis relying on orthology-based inference and predicted druggability (DG ≥ 3). (A) combined queries; (B) initial list of prioritized targets. (C) Any target list can be ‘transformed’ into a list of their associated drugs, using any of the available compound linking methods (see main text). (D) resulting list of bioactive compounds. (E) Example network sub-graph visualization from one selected compound, showing active and inactive bioactivity links. Compounds (green nodes) are connected to pathogen targets (orange) according to bioactivity records (green = active; red = inactive, see main text for activity thresholds). Targets, in turn, are connected (gray links) to functional affiliations (blue nodes). The sub-graph rendering provides visual hints on how the initial E. multilocularis target is connected with the selected compound in the network.
Exploration of orphan compounds
The activities of compounds extracted from the literature by curation appear in the form of target-based assays (direct link to target) or in the form of cell-based or whole-organism assays. In the absence of other information these latter classes of assays do not provide clues to the target or mechanism of action of compounds. During the process of chemical data updates in TDR6, we identified compounds with reported phenotypic effects on whole-organism or cell-based assays, based on their ChEMBL classifications. This information was used to identify ‘orphan’ compounds which are active against a particular pathogen in cell-based primary or secondary screenings, but for which there is no target-based assay.
Orphan compounds in TDR6 can be searched for any species with available phenotypic screening data, within the compounds search page. This enables a fast way of leveraging data from high-throughput assays, allowing users to start their prioritizations from compounds with known activity against a pathogen of interest.
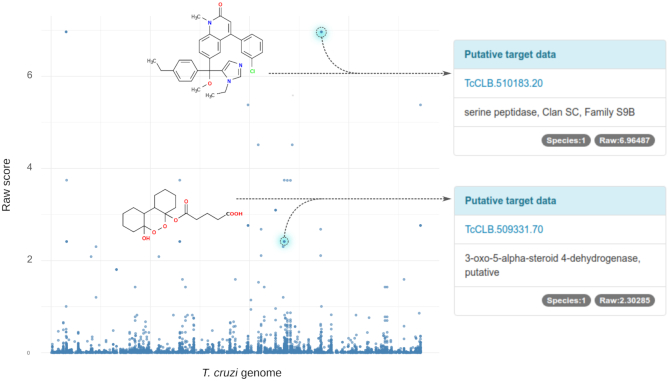
The integrated network model in TDR6 is also useful to identify candidate targets for orphan compounds. As described in the original publication (14), the computed chemical similarity neighborhood around a selected orphan compound can provide indirect links to one or more targets. Using this strategy we have performed target prioritizations for all orphan compounds in TDR6. These precomputed network-driven compound prioritizations are available for all organisms for which phenotypic screening data is available. Global summaries showing all orphan compounds for these organisms are linked from the ‘Data summary’ page (see https://tdrtargets.org/datasummary, and click on the species of interest). An example of orphan compound based prioritization for T. cruzi is shown in Figure 4. Whereas prioritizations starting from a single compound are available in each compound page.
Figure 4.
Exploration of candidate targets for Trypanosoma cruzi orphan compounds. The plot summarizes the network-driven target prioritization for orphan compounds active against T. cruzi. All protein-coding sequences in the genome of T. cruzi (candidate targets) are arranged on the x-axis. Data points in the plot correspond to target-drug associations scored by the algorithm (score plotted on the y-axis). As an example, we highlighted two putative targets for two different drugs (as displayed in the T. cruzi data summary page). Similar plots are available online for Tier 1 organisms in TDR6 (linked from the Data Summary page).
FUNCTIONALITY AND DATA UPDATES
New Genomic Data in TDR Targets v6.1
Since the previous publication of the TDR Targets database (13), several pathogen genomes have been added. A detailed list is provided in Table 3 as well as online at the TDR6 Data Summary Page (https://tdrtargets.org/datasummary).
Table 3.
Data availability summary for top tier pathogens. Summary of target data available for Tier 1 organisms in TDR Targets. CDS: Coding sequences; PFAM: number of proteins with mapped Pfam domain(s); GO: number of proteins with mapped Gene Ontology terms; EC: number of proteins with mapped Enzyme Commission (EC) numbers; Pathways: number of proteins mapped to KEGG pathway maps; Orthologs: number of sequences mapped to OrthoMCL ortholog groups. A more complete data summary table is available online at https://tdrtargets.org/datasummary
| Species | CDS | PFAM | GO | EC | Pathways | Orthologs |
|---|---|---|---|---|---|---|
| Plasmodium falciparum | 5349 | 3322 | 3551 | 750 | 1083 | 5166 |
| Plasmodium vivax | 5344 | 3264 | 2631 | 641 | 806 | 5207 |
| Toxoplasma gondii | 7946 | 4025 | 3795 | 772 | 967 | 6764 |
| Chlamydia trachomatis | 887 | 704 | 598 | 269 | 357 | 645 |
| Mycobacterium leprae | 1630 | 1236 | 929 | 628 | 611 | 1473 |
| Mycobacterium tuberculosis | 4004 | 2934 | 2001 | 1174 | 1145 | 3287 |
| Mycobacterium ulcerans | 4232 | 3602 | 2578 | 873 | 1002 | 3459 |
| Treponema pallidum | 1036 | 791 | 634 | 221 | 335 | 733 |
| Wolbachia endosymbiont of B. malayi | 805 | 628 | 577 | 308 | 382 | 688 |
| Brugia malayi | 11316 | 7042 | 6368 | 1278 | 1787 | 8424 |
| Echinococcus granulosus | 10249 | 6481 | 5432 | 854 | 1965 | 7109 |
| Echinococcus multilocularis | 10474 | 6817 | 5768 | 878 | 2079 | 7539 |
| Loa Loa (eye worm) | 16292 | 8071 | 6774 | 1539 | 2207 | 10484 |
| Onchocerca volvulus | 12224 | 3248 | 2178 | 246 | 563 | 4054 |
| Schistosoma mansoni | 12692 | 7818 | 7384 | 1218 | 1649 | 10386 |
| Leishmania major | 8280 | 4641 | 4415 | 1067 | 1162 | 8250 |
| Trypanosoma brucei | 10270 | 5665 | 5482 | 1019 | 1264 | 9259 |
| Trypanosoma cruzi | 18639 | 9908 | 8572 | 1495 | 1735 | 18140 |
| Entamoeba histolytica | 8211 | 4920 | 4087 | 645 | 1094 | 7692 |
| Giardia lamblia | 9665 | 2726 | 2263 | 326 | 514 | 5977 |
| Trichomonas vaginalis | 95600 | 35474 | 18435 | 843 | 1366 | 87303 |
Given the diversity of organisms integrated into TDR Targets and, consequently, the variety of data sources needed to cover all the genomes; substantial effort has been put into standardizing data retrieval and parsing of genome information from these organisms. Most of the complete genomes were obtained from EupathDB (18), GenBank (19), GeneDB (20), Wormbase Parasite (21), GenoList (22) or Mycobrowser (23). A full description on genome sources is given in Supplementary Table S1. To update the data for organisms present in previous version of TDR Targets protein coding genes from current release of genomes were either mapped to existing genes in TDR Targets, or otherwise entered as new records. The mapping algorithm uses a combination of conditions to track gene identifiers across releases and maintain the identity of genes: matching sequence checksums (using 128-bit hash values generated by the MD5 algorithm), gene names or identifiers and BLAST (24) if no perfect matches are found. After updating records, the pipeline calculates physicochemical properties using Pepstats (25), scans for transmembrane domains with TMHMM (26), signal peptides with SignalP (27), and glycosylphosphatidylinositol anchor points, using PredGPI (28). The algorithm dismisses all non coding sequences, as well as any pseudogenes, to avoid misleading annotations and minimizing false assumptions during prioritization workflows. As of TDR6, all tasks mentioned above for genome integration and update have been wrapped into an automated workflow to facilitate faster updates in future releases. A schematic of the update pipeline algorithm is shown in Supplementary Figure S1. The pipeline also automates the computation of annotations using ad hoc individual strategies for different annotations, relying on web services and APIs (such as the KAAS (29) service for mapping proteins to Metabolic Pathways and to the EC number classification of enzymes, or the OrthoMCL database and tool (30,31) for mapping proteins to ortholog groups. The pipeline also relies on computation against locally installed databases such as InterPro (32), using InterProScan (33) to identify protein domains (Pfam) and map terms to controlled vocabularies and classifications (GO terms). Additional resources such as 3D structures and structural models were retrieved from the Protein Data Bank (34) using web services and downloaded from the Modbase FTP site (35), respectively.
Also a number of key functional datasets were integrated in this release, including (i) transcriptomic datasets which provide evidence of gene expression in life cycle stages or experimental conditions which are relevant for drug discovery (36–47) and (ii) essentiality datasets derived from two Apicomplexan pathogens (P. berghei and T. gondii) (16,48), which provide vital information to assist prioritization strategies.
Updates of chemical data
For bioactive compounds also, the data update workflows have been automated for this release. The majority of the bioactive compounds were retrieved from ChEMBL 24th release (49), which contains some additional datasets such as those of pathogen specific chemical boxes – GSK Kinetoplastid Boxes (50), MMV Pathogen box (51). The integration process starts from molecule descriptions (2D) in SDF format, from which we calculated all necessary compound fingerprints (required for compound similarity/substructure searches) using CheckMol (52). The pipeline also calculates additional chemical properties such as the logP octanol/water partition coefficient and other structural descriptors using xLogp3 (53), and the Open Babel tools obprop and obrotamer (54). Other relevant data were obtained or calculated directly from the compound structure, such as the InChi and InChIKey (55) identifiers used for compound tracking; and other standard rules of thumb used in medicinal chemistry and drug discovery, such as Lipinski Rule of Five (56) and the related Rule of Three (57).
After integration into TDR Targets, all compounds were subject to an all vs all chemical similarity comparison calculation using ChemFP (58) which produces pairwise similarity measurements based on the Tanimoto index/distance (59). Also, we computed a global (all versus all) map of substructure relationships between compounds in the database (x is a substructure of y; y is a superstructure of x). Knowing that the problem of finding maximum common subgraphs between molecules is computationally hard, we applied a heuristic approach to find substructures. The algorithm first obtains a subset of possible candidate molecules by making use of previously calculated fingerprints. Candidates must have matching fingerprints with the subject molecule. Once a list of candidates is obtained, pairwise full atom-by-atom substructure determination is done using MatchMol (52). The data available for compounds and the queries that can be run on each data type are summarized in Table 2. The molecular weight (MW) and polar surface area (PSA) distribution for all compounds in the database is shown in Supplementary Figure S2.
Curation and integration of bioactivity data
As with chemical compounds, most bioactivities integrated into TDR Targets come directly from upstream data sources (e.g. ChEMBL). When integrating bioactivity data, we preserved both the annotation of the assay (e.g. ‘Motility reduction assay in vitro against Brugia malayi microfilariae at 10 μM’) and the numerical value and units associated with compound activities (e.g. ‘80% inhibition’, ‘1.5 μM IC50’, ‘10 nM MIC’), which are all searchable fields. In addition, and to facilitate user queries, the reported bioactivities were used to group assayed compounds into ‘active’, or ‘inactive’ classes. However, to minimize the effect of using hard boundaries around arbitrary thresholds and to increase separation between active/inactive classifications, we also defined an indeterminate grey area. Hence, compounds scoring just below an arbitrary threshold are not considered inactive for query and visualization purposes.
Not all activity types were amenable to classification, though. Despite efforts in standardization of these activity data, interpreting the activities of compounds at this scale is difficult, as they often depend on the particular assay type, reported units, and the particular conditions in which each assay was conducted. However, a significant set of assay types could be automatically classified into active/indeterminate/inactive categories based on activity thresholds. For this, all assay types with >100 000 reports (see Supplementary Figure S3 for an activity per assay type/per compound distribution plot) were considered for activity auditing, though only concentration based assays (such as IC50, Ki or Potency) were found robust enough for such determination, because percentage based assays (such as % Activity, % Residual activity or $ Inhibition) were ambiguous in bioactivity reports. The thresholds used to classify activities for each assay type can be found in Table 4, and the distribution of compounds in these activity classes is summarized in Figure 5.
Table 4.
Assay types and activity thresholds used for activity tag determination: only concentration based assays were used to determine activity tags. Activities reporting less than the maximum admitted value for positives were considered active (+) interactions, while those greater than the ‘minimum admitted value for negatives’ were considered inactive (−). Any activity reported in between these two values was considered as indeterminate (0)
| Assay type |
Standard unit |
Maximum admitted value for actives | Minimum admitted value for inactives |
|---|---|---|---|
| AC50 | nM | 20000 | 100000 |
| EC50 | nM | 20000 | 100000 |
| IC50 | nM | 20000 | 100000 |
| IC50 | ug ml−1 | 15 | 50 |
| K d | nM | 20000 | 100000 |
| K i | nM | 20000 | 100000 |
| Potency | nM | 20000 | 100000 |
Figure 5.
Activity tags distribution and evidence mixtures among data: (A) Venn diagram showing the distribution of bioactivity values in the active, inactive and indeterminate classes in TDR6 (see main text for details). Intersections count cases where the same drug has different activity outcomes against the same target. Examples of these cases are provided in panels B through F (compound IDs represent TDR6 identifiers). (B) Activity of C329137 (an hydroxy-benzamidine) against P. falciparum bifunctional dihydrofolate reductase-thymidylate synthase. (C) Example of positive records for Human acyltransferase inhibitors. (D) Example of negative and neutral activities for compounds Triphenylcarbinol and Benzohydrol, respectively. Finally, both positive (E) and negative (F) evidence may be mixed with indeterminate evidence, as shown for C2164865 tested against Horse cholinesterase and C306882 tested against recombinant P. falciparum deoxyuridine 5′-triphosphate nucleotide hydrolase, respectively.
The ChEMBL 24th release counts with over 15.2 million bioactivities reported, of which only about 6 million corresponded to relationships involving drugs and protein targets (either single proteins, protein families and protein complexes, with ∼ 93% being single proteins). Other remaining bioactivities in the database were reports for a wide variety of non-protein targets, such as whole-cells (3.6M), whole-organisms (2.2M), tissues (83K), and non-peptidic macromolecules (85K) or small molecules (<100). These were not used in network construction, because the network is protein (i.e. target) centric. Figure 5 also shows some example network visualizations that depict how TDR6 displays these bioactivities.
Integration of network-derived features: druggability and prioritizations
As mentioned above, genomic data, gene annotations, chemical compounds and gene–drug interactions were integrated into a complex network oriented to drug repurposing, as described in Berenstein et al. (14). The network was used to calculate a Network Druggability Score (NDS), for all targets in priority (Tier 1) pathogens. The NDS is related to the chance of finding bioactive compounds in the close vicinity of the network graph of a given target (range is 0 to 1). The algorithm has been previously described in detail (14), but briefly, based on an over-representation test of annotated known druggable proteins, it calculates a relevance score (RS) for every Pfam domain and Orthology group categories of the network. The NDS score for a given target results from a weighted cumulative sum over the RS’s of all affiliation contributions common to the target node, and neighbor proteins linked to active compounds.
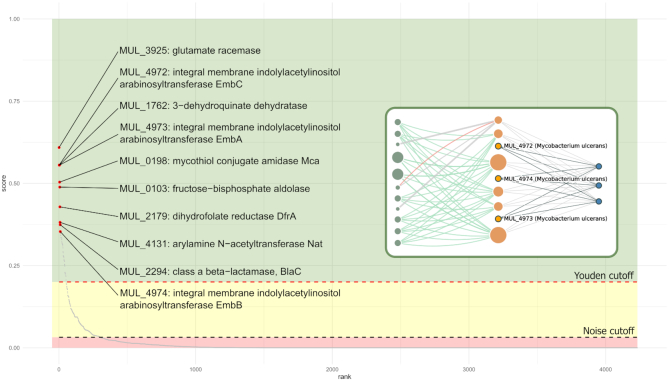
To facilitate interpretation of NDS scores we performed a statistical assessment to identify distinct Druggability Groups (DG) based on two types of thresholds that help classify druggability predictions into confidence zones. These are illustrated in Figure 6. On one hand, while all non-zero scoring targets have some degree of connectivity to known-druggable targets, a low NDS suggests these connections are not relevant for druggability assessment. Hence, a noise-cutoff (a baseline calculated as 5 times the value of 0.25 percentile from the complete NDS distribution) is considered to identify low scoring targets. The second threshold is derived from the Youden's J maximum index (60), which is calculated as the score at which both the specificity and the sensitivity are optimal (best sensitivity without compromising specificity, and vice versa). This value can only be calculated for pathogens with true positives (known druggable targets). An arbitrary minimum of 10 true positives was considered sufficient for Youden cutoff determination. For other pathogens lacking such information, a global Youden cutoff was used (calculated using all true positives in the network). The corresponding Druggability Groups are thus: DG1 for targets with NDS values ranging from 0 to the noise threshold; DG2 for targets with NDS values ranging from the noise threshold to the Youden's cutoff; and DGs 3, 4 and 5 with NDS values that are 1-, 10- and 100-fold above the Youden's cut-off. Accordingly, these latter groups make for the most likely druggable targets. Figure 6 shows a static example of a network-driven prioritization for Mycobacterium ulcerans (which lacks targets with known compounds in the current release). All prioritizations for TDR Targets priority organisms can be seen online at the data summary page for each species (see https://tdrtargets.org/datasummary, clicking on the species of interest). In this case, online plots are interactive and can be zoomed and exported. In cases where there are targets with known bioactive compounds for the species, these are shown distinctively in the plot.
Figure 6.
Network-driven whole-genome target prioritization for Mycobacterium ulcerans: Candidate targets in the M. ulcerans genome were ranked by their NDS (network druggability score, see main text). The plot depicts all genome targets (in the x axis) along with their corresponding NDS score (in the y axis). Red points correspond to the top-10 ranked targets, with labels indicating the gene name and product. Browsing whole-genome prioritization from the data summary, the user may access a gene page by clicking on it in the prioritization plot. A subgraph example from EmbA/EmbB/EmbC gene family is shown (as seen in their corresponding gene pages). The figure also displays confidence zones, DG1 (red): delimited by zero and noise cutoff; DG2 (yellow): between the noise and the Youden cutoff; and DG3–5: with scores higher than the Youden cutoff.
These network-driven prioritizations can work both ways. When starting from a compound of interest, the algorithm can prioritize targets, using the weighted similarity of chemical neighbors to initial candidate targets. And when starting from target of interest, it can prioritize compounds, using connected druggable neighbor targets and then following weighted links to candidate inhibitors/drugs. Precomputed scores for compounds and for targets are used internally by TDR6 and are at the core of network-based query transformations.
Network sub-graph visualizations and User Interface upgrade
The network sub-graphs for both compounds and targets (and their respective NDS scores) can be browsed from the web application using a drug or a target as a starting point to obtain hints for untested drugs or novel druggable targets, respectively. Through newly developed visualizations users can check out the network neighborhoods around drugs and targets in the corresponding pages. Lists of network derived putative interactions can also be explored in tabular format under the ‘Druggability’ (for targets) and ‘Known and predicted targets’ (for drugs) sections.
These visualizations are driven by D3.js (61) implementing forced layouts for sub-graph visualizations. Within the D3 subgraph panel, users can perform node searches within the graph (target identifiers), as well as toggle the visibility of targets on a species by species manner, and customize the opacity of nodes. Taken together these new features provide a clear and comprehensive visualization of the sub-network vicinity of targets and compounds, allowing users to manipulate the graphs while exploring the data.
The user interface (UI) and the available tools for drug repurposing and target prioritization have gone through a major upgrade. In the first place, the UI has been redesigned under W3C standards to achieve a healthier and more scalable application. We integrated the Bootstrap (https://getbootstrap.com/) and jQuery (https://jquery.com) frameworks in the development and design of the TDR6 web application and in the front-end functionality. For compound structure queries we have licensed and implemented the Marvin JS chemical drawing application from Chemaxon (https://chemaxon.com/products/marvin-js). Tabulated records within target and drug pages now use the DataTable javascript jquery plugin (https://datatables.net) to easily create paginations, filtering and sorting functionalities. Finally, compound 2D representations are now automatically generated using an implementation of the SmilesDrawer javascript module (62).
Commercial availability of compounds
One important aspect when prioritizing compounds for testing in the lab, is their availability. In TDR6 we are now displaying information on commercial availability of compounds. Currently we have started this feature by linking with Molport (a chemical online marketplace that sources compounds from major suppliers) and show users a visual clue on compound pages that give a fast indication of whether the compound is either in stock or can be made to order. Because commercial availability of compounds is currently implemented in TDR6 in the form of asynchronous queries against Molport, at this time this feature is only available in browsing mode (not in queries). However, users can prioritize compounds using any of the available query strategies in TDR6 and then finalize their compound selections by inspecting compounds manually for commercial availability.
DISCUSSION AND FUTURE DIRECTIONS
The new data, interface and functionality of TDR6 provides users with improved navigation and visualization of targets and compounds.
The current network model connects targets through affiliation of entities (proteins) to annotation concepts (Pfam domains, Ortholog groups). These have been selected based on their wide coverage and relative ease of calculation. Complementing these concepts with other important criteria for drug target validation (essentiality, expression in relevant life cycle stages) can be done by users with the tools and functionality provided by TDR6 but in the future they can be built into the underlying network model itself, at least for some organisms amenable to genome-wide experimental assessment.
Several key improvements are necessary to keep TDR Targets relevant for the community of scientists working on tropical diseases. Integration of natural metabolites, and connecting these small molecules to other bioactive compounds through shared substructures or by chemical similarity will be a major focus in the future. This will allow navigating the drug-targets graph using the concepts of biochemical reactions also, which naturally connect non-orthologous enzymes through their shared substrates/products and cofactors.
Finally, as already mentioned before (13), there is still a large curation gap that needs to be filled. Many bioactive compounds have been tested by the community of researchers working in Neglected Tropical Diseases. Yet many of these assays and outcomes are reported in journals outside the mainstream Medicinal Chemistry journals and thus are missed by large curation efforts such as the one led by ChEMBL (49). Curation and integration of these missing data (including negative data!) should be a priority for the community, as it would save valuable time and resources.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Matthew Berriman and Magdalena Zarowiecki (Wellcome Trust Sanger Institute) for sharing pre-release Echinococcus genome data and annotation for inclusion in TDR Targets; and Ben Webb and Andrej Sali (University of California San Francisco) for the calculation of 3D models for Tier 1 pathogen genomes in TDR Targets. L.U.L., A.B. and S.V. were or are supported by fellowships from the National Research Council (CONICET, Argentina). A.C. and F.A. are members of the Research Career of the National Research Council (CONICET, Argentina). P.M. would like to acknowledge a fellowship from University Grants Commission (UGC), India.
Notes
Present address: Ariel J Berenstein, Instituto Multidisciplinario de Investigaciones en Patologías Pediátricas (IMIPP), CONICET-GCBA, Laboratorio de Biología Molecular, División Patología, Hospital de Niños Ricardo Gutiérrez, Ciudad Autónoma de Buenos Aires, Argentina.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
GlaxoSmithKline Argentina and the National Agency for the Promotion of Science and Technology, Argentina (ANPCyT) [PICTO-Glaxo-2013-0067 to F.A.]; Indo-Argentina Bilateral Cooperation Project (Joint Funding from the Indian Department of Science and Technology (DST) and the Argentinian Ministry of Science and Technology (MINCyT) [IN-1405 to F.A. and D.S.]. Funding for open access charge: Fondo para la Investigación Científica y Tecnológica [PICT-2017-0175].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hotez P.J., Molyneux D.H., Fenwick A., Kumaresan J., Sachs S.E., Sachs J.D., Savioli L.. Control of neglected tropical diseases. N. Engl. J. Med. 2007; 357:1018–1027. [DOI] [PubMed] [Google Scholar]
- 2. Trouiller P., Olliaro P., Torreele E., Orbinski J., Laing R., Ford N.. Drug development for neglected diseases: a deficient market and a public-health policy failure. Lancet North Am. Ed. 2002; 359:2188–2194. [DOI] [PubMed] [Google Scholar]
- 3. Hughes J., Rees S., Kalindjian S., Philpott K.. Principles of early drug discovery. Br. J. Pharmacol. 2011; 162:1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adams C.P., Brantner V.V.. Estimating the cost of new drug development: is it really $802 million. Health Aff. (Millwood). 2006; 25:420–428. [DOI] [PubMed] [Google Scholar]
- 5. Wyatt P.G., Gilbert I.H., Read K.D., Fairlamb A.H.. Target validation: linking target and chemical properties to desired product profile. Curr. Top. Med. Chem. 2011; 11:1275–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farha M.A., Brown E.D.. Drug repurposing for antimicrobial discovery. Nat. Microbiol. 2019; 4:565–577. [DOI] [PubMed] [Google Scholar]
- 7. Hernandez H.W., Soeung M., Zorn K.M., Ashoura N., Mottin M., Andrade C.H., Caffrey C.R., de Siqueira-Neto J.L., Ekins S.. High throughput and computational repurposing for neglected diseases. Pharm. Res. 2018; 36:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wooller S.K., Benstead-Hume G., Chen X., Ali Y., Pearl F.M.G.. Bioinformatics in translational drug discovery. Biosci. Rep. 2017; 37:doi:10.1042/BSR20160180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Agüero F., Al-Lazikani B., Aslett M., Berriman M., Buckner F.S., Campbell R.K., Carmona S., Carruthers I.M., Chan A.W.E., Chen F. et al.. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat. Rev. Drug Discov. 2008; 7:900–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crowther G.J., Shanmugam D., Carmona S.J., Doyle M.A., Hertz-Fowler C., Berriman M., Nwaka S., Ralph S.A., Roos D.S., Van Voorhis W.C. et al.. Identification of attractive drug targets in neglected-disease pathogens using an in silico approach. PLoS Negl. Trop. Dis. 2010; 4:e804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lykins J.D., Filippova E.V., Halavaty A.S., Minasov G., Zhou Y., Dubrovska I., Flores K.J., Shuvalova L.A., Ruan J., El Bissati K. et al.. CSGID solves structures and identifies phenotypes for five enzymes in Toxoplasma gondii. Front. Cell. Infect. Microbiol. 2018; 8:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shanmugam D., Ralph S.A., Carmona S.J., Crowther G.J., Roos D.S., Agüero F.. Caffrey CR, Selzer PM. Integrating and Mining Helminth Genomes to Discover and Prioritize Novel Therapeutic Targets. Parasitic Helminths: Targets, Screens, Drugs and Vaccines. 2012; Wiley-Blackwell; 43–59. [Google Scholar]
- 13. Magariños M.P., Carmona S.J., Crowther G.J., Ralph S.A., Roos D.S., Shanmugam D., Van Voorhis W.C., Agüero F.. TDR Targets: a chemogenomics resource for neglected diseases. Nucleic. Acids. Res. 2012; 40:D1118–D1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berenstein A.J., Magariños M.P., Chernomoretz A., Agüero F.. A multilayer network approach for guiding drug repositioning in neglected diseases. PLoS Negl. Trop. Dis. 2016; 10:e0004300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim K., Weiss L.M.. Toxoplasma gondii: the model apicomplexan. Int. J. Parasitol. 2004; 34:423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sidik S.M., Huet D., Ganesan S.M., Huynh M.-H., Wang T., Nasamu A.S., Thiru P., Saeij J.P.J., Carruthers V.B., Niles J.C. et al.. A genome-wide CRISPR screen in Toxoplasma identifies essential apicomplexan genes. Cell. 2016; 166:1423–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gajria B., Bahl A., Brestelli J., Dommer J., Fischer S., Gao X., Heiges M., Iodice J., Kissinger J.C., Mackey A.J. et al.. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2007; 36:D553–D556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Warrenfeltz S., Basenko E.Y., Crouch K., Harb O.S., Kissinger J.C., Roos D.S., Shanmugasundram A., Silva-Franco F.. Kollmar M. EuPathDB: the eukaryotic pathogen genomics database resource. Eukaryotic Genomic Databases. 2018; 1757:NY: Springer; 69–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sayers E.W., Agarwala R., Bolton E.E., Brister J.R., Canese K., Clark K., Connor R., Fiorini N., Funk K., Hefferon T. et al.. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019; 47:D23–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hertz-Fowler C., Peacock C.S.. Introducing GeneDB: a generic database. Trends Parasitol. 2002; 18:465–467. [Google Scholar]
- 21. Bolt B.J., Rodgers F.H., Shafie M., Kersey P.J., Berriman M., Howe K.L.. Using wormbase parasite: an integrated platform for exploring helminth genomic data. Methods Mol. Biol. 2018; 1757:471–491. [DOI] [PubMed] [Google Scholar]
- 22. Lechat P., Hummel L., Rousseau S., Moszer I.. GenoList: an integrated environment for comparative analysis of microbial genomes. Nucleic Acids Res. 2007; 36:D469–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kapopoulou A., Lew J.M., Cole S.T.. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis. 2011; 91:8–13. [DOI] [PubMed] [Google Scholar]
- 24. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hancock J.M., Bishop M.J.. EMBOSS (The European Molecular Biology Open Software Suite). Dictionary of Bioinformatics and Computational Biology. 2004; Chichester: John Wiley & Sons, Ltd; dob0206. [Google Scholar]
- 26. Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001; 305:567–580. [DOI] [PubMed] [Google Scholar]
- 27. Almagro Armenteros J.J., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H.. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019; 37:420–423. [DOI] [PubMed] [Google Scholar]
- 28. Pierleoni A., Martelli P., Casadio R.. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008; 9:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M.. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007; 35:W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen F., Mackey A.J., Stoeckert C.J. Jr, Roos D.S.. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006; 34:D363–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fischer S., Brunk B.P., Chen F., Gao X., Harb O.S., Iodice J.B., Shanmugam D., Roos D.S., Stoeckert C.J. Jr. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinforma. 2011; doi:10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mitchell A.L., Attwood T.K., Babbitt P.C., Blum M., Bork P., Bridge A., Brown S.D., Chang H.-Y., El-Gebali S., Fraser M.I. et al.. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019; 47:D351–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G. et al.. InterProScan 5: genome-scale protein function classification. Bioinforma. Oxf. Engl. 2014; 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Costanzo L.D., Christie C., Duarte J.M., Dutta S., Feng Z. et al.. Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019; 47:D520–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pieper U., Webb B.M., Dong G.Q., Schneidman-Duhovny D., Fan H., Kim S.J., Khuri N., Spill Y.G., Weinkam P., Hammel M. et al.. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2014; 42:D336–D346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu L., Mok S., Imwong M., Jaidee A., Russell B., Nosten F., Day N.P., White N.J., Preiser P.R., Bozdech Z.. New insights into the Plasmodium vivax transcriptome using RNA-Seq. Sci. Rep. 2016; 6:20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smircich P., Eastman G., Bispo S., Duhagon M.A., Guerra-Slompo E.P., Garat B., Goldenberg S., Munroe D.J., Dallagiovanna B., Holetz F. et al.. Ribosome profiling reveals translation control as a key mechanism generating differential gene expression in Trypanosoma cruzi. BMC Genomics. 2015; 16:443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lasonder E., Rijpma S.R., van Schaijk B.C.L., Hoeijmakers W.A.M., Kensche P.R., Gresnigt M.S., Italiaander A., Vos M.W., Woestenenk R., Bousema T. et al.. Integrated transcriptomic and proteomic analyses of P. falciparum gametocytes: molecular insight into sex-specific processes and translational repression. Nucleic Acids Res. 2016; 44:6087–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Otto T.D., Wilinski D., Assefa S., Keane T.M., Sarry L.R., Böhme U., Lemieux J., Barrell B., Pain A., Berriman M. et al.. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 2010; 76:12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Otto T.D., Böhme U., Jackson A.P., Hunt M., Franke-Fayard B., Hoeijmakers W.A.M., Religa A.A., Robertson L., Sanders M., Ogun S.A. et al.. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 2014; 12:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zanghì G., Vembar S.S., Baumgarten S., Ding S., Guizetti J., Bryant J.M., Mattei D., Jensen A.T.R., Rénia L., Goh Y.S. et al.. A Specific PfEMP1 is expressed in P. falciparum sporozoites and plays a role in hepatocyte infection. Cell Rep. 2018; 22:2951–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fernandes M.C., Dillon L.A.L., Belew A.T., Bravo H.C., Mosser D.M., El-Sayed N.M.. Dual transcriptome profiling of leishmania-infected human macrophages reveals distinct reprogramming signatures. mBio. 2016; 7:e00027-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fritz H.M., Buchholz K.R., Chen X., Durbin-Johnson B., Rocke D.M., Conrad P.A., Boothroyd J.C.. Transcriptomic analysis of toxoplasma development reveals many novel functions and structures specific to sporozoites and oocysts. PLoS One. 2012; 7:e29998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hon C.-C., Weber C., Sismeiro O., Proux C., Koutero M., Deloger M., Das S., Agrahari M., Dillies M.-A., Jagla B. et al.. Quantification of stochastic noise of splicing and polyadenylation in Entamoeba histolytica. Nucleic Acids Res. 2013; 41:1936–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Siegel T.N., Hekstra D.R., Wang X., Dewell S., Cross G.A.M.. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010; 38:4946–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yeoh L.M., Goodman C.D., Mollard V., McFadden G.I., Ralph S.A.. Comparative transcriptomics of female and male gametocytes in Plasmodium berghei and the evolution of sex in alveolates. BMC Genomics. 2017; 18:734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hehl A.B., Basso W.U., Lippuner C., Ramakrishnan C., Okoniewski M., Walker R.A., Grigg M.E., Smith N.C., Deplazes P.. Asexual expansion of Toxoplasma gondii merozoites is distinct from tachyzoites and entails expression of non-overlapping gene families to attach, invade, and replicate within feline enterocytes. BMC Genomics. 2015; 16:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bushell E., Gomes A.R., Sanderson T., Anar B., Girling G., Herd C., Metcalf T., Modrzynska K., Schwach F., Martin R.E. et al.. Functional profiling of a Plasmodium genome reveals an abundance of essential genes. Cell. 2017; 170:260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., Magariños M.P., Mosquera J.F., Mutowo P., Nowotka M. et al.. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019; 47:D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peña I., Pilar Manzano M., Cantizani J., Kessler A., Alonso-Padilla J., Bardera A.I., Alvarez E., Colmenarejo G., Cotillo I., Roquero I. et al.. New compound sets identified from high throughput phenotypic screening against three kinetoplastid parasites: an open resource. Sci. Rep. 2015; 5:8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spangenberg T., Burrows J.N., Kowalczyk P., McDonald S., Wells T.N.C., Willis P.. The open access malaria box: a drug discovery catalyst for neglected diseases. PLoS One. 2013; 8:e62906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Haider N. Functionality pattern matching as an efficient complementary structure/reaction search tool: an open-source approach. Molecules. 2010; 15:5079–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheng T., Zhao Y., Li X., Lin F., Xu Y., Zhang X., Li Y., Wang R., Lai L.. Computation of octanol-water partition coefficients by guiding an additive model with knowledge. J. Chem. Inf. Model. 2007; 47:2140–2148. [DOI] [PubMed] [Google Scholar]
- 54. O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R.. Open Babel: an open chemical toolbox. J. Cheminformatics. 2011; 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Heller S.R., McNaught A., Pletnev I., Stein S., Tchekhovskoi D.. InChI, the IUPAC International Chemical Identifier. J. Cheminformatics. 2015; 7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J.. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001; 46:3–26. [DOI] [PubMed] [Google Scholar]
- 57. Congreve M., Carr R., Murray C., Jhoti H.. A ‘Rule of Three’ for fragment-based lead discovery. Drug Discov. Today. 2003; 8:876–877. [DOI] [PubMed] [Google Scholar]
- 58. Dalke A. chemfp - fast and portable fingerprint formats and tools. J. Cheminformatics. 2011; 3:P12. [Google Scholar]
- 59. Rogers D.J., Tanimoto T.T.. A computer program for classifying Plants. Science. 1960; 132:1115–1118. [DOI] [PubMed] [Google Scholar]
- 60. Youden W.J. Index for rating diagnostic tests. Cancer. 1950; 3:32–35. [DOI] [PubMed] [Google Scholar]
- 61. Bostock M., Ogievetsky V., Heer J.. D3 data-driven documents. IEEE Trans. Vis. Comput. Graph. 2011; 17:2301–2309. [DOI] [PubMed] [Google Scholar]
- 62. Probst D., Reymond J.-L.. Smilesdrawer: parsing and drawing SMILES-encoded molecular structures using client-side javascript. J. Chem. Inf. Model. 2018; 58:1–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.