Abstract
Bacterial viruses encode a vast number of ORFan genes that lack similarity to any other known proteins. Here, we present a 2.20 Å crystal structure of N4-related Pseudomonas virus LUZ7 ORFan gp14, and elucidate its function. We demonstrate that gp14, termed here as Drc (ssDNA-binding RNA Polymerase Cofactor), preferentially binds single-stranded DNA, yet contains a structural fold distinct from other ssDNA-binding proteins (SSBs). By comparison with other SSB folds and creation of truncation and amino acid substitution mutants, we provide the first evidence for the binding mechanism of this unique fold. From a biological perspective, Drc interacts with the phage-encoded RNA Polymerase complex (RNAPII), implying a functional role as an SSB required for the transition from early to middle gene transcription during phage infection. Similar to the coliphage N4 gp2 protein, Drc likely binds locally unwound middle promoters and recruits the phage RNA polymerase. However, unlike gp2, Drc does not seem to need an additional cofactor for promoter melting. A comparison among N4-related phage genera highlights the evolutionary diversity of SSB proteins in an otherwise conserved transcription regulation mechanism.
INTRODUCTION
The availability of sequencing technologies has led to a vast increase in available genome sequences of microorganisms. Consequently, the number of unknown open reading frames whose products are termed ‘ORFans’, continues to increase. These ORFans have no sequence similarity to known sequences, which makes it hard to predict their function (1). Research on ORFans often results in discovery of very diverged known proteins or sometimes entirely new classes and folds (2–5). As such, their analysis can vastly expand the set of known structural folds and domains, which in turn will allow improved structural and functional predictions. Bacterial viruses encode a vast array of ORFans (35% of their ORFs), of which about a third are shared by fewer than five different bacteriophages (6). Many phage ORFans are expressed immediately after infection and are likely required for host takeover (7). Indeed, several ORFans have been shown to mediate crucial host processes in very distinct and unique ways (8). Apart from the search for new functionalities, domains and folds, the study of these ORFans is key to understanding host reprogramming in the course of infection and can lead to novel antibacterial strategies (9) or discovery of new biotechnological tools (7,10–12). Multiple studies have been conducted to harness ORFans potential, mostly using screening-based techniques. These range from mapping the protein–protein interaction networks within the phage (13) or with the host (14–16), to observation of phage phenotypes upon gene deletions (17), or host phenotypes upon phage genes expression (9,18,19).
While these screens provide vital clues, further in-depth experimentation is required to fully unravel the function of an individual ORFan. Here we focus on a small ORFan protein (gp14) from Pseudomonas virus LUZ7, which was identified as toxic to the host in one such screen (18). LUZ7 gp14 causes filamentous growth of the host, but to date no host protein interaction partners have been found, despite an extensive yeast two-hybrid screen (18). We demonstrate that gp14 (here termed Drc, ssDNA-binding RNA Polymerase cofactor) interacts with single-strand DNA (ssDNA) by means of a unique ssDNA-binding protein (SSB) fold. Additionally, by showing interaction with the phage-encoded RNA Polymerase (RNAPII), we establish the biological role of Drc in the transcription regulation of the N4-related Pseudomonas virus LUZ7. Drc is likely involved in the same process as the coliphage N4-encoded gp2 protein, which mediates the recruitment of the viral RNAPII complex to its single-stranded promoters (20). Even though N4-related viruses have a generally conserved transcriptional progression scheme, further phylogenetic analysis unveils that there is diversity of transcription-activating SSB proteins across the N4-like phage genera.
MATERIALS AND METHODS
Construction of expression plasmids
Genomic LUZ7 DNA was used to amplify ORF14, ORF20 and ORF22. ORF14 was amplified using primers 5′-ATGGATCCATGGCACTCGTCAAGAAGAA-3′ and 5′-CTGAATTCTTACAGGTCGAGCGCG-3′ prior to restriction cloning in pGEX-6P-1 (GE Healthcare) with BamHI and EcoRI, which results in Drc having an N-terminal GST-tag fusion. ORF20 and ORF22 were cloned in MCS1 and MCS2 of pRSFDuet-1 (Novagen) using BamHI and SacI or NdeI and KpnI, respectively. Primer pairs 5′-GTAGGATCCTCACCCAACTCTAATGCTC-3′ and 5′-GACGAGCTCTCATTTGGTAATCCTCAGATG-3′ or 5′-GATCATATGAAGCGTTACACTGGCTTTG-3′ and 5′-CTTGGTACCTTAAGACAGGGCATACTCACTCTC-3′, were used for their PCR amplification. The resulting plasmid encodes an N-terminally 6xHis-tagged RNAP1 subunit and untagged RNAP2 subunit. Drc.Y23A was created by a modified inverse PCR (21). In short, two non-overlapping primers of which one contains the desired mutation (5′-CAACAAGGGCGCTTCGGCCGCCCTGAACTTCCACTTC-3′, 5′-TCGGTGGCTTGGGTGTTGCGAGCTTGGTTC-3′), were phosphorylated with PNK (Thermo Fisher Scientific) and used to amplify 150 pg of template plasmid. The amplicon was then purified by agarose gel electrophoresis followed by ligation, propagation and sequencing of the plasmid. The same process was used to add a C-terminal Strep-tag II to the RNAP2 subunit, using primers 5′-TAAGGTACCCTCGAGTCTGGTAAAGAAAC-3′ and 5′-TTTTTCGAACTGCGGGTGGCTCCAAGCGCTAGACAGGGCATACTCACTCTC-3′. Drc(Δ67–99) was cloned into pEXP-5-CT/TOPO (Thermo Fisher Scientific) after amplification from LUZ7 genomic DNA with primers 5′-ATGGCACTCGTCAAGAAGAAC-3′ and 5′-CAGCGGCTTGCCCTTGTC-3′, thus encoding a C-terminally 6xHis-tagged protein. ORF14 was also cloned into pEXP-5-CT/TOPO with primers 5′-ATGGCACTCGTCAAGAAGAAC-3′ and 5′-TTACAGGTCGAGCGCGCGCTC-3′, after which a Strep-tag II was added by inverse PCR with primers 5′-AGCGCTTGGAGCCACCCGCAGTTCGAAAAATAAAAGGGTCATCATCACCATC-3′ and 5′-CAGGTCGAGCGCGCGCTC-3′.
Expression and purification of GST-tagged proteins (Drc and Drc.Y23A)
All expressions were done with Escherichia coli BL21(DE3)pLysS expression strains. Cells were grown in Lysogeny Broth (LB) with 100 μg/ml ampicillin at 37°C to OD600 of 0.6. Expression was then induced by addition of 1 mM IPTG and cells were grown for an additional 4h at 37°C. The culture was pelleted by centrifugation (4600 g, 4°C). Cells were resuspended in Lysis buffer (150 mM NaCl, 100 mM Tris–HCl, pH 7.6, 100 μM PefaBloc SC (Merck), 2 mg/ml HEW Lysozyme (Sigma)), freeze-thawed three times, DNase I treated (Thermo Fisher Scientific) and pulse-sonicated on ice. The lysate was centrifuged at 60 000 g and filtered over a 0.22 μm filter (Merck). This cleared lysate was purified on an ÄKTA FPLC (GE Healthcare) by affinity chromatography using a 5 ml GSTrap HP column (GE Healthcare). Before loading, the column was equilibrated with wash buffer (140 mM NaCl, 10 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3). After loading, the column was washed again with the same buffer before elution with 10 mM reduced glutathione, 50 mM Tris–HCl (pH 8). After assessment by SDS-PAGE for presence of the correct protein, fractions were pooled and concentrated with a 3K Microsep Advance Centrifugal device (Pall) and then dialyzed in a Slide-A-Lyzer MINI dialysis device, 3.5K MWCO (Thermo Fisher Scientific) against digestion buffer (50 mM Tris–HCl, pH 7.3, 150 mM NaCl, 1 mM EDTA, 1 mM DTT). The GST-tag was cleaved off overnight at 4°C with PreScission Protease (2.5 U/mg; GE Healthcare) and removed by gel filtration on a HiLoad 16/600 SuperDex 75 pg column (GE healthcare) with running buffer (150 mM NaCl, 100 mM Tris–HCl, pH 7.6). Protein fractions were pooled as before, flash-frozen and stored at −80°C. Protein concentrations were determined by absorbance measurement at 280 nm on a nanodrop 1000 (Thermo Fisher Scientific). Concentrations for Drc and mutants thereof were calculated assuming protein dimer formation. The resulting proteins have a five amino acid scar at the N-terminus and are referred to in the assays as Drc or Drc.Y23A for the wild-type and Y23A mutant protein, respectively.
Expression and purification of His-tagged proteins (RNAPII, RNAPII.Strep and Drc(Δ67-99))
Proteins in the assays termed RNAPII, RNAPII.strep consist of two subunits, RNAP1 and RNAP2. In RNAPII there is a single N-terminal His-tag on RNAP1, whereas RNAPII.strep also contains a C-terminal strep-tag on the RNAP2 subunit. Drc(Δ67–99) has a C-terminal His-tag. Expression of these proteins was performed as described for GST-tagged proteins, with minor changes. Cells were grown in LB with 50 μg/ml kanamycin at 37°C and were transferred to 30°C overnight incubation after induction with 1 mM IPTG (0.5 mM for Drc(Δ67–99)). Lysates were created in a lysis buffer containing 50 mM Tris–HCl, pH 8, 0.5 M NaCl, 100 μM PefaBloc SC and 2 mg/ml HEW Lysozyme. Lysates were purified with a 1 ml Protino Ni-NTA column (Macherey-Nagel), which was pre-equilibrated in imidazole wash buffer (50 mM Tris–HCl, pH 8.0, 0.5 M NaCl, 30 mM imidazole). After loading this buffer was used to wash the column before eluting in the same buffer with 500 mM imidazole. Fractions were pooled and concentrated using a 10K Microsep Advance Centrifugal device (Pall), and then dialysed against running buffer and stored at −80°C. In case of Drc(Δ67–99) an additional gel filtration step was performed as was described for GST-tagged proteins.
Expression and purification of Strep-tag II proteins (Drc.strep)
Proteins in the assays termed Drc.strep have a C-terminal strep-tag and their expression was performed as described for GST proteins. Lysates were created similarly with a different lysis buffer (100 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.1% Nonidet P-40, 100 μM PefaBloc SC and 2 mg/ml HEW Lysozyme). Proteins were purified on a column loaded with 2 ml Strep-Tactin sepharose beads (IBA). Buffers W and E were used as wash and elution buffers according to the manufacturer's instructions. Before storage at −80°C, fractions were pooled and concentrated using a 3K Microsep Advance Centrifugal device (Pall) and dialysed to running buffer.
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed as described previously (22). In short, for EMSAs on agarose gel, protein dilutions were mixed with the DNA (PhiX174 virion DNA and/or PhiX174 RF I; NEB) in reaction buffer (20 mM Tris–HCl, pH 8.0, 1 mM MgCl2, 50 mM NaCl, 0.4 mM EDTA, 0.1 mM DTT, 12.5% glycerol) and incubated for 20 min at room temperature. Samples were mixed with 6× DNA loading dye (Thermo Fisher Scientific) and loaded on a 1% agarose gel pre-stained with ethidium bromide or stained afterwards with SYBR Gold (Thermo Fisher Scientific) or by Coomassie staining (GelCode Blue Safe Protein Stain, Thermo Fisher Scientific). The gel was run at 100 V for 30 min prior to visualization with UV light.
Sample preparation for EMSA with short, 5′ fluorescein amidite (6-FAM) labelled DNA was performed similarly. Labelled DNA (5′-6-FAM-TGAGTTTTTTTCATTTTTTGCGTAAATTTC-3′, used for both ssDNA and dsDNA, and 5′-GAAATTTACGCAAAAAATGAAAAAAACTCA-3′ for generation of dsDNA) was ordered from IDT and prepared as described by Peeters et al. (22). For the internally melted duplex test, 5′-6-FAM-GGGCGGCGGTTTTTTTTTTGCGGGGCGG-3′ was combined with either 5′-CCGCCCCGCTTTTTTTTTTCCGCCGCCC-3′ or 5′-CCGCCCCGCAAAAAAAAAACCGCCGCCC-3′ to create internally melted and full duplex, respectively. The prepared DNA was mixed with the protein dilutions in aforementioned reaction buffer to a 100 nM final concentration. After incubation, these samples were run on a 12% native gel with a TBE running buffer (90 mM Tris–HCl, 90 mM boric acid, 2 mM EDTA) at 150 V on ice for 35 min. Gels were imaged at an excitation wavelength of 470 nm with a G:BOX imager, equipped with an UltraBright-LED transilluminator (Syngene). All EMSAs were performed in duplicate and results show representative gels.
Mobility shift assay (MSA)
The assayed proteins were diluted in protein buffer to the required concentrations and incubated at room temperature for 20 min. Loading dye (50% (w/v) glycerol, 0.2% (w/v) bromophenol blue, 300 mM DTT) was added before loading the samples on a 10% native acrylamide gel. Gels were run on ice at 150 V in 25 mM Tris–HCl, 250 mM glycine buffer and proteins were visualized by Coomassie staining.
Enzyme-linked immunosorbent assay (ELISA)
ELISA was performed in Pierce Nickel coated plates (Thermo Fisher Scientific). 100 pmol RNAPII, diluted in PBS with 2% bovine serum albumin (BSA; Sigma), was added to each well. After a 1h incubation, wells were washed three times with PBST (140 mM NaCl, 10 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3, 0.1% Tween) and three times with PBS. Drc.strep was then added at 100 pmol in PBS with 2%BSA followed by a 1 h incubation. The wash steps were repeated before adding a 1:5000 dilution of StrepMAB-Classic, HRP conjugate (IBA) in PBS + 2%BSA. After a final 1 h incubation, the reaction by HRP was started with 1-step Slow TMB-ELISA substrate solution (Thermo Fisher Scientific). Stop Reagent for TMB Substrate (Sigma) was added after 25 min and colorimetric detection was done at OD450 with a Bio-Rad model 680 microplate reader. All ELISA binding assays were performed in triplicate, RNAPII.strep served as positive control and negative controls included wells where RNAPII, Drc.strep or both were replaced by their respective buffer solution.
Structure determination of Drc
Drc crystals were grown in sitting drops containing 1 μl of protein solution (7 mg/ml, 10 mM Tris–HCl pH 7.5) and 1 μl of precipitant solution equilibrated against 100 μl of precipitant in the reservoir. For the crystal used for the non-anomalous dataset, this precipitant consisted of 1.8 M monobasic ammonium phosphate and 0.1 M sodium acetate pH 4.6, while it consisted of 2.0 M ammonium citrate pH 7.0 and 0.1 M sodium acetate for the crystal used for anomalous data acquisition. The crystals grew for approximately two and a half years and were soaked in 40% PEG 400 before flash freezing in liquid nitrogen.
X-ray diffraction data was collected under a 100 K nitrogen stream at the X06DA beamline of the Swiss Light Source at the Paul Scherrer Institute (Villigen, Switzerland). After data acquisition using an X-ray wavelength of 1.00 Å (12.4 keV), seven datasets at different χ angles (0° to 30°, 5° increment) with a wavelength of 2.07 Å (5.98 keV) were collected to ensure a high multiplicity necessary for S-SAD phasing.
The first dataset was indexed and integrated using XDS v. 3 November 2014 (23) and scaled and merged using Aimless (24). Data collection and refinement statistics can be found in Table 1. The phase problem was solved using native S-SAD using the seven other datasets, as described in (25). These datasets were also processed using XDS v. 3 November 2014, but scaled using XSCALE v. 3 November 2014 (23), which resulted in an anomalous multiplicity of 83.8. Sulphur positions were found using SHELXC/D (26) and subsequent density modification and auto-tracing of the protein backbone was done using SHELXE (26). This model and the non-anomalous data were used to build the initial model with Phenix.Autobuild v. 1.9 (27). This model was further refined using Phenix.refine v. 1.11 (28) and COOT v.0.8.2 (29). Figures were generated using PyMOL.
Table 1.
Data collection and merging statistics for Drc
| Data collection statistics | ||
| Wavelength (Å) | 1.00 | 2.07 |
| Resolution range (Å) | 44.42–2.20 (2.27–2.20)a | 44.42–2.80 (2.95–2.80) |
| Space group | I 41 2 2 | I 41 2 2 |
| Unit cell (Å; °) | 88.84, 88.84, 63.19; 90, 90, 90 | 88.84, 88.84, 63.13; 90, 90, 90 |
| R merge (%) | 4.3 (43.4) | 6.7 (27.4) |
| R pim (%) | 1.8 (17.3) | 0.7 (3.1) |
| <I/σ(I)> | 31.3 (5.9) | 78.90 (19.62) |
| Total reflections | 86 244 (7801) | 505 914 (33 930) |
| Unique reflections | 6690 (571) | 5970 (444) |
| Completeness | 100.0 (100.0) | 99.9 (99.9) |
| Multiplicity | 12.9 (13.7) | 83.8 (77.1) |
| CCano (%) | NA | 50 (19) |
| SigAno | NA | 1.77 (0.99) |
| Refinement statistics | ||
| R work/Rfreeb (%) | 22.72/28.59 | |
| R.m.s.d. bond lengths (Å) | 0.007 | |
| R.m.s.d. bond angles (°) | 0.88 | |
| Average B-factors (Å2) | ||
| Main chain | 55.51 | |
| Side chain | 60.53 | |
| Waters | 64.60 | |
| Phosphate | 70.08 | |
| Ramachandran plotc (%) | ||
| Residues in favoured regions | 93.75 | |
| Outliers | 0.00 | |
aValues in parentheses are for the outermost resolution shell.
b R free is calculated using a random 5% of data excluded from the refinement.
cRamachandran analysis was carried out using Molprobity (55).
NA = not applicable.
Phylogenetic analysis
Phylogenetic analysis was performed by the maximum likelihood method and a Jones–Thornton–Taylor matrix-based model (30). The phylogenetic tree was generated based on the dataset and methods described in (31), using MUSCLE (32) for sequence alignment and MEGAX (33) for phylogram generation. A bootstrap test (1000) was performed for the percentages of replicate trees. Homologs of Drc and N4 gp2 were assigned based on a PSI-BLAST search (34) with up to three iterations using the default parameters and retaining all significant hits for the next iteration.
RESULTS AND DISCUSSION
LUZ7 gp14 (‘Drc’) is a ssDNA-binding protein
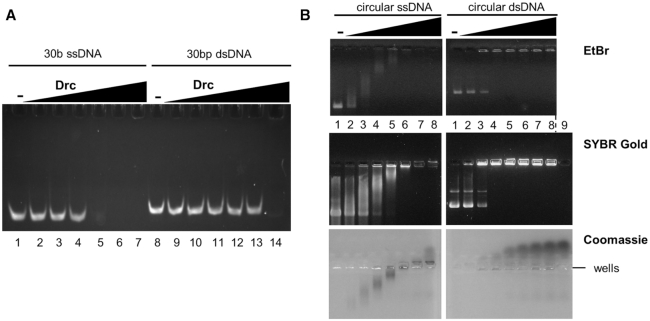
While LUZ7 gp14 has previously been found to be toxic upon expression in the bacterial host, Pseudomonas aeruginosa, no host protein interactions of gp14 have been observed by a high-throughput yeast two-hybrid (Y2H) screen (18). However, the same study suggested that this protein has a negative impact on host transcription, even though no interaction with the host RNA polymerase could be observed in pull-down experiments with this polymerase as bait. LUZ7 gp14 (further referred to as Drc, ssDNA-binding RNA Polymerase Cofactor, based on evidence provided below) could also impact transcription by direct binding of nucleic acids. To verify this hypothesis, nucleic acid binding activity of Drc was assessed by Electrophoretic Mobility Shift Assays (EMSAs), using short 5′-6-FAM labelled single-stranded and double-stranded oligonucleotides (ssDNA and dsDNA; Figure 1A). Drc caused a significant shift in migration of ssDNA and shifted nearly all DNA substrate at a 15:1 (Drc:DNA) molar ratio. A similar shift for dsDNA was only observed at the highest tested ratio of 500:1, thus demonstrating a clear preference for ssDNA. Notably, Drc prevented the ssDNA from entering the gel. This effect could be caused by the high pI of the native protein (9.16, estimated by Protparam (35)), which could result in the movement of bound DNA towards the negative pole. Alternatively, the protein could be forming larger DNA-bound complexes that are unable to enter the gel. Indeed, in some cases, DNA was clearly visible in wells, which would be consistent with higher molecular weight complexes/aggregate formation (Supplementary Figure S1). On other gels, like the one shown in Figure 1A, no DNA in the well was observed. It could be that the DNA–protein complex was washed away from the well during handling.
Figure 1.
(A) EMSA on native acrylamide gel with 100 nM of 5′-6-FAM-labeled ssDNA oligonucleotides (lanes 1–7) or dsDNA (lanes 8–14), each with increasing concentrations of Drc (left to right: 0, 0.25, 0.5, 0.75, 1.5, 3, 50 μM). (B) EMSAs on agarose gel with 57 nM phiX174 virion DNA (left) or with its double-stranded RFI form (right). Drc was used in increasing concentrations (lane 1 to 8; 0, 3, 6, 9, 12, 15, 18, 21 μM) for both conditions. Gels were either stained with ethidium bromide (top), SYBR Gold (middle) or Coomassie (bottom). A control with only 21 μM Drc, without DNA was loaded in lane 9 for SYBR Gold and Coomassie stains. Gels are representative of at least two replicates.
The DNA binding activity of Drc was also investigated using agarose gels with larger, circular DNA (Figure 1B). With these substrates, the preference for ssDNA was less pronounced, but a clear distinction in binding for either ssDNA and dsDNA was evident. For ssDNA, a gradual shift with increasing Drc concentrations was observed, whereas on dsDNA there was an all-or-none change from unbound to shifted DNA, without any intermediates. Moreover, ssDNA seemed to disappear from the gel at high Drc concentrations, while dsDNA remained clearly visible in the wells. To detect whether Drc-bound ssDNA is indeed migrating to the negative pole or remains in the wells like dsDNA, the more sensitive SYBR Gold stain was used (Figure 1B, middle). As for dsDNA, the shifted ssDNA was found in the wells, with no ssDNA shifted towards the negative pole. Similar to what was seen for EtBr-stained gels, the bands of ssDNA become increasingly faint at higher Drc concentrations, whereas dsDNA band intensity was unchanged. To gain additional insights into the observed binding effects and the localization of the protein, the SYBR gold agarose gel was subsequently stained with Coomassie (Figure 1B, bottom). Drc was found tightly associated with the gradually shifting ssDNA band at low to intermediate Drc concentrations. Only at the highest Drc concentrations the protein migrated towards the negative pole, where ssDNA was not observed. For dsDNA, on the other hand, Drc readily shifted to the negative pole without dsDNA, even at low protein concentrations.
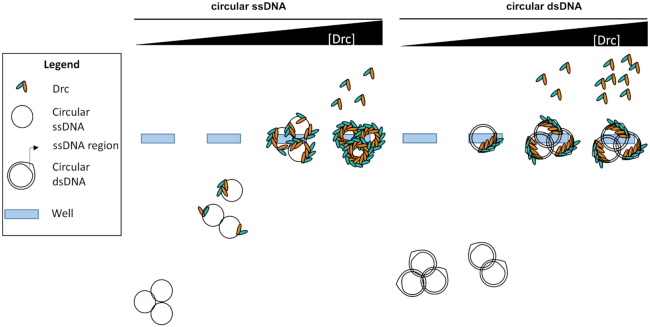
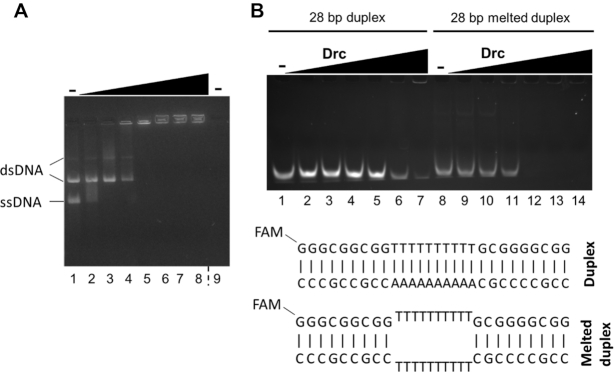
A model that accounts for these differences between ss and dsDNA binding by Drc is given in Figure 2. It assumes that Drc can (cooperatively) bind internally melted duplex DNA and that there is a preference for ssDNA. This preference is evident on small oligos (30 bases; Figure 1A) but not on larger circular DNA substrates (Figure 1B). To further demonstrate the ssDNA-binding preference, a binding assay was performed with a mixture of large circular ssDNA and dsDNA (Figure 3A). If Drc had a preference for one of the substrates, that substrate would be expected to be shifted at lower Drc concentrations than the other. As can be seen, dsDNA was only shifting after all of the ssDNA was shifted to the wells, indicating a clear preference for ssDNA. The ability of Drc to bind internally melted duplex DNA was assayed to further validate the model. Binding to small 28 bp duplex DNA, with a 10 bp mismatch in the center, was compared to a fully complementary duplex (Figure 3B). The full duplex DNA was barely bound by Drc. In contrast, the internally melted duplex was bound almost to the same extent as ssDNA, with a full shift occurring at a 5:1 Drc:DNA ratio. Thus, not only is Drc able to bind ssDNA, it can also bind internally unwound duplexes, which matches with the binding model presented in Figure 2.
Figure 2.
Model for DNA binding on long circular ssDNA versus dsDNA. Drc preferentially binds ssDNA, hence at low concentrations of Drc, it is spread across all ssDNA molecules, resulting in a gradual shift. In case of dsDNA it can only bind in ssDNA regions that can be formed on supercoiled dsDNA (56). This binding seems to be occurring in a cooperative manner. Hence, as soon as one region is bound, the other proteins will preferentially bind the same dsDNA molecule until its ssDNA region is filled. This final complex will be too large to migrate into the gel, resulting in a dsDNA molecule to be either fully shifted or not shifted at all. At higher concentrations of Drc, there will still be space to bind on ssDNA molecules but not on dsDNA molecules as it can only bind the ssDNA region. Therefore, unbound Drc is seen to shift upwards to the negative pole for samples with dsDNA molecules but not yet for ssDNA molecules. Only at very high concentrations unbound Drc is observed in ssDNA-bindng conditions. At this point, the ssDNA is fully coated by Drc, blocking it from being bound by DNA dyes. DsDNA will still be stained as Drc only binds the ssDNA region and the dye can easily reach the unbound dsDNA regions.
Figure 3.
(A) EMSAs on agarose gel with 57 nM of both phiX174 virion DNA and its double-stranded RFI form (lanes 1–8). Drc was used in increasing concentrations from left to right (0, 3, 6, 9, 12, 15, 18, 21 μM) for both conditions. A control with only 21 μM Drc, without DNA was loaded in lane 9. The gel was stained with EtBr. (B) EMSA on native acrylamide gel with Drc and 100 nM 5′-6-FAM-labeled dsDNA oligonucleotides (lanes 1–7) or internally melted duplex DNA (lanes 8–14). In both conditions, increasing concentrations of protein were used from left to right: 0, 0.5, 1, 1.5, 5, 20, 50 μM. Below, a representation of both oligonucleotide combinations is shown. Gels are representative of two replicates.
As is often the case with other SSB proteins, the binding by Drc does not seem to be sequence specific. It binds not only LUZ7 DNA but also non-cognate sequences used in the assays (Figures 1, 3 and Supplementary Figure S2). The ssDNA-binding activity of Drc, might explain its previously observed toxicity (18). Genome regions with higher levels of non-duplex DNA, like those that are undergoing replication or transcription, could be bound by Drc. This could hinder these crucial processes and cause toxicity. However, this effect is likely not related to the biological function, which is discussed later on and is a consequence of artificially increased Drc concentrations during screening.
Drc forms a stable homodimer with a novel ssDNA-binding fold
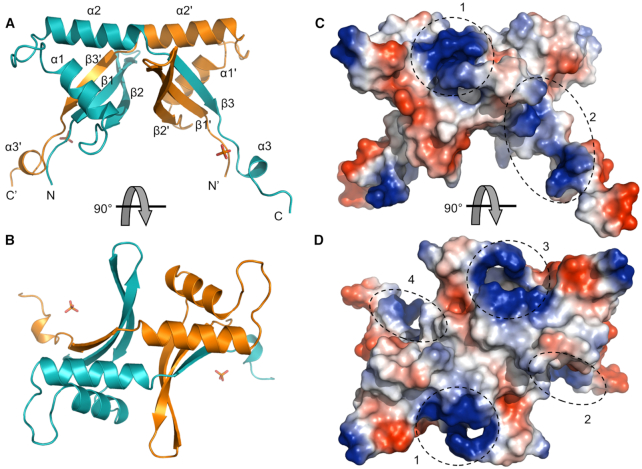
Most SSBs in dsDNA viruses bear one or more oligonucleotide-binding (OB) fold domains, which form a five-stranded β-barrel that binds the nucleic acid strand (36). Although Drc is a ssDNA-binding protein, it does not share any significant sequence similarity with the typical OB-fold or other SSB proteins. Therefore, the crystal structure of Drc was determined. The protein crystallized in space group I4122 and the structure was solved by single-wavelength anomalous dispersion of Sulphur atoms (S-SAD) using a dataset of 2.80 Å resolution. After density modification and auto-tracing of the protein backbone, the model was subsequently refined using another 2.20 Å resolution dataset, from a crystal of which no anomalous data was recorded, to Rwork and Rfree values of 22.72% and 28.59%, respectively. The asymmetric unit contains furthermore 30 ordered solvent molecules and one phosphate group. Additional data collection and refinement statistics are provided in Table 1. Wild-type Drc has 99 amino acids, of which the model spans residues 18–99. The N-terminal residues 1–17 of Drc and five amino acids that remained from the tag could not be resolved from the electron density maps. This region is predicted by ESpritz to be disordered (Supplementary Figure S3), which could explain the weak electron density (37). Although this part could not be resolved in the crystal structure, it cannot be excluded that the flexibility of this terminus is required for binding ssDNA and that it might adopt a more fixed conformation once the target is bound.
The crystal structure clearly shows two Drc monomers strongly interacting via 26 hydrogen bonds and six salt bridges to form a stable dimer with a solvation free energy gain upon interface formation (ΔGi) of –30.3 kcal/mol, calculated with PDBePISA (38) (Figure 4A, B and Supplementary Figure S4). On both sides of the dimer, two profound concave beta-sheets are present at the surface. Each β-sheet consists of three β-strands, one of which (β3′) is coming from the partnering monomer. The sheets are bridged by two α-helices (α1, α2) that are separated by a loop region. The C-terminus of the protein ends in a long tail structure containing a small helix (α3). The basic residues, which are abundantly present in Drc and result in the high pI of the protein, are mostly clustering in four patches on the dimeric surface (Figure 4C, D). The electrostatic surface potential shows two positively charged patches on the helical rim, formed by α2 and α2′, and one on and near each concave surface.
Figure 4.
(A) Crystal structure of the Drc dimer with indicated helices and strands. The two chains in the dimer are colored in cyan and orange (the orange chain is generated by symmetry code x, –y-1/2, –z-3/4). The bound phosphate group is shown as a stick model. (B) 90° rotated view of the Drc model. (C) Electrostatic surface potential of Drc (generated by PyMOL vacuum electrostatics) with positive charges in blue and negative in red. The different positively charged patches are indicated by dashed circles. (D) Rotated view of the surface potential.
As was seen at the sequence level, Drc does not share structural similarity with the canonical OB-fold. Moreover, when comparing the model to known structures in the PDB database using the DALI and PDBeFold servers (39,40), no significant matches with other structures can be found. Since Drc forms a dimer, which has larger β-sheets than the monomer, the search was broadened by using the dimeric form. This resulted in the retrieval of 19 hits with DALI (Supplementary Table S1) and three hits with PDBeFOLD (Supplementary Table S2), of which the top scoring include viral matrix proteins (seven occurrences in DALI), DNA topoisomerases of type IA (four occurrences in DALI) and cyclophilines (three occurrences in DALI, three in PBDeFOLD). Hence, only the cyclophilins are found by both servers and only when reducing the standard similarity cut-off values to 60% in PDBeFOLD, already indicating that the fit between the Drc dimer and the hit structures is rather poor. Indeed, the rmsd between Drc and the DNA topoisomerases are relatively high (6.6–7.4 Å). Moreover, the hit structures mainly match through similarly placed β-sheets, which upon closer inspection have a different build-up than those of Drc (i.e. amount, length and curvature of the strands) and the order of α-helices and β-sheets largely differs (Supplementary Figure S5). Also the function of these proteins strongly varies, ranging from viral packaging and budding (viral matrix proteins (41)) to regulation of DNA supercoiling (DNA topoisomerase type IA; (42)) and protein folding/trafficking (cyclophilins; (43)), further indicating these hits are coincidental. Given the lack of hits with the monomeric Drc structure and the rather weak similarity between the Drc dimer and hit structures, Drc seems to be a novel type of ssDNA-binding fold.
Honing in on the DNA-binding site of Drc
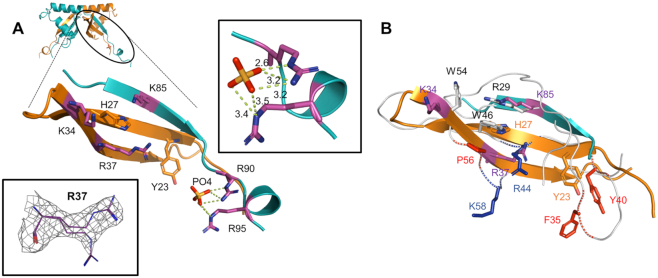
To gain insights in the ssDNA-binding mechanism of Drc, it's structure was manually compared and aligned with known ssDNA-binding folds. Interestingly, the β-sheet of Drc can be aligned well with part of the β-sheet of PC4-fold proteins (Supplementary Figure S6). This fold has been described for various proteins involved in different ssDNA-binding processes, primarily linked with DNA repair, replication or transcriptional regulation (44–47). Although the protein aligns well at a single β-sheet, the remainder of the protein does not. Nevertheless, provided that PC4-fold proteins generally bind ssDNA on their concave β-sheet surface, a comparison with this sheet could provide a good indication of the DNA-binding mechanism of Drc. PC4-fold proteins mainly bind ssDNA through aromatic amino acids that stack with the bases and basic residues that interact with the phosphate backbone (48). The Drc β-sheets might perform similarly as they have multiple lysine and arginine residues alongside the surface with two aromatic residues on the central β-strand (Figure 5A). The overlay of the Drc sheet with that PC4-fold protein of bacteriophage T5 (PDB ID: 4BG7) reveals several similarly placed aromatic and positively charged residues (Figure 5B; e.g. R29/K85, W46/H27). The most prominent of these is Drc's Y23, which localizes approximate to Y40 of the PC4-fold. This tyrosine is one of the residues reported to be critical in binding ssDNA by PC4-folds (46). This implies that Y23 of Drc could be involved in binding DNA, potentially through base stacking interactions. In addition, Y23 is flanked by positively charged residues that could bind the phosphate backbone. Two of these residues are arginines (R90 and R95) that are bound to a phosphate group in the crystal structure, which could reflect this backbone binding (Figure 5A). Another (R37), is located on the β-sheet surface near Y23 and is conserved among all Drc homologs from related phages. Moreover, the electron density of R37 was interpreted as two conformations with occupancies of 0.51 and 0.49 (Figure 5A). This might indicate a certain flexibility of this residue when Drc is not bound to DNA. It may adopt a fixed conformation when it interacts with the DNA backbone. Overall, Y23 and the basic residues that lie around it could form the main ssDNA-binding site of Drc.
Figure 5.
(A) Zoom-in on one of the Drc beta-sheets and the C-terminal tail. Aromatic residues and positively charged residues on the concave side are displayed in orange and purple respectively. Bottom left inset displays the two conformations of R37 within the electron density (2Fo– Fc map contoured at 0.5σ). Top right inset shows the polar contacts between the arginines R90 and R95 with the phosphate group (distances in ångström). (B) Overlay of the beta-sheets of bacteriophage T5 PC4-fold protein (PDB ID: 4BG7) and Drc. The T5 protein backbone is shown as a white ribbon with critical DNA-binding residues on the concave side as red or blue sticks for respectively hydrophobic/aromatic or positively charged residues. Non-critical residues with these properties are displayed as white sticks.
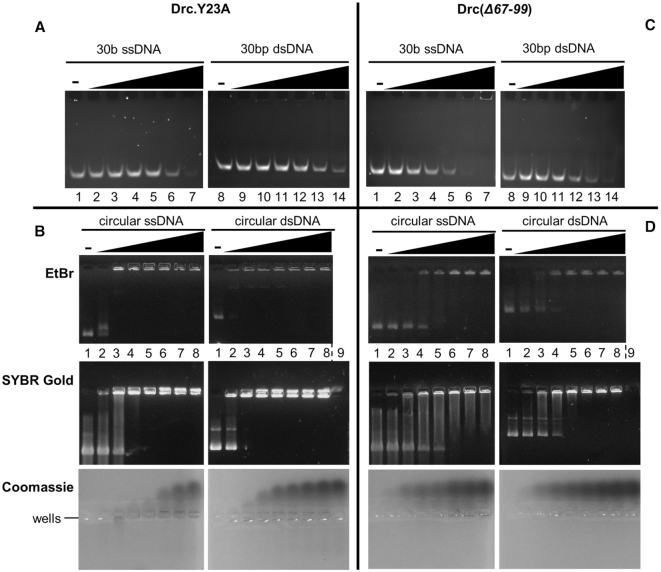
To confirm that Y23 is a core ssDNA-binding residue of Drc, a recombinant Y23A mutant was produced. During purification it behaved similarly to the wild type and eluted from a gel filtration column at the same time, indicating proper folding (Supplementary Figure S7A, B). Drc.Y23A was then tested for ssDNA binding and compared to its wild-type counterpart. On a small 30b DNA substrate, the mutant protein no longer caused a gel shift (Figure 6A); only at the highest concentration tested (50 μM) some residual shifting was visible. This is an almost 50-fold higher concentration than that of wild-type Drc to obtain a similar shift. Thus, this tyrosine residue is important for ssDNA binding by Drc. For small dsDNA, the results were similar: any shift that was seen with wild-type Drc (Figure 1A), was nearly completely abolished with the mutant. The Y23A mutant was also tested on larger circular DNA (Figure 6B). Intriguingly, compared to the wild type, both the ssDNA and dsDNA shifted at nearly the same or even lower mutant protein concentration. However, the manner in which the shift occurred for ssDNA was changed. The wild-type protein-DNA complex was moving gradually slower with increasing protein concentrations, while for the mutant the DNA band was either fully shifted or not at all. This hints at an increased cooperative binding of the mutant protein. In addition, the ssDNA remained visible in the wells even at high Drc.Y23A concentrations, while in the case of wild-type Drc it disappeared, likely due to a full coating of DNA by the protein. Taken together, while the EMSAs with short oligos demonstrate the importance of Y23 for binding smaller ssDNA sites, the ability of Drc.Y23A to bind large ssDNA means that, if the DNA is sufficiently long, other interactions can take place that compensate for the loss of binding at short substrates. These interactions seem to favour cooperative binding of the protein. This cooperative binding was also seen for wild-type Drc on dsDNA, likely in the ssDNA regions, but was less apparent on ssDNA (Figure 1B). Despite the cooperative binding, the mutant seems no longer capable of fully coating the ssDNA and parts of the DNA remain accessible for the DNA stains. This is presumably also why more unbound protein is seen upon Coomassie staining (Figure 6B).
Figure 6.
(A) EMSA on native acrylamide gel with 100 nM of 5′-6-FAM-labeled ssDNA oligonucleotides (lane 1–7) or dsDNA (lane 8–14), each with increasing concentrations of Drc.Y23A (left to right: 0, 0.5, 1, 1.5, 5, 20, 50 μM). (B) EMSAs on agarose gel with 57 nM phiX174 virion DNA (left) or with its double-stranded RFI form (right). Drc.Y23A was used in increasing concentrations (lane 1 to 8; 0, 3, 6, 9, 12, 15, 18, 21 μM) for both conditions. Gels were either stained with Ethidium Bromide (top), SYBR Gold (middle) or Coomassie (bottom). A control with only 21 μM Drc, without DNA was loaded in lane 9 for SYBR Gold and Coomassie stains. (C, D) Identical to A and B, respectively, but with mutant Drc(Δ67–99). Gels are representative for two replicates.
To verify if and which charged interactions between Drc and the DNA backbone play a role in binding, a Drc mutant lacking 33 C-terminal amino acids and fused to a C-terminal 6xHis-tag was produced. Due to this truncation, the mutant ‘Drc(Δ67–99)’ misses the phosphate binding residues R90 and R95 (Figure 5A), a lysine residue on β3 and three basic residues on α2. The protein elutes as a dimer during gel filtration (∼18 kDa, predicted size: 16.75 kDa) indicating correct protein folding (Supplementary Figure S7C). The mutant protein is able to bind DNA (Figure 6C, D). For short oligo binding, the truncation has less effect than the single Y23A substitution (Figure 6A, C). However, binding to ssDNA is not nearly as good as wild-type Drc (>10-fold decrease in the mutant). On longer DNA substrates, the impact of the truncation on DNA binding becomes more obvious (Figure 6D). The ssDNA binding is strongly impacted, whereas the effect is less severe for dsDNA binding. Even at the highest tested protein concentrations, not all ssDNA is bound, while a lot of unbound protein is already seen at low protein concentrations. Given that the truncated mutant Drc(Δ67–99) lacks many positively charged residues, this indicates that electrostatic interactions with the DNA backbone play a role in ssDNA binding by Drc.
Drc is part of the transcription complex responsible for middle transcription in LUZ7
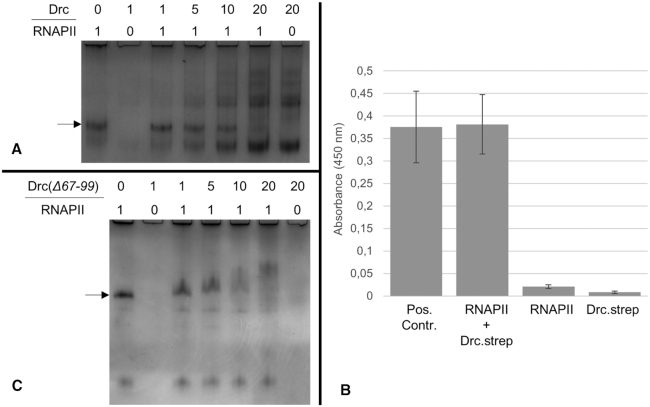
LUZ7 is an N4-related phage as it shares a distinct transcription pattern with coliphage N4. Immediately after infection these phages co-inject a virion-associated RNA polymerase (vRNAP) with their DNA, which guides early viral transcription aided by the host SSB; middle gene transcription is performed by the phage-encoded RNAPII in consort with N4 gp2, and the late viral genes are transcribed by the host RNA polymerase, guided by N4SSB (49). In LUZ7 and its host, homologs have been predicted for all these RNA polymerases and SSB proteins except N4 gp2, the SSB required for recruiting RNAPII during transcription of middle genes (20,50). To investigate whether Drc encodes a functional homolog of N4 gp2 in phage LUZ7, its interaction with the heterodimeric LUZ7 RNAPII polymerase was assessed. To this end, a Mobility Shift Assay (MSA) with the RNAPII complex and Drc was performed (Figure 7A). Upon the addition of twenty-fold excess of Drc to RNAPII, the RNAPII band was fully shifted, indicating the interaction between the two proteins. Consistent with EMSA results, a complete disappearance of shifted RNAPII is observed, most likely due to the high pI of Drc. Indeed, when the poles of the electrophoresis are reversed, Drc migrates into the gel and disappears upon addition of RNAPII, which does not migrate to this pole (Supplementary Figure S8).
Figure 7.
(A) MSA with Drc and the RNAPII complex on a native 10% acrylamide gel. At the top of each lane the ratio of Drc: RNAPII in each condition is indicated (with 1 being 2.25 μM and 0 being a buffer control). RNAPII was kept constant at 2.25 μM, while the concentration of Drc was increased from left to right. The arrow indicates the height of the RNAPII band in unbound state. (B) ELISA of Drc.strep and the RNAPII complex (with His-tagged RNAP1 subunit) at 100 pmol. The positive control, RNAPII.strep (bar 1) consists of the two RNAPII subunits, of which RNAP1 is His-tagged and RNAP2 strep-tagged. Bar two represents the interaction of RNAPII (only His-tagged at RNAP1 subunit) and Drc.strep at equimolar amounts. Bar three and four are negative controls with respectively Drc and RNAPII replaced by protein buffer. Colorimetric error bars are based on three replicates. detection was done at 450 nm. (C) MSA of Drc(Δ67–99) and the RNAPII complex on a 10% native acrylamide gel. Like in A, the ratios are given at the top of each lane and the arrow represents the unbound RNAPII complex.
The interaction of Drc with the RNAPII complex was independently validated by an Enzyme-Linked ImmunoSorbent Assay (ELISA) with equimolar amounts of strep-tagged Drc (Drc.strep) as prey and His-tagged RNAPII complex as bait. A strong signal was detected when both RNAPII and Drc.strep are present in the sample, thus demonstrating their interaction (Figure 7B). The interaction between RNAP1 and RNAP2, both subunits of the RNAPII complex, served as a positive control and reached similar signal intensities in the assay. This indicates that Drc interacts with the RNAPII complex with an affinity similar to that between the two subunits in this complex. Together with the binding of ssDNA discussed above, this interaction provides evidence of a shared biological function between non-related proteins, N4 gp2 and Drc.
To narrow down the domain of Drc that is required for protein binding, the truncated mutant of Drc was also subjected to the MSA. With the mutant, the mobility shift occurs around the same concentration as with wild-type Drc, but is a lot less pronounced (Figure 7C). While the less extreme shift might be due to charge and size differences of the truncated protein, the observed smearing of the band does suggest a reduced strength of the interaction. Binding to RNAPII is thus impaired for Drc(Δ67–99), suggesting that the C-terminus does play a role in RNAPII recognition in addition to its role in binding DNA. However, since Drc(Δ67–99) still causes some shifting of the RNAPII band, the C-terminus, while important, is not the only region that interacts with RNAPII. Since the C-terminus encompasses the positively charged patch on the helical rim, it could be involved in RNAPII binding through electrostatic interactions. Recently, Molodtsov & Murakami (2018) suggested that the specificity loop of N4 RNAPII, which is usually required for DNA recognition, is too negatively charged and too rigid due to self-interaction with its N-terminal domain (NTD) to do so (51). Moreover, the NTD is thought to be involved in recruitment of cofactors rather than promoter binding. Therefore, it could be that the Drc C-terminus binds either negatively charged patches on the RNAPII’s NTD or its negatively charged specificity loop, and thereby aids RNAPII recruitment to the promoter region, which is supposedly mostly bound through the Drc concave surface.
Both N4 gp2 and Drc share their ability to bind ssDNA and the phage-encoded RNAPII, implying a similar biological role in transcription activation. However, they presumably perform this function in a distinct manner. Drc is able to efficiently bind supercoiled plasmid DNA (Figure 1B, Supplementary Figure S2A), while gp2 is reported not to be able to do so (20). The difference could be explained by their strongly differing pI values (9.16 and 5.20, respectively for Drc and gp2, estimated by Protparam (35)). The higher positive charge of Drc would allow more interactions with the DNA backbone and increase its affinity for DNA. The differences between the two proteins can also be explained by the low sequence identity of their interacting RNA polymerases. The RNAP1 and RNAP2 subunits of LUZ7 RNAPII share only 36% and 31% with their respective subunits in N4 (determined by psiBLAST (34)). Moreover, the LUZ7 RNA polymerase is about 50 amino acids larger than the polymerase of N4. The major addition comes in at the N-terminal domain (NTD) on the RNAP1 subunit, which is proposed to be involved in cofactor recruitment (51). Since the NTD between the two RNA polymerases is dissimilar, it is likely that also the cofactors themselves should be sequentially and structurally different.
Proposed model for Drc function
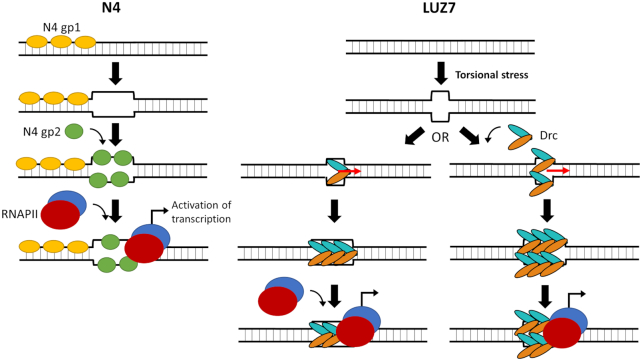
Combining the existing model of gp2-mediated RNAPII recruitment (49) with the EMSA and structural data for Drc reported here, a model for Drc-mediated recruitment can be proposed (Figure 8). In N4, an additional factor (gp1) is thought to be required for initial promoter unwinding before the SSB (gp2) can bind (49). LUZ7 does not encode any homologs of N4 gp1 and does not seem to require this additional factor, since Drc is capable of binding duplex/supercoiled DNA that is locally unwound due to torsional stress. Torsional stress is known to play an important role in activation of early promoters, hence it is not unlikely that also for middle promoters torsional stress is present and plays a role (52). After promoter melting, Drc can bind the ssDNA regions that become available. On short ssDNA molecules, Y23 of Drc is crucial to mediate binding. On longer DNA substrates, additional, likely electrostatic, interactions with the DNA backbone become more important. Given the two binding surfaces present on the Drc dimer, it could be that Drc binds both juxtaposed strands simultaneously, or each strand is bound separately by different Drc dimers that wrap the DNA around their surface. In addition to just binding, Drc might also destabilise and/or unwind dsDNA. Other SSB proteins that have two opposing ssDNA-binding surfaces (i.e. PC4-fold proteins) are reported of not only binding the juxtaposed strands, but also destabilizing the dsDNA and unwinding it (53). Whether Drc is also capable of this is yet unclear, but it might be a way for Drc to provide additional binding sites for cooperative recruitment of additional Drc proteins and make room for the positioning of RNAPII. RNAPII is guided to the DNA by direct protein-protein interaction with Drc. The exact nature of this interaction is still to be investigated, but our evidence suggests that it is in part mediated by the C-terminus of Drc. In N4, transcription from middle promoters is initiated by the recruitment of RNAPII. It is tempting to hypothesize that Drc-mediated RNAPII recruitment activates transcription of LUZ7 middle promoters. The proposed model together with the structural and functional data of this study, provide a strong basis to direct future research into these interactions and how or if it can result in activation of transcription.
Figure 8.
Comparison of RNAPII recruitment to middle promoters in N4 and LUZ7. (left) Current model of transcription activation by N4 (49). In N4 there is a need for initial unwinding of the promoter region, thought to be performed by gp1. The resulting ssDNA region can then be bound by gp2, which recruits the phage-encoded RNAPII and thus activates its transcription. (right) Proposed model of RNAPII recruitment in LUZ7. Drc can already bind ssDNA regions that become available by torsional stress. Therefore it might not require an additional DNA-unwinding cofactor. Due to its double binding surface it can either bind one strand on each surface, or it binds a single strand with both surfaces. Additional Drc proteins are recruited through cooperative binding. At this stage, Drc might also be destabilizing the duplex and allow additional Drc to be bound. Drc then recruits RNAPII through direct protein-protein interaction. Potentially, this will lead to transcription activation similar to how it occurs in N4.
Drc reveals an unexpected SSB diversity in an otherwise conserved transcription scheme
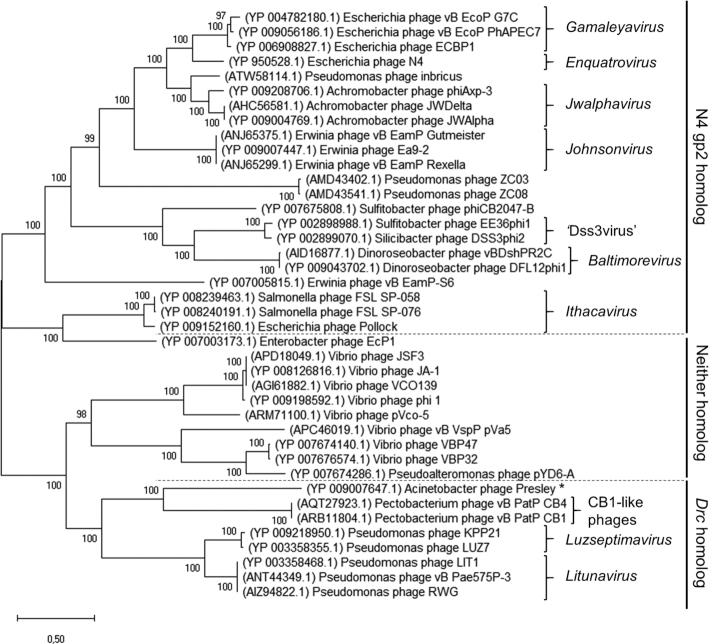
It is remarkable that a crucial process, which is conserved among most N4-like phages, is different in LUZ7. To see whether this is a distinct case or the usage of this SSB type has a broader occurrence, a phylogenetic tree was built based on a previously described dataset (31) which contains vRNAP sequences of various N4-like phages with representative phages for each currently classified genus (Figure 9). Members encoding a homolog of either Drc or N4 gp2 were then assigned based on a psiBLAST search (Supplementary Table S3) (34). From this analysis, it becomes apparent that the Drc mediated transcription scheme is spread across all phages within the Luzseptimavirus and Litunavirus genera, but also the recently described and more distant group of Pectobacterium-infecting CB1-like phages (31). Furthermore, a large clade consisting primarily of N4-related Vibrio phages contains homologs for neither N4 gp2 or Drc, hinting at the existence of a third distinct SSB protein. The most logical explanation for this difference would be that the host imposes a restriction in the SSB that can be used by the phage. Apart from the Vibrio-infecting clade, Luzseptimavirus and Litunavirus members all infect P. aeruginosa, whereas CB1-like phages have a Pectobacterium atrosepticum host. However, some N4-related Pseudomonas phages including ZC03 and ZC08 do encode a gp2 homolog (54), ruling out the host as the determining factor. Interestingly, nearly all phages without a gp2 homolog form a distinct branch with multiple clades, separated from the phages with clear gp2 homologs. This indicates that the distinction in the SSB that these phages use was made during the formation of the separate clades. Potentially, the splitting of the single subunit T7-like RNA polymerase into two subunits, which currently form RNAPII of these phages, has driven the formation of the different SSB proteins. The T7 RNA polymerase is capable of binding DNA without help of external factors, while RNAPII is unable to do so and requires an SSB protein (49). This splitting would thus have driven the acquisition of (SSB) cofactors to improve promoter binding. It could be that the crucial function of RNAPII requires a specific kind of SSB, which has led to the unique fold of this protein. It would be interesting to see whether N4 gp2 or the missing SSB in the Vibrio phages are structurally similar to the Drc fold or if they perform their role through an entirely different structure. The latter case would mean that the typical transcriptional progression that is shared in all N4-like phages is more diverse than originally thought. While looking similarly at the RNA polymerase level, these phages might be very distinct in the SSB they use for middle transcription.
Figure 9.
Evolutionary analysis by Maximum Likelihood, using the vRNAP protein sequences (log likelihood –174006.59) of the various N4-like phages in the dataset of Buttimer and colleagues (31). Taxonomically assigned groups are indicated by brackets with their corresponding genus in italics. The proposed group of ‘Dss3virus’ and suggested group of CB1-like phages are also indicated. Dashed lines delineate the phages that have N4 gp2 homologs, Drc homologs or neither of these two proteins. Phage Presley is indicated with asterisk as it's putative Drc homolog fell under the threshold of significance (Supplementary Table S3). Bootstrap values indicated at each branch point are derived from 1000 replicates.
DATA AVAILABILITY
ProtParam is a tool for the computation of physical and chemical parameters based on protein sequence and is available at https://web.expasy.org/protparam/.
PDBePISA is an interactive tool for the exploration of macromolecular interfaces and is available at http://www.ebi.ac.uk/pdbe/pisa/.
PDBeFOLD is the Protein structure comparison service at European Bioinformatics Institute and is available at http://www.ebi.ac.uk/msd-srv/ssm.
ESpritz is a protein disorder prediction server available at http://protein.bio.unipd.it/espritz/.
The Dali server is a network service for comparing protein structures in 3D available at http://ekhidna2.biocenter.helsinki.fi/dali/.
PyMOL is a molecular visualization system available at https://pymol.org/2/.
The PSI-BLAST suite allows for a position-specific iterated search in the protein databases using a protein query and is available at https://blast.ncbi.nlm.nih.gov/Blast.cgi.
The Protein Data Bank (PDB) is available at http://www.rcsb.org/.
Atomic coordinates and structure factors for Drc have been deposited with the Protein Data Bank under accession number 6QLC.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Abram Aertsen for his practical advice and valuable discussions and acknowledge the staff of beam line X06DA at the Swiss Light Source for their assistance during X-ray data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
KU Leuven as part of the Geconcerteerde Onderzoeksacties program [3E140356]; European Research Council Consolidator grant ‘BIONICbacteria’ [819800 to R.L.]; Research Foundation Flanders (FWO) [131620 to E.D.Z., 52318 to J.D.S.]. Funding for open access charge: KU Leuven [3E140356].
Conflict of interest statement. None declared.
REFERENCES
- 1. Fischer D., Eisenberg D.. Finding families for genomic ORFans. Bioinformatics. 1999; 15:759–762. [DOI] [PubMed] [Google Scholar]
- 2. Zhang R., Joachimiak G., Jiang S., Cipriani A., Collart F., Joachimiak A.. Structure of phage protein BC1872 from Bacillus cereus, a singleton with new fold. Proteins. 2006; 64:280–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hakim M., Ezerina D., Alon A., Vonshak O., Fass D.. Exploring ORFan Domains in giant viruses: structure of Mimivirus sulfhydryl oxidase R596. PLoS One. 2012; 7:e50649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rossi P., Barbieri C.M., Aramini J.M., Bini E., Lee H.-W., Janjua H., Xiao R., Acton T.B., Montelione G.T.. Structures of apo-and ssDNA-bound YdbC from Lactococcus lactis uncover the function of protein domain family DUF2128 and expand the single-stranded DNA-binding domain proteome. Nucleic Acids Res. 2013; 41:2756–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Van den Bossche A., Hardwick S.W., Ceyssens P.-J., Hendrix H., Voet M., Dendooven T., Bandyra K.J., De Maeyer M., Aertsen A., Noben J.-P. et al.. Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome. Elife. 2016; 5:e16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yin Y., Fischer D.. Identification and investigation of ORFans in the viral world. BMC Genomics. 2008; 9:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roucourt B., Lavigne R.. The role of interactions between phage and bacterial proteins within the infected cell: a diverse and puzzling interactome. Environ. Microbiol. 2009; 11:2789–2805. [DOI] [PubMed] [Google Scholar]
- 8. De Smet J., Hendrix H., Blasdel B.G., Danis-Wlodarczyk K., Lavigne R.. Pseudomonas predators: Understanding and exploiting phage-host interactions. Nat. Rev. Microbiol. 2017; 15:517–530. [DOI] [PubMed] [Google Scholar]
- 9. Liu J., Dehbi M., Moeck G., Arhin F., Bauda P., Bergeron D., Callejo M., Ferretti V., Ha N., Kwan T. et al.. Antimicrobial drug discovery through bacteriophage genomics. Nat. Biotechnol. 2004; 22:185–191. [DOI] [PubMed] [Google Scholar]
- 10. O’Sullivan L., Buttimer C., McAuliffe O., Bolton D., Coffey A.. Bacteriophage-based tools: recent advances and novel applications [version 1; referees: 3 approved]. F1000Research. 2016; 5:2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henry M., Debarbieux L.. Tools from viruses: Bacteriophage successes and beyond. Virology. 2012; 434:151–161. [DOI] [PubMed] [Google Scholar]
- 12. De Smet J., Zimmermann M., Kogadeeva M., Ceyssens P.-J., Vermaelen W., Blasdel B., Bin Jang H., Sauer U., Lavigne R.. High coverage metabolomics analysis reveals phage-specific alterations to Pseudomonas aeruginosa physiology during infection. ISME J. 2016; 10:1823–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berjón-Otero M., Lechuga A., Mehla J., Uetz P., Salas M., Redrejo-Rodríguez M.. Bam35 tectivirus intraviral interaction map unveils new function and localization of phage ORFan proteins. J. Virol. 2017; 91:e00870-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blasche S., Wuchty S., Rajagopala S.V., Uetz P.. The protein interaction network of bacteriophage lambda with its host, Escherichia coli. J. Virol. 2013; 87:12745–12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mehla J., Dedrick R.M., Caufield J.H., Wagemans J., Sakhawalkar N., Johnson A., Hatfull G.F., Uetz P.. Virus-host protein-protein interactions of mycobacteriophage Giles. Sci. Rep. 2017; 7:16514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van den Bossche A., Ceyssens P.-J., De Smet J., Hendrix H., Bellon H., Leimer N., Wagemans J., Delattre A.-S., Cenens W., Aertsen A. et al.. Systematic identification of hypothetical bacteriophage proteins targeting key protein complexes of Pseudomonas aeruginosa. J. Proteome Res. 2014; 13:4446–4456. [DOI] [PubMed] [Google Scholar]
- 17. Dedrick R.M., Marinelli L.J., Newton G.L., Pogliano K., Pogliano J., Hatfull G.F.. Functional requirements for bacteriophage growth: gene essentiality and expression in mycobacteriophage Giles. Mol. Microbiol. 2013; 88:577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wagemans J., Blasdel B.G., Van den Bossche A., Uytterhoeven B., De Smet J., Paeshuyse J., Cenens W., Aertsen A., Uetz P., Delattre A.-S. et al.. Functional elucidation of antibacterial phage ORFans targeting Pseudomonas aeruginosa. Cell. Microbiol. 2014; 16:1822–1835. [DOI] [PubMed] [Google Scholar]
- 19. Wagemans J., Delattre A.-S., Uytterhoeven B., De Smet J., Cenens W., Aertsen A., Ceyssens P.-J., Lavigne R.. Antibacterial phage ORFans of Pseudomonas aeruginosa phage LUZ24 reveal a novel MvaT inhibiting protein. Front. Microbiol. 2015; 6:1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carter R.H., Demidenko A.A., Hattingh-Willis S., Rothman-Denes L.B.. Phage N4 RNA polymerase II recruitment to DNA by a single-stranded DNA-binding protein. Genes Dev. 2003; 17:2334–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Silva D., Santos G., Barroca M., Collins T.. Domingues L. Inverse PCR for point mutation introduction. PCR. Methods in Molecular Biology. 2017; NY: Springer; 87–100. [DOI] [PubMed] [Google Scholar]
- 22. Peeters E., Boon M., Rollie C., Willaert R.G., Voet M., White M.F., Prangishvili D., Lavigne R., Quax T.E.F.. DNA-Interacting characteristics of the archaeal rudiviral protein SIRV2_Gp1. Viruses. 2017; 9:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kabsch W. XDS. Acta Crystallogr. 2010; D66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Evans P.R., Murshudov G.N.. How good are my data and what is the resolution. Acta Crystallogr. 2013; D69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weinert T., Olieric V., Waltersperger S., Panepucci E., Chen L., Zhang H., Zhou D., Rose J., Ebihara A., Kuramitsu S. et al.. Fast native-SAD phasing for routine macromolecular structure determination. Nat. Methods. 2015; 12:131–133. [DOI] [PubMed] [Google Scholar]
- 26. Sheldrick G.M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. 2010; D66:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adams P.D., Afonine P. V, Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 2010; D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Afonine P. V, Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D.. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. 2012; D68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. 2010; D66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones D.T., Taylor W.R., Thornton J.M.. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992; 8:275–282. [DOI] [PubMed] [Google Scholar]
- 31. Buttimer C., Hendrix H., Lucid A., Neve H., Noben J.-P., Franz C., O’Mahony J., Lavigne R., Coffey A.. Novel N4-Like Bacteriophages of Pectobacterium atrosepticum. Pharmaceuticals. 2018; 11:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumar S., Stecher G., Li M., Knyaz C., Tamura K.. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018; 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A.. Walker JM. Protein identification and analysis tools on the ExPASy server. The Proteomics Protocols Handbook. 2005; Totowa: Humana Press; 571–607. [Google Scholar]
- 36. Kazlauskas D., Krupovic M., Venclovas Č. The logic of DNA replication in double-stranded DNA viruses: insights from global analysis of viral genomes. Nucleic Acids Res. 2016; 44:4551–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walsh I., Martin A.J.M., Di Domenico T., Tosatto S.C.E.. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics. 2012; 28:503–509. [DOI] [PubMed] [Google Scholar]
- 38. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 39. Holm L., Laakso L.M.. Dali server update. Nucleic Acids Res. 2016; 44:W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krissinel E., Henrick K.. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D. 2004; 60:2256–2268. [DOI] [PubMed] [Google Scholar]
- 41. Radzimanowski J., Effantin G., Weissenhorn W.. Conformational plasticity of the Ebola virus matrix protein. Protein Sci. 2014; 23:1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang J.C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002; 3:430–440. [DOI] [PubMed] [Google Scholar]
- 43. Nigro P., Pompilio G., Capogrossi M.C.. Cyclophilin A: a key player for human disease. Cell Death Dis. 2013; 4:e888–e888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ge H., Roeder R.G.. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994; 78:513–523. [DOI] [PubMed] [Google Scholar]
- 45. Caldwell R.B., Braselmann H., Schoetz U., Heuer S., Scherthan H., Zitzelsberger H.. Positive Cofactor 4 (PC4) is critical for DNA repair pathway re-routing in DT40 cells. Sci. Rep. 2016; 6:28890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Steigemann B., Schulz A., Werten S.. Bacteriophage T5 encodes a homolog of the eukaryotic transcription coactivator PC4 implicated in recombination-dependent DNA replication. J. Mol. Biol. 2013; 425:4125–4133. [DOI] [PubMed] [Google Scholar]
- 47. Conesa C., Acker J.. Sub1/PC4 a chromatin associated protein with multiple functions in transcription. RNA Biol. 2010; 7:287–290. [DOI] [PubMed] [Google Scholar]
- 48. Werten S., Moras D.. A global transcription cofactor bound to juxtaposed strands of unwound DNA. Nat. Struct. Mol. Biol. 2006; 13:181–182. [DOI] [PubMed] [Google Scholar]
- 49. Lenneman B.R., Rothman-denes L.B.. Structural and biochemical investigation of bacteriophage N4-Encoded RNA polymerases. Biomolecules. 2015; 5:647–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ceyssens P.-J., Brabban A., Rogge L., Lewis M.S., Pickard D., Goulding D., Dougan G., Noben J.-P., Kropinski A., Kutter E. et al.. Molecular and physiological analysis of three Pseudomonas aeruginosa phages belonging to the ‘N4-like viruses’. Virology. 2010; 405:26–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Molodtsov V., Murakami K.S.. Minimalism and functionality: Structural lessons from the heterodimeric N4 bacteriophage RNA polymerase II. J. Biol. Chem. 2018; 293:13616–13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dai X., Rothman-Denes L.B.. Sequence and DNA structural determinants of N4 virion RNA polymerase-promoter recognition. Genes Dev. 1998; 12:2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Werten S., Langen F.W.M., van Schaik R., Timmers H.T.M., Meisterernst M., van der Vliet P.C.. High-affinity DNA binding by the C-terminal domain of the transcriptional coactivator PC4 requires simultaneous interaction with two opposing unpaired strands and results in helix destabilization. J. Mol. Biol. 1998; 276:367–377. [DOI] [PubMed] [Google Scholar]
- 54. Amgarten D., Martins L.F., Lombardi K.C., Antunes L.P., de Souza A.P.S., Nicastro G.G., Kitajima E.W., Quaggio R.B., Upton C., Setubal J.C. et al.. Three novel Pseudomonas phages isolated from composting provide insights into the evolution and diversity of tailed phages. BMC Genomics. 2017; 18:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen V.B., Arendall W.B. III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. 2010; D66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Adamcik J., Jeon J.H., Karczewski K.J., Metzler R., Dietler G.. Quantifying supercoiling-induced denaturation bubbles in DNA. Soft Matter. 2012; 8:8651–8658. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ProtParam is a tool for the computation of physical and chemical parameters based on protein sequence and is available at https://web.expasy.org/protparam/.
PDBePISA is an interactive tool for the exploration of macromolecular interfaces and is available at http://www.ebi.ac.uk/pdbe/pisa/.
PDBeFOLD is the Protein structure comparison service at European Bioinformatics Institute and is available at http://www.ebi.ac.uk/msd-srv/ssm.
ESpritz is a protein disorder prediction server available at http://protein.bio.unipd.it/espritz/.
The Dali server is a network service for comparing protein structures in 3D available at http://ekhidna2.biocenter.helsinki.fi/dali/.
PyMOL is a molecular visualization system available at https://pymol.org/2/.
The PSI-BLAST suite allows for a position-specific iterated search in the protein databases using a protein query and is available at https://blast.ncbi.nlm.nih.gov/Blast.cgi.
The Protein Data Bank (PDB) is available at http://www.rcsb.org/.
Atomic coordinates and structure factors for Drc have been deposited with the Protein Data Bank under accession number 6QLC.