Abstract
The ribosome is not only a protein-making machine, but also a regulatory element in protein synthesis. This view is supported by our earlier data showing that Arabidopsis mitoribosomes altered due to the silencing of the nuclear RPS10 gene encoding mitochondrial ribosomal protein S10 differentially translate mitochondrial transcripts compared with the wild-type. Here, we used ribosome profiling to determine the contribution of transcriptional and translational control in the regulation of protein synthesis in rps10 mitochondria compared with the wild-type ones. Oxidative phosphorylation system proteins are preferentially synthesized in wild-type mitochondria but this feature is lost in the mutant. The rps10 mitoribosomes show slightly reduced translation efficiency of most respiration-related proteins and at the same time markedly more efficiently synthesize ribosomal proteins and MatR and TatC proteins. The mitoribosomes deficient in S10 protein protect shorter transcript fragments which exhibit a weaker 3-nt periodicity compared with the wild-type. The decrease in the triplet periodicity is particularly drastic for genes containing introns. Notably, splicing is considerably less effective in the mutant, indicating an unexpected link between the deficiency of S10 and mitochondrial splicing. Thus, a shortage of the mitoribosomal S10 protein has wide-ranging consequences on mitochondrial gene expression.
INTRODUCTION
Mitochondria are semi-autonomous organelles with their own genetic material and a system of its expression. In plants, the expression of the mitochondrial genome is regulated mainly at the post-transcriptional and translational levels (1). The plant mitoribosomes are composed of a large and a small subunit, as are all ribosomes, but they are substantially divergent in terms of structure, length of rRNA and protein content from all other mitoribosomes and their bacterial counterparts (2). Recently, cryo-electron microscopy has revealed that the small ribosomal subunit of Arabidopsis mitoribosomes is in fact larger than the large subunit, bearing an additional RNA domain grafted onto the head (2). Out of the 81 protein species identified, 19 are unique to the plant mitoribosome. Ten of them are pentatricopeptide repeat (PPR) proteins. Moreover, some studies imply that the protein composition of the plant mitoribosome could vary at different developmental stages resulting in a heterogeneous population of mitoribosomes (3). It has also been shown that plant mitoribosomes interact extensively with the inner mitochondrial membrane but their mode of membrane association remains unknown (4).
To generate mature mRNAs, the primary transcripts in plant mitochondria undergo extensive and complex processing which includes maturation of the 3′ and 5′ termini, RNA editing as well as cis- and trans-splicing of introns (5,6). The mature protein-encoding mRNAs tend to have heterogeneous 5′ ends but generally well-defined 3′ ends (7). The ribosome-binding Shine-Dalgarno sequence present in prokaryotic and chloroplast mRNAs is absent in mitochondrial mRNAs from higher plants (8). Moreover, no obvious motifs have been found within the 5′ untranslated sequences (5′ UTRs) suggesting a gene-specific regulation of translation (9). In several cases it has been shown that functionally translated mRNAs lack any start or stop codon, indicating that the translational machinery in plant mitochondria has evolved ways to efficiently decode mRNAs without a conventional start or stop codon (10). These unique features suggest that trans-factors assist the ribosome in performing efficient translation in plant mitochondria. This role could be played by nuclear-encoded RNA-binding proteins, most of them belonging to the PPR family (11–13). The PPR6 protein stimulates translation of the mRNA encoding ribosomal protein S3 by taking part in its 5′ processing required for the generation of a translatable mature mRNA (14). The MTL1 protein is engaged in both splicing and translation of nad7 mRNA. So far, the molecular basis of its essentiality for translation is not understood, but it is not connected with the transcript processing (15). It has been shown that PPR proteins are also components of Arabidopsis mitoribosomes (2). As part of the mitoribosome, ribosomal PPR proteins (rPPRs) may help in the recruitment and docking of mRNAs to the small subunit or they may be involved directly in the process of translation, for example by stabilizing rRNAs. Thus, rPPR1, previously called PPR336, has been shown to be a generic translation factor required for optimal translation levels of most, if not all, mRNAs in Arabidopsis mitochondria (2).
The binding of PPR proteins to transcripts in organelles often leads to the formation of short RNA footprints that are indicators of their activity (16–18). A global analysis of sRNA-seq datasets has allowed identification of 222 such footprints named clustered organellar sRNAs (cosRNAs) (18). The largest fraction of cosRNAs was found in non-coding regions. Fifteen of them overlapped with previously mapped 3′ ends of mature mitochondrial transcripts. A typical mitochondrial cosRNA of this type is 15–50 nt long, with a sharp 3′ end and a fuzzy 5′ end. It is assumed that these cosRNAs are footprints of PPR proteins like MTSF1 (19), which binds to the 3′ region of a precursor mRNA and acts as a roadblock against the progression of a 3′ to 5′ exonuclease that would otherwise degrade the transcript. Thus, the 3′ end processing is associated with mRNA stabilization. Recently, it has been shown that the PPR protein-mediated protective mechanism blocking the exonucleolytic decay is not limited to the generation and stabilization of the 3′ ends of mature mitochondrial mRNAs but also concerns the stabilization of a nad1 precursor transcript containing only the second and third exon and defines the 3′ extremity of the first half of its trans-intron (20,21). In contrast, the 5′ ends of mature mitochondrial transcripts rarely overlap with a cosRNA. Instead, an efficient generation of mature 5′ termini of RNA requires an RNA stem-loop structure at the 5′ end (i.e. t-elements) and/or a PPR protein termed RNA processing factor bound upstream of the processing site (22,23). Notably, steady-state RNA analyses have demonstrated that cosRNAs accumulate differentially during the plant development suggesting their regulatory function (18).
Using two independent methods, in organello protein synthesis and polysomal profiling, we showed previously that silencing of the nuclear RPS10 gene encoding mitochondrial ribosomal protein S10 altered the mitoribosomes such that they oversynthesized ribosomal proteins and inefficiently synthesized some oxidative phosphorylation subunits (24). Here, we extend the analysis of the rps10 mutant to produce a genome-wide view of the Arabidopsis mitochondrial translatome at nucleotide resolution. This strategy combines sequencing of ribosome footprints (Ribo-seq) and of total RNA fragments (RNA-seq) isolated from the same starting material. In contrast to the profiling of mitoribosome footprints in Arabidopsis reported so far, which was based on footprints isolated from a total flower cell extract (2,10), we isolated ribosome-protected RNA fragments (RPFs) from purified mitochondria from leaves.
On the whole, the results reported here provide a comprehensive description of the alterations of the mitochondrial transcriptome and translatome in Arabidopsis with the mitochondria altered due to the silencing of the RPS10 gene. We found that shorter fragments of transcripts were protected by the rps10 mitoribosomes likely due to the distorted small subunit (SSU) structure, and these footprints showed a weaker 3-nt periodicity. The altered mitoribosomes in rps10 selectively affected translation of subsets of mRNAs, most notably by increasing the synthesis of ribosomal proteins, maturase (MatR) and the TatC protein also known as MttB. Unexpectedly, we also found that the splicing of mitochondrial transcripts was inefficient in the rps10 mutant. This finding reveals a previously unappreciated link between splicing and translational events in the mitochondria of plants.
MATERIALS AND METHODS
Plant material
Transgenic Arabidopsis thaliana with RNAi-silenced expression of the RPS10 gene (further referred to as rps10 or P2 and P3 phenotypes) (25) and wild-type plants, both Columbia-0 ecotype (Col-0), were used in this study. The growth conditions were the same as described previously (24). The youngest leaves of 9–10-week-old plants were the starting material for the experiments.
Isolation of mitochondrial fraction
Mitochondria were isolated from leaves as described in (26). Approximately 15 g (fresh weight) of leaves from rps10 and 30 g from wild-type plants was used per one isolation, yielding typically ∼1 mg of mitochondrial protein. Mitochondrial fractions were frozen in liquid nitrogen and stored at −80°C.
Construction of mtRNA-seq and mtRibo-seq libraries
Mitochondrial Ribo-seq and RNA-seq libraries were prepared using the ARTseq Ribosome Profiling Kit (Epicentre) dedicated for yeast, with slight modifications. Each library was prepared from 15 independent mitochondrial preparations. Immediately after removing mitochondrial preparations from the −80°C freezer chloramphenicol was added to a final concentration of 100 μg/ml and thawed mitochondria were pooled, suspended in polysome extraction buffer (0.2 M sucrose, 0.2 M KCl, 10 mM MgCl2, 2% polyoxyethylene 10 tridecyl ether, 1 mM dithiothreitol (DTT), 1% Triton X-100, DNase I (0.01 U/μl), 100 μg/ml chloramphenicol, 40 mM Tris-acetate, pH 8.0), incubated on ice for 10 min and centrifuged for 10 min at 28 154 × g at 4°C to obtain clarified lysate. RNA concentration in the lysate was determined as OD260 and aliquots of 100 (mtRNA-seq library) and 200 μl (mtRibo-seq library) were prepared. The 100 μl aliquot was supplemented with 10 μl of 10% sodium dodecyl sulphate (SDS) and total RNA was purified using the RNA Clean & Concentrator-25 kit according to the protocol for large RNAs >200 nt (Zymo Research). The 200 μl aliquot was treated with ARTseq nuclease (0.3 U nuclease per 1 OD260 of lysate) to degrade unprotected RNA following Epicentre instruction, and ribosome protected fragments (RPFs) were isolated using sucrose cushion ultracentrifugation at 294 000 × g for 4 h at 4°C. The pellet was suspended in 100 μl of nuclease-free water and 10 μl of 10% (w/v) SDS was added. Next, the RPFs were purified using RNA Clean & Concentrator-25 kit according to the protocol for small RNAs (17–200 nt). After purification the total RNA and RPFs preparations were depleted of rRNA using the Ribo-Zero rRNA Removal Plant Leaf kit (Illumina) and then were purified again using the RNA Clean & Concentrator-25 kit according to the protocol for large RNAs >200 nt for total RNA and the RNA Clean & Concentrator-5 kit according to the protocol for RNA >17 nt for RPFs. The total RNA preparation without ribosomal RNA was heat-fragmented by incubation at 94°C for 25 min. The RPFs without ribosomal RNA were purified by 15% Tris-borate-EDTA (TBE)-urea-polyacrylamide gel electrophoresis (PAGE) (Thermo Fisher Scientific) and the gel fragment containing RNA of 25–35 nt was excised. The RPFs were recovered from the excised gel slices following overnight elution as specified in the ARTseq Ribosome Profiling Kit. The fragmented total RNA and RPFs were end-polished, purified by RNA Clean & Concentrator-5 kit according to the protocol for >17 nt, ligated with 3′ adaptors, reverse-transcribed and purified as above. Purified cDNAs were separated on 10% TBE-urea PAGE, and gel fragments containing ssDNA of 80–100 nt for cDNA generated from total RNA fragments, and 70–80 nt for cDNA from RPFs, were excised. cDNA was recovered by overnight elution as specified in the ARTseq Ribosome Profiling Kit. The purified cDNA was then circularized and used for polymerase chain reaction (PCR) with index PCR primers for nine cycles. The libraries were purified by 8% Native TBE PAGE and AMPure XP beads (Beckman Coulter) and their quality and concentration was determined using an Agilent 2100 Bioanalyzer. High-throughput sequencing was performed on an Illumina MiSeq sequencer in the single-end mode for 50 nt at Wyzer Biosciences (Cambridge, MA, USA).
Bioinformatic analysis
mtRibo-seq and mtRNA-seq data preprocessing and alignment
3′ adapter sequences were trimmed from raw mtRibo-seq and mtRNA-seq data and low quality reads were removed using FastX-toolKit. Reads shorter than 25 nt were discarded. Alignments of cleaned reads were performed using Bowtie 2 with default ‘very-sensitive’ preset (27). The reads were aligned to the whole A. thaliana mitochondrial genome sequence (BK010421.1). All rRNA and tRNA sequences were filtered out by aligning the reads to their sequences in three steps, with unaligned reads from each step used as an input for the next round of alignment. In the first step mitochondrial tRNAs and rRNAs were removed, then chloroplast tRNAs and rRNAs, and subsequently nuclear tRNAs and rRNAs. The remaining reads were aligned to the A. thaliana mitochondrial genome with the split-aware aligner STAR (Spliced Transcripts Alignment to Reference) version 2.5.2b (28) with default settings allowing for up to five mismatches. We found that alignment with up to five mismatches allows better mapping of Arabidopsis mitochondrial genes with high editing frequency compared to more-stringent alignments. Next, the output reads were mapped to the chloroplast and subsequently to the nuclear genome allowing for up to two mismatches. This approach allowed determining the proportion of reads mapping, respectively, to the mitochondrial, chloroplast and nuclear genomes. All the reads mapped to the mitochondrial genome were sorted into two groups: one group with unique reads that mapped to a single location in the genome, and the other with reads that mapped to multiple locations in the genome. Only the unique alignments were used for further analysis. The sequence alignments were stored in indexed, sorted BAM files, which were browsed in the Integrative Genomics Viewer (IGV).
Ribosome P-site positioning and periodicity analysis
P-site offsets (i.e. the distances of the P-site from the 5′ and 3′ ends of the reads) were determined using riboWaltz v.1.0 (29). For these results RiboTaper software was used to determine the 3-nt periodic pattern of analyzed sequences in all three possible frames assuming P-value < 0.05 (30) using 5′ offsets of 12–21 nt for footprint lengths of 25–34 nt, respectively.
Transcriptional/translational output and translation efficiency analysis
To calculate the transcriptional output of a gene, mtRNA-seq reads of all lengths aligned to coding sequences (CDS) were used, whereas only 25–29 nt-long mtRibo-seq reads were applied to estimate translational output. Both types of reads were counted by featureCounts (assuming fracOverlap = 1) and then normalized for sequencing depth and length and expressed in TPM units, i.e. transcripts per million. Intron-containing genes were excluded from this analysis. Translational efficiency (TE) of a given CDS was calculated as the ratio of normalized transcript abundance determined by mtRibo-seq to the normalized abundance determined by mtRNA-seq.
Details of other bioinformatic analyses are provided in Supplementary Data.
Statistical analysis
Paired Wilcoxon signed rank test was applied to compare the distribution of fractions of footprints mapped to individual reading frames. We applied the Benjamini–Hochberg method for P-value correction to control the false discovery rate in multiple testing. The Kolmogorov–Smirnov and unpaired Wilcoxon–Mann–Whitney tests were used for comparison of the distribution of read lengths between the wild-type and the mutant. The statistical analyses were carried out in R package 3.5.1 (R Core Team 2018, http//www.R-project.org).
RT-qPCR analysis of pre-mRNA and mature mRNA levels
Total RNA was isolated using the GeneMATRIX Universal RNA purification kit (EURx). Reverse transcription was performed using up to 2 μg of total RNA, random hexamers and High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) according to the manufacturer's directions. The resulting cDNA was used as a template for quantitative real-time PCR analysis on a LightCycler480 instrument with dedicated software (Roche Applied Science) and with sets of primers for pre-mRNAs and mature mRNAs described in (19). Splicing efficiency was determined by calculating the ratio of spliced to unspliced transcripts as described in (15) and (21).
Northern blot analysis
Small RNAs (sRNAs) were purified using RNA Clean & Concentrator kit-5 (Zymo Research) according to the protocol for small RNAs (17–200 nt). They were isolated from polysomal fraction previously treated or not with ARTseq nuclease (0.3 U nuclease per 1 OD260 of lysate). The polysomal fraction was obtained by sucrose cushion ultracentrifugation at 294 000 × g at 4°C for 4 h from mitochondria lysed in polysome extraction buffer. sRNA gel blot hybridization was carried out as described in (31). For analysis of total mtRNA RNA was extracted from mitochondria using the GeneMATRIX Universal RNA purification kit (EURx), and blotting and hybridization were performed as described in (32). The following oligonucleotide probes biotinylated on the 5′ terminus were used: rps4 5′ [Btn]TCGTTGAATCAGTTTTTTAA GCAGCCACAT, rps12 5′ [Btn] TCGACCCGGAATTCCCATCAAATCCTTT, rpl5 5′ [Btn] ATCTTCGTAATGAAAATTGAGTGGAAACAT, nad3 5′ [Btn] GATAGAAATTGGTGCAAA TTCTGACATCAT, nad9 5′ [Btn] ATAGCATTTCCTATTGATTTGTCCCCTGGACTGG, atp1 5′ [Btn] AGGCAAGGAGGAATACCGCTTTAGAA, atp4 5′ [Btn] AGTAGTCTTGGTATTG GATCCGCTCTTCTGAGAGCA and atp9 5′ [Btn] GCTTGCTTTATGAGACTGAATGGA AACCTT. Hybridization was performed overnight in PerfectHyb Plus buffer (Sigma Aldrich) at 39°C and then the membrane was washed three times with 2× saline-sodium citrate (SSC) for 10 min at 39°C. The biotin-labeled probes were detected using the Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher Scientific). Data were analyzed using G:Box software (Syngene).
RESULTS
Experimental setup of mitoribosome profiling assay
To obtain mitochondrial ribosome protected fragments (mtRPFs), we used purified mitochondrial fractions rather than total cell extracts that were used by other authors (2,10,33) since several reports suggesting that transcripts derived from mitochondrial genes inserted in the nuclear genome could contribute to the transcriptome in plants (34,35). We performed in parallel RNA-seq and Ribo-seq analyses for mitochondria isolated from two phenotypes of the rps10 Arabidopsis hemizygous transformants, P2 and P3, differing by the onset of RPS10 silencing, and wild-type plants (25). The P2 and P3 plants exhibit a similar decrease in the RPS10 transcript level and in consequence similarly altered population of mitoribosomes (24). To obtain high-coverage sequencing data, we pooled several independent preparations of mitochondria (see ‘Materials and Methods’ section). The vast majority of reads in all the libraries obtained mapped to the mitochondrial genome, albeit in the wild-type libraries the non-mitochondrial reads were more frequent than in the rps10 ones (Supplementary Figure S1). This difference probably reflects the poorer purity of the wild-type mitochondrial preparations compared with rps10. We found that some of the reads aligning to nuclear sequences detected in our libraries came from mRNAs encoding mitochondria-destined proteins undergoing translation by cytosolic ribosomes at the mitochondrial outer membrane (36).
General characteristics of mitochondrial RNA-seq and Ribo-seq data
After trimming adapters and removal of low quality reads, the mtRibo-seq assay produced about 8 million mtRPFs for the WT and approximately 15 and 12 million for the P2 and P3 phenotypes, respectively (Supplementary Table S1). There were over 4, 8 and around 6 million mtRNA-seq reads for the WT, P2 and P3, respectively. The reads were processed to filter out all rRNA and tRNA sequences and then were aligned to the Arabidopsis mitochondrial reference genome, allowing for up to five mismatches. The vast majority of the initial mtRPFs mapped to mitochondrial rRNA and tRNA. At last, ca. 0.7 million and ca. 0.5 and 0.3 million of mtRibo-seq reads for WT, P2 and P3, respectively, could be aligned to the Arabidopsis mitochondrial genome outside the rDNA/tDNA sequences (see Supplementary Table S1 for exact numbers). Due to the high abundance of the rRNA and tRNA sequences, the mapping efficiency of the mtRibo-seq study was relatively low (∼9% for WT and ∼3% for P2 and P3). In contrast, the mapping efficiency of the mtRNA-seq reads was much higher (∼76% for WT, ∼79% for P2 and 81% for P3) because of the much lower content of the non-coding mitochondrial RNAs. It should be stressed that the normalized mtRNA-seq and mtRibo-seq results of the two independent experiments for the two phenotypes of the rps10 mutant, P2 and P3, were nearly identical, with the Pearson correlation coefficient of 0.99 for mtRNA-seq and 0.92 for the mtRibo-seq data (Supplementary Figure S2). This shows, on the one hand, the excellent reproducibility of the whole procedure, and on the other that the conclusions drawn from these data are not limited to a particular rps10 line but concern fundamental consequences of the S10 deficit.
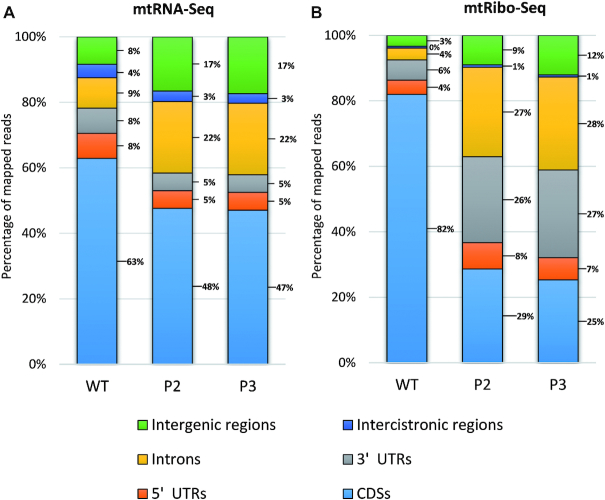
The reads mapping to the mitochondrial genome were then sorted into two subgroups: those that mapped to a single genome location only (unique match) and those mapping to more than one location (multi-position match) (Table 1). The first subgroup comprised ca. 89% of mtRibo-seq reads, the second approximately 11%. As shown in Figure 1B, the vast majority of the unique reads from wild-type mitochondria mapped to CDS (82%). Surprisingly, this percentage was substantially lower for the mutant (∼29 and 25% for P2 and P3, respectively), indicating that in rps10 the mtRibo-seq reads lie mostly in regions outside the annotated CDSs. This tendency was much weaker in the mtRNA-seq data and concerned mainly introns and intergenic regions (Figure 1A).
Table 1.
Quantitative characteristics of mtRNA-seq and mtRibo-seq reads mapping to Arabidopsis mitochondrial genome
| WT mtRNA-seq | P2 mtRNA-seq | P3 mtRNA-seq | WT mtRibo-seq | P2 mtRibo-seq | P3 mtRibo-seq | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Feature | number of reads | % of total mapped reads | number of reads | % of total mapped reads | number of reads | % of total mapped reads | number of reads | % of total mapped reads | number of reads | % of total mapped reads | number of reads | % of total mapped reads |
| Total reads mapped to mitochondrial genome | 3 266 398 | 100.00% | 6 643 268 | 100.00% | 4 598 122 | 100.00% | 665 159 | 100.00% | 461 960 | 100.00% | 327 439 | 100.00% |
| Unique match | 2 671 482 | 81.79% | 5 824 461 | 87.67% | 3 995 954 | 86.90% | 594 404 | 89.36% | 408 669 | 88.46% | 289 775 | 88.50% |
| Multi-position match | 594 916 | 18.21% | 818 807 | 12.33% | 602 168 | 13.10% | 70 755 | 10.64% | 53 291 | 11.54% | 37 664 | 11.50% |
Number and relative abundance of unique and multi-position match reads in WT and two rps10 phenotypes (P2 and P3) is shown.
Figure 1.
Percentage of unique (A) mtRNA-seq and (B) mtRibo-seq reads mapping to various regions of mitochondrial genome in P2 and P3 phenotypes of rps10 and wild-type.
Shorter mitoribosome footprints with a weaker triplet periodicity due to the S10 protein deficit
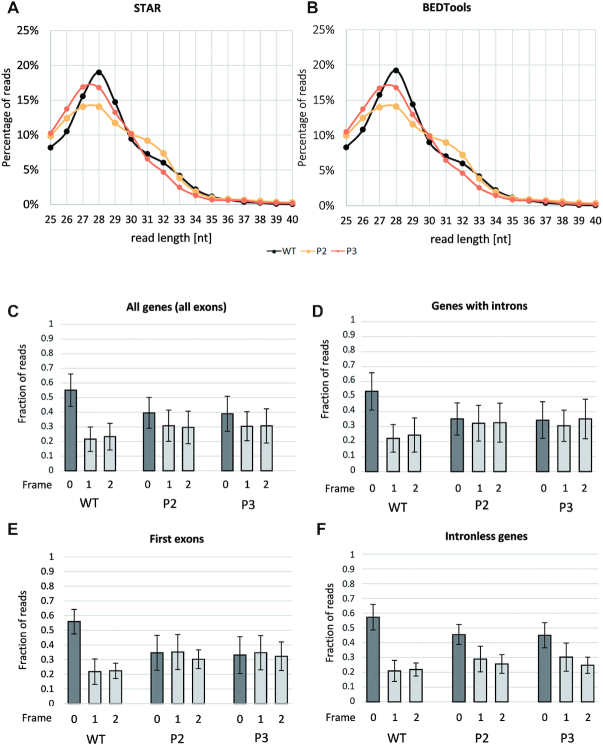
The size distribution of the ribosomal footprints derived from mitochondrial protein-CDS was estimated by two programs, STAR and BEDTools (Figure 2A and B). In both analyses mtRPFs of 28 nt were by far the most numerous in the WT, while in the both rps10 phenotypes the length distribution was shifted toward shorter reads and those of 27 and 28 nt were equally abundant. Notably, this minute difference in the distribution is statistically significant with P < 10−15. Also shorter reads (25 and 26 nt) were more abundant in rps10 than in the wild-type. In addition, a minor, broader peak was present at 31–32 nt for all samples examined. As in the maize and human mitochondria (33,37), shorter footprints were more numerous than longer ones.
Figure 2.
Characteristics of mtRibo-seq reads in P2 and P3 phenotypes of rps10 and wild-type. Length distribution of mtRibo-seq reads mapping to mitochondrial protein-CDS determined with (A) STAR and with (B) BEDTools software. (C) Average 3-nt periodicity of 25–29 nt long mtRibo-seq reads mapping to all mitochondrial protein-coding genes, (D) mapping exclusively to mitochondrial protein-coding genes containing introns, (E) mapping exclusively to first exons, (F) mapping exclusively to mitochondrial protein-coding intronless genes. Whiskers represent standard deviation.
True ribosome footprints representing actively translating ribosomes are expected to exhibit a 3-nt periodicity in the protein-coding regions. Crucial for determining such periodicity is the identification of the location of the ribosome P-site for each protected fragment. Using offsets from RiboWaltz (29), we analyzed the ribosome footprints on the CDSs for OXPHOS, ribosomal proteins, proteins involved in cytochrome c biogenesis, maturase (MatR) and the transport membrane protein TatC using the RiboTaper software (30). First, the wild-type data were analyzed for each footprint length separately to assess for which length the periodicity was the strongest (Supplementary Figure S3). The reads of 25–29 nt showed a substantially higher percentage of significant 3-nt periodicity and correct frames of CDSs as well as the median frequency of the correct frames than did the longer reads (30–34 nt) (Supplementary Figure S3). Therefore, we concluded that the 3-nt periodicity is clear-cut in the shorter reads, and they were selected for further analyses, unless noted otherwise. The reads of 30–34 nt may represent footprints of RNA-binding proteins or alternative ribosome conformations, probably inactive in translation.
Although the 25–29 nt long footprints had a statistically significant (P < 0.006) fraction of P-site positions in the correct frame rather than in the alternative frames, both in the wild-type and the two phenotypes of rps10, the fraction of P-site positions in the correct frame was statistically significantly (P < 10−12) higher in the wild-type (on average 55%) than in P2 or P3 (ca. 40%) (Figure 2C). Remarkably, for genes containing introns considered separately (Figure 2D) the footprints in all three possible frames showed similar abundancies in P2 and P3 (P > 0.45), whereas in the wild-type the correct frame was still significantly dominant (P < 0.00002). Interestingly, a similar distribution of footprints among frames was obtained when all exons or only the first exon were studied (Figure 2C and E). When only the intronless genes were analyzed, the proportion of correctly mapping reads increased, but still did not reach 50% in rps10 (Figure 2F).
Lower efficiency of intron splicing as a result of the S10 protein deficit
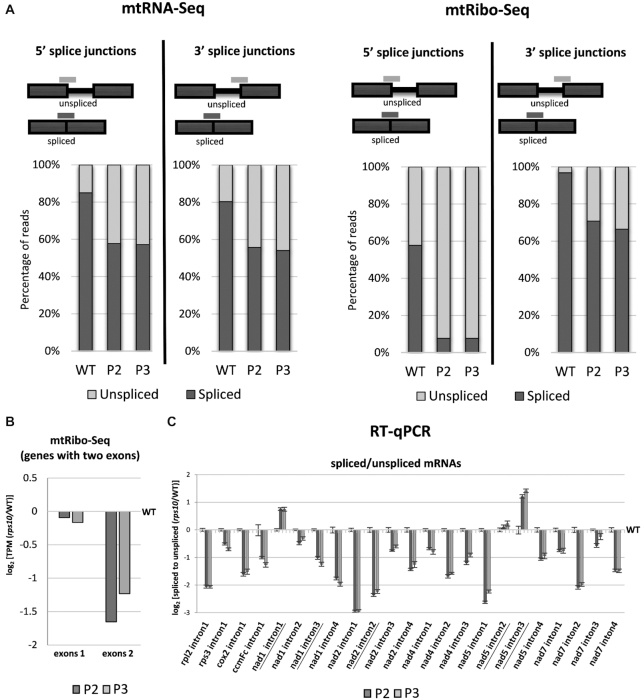
To evaluate the possible effects of the S10 deficit on the efficiency of mitochondrial splicing and/or translation of unspliced RNAs, all reads spanning spliced exon–exon junctions and all unspliced exon–intron junctions with the exception of junctions involving trans-spliced introns were quantified in the mtRNA-seq and mtRibo-seq reads (Figure 3A). This analysis was performed separately for the 5′ splice junctions (the 5′ end of an intron and the 3′ end of the preceding exon) and the 3′ ones. The percentage of unspliced reads was strikingly higher in the mutant than in the wild-type, both in mtRNA-seq and mtRibo-seq (Figure 3A). As could be expected, in the mtRNA-seq data the 5′ and 3′ junctions gave nearly identical results, being present in ca. 15–20% of reads in the wild-type and ca. 42–45% in rps10. Conversely, in the mtRibo-seq data the two junctions showed strikingly different frequencies. In the WT, the 5′ junction was found in ca. 42% of reads while the 3′ one was hardly detectable at ca. 3%. In the rps10 phenotypes these frequencies were many times higher, constituting ca. 92% and 30–35% of reads, respectively. The presence of the exon–intron junctions in the mtRNA-seq reads simply reflects the retention of introns in mRNAs, while in the mtRibo-seq reads it likely indicates that ribosomes actually carry out translation of unspliced mRNAs and that this is much more frequent in the mutant than in the wild-type. That the 5′ junctions are several-fold more frequent than the 3′ ones likely reflects the direction of translation and stalling of the ribosome at or downstream of the exon–intron junction and/or subsequent termination of translation somewhere along the intron sequence. This suggestion is supported by a lower abundance of ribosome footprints across exon 2 than exon 1 in the mutant compared with the wild-type (Figure 3B). Alternatively, the reads mapped to the exon–intron junctions could be not mitoribosome footprints but rather result from mRNA protection by RNA-binding proteins involved in splicing. Such proteins could be those engaged in the interactions between exon-binding sequences (EBSs) within the intron and intron-binding sequences (IBSs) in the flanking exons. The increased proportion of unspliced transcripts due to inefficient splicing observed in the mutant would be expected to increase the abundance of such reads.
Figure 3.
Abundance of spliced and unspliced mitochondrial mRNAs in P2 and P3 phenotypes of rps10 and wild-type. (A) Percentage of mtRNA-seq and mtRibo-seq reads spanning spliced exon–exon junctions and unspliced exon–intron junctions. The results are presented separately for 5′ and 3′ splice junctions. (B) Log2 ratios of mtRibo-seq TPM (reads per kilobase per million reads mapping to CDS) in exons 1 and exons 2 for genes with only two exons (rps3, rpl2, cox2 and ccmFc) in P2 and P3 phenotypes of rps10 compared with the wild-type. (C) Log2 ratios of spliced to unspliced transcripts for each intron estimated by RT-qPCR in P2 and P3 phenotypes of rps10 compared with the wild-type. The values are means of five biological replicates, with error bars representing standard errors. Trans-spliced introns are underlined.
These results prompted us to determine the efficiency of splicing for individual genes in the WT and rps10 by quantitative RT-PCR. For most genes, the level of both spliced and unspliced transcripts was elevated in the mutant compared with the wild-type, but noticeably more so for the unspliced ones (Supplementary Figure S4). As a result, the ratio of spliced to unspliced mRNAs turned out to be lower in the mutant than in the WT, ca. 2-fold for most transcripts, indicating a reduced splicing efficiency of all transcripts except for intron 1 in nad1, and introns 2 and 3 in nad5, which are all trans-spliced (Figure 3C). Thus, our results indicate that a deficit of the S10 protein upregulates mitochondrial transcripts and shifts the balance between spliced and unspliced transcripts toward the latter. These two effects are nearly universal, affecting most mRNAs containing introns to a similar degree. Whether these effects are due to dysfunctional mitoribosomes or are caused by compromised hypothetical non-ribosomal function of S10 remains to be established.
Most mtRibo-seq reads mapping to non-coding regions of mitochondrial transcripts represent putative footprints of RNA-binding proteins (mtRibo-seq-sRNAs)
The significant accumulation of mtRibo-seq reads mapping to regions outside the annotated CDSs in rps10 compared to the wild-type (Figure 1) prompted us to study their nature further. The lack of the 3-nt periodicity for the vast majority of them suggested that those reads are not footprints of mitoribosomes but rather originate from RNA-binding proteins (RBPs) like PPR proteins. Ruwe et al. have developed a software (sRNA miner) for identifying potential footprints of RBPs in sRNA sequencing datasets (18). We used this tool to find potential footprints of RBPs aligning to non-coding regions of mitochondrial mRNAs (5′ UTRs, introns, 3′ UTRs and intercistronic regions) in our mtRibo-seq data. The parameters in sRNA miner were set to detect clusters of sRNAs with one or both ends sharp. Forty such sRNAs, further referred to as mtRibo-seq-sRNAs, mainly 25–28 nt in length, were found in all three (WT, P2, P3) and 35 additional ones in two of the phenotypes examined (Supplementary Figure S5). This indicates that the detected fragments arise reproducibly and are not products of random decay. We focused on 58 unique-match (non-redundant) mtRibo-seq-sRNAs (9 in 3′ UTRs, 10 in 5′ UTRs, 37 in introns and 2 in intercistronic regions) (Supplementary Table S2) and their abundance was calculated relative to the library depth (Supplementary Figure S6A). In all the transcript regions investigated, except for the intercistronic ones, the mtRibo-seq-sRNAs reads were more abundant in the mutant than in the wild-type, most markedly so for the mtRibo-seq-sRNAs mapping to introns which were only rarely found in the wild-type. The result for 3′ UTR regions is presented in two versions. The case of the sRNA reads mapping to 3′ UTRs was unusual in that ca. 90% of them derived from a single transcript, rpl16. We therefore analyzed these reads in two ways—for all the reads, and after the dominating rpl16 reads had been removed. In both analyses the reads were several-fold less common in WT than in the mutant (compare relevant graphs in Supplementary Figure S6A).
The mtRibo-seq-sRNA reads constituted a substantial fraction of all mtRibo-seq reads mapping to particular regions of mitochondrial mRNA in both the WT and the mutant (Supplementary Figure S6B). This was most strongly pronounced for the UTRs, where the mtRibo-seq-sRNAs comprised 89% and ca. 97% of all reads from 3′ UTRs, and 85% and ca. 86% of those from 5′ UTRs in the wild-type and rps10, respectively. The corresponding numbers for introns and intercistronic regions were, respectively, 46 and 55%, and 17 and 14%. This result indicates that the vast majority of reads from the UTRs in the mtRibo-seq data actually represent mtRibo-seq-sRNAs. In contrast, among the reads from introns and intercistronic regions in the mutant only half or less represent mtRibo-seq-sRNAs and the rest could be bone fide mtRPFs.
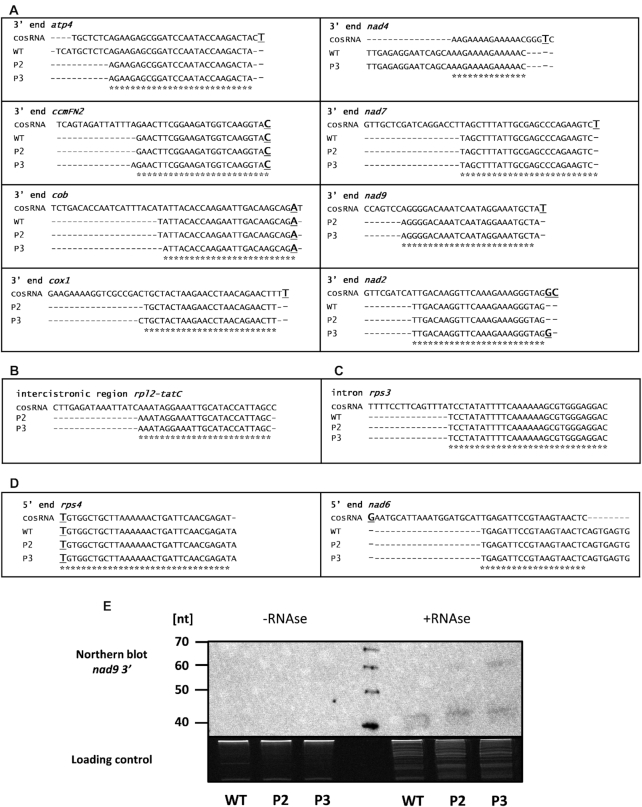
Some mtRibo-seq-sRNAs resemble cosRNAs (clustered organellar sRNAs)
Eight of the mtRibo-seq-sRNAs mapping to 3′ UTRs resembled the cosRNAs reported earlier that coincide with the 3′ termini of mature mitochondrial transcripts (18) (Figure 4A). The mitochondrial cosRNAs positioned at mRNA 3′ ends had strictly defined 3′ ends and relaxed 5′ ends. The mtRibo-seq-sRNAs identified by us, with the exception of the nad4 mtRibo-seq-sRNA partially overlapping the cosRNA representing the binding site of mitochondrial stability factor1 (MTSF1) (19), have identical or nearly identical 3′ ends (1 or 2 nt shorter) with cosRNAs and the 5′ ends recessed by several nucleotides. Similarly to the 3′ UTR-derived mtRibo-seq-sRNAs also the mtRibo-Seq-sRNA from the intercistronic region of rpl2-tatC transcripts and that from the rps3 intron had the 3′ ends nearly identical with the corresponding cosRNA and were trimmed at the 5′ end (Figure 4B and C). Only two mtRibo-seq-sRNAs mapping to 5′ UTRs matched cosRNAs fully or partially (Figure 4D). Notably, the rps4 mtRibo-seq-sRNA has an identical 5′ end with the corresponding cosRNA and is only 1 nt longer at the 3′ end. It is likely that it represents a footprint of the PPR protein involved in the formation of the atypical 5′ end of mature rps4 mRNA lacking the AUG start codon (38).
Figure 4.
mtRibo-seq-sRNAs identified in P2 and P3 phenotypes of rps10 and wild-type. Comparison of mtRibo-seq-sRNAs sequences identified in mtRibo-seq reads in rps10 and wild-type plants with previously published cosRNAs (A) from 3′ ends of mRNAs, (B) intercistronic regions, (C) introns and (D) 5′ ends of mRNAs. The cosRNA sequences are from (18). Asterisks mark sequences common to mtRibo-seq-sRNA and cosRNA. Major mRNA ends are in bold and underlined. (E) Detection of nad9 mtRibo-seq-sRNA in sRNA fractions of mitochondrial polysomal RNA isolated from rps10 and wild-type plants. Equal amounts (500 ng) of two sRNA fractions were analyzed: sRNAs isolated from polysomal fraction, and sRNAs isolated from polysomal fraction treated with RNase before isolation of sRNAs. The fractions were run on a denaturing 15% polyacrylamide gel, transferred to a nylon membrane and detected using 5′- biotinylated oligonucleotide probe. The image of the agarose gel stained with ethidium bromide is shown as a loading control in the bottom panel.
To test if the sRNAs identified among the mtRibo-seq reads exist in vivo, we studied them by Northern hybridization using the mtRibo-seq-sRNA from the 3′ UTR of the nad9 gene as a probe (Figure 4E). No signal was found when the sRNA fraction isolated from polysomal RNA was used, while clear signals were found for sRNAs purified from polysomes treated with nuclease, suggesting that the mtRibo-seq-sRNAs are not associated with ribosomes in vivo but are products of degradation of mRNAs undergoing translation by the nuclease added during the ribosomal profiling procedure. Thus, the difference in length between the mtRibo-seq-sRNAs identified by us and the corresponding cosRNAs is likely due to the specificity of the ribonuclease used in the ribosome profiling procedure.
Differential changes in transcriptional output and efficiency of mitochondrial translation caused by the S10 protein deficit
To determine how the S10 deficit affects mitochondrial transcription/translation, we compared the abundance of mtRNA-seq and mtRibo-seq reads aligning to protein-CDS of mitochondrial transcripts i.e. those widely known to be translated in the WT and rps10. We decided to include in the analysis only intronless transcripts since the unspliced transcripts, substantially more abundant in the mutant, could disturb the picture.
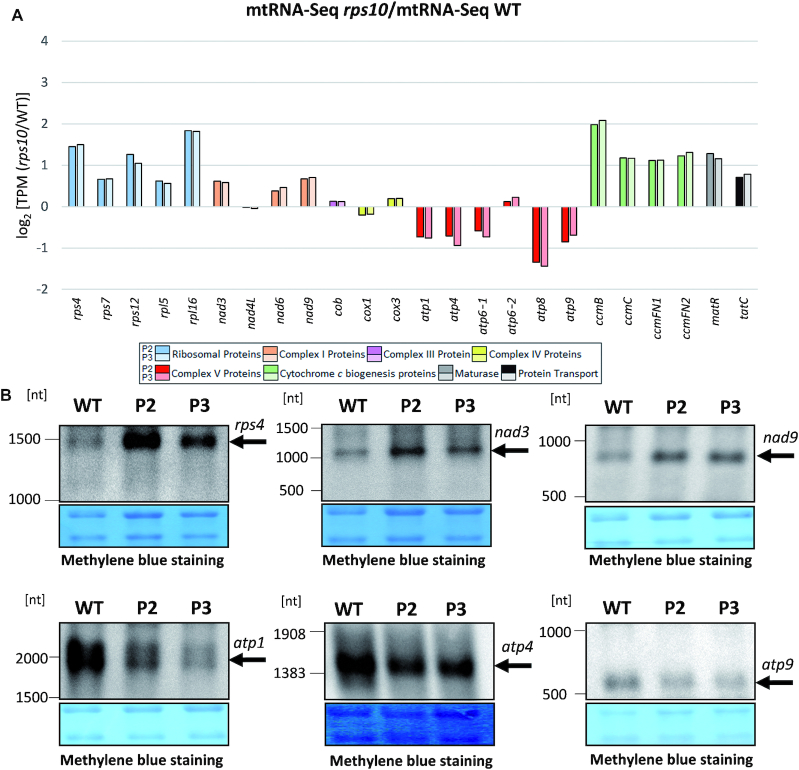
The transcriptional output (mtRNA-seq) of all ribosomal, cytochrome c maturation, matR and tatC genes was elevated markedly in the mutant (Figure 5A). In contrast, the transcriptional response of the OXPHOS genes was varied and overall markedly weaker. The genes encoding subunits of complex I were moderately upregulated in rps10 compared with the wild-type while those encoding subunits of complex V, except for atp6-2, moderately downregulated. Previously, basing on quantitative RT-PCR analysis, we reported that all mitochondrial transcripts were upregulated in rps10, including those encoding subunits of complex V, although their increase was noticeably lower than that of the other transcripts (24). To resolve this discrepancy, we performed Northern hybridization on mitochondrial RNA isolated from the wild-type and P2 and P3 plants using probes specific to several transcripts encoding subunits of complex V (atp1, atp4, atp9) and some other mitochondrial transcripts (rps4, nad3, nad9). As in the mtRNA-seq results, the level of the atp1, atp4 and atp9 transcripts was downregulated and that of rps4, nad3 and nad9 upregulated in rps10 (Figure 5B).
Figure 5.
Comparison of transcriptional output of mitochondrial intronless genes in P2 and P3 phenotypes of rps10 and wild-type. Transcriptional output is defined as mtRNA-seq TPM (reads per kilobase per million reads mapping to CDS). (A) Relative mRNA level defined as mtRNA-seq in rps10 compared with WT. Log2 of the ratio is shown. Transcripts are divided into groups encoding: ribosomal proteins, proteins of individual OXPHOS complexes, proteins involved in cytochrome c biogenesis, maturase (MatR) and transport membrane protein (TatC). (B) Level of selected mitochondrial transcripts in wild-type and rps10 analyzed by Northern hybridization. Total mitochondrial RNA was electrophoresed in denaturing conditions on 1% agarose, transferred to a nylon membrane and hybridized with 5′- biotinylated oligonucleotide probes for the rps4, nad3, nad9, atp1, atp4 and atp9 transcripts.
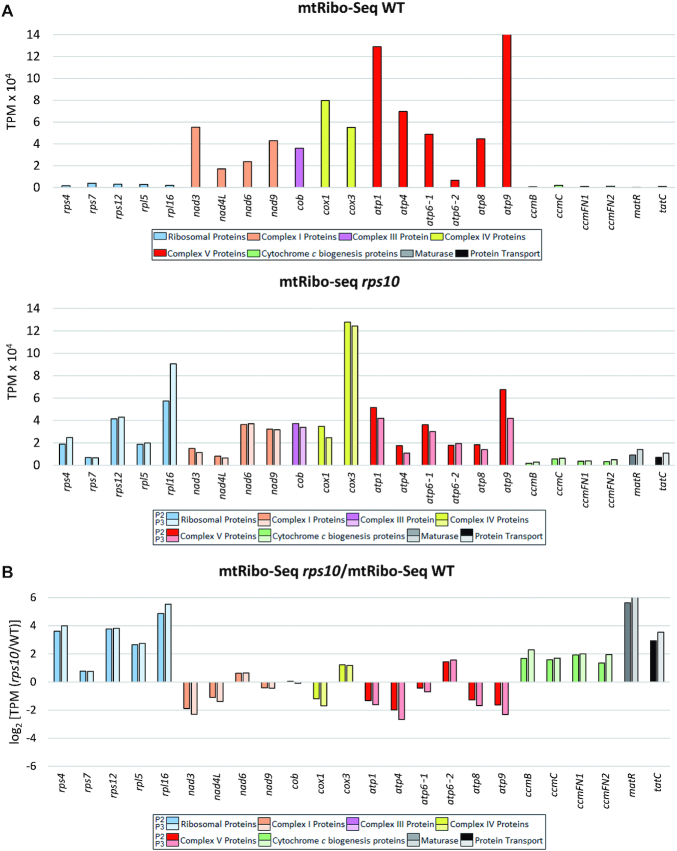
Considering the translation intensity reflected in mtRibo-seq data, in the wild-type mitochondria it was definitely much higher for the OXPHOS transcripts than for the remaining mitochondrial genes (Figure 6A). A similar conclusion has been reported recently based on ribosomal profiling of Arabidopsis mitochondrial transcripts in total cell extracts (10). In contrast, in rps10 the OXPHOS transcripts were no longer translated preferentially: the production of OXPHOS proteins was either reduced, unchanged or, for nad6, cox3 and atp6-2, modestly increased relative to the WT, while for all the other transcripts studied a clear-cut increase was observed, either moderate (cytochrome c maturation proteins) or several-fold (ribosomal proteins, MatR and TatC) (Figure 6B).
Figure 6.
Translational output of intronless mitochondrial genes in P2 and P3 phenotypes of rps10 and wild-type. Translational output is defined as mtRibo-seq TPM (reads per kilobase per million reads mapping to CDS). Genes are divided into groups as in Figure 5. (A) Translational output in wild-type (top) and rps10 (bottom). (B) Relative translational output in rps10 compared with WT. Log2 of the ratio is shown.
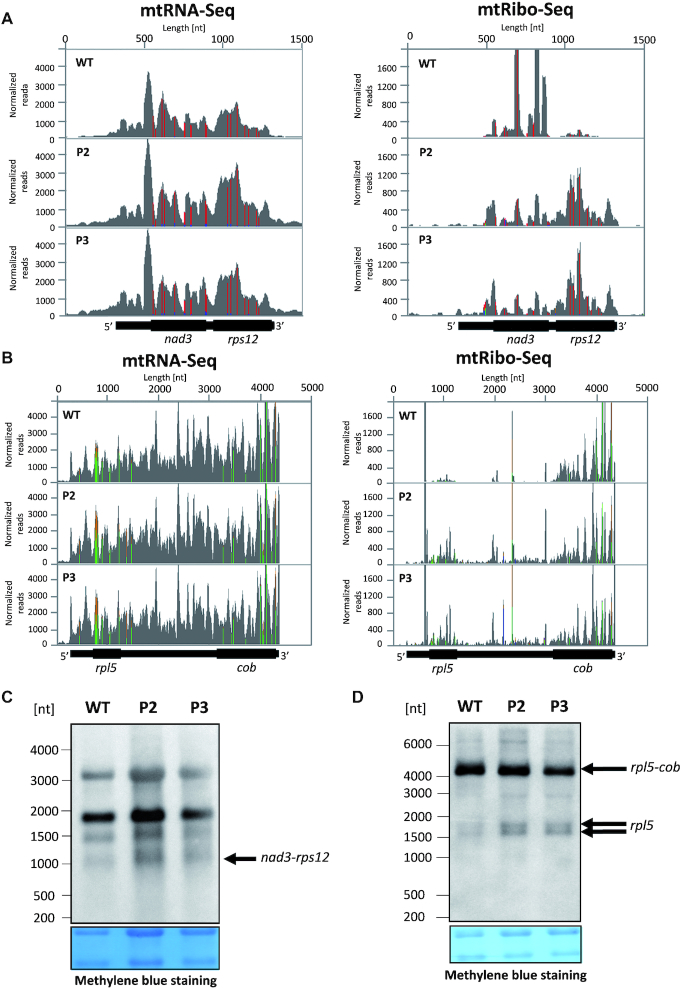
A striking illustration of the differential translation of proteins of different functions and its modulation by rps10 silencing is provided by the dicistronic nad3-rps12 transcript (Figure 7A). In the wild-type, the translational output of the OXPHOS protein (NAD3) is considerably higher than that of the ribosomal protein (RPS12). Conversely, in rps10 the ribosomal footprints for rps12 are more abundant than those for nad3. A similar tendency was observed for another dicistronic transcript comprised of genes encoding OXPHOS and ribosomal proteins, rpl5-cob (Figure 7B). Here the translation of rpl5 was clearly upregulated in rps10 while that of cob was rather unchanged. Thus, independently of their localization along the transcript, translation of the cistron encoding a ribosomal protein is markedly upregulated in the mutant compared to the wild-type. Next, we checked if the enhanced translation of non-OXPHOS proteins encoded by dicistronic transcripts could be associated with their intercistronic processing since a relationship between translation and such processing in chloroplasts has been shown (39). It was documented that monocistronic mRNAs were translated better than polycistronic ones. The Northern hybridization analysis clearly indicates that no monocistronic rps12 mRNA can be detected in wild-type or rps10 mitochondria (Figure 7C), in agreement with a previous report (7). Thus, only the dicistronic nad3-rps12 transcript is used as the template for the synthesis of S12, and intercistronic processing is not required for its efficient synthesis in the mutant. In contrast, the results of Northern hybridization using the rpl5 probe suggest that the dicistronic rpl5-cob transcript coexists with a monocistronic transcript coding for L5 (Figure 7D). The unprocessed dicistronic form accumulates in all samples at a comparable level, while the abundance of the putative monocistronic rpl5 is generally very low, but definitely higher in the mutant. This result suggests that the increased translation of L5 in rps10 could indeed be associated at least partially with a more efficient formation of the monocistronic rpl5 transcript.
Figure 7.
Transcriptional and translational status of dicistronic nad3-rps12 and rpl5-cob transcripts in P2 and P3 phenotypes of rps10 and wild-type. Coverage of total mRNA reads and ribosomal footprints on the dicistronic (A) nad3-rps12 and (B) rpl5-cob transcript. Reads aligning to the transcript were normalized against the number of all reads in mtRNA-seq and mtRibo-seq libraries and their distribution was visualized using Integrative Genomics Viewer (IGV). (C) Northern hybridization of total mitochondrial RNA using a 5′- biotinylated oligonucleotide probe specific for rps12. Size of dicistronic form of nad3-rps12 transcripts was inferred from transcript 5′ and 3′ ends defined in (7). (D) Northern hybridization of total mitochondrial RNA using a 5′-biotinylated oligonucleotide probe specific for rpl5. Sizes of monocistronic forms of rpl5 and dicistronic form of rpl5-cob were inferred from two 5′ ends and 3′end of rpl5-cob defined in (7) and from cosRNA identified in our analysis in the intercistronic region, putatively coinciding with 3′ end of rpl5. Arrows indicate discussed forms. The membranes were stained with methylene blue and are shown as loading control in the bottom panels.
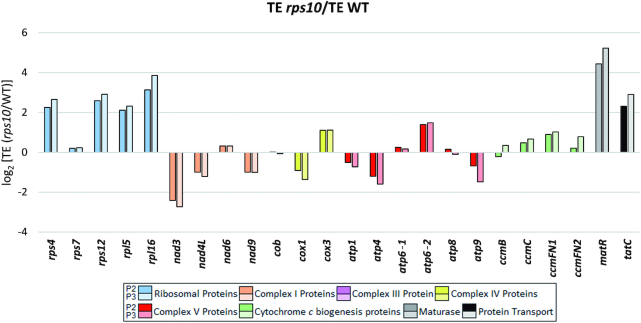
The translational output of a gene depends on both the level of its transcript and its translation efficiency (TE). To better understand the altered translational output of mitochondrial proteins in the rps10 mutant, the relative TE of individual genes in the rps10 and wild-type mitochondria was calculated (Figure 8). The TE was defined here as the ratio of ribosome footprint reads (mtRibo-seq) to mRNA reads (mtRNA-seq) mapping to the coding region of a gene. The largest positive TE difference between rps10 and WT was observed for a subset of genes with a markedly elevated transcript level in the mutant (rps4, rps12, rpl5, rpl16, matR and tatC), implying that these genes are under strong upregulation in rps10, both transcriptional and translational. The TE of another set of transcripts, those encoding proteins involved in cytochrome c biogenesis, was only slightly increased in rps10 suggesting that their enhanced translational output in the mutant is connected mainly with the greater transcriptional response. In contrast to these quite uniform group-specific alterations, the direction of the TE changes and their magnitude for the OXPHOS transcripts was varied. For most of them (nad3, nad4L, nad9, cox1, atp1, atp4 and atp9) the TE was moderately or slightly decreased in rps10. The case of the nad3 and nad4L genes encoding subunits of complex I is particularly interesting. The level of their mRNAs was markedly increased in rps10, but this was over-compensated by a decreased TE, giving an overall reduced level of expression at the protein level. This result suggests a dominance of the translational over transcriptional control in the expression of at least some subunits of complex I (NAD3 and NAD9) in the rps10 mutant.
Figure 8.
TE of intronless mitochondrial genes in P2 and P3 phenotypes of rps10 and wild-type. TE is defined as the ratio of mtRibo-seq TPM to mtRNA-seq TPM and expressed as log2 of (rps10/WT) ratios.
DISCUSSION
How the ribosome functions in translation is understood in considerable detail, but its role as a regulator of gene expression is only beginning to be appreciated. Previously we showed that mitoribosomes altered due to silencing of the nuclear RPS10 gene encoding a mitochondrial ribosomal protein differentially affected translation of several mitochondrial mRNAs in Arabidopsis (24). Here, we elaborate on that finding by characterizing in details the specific features of the altered translation in rps10 mitochondria by ribosomal profiling. We also uncover unexpected coupling between the deficit of the mitoribosomal protein S10 and the efficiency of the mitochondrial splicing machinery.
While the wild-type mitoribosomes preferentially synthesize OXPHOS proteins, the deficiency of the S10 protein substantially increases the translational output of ribosomal proteins, MatR, TatC and proteins involved in cytochrome c maturation, and only weakly and diversely alters OXPHOS protein synthesis with a tendency to reduction. The enhanced production of ribosomal proteins, MatR and TatC involves both transcriptional and translational upregulation with a dominance of the latter while the more intensive synthesis of cytochrome c biogenesis proteins mainly reflects an increased level of their mRNAs. In contrast, for some complex I subunits (NAD3 and NAD9) a negative correlation between mRNA level and TE is observed; the decreased TE overrides the accumulation of transcripts leading to a reduced level of these proteins. The present genome-wide examination shows that our previous report based on an analysis of several genes only oversimplified the effects of RPS10 downregulation on the expression of the mitochondrial genome (24). We previously claimed that the altered mitochondrial translation in rps10 is associated with a hyperaccumulation of mitochondrial transcripts encoding ribosomal and OXPHOS proteins. In the light of the current findings this conclusion still holds for the transcripts encoding ribosomal proteins and subunits of complex I, but not for the transcripts encoding subunits of complex V which, with the exception of the atp6-2 transcript, are actually less abundant in the mutant.
Given that the altered population of mitoribosomes deficient in S10 leads to reprogramming of mitochondrial gene expression by induction of changes at the transcript level and by gene-specific modulation of TE, we asked what specific characteristics of the rps10 mutant are responsible for these phenomena. We found a number of features, such as shorter fragments of mRNA protected by the mitoribosome, weaker 3-nt periodicity, accumulation of unspliced transcripts and increased production of mtRibo-seq-sRNAs by the footprinting procedure.
The length distribution of the ribosome-protected fragments was different in the wild-type and rps10. Reads of 28 nt dominated in the wild-type, while in the mutant reads of 27 and 28 nt were almost equally abundant. Also the shorter reads (25 and 26 nt) accumulate in rps10 compared with the wild-type. It should be kept in mind that the population of mitoribosomes in rps10 is heterogeneous and correctly assembled mitoribosomes coexist with those with partially assembled small subunit and with an excess of free large subunits (24). Despite the rps10 silencing the steady-state level of the S10 protein is nearly the same in the mutant and wild-type mitochondria, suggesting that the rps10 mutant does not suffer from an absolute insufficiency of functional mitoribosomes. However, defective complexes are also formed as indicated by an excess of another small ribosomal subunit component, S4, over the S10 protein (24). It is conceivable that the shorter ribosome-protected mRNA fragments in the rps10 mutant reflect distinct conformations of a fraction of the mitoribosomes due to their altered biogenesis. This conclusion is in agreement with reports showing that distinct ribosome conformations lead to different mRNA footprint lengths (40).
Both the wild-type and the rps10-specific population of mitoribosomes are engaged in active translation because the fraction of the 25–29 nt-long RPFs aligning with the correct reading frame of protein-CDS is larger than for the alternative frames. However, the fraction of correctly mapping footprints is significantly smaller in rps10 and the 3-nt periodicity is generally weaker in the mutant than in the wild-type. In this respect, the intron-containing genes represent an extreme because they show no significant difference between the fraction of footprints in the correct and the alternative frames in the rps10 mutant. The triplet periodicity reflects the specific stepwise movement of the ribosome associated with translation of genetic information. Given that the small ribosomal subunit is responsible for the decoding, the weaker periodicity observed in the mutant with an altered biogenesis of this subunit could be expected. The S10 protein is located in the head region of the small subunit in close proximity to the entrance site of mRNA and the decoding center (41). In this context it is of utmost interest that the bacterial S10 protein is involved in the binding of tRNA to the ribosomal P-site (42) and the translational frame is controlled mainly by the stability of the codon-anticodon interactions at the P-site (43). It is conceivable that a deficiency of S10 introduces structural distortions at the P-site and thereby affects the fidelity of the codon–anticodon interactions. This structural instability may lead to more frequent dissociation of tRNA from its codon at the P-site and promote interactions with out-of-frame transcript regions. Thus, the weaker 3-nt periodicity in rps10 could be associated with destabilization of the codon-anticodon contact at the P-site and consequent incorrect reading of transcripts. Apparently the frameshifting occurs already on translation initiation because the disturbance of the periodicity affects to a similar extent all exons, including the first one.
One of the most intriguing findings of this study is the strong negative impact of a deficit of the ribosomal S10 protein on the efficiency of mitochondrial splicing. The proportion of unspliced to spliced transcripts is clearly increased in the rps10 mutant, but the presence of mature correctly spliced transcripts indicates that the inhibition of splicing is only partial. The effect is general as it is observed for almost all genes having introns, except for intron 1 in nad1, and introns 2 and 3 in nad5. It seems that the efficiency of removal of these introns in the mutant is comparable to or even more efficient than in the wild-type since less unspliced transcripts are produced. Notably, these introns belong to the trans-spliced type, but the reason why they are exceptional is currently unknown. The association of both unspliced and spliced mRNAs with ribosomes in plant mitochondria has been reported earlier, suggesting that mitochondrial transcripts still containing introns could undergo translation (44). In fact, our mtRibo-seq data imply that mitoribosomes do synthesize polypeptides across exon–intron junctions and this trend is particularly strong in the mutant. Translation of unspliced RNAs could, in principle, generate nonfunctional or truncated proteins. Our attempts to confirm the presence of such aberrant proteins in rps10 using a targeted mass spectrometry-based approach, Multiple Reaction Monitoring, have unfortunately ended in failure.
It is currently impossible to precisely deduce how the silencing of RPS10 expression leads to the impairment of mitochondrial splicing but studies in yeast have revealed that mitoribosomes occur in higher-order complexes called MIOREX, which could couple protein synthesis with mRNA maturation processes including splicing (45). It is possible that the splicing repression is connected with the bifunctionality of PPR proteins which are components of mitoribosomes or interact with them and, like MTL1, are involved in both translation and splicing (15). We posit that the S10-depleted mitoribosomes could aberrantly interact with the bifunctional PPRs thereby preventing their participation in intron removal. Another possibility, not mutually exclusive with the above one, is that unproductive movement of mitoribosomes into introns inhibits splicing by disrupting the folded structure of the RNA, or that ribosomes lacking S10 protein are unable to act efficiently as RNA chaperones to facilitate proper excision of introns in accordance with an earlier study (46). While we prefer the hypothesis that the defect in splicing is associated with the alteration in the mitoribosomes due to the deficiency of S10, one cannot exclude an alternative explanation, namely that the splicing disorder stems from an additional, non-ribosomal function of the S10 protein. Such a function has been reported for the S10 homologue in E. coli, where it participates in transcription antitermination (47). In the context of a possible non-ribosomal function of the Arabidopsis S10 protein of a particular interest is our finding concerning the activation of intergenic processing of the dicistronic transcript rpl5-cob in rps10. The S10 protein could form a complex with RNA polymerase preventing transcription termination at otherwise functional terminators. Then, an S10 deficit in the rps10 mutant would weaker that mechanism, causing generation of monocistronic rpl5 transcripts. The identification of a mtRibo-seq-sRNA representing a putative footprint of a PPR protein defining the 3′terminus of rpl5 transcript led us to an alternative hypothesis. The S10 protein, likely with other regulatory proteins, could stabilize the dicistronic transcript while its deficiency would stimulate exonucleolytic degradation up to the position of the PPR protein bound to the intergenic region, resulting in the accumulation of monocistronic rpl5 transcripts.
Ample data document the involvement of RNA-binding proteins like PPR in post-transcriptional processes in plant mitochondria (11–13). One should bear in mind that the so-called ‘ribosome profiling’ is a footprinting assay that in fact detects both the ribosome-RNA and non-ribosomal protein–RNA complexes (48,49). Our results suggest that a considerable number of mtRibo-seq reads from untranslated regions, present in large quantities in the rps10 mutant, were in fact footprints of RNA binding proteins (mtRibo-seq-sRNAs). Furthermore, we showed that some of those mtRibo-seq-sRNAs resembled cosRNAs—short RNA fragments originating from mitochondrial mRNAs as a result of the protective action of PPR proteins against endogenous ribonucleases, i.e. footprints of PPR proteins (18). Our data suggest that mtRibo-seq-sRNAs mapping to 3′ UTRs, like the matching cosRNAs, are remnants of PPR protein targets but in contrast to cosRNAs they are not generated in vivo but are produced by the ribonuclease added during the ribosomal profiling procedure. The shorter length of the mtRibo-seq-sRNAs compared with the corresponding cosRNAs is likely due to the catalytic differences between the ribonucleases present in plant mitochondria and that used in ribosomal profiling. This makes our data useful in precise determination of the binding sites of the PPRs.
Taken together, our study shows that plant mitoribosomes do not synthesize proteins non-selectively but execute translational control. In contrast to the wild-type, where the OXPHOS transcripts show the highest TE, the S10-deficient mitoribosomes translate different transcripts with a more uniform efficiency. It appears that the mutant mitoribosomes are not only less selective toward individual mRNA species, but also less accurate because they exhibit a weaker 3-nt periodicity compared with the wild-type ones. Our data also indicate that a proper level of S10 protein is necessary not only for mitoribosome biogenesis but also for efficient splicing in mitochondria. Otherwise unspliced transcripts accumulate and compensatory changes at the transcript and protein levels are provoked to increase production of ribosomal proteins and maturase. Moreover, the level of S10 protein controls intergenic cleavage of the dicistronic rpl5-cob transcript. Thus, it is likely that the plant S10 also has an extraribosomal function(s). Further studies will be needed to verify this assumption.
DATA AVAILABILITY
The datasets supporting the conclusions of this article are included within the article and its Supplementary Files. Raw and processed mtRibo-seq and mtRNA-seq data have been deposited at NCBI Gene Expression Omnibus under the accession number GSE132441.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Bartosz Wojtas (Laboratory of Molecular Neurobiology, Neurobiology Center, Nencki Institute of Experimental Biology, Polish Academy of Sciences, Warsaw, Poland) for participation in bioinformatic analyses. We are very grateful to Fabio Lauria (Institute of Biophysics, CNR Unit at Trento, Trento, Italy), Lorenzo Calviello (Berlin Institute for Medical Systems Biology, Max Delbrück Center for Molecular Medicine, Berlin, Germany) and Hannes Ruwe (Molecular Genetics Group, Institute of Biology, Faculty of Life Sciences, Humboldt University of Berlin, Berlin, Germany) for fruitful discussion and explanations concerning their software. We are also very grateful to Hakim Mireau (Institut Jean-Pierre Bourgin INRA, AgroParisTech, CNRS, Université Paris-Saclay, Versailles, France) for his helpful comments.
Notes
Present address: Hanna Janska, Department of Cellular Molecular Biology, Faculty of Biotechnology, University of Wroclaw, F. Joliot-Curie 14A, 50-383 Wroclaw, Poland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Centre, Poland [2014/15/B/NZ2/01065 to H.J.]. Funding for open access charge: National Science Centre, Poland [2014/15/B/NZ2/01065].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hammani K., Giegé P.. RNA metabolism in plant mitochondria. Trends Plant Sci. 2014; 19:380–389. [DOI] [PubMed] [Google Scholar]
- 2. Waltz F., Nguyen T.T., Arrivé M., Bochler A., Chicher J., Hammann P., Kuhn L., Quadrado M., Mireau H., Hashem Y., Giegé P.. Small is big in Arabidopsis mitochondrial ribosome. Nat. Plants. 2019; 5:106–117. [DOI] [PubMed] [Google Scholar]
- 3. Robles P., Quesada V.. Emerging roles of mitochondrial ribosomal proteins in plant development. Int. J. Mol. Sci. 2017; 18:2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Delage L., Giegé P., Sakamoto M., Maréchal-Drouard L.. Four paralogues of RPL12 are differentially associated to ribosome in plant mitochondria. Biochimie. 2007; 89:658–668. [DOI] [PubMed] [Google Scholar]
- 5. Budar F., Mireau H.. Logan DC. The cross‐talk between genomes: How co‐evolution shaped plant mitochondrial gene expression. Annual Plant Reviews. 2018; 50:Chichester: John Wiley & Sons, Ltd; 33–66. [Google Scholar]
- 6. Zmudjak M., Ostersetzer-Biran O.. Logan DC. RNA Metabolism and Transcript Regulation. Annual Plant Reviews. 2018; 50:Chichester: John Wiley & Sons, Ltd; 143–184. [Google Scholar]
- 7. Forner J., Weber B., Thuss S., Wildum S., Binder S.. Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res. 2007; 35:3676–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hazle T., Bonen L.. Comparative analysis of sequences preceding Protein-Coding mitochondrial genes in flowering plants. Mol. Biol. Evol. 2007; 24:1101–1112. [DOI] [PubMed] [Google Scholar]
- 9. Kazama T., Yagi Y., Toriyama K., Nakamura T.. Heterogeneity of the 5′-end in plant mRNA may be involved in mitochondrial translation. Front. Plant Sci. 2013; 4:517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Planchard N., Bertin P., Quadrado M., Dargel-Graffin C., Hatin I., Namy O., Mireau H.. The translational landscape of Arabidopsis mitochondria. Nucleic Acids Res. 2018; 46:6218–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmitz-Linneweber C., Small I.. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008; 13:663–670. [DOI] [PubMed] [Google Scholar]
- 12. Barkan A., Small I.. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014; 65:415–442. [DOI] [PubMed] [Google Scholar]
- 13. Manna S. An overview of pentatricopeptide repeat proteins and their applications. Biochimie. 2015; 113:93–99. [DOI] [PubMed] [Google Scholar]
- 14. Manavski N., Guyon V., Meurer J., Wienand U., Brettschneider R.. An essential pentatricopeptide repeat protein facilitates 5′ maturation and translation initiation of rps3 mRNA in maize mitochondria. Plant Cell. 2012; 24:3087–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haïli N., Planchard N., Arnal N., Quadrado M., Vrielynck N., Dahan J., des Francs-Small C.C., Mireau H.. The MTL1 pentatricopeptide repeat protein is required for both translation and splicing of the mitochondrial NADH dehydrogenase subunit 7 mRNA in Arabidopsis. Plant Physiol. 2016; 170:354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ruwe H., Schmitz-Linneweber C.. Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins. Nucleic Acids Res. 2012; 40:3106–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhelyazkova P., Sharma C.M., Forstner K.U., Liere K., Vogel J., Borner T.. The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell. 2012; 24:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruwe H., Wang G., Gusewski S., Schmitz-Linneweber C.. Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms. Nucleic Acids Res. 2016; 44:7406–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haïli N., Arnal N., Quadrado M., Amiar S., Tcherkez G., Dahan J., Briozzo P., Colas des Francs-Small C., Vrielynck N., Mireau H.. The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res. 2013; 41:6650–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee K., Han J.H., Park Y.-I., Colas des Francs-Small C., Small I., Kang H.. The mitochondrial pentatricopeptide repeat protein PPR19 is involved in the stabilization of NADH dehydrogenase 1 transcripts and is crucial for mitochondrial function and Arabidopsis thaliana development. New Phytol. 2017; 215:202–216. [DOI] [PubMed] [Google Scholar]
- 21. Wang C., Aubé F., Planchard N., Quadrado M., Dargel-Graffin C., Nogué F., Mireau H.. The pentatricopeptide repeat protein MTSF2 stabilizes a nad1 precursor transcript and defines the 3′ end of its 5′-half intron. Nucleic Acids Res. 2017; 45:6119–6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hauler A., Jonietz C., Stoll B., Stoll K., Braun H.-P., Binder S.. RNA processing factor 5 is required for efficient 5′ cleavage at a processing site conserved in RNAs of three different mitochondrial genes in Arabidopsis thaliana. Plant J. 2013; 74:593–604. [DOI] [PubMed] [Google Scholar]
- 23. Binder S., Stoll K., Stoll B.. Maturation of 5′ ends of plant mitochondrial RNAs. Physiol. Plant. 2016; 157:280–288. [DOI] [PubMed] [Google Scholar]
- 24. Kwasniak M., Majewski P., Skibior R., Adamowicz A., Czarna M., Sliwinska E., Janska H.. Silencing of the nuclear RPS10 gene encoding mitochondrial ribosomal protein alters translation in arabidopsis mitochondria. Plant Cell. 2013; 25:1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Majewski P., Wołoszyńska M., Jańska H.. Developmentally early and late onset of Rps10 silencing in Arabidopsis thaliana: genetic and environmental regulation. J. Exp. Bot. 2009; 60:1163–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lister R., Carrie C., Duncan O., Ho L.H.M., Howell K.A., Murcha M.W., Whelan J.. Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell. 2007; 19:3739–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lauria F., Tebaldi T., Bernabò P., Groen E.J.N., Gillingwater T.H., Viero G.. riboWaltz: optimization of ribosome P-site positioning in ribosome profiling data. PLoS Comput. Biol. 2018; 14:e1006169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Calviello L., Mukherjee N., Wyler E., Zauber H., Hirsekorn A., Selbach M., Landthaler M., Obermayer B., Ohler U.. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods. 2016; 13:165–170. [DOI] [PubMed] [Google Scholar]
- 31. Pall G.S., Hamilton A.J.. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 2008; 3:1077–1084. [DOI] [PubMed] [Google Scholar]
- 32. Keren I., Abudraham S., Shaya F., Ostersetzer-Biran O.. An optimized method for the analysis of plant mitochondria RNAs by Northern-blotting. J. Endocyt. Cell Res. 2011; 21:34–42. [Google Scholar]
- 33. Chotewutmontri P., Barkan A.. Dynamics of chloroplast translation during chloroplast differentiation in maize. PLos Genet. 2016; 12:e1006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Michalovova M., Vyskot B., Kejnovsky E.. Analysis of plastid and mitochondrial DNA insertions in the nucleus (NUPTs and NUMTs) of six plant species: size, relative age and chromosomal localization. Heredity (Edinb). 2013; 111:314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang D., Qu Z., Adelson D.L., Zhu J.-K., Timmis J.N.. Transcription of nuclear organellar DNA in a model plant system. Genome Biol. Evol. 2014; 6:1327–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vincent T., Vingadassalon A., Ubrig E., Azeredo K., Srour O., Cognat V., Graindorge S., Salinas T., Maréchal-Drouard L., Duchêne A.-M.. A genome-scale analysis of mRNAs targeting to plant mitochondria: upstream AUGs in 5′ untranslated regions reduce mitochondrial association. Plant J. 2017; 92:1132–1142. [DOI] [PubMed] [Google Scholar]
- 37. Rooijers K., Loayza-Puch F., Nijtmans L.G., Agami R.. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat. Commun. 2013; 4:2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raczynska K.D., Le Ret M., Rurek M., Bonnard G., Augustyniak H., Gualberto J.M.. Plant mitochondrial genes can be expressed from mRNAs lacking stop codons. FEBS Lett. 2006; 580:5641–5646. [DOI] [PubMed] [Google Scholar]
- 39. Zoschke R., Bock R.. Chloroplast translation: structural and functional organization, operational control, and regulation. Plant Cell. 2018; 30:745–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lareau L.F., Hite D.H., Hogan G.J., Brown P.O.. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife. 2014; 3:e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Burmann B.M., Schweimer K., Luo X., Wahl M.C., Stitt B.L., Gottesman M.E., Rösch P.. A NusE:NusG complex links transcription and translation. Science. 2010; 328:501–504. [DOI] [PubMed] [Google Scholar]
- 42. Riehl N., Remy P., Ebel J.-P., Ehresmann B.. Crosslinking of N-Acetyl-phenylalanyl [s4U]tRNAPhe to Protein S10 in the Ribosomal P Site. Eur. J. Biochem. 2005; 128:427–433. [DOI] [PubMed] [Google Scholar]
- 43. Hansen T.M., Baranov P.V, Ivanov I.P., Gesteland R.F., Atkins J.F.. Maintenance of the correct open reading frame by the ribosome. EMBO Rep. 2003; 4:499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang A.J., Mulligan R.M.. Distribution of maize mitochondrial transcripts in polysomal RNA: evidence for non-selectivity in recruitment of mRNAs. Curr. Genet. 1993; 23:532–536. [DOI] [PubMed] [Google Scholar]
- 45. Kehrein K., Schilling R., Möller-Hergt B.V., Wurm C.A., Jakobs S., Lamkemeyer T., Langer T., Ott M.. Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Rep. 2015; 10:843–853. [DOI] [PubMed] [Google Scholar]
- 46. Semrad K., Schroeder R.. A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo. Genes Dev. 1998; 12:1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Luo X., Hsiao H.-H., Bubunenko M., Weber G., Court D.L., Gottesman M.E., Urlaub H., Wahl M.C.. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol. Cell. 2008; 32:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ji Z., Song R., Regev A., Struhl K.. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015; 4:e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ji Z. Rfoot: transcriptome-scale identification of RNA-protein complexes from ribosome profiling data. Curr. Protoc. Mol. Biol. 2018; 124:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its Supplementary Files. Raw and processed mtRibo-seq and mtRNA-seq data have been deposited at NCBI Gene Expression Omnibus under the accession number GSE132441.