Abstract
Genome-wide association studies (GWASs) have revolutionized the field of complex trait genetics over the past decade, yet for most of the significant genotype-phenotype associations the true causal variants remain unknown. Identifying and interpreting how causal genetic variants confer disease susceptibility is still a big challenge. Herein we introduce a new database, CAUSALdb, to integrate the most comprehensive GWAS summary statistics to date and identify credible sets of potential causal variants using uniformly processed fine-mapping. The database has six major features: it (i) curates 3052 high-quality, fine-mappable GWAS summary statistics across five human super-populations and 2629 unique traits; (ii) estimates causal probabilities of all genetic variants in GWAS significant loci using three state-of-the-art fine-mapping tools; (iii) maps the reported traits to a powerful ontology MeSH, making it simple for users to browse studies on the trait tree; (iv) incorporates highly interactive Manhattan and LocusZoom-like plots to allow visualization of credible sets in a single web page more efficiently; (v) enables online comparison of causal relations on variant-, gene- and trait-levels among studies with different sample sizes or populations and (vi) offers comprehensive variant annotations by integrating massive base-wise and allele-specific functional annotations. CAUSALdb is freely available at http://mulinlab.org/causaldb.
INTRODUCTION
From the first genome-wide association study (GWAS) on macular degeneration in 2005 (1) to the present, thousands of GWASs have been conducted to explore diverse quantitative traits and complex diseases, identifying numerous significant associations between genotypes and phenotypes. In particular, with the introduction of methodologies such as imputation (2), meta-analysis (3) and multi-trait test (4), and emergence of projects with a large sample size such as the UK Biobank (UKBB) (5), the number of identified significant trait/disease-associated loci is rapidly accumulating, covering most regions of the human genome. Because there are many restrictions on accessing the genotype and phenotype data of individuals, publicly available GWAS summary statistics together with derived statistical methods are of a great value to GWAS downstream studies and for applications such as potential causal variant fine-mapping and validation, causal relationship inference among traits, polygenic risk prediction, as well as drug target discovery (6,7).
Although some catalogs of GWAS, such as the NHGRI-EBI GWAS Catalog (8), GWASdb (9) and GRASP (10), have been curating statistically significant variants for years, most of these resources only focus on the reported GWAS signals in each publication, particularly those with the most significant P-value at the associated locus. However, given the complexity of the linkage disequilibrium (LD) structure in the investigated GWAS cohort and statistical fluctuations of association analysis, several GWAS leading variants may not necessarily be trait/disease causal variants (11). Therefore, previous curation projects inevitably missed many causal signals and only provided marker information on genetic associations. Fortunately, recent GWAS communities advocate the public release of GWAS summary statistics, and some pertinent data have been archived in existing databases including LD Hub (12), GWAS Catalog (8), PhenoScanner (13), MR-Base (14), Gene ATLAS (15) and GWAS ATLAS (16). Nevertheless, these resources have only collected limited or specific GWAS summary statistics to date and were not particularly designed to prioritize causal variants. However, statistical fine-mapping technologies were developed to identify underlying causality from GWAS summary information (17). Although some recent GWASs reported their associated signals along with the fine-mapping results, majority of existing GWASs did not point out potential causal variants in each significant locus. At present, there is no resource that follows a consistent procedure to systematically fine-map potential trait/disease causal variants at the genome-wide level. Moreover, online manipulation of genome-wide summary statistics involves intensive network data transmission load, and it is laborious for researchers to deploy fine-mapping pipeline on their own. Despite several web-based visualization tools, including LocusExplorer (18) and LocusZoom.js (19), that attempt to provide researchers with options for displaying potential causal variants, an online resource that can efficiently help visualize and operate genome-wide summary statistics and elucidate underlying causal signatures is still lacking. Finally, statistical fine-mapping usually fails to distinguish a true causal variant from extremely high LD (20); therefore, the integration of fine-scale functional annotation information is required to further prioritize fine-mapped variants.
To address the aforementioned issues, we herein developed a database called CAUSALdb, in which we curated and collected the majority of the published GWASs having complete summary statistics and performed statistical fine-mapping using three commonly used tools. CAUSALdb allows users to explore causal signatures across studies on variant-, gene- and trait-levels. By integrating comprehensive functional annotation resources, we constructed a highly interactive viewer to visualize and annotate potential causal variants for each trait/disease. CAUSALdb is free and open access: http://mulinlab.org/causaldb or http://mulinlab.tmu.edu.cn/causaldb.
MATERIALS AND METHODS
GWAS summary statistics curation and integration
We collected publicly available GWAS summary statistics from two major sources according to the investigated cohorts: UKBB and non-UKBB cohorts. The latter includes samples from other specific projects (including meta-analysis, which combines the UKBB cohort). GWAS summary statistics of the UKBB cohort were collected from three resources: Neale Lab UKBB v3 (http://www.nealelab.is/uk-biobank), Gene ATLAS (15) and GWAS ATLAS (16). Although they are all derived from the UKBB cohort, the incorporated samples, quality control (QC) processes, and association models are different. Consequently, the summary statistics among these datasets could be distinct (Supplementary Table S1). For Neale Lab's release data containing over 10 000 tests, to exclude low power results, we only included ICD10 binary traits with total sample size of >50 000 and number of cases >1000, and selected continuous traits with total sample size >50 000 tested by PHESANT (21). Besides, we integrated GWAS summary statistics of non-UKBB cohorts from several public databases, including GWAS Catalog (8), LD Hub (12), GRASP (10), PhenoScanner (13) and dbGaP (22). We also curated hundreds of summary statistics from websites of consortiums such as PGC (https://www.med.unc.edu/pgc), MAGIC (https://www.magicinvestigators.org), SSGAC (https://www.thessgac.org), and JENGER (http://jenger.riken.jp/en/). We only included studies for which the original publication was available and if population-related information and sample size were clearly recorded. To remove duplicate GWAS summary statistics among these resources, we identified redundancy by publication source and only retained the one with the most information. We extracted the sample size, population, and source information across these databases and the original study. GWAS population information was mapped to five super-populations (AFR, AMR, EAS, EUR and SAS) in the 1000 Genomes Project (1KGP). To ensure accurate fine-mapping using the 1KGP LD information, we did not include GWASs conducted on mixed populations.
Ontology mapping
We manually mapped the reported traits of included GWASs to Medical Subject Headings (MeSH) (23). To ensure the accuracy of trait mapping, we accounted for some auxiliary information within original studies and descriptions using MeSH terms. For UKBB cohort traits, we considered the descriptions in ICD10 (https://icd.who.int/browse10/2016/en) and related notes on UKBB Showcase. We also sought suggestions from the search function of MeSH (https://meshb.nlm.nih.gov/search). For example, let us consider the ICD10 term ‘insulin-dependent diabetes mellitus’ in UKBB; we searched this term in the MeSH Browser and got a match for ‘diabetes mellitus, type 1.’ For non-UKBB cohort traits, we took into account the Abstract and list of suggested MeSH terms on PubMed. For example, the reported trait of a recent GWAS was ‘primary sclerosing cholangitis’ (24); ‘cholangitis, sclerosing’ was determined to be the relevant MeSH term on PubMed. Therefore, an exact match could be determined. CAUSALdb presents a tree of traits that has the same architecture as Tree View in the MeSH Browser (https://meshb.nlm.nih.gov/treeView).
Pre-fine-mapping QC and summary statistics standardization
GWAS fine-mapping methods based on summary information usually require complete association statistics [such as variant coordinate, minor allele frequency (MAF), effect/non-effect allele, P, effect size (beta-coefficient; BETA), and standard error (SE)] and LD information on variants. To ensure that these curated GWAS summary statistics fit the input requirements of fine-mapping tools, we performed a series of QC steps on the raw downloaded data. First, we inspected the coordinates and dbSNP ID (rsID) for each variant and converted non-GRCh37 coordinates to GRCh37 (hg19) coordinates. When either the coordinate or rsID was missing, we extracted it from dbSNP 151. The statistics were excluded when both the coordinates and rsID were missing. Second, CAUSALdb only curated summary statistics with an explicitly defined effect allele. When only the effect allele was available, the non-effect allele was inferred from 1KGP biallelic sites, and we excluded variants if the non-effect allele could not be clearly determined. Third, MAF is required by certain fine-mapping tools, but it was at times unavailable in the raw data. In such cases, we converted other allele frequencies (such as reference allele frequency or effect allele frequency) to MAF or estimated it from matched 1KGP populations. Fourth, we discarded summary statistics that did not have P-value and effect size [BETA or odds ratio (OR)] for test variants. In some cases for which standard error (SE) of BETA or confidence intervals of OR was missing but effect size was available, we calculated SE using effect size, P, and sample size using the quantile function. In addition, if INFO metric of imputation was available in the raw data, variants with INFO <0.9 were filtered out. Thus, for all summary statistics in the Neale Lab UKBB cohort, we excluded variants with INFO <0.9.
GWAS fine-mapping
LD block partition
To perform fine-mapping on curated GWAS summary statistics, we partitioned the genetic variants with relatively independent LD blocks estimated using LDetect (25). We checked each file and extracted the variants in LD blocks (termed causal blocks) that had at least one genome-wide significant variant (P-value ≤ 5E−8). For studies without any genome-wide significant variants (P-value ≤ 5E−8), we only selected the LD block in which the variant with the lowest genome-wide P-value located as the potentially causal block for each trait. The GWAS summary information for each causal block was then reformatted into the format required by the fine-mapping tools.
LD estimation
We estimated the LD information of GWAS variants in each LD block using five super-populations (AFR, AMR, EAS, EUR and SAS) from the 1KGP reference panel. Since LDetect only contains LD block information for three continental populations, we assigned the five 1KGP super-populations to them (mapping EUR and AMR to European population, EAS and SAS to Asian population, and AFR to African population). For each 1KGP super-population, we only retained biallelic variants and discarded sites with MAF = 0. To accelerate the process, we further divided the VCF genotype file into LD block-wise files. We mapped test variants to corresponding variants in the reference panel according to identical coordinates and alleles, and harmonized complementary alleles for reverse strands. We used the PAINTOR (26) framework to calculate pairwise Pearson correlation coefficients between each variant in the LD block and block-wise LD matrix.
Fine-mapping
We performed fine-mapping based on summary statistics and matched LD matrix for each causal block of each trait using three commonly used tools, namely FINEMAP (27), PAINTOR (26) and CAVIARBF (28) (Supplementary Table S2). We assumed that there was only one causal variant in a causal block and used the recommended parameters of the tools. These fine-mapping tools can report the posterior probability (PP) of each variant being causal in the specific model. A credible set is the set of variants with a sum of PP of more than α, which means considering the cumulative sum of PPs from the largest to smallest until it is not smaller than α. In CAUSALdb, we reported potential causal variants within the credible set upon the adjustment of α. The code for reproducing the CAUSALdb GWAS fine-mapping procedure can be found at https://github.com/mulinlab/CAUSALdb-finemapping-pip.
Post-fine-mapping QC
Because some human genetic variants in 1KGP are not complete, there are variants without any LD information in some GWASs, which may markedly affect causal variant estimation in the process of fine-mapping. Thus, to avoid overestimation of causality for such variants, we excluded them (P-value > 5E−5) from the credible set, but still allowed users to inspect the original summary statistics. Also, for variants without LD information, we set the default PP value to −1. Since fine-mapping results may be inconsistent among three applied tools, we used rank product value to combine PP ranks for each credible set variant and prioritize the potential causal variants.
Variant-level potential pleiotropy estimation
We selected the most representative GWAS which contains maximum causal blocks for each MeSH term. Then we inspected the causal blocks in GWAS pairs and calculated the PP of a variant influencing both traits (potential pleiotropy) using gwas-pw (model 3) (29). Since the overlapping samples or comorbid samples were largely unreported accompanying released GWAS summary statistics, we empirically set the expected correlation (-cor in gwas-pw) between two traits as described in a recent simulation study (30). For the UKBB cohort studies we were certain that they contain overlapping samples and incorporate similar categories of traits, we varied the expected correlation of 0.09, 0.18, 0.27, 0.36, 0.45 according to the hierarchy of MeSH tree. For example, the tree numbers of Heart Failure and Atrial Fibrillation are C14.280.434 and C14.280.067.198 respectively which all belong to C14.280 parent node of Heart Diseases. We hence classified the relationship between these two diseases into Level 2, so the expected correlation in summary statistics between Heart Failure and Atrial Fibrillation was set to 0.18. For non-UKBB cohort studies which we cannot ascertain overlapping samples, the expected correlation was uniformly set to 0.
Functional annotation
Functional annotations integrated into CAUSALdb can be divided into four major categories according to the attributes and usage of the collected datasets: variant information, functional prediction, functional evidence, and trait association. Variant information annotations report basic information, including variant genomic features derived from dbSNP and CADD (31) and variant allele frequency derived from gnomAD (32) and 1KGP phase 3 (33). Functional prediction annotations incorporate several integrative variant function prediction results from frequently used resources, including aggregated conservation scores from CADD, aggregated non-coding variant prediction scores from regBase (34), aggregated missense mutation pathogenic scores from dbNSFP (35), aggregated splicing altering prediction scores from dbscSNV (36), aggregated miRNA-target altering prediction scores from dbMTS, and several function predictions from HaploReg (37), RegulomeDB (38) and InterVar (39). Functional evidence annotations integrate large-scale tissue/cell type-specific epigenomic profiling (e.g. histone modifications, transcription factor binding, open chromatin and nascent transcription) data from different resources, such as the Roadmap Epigenomics Project (40), CistromeDB (41) and FANTOM5 Project (42). Trait association annotations collect important disease/trait-associated information, including GTEx eQTLs (43), GWAS Catalog significant variants, ClinVar reported variants (44), DisGeNET recorded variants (45), and ICGC somatic mutation information (46) (Supplementary Table S3).
Database design
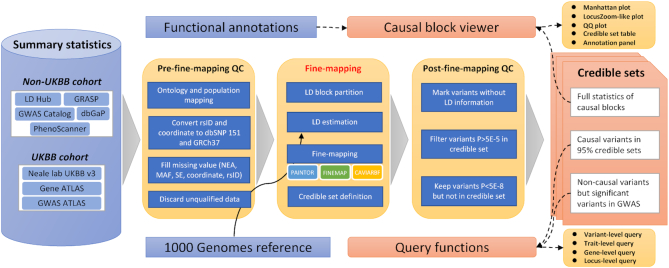
CAUSALdb was established using a JAVA-based web framework. We stored the partitioned summary statistics and fine-mapping results of causal blocks in flat files. The information pertaining to curated GWASs and potential causal variants was stored in MySQL for quick retrieval. The annotation information was indexed and stored using MySQL or Tabix (47). We generated highly interactive Manhattan and LocusZoom-like plots for users to inspect causal blocks using jQuery, D3.js, and related JavaScript modules. The overall architecture of CAUSALdb is shown in Figure 1.
Figure 1.
Data processing workflow and overall architecture of CAUSALdb.
RESULTS
CAUSALdb statistics
We started with the collection and curation of GWAS summary statistics from various resources and publications (details in Materials and Methods). After pre-fine-mapping QC, up to the latest update in July 2019, CAUSALdb curated 3052 fine-mappable GWAS summary statistics in total: 1237 belonged to non-UKBB cohorts and 1815 belonged to the UKBB cohort. In total, 2629 unique traits were identified that could be mapped to 855 MeSH terms. According to the ontology mapping, around two-thirds of the studies were based on common diseases such as cardiovascular diseases and neoplasms, while the remainder focused on quantitative traits of human phenotypes (Supplementary Figure S1). In the non-UKBB cohort data, 92.07% of studies were based on the EUR population, 6.91% on EAS population, and only seven, six, and two studies were based on AMR, AFR and SAS populations, respectively (Supplementary Figure S2), which indicates an unequal ancestry composition in the current GWASs. The average sample size in the non-UKBB cohort studies was 43 516, with a meta-analysis of atrial fibrillation (48) having the largest sample size (1 030 836). In the UKBB cohort data, we incorporated summary statistics from three independent sources; the collected data therefore included varied sample sizes and distinct summary statistics (Supplementary Table S1).
We performed systematic fine-mapping using three commonly used tools and observed highly concordant results in identifying credible set variants in each causal block (Supplementary Figure S3). Among all identified causal blocks in the CAUSALdb, only five of them show relatively low correlation between FINEMAP and other two tools (Spearman's Rank correlation coefficient <0.8, Supplementary Figure S3A and Table S4), which may due to the Shotgun Stochastic Search algorithm used in FINEMAP. Also, the credible set size of each causal block is largely similar across fine-mapping tools, especially between CAVIARBF and FINEMAP, probably due to they used similar statistical models (Supplementary Figure S3B). By pooling fine-mapped variants in the 95% credible set (α = 0.95), we built a dataset composed of 962 176 potential causal variants corresponding to 5 097 732 genotype–phenotype associations across the entire human genome (Supplementary Figure S4). The genomic distribution of these variants showed that 98.9% of them were located in non-coding genomic regions (Supplementary Figure S5), emphasizing the pivotal role of regulatory variants in the development of complex traits. Among these variants, ∼55% were identified by more than one study, which implies that shared genetic effects among traits could be very common. Notably, many test variants with genome-wide significance (P-value ≤ 5E−8) were not present in the credible set, which included 1 339 760 unique variants and 11 528 369 genotype-phenotype associations, demonstrating that fine-mapping can greatly narrow down potential causal hits. In 1703 relatively independent LD blocks of EUR population defined by LDetect (25), 1699 blocks were noted to contain potential causal variants. Among these blocks, 99.88% contained potential causal variants across multiple studies; 6p21.32 (HLA locus) was the top causal block, associated with 514 studies and 176 traits. To investigate the potential pleiotropic variants in CAUSALdb, we performed joint analysis in GWAS pairs using gwas-pw (29). We in total identified 24 286 potential variant-level pleiotropy (PP > 0.5 in gwas-pw model 3) for 219 880 GWAS pairs. Although this estimation cannot further distinguish vertical or horizontal pleiotropy, it may help users cautiously interpret the potential causal variants in the context of shared genetic effect.
Database usage and interface
Query entries
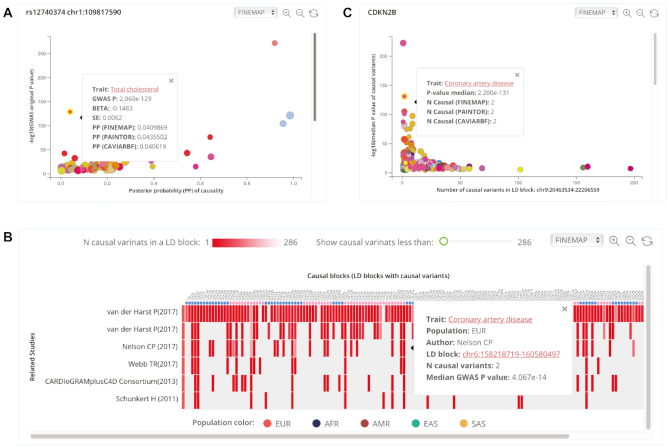
CAUSALdb allows users to explore variant causality across studies through querying variants, traits, genes, or chromosome loci of their choice. The general query result displays all matched GWASs and related study summaries, including trait name, sample size, population, number of cases/controls, number of variants with summary statistics, publication information, source link, mapped MeSH terms, as well as our QC notes (Supplementary Figure S6). In terms of specific functions, by searching for either rsID or variant chromosome position, users can visualize the PP of the causality versus the original GWAS P-value in a scatter plot (Figure 2A). The dot size in the plot represents the study sample size and the dot color represents the mapped MeSH term. Obviously, variants on the upper right corner are more likely to be causal. Users can inspect the summary table on the right of the scatter plot and switch the fine-mapping tools via a drop-down box. The link on the hover tip can directly guide users to the causal block viewer of a particular study. As fine-mapping would have narrowed down the significant variants to a smaller set, users sometimes may get no results when searching for rsID. We have included an additional query function for users to browse variants with genome-wide significance by clicking ‘Only show P-value’ in a phenome-wide-like plot (Supplementary Figure S7). In addition, we listed the traits associated with potential pleiotropy of queried variant in this plot. On searching for a trait name, auto-completion should help users select the potential trait from the mapped MeSH terms. In the search result, users can see all causal blocks across the studies related to the searched trait in a heatmap plot (Figure 2B). Each column represents an independent causal block and each row depicts a separate GWAS. The grid color in the heatmap represents the number of potential causal variants in a corresponding causal block. On the hover tips, users can find the median GWAS P-value of the credible set, and on clicking the block genomic position, they can navigate to the corresponding block view page. By searching for a gene name, considering that the average length of LD blocks is 1.6 Mb, the system will locate the target LD block in which the searched gene is present and display a block-wise causality plot (the number of potential causal variants in the LD block versus the median GWAS P-value of these variants) (Figure 2C). By searching for a chromosome region that is <10 Mb, users can investigate the causality of the most relevant LD block overlapped with the input region, and the results are similar to those when searching for a gene. By mapping the reported traits to MeSH, we established an ontology tree for users to browse the profile information of our collected GWASs, which further facilitates the navigation to traits of interest and the related causal block viewer (Supplementary Figure S8).
Figure 2.
Query results from CAUSALdb. (A) Scatter plot of −log10(P-value) and posterior probability for rs12740374. (B) Heatmap plot of the number of potential causal variants in all causal blocks across CAD GWASs. (C) Scatter plot of −log10(median P-value) and the number of potential causal variants for CDKN2B located causal block.
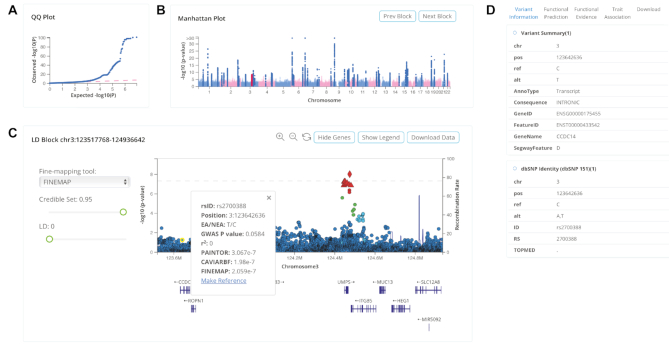
Causal block viewer
To ensure interactive visualization and seamless operation of genome-wide summary statistics in the web environment, we designed an optimized web architecture to reduce intensive network data transmission load and developed dynamic Manhattan and LocusZoom-like plots. We first introduced a causal block viewer that integrates QQ, Manhattan, and LocusZoom-like plots along with a table displaying credible set variants and a variant annotation panel into a single user-friendly web page (Figure 3). Specifically, QQ (Figure 3A) and Manhattan (Figure 3B) plots can be used to evaluate the quality of GWAS globally. By clicking the highlighted strip or ‘Prev Block’ and ‘Next Block’ buttons in the Manhattan plot, users can switch the causal block of interest. All GWAS variants in each causal block can be displayed in the LocusZoom-like plot (Figure 3C), and users can zoom in or zoom out smoothly using the buttons. Moreover, we added some glyphs to signify the credible set and leading variant. Triangles represent the variants in the credible set and the diamond represents the potential causal variant with leading GWAS signal. When the leading variant is not in the credible set, it will be marked with an inverted triangle. Users can click each variant in the plot to check the summary statistics and causality information, and even reset any variant as an LD proxy. By dragging the slider bar, users can adjust the fine-mapping tools, credible set threshold, and LD r2 to filter out variants in the LocusZoom-like plot. The bottom table displays summary statistics information and product rank values on potential causal variants as the change of credible set threshold. To further distinguish a true causal variant from an extremely high LD, users can select potential causal variants in credible set and compare functional prediction scores (e.g. CADD or FATHMM-MKL) or the number of overlapped epigenomic features (e.g. chromatin accessibility or transcription factor binding) in popup bar plots. Importantly, users can download complete summary statistics for the causal block for further analysis by clicking the ‘Download Data’ button. Also, the fine-mapping results of all causal blocks of each GWAS are stored in a compressed file, which can be downloaded in bulk.
Figure 3.
Causal block viewer in CAUSALdb. (A) QQ plot of selected GWAS. (B) Manhattan plot of selected GWAS, with highlighted blocks that are clickable. (C) LocusZoom-like plot of the selected causal block from Manhattan plot. (D) Functional annotation panel of a selected variant in LocusZoom-like plot.
Functional annotations
Statistical fine-mapping usually contains false positives and fails to distinguish a true causal variant from other variants in extremely high LD; therefore, it requires functional annotation information to further prioritize fine-mapped variants. CAUSALdb integrates and compiles over 22 annotations (Supplementary Table S3) from four major categories according to the attributes and usage of collected datasets: variant information, functional prediction, functional evidence, and trait association (details in Materials and Methods). All annotations can be inspected from the right panel of the causal block viewer and are downloadable (Figure 3D).
Application of CAUSALdb to identify potential causal variants
We investigated the reliability and practicality of CAUSALdb using GWAS results for coronary artery disease (CAD). By searching for the trait name ‘Coronary Artery Disease,’ we found six CAD GWAS summary statistics from the EUR population in the current version of CAUSALdb (Supplementary Table S5). Among them, the largest study, involving meta-analysis of the UKBB and CARDIoGRAMplusC4D samples (49), showed the highest number of causal blocks (n = 165). In the original publication, the authors performed GWAS fine-mapping on 161 CAD risk loci using PAINTOR (26). Although they only considered variants having r2 >0.1 with the leading variant and GWAS P-value of <0.01 as independent loci, we found highly consistent PP of potential causal variants between the original results and CAUSALdb (Pearson correlation coefficient: PAINTOR = 0.922, CAVIARBF = 0.920, FINEMAP = 0.921; Supplementary Figure S9A). For example, CAUSALdb recapitulated 100% (8/8) variants with PP equal to 1 and 92% (69/75) variants with PP >0.5. These variants with high PP were very easy to distinguish from others and were more likely to be causal. One of the eight variants with the highest PP, rs11556924, was also supported by four additional CAD GWASs in CAUSALdb, with average PP of 0.955 (Supplementary Figure S9B and Table S6). This missense variant has been reported to disrupt NIPA function and plays a critical role in cell cycle regulation (50).
Distinguishing a true causal variant from an extremely high LD is a challenging task, and fine-mapping usually generates a challenging credible set in which highly linked variants achieve a similar PP of causality. For example, at locus 15p22.3 in the CAD GWAS mentioned before, there were five variants in the credible set, with similar PP of ∼0.2 (Supplementary Figure S9C). CAUSALdb provides base-wise variant annotations and tissue/cell type-specific epigenome data that help researchers determine true causal variants. By inspecting the functional annotations from CAUSALdb, we found that although rs17293632 did not obtain the highest PP, it could be a causal CAD variant at 15p22.3, with substantial supporting evidence. First, according to our aggregated conservation scores, rs17293632 was noted to be more conserved than the other four variants (Supplementary Table S7). Second, this variant obtained significantly higher functional scores than the other variants according to our integrated non-coding variant functional prediction tool, namely regBase (34) (Supplementary Figure S9D). Third, rs17293632 was found to overlap with most epigenomics signals such as open chromatin, histone modification, and transcription factor binding (Supplementary Figure S9E), particularly in CAD-related tissues/cell types such as the endothelium and blood tissue. Finally, we found that rs17293632 was top-ranked in RegulomeDB and identified as the top causal variant in other complex traits/diseases, such as asthma and inflammatory bowel disease; this further supported its causal and potentially pleiotropic effects. Besides, two recent studies reported that this variant disrupts the binding of the AP-1 transcription factor (51,52).
As for quantitative traits, we illustrated the effectiveness of CAUSALdb fine-mapping results by taking body mass index (BMI) as example. In the 31 collected GWASs of BMI for EUR population, genetic locus 16q12.2 was found containing potential causal variants in most studies (29, 93.5%). The credible set variants of these studies at this locus all lie in the first intron of FTO gene. Notably two potential causal variants in perfect LD, rs1558902 and rs1421085, were found in credible sets of 18 and 14 BMI studies, respectively, which are far more than only six studies involving the third one rs9937053, indicating the significance of functional follow-up to dissect their biological mechanism. The variant rs1421085 has been proven to repress mitochondrial thermogenesis in adipocyte precursor cells by disrupting a conserved motif for the ARID5B repressor and affecting IRX3 and IRX5 enhancer activity (53). Taken together, CAUSALdb offers a comprehensive knowledgebase to fine-map potential causal variants that confer susceptibility to complex traits/diseases.
DISCUSSION
Identifying and interpreting the genetic causality of complex traits is a major task in the post-GWAS era. Although several statistical fine-mappings have been used in many recent GWASs to estimate potential causal variants, the complexity of data representation as well as discrepancies of the applied statistical methods have inhibited systematic and valuable curation. Nowadays, an increasing amount of summary-level GWAS data has become publicly available, which provides unprecedented opportunities to gain in-depth understanding of the genetic mechanisms of complex traits via integrative analysis. However, no resources have leveraged published GWAS summary statistics to comprehensively fine-map causal variants and annotate their potential mechanisms. Herein, we performed strict quality control process and finally selected 3,052 fine-mappable GWAS summary statistics. We developed a unique user-friendly platform called CAUSALdb that integrates a repository of high-quality GWAS summary statistics, identifies potential causal variants by three state-of-the-art fine-mapping tools, and offers comprehensive variant annotations.
We used several well-known GWAS loci as examples and found that CAUSALdb could identify potential causal variants that were verified or were about to be verified; moreover, it facilitated the identification of true causal variants in a difficult credible set. The query functions of CAUSALdb are very beneficial for cross-study and cross-trait comparisons of causality. For instance, rs17293632 shows notable PP of causality in multiple disease categories, including cardiovascular and autoimmune diseases, implying that this variant plays a role in pleiotropy. Furthermore, CAUSALdb provides fine-mapping results of the 95% credible set of all GWASs, which may be a useful resource for researchers in other fields, such as disease risk prediction and drug repositioning. The well-formatted summary statistics of each causal block are also downloadable, enabling specific downstream analysis such as Mendelian randomization and colocalization. Although we have established several novel online functions for trait causality investigation, there are still some points that can be further improved in future. Many complex genetic loci harbor multiple causal variants for a particular trait/disease (54,55); given the computational burden and relatively low accuracy of multi-causal variants inference, CAUSALdb only assumes a single causal signal in each independent LD block. However, it allows users to download block-wise summary statistics for customized fine-mapping. In addition, several fine-mapping approaches in trans-ethnics have been proposed (56); CAUSALdb excludes GWASs with mixed populations in the current version because of complex LD patterns. As the GWAS summary data in CAUSALdb are from worldwide populations and considering the restrictions on obtaining original individual genotypes or large LD references (57), we only extracted LD information from 1KGP, which may not reflect the actual LD pattern in the corresponding GWAS cohort. In future studies, we plan to address the aforementioned issues by adding new features to CAUSALdb. We also aim to perform monthly curation of newly available GWAS summary statistics and frequently update variant information and functional annotations. In conclusion, with the accumulation of summary-level GWAS data, we believe that CAUSALdb should considerably aid researchers to interrogate the genetic mechanisms underlying diseases, thereby creating a significant impact in the post-GWAS era.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31701143, 31871327]; Natural Science Foundation of Tianjin [18JCZDJC34700]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Narayanan R., Butani V., Boyer D.S., Atilano S.R., Resende G.P., Kim D.S., Chakrabarti S., Kuppermann B.D., Khatibi N., Chwa M. et al.. Complement factor H polymorphism in age-related macular degeneration. Ophthalmology. 2007; 114:1327–1331. [DOI] [PubMed] [Google Scholar]
- 2. Marchini J., Howie B.. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010; 11:499–511. [DOI] [PubMed] [Google Scholar]
- 3. Evangelou E., Ioannidis J.P.. Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet. 2013; 14:379–389. [DOI] [PubMed] [Google Scholar]
- 4. Solovieff N., Cotsapas C., Lee P.H., Purcell S.M., Smoller J.W.. Pleiotropy in complex traits: challenges and strategies. Nat. Rev. Genet. 2013; 14:483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M. et al.. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015; 12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tam V., Patel N., Turcotte M., Bosse Y., Pare G., Meyre D.. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019; 20:467–484. [DOI] [PubMed] [Google Scholar]
- 7. Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J.. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017; 101:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E. et al.. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019; 47:D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li M.J., Liu Z., Wang P., Wong M.P., Nelson M.R., Kocher J.P., Yeager M., Sham P.C., Chanock S.J., Xia Z. et al.. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2016; 44:D869–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eicher J.D., Landowski C., Stackhouse B., Sloan A., Chen W., Jensen N., Lien J.P., Leslie R., Johnson A.D.. GRASP v2.0: an update on the Genome-Wide Repository of Associations between SNPs and phenotypes. Nucleic Acids Res. 2015; 43:D799–D804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farh K.K., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J., Shishkin A.A. et al.. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015; 518:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zheng J., Erzurumluoglu A.M., Elsworth B.L., Kemp J.P., Howe L., Haycock P.C., Hemani G., Tansey K., Laurin C., Early G. et al.. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017; 33:272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kamat M.A., Blackshaw J.A., Young R., Surendran P., Burgess S., Danesh J., Butterworth A.S., Staley J.R.. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019; doi:10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hemani G., Zheng J., Elsworth B., Wade K.H., Haberland V., Baird D., Laurin C., Burgess S., Bowden J., Langdon R. et al.. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018; 7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Canela-Xandri O., Rawlik K., Tenesa A.. An atlas of genetic associations in UK Biobank. Nat. Genet. 2018; 50:1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Watanabe K., Stringer S., Frei O., Umicevic Mirkov M., de Leeuw C., Polderman T.J.C., van der Sluis S., Andreassen O.A., Neale B.M., Posthuma D.. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019; 51:1339–1348. [DOI] [PubMed] [Google Scholar]
- 17. Schaid D.J., Chen W., Larson N.B.. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 2018; 19:491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dadaev T., Leongamornlert D.A., Saunders E.J., Eeles R., Kote-Jarai Z.. LocusExplorer: a user-friendly tool for integrated visualization of human genetic association data and biological annotations. Bioinformatics. 2016; 32:949–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J.. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010; 26:2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spain S.L., Barrett J.C.. Strategies for fine-mapping complex traits. Hum. Mol. Genet. 2015; 24:R111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Millard L.A.C., Davies N.M., Gaunt T.R., Davey Smith G., Tilling K.. Software Application Profile: PHESANT: a tool for performing automated phenome scans in UK Biobank. Int. J. Epidemiol. 2017; doi:10.1093/ije/dyx204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tryka K.A., Hao L., Sturcke A., Jin Y., Wang Z.Y., Ziyabari L., Lee M., Popova N., Sharopova N., Kimura M. et al.. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014; 42:D975–D979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lipscomb C.E. Medical Subject Headings (MeSH). Bull. Med. Libr. Assoc. 2000; 88:265–266. [PMC free article] [PubMed] [Google Scholar]
- 24. Ji S.G., Juran B.D., Mucha S., Folseraas T., Jostins L., Melum E., Kumasaka N., Atkinson E.J., Schlicht E.M., Liu J.Z. et al.. Genome-wide association study of primary sclerosing cholangitis identifies new risk loci and quantifies the genetic relationship with inflammatory bowel disease. Nat. Genet. 2017; 49:269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berisa T., Pickrell J.K.. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics. 2016; 32:283–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kichaev G., Roytman M., Johnson R., Eskin E., Lindstrom S., Kraft P., Pasaniuc B.. Improved methods for multi-trait fine mapping of pleiotropic risk loci. Bioinformatics. 2017; 33:248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benner C., Spencer C.C., Havulinna A.S., Salomaa V., Ripatti S., Pirinen M.. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016; 32:1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen W., Larrabee B.R., Ovsyannikova I.G., Kennedy R.B., Haralambieva I.H., Poland G.A., Schaid D.J.. Fine Mapping Causal Variants with an Approximate Bayesian Method Using Marginal Test Statistics. Genetics. 2015; 200:719–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pickrell J.K., Berisa T., Liu J.Z., Segurel L., Tung J.Y., Hinds D.A.. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 2016; 48:709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. LeBlanc M., Zuber V., Thompson W.K., Andreassen O.A. Schizophrenia, Bipolar Disorder Working Groups of the Psychiatric Genomics, C. Schizophrenia, Bipolar Disorder Working Groups of the Psychiatric Genomics, C. Frigessi A., Andreassen B.K.. A correction for sample overlap in genome-wide association studies in a polygenic pleiotropy-informed framework. BMC Genomics. 2018; 19:494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014; 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B. et al.. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016; 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H. et al.. An integrated map of structural variation in 2,504 human genomes. Nature. 2015; 526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang S., He Y., Liu H., Zhai H., Huang D., Yi X., Dong X., Wang Z., Zhao K., Zhou Y. et al.. regBase: whole genome base-wise aggregation and functional prediction for human non-coding regulatory variants. Nucleic Acids Res. 2019; doi:10.1093/nar/gkz774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu X., Wu C., Li C., Boerwinkle E.. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat. 2016; 37:235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jian X., Boerwinkle E., Liu X.. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014; 42:13534–13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ward L.D., Kellis M.. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016; 44:D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. et al.. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012; 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Q., Wang K.. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am. J. Hum. Genet. 2017; 100:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roadmap Epigenomics C., Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. et al.. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zheng R., Wan C., Mei S., Qin Q., Wu Q., Sun H., Chen C.H., Brown M., Zhang X., Meyer C.A. et al.. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019; 47:D729–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Consortium F. the, R.P., Clst the, R.P., Clst Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T. et al.. A promoter-level mammalian expression atlas. Nature. 2014; 507:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Consortium G.T., Laboratory D.A. Coordinating Center -Analysis Working, G., Statistical Methods groups-Analysis Working, G., Enhancing, G.g., Fund, N.I.H.C., Nih/Nci, Nih/Nhgri, Nih/Nimh, Nih/Nida et al.. Genetic effects on gene expression across human tissues. Nature. 2017; 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. et al.. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018; 46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pinero J., Bravo A., Queralt-Rosinach N., Gutierrez-Sacristan A., Deu-Pons J., Centeno E., Garcia-Garcia J., Sanz F., Furlong L.I.. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017; 45:D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. International Cancer Genome, C. Hudson T.J., Anderson W., Artez A., Barker A.D., Bell C., Bernabe R.R., Bhan M.K., Calvo F., Eerola I. et al.. International network of cancer genome projects. Nature. 2010; 464:993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011; 27:718–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nielsen J.B., Thorolfsdottir R.B., Fritsche L.G., Zhou W., Skov M.W., Graham S.E., Herron T.J., McCarthy S., Schmidt E.M., Sveinbjornsson G. et al.. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 2018; 50:1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van der Harst P., Verweij N.. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ. Res. 2018; 122:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jones P.D., Kaiser M.A., Ghaderi Najafabadi M., McVey D.G., Beveridge A.J., Schofield C.L., Samani N.J., Webb T.R.. The coronary artery disease-associated coding variant in zinc finger C3HC-type containing 1 (ZC3HC1) affects cell cycle regulation. J. Biol. Chem. 2016; 291:16318–16327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Turner A.W., Martinuk A., Silva A., Lau P., Nikpay M., Eriksson P., Folkersen L., Perisic L., Hedin U., Soubeyrand S. et al.. Functional analysis of a novel genome-wide association study signal in SMAD3 that confers protection from coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2016; 36:972–983. [DOI] [PubMed] [Google Scholar]
- 52. Miller C.L., Pjanic M., Wang T., Nguyen T., Cohain A., Lee J.D., Perisic L., Hedin U., Kundu R.K., Majmudar D. et al.. Integrative functional genomics identifies regulatory mechanisms at coronary artery disease loci. Nat. Commun. 2016; 7:12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Claussnitzer M., Dankel S.N., Kim K.H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V. et al.. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 2015; 373:895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang J., Ferreira T., Morris A.P., Medland S.E. Genetic Investigation of, A.T.C. Genetic Investigation of, A.T.C. Replication D.I.G., Meta-analysis C., Madden P.A., Heath A.C., Martin N.G. et al.. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012; 44:369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dadaev T., Saunders E.J., Newcombe P.J., Anokian E., Leongamornlert D.A., Brook M.N., Cieza-Borrella C., Mijuskovic M., Wakerell S., Olama A.A.A. et al.. Fine-mapping of prostate cancer susceptibility loci in a large meta-analysis identifies candidate causal variants. Nat. Commun. 2018; 9:2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li Y.R., Keating B.J.. Trans-ethnic genome-wide association studies: advantages and challenges of mapping in diverse populations. Genome Med. 2014; 6:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K. et al.. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016; 48:1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.