Abstract
Thousands of frozen, archived tissue samples from the human central nervous system (CNS) are currently available in brain banks. As recent developments in RNA sequencing technologies are beginning to elucidate the cellular diversity present within the human CNS, it is becoming clear that an understanding of this diversity would greatly benefit from deeper transcriptional analyses. Single cell and single nucleus RNA profiling provide one avenue to decipher this heterogeneity. An alternative, complementary approach is to profile isolated, pre-defined cell types and use methods that can be applied to many archived human tissue samples that have been stored long-term. Here, we developed FIN-Seq (Frozen Immunolabeled Nuclei Sequencing), a method that accomplishes these goals. FIN-Seq uses immunohistochemical isolation of nuclei of specific cell types from frozen human tissue, followed by bulk RNA-Sequencing. We applied this method to frozen postmortem samples of human cerebral cortex and retina and were able to identify transcripts, including low abundance transcripts, in specific cell types.
INTRODUCTION
The human central nervous system (CNS) comprises an extremely diverse set of cell types. While this heterogeneity has been appreciated since the work of early anatomists, it was not until recently that different cell types of the CNS have begun to be defined at the molecular level (1–9). Two of the most well studied CNS areas, the cerebral cortex and retina, have been the subjects of some of the earliest molecular characterizations, leading to the identification of at least 16 neuronal subtypes in the adult human cerebral cortex (4) and 18 major cell types in the adult human retina (7). While these pioneering studies have started to highlight the heterogeneity of the adult human CNS, more fine-grained distinctions among cell types are likely present. These distinctions will become more apparent with an increased number of cells profiled, and/or greater depth in sequencing of individual cell types. Such studies will greatly enable our understanding of the development and function of cell types in health and disease.
Transcriptional profiling to define cell types among heterogeneous populations, or to define gene expression features among different cell types, are now frequently carried out using single cell RNA sequencing (10–13). Although a very powerful approach, single cell RNA sequencing does not provide a depth of coverage of rare cell types, unless a very large number of cells is sequenced. An alternative is to use bulk RNA sequencing of defined, potentially rare, cell types, to avoid sequencing a large number of more abundant cell types. The discovery of novel markers has facilitated the isolation of specific cell types from diverse tissues, with isolation based on genetic markers, dyes, or antibodies (14–19).
Most postmortem human tissue is preserved by fixation or flash-freezing. While whole-cell approaches are incompatible with flash-frozen CNS tissue, the nuclei from frozen tissue stay intact and can be profiled. In addition, nuclear RNA has been successfully used as a proxy for the cellular transcriptome (4,20–24). Single nucleus RNA sequencing has been used to profile neuronal subtypes from frozen human cerebral cortex tissue (4). Bulk sequencing of immunolabeled nuclei also has been used to characterize the transcriptome of specific cell types in frozen human postmortem cerebellum (25). This example provides encouragement to explore further the use of frozen samples for antibody-based FACS purification of specific cell populations and subsequent RNA profiling.
Thousands of frozen human postmortem brain tissue samples, including those with disease, are readily available through brain banks. These samples are a valuable resource that is immediately available. A significant number of samples are archived, which, given the wide genetic variation among humans, will be important for the interpretation of disease-specific changes. This resource has not been fully exploited due to technical limitations in the retrieval of cell type specific RNA from frozen specimens. It also has been unclear whether long term storage, over a period of decades, would lead to diminished RNA quality and/or antigen detection.
Here, we developed FIN-Seq (Frozen Immunolabeled Nuclei Sequencing), a technique that combines nuclear isolation, fixation, immunolabeling, FACS, and RNA sequencing from frozen, archived human CNS tissue. While some antibodies such as those against NeuN and SOX6 are known to work with fresh tissue (26), a simple method to apply a wider range of antibodies against cell-type specific markers in archived frozen tissue has not been available until recently (25). With FIN-Seq, we isolated and profiled specific excitatory and inhibitory neuronal subtypes from frozen human cerebral cortex tissue, some of which had been stored for over fifteen years. We also extended the method to a different part of the CNS by isolating and profiling cone photoreceptors from the frozen human retina. Successful isolation of cone photoreceptors, which constitute roughly 2% of retinal cells, signified that rare populations could be reliably profiled from a frozen tissue sample. Interestingly, we also found that the nuclear transcripts captured with FIN-Seq represented more of the whole-cell transcriptome than has been seen using single nuclei sequencing (24). FIN-Seq is cost-effective and provides access to the transcriptomes of user-defined cell types from widely-available frozen human CNS samples.
MATERIALS AND METHODS
Mouse brain samples
All animals were handled according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Harvard University. For each biological replicate, the neocortex of P30 or adult (1+ years old) CD1 mice were microdissected, flash-frozen in an isopentane/dry ice slurry, and stored at −80°C.
Frozen human CNS samples
Frozen Brodmann Area 4 (Primary Motor Cortex) samples of Patient 1569, 3529, 3589, 4340 and 5650 were obtained from Human Brain and Spinal Fluid Resource Center at University of California, Los Angeles through the NIH NeuroBioBank. Patient 1569 was a 61-year-old male with no clinical brain diagnosis and the postmortem interval was 9 h. Patient 3529 was a 58-year-old male with no clinical brain diagnosis and the postmortem interval was 9 h. Patient 3589 was a 53-year-old male with no clinical brain diagnosis and the postmortem interval was 15 h. Patient 4340 was a 47-year-old male with no clinical brain diagnosis and the postmortem interval was 12.5 h. Patient 5650 was a 55-year-old male with no clinical brain diagnosis and the postmortem interval was 22.6 h. This IRB protocol (IRB16-2037) was determined to be not human subjects research by the Harvard University-Area Committee on the Use of Human Subjects.
Frozen eyes were obtained from Restore Life USA (Elizabethton, TN) through TissueForResearch. Patient DRLU032618A was a 52-year-old female with no clinical eye diagnosis and the postmortem interval was 8 h. Patient DRLU041518A was a 57-year-old male with no clinical eye diagnosis and the postmortem interval was 5 h. Patient DRLU041818C was a 53-year-old female with no clinical eye diagnosis and the postmortem interval was 9 h. Patient DRLU051918A was a 43-year-old female with no clinical eye diagnosis and the postmortem interval was 5 h. This IRB protocol (IRB17-1781) was determined to be not human subjects research by the Harvard University-Area Committee on the Use of Human Subjects.
Nuclei Isolation, Immunolabeling, and FACS
Thawed tissue was minced and immediately incubated in 1% PFA (with 1 μl ml−1 RNasin Plus (Promega, Madison, WI)) for 5 min. Nuclei were prepared by Dounce homogenizing in 0.1% Triton X-100 homogenization buffer (250 mM sucrose, 25 mM KCl, 5 mM MgCl2, 10 mM Tris buffer, pH 8.0, 1 μM DTT, 1× Protease Inhibitor (Promega), Hoechst 33342 10 ng ml−1 (Thermo Fisher Scientific, Waltham, MA, USA), 0.1% Triton X-100, 1 μl ml−1 RNasin Plus). Sample was then overlaid on top of 20% sucrose bed (25 mM KCl, 5 mM MgCl2, 10 mM Tris buffer, pH 8.0) and spun at 500×g for 12 min at 4°C. The pellet was resuspended in 4% PFA (with 1 ul ml−1 RNasin Plus) and incubated for 15 min on ice. The sample was spun at 2000×g for 5 min at 4°C and the supernatant was discarded. The sample was then resuspended in blocking buffer (0.5% BSA in nuclease-free PBS, 0.5 μl ml−1 RNasin Plus) and incubated for 15 min. Sample was spun and the pellet was resuspended and incubated in primary antibody (1:50 SATB2 antibody (Abcam, Cambridge, UK), 1:100 BCL11B antibody (Abcam), 1:1,000 CAR antibody (kind gift from Dr Sheryl Craft) in blocking buffer) for 30 min at 4°C. After washing 1× with blocking buffer, the sample was incubated in secondary antibody (1:750 appropriate AlexaFluor secondary antibodies (Thermo Fisher Scientific)) for 30 min at 4°C. After 1× wash, the sample was passed through a 35μm filter (Corning, Corning, NY, USA) before proceeding to FACS using FACSAria (BD Biosciences, Franklin Lakes, NJ, USA). 2N nuclei were determined by a Hoechst histogram. From the 2N nuclei, those that ran along the diagonal were considered to be the negative population. This was evident from the secondary antibody-only control FACS plot (Supplementary Figure S1), where single nuclei events displayed a continuum of autofluorescence in a diagonal line with events showing high fluorescence in one laser channel (e.g. 647nm) that were also were high in another laser channel (e.g. 594 nm). Nuclei that were highly fluorescent in all laser channels were thus also considered to be negative. Traditional gating methods with four quadrants include the events that display high fluorescence in all channels. Using the diagonal line to assess the negative cells then allowed the selection of the positive population, i.e. those nuclei that were right-shifted or left-shifted compared to the nuclei on the diagonal. Isolated populations were sorted into blocking buffer. Sorted nuclei were spun at 3000×g for 7 min, and the supernatant was discarded. A step-by-step online protocol is available at https://www.protocols.io/view/fin-seq-frozen-immunolabeled-nuclei-sequencing-zxbf7in.
RNA isolation and library preparation
RNA was extracted using the RecoverAll Total Nuclear Isolation Kit (Thermo Fisher Scientific). Crosslinking was reversed by incubating the nuclear pellet in Digestion Buffer and Protease mixture (100 μl buffer and 4 μl protease) for 3 h at 50°C, which differs from the manufacturer's protocol. The downstream steps were according to the manufacturer's protocol. RNA-seq libraries were generated using the SMART-Seq v.4 Ultra Low Input RNA kit (Takara Bio, Kusatsu, Japan) and Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer's protocol. The number of cycles was determined based on the number of nuclei sorted, as indicated in the SMART-Seq v.4 protocol. However, given the reduced amount of RNA in nuclei, at least 2 cycles were added from the protocol recommendation. The Nextera XT Kit uses 150 pg total cDNA as the input after SMART-Seq v.4; therefore, SMART-Seq v.4 should generate in the range of 1–2 ng of cDNA. The cDNA library fragment size was determined by the BioAnalyzer 2100 HS DNA Assay (Agilent, Santa Clara, CA, USA). The libraries were sequenced as paired-end reads on HiSeq 2500 or NextSeq 500.
RNA-seq data processing
Quality control of RNA-seq reads were performed using fastqc version 0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). RNA-seq reads were clipped and mapped onto the mouse genome (Ensembl GRCm38.88) or human genome (Ensembl GRCh38.87) using STAR version 2.5 (27,28). Parameters used were as follows: –runThreadN 6 –readFilesCommand zcat –outSAMtype BAM SortedByCoordinate –outSAMunmapped Within –outSAMattributes Standard –clip3pAdapterSeq –quantMode TranscriptomeSAM GeneCounts
Alignment quality control was performed using Qualimap version 2.2.1 (29). Read counts were generated by HT-seq version 0.6.1 (30). Sample parameters used were as follows: -i gene_name -s no.
The resulting matrix of read counts were analyzed for differential expression by DESeq2 version 3.5 (31). Samples with non-neuronal cell contamination were discarded for analysis (BCL11B+ 3529 and BCL11B+ 3589). For the DE analysis of human retina samples, any genes with more than five samples with zero reads were discarded. The R scripts used for differential expression analysis are available in Supplementary Files.
Gene set enrichment analysis
GSEAPreranked analysis was performed on the All versus SATB2+ dataset using GSEA v3.0 (32). Gene set databases including markers that define neuronal subtypes identified by Darmanis et al. (2) and Lake et al. (4) were generated. Parameters used were as follows: Number of permutations: 1000; Enrichment statistic: classic; the ranked file was generated using log2FoldChange generated by DESeq2. To determine significance, we used the default FDR <0.25 for all gene sets.
RNAscope
P30 and adult (>1 year old) mouse brains were perfused with 4% PFA in PBS and sectioned on a cryostat at a thickness of 16 μm. Double in situ fluorescence hybridization was performed using the RNAscope Fluorescent Multiplex assay according to the manufacturer's protocol (Advanced Cell Diagnostics, Newark, CA, USA). The following probes were used for the mouse study: Satb2-C1 (Catalog #: 413261), Satb2-C2 (Catalog#: 420981-C2), Bcl11b-C3 (Catalog#: 413271-C3), Ddit4l-C1 (Catalog#: 468551), Kcnn2-C1 (Catalog#: 427971), Unc5d-C2 (Catalog#: 480461-C2) and Rprm-C2 (Catalog#: 466071-C2).
FFPE adult human cerebral cortex tissue from a 54-year-old female was obtained from Abcam (ab4296). Chromogenic double in situ hybridization was performed for the human brain tissue using the RNAscope 2.5 HD Duplex Assay (Advanced Cell Diagnostics) according to the manufacturer's protocol. A Fluorescent Multiplex assay was used for the human retina tissue according to the manufacturer's protocol (Advanced Cell Diagnostics). The following probes were used for the human study: SATB2-C2 (Catalog#: 420981-C2), RORB-C1 (Catalog#: 446061), UNC5D-C1 (Catalog#: 459991), CRYM-C2 (Catalog#: 460031-C2), GAD1-C1 (Catalog#: 404031), COL6A1-C1 (Catalog#: 482461), ANXA1-C1 (Catalog#: 465411), ARR3-C2 (Catalog#: 486461-C2), DHRS3-C1 (Catalog#: 504941), RAB41-C1 (Catalog#: 559561).
Immunohistochemistry
P30 and adult (>1 year old) mouse brains were perfused with 4% PFA in PBS and sectioned on a vibratome at a thickness of 40 μm. Immunohistochemistry was performed as previously described (14) with anti-SATB2 and anti-BCL11B antibodies.
FFPE adult human cerebral cortex tissue from a 54-year-old female was obtained from Abcam (ab4296). The brain tissue was deparaffinized by 2× xylene incubation (3 min each) followed by 1 × 100% ethanol (3 min), 1 × 95% ethanol (3 min), 1 × 70% ethanol (3 min) washes. Antigen retrieval was performed in a citrate buffer (10 mM citric acid, pH 6.0) in a rice cooker with boiling water for 20 min. Subsequently, immunohistochemistry was performed as described above with an additional step of incubation in TrueBlack (Biotium, Fremont, CA, USA) after incubation in blocking buffer to quench the lipofuscin autofluorescence.
For human eye immunohistochemistry, formalin-fixed human postmortem eyes were obtained from Restore Life USA. Patient DRLU101818C was a 54-year-old male with no clinical eye diagnosis and the postmortem interval was 4 h. Patient DRLU110118A was a 59-year-old female with no clinical eye diagnosis and the postmortem interval was 4 h. The retina was cryosectioned at 16 μm thickness. Immunohistochemistry was performed as previously described (14) with anti-CAR antibody (1:10,000).
Imaging
Fluorescent confocal images of the brain were acquired with Zeiss LSM 700 confocal microscope and analyzed with the ZEN Black software. Fluorescent confocal images of the retina were acquired with Nikon Ti inverted spinning disk microscope and analyzed with the NIS-Elements software. Brightfield color images of the human brain were acquired with AxioZoom V16 Zoom Microscope.
RESULTS
FACS isolation of immunolabeled nuclei from frozen mouse brain samples
To test whether sequencing of nuclear RNA from frozen tissue is feasible, specific nuclear populations from frozen mouse neocortex were isolated. To this end, modifications were made to protocols that use intracellular antibody staining to isolate specific cell types (17,33–36). Since these protocols were for intact cells, which cannot be dissociated from frozen tissue, we developed a protocol for the isolation of antibody-labeled nuclei (Figure 1A). Isolation of nuclei eliminates the need for enzymatic dissociation, which induces aberrant activation of immediate early genes (37). We added a 1% PFA step before the extraction of the nuclei to ensure the structural integrity of the nuclei. In addition, our method of RNA extraction using the modified Recoverall Kit protocol, which includes a three-hour protease step, outperformed other FFPE RNA extraction kits in terms of RNA recovery.
Figure 1.
Isolation and transcriptome sequencing of two neuronal subtypes from the frozen mouse neocortex. (A) Schematic of FIN-Seq for frozen adult mouse brain. Nuclei were extracted from the frozen mouse neocortex by Dounce homogenization. The nuclei were fixed and immunolabeled with anti-BCL11B and anti-SATB2 antibodies. Two nuclear populations were isolated by FACS based on expression level of these two proteins. The nuclei were reverse crosslinked by protease digestion, and the RNA was extracted. Sequencing libraries were generated and subsequently sequenced to obtain cell type specific transcriptomes. (B) Representative immunohistochemistry images using BCL11B and SATB2 antibodies in the P30 mouse neocortex showed SATB2 expression in the upper layers and BCL11B expression in the deep layers (Left image). In layer 5, there were sparse cells that express both SATB2 and BCL11B (bottom right image). (C) FACS plot of nuclei labeled with SATB2 and BCL11B antibodies showed a cluster of nuclei immunolabeled with BCL11B and a cluster of nuclei labeled with SATB2. (D) Isolated nuclei were counterstained by the Hoechst dye and either SATB2 or BCL11B in the SATB2+ population (top panels) or BCL11B+ population (bottom panels). (E) Representative quantification of read counts mapped by transcript position (5′ to 3′) for every gene. (F) Representative quantification of percentage of read counts mapped to exonic, intronic, or intergenic regions of the genome. (G) Heatmap of unbiased top 50 differentially expressed genes between SATB2+ and BCL11B+ populations. Known markers of callosal projection neurons (in red) were enriched in the SATB2+ population while known markers of corticofugal projection neurons (in green) were enriched in the BCL11B+ population. Scale bars; 100 μm (B, right panels, D), 500 μm (B, left panel).
From the flash-frozen neocortex of P30 mice, we sought to isolate two populations of projection neurons, Corticofugal Projection Neurons (CFuPN) and Callosal Projection Neurons (CPN). In the adult mouse brain, BCL11B (also known as CTIP2) is largely expressed in CFuPNs in layer 5b and 6 and in sparse populations of interneurons. SATB2 is expressed by CPNs in all layers (38) (Figure 1B). BCL11B and SATB2 expression are largely mutually exclusive, with a small population of layer 5 neurons expressing both markers (17,39) (Figure 1B, layer 5, inset). Upon isolation by homogenization, nuclei were fixed, immunolabeled with antibodies against BCL11B and SATB2, and separated into two populations by FACS: SATB2LOBCL11BHI (BCL11B+) and SATB2HIBCL11BLO (SATB2+) (n = 2 for each population) (Figure 1C, D, Supplementary Figure S1). On average, we collected 55,215 BCL11B+ nuclei and 102,016 SATB2+ nuclei per biological replicate. These results indicate that this protocol could result in the isolation of intact nuclei that are immunolabeled with specific intranuclear antibodies.
To determine whether the correct neuronal populations were isolated, and to test nuclear transcriptional profiling using these samples, SMART-Seq v.4 RNA-seq libraries were generated and sequenced on HiSeq 2500. For each sample, libraries were sequenced to a mean of 40 million 100 bp paired-end reads (range: 36–48 million reads per sample) to be able to reliably detect low-abundance transcripts. To determine the degree of RNA degradation, we measured the 3′ bias using Qualimap (29). The 3′ bias for P30 samples ranged from 0.65 to 0.69 (mean±SD: 0.685±0.02), which is comparable to RNA Integrity Number (RIN) of 2–4 (40) (Figure 1E). Consistent with the idea that nuclear transcripts are predominantly nascent RNA, we found that a substantial number of reads mapped to intronic regions (Exonic: 63.16±4.89%; Intronic: 32.49±4.48%; Intergenic: 4.36±0.56%) (Figure 1F) (4,22,24). Distribution of normalized read counts was virtually identical among samples (Supplementary Figure S2A). Unbiased hierarchical clustering showed that the samples of the same population clustered together (average Pearson correlation between samples within population: r = 0.98) (Supplementary Figure S2B). Subsequently, the two populations were analyzed for differential (gene) expression (DE). The frequency distribution of all P-values showed an even distribution of null P-values, thus allowing for calculation of adjusted P-value using the Benjamini-Hochberg procedure (Supplementary Figure S2C). Between populations, we found 2,698 differentially expressed genes (adjusted P-value < 0.05) out of 17,662 genes (Supplementary Figure S2D). The high number of genes detected suggests identification of low abundance transcripts.
From the DE analysis, we found an enrichment of known CPN and Layer 4 (L4) markers (e.g. Cux2, Unc5d and Rorb) in the SATB2+ population among the unbiased top 50 DE genes. Conversely, we found an enrichment of CFuPN markers (e.g. Fezf2, Foxp2, and Crym) in the BCL11B+ population (Figure 1G). BCL11B also labels interneurons in all layers of the mouse neocortex (14,41). Accordingly, we found an enrichment of some interneuron markers in the BCL11B+ population (e.g. Gad1 and Gad2) (Figure 1G). To confirm the molecular identities of the isolated neuronal populations, we also determined the relative expression levels of known CPN and CFuPN marker genes that were differentially expressed between CPN and CFuPN in previous studies (21 CPN markers and 22 CFuPN markers) (14,17,38). We found that all CPN markers were enriched in the SATB2+ population and all CFuPN markers were enriched in the BCL11B+ population (Supplementary Figure S3). To validate the differentially expressed genes, we chose four DE genes (Ddit4l, Unc5d, Kcnn2 and Rprm) for further analysis. Using RNAscope double fluorescent in situ hybridization (FISH), we localized the transcripts of these genes in specific neuronal populations. We found that Ddit4l and Unc5d were expressed in layers 2 through 4 and were localized to Satb2+ neurons (Supplementary Figure S4). Additionally, Kcnn2 and Rprm were expressed in layers 5 and 6, respectively, and they were specifically confined to Bcl11b+ neurons (Supplementary Figure S4). In addition, we successfully isolated and profiled the same neuronal populations from mature, adult (1+ years old) mouse neocortex (Supplementary Figures S5 and S6). These results indicate that FIN-Seq can be used to isolate CFuPN and CPN nuclei from flash-frozen mouse neocortex for downstream quantitative RNA-seq analysis of specific neuronal populations.
To determine the degree to which nuclear transcript abundance correlates to cellular transcript abundance, we sought to compare the transcriptional profiles of BCL11B+ nuclei and cells. For cells, we dissociated the brains of P7 mice using a protocol described previously (17). For nuclei, we performed the FIN-Seq protocol, starting with a fresh P7 brain instead of flash-freezing to keep the starting material consistent between cells and nuclei. We chose the P7 time point because dissociation of the adult mouse brain into single cells affects cell viability at later ages (42,43). Transcriptional analysis of BCL11B+ cells and nuclei showed a high degree of correlation (average Pearson correlation between cellular versus nuclear: r = 0.90; cellular versus cellular: r = 0.93; nuclear versus nuclear: r = 0.93) (Supplementary Figure S7). In contrast, previous comparison of single nucleus and single cell transcriptomes from the adult mouse brain showed a lower degree of correlation (r = 0.77) (24). These results indicate that bulk sequencing of isolated nuclei using FIN-Seq could more accurately represent the transcript abundance found within whole cells.
Specific neuronal subtypes can be isolated from frozen postmortem human brain samples
To determine whether this protocol is applicable to frozen postmortem samples of the human brain, we obtained five frozen postmortem brain samples (Brodmann area 4, primary motor cortex; ages: 47–61) from a tissue bank that had stored them long-term (for description of the samples, see Materials and Methods). Of note, the oldest frozen sample had been archived for over 25 years. We implemented the same FIN-Seq protocol as above to the frozen human cortical tissue (Figure 2A), in which we found BCL11B+/SATB2−, BCL11B+/SATB2+ and BCL11B−/SATB2+ nuclei (Figure 2B). Although the precise identity of these neurons remains unclear due to the lack of data on additional features, including their electrophysiological properties and patterns of connectivity, the transcriptomic data indicate that they are subtypes of excitatory and inhibitory neurons (4). We used FACS to isolate SATB2LOBCL11BHI and SATB2HIBCL11BLO nuclei as well as all cortical nuclei (henceforth called BCL11B+, SATB2+ and all, respectively) for comparison (BCL11B+: 26,616 nuclei/replicate, n = 5; SATB2+: 104,865 nuclei/replicate, n = 5; All: 67,580 nuclei/replicate, n = 5) (Supplementary Figure S8). These results indicate that nuclear isolation of specific neuronal subtypes from frozen postmortem human brain tissue is feasible using this technique.
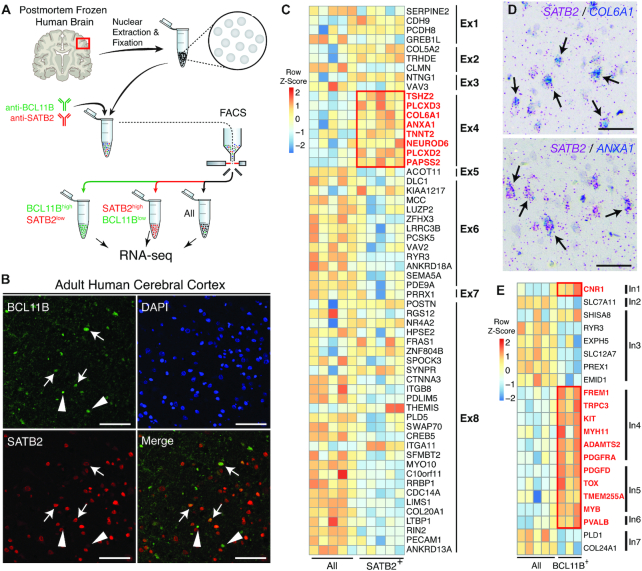
Figure 2.
Isolation and profiling of neuronal subtypes from the frozen human cerebral cortex. (A) Schematic of FIN-Seq for frozen human cerebral cortex. Nuclei were isolated and subsequently fixed in 4% PFA. They were immunolabeled with anti-BCL11B and anti-SATB2 antibodies, and FACS isolated into populations. RNA from the nuclei were sequenced to obtain a cell type specific transcriptome. (B) Representative immunohistochemistry of the adult human cerebral cortex using anti-BCL11B and anti-SATB2 antibodies. Some nuclei expressed both SATB2 and BCL11B (arrows), some nuclei expressed BCL11B but not SATB2 (arrowheads), and many nuclei expressed SATB2 but not BCL11B. (C) A heatmap representing relative expression levels of excitatory neuron markers previously identified by single nuclei RNA sequencing that are differentially expressed (adjusted P-value < 0.05) between SATB2+ and All populations. Markers of neuronal subtype Ex4 (outlined in red), which expresses SATB2, were enriched in the SATB2+ population. (D) Validation of Ex4 markers, COL6A1 (left panel) and ANXA1 (right panel) using RNAscope single molecule FISH. Both COL6A1 and ANXA1 were expressed in SATB2+ neurons (arrows). (E) A heatmap representing relative expression levels of inhibitory neuron markers previously identified by single nuclei RNA sequencing that are differentially expressed (adjusted P-value < 0.05) between BCL11B+ and All populations. Markers of neuronal subtypes, In1, In4, In5 and In6, all of which express BCL11B, were enriched in the BCL11B+ population. Scale bars: 100 μm (B), 50 μm (D).
To determine the molecular identity of the isolated neuronal populations, we performed RNA sequencing of each population (BCL11B+, SATB2+, and All, sequenced to a mean of 36 million paired-end 100 bp reads). The average RIN of the frozen human brain samples prior to FIN-Seq was 3.9. After FIN-Seq, the 3′ bias ranged from 0.69 to 0.78 (mean ± SD: 0.73 ± 0.02), which corresponds to a RIN of 2–4, indicating that the FIN-Seq protocol does not further decrease the integrity of the RNA (Supplementary Figure S9A). The human brain contains an increased number of nascent transcripts compared to other organs and organisms (44). Accordingly, we found that the proportion of intronic reads was higher in the human neuronal samples compared to that in mice (exonic: 47.76 ± 5.82%; intronic: 45.51 ± 5.04%; intergenic: 6.72 ± 1.13%) (Supplementary Figure S9B). Quality control of the sequencing reads and differential expression analysis indicated successful sample separation and differential expression analysis (Supplementary Figure S10). Interestingly, we did not observe an increase in 3′ sequencing bias with an increase in the number of years that the frozen specimens had been stored at −80°C (Supplementary Figure S10). Between SATB2+ and All populations, we found 4,917 differentially expressed genes (adjusted P-value < 0.05) out of 24,979 genes. Between BCL11B+ and All populations, we found 2,812 differentially expressed genes (adjusted P-value < 0.05) out of 24,477 genes (Supplementary Figure S10).
To determine the molecular identity of the SATB2+ and BCL11B+ populations, we first compared the gene expression levels of known markers of oligodendrocytes, astrocytes, and neurons. We found that neuronal markers were enriched in both SATB2+ and BCL11B+ populations. However, two of the BCL11B+ samples were discarded following this analysis due to enrichment of oligodendrocyte and astrocyte markers, indicating non-neuronal cell contamination. We also found an enrichment in the BCL11B+ population of PDGFRA, normally considered an oligodendrocyte marker, but also previously shown to be expressed by a subset of inhibitory neurons in the human cerebral cortex (Supplementary Figure S11) (4). The SATB2+ population highly expressed SLC17A7 (also known as VGLUT1) and did not express GAD1 or GAD2, while the BCL11B+ population expressed GAD1 and GAD2 at high levels, indicating that, while SATB2+ population contained mainly excitatory neurons, the BCL11B+ population contained also inhibitory neurons (Supplementary Figure S12).
We next sought to understand the identity of the SATB2+ and BCL11B+ populations at the neuronal subtype-level. Previously, single nucleus RNA-seq has identified eight excitatory neuronal subtypes (Ex1–Ex8) and eight inhibitory neuronal subtypes (In1–In8) in the adult human neocortex (4). SATB2 is expressed in all excitatory neurons, but it is most highly expressed in one of the neuronal subtypes referred to, in this prior study, as Ex4. BCL11B is highly expressed in In1, In4, In5, and In6. SATB2 and BCL11B are both expressed in Ex6 and Ex8, but we would not expect to see these subtypes in our populations as we did not collect the SATB2HIBCL11BHI population. For the SATB2+ population, we cross-referenced our DE gene set (adjusted P-value < 0.05) to the molecular signature genes that define the eight excitatory cortical neuronal subtypes (Ex1–Ex8). From this analysis, we observed a high level of expression of Ex4 markers in the SATB2+ population compared to the All population (Figure 2E). To confirm these results, we also ran the dataset through a gene set enrichment analysis (GSEA) against all marker genes that define Ex1–Ex8 (32). We found that Ex4 gene set was significantly enriched in the SATB2+ population while Ex6 and Ex8 gene sets were enriched in the All population (default significance at FDR < 0.25; Ex4: FDR = 0.139; Ex6: FDR = 0.043; Ex8: FDR = 0.005). Depletion of Ex6 and Ex8 from the SATB2+ population is likely due to the exclusion of SATB2HIBCL11BHI nuclei. We confirmed the expression of COL6A1 and ANXA1, two Ex4 markers, in SATB2+ neurons by single molecule FISH (Figure 2D). In the BCL11B+ population, we found that the markers for In1, In4, In5 and In6 were enriched compared to the All population (Figure 2E). To directly compare the BCL11B+ population and the SATB2+ population, we cross-referenced our DE gene set (adjusted P-value < 0.05) to the molecular signature genes that define the eight excitatory and eight inhibitory cortical neuronal subtypes (Ex1–Ex8 and In1–In8). From this analysis, we saw that the majority of Ex4 markers were enriched in the SATB2+ population while Ex6, In4 and In5 markers were enriched in the BCL11B+ population (Supplementary Figure S13). Furthermore, previous single cell sequencing of the fresh adult human brain identified seven neuronal communities (NC), of which SATB2 is highly expressed in neuronal community 4 (NC4) (2). Accordingly, we found that the markers for NC4 are highly expressed in the isolated SATB2+ population (Supplementary Figure S14). By GSEA analysis, we also found that NC4 gene set was significantly enriched in the SATB2+ population (FDR = 0.037). Taken together, our results show the FIN-Seq protocol can isolate neuronal subtypes defined by antibody staining for downstream transcriptional profiling from frozen postmortem human cortical samples.
Isolation and transcriptional profiling of cone photoreceptors from the human retina
To determine whether we could use FIN-Seq to isolate and profile specific cell types from another region of the human CNS, we chose to isolate cone photoreceptors from the retina. Cones comprise only ∼2–4% of the human retina (6,7) so this was also a test of the ability of FIN-Seq to allow for a deeper transcriptomic analysis of a relatively rare cell type. We obtained four freshly frozen postmortem eyes (age range: 40–60, see Materials and Methods for description of samples) from patients without known retinal disorders. Nuclei were extracted from the mid-peripheral retina, fixed, and immunostained by a human Cone Arrestin (CAR, also known as ARR3) antibody (Figure 3A). In human retinal cross-sections, we found CAR expression in the nuclei and cell bodies of cone photoreceptors, located in the outer nuclear layer where all photoreceptors reside (Figure 3B). CAR+ and CAR− nuclei were isolated by FACS, and the RNA was extracted for deep sequencing (CAR+: 8,500 nuclei/replicate, n = 4; CAR−: 180,000 nuclei/replicate, n = 4). On average, 1.97% of all nuclei were CAR+, a proportion similar to known percentage of cone photoreceptors in the human retina based on single cell RNA sequencing (6,7). To determine whether fixation was necessary for antibody penetration, we performed the FIN-Seq protocol with and without fixation. We found that the distinct CAR+ population was present only with fixation, suggesting that, unlike the NeuN antibody, fixation is necessary for optimal immunolabeling of CAR (Supplementary Figure S15). cDNA sequencing libraries were generated using SMART-Seq v.4 and sequenced to a mean depth of 43 million (range: 37–53 million reads/replicate) 75 bp paired-end reads. The sequencing reads were analyzed, and the quality control parameters indicated successful sample separation and differential expression analysis (Supplementary Figure S16). We found 5,260 DE genes (adjusted P-value < 0.05) out of 12,910 genes between CAR+ and CAR− nuclear populations.
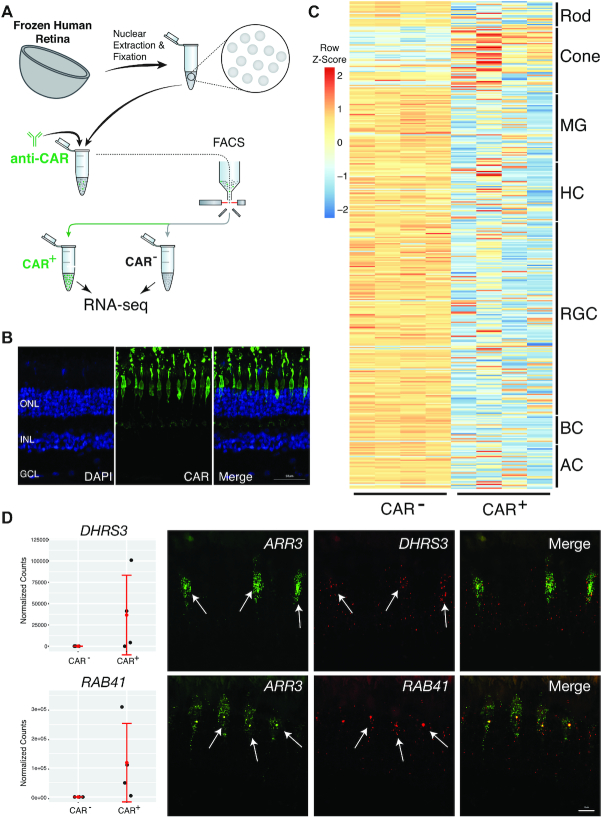
Figure 3.
Isolation and sequencing of cone photoreceptor nuclei from the frozen human retina. (A) Schematic of FIN-Seq for the frozen human retina. Nuclei were extracted from the frozen retina and subsequently fixed in 4% PFA. Nuclei were then immunolabeled with an anti-CAR antibody and sorted. CAR+ and CAR− populations were obtained and the nuclear RNA was sequenced. (B) Representative immunohistochemistry of an adult human retina section using the anti-CAR antibody (middle panel) and DAPI (left panel). CAR+ cone photoreceptors were localized to the uppermost layer of the ONL. (C) A heatmap representing relative expression levels of retinal cell type markers previously identified by single nuclei RNA sequencing (adjusted P-value < 1E−10) that are differentially expressed (adjusted P-value < 0.05) between CAR− and CAR+ populations. Cone markers were enriched in the CAR+ population. (D) Validation of new human cone photoreceptor markers by single molecule FISH. Expression levels for DHRS3 and RAB41 from the RNA-seq are indicated in the graphs (left panels). Both DHRS3 and RAB41 were expressed in the ARR3+ cone photoreceptors (arrows). ONL, Outer Nuclear Layer; INL, Inner Nuclear Layer; GCL, Ganglion Cell Layer; MG, Müller Glia; AC, Amacrine Cell; HC, Horizontal Cell; RGC, Retinal Ganglion Cell; BC, Bipolar Cell. Scale bars; 50 μm (B), 10 μm (D).
To determine the cellular identity of the CAR+ population, we examined the top 50 differential expressed genes between CAR+ and CAR− populations. Of the 16 genes enriched in the CAR+ population in the top 50 DE genes, eight are known markers of human cone photoreceptors, identified by previous single cell RNA sequencing experiments (Supplementary Figure S17, cone markers in red) (7). We then cross-referenced our DE gene set (adjusted P-value < 0.05) to the retinal cell type specific markers identified by single nucleus RNA sequencing (adjusted P-value < 1E−10) (6). We found that cone specific markers were upregulated in the CAR+ population while other cell-type specific markers were enriched in the CAR− population (Figure 3C). We also performed single molecule FISH for two previously uncharacterized cone markers, RAB41 and DHRS3, and found that they were specifically expressed in ARR3+ cone photoreceptors (Figure 3D). These results indicate that FIN-Seq enabled successful isolation and transcriptional profilng of cone photoreceptors from frozen postmortem human retinas.
DISCUSSION
Technologies for the profiling of RNA expression within the CNS are rapidly expanding. At the tissue level, analyses of gene expression within distinct regions of the fetal and adult human brain have been carried out (45–56). Despite progress, these tissue-level approaches cannot account for the cellular heterogeneity of the brain, an organ with tremendous cellular diversity. This is important especially when human CNS disorders are under study, as histological studies have underscored the cell type-specific nature of cellular dysfunction and degeneration (57–59). Insights into the mechanism of such diseases may result from the isolation and transcriptional profiling of the specific neuronal populations affected. These may be rare cell types within the tissue, further underscoring the need for technologies that allow enrichment of defined cell types.
Recent developments of single cell RNA-seq technology have enabled unbiased sampling of all cell types from a human CNS tissue sample (1–9). However, for some types of studies, it is impractical to assess the gene expression changes in all cell types. If the cell type of interest is known, bulk RNA-seq of isolated neuronal populations is a complementary approach to quantify gene expression more comprehensively in specific cells of interest. Here, we developed a method, FIN-Seq, to quantify gene expression in isolated neuronal populations from frozen archived postmortem human CNS tissue. Bulk RNA sequencing can lead to the detection of low abundance transcripts and rare splice variants, which are often not detected in single cell or single nucleus RNA sequencing (60,61). We also show that bulk nuclear sequencing can represent more of the transcriptome of the entire cell, compared to single nucleus sequencing. Moreover, as suggested by the data from cone photoreceptors, which comprise only 2% of retinal cells, FIN-seq may prove to be especially valuable for the deep profiling of rare cell types.
The challenge of applying FIN-Seq for some cell types is the availability of suitable nuclear antibodies. With the rapid progress of single cell sequencing, markers of molecularly distinct human neuronal subtypes are becoming available. For most of these markers, however, no antibody exists. FIN-Seq could greatly benefit from efforts to generate a validated antibody catalog such as the Protein Capture Reagents Program, in which over 700 validated monoclonal antibodies against human transcription factors have been produced (62). Interestingly, ER proteins also have been used to purify cell type-specific human nuclei using FACS, which could significantly increase the number of available cell type-specific antibodies (25). For molecular markers without an antibody, FIN-Seq could be further developed to isolate specific cell populations using nuclear RNA by FISH techniques such as RNAscope or SABER (63,64). Labeling specific nuclear transcripts of human neuronal nuclei for downstream FACS and transcriptome sequencing will enable FIN-Seq to capture any cell type of interest.
Taken together, FIN-Seq can enable transcriptional profiling of specific neuronal subtypes in the postmortem human CNS, without a need for genetic labeling. Counting only those from the NIH brain bank, over 16,000 postmortem samples are available, including those with neurological disorders, and many of them are stored long-term as flash-frozen samples. With FIN-Seq, we can start to interrogate the transcriptional changes that accompany specific neuronal subtypes in the adult human brain and identify molecular mechanisms underlying cell type specific pathology.
DATA AVAILABILITY
The mouse transcriptome data have been deposited to GEO (GEO Accession # GSE130143).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank former and current members of the Arlotta and Cepko Labs for the insightful discussion and feedback. We thank D. Richardson, Harvard Center for Biological Imaging, P.M. Llopis, R. Stephansky, and the MicRoN core at Harvard Medical School for their assistance with microscopy. We thank S.H. Sui, V. Barrera, and M. Ziller for their assistance with bioinformatics. We thank S. Craft for sharing of the hCAR antibody. We thank C. Araneo, F. Lopez, and the Flow Cytometry Core Facility for their assistance with flow cytometry.
Author contributions: R.A., E.Z., H.-H.C. and P.A. conceived the work and designed the experiments. R.A. performed the majority of the experiments and analyzed the data. E.Z. and P.A. conceived the method. E.Z. and H.-H.C. assessed feasibility in preliminary steps. R.A., P.A. and C.L.C. wrote the manuscript. E.Z. edited the manuscript. N.C.C. and H.-H.C. assisted with the FACS. S.K. assisted with the initial antibody screening for the FACS. P.A. and C.L.C. supervised the brain and retina aspects of the project, respectively.
Notes
Present address: Emanuela Zuccaro, Department of Biomedical Sciences, University of Padova, 35131 Padova, Italy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health [R01MH112940, R01MH09606]; Howard Hughes Medical Institute; Edward R. and Anne G. Lefler Postdoctoral Fellowship. Funding for open access charge: Howard Hughes Medical Institute; NIMH Grant.
Conflict of interest statement. A provisional US patent has been filed based on this work.
REFERENCES
- 1. Cherry T.J., Yang M.G., Harmin D.A., Tao P., Timms A.E., Bauwens M., Allikmets R., Jones E.M., Chen R., DeBaere E. et al.. Epigenomic profiling and single-nucleus-RNA-seq reveal cis-regulatory elements in human retina, macula and rpe and non-coding genetic variation. 2018; bioRxiv doi:08 September 2018, preprint: not peer reviewed 10.1101/412361. [DOI]
- 2. Darmanis S., Sloan S.A., Zhang Y., Enge M., Caneda C., Shuer L.M., Hayden Gephart M.G., Barres B.A., Quake S.R.. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hodge R.D., Bakken T.E., Miller J.A., Smith K.A., Barkan E.R., Graybuck L.T., Close J.L., Long B., Penn O., Yao Z. et al.. Conserved cell types with divergent features between human and mouse cortex. 2018; bioRxiv doi:05 August 2018, preprint: not peer reviewed 10.1101/384826. [DOI] [PMC free article] [PubMed]
- 4. Lake B.B., Ai R., Kaeser G.E., Salathia N.S., Yung Y.C., Liu R., Wildberg A., Gao D., Fung H.-L.L., Chen S. et al.. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science (New York, N.Y.). 2016; 352:1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lake B.B., Chen S., Sos B.C., Fan J., Kaeser G.E., Yung Y.C., Duong T.E., Gao D., Chun J., Kharchenko P.V. et al.. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 2018; 36:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liang Q., Dharmat R., Owen L., Shakoor A., Li Y., Kim S., Vitale A., Kim I., Morgan D., Wu N. et al.. Single-nuclei RNA-seq on human retinal tissue provides improved transcriptome profiling. 2019; bioRxiv doi:11 November 2018, preprint: not peer reviewed 10.1101/468207. [DOI] [PMC free article] [PubMed]
- 7. Lukowski S., Lo C., Sharov A., Nguyen Q., Fang L., Hung S., Zhu L., Zhang T., Nguyen T., Senabouth A. et al.. Generation of human neural retina transcriptome atlas by single cell RNA sequencing. 2018; bioRxiv doi:24 September 2018, preprint: not peer reviewed 10.1101/425223. [DOI]
- 8. Peng Y.-R.R., Shekhar K., Yan W., Herrmann D., Sappington A., Bryman G.S., van Zyl T., Do M.T.H.T.H., Regev A., Sanes J.R.. Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell. 2019; 176:1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Phillips M.J., Jiang P., Howden S., Barney P., Min J., York N.W., Chu L.-F.F., Capowski E.E., Cash A., Jain S. et al.. A novel approach to single cell rna-sequence analysis facilitates in silico gene reporting of human pluripotent stem cell-derived retinal cell types. Stem Cells. 2018; 36:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. et al.. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015; 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shekhar K., Lapan S.W., Whitney I.E., Tran N.M., Macosko E.Z., Kowalczyk M., Adiconis X., Levin J.Z., Nemesh J., Goldman M. et al.. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 2016; 166:1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tasic B., Menon V., Nguyen T.N., Kim T.K., Jarsky T., Yao Z., Levi B., Gray L.T., Sorensen S.A., Dolbeare T. et al.. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 2016; 19:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zeisel A., Muñoz-Manchado A.B., Codeluppi S., Lönnerberg P., La Manno G., Juréus A., Marques S., Munguba H., He L., Betsholtz C. et al.. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science (New York, N.Y.). 2015; 347:1138–1142. [DOI] [PubMed] [Google Scholar]
- 14. Arlotta P., Molyneaux B.J., Chen J., Inoue J., Kominami R., Macklis J.D.. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005; 45:207–221. [DOI] [PubMed] [Google Scholar]
- 15. Heiman M., Schaefer A., Gong S., Peterson J.D., Day M., Ramsey K.E., Suárez-Fariñas M., Schwarz C., Stephan D.A., Surmeier D.J. et al.. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008; 135:738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lobo M.K., Karsten S.L., Gray M., Geschwind D.H., Yang X.W.. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat. Neurosci. 2006; 9:443–452. [DOI] [PubMed] [Google Scholar]
- 17. Molyneaux B.J., Goff L.A., Brettler A.C., Chen H.-H.H., Brown J.R., Hrvatin S., Rinn J.L., Arlotta P.. DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron. 2015; 85:275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Siegert S., Cabuy E., Scherf B.G., Kohler H., Panda S., Le Y.-Z.Z., Fehling H.J., Gaidatzis D., Stadler M.B., Roska B.. Transcriptional code and disease map for adult retinal cell types. Nat. Neurosci. 2012; 15:487. [DOI] [PubMed] [Google Scholar]
- 19. Telley L., Govindan S., Prados J., Stevant I., Nef S., Dermitzakis E., Dayer A., Jabaudon D.. Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science (New York, N.Y.). 2016; 351:1443–1446. [DOI] [PubMed] [Google Scholar]
- 20. Barthelson R.A., Lambert G.M., Vanier C., Lynch R.M., Galbraith D.W.. Comparison of the contributions of the nuclear and cytoplasmic compartments to global gene expression in human cells. BMC Genomics. 2007; 8:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grindberg R.V., Yee-Greenbaum J.L., McConnell M.J., Novotny M., O'Shaughnessy A.L., Lambert G.M., Araúzo-Bravo M.J., Lee J., Fishman M., Robbins G.E. et al.. RNA-sequencing from single nuclei. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:19802–19807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Habib N., Avraham-Davidi I., Basu A., Burks T., Shekhar K., Hofree M., Choudhury S.R., Aguet F., Gelfand E., Ardlie K. et al.. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. methods. 2017; 14:955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krishnaswami S.R., Grindberg R.V., Novotny M., Venepally P., Lacar B., Bhutani K., Linker S.B., Pham S., Erwin J.A., Miller J.A. et al.. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat. Protoc. 2016; 11:499–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lake B.B., Codeluppi S., Yung Y.C., Gao D., Chun J., Kharchenko P.V., Linnarsson S., Zhang K.. A comparative strategy for single-nucleus and single-cell transcriptomes confirms accuracy in predicted cell-type expression from nuclear RNA. Sci. Rep. 2017; 7:6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu X., Stoyanova E.I., Lemiesz A.E., Xing J., Mash D.C., Heintz N.. Species and cell-type properties of classically defined human and rodent neurons and glia. eLife. 2018; 7:e37551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kozlenkov A., Li J., Apontes P., Hurd Y.L., Byne W.M., Koonin E.V., Wegner M., Mukamel E.A., Dracheva S.. A unique role for DNA (hydroxy)methylation in epigenetic regulation of human inhibitory neurons. Sci. Adv. 2018; 4:eaau6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J., Billis K., Carvalho-Silva D., Cummins C., Clapham P. et al.. Ensembl 2017. Nucleic Acids Res. 2017; 45:D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okonechnikov K., Conesa A., García-Alcalde F.. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016; 32:292–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anders S., Pyl P.T., Huber W.. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hrvatin S., Deng F., O’Donnell C.W., Gifford D.K., Melton D.A.. MARIS: method for analyzing RNA following intracellular sorting. PLoS One. 2014; 9:e89459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pechhold S., Stouffer M., Walker G., Martel R., Seligmann B., Hang Y., Stein R., Harlan D.M., Pechhold K.. Transcriptional analysis of intracytoplasmically stained, FACS-purified cells by high-throughput, quantitative nuclease protection. Nat. Biotechnol. 2009; 27:1038–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pan Y., Ouyang Z., Wong W.H., Baker J.C.. A new FACS approach isolates hESC derived endoderm using transcription factors. PLoS One. 2011; 6:e17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamada H., Maruo R., Watanabe M., Hidaka Y., Iwatani Y., Takano T.. Messenger RNA quantification after fluorescence activated cell sorting using intracellular antigens. Biochem. Biophys. Res. Commun. 2010; 397:425–428. [DOI] [PubMed] [Google Scholar]
- 37. Lacar B., Linker S.B., Jaeger B.N., Krishnaswami S.R., Barron J.J., Kelder M.J.E.J.E., Parylak S.L., Paquola A.C.M.C.M., Venepally P., Novotny M. et al.. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun. 2016; 7:11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Molyneaux B.J., Arlotta P., Menezes J.R., Macklis J.D.. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007; 8:427–437. [DOI] [PubMed] [Google Scholar]
- 39. Harb K., Magrinelli E., Nicolas C.S.S., Lukianets N., Frangeul L., Pietri M., Sun T., Sandoz G., Grammont F., Jabaudon D. et al.. Area-specific development of distinct projection neuron subclasses is regulated by postnatal epigenetic modifications. eLife. 2016; 5:e09531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sigurgeirsson B., Emanuelsson O., Lundeberg J.. Sequencing degraded RNA addressed by 3′ tag counting. PLoS One. 2014; 9:e91851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nikouei K., Muñoz-Manchado A.B., Hjerling-Leffler J.. BCL11B/CTIP2 is highly expressed in GABAergic interneurons of the mouse somatosensory cortex. J. Chem. Neuroanat. 2016; 71:1–5. [DOI] [PubMed] [Google Scholar]
- 42. Habib N., Li Y., Heidenreich M., Swiech L., Avraham-Davidi I., Trombetta J.J., Hession C., Zhang F., Regev A.. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science (New York, N.Y.). 2016; 353:925–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saxena A., Wagatsuma A., Noro Y., Kuji T., Asaka-Oba A., Watahiki A., Gurnot C., Fagiolini M., Hensch T.K., Carninci P.. Trehalose-enhanced isolation of neuronal sub-types from adult mouse brain. BioTechniques. 2012; 52:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ameur A., Zaghlool A., Halvardson J., Wetterbom A., Gyllensten U., Cavelier L., Feuk L.. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat. Struct. Mol. Biol. 2011; 18:1435–1440. [DOI] [PubMed] [Google Scholar]
- 45. Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L. et al.. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012; 489:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kang H.J., Kawasawa Y.I., Cheng F., Zhu Y., Xu X., Li M., Sousa A.M.M.M., Pletikos M., Meyer K.A., Sedmak G. et al.. Spatio-temporal transcriptome of the human brain. Nature. 2011; 478:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang X.-S.S., Simmons Z., Liu W., Boyer P.J., Connor J.R.. Differential expression of genes in amyotrophic lateral sclerosis revealed by profiling the post mortem cortex. Amyotroph. Lateral Scler. 2006; 7:201–210. [DOI] [PubMed] [Google Scholar]
- 48. Lederer C.W., Torrisi A., Pantelidou M., Santama N., Cavallaro S.. Pathways and genes differentially expressed in the motor cortex of patients with sporadic amyotrophic lateral sclerosis. BMC Genomics. 2007; 8:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dangond F., Hwang D., Camelo S., Pasinelli P., Frosch M.P., Stephanopoulos G., Stephanopoulos G., Brown R.H., Gullans S.R.. Molecular signature of late-stage human ALS revealed by expression profiling of postmortem spinal cord gray matter. Physiol. Genomics. 2004; 16:229–239. [DOI] [PubMed] [Google Scholar]
- 50. Offen D., Barhum Y., Melamed E., Embacher N., Schindler C., Ransmayr G.. Spinal cord mRNA profile in patients with ALS: comparison with transgenic mice expressing the human SOD-1 mutant. J. Mol. Neurosci. 2009; 38:85–93. [DOI] [PubMed] [Google Scholar]
- 51. Bossers K., Meerhoff G., Balesar R., van Dongen J.W., Kruse C.G., Swaab D.F., Verhaagen J.. Analysis of gene expression in Parkinson's disease: possible involvement of neurotrophic support and axon guidance in dopaminergic cell death. Brain Pathol. 2009; 19:91–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miller R.M., Kiser G.L., Kaysser-Kranich T.M., Lockner R.J., Palaniappan C., Federoff H.J.. Robust dysregulation of gene expression in substantia nigra and striatum in Parkinson's disease. Neurobiol. Dis. 2006; 21:305–313. [DOI] [PubMed] [Google Scholar]
- 53. Hauser M.A., Li Y.-J.J., Xu H., Noureddine M.A., Shao Y.S., Gullans S.R., Scherzer C.R., Jensen R.V., McLaurin A.C., Gibson J.R. et al.. Expression profiling of substantia nigra in Parkinson disease, progressive supranuclear palsy, and frontotemporal dementia with parkinsonism. Arch. Neurol. 2005; 62:917–921. [DOI] [PubMed] [Google Scholar]
- 54. Moran L.B., Duke D.C., Deprez M., Dexter D.T., Pearce R.K., Graeber M.B.. Whole genome expression profiling of the medial and lateral substantia nigra in Parkinson's disease. Neurogenetics. 2006; 7:1–11. [DOI] [PubMed] [Google Scholar]
- 55. Papapetropoulos S., Ffrench-Mullen J., McCorquodale D., Qin Y., Pablo J., Mash D.C.. Multiregional gene expression profiling identifies MRPS6 as a possible candidate gene for Parkinson's disease. Gene Expr. 2006; 13:205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dumitriu A., Latourelle J.C., Hadzi T.C., Pankratz N., Garza D., Miller J.P., Vance J.M., Foroud T., Beach T.G., Myers R.H.. Gene expression profiles in Parkinson disease prefrontal cortex implicate FOXO1 and genes under its transcriptional regulation. PLos Genet. 2012; 8:e1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hartong D.T., Berson E.L., Dryja T.P.. Retinitis pigmentosa. Lancet (London, England). 2006; 368:1795–1809. [DOI] [PubMed] [Google Scholar]
- 58. Mitchell J.D., Borasio G.D.. Amyotrophic lateral sclerosis. Lancet (London, England). 2007; 369:2031–2041. [DOI] [PubMed] [Google Scholar]
- 59. Sulzer D., Surmeier D.J.. Neuronal vulnerability, pathogenesis, and Parkinson's disease. Mov. Disord. 2013; 28:715–724. [DOI] [PubMed] [Google Scholar]
- 60. Arzalluz-Luque Á., Conesa A.. Single-cell RNAseq for the study of isoforms-how is that possible. Genome Biol. 2018; 19:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu S., Trapnell C.. Single-cell transcriptome sequencing: recent advances and remaining challenges [version 1; peer review: 2 approved]. F1000Research. 2016; 5:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Venkataraman A., Yang K., Irizarry J., Mackiewicz M., Mita P., Kuang Z., Xue L., Ghosh D., Liu S., Ramos P. et al.. A toolbox of immunoprecipitation-grade monoclonal antibodies to human transcription factors. Nat. Methods. 2018; 15:330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kishi J.Y., Lapan S.W., Beliveau B.J., West E.R., Zhu A., Sasaki H.M., Saka S.K., Wang Y., Cepko C.L., Yin P.. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat. Methods. 2019; 16:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Klemm S., Semrau S., Wiebrands K., Mooijman D., Faddah D.A., Jaenisch R., van Oudenaarden A.. Transcriptional profiling of cells sorted by RNA abundance. Nat. Methods. 2014; 11:549–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mouse transcriptome data have been deposited to GEO (GEO Accession # GSE130143).