Abstract
Population bottlenecks leading to a drastic reduction of the population size are common in the evolutionary dynamics of natural populations; their occurrence is known to have implications for genome evolution due to genetic drift, the consequent reduction in genetic diversity, and the rate of adaptation. Nevertheless, an empirical characterization of the effect of population bottleneck size on evolutionary dynamics of bacteria is currently lacking. In this study, we show that selective conditions have a stronger effect on the evolutionary history of bacteria in comparison to population bottlenecks. We evolved Escherichia coli populations under three different population bottleneck sizes (small, medium, and large) in two temperature regimes (37 °C and 20 °C). We find a high genetic diversity in the large in comparison to the small bottleneck size. Nonetheless, the cold temperature led to reduced genetic diversity regardless the bottleneck size; hence, the temperature has a stronger effect on the genetic diversity in comparison to the bottleneck size. A comparison of the fitness gain among the evolved populations reveals a similar pattern where the temperature has a significant effect on the fitness. Our study demonstrates that population bottlenecks are an important determinant of bacterial evolvability; their consequences depend on the selective conditions and are best understood via their effect on the standing genetic variation.
Keywords: experimental evolution, genetic drift, fitness
Introduction
Population bottlenecks impose a rapid and often drastic reduction on the population size. Such events are common and unavoidable during the evolutionary dynamics of natural populations. Fluctuations in the size of the population over time play a major role in the ecology and evolution of prokaryotic organisms (Fraser et al. 2009). Abiotic factors in the environment can lead to severe population size reduction. Examples are seasonal change (e.g., temperature fluctuations) or resource limitation that can result in only a small fraction of the population surviving a temporary selective event (e.g., via persistence or dormancy [Balaban et al. 2004]). Species interactions (i.e., biotic factors) may also lead to fluctuations in bacterial population size over time. For example, the life cycle of host-associated bacteria is often characterized by repeated population bottleneck events. For pathogenic bacteria, the transmission to a new host and the selection by the host immune system drastically reduce bacterial population size in every infection cycle (Didelot et al. 2016; Moxon and Kussell 2017). The life cycle of bacteria in mutualistic interactions (i.e., beneficial symbiosis) is also characterized by successive population bottlenecks, which typically occur at the initial stages of the host colonization due to founder effects. The effect of strong population bottlenecks is well recognized in vertically inherited bacterial symbionts, where only few bacterial cells are transferred to the next generation (e.g., as in aphid or beetle symbioses [Funk et al. 2001; McCutcheon and Moran 2012; Salem et al. 2017]). The evolution of horizontally transmitted symbionts can be characterized by strong population bottlenecks as well. Next to founder effects, priority effects in colonization may induce strong population bottlenecks for successively incoming colonizers, where the first colonizer restricts the habitat for the later incoming colonizers (Stephens et al. 2015; Wein et al. 2018). Finally, phage predation constitutes a major factor leading to repeated population bottlenecks and consequently fluctuating population size of bacterial populations (Avrani et al. 2011; Koskella and Brockhurst 2014). Notably, population bottlenecks may be either neutral due to random sampling of the bacterial population or selective—where the probability of surviving the bottleneck depends on the genotype.
The occurrence of population bottlenecks is known to have significant implications for bacterial genome evolution due to their potential to lead to genetic drift, which results in a reduction of the population genetic diversity. This is true for both types of bottlenecks, where in the adaptive bottleneck the effect of drift is limited to alleles in linkage with the selected genotype. The stochastic elimination of rare alleles from the population during genetic drift may also have consequences for the rate of adaption as they decrease the efficacy of natural selection (Brandvain and Wright 2016). Strong (or small) population bottlenecks are assumed to maintain slightly deleterious mutations in the population (Lynch et al. 1993; Elena and Lenski 2003) and thus decrease the rate of adaptation, whereas weak (or large) population bottlenecks are expected to maintain a higher rate of adaptation (Lachapelle et al. 2015; Vogwill et al. 2016). However, in large populations that experience weak neutral population bottlenecks, independently derived beneficial mutations are expected to compete in the population. The relative frequency of such beneficial mutations in the population has a direct effect on the dynamics of less beneficial mutations due to linkage disequilibrium. Under adaptive bottlenecks (i.e., strong selective conditions), this phenomenon—which has been termed Hill–Robertson effect or clonal interference (Hill and Robertson 1966; Gerrish and Lenski 1998)—can thus lead to bacterial adaptation that is driven by highly beneficial mutations and is characterized by a high probability of parallel evolution (Herron and Doebeli 2013). The dynamics of clonal interference may be nonetheless perturbed in the presence of population bottlenecks where strong bottlenecks are expected, in addition, to decrease the probability of parallel evolution (Wahl et al. 2002).
Population bottlenecks thus constitute an important determinant of allele dynamics that interferes with selective processes, including purifying selection of deleterious alleles as well as positive selection for beneficial alleles. Nonetheless, a quantification of the combined effect of population bottlenecks and selection regimes on allele frequency (AF) dynamics remains challenging due to the multiplicity of confounding factors. Here, we compare genome and phenotype evolution in Escherichia coli under three different population bottleneck sizes and two different selection regimes. Theory predicts that the population size, as imposed by the strength of the bottleneck events, has an effect on the genetic diversity depending on the environmental conditions (i.e., selection regime [Lanfear et al. 2014]). The level of adaptation is similarly expected to vary among the different combinations of population bottleneck size and selection regimes; the highest increase in fitness (i.e., rapid adaptation) is expected in the largest population bottleneck size and harshest selective conditions.
Materials and Methods
Experimental Evolution
The experiment was conducted with the E. coli K12 strain MG1655. Eight ancestral replicates were picked as single colonies from LB-agar plates and inoculates for overnight growth at 37 °C. Thereafter, the ancestral cultures were divided into two temperatures of 37 and 20 °C and three bottleneck sizes. The population bottleneck sizes were applied every serial transfer with a dilution of 1:100 (1%, large, L), 1:1,000 (0.1%, medium, M), and 1:10,000 (0.01%, small, S). The replicated populations were evolved in a total volume of 1 ml LB medium. The populations were propagated either every 12 h at 37 °C or every 24 h at 20 °C according to their growth dynamics for a total of 130 for L, 110 for M, and 90 for S transfers (i.e., bottlenecks) and ∼1,000 generations (as performed in Vogwill et al. [2016]; see supplementary fig. S1, Supplementary Material online, for a detailed calculation of generation time). Note that the experiment at 20 °C was conducted twice as long as the 37 °C experiment in order to adjust for an equal number of generations in all evolved populations.
Population Growth Analysis
The growth dynamics of E. coli populations under different population bottlenecks (S, M, and L) were measured by determining the optical density at 600 nm (OD600) using a photospectrometer (Thermo Fisher Scientific). The OD600 was measured at the onset of the experiment and was repeated every 2 h for 12 h at 37 °C and for 24 h at 20 °C with three replicates per bottleneck size and temperature. After measuring the growth, the R package growthcurver (Sprouffske and Wagner 2016) was used to fit a logistic growth model to the results and estimate the growth parameters (supplementary fig. S1, Supplementary Material online).
Sequence Analysis
Population sequencing was applied to enable the detection of variant alleles. Total DNA was isolated from 1 ml culture of the ancestor and all evolved populations using the Wizard Genomic DNA Purification Kit (Promega). Concentration and quality of the extracted DNA were assessed using the NanoDropTM (Thermo Fisher Scientific) and Qubit (Invitrogen by Life Technologies). The sample libraries for Illumina sequencing were prepared using the Nextera DNA Flex and the Nextera XT library kit (Illumina, Inc), and sequencing was performed with paired-end reads on the Hiseq system (Illumina, Inc). The median coverage in all populations ranged between 55× and 255×.
Sequencing reads were trimmed to remove Illumina specific adaptors and low-quality bases using the program Trimmomatic v.0.35 (Bolger et al. 2014) (parameters: NexteraPE-PE.fa: 2: 30: 10 CROP: 125 HEADCROP: 5 LEADING: 5 TRAILING: 5 SLIDINGWINDOW: 4: 20 MINLEN: 36). The sequencing reads were mapped to the reference genomes using BWA-MEM 0.7.16a-r1181 (Li and Durbin 2009). As the reference we used the E. coli MG1655 genome (GenBank accession number NC_000913.3). Mapping statistics were retrieved using Alfred v0.1.5 (Rausch et al. 2018). Subsequent indexing and local realignment of sequencing reads were performed using PICARD tools, SAMtools v1.6 (Li et al. 2009), and GATK v3.8-0-ge9d806836 (McKenna et al. 2010). Short indels and single nucleotide variants (SNVs) were called using LoFreq v.2.1.2 (Wilm et al. 2012). The annotation of evolved SNVs was performed with an in-house PERL script.
The analysis of the evolved population revealed cross contamination in four populations that were excluded from further analysis. Data analysis and statistical tests were performed with MatLab and R version 3.5.1. Data transformation for the statistical tests was performed with the ARTtool package (Wobbrock et al. 2011).
Nucleotide diversity (π) was calculated as in Schloissnig et al. (2013):
where G is the length of E. coli reference genome, and Xi,Bj is the count of a specific nucleotide Bj at a specific locus i with coverage Ci. The proportion of polymorphic nonsynonymous and synonymous SNVs (i.e., pN and pS) was calculated using a threshold of AF < 0.7 for polymorphic SNVs (supplementary table S1/S2, Supplementary Material online).
Competition Experiments
The relative fitness (w; Lenski et al. 1991) of the evolved versus the ancestral strain (marked; E. coli MG1655 Tmr) was estimated by direct pairwise competition experiments, with three replicates per three populations per bottleneck and temperature. Previously, we showed that the fitness difference between the marked E. coli MG1655 Tmr strain and the wild-type E. coli MG1655 is negligible (Wein et al. 2019). The starter cultures were transferred twice prior to the competition experiment. All competition experiments were initiated with a 1:1 mixture of 1:100 diluted evolved strain and ancestral strain from overnight cultures in a total volume of 1 ml of LB medium. The experiments were conducted at 20 °C for populations evolved at 20 °C and at 37 °C for populations evolved at 37 °C. The relative fitness of the evolved strains was calculated from viable cell counts at the time points 0, 24, and 48 h. The strains were distinguished through plating on nonselective (LB) and selective media (LB supplemented with trimethoprim 150 µg/ml).
Results
To test the theoretical predictions, we conducted an experimental evolution study of E. coli strain K-12 MG1655 in a serial transfer approach. The three bottleneck sizes were applied in every serial transfer with a dilution of 1:100 (1% of the total population, large, L), 1:1,000 (0.1% of the total population, medium, M), and 1:10,000 (0.01% of the total population, small, S). The populations were evolved in two environmental temperatures: 37 °C that is considered as optimal growth conditions and 20 °C that is considered suboptimal conditions for E. coli. The experiment was conducted with eight replicates for ∼1,000 generations (see Materials and Methods for details). Comparative genomics of the evolved and ancestral populations revealed 212 evolved single-nucleotide variants (SNVs; using a threshold of SNV AF >0.02), of which 122 (57%) are nonsynonymous, 27 (13%) are synonymous, and 63 (30%) are intergenic. A total of 66 (31%) SNVs were observed in more than one population (with 43 nonsynonymous and 8 synonymous parallel SNVs; supplementary table S1, Supplementary Material online).
High Genetic Diversity in the Large Bottleneck and Reduction of Diversity at 20 °C
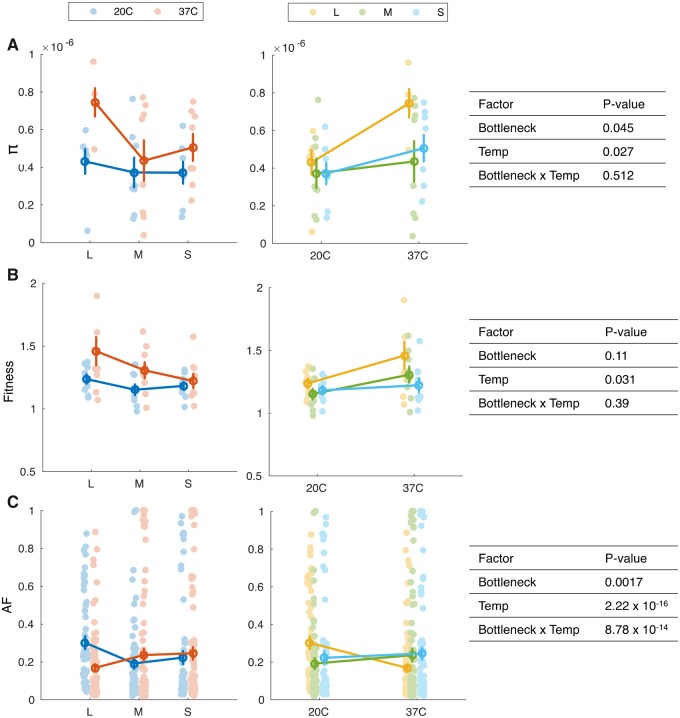
To examine the effect of bottleneck size and temperature on the degree of genetic polymorphism, we compared the nucleotide diversity among the evolved populations. Our results revealed a significant effect of the temperature and bottleneck size on the population nucleotide diversity (fig. 1A). Evolution in 20 °C resulted in overall low genetic diversity in all populations compared with populations evolved at 37 °C. The highest genetic diversity was observed in L populations and the lowest was observed in M populations in both temperature regimes (fig. 1A). Furthermore, our results show that there is no interaction between the effects of bottleneck size and temperature on the mean population nucleotide diversity; hence, the effect of the bottleneck was similar in both temperature regimes (fig. 1A). Consequently, we conclude that the population bottleneck size had an impact on the genetic diversity regardless the selection pressure imposed by the temperature in our experiment.
Fig. 1.
—The effect of bottleneck size and temperature on (A) nucleotide diversity (π), (B) relative fitness, and (C) AF. Bottleneck size is denoted as L: Large, M: Medium, or S: Small. The data are presented by dots; mean values ± SEM are marked by a circle with error bars. Note that π and fitness are presented per population, whereas AF is presented for all mutated loci in all populations. Tables on the right show the results of ANOVA two-way performed on transformed data (using aligned transformation [Wobbrock et al. 2011]). The quality of the transformation was validated by ANOVA; All transformations yielded F-values close to 0 as required. The statistical test of the effect on AF was performed including only parallel loci (i.e., evolved in ≥1 population) and the populations were considered as replicates. Fitness values >1 indicate a relative fitness increase of the evolved population compared with the ancestral population.
Significant Effect of Temperature on the Fitness of Evolved Populations
The majority of evolved SNVs are nonsynonymous, and furthermore the proportion of polymorphic synonymous sites is significantly lower than the expected by chance in most populations evolved at all conditions in our experiment (supplementary table S2 and fig. S2, Supplementary Material online). To test the effect of bottleneck size on adaptive evolution in our experiment, we compared the fitness of the evolved populations relative to the ancestral population. For that purpose, we conducted competition experiments between a marked ancestor and the evolved populations in the corresponding temperature regime. Thus, populations evolved at 20 °C were competed against the ancestor at 20 °C and populations evolved at 37 °C were competed against the ancestor at 37 °C. Our results revealed that the tested populations had an increased fitness relative to the ancestral population (fig. 1B). Our results further show that the effect of temperature on the evolved population fitness is significant. The relative fitness increase of the populations evolved at 37 °C was higher in comparison to the populations evolved at 20 °C (fig. 1B). In contrast, the bottleneck size had no significant effect on the evolved populations’ relative fitness (fig. 1B; but we note that a trend of increased fitness with population size bottleneck at 37 °C can be observed). Furthermore, there is no significant interaction between the effects of bottleneck size and temperature on the mean relative fitness; hence, the effect of bottleneck size was similar in both temperature regimes (fig. 1B). Notably, the effect of bottleneck size and temperature on the relative fitness was similar to what we observed in the comparison of genetic diversity among the evolved populations (fig. 1A).
AF Dynamics Depend on Both Bottleneck Size and Temperature
Comparing the AF of SNVs in the evolved populations, we found that both temperature and population bottleneck size had an effect on the distribution of SNV frequency in the population (fig. 1C). Furthermore, the effect of bottleneck size on the AF varied among the growth temperatures. Indeed, we found a significant interaction between temperature and bottleneck size; hence, the effect of bottleneck size on the AF depends on the growth temperature (and vice versa). Although the AF distribution in M and S populations was similar in both temperature regimes, the L populations evolved at 37 °C (L37) where characterized by lower AF in comparison to L populations evolved at 20 °C (L20; fig. 1C). A comparison of AF distribution among synonymous and nonsynonymous SNVs further showed that the observed differences in AFs are well explained by the allele dynamics of nonsynonymous rather than synonymous SNVs (supplementary fig. S3, Supplementary Material online). Notably, no SNVs reached fixation (i.e., AF > 0.9) in the L37 populations, whereas several SNVs reached a high frequency (AF > 0.9) in the M and S populations at both growth temperatures (fig. 1C;supplementary table S1, Supplementary Material online). This suggests that the effect of purifying selection varies among the bottleneck sizes.
The fixed mutations include several genetic variants that may be linked to osmotic stress (triggered by the salt concentration). Examples are fixed mutations observed in the sodium/glutamate symporter gene (gltS; M37) and the potassium uptake system. These include mutations in trkH or trkD (kup) that were fixed across the three bottleneck sizes at 37 °C (but not all replicates), as well as mutations in sapD, which were fixed only in the small bottleneck size of both temperature regimes. Further fixed mutations targeting transcriptional processes occurred across temperatures and bottleneck sizes in the DNA-dependent RNA polymerase gene (rpoC), whereas fixed mutations in rpoB only occurred in L20 populations. Mutations related to translational processes occurred mainly at 20 °C in genes encoding for ribosomal proteins, rpsG and rpsA. In addition, fixed mutations emerged in genes involved in nutrient abundance and starvation response (e.g., spoT and sspA in L20 populations) or regulation of anoxic metabolism (e.g., arcA in one M37 population). All of the fixed variants described above are nonsynonymous substitutions.
To further examine differences in AF dynamics depending on the evolutionary factors, we examined the fate of preexisting SNVs in the ancestral population. The ancestral E. coli strain in our population had four SNVs in comparison to the reference genome; of which two were at a high AF: a synonymous substitution in aroE (AFancestral = 0.98) and a synonymous substitution in yciM (AFancestral = 0.77) (supplementary table S1, Supplementary Material online). These two substitutions reached fixation (AF > 0.9) in all evolved populations (supplementary fig. S4, Supplementary Material online). One intergenic mutation remained either at a similar low frequency (AF ≈ 0.3) in some populations or slightly decreased in others regardless of the temperature conditions or bottleneck size. This intergenic ancestral variant is thus likely to be selectively neutral. An additional ancestral synonymous SNV in rpoD that was present with an AF = 0.2 decreased in frequency in almost all populations. RpoD is sigma factor that is involved in translation during exponential growth (e.g., ribosomal operons or rRNA and tRNA genes). The allele dynamics of the RpoD SNV indicate that this variant evolved under purifying selection.
Large Population Bottlenecks Show the Highest Degree of Parallel Evolution
The distribution of shared SNVs among replicate populations evolved under the same evolutionary factors revealed that more parallel SNVs were found among replicates in the L populations in comparison to the M and S populations at both temperature regimes (supplementary fig. S5, Supplementary Material online). The parallel SNVs in L populations were, however, specific to the temperature regime; this is true for the specific mutation position as well as for the gene in which the mutation occurred (fig. 2 and supplementary fig. S5, Supplementary Material online). Notably, the comparison among populations evolved under the same temperature did not reveal a strong signal of parallel evolution. Nevertheless, at 37 °C we observed more parallel variants across bottleneck sizes than at 20 °C (supplementary fig. S5, Supplementary Material online). Moreover, several of the variants detected as parallel in a specific bottleneck, for example, S20 or S37, did not occur in another bottleneck size and can thus be considered bottleneck size specific (fig. 2).
Fig. 2.
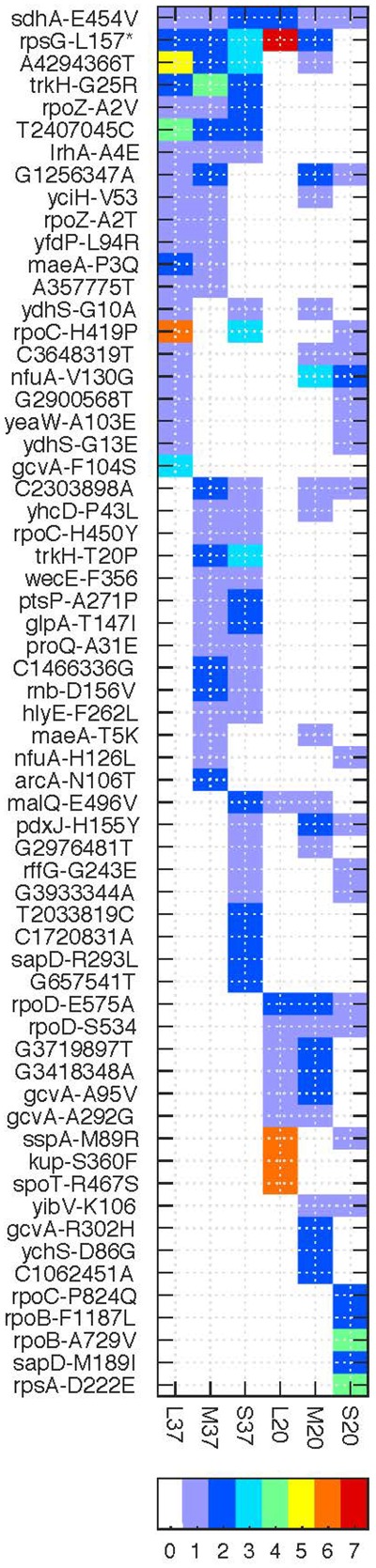
—The distribution and annotation of parallel SNVs in the evolved populations. SNVs that have been observed in >1 replicate population (AF ≥ 0.02) are presented with the number of replicated color coded according to the colorbar at the bottom. The variant annotation is listed on the left according to the following format: Intergenic variants are presented by [ancestral nucleotide][genomic position][evolved nucleotide]. Intragenic variants are presented with the gene symbol [ancestral amino acid][amino acid position][evolved amino acid]. For synonymous SVNs, the evolved amino acid is omitted; STOP codons are marked by *. Ancestral variants are excluded.
One variant was detected across all bottleneck sizes and both temperatures. The nonsynonymous SNV in sdhA was observed in eight replicate populations across all conditions (sdhA-E454V). SdhA is involved in the synthesis of fumarate from succinate and can switch function between aerobic and anaerobic metabolism (Ruprecht et al. 2009). The next most common mutation across all replicates is a nonsense mutation (premature stop codon) in the ribosomal gene rpsG that encodes for the 30S ribosomal subunit protein S7, which is involved in mRNA binding. A nonsense mutation in rpsG was previously described as advantageous in a study of E. coli adaptation to elevated salt concentrations (Wu et al. 2014).
Several SNVs are specific to the temperature regime (yet not across all bottleneck sizes or replicates). For example, at 37 °C, this includes variants in genes related to transcriptional processes such as the RNA-polymerase subunit gene rpoZ as well as the transcriptional regulator lrhA. At 20 °C, we observed two more variants in rpoD that did not emerge at 37 °C. Many parallel occurring variants include genes associated with growth (e.g., rpoB, rpoC, rpoZ); evolution of these genes has been frequently observed in E. coli in response to novel environmental conditions (Herring et al. 2006; Conrad et al. 2010; Bosshard et al. 2019). Additional parallel variants are associated with persistence in the extended stationary growth phase such as the anoxic regulation protein system arcA/B or proteins involved in starvation response, for example, spoT encoding for bifunctional (p)ppGpp synthase/hydrolase that is activated in response to nutritional changes and sspA encoding for the stringent starvation protein. Notably, only at 20 °C we observed fixed parallel substitutions in the gene encoding ribosomal protein RpsG that is involved in translation; the parallel substitutions were shared across the M and L bottleneck size indicating that the substitutions are associated with the temperature regime. Overall, most of the parallel SNVs were observed in the L populations regardless of the temperature; yet, parallel SNVs across bottlenecks were more frequent in populations evolved at 37 °C.
Discussion
The evolutionary trajectory of populations through time is influenced by the interplay of drift and selection that act on the standing genetic variation. In the context of sexually reproducing eukaryotes, the recognition that population bottlenecks reduce genetic variation was made long ago (Mayr 1963) whereas Nei et al. (1975) quantified the extent of loss in variation. Our results are in agreement with those observations, and extend their applicability to the prokaryotic domain, because populations evolved under a large bottleneck maintain the highest genetic diversity over time. Nonetheless, our results show that the differences between the medium and small bottlenecks are only marginal. The lack of difference between those bottlenecks suggests that the small population bottleneck in our experiment (105 cells) already includes a sufficient number of variants for maintaining a basic level of standing genetic variation and by that avoids a great loss of diversity within the population. Notably, the number of fixed mutations (i.e., AF ≥ 0.9) is highest in the S populations and lowest in the L populations (fig. 1C). The difference in the frequency of fixed SNVs may be explained by a strong effect of competing beneficial mutations in the L populations (i.e., clonal interference), and in addition, a strong impact of genetic drift in the S populations (fig. 1C).
The degree of standing genetic variation is expected to impose a fundamental constraint on the rate of adaptation. Our results indicate that both genetic variation and adaptation (i.e., fitness) are highest in the large bottleneck, where the differences between the population bottleneck sizes are more prominent at 37 °C. In the cold temperature regime, highly adaptive mutations may induce selective sweeps that reduce the diversity and increase the number of fixed mutations. Our results thus show that the genetic diversity, the frequency of nonsynonymous mutations and fitness are tightly linked and may be of assistance when predicting the degree of adaptation in other (natural) settings.
Additionally, we observed that the degree of parallel evolution is sensitive to the bottleneck size regardless of the environment, contrasting previous studies on the effect of serial bottlenecks (Vogwill et al. 2016). Theory predicts that the degree of parallel evolution is expected to be highest in large populations compared with small populations (Orr 2005). Thus, our findings for the effect of bottleneck size on parallel evolution are in agreement with theoretical predictions for the effect of constant population size.
The genes evolved in our experiment reveal a strong impact of our experimental batch culture system. The high abundance of variants related to growth (e.g., rpo genes) in the evolved populations indicates that the exponential growth phase, which is typical for serial transfer experiments, imposes a strong selection pressure on traits related to growth dynamics, regardless of the temperature or bottleneck size. Our results are in agreement with other experimental evolution studies of E. coli that observed evolution of growth-related genes in response to novel environmental conditions (Herring et al. 2006; Conrad et al. 2010; Tenaillon et al. 2012; Bosshard et al. 2019). In addition, genes related to persistence and nutrient starvation are important in the stationary growth phase of bacteria and thus may be important for growth in the extended stationary phase, especially in the L populations. Mutations in genes related to such adaptation seem to evolve under strong positive selection, and their allele dynamics may mask other low frequency variants that emerged during the experiment. We propose that the serial transfer approach may select for the mutations that reached the highest frequencies (i.e., of fast growers) and thus reduce the overall genetic diversity. Thus, upon the induction of the bottleneck effect (i.e., the transfer), high-frequency variants will increase whereas low-frequency variants may quickly disappear. This implies that the probability of fixation to occur in the population is not uniformly distributed across growth phases; mutations that emerge early in the growth phase have a higher probability of being fixed, whereas mutations that emerge at a later stage are a minority and therefore less likely to be fixed. This suggestion is in agreement with previously published mathematical models of the survival probability of mutations in bacterial populations grown in batch cultures (Wahl and Gerrish 2001; Wahl and Zhu 2015). We suggest that the differences in the growth dynamics between the large, and medium/small populations may have an effect on the allele dynamics within the population.
Our results indicate that the experimental conditions (i.e., selection) constitute an important factor governing the evolutionary dynamics in our experiment, and selective conditions may have a stronger effect on the evolutionary history of bacteria in comparison to repeated bottlenecks of various sizes. Exception are extremely small population bottlenecks of a single cell or only very few individuals leading to a strong genetic drift and fitness decline (Lynch et al. 1993). Nevertheless, serial bottleneck effects remain an important determinant in the evolution of bacterial populations, especially with regards to the rate of adaptation. An example for the importance of population bottlenecks in natural habitats is the impact of phage predation on the evolution of their bacterial hosts. It is well known that phage predation imposes a strong population bottleneck on the host and several studies showed that this may lead to rapid evolution of phage resistance (Paterson et al. 2010). Nonetheless, our results suggest that although the strong population bottleneck will lead to the fixation of specific genotypes (i.e., the resistant genotypes; as in our S bottleneck [fig. 1C]), at the same time, the bottleneck will lead to a purge of the host genetic diversity (as in the S bottleneck, fig. 1A). Taken together, we expect that the adaptability of the host population to other selection pressures (e.g., abiotic factors in the environment) will be significantly reduced due to repeated bottlenecks. Indeed, previous studies observed that the adaptability of bacterial populations is reduced when exposed to multiple stressors (e.g., phage and fast growth or predation and antibiotics [Avrani and Lindell 2015; Hiltunen et al. 2018]). Similarly, selection events for the dissemination of plasmids encoding for antibiotics (or metal) resistance may also impose a strong population bottleneck on the plasmid-carrying cells. Fluctuating selective conditions for the plasmid-encoded function (i.e., antibiotics) are parallel to serial population bottlenecks for the portion of the population that carries the plasmid (Wein et al. 2019). Also here, the strong selection for the plasmid presence may lead to the fixation of successful plasmid–host genotypes (De Gelder et al. 2008; Harrison et al. 2015), but, at the same time, to a reduction in the population genetic diversity, which is expected to decrease the rate of adaptation to alternative selection pressures in the environment. Thus, population bottlenecks induced by abiotic and biotic factors are expected to have a significant effect on the adaptability of bacterial population and their consequences are best understood via their effect on the standing genetic variation.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
With thank Giddy Landan, Anne Kupczok, Devani Romero Picazo, Elie Jami, Maxime Godfroid, Ana Garona, and Nils Hülter for fruitful discussions and critical comments on the manuscript. Genome sequencing was performed in the Centre for Genome Analysis Kiel that is funded by the German Research Foundation (DFG). The study was supported by the ZMB Young Scientist Grant 2018 (awarded to T.W.) and the DFG Focus Program 1819 (Grant No. DA1202/2-1; awarded to T.D.).
Data deposition: Sequence reads have been deposited at SRA under the accession PRJNA574586.
Literature Cited
- Avrani S, Lindell D.. 2015. Convergent evolution toward an improved growth rate and a reduced resistance range in Prochlorococcus strains resistant to phage. Proc Natl Acad Sci U S A. 112(17):E2191–E2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D.. 2011. Genomic island variability facilitates Prochlorococcus–virus coexistence. Nature 474(7353):604–608. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S.. 2004. Bacterial persistence as a phenotypic switch. Science 305(5690):1622–1625. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosshard L, Peischl S, Ackermann M, Excoffier L.. 2019. Mutational and selective processes involved in evolution during bacterial range expansions. Mol Biol Evol. 36(10):2313–2327. [DOI] [PubMed] [Google Scholar]
- Brandvain Y, Wright SI.. 2016. The limits of natural selection in a nonequilibrium world. Trends Genet. 32(4):201–210. [DOI] [PubMed] [Google Scholar]
- Conrad TM, et al. 2010. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci U S A. 107(47):20500–20505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gelder L, Williams JJ, Ponciano JEM, Sota M, Top EM.. 2008. Adaptive plasmid evolution results in host-range expansion of a broad-host-range plasmid. Genetics 178(4):2179–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ.. 2016. Within-host evolution of bacterial pathogens. Nat Rev Microbiol. 14(3):150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, Lenski RE.. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 4(6):457–469. [DOI] [PubMed] [Google Scholar]
- Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP.. 2009. The bacterial species challenge: making sense of genetic and ecological diversity. Science 323(5915):741–746. [DOI] [PubMed] [Google Scholar]
- Funk DJ, Wernegreen JJ, Moran NA.. 2001. Intraspecific variation in symbiont genomes: bottlenecks and the aphid-buchnera association. Genetics 157(2):477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish PJ, Lenski RE.. 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102–103:127–144. [PubMed] [Google Scholar]
- Harrison E, Guymer D, Spiers AJ, Paterson S, Brockhurst MA.. 2015. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr Biol. 25(15):2034–2039. [DOI] [PubMed] [Google Scholar]
- Herring CD, et al. 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 38(12):1406–1412. [DOI] [PubMed] [Google Scholar]
- Herron MD, Doebeli M.. 2013. Parallel evolutionary dynamics of adaptive diversification in Escherichia coli. PLoS Biol. 11(2):e1001490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A.. 1966. The effect of linkage on limits to artificial selection. Genet Res. 8(3):269–294. [PubMed] [Google Scholar]
- Hiltunen T, et al. 2018. Dual-stressor selection alters eco-evolutionary dynamics in experimental communities. Nat Ecol Evol. 2(12):1974–1981. [DOI] [PubMed] [Google Scholar]
- Koskella B, Brockhurst MA.. 2014. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 38(5):916–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachapelle J, Reid J, Colegrave N.. 2015. Repeatability of adaptation in experimental populations of different sizes. Proc R Soc B. 282(1805):20143033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Kokko H, Eyre-Walker A.. 2014. Population size and the rate of evolution. Trends Ecol Evol. 29(1):33–41. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC.. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 138(6):1315–1341. [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. ; 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Bürger R, Butcher D, Gabriel W.. 1993. The mutational meltdown in asexual populations. J Hered. 84(5):339–344. [DOI] [PubMed] [Google Scholar]
- Mayr E. 1963. Animal species and evolution. Cambridge, Massachusetts: Harvard University Press; p. 1362–4962. [Google Scholar]
- McCutcheon JP, Moran NA.. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10(1):13–26. [DOI] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon R, Kussell E.. 2017. The impact of bottlenecks on microbial survival, adaptation, and phenotypic switching in host-pathogen interactions. Evolution 71(12):2803–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Maruyama T, Chakraborty R.. 1975. The bottleneck effect and genetic variability in populations. Evolution 29(1):1.. [DOI] [PubMed] [Google Scholar]
- Orr HA. 2005. The probability of parallel evolution. Evolution 59(1):216–220. [PubMed] [Google Scholar]
- Paterson S, et al. 2010. Antagonistic coevolution accelerates molecular evolution. Nature 464(7286):275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Fritz M-Y, Korbel JO, Benes V.. 2018. Alfred: interactive multi-sample BAM alignment statistics, feature counting and feature annotation for long- and short-read sequencing. Bioinformatics 28:1530.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht J, Yankovskaya V, Maklashina E, Iwata S, Cecchini G.. 2009. Structure of Escherichia coli succinate: quinone oxidoreductase with an occupied and empty quinone-binding site. J Biol Chem. 284(43):29836–29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem H, et al. 2017. Drastic genome reduction in an herbivore's pectinolytic symbiont. Cell 171(7):1520–1531. [DOI] [PubMed] [Google Scholar]
- Schloissnig S, et al. 2013. Genomic variation landscape of the human gut microbiome. Nature 493(7430):45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouffske K, Wagner A.. 2016. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17(1):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens WZ, et al. 2015. Identification of population bottlenecks and colonization factors during assembly of bacterial communities within the zebrafish Intestine. mBio. 6(6):e01163-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, et al. 2012. The molecular divergence of adaptive convergence. Science 335(6067):457–461. [DOI] [PubMed] [Google Scholar]
- Vogwill T, Phillips RL, Gifford DR, Maclean RC.. 2016. Divergent evolution peaks under intermediate population bottlenecks during bacterial experimental evolution. Proc R Soc B. 283(1835):20160749.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl LM, Gerrish PJ.. 2001. The probability that beneficial mutations are lost in populations with periodic bottlenecks. Evolution 55(12):2606–2610. [DOI] [PubMed] [Google Scholar]
- Wahl LM, Gerrish PJ, Saika-Voivod I.. 2002. Evaluating the impact of population bottlenecks in experimental evolution. Genetics 162(2):961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl LM, Zhu AD.. 2015. Survival probability of beneficial mutations in bacterial batch culture. Genetics 200(1):309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein T, et al. 2018. Carrying capacity and colonization dynamics of Curvibacter in the Hydra host habitat. Front Microbiol. 9:443.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein T, Hülter NF, Mizrahi I, Dagan T.. 2019. Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nat Commun. 10(1):2595.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm A, et al. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 40(22):11189–11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobbrock JO, Findlater L, Gergle D, Higgens JJ.. 2011. The aligned rank transform for nonparametric factorial analyses using only ANOVA procedures. New York: ACM; p. 143–146. [Google Scholar]
- Wu X, Altman R, Eiteman MA, Altman E.. 2014. Adaptation of Escherichia coli to elevated sodium concentrations increases cation tolerance and enables greater lactic acid production. Appl Environ Microbiol. 80(9):2880–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.