Abstract
Charcot-Marie-Tooth 1A (CMT1A) is the most common inherited neuropathy without a known therapy, which is caused by a 1.4 Mb duplication on human chromosome 17, which includes the gene encoding the peripheral myelin protein of 22 kDa (PMP22). Overexpressed PMP22 protein from its gene duplication is thought to cause demyelination and subsequently axonal degeneration in the peripheral nervous system (PNS). Here, we targeted TATA-box of human PMP22 promoter to normalize overexpressed PMP22 level in C22 mice, a mouse model of CMT1A harboring multiple copies of human PMP22. Direct local intraneural delivery of CRISPR/Cas9 designed to target TATA-box of PMP22 before the onset of disease, downregulates gene expression of PMP22 and preserves both myelin and axons. Notably, the same approach was effective in partial rescue of demyelination even after the onset of disease. Collectively, our data present a proof-of-concept that CRISPR/Cas9-mediated targeting of TATA-box can be utilized to treat CMT1A.
INTRODUCTION
Charcot-Marie-Tooth disease (CMT), which affects 1 in 2500 people, is the most common hereditary peripheral neuropathy (1). The predominant subtype, CMT type 1A (CMT1A), accounts for an estimated 50% of CMT cases, and involves prominent demyelination of the peripheral nerves (2). CMT1A is associated with a duplication of the gene encoding the peripheral myelin protein of 22 kDa (PMP22) (3,4). PMP22 is an intrinsic membrane protein of myelin that is developmentally induced in Schwann cells when initiating myelination in the peripheral nerve, and PMP22 overexpression causes a dedifferentiated phenotype in Schwann cells. On the other hand, deletion of the same 1.4 Mb region is also associated with other neuropathy, hereditary neuropathy with liability to pressure palsy (HNPP) (5). Transgenic rodents harboring multiple copies of Pmp22 revealed that aberrant Pmp22 expression is linked with CMT1A-associated peripheral neuropathy (6–9) whereas heterozygous knockout of Pmp22 resembled some aspects of HNPP (10). These results indicate that proper regulation of PMP22 is required to maintain PNS myelin. Therefore, a rational therapeutic approach for CMT1A is to downregulate PMP22 expression in Schwann cells.
There have been attempts to reduce PMP22 expression for CMT1A in indirect manners. For example, effective downregulation of PMP22 by high-dose vitamin C in C22 mice led to clinical trials of vitamin C (11), which was ineffective in reducing PMP22 gene expression level in skin biopsies from treated patients (12–14). Furthermore, although, progesterone antagonist was suggested as a potential therapy as it downregulated Pmp22 gene in rodent models of Pmp22 overexpression (15,16). However, as these approaches can influence in other cellular function, directly targeting PMP22 itself would be more specific therapeutic approach. For direct targeting of PMP22, Zhao et al., recently showed that direct downregulation of PMP22 via antisense oligonucleotide (ASO) can ameliorate the clinical symptoms associated with peripheral neuropathy in animal models of CMT1A. CRISPR-based gene editing also has great potential for treating genetic diseases including CMT1A by directly targeting disease causing genes. Additionally, CRISPR-based gene editing has potential to be a single dose therapy.
There are two different promoters driving expression of PMP22, P1 and P2. Transcribed mRNAs from these promoters only differ in their 5′ non-coding region (17,18). Although, Pmp22 expression from P1 and P2 promoters are upregulated during myelination, Pmp22 expression from P1 promoter is Schwann cell specific as transcripts from P2 promoter is expressed in other non-PNS tissues. For this reason, to downregulate PMP22 in Schwann Cell, we targeted TATA-box of PMP22 P1 promoter as CRISPR-mediated TATA element disruption was shown to regulate mRNA expression (19).
Here, we tested the therapeutic efficacy of CRISPR/Cas9-mediated downregulation of PMP22 via targeting TATA-box of PMP22 P1 promoter region in the C22 mouse, a model of CMT1A by overexpressing human PMP22 gene (20), allowing for direct translation of the target sequence to the clinic.
MATERIALS AND METHODS
Preparation of sgRNA
sgRNAs were generated by in vitro transcription using T7 polymerase (New England BioLabs) according to the manufacturer's protocol.
Cell culture
A human Schwann-like cell line (ATCC crl-2885) and human Schwann cells (ScienCell) were maintained as described in the distributor's manual. Human Schwann-like cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) with high glucose (WelGene) supplemented with 10% fetal calf serum (WelGene) and 1× penicillin/streptomycin (WelGene). Human Schwann cells were maintained in the provided Schwann Cell medium (ScienCell) and cultured on 1× poly-D-Lysine coated culture dishes. For differentiation, cells were cultured with DMEM with low glucose (WelGene) supplemented with 1% fetal calf serum (WelGene) and myelination signals, consisting of 100 ng/ml Nrg1 (Peprotech) and 100 μM dbcAMP (Sigma-Aldrich), for 7 days. For transfection of CRISPR/Cas9 components, ribonucleoprotein (RNP) complexes were formed by incubating 4 μg of Cas9-HA protein (ToolGen) with 1 μg of sgRNA at room temperature for 15 min. Then, 2 × 105 cells were electroporated with the RNP complexes using a Neon electroporator (ThermoFisher) with 10 uL electroporation tips. For targeted deep sequencing, genomic DNA (gDNA) was collected from cells 72 h after transfection.
In vitro primary Schwann cell culture, CRISPR/Cas9 transfection and DNA/RNA analysis
About 3–4 weeks old C22 mice (6–10 mice/preparation) were sacrificed by CO2 gas chamber. The accompanying procedure requires a sterile environment, equipment and cell culture tools. Both sciatic nerves were exposed, dissected out. Then, the surrounding membranes and muscular tissue of isolated nerves were carefully removed under a stereomicroscope. The epineurium was stripped off with fine forceps. The remaining nerves were then transferred to tube containing ice-cold phosphate-buffered saline (PBS) and centrifuged at 1500 rpm for 10 min. For single cell dissociation, enzymatic digestion was performed with 0.05% collagenase-A solution (Sigma) for 30 min at 37°C. Enzymatic activity was stopped by fetal bovine serum (Welgene) and centrifuged for 5 min at 1500 rpm. Dissociated cells were then seeded onto poly-L-ornithine- (Sigma) and laminin (ThermoFisher) coated dishes and allowed to adhere overnight. To eliminate contaminating fibroblasts, 10 μM AraC (Sigma) was added to the medium. After 48 h, the medium was replaced by DMEM (Welgene) containing 3% FBS with 3 μM forskolin (Sigma) and 20 ng/ml neuregulin (R&D systems) to expand the cells. For transfection of CRISPR/Cas9 components, 2 × 105 cells were electroporated with the RNP complexes as described above using a Neon electroporator (ThermoFisher).
Real time PCR (qRT-PCR)
Human Schwann cells in vitro
mRNA was extracted from primary human Schwann cells using a RNeasy mini kit (Qiagen) according to the manufacturer's protocol. A total of 100 ng of mRNA was then reverse transcribed using a High-capacity cDNA reverse transcription kit (ThermoFisher). Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed with 100 ng of cDNA using Taqman Gene expression master mix according to the manufacturer's protocol (Thermo Fisher) on QuantStudio 3. The level of PMP22 expression was calculated using the CT value and GAPDH was utilized as an endogenous control. Taqman probes (Thermo Fisher) utilized in this study are listed in Supplementary Table S7.
Primary Schwann cells from C22 mice
For gene expression analysis, total RNA were extracted using RNeasy Mini Kit (QIAGEN) according to manufacturer's protocol, 5 days post transfection. cDNA was obtained using SuperScript II according to the manufacturer's protocol (Thermo Fisher) as total mRNA extracted in the manner mentioned above. qRT-PCRwas performed using Power SYBR® Green Master Mix (Thermo Fisher) protocol with the following primers:
Human P1-PMP22-F, 5′-CTTAGTCTGTCGGCTGCGGG-3′; Human P1-PMP22-R: 5′-GGCCAAACAGCGTAACCCCT-3′; Human P2-PMP22-F: 5′-CGTTAAAGGGGAACGCCAGGA-3′; Human P2-PMP22-R: 5′-CAGGGTGGCCTCAAACACAA-3′; Mouse Mpz-F: 5′-CGGACAGGGAAATCTATGGTGC-3′; Mouse Mpz-R: 5′-GCGCCAGGTAAAAGAGATGTCA-3′; Mouse P1-Pmp22-F: 5′-AGCTCCACCAGAGAACCTCTCA-3′; Mouse P1-Pmp22-R: 5′-TGAGGAGTAGCAGTGTTGGACGG-3′; Mouse P2-Pmp22-F: 5′- TGACCCGCAGCACAGCTGTCTTTG -3′; Mouse P2-Pmp22-R: 5′- TGAGGAGTAGCAGTGTTGGACGG 3′
Sciatic nerves in vivo
Total mRNA was extracted from sciatic nerves from WT and C22 mice 15 weeks post CRISPR/Cas9 administration, which had been treated with either PMP22-TATA or mRosa26 RNPs, using an RNeasy Mini Kit (Qiagen). A total of 200 ng of total mRNA was then reverse transcribed using SuperScript™ II reverse transcriptase (Thermo Fisher). The resulting cDNA (3 μl) then served as the template for PCR amplification using the following primers: human PMP22-F, 5′-CCTCAGGAAATGTCCACCAC-3′; human PMP22-R, 5′-AGCACTCATCACGCACAGAC-3′; mouse actin-F, 5′-GTGACGTTGACATCCGTAAAGA-3′; and mouse actin-R, 5′-GCCGGACTCATCGTACTCTCC-3′. Real-time PCR reactions were performed in a total volume of 10 μl that consisted of 3 μl cDNA as template, 5 μl of SYBR Green PCR master mix (Applied Biosystems) and 2.5 pmol of each primer listed above. PCR amplifications (50 cycles at 95°C for 30 s and 60°C for 60 s) were performed using an ABI QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems). To determine PMP22 expression, the average Ct values calculated from quadruplicate PCR reactions were normalized to the average Ct values for actin. These normalized values were then used to calculate PMP22 expression relative to the control according to formula 2–(meanΔΔCt).
Targeted deep sequencing
The on-target region was PCR amplified from gDNA extracted from transfected cells using Phusion polymerase (New England BioLabs). The resulting PCR amplicons were then subjected to paired-end deep sequencing using Mi-Seq (Illumina). Data from deep sequencing were analysed using the online Cas-Analyzer tool (www.rgenome.net) (21). Indels in the region 3 bp upstream from the protospacer-adjacent motif (PAM) sequence were considered to be mutations resulting from Cas9. Primer lists utilized in this study are listed in Supplementary Table S8.
In silico identification of off-target sites
Potential off-target sites were identified in silico using an on-line tool (www.rgenome.net) (21). Sequences containing up to 3 bp mismatches were considered to be off-target sites.
Digenome-seq
gDNA was purified from HeLa cells with a DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer's instructions. gDNA (10 μg) was mixed with the pre-incubated Cas9 protein (100 nM) and sgRNA (300 nM) RNP in a reaction volume of 1 ml (100 mM NaCl, 50 mM Tris–HCl, 10 mM MgCl2, 100 μg/ml bovine serum albumin, pH 7.9) and incubated at 37°C for 8 h. Digested gDNA was treated with RNase A (50 μg/ml) and purified again with a DNeasy Tissue Kit (Qiagen). A total of 1 μg of digested gDNA was fragmented using the Covaris system; the fragmented DNA was then ligated with adapters to produce libraries. These libraries were then subjected to whole genome sequencing using a HiSeq X Ten Sequencer (Illumina), at a sequencing depth of 30–40 × (Macrogen). An in vitro DNA cleavage score was calculated at each nucleotide position across the genome with a DNA cleavage scoring system described previously (22).
Mice and intraneural injection
All animals were tested in a blinded manner. After administration, all genotyping, NCS data acquisition, and histological analyses were performed separately by different researchers without information regarding the treatments. All animal studies were carried out following the guidelines set by the Samsung Animal Care and Use Committee in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care International regulations (SMC-20170206001).
The C22 mice [B6;CBACa-Tg(PMP22)C22Clh/H] were purchased from MRC Harwell (Oxfordshire, UK). Sciatic nerves of C22 and its wild-type littermate (WT) mice were treated with mRosa26 or PMP22-TATA targeting RNPs at postnatal day 6 (p6) and 21 (p21). Intraneural injection was performed by a single injection distal to the sciatic notch as previously described (23,24). A 10 μl Hamilton syringe connected to a 33-gauge needle was used for intraneural injection. RNP complexes containing 11 μg of Cas9-HA protein (ToolGen) and 2.75 μg of sgRNA per animal with Lipofectamine 3000 (Thermo Fisher) were administered. To analyze the efficiency of CRISPR/Cas9-mediated genome editing efficiency and off-target analysis in vivo, gDNAs from sciatic nerves of treated animals 11 weeks post CRISPR/Cas9 administration were collected and subjected for targeted deep-sequencing.
Immunohistochemistry and RNP distribution analysis in vivo
For immunostaining, mice were sacrificed 24 h post RNP administration. Sciatic nerves were isolated and fixed in 4% paraformaldehyde. These were then cryoprotected in 30% sucrose and processed for cryosection. Longitudinal sections were permeabilized with cold methanol and washed with PBS. Sections were then blocked and permeabilized with 10% normal goat serum and 0.3% Triton X-100 in PBS for 1 h at room temperature. Rabbit Anti-HA tag (Cell signaling Technology C29F4) were applied on sections for 24 h at 4°C followed by anti-rabbit Alexa-Fluor 488 (ThermoFisher Scientific) for 1 h at room termperatures. Sections were then mounted with ProLong Gold Antifade Mountant with DAPI (ThermoFisher Scientific). Images were taken under a Axio Scan.Z1 fluorescence microscope with Zen software (Carl Zeiss).
Electrophysiological study
To assess electrophysiological status, a nerve conduction study (NCS) was performed for WT and C22 mice 11 weeks post CRISPR/Cas9 administration as previously described (25). Briefly, mice were anesthetized with 1.5% isoflurane supplied using a nose cone for the duration of the procedure. The fur from the distal back to the hind limbs was completely removed. The NCS was performed using a Nicolet VikingQuest device (Natus Medical). For measurement of motor nerve conduction velocity, stimulating cathodes were placed at sciatic notch and 6 mm distal to the sciatic notch, while recording electrodes were placed on the muscle belly of the tibialis anterior muscle. And a ground electrode was placed at the animal's back, near the midline. Motor nerve conduction velocity (MNCV) and compound muscle action potential (CMAP) amplitudes were determined by an independent examiner who was blinded for the genotype and treatment groups. We measured CMAPs at supramaximal stimulation.
Nerve histology and imaging
The sciatic nerves were biopsied from both WT and C22 mice 11 weeks post CRISPR/Cas9 administration and pathological examinations of affected specimens were performed with microscopic analyses. Specimens were fixed overnight with 2.5% glutaraldehyde in 4% paraformaldehyde solution at 4 C. After incubation for 1 h in 1% OsO4, the specimens were dehydrated in an ethanol series, passed through propylene oxide, and embedded in epoxy resin (Epok 812, 02–1001, Oken, Japan). The tissues were cut into semi-thin (1 μm) sections (2 mm piece cut from sciatic notch where intraneural injection was performed) and stained with toluidine blue for 5 to 10 s. Semi-thin sections were imaged using Bx51 upright microscope (Olympus) and analyzed using Cell sense (Olympus). Ultrathin sections (65 nm) were collected on 200 mesh nickel grids and stained for 15 min in 2% uranyl acetate and 5 min lead citrate. The specimens were observed with a Hitachi HT7700 electron microscope at 80 kV. Electron microscope analysis was performed by the electron microscope facility of the Samsung Biomedical Research Institute. About 500 axons were counted per animal. The numbers of myelinated axons and unmyelinated axons (axons greater than 1 μm in diameter without a myelin sheath) were expressed as percentage of total counted axons. And, onion bulbs were also expressed as percentage of total counted axons. Axons with a diameter larger than 1 μm were counted from randomly selected 20–25 images from each animal (50 × 40 μm, n = 5). Determination of the g-ratio (axon diameter divided by fiber diameter) was performed by measuring the outer and inner diameter of the myelin sheath using the Zeiss Zen2 program (Carl Zeiss).
Gastrocnemius muscle examination and measurement
Biopsies were taken from the gastrocnemius muscles of WT or C22 mice 15 weeks post CRISPR/Cas9 treatments. The animals were sacrificed by CO2 inhalation. The gastrocnemius muscle was detached and weighed to compare the gastrocnemius muscle weight (mg) with the total body weight (g).
Statistical analysis
All values are expressed as mean ± SEM. The statistical significance of the data presented in Figure 1D was evaluated by one-way ANOVA with post-hoc Tukey's multiple comparison. The statistical significance of data presented in elsewhere were evaluated by student's t-test. Graphs and data generated in this study were analyzed using GraphPad Prism. The level of significance was set to 0.05.
Figure 1.
Efficient and specific downregulation of PMP22 via CRISPR/Cas9 targeting the TATA-box region of the human PMP22 gene in vitro. (A) The target sequence in the promoter region of the human PMP22 locus. TATA-box sequences are shown in red boxes and the PAM and sgRNA target sequences are shown in green and blue, respectively. (B) Indel frequencies measured by targeted deep sequencing in primary human Schawann cells. (C) Frequency of sequences with TATA-box mutations relative to the total number of sequences with indels (of (B)) as determined by targeted deep sequencing (n = 3). (D) Relative PMP22 mRNA levels in primary human Schwann cells with or without myelination signals (Nrg1, dbcAMP) and RNP complexes, measured by quantitative real-time PCR (qRT-PCR) (n = 3, ** P < 0.01). (E) Genome-wide Circos plot showing in vitro cleavage sites. Human genomic DNA is shown in gray and PMP22-TATA RNP digested genomic DNA is shown in blue. (F) Off-target sites validated in human Schwann cells by targeted deep sequencing. The mismatched nucleotides are shown in red and PAM sequences are shown in blue. Error bars indicate SEM.
RESULTS
CRISPR/Cas9-mediated genome editing of TATA-box of PMP22
To target TATA-box region of human PMP22, we searched for PAM sequences (NGG for Streptococcus pyogenes Cas9) near the TATA-box region of human PMP22 and designed eight different single guide RNAs (sgRNAs) to target this region (Supplementary Table S1). The activities of these sgRNAs were screened after transfection of sgRNA-Cas9 RNP complexes (26) into a human cell line derived from metastatic site of neurofibromatosis type 1 displaying Schwann cell-like characteristics (human Schwann-like cell line) (Supplementary Figure S1). Cas9-mediated insertion or deletion (indel) mutations at the desired sites were evaluated using targeted deep sequencing. We found that sgRNA1, gRNA4 and sgRNA7 showed high indel efficiency. From these sgRNAs we selected sgRNA1 as lead sgRNA as it showed greatest efficiency to disrupt the TATA-box from targeted deep sequencing (Data not shown), we selected this sgRNA (PMP22-TATA sgRNA hereafter) as the lead sgRNA. Bioinformatics analysis revealed that the selected PMP22-TATA sgRNA sequence is conserved in both human and mouse. The high efficiency of PMP22-TATA sgRNA in indel mutation formation was replicated in primary human Schwann cells (Figure 1A and B). Targeted deep sequencing analysis revealed that 89.54 ± 1.39% of the total indel reads corresponded to the intended TATA-box region of human PMP22 (Figure 1C and Supplementary Table S2).
Targeted in vitro deletion of TATA-box of PMP22 downregulates PMP22 expression level
To determine whether mutations in the PMP22 TATA-box lead to a reduction in human PMP22 gene expression, we performed qRT-PCR analysis. Because PMP22 is transcribed during the late stage of Schwann cell differentiation, we treated primary human Schwann cells with a known potent pro-myelination signal, consisting of Neuregulin-1 (Nrg1) and dibutryryl cyclic AMP (dbcAMP) (27), for 7 days, which resulted in an ∼9 fold induction of human PMP22 expression compared to cells without Nrg1 and dbcAMP treatment (Figure 1D). In contrast, the PMP22-TATA RNP treated culture showed an ∼6 fold induction of human PMP22 expression (Figure 1D). Schwann cells treated with a control AAVS1-targeting RNP before exposure to the differentiation signal showed no difference in human PMP22 gene expression when compared to a culture treated with the differentiation signal alone (Figure 1D).
As PMP22-TATA RNP targets PMP22 P1 transcript, we evaluated its targeting specificity. For this, we treated PMP22-TATA or mRosa26 targeting RNP in primary Schwann cells isolated from C22 mice. Successful targeting of TATA-box of P1 promoter of human and mouse PMP22 locus was confirmed by targeted deep sequencing (Supplementary Figure S2a). To determine specific targeting of P1 transcript of PMP22 we performed qRT-PCR using primers specific for human P1-PMP22 (P1-hPMP22), P2-PMP22 (P2-hPMP22), mouse P1-Pmp22 (P1-mPmp22) and mouse P2-Pmp22 (P2-mPmp22). In PMP22-TATA RNP treated culture, significant knockdown of P1-hPMP22 transcript and although not statistically significant, knockdown trend of P1-mPmp22 transcript were found whereas no changes in neither P2-hPMP22 nor P2-mPmp22 expression level when compared to mRosa26 RNP treated culture (Supplementary Figure S2b–e). These results indicate that PMP22-TATA RNP is highly specific to P1 transcript of PMP22.
No detectable off-target effects in genome edited cells
To analyze the specificity of the PMP22-TATA RNP, we performed in silico-based off-target analysis, which showed potential off-target sites containing up to 3 bp mismatches in the human genome. Targeted deep sequencing revealed no indel mutations above sequencing error rates (0.1%, on average) at the potential off-target sites that were found by in silico analysis (Supplementary Figure S3 and Table S3). Because in silico-based off-target analysis can be biased (28), we performed Digenome-seq (22), an unbiased whole genome sequencing-based off-target analysis. This approach revealed nine potential off-target sites cleaved by the PMP22-TATA RNP in vitro. However, re-analysis by targeted deep sequencing detected no aberrant indel mutations in these potential off-target sites in vivo (Figure 1E and F; Supplementary Table S4). Together, these results indicate that efficient and specific disruption of the PMP22 TATA-box by the PMP22-TATA RNP can modulate the level of PMP22 transcription in primary Schwann cells.
Establishment of intraneural CRISPR-RNP delivery to sciatic nerve and evaluation of its distribution
For PNS genome editing in vivo, we delivered CRISPR-RNP comprising sgRNA targeting mouse Rosa26 (mRosa26) and Cas9 with HA tag (Cas9-HA) encapsulated with liposome to left sciatic nerves (ipsilateral) of WT C57Bl/6 mice at postnatal day 6 (p6). The other uninjected sciatic nerves (contralateral) were utilized as control. Specifically, we injected RNP immediately distal to the sciatic notch as described before (Figure 2A) (23,24). As preassembled CRISPR-RNP was shown to degrade within 3 days in vivo (29), we sacrificed animals 24 h post-injection and prepared cryosections of injected sciatic nerves for RNP distribution. For this, we immunostained sections for HA and counterstained with DAPI to visualize nuclei. To determine RNP spread along the whole sciatic nerve, we divided the nerve into three segments (proximal, middle and distal) and evaluated HA-positive RNP containing DAPI+ nuclei (Figure 2B). We found that the percentages of HA-positive nuclei were ∼20% in the proximal, middle and distal segments (Figure 2C), indicating injected RNP was distributed evenly throughout the nerve and localized into the nucleus for gene editing.
Figure 2.
Intraneural CRISPR-RNP delivery results in efficient biodistribution of RNP in vivo. (A) Schematic diagram showing the intraneural injection into the sciatic nerve immediately distal to the sciatic notch along with the three segments (proximal, middle and distal) of the sciatic nerves utilized for analysis. (B) Representative immunostained images from contralateral (uninjected) and proximal, middle and distal segments of ipsilateral sciatic nerves (Scale bar = 50 μm). Higher magnification images of ipsilateral nerves showing nuclear localization of Cas9-HA were shown below (Scale bar = 10 μm). (C) Percentage of DAPI-positive nuclei containing HA-positive Cas9 (n = 3 nerves and mice).
Specific in vivo genome editing of TATA-box of PMP22
Having achieved efficient intraneural delivery of CRISPR-RNP, we then tested whether the PMP22-TATA RNP can regulate PMP22 transcription in vivo. We delivered RNP complexes encapsulated with liposome carrying sgRNAs targeting either mRosa26 as a control or PMP22-TATA into the left sciatic nerve (ipsilateral) of C22 mice or their wild-type littermates (WT) by intraneural injection at p6 (Figure 3A). The right sciatic nerve was used as an internal control (contralateral). We examined the gene editing efficiency of RNP complexes after intraneural delivery via targeted deep sequencing of genomic DNA collected from whole sciatic nerves 11 weeks post injection, which showed ∼11% indel formation at the appropriate locus in both mRosa26 and PMP22-TATA RNP treated sciatic nerves (Supplementary Figure S4a and Table S5). To ascertain whether the TATA-box mutations downregulated human PMP22 expression in vivo, we performed qRT-PCR analysis of mRNA extracted from whole PMP22-TATA RNP treated sciatic nerves 16 weeks post injection. In agreement with the in vitro analysis, we found that human PMP22 gene expression was significantly reduced (44.64% ± 11.41 knockdown) when compared to controls (Figure 3B).
Figure 3.
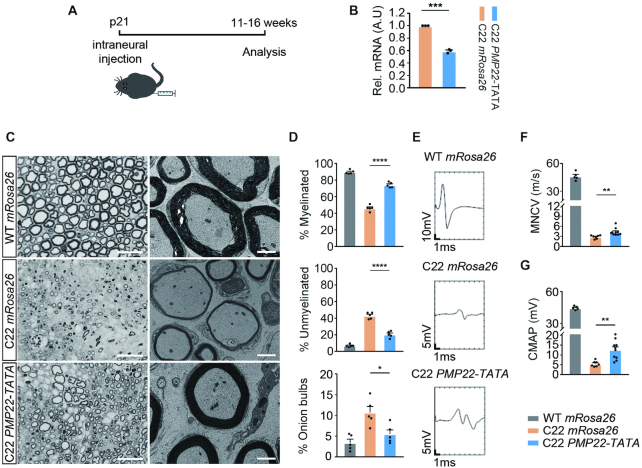
CRISPR/Cas9-mediated PMP22 downregulation prevented disease phenotypes in CMT1A mice. (A) Six-day-old C22 pups were injected with mRosa26 RNP or PMP22-TATA RNP intraneurally, and then culled at 11–16 weeks of age for analysis. (B) Relative PMP22 mRNA levels from mRosa26 or PMP22-TATA RNP treated sciatic nerves from C22 mice, measured by qRT-PCR (n = 3 for both treatments). (C) Representative semi-thin cross-section images stained with toluidine blue (left) and ultra-thin electron micrographs from same sections (right) from WT mice injected with mRosa26 RNP and C22 mice injected with mRosa26 or PMP22-TATA RNP. Scale bar = 20μm for semi-thin and for 2 μm ultra-thin sections. (D) Quantification of percentage of myelinated, unmyelinated and onion bulb axons (n = 5). (E) Representative electrophysiological trace, (F) measurement of motor nerve conduction velocity (MNCV) and (G) compound muscle action potentials (CMAP) from WT mice injected with mRosa26 RNP and C22 mice injected with mRosa26 or PMP22-TATA RNP. n = 7 for mRosa26 RNP; n = 11 PMP22-TATA RNP. n = 7 for mRosa26 RNP, n = 10 for PMP22-TATA RNP. *, P < 0.05, **, P < 0.01. ***, P < 0.005, **** P < 0.0001.
We then investigated whether the PMP22-TATA RNP caused any off-target mutations in vivo. To address this question, we performed in silico-based off-target analysis and identified eight potential off-target sites containing up to 3 bp mismatches in the mouse genome (Supplementary Table S6). Targeted deep sequencing revealed no comparable indel mutations at these sites in PMP22-TATA RNP treated ipsilateral nerves when compared to contralateral nerves (Supplementary Figure S5).
Prevention of neuropathological and electrophysiological deficits in C22 mice following targeted genome editing of TATA-box of PMP22 at p6
To investigate whether PMP22-TATA RNP-mediated reduction of PMP22 transcription prevents demyelination in C22 mice, we performed histological analysis of the semi-thin cross sections of the sciatic nerves treated with either PMP22-TATA or mRosa26 RNPs 11 weeks post injection and revealed significant reduction of unmyelinated axons and ‘onion bulbs’ (Supplementary Figure S6), a classic hallmark of repetitive segmental demyelination and remyelination of peripheral nerve (Figure 3C and D). We also measured the axon diameter and fiber diameter (axon with myelin) to determine the g-ratio, and found that it was reduced in PMP22-TATA compared to mRosa26 RNP treated nerve, which suggests that a thicker myelin sheath formed in PMP22-TATA compared to mRosa26 RNP treated nerve (Supplementary Figure S7). Furthermore, we also observed more large diameter axons in PMP22-TATA compared to mRosa26 RNP treated nerve (Figure 3C and Supplementary Figure S7).
Given the substantial improvements seen in histological analysis of myelination, we interrogated the electrophysiological profiles of both groups. We found a significantly increased motor nerve conduction velocity (MNCV; Figure 3E and F) in PMP22-TATA compared to mRosa26 treated sciatic nerves of C22 mice 10 weeks post injection, which correlates with the increment in myelin thickness and axon diameter in PMP22-TATA RNP nerves. Moreover, PMP22-TATA RNP treated nerve showed a significant increase in the amplitude of compound muscle action potential (CMAP; Figure 3E and G). Furthermore, along with histological and electrophysiological improvements, we found a significant muscle gain in PMP22-TATA compared to mRosa26 RNP treated C22 mice at 15 weeks post administration of RNPs (Supplementary Figure S8).
Neuropathological and electrophysiological rescue of C22 mice following targeted genome editing of TATA-box of PMP22 at p21
Although we have found a significant amelioration of demyelination-associated neuropathy in C22 mice by targeted mutagenesis of TATA-box of PMP22, administration of therapeutic CRISPR at p6 could be a prevention rather than treatment. To see whether our approach can modulate phenotypes after the onset of disease in C22 mice, we administered PMP22-TATA RNP at p21 intraneurally (Figure 4A) at which C22 mice exhibit neuropathological symptoms including histopathology and electrophysiology (30). In line with the prevention study (administration at p6), we achieved similar downregulation of human PMP22 measured by qRT-PCR analysis when compared to mRosa26 treated controls (40.67% ± 1.74 knockdown; Figure 4B). Downregulation of PMP22 by PMP22-TATA RNP partially rescued histopathological phenotypes in C22 mice where less number of unmyelinated axons and ‘onion bulbs’ whereas increased number of myelinated axons were found in sciatic nerve sections of PMP22-TATA RNP treated C22 mice when compared to mRosa26 treated C22 mice control (Figure 4C and D). Furthermore, improved histopathological features correlated with partial rescue of electrophysiological properties where increased MNCV and CMAP were found in sciatic nerves of PMP22-TATA RNP treated C22 mice when compared to mRosa26 treated C22 mice control (Figure 4E–G).
Figure 4.
CRISPR/Cas9-mediated PMP22 downregulation ameliorated disease phenotypes in CMT1A mice. (A) Twenty-one-day-old C22 mice were injected with mRosa26 RNP or PMP22-TATA RNP intraneurally, and then culled 11–16 weeks of age for analysis. (B) Relative PMP22 mRNA levels from mRosa26 or PMP22-TATA RNP-treated sciatic nerves from C22 mice, measured by qRT-PCR (n = 3 for both treatments). (C) Representative semi-thin cross-section images stained with toluidine blue (left) and ultra-thin electron micrographs from same sections (right) from WT mice injected with mRosa26 RNP and C22 mice injected with mRosa26 or PMP22-TATA RNP. Scale bar = 20 μm for semi-thin and for 2 μm ultra-thin sections. (D) Quantification of percentage of myelinated, unmyelinated and onion bulb axons (n = 5). (E) Representative electrophysiological trace, (F) measurement of motor nerve conduction velocity (NCV) and (G) compound muscle action potentials (CMAP) from WT mice injected with mRosa26 RNP and C22 mice injected with mRosa26 or PMP22-TATA RNP. n = 7 for mRosa26 RNP; n = 10 PMP22-TATA RNP. n = 7 for mRosa26 RNP, n = 10 for PMP22-TATA RNP. *, P < 0.05, **, P < 0.01. ***, P < 0.005, **** P < 0.0001
DISCUSSION
Overexpressed PMP22 in Schwann cells due to its duplication in CMT1A patients has been generally thought to be the cause of peripheral neuropathy including demyelination in affected patients. As heterozygous deletion of PMP22 (leads to downregulation of PMP22) cause HNPP (31), a rationale therapeutic approach for CMT1A would be normalizing PMP22 gene expression to physiological level.
Transcription of PMP22 is regulated by multiple elements such as EGR2 and SOX10 transcription factor responsive intronic enhancer region (32), the late myelination Schwann cell-specific element (LMSE) (33) and super-enhancer region located far distal from the PMP22 promoter (34,35). These multiple regulatory components of PMP22 gene transcription further support the importance of tight regulation of its expression.
It was shown that targeted TATA-box editing of c-myc gene by CRISPR/Cas9 reduced its mRNA and subsequently protein expression (19). We utilized similar approach and for specific downregulation of PMP22 in Schwann cells, TATA-box of PMP22 P1 promoter was targeted by CRISPR/Cas9. In contrast to targeting protein coding sequence of PMP22, TATA-box editing may prevent unwanted effects from knocking down PMP22 expression too low which may cause HNPP. PMP22-TATA targeting CRISPR/Cas9 treatment resulted in specific editing of TATA-box of PMP22 P1 promoter as validated by qRT-PCR-based gene expression analysis using specific primers for P1 and P2 transcripts of PMP22. Schwann cells were shown to express PMP22 driven by either P1 or P2 promoter whereas non-neural cells express PMP22 driven by P2 promoter specifically (18). Thereby, PMP22-TATA sgRNA would have reduced potential off-target in non-Schwann cells. Furthermore, unbiased whole genome sequencing followed by targeted deep sequencing revealed no genome-wide off-target cleavage, suggesting that selected lead sgRNA targeting TATA-box of PMP22 in this study is potentially translatable.
There are some intriguing results that need to be discussed. We observed ∼40% reduction in PMP22 transcripts in sciatic nerves but detected indels in only ∼10% of sequenced PMP22 transgenes. It is difficult to speculate the reason behind this without further study; however, CRISPR/Cas9-mediated in vivo gene editing studies also reported phenotypic changes that exceeds on-target indel frequency (36,37). In addition, it was suprising that the PMP22 downregulation effects appear to be quite consistent throughout the in vivo experiments. Although we achieved quite consistent indel efficiency in vivo, each CRISPR delivery thought to produce different indel formation in each cells. To explain the consistent downregulation of PMP22 in vivo, a clonal analysis with different indels of the TATA-box can be performed in the future to delineate quantitative downregulation of PMP22 upon different indels of the TATA-box. Furthermore, although PMP22-TATA RNP treatment showed phenotypic correction when compared to mRosa26 targeting RNP, this never reached wild-type level. To further enhance therapeutic efficacy, more efficient delivery vehicles are required for the PNS and intrathecal administration of certain viruses may pave the way for this (38,39).
Furthermore, although it varies, Schwann cells from CMT1A patients exhibit ∼1.7-fold PMP22 overexpression. Whereas, C22 mice harbors seven–eight copies of human PMP22, which display an ∼2.7-fold PMP22 mRNA overexpression when compared to endogenous mouse Pmp22 (8,11), which indicates that C22 mouse model is not a perfect model for CMT1A as it harbors high copy number of PMP22 and the expression level from transgene differs from CMT1A patient scenario. Therefore, transgenic animal models with low copy number of PMP22 such as C3 mice or Pmp22 transgenic rats which harbors four copies of human PMP22 or three copies of mouse Pmp22, respectively could be utilized to determine therapeutic efficacy of our strategy in more CMT1A-like in vivo scenario.
Notably, we observed therapeutic efficacy of TATA-box of PMP22 P1 promoter targeted CRISPR/Cas9 in both p6 and p21 timepoints. As previous studies observed a prominent neurological phenotype as early as p21 in c22 mice (30), our study demonstrates that targeting TATA-box of PMP22 P1 promoter by CRISPR/Cas9 not only applicable in preventative manner before demyelination (p6 timepoint), but also may applicable in treatment paradigm (p21 timepoint). Further investigations are required to test whether targeting TATA-box of PMP22 by CRISPR/Cas9 also has therapeutic effect in later stage of development, which would help determining therapeutic time window for our approach.
To our surprise, single dose of liposomal PMP22-TATA RNP delivery resulted in the partial phenotypic rescue of C22 mice. Through our analysis, we found that delivered RNPs were evenly distributed along the sciatic nerves, however only ∼20% of cells contained RNPs within their nucleus which may suggest relatively low indel efficiency (∼11% at p6) and partial not full rescue of phenotypes of C22 mice in vivo.
Although RNP-based delivery of CRISPR/Cas9 showed efficient gene editing in vivo, one should consider about immunogenicity and stability of the Cas9 protein in the host. To overcome potential immunogenicity against Cas9 protein, proper encapsulation of RNP complexes using nanoparticles required. On the other hand, the rational Cas9 engineering approach to eliminate T-cell epitopes showed promise to evade immunogenicity against Cas9 protein (40). To increase the stability of the Cas9 protein in the host, other variants of Cas9 such as Cas9 derived from a thermophilic bacterium, Geobacillus stearothermophilus (GeoCas9) can be utilized (41).
Moreover, to further evaluate therapeutic efficacy of targeting TATA-box of PMP22 promoter in vivo, systemic delivery approach to maximize PNS Schwann cell targeting efficiency such as intrathecal administration using viral vectors maybe required (42). Furthermore, this systemic delivery would allow us to investigate on behavioural improvement upon therapeutic vehicle delivery.
For successful translation of CRISPR/Cas9-based genome editing for therapy, potential unwanted off-target effects should be thoroughly considered. For this, we performed whole-genome sequencing based Digenome-seq to study off-target effects. However, this may not be enough to cover other off-target effects that are recently identified such as unintended large deletions and complex rearrangements (43). Although there are differences regarding developmental stage of target cells (Schwann cells versus mouse induced pluripotent stem cells) and modality of CRISPR/Cas9 tools (RNP versus piggybac) between current study and (43), meticulous analysis of off-target effects that could be caused by CRISPR/Cas9 should be evaluated.
While we were preparing this study, therapeutic approach involving ASO-based downregulation of PMP22 was reported to be effective in rodent models of CMT1A when delivered in repeated dose (44). In consistent with ASO-based approach, our results indicate that lowering PMP22 level has potential to be translated into the clinic. Moreover, as CRISPR/Cas9 can edit DNA, it has the advantage of potentially a single dose regimen.
In summary, our data demonstrate the proof-of-principle for PMP22 downregulation via CRISPR/Cas9 targeting TATA-box of the PMP22 promoter region. Non-viral delivery of PMP22-TATA RNP directly into the sciatic nerves of C22 mice ameliorates the neuropathological symptoms caused by PMP22 over-expression. Therefore, CRISPR/Cas9-mediated modification of the TATA-box of PMP22 promoter holds promise for the treatment of CMT1A. Furthermore, to our knowledge, apart from being the first study to demonstrate genome editing in the PNS, this is the first study of utilization of CRISPR/Cas9 to treat gene duplication mutation and further study warrants for the use of CRISPR/Cas9 for other gene duplication mutations.
DATA AVAILABILITY
The datasets generated or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions’: J.-S.L., J.Y.L., and D.W.S. designed and performed the experiments, interpreted the data, and wrote the manuscript. H.S.B., H.M.D., H.S.Y., and K.J.L. collected, analyzed, and interpreted data. H.K.K. and H.H. performed behaviour tests and statistical analysis. G.G. performed electrophysiological and pathological studies. D.K. performed Digenome-seq. J.Y.L., S.K., Y.H.B, J.M.L. and B.-O.C. supervised the project and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Korean Health Technology R&D Project, Ministry of Health & Welfare [HI14C3484, HI16C0426]; Korean Government, NRF Grants [2016R1A5A2007009, 2017R1A2B2004699, 2018R1A4A1024506, 2017M3A9B4061404, 2018M3A9H3020844]; National Institute of Health [RO1 NS094388]. Funding for open access charge: ToolGen Inc.
Conflict of interest statement. J.Y.L., D.W.S., J.M.L., H.S.B., H.S.Y., K.J.L. and S.K are employed by ToolGen. There are patent documents related to this work.
REFERENCES
- 1. Skre H. Genetic and clinical aspects of Charcot-Marie-Tooth's disease. Clin. Genet. 1974; 6:98–118. [DOI] [PubMed] [Google Scholar]
- 2. Boerkoel C.F., Takashima H., Garcia C.A., Olney R.K., Johnson J., Berry K., Russo P., Kennedy S., Teebi A.S., Scavina M. et al.. Charcot-Marie-Tooth disease and related neuropathies: mutation distribution and genotype-phenotype correlation. Ann. Neurol. 2002; 51:190–201. [DOI] [PubMed] [Google Scholar]
- 3. Lupski J.R., de Oca-Luna R.M., Slaugenhaupt S., Pentao L., Guzzetta V., Trask B.J., Saucedo-Cardenas O., Barker D.F., Killian J.M., Garcia C.A. et al.. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991; 66:219–232. [DOI] [PubMed] [Google Scholar]
- 4. Raeymaekers P., Timmerman V., Nelis E., De Jonghe P., Hoogendijk J.E., Baas F., Barker D.F., Martin J.J., De Visser M., Bolhuis P.A. et al.. Duplication in chromosome 17p11.2 in Charcot-Marie-Tooth neuropathy type 1a (CMT 1a). The HMSN Collaborative Research Group. Neuromuscul. Disord. 1991; 1:93–97. [DOI] [PubMed] [Google Scholar]
- 5. Chance P.F., Alderson M.K., Leppig K.A., Lensch M.W., Matsunami N., Smith B., Swanson P.D., Odelberg S.J., Disteche C.M., Bird T.D.. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993; 72:143–151. [DOI] [PubMed] [Google Scholar]
- 6. Sereda M., Griffiths I., Puhlhofer A., Stewart H., Rossner M.J., Zimmerman F., Magyar J.P., Schneider A., Hund E., Meinck H.M. et al.. A transgenic rat model of Charcot-Marie-Tooth disease. Neuron. 1996; 16:1049–1060. [DOI] [PubMed] [Google Scholar]
- 7. Magyar J.P., Martini R., Ruelicke T., Aguzzi A., Adlkofer K., Dembic Z., Zielasek J., Toyka K.V., Suter U.. Impaired differentiation of Schwann cells in transgenic mice with increased PMP22 gene dosage. J. Neurosci. 1996; 16:5351–5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huxley C., Passage E., Manson A., Putzu G., Figarella-Branger D., Pellissier J.F., Fontes M.. Construction of a mouse model of Charcot-Marie-Tooth disease type 1A by pronuclear injection of human YAC DNA. Hum. Mol. Genet. 1996; 5:563–569. [DOI] [PubMed] [Google Scholar]
- 9. Perea J., Robertson A., Tolmachova T., Muddle J., King R.H., Ponsford S., Thomas P.K., Huxley C.. Induced myelination and demyelination in a conditional mouse model of Charcot-Marie-Tooth disease type 1A. Hum. Mol. Genet. 2001; 10:1007–1018. [DOI] [PubMed] [Google Scholar]
- 10. Adlkofer K., Frei R., Neuberg D.H., Zielasek J., Toyka K.V., Suter U.. Heterozygous peripheral myelin protein 22-deficient mice are affected by a progressive demyelinating tomaculous neuropathy. J. Neurosci. 1997; 17:4662–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Passage E., Norreel J.C., Noack-Fraissignes P., Sanguedolce V., Pizant J., Thirion X., Robaglia-Schlupp A., Pellissier J.F., Fontes M.. Ascorbic acid treatment corrects the phenotype of a mouse model of Charcot-Marie-Tooth disease. Nat. Med. 2004; 10:396–401. [DOI] [PubMed] [Google Scholar]
- 12. Lewis R.A., McDermott M.P., Herrmann D.N., Hoke A., Clawson L.L., Siskind C., Feely S.M., Miller L.J., Barohn R.J., Smith P. et al.. High-dosage ascorbic acid treatment in Charcot-Marie-Tooth disease type 1A: results of a randomized, double-masked, controlled trial. JAMA Neurol. 2013; 70:981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nobbio L., Visigalli D., Radice D., Fiorina E., Solari A., Lauria G., Reilly M.M., Santoro L., Schenone A., Pareyson D. et al.. PMP22 messenger RNA levels in skin biopsies: testing the effectiveness of a Charcot-Marie-Tooth 1A biomarker. Brain. 2014; 137:1614–1620. [DOI] [PubMed] [Google Scholar]
- 14. Pareyson D., Reilly M.M., Schenone A., Fabrizi G.M., Cavallaro T., Santoro L., Vita G., Quattrone A., Padua L., Gemignani F. et al.. Ascorbic acid in Charcot-Marie-Tooth disease type 1A (CMT-TRIAAL and CMT-TRAUK): a double-blind randomised trial. Lancet. Neurol. 2011; 10:320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sereda M.W., Meyer zu Horste G., Suter U., Uzma N., Nave K.A.. Therapeutic administration of progesterone antagonist in a model of Charcot-Marie-Tooth disease (CMT-1A). Nat. Med. 2003; 9:1533–1537. [DOI] [PubMed] [Google Scholar]
- 16. Meyer zu Horste G., Prukop T., Liebetanz D., Mobius W., Nave K.A., Sereda M.W.. Antiprogesterone therapy uncouples axonal loss from demyelination in a transgenic rat model of CMT1A neuropathy. Ann. Neurol. 2007; 61:61–72. [DOI] [PubMed] [Google Scholar]
- 17. Bosse F., Zoidl G., Wilms S., Gillen C.P., Kuhn H.G., Muller H.W.. Differential expression of two mRNA species indicates a dual function of peripheral myelin protein PMP22 in cell growth and myelination. J. Neurosci. Res. 1994; 37:529–537. [DOI] [PubMed] [Google Scholar]
- 18. Suter U., Snipes G.J., Schoener-Scott R., Welcher A.A., Pareek S., Lupski J.R., Murphy R.A., Shooter E.M., Patel P.I.. Regulation of tissue-specific expression of alternative peripheral myelin protein-22 (PMP22) gene transcripts by two promoters. J. Biol. Chem. 1994; 269:25795–25808. [PubMed] [Google Scholar]
- 19. Slobodin B., Han R., Calderone V., Vrielink J., Loayza-Puch F., Elkon R., Agami R.. Transcription impacts the efficiency of mRNA translation via Co-transcriptional N6-adenosine methylation. Cell. 2017; 169:326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huxley C., Passage E., Robertson A.M., Youl B., Huston S., Manson A., Saberan-Djoniedi D., Figarella-Branger D., Pellissier J.F., Thomas P.K. et al.. Correlation between varying levels of PMP22 expression and the degree of demyelination and reduction in nerve conduction velocity in transgenic mice. Hum. Mol. Genet. 1998; 7:449–458. [DOI] [PubMed] [Google Scholar]
- 21. Park J., Lim K., Kim J.S., Bae S.. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics. 2017; 33:286–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.S.. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015; 12:237–243. [DOI] [PubMed] [Google Scholar]
- 23. Gonzalez S., Fernando R.N., Perrin-Tricaud C., Tricaud N.. In vivo introduction of transgenes into mouse sciatic nerve cells in situ using viral vectors. Nat. Protoc. 2014; 9:1160–1169. [DOI] [PubMed] [Google Scholar]
- 24. Sargiannidou I., Kagiava A., Bashiardes S., Richter J., Christodoulou C., Scherer S.S., Kleopa K.A.. Intraneural GJB1 gene delivery improves nerve pathology in a model of X-linked Charcot-Marie-Tooth disease. Ann. Neurol. 2015; 78:303–316. [DOI] [PubMed] [Google Scholar]
- 25. Lee J., Jung S.C., Joo J., Choi Y.R., Moon H.W., Kwak G., Yeo H.K., Lee J.S., Ahn H.J., Jung N. et al.. Overexpression of mutant HSP27 causes axonal neuropathy in mice. J. Biomed. Sci. 2015; 22:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim S., Kim D., Cho S.W., Kim J., Kim J.S.. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014; 24:1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arthur-Farraj P., Wanek K., Hantke J., Davis C.M., Jayakar A., Parkinson D.B., Mirsky R., Jessen K.R.. Mouse schwann cells need both NRG1 and cyclic AMP to myelinate. Glia. 2011; 59:720–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chuai G.H., Wang Q.L., Liu Q.. In silico meets in vivo: towards computational CRISPR-Based sgRNA Design. Trends Biotechnol. 2017; 35:12–21. [DOI] [PubMed] [Google Scholar]
- 29. Kim K., Park S.W., Kim J.H., Lee S.H., Kim D., Koo T., Kim K.E., Kim J.H., Kim J.S.. Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration. Genome Res. 2017; 27:419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Verhamme C., King R.H., ten Asbroek A.L., Muddle J.R., Nourallah M., Wolterman R., Baas F., van Schaik I.N.. Myelin and axon pathology in a long-term study of PMP22-overexpressing mice. J. Neuropathol. Exp. Neurol. 2011; 70:386–398. [DOI] [PubMed] [Google Scholar]
- 31. van Paassen B.W., van der Kooi A.J., van Spaendonck-Zwarts K.Y., Verhamme C., Baas F., de Visser M.. PMP22 related neuropathies: Charcot-Marie-Tooth disease type 1A and hereditary neuropathy with liability to pressure palsies. Orphanet. J. Rare Dis. 2014; 9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jones E.A., Lopez-Anido C., Srinivasan R., Krueger C., Chang L.W., Nagarajan R., Svaren J.. Regulation of the PMP22 gene through an intronic enhancer. J. Neurosci. 2011; 31:4242–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maier M., Castagner F., Berger P., Suter U.. Distinct elements of the peripheral myelin protein 22 (PMP22) promoter regulate expression in Schwann cells and sensory neurons. Mol. Cell Neurosci. 2003; 24:803–817. [DOI] [PubMed] [Google Scholar]
- 34. Lopez-Anido C., Poitelon Y., Gopinath C., Moran J.J., Ma K.H., Law W.D., Antonellis A., Feltri M.L., Svaren J.. Tead1 regulates the expression of Peripheral Myelin Protein 22 during Schwann cell development. Hum. Mol. Genet. 2016; 25:3055–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pantera H., Moran J.J., Hung H.A., Pak E., Dutra A., Svaren J.. Regulation of the neuropathy-associated Pmp22 gene by a distal super-enhancer. Hum. Mol. Genet. 2018; 27:2830–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Long C., McAnally J.R., Shelton J.M., Mireault A.A., Bassel-Duby R., Olson E.N.. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014; 345:1184–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. et al.. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015; 520:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kagiava A., Sargiannidou I., Theophilidis G., Karaiskos C., Richter J., Bashiardes S., Schiza N., Nearchou M., Christodoulou C., Scherer S.S. et al.. Intrathecal gene therapy rescues a model of demyelinating peripheral neuropathy. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E2421–E2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kagiava A., Karaiskos C., Richter J., Tryfonos C., Lapathitis G., Sargiannidou I., Christodoulou C., Kleopa K.A.. Intrathecal gene therapy in mouse models expressing CMT1X mutations. Hum. Mol. Genet. 2018; 27:1460–1473. [DOI] [PubMed] [Google Scholar]
- 40. Ferdosi S.R., Ewaisha R., Moghadam F., Krishna S., Park J.G., Ebrahimkhani M.R., Kiani S., Anderson K.S.. Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nat. Commun. 2019; 10:1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harrington L.B., Paez-Espino D., Staahl B.T., Chen J.S., Ma E., Kyrpides N.C., Doudna J.A.. A thermostable Cas9 with increased lifetime in human plasma. Nat. Commun. 2017; 8:1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kagiava A., Kleopa K.A.. Intrathecal delivery of viral vectors for gene therapy. Methods Mol. Biol. 2018; 1791:277–285. [DOI] [PubMed] [Google Scholar]
- 43. Kosicki M., Tomberg K., Bradley A.. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018; 36:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao H.T., Damle S., Ikeda-Lee K., Kuntz S., Li J., Mohan A., Kim A., Hung G., Scheideler M.A., Scherer S.S. et al.. PMP22 antisense oligonucleotides reverse Charcot-Marie-Tooth disease type 1A features in rodent models. J. Clin. Invest. 2018; 128:359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated or analyzed during the current study are available from the corresponding author on reasonable request.