Abstract
Norine, the unique resource dedicated to nonribosomal peptides (NRPs), is now updated with a new pipeline to automate massive sourcing and enhance annotation. External databases are mined to extract NRPs that are not yet in Norine. To maintain a high data quality, successive filters are applied to automatically validate the NRP annotations and only validated data is inserted in the database. External databases were also used to complete annotations of NRPs already in Norine. Besides, annotation consistency inside Norine and between Norine and external sources have reported annotation errors. Some can be corrected automatically, while others need manual curation. This new approach led to the insertion of 539 new NRPs and the addition or correction of annotations of nearly all Norine entries. Two new tools to analyse the chemical structures of NRPs (rBAN) and to infer a molecular formula from the mass-to-charge ratio of an NRP (Kendrick Formula Predictor) were also integrated. Norine is freely accessible from the following URL: https://bioinfo.cristal.univ-lille.fr/norine/
INTRODUCTION
Norine has been and remains the unique resource dedicated to nonribosomal peptides (NRPs) (1). These secondary metabolites are produced by bacteria and fungi and display a diverse spectrum of biological activity. They are called peptides because they are composed minimally of amino acids connected by peptide bonds and because their length is between two and 26 building blocks. In fact, >500 different building blocks, called monomers, are observed in these peptides, such as derivatives of the proteinogenic amino acids, rare amino acids, fatty acids or carbohydrates. In addition, various types of bonds connect their monomers such as disulfide or phenolic bonds. Some monomers can connect with up to five other monomers, making cycles or branches in the structure of the NRPs. This structural diversity leads to multiple biological functions, which can be further developed to yield pharmaceuticals, biocontrol agents, biocosmetics and bio-cleansing. These molecules have in common that they are synthesized by nonribosomal peptide synthetases. As their name suggests, they are not synthesized following DNA transcription through translation by ribosomes. Nonribosomal peptides synthetases form huge enzymatic complexes that select amino acids or other monomers and connect them with several types of bonds (2).
Norine is a platform dedicated to these compounds. The database stores only natural nonribosomal peptides and it is complemented with analysis tools. In recent years, Norine has been extended and improved to address the needs of distinct scientific communities (mainly biologists and biochemists but also pharmacists among others). In particular, we developed a new pipeline in order to (i) massively add new NRPs and new annotations from external databases and (ii) enhance the quality of these annotations via automatic validation procedures. We also developed new tools processing NRP chemical structures as well as mass spectra.
Furthermore, the web interface has been upgraded to ease the retrieval and the reading of the data. A powerful query builder now allows combining multiple criteria to optimize the database search. A query is built dynamically by the user that can search for any term in any NRP annotation. In the case of annotations with a limited number of possible terms, the term list is displayed and one or several values can be selected. In other cases, an auto-complete feature is provided. Several criteria can be combined using Boolean operators such as AND, OR and AND NOT.
The NRP description page has been refactored. The different categories of annotations are accessible by tabs, allowing more detailed annotation display. For example, the producing organisms are located in a taxonomic tree and a schema of the chemical structure identifies the monomers by different colors generated by in-house tools from the SMILES (Simplified Molecular Input Line Entry Specification) codes.
The pipeline to automate massive sourcing and enhance annotation as well as the new tools are described in the following sections.
AUTOMATIC AND MASSIVE SOURCING
In 2015, Norine was opened to crowd-sourcing through the creation of MyNorine (1) a tool that grants the scientific community access to Norine for submitting new NRPs or suggesting modifications of existing entries. NRP annotation is undoubtedly best achieved by experts. Submissions are manually verified and validated by the Norine team to ensure correctness and quality of data. About 50 contributors registered to MyNorine up to now, and ∼90 new NRPs or modification of annotations have been submitted and validated.
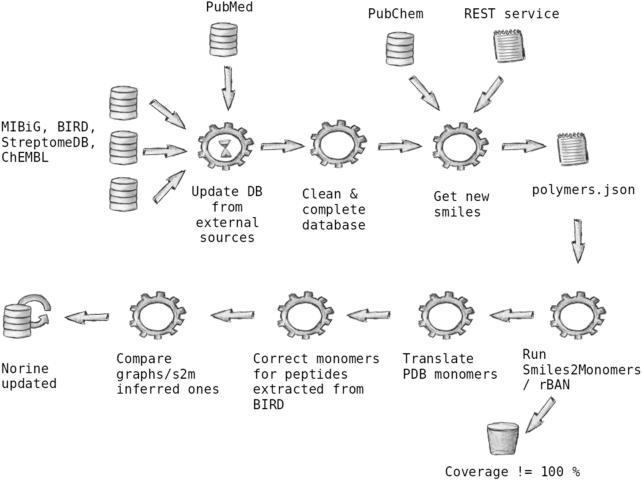
Our next aim was to automatically and massively fill the Norine database with new NRPs or to refine annotations using external resources, without quality loss. To reach this goal, we have developed a new pipeline (see Figure 1 and a detailed description in the supplementary material). Three main databases were targeted for NRP sourcing:
MIBiG (3) (Minimum Information about a Biosynthetic Gene Cluster) is a repository of known biosynthetic gene clusters of secondary metabolites. MIBiG provides community standards for annotations and metadata on biosynthetic gene clusters and their molecular products. A tarball with all entries in raw JSON format is freely available for download.
BIRD (4) (Biologically Interesting Molecule Reference Dictionary) is a resource that provides information about biologically interesting peptide-like antibiotics and inhibitor molecules in the PDB (5) archive. The entire BIRD resource can be downloaded from the wwPDB (World Wide Protein Data Bank) server in CIF format.
StreptomeDB (6) is a database of molecules produced by bacteria in the Streptomyces genus, well known for their prolific production of NRPs. Again, StreptomeDB provides a link to download a file containing all entries.
Figure 1.
Global view of the update process of the Norine database.
In the following paragraphs, we describe the pipeline through which data is fetched and selected for insertion in the Norine database. This pipeline is composed of Python scripts that are sequentially executed. For each database, the last version is downloaded and the files are checked before executing the update process. The scripts parse all entries to extract the putative nonribosomal peptides from the external sources. Keywords distinguish the NRPs from the other types of secondary metabolites stored in the external entries. Then, several checks verify if these NRPs are already in Norine or not (see annotation quality enhancement 1 section and supplementary material for more details). Moreover, the source field is automatically instantiated with the name of the source database, the status field is set to unreviewed value and a link to the source entry is recorded. These annotations associated with each newly and automatically added NRP guarantee data traceability.
MIBiG is the first external resource that is parsed. At this point in time, the archive contains ∼1800 entries. Thus, the first script parses all JSON entries and fills the database with the following annotations about the new NRPs: peptide name, family, synonyms, structure type, accession number of the corresponding MIBiG entry and PubChem ID when available, in order to reference MIBiG and PubChem (7) in the corresponding NRP entry of Norine. Finally, the SMILES are fetched for these new Norine entries but not necessarily added. The selection process is detailed in the annotation quality enhancement section 1. As MIBiG is more focused on gene clusters, some NRP structures are incomplete and do not reach Norine quality criteria.
A second script handles entries of BIRD. The CIF files contain information about the chemistry, the biology and structures of ∼1300 molecules. As for MIBiG entries, the script parses these files for finding new NRPs and annotations. The BIRD repository already references 12 Norine entries and, with the script, the cross-links between Norine and BIRD could be extended up to 171 entries. Classic annotation (name, type, synonyms, SMILES, etc.) is completed with formula, molecular weight and monomer composition generated by using an in-house conversion table. Indeed, Norine relies on its own monomer notation which can differ from that of PDB.
The addition of new NRPs from StreptomeDB follows the same process. Then, a dedicated script completes general NRP annotations with chemical activity, producing organisms and references to PubMed IDs (https://www.ncbi.nlm.nih.gov/pubmed/).
Once these scripts are executed, additional annotations are extracted from other sources. When PubMed IDs are fetched, the E-Utilities (Entrez Programming Utilities) API gives access to the entire references associated with NRPs (both existing and new ones) such as article title, full journal name and DOI. In the same way, the PubChem IDs give access to the SMILES of the compound. Also, links and chemical annotations from ChEMBL (8), a manually curated database of bioactive molecules with drug-like properties, are retrieved using a REST service (9). Unfortunately, not all SMILES of the three databases could be compared to the SMILES of Norine, because most NRPs entries in Norine did not contain SMILES information. In order to solve this discrepancy, all NRP entries lacking SMILES data were manually searched by name in chemical databases to retrieve the missing information. The execution of the pipeline led to the insertion of 539 new NRPs in Norine. Table 1 shows the contribution of the main external databases and the 666 external links that have been updated or added. Also, about 527 SMILES and 393 references were updated or added.
Table 1.
New data for Norine
| - | MIBiG | BIRD | StreptomeDB | ChEMBL |
|---|---|---|---|---|
| New NRPs | 293 | 171 | 75 | - |
| External links | ||||
| (NRPs updated/added) | 19/307 | - /171 | 2/80 | 49/38 |
| Associated references | ||||
| (NRPs updated/added) | 20/10 | - /1 | 8/286 | 65/3 |
ANNOTATIONS QUALITY ENHANCEMENT
The quality of NRP annotation in Norine has been and remains our top priority. Since its creation in 2006 (10), Norine NRP curation has relied on manual extraction from scientific literature as well as meticulous validation prior to insertion in the database. Of course, some errors or incorrect information may occur but these are removed through regular checks. As cited in the first section, the MyNorine tool has been created to boost precision. Nonetheless, automatic massive sourcing from external databases is needed to boost the number of entries, but it also increases the risk of introducing incorrect data. That is the reason why a strict validation pipeline was created. By strict validation, we mean that no entry is added if validation filters fail. Figure 1 shows some validation filters used during the execution process of the pipeline. The following section provides details of some of these filters.
First of all, we obviously verify that any external NRP or particular annotation (i.e. synonym, reference, access code, etc.) is not already present in Norine before adding it. If a peptide name is already in Norine, missing annotations for this NRP are added. When a graph (monomer composition of the NRP) is available, the Norine and external graphs are compared. If differences are detected, a failure report is generated. Various reports are created during the process to highlight inconsistencies or errors. These reports are intended to be manually analysed to curate the data. A second verification filter targets the new SMILES that were missing for many NRPs of Norine and that are fetched from external databases. For that purpose, we use Smiles2Monomers (s2m) (11) and rBAN (12), in-house tools that check the consistency of the graph. They infer the monomeric structure of an NRP from its SMILES with two distinct strategies (see next section for description of rBAN). Thus, the new SMILES is only added if the inferred graph is the same as the given NRP graph. s2m and rBAN are used to detect and correct potential errors in the SMILES or monomeric structures recorded in Norine. More than 50 structures were corrected in that way.
Another filter compares all SMILES to check if two NRPs are identical but registered with different names. SMILES are canonized prior to this comparison. Other filters not detailed in this article were developed to enhance the quality of annotation in order to rebuild the taxonomy tree of the producing organisms, remove duplicates, convert PDB monomers notation, etc.
NEW SOFTWARE
Norine has extended its range of NRP-dedicated software by including two additional tools.
rBAN (12) (retroBiosynthetic Analysis of Nonribosomal peptides) infers the monomeric structure of a NRP from a SMILES code. This process can also be qualified as simulating the retro-biosynthesis of NRPs. The first step in rBAN is the fragmentation of a molecule that is broken following a set of pattern bonds. Then, the resulting fragments are matched to Norine monomers. The tool can be run in a ‘discovery mode’ when a monomer cannot be matched to Norine. Then, missing substructure(s) are searched in PubChem so as to suggest potential new monomers. All results are displayed in a directed graph format highlighting the bond types between monomers. rBAN addresses some of the limitations of s2m cited above. URL: http://bioinfo.cristal.univ-lille.fr/rban
The Kendrick Formula Predictor is a tool that uses the Kendrick mass defect (KMD) (13) to predict the chemical formula from the mass-to-charge ratio of a NRP. The required data for the development of the method is extracted from Norine and PubChem. The software was tested with high resolution mass spectrometry data from the surfactin family and the results confirmed the capacity of the tool to successfully predict NRP molecular formulae. Note that this is the first tool in Norine specifically dedicated to the mass spectrometry of NRPs. URL: https://bioinfo.cristal.univ-lille.fr/kendrick-webapp/
Both tools have contributed to the curation/extension of the Norine database. A pipeline combining PubChem and rBAN led to the validation of 97.26% of the records in Norine, a two-fold extension of its SMILES and the introduction of 11 new monomers using the discovery mode (12). Kendrick Formula Predictor was used to add missing molecular formulas in Norine increasing the percentage of entries containing this information to 95%. From these formulas, the exact mono-isotopic masses of the peptides were calculated.
CONCLUSION
This paper describes a substantial update of the Norine database. Almost 500 new NRPs and hundreds of annotations for existing Norine NRPs (SMILES, chemical formulas, synonyms, references, external links, etc.) have been added. The quality of annotation was significantly enhanced. To achieve this goal, we developed a pipeline that relies on three main databases that potentially contain NRPs, namely, MIBiG, BIRD and StreptomeDB, and other resources such as PubMed, PubChem or ChEMBL to complement the fetched data. It should be noted that the status of these new NRPs is tagged as unreviewed. Our pipeline checks the data before insertion into the database. For example the monomeric composition is inferred from a SMILES using tools such as rBAN or s2m. If in doubt, a failure report is generated during the execution process of the pipeline to facilitate manual verification by experts. In that sense, the process also enhances the quality of manual annotation that is in turn validated or not by automatic checking. Finally, Norine benefits from two complementary data sources: expert annotations input through the MyNorine tool and automatic annotations produced with the pipeline. In a virtuous circle, data is entered manually by experts, is verified and possibly completed and corrected automatically. Alternatively, data is entered automatically by the pipeline, is verified and possibly completed and corrected manually. The history of all changes for each NRP is kept and easily available from the NRP description page for traceability.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all scientists that have contributed to Norine by inserting new peptides or new annotations related to an already known NRP.
AF is the main developer of Norine and supervises other developers. E.R. has developed rBAN, the Kendrick Formula Predictor, and parts of the pipeline in relation to peptide structures and also participates in data correction. C.C. participates in the development of Norine. M.C. participates in the design of the Kendrick Formula Predictor, and in data correction. Y.D. and J.M. participated in the development of the pipeline and Y.D. in data correction. P.J., C.F., F.L., V.L. and M.P. supervise Norine database and tool design as well as data curation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CPER Alibiotech project and the INTERREG V France-Wallonie-Vlaanderen Project SmartBioControl/BioScreen; Institut Français de Bioinformatique [ANR-11-INBS-0013 to J.M.]; SIB Swiss Institute of Bioinformatics Fellowship Programme (to E.R.); Mobility between the CRIStAL and PIG teams is supported by the Germaine de Stael French-Swiss cooperation programme [39517NH]. Funding for open access charge: University of Lille (SmartBioControl/BioScreen project).
Conflict of interest statement. None declared.
REFERENCES
- 1. Flissi A., Dufresne Y., Michalik J., Tonon L., Janot S., Noé L., Jacques P., Leclère V., Pupin M.. Norine, the knowledgebase dedicated to non-ribosomal peptides, is now open to crowdsourcing. Nucleic Acids Res. 2016; 44:D1113–D1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Süssmuth R.D., Mainz A.. Nonribosomal peptide synthesis-principles and prospects. Angew. Chem. Int. Ed. 2017; 56:3770–3821. [DOI] [PubMed] [Google Scholar]
- 3. Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K., de Bruijn I., Chooi Y.H., Claesen J., Coates R.C. et al.. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015; 11:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Young J.Y., Feng Z., Dimitropoulos D., Sala R., Westbrook J., Zhuravleva M., Shao C., Quesada M., Peisach E., Berman H.M.. Chemical annotation of small and peptide-like molecules at the protein data Bank. Database. 2013; 2013:bat079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Costanzo L.D., Christie C., Duarte J.M., Dutta S., Feng Z. et al.. Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019; 47:D520–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klementz D., Döring K., Lucas X., Telukunta K.K., Erxleben A., Deubel D., Erber A., Santillana I., Thomas O.S., Bechthold A. et al.. StreptomeDB 2.0-an extended resource of natural products produced by streptomycetes. Nucleic Acids Res. 2016; 44:D509–D514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B. et al.. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019; 47:D1102–D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gaulton A., Hersey A., Nowotka M., Bento A.P., Chambers J., Mendez D., Mutowo P., Atkinson F., Bellis L.J., Cibriàn-Uhalte E. et al.. The ChEMBL database in 2017. Nucleic Acids Res. 2017; 45:D945–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davies M., Nowotka M., Papadatos G., Dedman N., Gaulton A., Atkinson F., Bellis L., Overington J.P.. ChEMBL web services: streamlining access to drug discovery data and utilities. Nucleic Acids Res. 2015; 43:W612–W620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caboche S., Pupin M., Leclère V., Fontaine A., Jacques P., Kucherov G.. NORINE: a database of nonribosomal peptides. Nucleic Acids Res. 2008; 36:D326–D331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dufresne Y., Noé L., Leclère V., Pupin M.. Smiles2Monomers: a link between chemical and biological structures for polymers. J. Cheminform. 2015; 7:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ricart E., Leclère V., Flissi A., Mueller M., Pupin M., Lisacek F.. rBAN: retro-biosynthetic analysis of nonribosomal peptides. J. Cheminform. 2019; 11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kendrick E. A mass scale based on CH2 = 14.0000 for high resolution mass spectrometry of organic compounds. Anal. Chem. 1963; 35:2146–2154. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.