Abstract
Here, we presented an integrative database named DrLLPS (http://llps.biocuckoo.cn/) for proteins involved in liquid–liquid phase separation (LLPS), which is a ubiquitous and crucial mechanism for spatiotemporal organization of various biochemical reactions, by creating membraneless organelles (MLOs) in eukaryotic cells. From the literature, we manually collected 150 scaffold proteins that are drivers of LLPS, 987 regulators that contribute in modulating LLPS, and 8148 potential client proteins that might be dispensable for the formation of MLOs, which were then categorized into 40 biomolecular condensates. We searched potential orthologs of these known proteins, and in total DrLLPS contained 437 887 known and potential LLPS-associated proteins in 164 eukaryotes. Furthermore, we carefully annotated LLPS-associated proteins in eight model organisms, by using the knowledge integrated from 110 widely used resources that covered 16 aspects, including protein disordered regions, domain annotations, post-translational modifications (PTMs), genetic variations, cancer mutations, molecular interactions, disease-associated information, drug-target relations, physicochemical property, protein functional annotations, protein expressions/proteomics, protein 3D structures, subcellular localizations, mRNA expressions, DNA & RNA elements, and DNA methylations. We anticipate DrLLPS can serve as a helpful resource for further analysis of LLPS.
INTRODUCTION
In eukaryotes, cellular compartmentalization is a fundamental principle to dynamically and transiently organize complex biochemical reactions within distinct chemical environments, by forming membrane-bound compartments or membraneless organelles (MLOs) (1,2). Although the former have been well documented, the latter were poorly understood until recent advances in mechanistic analyses of protein phase separation, or liquid–liquid phase separation (LLPS) (1–7). LLPS provides a simple but critical mechanism to interpret how cells can spatiotemporally create MLOs, through condensing solutions of biomolecules such as proteins or nucleic acids into dense-phase liquid droplets that coexist with the dilute-phase cytoplasm (2,5–8). More formally, chemical potentials drive LLPS which in turn forms MLOs, and multiple factors including fusion and Ostwald ripening can contribute to droplet size (2,9,10). To date, a large number of MLOs have been discovered, including but not limited to stress granule, processing body (P-body), P granule, centrosome, spindle apparatus and nucleolus (1,5,11). Besides MLOs, LLPS also contributes to the formation of other subcellular structures such as heterochromatin, nuclear pore complex and receptor clusters, although conventional macromolecular assembly mechanisms are also important for nuclear pore formation (1,7,12–14). Collectively, MLOs assembled via LLPS were termed with a unique name, biomolecular condensates, which play critical roles in regulating a variety of biological processes such as stress response, RNA metabolism, DNA damage response and signal transduction (6,7,10,15,16). In living cells, the nucleation, formation and biological properties of biomolecular condensates are precisely regulated, while dysregulation or mutations of LLPS-associated proteins have been linked with human diseases such as neurodegeneration and cancer (4,5,15).
The identification of proteins undergoing LLPS is the foundation of understanding the molecular mechanisms of LLPS. Two types of proteins undergoing LLPS have been discovered, including structured proteins with multiple folded domains and intrinsically disordered proteins (IDPs) (7,17,18). Protein LLPS is mediated by weak multivalent interactions, such as electrostatic, cation–π, π–π and hydrophobic interactions, whereas protein–protein interactions (PPIs), protein–RNA interactions, post-translational modifications (PTMs), mutations and various cellular factors dynamically regulate the stability and state of protein condensates (3,4,7,19). For example, although either PGL-1 or PGL-3, two components of the P granule in germline cells of Caenorhabditis elegans, can form liquid droplets in vitro, mixing PGL-1 and PGL-3 produced larger droplets and lowered the critical concentration for LLPS occurrence (8). A receptor protein SEPA-1 interacts with PGL-3 to facilitate LLPS of PGL-1/-3, while the droplet size and mobility are modulated by EPG-2 (8). Although either SEPA-1 or EPG-2 fails to undergo LLPS alone, the four resident proteins interact with each other as scaffolds to drive LLPS-mediated assembly of PGL granules (8). Moreover, arginine methylation of PGL-1/–3 by the protein arginine N-methyltransferase 1 (PRMT1) homolog EPG-11 inhibits LLPS, which is prompted through the phosphorylation of PGL-1 by LET-363, the ortholog of mammalian target of rapamycin (mTOR) (8). EPG-11 and LET-363 are not resident molecules of PGL granules, but contribute in modulating LLPS (4,8). The characterization of these regulators is undoubtedly important for analyzing LLPS. To date, only a small proportion of MLO-associated components were identified as scaffold proteins. A large number of other remaining proteins in MLOs dispensable for condensate formation were named as clients, which might be selectively recruited into MLOs through interactions with scaffold proteins (7,10,20,21). Although great efforts have been taken on the discovery of new LLPS-associated proteins, an integrative data resource was still not available.
In this study, we first collected 9285 experimentally identified LLPS-associated proteins, including 150 scaffolds, 987 regulators and 8148 potential clients, from the literature (Figure 1, Supplementary Table S1). These proteins were classified into 40 distinct biomolecular condensates, and we computationally identified potential orthologs of these known proteins in other eukaryotes (Figure 1). In total, the data resource of LLPS (DrLLPS) contained 437 887 known and potential LLPS-associated proteins, including 7993 scaffolds, 72 300 regulators and 357 594 clients in 164 eukaryotic species. Rich annotations were provided for LLPS-associated proteins in eight model organisms especially in human, by compiling and integrating the knowledge that covered 16 aspects, including intrinsically disordered regions (IDRs), domain annotations, PTMs, genetic variations, cancer mutations, molecular interactions, disease-associated information, drug–target relations, physicochemical property, protein functional annotations, protein expressions/proteomics, protein 3D structures, subcellular localizations, mRNA expressions, DNA & RNA elements and DNA methylations from 110 widely used databases (Figure 1, Supplementary Table S2). With a data size of 203.11 GB, DrLLPS can be useful for further study of LLPS, and free for all users at: http://llps.biocuckoo.cn/.
Figure 1.
The experimental procedure for the construction of DrLLPS. First, we searched PubMed to collect and curate experimentally identified scaffolds, regulators and potential clients (Supplementary Table S1). We classified all collected proteins into 40 types of biomolecular condensates of five super-classes, including ‘In vitro droplet’, ‘Nucleus’, ‘Cytoplasm’, ‘Germ cell’ and ‘Others’. Then we computationally identified potential orthologs of these known proteins in 164 eukaryotes, including 68 animals, 50 plants and 46 fungi. Besides basic annotations, we further integrated annotations in 110 public data resources covering 16 aspects, including (i) IDRs, (ii) domain annotations, (iii) PTMs, (iv) genetic variations, (v) cancer mutations, (vi) molecular interactions, (vii) disease-associated information, (viii) drug–target relations, (ix) physicochemical properties, (x) protein functional annotations, (xi) protein expressions/proteomics, (xii) protein 3D structures, (xiii) subcellular localizations, (xiv) mRNA expressions, (xv) DNA & RNA elements and (xvi) DNA methylations (Supplementary Table S2).
CONSTRUCTION AND CONTENT
Data collection, classification and genome-wide identification
From PubMed, we first used a single keyword combination of ‘((phase separation) OR (phase transition)) AND (protein OR proteins)’ to search experimentally identified LLPS-associated proteins, by manually checking abstracts or full texts of the scientific papers published before 1 January 2019. To avoid missing any known proteins, we further used multiple keyword combinations to search proteins located in various biomolecular condensates. For example, a keyword combination ‘((Cajal body) OR (Cajal bodies)) AND ((formation) OR (protein OR proteins))’ was adopted to search constitutive proteins of Cajal bodies. We used this approach to collect proteins located in 40 types of biomolecular condensates.
As previously described (7,10,20,21), all collected proteins were classified as scaffolds, regulators or potential clients. Scaffolds were defined as the drivers of LLPS essential for the structural integrity of MLOs, and the major components which, alone or with co-scaffolds, undergo LLPS (7,10,20,21). For example, the human fused in sarcoma (FUS), a well-characterized RNA-binding protein undergoing LLPS involved in formation of multiple biomolecular condensates (16,22–24), forms liquid-like droplets both in cells and at near physiological conditions in vitro (25). Thus, the human FUS was annotated as a scaffold (Figure 2). The LLPS of scaffold proteins and the stability of MLOs can be modulated by PTMs and other proteins (3,4,7,19). For example, arginine methylation of FUS by PRMT1 or PRMT8 prevents the LLPS of FUS, and both PRMTs were classified as PTM regulators (Figure 2). Also, Buchan et al. conducted a microscopy-based genetic screen of >4000 gene deletions in Saccharomyces cerevisiae, and identified 125 genes to be involved in regulating the stability and formation of P-bodies and/or stress granules (26). None of these proteins have been characterized to undergo LLPS, and here we classified all the 125 proteins as MLO regulators. In addition, a large number of proteins were identified to be co-complexed with known MLO scaffolds by conventional biochemical assays or mass spectrometry, or co-localized with MLOs by immunofluorescence. For example, C14ORF166, FAM98A/B and RTCB were identified as candidate stress granule proteins, which form an RNA transport complex with the DEAD-box helicase DDX1, a known scaffold of multiple MLOs including the DDX1 body (27). It was not known whether C14ORF166, FAM98A, FAM98B and RTCB are indispensable for stress granule assembly, and the four proteins were annotated as potential clients (Figure 2). In total, we collected 9285 known LLPS-associated proteins, including 150 scaffolds, 987 regulators and 8148 potential clients, from 23 eukaryotes (Supplementary Table S1).
Figure 2.
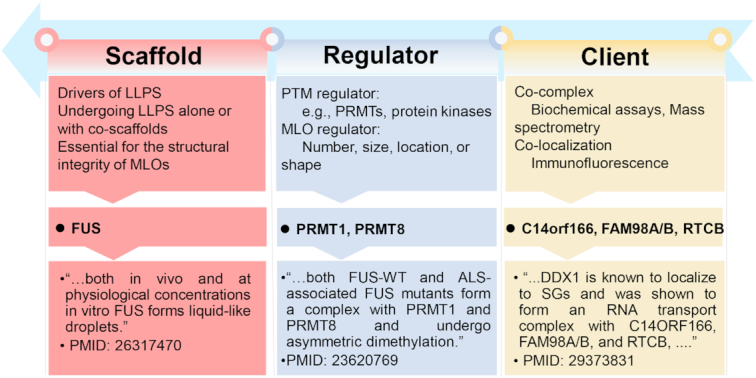
The classification of all known LLPS-associated proteins as scaffolds, regulators or potential clients (7,10,20,21). Scaffolds are the drivers of LLPS essential for the structural integrity of MLOs, as the major components which, alone or with co-scaffolds, undergo LLPS (7,10,20,21). The LLPS of scaffolds and the MLO dynamics such as number, size, location or shape can be affected by various PTM regulators and other proteins, which were collectively regarded as regulators (3,4,7,19). Potential clients were defined as proteins co-complexed or co-localized with known MLO scaffolds. Typical examples of scaffolds, regulators and potential clients were shown.
Then, we categorized these proteins into 40 biomolecular condensates belonged to five super-classes as: (i) in vitro droplet, containing scaffolds and regulators involved in the formation of liquid droplets in vitro; (ii) nucleus, including Cajal body, chromatin, cleavage body, DDX1 body, DNA damage foci, Gemini of Cajal body, Histone locus body, insulator body, nuclear pore complex, nuclear speckle, nuclear stress body, nucleolus, OPT domain, paraspeckle, PcG body, perinucleolar compartment, PML nuclear body and Sam68 nuclear body; (iii) cytoplasm, including centrosome/spindle pole body, microtubule, neuronal granule, P-body, pericentriolar matrix, siRNA body, spindle apparatus, stress granule, TAM body and U body; (iv) germ cell, including Balbiani body, chromatoid body, germ plasm/polar granule, nuage, P granule and sponge body; (v) others, including mitochondrial RNA granule, postsynaptic density, pyrenoid matrix and receptor cluster, as well as Others for unclassified proteins (Figure 1). It should be noted that several biomolecular condensates only existed in specific organisms, e.g. PML nuclear body, neuronal granule and postsynaptic density in animals, pyrenoid matrix in plants, centrosome in metazoans and spindle pole body in yeasts (1,3,6,7,10,18,28). Also, all Germ cell condensates were exclusively found in animals, whereas P granules only exist in nematodes (5,10,29).
Using these known LLPS-associated proteins, we performed a genome-wide detection of their orthologs in other species (Figure 1). We downloaded the complete proteome sets of 164 eukaryotes, including 68 animals from Ensembl (release version 95, http://www.ensembl.org/), 50 plants from EnsemblPlants (release version 42, http://plants.ensembl.org/) and 46 fungi from EnsemblFungi (release version 42, http://fungi.ensembl.org/), respectively (30). As previously described (31), low-quality protein sequences containing one or more ‘X’ characters were removed. The Ensembl Gene ID was chosen as the primary accession to avoid redundancy, since multiple isoform proteins can be derived from one gene. The longest protein and its corresponding nucleotide coding sequence (CDS) was reserved for each gene with multiple alternatively splicing isoforms. For each proteome set, we used a tool called CD-HIT to eliminate redundant proteins with 100% identity (32). Then, we adopted the classical method of reciprocal best hits (RBHs), which can pairwisely detect orthologous pairs, if two proteins in two different organisms reciprocally find each other as the best hit (33). For detection of potential orthologs, we used the blastall program in the software package of BLAST, with a stringent threshold of E-value ≤ 10−6 (34). In total, we additionally predicted 7843 scaffolds, 71 313 regulators and 349 446 clients in 164 eukaryotes, and the classification information of these proteins were determined based on their orthologous known cognates. If the condensate of a protein does not exist in a species, this protein was classified into the category of Others/Others. Both known and computationally identified LLPS-associated proteins were included into DrLLPS, and a software package of Heatmap Illustrator (HemI) (35) was adopted to illustrate the distribution of numbers of LLPS-associated proteins in 40 biomolecular condensates across the 164 species (Supplementary Figure S1). A detailed data statistics was available for known and potential scaffolds, regulators and potential clients in each eukaryotes (Supplementary Table S3).
A comprehensive annotation of LLPS-associated proteins
We constructed DrLLPS as a gene-centered database, and a variety of basic annotations, such as protein/gene names/aliases, Ensembl/UniProt/GeneBank/RefSeq accession numbers, functional descriptions, protein/nucleotide sequences, keywords, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (36) and Gene Ontology (GO) terms (37), were obtained from Ensembl (30) and UniProt (38) databases. For each known LLPS-associated protein, brief descriptions on its regulatory roles in LLPS or localizations in biomolecular condensates were present, and corresponding tissues or cell lines for experimental analyses were provided, as well as PMIDs of primary references. Recently, a minimum set of six experimental tests, including the assembly of spherical droplets, the observation of fusion events, and the identification of mutations that inhibit LLPS in vitro and in cells have been proposed for rigorous analysis of LLPS processes (7). For each known scaffold protein, descriptions on performed assays of the minimum set of experiments were presented on its gene page.
Next, we compiled and integrated the knowledge from 110 additionally public resources, and carefully annotated 28 024 known or potential LLPS-associated proteins in eight model species, including Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, C. elegans, Danio rerio, Arabidopsis thaliana and S. cerevisiae. These resources covered 16 distinct aspects: (i) computationally predicted IDRs in protein sequences by multiple tools; (ii) functional domain annotations; (iii) up to 42 types of PTM sites in proteins; (iv) genetic variations as non-synonymous single nucleotide polymorphisms (nsSNPs) in nucleotide CDS sequence; (v) cancer mutations detected in clinical samples; (vi) molecular interactions including PPIs and protein–RNA interactions; (vii) disease-associated SNPs, cancer mutations, PTMs and gene fusions; (viii) drug–target relations; (ix) physicochemical properties; (x) protein functional annotations; (xi) protein expressions derived from the proteomic data; (xii) protein 3D structures; (xiii) known or predicted subcellular localizations; (xiv) mRNA expressions; (xv) DNA & RNA elements; (xvi) DNA methylations (Figure 1, Supplementary Table S2). The details on processing each resource were present in Supplementary Methods. All annotations in DrLLPS were downloadable at http://llps.biocuckoo.cn/download.php.
USAGE
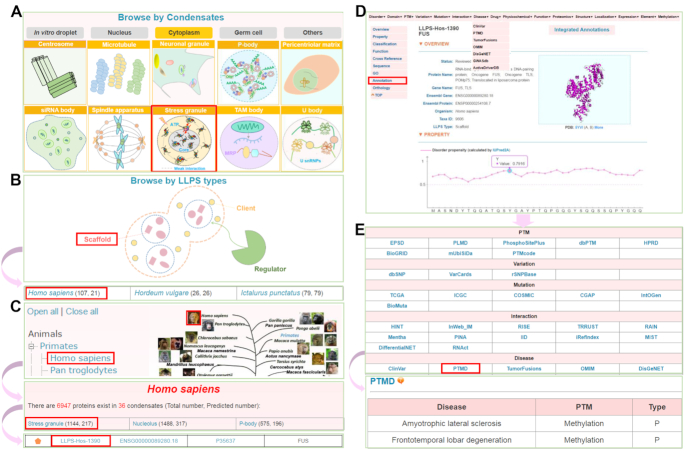
The online service of DrLLPS was developed in an easy-to-use manner. Here, we selected the human FUS protein as an example to describe the usage of DrLLPS. For browsing the data in DrLLPS, we implemented three options, including ‘Browse by Condensates’, ‘Browse by LLPS types’, and ‘Browse by species’ (Figure 3). In the option of ‘Browse by Condensates’, users can click the condensate of ‘Stress granule’ under the Cytoplasm super-class to browse all known and predicted LLPS-associated proteins involved in stress granules in eukaryotes (Figure 3A). Since human FUS is a known scaffold protein, users can directly click ‘Scaffold’ in the option of ‘Browse by LLPS types’, while the numbers of total and predicted human scaffolds were shown, respectively (Figure 3B). Then, users can click ‘Homo sapiens’ of the returned page to view all human scaffold proteins (Figure 3B). Moreover, in the option of ‘Browse by species’, the Ensembl taxonomic categories were shown in the left side, while the phylogenetic relations of the eukaryotes annotated in Ensembl were illustrated in the right side (Figure 3C). In DrLLPS, all known LLPS-associated proteins were designated as ‘Reviewed’ and marked with an orange pentagon, whereas computationally identified proteins were marked with a grey pentagon as ‘Unreviewed’ (Figure 3C). For each MLO, the numbers of total and predicted LLPS-associated proteins were present, respectively (Figure 3C). By clicking ‘Homo sapiens’, ‘Stress granule’ and ‘LLPS-Hos-1390’, the final page of human FUS will be shown (Figure 3D). In the gene page, basic annotations such as status, protein/gene names, Ensembl gene/protein IDs, Taxa ID and LLPS type can be viewed (Figure 3D). The disorder propensity of human FUS protein calculated by IUPred2A (39) and its domain structures were shown, as well as additional fundamental annotations (Figure 3D). For additional annotations, users can either click ‘Integrated Annotations’ on the gene page, or ‘Annotation’ in the left bar (Figure 3D). A specific type of annotation can be selected by clicking on its corresponding button (Figure 3E). For example, users can click ‘PTMD’ to view known PTM-disease associations of human FUS (Figure 3E). Besides the three options for browsing the database, we also provided several options, including ‘Simple Search’, ‘Batch Search’, ‘Advance Search’ and ‘BLAST Search’, for searching the data in DrLLPS (http://llps.biocuckoo.cn/advance.php) (Supplementary Figure S2).
Figure 3.
The browse options of DrLLPS. A well characterized scaffold protein, human FUS, was chosen as an example for describing the usage. (A) Browse by Condensates. All proteins classified in a specific biomolecular condensates can be viewed by clicking on the corresponding icon. (B) Browse by LLPS types. Known and predicted scaffolds, regulators or clients can be browsed by clicking on their corresponding names. (C) Browse by species. (D) The gene page of human FUS protein with basic annotations (E) The 16 aspects of additional annotations of FUS. From the PTMD database, it could be found that methylation of FUS protein is associated with human diseases.
DISCUSSION
Since the discovery of nematode germline P Granules as liquid droplets in 2009 (29), LLPS, a typical type of phase transitions, has emerged to be an intriguing mechanism to interpret how MLOs can be created to spatiotemporally organize complex biochemical reactions in living cells (1–7). Later in 2012, both structurally folded proteins and IDPs were found to undergo LLPS, and weak multivalent interactions among amino acid residues have been proposed as a major molecular signature of protein LLPS (7,17,18,40). To date, only a small proportion of constituents in various MLOs were characterized as scaffold proteins, which act as drivers essential for driving LLPS-mediated MLO assembly (4,6–8,20). Besides experimental efforts, the development of computational approaches for the prediction of protein LLPS has also emerged to be an intriguing challenge (3,19). Recently, Vernon et al. compiled a benchmark data set containing 30 human proteins known to be involved in LLPS, and carefully compared the prediction performance of seven predictors, which used multiple sequence features including prion-like amino acid composition (PLAAC), low-complexity aromatic-rich kinked segments (LARKS), the number of arginine and tyrosine residues (R + Y), DDX4-like sequences, sequence composition statistics in catGRANULE, pi–pi contacts in PScore, and non-specific protein interaction propensity in CRAPome (3). They found both catGRANULE and PScore were better than other tools, with an area under the curve (AUC) value as 0.87 (3). It was proposed that the combination of multiple features might be helpful for developing more accurate predictors.
Since more and more LLPS-associated proteins have been experimentally identified, a comprehensive database will be helpful for further analyses. In this study, we developed an integrative resource called DrLLPS, containing 7993, 72 300 and 357 594 known and potential scaffolds, regulators and clients of 40 biomolecular condensates from 164 eukaryotes, respectively. For known scaffold proteins, various experimental tests performed for analyzing their LLPS behaviors were summarized (7) (Supplementary Table S4). Besides LLPS, biomolecular condensates can also be converted into other forms such as gel-like and solid states, and various associated physical properties have been identified in several systems under different conditions (2,4,5,7,15). Indeed, transitions between these different states can be biologically important and attributed for disease-associated states. For example, EPG-2 promotes the liquid-to-gel-like transition of PGL granules to reduce their mobility and increase their salt resistance (8). Also, G156E and R244C, two disease mutations of FUS derived from amyotrophic lateral sclerosis (ALS) patients, can accelerate the liquid-to-solid transition to form aggregates (25). Descriptions on other forms that scaffolds could be involved in were also summarized on gene pages, if available. It should be noted that scaffold proteins can interact with various partners such as RNA, DNA or other proteins to drive LLPS in a much more facile manner. For example, SEPA-1 forms a complex with PGL-1 and PGL-3 to facilitate the LLPS of PGL granules (8). If available, co-complexed partners and corresponding descriptions were shown for known scaffolds on their gene pages.
Besides basic annotations, we further annotated 28 024 known and potential LLPS-associated proteins in eight species by integrating the knowledge from 110 public resources, and human FUS protein was chosen as an example to demonstrate the usefulness of rich annotations in DrLLPS (Figure 4). According to the prediction results of 11 tools integrated from three databases including D2P2 (41), IUPred2A (39) and MobiDB (42), averagely 84.15% of residues in FUS were located in IDRs, which are important for its LLPS through the multivalent interaction (17) (Figure 4 and Supplementary Figure S3A). From the domain annotations, it could be found that FUS harbors multiple distinct domains, including a prion-like domain (PLD), an RNA recognition motif (RRM) and a zinc finger (ZnF) domain. The former two are important for FUS LLPS (4,7,18), whereas a low complexity domain (LCD) is overlapped with PLD and ZnF (Figure 4 and Supplementary Figure S3B). For PTM information, there were eight types of PTMs with 116 sites integrated for the FUS protein. Among these known PTM sites, at least phosphorylation of S42, S54, S61, T68, S84 and/or S87 disrupts the formation of liquid droplets (22) (Figure 4 and Supplementary Figure S3B). In the nucleotide CDS sequence of FUS, 201 nsSNPs were retrieved from the database dbSNP (43), and six of them including rs387906627 (R495*), rs121909667 (H517Q), rs121909669 (R518K), rs121909667 (R521H), rs121909668 (R521C) and rs886041390 (P525L) were annotated to be associated with ALS from ClinVar (44) (Figure 4 and Supplementary Figure S3B). The R495* and P525L severely abrogate the interaction with transportin-1 (TNPO1) to block the nuclear importing of FUS, and all the six ALS-related nsSNPs promote the abnormal LLPS of FUS in cytoplasmic stress granules but not in nucleus (16,23,24). Furthermore, we obtained 269 missense cancer mutations of 26 cancer types from TCGA for FUS (45), which is highly mutated in uterine corpus endometrial carcinoma (UCEC), colon adenocarcinoma (COAD) and skin cutaneous melanoma (SKCM) (Figure 4, and Supplementary Figure S3C). There were 82 small chemicals annotated to target FUS and influence its protein expression, while its molecular weight and isoelectric point of FUS were calculated as 53 425.84 and 9.40, respectively (Figure 4). Human FUS has been annotated as an RNA-binding protein (22), with 13 3D structures maintained in PDB (46). FUS protein interacts with 2445 miRNAs, and tends to be localized in nucleus. In addition, FUS protein is highly expressed in fetal ovary, whereas its mRNA expression level is upregulated in thymoma (THYM), testicular germ cell tumors (TGCT) and lymphoid neoplasm diffuse large B-cell lymphoma (DLBC) (Figure 4 and Supplementary Figure S3D, E).
Figure 4.
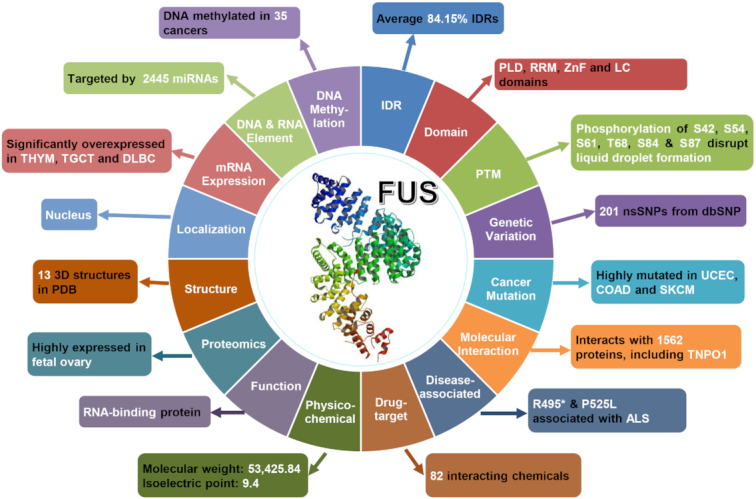
The overview of additional annotations for human FUS. A brief summary of the 110 data resources was shown in Supplementary Table S2. A detailed description on processing each resource was present in Supplementary Methods.
In the future, DrLLPS will be continuously maintained and updated to collect and annotate newly identified LLPS-associated proteins. It should be noted that new functions might be reported for existing proteins in DrLLPS, and their LLPS types and the classification information will also be refined. We anticipate DrLLPS can serve as a useful resource for further biophysical, biochemical, biological and bioinformatic analyses of LLPS.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Special Project on Precision Medicine under the National Key R&D Program [2017YFC0906600, 2018YFC0910500]; Natural Science Foundation of China [31970633, 31930021, 31671360, 81701567, 31671348]; Fundamental Research Funds for the Central Universities [2017KFXKJC001, 2019kfyRCPY043]; Changjiang Scholars Program of China, and the program for HUST Academic Frontier Youth Team.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bentley E.P., Frey B.B., Deniz A.A.. Physical chemistry of cellular liquid-phase separation. Chemistry. 2019; 25:5600–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hyman A.A., Weber C.A., Julicher F.. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014; 30:39–58. [DOI] [PubMed] [Google Scholar]
- 3. Vernon R.M., Forman-Kay J.D.. First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol. 2019; 58:88–96. [DOI] [PubMed] [Google Scholar]
- 4. Wang Z., Zhang H.. Phase separation, transition, and autophagic degradation of proteins in development and pathogenesis. Trends Cell Biol. 2019; 29:417–427. [DOI] [PubMed] [Google Scholar]
- 5. Gomes E., Shorter J.. The molecular language of membraneless organelles. J. Biol. Chem. 2019; 294:7115–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feng Z., Chen X., Zeng M., Zhang M.. Phase separation as a mechanism for assembling dynamic postsynaptic density signalling complexes. Curr. Opin. Neurobiol. 2019; 57:1–8. [DOI] [PubMed] [Google Scholar]
- 7. Alberti S., Gladfelter A., Mittag T.. Considerations and challenges in studying liquid–liquid phase separation and biomolecular condensates. Cell. 2019; 176:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang G., Wang Z., Du Z., Zhang H.. mTOR regulates phase separation of PGL Granules to modulate their autophagic degradation. Cell. 2018; 174:1492–1506. [DOI] [PubMed] [Google Scholar]
- 9. Bergeron-Sandoval L.P., Safaee N., Michnick S.W.. Mechanisms and consequences of macromolecular phase separation. Cell. 2016; 165:1067–1079. [DOI] [PubMed] [Google Scholar]
- 10. Banani S.F., Lee H.O., Hyman A.A., Rosen M.K.. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017; 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marnik E.A., Updike D.L.. Membraneless organelles: P granules in Caenorhabditis elegans. Traffic. 2019; 20:373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., Karpen G.H.. Phase separation drives heterochromatin domain formation. Nature. 2017; 547:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee K.H., Zhang P., Kim H.J., Mitrea D.M., Sarkar M., Freibaum B.D., Cika J., Coughlin M., Messing J., Molliex A. et al.. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016; 167:774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mugler A., Bailey A.G., Takahashi K., ten Wolde P.R.. Membrane clustering and the role of rebinding in biochemical signaling. Biophys. J. 2012; 102:1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun D., Wu R., Li P., Yu L.. Phase separation in regulation of aggrephagy. J. Mol. Biol. 2019; doi:10.1016/j.jmb.2019.06.026. [DOI] [PubMed] [Google Scholar]
- 16. Hofweber M., Hutten S., Bourgeois B., Spreitzer E., Niedner-Boblenz A., Schifferer M., Ruepp M.D., Simons M., Niessing D., Madl T. et al.. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell. 2018; 173:706–719. [DOI] [PubMed] [Google Scholar]
- 17. Li P., Banjade S., Cheng H.C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J.V., King D.S., Banani S.F. et al.. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012; 483:336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J. et al.. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012; 149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vernon R.M., Chong P.A., Tsang B., Kim T.H., Bah A., Farber P., Lin H., Forman-Kay J.D.. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife. 2018; 7:e31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., Rosen M.K.. Compositional control of phase-separated cellular bodies. Cell. 2016; 166:651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hyman A.A., Simons K.. Cell biology. Beyond oil and water–phase transitions in cells. Science. 2012; 337:1047–1049. [DOI] [PubMed] [Google Scholar]
- 22. Murray D.T., Kato M., Lin Y., Thurber K.R., Hung I., McKnight S.L., Tycko R.. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017; 171:615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo L., Kim H.J., Wang H., Monaghan J., Freyermuth F., Sung J.C., O’Donovan K., Fare C.M., Diaz Z., Singh N. et al.. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell. 2018; 173:677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoshizawa T., Ali R., Jiou J., Fung H.Y.J., Burke K.A., Kim S.J., Lin Y., Peeples W.B., Saltzberg D., Soniat M. et al.. Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell. 2018; 173:693–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M. et al.. A Liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015; 162:1066–1077. [DOI] [PubMed] [Google Scholar]
- 26. Buchan J.R., Kolaitis R.M., Taylor J.P., Parker R.. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013; 153:1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Markmiller S., Soltanieh S., Server K.L., Mak R., Jin W., Fang M.Y., Luo E.C., Krach F., Yang D., Sen A. et al.. Context-dependent and disease-specific diversity in protein Interactions within stress granules. Cell. 2018; 172:590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rai A.K., Chen J.X., Selbach M., Pelkmans L.. Kinase-controlled phase transition of membraneless organelles in mitosis. Nature. 2018; 559:211–216. [DOI] [PubMed] [Google Scholar]
- 29. Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Julicher F., Hyman A.A.. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009; 324:1729–1732. [DOI] [PubMed] [Google Scholar]
- 30. Cunningham F., Achuthan P., Akanni W., Allen J., Amode M.R., Armean I.M., Bennett R., Bhai J., Billis K., Boddu S. et al.. Ensembl 2019. Nucleic Acids Res. 2019; 47:D745–D751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo Y., Peng D., Zhou J., Lin S., Wang C., Ning W., Xu H., Deng W., Xue Y.. iEKPD 2.0: an update with rich annotations for eukaryotic protein kinases, protein phosphatases and proteins containing phosphoprotein-binding domains. Nucleic Acids Res. 2019; 47:D344–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fu L., Niu B., Zhu Z., Wu S., Li W.. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012; 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tatusov R.L., Koonin E.V., Lipman D.J.. A genomic perspective on protein families. Science. 1997; 278:631–637. [DOI] [PubMed] [Google Scholar]
- 34. Sayers E.W., Cavanaugh M., Clark K., Ostell J., Pruitt K.D., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2019; 47:D94–D99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deng W., Wang Y., Liu Z., Cheng H., Xue Y.. HemI: a toolkit for illustrating heatmaps. PLoS one. 2014; 9:e111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M.. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019; 47:D590–D595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019; 47:D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Consortium UniProt. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meszaros B., Erdos G., Dosztanyi Z.. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018; 46:W329–W337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chong P.A., Forman-Kay J.D.. Liquid-liquid phase separation in cellular signaling systems. Curr. Opin. Struct. Biol. 2016; 41:180–186. [DOI] [PubMed] [Google Scholar]
- 41. Oates M.E., Romero P., Ishida T., Ghalwash M., Mizianty M.J., Xue B., Dosztanyi Z., Uversky V.N., Obradovic Z., Kurgan L. et al.. D(2)P(2): database of disordered protein predictions. Nucleic Acids Res. 2013; 41:D508–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Piovesan D., Tabaro F., Paladin L., Necci M., Micetic I., Camilloni C., Davey N., Dosztanyi Z., Meszaros B., Monzon A.M. et al.. MobiDB 3.0: more annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Res. 2018; 46:D471–D476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. et al.. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018; 46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hutter C., Zenklusen J.C.. The Cancer Genome Atlas: creating lasting value beyond its data. Cell. 2018; 173:283–285. [DOI] [PubMed] [Google Scholar]
- 46. Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L., Christie C., Dalenberg K., Duarte J.M., Dutta S. et al.. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019; 47:D464–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.