Abstract
The common pheasant (Phasianus colchicus) in the order Galliformes and the family Phasianidae, has 30 subspecies distributed across its native range in the Palearctic realm and has been introduced to Europe, North America, and Australia. It is an important game bird often subjected to wildlife management as well as a model species to study speciation, biogeography, and local adaptation. However, the genomic resources for the common pheasant are generally lacking. We sequenced a male individual of the subspecies torquatus of the common pheasant with the Illumina HiSeq platform. We obtained 94.88 Gb of usable sequences by filtering out low-quality reads of the raw data generated. This resulted in a 1.02 Gb final assembly, which equals the estimated genome size. BUSCO analysis using chicken as a model showed that 93.3% of genes were complete. The contig N50 and scaffold N50 sizes were 178 kb and 10.2 Mb, respectively. All these indicate that we obtained a high-quality genome assembly. We annotated 16,485 protein-coding genes and 123.3 Mb (12.05% of the genome) of repetitive sequences by ab initio and homology-based prediction. Furthermore, we applied a RAD-sequencing approach for another 45 individuals of seven representative subspecies in China and identified 4,376,351 novel single nucleotide polymorphism (SNPs) markers. Using this unprecedented data set, we uncovered the geographic population structure and genetic introgression among common pheasants in China. Our results provide the first high-quality reference genome for the common pheasant and a valuable genome-wide SNP database for studying population genomics and demographic history.
Keywords: common pheasant, Phasianus colchicus, genome sequencing, RAD-sequencing, SNP, population genomics
Introduction
The common pheasant (Phasianus colchicus), belonging to the order Galliformes in the family Phasianidae, is a common gamebird with a worldwide distribution (Hill and Robertson 1988; Pfarr 2012). It is a well-known game bird with global importance for both research and wildlife management. First, as one of the world’s most widespread resident species, the common pheasant is native to the temperate zones in the Palearctic region, from the Russian Far East to eastern-southeastern Europe (east of the Black Sea), and southwards to Indochina and Afghanistan (Cramp 1980; Johnsgard 1999). Second, it is among the most subspecies-rich bird species with thirty described subspecies defined mainly by plumage characters in males and geographical range (Madge et al. 2002). These subspecies occupy substantially different environments and climatic zones, from isolated oases in semideserts, to montane regions, displaying unique phenotypes and genotypes (Hill and Robertson 1988; Pfarr 2012). The common pheasant also has great economic values, and have long history of being released for hunting or kept captive in bird farms in Western Europe, North America and Australia (Johnsgard 1999; Madge et al. 2002). This makes the common pheasant a promising model to investigate important questions about speciation, trait evolution, biogeography and local adaptation to various climatic conditions, as well as in wildlife management and conservation genetics.
Previous studies of the common pheasant have mainly focused on its ecology and biology. Several aspects of the species’ life history and reproduction, such as survival rate (Draycott et al. 2008), habitat selection (Long et al. 2007), and breeding biology (Robertson 1996) has been well quantified, which provide useful knowledge for sustainable management. The taxonomy and systematics of common pheasant have long been actively studied. For example, the thirty-recognized subspecies were hypothesized to be clustered into five subspecies-groups based on morphological resemblance and biogeographical affinity (Madge et al. 2002). However, given that clinal variation may explain connectivity and contiguous distributions, the validity of some subspecies has been questioned (Cramp 1980; Liu and Sun 1992; Johnsgard 1999; Madge et al. 2002). Molecular phylogenetic approaches have been applied to resolve these puzzles since the subspecies-groups have been identified as independent evolutionary lineages by mitochondrial fragments (Qu et al. 2009; Liu et al. 2010; Zhang et al. 2014). Recent works using multi-locus nuclear markers corroborate previous findings, and further showed that viscous boundaries between subspecies are probably due to extensive gene flow among contiguous populations (Kayvanfar et al. 2017; Wang et al. 2017). These works used a small number of genetic markers, which are capable of delineating population structure and subspecies relationships. However, due to the lack of genomic-level data, many questions related to demographic histories, for example (Nadachowska-Brzyska et al. 2013), population admixture and molecular genetic basis of phenotypes, for example, plumage color and pattern (Toews et al. 2016), remain undetermined.
Taking advantage of next-generation sequencing (NGS), it is now feasible to obtain unprecedented genomic-scale data, allowing direct quantification of genomic variation and identification of the genomic landscape associated with specific phenotypes in nonmodel organisms (Ellegren 2014; Toews et al. 2016; Bosse et al. 2017). Particularly in the study of speciation, it is of interest to understand the size and extent of regions of divergence across the genome among the related species (Wu 2001). Recent empirical studies have been uncovering a notable pattern that genomic regions with accumulated genetic differentiation between closely related avian species/subspecies pairs contain genes involved in divergence whereas gene flow and genetic drift homogenize other regions (Ellegren et al. 2012; Poelstra et al. 2014). Divergent heterogeneous regions have been coined “islands of divergence” (Turner et al. 2005; Harr 2006; Nosil et al. 2009) and divergent selection is considered to contribute to such differentiation islands, though it is not necessarily the only evolutionary force explaining this phenomenon (Cruickshank and Hahn 2014). Other processes, such as linked selection are evident to contribute divergent genomic landscapes as shown by some recent studies (Burri et al. 2015, Vijay et al. 2017). The common pheasant is distributed in a vast geographical range and provide several evolutionary contrasts to investigate complex evolutionary processes and demographic history that could contribute intraspecific divergence. Obviously, the availability of genomic information will help facilitate finer-scale characterization of the genomic-wide regions involved in divergence and adaptation of common pheasant.
In this study, we present the first genome assembly of the common pheasant with detailed description of its genetic architecture and population-level genomic polymorphisms in order to provide genomic resources toward studies of speciation, local adaptation, and conservation genomics of an ecologically important species.
Materials and Methods
Sampling and Sequencing
The sequenced sample was fresh blood from a male common pheasant, (the subspecies P.colchicus torquatus: NCBI taxonomy ID 9054; BioProject ID PRJNA449162; BioSample ID SAMN08888528). This individual was a captive individual with a wild origin from Beijing Wild Animal Park (Daxing) sampled in August 2015. All sampling procedures were performed in accordance with Chinese wildlife regulations and protocols.
Genomic DNA was extracted using a Qiagen DNA purification kit following the manufacturer’s instruction and the quality of extracted DNA was checked using gel electrophoresis (1% agarose gel/40 ng loading). We built four short insert libraries (two for 250 bp and two for 450 bp) and three mate pair libraries (2 kbp, 5 kbp, 10 kbp) following Illumina’s standard protocol. Briefly, the qualified genomic DNA was randomly sheared into short fragments by hydrodynamic shearing system (Covaris, Massachusetts, USA). Then, followed by end repairing, dA-tailing and further ligation with Illumina adapter, the required fragments (in 300–500 bp size) with both P5 and P7 sequences were PCR selected and amplified. After gel electrophoresis and subsequent purification, the required fragments were obtained. The constructed libraries were loaded on the Illumina HiSeq platform for paired-end sequencing, with the read length of 150 bp at each end.
Raw data obtained from sequencing also contain adapter contamination and low-quality reads. These sequence artifacts may complicate the downstream processing analysis. The raw data were thus filtered to reduce low quality bases and reads by the following strategies: 1) filtering our reads with adapters; 2) reads with N bases >5%; 3) the paired reads when single end sequencing reads contain low quality (<5) bases that exceed 10% of the read length.
Evaluation of Genome Size
The genome size was estimated according to a k-mer analysis with the formula: G = k-mer_number/k-mer_depth, where G is the genome size, k-mer_number is the total counts of kmers and k-mer_depth refers to the main peak in the k-mer distribution. In this study, we collected all reads in a short-insert library to conduct the 19-mer analysis with Jellyfish 2.0 (Marçais and Kingsford 2011). A total of 39,151,211,661 k-mers were produced and the peak k-mer depth was 38 (supplementary fig. S1, Supplementary Material online).
Genome Assembly and Assessment
To assemble the common pheasant genome, we firstly evaluated genome-wide heterozygosity using the above k-mer analysis. Double peaks suggested that this diploid genome was highly heterozygous. We therefore employed Platanus v1.2.4 (Kajitani et al. 2014), which is particularly designed for highly heterozygous genomes, to assemble the common pheasant genome. The first round included three steps: Contig assembly, scaffolding, and gap closing. Firstly, all filtered reads in short-insert libraries (250 bp, 450 bp) were input for contig assembly. After constructing de Brujin graphs, clipping tips, merging bubbles, and removing low coverage links with default parameters, assembled contigs and bubbles in the graphs were obtained in this step. In the scaffolding steps, the bubbles and reads from both short-insert library (250 bp, 450 bp) and long-insert library (2 kbp, 5 kbp, 10 kbp) were mapped onto contig sequences to build scaffolds with default parameters. Finally, the intrascaffold gaps were filled with reads from all libraries. After five rounds of gap closing, the gap rate in the scaffolds reached a plateau. To evaluate the completeness of the genome, we performed BUSCO v3 (Simão et al. 2015) with the representative chicken gene data set aves_odb9.
Genome Annotation
To identify genomic repeat elements in the assembly, both ab initio and homolog-based methods were used. For the homolog-based methods, we used RepeatMasker (www.repeatmasker.org; last accessed November 22, 2019) (Smit et al. 2016) to search against the Repbase library version 22.12 (Jurka 1998). In the ab initio method, a custom repeat library was constructed using RepeatModeler (www.repeatmasker.org) (Smit and Hubley 2008) with RECON (Bao and Eddy 2002), RepeatScout (Price et al. 2005), and Tandem repeats finder (TRF) (Benson 1999), which was then used in RepeatMasker to annotate repeats.
Gene annotation for the common pheasant genome assembly were conducted with the MAKER2 pipeline (Holt and Yandell 2011), which incorporates ab initio prediction and homology-based prediction. For the ab initio method, repeat regions were first masked based on the previous results of repeat annotation, and then Augustus (Stanke and Waack 2003) and GeneMark_ES (Lomsadze et al. 2005) were employed to generate gene structures. In addition, FGENESH (Salamov and Solovyev 2000) was also used to for ab initio prediction. For homology-based prediction, protein sequences from three different species, Chicken (Gallus gallus), Zebra finch (Taeniopygia guttata), Turkey (Meleagris gallopavo) (downloaded from Ensemble database 9.1 release), were mapped onto the genome assembly using tBlastN of the NCBI BLAST suite v2.7.1 (Madden 2013; Coordinators 2017) and Exonerate v2.2.0 (Slater and Birney 2005) was used to polish BLAST hits to get exact intron/exon position.
Population Genomics Analysis Using the Reference Genome of the Common Pheasant
To facilitate future population genomics studies of the common pheasant, we further sequenced an additional 45 male individuals from seven subspecies across China (fig. 1A) using restriction site-associated DNA sequencing (RAD sequencing) (Miller et al. 2007). The de novo common pheasant genome was used as a reference to facilitate mapping and SNP calling. We analyzed population structure among 45 individuals using ADMIXTURE 1.3 (Alexander et al. 2009) and principal component analysis (PCA). The detailed lab and sequencing procedures, and analyses are available in the supplementary appendix, Supplementary Material online.
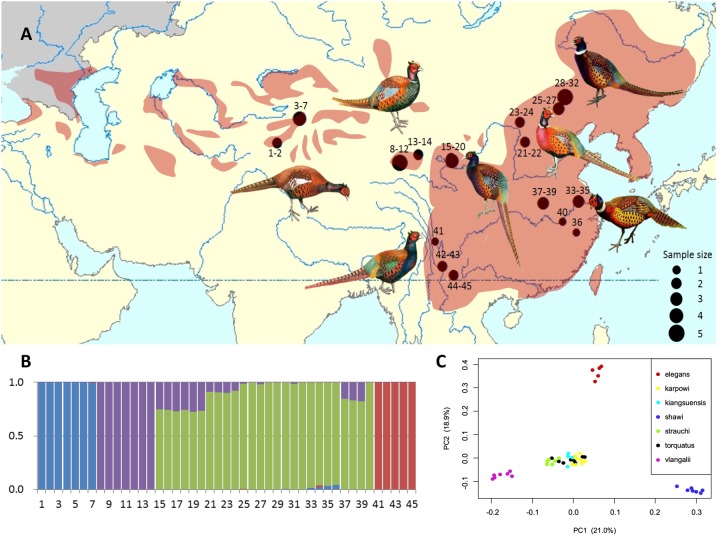
Fig. 1.
—Population genetic structure of common pheasant in China. (A) The distributions of samples of seven subspecies of common pheasant. Shown are males with divergent phenotypes (pheasant artworks were modified from Pfarr 2012). The size of the circles is proportional to the number of individuals. (B) Four genetic clusters were inferred by ADMIXTURE using 4,376,351 SNPs. The Y-axis represents the proportion of each ancestral genetic component and numbers in the X-axis represent sample locations (details in supplementary table S5, Supplementary Material online) and seven subspecies affinities: 1–7: shawii; 8–14: vlangalii; 15–20: strauchi; 21–24: kiangsuensis; 25–32: karpowi; 33–40: torquatus; 41–45: elegans. (C) Principal component analysis (PCA) of 45 individuals from seven subspecies of common pheasant using the same SNP data set with ADMIXTURE analysis. Color dots represent individuals of different subspecies.
Results and Discussion
We sequenced and assembled a reference genome of a male common pheasant. We obtained 94.88 Gb clean paired reads (supplementary table S1, Supplementary Material online). The assembled genome size of is 1.02 Gb (1,021,360,992 bp) in length with a genomic coverage = 93×. The assembled genome contains 58,369 contigs (contig N50 of 178 kb) and 39,677 scaffolds (scaffold N50 of 10.2 Mb) (table 1). The completeness of the common pheasant draft genome is high: We totally identified 4790 BUSCOs (97.5%) including 4,585 complete (93.3%) and 205 fragmented (4.2%) BUSCOs (supplementary table S2, Supplementary Material online).
Table 1.
Summary statistics of the genome assembly for the common pheasant
| Common Pheasant Genome | |
|---|---|
| Total length | 1,021,360,992 bp |
| Number of contigs | 58,369 |
| N50 of contigs | 178,013 bp |
| Number of scaffolds | 39,677 |
| N50 of scaffolds | 10,186,719 bp |
| Longest scaffolds | 42,030,034 bp |
| GC level | 41.24% |
| Number of BUSCOs | 4790 (97.5%) |
| Number of CDS | 165,367 |
| Number of mRNA | 16,485 |
| length of repeat elements | 123,323,121 bp |
We found that a total of 12.05% (123.3 Mb) repeats elements were identified in the genome assembly of common pheasant, with unclassified elements constituting the greatest proportion (supplementary table S3, Supplementary Material online). We used different prediction methods to produce a consensus gene set. In total 16,485 protein-coding genes were identified in the common pheasant genome using our described prediction methods (supplementary table S4, Supplementary Material online).
In addition, ∼114.91 Gb were generated (clean data) for all 45 samples (supplementary table S5, Supplementary Material online). After filtering and SNP callings, we obtained 4,376,351 SNPs overall, and we established a database including 328,473–999,733 SNPs for each individual (supplementary table S5, Supplementary Material online). We managed to identify 59,453 SNPs in exon regions, UTR regions and splice sites. 602,747 SNPs were identified in 5 kbp upstream/downstream regions, which can also be associated with phenotypes and functions. In addition, 1,260,753 SNPs and 3,092,463 SNPs were found in intron regions and intergenic regions, respectively. Intron regions are suggested to have genomic functions (Cech 1990) and we can further test this hypothesis using the common pheasant model. Since populations of common pheasants dwelling in contrasting environments and climatic zones, for example, monsoon regions, basins in the Qinghai-Tibetan plateau, semiarid zones, and deserts. These resources can be used to investigate population demography and genomic architectures associated with local adaptation of the common pheasant in the future (supplementary table S6, Supplementary Material online).
Our population structure results by ADMIXTURE clearly show that when four groups were inferred, the cross-validation error has the lowest value among the alternatives (supplementary fig. S2, Supplementary Material online). Given this, populations in western Xinjiang (subspecies shawii), Qinghai (subspecies vlanglii), Yunnan (subspecies elegans) and the remaining four populations in central and eastern China (subspecies strauchi, kiangsuensis, karpowi, and torquatus) form distinct genetic groups, respectively (fig. 1B). The results of PCA using the similar SNP data set were consistent with ADMIXTURE results, showing four genetic clusters, that is, Yunnan, western Xinjiang Qinghai, and the remaining subspecies (fig. 1C). Clearly, the subspecies of karpowi, kiangsuensis, and torquatus show varied magnitudes of genetic introgression with the subspecies of vlanglii from Qinghai, which corroborates previous results as shown by multilocus phylogeography studies (Liu et al. 2010; Kayvanfar et al. 2017). The resulting population structuring pattern is likely due to genetic drift of small and isolated populations in those regions, but consistent with genetic introgression caused by population expansion in contiguous populations in northern China (Liu et al. 2010; Kayvanfar et al. 2017). Obviously, the derived genomic-level polymorphism provides unprecedented power, which allows exclusive tests of specific hypotheses demographic history in common pheasant using model-based population genomic analysis (Luikart et al. 2003).
In conclusion, we report the first high quality assembled and annotated common pheasant genome. This reference genome facilitated us to obtain a high-resolution population genomic data. Such valuable resources allow unbiased downstream population genetic analysis that might be more robust than SNP sets called from de novo approaches (Shafer et al. 2017). Overall, our effort will offer an opportunity to future deep investigation of questions in evolutionary biology and wildlife management in the common pheasant, a bird species of long-standing interest in ecology, evolution, and economics.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 3157225 to Y.L. and No. 31702013 to B.W.). Partial computational machinery time work was granted by Special Program for Applied Research on Super Computation of the NSFC-Guangdong Joint Fund (2nd phase) under Grant No. U1501501 to Y.L. We also acknowledge support from Uppsala Multidisciplinary Center for Advanced Computational Science (SNIC-UPPMAX) for aiding in computational infrastructure. We thank the following persons who kindly provided samples or assisted with sampling: Xiaoju Niu, Ying Liu, and Jun Gou and we thank Dr Judith Mank for comments on an early version of the manuscript. We acknowledge Total Genomics Solution Institute, Shenzhen, China for bioinformatics advices.
Data deposition: This project has been deposited at the NCBI Sequence Read Archive under the accession (SRR6987054–SRR6987060), and the Whole Genome Shotgun project is deposited at NCBI GenBank under accession numbers BioProject PRJNA449162, BioSample SAMN08888240, and Genome assembly QCWP00000000. The SNPs are deposited at the European Bioinformatics Institute (EMBL-EBI) EVA database under accession number PRJEB26609.
Literature Cited
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Eddy SR.. 2002. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12(8):1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse M, et al. 2017. Recent natural selection causes adaptive evolution of an avian polygenic trait. Science 358(6361):365–368. [DOI] [PubMed] [Google Scholar]
- Burri R, et al. 2015. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 25(11):1656–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR. 1990. Conserved sequences and structures of group I introns: building an active site for RNA catalysis—a review In: RNA: catalysis, splicing, evolution. Amsterdam: Elsevier; p. 191–203. [DOI] [PubMed] [Google Scholar]
- Coordinators NR. 2017. Database resources of the national center for biotechnology information. Nucleic Acids Res. 45(Database issue):D12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramp S. 1980. Birds of the western Palearctic: volume 2, Hawks to Bustards. Oxford: Oxford University Press. [Google Scholar]
- Cruickshank TE, Hahn MW.. 2014. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 23(13):3133–3157. [DOI] [PubMed] [Google Scholar]
- Draycott RA, Hoodless AN, Woodburn MI, Sage RB.. 2008. Nest predation of common pheasants Phasianus colchicus. Ibis 150(s1):37–44. [Google Scholar]
- Ellegren H. 2014. Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol. 29(1):51–63. [DOI] [PubMed] [Google Scholar]
- Ellegren H, et al. 2012. The genomic landscape of species divergence in Ficedula flycatchers. Nature 491(7426):756.. [DOI] [PubMed] [Google Scholar]
- Harr B. 2006. Genomic islands of differentiation between house mouse subspecies. Genome Res. 16(6):730–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D, Robertson P.. 1988. The pheasant: ecology, management and conservation. Oxford: BSP Professional Books. [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12(1):491.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsgard P. 1999. The pheasant of the world: biology and natural history. Vol. 2 Washington (DC: ): Smithsonian Institute; p. 179–193. [Google Scholar]
- Jurka J. 1998. Repeats in genomic DNA: mining and meaning. Curr Opin Struct Biol. 8(3):333–337. [DOI] [PubMed] [Google Scholar]
- Kajitani R, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24(8):1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayvanfar N, Aliabadian M, Niu X, Zhang Z, Liu Y.. 2017. Phylogeography of the common pheasant Phasianus colchicus. Ibis. 159(2):430–442. [Google Scholar]
- Liu N, Sun H.. 1992. The geographic variance and classification of Phasianus colchicus strauchi. J Lanzhou Univ. 28(2):135–139. [Google Scholar]
- Liu Y, Zhan X, Wang N, Chang J, Zhang Z.. 2010. Effect of geological vicariance on mitochondrial DNA differentiation in Common Pheasant populations of the Loess Plateau and eastern China. Mol Phylogenet Evol. 55(2):409–417. [DOI] [PubMed] [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M.. 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33(20):6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S, Zhou C-Q, Wang W-K, Wei W, Hu J-C.. 2007. The habitat and nest-site selection of common pheasants in spring and summer in Nanchong, China . Zool Res. 28(3):249. [Google Scholar]
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. 2003. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 4(12): 981–994. [DOI] [PubMed]
- Madden T. 2013. “The BLAST sequence analysis tool.” The NCBI Handbook [Internet]. 2nd edition. National Center for Biotechnology Information (US).
- Madge S, McGowan PJ, Kirwan GM.. 2002. Pheasants, partridges and grouse: a guide to the pheasants, partridges, quails, grouse, guineafowl, buttonquails and sandgrouse of the world: A&C Black.
- Marçais G, Kingsford C.. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27(6):764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA.. 2007. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 17(2):240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadachowska-Brzyska K, et al. 2013. Demographic divergence history of pied flycatcher and collared flycatcher inferred from whole-genome re-sequencing data. PLoS Genet. 9(11):e1003942.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz‐Barrientos D.. 2009. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 18(3):375.. [DOI] [PubMed] [Google Scholar]
- Pfarr J. 2012. True Pheasants: a noble quarry. Surrey, BC: Hancock House Publishers. [Google Scholar]
- Poelstra JW, et al. 2014. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science 344(6190):1410–1414. [DOI] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA.. 2005. De novo identification of repeat families in large genomes. Bioinformatics 21(Suppl 1):i351–i358. [DOI] [PubMed] [Google Scholar]
- Qu J, Liu N, Bao X, Wang X.. 2009. Phylogeography of the ring-necked pheasant (Phasianus colchicus) in China. Mol Phylogenet Evol. 52(1):125–132. [DOI] [PubMed] [Google Scholar]
- Robertson PA. 1996. Does nesting cover limit abundance of ring-necked pheasants in North America? Wildlife Soc Bull. 24:98–106. [Google Scholar]
- Salamov AA, Solovyev VV.. 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10(4):516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer AB, et al. 2017. Bioinformatic processing of RAD‐seq data dramatically impacts downstream population genetic inference. Methods Ecol Evol. 8(8):907. [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Slater GSC, Birney E.. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6(1):31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R.. 2008. RepeatModeler Open-1.0. http://www.repeatmasker.org.
- Smit A, Hubley R, Green P.. 2016. RepeatMasker Open-4.0. http://www.repeatmasker.org.
- Stanke M, Waack S.. 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19(Suppl 2):ii215–ii225. [DOI] [PubMed] [Google Scholar]
- Toews DP, et al. 2016. Plumage genes and little else distinguish the genomes of hybridizing warblers. Curr Biol. 26(17):2313–2318. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV.. 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3(9):e285.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay N, et al. 2017. Genomewide patterns of variation in genetic diversity are shared among populations, species and higher‐order taxa. Mol Ecol. 26(16):4284. [DOI] [PubMed] [Google Scholar]
- Wang B, et al. 2017. Development and characterization of novel microsatellite markers for the Common Pheasant (Phasianus colchicus) using RAD-seq. Avian Res. 8(1):4. [Google Scholar]
- Wu CI. 2001. The genic view of the process of speciation. J Evol Biol. 14(6):851–865. [Google Scholar]
- Zhang L, An B, Backström N, Liu N.. 2014. Phylogeography-based delimitation of subspecies boundaries in the Common Pheasant (Phasianus colchicus). Biochem Genet. 52(1–2):38–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.