Abstract
Microbial secondary metabolism is a reservoir of bioactive compounds of immense biotechnological and biomedical potential. The biosynthetic machinery responsible for the production of these secondary metabolites (SMs) (also called natural products) is often encoded by collocated groups of genes called biosynthetic gene clusters (BGCs). High-throughput genome sequencing of both isolates and metagenomic samples combined with the development of specialized computational workflows is enabling systematic identification of BGCs and the discovery of novel SMs. In order to advance exploration of microbial secondary metabolism and its diversity, we developed the largest publicly available database of predicted BGCs combined with experimentally verified BGCs, the Integrated Microbial Genomes Atlas of Biosynthetic gene Clusters (IMG-ABC) (https://img.jgi.doe.gov/abc-public). Here we describe the first major content update of the IMG-ABC knowledgebase, since its initial release in 2015, refreshing the BGC prediction pipeline with the latest version of antiSMASH (v5) as well as presenting the data in the context of underlying environmental metadata sourced from GOLD (https://gold.jgi.doe.gov/). This update has greatly improved the quality and expanded the types of predicted BGCs compared to the previous version.
INTRODUCTION
Microorganisms have the remarkable ability to produce structurally complex secondary metabolites (SMs) serving important biological functions. SMs (also called natural products) are small molecules not critical for normal growth, but playing important roles in defense or signaling, nutrient acquisition or otherwise providing competitive advantage for the synthesizing organism (1–6). Owing to their bioactivity and structural diversity, SMs are used in a wide range of applications such as antibiotics, antitumor therapeutics, cholesterol-lowering agents, insecticides, fungicides, biofuels and more (7–12). However, there has been a recognized plateau in the rate of new SM discoveries (13), and renewed efforts are underway, seeking better understanding of the role of SMs in microbial interactions within complex communities, as well as novel SMs for biotechnological applications.
Genome sequence-based mining has revealed a previously undiscovered richness of biosynthetic potential via novel biosynthetic gene clusters (BGCs) (13–18). Advancements in high-throughput genome sequencing and assembly, combined with the development of novel bioinformatics pipelines (19–31) for systematic identification of BGCs offers renewed opportunity for the discovery of novel SMs. Importantly, these computational tools provide a culture-independent route to find new SMs where traditional laboratory-based approaches fail. Thus, integrating computational and experimental technologies together into a comparative platform that can address large-scale natural product characterization projects will enable further exploration of the natural products (32).
Among the computational BGC prediction software packages, antiSMASH (antibiotics and Secondary Metabolite Analysis SHell) has emerged as a popular tool widely used by the BGC research community (24–29). Its latest version, antiSMASH v5, incorporates an extensive set of manually curated and validated detection rules for more than 50 types of biosynthetic gene clusters including non-ribosomal peptide and polyketide synthases, many types of ribosomally synthesized and post-translationally modified peptides (RiPPs), as well as BGCs encoding biosynthesis of acyl-amino acids, beta-lactones, polybrominated diphenyl ethers, C-nucleosides, PPY-like ketones and lipolanthines (29).
The Integrated Microbial Genomes & Microbiomes (IMG/M) data management system is a system for comparative analysis of microbial genomes and metagenomes, which combines the largest collection of microbial genomes and assembled metagenomes with an extensive set of visualization and analytical tools (33). To further advance the discovery and analysis of BGCs and SMs in bacterial genomes and metagenomes, an IMG-ABC system (Atlas of Biosynthetic gene Clusters within IMG) was introduced in 2015 (34,35). Here we present an updated IMG-ABC system, which takes advantage of the improved performance of antiSMASH v5 to predict BGCs in IMG/M’s large collection of genomes including metagenome bins (sets of scaffolds originating from the same microbial population), which unveil the biosynthetic potential of uncultivated microbial lineages. In addition to content update, we also re-architected the IMG-ABC system in anticipation of future genome and metagenome data growth and extension of the suite of BGC prediction tools beyond antiSMASH. We expect that this new architecture will further benefit users by enabling a comparison of results from multiple BGC prediction methods and improving the overall sensitivity and specificity of BGC detection.
Data content
The new updated IMG-ABC v5 (as of September 2019) contains a total of 330,884 BGCs with 317,423 predicted clusters in 42,892 public isolate genomes and 12,176 predicted clusters from 4,944 metagenome-derived high-quality (HQ) scaffold bins, as well as 1,285 experimentally verified known clusters. All of these clusters were predicted by antiSMASH v5; clusters predicted by the previous versions of antiSMASH were discarded. Processing of additional scaffold bins and other uncultivated entities like single cells and metagenome-assembled genomes are currently underway and will be updated routinely. Compared to the previous versions of IMG-ABC, this update contains much fewer clusters owing to better specificity of antiSMASH v5, as well as an introduction of a hierarchy of BGC-related objects, which include core genes, protoclusters, candidate clusters and regions. As a result, an average ‘candidate cluster’ selected as the basic ‘BGC feature’ in IMG-ABC v5 is longer than an average BGC in the previous IMG-ABC versions.
In addition to antiSMASH ‘BGC type’ found in the previous IMG-ABC versions, IMG-ABC v5 schema captures important details of antiSMASH annotations, such as hits to antiSMASH Hidden Markov Models (HMMs), HMM hit type (e.g., biosynthetic, biosynthetic-accessory), specific rules used to predict BGCs, and assignment of genes to cluster core. For experimentally verified BGCs, links to MIBiG (v.1.4) (22) (a BGC community supported centralized database of experimentally verified BGCs) were added based on Genbank accessions and coordinates of experimental clusters.
Data analysis
IMG users can query the data and perform comparative analysis through the public IMG-ABC web user interface (https://img.jgi.doe.gov/abc-public/) The new IMG-ABC menu essentially follows the layout of the IMG/MER (33) menu, but with two biosynthetic gene cluster (BGC) related items: Search BGCs and Browse BGCs menus. The Search BGCs menu allows users to search BGCs and SMs by IDs or by a variety of attributes. ‘Search by BGC Attributes’ option has been updated to include 58 types of biosynthetic gene clusters predicted by antiSMASH v5. An IMG-ABC tool for identification of putative clusters based on co-located genes with Pfams of interest, ClusterScout (35), has been also moved into Search BGCs menu. Furthermore, the tool has been modified to allow search in specific genomes of interest, which is available to authenticated users in IMG/MER-ABC (https://img.jgi.doe.gov/abc/). Such users can select a genome set from their Genome Workspace and run ClusterScout only on these genomes rather than entire IMG collection of public isolate genomes, thereby offering a faster turnaround from a more focused search. The default option of searching in all public isolate genomes is available to all users.
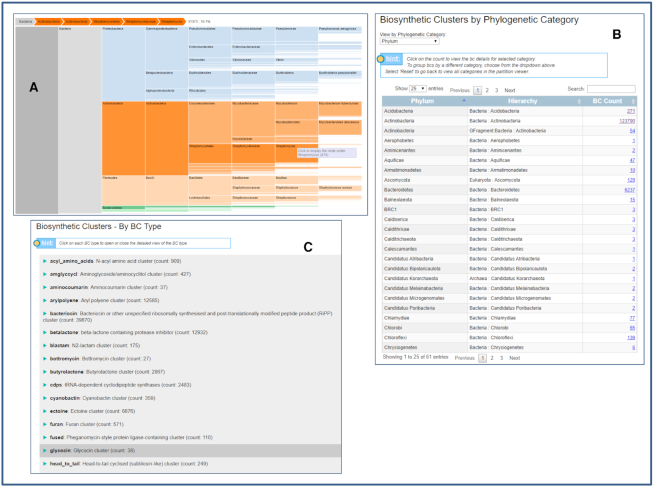
The Browse BGCs menu now provides seven options: Summary, -by Taxonomy, -by Ecosystem, -by BGC Type, -by SM Type, -by Gene Count and -by Pfam. The new ‘Browse by Taxonomy’ option allows users to view taxonomy of genomes, in which BGCs were predicted in a colorful tree map display as shown in Figure 1A. Users can zoom into any level of taxonomy by clicking on it. The ‘View by Phylogenetic Category’ dropdown list allows users to select a category (domain, phylum, class, etc.) to view cluster counts within each category in a tabular display (Figure 1B). The new ‘Browse by Ecosystem’ option displays ecosystem classification of genomes, similar to the ‘by Taxonomy’ display option. The new ‘Browse By BGC Type’ has 2 options: ‘All BGC Types’ shows all biosynthetic gene cluster types and the count of clusters associated with a particular BGC type (Figure 1C). Since antiSMASH v.5 predicts hybrid clusters, which may belong to multiple BGC types, users can click on each particular BGC type to see additional hybrid types. ‘BGC Types in Genomes’ allows users to select a set of genomes to view counts of BGC types of clusters in selected genomes in a clickable heatmap profile display. The rest of the options in ‘Browse BGCs’ menu are the same as in the previous version of IMG-ABC (35), but updated with the new data.
Figure 1.
Browse BGCs functions in IMG-ABC. (A) Browse By Taxonomy. (B) Biosynthetic Clusters by Phylogenetic Category. (C) Browse by BGC Type.
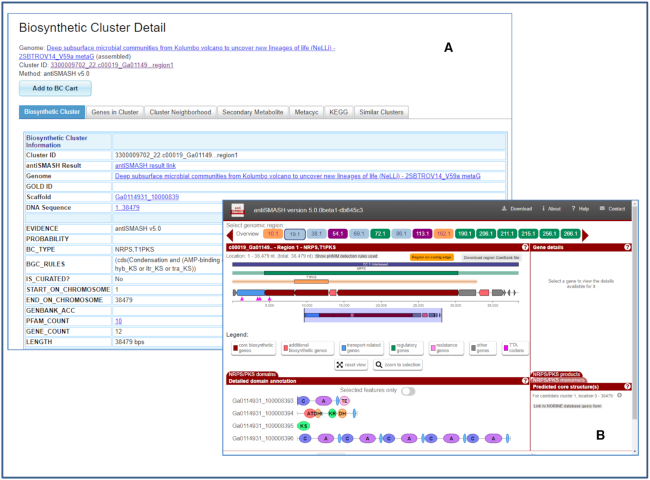
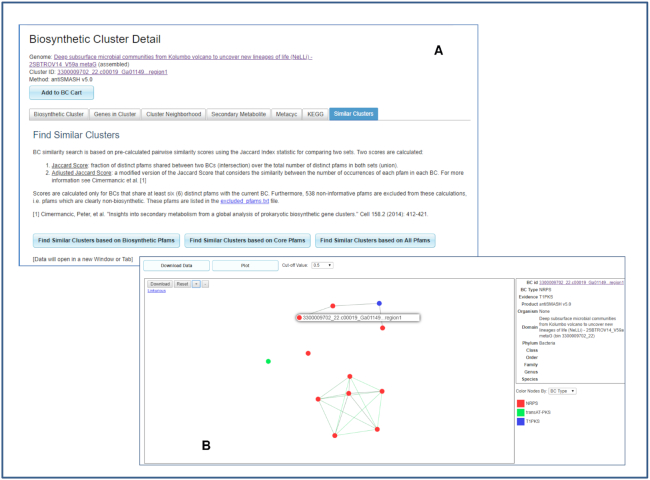
An updated Biosynthetic Cluster Detail page elaborates the BGC annotation details generated by antiSMASH v5. For example, cluster 3300009702_22.c00019_Ga01149…region1 detected in metagenome bin 3300009702_22 from Deep subsurface microbial communities from Kolumbo volcano to uncover new lineages of life (NeLLi) - 2SBTROV14_V59a metaG (IMG Taxon OID 3300009702, a public JGI metagenome) has been identified by antiSMASH v5 as a hybrid NRPS, T1PKS type (Figure 2A). In the ‘Biosynthetic Cluster’ tab, the ‘antiSMASH Result’ allows users to link out to the native display in antiSMASH (Figure 2B). ‘BGC_RULES’ displays the antiSMASH rule used to predict the cluster, while ‘Genes in Cluster’ tab shows IMG-generated Pfam, TIGRfam and KO annotations for all genes within the cluster. ‘Cluster Neighborhood’ tab displaying a gene cluster schematic has been updated to allow selection of a gene-coloring scheme based on COG, KEGG, Pfam or TIGRfam annotations. Other coloring options include GC content and taxonomy of top LAST hits (Phylo Distribution) (33) of genes encoded by the BGC. In addition, a search for potentially ‘Similar Clusters’ (tab) now has three options: use all Pfams assigned to genes in the BGC, use only ‘biosynthetic’ Pfams (i.e., Pfams assigned to genes annotated by antiSMASH as ‘biosynthetic’ or ‘biosynthetic-additional’) or use only ‘core’ Pfams (i.e., Pfams assigned to genes annotated by antiSMASH as ‘core’). While the Jaccard distance between BGCs is computed the same way as in previous IMG-ABC versions using a Jaccard score (20,35), these three options enable searches of different stringency. ‘Core Pfams’ option is the most stringent and requires that similar clusters have the same ‘Core Pfams’. On the other hand, ‘All Pfams’ option is the least stringent and retrieves similar clusters that share any Pfams, including those that are merely found within the neighborhood, but possibly not a bonafide member of the BGC (Figure 3A).
Figure 2.
A biosynthetic cluster example. (A) Detail view of cluster 3300009702_22.c00019_Ga01149…region1 in metagenome bin 3300009702_22 from Deep subsurface microbial communities from Kolumbo volcano to uncover new lineages of life (NeLLi) - 2SBTROV14_V59a metaG (B) antiSMASH v5 representation of the same cluster.
Figure 3.
IMG allows users to find similar clusters based on Pfam associations. (A) From the Similar Clusters tab, users can select to find similar clusters using only biosynthetic Pfams, only core Pfams or all Pfams. (B) Users can add similar clusters to Biosynthetic Cluster Cart to view similarities among them based on a selected cut-off value.
In order to enable seamless navigation between different IMG data marts, IMG now supports persistent Analysis Carts. For instance, users can use IMG/MER to find, select, filter and save a set of genomes, genes or functions to Analysis Carts, and then switch to IMG-ABC to perform BGC-related analyses using IMG-ABC specific tools and features. Several improvements have been made to the Biosynthetic Cluster Cart. All of the tools in the BGC Cart now support clusters from both isolate genomes and metagenome bins, as well as predicted BGCs generated by user-specified ClusterScout searches. An existing ‘Function HeatMap’ tab has been supplemented by ‘Function Profile’ tool, which enables selection of one function annotation source (COG, Pfam, TIGRfam, EC Numbers or KEGG KO Terms) and displays the counts of these functions in selected BGCs. The ‘Similarity Network’ tool has been updated to allow selection of cluster similarity cut-off value: user can adjust the default 0.5 cut-off to a different value between 0.2 and 0.9 and redisplay the results. For example, finding similar clusters of 3300009702_22.c00019_Ga01149…region1 based on biosynthetic Pfams returns a list of 100 similar clusters from both isolate genomes and metagenome scaffold bins. After saving the top finds into Biosynthetic Cluster Cart, ‘Similarity Network’ tab allows the user to adjust the cutoff value and plot similarity among selected BGCs as shown in Figure 3B.
USE CASES
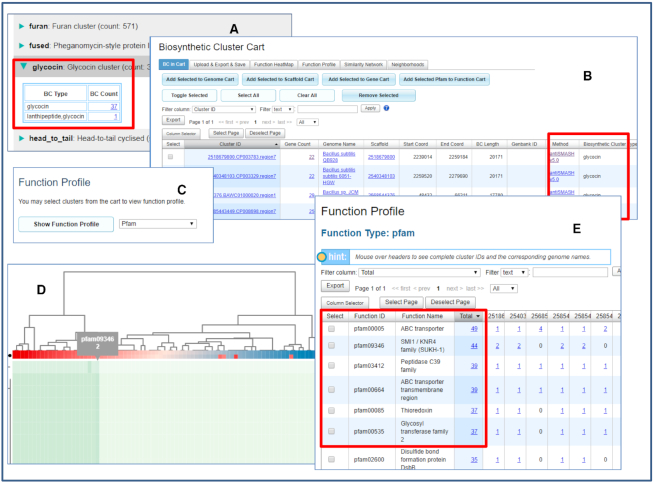
In order to demonstrate the capabilities of updated IMG-ABC v5, we chose an example of glycosylated bacteriocins called glycocins. Glycocins, such as sublancin (36) and glycocin F (37), are glycopeptide lantibiotics, which belong to class I bacteriocins. Several structurally characterized glycocins contain sugar moieties linked to side chains of cysteine residues via an unusual S-glycosidic bond (38). AntiSMASH v5 predicts glycocin biosynthesis gene clusters using sequence similarity to the precursor peptide, glycocin or sublancin, resulting in prediction of 38 glycocin BGCs in IMG isolate genomes, including one hybrid lantipeptide-glycocin type (Figure 4A). Taxonomy of the genomes harboring BGCs of glycocin type is restricted to the order Bacillales, which may be due to the limited sensitivity of sequence similarity search with short peptide sequence as query. IMG-ABC tools enable in-depth analysis of these clusters and suggest supplementary or alternative ways to predict putative glycocin-encoding BGCs and prioritize them for experimental characterization.
Figure 4.
(A) There are 38 predicted glycocin BGCs in IMG/ABC v5. (B) Users can add all 38 clusters to Biosynthetic Cluster Cart. (C) Biosynthetic Cluster Cart provides many analysis functions, including the new Function Profile. (D) Function Heatmap shows Pfam associations of the clusters in a heap map tree display. (E) Function Profile shows that most glycocin BGCs are associated with a few Pfams.
After adding all predicted glycocin BGCs to Biosynthetic Cluster Cart (Figure 4B), their function profiles can be analyzed using Function Profile (Figure 4C) and/or Function Heat Map (Figure 4D) panels leading to identification of the predominant protein families in these clusters. Function Profile tool with a TIGRfam and a Pfam option suggests two alternatives: using TIGRfams TIGR04196 and/or TIGR04195 family (glycopeptide, sublancin family and peptide S-glycosyltransferase, SunS family, respectively) or a combination of four Pfams (PF00005, PF00535, PF03412 and PF00664 (Figure 4E), representing ABC transporter, Glycosyl transferase family 2, Peptidase C39 family and ABC transporter transmembrane region, respectively).
Genes assigned to TIGR04195 and TIGR04196 can be retrieved using generic IMG tools (33) like Function Search in Find Functions menu, now available in IMG-ABC. This search retrieves the operons encoding biosynthesis of sublancin, thurandacin (39) and pallidocin (40). The genes assigned to these TIGRfams in all or a subset of IMG genomes can be selected, added to Gene Cart, and analyzed using both generic and IMG-ABC-specific Gene Cart displays. For instance, users can see whether these genes are part of antiSMASH-predicted BGCs. Interestingly, SunS family glycosyltransferase (TIGR04195), but not the precursor peptide, is found in two strains of a thermophilic firmicute Laceyella sacchari (41). Although putative glycocin-encoding operon in these genomes is not part of a prophage like in Bacillus subtilis, it still shows clear signs of horizontal gene transfer (GC content much lower than average and no similar genes in other genomes of Thermoactinomycetaceae). Unfortunately, the lack of precursor peptides in these genomes is owing to the errors of gene finding tools, which often fail to predict short peptides with unusual amino acid composition. IMG’s generic tools, such as ‘Sequence Viewer for Alternate ORF Search’ available on Gene Details pages enables prediction of putative precursor glycocin-like peptides based on their unique features, including the presence of double-glycine signal peptide (38) and enrichment of the C-terminus in serine and cysteine residues.
A much broader approach to predict glycocin-like BGCs is to use ClusterScout tool in Search BGCs menu in IMG-ABC (35), which allows users to find genomic regions of interest based on the co-occurrence of genes assigned to certain Pfam families, such as those identified as predominant through Function Profile analysis of antiSMASH-predicted glycocin clusters. A search using PF00535, PF03412, and PF00664 corresponding to glycosyltransferase and a fused peptidase/ABC transporter as ClusterScout hooks with a maximum distance of 900 nt between them and a minimum distance from scaffold/contig edge of 1000 nt results in 268 genomic regions (Figure 5). After selecting some or all of them and adding to Biosynthetic Cluster Cart (Figure 5B), users can visualize them in Neighborhoods tab (Figure 5C) and remove spurious genomic regions, such as those with disruptions by IS elements and frameshifts or those in which glycosyltransferase and peptidase/ABC transporter are convergent and therefore unlikely to be part of the same operon.
Figure 5.
(A) Use ClusterScout to search using PF00535, PF03412 and PF00664 corresponding to glycosyltransferase and a fused peptidase/ABC transporter as ClusterScout hooks with a maximum distance of 900 nt between them and a minimum distance from scaffold/contig edge of 1000 nt. (B) Results can be saved into Biosynthetic Cluster Cart. (C) User can use the ‘View Selected BGCs Neighborhoods’ to check neighborhood of selected BGCs. (D) Similarity Network of glycocin-like BGCs predicted in Bacillus subtilis, Bacillus cereus, and Bacillus thuringiensis genomes using cut-off value of 0.5 and coloring based on species. While all clusters predicted in B. subtilis fall into a single group based on the similarity of their overall Pfam composition, predicted BGCs in B. cereus and B. thuringiensis are split into 3 distinct clusters. (E) Alignment of putative glycocin precursors from Firmicutes genomes.
Review of ClusterScout search results shows that genomic regions include the majority of glycocin clusters predicted by antiSMASH, as well as glycocin BGCs selected for expression in (42). New and updated Biosynthetic Cluster Details and Biosynthetic Cluster Cart tools enable users to visualize the overall similarity between clusters based on their Pfam composition via Similarity Network and Function HeatMap in Biosynthetic Cluster Cart (limit of 50 BGCs). Figure 5D shows the Similarity Network of glycocin-like BCs predicted in B. subtilis, Bacillus cereus and Bacillus thuringiensis genomes. A cut-off value of 0.5 and coloring based on species is used in visualization, which shows that, while all clusters predicted in B. subtilis fall into a single group based on the similarity of their overall Pfam composition, predicted BGCs in B. cereus and B. thuringiensis are split into three distinct groups.
Function Profile tab in Biosynthetic Cluster Cart allows users to select protein families of interest for in-depth analysis. For instance, users can click the count of genes associated with PF00535 (glycosyltransferase), select some or all of them and add them to Gene Cart in order to perform multiple sequence alignment and tree reconstruction with Clustal Omega (43) and visualize the alignment using MSA Viewer (44). These analyses help delineating subfamilies of SunS-like S-glycosyltransferases, as well as O-/S-glycosyltransefases (39,45) and predict potential glycosylation patterns. At last, by adding all genes encoded by BGCs of interest to Gene Cart, and using Gene Cart table configuration and filtering options to remove longer proteins, as well as those assigned to Pfam, users can study the diversity of putative glycocin precursor peptides by performing multiple sequence alignment. For example, alignment of putative glycocin precursors found in ClusterScout-predicted genomic regions in Firmicutes genomes (Figure 5E) shows that in addition to previously characterized peptides with five Cys residues, such as sublancin, thurandacin and glycocin F, there is a peptide with seven Cys residues found in the genome of Bacillus plakortidis DSM 19153. Furthermore, there are peptides with three Cys residues found in Paenibacillus strains and even peptides with a single Cys residue found in multiple B. cereus and B. thuringiensis strains suggesting that the diversity of glycocin-like glycosylated peptides is far from being fully characterized even in Firmicutes, let alone other lineages.
The taxonomic origin of additional predicted glycocin BGCs can be explored by selecting BGCs of interest and adding their genomes to Genome Cart. The majority of predicted clusters are found in Firmicutes genomes, including several Streptococcus and Staphylococcus spp. However, they are also present in several Streptomyces genomes, and even in a few representatives of gram-negative Bacteroidetes phylum. Thus, an updated set of IMG-ABC tools enables users to review antiSMASH predictions, formulate their own BGC detection rules, compare the results of the two and identify the most interesting candidates for in-depth analysis of their diversity.
CONCLUSION
SMs are an incredible resource of useful bioactive molecules and there still remains significant untapped diversity. Access to cryptic BGCs resulting from high-throughput whole-genome and metagenome sequences has resulted in renewed interest in natural product discovery (4,10,12,13).
IMG-ABC is the largest publicly available database of predicted and experimental BGCs and resultant SMs (34,35). IMG-ABC relies on IMG’s comprehensive integrated structural and functional genomic data for the analysis of BGCs and their associated SMs, as well as environmental metadata from GOLD (46). SMs and BGCs serve as the two main classes of objects in IMG-ABC, each with a rich collection of attributes. A unique feature of IMG-ABC is the incorporation of both experimentally validated and computationally predicted BGCs in genomes, as well as metagenome-derived scaffold bins, revealing BGCs in uncultured populations and rare taxa. As illustrated in the case study example, the system also includes powerful search and analysis tools that are integrated with IMG’s extensive genomic/metagenomic data and analysis tool kits. An overlay of BGC data on a unique collection of IMG genome and metagenome data combined with an extensive collection of search, analysis and export tools enables users to perform large-scale BGC analyses at various granularity levels ranging from individual genes and proteins to whole ecosystems thereby complementing BGC-specific database, such as antiSMASH database (26). IMG-ABC strives to fill the niche among resources for integrated computational exploration of the secondary metabolism universe; its underlying scalable framework enables traversal of uncovered phylogenetic and chemical structure space, serving as a platform for the discovery of novel molecules. As new research on BGCs and SMs is published, and more genomes are sequenced, IMG-ABC will continue to expand, with the goal of becoming an essential component of any bioinformatic exploration of the secondary metabolism community. We will also seek community input on what features are most desirable to include in future releases of IMG-ABC.
ACKNOWLEDGEMENTS
We thank Daniel Udwary (DOE Joint Genome Institute) for his help with antiSMASH v5 BGC prediction of IMG genomes and his suggestions to improve the paper.
FUNDING
Director, Office of Science, Office of Biological and Environmental Research, Life Sciences Division, U.S. Department of Energy [DE-AC02–05CH11231]. Funding for open access charge: Joint Genome Institute; Lawrence Berkeley National Laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1. Traxler M.F., Kolter R.. Natural products in soil microbe interactions and evolution. Nat. Prod. Rep. 2015; 32:956–970. [DOI] [PubMed] [Google Scholar]
- 2. Donia M.S., Fischbach M.A.. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science. 2015; 349:1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guo C.J., Chang F.Y., Wyche T.P., Backus K.M., Acker T.M., Funabashi M., Taketani M., Donia M.S., Nayfach S., Pollard K.S. et al.. Discovery of reactive microbiota-derived metabolites that inhibit host proteases. Cell. 2017; 168:e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jensen P.R. Natural products and the gene cluster revolution. Trends Microbiol. 2016; 24:968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mendes R., Kruijt M., de Bruijn I., Dekkers E., van der Voort M., Schneider J.H., Piceno Y.M., DeSantis T.Z., Andersen G.L., Bakker P.A. et al.. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011; 332:1097–1100. [DOI] [PubMed] [Google Scholar]
- 6. Piel J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2009; 26:338–362. [DOI] [PubMed] [Google Scholar]
- 7. Ling L.L., Schneider T., Peoples A.J., Spoering A.L., Engels I., Conlon B.P., Mueller A., Schäberle T.F., Hughes D.E., Epstein S. et al.. A new antibiotic kills pathogens without detectable resistance. Nature. 2015; 517:455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Newman D.J., Cragg G.M.. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016; 79:629–661. [DOI] [PubMed] [Google Scholar]
- 9. Zhang F., Rodriguez S., Keasling J.D.. Metabolic engineering of microbial pathways for advanced biofuels production. Curr. Opin. Biotechnol. 2011; 22:775–783. [DOI] [PubMed] [Google Scholar]
- 10. Shen B. A new golden age of natural products drug discovery. Cell. 2015; 163:1297–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davies J. Specialized microbial metabolites: functions and origins. J. Antibiot. (Tokyo). 2013; 66:361–364. [DOI] [PubMed] [Google Scholar]
- 12. Baltz R.H. Gifted microbes for genome mining and natural product discovery. J. Ind. Microbiol. Biotechnol. 2017; 44:573–588. [DOI] [PubMed] [Google Scholar]
- 13. Baltz R.H. Natural product drug discovery in the genomic era: realities, conjectures, misconceptions, and opportunities. J. Ind. Microbiol. Biotechnol. 2018; 46:281–299. [DOI] [PubMed] [Google Scholar]
- 14. Tietz J.I., Schwalen C.J., Patel P.S., Maxson T., Blair P.M., Tai H.C., Zakai U.I., Mitchell D.A.. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 2017; 13:470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gomez-Escribano J.P., Alt S., Bibb M.J.. Next generation sequencing of actinobacteria for the discovery of novel natural products. Mar. Drugs. 2016; 14:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Katz L., Baltz R.H.. Natural product discovery: past, present, and future. J. Ind. Microbiol. Biotechnol. 2016; 43:155–176. [DOI] [PubMed] [Google Scholar]
- 17. Grubbs K.J., Bleich R.M., Santa Maria K.C., Allen S.E., Farag S., Shank E.A., Bowers A.A., Bowers A.A.. Large-scale bioinformatics analysis of Bacillus genomes uncovers conserved roles of natural products in bacterial physiology. mSystems. 2017; 2:e00040-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doroghazi J.R., Albright J.C., Goering A.W., Ju K.S., Haines R.R., Tchalukov K.A., Labeda D.P., Kelleher N.L., Metcalf W.W.. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 2014; 10:963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chavali A.K., Rhee S.Y.. Bioinformatics tools for the identification of gene clusters that biosynthesize specialized metabolites. Brief. Bioinform. 2017; 19:1022–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cimermancic P., Medema M.H., Claesen J., Kurita K., Wieland Brown L.C., Mavrommatis K., Pati A., Godfrey P.A., Koehrsen M., Clardy J. et al.. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014; 158:412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen R., Wong H.L., Burns B.P.. New approaches to detect biosynthetic gene clusters in the environment. Medicines. 2019; 6:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K., de Bruijn I., Chooi Y.H., Claesen J., Coates R.C. et al.. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015; 11:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Medema M.H., Fischbach M.A.. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015; 11:639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blin K., Kim H.U., Medema M.H., Weber T.. Recent development of antiSMASH and other computational approaches to mine secondary metabolite biosynthetic gene clusters. Brief. Bioinform. 2017; 20:1103–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Medema M.H., Blin K., Cimermancic P., de Jager V., Zakrzewski P., Fischbach M.A., Weber T., Takano E., Breitling R.. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011; 39:W339–W346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blin K., Medema M.H., Kazempour D., Fischbach M.A., Breitling R., Takano E., Weber T.. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013; 41:W204–W212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R., Lee S.Y., Fischbach M.A., Müller R., Wohlleben W. et al.. AntiSMASH 3.0-A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015; 43:W237–W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blin K., Wolf T., Chevrette M.G., Lu X., Schwalen C.J., Kautsar S.A., Suarez Duran H.G., de Los Santos E.L.C., Kim H.U., Nave M. et al.. antiSMASH 4.0- improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017; 45:W36–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S.Y., Weber T.. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019; 47:W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blin K., Medema M.H., Kottmann R., Lee S.Y., Weber T.. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2017; 45:D555–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Skinnider M.A., Dejong C.A., Rees P.N., Johnston C.W., Li H., Webster A.L., Wyatt M.A., Magarvey N.A.. Genomes to natural products PRediction Informatics for Secondary Metabo- lomes (PRISM). Nucleic Acids Res. 2015; 43:9645–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mouncey N.J., Otani H., Udwary D., Yoshikuni Y.. New voyages to explore the natural product galaxy. J. Ind. Microbiol. Biotechnol. 2019; 46:273–279. [DOI] [PubMed] [Google Scholar]
- 33. Chen I.A., Chu K., Palaniappan K., Pillay M., Ratner A., Huang J., Kyrpides N.C.. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019; 47:D666–D677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hadjithomas M., Chen I.M., Chu K., Ratner A., Palaniappan K., Szeto E., Huang J., Reddy T.B., Cimermancic P., Fischbach M.A. et al.. IMG–ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. Mbio. 2015; 6:e00932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hadjithomas M., Chen I.A., Chu K., Huang J., Ratner A., Palaniappan K., Andersen E., Markowitz V., Kyrpides N.C., Ivanova N.N.. IMG-ABC: new features for bacterial secondary metabolism analysis and targeted biosynthetic gene cluster discovery in thousands of microbial genomes. Nucleic Acids Res. 2017; 45:D560–D565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oman T.J., Boettcher J.M., Wang H., Okalibe X.N., van der Donk W.A.. Sublancin is not a lantibiotic but an S-linked glycopeptide. Nat. Chem. Biol. 2011; 7:78–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stepper J., Shastri S., Loo T.S., Preston J.C., Novak P., Man P., Moore C.H., Havlíček V., Patchett M.L., Norris G.E.. Cysteine S-glycosylation, a new post-translational modification found in glycopeptide bacteriocins. FEBS Lett. 2011; 585:645–650. [DOI] [PubMed] [Google Scholar]
- 38. Norris G.E., Patchett M.L.. The glycocins: in a class of their own. Curr. Opin. Struct. Biol. 2016; 40:112–119. [DOI] [PubMed] [Google Scholar]
- 39. Wang H., Oman T.J., Zhang R., Garcia De Gonzalo C.V., Zhang Q., van der Donk W.A.. The glycosyltransferase involved in thurandacin biosynthesis catalyzes both O- and S-glycosylation. J. Am. Chem. Soc. 2013; 136:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kaunietis A., Buivydas A., Čitavičius D.J., Kuipers O.P.. Heterologous biosynthesis and characterization of a glycocin from a thermophilic bacterium. Nat. Commun. 2019; 10:1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lacey J. Thermoactinomyces sacchari sp. nov., a thermophilic actinomycete causing bagassosis. J. Gen. Microbiol. 1971; 66:327–338. [DOI] [PubMed] [Google Scholar]
- 42. Ren H., Biswas S., Ho S., van der Donk W.A., Zhao H.. Rapid discovery of glycocins through pathway refactoring in Escherichia coli. ACS Chem. Biol. 2018; 13:2966–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Higgins D.G.. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011; 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yachdav G., Wilzbach S., Rauscher B., Sheridan R., Sillitoe I., Procter J., Goldberg T.. MSAViewer: interactive JavaScript visualization of multiple sequence alignments. Bioinformatics (Oxford, England). 2016; 32:3501–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nagar R., Rao A.. An iterative glycosyltransferase EntS catalyzes transfer and extension of O- and S-linked monosaccharide in enterocin 96. Glycobiology. 2017; 27:766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Katta H.Y., Mojica A., Reddy T.. Genomes OnLine database (GOLD) v.7: updates and new features. Nucleic acids Res. 2019; 47:D649–D659. [DOI] [PMC free article] [PubMed] [Google Scholar]