Abstract
BMVC is the first fluorescent probe designed to detect G-quadruplexes (G4s) in vivo. The MYC oncogene promoter forms a G4 (MycG4) which acts as a transcription silencer. Here, we report the high-affinity and specific binding of BMVC to MycG4 with unusual slow-exchange rates on the NMR timescale. We also show that BMVC represses MYC in cancer cells. We determined the solution structures of the 1:1 and 2:1 BMVC–MycG4 complexes. BMVC first binds the 5′-end of MycG4 to form a 1:1 complex with a well-defined structure. At higher ratio, BMVC also binds the 3′-end to form a second complex. In both complexes, the crescent-shaped BMVC recruits a flanking DNA residue to form a BMVC-base plane stacking over the external G-tetrad. Remarkably, BMVC adjusts its conformation to a contracted form to match the G-tetrad for an optimal stacking interaction. This is the first structural example showing the importance of ligand conformational adjustment in G4 recognition. BMVC binds the more accessible 5′-end with higher affinity, whereas sequence specificity is present at the weaker-binding 3′-site. Our structures provide insights into specific recognition of MycG4 by BMVC and useful information for design of G4-targeted anticancer drugs and fluorescent probes.
INTRODUCTION
G-quadruplexes are non-canonical four-stranded nucleic acid secondary structures that consist of stacked G-tetrads, which are formed by four guanines connected by Hoogsteen hydrogen bonding and stabilized by monovalent cations such as K+ and Na+ (1). Intramolecular DNA G-quadruplexes form in human guanine-rich sequences with functional significance, such as human telomeres (2), the promoter regions of human oncogenes (3–5), and 5′-UTRs (6). Significantly, G-quadruplexes formation were elevated in both human precancerous cells and cancer tissues (7), and G-quadruplex structures were enriched in the MYC promoter of highly transcribed cells (7,8).
The MYC proto-oncogene is a critical transcription factor, regulating genes in various normal cellular processes such as cell growth, proliferation, differentiation, as well as apoptosis (9,10). Overexpression of MYC is widely observed in most types of human malignancies (11–15). Transcriptional repression of MYC is an attractive strategy in modulating MYC expression (16). The nuclease hypersensitive element (NHE) III1 region of the MYC promoter controls 80–95% of MYC transcription (17,18) and can form a DNA G-quadruplex that is a transcription silencer (19,20). Compounds that stabilize the MYC promoter G-quadruplex repress MYC gene expression in vivo (21–24). Therefore, the MYC promoter G-quadruplex is a promising target for anticancer drug development (4,5,20).
The major G-quadruplex formed in the MYC promoter NHE III1 adopts a parallel-stranded folding topology in K+ solution (25,26). We have previously determined the molecular structure of this major MYC promoter G-quadruplex (27). It is a three-tetrad parallel structure with three propeller loops of 1-, 2- and 1-nt (MycG4, Figure 1A). Two solution structures of the ligand-complexes of MycG4 have been reported (28,29). Both ligands bind at intermediate or intermediate-to-fast exchange rate on the NMR time scale.
Figure 1.

(A) MycG4, the major G-quadruplex formed in the MYC promoter NHE III1 in K+ solution, a parallel-stranded structure with a 1:2:1 loop-length arrangement. Red box = guanine, green ball = adenine, blue ball = thymine. (B) Structure of BMVC molecule with numbering.
3,6-bis(1-Methyl-4-vinylpyridinium) carbazole diiodide (BMVC, Figure 1B) is a G-quadruplex-specific ligand and the first in vivo fluorescent probe to detect the G-quadruplex structures in human telomeres (30–32). However, the cellular localizations and effects of BMVC and its derivatives are not limited to telomeres (33,34). BMVC is also a promising fluorescent marker of cancer cells and a potential antitumor agent (35–41). However, the molecular basis of G-quadruplex recognition by BMVC is unknown.
Herein, we report the specific binding of BMVC to the MycG4. As shown by the slow-exchange binding on the NMR timescales, BMVC binds the MycG4 with greater affinity and specificity than the human telomeric G-quadruplexes. BMVC first binds the 5′-end of MycG4 to form a 1:1 complex; at higher ratio, BMVC also binds the 3′-end to form a second complex. The solution structures of the 1:1 and 2:1 BMVC–MycG4 complexes are determined by NMR. The molecular structures show that the specific binding of BMVC with MycG4 is achieved by the pairing recognition of the MycG4 flanking bases and the conformational adjustment of the BMVC molecule. Moreover, BMVC represses MYC expression as demonstrated by qRT-PCR and western blot results. Our structures provide molecular-level mechanism of MycG4 recognition by BMVC, and useful information for future design of G4-targeted anticancer drugs and fluorescent probes.
MATERIALS AND METHODS
Oligonucleotides
DNA oligonucleotides were synthesized using β-cyanoethylphosphoramidite solid-phase chemistry on an Expedite 8809 nucleic acid synthesis system (Applied Biosystems, Inc.) with dimethoxytrityl (DMT)-ON setting and were purified using MicroPure II Columns from BioSearch Technologies (Novato, CA, USA), as described previously (27,28).
NMR Experiments
The water NMR samples were prepared in 25 mM K-phosphate and 70 mM KCl buffer at pH 7 in D2O/H2O (10%/90%). The D2O samples were lyophilized and redissolved in 99.98% D2O two more times. Each sample was heated to 95 °C for 5 min and cooled slowly to room temperature. The final concentrations of DNA oligonucleotides were 0.1–2.5 mM. NMR experiments were performed on a Bruker DRX-600 MHz spectrometer Standard homonuclear 2D NMR experiments, including DQF-COSY, TOCSY and NOESY, were collected at temperatures of 5, 15, 25 and 35 °C for the BMVC and MycG4 complex samples in water and D2O solution in 95 mM K+ at pH 7. The mixing times were set from 50–250 ms for NOESY, and at 40 ms and 80 ms for TOCSY experiments. The NMR experiments for samples in water solution were performed with WATERGATE or jump-return (NOE11) water suppression techniques. Peak assignment and integration were achieved using Sparky (UCSF). Distances between protons were assigned based on the nuclear overhauser effect (NOE) cross peaks integrated at 200 ms mixing times, with the upper and lower boundaries assigned to ±20% of the estimated distances. The methyl base proton Me-H6 distance (2.99 Å) was used as a reference. The distances involving the unresolved protons, e.g. methyl protons, were assigned using pseudo-atom notation in X-PLOR.
NOE-Distance restrained molecular dynamics simulation
Structure calculations were performed using NOE-restrained molecular dynamics simulation in the program XPLOR (42) and Insight II/Discover (version 2000L, Accelrys, CA, USA). The starting model of the 1:1 and 2:1 BMVC–MycG4 complexes, respectively, were constructed in Insight II, with the conformations deduced from the NOE data. This starting model with two potassium cations in the tetrad core and 21 potassium counter-ions was then soaked into water solvent using 15 Å water layer in Insight II and CFF force field was used for the calculations. The complete molecular system was first subjected to energy minimization to the system convergence of 1 kcal/mol/Å in Insight II. Topology and parameter files were generated using PRODRG (43). The energy minimized molecular system was then subjected to a NOE distance-restrained molecular dynamic simulation at 300 K in XPLOR. A total of 481 and 516 distance restraints were used in the NOE-restrained dynamics calculations of the 1:1 and 2:1 complexes, respectively, of which 82 and 120 are from intermolecular NOEs between BMVC and MycG4. Hydrogen bond restraints were applied to the G-tetrads. NOE-restrained simulated annealing refinement calculations were performed as described previously (28). The 20 best molecules were selected based on their minimal energy terms and minimal number of NOE violations.
Fluorescence Kd measurement
Fluorescence spectra were acquired with a Jasco-FP8300 spectrofluorometer (Jasco Inc., Easton, MD, USA) equipped with a temperature-controlled circulator. Fluorescence was measured between 500 and 600 nm in a quartz cell with path length of 1 cm, using an excitation wavelength of 475 nm. The titration experiments were carried out at a BMVC concentration of 20 nM in presence of 25 mM K-phosphate and 70 mM KCl buffer or 25 mM Na-phosphate and 70 mM NaCl buffer at pH 7. Six different DNA sequences at the specific concentration were added into the BMVC solutions and the resulting solutions were incubated for 2 min before fluorescence was measured. The apparent Kd was calculated using GraphPad Prism software to fit the equation assuming a 1:1 model: F = Fmax [(MT + LT + Kd) – ((MT + LT + Kd)2 – (4MTLT))1/2]/(2LT), where F represents the BMVC fluorescence intensity at 563 nm. Fmax is fitted in addition to apparent Kd. The total BMVC (ligand) concentration, LT, was held constant and MT, the total DNA (Macromolecule) concentration is an independent variable, varying with each titration step. The binding isotherm could be fitted with the equation assuming a 1:1 model, suggesting that the apparent Kd is probably dominated by the stronger binding site at the 5′-end.
Circular dichroism (CD) spectroscopy experiments
Circular dichroism spectra were recorded using a Jasco-1100 spectropolarimeter (Jasco Inc., Easton, MD, USA) equipped with a temperature control cell holder. Samples were prepared in 0.5 mM K+-containing phosphate buffer at MycG4 DNA concentrations of 15 μM. CD measurements were taken through a quartz cell with a 1 mm path length. Spectra were attained using three averaged scans between 230 and 330 nm at 25 °C. The baseline was corrected by subtracting signal contributions from the buffer. Melting experiments were conducted by recording the CD intensity of the DNA or DNA–ligand complex at 264 nm as a function of temperature. The temperature ranged from 25 to 95 °C with 2 °C/min heating rate.
Electrophoretic mobility shift assay (EMSA)
Gel electrophoresis experiments were carried out with a 10 × 7 cm 1.5 mm thick native gel containing 16% acrylamide (acrylamide:bis-acrylamide = 29:1) in 1× TBE buffer, pH 8, supplemented with 12.5 mM KCl. Each sample contains 0.4 nmol DNA. DNA bands were visualized by ethidium bromide staining. The gels were photographed under UV light at 254 nm by a Bio-Rad imaging detector and recorded by a digital camera.
Western blot
After pelleting down, MCF7 cells were solubilized in 1× RIPA buffer supplemented with 1× protease inhibitor cocktail (Roche, USA). The nucleus and intracellular organelles were disrupted by sonication five times for 10 s each. After incubation for 20 min at 4 °C, the suspensions were separated by centrifugation at 13 000 rpm for 10 min at 4 °C and the supernatant was collected. 20 ng protein was used for Western Blot analysis. The protein bands were detected using anti-MYC (D84C12, Cell Signaling Technology, USA) and anti-GAPDH (FL-335, Santa Cruz Biotechnology, USA) antibodies.
Real-Time quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was isolated using TRIzol reagent. 1 μg of RNA was subjected to cDNA synthesis using qScript cDNA Supermix (Quanta BioSciences, USA) according to manufacturer's instructions. For each reaction, a mix of the following reaction components was prepared to the indicated end-concentration: 3.2 μl water, 0.8 μl of 1:3 diluted cDNA synthesis products, 0.25 μM of each primer for MYC (forward primer: 5′-GCTGCTTAGACGCTGGATT-3′; reverse primer: 5′-TCCTCCTCGTCGCAGTAGA-3′) and GAPDH (forward primer: 5′-GGTGGTCTCCTCTGACTTCAACA-3′, reverse primer: 5′-GTTGCTGTAGCCAAATTCGTTGT-3′) and 5 μl of IQ SYBR Green Supermix (Bio-Rad Laboratories, USA). Relative gene expression was calculated by using the 2−ΔΔCT, in which the amount of MYC mRNA was normalized to an endogenous reference (GAPDH). Melting curve analysis or agarose gel electrophoresis was carried out to confirm correct PCR products.
RESULTS
BMVC specifically binds the MycG4 with high affinity
We used NMR to investigate the interaction modes and dynamic binding of the MycG4 (Figure 1A) with BMVC (Figure 1B) under the physiologically relevant K+ conditions. In 1D 1H NMR spectra (Figure 2), upon addition of BMVC into the free MycG4 solution, a new set of sharp, distinct imino proton peaks (labeled in red) appeared immediately at lower ligand equivalence 0–0.7, while the imino proton peaks of free MycG4 remained, indicating a slow-exchange binding of BMVC to the MycG4 on the NMR timescale, characteristic of high-affinity binding. The new set of 12 well-resolved imino proton peaks suggests the formation of a well-defined BMVC−MycG4 complex. The presence of a second BMVC binding site was observed, as further addition of BMVC at ligand equivalence 0.7–1.8 showed another distinct set of imino proton peaks (labeled in blue). At BMVC/MycG4 ratio of 1.5, the imino proton spectrum showed the clear presence of two BMVC−MycG4 complexes. BMVC binds the higher-affinity binding site of MycG4 with an apparent Kd value of 36 nM (Supplementary Figure S1), stronger than most reported G4-ligands (44,45), and significantly increases the thermal stability of the MycG4 (Supplementary Figure S2). BMVC-bound MycG4 was visualized by electrophoretic mobility shift assay (EMSA) under native conditions (Supplementary Figure S3). The clear bands at the BMVC/MycG4 ratio of 0.5, 1 and 2, indicated stable complex formation between BMVC and MycG4 and that the complexes were observed at monomeric states.
Figure 2.
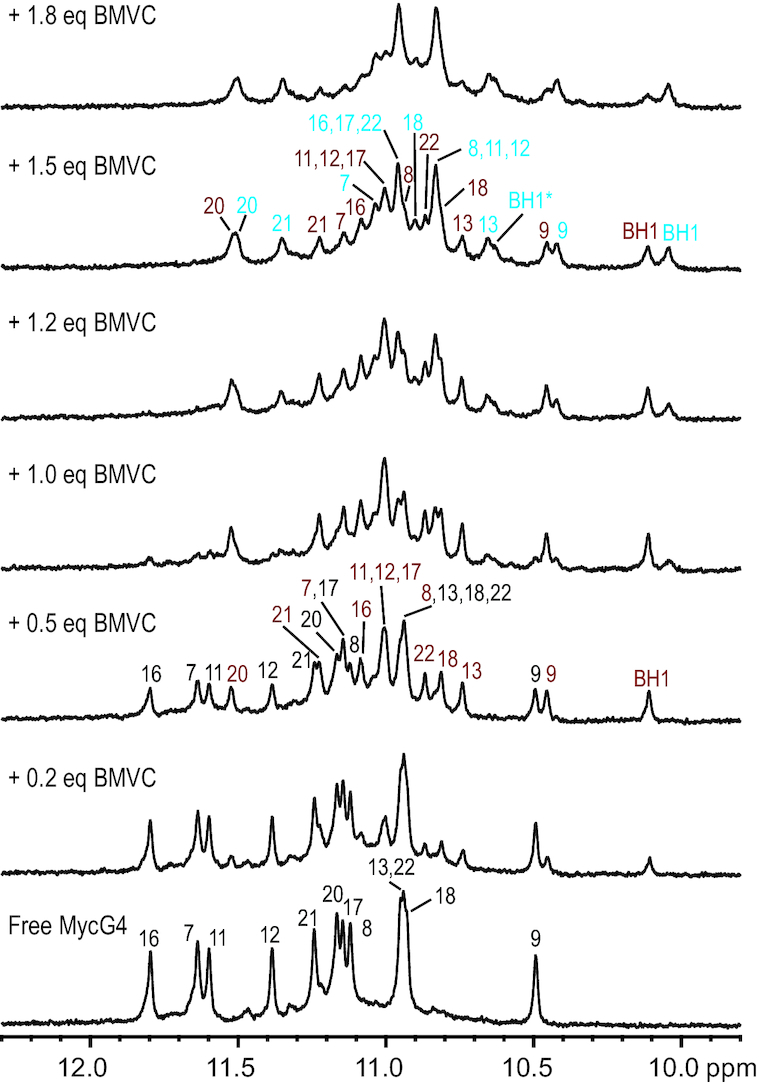
Imino proton region of the 1D 1H NMR titration spectrum of MycG4 with BMVC in pH 7, 95 mM K+ solution, at 25 °C. The assignments of imino protons from free MycG4 DNA are colored in black, 1:1 BMVC–MycG4 complex in red, 2:1 BMVC–MycG4 complex in blue. The H1 protons of BMVC molecules (BH1 in the 5'-end complex or BH1* in the 3'-end complex) are also shown in the spectra. The BMVC/MycG4 ratios are shown on the left side of spectra.
Using 1D 1H NMR, we titrated BMVC against various other G-quadruplex sequences, including basket-type and hybrid-type human telomeric G-quadruplexes, and parallel-stranded VEGF and Myc1234 G-quadruplexes (Supplementary Figure S4). BMVC showed no specific binding to basket-type or hybrid-type human telomeric G-quadruplexes, as observed by the broadened and not well-resolved imino proton peaks of those G-quadruplexes at ligand/DNA ratio 0.5–1.0, while better binding was observed for the parallel-stranded VEGF and Myc1234 G-quadruplexes. We also determined the apparent Kd values of BMVC binding to other G-quadruplexes using fluorescence titrations. The results showed the highest affinity of BMVC for MycG4, but the lowest affinity for the basket-type human telomeric G-quadruplex.
2D NMR studies of the 1:1 and 2:1 BMVC−MycG4 complexes
As the well-resolved 1D NMR spectra (Figure 2) indicated both BMVC−MycG4 complexes were suitable for high-resolution structural analysis, we decided to solve these two structures in order to understand how BMVC interacts with MycG4. The NMR structure determination of the BMVC−MycG4 complexes was not trivial, as BMVC did not form a single species of 1:1 or 2:1 complex at any ligand:DNA ratio. Instead, at BMVC/MycG4 ratio of 0.5, a mixture of the free DNA and 1:1 BMVC−MycG4 complex was observed, while at BMVC/MycG4 ratio of 1.5, a mixture of 1:1 and 2:1 BMVC−MycG4 complexes was observed. Therefore, the NMR spectral assignment and structural determination of the 1:1 and 2:1 complexes were both conducted in the co-presence of another molecular species. We collected sets of 2D-NOESY, TOCSY and COSY NMR data in both water and D2O at BMVC/MycG4 ratios of 0.5 and 1.5, at temperatures of 5, 15, 25 and 35 °C. Notably, the BMVC-H1 proton (BH1) was clearly observed for the 1:1 and 2:1 complex, respectively, and was an important indicator of complex formation (Figure 2). At BMVC/MycG4 ratio of 0.5, one BMVC-H1 signal was detected at chemical shift ∼10.1 ppm, indicating the formation of one BMVC−MycG4 complex at the 5′-end. At BMVC/MycG4 ratio of 1.5, three BMVC-H1 signals were observed (10, 10.1 and 10.6 ppm), with one from the 1:1 complex at the 5′-end and two from the 2:1 complex at the 5′- and 3′-end, indicating the formation of two BMVC-MycG4 complexes.
Complete 2D NMR proton assignment of the 1:1 BMVC−MycG4 complex showing BMVC binds to the 5′-end of MycG4 and stabilizes the MycG4
The imino and aromatic proton resonances of the bound MycG4 in the 1:1 complex were unambiguously assigned based on the exchange cross-peaks of the free and bound MycG4 at BMVC/MycG4 ratios of 0.5 (Figure 3A). The proton assignment of the free MycG4 G-quadruplex was based on the previous report (27), with corrections on G5 and A6 assignments (Supplementary Figure S5 and Table S1). Using the sequential assignment method, we were able to assign all MycG4 protons of the 1:1 complex (Figure 3B, Supplementary Figure S5 and Table S2). The similar characteristic guanine H1-H8 protons connectivity patterns observed for the two MycG4 molecules at BMVC/MycG4 ratio of 0.5 (Supplementary Figure S6) indicated that the MycG4 folding remained unchanged. Only the T4 residue adopted a syn glycosidic conformation as supported by the strong intraresidue H6-H1′ NOE (Figure 3B), which is different from the free MycG4. Other residues adopted anti glycosidic conformations as indicated by the weak intraresidue H6/H8-H1′ NOEs.
Figure 3.
(A) The expanded H1-H1 region of the 2D-NOESY spectrum at BMVC/MycG4 ratio of 0.5. Exchange cross-peaks of imino protons between free DNA and 1:1 BMVC–MycG4 complex are labeled with the corresponding residue numbers. (B) The expanded H8/H6-H1′ region of the 2D-NOESY spectrum of the 1:1 complex at BMVC/MycG4 ratio of 0.5. Complete sequential assignment pathway is shown. The missing connectivity is labeled with red asterisk. (C) The expanded 2D-NOESY spectra of the 1:1 complex, showing some examples of the intramolecular BMVC NOEs (left) and intermolecular NOEs between BMVC with the 5′-end of MycG4 (right). Conditions: pH 7, 95 mM K+, 25 °C, mixing time 200 ms.
The protons of bound BMVC were assigned using the NOESY (Figure 3C), COSY and TOCSY spectra (Supplementary Figure S7). Exchange cross-peaks of BMVC protons also could be found in NOESY spectra (Supplementary Figure S8), corresponding to the bound and non-bound BMVC in equilibrium. The H1 protons of bound BMVCs were confirmed by the intramolecular NOEs with BMVC protons H8, H9, H12 and H13.
Based on the complete proton assignment, the proton chemical shift differences between the free MycG4 and 1:1 BMVC−MycG4 complex were calculated (Δppm 1, Figure 4A). The large values of Δppm 1 were found in the 5′ G-tetrad residues of G7, G11, G16 and G20, indicating that BMVC bound at the 5′-end and stacked over the 5′ G-tetrad. Significant chemical shift differences were also observed for 5′-flanking residues of T4, G5 and A6, indicating a large conformational change of the 5′-flanking region. 82 intermolecular NOEs between the protons of BMVC and MycG4 5′-end were observed (Figure 4B and Supplementary Table S3), revealing a specific binding of BMVC with MycG4.
Figure 4.
(A) Δppm 1, the MycG4 proton chemical shift difference between the free MycG4 and 1:1 BMVC–MycG4 complex and (D) Δppm 2, the MycG4 proton chemical shift difference between the 1:1 and 2:1 BMVC−MycG4 complexes. The residue numbers of MycG4 are shown. Schematic diagrams of the inter- and intra-molecular NOEs of the 5'-end complex in the 1:1 complex (B) and of the 3′-end complex in the 2:1 complex (C). Thin, medium, and thick lines correspond to strong, medium and weak NOE interactions, respectively.
The NMR structure of the 1:1 BMVC−MycG4 complex showed BMVC induced large structural rearrangement at the 5′-end to form a well-defined binding pocket
The 1:1 BMVC−MycG4 complex structure was calculated using established molecular dynamics methods (28) based on the NOE data (Table 1). The 20 lowest-energy structures are superimposed and presented in Supplementary Figure S9, with a representative structure shown in Figure 5. As shown in the structure, BMVC binds to the 5′-end of MycG4 to form a well-defined binding pocket (Figure 5A and B). The 5′ flanking segments showed large rearrangement induced by BMVC binding. In the free MycG4, G5 and A6 at the 5′-end cover the G-tetrad, while T4 is extended toward the solvent (27). In the 1:1 BMVC−MycG4 complex, BMVC reorganized the capping structure and recruited A6 to form a BMVC–adenine plane stacking on the 5′ G-tetrad (Figure 5C). Interestingly, the BMVC molecule adopts a contracted conformation, which perfectly matches the external G-tetrad and covers the bases of G11, G16 and G20. Significant chemical shift differences of the protons of tetrad guanines (Figure 4A) and numerous NOEs between BMVC and G-tetrads were observed in the NMR spectra (Figure 4B and Supplementary Table S3), indicating specific ligand binding. Particularly, intermolecular NOEs between BMVC-H1 and the protons of G16 were observed (Figure 4B and Supplementary Figure S10D). A6 stacks over G7 and inserts into the arc of BMVC, which is consistent with the marked downfield shifted resonances of A6H2 and A6H8 and upfield shifted resonance of G7H1 (Figure 4A) as well as interresidue NOEs corresponding to A6H8/G20H1 and A6H2/G7H1 (Figure 4B, Supplementary Figure S6 and Supplementary Table S4). In addition, intermolecular NOEs were found between the protons of A6H8 and BMVC ring A, and between the protons of A6/H2 and BMVC ring E (Figure 4B and Supplementary Figure S10A), indicating the recruitment of A6 into the arc of BMVC which locks the BMVC orientation. Meanwhile, NOEs between the protons of G5 sugar with BMVC ring A and A6H8 (Figure 4B) suggested that the G5 sugar moiety stacked over the BMVC ring A and the H8 end of A6 (Figure 5D), in agreement with the significant upfield shifting of G5 sugar protons (Figure 4A). However, the G5 base appeared to be more flexible and did not show NOE interactions with BMVC (Figure 4B). The 5′ T4 with a syn glycosidic conformation was well-defined and stacked directly over the central carbazole segment of BMVC stabilizing the BMVC-A6 plane (Figure 5D). The well-stacked conformation of T4 was supported by the significant upfield shifting of T4 protons (Figure 4A) and the NOEs between the protons of T4 and A6/H2 (Figure 4B and Supplementary Figure S10B) and BMVC ring B, C and D (Figure 4B and Supplementary Figure S10C, D).
Table 1.
Structural statistics for the 1:1 and 2:1 BMVC–MycG4 complex structures
| Structure statistics | ||
|---|---|---|
| 1:1 complex | 2:1 complex | |
| Distance restraints | 481 | 516 |
| MycG4 | 399 | 396 |
| Intraresidue | 259 | 272 |
| Interresidue | 116 | 100 |
| Hydrogen bonds | 24 | 24 |
| Intermolecular | 82 | 120 |
| Deviations from standard geometry | ||
| Bond length (Å) | 0.01 ± 0.00 | 0.08 ± 0.02 |
| Bond angle (deg.) | 1.26 ± 0.00 | 1.22 ± 0.00 |
| NOE violations | ||
| Numbers (> 0.2 Å) | 0.01 ± 0.00 | 0.01 ± 0.00 |
| Pairwise RMSD of heavy atoms (Å) | ||
| G-tetrad core | 0.80 ± 0.17 | 0.60 ± 0.17 |
| MycG4 | 2.12 ± 0.48 | 2.02 ± 0.48 |
| All | 2.06 ± 0.47 | 2.00 ± 0.47 |
Figure 5.
(A-B) A representative model of the NMR-refined 1:1 BMVC–MycG4 complex structure (the 5'-end complex) shown in two different views. (PDB ID 6JJ0) (C) top and (D) side view of the BMVC-induced binding pocket at the 5′-end of MycG4. BMVC molecules are shown in yellow. Guanines are in green, adenines in magenta and thymines in cyan.
Complete 2D NMR proton assignment of the 2:1 BMVC−MycG4 complex showing the second BMVC binds to the 3′-end of MycG4
Similar to the assignment of the 1:1 complex, we unambiguously assigned the imino and aromatic protons, and subsequently completely assigned the protons of the bound-MycG4 in the 2:1 BMVC−MycG4 complex using the COSY, TOCSY and NOESY spectra (Figure 6, Supplementary Figures S11–S14 and Supplementary Tables S5–S8) at the BMVC/MycG4 ratio of 1.5. Similar to the 1:1 complex, the parallel folding of MycG4 was conserved in the 2:1 complex (Supplementary Figure S12) and all residues adopted anti glycosidic conformations except for the syn T4, which showed a strong H6-H1′ NOE cross-peak (Figure 6B).
Figure 6.
(A) The expanded H1-H1 region of the 2D-NOESY spectrum at BMVC/MycG4 ratio 1.5. Exchange cross-peaks of imino protons between the 1:1 BMVC–MycG4 complex and 2:1 BMVC–MycG4 complex are labeled with the corresponding residue numbers. Condition: pH 7, 95 mM K+, 25 °C. (B) The expanded H8/H6-H1′ region of the 2D-NOESY spectrum of the 2:1 BMVC–MycG4 complex at BMVC/MycG4 ratio of 1.5:1. The complete sequential assignment pathway is shown. The missing connectivity is labeled with red asterisks. The assignment of the 2:1 BMVC–MycG4 complex is labeled with superscript b. Condition: pH 7, 95 mM K+, 5 °C. (C) The expanded 2D-NOESY spectra of the 2:1 BMVC–MycG4 complex at BMVC/MycG4 ratio of 1.5:1, showing the intermolecular NOEs between BMVC and the 3′-end of MycG4. Condition: pH 7, 95 mM K+, 5 °C, mixing time 200 ms.
Based on the complete proton assignment, the MycG4 proton chemical shift differences between the 1:1 and 2:1 complexes (Δppm 2, Figure 4D) were calculated. In Δppm 2, 3′ G-tetrad residues of G9, G13, G18 and G22 and 3′ flanking residues of T23, A24 and A25 were most affected, indicating that the second BMVC stacks over the 3′-end G-tetrad and the 3′-capping region undergoes large conformational changes. The binding of BMVC with the MycG4 3′-end was also supported by 38 intermolecular NOEs between the protons of BMVC and 3′-end (Figure 4C and Supplementary Table S9). The same NOE interactions were observed between BMVC and the 5′-end in the 2:1 complex and the 1:1 complex, suggesting the same conformation of the 5′-end complex. However, the significant difference in total intermolecular NOEs between 5′-end (82 NOEs) and 3′-end (38 NOEs) of the BMVC−MycG4 complex further supported that the 5′-end complex was better defined as compared to the 3′-end complex (Figure 4B and C).
The NMR structure of the 2:1 BMVC−MycG4 complex showed 3′-end has a similar binding pattern to the 5′-end with a more dynamic conformation
The 3′ flanking region showed large rearrangement induced by BMVC binding (Supplementary Figure S15). In the 2:1 complex structure, the sequential NOE connectivity was interrupted at the T23-A24 and A24-A25 steps (Figure 6B), illustrating the fold-back stacking conformation in the free MycG4 (27) was disrupted and the 3′ flanking region was reoriented to form a new binding pocket at the 3′-end. T23 was recruited by BMVC to establish a new BMVC−thymine plane which stacks over the 3′ external G-tetrad (Figure 7C). As a result, the protons of T23 showed significant upfield shifting (Figure 4D) and NOE interactions with BMVC (Figure 4C). In addition, T23 stacks directly over G22 as supported by NOEs between T23Me/G22H8 and T23Me/G18H1 (Figure 4C and Supplementary Table S10). On the other hand, A24 and A25 were dynamic in conformation with few NOE interactions (Figure 4C) with BMVC and loss of interresidue NOE connectivity pattern (Figure 6B).
Figure 7.
(A, B) A representative model of the NMR-refined 2:1 BMVC–MycG4 complex structure shown in two different views. (PDB ID 6O2L) (C) Top and (D) side views of the BMVC-induced binding pocket at the 3′-end of MycG4. BMVC molecules are shown in yellow. Guanines are in green, adenines in magenta, and thymines in cyan.
The BMVC molecule adjusts its conformation to match three bases of external G-tetrads in both the BMVC−MycG4 complexes
In both the 1:1 and 2:1 BMVC-MycG4 complex structures, the BMVC orientations and positions are well-defined (Figures 5C and 7C). The BMVC molecule has a crescent shape and possesses two double-bond-linked pyridine moieties. BMVC can adopt two conformations, extended and contracted (Figure 8). Using OPLS3 force field from Schrödinger Inc., the conformational energy difference between the two free BMVC conformations was calculated to be 2.6 kcal/mol with the extended conformation being more stable. In the free form, BMVC adopts the extended conformation. However, upon binding to MycG4, BMVC molecules adjusted to a contracted conformation to perfectly match three bases of G-tetrads with an optimal stacking interaction in both complexes (Figure 8A). Each BMVC covers three bases of the G-tetrad at each end of MycG4, with A6 or T23 recruited into the BMVC arc. Specifically, at the 5′-end (Figures 4B and 5C), BMVC ring A covered the G20 base, as indicated by the NOEs such as G20H1/BH21 and G20H1/BH20. BMVC ring B, C and D cover G16 as indicated by NOEs such as G16H1/BH6 and G16H8/BH1. BMVC ring E covers G11 as indicated by the NOEs such as G11H8/BH26 and G11H8/BH25. Similarly, at the 3′-end (Figure 7C), the binding position of BMVC is also clearly defined by NOE interactions (Figure 4C), with BMVC ring A covering G9, ring B, C and D covering G13, and ring E covering G18.
Figure 8.

The stacking view of the 5′-end MycG4 G-tetrad with the contracted-form (A) or extended-form BMVC (B). BMVC is shown in magenta. (C) Proton distances between 5′-end G-tetrad with contracted-form or extended-form BMVC.
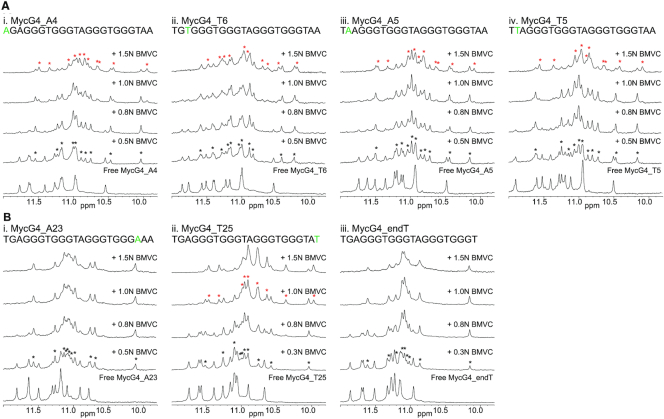
Mutational analysis showed the 3′-flanking sequence has more influence on the binding of BMVC
The mutational analysis with various modified MycG4 sequences (Figure 9) showed that the mutations at the 5′-flanking segment, i.e., T4, G5 and A6, of MycG4 (Figure 9A) do not affect BMVC’s binding at the 5′-end, as indicated by the similar 1D NMR titration spectra with two slow-exchanged binding stages. However, the mutations of the 3′-flanking residues (Figure 9B) showed a much greater effect on the 3′-end binding. BMVC failed to bind the 3′-end of MycG4 after T23 was mutated to A23 (Figure 9B-i), illustrating that residue T23 is critical to the specific binding of BMVC at the 3′-end. In addition, BMVC binding at the 3′-end was disrupted when the 3′-flanking segment was truncated (Figure 9B-iii), indicating that flanking residues are required for BMVC binding at the 3′-end. Mutational analysis of Myc1234 and VEGF G4 sequences also showed similar results (Supplementary Figure S16).
Figure 9.
Imino proton regions of the 1D 1H NMR titration spectra of BMVC with various 5′-end modified (A) or 3′-end modified (B) MycG4 sequences in pH 7, 95 mM K+ solution, at 25 °C. The modified sequences are shown above the spectra with modified residues colored in green. Imino protons arising from the 1:1 or 2:1 complex formation are marked with asterisks in black or red, respectively.
BMVC represses the MYC gene expression in cancer cells
Suppression of MYC transcription can be achieved by stabilization of MYC promoter G4 DNA structures (19,25,46). To understand the cellular effects of BMVC on MYC gene expression, we have carried out western blot and qRT-PCR assays using MCF-7 breast cancer cells. We found that MYC expression levels were significantly reduced after BMVC treatments, as shown by western blot (Figure 10A). Lower MYC gene expression was observed with increased BMVC concentration, as shown at 4 and 10 μM, and increased BMVC treatment time. We further performed qRT-PCR experiments and found that MYC mRNA levels were decreased below 50% in the presence of BMVC at both 4 and 10 μM (Figure 10B). This was in agreement with the western blot data that BMVC functions as a MYC repressor, likely by binding and stabilizing the MYC promoter G4 to repress transcription.
Figure 10.
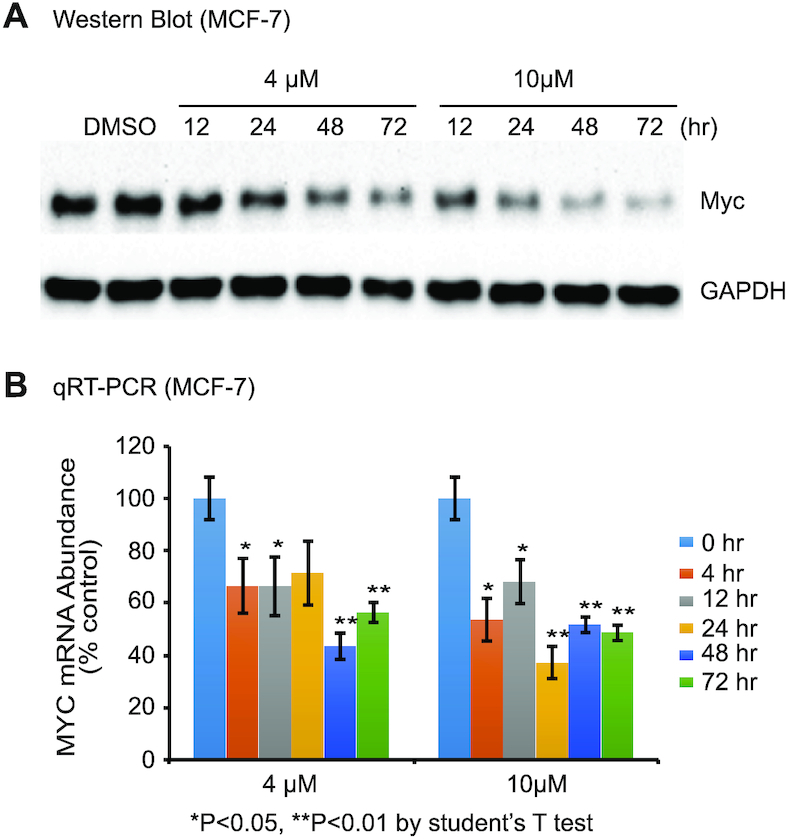
BMVC represses MYC expression in MCF-7 breast cancer cells. (A) BMVC lowers Myc protein levels but not glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels as shown by western blots. 0.1% DMSO is used as control. Cells treated with DMSO for 72 hr are shown in the left 2 lanes (replicates). Two concentrations of BMVC are added to different cell samples with difference treatment times (right 8 lanes). (B) qRT-PCR results showing BMVC decreases MYC mRNA levels. The BMVC-exposure time is shown to the right of the figure. (n = 3 biologically independent samples. Error bars represent mean ± s.d. *P < 0.05, **P < 0.01 by Student's unpaired t-test).
DISCUSSION
Solving the NMR structures of the 1:1 and 2:1 BMVC–MycG4 complexes allows us to understand the specific molecular recognition of the MYC G-quadruplex by BMVC. As shown by the complex structures (Figures 5 and 7), there are common features between the overall structures of the 5′-end and 3′-end complexes. At both ends, the BMVC molecule stacks over three guanine bases and recruits an adenine or thymine to form a new ligand-base plane covering the external G-tetrad. The crescent shape of BMVC allows it to recruit a flanking DNA base, which locks the BMVC orientation. Similar base recruitment recognitions have been reported for the MycG4 with the compounds quindoline (28) and DC-34 (29), and for the human telomeric G-quadruplex hybrid-2 with the protoberberine alkaloid epiberberine (47) and hybrid-1 with the platinum(II) compound Pt-tripod (48). This suggests that crescent shaped compounds such as BMVC can specifically recognize and engage a flanking DNA base to anchor its specific position, which provides a means to achieve sequence selectivity into the G4 recognition. The previously reported NMR complex structures of a modified MYC G-quadruplex with TMPyP4 (49) and Phen-DC3 (50), or the Myc1234 G-quadruplex with DAOTA-M2 (51) show the importance of extensive π–π stacking in the ligand binding with G-quadruplexes. However, in these complexes, information for specific recognition is limited.
Remarkably, upon binding of MycG4, BMVC adjusts its pyridine rings to adopt a contracted conformation to perfectly match the three bases of a G-tetrad. This gives rise to an optimized π-π stacking of BMVC with the external G-tetrad. Importantly, the rotatable double-bond linker (C7−C14 and C11−C22) of the BMVC molecule enables it to adjust its conformation. As shown in Figure 8, BMVC can adopt both the extended and contracted conformations. The contracted conformation of BMVC clearly exhibits better stacking with the G-tetrad (Figure 8A) as compared to the extended conformation (Figure 8B). The proton distances of contracted BMVC with the external G-tetrad are closer than those of extended BMVC (Figure 8C). This highly optimized stacking interaction with G-tetrad contributes to the high-affinity binding of BMVC to MycG4. This conformational adjustment upon G4 binding also provides a potential molecular mechanism for the large, G4-specific fluorescence enhancement of BMVC.
BMVC shows a rare case of slow-exchange binding on the NMR timescale, in particular for the parallel-stranded MycG4 with short-loops. Unlike hybrid-type telomeric G-quadruplexes that contain lateral loops, which can contribute to a more extensive binding pocket formation (47,48), the ligand binding pockets of a parallel G-quadruplex with short-loops are formed by the 5′ or the 3′ flanking segments. For example, quindoline binds MycG4 at an intermediate exchange rate (28), while DC-34 binds at an intermediate-to-fast exchange rate (29). In contrast, the slow-exchange rate suggests a higher-affinity binding of MycG4 by BMVC. Moreover, BMVC shows more preferred binding to parallel-stranded G-quadruplexes, which are common in the human promoter G-quadruplexes (4), than to basket- or hybrid-type telomeric G-quadruplexes. The reason is probably that the lateral and diagonal loops found in these topologies can sterically hinder the tetrad stacking of BMVC.
There are also significant differences between the 5′- and 3′-end complexes in the binding of BMVC to MycG4. BMVC binds tighter at the 5′-end to form a well-defined complex, whereas the 3′-end complex showing a more dynamic conformation. This difference in binding is confirmed by NMR and mutational analysis. As shown by the NMR titration data (Figure 2), BMVC binds to MycG4 in two stages. BMVC first binds to the 5′-end of MycG4, and then binds the 3′-end at higher ligand ratios. The binding affinity of BMVC at the 5′-end appears to be 1–2 orders of magnitude higher than that at the 3′-end (Figure 2). The more-accessible planar surface of the 5′ external tetrad allows a greater stacking interaction and thus a high-affinity binding, whereas the accessible planar surface of the 3′ external tetrad for stacking is limited by the protruding sugar moieties. The mutations on the 5′ flanking residues do not affect the binding of BMVC to the 5′-end (Figure 9A). In contrast, the 3′ flanking residues showed a clear effect on BMVC binding (Figure 9B), suggesting that the weaker binding site at the 3′-end is more sequence-specific.
CONCLUSION
In conclusion, we show that BMVC binds the major MYC promoter G-quadruplex with greater affinity and specificity than the human telomeric G-quadruplexes, and that BMVC represses MYC expression in cancer cells. The determined NMR solution structures of the 1:1 and 2:1 BMVC–MycG4 complexes show that BMVC first binds to the 5′-end with high affinity to form a very well-defined binding pocket, then binds to the 3′-end with a weaker affinity to form a more-dynamic complex. We find the crescent shape and the adjustable conformation of the BMVC molecule both play important roles in the specific and high-affinity binding to the MYC G-quadruplex. BMVC specifically recruits a flanking base of MycG4 to form a ligand-base pairing plane to anchor its position. Remarkably, upon binding of MycG4, BMVC undergoes a conformational adjustment to a contracted form to perfectly match the external G-tetrad for a highly optimized stacking interaction. This is the first structural evidence showing the importance of small molecule conformational adjustment in G-quadruplex recognition. This conformational adjustment upon G4 binding is likely responsible for the large, G4-specific fluorescence enhancement of BMVC. Our results provide the molecular mechanism for the specific recognition of G-quadruplexes by BMVC and important insights into rational design of G-quadruplex-targeting anticancer drugs and fluorescence probes.
DATA AVAILABILITY
The coordinates for the structures of the 1:1 (accession code: 6JJ0) and 2:1 (accession code: 6O2L) BMVC–MycG4 complexes have been deposited in the Protein Data bank.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Jonathan Dickerhoff for his help proofreading and revising the manuscript. We thank Dr Kaibo Wang for the Kd measurements.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01CA177585 to D.Y., P30CA023168 to Purdue Center for Cancer Research]; Ministry of Education of China [IRT-17R111 to W.L.]. Funding for open access charge: National Institutes of Health, National Cancer Institute Grant [R01CA177585].
Conflict of interest statement. None declared.
REFERENCES
- 1. Yang D., Okamoto K.. Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med. Chem. 2010; 2:619–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Neidle S., Parkinson G.. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002; 1:383–393. [DOI] [PubMed] [Google Scholar]
- 3. Qin Y., Hurley L.H.. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008; 90:1149–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Y., Yang D.Z.. Sequence, stability, and structure of G-Quadruplexes and their interactions with drugs. Curr. Protoc. Nucleic Acid Chem. 2012; 50:17.15.11–17.15.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy?. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bugaut A., Balasubramanian S.. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012; 40:4727–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Biffi G., Tannahill D., Miller J., Howat W.J., Balasubramanian S.. Elevated levels of G-quadruplex formation in human stomach and liver cancer tissues. PLoS One. 2014; 9:e102711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M. et al.. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 9. Marcu K.B., Bossone S.A., Patel A.J.. myc function and regulation. Annu. Rev. Biochem. 1992; 61:809–860. [DOI] [PubMed] [Google Scholar]
- 10. Pelengaris S., Khan M.. The many faces of c-MYC. Arch. Biochem. Biophys. 2003; 416:129–136. [DOI] [PubMed] [Google Scholar]
- 11. Kinzler K.W., Vogelstein B.. Lessons from hereditary colorectal cancer. Cell. 1996; 87:159–170. [DOI] [PubMed] [Google Scholar]
- 12. Nupponen N.N., Kakkola L., Koivisto P., Visakorpi T.. Genetic alterations in hormone-refractory recurrent prostate carcinomas. Am. J. Pathol. 1998; 153:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dang C.V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell Biol. 1999; 19:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nesbit C.E., Tersak J.M., Prochownik E.V.. MYC oncogenes and human neoplastic disease. Oncogene. 1999; 18:3004–3016. [DOI] [PubMed] [Google Scholar]
- 15. Schlagbauer-Wadl H., Griffioen M., van Elsas A., Schrier P.I., Pustelnik T., Eichler H.G., Wolff K., Pehamberger H., Jansen B.. Influence of increased c-Myc expression on the growth characteristics of human melanoma. J. Invest. Dermatol. 1999; 112:332–336. [DOI] [PubMed] [Google Scholar]
- 16. Prochownik E.V., Vogt P.K.. Therapeutic targeting of Myc. Genes Cancer. 2010; 1:650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sakatsume O., Tsutsui H., Wang Y., Gao H., Tang X., Yamauchi T., Murata T., Itakura K., Yokoyama K.K.. Binding of THZif-1, a MAZ-like zinc finger protein to the nuclease-hypersensitive element in the promoter region of the c-MYC protooncogene. J. Biol. Chem. 1996; 271:31322–31333. [DOI] [PubMed] [Google Scholar]
- 18. Tomonaga T., Levens D.. Activating transcription from single stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:5830–5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H.. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S A. 2002; 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brooks T.A., Hurley L.H.. Targeting MYC Expression through G-Quadruplexes. Genes Cancer. 2010; 1:641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ou T.M., Lu Y.J., Zhang C., Huang Z.S., Wang X.D., Tan J.H., Chen Y., Ma D.L., Wong K.Y., Tang J.C et al.. Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc by quindoline derivatives. J. Med. Chem. 2007; 50:1465–1474. [DOI] [PubMed] [Google Scholar]
- 22. Grand C.L., Han H., Munoz R.M., Weitman S., Von Hoff D.D., Hurley L.H., Bearss D.J.. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 2002; 1:565–573. [PubMed] [Google Scholar]
- 23. Liu J.N., Deng R., Guo J.F., Zhou J.M., Feng G.K., Huang Z.S., Gu L.Q., Zeng Y.X., Zhu X.F.. Inhibition of myc promoter and telomerase activity and induction of delayed apoptosis by SYUIQ-5, a novel G-quadruplex interactive agent in leukemia cells. Leukemia. 2007; 21:1300–1302. [DOI] [PubMed] [Google Scholar]
- 24. Kang H.J., Park H.J.. Novel molecular mechanism for actinomycin D activity as an oncogenic promoter G-Quadruplex binder. Biochemistry. 2009; 48:7392–7398. [DOI] [PubMed] [Google Scholar]
- 25. Seenisamy J., Rezler E.M., Powell T.J., Tye D., Gokhale V., Joshi C.S., Siddiqui-Jain A., Hurley L.H.. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc. 2004; 126:8702–8709. [DOI] [PubMed] [Google Scholar]
- 26. Phan A.T., Modi Y.S., Patel D.J.. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004; 126:8710–8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ambrus A., Chen D., Dai J., Jones R.A., Yang D.. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005; 44:2048–2058. [DOI] [PubMed] [Google Scholar]
- 28. Dai J., Carver M., Hurley L.H., Yang D.. Solution structure of a 2:1 quindoline-c-MYC G-quadruplex: insights into G-quadruplex-interactive small molecule drug design. J. Am. Chem. Soc. 2011; 133:17673–17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Calabrese D.R., Chen X., Leon E.C., Gaikwad S.M., Phyo Z., Hewitt W.M., Alden S., Hilimire T.A., He F., Michalowski A.M et al.. Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex. Nat. Commun. 2018; 9:4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chang C.C., Wu J.Y., Chien C.W., Wu W.S., Liu H., Kang C.C., Yu L.J., Chang T.C.. A fluorescent carbazole derivative: high sensitivity for quadruplex DNA. Anal. Chem. 2003; 75:6177–6183. [DOI] [PubMed] [Google Scholar]
- 31. Chang C.C., Kuo I.C., Ling I.F., Chen C.T., Chen H.C., Lou P.J., Lin J.J., Chang T.C.. Detection of quadruplex DNA structures in human telomeres by a fluorescent carbazole derivative. Anal. Chem. 2004; 76:4490–4494. [DOI] [PubMed] [Google Scholar]
- 32. Chang C.C., Chu J.F., Kao F.J., Chiu Y.C., Lou P.J., Chen H.C., Chang T.C.. Verification of antiparallel G-quadruplex structure in human telomeres by using two-photon excitation fluorescence lifetime imaging microscopy of the 3,6-Bis(1-methyl-4-vinylpyridinium)carbazole diiodide molecule. Anal. Chem. 2006; 78:2810–2815. [DOI] [PubMed] [Google Scholar]
- 33. Huang F.C., Chang C.C., Wang J.M., Chang T.C., Lin J.J.. Induction of senescence in cancer cells by the G-quadruplex stabilizer, BMVC4, is independent of its telomerase inhibitory activity. Br. J. Pharmacol. 2012; 167:393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kang C.C., Huang W.C., Kouh C.W., Wang Z.F., Cho C.C., Chang C.C., Wang C.L., Chang T.C., Seemann J., Huang L.J.. Chemical principles for the design of a novel fluorescent probe with high cancer-targeting selectivity and sensitivity. Integr. Biol. (Camb.). 2013; 5:1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang C.C., Kuo I.C., Lin J.J., Lu Y.C., Chen C.T., Back H.T., Lou P.J., Chang T.C.. A novel carbazole derivative, BMVC: a potential antitumor agent and fluorescence marker of cancer cells. Chem. Biodivers. 2004; 1:1377–1384. [DOI] [PubMed] [Google Scholar]
- 36. Kang C.-C., Chang C.-C., Cheng J.-Y., Chang T.-C.. Simple method in diagnosing cancer cells by a novel fluorescence probe BMVC. J. Chinese Chem. Soc. 2005; 52:1069–1072. [Google Scholar]
- 37. Kang C.C., Chang C.C., Chang T.C., Liao L.J., Lou P.J., Xie W., Yeung E.S.. A handheld device for potential point-of-care screening of cancer. Analyst. 2007; 132:745–749. [DOI] [PubMed] [Google Scholar]
- 38. Huang F.C., Chang C.C., Lou P.J., Kuo I.C., Chien C.W., Chen C.T., Shieh F.Y., Chang T.C., Lin J.J.. G-quadruplex stabilizer 3,6-bis(1-methyl-4-vinylpyridinium)carbazole diiodide induces accelerated senescence and inhibits tumorigenic properties in cancer cells. Mol. Cancer Res. 2008; 6:955–964. [DOI] [PubMed] [Google Scholar]
- 39. Yang T.L., Lin L., Lou P.J., Chang T.C., Young T.H.. Detection of cell carcinogenic transformation by a quadruplex DNA binding fluorescent probe. PLoS One. 2014; 9:e86143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tseng T.-Y., Chang C.-C., Lin J.-J., Chang T.-C.. A Fluorescent Anti-Cancer Agent, 3,6-bis(1-methyl-4-vinylpyridinium) carbazole diiodide, stains G-Quadruplexes in cells and inhibits tumor growth. Curr. Top. Med. Chem. 2015; 15:1964–1970. [DOI] [PubMed] [Google Scholar]
- 41. Tseng T.Y., Chen W.W., Chu I.T., Wang C.L., Chang C.C., Lin M.C., Lou P.J., Chang T.C.. The G-quadruplex fluorescent probe 3,6-bis(1-methyl-2-vinyl-pyridinium) carbazole diiodide as a biosensor for human cancers. Sci. Rep. 2018; 8:16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brünger A.T. X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. 1993; New Haven, CT: Yale University Press. [Google Scholar]
- 43. Schuttelkopf A.W., van Aalten D.M.. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica. D, Biol. Crystallogr. 2004; 60:1355–1363. [DOI] [PubMed] [Google Scholar]
- 44. Murat P., Bonnet R., Van der Heyden A., Spinelli N., Labbe P., Monchaud D., Teulade-Fichou M.P., Dumy P., Defrancq E.. Template-assembled synthetic G-quadruplex (TASQ): a useful system for investigating the interactions of ligands with constrained quadruplex topologies. Chemistry. 2010; 16:6106–6114. [DOI] [PubMed] [Google Scholar]
- 45. Dash J., Shirude P.S., Hsu S.T., Balasubramanian S.. Diarylethynyl amides that recognize the parallel conformation of genomic promoter DNA G-quadruplexes. J. Am. Chem. Soc. 2008; 130:15950–15956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gonzalez V., Guo K.X., Hurley L., Sun D.. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009; 284:23622–23635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lin C., Wu G., Wang K., Onel B., Sakai S., Shao Y., Yang D.. Molecular recognition of the Hybrid-2 human telomeric G-Quadruplex by epiberberine: insights into conversion of telomeric G-Quadruplex structures. Angew. Chem. Int. Ed. Engl. 2018; 57:10888–10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu W.T., Zhong Y.F., Liu L.Y., Shen C.T., Zeng W.J., Wang F.Y., Yang D.Z., Mao Z.W.. Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding. Nat. Commun. 2018; 9:3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Phan A.T., Kuryavyi V., Gaw H.Y., Patel D.J.. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005; 1:167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chung W.J., Heddi B., Hamon F., Teulade-Fichou M.P., Phan A.T.. Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC(3). Angew. Chem. Int. Ed. Engl. 2014; 53:999–1002. [DOI] [PubMed] [Google Scholar]
- 51. Kotar A., Wang B., Shivalingam A., Gonzalez-Garcia J., Vilar R., Plavec J.. NMR structure of a triangulenium-based Long-lived fluorescence probe bound to a G-quadruplex. Angew. Chem. Int. Ed. Engl. 2016; 55:12508–12511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates for the structures of the 1:1 (accession code: 6JJ0) and 2:1 (accession code: 6O2L) BMVC–MycG4 complexes have been deposited in the Protein Data bank.