Abstract
PUF proteins, named for Drosophila Pumilio (PUM) and Caenorhabditis elegans fem-3-binding factor (FBF), recognize specific sequences in the mRNAs they bind and control. RNA binding by classical PUF proteins is mediated by a characteristic PUM homology domain (PUM-HD). The Puf1 and Puf2 proteins possess a distinct architecture and comprise a highly conserved subfamily among fungal species. Puf1/Puf2 proteins contain two types of RNA-binding domain: a divergent PUM-HD and an RNA recognition motif (RRM). They recognize RNAs containing UAAU motifs, often in clusters. Here, we report a crystal structure of the PUM-HD of a fungal Puf1 in complex with a dual UAAU motif RNA. Each of the two UAAU tetranucleotides are bound by a Puf1 PUM-HD forming a 2:1 protein-to-RNA complex. We also determined crystal structures of the Puf1 RRM domain that identified a dimerization interface. The PUM-HD and RRM domains act in concert to determine RNA-binding specificity: the PUM-HD dictates binding to UAAU, and dimerization of the RRM domain favors binding to dual UAAU motifs rather than a single UAAU. Cooperative action of the RRM and PUM-HD identifies a new mechanism by which multiple RNA-binding modules in a single protein collaborate to create a unique RNA-binding specificity.
INTRODUCTION
RNA-binding proteins (RBPs) regulate every step in the life of an mRNA, from processing to translation and destruction. As a result, they are critical in determining the level of protein that a gene produces. RBP dysfunction, due to mutations, incorrect protein levels or inappropriate localization, leads to aberrant gene expression. Dysfunctional RBPs are linked with human neurological disorders and cancer (1–4). mRBPs bind mature mRNAs, often in their 3′-untranslated regions (3′-UTRs), to control stability, translation and localization of their mRNA targets (5). The PUF family proteins, named after founding members, Drosophila melanogaster Pumilio (PUM) and Caenorhabditis elegans fem-3 binding factor (FBF) (6,7), are exemplary mRBPs, and their roles in mRNA control have been studied extensively (8–11).
PUF proteins are evolutionarily conserved throughout eukaryotes. In metazoa, PUF proteins regulate mRNAs that encode proteins involved in embryonic development, stem cell control and neurogenesis, among other roles (12–14). In Saccharomyces cerevisiae, each PUF subfamily binds multiple mRNAs, often with functional and cytotopic relatedness (15). Their roles are widespread, and include controls of mating type, cell wall integrity, mitochondrial functions, ion sensitivity, and other cellular events (15–21).
A single eukaryotic species can encode multiple PUF proteins that comprise as many as four subgroups: classical PUF proteins, Puf6 proteins, Nop9 proteins and the fungal-specific Puf1/Puf2 proteins. PUF proteins are characterized by RNA-binding domains composed of multiple α-helical PUM repeats, and the number and arrangement of the PUM repeats varies for each of the subgroups. Although crystal structures have revealed how the PUM repeats of classical, Puf6 and Nop9 PUF proteins assemble for specific RNA recognition (22–32), no structural information is available for the Puf1/Puf2 proteins.
Classical PUF proteins, including yeast Puf3, Puf4 and Puf5, are the most widely characterized subgroup, and are marked by a sequence-specific RNA-binding domain called the Pumilio-homology domain (PUM-HD) (6,7). Crystal structures of the PUM-HD from classical PUF proteins reveal eight PUM repeats arranged in a crescent shape (22–23,25–28,30). Single-stranded RNA binds to the inner concave face of the protein. Typically, one PUM repeat comprising three α helices contacts one RNA base. A five-residue motif of X1X2øøX3 (ø, hydrophobic residue) in the second α helix of a PUM repeat defines the sequence specificity of RNA base recognition (33,34). Residues X1 and X3 make edge-on hydrogen bond or Van der Waals interactions with the RNA base, whereas residue X2 stacks with and often between bases. This X1X2øøX3 motif is termed the tripartite recognition motif (TRM), and the RNA recognition residues are denoted X1X3/X2 (35). For example, a uracil-specific TRM, NYøøQ, is denoted NQ/Y. Yeast Puf3, Puf4 and Puf5 proteins have diverged in the length and sequence of the RNAs they bind with their eight PUM repeats (15,36). For example, yeast Puf4 recognizes nucleotides 1–6 and 8–9 of its 9-nt recognition motif, but nucleotide 7 is flipped away from the RNA-binding surface. Flipping of a specific nucleotide is a key feature of the Puf4 RNA recognition pattern (23).
Yeast Puf6 and Nop9 define two atypical PUF protein subgroups, distinct from each other and the classical PUF proteins. Both proteins are nucleolar localized and involved in pre-rRNA processing (37,38,39). In contrast to the classical PUF proteins, Puf6 and Nop9 both comprise 11 PUM repeats, arranged in an ‘L’ or ‘C’-like shape, respectively (24,29). Puf6 and human homolog Puf-A bind to single- or double-stranded RNA with no apparent sequence specificity, and most of their PUM repeats lack the characteristic RNA base recognition residues that are found in the classical PUF proteins. Nop9 utilizes a combination of classical PUF protein RNA sequence recognition and structured RNA recognition to bind to its target RNAs (29,31,39).
Puf1 and Puf2 comprise a subgroup of PUF proteins unique to fungi, yet conserved for hundreds of millions of years. They differ from other PUF proteins in containing two types of RNA-binding domain: a divergent PUM-HD and an RNA recognition motif (RRM) domain (Figure 1A). Only six PUM repeats are predicted in the PUM-HD, and some of their RNA-interacting TRMs differ from classical PUF proteins. S. cerevisiae Puf1 and Puf2 interact preferentially with mRNAs encoding cell periphery proteins (15), and Puf1/Jsn1 has been shown to be localized to the cell perimeter (40). Puf1 also co-localizes with mitochondria and associates with the Arp2/3 complex (41). Deletion of Puf1 causes defects in mitochondrial morphology and motility, but it is not known whether Puf1 RNA regulatory activity is involved. Puf1 and Puf2 mediate responses to environmental stresses (19,21) and do so via control of specific mRNAs (19). They also appear to control some target RNAs in conjunction with the yeast classical PUF proteins: Puf3, Puf4 and Puf5 (20–21,42).
Figure 1.
Two molecules of Puf1 recognize a dual UAAU motif. (A) Schematic drawing of the structural domains of Puf1 protein and the sequence of a dual UAAU motif RNA. Puf1 contains an N-terminal RRM (blue) and C-terminal PUM-HD (purple and gray ovals) beginning with an α-helical region (αN, green). Disordered regions are predicted between the RRM and the αN domain and after the PUM-HD. Puf1 protein constructs used in our studies are indicated. The two UAAU motifs in the RNA sequence are colored as in (B), and the nucleotide numbering used in the text is shown. (B) Crystal structure of the Puf1 PUM-HD in complex with a dual UAAU motif. Each of the two Puf1 PUM-HD proteins recognizes one of the UAAU sequences. We designate molecule A as the Puf1 PUM-HD interacting with the 5′-UAAU and molecule B as that interacting with the downstream UAAU-3′. The proteins are shown as ribbon diagrams with the N-terminal α-helical repeats (R1′ and R2-R5) colored alternately purple and light blue and non-RNA-binding repeats (R6-R8) colored gray. The N- and C-termini of the Puf1 PUM-HD are indicated by blue and red spheres, respectively. The αN region is colored green, and a helix encoded by the vector sequence is shown in beige. The RNA is shown as a cartoon highlighting the two UAAU motifs. (C) Conserved surfaces of the Puf1 PUM-HD interact with dual UAAU motif RNA. The two Puf1 molecules are shown as surface representations colored by degree of sequence conservation calculated using the ConSurf server (55). Highly conserved residues are colored maroon and weakly conserved residues are colored cyan.
Analysis of S. cerevisiae Puf2 mRNA targets from RNA immunopurification (RIP)-microarray and CLIP-seq (UV crosslinking and immunoprecipitation with deep sequencing) identified the UAAU tetranucleotide sequence as its binding motif (43–45), in contrast to the 5′-UGUA sequence motif found within the target RNAs of classical PUF proteins. The presence of more than two UAAU sequences is predominant in the top mRNA targets of Puf2, although one Puf2 molecule binds one UAAU sequence (45). The sequence and length of the linker region between the two UAAU sequences have little effect on Puf2 binding, based on analysis by yeast three-hybrid assay (44). CLIP analysis of Puf2 mutants lacking the RRM domain results in a lower percentage of RNA targets containing the UAAU motif, indicating that the RRM is not required to bind a UAAU motif but it enhances target selection in vivo (45). Although their sequence specificity has been examined, the Puf1/Puf2 proteins are the only remaining PUF family for which we have no structural information.
We sought to determine a crystal structure of a Puf1 protein to determine how the six-repeat Puf1/Puf2 PUM-HD recognizes UAAU motifs and how their single RRM domain influences RNA-binding activity. Here, we present a crystal structure of the PUM-HD of Schizosaccharomyces pombe Puf1 in complex with target RNA containing dual UAAU motifs. The structure reveals an extended flat PUF scaffold comprising seven PUM repeats. Unlike all other PUF-RNA complexes, two Puf1 molecules bind one RNA. PUM repeats R2 to R5 form both sequence-specific and sequence-independent protein–RNA interactions. Two crystal structures of the RRM domain of Puf1 reveal a homodimer. The RRM appears to lack RNA-binding activity in vitro, but when together with the PUM-HD, it enables Puf1 to preferentially bind to RNAs with dual UAAU sites over single sites. Dimerization via the RRM is required for high-affinity binding to the dual UAAU motif. This example of cooperative action of the RRM and PUM-HD identifies a new mechanism by which multiple RNA-binding modules within an RBP may collaborate for RNA target selection.
MATERIALS AND METHODS
Protein expression and purification
A cDNA fragment encoding the PUM-HD of Puf1 (residues 109–485) was amplified by polymerase chain reaction (PCR) from an S. pombe cDNA library pTN-RC5 (obtained from the Yeast Genetic Resource Center in Japan). Attempts to amplify fragments encoding the Puf1 RRM domain (residues 1–79) and the sequence containing both the RRM and PUM-HD domains (RP, residues 1–485) from the same cDNA library were unsuccessful. Therefore, a cDNA fragment corresponding to nucleotides 1–487 of the Puf1 gene sequence was synthesized (Thermo Fisher). The synthesized DNA was used as a PCR template to amplify a cDNA encoding the RRM domain. The RP fragment was constructed by ligating the cDNAs encoding the RRM domain and the PUM-HD through a BglII restriction site. The cDNAs were cloned into the pSMT3 vector that encodes an N-terminal His6-SUMO tag (kindly provided by Dr Christopher Lima) through NotI and XhoI restriction sites (46). The usage of the NotI site introduces 12 additional amino acid residues at the N terminus generated from the vector sequence that remain after Ulp1 cleavage to remove the His6-SUMO tag. The recombinant plasmids were transformed into BL21-CodonPlus (DE3)-RIL competent cells (Agilent) for protein expression.
Puf1 proteins were overexpressed by inducing cultures in log phase with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 16°C for ∼18 h for the PUM-HD and RP or 0.4 mM IPTG at 37°C for 3 h for the RRM. Bacteria were disrupted by sonication in lysis buffer containing 20 mM Tris (pH 8.0), 0.5 M NaCl, 20 mM imidazole, 5% (v/v) glycerol and 0.1% (v/v) β-mercaptoethanol. The proteins were purified by three sequential chromatography columns: a gravity-flow Ni-NTA chelating column (Qiagen), a Hitrap Q or a Heparin column (GE Healthcare) and a Superdex 75 column (GE Healthcare). The Ni-column elution buffer contained 20 mM Tris (pH 8.0), 50 mM NaCl, 0.2 M imidazole and 1 mM dithiothreitol (DTT). The Ni-column eluent was incubated with Ulp1 protease for 2 h (RRM or RP) or overnight (PUM-HD) to remove the His6-SUMO tag before loading onto the next column. Buffer A for the Hitrap Q or Heparin column was 20 mM Tris (pH 8.0), 1 mM DTT and buffer B contained an additional 1 M NaCl. With a linear gradient from 5–100% buffer B, the RRM protein was eluted off the Hitrap Q column with ∼15% buffer B. The PUM-HD protein was eluted off the Heparin column with ∼40% buffer B. The RP protein did not bind the Q resin but passing the solution over the column removed contaminating proteins. The protein solutions were concentrated before loading onto a Superdex 75 column equilibrated in 20 mM HEPES (pH 7.4), 0.15 M NaCl and 2 mM DTT. Peak fractions were pooled and concentrated for crystallization or RNA-binding assays.
To prepare selenomethionine (SeMet)-substituted PUM-HD protein, the plasmid was transformed into B834(DE3) Escherichia coli cells. The cells were grown in a media made from SeMet medium base (Molecular Dimensions) supplemented with 10 mg/ml SeMet. The protein was expressed and purified as the native protein. Single amino-acid substitution mutants of Puf1 PUM-HD (R315E or R318E) and Puf1 RP (Y3A/I39A/K73A) were generated using site-directed mutagenesis. The mutants were expressed and purified in the same way as the wild-type protein.
Crystallization
Concentrated Puf1 PUM-HD protein (OD280 = 3.0, ∼105 μM) was mixed with synthetic RNA purchased from GE Dharmacon (5′-UUAAUAACUUAAU-3′) with a molar ratio of 1:1 and incubated on ice for two hours prior to crystallization screening. Crystals were grown in a crystallization solution of 1.4 M MgSO4, 0.1 M MES (pH 5.6) with a 1:1 ratio of sample:reservoir solution by hanging drop vapor diffusion at 20°C. SeMet derivative crystals were obtained in the same conditions as native crystals. Crystals were cryoprotected in the crystallization solution supplemented with 20% (v/v) glycerol and flash frozen in liquid nitrogen.
The Puf1 RRM protein crystallized readily, however, the needle-shaped crystals were soft and did not diffract. To improve the crystal quality, we first deleted vector sequence encoding 12 N-terminal residues that remain after Ulp1 cleavage but this produced a fusion protein where the His6-SUMO tag could not be removed by proteolytic cleavage. We therefore introduced a 3-aa linker (GGS) between the His6-SUMO tag and the RRM that restored Ulp1 cleavage. The new RRM construct was purified in the same way as the initial construct. The protein was concentrated to 15 mg/ml. Numerous conditions from sparse-matrix crystallization screens yielded crystals. Two different conditions were subjected to optimization. One condition contained 0.3 M NaCl, 0.1 M citric acid (pH 3.6), 20% (v/v) glycerol. Crystals were directly looped out of the drop and flash frozen in liquid nitrogen. The other condition contained 15% (w/v) PEG 3350, 0.1 M MES (pH 5.6). Crystals were cryoprotected in the crystallization solution supplemented with 20% (v/v) glycerol and flash frozen in liquid nitrogen.
Data collection and structure determination
X-ray diffraction data were collected at beamline 22-ID of the Advanced Photon Source (APS) or using our in-house system, a 007HF Rigaku X-ray generator equipped with a Saturn944 CCD detector. A single-wavelength anomalous dispersion (SAD) dataset was collected on a SeMet derivative PUM-HD:RNA crystal at the wavelength of 0.97942 Å. All datasets were integrated and scaled with HKL2000 (47). For the PUM-HD:RNA structure, phases were determined from the 3.6 Å-resolution SAD dataset. Phenix AutoSol (48) found all 18 selenium sites from two protein molecules. Iterative AutoBuild and manual model building with COOT (49) were performed. The final structure was refined in Phenix to Rwork/Rfree of 0.211/0.263 at 3.0 Å resolution using a dataset from a native crystal. The crystals belong to the I4122 space group, and the asymmetric unit contains a complex of two Puf1 PUM-HD molecules bound to one RNA molecule. Protein molecule A contains residues 109–482 as well as the twelve residues encoded by the vector. Only the three C-terminal residues 483–485 are not modeled in molecule A due to missing electron density. Residues 154–163, 443–447 and 479–485 are not modeled in protein molecule B. All 13 nt in the RNA molecule are modeled based on clear electron density. The Puf1 PUM-HD protein molecules A and B can be aligned with an rmsd of 1.27 Å over 363 Cα.
The Puf1 RRM crystals that were grown in two different conditions belong to different space groups. The crystals from the PEG condition belong to the P21 space group, and the crystals from the NaCl condition belong to the P212121 space group. Molecular replacement (MR) with Phaser was attempted using several different RRM structures as the search model. The structure of Sex-lethal (Sxl) protein RRM1 (PDB ID: 4QQB) (50) with loops deleted led to a solution using the P21 dataset. The P212121 structure was solved using the P21 structure as the MR search model. In both structures there are four RRM molecules in an asymmetric unit. The final P21 structure was refined to Rwork/Rfree of 0.193/0.231 at 1.55 Å resolution. The relatively high Rfree is due to low completeness in the low-resolution shell. Molecule A contains residues 1–79 as well as the N-terminal linker sequence, SGGS. Four or five C-terminal residues are not modeled in molecules B, C and D. The final P212121 structure was refined to Rwork/Rfree of 0.172/0.225 at 2.05 Å resolution. X-ray data and refinement statistics for all structures are listed in Table 1.
Table 1.
Data collection and refinement statistics
| PUM-HD with RNA | RRM (PEG) | RRM (NaCl) | ||
|---|---|---|---|---|
| Data collection | ||||
| Wavelength (Å) | 1.54 | 1.0 | 1.54 | |
| Space group | I4122 | P21 | P212121 | |
| Cell dimensions | a, b, c (Å) | 159.68 159.68 215.53 | 46.85 37.59 73.93 | 54.69 63.90 79.54 |
| α, β, γ (°) | 90 90 90 | 90 94.2 90 | 90 90 90 | |
| Resolution (Å) | 50.0-3.0 (3.05-3.0) | 50.0-1.55 (1.58-1.55) | 50.0-2.06 (2.1-2.06) | |
| R merge | 0.062 (0.526) | 0.061 (0.549) | 0.08 (0.206) | |
| I / σI | 34.2 (4.0) | 20.1 (2.4) | 13.4 (4.0) | |
| Completeness (%) | 99.9 (100.0) | 98.1 (90.9) | 97.9 (95.1) | |
| Redundancy | 8.1 (8.1) | 5.5 (5.0) | 3.6 (2.2) | |
| Refinement | ||||
| Resolution (Å) | 32.8-3.0 | 38.2-1.55 | 24.5-2.05 | |
| No. reflections | 27506 | 33403 | 16888 | |
| R work / Rfree | 0.211 / 0.263 | 0.193 / 0.231 | 0.172 / 0.225 | |
| No. atoms | ||||
| Protein | 6099 | 2378 | 2309 | |
| RNA | 272 | |||
| Solvent | 51 | 234 | 215 | |
| B-factors | ||||
| Wilson B | 77.8 | 11.9 | 16.8 | |
| Protein | 92.9 | 23.2 | 17.7 | |
| RNA | 98.2 | |||
| Solvent | 98.8 | 29.4 | 22.9 | |
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.003 | 0.006 | 0.002 | |
| Bond angles (°) | 0.554 | 0.822 | 0.456 | |
Statistics for the highest-resolution shell are shown in parentheses.
Electrophoretic mobility shift assays
Synthetic RNAs (GE Dharmacon) were labeled with 32P- γ-ATP by T4 polynucleotide kinase for 1 h at 37°C. Unincorporated 32P- γ-ATP was removed using Illustra MicroSpin G-25 columns. For the PUM-HD and RP proteins, radiolabeled RNAs (100 pM) were incubated with 2-fold serially diluted protein samples from 4000 to 0.49 nM at 4°C for 1 h in 10 mM HEPES (pH 7.4), 50 mM NaCl, 0.01% (v/v) Tween-20, 0.1 mg/ml BSA, 10 μg/ml yeast tRNA and 2 mM DTT. For the RRM protein, the binding buffer was simpler with 10 mM HEPES (pH 7.4), 50 mM NaCl, and 0.01% (v/v) Tween-20. The samples were resolved on 10% polyacrylamide native TBE gels run at constant voltage (100 V) with 1× TBE buffer at 4°C for 35 min. The gels were dried and visualized using a Typhoon PhosphorImager (GE Healthcare). Band intensities were quantified with ImageQuant 5.1. The data were fit using the Hill equation with GraphPad Prism 7. EMSAs were performed three times, and mean Kd’s and standard error of the mean are reported (Table 2 and Supplementary Table S1).
Table 2.
In vitro RNA-binding affinities of Puf1 determined by EMSA
| RNA variant | Sequencea | PUM-HD, Kd | K rel b | Puf1 RP, Kd | K rel b |
|---|---|---|---|---|---|
| Dual UAAU | UUAAUAACUUAAU | 50.1 ± 1.9 | 1 | 29.5 ± 1.3 | 1 |
| Dual UAAU-L2 | UUAAUA - - UUAAU | 57.3 ± 0.3 | 1.1 (0.02) | N.D.c | |
| Dual UAAU-L0 | UUAAU - - - - UAAU | 90.5 ± 8.7 | 1.8 (0.01) | 43.5 ± 1.3 | 1.5 (0.0002) |
| Dual UAAU-L11 | CUAAUGAUAUUUUGACUAAU | 48.5 ± 0.8 | 1 (0.48) | 25.9 ± 2.4 | 0.9 (0.23) |
| Mono UAAU (Site 1) | UUAAUAACUACAG | 71.9 ± 0.5 | 1.4 (0.0004) | 169 ± 48 | 5.7 (0.007) |
| Mono UAAU (Site 2) | UACAGAACUUAAU | 92.8 ± 3.5 | 1.9 (0.008) | 125 ± 6.8 | 4.2 (0.00002) |
| U1G, U9G | UGAAUAACUGAAU | 435 ± 43 | 9 (0.0008) | 491 ± 8.5 | 17 (<0.00001) |
| A2G, A10G | UUGAUAACUUGAU | 484 ± 6.9 | 10 (<0.00001) | 423 ± 50 | 14 (0.0002) |
| A3G, A11G | UUAGUAACUUAGU | 133 ± 8.0 | 2.6 (0.0006) | 98 ± 7.3 | 3.3 (0.00009) |
| U4G, U12G | UUAAGAACUUAAG | 1191 ± 167 | 24 (0.002) | 745 ± 189 | 25 (0.003) |
| U1G | UGAAUAACUUAAU | 93 ± 6.0 | 1.8 (0.003) | ||
| A2G | UUGAUAACUUAAU | 80 ± 2.0 | 1.6 (0.0004) | ||
| U4G | UUAAGAACUUAAU | 112 ± 5.0 | 2.2 (0.0003) | ||
| U9G | UUAAUAACUGAAU | 91 ± 9.1 | 1.9 (0.012) | ||
| A10G | UUAAUAACUUGAU | 106 ± 5.5 | 2.1 (0.0007) | ||
| U12G | UUAAUAACUUAAG | 234 ± 23 | 4.7 (0.0013) |
aUAAU motifs are underlined, and altered nucleotides are in boldface.
b K rel values are calculated relative to the Kd for binding to the dual UAAU RNA (top row). P-values for comparison with the Kd for binding to the dual UAAU RNA (unpaired two-tailed t-tests) are in parentheses.
cN.D., not determined.
Differential scanning fluorimetry assay
Reaction mixtures (20 μl) contained ∼0.2 mg/ml Puf1 PUM-HD proteins (wild-type, R315E or R318E) and Sypro Orange dye (1:1000 dilution) (Invitrogen). Fluorescent intensity was collected from 25°C to 95°C (3°C increment/min) with a real time PCR instrument (Applied Biosystems™ QuantStudio 7 Flex System) using excitation and emission wavelengths of 470 and 586 nm, respectively. The Protein Thermal Shift software (Applied Biosystems) was used to analyze protein melting curves and calculate melting temperatures (Tm).
Size exclusion chromatography-multiangle light scattering (SEC-MALS)
Molecular masses of Puf1 proteins were assessed by SEC-MALS using an AKTA FPLC system (GE Healthcare) coupled to miniDawn TREOS and Optilab rEX detectors (Wyatt Technology). The PUM-HD and RP proteins, with or without RNA, were run on a Superdex-200 10/300 GL column (GE Healthcare) in a buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl and 2 mM DTT. A Superdex 75 10/300 column with the same buffer was used for the RRM protein. All data were analyzed using ASTRA 7.1.4 software (Wyatt Technology).
CLIP-seq analyses
CLIP-seq data for S. cerevisiae Puf2 protein variants from Porter et al., (45) were reanalyzed. The S. cerevisiae Puf2 proteins lacked the polyasparagine prion region at the C-terminus of the protein, and we refer to the Puf2 construct lacking only the polyasparagine region as the wild-type protein. Yeast strains had been UV-irradiated to covalently cross-link protein with directly bound RNA. The Puf2 protein was then affinity purified, and the attached segments of RNA had been identified by deep sequencing (45,51–53). The Puf2 CLIP-seq FASTQ files with NCBI accession number GSE73273 were downloaded. Adapter sequences (TGGAATTCTCGGGTGCCAAGG) and duplicates were removed using fastq-mcf with the parameter -D 35 (https://github.com/ExpressionAnalysis/ea-utils). Reads were mapped to the S. cerevisiae genome (EF4 release 70) using Bowtie2 (54) with the parameters ‘-5 5 –local.’ Peaks were obtained as reported in Porter et al., (45). Peak regions were defined by first merging the peak lists from yeast expressing Puf2 wild-type, PUM-HD only and ΔRRM. For overlapping peaks in these data sets, the highest peak for each region was retained. Peaks that corresponded to snoRNAs and tRNAs were discarded. Peaks with widths >40 nt were also discarded, as it was difficult to differentiate a widely spaced UAAU motif cluster from independent monomeric motifs. This retained 1614 peaks. UAAU motifs in peaks were counted, and overlapping motifs (e.g. UAAUAAU) were counted as one motif. Binding at a peak was defined as the maximum read depth in the peak region. For each dataset, we normalized binding to the median peak height for that particular Puf2 protein variant.
RESULTS
Two molecules of Puf1 recognize the dual UAAU motif in target mRNA
To provide a structural basis for understanding target RNA recognition by the yeast Puf1 and Puf2 protein family, we determined a crystal structure of S. pombe Puf1 in complex with RNA. We attempted to crystallize S. cerevisiae Puf1 and Puf2, but many different expression constructs yielded only insoluble or aggregated protein in E. coli. As a result, we turned to S. pombe Puf1 whose PUM-HD shares ∼40% sequence identity (61% similarity) with S. cerevisiae Puf1 and ∼38% sequence identity (59% similarity) with S. cerevisiae Puf2. Here we present a 3.0-Å resolution crystal structure of the PUM-HD of S. pombe Puf1 (residues 109–485) in complex with a model RNA binding element, derived from ARF1 mRNA and analyzed previously in three-hybrid assays (45) (5′-UUAAUAACUUAAU-3′, UAAU motifs underlined) (Figure 1A and Table 1). This RNA contains a dual UAAU motif, as is common in targets of the Puf1/Puf2 family. We define the first UAAU motif as positions 1–4 and the second motif as positions 9–12.
The crystal structure of Puf1 reveals a unique PUF protein scaffold formed by one pseudo-repeat R1′, consisting of two α helices, and followed by seven PUM repeats with three α helices. Although only six PUM repeats were predicted in sequences of Puf1/Puf2 family members, the structure identified seven repeats that together form an extended flat structure whose overall shape deviates from the curved crescent shape of classical PUF proteins like Puf3 (Figure 1B and Supplementary Figure S1). Unlike the highly regular α-helical structural repeats of human PUM1, Puf1 repeats are variable, especially in the loop between the second and third α helices (Supplementary Figure S1C). Repeats R3 and R8 are most distinct; their third α helices do not align well with those of the other repeats (Supplementary Figure S1D). A sequence alignment of Puf1/Puf2 homologs and yeast Puf3 indicates that the TRMs of the Puf1 N-terminal PUM repeats align best with repeats R2-R4 of Puf3. We designated the seven Puf1 PUM repeats as R2-R8 (Figure 1B and Supplementary Figure S2). These repeat designations thus are renumbered relative to those for S. cerevisiae Puf2 in Porter et al. (45). Repeat R2 here corresponds to Repeat R1 in their analyses. In addition to the tandem α-helical repeats, the PUM-HD of Puf1 contains an N-terminal extension (αN, residues 109–159) (Supplementary Figure S2). Residues 109–144 form a small structural subunit made of three α helices that pack against the non-RNA binding surfaces of repeats R4-R6 in the center of the structure, and residues 145–159 form a long coil that packs along the junctions between repeats R3-R4, R2-R3 and R1′-R2 and then leads into the pseudo-repeat R1′ at one end of the extended structure (Supplementary Figure S3A).
The structure of the Puf1 PUM-HD:RNA complex reveals a novel stoichiometry of PUF protein RNA recognition with two Puf1 protein molecules bound to the one RNA molecule containing two UAAU sites (Figure 1B). The presence of one protein per UAAU was predicted from compensatory mutant analyses of S. cerevisiae Puf2 (45), but is seen here for the first time. The two Puf1 molecules (A and B) are arranged in a V-like shape. The RNA is bent between U8 and U9, which places the two Puf1 N-terminal pseudo repeats near each other. This V-shaped arrangement is distinctive but may not be essential, as it could vary depending on the length of the linker sequence between the two UAAU motifs (see ‘Discussion’ section). Each UAAU tetranucleotide is bound by one protein molecule via PUM repeats R5 to R2. Repeats R6 to R8 do not contact RNA. Puf1 molecule A binds to U1-A2-A3-U4, and molecule B binds to U9-A10-A11-U12. The protein–RNA contacts are nearly identical between the two proteins and the two UAAU binding sites, and below we describe in detail protein–RNA interactions based on protein molecule A and RNA nucleotides U1-A2-A3-U4. The 4 nt in the RNA linker region, A5-A6-C7-U8, stack with each other. A5 stacks with the phenol ring of Tyr203 (Figure 1B and Supplementary Figure S3B). Nucleotides A6-C7-U8 and -1U make no contact with the proteins.
Sequence conservation analysis using the ConSurf server (55) indicates that the RNA-binding surface is highly conserved across fungal Puf1/Puf2 proteins (Figure 1C). We therefore predict that RNA recognition by other fungal Puf1/Puf2 proteins is similar to that of S. pombe Puf1. The strongest conservation is focused at the second α helices of PUM repeats, which include the RNA base-interacting TRM motifs. Repeat R8, which is not involved in RNA recognition, is less conserved than the other repeats.
Puf1 recognizes the UAAU motif sequence and flanking structural features
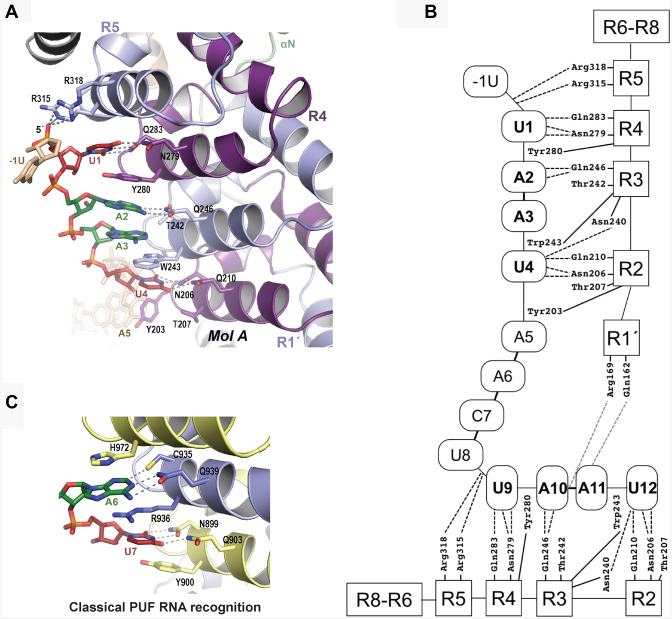
Puf1 recognizes RNA using a combination of classical PUF protein RNA base contacts and atypical interaction between divergent PUM repeats and RNA structural features. Only repeats R2-R4 in Puf1 bear RNA interacting residues similar to the classical sequence-specific PUM repeats (Figure 2A and B; Supplementary Figure S2), whereas other repeats display divergent sequences. In classical PUF proteins the set of residues in the TRM dictates the RNA base recognized by that repeat. Typically, NQ/Y selects U and CQ/R or SQ/R selects A (Figure 2C) (34). Puf1 repeats R4, R3 and R2 recognize U1, A2/A3 and U4, respectively. U1 is recognized sequence-specifically by the typical U-recognizing TRM NQ/Y of repeat R4 (Asn279, Tyr280 and Gln283) (Figure 2B). Additionally, the atypical TRM (AR/R) in repeat R5 contributes electrostatic interactions between Arg315 and Arg318 and the U1 phosphate group (Figure 2A). U4 is recognized by the TRM NQ/T in repeat R2, as had been predicted and confirmed for S. cerevisiae Puf2 by Porter et al. (45). The edge-interacting residues Asn206 and Gln210 form typical interactions with U4, but Thr207 fails to stack with the base due to its small side chain. There is an additional interaction between the O2 atom of U4 and Asn240 of repeat R3 (Figure 2B).
Figure 2.
Puf1 repeats R2-R4 recognize the UAAU motifs. (A) Interaction between Puf1 molecule A and 5′UAAU sequence. RNA-binding TRM residues from Puf1 are shown as stick models and gray dashed lines indicate atoms within hydrogen bonding distance of the RNA. The structure is colored as in Figure 1. (B) Schematic drawing of the interactions between Puf1 and the dual UAAU motif RNA. PUM repeats are represented by rectangles and RNA nucleotides are represented by ovals. (C) A classical PUF protein RNA interaction for comparison. RNA-binding TRM residues from human Pumilio1 are shown as stick models and gray dashed lines indicate interactions between the amino acid side chains and RNA nucleotides. The RNA is colored as in panel A.
In contrast to the more typical interactions of repeats R4 and R2 with U1 and U4, respectively, the A2 and A3 bases are stacked with one another and together are bound by the A-recognizing TRM, TQ/W, of repeat R3 (Figure 2). The edge-interacting residue Gln246 of repeat R3 forms two hydrogen bonds with A2 while the base-stacking residue Trp243 of repeat R3 stacks with A3. A2 is also stacked with Tyr280 from repeat R4. The flat RNA-binding surface formed by the Puf1 PUM repeats appears to accommodate the interaction of the single R3 repeat with the directly stacked A2 and A3 bases. The second UAAU motif (U9-A10-A11-U12) is recognized by molecule B just as molecule A contacts the first UAAU motif (Supplementary Figure S3B). In addition to recognition by molecule B, A11 of the second UAAU motif is bound by residues in the N-terminal pseudo-repeat of molecule A. Gln162 of molecule A forms a hydrogen bond with the N6 atom of A11 and Arg169 of molecule A forms a salt bridge with the A11 phosphate group (Figure 2B and Supplementary Figure S3B).
Puf1 PUM-HD RNA-binding activity depends upon UAAU motif and RNA backbone recognition
We probed the RNA-binding activity of the Puf1 PUM-HD and found that it binds with high affinity and specificity to the UAAU motif. We measured in vitro RNA-binding affinity of purified S. pombe Puf1 PUM-HD protein by electrophoretic mobility shift assay (EMSA) (Table 2, Supplementary Figure 4A). We used the dual UAAU motif RNA that was crystallized, which contains two UAAU sites separated by a 4-nt linker. Puf1 bound the dual UAAU motif RNA with an apparent Kd of 50.1 nM. This tight binding is similar to the in vitro affinity of S. cerevisiae Puf2 for a dual UAAU motif in PMP2 mRNA (Kd = 53 nM) (44). Our crystal structure revealed sequence-specific recognition of U1/U9, A2/A10 and U4/U12 by the TRMs of repeats R2-R4, but recognition of A3/A11 appeared to be sequence independent. We substituted A with G in the third positions of both UAAU motifs of the dual UAAU RNA (A3G, A11G). These sequence changes resulted in a modest 2.6-fold reduction in affinity. In contrast, substituting A with G in the second positions of both UAAU motifs (A2G, A10G) reduced binding affinity 10-fold relative to the base dual UAAU RNA sequence (Table 2). Therefore, as suggested by the crystal structure, an A at the second position of the UAAU motif is critical while the identity of the nucleotide at the third position is less important.
Figure 4.
The Puf1 RRM promotes binding of the PUM-HD to dual UAAU motif RNA. Bar graphs illustrate binding affinities relative to that of the corresponding protein with dual UAAU motif RNA (Krel). (A) Relative binding affinities of the Puf1 PUM-HD (gray) or RP (black) for RNA variants with dual or mono UAAU motifs, showing the effects of motif number and linker length. ‘Dual UAAU’ denotes an RNA with a 4-nt linker between the two UAAU motifs, ‘Dual UAAU-L0’ denotes no linker nucleotides and ‘Dual UAAU-L11’ denotes an 11-nt linker. RNA sequences and Kd values are shown in Table 2. Error bars in all graphs are the standard error of the mean of three replicate experiments. The measured Kd for binding to the dual UAAU motif RNA is shown above the bar. The Krel is the Kd for binding to a particular RNA divided by the Kd for binding of the same protein to the dual UAAU motif RNA. (B) Relative binding affinities of the Puf1 PUM-HD (gray) or RP (black) for RNAs bearing UAAU sequence variants, showing the effects of disrupting the UAAU motif sequences at different positions. (C) A common dimerization interface in crystal structures of the Puf1 RRM. Backbone traces of non-crystallographic dimers in crystals of the Puf1 RRM are superimposed (space group P21, blue and cyan; space group P212121, gray). Side chains for residues that were mutated to alanine in Puf1 RP TM are shown as stick models. (D) Relative binding affinities of the Puf1 RP (black) or RP TM (dark gray) for RNAs bearing dual or mono UAAU motifs.
The RNA-interacting TRM motif in repeat R4 of Puf1 (AR/R) is distinct from classical PUM repeats, and we found that the two arginine residues that interact with the phosphate backbone are essential for RNA complex formation. Our crystal structure revealed that Arg315 and Arg318 electrostatically interact with the phosphate groups of U1 (molecule A) and U9 (molecule B) (Figure 2 and Supplementary Figure S3B). We mutated Arg315 or Arg318 to glutamic acid and measured binding of the mutant proteins to the dual UAAU RNA (Supplementary Table S1). No protein–RNA complex was detected for the R315E mutant with the dual UAAU RNA, even at a protein concentration as high as 1.5 μM. We detected binding of the R318E mutant to the dual UAAU RNA, but the mutant protein bound 4-fold more weakly than wild-type Puf1. To ensure that these binding defects were not due to misfolding of the mutant proteins, we checked the protein stability by differential scanning fluorimetry. The R315E mutant displayed a slightly lower melting temperature (Tm) than wild-type (49.3°C versus 52.2°C), whereas R318E displayed a slightly higher Tm of 54.6°C. The small difference in protein stability of the R315E mutant does not account for the loss of RNA-binding activity. Therefore, Arg315 is a critical residue for RNA binding. Together, the interaction of Arg315 and Arg318 with the RNA backbone may support the RNA conformation at the 5′ ends of the UAAU motifs that is observed in the crystal structure.
S. cerevisiae Puf2 binds to dual UAAU motifs separated by linkers of a range of lengths and sequences, and we found that for S. pombe Puf1 the length of the linker between UAAU sites also plays a minor role in affinity. We measured binding of the S. pombe Puf1 PUM-HD to RNAs with a 2-nt linker (dual UAAU-L2) or an 11-nt linker (dual UAAU-L11, corresponding to a sequence in the 3′-UTR of S. pombe TPO1 mRNA) and found that Puf1 bound to the dual UAAU-L2 or L11 RNA with similar affinity as to the dual UAAU motif with a 4-nt linker (dual UAAU, Table 2). When we removed the entire linker sequence (dual UAAU-L0), Puf1 binding was only 2-fold weaker than binding to the dual UAAU RNA with a 4-nt linker (Table 2).
In contrast to the importance of a dual UAAU motif for S. cerevisiae Puf2 binding, we found that a single UAAU sequence is sufficient for S. pombe Puf1 PUM-HD recognition. Although many mRNAs contain a dual UAAU motif, other yeast Puf1/2 target mRNAs contain only one UAAU site. However, previous yeast 3-hybrid analyses of S. cerevisiae Puf2 indicated that its minimal recognition site was a dual UAAU motif and a mono UAAU site did not support binding by S. cerevisiae Puf1 or Puf2 (44). We therefore tested whether the S. pombe Puf1 PUM-HD binds to RNAs with a single UAAU motif. We mutated either the first or second UAAU site in the dual UAAU RNA to ACAG. Puf1 bound to either RNA with a single UAAU site with only slightly reduced affinity (Table 2, 1.4- to 1.9-fold weaker than to the dual UAAU motif). Consistent with this finding, Puf1 bound no more than 2-fold weaker to dual UAAU RNAs that retained one UAAU motif and bore a single-base substitution in the other motif (A2G, A10G, U1G or U9G).
The Puf1 RRM domain favors dual UAAU motif binding
An N-terminal RRM domain is unique to the fungal Puf1/Puf2 subgroup of proteins, which led us to investigate its role, if any, in RNA recognition. Our crystal structures of the Puf1 RRM suggested it is similar to other RNA-binding RRMs, as it possesses features that typically contribute to RNA binding. We determined crystal structures of the Puf1 RRM by molecular replacement using a crystal structure of the RRM1 domain of D. melanogaster Sex-lethal (Sxl, PDB ID: 4QQB) (50) as the search model (Table 1). The Puf1 RRM domain adopts the classical RRM fold comprising a four-stranded antiparallel β-sheet packed against two α helices (Figure 3A). Its structure aligned with the Sxl RRM1 molecular replacement search model with an RMSD of 1.06 Å over 70 Cα atoms, although the sequence identity is <30% (Figure 3B and C). Canonical RRM domains are characterized by two conserved ribonucleoprotein (RNP) sequences that are important for RNA binding: eight-residue RNP1 located on strand β3 and six-residue RNP2 located on strand β1. Typically the hydrophobic side chains at position 5 in RNP1 and position 2 in RNP2 play key roles in interacting with single-stranded RNA bases (56). The sequence of the S. pombe Puf1 RNP2 matches the consensus motif with a tyrosine residue at position 2 (Y3), which could form a stacking interaction with an RNA base. The sequence of RNP1 deviates considerably from the consensus motif, yet it retains an isoleucine residue at position 5 (I39) that could form a hydrophobic interaction with an RNA base (Figure 3D).
Figure 3.
Crystal structure of the Puf1 RRM identifies potential RNA-interacting residues. (A) Ribbon diagram of a crystal structure of the S. pombe Puf1 RRM. Residues at positions on the β sheet that typically bind RNA are shown as stick models. (B) Structural superposition of the Puf1 RRM with RRM1 of D. melanogaster Sxl protein (PDB ID: 4QQB). The structures are shown as Cα traces (Puf1, blue; Sxl, mauve). (C) Ribbon diagram of a crystal structure of D. melanogaster Sxl RRM1 in complex with msl2 RNA. Selected residues on the β sheet that bind the RNA are shown as stick models. (D) Amino acid sequence alignment of the RRM domains of Puf1 homologs and D. melanogaster Sxl protein. Secondary structural elements of the Puf1 RRM are shown above the sequences, and the RNP1 and RNP2 motif sequences are boxed. Residues shown as stick models in (A), (B) and (C) are in boldface. Consensus sequences for the RNP1 and RNP2 motifs are shown below the sequences.
Since the mode of RNA recognition by RRMs varies from protein to protein and some RRMs do not bind RNA, we cannot predict whether or not the Puf1 RRM domain binds to RNA based solely on the apo-RRM crystal structure. We therefore tested whether purified Puf1 RRM protein bound to RNA in vitro. Reasoning that the RRM could bind to sequences outside the dual UAAU motif, we used an RNA similar to the dual UAAU RNA, but with additional flanking sequence (5′-ACAUUAAUAACUUAAUA-3′) and measured RNA binding by EMSA. The Puf1 RRM did not bind to this RNA, even at a protein concentration of 10 μM. We also tested binding to two additional RNA sequences, a 35 nt sequence from S. pombe TPO1 (5′-UUUUUUUCUAAUGAUAUUUUGACUAAUACGGAUUA-3′), the UAAU-L11 sequence with 5′ and 3′ extensions (Table 2), and a 30 nt sequence modified from S. cerevisiae PMP2* (5′-AAUUUCUAAUAAUUAAUACAUUUUUCCUCU-3′) (44). The Puf1 RRM domain did not bind to either of these RNAs, even at a protein concentration of 20 μM. Therefore, although the Puf1/2 RRM enhances UAAU motif recognition in vivo, it lacked detectable in vitro RNA-binding activity on its own.
We hypothesized that the RRM domain could alter the RNA-binding selectivity of the PUM-HD and found that addition of the Puf1 RRM to the PUM-HD surprisingly favored binding to a dual UAAU motif over a mono UAAU site, but had little effect on overall affinity and no effect on UAAU motif sequence specificity. We measured RNA-binding affinities of a Puf1 protein containing both the RRM and PUM-HD (Puf1 RP, residues 1-485). We found that Puf1 RP bound the dual UAAU RNA with a Kd of 29.5 nM, which is similar to the value we measured for the PUM-HD alone (Table 2 and Supplementary Figure S4B). We also found that the binding affinity was not substantially affected by the length of the linker separating the two UAAU motifs: Puf1 RP binding affinity for dual UAAU-L11 RNA with an 11-nt linker was unchanged and for the dual UAAU-L0 RNA with no linker was only 1.5-fold weaker than to the RNA with a 4-nt linker (Figure 4A and Table 2). Sequence substitutions in the UAAU motifs had the same effects as for the Puf1 PUM-HD: mutations of the first (U1G/U9G), second (A2G/A10G) and fourth (U4G/U12G) nucleotides weakened Puf1 RP binding ∼14-fold, but mutations of the third nucleotides (A3G/A11G) only weakened binding 3-fold (Figure 4B and Table 2). These results indicate that the PUM-HD is the main contributor to sequence-specific RNA recognition by Puf1. Lastly, we tested binding affinity of Puf1 RP for RNAs bearing a single UAAU motif and found that mono UAAU RNAs bound ∼5-fold weaker than dual UAAU RNAs (Figure 4A). In contrast, the PUM-HD bound to the same RNAs with less than 2-fold difference in affinity. We conclude that the RRM domain together with the PUM-HD leads to preferential Puf1 binding to target RNAs with dual UAAU motifs.
The Puf1 RRM promotes dimerization and stable dual UAAU motif recognition
In addition to binding RNA, RRM domains can mediate protein–protein interactions by forming homodimers, making intramolecular contacts with flanking regions, and binding other proteins (56). We therefore examined whether the Puf1 RRM domain promotes dimerization, which might favor dual UAAU recognition. We determined molecular masses of Puf1 proteins and protein–RNA complexes by SEC-MALS and found that the Puf1 RP protein forms a stable 2:1 complex with a dual UAAU RNA in solution (Table 3). The apparent molecular mass of the PUM-HD was 43.8 kDa, similar to the calculated protein molecular weight (MW) of 44.6 kDa, indicating that the protein is monomeric in solution. In contrast, the apparent molecular masses of the Puf1 RRM and RP were ∼1.6–1.7 times their respective calculated MWs: 15.1 kDa for the RRM versus 8.9 kDa (calculated) and 86.1 kDa for Puf1 RP versus 54.6 kDa (calculated). These intermediate molecular masses suggest that monomer and dimer forms of the Puf1 RRM and RP proteins in the absence of RNA are in a fast equilibrium, because we observed single peaks that appear to be one species (Supplementary Figure S5). When we analyzed complexes of the Puf1 PUM-HD with dual UAAU RNAs, we found that the molecular masses were similar for RNAs with linker lengths of 0, 2 and 4 nts, about 1.5 times the monomer size (Table 3). Therefore, the PUM-HD is in a rapid equilibrium of one or two protein molecules bound to the dual UAAU motif RNA. The molecular mass of a complex of the PUM-HD with a mono UAAU RNA bearing only one intact UAAU motif (site 1) was 44.5 kDa, corresponding to a stable 1:1 complex. In contrast, the molecular mass for Puf1 RP in complex with the dual UAAU RNA was 110.6 kDa, matching the calculated MW of 113.2 kDa for two Puf1 RP molecules bound to an RNA. Thus the RRM appears to stabilize the 2:1 Puf1 RP-dual UAAU motif RNA complex in solution.
Table 3.
SEC-MALS molecular weight (kDa)
| Calc MW | No RNA | Dual UAAUa | Dual UAAU-L2 | Dual UAAU-L0 | Mono UAAU | |
|---|---|---|---|---|---|---|
| PUM-HD | 44.6 | 43.8 | 68.5 | 68.3 | 67.0 | 44.5 |
| RRM | 8.9 | 15.1 | ||||
| Puf1 RP | 54.6 | 86.1 | 110.6 | 97.3b | ||
| Puf1 RP TM | 54.4 | 53.1 | 106.9 | 58.6 |
aRNA MW = ∼4 kDa.
bWhen only one UAAU motif is available for binding, the Puf1 RP appears to be in an equilibrium of various species of monomer/dimer bound to the mono UAAU RNA.
We obtained two different types of crystals of the RRM domain of S. pombe Puf1: one was grown in a crystallization condition with PEG (P21 space group) and the other was grown in a condition with NaCl (P212121 space group). Both models contain four molecules (labeled A, B, C, D) in an asymmetric unit, and all eight molecules are highly similar (RMSD 0.2-1.4 Å over 75–78 Cα atoms). The four molecules in both structures are related by non-crystallographic symmetry, with the A and B pair equivalent to the C and D pair. Molecules A and B are arranged similarly with respect to each other in the two structures, despite different space groups (Figure 4C). The protein molecules contact each other through hydrophobic and H-bonding interactions via residues in the β-sheet and the C-terminal loop. Although RRM homodimers have been observed previously (56), a search for similar homodimers in the Protein Data Bank did not identify a similar RRM-RRM interface. The PDBePISA server (57) calculated a buried surface area of 1360–1900 Å2 with a Complexation Significance Score of 1.0. This indicates that the observed homodimer AB is likely to be biologically relevant, rather than a crystal-packing artifact.
To determine the importance of the RRM homodimer, we mutated to alanine three residues located at the interface (Tyr3 in the RNP2 motif, Ile39 in the RNP1 motif and Lys73 in the C-terminal loop, Figure 4C) and found that dimerization is critical for dual UAAU motif preference. By SEC-MALS analysis, the apo Puf1 RP tri-mutant (TM) protein now exhibited the MW of a monomer, 58.6 kDa, indicating that the interface is important for dimer formation (Table 3). Nevertheless, the Puf1 RP TM protein bound tightly to the dual UAAU RNA (Supplementary Table S1) and retained the ability to form a stable 2:1 complex with dual UAAU RNA with a MW of 106.9 kDa (Table 3). Therefore, the alanine substitutions that affect Puf1 RP dimerization do not disrupt the RRM’s function to stabilize complex formation on a dual UAAU motif RNA. Instead RRM dimerization is necessary for preferential binding to a dual UAAU motif over a single UAAU motif. The Puf1 RP TM protein behaved like the PUM-HD alone and bound to a mono UAAU RNA (site 1) only 1.5-fold weaker than binding to a dual UAAU motif rather than the 6-fold stronger binding affinity of the wild-type protein for the dual UAAU motif RNA (Figure 4D). We conclude that the RRM imposes a preference in vitro for dual- versus mono-UAAU elements on the PUM-HD. Past work has shown that members of the Puf1/Puf2 family prefer multiple UAAU elements (45). We predict that in vivo preference relies on the RRM.
In vivo studies support collaboration between the Puf1/2 RRM and PUM-HD
To examine whether the Puf1/2 RRM influences RNA-binding specificity in living cells, we re-analyzed our published CLIP-seq studies of S. cerevisiae Puf2 (45) and found that Puf2 lacking the RRM fails to preferentially bind to sites with multiple UAAU elements. In S. cerevisiae and S. pombe, Puf2 is closely related to Puf1, with the same overall protein architecture (15,43,58). In our CLIP-seq studies we had identified in vivo RNA interactions for three forms of Puf2: wild-type protein containing both the RRM and PUM-HD, a mutant protein that contained essentially the PUM-HD alone and a second mutant that lacked the RRM (ΔRRM) (45). In light of our in vitro data indicating the influence of the RRM on binding to sites with dual UAAU motifs, we assessed the preferences of the wild-type and mutant proteins for single versus multiple UAAU motifs in vivo. We first compared the number of UAAU motifs present in binding regions (‘peaks’) for the three Puf2 forms. We sorted peaks into those containing 0, 1, 2, 3 or >3 UAAU motifs and assessed the ‘peak heights,’ defined as the maximum read depth in each peak and normalized to the median peak height for the proteins. For the wild-type protein, we found that the peak heights, which serve as a surrogate for protein binding in vivo, increased progressively with increasing number of UAAU motifs in the peak (Figure 5A left, ‘Wild-type’). This progressive increase disappeared with a protein that possessed only the PUM-HD (Figure 5A middle, ‘PUM-HD’). Moreover, the Puf2p ΔRRM variant (which lacks only the RRM domain) also failed to exhibit increased binding to RNA sites with two or more UAAU motifs (Figure 5A right, ‘ΔRRM’). Therefore, the Puf2 mutant proteins lacking the RRM bind equally well to sites with one or multiple UAAU motifs.
Figure 5.
CLIP-seq analyses support a role for the Puf1/2 RRM in binding site selection in vivo. (A) S. cerevisiae Puf2 protein shows increasing binding to sites with 2, 3 or >3 UAAU motifs, which disappears in the absence of an RRM domain. Peak heights of CLIP-seq peaks for the indicated proteins sorted by the number of UAAU motifs. The log2 peak heights normalized to the median peak height for each protein are shown. Box plots show median values and the interquartile range (IQR) from the 25th to 75th percentile. The whiskers extend 1.5 × IQR, and outliers are shown as diamonds. (B) Puf2 protein binding to sites with multiple UAAU motifs is greater for wild-type protein than for mutants lacking an RRM domain. Peak heights of CLIP-seq peaks for the indicated proteins sorted by the number of UAAU motifs. The log2 ratio of mutant protein peak heights to wild-type protein are shown. The ratio of PUM-HD binding to wild-type Puf2 binding is lower for sites with multiple UAAU motifs versus one motif (P< 10−3). The same is true for the ΔRRM protein (P< 10−8).
We next determined, peak-by-peak, the ratio of peak heights for the mutant proteins relative to that of wild-type protein. For binding regions with 1 UAAU motif, the peak heights were similar for wild-type and mutant proteins (median log2 ratio of peak heights near 0), suggesting they bind equally well to sites with a single motif (Figure 5B). In contrast, the peak heights for binding of the PUM-HD and ΔRRM proteins to regions with multiple UAAUs were lower than for the wild-type protein (negative values for median log2 ratio of peak heights), reflecting the favorable binding of the wild-type protein to sites with multiple UAAUs (Figure 5B) and supporting our prediction that in vivo, the RRM imposes preferential binding to RNAs with multiple UAAU motifs.
DISCUSSION
The seven yeast PUF proteins fully represent the diversity of the PUF protein family across eukaryotes. Puf3, Puf4 and Puf5 are classical sequence-specific RBPs, and Nop9 and Puf6 represent the two atypical PUF proteins common among all eukaryotes. The Puf1/Puf2 subgroup is specific to fungi and has been conserved over hundreds of millions of years. Its defining characteristics are a divergent PUM-HD, an RRM RNA-binding domain, distinctive RNA sequence specificity, and novel 2:1 protein-to-RNA stoichiometry. Our crystal structure of S. pombe Puf1 reveals that the PUM-HD directly binds the UAAU motif using PUM repeat RNA recognition residues for both classical RNA base recognition and interaction with backbone phosphate groups. The RRM also has a key role, in that it promotes preferential Puf1 binding to dual UAAU motifs over single UAAU sequences (Figure 6). Dual UAAU motifs are present frequently in high-ranked target RNAs of S. cerevisiae Puf2, while mono UAAU motifs are common in low-ranked targets (45), strongly suggesting that this function of the RRM is biologically significant.
Figure 6.
Model for Puf1 UAAU RNA recognition selectivity. Schematic representation of RNA recognition by the Puf1 PUM-HD alone or in combination with the RRM. The PUM-HD alone binds with similar affinity to either dual or single UAAU motif RNA (top). In contrast, the RRM + PUM-HD (RP) forms a stable 2:1 complex with dual UAAU motif RNA and binds 6-fold weaker to an RNA with a single UAAU than to a dual site (bottom).
The specificity of Puf1/Puf2 proteins for their RNA targets in vivo reflects the interplay of multiple structural features in the proteins. The Puf1 PUM-HD recognizes UAAU motif RNA by bringing together the sequence-specific RNA-binding mode adopted by classical PUF proteins with the electrostatic interaction mode of Puf6 proteins. Of the seven PUM repeats in Puf1, only repeats R2-R4 bear RNA base-interacting TRMs similar to the classical sequence-specific PUM repeats: Repeats R4, R3 and R2 recognize U1, A2/A3 and U4, respectively. In contrast, Puf1 PUM repeats R5-R8 display divergent TRM sequences. Repeats R6 to R8 of Puf1/Puf2 protein do not contact RNA, but repeat R5 uses two arginine residues in its TRM to make electrostatic contacts with the U1 and U9 phosphate groups. This interaction by repeat R5 is reminiscent of that observed in the atypical PUF protein subgroup containing Puf6 and its human ortholog Puf-A (24). Conserved basic residues within multiple TRMs interact with the phosphate backbone in a crystal structure of Puf-A in complex with double-stranded DNA.
Dual UAAU motifs contain linkers separating the two UAAU motifs ranging from 0 to 10 nt, with 3 nt being the most prevalent (44). Our structure of Puf1 bound to a dual UAAU RNA with a 4-nt linker revealed that each UAAU sequence is recognized by one Puf1 molecule, and RNA-binding assays showed that binding by the Puf1 PUM-HD with or without the RRM is tight regardless of the linker length. In our crystal structure, the RNA makes a sharp bend between U8 and U9, which are the last nucleotide of the linker and the first nucleotide of the second UAAU motif, respectively. The intervening phosphate group is contacted by arginine side chains in the repeat R5 TRM and a Mg2+ ion supports the RNA conformation. We speculate that the relative orientation of the two RNA-bound protein molecules must change to accommodate different length linkers. For example, with no linker nucleotides, the observed relative orientation of the two Puf1 molecules is impossible, and with fewer than four intervening nucleotides, the observed interactions at both ends of the linker sequence could not be made. With sequences longer than 4 nt, the linker RNA may adopt different conformations. We did not observe protein-protein interactions between the two Puf1 PUM-HD molecules in our crystal structure or in solution measurements, but intermolecular interactions might occur between the two protein molecules when bound to RNAs with longer or shorter linker lengths.
The Puf1/Puf2 RRM domain enhances target RNA selection with dimerization favoring binding of Puf1 to the dual UAAU motif, and this finding is reflected in the mRNA targets of S. cerevisiae Puf2 protein in vivo. A single UAAU motif is relatively low in sequence complexity compared to the 8–12 nt sequences recognized by classical PUF proteins. When both the Puf1 RRM and PUM-HD are present, the binding affinity is now sensitive to loss of one of the two UAAU motifs. The highest ranked mRNA targets of S. cerevisiae Puf2 in vivo, as defined by CLIP experiments, possess two UAAU motifs, which progressively declines to a plateau of an average of ∼1.5 motifs in the weakest binding RNAs (45). The RRM does not appear to affect sequence specificity of the PUM-HD for a UAAU motif in vitro, but it does affect target mRNA selection in vivo. A UAAU motif is found in 81% of target RNAs of Puf2 with both RRM and PUM-HD. In contrast, only 48% of target RNAs of the Puf2 PUM-HD alone contain a UAAU motif (45). Furthermore, as shown here via in vivo CLIP studies, the RRM imposes a systematic preference for binding sites with 2, 3 or >3 UAAU elements. Collectively, these data strongly support the view that the Puf1/Puf2 PUM-HD directs sequence specificity and that the RRM enhances recognition of multiple UAAU motifs.
The presence of both an RRM and PUM-HD, though unique to the fungal Puf1/Puf2 PUF subgroup, emphasizes the broad principle that distinct RNA-binding modules within a single protein can alter RNA recognition even without both domains interacting with RNA. RRMs previously have been shown to collaborate with other RBPs. For example, the RRM of the 68-kDa subunit of Cleavage Factor Im (CFIm68) has been reported to alter the RNA-binding properties of the 25-kDa Nudix domain subunits (CFIm25). Similar to Puf1, the CFIm68 RRM does not affect sequence specificity of CFIm25, as CFIm25 recognizes a UGUA motif in the absence of CFIm68 (59). Instead, it appears that the RRM of CFIm68 restricts binding of the complex to RNAs with two UGUA motifs separated by >9 nt. RRMs are often combined with other RNA-binding modules in RBPs. Given our findings with Puf1, we expect that some of these RRMs might collaborate with neighboring RNA-binding domains to modulate RNA recognition. This also applies to RBPs with multiple RRMs that together determine the specificity of RNA recognition. The RBP HuR contains two N-terminal RRMs in tandem and a third C-terminal RRM3. The N-terminal RRMs comprise a module that is sufficient for AU-rich or U-rich RNA element recognition, but the C-terminal RRM3 has been shown to enhance RNA-binding affinity through both dimerization and direct RNA recognition (60,61). The dimerization interface of HuR RRM3 does not obstruct its RNA-binding surface, which is different from what we observed for the Puf1 RRM. Dimerization of the Puf1 RRM involves the β-sheet RNA-binding surface and we could not demonstrate RNA binding by the Puf1 RRM, suggesting that it may not have a dual function like that of HuR RRM3. However, the apparent fast equilibrium between monomer and dimer forms of the Puf1 RRM in the absence of RNA suggests that its β-sheet surface could become available for RNA binding. A crystal structure of a Puf1 RP–RNA complex is needed to enhance understanding of the cooperative activities of the RRM and PUM-HD, but our efforts have not yet yielded crystals.
As illustrated here, deep studies of model RNA-binding proteins produce new principles of RNA recognition, in this case how two RNA-binding domains within a protein collaborate to produce a distinct regulatory motif. Encyclopedias of RNA-binding proteins continue to expand along with glimpses into their RNA recognition specificities and target networks through powerful high-throughput, genome-wide probing (62–64). The discovery of cooperative modes of RNA recognition can drive genomic studies forward by suggesting new parameters for analyzing existing data sets and refinements for generating new data.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 6NY5 (Puf1 PUM-HD and RNA), 6NX5 (Puf1 RRM) and 6NWW (Puf1 RRM).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to our colleagues Lars Pedersen and Monica Pillon for critical reading of our manuscript. We thank John Gonczy for assistance with data collection at SER-CAT beamlines 22-ID and 22-BM at the Advanced Photon Source, Argonne National Laboratory, Lars Pedersen and Juno Krahn for crystallographic and data collection support at NIEHS, and Geoffrey Mueller for preliminary NMR characterization.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences [1ZIA50165 to T.M.T.H., in part]; National Institutes of Health, National Institute of General Medical Sciences [GM050942 to M.W.]; U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences [W-31-109-Eng-38]. Funding for open access charge: National Institutes of Health, National Institute of Environmental Health Sciences [ZIA50165].
Conflict of interest statement. None declared.
REFERENCES
- 1. Coppin L., Leclerc J., Vincent A., Porchet N., Pigny P.. Messenger RNA life-cycle in cancer cells: emerging role of conventional and non-conventional RNA-binding proteins. Int. J. Mol. Sci. 2018; 19:E650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang E.T., Taliaferro J.M., Lee J.A., Sudhakaran I.P., Rossoll W., Gross C., Moss K.R., Bassell G.J.. Dysregulation of mRNA localization and translation in genetic disease. J. Neurosci. 2016; 36:11418–11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lenzken S.C., Achsel T., Carri M.T., Barabino S.M.. Neuronal RNA-binding proteins in health and disease. Wiley Interdiscip. Rev. RNA. 2014; 5:565–576. [DOI] [PubMed] [Google Scholar]
- 4. Ravanidis S., Kattan F.G., Doxakis E.. Unraveling the pathways to neuronal homeostasis and disease: mechanistic insights into the role of RNA-binding proteins and associated factors. Int. J. Mol. Sci. 2018; 19:E2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mitchell S.F., Parker R.. Principles and properties of eukaryotic mRNPs. Mol. Cell. 2014; 54:547–558. [DOI] [PubMed] [Google Scholar]
- 6. Zamore P.D., Williamson J.R., Lehmann R.. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997; 3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., Kimble J., Wickens M.P.. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997; 390:477–484. [DOI] [PubMed] [Google Scholar]
- 8. Wickens M., Bernstein D.S., Kimble J., Parker R.. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002; 18:150–157. [DOI] [PubMed] [Google Scholar]
- 9. Wharton R.P., Aggarwal A.K.. mRNA regulation by Puf domain proteins. Sci. STKE. 2006; 2006:pe37. [DOI] [PubMed] [Google Scholar]
- 10. Miller M.A., Olivas W.M.. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip. Rev. RNA. 2011; 2:471–492. [DOI] [PubMed] [Google Scholar]
- 11. Wang M., Oge L., Perez-Garcia M.D., Hamama L., Sakr S.. The PUF protein family: overview on PUF RNA targets, biological functions, and post transcriptional regulation. Int. J. Mol. Sci. 2018; 19:E410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Forbes A., Lehmann R.. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998; 125:679–690. [DOI] [PubMed] [Google Scholar]
- 13. Crittenden S.L., Bernstein D.S., Bachorik J.L., Thompson B.E., Gallegos M., Petcherski A.G., Moulder G., Barstead R., Wickens M., Kimble J.. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002; 417:660–663. [DOI] [PubMed] [Google Scholar]
- 14. Goldstrohm A.C., Hall T.M.T., McKenney K.M.. Post-transcriptional regulatory functions of mammalian pumilio proteins. Trends Genet. 2018; 34:972–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gerber A.P., Herschlag D., Brown P.O.. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004; 2:E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lapointe C.P., Stefely J.A., Jochem A., Hutchins P.D., Wilson G.M., Kwiecien N.W., Coon J.J., Wickens M., Pagliarini D.J.. Multi-omics reveal specific targets of the RNA-binding protein Puf3p and its orchestration of mitochondrial biogenesis. Cell Syst. 2018; 6:125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stewart M.S., Krause S.A., McGhie J., Gray J.V.. Mpt5p, a stress tolerance- and lifespan-promoting PUF protein in Saccharomyces cerevisiae, acts upstream of the cell wall integrity pathway. Eukaryot. Cell. 2007; 6:262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Viet N.T.M., Duy D.L., Saito K., Irie K., Suda Y., Mizuno T., Irie K.. Regulation of LRG1 expression by RNA-binding protein Puf5 in the budding yeast cell wall integrity pathway. Genes Cells. 2018; 23:988–997. [DOI] [PubMed] [Google Scholar]
- 19. Haramati O., Brodov A., Yelin I., Atir-Lande A., Samra N., Arava Y.. Identification and characterization of roles for Puf1 and Puf2 proteins in the yeast response to high calcium. Sci. Rep. 2017; 7:3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fischer A.D., Olivas W.M.. Multiple Puf proteins regulate the stability of ribosome biogenesis transcripts. RNA Biol. 2018; 15:1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Russo J., Olivas W.M.. Conditional regulation of Puf1p, Puf4p, and Puf5p activity alters YHB1 mRNA stability for a rapid response to toxic nitric oxide stress in yeast. Mol. Biol. Cell. 2015; 26:1015–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Edwards T.A., Pyle S.E., Wharton R.P., Aggarwal A.K.. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001; 105:281–289. [DOI] [PubMed] [Google Scholar]
- 23. Miller M.T., Higgin J.J., Hall T.M.T.. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat. Struct. Mol. Biol. 2008; 15:397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qiu C., McCann K.L., Wine R.N., Baserga S.J., Hall T.M.T.. A divergent Pumilio repeat protein family for pre-rRNA processing and mRNA localization. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:18554–18559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang X., McLachlan J., Zamore P.D., Hall T.M.T.. Modular recognition of RNA by a human Pumilio-homology domain. Cell. 2002; 110:501–512. [DOI] [PubMed] [Google Scholar]
- 26. Wang X., Zamore P.D., Hall T.M.T.. Crystal structure of a Pumilio homology domain. Mol. Cell. 2001; 7:855–865. [DOI] [PubMed] [Google Scholar]
- 27. Wang Y., Opperman L., Wickens M., Hall T.M.T.. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:20186–20191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wilinski D., Qiu C., Lapointe C.P., Nevil M., Campbell Z.T., Hall T.M.T., Wickens M.. RNA regulatory networks diversified through curvature of the PUF protein scaffold. Nat. Commun. 2015; 6:8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang J., McCann K.L., Qiu C., Gonzalez L.E., Baserga S.J., Hall T.M.T.. Nop9 is a PUF-like protein that prevents premature cleavage to correctly process pre-18S rRNA. Nat. Commun. 2016; 7:13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu D., Stumpf C.R., Krahn J.M., Wickens M., Hall T.M.T.. A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:20192–20197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bao H., Wang N., Wang C., Jiang Y., Liu J., Xu L., Wu J., Shi Y.. Structural basis for the specific recognition of 18S rRNA by APUM23. Nucleic Acids Res. 2017; 45:12005–12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weidmann C.A., Qiu C., Arvola R.M., Lou T.F., Killingsworth J., Campbell Z.T., Hall T.M.T., Goldstrohm A.C.. Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio. Elife. 2016; 5:e17096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cheong C.G., Hall T.M.T.. Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:13635–13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hall T.M.T. Expanding the RNA-recognition code of PUF proteins. Nat. Struct. Mol. Biol. 2014; 21:653–655. [DOI] [PubMed] [Google Scholar]
- 35. Campbell Z.T., Valley C.T., Wickens M.. A protein-RNA specificity code enables targeted activation of an endogenous human transcript. Nat. Struct. Mol. Biol. 2014; 21:732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lapointe C.P., Preston M.A., Wilinski D., Saunders H.A.J., Campbell Z.T., Wickens M.. Architecture and dynamics of overlapped RNA regulatory networks. RNA. 2017; 23:1636–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Z., Lee I., Moradi E., Hung N.J., Johnson A.W., Marcotte E.M.. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009; 7:e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thomson E., Rappsilber J., Tollervey D.. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA. 2007; 13:2165–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang B., Ye K.. Nop9 binds the central pseudoknot region of 18S rRNA. Nucleic Acids Res. 2017; 45:3559–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Machin N.A., Lee J.M., Barnes G.. Microtubule stability in budding yeast: characterization and dosage suppression of a benomyl-dependent tubulin mutant. Mol. Biol. Cell. 1995; 6:1241–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fehrenbacher K.L., Boldogh I.R., Pon L.A.. A role for Jsn1p in recruiting the Arp2/3 complex to mitochondria in budding yeast. Mol. Biol. Cell. 2005; 16:5094–5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ulbricht R.J., Olivas W.M.. Puf1p acts in combination with other yeast Puf proteins to control mRNA stability. RNA. 2008; 14:246–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hogan D.J., Riordan D.P., Gerber A.P., Herschlag D., Brown P.O.. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008; 6:e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yosefzon Y., Koh Y.Y., Chritton J.J., Lande A., Leibovich L., Barziv L., Petzold C., Yakhini Z., Mandel-Gutfreund Y., Wickens M. et al.. Divergent RNA binding specificity of yeast Puf2p. RNA. 2011; 17:1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Porter D.F., Koh Y.Y., VanVeller B., Raines R.T., Wickens M.. Target selection by natural and redesigned PUF proteins. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:15868–15873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mossessova E., Lima C.D.. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000; 5:865–876. [DOI] [PubMed] [Google Scholar]
- 47. Otwinowski Z., Minor W.. [20]Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 48. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 50. Hennig J., Militti C., Popowicz G.M., Wang I., Sonntag M., Geerlof A., Gabel F., Gebauer F., Sattler M.. Structural basis for the assembly of the Sxl-Unr translation regulatory complex. Nature. 2014; 515:287–290. [DOI] [PubMed] [Google Scholar]
- 51. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr, Jungkamp A.C., Munschauer M. et al.. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W., Clark T.A., Schweitzer A.C., Blume J.E., Wang X. et al.. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008; 456:464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ule J., Jensen K., Mele A., Darnell R.B.. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005; 37:376–386. [DOI] [PubMed] [Google Scholar]
- 54. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N.. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010; 38:W529–W533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Muto Y., Yokoyama S.. Structural insight into RNA recognition motifs: versatile molecular Lego building blocks for biological systems. Wiley Interdiscip. Rev. RNA. 2012; 3:229–246. [DOI] [PubMed] [Google Scholar]
- 57. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 58. Hogan G.J., Brown P.O., Herschlag D.. Evolutionary conservation and diversification of Puf RNA binding proteins and their mRNA targets. PLoS Biol. 2015; 13:e1002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang Q., Coseno M., Gilmartin G.M., Doublie S.. Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure. 2011; 19:368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pabis M., Popowicz G.M., Stehle R., Fernandez-Ramos D., Asami S., Warner L., Garcia-Maurino S.M., Schlundt A., Martinez-Chantar M.L., Diaz-Moreno I. et al.. HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs. Nucleic Acids Res. 2018; 47:1011–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ripin N., Boudet J., Duszczyk M.M., Hinniger A., Faller M., Krepl M., Gadi A., Schneider R.J., Šponer J., Meisner-Kober N.C. et al.. Molecular basis for AU-rich element recognition and dimerization by the HuR C-terminal RRM. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:2935–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Campbell Z.T., Wickens M.. Probing RNA-protein networks: biochemistry meets genomics. Trends Biochem. Sci. 2015; 40:157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dominguez D., Freese P., Alexis M.S., Su A., Hochman M., Palden T., Bazile C., Lambert N.J., Van Nostrand E.L., Pratt G.A. et al.. Sequence, structure, and context preferences of human RNA binding proteins. Mol. Cell. 2018; 70:854–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gerstberger S., Hafner M., Tuschl T.. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014; 15:829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession numbers 6NY5 (Puf1 PUM-HD and RNA), 6NX5 (Puf1 RRM) and 6NWW (Puf1 RRM).