Abstract
Malvaceae is a family of flowering plants containing many economically important plant species including cotton, cacao and durian. Recently, the genomes of several Malvaceae species have been decoded, and many omics data were generated for individual species. However, no integrative database of multiple species, enabling users to jointly compare and analyse relevant data, is available for Malvaceae. Thus, we developed a user-friendly database named MaGenDB (http://magen.whu.edu.cn) as a functional genomics hub for the plant community. We collected the genomes of 13 Malvaceae species, and comprehensively annotated genes from different perspectives including functional RNA/protein element, gene ontology, KEGG orthology, and gene family. We processed 374 sets of diverse omics data with the ENCODE pipelines and integrated them into a customised genome browser, and designed multiple dynamic charts to present gene/RNA/protein-level knowledge such as dynamic expression profiles and functional elements. We also implemented a smart search system for efficiently mining genes. In addition, we constructed a functional comparison system to help comparative analysis between genes on multiple features in one species or across closely related species. This database and associated tools will allow users to quickly retrieve large-scale functional information for biological discovery.
INTRODUCTION
The Malvaceae is a family of flowering plants containing 243 genera with 4225 known species, including many economically important crop plants (1). These include cotton (Gossypium spp.), a principal natural fibre source for the textile industry; cacao (Theobroma spp.), yielding millions of tonnes of cocoa for making chocolate; durian (Durio spp.), a major Southeast Asian food known as the ‘king of fruits’; and okra (Abelmoschus spp.), which is a nutritious vegetable. These species exhibit wide phenotypic diversity in life forms (from herbaceous to woody), flowers, fruits and seeds. The underlying functional genomic elements need to be deciphered to enhance their traits in productivity and quality.
In recent years, genome sequencing data have rapidly accumulated for Malvaceae species, including Theobroma cacao (2), four species of cotton (3–9), Durio zibethinus (10), and Bombax ceiba (11). Many transcriptomic, epigenomic and proteomic studies have uncovered the gene expression profiles and genomic features in individual species. However, an integrative functional genomic database of multiple species, enabling users to jointly examine and utilize relevant data, is missing for the Malvaceae, probably due to difficulties in collecting, analyzing, storing, and visualizing large-scale data. The previously published CottonGen (12), CottonFGD (13) and ccNET (14) databases are useful, but are cotton-specific and lack functions for comparative genomics.
To provide a functional genomics hub for the Malvaceae and the plant community, we developed a user-friendly database named the MaGenDB (http://magen.whu.edu.cn). We collected the genomes of 13 species which coverage all genera in Malvaceae with whole genome sequence in NCBI genome database, and comprehensively analysed the functional features of genes including various RNA/protein elements, gene ontology (GO), KEGG orthology (KO), enzyme characterization, protein 3D structure and gene families. To date, we processed 374 sets of diverse omics data with standard pipelines and integrated them into MaGenDB, including ChIP-seq, DNase-seq, BS-seq, RNA-seq, small RNA-seq, PacBio long-read Iso-seq, CAGE-seq, polyA-seq and mass spectra data. Based on these comprehensive data analyses, we constructed a customised genome browser to visualize these high-throughput genomic data, and designed multiple dynamic charts to present gene/RNA/protein-level knowledge such as dynamic expression profiles, functional elements, and interacting protein networks. Furthermore, we also implemented a smart search system for efficiently mining genes by functional element, ontology annotation, and gene family. In addition, we constructed a functional comparison system to help comparative analysis between genes on multiple features in one species or across closely related species.
MATERIALS AND METHODS
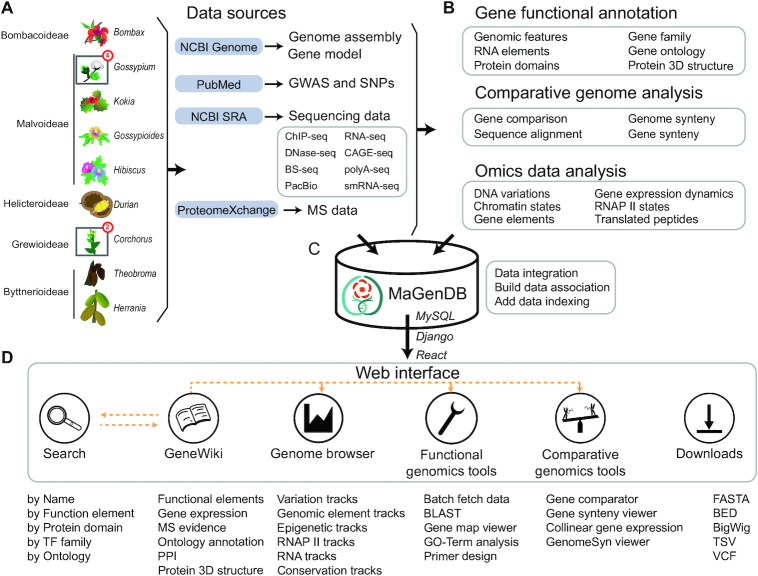
All data sources, data processing, and web interface features are summarized in Figure 1. The MaGenDB consists of four parts: data collection, gene functional annotation and omics data processing, database construction and web interface and toolkit development. The key steps, tools, and generated data are summarized in Supplementary Figure S1 and described below.
Figure 1.
Schematic of MaGenDB. (A) Data sources included 13 species from five subfamilies in Malvaceae and associated gene models, GWAS, eight types of sequencing data, and MS data. (B) Summary of data processing and obtained biological knowledge. (C) All data are stored in a MySQL relationship database with additional indexes. Django and React frameworks are used for interactive queries between frontend and backend. (D) Overview of the web interface and usage of MaGenDB. Main functions or data are listed under the first-level menu. The dashed lines indicate the linkages between different pages.
Data sources
All species and genome assemblies used in this study were collected from public databases (Supplementary Table S1). The genome-wide association study (GWAS) and single nucleotide polymorphism (SNP) data for genomes were manually extracted from the NCBI PubMed database. Diverse types of omics data—including RNA-seq, small RNA-seq (smRNA-seq), CAGE-seq, polyA-seq, ChIP-seq, DNase-seq, Bisulfite-seq (BS-seq) and PacBio long-read sequencing—were downloaded from the NCBI SRA database (www.ncbi.nlm.nih.gov/sra). Mass spectrometry data were downloaded from the ProteomeXchange (15) database (Figure 1A). All metadata of these omics data are summarised in Supplementary Table S2.
Gene functional annotation
All functional elements in Malvaceae genomes were thoroughly annotated with a unified and standard procedure (Figure 1B). For genomes without gene annotations, gene models were reconstructed using StringTie (16) with default parameters from RNA-seq data, except the IGIA gene annotation for Gossypium arboretum was recently assembled from integrative multi-strategic RNA-seq data (17). The tRNA and various kinds of ncRNA genes were predicted using tRNAscan-SE 2.0 (18) and rfam_scan with Rfam models (19), respectively. The tandem repeats were identified using TRF program (20). The protein-coding potential scores were computed using CPAT (21). Three local BLAST databases were built, including the NCBI Nucleotide (NT), non-redundant protein, and the Arabidopsis Information Resource (TAIR) databases (22), to which the DNA and protein sequences of all gene models were compared using blastn and blastp programs (23) with E-value cutoff 1e–6. The GO terms and EC numbers for genes were assigned using Blast2GO (24). The KO identifiers were annotated using the KAAS web server (25), and homologous genes were identified using the eggNOG web server (26). Potential transcription-factor (TF) families of protein-coding genes were mapped to the PlantTFDB database (27).
Various functional protein domains and RNA elements were also identified. InterPro, Pfam, and conserved protein domains were predicted using InterProScan (28) and InterPro database (29), PfamScan (30) and Pfam database (31), and NCBI conserved domains (CDD) database (32), respectively. Signal peptides, transmembrane helices and disorder regions were predicted using signalP (33), TMHMM (34) and IUPred2A (35), respectively. The RNA G-quadruplexes (RG4) were predicted using QGRS (36) and miRNA target sites were predicted by psRNATarget web server (37). Protein 3D structures were modelled using the SWISS-MODEL web server (38).
Comparative genome analysis
The plant genomes for comparative analysis with Malvaceae were downloaded from PLAZA (39). Gene synteny clusters between any two genomes in MaGenDB were predicted using MCScanX (40). Multiple sequence alignments between collinear genes were built using Clustal Omega (41).
Protein–protein interactions (PPI) were extracted from the STRING database (42) for two Malvaceae genomes (Gossypium raimondii and Theobroma cacao) and Arabidopsis thaliana. A similar but stricter strategy to that used in STRING, transferring PPI score by gene collinearity, was used to predict the PPI network for other genomes in MaGenDB which were highly collinear. For two proteins A and B, the ‘combined score’ from one genome was calculated as the average interaction scores between collinear gene pairs of A and B. The ‘combined score’ from multiple genomes were averaged as the predicted interaction strength.
Omics data analysis
High-throughput sequencing data passing the fastQC quality control (www.bioinformatics.babraham.ac.uk/projects/fastqc/) were used in this study. The raw reads were filtered to remove the adaptors and low-quality bases using cutadapt (43). For the omics data of RNA-seq, smRNA-seq, ChIP-seq, DNase-seq and BS-seq, the data processing followed the recommendations of the ENCODE pipelines (44). Briefly, RNA-seq and smRNA-seq reads were mapped to the genome using STAR (45). The ChIP-seq and DNase-seq reads were mapped by BWA (46), and the BS-seq reads were processed by Bismark (47). The gene expression quantification in FPKM (Fragments Per Kb of exon per Million mapped fragments) was computed by StringTie (16). The peaks for ChIP-seq and DNase-seq were identified by MACS2 (48) and Hotspot2 (newer version of Hotspot) (49), respectively.
For CAGE-seq and polyA-seq, the 5′ random barcodes were extracted and trimmed before mapping. The potential rRNAs and PCR duplicate reads were removed, and the cleaned reads were mapped to the genome using STAR in End-to-End mode.
For PacBio data, the long subreads were corrected with RNA-seq data using proovread (50) to reduce the sequencing errors. The corrected reads were aligned to the genome using STARlong with the parameters recommended by the PacBio official documentation.
The raw mass spectra data were converted to mgf format using ProteoWizard (51) before peptide searching. The programs SearchGUI (52) and peptideShaker (53) were used to match the MS/MS fragmentation spectra to peptide sequences with default parameters. The genomic positions of matched peptides were recovered using custom Python scripts.
Data integration
We integrated functional annotations at gene, transcript, and protein levels for different categories separately in the MySQL database. Tables for unique genes, transcripts, and proteins were created by genomic positions. For one specific type annotation, the functional information was mapped to the unique records to remove redundancy. For gene-level element annotations, we merged genomic blocks to build ‘dense’ format track in the GeneWiki page, while for functional annotations (e.g. GO-term), the information from the longest transcript was used.
Database construction
All pre-processed data were integrated into the MaGenDB MySQL database, in which different tables were associated together by the functional element, ontology, and genomic position (Figure 1C). Django framework was used to provide query and computation supports from the database backend.
Web interface and toolkit development
A user-friendly web interface was developed by React framework (Figure 1D), and multiple customised dynamic charts were implemented using BizCharts library. All genomic features and omics data were visualised using JBrowse (54) and its plugins. The BLAST server was driven by Sequenceserver (55). The PCR primer design was implemented by Primer3 (56) with customization to accommodate support for alternative splicing events. The GO enrichment analysis was provided using R package topGO. Protein 3D structure was presented using NGL viewer (57). GenomeSyn viewer was developed to visualize gene synteny clusters between two genomes. All programs and packages used in this study are listed in Supplementary Table S3.
RESULTS
Overview of MaGenDB
MaGenDB (http://magen.whu.edu.cn) is a family-level functional genomics hub for Malvaceae plants, which contains 367 available deep-sequencing data of eight types for 13 species (Figure 1A). For each genome, the database provides multiple functional annotations, DNA variations, chromatin and RNAP II states, and RNA landscape including gene expression across different tissues (Figure 1B). The GO/KO/EC annotations, gene family, and TF family for 759 700 transcripts are stored in MaGenDB. About 370 million BLAST annotations to the NT, NCBI non-redundant protein database, and TAIR are saved in MaGenDB. More than 24 million functional elements are annotated, including InterPro domains, Pfam domains, CDD, signal peptides, transmembrane helices, protein disorder regions and RG4. The database contains 170 660 615 PPIs and 98 831 protein 3D structures from prediction (Table 1). The coding-potential scores for all transcripts and the proteins with mass spectrometry evidence were recorded. For comparative genome analysis, 51 other plant genomes were collected (Supplementary Table S4) and a total of 28 976 317 gene syntenies were annotated (Table 1).
Table 1.
Summary of MaGenDB data
| Data | Statistics |
|---|---|
| Subfamily/Genus/Species | 5/9/13 |
| Transcripts/Proteins | 759 700/728 366 |
| Types of omics data | 9 |
| Omics data samples | 374 |
| GO/KO/EC annotation | 494 364/254 551/85 157 |
| Gene/TF families | 656 516/280 976 |
| BLAST annotation (NT/NR/TAIR) | 63 313 713/307 542 675/635 804 |
| Types of functional elements | 7 |
| Functional elements | 24 855 369 |
| Synteny gene pairs | 28 976 317 |
| Protein-protein interactions | 170 660 615 |
| Protein 3D structures | 98 831 |
The MaGenDB is aimed to be a user-friendly comparative functional genomics database, in which multiple dynamic charts and hyperlinks are generated. The web pages were well-designed, and several useful tools were developed in MaGenDB for researchers (Figure 1D). A detailed user guide was provided to help use MaGenDB efficiently. The database was designed to also be compatible with tablet and phone devices. All data stored in the MaGenDB are freely accessible and downloadable for academic purposes.
Smart search system
For users to quickly find the data of interest, a smart search system was designed for genes, transcripts, and proteins. The supported queries include the gene symbol; the identifiers of gene, transcript, and protein; the name and identifier of KO, EC, KEGG pathway, KEGG model, KEGG disease, InterPro domain and Pfam domain; the identifier of GO and CDD domain; the name of TF family; and the name of eggNOG gene family (Table 2).
Table 2.
Query fields and examples in smart search system
| Query field | Query content | Query example |
|---|---|---|
| Name | Locus/Transcript/Protein name | GhRDL1 |
| Name | Locus/Transcript/Protein ID | Ga14g01129.F1 |
| Name | TAIR homolog gene symbol | ACX3 |
| GO | GO accession ID | GO:0000032 |
| KO | KO accession ID | K11091 |
| KO | KEGG ontology name | ATPF1B, atpD |
| EC | Enzyme Commission ID | 1.17.7.2 |
| EC | Enzyme Commission name | Very-long-chain 3-oxoacyl-CoA synthase |
| KEGG pathway | KEGG pathway ID | ko04024 |
| KEGG pathway | KEGG pathway name | cAMP signaling pathway |
| KEGG model | KEGG model ID | M00087 |
| KEGG model | KEGG model name | Fe-S protein |
| KEGG disease | KEGG disease ID | H00254 |
| KEGG disease | KEGG disease name | Lysosomal acid lipase deficiency |
| InterPro domain | InterPro ID | IPR001736 |
| InterPro domain | InterPro domain name | cd01254 |
| Pfam domain | Pfam ID | PF00614.22 |
| Pfam domain | Pfam domain name | PLDc |
| CDD domain | NCBI CDD ID | 197200 |
| TF family | TF family name | C2H2 |
| eggNOG gene family | eggNOG root name | EOY06593 |
| eggNOG gene family | eggNOG hit gene | COG1502 |
For example, the gene name ‘Pgam5’, TAIR gene symbol, or gene ID ‘Ga01g00214.F3’ can be used to query the same gene. Fuzzy matching is supported, for example, ‘cAMP’ instead of ‘cAMP signalling pathway’. Additionally, the search box can trigger the appearance of a list of suitable queries that is filtered as the user types.
GeneWiki
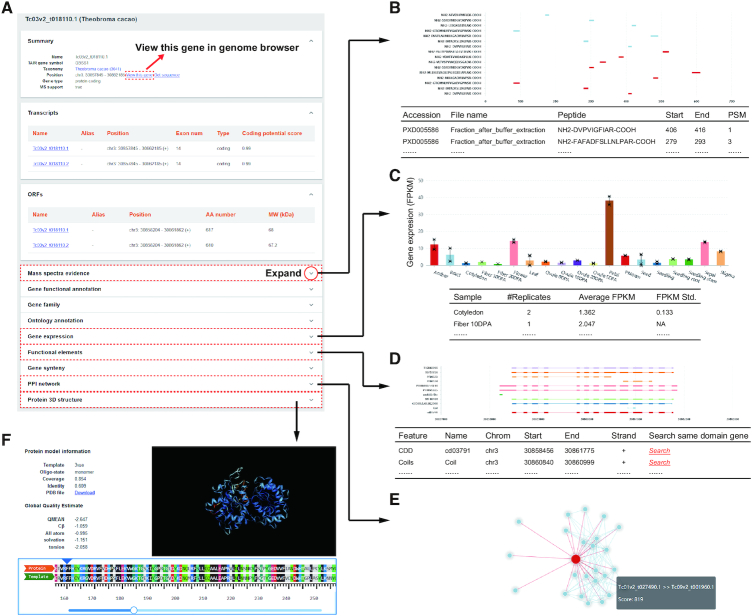
GeneWiki is the page where MaGenDB displays all functional annotations for gene, transcript, and protein (Figure 2), such as gene structure, ontology annotation, gene expression, mass spectra evidence, functional domains, gene synteny and predicted protein 3D structure. Users land in this page from the ‘Search’ page or other hyperlinks. Detailed data can be viewed as organised sections by clicking the drop-down buttons (Figure 2A).
Figure 2.
Functional annotations in GeneWiki. (A) Different types of functional annotations in GeneWiki page. (B) Custom figure and table showing the positions and details of MS evidence for a protein. (C) Bar plot and table showing the gene expression level in FPKM across different tissues. (D) Custom dynamic charts and tables of functional elements annotated for a protein. (E) An interactive view of the PPI network for the query protein. (F) Dynamic visualization of the predicted 3D structure and the model details for a protein.
For a gene locus, the information of gene symbol, taxonomy, genomic position and gene type are shown in the ‘Summary’ section. Users can view the taxonomy, jump to the genome browser, or download DNA sequences via hyperlinks. All unique transcripts and proteins are listed in the ‘Transcript information’ and ‘ORF information’ sections, respectively (Figure 2A).
The ‘Functional annotation’ section contains the NT annotation and Arabidopsis thaliana homolog gene predicted by BLAST. The annotation of eggNOG gene family and TF genes from PlantTFDB are arranged in the sections ‘Gene family’ and ‘TF family’. The GO, KEGG ontology annotation, pathway, model and disease annotation, and enzyme commission annotation are listed in the ‘Ontology annotation’ section (Figure 2A).
The mass spectra evidence from the different sources is shown using dynamic charts and tables (Figure 2B). The detailed information includes data source, matched peptide, and peptide-spectrum match (PSM). The elements can be viewed at multiple linear scales including gene locus, mRNA transcript, and amino acid sequence of protein.
The ‘Gene expression’ section shows the expression profiles in different tissues, with error bars indicating the standard deviation among different datasets (Figure 2C). The expression values in FPKM of each data set are shown in the bottom table.
The ‘Functional element’ section includes InterPro domain, NCBI CDD domain, Pfam domain, signal peptide, transmembrane helices, disorder region, and RG4, displayed by dynamic charts in different colours (Figure 2D). The protein network interacting with the query gene is shown in an interactive chart (Figure 2E). The combined scores of interactions are shown and can be used to filter the network. The information for a specific gene appears when clicking the corresponding node. The 3D structure model of protein predicted using SWISS-MODEL is displayed as an interactive figure in the ‘Protein 3D structure’ section (Figure 2F). The sequence alignment between the protein and its template can be dynamically explored.
Genome browser and multi-omics data integration
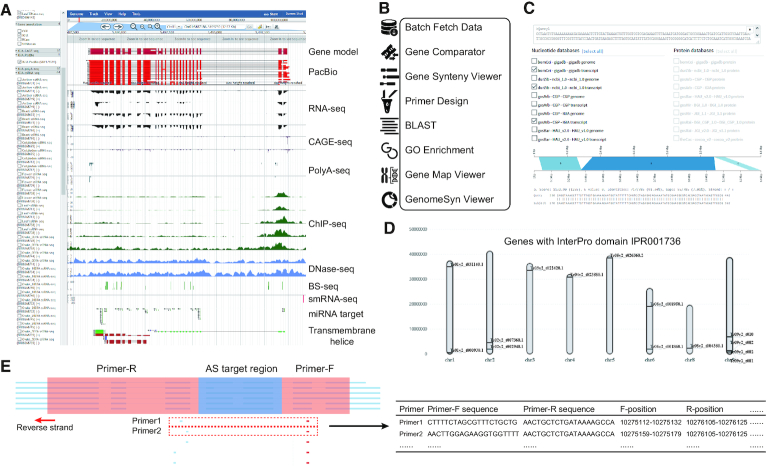
All gene models, processed omics data, annotated functional elements in MaGenDB are visualised using customised JBrowse with suitable default settings (Figure 3A). A set of plugins were configured to enhance the functionality and usability of the browser including generating high-resolution figures (58). The user can conveniently explore the gene of interest by entering the genomic location or following hyperlinks in other pages like GeneWiki. Similar tracks are organised into track groups. The tracks can be shown or hidden by ticking on or off. All functional data are unified in the same coordinate system, and hypotheses are often proposed from jointly exploring multiple tracks simultaneously.
Figure 3.
Genome browser and functional genomics tools in MaGenDB. (A) Genome browser view of processed omics data for a gene example. For any genome, the available data are organised in track groups and can be dynamically selected/unselected. (B) Overview of genomics tools provided in MaGenDB. (C) The interface of BLAST service for the different databases. (D) Example of genome map viewer of multiple genes. (E) Primer design page considering alternative splicing (AS) events.
Functional genomics tools in MaGenDB
There are many useful genomics tools in MaGenDB that can aid researchers to explore and analyse the data (Figure 3B). Users can get annotation data in batches, perform different BLAST operations (Figure 3C), dynamically visualize multiple genes (in the same pathway or complex, or with same functional domain) in chromosomes simultaneously (Figure 3D), and design primers with options for alternative splicing (AS) events (Figure 3E). A topGO-based GO enrichment service is also provided online.
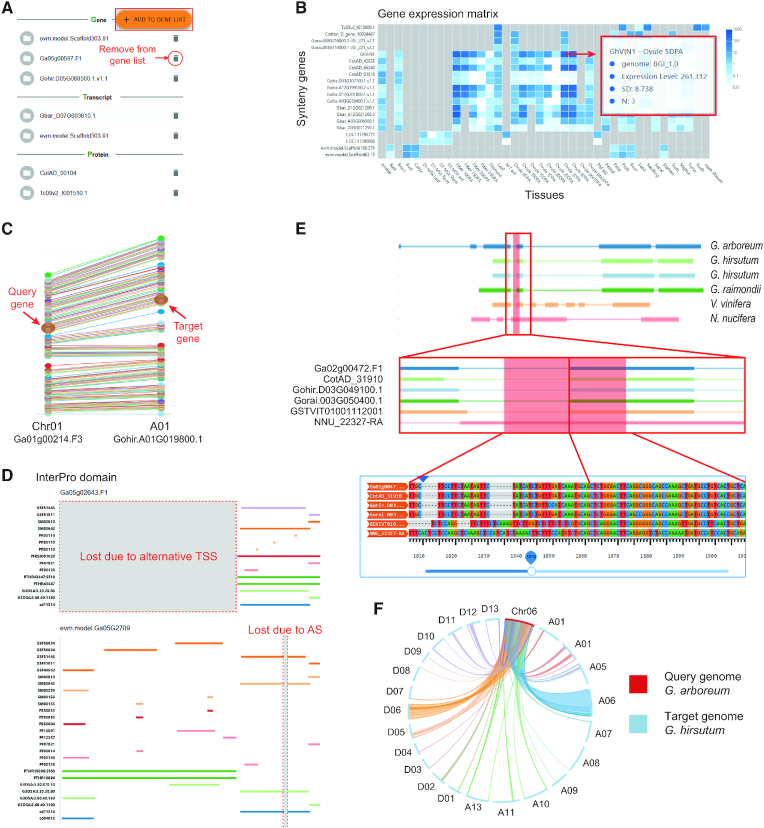
Comparative genomics tools in MaGenDB
The MaGenDB includes several ingenious comparative genomics analysis tools. The genes of interest are managed by adding or removing gene from the ‘gene list’, a comparative analysis framework (Figure 4A). Because the functions of collinear homologous genes tend to be similar (59), their expression patterns in different plant tissues often have similar characteristics (60). The MaGenDB can automatically generate gene expression heatmaps for all genes in the synteny block of the query gene, as shown in a fiber and ovule early stage specific gene example, of which the collinear genes have same expression pattern (Figure 4B). Furthermore, the chromosomal locations of collinear gene clusters can be visualized interactively (Figure 4C).
Figure 4.
Comparative genomics tools in MaGenDB. (A) Management of gene list of interest for comparative analysis. (B) Gene expression heatmap across different tissues for collinear genes to a query gene from the GeneWiki page. (C) Genomic position mapping of collinear genes. (D) Comparison of functional domains between two proteins. Grey box marks the missing region in one vs the other. (E) An interactive view of multiple alignments of collinear genes from the gene structure perspective. (F) A circular view of the gene synteny clusters between G. arboreum Chr06 with G. hirsutum chromosomes.
To intuitively evaluate the effects on proteins of alternative RNA processing, we developed the gene comparator tool to compare biological annotations and functional structures between two gene loci, transcripts, or proteins. An example is shown in Figure 4D, in which the protein Ga05g02643.F1 lost almost two-thirds of the amino acid sequence compared to the protein evm.model.Ga05G2709 due to alternative transcription start sites, and so the Ga05g02643.F1 protein is missing a significant amount of InterPro domains. The Gene Synteny viewer was developed to compare gene structures between collinear genes. The multi-alignments are dynamically presented, which facilitates discovery of conserved elements, as shown in Figure 4E. The gene synteny clusters between two genomes can be dynamically presented with GenomeSyn viewer as circular plot. As shown in Figure 4F, the Chr06 from diploid G. arboretum and the two homologous chromosomes from tetraploid G. hirsutum have the largest synteny blocks, indicating good quality of our results.
DISCUSSION
The MaGenDB fills the gap for an important plant family, integrates large-scale diverse omics data, implements comprehensive data visualization methods and constructs a new functional comparison system. A total of 374 processed omics data of nine techniques, 18 types of annotation and >24 million functional elements are stored in MaGenDB and presented in a user-friendly way using well-designed custom dynamic charts. The large set of curated data sets and several powerful tools provide comparative functional genomics resources and services. Thus, MaGenDB will be useful for plant and evolution scientists in both experimental and computational directions. The differences between MaGenDB and other cotton-specific databases are also summarized in Supplementary Table S5. In the future, we will continuously update MaGenDB as new Malvaceae genomes and omics data become available, and will add more annotation and functionalities to the database including whole genome alignments in the context of comparative genome analysis, and other types of omics data such as Hi-C, ChIA-PET and CLIP-seq data.
DATA AVAILABILITY
The MaGenDB database can be accessed through the web server at http://magen.whu.edu.cn. The codes for the omics data analysis are available in the GitHub repository at https://github.com/zhouyulab/magendb.
Supplementary Material
ACKNOWLEDGEMENTS
Part of the computation in this work was done on the supercomputing system in the Supercomputing Center of Wuhan University.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Innovation Team Program from Wuhan University [2042017kf0233 to Y.Z., Y.X.Z., K.W.]; National Natural Science Foundation of China [91640115 and 31670827 to Y.Z., 31690090 to Y.X.Z., 31770310 to K.W.]; National Key R&D Program of China [2017YFA0504400 to Y.Z.]. Funding for open access charge: Innovation Team Program from Wuhan University [2042017kf0233].
Conflict of interest statement. None declared.
REFERENCES
- 1. Group The Angiosperm Phylogeny. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016; 181:1–20. [Google Scholar]
- 2. Argout X., Salse J., Aury J.-M., Guiltinan M.J., Droc G., Gouzy J., Allegre M., Chaparro C., Legavre T., Maximova S.N. et al.. The genome of Theobroma cacao. Nat. Genet. 2011; 43:101–108. [DOI] [PubMed] [Google Scholar]
- 3. Paterson A.H., Wendel J.F., Gundlach H., Guo H., Jenkins J., Jin D., Llewellyn D., Showmaker K.C., Shu S., Udall J. et al.. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012; 492:423–427. [DOI] [PubMed] [Google Scholar]
- 4. Wang K., Wang Z., Li F., Ye W., Wang J., Song G., Yue Z., Cong L., Shang H., Zhu S. et al.. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012; 44:1098–1103. [DOI] [PubMed] [Google Scholar]
- 5. Li F., Fan G., Wang K., Sun F., Yuan Y., Song G., Li Q., Ma Z., Lu C., Zou C. et al.. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014; 46:567–572. [DOI] [PubMed] [Google Scholar]
- 6. Li F., Fan G., Lu C., Xiao G., Zou C., Kohel R.J., Ma Z., Shang H., Ma X., Wu J. et al.. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015; 33:524–530. [DOI] [PubMed] [Google Scholar]
- 7. Du X., Huang G., He S., Yang Z., Sun G., Ma X., Li N., Zhang X., Sun J., Liu M. et al.. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 2018; 50:796–802. [DOI] [PubMed] [Google Scholar]
- 8. Wang M., Tu L., Yuan D., Zhu D., Shen C., Li J., Liu F., Pei L., Wang P., Zhao G. et al.. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019; 51:224–229. [DOI] [PubMed] [Google Scholar]
- 9. Hu Y., Chen J., Fang L., Zhang Z., Ma W., Niu Y., Ju L., Deng J., Zhao T., Lian J. et al.. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019; 51:739–748. [DOI] [PubMed] [Google Scholar]
- 10. Teh B.T., Lim K., Yong C.H., Ng C.C.Y., Rao S.R., Rajasegaran V., Lim W.K., Ong C.K., Chan K., Cheng V.K.Y. et al.. The draft genome of tropical fruit durian (Durio zibethinus). Nat. Genet. 2017; 49:1633–1641. [DOI] [PubMed] [Google Scholar]
- 11. Gao Y., Wang H., Liu C., Chu H., Dai D., Song S., Yu L., Han L., Fu Y., Tian B. et al.. De novo genome assembly of the red silk cotton tree (Bombax ceiba). GigaScience. 2018; 7:giy051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu J., Jung S., Cheng C.-H., Ficklin S.P., Lee T., Zheng P., Jones D., Percy R.G., Main D.. CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2014; 42:D1229–D1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu T., Liang C., Meng Z., Sun G., Meng Z., Guo S., Zhang R.. CottonFGD: an integrated functional genomics database for cotton. BMC Plant Biol. 2017; 17:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. You Q., Xu W., Zhang K., Zhang L., Yi X., Yao D., Wang C., Zhang X., Zhao X., Provart N.J. et al.. ccNET: Database of co-expression networks with functional modules for diploid and polyploid Gossypium. Nucleic Acids Res. 2017; 45:D1090–D1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deutsch E.W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D.S., Bernal-Llinares M., Okuda S., Kawano S. et al.. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017; 45:D1100–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L.. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016; 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang K., Wang D., Zheng X., Qin A., Zhou J., Guo B., Chen Y., Wen X., Ye W., Zhou Y. et al.. Multi-strategic RNA-seq analysis reveals a high-resolution transcriptional landscape in cotton. Nat. Commun. 2019; 10:4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lowe T.M., Chan P.P.. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016; 44:W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kalvari I., Argasinska J., Quinones-Olvera N., Nawrocki E.P., Rivas E., Eddy S.R., Bateman A., Finn R.D., Petrov A.I.. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018; 46:D335–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999; 27:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang L., Park H.J., Dasari S., Wang S., Kocher J.-P., Li W.. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013; 41:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berardini T.Z., Reiser L., Li D., Mezheritsky Y., Muller R., Strait E., Huala E.. The arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome: Tair: making and Mining the “Gold Standard” Plant Genome. Genesis. 2015; 53:474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Conesa A., Götz S.. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics. 2008; 2008:619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M.. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007; 35:W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S.K., Cook H., Mende D.R., Letunic I., Rattei T., Jensen L.J. et al.. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019; 47:D309–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jin J., Tian F., Yang D.-C., Meng Y.-Q., Kong L., Luo J., Gao G.. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017; 45:D1040–D1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G. et al.. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014; 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mitchell A.L., Attwood T.K., Babbitt P.C., Blum M., Bork P., Bridge A., Brown S.D., Chang H.-Y., El-Gebali S., Fraser M.I. et al.. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019; 47:D351–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mistry J., Bateman A., Finn R.D.. Predicting active site residue annotations in the Pfam database. BMC Bioinformatics. 2007; 8:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A. et al.. The Pfam protein families database in 2019. Nucleic Acids Res. 2019; 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R. et al.. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017; 45:D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nielsen H. Predicting Secretory Proteins with SignalP. Methods Mol. Biol. 2017; 1611:59–73. [DOI] [PubMed] [Google Scholar]
- 34. Sonnhammer E.L., von Heijne G., Krogh A.. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998; 6:175–182. [PubMed] [Google Scholar]
- 35. Mészáros B., Erdős G., Dosztányi Z.. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018; 46:W329–W337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kikin O., D’Antonio L., Bagga P.S.. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006; 34:W676–W682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dai X., Zhao P.X.. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011; 39:W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. et al.. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018; 46:W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Bel M., Diels T., Vancaester E., Kreft L., Botzki A., Van de Peer Y., Coppens F., Vandepoele K.. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 2018; 46:D1190–D1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T. -h., Jin H., Marler B., Guo H. et al.. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012; 40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D. et al.. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019; 47:W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. et al.. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45:D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10. [Google Scholar]
- 44. Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K. et al.. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018; 46:D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Krueger F., Andrews S.R.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nussbaum C., Myers R.M., Brown M., Li W. et al.. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. John S., Sabo P.J., Thurman R.E., Sung M.-H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A.. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011; 43:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hackl T., Hedrich R., Schultz J., Förster F.. proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics. 2014; 30:3004–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kessner D., Chambers M., Burke R., Agus D., Mallick P.. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008; 24:2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vaudel M., Barsnes H., Berven F.S., Sickmann A., Martens L.. SearchGUI: an open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics. 2011; 11:996–999. [DOI] [PubMed] [Google Scholar]
- 53. Vaudel M., Burkhart J.M., Zahedi R.P., Oveland E., Berven F.S., Sickmann A., Martens L., Barsnes H.. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 2015; 33:22–24. [DOI] [PubMed] [Google Scholar]
- 54. Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H.. JBrowse: a next-generation genome browser. Genome Res. 2009; 19:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Priyam A., Woodcroft B.J., Rai V., Moghul I., Munagala A., Ter F., Chowdhary H., Pieniak I., Maynard L.J., Gibbins M.A. et al.. Sequenceserver: a modern graphical user interface for custom BLAST databases. Mol. Biol. Evol. 2019; msz185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G.. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012; 40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rose A.S., Bradley A.R., Valasatava Y., Duarte J.M., Prlić A., Rose P.W.. NGL viewer: web-based molecular graphics for large complexes. Bioinformatics. 2018; 34:3755–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., Goodstein D.M., Elsik C.G., Lewis S.E., Stein L. et al.. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016; 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Koonin E.V. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005; 39:309–338. [DOI] [PubMed] [Google Scholar]
- 60. Patel R.V., Nahal H.K., Breit R., Provart N.J.. BAR expressolog identification: expression profile similarity ranking of homologous genes in plant species: Expression profile similarity ranking of homologous genes. Plant J. 2012; 71:1038–1050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MaGenDB database can be accessed through the web server at http://magen.whu.edu.cn. The codes for the omics data analysis are available in the GitHub repository at https://github.com/zhouyulab/magendb.