Abstract
The Fragile X Mental Retardation Protein (FMRP) is an RNA binding protein that regulates translation and is required for normal cognition. FMRP upregulates and downregulates the activity of microRNA (miRNA)-mediated silencing in the 3′ UTR of a subset of mRNAs through its interaction with RNA helicase Moloney leukemia virus 10 (MOV10). This bi-functional role is modulated through RNA secondary structures known as G-Quadruplexes. We elucidated the mechanism of FMRP’s role in suppressing Argonaute (AGO) family members’ association with mRNAs by mapping the interacting domains of FMRP, MOV10 and AGO and then showed that the RGG box of FMRP protects a subset of co-bound mRNAs from AGO association. The N-terminus of MOV10 is required for this protection: its over-expression leads to increased levels of the endogenous proteins encoded by this co-bound subset of mRNAs. The N-terminus of MOV10 also leads to increased RGG box-dependent binding to the SC1 RNA G-Quadruplex and is required for outgrowth of neurites. Lastly, we showed that FMRP has a global role in miRNA-mediated translational regulation by recruiting AGO2 to a large subset of RNAs in mouse brain.
INTRODUCTION
The Fragile X Mental Retardation Protein (FMRP) is an RNA binding protein that binds ∼4% of mRNAs in the brain (1,2). Loss of FMRP expression causes Fragile X Syndrome (FXS), the most common inherited form of intellectual disability (3,4). Loss of FMRP contributes to an altered proteome (5), but the critical open question in the field is how does FMRP binding affect translation of its bound mRNAs?
FMRP was first implicated in miRNA-mediated regulation in two independent studies using the Drosophila ortholog dFmr1 (6,7). These results were extended to mammalian cells when FMRP was shown to associate with endogenous miRNAs, DICER activity and AGO1 (7). miRNA-mediated regulation by FMRP was explored in brain when FMRP was shown to co-immunoprecipitate with a number of miRNAs important in neuronal function (8). CLIP-seq analysis of brain FMRP showed that FMRP bound primarily in the coding sequence of its mRNA targets (9). However, a subsequent study in HEK293 cells showed that the FMRP CLIP sites were comparably distributed between coding sequence and 3′UTR (10). Recently, eCLIP identification of FMRP targets in human postmortem frontal cortex showed FMRP binding primarily in the 3′UTR (11).
In this work, we map the interaction domains in the FMRP RiboNucleoProtein complex formed by FMRP and associated mRNAs (mRNP). FMRP contains two putative RNA binding domains, the K-homology domains KH1 and KH2 (12,13), and an arginine-glycine-glycine (RGG) box that binds G-Quadruplex RNA structures (hereafter referred to as rG4s) (14–18). FMRP’s KH0 domain is thought to be a protein-binding domain (19–21). We hypothesized that FMRP associates with other proteins that participate in translation of its bound mRNAs and identified the RNA helicase MOV10 as functionally associating with FMRP (22). We found that FMRP exhibits a bifunctional role in regulating subsets of mRNAs modulated through its interaction with MOV10 (23), meaning that it both blocks and facilitates translation. MOV10’s recruitment by FMRP facilitates miRNA-mediated translational suppression, likely by resolving RNA secondary structure and exposing miRNA recognition elements (MREs) within the 3′ UTR. However, FMRP also blocks association of AGO family members (AGO) in a separate subset of mRNAs, resulting in the inhibition of translational suppression. How FMRP dynamically functions to translationally regulate its bound mRNAs is poorly understood. Here we determine the mechanism in which FMRP association with mRNAs is modulated by interacting with MOV10 at rG4s. We identify the interacting domains in the FMRP/MOV10/AGO complex and show how their association modulates translation regulation. By comparing AGO2 eCLIP data from Fmr1 KO (knock out) mouse brain to C57BL6/J wild-type (WT) mouse brain, we show that AGO2’s association with a large subset of neuronal mRNAs is greatly reduced in the absence of FMRP, suggesting that FMRP recruits AGO2 to specific MREs and has a global role in the miRNA pathway.
MATERIALS AND METHODS
Plasmids
WT FMRP, ΔRGG and I304 mutants were generous gifts from Dr Jennifer Darnell (The Rockefeller University). The FMRP KH1 and KH2 mutants were generous gifts from Dr Edouard Khandjian (Universite Laval) (24). The N-terminus and C-terminus of MOV10 were generous gifts from Dr Unutmaz (25). N-terminal FMRP (aa 1–404) and C-terminal FMRP (aa 216–632) were cloned into the pEGFP-C1 vector (BD Biosciences, Catalog #6084–1) using the EcoRI and NotI recognition sites. The N-terminus of MOV10 and the C-terminus of MOV10 were cloned into the pmCherry-C1 vector (TakaRa, Catalog #632524) using the EcoRI and XhoI recognition sites. The iSpinach sequence was provided by Dr Michael Ryckelynck, University of Strasbourg (26).
Animals
Experiments were performed on newly born (P0) C57BL6/J WT and Fmr1 KO mice from both sexes. Animals were kept on a 12/12 h light/dark cycle with food and water ad libitum. All experiments were performed during the light phase (7 AM–7 PM). Animals were treated in accordance with Institutional Animal Care and Use Committee guidelines. All work with animals was done in compliance with the Institutional Animal Care and Use Committee, IACUC protocols 17 164 and 17 000.
Western blot
Samples from at least three biological replicates were prepared for immunoblotting after quantification by Bradford assay and suspension in 1× Laemmli sample buffer, resolved by sodium dodecyl sulphate-polyacrylamide gelelectrophoresis and analyzed by western/immunoblotting. Briefly, membranes were blocked with 5% non-fat dry milk in phosphate-buffered saline (PBS) containing 1% TWEEN-20 for 1 h at room temperature. Primary antibody was applied for 1 h at room temperature or overnight at 4°C followed by a brief wash in 1% non-fat milk PBS containing 1% TWEEN-20 wash buffer. Horseradish peroxidase (HRP)-conjugated secondary antibody was applied at 1:5000 dilution for 1 h at room temperature and washed 4 × 15 min using wash buffer. The HRP signal was detected using an enhanced chemiluminescent (ECL) substrate and exposed to film. The following antibodies were used: anti-MOV10 (A301–571A; Bethyl Laboratories) at 1:1000, anti-mCherry (ab125096; Abcam) at 1:1000, anti-USP22 (ab4812; Abcam) at 1:1000, anti-HN1L (ab200587; Abcam) at 1:1000, anti-RPL5 (ab157099; Abcam) at 1:1000, anti-NCS1 (ab157099; Abcam) at 1:500, anti-WHSC1 (ab75359; Abcam) at 1:1000, anti-LETM1 (ab; Abcam) at 1:1000, anti-PHACTR2 (ab85262; Abcam) at 1:1000, anti-GFP (A-11122, ThermoFisher Scientific) at 1:200, anti-Fus/TLS (sc-47711, Santa Cruz Biotechnology) at 1:250, anti-eIF5 (sc-282, Santa Cruz Biotechnology) at 1:5000, anti-MAZ (sc-28745, Santa Cruz Biotechnology) at 1:1000, anti-HA (HA.11, 901501, Biolegend) at 1:1000, anti-Neurod2 (LS-C352562, LifeSpan Biosciences Inc.) at 1:1000, anti-SOX4 (LS-C499849m, LifeSpan Biosciences Inc.) at 1:1000, anti-eIF2B2 (LS-C409107, LifeSpan Biosciences Inc) at 1:1000, anti-CELF1 (LS-C408897, LifeSpan Biosciences Inc.) at 1:1000. HRP-conjugated anti rabbit and anti-mouse antibodies from Amersham and Jackson laboratories, respectively. A total of 30 μg of overall protein in lysate was used as the input sample. The level of significance and tests performed are described in the figure legends for each experiment.
Quantification: Densitometric quantification of western blot bands was performed using imageJ following NIH protocol: https://imagej.nih.gov/ij/docs/menus/analyze.html#gels
Bands were normalized to an eIF5B loading control.
Immunoprecipitations (IP’s) for cobound mRNAs or associated proteins
Bead preparation (AGO): 25 μl of Protein A Sepharose (PAS) beads (GE Healthcare Biosciences CL-48) were washed with 0.1 M sodium phosphate (pH 8.0) and 0.12 mg of rabbit anti-mouse IgG (Jackson Immunoresearch) was added to beads, rinsed, followed by addition of 10 μg of the pan-AGO 2A8 antibody, which recognizes AGO family members (#MABE56, Millipore Sigma Corp).
HEK293 cells (∼1.5 × 107) were transfected with 100 μg of the FMRP-eGFP, FMRP-eGFP ΔRGG or the N-terminal MOV10 constructs. Cells (∼1.5 × 107) were lysed in 0.5 ml lysis buffer (20 mM Tris–HCl pH 7.5, 200 mM sodium chloride, 2.5 mM magnesium chloride, 0.5% Triton X-100) with protease Inhibitor (1 tablet per 10 ml lysis buffer, Complete Mini, ethylenediaminetetraacetic acid (EDTA) free, 35440400, Roche) and RNase Inhibitor (80 U/ml, RNasin, N2511, Promega), and immunoprecipitated with the prepared beads at 4°C for 12 h. For all RNA IPs, an IgG control was run, which is the bridging rabbit anti-mouse antibody. After IP, the beads were washed in 0.1 M sodium phosphate, pH 8.0 and treated with 20 U DNase I (M0303S, NEB) at 37°C for 10 min followed by 200 μl proteinase K (10 μg/ul, P1078S, NEB) at 37°C for 10 min to digest the proteins and release the RNAs from the beads.
Bead preparation (FMRP): 25 μl Protein A Sepharose beads (GE Healthcare Biosciences CL-48) were washed with lysis buffer (20 mM Tris–HCl pH 7.5, 200 mM sodium chloride, 30 mM EDTA, 2.5 mM magnesium chloride, 0.5% Triton X-100) and 5 μg of 7G1–1 antibody [1] was added to beads.
WT and Mov10 KO N2a cells were transfected with 100 μg of plasmids encoding MOV10, KH1 peptide, or control vector DNA. Cells (∼1.5 × 107) were lysed with 0.5 ml lysis buffer (20 mM Tris–HCl pH 7.5, 200 mM sodium chloride, 30 mM EDTA, 2.5 mM magnesium chloride, 0.5% Triton X-100) with protease Inhibitor (1 tablet per 10 ml Lysis buffer, Complete Mini, EDTA free, 35440400, Roche), and RNase Inhibitor (80 U/ml, RNasin, N2511, Promega), and immunoprecipitated with prepared beads at 4°C for 12 h. The beads were then washed in lysis buffer and treated with 20 U DNase I (M0303S, NEB) at 37°C for 10 min followed by 200 μl proteinase K (10 μg/μl, P1078S, NEB) at 37°C for 10 min to isolate RNAs from the beads.
For RNA immunoprecipitation: Samples were treated with 200 μl proteinase K (10 μg/μl, P1078S, NEB) at 37°C for 10 min to isolate RNA from the PAS beads. RNA was then purified from the supernatant by following RNA purification/ RT-qPCR experiment procedure (below).
For protein co-immunoprecipitations: after immunoprecipitation, samples were treated with 1× Laemmli buffer and analyzed via western blot protocol (above).
RNA purification/ RT-qPCR experiments
The RNA was extracted from brain and cell lines with TriZOL reagent (15596018, ThermoFisher Scientific) per ThermoFisher protocol, precipitated from 100% isopropanol and washed in 70% ethanol. RNA was quantified and purity assessed (260 nm: 280 nm value between 1.8 and 2.2) by UV-Vis spectroscopy (Beckman Coulter DU 640 Spectrophotometer). cDNA was synthesized using 2.5 μM Oligo dT 20 primer (18418020, ThermoFisher Scientific) and Superscript III Reverse Transcriptase (18080044, ThermoFisher Scientfic) per ThermoFisher published protocol. RT-qPCR was performed with iQ SYBR Green Supermix (Bio-Rad) using a StepOnePlus RT-qPCR machine (Applied Biosystems) with gene specific primers (Supplementary Table S1). mRNA levels were normalized to GAPDH. Relative levels of mRNAs IP’ed were then normalized to overall mRNA levels in lysate. Ct values obtained were verified to be within the linear concentration range obtained for primers (see MIQE in Supplementary Files). A sample lacking reverse transcriptase (RT-) and a non-template control (NTC) were run for each sample/ primer set and verified to have non-detectable fluorescence. Melt curves were analyzed to verify a single peak at the appropriate melt temperature. Experiments were performed at least three times and a two-tailed T-test was performed to determine significance.
MOV10 purification
Myc-tagged murine MOV10 (27) construct DNA was transfected using PEI (polyethylenimine, # 408727, Sigma-Aldrich) in Freestyle HEK 293F cells (Invitrogen) and cultured according to the manufacturer's protocol and as described (22). Cells were harvested after 48 h and lysed in Lysis buffer (50 mM Tris–HCl pH 7.5, 300 mM NaCl, 30 mM EDTA, 0.5% Triton) containing Protease inhibitors (Roche, Indianapolis, IN, USA) and spun at 8000 rpm for 10 min at 4°C. The supernatant was immunoprecipitated overnight with anti-myc agarose (A7470, Sigma-Aldrich). After four washes, myc-MOV10 was eluted with myc peptide 2 mg/ml, (synthesized by the Protein Sciences, Roy J. Carver Biotech Center, UIUC) for 2 h at 4°C and approximated concentration by comparing band using Coomassie Brilliant Blue staining.
Mass spectrometry
Protocol for MOV10 purification was followed (above). A band at approximately 75 kDa was excised from the gel and sent for analysis. Samples were cleaned up using Perfect Focus (G-Biosciences, St Louis, MO, USA) and digested using Trypsin (Proteomics Grade, G-Biosciences) at a ratio of 1:50 w/w in 25 mM Ammonium bicarbonate buffer in a CEM (Matthews, NC, USA) Discover Focused Microwave digestor for 30 min at 55 C (50 Watts max power). The digested peptides were lyophilized and suspended in 5% acetonitrile + 0.1% formic acid. Nano-ESI LC/MS was performed in a system consists of a Waters (Milford, MA, USA) NanoAquity UPLC connected to a Waters Q-ToF mass spectrometer. For chromatography, the column used was Waters Atlantis C-18 3 μm 75 μm × 150 mm. The flow rate was set at 300 nanoliters using a gradient of water + 0.1% formic acid to 60% acetonitrile + 0.1% formic acid in 60 min. Mass spec results were filtered and sorted by Waters ProteinLynx Global Server to PKL format and further analyzed using Mascot (Matrix Sciences, London, UK) and searched against NCBI NR Human database.
MOV10 unwinding assay
In vitro transcription of iSpinach was performed using Megashortscript T7 Transcription Kit (AM1354, ThermoFisher Scientific). The iSpinach RNA was then cleaned and concentrated using a Zymo Research Kit (R1013, Zymo Research). A total of 200 nM recombinant MOV10 was added to a solution containing 2 μM of iSpinach in 2 μM of DHFBI, 80 mM Tris–HCl, pH 7.5, 2 mM MgCl2, 2 μM DHFBI, 200 mM KCl, 2 mM DTT, 0.01% RNasin with or without 4 mM adenosinetriphosphate (ATP). Fluorescence was measured using a SpectraMax M2 microplate reader (ex: 492 nm/em: 516 nm), according to protocol in (26), at the Roy J Carver Biotech Center, UIUC, every 30 s.
Neurite outgrowth
WT and Mov10 KO N2a cells were plated in triplicate (density of 1.5 × 104 cells/well) and incubated at 37°C in Dulbecco's modified Eagle's medium (DMEM, 10% fetal bovine serum). Mov10 KO N2a cells were transfected with the N- or C-terminal domains of MOV10 (25) or the helicase mutant (K531A) DNA using Lipofectamine 2000 (ThermoFisher Scientific). Cells were differentiated 24 h post-transfection. and imaged under transmitted light using an EVOS cell-imaging microscope. The images were anonymized and analyzed by an experimenter blinded to the conditions using the AxioVision image analysis software. About 500–800 differentiated neurons were identified and their processes measured from triplicate experiments, and a total of 11 images were counted per condition.
AGO2 eCLIP
Brains from newly born mice (P0) Jax WT C57BL/6 and Fmr1 KO mice were flash-frozen and sent to Eclipse BioInnovations. eCLIP was performed per (28), using anti-AGO2 antibody (EAG009, Eclipse BioInnovations). Single-end (75nt) sequencing was performed on the HiSeq 4000 platform. The first 10 nt of each read contains a unique molecular identifier (UMI) which was extracted from each read with UMI tools (version 5.2) (29) and appended to the end of the read name. Next, sequencing adapters were trimmed from the 3′ end of each read using Cutadapt (version 1.15) (30). Reads were then mapped to a database of mouse repeats using STAR (version 2.6.0c) (31). Reads that mapped to the repeats were removed. The remaining reads were mapped to the mouse genome (mm10) using STAR (version 2.6.0c). Polymerase chain reaction (PCR) duplication removal was performed using UMI tools (version 5.2). CLIPper (version 1.4) was then used to identify clusters within the IP samples, and read density within clusters was compared against the size matched input sample using a custom perl script to identify peaks enriched in the CLIP sample versus the input sample.
SC1 RNA capture experiments
In-vitro transcription of SC1 and SC1 mutant was performed using Megashortscript T7 Transcription Kit (AM1354, ThermoFisher Scientific). The RNA was then cleaned and concentrated using a Zymo Research Kit (R1013, Zymo Research).
Bead preparation for FMRP IP was as described above.
WT and Mov10 KO N2a cells were transfected with 100 μg of MOV10 or a control vector DNA. Approximately 1.5 × 107 cells were lysed (20 mM Tris–HCl pH 7.5, 200 mM sodium chloride, 2.5 mM magnesium chloride, 0.5% Triton X-100), with protease Inhibitor (1 tablet per 10 ml Lysis buffer (Complete Mini, EDTA free, 35440400, Roche) and RNase Inhibitor (80 U/ ml) and immunoprecipitated with prepared beads at 4°C for 12 h. The beads were then washed with lysis buffer and 1 μg of SC1 or SC1 mutant was added and immunoprecipitated for 4 h and washed with lysis buffer. Samples were then treated with DNAse at 37°C for 10 min followed by proteinase K at 37°C for 20 min. RNA was extracted and analyzed by RT-qPCR as described above using a modified poly dT primer (Supplementary Table S1) and the primers described Supplementary Table S1.
Luciferase reporter assay
Luciferase assay constructs were obtained by cloning a truncated MAZ 3′UTR (WT), or with a point mutation in the miR-328 MRE, or with three point mutations disrupting the rG4 containing the miR-328 MRE site (Supplementary Table S1). The amplified fragment was cloned into the Psicheck2 vector using the NotI and Xho restriction sites. HEK293 and N2a cells were seeded at 5 × 104 cells into a 24-well plate for 24 h and transfected with 500 ng of the luciferase construct along FMRP-eGFP, FMRP-eGFP ΔRGG, N-terminal MOV10, using PEI (HEK293 cells) or Lipofectamine 2000 reagent (N2a cells) to a total transfected DNA concentration of 1 μg, luciferase activity was measured using a dual-luciferase reporter assay kit (Promega) on a SynergyTM HT Multi-detection plate reader 24 h post-transfection. Renilla Luciferase activity was normalized to Firefly Luciferase activity, then normalized to the luciferase activity of the Δ328 control.
RESULTS
FMRP directly interacts with MOV10 and AGO
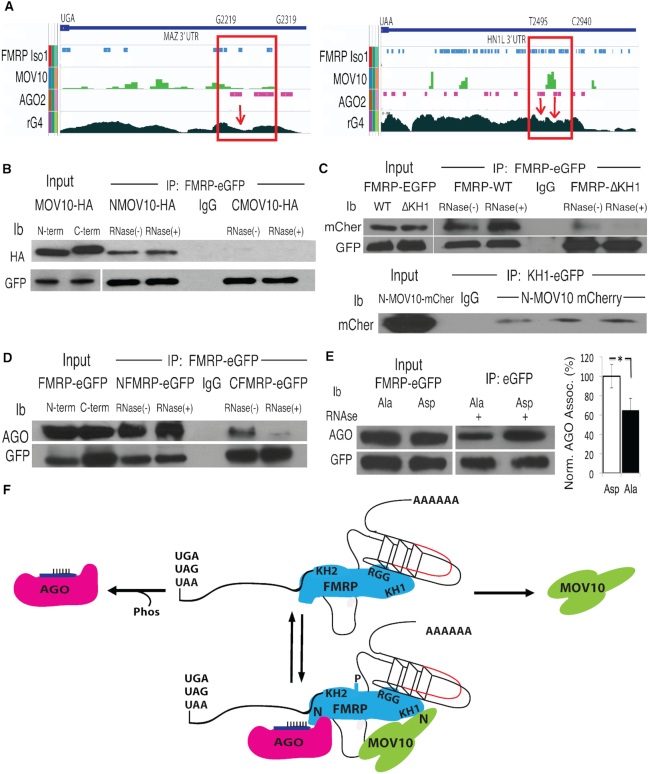
In previous work, we showed that FMRP binding in the 3′ UTR of target mRNAs either upregulated or downregulated luciferase activity (22). Comparing CLIP-seq binding sites of FMRP (9,10), the RNA helicase MOV10 (22) and AGO2 as a proxy for active MREs (32), we found that, in most cases, FMRP recruited MOV10 to co-bound mRNAs and facilitated AGO-associated translational suppression, possibly by resolving RNA secondary structure (22). However, we also found that a subset of mRNAs was upregulated by FMRP–MOV10 interaction, suggesting that in these mRNAs, this complex was protecting the mRNA from AGO-associated translational suppression (22). The upregulated mRNAs (hereafter referred to as protected mRNAs, examples are MAZ and HN1L), used in this study were selected from previous work (22) and exhibited regions in the 3′ UTR in which the binding sites for all three proteins, FMRP (10), MOV10 (22) and AGO2 (32), were proximal to a rG4, obtained from rG4-seq data, which are indicated as valleys in the plot (Figure 1A). rG4-seq is a method to determine real time rG4s in mRNAs via a loss in read coverage due to the stalling of reverse transcriptase at the rG4 during cDNA synthesis (33,34). We conclude that assembly of the FMRP–MOV10 complex on an rG4 blocked AGO association.
Figure 1.
FMRP binds the N-terminus of MOV10 through its KH1 domain and to AGO through its N-terminus in a phosphorylation dependent manner. (A) Integrated Genomics Viewer (IGV) screen shot of the 3′UTRs (stop codons indicated) of two protected mRNAs, MAZ and HN1L. CLIP regions in which FMRP, MOV10 and AGO2 binding are coincident with rG4s are shown in red boxes, indicated by nucleotide position at the top. rG4s, determined by rG4-seq, are troughs (indicated by red arrows). (B) A GFP-tagged FMRP construct was co-transfected with an HA-tagged N-terminal MOV10 (NMOV10-HA) or HA-tagged C-terminal MOV10 (CMOV10-HA) construct into HEK293 and immunoprecipitated (IP’ed) with an anti-eGFP antibody and examined by immunoblot (ib): antibody indicated on the left. IgG indicates the IP’ing antibody alone. RNase indicates treatment (+) or not (−) with RNase A. (C) (Top) eGFP-tagged KH1 domain of FMRP was IP’ed and examined for mCherry tagged N-terminal MOV10 (mCher). (Bottom) eGFP-tagged FMRP WT or ΔKH1 was IP’ed with anti-eGFP and examined for association with N-terminal MOV10 (mCher), three biological replicates are shown, (n = 3). (D) N-terminal half of FMRP (N-term through KH2 domain) or C-terminal half of FMRP (KH1 through C-terminus) was IP’ed and AGO association examined with the pan-AGO antibody 2A8, which recognizes the four AGO family members in the presence or absence of RNase A. (E) The primary phosphorylation site of murine FMRP (S499) was substituted to alanine (Ala) or aspartic acid (Asp) and association with AGO examined by eGFP-FMRP-IP. AGO co-IP was quantified by normalizing to AGO and transgene protein levels, (n = 3), error bars represent SD, *P < 0.05, Student t-test. (F) Based on their respective interacting domains, a representation of a dynamic RNP complex involving FMRP, MOV10 and AGO proteins in the 3′ UTR of an mRNA (stop codons indicated) bearing an RNA G-Quadruplex (rG4) with an MRE site (red). N denotes N-terminus. All experiments were performed in triplicate, representative immunoblots are shown.
To determine how FMRP and MOV10 assemble to protect co-bound mRNAs from AGO-mediated translational suppression, we mapped the interaction domains of FMRP, MOV10 and AGO by using tagged truncation or deletion constructs as indicated. FMRP associated with the N-terminal domain of MOV10 (aa 1–495) but not with the C-terminal domain of MOV10 (aa 495–1003) (Figure 1B). To identify the region of FMRP that interacts with the N-terminus of MOV10, we expressed N-terminal FMRP (aa 1–404) or C-terminal FMRP (aa 216–632) and found that the N-terminus of MOV10 bound both, suggesting that the interacting domain of FMRP was either the KH1 or KH2 domain shared by both constructs (Supplementary Figure S1A). To identify the FMRP domain that is recognized by the N-terminus of MOV10, we used FMRP-ΔKH1 or FMRP-ΔKH2 domain deletion constructs (Supplementary Figure S1B, top and center) along with a N-terminal MOV10 construct and found that the FMRP–MOV10 interaction was disrupted in the FMRP-ΔKH1 mutant (Figure 1C, top) but remained associated in the FMRP-ΔKH2 mutant (Supplementary Figure S1C). To verify that FMRP’s KH1 domain directly interacts with MOV10, we transfected the FMRP-KH1 construct (Supplementary Figure S1B, bottom) along with an N-terminal MOV10 construct and found that we were able to co-immunoprecipitate N-terminal MOV10 with the FMRP-KH1 (Figure 1C, bottom). We conclude that the N-terminal domain of MOV10 interacts with the KH1 domain of FMRP.
FMRP has been shown to directly interact with AGO1 and AGO2 (35), specifically through AGO2’s MID domain (36). Interestingly, recent work suggests that assembly of the FMRP–AGO2 complex is also mediated through miRNA binding to the KH1/KH2 domains of FMRP (37). To identify the region of FMRP that interacts with AGO, we expressed the N- and C-terminal halves of FMRP (Supplementary Figure S1A, top and bottom, respectively) and found that only the N-terminal half bound AGO in the absence of RNA but not the C-terminal half (Figure 1D), suggesting that AGO proteins bind the N-terminal half of FMRP.
Phosphorylation of FMRP has been shown to regulate association with AGO (38). To determine the role of FMRP phosphorylation in this interaction, we expressed two eGFP-FMRP mutants, FMRP-Asp (S499D), a phosphomimic of FMRP (39) and FMRP-Ala (S499A), constitutively unphosphorylated FMRP, and found significantly less S499A associated with AGO compared to S499D (Figure 1E), in an RNA-independent manner, in contrast to (38). The phosphorylation status of FMRP did not affect its ability to bind MOV10 (Supplementary Figure S1D). In sum, we propose a dynamic FMRP RNP complex in which the proteins change their association based on conformational changes—some of which may be driven by binding other proteins or by post-translational modifications like phosphorylation or arginine methylation (40–43). Based on our interactive domain mapping data and what is currently known about these proteins, FMRP could act as a scaffolding protein at rG4s, where these three components of the RNP complex (rG4s, MOV10 and AGO) may dynamically interact as their respective binding sites are revealed (FIgure 1F, top) and how they may assemble as shown (Figure 1F, bottom).
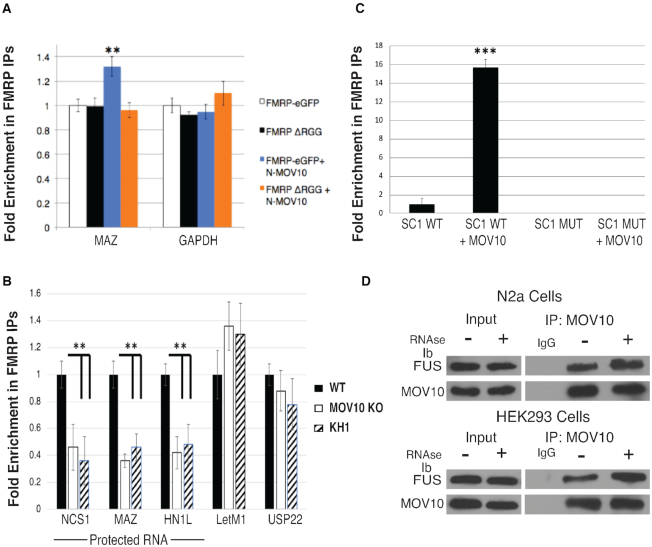
FMRP inhibits AGO association with a subset of mRNAs through its RGG box
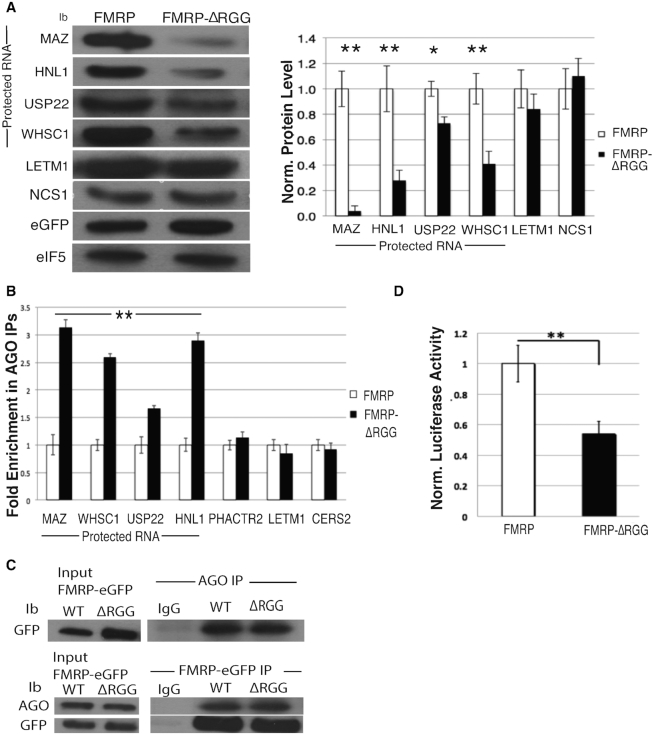
FMRP binds RNA rG4s through its RGG box and stabilizes them (16,17,44,45), suggesting a role for FMRP in modulating regulatory elements, such as MREs, within rG4 structures. To elucidate FMRP’s role in protecting mRNAs from miRNA-mediated translational suppression, we focused on understanding the role of FMRP’s RGG box in modulating AGO association with mRNAs. We first overexpressed WT FMRP or an RGG box deletion (FMRP-ΔRGG) and compared protein levels from mRNAs that were protected by the FMRP–MOV10 complex to those mRNAs that were translationally suppressed by the FMRP–MOV10 complex [22] (Figure 2A). In the presence of FMRP-ΔRGG, protein levels associated with protected mRNAs were significantly reduced compared to the protein levels of the unprotected RNAs that are suppressed by AGO [Figure 2A, compare protein levels of protected mRNAs MAZ, HNL1, USP22, WHSC1 to non-protected mRNAs LETM1 and NSC1 per (22)], suggesting that the RGG box prevents degradation of those RNAs. As a control, we verified levels of FMRP by measuring eGFP in each sample and normalized loading to eIF5B protein levels.
Figure 2.
RGG box of FMRP attenuates AGO association and subsequent degradation of MOV10-FMRP co-bound mRNAs. (A) (Left) Immunoblot (ib) of endogenous proteins encoded by mRNAs co-bound by FMRP–MOV10 in HEK293 cells expressing FMRP or FMRP-ΔRGG. eIF5B is the loading control. Vertical line denotes the proteins that are ‘protected’. (Right) Quantification of immunoblots (n = 3), error bars represent SD, *P < 0.05, ** P < 0.001 Student t-test. (B) RT-qPCR of IP’ed AGO- associated mRNAs in cells expressing WT FMRP or FMRP-ΔRGG, (n = 3), error bars represent SD, ** P < 0.001 Student t-test. Statistical significance is for all four protected RNAs, MAZ through HN1L. (C) (Top) AGO was IP’ed and examined for GFP-tagged FMRP and FMRP-ΔRGG (GFP). (Bottom) GFP tagged FMRP and FMRP-ΔRGG were IP’ed and examined for AGO. (D) Luciferase activity of the MAZ 3′ UTR in cells expressing the WT FMRP or FMRP-ΔRGG constructs, (n = 3), error bars represent SD, ** P < 0.001 Student t-test. All experiments were performed in triplicate, representative immunoblots are shown.
To directly test this hypothesis, we immunoprecipitated AGO from cells expressing WT FMRP or FMRP-ΔRGG in HEK293 cells and quantified AGO-associated mRNAs using RT-qPCR. AGO association with protected mRNAs (MAZ, HN1L, USP22 and WHSC1) increased significantly with the loss of FMRP’s RGG box, suggesting that the RGG box blocks AGO association (Figure 2B). However, AGO’s association with the mRNAs that are unprotected [PHACTR2 and LETM1 per (22)] was unchanged in cells containing either FMRP or FMRP-ΔRGG (Figure 2B). We verified that AGO association with FMRP was not affected by a loss of FMRP’s RGG box (Figure 2C), ruling out the possibility that the RGG box itself affected AGO association. We normalized to overall mRNA levels in each sample and used CERS2 as an immunoprecipitation control because CERS2 is bound by AGO but is not a target of FMRP. To verify that this reduction in AGO association observed in protected RNAs is not due to a loss in mRNA binding by FMRP-ΔRGG, we immunoprecipitated FMRP and FMRP-ΔRGG from HEK293 cells and quantified associated mRNAs using RT-qPCR. We found that FMRP’s ability to bind this subset of mRNAs was not affected by a loss of the RGG box (Supplementary Figure S2A) but was affected by loss of the KH2 domain as well as the I304N point mutation located within the KH2 domain (24) (Supplementary Figure S2B, left and right). Thus, FMRP-ΔRGG is able to bind normal amounts of RNA; however, the RGG box could have an effect on the translational fate of protected RNAs.
We verified that the RGG box is required for maintaining the level of protein production encoded by FMRP-protected mRNAs by cloning the 3′ UTR of the MAZ gene into a luciferase reporter in cells over-expressing the FMRP-ΔRGG mutant. We found a significant decrease in luciferase activity compared to WT (Figure 2D), suggesting that the RGG box is required to protect expression from the rG4-rich MAZ 3′UTR. These data demonstrate that FMRP’s ability to inhibit AGO association with the protected MAZ RNA is modulated through FMRP’s RGG box.
The N-terminus of MOV10 is necessary to block AGO association with protected mRNAs
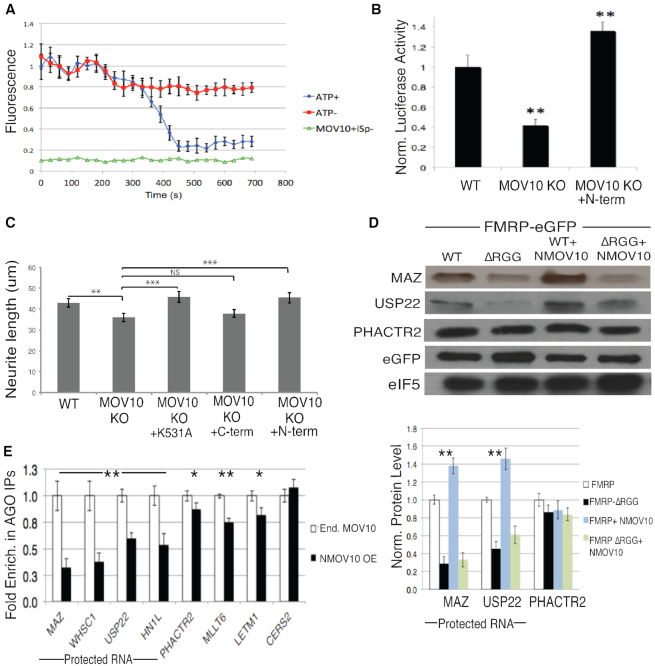
RNA helicases remodel the secondary structural landscape of target RNAs. MOV10 is an RNA helicase that translocates in the 5′ to 3′ direction (46) and its binding sites are highly enriched in the 3′ UTR’s of target mRNAs (22,46). Although originally identified as functionally associating with AGO2 (47), MOV10 has been implicated in facilitating nonsense-mediated decay through its direct association with UPF1 (46) and inhibiting retrotransposition during reverse transcription (48,49). MOV10 binds G-rich structures and facilitates miRNA-mediated translational suppression (22). Although it has been shown to unwind short double stranded RNA in vitro (46), it was unknown if MOV10 could resolve more thermodynamically stable nucleic acid structures, such as rG4s. To answer this question, we employed the iSpinach aptamer to develop an unwinding assay. iSpinach is a modified version of the Spinach aptamer (50) that is optimized for in vitro analysis (26). The iSpinach aptamer contains a rG4 structure that is necessary to bind the DHFBI fluorophore. Upon binding of the fluorophore, the complex fluoresces as a green fluorescent mimic (50–52). We modified the iSpinach aptamer to incorporate a 5′ overhang known to be bound by MOV10 (46), and measured the ability of MOV10 to unwind the rG4 by measuring the loss of fluorescence as a function of time. We found that MOV10 is able to unwind the iSpinach aptamer in an ATP-dependent manner (Figure 3A), suggesting that MOV10 can resolve rG4s to reveal proximal MREs.
Figure 3.
MOV10 unwinds rG4, enhances rG4 luciferase activity from a reporter and enhances FMRP-mediated translation of a subset of RNAs through its N-terminus. (A) Purified murine MOV10 unwinds a synthesized rG4 reporter (iSpinach). Red: iSpinach in the presence of MOV10, DHFBI, in the absence of ATP. Blue: iSpinach in the presence of MOV10, DHFBI, and ATP. Green: MOV10 and ATP, absence of DFHBI. (B) Luciferase activity of MAZ 3′ UTR reporter in WT N2a, Mov10 KO N2a and Mov10 KO N2a transfected with the N-terminal MOV10 construct, (n = 3), error bars represent SD, ** P < 0.001 Student t-test. (C) Quantification of neurite length (μm) in WT or Mov10 KO N2a cells post differentiation transfected with null or N-,C-terminal MOV10, or a translocation mutant (K531A), error bars represent SEM, one way ANOVA (F(4.94) = 3.964, P = 0.0052, **P< 0.05,***P< 0.01. (D) Immunoblot of endogenous proteins encoded by mRNAs co-bound by FMRP–MOV10 in HEK293 cells expressing WT FMRP or FMRP-ΔRGG transfected with N-terminal MOV10. (Bottom) Quantification of protein levels, (n = 3), error bars represent SD, **P < 0.001 Student t-test. (E) RT-qPCR of IP’ed AGO-associated mRNAs in HEK293 cells expressing endogenous levels of MOV10 or over-expressing N-terminal MOV10, (n = 3), error bars represent SD, *P < 0.05, ** P < 0.001 Student t-test.
To elucidate MOV10’s role in protecting FMRP–MOV10 co-bound mRNAs, we first identified the functional domain of MOV10 that is necessary for this activity. The N-terminus of MOV10, which directly interacts with FMRP, contains no known structure, while all of its helicase activity is within its two C-terminal recA domains (53). In addition, the N-terminus of MOV10 was found to block HIV infection, suggesting an independent functional role (53). We cloned the 3′ UTR of the protected mRNA of the MAZ gene into a luciferase reporter and transfected the construct into Mov10 KO N2a cells that were expressing the N-and C- terminal regions of MOV10 (53). As expected, MAZ expression was reduced in the absence of MOV10 (Figure. 3B) that we described in (22). In contrast, expression of the N-terminus of MOV10 was sufficient to restore activity of the MAZ reporter, suggesting that this process is independent of MOV10’s helicase activity and that the N-terminus is able to form the protective complex that may include FMRP (Figure 3B).
In an earlier study, we provided evidence that FMRP and MOV10 function in the same pathway for neurite outgrowth (49). To determine if the N-terminus of MOV10 has a functional role in neuronal process formation, we used the murine Mov10 KO N2a cell line (49). MOV10 is highly expressed in neurites (54) and, similar to loss of FMRP (55), the absence of MOV10 results in significantly shorter neurites (49). We measured neurite length in WT N2a, Mov10 KO N2a cells, and Mov10 N2a KO cells transfected with transgenes encoding the N- and C- terminus of MOV10 along with a MOV10 helicase mutant (K531A) that is unable to translocate (46). Upon verifying the overexpression of these constructs by Western Blot (Supplementary Figure S3A), we found that N-terminal MOV10, unlike C-terminal MOV10, was able to rescue neurite outgrowth (Figure 3C). Interestingly, the full length K531A mutant also rescued the neurite length, suggesting that the presence of the N-terminus was sufficient in restoring neurite length and did not require helicase activity (Figure 3C). This result suggests that the N-terminal domain of MOV10 has a unique and important function.
Our hypothesis is that the N-terminus of MOV10 binds and modulates FMRP association with rG4s. To determine if the N-terminus of MOV10, along with FMRP’s RGG box, acted in conjunction to regulate translation, we over-expressed the N-terminus of MOV10 with either WT FMRP or FMRP-ΔRGG and quantified the endogenous protein levels of protected mRNAs compared to unprotected mRNAs. We verified that loss of FMRP’s RGG box did not affect its ability to bind MOV10 (Supplementary Figure S3B). Loss of FMRP’s RGG box resulted in a decrease in MAZ and USP22 protein levels (Figure 3D). However, the addition of the N-terminus of MOV10 in the presence of FMRP’s RGG box resulted in increased protein levels of the protected mRNAs compared to the non-protected mRNA, PHACTR2 (Figure 3D, top and bottom). This result suggests that association of the N-terminus of MOV10 leads to increased translation of protected RNAs mediated through the FMRP’s RGG box. We verified that a loss of FMRP’s RGG box did not affect MOV10 protein levels (Supplementary Figure S3C) and that FMRP protein levels were not affected by the absence of MOV10 (Supplementary Figure S3D).
To determine if MOV10 affected AGO association with these protected mRNAs, we immunoprecipitated AGO from HEK293 cells containing endogenous levels of MOV10 and in cells in which we had overexpressed N-terminal MOV10, verified by Western Blot (Supplementary Figure S3E) and quantified AGO-associated mRNAs using RT-qPCR. In cells expressing N-terminal MOV10, AGO association with protected mRNAs was significantly reduced (Figure 3E). There were significant yet smaller increases in AGO association with unprotected mRNAs, most likely because we have disrupted the interaction of FMRP with full length MOV10 so there is reduced helicase activity to reveal MREs for AGO association. As a control, we quantified the AGO association of CERS2 mRNA, which was bound by AGO but is not a target of FMRP or MOV10 (Figure 3E). These data suggest that the N-terminus is sufficient in forming the FMRP–MOV10 RNP complex that acts to protect a subset of mRNAs from miRNA-mediated translational suppression.
FMRP and MOV10 interact to regulate MREs embedded within RNA rG4s
RNA secondary structure can modulate translational regulation intrinsically (56,57) or through association of RBPs (22,32,58). The rG4 is a stable RNA secondary structure formed by the stacking of co-planar arrays known as G-quartets that are stabilized by Hoogsteen-type hydrogen bonds (59,60) to form the most stable nucleic acid structures in nature (61,62). rG4s are highly enriched in key neuronal genes and have been implicated in facilitating mRNA localization, regulating neurite outgrowth (63), facilitating alternative splicing (64), as well as increasing mRNA stability (65). rG4s also obstruct RISC effector proteins from accessing MREs (22,56). In support of this function, global identification of rG4s using rG4-seq have shown that rG4s are predominantly in the 3′ UTR of mRNAs and are enriched at MREs (33), suggesting a role in miRNA regulation. FMRP binds to rG4 structures through its RGG box and stabilizes them, (14,16), suggesting a role for FMRP in regulating MREs embedded within rG4 structures.
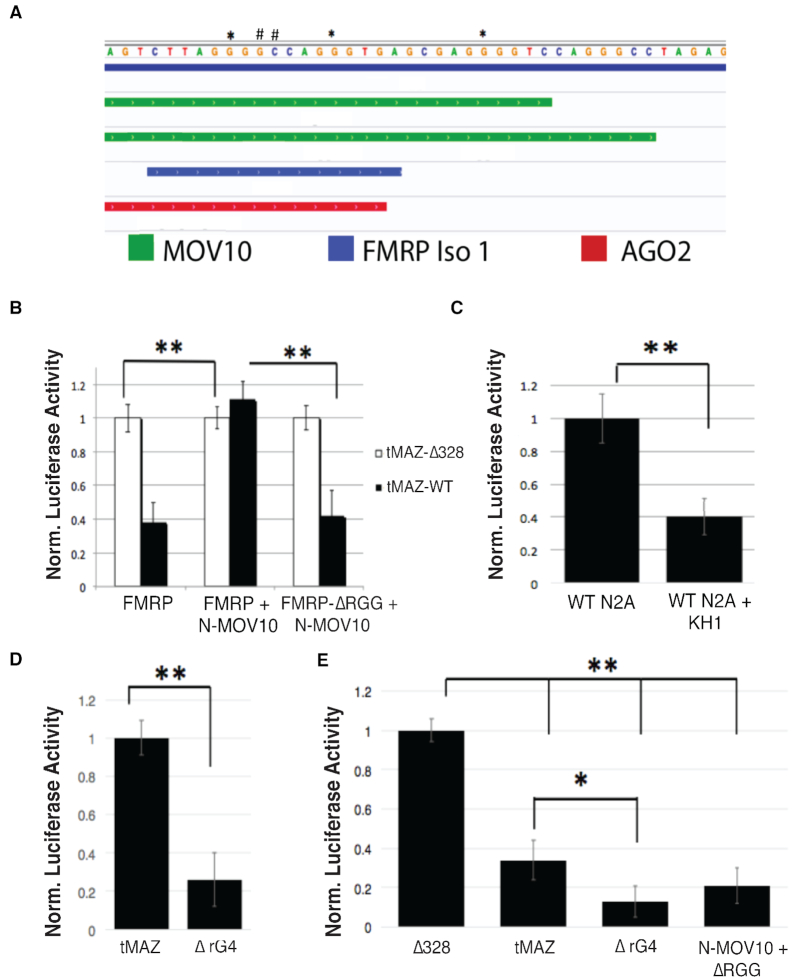
In previous work, we showed that the MRE site for miR-328 overlaps with a predicted rG4 structure in the 3′ UTR of MAZ mRNA (22). To determine if FMRP and MOV10 regulate MREs within rG4s, we expressed a truncated version of the MAZ 3′ UTR (tMAZ-WT) containing the miR-328 MRE and the rG4 (Supplementary Table S2). We also introduced two point mutations (G2277A, C2278T) (Figure 4A, denoted with pound signs) that obliterated the miR-328 MRE site (tMAZ-Δ328) (Supplementary Table S2). The truncated region also contained CLIP binding sites for FMRP, MOV10 and AGO (Figure 4A). We first verified that this region was modulated by FMRP by quantifying luciferase activity in WT N2a cells expressing FMRP or FMRP-ΔRGG (Supplementary Figure S4A). We found that the RGG box of FMRP was required for normal luciferase activity, indicating that this region is protected by FMRP through the RGG box (Supplementary Figure S4A). We next examined the role of FMRP on expression of the reporters when only the N-terminus of MOV10 was present. We expressed tMAZ-WT and tMAZ-Δ328 in Mov10 KO N2a cells, as well as in Mov10 KO N2a cells transfected with FMRP or FMRP-ΔRGG constructs and with or without the N-terminus of MOV10, as indicated in Figure 4B. Cells expressing the tMAZ-Δ328 construct were used as a normalization control since it has a mutated miR-328 MRE and luciferase activity is independent of miRNA regulation. As expected, cells expressing WT FMRP in the absence of MOV10 led to a decrease in luciferase activity of the tMAZ-WT construct compared to the tMAZ-Δ328 (Figure 4B, left). Introduction of the N-terminus of MOV10 significantly increased tMAZ-WT luciferase activity—up to the activity levels observed in tMAZ-Δ328 (Figure 4B, center), suggesting that the N-terminus of MOV10 inhibits miRNA-mediated translational regulation and restores luciferase activity. In contrast, the N-terminus of MOV10 was not able to rescue tMAZ-WT luciferase activity in the absence of the RGG box (Figure 4B, right), indicating that protection by the N-terminus of MOV10 requires the RGG box of FMRP. As a control, the C-terminus of MOV10 was transfected into Mov10 KO N2a cells expressing FMRP or FMRP-ΔRGG and did not rescue reporter activity (Supplementary Figure S4B). We verified that over-expressing the N- or C-terminal domains of MOV10 in WT and Mov10 KO N2a cells expressing FMRP or FMRP-ΔRGG did not affect the luciferase activity of the empty vector (Supplementary Figure S4C). These data suggest that FMRP and MOV10 cooperatively function to regulate the miR-328 MRE site, inhibiting translational suppression of the tMAZ WT reporter.
Figure 4.
FMRP and N-terminal MOV10 function co-operatively to regulate MREs contained within rG4 in the 3′ UTR of MAZ. (A) IGV screen shot of the region in the 3′ UTR of the MAZ mRNA cloned into the psicheck 2 dual luciferase reporter. ** mutations in the Gs required to form an rG4. #- mutations in the miR-328 MRE site. Top two bars (green): MOV10 CLIP sites. Third bar (blue): FMRP CLIP site. Bottom bar (red): AGO2 CLIP site. (B) Luciferase activity in Mov10 KO N2a cells transfected with the transgenes indicated below, (n = 3), error bars represent SD, **P < 0.001 Student t-test. (C) Luciferase activity in WT N2a cells and WT N2a cells transfected with a construct encoding the KH1 peptide, n = 3, ** P < 0.001, Student t-test. (D) Luciferase activity in WT N2a cells expressing the MAZ 3′ UTR rG4 mutant, (n = 3), error bars represent SD, ** P < 0.001 Student t-test. (E) Luciferase activity in Mov10 KO N2a cells transfected with the MAZ 3′ UTR MRE site mutation (Δ328), the WT MAZ 3′ UTR (tMAZ), the MAZ 3′ UTR rG4 mutant (ΔrG4) and the MAZ 3′ UTR rG4 mutant expressing N-terminal MOV10 and FMRP-ΔRGG, (n = 3), error bars represent SD, *P < 0.05, ** P < 0.001 Student t-test.
To identify the relevance of the rG4 in regulating the MAZ MRE, we utilized the same reporter, but introduced three new point mutations (G2275A, G2282A, G2292A) (Supplementary Table S2) that disrupt the rG4 structure per the in-silico rG4 predictive analysis tool, QGRS Mapper (66) (Figure 4A, denoted with asterisks), leaving the miR-328 MRE intact. We expressed this construct and tMAZ-WT in WT N2a cells and found that luciferase activity significantly decreased, suggesting that a functional rG4 is required for the FMRP–MOV10 complex to protect mRNAs (Figure 4D). In support of this result, expression of the construct encoding the KH1 peptide, which was identified as the MOV10 interacting domain of FMRP in Figure 1, should competitively disrupt FMRP–MOV10 interaction. In Figure 4C, we show that expression of the KH1 peptide reduced luciferase activity.
We hypothesized that FMRP binds the rG4 and recruits MOV10 to form a stabilizing complex that blocks AGO association. To test this, we transfected the rG4 mutant into Mov10 KO N2a cells and normalized to the mutated 328 MRE site (Δ328), which cannot be bound by AGO (Figure 4E). In the absence of MOV10, there is a significant decrease in WT tMaz reporter activity. Interestingly, loss of the rG4 structure led to a further reduction in reporter activity, suggesting that FMRP may act to protect the mRNA to a lesser degree than in the presence of the FMRP–MOV10 complex (Figure 4E). In the presence of N-terminal MOV10 but in the absence of the RGG box, reporter activity of the rG4 mutant was not rescued, suggesting the FMRP–MOV10 complex at the miR-328 MRE no longer has a protective role in the absence of the rG4 (Figure 4E). These data suggest that FMRP and MOV10 cooperatively act to inhibit AGO association with the rG4 embedded miR-328 MRE.
The N-terminus of MOV10 enhances FMRP’s ability to bind rG4s
We have shown that the N-terminus of MOV10 modulates AGO’s access to mRNAs (Figure 3). To examine the effect of the N-terminus of MOV10 on FMRP’s ability to bind mRNA, we immunoprecipitated exogenously expressed FMRP in Fmr1 KO STEK cells (67) that had been transfected with either WT FMRP, FMRPΔ-RGG and/or the N-terminus of MOV10 as indicated in Figure 5A. We then quantified FMRP-associated MAZ mRNA using RT-qPCR. In the presence of the N-terminus of MOV10, FMRP association with MAZ mRNA increased significantly (Figure 5A), suggesting that the N-terminus of MOV10 bound and stabilized the interaction of the RGG box with the rG4-rich MAZ mRNA. We expanded upon this observation by testing the ability of MOV10 to modulate FMRP’s association with additional protected mRNAs in N2a cells. We immunoprecipitated FMRP in the presence of MOV10 (WT N2a cells), in the absence of MOV10 (Mov10 KO N2a cells), or in the presence of MOV10 (WT N2a cells) expressing FMRP’s KH1 peptide. We found that introduction of the KH1 peptide had the same effect as complete loss of MOV10 (Figure 5B). Thus, in the presence of FMRP’s KH1 domain, FMRP binding to protected mRNAs decreased, similar to what was observed in the absence of MOV10, suggesting that MOV10’s direct interaction with FMRP is required to modulate its affinity for these mRNAs (Figure 5B).
Figure 5.
MOV10 increases FMRP association with rG4s, which is mediated through the KH1 domain and directly interacts with FUS. (A) RT-qPCR of FMRP-associated mRNAs in murine Fmr1 KO STEK cells exogenously expressing WT FMRP or FMRP-ΔRGG in the presence or absence of N-terminal MOV10, (n = 3), error bars represent SD, ** P < 0.001 Student t-test. (B) RT-qPCR of FMRP-associated mRNAs in WT and Mov10 KO N2a cells in the presence or absence of the FMRP’s KH1 peptide, (n = 3), error bars represent SD, ** P < 0.001 Student t-test. (C) RT-qPCR of IP’ed FMRP-associated SC1 rG4 in the presence or absence of MOV10, (n = 3), error bars represent SD, ** P < 0.001 Student T-test. (D) Representative immunoblots of MOV10-associated FUS in the presence and absence of RNase in N2a cells and in HEK293 cells. Experiments were performed in triplicate.
FMRP, through its RGG box, binds the SC1 rG4 with nanomolar affinity (14,68). To determine if MOV10 increases FMRP’s association with rG4 structures, FMRP isolated from Mov10 KO N2a cells was incubated with or without purified MOV10. After washing, we added SC1 rG4 RNA or SC1 Mut (Supplementary Table S2), a mutant SC1 that could not form a rG4 structure and quantified FMRP associated SC1 (or SC1 MUT). In the presence of MOV10, FMRP’s association with SC1 was significantly increased (Figure 5C). In contrast, FMRP did not associate with the SC1 mutant in the presence or absence of MOV10. Taken together, our data suggest that MOV10’s association with FMRP increases the ability of FMRP to bind rG4s.
MOV10 directly associates with FUS, another RGG box containing RBP
We hypothesized that MOV10 may be interacting with other RBPs that contain RGG box domains and possibly modulating their association with mRNA. MOV10 has been shown to directly interact with DHX9 (46), an RGG box protein containing 3′ to 5′ RNA/DNA helicase activity known to regulate transcription and translation (69). To identify MOV10-associated proteins, we performed a mass spectrometry screen and found Fused in Sarcoma (FUS) associates with MOV10 (Supplementary Figure S5A). FMRP has also been found to directly interact with FUS (70) and like MOV10, FUS has been shown to interact with AGO2 to facilitate miRNA-translational regulation (71) as well as modulate NMD factors in RNA granules (72). Interestingly, much like FMRP, FUS contains a low complexity domain (LCD), two RGG boxes and associates with FMRP in RNA granules (73–75). We found that MOV10 directly interacts with FUS (Figure 5D) in mouse N2a cells as well as HEK293 cells, suggesting that there may be a wider scope of MOV10-modulated regulation of gene expression.
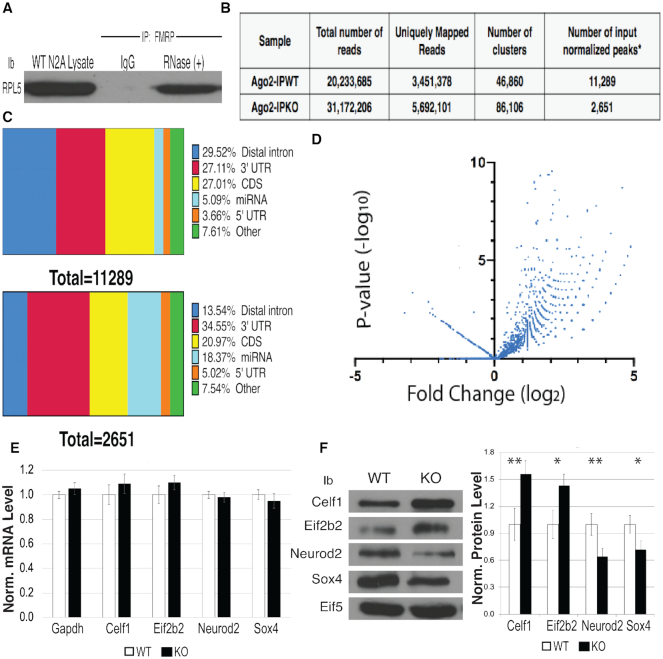
FMRP facilitates AGO association with target mRNAs
FMRP binding is enriched in coding sequences and has been shown to translationally suppress bound mRNAs by stalling ribosomes (9). This function likely occurs by FMRP’s direct association with the large subunit ribosomal protein RPL5 in Drosophila (76). Here we show that FMRP also directly associates with RPL5 in N2a cells (Figure 6A), suggesting that ribosome stalling may occur similarly in mice. In addition, there is a great deal of evidence, including from our lab, that FMRP binds AGO and functions in the 3′UTR to regulate translation. To understand FMRP’s global role in regulating AGO association with mRNAs, we performed enhanced Cross-linking Immunoprecipitations (eCLIP) of AGO2 in the presence or absence of FMRP in P0 mouse brain (28). An AGO2-specific antibody developed by Eclipse BioInnovations (EAG005) was used for the eCLIP. This antibody immunoprecipitates comparably to the pan-AGO antibody 2A8 (Supplementary Figure S6A). Immunoprecipitated samples were normalized to input to obtain the identity and relative enrichment of AGO2 bound mRNAs. We chose age P0 because FMRP is elevated in all brain cell types at this time (77). As shown in Figure 6B, we obtained 20 233 685 reads from the WT AGO2 IP and 31 172 206 reads from the Fmr1 KO AGO2 IP. A total of 3 451 378 of the WT reads and 5 692 101 of the Fmr1 KO reads mapped to the mouse genome. As shown by others, loss of FMRP does not have a large effect on target transcript levels (10,78,79), allowing for the comparison of AGO2 binding to mRNAs in the presence or absence of FMRP. After normalizing to input, the AGO WT IP had 11 289 peaks and the AGO Fmr1 KO IP had 2651 peaks—a significant reduction in AGO association with mRNAs in the absence of FMRP (Figure 6B: a peak is defined as a cluster with log2 fold enrichment greater than or equal to 3 and P-value ≥ to 0.001). This result suggests that FMRP participates in the recruitment of AGO2 to mRNAs because there was an approximately four-fold reduction in peaks in the absence of FMRP. We conclude that FMRP is required to recruit AGO2 to a subset of RNAs.
Figure 6.
FMRP facilitates AGO binding in a large subset of brain mRNAs. (A) IP of FMRP treated with RNase A and probed for RPL5. Experiment performed in triplicate, representative immunoblot is shown. (B) Number of normalized peaks from AGO2 eCLIP in WT versus Fmr1 KO mouse brains. (C) Distribution of enriched AGO2 binding sites across transcript regions determined by eCLIP: WT mouse brain normalized to input (top), Fmr1 KO brains normalized to input (bottom). (D) Volcano plot of AGO2 enriched peaks in WT and Fmr1 KO mouse brain. (E) RT-qPCR of statistically differentially bound targets of AGO in the absence of FMRP (n = 3), error bars represent SD. (F) Immunoblot of protein levels of differentially bound AGO targets from WT and Fmr1 KO mouse brain. eIF5B is a loading control, (n = 3), error bars represent SD, *P < 0.05, ** P < 0.001 Student t-test.
We next evaluated the relative frequency of the peaks that map to each RNA region (Distal intron, 3′UTR, CDS, miRNA and 5′UTR). The overall range of AGO2 binding across specific regions of the mRNA, like the distal intron and coding sequence, as denoted by horizontal slice charts (Figure 6C), supports recent work demonstrating AGO2’s function in the nucleus and modulating microRNA-mediated translational regulation within the coding regions of transcripts, respectively (80). In the AGO2 IP from WT brain, 27.1% of the peaks mapped to the 3′UTR (Figure 6C, top), comparable to the AGO2 IP from Fmr1 KO P0 brain in which 34.6% of the peaks mapped to the 3′UTR (Figure 6C, bottom). These percentages are similar to previously published AGO HITS-CLIP data of P13 brain where 32% of the reads mapped to the 3′UTR (81). Interestingly, the distribution of AGO binding in distal introns and miRNAs varied largely between the samples, and could reflect AGO2’s inability to localize to the nucleus via FMRP interaction (82) and FMRP’s ability to bind miRNAs and modulate the AGO–miRNA complex (37). This needs to be studied further.
When analyzing differential binding of AGO2 in WT versus Fmr1 KO mouse brain in the 3′ UTR of target mRNA, we found that 2505 peaks representing 1648 genes had significantly more AGO2 binding in WT compared to the Fmr1 KO brain (Supplementary File 1). Eif2b2 had the highest fold change between WT and Fmr1 KO [log2 fold-change 5.19, P-value (6.9 × 10−8)] and had no eCLIP sites in the absence of FMRP (Supplementary Figure S6B, top), suggesting that FMRP is required for AGO2 recruitment to that mRNA. We also found a number of significantly changed genes that had the same AGO2 eCLIP sites in the 3′UTR in both WT and Fmr1 KO but there were less peaks in the absence of FMRP, an example being Celf1 [log2 fold change 4.8, P-value (5.2 × 10−5) (Supplementary Figure S6B, bottom). Interestingly, transcript levels of Eif2b2 and Celf1 were not significantly changed between genotypes (Figure 6E), though the protein levels of these two genes increased significantly in the absence of FMRP (Figure 6F). This result suggests that FMRP recruits AGO2 to a large subset of mRNAs and is required for AGO2 association with the 3′UTR.
In support of FMRP blocking AGO2 association with some targets, we identified 32 peaks (P < 0.05) representing 27 genes (Supplementary File 1) in which AGO2 bound more highly in the Fmr1 KO mouse brain than in WT (Figure 6D). In this case, we found significantly more AGO2 CLIP sites in the 3′ UTRs of the Fmr1 KO samples as compared to WT. This was observed for Sox4 and Neuro2d (Supplementary Figure S6C, left and right). Transcript levels of Sox4 and Neuro2d were not significantly changed between genotypes (Figure 6E), though protein levels decreased significantly (Figure 6F). We hypothesize that these sites correlate to genes in which FMRP blocks AGO2 association. Sox4 and Neurod2 are transcription factors that are critical for neuronal differentiation and survival. Also included in this group of genes are Ntm, and DCX, among others, that are involved in establishing neuronal structure. In summary, this work establishes a global role for FMRP in the 3′UTR where it is required for AGO2 recruitment.
DISCUSSION
In this study, we characterize the FMRP-containing mRNP that regulates translation of bound mRNAs at the molecular level. Although RBPs are studied individually as recombinant proteins, it seems unlikely that they exist in cells as monomers. In fact, FMRP does not exist as a free protein in cells (83), rather it is present in large complexes as has been described for other RNA binding proteins in neuronal granules (84,85). This is one of the first studies to examine the function of a complex of RNA binding proteins, namely FMRP and MOV10 and how their interaction modulates association with rG4s and the consequence on AGO access to proximal MREs. Since MOV10 association with FMRP increases association with rG4s, the levels of MOV10 in brain and its availability for association with FMRP could explain why FMRP association with rG4s varies in brain samples (14). In fact, an FMRP HITS-CLIP experiment performed on specific brain regions of postnatal day 13 mice found an enrichment of rG4 in FMRP brain CLIP targets (44). This body of work, along with recent data showing that FMRP binding is enriched in the 3′ UTR in specific regions of the human brain (11), suggest a global role for FMRP in dynamically regulating translation through the miRNA pathway.
As previously described (38), we found that phosphorylation of FMRP affects association with AGO, though we found this to be an RNA-independent interaction. Often, the phosphorylation of a protein facilitates the necessary conformational changes–proximal or remotely to the phosphorylation site–to confer association with other proteins (40). The phosphorylation of FMRP (S500 human, S499 mouse) occurs in the C-terminal region, suggesting that phosphorylation initiates a conformational change in the molecule, allowing it to interact more strongly with AGO in the N-terminus. Likewise, the binding of the N-terminus of MOV10 to FMRP’s KH1 domain most likely changes the conformation of FMRP to facilitate rG4 binding by FMRP’s RGG box.
Our model is that FMRP recruits MOV10 to mRNAs for unwinding; however, if FMRP is bound to an rG4 first, the N-terminus of MOV10 binds FMRP to stabilize this association. It was somewhat surprising to us that removal of the RGG box of FMRP did not result in an overall reduction in the quantity of the mRNAs bound (Supplementary Figure S2A)—some of which contain rG4s. We believe that this is because the KH2 domain is the primary RNA binding domain in the coding region of the FMRP CLIP targets. For example, the MAZ mRNA contains 21 FMRP CLIP sites (10). Of those, only 3 (14%) were found in G-rich sequences and confirmed to be rG4s via rG4-seq mapping (33). We believe that the FMRP-rG4 sites in the 3′UTR are important translation regulation regions that are regulated by MOV10 binding, though these regions may minimally affect FMRP’s overall binding to specific mRNAs.
One future direction is to understand how this complex is resolved. One method is by RGG box methylation (42,43), which we showed disrupts FMRP’s rG4 binding. Another way to resolve this association is by the degradation of FMRP (86,87), which would release MOV10 to unwind the rG4 and reveal the MRE for AGO association.
A recent paper provides compelling evidence that FMRP binds some miRNAs, specifically, miR-125a and miR-125b, in regions outside of the seed sequence (37). This would be an intriguing way for FMRP to recruit specific AGO–miRNA complexes to its bound mRNAs. FMRP binding to miRNAs was shown to occur through the KH1–KH2 domain, thus it would be interesting to know if competitive binding of MOV10 to the KH1 domain competes with miRNA binding, therefore freeing the AGO–miRNA complex to bind its MRE in the 3′UTR. In addition, it is possible that the absence of the RGG box in Figure 2B could affect FMRP’s binding affinity for the RISC-associated miRNAs, which in turn would affect the RISC association with the mRNA.
Furthermore, we would like to understand how the FMRP–MOV10–AGO mRNP is regulated. A loss of MOV10 negatively affects neurite outgrowth in N2a cells and correlates to an overall decrease in gene expression (49), suggesting that MOV10 may act to inhibit translational suppression in these cells. In HEK293 cells we saw an opposite effect, where a loss in MOV10 expression correlated to an overall increase in global gene expression levels, suggesting that MOV10 acted to translationally suppress bound RNA via its helicase activity (22). Thus, it would be interesting to understand if FMRP’s bifunctional role in translational regulation is cell type-dependent and importantly, which cell type specific interactors lead to these different outcomes.
DATA AVAILABILITY
All eCLIP data files are available from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession numbers GSE129885 (AGO eCLIP-SEQ).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the following individuals for sharing reagents and for their invaluable advice: Dr Unutmaz for the N- and C-terminal MOV10 constructs, Dr Michael Ryckelynck for iSpinach sequences and advice, Dr Jennifer Darnell for FMRP and FMRP I304N and RGG mutant constructs, Dr Edouard W. Khandjian for the FMRP KH2 mutant and Dr Auinash Kalsotra for critical reading of this manuscript.
Author contributions’: S.C. and P.J.K conceived the project, performed experiments and wrote the manuscript. M.K., G.S. and J.N. performed experiments. M.C.L performed animal husbandry and isolated P0 brains from the mice.
Notes
Present address: Miri Kim, Loyola University Medical Center, Department of Neurosurgery, Maywood, IL 60153, USA.
Present address: Geena Skariah, University of Michigan, Department of Neurology, Ann Arbor, MI 48109, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health/National Institutes of Mental Health [MH093661]; Kiwanis Neuroscience Research Foundation; National Science Foundation (NSF) [1855474]. Funding for open access charge: Kiwanis Neuroscience Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Brown V., Jin P., Ceman S., Darnell J.C., O’Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D. et al.. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001; 107:477–487. [DOI] [PubMed] [Google Scholar]
- 2. Ashley C.T.J., Wilkinson K.D., Reines D., Warren S.T.. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993; 262:563–566. [DOI] [PubMed] [Google Scholar]
- 3. Kelleher R.J. 3rd, Bear M.F.. The autistic neuron: troubled translation?. Cell. 2008; 135:401–406. [DOI] [PubMed] [Google Scholar]
- 4. Lozano R., Summers S., Lozano C., Mu Y., Hessl D., Nguyen D., Tassone F., Hagerman R.. Association between macroorchidism and intelligence in FMR1 premutation carriers. Am. J. Med. Genet. A. 2014; 164A:2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bowling H., Bhattacharya A., Zhang G., Alam D., Lebowitz J.Z., Bohm-Levine N., Lin D., Singha P., Mamcarz M., Puckett R. et al.. Altered steady state and activity-dependent de novo protein expression in fragile X syndrome. Nat. Commun. 2019; 10:1710–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caudy A.A., Myers M., Hannon G.J., Hammond S.M.. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002; 16:2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ishizuka A., Siomi M.C., Siomi H.. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002; 16:2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edbauer D., Neilson J.R., Foster K.A., Wang C.-F., Seeburg D.P., Batterton M.N., Tada T., Dolan B.M., Sharp P.A., Sheng M.. Regulation of synaptic structure and function by FMRP-Associated MicroRNAs miR-125b and miR-132. Neuron. 2010; 65:373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W. et al.. FMRP Stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011; 146:247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ascano M., Mukherjee N., Bandaru P., Miller J.B., Nusbaum J.D., Corcoran D.L., Langlois C., Munschauer M., Dewell S., Hafner M. et al.. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012; 492:382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tran S.S., Jun H.-I., Bahn J.H., Azghadi A., Ramaswami G., Van Nostrand E.L., Nguyen T.B., Hsiao Y.-H.E., Lee C., Pratt G.A. et al.. Widespread RNA editing dysregulation in brains from autistic individuals. Nat. Neurosci. 2019; 22:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Siomi H., Matunis M.J., Michael W.M., Dreyfuss G.. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nuclic Acids Res. 1993; 21:1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Darnell J.C., Fraser C.E., Mostovetsky O., Stefani G., Jones T.A., Eddy S.R., Darnell R.B.. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005; 19:903–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B.. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001; 107:489–499. [DOI] [PubMed] [Google Scholar]
- 15. Ramos A., Hollingworth D., Pastore A.. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA. 2003; 9:1198–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phan A.T., Kuryavyi V., Darnell J.C., Serganov A., Majumdar A., Ilin S., Raslin T., Polonskaia A., Chen C., Clain D. et al.. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat. Struct. Mol. Biol. 2011; 18:796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vasilyev N., Polonskaia A., Darnell J.C., Darnell R.B., Patel D.J., Serganov A.. Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP. PNAS. 2015; 112:E5391–E5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alpatov R., Lesch Bluma J., Nakamoto-Kinoshita M., Blanco A., Chen S., Stützer A., Armache Karim J., Simon Matthew D., Xu C., Ali M. et al.. A Chromatin-Dependent role of the fragile X mental retardation protein FMRP in the DNA damage response. Cell. 2014; 157:869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Myrick L.K., Hashimoto H., Cheng X., Warren S.T.. Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain. Hum. Mol. Genet. 2015; 24:1733–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schenck A., Bardoni B., Moro A., Bagni C., Mandel J.L.. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8844–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu Y., Chen Z., Fu Y., He Q., Jiang L., Zheng J., Gao Y., Mei P., Chen Z., Ren X.. The amino-terminal structure of human fragile X mental retardation protein obtained using precipitant-immobilized imprinted polymers. Nat. Commun. 2015; 6:6634–6645. [DOI] [PubMed] [Google Scholar]
- 22. Kenny P.J., Zhou H., Kim M., Skariah G., Khetani R.S., Drnevich J., Arcila M.L., Kosik K.S., Ceman S.. MOV10 and FMRP regulate AGO2 association with microRNA recognition elements. Cell Rep. 2014; 9:1729–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kenny P., Ceman S.. RNA Secondary structure modulates FMRP’s Bi-Functional role in the MicroRNA pathway. Int. J. Mol. Sci. 2016; 17:985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mazroui R., Huot M.E., Tremblay S., Boilard N., Labelle Y., Khandjian E.W.. Fragile X Mental Retardation protein determinants required for its association with polyribosomal mRNPs. Hum. Mol. Genet. 2003; 12:3087–3096. [DOI] [PubMed] [Google Scholar]
- 25. Furtak V., Mulky A., Rawlings S.A., Kozhaya L., Lee K., KewalRamani V.N., Unutmaz D.. Perturbation of the P-Body component Mov10 inhibits HIV-1 infectivity. PLoS One. 2010; 5:e9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Autour A., Westhof E., Ryckelynck M.. iSpinach: a fluorogenic RNA aptamer optimized for in vitro applications. Nucleic Acids Res. 2016; 44:2491–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mooslehner K., Muller U., Karls U., Hamann L., Harbers K.. Structure and expression of a gene encoding a putative GTP-binding protein identified by provirus integration in a transgenic mouse strain. Mol. Cell Biol. 1991; 11:886–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K. et al.. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods. 2016; 13:508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith T., Heger A., Sudbery I.. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017; 27:491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. J. EMBnet. 2011; 17:10–12. [Google Scholar]
- 31. Rivals E., Delgrange O.. STAR: an algorithm to Search for Tandem Approximate Repeats. Bioinformatics. 2004; 20:2812–2820. [DOI] [PubMed] [Google Scholar]
- 32. Xue Y., Ouyang K., Huang J., Zhou Y., Ouyang H., Li H., Wang G., Wu Q., Wei C., Bi Y. et al.. Direct conversion of fibroblasts to neurons by reprogramming PTB-Regulated MicroRNA circuits. Cell. 2013; 152:82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kwok C.K., Marsico G., Sahakyan A.B., Chambers V.S., Balasubramanian S.. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Meth. 2016; 13:841–844. [DOI] [PubMed] [Google Scholar]
- 34. Guo J.U., Bartel D.P.. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016; 353:aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee E.K., Kim H.H., Kuwano Y., Abdelmohsen K., Srikantan S., Subaran S.S., Gleichmann M., Mughal M.R., Martindale J.L., Yang X. et al.. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat. Struct. Mol. Biol. 2010; 17:732–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Y., Tang W., Zhang L.-R, Zhang C.-Y.. FMRP regulates miR196a-mediated repression of HOXB8 via interaction with the AGO2 MID domain. Mol. Biosyst. 2014; 10:1757–1764. [DOI] [PubMed] [Google Scholar]
- 37. DeMarco B., Stefanovic S., Williams A., Moss K.R., Anderson B.R., Bassell G.J., Mihailescu M.R.. FMRP - G-quadruplex mRNA - miR-125a interactions: Implications for miR-125a mediated translation regulation of PSD-95 mRNA. PLoS One. 2019; 14:e0217275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muddashetty R.S., Nalavadi V.C., Gross C., Yao X., Xing L., Laur O., Warren S.T., Bassell G.J.. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell. 2011; 42:673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ceman S., O’Donnell W.T., Reed M., Patton S., Pohl J., Warren S.T.. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum. Mol. Genet. 2003; 12:3295–3305. [DOI] [PubMed] [Google Scholar]
- 40. Johnson L.N., Barford D.. The effects of phosphorylation on the structure and function of proteins. Annu. Rev. Biophys. Biomol. Struct. 1993; 22:199–232. [DOI] [PubMed] [Google Scholar]
- 41. Patton E., Stetler A., Ceman S.. Sung YJ, Denman RB. The role of phosphorylation in regulating FMRP function. The Molecular Basis of Fragile X Syndrome. 2005; Kerala: Research Signpost; 229–243. [Google Scholar]
- 42. Stetler A., Winograd C., Sayegh J., Cheever A., Patton E., Zhang X., Clarke S., Ceman S.. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum. Mol. Genet. 2006; 15:87–96. [DOI] [PubMed] [Google Scholar]
- 43. Blackwell E., Zhang X., Ceman S.. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum. Mol. Genet. 2010; 19:1314–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maurin T., Lebrigand K., Castagnola S., Paquet A., Jarjat M., Popa A., Grossi M., Rage F., Bardoni B.. HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic Acids Res. 2018; 46:6344–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zanotti K.J., Lackey P.E., Evans G.L., Mihailescu M.-R.. Thermodynamics of the fragile X mental retardation protein RGG box interactions with G quartet forming RNA. Biochemistry. 2006; 45:8319–8330. [DOI] [PubMed] [Google Scholar]
- 46. Gregersen L.H., Schueler M., Munschauer M., Mastrobuoni G., Chen W., Kempa S., Dieterich C., Landthaler M.. MOV10 Is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol. Cell. 2014; 54:573–585. [DOI] [PubMed] [Google Scholar]
- 47. Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T.. Identification of novel argonaute-associated proteins. Curr. Biol. 2005; 15:2149–2155. [DOI] [PubMed] [Google Scholar]
- 48. Goodier J.L., Cheung L.E., Kazazian H.H. Jr. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLos Genet. 2012; 8:e1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Skariah G., Seimetz J., Norsworthy M., Lannom M.C., Kenny P. J., Elrakhawy M., Forsthoefel C., Drnevich J., Kalsotra A., Ceman S.. Mov10 suppresses retroelements and regulates neuronal development and function in developing brain. BMC Biol. 2017; 15:54–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Paige J.S., Wu K., Jaffrey S.R.. RNA mimics of green fluorescent protein. Science. 2011; 333:642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Warner K.D., Chen M.C., Song W., Strack R.L., Thorn A., Jaffrey S.R., Ferré-D’Amaré A.R.. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat.Struct. Mol. Biol. 2014; 21:658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang H., Suslov N.B., Li N.-S., Shelke S.A., Evans M.E., Koldobskaya Y., Rice P.A., Piccirilli J.A.. A G-quadruplex–containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014; 10:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang X., Han Y., Dang Y., Fu W., Zhou T., Ptak R.G., Zheng Y.-H.. Moloney leukemia virus 10 (MOV10) protein inhibits retrovirus replication. J. Biol. Chem. 2010; 285:14346–14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zappulo A., van den Bruck D., Ciolli Mattioli C., Franke V., Imami K., McShane E., Moreno-Estelles M., Calviello L., Filipchyk A., Peguero-Sanchez E. et al.. RNA localization is a key determinant of neurite-enriched proteome. Nat. Commun. 2017; 8:583–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nolze A., Schneider J., Keil R., Lederer M., Hüttelmaier S., Kessels M.M., Qualmann B., Hatzfeld M.. FMRP regulates actin filament organization via the armadillo protein p0071. RNA. 2013; 19:1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stefanovic S., Bassell G.J., Mihailescu M.R.. G quadruplex RNA structures in PSD-95 mRNA: potential regulators of miR-125a seed binding site accessibility. RNA. 2015; 21:48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rouleau S., Glouzon J.-P.S., Brumwell A., Bisaillon M., Perreault J.-P.. 3′ UTR G-quadruplexes regulate miRNA binding. RNA. 2017; 23:1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kedde M., van Kouwenhove M., Zwart W., Oude Vrielink J.A.F., Elkon R., Agami R.. A Pumilio-induced RNA structure switch in p27-3′UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010; 12:1014–1020. [DOI] [PubMed] [Google Scholar]
- 59. Arthanari H., Bolton P.H.. Functional and dysfunctional roles of quadruplex DNA in cells. Chem. Biol. 2001; 8:221–230. [DOI] [PubMed] [Google Scholar]
- 60. Kostadinov R., Malhotra N., Viotti M., Shine R., D’Antonio L., Bagga P.. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006; 34:D119–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Creacy S.D., Routh E.D., Iwamoto F., Nagamine Y., Akman S.A., Vaughn J.P.. G4 Resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2008; 283:34626–34634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Beaudoin J.-D., Perreault J.-P.. 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010; 38:7022–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Subramanian M., Rage F., Tabet R., Flatter E., Mandel J.-L., Moine H.. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011; 12:697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huang H., Zhang J., Harvey S.E., Hu X., Cheng C.. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017; 31:2296–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pandey S., Agarwala P., Maiti S.. Effect of loops and G-Quartets on the stability of RNA G-Quadruplexes. J. Phys. Chem. B. 2013; 117:6896–6905. [DOI] [PubMed] [Google Scholar]
- 66. Kikin O., D’Antonio L., Bagga P.. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006; 34:W676–W682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mazroui R., Huot M.E., Tremblay S., Filion C., Labelle Y., Khandjian E.W.. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 2002; 11:3007–3017. [DOI] [PubMed] [Google Scholar]
- 68. Schaeffer C., Bardoni B., Mandel J.L., Ehresmann B., Ehresmann C., Moine H.. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001; 20:4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lee T., Pelletier J.. The biology of DHX9 and its potential as a therapeutic target. Oncotarget. 2016; 7:42716–42739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. He Q., Ge W.. The tandem Agenet domain of fragile X mental retardation protein interacts with FUS. Sci. Rep. 2017; 7:962–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang T., Wu Y.-C., Mullane P., Ji Y.J., Liu H., He L., Arora A., Hwang H.-Y., Alessi A.F., Niaki A.G. et al.. FUS regulates activity of MicroRNA-Mediated gene silencing. Mol. Cell. 2018; 69:787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kamelgarn M., Chen J., Kuang L., Jin H., Kasarskis E.J., Zhu H.. ALS mutations of FUS suppress protein translation and disrupt the regulation of nonsense-mediated decay. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E11904–E11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P.. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005; 169:871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yasuda K., Zhang H., Loiselle D., Haystead T., Macara I.G., Mili S.. The RNA-binding protein Fus directs translation of localized mRNAs in APC-RNP granules. J. Cell Biol. 2013; 203:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sephton C.F., Yu G.. The function of RNA-binding proteins at the synapse: implications for neurodegeneration. Cell Mol. Life Sci. 2015; 72:3621–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chen E., Sharma M.R., Shi X., Agrawal R.K., Joseph S.. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell. 2014; 54:407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gholizadeh S., Halder S.K., Hampson D.R.. Expression of fragile X mental retardation protein in neurons and glia of the developing and adult mouse brain. Brain Res. 2015; 1596:22–30. [DOI] [PubMed] [Google Scholar]
- 78. Das Sharma S., Metz J.B., Li H., Hobson B.D., Hornstein N., Sulzer D., Tang G., Sims P.A.. Widespread alterations in translation elongation in the brain of juvenile Fmr1 knockout mice. Cell Rep. 2019; 26:3313–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Filippini A., Bonini D., Lacoux C., Pacini L., Zingariello M., Sancillo L., Bosisio D., Salvi V., Mingardi J., La Via L. et al.. Absence of the Fragile X Mental Retardation Protein results in defects of RNA editing of neuronal mRNAs in mouse. RNA Biol. 2017; 14:1580–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sarshad A.A., Juan A.H., Muler A.I.C., Anastasakis D.G., Wang X., Genzor P., Feng X., Tsai P.-F., Sun H.-W., Haase A.D. et al.. Argonaute-miRNA complexes silence target mRNAs in the nucleus of mammalian stem cells. Mol. Cell. 2018; 71:1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chi S.W., Zang J.B., Mele A., Darnell R.B.. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009; 460:479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kim M., Bellini M., Ceman S.. Fragile X mental retardation protein FMRP binds mRNAs in the nucleus. Mol. Cell Biol. 2009; 29:214–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Feng Y., Absher D., Eberhart D.E., Brown V., Malter H.E., Warren S.T.. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1997; 1:109–118. [DOI] [PubMed] [Google Scholar]
- 84. Barbee S.A., Estes P.S., Cziko A.M., Hillebrand J., Luedeman R.A., Coller J.M., Johnson N., Howlett I.C., Geng C., Ueda R. et al.. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006; 52:997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kiebler M.A., Bassell G.J.. Neuronal RNA Granules: movers and makers. Neuron. 2006; 51:685–690. [DOI] [PubMed] [Google Scholar]
- 86. Hou L., Antion M.D., Hu D., Spencer C.M., Paylor R., Klann E.. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-Dependent Long-Term depression. Neuron. 2006; 51:441–454. [DOI] [PubMed] [Google Scholar]
- 87. Bassell G.J., Warren S.T.. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008; 60:201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All eCLIP data files are available from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession numbers GSE129885 (AGO eCLIP-SEQ).