Abstract
Ancient duplication events and retained gene duplicates have contributed to the evolution of many novel plant traits and, consequently, to the diversity and complexity within and across plant lineages. Although mounting evidence highlights the importance of whole-genome duplication (WGD; polyploidy) and its key role as an evolutionary driver, gene duplication dynamics and mechanisms, both of which are fundamental to our understanding of evolutionary process and patterns of plant diversity, remain poorly characterized in many clades. We use newly available transcriptomic data and a robust phylogeny to investigate the prevalence, occurrence, and timing of gene duplications in Lamiaceae (mints), a species-rich and chemically diverse clade with many ecologically, economically, and culturally important species. We also infer putative WGDs—an extreme mechanism of gene duplication—using large-scale data sets from synonymous divergence (KS), phylotranscriptomic, and divergence time analyses. We find evidence for widespread but asymmetrical levels of gene duplication and ancient polyploidy in Lamiaceae that correlate with species richness, including pronounced levels of gene duplication and putative ancient WGDs (7–18 events) within the large subclade Nepetoideae and up to 10 additional WGD events in other subclades. Our results help disentangle WGD-derived gene duplicates from those produced by other mechanisms and illustrate the nonuniformity of duplication dynamics in mints, setting the stage for future investigations that explore their impacts on trait diversity and species diversification. Our results also provide a practical context for evaluating the benefits and limitations of transcriptome-based approaches to inferring WGD, and we offer recommendations for researchers interested in investigating ancient WGDs in other plant groups.
Keywords: Lamiaceae, mints, phylotranscriptomics, gene duplication, whole-genome duplication, ancient polyploidy
Introduction
Duplicated genes are abundant in plant genomes and can arise via multiple mechanisms, including tandem duplications, chromosomal segmental duplications, or whole-genome duplications (WGDs; polyploidy). On average, 64.5% of annotated genes in plant genomes have a duplicate copy, and comparative studies suggest that most are derived from WGD (Panchy et al. 2016).
In angiosperms, the prevalence of WGD and its role as a key driver in the diversification of species and traits have become increasingly evident. An estimated 35% of all extant angiosperm species are considered recent polyploids, yet past polyploidy events, accompanying the divergence of many extant lineages, are also found throughout angiosperm phylogeny (Wood et al. 2009; Jiao et al. 2011, 2014; Husband et al. 2012; Tank et al. 2015; McKain et al. 2016; Landis et al. 2018). Ancient polyploidy has preceded the diversification of all seed plants, angiosperms, eudicots, and monocots (Tuskan et al. 2006; Tang et al. 2010; Jiao et al. 2011, 2012; Amborella Genome Project 2013; Li et al. 2015). Furthermore, recent evidence indicates that WGD has occurred much more frequently than traditionally appreciated both within angiosperms (Landis et al. 2018) and in most land plant lineages (One Thousand Plant Transcriptomes Initiative forthcoming).
WGD can arise through multiple avenues in plants (Ramsey and Schemske 1998; Comai 2005), and it is a common feature of many species-rich clades (Soltis et al. 2009). The novel genetic material resulting from WGD events (e.g., Comai 2005; Freeling and Thomas 2006; Van de Peer et al. 2009; Rensing 2014; Soltis et al. 2014, 2015) may contribute to key evolutionary innovations that are central to rapid radiations (e.g., Edger et al. 2015; Soltis and Soltis 2016; Green Plant Consortium, unpublished). For instance, ancient WGDs contributed to the proliferation of novel genes and gene interactions in Brassicales, vastly increasing specialized metabolite diversity in the clade, particularly within Brassicaceae (mustards). Importantly, this expansion of chemical diversity was accompanied by an increase in species diversification rates (Edger et al. 2015), highlighting the importance of WGD as a driver of both chemical innovation and speciation in plants (Soltis et al. 2018).
Despite increasing evidence for the prevalence of WGDs and their impacts on trait evolution (reviewed in Van de Peer et al. [2009]), gene duplication dynamics and mechanisms remain underexplored in most plant clades. Lamiaceae (mints) are one of the most species-rich angiosperm families, with more than 7,000 species, and possess a high degree of chemical diversity and many ecologically, economically, and culturally important species appreciated by peoples worldwide. Beyond differences in mint genome sizes (MEGC 2018) and well-documented examples of auto- and allo-polyploidy, the latter of which is common throughout the clade (Harley et al. 2004), little is known about gene duplication dynamics and mechanisms in Lamiaceae. For example, it is unclear how often WGD events have occurred throughout mint evolutionary history and whether gene or WGD events have occurred with greater frequency in species-rich mint lineages such as Nepetoideae (>3,300 species) as compared with other lineages with much lower species richness.
Recent WGDs are evident through comparison of chromosome numbers, but evidence of ancient WGD is often erased through postpolyploid diploidization, making detection from chromosome numbers difficult or impossible. Detection of ancient WGD has relied largely on estimates of synonymous substitutions per synonymous site (KS) among paralogous sequence pairs present in EST or transcriptome data sets, which are subsequently used to infer the ages of gene duplications within individual species (Maere et al. 2005). At the genome level, KS distributions representing all paralogous gene pairs allow for identification of putative WGD events, which appear as punctuated episodes of large-scale gene duplication in time and stand out against background levels of gene duplication arising from other processes (Lynch and Conery 2000; Cui et al. 2006; Barker et al. 2008). Recently, researchers have used phylotranscriptomic approaches to infer and place putative WGDs in a multispecies phylogenetic context. Two of these approaches consider gene or gene family trees and reconcile gene duplications using a species tree reference (Li et al. 2015; McKain et al. 2016), whereas another method estimates the ages of gene duplication events and places them within the context of a dated species tree (Jiao et al. 2011, 2014).
To better understand the prevalence, occurrence, and timing of gene duplications in Lamiaceae, we use a combination of KS, phylotranscriptomic, and divergence time approaches to document and characterize gene duplication dynamics and to infer ancient polyploidy in the clade, taking advantage of recently available transcriptomic resources for mints and a robust phylogeny based on data from 520 nuclear genes (MEGC 2018). We compare patterns of putative ancient WGD events across 11 major mint subclades (=traditional subfamilies) and correlate these patterns with their species-level diversity. We also compare and discuss the congruence of results across different analyses as well as the efficacy of these approaches. Most importantly, we provide a set of hypotheses for gene duplication dynamics and ancient polyploidy that can be used in future investigations that explore their impacts on mint diversity and diversification.
Materials and Methods
Species Tree and Transcriptome Data Used in This Study
We sourced a robust species tree hypothesis (fig. 1a) and available leaf transcriptomes from 48 mint species and four outgroup species from Lamiales (supplementary table S1, Supplementary Material online) generated by the Mint Evolutionary Genomics Consortium (2018) from the Dryad Digital Repository (doi:10.5061/dryad.tj1p3). The evolutionary sampling scheme from the Mint Evolutionary Genomics Consortium (2018) study reflects our current understanding of Lamiaceae phylogeny and includes species from 11 of 12 recognized subclades, thereby representing very well the phylogenetic diversity of mints and providing a suitable data set for investigation of gene duplication dynamics and ancient WGD in the clade. All transcriptome data sets were prefiltered and included both Transdecoder-predicted (Haas et al. 2013) peptides and coding sequences from representative transcriptome assemblies (RTAs)—that is, filtered transcriptomes that included only the longest assembled isoform for each gene (see Mint Evolutionary Genomics Consortium [2018] for methods and details).
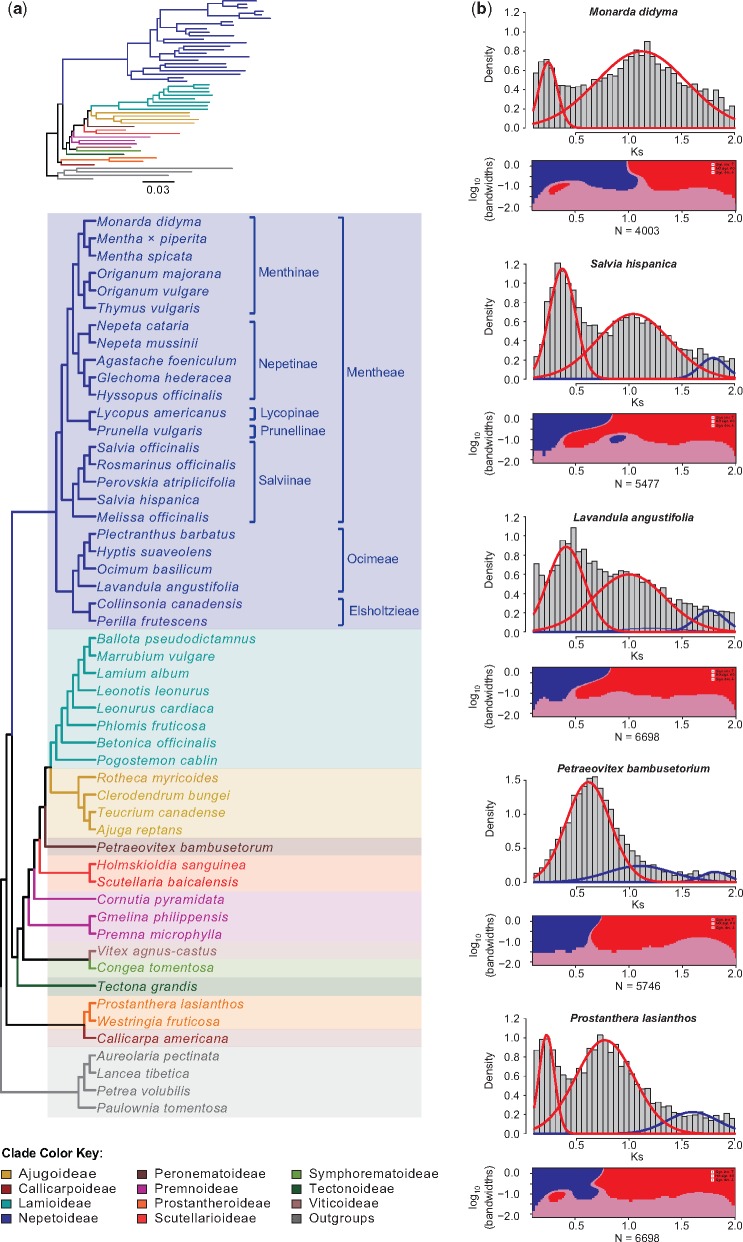
Fig. 1.
—(a) Maximum likelihood phylogeny of Lamiaceae inferred from 520 single-copy nuclear genes by the Mint Evolutionary Genomics Consortium (2018). The tree topology is shown as a cladogram, with a phylogram illustrating the tree shape as an inset. Subfamilial classifications (Li et al. 2016; Li and Olmstead 2017) are color coded according to the provided key, and tribal and subtribal classifications for Nepetoideae (Harley et al. 2004; Drew and Sytsma 2012) are indicated with brackets. (b) KS (top) and SiZer (bottom) plots for a selection of five species sampled by our study. Gaussian distributions produced by mixture models are shown as overlays on each KS distribution, with red or blue color-coded peaks representing putative WGD events that were either corroborated or not corroborated, respectively, by a SiZer analysis (Chaudhuri and Marron 1999). As shown in plots below each KS distribution, SiZer tests for significant increases (blue) or decreases (red), or no significant changes (pink) across a distribution at various (log transformed) bandwidths to distinguish true data features from noise.
Inferring WGDs from Estimates of Synonymous Divergence
The fraction of synonymous substitutions per synonymous site (KS) between a given pair of paralogous sequences can be used to estimate their time of duplication (Lynch and Conery 2000; Blanc and Wolfe 2004; Maere et al. 2005; Cui et al. 2006). At the genome level, KS distributions representing all paralogous gene pairs can reveal temporal patterns of gene duplication that may reflect ancient WGD events (Lynch and Conery 2000). This methodology has been widely used to assess putative ancient WGDs. We analyzed RTAs from 52 species (48 ingroup and 4 outgroup) with DupPipe (Barker et al. 2010). DupPipe uses a reciprocal best-BLAST-hit approach to identify pairs of putatively paralogous transcripts (gene duplicates) and estimates their pairwise synonymous site divergence (KS) from protein-guided DNA alignments with PAML and the F3 × 4 model (Yang 2007). The resulting KS estimates from each species were plotted as a histogram (KS plot) with age distributions from 0.1 to 2; all gene pairs with KS values below and above these upper and lower values, respectively, were removed to enable reliable inference of WGD events (e.g., Vanneste et al. 2013). Significant peaks in each observed KS distribution were inferred with Gaussian mixture models, as implemented in the mixtools R package (Benaglia et al. 2009) with the expectation-maximization (EM) algorithm (McLachlan and Peel 2000), and the most likely number of Gaussian components that fit each distribution was tested with parametric bootstrap analyses (100 bootstraps) of the likelihood ratio statistic using the “boot.comp” function of mixtools. In cases where Gaussian peaks did not align with KS peaks or multiple peaks were placed in approximately the same position, the number of components was manually adjusted to better fit the KS distribution. We further compared all mixture model components with results from a SiZer analysis (Chaudhuri and Marron 1999). SiZer tests for significant increases or decreases or no significant changes across a distribution at various bandwidths to distinguish true data features from noise. Values of KS ≤ 2 and bandwidths ranging from KS = 0.01 to KS = 2 were used to identify significant (α = 0.05) features in each observed KS distribution. Shifts from significant increases to significant decreases identified by SiZer were interpreted as true peaks, and all peaks corroborating Gaussian mixture models were inferred as WGD events.
Inferring and Placing WGDs in a Phylogenetic Context
Although KS plots are useful for documenting putative WGD events that occurred within the evolutionary histories of individual species, phylotranscriptomic approaches utilize transcriptome data from multiple species and test for the occurrence of putative WGDs across all nodes of a given species tree to facilitate detection of shared WGDs across lineages. To characterize patterns of ancestral polyploidy in Lamiaceae, we inferred and placed putative WGD events in the context of our best species tree hypothesis (fig. 1a) using two available pipelines: Multi-tAxon Paleopolyploidy Search (MAPS) (Li et al. 2015) and Phylogenetic Placement of Polyploidy Using Genomes (PUG) (McKain et al. 2016).
Guide Tree Preparation
The MAPS algorithm searches gene family tree topologies and identifies where paralogs coalesce within the context of a user-provided species tree with stepwise branching. Because our species tree topology for Lamiaceae (fig. 1a) does not have a stepwise branching pattern, we followed Li et al. (2015, 2018) and systematically pruned taxa from the tree in R using the APE package (Paradis et al. 2004), producing 12 stepwise guide trees for use with MAPS (supplementary fig. S1, Supplementary Material online). Each guide tree contained a focal clade (i.e., a subclade) from the species tree, and the taxon representation in its sister clade and in all other subclades in the species tree was reduced to a single individual to produce a tree with a stepwise branching pattern. Collectively, these guide trees enabled investigations of WGD events across most nodes in our (unpruned) species tree. Preparation of a guide tree was not necessary for use of PUG.
Gene Family Circumscription
We precomputed reciprocal protein BLAST (BlastP) searches among RTAs comprising Transdecoder-predicted peptide sequences from all 52 species using NCBI BLAST version 2.2.31 (Altschul et al. 1990) with an e-value threshold of 0.001. Gene families (i.e., orthogroups comprising gene orthologs and close paralogs) were circumscribed from the BLAST results with OrthoFinder (Emms and Kelly 2015) version 0.7.1 using default parameters. The resulting gene family data were processed directly in preparation for the PUG analysis. However, additional OrthoFinder processing was necessary to prepare individual data sets for each of the MAPS analyses. For the latter, we systematically reclustered BLAST output using the corresponding taxon sampling in each MAPS guide tree (supplementary fig. S1, Supplementary Material online), producing new gene family circumscriptions for each MAPS data set. In all, 13 independent gene family data sets with partial or complete taxon sampling (i.e., 12 for MAPS and one for PUG, respectively) were produced for downstream filtering, multiple sequence alignment, tree estimation, and analysis.
Filtering, Multiple Sequence Alignment, and Tree Estimation
Amino acid sequences from gene families occupied by at least one sequence per species represented in a corresponding guide tree were parsed and passed to PASTA (Mirarab et al. 2015) for automated multiple sequence alignment and phylogenetic inference; all remaining data were discarded. Sequences were divided into subsets and aligned using MAFFT (Katoh et al. 2002), with MUSCLE (Edgar 2004) and RAxML (Stamatakis 2014) used for pairwise merger of aligned subsets and tree estimation, respectively, according to the default parameters set by PASTA for each tool. For each gene family, we ran PASTA until no improvement in the likelihood score was reached after three iterations using a centroid break strategy. We followed Li et al. (2015) and compiled the best-scoring amino acid-based PASTA trees for each data set for use as tree input in their respective MAPS analyses. To prepare tree input for PUG, we followed McKain et al. (2016). Corresponding coding sequences were forced onto the amino acid alignments from PASTA using PAL2NAL (Suyama et al. 2006), and gene family trees were estimated from the resulting nucleotide alignments. All phylogenetic analyses with nucleotide sequence data were conducted using RAxML version 8.2.10 and the GTRGAMMA model, and topological uncertainty in each of the resulting maximum likelihood (ML) trees was assessed with 100 rapid bootstraps. The resulting bootstrapped (nucleotide-based) ML trees were compiled into a single file for analysis with PUG.
MAPS and PUG Analyses
We ran 12 MAPS analyses to infer and place putative WGD events in Lamiaceae in a phylogenetic context. A compiled set of multispecies gene family phylogenies and corresponding guide tree were used as input for each analysis. Using MAPS output, we identified nodes in each guide tree that exhibited shared duplications among descendent taxa in ≥40% of gene family trees with congruent topologies and had higher levels of duplication relative to neighboring nodes. We interpreted nodes at or above this threshold as putative WGD nodes, following Li et al. (2015) and Barker et al. (2016). We summarized results across all MAPS analyses by resolving WGD nodes in each guide tree to our species tree topology. For comparison, we also ran an analysis with PUG to infer and place WGD events in the context of our species tree. The “estimate_paralogs” option in PUG was used to identify all putative gene paralog pairs in each gene family phylogeny, whose coalescence nodes were subsequently identified and resolved to our species tree topology. We accounted for possible phylogenetic uncertainty in each gene family tree topology and applied a stringent minimum bootstrap threshold to filter our results and confidently place gene duplication events within the context of our species tree; following McKain et al. (2016) and Unruh et al. (2018), only paralog pair coalescence nodes with ML bootstrap values ≥80 in each gene family tree were retained and interpreted as part of the final data set. We used an R script, PUG_Figure_Maker.R (Unruh et al. 2018), to identify putative WGDs in our species tree. The script identifies the maximum number of paralog pairs mapped to any given node in the species tree and labels the stem lineage subtending this and all other nodes with ≥10% of the maximum number as having putative WGD events.
Inferring and Placing WGDs in a Temporal Context
Although the phylotranscriptomic approaches described above are useful for placing gene duplication events, they do not account for factors such as the phylogenetic timing and ages of gene duplications, both of which are useful for identifying putative shared WGD events. We investigated both factors and placed gene duplications within a time-calibrated phylogeny to identify temporal patterns of punctuated gene duplication that are indicative of putative ancient WGD events (Lynch and Conery 2000). We also compared these results with putative WGD events inferred with KS and other phylotranscriptomic approaches.
Divergence Time Estimations
We extracted a set of gene family trees for molecular dating that could be reliably rooted with all outgroup species (using the APE package in R) and contained putative gene paralog pairs identified by PUG. Divergence times were estimated for species in our species tree for Lamiaceae (fig. 1a) and for paralog pairs in each gene family tree assuming a relaxed molecular clock, as implemented in treePL (Smith and O’Meara 2012) with a penalized likelihood approach (Sanderson 2002). We used the “prime” option in treePL to identify the best optimization parameters for each analysis, which was followed by a “thorough” analysis using those parameters. The best smoothing parameter for each analysis was determined by cross-validation. We used the following age constraints for the estimation procedure: a minimum and maximum age of 59.99 and 70.94 Myr, respectively, for the crown node of Lamiaceae, and a maximum age of 107 Myr for the root node (Lamiales crown). These constraints correspond to the lower and upper bounds of the 95% highest posterior density interval of estimated ages reported for the Lamiaceae crown (Yao et al. 2016) and the oldest estimated age (upper confidence interval) reported for the Lamiales crown (Janssens et al. 2009), respectively. All divergence times for paralog pair coalescence nodes inferred with PUG were extracted with the APE R package and compiled for downstream analyses; Estimated ages for constrained nodes were not considered.
Statistical Analyses
We extracted and analyzed the estimated divergence times for paralog pairs representing unique duplication events resolved by PUG to individual nodes in our species tree and used these ages to place duplications within the context of our time-calibrated mint phylogeny. We also assessed whether gene duplication events at each node were clustered in time (i.e., representing ≥1 putative WGD event). For the latter, we followed Cui et al. (2006) and compared each observed distribution of gene duplication ages with an expected (null) distribution that was generated under a constant-rate birth–death model. The null model forms a declining exponential distribution whose decay rate was informed by an observed age distribution. The null distribution differs from possible WGD scenarios, where observed distributions are expected to show temporal patterns of punctuated gene duplication. We estimated a decay parameter for nodes in our species tree that had observed age distributions with samples sizes ≥5 and used it to randomly generate a null distribution that was subsampled to reflect the range and size of the observed distribution. The decay parameter for each test node corresponded to its expected gene death rate, which was estimated from the sample mean of its observed age distribution. Unlike Cui et al. (2006), who incorporated error into their analyses (with KS data), we accounted for possible outliers in our observed age distributions and removed the 1.5th and 99.5th percentiles prior to estimating decay parameters and generating null distributions. We also accounted for possible subsampling artifacts when simulating null distributions and repeated the process 5,000 times. To compare the observed distribution of gene duplication ages at each test node with each simulated null model, we used the bootstrap version of the univariate Kolmogorov–Smirnov (KS) test, as implemented with 1,000 replicates in R with the “ks.boot” function (http://sekhon.polisci.berkeley.edu/matching/ks.boot.html).
Our strategy produced a distribution of 5,000 bootstrapped P values for each test node, which we examined to identify observed age distributions that bore signatures of putative ancient WGDs. If KS tests with an observed age distribution yielded a relatively uniform distribution of P values (0 ≤ P ≤ 1), we concluded that the distribution was largely consistent with the null model (Breheny et al. 2018). Conversely, if the P value distributions showed a large skew toward marginal or significant P values (P < 0.05), we concluded that the distribution was inconsistent with the null model and performed additional tests to characterize possible age clustering. For the latter, we inferred data features within each observed distribution using finite Gaussian mixture models, as implemented using the Mclust function of the “mclust” R package with the EM algorithm (Fraley and Raftery 2002). For the univariate data used here, the EM algorithm identifies data features under variable variance models and tests a range of possible numbers of mixture components representing individual gene duplication age clusters; the optimal number of components is selected using the Bayesian Information Criterion. We tested a range of one to three possible mixture components per distribution and inferred the optimal number of components within each. Given that mixture models could incorrectly infer one or more components within some broad age distributions (Johnson et al. 2016), we performed an additional test to assess whether mixture model components represented “true” data features (i.e., clusters of gene duplication in time). We conducted a SiZer analysis to visualize the data and test for significant data features in each distribution, as described above for KS analyses. Clusters of gene duplication ages inferred with mixture models that were corroborated by our SiZer results were interpreted as putative WGD events.
Inferring Associations between Species Richness and Cumulative WGDs
Associations between lineage-specific species richness and cumulative levels of ancient polyploidy were examined using Kendall’s tau-b (τb) correlation coefficient, which provides a nonparametric measure of the strength and direction of association between these variables and enables adjustments for ties within the rankings. Plant name records from The Plant List (www.theplantlist.org) were downloaded and curated, and numbers of currently recognized species were summed for each of the following clades (=traditional subfamilies) to produce species richness estimates: Ajugoideae, Callicarpoideae, Lamioideae, Nepetoideae, Peronematoideae, Premnoideae, Prostantheroideae, Scutellarioideae, Symphorematoideae, Tectonoideae, and Viticoideae (see fig. 1a). Cumulative levels of ancient polyploidy were compiled for each clade by summing the number of putative ancient WGD events inferred from MAPS, PUG, and divergence times results, respectively, and placed at or near nodes within each clade and all ancestral nodes through the most recent common ancestor (MRCA) of Lamiaceae. All statistical analyses were conducted in R version 3.5.3 (R-Core-Team 2019).
Results
WGDs Inferred from KS Distributions
We observed peaks of gene duplication consistent with WGD events in the KS distributions of all study species (fig. 1b and supplementary fig. S2, Supplementary Material online). Our mixture modeling identified two to four significant peaks per KS distribution, but only one to three of these were corroborated by our SiZer results (supplementary fig. S2 and table S2, Supplementary Material online). At least 1 putative WGD event per each of the 52 species analyzed was inferred. However, 17 species showed evidence for 2 putative WGDs, and 1 species, Paulownia tomentosa (Thunb.) Steud., had significant peaks corresponding to 3 putative WGDs. The range of mean KS values estimated by mixtools for peaks confirmed by SiZer ranged from 0.21 to 1.657 (supplementary table S2, Supplementary Material online).
Phylogenetic Analyses and Placement of WGDs
Gene Family Circumscription, Multiple Sequence Alignment, and Tree Estimation
The number of transcriptomes (RTAs) and genes comprising each MAPS data set varied considerably (supplementary fig. S1 and table S3, Supplementary Material online), with 5–18 species and 227,344–830,078 genes represented in each set, respectively. OrthoFinder circumscribed ∼67% of the genes in each data set into gene families (orthogroups) and yielded a range of 25,731–46,731 gene families per data set (mean = 34,124; median = 33,319). More than 8,000 gene families in each MAPS data set had full species representation, and sequences from these families were filtered for phylogenetic analysis (supplementary table S3, Supplementary Material online). We successfully generated amino acid sequence alignments and corresponding ML trees with PASTA for gene families comprising each filtered MAPS data set, albeit with a few exceptions. PASTA failed to complete analysis for 18 gene families distributed among 4 data sets, and we excluded these data from downstream analyses. The OrthoFinder analysis of sequence data comprising our PUG data set, which included RTAs from all 52 species and over 2 million gene sequences, yielded 89,266 gene families containing two-thirds of the total genes analyzed (supplementary table S3, Supplementary Material online). In all, 6,336 gene families had full species representation, and multiple sequence alignments and ML trees were successfully generated for ∼97% of these families and used for the PUG analysis; PASTA alignments or RAxML trees were not obtained for 174 gene families.
MAPS Results
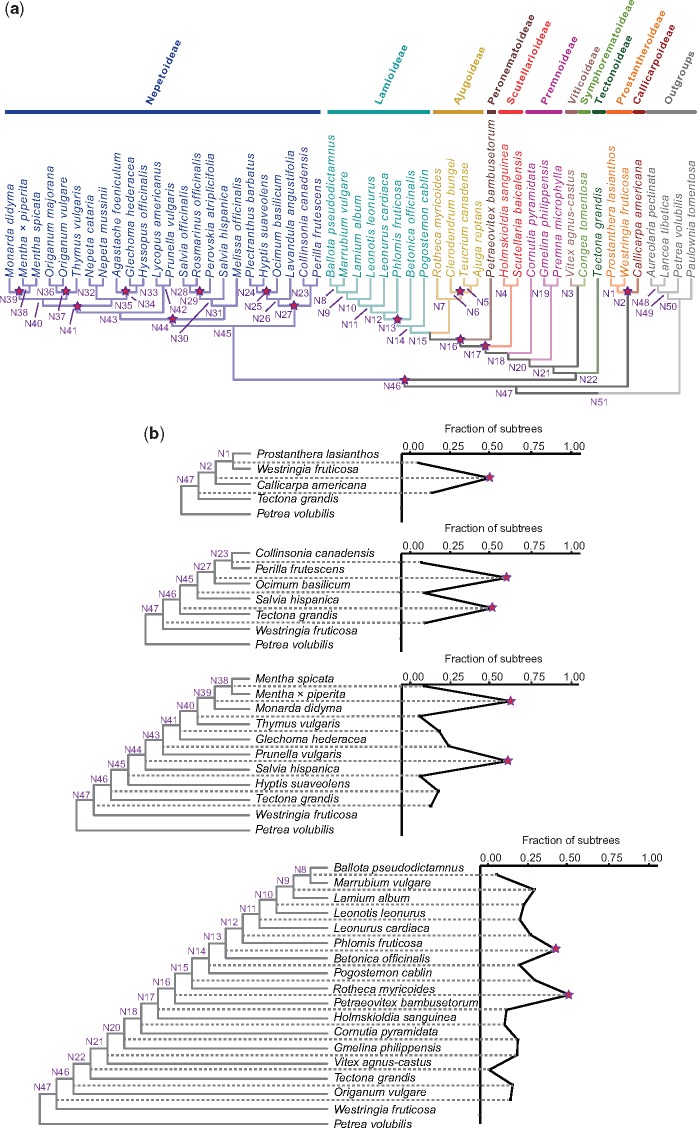
Analyses of gene family tree data and guide trees with MAPS identified putative WGD events in 10 of 12 data sets (supplementary fig. S1, Supplementary Material online). As summarized on our best species tree hypothesis for Lamiaceae, we identified pronounced gene duplication levels consistent with WGDs at 14 nodes (fig. 2a). Each of these nodes showed a characteristic upsurge in gene duplication levels relative to neighboring nodes and had a high proportion of gene family trees with shared gene duplications observed across descendent taxa (i.e., ≥40% of the gene trees analyzed were topologically congruent with respect to the guide tree and showed evidence for duplication at that node) (supplementary fig. S1 and table S4, Supplementary Material online). Because the species tree was subsampled to generate stepwise guide trees for MAPS, six species tree nodes (fig. 2a:N16, N17, N27, N41, N44, and N46) were represented more than once in our set of guide trees and, consequently, were tested more than once across individual MAPS analyses. MAPS consistently inferred putative WGD events at only three of these nodes (fig. 2a:N17, N41, and N46). As for the remaining nodes with putative WGDs, eight were tested only once (fig. 2a:N2, N6, N13, N25, N29, N34, N37, and N39). The total number of putative WGDs inferred by MAPS as well as the numbers and proportions of gene family trees showing shared gene duplications across MAPS data sets were most pronounced within Nepetoideae, especially within the species-rich subclade Mentheae. We inferred putative WGDs at the node representing the MRCA of Mentheae (fig. 2a:N44) and at five additional nodes within most major subclades comprising this clade (fig. 2a:N29, N34, N37, N39, and N41). Two additional WGDs were identified by MAPS within Nepetoideae—one at the node representing the MRCA of Elscholzieae + Ocimeae (fig. 2a:N27) and another at the MRCA of Ocimeae (fig. 2a:N25)—for a sum of eight WGD events in Nepetoideae. Outside Nepetoideae, MAPS inferred six WGDs. These events were placed at the MRCA of Callicarpoideae and Prostantheroideae (fig. 2a:N2), the MRCA of Nepetoideae and a grade of Tectonoideae + Symphorematoideae + Viticoideae s.s., Premnoideae + Scutellarioideae + Peronematoideae + Ajugoideae + Lamoideae (fig. 2a:N46), the MRCA of Scutellarioideae and Premnoideae + Scutellarioideae + Preonematoideae + Ajugoideae + Lamoideae (fig. 2a:N17), the MRCA of Peronematoideae and Ajugoideae + Lamoideae (fig. 2a:N16), and at a single node each within Lamioideae (fig. 2a: N13) and Ajugoideae (fig. 2a:N6).
Fig. 2.
—(a) Putative WGD events in Lamiaceae inferred and placed with the MAPS pipeline (Li et al. 2015). Summarized here on our best species tree hypothesis (see fig. 1) are 14 ancient polyploidy events. Red stars denote nodes with pronounced gene duplication levels consistent with WGDs that were identified from at least one of 12 independent MAPS analyses. Node numbers correspond to PUG output (see fig. 3) and are included here to facilitate results comparisons across analyses. (b) Examples of guide trees used with MAPS (left), with plots (right) showing the percentage of subtrees in each analysis that contained a gene duplication. Red stars denote putative WGD nodes, which showed duplications in ≥40% of the gene trees analyzed and pronounced gene duplication levels relative to neighboring nodes. Refer to supplementary figure S2, Supplementary Material online, for the full set of MAPS guide trees and corresponding plots.
PUG Results
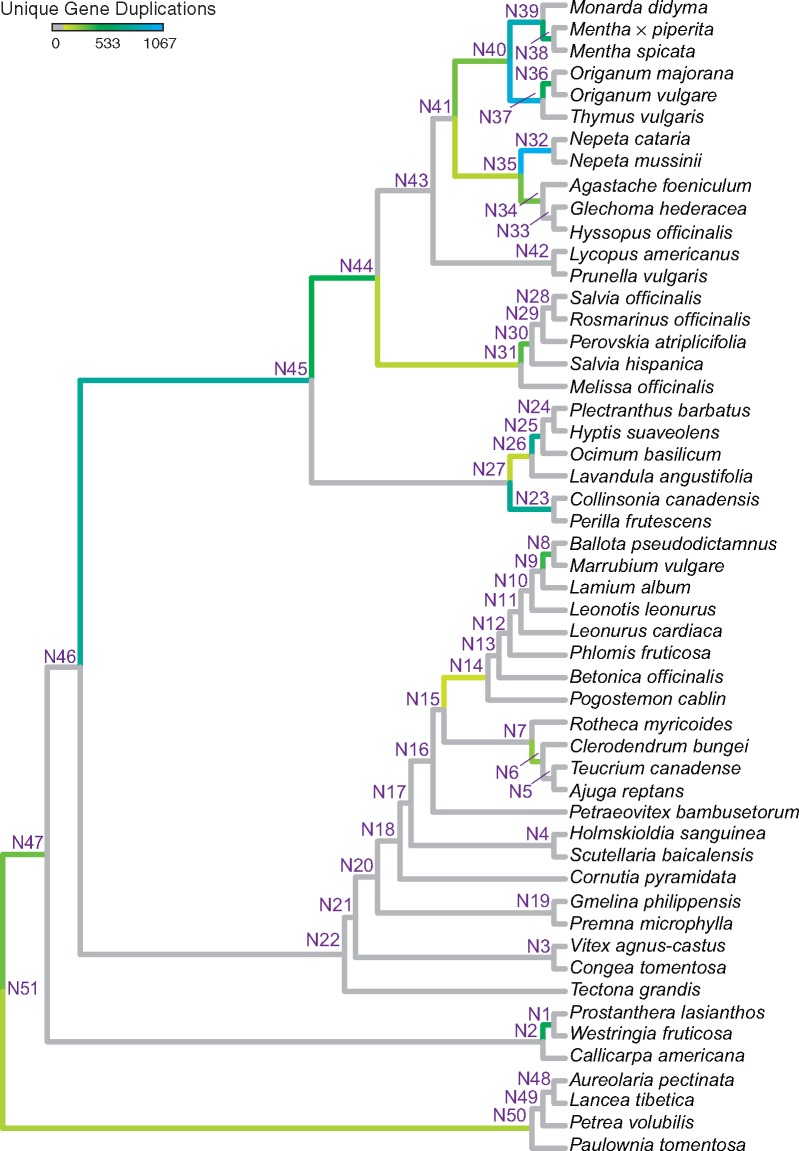
Our PUG analysis identified 168,547 putative paralog pairs within the filtered set of 6,162 gene family trees. However, only 10,690 of these pairs resolved to nodes supporting our species tree topology (fig. 1a) and surpassed our minimum bootstrap threshold (ML BS ≥ 80) for inclusion in the final data set (supplementary table S5, Supplementary Material online). These pairs were interpreted as uniquely mapped duplication events and provided evidence for 21 putative WGD events distributed across the species tree (fig. 3), including one WGD within the outgroup stem lineage (201 duplication events; fig. 3: N50), a second WGD within the Lamiaceae stem lineage (332 duplication events; fig. 3: N47), and 19 additional WGDs within Lamiaceae (fig. 3: descendants of N47). Regarding the latter, it is noteworthy that the greatest number of uniquely mapped duplications and inferred WGD events were observed for Nepetoideae (i.e., 15 WGDs and 8,397 duplication events in total, including the Nepetoideae stem lineage [fig. 3: N45]), with pronounced levels of duplication observed within Mentheae. Outside Nepetoideae, additional WGDs were inferred within the stem lineages of Prostantheroideae and Lamioideae (fig. 3: N1 and N14, respectively), within the stem lineage of Ballota + Marrubium (Lamioideae) (fig. 3: N8), and within the stem lineage of Clerodendrum + Teucrium + Ajuga (Ajugoideae) (fig. 3: N6).
Fig. 3.
—Putative WGD events in Lamiaceae inferred and placed with the PUG pipeline (McKain et al. 2016). Twenty-one stem lineages with putative ancient polyploidy events are shown in color, with total numbers of unique duplication events color coded according to the heat map key provided. Node numbers correspond to PUG output and are included to facilitate comparisons across analyses.
Divergence Time Results
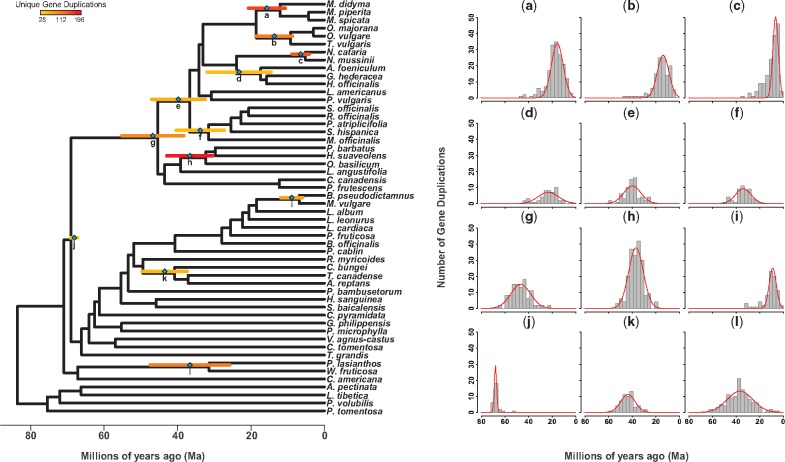
Of the 6,181 gene family trees comprising the PUG data set, 4,021 trees contained well-supported (ML BS ≥ 80) nodes representing gene duplication events in our species tree (supplementary table S5, Supplementary Material online). However, only 910 of these trees could be reliably rooted with all outgroup species and were filtered for phylogenetic dating. The final dated tree set included estimated ages for 1,868 uniquely mapped gene duplication events corresponding to 43 species tree stem lineages (supplementary tables S5 and S6, Supplementary Material online), and significant (P < 0.05) KS test results revealed that 12 of these had nonuniform data distributions with clustered gene duplication ages (supplementary figs. S3 and S4, Supplementary Material online). Mixture modeling (supplementary table S7, Supplementary Material online) identified as many as 2 age clusters in 6 of the 12 distributions, but only 1 “true” cluster (or peak) per distribution was validated by SiZer analyses (supplementary fig. S3 and table S7, Supplementary Material online). Following all validation procedures, we identified 12 stem lineages in Lamiaceae with gene duplication age distributions consistent with WGD (fig. 4a–l). Many of these WGDs were placed in Nepetoideae (eight total), including one event (fig. 4g) in the stem lineage of the clade at ∼46.72 Ma and within seven sublineages with estimated ages from ∼6.47 to 39.76 Ma (fig. 4a–f and h; supplementary table S7, Supplementary Material online). Only four events were inferred outside Nepetoideae. Of these, one putative WGD (fig. 4j) was placed at ∼68.12 Ma within a stem lineage that includes Nepetoideae and a grade of lineages comprising Tectonoideae, Symphorematoideae, Viticoideae s.s., Premnoideae, Scutellarioideae, Peronematoideae, Ajugoideae, and Lamioideae. The remaining events were placed within the following stem lineages: Prostantheroideae (fig. 4l) at ∼36.6 Ma; Clerodendrum + (Teucrium + Ajuga) at ∼43.52 Ma; and Ballota + Marrubium (fig. 4i) at ∼8.91 Ma (supplementary table S7, Supplementary Material online).
Fig. 4.
—Phylogenetic distribution and timing of putative WGD events in Lamiaceae. Shown here as labeled bars (a–l) on a species tree chronogram are 12 putative ancient polyploidy events inferred from analyses of divergence times corresponding to 1,868 uniquely mapped gene duplication events. The mean age of each event is indicated with a teal diamond, with error bars representing ± one standard deviation the mean (i.e., ∼68% of each observed age distribution). Color coding corresponds to the number of dated gene duplicates associated with each event, as defined in the provided key. Gene duplication age distributions corresponding to each event are also shown as histograms, with means and standard deviations provided by mixture models.
Associations between Species Richness and Cumulative WGDs
After records filtering and taxonomic curation, we compiled a list of 7,193 accepted species names that could be assigned to one of 12 major mint clades (=traditional subfamilies). Of the 11 clades represented in our study (Cymarioideae not sampled), Nepetoideae (3,302 species), Lamioideae (1,488 species), and Ajugoideae (947 species) had high species richness and higher cumulative levels of ancient polyploidy than the other mint clades (supplementary table S8, Supplementary Material online). Across Lamiaceae, Kendall’s τb correlation tests revealed strong, positive relationships between lineage-specific species richness and cumulative levels of ancient polyploidy (τb ≥ 0.58, P < 0.02), regardless of whether putative ancient WGD events were inferred from MAPS, PUG, or divergence time results (supplementary fig. S5, Supplementary Material online).
Discussion
Ancient Polyploidy in the Mints
Ancient polyploidy events have been identified in Lamiales by a number of studies (Wang et al. 2014; He et al. 2016; Edger et al. 2017; Van de Peer et al. 2017; Ren et al. 2018), but sparse sampling within this species-rich lineage (∼45,000 species) has limited characterization and placement of putative ancient WGD events within internal clades such as Lamiaceae, which was either unsampled or represented by one or two species in previous studies. Phylogenetic evidence from an investigation of floral regulatory gene duplications (Aagaard et al. 2005) suggests that at least one lineage-specific ancient WGD event occurred within the mints. However, until now, ancient polyploidy has never been formally investigated in mints and with taxon sampling spanning the phylogenetic diversity of the clade.
Our study is the first to characterize patterns of gene duplication broadly in Lamiaceae, and we make use of available transcriptome data as well as KS, phylotranscriptomic, and divergence time analyses to infer and place putative ancient WGD events within the context of our best estimate of mint phylogeny. Here, we present several lines of evidence revealing widespread but asymmetrical levels of gene duplication and ancient polyploidy in Lamiaceae—evidence derived from multiple, large-scale data sets (nearly 127,000 gene trees).
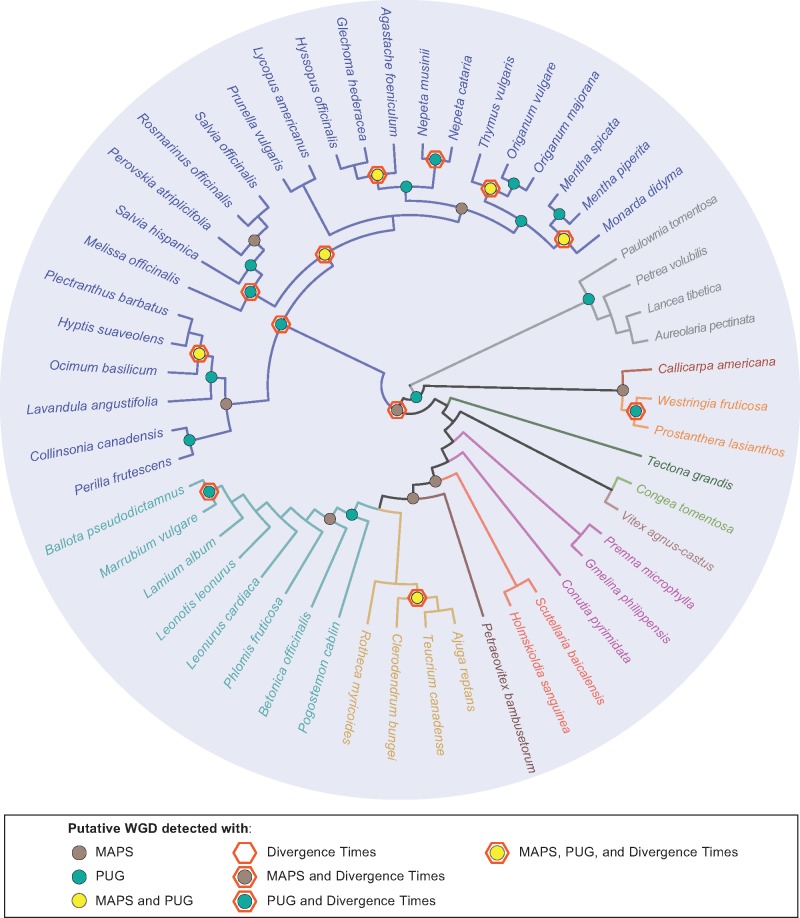
The prevalence of putative WGDs in Lamiaceae is striking, as emphasized by the one or more events detected in each of our sampled mint transcriptomes (supplementary fig. S2 and table S2, Supplementary Material online) and the large numbers of putative ancient WGDs inferred and placed on the species tree by phylotranscriptomic and divergence time analyses (figs. 2–5). Across all hypotheses generated by our study, we uniquely inferred and placed as many as 28 putative ancient WGDs in the mint family (fig. 5), exceeding maximum estimated numbers of events reported in previous studies for larger clades such as Malpighiales (>16,000 species), Poales (∼21,000 species), and Asteraceae (24,000–35,000 species) (i.e., 22–24 [Cai et al. 2019], 9–14 [McKain et al. 2016], and 6–17 [Huang et al. 2016] ancient WGD events, respectively). However, we also caution that WGD hypotheses were not consistent across analyses. Of the 14 and 21 putative WGDs inferred with MAPS and PUG, respectively, only 6 were consistent between the two and with divergence time-derived hypotheses. Nearly all of these consistent WGDs were placed within Nepetoideae, providing some of the most compelling evidence for ancient polyploidy in the family. However, in all, 12 of 29 WGD hypotheses were consistent across 2 or more analyses, supporting at least 8 events in Nepetoideae and 4 additional events outside the clade.
Fig. 5.
—Comparison of putative WGD events inferred across MAPS, PUG, and divergence times analyses. As illustrated with colored symbols, our results reveal signatures of widespread ancient polyploidy in Lamiaceae, including 29 uniquely placed WGD events in the family and pronounced levels of WGDs in the species-rich Nepetoideae (blue). Twelve of these events were inferred by more than one analysis, whereas six were consistently inferred across all analyses.
We consistently recovered evidence for widespread WGD in Nepetoideae. Of the WGD hypotheses corroborated by 1 or more analyses, most (8/12 events) were placed within this species-rich clade, including 1 event each within the stem lineages (or near their MRCA) of Nepetoideae, Mentheae, Salviinae, and Ocimeae, respectively, and 4 additional events within internal stem lineages or nodes in Menthinae (fig. 5). Notably, we observed pronounced levels of gene duplications or large numbers of putative WGDs (or both) in Nepetoideae across analyses, emphasizing the asymmetry of duplication dynamics among major mint subclades. These results suggest that gene duplication events—whether or not from WGDs—may be an important evolutionary process in this clade. Given its large number of species (3,300+ species) and extraordinary phenotypic diversity (including chemodiversity), it is possible that gene duplications or WGDs (or both) may be driving key innovations and subsequent species diversification in Nepetoideae.
Caveats to Interpretations of Results
The KS and phylotranscriptomic results presented here provide needed estimates of and working hypotheses for gene duplication dynamics and ancient polyploidy in mints, but a few important caveats should be considered when interpreting results (see also discussion of approaches below). First, although our taxon sampling scheme is broadly representative of the phylogenetic diversity in Lamiaceae, we acknowledge that it is not representative of the phylogenetic diversity within all major mint lineages (e.g., within some subclades [=traditional subtribes]). Phylotranscriptomic analyses with broader taxon sampling may improve resolution and placement of putative ancient WGD events in some lineages. Second, in the absence of available genomes spanning the phylogenetic diversity of mints, transcriptomes are useful as proxies for exploring Lamiaceae-wide genome-level duplication dynamics. However, we acknowledge that inferences solely from transcriptome data sets, particularly those generated from a single tissue, could contribute to underestimates of gene duplication because of incomplete sampling of both nonfunctional (e.g., absence of silenced paralogs and missing pseudogenes removed by transcript filtering procedures) and functional (e.g., nonexpressed stress-related or tissue-specific genes, and poor representation of transcripts from lowly expressed genes) genes. These issues are not specific to this study but are common to all analyses of this type; in fact, the original description of the KS method (Lynch and Conery 2000) used genomes rather than transcriptomes, so all applications using transcriptomes (e.g., Li et al. 2015, 2018; McKain et al. 2016; Unruh et al. 2018) suffer from the same set of concerns. More detailed analyses based on whole-genome sequences are needed to provide further resolution of ancient polyploidy events in mints as well as other organisms.
Ancient Polyploidy and Mint Species Richness
Recent studies have examined the evolutionary role of ancient polyploidy in angiosperm diversification, associating lineage-specific WGDs with species richness or diversification rates (e.g., Edger et al. 2015; Tank et al. 2015; Smith et al. 2018; Landis et al. 2018; Ren et al. 2018). Many angiosperm clades with early putative WGD events had higher species richness than their sister lineages (Landis et al. 2018, Ren et al. 2018), supporting a possible relationship between polyploidy and net diversification. However, some small angiosperm clades were also marked by these events (Landis et al. 2018; Ren et al. 2018), suggesting that WGDs do not always promote diversification. Our results for Lamiaceae mirrored previous angiosperm-wide patterns and revealed putative WGD events that were distributed across clades with varying levels of high and low species richness (fig. 5). However, when accounting for cumulative levels of WGD within the ancestry of each major clade, we found evidence for a strong, positive association between ancient polyploidy and lineage-specific species richness (supplementary fig. S5, Supplementary Material online). Thus, our results suggest that ancient polyploidy may have played an important role in mint diversification. Future analyses of mint diversification rates may shed new light on this role, perhaps by linking WGDs with increases in net diversification and revealing patterns of nested radiations that correspond to WGD events.
Approaches Used
KS Analyses
Although KS plots were useful for detecting signatures of ancient polyploidy within the transcriptomes of each of our 52 species, we found it challenging (for a number of reasons) to compare KS plots across samples and use these results to corroborate our interpretations of results from other phylotranscriptomic analyses. Analyses of KS plots are prone to many pitfalls that prevent accurate detection of ancient and recent WGDs (reviewed in Tiley et al. [2018]). We observed evidence for numerous recent WGD events in our phylotranscriptomic results that we also visualized in some KS plots. For example, we visualized peaks consistent with recent WGDs in Monarda and Mentha (KS = 0.24–0.39; supplementary fig. S2, Supplementary Material online), which may correspond to the putative WGD predicted in the stem lineage or near the MRCA of these species by MAPS, PUG, and divergence time analyses (fig. 5). However, in this and other cases, we lacked the ability to statistically discern these patterns across the full range of observed KS values due to documented limitations associated with mixture modeling (Tiley et al. 2018). It is also plausible that some peaks identified by our analyses were artifactual, confounding our ability to interpret shared patterns of WGD within Lamiaceae. Mixture models are sensitive to data filtration and, in some cases, can overfit Gaussian components onto KS distributions (Johnson et al. 2016), producing artificial peaks that are erroneously inferred as ancient WGDs (Tiley et al. 2018). Although we used SiZer analyses (Vanneste et al. 2013, 2015) to mitigate the propensity for these errors, our comparisons of KS values across species remained problematic for other reasons. For example, differences in retention rates of gene duplicates across lineages may have hindered our ability to identify some ancient WGD events that were shared across lineages. Moreover, because it was not possible to estimate the ages of these events accurately (i.e., due to possible differences in substitution rates across lineages), we were not able to place and interpret putative WGD events inferred from KS plots within the context of our dated Lamiaceae phylogeny. For all of these reasons, we concluded that KS plots had limited utility for documenting shared patterns of ancient polyploidy in Lamiaceae.
Phylotranscriptomic and Divergence Time Investigations
As described above, we observed inconsistent numbers and phylogenetic placements of putative ancient WGDs across the analyses employed. These inconsistencies forced us to consider possible benefits and limitations of MAPS, PUG, and divergence time approaches that have implications for accurate detection and placement of WGDs in Lamiaceae, as well as other plant clades. Although our discussion of these benefits and limitations is not exhaustive, we highlight some important differences among these approaches and make recommendations to researchers planning investigations of ancient WGD in other lineages.
Unlike PUG or divergence time approaches, which accommodated use of our best estimate of mint phylogeny, the MAPS pipeline required use of multiple stepwise guide trees to identify and place gene duplicates on our species tree. From a practical standpoint, this requirement was extremely costly in terms of time, requiring 12 independent analyses and considerably more data processing (e.g., orthogroup clustering based on 12 subsampled taxon sets and alignment and tree inference for ∼120,000+ orthogroups), and necessitated post hoc reconciliation of independent MAPS results. From a theoretical standpoint, taxon subsampling was an important concern, primarily because all descendent lineages could not be included in a single analysis. This practice may have introduced several sources of potential bias that affected placement of gene duplications within individual guide trees and, ultimately, our interpretations of WGDs—including both total numbers and placements of events resolved on our fully sampled mint tree. Taxon sampling is a known factor affecting accuracy in phylogenetic estimations (reviewed in Nabhan and Sarkar [2012]). In our case, this subsampling may have introduced errors into gene tree estimates that were subsequently used by MAPS to identify and place gene duplications on corresponding guide trees. Errors in gene trees tend to increase numbers of gene duplicates placed toward the root of the species tree and decrease numbers toward the tips, potentially skewing our interpretations of WGD events (Hahn 2007). Nevertheless, even in the absence of these errors, reconciling results across independent MAPS analyses remained problematic. We noted that placements of gene duplications or WGD inferences (or both) differed across some MAPS results, in spite of our use of overlapping taxon sampling. For example, we observed elevated levels of gene duplications (i.e., relative to neighboring nodes) near the MRCAs of Tectona and Nepetoideae in our Elscholtzieae-focused analysis (supplementary fig. S1, Supplementary Material online) and inferred putative ancient WGDs at these nodes, but these events were not recovered by other MAPS analyses. It is reasonable to assume that some duplications were resolved by MAPS to nodes in our sparsely sampled guide trees that corresponded to deep-level nodes in our species tree, but these events may have represented shared events observed in other densely sampled guide trees that were placed at alternative nodes in the species tree. In consideration of these limitations and others described below, we were cautious with our final interpretations of putative WGDs reconciled to our species tree. Our summary of MAPS results for Lamiaceae likely represented an overestimation of putative ancient WGD events, and consideration of other evidence was needed to corroborate individual hypotheses.
Compared with MAPS, we observed more benefits and fewer limitations for PUG and divergence time approaches for investigating ancient WGDs. Although all approaches incorporated information from large numbers of gene trees that may have contained phylogenetic error that could potentially bias placements of gene duplicates or WGD inferences, PUG and divergence time approaches were appealing because they accommodated use of a single ML tree hypothesis for Lamiaceae and gene trees with full taxon sampling. Available research suggests that increasing the density of taxon sampling within an organismal phylogeny can improve precision in estimates of the timing of WGD events (McKain et al. 2016). Thus, the ability to analyze complete data sets was a clear benefit of both PUG and divergence time approaches. Moreover, PUG provided options for results filtering based on clade support values (e.g., ML BS ≥ 80 and ML BS ≥ 50), facilitating exclusion of poorly supported gene tree results that may have confounded our interpretations of gene duplication patterns. Because our divergence time analysis was based on paralog data exported by PUG, this approach also benefited from the filtering feature.
From a theoretical standpoint, one major limitation of the PUG approach was that it did not consider the timing of gene duplication events placed in the species tree. As a result, we were not able to establish solely from PUG results whether gene duplications were produced by ancient WGDs or other duplication processes. Moreover, where we inferred putative ancient WGDs with PUG, we could not discern whether one or more WGD events had occurred within a stem lineage. If WGD hypotheses inferred from PUG results are real events, we would expect that the estimated dates for duplication events would be similar in independent gene trees (i.e., duplication ages are clustered in time) (Jiao et al. 2011). Our divergence time approach, which represents a possible extension of PUG here, provides a clear benefit in this regard by facilitating characterization of gene duplication patterns in a temporal context. We corroborated over half of the events inferred with PUG (i.e., 11 of 21 WGDs) with divergence time data and inferred an additional event inferred by MAPS at a deep node within the family (fig. 2a:N46 and fig. 5). Our application of a gene birth–death process to observed distributions of gene duplication ages may have facilitated detection of the latter (MAPS) event because we were able to treat nodes of different ages independently. As a result, we avoided one potential pitfall of previous strategies employing PUG, which identifies putative WGD events by applying the same threshold of gene duplication across nodes of different ages, in spite of expectations that older nodes may have fewer surviving paralogs than younger nodes (Lynch and Conery 2000). Strategies employing MAPS are also prone to this same pitfall, especially because a pronounced level or “burst” of gene duplication at a given node relative to its neighbors is required to infer a putative WGD event. In some cases with MAPS, we may have failed to detect putative WGDs at older nodes with low to moderate, but similar, levels of gene duplication.
Despite its value, our divergence time approach was not without limitations. First, our ability to corroborate only some MAPS or PUG WGDs may have been limited because we were conservative and employed only a small percentage of our total gene trees that could be confidently rooted with all designated outgroups. Other researchers are working on new strategies for rooting gene trees for dating analyses (S. Smith, personal communication), and investigators of WGD may be able to take advantage of these in the near future. Use of additional time-calibrated gene trees may facilitate corroboration of additional WGD events. Second, uncertainty in molecular dating estimates may have affected some of our downstream analyses and interpretations of observed age distributions identified as putative WGDs. We observed a number of broad distributions of gene ages, but we were not able to assess whether these resulted from a series of gene duplications or reflected error in phylogenetic dating estimates across independent gene trees; we did not account for uncertainty in either our time-calibrated species tree or gene trees. Future research should consider bootstrapping approaches to estimate confidence intervals for divergence time estimates (e.g., Sanderson 2003) and incorporate these into analysis pipelines.
Conclusions and Future Directions
This study presents compelling evidence revealing widespread ancient polyploidy in Lamiaceae, particularly within the species-rich and chemically diverse Nepetoideae. Within Nepetoideae, we show extraordinarily high levels of gene duplication events relative to other mint subclades. These duplication events may have resulted from single, large-scale duplications such as segmental duplications or WGDs (as inferred here), or from many individual tandem duplications. Although we have no reason to believe that any of these processes are mutually exclusive, it seems clear from our results and from other recent studies of Lamiaceae (e.g., Mint Evolutionary Genomics Consortium 2018; Zhao et al. 2019; Benjamin R. Lichman, Grant T. Godden, John P. Hamilton, Lira Palmer, Mohamed O. Kamileen, Dongyan Zhao, Brieanne Vaillancourt, Joshua Wood, Miao Sun, Taliesin J. Kinser, Laura K. Henry, Carlos Rodriguez Lopez, Natalia Dudareva, Douglas E. Soltis, Pamela S. Soltis, C. Robin Buell, Sarah E. O'Connor, in preparation) that gene duplications—regardless of the processes that produced them—are potentially important drivers of evolution in this ecologically, economically, and culturally important clade. However, the extent to which retained gene duplicates produced by WGDs or other mechanisms contribute to specific trait innovations, particularly novelty in specialized metabolites, and subsequent diversification in Nepetoideae remains poorly known and represents an important avenue for future research that can build upon the foundation established here. Although these questions are beyond the scope of this study, we plan to test for correlations among these features as part of future investigations. Lastly, for researchers interested in exploring gene duplication dynamics and ancient polyploidy in other plant groups, we hope the observations and recommendations described here prove useful for planning analyses. For reasons described above, we strongly encourage integration of divergence time approaches into research designs.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation Plant Genome Research Program (IOS-1444499). We thank members of the Mint Evolutionary Genomics Consortium for their contributions to this manuscript and to Stephen Smith (and lab members) and Yuannian Jiao for consultations regarding divergence time analyses.
Data deposition: This project has been deposited at Dryad under the accession doi: 10.5061/dryad.qbzkh18cr.
Literature Cited
- Aagaard JE, Olmstead RG, Willis JH, Phillips PC.. 2005. Duplication of floral regulatory genes in the Lamiales. Am J Bot. 92(8):1284–1293. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Amborella Genome Project. 2013. The Amborella genome and the evolution of flowering plants. Science 342:1241089. [DOI] [PubMed] [Google Scholar]
- Barker MS, et al. 2008. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol Biol Evol. 25(11):2445–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, et al. 2010. EvoPipes.net: bioinformatic tools for ecological and evolutionary genomics. Evol Bioinform Online 6:143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, et al. 2016. Most Compositae (Asteraceae) are descendants of a paleohexaploid and all share a paleotetraploid ancestor with the Calyceraceae. Am J Bot. 103(7):1203–1211. [DOI] [PubMed] [Google Scholar]
- Benaglia T, Chauveau D, Hunter D, Young D.. 2009. mixtools: an R package for analyzing finite mixture models. J Stat Softw. 32:1–29. [Google Scholar]
- Blanc G, Wolfe KH.. 2004. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16(7):1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breheny P, Stromberg A, Lambert J.. 2018. p-Value histograms: inference and diagnostics. High-Throughput 7(3):23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, et al. 2019. Widespread ancient whole-genome duplications in Malpighiales coincide with Eocene global climatic upheaval. New Phytol. 221(1):565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri P, Marron J.. 1999. SiZer for exploration of structures in curves. J Am Stat Assoc. 94:907–823. [Google Scholar]
- Comai L. 2005. The advantages and disadvantages of being polyploid. Nat Rev Genet. 6(11):836.. [DOI] [PubMed] [Google Scholar]
- Cui L, et al. 2006. Widespread genome duplications throughout the history of flowering plants. Genome Res. 16(6):738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew BT, Sytsma KJ.. 2012. Phylogenetics, biogeography, and staminal evolution in the tribe Mentheae (Lamiaceae). Am J Bot. 99(5):933–953. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, et al. 2015. The butterfly plant arms-race escalated by gene and genome duplications. Proc Natl Acad Sci U S A. 112(27):8362–8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, et al. 2017. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year-old naturally established neo-allopolyploid monkeyflower. Plant Cell 29(9):2150–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16(1):157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley C, Raftery AE.. 2002. Model-based clustering, discriminant analysis, and density estimation. J Am Stat Assoc. 97(458):611–631. [Google Scholar]
- Freeling M, Thomas BC.. 2006. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 16(7):805–814. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. 2007. Bias in phylogenetic tree reconciliation methods: implications for vertebrate genome evolution. Genome Biol. 8(7):R141.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley RM, et al. 2004. Labiatae In: Kadeiret JW, editor. The families and genera of vascular plants: flowering plants—dicotyledons. Vol. VII Berlin: Springer-Verlag; p. 167–275. [Google Scholar]
- He Y, et al. 2016. Survey of the genome of Pogostemon cablin provides insights into its evolutionary history and sesquiterpenoid biosynthesis. Sci Rep. 6:26405.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C-H, et al. 2016. Multiple polyploidization events across Asteraceae with two nested events in the early history revealed by nuclear phylogenomics. Mol Biol Evol. 33(11):2820–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husband BC, Baldwin SJ, and Suda J. 2013. The incidence of polyploidy in natural plant populations: Major patterns and evolutionary processes. In: J. Greilhuber, Dolezel J, Wendel JF, editors, Plant genome diversity, Austria: Springer Vienna. 255–276. [Google Scholar]
- Janssens SB, Knox EB, Huysmans S, Smets EF, Merckx VS. 2009. Rapid radiation of Impatiens (Balsaminaceae) during Pliocene and Pleistocene: result of a global climate change. Mol Phylogenet Evol, 52(3): 806–824. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Li J, Tang H, Paterson AH.. 2014. Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 26(7):2792–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473(7345):97–100. [DOI] [PubMed] [Google Scholar]
- Jiao Y, et al. 2012. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 123:R3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MG, Malley C, Goffinet B, Shaw AJ, Wicket NJ.. 2016. A phylotranscriptomic analysis of gene family expansion and evolution in the largest order of mosses (Hypnales, Bryophyta). Mol Phylogenet Evol. 98:29–40. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma KI, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis JB, et al. 2018. Impact of whole-genome duplication events on diversification rates in angiosperms. Am J Bot. 105(3):348–363. [DOI] [PubMed] [Google Scholar]
- Li B, Olmstead RG.. 2017. Two new subfamilies in Lamiaceae. Phytotaxa 313(2):222–226. [Google Scholar]
- Li B, et al. 2016. A large-scale chloroplast phylogeny of the Lamiaceae sheds new light on its subfamilial classification. Sci Rep. 6:34343.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. 2015. Early genome duplications in conifers and other seed plants. Sci Adv. 1(10):e1501084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. 2018. Multiple large-scale gene and genome duplications during the evolution of hexapods. Proc Natl Acad Sci U S A. 115(18):4713–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2000. The evolutionary fate and consequences of duplicated genes. Science 290(5494):1151–1155. [DOI] [PubMed] [Google Scholar]
- Maere S, et al. 2005. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci U S A. 102(15):5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan G, Peel DA. 2000. Finite Mixture Models. New York: Wiley. [Google Scholar]
- McKain MR, et al. 2016. A phylogenomic assessment of ancient polyploidy and genome evolution across the Poales. Genome Biol Evol. 8(4):1150–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mint Evolutionary Genomics Consortium. 2018. Phylogenomic mining of the mints reveals multiple mechanisms contributing to the evolution of chemical diversity in Lamiaceae. Mol Plant 11:1084–1096. [DOI] [PubMed] [Google Scholar]
- Mirarab S, et al. 2015. PASTA: ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J Comput Biol. 22(5):377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan AR, Sarkar IN.. 2012. The impact of taxon sampling on phylogenetic inference: a review of two decades of controversy. Brief Bioinform. 13(1):122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- One Thousand Plant Transcriptomes Initiative. 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574:679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu S-H.. 2016. Evolution of gene duplication in plants. Plant Physiol. 171(4):2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K.. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20(2):289–290. [DOI] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW.. 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst. 29(1):467–501. [Google Scholar]
- R-Core-Team. 2019. R: a language and environment for statistical computing, version 3.5.3. Vienna (Austria: ): R Foundation for Statistical Computing; Available from: https://www.R-project.org/ (accessed March 11, 2019). [Google Scholar]
- Ren R, et al. 2018. Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol Plant 11(3):414–428. [DOI] [PubMed] [Google Scholar]
- Rensing SA. 2014. Gene duplication as a driver of plant morphogenetic evolution. Curr Opin Plant Biol. 17:43–48. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol Biol Evol. 19(1):101–109. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. 2003. rR8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19(2):301–302. [DOI] [PubMed] [Google Scholar]
- Smith SA, O’Meara BC.. 2012. treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28(20):2689–2690. [DOI] [PubMed] [Google Scholar]
- Smith SA, et al. 2018. Disparity, diversity, and duplications in the Caryophyllales. New Phytol. 217(2):836–854. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Liu X, Marchant DB, Visger CJ, Soltis DE.. 2014. Polyploidy and novelty: Gottlieb’s legacy. Philos Trans R Soc B 369(1648):20130351.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Marchant DB, Van de Peer Y, Soltis DE.. 2015. Polyploidy and genome evolution in plants. Curr Opin Genet Dev. 35:119–125. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE.. 2016. Ancient WGD events as drivers of key innovations in angiosperms. Curr Opin Plant Biol. 30:159–165. [DOI] [PubMed] [Google Scholar]
- Soltis DE, et al. 2009. Polyploidy and angiosperm diversification. Am J Bot. 96(1):336–348. [DOI] [PubMed] [Google Scholar]
- Soltis DE, et al. 2018. Phylogeny and evolution of the Angiosperms: revised and updated edition. Chicago: University of Chicago Press. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Paterson AH.. 2010. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci U S A. 107(1):472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank DC, et al. 2015. Progressive radiations and the pulse of angiosperm diversification. New Phytol. 207(2):454–467. [DOI] [PubMed] [Google Scholar]
- Tiley GP, Barker MS, Burleigh JG.. 2018. Assessing the performance of KS plots for detecting ancient whole genome duplications. Genome Biol Evol. 10(11):2882–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313(5793):1596–1604. [DOI] [PubMed] [Google Scholar]
- Unruh SA, et al. 2018. Phylotranscriptomic analysis and genome evolution of the Cypripedioideae (Orchidaceae). Am J Bot. 105(4):631–640. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A.. 2009. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 10(10):725–732. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K.. 2017. The evolutionary significance of polyploidy. Nat Rev Genet. 18(7):411–424. [DOI] [PubMed] [Google Scholar]
- Vanneste K, Van de Peer Y, Maere S.. 2013. Inference of genome duplications from age distributions revisited. Mol Biol Evol. 30(1):177–190. [DOI] [PubMed] [Google Scholar]
- Vanneste K, Sterck L, Myburg AA, Van de Peer Y, Mizrachi E. 2015. Horsetails are ancient polyploids: evidence from Equisetum giganteum. Plant Cell 27(6):1567–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, et al. 2014. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 15(2):R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TE, et al. 2009. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci U S A. 106(33):13875–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yao G, et al. 2016. Phylogenetic relationships, character evolution and biogeographic diversification of Pogostemon s.l. (Lamiaceae). Mol Phylogenet Evol. 98:184–200. [DOI] [PubMed] [Google Scholar]
- Zhao D, et al. 2019. A chromosomal-scale genome assembly of Tectona grandis reveals the importance of tandem gene duplication and enables discovery of genes in natural product biosynthetic pathways. Gigascience 8(3):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.