SUMMARY
Metazoan retromer (VPS26/VPS35/VPS29) associates with sorting nexins on endosomal tubules to sort proteins to the trans-Golgi network or plasma membrane. Mechanisms of metazoan retromer assembly remain undefined. We combine single-particle cryoelectron microscopy with biophysical methods to uncover multiple oligomer structures. 2D class averages reveal mammalian heterotrimers; dimers of trimers; tetramers of trimers; and flat chains. These species are further supported by biophysical solution studies. We provide reconstructions of all species, including key sub-structures (~5 Å resolution). Local resolution variation suggests that heterotrimers and dimers adopt multiple conformations. Our structures identify a flexible, highly conserved electrostatic dimeric interface formed by VPS35 subunits. We generate structure-based mutants to disrupt this interface in vitro. Equivalent mutations in yeast demonstrate a mild cargo-sorting defect. Our data suggest the metazoan retromer is an adaptable and plastic scaffold that accommodates interactions with different sorting nexins to sort multiple cargoes from endosomes their final destinations.
Graphical Abstract
In Brief
Retromer is an important trafficking complex linked to human brain disease, but structures and mechanisms of metazoan retromer assembly remain elusive. Kendall et al. combine single-particle cryo-EM with biophysical studies to demonstrate that retromer forms multiple flexible oligomers. Mutagenesis confirms a key electrostatic dimer interface mediated by VPS35 subunits.
INTRODUCTION
Retromer is a multi-subunit protein complex that forms coats on tubules emerging from endosomes (reviewed in Burd and Cullen, 2014; Trousdale and Kim, 2015). The core heterotrimer, formerly called cargo-selective complex, contains VPS26, VPS35, and VPS29 subunits that together form a ~150-kDa heterotrimer; for clarity, we will refer to the VPS26/35/29 heterotrimer as “retromer” in this manuscript. Retromer associates with sorting nexin (SNX) proteins that help recruit it to endosomal membranes enriched in phosphatidylinositol-3-phosphate (PI3P). In yeast, retromer forms a pentamer with two SNX-BAR proteins, Vps5 and Vps17, which associate to form heterodimers (Seaman et al., 1998). Retromer/Vps5/Vps17 coats sort acid hydrolase receptors, including Vps10, from endosomes to the trans-Golgi network (TGN) (Seaman et al., 1998; Suzuki et al., 2019). Yeast retromer also interacts with SNX3 (Bean et al., 2017; Harrison et al., 2014; Strochlic et al., 2007), a sorting nexin that lacks a BAR domain. In metazoans, retromer interacts with multiple sorting nexin proteins (SNX-BARs, SNX3, and SNX27) to further diversify its cargo repertoire. Hydrolase receptors, including mannose 6-phosphate receptors (Arighi et al., 2004) are sorted in a retrograde pathway to the TGN, while other transmembrane receptor cargoes (e.g., β2-adrenergic receptor, GLUT1) are recycled directly from endosomes to the plasma membrane (Steinberg et al., 2013; Temkin et al., 2011). Mammalian retromer uses additional SNX proteins, including SNX3 (Strochlic et al., 2007) and SNX27 (Clairfeuille et al., 2016; Gallon et al., 2014), as cargo adaptors. SNX3/retromer is implicated in Wntless retrograde sorting (Belenkaya et al., 2008; Franch-Marro et al., 2008; Pan et al., 2008; Port et al., 2008; Yang et al., 2008), while SNX27 sorts many transmembrane cargoes bearing specific PDZ-binding motifs to the plasma membrane (Steinberg et al., 2013). SNX-BAR proteins have recently emerged as cargo adaptors (Kvainickas et al., 2017a; Simonetti et al., 2017). Biochemical evidence suggests VPS26 itself may directly bind one cargo (SorLA) in mammalian cells (Fjorback et al., 2012).
Mechanisms governing the assembly and structures of coated tubules are now starting to emerge (recently reviewed in Chen et al., 2019; Simonetti and Cullen, 2018). Multiple crystal structures of mammalian retromer subunits (Collins et al., 2005, 2008; Shi et al., 2006; Wang et al., 2005) and sub-complexes (Hierro et al., 2007; Lucas et al., 2016) have been determined. Two groups have proposed that mammalian retromer forms a “dimer of trimers” (Lucas et al., 2016; Norwood et al., 2010) based on low-resolution small-angle X-ray scattering data, and a structure has been proposed for the budding yeast heterotrimer from electron microscopy (EM) data (Purushothaman et al., 2017). A recent cryoelectron tomography (cryo-ET) reconstruction reveals the overall architecture of reconstituted thermophilic yeast retromer with a Vps5 homodimer (Kovtun et al., 2018). In this structure, Vps5 dimers on the membrane organize VPS26 into dimers, while VPS35 subunits form a second dimer at the apex of retromer arches. However, immunoprecipitation data from budding yeast (Suzuki et al., 2019) do not fully support this structural model.
How metazoan retromer assembles, and whether it assembles like thermophilic yeast retromer, remains an open question. Modeling crystal structures into the cryo-ET reconstruction suggests that SNX3 could interact with retromer, but SNX-BAR and SNX3 binding would be mutually exclusive (Chen et al., 2019; Kovtun et al., 2018; Simonetti and Cullen, 2018). It also remains unclear how VPS26 would assemble dimers in the absence of SNX-BAR proteins. Furthermore, the established SNX27 PDZ cargo-binding site on VPS26 is located far from the membrane when crystal structures (Gallon et al., 2014) are docked into the yeast cryo-ET model. SNX proteins lacking BAR domains altogether may require different retromer assemblies to sort distinct cargoes.
Here, we present structural and biophysical data to reveal how murine retromer forms a variety of oligomers. We find retromer forms dimers of trimers; a tetramer of trimers; and longer chains. Our structures suggest that retromer contains a key electrostatic yet flexible assembly interface mediated by the backside of VPS35 subunits. We test the proposed interface biochemically by introducing structure-based point mutations and determining whether retromer can form oligomers in vitro. Finally, we introduce equivalent mutations into S. cerevisiae and show that disrupting the conserved interface partially impedes cargo sorting. Disruption of this interface with specific point mutations suggests that the interface may be relevant for assembling higher-order structures and provides an important molecular tool for the community. Based on our structures, we hypothesize that retromer can function as a plastic and adaptable scaffold, and we present possible assembly models in the presence of mammalian sorting nexins on endosomal membranes.
RESULTS
Structural and Biophysical Studies of Mammalian Retromer
In preliminary studies, we analyzed whether purified recombinant retromer was suitable for single-particle cryoelectron microscopy (cryo-EM) studies. We first obtained 2D class averages and produced random conical tilt reconstructions from negatively stained samples (data not shown). 2D class averages from untilted particles revealed heterotrimers along with multiple oligomeric species, including dimers and tetramers of trimers (data not shown). We next undertook single-particle cryo-EM studies (Figure 1) to determine structures of mammalian retromer heterotrimers (Figure 1A) and oligomers (Figures 1B–1E). In vitrified ice, we clearly observe the retromer heterotrimer; dimers of trimers; and tetramers of trimers both in micrographs (Figure S1) and in 2D class averages (Figure 1). We also observe longer, flat chains of retromer (Figures 1C and 1D). We used 2D classification to separate each biochemical species and generated reconstructions for each (workflow in Figure S2; full details in the STAR Methods). We independently confirmed that retromer forms oligomers in solution using size-exclusion chromatography with multi-angle laser light scattering (Figure 2A) and dynamic light scattering (Figure 2B). We find oligomer formation depends on salt concentration; the retromer heterotrimer forms both dimers and tetramers near physiological salt concentrations (50–100 mM NaCl) but exists as a heterotrimer at higher concentrations (150–500 mM NaCl). We describe each structure and its key features below.
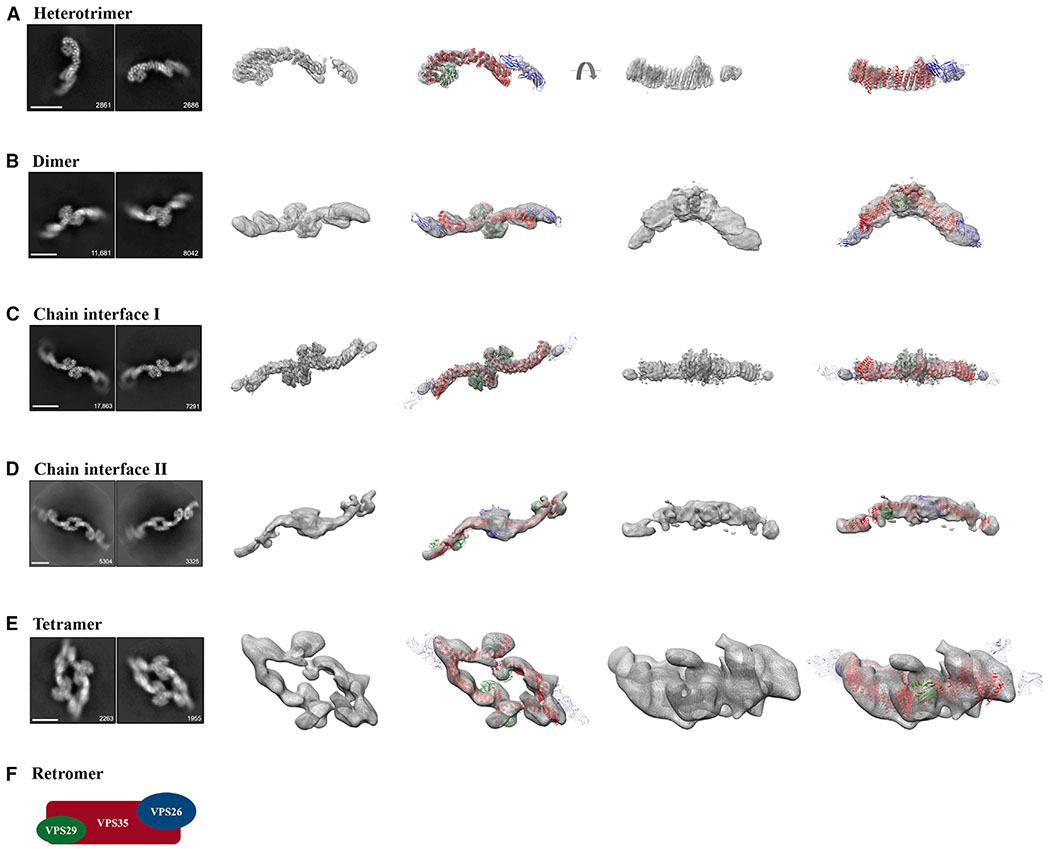
Figure 1. Single-Particle Cryo-EM Reconstructions of Mammalian Retromer.
Four retromer species were resolved between 27 and 6 Å: (A) the retromer heterotrimer; (B) a dimer of trimers; (C and D) retromer chains; and (E) a tetramer of trimers. For each row (A–E), the first column shows two representative 2D class averages for the species, including particle numbers. Scale bars represent 10 nm. The middle two columns show 3D reconstructions (see Figure S2 for contour details) with and without a fitted model. The last two columns show an additional view (rotated by 90°) of 3D reconstructions with and without fitted models. Initial models were generated from partial crystal structures (PDB: 2R17, 5F0J). VPS29 is shown in green, VPS35 in red, and VPS26 in blue or transparent blue (when averaged out in a reconstruction). A schematic of the retromer heterotrimer is shown in (F). See also Figures S1 and S2, Tables S1 and S2.
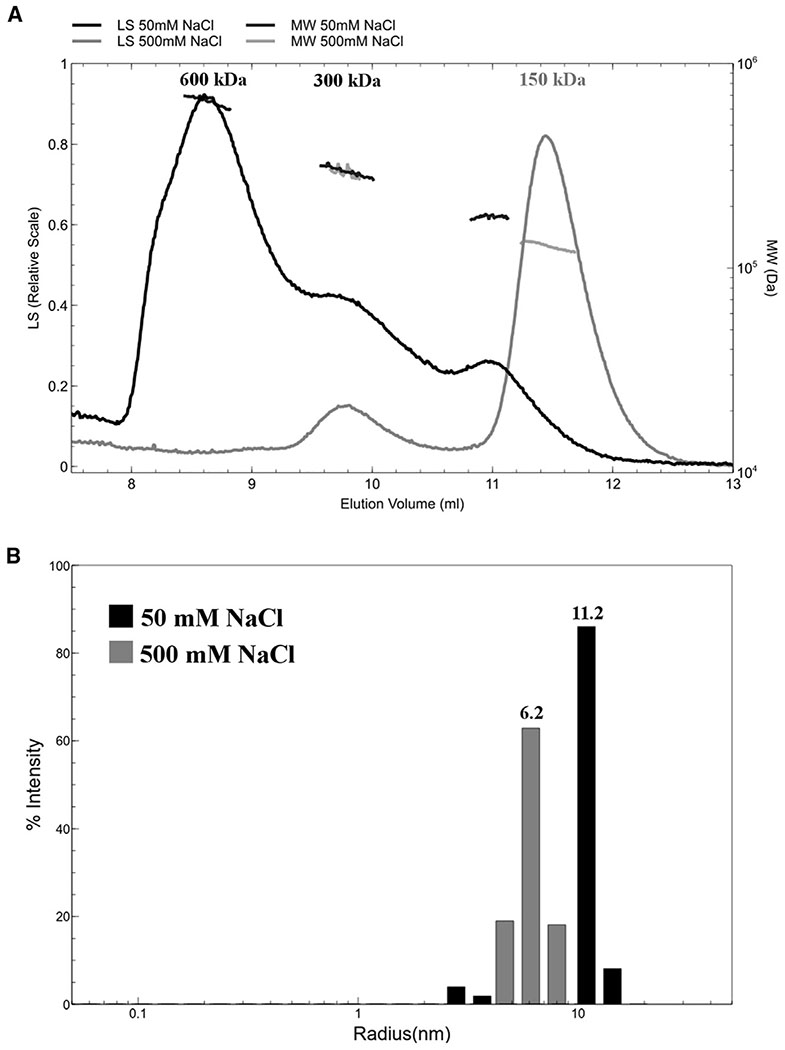
Figure 2. Mammalian Retromer Forms Oligomers in Solution in a Salt-Dependent Manner.
(A) At 500 mM NaCl (gray trace), the main retromer peak, elutes from a size-exclusion column at a volume consistent with one copy of the VPS26/VPS35/VPS29 heterotrimer (~150 kDa). A small population of retromer elutes in a second peak as a “dimer of trimers” (~300 kDa). At 50 mM NaCl (black trace)the peak profile shifts: the predominant peak is now consistent with four copies of the heterotrimer (600 kDa); a second peak or shoulder is consistent with two copies, and the third is consistent with one copy.
(B) Dynamic light scattering reveals that retromer particles in low salt (50 mM NaCl, gray bars) have approximately double the average radius compared with retromer in high salt (500 mM NaCl, black bars). This is consistent with size-exclusion chromatography with multi-angle laser light scattering data in (A) indicating retromer forms oligomers in solution.
Heterotrimer
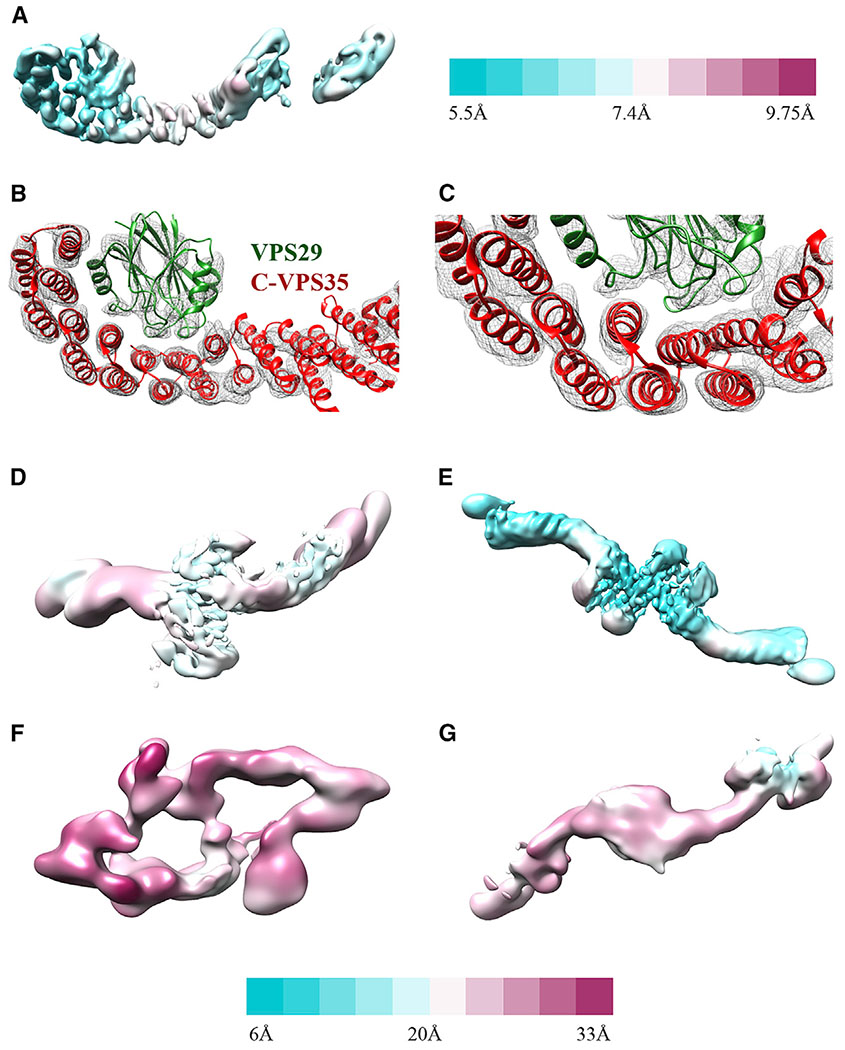
The structure of full-length mammalian retromer is directly observed here for the first time (5.7 Å average resolution from 26,369 particles). 3D reconstructions reveal a well-resolved interface between the VPS35 C terminus (C-VPS35) and VPS29. This interface is nearly identical to an X-ray crystal structure comprising the C-VPS35 and full-length VPS29 (PDB: 2R17), which we used as a model for fitting into reconstructions. The resolution of the map corresponding to C-VPS35/VPS29 is ~5.5 Å (Figure 3A), and α-helical density for the VPS35 solenoid is clearly distinguishable and can be fit well (Figures 3B and 3C). In contrast, the VPS35 N terminus exhibits substantial flexibility. The VPS35 N-terminal interface that binds VPS26 is not well ordered, and this part of the map is less ordered (~8–9 Å; Figure 3A). VPS26 may not be well resolved for two reasons. First, VPS26 is an all β sheet protein, and at intermediate resolution it is more difficult to assign a model for β sheets than for α helices. Second, both N-VPS35 and VPS26 appear to be flexible, based on variation in local resolution estimates (Figure 3A). Finally, VPS35 seems to be flexible along the length of the solenoid based on local resolution estimates, which suggest there may be a hinge point in the middle of VPS35 (Figure 3A).
Figure 3. Local Resolution and Features.
(A) Retromer heterotrimer resolution range across the model with legend shown to right (5.5–9.8 Å).
(B) View of C-VPS35/VPS29 interface to show model fitting into α helices (contoured at 6σ).
(C) Close-up view of VPS35 C-terminus to show model fitting into α helices (contoured at 6σ).
(D–G) Resolution range for dimer (D); VPS35-centered chain I interface (E); tetramer (F); and VPS26-centered chain II interface (G). Legend shown below from 6 (cyan) to 33 (magenta) Å.
Retromer Dimer of Trimers
Retromer forms dimers of trimers in both negatively stained (data not shown) and vitrified samples (Figure 1B). The dimeric interface between two heterotrimers is immediately clear in 2D class averages, because we can distinguish between the VPS35 and VPS26 ends of retromer. Initial reconstructions (18 Å average resolution from 31,022 particles) reveal that the C termini of VPS35 subunits mediate dimer formation. Although the dimer structure underwent postprocessing in RELION, the reported resolution of ~9 Å seems overly optimistic based on map quality. The map is better resolved at the dimer interface mediated by VPS35 (14 Å resolution; Figure 3D). The mammalian retromer dimer is wide, fairly flat, and does not exhibit 2-fold symmetry. Instead, one heterotrimer twists and rotates relative to its partner, giving rise to a dimer with an angle of ~150° between two heterotrimer legs. The retromer dimer is a stable biochemical species (Figure 2), and, like the heterotrimer, it exhibits substantial conformational flexibility. Local resolution across the dimer varies from 14 to 26 Å (Figure 3D). The VPS35-mediated interface is slightly different across the population of dimers, giving rise to slightly different dimers with different angles between heterotrimer legs. Furthermore, the presence of VPS26 seems to affect dimer formation and overall retromer structure. Class averages of negatively stained VPS35/VPS29 sub-complex (Figure S4A) reveal these dimers are more flexible than dimers formed by intact retromer. VPS26 appears to impart rigidity to the VPS35 solenoid, even though structural data from heterotrimers and dimers reveal flexibility in the N-terminal VPS35/VPS26 interface. Overall, these data suggest that retromer is plastic and adaptable at multiple interfaces.
Retromer Chains
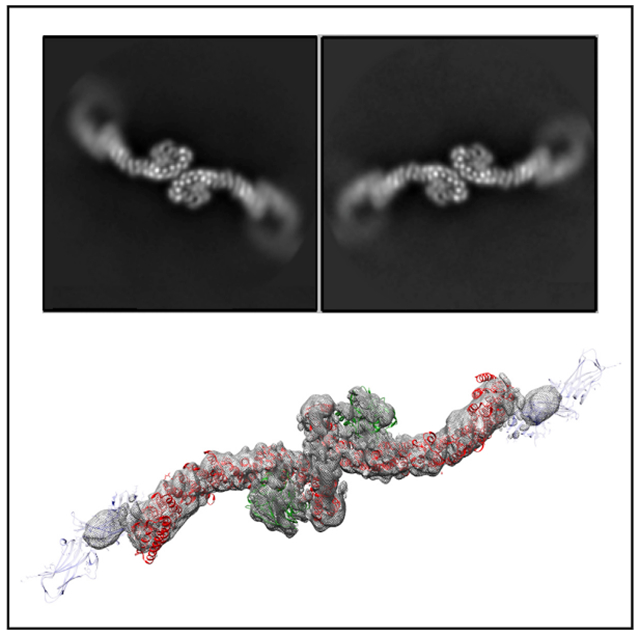
A surprising result from our structural studies was the existence of longer retromer chains in vitrified ice (Figures S1C, S1D, 1C, and 1D). We did not observe chain structures in negative stain, and they may occur only at higher concentrations present in vitrified samples and when retromer is concentrated on membranes. We also cannot exclude the possibility that chains form as a result of interactions with the air-water interface or carbon surface. Chain structures arise when retromer links together at both the C-VPS35 (Figure 1C) and VPS26 ends (Figure 1D); VPS26-mediated interfaces resemble a chain link. We observed chains as short as 3 links (Figure S1C) and up to ~20 links, because chains can span an entire micrograph (data not shown). To analyze the structure of chains computationally, we used masks to separate chains into two different interfaces: the first interface was centered on VPS35-mediated dimers (chain interface I; Figure 1C), while the second was centered on VPS26 chain links (chain interface II; Figure 1D). We generated 3D reconstructions of the VPS35 dimer from chains (6.9 Å average resolution from 75,790 particles); the best-resolved portion occurs at the dimer interface (Figure 3E). This VPS35-mediated interface appears flat and exhibits 2-fold symmetry.
We also generated reconstructions of the VPS26-mediated interface (Figures 1D and 3G) at 17 Å average resolution from 13,782 particles. Retromer chains exhibit preferred orientation (Figure S2), which limits the resolution of our model. The data do not allow us to build an accurate model that captures how VPS26 subunits pack together. However, we clearly see a symmetrical 2-fold interface mediated by VPS26 in 2D classes. These links allow adjacent VPS35 subunits to curve out and away from each other, and we clearly observe the curve of VPS35 solenoids in both 2D classes and 3D reconstructions. Alternating heterotrimer units in this way would allow retromer to form an elongated repeating structure. Overall, the existence of retromer chains suggests that the heterotrimer alone may encode assembly information to build a flexible scaffold to accommodate various binding partners.
Retromer Tetramer of Trimmers
A second surprising result from EM studies was the presence of stable biochemical tetramers (Figure 1E). We observe tetramers in both micrographs and 2D class averages obtained from negatively stained (data not shown) or vitrified retromer (Figure S1B). Their presence is further supported by biophysical data (Figure 2). Tetramers are the rarest species in our samples (6,015 particles) but the overall architecture is relatively clear. Tetramers assemble using two interfaces: the first is a curved VPS35-mediated dimer interface similar to that observed in the dimer species. The second interface is mediated by VPS26, although this interface is very poorly resolved. Most of our views are “top-down” views, and limited side views suggest a curved particle shaped like a shallow boat or bowl. However, the low particle numbers, inherent VPS26 flexibility, and limited views impede our ability to generate reconstructions beyond 26 Å (Figures 1E and 3F).
VPS35 Contains a Conserved Electrostatic Interface to Mediate Dimer Assembly
We observed two different dimers mediated by C-VPS35 subunits in our retromer oligomers (Figures 1B and 1C). We specifically set out to compare the VPS35-mediated dimer found in chains with the curved VPS35 interface observed in dimers (discussed above). Dimer formation seems to be an inherent property of VPS35, since dimers form in solution (Figure 2) and on grids in the absence of sorting nexins or other binding partners. VPS35 α-solenoid regions consistently produced the best-resolved portions of 3D reconstructions (Figures 3A, 3B, and 3E). We therefore pursued sub-structures of VPS35-mediated interfaces observed in retromer dimers and in chain interface I (Figures 4 and S3). In both species, we masked the VPS35 N terminus and VPS26 subunits in order to focus on C-VPS35/VPS29 (masks shown in Figure S3). We produced reconstructions of each dimer sub-structure (Figures 4C and 4D) using the C-VPS35/VPS29 crystal structure (PDB: 2R17) as an initial model for rigid body fitting in Chimera.
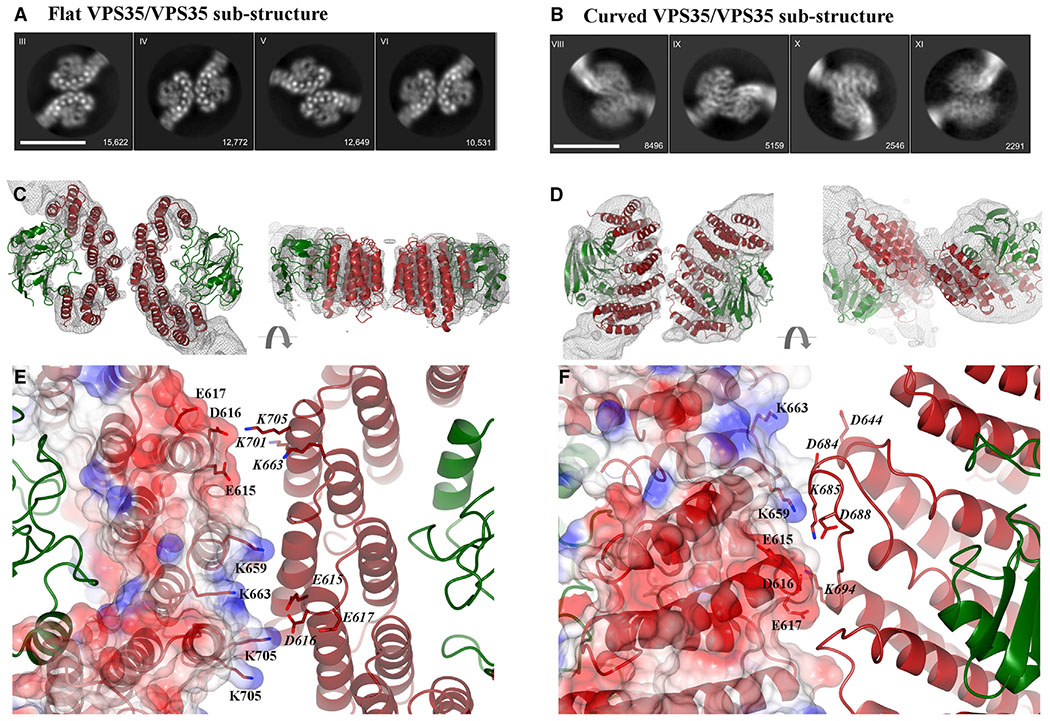
Figure 4. Conserved Electrostatic Patches Mediate Key VPS35 Dimer Interfaces.
(A and B) Representative 2D class averages of (A) the VPS35/VPS35 flat dimer sub-structure generated from retromer chains and (B) the VPS35/VPS35 curved sub-structure generated from retromer dimers. Scale bars represent 10 nm.
(C) Two different views (rotated 90°) of flat sub-structure reconstruction at ~5 Å shown as gray mesh (contoured at 4σ) with fitted model overlaid (PDB: 2R17). VPS35 is shown in red, VPS29 in green.
(D) Two different views of curved sub-structure reconstruction at ~6 Å with fitted model. Reconstructions are shown as gray mesh (contoured at 2σ).
(E) Inset shows detailed view of the flat dimer interface, which exhibits 2-fold symmetry. One VPS35 copy is shown as an electrostatic surface with residues marked in black text; the second copy is shown as a ribbon diagram with the same residues shown in italics. Key acidic residues in one copy (E615, D616, E617) interact with basic residues in the second copy (K659, K663, K701, K705).
(F) Inset shows detailed view of curved dimer interface, which does not exhibit 2-fold symmetry. The interface is shown as described in (E). Many of the same residues are observed in both dimer interfaces, including E615, D616, E617, K659, and K663.
See also Figure S3.
The sub-structure reconstruction from chain interface I (Figures 4A, 4C, and S3; 69,195 particles at 4.9 Å average resolution in RELION) revealed a symmetrical and flat 2-fold interface. In contrast, the sub-structure reconstruction from dimers (Figures 4B, 4D, and S3; 32,435 particles at 5.3 Å average resolution) revealed a curved orientation between VPS35 subunits, which does not exhibit 2-fold symmetry. We clearly observe α helices in 2D classes (Figure 4A) and 3D reconstructions (Figure 4C) of flat dimers. The curved sub-structure was more challenging because of its inherent flexibility, but we can observe overall architecture of the dimer following a rigid body fit in Chimera (Figure 4D).
Overall, improved reconstructions from sub-structures allowed us to generate models that reveal dimer interfaces located on the back side of VPS35 subunits, on the opposite face from where VPS35 binds VPS29 (Figures 4C and 4D). The flat dimer exhibits 2-fold symmetry, while the curved dimer shows one C-VPS35/VP29 has rotated away from its partner. In both dimers, VPS29 subunits point out and away from VPS35. VPS29 residues known to mediate interactions with key regulators, including VARP (Hesketh et al., 2014b) and TBC1D5 (Jia et al., 2016; Seaman et al., 2009), are fully exposed in each dimer. Maps from the flat sub-structure were well resolved (Figure 4C) and α helices are easily assigned. The structure is symmetrical and is not flexible, so we refined the model using real space refinement in PHENIX (Liebschner et al., 2019) (Table S2; STAR Methods). Maps from the curved substructure are poorer due to high flexibility and lack of symmetry, so we could not justify further refinement. However, reconstructions of the curved sub-structure allow us to observe overall arrangement of secondary and tertiary structural elements in order to compare the two dimer interfaces.
Both dimer interfaces contain multiple electrostatic residues (Figures 4E and 4F). The structures are further supported by biophysical data, which show that retromer oligomer formation depends on salt concentration (Figure 2). Many of the same residues are observed in both interfaces, including E615, D616, E617, K659, and K653. In the flat dimer, acidic residues (E615, D616, and E617) in one copy associate with basic residues (K663, K701, and K703) in its symmetry copy (Figure 4E). In the curved dimer, we observe the same acidic residues (E615, D616, and E617), but they appear to mediate different contacts with K685 and K694. The resolution of our sub-structures is insufficient to measure distances accurately. However, the model of the flat substructure suggests these electrostatic residues are located within 7–8 Å, which would be close enough to mediate salt bridge formation.
Many key residues in both interfaces are highly or absolutely conserved from yeast to humans (see next section), suggesting that they play an important biological role. We compared our interfaces with other proposed retromer dimers. Our interfaces differ from mammalian dimers proposed from SAXS data (Lucas et al., 2016; Norwood et al., 2010), which suggest an end-to-end VPS35 dimer. Our structures are more similar to a recent model proposed from C. thermophilum retromer (Kovtun et al., 2018) reconstituted on membranes. The cryo-ET model at 9 Å cannot resolve side chains, but our proposed residues are both conserved in C. thermophilum and are located in the VPS35 dimer interface proposed to form arch-like structures (Figure 6B). Based on available sequence and structural data, these electrostatic residues make excellent candidates for testing structure-based models (next section).
Figure 6. The VPS35 C Terminus Is Conserved across Species.
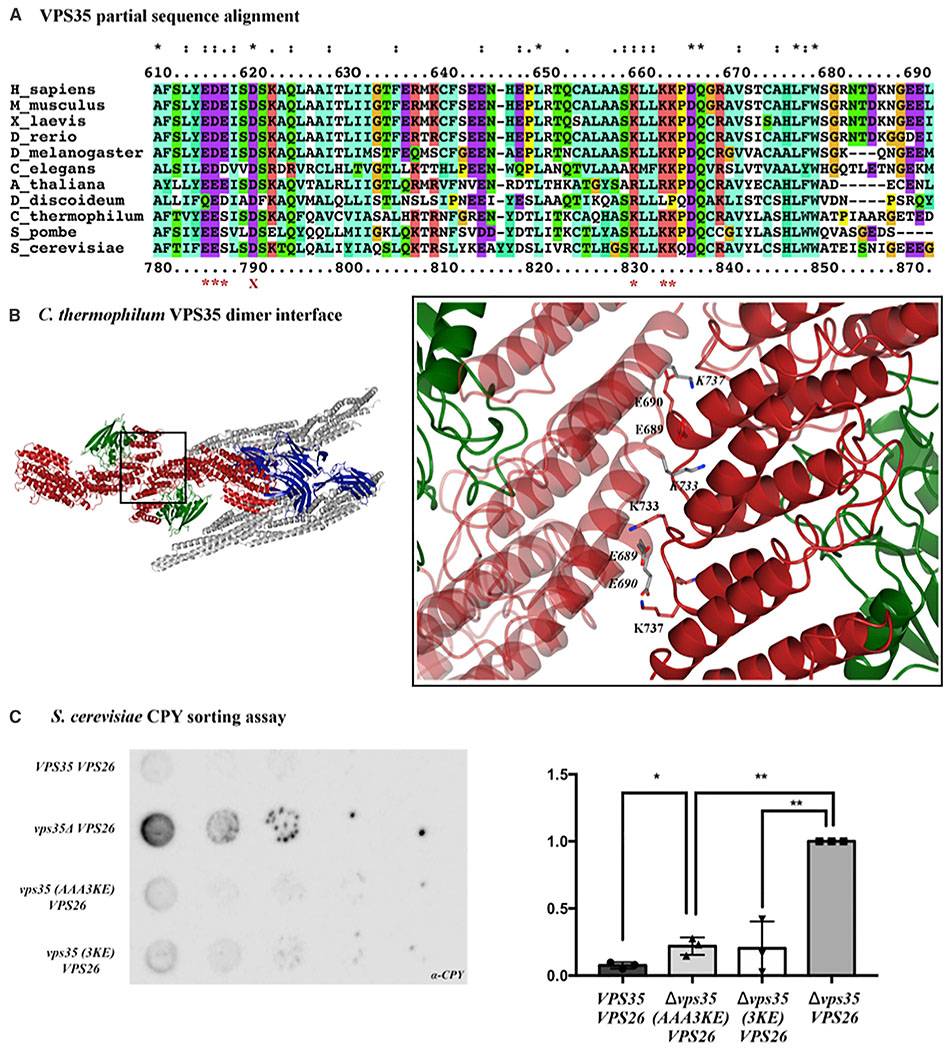
(A) Sequence alignments were generated in Clustal Omega; a partial alignment corresponding to M. musculus amino acid residues 610–690 are shown here for clarity. Human and yeast amino acid numbers are shown above and below the alignment. The following species were used to represent diversity across eukaryotes in the full-length alignment: S. cerevisiae, S. pombe, C. thermophilum, D. discoideum, A. thaliana, C. elegans, D. melanogaster, D. rerio, X. laevis, M. musculus, and H. sapiens. Red asterisks below the alignment mark conserved residues; these residues were mutated to generate the electrostatic mutant described in the main text. The red X marks residue D620, a residue implicated in late-onset Parkinson disease when mutated to asparagine.
(B) Conserved acidic and basic residues are found in the VPS35/VPS35 dimer interface of the C. thermophilum cryo-ET structure. Left-hand view shows top-down view of Vps5/retromer arches. TheVPS35 dimer interface shown in the inset. E689, E690, K733, and K737 are the thermophilic yeast equivalents of mouse residues E615, D616/E617, K659, and K662.
(C) One representative blot from a CPY secretion experiment in the ΔVPS35ΔVPS26 retromer strain at 26°C (left) with quantification from three biological replicates for each genotype in CPY secretion experiments (right). Data were analyzed using a one-way ANOVA with Tukey-Kramer post-hoc test. CPY secretion was normalized to the ΔVPS35 yeast strain (set at 1.0). The yeast retromer 3KE and electrostatic AAA3KE mutants secrete about twice as much CPY compared with wild-type retromer. Raw western blots from all three replicates are shown; see also Figures 3 and S5, and Table S3.
Mutating the Conserved Interface Disrupts Assembly In Vitro
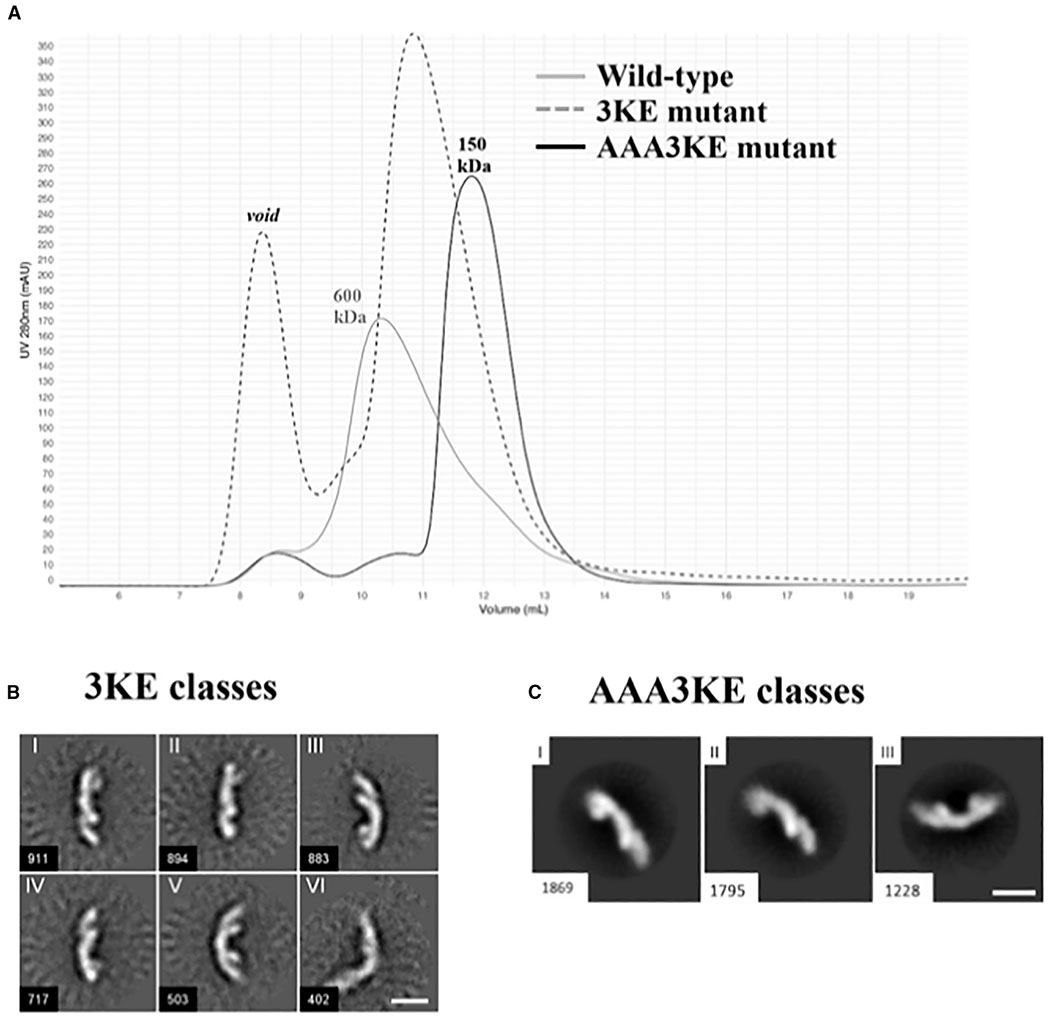
We hypothesized that key electrostatic residues in our proposed VPS35/VPS35 interfaces should promote assembly. To test this explicitly, we introduced point mutations in VPS35 subunits predicted to break apart the electrostatic interface and generated recombinant purified retromer protein containing mutant VPS35 subunits with wild-type VPS26 and VPS29. To test assembly in vitro, we combined size-exclusion chromatography with negative stain EM to test whether point mutations affect whether retromer forms higher-order oligomer structures. We identified five candidate residues conserved from yeast to humans (Figure 6A): E615, E617, K659, K662, and K663. We also included D616, which is conserved from insects to vertebrates. Initially, we attempted a charge swap in which we mutated three conserved lysine residues (K659, K662, and K663) to glutamates. We predicted that this should prevent dimer formation by causing VPS35 subunits to repel each other. This mutant gave a partial phenotype (Figures 5A and 5B) and exhibited a shift over gel filtration. The main wild-type retromer peak elutes predominantly at volumes consistent with tetramers over gel filtration, while the 3KE mutant elutes predominantly as a heterotrimer. This suggests that we successfully mutated residues that mediate assembly. However, we still observed dimers in negatively stained samples (Figure 5B), although at lower frequency than in wild-type samples (Table S4).
Figure 5. Mutating the Conserved VPS35 Interface Disrupts Assembly In Vitro.
(A) Gel filtration profiles of wild-type mammalian retromer (gray trace); the partial 3KE mutant (E615A/D616A/E617A; dashed trace) and the electrostatic AAA3KE mutant (E615A/D616A/E617A/K659E/K662E/K663E; black trace). The main wild-type peak elutes at a volume consistent with larger oligomeric assemblies (~600 kDa). The 3KE mutant is intermediate, while the electrostatic mutant peak elutes at a volume consistent mostly with heterotrimer (~150 kDa).
(B) Representative 2D class averages of the 3KE electrostatic mutant in negative stain. Most retromer particles exist as heterotrimers, but a population of dimers remains (class VI). Scale bar represents 10 nm.
(C) Representative 2D class averages of the AAA3KE mutant show most protein exists as heterotrimers. For both mutants, the overall 3D structure of the heterotrimer is preserved. Scale bar represents 10 nm.
Therefore, we introduced three additional mutations to generate the retromer electrostatic mutant, AAA3KE (E615A/D616A/E617A/K659E/K662E/K663E). This mutant exhibited a clear shift in gel filtration profile (Figure 5A). We compared negatively stained samples of wild-type, 3KE, and AAA3KE mutant retromer at the same protein concentration (Table S4). We selected five representative micrographs for each sample; in total, micrographs contained ~2,100 copies of retromer. We then counted observed structures in all samples. We found 79% of wild-type retromer exists as heterotrimers, 19% as dimers, and 2% as tetramers. For the 3KE mutant, 86% exists as heterotrimers; 13% as dimers; and less than 1% as tetramers. For the AAA3KE mutant, 98% of observed structures are heterotrimers. We identified 30 (of 2,250) particles that might be dimers or monomers located very close to one another. We observed no tetramers. Finally, representative 2D class averages of negatively stained mutant 3KE or AAA3KE protein revealed stable heterotrimers (Figures 5B and 5C; cf. S4B); this confirms that we did not disrupt overall heterotrimer fold. These data suggest that we successfully disrupted a key interface mediating formation of oligomers formed by mammalian retromer in vitro.
Mutating the Conserved Interface Partially Impedes Cargo Sorting in Budding Yeast
We further tested whether mutants based on our structural models would disrupt retromer assembly in vivo. Reconstituted Vps5/retromer from thermophilic yeast assembles VPS35-mediated dimers containing the same conserved residues (Figures 6A and 6B), so we tested whether this interface is conserved across eukaryotes. We undertook a functional cargo-sorting assay in budding yeast, S. cerevisiae, to determine whether breaking the conserved assembly interface has a functional consequence. We used the well-characterized carboxypeptidase Y (CPY) secretion assay (Paravicini et al., 1992). In brief, budding yeast sort CPY to the vacuole when retromer is present and functional. Deletion of retromer genes instead drives CPY secretion through mis-sorting of Vps10, and CPY secretion is measured and quantified at the cell surface using an immunoblotting assay.
We generated a Δvps35Δvps26 deletion strain from a well-established Δvps35 strain (Paravicini et al., 1992) (kindly provided by Scott Emr’s lab, Cornell). We then re-introduced either wild-type or mutant retromer VPS35, together with wild-type VPS26, on plasmids under control of endogenous promoters and measured how much CPY was secreted in cells containing wild-type or mutant retromer. We normalized our data against a strain lacking VPS35 by arbitrarily setting its CPY secretion at 1.0. The recombinant wild-type retromer strain, in which both VPS35 and VPS26 genes were re-introduced, secreted CPY at ~10% of Δvps35 levels (Figures 6C and S5). Both the vps35 3KE or AAA3KE mutants consistently secreted about twice as much (~20%) CPY as compared with wild-type, across three biological replicates (Figures 6C and S5).
Mutating key conserved residues in the VPS35 dimer interface thus causes a reproducible and measurable cargo-sorting defect, but loss of these residues does not preclude cargo sorting in budding yeast. The heterotrimer has long been proposed to serve as the cargo-binding module for yeast receptors, while recent data from mammalian systems (Kvainickas et al., 2017b; Simonetti et al., 2017) suggests that sorting nexins act as cargo adaptors. Our data may support the idea that either VPS26 or SNX proteins influence cargo sorting more directly in yeast. Mutating the heterotrimer scaffold at the VPS35 interface may partially destabilize the coat, but it is insufficient to block cargo sorting. The data may also be explained by the adaptability of the retromer scaffold (discussed further below).
DISCUSSION
We have shown here how the conserved retromer heterotrimer (VPS26/VPS35/VPS29 subunits) forms stable oligomers. Single-particle cryo-EM reconstructions provide the first structural snapshots of multiple mammalian retromer oligomers. Previous reports (Lucas et al., 2016; Norwood et al., 2010) identified a mammalian dimer based on very-low-resolution SAXS data, in which two groups proposed dimers mediated by the very C-terminal tips of VPS35 subunits. Our structural data at improved resolution instead support an interface mediated by key conserved electrostatic residues located on the backside of VPS35 C termini. One of our dimer models is similar to a published thermophilic yeast cryo-ET model (Kovtun et al., 2018) (discussed further in next section). Biochemical data suggest that this conserved interface mediates retromer dimer assembly. Identification and verification of specific electrostatic residues in the interface provides a useful molecular tool to the community for testing retromer function more precisely with different cargoes across multiple model systems.
Implications for Retromer Assembly and Regulation
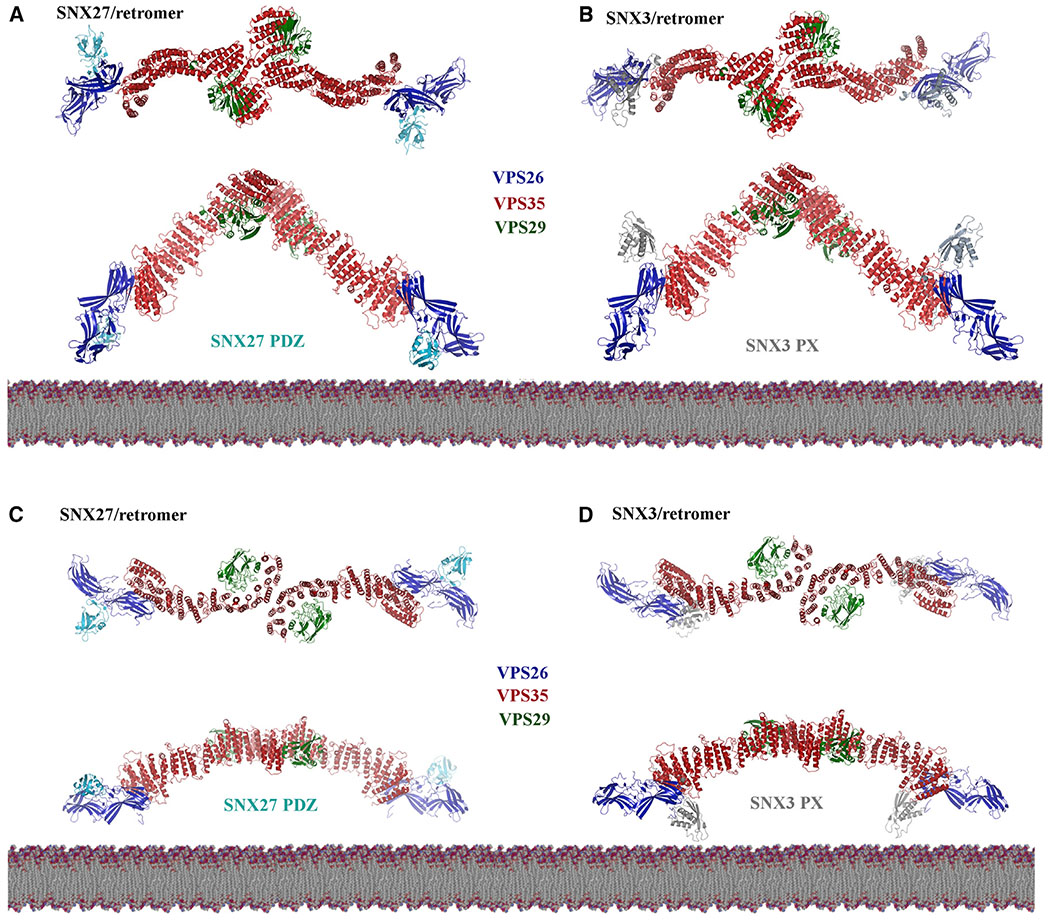
Our structural data suggest possible models for retromer assembly (Figure 7). We generated models for structures with SNX27 and SNX3 based on the curved and flat VPS35-mediated dimer interfaces by overlaying crystal structures (PDB: 4P2A and 5F0J) onto cryo-EM reconstructions. The considerable VPS26 flexibility (Figure 3) makes it challenging to depict the orientation of VPS26, but its position seems consistent with that observed in a crystal structure (Lucas et al., 2016) and cryo-ET reconstructions (Kovtun et al., 2018). We modeled VPS26 based on the crystal structure here, but our data explicitly show substantial flexibility at the VPS26/N-VPS35 interface, implying that VPS26 may adopt multiple conformations.
Figure 7. Retromer Is a Plastic Scaffold for Cargo Sorting on Membranes.
Possible models of assembled retromer dimers (based on this work) shown with mammalian sorting nexins, SNX27 and SNX3. The left-hand column shows putative models of curved (A) or flat (C) retromer dimers with the SNX27 PDZ domain; models were generated by overlaying dimers with the VPS26/SNX27 PDZ crystal structure (PDB: 4P2A). Both retromer dimers place the SNX27 PDZ domain close to the membrane, where it could engage cargoes containing a PDZ-binding motif. The SNX27 PX and FERM domains are not shown, because there are no structures of full-length SNX27. The right-hand column shows models of curved (B) or flat (D) retromer dimers with the SNX3 PX domain; models were generated by overlaying dimers with the N-VPS35/VPS26/SNX3 PX crystal structure (PDB: 5F0J). Only flat retromer dimers (D) place the SNX3 PX domain close to the membrane in an orientation where known PX residues could interact with PI3P. See also Figures S6 and S7.
Overall, extended arch-like retromer dimers seem more likely to be partial retromer assemblies in solution or the cytoplasm. Dimer models (Figures 7A and 7B) position the SNX27 PDZ cargo-binding domain close to the membrane (Figure 7A). Retromer could sit in this orientation, but it remains unclear how retromer would form a repeating unit. The SNX3/retromer model positions the PX domain relatively far from the membrane (Figure 7B), another unlikely scenario for coat assembly. We predict that encountering specific SNXs or cargoes in PI3P-enriched membranes will correctly orient and position the retromer scaffold depending on biological need (i.e., what cargoes require sorting).
In contrast, the existence of flat chains may suggest a new assembly mechanism for mammalian retromer. Formation of flat chains could position both SNX27 PDZ (Figure 7C) and SNX3 PX domains (Figure 7D) close to the membrane. SNX27/retromer coats would presumably be further oriented through binding PI3P via the PX domain and NPxY cargoes via the FERM domain. The PI3P-binding pocket for SNX3/retromer would also be apposed to the membrane. Chains further suggest a way for VPS26 subunits to form interfaces that extend retromer into repeating units (Figure S6). These chains would form very elongated structures with shallow and gently curving arches. These chains position VPS29 subunits at arch apexes located closer to the membrane, which in turn may have regulatory implications. Both VARP (Hesketh et al., 2014a) and TBC1D5 (Jia et al., 2016; Seaman et al., 2009) interact with VPS29, and key VPS29 residues are exposed and available in all of our models. VARP contains two cysteine-rich repeats, and both have been shown to bind the same patch (L152) on VPS29 (Hesketh et al., 2014a). Together with structural data, this implies that one VARP could “bridge” a retromer VPS35-mediated dimer. VARP also binds endosomal Rab proteins and the R-SNARE, VAMP7. We can speculate VARP may favor binding a flatter dimer located closer to the membrane, or, alternatively, VARP could prompt a conformational change as part of its regulatory role.
Comparison with Yeast Vps5/Retromer Structure
The cryo-ET reconstruction of thermophilic yeast Vps5/retromer revealed how the retromer heterotrimer forms two types of dimers when assembled in vitro on membranes in the presence of an SNX-BAR protein (Kovtun et al., 2018). Overall, we observe no major differences between heterotrimer architecture (Figure S7A), although our local resolution data suggest a “hinge point” located approximately in the middle of the VPS35 solenoid. In the yeast structure, a symmetrical VPS26-mediated dimer is organized directly through interactions with Vps5 homodimers. VPS35 subunits mediate formation of a second arch-shaped dimer. We attempted to address two questions arising from the cryo-ET model here. Does the VPS35-mediated dimer exist only in the presence of SNXs, and is this dimer conserved across evolution? Our biochemical and structural data agree with previously published reports (Lucas et al., 2016; Norwood et al., 2010) indicating that mammalian retromer forms dimers on its own in solution. We have now visualized those dimers, as well as new chain and tetramer structures, for the first time. Together, these data imply that the heterotrimer acts as a scaffold with intrinsic assembly capabilities. We further show how VPS35 contains a specific electrostatic interface, because we can disrupt dimer assembly in vitro with targeted point mutations (Figure 5).
We note that interfaces observed in flat VPS35 dimers from mammalian chains look similar to the VPS35 dimer observed at the top of thermophilic yeast arches; the mammalian dimer is just flatter (Figure S7B). This could place mammalian arch apexes closer to the membrane and cause the rest of the heterotrimer to adopt a different conformation, which leads to formation of flat chains instead of tall arches. Local resolution estimates in our structures revealed that retromer exhibits inherent flexibility at multiple points, including a hinge point in the VPS35 solenoid; at VPS35/VPS35 dimer interfaces; and at VPS26/ N-VPS35 interfaces. These data support the idea that retromer is an adaptable and plastic scaffold that can adopt a range of conformations. Retromer may interact with SNX-BAR proteins by forming arches (Kovtun et al., 2018); SNX-BAR binding could promote or induce arch formation by organizing the scaffold. Modeling suggests that flat chains could bind either SNX27 or SNX3, both of which lack BAR domains, in orientations placing lipid- or cargo-binding domains close to the membrane. Finally, although they are not well resolved, the VPS26/VPS26 dimer observed in chain links (Figure 1D) is very different from the yeast VPS26 dimer observed in the presence of Vps5 (Kovtun et al., 2018). Chains seem to form by tip-to-tip interactions between adjacent VPS26 subunits, as opposed to back-to-back interactions between N-VPS26 subdomains. Higher-resolution structures for both binding modes will be required to understand these structures in more detail and to test assembly interfaces both in vitro and in vivo.
Why might retromer scaffold plasticity be advantageous for cells? Yeast retromer often occurs as a pentamer, in which the heterotrimer assembles with Vps5/Vps17 dimers containing BAR domains that recognize or drive membrane curvature. However, both yeast and mammalian retromer also assemble with SNXs that lack BAR domains. Mammalian retromer also sorts cargoes to different destinations from a common origin. Thus, retromer may require plasticity to assemble distinct retromer complexes with specific sorting nexins to sort cargoes to different destinations.
An unresolved question is whether the tetramers we observe are relevant to retromer assembly. Tetramers are observed in micrographs (Figure S1) and in solution (Figure 2). Tetramers must interact via both VPS35- and VPS26-mediated interfaces. There are at least three possibilities that explain our observation of tetramers. First, they may be a biochemical artifact. We do not favor this interpretation, because we can specifically disrupt their formation with point mutations, but it formally remains a possibility. Second, tetramers may represent a cytosolic species that provides a retromer pool for rapid assembly on membranes in the presence of sorting nexins and cargo. Clathrin has been shown to assemble on membranes from pre-existing cytosolic pools (Avinoam et al., 2015; Heuser, 1980), and we can conceptually consider retromer as a scaffold analogous to the clathrin cage. Finally, tetramers may represent another way to interact with membranes to sort cargoes; perhaps retromer could interact with specific SNX proteins in this way. We cannot ascertain the orientation of VPS26 in our current tetramer reconstructions. We will require many more particles to generate higher-resolution structures and ascertain tetramer architecture.
Functional Consequences in Yeast
Similarities between yeast and mammalian retromer assembly and function remain a major question in the retromer field. We explicitly tested for functional conservation in the electrostatic interface using the CPY secretion assay in budding yeast (Figure 6C). Disrupting conserved residues in the retromer scaffold has a modest but reproducible effect on CPY secretion. There are multiple possible reasons for the modest effect in budding yeast. First, the well-established CPY secretion assay is based on deleting retromer genes, which causes substantial Vps10 sorting defects (Paravicini et al., 1992). Our point mutations instead target specific residues in a specific interface. Second, yeast retromer is thought to exist as a relatively stable pentamer, in contrast to mammalian retromer. Multiple binding interfaces remain intact in our mutants, including the Vps5/Vps17 and VPS26 dimers (Kovtun et al., 2018). Therefore, targeted mutations in the VPS35 interface alone may be insufficient to drive a strong defect. Finally, the biochemical basis for Vps10 cargo binding to retromer remains unclear in yeast. In mammalian cells, the Vps10 homologue (SorLA) has been shown to depend upon VPS26 (Fjorback et al., 2012). However, other key receptors, such as CI-MPR bind SNX proteins (Kvainickas et al., 2017a; Simonetti et al., 2017). It will be important to test other cargoes and to establish the molecular basis for cargo binding by retromer in yeast to more fully understand these phenotypes.
Implications for Human Disease
The VPS35/VPS35 interfaces observed here and in the yeast cryo-ET structure have relevance to human disease. The VPS35 D620N mutation is linked to an autosomal dominant form of Parkinson disease (Vilariño-Güell et al., 2011; Zimprich et al., 2011). Our structural data suggest that D620 is located close to the acidic patch formed by E615, D616, and E617. This may suggest that D620 plays an indirect role in maintaining the acidic surface required at the interface. Its mutation to asparagine may alter overall charge distribution and destabilize retromer assembly. This mutation has been proposed to interfere with the ability of VPS35 to associate with the WASH complex (McGough et al., 2014; Zavodszky et al., 2014). The WASH complex may favor interacting with a retromer dimer, and thus destabilizing the VPS35/VPS35 dimer interface would indirectly affect the ability of retromer to engage WASH. Alternatively, because this residue is located underneath the VPS35 dimer in the arch, it may play a role in engaging other binding partners.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
All resources and reagents, including plasmids, are available from the Lead Contact, Lauren P. Jackson (lauren.p.jackson@vanderbilt.edu) with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains are listed in Table S3. Standard media and techniques for growing and transforming S. cerevisiae were used. Yeast gene knockout was performed using a PCR toolbox (Janke et al., 2004). The VPS26/VPS35 double knockout strains were generated from a well-characterized ΔVPS35 strain (Paravicini et al., 1992). The double ΔVPS35/ ΔVPS26 knockout strain was confirmed by PCR-based genotyping. Details on yeast strains are provided in Table S3. Plasmids were generated using Gibson assembly methods. Wild-type and mutant VPS35 was inserted into pRS315, and wild-type VPS26 into pRS416. Both genes remain under control of the endogenous promoter and are expressed at wild-type levels.
METHOD DETAILS
Reagents
Unless otherwise noted, all chemicals were purchased from Sigma (St. Louis, MO, USA).
Molecular Biology and Cloning
Retromer constructs were generated in the labs of David Owen and Brett Collins and have been published previously (Collins et al., 2005, 2008). We used a two-stage quick-change mutagenesis protocol adapted from Wang and Malcolm (Biotechniques 1999) to introduce point mutations into retromer plasmids to generate retromer electrostatic mutants. Briefly, mutagenic primers (Sigma) were created for the desired mutations. In the first step, two polymerase chain reactions (PCRs), with either the mutagenic 5’ or 3’ primer, were amplified around the plasmid. The two reactions were then combined in an additional PCR step, and the product was digested using Dpn I. Digested product was used to transform XL1 Blue (Agilent) competent cells, and colonies were sequenced using Sanger methods (Genewiz).
Protein Expression and Purification
We expressed and purified recombinant retromer protein (wild-type and mutants) from E. coli as previously described (Collins et al., 2008). Briefly, retromer plasmids were transformed into BL21(DE3) Rosetta2 pLysS cells (Millipore). Cells were grown to OD600 between 0.8-1.0 and induced for 16-20 hours at 22°C with 0.4 mM IPTG. Cells were lysed by a disruptor (Constant Systems Limited). Protein was purified in 10 mM Tris-HCl (pH 8.0), 200 mM NaCl, 2 mM βME using glutathione sepharose (GE Healthcare). Protein was cleaved overnight using thrombin (Recothrom, The Medicines Company) at room temperature and batch eluted in buffer. Retromer was further purified by gel filtration on a Superdex S200 analytical column (GE Healthcare) into 10 mM Tris-HCl (pH 8.0), 200 mM NaCl, or dialysis into 20 mM HEPES pH 8.2, 50 mM NaCl, 2 mM DTT for cryoEM experiments.
Negative Stain Grid Preparation & Screening
For screening of negatively stained retromer samples, 10 μl of retromer at concentrations between 5 and 10 μg/mL were applied to continuous carbon film on 400 square mesh copper EM grids (Electron Microscopy Sciences, Hatfield, PA) and washed twice with water. The grids were stained with 2% uranyl formate and 1% uranyl acetate and air dried overnight. The grids were screened on a ThermoFisher FEI Morgagni microscope operating at 100kV with a AMT 1k×1k CCD camera (Vanderbilt University CryoEM Facility) to verify protein quality. The grids were imaged on a ThermoFisher FEI Tecnai F20 operating at 200 kV with a 4kx4k CCD camera (Vanderbilt University CryoEM Facility).
Cryo-EM Grid Preparation and Data Collection
For cryo-electron microscopy, retromer at a concentration of 100 μg/ml was applied to freshly glow discharged CF-2/2-2C C-Flat grids (Protochips, Morrisville, NC), and the grids were vitrified in liquid ethane using either a ThermoFisher FEI MarkIII or MarkIV Vitrobot. Data were collected from 2779 micrographs at the National Resource for Automated Molecular Microscopy (NRAMM) in two different data collection sessions using ThermoFisher FEI Titan Krios microscopes operating at 300keV. The first data collection used a Titan Krios equipped with a spherical aberration (Cs) corrector; a Volta phase plate; a Gatan BioQuantum energy filter; and a Gatan K2 Summit camera. The second data collection used a similar Krios instrument but without a Cs corrector. The nominal magnification used during each data collection was 105,000x and 130,000x, respectively. The effective pixel size was 1.096Å/pix for the first data collection and 1.06Å/pix for the second data collection. The total electron dose during the two data collection sessions was between 69 and 74 e−/A2. Table S1 contains a data collection summary.
Single Particle Cryo-EM Image & Data Processing
All images were motion corrected using MotionCor2 (Zheng et al., 2017). Because we used different Krios microscopes in two data collection sessions, micrographs from the second data collection were rescaled to match the 1.096Å/pix pixel size from the first data collection using an NRAMM script written for MotionCor2. The CTF of each micrograph was determined using Gctf (Zhang, 2016); defocus values for the data varied between −0.7 and −4.4μm. RELION-2 (Scheres, 2012) and RELION-3 (Zivanov et al., 2018) were used for all image processing unless otherwise indicated.
For each dataset, we manually selected several thousand particles and performed an initial 2D classification to produce templates for autopicking. Autopicking for the heterotrimer, dimer, chain interface I, tetramer, and curved and flat substructures identified 249,562 particles from the first Krios dataset and 190,084 particles from the second dataset. Multiple rounds of 2D classification were performed to separate the different species into distinct particle sets. Initial models for 3D classification were generated from models generated by earlier experiments, filtered to 60 A resolution. Detailed methods are provided below; image processing work-flow diagrams are provided in Figures S2 and S5; and statistics are provided in Table S2.
Heterotrimer
2D classification of the heterotrimer particle began with 29,771 particles at a box size of 252x252 Å. Multiple rounds of 2D classification and re-centering of these particles yielded 29,702 particles suitable to continue to 3D classification. A preliminary 3D model generated using only particles from the first dataset was filtered to 60 Å and used as an input model for 3D classification; 6 output classes were used. 26,369 particles from 3D classification were selected to continue to 3D refinement and postprocessing. The final masked heterotrimer model had a resolution of 5.7 Å and a Relion-determined B-factor of −212. Directional resolution was determined using the 3D FSC remote server (Tan et al., 2017). The heterotrimer was refined in PHENIX (Liebschner et al., 2019) with phenix.real_space_refine using global minimization and rigid body parameters (Table S2).
Dimer
2D classification of the dimer particle began with 34,371 particles at a box size of 395x395 Å. Numerous rounds of 2D classification were required to separate the dimer structure from the flat chain (chain interface I), and 31,147 dimer particles were selected to continue to 3D classification. A preliminary 3D model generated using only particles from the first dataset was filtered to 60 Å and used as an input model for 3D classification; 6 output classes were used. 31,022 particles from 3D classification were selected to continue to 3D refinement and postprocessing. The final post-processed dimer model had a resolution of 9.3 Å and a user-imposed B-factor of −100 to optimize contrast and minimize noise (Jakobi et al., 2017). Directional resolution was determined using the 3D FSC remote server (Tan et al., 2017).
Chain Interface I
For computational reasons, chain particles were separated into two units (called chain interface I and chain interface II) for data processing; the box size for a longer chain encompassing both interfaces is impractical. 2D classification of the chain interface I particle (centered on the VPS35/VPS35 interface) began with 79,104 particles at a box size of 395x395 Å. Numerous rounds of 2D classification were required to separate the chain interface I structure from the dimer. 76,852 particles were selected to continue to 3D classification using C2 symmetry, and a preliminary 3D model generated using only particles from the first dataset was filtered to 60 Å and used as input; 6 output classes were used. 75,790 particles were selected for 3D refinement and postprocessing. The final masked model had a resolution of 7.1 Å and a Relion-determined B-factor of −189. Directional resolution was determined using the 3D FSC remote server (Tan et al., 2017).
Chain Interface II
A single class (class XVI) from the original 2D classification of all autopicked particles was used for a new round of autopicking. This autopick job identified 301,950 particles from the first dataset and 141,167 particles from the second dataset. These 443,117 particles, with a box size of 482x482 Å, were then subjected to 2D classification. 14,592 particles were selected to continue to 3D classification. A preliminary 3D model of this chain was built in Chimera using two heterotrimers interacting at their VPS26 ends; this preliminary model was then filtered to 60 Å and used as input for 3D classification; 6 output classes were used. 13,782 particles were selected for 3D refinement and postprocessing. The final masked chain interface II model had a resolution of 16.9 Å and a Relion-determined B-factor of −212. Directional resolution was determined using the 3D FSC remote server (Tan et al., 2017).
Flat Substructure
Processing of the flat substructure began with re-extraction of 79,104 chain interface I particles at a box size of 180x180Å. Repeated rounds of 2D classification with a mask of 149 Å were performed. 76,927 particles were selected to continue to 3D classification using C2 symmetry, and a preliminary 3D model generated using only particles from the first dataset was filtered to 60 Å and used as input; 6 output classes were used. 69,195 particles selected from 3D classification were subjected to CTF refinement, then continued to 3D refinement and postprocessing. The final masked flat substructure model had a resolution of 5.0 Å and a Relion-determined B-factor of −56. Directional resolution was determined using the 3D FSC remote server (Tan et al., 2017). The flat sub-structure was refined in PHENIX (Liebschner et al., 2019) using phenix.real_space_refine with the global minimization and rigid body parameters (Table S2).
Curved VPS35/VPS35 Substructure
Processing of the curved sub-structure began with re-extraction of 34,371 dimer particles at a box size of 180x180 Å. Repeated rounds of 2D classification with a mask of 149 Å were performed. 33,883 particles were selected to continue to 3D classification, and a preliminary 3D model generated using only particles from the first dataset was filtered to 60 Å and used as input; 6 output classes were used. 32,435 particles from 3D classification were selected to continue to 3D refinement and postprocessing. The final masked curved substructure model had a resolution of 5.3 Å and a Relion-determined B-factor of −58. Directional resolution was determined using the 3D FSC remote server (Tan et al., 2017).
Tetramer
2D classification of the tetramer particle began with 6,127 particles at a box size of 329x329 Å. 6101 particles continued to 3D classification, which used an initial model generated in cryoSPARC (Punjani et al., 2017) and filtered to 60 Å; 6 output classes were used. 6015 particles from 3D classification were selected to continue to 3D refinement and postprocessing. The final masked tetramer model had a resolution of 27.4 Å; no B-factor was determined by Relion in postprocessing. Directional resolution was determined using the 3D FSC remote server (Tan et al., 2017).
Model Building & Docking
An initial model of mammalian retromer heterotrimer was produced by combining X-ray crystallographic structures of retromer subunits (PDB: 2R17, 5F0J). Models for VPS29 and the VPS35 C-terminus were obtained from PDB 2R17, while models for N-VPS35 and VPS26A were obtained from PDB 5F0J. We omitted a flexible unstructured loop (amino acids 470–482) that is absent in all crystal structures. Rigid-body docking and map visualization were performed using Chimera (Pettersen et al., 2004) using Chimera Fit in Map routine.
SEC MALS
Size-exclusion chromatography coupled to multi-angle laser light scattering (SEC-MALLS) was performed on an ÄKTA Purifier FPLC system (GE Healthcare) in-line with an Agilent Technologies 1200 Series refractive index Detector and a Dawn Helios 8+ MALS unit (Wyatt). 100 μL of sample at 15 μM was injected from a static loop beginning at the 0 ml point of each run onto a Superdex 200 increase 10/300 GL column (GE Healthcare) at a flow rate of 0.5 ml/min. Samples were assayed at 25°C in 20 mM Tris-HCl (pH 8.5), 2mM DTT, and 8 mM NaN3 at two different salt concentrations (50 mM NaCl and 500 mM NaCl). Refractive index and light scattering data were recorded and analyzed using ASTRA 6.1 (Wyatt) software. Molecular masses were calculated across distinct eluted peaks using the Zimm algorithm with a dn/dc value of 0.185 ml/g.
Dynamic Light Scattering
Dynamic light scattering experiments were performed using a Wyatt Technology DynaPro NanoStar instrument. Retromer samples were assayed in 20 mM Tris-HCl (pH 8.5), 2 mM DTT at two different salt concentrations (50 mM NaCl and 500 mM NaCl). 10 μL of sample at 7 μM was equilibrated at 25°C for 5 minutes prior to data acquisition in a disposable 4 μL cyclic olefin copolymer cuvette. Twenty acquisitions at 5s intervals were recorded for each sample and fitted with the coils shape model using the Wyatt Dynamics software.
Yeast Plasmids
Plasmids were generated using Gibson assembly methods. Wild-type and mutant versions of VPS35 (3KE or AAA3KE) were inserted into pRS315, and wild-type VPS26 into pRS416. Both genes remain under control of the endogenous promoter and are expressed at wild-type levels.
Carboxypeptidase Y Secretion Assay
Retromer mutant yeast cells were 10-fold serial diluted and spotted on YPD plates at 26°C. The cells were grown for 24 hr prior to being overlaid with a nitrocellulose membrane. The operation is carefully performed to prevent air bubbles from being trapped under the membrane. The plates were grown for an additional day. The nitrocellulose membranes were washed several times with a gentle flow of deionized water and then placed in PBS-T buffer (standard phosphate-buffered saline with 0.1% (v/v) Tween-20) until ready to use. The membrane was blocked in Odyssey Blocking Buffer (LI-COR Biosciences) for 1 hr at room temperature then incubated with anti-CPY antibodies (Invitrogen) at1:1000 in blocking buffer overnight. After washing with PBS-T, the membrane was incubated with HRP-goat anti-mouse IgG antibody (Invitrogen) (1:10,000 in PBS-T +5% non-fat milk) for 1 hr at room temperature. The membrane was washed by PBS-T and then developed by Amersham ECL Western Blotting Detection Reagent (GE Healthcare).
QUANTIFICATION AND STATISTICAL ANALYSIS
Carboxypeptidase Y Secretion Assay
Yeast blots were imaged with the AI600 Chemiluminescent Imager system (GE Life Sciences). The dot intensity was quantified by ImageStudio (GE Life Sciences). Statistical differences were determined using a one-way ANOVA on the means of at least three independent experiments using GraphPad Prism (GraphPad Software). Probability values of less than 0.05, 0.01 and 0.001 were used to show statistically significant differences and are represented with *, ** or *** respectively.
DATA AND CodeAvailability
EM electron density maps were deposited in the EMDB with accession numbers EMD-21136, EMD-21117, EMD-21116, EMD-21118, and EMD-21011 corresponding to the retromer heterotrimer, dimers, chain interface I, chain interface II, and tetramer maps. Sub-structure maps for the flat and curved VPS35 dimers were deposited as EMD-21135 and EMD-21119. Coordinates for the heterotrimer and flat VPS35/VPS35 sub-structures were deposited with PDB accession codes 6VAC and 6VAB.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CPY | Invitrogen | A-6428; RRID: AB_2536203 |
| Deposited Data | ||
| Heterotrimer map | This paper | EMDB- EMD-21136 |
| Dimer map | This paper | EMDB: EMD-21117 |
| Tetramer map | This paper | EMDB: EMD-21101 |
| Chain interface I map | This paper | EMDB: EMD-21116 |
| Chain interface II map | This paper | EMDB: EMD-21118 |
| Flat VPS35/VPS35 sub-structure map | This paper | EMDB: EMD-21135 |
| Curved VPS35/VPS35 sub-structure map | This paper | EMDB: EMD-21119 |
| Heterotrimer model | This paper | PDB: 6VAC |
| Flat VPS35/VPS35 sub-structure model | This paper | PDB: 6VAB |
| VPS35 C-terminus/VPS29 structure | (Hierro et al., 2007) | PDB: 2R17 |
| N-VPS35/VPS26/SNX3 structure | (Lucas et al., 2016) | PDB: 5F0J |
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae; SEY6210.1 MATa leu2-3, 112 ura3-52 his3-Δ 200 trp1-Δ901 lys2-801 suc2- Δ9 vps35D::KanMX6 | This paper | N/A |
| Recombinant DNA | ||
| GST-VPS35-VPS29 in pGEX4T2 (mouse) | (Collins et al., 2005, 2008) | N/A |
| H6-VPS26 in pMWKan (mouse) | (Collins et al., 2005, 2008) | N/A |
| GST-VPS35 (3KE)-VPS29 in pGEX4T2 (mouse) | This paper | N/A |
| GST-VPS35 (AAA3KE)-VPS29 in pGEX4T2 (mouse) | This paper | N/A |
| VPS35 in pRS315 (S. cerevisiae) | This paper | N/A |
| vps35 3KE in pRS315 (S. cerevisiae) | This paper | N/A |
| vps35 AAA3KE in pRS315 (S. cerevisiae) | This paper | N/A |
| VPS26 in pRS416 (S. cerevisiae) | This paper | N/A |
| Software and Algorithms | ||
| RELION-2 | (Scheres, 2012) | N/A |
| RELION-3 | (Zivanov et al., 2018) | N/A |
| MotionCor2 | (Zheng etal., 2017) | N/A |
| Gctf | (Zhang, 2016) | N/A |
| Chimera | (Pettersen et al., 2004) | N/A |
| PHENIX | (Liebschner et al., 2019) | N/A |
| 3DFSC | (Tan et al., 2017) | N/A |
| cryoSPARC | (Punjani et al., 2017) | N/A |
Highlights.
The metazoan retromer heterotrimer forms dimers, tetramers, and chains
Cryo-EM and biophysical studies reveal an electrostatic VPS35 dimer interface
Targeted structure-based point mutations disrupt the VPS35 interface in vitro
Structural data suggest that retromer is an adaptable scaffold
ACKNOWLEDGMENTS
We sincerely thank Bridget Carragher and Clint Potter for their generosity in providing access to excellent instrumentation. Electron microscopy data collection was performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (349247) and the NIH National Institute of General Medical Sciences (GM103310). We thank Michael Sheedlo, Rick Baker, and Mike Cianfrocco for helpful feedback and discussions on data processing; and Brett Collins and David Owen for helpful discussions and sharing unpublished data. A.K.K., B.X., R.B., M.N.F., and L.P.J. are supported by NIH R35GM119525. L.P.J. is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. Initial screening for EM data collection was undertaken on a ThermoFisher FEI Polara and TF20 microscopes at the Center for Structural Biology CryoEM facility (V-CEM) at Vanderbilt University. We thank Dr. Scott Collier for his support at the facility. P.X. and T.R.G. are supported by NIH R01GM118452.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.str.2020.01.009.
DECLARATION OF INTERESTS
The authors have no competing interests to declare.
REFERENCES
- Arighi CN, Hartnell LM, Aguilar RC, Haft CR, and Bonifacino JS (2004). Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol 165, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avinoam O, Schorb M, Beese CJ, Briggs JAG, and Kaksonen M (2015). Endocytosis. Endocytic sites mature by continuous bending and remodeling of the clathrin coat. Science 348, 1369–1372. [DOI] [PubMed] [Google Scholar]
- Bean BDM, Davey M, and Conibear E (2017). Cargo selectivity of yeast sorting nexins. Traffic 18, 110–122. [DOI] [PubMed] [Google Scholar]
- Belenkaya TY, Wu Y, Tang X, Zhou B, Cheng L, Sharma YV, Yan D, Selva EM, and Lin X (2008). The retromer complex influences Wnt secretion by recycling wntless from endosomes to the trans-Golgi network. Dev. Cell 14, 120–131. [DOI] [PubMed] [Google Scholar]
- Burd CG, and Cullen PJ (2014). Retromer: a master conductor of endosome sorting. Cold Spring Harb. Perspect. Biol 6, 10.1101/cshperspect.a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K-E, Healy MD, and Collins BM (2019). Towards a molecular understanding of endosomal trafficking by Retromer and Retriever. Traffic 20, 465–478. [DOI] [PubMed] [Google Scholar]
- Clairfeuille T, Mas C, Chan ASM, Yang Z, Tello-Lafoz M, Chandra M, Widagdo J, Kerr MC, Paul B, Mérida I, et al. (2016). A molecular code for endosomal recycling of phosphorylated cargos by the SNX27-retromer complex. Nat. Struct. Mol. Biol 23, 921–932. [DOI] [PubMed] [Google Scholar]
- Collins BM, Skinner CF, Watson PJ, Seaman MN, and Owen DJ (2005). Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly. Nat. Struct. Mol. Biol 12, 594–602. [DOI] [PubMed] [Google Scholar]
- Collins BM, Norwood SJ, Kerr MC, Mahony D, Seaman MNJ, Teasdale RD, and Owen DJ (2008). Structure of Vps26B and mapping of its interaction with the retromer protein complex. Traffic 9, 366–379. [DOI] [PubMed] [Google Scholar]
- Fjorback A, Seaman M, Gustafsen C, Mehmedbasic A, Gokool S, Wu C, Militz D, Schmidt V, Madsen P, Nyengaard JR, et al. (2012). Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J. Neurosci 32, 1467–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franch-Marro X, Wendler F, Guidato S, Griffith J, Baena-Lopez A, Itasaki N, Maurice MM, Vincent J-P (2008). Wingless secretion requires endosome-to-Golgi retrieval of Wntless/Evi/Sprinter by the retromer complex. Nat. Cell Biol 10, 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallon M, Clairfeuille T, Steinberg F, Mas C, Ghai R, Sessions RB, Teasdale RD, Collins BM, and Cullen PJ (2014). A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer. Proc. Natl. Acad. Sci. U S A 111, E3604–E3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MS, Hung C-S, Liu T, Christiano R, Walther TC, Burd CG (2014). A mechanism for retromer endosomal coat complex assembly with cargo. Proc. Natl. Acad. Sci. U S A 111, 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh G, Perez-Dorado I, Jackson LP, Wartosch L, Schafer I, Gray S, McCoy A, Zeldin O, Garman E, Harbour M, et al. (2014a). VARP is recruited onto endosomes by direct interaction with retromer where together they function in export to the cell surface. Dev. Cell 29, 591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh GG, Pérez-Dorado I, Jackson LP, Wartosch L, Schäfer IB, Gray SR, McCoy AJ, Zeldin OB, Garman EF, Harbour ME, et al. (2014b). VARP is recruited on to endosomes by direct interaction with retromer, where together they function in export to the cell surface. Dev. Cell 29, 591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser J (1980). Three-dimensional visualization of coated vesicle formation in fibroblasts. J. Cell Biol 84, 560–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro A, Rojas AL, Rojas R, Murthy N, Effantin G, Kajava AV, Steven AC, Bonifacino JS, and Hurley JH (2007). Functional architecture of the retromer cargo-recognition complex. Nature 449, 1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobi AJ, Wilmanns M, and Sachse C (2017). Model-based local density sharpening of cryo-EM maps. Elife 6, 10.7554/eLife.27131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. (2004). A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962. [DOI] [PubMed] [Google Scholar]
- Jia D, Zhang J-S, Li F, Wang J, Deng Z, White MA, Osborne DG, Phillips-Krawczak C, Gomez TS, Li H, et al. (2016). Structural and mechanistic insights into regulation of the retromer coat by TBC1d5. Nat. Commun 7, 13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun O, Leneva N, Bykov YS, Ariotti N, Teasdale RD, Schaffer M, Engel BD, Owen DJ, Briggs JAG, and Collins BM (2018). Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature 561, 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvainickas A, Jimenez-Orgaz A, Nägele H, Hu Z, Dengjel J, and Steinberg F (2017a). Cargo-selective SNX-BAR proteins mediate retromer trimer independent retrograde transport. J. Cell Biol 216, 3677–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvainickas A, Jimenez-Orgaz A, Nägele H, Hu Z, Dengjel J, and Steinberg F (2017b). Cargo-selective SNX-BAR proteins mediate retromer trimer independent retrograde transport. J. Cell Biol 216, 3677–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Gershlick DC, Vidaurrazaga A, Rojas AL, Bonifacino JS, and Hierro A (2016). Structural mechanism for cargo recognition by the retromer complex. Cell 167, 1623–1635.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough IJ, Steinberg F, Jia D, Barbuti PA, McMillan KJ, Heesom KJ, Whone AL, Caldwell MA, Billadeau DD, Rosen MK, et al. (2014). Retromer binding to FAM21 and the WASH complex is perturbed by the Parkinson disease-linked VPS35(D620N) mutation. Curr. Biol 24,1670–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norwood SJ, Shaw DJ, Cowieson NP, Owen DJ, Teasdale RD, and Collins BM (2010). Assembly and solution structure of the core retromer protein complex. Traffic 12, 56–71. [DOI] [PubMed] [Google Scholar]
- Pan C-L, Baum PD, Gu M, Jorgensen EM, Clark SG, and Garriga G (2008). C. elegans AP-2 and retromer control Wnt signaling by regulating mig-14/Wntless. Dev. Cell 14, 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paravicini G, Horazdovsky BF, and Emr SD (1992). Alternative pathways for the sorting of soluble vacuolar proteins in yeast: a vps35 null mutant missorts and secretes only a subset of vacuolar hydrolases. Mol. Biol. Cell 3, 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Port F, Kuster M, Herr P, Furger E, Bänziger C, Hausmann G, and Basler K (2008). Wingless secretion promotes and requires retromer-dependent cycling of Wntless. Nat. Cell Biol 10, 178–185. [DOI] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Purushothaman LK, Arlt H, Kuhlee A, Raunser S, and Ungermann C (2017). Retromer-driven membrane tubulation separates endosomal recycling from Rab7/Ypt7-dependent fusion. Mol. Biol. Cell 28, 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, McCaffery JM, and Emr SD (1998). A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol 142,665–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ, Harbour ME, Tattersall D, Read E, and Bright N (2009). Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J. Cell Sci 122, 2371–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Rojas R, Bonifacino JS, and Hurley JH (2006). The retromer subunit Vps26 hasan arrestin fold and binds Vps35 through its C-terminal domain. Nat. Struct. Mol. Biol 13, 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti B, and Cullen PJ (2018). Endosomal sorting: architecture of the retromer coat. Curr. Biol 28, R1350–R1352. [DOI] [PubMed] [Google Scholar]
- Simonetti B, Danson CM, Heesom KJ, and Cullen PJ (2017). Sequence-dependent cargo recognition by SNX-BARs mediates retromer-independent transport of CI-MPR. J. Cell Biol 216, 3695–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg F, Gallon M, Winfield M, Thomas EC, Bell AJ, Heesom KJ, Tavaré JM, and Cullen PJ (2013). A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell Biol 15, 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, Setty TG, Sitaram A, and Burd CG (2007). Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J. Cell Biol 177, 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SW, Chuang Y-S, Li M, Seaman MNJ, and Emr SD (2019). A bipartite sorting signal ensures specificity of retromer complex in membrane protein recycling. J. Cell Biol 218, 2876–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YZ, Baldwin PR, Davis JH, Williamson JR, Potter CS, Carragher B, and Lyumkis D (2017). Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temkin P, Lauffer B, Jäger S, Cimermancic P, Krogan NJ, and von Zastrow M (2011). SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat. Cell Biol 13, 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trousdale C, and Kim K (2015). Retromer: structure, function, and roles in mammalian disease. Eur. J. Cell Biol 94, 513–521. [DOI] [PubMed] [Google Scholar]
- Vilariño-Güell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, Soto-Ortolaza AI, Cobb SA, Wilhoite GJ, Bacon JA, et al. (2011). VPS35 mutations in Parkinson disease. Am. J. Hum. Genet 89, 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Guo M, Liang Z, Fan J, Zhu Z, Zang J, Li X, Teng M, Niu L, Dong Y, et al. (2005). Crystal structure of human vacuolar protein sorting protein 29 reveals a phosphodiesterase/nuclease-like fold and two protein-protein interaction sites. J. Biol. Chem 280, 22962–22967. [DOI] [PubMed] [Google Scholar]
- Yang P-T, Lorenowicz MJ, Silhankova M, Coudreuse DYM, Betist MC, and Korswagen HC (2008). Wnt signaling requires retromer-dependent recycling of MIG-14/Wntless in Wnt-producing cells. Dev. Cell 14, 140–147. [DOI] [PubMed] [Google Scholar]
- Zavodszky E, Seaman MNJ, Moreau K, Jimenez-Sanchez M, Breusegem SY, Harbour ME, and Rubinsztein DC (2014). Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat. Commun 5, 3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: real-time CTF determination and correction. J. Struct. Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Benet-Pagés A, Struhal W, Graf E, Eck SH, Offman MN, Haubenberger D, Spielberger S, Schulte EC, Lichtner P, et al. (2011). A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet 89, 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.