Abstract
Despite recent improvements in sequencing methods, there remains a need for assays that provide high sequencing depth and comprehensive variant detection. Current methods1-4 are limited by the loss of native modifications, short read length, high input requirements, low yield, or long protocols. Here, we describe nanopore Cas9-targeted sequencing (nCATS), an enrichment strategy that uses targeted cleavage of chromosomal DNA with Cas9 to ligate adaptors for nanopore sequencing. We show that nCATS can simultaneously assess haplotype-resolved single-nucleotide variants (SNVs), structural variations (SVs) and CpG methylation. We apply nCATS to four cell lines, a cell-line-derived xenograft, and normal and paired tumor/normal primary human breast tissue. Median sequencing coverage was 675X using a minION flow cell and 34X using the smaller flongle flow cell. nCATS requires only ~3μg of genomic DNA and can target a large number of loci in a single reaction. The method will facilitate the use of long-read sequencing in research and in the clinic.
Editorial summary
Point mutations, structural variants and DNA methylation at target loci are assessed by nanopore sequencing.
Targeted sequencing allows investigators to enrich for loci of interest, reducing sequencing costs and labor to achieve high coverage data at desired genomic regions. This approach is critical for interrogation of methylation patterns or mutation frequency in heterogeneous clinical samples. For next-generation sequencing, leading strategies are amplification or hybridization capture5, but these do not take advantage of the benefits of newer long-read sequencing technologies, as amplification would lose any base modifications present and hybridization capture has yet to be fully optimized for long fragments. Some approaches for long-read enrichment have used PCR to amplify regions of interest and then either sequenced the amplicons directly1 or cloned into expression plasmids2 prior to sequencing, but both of these strategies can be affected by amplification bias and lose any information about modified nucleotides. Another method described for target enrichment with nanopore sequencing is CATCH-seq3, wherein regions of interest are excised by dual Cas9 cleavage then enriched for by size selection, but the low recovery from this method required amplification for enrichment from the human genome. Most recently, the enrichment field has had a burgeoning interest in ligating sequencing adaptors to cuts with Cas endonucleases, as demonstrated by others for enrichment and methylation status evaluation of the C9orf72 locus4.
For this work, we enriched by selectively ligating sequencing adaptors to fresh cut sites created by active Cas9/guide RNA ribonucleoprotein complex (RNP). By dephosphorylating pre-existing DNA ends before cutting with Cas9, we preferentially ligate to the newly produced DNA ends at Cas9 cleavage sites (Figure 1A). We first validated and tested the nCATS method by comparing our data on the well-characterized GM12878 cell line to both annotated variants6 and whole-genome bisulfite methylation data7. We then applied the enrichment strategy to assess genetic and epigenetic changes in breast cell lines, a breast cancer cell line xenograft, and primary patient tissue.
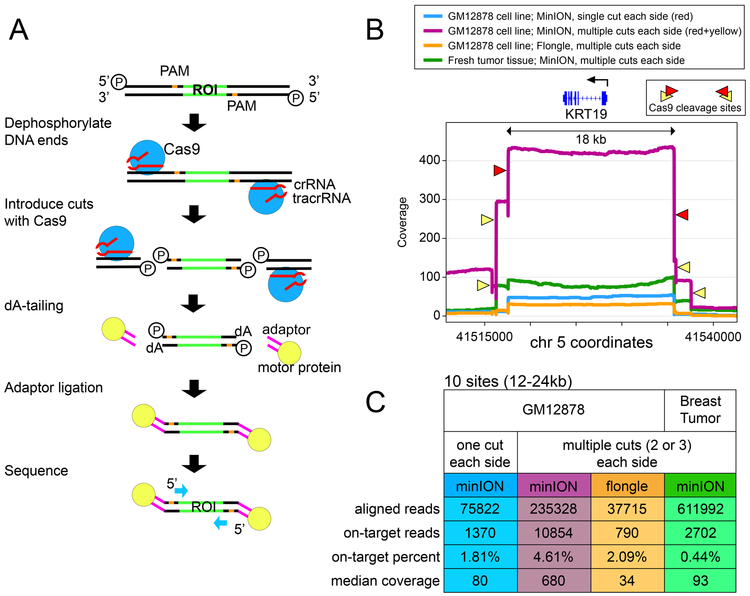
Figure 1 -. Method schematic and coverage data.
(A) Schematic of Cas9 enrichment operation. ROI = region of interest. First, DNA ends are dephosphorylated, then new cuts introduced with Cas9/guideRNA complex, and nanopore sequencing adaptors are ligated to cuts around the ROI prior to loading the sample on the nanopore sequencer. (B) Coverage plots at the KRT19 gene (enriched area 18kb) in four separate enrichment experiments: GM12878 with a single gRNA on each side (minION); GM12878 with three gRNA on each side (minION); GM12878 with three gRNA on each side (flongle); and fresh tumor tissue with three guideRNAs on each side (minION). (C) Table showing total aligned read count, on-target reads (within 20kb of a guideRNA site), on-target percentage, and median coverage at each of the ten enriched regions.
After cleavage with Cas9, the enzyme remains bound to the DNA on the 5’ side of the gRNA8, resulting in preferential ligation of adaptors onto the 3’ side of the cut. We introduced cuts flanking regions of interest to achieve coverage on both strands (Figure 1A). We targeted 10 sites in our initial panel, with sizes ranging from 12–24kb (Supplementary Table 1). For evaluating single nucleotide mutations we selected three cancer-associated genes (TP53, KRAS, and BRAF) with annotated mutations in the MDA-MB-231 cell line9. Five regions for methylation studies (KRT19, SLC12A4, GSTP1, TPM2, and GPX1) and two candidate large deletions (6–8kb) were selected based on previous whole-genome nanopore data from our lab10 as well as existing expression data in these breast cell lines11. An 11th region, the BRCA1 locus, was included in some sequencing runs (Supplementary Table 1) to test our ability to capture larger regions (>80kb), and to evaluate this method for sequencing repetitive regions12.
In our initial experiments, we used one guideRNAs on either side of each region. We applied this to four cell lines: the well-characterized GM12878 lymphoblast cell line and three breast cell lines (MCF-10A, MCF-7, and MDA-MB-231). Libraries were prepared from 3ug of starting DNA and run on a minION flow cell, resulting in coverage ranging from 18X to 846X (Supplementary Table 2). When libraries were run on the smaller flongle flow cell, we measured coverage between 8X and 65X (Supplementary Table 2).
We attributed the variable coverage between regions to differing on-target cutting efficiency and off-target binding of the different guideRNAs. Subsequently, we experimented with a combination of multiple guideRNAs at the same locus in GM12878 and found this significantly improved median coverage. For example, at the KRT19 locus, coverage with multiple guides increased to 407X versus 47X with single guides (Figure 1B). Using multiple guides at all loci yielded greater than 400X at all sites from the MinION flow cell (Median 680X) and greater than 25X at all sites from a flongle flow cell (Median 34X) (Figure 1C; Supplementary Table 2).
From GM12878 minION data, the percentage of ‘on-target’ reads, was 1.8% with the single guideRNA panel and 4.6% with the multi-guideRNA panel (Supplementary Table 2). Genome-wide coverage analysis found the off-target reads to be distributed randomly across the genome, indicating they result primarily from ligation of nanopore adaptors to random breakage points. For example, in the GM12878 cell line with single guideRNAs flanking each site, after quality filtering alignments (MAPQ > 30) there were only 2 genomic sites outside target regions where coverage reached 25X. Both of these are at repetitive peri-centromeric sites and contain reads with lower mapping quality (MAPQ 30–50), suggesting the increased coverage to be the result of alignment errors in these poorly mappable regions. We did note the occurrence of some off-target cleaving with the inclusion of guideRNAs designed to flank the BRCA1 locus (Supplementary Figure 1, Supplementary Table 3), which we attribute to the abundance of repetitive regions12 at this locus resulting in increased homology with other genomic loci.
With this new panel of guides in hand, we tested the assay’s performance in tissue samples: normal human breast tissue, a breast cancer cell-line-derived xenograft, and a human breast tumor/normal pair. In tissue from a reduction mammoplasty (normal) and cell-line-derived mouse xenograft we measured a median coverage of 162X/312X; and from the paired primary tumor/normal sample with limited input we achieved median coverage of 93X/70X (Figure 1C, Supplementary Table 2).
Nanopore sequencing still has intrinsically high error rates (~5–10%) due to the inability of the basecaller to distinguish between some k-mers and the difficulty in discriminating signal events in repetitive regions (e.g. homopolymers). We explored how the increased coverage data from the nCATS protocol would affect the ability to call variants from nanopolish data. To simplify analysis, we limited variant calls to single nucleotide substitutions. There are numerous tools that currently exist for calling variants, and we selected four for comparison: (1) the Samtools/Bcftools package13, which generates genotype likelihoods from alignment data (2) Clair 14, which uses a deep neural network for variant calling from alignment data, (3) Medaka, a tool from Oxford Nanopore which also uses a neural network algorithm, and (4) Nanopolish15, which uses a hidden Markov model to interrogate the raw electrical data as well as alignment data.
For initial validation, we used the GM12878 cell line and the platinum genome dataset6 as ground truth for single nucleotide variants (SNVs). We benchmarked SNVs over the 8 loci without large deletions (total enriched area of 140kb) which have a total of 174 annotated SNVs. To explore the relationship between coverage and variant calling efficiency, we subsampled the aligned data to coverage of 300X, 200X, 100X, 50X and 25X (see methods). During filtering we selected for reads spanning the region, and maintained balanced coverage between both DNA strands.
We found that at lower coverage data (25X and 50X) Clair had the greatest sensitivity (0.98). However, the current model for Clair was trained and assessed on whole genome data only up to 100X coverage; and above this coverage it no longer functioned. Medaka showed peak sensitivity of 0.93 at both 50X and 100X coverage, with sensitivity remaining robust at higher coverage. Samtools variant calling and Nanopolish variant calling both increased in sensitivity up to 200X coverage, at which point they plateaued with sensitivities of 0.97 and 0.98, respectively (Figure 2A, Supplementary Table 4).
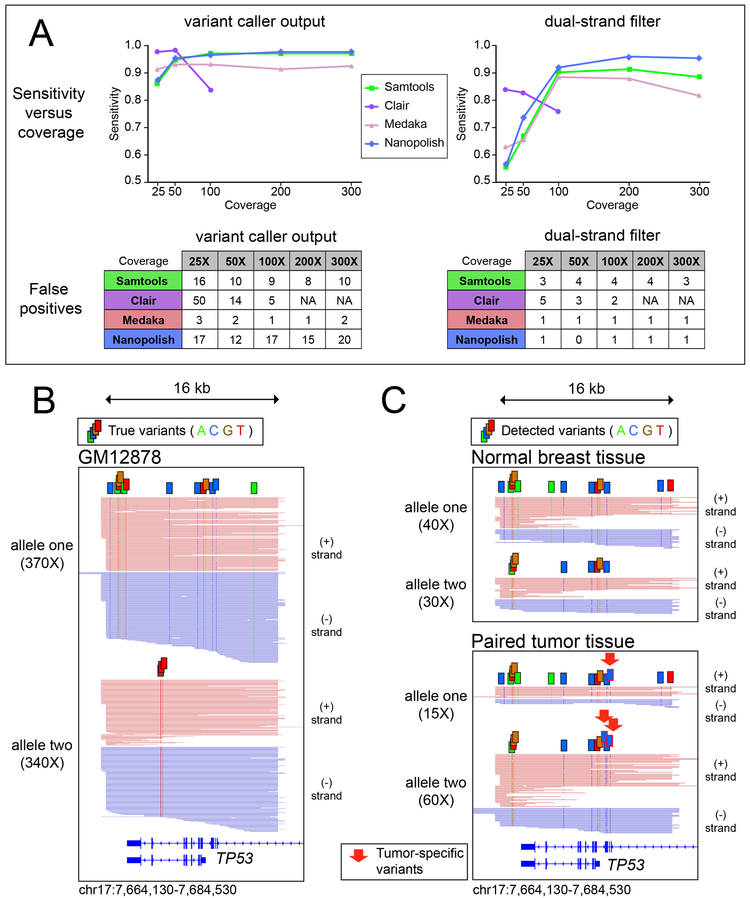
Figure 2 -. Single Nucleotide Variants.
(A) Plot of sensitivity versus coverage using four tools to call single nucleotide variants from enrichment data in GM12878 for a 140kb region containing 174 annotated SNVs (B) Visual representation of high-confidence variants detected by nanopolish in the MinION data from GM12878 for the captured region around TP53, reads phased into homologous alleles using WhatsHap. (C) High-confidence variants identified in primary tissue from a tumor/normal pair, red arrows used to demarcate tumor-specific variants.
One important caveat of the raw output of these variant caller pipelines is the persistence of false positives, limiting the use of this method for de novo SNV discovery (Figure 2A). On inspection, we noted many false positives to occur on only one strand (Supplementary Figure 2), suggesting the basecaller has systematic issues with the sequence of k-mers on one strand but not on the other. Thus, we implemented a filter requiring variants to be supported by reads from both strands (“dual-strand filter”). This filter caused a decrease in sensitivity, especially at lower coverage. But strikingly this filter eliminated nearly all false positive variant calls (Supplementary Table 4), yielding a set of high-confidence variants. The dual-strand filter performed best with 200X coverage using nanopolish variant calling (Sensitivity: 0.96, F1score: 0.97), with the sole false positive variant existing in a thymidine-dense homopolymer region (Supplementary Figure 3). We then applied WhatsHap16, a weighted haplotype assembler that uses statistical information as well as coverage depth to assign reads into parental haplotypes based on SNVs detected in long-read data. A graphical depiction of detected variants is shown in Figure 2B, highlighting the identification and phasing of variants in the captured region of TP53 in GM12878. All 17 of the annotated SNVs in this region were detected by the dual-strand filtered data with no false positives.
We then applied this variant caller pipeline to our data from the MDA-MB-231 cell line to detect cancer-associated mutations. Across the captured regions of three cancer-associated genes (BRAF, KRAS, and TP53) nanopolish called 42 high-confidence SNVs (Supplementary Table 5), including 2 of the 3 annotated in the COSMIC database for MDA-MB-23117. The third variant was detected, but at a lower frequency in this aneuploid line and thereby did not pass dual-strand filtering. Finally, we applied this variant calling pipeline to the paired tumor/normal breast tissue sample and phased the reads using WhatsHap16. We noticed a strong variation in the number of reads per haplotype in the TP53 region, implying an imbalanced copy number in tumor cells (Figure 2C). We examined two other captured regions on the same chromosome and observed similar chromosomal imbalance with additional mutations in tumor samples (Supplementary Figure 4).
We next evaluated CpG methylation, which can be measured from nanopore electrical data15. Sites for methylation studies were selected by searching whole-genome nanopore data10 for differentially methylated promoters between the non-tumorigenic breast cell line MCF-10A and the tumorigenic breast cell lines MCF-7 and MDA-MB-231. Candidate loci were further filtered by comparing to existing RNA-seq data11 and genes with prognostic implications in human cancer18–20.
We use read-level methylation plots to display methylation information from both a minION sequencing run and a flongle sequencing run (Figure 3A; Supplementary Figure 5). Methylation data for one locus (KRT19) is shown in Figure 3A, with four additional genes (GSTP1, GPX1, SLC12A4, and TPM2) in Supplementary Figure 5. We compared nanopore methylation patterns with existing whole genome bisulfite sequencing (WGBS) data in GM128787 using smoothed (loess) line plots (Figure 3B, Supplementary Figure 5). Directly comparing per-CpG methylation (Supplementary Figure 5) at each locus, we observed per-CpG methylation largely clustered at points reflecting completely methylated or unmethylated sites, with an aggregate per-CpG correlation of 0.81 (Pearson).
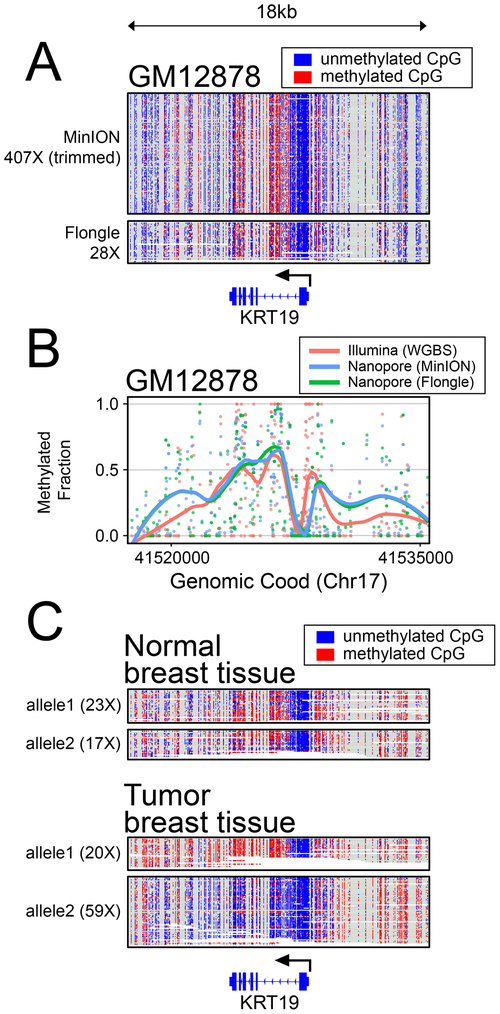
Figure 3 -. Methylation Analysis.
(A) Read-level plots showing methylation patterns in GM12878 from minION and flongle data at the KRT19 locus. (B) Methylation calls (points) and line plots at the same locus as in (A) showing smoothed (loess) methylation calls from whole genome bisulfite sequencing on the Illumina platform7, compared with methylation calls from minION and flongle targeted nanopore sequencing. (C) Haplotype phased methylation calls in primary patient tissue and paired tumor at the KRT19 locus.
We applied this strategy to our data for breast cell lines, looking for regions with differential methylation at these loci. One gene where we observed differential methylation in breast cell lines is the keratin family member gene: KRT19. KRT19 is known to be upregulated in breast cancer19, and detection of KRT19 mRNA has been used to identify micrometastasis of breast cancer to lymph nodes21 and to detect circulating tumor cells20. We observed that KRT19 remains largely methylated in the non-tumorigenic MCF-10-A cell line, but becomes hypomethylated in both of the transformed cell lines, MCF-7 and MDA-MB-231 (Supplementary Figure 6). This is correlated with an observed increased transcript level for KRT19 in the transformed cell lines (Supplementary Figure 7, GEO: GSE75168). Further, we note the observed pattern of methylation is largely maintained in mouse xenografts derived from the MDA-MB-231 cell line (Supplementary Figure 8). In evaluation of the paired tumor/normal patient sample, we found that the primary patient tumor had a dramatic allele-specific hypomethylation of KRT19 on the haplotype with increased copy number (Figure 3C, Supplementary Figure 8), in line with evidence suggesting increased expression in tumor cells19-21. This unveils nuance about allele-specific methylation and copy number changes that would be difficult to query without the high-coverage long-read data as achieved by this methodology.
We next applied this method to evaluate structural variations by confirming the presence of candidate deletions from whole genome nanopore sequencing data10. We selected two deletions present in the MDA-MB-231 and MCF-7 breast cancer lines and absent in the MCF-10-A cell line, and designed guideRNAs to flank breakpoints by ~5kb. Plotting reads in IGV showed both deletions as heterozygous in MDA-MB-231 and homozygous in MCF-7 (Figure 4A, Supplementary Figure 9). The alignment data was passed to the Sniffles variant caller22, which identified the breakpoints and zygosity of both deletions in line with our observations (Supplementary Table 6). We also performed methylation studies on these regions but did not note any difference in methylation patterns between the deleted and intact allele (Supplementary Figure 10).
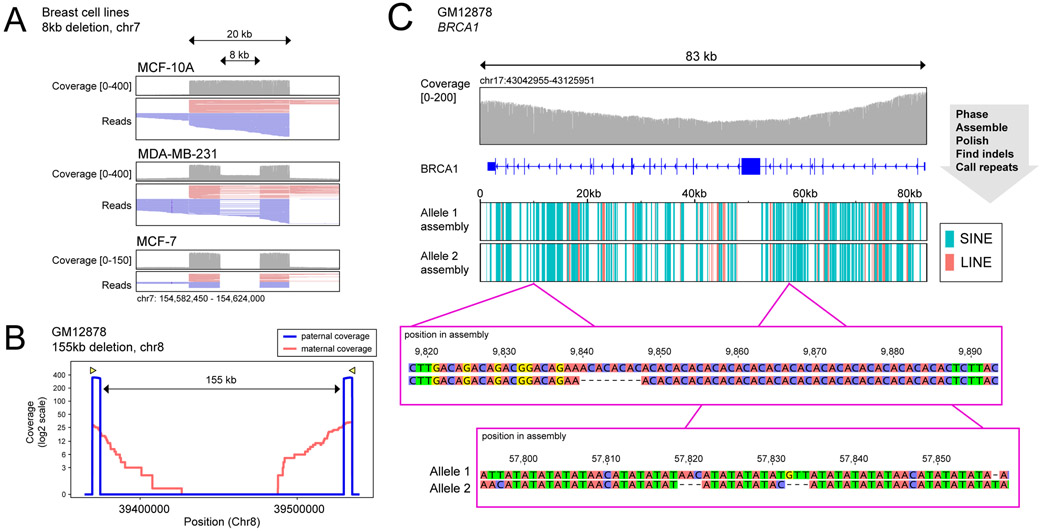
Figure 4 -. Structural Variation.
(A) Reads around an ~8kb deletion in chromosome 7 present in MCF-7 and MDA-MB-231, and absent in MCF-10A. (B) Coverage on each parental allele in the region of a large (155kb) heterozygous deletion in GM12878. (C) Top: Coverage at the BRCA1 locus from DNA extracted using Circulomics CBB kit. Middle: LINE and SINE components identified by RepeatMasker on each of the BRCA1 allele assemblies. Bottom: Three indels discovered between BRCA1 assemblies not annotated in platinum genome data set for GM128789.
To explore the use of this method for targeting larger fragments of DNA, we enriched for regions harboring large (>70kb) heterozygous chromosomal deletions. We identified three large heterozygous deletions in GM12878 from available 10X Genomics data through the Genome In a Bottle (GIAB) Consortium23, two with sizes of ~70kb and one ~155kb. Guide RNAs were designed to flank the deletion breakpoints by 5kb, resulting in reads of ~10 kb on the deleted allele, and spanning the region between cut sites (80kb/165kb) on the non-deleted allele. We again phased the reads into parental alleles16 and compared read lengths and read counts achieved from each allele. Interestingly, we found that the allele containing the deletion, with the correspondingly shorter distance between the cut sites, demonstrated an order of magnitude higher number of reads (Figure 4B, Supplementary Figure 11). This reflects a bias against achieving reads >50kb, likely introduced during DNA purification, library preparation, or delivery to the pore. To confirm this size-bias, we performed similar parental-allele segregation on sites without SVs and did not observe bias towards either parental allele (Supplementary Figure 12). The alignment data was passed to the Sniffles variant caller22, which identified all 3 of the deletions within 10nt from the annotated breakpoints in existing GIAB data (Supplementary Table 7). We adjusted Sniffles parameters to call SVs as heterozygous if an allele was supported by even a very low amount (0.1%) of reads, as the imbalance of reads from the two alleles caused the software to initially identify these deletions as homozygous (see Methods).
Finally, we targeted the BRCA1 gene, because of the well-documented association of this gene with familial breast cancer12. BRCA1 is an attractive target for long-read sequencing because of the abundance of hard to map repetitive Alu elements. To capture the entire BRCA1 gene (distance between flanking guideRNAs: 83kb) further adjustments to DNA extraction were needed. Initial MinION sequencing runs from 3ug extracted GM12878 gDNA resulted in only 10 sequencing reads spanning the entire region with many smaller fragments (Supplementary Figure 13). We found that using the Circulomics NanoBind kit for DNA extraction resulted in an increase to nearly 30 reads completely spanning the BRCA1 gene, with coverage between guideRNAs ranging between 100X and 200X (Figure 4C, Supplementary Figure 13). We phased the BRCA1 reads into haplotypes16 with de novo called high-confidence variants found with nanopolish. We then built an assembly for each of the two alleles using the Flye assembler24 and polished the assemblies using Racon25 and Medaka (see Methods). This resulted in full length assemblies for each of the two alleles, within which we identified the presence of SINEs (e.g. Alu-elements) and LINEs using RepeatMasker. We compared the two assemblies for variants differing between alleles and found numerous indels and single nucleotide changes (Supplementary Table 8) using the minimap2 suite26. After filtering for homopolymer regions (which nanopore sequencing is known to still have difficulty resolving15), we found 10 indels of at least 3 nucleotides between the two assemblies (Supplementary Table 8). Seven of these ten were annotated in the platinum genome data set for GM128786. The remaining three unannotated indels are within large repetitive regions (Figure 4C), making it difficult to map reads from conventional short read sequencing. To validate these indels, we compared against recently released whole genome PacBio data for GM12878 (SRA: SRR9001768-SRR9001773)23 which confirmed each of these three small unannotated indels (Supplementary Figure 14). Further, because we used nanopore sequencing, we were also able to call CpG methylation on this complex region, and compare methylation profiles between BRCA1 alleles (Supplementary Figure 15).
Because of the low cost to entry and small footprint of the instrument, this assay has the potential to be widely utilized as a tool for identifying single nucleotide changes, evaluating DNA methylation, and studying structural variation. We were even able to apply this to clinical tissue despite the relatively high DNA input requirements (3μg). We show that single nucleotide variants in regions of interest can be queried with the nCATS protocol, although there are persisting limitations, as evinced by the few SNVs not detected by this approach. We found that by using only high-confidence variants, we were able to phase nanopore sequencing reads into parental alleles using WhatsHap16, permitting haplotype resolution of high-coverage nanopore data. As basecalling and variant-calling algorithms continue to improve we anticipate higher future performance for surveillance and identification of mutations. We also highlight the use of nCATS to detect and validate structural variants. It is only with the advent of long-read sequencing that the great diversity of structural variation in human genomes has been appreciated27,28, and this method provides a dynamic tool to evaluate genomic rearrangements, including large structural variants and hard-to-map repetitive regions29. Importantly, because nanopore sequencing interrogates the DNA strand rather than sequencing ”by-synthesis”, we can simultaneously profile methylation in these loci, providing biological as well as diagnostic insight into the epigenome, which is commonly disrupted in human neoplasia30. In fact, as we show long reads allow easy phasing of methylation into different alleles, allowing careful exploration of allele specific epigenetic changes. The high sequencing depth granted by this method is especially useful to characterize genetically and epigenetically heterogeneous samples typically obtained from clinical samples; giving us insight into the frequency of different mutations and epigenetic changes present.
ONLINE METHODS
Cell culture and DNA prep
Cell lines were obtained from ATCC: MCF10A (CRL-10317), MCF7 (HTB-22), MDA-MB-231 (HTB-26); or Coriell institute: CEPH/UTAH Pedigree 1463 (GM12878). Cells were cultured according to recommended protocols. Briefly, all cell lines were maintained at 37°C in 5% CO2. The GM12878 cell line was grown in high-glucose RPMI media supplemented with 10% fetal calf serum (FCS), penicillin-streptomycin antibiotics (pen-strep), and L-glutamine. MCF-7 and MDA-MB-231 were grown hi-glucose DMEM media supplemented with 10% FCS, pen-strep, and L-glutamine. MCF-10A cells were grown in hi-glucose DMEM media supplemented with 5% horse serum, pen-strep, L-glutamine, epidermal growth factor, insulin, hydrocortisone, and cholera toxin. DNA was extracted from cells, using either the MasterPure kit (Lucigen, MC85200), or the Nanobind kit (Circulomics, NB-900–001-01) and stored at 4°C until use. DNA was quantified using the Qubit fluorometer (Thermo) immediately before performing the assay.
Patient Tissue and Mouse Xenograft
All human samples were collected with appropriate approval from the Johns Hopkins institutional review board. The primary breast tumor was identified as ER/PR+ by immunohistochemistry and snap frozen. Mouse experiments were conducted with prior approval from JH-ACUC. Mouse xenografts were generated by injecting 106 ER/PR/HER2-negative MDA-MB-231 breast cancer cells into the mammary fat pad of athymic mice. Tumors were collected 6–8 weeks later and frozen immediately as small chunks. The snap frozen tissue was ground under liquid nitrogen using a CryoMill (Retch) and DNA extracted using MasterPure kit (Lucigen, MC85200).
Guide RNA design
Guide RNAs were assembled as a duplex from synthetic crRNAs (IDT, custom designed) and tracrRNAs (IDT, 1072532). Sequences are provided in Supplementary Table 1. The crRNAs were designed using IDT’s design tool and selected for the highest predicted on-target performance with minimal off-target activity. The gRNA duplex was designed to introduce cuts on complementary strands flanking the region of interest. For methylation studies and SNV studies, the target size between gRNAs was 12–24 kb; for deletions, the gRNAs were designed to flank the suspected breakpoints by ~5kb.
Ribonucleoprotein Complex Assembly
Prior to guide RNA assembly, all crRNAs were pooled into an equimolar mix, with a total concentration of 100uM. The crRNA mix and tracrRNA were then combined such that the tracrRNA concentration and total crRNA concentration were both 10uM. The gRNA duplexes were formed by denaturation for 5 minutes at 95°C, then allowed to cool to room temp for 5 minutes on a benchtop. Ribonucleoprotein complexes (RNPs) were constructed by combining 10pmol of gRNA duplexes with 10pmol of HiFi Cas9 Nuclease V3 (IDT, 1081060) in 1X CutSmart Buffer (NEB, B7204) at a final volume of 30μL (conc: 333nM), incubated 20 minutes at room temperature, then stored at 4°C until use, up to 2 days.
Cas9 Cleavage and Library Prep
3ug of input DNA was resuspended in 30uL of 1X CutSmart buffer (NEB, B7204), and dephosphorylated with 3uL of Quick CIP enzyme (NEB, M0508) for 10 min at 37C, followed by heating for 2 minutes at 80C for CIP enzyme inactivation. After allowing the sample to return to room temp, 10uL of the pre-assembled 333nM Cas9/gRNA complex was added to the sample. In the same tube, 1uL of 10mM dATP (Zymo, D1005) and 1uL of Taq DNA polymerase (NEB, M0267) were added for A-tailing of DNA ends. The sample was then incubated at 37C for 20min for Cas9 cleavage followed by 5 minutes at 72C for A-tailing. Sequencing adaptors and ligation buffer from the Oxford Nanopore Ligation Sequencing Kit (ONT, LSK109) were ligated to DNA ends using Quick Ligase (NEB, M2200) for 10 min at room temp. The sample was cleaned up using 0.3X Ampure XP beads (Beckman Coulter, A63881), washing twice on a magnetic rack with the long-fragment buffer (ONT, LSK109) before eluting in 15uL of elution buffer (ONT, LSK109). Sequencing libraries were prepared by adding the following to the eluate: 25uL sequencing buffer (ONT, LSK109), 9.5uL loading beads (ONT, LSK109), and 0.5uL sequencing tether (ONT, LSK109). A detailed step-wise description of the enrichment method is available on protocols.io (https://www.protocols.io/view/cas9-enrichment-for-nanopore-sequencing-68ihhue)
Sequencing
Samples were run on a MinION (ver 9.4.1) flow cell or Flongle flow cell (ver 9.4.1 pore), using the MK1B or GridION sequencer. Sequencing runs were operated using the MinKNOW software (v19.2.2). A detailed description of runs (flow cell, guideRNAs, sequencer) is provided in Supplementary Table 1.
Analysis
Basecalling was performed using GUPPY (Version 3.0.3) to generate FASTQ sequencing reads from electrical data. Reads were aligned to the human reference genome (Hg38) using Minimap2 (v2.17)26. Per-nucleotide coverage was determined using samtools, and clustered using the ‘bincov’ script of the SURVIVOR (v1.0.7) software package31. On-target reads were defined as those which aligned within 20kb of a guideRNA site. Average coverage per region is the average of coverage of all bases between the innermost guideRNA sites, using coverage found by samtools.
De novo variant calling was performed using samtools (v1.9)13, Clair (v2.0.0)14, Medaka (v0.10.0) or nanopolish (v0.11.1)15. For validation, we compared SNV calls to those annotated for GM12878 as part of the platinum genome dataset6. To achieve different coverage values for validation of GM12878 data, each region was subsampled at random using samtools to achieve 300X coverage with balanced read counts on each strand. The reads were then further subsampled to achieve the lower coverage values of 200X, 100X, 50X and 25X. Sensitivity was calculated as correctly called SNVs (true positives) out of all true SNVs (true positives plus false negatives). The F1 score is included as a measure of overall test accuracy, calculated as the harmonic mean of precision and recall.
High-confidence variants were generated by an additional filter requiring variants to be supported by reads from both strands. Bam alignment files were split into reads aligning to forward strand and reverse strand, and variant calls performed were performed on each set of reads separately. Variants were only included in the high-confidence set if they were called in forward strand reads alone, reverse strand reads alone, and the complete data set.
Segregation of reads into parental alleles was performed with WhatsHap (v0.18)16, using only de novo called high-confidence variants. For patient tumor tissue, reads phased into haplotypes using only the variants identified from paired normal tissue.
CpG methylation calling on nanopore data was performed using nanopolish (v0.11.1)15. Methylation calling on existing WGBS GM12878 data (GEO: GSE86765)7 was performed using the Bismark (v0.18.2) software tool32. The bismark output files were processed using the bsseq R package (v3.9)33, and a Pearson correlation coefficient was calculated using base R. RNA-seq data of MCF-10A, MCF-7, and MDA-MB-231 were downloaded from GEO (Accession: GSE75168) in the form of RNA counts.
Deletions were called using the structural variant caller Sniffles (v1.0.11)22, set to find deletions with a minimum size of 100bp. In the instance of the very large (>70kb) heterozygous deletions in GM12878, the allelic size bias caused the ploidy to be incorrectly called as homozygous. To correct this, we used the option “--min_homo_af” set to 99.9, which ensured a deletion was called as heterozygous if supporting reads for an allele were present at a rate as low as one in one thousand.
For assembly of the BRCA1 region, reads were first split into haplotypes with WhatsHap16. A draft assembly for each allele was built using the Flye (v2.4.2) assembly tool24, with default parameters for nanopore reads. Draft assemblies were then corrected by using four iterative rounds of polishing with the Racon error-correction software (v1.3.3)25, with the score for matching bases (“-m”) increased to 8 and the score for mismatching bases (“-x”) decreased to −6. A final round of polishing was performed using the Medaka consensus tool with default parameters. The assemblies were surveilled for indels using the paftools helper script of the Minimap2 suite (v2.17)26.
DATA AVAILABILITY
Sequencing data from all non-primary patient samples for this study can be retrieved from the Sequence Read Archive (SRA), under the BioProject ID PRJNA531320
CODE AVAILABILITY
The computational code used in all of the analysis is hosted on GitHub (see https://github.com/timplab/Cas9Enrichment, https://github.com/isaclee/nanopore-methylation-utilities).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by funding from NIH R01 HG009190 (NHGRI).
Footnotes
COMPETING FINANCIAL INTERESTS
JG, ER, RB, and AH are employees of ONT. WT has two patents licensed to ONT (US Patent 8,748,091 and US Patent 8,394,584). TG, IL, and WT have received travel funds to speak at symposia organized by Oxford Nanopore Technologies.
REFERENCES
- 1.Karamitros T & Magiorkinis G Multiplexed Targeted Sequencing for Oxford Nanopore MinION: A Detailed Library Preparation Procedure. Methods Mol. Biol 1712, 43–51 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Leija-Salazar M et al. Evaluation of the detection of GBA missense mutations and other variants using the Oxford Nanopore MinION. Mol Genet Genomic Med 7, e564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabrieli T et al. Selective nanopore sequencing of human BRCA1 by Cas9-assisted targeting of chromosome segments (CATCH). Nucleic Acids Res. 46, e87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giesselmann P et al. Analysis of short tandem repeat expansions and their methylation state with nanopore sequencing. Nat. Biotechnol. 37, 1478–1481 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Kozarewa I, Armisen J, Gardner AF, Slatko BE & Hendrickson CL Overview of Target Enrichment Strategies. Curr. Protoc. Mol. Biol 112, 7.21.1–7.21.23 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Eberle MA et al. A reference data set of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res. (2016) doi: 10.1101/gr.210500.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sternberg SH, Redding S, Jinek M, Greene EC & Doudna JA DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forbes SA et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 45, D777–D783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee I et al. Simultaneous profiling of chromatin accessibility and methylation on human cell lines with nanopore sequencing. bioRxiv 504993 (2018) doi: 10.1101/504993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Messier TL et al. Histone H3 lysine 4 acetylation and methylation dynamics define breast cancer subtypes. Oncotarget 7, 5094–5109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welcsh PL & King MC BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. 10, 705–713 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Li H A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo R et al. Clair: Exploring the limit of using a deep neural network on pileup data for germline variant calling. bioRxiv 865782 (2019) doi: 10.1101/865782. [DOI] [Google Scholar]
- 15.Simpson JT et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Martin M et al. Whatshap: fast and accurate read-based phasing. bioRxiv. 2016. [Google Scholar]
- 17.Tate JG et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martignano F et al. GSTP1 Methylation and Protein Expression in Prostate Cancer: Diagnostic Implications. Dis. Markers 2016, 4358292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabir NN, Rönnstrand L & Kazi JU Keratin 19 expression correlates with poor prognosis in breast cancer. Mol. Biol. Rep 41, 7729–7735 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Wang X-M, Zhang Z, Pan L-H, Cao X-C & Xiao C KRT19 and CEACAM5 mRNA-marked circulated tumor cells indicate unfavorable prognosis of breast cancer patients. Breast Cancer Res. Treat (2018) doi: 10.1007/s10549-018-05069-9. [DOI] [PubMed] [Google Scholar]
- 21.Noguchi S et al. Detection of breast cancer micrometastases in axillary lymph nodes by means of reverse transcriptase-polymerase chain reaction. Comparison between MUC1 mRNA and keratin 19 mRNA amplification. Am. J. Pathol 148, 649–656 (1996). [PMC free article] [PubMed] [Google Scholar]
- 22.Sedlazeck FJ et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zook JM et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Scientific data vol. 3 160025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolmogorov M, Yuan J, Lin Y & Pevzner PA Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol 37, 540–546 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Vaser R, Sović I, Nagarajan N & Šikić M Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaisson MJP et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. bioRxiv 193144 (2018) doi: 10.1101/193144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Audano PA et al. Characterizing the Major Structural Variant Alleles of the Human Genome. Cell 176, 663–675.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dixon JR et al. Integrative detection and analysis of structural variation in cancer genomes. Nat. Genet 50, 1388–1398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timp W & Feinberg AP Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer 13, 497–510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 31.Jeffares DC et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun 8, 14061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krueger F & Andrews SR Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen KD, Langmead B & Irizarry RA BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 13, R83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data from all non-primary patient samples for this study can be retrieved from the Sequence Read Archive (SRA), under the BioProject ID PRJNA531320