Abstract
Tissue sections have long been the subject matter for the application of imaging mass spectrometry, but recently the technique has been adapted for many other purposes including bacterial colonies and 3D cell culture. Here we present a simple preparation method for unsectioned invertebrate tissue without the need for fixing, embedding, or slicing. The protocol was used to successfully prepare a Hawaiian bobtail squid hatchling for analysis, and the resulting data detected ions that correspond to compounds that present in the host only during its symbiotic colonization by Vibrio fischeri.
Keywords: Vibrio fischeri, Euprymna scolopes, symbiosis, microbiology, unsectioned organism imaging
Introduction
Imaging mass spectrometry (IMS) of tissue from animals has long been used to map the molecular information of molecules.1 Originally adapted for analysis of thin tissue sections, IMS has been expanded to a wide range of applications,2–6 with thin tissue sectioning remaining the most common preparation method for the technology. In an effort to expand upon the original methods to image organs or tumors, some recent achievements in unsectioned organism imaging include evaluation of tissue sections of mice,7 zebrafish8 and whole flies.9 While a single tissue section 10–20 μm in thickness generates a wealth of data, it requires significant manipulation of the original host, which can include dissection and cryopreservation of specific tissues, fixation of the tissue,10 embedding of tissues in specific substrates such as gelatin,11,12 or expertise in sectioning an organism or tissue that is primarily composed of water to reduce carry over between sections. Fixing and embedding have been successfully performed for small cephalopods for fluorescence microscopy and histology,10,11,13 but the materials required for such protocols interfere with ionization in a matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS system. There are also spatial limitations of the target plates which may limit how many time points can be integrated within one analysis, which can limit spatiotemporal applications. Imaging of concurrent tissue sections has been performed to construct a 3D rendering of the spatial distribution of biomolecules,14,15 an approach realized only recently.16,17 Tissue manipulation may adulterate the type of molecular information that is ultimately available from the resulting data. While some organisms and systems can only be visualized using this approach, due to size and tissue composition, it is feasible that some invertebrate animals are small enough to be imaged with intact organs or structures, without any embedding, fixing, or sectioning.
A system that has yet to see success with embedding and sectioning is the Hawaiian bobtail squid, Euprymna scolopes, because of its water-based, gelatinous body and its small size, usually about 2 × 4 mm as a hatchling. E. scolopes is most well-known for its symbiotic relationship with Vibrio fischeri; the squid is born free of any bacterial symbiont and within hours acquires V. fischeri exclusively from the water column, even though the bacteria comprises less than 0.02% of the bacterial population in the water column.18 The microbial colonies are housed inside the light organ, which measures a mere 500 μm across and sits under the ink sac inside the mantle cavity that covers the dorsal half of the body.19 The early, specific microbial colonization factors in this symbiosis have remained elusive and, given the small size of the organ, would be a model subject for imaging in situ pre- and post-colonization.20,21 Because the bacteria exist only in a small organ, disturbing that organ may result in loss of material or signal, which motivated the development of an approach where the light organ is not disrupted. Efforts to apply MALDI-TOF MS analysis to squid tissue has inspired the development of a minimal sample preparation method presented here. Several optimization attempts were required to reach the simple, 3-step protocol for unsectioned E. scolopes tissue preparation. Removal of several biological elements that generated height differences after desiccation and exposure of the light organ resulted in the optimal preparation procedure to begin to investigate the chemical communication that drives this inter-kingdom symbiosis in situ.
Materials and Methods
Squid colonization.
The wild-type V. fischeri strain MJM1100 (ES114) was grown overnight with aeration at 25°C in LBS. The overnight culture was diluted 1:80 in LBS and grown at 25°C again to OD600 of approximately 0.2. E. scolopes that hatched within the 24 h were placed in cups containing filter-sterilized instant ocean (FSIO) and inoculated with 3.3 × 103 CFU/ml.22 At 4 h post-inoculation, squid were removed from cups, washed in FSIO and transferred with 750 mL FSIO to microcentrifuge tubes to be euthanized by storage at −80°C. For the 48 h time point, samples were prepared as follows: at 3 h post-inoculation, squid were washed in FSIO and transferred to individual Drosophila vials with FSIO and kept in day/night cycling room for the additional time (water was changed at 24 h post-inoculation). At 48 h they were euthanized and any surface-associated V. fischeri were killed by storage at −80°C.22 Luminescence of these squid was checked before freezing to ensure colonization.22 Aposymbiotic hatchlings were not inoculated with any bacteria, were confirmed to not contain significant numbers of symbionts by luminescence, and were processed side by side with colonized hatchlings for the two time points. Four specimens were analyzed: 1) aposymbiotic hatchlings euthanized at 4 h, 2) aposymbiotic hatchlings euthanized at 48 h, 3) hatchlings colonized with ES114 euthanized at 4 h, and 4) hatchlings colonized with ES114 euthanized at 48 h.
Microscopy and organism dissection.
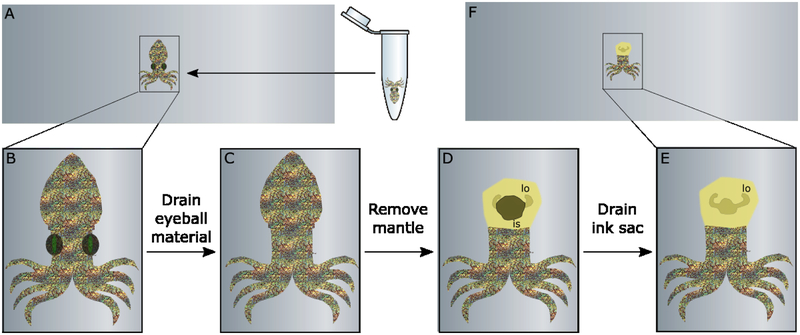
Prior to dissection, all of the tools used were sterilized and the squid was dissected on the same slide which it was imaged on, to minimize transfers and artifacts. A squeeze pipette was used to transfer the hatchling onto an ITO-coated glass slide (Figure 1A–B). Dissection was performed using an Imaging Leica MZ16 dissecting microscope equipped with an RTKE spot camera. Using a PrecisionGlide® needle (21G 1 ½, BD), the eyeballs were completely removed from the eye sockets. Forceps were used to remove the entire mantle by pulling upward and away from the body, exposing the ink sac and the light organ. Ink was removed using the same PrecisionGlide® needle. The ink sac was pierced and continually prodded until the majority of the ink had drained. The squid was rinsed fully in fresh DI water and the slide gently wiped free of excess material. A small aliquot of the squid ink was preserved to use as a negative control during IMS. The squid was then returned to the slide with minimal water and manually arranged so that no tissue was overlapping.
Figure 1.
Preparation of E. scolopes tissue requires a few simple steps. A. Frozen hatchling is transferred to ITO-coated slide. B. Slide is transferred to microscope for easy visualization. C. Squid eyeballs are removed by needle. C. Mantle is completely detached using forceps. E. Ink sac (is) is drained so light organ (lo) is visible. F. Squid is rinsed and returned to slide for dehydration.
Imaging mass spectrometry.
The slide containing one or multiple squids was put into an oven at 37°C for 2–4 h to dehydrate the squid body completely. Following desiccation, the samples were prepared for MALDI-TOF IMS. A 1:1 mixture of 2,5-dihydroxybenzoic acid (DHB (98%), Sigma) and α-cyano-4-hydroxycinnamic acid (CHCA (98%), Sigma) was applied via an HTX TM Sprayer. The solution for the sprayer was prepared as follows: 5 mg/mL of a 1:1 mixture of DHB:CHCA was dissolved in 90:10 ACN:H2O + 0.1% TFA and sonicated to ensure solubility. The matrix mixture was recrystallized in house as previously reported.4 The following instrument parameters were used to apply the matrix: flow rate = 0.2 mL/min, velocity = 1100 mm/min, temperature: 30°C, track spacing = 3 mm, passes = 8, nitrogen pressure = 10 psi, spray pattern = CC, drying time = 0 sec, and nozzle height = 40 mm.
Phosphorus red was used as a calibrant; 0.5 μL of a 1 mg/μL solution in MeOH was spotted onto the glass directly adjacent to the tissue on the glass. The ink control was mixed with matrix (1:1 CHCA:DHB in 78:22 ACN:H2O + 0.1% TFA) in a 1:1 ratio and 1 μL was spotted. ‘X’ marks were drawn onto the four corners of the glass slide for use as teach points when assigning the regions of interest for IMS. The full slide was scanned at 1200 dpi using a Hewlett Packard ScanJet 5550c and imported into the imaging software. IMS data was acquired using flexControl v 3.4 at 20 μm spatial resolution on an Autoflex Speed LRF MALDI-TOF mass spectrometer (Bruker Scientific, Billerica, MA) over the mass range 40−1000 Da. In positive reflectron mode, laser power was set to 40%, laser width to 3 and reflector gain to 10X. For each raster point 500 laser shots at 2000 Hz were shot in a random walk method.
Data and Statistical Analysis.
Data was analyzed in flexImaging v 4.1 × 64 (Bruker Scientific, Billerica, MA). All spectra were normalized to root mean square (RMS). The statistical analysis software SCiLS (Bruker Scientific, Billerica, MA) was used to determine which mass features were statistically more abundant in certain conditions. The co-localization algorithm was employed to detect mass features that were more significant in the light organ region of the colonized 48 h sample compared to the light organ region of all of the other colonization conditions, with N=4. Segmentation in SCiLS generates areas of analyzed sample that are defined by shared m/z’s. Co-localization and Segmentation workflows use the following parameters: Normalization = TIC, Interval width = 0.1 Da, Denoising = Weak, and algorithms work on individual spectra.
Light Organ Extraction and LC-MS/MS Analysis.
Whole light organs were extracted from the four E. scolopes samples that were not used for IMS analysis. Light organs were dissected by removing the mantle and cutting the remainder of the body from the light organ and ink sac. The ink sac was drained and the light organs were rinsed in DI water to remove residual ink. Each light organ was submerged in 1 mL EtOAc in a 4-mL vial for at least 12 h, then sonicated for 10 min and 1 mL transferred to microcentrifuge tubes. Samples were spun in a microcentrifuge for 2 min at 112 RCF and the supernatant transferred to new microcentrifuge tubes. EtOAc was dried under a steady stream of air for 15 min or until dryness, and then resuspended in 10 μL of MeOH for LC-MS/MS.
Organic and polar resin extractions of large-scale WT V. fischeri (ES114) cultures were generated for LC-MS/MS. A frozen stock of ES114 was revived in 5 mL LBS, after 24 h was transferred into 30 mL LBS, and after 24 h was transferred into 1 L LBS. Bacteria were grown in an incubator at 25°C shaking at 225 rpm for 48 h. 20 g XAD16 resin was added and shaken at 225 rpm for 1 h. The resin was filtered and back extracted with 1:1 DCM:MeOH and dried in vacuo to generate the organic extract. In the spent media from the XAD16 resin, 20 g of a 1:1:1 mixture of XAD2:XAD4:XAD7 resin was added and shaken at 225 rpm for 1 h, then back extracted in 1:1 ACN:H2O + 0.1% TFA, shaken at 225 rpm for 1 h. The XAD2:4:7 resin mixture was collected by filtration and dried in vacuo to generate the ‘polar’ extraction. Both extracts were resuspended in MeOH at mg/mL.
LC-MS/MS data were collected on a Bruker impact II qTOF in positive mode with the detection window set from 50 to 1500 Da. Samples (10 μL each) were subjected to a UPLC gradient of 10–100% B over 16 minutes (Solvent A: H2O + 0.02% formic acid, Solvent B: ACN : 0.02% formic acid). The ESI conditions were set with the capillary voltage at 4.5 kV. Dynamic exclusion was used, and the top nine precursor ions from each MS1 scan were subjected to collision energies according to mass and charge state (see Table S1). The m/z 806 was fragmented at 44.2 eV. All LC-MS/MS data sets were converted to mzXML by the export function from Compass DataAnalysis (Bruker) and imported into GNPS. The following parameters were used to generate a molecular network: Parent mass error = 0.02 Da, Fragment mass error = 0.02 Da, Minimum matching peaks = 3, Minimum cosine score = 0.7, with all other parameters at default value.
Results and Discussion
Investigation of communication that drives symbiosis in E. scolopes has been challenging due to the size and location of the light organ that harbors V. fischeri. In order to facilitate extension of this symbiosis to be compatible with MALDI-based imaging platforms, a method for dissection is done microscopically and only requires three main steps: removal of the eyeball material (Figure 1C), separation of the mantle from the body (Figure 1D), and drainage of the ink sac (Figure 1E). Removal of the eyeball material was required to achieve uniform height after dehydration of the tissue. Removal of the mantle also results in decreased height overall upon desiccation which allows for more efficient ionization from the light organ below. When the mantle is kept in place, it is possible to generate ions from the surface of the squid (Figure S1), and segmentation of the IMS data does not reveal an area specific to the light organ, because ions cluster by height. Similarly, when the mantle is flayed open but not entirely removed, ionization only occurs in hot spots across the tissue (Figure S2). The light organ area becomes more distinctly defined by ions here, but the differences in height still affect the ionization. Figure S3 shows that complete removal of the mantle and ink sac results in a region surrounding the light organ that is detected and segmented based on the unique localization of these signals.
While the idea of unsectioned body imaging is not novel,23 height heterogeneity and obstructing tissues limit the application of unsectioned imaging, and we sought to solve this problem in a group of biologically-informative cephalopods. The approach of maintaining an unsectioned structure of an organism with its microbial symbiont relatively undisturbed can enhance our understanding of the specialized metabolites and their role in maintenance or colonization. This is especially true in a system such as the squid-Vibrio symbiosis, where tissue sectioning is a challenging task (data not shown). Several aspects of the minimal preparation approach are critical for adaptation to another organism. The first is that any feature of an invertebrate that contributes to height in a dried sample must be removed prior to desiccation unless it is completely necessary for analysis. As demonstrated in this preparation of squid tissue, it is possible to remove many anatomical features that interfere with height uniformity while leaving important internal regions (namely the light organ) relatively undisturbed.
Another aspect for consideration during preparation is the removal of interfering chemical signals. For example, it is known that the squid ink sac holds a rich collection of melanin24 and almost entirely obscures visual observation of the light organ. If the purpose of the analysis is to evaluate biomolecular spatial distribution in the entire organism, as opposed to one single organ, then removal of chemical-rich areas is not necessary. An advantage of using the unsectioned organism is that the entire sample can be manipulated manually to create a pseudo 3D viewpoint of the organism for IMS. For example, two hatchlings were prepared and were mounted on opposite sides: one with its anterior side up and one with its posterior side up (Figure 2). Because both light organs in these hatchlings were colonized by ES114, it was not expected that the IMS analysis would detect differences in metabolite production, and that is what was observed. Both m/z’s 192 and 329 were detected in the light organ of E. scolopes regardless of the mounted orientation of the squid, which also lends further confidence in the data as these were two distinct hatchlings. The only significance of the two m/z’s was that they can be easily detected from both the anterior and posterior side of the organism, indicating that much attention need not be paid to orienting the squid in a specific way when preparing them for IMS analysis. It also is clear evidence that even analysis of the entire squid results in several signals that are specifically localized to the light organ. Isolation of these signals is ongoing for further experimentation and structure elucidation.
Figure 2.
Orientation of squid tissue does not affect ionization of metabolites. Both anterior and posterior orientation of E. scolopes tissue generated similar signals, m/z’s 192 and 329, localized to the light organ region. An ink spot is included as a negative control.
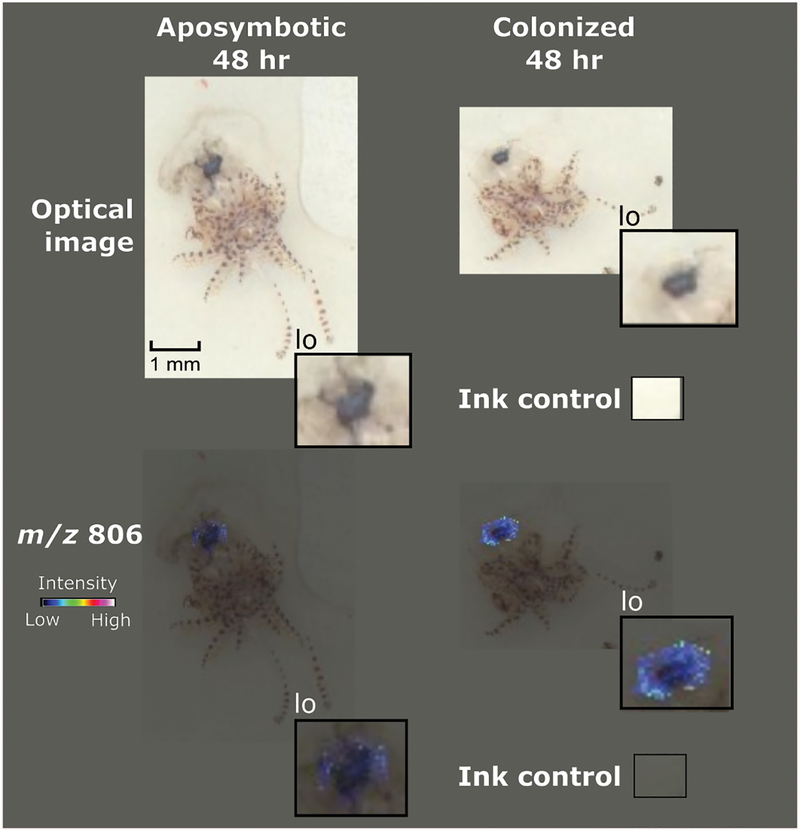
Once it was determined that the analysis of the desiccated unsectioned organism at 20 μm spatial resolution resulted in mass signals that localized to the light organ (Figure S3), we explored the spatiotemporal presence of specific ions during colonization. Four E. scolopes samples (Described in Materials and Methods) were evaluated. At 4 hours, bacteria have aggregated on the outside of the light organ in host mucus and are about to begin the process of light organ entry. At 48 hours, the light organ has been sufficiently colonized and long-term symbiosis is beginning to be established. Although statistical analysis using SCiLS was not able to detect any differences in signals with a default value of p < 0.05, the ion that presented the most statistical significance when the value was raised to a threshold of p < 0.1 was identified as m/z 806. Therefore, the actual p value of this ion lies between 0.1< p < 0.05 (Figure 3). This signal was detected with high abundance in the 48 h colonized squid and was undetectable in the 48 h aposymbiotic squid.
Figure 3.
Preparation of squid tissue facilitated detection of a colonization signal. Comparison between a hatchling at 48 hours that had not been exposed to any V. fischeri and a hatchling that was colonized displayed a metabolite, m/z 806 that is believed to be a bacterial metabolite.
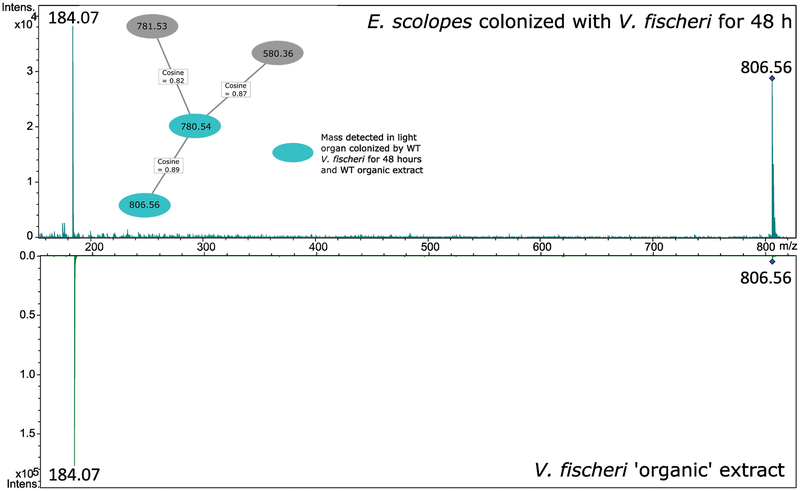
Because the biological origin (E. scolopes or V. fischeri) of m/z 806 cannot be determined by IMS analysis, LC-MS/MS of light organ extractions of all four samples was conducted orthogonally. The four light organ extracts were run alongside two WT V. fischeri extracts (Materials and Methods) and the tandem data were analyzed using Global Natural Products Social molecular networking (GNPS)25 (Figure S4). Molecular networking allows for the rapid identification of similarities in ions across samples based on fragmentation patterns as well as the dereplication of known unknowns compared to the library spectra.26 These data generated a small network that indicated m/z 806 is only present in the E. scolopes light organ sample that had been colonized with V. fischeri for 48 hours, but is also a product in the organic extract of the large-scale WT V. fischeri culture (Figure 4). Comparison of the accurate mass and fragmentation patterns of the light organ metabolite and the V. fischeri metabolite indicate that m/z 806 is likely of microbial origin. A putative identification in GNPS suggests that the ion is a phosphocholine analog (Figure S5), but further structure elucidation is required to verify this assignment and if there is a biological relevance to colonization. The TICs of all LC-MS/MS analyses can be found in Figure S6, and the TIC of the IMS run from Figure 3 can be found in Figure S7.
Figure 4.
GNPS Molecular Networking generated a network of phosphocholine analogs that may help to identify m/z 806. High-resolution LC-MS/MS data is compared to a database of natural products and fragmentation to assign putative identifications. Inset is network containing m/z 806 and three other metabolites, including their relatedness via cosine score (1.0 is identical). The m/z 806 node is a consensus spectrum of all m/z 806.56 scans detected, and two representative spectra are expanded. Two major fragments match between the colonized hatchling light organ and the V. fischeri organic extraction.
Ultimately, as long as the organism in question can be manipulated such that the region to be imaged dries to uniform flatness upon desiccation, the simple preparation method can be applied to any invertebrate. Even more directly impactful are the questions in the squid-Vibrio research community that may be answered by this approach. Studying any small organ in situ without the need for labelling or fully dissecting the animal tissue allows researchers to gain perspective on the chemical communication patterns that exist between two symbiotic partners both during colonization and throughout the partnership. The discovery of m/z 806 in the colonized hatchling may indicate a role for the metabolite during bacterial establishment in the light organ. While orthogonal methods (LC-MS/MS) were required to verify both the significance and structural features of this metabolite, IMS was needed to assign biological significance to this compound in situ.
Conclusions
Optimization of unsectioned body tissue imaging resulted in a minimal sample preparation protocol of the invertebrate E. scolopes for a MALDI-TOF MS system. The protocol enabled the successful preparation of unsectioned E. scolopes hatchlings for IMS of small molecules (<1000 Da) and ultimately facilitated rapid comparison of metabolites between hatchlings in colonization states. This method may provide a solution to those tissues that cannot be prepared in the traditional cryosectioning approach, but still require spatial orientation when mapping metabolites. In the case of E. scolopes, an intact light organ was required to assign particular molecules to the organ, which cannot be accomplished with a homogenized tissue sample. This protocol can be applied to any invertebrate organism as long as its size and desiccation capability can be aligned with the parameters for MALDI-TOF MS.
Supplementary Material
Acknowledgements
This publication was funded in part by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust (L.M.S. and M.J.M) and UIC startup funds (L.M.S.). Studies in the lab of M.J.M are supported by NIH grants R35GM119627 and R21AI117262 and NSF grant IOS-1757297. This work was supported in part by the National Cancer Institute of the National Institutes of Health under Award Number F31CA236237 (K.E.Z.).
References
- (1).Caprioli RM; Farmer TB; Gile J Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Anal. Chem 1997, 69 (23), 4751–4760. [DOI] [PubMed] [Google Scholar]
- (2).Yang JY; Phelan VV; Simkovsky R; Watrous JD; Trial RM; Fleming TC; Wenter R; Moore BS; Golden SS; Pogliano K; et al. Primer on Agar-Based Microbial Imaging Mass Spectrometry. J. Bacteriol 2012, 194 (22), 6023–6028. 10.1128/JB.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gemperline E; Horn HA; DeLaney K; Currie CR; Li L Imaging with Mass Spectrometry of Bacteria on the Exoskeleton of Fungus-Growing Ants. ACS Chem. Biol 2017, 12 (8), 1980–1985. 10.1021/acschembio.7b00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Zink KE; Dean M; Burdette JE; Sanchez LM Imaging Mass Spectrometry Reveals Crosstalk between the Fallopian Tube and the Ovary That Drives Primary Metastasis of Ovarian Cancer. ACS Cent Sci 2018, 4 (10), 1360–1370. 10.1021/acscentsci.8b00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Li H; Hummon AB Imaging Mass Spectrometry of Three-Dimensional Cell Culture Systems. Anal. Chem 2011, 83 (22), 8794–8801. 10.1021/ac202356g. [DOI] [PubMed] [Google Scholar]
- (6).ković D; Chu RK; Carrell AA; Thomas M; Paŝa-Tolić L; Weston DJ; Anderton CR Multimodal MSI in Conjunction with Broad Coverage Spatially Resolved MS2 Increases Confidence in Both Molecular Identification and Localization. Anal. Chem 2017, 90 (1), 702–707. [DOI] [PubMed] [Google Scholar]
- (7).Paine MRL; Kim J; Bennett RV; Parry RM; Gaul DA; Wang MD; Matzuk MM; Fernández FM Whole Reproductive System Non-Negative Matrix Factorization Mass Spectrometry Imaging of an Early-Stage Ovarian Cancer Mouse Model. PLoS One 2016, 11 (5), e0154837 10.1371/journal.pone.0154837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Perez CJ; Tata A; de Campos ML; Peng C; Ifa DR Monitoring Toxic Ionic Liquids in Zebrafish (Danio Rerio) with Desorption Electrospray Ionization Mass Spectrometry Imaging (DESI-MSI). J. Am. Soc. Mass Spectrom 2017, 28 (6), 1136–1148. 10.1007/s13361-016-1515-9. [DOI] [PubMed] [Google Scholar]
- (9).Yang E; Gamberi C; Chaurand P Mapping the Fly Malpighian Tubule Lipidome by Imaging Mass Spectrometry. J. Mass Spectrom 2019. 10.1002/jms.4366. [DOI] [PubMed] [Google Scholar]
- (10).Kingston ACN; Wardill TJ; Hanlon RT; Cronin TW An Unexpected Diversity of Photoreceptor Classes in the Longfin Squid, Doryteuthis Pealeii. PLoS One 2015, 10 (9), e0135381 10.1371/journal.pone.0135381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Andouche A; Bassaglia Y; Baratte S; Bonnaud L Reflectin Genes and Development of Iridophore Patterns in Sepia Officinalis Embryos (Mollusca, Cephalopoda). Dev. Dyn 2013, 242 (5), 560–571. 10.1002/dvdy.23938. [DOI] [PubMed] [Google Scholar]
- (12).Mainini V; Lalowski M; Gotsopoulos A; Bitsika V; Baumann M; Magni F MALDI-Imaging Mass Spectrometry on Tissues In Clinical Proteomics: Methods and Protocols; Vlahou A, Makridakis M, Eds.; Springer New York: New York, NY, 2015; pp 139–164. 10.1007/978-1-4939-1872-0_8. [DOI] [PubMed] [Google Scholar]
- (13).Nilsson A; Goodwin RJA; Shariatgorji M; Vallianatou T; Webborn PJH; Andrén PE Mass Spectrometry Imaging in Drug Development. Anal. Chem 2015, 87 (3), 1437–1455. 10.1021/ac504734s. [DOI] [PubMed] [Google Scholar]
- (14).León M; Ferreira CR; Eberlin LS; Jarmusch AK; Pirro V; Rodrigues ACB; Favaron PO; Miglino MA; Cooks RG Metabolites and Lipids Associated with Fetal Swine Anatomy via Desorption Electrospray Ionization - Mass Spectrometry Imaging. Sci. Rep 2019, 9 (1), 7247 10.1038/s41598-019-43698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Giordano S; Morosi L; Veglianese P; Licandro SA; Frapolli R; Zucchetti M; Cappelletti G; Falciola L; Pifferi V; Visentin S; et al. 3D Mass Spectrometry Imaging Reveals a Very Heterogeneous Drug Distribution in Tumors. Sci. Rep 2016, 6, 37027 10.1038/srep37027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Andersson M; Groseclose MR; Deutch AY; Caprioli RM Imaging Mass Spectrometry of Proteins and Peptides: 3D Volume Reconstruction. Nat. Methods 2008, 5 (1), 101–108. 10.1038/nmeth1145. [DOI] [PubMed] [Google Scholar]
- (17).Seeley EH; Caprioli RM 3D Imaging by Mass Spectrometry: A New Frontier. Anal. Chem 2012, 84 (5), 2105–2110. 10.1021/ac2032707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mandel MJ Models and Approaches to Dissect Host–symbiont Specificity. Trends Microbiol. 2010, 18 (11), 504–511. 10.1016/j.tim.2010.07.005. [DOI] [PubMed] [Google Scholar]
- (19).Ruby EG; Lee KH The Vibrio Fischeri-Euprymna Scolopes Light Organ Association: Current Ecological Paradigms. Appl. Environ. Microbiol 1998, 64 (3), 805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Cleary JL; Condren AR; Zink KE; Sanchez LM Calling All Hosts: Bacterial Communication in Situ. Chem 2017, 2 (3), 334–358. 10.1016/j.chempr.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Galey MM; Sanchez LM Spatial Analyses of Specialized Metabolites: The Key to Studying Function in Hosts. mSystems 2018, 3 (2). 10.1128/mSystems.00148-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Naughton LM; Mandel MJ Colonization of Euprymna Scolopes Squid by Vibrio Fischeri. J. Vis. Exp 2012, No. 61, e3758 10.3791/3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Khatib-Shahidi S; Andersson M; Herman JL; Gillespie TA; Caprioli RM Direct Molecular Analysis of Whole-Body Animal Tissue Sections by Imaging MALDI Mass Spectrometry. Anal. Chem 2006, 78 (18), 6448–6456. 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- (24).McFall-Ngai MJ Consequences of Evolving with Bacterial Symbionts: Insights from the Squid-Vibrio Associations. Annu. Rev. Ecol. Syst 1999, 3, 235–236. [Google Scholar]
- (25).Wang M; Carver JJ; Phelan VV; Sanchez LM; Garg N; Peng Y; Nguyen DD; Watrous J; Kapono CA; Luzzatto-Knaan T; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol 2016, 34 (8), 828–837. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Yang JY; Sanchez LM; Rath CM; Liu X; Boudreau PD; Bruns N; Glukhov E; Wodtke A; de Felicio R; Fenner A; et al. Molecular Networking as a Dereplication Strategy. J. Nat. Prod 2013, 76 (9), 1686–1699. 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.