Abstract
Antidepressants are widely prescribed, but their efficacy relative to placebo is modest, in part because the clinical diagnosis of major depression encompasses biologically heterogeneous conditions. Here, we sought to identify a neurobiological signature of response to antidepressant treatment as compared to placebo. We designed a latent-space machine learning algorithm tailored for resting-state electroencephalography (rsEEG) and applied it to data from the largest imaging-coupled, placebo-controlled antidepressant study (n=309). Symptom improvement was robustly predicted in a manner both specific for the antidepressant sertraline (versus placebo) and generalizable across different study sites and EEG equipment. This sertraline-predictive EEG signature generalized to two depression samples, wherein it reflected general antidepressant medication responsivity, and related differentially to repetitive transcranial magnetic stimulation (rTMS) treatment outcome. Furthermore, we found that the sertraline rsEEG signature indexed prefrontal neural responsivity, as measured by concurrent TMS/EEG. Our findings advance the neurobiological understanding of antidepressant treatment through an EEG-tailored computational model and provide a clinical avenue for personalized treatment of depression.
Major depression is currently defined based on clinical criteria and encompasses a heterogeneous mix of neurobiological phenotypes 1. This heterogeneity may account for the modest superiority of antidepressant medication over placebo (Cohen’s d ~ 0.3) 2-6. Work over the past two decades has suggested that resting-state electroencephalography (rsEEG) may be able to identify treatment-predictive heterogeneity in depression 7-10. Specific attention has been paid to prefrontal and parietal signals carried by the theta (4-7Hz) and alpha (8-12Hz) frequency bands 7-13. However, due to lack of cross-validation and small sample sizes, prior studies have identified either non-specific predictors that do not differentiate between response to drug versus placebo, such as rostral anterior cingulate theta current density 11-13, or failed to yield robust (i.e., generalizable) and reproducible neural signatures that are predictive at the individual patient level 8. As such, we still lack a robust neurobiological signature for an antidepressant-responsive phenotype that could identify which patients will derive a large benefit from medication. Delineating such a signature would advance both a neurobiological understanding of treatment response and yield important clinical implications.
In order to identify a robust antidepressant-responsive depression phenotype, machine learning can be used to combine across the complex multivariate relationships existing within rsEEG data. An effective predictive rsEEG computational model, however, must deal with three critical challenges. First, smearing of signal and noise results from volume conduction due to each electrode picking up neural signals from multiple sources and adjacent electrodes detecting neural signals from the same sources 14, Second, there is a risk of overfitting the model given the high spatio-temporal dimensionality and noisiness of EEG data 15, 16. Third, there are challenges in simultaneously optimizing feature identification and fitting of predictive regression models due to the nonlinearity of the error function with respect to the model parameters 17.
To address each of these challenges, we developed a machine learning algorithm that we call Sparse EEG Latent SpacE Regression (SELSER). The data were derived from four studies. First, we established the rsEEG predictive signature by training SELSER on data derived from the largest neuroimaging-coupled placebo-controlled randomized clinical study of antidepressant efficacy (n = 309). We then used three additional data sets to validate our findings. Together, these efforts aimed to reveal a treatment-responsive phenotype in depression, dissociate between medication and placebo response, establish its mechanistic significance, and provide initial evidence for the potential for treatment selection based on a rsEEG signature.
Results
Development of SELSER
We developed SELSER (Fig. 1, Methods and Supplementary Code) to address the challenges mentioned above and identify a robust signature from EEG that would predict response to antidepressants. Signal identification and mitigation of volume conduction are accomplished by amplifying signal-to-noise ratio through use of spatial filters 18. Each spatial filter transforms the multi-channel EEG data into a single latent signal, the power of which is used as a feature for the machine learning algorithm. Since model fitting is done under a sparse constraint on the number of spatial filters, this also serves to reduce dimensions of the underlying latent signals and thus decrease the chance of overfitting. Finally, we mathematically formulated the outcome prediction framework as solving a convex problem that related EEG time series to the treatment outcome directly, yielding a single and globally optimal solution 19. This approach can be contrasted, for example, with regression models being applied to channel-level rsEEG power measures, which does little to mitigate volume conduction. Likewise, conventional latent space methods such as Independent Component Analysis (ICA) 20 and Principal Component Analysis (PCA) are not optimal in that they are unsupervised approaches not directly related to optimizing for the treatment outcome prediction target. In light of prior rsEEG work 7-9, we predicted that SELSER-established neural signals drawn from theta and alpha frequency bands would most strongly predict treatment outcome.
Fig. 1.
End-to-end prediction of the treatment outcome with a latent space model. The model consists of three stages: 1) Spatial filtering that linearly transforms the EEG signals to the latent signals; 2) Band power feature extraction that computes the band power of each latent signal; 3) Linear regression that uses the band powers to predict the treatment outcome. By solving a convex optimization problem, all the unknown parameters (spatial filters and linear regression weight coefficients) are optimized in conjunction under a unified objective function that trades off between the prediction error and dimensionality of the latent signals. S1, S2, and SN refer to Subject 1, Subject 2, and the Nth Subject, respectively. C1, C2, F1, F2 and Pz refer to electrode locations according to the 10/10 international system. (·)2 denotes the square operator, and ∫t denotes the average of a time series over time.
Data were drawn from four studies. Establishment of the treatment-predictive rsEEG signature was accomplished with data from the Establishing Moderators and Biosignatures of Antidepressant Response in Clinic Care (EMBARC) study 21. EMBARC is the largest neuroimaging-coupled placebo-controlled randomized clinical trial (RCT) in depression to date and involved randomization of 309 medication-free depressed outpatients (n = 228 with high quality rsEEG data) to receive either the selective serotonin reuptake inhibitor (SSRI) sertraline or placebo for eight weeks (Supplementary Fig. 1). Eyes-open and eyes-closed EEG data were collected prior to randomization at four sites in the U.S., each of which used a different high-density EEG system and/or electrode montage (60 or more electrodes). Clinical outcome was assessed on the 17-item clinician-administered Hamilton Depression Rating Scale (HAMD17).
The generalizability of the antidepressant-predictive signature was then tested in a second independent sample of depressed patients (n = 72), for whom we had historical information about treatment response during the current depressive episode and rsEEG data. A third independent sample of depressed patients (n = 24) was used to assess two features of the treatment-predictive rsEEG signature: convergent validity and neurobiological significance. Specifically, we tested whether expression of our rsEEG signature correlated with another machine learning signature we developed based on task-based fMRI activation in EMBARC 22, as this would provide further convergent validation of the rsEEG signature identified here. We also tested in the third sample whether regions that were prominent in the rsEEG signature reflected individual differences in cortical responsivity, as directly assessed through single pulse transcranial magnetic stimulation (TMS) during concurrent EEG recording.
Finally, in a fourth depressed sample (n = 152) that was treated with either 10Hz left dorsolateral prefrontal repetitive TMS (rTMS) or 1Hz right dorsolateral prefrontal rTMS (both with concurrent psychotherapy), we tested whether the strength of the EMBARC-trained rsEEG signature predicted outcome with an antidepressant treatment that has a putatively different mechanism of action. This allowed us to test the generalizability of our results and open up the potential for treatment selection by defining the neural predictors of antidepressant response.
Treatment prediction from pretreatment resting EEG using SELSER
We built prediction models using pretreatment rsEEG by applying SELSER to each of four canonical EEG frequency bands (theta: 4-7 Hz, alpha: 8-12 Hz, beta: 13-30 Hz, gamma: 31-50 Hz) in each resting condition (two 2-minute blocks each of eyes open or eyes closed). Treatment outcome was quantified as the pre- minus-post-treatment difference in HAMD17 scores, with missing endpoint values imputed to maintain an intent-to-treat framework. Model performance was tested using 10-fold cross-validation (Fig. 1 and Supplementary Fig. 2, Methods).
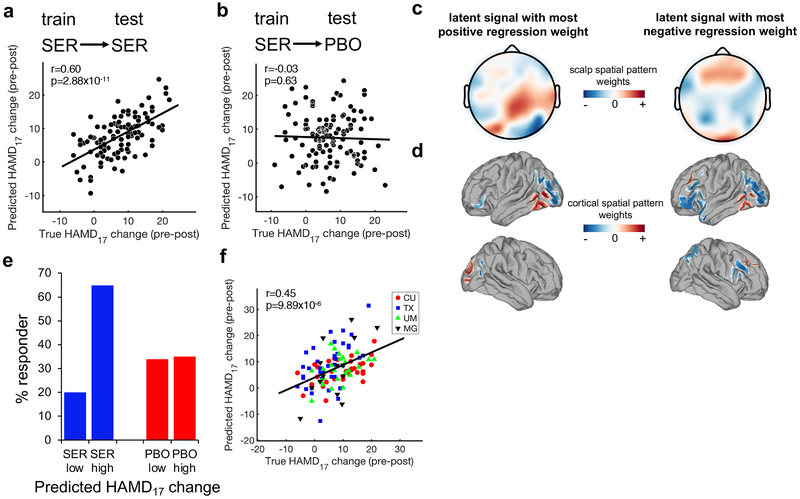
For the sertraline arm, only alpha signals from the resting eyes open (REO) condition were significantly predictive of the observed treatment score changes during cross-validation (Fig. 2a; Pearson’s r = 0.60, root mean square error (RMSE) = 5.68, Bonferroni-corrected p = 2.88x10−11; permutation test-verified using 1000 permutations p < 10−3). When the sertraline-trained model was applied to the placebo arm, however, outcome could not be predicted (Fig. 2b, Pearson’s r = −0.03, RMSE = 9.77, p = 0.63), thus demonstrating the specificity of this model for sertraline efficacy prediction (Fisher’s z test: z = 4.94, p = 8x10−7). Application of SELSER to alpha-frequency REO EEG signals could not predict baseline HAMD17 scores (Pearson’s r = 0.06, RMSE = 6.10, p = 0.27), illustrating that the treatment-predictive model was not related to baseline depression severity.
Fig. 2.
Prediction of outcome specific to sertraline using SELSER on resting eyes open alpha-frequency range data. (a) 10x10 stratified cross-validation prediction of HAMD17 change in the sertraline arm (n = 109) using SELSER. Pearson’s r = 0.60, Bonferroni-corrected p = 2.88x10−11 based on the one-sided test against the alternative hypothesis that r > 0. (b) Application of the sertraline-trained model to the placebo arm (n = 119) failed to predict outcome, demonstrating specificity of the model for sertraline prediction. Pearson’s r = −0.03, p = 0.63 based on the one-sided test against the alternative hypothesis that r > 0. (c) Scalp spatial patterns of the SELSER latent signals, with the most positive (β = 759.31; left) and negative (β = −853.13; right) regression weights, respectively. n =109. (d) Cortical spatial patterns of the SELSER latent signals, with the most positive (β = 759.31; left) and negative (β = −853.13; right) regression weights, respectively. n = 109. (e) Purely for the purpose of visualizing the utility of the rsEEG predictive signature, patients in each arm were partitioned into the low and high groups by applying a median split on the cross-validated predicted HAMD17 score changes for sertraline response. The response rate was then calculated for each group (defined as a 50% or greater decrease in symptoms from baseline). SER = sertraline (blue), PBO = placebo (red). (f) Treatment prediction across study sites in a leave-study-site-out cross-validation on the alpha REO sertraline model (n = 109). Study sites were Columbia University (CU), University of Texas Southwestern Medical Center (TX), University of Michigan (UM) and Massachusetts General Hospital (MG). Site effect was corrected for by removing mean of the covariance matrix from each study site prior to the SELSER analysis. Pearson’s r = 0.45, Bonferroni-corrected p = 9.89x10−6 based on the one-sided test against the alternative hypothesis that r > 0.
As a result of the algorithm-enforced low-dimensionality constraint on the latent signals in SELSER, only a few latent signals were obtained in each model (Supplementary Fig. 3). For the sertraline alpha REO model, the scalp and cortical spatial maps of the two latent signals with the most positive and negative regression weights are shown in Figs 2c and 2d, respectively. The spatial pattern of the latent signal with the most positive regression weight was mainly centered around the right parietal-occipital regions, in line with prior work 23. By contrast, the spatial pattern of the latent signal with the most negative regression weight was heavily concentrated in both the lateral prefrontal and parieto-occipital regions.
We also tested the effects of data amount on model fitting, and found that performance began degrading once less than two blocks of 1.5 minutes each per patient were used (Supplementary results and Supplementary Fig. 4 and 5).
For the placebo arm, both alpha signals from the REO and resting eyes closed (REC) conditions significantly predicted the HAMD17 score change (REO Fig. 3a, Pearson’s r = 0.41, RMSE = 6.34, Bonferroni-corrected p = 2.73x10−5; and REC Fig. 3c, Pearson’s r = 0.31, RMSE = 7.60, Bonferroni-corrected p = 4.13x10−3; permutation test-validated p < 10−3). The spatial maps of the two latent signals with the most positive and negative regression weights are shown in Supplementary Fig. 6 for the REO and REC conditions. For the REO condition, the spatial patterns of the latent signals were predominantly in the temporal and occipital regions, whereas for the REC condition, frontal-parietal and frontal regions were the most prominent. When applied to the sertraline arm, both regression models failed to predict outcome (Fig. 3b and 3d, Fisher’s z test: z > 2.98, p < 3x10−3), demonstrating the specificity of these models for placebo outcome prediction and distinction from the sertraline-predictive model above.
Fig. 3.
Prediction of outcome specific to placebo using SELSER on alpha-frequency range data. (a, c) 10x10 stratified cross-validation prediction of HAMD17 change in the placebo arm (n = 119) using SELSER on resting eyes open (a; Pearson’s r = 0.41, Bonferroni-corrected p = 2.73x10−5 based on the one-sided test against the alternative hypothesis that r > 0) and resting eyes closed (c; Pearson’s r = 0.31, Bonferroni-corrected p = 4.13x10−3 based on the one-sided test against the alternative hypothesis that r > 0) alpha-frequency range data, respectively. (b, d) Application of the resting eyes open (b; Pearson’s r = −0.13, p = 0.91 based on the one-sided test against the alternative hypothesis that r > 0) and resting eyes closed (d; Pearson’s r = −0.08, p = 0.79 based on the one-sided test against the alternative hypothesis that r > 0) placebo-trained models to the sertraline arm (n = 109) failed to predict outcome, demonstrating specificity of the model for placebo prediction.
To visualize how the SELSER predictions in Fig. 2a could be used for treatment stratification, we partitioned the patients in each arm by applying an arbitrary median split on the cross-validated predicted HAMD17 score changes derived from the sertraline alpha REO model. We then calculated the rates of treatment response (≥ 50% reduction in symptoms) for the portion of patients above the median (“high”) and below the median (“low”), respectively, based on the model-predicted HAMD17 change scores (Fig. 2e and Supplementary Fig. 7). For the sertraline arm, the high group reached a response rate of 65%, which more than tripled the response rate (20%) in the low group and was considerably higher than the response rates in the placebo arm (35% and 34% for the high and low groups, respectively).
Treatment prediction across study sites
To further assess the generalizability of the prediction models to unseen data collected with different EEG amplifiers, a leave-study-site-out analysis was performed by iteratively using three study sites’ data to train the model, and the fourth site’s data for testing the model. Since the four study sites’ EEG data were acquired with different EEG amplifiers and/or electrode montages (Supplementary Table 1), marked variability of prediction performance was observed across study sites (Supplementary Fig. 8). To mitigate this site effect, the mean of the covariance matrix was removed from each study site prior to the SELSER analysis. For the sertraline arm, only the alpha REO model was significantly predictive of the treatment outcome when doing leave-study-site-out cross-validation (Fig. 2f; Pearson’s r = 0.45, RMSE = 7.02, Bonferroni-corrected p = 9.89x10−6; permutation test-validated using 1000 permutations p < 10−3).This demonstrates the robustness of our model for unseen data from a different EEG amplifier – arguably a worst-case scenario with respect to testing through cross-validation. An equivalent 4-fold cross-validation model sampling across all sites remained strongly predictive (Pearson’s r = 0.58, RMSE = 5.63, p = 1.59x10−11; permutation test-validated using 1000 permutations p < 10−3). Further restricting the sample size via a 2-fold cross-validation across all sites yielded a lower yet still highly significant predictive performance (Pearson’s r = 0.38, RMSE = 6.32, p = 2.25 x 10−5). For the placebo arm, none of the REO and REC models were predictive of the treatment outcome when cross-validating between study sites (Pearson’s r’s < 0.22, RMSE > 7.90, Bonferroni-corrected p > 0.07). The sertraline leave-study-site-out alpha REO predictive model also still showed significantly greater specificity in predicting sertraline over placebo response (Fisher’s z test: z = 3.83, p = 10−4).
Comparison of SELSER predictions to prior methods
In order to benchmark SELSER against conventional machine learning approaches that do not use latent space modeling, we also trained linear regression models on eyes open rsEEG data by using the Relevance Vector Machine (RVM) 24 on channel-level alpha band power 25, 26, theta band power 27, 28, and theta cordance 13, 23 (Online Methods and Supplementary Fig. 9 and 10). However, none of the models were predictive of the treatment outcome on cross-validation (Pearson’s r’s < −0.06, RMSE > 8.20, p > 0.74). Moreover, to demonstrate the improvement of SELSER over conventional latent space modeling approaches, we also trained the RVM on alpha band power of the latent signals extracted with PCA 29 (Supplementary Fig 9d) or ICA 25 (Supplementary Fig 9e), which are among the most popular unsupervised methods to derive spatial filters from EEG (Online Methods and Supplementary Fig. 10). Here as well, both models failed to predict the treatment outcome (Pearson’s r’s < 0.15, RMSE > 7.17, p > 0.09).
Treatment prediction from symptoms
Assessing brain activation for defining an individual’s sertraline responsive phenotype may not be relevant in practice if lower-cost measures such as clinical severity scores, demographic variables or historical factors like childhood trauma exposure could usefully predict outcome. This did not prove to be the case, however, as RVM trained with all of these was only modestly predictive (Supplementary Fig. 11; Pearson’s r = 0.26, RMSE = 7.93, p = 3x10−3 for sertraline, and Pearson’s r = 0.16, RMSE = 9.56, p = 0.05 for placebo), and worse if using only the Quick Inventory of Depressive Symptomatology (QIDS) scale (Pearson’s r = 0.12, RMSE= 6.85, p = 0.12 for sertraline, and Pearson’s r = 0.06, RMSE = 6.72, p = 0.26 for placebo). Sertraline outcome prediction with clinical measures was also significantly weaker than the alpha REO rsEEG model above (Fisher’s z test; z = 3.11, p = 0.0019).
Testing the generalization of the rsEEG sertraline-predictive signature
We next tested the generalizability of the SELSER rsEEG sertraline-predictive signature from EMBARC in a second independent cohort of patients with depression. This cohort of patients were drawn from a naturalistic, longitudinal depression study in which rsEEG data were recorded at the baseline visit 30. Patients also completed the Antidepressant Treatment Response Questionnaire (ATRQ), which provided historical information about the number of adequate antidepressant medication trials in the current episode, as well as whether patients responded to them or not. Following conventional groupings, patients were categorized as either treatment-resistant (two or more failed antidepressant trials; n = 21), or as partial responders (partial response to at least one medication; n = 51). The mean-removal site correction procedure was performed for the rsEEG data, as in the leave-study-site-out analysis of the EMBARC study. We then applied the EMBARC-trained sertraline alpha-band rsEEG model to each patient, yielding a predicted HAMD17 change for each individual, which reflects their strength of expression of the sertraline-predictive rsEEG signature. As expected, the predicted HAMD17 change was higher for the partial responder group than the treatment resistant group (Fig. 4), demonstrating the generalizability of the EMBARC rsEEG sertraline signature to the broader construct of treatment responsiveness/resistance to antidepressant medication. Moreover, information on the number of within-episode failed antidepressant trials was available for 45 of the 72 patients. Also as expected, we found a negative correlation between the number of failed trials and the magnitude of the rsEEG sertraline signature-predicted HAMD17 improvement (Pearson’s r = −0.34, p=0.023).
Fig. 4.
Prediction of treatment outcome by the EMBARC-trained sertraline rsEEG model, applying to baseline eyes open rsEEG of the second depression study cohort. The plot shows the predicted HAMD17 change for patients who are partial responder (n = 51) or treatment resistant (n = 21). These data demonstrate the predicted HAMD17 change is significantly larger in patients whose are partial responders than in those who are treatment resistant (two-sample and two sided t-test p = 0.016). Error bars depict s.e.m.
Convergence between rsEEG- and task-fMRI-derived machine learning predictions
To test the convergent validity of the sertraline rsEEG model from EMBARC, we examined a separate dataset of 24 patients with depression who were assessed in a cross-sectional manner (i.e., without treatment) using both rsEEG and task-based fMRI with the emotional conflict task 22. The reason for doing so is that we could test whether the predicted HAMD17 change based on the EMBARC-trained sertraline rsEEG model correlated with the predicted HAMD17 change based on an fMRI emotional conflict task-based machine learning model we established in a separate analysis of EMBARC data 22 (Methods). Since the EEG data were recorded with yet another amplifier distinct from those used in EMBARC, the mean-removal site correction procedure was performed. The rsEEG and task-fMRI predictions were significantly correlated with each other in these independent data (Fig. 5a; Pearson’s r = 0.44, p = 0.02). This finding provides convergent support, across fMRI and EEG, for the existence of a treatment-responsive neurobiological phenotype in major depressive disorder across populations, and across assessment modalities.
Fig. 5.
Alignment of predicted HAMD17 change calculated by the rsEEG model and predicted HAMD17 change calculated by a machine learning model trained on task-based fMRI activation from a separate analysis on EMBARC data, as well as neural responsivity assessed through spTMS/EEG. n = 24. a) The EMBARC-trained rsEEG and task fMRI models were applied to an independent major depressive disorder data set that had both data types, and the ensuing predicted HAMD17 changes from both models were correlated with each other. spTMS/EEG correlates of the rsEEG phenotype in the independent depressed data set. Pearson’s r = 0.44, p = 0.02 based on the one-sided test against the alternative hypothesis that r > 0. (b) TMS was delivered to bilateral posterior dorsolateral prefrontal cortices (pDLPFC, part of the fronto-parietal control network), anterior DLPFC (aDLPFC, part of the ventral attention network), primary motor cortex (M1), and primary visual cortex (V1). These sites were identified based on independent components analyses on resting-state fMRI data from a separate cohort. (c) A significance plot of the correlation between the spTMS/EEG responses and rsEEG phenotype, as indexed by the leave-one-out cross-validated Pearson’s correlation coefficients between the SELSER-predicted rsEEG phenotype and true rsEEG phenotype, for each of the stimulation sites. n = 24. The SELSER analysis was performed separately for the same set of frequency bands as used in the rsEEG prediction analysis (θ: theta, α: alpha, β: beta, γ: gamma), and for three time windows relative to the TMS pulse (0 – 200ms, 200 – 400ms, 400 – 600ms), followed by a false discovery rate correction (FDR) across all of these tests. Only right aDLPFC stimulation (alpha band, 200 – 400ms: Pearson’s r = 0.60, p = 5.5x10−4 based on the one-sided test against the alternative hypothesis that r > 0), left pDLPFC stimulation (gamma band, 200 – 400ms: Pearson’s r = 0.58, p = 8x10−4 based on the one-sided test against the alternative hypothesis that r > 0) and right pDLPFC stimulation (beta band, 0 – 200ms: Pearson’s r = 0.60, p = 4.6x10−4 based on the one-sided test against the alternative hypothesis that r > 0) survived FDR correction (denoted by asterisks). The plot shows −log10(p) of the correlation of the SELSER-predicted rsEEG phenotype with true rsEEG phenotype.
TMS/EEG correlates of rsEEG phenotype
Next, to provide further insight into the neural signals driving our sertraline-predictive rsEEG-defined phenotype, we analyzed concurrent single-pulse TMS/EEG (spTMS/EEG) 31 data from the third independent depression sample as used above. Specifically, we sought to test whether cortical responsivity, as assessed by direct stimulation using spTMS/EEG to regions either prominent or minimal within the spatial patterns of the rsEEG latent signals, induced neural responses that correlated with the rsEEG-defined treatment-predictive phenotype. The stimulated regions were the bilateral posterior dorsolateral prefrontal cortices (pDLPFC), anterior DLPFC (aDLPFC), along with primary visual cortex (V1) and bilateral primary motor cortices (M1) as the control regions (Fig. 5b). We localized pDLPFC and aDLPFC using neuronavigation based on their being nodes within the frontoparietal and salience resting-state networks, respectively, as we have done in our prior work 32. To quantify the correlation between the spTMS/EEG responses and sertraline-predictive rsEEG phenotype, we again employed SELSER to relate spTMS/EEG data to the sertraline rsEEG signature (Fig. 5c). Correlations between the sertraline rsEEG phenotype and spTMS/EEG responses to stimulation at three of four prefrontal cortical regions survived correction for multiple comparisons. This included right aDLPFC stimulation (alpha band, 200 – 400ms: Pearson’s r = 0.60, false discovery rate (FDR)-corrected p = 5.5x10−4), left pDLPFC stimulation (gamma band, 200 – 400ms: Pearson’s r = 0.58, FDR-corrected p = 8x10−4) and right pDLPFC stimulation (beta band, 0 – 200ms: Pearson’s r = 0.60, FDR-corrected p = 4.6x10−4). Correlation of responses to stimulation of primary motor or visual cortices did not survive correction.
Sertraline signature assessment in a combined repetitive TMS and psychotherapy treatment study
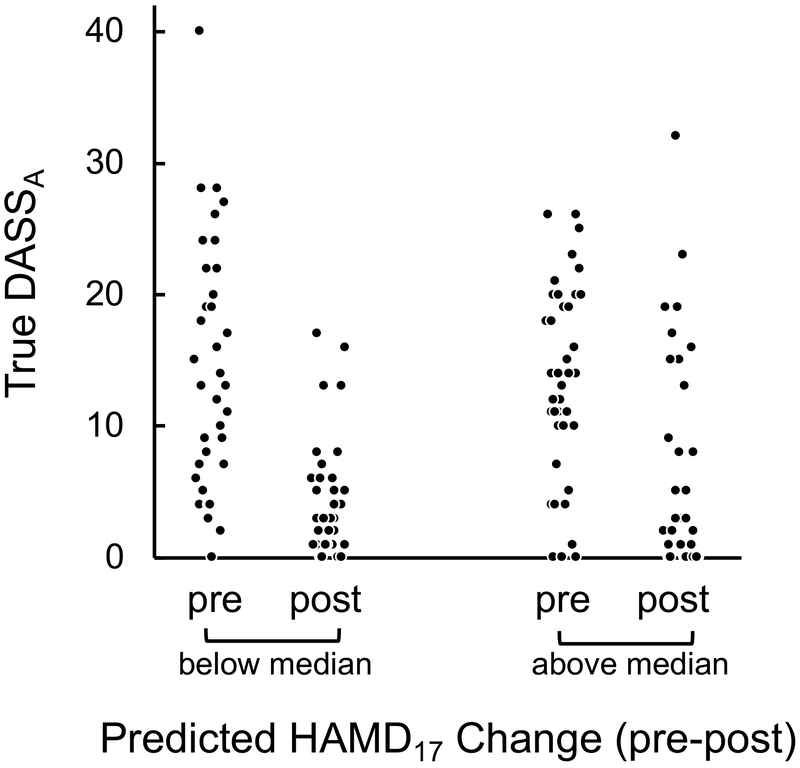
In light of the sertraline rsEEG phenotype indexing cortical responsivity in response to stimulation at several DLPFC locations, we next considered whether the strength of this phenotype could predict outcome with repetitive TMS (rTMS) treatment in depression. Analyses were performed on a fourth previously-reported dataset of patients with depression and pre-treatment rsEEG recordings, who received at least 10 sessions of simultaneous rTMS and psychotherapy and on stable medication regiments 33, 34. Treatment involved rTMS applied with either a 10 Hz protocol over the left DLPFC (n = 64) or a 1 Hz protocol over the right DLPFC (n = 88; Online Methods). We computed each patient’s expression of the EMBARC-trained sertraline rsEEG model (expressed as predicted HAMD17 change) using the same mean site removal procedure as above. Symptoms were assessed with the Beck Depression Inventory (BDI) and the three subscales of the Depression, Anxiety and Stress Scale (DASS), separately by rTMS protocol, using linear mixed models and a Bonferroni-correction for eight comparisons (two frequencies, four outcome measures). One relationship survived, wherein less rsEEG-predicted HAMD17 change with sertraline was associated with greater response to 1 Hz rTMS on the DASS anxiety scale (rsEEG predicted HAMD17 sertraline change x Time interaction: F(1,128) = 9.02, p = 4x10−3; Fig. 6). This suggests that patients who fail to respond to sertraline may be more amenable to 1 Hz right DLPFC rTMS, providing a potential evidence-based treatment selection approach for depression. This relationship was also specific to 1Hz right DLPFC rTMS, as we found a treatment protocol x predicted HAMD17 change x Time interaction when including both arms in a linear regression (F(1,126) = 4.54, p = 0.035).
Fig. 6.
Prediction of treatment outcome with right DLPFC 1Hz rTMS treatment by the EMBARC-trained sertraline rsEEG model, applying to pre-rTMS eyes open rsEEG. The scatterplot shows the pre and post-treatment scores for patients on the Anxiety subscale of the Depression, Anxiety and Stress Scale (DASSA). In order to visualize the linear mixed model relating rsEEG-predicted HAMD17 change to observed changes in DASSA scores, shown here is a median split on the predicted HAMD17 change values. These data demonstrate that the degree of pre-to-post change in DASSA symptoms due to 1Hz rTMS treatment is greater in those patients with smaller expected HAMD17 change scores using the EMBARC-trained sertraline rsEEG model.
Discussion
Here we developed a rsEEG-optimized latent space computational model, called Sparse EEG Latent SpacE Regression (SELSER), with which we obtained robust prediction of antidepressant outcome, and moderation (i.e. differential prediction) between outcome with an antidepressant versus placebo in a large placebo-controlled study. The antidepressant-predictive signature identified using SELSER on alpha frequency range eyes-open rsEEG data was superior to conventional machine learning models, or latent modeling methods such as ICA or PCA. This signature was furthermore superior to a model trained on clinical data alone, was able to predict outcome on rsEEG data acquired at a study site not included in the model training set and which used a different EEG amplifier and/or electrode montage, and related to general antidepressant responsiveness/resistance in a separate depression sample. These attributes of our SELSER model support its potential utility in the context of real-world clinical care and with regard to stratification of future depression studies based on the expected antidepressant-specific treatment outcome for that individual. Critically, though promising and highly influential signals for antidepressant prediction with rsEEG have been reported for the past two decades 7-9, 35, prior signals neither moderated between outcome with an antidepressant versus placebo, nor provided robust individual patient-level prediction. Our present work is thus distinguished from prior findings on both fronts.
We also found evidence of multi-modal convergent validity for our rsEEG antidepressant-response signature in a third depression data set by virtue of its correlation with expression of a task-based fMRI signature we recently identified using EMBARC data 22. The strength of our rsEEG signature also correlated with prefrontal neural responsivity, as indexed by direct stimulation with spTMS/EEG. This led us to test the relationship between the sertraline rsEEG model and treatment outcome with rTMS in a fourth sample. There, we found an opposite relationship between sertraline-predicted improvement and observed treatment outcome with 1Hz right DLPFC rTMS. This finding also opens an exciting avenue for neural signature-driven treatment selection in depression.
From a neural mechanism perspective, we note that the sertraline SELSER model revealed both positively and negatively-weighted posterior cortical eyes open rsEEG signals, but only heavily negatively-weighted prefrontal signals. Considering suggestions that resting alpha power reflects inhibitory tone in a brain region 36, 37, the negative weighting of prefrontal alpha in our sertraline model suggests that the prefrontal cortices of better treatment responders are more active or excitable than those of poor responders. The positive and negative weights for posterior signals suggests that optimizing the balance of distinct posterior predictive signals may be what is critical to establishing a robust computational model. Prediction analysis using electrodes exclusively from posterior regions corroborates this claim (Supplementary Fig. 12). Our results are broadly in line with prior reports of better outcome being associated with greater posterior cortical alpha power 23. However, a large-scale study that lacked a placebo control arm failed to replicate these findings 38. Thus, as our RVM model trained on channel-level alpha power failed to predict outcome, the critical element in attaining individual level-robust outcome prediction may be the use of a latent space computational model .
Perhaps somewhat surprising is that only eyes open alpha rsEEG, but not eyes closed, was predictive of the treatment outcome, given that the alpha rhythm is more strongly present in the eyes closed condition. One explanation is that the increasing alpha rhythm during the eyes closed condition is indicative of cortical areas being deactivated 39, and thus may contribute to background noise rather than predictive signal. This view is consistent with motor intention decoding work, wherein the goal has been to enhance the difference between the mu motor rhythm while suppressing the more broadly-distributed alpha rhythm 40,41.
The present work does not directly inform the cognitive and emotional information processing relevance of the antidepressant-response rsEEG signature. However, the relationship between the rsEEG signature and one derived from task-based fMRI data from EMBARC22 suggests that individuals with stronger expression of the rsEEG signature may be better able to regulate emotional conflict.
We utilized a range of cross-validation methods (including on data from entirely unseen study sites collected on different EEG equipment), compared prediction of outcome with sertraline versus placebo across cross-validation methods, and tested for generalization of the signals across a number of complimentary datasets. As such, our data provide neurobiological evidence that an antidepressant-responsive phenotype exists within the biological heterogeneity characteristic of the broader clinical diagnosis of depression. These findings thereby not only advance a “personalized” approach to depression 1, 5, 42, but also demonstrate that antidepressants only appear to be modestly more effective than placebo because they are typically given to an unselected sample of depression patients.
If replication and extension of this phenotype to other antidepressant medications is successful in progressively larger and more diverse data sets, the rsEEG signature we identified may be helpful in deciding whether a patient should continue further medication trials after an initial failure with an antidepressant medication, or switch to treatments with putatively different mechanisms of action (e.g. rTMS, electroconvulsive therapy (ECT), or psychotherapy). Indeed, depressed patients often undergo many medication trials before advancing to other treatments such as rTMS 43, 44, which are effective for some medication-resistant patients45, 46. This could result in potentially avoidable morbidity and economic cost if they are switched to another intervention earlier based on evidence of little expected benefit with an antidepressant using our rsEEG signature.
Our finding of an opposite relationship between predicted change with sertraline and treatment outcome with 1Hz rTMS and concurrent psychotherapy directly support the potential for these findings to guide treatment selection in depression, pending further replication. Furthermore, as the sertraline-predictive rsEEG signature did not predict outcome with 10Hz rTMS during concurrent psychotherapy, this suggests that it is the effect of the specific rTMS protocol that is being predicted rather than the effects of psychotherapy, though further investigation with rTMS treatment without psychotherapy is needed for this to be conclusive. Interestingly, this opposite direction prediction between antidepressants and rTMS is consistent with our prior work as well as that of others. More intact default mode network connectivity in the iSPOT-D study predicted better treatment outcome with antidepressant treatment 47, while more disrupted default mode network connectivity predicted outcome with rTMS in a clinic-based cohort 48.
There are also several limitations to consider. First, our specific prediction findings remain to be replicated in an entirely independent sample, nor is it known whether the sertraline signature predicts outcome with the broader class of selective serotonin reuptake inhibitors, or antidepressant medication more generally. Second, EMBARC assessed all patients while medication-free at baseline. Thus, while this removes potential confounds from the experimental design, it also limits generalizability to typical outpatients, who are often already on an antidepressant. Finally, SELSER is a static latent space modeling approach that does not consider resting EEG dynamics. Dynamic latent space models have been recently developed to predict mood from multisite intracortical human brain signals 49.
In summary, we developed a rsEEG-optimized latent space computation model that was capable of robustly predicting treatment outcome with the antidepressant sertraline and distinguishing between response to sertraline versus placebo at the individual patient level, and which may furthermore support treatment selection between medication and rTMS. Together, these findings ground in individual-level neurobiology a treatment-responsive phenotype obscured within the broader clinical diagnosis of depression and its associated biological heterogeneity and lay a path towards machine learning-driven personalized approaches to treatment in depression.
Online Methods
EMBARC study
Trial Registration
Establishing Moderators and Biosignatures of Antidepressant Response for Clinical Care for Depression (EMBARC), NCT01407094
Participants and treatment
Written informed consent was obtained from each participant under institutional review board-approved protocols at each of the four study sites: University of Texas Southwestern Medical Center (TX), Massachusetts General Hospital (MG), Columbia University (CU), and University of Michigan (UM). Data reported here are based on EMBARC participants who were randomly assigned to sertraline or placebo during stage 1 of the trial (N=309 total). Key eligibility for the study included the following: being 18-65 years old; having major depression as a primary diagnosis by the Structured Clinical Interview for DSM-IV Axis I Disorders (SCID); at least moderate depression severity with a score ⩾14 on the Quick Inventory of Depressive Symptomatology-Self Report (QIDS-SR) at screening and randomization; a major depressive episode beginning before age 30; either a chronic recurrent episode (duration ⩾ 2 years) or recurrent major depressive disorder (at least 2 lifetime episodes); no antidepressant failure during the current episode. Exclusion criteria included the following: current pregnancy, breastfeeding, no use of contraception; lifetime history of psychosis or bipolar disorder; substance dependence in the past six months or substance abuse in the past two months; unstable psychiatric or general medical conditions requiring hospitalization; study medication contraindication; clinically significant laboratory abnormalities; history of epilepsy or condition requiring an anticonvulsant; electroconvulsive therapy (ECT), vagal nerve stimulation (VNS), transcranial magnetic stimulation (TMS) or other somatic treatments in the current episode; medications (including but not limited to antipsychotics and mood stabilizers); current psychotherapy; significant suicide risk; or failure to respond to any antidepressant at adequate dose and duration in the current episode.
Clinical trial
EMBARC used a double-blind design, wherein participants were randomized to an 8-week course of sertraline or placebo. Randomization was stratified by site, depression severity, and chronicity using a block randomization procedure. Sertraline dosing began at 50mg using 50mg capsules and was increased as tolerated if the patient did not respond until a maximum of 200mg. A similar dosing approach was used for placebo capsules.
Clinical outcome measure
Our primary outcome was the Hamilton Depression Rating Scale (HAMD17). For participants lacking an endpoint HAMD17, multiple imputation by chained equations was conducted in R using the package mice 50. The following observed variables were utilized in order to impute endpoint HAMD17 values for missing data via Bayesian regression: baseline HAMD17, week 1 HAMD17, week 2 HAMD17, week 3 HAMD17, week 4 HAMD17, week 6 HAMD17, baseline Quick Inventory of Depressive Symptoms (QIDS) total score, baseline Mood and Symptom Questionnaire subscale scores for Anxious Arousal, Anhedonic Depression, and General Distress, Snaith-Hamilton Pleasure Scale (SHAPS) total score, age, years of education, gender, and Wechsler Abbreviated Scale of Intelligence (WASI) t-scores for Vocabulary and Matrix Reasoning.
Resting-state EEG acquisition
Resting-state EEG (rsEEG) were recorded from each of the four study sites. The EEG amplifier settings are summarized in SupplementaryTable 1. At all study sites, amplifier calibrations were performed. Experimenters were certified by the Columbia EEG team after demonstrating accurate EEG cap placement and delivery of task instructions via video conference, and then submitting satisfactory EEG data from a pilot subject.
rsEEG were recorded during four 2-minute blocks (2 blocks for eyes-closed, and 2 blocks for eyes open) in a counterbalanced order. Participants were instructed to remain still and minimize blinks or eye movements, and to fixate on a centrally presented cross during the eyes-open condition.
Resting-state EEG preprocessing
The recorded rsEEG data were cleaned offline with our in-house fully automated artifact rejection pipeline, thereby minimizing the biases in preprocessing possible with manual rejection of artifacts. The steps are briefly described as follows: 1) The EEG data were resampled to 250 Hz; 2) The 60 Hz AC line noise artifact was removed using CleanLine (https://www.nitrc.org/projects/cleanline). 3) Non-physiological slow drifts in the EEG recordings were removed using a 0.01 Hz high-pass filter; 4) The spectrally filtered EEG data were then re-referenced to the common average; 5) Bad epochs were rejected by thresholding the magnitude of each epoch. Bad channels were rejected based on thresholding the spatial correlations among channels. Subjects with more than 20% bad channels were discarded. The rejected bad channels were then interpolated from the EEG of adjacent channels via the spherical spline interpolation; 6) Remaining artifacts were removed using independent component analysis (ICA). Independent components (ICs) related to the scalp muscle artifact, ocular artifact, ECG artifact, were automatically rejected using a pattern classifier trained on expert-labeled ICs from another independent EEG data set; 7) EEG data were re-referenced to the common average. After artifact rejection, 54 EEG channels common to all four study sites were identified and extracted for each subject. Subjects whose total powers across all the channels were beyond three standard deviations of the mean total power were discarded. Consequently, of the 266 patients with pretreatment EEG recordings, 228 had usable EEG data for analyses. The baseline sociodemographic and clinical information of these 228 patients is provided in Supplementary Table 1. The 38 patients with unusable EEG recordings were mainly had too many bad EEG channels and exceedingly large total power across channels.
Second depression study cohort (validating rsEEG antidepressant predictive signature)
Participants and treatment
The second depression study was carried out at University of Texas Southwestern Medical Center (UTSW), which is one of the four study sites in EMBARC 30. Written informed consent was obtained under institutional review board-approved protocol at UTSW. Individuals were eligible for the study if they were aged 10 years or older, and had the ability to speak, read, and understand English. To be included, participants needed to have a lifetime or current diagnosis of a mood disorder (i.e., major depressive disorder, persistent depressive disorder, bipolar I/II/NOS, bipolar/mood disorder with psychotic features, depressive disorder other specified (i.e., subthreshold)) based upon a semi-structured diagnostic interview.
Exclusion criteria included the following: history of schizophrenia, schizoaffective disorders or chronic psychotic disorders based upon a semi-structured diagnostic interview; inability to provide a stable home address and contact information; having had any condition for which, in the opinion of the investigator or designee, study participation would not be in their best interest (including but not limited to significant cognitive impairment, unstable general medical condition, intoxication, active psychosis) or that could prevent, limit, or confound the protocol-specified assessments; or requirement for immediate hospitalization for psychiatric disorder or suicidal risk as assessed by a licensed study clinician.
Screening and Baseline assessments
Whenever possible, screening and baseline assessments took place on the same day and began after informed consent was obtained. A potential participant’s eligibility was determined following review of the inclusion/exclusion criteria and assessment with the Mini-International Neuropsychiatric Interview (MINI) diagnostic interview. In addition to a number of self-report symptom measures, which are outside the scope of the current analysis, patient completed the Antidepressant Treatment Response Questionnaire (ATRQ) at the initial visit, which is a self-rated scale for determining treatment resistance in major depressive disorder, within the current depressive episode.
Once eligibility requirements were met, baseline procedures were completed over two visits depending on the needs of the individual participant. In the event of a split visit, neuroimaging and EEG procedures might be performed on separate days. This study captured a range of information including socio-demographics, general clinical data, physical exam, blood and stool samples, behavioral testing, neuroimaging and EEG, though here we focus only on the EEG data given the scope of the present study.
Resting-state EEG acquisition
EEG signals were acquired with two EEG amplifiers, each for a different portion of the participants. The first amplifier was the same 62-channel NeuroScan SynAmps amplifier (NeuroScan Inc., USA) used in EMBARC with identical acquisition parameters. The second amplifier was the Net Amps 300 amplifier with the high-density 256-channel HydroCel Geodesic Sensor Net (Electrical Geodesic Inc., USA). Cz was used as the reference electrode and the sampling rate was set at 1,000 Hz. Electrode impedances were kept below 50 kΩ. A total of 35 and 37 participants’ EEG data were collected using the first and second amplifiers, respectively. Participants were seated on a comfortable reclining chair and were instructed to remain awake and let their mind naturally wander in the eyes closed paradigm and then fixate a given point in the eyes open paradigm, each paradigm for two 2-minute blocks.
Resting-state EEG preprocessing and analysis
The recorded rsEEG data were cleaned offline with the identical fully automated artifact rejection pipeline as used in EMBARC. After artifact rejection, 54 EEG channels common to all four study sites in EMBARC were identified and extracted for each subject. Subjects whose total powers across all the channels were beyond three standard deviations of the mean total power were discarded.
Third depression study cohort (correlation between rsEEG antidepressant predictive signature and fMRI signature and with TMS/EEG response to probe stimulation)
Participants
Written informed consent was obtained from each participant under institutional review board-approved protocol at Stanford University. Participants in the study underwent several assessments during a clinical intake interview to determine eligibility and classification for the study. The diagnosis of depression (and comorbid conditions) was assessed by a clinician using the SCID, as for EMBARC. Key eligibility for the study included the following: age 18-50 years old; no current psychotherapy; free of metal or ferrous implant; good English comprehension and non-impaired intellectual abilities to ensure understanding of task instructions; no history of neurological disorders, brain surgery, electroconvulsive or radiation treatment, brain hemorrhage or tumor, stroke, epilepsy, hypo- or hyperthyroidism; no daily use of PRN benzodiazepines or opiates (max: 3x/week), or daily thyroid medications, and no antidepressant, anticonvulsant or antipsychotic medications for > 2 weeks (fluoxetine > 6 weeks). Exclusion criteria included the following: left-handed; did not graduate from an English-speaking high school and English was not their first language; psychiatric medications (including but not limited to antipsychotics and mood stabilizers) and hormonal and/or cancer medications; current psychotherapy; current repetitive transcranial magnetic stimulation (rTMS) treatment or electroconvulsive therapy treatment; loss of consciousness greater than 30 minutes and/or a loss of memory greater than 24 hours; lifetime evidence of psychosis, mania, hypomania, or bipolar disorders and/or manic episodes on the SCID; diagnosis of substance dependence within the past 3 months (but not abuse). 24 subjects for whom resting EEG, single-pulse TMS/EEG, and task fMRI data were all acquired were considered in the subsequent analyses.
Resting EEG acquisition
EEG recordings were acquired with a BrainAmp DC amplifier (sampling rate: 5 kHz; measurement range: ± 16.384 mv; cut-off frequencies of the analog high-pass and low-pass filters: 0 and 1 kHz) and the Easy EEG cap with 64 extra-flat, freely rotatable, sintered Ag-AgCl electrodes (Brain Products GmbH, Germany). The electrode montage followed an equidistant arrangement extending from below the cheekbone back to below the inion. Electrode impedances were kept below 5 kΩ. An electrode attached to the tip of the nose was used as the reference. Participants were seated on a comfortable reclining chair and were instructed to remain awake and let their mind wander in the eyes closed paradigm and then fixate a given point in the eyes open paradigm, each 3 minutes. Recordings were immediately assessed for quality using a custom MATLAB (R2014b, The Mathworks Inc., MA) script and rerun if necessary.
Resting-state EEG preprocessing
The recorded rsEEG data were cleaned offline with the identical fully automated artifact rejection pipeline as used in EMBARC.
Emotional conflict task
This well-characterized paradigm assesses both emotional conflict and emotional conflict regulation 51. Each trial involved presentation of an emotional face with either a fearful or happy expression, drawn from the set of Ekman & Friesen, with an overlaid emotion word (“FEAR” or “HAPPY”). Participants were instructed to identify the facial emotion with a key press, while trying to ignore the emotion word. The task consisted of 148 trials, with stimuli presented for 1000 milliseconds (ms) in a fast event-related design. Interstimulus intervals were 3000-5000ms in a pseudo-randomized order counterbalanced for facial expression, gender, word, and response button. Stimuli were either congruent (e.g. fearful face with “FEAR”) or incongruent (e.g. fearful face with “HAPPY”), and stimuli were furthermore balanced to achieve an equal fraction of current and prior trial congruency, while ensuring no direct stimulus repetitions. Prior to performance of the task during neuroimaging, all participants underwent a practice version to ensure task proficiency was reached (minimum 80% accuracy) and the task instructions were understood. The neuroimaging task lasted 13 minutes and 14 seconds.
Regulation in the emotional conflict task occurs via an implicit process when conflict trials are preceded by other conflict trials 51, 52. That is, while emotional conflict results in slowing of reaction times, this effect can be mitigated in incongruent trials that follow incongruent trials (iI trials), compared to incongruent trials that follow congruent trials (cI trials). This trial-to-trial adaptive regulation of emotional conflict reflects an active process by which the brain increases emotional control in response to prior trial conflict, which then benefits regulation of emotional conflict on the subsequent trial (captured by the iI-cI contrast). This regulation effect, captured through the same contrast, has also been extensively described for non-emotional conflict stimuli 53. Critically, this contrast between post-incongruent incongruent and post-congruent incongruent trial compares brain responses to physically identical stimuli (i.e. incongruent trials) that differ only on the relative emotional conflict regulatory context in which they come due to prior trial congruency, and is furthermore independent of the incongruent versus congruent trial (I-C) conflict response contrast. Neuroimaging acquisition parameters are shown in SupplementaryTable 3.
fMRI preprocessing and first-level modeling
FSL tools were used to preprocess imaging data 54. Functional images were first realigned to structural images using an affine registration matrix and boundary-based registration based upon tissue segmentation as implemented in FSL’s FLIRT, which was concatenated with a non-linear normalization of each participant’s T1 image to the Montreal Neurological Institute (MNI) 152-person 1 mm3 T1 template using FNIRT from FSL 5.0 to result in a single transformation step from individual native functional space to structurally-aligned and spatially-normalized template space. Functional images were realigned to the middle volume of the run. Nuisance signals corresponding to segmented white matter and CSF were regressed out of motion-corrected functional images. A 6 mm full-width half max (FWHM) isotropic smoothing kernel was then applied to preprocessed time series images to account for individual anatomical variability. In order to ensure the quality of imaging measures, we instituted cutoffs for absolute level of motion (root mean square of the absolute level of movement < 4mm across the mean of the squared maximum displacements in each of the 6 translational and rotational parameters estimated during realignment). In addition, in order to ensure brain activation measures reflect task-relevant metrics, we also instituted a minimum level of behavioral accuracy during completion of the emotional conflict task as an additional quality control metric (total accuracy ⩾ 70% of trials correct). Functional runs displaying motion higher than our cutoff OR accuracy below the minimum cutoff were excluded from further analyses.
For individual-level analyses for each participant, regressors modeling trials of interest were convolved with the hemodynamic response function. First-level general linear models were estimated in SPM 8 55. Regressors corresponded to zero-duration markers set at the onset of stimuli, which were explicitly categorized by congruency (Incongruent or Congruent) and prior trial type (Post-incongruent or Post-congruent) in order to model conflict response and regulation effects. This resulted in 4 different trial types in total, in addition to nuisance regressors for error trials and post-error trials (when applicable) and six motion parameters.
Single-pulse TMS/EEG acquisition
Following an anatomical MRI (T1-weghted, 3T) to determine MRI-guided single-pulse TMS (spTMS) targets, subjects received spTMS using a Cool-B B6 5 butterfly coil and a MagPro X100 TMS stimulator (MagVenture, Denmark). Stimulations were delivered to middle primary visual cortex (V1), bilateral primary motor cortices (M1), bilateral dorsolateral prefrontal cortices (pDLPFC), and bilateral anterior DLPFC (aDLPFC) in a randomized order for each subject. Among these stimulation sites, M1 was defined as the hand knob in the standard Montreal Neurological Institute (MNI) space (MNI coordinate: (−38, −18, 64) for left M1 and (40, −18, 64) for right M1). V1 was defined by its MNI anatomical target (MNI coordinate: (0, −100, 2)). For pDLPFC and aDLPFC, the stimulation sites were targeted based on the location of the frontoparietal (executive) control network and ventral attention (salience) network in separate resting-state fMRI data (MNI coordinate: (−32, 42, 34) for left aDLPFC, (30, 50, 26) for right aDLPFC, (−38, 22, 38) for left pDLPFC, and (46, 26, 38) for right pDLPFC). Following our previous work, these targets were established using a group ICA on a separate cohort of 38 participants, with the pDLPFC and aDLPFC targets representing peak voxels within the middle frontal clusters of these two networks 22, 32. Coordinates for the pDLPFC and aDLPFC stimulation targets were then transformed to individual subject native space using non-linear spatial normalization with FSL (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki) and used for TMS targeting. The resting motor threshold (rMT) was determined as the minimum stimulation intensity that produced visible finger movement of the right hand at least 50% of the times when the subject’s left M1 is stimulated. TMS coil placement was guided by Visor2 LT 3D neuro-navigation system (ANT Neuro, Netherlands) based on co-registration of the functionally defined target to each participant’s structural MRI (T1 weighted, slice distance 1mm, slice thickness 1mm, sagittal orientation, acquisition matrix 256x256) acquired with a 3T GE DISCOVERY MR750 scanner. The TMS coil was placed in a posterior to anterior direction, with an angle of 45 degrees to the nasion-inion axis (studying the optimal coil angles is beyond the scope of this paper). Each target site was stimulated with 60 pulses (biphasic TMS pulses, 280 μs pulse width, 120% rMT, 1500 ms recharge delay), interleaved at a random interval of 3 s ± 300 ms. A thin foam pad was attached to the surface of the TMS coil to decrease electrode movement. The subjects were instructed to relax and to fixate at a cross located on the opposing wall while stimulations were administered by a research assistant.
The same TMS-compatible 64-channel BrainAmp DC amplifier as for rsEEG recordings was used to record spTMS/EEG data. Electrode impedances were kept below 5 kΩ. An electrode attached to the tip of the nose was used as the reference. DC correction was manually triggered at the end of the stimulations at each site to prevent the saturation of the amplifier due to the DC drift.
Single-pulse TMS/EEG preprocessing
The recorded spTMS/EEG data were cleaned offline with ARTIST, which is a fully automated artifact rejection algorithm for spTMS/EEG 56: 1) The initial 10 ms data segment following TMS pulses were discarded to remove the large stimulation-induced electric artifact. 2) The EEG data were downsampled to 1 kHz. 3) Big decay artifacts were automatically removed using ICA based on thresholding. 4) The 60 Hz AC line noise artifact was removed by a notch filter. 5) Non-physiological slow drifts in the EEG recordings were removed using a 0.01 Hz high-pass filter, and high-frequency noise was removed by using a 100 Hz low-pass filter. 6) The spectrally filtered EEG data was then re-referenced to the common average and epoched with respect to the TMS pulse (−500 ~ 1500 ms). 7) Bad trials were rejected by thresholding the magnitude of each trial. Bad channels were rejected based on the spatial correlations among channels. The rejected bad channels were then interpolated from the EEG of adjacent channels. 8) Remaining artifacts were automatically removed using ICA. ICs related to the scalp muscle artifact, ocular artifact, ECG artifact, were rejected using a pattern classifier trained on expert-labeled ICs from other TMS/EEG data sets.
Fourth depression study cohort (prediction of outcome with rTMS treatment)
Participants
This study was a naturalistic open-label clinical study, and has been previously reported in greater detail elsewhere 33. Briefly, patients were drawn from three outpatient mental health care clinics in the Netherlands (neuroCare Clinic Nijmegen, neuroCare Clinic The Hague, and Psychologenpraktijk Timmers Oosterhout) between May 2007 and November 2016. Inclusion criteria included: 1) a primary diagnosis of non-psychotic major depressive disorder or dysthymia, 2) Beck Depression Inventory, second edition, Dutch version (BDI-II-NL)≥14 at baseline, 3) treatment with at least 10 sessions of rTMS over the DLPFC or response within these 10 sessions. All participants signed an informed consent under an approved IRB protocol. Additional exclusion criteria included: previous ECT treatment, epilepsy, traumatic brain injury, current psychotic disorder, wearing a cardiac pacemaker or metal parts in the head, and current pregnancy.
Treatment
All patients were treated with either a high frequency (10 Hz) protocol over the left DLPFC or a low frequency (1 Hz) protocol over the right DLPFC, or both sequentially. The rTMS data included a long time-span and the rTMS protocol applied was never based on clinical symptomatology. In the beginning (2006-2012) the standard protocol applied was 10 Hz left DLPFC rTMS, and only in some cases 1 Hz right DLPFC rTMS was applied (when there were concerns for safety e.g. paroxysmal activity, seizure risk) due to 1 Hz rTMS being considered a safer protocol. On first inspection of those data 28, it was found that the clinical benefits for 10 Hz and 1 Hz were indistinguishable, after which time period the standard protocol became 1 Hz right DLPFC 33. The analyses reported here focus only on patients that received only 10 Hz or 1 Hz rTMS as too few data sets were available on patients who received both treatments or switched treatments mid-way. There were 73 patients in the 10 Hz arm, of which 64 had high quality EEG data, while in the 1 Hz arm there were 104 patients, of which 88 had high quality EEG data. Selection of the treatment protocol was not done in a randomized manner, but rather in the context of clinical care, and thus each arm is analyzed separately. rTMS was performed using a Magstim Rapid2 (Magstim Company, Spring Gardens, UK) or a Deymed DuoMag XT-100 stimulator with a Fig.-of-8 coil, 70 mm diameter. For the 10 Hz protocol, rTMS was administered at 10 Hz over the left DLPFC, 110-120% of the resting motor threshold (MT), 30 trains of 5s duration, inter-train interval (ITI) 30s, 1500 pulses per session. The 1Hz protocol consisted of rTMS at 1 Hz over the right DLPFC, 110-120% MT, 120 trains of 10s duration, ITI 1s, 1200 pulses per session. In case of both protocols, the LF protocol was administered first with a shorter duration of 1000 pulses per session and subsequently the HF protocol at full length. The DLPFC was localized using either the 5-cm rule or the Beam F3/F4 method. Furthermore, rTMS treatment was complemented with cognitive behavioral psychotherapy by a trained psychologist. Psychotherapy was performed concurrent with the rTMS treatment in 45-minute sessions (the rTMS lasting 20 minutes). Sessions took place with a minimum frequency of two to three times per week and a maximum frequency of two per day, as per the patient’s availability.
As these data were drawn from naturalistic clinical care, the total number of sessions depended on clinical decisions, and thus varied across patients. Decisions to continue treatment were based on response to treatment, clinical evaluation of symptom severity, and the patient's own request. Decisions followed several rules: if a BDI decrease was observed of at least 20% from baseline 10 sessions, the treatment was continued, and re-evaluated every five sessions. If no response occurred by session 20-25, treatment was recommended to be terminated unless the patient requested to extend it. If BDI scores reached 12 or below for five sessions, which indicated remission, the patient was given the option of ending or tapering treatment, with an option to extend into maintenance sessions (one session every 6-8 weeks). However, if the threshold of BDI=12 was reached, but symptom improvement continued, treatment was continued until BDI scores ceased improving.
Clinical outcome measures
Clinical outcome was assessed on the BDI (which was the primary outcome measure for the decision rules above) as well as the Depression, Anxiety and Stress Scale (DASS) 57. The DASS is a self-report questionnaire and consists of three subscales: depression (DASSD), anxiety (DASSA), and stress (DASSS). Each scale consists of 14 items with a 4-point severity score, with a maximum total score of 42 on each scale. The patient is asked to fill in the items based on experiences in the previous week.
Resting EEG acquisition
EEG data were acquired from 26 channels according to the 10–20 electrode international system (Quickcap; NuAmps). Data were referenced to averaged mastoids with a ground at Fpz. The sampling rate of all channels was 500 Hz. A low pass filter with attenuation of 40dB per decade above 100Hz was employed prior to digitization. Subjects were asked to rest quietly with their eyes open and eyes closed for 3 minutes each.
Preprocessing of resting-state EEG and clinical outcome metrics
The recorded rsEEG data were cleaned offline with the identical fully automated artifact rejection pipeline as used in EMBARC. Missing data in the clinical metrics were imputed in the same manner as in the EMBARC data, separately by treatment arm.
Machine learning analysis
Sparse EEG latent space regression
We developed a novel end-to-end machine learning algorithm for predicting the treatment outcome from the baseline resting EEG. This new algorithm, referred to as Sparse EEG Latent SpacE Regression (SELSER), optimizes a latent space model that maps the resting EEG data to the treatment outcome by minimizing the prediction error, subject to a constraint on the dimensionality of the latent signals. The band powers in each of the four canonical EEG frequency bands (theta: 4-7 Hz, alpha: 8-12 Hz, beta: 13-30 Hz, gamma: 31-50 Hz; Filtered using zero-phase FIR filters) are employed as the features. Due to the volume conduction, these band power features are best captured in a latent space rather than in the sensor space. For this purpose, SELSER optimizes a set of spatial filters to linearly transform the multi-channel EEG signals in the sensor space to low-dimensional latent signals. A linear regression model is then built to relate the band powers of the latent signals to the treatment outcome.
More formally, SELSER models the treatment outcome yi for the i-th subject as follows (i = 1, …, M):
| (1) |
where denotes the filtered EEG data for the i-th subject, C is the number of channels, and N is the number of sampled time points. denotes the predicted treatment outcome for the i-th subject. is the k-th spatial filter (k = 1, …, L), βk is the k-th weight coefficient of a linear regression model, and b is the intercept of the linear regression model. As can be seen in Supplementary Fig. 10a, prediction is carried out in three phases: (1) The multi-channel EEG signals is transformed to L latent signals via L spatial filters . (2) The band powers of the L latent signals are calculated: . (3) A linear regression model is used to combine the band powers of the latent signals to predict the treatment outcome: . It is expected that each latent signal captures a certain portion of information predictive of the treatment outcome, quantitated by the band power of the rhythmic EEG activity.
Unlike conventional approaches where the unknown parameters in the spatial filters and regression model, namely , were optimized separately under distinct objective functions which may or may not be directly associated with the treatment outcome, we proposed a computationally efficient algorithm (see next section) for optimizing all the model parameters by directly minimizing the mean-squared prediction error while preventing L from getting too large to guard against overfitting.
Parameter optimization in SELSER
Let denote the EEG spatial covariance matrix. The predicted treatment outcome can be represented alternatively as the following 58:
| (2) |
where is a symmetric matrix. Tr(·) stands for the trace operator, which takes the sum of the diagonal elements of a matrix. Suppose the spatial filters are orthogonal to each other, then and are the eigenvectors and eigenvalues of W, respectively. As a result, optimizing and amounts to optimizing W, after which and can be obtained by performing eigendecomposition of W.
However, the number of unknown parameters in W, C(C + 1)/2 is typically much larger than the number of training samples, hence simply minimizing the prediction error is prone to model overfitting. To address this issue, in addition to minimizing the prediction error, we added the rank of W, which is equal to L, as a penalty term into the objective to limit the dimensionality of the latent signals, yielding the following optimization problem:
| (3) |
where ∥W∥0 denotes the rank of W. However, Problem (3) is NP-hard and the rank penalty is non-smooth. Alternatively, the following nuclear norm has been widely used as a convex surrogate of the rank of matrices in a wide range of applications in signal processing and machine learning 58-60:
| (4) |
where are the singular values of W. Consequently, replacing ∥W∥0 with ∥W∥*, yields
| (5) |
Problem (5) is a convex optimization problem since the objective function is a convex function of W and b. The global minimum solution can be obtained with the accelerated proximal gradient method 61.
To remove the inter-subject variability due to the overall power variation, for each subject Ci is normalized by dividing its trace prior to the SELSER analysis: Ci = Ci/Tr(Ci).
Relevance Vector Machine
To compare with SELSER, Relevance Vector Machine (RVM) 24 with the linear kernel was used to build sparse linear regression models for treatment prediction from non-SELSER-optimized features. By leveraging a sparse prior to penalize overly complex models under the sparse Bayesian learning framework, RVM is able to automatically select relevant features for prediction via marginal likelihood maximization. Hence, RVM obviates the need of additional validation data to determine the hyperparameters.
Treatment prediction using SELSER
We applied SELSER to each canonical EEG frequency band to predict the treatment outcome. In order to increase the sample size, each of the two blocks of EEG in each resting-state condition (eyes open or eyes closed) was treated as a separate sample for training the model. However, during the leave-subject-out cross-validation, the two blocks from each subject were always grouped together such that if one block was included in the training set, the other was included as well. The predicted outcome of each subject was the average of the predicted outcomes of the two EEG blocks from the subject.
Visualizing spatial maps of latent signals
Each latent signal has a spatial map that could be visualized on both the scalp and cortical surface to facilitate neurophysiological interpretation. The scalp spatial maps could be calculated as follows 18:
| (11) |
where contains the scalp spatial maps of the latent signals as columns, is the mean spatial covariance matrix of the EEG signals across subjects, , and is the mean covariance matrix of the latent signals across subjects.
In order to obtain the cortical spatial maps of the latent signals, a three-layer (scalp, skull, and cortical surface) boundary element head model was computed with the OpenMEEG 62 based on the FreeSurfer average brain template 63. A total of M (M = 15,002) fixed-orientation dipoles whose orientations were normal to the cortical surface were generated. The lead-field matrix relating the dipole activities to the EEG was obtained as a result of the boundary element modeling. A linear inverse matrix that maps the EEG signals to the cortical sources was then computed via the sLORETA algorithm 64. The cortical spatial maps could subsequently be calculated as follows:
| (12) |
where contains the cortical spatial maps of the latent signals as columns.
The spatial maps used for visualization were obtained by training SELSER on the entire EMBARC sample.
Treatment prediction using the channel-level alpha band power or theta cordance features
To contrast with prediction approaches based on channel-level measures, we trained the RVM using the channel-level alpha band power (Supplementary Fig. 10c) and theta cordance features, respectively. Cordance is a quantitative EEG (QEEG) measure that has been implicated as a predictive biomarker for antidepressant treatment 13. In particular, it was reported that lower prefrontal theta cordance during the placebo lead-in phase predicted better antidepressant (fluoxetine, which is an SSRI, and venlafaxine, which is an SNRI) treatment outcome 65. Theta cordance was calculated for each participant via the following steps: 1) EEG power re-attribution: Absolute re-attributed theta power of each electrode was calculated as the average of the theta band (4-7Hz) power of all bipolar neighboring electrode pairs that share that electrode, and absolute re-attributed total power of each electrode was calculated as the average of the total (1-50Hz) power of all bipolar neighboring electrode pairs that share that electrode. Relative re-attributed power of each electrode was calculated as the absolute re-attributed theta power divided by the average of the absolute re-attributed total power. 2) Spatial normalization: The absolute and relative re-attributed theta power values were each normalized by their average across channels. 3) Combination of absolute and relative power: Theta cordance was calculated as the sum of the spatially normalized absolute and relative re-attributed theta power.
Treatment prediction using band power features of the latent signals extracted with independent component analysis or principal component analysis
SELSER was also benchmarked against prediction approaches using band power features of the latent signals estimated by Independent Component Analysis (ICA) and Principal Component Analysis (PCA), respectively. ICA and PCA are widely used unsupervised approaches for estimating latent signals from EEG signals 66, 67, based on different statistical criteria - statistical independence is maximized among latent signals in ICA, while variances of the latent signals are maximized in PCA. Over the years, a multitude of ICA algorithms have been developed as statistical independence could be quantified in a variety of ways 68. In this work, we used the information maximization (Infomax) algorithm 20 for performing ICA.
To align with the SELSER analyses, the prediction framework based on ICA/PCA followed the same workflow as in SELSER (Supplementary Fig. 10b). Each subject’s EEG signal was first normalized by dividing the square root of its total power across channels. The latent signals were then estimated by applying ICA or PCA to temporally concatenated EEG signals across subjects. After that, the band powers of the full set of latent signals were computed as the features, followed by an RVM with the linear kernel that related the band power features to the HAMD17 score changes.
Performance evaluation using cross-validation
Stratified 10-fold leave-subject-out cross-validation 69 (Supplementary Fig. 2) was employed to assess the predictive performance of each prediction approach. More specifically, the data were randomly partitioned into 10 subsets, such that each subset containing an approximately equal number of subjects from each of the four study sites. A subset was left out as the test data, and the remaining 9 subsets were used as the training data. For SELSER, the regularization parameter λ was determined using an inner 10-fold cross-validation on the training data. The process was then repeated 10 times, where each of the 10 subsets was used exactly once as the test data. As a result, each subject had a predicted HAMD17 score change. To enhance the stability of the prediction, the data were randomized 10 times and the stratified 10-fold cross-validation was run on each randomized data. The median of the resulting 10 predicted HAMD17 score changes of each subject was used as the final prediction. The prediction performance was then quantified by the Pearson’s correlation coefficient and root mean square error (RMSE) between the cross-validated prediction of the HAMD17 score change and the true HAMD17 score change. The P value for the one-tailed alternative hypothesis that the Pearson’s correlation coefficient was greater than 0 was also reported.
Specificity of the prediction was tested by applying, at each fold of stratified 10x10 cross-validation, the prediction model to the data from the other treatment arm, which was summarized for each participant by taking the median of the 100 folds of cross-validation.
Statistical test
A non-parametric permutation test was used to assess the statistical significance of the treatment prediction results. The observed HAMD17 score changes were randomly shuffled across subjects 1000 times. Each time the cross-validated prediction procedure was repeated, resulting in a distribution of the Pearson’s correlation coefficient. The P value was then defined as the proportion of cross-validated correlation coefficients that were greater than the cross-validated correlation coefficient without permutation.
Application of machine learning models to independent major depressive disorder data
Calculating rsEEG predictions in the second major depressive disorder study
We applied the result of the alpha SELSER model trained on the sertraline arm of the EMBARC sample to data from the 2nd major depressive disorder study in which rsEEG data were collected from 72 patients with depression who were assessed in cross-sectionally at the baseline visit. Since 37 patients’ EEG data were recorded with a new amplifier (i.e., the EGI Net Amps 300 amplifier) distinct from those used in EMBARC, the mean-removal site correction procedure was performed on both the EMBARC data to train the model, and the 37 patients’ EEG data in the 2nd study, as in the leave-study-site-out analysis. The rsEEG were fed into Equation (1) to yield predictions with the SELSER model trained at each fold of cross-validation in EMBARC. The prediction was summarized for each participant by taking the median of the predictions from the 100 folds of cross-validation. This yielded a measure of the EMBARC SELSER model expression strength for each individual in the second major depressive disorder study, expressed as a predicted HAMD17 change for each patient. Given the focus in the present study on testing the generalizability of the EMBARC sertraline-predictive signature in an independent data set, we compared the SELSER model-predicted HAMD17 change scores between patients with a treatment-resistant profile on the ATRQ (failed two or more medications in the current episode) versus those who showed a partial response to antidepressant treatment within-episode.
Calculating rsEEG predictions in the third MDD study
We applied the result of the alpha SELSER model trained on the sertraline arm of the EMBARC sample to data from the 3rd major depressive disorder study in which rsEEG data were collected from 24 patients with depression who were assessed in a cross-sectional manner (i.e. without treatment). Since the EEG data were recorded with yet another amplifier distinct from those used in EMBARC, the mean-removal site correction procedure was performed, as in the leave-study-site-out analysis. Due to the difference of electrode montages between this study and EMBARC, the SELSER model trained on EMBARC cannot be directly applied to this study. To address this issue, we source localized the rsEEG from the study based on the linear inverse matrix obtained similarly as in EMBARC, and then mapped the source activity to the EMBARC electrodes via the lead-field matrix from EMBARC. Next, the mapped rsEEG at EMBARC electrodes were fed into Equation (1) to yield predictions with the SELSER model trained at each fold of cross-validation in EMBARC. The prediction was summarized for each participant by taking the median of the predictions from the 100 folds of cross-validation.
Calculating task fMRI predictions in the third major depressive disorder study
We also applied a previously-described RVM model trained on emotional conflict task fMRI data from EMBARC to data from the 3rd major depressive disorder study 22. Development of the model on EMBARC data is described in brief below. Extractions were conducted on cortical regions of interest (ROIs), defined based on a recently-published cortical parcellation derived from applying a combination of local gradient analysis and global signal similarity on an independent resting-state fMRI cohort 70. Since functional parcellations typically rely on resting-state connectivity patterns, which may or may not adequately describe activity patterns in the emotional conflict task, we pooled ROIs from the 200, 400, and 600 region parcellations in order to limit parcellation-related specificity. ROIs were mapped to seven previously identified functional networks based on the spatial overlap between each ROI and each network 70. In addition to these cortical ROIs, subcortical ROIs included striatal and cerebellar parcellations based on the same seven functional networks, amygdala ROIs, anterior and posterior hippocampal ROIs and the thalamus. We then regressed imaging site out of these data using multiple linear regression within the training set at each run of the RVM model, and the residualized brain signals were then used for predicting the HAMD17 score change with the RVM model trained at each fold of cross-validation in EMBARC. The prediction was summarized for each participant by taking the median of the predictions from the 100 folds of cross-validation. fMRI data from our third major depressive disorder study were preprocessed in the same manner, and the EMBARC-derived weight vector was applied to the extracted ROI data to determine each participant’s strength of expression of the EMBARC fMRI RVM model.
Correlating spTMS/EEG with rsEEG predictions in the third major depressive disorder study
To quantify in the 3rd major depressive disorder study the correlation between the spTMS/EEG responses and the EMBARC-defined rsEEG phenotype, we employed SELSER to learn predictive models from the spTMS/EEG data to the rsEEG predictions, and calculated the leave-one-out cross-validated Pearson’s correlation coefficients between the predicted rsEEG predictions and true rsEEG predictions. The SELSER analysis was performed separately for the seven stimulation sites (bilateral pDLPFC, bilateral aDLPFC, bilateral M1, and V1), the same set of frequency bands as used in the rsEEG prediction analysis (theta, alpha, beta, and gamma), and for three time windows relative to the TMS pulse (0 – 200ms, 200 – 400ms, 400 – 600ms). For each SELSER analysis, the spTMS/EEG data were concatenated across trials. Significance was evaluated after correcting for the false discovery rate (p < 0.05) across all SELSER models (i.e. encompassing stimulation sites x frequency bins x time windows).
Testing the relationship between rsEEG predictions and treatment outcome in the fourth major depressive disorder study
We computed each patient’s expression of the EMBARC-trained SELSER rsEEG model (expressed as predicted HAMD17 change) using the same mean site removal procedure as above. We then conducted linear mixed models (SPSS version 25, IBM Corporation) between the SELSER rsEEG-generated predicted HAMD17 change and outcome on the BDI as well as each of the DASS subscales, separately by rTMS protocol. Terms were time, predicted HAMD17 change and predicted HAMD17 change x time, using a random intercept and fixed slope. A Bonferroni correction for eight comparisons (two stimulation frequencies, four outcome measures) was then conducted on the predicted HAMD17 change x time results.
Life Sciences Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
Code for SELSER is available as Supplementary Code for non-commercial use only at xxxxxxxx. For commercial use, please contact Alto Neuroscience at info@altoneuroscience.com.
Data availability
The EMBARC data are publicly available through the NIMH Data Archive (NDAR) (https://nda.nih.gov/edit_collection.html?id=2199).
Supplementary Material
Acknowledgments:
We thank Corey J. Keller and Sanggyun Kim. The EMBARC study was supported by the National Institute of Mental Health of the National Institutes of Health under award numbers U01MH092221 (Trivedi, M.H.) and U01MH092250 (McGrath, P.J., Parsey, R.V., Weissman, M.M.). Data from the second depressed cohort were acquired under R01MH103324 (Etkin, A) and Big Idea in Neuroscience research funds from the Stanford Neurosciences Institute (Etkin A). This work was also funded in part by the Hersh Foundation (Trivedi MH PI). Etkin, A and Wu, W were additionally funded by NIH grant DP1 MH116506. Wu, W was also funded by National Key Research and Development Plan of China (No. 2017YFB1002505) and National Natural Science Foundation of China (No. 61876063 and No. 61836003).
Footnotes
Competing interest: Dr. Etkin (lifetime disclosure) receives salary and equity from Alto Neuroscience since November 1, 2019, to which the pending patent for SELSER has been licensed from Stanford. He holds equity in Mindstrong Health, Akili Interactive and Sizung for unrelated work, has received research funding from the National Institute of Mental Health, Department of Veterans Affairs, Cohen Veterans Bioscience, Brain and Behavior Research Foundation, Dana Foundation, Brain Resource Inc, and the Stanford Neurosciences Institute, and consulted for Cervel, Takaeda, Posit Science, Acadia, Otsuka, Lundbeck and Janssen. Over the past three years, Dr. Pizzagalli has received consulting fees from Alkermes, BlackThorn Therapeutics, Boehreinger Ingelheim, Posit Science, and Takeda Pharmaceuticals. He has received funding from NIMH, the Dana Foundation, and Brain and Behavior Research Foundation. Dr. Deckersbach’s research has been funded by NIH, NIMH, NARSAD, TSA, IOCDF, Tufts University, DBDAT and Otsuka Pharmaceuticals, he has received honoraria, consultation fees and/or royalties from the MGH Psychiatry Academy, BrainCells Inc., Clintara, LLC, Inc., Systems Research and Applications Corporation, Boston University, the Catalan Agency for Health Technology Assessment and Research, the National Association of Social Workers Massachusetts, the Massachusetts Medical Society, Tufts University, NIDA, NIMH, Oxford University Press, Guilford Press, and Rutledge. He has also participated in research funded by DARPA, NIH, NIA, AHRQ, PCORI, Janssen Pharmaceuticals, The Forest Research Institute, Shire Development Inc., Medtronic, Cyberonics, Northstar, and Takeda. Dr. Pat McGrath has received funding from the National Institute of Mental Health, New York State Department of Mental Hygiene, Research Foundation for Mental Hygiene (New York State), Forest Research Laboratories, Sunovion Pharmaceuticals, and Naurex Pharmaceuticals (now Allergan). In the past two years, Dr. Myrna Weissman received funding from the National Institute of Mental Health (NIMH), the National Institute on Drug Abuse (NIDA), the National Alliance for Research on Schizophrenia and Depression (NARSAD), the Sackler Foundation, the Templeton Foundation; and receives royalties from the Oxford University Press, Perseus Press, the American Psychiatric Association Press, and MultiHealth Systems. Dr. Fava has received research support from Abbot Laboratories; Alkermes, Inc.; American Cyanamid; Aspect Medical Systems; AstraZeneca; Avanir Pharmaceuticals; BioResearch; BrainCells Inc.; Bristol-Myers Squibb; CeNeRx BioPharma; Cephalon; Clintara, LLC; Cerecor; Covance; Covidien; Eli Lilly and Company; EnVivo Pharmaceuticals, Inc.; Euthymics Bioscience, Inc.; Forest Pharmaceuticals, Inc.; Ganeden Biotech, Inc.; GlaxoSmithKline; Harvard Clinical Research Institute; Hoffman-LaRoche; Icon Clinical Research; i3 Innovus/Ingenix; Janssen R&D, LLC; Jed Foundation; Johnson & Johnson Pharmaceutical Research & Development; Lichtwer Pharma GmbH; Lorex Pharmaceuticals; Lundbeck Inc.; MedAvante; Methylation Sciences Inc.; National Alliance for Research on Schizophrenia & Depression (NARSAD); National Center for Complementary and Alternative Medicine (NCCAM); National Institute of Drug Abuse (NIDA); National Institute of Mental Health (NIMH); Neuralstem, Inc.; Novartis AG; Organon Pharmaceuticals; PamLab, LLC.; Pfizer Inc.; Pharmacia-Upjohn; Pharmaceutical Research Associates., Inc.; Pharmavite® LLC; PharmoRx Therapeutics; Photothera; Reckitt Benckiser; Roche Pharmaceuticals; RCT Logic, LLC (formerly Clinical Trials Solutions, LLC); Sanofi-Aventis US LLC; Shire; Solvay Pharmaceuticals, Inc.; Stanley Medical Research Institute (SMRI); Synthelabo; Tal Medical; Wyeth-Ayerst Laboratories; he has served as advisor or consultant to Abbott Laboratories; Acadia; Affectis Pharmaceuticals AG; Alkermes, Inc.; Amarin Pharma Inc.; Aspect Medical Systems; AstraZeneca; Auspex Pharmaceuticals; Avanir Pharmaceuticals; AXSOME Therapeutics; Bayer AG; Best Practice Project Management, Inc.; Biogen; BioMarin Pharmaceuticals, Inc.; Biovail Corporation; BrainCells Inc; Bristol- Myers Squibb; CeNeRx BioPharma; Cephalon, Inc.; Cerecor; CNS Response, Inc.; Compellis Pharmaceuticals; Cypress Pharmaceutical, Inc.; DiagnoSearch Life Sciences (P) Ltd.; Dinippon Sumitomo Pharma Co. Inc.; Dov Pharmaceuticals, Inc.; Edgemont Pharmaceuticals, Inc.; Eisai Inc.; Eli Lilly and Company; EnVivo Pharmaceuticals, Inc.; ePharmaSolutions; EPIX Pharmaceuticals, Inc.; Euthymics Bioscience, Inc.; Fabre-Kramer Pharmaceuticals, Inc.; Forest Pharmaceuticals, Inc.; Forum Pharmaceuticals; GenOmind, LLC; GlaxoSmithKline; Grunenthal GmbH; i3 Innovus/Ingenis; Intracellular; Janssen Pharmaceutica; Jazz Pharmaceuticals, Inc.; Johnson & Johnson Pharmaceutical Research & Development, LLC; Knoll Pharmaceuticals Corp.; Labopharm Inc.; Lorex Pharmaceuticals; Lundbeck Inc.; MedAvante, Inc.; Merck & Co., Inc.; MSI Methylation Sciences, Inc.; Naurex, Inc.; Nestle Health Sciences; Neuralstem, Inc.; Neuronetics, Inc.; NextWave Pharmaceuticals; Novartis AG; Nutrition 21; Orexigen Therapeutics, Inc.; Organon Pharmaceuticals; Osmotica; Otsuka Pharmaceuticals; Pamlab, LLC.; Pfizer Inc.; PharmaStar; Pharmavite® LLC.; PharmoRx Therapeutics; Precision Human Biolaboratory; Prexa Pharmaceuticals, Inc.; Puretech Ventures; PsychoGenics; Psylin Neurosciences, Inc.; RCT Logic, LLC Formerly Clinical Trials Solutions, LLC; Rexahn Pharmaceuticals, Inc.; Ridge Diagnostics, Inc.; Roche; Sanofi-Aventis US LLC.; Sepracor Inc.; Servier Laboratories; Schering-Plough Corporation; Solvay Pharmaceuticals, Inc.; Somaxon Pharmaceuticals, Inc.; Somerset Pharmaceuticals, Inc.; Sunovion Pharmaceuticals; Supernus Pharmaceuticals, Inc.; Synthelabo; Taisho Pharmaceutical; Takeda Pharmaceutical Company Limited; Tal Medical, Inc.; Tetragenex Pharmaceuticals, Inc.; TransForm Pharmaceuticals, Inc.; Transcept Pharmaceuticals, Inc.; Vanda Pharmaceuticals, Inc.; VistaGen; he has received speaking or publishing fees from Adamed, Co; Advanced Meeting Partners; American Psychiatric Association; American Society of Clinical Psychopharmacology; AstraZeneca; Belvoir Media Group; Boehringer Ingelheim GmbH; Bristol-Myers Squibb; Cephalon, Inc.; CME Institute/Physicians Postgraduate Press, Inc.; Eli Lilly and Company; Forest Pharmaceuticals, Inc.; GlaxoSmithKline; Imedex, LLC; MGH Psychiatry Academy/Primedia; MGH Psychiatry Academy/Reed Elsevier; Novartis AG; Organon Pharmaceuticals; Pfizer Inc.; PharmaStar; United BioSource, Corp.; Wyeth-Ayerst Laboratories; he has equity holdings in Compellis and PsyBrain, Inc.; he has a patent for Sequential Parallel Comparison Design (SPCD), which are licensed by MGH to Pharmaceutical Product Development, LLC (PPD); and patent application for a combination of Ketamine plus Scopolamine in Major Depressive Disorder (MDD), licensed by MGH to Biohaven; and he receives copyright royalties for the MGH Cognitive & Physical Functioning Questionnaire (CPFQ), Sexual Functioning Inventory (SFI), Antidepressant Treatment Response Questionnaire (ATRQ), Discontinuation-Emergent Signs & Symptoms (DESS), Symptoms of Depression Questionnaire (SDQ), and SAFER; Lippincott, Williams & Wilkins; Wolkers Kluwer; World Scientific Publishing Co. Pte.Ltd. Dr. Trivedi, is or has been an advisor/consultant and received fee from (lifetime disclosure): Abbott Laboratories, Inc., Abdi Ibrahim, Akzo (Organon Pharmaceuticals Inc.), Alkermes, AstraZeneca, Axon Advisors, Bristol-Myers Squibb Company, Cephalon, Inc., Cerecor, CME Institute of Physicians, Concert Pharmaceuticals, Inc., Eli Lilly & Company, Evotec, Fabre Kramer Pharmaceuticals, Inc., Forest Pharmaceuticals, GlaxoSmithKline, Janssen Global Services, LLC, Janssen Pharmaceutica Products, LP, Johnson & Johnson PRD, Libby, Lundbeck, Meade Johnson, MedAvante, Medtronic, Merck, Mitsubishi Tanabe Pharma Development America, Inc., Naurex, Neuronetics, Otsuka Pharmaceuticals, Pamlab, Parke-Davis Pharmaceuticals, Inc., Pfizer Inc., PgxHealth, Phoenix Marketing Solutions, Rexahn Pharmaceuticals, Ridge Diagnostics, Roche Products Ltd., Sepracor, SHIRE Development, Sierra, SK Life and Science, Sunovion, Takeda, Tal Medical/Puretech Venture, Targacept, Transcept, VantagePoint, Vivus, and Wyeth-Ayerst Laboratories. In addition, he has received grants/research support from: Agency for Healthcare Research and Quality (AHRQ), Cyberonics, Inc., National Alliance for Research in Schizophrenia and Depression, National Institute of Mental Health and National Institute on Drug Abuse. Dr. Trombello currently owns stock in Merck and Gilead Sciences and within the past 36 months previously owned stock in Johnson & Johnson. Dr. Arns holds options from Brain Resource (Sydney, Australia); is director and owner of Research Institute Brainclinics, equity in neuroCare Group (Munich, Germany), and a coinventor on 4 patent applications (A61B5/0402; US2007/0299323, A1; WO2010/139361 A1) related to EEG, neuromodulation and psychophysiology, but does not own these nor receives any proceeds related to these patents; Research Institute Brainclinics funding from Brain Resource (Sydney, Australia) and neuroCare Group (Munich, Germany), and equipment support from Deymed, neuroConn and Magventure. Drs. Wu, Zhang, Jiang, Fonzo, Cooper, Chin-Fatt, Jha, Krepel and Adams report no competing interests.
References
- 1.Drysdale AT et al. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nature medicine 23, 28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cipriani A et al. Comparative efficacy and acceptability of 21 antidepressant drugs for the acute treatment of adults with major depressive disorder: a systematic review and network meta-analysis. Lancet (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fournier JC et al. Antidepressant drug effects and depression severity: a patient-level meta-analysis. JAMA 303, 47–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan A & Brown WA Antidepressants versus placebo in major depression: an overview. World Psychiatry 14, 294–300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirsch I The Emperor’s New Drugs: Exploding the Antidepressant Myth. (Random House, 2009). [Google Scholar]
- 6.Kirsch I et al. Initial severity and antidepressant benefits: a meta-analysis of data submitted to the Food and Drug Administration. PLoS Med 5, e45 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wade EC & Iosifescu DV Using electroencephalography for treatment guidance in major depressive disorder. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging 1, 411–422 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Widge AS et al. Electroencephalographic Biomarkers for Treatment Response Prediction in Major Depressive Illness: A Meta-Analysis. American Journal of Psychiatry, appi. ajp. 2018.17121358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olbrich S & Arns M EEG biomarkers in major depressive disorder: discriminative power and prediction of treatment response. International Review of Psychiatry 25, 604–618 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Jaworska N, de la Salle S, Ibrahim M-H, Blier P & Knott V Leveraging machine learning approaches for predicting antidepressant treatment response using electroencephalography (EEG) and clinical data. Frontiers in psychiatry 9, 768 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pizzagalli DA et al. Pretreatment Rostral Anterior Cingulate Cortex Theta Activity in Relation to Symptom Improvement in Depression: A Randomized Clinical Trial. JAMA psychiatry 75, 547–554 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korb AS, Hunter AM, Cook IA & Leuchter AF Rostral anterior cingulate cortex theta current density and response to antidepressants and placebo in major depression. Clinical Neurophysiology 120, 1313–1319 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leuchter AF, Cook IA, Witte EA, Morgan M & Abrams M Changes in brain function of depressed subjects during treatment with placebo. American Journal of Psychiatry 159, 122–129 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Nunez PL & Srinivasan R Electric fields of the brain: the neurophysics of EEG. (Oxford University Press, USA, 2006). [Google Scholar]
- 15.Müller K-R et al. Machine learning for real-time single-trial EEG-analysis: from brain–computer interfacing to mental state monitoring. Journal of neuroscience methods 167, 82–90 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Wu W, Nagarajan S & Chen Z Bayesian Machine Learning: EEG/MEG signal processing measurements. IEEE Signal Processing Magazine 33, 14–36 (2016). [Google Scholar]
- 17.Schirrmeister RT et al. Deep learning with convolutional neural networks for EEG decoding and visualization. Human brain mapping 38, 5391–5420 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haufe S et al. On the interpretation of weight vectors of linear models in multivariate neuroimaging. Neuroimage 87, 96–110 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Boyd S & Vandenberghe L Convex optimization. (Cambridge university press, 2004). [Google Scholar]
- 20.Bell AJ & Sejnowski TJ An information-maximization approach to blind separation and blind deconvolution. Neural computation 7, 1129–1159 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Trivedi MH et al. Establishing moderators and biosignatures of antidepressant response in clinical care (EMBARC): Rationale and design. J Psychiatr Res 78, 11–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fonzo GA, Etkin A, Zhang Y, Wu W, Cooper C, Chin-Fatt C, Jha MK, Trombello J, Deckersbach T, Adams P, McInnis M, McGrath PJ, Weissman MM, Fava M, Trivedi MH Brain Regulation of Emotional Conflict Differentiates Response to Antidepressants Versus Placebo in Depression. Nature Human Behaviour (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruder GE et al. Electroencephalographic alpha measures predict therapeutic response to a selective serotonin reuptake inhibitor antidepressant: pre-and post-treatment findings. Biological psychiatry 63, 1171–1177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tipping ME Sparse Bayesian learning and the relevance vector machine. Journal of machine learning research 1, 211–244 (2001). [Google Scholar]
- 25.Grin-Yatsenko VA, Baas I, Ponomarev VA & Kropotov JD Independent component approach to the analysis of EEG recordings at early stages of depressive disorders. Clinical Neurophysiology 121, 281–289 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Pozzi D, Golimstock A, Petracchi M, García H & Starkstein S Quantified electroencephalographic changes in depressed patients with and without dementia. Biological psychiatry 38, 677–683 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Iosifescu DV et al. Frontal EEG predictors of treatment outcome in major depressive disorder. European Neuropsychopharmacology 19, 772–777 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Arns M, Drinkenburg WH, Fitzgerald PB & Kenemans JL Neurophysiological predictors of non-response to rTMS in depression. Brain stimulation 5, 569–576 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Tipping ME & Bishop CM Probabilistic principal component analysis. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 61, 611–622 (1999). [Google Scholar]
- 30.Trivedi MH et al. Comprehensive phenotyping of depression disease trajectory and risk: Rationale and design of Texas Resilience Against Depression study (T-RAD). Journal of Psychiatric Research (In Press). [DOI] [PubMed] [Google Scholar]
- 31.Hill AT, Rogasch NC, Fitzgerald PB & Hoy KE TMS-EEG: A window into the neurophysiological effects of transcranial electrical stimulation in non-motor brain regions. Neuroscience & Biobehavioral Reviews 64, 175–184 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Chen AC et al. Causal interactions between fronto-parietal central executive and default-mode networks in humans. Proceedings of the National Academy of Sciences 110, 19944–19949 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donse L, Padberg F, Sack AT, Rush AJ & Arns M Simultaneous rTMS and psychotherapy in major depressive disorder: Clinical outcomes and predictors from a large naturalistic study. Brain stimulation 11, 337–345 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Krepel N et al. Non-replication of neurophysiological predictors of non-response to rTMS in depression and neurophysiological data-sharing proposal. Brain Stimulation: Basic, Translational, and Clinical Research in Neuromodulation 11, 639–641 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Leuchter AF et al. Comparative effectiveness of biomarkers and clinical indicators for predicting outcomes of SSRI treatment in Major Depressive Disorder: results of the BRITE-MD study. Psychiatry Research 169, 124–131 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Klimesch W, Sauseng P & Hanslmayr S EEG alpha oscillations: the inhibition–timing hypothesis. Brain research reviews 53, 63–88 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Jensen O & Mazaheri A Shaping functional architecture by oscillatory alpha activity: gating by inhibition. Frontiers in human neuroscience 4, 186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arns M et al. EEG alpha asymmetry as a gender-specific predictor of outcome to acute treatment with different antidepressant medications in the randomized iSPOT-D study. Clinical Neurophysiology 127, 509–519 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Lehtonen J & Lehtinen I Alpha rhythm and uniform visual field in man. Electroencephalography and clinical neurophysiology 32, 139–147 (1972). [DOI] [PubMed] [Google Scholar]
- 40.Hari R & Salmelin R Human cortical oscillations: a neuromagnetic view through the skull. Trends in neurosciences 20, 44–49 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Ramoser H, Muller-Gerking J & Pfurtscheller G Optimal spatial filtering of single trial EEG during imagined hand movement. IEEE transactions on rehabilitation engineering 8, 441–446 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Kraemer HC Messages for Clinicians: Moderators and Mediators of Treatment Outcome in Randomized Clinical Trials. Am J Psychiatry 173, 672–679 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Nguyen KH & Gordon LG Cost-Effectiveness of Repetitive Transcranial Magnetic Stimulation versus Antidepressant Therapy for Treatment-Resistant Depression. Value Health 18, 597–604 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Voigt J, Carpenter L & Leuchter A Cost effectiveness analysis comparing repetitive transcranial magnetic stimulation to antidepressant medications after a first treatment failure for major depressive disorder in newly diagnosed patients - A lifetime analysis. PLoS One 12, e0186950 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Reardon JP et al. Efficacy and safety of transcranial magnetic stimulation in the acute treatment of major depression: a multisite randomized controlled trial. Biological psychiatry 62, 1208–1216 (2007). [DOI] [PubMed] [Google Scholar]
- 46.George MS et al. Daily left prefrontal transcranial magnetic stimulation therapy for major depressive disorder: a sham-controlled randomized trial. Archives of general psychiatry 67, 507–516 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Williams LM, Debattista C, Duchemin A, Schatzberg A & Nemeroff C Childhood trauma predicts antidepressant response in adults with major depression: data from the randomized international study to predict optimized treatment for depression. Translational psychiatry 6, e799 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liston C et al. Default mode network mechanisms of transcranial magnetic stimulation in depression. Biological psychiatry 76, 517–526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sani OG et al. Mood variations decoded from multi-site intracranial human brain activity. Nature biotechnology 36, 954 (2018). [DOI] [PubMed] [Google Scholar]
Methods-only references
- 50.van Buuren S & Groothuis-Oudshoorn K mice: Multivariate Imputation by Chained Equations in R. 2011 45, 67 (2011). [Google Scholar]
- 51.Etkin A, Egner T, Peraza DM, Kandel ER & Hirsch J Resolving emotional conflict: a role for the rostral anterior cingulate cortex in modulating activity in the amygdala. Neuron 51, 871–882 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Etkin A, Buchel C & Gross JJ The neural bases of emotion regulation. Nat Rev Neurosci 16, 693–700 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Egner T & Hirsch J Cognitive control mechanisms resolve conflict through cortical amplification of task-relevant information. Nat Neurosci 8, 1784–1790 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW & Smith SM FSL. Neuroimage 62, 782–790 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Friston KJ et al. Statistical parametric maps in functional imaging: a general linear approach. Hum Brain Mapp 2, 189–210 (1995). [Google Scholar]
- 56.Wu W et al. ARTIST: A fully automated artifact rejection algorithm for single-pulse TMS-EEG data. Human brain mapping 39, 1607–1625 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lovibond PF & Lovibond SH The structure of negative emotional states: Comparison of the Depression Anxiety Stress Scales (DASS) with the Beck Depression and Anxiety Inventories. Behaviour research and therapy 33, 335–343 (1995). [DOI] [PubMed] [Google Scholar]
- 58.Tomioka R & Müller K-R A regularized discriminative framework for EEG analysis with application to brain–computer interface. NeuroImage 49, 415–432 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Srebro N & Jaakkola T in Proceedings of the 20th International Conference on Machine Learning (ICML-03) 720–727 (2003). [Google Scholar]
- 60.Candès EJ, Li X, Ma Y & Wright J Robust principal component analysis? Journal of the ACM (JACM) 58, 11 (2011). [Google Scholar]
- 61.Parikh N & Boyd S Proximal algorithms. Foundations and Trends® in Optimization 1, 127–239 (2014). [Google Scholar]
- 62.Gramfort A, Papadopoulo T, Olivi E & Clerc M OpenMEEG: opensource software for quasistatic bioelectromagnetics. Biomedical engineering online 9, 45 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fischl B FreeSurfer. Neuroimage 62, 774–781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pascual-Marqui RD Standardized low-resolution brain electromagnetic tomography (sLORETA): technical details. Methods Find Exp Clin Pharmacol 24, 5–12 (2002). [PubMed] [Google Scholar]
- 65.Hunter AM, Leuchter AF, Morgan ML & Cook IA Changes in brain function (quantitative EEG cordance) during placebo lead-in and treatment outcomes in clinical trials for major depression. American Journal of Psychiatry 163, 1426–1432 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Makeig S, Bell AJ, Jung T-P & Sejnowski TJ in Advances in neural information processing systems 145–151 (1996).
- 67.Ghosh-Dastidar S, Adeli H & Dadmehr N Principal component analysis-enhanced cosine radial basis function neural network for robust epilepsy and seizure detection. IEEE Transactions on Biomedical Engineering 55, 512–518 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Cichocki A & Amari S.-i. Adaptive blind signal and image processing: learning algorithms and applications, Vol. 1 (John Wiley & Sons, 2002). [Google Scholar]
- 69.Witten IH, Frank E, Hall MA & Pal CJ Data Mining: Practical machine learning tools and techniques. (Morgan Kaufmann, 2016). [Google Scholar]
- 70.Schaefer A et al. Local-Global Parcellation of the Human Cerebral Cortex from Intrinsic Functional Connectivity MRI. Cereb Cortex, 1–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The EMBARC data are publicly available through the NIMH Data Archive (NDAR) (https://nda.nih.gov/edit_collection.html?id=2199).