Abstract
Lung adenocarcinoma (LUAD) is the second most common cancer, affecting both men and women. Fibrosis is a hallmark of LUAD occurring throughout progression with excess production of extracellular matrix (ECM) components that lead to metastatic cell processes. Understanding the ECM cues that drive LUAD progression has been limited due to a lack of tools that can access and report on ECM components within the complex tumor microenvironment. Here, we test whether low grade LUAD can be distinguished from normal lung tissue using a novel extracellular matrix imaging mass spectrometry (ECM IMS) approach. ECM IMS analysis of a tissue microarray with 20 low-grade LUAD tissues and 20 normal lung samples from 10 patients revealed 25 peptides that could discriminate between normal and low-grade LUAD using area under the receiver (AUC) operating curve ≥0.7, p-value ≤0.001. Principal component analysis demonstrated that 62.4% of the variance could be explained by sample origin from normal or low-grade tumor tissue. Additional work performed on a wedge resection with moderately differentiated LUAD demonstrated that the ECM IMS analytical approach could distinguish LUAD spectral features from spectral features of normal adjacent lung tissue. Conventional LC-MS/MS proteomics demonstrated that specific sites of hydroxylation of proline (HYP) were a main collagen post-translational modification that was readily detected in LUAD. A distinct peptide from collagen 3A1 modified by HYP was increased 3.5 fold in low grade LUAD compared to normal lung tissue (AUC 0.914, p-value <0.001). This suggests that regulation of collagen proline hydroxylation could be an important process during early LUAD fibrotic deposition. ECM IMS is a useful tool that may be used to define fibrotic deposition in low-grade LUAD.
Keywords: lung cancer, adenocarcinoma, extracellular matrix, imaging mass spectrometry, proteomics, peptide imaging, formalin-fixed, paraffin-embedded tissue imaging, tissue imaging, MALDI imaging mass spectrometry, collagen
1. Introduction
Lung cancer (both small cell and non-small cell) accounts for 13% of all new cancer cases every year and is the second most common cancer in men and women [1, 2]. LUAD is a very heterogeneous cancer with many variations in genetic, epigenetic and protein expression contributing to the resultant pathology[3, 4]. In spite of increasing knowledge of targetable components in LUAD progression, the 5-year survival rate remains low, around 16% at 5 years [5]. The major reason for low survival rate is attributed to highly variable symptoms and signs depending on where the tumor forms in the lung. It is estimated that around 70% of all patients diagnosed with LUAD present with advanced lung cancer [4, 6]. Lung cancer screening in high risk adults age 55–80 years has been shown to reduce 20% lung cancer-related mortality [7]. However, it is acknowledged that current screening, which include both invasive and noninvasive approaches, results in high false positive rates and contributes to over diagnoses[8]. A main challenge in LUAD is defining new molecules can act as a prognostic or diagnostic indicator at earlier time points of progression.
Fibrosis is a hallmark of lung adenocarcinoma. The cancer-associated fibroblasts (CAF) and the extracellular matrix (ECM) components that CAFs secrete have become a focus of research [1, 9, 10]. ECM is a dynamic component of all tissues that forms a scaffold for cells, but also mediates important functions such as cell adhesion and communication. It consists of proteins including collagen, fibronectin, tenascin, elastin, laminin, proteoglycans, and a variety of growth factors, all of which are important components of the ECM microenvironment that provide cues to the cells. Increased production and deposition of ECM components such as collagen and fibronectin as well as their cross-linking are shared features of non-tumor fibrosis and the tumor microenvironment. The role of the ECM in regulating tumor cell behavior was originally proposed by Bissell et al[11]. Since then, some progress has been made in understanding the role of the ECM in tumors. For example, Paszek et al showed that increased ECM production drives tumor progression and promotes tumor cell migration[12]. Further, tumor cell metastatic behavior is shaped by the cross-linked matrix produced by cancer associated fibroblasts[13]. In non-small cell lung cancer, high stroma-tumor ratios (≥50% stroma) correlate with decreased survival [14]. Culture of cells from LUAD diagnosed patients showed that the presence of collagen type 1, a main protein deposited in fibrosis, resulted in significantly shorter progression-free survival time after therapy[15]. Since cancer-related mortality is due to tumor cell metastasis, which is driven by tumor-associated fibroblasts and fibrosis, approaches to mitigate fibrotic initiation in the setting of solid tumors could reduce metastasis and improve survival. Furthermore, detecting these changes in low-grade adenocarcinoma could uncover useful targets that limit advancement to metastasis.
New tools for noninvasive imaging of ECM in fibrotic processes have recently been developed [16, 17], yet there is still a gap in defining disease-specific ECM deposition for molecular targeting. ECM proteins are challenging for mass spectrometry analyses due to lower basic amino acid content and extensive post-translational modifications that include crosslinking. Recently, we reported parallel chromatographic and tissue imaging proteomic workflows that access ECM proteins from formalin-fixed, paraffin-embedded clinical specimens [18, 19]. This novel method leverages collagenase type enzymes to specifically target ECM proteins and produce peptides useful for ECM structure evaluation. Specifically, collagenase type III was shown to target collagen type proteins and other ECM proteins including fibronectin, tenascin X, tenascin C, laminin, lumican, and proteoglycans such as basement membrane-specific heparin sulfate proteoglycan core protein (HSPG2). The target list suggests that this approach would be ideal for evaluating the emerging patterns of fibrosis and predicting new molecular targets for therapy.
In the current work, we explore the use of extracellular matrix imaging mass spectrometry (ECM IMS) as an approach to determine molecular differences in ECM between normal lung and low grade (0–1) lung adenocarcinoma (LUAD) compared to normal tissue. As a proof of concept, we evaluate a tissue microarray (TMA) containing 20 patient cores of early lung adenocarcinoma compared to 10 normal tissues. We compare this signature pattern to tissue sections containing advanced lung adenocarcinoma characterized by excessive stroma composition. Interestingly, this proof of concept study suggests that a primary difference between normal and early grade lung adenocarcinomas is the presence of collagen peptides containing hydroxylated prolines. This approach provides a new tool for investigations on fibrotic progression during progression of adenocarcinoma.
Experimental
2.1. Materials and Reagents
Acetonitrile, α-Cyano-4-hydroxycinnamic acid (CHCA), trifluoroacetic acid (TFA), ammonium bicarbonate, ammonium phosphate monobasic, calcium chloride, and Trizma® base were purchased from Sigma-Aldrich (St. Louis, MO, USA). Collagenase type III (COLase3) (C. histolyticum) was purchased from Worthington (Lakewood, NJ, USA). Xylenes, 200 proof ethanol, methanol, HPLC grade water were purchased from Fisher Scientific (Pittsburgh, PA, USA).
2.2. Tissue Procurement
Tissue use was in accordance with protocols approved by the Medical University of South Carolina Institutional Review Board. The tissue microarray (TMA) was purchased from Biomax (Rockville MD, USA) as defined primarily low grade lung adenocarcinoma pathology (20 cases) and 10 cases of normal lung tissue with two independent collections per normal lung (a total of 20 normal lung tissue cores). The TMA was created in 2015 and used the WHO 2015 guidelines for grading. Tissue sections used for proteomics were procured via the Biorepository & Tissue Analysis Shared Resource at the Hollings Cancer Center, Medical University of South Carolina with Institutional Review Board (IRB) approval.
2.3. Tissue Preparation
The formalin-fixed, paraffin-embedded (FFPE) tissues and TMA were prepared as previously described.[18, 19] Briefly, tissues were heated, dewaxed, antigen retrieved at pH 9 using 10 mM tris buffer, and sprayed with COLase3 using an automated sprayer (M3 TM-Sprayer, HTXImaging, Chapel Hill, NC, USA). COLAse3 was sprayed onto tissues using parameters of 45°C, 10 psi, 25 μL/min, 1200 velocity, and 15 passes with a 3.0 mm offset. Samples were digested in high humidity at 37.5°C for five hours followed by automated spraying of CHCA matrix prepared as 7 mg/mL in 50% acetonitrile, 1% TFA with a spiked standard of 200 femtomole/microliter [Glu1]-Fibrinopeptide B human (Glufib) (Sigma-Aldrich), St. Louis, MO, USA). CHCA was applied by automated sprayer with parameters of 77°C, 10 psi, 100 μL/min, 1300 velocity, and 10 passes with a 2.5 mm offset. Slides were rapidly dipped (<1 s) in cold 5 mM ammonium phosphate, monobasic and dried in a desiccator prior to imaging mass spectrometry data acquisition.
2.4. Imaging Mass Spectrometry Acquisition and Analyses
Lung TMA and tissues were analyzed by MALDI-FT-ICR (solariX™ Legacy 7.0 Tesla, Bruker, Bremen, Germany) in positive ion mode, collecting 200 laser shots per pixel with stepsizes of 100 (TMAs) to 150 (tissue) μm between pixels. Transients of 512 kilowords were acquired in broadband mode over m/z 600–5000, with a calculated on-tissue mass resolution at full width half maximum of 81,000 at m/z 1400. Lockmass (GluFib peptide m/z 1570.6768) was maintained at 5 ppm during tissue imaging. Data were analyzed using SCiLS Lab software 2019 Pro Build 7.02.10901 (Bruker Scientific, LLC, Bremen, Germany). All images are shown normalized to total ion current. Image segmentation used the Manhattan metric for segmentation without denoising. Extracted peak areas were exported from SCiLs and are shown visualized as heat maps with clusterVis[20]. Area under the receiver-operating curve (AUC) was used to evaluate for peaks that discriminated between cancer types, subtypes and staging. Clustervis[20] (https://biit.cs.ut.ee/clustvis/) was used to visualize peptide intensity data and perform multivariate analyses based on selected peptides.
2.5. Chromatographic Proteomics
Lung tissue sections were treated as described previously[18], digesting with COLase3 overnight. From these, COLAse3 peptides was dried down, cleaned by solid phase extraction by C18 Ziptip (EMD Millipore, Darmstadt, Germany). The peptides were loaded on a trap column and separated on a 30 cm × 75 μm C18RP analytical column (Dr. Maisch; 1.9 μm particle size) at 50°C with a gradient of 5–40% in 180 min at a flow rate of 200 nL/min. Solvents A (2% acetonitrile) and B (98% acetonitrile) contained 0.2% formic acid. Peptides were analyzed by data dependent acquisition on an Orbitrap Elite mass spectrometer equipped with a U3000 nanoLC system (Thermo Scientific). Survey MS scans were acquired in the orbitrap with an automatic gain control target value of 106 at a resolution of 60,000 at m/z 400. Collision-induced dissociation MS/MS was performed on the top 10 most intense ions with dynamic exclusion enabled with a repeat count of 3, repeat duration of 30 sec, exclusion list of 50, and exclusion duration of 100 sec. Tandem mass spectra acquired in the ion trap were searched using both MASCOT (Version 2.4.01) and SEQUEST HT via Proteome Discoverer 1.4 (Thermo Scientific, Waltham, MA, USA) against a subset of human protein sequences downloaded on May 7, 2017 from UniProtKB (SwissProt) containing 1,783 entries (keywords used: collagen, elastin, aggrecan, gelatin, osteonectin, perlecan, plasminogen, and fibronectin). Search parameters were unspecified enzyme, precursor mass tolerances of ± 20 ppm, and fragment mass tolerances ± 0.8 Da. Methionine oxidation, asparagine and glutamine deamidation were used as variable modifications. Additional post-translational modifications were searched using Byonic[21] and including a wild card search. Data were uploaded into Scaffold v4.8.1 (Proteomesoftware, Portland OR, USA) and a peptide probability of 99% was used to report peptide identifications. Hydroxyproline site modification probabilities are reported using MaxQuant and Perseus[22, 23]. Protein localization and function was determined using Uniprot[24] via incorporated databases (Reactome) and primary publications. Accurate mass comparisons between image data and proteomic data were done by calculating accurate mass of a peptide using Protein Prospector version 5.22.0 (Baker, P.R. and Clauser, K.R. http://prospector.ucsf.edu, University of California at San Francisco, San Francisco CA, USA).
3. Results and Discussion
3.1. Heuristic comparison of tissue microarray data demonstrates aggregate changes in collagenase type III peptide data based on normal or adenocarcinoma status.
Our previously published approach to targeting ECM peptides from FFPE tissue was used to determine if we could detect differences in normal versus low grade lung adenocarcinoma[18, 19]. A commercial tissue microarray (TMA) was used to explore differences between low grade lung adenocarcinoma and normal lung tissue (Figure 1A). The TMA contained 20 cores with primarily low stage (I-II) low grade (G1 and G2–3) tumors that had not spread to lymph nodes (Table 1, Supplemental Table 1). Normal lung tissue was from 10 patients with two sites of collection per patient lung. Hematoxylin and eosin stain demonstrated increased stroma in low grade LUAD (Figure 1B). By ECM IMS, total of 2,190 peaks were reported across various cores. Image segmentation was used as a heuristic approach to compute spectra assigned by histological region (Figure 1C). Segmentation showed distinct clustering between LUAD and normal lung tissue cores. A total of 756 spectra clustered with normal cores as compared to LUAD cores (Figure 1D). Adenocarcinoma specific signatures appeared split into two primary groups of spectral features with a combined total of 1168 spectra. The greater number of spectra found in cancer cores was expected because these cores contain dense stroma as compared to normal lung tissue. Extraction of each segmented region highlights that spectral signatures separate between lung adenocarcinoma and normal lung tissue (Figure 1E). Overall, the analysis of peptides generated by ECM IMS across the TMA demonstrated that at an aggregate level there are distinct molecular differences between low grade lung adenocarcinoma and normal lung tissue.
Figure 1.
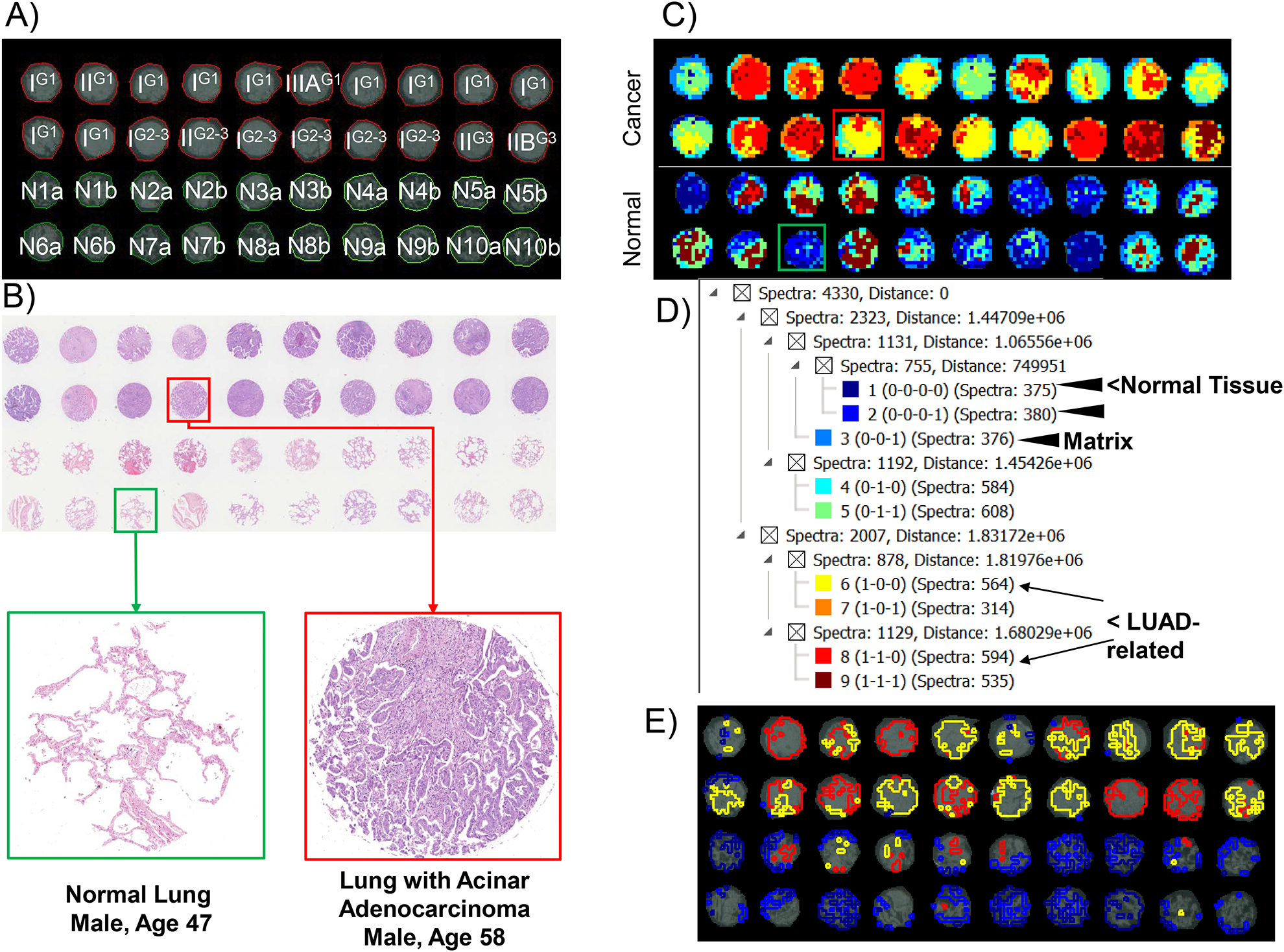
Tissue microarray containing lung adenocarcinoma (LUAD; n=20, one core from each patient tissue) and normal lung tissue (n=10, two different cores from each normal lung). A. 2015 WHO classification of lung cancer. Stage and grading of the TMA stage is indicated in capital I, II, IIB or IIIA. Grade is superscripted. Normal cores are marked N for normal, number is the normal lung core number, a or b reports different locations. B) hematoxylin and eosin staining. Red highlights lung adenocarcinoma core while green highlights a normal core, shown below in higher magnification. Cores are 1 mm in diameter. C) Image segmentation done on 2,190 peaks highlighting overall differences in spectral composition between normal and adenocarcinoma. D) Hierarchical tree from image segmentation reporting clusters per tissue type. E) Extracted regions enriched in normal or LUAD.
Table 1.
Clinical characteristics of the tissue microarray.
| Normal | Adenocarcinoma | |
|---|---|---|
| Age, years [Median, 95% CI] | 38.5 [30.3, 46.7] | 60 [55.9, 64.1] |
| Female | 2/10 (20%) | 4/20 (20% |
| Male | 8/10 (80%) | 16/20 (80%) |
| Grade 1 | 12 (60%) | |
| Grade 2–3 | 6 (30%) | |
| Grade 3 | 2 (10%) | |
| Stage I | 15 (75%) | |
| Stage II (A&B) | 4 (20%) | |
| Stage III (A) | 1 (5%) | |
| Papillary | ||
| adenocarcinoma | 6 (30%) | |
| Acinar adenocarcinoma | 12 (60%) | |
| Solid adenocarcinoma | 2 (10%) | |
3.2. Distinct collagenase type III peptides differentiate low-grade lung adenocarcinoma from normal lung tissue.
Multivariate analyses was used to compare spectral features between the two sets of data. Area under the receiver-operating curve (AUC) was used to determine how well each spectral feature could distinguish between adenocarcinoma and normal tissue. Figure 2A demonstrates 25 peptides that could distinguish between normal and disease. Interestingly, even though lung adenocarcinoma involves a clear increase in stroma, there were at least 10 peptides that were higher in normal tissue. This could potentially indicate a change in post-translational modification of the peptide. Principal Component Analysis was used to determine variation of the peptide set per sample. Using the collective 25 peptides as variables, 62.5% of the variance could be explained by spectral origin from adenocarcinoma or from normal tissue (Figure 2A). Intriguingly, cores from normal tissue (samples # 21–40) showed a broad distribution pattern and duplicate cores from the same patient did not necessarily cluster together. This could be due to duplicate cores being from different lung regions. Adenocarcinoma on the other hand, formed a tighter cluster, regardless of the fact that each core came from a different patient. An exception was core adenocarcinoma core #1, which separated apart from any other core. Correlation with aggregate data showed that this core had a minimal spectral signature unlike adenocarcinoma or normal tissue; however, close inspection of the pathology did not report anything unique about this core compared to other adenocarcinoma cores. Examination of peptide distribution revealed variation in localization to histology (Fig 2C–D). However, expression levels from each core were reproducible and reported significant alteration between normal and adenocarcinoma and showed significant fold changes between normal and disease states. Altogether, analysis of individual peptide peaks generated by ECM IMS revealed substantial changes in peptide expression level between low grade lung adenocarcinoma and normal lung tissue.
Figure 2.
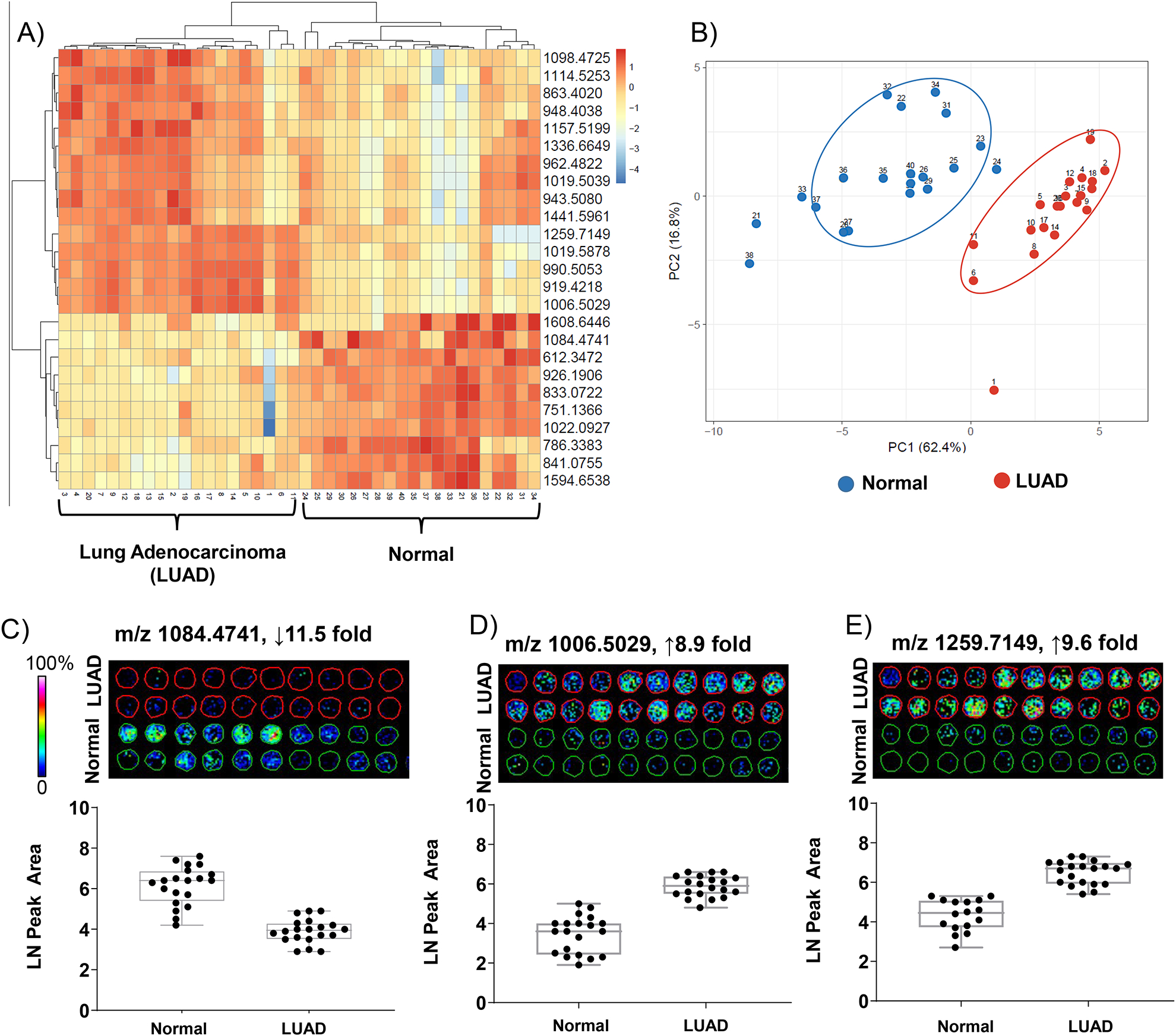
ECM IMS differentiates low-grade lung adenocarcinoma from normal lung tissue. A) Peptides differentiated by area under the receiver operating curve >0.7. B) Principal component analysis demonstrates separation based on origin of spectral data from normal or adenocarcinoma cores. Component 1, differences between samples types, explains 62% of the variance. C-D) Example peptides shown as images and quantitative data points per core. Peptide changes are greater than 8 fold.
3.3. Collagenase type III peptides define the lung adenocarcinoma tumor microenvironment.
A wedge resection of LUAD was characterized by ECM IMS to understand if collagenase type III generated peptides aligned with tumor composition. Detection of peptides in tumor tissue and not normal tissue could increase the potential for a particular peptide to act as a marker of LUAD status. Here, the wedge resection was defined as having moderately differentiated adenocarcinoma and was staged as T2 (Figure 3A). The tissue section represented a large proportion of tumor compared to a smaller region of normal adjacent tumor (NAT; approximately 10% of area compared to tumor based on pathology). The wedge resection was further differentiated by the presence of mixed tumor morphologies including mucinous and minor clear cell components. The tumor stroma had a significant lymphoplasmacytic inflammatory infiltrate including scattered lymphoid aggregates (Figure 3B). Figure 3C demonstrates that hierarchical clustering of image data reports the majority of spectra associated with the tumor (7,056/12,253; 57% of total spectra). NAT tissue represented a distinct region with 859/12,253; 7% of total spectra). Extraction of spectral regions highlighted that molecular patterning followed pathologically defined regions of normal and tumor within the wedge resection (Figure 3D). Like the TMA data, principal component analysis reported that the main variance could be explained by localization to normal or tumor tissue. Component 1 reported that 60.8% of the variance could be explained by spectral origin from adenocarcinoma or from normal tissue (Figure 3F). To conclude, our analysis of lung tissue with moderately differentiated adenocarcinoma and normal adjacent tissue demonstrated that ECM IMS could distinguish between tumor and normal features within a heterogeneous tumor microenvironment.
Figure 3.
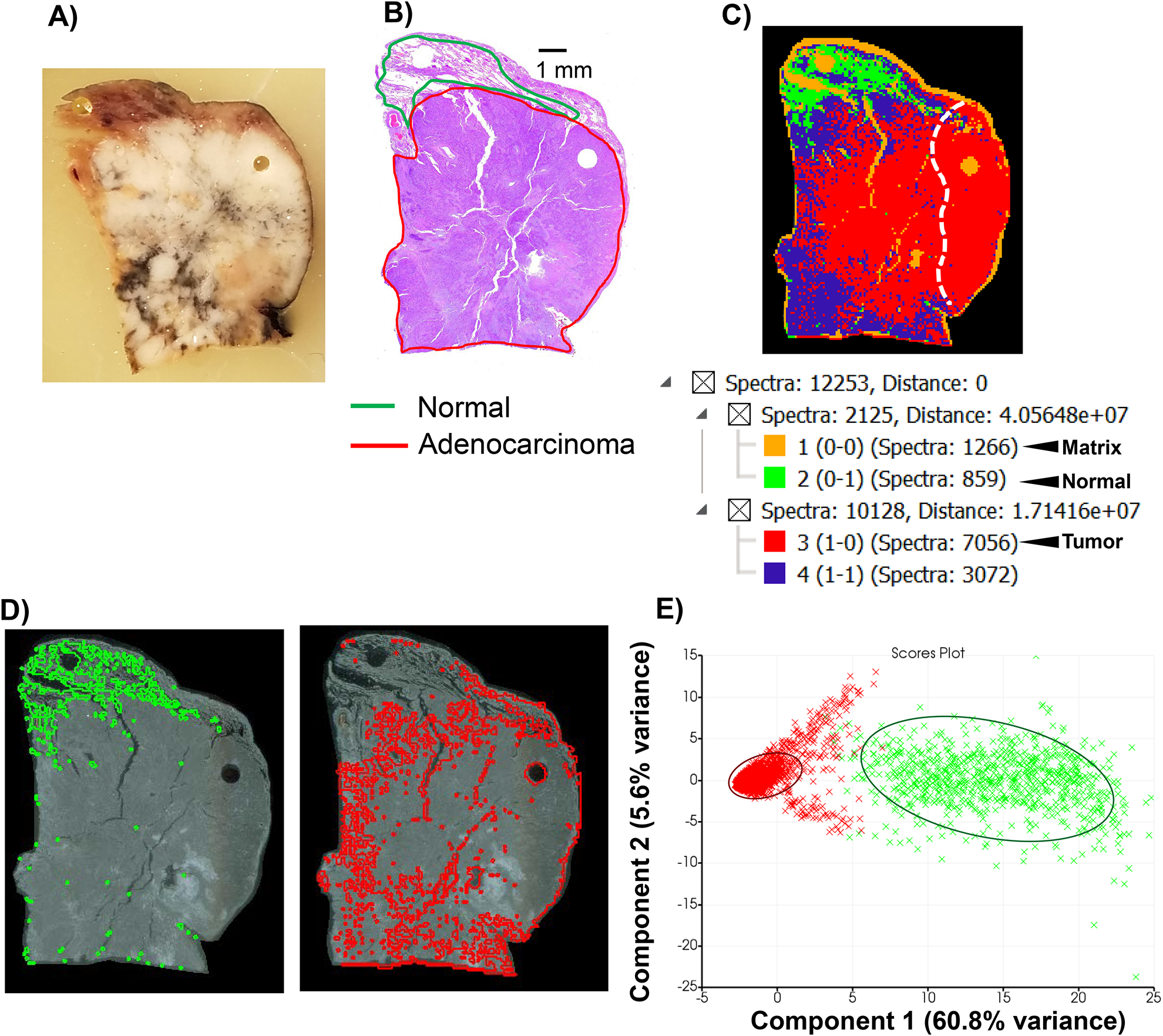
ECM IMS on lung tissue with moderately differentiated T2 adenocarcinoma. A) Block face of wedge resection. Gray/black discoloration is anthracotic pigmentation caused by accumulation of carbon in the lungs. This could be inhalation of wood smoke, coal dust or from air pollution. B) Hematoxylin and eosin (H&E) stain on section neighboring to section used for IMS. C) Image segmentation on 4,342 peaks from imaging data. C) Spectral hierarchical clustering defines normal and tumor region and correlates with pathology reading. D) Extracted regions from image segmentation normal (green, 859 spectra; 7% of total spectra) and tumor (red, 7,056 spectra; 57% of total spectra). E) Principal component analysis (PCA) of spectra from normal versus tumor. Circled regions highlight main clusters of tumor spectra (red) or normal spectra (green). Separation of component 1 is based on localization to tumor or normal tissue and accounts for 60.8% of the variance.
3.4. Collagenase type III peptides from the tumor microarray show unique molecular gradients within the adenocarcinoma microenvironment.
TMA cores are selected as being most representative of the pathology grade. This contrasts with tissue sections where molecular gradients might be present from normal adjacent to tumor tissue within a single section. Peptides that discriminated (Figure 2A) between pathology grading in the TMA were investigated to understand if ECM IMS reported molecular gradients for these peptides in the complex tumor microenvironment. Peptides elevated in the TMA tumor tissues across many patients also corresponded to tumor localization in the wedge resection (Figure 4A). For instance m/z 943.5080 (m/z 943) had an intense distribution at the bottom of the primary tumor region and varied in intensity throughout the tumor. The m/z 1019.5878 (m/z 1019.58) localized to the primary tumor region and in the TMA had significantly higher expression in LUAD cores (9.5 fold LUAD/normal, Mann-Whitney t-test p-value <0.001). The m/z 1336.6709 (m/z 1336) demonstrated uniform expression throughout the entire characterized tumor. Examples of peptides with lower expression in the TMA tumor were also examined (Figure 4B). The peptide m/z 1019.5039 (m/z 1019.50) was found in the TMA in both tumor and cores but higher in tumor. In the tissue section, 1019.50 was localized to blood vessels. A peptide m/z 1125.5829 (m/z 1125) showed higher expression levels in regions of stroma found in normal and tumor; this peptide mapped to both normal and tumor in the TMA. The peptide m/z 1399.6566 (m/z 1399) showed lower intensity in the regions with a low amount of clear-cell differentiation within the tumor, appearing to align with inflammatory aggregate and desmoplastic tumor stroma. However, m/z 1399 was also present in bronchioles that were found in both normal and tumor tissue. Spatial correlation coefficient (R2s) was further used to determine if peaks significantly elevated in healthy lung TMA cores (10 peaks, Figure 2A) correlated to NAT regions from the wedge resection. A high correlation coefficient (≥0.6) was used to indicate a positive association between the map of the indicated peptide and the tissue region graded as normal adjacent to tumor. Many peaks detected in normal tissue from the TMA were not present, or below noise threshold, in the normal adjacent tissue. This is not surprising, as normal adjacent tissue may be influenced by the nearby tumor, which is believed to contribute to the spread of cancer, known as the “field effect”[25]. A single peak, m/z 1022.0927, was detected that showed some spatial correlation to the defined normal adjacent tumor region (R2s = 0.54, Supplemental Figure 1). Expression was higher in the NAT region, but with low expression throughout the tissue. Within the wedge resection, expression ratio of NAT/Tumor for m/z 1022 was 4.32, AUC =0.972, suggesting that this peak could potentially discriminate normal type regions. Finally, the TMA peak data that was significantly higher in LUAD (15 peaks, Figure 2A) was compared to the tumor region of the wedge resection. The correlation factor R2s ≥0.6 was used to indicate a positive association with the defined tumor region. A total of three peaks m/z 919.4218 (m/z 919), m/z 943.5080 (m/z 943), and m/z 1019.5878 (m/z 1019.58) showed positive correlation with the LUAD tumor from the wedge resection (spatial correlation coefficient R2s 1.00, 0.568, and 0.686, respectively). It should be noted that a positive correlation coefficient does not depict variation due heterogeneity of the tumor microenvironment; this is best demonstrated with m/z 1019.58, which is positively associated with the overall tumor region but localizes to the primary tumor. These data demonstrated that ECM IMS is reproducible in obtaining signatures from lung adenocarcinoma tissue types and that the resulting peptides report on pathological molecular gradients within the complex tumor microenvironment.
Figure 4.
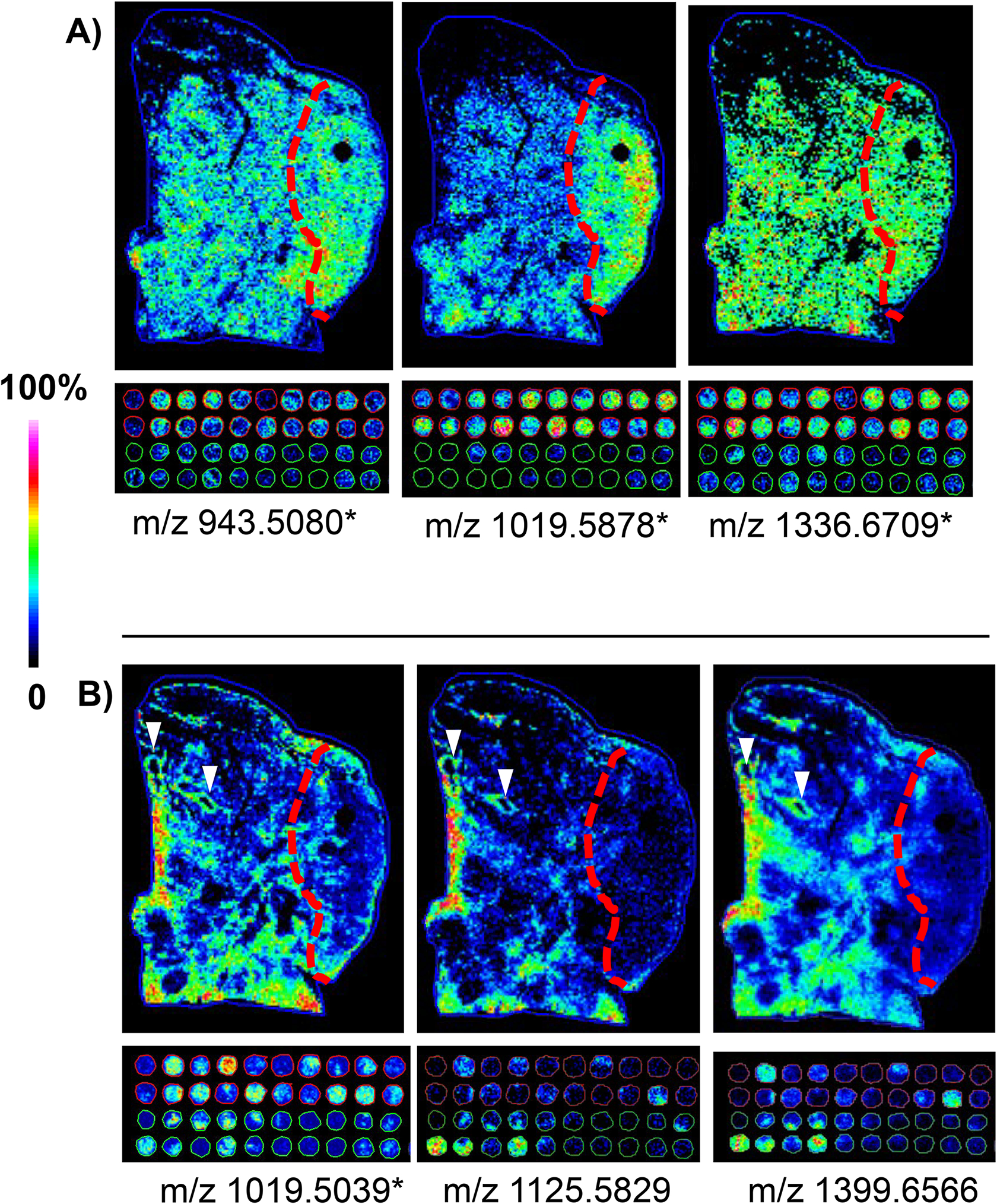
Comparison of peak distributions across a tumor tissue microenvironment and the tissue microarray representing low-grade adenocarcinoma. * - peaks detected as significantly different when compared normal tissue to lung adenocarcinoma in the TMA data using area under the receiver operating curve ≥ 0.7. A) Peptides elevated in tumor regions in the wedge resection also elevated in tissue microarray cores of low grade lung adenocarcinoma. B) Peptides decreased in the tumor regions of the wedge resection with low detection in tumor tissue microarray cores of low grade lung adenocarcinoma. White arrows indicate blood vessels.
3.5. Collagenase type III proteomics identifies hydroxylated proline modified collagen type peptides from lung adenocarcinoma.
One challenge with TMA analyses is that it is difficult to obtain multiple peptide identifications from the small tissue cores. Parallel chromatographic workflows on wedge sections bracketing the sections used for ECM IMS were performed to further understand the adenocarcinoma ECM proteome. The workflows use the same enzymes and antigen retrieval prior to digesting the tissue section in solution [18, 19]. A total of 40 proteins were identified by the approach (Supplemental Table 2). The majority of the proteins mapped to ECM (67%; Figure 5A). Some membrane proteins were detected that have been previously reported by the method [18]. These include basement membrane-specific heparan sulfate proteoglycan core protein (HSPG2) which controls cell adhesion to collagen type proteins. Bioinformatics done on non-collagen ECM type proteins reported main functions of collagen binding (8 proteins, 20%), collagen regulation (5 proteins, 12%) and collagen fibril formation (4 proteins, 10%) (Figure 5B; Supplemental Table 2). By conventional proteomics and antibody staining, type 1 collagen, but not type III or IV, has been reported the major collagen produced in lung tumor cells, xenografts and patient derived cancer cells [26–28]. Our data supported this, demonstrating that COL1A1 and COL1A2 were the major collagens in the wedge resection (6.1% and 18.6% of total spectral counts, respectively). Comparison of the FFPE collagenase digest to recent ECM proteomic papers using trypsin digestion on primary human lung cells from endstage idiopathic pulmonary fibrosis [27] and a mouse model of lung fibrosis to LUAD [28] demonstrated good overlap in protein identifications 60% (24/40 proteins) and 50% (20/40 proteins). The cited studies utilized trypsin digestion on four biological replicates of ECM from 25,000 cells (initial plating) or on 50–100 mg of fresh tissue[28] as compared to our approach on a single section of FFPE tissue in duplicate. Interestingly, comparison to a collagenase type III digest on human breast tissue[19] demonstrated 50% overlap in proteins identified. A protein unique to the FFPE lung wedge resection was dystonin, a cytoskeletal protein having multiple helical bundles with a role in anchoring fibril formation[29]. Additionally, two redox sensitive proteins were identified, protein disulfide-isomerase A6 (PDIA6), which plays a role in regulating the unfolded protein response, and peroxiredoxin-5 (PRDX5), which plays a role in regulating redox homeostasis. Protein disulfide-isomerase was also identified, a protein that may act as a chaperone to inhibit aggregation of misfolded proteins, but also acts as a subunit for prolyl 4-hydroxylase (P4HA). P4HA catalyzes the formation of 4-hydroxyproline in collagen. Hydroxylation of proline (HYP) is a major collagen post-translational modification that can occur on variable prolines to regulate collagen stability [30, 31]. Collagen stability controls cell processes of migration and invasion and also controls drug access to tumor [32–34]. Although not detected here, metastasis of non-small cell lung cancer has been positively correlated with increased expression of hypoxia-inducible factor prolyl hydroxylase 3 (HIFPH3) [35]. In total, protein identification from FFPE adenocarcinoma tissue reported significant ECM interactions associated with collagen, including several proteins that guide protein folding and aggregation.
Figure 5.
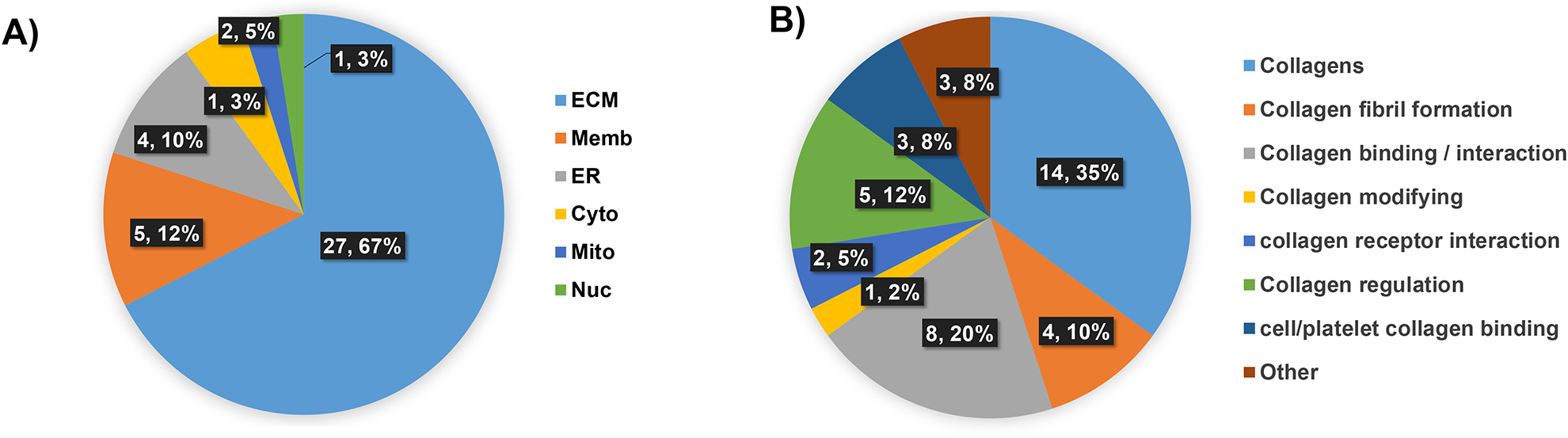
Proteomics data on lung tissue sections bracketing the section used for ECM IMS (40 proteins). A) Cellular localization of the proteome is enriched for ECM type proteins. B) Proteins organized by collagen or collagen type interactions. Almost all proteins are involved in collagen fibril formation, regulation, binding or modification.
3.6. Collagenase type III proteomics identifies hydroxylated proline modified collagen type peptides from lung adenocarcinoma.
Previous work done on carefully produced tryptic peptide libraries has identified that collagenase type II has a strong cleavage preference for Gly-Pro-X and Gly-X-HYP[36], where HYP is a hydroxylated proline residue. Crystal structure data supports that the active site of bacterial collagenases recognizes triple helix strands through flexibility in the binding site that accommodates modified and unmodified prolines [37]. To our knowledge, collagenase type III produced peptides have not been investigated for post translational modifications. Based on these data, additional work was done to investigate for hydroxylated lysine and proline residues from collagens detected in adenocarcinoma. A total of 5 peptides were confidently identified with hydroxylated lysine residues (Supplemental Table 3). We attribute this to limited material and potential crosslinking in formalin fixed tissue sections; studies on ECM secreted by fibrotic lung cells have identified 14 hydroxylated lysine residues on ECM [27]. However, both the current study and previous study identified that lysine residue 277 on COL1A1 was hydroxylated. Hydroxylation of this particular lysine is reduced by TGFβ1 stimulation, suggesting a role in the development of fibrosis[27]. Investigation on hydroxylated proline modifications reported that 8.7% of all identified peptides were HYP modified (Figure 6A). Multiple HYP peptides were identified from 9 of the 14 identified collagens (Figure 6B). Site probability analysis of hydroxylated prolines reported that very specific prolines were modified (Supplemental Table 3). HYP peptide lists were compared to imaging runs using high mass accuracy matches to test if HYP peptides were involved in low grade adenocarcinoma. In the tissue section, COL3A1 peptide VAVGGLAGYPx at m/x 919.4884 (m/z 919), was intense in the primary tumor and appeared throughout the tumor region (Figure 6D). Expression levels of m/z 919 in the tumor tissue section compared to normal showed a 5.3-fold increase in tumor. In the TMA, m/z 919 was able to discriminate between tumor cores compared to normal tissue cores (3.5 fold change LUAD/normal; AUC 0.914, p-value <0.001). Importantly, detection of HYP modified peptides correlates with LC-MS/MS identification of the P4HA, an enzyme that hydroxylates proline residues on collagen. Together, this data suggests that regulation of the collagen structure by hydroxylated proline-modified may be an important component of ECM deposition related to adenocarcinoma.
Figure 6.
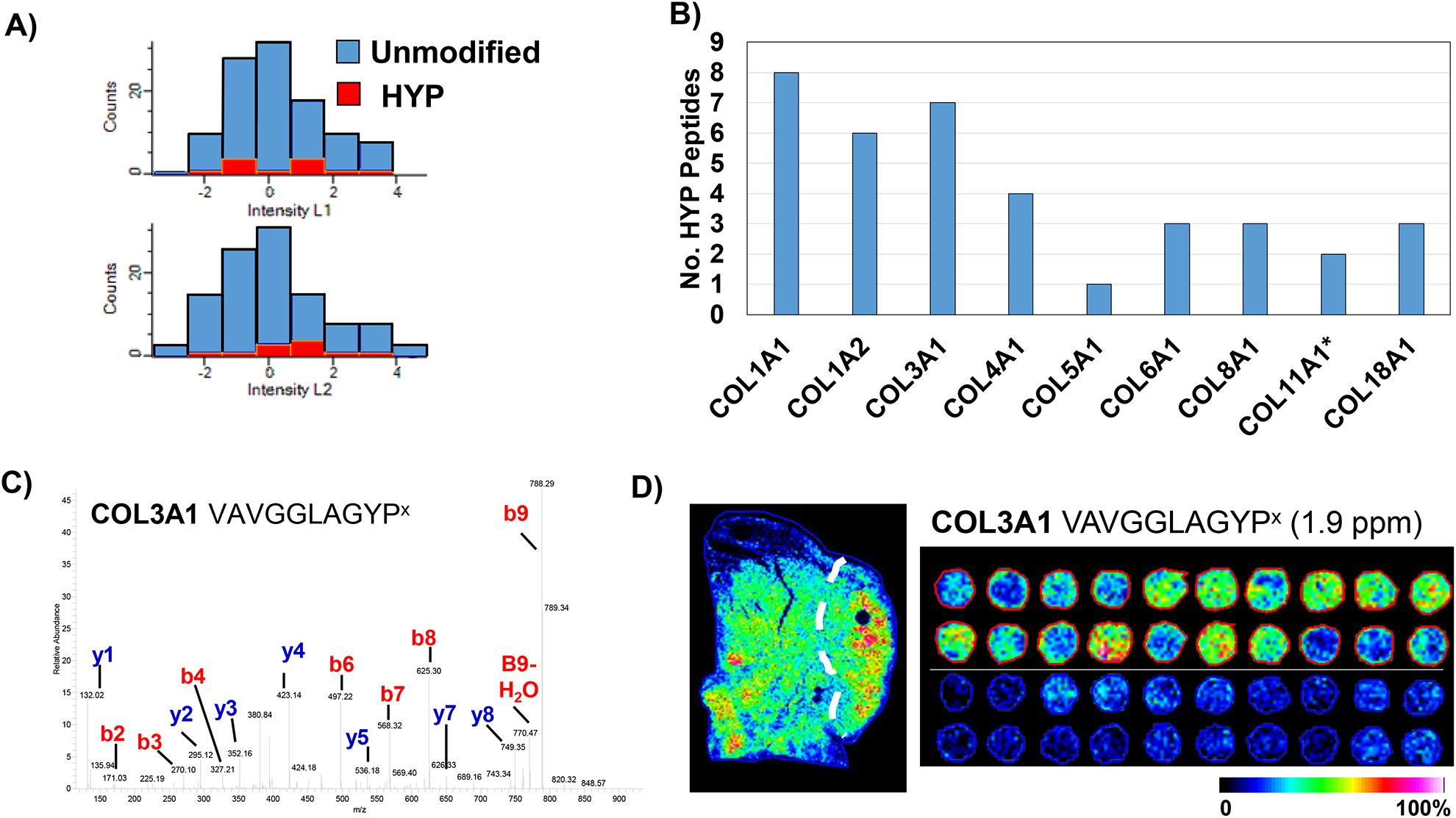
Analyses of hydroxylated proline residues, which stabilize the collagen triple helix. A) Collagenase proteomics reported approximately 8.7% of peptides (46/525 unique peptides) modified by hydroxylated proline. Data is from duplicate LC-MS/MS analyses of tissue sections bracketing the imaging data. B) Comparison of hydroxylated proline residues in collagen peptide data. C) Example MS/MS of hydroxylated proline peptide fragmentation. D) Collagen 3A1 hydroxylated proline peptide matched by accurate mass demonstrates high signature in the tumor region of the lung tissue section. Comparison of the same putative peptide in the imaging data by accurate mass demonstrates the modified COL3A1 peptide present almost exclusively in lung adenocarcinoma patients, not in normal tissue.
Conclusion
Extracellular matrix has key regulatory roles in the course of lung adenocarcinoma; functions of ECM that promote progression are an emerging topic for therapeutic strategies [38]. The ability to explore ECM components as a therapeutic route has been challenging due to lack of available tools for detecting molecular features of ECM components. Tools for investigation of ECM in formalin-fixed paraffin-embedded samples, used for preserving clinical specimens, are especially limited due to the additional complication of formalin crosslinking. This preservation excludes options for examining the decellularized tissue structure. In the current work we explored low-grade lung adenocarcinoma compared to normal lung tissue using our novel workflow that targets ECM proteins. We believe the approach targets mainly extracellular ECM compared to intracellular ECM due to the size of the laser spot and comparison of stained tissue to correlative ECM peptide images shown here and in work previously published by our group [18, 19]. The imaging mass spectrometry data reported that there are many peptides from collagenase type III digest that can discriminate between normal and low-grade lung adenocarcinoma. This data was demonstrated in both a tissue microarray and a wedge resection representative of the complex tumor microenvironment. In-solution proteomics on single tissue sections confirmed that the ECM IMS workflow identifies several types of collagens and their interacting proteins. Comparison with conventional tryptic digestion proteomics on ECM from cell culture and fresh tissue demonstrated good overlap in protein identifications. This included identifying a common hydroxylated lysine residue (K277) in COL1A1 sensitive to TGFβ1 stimulation[27]. Proteins identified associated with cell-collagen adhesion, which could be useful for understanding immune cell recruitment throughout the tumor and linked to oxidative stress, including the unfolded protein response, could be leveraged to understand mechanisms of ECM deposition in emergent cancer. An additional finding was that collagen peptides with hydroxylated proline residues were sequenced and mapped from the wedge resection proteome. By accurate mass matching, a hydroxylated proline peptide from COL3A1 could distinguish between the tumor region and normal region. Hydroxylated proline type residues have a critical role in stabilizing the collagen structure which feeds sensory information to cells [32–34] and thus could represent new research avenues for lung adenocarcinoma. More work needs to be done over many tissue types to evaluate detection of hydroxylated proline type residues when using collagenase type enzymes for proteomics.
In conclusion, ECM IMS, including the associated in-solution workflows, is an important advance towards understanding low-grade adenocarcinoma represented in clinically preserved FFPE specimens. Non-invasive imaging probes have been developed that target collagen 1[16] or contrast against collagen type I [17]. We suggest that the molecular data produced by ECM IMS is useful in pinpointing specific peptide components of collagen altered by emergent adenocarcinoma. Studies on larger cohorts could be used to develop new tests that improve detection and accuracy of low-grade lung adenocarcinoma.
Supplementary Material
Acknowledgements
PMA is supported by P20GM103542 (NIH/NIGMS), HL007260 (NHLBI), and in part by pilot research funding, Hollings Cancer Center Support Grant P30 CA138313 at the Medical University of South Carolina. Additional support was provided by the South Carolina Centers of Economic Excellence SmartState program to RRD and CFB. The Mass Spectrometry Facility and Redox Proteomics Core is supported by the University and P20GM103542 (NIH/NIGMS) with shared instrumentation S10 OD010731 and S10 OD025126 (NIH/OD) to LEB.
Abbreviations:
- COLase3
collagenase type III
- ECM
extracellular matrix
- ECM IMS
extracellular matrix imaging mass spectrometry
- LUAD
lung adenocarcinoma
- MALDI IMS
matrix-assisted laser desorption / ionization imaging mass spectrometry
- TMA
tissue microarray
References
- 1.American Cancer Society. Facts & Figures 2019. American Cancer Society; Atlanta, G. [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A: Cancer Statistics, 2016. CA: a cancer journal for clinicians; 66, 7–30 (2016) [DOI] [PubMed] [Google Scholar]
- 3.Testa U, Castelli G, Pelosi E: Lung cancers: molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers. 10, 248 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemjabbar-Alaoui H, Hassan OUI, Yang Y-W, Buchanan P: Lung cancer: Biology and treatment options. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 1856, 189–210 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alberg AJ, Ford JG, Samet JM: Epidemiology of lung cancer: ACCP evidence-based clinical practice guidelines. Chest. 132, 29S–55S (2007) [DOI] [PubMed] [Google Scholar]
- 6.Borczuk AC: Prognostic considerations of the new World Health Organization classification of lung adenocarcinoma. European Respiratory Review. 25, 364–371 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Lung Screening Trial Research, T.: Reduced lung-cancer mortality with low-dose computed tomographic screening. New England Journal of Medicine. 365, 395–409 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabrikant MS, Wisnivesky JP, Marron T, Taioli E, Veluswamy RR: Benefits and challenges of lung cancer screening in older adults. Clinical Therapeutics. 40, 526–534 (2018) [DOI] [PubMed] [Google Scholar]
- 9.Walker C, Mojares E, del Río Hernández A: Role of extracellular matrix in development and cancer progression. International Journal of Molecular Sciences. 19, 3028 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox TR, Erler JT: Molecular pathways: connecting fibrosis and solid tumor metastasis. Clinical Cancer Research. 20, 3637–3643 (2014) [DOI] [PubMed] [Google Scholar]
- 11.Bissell MJ, Hall HG, Parry G: How does the extracellular matrix direct gene expression? Journal of Theoretical Biology. 99, 31–68 (1982) [DOI] [PubMed] [Google Scholar]
- 12.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D: Tensional homeostasis and the malignant phenotype. Cancer Cell. 8, 241–254 (2005) [DOI] [PubMed] [Google Scholar]
- 13.Yamauchi M, Barker TH, Gibbons DL, Kurie JM: The fibrotic tumor stroma. The Journal of Clinical Investigation. 128, 16–25 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xi K-X, Wen Y-S, Zhu C-M, Yu X-Y, Qin R-Q, Zhang X-W, Lin Y-B, Rong T-H, Wang W-D, Chen Y-Q: Tumor-stroma ratio (TSR) in non-small cell lung cancer (NSCLC) patients after lung resection is a prognostic factor for survival. Journal of Thoracic Disease. 9, 4017 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamazaki S, Higuchi Y, Ishibashi M, Hashimoto H, Yasunaga M, Matsumura Y, Tsuchihara K, Tsuboi M, Goto K, Ochiai A: Collagen type I induces EGFR-TKI resistance in EGFR‐mutated cancer cells by mTOR activation through Akt-independent pathway. Cancer Science. 109, 2063–2073 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrar CT, Gale EM, Kennan R, Ramsay I, Masia R, Arora G, Looby K, Wei L, Kalpathy-Cramer J, Bunzel MM: CM-101: type I collagen–targeted MR imaging probe for detection of liver fibrosis. Radiology. 287, 581–589 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caravan P, Das B, Dumas S, Epstein FH, Helm PA, Jacques V, Koerner S, Kolodziej A, Shen L, Sun WC: Collagen-targeted MRI contrast agent for molecular imaging of fibrosis. Angewandte Chemie International Edition. 46, 8171–8173 (2007) [DOI] [PubMed] [Google Scholar]
- 18.Angel PM, Comte-Walters S, Ball LE, Talbot K, Brockbank KGM, Mehta AS, Drake RR: Mapping Extracellular Matrix Proteins in Formalin-Fixed, Paraffin-embedded Tissues by MALDI Imaging Mass Spectrometry. Journal of Proteome Research. 17, 635–646. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angel PM, Schwamborn K, Comte-Walters S, Clift C, Ball LE, Mehta AS, Drake RR: Extracellular Matrix Imaging of Breast Tissue Pathologies by MALDI Imaging Mass Spectrometry. Proteomics Clinical Applications. 13, (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metsalu T, Vilo J: ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Research. 43, W566–W570 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bern M, Kil YJ, Becker C: Byonic: advanced peptide and protein identification software. Current Protocols in Bioinformatics. 13–20. (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyanova S, Temu T, Cox J: The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature Protocols. 11, 2301–2319 (2016) [DOI] [PubMed] [Google Scholar]
- 23.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J: The Perseus computational platform for comprehensive analysis of (prote) omics data. Nature methods. 13, 731 (2016) [DOI] [PubMed] [Google Scholar]
- 24.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M: UniProt: the universal protein knowledgebase. Nucleic Acids Research. 47, D506–515 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lochhead P, Chan AT, Nishihara R, Fuchs CS, Beck AH, Giovannucci E, Ogino S: Etiologic field effect: reappraisal of the field effect concept in cancer predisposition and progression. Modern Pathology. 28, 14 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang S, Dai Y, Mei Y, Yang M, Hu L, Yang H, Guan X, Li J: Clinical significance and biological role of cancer-derived Type I collagen in lung and esophageal cancers. Thoracic Cancer. 10, 277–288 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merl-Pham J, Basak T, Knüppel L, Ramanujam D, Athanason M, Behr J, Engelhardt S, Eickelberg O, Hauck SM, Vanacore R: Quantitative proteomic profiling of extracellular matrix and site-specific collagen post-translational modifications in an in vitro model of lung fibrosis. Matrix Biology Plus. 1, 100005 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gocheva V, Naba A, Bhutkar A, Guardia T, Miller KM, Li CM-C, Dayton TL, Sanchez-Rivera FJ, Kim-Kiselak C, Jailkhani N: Quantitative proteomics identify Tenascin-C as a promoter of lung cancer progression and contributor to a signature prognostic of patient survival. Proceedings of the National Academy of Sciences. 114, E5625–E5634 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrier A, Boyer JG, Kothary R: Cellular and molecular biology of neuronal dystonin. Elsevier, (2013) [DOI] [PubMed] [Google Scholar]
- 30.Shoulders MD, Raines RT: Collagen structure and stability. Annual Review of Biochemistry. 78, 929–958. (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gelse K, Pöschl E, Aigner T: Collagens—structure, function, and biosynthesis. Advanced Drug Delivery Reviews. 55, 1531–1546 (2003) [DOI] [PubMed] [Google Scholar]
- 32.Aikawa E, Whittaker P, Farber M, Mendelson K, Padera RF, Aikawa M, Schoen FJ: Human Semilunar Cardiac Valve Remodeling by Activated Cells From Fetus to Adult: Implications for Postnatal Adaptation, Pathology, and Tissue Engineering. Circulation. 113, 1344–1352. (2006) [DOI] [PubMed] [Google Scholar]
- 33.Vellinga TT, den Uil S, Rinkes IHB, Marvin D, Ponsioen B, Alvarez-Varela A, Fatrai S, Scheele C, Zwijnenburg DA, Snippert H: Collagen-rich stroma in aggressive colon tumors induces mesenchymal gene expression and tumor cell invasion. Oncogene. 35, 5263–5271. (2016) [DOI] [PubMed] [Google Scholar]
- 34.Lu P, Takai K, Weaver VM, Werb Z: Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harbor Perspectives in Biology. 3, a005058 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu X, Zhu C-C, Liu H, Wang J-C: Expression of hypoxia-inducible factor prolyl hydroxylase 3 HIFPH3 in human non-small cell lung cancer (NSCLC) and its correlation with prognosis. Asian Pacific Journal of Cancer Prevention. 15, 5819–5823 (2014) [DOI] [PubMed] [Google Scholar]
- 36.Eckhard U, Huesgen PF, Brandstetter H, Overall CM: Proteomic protease specificity profiling of clostridial collagenases reveals their intrinsic nature as dedicated degraders of collagen. Journal of proteomics. 100, 102–114. (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckhard U, Schönauer E, Nüss D, Brandstetter H: Structure of collagenase G reveals a chew-and-digest mechanism of bacterial collagenolysis. Nature Structural & Molecular Biology. 18, 1109 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Götte M, Kovalszky I: Extracellular matrix functions in lung cancer. Matrix Biology. 73, 105–121 (2018) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


