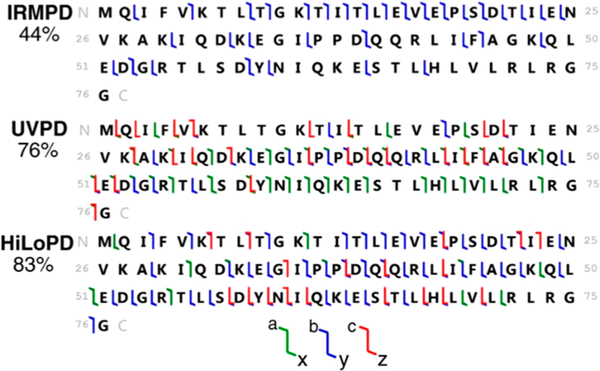Abstract
The development of new ion-activation/dissociation methods continues to be one of the most active areas of mass spectrometry owing to the broad applications of tandem mass spectrometry in the identification and structural characterization of molecules. This Review will showcase the impact of ultraviolet photodissociation (UVPD) as a frontier strategy for generating informative fragmentation patterns of ions, especially for biological molecules whose complicated structures, subtle modifications, and large sizes often impede molecular characterization. UVPD energizes ions via absorption of high-energy photons, which allows access to new dissociation pathways relative to more conventional ion-activation methods. Applications of UVPD for the analysis of peptides, proteins, lipids, and other classes of biologically relevant molecules are emphasized in this Review.
Graphical Abstract

1. INTRODUCTION
1.1. Scope of Review
Tremendous advances in the scope and applications of mass spectrometry have catalyzed interest in exploring and developing new ion-activation methods. The success of tandem mass spectrometry (MS/MS) is largely driven by the ability to generate fragmentation patterns of ions that allow confirmation of their identities or characterization of new structural features. MS/MS spectra afford diagnostic molecular fingerprints that have been used to decipher sequences of biopolymers such as peptides, proteins, and nucleic acids as well as a myriad of other classes of molecules, ranging from small molecules like drugs, pesticides, and metabolites to other types of biological molecules, such as lipids and polysaccharides. There have been many past reviews that have focused on tandem mass spectrometry for analysis of different classes of molecules, and readers are directed to these reviews.1–14 Fragmentation of molecular ions requires deposition of internal energy; this frames the basis of the ion-activation process and can be implemented in a number of ways. Traditional collisional-based methods (commonly known as collision-induced dissociation (CID) or collisional-activation dissociation (CAD)) are the most widely used and most successful methods for tandem mass spectrometry.15 The quest for higher-energy deposition and alternative activation mechanisms to access new fragmentation pathways and overcome limitations of collision-based methods has led to the development of many alternative techniques, including electron-based methods (electron-capture dissociation (ECD) and electron-transfer dissociation (ETD)),16 surface-induced dissociation (SID),17 and ion–ion reactions,18 as well as newly emerging hybrid approaches19 that combine two activation methods, such as electron-transfer higher-energy collision dissociation (EThcD).20 Another type of activation method, termed photodissociation, relies on the absorption of photons for energy deposition. Photodissociation methods have been developed using photons spanning a range of energies, from low-energy infrared photons (i.e., infrared multiphoton dissociation (IRMPD)21) to the extreme ultraviolet range (40–80 nm),22 which leads to dissociative ionization. New photon-based ion-activation methods continue to emerge owing to the diverse array of light sources (lasers, light-emitting diodes (LEDs), lamps, and synchrotron radiation sources) and wavelength ranges that mediate ion-activation/dissociation mechanisms.
This Review will focus on applications of ultraviolet photodissociation (UVPD), a method that has garnered significant attention over the past decade and couples well with mass spectrometry. The type of photodissociation described here is initiated using a single wavelength for activation, such as the most commonly used 157, 193, or 266 nm wavelengths. This Review will primarily highlight new developments of UVPD for the analysis of biological molecules over the past 5 years, ranging from peptides, proteins, and protein complexes to lipids, nucleic acids, and carbohydrates (Figure 1). Readers are directed to other previous reviews that have covered work prior to 2014.23–26 Owing to the large number of applications that have used 193 nm photons for UVPD, this wavelength is showcased in the present Review. Advances in ion spectroscopy, in which ion fragmentation is monitored as a function of wavelength, have been covered in a number of excellent reviews and will not be addressed here.27–35 Moreover, this Review will not discuss the numerous studies of UV photodissociation for small molecules.
Figure 1.
UVPD has been used to characterize numerous classes of biological molecules.
Photodissociation entails the exposure of ions to photons, generally of a specific wavelength characteristic of a particular light source. For UVPD, ion-activation occurs via the absorption of one or more high-energy UV photons. UV radiation may be subdivided based on the range of wavelength: vacuum-UV or far-UV is 10–200 nm, middle-UV extends from 200 to 300 nm (UV-C is 200–280 nm, and UV-B is 280–315 nm), and near-UV or UV-A spans 300–400 nm. UVPD is most commonly implemented using a pulsed laser to deliver a high flux of photons in a short period, often just a few ns. The number of pulses, the pulse energy, and the wavelength are key parameters that modulate energy deposition and affect the outcome of UVPD in terms of the mechanism of photoactivation, fragmentation efficiency, and both type and distribution of fragment ions. UV photoabsorption results in excitation of ions to excited electronic states, a process that allows access to higher-energy dissociation pathways not typically observed upon low-energy collisional activation. The detailed mechanism of UVPD is not fully resolved.36 After electronic excitation, ions may dissociate directly from the excited states; these are the fragmentation channels that are most unique to UVPD.29 In other cases, relaxation via internal conversion to the ground electronic state along with intramolecular vibrational energy redistribution (IVR) results in other lower-energy fragmentation pathways that are more akin to ones induced by conventional collisional-activation methods.
Integrating UV lasers with mass spectrometry for photodissociation was first reported over 30 years ago37 and followed with a limited number of studies,38–41 but the strategy was not widely adopted for characterization of biological molecules owing to the low signal-to-noise of mass spectra and limited availability of ion-trapping mass spectrometers (which are arguably more well-suited for UVPD than other types of MS platforms). The more recent resurgence and success of UVPD that began in 200442 and expanded over the past 15 years has capitalized on substantial improvements in the performance metrics of mass spectrometers and significant advances in ionization methods that facilitate analysis of biological molecules. High-resolution/high-accuracy mass measurements are especially instrumental in allowing dense MS/MS spectra, like ones commonly generated by UVPD, to be deciphered.
Another variation of photon-based activation method is electron-photodetachment dissociation (EPD).35 In this process, electrons are detached from multicharged anions after absorption of UV (or visible) photons, resulting in the production of charge-reduced radical anions. Subsequent fragmentation of the radical anions is frequently promoted by collisional activation, and this combined EPD/collisional-activation process is known as activated-electron-photodetachment dissociation (a-EPD). EPD or a-EPD methods have been used for analysis of oligonucleotides, peptides, proteins, oligosaccharides, lipids, and polymers. The mechanisms and applications of electron photodetachment have been reviewed elsewhere.35
1.2. Instrumentation
UVPD has been implemented predominantly on ion-trap mass spectrometers,43,44 and to a lesser extent on time-of-flight (TOF)42,45 and quadrupole TOF (Q-TOF)46 systems. The TOF and Q-TOF platforms have been less popular for UVPD for practical reasons, such as more difficult synchronization of laser pulses with ion-flight times (in the case of TOF-based systems) or more challenging optical access (for TOF and Q-TOF instruments). However, some of the commercial Q-TOF systems have often been equipped with ion-mobility capabilities and thus offer an additional experimental feature not routinely available on ion-trapping systems (linear ion trap, Orbitrap, Fourier-transform ion cyclotron resonance (FTICR)).46 UVPD has been integrated with 3D quadrupole ion traps and 2D linear ion traps, as well as more recently with Orbitrap systems.47–50 For the Orbitrap platforms, UVPD does not occur in the Orbitrap analyzer but rather in an higher-energy C-trap dissociation (HCD) cell or in a linear ion trap prior to mass analysis in the Orbitrap analyzer (Figure 2). The versatility of UVPD for applications in the fields of lipidomics, glycomics, and proteomics has motivated adaptation of UVPD to other platforms, including FTICR mass spectrometers51 and Q-TOFs equipped with ion-mobility capabilities.52
Figure 2.
Implementation of UVPD on (a) an Orbitrap Lumos mass spectrometer50 and (b) Synapt Q-TOF mass spectrometer.52 Adapted with permission from refs 50 and 52. Copyright 2016 and 2018 American Chemical Society, respectively.
All of the ion-trapping systems are naturally well-suited for UVPD because ions can be accumulated and stored during the photoactivation event, allowing more facile control of energy deposition, fragmentation efficiency, and S/N of the resulting spectra. For the aforementioned developments, UVPD has been primarily implemented in a semicustomized, in-house fashion in which lasers or other light sources have been integrated with commercial mass spectrometers by individual research groups, often with technical assistance or software modifications provided by the manufacturer of the mass spectrometer. The primary instrument modifications required for implementation of UVPD include addition of an optical window, inclusion of optics (lens and/or mirrors) as needed to direct the photons to the ions, and a triggering method to synchronize the photoirradiation with the ion-analysis events. The experimental parameters for enabling UVPD are often modulated to prevent excessive energy deposition, which can lead to decomposition of precursor ions into overly small fragments that lack diagnostic value as well as annihilation of primary product ions. In 2017, the first fully integrated commercial version of UVPD using 213 nm photons from a solid-state laser was launched on an Orbitrap mass spectrometer, and publications have started to emerge based on utilization of this type of fully commercial UVPD-MS system.53–55
A variety of light sources have been used for UVPD, including excimer lasers (e.g., F2: 157 nm, 7.9 eV per photon;42 ArF: 193 nm, 6.4 eV per photon;47 and XeF: 351 nm, 3.5 eV per photon56), solid state Nd:YAG lasers (213 nm, 5.8 eV per photon;52 266 nm, 4.7 eV per photon;44 or 355 nm, 3.5 eV per photon43), gas-discharge lamps,22 synchrotron radiation,57 and even light-emitting diodes (LEDs) in one report.58 More powerful lasers or ones with high repetition rates enable UVPD on a fast time scale and are particularly beneficial for higher-throughput LC-MS applications. Tunable lasers have also been employed, affording the ability to evaluate UVPD over a broader range of wavelengths to enable ion spectroscopy experiments.27–29
2. UVPD FOR PEPTIDES
2.1. Mechanistic Studies of Peptides
Integral to the effective application of UVPD for characterization of biomolecules is the development of a mechanistic understanding of photodissociation in model systems, particularly peptides because of the vast interest in proteomics. In fact, most of the mechanistic studies to date have focused on peptides, in large part owing to their linear structures and well-developed fragmentation nomenclature (Figure 3) or even individual amino acids.
Figure 3.
Fragmentation nomenclature for peptides and proteins: a/x, b/y, and c/z ions according to cleavage of backbone bonds. The subscript indicates the number of residues contained in the product, as illustrated for a tetrapeptide.
To this end, several groups have actively pursued fundamental studies focused on UVPD of peptides using 157, 193, 213, and 266 nm photons to induce fragmentation.29,36,59–76 Readers are directed to these studies for a deeper mechanistic understanding as only a brief overview of key findings and milestone studies is summarized in this section. The diversity of fragments produced upon UVPD has generally been observed to be greater for wavelengths below 266 nm; consequently, a number of studies using 157, 193, and 213 nm photons have examined peptide fragmentation as a function of peptide size and sequence, charge state, charge location, and ion polarity. Examination of UVPD as a function of charge state and charge site has also been undertaken to understand fragmentation pathways of peptides.60,66,54,73,76 The peptide backbone has a number of chromophores, and irradiation with 157, 193, and 220 nm photons aligns well with numerous predicted electronic transitions (e.g., π-to-π* involving π orbitals of amide group and n-to-π* involving nonbonding electrons of oxygen of carbonyl) occurring around 160, 190, or 220 nm.77
The Reilly group was the first to examine the mechanism of UVPD and presented a number of studies demonstrating that 157 nm UVPD of peptides having a C-terminal arginine resulted in the production of abundant x- (and x + 1) and y-type ions in addition to v- and w-type ions, which result from side-chain cleavage at particular amino acids.59–61 In contrast, peptides protonated with an N-terminal arginine were found to produce predominantly a- (and a + 1) and d-type ions upon 157 nm UVPD (Figure 4).59 On the basis of the production of x + 1 and a + 1 radical ions and the absence of thermal b- and y-type ions, Reilly proposed fragmentation to occur via a Norrish type I reaction in which 157 nm photoactivation resulted in direct access to a dissociative state.42
Figure 4.
Production of a + 1/a and x + 1/x ions from UVPD of a protonated peptide. Adapted with permission from ref 59. Copyright 2005 Elsevier.
This was later confirmed in a density functional theory (DFT) study of ArgAla and AlaArg dipeptides that demonstrated that 157 nm UVPD accesses a high-lying Rydberg state.61 Irradiation of peptides presumably protonated at terminal lysine residues rather than arginine was observed to produce more abundant b- and y-type ions, in addition to x-, a-, v-, and d-type ions, suggesting that vibrational-energy redistribution contributes to secondary processes such as hydrogen or side-chain loss as well as production of b- and y-type ions.61 Other studies have examined secondary dissociation processes rigorously, and it was demonstrated through hydrogen/deuterium exchange that β-hydrogen and amide hydrogen elimination occurred from a + 1 radical ions to result in even electron a ions, explaining the diversity of a + 1, a, x + 1, and x ions commonly observed upon UVPD.62 A more recent study further explored hydrogen elimination observed in fragment ions produced by 193 nm UVPD using a statistical analysis of a large number of peptides, all terminated in Lys at the first residue.78 It was deduced that hydrogen elimination varied as a function of the identity of the amino acid situated N-terminal to the backbone-cleavage site, presumably due to the structure of the side chain, and the presence or absence of hydrogen bonding adjacent to the cleaved residue.78
The Reilly group has also undertaken a comparison of the dissociation of singly charged peptides upon 157 and 193 nm UVPD in both a MALDI-TOF and linear ion-trap instrument.60 Fragment ions produced upon 157 and 193 nm activation were found to be remarkably similar in the ion-trap and TOF instruments, although higher mass fragments from 157 nm vs 193 nm UVPD were found to be more abundant in the TOF.60 Data collected using 193 nm dissociation on TOF instruments using short 1 and 10 μs analysis time scales echoed these results.41,45,60,64 It has been previously noted that 193 nm UVPD results in more-abundant thermal fragments than does activation with 157 nm, an effect undoubtedly related to excitation of different electronic transitions at the two wavelengths.59 Interestingly, this effect was not strongly evident in a comparison of 157 and 193 nm UVPD of singly charged peptides.60 A systematic study of the 193 and 266 nm fragmentation of six adrenocorticotropic hormone (ACTH) peptide variants as a function of charge state demonstrated that thermal b- and y-type ions were far more prevalent for higher charge states of the two smallest ACTH variants (residues 1–10 and 1–14), suggesting that vibrational-deactivation pathways are more competitive in smaller, more highly charged peptides.65 Fast and slow dissociation channels were explored using time-resolved photodissociation studies on a matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) platform with 193 and 266 nm UVPD of peptides containing C-terminal arginine residues.66 Similar spectra were obtained using 266 and 193 nm photoactivation, featuring dominant b-type ions and lower-abundance v-, w-, x-, y-, and z-type ions.66 The similarity of the spectra regardless of activation wavelength led to the unique consideration of reaction kinetics as a function of total energy deposited by the two wavelengths rather than the different electronic states accessed. Using a photodissociation cell floated at 2 keV, the authors were able to distinguish ions produced inside the cell from ions produced outside of the cell.66 High-energy product ions were found to arise from a common intermediate, the x + 1 ion.66 Interestingly, high- (x-, w-, y-, a-, and d-type) and low-energy (b and y) products were found to evolve from pathways with separate rate constants, from which the authors concluded that the high- and low-energy pathways were not in direct competition.66 Several possibilities were explored to account for this, including multiphoton effects, internal conversions, thermal energy differences, and conformational differences in the ion population. Although none of these studies were completely conclusive, conformational differences could account for some of the observed differences.
The third peptide backbone chromophore features an absorption maximum around 220 nm. Peptide fragmentation has been explored using a tunable laser27,29,33 and more recently has been explored via UVPD of peptides using the fifth harmonic of Nd:YAG lasers (213 nm). Mirroring an earlier study by the Reilly group in which bn + 2 and an + 2 fragment ions were observed in proline-containing peptides,67 213 nm UVPD was used in conjunction with molecular dynamics and DFT calculations to probe the electronic states accessed for photoactivation of proline-containing peptides.72 Upon 213 nm photoactivation of several peptides, yn − 2, bn + 2, and an + 2 products were found to exclusively occur from cleavage N-terminal to proline residues, from which it was concluded that a novel mechanism contributed to the formation of these ions, which was linked to the structure of the prolyl ring.72 Homolytic cleavage of the C–N bond was found to proceed with simultaneous rearrangement and elimination of both CO and two radical hydrogens to generate yn − 2 ions for C-terminal protonation.72 Interestingly, a different mechanism was found to account for fragmentation for N-terminally protonated peptides, in which C–C activation and subsequent proton rearrangements result in production of bn + 2 and an + 2 fragments; this is in agreement with a study using 193 nm UVPD in which different fragment types were observed for peptides featuring N-terminal vs C-terminal protonation sites.73
The fragmentation of peptide anions has also been explored. Photoactivation of negatively charged peptides typically results in electron photodetachment to generate hydrogen-deficient radicals, from which additional collisional activation can be used to induce dissociation, a process termed EPD.35 Many unique ion types were identified, including radical an + 1, an, an + 2, xn, radical xn + 1, xn + 2, and xn − 1,75, 75 as well as numerous side-chain-loss fragments and b/y, c/z, yn − 1, yn − 2, bn + 2, among others.75
In summary, numerous investigations of the dissociation pathways of peptides upon irradiation by 157, 193, and 213 nm photons indicate production of diverse ion types as a consequence of electronic excitation. Evidence points to fragmentation pathways consistent wtih direct dissociation from the excited electronic states in addition to ones that spring from internal conversion and intramolecular vibrational energy redistribution, with the latter ultimately resulting in b/y-type ions commonly created upon collisional activation.
2.2. UVPD for Peptide Sequencing and Bottom-Up Proteomics
The impressive performance of UVPD for fragmentation of peptides motivated the utilization of UVPD for more elaborate applications in proteomics. Proteomics entails the larger-scale study of proteins in a biological context, a challenging, multilevel problem for which mass spectrometry has proven to be the preeminent analytical strategy.1 Implementation of UVPD for proteomics has ranged from bottom-up approaches entailing analysis of peptides generated from enzymatic digestion of proteins to top-down strategies that focus on characterization of intact proteins.1 In the context of high-throughput proteomics, UVPD is particularly compelling because its fast time scale, high-energy deposition, and unique mechanisms afford extensive fragmentation of peptides and proteins and allow retention of modifications, an outcome that is critical for localization of post-translational modifications. A number of clever tagging strategies have also been developed to incorporate chromophores that allow a large degree of fragmentation selectivity, either via incorporation of photocleavable motifs or highly specific photoabsorption properties. De novo sequencing based on UVPD has also emerged as a promising approach owing to the ability to modulate the extent of peptide fragmentation and types of product ions based on the wavelength utilized for photoactivation.
Applications of UVPD for bottom-up proteomics began to surface over a decade ago, primarily using 193 nm photons.79–81 The high sequence coverage offered by UVPD and its lack of dependence on charge state were recognized as advantageous attributes of 193 nm UVPD for analysis of peptides using both positive and negative ionization modes.79–81 UVPD (193 nm) has also been used to characterize cyclic peptides, including ones representative of stapled peptides,82 developed as a new type of biostable therapeutic; antimicrobial cyclotides,83 disulfide-rich peptides derived from plants; and microcystins,84 nonribosomal peptides generated as metabolites in cyanobacteria. For characterization of nonconventional peptides like the aforementioned cyclic ones, CID and 193 nm UVPD have typically been used in a comparative manner, resulting in the general outcome that the two methods often yielded orthogonal information that could be used in a complementary fashion to enhance characterization of the most intractable structures.
The versatility of UVPD for proteomic analysis was examined more extensively by incorporating alternative proteases into the bottom-up workflow85 and by integrating both positive-mode and negative-mode data in a complementary manner.86 The diverse array of fragment ions created by 193 nm UVPD is illustrated in Figure 5 for identification of proteins from a bacterial cell lysate using a bottom-up approach, with both the types of ions and the distribution of N-terminal (a, b, c) and C-terminal (x, y, z) varying considerably for UVPD versus higher-energy collisional dissociation (HCD).85 In addition, side-chain losses were only consistently observed for UVPD.85
Figure 5.
Distribution of 193 nm UVPD and HCD fragment ions for different proteases (GluC, LysC, trypsin, and chymotrypsin) used for Halobacterium cell lysate. The abundances of the various fragment ion types were compiled for the 25 most confidently identified peptides (based on XCorr scores) from each digest. Reproduced with permission from ref 85. Copyright 2015 American Chemical Society.
Deprotonated peptides produce a/x-type ions upon absorption of 193 nm photons, representing one of the unique signatures of UVPD. Significant differences in the fragmentation patterns of protonated and deprotonated peptides were observed upon UVPD, with UVPD favoring the production of a/x ions for deprotonated peptides in comparison to b/y ions for protonated peptides (Figure 6).86 Although using the negative polarity allows analysis of classes of peptides not readily detected in the positive mode, generating peptides as anions may be impeded by low ionization efficiencies.
Figure 6.
(A) HCD (3+), (B) 193 nm UVPD (3+), and (C) 193 nm UVPD (2−) spectra of tryptic peptide LNDGHFMPVLGFGTYAPPEVPR showing complementary information from each technique. Combining all methods yields 70% coverage by a- and b-type ions and 70% coverage by x- and y-type ions. Reproduced with permission from ref 86. Copyright 2018 American Chemical Society.
To circumvent this issue, a workflow incorporating carbamylation of lysine resides of peptides was undertaken prior to 193 nm UVPD.87 Carbamylation was employed to convert basic amine groups to nonbasic amide groups, thus reducing the basicities of the peptides and enhancing their ionization in the negative mode. This strategy was used for characterization of the acidic proteome of H. salinarum.87 The increased charge state of the precursor ions in the negative mode afforded by carbamylation of lysine residues (and the peptide N-terminus) provided deeper characterization of the proteome in the negative mode (Figure 7). Combining carbamylation with 193 nm UVPD resulted in identification of 1086 peptides corresponding to 430 proteins, outperforming UVPD of an unmodified digest by 25%.87
Figure 7.
UVPD (193 nm) mass spectra of GEEVTAEVADGPQSVIFDQAENR from H. salinarum: (A) unmodified peptide (2−) and (B) carbamylated peptide (2−). # indicates the loss of water; Δ represents carbamylation. Combined number of unique (C) peptide and (D) protein identifications from LC/UVPD-MS analyses of carbamylated and unmodified tryptic peptides from H. salinarum in the negative mode in triplicate. Reproduced with permission from ref 87. Copyright 2014 American Chemical Society.
2.3. UVPD for Identification of Post-Translational Modifications
Post-translational modification (PTM) of proteins plays an enormous role in modulating the structural and functional diversity of the proteome, and the large array of potential modifications mediate protein interactions, signaling, catalytic activity, and numerous other processes.88 The comprehensive mapping of post-translational modifications is impeded by the large range of PTMs, their dynamic nature, and the critical impact of their locations. Despite the significant advances in MS/MS methods for profiling and quantifying proteins, pinpointing PTMs remains challenging.89 Many conventional collisional activation methods cause preferential cleavages of labile PTMs, mitigating the ability to localize their sites.89 In addition, the most confident localization demands that the sites be bracketed by backbone cleavages adjacent to the sites of PTMs, thus requiring activation methods that provide extensive fragmentation. This latter issue motivated the exploration of UVPD for mapping PTMs.
Modulation of phosphorylation in proteins is integral to cell signaling, and characterization of individual and combinations of phosphorylation sites has become an important aspect of understanding cellular communication and monitoring the progression of a variety of diseases.90 Localization of phosphorylation sites through MS/MS approaches is one of the most common goals in proteomics. Electron-transfer dissociation, useful for sequencing and localization because phosphate groups remain appended to the peptides, and collision-induced dissociation, in which the production of diagnostic phosphate loss ions allow facile identification of phosphorylated peptides, are often used in combination.89 A number of studies have previously investigated the utility of photodissociation as an alternative to the electron- and collision-based activation methods, utilizing the diagnostic fragment ions to localize the phosphate groups.48,73,79,91–96 In one of the first applications of UVPD for phosphopeptide analysis, singly protonated phosphopeptides were activated using 193 nm photons, producing unusual an − 97 ions exclusively for cleavages C-terminal to the residue bearing the phosphate moiety.94 This unique fragmentation pathway provided a direct means to localize the phosphate moiety, although the formation of the diagnostic an − 97 ions was restricted to peptides bearing a proton-sequestering residue at the N-terminus.94 UVPD (193 nm) was implemented in an LC-MS mode to allow higher-throughput analysis of peptides, including phosphopeptides, for proteomics applications.79 A follow-up study explored a hybrid MS/MS method combining electron-transfer reactions to generate radical ions and UVPD to create fragmentation patterns dominated by z ions for identification of new phosphorylation sites in the protein TrpM7.73
Application of UVPD to phosphoproteomics was further extended using 193 nm UVPD implemented on a Q Exactive Orbitrap mass spectrometer.48 The number of peptide spectral matches (PSMs) from a phosphoenriched HeLa lysate was compared for UVPD and HCD, and the degree of phosphate retention was monitored for both activation techniques.48 Examples of the HCD and UVPD mass spectra of a phosphopeptide are shown in Figure 8. While the total number of peptide spectral matches was higher for HCD than UVPD by 20%, the prevalence of phosphorylation loss from diagnostic product ions was 45% lower for UVPD, suggesting better retention of the modifications and presumably better capacity to localize the phosphate moieties.48
Figure 8.
Comparison of (A) HCD and (B) 193 nm UVPD for analysis of phosphopeptide APPDNLPSPGGSR. The modification losses are encoded as (1) H3PO4 + H2O, (2) H3PO4, and (3) full modification or HPO3. The insets show the contribution of neutral losses of phosphomodifications (red pie segments) to the total ion current. (C) Percentage of neutral losses of phosphorylation modifications caused by HCD (red bars) versus UVPD (blue bars) for mono-, bi-, and triphosphorylated peptides, expressed as a percentage of total ion current. Reproduced with permission from ref 48. Copyright 2016 American Chemical Society.
The performance of UVPD for analysis of phosphopeptides from HeLa and HCC70 primary ductal carcinoma lysates was evaluated on an Orbitrap Fusion mass spectrometer in both positive and negative ionization modes with the latter being particularly useful for multiply phosphorylated peptides.95 In comparison to HCD, the richer fragmentation patterns generated by 193 nm UVPD enhanced the ability to pinpoint the phosphorylation sites with confidence, although again it was found that HCD identified a larger total number of phosphopeptides.95 UVPD was also applied to characterize phosphorylation sites of the C-terminal domain (CTD) of RNA polymerase II, a protein that is important in the transcription process. The ability to map the phosphorylation of CTD is impeded by its repeating heptad motifs for which the pattern of phosphorylation creates a complicated code that modulates the specific function of the protein.96 Phosphorylation of CTD by two different kinases, TFIIH and Erk2, was elucidated in detail using the diagnostic fragmentation patterns produced by UVPD.96 UVPD bracketed each potential phosphorylation site via backbone cleavages adjacent to each of the Ser, Tyr, and Thr residues in the heptads, as illustrated in Figure 9. Examples of the 193 nm UVPD spectra acquired for one representative CTD peptide in both positive and negative modes are shown in Figure 9, along with a map of the phosphorylation sites derived for a panel of CTD peptides. The large array of phosphopeptides deciphered by UVPD allowed construction of a “rule book” summarizing the sequence motifs that guided phosphorylation, as illustrated in Figure 9D.96
Figure 9.
Phosphorylations identified in Drosophila melanogaster C-terminal domain (CTD) of RNA polymerase II following treatment with Erk2 using LC-UVPD-MS. (A) Sites of phosphorylation in Drosophila melanogaster CTD, where confirmed sites are highlighted in green. One peptide (repeat 28) shows a single phosphorylation, but the position of the phosphate could not be distinguished among three sites (shown in gold). Regions of the protein in gray were not detected in the Erk2-treated or control CTD samples. The phosphorylation map is the composite of sites identified using positive-mode and negative-mode LC-UVPD-MS. Representative 193 nm UVPD mass spectra from positive-mode (B) and negative-mode (C) analyses are shown for the chymotryptic peptide SPTpSPVYSPTVQF, which covers heptads 11 and 12. In both polarities, the doubly charged ions of m/z 745.3 (for positive mode) and m/z 743.3 (for negative mode) were activated using 2 laser pulses at 2 mJ. Ions that are detected following phosphate neutral loss are denoted by “-P”. (D) Graphical schematic of the rule book for CTD phosphorylation by Erk2 derived from the UVPD mass spectra for numerous phosphopeptides. SP motifs are recognized with the strict requirement for proline (blue) following serine/threonine (orange and red). Modification of Ser5 (S, red) is favored over Ser2 (S, orange) during phosphorylation by Erk2. Thr4 (T) and Ser7 (S) shown in dashed font had little impact on the phosphorylation outcome. An aromatic residue such as tyrosine (Y) or phenylalanine (F) is required (colored green) for phosphorylation. Reproduced with permission from ref 96. Copyright 2017 American Chemical Society.
The application of UV photodissociation to sulfated97–101 and nitrated102 tyrosine residues has been explored to evaluate the utility of photoactivation methods for identification and characterization of other PTMs. Tyrosine sulfation mediates protein–protein interactions in the intercellular space and has been implicated in the inflammatory response.103 The lability of the sulfoester bond prevents retention of the modification during CID. UVPD (193 nm) of deprotonated sulfopeptides was demonstrated to circumvent these problems; the resulting spectra typically featured nearly full sequence coverage with consistent retention of the sulfate goups.97 Detection limits were found to be ~100 fmol for sulfated peptides based on evaluation of caerfulein, cionin, and leu-enkephalin, and the sulfation site of bovine fibrinogen was confidently identified based on characterization of the tryptic peptide GlpFPTDsYDEGQDDRPK.97 This UVPD method was expanded by integrating an enrichment method, weak anion exchange, with carbamylation to reduce the basicities and ionizabilities of primary amines.99 This multistep method allowed the sulfate modification of bovine coagulation factor V to be localized to a single site, Tyr 1513, found on tryptic peptide DDGDGsYIEIIPR.99
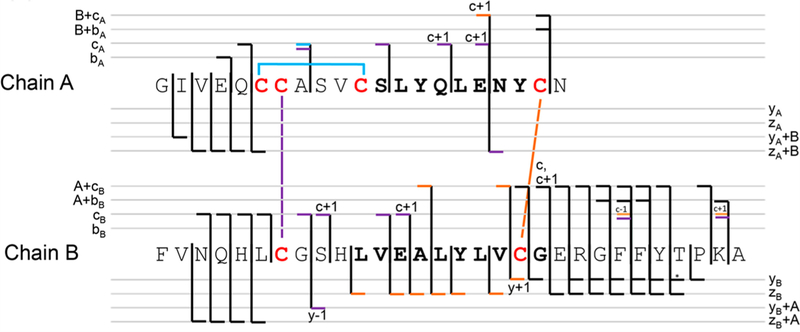
The formation of disulfide bonds connecting cysteine residues modulates the activities and structures of proteins.88 This type of post-translational modification is particularly challenging to determine by mass spectrometry owing to the high degree of complexity from the presence of the extra intra- and intermolecular bonds that stymie database search methods and thus require manual interpretation of spectra. By implementing a partial reduction and digestion process to generate peptide products containing a single disulfide bond prior to MS/MS analysis, 193 nm UVPD afforded homolytic cleavage of disulfide bonds and facilitated elucidation of complicated patterns of disulfide bonds in proteins.104 An example of an annotated UVPD mass spectrum for a 30-residue segment of lysozyme linked via an intermolecular disulfide bond is shown in Figure 10. UVPD resulted in extensive cleavages of the backbone, resulting in a broad array of b/y ions, including those that retained the second chain. As one demonstration of the potential utility of partial reduction combined with UVPD, all 19 disulfide bonds of serotransferrin were mapped.104
Figure 10.
Deconvoluted 193 nm UVPD spectrum of partially reduced peptide pair from lysozyme digest (4+). For this segment of the protein, the disulfide bond linking Cys76 and Cys94 (shown with blue stars) was reduced and alkylated, whereas the disulfide bond between Cys64 and Cys80 (connecting the red and blue chains) remained intact prior to UVPD. Fragment ions denoted with a blue dot contain the intact mass of the B-chain. Fragment ions denoted with a red dot contain the intact mass of the A-chain. Spectrum magnified 5×. Reproduced with permission from ref 104. Copyright 2018 American Chemical Society.
2.4. PTM Analysis Using Other Wavelengths
Although most of the recent applications of UVPD for peptide analysis have entailed activation using 193 nm photons, other wavelengths, particularly 266, 351, or 355 nm, and more recently 213 nm, have also been utilized for elucidation of PTMs. For example, nitration of tyrosine influences protein activity and has been associated with a number of diseases.105 MALDI/in-source photodissociation, in which peptides were ionized by MALDI and irradiated by a 355 nm beam oriented parallel to the MALDI plate, was used to identify nitrated peptides.102 Peptides lacking the 3-nitrotyrosine chromophore failed to undergo photoabsorption at 355 nm, thus allowing selective UVPD of only the peptides containing 3-nitrotyrosine.102 The targeted peptides containing 3-nitrotyrosine underwent charge-directed cleavage, resulting in prominent neutral losses of the nitrotyrosine side chain, as well as production of abundant b/y and immonium ions, the latter of which were thought to arise from multiphoton absorption events.102 The extent of photodepletion of the precursor ions was used to differentiate nitrated peptides from other peptides in a tryptic digest of bovine serum albumin (BSA); two nitropeptides were spiked into the digest and MALDI/in-source PD was found to significantly improve the confidence of identification of nitrated peptides.102
A clever two-stage ETD/UVPD method was designed to enhance the characterization of phosphopeptides in which radical z-type ions generated from ETD were subjected to 355 nm UVPD.92 The resulting fragmentations allowed confident localization of the phosphorylation of serine in a series of isomeric peptides: AAApSR, AApSAR, ApSAAR, and pSAAAR.92 For comparison, collision-induced dissociation of the same z ion resulted exclusively in loss of the phosphorylation modification, thus preventing determination of the site of the modification. Via DFT calculations, it was shown that the variations in fragmentation patterns that allowed differentiation of the isomer depended on access to different electronic transitions of the isomeric z ions.92
Exposure of peptides or proteins to 266 nm photons promoted unusually selective cleavage of disulfide bonds, allowing elucidation of disulfide-bonding patterns in peptides and proteins.106,107 Selective homolytic cleavage of the disulfide bond resulted in the production of the two constituent peptides connected by the linkage.106 In a subsequent study that focused on improving MS/MS strategies for top-down characterization of intact proteins, 266 nm photons were used to selectively cleave up to three disulfide bonds in proteins prior to sequencing by electron-capture dissociation.106 By cleaving the disulfide linkages, enhanced sequence coverage was obtained for the proteins.107 A similar selectivity for cleavage of disulfide linkages was found using 213 nm photons. Upon photoactivation using 213 nm photons, it was found that both disulfide and the adjacent carbon–sulfur bonds were selectively cleaved, yielding characteristic trios of fragment ions that allowed facile identification of the disulfide bonds.108,109 This strategy was used as a first step for disulfide mapping in a biotherapeutic antibody.109
2.5. Improving S/N of UVPD
In parallel with the numerous applications of UVPD for peptide analysis in bottom-up proteomics, there have also been efforts dedicated to improving fragmentation efficiency and ultimately increasing the signal-to-noise (S/N) of the resulting spectra. The S/N of UVPD mass spectra are often lower than the S/N observed in other types of MS/MS spectra, such as those generated by collisional-activation methods, for several primary reasons. First, the large array of fragment ions produced by UVPD results in dispersion of the ion current across many types of fragment ions, resulting in information-rich spectra at the expense of lower-abundance ions. Second, owing to the fact that UV photoirradiation may activate precursor ions and fragment ions, the use of multiple laser pulses to increase fragmentation efficiency may simultaneously result in extensive secondary dissociation and depletion of fragment ions. This means that UVPD is frequently implemented using lower photon fluxes (e.g., lower laser power or fewer laser pulses) to mitigate secondary dissociation processes, thus limiting the overall efficiency. Third, the successful implementation of UVPD requires optimal overlap of photons with the ion population, whether spatially (for ion-trapping instruments) or temporally (for nontrapping systems like time-of-flight platforms), and many instrument-design and method-implementation factors influence the ability to achieve this overlap. Fragmentation efficiencies can be enhanced by attaching chromophores that amplify the photoabsorption cross sections of the analyte ions, for example, by tagging peptides with aromatic groups.110 Alternatively, strategies to overcome secondary fragmentation and/or increase the population of ions subjected to UVPD have yielded promising dividends. Two approaches have exploited the application of supplemental waveforms to manipulate ions during acquisition of UVPD spectra in ion-trap mass spectrometers.111,112 In one case, selective ejection of nondissociated precursor ions after UV photoactivation offered a means to mitigate space charge problems that decreased S/N of the UVPD spectra.111 This strategy, termed precursor-ejection UVPD (PE-UVPD), alleviated mass shifts of fragment ions caused by space charge in the ion trap originating from accumulating an excessive number of ions as illustrated in Figure 11. Ejection of the nondissociated precursor, typically the most abundant ion during and after UVPD, significantly reduced mass errors in the resulting spectra. Demonstration of this strategy for analysis of peptides in an E. coli lysate resulted in better scores owing to the higher S/N values of the spectra.111 A complementary method utilized supplemental waveforms to manipulate the fragment ions, not the precursor ions, to protect them from UV irradiation.112 This latter strategy was applied for characterization of intact proteins, resulting in higher S/N of the resulting spectra, as described in section 3.2.112
Figure 11.
Precursor-ejection UVPD (PE-UVPD). (A) Schematic representation of a sequence of events to relieve space charge effects beginning with ion accumulation, then 193 nm UVPD, followed by resonance ejection applied to the remaining undissociated precursor. UVPD mass spectra (1 pulse, 2 mJ) of angiotensin (3+) obtained using an AGC target of 3 × 104 (B) without PE-UVPD and (C) with PE-UVPD. Expansions of the m/z 125–350 range are also shown without and with PE-UVPD, illustrating the mass shifts in the fragment ion assignments. (D) Mass error (ppm) of fragment ions ranging from m/z 130–1030 using AGC target 5 × 104 without PE-UVPD (light red line) and with PE-UVPD (red dashed line), as well as 3 × 104 without PE-UVPD (light blue line) and with PE-UVPD (blue dashed line). The m/z value of the precursor ion is indicated by a solid black vertical line. The zero ppm error value is shown with a horizontal gray line. Reproduced with permission from ref 111. Copyright 2017 American Chemical Society.
2.6. UVPD and Derivatization Strategies
Strategic chemical modification of peptides and subsequent photoactivation has emerged as an attractive approach for a variety of peptide analysis and sequencing-based proteomic applications. For example, addition of chromogenic tags to peptides has been used to facilitate photoabsorption using wavelengths at which peptides would not otherwise absorb or dissociate (such as in the range of 350 nm). This strategy permits selective photoactivation and dissociation of exclusively the tagged peptides or alternatively endows peptides with uniquely photocleavable tags, thus improving the specificity of proteomic approaches or affording insight into structural features not targeted by more universal ion-activation methods like CID. Other derivatization strategies have utilized covalent modification of peptides to target other inherent challenges of proteomics or UVPD, including hurdles associated with the negative ionization mode (ionization efficiency and ineffective collisional-activated dissociation of deprotonated peptides) or overcoming low fragmentation efficiency of some UVPD methods.
2.6.1. Radical-Directed Dissociation.
One strategy coupling derivatization with 266 nm UVPD has emerged as a versatile method to promote site-selective bond cleavages via a process termed radical-directed dissociation (RDD).113 RDD is a hybrid MS3 strategy that typically uses 266 nm photoactivation to cause specific homolytic bond cleavages, producing radical ions that are subsequently subjected to collisional activation to generate diagnostic fragment ions.113 A frequency-quadrupled solid-state Nd:YAG laser provides a convenient way to generate 266 nm photons and hence has been the most commonly used laser for RDD applications. The characteristic RDD fragmentation pathways result in a/x and c/z ions in addition to products from side-chain losses. To enable RDD, the analyte molecules of interest, often peptides, are derivatized to install a photocleavable bond that facilitates the production of radical ions. For example, iodine-containing aromatic compounds have proven to have ideal properties for initiating RDD because the aromatic group affords a good chromophore for absorption of 266 nm photons and the photolabile C–I bond is homolytically cleaved with both high efficiency and high selectivity to generate radical species. This RDD process, pioneered by the Julian group in 2008,113 has been utilized in an enormous array of applications to characterize key structural features of peptides and proteins, as well as other types of biomolecules discussed in later sections. Some of the mechanistic details of the RDD process, including the propensity for radical migration, were deciphered in a series of more fundamental studies involving model peptides.114,115 RDD has been utilized to facilitate the localization of phosphorylation sites93,116,117 and iodotyrosine residues of peptides,118 the quantification of the number and type (reduced or oxidized) of cysteines in peptides and proteins,119 and the determination of charge state of proteins and fragment ions.120 Different methods for creating appropriate radical sites for RDD based on installing different types of photocleavable bonds have also been reported.116,119,121,122
One of the more recent applications of RDD focused on the elucidation of the antioxidant regions of proteins based on evaluation of the extent of RDD of tryptic peptides created from proteolysis of the targeted proteins.123,124 The peptides were modified using 4-iodobenzoic acid to install a cleavable C–I site, then subjected to the RDD process using 266 nm photons for photocleavage, followed by collisional activation to generate diagnostic fragmentation patterns (Figure 12). Those peptides with high antioxidant capacity exhibited little of the expected RDD pathways (production of a/x ions and side-chain losses) and instead dissociated primarily via production of b/y-type ions characteristic of mobile-proton pathways. Generation of b/y-type ions suggested that the radical site created by 266 nm photocleavage of the C–I bond was sequestered, thus inactivating the subsequent radical-initiated fragmentation pathways.123,124 The ratio of b/y-type ions to fragments associated with radical-directed pathways provided a means to categorize the antioxidant properties of peptides in a quantitative manner via a radical sequestering score (RSS). This strategy demonstrated the strong correlation between radical sequesteration and antioxidant potential, with examples drawn from analysis of human serum albumin, β-lactoglobulin, αA-crystallin, αB-crystallin, and myoglobin.123,124
Figure 12.
Radical-directed dissociation (RDD) entails derivatization of peptides with 4-iodobenzoic acid to install a C–I bond that is selectively cleaved by 266 nm UVPD. Collisional activation of the resulting product yields fragmentation patterns that reflect the antioxidant properties of the peptides. MS/MS spectra are shown for two peptides derived from β-lactoglobulin. (a) CID of IPAVFK4IB radical (1+). (b) CID of VYVEELK4IBPTPEGDLEILLQK radical (3+). Fragments derived from radical-initiated pathways are labeled in red; fragments from proton-driven pathways are labeled in green. The radical sequestering score (RSS) shown on each spectrum represents the ratio of summed fragment ion abundances originating from b/y-type ions (labeled in green font) to those originating from radical-directed fragmentation (labeled in red font). Peptides exhibiting antioxidant behavior have RSS scores greater than 1.4, indicating that the radical is sequestered. Reproduced with permission from ref 124. Copyright 2015 Royal Society of Chemistry.
Chirality inversion of single amino acids (with l being the natural chirality and d representing the rare chirality) in peptides or proteins is a subtle PTM that has a profound impact on the function of the molecules in eukaryotic systems. Isomeric modifications of this kind, however, are extremely difficult to detect using conventional MS/MS strategies and require discrimination of d-amino-acid-containing peptides in diastereomeric mixtures. Characterization of such peptide epimers was recently demonstrated using RDD.125–128 The C–I motifs that promote RDD were incorporated into the peptides by direct iodination of tyrosine residues (for those peptides that contained tyrosine) or coupling of an iodobenzoic acid moiety to the N-terminus or lysine side chains. Radical migration during the collisional-activation step has proven to be highly dependent on peptide/protein structure; consequently, facile discrimination of peptide epimers was achieved for eight peptides containing d-alanine, d-serine, or d-aspartic acid.126 Examples of the UVPD mass spectra are shown in Figure 13 for the peptide IQTGLDATHAER, which features racemization of the Asp residue. Photodissociation of the protonated d- and l-peptides resulted in different fragmentation patterns; the appearance of a product ion attributed to loss of 56 Da from the side chain of Leu was shown to be unique to the d-Asp epimer and allowed confident differentiation of the epimeric forms. A similar RDD strategy was used to quantify four isomeric peptides containing l-Asp, d-Asp, l-isoAsp, or d-isoAsp (peptide 4IB-VKLNXG where X = G, H, S, or A and 4IB is the para-iodobenzoic acid tag used for the C–I bond cleavage) as a function of solution conditions used to modulate deamidation. The use of bicarbonate buffer or high temperature was found to cause significant racemization, yielding increased amounts of d-Asp and d-isoAsp. Variation of the pH of the solution caused a far more limited impact on racemization.126
Figure 13.
UV photodissociation of two epimeric peptides: (a) [IQTGLDATHAER + H]+ and (b) [IQTGLDATHAER + 2H]2+. The d-form shows abundant −56 Da loss from Leu for both charge states. The downward arrow indicates the selected precursor ion. Reproduced with permission from ref 125. Copyright 2012 American Chemical Society.
This RDD method was subsequently utilized for several biologically relevant applications involving monitoring d/l epimerization and isomerization of aspartic acid to isoaspartic acid in proteins.127,128 In one application, RDD was integrated into an LC-MS mode to allow characterization of amino acid epimerization in crystallin proteins from mammalian eye lenses.127,128 Tryptic digests of the targeted proteins were subjected to the iodobenzoic acid conjugation procedure prior to LC-MS analysis and 266 nm UV-initiated RDD (e.g., 266 nm photoactivation for C–I bond cleavage, followed by collisional activation to generate informative fragmentation patterns from the precursor peptide radical ions). The identification of the peptides was facilitated by incorporating the +230 Da shift, corresponding to addition of the 4-iodobenzoic acid tag, as a variable modification at the N-terminus or lysine side chains of each peptide in the database search. A number of new sites of isomerization involving serine and aspartic acid residues were discovered, with a high frequency of these modifications found in the more disordered regions of the proteins.127,128 The extent of epimerization/isomerization was based on the ratios of key diagnostic b/y fragment ions that differed for l- versus d-enantiomers of each peptide. For the sheep crystallin protein in Figure 14, the degree of isomerization was found to be substantially greater for the N- and C-terminal regions than in the crystallin domain.128
Figure 14.
Percent aspartic acid isomerization of water-soluble (WS, upper bars) αA sheep crystallin proteins versus water-insoluble (WI, lower bars) αA sheep crystallin proteins determined from RDD mass spectra using 266 nm photons and CID. The color-coded bars correspond to three structural regions: orange, disordered N-terminus; blue, structured α-crystallin domain; purple, disordered C-terminus. Three separate digests were performed; error bars represent standard deviations. Number ranges above the bars represent peptide sequences. Peptide 164–173 does not contain error bars because it only appeared baseline-resolved in one digest. The full protein sequence is given below the plot, with aspartic acid residues in bolded/black and serine residues in underlined/black. Asp105 and Asp106 are in bold red text in the amino acid sequence and are shown explicitly in the crystal structure (PDB 3L1F) to highlight an important region of isomerization. Stars indicate isomerized regions where isomerization was identified but quantitation was not possible due to incomplete chromatographic separation. Reproduced with permission from ref 128. Copyright 2017 American Chemical Society.
UVPD using 266 nm photons for the radical-directed dissociation (RDD) strategy described earlier has also been used to characterize protein structure.113,129 In the founding RDD study of proteins, ubiquitin was iodinated at Tyr59 in solution, thus installing a C–I bond that was susceptible to cleavage upon exposure to 266 nm photons and that created a reactive radical site.113 Because radical migration depends intrinsically on tertiary structure, fragmentation was observed at remote locations from the Tyr59 residue for the compact, folded charge states of ubiquitin (4+ to 6+). In contrast, fragmentation of the higher charge states (8+ to 10+), which were thought to be helical or unfolded,130–132 occurred primarily at residues immediately adjacent to Tyr59.113 Deeper analysis of the tertiary contacts of ubiquitin based on constrained molecular dynamics simulations suggested that significant structural reorganization occurred for ubiquitin in the gas phase.113 This strategy was extended to another three-helix protein domain, KIX, from the KREB protein for the 6+ to 16+ charge states.129 Tyr50 was the primary iodination site for this protein, and subsequent RDD of the low charge states resulted in fragmentation at Asn46, located in an adjacent turn of the α-helix at Tyr50, as well as even more distant sites such as Trp10 and His21. These findings suggested the existence of two different conformations that were similar to one identified by NMR.129 In general, the RDD method exhibited significant sensitivity to the secondary and tertiary features of proteins that modulated migration of radicals through space in the gas phase, even for residues not close in the primary sequence.
2.6.2. Photoelectron-Transfer Dissociation.
Another approach using 266 nm UVPD, termed photoelectron-transfer dissociation (PETD), was aimed at deciphering zwitterionic pairs (i.e., salt bridges, a key structural motif) in peptides and proteins.133 PETD is not a new type of method but rather an intramolecular process that results in production of c/z fragment ions upon exposure to 266 nm photon. Those peptides or proteins that produced c/z fragment ions were determined to have anionic sites characteristic of zwitterions. The lack of c/z ions indicated the absence of anionic sites among the acidic functional groups of the peptides or proteins, thus representing nonzwitterionic structures. This method was used to categorize a series of peptides and small proteins, including the beta chain of insulin and bovine ubiquitin, as zwitterionic or not.133 An example is shown in Figure 15 for ubiquitin.133 On the basis of the prevalent formation of c-type ions upon 266 nm UVPD, it was determined that ubiquitin, with 13 basic sites, could adopt zwitterionic states via engagement of salt bridges and protonation at nonbasic sites, for all charge states from 5+ to 14+, with greater dominance of zwitterionic contributions for the lower charge states.133 The production of c/z ions decreased and the generation of b/y ions increased with charge state, an outcome reflecting a shift to more nonzwitterionic structures.
Figure 15.
(a) Photoelectron-transfer dissociation (PETD) mass spectrum for ubiquitin (5+) and (b) its corresponding c/z fragmentation map. CO2 losses are marked by an asterisk. Reproduced with permission from ref 133. Copyright 2017 American Chemical Society.
2.6.3. UVPD (266 nm) for Selective Bond Cleavages.
In addition to a large array of strategies that have exploited 266 nm photons for radical-directed dissociation (RDD) described earlier, 266 nm UVPD has also been paired with other derivatization strategies for specialized applications.134,135 For example, 266 nm photoactivation promotes selective S–Se bond cleavage and thus has been exploited for a cysteine-selective peptide-analysis method.134 This strategy was designed to enhance the level of selectivity in a bottom-up proteomics workflow in order to mitigate the common problem of chimeric spectra.134 Chimeric spectra are MS/MS spectra containing fragment ions from multiple isobaric precursors, thus confounding interpretation. For this 266 nm UVPD method, N-(phenylseleno)phthalimide (NPSP) was used to alkylate thiols of cysteine residues, thus installing a chromogenic benzeneselenol tag. The S–Se bond was selectively cleaved upon photoactivation using 266 nm photons, resulting in a trackable neutral loss of 156 Da that was used as a means to target cysteine-containing peptides by CID. An example of the highly selective cleavage of the S–Se bond upon exposure of the modified peptides to 266 nm UV photons is shown in Figure 16.134 The CID spectrum acquired for the targeted cysteine-containing peptide allows facile identification based on the generation of b/y ions. Data-dependent acquisition based on the cysteine screening method along with a filtered cysteine database search provided a streamlined bottom-up workflow.134 Another strategy implemented the N-terminal modification of peptides using 4-acetamido-4-isothiocyanatostilbene-2,2-disulfonic acid (SITS) to improve the photoabsorption cross sections of peptides at 266 nm. The SITS-tagged peptides exhibited higher fragmentation efficiencies and greater numbers of C-terminal (x,y,z) sequence ions than untagged peptides upon 266 nm UVPD.135
Figure 16.
(a) Electrospray ionization (ESI) mass spectrum of a tryptic peptide CAQCHTVEK containing two benzeneselenol tags, (b) 266 nm UVPD mass spectrum of C*AQC*HTVEK (2+), and (c) UVPDnLossCID spectrum. The asterisks represent incorporation of benzeneselenol tag. Reproduced with permission from ref 134. Copyright 2016 American Chemical Society.
2.6.4. UVPD Using 351/355 nm Photons.
Utilization of chromogenic tags to endow peptides with photoreactivity has emerged as a powerful approach for selective fragmentation using ~350 nm photons.56,136–143 Typically only peptides that are tagged with a specific chromophore absorb 350 nm photons, thus enabling selective activation and dissociation. In essence, modification of peptides via attachment of a chromophore converts nonabsorbing peptides to highly chromogenic ones whereas nontagged peptides remain silent. This strategy was originally developed for peptides using 355 nm photons produced by a frequency-tripled Nd:YAG laser136,137 and subsequently extended by using 351 nm photons generated by a XeF excimer laser.56,142,143 The chromophore-tagging method was used to map solvent accessibility of proteins based on UVPD analysis using a chromogenic chemical probe to react selectively with the side chains of lysine residues.138,139 It was also used to characterize antigen-binding regions of antibodies by analysis of chromophore-tagged cysteine-containing peptides.140 Selective tagging of histidine and tyrosine was more recently demonstrated in a workflow utilizing 351 nm UVPD.141 For this application, peptides were labeled with a diazonium chromophore at tyrosine and histidine residues, a strategy that exploited the relative low abundance of these amino acids to streamline protein identification. Irradiation at 351 nm resulted in the detection of b- and y-type ions produced only from the tagged peptides.141 A customized database in which exclusively Tyr- and His-containing peptides were included reduced the search space and thereby improved the confidence of the identifications.141
Although in silico database searching has been a more popular peptide-identification strategy than de novo methods, de novo sequencing has remained a compelling approach because of its unique capacity to identify peptides from proteins lacking sequenced genomes or those with unusual modifications.144–146 De novo methods utilize differences in m/z between adjacent fragments of an ion series to predict peptide sequences.144–146 To date, the performance of de novo methods has been hampered by incomplete fragment ion series, the presence of isomeric residues (leucine and isoleucine), and difficulties discerning the directionality of ion series, among others.144–146 The excellent performance metrics offered by UVPD provides a means to overcome gaps in sequence coverage of peptides as demonstrated previously using 157 nm UVPD147,148 and 193 nm UVPD.149 The production of v- and w-type side-chain loss ions has even allowed differentiation of leucine and isoleucine.147,148 Along with the development of improved ion-activation modes, peptide-derivatization methods have been implemented for a number of years to improve de novo sequencing.150–153 Derivatization methods that enhance the selective and exclusive production of either C- or N-terminal ion series can reduce the redundant formation of the opposite series of fragment ions (N- or C-terminal), which confounds fragment ion assignment.150–153 One new UVPD-based strategy capitalized on a peptide-derivatization method to ameliorate some of the other challenges traditionally associated with de novo sequencing.56,142,143 Derivatization of peptides with sulfosuccinimydyl-7-amino-4-methyl-coumarin-3-acetic acid (sulfo-NHS AMCA) appended a 350 nm chromophore to the N-terminus of tryptic peptides as illustrated using variants of green fluorescent protein (GFP).56 UVPD using 351 nm photons from an excimer laser resulted in selective dissociation of the chromophore-labeled peptides and fragments.56,142 Utilization of a high number of laser pulses (15) resulted in annihilation of N-terminal fragment ions, leaving a clean y-ion series from which de novo analysis was performed using a de novo algorithm.56,142 An example of the MS/MS spectrum resulting from 351 nm UVPD of an AMCA-tagged peptide is shown in Figure 17. The scoring of the UVPD spectra suffered, however, owing to the absence of b ions, which were expected to be found and scored using commercially available de novo search algorthims. Thus, a custom de novo algorithm, UVnovo, was developed to mitigate this problem and was used to identify 2 983 peptides from an E. coli lysate with excellent precision.142 This method was extended to utilize matched pairs of CID and 351 nm UVPD spectra in which a machine learning process was developed to facilitate interpretation of the fragmentation patterns.143 This more advanced strategy was used for de novo sequencing of proteins from an E. coli lysate in which the complementary nature of the UVPD spectra (containing only y ions) and the CID spectra (b/y ions) provided even higher confidence in assigning the ion series.143 For example, on the basis of utilizing only the CID mass spectra, 2617 peptides were identified from de novo sequencing of the AMCA-tagged peptides, and 3422 peptides were identified using the UVPD mass spectra. Combining the CID and UVPD spectra facilitated differentiation and assignment of b and y ions, ultimately resulting in 3835 peptide identifications.143 The UVnovo algorithm successfully identified peptides corresponding to 83% of the matched CID/UVPD spectra in a high-throughtput workflow.143 In general, these UVPD-based methods offered a clever solution to some of the problems associated with conventional de novo sequencing. A combined derivatization/selective UVPD strategy might be envisioned for de novo sequencing of proteins, but this concept is impeded by the likelihood of substoichiometric derivatization reactions of intact proteins that would result in an overly heterogeneous population of tagged proteins, complicating interpretation.
Figure 17.
UVPD (351 nm) (3 mJ/pulse, 15 pulses) mass spectrum of elongation factor G peptide V[AMCA]YSGVVNSGDTVLNSVK[carbamyl]AAR (2+) from a tryptic digest of E. coli lysate. AMCA (7-amino-4-methylcoumarin-3-acetic acid) is the chromogenic tag appended to the peptide at the N-terminus. The lysine residue is carbamylated. The precursor is labeled with an asterisk. The inset shows the number and overlap of spectra that were identified correctly from an E. coli data set using the UVnovo algorithm. UVnovo identifications from the paired CID + UVPD, CID-only, and UVPD-only mass spectra are shown. Adapted with permission from refs 142 and 143. Copyright 2016 and 2017 American Chemical Society, respectively.
2.6.5. UVPD with Online Reactions and Hydrogen/Deuterium Exchange.
An alternative type of online peptide tagging strategy was implemented in conjunction with 193 nm UVPD in order to enhance UVPD fragmentation efficiency while taking advantage of the benefits of rapid microdroplet chemistry.154 A dual-spray reactor was built to enable simultaneous introduction and interaction of 4-formyl-1,3-benzenedisulfonic acid (FBDSA) with peptides, thus resulting in the noncovalent adduction of an anionic aromatic chromophore to a cationic peptide via a type of ion–ion interaction occurring at the intersection of two liquid sprays.154 The dual-spray setup and reaction chemistry is illustrated in Figure 18A. Schiff base formation was subsequently promoted in the gas phase by collisional activation of the noncovalent adducts in a linear ion trap, and then 193 nm UVPD was used to generate diagnostic fragmentation patterns of the resulting peptide complexes.154 The UVPD mass spectra exhibited improved sequence coverage relative to mass spectra generated by CID and with greater peptide fragmentation efficiency relative to UVPD of the untagged peptides, as exemplified by the spectra shown in Figure 18B.154 The higher fragmentation efficiency was attributed to the chromogenic properties of FBDSA, thus enhancing the UV photoabsorption cross sections of peptides.154 This strategy was successfully implemented on an LC time scale for analysis of a peptide mixture using a data-dependent neutral loss strategy, from which the improvements in sequence coverage and UVPD efficiency were evident.154
Figure 18.
(a) Setup used for dual-spray-initiated bioconjugation of a peptide cation with an FBDSA anion for enhanced ultraviolet photodissociation. (b) Comparison of 193 nm UVPD efficiencies before and after Schiff base modification of KLVANNTRL (1+): (1) MS2 UVPD mass spectrum for unlabeled peptide and (2) MS3 UVPD mass spectrum following online derivatization using the dual-spray reactor. Reproduced with permission from ref 154. Copyright 2015 American Chemical Society.
Hydrogen/deuterium-exchange reactions represent another type of solution chemistry commonly coupled with mass spectrometry analysis, primarily for mapping dynamics and structural features of proteins.155 UVPD (213 nm) was used to characterize peptides subjected to hydrogen/deuterium-exchange reactions, as illustrated for deuterium-labeled peptide HHHHHHIIHIIK in Figure 19. On the basis of mass analysis of the array of fragment ions, it was found that UVPD caused little H/D scrambling among the backbone amides, consistent with amounts observed by ETD.52 The absence of significant scrambling provided compelling evidence that UVPD could be successfully used to improve the spatial resolution of hydrogen/deuterium-exchange (HDX) strategies for conformational studies of proteins.52
Figure 19.
Comparison of H/D scrambling in the regioselective deuterium-labeled peptide HHHHHHIIHIIK following 213 nm UVPD, ETD, or CID using mild ion source conditions. The degree of H/D scrambling was quantitated for all fragment ion types present at suitable ion abundance. Experiments were done in at least three replicates, and the error bars indicate single standard deviations. Reproduced with permission from ref 52. Copyright 2018 American Chemical Society.
2.6.6. Derivatization and Photodissociation Using Visible Wavelengths.
Although this Review focuses on the ultraviolet range, many of the same concepts related to design of specialized selective photodissociation applications using chromogenic tags are readily adapted to other regions of the electromagnetic spectrum. Perhaps unsurprisingly, tagging strategies and selective photodissociation have also been extended to the visible range. The Dugourd group developed a cysteine tagging approach using a cysteine-selective dabcyl maleimide reagent in conjunction with single reaction monitoring (SRM) for profiling plasma proteins.156–158 Cysteine-containing peptides modified with the dabcyl reagent underwent efficient photoabsorption at 473 nm, a wavelength not commonly absorbed by unmodified peptides. Owing to the low frequency of cysteine in peptides and proteins, the net outcome was a reduction in interferences of the photo-SRM mode relative to conventional SRM. The targeted photo-SRM method resulted in lower limits of detection than conventional SRM.158 These types of innovative approaches illustrate the versatility and analytical attributes of coupling tagging/derivatization methods to wavelength-specific photodissociation across the electromagnetic spectrum.
2.7. UVPD for Middle-Down Proteomics
An emerging strategy for proteomics is termed “middle-down”, a scaled variation of bottom-up methods.159,160 The middle-down approach represents an intermediate between conventional bottom-up methods, which use proteases to decompose proteins into small, easily analyzed peptides, and top-down methods, which directly analyze intact proteins. In practice, middle-down methods use limited proteolysis to generate large peptides, ones that cover longer sections of proteins.159,160 The potential benefits of this strategy include the reduction in the number of overly short peptides that do not uniquely identify any specific protein and an enhancement in the ability to characterize larger sequence stretches that might contain multiple modification sites. At the same time, the challenges of analyzing large proteins as intact species, both in terms of chromatographic separation and mass spectrometric analysis, are mitigated by using middle-down approaches. Successful implementation of middle-down methods require MS/MS techniques that offer extensive fragmentation of very large peptides (>3 kDa), and this has motivated the exploration of UVPD for middle-down applications in several recent studies, all utilizing 193 nm photoactivation for analysis of large peptides from proteins.161–164
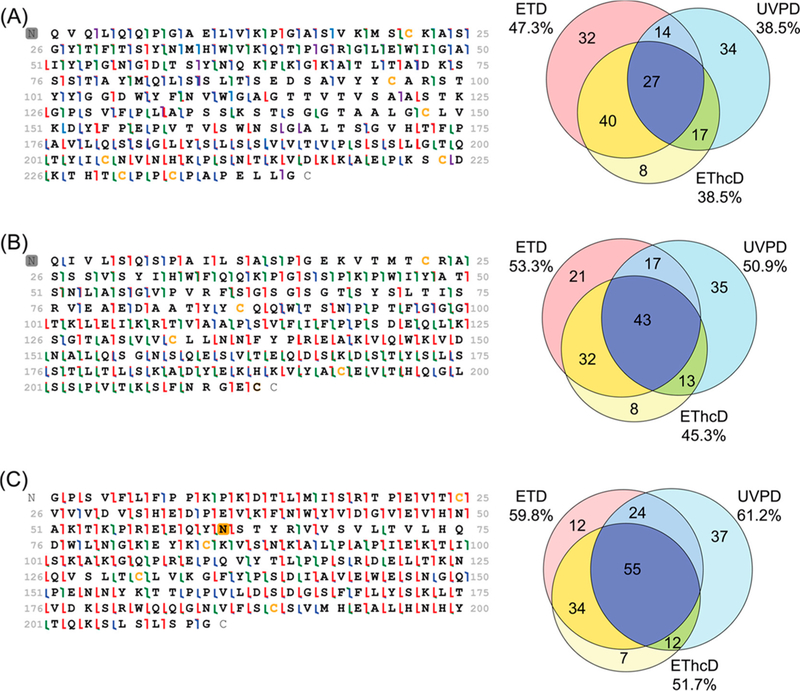
The development of analytical methods for characterization of antibodies has exploded in recent years owing to the growing understanding of the adaptive immune response and the quest for new vaccines and immunotherapies for disease treatment.165 Antibodies are glycoproteins that pose significant analytical challenges owing to their large size and often high degree of sequence similarity. For one middle-down UVPD-MS application, the use of limited Lys-C proteolysis allowed production of larger peptides of immunoglobulin G.161 Using 193 nm UVPD for MS/MS characterization of the peptides improved the differentiation of antibodies by affording up to 100% sequence coverage of the most diagnostic complementarity-determining regions (CDRs). For this strategy, Lys-C protease cleaved the antibodies at the lysines, yielding unique peptides larger than 4.5 kDa. The number of Lys-C peptides identified by 193 nm UVPD, HCD, and ETD for trastuzumab are shown in Figure 20, along with E-scores (a confidence metric that describes the probability at which a set of fragment ions match a given sequence by random chance alone) used to assess scoring confidence for 29 peptides found in common by all three MS/MS methods.161 The E-scores were consistently higher for UVPD, an outcome attributed to the richer fragmentation patterns promoted by UVPD as illustrated by the sequence maps shown for one CDR peptide in Figure 20.161 UVPD offered especially informative fragmentation of the key hypervariable CDR sequences that are flanked by conserved lysine residues.161 The performance of a customized LysC/UVPD/database search strategy, coupled with a library containing 14 515 VH genes (heavy-chain sequences), was applied successfully for the analysis of a mixture of anti-influenza monoclonal antibodies.161
Figure 20.
Comparison of 193 nm UVPD, HCD, and ETD for middle-down analysis of Lys-C digested Trastuzumab in terms of (a) overlap in identified peptide populations and (b) average peptide mass and sequence coverage obtained by each activation method. (c) Relative E-scores for the 29 overlapping peptides identified by all three activation methods and (d) ion-cleavage maps obtained for an 8.2 kDa CDR-H1 and CDR-H2-containing peptide. The hypervariable regions are shaded in gray. Reproduced with permission from ref 161. Copyright 2017 American Chemical Society.
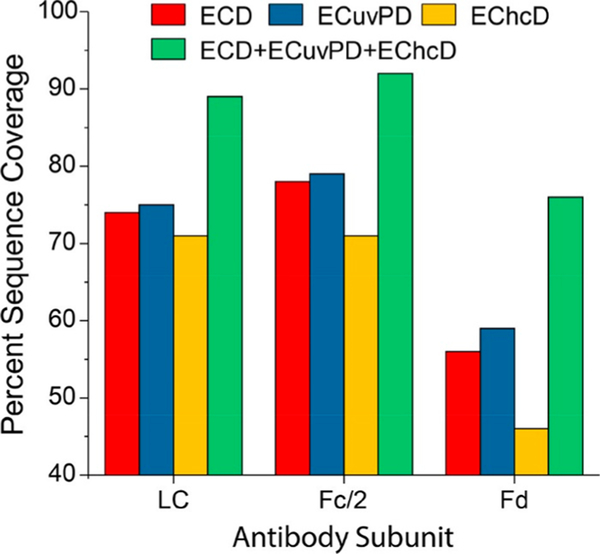
In a second middle-down application, UVPD was used to characterize therapeutic monoclonal antibodies, trastuzumab and adalimumab, using a targeted approach that utilized IdeS, a hinge-selective G-degrading enzyme, to generate ~25 kDa Fc/2, Lc, and Fd antibody subunits.162 Combining the fragmentation obtained from UVPD using different numbers of laser pulses and laser energies resulted in 85% sequence coverage, including extensive characterization of the antigen-binding CDRs as well as unambiguous localization of the glycosylation site of the Fc/2 subunit.162 The latter finding confirmed that labile post-translational modifications, such as those added via glycosylation, were not preferentially cleaved during UVPD. Instead, glycans remained attached to fragment ions that facilitated their localization. Figure 21 displays an expanded region of the deconvoluted UVPD mass spectrum acquired for the Fc/2 subunit of antibody trastuzmab. In addition to an array of diagnostic a, x, y, and z sequence ions, the mass difference between the a60 and a61 ions is consistent with the addition of one amino acid (Asn, N) plus an octasaccharide.162 The production and assignment of a continuous series of fragment ions (e.g., a59, a60, a61, a62, etc.) is what facilitated the localization of the octasaccharide based on incorporation of the mass shift associated with the PTM.
Figure 21.
MS1 spectrum of the Fc/2 subunit of trastuzumab showing mass shifts consistent with glycoform microheterogeneity (top). The inset demonstrates consecutive saccharide additions to the core N-linked glycan structure. The expanded region of the deconvoluted 193 nm UVPD mass spectrum (middle) of the G0F glycoform (25+) shows a mass shift consistent with the intact glycan between consecutive a ions (a60 and a61). An abrupt stop in matched N-terminally derived ions allows unambiguous glycan site localization at a single Asn (N61) residue in the sequence map (bottom). Reproduced with permission from ref 162. Copyright 2016 American Chemical Society.
Another middle-down strategy combined a protein-derivatization method with UVPD to yield high sequence coverage of large peptides.163 Carbamylation modified the side chains of lysines, making them unrecognized by trypsin (which normally cleaves at both lysine and arginine residues) and thus restricting the proteolysis to only Arg residues.163 Large peptides were subsequently generated by trypsin proteolysis of the carbamylated proteins. UVPD of the resulting peptides yielded high sequence coverage, and the carbamylation/trypsin/UVPD workflow was readily adapted for analysis of proteins from E. coli.163
Middle-down strategies are also effective for characterization of proteins containing multiple post-translational modifications. Mapping combinatorial modifications is a particularly vexing problem for traditional bottom-up methods as it is difficult or even impossible to reconstruct the patterns of multiple modifications based on analysis of short peptides that do not retain the integrity of the original pattern of modifications. For example, histones are not particularly large proteins, often <15 kDa, but they may be heavily modified along the first 50–60 residues adjacent to the N-terminus.166 UVPD (193 nm) was used to analyze large histone peptides generated by GluC proteolysis and isolated by weak cation-exchange–hydrophilic interaction chromatography.164 The performance of 193 nm UVPD remained robust irrespective of the size and charge state of the peptides generated from H3 and H4 histones. For example, 90% sequence coverage was obtained from UVPD of the N-terminal peptide of hepta-modified histone H3K4me1K9-me2K14acK18acK23acK27acK36me3 (8+), as illustrated in Figure 22.164 The characterization of this histone tail was challenged by the number (7) and diversity of modifications (acetylation, methylation, dimethylation, and trimethylation). All of the modifications were bracketed by N- and C-terminal fragment ions corresponding to backbone cleavages on each side of the modified residues, thus offering confidence in their localization. For this middle-down/UVPD study, over 300 modified histones were identified by UVPD.164
Figure 22.
Comparison of deconvoluted (A) ETD and (B) 193 nm UVPD mass spectra of the N-terminal tail of H3K4me1K9me2K14acK18-acK23acK27acK36me3 (50 residues, 8+ charge state) showing fragment ion maps, P-score, sequence coverage, and labeled PTM site-localizing ions. Residues are shaded to indicate modifications: red indicates acetylation, blue indicates dimethylation, green indicates methylation, and purple indicates trimethylation. Reproduced with permission from ref 164. Copyright 2018 American Chemical Society.
3. UVPD FOR INTACT PROTEINS
In the field of mass spectrometry, the examination of intact proteins is called top-down analysis, and historically it has been one of the most challenging approaches for proteomics.167–169 More conventional MS/MS methods used for the characterization of intact proteins, such as collision-induced dissociation (CID), typically favor cleavage of the most labile backbone bonds close to the N- and C-termini of the protein, which limits sequence coverage. The fragmentation patterns of intact proteins are congested and complicated, requiring high-performance deconvolution, ion assignment, and scoring algorithms. Advances in informatics over the past decade have overcome many of these technical difficulties.170,171 In addition, the growing accessibility to mass spectrometer platforms with exceptionally high mass accuracy and resolution, like Orbitrap platforms and Fourier transform ion cyclotron resonance mass spectrometers, have catalyzed new growth in the development of top-down proteomic methods.167–170 The large photoabsorption cross section of proteins at 193 nm and the resulting high internal energy deposition have spurred the development of UVPD for characterization of intact proteins. The first successful application of 193 nm UVPD for top-down analysis of proteins was reported in 2013, in which an excimer laser was integrated with an Orbitrap mass spectrometer for fragmentation and characterization of intact proteins.47 Extensive dissociation of proteins, including myoglobin (17 kDa) and carbonic anhydrase (29 kDa), was achieved via photoirradiation using 193 nm photons, and the resulting fragment ions were transferred to the Orbitrap analyzer for high-accuracy analysis.47 A sequence coverage of 87% was obtained for carbonic anhydrase using a single laser pulse, and this represented a significant improvement over that obtained by collisional activation on the same MS platform (30%).47
This landmark study opened a new frontier of top-down analysis using 193 nm UVPD, including adoption of the method for detailed characterization of proteins containing post-translational modifications,47,172,173 engineered mutations,174 unnatural amino acids,175,176 solvent-accessibility tags,177 and cross-linked sites.178 The successful characterization of proteins using 193 nm photoactivation also motivated the implementation of UVPD on other high-performance mass spectrometers including a 15 T FTICR platform51 and other variations of Orbitrap systems,48,179,180 which offer sufficiently high mass accuracy and mass resolution to allow assignment of fragment ions in the dense spectra created by UVPD of intact proteins. UVPD (193 nm) has also been used to characterize native-like folded proteins in the context of structural biology applications, as described in the next section. This array of top-down protein studies was motivated in large part by the rich fragmentation patterns provided by 193 nm UVPD that yielded high coverage (meaning a substantial number of backbone cleavages throughout the protein) for both confirmation of protein sequences and localization of modifications.47,172–176 In particular, the ability to localize modifications with confidence depends on generating fragment ions that bracket the modifications via backbone cleavages that generate fragment ions that retain the mass shift associated with the modified residue. Tracking the pattern of fragment ions that contain no mass shift versus those that incorporate a mass shift alleviates ambiguity about the specific locations of the modified residues in a protein sequence.
To demonstrate the attributes of UVPD for mapping post-translational modifications, 193 nm UVPD was applied to the characterization of histones, proteins which are known to be among the most heavily modified.173 The extensive dynamic variation of modifications creates the histone code that regulates transcription and DNA-damage repair.166 Mapping the modifications of histones is particularly challenging owing to the complexity of the combinatorial pattern, often resulting in acetylation and methylation of multiple lysines and/or arginines of the N-terminal tail. An example of the sequence maps and scoring metrics obtained by 193 nm UVPD for one H4 proteoform is shown in Figure 23 in comparison to HCD and EThcD.173 For this particular proteoform, UVPD offered the highest sequence coverage, promoting backbone cleavage of nearly every inter-residue position, which allowed the three modifications to be localized and the sequence to be well-characterized. From a mixture of histones, over 500 modified histoforms and 288 unique proteoforms spanning H1, H2, H3, and H4 variants were elucidated by 193 nm UVPD, in many cases pinpointing multiple modification sites based on backbone cleavages that flanked the methylation or acetylation sites and allowed them to be confidently bracketed.173 UVPD returned better scoring metrics for 80% of histoforms found in common with HCD.173 The same type of bracketing strategy, which depends on the ability of UVPD to promote cleavage of nearly every backbone position, was used to confirm the incorporation of selenocysteines into proteins175,176 and to facilitate the localization of cross-linking sites in ubiquitin and insulin.178 Data processing for the latter application required extensive iterative analysis to localize the cross-linker attachment sites based on determination of the transition points from unmodified fragment ions to mass-shifted (e.g., containing the cross-linker) fragment ions.178
Figure 23.
Sequence maps of the H4 proteoform acNacK16me2K20 showing coverage and scoring metrics by (a) HCD, (b) EThcD, and (c) 193 nm UVPD. Reproduced with permission from ref 173. Copyright 2018 American Chemical Society.
The amide group serves as a chromophore at 193 nm, meaning that proteins may have hundreds of suitable chromophores to ensure efficient photoabsorption (π-to-π* transition).181 As the number of amino acids in a protein increases, its photoabsorption cross section also increases, meaning that the laser power or number of pulses used to cause photodissociation can be decreased as the size of the protein increases. Owing to the high repetition rate of excimer lasers and the need for as little as one pulse for satisfactory dissociation, UVPD was readily integrated into an LC-MS workflow for high-throughput proteomics.182,183 Top-down analysis using UVPD has been demonstrated for mixtures of ribosomal proteins182 and more elaborate mixtures of proteins extracted from HeLa cells.183 In the latter study, 153 proteins and 489 proteoforms were assigned by UVPD.183 A greater number of proteins and proteoforms were identified by higher-energy collisional dissociation (HCD), but the scoring metrics that enable confident characterization of sequences were higher for UVPD.183 For example, UVPD afforded up to 74% higher sequence coverage for specific proteins than the same ones identified by HCD, and UVPD characterized 203 unique proteoforms. The results of the two comparative studies demonstrated that UVPD offered a complementary strategy to HCD, in terms of identification of a number of unique proteoforms not found by HCD or improving characterization of a number of specific proteins identified by both HCD and UVPD, for high-throughput top-down proteomics.183
3.1. Fundamental Aspects
Owing to the successful application of UVPD for both characterization of individual proteins and for high-throughput top-down proteomics, efforts to better understand the mechanism of UVPD and the impact of charge state have been pursued.184–187 In one extensive study, the influence of charge state and proton mobility on the UVPD of various proteins was examined with carbamylation to modify the basicity of primary amines (Lys side chains and N-terminus) and suppress protonation of those sites.184 Although carbamylation substantially reduced the sequence coverage obtained upon collisional activation of proteins, the same effect was not observed upon UVPD, confirming that the mechanism of UVPD displayed little dependence on charge state.184 A related study generated proteins in low charge states by a number of different methods, including modulating the pH or ionic strength of the protein solution prior to electrospray ionization.185 The results from this latter study demonstrated that UVPD consistently favored nonselective backbone cleavages, meaning that UVPD exhibited little preference for cleavages adjacent to specific amino acids.185 The latter phenomenon is prevalent upon collisional activation depending on the charge state of the precursor ion and is thought to be one of the factors that contributes to limited sequence coverage of intact proteins by CID.
UVPD of multicharged proteins results predominantly in production of a/x-type fragment ions arising from cleavages of the C–Ca backbone positions. The pattern of a/x ions was used in an innovative way to derive the locations of charge sites in proteins.186 Each sequential protonation site from N-terminus to C-terminus of the protein was assigned by monitoring the transitions in the charge states of a/x ions generated upon UVPD.186 This strategy focused on analysis of a/x ions owing to their origination from direct fragmentation of ions in excited electronic states for which proton mobility does not play a signifciant role. The localization of charge sites was based on the assumption that each transition reflected the retention of an additional proton localized at or near the terminal residue of a more highly charged fragment ion containing N residues relative to a lower-charged fragment ion containing N-1 residues. An example of the distribution of the charge states of a/x ions generated upon UVPD of ubiquitin is shown in Figure 24.186 The shifts in the charge states of the fragment ions are clearly demarcated, thus facilitating prediction of charge site locations. The prediction of charge sites was also demonstrated for β-lactoglobulin.186
Figure 24.
Fractional abundance per charge state of the a and x ions of ubiquitin for 5+ (a, b), 6+, (c, d), 7+ (e, f), and 8+ (g, h). (i) Charge sites of ubiquitin elucidated based on the a/x ions from UVPD are highlighted in bold red font for the 5+, 6+, 7+, and 8+ charge states. The arrows are shown to highlight the changes in protonation sites as a function of increasing charge state. (j) The secondary and tertiary structures of ubiquitin (5+ and 8+) are represented with salt bridges denoted by hashed lines. The amino acids that are protonated for the 5+ and 8+ charge states are highlighted in red font. Reprinted with permission from ref 186. Copyright 2016 Royal Society of Chemistry.
3.2. Hybrid Methods and New Concepts for Top-Down Analysis
To enhance the array of fragment ions and expand the sequence coverage obtained by UVPD, several new strategies have been developed.187–189 One study reported the hybridization of ETD and UVPD for analysis of intact proteins.187 This hybrid ETD/UVPD method accomplished two goals: (i) allowing modulation of the distribution of a/x, b/y, and c/z ions and (ii) reducing the density of key regions of the MS/MS spectra by affording greater dispersion of fragment ions among different charge states over a broader m/z range. These two features improved characterization of intact proteins by yielding new fragment ions that filled in sparse regions of coverage and by decreasing spectral crowding in regions of the MS/MS spectra more densely congested with fragment ions of similar m/z values.187 An example of UVPD and two hybrid methods, ET/UVPD (in which UVPD is used to activate one specific product from the initial ET process) and ET/broadband UVPD (in which all fragment ions created by ETD are simultaneously subjected to UVPD), employed for analysis of ubiquitin is shown in Figure 25.187 The blue-shaded sections of the spectra illustrate regions where new fragment ions emerge upon adoption of the hybrid methods. The hybrid ET/UVPD method yielded a different distribution of fragment ions compared to UVPD alone.187
Figure 25.
Portion of MS/MS spectra for ubiquitin acquired using three UVPD methods: (A) 193 nm UVPD (one 2.5 mJ laser pulse) of ubiquitin (12+), (B) ET/UVPD (8 ms ETD in LIT) of ubiquitin (13+) followed by one 1.8 mJ laser pulse of ubiquitin (12+·) (MS3), (C) ET/broadband UVPD (15 ms ETD) of ubiquitin (13+) in HCD cell followed by one 2.5 mJ laser pulse of all product ions. All spectra are shown on the same scale. (D) Segmented bar graph showing percentages of ion-type pairs identified by UVPD, ETD, and ET/UVPD for ubiquitin with all activation events performed in the HCD cell (UVPD, one 2.5 mJ laser pulse; ETD, 15 ms; ET/UVPD, 4 ms ETD, and one 1.0 mJ laser pulse). Reproduced with permission from ref 187. Copyright 2014 American Chemical Society.
The strategic use of multiple MS/MS modes was further extended by employing ECD, UVPD, HCD, and hybrid combinations (ECuvPD and EChcD) in order to expand the sequence coverage obtained for intact proteins, including subunits of monoclonal antibodies.190 A comparison of the sequence coverages obtained for the three subunits of an antibody in Figure 26 shows that combining MS/MS methods yielded the highest sequence coverage among all of the MS/MS methods evaluated.190 In another study, charge-reducing proton-transfer reactions were used as a sophisticated means to modulate protein charge states and to focus broad arrays of charge states into individual charge states, both for denatured proteins in high-charge states188 and for native-like proteins in low-charge states.189 By manipulating charge states of proteins, mass spectra could be decongested via charge-state separation of overlapping species, as demonstrated for monomeric and dimeric forms of superoxide dismutase in which, for example, the 5+ state of the monomer had an identical m/z value as the 10+ state of the dimeric protein.189
Figure 26.
Sequence coverages obtained from a single LC/MS/MS experiment using ECD, ECuvPD (1 pulse at 0.5 mJ, 193 nm), EChcD (CE = 12 eV), and combined coverage for the LC, Fc/2, and Fd subunits of SigmaMAB (MSQC4). The reported sequence coverage for the Fc/2 subunit is for the G0F glycoform. Analyses were performed for the 20+, 25+, and 24+ charge states of the LC, Fc/2, and Fd subunits, respectively. Reproduced with permission from ref 190. Copyright 2018 American Chemical Society.
The development of hybrid activation methods and combined utilization of multiple activation method have offered significant inroads for more comprehensive characterization of intact proteins by top-down mass spectrometry methods. However, these approaches require more elaborate optimization of experimental parameters for multiple MS/MS modes and increase the burden of data analysis. An algorithm, topdownr, was developed to address the hurdles of managing workflows that deploy multiple MS/MS methods such as CID, HCD, ETD, ETciD, EThcD, and UVPD.191 As demonstrated for a series of five proteins spanning a range of molecular weights, the software package accomplished curation of thousands of MS/MS spectra, designed guidelines for optimizing protein fragmentation, and streamlined development of multimodal MS/MS approaches.191 By strategic combination of MS/MS spectra obtained using different activation methods, unsurpassed levels of sequence coverage were obtained while minimizing data acquisition time from excessive optimization of ion-activation parameters.191
Consistent with the prevailing understanding of the impact of charge state on the fragmentation of proteins, the total sequence coverage obtained upon activation/dissociation of an intact protein was significantly more dependent on the charge state of the protein for HCD compared to UVPD.184 The efficiency and extent of fragmentation promoted by UVPD depend on the photon flux, a factor that is modulated by the laser power and the number of laser pulses. Application of a greater number of pulses offers one way to improve the conversion of precursor ions into diagnostic product ions. However, owing to the nonselective activation mechanism of UVPD, both fragment ions and precursor ions that pass through the path of the laser beam may undergo photoabsorption.192 Although often this is beneficial for increasing the conversion of precursor ions into product ions, it also increases the probability that fragment ions will undergo secondary dissociation.192 The latter process has several drawbacks. For one, it enhances the formation of small, uninformative fragment ions and internal ions (ones that retain neither C-terminus nor N-terminus) that are challenging to identify.192 Furthermore, secondary dissociation can disperse the fragment ion current into more channels, thus decreasing the signal-to-noise of the resulting spectra. A strategy, termed fragment ion protection, was developed to utilize dipolar excitation waveforms to displace the fragment-ion population outside of the laser beam path and thereby arrest undesirable secondary photoactivation/photodissociation processes.112 The concept and examples of results are illustrated in Figure 27. The selective ion-displacement method prevented photoabsorption and secondary dissociation of primary fragment ions (see Figure 27A) while allowing the use of multiple laser pulses to increase conversion of the targeted precursor ions into meaningful fragment ions.112 For UVPD of intact proteins, the production of larger fragment ions via backbone cleavages in the midsection of the protein was enhanced. The net outcome was an increase in the S/N of certain fragment ions by a factor >3 and a doubling in the total abundances of larger fragment ions that are critical for covering the midsections of protein sequences.112 Examples displaying the improvement in S/N for four diagnostic fragment ions (y5810+, a7010+, z639+, and a7110+) from UVPD of ubiquitin are shown in Figure 27B.
Figure 27.
(A) Concept of UVPD with fragment ion protection (FIP) showing resonant excitation to move fragment ions away from the laser beam. (B) Sequence coverage map of UVPD of ubiquitin (12+) using eight 0.25 mJ pulses and expansions of ion profiles and signal-to-noise levels for four large fragment ions without FIP (upper profiles) and with FIP (lower profiles), including y5810+ (m/z 654), a7010+(m/z 787), z639+ (m/z 788), and a7110+ (m/z 798). Results represent the average of three spectra (each consisting of 6 μscans) generated by Thermo Xcalibur Qual Browser v2.2. Reproduced with permission from ref 112. Copyright 2018 American Chemical Society.
3.3. Other Wavelengths for Top-Down Analysis
Although the great majority of studies that have reported UVPD of intact proteins have utilized 193 nm photons, there have been a few other studies that used 266 nm193,194 or 213 nm photons.54,195–197 In one study, 266 nm UVPD was integrated with electron-capture dissociation (ECD) to increase the sequence coverage obtained upon top-down analysis of intact proteins.193 Proteins containing disulfide bonds typically display significantly suppressed fragmentation in those regions of the sequence anchored by disulfide bonds. Upon exposure to 266 nm photons, disulfide bonds in the proteins were cleaved, thus facilitating the subsequent fragmentation of the protein by ECD.193 Evidence suggested that up to three or four disulfide bonds could be selectively cleaved by 266 nm UVPD, thus offering an intriguing opportunity for overcoming one of the hurdles of MS/MS analysis of intact proteins.193 An example illustrating the cleavage of three disulfide bonds of insulin using a hybrid 266 nm UVPD/ECD method is shown in Figure 28. Another study reported an innovative derivatization strategy with 266 nm UVPD to determine the number of free and bound cysteines in proteins.194 Proteins were derivatized using N-(phenylseleno)phthalimide (NPSP) to create Se–S bonds that selectively cleaved upon exposure to 266 nm photons. The detachment of SePh groups, resulting in consecutive mass shifts of 156 Da, reflected the number of cysteines per protein. The cysteine content was determined for proteins containing up to eight cysteine residues, such as lysozyme and α-lactalbumin, by this 266 nm UVPD method.194
Figure 28.
Cleavage pattern of insulin by 266 nm UV-activation/ECD showing three disulfide bonds cleaved (blue, purple, and gold). Fragment ions are color-coded according to the corresponding cleaved disulfide bonds. Reprinted with permission from ref 193. Copyright 2015 Elsevier.
The integration of two lasers (IR and Nd:YAG) on a quadrupole-Orbitrap mass spectrometer was the first strategy allowing simultaneous or consecutive irradiation of intact ubiquitin ions with low-energy (10.6 μm from a 50 W cw CO2 laser) and higher-energy (213 nm from a pulsed Nd:YAG laser) photons.195,197 This dual-photactivation process, termed HiLoPD, was compared to IRMPD or 213 nm UVPD alone. Strategic control of the photoactivation process undertaken in the HCD cell allowed modulation of the distribution of fragment ions as well as production of diagnostic side-chain loss ions via secondary fragmentation processes that aided the differentiation of isomeric amino acids and localization of post-translational modifications of peptides.195 Examples of the number and distributions of fragment ions generated by IRMPD, 213 nm UVPD, and HiLoPD are shown in Figure 29 for two peptides containing PTMs, one a phosphopeptide and one a glycosylated peptide.197 HiLoPD afforded excellent sequence coverage and allowed ample retention of PTMs, including phosphorylation, sulfation, and glycyoslation, which facilitated their characterization. For analysis of intact proteins, the combination HiLoPD method provided greater sequence coverage than IRMPD or UVPD alone, as demonstrated in Figure 30 for the analysis of ubiquitin.195 HiLoPD truly offers a hybrid mixture of the types of fragment ions generated by IRMPD and UVPD.
Figure 29.
Number of fragment ions (without PTM loss) detected in IRMPD (using a 50 W cw CO2 laser, 50–200 ms), UVPD (using 1–2 pulses from a 213 nm Nd:YAG laser), and HiLoPD (using simultaneous IR and UV irradiation) mass spectra of (a) the +3 charge state precursor ion (m/z 665.3544) of FFKNIVTPRT(H2PO4)-PPPSQGK peptide and (b) the +3 charge state precursor ion (m/z 556.9529) of EAISPPDAAS(GalNAc)AAPLR peptide. Reprinted with permission from ref 197. Copyright 2018 Springer.
Figure 30.
Sequence coverage of the +13 charge state precursor ion (m/z = 659.8249) of ubiquitin observed by IRMPD, UVPD (four pulses, 1 mJ/pulse of 213 nm from a Nd:YAG laser), and HiLoPD (simultaneous introduction of UV and IR lasers using 1 s IR irradiation and four pulses, 1 mJ/pulse of 213 nm from a Nd:YAG laser). Reprinted with permission from ref 195. Copyright 2016 Springer.
With the recent commercialization of 213 nm UVPD on an Orbitrap mass spectrometer using a small solid-state laser, two studies have presented results of UVPD for analysis of intact proteins.54,196 One study reported the use of 213 nm UVPD and ETD for combined top-down and middle-down analysis of a 150 kDa monoclonal antibody.54 Coverage of 40% was achieved by combining the results from ETD and 213 nm UVPD, representing the highest performance achieved for an intact antibody. Even greater coverage was obtained by integrating the fragmentation provided by ETD, 213 nm UVPD, and EThcD methods for subunits created by IdeS digestion of intact antibodies. Examples of the sequence maps showing total coverages of 72.0% (Fd subunit) to 86.6% (Fc/2 subunit) for rituximab and Venn diagrams displaying the levels of unique coverage and overlapping coverage among the three MS/MS methods are shown in Figure 31.54 The other study described the application of 213 nm UVPD in conjunction with hydrogen–deuterium exchange for analysis of myoglobin.196 UVPD (213 nm) provided fragmentation deeper into the middle region of the protein, thus enhancing characterization of the protein.196
Figure 31.
LC–MS characterization of the three ~25 kDa subunits generated from IdeS proteolysis of rituximab. The left sides of panels A–C show the fragmentation maps of Fd, Lc, and Fc/2, respectively, obtained by summing data from three ETD runs, two 213 nm UVPD runs, and one EThcD run (total of six LC–MS2 runs). The presence of the N-linked glycan, mapped using diagnostic fragment ions that include the mass of the sugar chains G0F, G1F, and G2F, is highlighted by an orange rectangle. For each panel, the right side shows the corresponding Venn diagram of shared/unique matched fragment ions for each of the three ion fragmentation techniques. Reproduced with permission from ref 54. Copyright 2018 American Chemical Society.
4. UVPD FOR NATIVE MASS SPECTROMETRY AND STRUCTURAL BIOLOGY APPLICATIONS
An array of mass spectrometry-based approaches have been developed to characterize composition, stoichiometry, topology, higher-order structure, binding affinities, dynamics, and thermodynamics of proteins and protein complexes.198–202 Some of these methods have relied on traditional chemical reaction strategies to probe proteins in solution, followed by analysis of the products by mass spectrometry. These reaction strategies include hydrogen/deuterium exchange (HDX), cross-linking, and other chemical-probe methods used to evaluate solvent-accessible regions of folded proteins and protein complexes.155,199,200,203 The workflow for these methods typically relies on conventional bottom-up MS/MS analysis to analyze the resulting peptides and relate mass shifts or abundances to structural features of the targeted proteins. Another suite of strategies has entailed transfer of intact native-like proteins or protein complexes into the gas phase, followed by direct MS1 and/or MS/MS analysis, occasionally with ion mobility, to glean information about secondary, tertiary, and quaternary structural features in a top-down manner.201,202 Both the bottom-up and top-down approaches rely on comprehensive characterization of the peptide or protein products, so the performance of the MS/MS method is critical and has catalyzed the use of alternatives, such as UVPD, to the more conventional collisional-activation methods.
4.1. Chemical-Probe Methods
Chemical-probe strategies generally entail exposing a protein or protein complex to a reactive agent in solution, causing covalent modification of residues based on a combination of site-specific reactivity and solvent accessibility.203 It is presumed that the reactivity of the various amino acids correlates with their accessibility, further modulated by local pKa environment and participation of side chains in hydrogen bonds and salt bridges. In essence, less-reactive sites are typically those that reside in the interior or are engaged in interactions within the protein. The resulting proteins are typically proteolytically digested prior to analysis of the constituent peptides.203 The abundances of modified and unmodified peptides reflect the reactivity or accessibility of the protein in those regions mapped by the identified peptides. Analysis of the proteolytic mixtures is complicated by the heterogeneous array of both modified and unmodified peptides, posing problems for both identification and quantitation. A number of strategies using chemical probes and cross-linkers that are selectively cleavable or enrichable have been designed to address these challenges.199,203 In addition, more sophisticated or selective MS/MS methods have been developed to better target the often low abundances of the key modified peptides in mixtures or to improve the localization of modified sites. In this context, the attributes of UVPD, particular with respect to its potential for high selectivity and the production of rich, highly informative fragmentation patterns, have been explored for MS-based chemical-probe applications. In one approach, the selectivity of absorption of 351 nm photons was exploited as a means to streamline the identification of probe-modified peptides in complex mixtures.138,139,204 An amine-specific chromogenic chemical probe was developed to react with lysine residues of proteins in a way that correlated with their solvent accessibility.138,139 The chemical probe contained a chromophore that endowed all modified peptides with high photoabsorption cross sections at 351 nm, whereas all unmodified peptides would be “silent”.138,139 Using 351 nm photodissociation as the MS/MS method in the bottom-up LC-MS workflow facilitated differentiation of probe-modified and unmodified peptides.138,139 UVPD analysis of the probe-modified peptides allowed details of the structures of several proteins, such as eIF4E, to be unravelled, such as differences related to complexation with a cognate peptide or guanosine-5′-triphosphate (GTP) ligand as well as the impact of the redox state of key disulfide bonds.139
To overcome some of the limitations of bottom-up methods used in conjunction with chemical-probe methods, top-down strategies that involve activation and dissociation of intact proteins have gained attention. The MS1 analysis of an intact protein allows unambiguous determination of the mass of the protein in addition to the number of covalent labels attached from a chemical-probe method. In addition to alleviating the potential need to use multiple proteases to generate panels of peptides that offer broad protein coverage, MS/MS analysis of intact proteins is ideally suited for elucidation of all sites of modifications if sufficient sequence coverage is obtained. The latter feature is a particular attribute of UVPD and has motivated the application of UVPD for chemical-probe and cross-linking methods.138,139,177 In one study, UVPD was used for top-down characterization of myoglobin and C-domain of poly(ADP-ribose) polymerase 1 (PARP-1) labeled with an amine-reactive chemical probe, S-ethylacetimidate (SETA).177 The goal was to evaluate top-down UVPD-MS as a means to monitor conformational changes and the engagement of polar contacts of proteins as a function of denaturation in acetonitrile and to elucidate changes in the nature of tertiary polar contacts. Each protein was reacted with SETA in different solution compositions ranging from 0% to 50% acetonitrile, followed by analysis by ESI-UVPD-MS.177 The degree of SETA incorporation at different lysine side chains was expected to correlate with the degree of solvent accessibility. Envelopes of fragment ions containing a specific diagnostic ion (unmodifed) plus the SETA-modified analogues were used to estimate SETA incorporation at each lysine residue in the protein, as illustrated in histogram format for 16 lysine residues of PARP-C (16 kDa) in Figure 32.177 For the lysine side chains, the SETA reactivity values ranged from near 0 (i.e., no reactivity) to 0.5 (high reactivity). It was found that the level of acetonitrile in the initial protein solution modulated the reactivity of different lysine residues (measured based on the attachment of the chemical probe) in a way that implied specific residues that shared topological orientations exhibited similar responses to denaturation, such as an increase in reactivity for buried residues that became more exposed upon protein unfolding.177 The high sequence coverage afforded by UVPD, ranging from 72% to 82%, proved to be an important metric for this type of top-down/chemical-probe study as it dictated the number of residues that could be monitored in a quantitative manner.177
Figure 32.
(A) Absolute SETA probe-incorporation values (n = 3) of PARP-C per lysine for the native (0% acetonitrile (ACN)), 25% ACN, and 50% ACN solution conditions based on 193 nm UVPD data. K324 was not quantified in the 50% ACN solution due to lack of sequence coverage beyond K324. K305 and K320 were not observed. K347 and K349 were averaged together. (B) PARP-C with color-coded lysine residues that represent their SETA incorporation trend as a function of acetonitrile solution composition with two views rotated by 180°. Pink = increasing SETA incorporation, blue = decreasing SETA incorporation, light brown = no change in SETA incorporation, and orange = variable SETA incorporation. NMR structure PDB ID 2JVN, state 1 shown. Reproduced with permission from ref 177. Copyright 2014 American Chemical Society.
Protein cross-linking integrated with mass spectrometry is another pivotal strategy used to profile conformational changes of proteins and map networks of protein–protein interactions.205,206 Efforts to advance cross-linking methodology have focused on developing ways to extract more information from MS/MS spectra of cross-linked species and enhance interpretation of data sets that may be saturated with false positives. Two of the ongoing hurdles include (1) differentiation of cross-linked peptides from unmodified ones in enzymatic digests of cross-linked proteins and (2) the identification of specific cross-link locations using tandem MS.205,206 To circumvent these limitations, extensive effort has been devoted to designing new cross-linkers that incorporate isotope labels or fluorophores, or ones that have preferentially cleavable bonds to simplify identification of low-abundance cross-linked species in complex mixtures containing many unmodified peptides.205,206 UVPD has been integrated with bottom-up analysis of cross-linked proteins to facilitate characterization of peptides based on the dense information-rich fragmentation patterns created by UVPD.207 UVPD enhanced the identification of larger cross-linked peptides found in higher charge states and those involving acidic residues compared to those determined from HCD.207 At this stage, UVPD should be considered a complementary MS/MS option, not a replacement, for conventional collision- and electron-based methods in the workflow for the bottom-up analysis of cross-linked peptides.
Another alternative approach that has been far less widely explored in the arena of cross-linking is top-down MS/MS analysis of intact cross-linked proteins. Few top-down studies have been reported owing to the challenging interpretation of MS/MS spectra produced for proteins that contain a heterogeneous set of cross-linked sites, in addition to insufficient sequence coverage to allow confident localization of those sites. UVPD (193 nm) was used in one study that focused on characterizing both intramolecular and intermolecular cross-links in two protein systems, ubiquitin and hexameric insulin.178 In each case, nanoflow liquid chromatography allowed separation of the cross-linked proteins from unmodified or dead-end modified proteins and even provided a degree of separation of isomeric cross-linked proteins. Cross-linking sites were identified by monitoring cross-link-specific mass shifts of fragment ions produced by UVPD. For ubiquitin, three pairs of cross-linked amines were determined by UVPD, K11/K22, N-terminus/K29, and N-terminus/K63.178 Three intermolecular cross-links were identified upon UVPD of hexameric insulin (N-terminus of A-chain/N-terminus of B-chain, N-termini of two B-chains, and N-terminus of A-chain/Lys29 of B-chain).178 Although using UVPD for top-down analysis of cross-linked proteins is a sophisticated strategy that solves some of the shortcomings of bottom-up approaches, the limited chromatographic resolution obtained for intact proteins and incomplete sequence coverage remain obstacles that have deterred greater adoption of the method.
4.2. Native MS
In lieu of chemical-probe, HDX, and cross-linking methods, the newest paradigm in mass spectrometry-based structural biology entails direct analysis of intact protein complexes by mass spectrometry.198–202 Advances in ionization methods have facilitated the transfer of proteins in native-like conformations as well as noncovalent protein complexes into the gas phase, thus founding a new frontier for mass spectrometry in the field of structural biology. Native mass spectrometry (native-MS) typically uses nanoelectrospray ionization (nanoESI) for analysis of solutions of high ionic strength, thus preserving noncovalent interactions of macromolecular complexes.200 The degree to which true native structures and conformational integrity of proteins are retained in the solvent-free environment of the gas phase remain a topic of debate, but a growing number of studies have demonstrated evidence for compelling correlations between proteins in solution and those ejected from droplets in the ESI process.200,208 Integrating this native ESI method with high-performance mass spectrometry allows high-mass-accuracy/high-resolution assignment of ions, often coupled with structural analysis by MS/MS. These MS-based strategies have been used to analyze incredibly large and complex assemblies, such as 18 MDa capsids of bacteriophage HK97.209 The unique capacity of mass spectrometry to discriminate proteoforms and different complexes on the basis of m/z allows investigation of highly heterogeneous populations of proteins. Additional biophysical information can be obtained by separating proteins by ion mobility,199,201 thus providing information about different conformations, or by dissociating the proteins and protein complexes to afford sequence information that may reflect the impact of ligand binding and both intramolecular and intermolecular interactions.202
4.2.1. Protein–Ligand Complexes.
A number of ion-activation methods, including collision-induced dissociation, electron-activation methods (electron-capture dissociation and electron-transfer dissociation), surface-induced dissociation, and photoactivation, have been implemented to disassemble protein complexes and characterize the constituents.210 Collisional activation of protein complexes frequently causes cleavage of noncovalent electrostatic interactions as the dominant process, resulting in unfolding of proteins and asymmetric charge redistribution prior to separation of the components.210 This outcome has limited the application of collisional activation for conformational investigation of protein complexes. Electron-capture dissociation (ECD) has been particularly useful for characterization of protein conformational features and protein–ligand complexes.211 Noncovalent interactions are maintained during the electron-activation process, thus allowing dissection of the protein backbone to generate informative sequence ions with retention of bound ligands in a manner that allows elucidation of binding sites. ECD favors backbone cleavages in regions of proteins that are more flexible, disordered, and dynamic, thus demonstrating a correlation with B-factors from crystallography meaurements.211 Similar findings have arisen from the use of electron-transfer dissociation (ETD), an analogue of ECD that uses reagent anions to provide the source of electrons for the electron-activation process. ETD has thus been used for a number of fundamental studies of native-like proteins.212 The ability to derive structurally meaningful information from ECD and ETD spurred others to consider the underpinnings of the method and whether UVPD, owing to its fast activation process and high-energy deposition, might offer similar attributes for characterization of protein complexes.
A series of investigations emerged beginning in 2014 to explore the use of 193 nm UVPD to characterize protein complexes.213–222 One of the primary findings was that UVPD, in addition to causing cleavages of backbone bonds of the proteins to generate sequence-type a/b/c/x/y/z ions, also generated products that contained bound ligands (termed “holo product ions”).213 These latter products presumably signified that noncovalent interactions were retained even as covalent bonds of the protein backbone were cleaved. This interesting outcome signaled the ability to use UVPD to characterize structures of native-like protein complexes, including localization of ligand-binding sites and examination of conformational variations in protein structures as a function of ligand binding. The first study in this body of work demonstrated the use of UVPD both to characterize the sequences of proteins in native-like protein complexes and to shed light on the binding sites of the ligands and the nature of the protein–protein interfaces.213 The featured protein–ligand complexes included myoglobin/heme, eIF4E/m7GTP, and the complex containing human peptidyl-prolyl cis–trans isomerase 1 (Pin1) and a peptide derived from the C-terminal domain of RNA polymerase II (CTD).213 For example, C-terminal (x, y, z) fragment ions of Pin1 that retained the CTD peptide were those that contained the key WW domain, and the only N-terminal (a, b, c) sequence ions that retained the CTD peptide were those that covered the Pin1 PPIase domain.213 The pattern reflected the key outcome that retention of noncovalently bound ligands typically correlated with stabilization provided by the known binding domains of the protein. Some fragment ions produced upon UVPD of oligomeric β-lactoglobulin dimers and hexameric insulin complexes were consistent with ones containing portions of multiple subunits, reinforcing the idea that the high-energy deposition that accrued during absorption of 193 nm photons allowed retention of noncovalent protein–protein interactions even during cleavage of covalent backbone bonds.213 The intensive data analysis required for interpretation of the UVPD mass spectra motivated the development of a portfolio of data-processing tools collectively termed UV-POSIT.214 These data-processing utilities facilitated the evaluation of fragmentation efficiencies throughout the protein backbone, assessment of locations of charge sites, analysis of hydrogen atom migration during fragmentation, and localization of mass shifts associated with bound ligands.214
Deciphering secondary structural features of proteins remains a significant challenge. A strategy that dissected the distribution of a-type ions generated upon 193 nm UVPD was recently reported as a means to decipher hydrogen-bonding interactions, ones important in the construction of secondary structure motifs of native-like proteins.78,215 Termed “hydrogen-elimination monitoring” (HEM), the method focused on tracking the formation of a versus a + 1 ions upon cleavage of each interresidue backbone bond of a protein. The concept of hydrogen-elimination monitoring arose from the hypothesis that a ions originated via hydrogen loss from a + 1 ions, a pathway modulated by the hydrogen-bonding motifs localized to the amino acids linked by the backbone bond undergoing cleavage.78 The preferential production of a + 1 ions over a ions correlated with the engagement of the protein backbone in hydrogen-bonding interactions. In essence, the ratio of a to a + 1 ions proved to be sensitive to the identity of the amino acid and the presence or absence of hydrogen bonds at the amide carbonyl oxygen atoms.78,215 The HEM UVPD method was applied for characterization of hydrogen-bonding motifs in structured proteins like Scp1 and calmodulin as well as disordered proteins akin to α-synuclein.215
Another study used 193 nm UVPD to examine the structures of apo- and holo-myoglobin in the gas phase.216 The UVPD fragmentation efficiencies correlated with the B-factors of myoglobin derived from crystallographic measurements, with the highest values being for the most disordered loop regions and the frayed helical termini. Both holo- and apo-myoglobin displayed low protein backbone fragmentation efficiences in the region spanning the AGH helical core.216 Moreover, backbone cleavages were suppressed in regions of myoglobin that interacted with heme. In general, it was found that the fragmentation patterns obtained from UVPD exhibited little dependence on side-chain interactions; however, backbone cleavages were suppressed in regions where amides were involved in hydrogen-bonding interactions. Moreover, as the charge state increased, the UVPD fragmentation yields increased for the N-terminus region of the G-helix, the F-helix, and the FG loop, suggesting a degree of conformational reorganization.216
Analysis of UVPD fragmentation efficiency of the protein backbone was also used to examine the structure of dihydrofolate reductase (DHFR) and its complexes with cofactor NADPH, inhibitors methotrexate (MTX), and several propargyl-linked antifolates.217,218 Changes in fragmentation yield were observed between DHFR and the binary and ternary complexes; for example, fragmentation of the ternary complex was enhanced relative to apo-DFHR in two different loop regions (M20 and loop domain II) and suppressed in the adenosine-binding domain. Suppression of fragmentation in the adenosine-binding domain was found to be consistent with an expected reduction in conformational flexibility of this region upon ligand binding.217 Shown in Figure 33 are color-coded crystal structures of DHFR that indicate backbone sites where 193 nm UVPD is enhanced or suppressed upon binding of NADPH, MTX, or both ligands relative to the apo protein.
Figure 33.
Color-coded residues that had an enhanced (positive) or suppressed (negative) change in 193 nm UVPD fragmentation upon comparison of apo-dihydrofolate reductase (DHFR) and (a) DHFR·NADPH, (b) DHFR·MTX, and (c) DHFR·MTX·NADPH as well as comparison of (d) DHFR·NADPH to DHFR·MTX·NADPH are highlighted according to the colored ranges. In each case, the increase (enhancement: yellow, orange, and red) or decrease (suppression: turquoise, royal blue, and purple) in UVPD fragmentation yield is shown as a percentage representing the change in ion abundance (based on total ion current) and superimposed on the crystal structures. Crystal structures 1RX1, 1RG7, and 1RX3 were used to represent the DHFR·NADPH (a), DHFR·MTX (b), and DHFR·MTX·NADPH (c, d) complexes, respectively. Two 45° rotations are shown for each complex. MTX is methotrexate. Reproduced with permission from ref 217. Copyright 2015 American Chemical Society.
Enhancement or suppression of backbone cleavages are modulated depending on the nature of the ligand (NADPH versus MTX) and its binding location (adenosine-binding domain for NADPH versus loop domain for MTX). For these studies, 193 nm UVPD provided over 80% sequence coverage of DHFR and led to production of fragment ions that contained one or both bound ligands (e.g., NADPH, methotrexate, or both NADPH and methotrexate).217 The binding sites of the NADPH and MTX ligands were also reflected in the patterns of apo and holo fragment ions, showing NADPH affiliated with the adenosine-binding domain and methotrexate bound to the B loop.217 A subsequent study focused on two clinically relevant mutations, P21L and W30R, of DHFR that are found in trimethoprim-resistant strains of E. coli that cause urinary tract infections.218 It was found that both P21L and W30R mutants produced less-stable complexes with trimethoprim in the presence of cofactor NADPH compared to wild-type (WT)-DHFR. Examination of the fragmentation patterns obtained by UVPD of the various DHFR complexes indicated that structural changes were localized to the substrate-binding pocket for W30R and to the M20 loop region and the C-terminal portion containing the G-H functional loop for the P21L mutant, therefore suggesting that the single-point P21L and W30R mutations caused different structural changes of DHFR.218
UVPD was also used to evaluate the impact of mutations on the conformations of another biologically important GTPase enzyme, K-Ras, based on variations in its backbone-cleavage efficiencies.219 K-Ras is a member of the sarcoma family of proteins that play critical roles in cell cycle progression and proliferation.223 K-Ras is frequently mutated in tumors, in up to 90% of pancreatic tumors, for example. For this study, the fragmentation patterns of complexes containing K-Ras (wild-type or G12C, G12S, or G12 V variants) and guanosine diphosphate (GDP) or a GTP analogue were compared to decipher the structural changes in the protein as a function of the single-point mutations.219 The most significant changes in fragmentation efficiency for the GDP- versus GTP-containing complexes correlated with the phosphate- and guanine-binding regions and were consistent with modulation of the binding motif upon replacement of the ligand (GDP versus GTP analogue).219 Moreover, variations in the 193 nm UVPD fragmentation patterns were observed for each G12X mutant. For example, the G12S mutant displayed higher levels of backbone cleavages that spanned the P-loop (residues 8–15), switch I (residues 29–33), and beta strand B (residues 37–45) regions, implying an increase in flexibility at the beta strand region in comparison to the G12V and G12C proteins. This study demonstrated that variations in backbone-cleavage efficiencies likely arose from variations in intra- or intermolecular interactions that inhibited separation of fragment ions during UVPD, making them sensitive to changes in single amino acids.219 A subsequent study extended the investigation of K-Ras to its dimer complexes with Raf, a downstream effector protein.220 The resulting K-Ras + Raf heterocomplexes were characterized using 193 nm UVPD to decipher structural changes that correlated with the formation of the dimeric complexes for wild-type K-Ras and the G12C, G12V, and G12S mutants. The relative fragmentation efficiencies of the dimeric K-Ras·GppNHp + RafRBD complexes suggested that hydrogen-bonding amino acid substitutions (G12C, G12S) stabilized the β-interface interactions with Raf, whereas the larger, hydrophobic G12V mutation destabilized this interface.220
Another study employed 193 nm UVPD to map the conformational variations of adenylate kinase during its entire reaction cycle via characterization of its binary and ternary complexes with adenosine monophosphate (AMP), adenosine diphosphate (ADP), adenosine triphosphate (ATP), and P1,P4-di(adenosine-5′)tetraphosphate (AP4A, an inhibitor).221 Another recent study compared electron-induced dissociation (EID) to 193 nm UVPD for characterization of human carbonic anhydrase I (HCA-I) and apo-, Zn-, and Cu,Zn-superoxide dismutase (SOD1) dimers.222 EID is a higher-energy version of electron-capture dissociation (ECD) in which the interaction of multicharged ions with 30 eV electrons causes simultaneous electron detachment and electronic excitation via a fast activation process that has similarities to UVPD. Examples of the backbone-cleavage maps generated from EID and 193 nm UVPD for human carbonic anhydrase and superoxide dismutase dimer are displayed in Figure 34.222 It was found that the regions from which backbone cleavages occurred were similar for the two activation methods, although EID resulted in mainly c/z ions in contrast to the a/x ions predominantly produced by UVPD.222 Additional comparisons will provide more insight into the similarities and differences between these two higher-energy activation methods (EID and UVPD) for characterization of protein complexes.
Figure 34.
Plot of backbone-cleavage sites with the respect to ion types (a/x, b/y, and c/z) obtained via electron-induced dissociation (EID) with in-source decay (ISD) or via UVPD. (A) ISD (120 V)-EID of human carbonic anhydrase I (HCA-I) with a/x, b/y, and c/z ions plotted separately; (B) 193 nm UVPD of HCA-I; (C) EID of apo-WT superoxide dismutase (SOD1) dimer; (D) UVPD of apo-WT SOD1 dimer (each spectrum was acquired from 200 scans). Reproduced with permission from ref 222. Copyright 2017 American Chemical Society.
The studies summarized earlier demonstrated that the abundances of holo (ligand-bound) and apo (ligand-free) fragment ions produced by UVPD correlated wtih secondary and tertiary structural features of the proteins. The basis for the correlation derived from the suppression or enhancement of UVPD at each position along the backbone, a process that appeared to be modulated by survival of noncovalent interactions that prevented separation of fragments after backbone bond cleavage.
UVPD using wavelengths other than 193 nm has also been explored for characterization of protein complexes. For example, synchotron radiation (16 eV energy) was used to activate a small protein (IB5)/tannin complex in a linear ion-trap mass spectrometer.224 IB5 posed a unique challenge because it is an intrinically disordered protein that is intractable by more conventional X-ray crystallography and NMR methods. The patterns of backbone cleavages leading to predominantly C-terminal x, y, and z fragment ions are illustrated in Figure 35 for IB5 bound to tannin B2 3′OG. The tannin binding site could be localized by detection of holo product ions that contained a portion of the IB5 protein and retained the tannin ligand.224
Figure 35.
Localization of the tannin (B2 3′OG) binding site on IB5. Patterns of the fragments linked to B2 3′OG for [IB5a·B2 3′OG + 7H]7+ after 16 eV dissociative photoionization. The binding site of B2 3′OG is highlighted by the dashed rectangle. Reprinted with permission from ref 224. Copyright 2013 Wiley.
4.2.2. UVPD for Multimeric Protein Complexes.
Collisional activation offers a strategy for disassembling macromolecular complexes into subunits via disruption of noncovalent interactions between proteins, but this process also causes the proteins to unfold and does not allow evaluation of the protein architecture.225–231 Electron-activation methods (ECD, ETD) have also been employed to characterize multimeric protein complexes, affording some sequence ions and exhibiting retention of noncovalent interactions.211,222,231–237 To date, surface-induced dissociation (SID) has emerged as the best activation method for disassembly of multimeric protein complexes in a manner that discerns subunit architecture.238–243 However, SID does not yield extensive sequence ions to allow identification of constituent proteins within the complexes. UVPD has more recently been applied as a new means to characterize multimeric protein complexes.179,244–246 UVPD (193 nm) has been explored as an alternative activation method in the context of probing architectures of protein complexes, evaluating charge partitioning among the subunits, and characterizing the sequences of proteins comprising the multimeric assemblies.179,244–246
For analysis of protein complexes, the dissociation pathways of UVPD were found to depend on the laser power or energy (e.g., photon flux).179,244 One study examined 193 nm UVPD of tetrameric streptavidin, transthyretin, and hemoglobin, all three with dimer-of-dimer architectures;244 the other investigation focused on dimeric β-lactoglobulin and superoxide dismutase, tetrameric concanavilin A, and heptameric GroES and Gp31.179 Using lower laser-pulse energies (≤1.5 mJ per pulse) resulted in CID-like disassembly of protein complexes with disruption of noncovalent interactions, separation of intact subunits, and asymmetrical partitioning of charge relative to the masses of the released proteins. In contrast, UVPD using higher powers (≥1.5 mJ per pulse) promoted disassembly of the complexes in a manner that revealed the architecture of the assemblies and resulted in production of sequence ions from the individual proteins.179,244 An example of the UVPD mass spectra obtained for tetrameric streptavidin (13+) using two different laser powers is shown in Figure 36.244 The distribution of charges among the released proteins was more symmetrical using the higher laser powers compared to the lower laser powers, thus indicating a more-symmetrical partitioning of charges based on the masses of the released subunits (i.e., monomer versus dimer versus trimer from a tetrameric presursor complex). Diagnostic fragment ions characteristic of the protein sequence are observed in the lower m/z range of the mass spectrum in Figure 34.244 In general, the efficiency of UVPD displayed little dependence on charge state, making it a robust method for characterization of protein complexes, especially given the higher level of sequence coverage obtained for the proteins relative to collisional activation.
Figure 36.
UVPD (193 nm) of tetrameric streptavidin (13+) using a single (a) 1.0 mJ or (b) 3.0 mJ laser pulse. Reproduced with permission from ref 244. Copyright 2016 American Chemical Society.
A systematic study of UVPD and the impact of laser power and charge state was reported for an array of multimeric protein complexes, including dimeric superoxide dismutase, tetrameric streptavidin, hemoglobin, transthyretin, and pentameric C-reactive protein.246 The results demonstrated that higher photon fluxes (3 mJ per pulse) resulted in release of low-charged monomeric ions presumed to be in compact conformations as evidenced from their low-charge states. In contrast, using lower photon fluxes (e.g., lower laser powers) alternatively yielded unfolded monomers in higher charge states and complementary charge-stripped subcomplexes.246 The differences in the results obtained using low and high laser energies was attributed to the prevalence for single-photon versus multiphoton processes occurring during a single pulse. The relative interfacial strengths of the protein complexes correlated with the degree of symmetry in the distibution of charges during disassembly of the complexes, much akin to the outcome of SID.246
Another emerging strategy for analysis of protein complexes by mass spectrometry entails the integration of native MS methods (i.e., gentle transfer of intact native-like protein assemblies into the gas phase from solutions of high ionic strength) with multistage MS/MS methods to deliberately disassemble and characterize the constituents of the complexes.230,231,245,247,248 Measurement of the masses of the intact complexes allows determination of the stoichiometries of complexes. Multiprotein complexes can be disassembled into the constituent subunits and individual proteins via low-energy collisional activation prior to or after isolation of specific complexes. Subsequent stages of collisional or electron activation of the subunits or individual proteins allow more detailed dissection of the complexes, thus allowing identification of the proteins and even characterization of specific proteoforms.230,231,245,247,248 This impressive multitier MS strategy has been applied to even larger-scale proteomic characterization of endogenous complexes in human cancer cells.248 Recently, this approach was modified to incorporate 193 nm UVPD as the means to characterize the constituent proteins comprising the complexes, as demonstrated for branched-chain amino acid transferase 2 (BCAT2), a human mitochondrial enzyme.245 For this study, the multistage native UVPD-MS strategy was implemented on an extended mass range Orbitrap mass spectrometer (prototype UHMR), and in-source trapping (low-energy collisional activation) was employed to break down the complexes prior to isolation and activation. The method allowed analysis of the oligomeric state of the complexes, and the use of UVPD to characterize the proteins facilitated localization of amino acid mutations and identification of the cofactors (pyridoxal 5′-phosphate) for two variants of BCAT2, each containing a single amino acid mutation.245 Examples of the MS/MS spectra obtained from this type of multistage approach are shown in Figure 37 for wild-type BCAT2 and its single-point variant T186R.245 Beneath the MS/MS spectra are sequence maps summarizing the patterns of backbone cleavages that lead to the sequence ions observed in the spectra. The color-coded cleavages in the sequence maps illustrate that UVPD creates a more diverse variety of fragment ions (a/x, b/y, c/z) than HCD (b/y). UVPD provided higher sequence coverage (37–45%) than HCD (13–18%) for characterization of each of the two proteoforms. By mapping the holo (pyridoxal 5-phosphate-containing) fragment ions produced upon UVPD, it was determined that the cofactor interacted with a specific loop containing residue K229. The extensive array of fragments generated by UVPD was pivotal for localizing the cofactor.245
Figure 37.
HCD and 193 nm UVPD mass spectra of the 12+ charge state monomer of (A) WT and (B) T186R BCAT2 observed after in-source trapping (IST). Expanded views (insets) in panel (A) show selected fragment ions labeled. Holo fragment ions that contain the pyridoxal phosphate (PLP) cofactor are denoted by a diamond. The precursors are indicated with a filled star in the MS/MS spectra. The unfilled star denotes the charge-reduced precursor (11+) as the PLP cofactor is ejected during HCD. Sequence coverage maps for (1) HCD and (2) UVPD of (A) WT and (B) T186R BCAT2 are shown beneath the spectra, with cleavages leading to a/x ions colored in green, b/y ions in blue, and c/z ions in red. The UVPD sequence maps represent the combined coverage for identified apo and holo fragment ions for the spectra collected using 1 and 3 mJ per pulse. Sequence coverages were 13 and 18% from HCD and 45 and 37% from UVPD for WT and T186R BCAT2, respectively. The Thr residue mutated to an Arg is shaded in gold in panel (B). Reproduced with permission from ref 245. Copyright 2018 American Chemical Society.
4.3. UVPD and Ion Mobility
UVPD has also been combined with ion mobility, an orthogonal approach that allows conformationally selective MS/MS characterization of ions.46,249–254 Ion mobility separates molecules in the gas phase on the basis of charge and shape.201,228 Although the application of ion mobility integrated with UVPD is still in its infancy, this hybrid strategy offers enormous potential in the field of structural biology. UVPD (193 nm) was first integrated with ion mobility to characterize the fragmentation patterns of different conformations of ubiquitin.249,250 An electrostatic gate was used to isolate the mobility-separated conformers prior to irradiation with 193 nm photons and mass analysis. Although mass resolution of this time-of-flight mass analyzer was not sufficient to resolve many of the lowest-abundance fragment ions, several prominent fragment ions resulting from cleavage of N-terminal to proline were found to vary in abundance for the different conformations of ubiquitin.249,250 The majority of the differences resulted from backbone cleavages adjacent to Pro19, and the same proline-mediated effect was not observed for cleavages adjacent to Pro37 or Pro38.249 Evaluation of molecular dynamics simulations supported the hypothesis that cis isomerization of Pro19 resulted in conformational changes of ubiquitin, which accounted for the changes in the shape that allowed the different conformations to be separated.249 In essence, the differences in ion mobility and UVPD fragmentation of ubiquitin were consistent with cis/trans isomerization of a proline peptide bond.
UVPD has also been implemented on a commercial ion-mobility mass spectrometer (Synapt G2) to explore peptide and protein structures using 266 or 213 nm photons for UVPD.46,251,252 In one case, UVPD was implemented in the transfer cell region to enable 266 nm UVPD after separation of different conformations of gramicidin and melittin by ion mobility.251 Variations in the fragmentation patterns of the mobility-separated ions confirmed that UVPD was sensitive to differences in conformations of the peptides.251 Examples of the UVPD produced for two conformers of melittin (5+ charge state), a 26-residue peptide, are shown in Figure 38.251 The 266 nm UVPD patterns are visibly different for the two conformers, with more extensive fragmentation for the elongated peptide. In a complementary approach, proteins were subjected to collisional unfolding by modulation of the cone voltage of the ion source region prior to 213 nm UVPD.252 The fragmentation patterns varied as a function of the degree of unfolding, thus providing a strategy to monitor conformational reorganization by UVPD.252
Figure 38.
Drift-time selected 266 nm UVPD of melittin (5+) with an irradiation time of 2 s (20 laser shots). (a) UVPD of the compact conformer C produces cleavage between seven residues producing b and y fragments while (b) UVPD of the more extended conformer E yields more extensive fragmentation with cleavage between 17 residues. The black trace represents the trapped-only spectrum without laser irradiation; the red dotted trace represents laser on. The main fragments produced or enhanced by UVPD are labeled on the spectra. The insets show the arrival-time distributions (ATD) of the two separated conformers (shaded in blue as C and E, which corresponds with the UVPD spectra). Reproduced with permission from ref 251. Copyright 2016 American Chemical Society.
Using a dual drift tube Q-TOF system, the Dugourd group also successfully coupled UVPD and ion mobility in a design that allowed selection of conformations based on ion mobility prior to UVPD and then examination of the ion mobilities of the resulting product ions.253 This approach was refined to incorporate a tunable laser to allow photodissociation measurements as a function of wavelength.253 Three conformations of the 4− charge state of an eosin Y tagged peptide were mobility-separated and individually activated.254 Photofragmentation yield as a function of irradiation wavelength allowed discrimination of three conformations, an outcome correlated with variation of the deprotonation site on the eosin chromophore.254
5. UVPD FOR LIPIDS
Lipids constitute one of the four major classes of biological molecules.255 These fatty acid molecules and their derivatives encompass extensive diversity, varying in headgroup chemistry, number and length of acyl chains, branching, chirality, and position and number of sites of unsaturation.255 Tandem mass spectrometry has emerged as an increasingly popular means to characterize all classes of lipids; however, a universal approach that deciphers the complete structures of lipids remains elusive. Because of the tremendous inroads in mass spectrometry analysis of lipids, readers are directed to several comprehensive reviews that cover other MS/MS approaches in greater detail.10–12 UVPD has been utilized for analysis of a number of classes of lipids owing to its ability to generate very rich fragmentation patterns that have successfully allowed elucidation of key structural features. As described in the following section, UVPD has been used to decipher locations of double bonds and cyclopropane rings within acyl chains, as well as sn positions of acyl chains of phospholipids, and to assign a number of modifications of saccharolipids.
Because of the variety of linkages, double bonds, and chiral centers found in phospholipids, sphingolipids, gangliosides, and other classes of biological lipids, differentiation of isomers is among the primary objectives of mass spectrometry-based methods and is a far-reaching goal of grander-scale lipidomics.10–12 Integral to this challenge is the fact that although collisional- and electron-based activation strategies yield information about acyl-chain lengths and degree of unsaturation, they often fail to provide structurally diagnostic fragments that allow determination of chiral linkages, branching, and other features that play vital roles in structure and function.10–12 For example, for many glycolipids the loss of the glycan is a dominant process upon collisional activation, consequently providing little information regarding the glycan or the stereochemistry of double bonds and linkages. The quest to characterize these more challenging features motivated the development and application of photon-based strategies, particularly radical-directed dissociation and ultraviolet photodissociation (UVPD). Other innovative nonphoton-based MS/MS methods have also been reported in the past decade, and these other methods (e.g., OzID, EIEIO, Paterno-Buchi reactions, EID) have also expanded the scope of mass spectrometry for characterization of features like double-bond positions.256–259
5.1. Radical-Directed Dissociation
One of the UV photodissociation methods applied for the analysis of lipids is radical-directed dissociation.260–265 As described earlier, radical-directed dissociation couples 266 nm photoactivation (to generate radical ions) with collisional activation to produce diagnostic fragment ions.113 The analyte of interest is typically derivatized to incorporate a tag with a photocleavable bond that initiates the RDD process. The C–I bond, for example, is homolytically cleaved upon exposure to 266 nm photons and, thus, has been widely used for RDD applications. One of the first approaches combined complexation of fatty acids with 4-iodoaniline or 4-iodobenzoic acid with RDD to differentiate branching positions and double-bond positional isomers in glycerophospholipids, sphingomyelins, and triacylglycerols.260 Upon exposure to 266 nm photons, cleavage of the carbon–iodine bonds in the fatty acid/iodobenzoate complexes yielded reactive phenyl radical-containing products. Subsequent collisional activation of the products generated an array of fragment ions derived from carbon–carbon bond cleavages along the alkenyl chains. In addition to neutral losses of the fatty acid chains that allow elucidation of the acyl group, the RDD fragments revealed the intrachain bonding patterns of the fatty acyl substituents.260 This RDD approach was also utilized for rapid characterization of an array of fatty acids, both saturated straight-chain and branched ones, including mono- and polyunsaturated, proving to be successful for localizing points of unsaturation, chain branching, and hydroxylation.261 Representative examples of the RDD spectra generated for three fatty acid isomers are shown in Figure 39. One unique fragment ion originating from C–C bond cleavage close to the double bond is observed for each isomer, thus allowing the position of the double bond to be pinpointed.261 More recently, this RDD method was adapted for the characterization of (O-acyl)-ω-hydroxy fatty acids possessing ultralong acyl chains.265 In this latter case, the lipids were derivatized using 1-(3-(aminomethyl)-4-iodophenyl)-pyridin-1-ium to facilitate production of radical ions upon 266 nm photoabsorption. RDD created extensive arrays of product ions separated by 14 Da that originated from C–C bond cleavage along the entire acyl chain of the lipid. The lack of any gaps in the series of ions supported the absence of methyl chain branching.265
Figure 39.
Photodissociation (266 nm) mass spectra of monounsaturated isomers [fatty acid 4-iodobenzyl esters (18:1) + Na]+ derivatized from (a) fatty acid (11Z-18:1), (b) fatty acid (9Z-18:1), and (c) fatty acid (6Z-18:1). The asterisk indicates the diagnostic product ion formed selectively from each double-bond positional isomer, thus allowing differentiation of double-bond positional isomers. Reprinted with permission from ref 261. Copyright 2013 Wiley.
RDD has also been applied to the localization of double bonds in phospholipids possessing phosphoethanolamine (PE) or phosphoserine (PS) head groups via a strategy that incorporated adduction of 4-iodobenzyl 18-crown-6 onto the amine functionality of the lipids of interest.262 Adduction of the crown ether proved to be highly efficient and boosted the ionization efficiencies of PEs. Moreover, the mass shift caused by the adduction of the crown ether provided mass separation of the PEs from other lipids of similar mass, such as phosphatidylcholine, that did not undergo adduction.262 The mass shift offered an additional advantage for analysis of lipids in complex mixtures that contained a variety of isobaric species. As reported for the RDD studies of other lipids, RDD of the PE/crown ether complexes allowed localization of double bonds and discernment of branching patterns of carbon chains.262 In two related applications, covalent attachment of iodobenzoyl groups and/or noncovalent adduction of iodobenzoyl crown ethers to lipids were explored in the context of developing RDD for structural characterization of glycerophospholipids and glycosphingolipids, including those with lyso modifications.263,264 Unique fragments were found for the different types of lipids, again permitting the localization of double bonds along the fatty acid tails, as well as identification of the lipid headgroups and the linkages to the glycerol backbone. The 4-iodobenzyl 18-crown-6 adduction method also enhanced RDD for differentiation of epimeric lipids.264 For the analysis of chiral glycosphingolipids, the ratios of pairs of fragment ions were used to differentiate glucose- versus galactose-containing epimers and permitted relative quantitation based on the abundances of the diagnostic fragments.264 One other related RDD application focused on characterization of triglycerol lipids derived from plants via interrogation of noncovalent complexes between the lipids and protonated diiodoanilines, ultimately yielding cross-linked products initiated by 266 nm UV irradiation.266 In all of these RDD applications, the absorption of 266 nm photons proved to be highly efficient and resulted in very specific cleavage of the C–I bond installed in the various lipids. The selective C–I bond cleavage was crucial for producing the reactive radical species that generated highly diagnostic fragmentation patterns upon collisional activation.
5.2. UVPD (193 nm)
UVPD (193 nm) has proven to be a versatile strategy for characterization of many classes of lipids, ranging from phospholipids to gangliosides to lipopolysaccharides.50,55,267–286 For many of these applications, UVPD has been used as a standalone MS/MS strategy to create rich fragmentation patterns of the lipids. In other cases, UVPD has been used in conjunction with collisional activation to develop powerful hybrid MSn methods or to offer complementary fragmentation information. Such hybrid methods provided additional layers of structural information, allowed more selective dissection of substructures, and facilitated automated interpretation of MS/MS spectra. Particularly in the context of structural analysis of phospholipids, UVPD filled in key details not elucidated by conventional MS/MS methods. For example, collisional activation of phospholipids gives insight into headgroup and acyl-chain composition but provides insufficient information to delineate isomeric structures that differ in double-bond and acyl-chain positions.10–12 UVPD (193 nm) of glycerophospholipids, such as phosphatidylcholines, allowed localization of double bonds of the acyl chains based on formation of pairs of products differing in mass by 24 Da that bracketed the double-bond positions.283 In essence, UVPD uniquely promoted cleavage of the carbon–carbon bonds adjacent to the double bonds, thus enabling differentiation of positional isomers.283 Examples of the UVPD and comparative collision activated dissociation (HCD) are shown in Figure 40 for a pair of isomeric phosphatidylcholines.283 The dominant ions generated by HCD originate from loss of the headgroup or acyl-chain cleavages, whereas UVPD produces diagnostic pairs of fragment ions separated by 24 Da that confidently localize the positions of the double bonds.283
Figure 40.

(a) HCD (NCE 25) and (b) 193 nm UVPD (10 pulses, 6 mJ) spectra of protonated phosphatidylcholine (PC) 18:1(9Z)/18:1(9Z) ([M + H]+, m/z 786.60). (c) HCD (NCE 25) and (d) UVPD (10 pulses, 6 mJ) spectra of protonated PC 18:1(6Z)/18:1(6Z) ([M + H]+, m/z 786.60). Reproduced with permission from ref 283. Copyright 2017 American Chemical Society.
The subsequent development of a hybrid CID/UVPD method expanded the standalone UVPD method and allowed successful elucidation of the sn-positions (acyl-chain assignments) while also permitting distinction of double-bond positional isomers.284 For the hybrid strategy, collisional activation of a sodium-cationized phosphatidylcholine (PC) resulted in the production of dioxalane-type fragment ions via loss of the lipid headgroup.284 Representative HCD/UVPD spectra are shown in Figure 41 for two regioisomeric phosphatidylcholines (PC 16:0/18:1(n-9) and PC18:1(n-9)/16:0). UVPD of the headgroup loss ions promoted diagnostic cross-ring cleavages that allowed confident localization of the acyl chains to the sn-1 or sn-2 position.284 This HCD/UVPD MS3 method provided multiple levels of information, ranging from the composition of the lipid, acyl-chain assignment (i.e., sn-position), to the site-specific assignment of double bonds in the acyl chains. This approach allowed both sn-positional and double-bond-positional isomers to be distinguished, including regioisomers such as the phosphatidylcholines PC 16:0/18:1(n-9) and PC 18:1(n-9)/16:0 shown in Figure 41.284 The method was extended for the quantification of phosphatidylcholine sn-isomers in an FTICR mass spectrometer based on 213 nm UVPD of Fe2+ complexes.287
Figure 41.
HCD/UVPD spectra of two sodium-cationized sn-regioisomeric phosphatidylcholines (PC): (A) PC 16:0/18:1(n-9) and (B) PC 18:1(n-9)/16:0. These spectra were obtained by isolating the headgroup loss ions (m/z 599.5) generated by HCD and subjecting them to 10 laser pulses (193 nm) with 4 mJ per pulse. Reproduced with permission from ref 284. Copyright 2017 American Chemical Society.
5.3. UVPD (213 nm)
With the commercialization of UVPD on a commercial Orbitrap mass spectrometer in 2017,53,54 efforts to adapt UVPD using 213 nm photons for characterization of lipids have begun to emerge.55 The first study of this kind focused on the characterization of cyclopropane rings in bacterial phospholipids, particularly cyclopropane fatty acids in glycerophospholipids and mycolic acids.55 UVPD resulted in cross-ring cleavages of carbon–carbon bonds comprising the cyclopropane rings, thus generating pairs of fragment ions separated by 14 Da that proved highly diagnostic for the location of the rings. When integrated into an LC-MS workflow, this UVPD-MS strategy allowed characterization of mycolic acids containing multiple cyclopropane rings in Mycobacterium bovis and M. tuberculosis as well as cyclopropyl glycerophospholipids in Escherichia coli.55 The strategy is illustrated in Figure 42, showing a typical LC trace along with the HCD and 213 nm UVPD mass spectra obtained for one mycolic acid eluting around 13 min. The fragment ions generated from HCD and UVPD are mapped onto the structure of the fatty acid, and the unique ions from UVPD allow the positions of the cyclopropane rings to be deciphered.55
Figure 42.
(a) Negative-mode LC-MS base peak chromatogram and m/z 1136 extracted ion chromatogram of m/z 1136 from M. bovis lipid extract; representative (b) HCD and (c) 213 nm UVPD mass spectra of m/z 1136; and (d) fragment ion map. Reproduced with permission from ref 55. Copyright 2019 American Chemical Society.
Given the success of UVPD for phospholipid analysis, 193 nm UVPD was also used to characterize gangliosides and glycosphingolipids, and the resulting spectra displayed more unique products arising from cross-ring cleavages than observed upon CID.282 The classes of lipids characterized by 193 nm UVPD-MS were further expanded to include sphingosines, sphingadienes, and sphingomyelins, among others,286 thus recapitulating the broad versatility for analysis of many diverse subclasses of lipids.
5.4. UVPD and DESI
The success of the UVPD method for localizing double bonds in lipids motivated the integration of UVPD with desorption electrospray ionization (DESI) for tissue imaging based on profiling specific phospholipids that are biomarkers of disease states.285 The combination of DESI, an ambient ionization method developed for spatial mapping of chemical signatures of tissue,288 with MS/MS analysis by 193 nm UVPD offered the ability to characterize isomeric phospholipids directly in tissue.285 The extra structural details provided by the UVPD fragmentation patterns allowed isomeric phospholipids to be spatially resolved in tissue.285 For example, differences in the distribution of isomeric phosphatidylcholines 16:0_18:1(9Δ) and 16:0_18:1(11Δ) confirmed by UVPD mass spectra were shown to vary in gray brain matter versus white brain matter of mice as well as in normal and cancerous regions of human lymph nodes.285 Examples of the images of mouse brain tissue created by monitoring the unique fragment ions generated by UVPD are shown in Figure 43. UVPD allowed the double-bond positions to be identified, and the spatial variations in the lipids were mapped based on the ratios of the key diagnostic ions.
Figure 43.
Optical images of the H&E stained (a) mouse brain tissue section, (e) human brain tissue section, and (i) lymph node tissue section with thyroid cancer metastasis. (b) DESI-MS ion image of m/z 798, (c) DESI-UVPD ratio image of the ratio of the summed intensities of the 193 nm UVPD double-bond diagnostic ions (Im/z 660 + 684)/(Im/z 688 + 712), (d) expanded region of UVPD mass spectra of the white and gray matter, (f) DESI-MS ion image of m/z 798, (g) DESI ratio image of the ratio of the summed intensities of the UVPD double-bond diagnostic ions (Im/z 660 + 684)/(Im/z 688 + 712), (h) expanded region of UVPD mass spectra of the white and gray matter, (j) DESI-MS ion image of m/z 798, (k) DESI ratio image of the intensity m/z 684 divided by the intensity m/z 712, and (l) expanded regions of UVPD mass spectra of cancerous and normal parts of tissue. Representative spectra were taken from areas of tissue marked with a white arrow. Reproduced with permission from ref 285. Copyright 2018 American Chemical Society.
5.5. Lipid A and Lipopolysaccharides
UVPD has also been implemented for the analysis of lipopolysaccharides (LPSs), arguably one of the most structurally complex classes of lipids for which many analytical methods have fallen short.289 LPSs are lipoglycans that stabilize the outer membrane of Gram-negative bacteria.290,291 LPS has three structural domains: an O-antigen polyglycan, an oligosaccharide core, and a hydrophobic lipid A.290,291 LPSs are further decorated with various modifications, such as phosphorylation and additional sugars that modulate the toxicity of LPSs and magnify the complexity of their structures. Among all the classes of lipids, LPSs are among the most challenging for mass spectrometry analysis as a result of their low solubilities, their low ionization efficiencies in both the positive and negative modes, and their intrinsic complexity arising from their composite structures consisting of multiple acyl chains appended to hydrophilic, branched oligosaccharides. UVPD (193 nm) has emerged as a powerful approach for characterization of both the key lipid A domain and intact LPS.267–281 As lipid A is largely responsible for the toxicity of lipopolysaccharides, it is the most frequently studied subdomain and is separated from intact LPS by mild acid hydrolysis and isolated by solvent extraction. The lipid A region has a variable number of acyl chains of varying length, some of which are hydroxylated, as well as the presence of modifications of the phosphate groups, among other features.
The utility of 193 nm UVPD applied to structural analysis of lipid A was originally reported in 2011;267 production of a variety of cross-ring and glycosidic fragments was not typically observed from CID-enabled characterization of the connectivity of the 3–8 acyl tails appended to lipid A. In general, UVPD yielded an array of C–O cleavages of both primary and secondary acyl chains and also promoted many diagnostic C–N amide cleavages, which allowed identification of the chains at the 2 and 2′ positions.267 Cleavage of the secondary acyl chain located at the 3′ position was one of the most prevalent pathways upon UVPD, along with less-dominant cleavages at the 3′ and 3 acyl positions.267 The original benchmark study267 established the efficacy of 193 nm UVPD for detailed elucidation of lipid A and motivated a number of subsequent studies aimed at mapping critical details of lipid A structures.268–280 For example, the extensive fragmentation of lipid A caused by 193 nm UVPD generated unique fragment ions that identified and localized myristate (C14:0) and 3-hydroxylaurate (3-OH C12:0) acyl chains,268 in addition to glycine and diglycine modifications,269 of lipid A from Vibrio Cholera. These structural modifications of lipid A augmented resistance to antimicrobial peptides, an outcome attributed to modulation of the negative charge of the surfaces of the bacteria and making the ability to identify and pinpoint such modifications pivotal in unravelling mechanisms of antibiotic resistance. Other unique and biological relevant modifications, such as addition of phosphoethanolamine to lipid A of Campylobacter jejuni,271 multiple phosphate groups to lipid A of Pseudomonas aeruginosa,275 or heptaacylation of lipid A of Acinetobacter baumannii,277 were similarly deciphered by 193 nm UVPD.
To further extend the range of applications accessible to UVPD-MS, UVPD was integrated into a liquid chromatography workflow to allow higher-throughput analysis of complex mixtures of lipid A.272 Using a reversed-phase column (C8) and a mixed organic/aqueous mobile phase offered satisfactory separation of lipid A molecules. This strategy proved to be particularly useful for characterization of lipid A from strains of E. coli engineered for development of adjuvants for vaccines.272 The resulting lipid A species displayed combinatorial patterns of modifications, and the integrated HPLC-UVPD-MS approach aided the identification of the structural mutants of lipid A present in the mixtures.272
Akin to informatics bottlenecks that have been problematic in the realm of proteomics, spectral analysis of lipids is typically time-consuming and labor-intensive. Moreover, UVPD creates a greater array of fragment ions than conventional collisional activation, many of which are not incorporated in algorithms designed for interpretation of CID mass spectra.292 Consequently, a de novo approach for characterizing lipid A structures was developed using an MSn strategy.278 Diagnostic fragments produced upon UVPD and CID were used to deduce general structural features such as the presence or absence of a secondary acyl chain at the 2′ position of the glucosamine backbone of the lipid A at the MS2 and MS3 levels.278 UVPD was utilized to generate glycosidic fragments containing only a single glycan and up to four acyl tails. Subsequent CID of the resulting glycosidic fragments enabled comprehensive elucidation of the compositions without the need for preexisting knowledge of the structures. Using this approach, data collection and analysis were automated to streamline structural assignments in higher-throughput applications.278
An alternate strategy for automated structural assignments employed a hybrid CID/UVPD MS3 approach.280 Using collisional activation as the first step not only targeted a prominent acyl-chain loss but also filtered out isobaric contaminants frequently found in biological samples. In essence, only those ions that exhibited a characteristic acyl-chain loss were selected for UVPD, thus excluding nonlipid A species from analysis. This approach diminished the prevalence of chimeric spectra (ones composed of fragment ions originating from different isobaric precursors) that often confound interpretation of MS/MS spectra.280 Applying UVPD in the second stage of the MS3 method allowed localization of substituents of the lipid A via diagnostic cross-ring cleavages and other glycosidic cleavages that confirmed the location and identity of the phosphate modifications.280 Key modifications mapped using this MS3 strategy included phosphoethanolamine, aminoarabinose, and galactosamine, each attached via an ester bond to the phosphate group on either the reducing or nonreducing end of lipid A.280
UVPD (193 nm) has also been applied to analysis of lipooligosaccharides (LOS), lipid A structures bearing the core glycans, by using both LC-MS/MS and shotgun MS3 approaches.50,274,281 Collisional activation of LOS generally results in cleavage of the most labile bonds, such as the C–O glycoside bond between the glycan and lipid A segments, and does not provide adequate information about the other acyl chains or sugars.274 UVPD generated richer fragmentation patterns, including complementary pairs of fragment ions: b/y, c/z, and a/x, with the latter from cross-ring cleavages that are particularly useful for characterization of oligosaccharide composition and elucidation of branching patterns.274 UVPD also promoted specific C–C bond cleavages that were critical for mapping acyl-chain modifications.274 Electron photodetachment is also a prominent process upon 193 nm UVPD of deprotonated lipids (i.e., lipids analyzed as anions in the negative mode), resulting in radical-type ions that were not informative as they represented the same intact charge-reduced lipids, not fragment ions. In an effort to capitalize on the utility of these prevalent radical-type ions, a complementary strategy was devised to convert these types of charge-reduced ions to informative ones by applying collisional activation during UVPD.274 The fragmentation efficiency of this hybrid UVPD/HCD method was greater than that of UVPD alone, thus making the hybrid process an appealing option for characterization of LOS. The hybrid method was also readily integrated into an LC-MS workflow to allow high-throughput analysis of LOS extracted from E. coli.274
An alternative MS3 approach was designed to exploit the lability of the bond connecting the glycan region to lipid A of the lipopolysaccharide, using CID as a first step to separate the core glycan region from the lipid A region of lipooligosaccharide (Figure 44).50 Subsequent isolation and UVPD (MS3) of each of the two sections in a second MS/MS step resulted in the production an array of fragments from cross-ring cleavages of the core oligosaccharide and lipid A. This MS3 strategy enabled full characterization of the LOS in a single multistep experiment.50 Examples of the rich MS/MS spectra created for the oligosaccharide and lipid A portions of a lipooligosaccharide from S. enterica are shown in Figure 43. The fragmentation map below the series of MS3 spectra exhibits the compilation of acyl-chain, glycosidic, and cross-ring cleavages that allow comprehensive reconstruction of the structure, including localization of modifications.50 The level of detail is impressive and attests to the merits of combining activation methods for high-level structural characterization of amphipathic molecules.
Figure 44.
(a) CID–CID (NCE 18–NCE 30), (b) CID–HCD (NCE 18–NCE 35), and (c) CID–UVPD (NCE 18–5 pulses, 2 mJ) of the core oligosaccharide substructure of tetra-acylated S. enterica Rb containing a pyrophosphoethanolamine modification (m/z 851.81, charge state 2−). (d) CID–CID (NCE 18–NCE 30), (e) CID–HCD (NCE 18–NCE 35), and (f) CID–UVPD (NCE 18–5 pulses, 2 mJ) of the lipid A substructure of S. enterica Rb containing a pyrophosphoethanolamine modification (m/z 679.41, charges state 2−). A yellow star is used to denote the precursor ion. Only select neutral losses are labeled on the spectra to avoid congestion: ▼ = loss of pyrophosphethanolamine; Δ = loss of phosphoethanolamine; ■ = loss of HPO3; □ = loss of H2PO4 and indicates that both of the indicated cleavages occur to generate a particular fragment ion. The acyl chains are numbered, and the loss of a particular acyl chain is denoted as M – N, where M represents the LPS and N represents the acyl chain that is lost. Reproduced with permission from ref 50. Copyright 2016 American Chemical Society.
The types of hybrid MS/MS and hierarchical MS3 workflows described earlier facilitated assignment of complex glycolipid structures, and the structural details uniquely provided by UVPD are paramount to allowing deeper analysis of very complicated lipids. With continued development of LC-MS/MS strategies, these efforts will likely build into workflows that enable full profiling of complex lipid structures in heterogeneous mixtures.
6. UVPD FOR NUCLEIC ACIDS
Nucleic acids, including deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), are responsible for storing genetic information.293 They comprise nucleotides, molecules composed of a base, a sugar, and a phosphate group. To determine the structure and function of nucleic acids, it is important to determine the sequence, identify the presence of modifications, and characterize the interactions of nucleic acids with other molecules. Mass spectrometry has not yet assumed as important a role in genomics as it has for proteomics.2–5,294–298 This contrast in impact likely originates from three main reasons. (1) Traditional technologies such as polymerase chain reaction (PCR) and next-generation sequencing are extremely powerful, robust, and sensitive for both the qualitative and quantitative analysis of DNA. (2) Owing to the phosphodiester linkages between nucleotides, the backbones of nucleic acids are negatively charged, thus making them more amenable to ionization as anions. The negative polarity is inherently a more difficult ionization mode for mass spectrometry. (3) Nucleic acid sequencing poses an analytical challenge owing to the structural properties of these biopolymers. In contrast to proteins, nucleic acids are composed of only four nucleotides. For analysis by MS/MS, the formation of isobaric fragment ions from nucleic acids is commonplace, exacerbating the ability to derive unique sequences for nucleic acids. In addition, the mass of the nucleic acid increases dramatically with the size of the biopolymer (e.g., number of nucleotides). The development of new activation methods and multistage MSn strategies have provided some framework for using mass spectrometry to characterize nucleic acids2–5 as well as complexes containing nucleic acids, either DNA or RNA, with noncovalently bound ligands5,295–297 or covalent adducts298 of nucleic acids. In general, collisional activation of oligonucleotides results in base loss and cleavage of the internucleoside backbone bonds. These types of fragmentation pathways299 predominantly result in a-B and w ions as demarcated in Figure 45.
Figure 45.
Fragmentation nomenclature for nucleic acids.
The majority of mass spectrometry studies that have focused on characterization of nucleic acids have used collision-induced dissociation (CID).299 The quest for alternative activation methods to provide more-extensive fragmentation and facilitate the characterization of modifications motivated the development and application of photoactivation techniques, mainly electron photodetachment (EPD) and UVPD.300–305 Upon UV photoabsorption by a multideprotonated nucleic acid, the loss of one electron is a common process known as electron photodetachment (EPD), which results in an oxidized species.300 As these charge-reduced ions frequently have little excess internal energy, they do not dissociate into meaningful product ions. Fragmentation is enhanced by coupling EPD with a subsequent stage of collisional activation. The combined process is termed activated-electron photodetachment (a-EPD) and has been used extensively for characterization of nucleic acids (as well as other biopolymers).4,301,303 Activation of odd-electron precursor ions produced by electron photodetachment accesses different pathways, improving the fragment yield and consequently sequence coverage for nucleic acids. Many of the previous studies have shown that electron photodetachment of guanine-containing nucleic acids is particularly enhanced using 260 nm photons.4,301,303 However, 193 nm has also been used successfully for a-EPD of nucleic acids,302 resulting in production of a, (a-B), w, and d/z fragment ions.
UVPD (193 nm) has also been used for several hybrid MS/MS strategies to characterize oligodeoxynucleotides and oligodeoxyribonucleotides, including ones containing platinated, phosphothioated, and 2′-O-methylated modification sites.303 For example, in one hybrid method termed ET-UVPD, modified nucleic acids were ionized in the positive mode and charge-reduced using electron-transfer reactions, and then the radical cations were dissociated using 193 nm UVPD.303 ET-UVPD resulted in an elaborate array of w, a-B, a, b, c, d, x, y, and z ions and exhibited lower portions of base loss compared to conventional CID. In addition, the detection of very low mass sequence ions (w1, z1, a1), as well as individual nucleotides, facilitated characterization of modifications at the termini. Cisplatin-modified oligodeoxynucleotides were characterized by 193 nm UVPD in both positive and negative modes to map the cisplatin-binding sites.304 UVPD resulted in production of several types of diagnostic fragment ions (a-B, w, b/y, and c/x), including many that retained the cisplatin modification and thus allowed confident localization of the sites of modifications.304
Most recently, a-EPD was used to localize the binding sites of metal cations and ligands to G-quadruplexes.305 The DNA was supercharged in order to increase the abundance of higher charge states and improve sequence coverage while minimizing the impact on the original G-quadruplex topology. For this application, a-EPD was undertaken using 240 nm photons for the electron-photodetachment step, followed by collisional activation. a-EPD of a quadruplex consisting of GGGTTAGGGTTAGGGTTAGGGT without potassium (unfolded form) yielded comprehensive sequence coverage (Figure 46A), whereas CID alone resulted in missed cleavages on the 3′ side of thymines.305 For the potassium complexes, a portion of the sequence around the central loop (from G8/9 to G14) exhibited no fragmentation (Figure 46C and D). This outcome corresponded to the region where potassium ions remained bound during activation, as confirmed by the presence of holo ions.
Figure 46.
(A) EPD analysis of 22GT without potassium ion bound (M6−). (B) EPD of the nonspecific MK6− complex of a control strand not forming G-quadruplex. EPD of 22GT G-quadruplex with one (C) potassium (MK6−) or with (D) two potassium ions bound (MK26−). Reprinted with permission from ref 305. Copyright 2019 Royal Society of Chemistry.
Hybrid MS/MS methods such as ET-UVPD and a-EPD have demonstrated the ability to generate highly informative fragmentation patterns for modified nucleic acids; nevertheless, in the past decade, there have been few studies expanding the use of UVPD for this class of biomolecule. With advances in the capabilities of high-performance tandem mass spectrometers and with the eventual development of algorithms to assist interpretation of mass spectra, there are compelling opportunities to continue the exploration of UVPD and a-EPD for the characterization of larger nucleic acids, including examination of secondary and tertiary structural features.
7. UVPD FOR CARBOHYDRATES
Carbohydrates engage in an array of critical functions in biological processes, both on their own and when conjugated to other molecules like proteins and lipids.306,307 The structural complexity and heterogeneity of polysaccharides makes the difficulty of their characterization nearly unrivalled by any other class of biopolymer. Unlike proteins, which are expressed in a linear manner, many carbohydrates have branched structures. In addition, the numerous hydroxyl groups of carbohydrates may be heavily and diversely modified. Comprehensive characterization requires elucidation of many levels of details ranging from the identities of each monosaccharide, including stereochemistry and modifications, the anomeric configurations of the glycosidic bonds, both the linkage and branching locations, to the arrangement of the monosaccharides in the oligomer.306,307 When attached to proteins, the saccharide portion (glycan) is one type of post-translational modification and is typically linked to the side chain of asparagine (N-linked) or through the hydroxyl groups on side chains of serine or threonine (O-linked).
Considering the numerous challenges posed by carbohydrates, MS/MS is in many ways an ideal tool for their analysis due to its sensitivity and the potential array of structural information provided.6–8 The accepted nomenclature for carbohydrate fragmentation is the one introduced by Costello308 as shown in Figure 47. The ions that retain the charge at the reducing end are X (cross-ring fragmentation), Y, and Z (glycosidic fragmentation), and those that retain the charge on the nonreducing end are A (cross-ring fragmentation), B, and C (glycosidic fragmentation). Glycosidic fragments give information about the sequence, monosaccharide composition, and branching pattern, whereas cross-ring cleavages aid in the localization of modifications and facilitate deciphering the positions of glycosidic linkages between residues. Collision-induced dissociation has been the most prevalent activation method used for analysis of carbohydrates, and its performance continues to be considered the gold standard against which all alternative activation methods are compared.6–8
Figure 47.
Fragmentation nomenclature for carbohydrates.
Reilly’s founding work just over a decade ago established UVPD as a promising method for analysis of oligosaccharides.309–311 This original trio of seminal studies demonstrated the diverse production of B, Y, Z, and X ions using 157 nm photons, thus facilitating analysis of the sequence and branching pattern of sugars via the array of fragment ions from both glycosidic and cross-ring cleavages.309–311 A subsequent study focused on applying 157 nm UVPD for characterization of glycopeptides, demonstrating the formation of diagnostic peptide sequence ions that retained the glycan in addition to cross-ring cleavage products that provide glycan linkage insight.312 The promising results obtained using 157 nm UVPD spurred interest in evaluating the feasibility of using 193 nm photons for analysis of oligosaccharides, and this goal was successfully accomplished for highly branched sialylated oligosaccharides in 2011.313 The dense fragmentation patterns generated upon 193 nm UVPD of branched sialylated oligosaccharides containing up to 13 sugars demonstrated the same diversity of fragment ions observed using 157 nm photons.313
UVPD (193 nm) was extended to the analysis of O-linked acidic glycopeptide species from A. baumannii OmpA/MotB.314 A procedure combining enzymatic glycoprotein digestion, glycopeptide enrichment, and reversed phase LC-MS with UVPD as the MS/MS method allowed characterization of both the glycan and peptide moieties and pinpointed the modification sites. The a/x peptide sequence ions retained the glycan, and B, Y, C, Z, and cross-ring cleavage ions allowed elucidation of the glycan moiety.314 Additionally, UVPD proved critical for uncovering site-specificity of O-linked acidic glycopeptides from the pathogenic bacteria Acinetobacter baumannii.314 A follow-up study compared the fragmentation patterns of protonated and deprotonated O-linked glycopeptides by 193 nm UVPD in order to evaluate the impact of charge state on the fragmentation pathways.315 Deprotonated O-glycopeptides afforded significantly more extensive series of fragment ions, and in fact the protonated O-glycopeptides preferentially produced only glycan-type ions upon UVPD.315
Another application of 193 nm UVPD focused on characterization of oligosaccharides dissected from lipopolysaccharides of Gram-negative bacteria.316 Lipooligosaccharides are the amphipathic species found on the outer membranes, typically composed of hydrophilic lipid A and hydrophilic oligosaccharides.291 The structures of the core oligosaccharides were elucidated by activated-electron photodetachment via a hybrid UVPD-CID strategy.316 As the first step, 193 nm UVPD was used to produce charge-reduced precursor ions of high abundance, followed by collisional activation to cause glycosidic and cross-ring cleavages throughout the oligosaccharide, which revealed the composition of saccharides and branching patterns. For these complex oligosaccharides, a-EPD yielded more diverse fragmentation patterns than CID yet less complicated spectra than UVPD. This hybridized a-EPD approach was employed to characterize the structure and branching pattern of core oligosaccharides from lipopolysaccharides of E. coli.316 Examples of the a-EPD spectra generated for two isobaric tridecasaccharides from E. coli are shown in Figure 48 along with their fragmentation maps that show the unique fragment ions that allow elucidation of the differences in the nonreducing ends of the oligosaccharides.316
Figure 48.
(a) a-EPD (UVPD (4 mJ per pulse and 5 pulses) of m/z 855.70 (3−) followed by CID (15 NCE) of m/z 1283.53 (2−)) of E. coli R1 tridecasaccharide. The precursor is labeled with a star. The red asterisk denotes neutral phosphate loss from the precursor ion. a-EPD reveals diagnostic product ions differentiating the branching pattern and substitution of a glucosamine residue for a glucose or galactose. (b) The identified product ions are mapped onto the structure of E. coli R1 tridecasaccharide. (c) a-EPD (UVPD (4 mJ per pulse and 5 pulses) of m/z 855.36 (3−) followed by CID (15 NCE) of m/z 1283.04 (2−)) of E. coli R3 tridecasaccharide. (d) The identified product ions are mapped onto the structure of E. coli R3 tridecasaccharide. Reproduced with permission from ref 316. Copyright 2019 American Chemical Society.
UVPD has also been reported for the characterization of glycosaminoglycans, sulfated polysaccharides constructed of hexosamine and uronic acid disaccharides.317 Analysis of this class of oligosaccharide is particularly challenging owing to the facile decomposition of sulfates using collisional-activation methods. UVPD resulted in both glycosidic and cross-ring cleavages without disruption of sulfate modifications. The attributes of UVPD were demonstrated for characterization of fondaparinux, a highly sulfated heparin-like anticoagulant drug.317
Uniting the rich fragmentation patterns generated upon UVPD with the capabilities of ion mobility provides a means to separate and characterize isomeric oligosaccharides.318–320 Analyses of mixtures of isobaric disaccharides and trisaccharides using 157 nm photoactivation were reported using this ion-mobility/UVPD strategy.318,319 To increase the UV photoabsorption cross sections and improve fragmentation efficiencies of oligosaccharides, cobalt adduction was employed.320 Upon 193 nm UVPD, the cobalt-cationized complexes of branched and linear isomeric tetrasaccharides produced more distinctive arrays of fragment ions when compared to UVPD of sodium adducts. UVPD also generated a greater number of fragments when compared to CID. The mobility method allowed an isobaric pair of tetrasaccharides to be separated, and the branched tetrasaccharide yielded a larger number of fragment ions upon UVPD relative to the linear glycan.320
Tunable synchrotron radiation in the vacuum UV (VUV) range has also been used to explore UVPD of oligosaccharides.321–323 The structures of 35 oligogalacturonans originating from citrus fruit were characterized using 69 nm photons, with each photon depositing 18 eV of energy and resulting in extensive cross-ring and glycosidic bond cleavages that allowed differentiation of isomers.321 This UVPD method was also used to characterize highly sulfated oligosaccharides known as oligoporphyrans in red algae Porphyra umbilicalis.322,323 An example of the rich spectrum generated for a pair of coisolated porphyrin isomers is shown in Figure 49. The representative porphyrin is composed of repeated, sulfated units (α-l-Galp-6-sulfate (1 → 3) β-d-Galp (L6S-G)) along with agarose motifs (4-linked 3,6-anhydro-α-l-Galp(1 → 3) ß-d-Galp (LA-G)). An important finding was that the sulfate groups, typically some of the most labile modifications of biopolymers, were retained in the fragment ions generated by UVPD.322
Figure 49.
VUV-photoactivation spectrum of a porphyran using 18 eV photons from a synchrotron radiation source. The precursor ion of the L6S-G-LA-G-L6S-G oligoporphyran with one methyl group was isolated as the [M + 3Na − 2H]+ species (m/z 1213.4, positive mode). Two isomers ((1) L6S-G6Me-LA-G-L6S-G6 and (2) L6S-G-LA-G6Me-L6S-G6) could be unequivocally differentiated from the spectrum. Masses marked with ▼ showed a complete shift upon 18O-labeling, while masses marked with ▽ contain two fragments, one of which shifted and one of which did not shift upon 18O-labeling. Several ions, indicated with ⧧, correspond to doubly charged fragments. Ions indicated with ★ correspond to an internal rearrangement. Reproduced with permission from ref 322. Copyright 2015 American Chemical Society.
Several other UVPD methods have been developed for analysis of oligosaccharides, each using a different UV wavelength. Activated-electron photodetachment was used to analyze di- and tetrasaccharides from heparin.324 For this approach, UV irradiation using 220 nm photons caused ejection of one electron from each deprotonated oligosaccharide, and collisional activation of the charge-reduced ions yielded informative cross-ring and glycoside cleavages that provided key insights into the structures.324 Another UVPD strategy developed for characterization of oligosaccharides entailed photocleavage of carbon–iodine bonds in oligosaccharides derivatized to incorporate 2-iodoaniline tags.325 For this application, 266 nm photons were used to induce the highly selective and efficient cleavage of the carbon–iodine bonds. The radical species created upon the carbon–iodine bond cleavage were subsequently subjected to collisional activation, resulting in production of diagnostic cross-ring cleavage ions, primarily X-type.325 This radical-directed dissociation method was used to differentiate isomeric pentasaccharides and hexasacharides.325 A variation of RDD used 213 nm photons to break the photocleavable C–I bond incorporated into six isomeric trisaccharides.326 In this case, oligomers were modified with para-iodobenzoate via amide bond formation between the linker amine and the carboxylic acid of the iodine-containing reagent, thus installing a photocleavable C–I bond in the oligosaccharides.326 Examples of the UVPD spectra, showing the characteristic loss of I· from the C–I bond scission, and the more informative RDD spectra acquired upon collisional activation of the selected radical precursor are shown in Figure 50. The RDD spectra allowed differentiation of the isomeric trisaccharides based on the unique patterns of cross-ring and glycosidic bond cleavages demarcated on the structures to the right of the spectra.326
Figure 50.
Radical fragmentation spectra afforded by UVPD and RDD. The dot indicates radical products; the dagger indicates that the product forms a double bond. The [4IB M + Na]+ precursors of m/z 842 during UVPD and the −I• radical precursors (m/z 715) during RDD are indicated with arrows. Reproduced with permission from ref 326. Copyright 2018 American Chemical Society.
Owing to the array of diagnostic cross-ring and glycosidic cleavages induced by photoactivation, UVPD has proven to be a promising strategy for characterization of oligosaccharides and glycans, and it is anticipated to be incorporated into hybrid and multistage MS/MS methods for analysis of increasingly complex glycosylated molecules.327 Glycolipids and glycoproteins, other important classes of glycosylated molecules, have also been characterized using UVPD. However, these studies are covered in earlier sections and are not described again here.
8. CONCLUSIONS AND OUTLOOK
Ultraviolet photodissociation has evolved into a versatile ion-activation method, offering high-energy deposition and access to new fragmentation pathways not observed upon conventional collisional activation. UVPD has been utilized for a tremendous array of analytical problems, ranging from analysis of small molecules like peptides and lipids to characterization of large molecules like proteins. More recent studies have reported promising applications in the area of structural biology based on dissection of protein–ligand and multimeric protein complexes by UVPD. The fast activation process of UVPD has allowed it to be integrated into high-throughput strategies incorporating chromatographic separations, capillary electrophoresis, and imaging.
This collection of features of UVPD will continue to expand its adoption for many applications, extend its integration into new mass spectrometry platforms, and spur continuing interest in the search for alternative light sources. There has been recent headway with the latter, mainly motivated to develop lower-cost, user-friendly methods for UVPD. For example, LEDs (emission wavelengths of 255, 265, and 275 nm) were inserted directly into a trapping cell to allow UVPD and mass analysis using an Orbitrap mass spectrometer, as illustrated in Figure 51.58 Although the fragmentation efficiency was modest owing to the low photon flux of the LEDs, the strategy demonstrated the ability to use inexpensive light sources for photodissociation in a manner that required no external optics.58 Even more recently, a low-cost helium-discharge lamp was interfaced to a commercial Q-TOF mass spectrometer, offering another means to activate ions and promote photodissociation using very high energy photons.22 In this case, UV irradiation of multicharged ions resulted in electron detachment and radical-directed fragmentation.22 In addition, a deuterium lamp with output from 115 to 160 nm has been used to provide polychromatic irradiation of ions.328 Adapting UVPD to use lower-cost, user-friendly light sources should accelerate more widespread adoption and implementation on many mass spectrometry platforms.
Figure 51.
LED-HCD cell with housing and trapping rods removed. Red lines represent the axis of fixed alignment of each LED. The inset shows the cell assembled with planar trapping rods and the cover installed. Reprinted with permission from ref 58. Copyright 2016 Wiley.
The activation/dissociation processes that occur upon photoabsorption depend on the wavelength of light, meaning that there remains an additional need for deciphering the mechanistic details of UVPD and correlation of wavelength with fragmentation modes. There are also many opportunities for development of new wavelength-selective UVPD applications. Methods that tag molecules with chromophores or exploit intrinsic chromophores offer some of the most intriguing avenues for designing innovative approaches to target specific classes of molecules or promote bond-selective cleavages.
The outlook for continued use and development of UVPD coupled to other auxiliary methods such as ion mobility is bright. Moreover, as mass spectrometry becomes more established as a biophysical tool, its use for structural characterization of macromolecules will inspire other new applications of UVPD. At the same time, more advanced computational algorithms will be needed to deconvolve and interpret the increasingly complicated fragmentation patterns created upon UVPD of complex supramolecular assemblies.
ACKNOWLEDGMENTS
Funding from the NIH (RO1 GM103655 and RO1 GM121714), the National Science Foundation (CHE-1402753), and the Welch Foundation (F-1155) is gratefully acknowledged.
ABBREVIATIONS
- a-EPD
activated electron photodetachment
- AGC
automatic gain control
- CAD
collisional-activated dissociation
- CID
collision-induced dissociation
- CTD
C-terminal domain
- DESI
desorption electrospray ionization
- ECD
electron-capture dissociation
- ETD
electron-transfer dissociation
- FIP
fragment ion protection
- HCD
higher-energy collisional dissociation
- PETD
photoelectron-transfer dissociation
- PTM
post-translational modification
- RDD
radical-directed dissociation
- UVPD
ultraviolet photodissociation
Biographies
Jennifer S. Brodbelt earned her B.S. in chemistry from the University of Virginia and her Ph.D. from Purdue University. After a postdoctoral stint at the University of California at Santa Barbara, she joined the faculty at the University of Texas at Austin, where she is currently a professor and the Rowland Pettit Centennial Chair in Chemistry. She serves as an associate editor for Journal of the American Society for Mass Spectrometry. Her research focus is on the development of ultraviolet photodissociation and ion-trap mass spectrometry for characterization of peptides, proteins, protein complexes, and lipids.
Lindsay Morrison is currently a senior applications scientist with Waters Corporation. She received her Ph.D. from the Ohio State University in 2014 in biological mass spectrometry, focused specifically on the fragmentation of peptides and proteins for structural biology applications. She then moved to the University of Texas at Austin to work with Professor Jennifer Brodbelt on the ultraviolet photon dissociation of proteins. In her current role, Lindsay is responsible for structural biology applications associated with mass spectrometry, including native MS, top-down proteomics, and hydrogen–deuterium exchange MS.
Inês C. Santos is a postdoctoral fellow in the Department of Chemistry at the University of Texas at Austin. She obtained her Ph.D. degree in Biotechnology with specialization in Chemistry at the Faculty of Biotechnology of the Portuguese Catholic University (ESB-UCP) in 2015. She has a M.Sc. and a bachelor’s degree from ESB-UCP on Microbiology. In the past two years she worked on the development of new gas chromatography–vacuum ultraviolet absorption detection and matrix-assisted laser desorption/ionization–mass spectrometry methods to study the microbiome in environmental samples. Her research is currently focused on the mass spectrometric analysis of biological molecules such as proteins, protein complexes, and nucleic acids.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Zhang Y; Fonslow BR; Shan B; Baek MC; Yates JR 3rd Protein analysis by shotgun/bottom-up proteomics. Chem. Rev 2013, 113, 2343–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Banoub JH; Newton RP; Esmans E; Ewing DF; Mackenzie G Recent development in mass spectrometry for the characterization of nucleosides, nucleotides, oligonucleotides, and nucleic acids. Chem. Rev 2005, 105, 1869–1915. [DOI] [PubMed] [Google Scholar]
- (3).Fabris D A role for the MS analysis of nucleic acids in the post-genomics age. J. Am. Soc. Mass Spectrom 2010, 21, 1–13. [DOI] [PubMed] [Google Scholar]
- (4).Schürch S Characterization of nucleic acids by tandem mass spectrometry - The second decade (2004–2013): From DNA to RNA and modified sequences. Mass Spectrom. Rev 2016, 35, 483–523. [DOI] [PubMed] [Google Scholar]
- (5).Beck JL; Colgrave MO; Ralph SF; Sheil MM Electrospray ionization mass spectrometry of oligonucleotide complexes with drugs, metals, and proteins. Mass Spectrom. Rev 2001, 20, 61–87. [DOI] [PubMed] [Google Scholar]
- (6).Kailemia MJ; Ruhaak LR; Lebrilla CB; Amster IJ Oligosaccharide analysis by mass spectrometry: a review of recent developments. Anal. Chem 2014, 86, 196–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Mirgorodskaya E; Karlsson NG; Sihlbom C; Larson G; Nilsson CL Cracking the sugar code by mass spectrometry. J. Am. Soc. Mass Spectrom 2018, 29, 1065–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ruhaak LR; Xu G; Li Q; Goonatilleke E; Lebrilla CB Mass spectrometry approaches to glycomic and glycoproteomic analyses. Chem. Rev 2018, 118, 7886–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Li L; Han J; Wang Z; Liu J; Wei J; Xiong S; Zhao Z Mass spectrometry methodology in lipid analysis. Int. J. Mol. Sci 2014, 15, 10492–10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Han X Lipidomics: Comprehensive mass spectrometry of lipids; Wiley & Sons: Hoboken, NJ, 2016. [Google Scholar]
- (11).Hu T; Zhang J-L Mass-spectrometry-based lipidomics. J. Sep. Sci 2018, 41, 351–372. [DOI] [PubMed] [Google Scholar]
- (12).Rustam YH; Reid GE Analytical challenges and recent advances in mass spectrometry based lipidomics. Anal. Chem 2018, 90, 374–397. [DOI] [PubMed] [Google Scholar]
- (13).Leurs U; Mistarz UH; Rand KD Applications of mass spectrometry in drug development science In Analytical Techniques in the Pharmaceutical Sciences. Advances in Delivery Science and Technology; Mullertz A, Perrie Y, Rades T, Eds.; Springer: New York, 2016. [Google Scholar]
- (14).Ren JL; Zhang A-H; Kong L; Wang X-J Advances in mass spectrometry-based metabolomics for investigation of metabolites. RSC Adv. 2018, 8, 22335–22350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).McLuckey SA; Mentinova M Ion/neutral, ion/electron, ion/photon, and ion/ion interactions in tandem mass spectrometry: do we need them all? Are they enough? J. Am. Soc. Mass Spectrom 2011, 22, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zhurov KO; Fornelli L; Wodrich MD; Laskay UA; Tsybin YO Principles of electron capture and transfer dissociation mass spectrometry applied to peptide and protein structure analysis. Chem. Soc. Rev 2013, 42, 5014–5030. [DOI] [PubMed] [Google Scholar]
- (17).Zhou M; Wysocki VH Surface induced dissociation: dissecting noncovalent protein complexes in the gas phase. Acc. Chem. Res 2014, 47, 1010–1018. [DOI] [PubMed] [Google Scholar]
- (18).McLuckey SA; Huang TY Ion-ion reactions: new chemistry for analytical MS. Anal. Chem 2009, 81, 8669–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Brodbelt JS Ion activation methods for peptides and proteins. Anal. Chem 2016, 88, 30–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Frese CK; Zhou HJ; Taus T; Altelaar AFM; Mechtler K; Heck AJR; Mohammed S Unambiguous phosphosite localization using electron-transfer/higher-energy collision dissociation (EThcD). J. Proteome Res 2013, 12, 1520–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Brodbelt JS; Wilson JJ Infrared multiphoton dissociation in quadrupole ion traps. Mass Spectrom. Rev 2009, 28, 390–424. [DOI] [PubMed] [Google Scholar]
- (22).Giuliani A; Williams JP; Green MR Extreme ultraviolet radiation: a means of ion activation for tandem mass spectrometry. Anal. Chem 2018, 90, 7176–7180. [DOI] [PubMed] [Google Scholar]
- (23).Reilly JP Ultraviolet photofragmentation of biomolecular ions. Mass Spectrom. Rev 2009, 28, 425–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ly T; Julian RR Ultraviolet photodissociation: developments towards applications for mass-spectrometry-based proteomics. Angew. Chem., Int. Ed 2009, 48, 7130–7137. [DOI] [PubMed] [Google Scholar]
- (25).Brodbelt JS Shedding light on the frontier of photodissociation. J. Am. Soc. Mass Spectrom 2011, 22, 197–206. [DOI] [PubMed] [Google Scholar]
- (26).Brodbelt JS Photodissociation mass spectrometry: New tools for characterization of biological molecules. Chem. Soc. Rev 2014, 43, 2757–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Rizzo TR; Stearns JA; Boyarkin OV Spectroscopic studies of cold, gas-phase biomolecular ions. Int. Rev. Phys. Chem 2009, 28, 481–515. [Google Scholar]
- (28).Baer T; Dunbar RC Ion spectroscopy: Where did It come from; where is it now, and where is it going? J. Am. Soc. Mass Spectrom 2010, 21, 681–693. [DOI] [PubMed] [Google Scholar]
- (29).Antoine R; Dugourd P Visible and ultraviolet spectroscopy of gas phase protein ions. Phys. Chem. Chem. Phys 2011, 13, 16494–16509. [DOI] [PubMed] [Google Scholar]
- (30).Czar MF; Jockusch RA Sensitive probes of protein structure and dynamics in well-controlled environments: combining mass spectrometry with fluorescence spectroscopy. Curr. Opin. Struct. Biol 2015, 34, 123–134. [DOI] [PubMed] [Google Scholar]
- (31).Hendricks NG; Julian RR Leveraging ultraviolet photodissociation and spectroscopy to investigate peptide and protein three-dimensional structure with mass spectrometry. Analyst 2016, 141, 4534–4540. [DOI] [PubMed] [Google Scholar]
- (32).Cismesia AP; Bailey LS; Bell MR; Tesler LF; Polfer NC Making mass spectrometry see the light: the promises and challenges of cryogenic infrared ion spectroscopy as a bioanalytical tool. J. Am. Soc. Mass Spectrom 2016, 27, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Gunzer F; Kruger S; Grotemeyer J Photoionization and photofragmentation in mass spectrometry with visible and UV lasers. Mass Spectrom. Rev 2019, 38, 202–217. [DOI] [PubMed] [Google Scholar]
- (34).Soorkia S; Jouvet C; Gregoire G UV photoinduced dynamics of conformer-resolved aromatic peptides. Chem. Rev 2019, DOI: 10.1021/acs.chemrev.9b00316. [DOI] [PubMed] [Google Scholar]
- (35).Antoine R; Lemoine J; Dugourd P Electron photodetachment dissociation for structural characterization of synthetic and bio-polymer anions. Mass Spectrom. Rev 2014, 33, 501–522. [DOI] [PubMed] [Google Scholar]
- (36).Julian RR The mechanism behind top-down UVPD experiments: making sense of apparent contradictions. J. Am. Soc. Mass Spectrom 2017, 28, 1823–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Bowers WD; Delbert SS; Hunter RL; McIver RT Fragmentation of oligopeptide ions using ultraviolet laser radiation and Fourier transfer mass spectrometry. J. Am. Chem. Soc 1984, 106, 7288–7289. [Google Scholar]
- (38).Hunt DF; Shabanowitz J; Yates JR Peptide sequence analysis by laser photodissociation Fourier transform mass spectrometry. J. Chem. Soc., Chem. Commun 1987, 548–550. [Google Scholar]
- (39).Tecklenburg RE; Miller MN; Russell DH Laser ion beam photodissociation studies of model amino acids and peptides. J. Am. Chem. Soc 1989, 111, 1161–1171. [Google Scholar]
- (40).Guan Z; Kelleher NL; O’Connor PB; Aaserud DJ; Little DP; McLafferty FW 193 nm photodissociation of larger multiply-charged biomolecules. Int. J. Mass Spectrom. Ion Processes 1996, 157–158, 357–364. [Google Scholar]
- (41).Barbacci DC; Russell DH Sequence and side-chain specific photofragment (193 nm) ions from protonated substance P by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J. Am. Soc. Mass Spectrom 1999, 10, 1038–1040. [Google Scholar]
- (42).Thompson MS; Cui W; Reilly JP Fragmentation of singly charged peptide ions by photodissociation at lambda = 157 nm. Angew. Chem., Int. Ed 2004, 43, 4791–4794. [DOI] [PubMed] [Google Scholar]
- (43).Wilson JJ; Brodbelt JS MS/MS Simplification by 355 nm ultraviolet photodissociation of chromophore-derivatized peptides in a quadrupole ion trap. Anal. Chem 2007, 79, 7883–7892. [DOI] [PubMed] [Google Scholar]
- (44).Lai CK; Ng DCM; Pang HF; Le Blanc JCY; Hager JW; Fang DC; Cheung ASC; Chu IK Laser-induced dissociation of singly protonated peptides at 193 and 266 nm within a hybrid linear ion trap mass spectrometer. Rapid Commun. Mass Spectrom 2013, 27, 1119–1127. [DOI] [PubMed] [Google Scholar]
- (45).Morgan JW; Russell DH Comparative Studies of 193-nm photodissociation and TOF-TOFMS analysis of bradykinin analogues: the effects of charge site(s) and fragmentation timescales. J. Am. Soc. Mass Spectrom 2006, 17, 721–729. [DOI] [PubMed] [Google Scholar]
- (46).Bellina B; Brown JM; Ujma J; Murray P; Giles K; Morris M; Compagnon I; Barran PE UV photodissociation of trapped ions following ion mobility separation in a Q-ToF mass spectrometry. Analyst 2014, 139, 6348–6351. [DOI] [PubMed] [Google Scholar]
- (47).Shaw JB; Li W; Holden DD; Zhang Y; Griep-Raming J; Fellers RT; Early BP; Thomas PM; Kelleher NL; Brodbelt JS Complete protein characterization using top-down mass spectrometry and ultraviolet photodissociation. J. Am. Chem. Soc 2013, 135, 12646–12651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Fort KL; Dyachenko A; Potel CM; Corradini E; Marino F; Barendregt A; Makarov AA; Scheltema RA; Heck AJR Implementation of ultraviolet photodissociation on a benchtop Q exactive mass spectrometer and its application to phosphoproteomics. Anal. Chem 2016, 88, 2303–2310. [DOI] [PubMed] [Google Scholar]
- (49).Gargano AFG; Shaw JB; Zhou M; Wilkins CS; Fillmore TL; Moore RJ; Somsen GW; Pasa-Tolic L Increasing the separation capacity of intact histone proteoforms chromatography coupling online weak cation exchange-HILIC to reversed phase LC UVPD-HRMS. J. Proteome Res 2018, 17, 3791–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Klein D; Holden DD; Brodbelt JS Shotgun analysis of rough-type lipopolysaccharides using UVPD. Anal. Chem 2016, 88, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Shaw JB; Robinson EW; Pasa-Tolic L Vacuum ultraviolet photodissociation and Fourier Transform-ion cyclotron resonance (FT-ICR) mass spectrometry: revisited. Anal. Chem 2016, 88, 3019–3023. [DOI] [PubMed] [Google Scholar]
- (52).Mistarz UH; Bellina B; Jensen PF; Brown JM; Barran PE; Rand KD UV photodissociation mass spectrometry accurately localize sites of backbone deuteration in peptides. Anal. Chem 2018, 90, 1077–1080. [DOI] [PubMed] [Google Scholar]
- (53).Brodie NI; Huguet R; Zhang T; Viner R; Zabrouskov V; Pan J; Petrotchenko EV; Borchers CH Top-down hydrogen-deuterium exchange analysis of protein structures using ultraviolet photodissociation. Anal. Chem 2018, 90, 3079–3082. [DOI] [PubMed] [Google Scholar]
- (54).Fornelli L; Srzentic K; Huguet R; Mullen C; Sharma S; Zabrouskov V; Fellers RT; Durbin KR; Compton PD; Kelleher NL Accurate sequence analysis of a monoclonal antibody by top-down and middle-down orbitrap mass spectrometry applying multiple ion activation techniques. Anal. Chem 2018, 90, 8421–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Blevins M; Klein D; Brodbelt JS Localization of cyclopropane modifications in bacterial lipids via 213 nm ultraviolet photodissociation mass spectrometry. Anal. Chem 2019, 91, 6820–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Robotham SA; Kluwe C; Cannon JR; Ellington A; Brodbelt JS De novo sequencing of peptides using selective 351 nm ultraviolet photodissociation mass spectrometry. Anal. Chem 2013, 85, 9832–9838. [DOI] [PubMed] [Google Scholar]
- (57).Canon F; Milosavljevic AR; van der Rest G; Refregiers M; Nahon L; Sarni-Manchado P; Cheynier V; Giuliani A Photodissociation and dissociative photoionization mass spectrometry of proteins and noncovalent protein-ligand complexes. Angew. Chem., Int. Ed 2013, 52, 8377–8381. [DOI] [PubMed] [Google Scholar]
- (58).Holden DD; Makarov A; Schwartz JC; Sanders JD; Zhuk E; Brodbelt JS Ultraviolet photodissociation using light emitting diodes (LEDs) in a planar ion trap. Angew. Chem., Int. Ed 2016, 55, 12417–12421. [DOI] [PubMed] [Google Scholar]
- (59).Cui W; Thompson MS; Reilly JP Pathways of peptide ion fragmentation induced by vacuum ultraviolet light. J. Am. Soc. Mass Spectrom 2005, 16, 1384–1398. [DOI] [PubMed] [Google Scholar]
- (60).Thompson MS; Cui W; Reilly JP Factors that impact the vacuum ultraviolet photofragmentation of peptide ions. J. Am. Soc. Mass Spectrom 2007, 18, 1439–1452. [DOI] [PubMed] [Google Scholar]
- (61).Parthasarathi R; He Y; Reilly JP; Raghavachari K New insights into the vacuum UV photodissociation of peptides. J. Am. Chem. Soc 2010, 132, 1606–1610. [DOI] [PubMed] [Google Scholar]
- (62).Zhang L; Cui W; Thompson MS; Reilly JP Structures of α-type ions formed in the 157 nm photodissociation of singly-charged peptide ions. J. Am. Soc. Mass Spectrom 2006, 17, 1315–1321. [DOI] [PubMed] [Google Scholar]
- (63).Morgan JW; Russell DH Comparative studies of 193-nm photodissociation and TOF-TOFMS analysis of bradykinin analogues: the effects of charge site(s) and fragmentation timescales. J. Am. Soc. Mass Spectrom 2006, 17, 721–729. [DOI] [PubMed] [Google Scholar]
- (64).Moon J-H; Yoon S-H; Kim M-S Construction of an improved tandem time-of-flight mass spectrometer for photodissociation of ions generated by matrix-assisted laser desorption ionization (MALDI). Bull. Korean Chem. Soc 2005, 26, 763–768. [Google Scholar]
- (65).Robotham SA; Brodbelt JS Comparison of ultraviolet photodissociation and collision induced dissociation of adrenocorticotropic hormone peptides. J. Am. Soc. Mass Spectrom 2015, 26, 1570–1579. [DOI] [PubMed] [Google Scholar]
- (66).Yoon SH; Moon JH; Kim MS Dissociation mechanisms and implication for the presence of multiple conformations for peptide ions with arginine at the C-terminus: time-resolved photodissociation study. J. Mass Spectrom 2010, 45, 806–814. [DOI] [PubMed] [Google Scholar]
- (67).Kim TY; Valentine SJ; Clemmer DE; Reilly JP Gas-phase conformation-specific photofragmentation of proline-containing peptide ions. J. Am. Soc. Mass Spectrom 2010, 21, 1455–1465. [DOI] [PubMed] [Google Scholar]
- (68).Tabarin T; Antoine R; Broyer M; Dugourd P Specific photodissociation of peptides with multi-stage mass spectrometry. Rapid Commun. Mass Spectrom 2005, 19, 2883–2892. [DOI] [PubMed] [Google Scholar]
- (69).Antoine R; Broyer M; Chamot-Rooke J; Dedonder C; Desfrancois C; Dugourd P; Gregoire G; Jouvet C; Onidas D; Poulain P; Tabarin T; van der Rest G Comparison of the fragmentation pattern induced by collisions, laser excitation and electron capture. Influence of the initial excitation. Rapid Commun. Mass Spectrom 2006, 20, 1648–1652. [DOI] [PubMed] [Google Scholar]
- (70).Lemoine J; Tabarin T; Antoine R; Broyer M; Dugourd P UV photodissociation of phospho-seryl-containing peptides: laser stabilization of the phospho-seryl bond with multistage mass spectrometry. Rapid Commun. Mass Spectrom 2006, 20, 507–511. [DOI] [PubMed] [Google Scholar]
- (71).Joly L; Antoine R; Broyer M; Dugourd P; Lemoine J Specific UV photodissociation of tyrosyl-containing peptides in multistage mass spectrometry. J. Mass Spectrom 2007, 42, 818–824. [DOI] [PubMed] [Google Scholar]
- (72).Girod M; Sanader Z; Vojkovic M; Antoine R; MacAleese L; Lemoine J; Bonacic-Koutecky V; Dugourd P UV photodissociation of proline-containing peptide ions: insights from molecular dynamics. J. Am. Soc. Mass Spectrom 2015, 26, 432–443. [DOI] [PubMed] [Google Scholar]
- (73).Madsen JA; Cheng RR; Kaoud TS; Dalby KN; Makarov DE; Brodbelt JS Charge-site-dependent dissociation of hydrogenrich radical peptide cations upon vacuum UV photoexcitation. Chem. -Eur. J 2012, 18, 5374–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Shaw JB; Madsen JA; Xu H; Brodbelt JS Systematic comparison of ultraviolet photodissociation and electron transfer dissociation for peptide anion characterization. J. Am. Soc. Mass Spectrom 2012, 23, 1707–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Halim MA; Girod M; MacAleese L; Lemoine J; Antoine R; Dugourd P 213 nm Ultraviolet photodissociation on peptide anions: radical-directed fragmentation patterns. J. Am. Soc. Mass Spectrom 2016, 27, 474–486. [DOI] [PubMed] [Google Scholar]
- (76).Holden D; Pruet JM; Brodbelt JS Ultraviolet photodissociation of protonated, fixed charge, and charge-reduced peptides. Int. J. Mass Spectrom 2015, 390, 81–90. [Google Scholar]
- (77).Brown SB An introduction to spectroscopy for biochemists; Academic Press: London, 1980. [Google Scholar]
- (78).Morrison LJ; Rosenberg JA; Singleton JP; Brodbelt JS Statistical examination of the a and a+1 fragment ions from 193 nm UVPD reveals local hydrogen bonding interactions. J. Am. Soc. Mass Spectrom 2016, 27, 1443–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Madsen JA; Kaoud TS; Dalby KN; Brodbelt JS 193-nm photodissociation of singly and multiply charged peptide anions for acidic proteome characterization. Proteomics 2011, 11, 1329–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Madsen JA; Boutz DR; Brodbelt JS Ultrafast ultraviolet photodissociation at 193 nm and its applicability to proteomic workflows. J. Proteome Res 2010, 9, 4205–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Madsen JA; Xu H; Robinson MR; Horton AP; Shaw JB; Giles DK; Kaoud TS; Dalby KN; Trent MS; Brodbelt JS High-throughput database search and large-scale negative polarity LC-MS/MS with ultraviolet photodissociation for complex proteomics. Mol. Cell. Proteomics 2013, 12, 2604–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Crittenden CM; Parker WR; Jenner ZB; Bruns KA; Akin LD; McGee WM; Ciccimaro E; Brodbelt JS Exploitation of the ornithine effect enhances characterization of stapled and cyclic peptides. J. Am. Soc. Mass Spectrom 2016, 27, 856–863. [DOI] [PubMed] [Google Scholar]
- (83).Parsley NC; Kirkpatrick CL; Crittenden CM; Rad JG; Hoskin DW; Brodbelt JS; Hicks LM PepSAVI-MS reveals anticancer and antifungal cycloviolacins in Viola odorata. Phytochemistry 2018, 152, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Attard TJ; Carter MD; Fang MX; Johnson RC; Reid GE Structural characterization and absolute quantification of microcystin peptides using collision-induced and ultraviolet photodissociation tandem mass spectrometry. J. Am. Soc. Mass Spectrom 2018, 29, 1812–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Greer SM; Parker WR; Brodbelt JS Impact of protease on ultraviolet photodissociation mass spectrometry for bottom-up proteomics. J. Proteome Res 2015, 14, 2626–2632. [DOI] [PubMed] [Google Scholar]
- (86).Greer SM; Bern M; Becker C; Brodbelt JS Extending proteome coverage by combining MS/MS methods and a modified bioinformatics platform adapted for database searching of positive and negative polarity 193 nm ultraviolet photodissociation mass spectra. J. Proteome Res 2018, 17 (4), 1340–1347. [DOI] [PubMed] [Google Scholar]
- (87).Greer SM; Cannon JR; Brodbelt JS Improvement of shotgun proteomics in the negative mode by carbamylation of peptides and ultraviolet photodissociation mass spectrometry. Anal. Chem 2014, 86, 12285–12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Spoel SH Orchestrating the proteome with post-translational modifications. J. Exp. Bot 2018, 69, 4499–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Doll S; Burlingame AL Mass spectrometry-based detection and assignment of protein posttranslational modifications. ACS Chem. Biol 2015, 10, 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Grimsrud PA; Swaney DL; Wenger CD; Beauchene NA; Coon JJ Phosphoproteomics for the Masses. ACS Chem. Biol 2010, 5, 105–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Smith SA; Kalcic CL; Cui L; Reid GE Femtosecond laser-induced ionization/dissociation tandem mass spectrometry (fsLID-MS/MS) of deprotonated phosphopeptide anions. Rapid Commun. Mass Spectrom 2013, 27, 2807–2817. [DOI] [PubMed] [Google Scholar]
- (92).Shaffer CJ; Slováková K; Turečcek F Near-UV photodissociation of phosphopeptide cation-radicals. Int. J. Mass Spectrom 2015, 390, 71–80. [Google Scholar]
- (93).Diedrich JK; Julian RR Facile identification of phosphorylation sites in peptides by radical-directed dissociation. Anal. Chem 2011, 83, 6818–6826. [DOI] [PubMed] [Google Scholar]
- (94).Shin YS; Moon JH; Kim MS Observation of phosphorylation site-specific dissociation of singly protonated phosphopeptides. J. Am. Soc. Mass Spectrom 2010, 21, 53–59. [DOI] [PubMed] [Google Scholar]
- (95).Robinson MR; Taliaferro JM; Dalby KN; Brodbelt JS 193 nm Ultraviolet photodissociation mass spectrometry for phosphopeptide characterization in the positive and negative ion modes. J. Proteome Res 2016, 15, 2739–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Mayfield JE; Robinson MR; Cotham VC; Irani S; Matthews WL; Ram A; Gilmour DS; Cannon JR; Zhang YJ; Brodbelt JS Mapping the phosphorylation pattern of Drosophila melanogaster RNA polymerase II carboxyl-terminal domain using ultraviolet photodissociation mass spectrometry. ACS Chem. Biol 2017, 12, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Robinson MR; Moore KL; Brodbelt JS Direct identification of tyrosine sulfation by using ultraviolet photodissociation mass spectrometry. J. Am. Soc. Mass Spectrom 2014, 25, 1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Brunet C; Antoine R; Dugourd P; Canon F; Giuliani A; Nahon L Formation and fragmentation of radical peptide anions: insights from vacuum ultra violet spectroscopy. J. Am. Soc. Mass Spectrom 2012, 23, 274–281. [DOI] [PubMed] [Google Scholar]
- (99).Robinson MR; Brodbelt JS Integrating weak anion exchange and ultraviolet photodissociation mass spectrometry with strategic modulation of peptide basicity for the enrichment of sulfopeptides. Anal. Chem 2016, 88, 11037–11045. [DOI] [PubMed] [Google Scholar]
- (100).Han S-W; Lee S-W; Bahar O; Schwessinger B; Robinson MR; Shaw JB; Madsen JA; Brodbelt JS; Ronald PA Tyrosine sulfation in gram-negative bacterium. Nat. Commun 2012, 3, 1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Pruitt RN; Schwessinger B; Joe A; Thomas N; Liu F; Albert M; Robinson MR; Chan LJG; Luu DD; Chen H; Bahar O; Daudi A; De Vleesschauwer D; Caddell D; Zhang W; Zhao X; Li X; Heazlewood JL; Ruan D; Majumder D; Chern M; Kalbacher H; Midha S; Patil PB; Sonti RV; Petzold CJ; Liu C; Brodbelt JS; Felix G; Ronald PC The rice immune receptor XA21 recognizes a tyrosine-sulfated peptide from a Gram-negative bacterium. Sci. Adv 2015, 1, No. e1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Shin YS; Moon JH; Kim MS Selective screening of tyrosine-nitrated peptides in tryptic mixtures by in-source photodissociation at 355 nm in matrix-assisted laser desorption ionization. Anal. Chem 2011, 83, 1704–1708. [DOI] [PubMed] [Google Scholar]
- (103).Kehoe JW; Bertozzi CR Tyrosine sulfation: a modulator of extracellular protein–protein interactions. Chem. Biol 2000, 7, R57–R61. [DOI] [PubMed] [Google Scholar]
- (104).Quick MJ; Crittenden CM; Rosenberg JA; Brodbelt JS Characterization of disulfide linkages in proteins by 193 nm ultraviolet photodissociation (UVPD) mass spectrometry. Anal. Chem 2018, 90, 8523–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Turko IV; Murad F Protein nitration in cardiovascular diseases. Pharmacol. Rev 2002, 54, 619–634. [DOI] [PubMed] [Google Scholar]
- (106).Agarwal A; Diedrich JK; Julian RR Direct elucidation of disulfide bond partners using ultraviolet photodissociation mass spectrometry. Anal. Chem 2011, 83, 6455–6458. [DOI] [PubMed] [Google Scholar]
- (107).Wongkongkathep P; Li HL; Zhang X; Ogorzalek Loo RR; Julian RR; Loo JA Enhancing protein disulfide bond cleavage by UV excitation and electron capture dissociation for top-down mass spectrometry. Int. J. Mass Spectrom 2015, 390, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Talbert LE; Julian RR Directed-backbone dissociation following bond-specific carbon-sulfur UVPD at 213 nm. J. Am. Soc. Mass Spectrom 2018, 29, 1760–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Bonner J; Talbert LE; Akkawi N; Julian RR Simplified identification of disulfide, trisulfide, and thioether pairs with 213 nm UVPD. Analyst 2018, 143, 5176–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Vasicek L; Brodbelt JS Enhancement of ultraviolet photodissociation efficiencies through attachment of aromatic chromophores. Anal. Chem 2010, 82, 9441–9446. [DOI] [PubMed] [Google Scholar]
- (111).Holden DD; Brodbelt JS Improving performance metrics of ultraviolet photodissociation mass spectrometry by selective precursor ejection. Anal. Chem 2017, 89, 837–846. [DOI] [PubMed] [Google Scholar]
- (112).Holden DD; Sanders JD; Weisbrod CR; Mullen C; Schwartz JC; Brodbelt JS Implementation of fragment ion protection (FIP) during ultraviolet photodissociation (UVPD) mass spectrometry. Anal. Chem 2018, 90, 8583–8591. [DOI] [PubMed] [Google Scholar]
- (113).Ly T; Julian RR Residue-specific radical-directed dissociation of whole proteins in the gas phase. J. Am. Chem. Soc 2008, 130, 351–358. [DOI] [PubMed] [Google Scholar]
- (114).Ly T; Julian RR Tracking radical migration in large hydrogen deficient peptides with covalent labels: facile movement does not equal indiscriminate fragmentation. J. Am. Soc. Mass Spectrom 2009, 20, 1148–1158. [DOI] [PubMed] [Google Scholar]
- (115).Liu ZJ; Julian RR Deciphering the peptide iodination code: influence on subsequent gas-phase radical generation with photodissociation ESI-MS. J. Am. Soc. Mass Spectrom 2009, 20, 965–971. [DOI] [PubMed] [Google Scholar]
- (116).Diedrich JK; Julian RR Site-specific radical-directed dissociation of peptides at phosphorylated residues. J. Am. Chem. Soc 2008, 130, 12212–12218. [DOI] [PubMed] [Google Scholar]
- (117).Sun Q; Julian RR Probing sites of histidine phosphorylation with iodination and tandem mass spectrometry. Rapid Commun. Mass Spectrom 2011, 25, 2240–2246. [DOI] [PubMed] [Google Scholar]
- (118).Sun QY; Yin S; Loo JA; Julian RR Radical-directed dissociation for facile identification of iodotyrosine residues using electrospray ionization mass spectrometry. Anal. Chem 2010, 82, 3826–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Diedrich JK; Julian RR Site-selective fragmentation of peptides and proteins at quinone-modified cysteine residues investigated by ESI-MS. Anal. Chem 2010, 82, 4006–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Lyon YA; Julian RR Photolytic determination of charge state for large proteins and fragments in an ion trap mass spectrometer. Rapid Commun. Mass Spectrom 2015, 29, 322–326. [DOI] [PubMed] [Google Scholar]
- (121).Ly T; Zhang X; Sun QY; Moore B; Tao YQ; Julian RR Rapid, quantitative, and site specific synthesis of biomolecular radicals from a simple photocaged precursor. Chem. Commun 2011, 47, 2835–2837. [DOI] [PubMed] [Google Scholar]
- (122).Agarwal A; Diedrich JK; Julian RR Direct elucidation of disulfide bond partners using ultraviolet photodissociation mass spectrometry. Anal. Chem 2011, 83, 6455–6458. [DOI] [PubMed] [Google Scholar]
- (123).Hamdy OM; Lam S; Julian RR Identification of inherently antioxidant regions in proteins with radical-directed dissociation mass spectrometry. Anal. Chem 2014, 86, 3653–3658. [DOI] [PubMed] [Google Scholar]
- (124).Hamdy OM; Alizadeh A; Julian RR The innate capacity of proteins to protect against reactive radical species. Analyst 2015, 140, 5023–5028. [DOI] [PubMed] [Google Scholar]
- (125).Tao Y; Quebbemann NR; Julian RR Discriminating D-amino acid-containing peptide epimers by radical-directed dissociation mass spectrometry. Anal. Chem 2012, 84, 6814–6820. [DOI] [PubMed] [Google Scholar]
- (126).Riggs DL; Gomez SV; Julian RR Sequence and solution effects on the prevalence of D-isomers produced by deamidation. ACS Chem. Biol 2017, 12, 2875–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Tao Y; Julian RR Identification of amino acid epimerization and isomerization in Crystallin proteins by tandem LC-MS. Anal. Chem 2014, 86 (19), 9733–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Lyon YA; Sabbah GM; Julian RR Identification of sequence similarities among isomerization hotspots in Crystallin proteins. J. Proteome Res 2017, 16, 1797–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Zhang X; Julian RR Investigating the gas phase structure of KIX with radical-directed dissociation and molecular dynamics: Retention of the native structure. Int. J. Mass Spectrom 2011, 308, 225–231. [Google Scholar]
- (130).Shi H; Pierson NA; Valentine SJ; Clemmer DE Conformation types of ubiquitin [M+8H]8+ ions from water:methanol solutions: evidence for the N and A states in aqueous solution. J. Phys. Chem. B 2012, 116, 3344–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Shi H; Clemmer DE Evidence for two new solution states of ubiquitin by IMS-MS analysis. J. Phys. Chem. B 2014, 118, 3498–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Ridgeway ME; Silveira JA; Meier JE; Park MA Microheterogeneity within conformational states of ubiquitin revealed by high resolution trapped ion mobility spectrometry. Analyst 2015, 140, 6964–6972. [DOI] [PubMed] [Google Scholar]
- (133).Bonner J; Lyon YA; Nellessen C; Julian RR Photoelectron transfer dissociation reveals surprising favorability of zwitterionic states in large gaseous peptides and proteins. J. Am. Chem. Soc 2017, 139, 10286–10293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Parker WR; Holden DD; Cotham VC; Xu H; Brodbelt JS Cysteine-selective peptide identification: selenium-based chromophore for selective S-Se bond cleavage with 266 nm ultraviolet photodissociation (UVPD). Anal. Chem 2016, 88, 7222–7229. [DOI] [PubMed] [Google Scholar]
- (135).Quick MM; Mehaffey MR; Johns RW; Parker WR; Brodbelt JS SITS derivatization of peptides to enhance 266 nm ultraviolet photodissociation (UVPD). J. Am. Soc. Mass Spectrom 2017, 28, 1462–1472. [DOI] [PubMed] [Google Scholar]
- (136).Wilson JJ; Brodbelt JS MS/MS Simplification by 355 nm ultraviolet photodissociation of chromophore-derivatized peptides in a quadrupole ion trap. Anal. Chem 2007, 79 (20), 7883–7892. [DOI] [PubMed] [Google Scholar]
- (137).Gardner MW; Brodbelt JS Ultraviolet Photodissociation Mass Spectrometry of bis-aryl hydrazone conjugated peptides. Anal. Chem 2009, 81, 4864–4872. [DOI] [PubMed] [Google Scholar]
- (138).O’Brien JP; Pruet JM; Brodbelt JS Chromogenic chemical probe for protein structural characterization via ultraviolet photodissociation mass spectrometry. Anal. Chem 2013, 85 (15), 7391–7397. [DOI] [PubMed] [Google Scholar]
- (139).O’Brien JP; Mayberry LK; Murphy PA; Browning KS; Brodbelt JS Evaluating the conformation and binding interface of capbinding proteins and complexes via ultraviolet photodissociation mass spectrometry. J. Proteome Res 2013, 12, 5867–5877. [DOI] [PubMed] [Google Scholar]
- (140).Cotham VC; Wine Y; Brodbelt JS Selective 351 nm photodissociation of cysteine-containing peptides for improved sequencing of antigen-binding regions of IgG fragments in bottom-up LC-MS/MS workflows. Anal. Chem 2013, 85, 5577–5585. [DOI] [PubMed] [Google Scholar]
- (141).Aponte JR; Vasicek L; Swaminathan J; Xu H; Koag MC; Lee S; Brodbelt JS Streamlining bottom-up protein identification based on selective ultraviolet photodissociation (UVPD) of chromophore-tagged histidine- and tyrosine-containing peptides. Anal. Chem 2014, 86, 6237–6244. [DOI] [PubMed] [Google Scholar]
- (142).Robotham SA; Horton AP; Cannon JR; Cotham VC; Marcotte EM; Brodbelt JS UVnovo: A de novo sequencing algorithm using single series of fragment ions via chromophore tagging and 351 nm ultraviolet photodissociation mass spectrometry. Anal. Chem 2016, 88, 3990–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Horton A; Robotham SA; Cannon J; Holden DD; Marcotte E; Brodbelt JS Comprehensive de novo peptide sequencing from MS/MS pairs generated through complementary collision induced dissociation and 351 nm ultraviolet photodissociation. Anal. Chem 2017, 89, 3747–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Yan Y; Kusalik AJ; Wu F-X Recent developments in computational methods for de novo peptide sequencing from tandem mass spectrometry (MS/MS). Protein Pept. Lett 2015, 22, 983–991. [DOI] [PubMed] [Google Scholar]
- (145).Medzihradszky KF; Chalkley RJ Lessons in de novo peptide sequencing by tandem mass spectrometry. Mass Spectrom. Rev 2015, 34, 43–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).Ma B; Johnson R De novo sequencing and homology searching. Mol. Cell. Proteomics 2012, 11 (2), 014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Zhang L; Reilly JP De novo sequencing of tryptic peptides derived from deinococcus radiodurans ribosomal proteins using 157 nm photodissociation MALDI TOF/TOF mass spectrometry. J. Proteome Res 2010, 9, 3025–3034. [DOI] [PubMed] [Google Scholar]
- (148).Zhang L; Reilly JP Peptide de novo sequencing using 157 nm photodissociation in a tandem time-of-flight mass spectrometer. Anal. Chem 2010, 82, 898–908. [DOI] [PubMed] [Google Scholar]
- (149).Robinson MR; Madsen JA; Brodbelt JS 193 nm ultraviolet photodissociation of imidazolinylated lys-N peptides for de novo sequencing. Anal. Chem 2012, 84, 2433–2439. [DOI] [PubMed] [Google Scholar]
- (150).Keough T; Lacey MP; Youngquist RS Derivatization procedures to facilitate de novo sequencing of lysine-terminated tryptic peptides using postsource decay matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom 2000, 14, 2348–2356. [DOI] [PubMed] [Google Scholar]
- (151).Kim J-S; Song J-S; Kim Y; Park SB; Kim H-J De novo analysis of protein N-terminal sequence utilizing MALDI signal enhancing derivatization with Br signature. Anal. Bioanal. Chem 2012, 402, 1911–1919. [DOI] [PubMed] [Google Scholar]
- (152).Samgina TY; Kovalev SV; Gorshkov VA; Artemenko KA; Poljakov NB; Lebedev AT N-terminal tagging strategy for de novo sequencing of short peptides by ESI-MS/MS and MALDI-MS/MS. J. Am. Soc. Mass Spectrom 2010, 21, 104–111. [DOI] [PubMed] [Google Scholar]
- (153).Hennrich ML; Mohammed S; Altelaar AFM; Heck AJR Dimethyl isotope labeling assisted de novo peptide sequencing. J. Am. Soc. Mass Spectrom 2010, 21, 1957–1965. [DOI] [PubMed] [Google Scholar]
- (154).Cotham VC; Shaw JB; Brodbelt JS High-throughput bioconjugation for enhanced 193 nm photodissociation via droplet phase initiated ion/ion chemistry using a front-end dual spray reactor. Anal. Chem 2015, 87, 9396–9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (155).Engen JR; Wales TE Analytical aspects of hydrogen exchange mass spectrometry. Annu. Rev. Anal. Chem 2015, 8, 127–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (156).Enjalbert Q; Girod M; Simon R; Jeudy J; Chirot F; Salvador A; Antoine R; Dugourd P; Lemoine J Improved detection specificity for plasma proteins by targeting cysteine-containing peptides with photo-SRM. Anal. Bioanal. Chem 2013, 405, 2321–2331. [DOI] [PubMed] [Google Scholar]
- (157).Girod M; Biarc J; Enjalbert Q; Salvador A; Antoine R; Dugourd P; Lemoine J Implementing visible 473 nm photodissociation in a Q-Exactive mass spectrometer: towards specific detection of cysteine-containing peptides. Analyst 2014, 139, 5523–5530. [DOI] [PubMed] [Google Scholar]
- (158).Garcia L; Girod M; Rompais M; Dugourd P; Carapito C; Lemoine J Data-independent acquisition coupled to visible laser-induced dissociation at 473 nm (DIA-LID) for peptide-centric specific analysis of cysteine-containing peptide subset. Anal. Chem 2018, 90, 3928–3935. [DOI] [PubMed] [Google Scholar]
- (159).Cannon J; Lohnes K; Wynne C; Wang Y; Edwards N; Fenselau C High-throughput middle-down analysis using an Orbitrap. J. Proteome Res 2010, 9, 3886–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (160).Cristobal A; Marino F; Post H; van den Toorn HW; Mohammed S; Heck AJR Toward an optimized workflow for middle-down proteomics. Anal. Chem 2017, 89, 3318–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Cotham V; Horton A; Lee J; Georgiou G; Brodbelt JS Middle-down 193 nm ultraviolet photodissociation for unambiguous antibody identification and its implications for immunoproteomic analysis. Anal. Chem 2017, 89, 6498–6504. [DOI] [PubMed] [Google Scholar]
- (162).Cotham VC; Brodbelt JS Characterization of therapeutic monoclonal antibodies at the subunit-level using middle-down 193 nm ultraviolet photodissociation. Anal. Chem 2016, 88, 4004–4013. [DOI] [PubMed] [Google Scholar]
- (163).Sanders JD; Greer SM; Brodbelt JS Integrating carbamylation and ultraviolet photodissociation mass spectrometry for middle down proteomics. Anal. Chem 2017, 89, 11772–11778. [DOI] [PubMed] [Google Scholar]
- (164).Greer SM; Sidoli S; Coradin M; Schack Jespersen M; Schwammle V; Jensen ON; Garcia BA; Brodbelt JS Extensive characterization of heavily modified histone tails by 193 nm ultraviolet photodissociation via a middle-down strategy. Anal. Chem 2018, 90, 10425–10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Tiller KE; Tessier PM Advances in antibody design. Annu. Rev. Biomed. Eng 2015, 17, 191–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Yuan Z-F; Arnaudo AM; Garcia BA Mass spectrometric analysis of histone proteoforms. Annu. Rev. Anal. Chem 2014, 7, 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (167).Toby TK; Fornelli L; Kelleher NL Progress in top-down proteomics and the analysis of proteoforms. Annu. Rev. Anal. Chem 2016, 9, 499–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (168).Catherman AD; Skinner OS; Kelleher NL Top down proteomics: facts and perspectives. Biochem. Biophys. Res. Commun 2014, 445, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Chen B; Brown KA; Lin Z; Ge Y Top-down proteomics: ready for prime time? Anal. Chem 2018, 90, 110–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (170).Patrie SM Top-down mass spectrometry: proteomics to proteoforms. Adv. Exp. Med. Biol 2016, 919, 171–200. [DOI] [PubMed] [Google Scholar]
- (171).LeDuc RD; Fellers RT; Early BP; Greer JB; Shams DP; Thomas PM; Kelleher NL Accurate estimation of context-dependent false discovery rates in top-down proteomics. Mol. Cell. Proteomics 2019, 18, 796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (172).Cannon JS; Martinez Fonts K; Robotham SA; Matouschek A; Brodbelt JS Top down 193-nm ultraviolet photodissociation mass spectrometry for simultaneous determination of polyubiquitin chain length and topology. Anal. Chem 2015, 87, 1812–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Greer SM; Brodbelt JS Top-down characterization of heavily modified histones using 193 nm ultraviolet photodissociation (UVPD) mass spectrometry. J. Proteome Res 2018, 17, 1138–1145. [DOI] [PubMed] [Google Scholar]
- (174).Cannon JR; Kluwe C; Ellington A; Brodbelt JS Characterization of green fluorescent proteins by 193 nm ultraviolet photodissociation mass spectrometry. Proteomics 2014, 14, 1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (175).Thyer R; Robotham SA; Brodbelt JS; Ellington A Evolving tRNASec for efficient canonical incorporation of selenocysteine. J. Am. Chem. Soc 2015, 137, 46–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (176).Thyer R; Shroff R; Klein DR; d’Oelsnitz S; Cotham VC; Byrom M; Brodbelt JS; Ellington AD Custom selenoprotein production enabled by laboratory evolution of recoded bacterial strains. Nat. Biotechnol 2018, 36, 624–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (177).Cammarata M; Lin K-Y; Pruet J; Liu H-W; Brodbelt JS Probing the unfolding of myoglobin and domain C of PARP-1 with covalent labeling and top-down 193 nm ultraviolet photodissociation mass spectrometry. Anal. Chem 2014, 86, 2534–2542. [DOI] [PubMed] [Google Scholar]
- (178).Cammarata MB; Brodbelt JS Characterization of intra- and intermolecular protein crosslinking by top down ultraviolet photodissociation mass spectrometry. ChemistrySelect 2016, 1 (3), 590–593. [Google Scholar]
- (179).Tamara S; Dyachenko A; Fort KL; Makarov AA; Scheltema RA; Heck AJR Symmetry of charge partitioning in collisional and UV photon-induced dissociation of protein assemblies. J. Am. Chem. Soc 2016, 138, 10860–10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (180).Dilillo M; de Graaf EL; Yadav A; Belov ME; McDonnell LA Ultraviolet photodissociation of ESI- and MALDI-generated protein ions on a Q-Exactive mass spectrometer. J. Proteome Res 2018, 18, 557–564. [DOI] [PubMed] [Google Scholar]
- (181).Jiang J; Abramavicius D; Bulheller BM; Hirst JD; Mukamel S Ultraviolet spectroscopy of protein backbone transitions in aqueous solution: combined QM and MM simulations. J. Phys. Chem. B 2010, 114, 8270–8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (182).Cannon JR; Cammarata MB; Robotham SA; Cotham VC; Shaw JB; Fellers RT; Early BP; Thomas PM; Kelleher NL; Brodbelt JS Ultraviolet photodissociation for characterization of whole proteins on a chromatographic time scale. Anal. Chem 2014, 86, 2185–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Cleland TP; DeHart CJ; Fellers RT; VanNispen AJ; Greer JB; LeDuc RD; Parker WR; Thomas PM; Kelleher NL; Brodbelt JS High throughput analysis of intact human proteins using UVPD and HCD on an Orbitrap mass spectrometer. J. Proteome Res 2017, 16, 2072–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (184).Greer SM; Holden DD; Fellers R; Kelleher NL; Brodbelt JS Modulation of protein fragmentation through carbamylation of primary amines. J. Am. Soc. Mass Spectrom 2017, 28, 1587–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (185).Bashyal A; Sanders JD; Holden DD; Brodbelt JS Top-down analysis of proteins in low charge states. J. Am. Soc. Mass Spectrom 2019, 30, 704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (186).Morrison LJ; Brodbelt JS Charge site assignment in native proteins by ultraviolet photodissociation (UVPD) mass spectrometry. Analyst 2016, 141, 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (187).Cannon JR; Holden DD; Brodbelt JS Hybridizing ultraviolet photodissociation with electron transfer dissociation for intact protein characterization. Anal. Chem 2014, 86, 10970–10977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (188).Holden DD; McGee W; Brodbelt JS Integration of ultraviolet photodissociation with proton transfer reactions and ion parking for analysis of intact proteins. Anal. Chem 2016, 88, 1008–1016. [DOI] [PubMed] [Google Scholar]
- (189).Holden DD; Brodbelt JS Ultraviolet photodissociation of native proteins following proton transfer reactions in the gas phase. Anal. Chem 2016, 88, 12354–12362. [DOI] [PubMed] [Google Scholar]
- (190).Shaw JB; Malhan N; Vasil’ev YV; Lopez NI; Makarov A; Beckman JS; Voinov VG Sequencing grade tandem mass spectrometry for top-down proteomics using hybrid electron capture dissociation methods in a benchtop orbitrap mass spectrometer. Anal. Chem 2018, 90, 10819–10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (191).Shliaha PV; Gibb S; Gorshkov V; Jespersen MS; Andersen GR; Bailey D; Schwartz J; Eliuk S; Schwammle V; Jensen ON Maximizing sequence coverage in top-down proteomics by automated multimodal gas-phase protein fragmentation. Anal. Chem 2018, 90, 12519–12526. [DOI] [PubMed] [Google Scholar]
- (192).Lyon YA; Riggs D; Fornelli L; Compton PD; Julian RR The ups and downs of repeated cleavage and internal fragment production in top down proteomics. J. Am. Soc. Mass Spectrom 2018, 29, 150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (193).Wongkongkathep P; Li H; Zhang X; Ogorzalek Loo RR; Julian RR; Loo JA Enhancing protein disulfide bond cleavage by UV excitation and electron capture dissociation for top-down mass spectrometry. Int. J. Mass Spectrom 2015, 390, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (194).Parker WR; Brodbelt JS Characterization of the cysteine content in proteins utilizing cysteine selenylation with 266 nm ultraviolet photodissociation (UVPD). J. Am. Soc. Mass Spectrom 2016, 27 (8), 1344–1350. [DOI] [PubMed] [Google Scholar]
- (195).Halim MA; Girod M; MacAleese L; Lemoine J; Antoine R; Dugourd P Combined infrared multiphoton dissociation with ultraviolet photodissociation for ubiquitin characterization. J. Am. Soc. Mass Spectrom 2016, 27, 1435–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (196).Brodie NI; Huguet R; Zhang T; Viner R; Zabrouskov V; Pan J; Petrotchenko EV; Borchers CH Top-down hydrogen-deuterium exchange analysis of protein structures using ultraviolet photodissociation. Anal. Chem 2018, 90, 3079–3082. [DOI] [PubMed] [Google Scholar]
- (197).Halim MA; MacAleese L; Lemoine J; Antoine R; Dugourd P; Girod M Ultraviolet, Infrared, and High-Low Energy Photodissociation of Post-Translationally Modified Peptides. J. Am. Soc. Mass Spectrom 2018, 29, 270–283. [DOI] [PubMed] [Google Scholar]
- (198).Chait BT; Cadene M; Olinares PD; Rout MP; Shi Y Revealing higher order protein structure using mass spectrometry. J. Am. Soc. Mass Spectrom 2016, 27, 952–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (199).Allison TM; Bechara C Structural mass spectrometry comes of age: new insight into protein structure, function and interactions. Biochem. Soc. Trans 2019, 47, 317. [DOI] [PubMed] [Google Scholar]
- (200).Leney AC; Heck AJR Native mass spectrometry: what is in the name? J. Am. Soc. Mass Spectrom 2017, 28, 5–13. [DOI] [PubMed] [Google Scholar]
- (201).Konijnenberg A; Butterer A; Sobott F Native ion mobility-mass spectrometry and related methods in structural biology. Biochim. Biophys. Acta, Proteins Proteomics 2013, 1834, 1239–1256. [DOI] [PubMed] [Google Scholar]
- (202).Mehmood S; Allison TM; Robinson CV Mass spectrometry of protein complexes: from origins to applications. Annu. Rev. Phys. Chem 2015, 66, 453–474. [DOI] [PubMed] [Google Scholar]
- (203).Mendoza VL; Vachet RW Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom. Rev 2009, 28, 785–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (204).Thomas PW; Cammarata M; Brodbelt JS; Fast W Covalent inhibition of new delhi metallo-beta-lactamase-1 (NDM-1) by cefaclor. ChemBioChem 2014, 15, 2541–2548. [DOI] [PubMed] [Google Scholar]
- (205).O’Reilly FJ; Rappsilber J Cross-linking mass spectrometry: methods and applications in structural, molecular and systems biology. Nat. Struct. Mol. Biol 2018, 25, 1000–1008. [DOI] [PubMed] [Google Scholar]
- (206).Sinz A Cross-linking/mass spectrometry for studying protein structures and protein-protein interactions: where are we now and where should we go from here? Angew. Chem., Int. Ed 2018, 57, 6390–6396. [DOI] [PubMed] [Google Scholar]
- (207).Cammarata MB; Rosenberg J; Macias LA; Bolufer A; Brodbelt JS Expanding the scope of crosslink identifications by incorporating collisional activated dissociation and ultraviolet photodissociation methods. Anal. Chem 2018, 90, 6385–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (208).Bakhtiari M; Konermann L Protein ions generated by native electrospray ionization: comparison of gas phase, solution, and crystal structures. J. Phys. Chem. B 2019, 123, 1784–1796. [DOI] [PubMed] [Google Scholar]
- (209).Snijder J; Rose RJ; Veesler D; Johnson JE; Heck AJR Studying 18 mega dalton virus assemblies with native mass spectrometry. Angew. Chem., Int. Ed 2013, 52, 4020–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (210).Sharon M How far can we go with structural mass spectrometry of protein complexes? J. Am. Soc. Mass Spectrom 2010, 21, 487–500. [DOI] [PubMed] [Google Scholar]
- (211).Zhang H; Cui W; Wen J; Blankenship RE; Gross ML Native electrospray and electron-capture dissociation FTICR mass spectrometry for top-down studies of protein assemblies. Anal. Chem 2011, 83, 5598–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (212).Boeri Erba E; Petosa C The emerging role of native mass spectrometry in characterizing the structure and dynamics of macromolecular complexes. Protein Sci 2015, 24, 1176–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (213).O’Brien JP; Li W; Zhang Y; Brodbelt JS Characterization of native protein complexes using ultraviolet photodissociation mass spectrometry. J. Am. Chem. Soc 2014, 136, 12920–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (214).Rosenberg J; Parker WR; Cammarata MB; Brodbelt JS UV-POSIT: Web-based tools for rapid and facile structural interpretation of ultraviolet photodissociation (UVPD) mass spectra. J. Am. Soc. Mass Spectrom 2018, 29, 1323–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (215).Morrison LJ; Chai W; Rosenberg JA; Henkelman G; Brodbelt JS Characterization of hydrogen bonding motifs in proteins: hydrogen elimination monitoring by ultraviolet photodissociation mass spectrometry. Phys. Chem. Chem. Phys 2017, 19, 20057–20074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (216).Cammarata M; Brodbelt JS Structural characterization of apo-myoglobin by ultraviolet photodissociation. Chem. Sci 2015, 6, 1324–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (217).Cammarata M; Thyer R; Rosenberg J; Ellington A; Brodbelt JS Structural characterization of dihydrofolate reductase complexes by top-down ultraviolet photodissociation mass spectrometry. J. Am. Chem. Soc 2015, 137, 9128–9135. [DOI] [PubMed] [Google Scholar]
- (218).Cammarata M; Thyer R; Lombardo M; Anderson A; Wright D; Ellington A; Brodbelt JS Characterization of trimethoprim resistant E. coli dihydrofolate reductase mutants by mass spectrometry and P21L inhibition by propargyl-linked antifolates. Chem. Sci 2017, 8, 4062–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (219).Cammarata MB; Schardon CL; Mehaffey MR; Rosenberg J; Singleton J; Fast W; Brodbelt JS Impact of G12C mutation on conformation of GTPase K-Ras probed by ultraviolet photodissociation mass spectrometry. J. Am. Chem. Soc 2016, 138, 13187–13196. [DOI] [PubMed] [Google Scholar]
- (220).Mehaffey MR; Schardon CL; Novelli ET; Cammarata MB; Webb LJ; Fast W; Brodbelt JS Investigation of GTP-dependent dimerization of G12X K-ras variants using ultraviolet photodissociation mass spectrometry. Chem. Sci 2019, 10, 8025–8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (221).Mehaffey MR; Cammarata MB; Brodbelt JS Tracking the catalytic cycle of adenylate kinase by ultraviolet photodissociation mass spectrometry. Anal. Chem 2018, 90, 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (222).Li H; Sheng Y; McGee W; Cammarata M; Holden D; Loo JA Structural characterization of native proteins and protein complexes by electron ionization dissociation-mass spectrometry. Anal. Chem 2017, 89, 2731–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (223).Prior IA; Lewis PD; Mattos C A comprehensive survey of Ras mutations in cancer. Cancer Res 2012, 72, 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (224).Canon F; Milosavljevic AR; van der Rest G; Refregiers M; Nahon L; Sarni-Manchado P; Cheynier V; Giuliani A Photodissociation and dissociative photoionization mass spectrometry of proteins and noncovalent protein-ligand complexes. Angew. Chem., Int. Ed 2013, 52, 8377–8381. [DOI] [PubMed] [Google Scholar]
- (225).Jurchen JC; Williams ER Origin of asymmetric charge partitioning in the dissociation of gas-phase protein homodimers. J. Am. Chem. Soc 2003, 125, 2817–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Pagel K; Hyung S-J; Ruotolo BT; Robinson CV Alternate dissociation pathways identified in charge-reduced protein complex ions. Anal. Chem 2010, 82, 5363–5372. [DOI] [PubMed] [Google Scholar]
- (227).Sciuto SV; Liu J; Konermann L An electrostatic charge partitioning model for the dissociation of protein complexes in the gas phase. J. Am. Soc. Mass Spectrom 2011, 22, 1679–1689. [DOI] [PubMed] [Google Scholar]
- (228).Hall Z; Politis A; Bush MF; Smith LJ; Robinson CV Charge-state dependent compaction and dissociation of protein complexes: insights from ion mobility and molecular dynamics. J. Am. Chem. Soc 2012, 134, 3429–3438. [DOI] [PubMed] [Google Scholar]
- (229).Benesch JLP; Ruotolo BT; Sobott F; Wildgoose J; Gilbert A; Bateman R; Robinson CV Quadrupole-time-of-flight mass spectrometer modified for higher-energy dissociation reduces protein assemblies to peptide fragments. Anal. Chem 2009, 81, 1270–1274. [DOI] [PubMed] [Google Scholar]
- (230).Belov ME; Damoc E; Denisov E; Compton PD; Horning S; Makarov AA; Kelleher NL From protein complexes to subunit backbone fragments: A multi-stage approach to native mass spectrometry. Anal. Chem 2013, 85, 11163–11173. [DOI] [PubMed] [Google Scholar]
- (231).Li H; Nguyen H; Ogorzalek Loo RR; Campuzano IDG; Loo JA An integrated native mass spectrometry and top-down proteomics method that connects sequence to structure and function of macromolecular complexes. Nat. Chem 2018, 10, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (232).Clarke DJ; Murray E; Hupp T; Mackay CL; Langridge-Smith PRR Mapping a noncovalent protein–peptide interface by top-down FTICR mass spectrometry using electron capture dissociation. J. Am. Soc. Mass Spectrom 2011, 22, 1432–1440. [DOI] [PubMed] [Google Scholar]
- (233).Zhang H; Cui W; Gross ML Native electrospray ionization and electron-capture dissociation for comparison of protein structure in solution and the gas phase. Int. J. Mass Spectrom 2013, 354–355, 288–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (234).Schennach M; Breuker K Proteins with highly similar native folds can show vastly dissimilar folding behavior when desolvated. Angew. Chem., Int. Ed 2014, 53, 164–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (235).Li H; Wolff JJ; Van Orden SL; Loo JA Native top-down electrospray ionization-mass spectrometry of 158 kDa protein complex by high-resolution Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem 2014, 86, 317–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Lermyte F; Sobott F Electron transfer dissociation provides higher-order structural information of native and partially unfolded protein complexes. Proteomics 2015, 15, 2813–2822. [DOI] [PubMed] [Google Scholar]
- (237).Skinner OS; McAnally MO; Van Duyne RP; Schatz GC; Breuker K; Compton PD; Kelleher NL Native electron capture dissociation maps to iron-binding channels in horse spleen ferritin. Anal. Chem 2017, 89, 10711–10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (238).Zhou M; Dagan S; Wysocki VH Impact of charge state on gas-phase behaviors of noncovalent protein complexes in collision induced dissociation and surface induced dissociation. Analyst 2013, 138, 1353–1362. [DOI] [PubMed] [Google Scholar]
- (239).Zhou M; Jones CM; Wysocki VH Dissecting the large noncovalent protein complex GroEL with surface-induced dissociation and ion mobility-mass spectrometry. Anal. Chem 2013, 85, 8262–8267. [DOI] [PubMed] [Google Scholar]
- (240).Quintyn RS; Yan J; Wysocki VH Surface-induced dissociation of homotetramers with D2 symmetry yields their assembly pathways and characterizes the effect of ligand binding. Chem. Biol 2015, 22, 583–592. [DOI] [PubMed] [Google Scholar]
- (241).Harvey SR; Yan J; Brown JM; Hoyes E; Wysocki VH Extended gas-phase trapping followed by surface-induced dissociation of noncovalent protein complexes. Anal. Chem 2016, 88, 1218–1221. [DOI] [PubMed] [Google Scholar]
- (242).Quintyn RS; Harvey SR; Wysocki VH Illustration of SID-IM-SID (Surface-Induced Dissociation-Ion Mobility-SID) mass spectrometry: Homo and hetero model protein complexes. Analyst 2015, 140, 7012–7019. [DOI] [PubMed] [Google Scholar]
- (243).Van Aernum ZL; Gilbert JD; Belov ME; Makarov AA; Horning SR; Wysocki VH Surface-induced dissociation of noncovalent protein complexes in an extended mass range orbitrap mass spectrometer. Anal. Chem 2019, 91, 3611–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (244).Morrison LJ; Brodbelt JS 193 nm ultraviolet photodissociation mass spectrometry of tetrameric protein complexes reflects quaternary and secondary protein topology. J. Am. Chem. Soc 2016, 138 (4), 10849–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (245).Mehaffey MR; Sanders JD; Holden DD; Nilsson CL; Brodbelt JS Multi-stage ultraviolet photodissociation mass spectrometry to characterize single amino acid variants of human mitochondrial BCAT2. Anal. Chem 2018, 90, 9904–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (246).Sipe SN; Brodbelt JS Impact of charge state on 193 nm ultraviolet photodissociation of protein complexes. Phys. Chem. Chem. Phys 2019, 21, 9265–9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (247).Ben-Nissan G; Belov ME; Morgenstern D; Levin Y; Dym O; Arkind G; Lipson C; Makarov AA; Sharon M Triple-stage mass spectrometry unravels the heterogeneity of an endogenous protein complex. Anal. Chem 2017, 89, 4708–4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (248).Skinner OS; Haverland NA; Fornelli L; Melani RD; Do Vale LHF; Seckler HS; Doubleday PF; Schachner LF; Srzentić K; Kelleher NL; Compton PD Top-down characterization of endogenous protein complexes with native proteomics. Nat. Chem. Biol 2018, 14, 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (249).Warnke S; Baldauf C; Bowers MT; Pagel K; von Helden G Photodissociation of conformer-selected ubiquitin ions reveals site-specific cis/trans isomerization of protein peptide bonds. J. Am. Chem. Soc 2014, 136, 10308–10314. [DOI] [PubMed] [Google Scholar]
- (250).Warnke S; von Helden G; Pagel K Analyzing the higher order structure of proteins with conformer-selective ultraviolet photodissociation. Proteomics 2015, 15, 2804–2812. [DOI] [PubMed] [Google Scholar]
- (251).Theisen A; Yan B; Brown JM; Morris M; Bellina B; Barran PE Use of ultraviolet photodissociation coupled with ion mobility mass spectrometry to determine structure and sequence from drift time of selected peptides and proteins. Anal. Chem 2016, 88, 9964–9971. [DOI] [PubMed] [Google Scholar]
- (252).Theisen A; Black R; Corinti D; Brown JM; Bellina B; Barran PE Initial protein unfolding events in ubiquitin, cytochrome c, and myoglobin are revealed with the use of 213 nm UVPD coupled to IM-MS. J. Am. Soc. Mass Spectrom 2019, 30, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (253).Simon A-L; Chirot F; Choi CM; Clavier C; Barbaire M; Maurelli J; Dagany X; MacAleese L; Dugourd P Tandem ion mobility spectrometry coupled to laser excitation. Rev. Sci. Instrum 2015, 86 (9), 094101. [DOI] [PubMed] [Google Scholar]
- (254).Choi CM; Simon AL; Chirot F; Kulesza A; Knight G; Daly S; MacAleese L; Antoine R; Dugourd P Charge, color, and conformation: spectroscopy on isomer-selected peptide ions. J. Phys. Chem. B 2016, 120, 709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (255).Fahy E; Cotter D; Sud M; Subramaniam S Lipid classification, structures, and tools. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2011, 1811, 637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (256).Mitchell TW; Pham H; Thomas MC; Blanksby SJ Identification of double bond position in lipids: From GC to OzID. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci 2009, 877, 2722–2735. [DOI] [PubMed] [Google Scholar]
- (257).Yoo HJ; Håkansson K Determination of double bond location in fatty acids by manganese adduction and electron induced dissociation. Anal. Chem 2010, 82, 6940–6948. [DOI] [PubMed] [Google Scholar]
- (258).Baba T; Campbell JL; Le Blanc JCY; Baker PRS; Ikeda K Quantitative structural multiclass lipidomics using differential mobility: electron impact excitation of ions from organics (EIEIO) mass spectrometry. J. Lipid Res 2018, 59, 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (259).Zhang W; Zhang D; Chen Q; Wu J; Ouyang Z; Xia Y Online photochemical derivatization enables comprehensive mass spectrometric analysis of unsaturated phospholipid isomers. Nat. Commun 2019, 10, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (260).Pham HT; Ly T; Trevitt AJ; Mitchell TW; Blanksby SJ Differentiation of complex lipid isomers by radical-directed dissociation mass spectrometry. Anal. Chem 2012, 84, 7525–7532. [DOI] [PubMed] [Google Scholar]
- (261).Pham HT; Trevitt AJ; Mitchell TW; Blanksby SJ Rapid differentiation of isomeric lipids by photodissociation mass spectrometry of fatty acid derivatives. Rapid Commun. Mass Spectrom 2013, 27, 805–815. [DOI] [PubMed] [Google Scholar]
- (262).Pham HT; Julian RR Mass shifting and radical delivery with crown ether attachment for separation and analysis of phosphatidylethanolamine lipids. Anal. Chem 2014, 86, 3020–3027. [DOI] [PubMed] [Google Scholar]
- (263).Pham HT; Julian RR Radical delivery and fragmentation for structural analysis of glycerophospholipids. Int. J. Mass Spectrom 2014, 370, 58–65. [Google Scholar]
- (264).Pham HT; Julian RR Characterization of glycosphingolipid epimers by radical-directed dissociation mass spectrometry. Analyst 2016, 141, 1273–1278. [DOI] [PubMed] [Google Scholar]
- (265).Hancock SE; Ailuri R; Marshall DL; Brown SHJ; Saville JT; Narreddula VR; Boase NR; Poad BLJ; Trevitt AJ; Willcox MDP; Kelso MJ; Mitchell TW; Blanksby SJ Mass spectrometry-directed structure elucidation and total synthesis of ultralong chain (O-acyl)-omega-hydroxy fatty acids. J. Lipid Res 2018, 59, 1510–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (266).Nie S; Pham HT; Blanksby SJ; Reid GE Photoinduced intermolecular cross-linking of gas phase triacylglycerol lipid ions. Eur. J. Mass Spectrom 2015, 21, 287–296. [DOI] [PubMed] [Google Scholar]
- (267).Madsen JA; Cullen TW; Trent MS; Brodbelt JS IR and UV photodissociation as analytical tools for characterizing lipid A structures. Anal. Chem 2011, 83, 5107–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (268).Hankins JV; Madsen JA; Giles DK; Childers BM; Klose KE; Brodbelt JS; Trent MS Elucidation of a novel Vibrio Cholera lipid A secondary hydroxyl-acyltransferase and its roles in pathogenesis. Mol. Microbiol 2011, 81, 1313–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (269).Hankins JV; Madsen JA; Giles DK; Brodbelt JS; Trent MS Amino acid addition to the bacterial surface confers polymyxin B resistance in V. cholerae. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 8722–8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (270).Hankins JV; Madsen JA; Needham BD; Brodbelt JS; Trent MS The outer membrane of gram-negative bacteria: lipid A isolation and characterization. Methods Mol. Biol 2013, 966, 239–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (271).Cullen TW; O’Brien JP; Hendrixson DR; Giles DK; Hobb RI; Thompson SA; Brodbelt JS; Trent MS EptC of Campylobacter jejuni mediates phenotypes involved in host interactions and virulence. Infect. Immun 2013, 81, 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (272).O’Brien JP; Needham BD; Henderson JC; Nowicki EM; Trent MS; Brodbelt JS 193 nm Ultraviolet photodissociation mass spectrometry for the structural elucidation of lipid a compounds in complex mixtures. Anal. Chem 2014, 86, 2138–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (273).Rubin EJ; O’Brien JP; Ivanov PL; Brodbelt JS; Trent MS Identification of a broad family of lipid A late acyltransferases with non-canonical substrate specificity. Mol. Microbiol 2014, 91, 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (274).O’Brien JP; Needham BD; Brown DB; Trent MS; Brodbelt JS Top-down strategies for the structural elucidation of intact lipopolysaccharides. Chem. Sci 2014, 5, 4291–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (275).Nowicki E; O’Brien JP; Brodbelt JS; Trent MS Characterization of Pseudomonas aeruginosa LpxT reveals dual positional lipid A kinase activity and coordinated control of outer membrane modification. Mol. Microbiol 2014, 94, 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (276).Nowicki EM; O’Brien JP; Brodbelt JS; Trent MS Extracellular zinc induces phophoethanolamine addition to Pseudomonas aeruginosa lipid A via the CoIR response regulator. Mol. Microbiol 2015, 97, 166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (277).Boll JM; Tucker AT; Klein DR; Beltran AM; Brodbelt JS; Davies BW; Trent MS Reinforcing the cell surface, lipid A acylation in Acinetobacter baumannii promotes CAMP resistance and desiccation survival. mBio 2015, 6, e00478–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (278).Morrison LJ; Parker WR; Holden DD; Henderson JC; Boll J; Trent MS; Brodbelt JS UVliPiD: A UVPD-based hierarchical approach for de novo characterization of lipid A structures. Anal. Chem 2016, 88, 1812–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (279).Crittenden CM; Akin LD; Morrison LJ; Trent MS; Brodbelt JS Characterization of lipid A variants by MS/MS: impact of acyl chains. J. Am. Soc. Mass Spectrom 2017, 28, 1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (280).Crittenden CM; Herrera CM; Williams PE; Ricci DP; Swem LR; Trent MS; Brodbelt JS Mapping phosphate modifications of substituted lipid a via a targeted MS3 CID/UVPD strategy. Analyst 2018, 143, 3091–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (281).Klein DR; Powers MJ; Trent MS; Brodbelt JS Top-down characterization of lipooligosaccharides from antibiotic-resistant bacteria. Anal. Chem 2019, 91, 9608–0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (282).O’Brien JP; Brodbelt JS Structural characterization of gangliosides and glycolipids via ultraviolet photodissociation mass spectrometry. Anal. Chem 2013, 85, 10399–10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (283).Klein DR; Brodbelt JS Structural characterization of phosphatidylcholines using 193 nm ultraviolet photodissociation mass spectrometry. Anal. Chem 2017, 89, 1516–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (284).Williams PE; Klein DR; Greer SM; Brodbelt JS Pinpointing double bond and sn-positions in glycerophospholipids via hybrid 193 nm ultraviolet photodissociation mass spectrometry. J. Am. Chem. Soc 2017, 139, 15681–15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (285).Klein DR; Feider CL; Garza KY; Lin JQ; Eberlin LS; Brodbelt JS Desorption electrospray ionization coupled with ultraviolet photodissociation for characterization of phospholipid isomers in tissue sections. Anal. Chem 2018, 90, 10100–10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (286).Ryan E; Nguyen CQN; Shiea C; Reid GE Detailed structural characterization of sphingolipids via 193 nm ultraviolet photodissociation and ultra high resolution tandem mass spectrometry. J. Am. Soc. Mass Spectrom 2017, 28 (7), 1406–1419. [DOI] [PubMed] [Google Scholar]
- (287).Becher S; Esch P; Heiles S Relative quantification of phosphatidylcholine sn-isomers using positive doubly charged lipid-metal ion complexes. Anal. Chem 2018, 90, 11486–11494. [DOI] [PubMed] [Google Scholar]
- (288).Laskin J; Lanekoff I Ambient mass spectrometry imaging using direct liquid extraction techniques. Anal. Chem 2016, 88, 52–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (289).Novikov A; Breton A; Caroff M Micromethods for isolation and structural characterization of lipid A and polysaccharide regions of bacterial lipopolysaccharides. Methods Mol. Biol 2017, 1600, 167–186. [DOI] [PubMed] [Google Scholar]
- (290).Steimle A; Autenrieth IB; Frick J-S Structure and function: Lipid A modifications in commensals and pathogens. Int. J. Med. Microbiol 2016, 306, 290–301. [DOI] [PubMed] [Google Scholar]
- (291).Henderson JC; Zimmerman SM; Crofts AA; Boll JM; Kuhns LG; Herrera CM; Trent MS The power of asymmetry: architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu. Rev. Microbiol 2016, 70, 255–278. [DOI] [PubMed] [Google Scholar]
- (292).Ting YS; Shaffer SA; Jones JW; Ng WV; Ernst RK; Goodlett DR Automated lipid A structure assignment from hierarchical tandem mass spectrometry data. J. Am. Soc. Mass Spectrom 2011, 22, 856–866. [DOI] [PubMed] [Google Scholar]
- (293).Tinoco I Physical chemistry of nucleic acids. Annu. Rev. Phys. Chem 2002, 53, 1–15. [DOI] [PubMed] [Google Scholar]
- (294).Limbach PA; Crain PF; McCloskey JA Characterization of oligonucleotides and nucleic acids by mass spectrometry. Curr. Opin. Biotechnol 1995, 6, 96–102. [DOI] [PubMed] [Google Scholar]
- (295).Hofstadler SA; Griffey RH Analysis of noncovalent complexes of DNA and RNA by mass spectrometry. Chem. Rev 2001, 101, 377–390. [DOI] [PubMed] [Google Scholar]
- (296).Brodbelt JS Evaluation of DNA/ligand interactions by electrospray ionization mass spectrometry. Annu. Rev. Anal. Chem 2010, 3, 67–87. [DOI] [PubMed] [Google Scholar]
- (297).Brodbelt JS; Xu Z Ligand binding to nucleic acids In Nucleic Acids in the Gas Phase; Springer Verlag: 2014; DOI: 10.1007/978-3-642-54842-0_9. [DOI] [Google Scholar]
- (298).Silvestri C; Brodbelt JS Tandem mass spectrometry for characterization of covalent adducts of DNA with anticancer therapeutics. Mass Spectrom. Rev 2013, 32, 247–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (299).Wu J; McLuckey SA Gas-phase fragmentation of oligonucleotide ions. Int. J. Mass Spectrom 2004, 237, 197–241. [Google Scholar]
- (300).Gabelica V; Rosu F; De Pauw E; Antoine R; Tabarin T; Broyer M; Dugourd P Electron photodetachment dissociation of DNA anions with covalently or noncovalently bound chromophores. J. Am. Soc. Mass Spectrom 2007, 18, 1990–2000. [DOI] [PubMed] [Google Scholar]
- (301).Antoine R; Lemoine J; Dugourd P Electron photodetachment dissociation for structural characterization of synthetic and bio-polymer anions. Mass Spectrom. Rev 2014, 33, 501–522. [DOI] [PubMed] [Google Scholar]
- (302).Smith SI; Brodbelt JS Characterization of oligodeoxynucleotides and modifications by 193 nm photodissociation and electron photodetachment. Anal. Chem 2010, 82, 7218–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (303).Smith SI; Brodbelt JS Hybrid activation methods for elucidating nucleic acid modifications. Anal. Chem 2011, 83, 303–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (304).Xu Z; Shaw JB; Brodbelt JS Comparison of MS/MS methods for characterization of DNA/cisplatin adducts. J. Am. Soc. Mass Spectrom 2013, 24, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (305).Paul D; Marchand A; Verga D; Teulade-Fichou M-P; Bombard S; Rosu F; Gabelica V Probing ligand and cation binding sites in G-quadruplex nucleic acids by mass spectrometry and electron photodetachment dissociation sequencing. Analyst 2019, 144, 3518–3524. [DOI] [PubMed] [Google Scholar]
- (306).Varki A Biological roles of glycans. Glycobiology 2017, 27, 3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (307).Raman R; Raguram S; Venkataraman G; Paulson JC; Sasisekharan R Glycomics: an integrated systems approach to structure-function relationships of glycans. Nat. Methods 2005, 2, 817–824. [DOI] [PubMed] [Google Scholar]
- (308).Costello CE; Vath JE Tandem mass spectrometry of glycolipids. Methods Enzymol 1990, 193, 738–768. [DOI] [PubMed] [Google Scholar]
- (309).Devakumar A; Thompson MS; Reilly JP Fragmentation of oligosaccharide ions with 157 nm vacuum ultraviolet light. Rapid Commun. Mass Spectrom 2005, 19, 2313–2320. [DOI] [PubMed] [Google Scholar]
- (310).Devakumar A; Mechref Y; Kang P; Novotny MV; Reilly JP Laser-induced photofragmentation of neutral and acidic glycans inside an ion-trap mass spectrometer. Rapid Commun. Mass Spectrom 2007, 21, 1452–1460. [DOI] [PubMed] [Google Scholar]
- (311).Devakumar A; Mechref Y; Kang P; Novotny MV; Reilly JP Identification of isomeric N-glycan structures by mass spectrometry with 157 nm laser-induced photofragmentation. J. Am. Soc. Mass Spectrom 2008, 19, 1027–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (312).Zhang L; Reilly JP Extracting both peptide sequence and glycan structural information by 157 nm photodissociation of nlinked glycopeptides. J. Proteome Res 2009, 8, 734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (313).Ko BJ; Brodbelt JS 193 nm Ultraviolet photodissociation of deprotonated sialylated oligosaccharides. Anal. Chem 2011, 83, 8192–8200. [DOI] [PubMed] [Google Scholar]
- (314).Madsen JA; Ko BJ; Xu H; Iwashkiw JA; Robotham SA; Shaw JB; Feldman MF; Brodbelt JS Concurrent automated sequencing of the glycan and peptide portions of O-linked glycopeptide anions by ultraviolet photodissociation mass spectrometry. Anal. Chem 2013, 85, 9253–9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (315).Ko BJ; Brodbelt JS Comparison of glycopeptide fragmentation by collision induced dissociation and ultraviolet photodissociation. Int. J. Mass Spectrom 2015, 377, 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (316).Crittenden CM; Escobar EE; Williams PE; Sanders JD; Brodbelt JS Characterization of antigenic oligosaccharides from gram-negative bacteria via activated electron photodetachment mass spectrometry. Anal. Chem 2019, 91, 4672–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (317).Klein D; Leach FE III; Amster IJ; Brodbelt JS Structural characterization of glycosaminoglycan carbohydrates using 193 nm ultraviolet photodissociation. Anal. Chem 2019, 91, 6019–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (318).Zucker SM; Lee S; Webber N; Valentine SJ; Reilly JP; Clemmer DE An ion mobility/ion trap/photodissociation instrument for characterization of ion structure. J. Am. Soc. Mass Spectrom 2011, 22, 1477–1485. [DOI] [PubMed] [Google Scholar]
- (319).Lee S; Valentine SJ; Reilly JP; Clemmer DE Analyzing a mixture of disaccharides by IMS-VUVPD-MS. Int. J. Mass Spectrom 2012, 309, 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (320).Morrison KA; Clowers BH Differential fragmentation of mobility-selected glycans via ultraviolet photodissociation and ion mobility-mass spectrometry. J. Am. Soc. Mass Spectrom 2017, 28, 1236–1241. [DOI] [PubMed] [Google Scholar]
- (321).Ropartz D; Lemoine J; Giuliani A; Bittebiere Y; Enjalbert Q; Antoine R; Dugourd P; Ralet MC; Rogniaux H Deciphering the structure of isomeric oligosaccharides in a complex mixture by tandem mass spectrometry: Photon activation with vacuum ultra-violet brings unique information and enables definitive structure assignment. Anal. Chim. Acta 2014, 807, 84–95. [DOI] [PubMed] [Google Scholar]
- (322).Ropartz D; Giuliani A; Herve C; Geairon A; Jam M; Czjzek M; Rogniaux H High-energy photon activation tandem mass spectrometry provides unprecedented insights into the structure of highly sulfated oligosaccharides extracted from macroalgal cell walls. Anal. Chem 2015, 87, 1042–1049. [DOI] [PubMed] [Google Scholar]
- (323).Ropartz D; Giuliani A; Fanuel M; Herve C; Czjzek M; Rogniaux H Online coupling of high-resolution chromatography with extreme UV photon activation tandem mass spectrometry: Application to the structural investigation of complex glycans by dissociative photoionization. Anal. Chim. Acta 2016, 933, 1–9. [DOI] [PubMed] [Google Scholar]
- (324).Racaud A; Antoine R; Dugourd P; Lemoine J Photoinduced dissociation of heparin-derived oligosaccharides controlled by charge location. J. Am. Soc. Mass Spectrom 2010, 21, 2077–2084. [DOI] [PubMed] [Google Scholar]
- (325).Zhang X; Julian RR Radical mediated dissection of oligosaccharides. Int. J. Mass Spectrom 2014, 372, 22–28. [Google Scholar]
- (326).Riggs DL; Hofmann J; Hahm HS; Seeberger PH; Pagel K; Julian RR Glycan isomer identification using ultraviolet photodissociation initiated radical chemistry. Anal. Chem 2018, 90, 11581–11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (327).Reiding KR; Bondt A; Franc V; Heck AJR The benefits of hybrid fragmentation methods for glycoproteomics. TrAC, Trends Anal. Chem 2018, 108, 260–268. [Google Scholar]
- (328).Ickert S; Beck S; Linscheid MW; Riedel J VUV photodissociation induced by a deuterium lamp in an ion trap. J. Am. Soc. Mass Spectrom 2019, 30, 2114–2122. [DOI] [PubMed] [Google Scholar]