Abstract
Successful cancer treatment continues to elude modern medicine and its arsenal of therapeutic strategies. Therapy resistance is driven by significant tumor heterogeneity, complex interactions between malignant, microenvironmental and immune cells and cross talk between signaling pathways. Advances in molecular characterization technologies such as next generation sequencing have helped unravel this network of interactions and have vastly affected how cancer is diagnosed and treated. However, the translation of complex genomic analyses to pathological diagnosis remains challenging using conventional immunofluorescence (IF) staining, which is typically limited to 2–5 antigens. Numerous strategies to increase distinct antigen detection on a single sample have been investigated, but all have deleterious effects on the tissue limiting the maximum number of biomarkers that can be imaged on a single sample and none can be seamlessly integrated into routine clinical workflows. To facilitate ready integration into clinical histopathology, we have developed a novel cyclic IF (cycIF) technology based on antibody conjugated oligonucleotides (Ab-oligos). In situ hybridization of complementary oligonucleotides (oligos) facilitates biomarker labeling for imaging on any conventional fluorescent microscope. We have validated a variety of oligo configurations and their respective signal removal strategies capable of diminishing fluorescent signal to levels of autofluorescence before subsequent staining cycles. Robust signal removal is performed without the employment of harsh conditions or reagents, maintaining tissue integrity and antigenicity for higher dimensionality immunostaining of a single sample. Our platform Ab-oligo cycIF technology uses conventional fluorophores and microscopes, allowing for dissemination to a broad audience and congruent integration into clinical histopathology workflows.
Keywords: cyclic immunofluorescence, fluorescence microscopy, oligonucleotide, antibody conjugation, restriction enzyme, photocleavable linker, tumor heterogeneity, biomarker distribution
1. INTRODUCTION
Significant tumor heterogeneity is driven by complex interactions between malignant, microenvironmental and immune cells, as well as cross talk between signaling pathways. This heterogeneity has largely foiled modern medicine’s efforts towards successful cancer cures. Advances in molecular characterization technologies, such as next generation sequencing, have vastly affected how cancer is diagnosed and treated. For example, genetic analysis has led to the identification of subtypes within breast cancer [1,2], all of which have unique responses to therapy. Genomic analyses are a therapeutic tool, but at the cost of cellular and subcellular context of biomarker distribution. Recent discoveries, have revealed the significance of the spatial relationships of cancer, immune, and microenvironmental cells [3,4,5]. To further understand the diagnostic and prognostic implications of these relationships, a molecular profiling technology to measure both expression and spatial context of biomarkers, while preserving both will be required.
Histopathology, most commonly performed on formalin-fixed paraffin-embedded (FFPE) tissue, is one of the most important tools for pathological diagnosis. Conventional methods of immunohistochemical (IHC) and immunofluorescent (IF) staining are typically limited to labeling 2–5 antigens on a single sample [6], making translation of genomic analyses to conventional pathological diagnosis impossible. There have been a number of strategies proposed to increase specific antigen labeling on a single sample for both IHC and IF, all of which are unable to be integrated into routine clinical histopathology [6–12]. To this end, it is important to make note that IHC has lower dimensionality simultaneous immunostaining than IF due to detection of spectrally unique chromogens employed in antigen labeling. While IF labeling allows increased multiplexing, bandpass filtration allows visualization of a maximum of five fluorophores on a single sample while spectral unmixing techniques with specialized camera and software increases this limit to seven [6,7,13]. This limitation has led to the development of a variety of techniques using serial cycles of fluorescent tagging, imaging, and bleaching [8,9,14] or dissociation of affinity tags [6,10,11] to vastly increase imaging dimensionality on a single sample. Similar serial techniques have been proposed for IHC staining [12], but IHC is primarily a single channel method and utilize precipitates such as brown diamino-benzidine (DAB) which are less quantitative than fluorescence [15]. These techniques have highly multiplexed capabilities with reported limits of registration of up to 61 immunostained targets in a single sample [8]. These technologies greatly improve the multiplexing capabilities of IF, but will be difficult to integrate into clinical histopathology because (1) antigenicity of the tissue is affected by the bleaching and destaining methods [16], (2) steric hindrance issues can occur as cycle number increases, and (3) tissue integrity is compromised due to the extensive tissue processing, causing loss of tissue as cycle number increases [17,18].
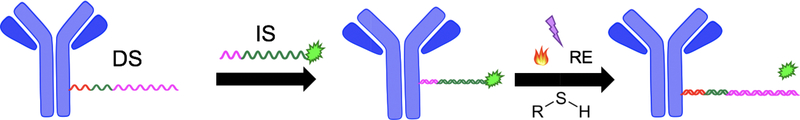
The shortcomings described above were the catalyst for development of our improved highly-multiplexable, cyclic IF (CycIF) technique capable of generating extreme multi-parametric images for quantifying biomarker expression and distribution. It was imperative that our technique required minimal adjustments to current clinical workflows for easy adoption and mild labeling conditions for robust staining cycles. Our technique, termed antibody conjugated oligonucleotide (Ab-oligo) cycIF, preserves tissue antigenicity and lends itself to ready integration into clinical workflows. Ab-oligo cycIF exploits in situ hybridization of complementary oligonucleotides for biomarker labeling and the modifiable nature of oligos to facilitate signal removal for sequential rounds of fluorescent tagging. In our technology, a single stranded oligo (docking strand, DS) is conjugated to the primary antibody. Then the introduction of a complementary single stranded oligo (imaging strand, IS) conjugated to a conventional fluorophore, facilitates specific on tissue fluorescence labeling through hybridization which can be imaged using any conventional fluorescence microscope. Signal removal can be completed using a variety of strategies, which result in full signal removal to levels of autofluorescence prior to subsequent staining cycles (Fig. 1).
Figure 1. Ab-oligo cycIF scheme.
Primary antibodies were conjugated to single stranded oligos (DS = docking strand). After Ab-oligo tissue labeling, the complementary oligo conjugated to a fluorophore (IS = imaging strand) was used for in situ hybridization. After imaging, signal removal was completed using thermal melting, UV light exposure, RE-based oligo cleavage or introduction of thiol for cleavage of a disulfide bond.
Herein, we describe the various modifications to the IS and/or signal removal procedures employed to achieve complete signal removal without the negative trade-offs in previously proposed cyclic immunostaining methods. Ab-oligo cycIF preserves tissue antigenicity and integrity allowing for a greater theoretical limit of labeled targets on a single sample for enhanced understanding of the heterogeneous nature of cancers.
2. METHODOLOGY
2.1. Ab-oligo conjugation
Monoclonal, IgG antibodies were purchased from Thermo Fisher Scientific (Waltham, MA), AbCam (Cambridge, UK), Biolegend (San Diego, CA), or Agilent Technologies (Santa Clara, CA) for the following targets: human epidermal growth factor 2 (HER2), cytokeratin 19 (CK19), lamin B1, estrogen receptor (ER), and programmed cell death protein 1 (PD-1). A unique 28 mer oligonucleotide (docking strand, DS) for each target was purchased from Integrated DNA Technologies (IDT, Skokie, IL). Antibody modification and ensuing conjugation with oligonucleotides (oligos) was performed with either the Solulink™ antibody modification kit (TriLink Biotechnologies, San Diego, CA) or the SiteClick™ Antibody Azido modification kit (Thermo Fisher Scientific) abiding to the manufacturers’ instructions. Excess, unconjugated oligo was filtered from Ab-oligo conjugates using molecular weight cut off (MWCO) spin columns.
2.2. Formalin Fixed Paraffin Embedded (FFPE) Tissue for Antibody Staining
After baking, the FFPE tissue slides were deparaffinized at room temperature (RT) in xylenes (2 × 10 minutes (min) washes). To rehydrate the tissue, a series of washes in ethanol and water solutions was performed: 100% ethanol (2 × 10 min), 95% ethanol (1 × 5 min), 70% ethanol (1 × 5 min), and 50% (1 × 5 min). The tissue was then rinsed in 100% deionized water (diH2O) and washed in PBS, pH 7.4 (1 × 10 min). Antigen retrieval was performed in a Pascal Pressure Cooker (Dako, Santa Clara, CA). RT solutions of 10 mM, pH 6.0 sodium citrate, diH2O, and pH 8.0, 1x tris hydrochloride (HCl) were placed in their respective containers in the basin of the chamber containing 500 mL of diH2O. The deparaffinized tissue slides were immersed in the container with sodium citrate buffer and the lid of the pressure cooker locked. The temperature was increased to 125 °C for a ~15-minute period where the pressure rose to ~15 psi. The pressure cooker then cooled to 90 °C and pressure decreased to 0 psi during a ~25 min interval. The lid was rem ved, the slides rinsed in the hot diH2O, placed in the hot tris-HCl buffer, and lid replaced for a 10 min incubation time. The slides were then transferred back to the hot diH2O and brought back to RT by slowly adding RT diH2O to the container. The slides remained in RT diH2O for 5 min and then placed in RT, pH 7.4 PBS for 5 minutes.
The slides were subsequently blocked for 30 min at RT in a solution comprised of 2% bovine serum albumin (BSA, bioWORLD, Dublin, OH), 0.5 mg/mL sheared salmon sperm DNA (Thermo Fisher Scientific), and 0.5% dextran sulfate (Sigma-Aldrich, St. Louis, MO) in 1X PBS, pH 7.4. Ab-oligo conjugates were diluted in the same blocking buffer to a final protein concentration of 15 μg/mL. Tissue sections were covered with adequate volume of diluted Ab-oligo conjugate, 40–100 μl depending on tissue size, and incubated at 4 °C overnight. The following day, the tissue sections were washed in pH 7.0 2X saline-sodium citrate (SSC) buffer (VWR, Radnor, PA) for 3 × 5 min. The tissue was then fixed with 2% paraformaldehyde (PFA, Sigma-Aldrich) for 15 min at RT and washed again in 2X SSC (3 × 5 min).
Procedures for the addition of the complementary fluorophore conjugated imaging strand (IS) can be stratified based on signal removal method as follows.
Restriction Enzyme/Thermal/PCL
Complementary IS labeled with either Alexa Fluor 488 (AF488), Alexa Fluor 546 (AF546), or Alexa Fluor 647 (AF647) were diluted to 350 nM in IS dilution buffer containing 2% BSA, 0.5 mg/mL sheared salmon sperm DNA, and 0.5% dextran sulfate in 2X SSC buffer. The sections were incubated with 40–100 μl of IS at RT for 45 min protected from light. Unbound IS was removed by washing with 2X SCC (3 × 5 min). DAPI (Thermo Fisher Scientific) was applied to the stained tissue sections at 300 nM for 10 min at RT and then washed with 2X SSC buffer (2 × 5 min). The stained slides were mounted in Fluoromount G (Southern Biotech, Birmingham, AL) and coverslipped for imaging.
Thiol
Adapted from Goltsev, et al. [19]. after fixation, the tissue slides were washed with buffer 405 containing 10 mM Tris 7.5, 650 mM NaCl (2 × 5 min). The tissue was then incubated for 4 min at RT with 50 mM Tris(2-carboxyethyl)phosphine hydrochloride solution (TCEP, Sigma-Aldrich) diluted in buffer 405 and subsequently washed with buffer 405 (3 × 5 min). The sections were blocked with 100 mM iodoacetamide (Fisher Scientific) in buffer 405, adjusted to pH 8.0 with sodium hydroxide (NaOH), for 1 hour (h) at RT. Complementary IS labeled with AF647 was diluted in buffer 4 containing 10 mM Tris 7.5, 10 mM MgCl2, 150 mM NaCl. The tissue was covered with diluted IS solution for 45 min at RT protected from light before being washed with buffer 405 (3 × 5 min). The procedure for application of DAPI, washing, and coverslip mounting was the same as the previous group.
2.3. Fluorescence Microscopy, Image Visualization
Antibody stained slides were imaged on a Zeiss AxioImager.M2 with motorized XY scanning stage (Carl Zeiss AG, Oberkochen, Germany) equipped with a CoolSNAP HQ2 14-bit CCD camera (Photometrics, Tucson, AZ). Fluorescence images were collected using three channels for IS visualization. Depending on the fluorophore used for IS labeling, the following Zeiss filter sets were used: 38HE, 43HE, and 50. DAPI was imaged using Zeiss filter set 49. Images acquired with this system were collected using an X-Cite 120Q (Excelitas Technologies Corporation, Waltham, MA) light source at 20X (Plan-Apochromat, 0.95NA) magnification. Images acquired both pre-signal removal treatment (pre-Tx) and post-signal removal treatment (post-Tx) were of the same region of interest (ROI). Each marker had an optimal exposure time, used for pre-Tx and post-Tx image acquisition, set using the auto-expose tool in Zen software (Zeiss). Quantitative comparison of pre-Tx and post-Tx image fluorescence signal intensity was performed in ImageJ with the f llowing processing pipeline. Pre-Tx and post-Tx images of the same target were opened and the auto-contrast tool applied to the pre-Tx image to set a dynamic range which was then applied to the post-Tx image to normalize the fluorescent signal between the two images. The merge channels tool was then employed to merge the pre-Tx images and post-Tx images to their respective DAPI channel images that were acquired at the same time point as the target channel images.
2.4. Fluorescence Signal Removal
The methods employed for Ab-oligo fluorescent signal removal after imaging are detailed below:
Restriction Enzyme
The coverslip was removed and the mounting media was washed from the slide with 0.1X SSC (3 × 5 min). The restriction enzyme dilution was made fresh to concentrations optimal for each Ab-oligo conjugate ranging from 5U to 200U with 1X cut smart buffer acting as the diluent. The restriction enzyme and diluent were kept on ice throughout the process. The 0.1X SSC was removed, then the tissue was incubated with 1X cut smart buffer (2 × 5 min). The 1X cut smart buffer was removed and the restriction enzyme dilution was added to the tissue and incubated at 37 °C for 30 min. The incubation was followed by 3 quick washes with 1X cut smart buffer before restriction enzyme dilution was reapplied and allowed to incubate for an additional 30 min at 37 °C. The restriction enzyme was again washed quickly 3 times with 1X cut smart buffer before a coverslip was reapplied with Fluoromount G as the mounting media.
Thermal
The coverslip was removed and tissue slides immersed in 0.1X SSC buffer heated to 70 °C. The slides were incubated for 3 × 5 mins with the buffer being replaced after each incubation. A coverslip was applied to the slide with Fluoromount G mounting media. The tissue was imaged to confirm full fluorescent signal removal.
Photocleavable Linker
The tissue slides were exposed to ultraviolet (UV) light through the cover glass for 15 minutes on a UVGL-55 Handheld UV lamp (UVP, Upland, CA). After the UV treatment, the coverslips were removed and the slides immersed in 0.1X SSC for 5 min. The slides were then removed from the bath and 0.1X SSC pipetted 10x to remove any residual cleaved fluorophore. A coverslip was then mounted again with Fluoromount G mounting media for imaging to confirm signal removal.
Thiol
The coverslip was removed and the mounting media washed off the slide with 0.1X SSC (3 × 5 min). The tissue was then incubated for 2 min at RT with 50 mM TCEP in buffer 405 and then washed with buffer 405 (2 × 5 min). The tissue was then incubated with freshly made 100 mM iodoacetamide in buffer 405 for 1 min. The tissue sections were washed with 2X SSC (3 × 3 min) and a coverslip mounted with Fluoromount G mounting media. The tissue was then imaged again to verify complete signal removal.
3. RESULTS & DISCUSSION
3.1. Restriction enzyme (RE) signal removal
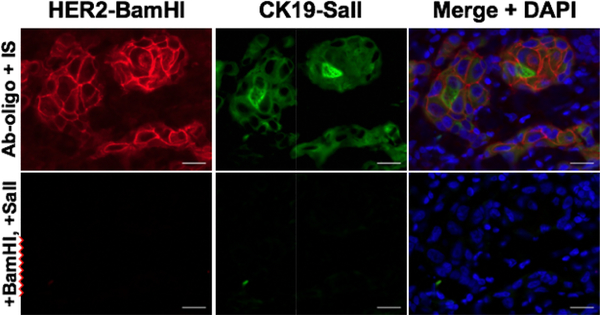
The incorporation of IS with engineered RE sites resulted in successful multiplexed Ab-oligo cycIF tissue labeling. Following confirmed antigen specific staining, signal removal facilitated by the use of RE specific to each Ab-oligo conjugate was performed. Imaging and image normalization succeeding RE signal removal confirmed reduction of fluorescent signal to autofluorescent levels (Fig. 2). The RE variant of the IS in the Ab-oligo platform enabled multiplexed in situ antigen labeling. There was no evidence of the incorporation of RE site causing loss of labeling functionality or specificity during the hybridization period between the DS and IS. To date, we have not observed tissue degradation after exposure to RE giving promise to this technique. More investigation into optimizing RE cleavage conditions is necessary because of the high concentration of RE necessary for signal removal of some conjugates. The costs of RE cleavage reagents as well as modifications to IS design weigh against the encouraging results reported here. Additionally, while the RE cleavage protocol is not laborious, it is time-consuming (~1 h) compared to other methods as discussed below.
Figure 2. RE signal removal.
Ab-oligo tissue labeling for HER2 and CK19 was performed with DS/IS pairs containing RE cleavage sites. Upon RE incubation, complete fluorescent signal was achieved.
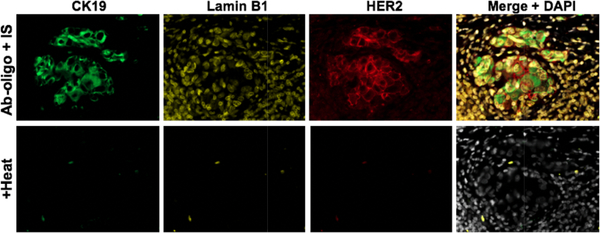
3.2. Thermal melting signal removal
Multiplexed Ab-oligo cycIF staining was performed to generate a three-target image with antigen specific labeling achieved. The observed staining patterns validated antigen specific labeling of Ab-oligo conjugates as CK19 and HER2 localize in similar tissue structures. Lamin B1 positively stained all nuclei as a major structural component found in all cells [20]. Exposure to optimal thermal melting conditions followed by imaging confirmed melting of all IS from their DS resulting in only autofluorescence following heating (Fig. 3). Utilization of the thermal melting permutation of Ab-oligo IF labeling does not require any expensive modifications to IS design. This technique is also simple and would be readily incorporated into a clinical histopathology workflow by employing equipment commonplace in histology laboratories. It is also important to emphasize thermal melting takes just 25% of the time required for RE cleavage. To date there have been no observations of loss of tissue antigenicity during cyclic staining. Further investigation into the upper tolerances of the tissue for exposure to heat at varied temperatures is necessary.
Figure 3. Thermal melting signal removal.
Ab-oligo conjugates for CK19, Lamin B1, and HER2 achieved specific tissue labeling. The samples underwent the thermal melting protocol, facilitating complete signal diminishment without any observed tissue damage.
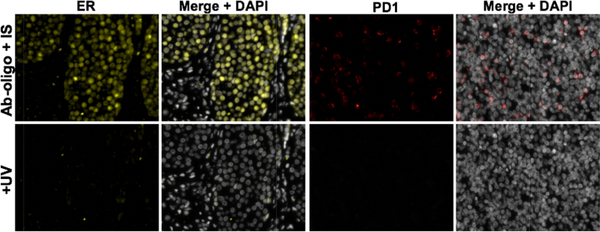
3.3. Photocleavable linker signal removal
Ab-oligo cycIF staining was performed in two separate tissues, one for estrogen receptor (ER) and one for immune marker PD-1. ER was labeled in ER positive BC tumor tissue and PD-1 was labeled in normal tonsil tissue with both demonstrating antigen specific tagging. There was no loss of functionality or specificity of the IS after engineering to include a PCL region. Under optimal PCL cleavage conditions, the functionality of UV light cleaving the PCL on the IS induced the release of fluorophores. The release of fluorophores was significant with resultant fluorescence levels being undetectable after image normalization (Fig. 4). As with RE cleaving and thermal melting, the incorporation of the PCL did not alter the standard Ab-oligo cycIF staining protocol, lending it to be integrated into the clinical lab setting. Similar to thermal melting, PCL cleavage was rapid requiring only 15 minutes. The potential for further optimization of PCL cleavage conditions could shorten the time required. PCL cleavage conditions, including buffers used and UV light exposure, are mild and to date have not caused observable damage to tissue integrity or antigenicity.
Figure 4. PCL cleavage facilitates signal removal.
Ab-oligo tissue labeling was performed for taER and PD1 on independent samples with each IS containing a PCL cleavage site. After imaging, UV light was used to induce cleavage at PCL sites and total signal removal achieved.
3.4. Thiol signal removal
The generic IS design was modified to include a disulfide region to perform Ab-oligo CycIF of HER2 in HER2 positive breast cancer (BC). This IS modification had no impact on antigen specific labeling. Under thiol removal conditions, with TCEP as the thiol of choice, fluorescent signal was completely removed (Fig. 5). It is also of note the TCEP and other buffers used in signal removal do not raise autofluorescence levels. The process by which a thiol is employed to cleave the disulfide bond containing IS was the shortest of all signal removal techniques studied herein. Initial testing does not suggest the thiol has any detrimental impact on tissue antigenicity, but more extensive testing would be for full evaluation. Thiol signal removal differs from the previously described techniques as it does require changes to the standard Ab-oligo cycIF labeling protocol. The modifications required to the protocol require less commonly found buffers that also must be made fresh before each staining. It also adds 1 h to the total staining time before initial imaging which is to the detriment of the potential of integrating such a technique into the clinical laboratory setting focused on both accuracy and efficiency to best serve clinicians and their patients.
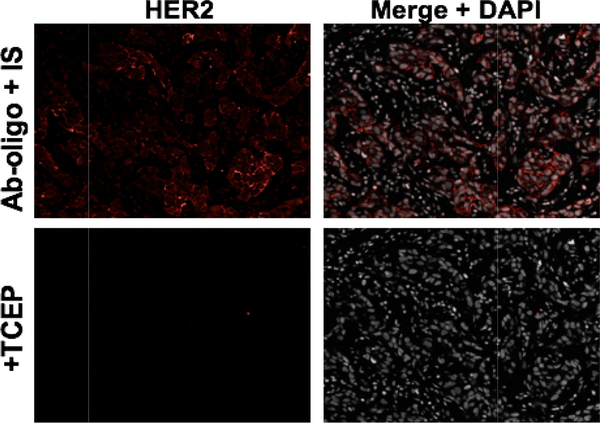
Figure 5. Thiol cleavage.
Ab-oligo HER2 antigen detection was achieved after adherence to the described thiol antibody staining procedure. Short incubation of TCEP induced cleavage of the disulfide bond sites on the IS, resulting in complete signal removal
4. CONCLUSION
Conventional methods of in situ IHC and IF staining of biomarker expression in tissue are limited to detecting of only 2–5 antigens in a single sample. This is insufficient to translate complex genomic analyses to pathological diagnosis. There have been many strategies proposed to increase the maximum antigens detected on a single sample, none of which are without drawbacks centered around their respective methods of signal removal. This has been a significant limiting factor in the maximum number of distinct antigens detected and adoption of such techniques into the clinical laboratory setting.
Ab-oligo cycIF is a technique capable of highly multiplexed immunolabeling and ready integration into the clinical workflow. Paired with Ab-oligo’s fluorescent biomarker labeling are a variety of robust signal removal strategies capable of complete signal removal between staining cycles. While robust, these signal erasing strategies do not require harsh conditions or reagents, ensuring tissue integrity is maintained for greater antigen detection. The generation of high dimensionality biomarker expression and spatial image data will further improve our understanding of tumor heterogeneity and in turn facilitate improved therapeutic strategies.
References
- [1].“Comprehensive molecular portraits of human breast tumors.”, Nature 490(7418), 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE and Børresen-Dale A-L, “Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications,” Proc. Natl. Acad. Sci. U. S. A 98(19), 10869–10874 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mezheyeuski A, Bergsland CH, Backman M, Djureinovic D, Sjöblom T, Bruun J and Micke P, “Multispectral imaging for quantitative and compartment-specific immune infiltrates reveals distinct immune profiles that classify lung cancer patients,” J. Pathol 244(4), 421–431 (2018). [DOI] [PubMed] [Google Scholar]
- [4].Galon J, Pagès F, Marincola FM, Angell HK, Thurin M, Lugli A, Zlobec I, Berger A, Bifulco C, Botti G, Tatangelo F, Britten CM, Kreiter S, Chouchane L, Delrio P, Arndt H, Asslaber M, Maio M, Masucci GV, et al. , “Cancer classification using the Immunoscore: a worldwide task force,” J. Transl. Med 10, 205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fridman WH, Pagès F, Sautès-Fridman C and Galon J, “The immune contexture in human tumours: impact on clinical outcome,” Nat. Rev. Cancer 12(4), 298–306 (2012). [DOI] [PubMed] [Google Scholar]
- [6].Stack EC, Wang C, Roman KA and Hoyt CC, “Multiplexed immunohistochemistry, imaging, and quantitation: A review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis,” Methods 70(1), 46–58 (2014). [DOI] [PubMed] [Google Scholar]
- [7].Tsurui H, Nishimura H, Hattori S, Hirose S, Okumura K and Shirai T, “Seven-color Fluorescence Imaging of Tissue Samples Based on Fourier Spectroscopy and Singular Value Decomposition,” J. Histochem. Cytochem 48(5), 653–662 (2000). [DOI] [PubMed] [Google Scholar]
- [8].Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, Can A, Corwin A, Dinn S, Filkins RJ, Hollman D, Kamath V, Kaanumalle S, Kenny K, Larsen M, Lazare M, Li Q, Lowes C, McCulloch CC, et al. , “Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue,” Proc. Natl. Acad. Sci. U. S. A 110(29), 11982–11987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lin J-R, Fallahi-Sichani M, Chen J-Y and Sorger PK, “Cyclic Immunofluorescence (CycIF), A Highly Multiplexed Method for Single-cell Imaging,” Curr. Protoc. Chem. Biol 8(4), 251–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zrazhevskiy P, True LD and Gao X, “Multicolor multicycle molecular profiling (M3P) with quantum dots for single-cell analysis,” Nat. Protoc. 8(10), 1852–1869 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].“Quantum dot imaging platform for single-cell molecular profiling | Nature Communications.”, <https://www.nature.com/articles/ncomms2635> (17 January 2019. ). [DOI] [PMC free article] [PubMed]
- [12].Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, Balter A, Kawashima R, Choe G, Sauer D, El Rassi E, Clayburgh DR, Kulesz-Martin MF, Lutz ER, Zheng L, Jaffee EM, Leyshock P, Margolin AA, Mori M, et al. , “Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis,” Cell Rep. 19(1), 203–217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Feng Z, Jensen SM, Messenheimer DJ, Farhad M, Neuberger M, Bifulco CB and Fox BA, “Multispectral Imaging of T and B Cells in Murine Spleen and Tumor,” J. Immunol. Author Choice 196(9), 3943–3950 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lin J-R, Fallahi-Sichani M and Sorger PK, “Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method,” Nat. Commun 6, 8390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rimm DL, “What brown cannot do for you,” Nat. Biotechnol 24, 914–916 (2006). [DOI] [PubMed] [Google Scholar]
- [16].“SIMPLE: A Sequential Immunoperoxidase Labeling and Erasing Method.”, <https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2746723/> (17 January 2019. ). [DOI] [PMC free article] [PubMed]
- [17].Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, Natkunam Y and Nolan GP, “Multiplexed ion beam imaging (MIBI) of human breast tumors,” Nat. Med 20(4), 436–442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schubert W, Dress A, Ruonala M, Krusche A, Hillert R, Gieseler A and Walden P, “Imaging cycler microscopy,” Proc. Natl. Acad. Sci. U. S. A 111(2), E215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, Black S and Nolan G, “Deep profiling of mouse splenic architecture with CODEX multiplexed imaging.,” bioRxiv (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shimi T, Butin-Israeli V, Adam SA, Hamanaka RB, Goldman AE, Lucas CA, Shumaker DK, Kosak ST, Chandel NS and Goldman RD, “The role of nuclear lamin B1 in cell proliferation and senescence,” Genes Dev. 25(24), 2579–2593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]