Abstract
Timely sister chromatid separation, promoted by separase, is essential for faithful chromosome segregation. Separase is a member of the CD clan of cysteine proteases, which also includes the pro-apoptotic enzymes known as caspases. We report a role for the C. elegans separase SEP-1, primarily known for its essential activity in cell division and cortical granule exocytosis, in developmentally programmed cell death when the predominant pro-apoptotic caspase CED-3 is compromised. Loss of SEP-1 results in extra surviving cells in a weak ced-3(-) mutant, and suppresses the embryonic lethality of a mutant defective for the apoptotic suppressor ced-9/Bcl-2 implicating SEP-1 in execution of apoptosis. We also report apparent non-apoptotic roles for CED-3 in promoting germ cell proliferation, meiotic chromosome disjunction, egg shell formation, and the normal rate of embryonic development. Moreover, loss of the soma-specific (CSP-3) and germline-specific (CSP-2) caspase inhibitors result in CED-3-dependent suppression of embryonic lethality and meiotic chromosome non-disjunction respectively, when separase function is compromised. Thus, while caspases and separases have evolved different substrate specificities associated with their specialized functions in apoptosis and cell division respectively, they appear to have retained the residual ability to participate in both processes, supporting the view that co-option of components in cell division may have led to the innovation of programmed cell suicide early in metazoan evolution.
Subject terms: Chromosome segregation, Cell proliferation
Introduction
Accurate segregation of chromosomes is essential for faithful transmission of the genome during somatic and germline mitotic proliferation and meiotic divisions associated with gametogenesis. Aneuploidy resulting from defective chromosome segregation can lead to a wide variety of genetic syndromes or embryonic lethality and is also associated with most malignant cells types, in some case conferring a growth advantage during cancer progression1. Prior to their segregation, chromosomes become aligned on the metaphase plate, where sister chromatids are held together until the onset of anaphase by the action of a ring-like structure, the cohesin complex, consisting of Scc1, Scc3, Smc1, and Smc32. Sister chromatid separation at the metaphase-to-anaphase transition is initiated when Scc1 (also called Mcd1 or Rad21) is cleaved by the enzyme separase3. Separase is activated at the metaphase-to-anaphase transition as a result of degradation of its inhibitors, securin and cyclin B, by a ubiquitin protein ligase, the “anaphase promoting complex” (APC). In C. elegans, a single separase, encoded by the sep-1 gene, functions during both meiosis and mitosis to promote sister chromatid separation4. In addition to its role in chromosome segregation, studies in C. elegans, Drosophila, and mammalian cells have revealed a role for separases in membrane trafficking5–9. Loss of separase activity in early C. elegans embryos also results in embryonic lethality owing to osmotic sensitivity that arises from defects in cortical granule exocytosis, as well as failure of cytokinesis, two processes that are separable from chromosome segregation8,10,11.
Separase is a member of the CD clan of cysteine proteases3,12. The proteases within this clan share conserved tertiary structures, arrangement of catalytic residues, and conserved motifs surrounding the catalytic residues, and apparently arose from a single evolutionary origin13 (Fig. 1A) The CD clan includes six distinct cysteine proteases, each that carries out unique cellular functions. These include the caspases, which are critical executors of apoptosis, or programmed cell death (PCD). When activated in cells destined to die, caspases cleave various cellular substrates, leading to the orderly dismantling of a dying cell. CED-3 is the predominant caspase in C. elegans responsible for nearly all of the 131 somatic and most of germline PCD during development. Three additional caspase-encoding genes encode six isoforms14,15, only one of which, CSP-1B, has been shown to possess proteolytic activity14,16. CSP-2 and CSP-3 instead act as negative regulators of CED-3 in the germline17 and soma18 respectively. In cells undergoing apoptosis, CED-3 is activated by trans-autoproteolysis through induced proximity mediated by the Apaf-1 homolog, CED-419–22.
Figure 1.
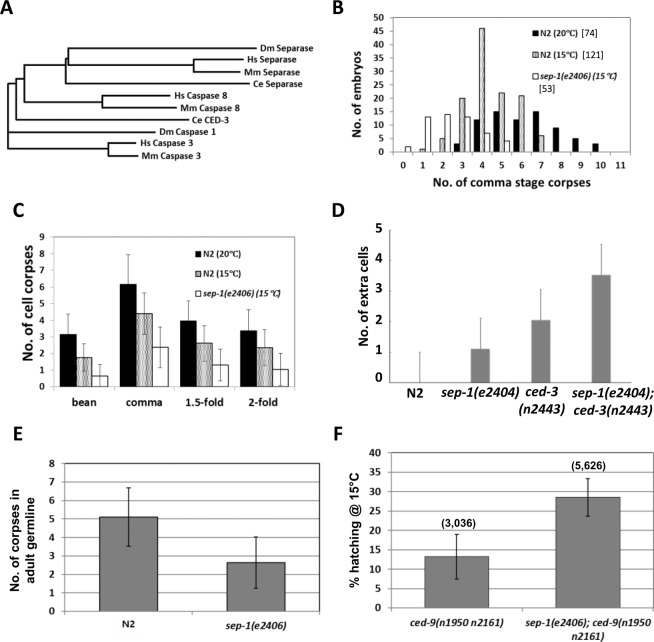
Pro-apoptotic action of separase in embryos and the germline. (A) Phylogenetic tree showing relationships of metazoan caspases and separases. Dm, Drosophila melanogaster; Hs, Homo sapiens; Mm, Mus musculus. Ce, C. elegans. (B) and (C) Subnormal numbers of cell corpses in sep-1(ts) embryos. In (B), cell corpses were scored at the “comma” stage at the indicated temperature. Total number of embryos scored is shown in brackets. (C) average number of cell corpses in N2 and sep-1(e2406) at the indicated stage and temperature. Error bars are ±SD. At least 16 embryos were scored for each stage. (D) Extra surviving cells in the anterior pharynx of sep-1(e2406ts) and ced-3(n2443), a weak allele of ced-3, L1 larvae. Mid-stage embryos were raised to 21 °C after completion of early divisions at the permissive temperature (15 °C) to prevent developmental arrest. (E) Decreased germ cell apoptosis in sep-1(e2406) mutants. Germ cell corpses were scored in both gonad arms of young adult N2 and sep-1(e2406) animals (50 worms were scored for each genotype) at 15 °C. (F) sep-1(e2406) suppresses ced-9(If) lethality. The hatching rate of embryos produced by sep-1(e2406); ced-9(n1950 n2161) animals and ced-9(n1950 n2161) animals are shown. Total number of embryos scored is shown above each bar. Values are expressed as mean ± SD.
Mounting evidence points to regulated caspase activation in mediating other process in addition to apoptosis, including cell proliferation and differentiation23–27. Some caspase targets are cell cycle regulators, including the negative regulators wee1 kinase, p27kip1, p21Waf1, the tumor suppressor Rb, and c-Abl, a kinase involved in cell cycle arrest28, and there is increasing evidence that proliferation and apoptosis are coupled29–31. Caspase activity is also required for T and B cell proliferation32,33. The apoptotic machinery in Drosophila is also involved in spermatogenesis and oogenesis34–37, and the Drosophila apical caspase dronc performs functions in cell migration38 and proliferation31. In C. elegans, the CED-3 caspase has been shown to cleave Dicer-1, a type III ribonuclease involved gene silencing processes by cleaving dsRNA39. CED-3 was also shown to function partially redundantly with the miRNA machinery to regulate gene expression in the course of normal development by cleaving, among other proteins, LIN-28, a key regulator of developmental timing40,41.
Here we present evidence that two CD clan proteases in C. elegans, the SEP-1 separase and the CED-3 caspase, perform shared roles in cell division and PCD. We found that sep-1 mutants contain subnormal numbers of cell corpses, enhance the PCD defect of weak ced-3 mutants, and partially rescue the embryonic lethality of mutants lacking the PCD inhibitor CED-9/BCL-2, suggesting an involvement in PCD. We further report that CED-3 is required for the normal rate of embryonic development as well as germ cell proliferation in the hermaphrodite germline. Moreover, loss of ced-3 function enhances the egg shell abnormalities, cell division defects, and meiotic chromosome non-disjunction phenotypes of a sep-1(-) mutant. Our results reveal unexpected promiscuity of clan CD cysteine proteases in the processes of cell division and PCD and raise the possibility that the machinery for PCD may have arisen from components originally required for cell division.
Results and Discussion
SEP-1 separase is required for developmental PCD in C. elegans
Caspases, best known for their roles in apoptosis, and separase, an essential mitotic regulator, are members of a clade of cysteine proteases, the CD clan (Fig. 1A), and are presumed to descend from a common ancient progenitor. Caspases and separase both cleave cohesins, albeit during the very distinct cellular events of chromosome separation and apoptosis3,28,42–44. We sought to investigate whether their potential functional overlap might be more than superficial by assessing whether SEP-1 separase is discernibly required for normal PCD. Loss of SEP-1 results in early embryonic lethality prior to onset of the developmental PCD as a result of osmotic sensitivity stemming from defects in eggshell formation as well as failure of cytokinesis and proper chromosome segregation4,8,10,45 To assess later roles of SEP-1, we took advantage of the temperature-sensitive sep-1(e2046) mutation, a missense allele (C450Y) that primarily affects vesicle trafficking and cortical granule exocytosis and that has minimal effect on chromosome segregation8. sep-1(e2046) animals show fully penetrant embryonic lethality at 22 °C, but are viable at 15 °C. We found that the sep-1(ts) mutant grown at permissive temperature showed a highly significant reduction in the number of apoptotic corpses (average of 2.4 ± 1.2 corpses; n = 53; p = 6.4 × 10−15) at the “comma” stage of embryogenesis compared to that in wild type (N2) embryos (average of 4.4 ± 1.2; n = 121; Fig. 1B). This decrease in the number of cell corpses does not appear to result from a delay in the execution of embryonic PCD, as we observed significantly fewer corpses throughout all late stages of sep-1(ts) embryos (Fig. 1C).
If the reduction in cell corpses reflects a bona fide block to PCD in some cells, it would be expected to result in supernumerary surviving cells, as is conveniently assayed by quantifying nuclei in the anterior pharynx46. Consistent with the decrease in cell corpses, we found, in comparison to N2, extra surviving nuclei in the anterior pharynx of sep-1(ts) L1 larvae (average of 1.1 ± 0.66 extra cells; n = 19; p = 8.42 × 10−7) resulting from mid-stage embryos that were shifted to 21 °C after completion of early divisions at the permissive temperature of 15 °C to prevent developmental arrest (Fig. 1D, Suppl. Figure 1). This value is similar to that seen in some weak ced-3(-) mutants47. In addition, we found that sep-1(e2046) grown at 21 °C significantly enhances the cell death defect of ced-3(n2443), a weak reduction-of-function allele, resulting in an average of 3.53 ± 0.91 extra nuclei in the anterior pharynx (n = 19; p = 2.86 × 10−6), compared to an average of 2.05 ± 0.71 in the ced-3(n2443) single mutant. Further supporting a role for SEP-1 in PCD, we found that the number of cell corpses in the adult germline is reduced approximately two-fold (2.6 ± 1.4 corpses; p = 6 × 10−11) in the sep-1(ts) mutant grown at permissive temperature of 15 °C compared to that in wild-type (Fig. 1E).
The observation that a sep-1 mutation even at permissive temperature appears to suppress the death of individual cells that are normally developmentally programmed to die raises the question of whether SEP-1 might be broadly involved in regulating PCD. The loss of CED-9/Bcl-2, a major apoptotic suppressor, results in maternal-effect lethality caused by massive ectopic PCD in embryos48. We found that the sep-1(e2046) mutation suppresses the maternal-effect lethality of ced-9(n1950 n2161) loss-of-function mutant: while 13.2 ± 5.8% (n = 3,036 total embryos from 39 worms) of ced-9(n1950 n2161) loss-of-function mutant embryos cultured at 15°C hatch, a significantly higher number of sep-1(e2046); ced-9(n1950 n2161) double mutant survive to hatching (28.6 ± 4.8%; n = 5,626 total embryos from 40 worms; p = 1.5 × 10−13)(Fig. 1F). Further, while the small number of hatching ced-9(-) mutants never developed past the early L1 stage, we found that approximately half of the arrested sep-1(-); ced-9(If) double mutant larvae grew to a larger size and appeared to have progressed further (late L1 or L2) before arresting.
While CED-3 is the predominant caspase in mediating apoptosis, these findings suggest that SEP-1 might also have a role in PCD. We note that these experiments were performed under conditions in which SEP-1 separase is able to execute its functions in cortical granule exocytosis, cytokinesis, and chromosome segregation. The early embryonic lethality in strongly affected sep-1(-) mutants precluded us from scoring PCD and extra cells in animals at the non-permissive temperature for sep-1(ts). Although the effects we have observed for the sep-1(-) are relatively mild, it is conceivable, therefore, that separase might actually play a larger role in the execution of PCD that was heretofore masked owing to its essential requirement in mitosis and early embryonic viability.
CED-3, but not PCD, is required for the normal rate of embryonic development
Given that separase and apoptotic caspases belong to same clade, the CD clan cysteine proteases, coupled with the evidence presented above that separase may possess pro-apoptotic activity, it was of interest to ask whether the predominant pro-apoptotic caspase in C. elegans, CED-3, might function in essential non-apoptotic roles performed by separase. The loss of ced-3 caspase function eliminates nearly all PCD in C. elegans;15 however, it does not lead to any debilitating or other overt phenotypes. This finding originally prompted the view that CED-3 caspase functions primarily or exclusively in PCD. While CED-3 is not essential for viability or fertility in C. elegans, we found that the ced-3(n717) mutation results in substantially slower embryonic development (Fig. 2A), and hence an increased time to hatching by ~20% or ~120 min, compared to wildtype N2 (p = 0.003), suggestive of a broader role for CED-3. To rule out specific effects of the ced-3(n717) mutant background on developmental rate, we analyzed six additional alleles of ced-3, of which three (n718, n2454 and n2442) show a strong defect in PCD, and the remainder (n1040, n2877 and n2921), a more moderate defect in PCD. Irrespective of their effect on PCD, all six alleles cause a delay in embryonic development similar to that seen in ced-3(n717) mutants (Fig. 2A). This slower developmental rate is not attributable to the absence of PCD, as a mutation in ced-4, the gene encoding C. elegans pro-apoptotic Apaf149, which also abolishes PCD, had no discernible effect on the rate of embryogenesis. These observations suggest that ced-3 performs functions that are important in normal progression through embryogenesis distinct from its function in activation of PCD.
Figure 2.
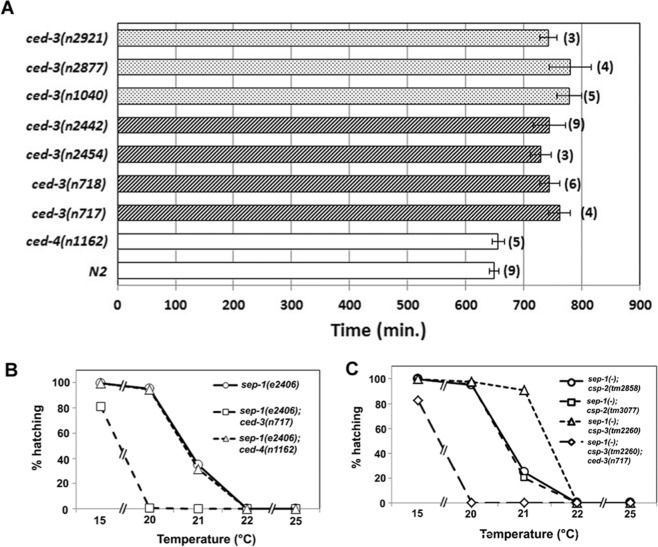
Delayed embryonic development in ced-3 mutants. (A) Slowed embryonic development in ced-3 mutants. Embryos of the indicated genotype were allowed to develop at 22.5 ± 0.5 °C and the time from 2-cell to hatching was measured. Stippled bars: ced-3 alleles that show a moderate defect in PCD (33). Striped bars: ced-3 alleles that show a strong defect in PCD. (B) Embryonic survival. Embryos of the indicated genotype and temperature were scored for hatching as the measure of embryonic viability. Each data point was obtained from >154 embryos. (C) Partial suppression of sep-1(-) embryonic lethality by csp-3(-). Embryos of the indicated genotype were scored as in A. Each data point was obtained from>1000 embryos.
Vital embryonic role of CED-3 is revealed when SEP-1 function is reduced
Mutations in sep-1 affect egg shell formation, cause defects in cytokinesis and chromosome separation, and block mitosis; in all cases leading to embryonic lethality4,6,8,10,11,50. We found that ced-3(-) mutations greatly enhance the embryonic lethality of sep-1(ts) mutants grown at a fully permissive or semi-permissive temperature range of 15–22°C (Fig. 2B). This effect was most striking at an intermediate temperature of 20 °C: while sep-1(ts) single-mutant embryos nearly always (~95%; n = 2,564) survive, those also lacking CED-3 function invariably die (<1% viable; n = 733). Given that sep-1(e2406) is a phenotypically complicated allele, we also compromised SEP-1 function by RNAi using HT115 E. coli strain expressing dsRNA targeting sep-1 from the Ahringer RNAi strain collection. RNAi of sep-1 in wild type (N2) background results in high lethality. While the embryonic lethality progressively decreases with increasing dilution in N2 background it remains higher in the ced-3 mutant background (Figure S1A). In addition to CED-3, the CSP-1B caspase, one of three isoforms encoded by csp-1, has been implicated in regulating a subset of embryonic cells destined to die16. We found that knockdown of csp-1 by RNAi had no effect on viability of sep-1(ts) mutant embryos (not shown); hence this effect is not general to caspases. More importantly, the greatly enhanced lethality is not related to abrogation of PCD per se, as a ced-4(-) mutation that eliminates virtually all PCD showed no enhancement of lethality in the sep-1(ts) mutant (Fig. 2B).
A non-catalytic caspase homolog, CSP-3, functions to buffer caspase activity in living cells by inhibiting CED-3 in the soma, raising the possibility that its removal might enhance the ability of CED-3 to provide separase-like activity in the absence of SEP-1. Indeed, we found that removal of CSP-3, the soma-specific caspase inhibitor18, suppresses the high embryonic lethality seen with the sep-1(ts) mutant grown at a semi-permissive temperature of 21°C from 65% to 9% lethality (p < 10−4) (Fig. 2C). We found that this strong suppression of lethality by csp-3(-) requires CED-3 function: the sep-1(ts); csp-3(-); ced-3(-) triple mutant shows 100% lethality at 20°C, a temperature that we found is nearly fully permissive for the sep-1(ts) mutation alone (Fig. 2C). In contrast, loss of the germline-specific CED-3 inhibitor CSP-217 does not suppress this lethality (Fig. 2C), as expected, since it does not function in the soma.
Our results showing that sep-1(e2406) shows low/impenetrant lethality at 20°C differs from earlier published studies in which it was reported that sep-1(e2406) carried in strain WH216 showed fully penetrant embryonic lethality at 20°C. In contrast to published findings, we found that embryos from fertile WH216-derived sep-1(e2406) homozygotes grown at 20 °C (in which temperature was closely monitored with three thermometer, <0.2°C variance), showed substantial viability (72 ± 8.1% lethality; n = 222), albeit lower than in our 5x backcrossed strain. In addition, we observed low lethality (5 ± 2.7%, n = 2444) in an independently constructed strain (JR3388 sep-1(e2406); ruIs32) at 20°C, as well as in three other independent strains in which lethality would not be expected to be suppressed (Fig. 2B,C). Thus, given the high survival rates we observed at 20°C in five independent sep-1(e2406) strains, our findings suggest that enhanced viability of the sep-1(e2406); csp-3(tm2260) at 21 °C is not likely the result of a background suppressor. The very sharp transition from permissive to non-permissive conditions for the sep-1(e2406) allele (see below) may account for exquisite sensitivity to genetic background at the transitional temperature of 20°C. We cannot rule out the presence of a tightly linked suppressor in our strains or, alternatively, a sep-1(e2406) enhancer in the WH216 background. Nonetheless, our tightly controlled comparisons strongly suggest that sep-1(e2406) lethality is enhanced by ced-3(-) and suppressed by csp-3(-).
Synergistic requirement for CED-3 and SEP-1 in osmotic integrity and chromosome segregation
The apparently shared requirement for CED-3 and SEP-1 in PCD and viability raises the possibility that CED-3 might also affect chromosome segregation when separase activity is attenuated. To follow chromosome separation in living embryos, we observed fluorescently tagged chromosomes during cleavage of the zygote (Fig. 3). The sep-1(ts) strain shows defects in vesicle trafficking and exocytosis that result in cytokinetic failure, impaired egg shell resulting in osmotic sensitivity, and chromosome segregation and centriole disengagement, defects which are separable from each other8–10,50. To ameliorate effects on osmotic sensitivity we analyzed embryos in osmotically balanced egg salt buffer. We found that nearly all mutant embryos carrying a temperature-sensitive mutation in sep-1 progress beyond the first division when incubated at 20°C. Under these conditions, the average time required for the transition from maximum alignment of metaphase chromosomes to initiation of chromosome segregation in wild-type was ~101 ± 14 sec (n = 20; Fig. 3A,B). While chromosome separation also succeeded in nearly all (95%; n = 21) sep-1(e2406ts) mutants at this temperature, the average time from maximum metaphase alignment to initiation of separation increased significantly, by nearly 2-fold, to ~3 min (p = 3.2 × 10−7) (Fig. 3A,B). As expected, chromosome separation and cytokinesis were always successful in ced-3(-) single mutants and the time for this event was virtually indistinguishable from that in wild-type (105 ± 18 sec; n = 20; Fig. 3B). In contrast, we found that nearly half (43%; n = 31) of sep-1(ts); ced-3(-) double mutant zygotes completely failed to undergo chromosome separation or cytokinesis (Fig. 3A). In the profoundly defective embryos, no prominent pseudocleavage furrow was observed and the embryo filled the eggshell. This suggests that the osmotic integrity of the eggshell in the double mutant was compromised under the same conditions in which the sep-1(e2406ts) single mutant embryos were able to complete zygotic cell division. We also found that chromatin condensation was affected, multipolar chromatin bridges were observed, and chromosomes remained in the center of the cell indefinitely (>40 min).
Figure 3.
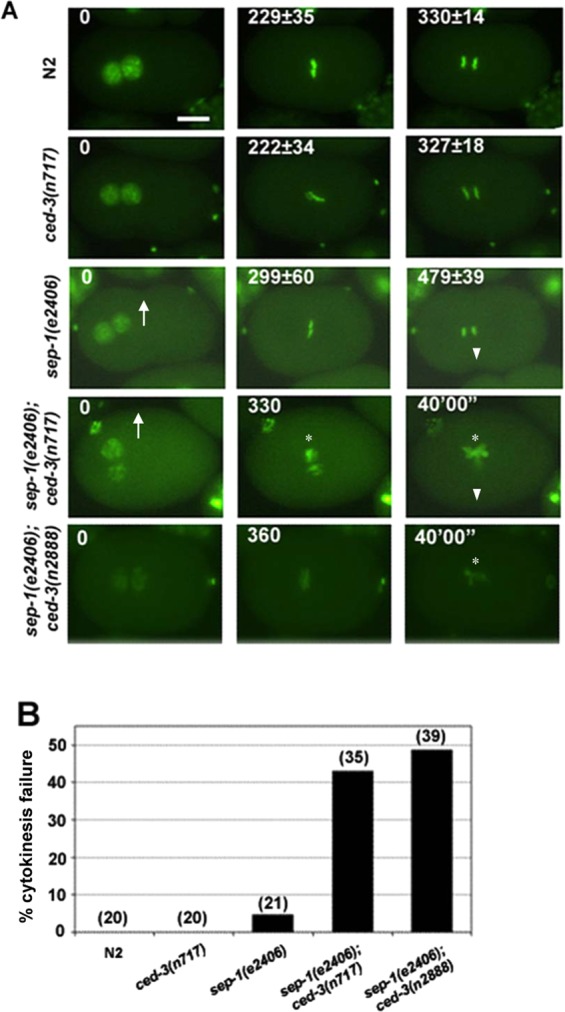
CED-3 enhances early cell division defects in separase mutants. (A) Progression of GFP-labeled mitotic chromosomes during zygotic cleavage. One-cell embryos of the indicated genotype, carrying the HIS-2B::GFP marker, are shown at pronuclear meeting (defined as t = 0; left panels), maximum metaphase alignment (middle panels), and immediately following separation of chromosomes (right panels, with the exception of the sep-1(e2406); ced-3(n717) embryo, which failed in chromosome separation). Embryos were dissected from young adults that had been pre-incubated at 21 °C for 90 mins and allowed to undergo cleavage at the same temperature. The time following pronuclear meeting is indicated in seconds, with the standard deviation reported for all embryos (see B) that underwent chromosome segregation with the exception of the last sep-1(-); ced-3(-) panel, in which time in indicated in minutes. Scale bar, 10 µm. The arrow and arrowhead indicate pseudocleavage and cleavage furrows respectively which are absent in the double mutant embryos. The asterisks show the chromatin condensation and separation defects in the last two panels of the double mutant embryos. (B) Average time for chromosome segregation in all embryos of the indicated genotype (n = 20 per genotype) in which separation of chromosomes was successful. The time from metaphase alignment to the start of chromosome segregation, with standard deviation, is indicated. For all experiments, embryos were imaged every 30 sec with a 500 ms exposure time.
Given that the sep-1; ced-3 mutant embryos are extremely sensitive to the osmolarity of the mounting medium, and to ensure the different mutant strains were subjected to identical environmental conditions, we also imaged early embryonic divisions of the sep-1(-) and ced-3(-) single and double mutants in utero on the same slide at ~19.5 °C. While, sep-1 and ced-3 single mutant embryos were able to complete meiotic and zygotic divisions (Suppl. movie 1 and 2), we observed that sep-1; ced-3 double mutant embryos were severely deformed and elongated. Some embryos exhibited defects in chromosome segregation with a large mass of DNA in the center and multipolar chromatin bridges (Suppl. movie 3). To assess whether the egg shell defects were separable from the chromosome segregation defects, we imaged the double mutant strain at a slightly lower temperature of ~18.2 °C (Suppl. movie 4). Under these conditions both the egg shell and chromosome separation appeared normal without any of the defects observed at a temperature that is ~1°C warmer. The profound enhancement of the chromosome segregation defects is not attributable to the block in germline PCD in the ced-3(-) mothers of the embryos per se, as mutations in ced-4, which also block virtually all PCD, do not enhance, and in some cases actually suppress, chromosome segregation phenotypes associated with loss of sep-1 function (see below). These findings implicate a partially redundant role for the pro-apoptotic caspase CED-3 in conjunction with separase during eggshell formation, cytokinesis, and chromosome segregation.
Requirement for CED-3 activity in germ cell proliferation
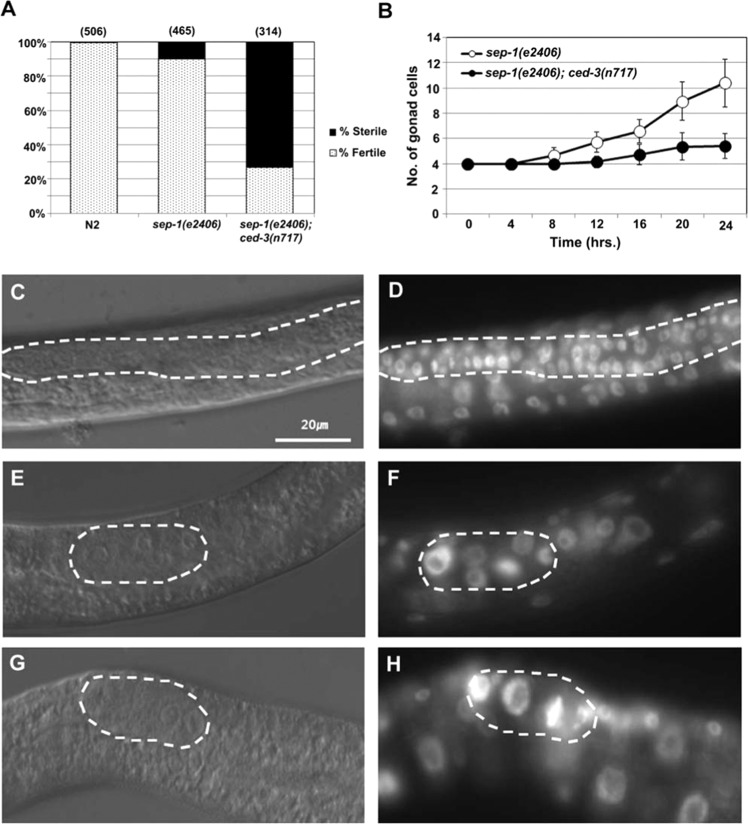
The mechanisms that regulate proliferation in the C. elegans germline are distinct from those acting in the embryo51,52. We therefore asked whether the requirement for CED-3 extends to germ cell proliferation. sep-1(e2406ts) mutants show a low level of sterility at a permissive temperature of 15°C (9.9%; n = 465). We found that this sterility is dramatically enhanced (to 72.9%; n = 314) by simultaneous removal of ced-3 activity (Fig. 4A). Examination of the sterile adult animals showed that they contained underproliferated germlines and frequent abnormal oocytes (not shown). Analysis of post-embryonic germline development in synchronized worms cultured at a non-permissive temperature of 25 °C starting immediately after hatching revealed that the block in germ cell proliferation resulting from loss of SEP-1 is greatly exacerbated by removal of CED-3 (Fig. 4). At hatching, the gonad primordium of wild-type worms contains 4 nuclei, which increases to an average of 44.2 ± 3.6 (n = 30) nuclei per gonad after 24 hours, (Fig. 4B-D). Under these conditions, sep-1(ts) animals contained an average of 10.4 nuclei per gonad (n = 50 animals), all of which were somewhat larger and stained more intensely with DAPI, revealing that removal of SEP-1 function strongly blocks cell proliferation in the gonad (Fig. 4B,E,F). We found that elimination of CED-3 function in the sep-1(-) mutant virtually completely blocks all proliferation of the initial 4 gonadal cells (the two somatic gonad precursor cells as well as the germline precursor cells) at hatching, with an average of only 5.4 nuclei/gonad (n = 50) (Fig. 4B,G,H). The very strong DAPI staining and increased size of the chromosome aggregates seen in sep-1(ts); ced-3(-) double mutants suggest that gonadal nuclei have apparently continued to undergo multiple rounds of DNA replication without karyokinesis (Fig. 4H).
Figure 4.
Synergy of CED-3 and SEP-1 in gonadal cell proliferation. (A) % sterility of adults grown at 15 °C. (B) Quantitation of gonadal (both somatic and germline) nuclei in larvae of the indicated genotype. Synchronized L1 worms (n = 50 for each data point) were grown at 25°C, and samples collected every 4 hrs. Larvae were fixed in Carnoy’s solution, stained with DAPI, and gonadal cells counted. (C), (E) and (G), DIC images of N2, sep-1(e2406) and sep-1(e2406); ced-3(n717), respectively, 24 hours after initiation of feeding of synchronized L1 larvae grown at 25oC. (D), (F) and (H), corresponding DAPI images of the same animals.
CED-3 is required for normal X-chromosome segregation in the germline
In C. elegans, sex is determined by the number of X-chromosomes relative to the number of autosomes, such that XX embryos develop into hermaphrodites and XO embryos become males53. A number of mutants that show X-chromosome nondisjunction result in a high incidence of males (Him) phenotype54. We found that sep-1(ts) single mutant animals grown at the permissive temperature exhibit a nearly nine-fold increased rate of male production (1.1%; p < 10−4) compared to wild-type, consistent with the role of SEP-1 in chromosome segregation. Similarly, we found that two different alleles of ced-3 result in a statistically significant (p < 0.01) increase in male production at 15 °C by as much as four-fold (0.48% males) compared to the wildtype (0.12%) (Table 1), implicating the CED-3 caspase in proper chromosome disjunction during meiosis. Further, we found that eliminating both ced-3 and sep-1 function together results in a strongly synergistic effect: sep-1(ts); ced-3(-) animals show a > 50-fold enhancement (6.2%; p < 10−4) in the rate of male production over wild-type. These results suggest that CED-3 and SEP-1 might cooperate in meiotic chromosome segregation.
Table 1.
Quantitation of X-chromosome non-disjunction.
| Genotype | % male ± SD (N) | p |
|---|---|---|
| N2 | 0.12 ± 0.04 (3433) | |
| sep-1(e2406) | 1.10 ± 0.08 (4267) | <0.0001a |
| ced-3(n717) | 0.48 ± 0.15 (4356) | <0.0001a |
| sep-1(e2406); ced-3(n717) | 6.15 ± 0.27 (3384) | <0.0001b |
| ced-3(2454) | 0.41 ± 0.02 (4115) | <0.01a |
| sep-1(e2406); ced-3(n2454) | 3.53 ± 0.20 (2949) | <0.0001b |
| ced-4(n1162) | 0.03 ± 0.03 (5799) | 0.14a |
| sep-1(e2406); ced-4(n1162) | 0.07 ± 0.02 (4323) | <0.0001b |
| sep-1(e2406); ced-4(n1162); ced-3(n717) | 2.18 ± 0.52 (2022) | <0.0001c |
| csp-2(tm2858) | 0.03 ± 0.03 (11930) | 0.06a |
| sep-1(e2406); csp-2(tm2858) | 0.06 ± 0.02 (10829) | <0.0001b |
| csp-2(tm2858) ced-3(n717) | 0.37 ± 0.08 (46185) | <0.0001d |
| csp-3(tm2260) | 0.28 ± 0.07 (33651) | 0.105a |
| sep-1(e2406) csp-3(tm2660) | 1.05 ± 0.10 (10558) | 0.862b |
| csp-3(tm2260); ced-3(n717) | 0.54 ± 0.03 (45007) |
All strains were maintained at 15 °C. Number of animals scored is indicated in parentheses. p value is based on Chi- square test. The genotypes compared for independence are as follows: a - comparison against against N2, b - comparison against sep-1(e2406), c - compared against sep-1(e2406); ced-4(n1162); d – compared against csp-2(tm2858).
Just as CSP-3 functions to buffer CED-3 pro-apoptotic activity in the soma, CSP-2 performs an analogous role in the germline. We found that removal of CSP-2, but not CSP-3, suppresses the Him phenotype of sep-1(-) mutants (0.06% versus 1.1% male production; p < 10−4; Table 1), consistent with an increase in CED-3 activities that might compensate for the lack of SEP-1. In contrast, the increased production of males in ced-3(-) single mutants is not significantly suppressed by elimination of CSP-2, as seen in the csp-2(-); ced-3(-) double mutant (Table 1), implying that CSP-2 functions through CED-3 in this process. Further, elimination of CSP-2 appears to decrease the naturally observed Him phenotype compared to that of wild-type (0.03% vs. 0.12%; p = 0.06), suggesting that elevated CED-3 levels resulting from the absence of CSP-2 may be sufficient to diminish the natural X non-disjunction seen in wild-type animals containing normal CED-3 and SEP-1.
In sharp contrast, a ced-4(-) mutation, which blocks PCD as effectively as a strong ced-3(-) mutation, has no effect on, or perhaps even slightly reduces chromosome non-disjunction compared to wild-type (0.03%; p = 0.14). Further, the increased X chromosome non-disjunction seen in the sep-1(ts) mutant is suppressed by the ced-4 mutation (0.07%; p < 10−4 for sep-1(ts) vs. sep-1(ts); ced-4(-)); i.e., removal of CED-4 counteracts the effect of debilitating separase function. CED-4/Apaf1 functions in apoptosis by binding to and directing CED-3 to the apoptotic pathway, a process that is very active in the germline55. We posit that removal of CED-4 may result in elevated levels of uncomplexed CED-3 that becomes available to participate in chromosome separation. Supporting this notion, we found that the suppression of meiotic non-disjunction by ced-4(-) is eliminated by removal of ced-3(+) activity (compare sep-1(e2406); ced-4(n1162) to sep-1(e2406); ced-4(n1162); ced-3(n717); p < 10−4; Table 1). Collectively, these results implicate the CED-3 caspase in meiotic chromosome segregation and suggest that CSP-2 and CED-4 antagonize its action in this process.
Discussion
This study and other recent reports reveal multiple functions of proteins that regulate cell death and also perform other vital functions in surviving cells24,27,56,57. Pro-apoptotic CED-4/Apaf1 has been reported to mediate DNA-damage-induced cell-cycle arrest at S phase27. CED-4 has also been shown to be required for regulation of cell size24. Caspases have been implicated in multiple cellular processes not related to cell death, including terminal differentiation, activation, proliferation, and cytoprotection56. Caspase-2 was also found to be involved in maintenance of the G2/M DNA damage checkpoint and DNA repair57. We note that caspase-2 is the only caspase that is constitutively present in the cell nucleus and is closely related to the C. elegans CED-347. In the worm, a consequence of abrogation of programmed germ cell death by ced-3 has been a reduction in quality of oocytes in aging worms due to a reduction in allocation of resources to developing oocytes58. CED-3 proteolytic activity, in conjunction with the miRNA silencing machinery40 and the N-end rule degradation machinery41, plays a role in regulating the robust expression of many genes during development. Dicer, a type-III ribonuclease is converted into a pro-apoptotic DNAase by CED-339 and consequentially might affect gene silencing by non-coding RNAs.
Like caspases, separase is predicted to have multiple cleavage substrates based on a consensus cleavage sequence43,59. Separase has been reported to cleave a kinetochore/spindle protein called Slk1960. Some of the potential separase substrates do not have mitotic functions and may provide clue to its role in other processes. Multiple studies have raised the possibility of a role for separase in apoptosis-related functions. Separase has been reported to cleave the yeast Scc1 to amplify of apoptotic signal initiated by H2O261. In another study, separase inhibition was found to be required for the survival of murine embryonic cells62,63. A constitutively active separase mutant leads to apoptotic cell death during embryonic development62,63. These results, coupled with our findings, suggest that separase may play a caspase-like executioner role and cleave multiple protein substrates during apoptosis.
The interplay between the cell death machinery and essential cellular activities is consistent with the notion that some of the molecular components regulating normal cellular processes may have been recruited into the complex apparatus that orchestrates the demise of cells64. The action of separase in mitosis is likely to be among its most ancient functions, as essentially all eukaryotes require this enzyme to direct chromosome segregation at mitosis2,4. Apoptosis was a major innovation associated with the emergence of multicellularity in metazoans64,65. The essential role of a cysteine protease that is required for cell division in developmentally programmed cell death supports the view that the apoptotic machinery arose as a consequence of co-option of basic cellular machinery involved in mitosis. Such co-option of essential cellular proteases as cellular executioners would presumably have been accompanied by the coevolution of buffering mechanisms that allow fine-tuning of enzymatic activity required to mediate essential cellular functions. Consistent with such a possibility, abrogation of buffering by CSP-2 and CSP-3 might allow for increased basal activity of CED-3 associated with its non-apoptotic functions in the germline and soma respectively, thereby suppressing the effects of reduced separase activity. Discovering the contribution of essential genes to PCD is complicated by the fact that their depletion often leads to lethality. The Janus-like behavior of SEP-1 might be representative of many other unexplored apoptotic functions of the essential components in the cellular toolkit.
The tight regulation of cell proliferation and cell survival is of fundamental importance to normal development and homeostasis. The notion of a connection between cell death and proliferation derives in part from evidence that apoptosis is a consequence of dysregulation of the cell-cycle machinery29. Disrupting or bypassing cell-cycle checkpoints frequently leads to increased apoptotic potential instead of increased proliferation. The involvement of common effector molecules CED-3 and SEP-1 at the interface between mitosis and apoptosis points to a further link between these two key cellular processes and provides a framework for understanding the complex signaling pathways that coordinate cell proliferation and cell death. These findings also raise a note of caution with the design of anti-tumor drug therapies targeted to separase activity; as we have observed, inhibition of this enzyme with the goal of blocking its mitosis-directing function might simultaneously prevent the major anti-tumor defensive mechanism, apoptosis, thereby potentially confounding such a strategy.
Materials and methods
Strains and culturing
C. elegans was cultured at 20°C by standard procedures66, unless otherwise noted. Temperature-sensitive strains were maintained at 15 °C. Strain N2 Bristol variety was used as the wild-type. sep-1(e2406) was isolated from strain WH216 sep-1(e2406)/hT2[bli-4(e937) let-?(q782) qIs48] (I;III), obtained from the Caenorhabditis Genetics Center (CGC), by backcrossing five times to generate JR3387. AZ212 unc-119(ed3);ruIs32[unc-119(+) pie-1::GFP::H2B] was obtained from the CGC and used to construct JR3304 ruIs32; ced-3(n717), JR3388 sep-1(e2406); ruIs32, JR3389 sep-1(e2406); ruIs32; ced-3 (n717) by standard methods. csp-2(tm2858), csp-2(tm3077), csp-3(tm2260), and csp-3(tm2486);csp-2(tm3077) were gifts from Dr. Ding Xue17,18. The various ced-3 alleles were a gift from Dr. H. Robert Horvitz47.
Time-lapse analysis of chromosome segregation
sep-1(e2406) and sep-1(e2406); ced-3(n717) embryos expressing GFP-tagged histone 2B (ruls32) were obtained from adult worms that had been pre-incubated at 21 °C for 90 min. Early one-cell embryos collected from dissected worms were mounted on agarose pads in egg salts buffer. Fluorescence time-lapse images were acquired at 24 ± 1 °C using a Nikon Eclipse Ti microscope controlled by NIS Elements AR software. Specimens were illuminated with an X-cite light source attenuated to 20% using a GFP filter (480/40 bandpass excitation filter). All images were obtained with a Hamamatsu CMOS sensor using a 20X objective. Images were acquired every 30 sec. by scanning through five to eight focal planes per time point.
Live in utero imaging
sep-1(e2406) and sep-1(e2406); ced-3(n717) and ced-3((n717) worms expressing GFP-tagged histone 2B (ruls32) were imaged simultaneously on the same slide to ensure that all three strains were subject to the same environmental conditions. Worms were picked at the mid-L4 stage and grown at 15 °C overnight. A 4% agarose pad was cut into three sections and gravid adult worms picked into a drop of Meiosis media (0.6 × Leibowitz-15 media, 0.2 × heat-inactivated fetal bovine serum, 25 mM HEPES pH 7.4, 5 mg/ml inulin)67 supplemented with 1 mM levamisole to immobilize the worms. The worms were imaged at 19.5 ± 0.2 °C or at 18.2 ± 0.2 °C. Temperature on the microscope stage was controlled using a Peltier thermocontroller (Brook Industries). Fluorescence time-lapse images were acquired every minute as described above.
Supplementary information
Acknowledgements
We thank S. Shaham for the csp-2 and -3 mutants, H. R. Horvitz for the ced-3 alleles, and S. Martin for the pGEX-CED-3 expression vector. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). This work was supported by grants from the NIH (#HD081266 and #HD082347) to J.H.R.
Author contributions
P.Y.J. was involved in the design of all experiments and obtained and analyzed the data presented in the manuscript. A.K. performed initial experiments implicating a role for SEP-1 in PCD. P.J. was involved in the concept, experimental design, data acquisition, data analysis, writing and revision of the manuscript. J.H.R. directed the project and was involved in the concept, experimental design, and data analysis of the project and writing of the manuscript.
Data availability
Strains reported in this manuscript will be made readily available upon request
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-63081-w.
References
- 1.Gordon, D. J., Resio, B. & Pellman, D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet (2012). doi:nrg3123 [pii]10.1038/nrg3123 [DOI] [PubMed]
- 2.Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science (80-.). 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- 3.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/S0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 4.Siomos MF, et al. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr. Biol. 2001;11:1825–1835. doi: 10.1016/S0960-9822(01)00588-7. [DOI] [PubMed] [Google Scholar]
- 5.Bard F, et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- 6.Richie CT, et al. Protein phosphatase 5 is a negative regulator of separase function during cortical granule exocytosis in C. elegans. J Cell Sci. 2011;124:2903–2913. doi: 10.1242/jcs.073379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bacac M, et al. Securin and separase modulate membrane traffic by affecting endosomal acidification. Traffic. 2011;12:615–626. doi: 10.1111/j.1600-0854.2011.01169.x. [DOI] [PubMed] [Google Scholar]
- 8.Bembenek JN, et al. Cortical granule exocytosis in C. elegans is regulated by cell cycle components including separase. Development. 2007;134:3837–3848. doi: 10.1242/dev.011361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schvarzstein M, Pattabiraman D, Bembenek JN, Villeneuve AM. Meiotic HORMA domain proteins prevent untimely centriole disengagement during Caenorhabditis elegans spermatocyte meiosis. Proc Natl Acad Sci USA. 2013;110:E898–907. doi: 10.1073/pnas.1213888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bembenek JN, White JG, Zheng Y. A Role for Separase in the Regulation of RAB-11-Positive Vesicles at the Cleavage Furrow and Midbody. Curr. Biol. 2010;20:259–264. doi: 10.1016/j.cub.2009.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai, X. & Bembenek, J. N. Protease dead separase inhibits chromosome segregation and RAB-11 vesicle trafficking. Cell Cycle (2017). doi:10.1080/15384101.2017.1363936 [DOI] [PMC free article] [PubMed]
- 12.Mottram JC, Helms MJ, Coombs GH, Sajid M. Clan CD cysteine peptidases of parasitic protozoa. Trends Parasitol. 2003;19:182–187. doi: 10.1016/S1471-4922(03)00038-2. [DOI] [PubMed] [Google Scholar]
- 13.Barrett AJ, Rawlings ND. Evolutionary lines of cysteine peptidases. Biol Chem. 2001;382:727–733. doi: 10.1515/bchm.2001.382.5.727. [DOI] [PubMed] [Google Scholar]
- 14.Shaham, S. & Shaham S. Identification of multiple Caenorhabditis elegans caspases and their potential roles in proteolytic cascades. 273, 35109–35117 (1998). [DOI] [PubMed]
- 15.Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 16.Denning DP, Hatch V, Horvitz HR. Both the caspase CSP-1 and a caspase-independent pathway promote programmed cell death in parallel to the canonical pathway for apoptosis in Caenorhabditis elegans. PLoS Genet. 2013;9:e1003341. doi: 10.1371/journal.pgen.1003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng X, et al. Caenorhabditis elegans caspase homolog CSP-2 inhibits CED-3 autoactivation and apoptosis in germ cells. Cell Death Differ. 2009;16:1385–1394. doi: 10.1038/cdd.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geng X, et al. Inhibition of CED-3 zymogen activation and apoptosis in Caenorhabditis elegans by caspase homolog CSP-3. Nat Struct Mol Biol. 2008;15:1094–1101. doi: 10.1038/nsmb.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conradt, B., Wu, Y. C. & Xue, D. Programmed cell death during Caenorhabditis elegans development. Genetics 203, (2016). [DOI] [PMC free article] [PubMed]
- 20.Qi S, et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 21.Huang, W. et al. Mechanistic insights into CED-4-mediated activation of CED-3. Genes Dev. (2013). 10.1101/gad.224428.113 [DOI] [PMC free article] [PubMed]
- 22.Yan, N. et al. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature (2005). 10.1038/nature04002 [DOI] [PubMed]
- 23.Algeciras-Schimnich A, Barnhart BC, Peter ME. Apoptosis-independent functions of killer caspases. Curr Opin Cell Biol. 2002;14:721–726. doi: 10.1016/S0955-0674(02)00384-8. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, McCloskey T, Joshi PM, Rothman J. H. ced-4 and proto-oncogene tfg-1 antagonistically regulate cell size and apoptosis in C. elegans. Curr Biol. 2008;18:1025–1033. doi: 10.1016/j.cub.2008.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrido C, Kroemer G. Life’s smile, death’s grin: vital functions of apoptosis-executing proteins. Curr. Opin. Cell Biol. 2004;16:639–646. doi: 10.1016/j.ceb.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Schwerk C, Schulze-Osthoff K. Non-apoptotic functions of caspases in cellular proliferation and differentiation. Biochem Pharmacol. 2003;66:1453–1458. doi: 10.1016/S0006-2952(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 27.Zermati Y, et al. Nonapoptotic role for Apaf-1 in the DNA damage checkpoint. Mol Cell. 2007;28:624–637. doi: 10.1016/j.molcel.2007.09.030. [DOI] [PubMed] [Google Scholar]
- 28.Fischer U, Janicke RU, Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abrams JM, White MA. Coordination of cell death and the cell cycle: linking proliferation to death through private and communal couplers. Curr. Opin. Cell Biol. 2004;16:634–638. doi: 10.1016/j.ceb.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 30.Greene LA, Biswas SC, Liu DX. Cell cycle molecules and vertebrate neuron death: E2F at the hub. Cell Death Differ. 2004;11:49–60. doi: 10.1038/sj.cdd.4401341. [DOI] [PubMed] [Google Scholar]
- 31.Huh JR, Guo M, Hay BA. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr Biol. 2004;14:1262–1266. doi: 10.1016/j.cub.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 32.Salmena L, et al. Essential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunity. Genes Dev. 2003;17:883–895. doi: 10.1101/gad.1063703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo M, et al. Caspase-3 regulates cell cycle in B cells: a consequence of substrate specificity. Nat Immunol. 2003;4:1016–1022. doi: 10.1038/ni976. [DOI] [PubMed] [Google Scholar]
- 34.Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell. 2003;4:687–697. doi: 10.1016/S1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- 35.Huh JR, et al. Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol. 2004;2:E15. doi: 10.1371/journal.pbio.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCall K. Eggs over easy: cell death in the Drosophila ovary. Dev. Biol. 2004;274:3–14. doi: 10.1016/j.ydbio.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 37.Peterson JS, Barkett M, McCall K. Stage-specific regulation of caspase activity in drosophila oogenesis. Dev Biol. 2003;260:113–123. doi: 10.1016/S0012-1606(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 38.Geisbrecht ER, Montell DJ. A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell. 2004;118:111–125. doi: 10.1016/j.cell.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa A, Shi Y, Kage-Nakadai E, Mitani S, Xue D. Caspase-dependent conversion of Dicer ribonuclease into a death-promoting deoxyribonuclease. Science (80-.). 2010;328:327–334. doi: 10.1126/science.1182374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weaver BP, et al. CED-3 caspase acts with miRNAs to regulate non-apoptotic gene expression dynamics for robust development in C. elegans. Elife. 2014;3:e04265. doi: 10.7554/eLife.04265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weaver BP, Weaver YM, Mitani S, Han M. Coupled Caspase and N-End Rule Ligase Activities Allow Recognition and Degradation of Pluripotency Factor LIN-28 during Non-Apoptotic Development. Dev Cell. 2017;41:665–673 e6. doi: 10.1016/j.devcel.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen F, et al. Caspase proteolysis of the cohesin component RAD21 promotes apoptosis. J Biol Chem. 2002;277:16775–16781. doi: 10.1074/jbc.M201322200. [DOI] [PubMed] [Google Scholar]
- 43.Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410:955–959. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 44.Azzopardi M, Farrugia G, Balzan R. Cell-cycle involvement in autophagy and apoptosis in yeast. Mech Ageing Dev. 2017;161:211–224. doi: 10.1016/j.mad.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 45.Lopez-Aviles S, Uhlmann F. Cell cycle: the art of multi-tasking. Curr Biol. 2010;20:R101–3. doi: 10.1016/j.cub.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Wu YC, Wang X, Xue D. Methods for studying programmed cell death in C. elegans. Methods Cell Biol. 2012;107:295–320. doi: 10.1016/B978-0-12-394620-1.00010-2. [DOI] [PubMed] [Google Scholar]
- 47.Shaham S, Reddien PW, Davies B, Horvitz HR. Mutational analysis of the Caenorhabditis elegans cell-death gene ced-3. Genetics. 1999;153:1655–1671. doi: 10.1093/genetics/153.4.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hengartner MO, Ellis RE, Horvitz HR. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature. 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- 49.Yuan JY, Horvitz HR. The Caenorhabditis elegans genes ced-3 and ced-4 act cell autonomously to cause programmed cell death. Dev Biol. 1990;138:33–41. doi: 10.1016/0012-1606(90)90174-H. [DOI] [PubMed] [Google Scholar]
- 50.Kimura K, Kimura A. Rab6 is required for the exocytosis of cortical granules and the recruitment of separase to the granules during the oocyte-to-embryo transition in Caenorhabditis elegans. J Cell Sci. 2012;125:5897–5905. doi: 10.1242/jcs.116400. [DOI] [PubMed] [Google Scholar]
- 51.Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- 52.White J, Strome S. Cleavage plane specification in C. elegans: how to divide the spoils. Cell. 1996;84:195–198. doi: 10.1016/S0092-8674(00)80974-5. [DOI] [PubMed] [Google Scholar]
- 53.Madl JE, Herman RK. Polyploids and sex determination in Caenorhabditis elegans. Genetics. 1979;93:393–402. doi: 10.1093/genetics/93.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hodgkin J, Horvitz HR, Brenner S. Nondisjunction Mutants of the Nematode CAENORHABDITIS ELEGANS. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 56.Galluzzi L, et al. No death without life: vital functions of apoptotic effectors. Cell Death Differ. 2008;15:1113–1123. doi: 10.1038/cdd.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi M, et al. DNA-PKcs-PIDDosome: a nuclear caspase-2-activating complex with role in G2/M checkpoint maintenance. Cell. 2009;136:508–520. doi: 10.1016/j.cell.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Andux S, Ellis RE. Apoptosis maintains oocyte quality in aging Caenorhabditis elegans females. PLoS Genet. 2008;4:e1000295. doi: 10.1371/journal.pgen.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pellman D, Christman MF. Separase anxiety: dissolving the sister bond and more. Nat Cell Biol. 2001;3:E207–9. doi: 10.1038/ncb0901-e207. [DOI] [PubMed] [Google Scholar]
- 60.Sullivan M, Lehane C, Uhlmann F. Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat Cell Biol. 2001;3:771–777. doi: 10.1038/ncb0901-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang H, Ren Q, Zhang Z. Cleavage of Mcd1 by caspase-like protease Esp1 promotes apoptosis in budding yeast. Mol Biol Cell. 2008;19:2127–2134. doi: 10.1091/mbc.e07-11-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang X, et al. Inhibitory phosphorylation of separase is essential for genome stability and viability of murine embryonic germ cells. PLoS Biol. 2008;6:e15. doi: 10.1371/journal.pbio.0060015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang X, et al. Preimplantation mouse embryos depend on inhibitory phosphorylation of separase to prevent chromosome missegregation. Mol Cell Biol. 2009;29:1498–1505. doi: 10.1128/MCB.01778-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ameisen JC. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ. 2002;9:367–393. doi: 10.1038/sj.cdd.4400950. [DOI] [PubMed] [Google Scholar]
- 65.Ratcliff WC, Denison RF, Borrello M, Travisano M. Experimental evolution of multicellularity. Proc Natl Acad Sci USA. 2012;109:1595–1600. doi: 10.1073/pnas.1115323109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dorn, J. F. et al. Actomyosin tube formation in polar body cytokinesis requires anillin in C. elegans. Curr. Biol. (2010). 10.1016/j.cub.2010.10.030 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains reported in this manuscript will be made readily available upon request