Abstract
Background
Growing evidence shows that the tumor microenvironment plays a crucial role in the pathogenesis of hepatocellular carcinoma (HCC). The present work aimed to screen tumor microenvironment-related genes strongly related to prognosis and to construct a prognostic gene expression model for HCC.
Material/Methods
We downloaded gene expression data of 371 HCC patients in The Cancer Genome Atlas (TCGA). A novel ESTIMATE algorithm was applied to calculate immune scores and stromal scores for each patient. Then, the differentially-expressed genes (DEGs) were detected according to the immune and stromal scores, and tumor microenvironment-related genes were further explored. Univariate, Lasso, and multivariate Cox analyses were performed to build the tumor microenvironment-related prediction model.
Results
Stromal and immune scores were calculated and were found to be correlated with the 3-year prognosis of HCC patients. DEGs were detected according to the stromal and immune scores. There were 49 genes with prognostic value in both TCGA and ICGC (International Cancer Genome Consortium) considered as prognostic tumor microenvironment-related genes. Univariate, Lasso, and multivariate Cox analyses were conducted. A novel 2-gene signature (IL18RAP and GPR182) was built for HCC 3-year prognosis prediction. The 2-gene signature was regarded as an independent prognostic predictor that was correlated with 3-year survival rate, as shown by Cox regression analysis.
Conclusions
This study offers a novel 2-gene signature to predict overall survival of patients with HCC, which has the potential to be used as an independent prognostic predictor. Overall, this study reveals more details about the tumor microenvironment in HCC and offers novel candidate biomarkers.
MeSH Keywords: Carcinoma, Hepatocellular; Prognosis; Tumor Microenvironment
Background
The liver is an organ with a high regenerative capacity, and liver damage can eventually induce substantial cell proliferation. When high cell proliferation is combined with DNA mutation, liver cancer morbidity significantly increases [1]. Most hepatocellular carcinoma (HCC) is diagnosed in at an advanced stage at which surgery is the only treatment option, with a survival rate below 30% [2]. In advanced HCC stages, resistance almost always occurs [3]. The various management guidelines for HCC unanimously suggest that preserved liver function is a cardinal pre-requisite to provide effective treatment to patients [2]. The HCC fatality rate remains high worldwide [4]. Consequently, new therapeutic strategies based on tumor biology characteristics are urgently needed to improve the prognosis of HCC.
The tumor microenvironment contributes greatly to the pathogenesis of various cancers [5]. The tumor microenvironment is a dynamic system, which largely consists of stromal cells, infiltrating immune cells, and surrounding extracellular matrix (ECM) [6]. Hyaluronic acid, a major ECM component, plays a crucial role in cancer progression [7]. A previous study reported that hyaluronic acid promotes the spread of hepatocellular carcinoma cells, focal adhesion, and membrane tension by phosphoinositide signaling [8]. ECM physical characteristics influence cell function and migration. Substrate viscosity may play an important role in this process [9]. A more recent study demonstrated that normal hepatocytes and hepatocellular carcinoma cells have different spreading dynamics, which were influenced by the substrate viscosity [10]. In recent years, targeting the tumor microenvironment has been proposed as a therapeutic strategy for HCC [11]. Improving knowledge about the tumor microenvironment might be important for identifying novel therapeutic and chemotherapy preventive targets.
The ESTIMATE (Estimation of STromal and Immune cells in MAlignant Tumor tissues using expression data) method was developed by Yoshihara et al. to calculate stromal and immune scores in tumor tissues [12]. A study reported that ESTIMATE can be used as a metric to evaluate the prognosis of cancer patients [13]. Recently, numerous studies have used the ESTIMATE method to assess various tumors, indicating successful uses of the method when applied to genomic data. For instance, Jia et al. and Yang et al. identified many potential prognostic tumor microenvironment-related genes for predicting outcomes in glioblastoma and cutaneous melanoma patients [14,15]. However, there are no studies elucidating the prognostic value of tumor microenvironment-related genes in HCC. The present work aimed to analyze the tumor microenvironment and detect prognostic tumor microenvironment-related biomarkers for HCC.
Material and Methods
Data collection
Hepatocellular carcinoma gene expression data were retrieved from the TCGA (up to June 1, 2019). Clinical data including gender, age, histologic grade, pathologic stage, survival and outcome were also retrieved from TCGA. For validation, gene expression data and the corresponding clinical data were retrieved from the ICGC data portal. Since the data were retrieved from public datasets and we followed the publishing guidelines offered by TCGA, there was no need for ethics approval.
Differential analysis of expressed genes
To calculate immune scores and stromal scores, a novel ESTIMATE algorithm implemented as an “ESTIMATE” package in R was used [12]. ESTIMATE is a tool used to predict stromal/immune cell infiltration. The ESTIMATE algorithm was constructed based on single-sample gene set enrichment analysis and generated a stromal score and an immune score. We divide patients into high/low immune score groups and high/low stromal score groups according to the median value of immune score and stromal score. |log2foldchange| >1 and FDR (False discovery rate) <0.05 was used as the threshold value to screen for differentially-expressed genes. Heatmaps were created using the pheatmap package of R. According to the upregulated and downregulated DEGs identified in immune score groups and stromal score groups, the VennDiagram R package was used to detect the intersecting DEGs.
Functional enrichment analysis
To better understand the potential biologic functions of intersecting DEGs, KEGG pathway enrichment analysis [16] and GO analysis [17] covering biological processes (BP), molecular functions (MF), and cellular components (CC) were conducted using the cluster Profiler package [18] in R. An adjusted p value less than 0.05 were regarded as significant, and the results were visualized with the ggplot2 package.
Survival analysis
Survival analyses were confined to patients with detailed follow-up times only. To identify the prognostic value of intersecting DEGs in HCC patients from TCGA, Kaplan-Meier plots were generated using the survival package in R. Genes with P value <0.05 were validated in ICGC databases using the survival package. Only those genes with P value less than 0.05 in both TCGA and ICGC were considered as candidate biomarkers associated with prognosis. The cBio Cancer Genomics Portal (cBioPortal) is a valuable tool for online exploration, visualization, and analysis of multidimensional cancer genomics data [19,20]. cBioPortal, which is the database used in this research, explores genetic alterations of prognostic genes in TCGA HCC patients. We investigated the cancer medicines affecting those genes. All operations were performed strictly following the cBioPortal’s online instructions.
Identification of the HCC-specific prognostic signatures
To construct the prognostic signature, univariate, Lasso, and multivariate Cox regression analyses were used to investigate the associations between the potential genes and 3-year survival. We built the HCC-specific prognostic signatures in 3 steps: (1) All prognostic genes from the survival analysis were selected. We conducted univariate Cox regression analysis using the survival package in R. Genes with P<0.05 were regarded as statistically significant and were chosen for Lasso Cox regression analysis. (2) The Lasso method can examine all the independent variables concurrently and can help select the most influential variables. Here, Lasso Cox regression analysis was utilized to remove highly correlated genes and prevent over-fitting. Lambda is an important parameter in Lasso regression, which can control the adjustment of Lasso regression complexity. When the partial likelihood deviance is the smallest, the most appropriate lambda can be taken. Lasso Cox regression analysis was implemented with glmnet and survival packages. (3) We performed ultivariate Cox regression analysis via the survminer and survival packages of R software. The linear combination of genes’ expression levels multiplies the regression coefficient determined from the multivariate Cox regression analysis, and risk scores were then calculated. Patients were stratified into a high-risk group and a low-risk group based on median risk scores. The Kaplan-Meier method was utilized to compare 3-year survival between high-risk group and low-risk groups through use of the survival package. To analyze factors affecting 3-year survival, univariate and multivariate analyses were completed on risk scores and several clinicopathological features.
Statistical analysis
All analyses were implemented with R version 3.5.1. We used gene expression data of 371 HCC samples from TCGA. CBioPortal was used to analyze the gene mutation information of the same 371 HCC samples. |log2foldchange|>1 and FDR (false discovery rate) <0.05 was used as the threshold value to screen for differentially-expressed genes. In survival analysis, genes with P values less than 0.05 were considered statistically significant. In univariate Cox regression analysis, genes with P values less than 0.05 were regarded as statistically significant.
Results
Stromal and immune scores correlated with 3-year prognosis in HCC patients
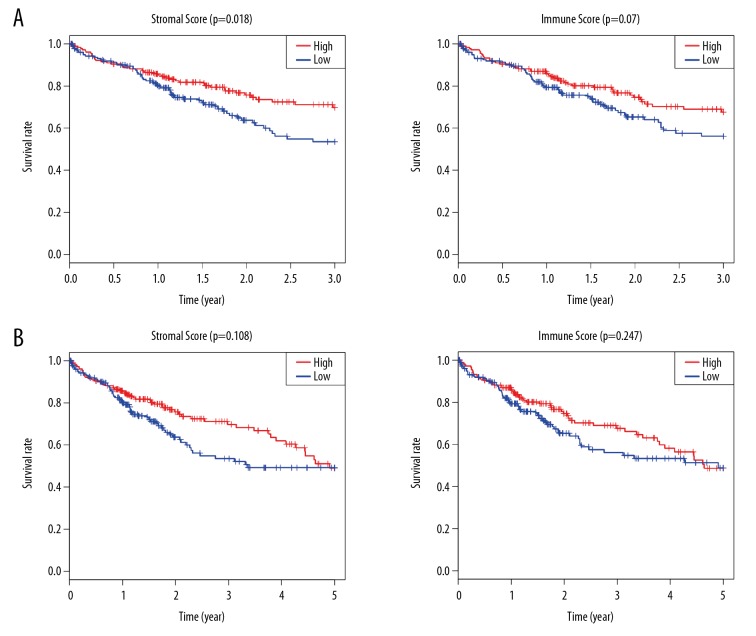
Gene expression profiles and clinical information of 371 HCC samples were acquired from TCGA. Patients were 59.44±13.52 years old and 67.39% were male. On the basis of ESTIMATE method, stromal scores varied from −1625.38 to 1171.12 and immune scores from −866.31 to 3146.06. To evaluate the association of immune/stromal scores with prognosis, HCC patients were categorized into high- and low-score groups based on their median score. The 3-year and 5-year survival rate were plotted with Kaplan-Meier survival curves. The 3-year survival rate of patients with high stromal scores was higher than in patient groups with low scores (p=0.018), while patients with high immune scores had a slightly higher 3-year survival rate (p=0.07; Figure 1A). Stromal scores were positively correlated with 3-year survival rate, and immune scores were weakly positively correlated with 3-year survival rate, suggesting that both were positive factors for 3-year prognosis of patients. However, there was no significant correlation between immune/stromal scores and 5-year survival rate (Figure 1B). The subsequent analysis in this study was based on the 3-year survival rate.
Figure 1.
The relationship between stromal/Immune scores and survival rate in HCC. (A) The analysis of patient 3-year survival rate based on stromal scores and immune scores. (B) The analysis of patient 5-year survival rate based on stromal scores and immune scores.
Identification of tumor microenvironment-related DEGs
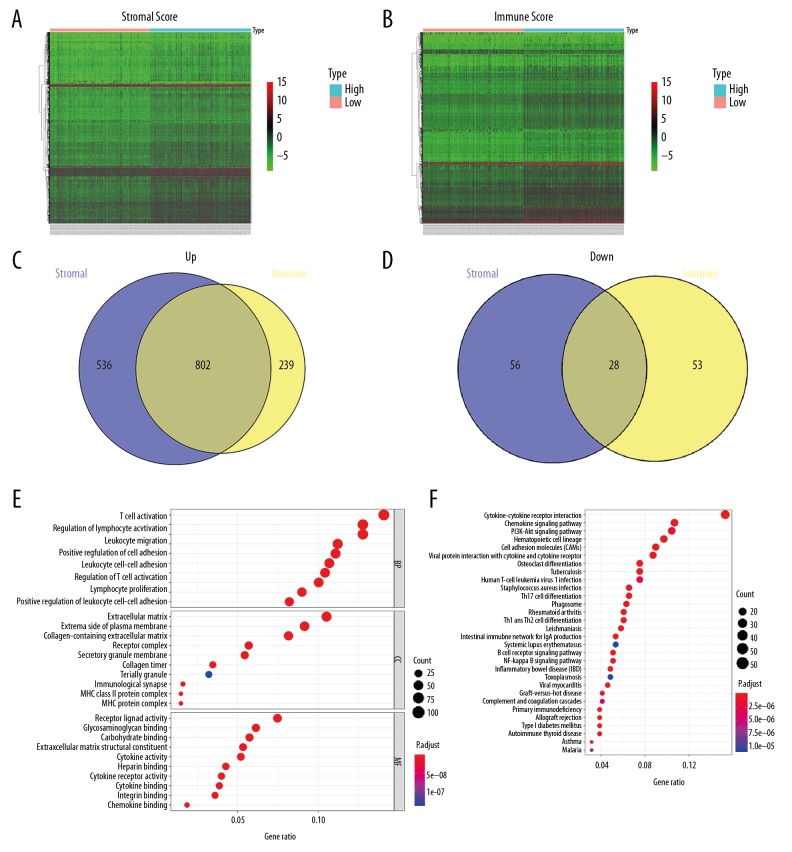
Tumor microenvironment-related genes were acquired via comparing high-score versus low-score groups. For the group with high immune scores, a total of 1122 DEGs – 1041 upregulated and 81 downregulated genes – were screened out. A total of 1422 DEGs – 1338 upregulated and 84 downregulated genes – were detected in the group with high stromal scores. The expression of DEGs in the high-score group versus low-score group are represented by heat maps (Figure 2A, 2B). Intersection analysis of DEGs in the high-score group showed 830 intersection DEGs, including 802 commonly upregulated and 28 commonly downregulated tumor microenvironment-related genes (Figure 2C, 2D). Moreover, the biological terms related to these intersection genes were explored. GO analysis illustrated that the main BP ontology were T cell activation, regulation of lymphocyte activation, and leukocyte migration. In terms of CC, the enriched terms included extracellular matrix and collagen-containing extracellular matrix. For MF, the major GO terms were receptor ligand activity, glycosaminoglycan binding, and carbohydrate binding. KEGG pathway analysis suggested that the intersection DEGs were particularly involved in cytokine-cytokine receptor interaction and chemokine signaling pathway (Figure 2E, 2F).
Figure 2.
Comparison of gene expression profile with stromal/immune scores in HCC. (A, B) Heatmap of significantly DEGs on the basis of stromal and immune scores. Red indicates higher expression and green indicates lower expression. (C, D) Venn diagrams displaying the intersection upregulated or downregulated DEGs in high-score groups. (E) Gene ontology analysis of the tumor microenvironment-related genes. (F) Top 30 enriched KEGG pathways for the tumor microenvironment-related genes.
Identification of prognostic tumor microenvironment-related genes
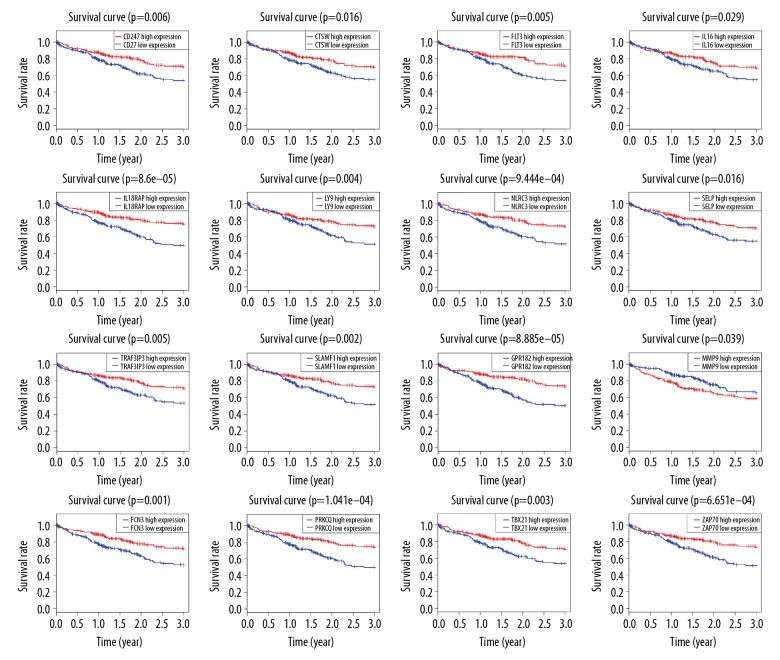
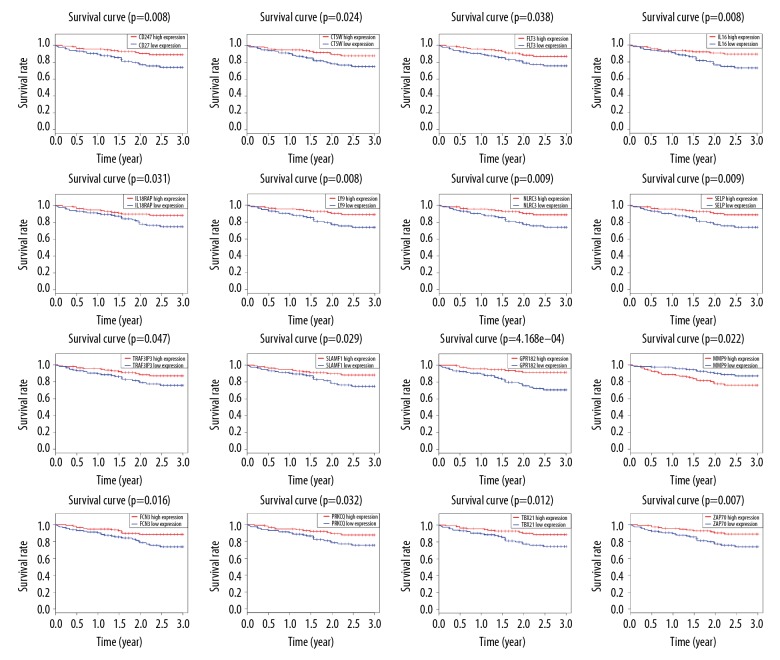
We chose 370 patients with detailed follow-up times for subsequent survival analysis. Kaplan-Meier graphs were fitted to investigate the relationship between the intersection genes and 3-year survival rate in HCC patients, showing that 166 intersection genes were significantly associated with 3-year survival in the TCGA database. Selected results are shown in Figure 3. To investigate whether prognostic genes in TCGA have prognostic value in other HCC cases, we collected and analyzed the data of 232 HCC patients from ICGC. There are 49 genes appearing to be correlated with 3-year survival rate (Figure 4), the majority of which had not been proved to be correlated with the prognosis of HCC patients (Table 1). These 49 genes with prognostic value in both TCGA and ICGC were regarded as prognostic tumor microenvironment-related genes.
Figure 3.
Kaplan-Meier survival curves of prognostic tumor microenvironment-related genes for 3-year survival of TCGA patients. (horizontal axis: 3-year of overall survival time; vertical axis: survival function).
Figure 4.
Validation of the TCGA results in ICGC cohort. Kaplan-Meier curve analysis of prognostic tumor microenvironment-related genes and 3-year survival rate in HCC patients. (horizontal axis: 3-year of overall survival time; vertical axis: survival function).
Table 1.
Genes significant in hepatocellular carcinoma 3-year overall survival identified in both TCGA and ICGC.
| Categories | Gene symbols |
|---|---|
| Cell surface | CD247, CD40LG, CD3E, CD3G, CLEC9A, GPM6A,LY9, SLAMF1, GGT5 |
| Cell adhesion molecules | CD2, CD6, PCDH7, SELP, SIRPG |
| Complements | C7, FCN3 |
| Extracellular matrices | CRISPLD2, DCN, MMP9 |
| G-protein coupled receptor 1 family | GPR171, GPR182, XCR1, PTGER2 |
| Interleukin-related Genes | IL16, IL18RAP |
| protein kinase superfamily | LCK, ITK, ZAP70, MAP4K1, PRKCQ, FLT3 |
| T-box transcription factors | TBX21, EOMES |
| Chromosome Open Reading Frame Genes | C11orf21, C11orf96, FAM163B, ZBP1 |
| Others | NTF3, CTSW, KCNA3, TNNT3, NLRC3, ACAP1, TRAF3IP3, SH2D1A, SHISA3, SIT1, CRHBP, INMT |
The prognostic value of bold type genes in hepatocellular carcinoma patients have not been studied previously.
Genomic changes and biological functions of prognostic tumor microenvironment-related genes
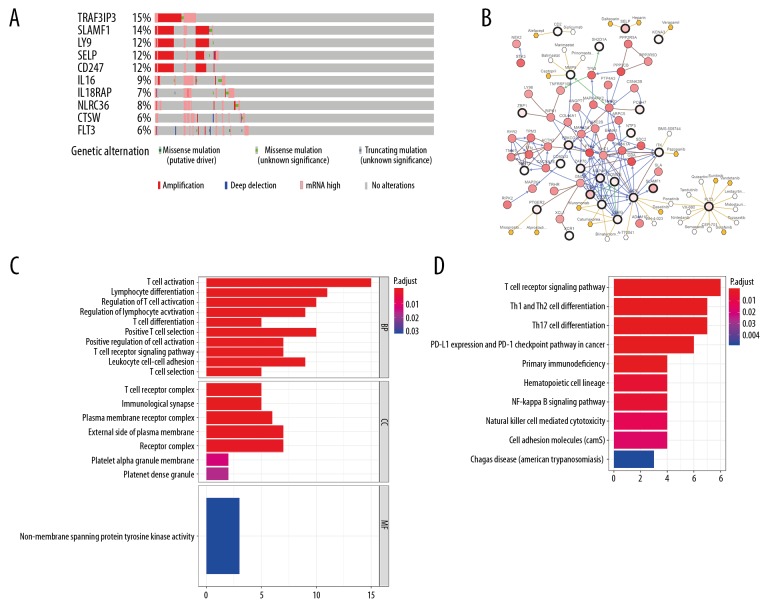
We used cBioPortal assay to investigate genomic alterations connected with these prognostic tumor microenvironment-related genes. Amplification and mRNA upregulation were the main genetic alterations, and 49 prognostic tumor microenvironment-related genes were altered in 229 (62%) of 371 patients. The mutations status of the top 10 genes with higher alteration frequencies are presented in Figure 5A, in which TRAF3IP3 and SLAMF1 were the most commonly mutated genes. Figure 5B shows the network established by 49 prognostic tumor microenvironment-related genes, and the 50 most frequently altered neighbor genes. Cancer drugs targeting those genes are displayed. Among them, 11 genes are currently regarded as anticancer drug targets. We considered that other genes may serve as new therapeutic targets. Additionally, the biological terms associated with 49 prognostic tumor microenvironment-related genes were investigated. Top GO terms included T cell activation and lymphocyte differentiation (Figure 5C). KEGG pathway analysis suggested that these genes were particularly involved in the T cell receptor signaling pathway (Figure 5D).
Figure 5.
Genomic changes and biological functions of prognostic tumor microenvironment-related genes. (A) The genetic alteration of the top 10 tumor microenvironment-related altered genes using the cBioPortal database. (B) The network included 99 nodes, including 49 query genes and the 50 most frequently altered neighbor genes (23 out of 49 were related to the 50 genes). The associations between tumor microenvironment-related genes and anticancer drugs are shown. (C) Gene ontology analysis of the prognostic tumor microenvironment-related genes. (D) Top 10 enriched KEGG pathways for the prognostic tumor microenvironment-related genes.
Construction of tumor microenvironment-related genes prognostic signatures
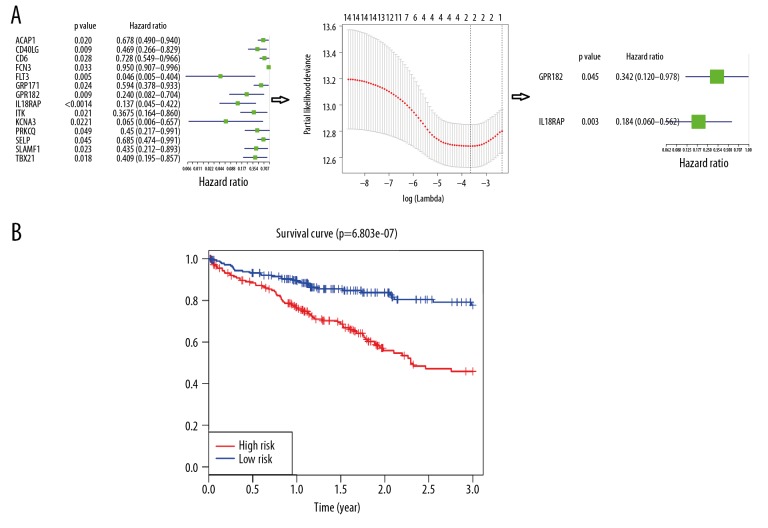
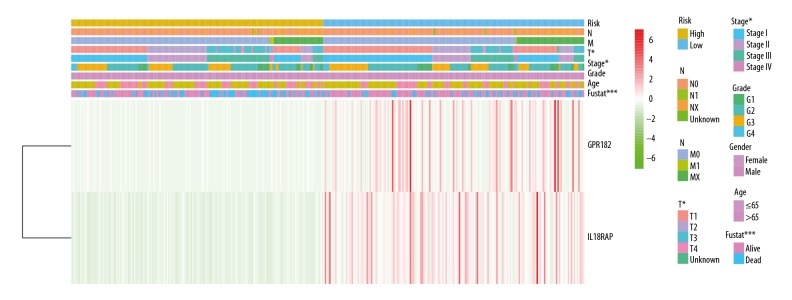
To construct tumor microenvironment-related genes prognostic signatures, expression values of 49 genes in 327 HCC patients were analyzed by univariate Cox regression analysis. As presented in Figure 5A, univariate Cox regression screened 14 genes significantly associated with 3-year survival (P<0.05). All the 14 genes were protective genes with HR <1. Next, Lasso Cox regression analysis was used to narrow the obtained genes (Figure 6A). Three genes were identified and were then subjected to multivariate Cox regression analysis. Only 2 genes – IL18RAP and GPR182 – were used to construct a prognostic model (Figure 6A). A risk score for each individual was generated and ranked based on the risk score. Then, patients were categorized into 2 groups: high-risk (n=185) and low-risk (n=185). Patients in the high-risk group had obviously poorer 3-year survival outcomes than patients in the low-risk group, as shown by the Kaplan-Meier curves (p=6.803e-07; Figure 6B). Figure 7 reveals the relationship between risk score and clinical characteristics, showing that risk score was obviously associated with pathological stage, T classification, and 3-year survival status.
Figure 6.
Establishment and verification of a prognostic gene signature for HCC. (A) The process of constructing the prognostic signature including 2 tumor microenvironment-related genes. First, univariate Cox regression screened 14 genes significantly associated with 3-year survival. Next, 3 genes were identified by Lasso Cox regression analysis. Then, only 2 genes were chosen to build a prognostic signature by multivariate Cox regression analysis. (B) The Kaplan-Meier curve of the 3-year survival rate between the high-risk and low-risk group was plotted according to the median risk score in TCGA.
Figure 7.
Heatmap represents the expression profiles of 2 tumor microenvironment-related gene signature in the high- and low-risk groups. The relationship between risk score and clinical characteristics is shown (* P<0.05, ** P<0.01, *** P<0.001).
Prognostic value of the 2 tumor microenvironment-related gene signatures in HCC
The independent predictive power of the 2-tumor microenvironment-related genes signature was investigated. Univariate analysis showed that pathological stage (P<0.001), T classification (P<0.001), and risk score (P<0.001) were significantly related to 3-year survival rate of TCGA HCC patients. After multivariate analysis, only risk score was an independent prognostic predictor correlated with 3-year survival rate (P<0.001; Table 2).
Table 2.
Univariate and multivariate analyses of 3-year survival in hepatocellular carcinoma patients of TCGA.
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | P-value | Hazard ratio (95% CI) | P-value | |
| Age | 1.004 (0.988–1.020) | 0.631 | 1.006 (0.990–1.022) | 0.490 |
| Gender | 0.839 (0.547–1.287) | 0.421 | 0.939 (0.606–1.454) | 0.776 |
| Histologic grade | 1.165 (0.883–1.538) | 0.280 | 1.187 (0.887–1.589) | 0.248 |
| Pathologic stage | 1.747 (1.394–2.191) | <0.001 | 1.030 (0.466–2.276) | 0.941 |
| T classification | 1.714 (1.386–2.120) | <0.001 | 1.596 (0.759–3.354) | 0.217 |
| Prognostic model | 3.319 (1.976–5.576) | <0.001 | 2.871 (1.690–4.878) | <0.001 |
Discussion
An increasing number of studies have suggested that the tumor microenvironment is involved in the biological processes of cancers [21]. The significance of the tumor microenvironment in HCC prognostic classification has also been noted [22]. The prognostic value of tumor microenvironment-related biomarkers for HCC patients has not yet been explored. Consequently, we performed bioinformatics analysis to screen tumor microenvironment-related genes that were correlated with the prognosis of HCC patients.
Using the TCGA cohort, we applied the ESTIMATE method to generate stromal and immune scores. Our findings suggested that patients with high stromal/immune scores had higher 3-year survival rates, indicating both were positive factors for 3-year prognosis of HCC patients. We screened tumor microenvironment-related genes by stratifying all samples into high immune/stromal score and low immune/stromal score groups, showing that immune/stromal scores and 5-year survival rate were not obviously correlated. Functional analysis based on these tumor microenvironment-related genes indicated that many molecular pathways were closely associated with tumor microenvironment, such as extracellular matrix, collagen-containing extracellular matrix, T cell activation, and cytokine-cytokine receptor interaction. These results were in accordance with previous reports that the roles of immune cells and extracellular matrix in the building of HCC tumor microenvironment are interlinked [23].
To screen prognostic tumor microenvironment-related genes, we conducted 3-year survival analysis of those 830 genes and screened 166 genes that were correlated with 3-year outcomes in HCC patients from TCGA. Validation in the ICGC database showed that 49 genes were associated with 3-year outcomes. The 49 genes with prognostic value in both TCGA and ICGC were considered as prognostic tumor microenvironment-related genes, and the potential functions were investigated. Functional analysis indicated prognostic genes particularly involved in T cell activation, regulation of T cell activation, T cell differentiation, and positive T cell selection. Most of these pathways were associated with T cell-mediated immune response. These results were consistent with our finding that most of the prognostic genes were protective genes. Additionally, these studies have revealed that T cells play an important role in the pathogenesis of HCC, and high levels of T cells were associated with significantly better prognosis, in accordance with pervious results.
In agreement with our findings, previous studies have revealed that T cells play a major role in the pathogenesis of HCC, and high levels of T cells are associated with considerably better prognosis [24,25]. Of these 49 genes identified, only 6 genes had been reported to be correlated with the prognosis of HCC patients. The other 43 genes had not been found to be associated with HCC outcomes, and may act as valuable biomarkers for prognosis of HCC in the future. We used the cBioPortal tool to investigate genomic alterations connected with these 49 prognostic tumor microenvironment-related genes. All prognosis genes were mutated in HCC samples appear to play a significant role in HCC pathogenesis. A gene network containing 49 prognostic tumor microenvironment-related genes and the 50 most frequently altered neighbor genes was established. Among the 49 genes, 11 (e.g., LCK, FLT3, and MMP9) were considered as anticancer drug targets. We are particularly interested in MMP9. MMP9 is an important member of the matrix metalloproteinases family and participates in extracellular matrix remodeling [26]. A growing number of studies indicate thatMMP9 plays a crucial role in mediating the tumor microenvironment [27,28]. In this study, MMP9 is the only gene negatively associated with prognosis among the 49 prognostic tumor microenvironment-related genes. Consistent with our results, numerous studies have demonstrated that overexpression of MMP9 is a predictor of worse prognosis in HCC patients [29]. As can be seen from the network diagram, captopril may exert its anti-tumor role via acting on MMP9. Previous studies have reported captopril may be an inhibitor of tumor growth in several tumors, such as lung cancer and glioblastoma [30,31]. However, no study has investigated the role of captopril in HCC patients. More work is necessary to explore its tumor-suppressor role in HCC.
Cox analyses were performed to establish a prognostic signature to predict HCC 3-year prognosis. We developed a novel 2-gene prognostic signature (IL18RAP and GPR182) using data derived from the TCGA database. The risk score of the 2-gene signature was calculated and then patients were categorized into high-risk and low-risk groups. Patients in the high-risk group had a worse 3-year survival rate than patients in the low-risk group. Moreover, Univariate Cox and multivariate Cox analyses suggested that the 2-gene signature is an independent factor predicting 3-year HCC prognosis. IL18RAP (interleukin 18 receptor accessory protein), a member of the interleukin-1 receptor family, is expressed by various cell types, including immune cells and stromal cells [32]. It has been demonstrated that IL18RAP plays a vital role in the IL-18-mediated signaling pathway by producing interferon gamma [33]. A study has shown that in renal cell carcinoma, high expression of IL18RAP was markedly correlated with poor prognosis based on TCGA datasets [34]. IL18RAP region polymorphisms may be related to the development of esophageal cancer [35]. A previous study investigated the expression of IL18RAP in human HCC patients infected with hepatitis C virus, indicating that IL18RAP and IL18R1 were coexpressed in HCC cell lines [36]. To date, however, few studies have reported the relationship of IL18RAP with HCC. Further research is needed to elucidate the exact role and prognostic value of IL-18RAP in HCC patients. GPR182 (G protein-coupled receptor 182, ADMR) is evidently involved in the G protein-coupled receptor 1 family. A recent study revealed that GPR182 was markedly downregulated in HCC tumorous compared with peritumoral tissues [37]. GPR182 was also reported to be downregulated in liver fibrosis and cirrhosis [37]. A previous study reported silencing GPR182 remarkably reduced the growth and metastasis of pancreatic cancer, without excessive harmful effects, which indicated GPR182 might be a valuable therapeutic target in pancreatic cancer [38]. So far, little work had been done to clarify the prognostic value and therapeutic target role of GPR182 in HCC. Further research is required to determine the prognostic and therapeutic significance of GPR182 in HCC.
Several shortcomings of the present study needed to be noted. Firstly, the work was entirely performed using public databases, and our findings need to be confirmed. Secondly, our survival analysis and Cox regression analysis were based on 3-year survival, not 5-year or overall survival times. Thirdly, further studies are required to clarify the biological roles of these prognostic tumor microenvironment-related genes in HCC. Additionally, the prognostic role of the 2-gene prognostic signature needs to be validated in more independent cohorts by clinical experiments.
Conclusions
We found that numerous tumor microenvironment-related genes were markedly associated with prognosis of HCC. In addition, we established a novel 2-gene signature as an independent prognostic predictor. The present work offers novel insights into the tumor microenvironment in HCC. Nonetheless, further research is required to validate these results.
Acknowledgments
We thank the TCGA and ICGC databases for the availability of the data. We gratefully appreciate Xiaochun Han for providing guidance with statistics.
Footnotes
Conflict of interests
None.
Source of support: Departmental sources
References
- 1.Yang YM, Kim SY, Seki E. Inflammation and Liver Cancer: Molecular mechanisms and therapeutic targets. Semin Liver Dis. 2019;39:26–42. doi: 10.1055/s-0038-1676806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taghizadeh S, Alimardani V, Roudbali PL, et al. Gold nanoparticles application in liver cancer. Photodiagnosis Photodyn Ther. 2019;25:389–400. doi: 10.1016/j.pdpdt.2019.01.027. [DOI] [PubMed] [Google Scholar]
- 3.Nault JC, Ningarhari M, Rebouissou S, Zucman-Rossi J. The role of telomeres and telomerase in cirrhosis and liver cancer. Nature reviews. Gastroenterol Hepatol. 2019;16:544–58. doi: 10.1038/s41575-019-0165-3. [DOI] [PubMed] [Google Scholar]
- 4.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–14. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 5.Chung HW, Lim JB. Role of the tumor microenvironment in the pathogenesis of gastric carcinoma. World J Gastroenterol. 2014;20:1667–80. doi: 10.3748/wjg.v20.i7.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leonardi GC, Candido S, Cervello M, et al. The tumor microenvironment in hepatocellular carcinoma (review) Int J Oncol. 2012;40:1733–47. doi: 10.3892/ijo.2012.1408. [DOI] [PubMed] [Google Scholar]
- 7.Wu R-L, Huang L, Zhao H-C, Geng X-P. Hyaluronic acid in digestive cancers. J Cancer Res Clin Oncol. 2017;143:1–16. doi: 10.1007/s00432-016-2213-5. [DOI] [PubMed] [Google Scholar]
- 8.Mandal K, Raz-Ben Aroush D, Graber ZT, et al. Soft hyaluronic gels promote cell spreading, stress fibers, focal adhesion, and membrane tension by phosphoinositide signaling, not traction force. ACS Nano. 2019;13:203–14. doi: 10.1021/acsnano.8b05286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaman MH, Trapani LM, Sieminski AL, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci USA. 2006;103:10889–94. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandal K, Gong Z, Rylander A, et al. Opposite responses of normal hepatocytes and hepatocellular carcinoma cells to substrate viscoelasticity. Biomater Sci. 2020;8(5):1316–28. doi: 10.1039/c9bm01339c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez-Gea V, Toffanin S, Friedman SL, Llovet JM. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology. 2013;144:512–27. doi: 10.1053/j.gastro.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W, Ye H, Liu YF, et al. Transcriptome-derived stromal and immune scores infer clinical outcomes of patients with cancer. Oncol Lett. 2018;15:4351–57. doi: 10.3892/ol.2018.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia D, Li S, Li D, Xue H, et al. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging. 2018;10:592–605. doi: 10.18632/aging.101415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S, Liu T, Nan H, et al. Comprehensive analysis of prognostic immune-related genes in the tumor microenvironment of cutaneous melanoma. J Cell Physiol. 2020;235(2):1025–35. doi: 10.1002/jcp.29018. [DOI] [PubMed] [Google Scholar]
- 16.Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–61. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Gene Ontology (GO) project in 2006. Nucleic acids research. 2006;34:D322–26. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–87. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6 doi: 10.1126/scisignal.2004088. pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pottier C, Wheatherspoon A, Roncarati P, et al. The importance of the tumor microenvironment in the therapeutic management of cancer. Expert Rev Anticancer Ther. 2015;15:943–54. doi: 10.1586/14737140.2015.1059279. [DOI] [PubMed] [Google Scholar]
- 22.Yang JD, Nakamura I, Roberts LR. The tumor microenvironment in hepatocellular carcinoma: Current status and therapeutic targets. Semin Cancer Biol. 2011;21:35–43. doi: 10.1016/j.semcancer.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Chen L. Tumor microenvironment and hepatocellular carcinoma metastasis. J Gastroenterol Hepatol. 2013;28(Suppl 1):43–48. doi: 10.1111/jgh.12091. [DOI] [PubMed] [Google Scholar]
- 24.Yao W, He JC, Yang Y, et al. The prognostic value of tumor-infiltrating lymphocytes in hepatocellular carcinoma: A systematic review and meta-analysis. Sci Rep. 2017;7:7525. doi: 10.1038/s41598-017-08128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X, Tan Y, Qian Y, et al. Clinicopathologic and prognostic significance of tumor-infiltrating CD8+ T cells in patients with hepatocellular carcinoma: A meta-analysis. Medicine. 2019;98:e13923. doi: 10.1097/MD.0000000000013923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagase H, Visse R, Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res. 2006;69:562–73. doi: 10.1016/j.cardiores.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Candido S, Abrams SL, Steelman LS, et al. Roles of NGAL and MMP-9 in the tumor microenvironment and sensitivity to targeted therapy. Biochim Biophys Acta. 2016;1863:438–48. doi: 10.1016/j.bbamcr.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Huang H. Matrix metalloproteinase-9 (MMP-9) as a cancer biomarker and MMP-9 biosensors: Recent advances. Sensors (Basel) 2018;18(10) doi: 10.3390/s18103249. pii: E3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Attoub S, Gaben AM, Al-Salam S, et al. Captopril as a potential inhibitor of lung tumor growth and metastasis. Ann NY Acad Sci. 2008;1138:65–72. doi: 10.1196/annals.1414.011. [DOI] [PubMed] [Google Scholar]
- 31.Kast RE, Halatsch ME. Matrix metalloproteinase-2 and -9 in glioblastoma: A trio of old drugs-captopril, disulfiram and nelfinavir-are inhibitors with potential as adjunctive treatments in glioblastoma. Arch Med Res. 2012;43:243–47. doi: 10.1016/j.arcmed.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev. 2008;223:20–38. doi: 10.1111/j.1600-065X.2008.00624.x. [DOI] [PubMed] [Google Scholar]
- 33.Cheung H, Chen NJ, Cao Z, et al. Accessory protein-like is essential for IL-18-mediated signaling. Journal of immunology (Baltimore, MD: 1950) 2005;174:5351–57. doi: 10.4049/jimmunol.174.9.5351. [DOI] [PubMed] [Google Scholar]
- 34.Yamada Y, Arai T, Kojima S, et al. Regulation of antitumor miR-144-5p targets oncogenes: Direct regulation of syndecan-3 and its clinical significance. Cancer Sci. 2018;109:2919–36. doi: 10.1111/cas.13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu J, Liu C, Teng X, et al. Association of the interleukin-18 receptor 1 and interleukin-18 receptor accessory protein polymorphisms with the risk of esophageal cancer. Biomed Rep. 2016;4:227–35. doi: 10.3892/br.2015.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asakawa M, Kono H, Amemiya H, et al. Role of interleukin-18 and its receptor in hepatocellular carcinoma associated with hepatitis C virus infection. Int J Cancer. 2006;118:564–70. doi: 10.1002/ijc.21367. [DOI] [PubMed] [Google Scholar]
- 37.Schmid CD, Schledzewski K, Mogler C, et al. GPR182 is a novel marker for sinusoidal endothelial differentiation with distinct GPCR signaling activity in vitro. Biochem Biophys Res Commun. 2018;497:32–38. doi: 10.1016/j.bbrc.2018.01.185. [DOI] [PubMed] [Google Scholar]
- 38.Ramachandran V, Arumugam T, Hwang RF, et al. Adrenomedullin is expressed in pancreatic cancer and stimulates cell proliferation and invasion in an autocrine manner via the adrenomedullin receptor, ADMR. Cancer Res. 2007;67:2666–75. doi: 10.1158/0008-5472.CAN-06-3362. [DOI] [PubMed] [Google Scholar]